Abstract
Background
In computational analysis, the RING-finger domain is one of the most frequently detected domains in the Arabidopsis proteome. In fact, it is more abundant in Arabidopsis than in other eukaryotic genomes. However, computational analysis might classify ambiguous domains of the closely related PHD and LIM motifs as RING domains by mistake. Thus, we set out to define an ordered set of Arabidopsis RING domains by evaluating predicted domains on the basis of recent structural data.
Results
Inspection of the proteome with a current InterPro release predicts 446 RING domains. We evaluated each detected domain and as a result eliminated 59 false positives. The remaining 387 domains were grouped by cluster analysis and according to their metal-ligand arrangement. We further defined novel patterns for additional computational analyses of the proteome. They were based on recent structural data that enable discrimination between the related RING, PHD and LIM domains. These patterns allow us to predict with different degrees of certainty whether a particular domain is indeed likely to form a RING finger.
Conclusions
In summary, 387 domains have a significant potential to form a RING-type cross-brace structure. Many of these RING domains overlap with predicted PHD domains; however, the RING domain signature mostly prevails. Thus, the abundance of PHD domains in Arabidopsis has been significantly overestimated. Cluster analysis of the RING domains defines groups of proteins, which frequently show significant similarity outside the RING domain. These groups document a common evolutionary origin of their members and potentially represent genes of overlapping functionality.
Background
The recent completion of the Arabidopsis thaliana genome sequence and its accessibility in annotated form [1] marks an essential breakthrough for basic and applied plant science. Extensive bioinformatics analysis, using both extrinsic and intrinsic data, initially detected 25,498 genes within the Arabidopsis genome. Around 69% of the corresponding proteins could be classified according to their sequence similarity to proteins of known function in plants and other organisms. Approximately 51% of the genes contain a functional domain detectable by InterPro. InterPro has proven to be especially powerful for functional domain detection [1,2,3].
One of the most abundant domains detected in the Arabidopsis proteome is the RING-finger domain, which was found 365 times in the initial characterization of the genome [1]. In fact, 1.42% of Arabidopsis proteins contain a RING-domain signature. Thus, it is overrepresented in Arabidopsis as compared to other complete eukaryotic genomes (Drosophila melanogaster, Caenorhabditis elegans and Saccharomyces cerevisiae), which contain 0.7-0.75% of RING-domain proteins. To date, the significance of this observation remains unclear.
The RING domain was originally named after the acronym for the first protein it was found in, encoded by the Really Interesting New Gene [4,5]. The motif is related to the zinc-finger domain; however, zinc fingers consist of two pairs of zinc ligands coordinately binding one zinc ion, whereas RING fingers consist of four pairs of ligands binding two ions. Two other motifs that consist of four metal-ligand pairs binding two zinc ions are related to the RING finger - the PHD (plant homeodomain) [6] and LIM (Lin11/Isl-1/Mec-3) [7] domains (Figure 1).
Figure 1.
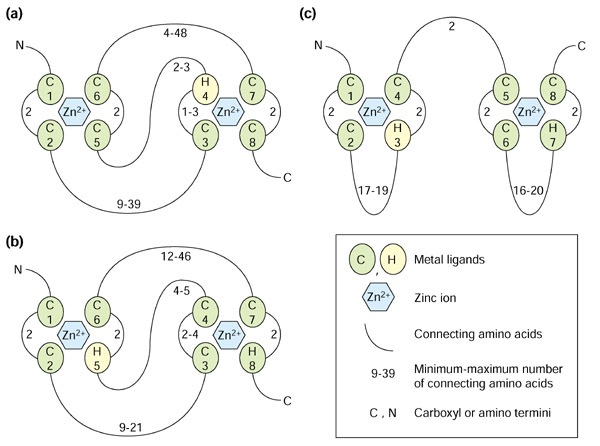
Schematic presentation of the structure of prototypical RING, PHD and LIM domains. The metal-ligand residues, either cysteine (C) or histidine (H), are shown as numbered spheres. Two pairs of metal ligands coordinate one zinc ion (hexagon). The numbers next to the loops connecting the metal-ligand residues indicate the minimum and maximum number of loop residues. (a) The structure of a RING domain (RING-HC type). The metal-ligand pairs 1 and 3 coordinate one zinc ion, while pairs 2 and 4 coordinate the second one in a so-called cross-brace arrangement. (b) The structure of a PHD domain reveals a cross-brace arrangement similar to the RING domain. (c) The LIM domain structure is distinct in its consecutive zinc ligation scheme: the first zinc ion is coordinated by the metal-ligand pairs 1 and 2, while the second ion is coordinated by pairs 3 and 4.
The RING-domain structure has been resolved at atomic resolution for three proteins - promyelotic leukemia protein (PML), equine herpes virus protein (IEEVH) and the human recombination protein RAG1 [8,9,10]. These studies revealed that the RING domain forms a distinct so-called cross-brace structure, in which metal-ligand pairs 1 and 3 coordinate to bind one zinc ion, and pairs 2 and 4 bind the second one (Figure 1a). Also, the structures of the LIM-domain proteins CRP1, CRIP and CRP2 [11,12,13] have been resolved and indicate that LIM domains behave like a double zinc finger, coordinating two zinc ions with the two consecutive pairs of ligands (Figure 1c). Finally, the recent solution of the structure of the PHD domain of KAP-1 [14] demonstrates that the zinc-ligation scheme of the PHD domain is similar to the RING domain; that is, it folds into a cross-brace structure (Figure 1b).
In functional terms, the RING domain can basically be considered a protein-interaction domain, and RING-finger proteins have been implicated in a range of diverse biological processes and biochemical activities, from transcriptional and translational regulation to targeted proteolysis [15,16,17]. For several RING proteins a biochemical ubiquitin ligase activity has been observed [18,19]. Thus it has been suggested that the abundance of RING proteins in Arabidopsis might reflect a bias towards target-specific proteolysis as a means of controlling gene activity. However, in most cases a RING domain by itself is not sufficient for ubiquitin ligase activity and the additional structural features required are not known. Thus, to date it remains unclear how many of the RING proteins encoded in the Arabidopsis genome could indeed be involved in protein degradation.
In this in-depth study, we evaluate and classify RING domain proteins of Arabidopsis by computational analyses as well as manual curation. We present a set of Arabidopsis RING domains, which we classify into related clusters and sort according to their potential to form a RING-type cross-brace structure on the basis of recent results from structural analyses.
Results and discussion
InterPro analysis of the Arabidopsis proteome for RING-domain proteins
In an initial characterization of the Arabidopsis proteome [1] by InterPro analysis (release 1.0) [2] a total of 365 RING domains were detected. We searched the proteome for RING domains again, using an updated version of InterPro (release 3.1). In total, our analysis retrieved 446 domains. An overall percentage of 10-15% of erroneously assigned exons has been estimated for the proteome. However, exons that contain functional domains, and which are therefore detectable and adjustable via similarity-based methods, are expected to have significantly fewer wrong assignments. We therefore have confidence in the detected number of RING-finger-containing proteins, although a deviation in the lower single-digit percentage range cannot be excluded.
We evaluated the first-pass computational analysis and manually annotated all detected domains according to a set of criteria that would qualify them as RING domains. All our results are organized on two websites that cross-reference each other [20,21]. Here we present an overview and a summary of the results; corresponding details and supplementary materials can be found on the websites.
Criteria for RING domains
Several of the detected domains did not represent a full-length RING domain. We inspected them in detail and attempted to complete them using the adjacent sequences in the respective proteins. This proved successful in a number of cases; in several others, however, it was not possible. We also noticed that in some domains the conserved spacing of the metal ligands was lacking, indicative of probable false-positive detection. In fact, in addition to programs relying on defined strings, modules not based on defined patterns (such as PfamHMM, which is based on hidden Markov models) are part of InterPro. As a result, domains not containing the defined patterns might have been detected by InterPro as RING domains. Thus, we inspected each of the 446 initially detected RING domains in detail, in order to eliminate false positives from the analysis set.
To verify the RING domains, we defined a set of criteria based on well characterized examples of RING-domain proteins. First, we required domains to contain at least seven of the full complement of eight metal ligands. Second, the metal-ligand residues had to correspond to the RING pattern, either to the prototypical RING pattern (metal ligand 4 is histidine, all others are cysteine, 'RING-HC', see below) or the frequent RING-H2 pattern (metal ligands 4 and 5 histidine, all others cysteine). Third, the spacing of the core residues (metal ligands 3 to 6) had to be conserved. These criteria leave room for subgroups of RING-finger structures in which the spacing between positions 7 and 8 is different from the generally conserved two residues [22], and for cases in which a metal ligand is missing, mostly at the seventh or eighth position. Finally, we also allowed additional deviations from the canonic criteria, which ensure that known variant-type RING domains [23] are included in our set. These modifications are metal ligand substitutions that are observed in a few well characterized RING domains, such as those in MDM2, mouse c-Cbl, Rbx1 and CART1. They include threonine for cysteine substitutions at metal ligand positions 1 or 3 and aspartate for cysteine substitution at metal ligand 8.
We did not allow for similar substitutions at other positions, as the ligands might not be interchangeable in every position [14]. However, we carried out additional computational analyses with more relaxed patterns. For instance, if metal ligand positions 1, 3, 5 and 8 are simultaneously allowed to be encoded by either cysteine, histidine, threonine or aspartate (see above), a total of 81 additional domains in 79 proteins are found. Although these domains are not detectable by InterPro analysis, they might have the potential to encode novel RING-domain variants. However, several LIM domains are included in this set. Moreover, the corresponding RING arrangement was unclear in most other cases and might represent distantly related motifs, such as the U-box [24]. Thus we did not include these additional domains in our proper set.
Other domains in RING proteins
A complete InterPro analysis on the protein set retrieved with the RING motif revealed other domains present in these proteins. The RING domain is closely related to the LIM and PHD domains, and 141 of the RING-domain proteins are also reported to carry a PHD domain. We inspected these predicted PHD domains and the vast majority of the respective metal ligands overlap with the detected RING domains. However, in most cases the RING-domain signature prevails, as some highly conserved residues characteristic of PHD domains are mostly missing. In fact, only in three cases did we clearly favor a PHD-domain architecture, and these domains were eliminated from our set. This means that the Arabidopsis proteome possibly contains 138 fewer PHD domains than detected. Thus, because of the overlap in the RING- and PHD-domain signatures, the frequency of the PHD domain has initially been significantly overestimated.
After the PHD domain, bipartite nuclear-localization signals were the second most frequently detected domain (63 times) in our RING-domain protein set. Most other domains are much less frequent however (< 10 times), and some combinations are obviously absent. For instance, the RING/B-box/coiled-coil protein family found in several eukaryotes seems to be absent from Arabidopsis.
Elimination of false positives
On the basis of our criteria above, the metal-ligand arrangement of every one of the initial 446 domains was reinspected. We noted that some proteins with a generally high content of cysteine and histidine residues represent false positives. Indeed, that is the case for a group of five cellulose synthases, which contain several zinc-finger domains. We eliminated these from our set.
We also eliminated 41 additional domains in which at least two metal ligands or possible substituting residues are missing, and 10 domains in which the spacing of the core residues (metal ligands 3 to 6) did not satisfy the criteria. Thus, in total, 59 probable false-positive domains have been eliminated from our initial set (Table 1).
Table 1.
Proteins with false-positive detected RING domains that were eliminated from the dataset
| Protein ID | Reason | Remarks |
| At1g02860 | Wrong ligand spacing | Spacer between metal ligands 2 and 3 too long |
| At1g05890 | Wrong ligand spacing | Protein contains one RING-variant domain |
| At1g09060 | Incomplete | |
| At1g10170 | PHD domain | |
| At1g55530 | Incomplete | Protein contains one RING-H2 domain |
| At1g60360 | Incomplete | Protein contains one RING-H2 domain |
| At1g62310 | Incomplete | |
| At1g74870 | Incomplete | |
| At2g28530 | Incomplete | |
| At2g31770 | Wrong ligand spacing | Either wrong spacing or incomplete |
| Protein contains one RING-variant domain | ||
| At2g31780 | Wrong ligand spacing | Either wrong spacing or incomplete |
| Protein contains one RING-variant domain | ||
| At2g39720 | Incomplete | Protein contains one RING-H2 domain |
| At2g42170 | Wrong ligand spacing | Pseudogene |
| At2g44330 | Incomplete | Two false-positive motifs |
| Protein contains one RING-H2 domain | ||
| At3g05870 | Incomplete | Protein contains one RING-H2 domain |
| At3g06330 | Wrong ligand spacing | |
| At3g07610 | Incomplete | |
| At3g08020 | PHD domain | |
| At3g18290 | Incomplete | Protein contains one RING-H2 domain |
| At3g19910 | Incomplete | |
| At3g19950 | Incomplete | Protein contains one RING-H2 domain |
| At3g23060 | Incomplete | Two false-positive motifs |
| At3g42830 | Incomplete | Rbx1-like |
| Protein contains one RING-variant domain | ||
| At3g45470 | Wrong ligand spacing | Protein contains one RING-variant domain |
| At3g45540 | Incomplete | Protein contains one RING-variant domain |
| At3g45630 | Incomplete | |
| At3g48070 | Wrong ligand spacing | |
| At3g52100 | PHD domain | |
| At3g60080 | Incomplete | Protein contains one RING-H2 domain |
| At3g62970 | Incomplete | Protein contains one RING-H2 domain |
| At4g00070 | Incomplete | |
| At4g01020 | Incomplete | Protein contains one RING-HC and |
| One RING-variant domain | ||
| At4g10940 | Incomplete | |
| At4g12150 | Incomplete | Protein contains one RING-variant domain, |
| Which is part of a RING-H2 cluster | ||
| At4g26400 | Incomplete | Protein contains one RING-H2 domain |
| At4g37880 | Incomplete | |
| At4g39350 | Wrong ligand spacing | Cellulose synthase |
| At5g05170 | Wrong ligand spacing | Cellulose synthase |
| At5g09870 | Wrong ligand spacing | Cellulose synthase |
| At5g10370 | Incomplete | Protein contains one RING variant domain |
| At5g15790 | Incomplete | Protein contains one RING-H2 domain |
| At5g17420 | Wrong ligand spacing | Cellulose synthase |
| At5g23110 | Incomplete | Protein contains one RING-HC domain |
| At5g25560 | Incomplete | Protein contains one RING-H2 domain |
| At5g28340 | Incomplete | |
| At5g38070 | Wrong ligand spacing | |
| At5g45290 | Incomplete | |
| At5g47430 | Incomplete | |
| At5g52140 | Incomplete | |
| At5g52150 | Incomplete | |
| At5g56340 | Incomplete | Protein contains one RING-H2 domain |
| At5g58410 | Wrong ligand spacing | |
| At5g62910 | Wrong ligand spacing | |
| At5g63740 | Incomplete | |
| At5g63750 | Incomplete | Drosophila Ariadne-like |
| Protein contains one RING-variant domain | ||
| At5g63970 | Incomplete | |
| At5g64740 | Wrong ligand spacing | Cellulose synthase |
To eliminate false-positive RING domains detected through computational analysis, the complete dataset was curated and false positives were eliminated. The first column shows proteins in which false-positive RING domains were detected. The principal reason for their elimination from the dataset is given in the second column. 'Incomplete', at least two metal ligands are missing; 'wrong ligand spacing', the spacing between the prospective metal ligands is not conserved; 'PHD domain', the domain has a PHD rather than a RING structure. See text for additional criteria for RING domains. Alternative gene names and other remarks are given in the third column. For instance, many of the proteins contain an independent, verified RING domain and are thus also listed in Table 3.
Classification of RING domains
We classified the remaining set of 387 domains according to metal-ligand arrangement. The originally described RING domains were characterized by a histidine at metal-ligand position 4. We have termed the domains in our set with a corresponding arrangement RING-HC domains. We found 118 domains of this type in 111 different proteins. However, the cysteine usually present at metal-ligand position 5 is frequently substituted by a histidine as well, and we identify these domains as RING-H2 domains. Of this type of domain, 215 were found in 214 proteins. The remaining 54 domains, in which not all of the metal ligands were either cysteine or histidine, but where one metal ligand is missing or substituted according to our criteria described above, were classified as RING-variant domains [23].
Derivation of additional patterns for computational analysis
An inherent problem with the computational detection of RING-finger domains is their relatedness to the PHD domain (see above). This ambiguity seems to be due to a lack of structural determinants that separate a given domain in one group or the other. Recently, the first solution structure for a PHD domain has been obtained [14]. Its comparison with related structures revealed some key features that separate LIM, PHD and RING domains.
The LIM domain is clearly set apart from the two others, with a more conserved spacing and conserved hydrophobic residues not found in RING or PHD domains. Among the conserved hydrophobic residues, one is located in front of metal ligand 3 and one after metal ligand 4, and these features result in a zinc ligation by consecutive metal ligand pairs. By contrast, hydrophobic residues in front of metal ligand 5 and after metal ligand 6 seem to result in a cross-brace arrangement, which is observed in RING and PHD domains. Despite this commonality, two features separate RING and PHD domains. First, the loop between metal ligands 4 and 5 can be up to five residues in PHD domains, rather than only up to three in RING domains. Second, in PHD domains the residue two positions in front of metal ligand 7 is an aromatic residue, which alters the hydrophobic core of the domain and thus its structural characteristics.
On the basis of the above data, we defined four patterns that specify a RING domain with increasing stringency (Table 2). In our first pattern (Stringent 1) we required that all metal ligands are present, according to our criteria outlined above, and that the position two residues in front of metal ligand 7 is not an aromatic amino acid. These criteria were satisfied by 324 domains, whereas it was not the case for 63 domains. Failure to match the criteria is mainly the result of a missing or substituted metal ligand; that is, these domains are classified as RING variants. However, among those 63 domains, 13 carry an aromatic residue two positions in front of metal ligand 7 and we thus consider them unlikely to form a RING domain. Rather, they are structurally more similar to PHD domains. Therefore, of the 141 PHD domains predicted in our initial RING-domain set (see above) as few as 16 might indeed represent a PHD structure.
Table 2.
Conventional and novel motif signatures used to identify RING domains in this study
| InterPro, RING-HC | C3HC4 long: C-X2-C-X9,39-C-X1,3-H-X2,3-C-X2-C-X4,48-C-X2-C and short: C-X-H-X-[LIVMFY]-C-X2-C-[LIVMYA] |
| InterPro, RING-HC | C3H2C3 long: C-X2-C-X9,39-C-X1,3-H-X2,3-H-X2-C-X4,48-C-X2-C and short: C-X-H-X-[LIVMFY]-H-X2-C-[LIVMYA] |
| Stringent 1 | [CT]-X2-C-X9,39-[CT]-X1,3-H-X2,3-[CH]-X2-C-X2,46-[KRCHDESTQNGPLVIMA]-X-C-X2-[CD] |
| Stringent 2 | [CT]-X2-C-X9,39-[CT]-X1,3-H-X1,2-[WFYLVIMAST]-[CH]-X2-C-[WFYLVIMAST]-X1,45-[KRCHDESTQNGPLVIMA]-X-C-X2-[CD] |
| Stringent 3 | [CT]-X2-C-X9,39-[CT]-X1,3-H-X1,2-[WFYLVIMAST]-[CH]-X2-C-[WFYLVIMA]-X1,45-[KRCHDESTQNGPLVIMA]-X-C-X2-[CD] |
| Stringent 4 | [CT]-X2-C-X9,39-[CT]-X1,3-H-X1,2-[WFYLVIMAST]-[CH]-X2-C-[WFYLVIMA]-X1,45-[KRCHDESTQNGP]-X-C-X2-[CD] |
Domain signatures used to identify RING domains in the Arabidopsis proteome. The signatures used by the InterPro package as well as the novel patterns defined by us (Stringent 1-Stringent 4) on the basis of recent structural results are given. Metal ligands are in bold. For details see text.
In our second pattern (Stringent 2) we added another criterion; we required the positions in front of metal ligand 5 and after metal ligand 6 to be hydrophobic or serine or threonine, which have been observed in some well characterized RING proteins [14]. Hydrophobic residues at these positions are critical for cross-brace structure formation. Thirty more domains failed this test. However, the remaining 294 domains can be considered fairly certain to form a RING structure.
Next (Stringent 3), we abolished the acceptance of threonine or serine after metal ligand 6, as these residues are rarely found in this position in well characterized RING proteins. Ten more domains did not comply with this requirement.
Finally, in our most stringent pattern (Stringent 4) we not only excluded aromatic residues from position 2 in front of metal ligand 7, but also other hydrophobic residues. An additional 55 domains from our set did not match this criterion. However, the 229 domains fulfilling these criteria can be considered to form a RING structure with near certainty.
Clustering of related RING domains
We sought to define groups of related RING-domain proteins beyond their classification by metal-ligand arrangement. However, it turned out that, at least in part, RING-domain proteins strongly deviate from each other outside the conserved domain. Thus it is not feasible to relate them using conventional phylogenetic methods. To circumvent this problem, the isolated RING-finger domains were used instead for further analysis. However, bootstrap values were again too low to reliably relate RING domains using phylogenetic methods. Therefore, a single-linkage-clustering method was applied to obtain clusters of related RING domains. We sorted our set on the basis of similarity restricted to the RING domains and excluding non-conserved amino- and carboxy-terminal parts of the respective proteins. A BLAST analysis [25] using a cut-off value of 10-15 was chosen to define meaningful similarities (Table 3). This analysis resulted in the definition of 54 clusters of RING domains (that is, two or more similar domains), in which 295 domains are grouped. Notably, with only one exception (cluster 2.8), all the RING-domain clusters only contain members from the same respective class of metal-ligand arrangement; that is, prototypical RING-HC domains are only found in clusters with other RING-HC domains. This finding confirms both the significance of our clustering and of the group definition described above. Of the 54 clusters, 28 consist of a pair of domains, whereas clusters with multiple domains contain up to 75 different domains. However, with the exception of the large clusters 2.1 (75 domains) and 2.2 (26 domains) found in the prototypical RING-H2 class, most clusters contain fewer than ten domains.
Table 3.
List of Arabidopsis RING domains
| Protein-ID | S1 | S2 | S3 | S4 | Alternative names and remarks | |
| (1) RING-HC: 118 domains in 111 proteins | ||||||
| Cluster 1.1 | At1g19310 | + | + | + | + | |
| At1g74990 | + | + | + | + | ||
| At2g23780 | + | + | + | + | ||
| At2g42030 | + | + | + | + | ||
| AT3g58030 | + | + | + | + | ||
| AT4g03510 | + | + | + | + | RMA1, ubiquitin ligase activity [29] | |
| AT4g27470 | + | + | + | + | ||
| AT4g28270 | + | + | + | + | ||
| Cluster 1.2 | At1g62370 | + | + | - | - | |
| AT3g07120 | + | + | - | - | ||
| AT3g25030 | + | + | - | - | ||
| AT4g03960 | + | + | - | - | ||
| AT4g13100 | + | + | - | - | ||
| AT4g22250 | + | + | - | - | ||
| Cluster 1.3 | At1g57800 | + | + | + | + | Second motif in protein |
| At1g57820 | + | + | + | + | Second motif in protein | |
| At1g66040 | + | + | + | + | Second motif in protein | |
| At1g66050 | + | + | + | + | Second motif in protein | |
| AT4g08590 | + | + | + | + | ||
| AT5g39550 | + | + | + | + | Second motif in protein | |
| Cluster 1.4 | At1g57800 | + | + | + | - | Second motif in protein |
| At1g57820 | + | + | + | - | Second motif in protein | |
| At1g66040 | + | + | + | - | Second motif in protein | |
| At1g66050 | + | + | + | - | Second motif in protein | |
| AT5g39550 | + | + | + | - | Second motif in protein | |
| Cluster 1.5 | AT3g06140 | + | + | + | + | |
| AT3g09770 | + | + | + | + | ||
| AT3g53410 | + | + | + | + | ||
| AT5g03200 | + | + | + | + | ||
| AT5g19080 | + | + | + | + | ||
| Cluster 1.6 | At1g24440 | + | + | + | + | |
| AT3g47160 | + | + | + | + | Potential RNA-binding protein | |
| AT5g01520 | + | + | + | + | ||
| AT5g58790 | + | + | + | + | ||
| Cluster 1.7 | At1g66620 | + | - | - | - | |
| At1g66630 | - | - | - | - | Likely not to be a RING | |
| AT5g37870 | + | - | - | - | ||
| AT5g37910 | + | - | - | - | ||
| Cluster 1.8 | At2g41980 | + | - | - | - | |
| AT3g58040 | + | - | - | - | ||
| AT3g61790 | + | - | - | - | Seven in absentia-like | |
| AT4g27880 | + | - | - | - | ||
| Cluster 1.9 | At1g69330 | + | + | + | - | |
| At1g74370 | + | + | + | - | ||
| AT3g29270 | + | + | + | - | ||
| Cluster 1.10 | At1g01350 | + | + | + | + | |
| AT4g01020 | + | + | + | + | Second motif in protein | |
| AT5g06420 | + | + | + | + | ||
| Cluster 1.11 | At1g30860 | + | + | + | + | |
| At2g34920 | + | + | + | + | ||
| AT5g44690 | + | + | + | - | ||
| Cluster 1.12 | AT5g07270 | + | + | + | - | |
| AT5g57740 | + | + | + | - | ||
| Cluster 1.13 | At1g59560 | + | - | - | - | |
| At1g63900 | + | - | - | - | ||
| Cluster 1.14 | At1g79110 | + | - | - | - | |
| AT3g12920 | + | - | - | - | ||
| Cluster 1.15 | AT5g22750 | + | + | + | + | |
| AT5g43530 | + | + | + | + | Putative DNA-repair protein | |
| Cluster 1.16 | At1g03370 | + | - | - | - | |
| AT4g03000 | + | - | - | - | ||
| Cluster 1.17 | AT3g01650 | + | - | - | - | |
| AT5g14420 | + | - | - | - | ||
| Cluster 1.18 | At1g03770 | + | + | + | + | |
| AT5g44280 | + | + | + | + | ||
| Cluster 1.19 | At1g32530 | + | + | - | - | |
| At2g35330 | + | + | - | - | ||
| Cluster 1.20 | At1g10650 | + | - | - | - | Apoptosis inhibitor-like |
| At1g60610 | + | - | - | - | ||
| Cluster 1.21 | At2g38190 | + | - | - | - | |
| AT5g01450 | + | - | - | - | ||
| Cluster 1.22 | At2g47090 | + | + | + | + | |
| AT3g62240 | + | + | + | + | ||
| Cluster 1.23 | AT4g19700 | + | - | - | - | |
| AT5g45100 | + | - | - | - | ||
| Cluster 1.24 | AT3g23280 | - | - | - | - | Likely not to be a RING |
| AT4g14360 | + | - | - | - | Ankyrin-like protein | |
| Unique | At1g05050 | + | - | - | - | Putative transcription factor |
| At1g05120 | + | + | + | - | ||
| At1g18660 | + | + | + | + | ||
| At1g21650 | + | + | + | - | Putative SecA-type plastid protein transport factor | |
| At1g32740 | + | - | - | - | ||
| At1g55250 | + | + | + | + | ||
| At1g61620 | + | + | + | + | ||
| At1g67180 | + | + | + | + | ||
| At1g67800 | + | - | - | - | ||
| At1g68820 | + | - | - | - | ||
| At1g79380 | + | - | - | - | ||
| At1g79810 | - | - | - | - | Likely not to be a RING | |
| At2g22010 | + | + | + | + | ||
| At2g25380 | + | + | + | + | ||
| At2g26350 | + | + | + | + | Putative peroxisome membrane protein | |
| At2g28840 | + | + | - | - | Ankyrin repeat protein | |
| At2g30580 | + | + | + | + | ||
| At2g32950 | + | + | + | + | COP1, putative ubiquitin ligase [39] | |
| At2g39100 | + | + | + | + | ||
| At2g40770 | + | + | + | - | SNF2/SWI2-like protein | |
| At2g44410 | + | + | + | + | ||
| At2g44950 | + | + | + | + | ||
| AT3g05250 | + | + | + | - | ||
| AT3g05670 | + | + | + | + | ||
| AT3g07200 | + | + | + | - | ||
| AT3g24800 | + | + | + | + | PRT1, N-end rule ubiquitin ligase [35] | |
| Second motif in protein | ||||||
| AT3g24800 | + | + | + | - | PRT1, N-end rule ubiquitin ligase [35] | |
| Second motif in protein | ||||||
| AT3g26730 | + | + | + | + | ||
| AT3g27330 | + | + | + | + | ||
| AT3g54360 | + | + | + | + | ||
| AT4g01740 | + | + | + | + | ||
| AT4g17680 | + | + | + | + | ||
| AT4g21070 | + | + | + | - | ||
| AT4g33940 | + | + | + | + | ||
| AT5g01160 | + | + | + | + | ||
| AT5g05130 | + | + | + | + | Helicase-like protein | |
| AT5g13530 | + | + | + | - | Ankyrin repeat protein | |
| AT5g19430 | + | + | + | + | ||
| AT5g23110 | + | + | - | - | ||
| AT5g47050 | + | - | - | - | ||
| AT5g63700 | + | + | + | + | ||
| (2) RING-H2: 215 domains in 214 proteins | ||||||
| Cluster 2.1 | At1g04360 | + | + | + | - | |
| At1g20810 | + | + | + | + | ||
| At1g22500 | + | + | + | + | ||
| At1g23980 | + | + | + | + | ||
| At1g28040 | + | + | + | + | ||
| At1g32360 | + | + | + | + | ||
| At1g33480 | + | + | + | + | ||
| At1g35330 | + | + | + | + | ||
| At1g49200 | + | + | + | + | ||
| At1g49210 | + | + | + | + | ||
| At1g49220 | + | + | + | + | ||
| At1g49230 | + | + | + | + | ||
| At1g53820 | + | + | + | + | ||
| At1g72200 | + | + | + | + | ||
| At1g72220 | + | + | + | + | ||
| At1g72310 | + | + | + | + | ATL3 | |
| At1g74410 | + | + | + | + | ||
| At1g76410 | + | + | + | + | ||
| At2g17460 | + | + | + | + | Pseudogene | |
| At2g17730 | + | + | + | - | ||
| At2g18650 | + | + | + | + | ||
| At2g20030 | + | + | + | - | ||
| At2g27940 | + | + | + | - | ||
| At2g34990 | + | + | + | + | ||
| At2g35000 | + | + | + | + | ||
| At2g35420 | + | + | + | + | ||
| At2g35910 | + | + | + | + | ||
| At2g37580 | + | + | + | + | ||
| At2g42350 | + | + | + | + | ||
| At2g42360 | + | + | + | + | ||
| At2g46160 | + | + | + | + | ||
| At2g47560 | + | + | + | + | ||
| AT3g03550 | + | + | + | + | ||
| AT3g05200 | + | + | + | - | ATL6 | |
| AT3g10910 | + | + | + | + | ||
| AT3g11110 | + | + | + | + | ||
| AT3g14320 | + | + | + | + | ||
| AT3g16720 | + | + | + | + | ||
| AT3g18930 | + | + | + | + | ||
| AT3g19140 | - | - | - | - | Likely not to be a RING | |
| AT3g48030 | + | + | + | + | ||
| AT3g60220 | + | + | + | + | ATL4 | |
| AT3g61550 | + | + | + | + | ||
| AT3g62690 | + | + | + | + | ATL5 | |
| AT4g09100 | + | + | + | + | ||
| AT4g09110 | + | + | + | + | ||
| AT4g09120 | + | + | + | + | ||
| AT4g09130 | + | + | + | + | ||
| AT4g10150 | + | + | + | + | ||
| AT4g10160 | + | + | + | + | ||
| AT4g15970 | + | + | + | + | ||
| AT4g17910 | + | + | + | + | ||
| AT4g17920 | + | + | + | + | ||
| AT4g28890 | + | + | + | - | ||
| AT4g30400 | + | + | + | + | ||
| AT4g35480 | + | + | + | + | RHA3b | |
| AT4g35840 | + | + | + | - | ||
| AT4g38140 | + | + | + | - | ||
| AT4g40070 | + | + | + | - | ||
| AT5g01880 | + | + | + | + | ||
| AT5g05280 | + | + | + | + | ||
| AT5g05810 | + | + | + | + | ||
| AT5g06490 | + | + | + | + | ||
| AT5g07040 | + | + | + | - | ||
| AT5g10380 | + | + | + | + | ||
| AT5g17600 | + | + | + | + | ||
| AT5g27420 | + | + | + | - | ||
| AT5g40250 | + | + | + | + | ||
| AT5g42200 | + | + | + | + | ||
| AT5g43420 | + | + | + | - | ||
| AT5g46650 | + | + | + | + | ||
| AT5g47610 | + | + | + | + | ||
| AT5g57750 | + | + | + | + | ||
| AT5g58580 | + | + | + | + | ||
| AT5g66070 | + | + | + | - | ||
| Cluster 2.2 | At1g14200 | + | + | + | - | |
| At1g26800 | + | + | + | + | ||
| At1g55530 | + | + | + | + | ||
| At1g60360 | + | + | + | + | ||
| At1g68180 | + | + | + | + | ||
| At2g03000 | + | + | + | + | ||
| At2g39720 | + | + | + | + | ||
| At2g40830 | + | + | + | + | ||
| At2g44330 | + | + | + | + | ||
| AT3g02340 | + | + | + | + | ||
| AT3g10810 | + | + | + | + | ||
| AT3g13430 | + | + | + | + | ||
| AT3g19950 | + | + | + | + | ||
| AT3g30460 | + | + | + | + | ||
| AT3g46620 | + | + | + | + | ||
| AT3g60080 | + | + | + | + | ||
| AT4g26400 | + | + | + | + | ||
| AT5g01980 | + | + | + | + | ||
| AT5g02750 | + | + | + | + | ||
| AT5g08140 | + | + | + | + | ||
| AT5g15820 | + | + | + | + | ||
| AT5g20910 | + | + | + | + | AIP2 | |
| AT5g56340 | + | + | + | + | ||
| AT5g59550 | + | + | + | + | ||
| AT5g60820 | + | + | + | + | ||
| AT5g64920 | + | + | + | + | CIP8, ubiquitin ligase activity [32] | |
| Cluster 2.3 | At1g17970 | + | + | + | + | |
| At1g45180 | + | + | + | + | ||
| At1g53190 | + | + | + | + | ||
| At1g73760 | + | + | + | + | ||
| At2g15530 | + | + | + | + | ||
| AT3g15070 | + | + | + | + | ||
| AT4g31450 | + | + | + | + | ||
| AT4g34040 | + | + | + | + | ||
| AT5g10650 | + | + | + | + | ||
| AT5g24870 | + | + | + | + | ||
| AT5g42940 | + | + | + | + | ||
| Cluster 2.4 | At1g12760 | + | + | + | - | |
| At1g63170 | + | + | + | - | ||
| At1g68070 | + | + | + | - | ||
| At1g80400 | + | + | + | - | ||
| AT3g61180 | + | + | + | - | ||
| AT4g11680 | + | + | + | + | ||
| AT4g26580 | + | + | + | + | ||
| AT4g32600 | + | + | + | - | ||
| Cluster 2.5 | At1g63840 | + | + | + | - | |
| AT3g43430 | + | + | + | + | ||
| AT3g61460 | + | + | + | + | ||
| AT4g11360 | + | + | + | - | RHA1b | |
| AT4g11370 | + | + | + | - | ||
| AT5g20880 | + | + | + | + | ||
| AT5g41400 | + | + | + | - | ||
| Cluster 2.6 | AT5g37200 | + | + | + | + | |
| AT5g37230 | + | + | + | + | ||
| AT5g37250 | + | + | + | + | ||
| AT5g37270 | + | + | + | + | ||
| AT5g37280 | + | + | + | + | ||
| Cluster 2.7 | AT3g02290 | + | + | + | + | |
| AT4g23450 | + | + | + | + | ||
| AT5g15790 | + | + | + | + | ||
| AT5g38890 | + | + | + | + | ||
| AT5g41350 | + | + | + | + | ||
| Cluster 2.8 | AT4g05350 | + | + | + | + | |
| AT4g12140 | + | + | + | + | ||
| AT4g12150 | - | - | - | - | Variant, ligand missing! | |
| AT4g12190 | + | + | + | + | ||
| AT4g12210 | + | + | + | + | ||
| Cluster 2.9 | At1g19680 | + | + | + | + | |
| At1g75400 | + | + | + | + | ||
| At2g21500 | + | + | + | + | ||
| AT4g39140 | + | + | + | + | ||
| AT5g18260 | + | + | + | + | ||
| Cluster 2.10 | At1g22670 | + | + | + | + | |
| At1g35630 | + | + | + | + | ||
| At1g71980 | + | + | + | + | RMRJR702 | |
| AT4g09560 | + | + | + | + | ||
| AT5g66160 | + | + | + | + | RMRJR700 | |
| Cluster 2.11 | At1g51930 | + | + | + | + | |
| AT5g41430 | + | + | + | + | ||
| AT5g41440 | + | + | + | + | ||
| AT5g41450 | + | + | + | - | ||
| Cluster 2.12 | AT3g62970 | - | - | - | - | |
| AT5g18650 | - | - | - | - | ||
| AT5g22920 | - | - | - | - | PGPD14-like protein | |
| AT5g25560 | - | - | - | - | ||
| Cluster 2.13 | At1g08050 | + | + | + | + | |
| At2g38970 | + | + | + | + | Putative pol polyprotein | |
| AT3g54780 | + | + | + | + | ||
| AT5g60710 | + | + | + | + | ||
| Cluster 2.14 | At1g18910 | - | - | - | - | Likely not to be a RING |
| At1g74760 | - | - | - | - | Likely not to be a RING | |
| AT3g18290 | - | - | - | - | Likely not to be a RING | |
| Cluster 2.15 | At2g24480 | + | + | + | + | |
| AT3g28620 | + | + | + | + | ||
| AT5g43200 | + | + | + | + | ||
| Cluster 2.16 | At1g15100 | - | - | - | - | Likely not to be a RING |
| At2g01150 | - | - | - | - | RHA2b, likely not to be a RING | |
| Cluster 2.17 | At1g65040 | + | + | + | + | |
| AT3g16090 | + | + | + | + | ||
| Cluster 2.18 | AT3g14970 | + | - | - | - | |
| AT3g15740 | + | - | - | - | ||
| Cluster 2.19 | At2g44580 | + | + | + | + | Second nearly identical motif in protein |
| At2g44580 | + | + | + | + | Second nearly identical motif in protein | |
| Cluster 2.20 | At2g22680 | + | + | + | - | Putative pol polyprotein |
| AT4g37890 | + | + | + | + | ||
| Cluster 2.21 | At2g18670 | + | + | + | + | |
| AT4g30370 | + | + | + | + | ||
| Cluster 2.22 | AT4g25230 | - | - | - | - | Likely not to be a RING |
| AT5g51450 | - | - | - | - | Likely not to be a RING | |
| Unique | At1g04790 | + | + | + | + | |
| At1g24580 | + | + | + | + | ||
| At1g27010 | + | + | + | + | ||
| At1g49850 | + | + | + | + | RHY1a | |
| At1g53010 | + | + | + | - | ||
| At1g55410 | - | - | - | - | Likely not to be a RING | |
| At1g57730 | + | + | + | + | ||
| At1g70910 | + | + | + | + | ||
| At1g74620 | + | + | + | + | ||
| At2g04240 | + | + | + | - | ||
| At2g05170 | + | + | + | + | ||
| At2g15260 | + | + | + | - | ||
| At2g15580 | + | + | + | - | ||
| At2g20650 | + | + | + | - | ||
| At2g26000 | + | + | + | + | ||
| At2g28920 | + | + | + | - | ||
| At2g47700 | + | + | + | - | ||
| AT3g05870 | + | + | + | - | ||
| AT3g47180 | + | + | + | + | ||
| AT3g47990 | + | + | + | - | ||
| AT3g55530 | + | + | + | + | ||
| AT3g58720 | + | + | + | + | ||
| AT4g01270 | + | + | + | + | ||
| AT4g13490 | + | + | + | - | ||
| AT4g14220 | + | + | + | + | RHF1a | |
| AT4g18110 | + | + | + | + | ||
| AT5g05530 | + | + | + | + | ||
| AT5g05910 | + | + | + | + | ||
| AT5g54990 | + | + | + | + | ||
| AT5g57820 | + | + | + | + | ||
| AT5g67120 | + | + | + | + | ||
| RING variants | ||||||
| (3) RING-HC, C6 or C7 position substituted or missing: 41 domains in 39 proteins | ||||||
| Cluster 3.1 | At2g26130 | - | - | - | - | Second motif in protein |
| At2g26130 | - | - | - | - | Second motif in protein | |
| AT3g43180 | + | + | + | + | ||
| AT3g43750 | - | - | - | - | ||
| AT3g45510 | - | - | - | - | ||
| AT3g45540 | - | - | - | - | ||
| AT3g45560 | - | - | - | - | ||
| AT3g45570 | - | - | - | - | ||
| AT3g45580 | - | - | - | - | ||
| AT5g60250 | + | + | + | + | Second motif in protein | |
| Cluster 3.2 | At1g05890 | - | - | - | - | |
| At1g65430 | - | - | - | - | ||
| At2g31510 | - | - | - | - | Dm Ariadne-like | |
| At2g31760 | - | - | - | - | Dm Ariadne-like | |
| At2g31770 | - | - | - | - | Dm Ariadne-like | |
| At2g31780 | - | - | - | - | Dm Ariadne-like | |
| Cluster 3.3 | At1g50410 | + | + | + | + | Putative DNA-binding protein |
| AT3g16600 | + | + | + | + | Putative DNA-binding protein | |
| AT3g20010 | + | + | + | + | RUSH1-alpha-like protein | |
| Cluster 3.4 | At2g16090 | - | - | - | - | |
| AT3g27710 | - | - | - | - | ||
| AT4g34370 | - | - | - | - | ||
| Cluster 3.5 | AT5g63750 | - | - | - | - | Dm Ariadne-like |
| AT5g63760 | - | - | - | - | ||
| Cluster 3.6 | AT4g01020 | - | - | - | - | Second motif in protein |
| AT5g10370 | - | - | - | - | ||
| Cluster 3.7 | At2g25360 | - | - | - | - | |
| AT3g45470 | - | - | - | - | ||
| Unique | At1g11100 | - | - | - | - | RUSH1 alpha-like protein |
| At1g32340 | - | - | - | - | ||
| At1g61140 | - | - | - | - | ||
| At2g19610 | - | - | - | - | ||
| AT3g14250 | - | - | - | - | ||
| AT3g45480 | + | + | + | + | ||
| AT3g45580 | - | - | - | - | Second motif in protein | |
| AT3g53690 | - | - | - | - | ||
| AT3g54460 | - | - | - | - | ||
| AT4g19670 | - | - | - | - | Second motif in protein | |
| AT4g19670 | - | - | - | - | Second motif in protein | |
| AT5g07640 | - | - | - | - | ||
| At5g60250 | - | - | - | - | Second motif in protein | |
| At5g60250 | - | - | - | - | Second motif in protein | |
| (4) RING-H2, C7 position substituted or missing: two domains in two proteins | ||||||
| AT3g42830 | + | + | + | + | Rbx1-like | |
| AT5g37220 | - | - | - | - | ||
| (5) C4 or C5 position substituted or missing: nine domains in nine proteins | ||||||
| Cluster 5.1 | At1g18760 | - | - | - | - | |
| At1g18770 | - | - | - | - | ||
| At1g18780 | - | - | - | - | ||
| At1g21960 | - | - | - | - | ||
| At2g29840 | - | - | - | - | ||
| AT5g53910 | - | - | - | - | Likely not to be a RING | |
| At1g77830 | - | - | - | - | RHA1b-like | |
| Unique | At1g36950 | - | - | - | - | |
| At2g34000 | - | - | - | - | ||
| (6) C2 position substituted or missing: two domains in two proteins | ||||||
| At2g37150 | - | - | - | - | ||
| AT5g63760 | - | - | - | - | Second motif in protein | |
List of Arabidopsis proteins (Protein-ID) that contain a manually curated RING domain according to the criteria described in the text. The table is divided into three principal sections: RING domains of the RING-HC type; RING domains of the RING-H2 type; and RING-variant domains. In addition, the domains are ordered in numbered clusters according to a single-linkage-clustering method with a 10-15 threshold as described in the text. If an individual domain meets the requirements of one of the stringent patterns 1 to 4 (S1-S4) (see Table 2), it is indicated by a plus sign, if not, it is indicated by a minus sign. Additional remarks such as previously assigned gene names and significant similarities are given in the last column, The remark 'likely not to be a RING' refers to the presence of an aromatic residue two positions in front of metal-ligand position 7 as outlined in the text.
Redundancy of clustered proteins
To investigate overall protein similarities and reveal potential functional redundancy between proteins of a given cluster, we decided to produce alignments of the full-length proteins by ClustalW analysis [26]. Links to these alignments are provided with each cluster on our web page. Overall similarities vary between clusters. For instance, the 75 domains of cluster 2.1 contain several members of a RING-H2 family that has been described [27,28]. The proteins in this cluster are generally short, with only very little additional sequence outside the RING domain, and are highly similar to each other. Thus, any functional overlaps could already be contained in the RING domain itself. Other clusters, for example 2.6 and 2.8, consist of genes that are derived from tandem duplication.
In numerous other cases, however, the full-length alignments frequently reveal additional similarities outside the RING domain. For instance, cluster 1.1 includes the RMA1 protein, which has been shown to be a membrane-bound ubiquitin ligase [29]. The proteins in this cluster show some sequence similarity besides the RING domain and share additional features, such as a transmembrane anchor. Thus, it seems likely that these other proteins might also be membrane-bound ubiquitin ligases. Another interesting group is cluster 2.2, which comprises 26 domains. The corresponding proteins also contain significant similarity in a stretch amino-terminal to the RING domain. Moreover, subgroups of proteins within the cluster even display additional similarity in the more distal amino-terminal regions. Two of the proteins in this cluster, AIP2 and CIP8, have been described [30,31]. For CIP8, a ubiquitin ligase activity has been demonstrated [32] and the same might be true for a subgroup of the cluster, which has a high structural similarity with CIP8 (C.H., unpublished data). Thus, this cluster might represent proteins that are functionally redundant to some extent. Notably, the similarity among members of cluster 2.2 also extends to their genomic organization: 22 of the 26 proteins are encoded by a single exon, underpinning their close relatedness and probable common evolutionary origin.
Conclusions
Gene redundancy in Arabidopsis has previously been shown to limit the number of mutants detectable by phenotype [33]. The completed genome sequence shows that a high degree of redundancy might indeed obscure the quest for many phenotypes. Accordingly, we suggest that there probably exists a high degree of functional redundancy among Arabidopsis RING-domain proteins. This would also correlate with the fact that surprisingly few genes in the complete set are characterized as mutants. To our knowledge, this is the case for only two of them, COP1 and PRT1 [34,35]. Notably, for both these proteins, a functional requirement for the RING domain has been demonstrated, and both are unique with respect to their RING domains.
In this study, we present an ordered set of manually curated RING domains of Arabidopsis. In summary, our set includes all bona fide RING domains, as well as common RING-variant domains. Notably, additional Arabidopsis proteins might have potential to form variant RING-finger domains, as has been suggested, for instance, for the HOS1 protein [36]. However, their primary sequences do not support this notion unambiguously and we chose not to include any RING-domain variants in our analysis for which no structural experimental evidence is yet available. Clearly, our findings show that predictions of cysteine-rich domains have to be met with skepticism. On a proteomics level, they can be misleading in drawing general conclusions, as is amply demonstrated by the overestimation of the abundance of the PHD domain owing to their overlapping classification with RING domains. Additional structural data are needed and have to be taken into account in computational analyses to resolve these issues. Our curated set of RING domains in Arabidopsis will serve as a vital starting point for further genome analysis in this field.
Materials and methods
The non-redundant Arabidopsis genome protein set available at MIPS [37] was screened for proteins containing RING-finger domains. Detailed results of this analysis are available on the web [20,21]. Analysis was undertaken using several discrete steps described in detail below.
Whole-genome analysis for proteins containing RING-finger motifs proteins
For initial analysis the InterPro system [2] (iprscan version 3.2) was used to calculate protein domains for all Arabidopsis proteins. The results were filtered for RING-finger domains matching the InterPro domains PF00097, PS00518 or PS50089 (corresponding to domain names ZF-C3HC4, ZINC_FINGER_C3HC4 and ZF_RING, respectively). Arabidopsis proteins containing one or more RING-finger domains were analyzed further.
Frequently detected overlapping domains, for example detected patterns with overlapping localization, were unified and only the domain with the most amino-terminal starting point was used for further analysis.
Prediction of additional domains
Proteins containing a RING-finger domain were subjected to an additional screen for the presence of additional domains using the InterPro package (see above).
Classification of RING-finger domains
The RING-finger domain summarizes different types of subdomains, namely the C3HC4-type and C3H2C3-type. We refer to these types as RING-HC and RING-H2, respectively. To differentiate between these two subtypes an additional fine analysis was carried out: RING-finger-containing genes were classified as C3HC4-type (RING-HC) for the patterns C-x-H-x-[LIVMFY]-C-x(2)-C-[LIVMYA] or C-x(2)-C-x(9-39)-C-x(1-3)-H-x(2-3)-C-x(2)-C-x(4-48)-C-x(2)-C and as C3H2C3-type (RING-H2) for the patterns C-x-H-x-[LIVMFY]-H-x(2)-C-[LIVMYA] or C-x(2)-C-x(9-39)-C-x(1-3)-H-x(2-3)-H-x(2)-C-x(4-48)-C-x(2)-C. RING-finger domains detected by InterPro that did not match these patterns were marked as 'others/unclear type'. Novel patterns for the evaluation of RING domains were defined as described in the text and Table 2.
Clustering of RING-finger domains
The isolated RING domains were related using BLASTP [25] (version 2.1.2) by testing the isolated RING-finger domains against a database containing all RING-finger domains assembled during the previous analysis steps. Domains below a threshold of 10-15 were united into clusters of related domains. This procedure is 'greedy'; for example, although domain A relates to domain B and B relates to C, A and C are not necessarily closely related enough to exceed the given threshold. Nevertheless, this procedure in general succeeded in grouping and/or separating individual subfamilies.
Multiple alignments
For RING-domain clusters with two or more members, multiple alignments of the respective complete protein sequences were done using the ClustalW program [26,38] with default parameter settings.
Manual expert curation
The individual RING-finger domains and clusters underwent manual inspection. Manual adjustments to clusters and rejections of individual domains and clusters on the basis of expert knowledge were carried out as explained in Results and discussion.
Additional data files
The curated set of clustered Arabidopsis RING domains, with their sequences and metal ligands, are provided in a supplemental table. Links to the individual genes and ClustalW analyses are included.
Supplementary Material
The curated set of clustered Arabidopsis RING domains, with their sequences and metal ligands
Acknowledgments
Acknowledgements
Our work was supported by the GABI project of the Bundesministerium für Bildung, Forschung und Technologie.
References
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. InterPro - an integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16:1145–1150. doi: 10.1093/bioinformatics/16.12.1145. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont PS, Hanson IM, Trowsdale J. A novel cysteine-rich sequence motif. Cell. 1991;64:483–484. doi: 10.1016/0092-8674(91)90229-r. [DOI] [PubMed] [Google Scholar]
- Lovering R, Hanson IM, Borden KL, Martin S, O'Reilly NJ, Evan GI, Rahman D, Pappin DJ, Trowsdale J, Freemont PS. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc Natl Acad Sci USA. 1993;90:2112–2116. doi: 10.1073/pnas.90.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- Borden KL, Boddy MN, Lally J, O'Reilly NJ, Martin S, Howe K, Solomon E, Freemont PS. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow PN, Luisi B, Milner A, Elliott M, Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol. 1994;237:201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- Perez-Alvarado GC, Miles C, Michelsen JW, Louis HA, Winge DR, Beckerle MC, Summers MF. Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat Struct Biol. 1994;1:388–398. doi: 10.1038/nsb0694-388. [DOI] [PubMed] [Google Scholar]
- Perez-Alvarado GC, Kosa JL, Louis HA, Beckerle MC, Winge DR, Summers MF. Structure of the cysteine-rich intestinal protein, CRIP. J Mol Biol. 1996;257:153–174. doi: 10.1006/jmbi.1996.0153. [DOI] [PubMed] [Google Scholar]
- Konrat R, Weiskirchen R, Krautler B, Bister K. Solution structure of the carboxyl-terminal LIM domain from quail cysteine-rich protein CRP2. J Biol Chem. 1997;272:12001–12007. doi: 10.1074/jbc.272.18.12001. [DOI] [PubMed] [Google Scholar]
- Capili AD, Schultz DC, Rauscher FJ, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ. Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- Kentsis A, Dwyer EC, Perez JM, Sharma M, Chen A, Pan ZQ, Borden KL. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J Mol Biol. 2001;312:609–623. doi: 10.1006/jmbi.2001.5003. [DOI] [PubMed] [Google Scholar]
- Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- Analysis of RING Finger domain proteins encoded by the Arabidopsis thaliana genome http://mips.gsf.de/proj/thal/ring/index.html
- Evaluation and classification of RING finger domains encoded by the Arabidopsis thaliana genome http://www.mcgill.ca/Biology/faculty/hardtke/ARARINGS/AraRINGindex.html [DOI] [PMC free article] [PubMed]
- Gervais V, Busso D, Wasielewski E, Poterszman A, Egly JM, Thierry JC, Kieffer B. Solution structure of the N-terminal domain of the human TFIIH MAT1 subunit: new insights into the RING finger family. J Biol Chem. 2001;276:7457–7464. doi: 10.1074/jbc.M007963200. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Borden KL, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Azevedo C, Santos-Rosa MJ, Shirasu K. The U-box protein family in plants. Trends Plant Sci. 2001;6:354–358. doi: 10.1016/s1360-1385(01)01960-4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Jensen KL, Jespersen HM, Skriver K. Widespread occurrence of a highly conserved RING-H2 zinc finger motif in the model plant Arabidopsis thaliana. FEBS Lett. 1998;436:283–287. doi: 10.1016/s0014-5793(98)01143-0. [DOI] [PubMed] [Google Scholar]
- Salinas-Mondragon RE, Garciduenas-Pina C, Guzman P. Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Mol Biol. 1999;40:579–590. doi: 10.1023/a:1006267201855. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Suzuki T, Tanaka K, Nakano A. Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J Cell Sci. 2001;114:1949–1957. doi: 10.1242/jcs.114.10.1949. [DOI] [PubMed] [Google Scholar]
- Kurup S, Jones HD, Holdsworth MJ. Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 2000;21:143–155. doi: 10.1046/j.1365-313x.2000.00663.x. [DOI] [PubMed] [Google Scholar]
- Torii KU, Stoop-Myer CD, Okamoto H, Coleman JE, Matsui M, Deng XW. The RING finger motif of photomorphogenic repressor COP1 specifically interacts with the RING-H2 motif of a novel Arabidopsis protein. J Biol Chem. 1999;274:27674–27681. doi: 10.1074/jbc.274.39.27674. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Okamoto H, Stoop-Myer C, Deng XW. Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 2002. [DOI] [PubMed]
- Bouche N, Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol. 2001;4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Potuschak T, Stary S, Schlogelhofer P, Becker F, Nejinskaia V, Bachmair A. PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc Natl Acad Sci USA. 1998;95:7904–7908. doi: 10.1073/pnas.95.14.7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIPS Arabidopsis thaliana genome project http://mips.gsf.de/proj/thal
- ClustalW http://www.ebi.ac.uk/clustalw/
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The curated set of clustered Arabidopsis RING domains, with their sequences and metal ligands


