Abstract
Collagenous connective tissue, found throughout the bodies of metazoans, plays a crucial role in maintaining structural integrity. This versatile tissue has the potential for numerous biomedical applications, including the development of innovative collagen-based biomaterials. Inspiration for such advancements can be drawn from echinoderms, a group of marine invertebrates that includes sea stars, sea cucumbers, brittle stars, sea urchins, and sea lilies. Through their nervous system, these organisms can reversibly control the pliability of their connective tissue components (i.e., tendons and ligaments) that are composed of mutable collagenous tissue (MCT). The variable tensile properties of the MCT allow echinoderms to perform unique functions, including postural maintenance, reduction of muscular energy use, autotomy to avoid predators, and asexual reproduction through fission. The changes in the tensile strength of MCT structures are specifically controlled by specialized neurosecretory cells called juxtaligamental cells. These cells release substances that either soften or stiffen the MCT. So far, only a few of these substances have been purified and characterized, and the genetic underpinning of MCT biology remains unknown. Therefore, we have conducted this research to identify MCT-related genes in echinoderms as a first step towards a better understanding of the MCT molecular control mechanisms. Our ultimate goal is to unlock new biomaterial applications based on this knowledge. In this project, we used RNA-Seq to identify and annotate differentially expressed genes in the MCT structures of the brittle star Ophiomastix wendtii. As a result, we present a list of 16 putative MCT modulator genes, which will be validated and characterized in forthcoming functional analyses.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10926-7.
Keywords: Echinoderm, Mutable collagenous tissue, Gene annotation, Histology, High-throughput sequencing
Introduction
Collagenous connective tissues are ubiquitous in metazoans, including humans. These tissues are found in all body parts, as they maintain body integrity by providing structural support and managing applied mechanical forces through dissipation, transmission, and resistance. Additionally, connective tissue carries out regulatory functions by affecting cell adhesion, differentiation, survival, and intercellular signaling via both mechanotransduction and biochemical events [1, 2]. The critical roles that connective tissue plays are underscored by a diverse array of collagen-related human diseases such as osteogenesis imperfect and Ehlers-Danlos syndrome that range in their manifestation from a moderately increased susceptibility to other ailments to devastating lethal outcomes [2–5].
In most animals, the mechanical properties of collagenous tissues either remain stable with time or change very slowly due to developmental processes, aging, exercise, or disease (e.g., fibrosis) [2, 6]. The only known animals in which collagenous connective tissues undergo rapid (within seconds) drastic changes in tensile strength and elastic stiffness are echinoderms. Echinoderms are a phylum of deuterostome marine invertebrates, which includes asteroids (sea stars), holothuroids (sea cucumbers), ophiuroids (brittle stars and basket stars), echinoids (sea urchins and sand dollars), and crinoids (sea lilies). The connective tissue capable of such rapid stiffening or softening in echinoderms is called mutable collagenous tissue (MCT) [6–8]. MCT appears to be unique to echinoderms, likely representing a synapomorphy of the phylum which is supported by indirect evidence of MCT in fossil echinoderms, including Middle Cambrian stylophorans and other Paleozoic species [9].
The unique dynamic nature of the echinoderm MCT allows it to perform additional functions, along with the standard roles that the collagenous tissue plays in all metazoan phyla. One such echinoderm-specific function is autotomy (i.e., defensive self-detachment of arms in stellate echinoderms and viscera in holothuroids). This process involves irreversible softening of otherwise mechanically stable collagenous structures at pre-determined anatomical locations in response to various mechanical, physical, and chemical stimuli [10–13]. The same mechanism of irreversible MCT destabilization is also thought to facilitate asexual reproduction through body fission [14, 15]. Reversible stiffening and de-stiffening of the MCT are the main mechanisms behind the energy-saving acquisition and maintenance of posture in echinoderms [16, 17].
As an echinoderm synapomorphy, the MCT has contributed to the evolutionary success of the phylum over the last 500 million years. A better understanding of the molecular physiology of echinoderm collagenous structures will yield valuable insights into the echinoderm evolutionary biology, development, regeneration, and other aspects of the biology of this taxon. Aside from the value in basic biology, the echinoderm MCT has the potential to inspire the development of new biomaterials with dynamic properties [8]. For example, one of the outstanding challenges in the field of tissue engineering is to design implantable extracellular materials with time-varying mechanics that mimic native tissue development that progresses in the direction of increasing stiffness. Moreover, the mechanical properties of such materials need to be amenable to manipulation at the fine scale to closely recapitulate natively non-uniform structures and anisotropic environments [2]. The echinoderms’ mutable collagenous structures have the potential to solve these challenges. In this paper, we execute the first steps to understand the genetic underpinnings of the molecular mechanisms that control the dynamic properties of MCT.
The tensile properties of the MCT are under nervous control [12]. Echinoderms have evolved a unique anatomical relationship between their nervous system and the collagenous connective tissue. The MCT is associated with specialized neurosecretory cells called juxtaligamental cells (JLCs), which receive motor nervous input and extend their long processes into the extracellular matrix [12, 18, 19]. When stimulated by motor neurons, the JLCs are thought to release secretory substances that modulate the strength of cohesive forces between collagen fibrils [12]. Some of those secretory substances enhance interfibrillar cohesion, thus locking the collagen fibrils in place and increasing stiffness. In contrast, other substances decrease the strength of the interfibrillar bonds, resulting in a more pliable state of the connective tissue [9].
Several protein modulators presumably released by the JLCs were purified with classical biochemical techniques [9, 20, 21]. In physiological assays, these protein modulators had either stiffening or softening effects on the MCT. However, except for one protein called tensilin [20, 22, 23], the corresponding protein-coding genes remain to be identified at the sequence level.
Furthermore, little is known about how protein modulators induce changes in the MCT. Therefore, the identification and characterization of MCT modulator genes remain a major bottleneck in better understanding the molecular physiology of the echinoderm collagenous tissues.
This study aims to identify genes that are differently expressed in tissues rich in JLCs and, thus, are potentially associated with MCT modulation. The identification of these putative MCT modulators will inform future cell-level gene expression assays and functional studies that will be able to confirm the involvement of key genes in MCT modulation. To this end, we employed a transcriptome-wide high-throughput gene expression analysis approach (RNA-Seq) to quantify gene expression in the inner arm core region (enriched in JLCs) of the brittle star Ophiomastix wendtii compared to the whole arm (containing the basal level (i.e., neither enriched nor depleted) of the JLCs) and stomach (which is devoid of JLCs).
Methods
Animal collection, maintenance, and tissue sample harvesting
Adult individuals of O. wendtii were collected at the Keys Marine Laboratory (Layton, FL) and transported in the natural seawater chilled in a box with water ice to the University of North Carolina at Charlotte. Once in the laboratory, the animals were allowed to acclimate gradually to the ambient room temperature overnight. The next day, the brittle stars were transferred to a tank with artificial aerated seawater (Instant Ocean, by Spectrum Brands, Blacksburg, VA), whose salinity was adjusted to the natural seawater from the animal habitat (40 ppt). The animals were allowed to acclimate for five days.
The animals were anesthetized in 0.1% solution of 1-phenoxy-2-propanol (Millipore Sigma, cat. no. 484423) in seawater until they stopped responding to touch. Tissue samples for both RNA-Seq and microscopic analysis were harvested from three individuals, including one male and two females (Fig. 1).
Fig. 1.
Ophiomastix wendtii individuals, from which tissue samples were collected. Animal 1 is male, and animals 2 and 3 are female
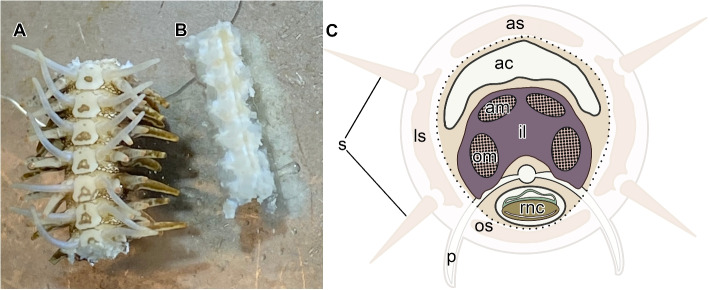
Three types of tissue samples were collected from each animal. (A) Since the MCT in brittle stars is known to be most prominent in the arm [18, 19, 24], we collected whole arm segments with all anatomical components being intact (Fig. 2A). (B) Within the arm, the mutable collagenous structures (ligaments and tendons) and the associated system of juxtaligamental nodes (ganglia) are mostly enriched in the inner core, which is surrounded by the peripheral skeletal plates. Therefore, we removed the peripheral plates to obtain MCT and JLC-enriched samples for analysis (Fig. 2B, C). (C) Stomach samples, which are devoid of MCT and JLCs (see the Results section), were used as a negative control.
Fig. 2.
Two types of tissue samples harvested from the arm. A “Whole arm segments” with all anatomical components left intact. B The “inner arm core” with the peripheral arm plates and associated structures removed. The oral side is shown for both specimens in A and B. C Diagram of the cross-section through the arm. The faded region indicates the peripheral tissues that are surgically removed when the inner arm core samples are prepared for RNA extraction. The dotted line indicates where the peripheral and inner arm core tissues are separated. Abbreviations: ac – arm coelom; am – aboral muscle; as – aboral skeletal shield; il – intervertebral ligament; ls – lateral skeletal shield; om – oral muscle; os – oral skeletal shield; p – podium; rnc – radial nerve cord; s – spine
RNA isolation
The tissue samples for RNA isolation (13–95 mg) were homogenized in 500 L of ice-cold TRI Reagent (Millipore Sigma, cat. no. T9424) using sterile closed-system disposable microtissue homogenizers (Kimble, cat. no. K7496250030). The homogenized samples were briefly vortexed, and an additional 500 L of the TRI Reagent was added to each sample. The samples were frozen at -80C for one week before RNA extraction. Upon retrieval from storage, the samples were thawed on ice, the insoluble material was precipitated by brief centrifugation, and the clear supernatant was transferred into fresh tubes. After a 5-minute incubation at room temperature, 100 L of 1-bromo-2-chloropropane (BCP) was added to each tube. The samples were then emulsified by vigorous shaking and allowed to sit at room temperature for 10 minutes. The organic and aqueous phases were separated by centrifugation at 16,000g for 45 min at 4C, and the aqueous phase was carefully aspirated and transferred into a fresh tube. The RNA was precipitated by adding 500 L of isopropanol to each sample, incubating at -20C for 1 hour, and centrifugation. The pellet was washed twice with ice-cold 70% ethanol and dissolved in 50 L of RNase-free water. The quantity of total RNA was determined with a NanoDrop Spectrophotometer (Thermo Scientific, USA) and a Qubit Fluorometer (Invitrogen, USA).
Microscopy
Tissue samples were immersion fixed overnight at 4C in 2.5% glutaraldehyde prepared in 0.05 M cacodylate buffer, pH 7.8, 1030 mOsm. The fixed samples were extensively washed and then postfixed in 1% OsO in the same buffer for 1 hour. The hard calcite endoskeleton was dissolved by incubation in a decalcifying solution composed of 1% ascorbic acid and 0.15M NaCl for seven days at 4C. The decalcifying solution was changed every 12 hours and freshly prepared each time.
The decalcified specimens were dehydrated in a graded series of ethanol and propylene oxide and embedded in Araldite 502/DDSA. Semithin (0.8 m) and ultrathin (70 – 80 nm) sections were cut with a PowerTome ultramicrotome (RMC Products) using glass knives. The semithin sections were collected on gelatin-covered slides, stained with toluidine blue, and coverslipped with the DPX mounting medium (Electron Microscopy Sciences). The ultrathin sections were collected on pioloform-covered copper slot grids and sequentially stained with 1% uranyl acetate in 10% ethanol, followed by Reynold’s lead citrate.
The semithin sections were photographed with an Olympus IX83 compound microscope. The ultrathin sections were imaged with the JEOL JEM 2100 LaB transmission electron microscope at 80 kV.
We note that our experimental design and the results presented in the manuscript do not include enough details to identify and categorize different types of JLCs in our tissues. Such categorization is beyond the scope of the manuscript, and the interested reader may find additional information in numerous studies published elsewhere [24, 25, 18].
Library preparation and sequencing
At the time we conducted our experiments, protocols for single-cell or single-nuclei sequencing optimized for brittle stars were not commercially available. The RNA-Seq libraries were prepared using the Illumina® Stranded mRNA Prep kit (Illumina, USA), following the “Illumina Stranded mRNA Prep Reference Guide” [26]. The fragmentation was enzyme-based. We conducted quality control on the final libraries to assess RNA concentration with a Qubit dsDNA BR (broad range) assay kit and quality by Tapestation 4200 analysis using the High Sensitivity D1000 Screentape (Agilent, USA). We prepared 16 Illumina paired-end read sequencing libraries to account for both the biological and technical replicates (Table 1). Sequencing was performed on an Illumina NextSeq 2000 (Illumina, USA). The adapter trimming feature in the Illumina FASTQ file generation pipeline was used to eliminate the library adapters and avoid problems with subsequent alignment and assembly steps [27].
Table 1.
A list of 16 Illumina paired-end read libraries prepared from three individuals and three sample types
| Animal | Whole arm segments | Inner arm core | Stomach |
|---|---|---|---|
| 1 | 2 | 2 | 2 |
| 2 | 1 | 2 | 2 |
| 3 | 1 | 2 | 2 |
Transcriptome assembly
After acquiring the raw transcriptome data, a series of quality preprocessing steps were executed. The quality of the raw sequencing data was assessed with FastQC v0.11.9, focusing on identifying and correcting common issues, such as low-quality bases and leftover adapter sequences. Next, the de novo transcriptome assembly [28, 29] was performed with Trinity v2.14.0 [30] installed as part of the OmicsBox v3.3 [31] bioinformatics software platform on an iMac computer. The computer ran the macOS 13.3.1 (22E772610a) operating system and was equipped with a 2.3 GHz 18-core Intel Xeon W processor, Radeon Pro Vega 64 16 GB graphics, and 128 GB of 2666 MHz DDR4 memory.
The following Trinity parameters were chosen to ensure an accurate and efficient assembly while accommodating the unique characteristics of the sequencing data. The assembly was not strand-specific, with a minimum contig length of 200 base pairs. Our approach included a thorough assessment of the read content without read normalization to maintain the original sequence data integrity. For paired-end reads, a pair distance of 400 base pairs was chosen. The minimum k-mer coverage was set to 1 during the Inchworm stage, allowing all k-mers to be included. In the Chrysalis stage, we limited the maximum reads per graph to 200,000 and set the minimum glue (i.e., the minimum number of reads required to join the Inchworm contigs) to 2 and the maximum cluster size to 25, thus balancing the complexity and manageability of the graphs. Finally, at the Butterfly stage, the default assembly algorithm was used. The path reinforcement distance was set to 25, with a minimum percent identity of 98%, a maximum of 2 allowed differences, and a maximum internal gap of 10 base pairs.
To evaluate the assembly quality, we assessed the number of full-length or nearly full-length (>80% of the length) protein-coding transcripts that were recovered in the de novo assembly by aligning our contigs to the reference proteome of the sea urchin S. purpuratus [32].
The completeness of the protein-coding gene representation in the assembled transcriptome was assessed with BUSCO v5.4.5 [33], which was run in the “transcriptome” mode against the OrthoDB v10 database [34] for the “Eukaryota” lineage. The Blast E-value threshold was set at to balance sensitivity and specificity while identifying orthologous genes.
Differential gene expression analysis and gene annotation
We quantified transcript and gene abundance across three different sample types (Fig. 2, Table 1): the whole arm segments, the inner arm core, and the stomach (control). The count tables were generated by the RSEM v1.3.3 [35] and Bowtie v2.5.1 [29] tools in OmicsBox and used as input for a comprehensive differential expression analysis with edgeR v3.18 [36].
The differentially expressed genes (DEGs) were identified in two pairwise comparisons among the sample types: whole arm segments vs stomach and inner arm core vs whole arm segments. A gene was considered differentially expressed in each comparison if the adjusted p-value was less than 0.05 and the fold-change was greater than 2 in either direction.
Following the DEG analysis, we combined the findings from the two comparisons to refine the list of candidate MCT-related genes. We kept only the genes that consistently exhibited upregulation in both scenarios. This strategy allowed us to filter out the genes that were less likely to be relevant.
The target genes were annotated using a suite of specialized tools in OmicsBox. We used InterProScan v5.63-95.0 [37] including searches against structural domains (Superfamily and Gene3D), other sequencing features (Phobius, SignalP, Coils, and TMHMM), and other categories (AntiFam, FunFam, and PIRSIR). Furthermore, the InterProScan parameters included searches against the following families, domains, sites, and repeats: NCBIfam, SFLD, Panther, HAMAP, ProSiteProfiles, ProSitePatterns, SMART, CDD, PRINTS, PfamA, and PIRSF.
Additionally, we used Diamond BLAST v2.1.9 [38] in the BLASTX mode. We set the E-value threshold at for Diamond BLAST and examined each top 5 hits with a 33 HSP length cut-off for a quick alignment against the NCBI nr (2023-02-01) database. Diamond Blast matches the sensitivity of NCBI BLAST but demonstrates a much higher throughput.
Gene Ontology (GO) mapping and annotation were performed to assign terms to the sequences and facilitate functional biological interpretation of the sequencing data [39–41]. We selected the latest database version (Goa version 2023.08) and performed fast GO mapping of the protein Blast hits against extensively curated GO annotated proteins to obtain functional labels. The reference data were retrieved from the Gene Annotation Files and by UniProt ID mapping. The annotation rule parameters were set as follows: the annotation cutoff to 55, the GO weight to 5, the E-Value-Hit-Filter to , the HSP-Hit Coverage cutoff to 0, and the Hit Filter to 500. Redundant terms for each sequence were removed, thus ensuring the assignment of only the most specific GO terms to a given sequence and avoiding the simultaneous assignment of multiple GO terms from the same branch to the same sequence. The individual GO term annotations were organized and summarized into higher-level GO slim sets.
Since the molecules that modulate the mechanical state of the MCT are secreted proteins that are produced and released by the JLCs [12], we further narrowed down the pool of the candidate genes by incorporating two advanced prediction tools that identify the proteins that are targeted for the extracellular or cell membrane localization. SignalP 6.0 was employed to predict the presence of signal peptides that direct candidate proteins toward the secretory pathway [42, 43]. The tool was run in the “fast” prediction mode with “Eukarya” specified as the “organism of origin”. Additionally, DeepLoc 2.0 was run in the “high-quality” mode to predict if the proteins are extracellular or can be in one of nine locations inside the cell [44].
For the final list of putative MCT modulators, we used Emboss v6.6.0.0 to retrieve the longest open reading frame (ORF) for one isoform per gene. We then used Expasy to calculate the molecular weight and isoelectric point for each corresponding amino acid sequence [45–48].
GO enrichment analysis
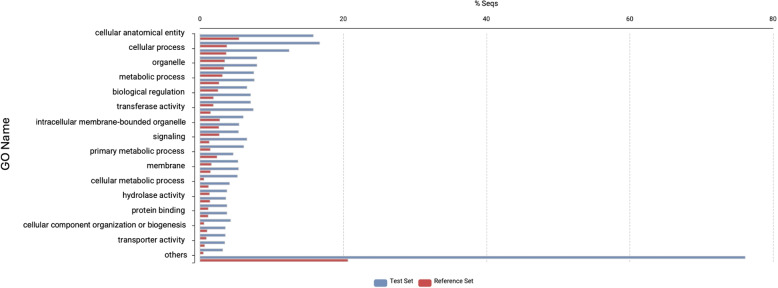
We ran Fisher’s exact test for over-representation to identify which GO terms are enriched among the candidate MCT-related genes, as implemented in the OmicsBox platform [31, 49]. The test set comprised the transcripts that exhibited upregulation in both contrasts (whole arm vs. stomach and inner arm core vs. whole arm), whereas the reference set contained all annotated transcripts. The p-value of the test was adjusted for multiple tests with the Benjamini-Hochberg method, and the cutoff value was set at 0.05. The enrichment results were represented by the most specific GO terms (i.e., the lowest possible level in the GO direct acyclic graph) and plotted as GO charts (Fig. 6).
Fig. 6.
Bar plot showing the significantly enriched GO terms in the shared set of 94 transcripts (blue bars) that were upregulated in both the whole arm vs stomach and inner arm core vs whole arm comparisons. Red bars represent the reference gene set (the entire annotated transcriptome)
Results
Microscopic analysis
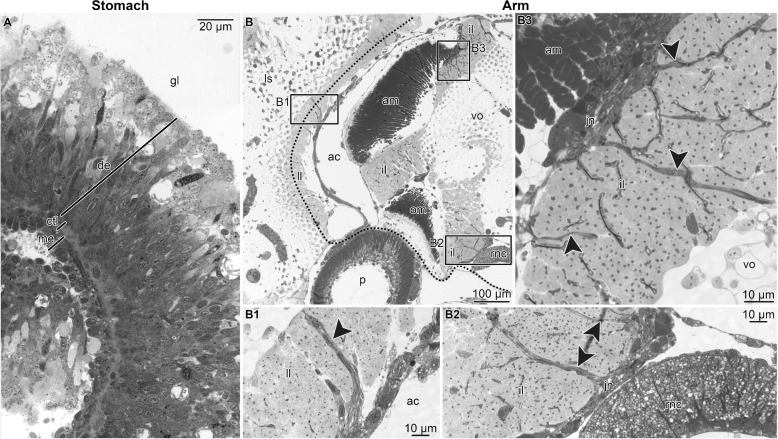
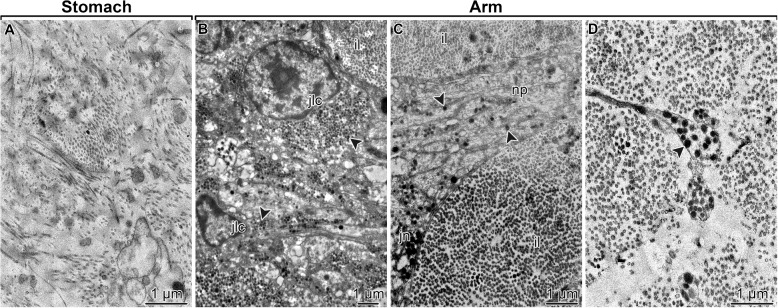
We implemented light and electron microscopy to demonstrate the absence of MCT structures in the stomach and their abundance in the inner arm core region of the brittle star O. wendtii. The stomach wall in O. wendtii, as in other echinoderms [50, 51], is composed of the inner digestive epithelium, outer mesothelium, and a connective tissue layer that separates the two epithelia (Fig. 3A). The connective tissue partition contained loosely organized bundles of collagen fibrils, which were never associated with neurosecretory cells (Fig. 4A). In contrast, MCT structures were abundant in the arm, including the centrally located intervertebral ligament, which connects adjacent vertebral ossicles, and the ligaments attaching the peripheral skeletal shields to the inner arm core (Fig. 3B – B2). The ligaments in the arm are made of densely packed collagen fibrils and are abundantly supplied with processes of neurosecretory JLCs (Fig. 4B – D). These processes contain abundant characteristic dense-core vesicles and originate from perikarya clustered in ganglion-like structures called juxtaligamental nodes (Fig. 3B2 and B3; Fig. 4B and C). Such direct association between the JLCs and the extracellular collagenous matrix is a typical microanatomical feature of the echinoderm MCT [18, 52]. When the peripheral arm shields were surgically removed (Fig. 2B), the majority of the MCT structures remained in the inner arm core samples (note the dotted line in Figs. 2C and 3B which indicates where the separation between the peripheral tissue from the inner arm core took place).
Fig. 3.
Representative micrographs showing the microanatomical and histological organization of the stomach (A) and the whole arm segments (B – B3) tissue samples used as a starting material for RNA-Seq. A High-magnification view of the stomach wall. B A general view of the whole arm segments in a cross-section. Boxed areas indicate the regions that are shown at a higher magnification in B1 – B3. B1 – lateral ligament. B2 – oral part of the intervertebral ligament. B3 – aboral part of the intervertebral ligament. Abbreviations: ac – arm coelom; am – aboral intervertebral muscle; ctl – connective tissue layer; de – digestive epithelium; gl – gut lumen; il – intervertebral ligament; jn – juxtaligamental node; ll – lateral ligament; ls – lateral shield; me – mesothelium; om – oral intervertebral muscle; rnc – radial nerve cord; vo – vertebral ossicle. Dotted line in B indicates where the separation of the peripheral tissues took place when preparing the inner core arm tissue samples for RNA-Seq. Arrowheads in B1 – B3 indicate bundles of neurites extending from juxtaligamental nodes into the extracellular matrix of ligaments. Plastic semithin sections stained with toluidine blue
Fig. 4.
Representative TEM micrographs showing the ultrastructural organization of the connective tissue compartments in the stomach (A) and whole arm segments (B – D) tissue samples used as a starting material for RNA-Seq. A Connective tissue layer of the stomach wall. B Juxtaligamental cells in the aboral juxtaligamental node. C A bundle of neurites projecting from the aboral juxtaligamental node into the aboral part of the intervertebral ligament. D Neurites of the juxtaligamental cells in the aboral part of the intervertebral ligament. Abbreviations: il – intervertebral ligament; jn – aboral juxtaligamental node; np – neurites. Arrowheads in B – D indicate secretory granules in juxtaligamental cells
RNA sequencing and de novo transcriptome assembly
The sequencing of the 16 libraries (Table 1) representing three sample types (whole intact arm segment, inner arm core, and stomach) yielded a total of 116,785,071 paired-end reads with an insert size of 160 bp and the maximum and minimum length of 76 bp and 35 bp, respectively.
All quality-processed reads were pooled together for a de novo assembly with Trinity [30] into 586,658 contigs corresponding to 325,320 Trinity “genes” with the average/median contig length of 713.25/412 bp, N50 of 1,067 bp, and L50 of 98,061 (Table 2).
Table 2.
Summary metrics of the de novo transcriptome assembly
| Metric | Value |
|---|---|
| Total assembled bases | 418,431,402 |
| Number of contigs | 586,658 |
| Number of Trinity “genes” | 325,320 |
| Average contig length, nt | 713.25 |
| Median contig length, nt | 412 |
| Contig N50, nt | 1,067 |
| L50 | 98,061 |
As one of the assembly quality metrics, we assessed the representation of full-length and near full-length (>80% length) protein-coding transcripts. To this end, we aligned our contigs to the reference proteome of the purple sea urchin S. purpuratus, the echinoderm species with the best-annotated genome to date. This analysis showed that 6,745 (out of 41,631) sea urchin orthologs were represented in the newly assembled O. wendtii transcriptome as complete/nearly-complete transcripts.
The quality of the de novo transcriptome assembly was further evaluated by assessing the representation of conserved eukaryote protein-coding genes with BUSCO [33]. Overall, 98.82% of these genes were recovered in the O. wendtii transcriptome (Table 3), of which the majority were “complete” (i.e., their length was within the length threshold established for the BUSCO dataset).
Table 3.
BUSCO analysis results for assessing the de novo transcriptome assembly
| Type | Number of BUSCOs | Percentage |
|---|---|---|
| Complete Single-Copy | 60 | 23.53% |
| Missing | 3 | 1.18% |
| Fragmented | 11 | 4.31% |
| Complete Duplicated | 181 | 70.98% |
Differential gene expression analysis: identification and annotation of the candidate MCT-related genes
Clean isolation of MCT structures and associated juxtaligamental nodes from brittle stars for the subsequent, direct analysis of molecular components remains a challenge. We thus implemented an alternative strategy by comparing gene transcript abundance among the three tissue samples with progressively higher levels of MCT abundance: the stomach, the whole intact arm segment, and the microsurgically isolated inner arm core. Microscopic analysis (see above) demonstrated the absence of MCT and associated JLCs in the stomach, while both structural elements are abundant in the arm, particularly in the central arm core. Also, we estimated the enrichment of JLCs in the inner arm core samples based on sample weight and assumed a consistent quantity of JLCs within arm tissues. Specifically, inner arm core samples from animals 1, 2, and 3 had 2.3, 2.5, and 2.56 times more JLCs than the corresponding whole arm samples, respectively.
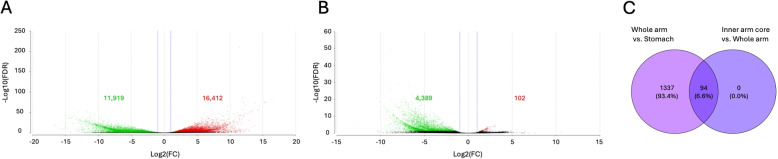
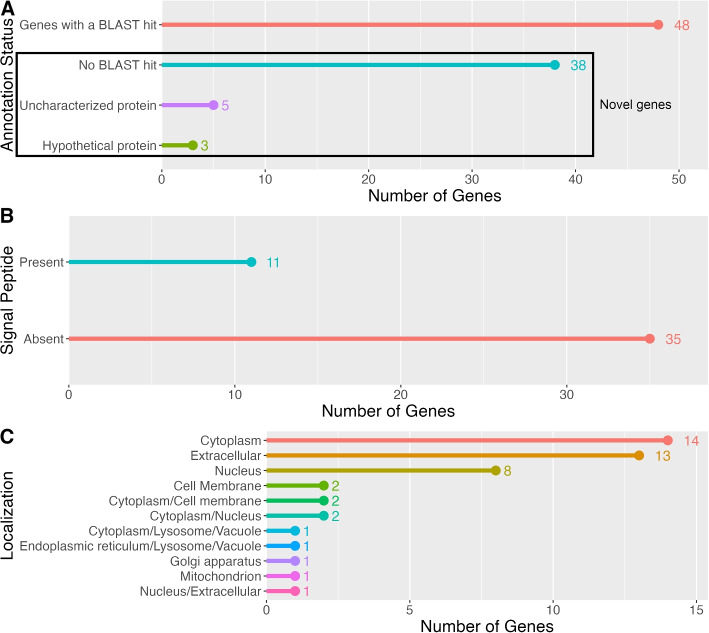
To identify the genes that are preferentially expressed in the MCT-rich inner arm core, two pairwise comparisons were made: whole arm vs stomach and inner arm core vs whole arm (Fig. 5A and B, respectively). Given this study design, the MCT-related genes are expected to be recovered as upregulated in both those contrasts (Fig. 5C). We identified 94 such genes (Additional File 1), which were then subjected to further annotation and analysis (Figs. 6 and 7). A significant proportion (46 out of 94, or 49%) of these genes are novel as they have not been previously studied. Of these 46 novel genes, 38 did not have a BLAST match, 8 genes mapped to “uncharacterized” or “hypothetical” proteins in all of their top 5 BLAST hits (Fig. 7A, Additional File 1). The vast majority of the annotated genes with a mapped BLAST hit encoded muscle-specific products, including cytoskeletal proteins, regulatory proteins, and proteins responsible for ion transport (Additional File 1). Other genes were predicted to be involved in regulating cell proliferation, differentiation, and migration, as well as in stress response (Additional File 1). Except for two genes that mapped to “uncharacterized” proteins, none of the BLAST-annotated transcripts in the 94 gene subset had a signal peptide that would direct them to the secretory pathway (Additional File 1) [53]. Likewise, the GO enrichment analysis (against the background set of all annotated genes in the transcriptome) of those BLAST-annotated genes did not reveal any functions specific to the connective tissue or extracellular matrix (Fig. 6; also see Additional File 2). Detailed annotations of all transcripts are provided in Additional File 3.
Fig. 5.
Differential gene expression analysis in A whole arm segments vs stomach and in B the inner arm core vs whole arm in O. wendtii. Volcano plots show the fold difference in expression (X-axis) plotted against (Y-axis). The significantly upregulated transcripts (red) are to the right, and the significantly downregulated transcripts (green) are to the left. The differentially expressed transcripts were defined as those with at least a two-fold difference in expression ( or ) and the associated adjusted p-value (FDR) below 0.05. Red dots indicate significantly upregulated genes (FDR , 1.0), and green dots represent significantly downregulated genes (FDR < 0.05, ). Black dots represent transcripts whose expression was not statistically different in the respective pairwise comparisons. The numbers on the plots refer to the number of up or downregulated transcripts. C Venn diagram showing the common set of 94 upregulated genes in both comparisons
Fig. 7.
Identification and annotation of the putative MCT-related candidate gene set. A BLAST annotation status of the whole set of the 94 genes that were identified as overexpressed in both the whole arm (in comparison to the stomach) and in the inner arm core (in comparison to the whole arm). The box indicates novel genes with unknown function. B Signal peptide presence (as predicted by the SignalP tool [43]) in a subset of 46 novel genes (those that had no associated BLAST hit or were recovered as either uncharacterized proteins or hypothetical proteins boxed are in (A)). C Cellular/extracellular localization of the protein products encoded by the subset of 46 novel candidate genes with unknown function (i.e., the same subset as in (B) and boxed area in (A)), as predicted by the DeepLoc tool [44]
Unlike the genes with mapped annotations, the 46 novel genes (boxed area in Fig. 7A) overexpressed in the inner arm core appear more relevant to the MCT function. SignalP [43] identified the presence of N-terminal signal peptides in 11 of those genes (Fig. 7B, Additional File 1), suggesting that they code for proteins targeted for a secretory pathway [53]. Similarly, DeepLoc [44] predicted extracellular localization for 14 proteins in the novel gene cohort (Fig. 7C, Additional File 1). The total set of novel proteins that contained a predicted signal peptide by SignalP or were identified as extracellular by DeepLoc constitutes the final dataset of interest for 16 putative MCT modulators (Additional File 1 and Table 4).
Table 4.
Summary of the main characteristics of 16 putative modulators of mutable collagenous tissue (MCT): top BLAST hit, the status of the open reading frame, number of amino acids (Size), molecular weight in Daltons (Weigth), the isoelectric point (pI), the presence of a signal peptide, as predicted by SignalP, and localization of the protein product, as predicted by DeepLoc. Sequences are identified by their suffix name, and all of them have the prefix “TRINITY_.” The corresponding amino acid sequences are available on Zenodo under DOI 10.5281/zenodo.13830943
| Sequence | Top BLAST hit | ORF status | Size, aa | Weight (Da) | pI | SignalP | DeepLoc |
|---|---|---|---|---|---|---|---|
| DN1888_c0_g1 | KAF6027037.1 hypothetical protein EB796_014653 (Bugula neritina) | 3’ partial | 872 | 87914.35 | 9.02 | No SP | Extracellular |
| DN9695_c0_g1 | XP_038071724.1 uncharacterized protein LOC119740469 (Patiria miniata) | 5’ partial | 941 | 105049.37 | 5.12 | SP | Cell Membrane |
| DN894_c1_g2 | XP_022109156.1 uncharacterized protein LOC110989228 (Acanthaster planci) | Complete | 574 | 62324.95 | 5.25 | SP | Extracellular |
| DN819_c1_g1 | No hit | Complete | 45 | 5243.09 | 4.12 | SP | Extracellular |
| DN81063_c0_g1 | XP_022104884.1 uncharacterized protein LOC110986900 (Acanthaster planci) | Internal | 525 | 58241.64 | 7.18 | No SP | Extracellular |
| DN1190_c0_g1 | No hit | Internal | 366 | 36461.54 | 11.07 | No SP | Extracellular |
| DN722_c0_g2 | No hit | Complete | 86 | 9508.15 | 8.4 | SP | Extracellular |
| DN3456_c0_g2 | No hit | 3’ partial | 109 | 11956.31 | 3.74 | SP | Extracellular |
| DN4715_c0_g1 | No hit | Complete | 176 | 18570.72 | 7.35 | SP | Cell membrane |
| DN130227_c0_g1 | No hit | 3’ partial | 47 | 5507.32 | 3.91 | SP | Extracellular |
| DN2540_c0_g1 | No hit | Complete | 451 | 45656.43 | 5.69 | SP | Extracellular |
| DN1888_c0_g2 | No hit | 3’ partial | 104 | 9040.43 | 10.86 | No SP | Nucleus/ extracellular |
| DN4472_c0_g1 | No hit | 3’ partial | 50 | 5673.68 | 4.38 | SP | Extracellular |
| DN24708_c1_g1 | No hit | Complete | 86 | 9219.84 | 8.39 | SP | Extracellular |
| DN2166_c2_g1 | No hit | Complete | 23 | 2274.83 | 8.34 | No SP | Extracellular |
| DN107250_c1_g1 | No hit | Complete | 65 | 7394.73 | 7.56 | SP | Extracellular |
The authors did not conduct an in silico analysis to predict the 3D structures of the candidate MCT modulators due to concerns about the accuracy of predictions when closely related homologous sequences were not used to train current methods. Such an analysis would require careful consideration and more space than is feasible in the current manuscript. While this is beyond the scope of our manuscript, we hope that the amino acid sequences provided in our supplementary digital materials will be valuable to researchers interested in exploring properties not covered here.
Other specific characteristics of the 16 putative MCT modulator
The annotation of the longest and most complete open reading frames (spanning from start to stop codon) from 16 representative isoforms of the putative MCT modulators reveals several notable patterns. While their top five BLAST hits, when available, primarily correspond to uncharacterized or hypothetical proteins, the results obtained from InterProScan analysis display greater diversity. These findings should be interpreted with caution, as the genetic distances between our sequences and those in the annotation databases may result in misleading or spurious matches. Nevertheless, in the interest of providing a comprehensive account of our results, we present the most salient patterns observed below.
Together, these sequences accumulate 62 putative topological domains that are either extracellular or cytoplasmatic, 48 signal cleavage regions, 11 different SignalP hits, and 7 THHMM hits. They also contain 186 putative antigenic regions that could indicate parts of a protein likely to be recognized by the immune system.
Three sequences (TRINITY_DN1888_c0_g1_i1, TRINITY_DN1888_c0_g2_i1, and TRINITY_DN3456_c0_g2_i1) contained predicted regions within proteins that do not fold into a fixed 3D structure, known as intrinsically disordered regions (IDRs), as categorized by MobiDB-lite.
Only one sequence (TRINITY_DN9695_c0_g1_i1) had a PANTHER hit to transforming growth factor beta receptor 3 (TGFBR3). The encoded receptor is a membrane proteoglycan that often functions as a co-receptor with other TGF-beta receptor superfamily members. Ectodomain shedding produces soluble TGFBR3, which may inhibit TGFB signaling. Decreased expression of this receptor has been observed in various cancers. Alternatively, spliced transcript variants encoding different isoforms have been identified for this gene.
In the same sequence (TRINITY_DN9695_c0_g1_i1), the TGFBR3 annotation overlaps with a Pfam annotation for the Zona pellucida (ZP) domain. This ZP domain hit matched a zp_4 SMART annotation. The ZP domain is a 260 amino acid polymerization module that is found at the C terminus of many secreted eukaryotic glycoproteins that play fundamental roles in development, hearing, immunity, and cancer. In this sequence, the domain is between positions 566 and 816 of a sequence that has 941 amino acids. None of the other remaining sequences had a Pfam hit. This sequence also has a hit on the PROSITE database, which is related to the ZP domain (ZP_2) between positions 565 and 823.
Phobius predicted 43 domains in 9 of these protein sequences, including the cytoplasmic domain, non-cytoplasmic domain, transmembrane region, and signal peptides. A separate file with detailed annotations for the amino acid sequences of proteins encoded by the 16 candidate genes is provided in Zenodo at https://doi.org/10.5281/zenodo.13830943.
Discussion
The aim of the present study was to address an important gap in our knowledge of the molecular constituents of the echinoderm MCT. The phenomenon of the variable tensile strength of echinoderm collagenous tissues has been known for decades [6, 8]. Its importance in echinoderm biology has been studied in the context of defensive autotomy, asexual reproduction, and energy-saving posture maintenance [10–17]. Careful and extensive microscopic surveys identified a unique population of specialized juxtaligamental neurosecretory cells as a potential source of secretory regulators of the MCT tensile strength [12, 18, 19]. Some of those modulatory proteins were purified biochemically, and their effect on the MCT properties has been tested in physiological assays [9, 20, 21]. Although those proteins were assigned names and their role in the MCT function was confirmed, they have remained largely unidentified at the sequence level, and the genes that code for them remain unknown.
Here, we performed high-throughput transcriptome sequencing and differential gene expression analysis to identify the genes that are preferentially expressed at high levels in the inner arm core of the brittle star O. wendtii. The inner arm core is enriched in MCT structures, including the large intervertebral ligament. Therefore, the genes that are overexpressed in this anatomical location are expected to contain MCT-specific genes.
The BUSCO results for the transcriptome assembly show that only 3% of genes, are missing, while gene duplication levels exceed 70%. This gene duplication level is expected given our experimental design. RNA-seq inherently amplifies regions with higher expression, leading to increased duplication levels. This effect can be further intensified in experiments that compare multiple tissue types, resulting in higher redundancy in the reads (for example, see [54] ). However, we found no indication that this is impacting our downstream analysis.
The differential gene expression analysis initially identified 94 genes that were overexpressed in the inner arm core compared to the whole arm and the stomach. Further analysis allowed us to refine this list of potential candidates by excluding genes with known functions in unrelated tissues. For example, many excluded genes code for proteins specific to muscle cells. The detection of overexpressed muscle genes in the inner arm core was expected, as it was impossible to isolate pure intervertebral ligaments (i.e., the MCT), and the samples used for RNA-Seq inevitably included intervertebral muscles.
Another filtering strategy involved identifying extracellular proteins with two state-of-the-art bioinformatic tools, SignalP [43] and DeepLoc [44]. This approach led to the identification of the 16 gene products with predicted extracellular or cell membrane localization, forming our final target candidate gene set. These genes will be further investigated in subsequent experiments aimed at (a) validating their expression in MCT-specific cell types, particularly JLCs; (b) functionally validating their involvement in the regulation of the MCT mechanical properties through expression perturbation (e.g., via RNA interference-mediated knockdown); and (c) isolating or synthesizing the protein products and characterizing their direct physiological effects.
The fact that no single of the 16 genes in the final candidate gene set has an ortholog with known functions underscores two key points: (a) the MCT phenomenon may be unique to echinoderms, with no conserved ubiquitous extracellular matrix proteins being co-opted in regulating the mechanical properties of the mutable collagenous structures; and (b) how much there is to learn about echinoderm MCT biology.
Confirming the role of the identified candidate genes in controlling MCT tensile strength will open up a wide range of new possibilities for both fundamental biology and biomedicine. The confirmed molecular and genomic identity of the MCT modulators will enable studies that would facilitate a better understanding of the evolution and molecular mechanisms of the echinoderm MCT. This enhanced fundamental understanding will, in turn, inform the design of new collagen-based biomaterials with dynamic, tunable mechanical properties for tissue engineering and regenerative medicine.
Conclusion
The present study is the first attempt at discovering novel genes specific to the echinoderm MCT using state-of-the-art sequencing, differential gene expression, and annotation approaches. We identified 16 putative candidate genes overexpressed in the brittle star inner arm core, an anatomical region enriched with MCT structures. All these genes displayed sequence features suggesting the localization of their protein products in the extracellular matrix (mostly) or cell membrane (in two cases) and represent novel proteins. This study enables further in silico and wet lab experiments in which the candidate genes will be probed for expression analysis at the cellular level and in which their molecular functions and physiological significance will be elucidated.
Supplementary information
The raw sequencing data were deposited at the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA1095919.
This Transcriptome Shotgun Assembly project has been deposited at DDBJ, ENA, and GenBank under accession GKWM00000000. This paper describes the first version, GKWM01000000. Also, the assembled transcriptome is available in EchinoDB (under “Ophiomastix wendtii”). The gene expression data were also submitted to the GEO database under accession GSE278778. Additional digital material is available in Zenodo at https://doi.org/10.5281/zenodo.13830943.
Supplementary information
Acknowledgements
We acknowledge several entities at the University of North Carolina at Charlotte for their support. This project was made possible through the collaboration and resources provided by the College of Computing and Informatics, the Department of Bioinformatics and Genomics sequencing facility, the University Research Computing, and the Center for Computational Intelligence to Predict Health and Environmental Risks (CIPHER). We also acknowledge the Wake Forest Institute of Regenerative Medicine for their contributions.
We would like to thank Sarah Calaway, Ph.D., Senior Field Applications Scientist from Illumina, for her exceptional technical and bioinformatic expertise and assistance during sequencing experiments. We are equally grateful to Lauren Roppolo Brazell for her dedicated help and support during sequencing.
Authors' contributions
R.N.: Methodology, validation, formal analysis, investigation, writing (original draft, review, and editing), visualization, and project administration. V.M.: Conceptualization, methodology, validation, formal analysis, investigation, writing (original draft, review, and editing), visualization, and project administration. A.H., G.N., and W.T.: Methodology, formal analysis, writing (review and editing). D.J. and R.R.: Writing (review and editing). D.J.M.: Conceptualization, methodology, validation, formal analysis, writing (original draft, review, and editing), supervision, project administration, and funding acquisition. All authors have reviewed the manuscript and agreed to submit it. The authors declare no conflict of interest.
Funding
Funding for this project was generously provided by Startup funds and the Faculty Research Grant (FRG) from UNC Charlotte.
Data availability
The raw sequencing data were deposited at the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA1095919 and in EchinoDB (accession number PENDING). The gene expression data were submitted to the GEO database under the accession number PENDING. Additional digital material is available in Zenodo at DOI: 10.5281/zenodo.13830943. Authors have started uploading the files to public databases. Files will be published upon manuscript acceptance. All data is currently available upon request.
Declarations
Ethics approval and consent to participate
The authors have complied with all ethical standards required for conducting this research. Consents and approvals are not applicable to this research.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gelse K, Pöschl E, Aigner T. Collagens-structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55(12):1531–46. [DOI] [PubMed] [Google Scholar]
- 2.Guimarães CF, Gasperini L, Marques AP, Reis RL. The stiffness of living tissues and its implications for tissue engineering. Nat Rev Mater. 2020;5:351–70. [Google Scholar]
- 3.Myllyharju J, Kivirikko KI. Collagens and collagen-related diseases. Ann Med. 2001;33:21–7. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez AP. Connective tissue disease: current concepts. Dermatol Clin. 2019;37(1):37–48. [DOI] [PubMed] [Google Scholar]
- 5.Asif MI, Kalra N, Sharma N, Jain N, Sharma M, Sinha R. Connective tissue disorders and eye: a review and recent updates. Indian J Ophthalmol. 2023;71(6):2385–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkie IC. Mutable collagenous tissue: overview and biotechnological perspective. Prog Mol Subcell Biol. 2005;39:221–50. 10.1007/3-540-27683-1_10. [DOI] [PubMed] [Google Scholar]
- 7.Goh KL, Holmes DF. Collagenous extracellular matrix biomaterials for tissue engineering: lessons from the common sea urchin tissue. Int J Mol Sci. 2017;18(5). 10.3390/ijms18050901. [DOI] [PMC free article] [PubMed]
- 8.Candia Carnevali MD, Sugni M, Bonasoro F, Wilkie IC. Mutable collagenous tissue: a concept generator for biomimetic materials and devices. Mar Drugs. 2024;22(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkie IC, Sugni M, Gupta H, Carnevali MC, Elphick M. 1. In: The mutable collagenous tissue of echinoderms: from biology to biomedical applications. Cambridge: Royal Society of Chemistry; 2021. 10.1039/9781839161124-00001.
- 10.Wilkie IC, Emson RH. The tendons of Ophiocomina nigra and their role in autotomy (Echinodermata, Ophiuroida). Zoomorphology. 1987;107:33–44. [Google Scholar]
- 11.Wilkie IC. Autotomy as a prelude to regeneration in echinoderms. Microsc Res Tech. 2001;55:369–96. 10.1002/jemt.1185. [DOI] [PubMed]
- 12.Wilkie IC, Daniela M, Carnevali C. Morphological and Physiological Aspects of Mutable Collagenous Tissue at the Autotomy Plane of the Starfish Asterias rubens L. (Echinodermata, Asteroidea): An Echinoderm Paradigm. Mar Drugs. 2023;21:138. 10.3390/md21030138. [DOI] [PMC free article] [PubMed]
- 13.Byrne M. Morphological, Physiological and Mechanical Features of the Mutable Collagenous Tissues Associated with Autotomy and Evisceration in Dendrochirotid Holothuroids. Mar Drugs. 2023;21:134. 10.3390/md21030134. [DOI] [PMC free article] [PubMed]
- 14.Motokawa T, Sato E, Umeyama K. Energy Expenditure Associated With Softening and Stiffening of Echinoderm Connective Tissue. Biol Bull. 2012;222:150–7. [DOI] [PubMed] [Google Scholar]
- 15.Dolmatov IY, Afanasyev S, Boyko AV. Molecular mechanisms of fission in echinoderms: Transcriptome analysis. PLoS ONE. 2018;13:e0195836. 10.1371/journal.pone.0195836. [DOI] [PMC free article] [PubMed]
- 16.Motokawa T, Shintani O, Birenheide R. Contraction and Stiffness Changes in Collagenous Arm Ligaments of the Stalked Crinoid Metacrinus rotundus (Echinodermata). Biol Bull. 2004;206:4–12. [DOI] [PubMed] [Google Scholar]
- 17.Takemae N, Nakaya F, Motokawa T. Low Oxygen Consumption and High Body Content of Catch Connective Tissue Contribute to Low Metabolic Rate of Sea Cucumbers. Biol Bull. 2009;216:45–54. [DOI] [PubMed] [Google Scholar]
- 18.Wilkie DIC. The juxtaligamental cells of Ophiocomina nigra (Abildgaard) (Echinodermata: Ophiuroidea) and their possible role in mechano-effector function of collagenous tissue. Cell Tissue Res. 1979;197:515–30. [DOI] [PubMed] [Google Scholar]
- 19.Zueva O, Khoury M, Heinzeller T, Mashanova D, Mashanov V. The complex simplicity of the brittle star nervous system. Front Zool. 2018;15(1):1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tipper JP, Lyons-Levy G, Atkinson MA, Trotter JA. Purification, characterization and cloning of tensilin, the collagen-fibril binding and tissue-stiffening factor from Cucumaria frondosa dermis. Matrix Biol. 2003;21(8):625–35. [DOI] [PubMed] [Google Scholar]
- 21.Trotter JA, Lyons-Levy G, Luna D, Koob TJ, Keene DR, Atkinson MA. Stiparin: a glycoprotein from sea cucumber dermis that aggregates collagen fibrils. Matrix Biol. 1996;15(2):99–110. [DOI] [PubMed] [Google Scholar]
- 22.Demeuldre M, Hennebert E, Bonneel M, Lengerer B, Van Dyck S, Wattiez R, et al. Mechanical adaptability of sea cucumber Cuvierian tubules involves a mutable collagenous tissue. J Exp Biol. 2017;220(11):2108–19. [DOI] [PubMed] [Google Scholar]
- 23.Mittal V, Reid RW, Machado DJ, Mashanov V, Janies DA. EchinoDB: An update to the web-based application for genomic and transcriptomic data on Echinoderms. BMC Genomic Data. 2022;23:75. 10.1186/s12863-022-01090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mashanov V, Charlina NA, Dolmatov IY, Wilkie IC. Juxtaligamental cells in the arm of the brittlestar Amphipholis kochiif Lütken, 1872 (Echinodermata: Ophiuroidea). Russian Journal of Marine Biology. 2007;33:110–7. [Google Scholar]
- 25.Charlina NA, Dolmatov IY, Wilkie IC. Juxtaligamental system of the disc and oral frame of the ophiuroid Amphipholis kochiif (Echinodermata: Ophiuroidea) and its role in autotomy. Invertebr Biol. 2009;128(2):145–56. [Google Scholar]
- 26.Illumina Stranded mRNA Prep Reference Guide. 2021. https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/illumina_prep/RNA/illumina-stranded-mrna-reference-guide-1000000124518-02.pdf.
- 27.Adapter trimming: Why are adapter sequences trimmed from only the 3’ ends of reads. https://knowledge.illumina.com/software/general/software-general-reference_material-list/000002905. Accessed 30 May 2024.
- 28.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2011;29(7):644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.BioBam Bioinformatics. OmicsBox – Bioinformatics Made Easy. 2019. https://www.biobam.com/omicsbox. Accessed 29 Jan 2024.
- 32.The NCBI collection of predicted protein sequences of the sea urchin Strongylocentrotus purpuratus. https://www.ncbi.nlm.nih.gov/protein/?term=txid7668%5Borganism:exp%5D. Accessed 30 June 2023.
- 33.Seppey M, Manni M. Zdobnov EM. BUSCO: Assessing Genome Assembly and Annotation Completeness. Gene Prediction Methods Protoc; 2019. pp. 227–45. 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 34.Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simão FA, et al. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019;47(D1):D807–11. 10.1093/nar/gky1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18(4):366–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 2023;51(D1):D523–31. [DOI] [PMC free article] [PubMed]
- 42.Nielsen HB. Predicting Secretory Proteins with SignalP. Methods Mol Biol. 2017;1611:59–73. [DOI] [PubMed] [Google Scholar]
- 43.Teufel F, Almagro Armenteros JJ, Johansen AR, Gíslason MH, Pihl SI, Tsirigos KD, et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022;40(7):1023–5. [DOI] [PMC free article] [PubMed]
- 44.Thumuluri V, Almagro Armenteros JJ, Johansen AR, Nielsen H, Winther O. DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022;50(W1):W228–34. [DOI] [PMC free article] [PubMed]
- 45.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite; 2000. Trends Genet. 16 (6): 276–7. [DOI] [PubMed]
- 46.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Totowa: Humana Press; 2005. pp. 571–607. Full text - Copyright Humana Press.
- 47.Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–31. MEDLINE: 8125050. [DOI] [PubMed]
- 48.Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–39. MEDLINE: 8055880. [DOI] [PubMed]
- 49.Al-Shahrour F, Díaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20(4):578–80. [DOI] [PubMed] [Google Scholar]
- 50.Ophiuroidea Byrne M. In: Harrison F, Chia F, editors. Microscopic Anatomy of Invertebrates, vol. 14. New York: Wiley-Liss; 1994. pp. 247–344. [Google Scholar]
- 51.Mashanov V, Frolova L, Dolmatov IY. Structure of the digestive tube in the holothurian Eupentacta fraudatrix (Holothuroidea: Dendrochirota). Russ J Mar Biol. 2004;30:314–22. [Google Scholar]
- 52.Wilkie IC, Carnevali MDC. The juxtaligamental cells of echinoderms and their role in the mechano-effector function of connective tissue. In: Frontiers in Invertebrate Physiology: A Collection of Reviews. Palm Bay: Apple Academic Press; 2024. pp. 345–432.
- 53.Owji H, Nezafat N, Negahdaripour M, Hajiebrahimi A, Ghasemi Y. A comprehensive review of signal peptides: structure, roles, and applications. Eur J Cell Biol. 2018;97(6):422–41. 10.1016/j.ejcb.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 54.Li W, Lan Y, Wang L, He L, Tang R, Price M, et al. Comparative transcriptomes of nine tissues for the Heilongjiang brown frog (Rana amurensis). Sci Rep. 2022;12(1):20759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data were deposited at the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA1095919 and in EchinoDB (accession number PENDING). The gene expression data were submitted to the GEO database under the accession number PENDING. Additional digital material is available in Zenodo at DOI: 10.5281/zenodo.13830943. Authors have started uploading the files to public databases. Files will be published upon manuscript acceptance. All data is currently available upon request.