ABSTRACT
Pseudomonas alloputida KT2440 is a ubiquitous, soil-dwelling bacterium that metabolizes recalcitrant and volatile carbon sources. The latter is utilized by two redundant, Ca- and lanthanide (Ln)-dependent, pyrroloquinoline quinone-dependent alcohol dehydrogenases (PQQ ADH), PedE and PedH, whose expression is regulated by Ln availability. P. alloputida KT2440 is the best-studied non-methylotroph in the context of Ln-utilization. Combined with microfluidic cultivation and single-cell elemental analysis, we studied the impact of light and heavy Ln on transcriptome-wide gene expression when growing P. alloputida KT2440 with 2-phenylethanol as the carbon and energy source. Light Ln (La, Ce, and Nd) and a mixture of light and heavy Ln (La, Ce, Nd, Dy, Ho, Er, and Yb) had a positive effect on growth, whereas supplementation with heavy Ln (Dy, Ho, Er, and Yb) exerted fitness costs. These were likely a consequence of mismetallation and non-utilizable Ln interfering with Ln sensing and signaling. The measured amounts of cell-associated Ln varied between elements. Gene expression analysis suggested that the Ln sensing and signaling machinery, the two-component system PedS2R2 and PedH, responds differently to (non-)utilizable Ln. We expanded our understanding of the lanthanide (Ln) switch in P. alloputida KT2440, demonstrating that it adjusts the levels of pedE and pedH transcripts based on the availability of Ln. We propose that the usability of Ln influences the bacterium’s response to different Ln elements.
IMPORTANCE
The Ln switch, the inverse regulation of Ca- and Ln-dependent PQQ ADH in response to Ln availability in organisms featuring both, is central to our understanding of Ln utilization. Although the preference of bacteria for light Ln is well known, the effect of different Ln, light and heavy, on growth and gene expression has rarely been studied. We provide evidence for a fine-tuning mechanism of Ca- and Ln-dependent PQQ ADH in P. alloputida KT2440 on the transcriptome level. The response to (non-)utilizable Ln differs depending on the element. Ln commonly co-occur in nature. Our findings underline that Ln-utilizing microbes must be able to discriminate between Ln to use them effectively. Considering the prevalence of Ln-dependent proteins in many microbial taxa, more work addressing Ln sensing and signaling is needed. Ln availability likely necessitates different adaptations regarding Ln utilization.
KEYWORDS: lanthanides, lanthanome, RNAseq, microfluidics, single-cell ICP-MS
INTRODUCTION
Together with the chemically similar elements scandium (Sc) and yttrium (Y), the lanthanides (elements 57–71 [La–Lu], Ln) are commonly known as “rare-earth elements” or more precisely, “rare-earth metals” (REM). Based on their electron configuration, which governs interactions with other elements, Ln are grouped into light (La–Eu, Sc; LREM) and heavy (Gd–Yb, Y; HREM) REM (1, 2). Ln are key resources for numerous high-tech applications and are in increasing demand for the ongoing green energy transition (3). The lack of sustainable mining and recycling methods puts a spotlight on understanding Ln-dependent metabolism as a foundation for biology-inspired Ln recovery strategies.
Our knowledge about Ln-dependent metabolism is primarily derived from methylotrophic bacteria, which utilize C1 compounds as carbon and energy sources (4, 5). The first discovered group of Ln-dependent enzymes, Xox-type methanol dehydrogenases (MDHs), catalyzes the oxidation of methanol to formaldehyde. The role of Xox-type MDH was long unclear, and calcium-dependent Mxa-type MDHs were considered the de facto key enzymes for methanol oxidation in methylotrophs (6, 7). Xox- and Mxa-type MDHs belong to the diverse group of pyrroloquinoline quinone (PQQ)-dependent alcohol dehydrogenases (ADH) (8). Genes encoding Xox-type MDHs were detected in many environments and also in non-canonical methylotrophs (9, 10), suggesting that methylovory (the supplemental use of C1 compounds as energy sources) is more common than anticipated (11). The PQQ ADH family includes over a dozen subclades (8). Most are assumed to rely on Ln. In bacteria possessing pairs of Ca- and Ln-dependent ADH, the expression of the corresponding genes is inversely regulated by the “Ln switch,” which is rooted in two-component systems and auxiliary proteins (12–17).
Microbes prefer LREM (5). Mechanisms underlying Ln mobilization and uptake likely differ depending on local Ln bioavailability. Work in the model methylotroph Methylorubrum extorquens identified two gene clusters, the lut-cluster (lanthanide utilization and transport) and the mll-/mlu-cluster (methylolanthanin uptake), which encode uptake machinery centered around a TonB-dependent receptor and an ABC transporter (18), and biosynthetic machinery for a Ln-binding metal chelator (19). Ln commonly co-occur in the environment. It is not clear how Ln-utilizing microbes distinguish between them. Using Beijerinckiaceae bacterium RH AL1, we could show that Ln are selectively taken up and that supplementation with different Ln elements had only minor effects on growth but substantial effects on gene expression (20).
Pseudomonas alloputida KT2440 is the best-studied, non-methylotroph in the context of microbial Ln utilization (15, 21–23). Strain KT2440 is characterized by a broad metabolic versatility, including the potential to use volatile organic compounds (VOC) that carry alcohols or aldehydes as functional groups (24–26). The utilization of VOC is facilitated by two inversely regulated, broad-range PQQ ADH PedH (Ln-dependent) and PedE (Ca-dependent). In P. alloputida KT2440, the Ln switch is rooted in the interplay between the Ca-dependent PQQ ADH PedE and the Ln-dependent PQQ ADH PedH (21). PedH also functions as a sensory module and is, together with the PedS2R2 (PP_2671, PP_2672) two-component system (15), integral for Ln signaling. It was previously postulated that PedS2 phosphorylates its LuxR-type response regulator PedR2 in the absence of La, which in turn activates pedE and limits pedH expression (15). If La is available, PedS2 kinase activity is reduced, pedE expression is decreased, and pedH transcription is activated (15).
In this study, we investigated the effect of Ln supplementation on growth, cell-associated Ln, and gene expression. Our findings indicate that the Ln switch fine-tunes the pedE and pedH transcript pools in P. alloputida KT2440. Differences in the response to (non-)utilizable elements suggest that P. alloputida KT2440 can distinguish Ln elements to effectively use them for its metabolism.
RESULTS
Effect of La concentration and Ln elements on growth
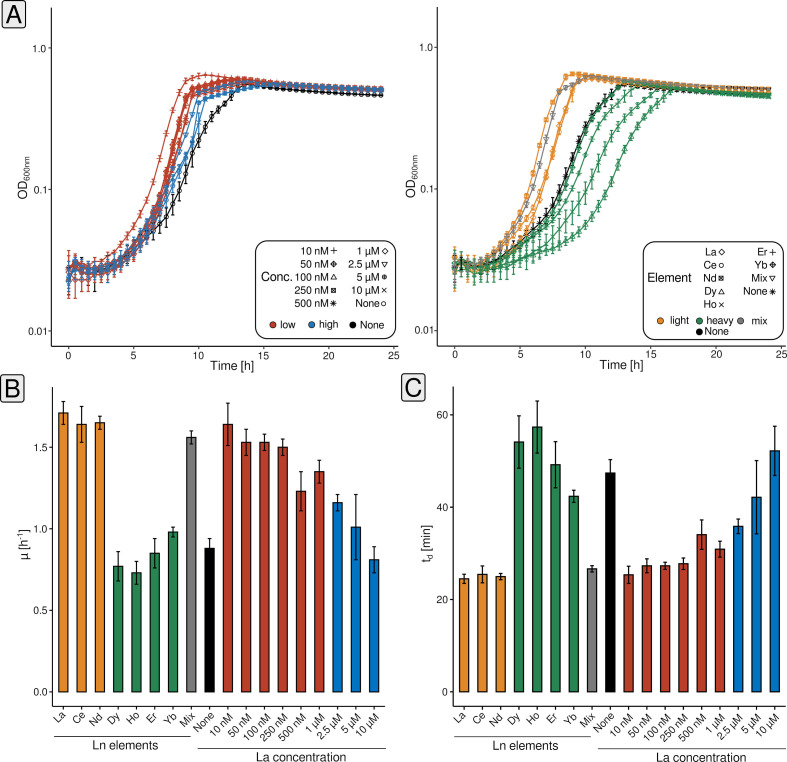
We first grew P. alloputida KT2440 with different La concentrations (10, 50, 100, 250, and 500 nM, 1, 2.5, 5, and 10 µM; and without added Ln) (Fig. 1A, left panel). The lag phase was shortest with 10 and 50 nM La. A maximum growth rate (r) of 1.64 ± 0.13 h−1 for the corresponding cultures was estimated, almost twice as high as for those without added Ln (Table S1). The shortest estimated doubling time (td) has been 0.42 ± 0.03 h in the case of the 10 nM cultures (Fig. 1B). Except for 10 µM, all tested concentrations had a positive effect on r and td (Table S1).
Fig 1.
Assessing the growth of P. alloputida KT2440 with different concentrations of La (10 nM - 10 µM) and different Ln elements (50 nM). Cultivations have been carried out in 96-well plates (n = 4 biological replicates) in 300 µL MP medium, and growth was tracked through spectrophotometry (OD600) (A). The symbols and color codes refer to different La concentrations, and different Ln elements, respectively (red = low concentrations, 10 nM–1 µM; blue = high concentrations, 2.5–10 µM; orange = light Ln, La–Nd; green = heavy Ln, Dy–Yb; grey = Ln mix [Ce, Nd, Dy, Ho, Er, and Yb]; black = no added Ln). Growth rates and doubling times were calculated based on averaged data using growthcurver (v0.3.1) (27) (B). Obtained growth curve data were fitted based on the standard form of the logistic equation in ecology and evolution, which is rooted in growth rate, initial population size, and carrying capacity (28, 29).
We compared the effect of 50 nM of different Ln elements on growth (La, Ce, Nd, Dy, Ho, Er, Yb, and Mix [equimolar mix of Ce, Nd, Dy, Ho, Er, and Yb; 50 nM total Ln]) (Fig. 1A, right panel). The lag phase was the shortest for Nd cultures. The lighter Ln (La, Ce, and Nd) and the Ln mix had a positive effect on growth when compared with cultures without added Ln (Fig. 1B and C). The estimated growth rates were between 1.71 ± 0.07 (La) and 1.56 ± 0.04 h−1 (Ln mix), with doubling times around 0.4 h (Table S1). The growth rate was 0.88 ± 0.06 h−1 without added Ln, and the doubling time was 0.79 ± 0.05 h. Heavier Ln impaired growth. Lag phases and doubling times were longer, and growth rates were decreased. Yb cultures almost behaved like those without added Ln.
Microfluidic one- and two-dimensional screenings with light and heavy Ln
We established microfluidic cultivation (Fig. S1) using segmented flow, based on a previously validated setup (30) (Fig. S2), for P. alloputida KT2440 in 500 nL droplets in PP9 (perfluoromethyldecalin) carrier medium, stored, and incubated in polytetrafluoroethylene (PTFE) tubing. The composition concerning added Ln was manipulated, yielding either a series of droplets characterized by increasing Ln concentration (0–75 nM) (one-dimensional screenings, Fig. 2A), or a series of droplets featuring different concentration ratios of two different Ln elements (0–75 nM, two-dimensional screenings, Fig. 2B).
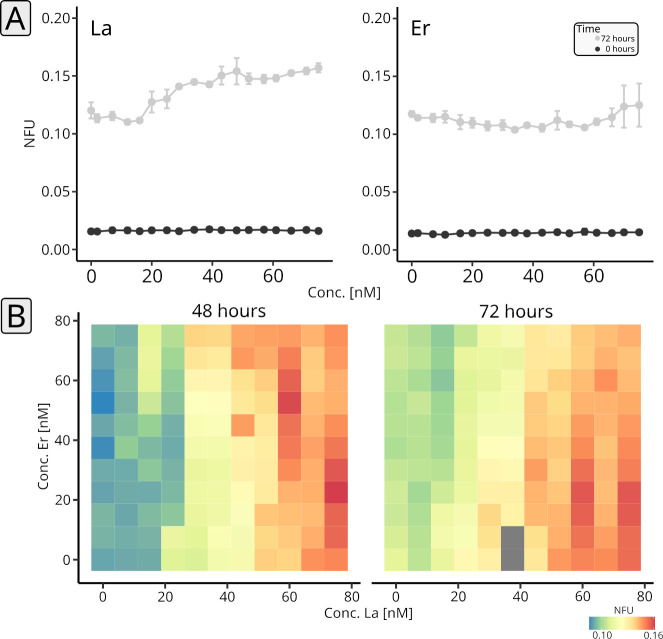
Fig 2.
One- and two-dimensional microfluidic cultivation of P. alloputida KT2440 with different Ln elements. One-dimensional screenings have been carried out for La and Er ramping from 0 to 75 nM, over 18 steps (n = 3–5 microfluidic segments per concentration, technical replicates). Per Ln element, two or three tube coils were incubated (n = 2–3, biological replicates). Incubations were done for 72 h. Plotted are normalized autofluorescence units (NFU) and standard deviations at the beginning (black) and end of the incubations (gray) (A). A two-dimensional screening has been done for La vs Er (B). We tested 121 combinations of concentrations (0–75 nM) against each other and incubated the corresponding microfluidic segments for three days. Per combination, between three and five microfluidic segments were generated (technical replicates). Plotted are NFU values and standard deviations after two and three days. For the measurements, complete sequences were moved through the sensors. Especially due to repeated measurements, it can happen that some droplets merge (see supplemental material). For the evaluation, only droplets that have not merged have been considered. Grayed-out tiles refer to droplets that suffered from droplet fusion.
P. alloputida KT2440 reached the stationary phase in the droplets after 72 h (data not shown). Growth was monitored by measuring the autofluorescence (405 nm excitation and 425 nm emission). In the case of La, the maximum attained autofluorescence after 3 days of incubation increased with La concentration, but only marginally for concentrations above 50 nM (Fig. 2A, left panel). Er had no impact on attained autofluorescence values, but higher concentrations were reflected in a higher observed variability (Fig. 2A, right panel).
We carried out a two-dimensional screening (La vs. Er) to study how the interplay between light and heavy Ln affects growth (Fig. 2B; Table S2). After 48 h, the highest autofluorescence values were observed for the combinations of 60 nM La and 52.5 nM Er, and 75 nM La and 30 nM Er, respectively (Fig. 2B). Overall, autofluorescence values tended to be higher with increasing La concentration. We did not see any interaction between the two effectors, La and Er. Additionally, added Er did not, or only marginally, impact growth.
Analysis of cell-bound Ln using single-cell elemental analysis
The observed effects of different Ln elements on growth prompted us to probe P. alloputida KT2440 for Ln accumulation and uptake. Using transmission electron microscopy (TEM) and energy-dispersive X-ray spectroscopy (EDX), we could neither identify extra- nor intracellular Ln accumulations when 50 nM or 1 µM Ln was added. Inspecting and measuring cell aggregates instead of individual cells did not yield any Ln-positive signals (Fig. S3). To assess cell-associated Ln, we made use of single-cell inductively coupled mass spectrometry (scICP-MS). We measured La, Nd, and Er starting from fixed biomass from incubations supplemented with an excess of Ln (1 µM of the respective element), and measured Ce, Nd, and Er in the case of the Ln mix.
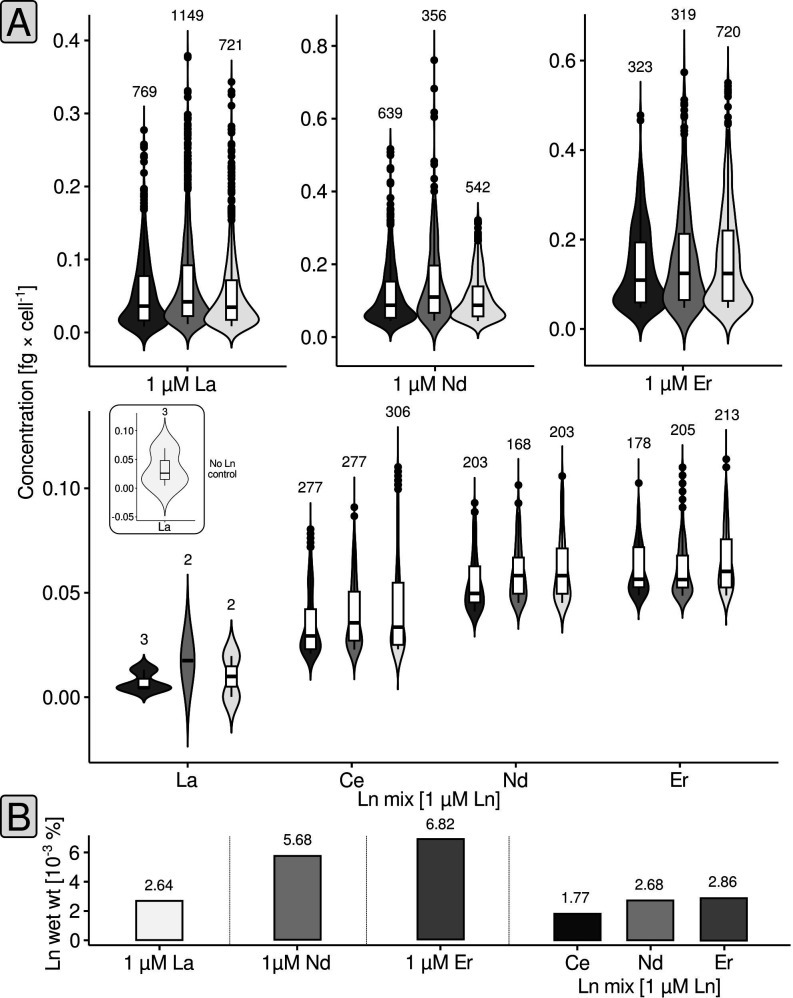
For each sample, several hundred events (= cells) have been analyzed (Fig. 3A). Cells grown with 1 µM La featured on average 0.058 ± 0.055 fg La cell−1. The amount of cell-associated Ln for cells grown with either Nd or Er was higher and around 0.125 ± 0.086 fg Nd cell−1 and 0.152 ± 0.106 fg Er cell−1. Assuming an average wet weight per cell of 2.2 pg (see Materials and Methods), between 2.64 × 10−3 (La) and 6.82 × 10−3% (Er) of the wet weight were made up by cell-associated Ln (Fig. 3B). For the Ln mix samples, we observed that Ln made up a slightly higher proportion of the wet weight (7.31 × 10−3%), and we could see that the amount of cell-associated Ln differed between Ln elements. We detected on average 0.039 ± 0.018 fg Ce cell−1 (1.77 × 10−3% cellular wet weight), 0.059 ± 0.037 fg Nd cell−1 (2.68 × 10−3%), and 0.063 ± 0.014 fg Er cell−1 (2.86 × 10−3%).
Fig 3.
Single-cell elemental analysis. The Ln concentration per cell is shown through combined violin and box plots (A). The different shades of gray indicate biological replicates (n = 3). The box plots indicate lower and upper quartiles, as well as median values. Black circles represent cells with concentrations above (or below) the whisker limits. The numbers above the violin plots indicate the number of considered events (= cells). The small subpanel shows the results of the control sample without any added Ln. Sporadically detected La events in the case of the control, and Ln mix samples represent background noise of the measurement. The %Ln of the wet weight (B) was calculated as outlined in Materials and Methods.
Overall effects of ln elements on gene expression
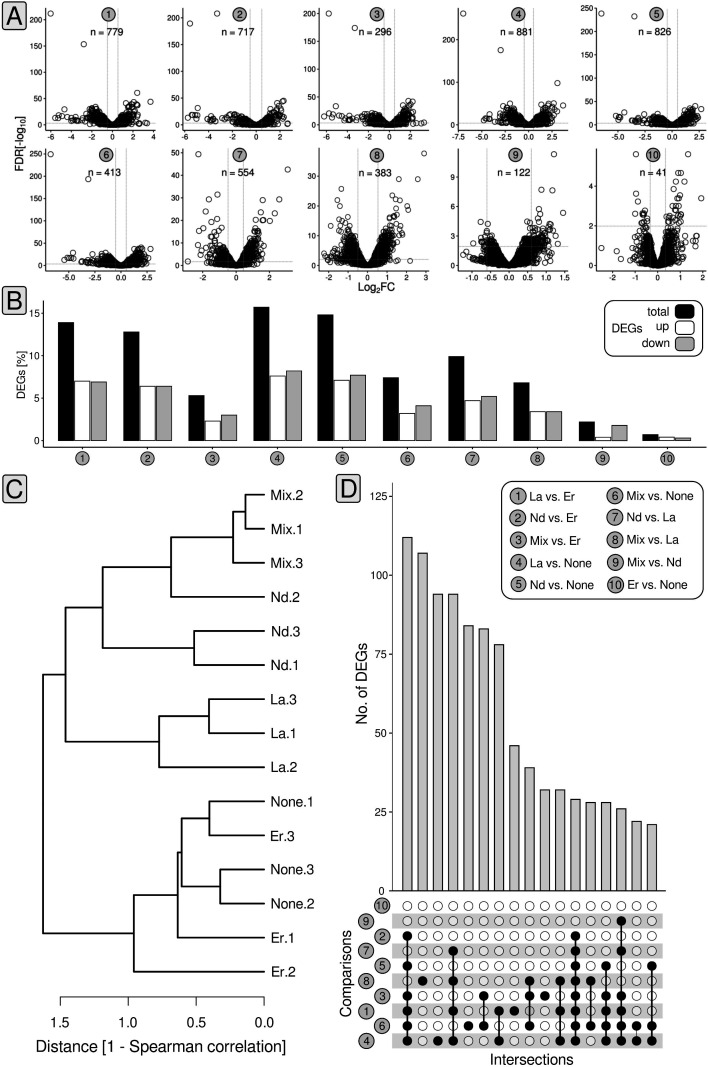
Starting from mid-exponential phase biomass samples (t1 + t2, Fig. S4) originating from cultures grown with 50 nM of different Ln elements (La, Nd, Er, Mix [equimolar mix of Ce, Nd, Dy, Ho, Er, Yb; 50 nM total Ln]) or without Ln, we performed RNAseq-based (Table S3) differential gene expression analysis (DGEA). We considered genes with changes in the expression above |0.58| log2fold change (FC; a log2FC > |0.58| is equivalent to gene expression changes > 50%), expression values higher than four log2CPM (counts per million), and false-discovery rate (FDR)-adjusted P-values < 0.05 as differentially expressed (Fig. 4A; Tables S4 to S13). We determined differentially expressed genes (DEGs) for a total of 10 different comparisons: (1) La vs. Er, (2) Nd vs. Er, (3) mix vs. Er, (4) La vs. none, (5) Nd vs. none, (6) mix vs. none, (7) Nd vs. La, (8) mix vs. La, (9) mix vs. Nd, and (10) Er vs. none (Fig. 4A).
Fig 4.
Statistics and summary of gene expression analysis. Volcano plots summarize the number of differentially expressed genes (DEGs) and the breadth of observed fold changes for the different comparisons (1–10). We defined differential gene expression based on changes in expression (> |0.58| log2FC [fold change]), relative gene expression (> 4 log2[counts per million [CPM]), and FDR-corrected P-values (<0.5). Gray dashed lines indicate the log2FC threshold of |0.58| (A). The proportion of DEGs was summarized as bar charts. The color code refers to the proportion of differentially expressed (black), upregulated (white), and downregulated genes (gray) (B). Based on the counts per million (CPM) matrix, inter-sample relationships have been determined by calculating Spearman distances (C). Shared genes between the different comparisons were highlighted using UpSet plots (D).
The number of DEGs ranged between 41 (Er vs. none) and 881 (La vs. none), which translates into 0.7% and 15.7% of the P. alloputida KT2440 protein-encoding genes (Fig. 4B). Replicate data sets from the incubations with Er and without added Ln clustered together. The same was true for the La, Nd, and Ln mix replicates (Fig. 4C).
We determined the overlap between individual comparisons (Fig. 4D) and noted the biggest overlap of 112 DEGs when looking at comparisons 1–6. Subsequently, 94 shared DEGs were observed in the case of comparisons 1, 4, 7, and 8, and 28 genes were differentially expressed in all comparisons except 9 and 10. There were no DGEs that were shared by all comparisons.
Differential gene expression and cluster analysis
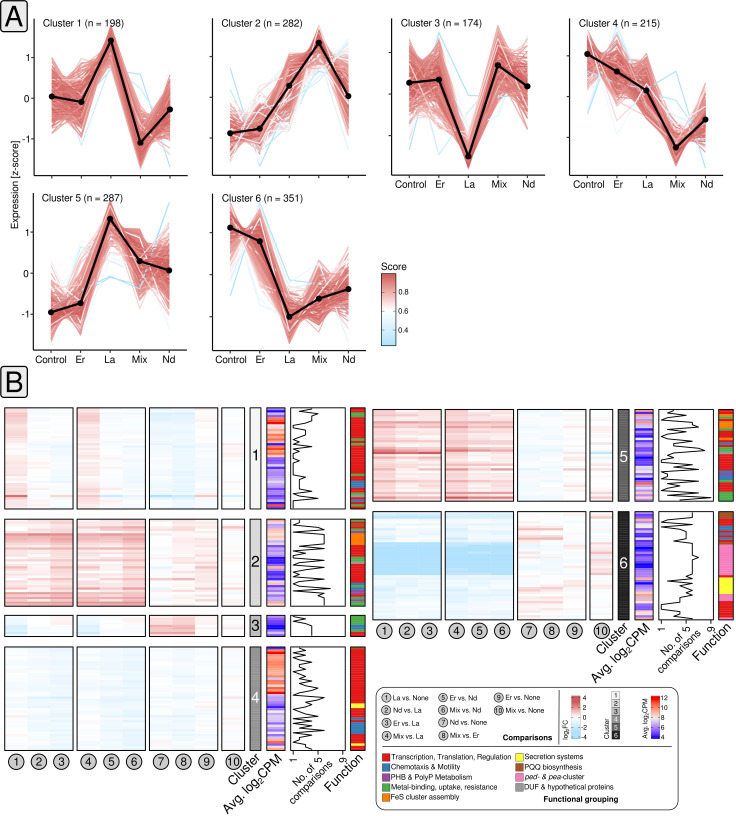
We used K-means clustering (31) for grouping differentially expressed genes based on co-expression. We evaluated different algorithms (Fig. S5) for estimating K, and ultimately settled on K = 6. Inspecting gene expression by determined z-scores, revealed pairs of clusters characterized by inversed gene expression patterns (clusters 1 and 3, 2 and 4, 5 and 6) (Fig. 5A).
Fig 5.
Cluster and differential gene expression analysis. K-means clustering (31) was used to identify clusters of genes with matching gene expression patterns (A). The number of clusters was estimated a priori through Spearman distances, the Calinski–Harabasz index (32), the sum of squared errors, the average silhouette width (33), and the gap statistic (34) (Fig. S5). We ultimately settled on K = 6. The color code indicates the Pearson correlation between each gene of the respective cluster and the cluster centroid (black). Changes in gene expression of cluster genes for each comparison (1–10) are shown for selected subsets of genes (B). The heatmap color codes refer to log2FC and average log2CPM values. The functional annotation of the genes is color-coded as well. The line plot indicates how often the genes were considered as differentially expressed based on the different comparisons.
We noted that cluster 1 genes not only responded to La supplementation by increased gene expression compared with Er and cultures without Ln, but also reduced gene expression when comparing La to Nd and the Ln mix. Cluster 1 included ribosomal protein (PP_0445–46) and transcriptional regulator (PP_4476) genes (Fig. 5B). We also noticed genes linked to spermidine/putrescine transport (PP_0412–13) and a gene coding for a DUF2790 domain-containing protein. The latter was strongly downregulated when comparing La supplemented cultures with those grown with Er or without Ln (log2FC −2.89,–2.62).
When looking at cluster 2, gene expression was elevated in all comparisons between cultures grown with La, Nd, or the Ln mix; and cultures supplemented with Er or no Ln. A weaker response was visible when Ln mix cultures were checked against, La, Nd, or Er. Cluster 2 comprised transcriptional regulator genes (PP_1683, PP_3756), genes encoding (heavy) metal sensor and transport proteins (PP_5139, PP_5383–84), a phasin gene (PP_5008), but also genes involved in Fe–S cluster assembly (PP_0843–46) (Fig. 5B).
Cluster 3 included two gene copies encoding the chemotaxis-related adaptor protein CheW (PP_1489, PP_1491). It featured reduced expression in the two comparisons between La and Er and between La and the control cultures, whereas expression was higher when contrasting Nd, and the Ln mix, against La. The genes belonging to cluster 4 included multiple TetR/Acr transcriptional regulator genes (PP_1961, PP_1968), as well as various ribosomal protein genes. We noted lower gene expression when comparing Nd and Ln mix cultures with Er and cultures without added Ln.
The addition of light/mixed Ln caused elevated gene expression of cluster 5 genes compared with the control set-up and cultures grown with Er. Genes for FeS-cluster assembly proteins were sensitive to Ln supplementation (log2FC −1.78 to 1.76), whereas the most responsive cluster 5 genes encoded metal-sensing, metal-binding proteins, and another DUF2790 domain-containing protein (PP_2969, PP_5732, PP_3494; log2FC −3.34 to 3.04). Cluster 6, covered in more detail in the next section, included lanthanome-related genes.
Differential gene expression of lanthanome-associated genes
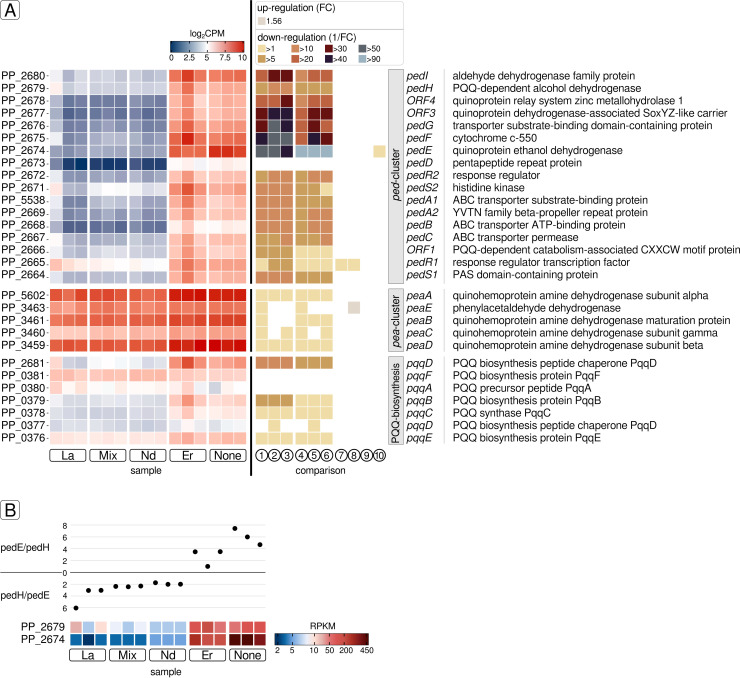
The lanthanome comprises all biomolecules, primarily proteins, directly involved in Ln utilization (35). In P. alloputida KT2440, Ln utilization is linked to the ped-cluster (PP_2664 – PP_2680), which is associated with 2-phenylethanol uptake and its conversion to phenylacetic acid (36). It includes pedE, pedH, as well as genes coding for the Ln-sensing two-component system PedS2/PedR2 (PP_2671/PP_2672), and an ABC transporter PedA1A2BC (PP_5538, PP_2669, PP_2668, PP_2667) facilitating cytoplasmic Ln uptake. Except for Er addition, the ped-cluster, including pedE and pedH, was downregulated upon Ln supplementation (Fig. 6A; Tables S4 to S13) (log2FC values between −1.29 and −6.98). Only pedE (PP_2674) was differentially expressed when comparing incubations with Er and without added Ln (log2FC −0.96). Depending on the added Ln element, pedE gene expression differed by more than two orders of magnitude (fold changes of up to 126). Its highest gene expression was detected in samples without Ln (reads per kilobase per million [RPKM] 410.28 ± 56.55), followed by Er (RPKM 210.45 ± 27.53). Significantly lower gene expression was observed in samples supplied with light Ln or the Ln mix. Gene expression ranged between 4.45 ± 0.29 (Nd) and 3.25 ± 0.57 (La) RPKM. The gene expression ratio of pedE and pedH differed depending on the Ln supply (Fig. 6B). In samples without Ln or with added Er, the pedE:pedH gene expression ratio was six and two, respectively. The pedE:pedH ratio shifted in favor of pedH in response to La, Nd, and the Ln mix. The expression of pedH was between 1.9-fold (Nd) and 4.2-fold (La) higher. The gene expression of pedE and pedH was additionally investigated via reverse transcription qPCR (RT-qPCR) from total RNA samples and making use of the 2-ΔΔCt method (37). For this purpose, cultivation was repeated for selected Ln elements (La, Er, controls without Ln), and samples for total RNA extraction were taken from the mid-exponential growth phase (OD600 between 0.29 ± 0.06 [no Ln] and 0.31 ± 0.01 [Er]). Compared with samples without added Ln (controls), La samples showed a 2.4-fold higher expression of pedH (Fig. S6). Samples with Er or without Ln, featured comparable pedH expression. Er samples showed higher gene expression of pedE compared with controls without Ln.
Fig 6.
Gene expression analysis of lanthanome-related genes. We considered the ped-cluster, the pea-cluster, and genes involved in PQQ biosynthesis as part of the lanthanome. Gene expression is plotted based on log2CPM values (A, left panel). Changes in gene expression are shown for each comparison (1 to 10 as described in Fig. 5) based on the respective fold change (FC) or its reciprocal (1/FC) in case of downregulation (A, right panel). The latter value describes the factor by which the expression of the respective gene is reduced. The Ln element-dependent gene expression ratios of the PQQ ADH-encoding genes pedE (PP_2674) and pedH (PP_2679) were calculated based on RPKM values (B).
Besides the ped-cluster, we also considered PQQ biosynthesis as part of the lanthanome. We noted a downregulation of PQQ biosynthesis-related genes (pqqBCDE [PP_0379, PP_0378, PP_0376, PP_2681]) when comparing La, Nd, and the Ln mix against Er and samples without Ln addition. Differential gene expression was not detected when light Ln samples were compared with each other or the Ln mix. There was no difference between the Er and no Ln samples. In addition, we looked at the pea-cluster (encoding genes of the 2-phenylethylamine pathway) since encoded proteins include quinoprotein dehydrogenases. So far, all known Ln-dependent enzymes are quinoprotein dehydrogenases, and we were wondering if genes belonging to the pea-cluster are responding to Ln supplementation. Genes associated with the pea-cluster (PP_3459–PP_3461, PP_5602) featured expression values between 6.21 ± 0.06 and 10.06 ± 0.09 log2CPM. Downregulation was observed when light Ln or the Ln mix was supplied (log2FC between −0.60 and −1.29).
DISCUSSION
Our findings suggest that the Ln sensing and signaling machinery in P. alloputida KT2440, encoded by pedE, pedH, and pedS2R2, responds differently to utilizable and non-utilizable Ln. Utilizable Ln reduce Ln signaling-related gene expression. A smaller pool of pedS2R2 transcripts presumably results in a reduction of PedS2 kinase activity, less phosphorylated PedR2, and ultimately reduced pedE expression. It is not clear if PedS2 binds heavier Ln, if heavier Ln reduce its activity, or if any other sensory elements are acting upstream of PedS2R2. Compared with cultures without added Ln, we did not observe an upregulation of pedH in response to Ln supplementation in RNAseq data sets. Except for Er, we noted the simultaneous downregulation of pedE and pedH in the presence of Ln, but also the reversion of their expression ratio. We also assessed changes in pedE and pedH expression by means of RT-qPCR, but had to use sample material from an independent incubation, due to a lack of original sample material. RT-qPCR revealed that pedH was upregulated in response to La, whereas its expression was stable, but not reduced, with added Er. The opposite was true for pedE. Expression was higher when Er was added and did not change significantly in the presence of La. Comparing RT-qPCR and RNAseq-derived data has inherent shortcomings. Different sets of incubations introduce biological variability. Both methods differ regarding sample processing. Factors, such as RNA quality, reverse transcription, and PCR efficiency add further uncertainty. Nonetheless, our data collectively show that the Ln switch is not acting as a simple on-off switch on the transcriptional level. Previously carried-out promoter activity assays (21) showed an increase of pedH and a decrease of pedE promoter activity in the presence of La.
Modulating the pool of PQQ ADH depending on Ln availability might be a mechanism to optimize substrate utilization. PedH features a higher Vmax and specific activity compared with PedE. At the same time, PedH has a much higher affinity for Ln than PedE for Ca (21). The downregulation of pedE, while pedH expression was unaltered when P. alloputida KT2440 was supplemented with Er, supports the idea that non-utilizable Ln might interfere with PedSR2 activity and the modulation of the PQQ ADH pool. Enzymatic activity assays have shown that PedH activity is highest with LREM and absent with heavy Ln, such as Er or Yb (21). Ln-dependent PQQ ADH can accommodate LREM and HREM with only minor changes in overall protein structure. However, differences in ionic radii and coordination number affect the catalytic efficiency of PQQ ADH and are potential reasons for bacteria preferring the larger and more abundant LREM (38). Accidental mismetallation of PedH with non-utilizable Ln and the reduced transcript pool of the second PQQ ADH, PedE, might explain the impaired growth observed with Er.
Droplet-based microfluidics combined with segmented flow is ideally suited for high-throughput cultivation and has been used for cultivating and characterizing various microbes, for instance, soil microorganisms, such as Rhodococcus sp. and Chromobacterium (39), cyanobacteria (40), as well as plant stem cells (41). Our one-dimensional microfluidic screenings suggested higher fitness with increasing La concentration, which matched the observed growth efficiency when using conventional cultivation. Considering that PedH is not active with Er (21), and changes in gene expression, the observed increase in autofluorescence variability for higher Er concentrations did not reflect higher fitness. Instead, it might be an indicator of increased oxidative stress, indicating a reduced overall fitness. Cellular autofluorescence in bacteria is primarily linked to NADH (NADH is fluorescent, emission spectrum 420–550 nm; NAD is not fluorescent) and, to a lesser extent, flavins. More noisy autofluorescence values suggest an effect on the NADH pool, which is easily affected by oxidative stress (42, 43).
Conventional cultivation in bottles has shown that supplementation with Er has a slight negative effect on growth. The performed two-dimensional screening suggested that potential negative effects mediated by Er were masked and compensated by the added La when strain KT2440 was exposed to both. This was in line with the positive effect seen for bottles supplemented with the Ln mix containing light and heavy Ln. For now, it remains unanswered how P. alloputida KT2440 recognizes and distinguishes between utilizable and non-utilizable Ln. In the future, coupling high-throughput microfluidic cultivation with downstream analyses, such as examining gene expression through RNAseq, might be a powerful strategy for studying Ln-related, phenotypic differences on multiple levels.
Observed growth patterns indicated that P. alloputida KT2440 interacts not only with light but also with heavy Ln. Intracellular Ln accumulation was previously shown for M. extorquens AM1 (18, 44) and Beijerinckiaceae bacterium RH AL1 (45). In the case of M. extorquens AM1, Ln are accumulated in the cytoplasm, whereas strain RH AL1 deposits Ln in the periplasm. In M. extorquens AM1 and Beijerinckiaceae bacterium RH AL1, Ln uptake into the peri- and cytoplasm is enabled by TonB-ABC transporter systems. The TonB-dependent receptor is in charge of uptake into the periplasm, and an ABC transporter facilitates translocation into the cytoplasm (18, 45, 46).
For P. alloputida KT2440, it was proposed that cytoplasmic Ln uptake relies on the PedA1A2BC (PP_2267–69, PP_5538) ABC transporter (22). We could neither identify extra- nor intracellular Ln accumulations in P. alloputida KT2440 through electron microscopy. Single-cell ICP-MS (47) enabled us to quantify cell-associated Ln. Cell-associated Lns in Ln-utilizing microbes have so far not been analyzed on the single-cell level. Despite only one known Ln-dependent protein, PedH, the determined values of cell-associated Ln (0.0025%–0.0068 % wet weight) are comparable to the total Fe content of a typical bacterial cell (48). We cannot distinguish between intra- and extracellular cell-associated Ln. Differences in the amount of cell-associated Ln are likely due to unspecific binding, governed by the intrinsic differences between Ln elements. Ln adsorption onto cell surfaces, with a preference for HREM, was previously shown for different organisms (49–51). We also showed this for Beijerinckiaceae bacterium RH AL1 (45). Unspecific extracellular binding, primarily through carboxyl and phosphoryl groups, skewed towards HREM, might facilitate the uptake of utilizable LREM.
Previous studies addressing Ln-utilizing methylotrophs revealed that Ln supplementation affected primarily xox- and mxa-cluster genes (52–55). Methylobacterium aquaticum 22A is the only microorganism featuring the Ln switch for which the effect of light (La) and heavy Ln (Ho, Lu) on gene expression was investigated over the whole transcriptome (53). Methylotrophy-related genes were sensitive to La addition, whereas Ho and Lu had no noticeable effect on gene expression. We could show that Ln have a tremendous effect on gene expression in Beijerinckiaceae bacterium RH AL1, which does not possess Ca-dependent PQQ ADH and lacks the Ln switch. Up to 41% of the encoded genes were differentially expressed in strain RH AL1 dependent on Ln supplementation (20).
We observed a clear difference between utilizable and non-utilizable Ln regarding gene expression. The upregulation of ribosomal protein genes L10 and L7/12; involved in ribosome stalk formation, translation factor recruitment, and GTP hydrolysis (56); when comparing La with Er and no Ln supplementation, suggested an overall effect on protein biosynthesis. The differential expression of numerous transcriptional regulator genes hinted towards dedicated mechanisms for transmitting changes in Ln availability. Genes encoding DUF2790-containing proteins were among the most differentially expressed genes dependent on Ln supplementation. This family of proteins of unknown function is conserved within the Pseudomonadaceae. The genome of P. alloputida KT2440 includes seven genes coding for DUF2790-containing proteins, of which four were responsive to Ln supplementation.
Some of the observed gene expression changes point towards Ln affecting the redox state of P. alloputida KT2440. Examples include the upregulation of genes linked to a spermidine/putrescine transport system when comparing La against Er and no Ln addition. Polyamines, such as spermidine and putrescine, function as antioxidants and signaling molecules (57, 58). When grown with La, Nd, or the Ln mix, genes associated with FeS cluster assembly were significantly more expressed, indicating higher demand, compared with cultures grown with Er or without Ln. Oxidative stress can cause an increased demand for functional FeS clusters (59). FeS cluster proteins are central to many forms of metabolism, including respiration, photosynthesis, as well as nitrogen fixation. FeS proteins can also function as sensors for intracellular oxygen and iron concentration (60).
Several genes for metal transporters showed differential gene expression when P. alloputida KT2440 was grown with Ln. Metalloproteins can accommodate different metals based on similar coordination geometry and the structure of the metal-binding site. Wehrmann and colleagues (22) could show that micromolar concentrations of Cu and Zn prevent the growth of a P. alloputida KT2440 pedE mutant when grown with nanomolar concentrations of Ln. Cu and Zn are known to strongly bind non-cognate metal-binding sites (61, 62).
P. alloputida KT2440 features two genes for phasin proteins (PP_5007, PP_5008), and one of them was significantly upregulated upon Ln supplementation. Scaffolds of phasin proteins surround polyhydroxybutyrate (PHB, a polyhydroxyalkanoate [PHA]) vacuoles (63). Besides from being linked to carbon storage, the presence of numerous proteins on their surface and their designation as carbonosomes highlights their assumed multifunctionality (64). PHB and related PHA are of economic relevance in the context of bioplastic production (65). For Beijerinckiaceae bacterium RH AL 1, we proposed that PHB plays a role in Ln uptake, periplasmic storage, and overall Ln homeostasis (20).
Concluding remarks
We provide evidence that Ln sensing in P. alloputida KT2440 is tuned towards utilizable Ln. Our findings support the idea that the Ln switch operates as a fine-tuning switch, modulating the pool of Ca- and Ln-dependent PQQ dependent on Ln availability. Ln-dependent gene expression changes have been opaque but were arguably skewed towards metal homeostasis and changes in the overall cellular redox state. The observed differences in cell-associated Ln demand a deeper investigation to clarify how P. alloputida KT2440 interacts with and discriminates between Ln. The continued and in-depth study of Ln-utilization in organisms beyond methylotrophy is needed to fully unravel the relevance of Ln in microbial physiology and thus its potential impact on microbial ecology and potential biotechnological applications.
MATERIALS AND METHODS
Cultivation
Pseudomonas alloputida KT2440 (DSM #6125) was maintained on solid MP (Methylobacterium PIPES) medium (66) supplemented with 2-phenylethanol (5 mM) at 30°C. Ln used for cultivation were supplemented as trichloride salts (retrieved in analytical grade from Carl Roth [Karlsruhe, Germany] and Sigma-Aldrich [Taufkirchen, Germany]). Incubation experiments were carried out with MP medium either in 96-well plates (BRANDplates, Brand + CO. KG, Wertheim, Germany) (cultivation volume 300 µL) or acid-washed 150 mL Erlenmeyer flasks (cultivation volume 50 mL). Pre-cultures were grown with succinate as the carbon source (25 mM). More information is given in the supplemental material.
Microfluidic cultivation
The setup used for microfluidic cultivation (Fig. S1) as well as its validation for two-dimensional screenings (Fig. S2) have been described in detail previously (67, 68). We used a syringe pump with six dosing units and a six-port droplet generator from Cetoni GmbH (Korbußen, Germany) to generate droplets with different compositions. Growth was tracked by recording the autofluorescence intensity It, which was normalized according to the initial measurement intensity It0. The background signal was determined based on the fluorinated-ethylene propylene (FEP) tubing filled with carrier liquid. Normalized autofluorescence units (NFU) were calculated as follows:
After droplet formation, droplets were passed through the transparent FEP tubes with an outer diameter (OD) of 1.6 mm and an inner diameter (ID) of 0.5 mm for photofluorimetric analysis (t = 0) and subsequently collected in connected PTFE tube coils (ID 0.5 mm and OD 1.0 mm) for incubation. Additional details, including information about the validation of the setup, can be found in the supplemental material.
Single-cell (sc) ICP-MS (inductively coupled plasma-mass spectrometry) analysis
Incubations were run as outlined before in 96-well plates, with three biological and six technical replicates. Incubations without Ln addition served as a control. Biomass was collected after the incubations reached OD600 values between 0.25 and 0.50. Technical replicates were pooled, harvested, and fixed with 500 µL of 2.5% (vol/vol) glutaraldehyde (in 100 mM cacodylate buffer) overnight at 4°C. Fixed and washed pellets were resuspended in 600 µL cacodylate buffer, and cell numbers were determined using a counting chamber. scICP-MS measurements were performed using an 8900 inductively coupled plasma-mass spectrometer (ICP-MS/MS) (Agilent Technologies, Inc., Santa Clara, CA, USA) equipped with a high-efficiency sample introduction system (Glass Expansion, Port Melbourne, Australia) composed of a total consumption spray chamber and a microconcentric nebulizer (47). Data acquisition is further outlined in the supplemental material. For determining the wet weight portion of Ln, we calculated with wet and dry weights of 1.7 g × L−1, and 0.4 g × L−1 per unit OD600 (69). Assuming a cell number of 7.8 × 108 × (mL × OD600)−1 (70), gave us a wet weight per cell of approximately 2.2 pg.
Electron microscopy
Transmission electron microscopy (TEM), and energy-dispersive X-ray spectroscopy (EDX) analyses were done as described previously (20, 45).
RNA extraction, mRNA enrichment, and sequencing library preparation
RNA extraction and subsequent mRNA enrichment were done based on previously described methods (45, 46, 71). Sequencing libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs, Germany). The size distribution of the libraries was checked by high-resolution gel electrophoresis with a Bioanalyzer instrument using the DNA 7500 Pico kit (Agilent Technologies). Libraries were quantified through fluorometry with a Qubit fluorometer and dsDNA HS reagents (Thermo Fisher Scientific, Darmstadt, Germany).
Sequencing and data pre-processing
An equimolar pool of the prepared libraries was sequenced with a NovaSeq 6000 instrument (Illumina, San Diego, California, USA) in paired-end mode (2 × 100 bp). Sequencing was carried out by the sequencing core facility of the Leibniz Institute on Aging-Fritz Lipmann Institute (Jena, Germany). We assessed the quality of raw and trimmed sequences with FastQC (v0.11.9) (72). Adaptor- and quality-trimming (settings: minlen = 75, qtrim = rl, ktrim = rl, k = 25, mink = 11, trimq = 20) were carried out with bbduk (v38.26) (73) using its included database of common sequence contaminants and adapters. The filtering of rRNA-derived reads, the mapping of mRNA-derived reads, and the generation of the read count table for subsequent differential gene expression analysis are detailed in the supplemental material. The read count data were assessed based on pseudocount distributions, determined biological coefficients of variation, as well as P-value distributions obtained from testing for differential gene expression between replicate groups of RNAseq data sets (Fig. S7 to S9).
Differential gene expression and cluster analysis
The R software framework for statistical computing (v4.2.1) and the package edgeR (v3.36.0) (74, 75) have been used to identify differentially expressed genes. We considered genes with changes in gene expression above |0.58| log2FC (FC), gene expression values higher than 4 log2counts per million (CPM), and FDR (false discovery rate) values smaller than 0.05 for differential gene expression analysis. Data exploration including cluster analysis is explained in more depth in the supplemental material.
Quantitative reverse transcription-PCR (RT-qPCR)
RT-qPCR was used to assess gene expression changes regarding pedE and pedH independent from RNAseq-based analysis using the 2-ΔΔCT method (37). The 16S rRNA gene was used as a constitutively expressed reference gene. Biomass samples originating from repeated incubations, set up the same way as outlined above, were subjected to the previously described extraction procedure for recovering total RNA. Details regarding the reverse transcription, subsequent qPCR, used primers, and cycling conditions can be found in the supplemental material.
Figure generation
We used the R software framework (v4.2.1) (76) for plotting and made use of the packages ggplot2 (v3.3.6) (77), gplots (v3.1.3) (78), ggpubr (v0.4.0) (79), cowplot (v1.1.1) (80), and upsetR (v1.4.0) (81), including their respective dependencies. Figures have been finalized with inkscape (https://inkscape.org/).
ACKNOWLEDGMENTS
The authors thank Josefine Bach, and Stefan Riedel (Friedrich Schiller University Jena, Aquatic Geomicrobiology Group) for help with cultivation work, especially growth monitoring. C.-E.W thanks Lena J. Daumann and Sophie Gutenthaler-Tietze (Heinrich Heine University Düsseldorf) for helpful discussions while preparing the manuscript. Sequencing was carried out by the sequencing core facility of the Leibniz Institute on Aging-Fritz Lipmann Institute (Jena, Germany).
J.C. gratefully acknowledges the financial support provided by a habilitation scholarship from the Technische Universität Ilmenau. This research was supported by the Deutsche Forschungsgemeinschaft (DFG) (grant: WE6579/4-1, granted to C.-E.W.; INST 275/441-1 FUGG granted to T.S. and S.H.), through the SFB 1127 ChemBioSys (project number 239748522), and by the de.NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI) and ELIXIR-DE (Forschungszentrum Jülich and W-de.NBI-001, W-de.NBI-004, W-de.NBI-008, W-de.NBI-010, W-de.NBI-013, W-de.NBI-014, W-de.NBI-016, W-de.NBI-022). Open access publishing was facilitated by the DFG (project number 512648189) and the Open Access Publication Fund of the Thueringer Universitaets- und Landesbibliothek Jena.
Contributor Information
Carl-Eric Wegner, Email: carl-eric.wegner@hhu.de.
Craig D. Ellermeier, The University of Iowa, Iowa City, Iowa, USA
DATA AVAILABILITY
RNAseq data sets are available via EBI/ENA ArrayExpress (accession: E-MTAB-13102) (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-13102). Details about data processing and analysis are also provided via the Open Science Framework (https://osf.io/ynsdc/). We also provide a reproducible, snakemake-based (82, 83) workflow for RNAseq data processing (https://github.com/wegnerce/smk_rnaseq, release v0.1).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msphere.00685-24.
Additional details regarding used methods, supplemental figures, and legends for supplemental tables.
Tables S1 to S15.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Gonzalez V, Vignati DAL, Leyval C, Giamberini L. 2014. Environmental fate and ecotoxicity of lanthanides: are they a uniform group beyond chemistry? Environ Int 71:148–157. doi: 10.1016/j.envint.2014.06.019 [DOI] [PubMed] [Google Scholar]
- 2. Kotelnikova AD, Rogova OB, Stolbova VV. 2021. Lanthanides in the soil: routes of entry, content, effect on plants, and genotoxicity (a review). Eur Soil Sci 54:117–134. [Google Scholar]
- 3. . Rare earths in the energy transition: what threats are there for the “vitamins of modern society”? IFPEN. Available from: https://www.ifpenergiesnouvelles.com/article/les-terres-rares-transition-energetique-quelles-menaces-les-vitamines-lere-moderne. Retrieved 1 Apr 2022.
- 4. Cotruvo JA. 2019. The chemistry of lanthanides in biology: recent discoveries, emerging principles, and technological applications. ACS Cent Sci 5:1496–1506. doi: 10.1021/acscentsci.9b00642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daumann LJ. 2019. Essential and ubiquitous: the emergence of lanthanide metallobiochemistry. Angew Chem Int Ed Engl 58:12795–12802. doi: 10.1002/anie.201904090 [DOI] [PubMed] [Google Scholar]
- 6. Anthony C, Zatman LJ. 1967. The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. Biochem J 104:960–969. doi: 10.1042/bj1040960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anthony C, Zatman LJ. 1967. The microbial oxidation of methanol. Purification and properties of the alcohol dehydrogenase of Pseudomonas sp. M27. Biochem J 104:953–959. doi: 10.1042/bj1040953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keltjens JT, Pol A, Reimann J, Op den Camp HJM. 2014. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol 98:6163–6183. doi: 10.1007/s00253-014-5766-8 [DOI] [PubMed] [Google Scholar]
- 9. Wilson MC, Mori T, Rückert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UAE, Heycke N, Schmitt S, Rinke C, Helfrich EJN, Brachmann AO, Gurgui C, Wakimoto T, Kracht M, Crüsemann M, Hentschel U, Abe I, Matsunaga S, Kalinowski J, Takeyama H, Piel J. 2014. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506:58–62. doi: 10.1038/nature12959 [DOI] [PubMed] [Google Scholar]
- 10. Butterfield CN, Li Z, Andeer PF, Spaulding S, Thomas BC, Singh A, Hettich RL, Suttle KB, Probst AJ, Tringe SG, Northen T, Pan C, Banfield JF. 2016. Proteogenomic analyses indicate bacterial methylotrophy and archaeal heterotrophy are prevalent below the grass root zone. PeerJ 4:e2687. doi: 10.7717/peerj.2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chistoserdova L, Kalyuzhnaya MG. 2018. Current trends in methylotrophy. Trends Microbiol 26:703–714. doi: 10.1016/j.tim.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 12. Chu F, Lidstrom ME. 2016. XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J Bacteriol 198:1317–1325. doi: 10.1128/JB.00959-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu F, Beck DAC, Lidstrom ME. 2016. MxaY regulates the lanthanide-mediated methanol dehydrogenase switch in Methylomicrobium buryatense. PeerJ 4:e2435. doi: 10.7717/peerj.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu Z, Chistoserdova L. 2017. Communal metabolism of methane and the rare earth element switch. J Bacteriol 199:e00328-17. doi: 10.1128/JB.00328-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wehrmann M, Berthelot C, Billard P, Klebensberger J. 2018. The PedS2/PedR2 two-component system is crucial for the rare earth element switch in Pseudomonas putida KT2440. mSphere 3:e00376-18. doi: 10.1128/mSphere.00376-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Groom JD, Ford SM, Pesesky MW, Lidstrom ME. 2019. A mutagenic screen identifies a TonB-dependent receptor required for the lanthanide metal switch in the type I methanotroph “Methylotuvimicrobium buryatense” 5GB1C. J Bacteriol 201:e00120-19. doi: 10.1128/JB.00120-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crombie AT. 2022. The effect of lanthanum on growth and gene expression in a facultative methanotroph. Environ Microbiol 24:596–613. doi: 10.1111/1462-2920.15685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roszczenko-Jasińska P, Vu HN, Subuyuj GA, Crisostomo RV, Cai J, Lien NF, Clippard EJ, Ayala EM, Ngo RT, Yarza F, Wingett JP, Raghuraman C, Hoeber CA, Martinez-Gomez NC, Skovran E. 2020. Gene products and processes contributing to lanthanide homeostasis and methanol metabolism in Methylorubrum extorquens AM1. Sci Rep 10:12663. doi: 10.1038/s41598-020-69401-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zytnick AM, Gutenthaler-Tietze SM, Aron AT, Reitz ZL, Phi MT, Good NM, Petras D, Daumann LJ, Martinez-Gomez NC. 2024. Identification and characterization of a small-molecule metallophore involved in lanthanide metabolism. Proc Natl Acad Sci U S A 121:e2322096121. doi: 10.1073/pnas.2322096121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gorniak L, Bechwar J, Westermann M, Steiniger F, Wegner C-E. 2023. Different lanthanide elements induce strong gene expression changes in a lanthanide-accumulating methylotroph. Microbiol Spectr 11:e0086723. doi: 10.1128/spectrum.00867-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J. 2017. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. MBio 8:e00570-17. doi: 10.1128/mBio.00570-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wehrmann M, Berthelot C, Billard P, Klebensberger J. 2019. Rare earth element (REE)-dependent growth of Pseudomonas putida KT2440 relies on the ABC-transporter PedA1A2BC and is influenced by iron availability. Front Microbiol 10:2494. doi: 10.3389/fmicb.2019.02494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wehrmann M, Toussaint M, Pfannstiel J, Billard P, Klebensberger J. 2020. The cellular response to lanthanum is substrate specific and reveals a novel route for glycerol metabolism in Pseudomonas putida KT2440. MBio 11:e00516-20. doi: 10.1128/mBio.00516-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiménez JI, Miñambres B, García JL, Díaz E. 2002. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4:824–841. doi: 10.1046/j.1462-2920.2002.00370.x [DOI] [PubMed] [Google Scholar]
- 25. Dos Santos V, Heim S, Moore ERB, Strätz M, Timmis KN. 2004. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ Microbiol 6:1264–1286. doi: 10.1111/j.1462-2920.2004.00734.x [DOI] [PubMed] [Google Scholar]
- 26. Belda E, van Heck RGA, José Lopez-Sanchez M, Cruveiller S, Barbe V, Fraser C, Klenk H-P, Petersen J, Morgat A, Nikel PI, Vallenet D, Rouy Z, Sekowska A, Martins Dos Santos VAP, de Lorenzo V, Danchin A, Médigue C. 2016. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ Microbiol 18:3403–3424. doi: 10.1111/1462-2920.13230 [DOI] [PubMed] [Google Scholar]
- 27. Sprouffske K, Wagner A. 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17:172. doi: 10.1186/s12859-016-1016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crow JF, Kimura M.. An introduction to population genetics theory. Harper & Row. New York, USA. [Google Scholar]
- 29. Rockwood LL. 2015. Introduction to population ecology. John Wiley & Sons. New York, USA. [Google Scholar]
- 30. Cao J, Richter F, Kastl M, Erdmann J, Burgold C, Dittrich D, Schneider S, Köhler JM, Groß GA. 2020. Droplet-based screening for the investigation of microbial nonlinear dose-response characteristics system, background and examples. Micromachines (Basel) 11:577. doi: 10.3390/mi11060577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin X, Han J. 2010. K-means clustering, p 563–564. In Sammut C, Webb GI (ed), Encyclopedia of machine learning. Springer US, Boston, MA. [Google Scholar]
- 32. Caliński T, Harabasz J.. 1974. A dendrite method for cluster analysis. Commun Stat Simul Comput 3:1–27. [Google Scholar]
- 33. Rousseeuw PJ. 1987. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65. [Google Scholar]
- 34. Tibshirani R, Walther G, Hastie T. 2001. Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Series B Stat Methodol 63:411–423. [Google Scholar]
- 35. Mattocks JA, Ho JV, Cotruvo JA. 2019. A selective, protein-based fluorescent sensor with picomolar affinity for rare earth elements. J Am Chem Soc 141:2857–2861. doi: 10.1021/jacs.8b12155 [DOI] [PubMed] [Google Scholar]
- 36. Arias S, Olivera ER, Arcos M, Naharro G, Luengo JM. 2008. Genetic analyses and molecular characterization of the pathways involved in the conversion of 2-phenylethylamine and 2-phenylethanol into phenylacetic acid in Pseudomonas putida U. Environ Microbiol 10:413–432. doi: 10.1111/j.1462-2920.2007.01464.x [DOI] [PubMed] [Google Scholar]
- 37. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 38. Jahn B, Pol A, Lumpe H, Barends TRM, Dietl A, Hogendoorn C, Op den Camp HJM, Daumann LJ. 2018. Similar but not the same: first kinetic and structural analyses of a methanol dehydrogenase containing a europium ion in the active site. Chembiochem 19:1147–1153. doi: 10.1002/cbic.201800130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cao J, Pliquett U, Yang L, Wiedemeier S, Cahill B, Michael Köhler J. 2022. Contactless optical and impedimetric sensing for droplet-based dose-response investigations of microorganisms. Sens Actuat B Chem 372:132688. doi: 10.1016/j.snb.2022.132688 [DOI] [Google Scholar]
- 40. Cao J, Russo DA, Xie T, Groß GA, Zedler JAZ. 2022. A droplet-based microfluidic platform enables high-throughput combinatorial optimization of cyanobacterial cultivation. Sci Rep 12:15536. doi: 10.1038/s41598-022-19773-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Richter F, Chen M, Schaub P, Wüst F, Zhang D, Schneider S, Groß GA, Mäder P, Dovzhenko O, Palme K, Köhler JM, Cao J. 2022. Induction of embryogenic development in haploid microspore stem cells in droplet-based microfluidics. Lab Chip 22:4292–4305. doi: 10.1039/d2lc00788f [DOI] [PubMed] [Google Scholar]
- 42. Surre J, Saint-Ruf C, Collin V, Orenga S, Ramjeet M, Matic I. 2018. Strong increase in the autofluorescence of cells signals struggle for survival. Sci Rep 8:12088. doi: 10.1038/s41598-018-30623-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schaefer PM, Kalinina S, Rueck A, von Arnim CAF, von Einem B. 2019. NADH autofluorescence-a marker on its way to boost bioenergetic research. Cytometry A 95:34–46. doi: 10.1002/cyto.a.23597 [DOI] [PubMed] [Google Scholar]
- 44. Good NM, Lee HD, Hawker ER, Su MZ, Gilad AA, Martinez-Gomez NC. 2022. Hyperaccumulation of gadolinium by Methylorubrum extorquens AM1 reveals impacts of lanthanides on cellular processes beyond methylotrophy. Front Microbiol 13:820327. doi: 10.3389/fmicb.2022.820327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wegner C-E, Westermann M, Steiniger F, Gorniak L, Budhraja R, Adrian L, Küsel K. 2021. Extracellular and intracellular lanthanide accumulation in the methylotrophic Beijerinckiaceae bacterium RH AL1. Appl Environ Microbiol 87:e0314420. doi: 10.1128/AEM.03144-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wegner C-E, Gorniak L, Riedel S, Westermann M, Küsel K. 2019. Lanthanide-dependent methylotrophs of the family Beijerinckiaceae: physiological and genomic insights. Appl Environ Microbiol 86:e01830-19. doi: 10.1128/AEM.01830-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hellmann S, García-Cancela P, Alonso-Fernández S, Corte-Rodríguez M, Bettmer J, Manteca A, Merten D, Gil-Díaz T, Schäfer T, Montes-Bayón M. 2024. Single cell ICP-MS to evaluate the interaction behaviour for Cd, Ce and U with Streptomyces coelicolor spores. Chemosphere 347:140633. doi: 10.1016/j.chemosphere.2023.140633 [DOI] [PubMed] [Google Scholar]
- 48. Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol 181:1415–1428. doi: 10.1128/JB.181.5.1415-1428.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Texier A-C, Andrès Y, Le Cloirec P. 1999. Selective biosorption of lanthanide (La, Eu, Yb) Ions by Pseudomonas aeruginosa. Environ Sci Technol 33:489–495. doi: 10.1021/es9807744 [DOI] [Google Scholar]
- 50. Texier A-C, Andrès Y, Illemassene M, Le Cloirec P. 2000. Characterization of lanthanide ions binding sites in the cell wall of Pseudomonas aeruginosa. Environ Sci Technol 34:610–615. doi: 10.1021/es990668h [DOI] [Google Scholar]
- 51. Maleke M, Valverde A, Vermeulen J-G, Cason E, Gomez-Arias A, Moloantoa K, Coetsee-Hugo L, Swart H, van Heerden E, Castillo J. 2019. Biomineralization and bioaccumulation of europium by a thermophilic metal resistant bacterium. Front Microbiol 10:81. doi: 10.3389/fmicb.2019.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gu W, Farhan Ul Haque M, DiSpirito AA, Semrau JD. 2016. Uptake and effect of rare earth elements on gene expression in Methylosinus trichosporium OB3b. FEMS Microbiol Lett 363:1–6. doi: 10.1093/femsle/fnw129 [DOI] [PubMed] [Google Scholar]
- 53. Masuda S, Suzuki Y, Fujitani Y, Mitsui R, Nakagawa T, Shintani M, Tani A. 2018. Lanthanide-dependent regulation of methylotrophy in Methylobacterium aquaticum strain 22A. mSphere 3. doi: 10.1128/mSphere.00462-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Good NM, Moore RS, Suriano CJ, Martinez-Gomez NC. 2019. Contrasting in vitro and in vivo methanol oxidation activities of lanthanide-dependent alcohol dehydrogenases XoxF1 and ExaF from Methylobacterium extorquens AM1. Sci Rep 9:4248. doi: 10.1038/s41598-019-41043-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ochsner AM, Hemmerle L, Vonderach T, Nüssli R, Bortfeld-Miller M, Hattendorf B, Vorholt JA. 2019. Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol Microbiol 111:1152–1166. doi: 10.1111/mmi.14208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Diaconu M, Kothe U, Schlünzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121:991–1004. doi: 10.1016/j.cell.2005.04.015 [DOI] [PubMed] [Google Scholar]
- 57. Tabor CW, Tabor H. 1985. Polyamines in microorganisms. Microbiol Rev 49:81–99. doi: 10.1128/mr.49.1.81-99.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rhee HJ, Kim E-J, Lee JK. 2007. Physiological polyamines: simple primordial stress molecules. J Cell Mol Med 11:685–703. doi: 10.1111/j.1582-4934.2007.00077.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mettert EL, Kiley PJ. 2015. How is Fe-S cluster formation regulated? Annu Rev Microbiol 69:505–526. doi: 10.1146/annurev-micro-091014-104457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Braymer JJ, Freibert SA, Rakwalska-Bange M, Lill R. 2021. Mechanistic concepts of iron-sulfur protein biogenesis in biology. Biochim Biophys Acta Mol Cell Res 1868:118863. doi: 10.1016/j.bbamcr.2020.118863 [DOI] [PubMed] [Google Scholar]
- 61. Irving H, Williams RJP. 1953. The stability of transition-metal complexes. J Chem Soc:3192. doi: 10.1039/jr9530003192 [DOI] [Google Scholar]
- 62. Foster AW, Osman D, Robinson NJ. 2014. Metal preferences and metallation. J Biol Chem 289:28095–28103. doi: 10.1074/jbc.R114.588145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bresan S, Sznajder A, Hauf W, Forchhammer K, Pfeiffer D, Jendrossek D. 2016. Polyhydroxyalkanoate (PHA) granules have no phospholipids. Sci Rep 6:26612. doi: 10.1038/srep26612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191:3195–3202. doi: 10.1128/JB.01723-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McAdam B, Brennan Fournet M, McDonald P, Mojicevic M. 2020. Production of polyhydroxybutyrate (PHB) and factors impacting its chemical and mechanical characteristics. Polymers (Basel) 12:2908. doi: 10.3390/polym12122908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Delaney NF, Kaczmarek ME, Ward LM, Swanson PK, Lee M-C, Marx CJ. 2013. Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS One 8:e62957. doi: 10.1371/journal.pone.0062957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cao J, Kürsten D, Krause K, Kothe E, Martin K, Roth M, Köhler JM. 2013. Application of micro-segmented flow for two-dimensional characterization of the combinatorial effect of zinc and copper ions on metal-tolerant Streptomyces strains. Appl Microbiol Biotechnol 97:8923–8930. doi: 10.1007/s00253-013-5147-8 [DOI] [PubMed] [Google Scholar]
- 68. Xie T, Köhler JM, Heyder S, Günther PM, Cao J. 2022. Microfluidically-assisted isolation and characterization of Achromobacter spanius from soils for microbial degradation of synthetic polymers and organic solvents. Environments 9:147. doi: 10.3390/environments9120147 [DOI] [Google Scholar]
- 69. Glazyrina J, Materne E-M, Dreher T, Storm D, Junne S, Adams T, Greller G, Neubauer P. 2010. High cell density cultivation and recombinant protein production with Escherichia coli in a rocking-motion-type bioreactor. Microb Cell Fact 9:42. doi: 10.1186/1475-2859-9-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Volkmer B, Heinemann M. 2011. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS ONE 6:e23126. doi: 10.1371/journal.pone.0023126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wegner C-E, Richter-Heitmann T, Klindworth A, Klockow C, Richter M, Achstetter T, Glöckner FO, Harder J. 2013. Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar Genomics 9:51–61. doi: 10.1016/j.margen.2012.12.001 [DOI] [PubMed] [Google Scholar]
- 72. Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 73. Bushnell B. 2016. BBMap short read aligner. Available from: https://www.sourceforge.net/projects/bbmap/
- 74. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. doi: 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Team R Core. 2022. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 77. Wickham H. 2016. Ggplot2: Elegant Graphics for Data Analysis. Springer, New York. https://ggplot2.tidyverse.org. [Google Scholar]
- 78. Warnes GR, Bolker B, Bonebakker L, Genteman R, Liaw WHA, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. 2012. Gplots: various R programming tools for plotting data. R package version 3.1.3. Available from: http://cran.r-project.org/package=gplots
- 79. Kassambara A. 2018. ggpubr: 'ggplot2' based publication ready plots. R package version 0.4.0. Available from: https://CRAN.R-project.org/package=ggpubr
- 80. Wilke CO. 2019. cowplot: streamlined plot theme and plot annotations for 'ggplot2' R package version 1.0. 0. Available from: https://CRAN.R-project.org/package=cowplot
- 81. Conway JR, Lex A, Gehlenborg N. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33:2938–2940. doi: 10.1093/bioinformatics/btx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Köster J, Rahmann S. 2012. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics 28:2520–2522. doi: 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- 83. Mölder F, Jablonski KP, Letcher B, Hall MB, Tomkins-Tinch CH, Sochat V, Forster J, Lee S, Twardziok SO, Kanitz A, Wilm A, Holtgrewe M, Rahmann S, Nahnsen S, Köster J. 2021. Sustainable data analysis with snakemake. F1000Res 10:33. doi: 10.12688/f1000research.29032.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional details regarding used methods, supplemental figures, and legends for supplemental tables.
Tables S1 to S15.
Data Availability Statement
RNAseq data sets are available via EBI/ENA ArrayExpress (accession: E-MTAB-13102) (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-13102). Details about data processing and analysis are also provided via the Open Science Framework (https://osf.io/ynsdc/). We also provide a reproducible, snakemake-based (82, 83) workflow for RNAseq data processing (https://github.com/wegnerce/smk_rnaseq, release v0.1).