Abstract
PURPOSE
In lung squamous cell carcinoma (LUSC), Black patients show significantly higher incidence and lower overall survival than White patients. Although socioeconomic factors likely contribute to this survival disparity, genomic factors have yet to be elucidated in LUSC.
METHODS
Using 416 LUSC tumor samples in the Cancer Genome Atlas (TCGA), we assessed genomic and transcriptomic profiles by ancestry. We replicated our analyses in pan-cancer data from TCGA, the American Association of Cancer Research (AACR) Genomics Evidence Neoplasia Information Exchange (GENIE), and Columbia University Medical Center.
RESULTS
We found increased MYC amplification, LUSC-specific MYC enhancer amplification, and chromosome arm 8q (chr8q) gain to be significantly associated with genetic AFR (African) ancestry in LUSC in TCGA. Furthermore, expression of MYC target genes was significantly enriched in AFR samples. Local ancestry analysis identified correlation of chr8q gain with AFR ancestry at the MYC locus in TCGA. We also found a significant correlation between chr8q and AFR ancestry in multiple cancer types and pan-cancer in TCGA. Similarly, in a pan-cancer subset of AACR GENIE data, we found a significant correlation between chr8q gain and race.
CONCLUSION
Together, our data suggest that ancestry may influence amplification of not only MYC but also its enhancer in LUSC. They also suggest a role for genetic ancestry in chr8q aneuploidy in cancer. These studies further define and expand patients who may benefit from future anti-MYC therapeutic approaches.
Amplification of a squamous-specific chr8q MYC enhancer is associated with African ancestry in lung squamous cell carcinoma
INTRODUCTION
The morbidity and mortality of lung cancer disproportionately affect racial and ethnic minorities, particularly Black/African American patients.1-3 This disparity is thought to be largely attributable to socioeconomic and environmental factors.1-10 However, germline risk variants and differential somatic alterations have been identified in Black populations for colorectal, breast, and prostate cancers11-15; this has informed population-specific risk stratification and screening paradigms.5,16-18 Cancer sequencing databases feature disproportionately low samples from Black patients and other racial/ethnic minorities.19
CONTEXT
Key Objective
Lung cancer incidence and mortality remain disproportionately higher in Black patients in the United States, despite a population-level decline in smoking comparable with that of White patients. We sought to characterize genomic hallmarks of lung squamous cell tumors that correlate with genetic ancestry to investigate the presence of targetable, ancestry-specific mechanisms of tumor progression.
Knowledge Generated
We show that amplification of the oncogene MYC, a lung squamous cell carcinoma (LUSC) MYC enhancer locus, and chromosome 8q is significantly associated with African ancestry in LUSC. We also show that across all cancers, chr8q gain is significantly associated with African ancestry.
Relevance
As approaches to target MYC therapeutically are developed, these findings emphasize the importance of including patients of African ancestry in translational and clinical studies in LUSC.
Many recent studies have leveraged genetic ancestry to overcome limitations of self-reported race-based analyses and elucidate ancestry-specific patterns in tumor biology.20,21 A comprehensive analysis of ancestry-related correlates in cancer found many tissue-specific effects and identified ancestry-specific mutation rates and genomic hallmarks.20
Here, in 416 lung squamous cell carcinoma (LUSC) tumors from the Cancer Genome Atlas (TCGA),22 we find that copy number gains of MYC, a LUSC-specific enhancer on chr8q24.21, and chromosome arm 8q (chr8q) all correlate with African (AFR) ancestry. We replicate our findings in pan-cancer cohorts, including TCGA, AACR GENIE, and a clinical cohort at the Columbia University Irving Medical Center (CUIMC). Our results suggest ancestry-specific patterns of oncogenic alterations that may modulate treatment response and survival in LUSC.
METHODS
Data Sets
TCGA
A total of 10,522 TCGA samples were assigned consensus genetic ancestry calls20 and filtered for consensus genetic ancestry assignments of predominantly European (EUR), AFR, or AFR plus other genetic ancestry (AFR_admix), comprising 9,897 TCGA samples. Data used include DNA mutation, DNA copy number, and gene expression (see below for data availability). Additional analyses were performed for the 416 LUSC samples, of which 24 were of AFR ancestry and 392 were of EUR ancestry. These numbers provide over an 80% power to detect three-fold changes for any event occurring at least 10% of the time. Clinical annotations, including overall survival status, overall survival months, progression-free survival status, progression-free months, TNM staging, age, sex, and race, were obtained from cBioPortal.23 Aneuploidy calls in TCGA were previously described.24
AACR GENIE
A total of 7,344 pan-cancer Memorial Sloan Kettering Cancer Center (MSKCC) samples from the AACR Project GENIE data set, filtered for Black or White self-identified race (MSKCC), had available segmented copy number files, as generated by FACETS from panel sequencing.25
Columbia University
A total of 1,040 deidentified pan-cancer primary clinical samples at the CUIMC, filtered for Black or White self-identified race, had been sequenced using FoundationOne for solid tumors.26 Only MYC amplification data were assessed.
Statistical Methods
All t-tests were performed using Welch's unpaired two-tailed t-test, using the t.test() function in R with default parameters. All simple correlations were performed using Pearson's method, using the cor.test() function in R. Chi-squared analysis was performed using Pearson's chi-square method, using the chi.sq() function in R. For local ancestry analysis, we used the analysis of variance (ANOVA) test to assess continuous data across three categorical genotypes, using the aov() function in R. This was followed by post hoc pairwise t-tests. Linear modeling was performed in R version 4.0.5 using the lm() function. The HiChIP heatmap was made using gTrack R package.
P values were adjusted for multiple corrections for the mutation-ancestry correlations, differential gene expression analysis, and aneuploidy correlations. These were adjusted by either a false discovery rate (FDR) of <0.1 or a Bonferroni cutoff of <0.1, specified in the text. P values for copy number variation (CNV)-ancestry correlations were not adjusted, as described in the text.
Ethics Approval and Consent to Participate
Studies involved data from samples from patients that were deidentified, namely, in the three data sets used, TCGA, AACR GENIE, and CUIMC panel data. TCGA and AACR GENIE data are publicly available.
RESULTS
AFR Ancestry Correlates With Amplification of MYC, Its Enhancer on Chr8q24.21, and Chr8q in LUSC
We first wanted to characterize any ancestry-associated genomic differences. We analyzed 416 LUSC tumors with either EUR consensus ancestry or AFR/AFR admix consensus ancestry (Table 1).20 We observed comparable tumor mutation burden (TMB), fraction of genome altered (FGA), and aneuploidy between AFR and EUR tumors (Data Supplement, Fig S1).
TABLE 1.
Summary of Clinical Information for All Patients With LUSC in TCGA
| Clinical Subsets Within LUSC TCGA | No. of Patients |
|---|---|
| Total | 416 |
| Self-identified race | |
| Black or African American | 24 |
| White | 288 |
| Not annotated | 104 |
| Consensus genetic ancestry | |
| AFR or AFR-admix | 24 |
| EUR | 392 |
| Sex | |
| Male | 307 |
| Female | 107 |
| Not annotated | 2 |
| Disease stage | |
| Stage I | 2 |
| Stage IA | 75 |
| Stage IB | 123 |
| Stage II | 1 |
| Stage IIA | 51 |
| Stage IIB | 82 |
| Stage III | 3 |
| Stage IIIA | 55 |
| Stage IIIB | 16 |
| Stage IV | 5 |
| Not annotated | 3 |
| Tumor stage | |
| T1 | 40 |
| T1A | 20 |
| T1B | 35 |
| T2 | 143 |
| T2A | 67 |
| T2B | 26 |
| T3 | 63 |
| T4 | 20 |
| Not annotated | 2 |
| Lymph node stage | |
| N0 | 262 |
| N1 | 108 |
| N2 | 35 |
| N3 | 5 |
| NX (unknown) | 4 |
| Not annotated | 2 |
| Metastatic stage | |
| M0 | 345 |
| M1 | 5 |
| MX (unknown) | 62 |
| Not annotated | 4 |
Abbreviations: AFR, African; EUR, European; LUSC, lung squamous cell carcinoma; TCGA, the Cancer Genome Atlas.
Although TMB did not correlate with ancestry, we wanted to discern whether any individual mutations occurred at significantly differential frequencies in association with ancestry in LUSC. We correlated percent AFR ancestry with mutation frequency across 21,387 genes, ranking mutations by significance with a cutoff of Bonferroni-adjusted P <.1. Although no mutations present at a frequency of 5% or greater were significantly associated with AFR ancestry, we found a subset of rare mutations occurring either significantly more frequently or exclusively in the AFR ancestry group that requires further replication in a larger cohort (Data Supplement, Fig S2).
We next wanted to identify whether individual somatic copy number alterations (SCNAs) correlated with genetic ancestry. We correlated amplified and deleted loci identified by GISTIC27 with percent AFR admixture, using a cutoff of unadjusted P = .05 to identify significant ancestry-associated SCNAs (Data Supplement, Table S1). Amplification of chr8q24.21 (Pearson's coefficient, 0.49; P = .028) and amplification of chr8q11.21 (Pearson's coefficient, 0.64; P = .005), among other events, were significantly associated with percentage of AFR ancestry (Fig 1A). Chr8q24.21 is amplified in two thirds of LUSC tumors, and its copy number was 1.43-fold higher in samples of AFR ancestry (Welch's t-test P = .007; Fig 1B). Copy number of chr8q24.21 significantly correlated with percent AFR admixture after correcting for FGA and tumor stage (coefficient, 0.420; P = .0012; Fig 1C). The amplicon on chr8q24.21 overlaps with an H3K27ac ChIP-seq peak identified in the LUSC cell line HARA; this region also interacts with MYC by HiCHIP28,29 (Fig 1D). These data suggest that a LUSC-specific MYC enhancer is located in the ancestry-correlated amplification peak.
FIG 1.
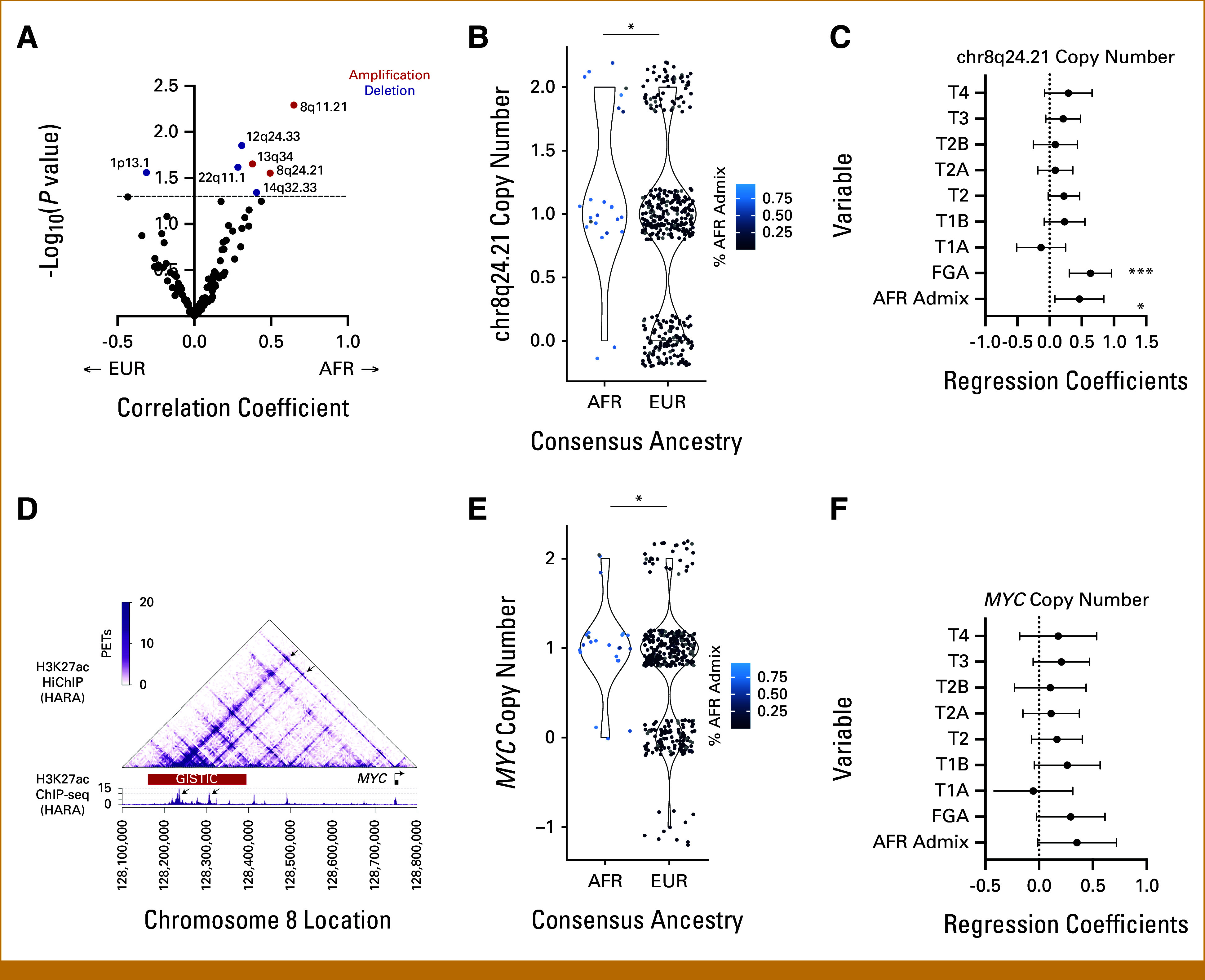
SCNAs associated with ancestry in AFR and EUR samples in LUSC in TCGA. (A) Volcano plot of focal SCNA frequency. Correlation with percent AFR admixture (X-axis) is plotted against log-transformed unadjusted P values (Y-axis). The dotted line marks the significance threshold of P = .05. Significant amplification events are colored red, and significant deletion events are colored blue. Copy number of (B) chr8q24.21 and (E) MYC gene by ancestry. Gradient color scale of data points represents percent AFR admixture, from black at 0% to blue at 100% (*P < .05; **P < .01; ***P < .005). Forest plots depicting multivariate linear regression models of (C) chr8q24.21 and (F) MYC copy number as a function of tumor stage, fraction genome altered, and AFR admixture, with variables listed along the Y-axis. Regression coefficients are plotted on the x-axis with a dotted line at x = 0. (D) Top: H3K27ac HiChIP data presenting the chromatin interactions at the MYC locus in the LUSC cell line HARA. Bottom: H3K27ac ChIP-seq data indicating two strong enhancers present in the GISTIC copy number peak (AFR). Arrows in the HiChIP heatmap indicate the interactions between the MYC promoter and the two highlighted enhancers within the GISTIC peak. AFR, African; EUR, European; LUSC, lung squamous cell carcinoma; SCNAs, somatic copy number alterations; TCGA, the Cancer Genome Atlas.
We also found copy number of the MYC gene to be significantly increased in AFR samples compared with EUR (Welch's t-test P = .018; Fig 1E). Though trending, MYC copy number did not significantly correlate with percent AFR admixture after correcting for FGA and tumor stage (Fig 1F). Copy number of MYC (Pearson's coefficient, 0.239; P = 7.8e-07; Data Supplement, Fig S3A) and the chr8q24.21 enhancer peak (Pearson's coefficient, 0.234; P = 1.4e-06; Data Supplement, Fig S3B) both correlated significantly with MYC gene expression.
We used a similar approach to identify significant aneuploidy events, correlating chromosome arm calls24 with percent AFR admixture. We found chr8q copy number to be significantly associated with percentage AFR ancestry (Pearson's coefficient, 0.115; P = .02; Fig 2A) and consensus ancestry (fold change, 1.79; P = .002; Fig 2B). This was still significant when correcting for FGA and tumor stage (coefficient, 0.422; P = .0248; Fig 2C). Overall, we find increased copy number of chr8q, amplification of an enhancer region of chr8q24.21, and amplification of the MYC gene to be associated with global AFR ancestry in LUSC.
FIG 2.
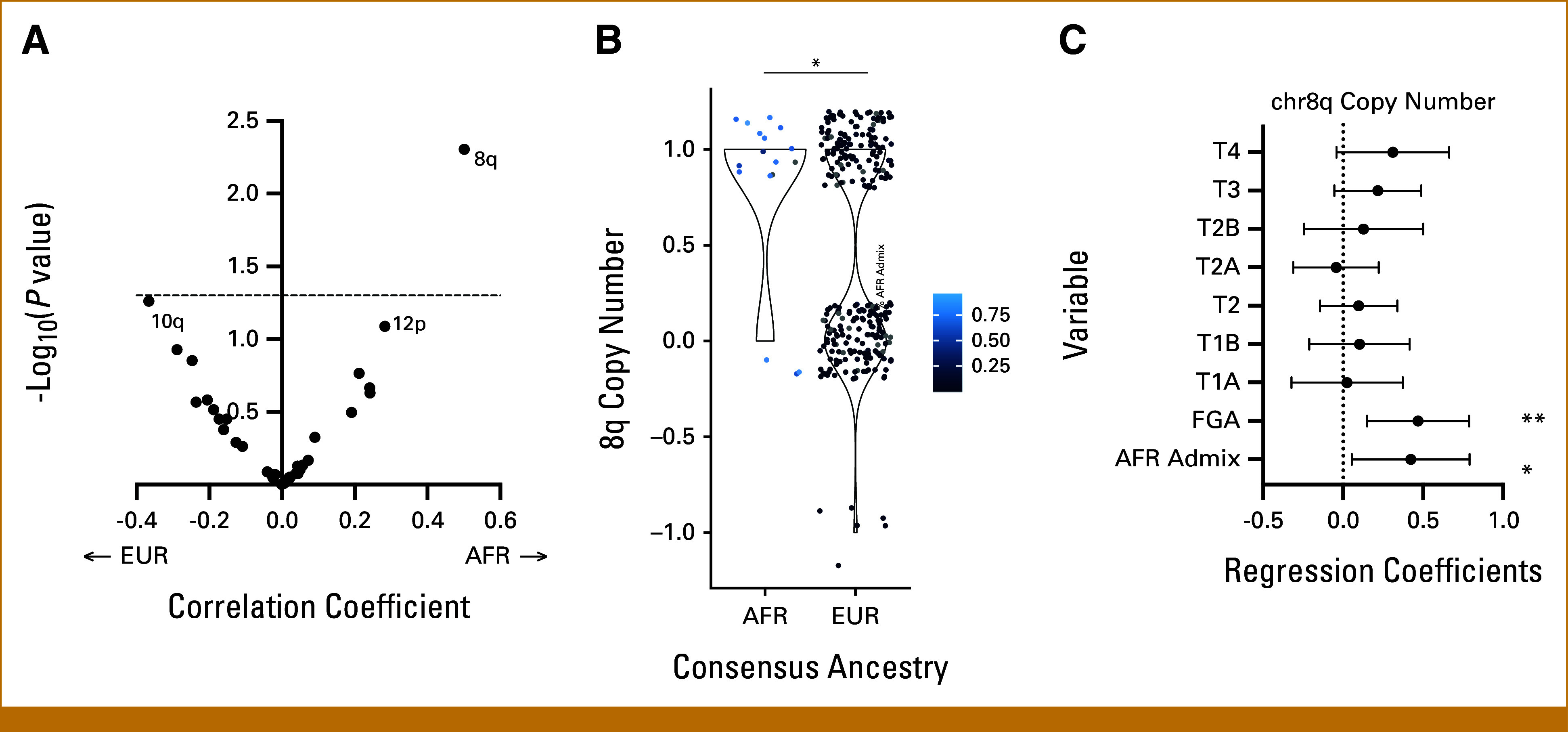
Aneuploidy alterations associated with ancestry in AFR and EUR samples in LUSC in TCGA. (A) Volcano plot of chromosome arm aneuploidy frequency. Correlation with percent AFR admixture (X-axis) is plotted against log-transformed Bonferroni-adjusted P values (Y-axis). The dotted line marks the significance threshold of Bonferroni-adjusted P = .05. (B) Copy number of chromosome 8q by ancestry. Gradient color scale of data points represents percent AFR admixture, from black at 0% to blue at 100% (*P < .05; **P < .01; ***P < .005). (C) Forest plots depicting multivariate linear regression models of chr8q copy number as a function of tumor stage, fraction genome altered, and AFR admixture, with variables listed along the Y-axis. Regression coefficients are plotted on the x-axis with a dotted line at x = 0. AFR, African; EUR, European; LUSC, lung squamous cell carcinoma; TCGA, the Cancer Genome Atlas.
Thus far, our analyses used global ancestry assigned to samples in TCGA.22 We next wanted to assess the relationship between local ancestry at the chr8q24.21 locus and its copy number. Global ancestry and local ancestry were 95% concordant (Data Supplement, Table S3). Homozygous AFR local ancestry was significantly associated with a 1.4-fold increase in chr8q24.21 copy number compared with homozygous EUR local ancestry (Welch's t-test P = .0257; Fig 3A). Homozygous AFR local ancestry was also significantly associated with a 1.64-fold increase in chr8q copy number compared with homozygous EUR (Welch's t-test P = .048; Fig 3B). ANOVA of all AFR/EUR local ancestry genotypes was significant for differences in means of chr8q24.21 (P = .0332; Fig 3A) and chr8q (P = .043; Fig 3B). A significant difference was observed in mean chr8q between local ancestry AFR/EUR heterozygotic loci and EUR homozygotic loci (fold change, 2.2; Welch's t-test P < 2.2E-16; Fig 3B). All other post hoc pairwise t-test comparisons were not significant (Figs 3A and 3C), likely because of a low sample size of heterozygotic loci. AFR local ancestry did not correlate significantly with MYC copy number (Welch two-sample t-test P = .096; Fig 3C). ANOVA of AFR/EUR local genotypes was also not significant for differences in means of MYC copy number (P = .285; Fig 3C). Overall, these results suggest that locus-specific ancestry at chr8q24.21 may play a role in its amplification in LUSC.
FIG 3.
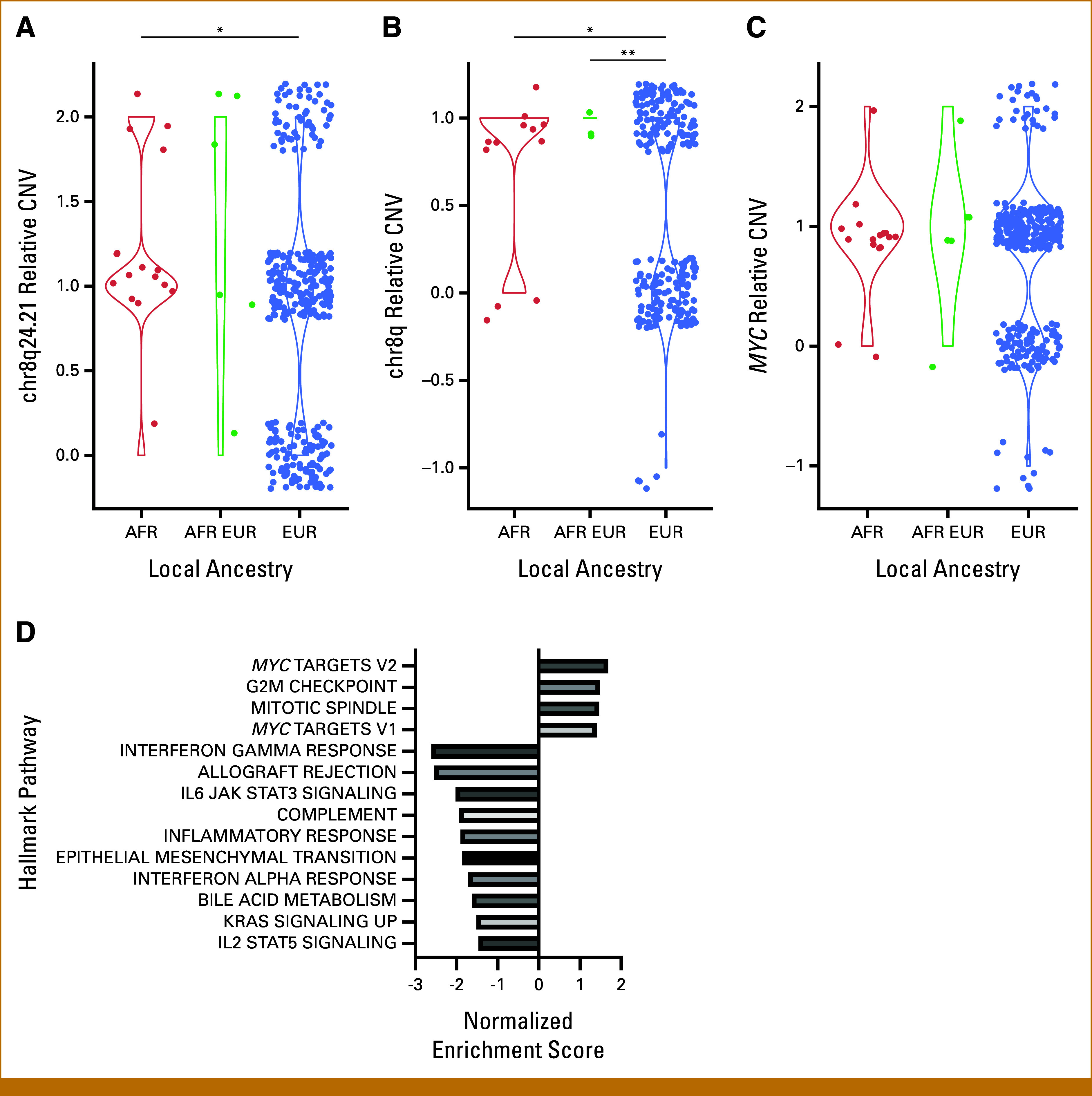
Relative CNV by local ancestry genotype and gene sets enriched with percent AFR global ancestry in LUSC. (A) Chr8q24.21 relative CNV, (B) chr8q relative CNV, and (C) MYC relative CNV (*P < .05; ** P < 1.0E-3, for post hoc pairwise t-tests). (D) Gene expression data were assessed for pathway enrichment using GSEA (preranked). All gene sets listed were significant (P < .05). Marked gene sets are significant by multiple corrections with a false discovery rate of <0.05 (*FDR < .05, **FDR < .01). AFR, African; CNV, copy number variation; EUR, European; FDR, false discovery rate; GSEA, Gene Set Enrichment Analysis; LUSC, lung squamous cell carcinoma.
AFR Ancestry Is Associated With Increased Expression of MYC Target Genes in LUSC
We next sought to discern whether the observed genomic differences associated with ancestry give rise to distinct ancestry-related gene expression profiles. For each gene, we correlated normalized expression with percentage admixture AFR. We then generated a list of genes ranked by fold-change and ran preranked Gene Set Enrichment Analysis (GSEA) to identify enriched pathways. Most significantly positively enriched in the AFR direction were MYC target genes (FDR q = .036; Fig 3D), consistent with increased MYC expression.
Notably, GSEA results also demonstrated significant negative enrichment of many immune pathways in association with AFR ancestry, including interferon-gamma response, allograft rejection, IL6-JAK-STAT3 signaling, and complement, among others (Fig 3D; adj P < .001). Interestingly, overall immune infiltrate was not significantly different between AFR and EUR samples, nor did it significantly correlate with percent AFR admixture (Data Supplement, Fig S4). This suggests a correlation between AFR ancestry and decreased immune signaling that is not explained by lower immune infiltrate.
AFR Ancestry Is Associated With Increased Chr8q Gain in Additional Cancer Types in TCGA
Having observed associations between AFR ancestry and MYC copy number, chr8q24.21 copy number, and chr8q copy number in LUSC, we next wanted to discern whether these trends are exclusive to LUSC or present more broadly across non–small cell lung cancer (NSCLC), particularly with the recent finding of MYC amplification associated with AFR ancestry in NSCLC in MSKCC and Foundation panel sequencing cohorts.30 We analyzed 498 lung adenocarcinoma (LUAD) samples in TCGA (EUR [n = 437], AFR [n = 35], AFR admixture [n = 17]) tumors. In either LUAD alone or the combined NSCLC cohort, we found no significant associations between AFR ancestry and either MYC or chr8q24.21 (Fig 4A). However, we found a significant trend of increased mean chr8q in AFR samples in NSCLC (fold change, 1.74; Welch's t-test P = 5E-04; Fig 4A) and a near-significant trend in LUAD alone (fold change, 1.46; Welch's t-test P = .067; Fig 4A). In summary, in TCGA, we see that ancestry-associated aneuploidy of chr8q can be generalized to NSCLC, but focal differences at this MYC enhancer are limited to LUSC.
FIG 4.
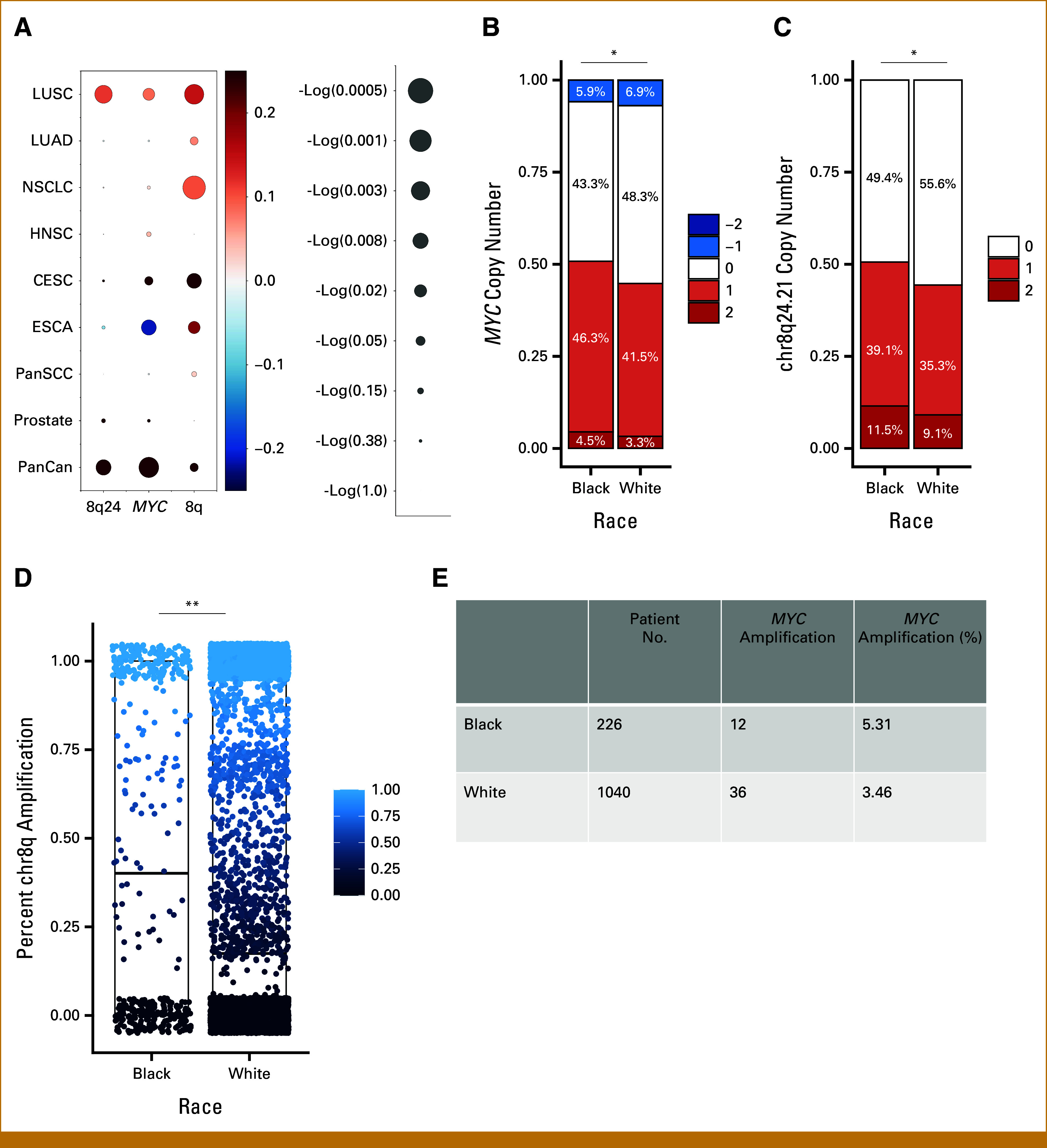
CNV associated with AFR ancestry in other TCGA cancer types, MSKCC GENIE, and CUIMC primary tumor sample cohorts. (A) For MYC, chr8q24.21, and chr8q, correlations with AFR ancestry are shown in listed cohorts in TCGA. Color indicates the strength of and direction of correlation per gradient scale on the left, with circle size inversely corresponding to P value. Scale for log(P value) scale and circle size shown on left. Proportional stacked bar plots of (B) MYC and (C) chr8q24.21 enhancer locus CNV by race in MSKCC GENIE samples (White [n = 5,910]; Black [n = 425]; *P < .05). (D) Percent chr8q amplification by race in MSKCC GENIE samples (mean in Black = 0.47, mean in White = 0.41; P = .008). (E) Proportion of samples with MYC amplification in the CUIMC clinical cohort by race. AFR, African; CESC, cervical squamous cell carcinomas; CNV, copy number variation; CUIMC, Columbia University Irving Medical Center; ESCA, esophageal squamous cell carcinomas; HNSC, head and neck squamous cell; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; NSCLC, non–small cell lung cancer; PanCan, pan-cancer; PanSCC, pan-squamous cell carcinoma; TCGA, the Cancer Genome Atlas.
We previously found that aneuploidy events correlate strongly with tumor type, including squamous tumors across lung, head and neck, esophagus, and cervix.24,31 In addition, we previously found increased MYC expression in head and neck squamous cell carcinoma (HNSCC) in Black patients.32 Since our analyses in lung suggest that squamous pathology shows a stronger correlation between ancestry and MYC amplification, we next wanted to look at squamous cancers in other tissues. We performed these correlations in 1386 squamous cell carcinoma (SCC) tumors (EUR [n = 1,089], AFR [n = 77], AFR admix [n = 39]), both across and within squamous cancer types. These included 89 esophageal squamous cell carcinomas, 210 cervical squamous cell carcinomas (CESC; EUR [n = 153], AFR [n = 16], AFR admix [n = 11]), and 498 HNSCC tumors (EUR [n = 426], AFR [n = 38], AFR admix [n = 11]). Within all individual cancer types, MYC and chr8q24.21 copy number trended higher in AFR samples but were not significantly different (Fig 4A). This could be attributed to (1) decreased frequency of this amplification in different cancer types and/or (2) different GISTIC peak locations by tumor type. We also observed a trend of increased mean chr8q associated with AFR ancestry in each analysis, with statistically significant differences in CESC (Welch's t-test P = .048) and near-significant differences in pan-squamous cell carcinoma (Pan-SCC) (Welch's t-test P = .059; Fig 4A). Overall, we did not see MYC or chr8q24.21 copy number significantly associated with AFR ancestry in other squamous cancers (Fig 4A).
AFR Ancestry Correlates With MYC and Chr8q24.21 Enhancer Copy Number Across All Cancers in TCGA
Underpowered for significance in individual cancer types, we performed a pan-cancer analysis across 9,897 tumors in TCGA (PANCAN) designated as EUR (n = 8,593), AFR (n = 636), or AFR-admix (n = 336) consensus ancestry, using linear models to correct for tumor type. We found both chr8q24.21 and MYC amplification to be approximately 1.1-fold higher in consensus AFR samples compared with consensus EUR samples in PANCAN (Welch's t-tests P = .0436, P = .016, respectively). It is important to distinguish that the peak identified for chr8q24.21 copy number amplification in GISTIC across cancers includes MYC, whereas the peaks for individual cancer types excluded MYC and are limited to enhancer sequences (Data Supplement, Table S4). Notably, the positive correlation between percentage AFR ancestry and chr8q24.21 copy number in PANCAN was preserved when correcting for tumor type (coefficient, 0.066; P = .038; Fig 4A). By contrast, the positive correlation between percent admixture AFR and MYC amplification in PANCAN was maintained but no longer statistically significant when correcting for tumor type (coefficient, 0.053; P = .083; Fig 4A). PANCAN analysis did not show a significant association between AFR ancestry and chr8q gain (Fig 4A).
Black Race Correlates With Amplification of MYC, Its Enhancer on Chr8q24.21, and Chr8q Across Cancers From MSKCC in AACR Project GENIE
We sought further independent data sets to validate our pan-cancer findings as we were limited by power to assess differences in LUSC and NSCLC in validation cohorts. We analyzed all 7,344 cancers in the MSKCC subset of the AACR Project GENIE data set with self-identified race designated as Black/African American (n = 425) or White (n = 5,910; there are no ancestry annotations available for this data set). We used GISTIC to identify peaks containing significant SCNAs and compared frequencies of these events in self-identified Black and White patients. We found 1.20-fold higher MYC copy number and 1.16-fold higher chr8q24.21 copy number in Black patient samples compared with White patient samples (Welch t-tests P = .012; P = .011, respectively; Figs 4B and 4C). Furthermore, when we corrected for tumor type, we found MYC copy number to be significantly correlated with Black race (coefficient, 0.064; P = .047). The correlation between Black race and chr8q24.21 neared significance (coefficient, 0.060; P = .058). As with the analysis across cancers in TCGA, the peak identified on chr8q24.21 in GISTIC for this cohort includes MYC. Assuming that race and genetic ancestry are strongly correlated,22 these findings independently validate the significant positive correlation between MYC copy number and AFR admixture that we found across all TCGA samples with tumor type correction. They also raise the possibility that, given sufficient power, an association between these copy number changes and genetic ancestry might be observed within other individual cancer types.
While we did not have aneuploidy calls from AACR GENIE data, we were able to aggregate copy number alterations of genes on chr8q to calculate percent amplification of chr8q. Notably, we found a significant 1.2-fold increase in percent amplification of chr8q associated with Black race (Welch's t-test P = .008; Fig 4D).
MYC Copy Number Trends Higher Across Cancers From a Clinical Cohort at CUIMC
To validate the findings of increased MYC in tumors from Black patients in an independent cohort, we looked at panel sequencing data of MYC in primary solid tumor samples from patients at CUIMC. We analyzed 1,266 samples across cancers, with 1,040 from self-identified White patients and 226 from self-identified Black patients. A total of 3.46% of samples from White patients featured MYC amplification, compared with 5.43% of samples from Black patients (Fig 4E). This trend is consistent with that observed in TCGA, though not statistically significant.
DISCUSSION
In this study, we found chr8q24.21 amplification (containing a MYC enhancer sequence), MYC amplification, and chr8q gain to be positively and significantly associated with AFR ancestry in LUSC. Gene expression and pathway analysis demonstrated that the top differentially expressed pathway by ancestry was MYC. These data converge on MYC and together suggest a previously undescribed role for ancestry-associated differences in MYC signaling in LUSC. We also described positive correlations between MYC/chr8q24.21 and AFR ancestry across all cancers in TCGA, correcting for tumor type. That the GISTIC peak identified in pan-cancer analyses contains MYC, whereas that in LUSC excludes MYC, also suggests a unique importance to this MYC enhancer in LUSC that is not found at a pan-cancer level. We found chr8q gain to be significantly associated with AFR ancestry in multiple other cancer types in TCGA. We independently validated increased MYC/chr8q24.21 copy gain in the MSKCC subcohort of the AACR Project GENIE in self-identified Black patients and also noted a significant increase in percent amplification of chr8q in these patients. We found MYC amplification to be more frequent among Black patients in a third data set obtained from samples at CUIMC. That the observed trends uphold in multiple pan-cancer data sets strengthens the generalizability of MYC in ancestry-specific cancer changes. This relationship may also be mediated by chr8q arm gain in specific cancers where we have shown aneuploidy patterns associated with AFR ancestry.
A particular strength of our approach was analysis by not only self-identified race or ancestry designation but also percent ancestry admixture. Compared with consensus ancestry, percent AFR admixture provides a more accurate approximation of the contribution of AFR ancestry to the patient's genetic makeup. It is a continuous variable, allowing us to correlate percent AFR admixture with genomic event frequencies to identify events of interest.
MYC is a transcription factor and well-characterized oncogene; relatively subtle (approximately 2-fold) changes in MYC expression affect cell proliferation and transformation.33-35 This imparts biologic significance to the 1.2-1.4-fold increase in MYC copy number observed in AFR samples compared with EUR, which could contribute to a more aggressive tumor phenotype. Indeed, we see enrichment of MYC target gene expression with AFR ancestry. In addition, we observe correlations between copy number of the chr8q24.21 peak and MYC gene expression, suggesting that copy number amplifications at MYC or its enhancer result in increased MYC activity in association with AFR ancestry. Recently, MYC amplification was found to be enriched with AFR ancestry in NSCLC, breast, and prostate cancers in a cohort of 333,908 tumor samples obtained for targeted panel sequencing.30 In our study, we establish an association not only between AFR ancestry and MYC but also with the lung squamous MYC enhancer on chr8q24.21.
The significant correlations observed between local ancestry and chr8q aneuploidy also suggest that arm-level events might drive differential chr8q24.21 copy number rather than focal amplifications. It is important to note that while local ancestry gives insight into chr8q24.21 loci heterozygous for ancestry, local ancestry was 95% concordant with global ancestry and therefore would be expected to correlate with 8q, MYC, and chr8q24.21 by association.
We observed a significant approximately two-fold increase in mean chr8q in association with AFR ancestry in LUSC and CESC, with near-significant increases in similar magnitude in pan-SCC and LUAD. We also observed a significant increase in the frequency of chr8q amplification in tumors from Black patients in MSKCC cancers in AACR GENIE. Previous studies have looked at aneuploidy alterations associated with genetic ancestry22,24,31; however, no findings of chr8q gain have been reported in association with AFR ancestry. The relationship between ancestry and the frequency of chr8q gain may underlie or amplify differences in chr8q24.21 and/or MYC dosage in these cancer types. Further studies describing aneuploidy events in diverse tumor samples are needed to explore this hypothesis.36
Targeted MYC inhibitors are now entering phase I/II clinical trials, and candidates for direct and indirect inhibition continue to emerge.36 Our findings suggest that MYC inhibition would particularly benefit these patients. Future preclinical and early-phase clinical trials that study MYC should robustly sample tumors from patients of AFR ancestry to (1) validate these findings, (2) describe clinic-pathologic correlates of MYC amplification in these patients, and (3) explore pharmacogenomic characteristics associated with ancestry. We hope that the advancement of precision medicine for minority populations can help mitigate persisting survival disparities.
SUPPORT
Supported in part by NIH/NIDCR R21 DE031112-02 (A.M.T. and F.M.-H) and NCI 1 R21 CA280577-01 (A.M.T. and J.C.-Z.).
DATA SHARING STATEMENT
The raw TCGA data, processed data, and clinical data can be found at the legacy archive of the GDC (https://portal.gdc.cancer.gov/legacy-archive/search/f) and the PanCanAtlas publication page (https://gdc.cancer.gov/about-data/publications/pancanatlas). The copy number data can be found at https://gdc.cancer.gov/about-data/publications/pancan-aneuploidy. The mutation data can be found at https://gdc.cancer.gov/about-data/publications/mc3-2017. The gene expression data can be found at https://gdc.cancer.gov/about-data/publications/pancanatlas. TCGA data can also be explored through the Broad Institute FireBrowse portal (http://gdac.broadinstitute.org) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org). The raw TCGA data, processed data, and clinical data can be found at the legacy archive of the GDC (https://portal.gdc.cancer.gov/legacy-archive/search/f) and the PanCanAtlas publication page (https://gdc.cancer.gov/about-data/publications/pancanatlas). The copy number data (including aneuploidy score and fraction of genome altered) can be found at https://gdc.cancer.gov/about-data/publications/pancan-aneuploidy. The mutation data can be found at https://gdc.cancer.gov/about-data/publications/mc3-2017. The gene expression data can be found at https://gdc.cancer.gov/about-data/publications/pancanatlas. TCGA data can also be explored through the Broad Institute FireBrowse portal (http://gdac.broadinstitute.org) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org).
AUTHOR CONTRIBUTIONS
Conception and design: Sejal Jain, Alison M. Taylor
Collection and assembly of data: Sejal Jain, Benjamin May, Alexandria G. Yao
Data analysis and interpretation: Sejal Jain, Xuechun Bai, Samyukta Mallick, Branden Kinghorn, Alexandria G. Yao, Diane Allen-Gipson, Xiaoyang Zhang, Brian S. Henick, Fatemeh Momen-Heravi, Jian Carrot-Zhang, Alison M. Taylor
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Brian S. Henick
This author is a member of the JCO Precision Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Honoraria: OncLive/MJH Life Sciences, DAVA Oncology
Consulting or Advisory Role: AstraZeneca, IDEAYA Biosciences, Jazz Pharmaceuticals, Sorrento Therapeutics, Genentech/Roche, Regeneron, Bristol Myers Squibb
Research Funding: NexImmune (Inst), Genentech/Roche (Inst), Johnson & Johnson/Janssen (Inst), Stand Up 2 Cancer (Inst), V Foundation (Inst)
Jian Carrot-Zhang
Travel, Accommodations, Expenses: Caris
Alison M. Taylor
Research Funding: Ono Pharmaceutical
No other potential conflicts of interest were reported.
REFERENCES
- 1.Haque AT, Berrington de Gonzalez A, Chen Y, et al. : Cancer mortality rates by racial and ethnic groups in the United States, 2018-2020. J Natl Cancer Inst 115:822-830, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aizer AA, Wilhite TJ, Chen MH, et al. : Lack of reduction in racial disparities in cancer-specific mortality over a 20-year period. Cancer 120:1532-1539, 2014 [DOI] [PubMed] [Google Scholar]
- 3.Giaquinto AN, Miller KD, Tossas KY, et al. : Cancer statistics for African American/Black people 2022. CA Cancer J Clin 72:202-229, 2022 [DOI] [PubMed] [Google Scholar]
- 4.Beyer KMM, Laud PW, Zhou Y, et al. : Housing discrimination and racial cancer disparities among the 100 largest US metropolitan areas. Cancer 125:3818-3827, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi E, Ding VY, Luo SJ, et al. : Risk model-based lung cancer screening and racial and ethnic disparities in the US. JAMA Oncol 9:1640-1648, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cronin KA, Scott S, Firth AU, et al. : Annual report to the nation on the status of cancer, part 1: National cancer statistics. Cancer 128:4251-4284, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Clark LP, Bechle MJ, et al. : Disparities in air pollution exposure in the United States by race/ethnicity and income, 1990-2010. Environ Health Perspect 129:127005, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel RL, Miller KD, Wagle NS, et al. : Cancer statistics, 2023. CA Cancer J Clin 73:17-48, 2023 [DOI] [PubMed] [Google Scholar]
- 9.Zahnd WE, Murphy C, Knoll M, et al. : The intersection of rural residence and minority race/ethnicity in cancer disparities in the United States. Int J Environ Res Public Health 18:1384, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zavala VA, Bracci PM, Carethers JM, et al. : Cancer health disparities in racial/ethnic minorities in the United States. Br J Cancer 124:315-332, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanetti KA, Wang Z, Aldrich M, et al. : Genome-wide association study confirms lung cancer susceptibility loci on chromosomes 5p15 and 15q25 in an African-American population. Lung Cancer 98:33-42, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naab TJ, Gautam A, Ricks-Santi L, et al. : MYC amplification in subtypes of breast cancers in African American women. BMC Cancer 18:274, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha S, Mitchell KA, Zingone A, et al. : Higher prevalence of homologous recombination deficiency in tumors from African Americans versus European Americans. Nat Cancer 1:112-121, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y, Rand KA, Hazelett DJ, et al. : Prostate cancer susceptibility in men of African ancestry at 8q24. J Natl Cancer Inst 108:djv431, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber TK, Chin HM, Rodriguez-Bigas M, et al. : Novel hMLH1 and hMSH2 germline mutations in African Americans with colorectal cancer. JAMA 281:2316-2320, 1999 [DOI] [PubMed] [Google Scholar]
- 16.Wolf AMD, Fontham ETH, Church TR, et al. : Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin 68:250-281, 2018 [DOI] [PubMed] [Google Scholar]
- 17.Montminy EM, Zhou M, Maniscalco L, et al. : Trends in the incidence of early-onset colorectal adenocarcinoma among Black and White US residents aged 40 to 49 years, 2000-2017. JAMA Netw Open 4:e2130433, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conti DV, Darst BF, Moss LC, et al. : Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet 53:65-75, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spratt DE, Chan T, Waldron L, et al. : Racial/ethnic disparities in genomic sequencing. JAMA Oncol 2:1070-1074, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network : Comprehensive genomic characterization of squamous cell lung cancers. Nature 489:519-525, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wojcik GL, Graff M, Nishimura KK, et al. : Genetic analyses of diverse populations improves discovery for complex traits. Nature 570:514-518, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrot-Zhang J, Chambwe N, Damrauer JS, et al. : Comprehensive analysis of genetic ancestry and its molecular correlates in cancer. Cancer Cell 37:639-654.e6, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, et al. : The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401-404, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor AM, Shih J, Ha G, et al. : Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33:676-689.e3, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen R, Seshan VE: FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44:e131, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milbury CA, Creeden J, Yip WK, et al. : Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One 17:e0264138, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mermel CH, Schumacher SE, Hill B, et al. : GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12:R41, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Guo B, Aguilera-Jimenez E, et al. : Chromatin looping shapes KLF5-dependent transcriptional programs in human epithelial cancers. Cancer Res 80:5464-5477, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Choi PS, Francis JM, et al. : Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet 48:176-182, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiagge E, Jin DX, Newberg JY, et al. : Tumor sequencing of African ancestry reveals differences in clinically relevant alterations across common cancers. Cancer Cell 41:1963-1971.e3, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell JD, Yau C, Bowlby R, et al. : Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep 23:194-212 e6, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mezghani N, Yao A, Vasilyeva D, et al. : Molecular subtypes of head and neck cancer in patients of African ancestry. Clin Cancer Res 29:910-920, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bazarov AV, Adachi S, Li SF, et al. : A modest reduction in c-myc expression has minimal effects on cell growth and apoptosis but dramatically reduces susceptibility to Ras and Raf transformation. Cancer Res 61:1178-1186, 2001 [PubMed] [Google Scholar]
- 34.Hofmann JW, Zhao X, De Cecco M, et al. : Reduced expression of MYC increases longevity and enhances healthspan. Cell 160:477-488, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy DJ, Junttila MR, Pouyet L, et al. : Distinct thresholds govern Myc's biological output in vivo. Cancer Cell 14:447-457, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llombart V, Mansour MR: Therapeutic targeting of “undruggable” MYC. EBioMedicine 75:103756, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw TCGA data, processed data, and clinical data can be found at the legacy archive of the GDC (https://portal.gdc.cancer.gov/legacy-archive/search/f) and the PanCanAtlas publication page (https://gdc.cancer.gov/about-data/publications/pancanatlas). The copy number data can be found at https://gdc.cancer.gov/about-data/publications/pancan-aneuploidy. The mutation data can be found at https://gdc.cancer.gov/about-data/publications/mc3-2017. The gene expression data can be found at https://gdc.cancer.gov/about-data/publications/pancanatlas. TCGA data can also be explored through the Broad Institute FireBrowse portal (http://gdac.broadinstitute.org) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org). The raw TCGA data, processed data, and clinical data can be found at the legacy archive of the GDC (https://portal.gdc.cancer.gov/legacy-archive/search/f) and the PanCanAtlas publication page (https://gdc.cancer.gov/about-data/publications/pancanatlas). The copy number data (including aneuploidy score and fraction of genome altered) can be found at https://gdc.cancer.gov/about-data/publications/pancan-aneuploidy. The mutation data can be found at https://gdc.cancer.gov/about-data/publications/mc3-2017. The gene expression data can be found at https://gdc.cancer.gov/about-data/publications/pancanatlas. TCGA data can also be explored through the Broad Institute FireBrowse portal (http://gdac.broadinstitute.org) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org).


