Abstract
Vitamin B12 is critical for hematopoiesis and myelination. Deficiency can cause neurologic deficits including loss of coordination and cognitive decline. However, diagnosis relies on measurement of vitamin B12 in the blood, which may not accurately reflect the concentration in the brain. Using programmable phage display, we identified an autoantibody targeting the transcobalamin receptor (CD320) in a patient with progressive tremor, ataxia, and scanning speech. Anti-CD320 impaired cellular uptake of cobalamin (B12) in vitro by depleting its target from the cell surface. Despite a normal serum concentration, B12 was nearly undetectable in her cerebrospinal fluid (CSF). Immunosuppressive treatment and high-dose systemic B12 supplementation were associated with increased B12 in the CSF and clinical improvement. Optofluidic screening enabled isolation of a patient-derived monoclonal antibody that impaired B12 transport across an in vitro model of the blood-brain barrier (BBB). Autoantibodies targeting the same epitope of CD320 were identified in seven other patients with neurologic deficits of unknown etiology, 6% of healthy controls, and 21.4% of a cohort of patients with neuropsychiatric lupus. In 132 paired serum and CSF samples, detection of anti-CD320 in the blood predicted B12 deficiency in the brain. However, these individuals did not display any hematologic signs of B12 deficiency despite systemic CD320 impairment. Using a genome-wide CRISPR screen, we found that the low-density lipoprotein receptor serves as an alternative B12 uptake pathway in hematopoietic cells. These findings dissect the tissue specificity of B12 transport and elucidate an autoimmune neurologic condition that may be amenable to immunomodulatory treatment and nutritional supplementation.
INTRODUCTION
Vitamin B12 deficiency is a prevalent but reversible condition. Up to 25% of individuals in the United States and up to 40% of vulnerable populations in other parts of the world have low serum B12 concentrations (1), leading to hematologic impairment (megaloblastic anemia) or neurologic deficits (loss of coordination, memory loss, and psychosis) (2). Comorbid B12 deficiency is associated with worse neurologic outcomes in patients with Alzheimer’s disease and Parkinson’s disease, and a randomized trial of high-dose B12 slowed functional decline in patients with amyotrophic lateral sclerosis (3–5). Therefore, improvements in the diagnosis and treatment of B12 deficiency, either as a primary disease or a comorbid condition, have the potential to improve brain health at a population level.
Chronic gastritis, pernicious anemia, and insufficient dietary intake are among the most common causes of vitamin B12 deficiency (1). However, diagnosis relies on vitamin B12 measurement in the blood, which may not accurately reflect the concentration in the brain. On its journey from the gut to the brain, B12 must pass through several barriers with the aid of carrier proteins and membrane transporters (6). Intrinsic factor binds B12 in the duodenum, and the cubam receptor shuttles this complex across the luminal surface of the distal ileum via receptor-mediated endocytosis (6). In the bloodstream, B12 is bound by either haptocorrin (biologically inert) or transcobalamin (biologically active). Transcobalamin-bound B12 (holotranscobalamin) enters cells via the transcobalamin receptor (CD320), the canonical pathway for tissue delivery of B12 (6). Although CD320 is ubiquitously expressed, CD320 knockout (KO) mice are selectively B12 deficient in the brain, and humans with CD320 mutations are hematologically normal despite methylmalonic aciduria (7–10). Moreover, individuals with systemic B12 deficiency frequently present with isolated neuropsychiatric symptoms without anemia (11). Thus, the mechanisms governing tissue-specific transport of vitamin B12 are poorly understood. Starting with a deep immunologic investigation of a single patient, we found an autoimmune cause of vitamin B12 deficiency restricted to the central nervous system (CNS), termed autoimmune B12 central deficiency (ABCD), and uncovered an alternative pathway for B12 uptake in hematopoietic cells.
RESULTS
Clinical presentation of the index patient
A 67-year-old woman with a history of type 2 diabetes, immune thrombocytopenia (ITP), pleuritis, and recent knee surgery presented with 5 weeks of progressive bilateral lower extremity pain, difficulty speaking, ataxia, and tremor (case 1; Fig. 1A). Her neurologic exam was notable for scanning speech, a rubral tremor of the head and hands, diminished vibratory sensation in her feet, dysmetria, and truncal ataxia. Magnetic resonance imaging (MRI) of the brain revealed bilateral symmetric T2-weighted hyperintensities in the dorsal brainstem involving the midbrain tegmentum, dentate nuclei, chief sensory nuclei of cranial nerve V, and rubrospinal tracts with gadolinium enhancement of the superior cerebellar peduncles (Fig. 1B). There were no lesions on MRI of the cervical spine. Lumbar puncture yielded noninflammatory cerebrospinal fluid (CSF) [two white blood cells, two red blood cells, total protein of 54 mg/dl, glucose of 91 mg/dl, zero oligoclonal bands, and a normal immunoglobulin G (IgG) index of 0.6; Fig. 1C and table S1]. Clinical autoimmune encephalopathy antibody testing was negative, including testing for aquaporin 4 (AQP4) antibodies. Myelin oligodendrocyte glycoprotein (MOG) antibody testing 4 years later was also negative. A rheumatologic work-up demonstrated elevated titers of antinuclear, antiphospholipid, and anti–double-stranded DNA antibodies. A repeat MRI 11 days later showed progression of disease, and she was empirically treated with 1 g of intravenous methylprednisolone for 5 days.
Fig. 1. Clinical course and initial diagnostic investigation of the index patient.
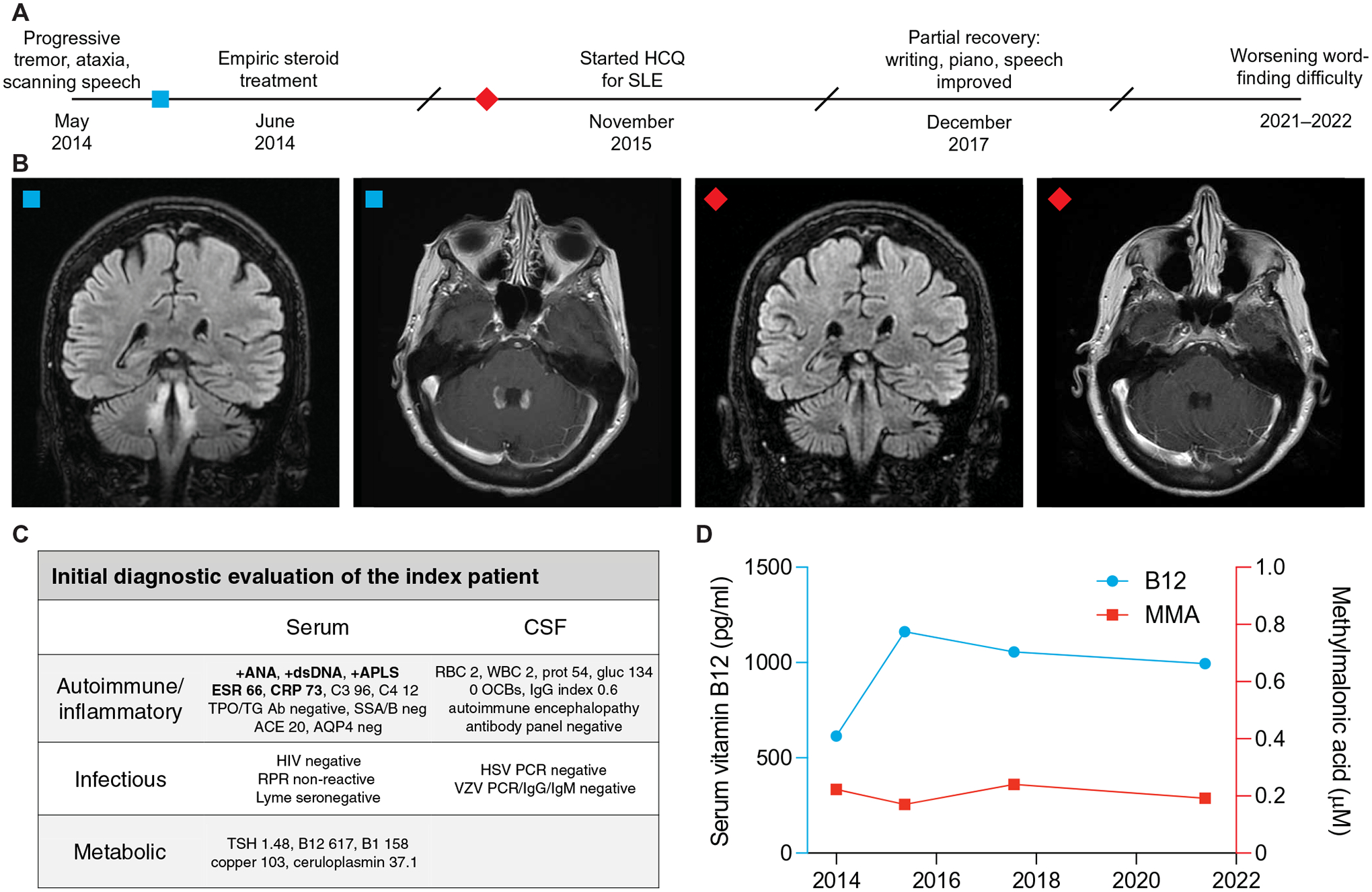
(A) Timeline of evaluation and treatment of the index patient. (B) Coronal slices of T2-weighted fluid-attenuated inversion recovery (FLAIR) MRI and axial slices of T1-weighted post-gadolinium MRI before (left two images) and 9 months after (right two images) steroid treatment. (C) Initial diagnostic evaluation of serum and CSF samples. Bolded items were found to be abnormal. (D) Serum vitamin B12 (blue) and MMA (red) concentrations over the clinical timecourse.
Her neurologic exam incrementally improved over the subsequent 3 months, and a repeat brain MRI showed an interval decrease in gadolinium enhancement. Eighteen months after her initial presentation, she was formally diagnosed with systemic lupus erythematosus (SLE) on the basis of her history of pleuritis and ITP and treated with hydroxychloroquine. Her functional status slowly recovered over the next 3 years: Her speech improved, her handwriting became legible, and she returned to playing the piano. Surveillance brain MRI showed resolution of contrast-enhancement but persistent T2 hyperintensities. Although her initial neurologic deficits stabilized, she experienced gradually worsening word-finding difficulty. Formal neuropsychologic assessment demonstrated mild impairments in processing speed, fluency, and naming. Blood tests for reversible causes of dementia were normal, including serum B12 concentrations between 614 and 1162 pg/ml and methylmalonic acid (MMA; a metabolite that accumulates downstream of B12 deficiency) concentrations between 0.17 and 0.24 μM (Fig. 1D).
Discovery of a functional transcobalamin receptor autoantibody
The patient was enrolled in a research study to detect autoantibodies in suspected neuroinflammatory disease. Programmable phage immunoprecipitation sequencing (PhIP-seq) (12) was used to screen for autoantibodies against a library containing ~730,000 peptide sequences tiling approximately 50,000 human proteoforms (13). CSF was incubated with the phage display library, and antibody-bound phage was immunoprecipitated, sequenced, and compared with mock immunoprecipitations. Among enriched antigens meeting a false discovery rate cutoff of <5%, CD320 was the top cell surface hit, a characteristic consistent with a pathogenic autoantibody (table S2) (14). Antibodies in the patient’s CSF selectively immunoprecipitated overlapping peptides in the extracellular domain of CD320, distant from the low-density lipoprotein receptor type A (LDLR-A) motifs that form the transcobalamin binding interface (Fig. 2A) (15, 16). Sequential alanine mutagenesis of the peptide further narrowed the critical region for autoantibody binding to a 15–amino acid epitope (fig. S1A). Overexpression of CD320 in a human embryonic kidney (HEK) 293T cell–based assay increased CSF antibody binding (Fig. 2B and fig. S1B), and Western blot confirmed the presence of anti-CD320 in serum and CSF (fig. S1C). Conversely, CRISPR-Cas9–mediated KO of CD320 in primary human brain endothelial cells eliminated CSF antibody binding at the live cell surface (Fig. 2C and fig. S1D). These findings confirmed that CD320 was the cell-surface target of an autoantibody present in the patient’s serum and CSF.
Fig. 2. Discovery and validation of a functional transcobalamin receptor autoantibody.
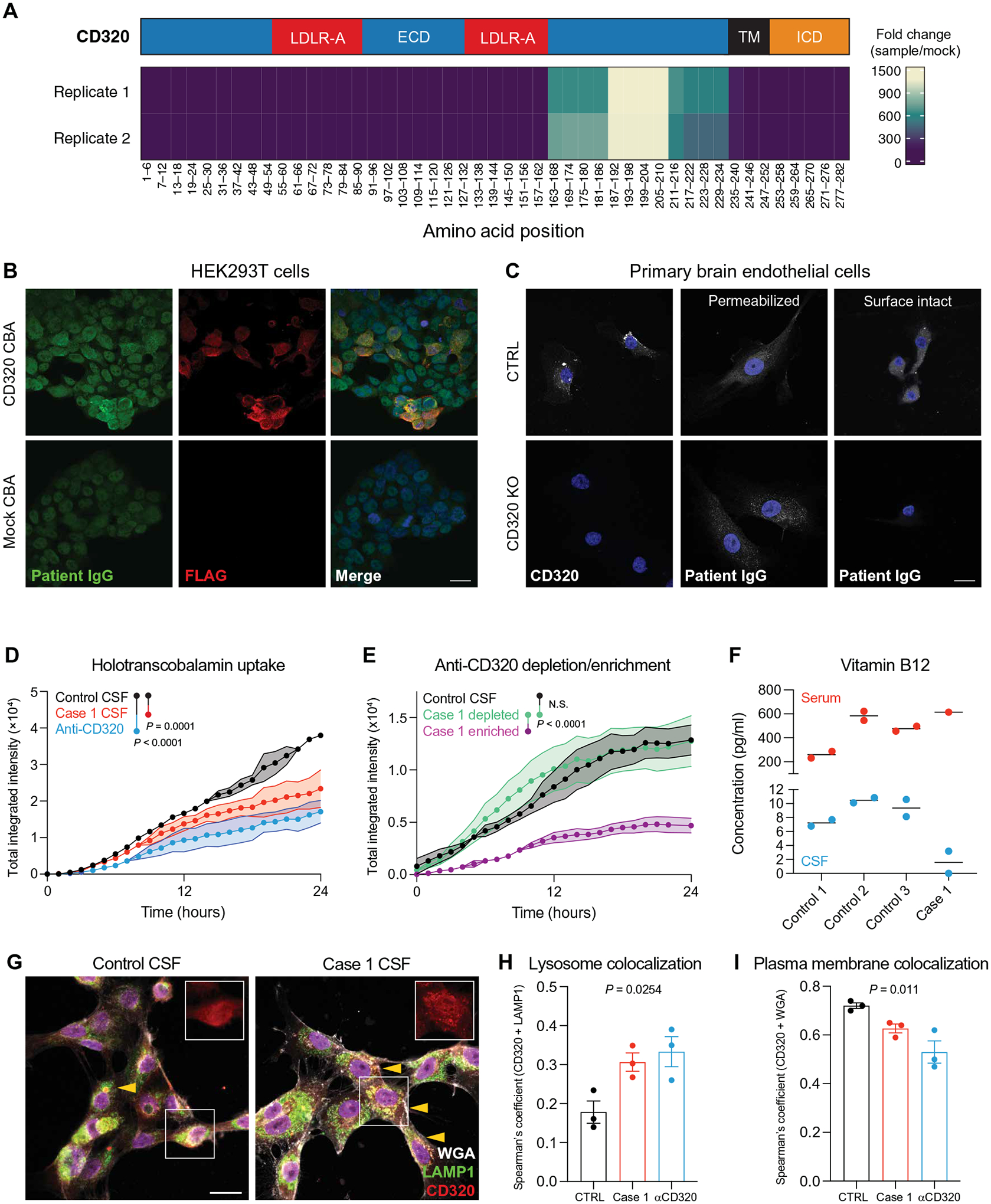
(A) Epitope map of CD320 peptides enriched by patient CSF antibodies. Coverage is divided into five amino acid bins and aligned to the full-length protein (NP_057663.1). The blue region represents the extracellular domain (ECD), the red regions represent LDLR-A domains within the ECD, the black region represents the transmembrane (TM) domain, and the orange region represents the intracellular domain (ICD). (B) Immunore-activity of patient CSF IgG (green) to HEK293T cells overexpressing a FLAG-tagged CD320 construct (red). Staining dilution = 1:25. Scale bar, 20 μm. (C) Control and CD320 KO primary human brain endothelial cells stained with a commercial CD320 antibody (left column) or patient CSF (middle and right columns) and DAPI (blue) to label nuclei. Scale bar, 20 μm. (D) Holotranscobalamin uptake in cells treated with case 1 CSF (red) or a commercial CD320 antibody (blue) compared with cells treated with healthy control CSF (black) (n = 2, paired one-way ANOVA with Tukey’s multiplicity correction, means ± SE). (E) Holotranscobalamin uptake in cells treated with control CSF (black) or case 1 CSF-enriched (magenta) or CSF-depleted (green) of anti-CD320 by affinity purification (n = 3, one-way ANOVA with Tukey’s multiplicity correction, means ± SE). (F) Vitamin B12 concentration in serum (red) and CSF (blue) of three healthy controls and case 1. (G) Representative images of brain endothelial cells treated with control or patient CSF, followed by staining for CD320 (red), wheat germ agglutinin (WGA, a plasma membrane marker; gray), and lysosomal-associated membrane protein (LAMP1; green). Arrows indicate yellow puncta where CD320 (red) colocalizes with lysosomes (LAMP1, green; scale bar = 20 μm). Quantification of CD320 colocalization with the LAMP1 (H) or WGA (I) in cells treated with control CSF (black), case 1 CSF (red), or a commercial anti-CD320 antibody (blue) (n = 3; one-way ANOVA; means ± SE).
CD320 is enriched in endothelial cells at the blood-brain barrier (BBB) (fig. S2, A to C) and mediates the cellular uptake and transcytosis of holotranscobalamin into the CNS (17–19). Incubation of HEK293T cells with patient CSF, but not antibody-negative healthy control CSF, inhibited holotranscobalamin uptake (Fig. 2D and fig. S3, A to D). Immunoglobulin depletion from patient CSF eliminated this inhibitory effect, and patient CSF had no effect on holotranscobalamin uptake in CD320-deficient cells (fig. S3, E to H). Using a biotinylated peptide mapping to the CD320 target epitope as an affinity reagent, we purified anti-CD320 from patient CSF (fig. S3I). Whereas the depleted supernatant had no effect on B12 uptake, purified anti-CD320 recapitulated the inhibitory effect of raw CSF (Fig. 2E). These findings demonstrated that anti-CD320 was a functional autoantibody capable of modulating B12 transport in vitro.
To interrogate a possible anti-CD320–mediated BBB transport defect in vivo, we measured B12 in paired serum and CSF samples. In three healthy controls, the mean serum concentration of B12 was 439 pg/ml, and the mean CSF concentration was 9.0 pg/ml, yielding a CSF to serum B12 ratio of 0.02, concordant with previous studies (Fig. 2F) (20, 21). In contrast, the patient’s serum B12 concentration was 617 pg/ml and CSF concentration was 1.6 pg/ml, yielding a CSF to serum B12 ratio of 0.003. Treatment of human brain endothelial cells with patient CSF increased lysosomal colocalization and decreased plasma membrane colocalization of CD320 (Fig. 2, G to I). Furthermore, immunostaining of postmortem human brain tissue with patient CSF demonstrated immunoglobulin binding to CD31+ endothelial cells (fig. S3J). These findings suggested that anti-CD320 may block B12 transport across the BBB by depleting cell-surface receptor availability. High-dose oral vitamin B12 supplementation was initiated, and the patient’s CSF B12 level increased to 4.8 pg/ml (fig. S3K) despite a persistently low CSF-to-serum ratio. After 9 months of treatment, she reported subjective improvements in mood and cognitive function.
Isolation of a patient-derived monoclonal autoantibody
We set out to identify the source of CD320 autoimmunity in the index patient using the Beacon optofluidic system, which uses optical tweezers and nanochambers to simultaneously analyze tens of thousands of single B cells for antigen-specific antibody secretion (Fig. 3A). Memory B cells were isolated by magnetic separation from the patient’s peripheral blood mononuclear cells (PBMCs), activated for antibody secretion, and loaded onto an optofluidic chip. Using fluorescent CD320-peptide tetramers, we identified and sequenced five CD320-specific clones from 23,205 IgG-secreting B cells (hit rate 0.02%). All five clones contained the same B cell receptor variable sequence, indicative of clonal expansion (fig. S4A). Next, we recombinantly expressed this patient-derived monoclonal antibody and confirmed antibody binding to full-length CD320 in a cell-based assay (Fig. 3B) and to CD320 peptide using biolayer interferometry (Fig. 3C). Sequencing of the heavy chain constant region demonstrated IgG1 subclass (Fig. 3D). Anti-CD320 impaired B12 uptake (Fig. 3E) without inducing cell death (Fig. 3F).
Fig. 3. Isolation of a patient-derived monoclonal autoantibody.
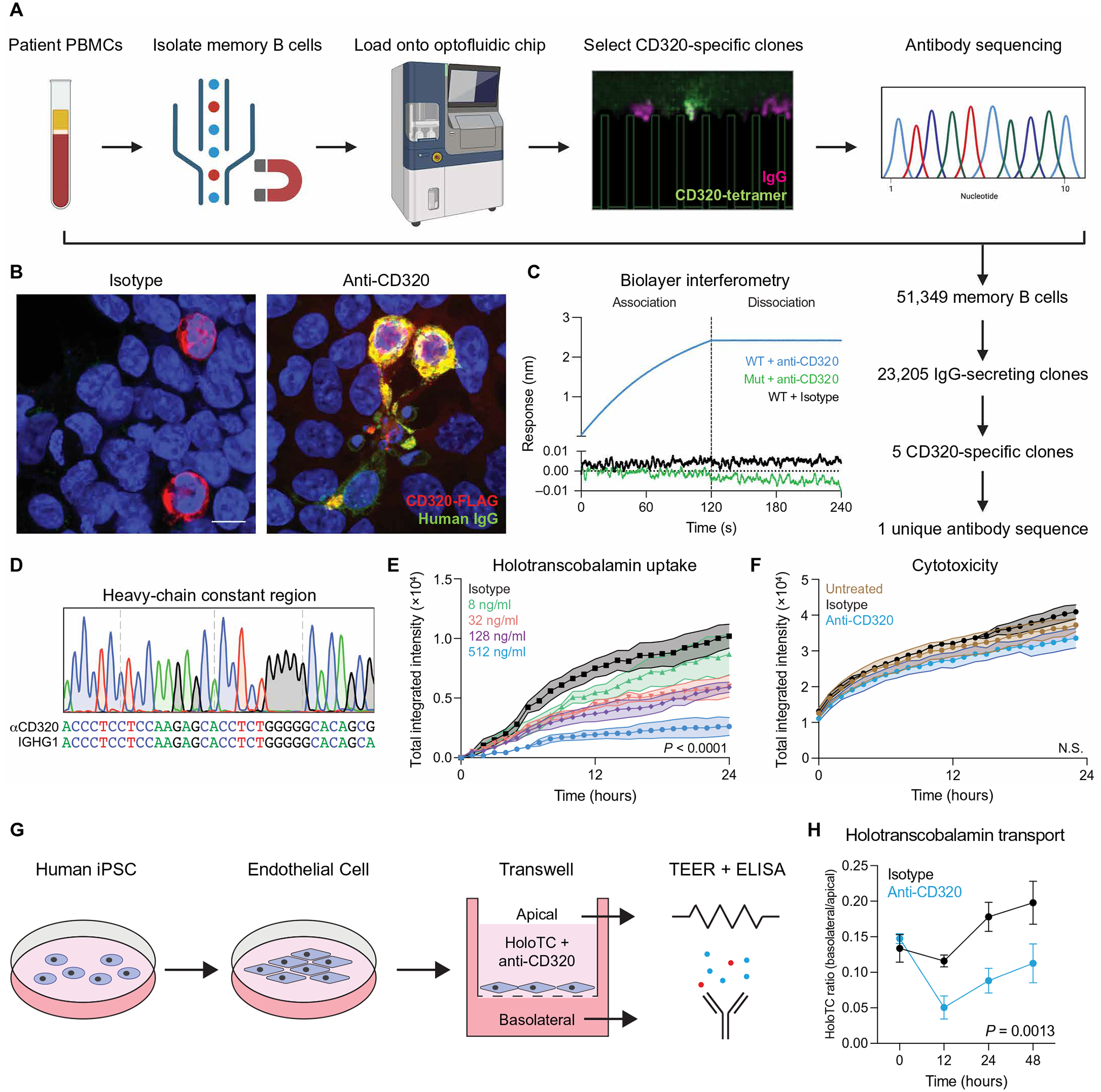
(A) Schematic of the Beacon optofluidic autoantibody discovery workflow. Memory B cells were magnetically isolated from PBMCs, loaded onto an optofluidic chip, and screened for antibody secretion (IgG, magenta) and antigen specificity (CD320-tetramers, green). CD320-reactive clones were exported for cDNA synthesis and immunoglobulin sequencing. (B) HEK293T cells transfected with CD320-FLAG (red) were stained with an isotype control antibody (left) or the patient-derived monoclonal antibody (green, right). Yellow signal represents colocalization of human IgG with the CD320-FLAG construct. Scale bar, 20 μm. (C) Kinetic analysis of anti-CD320 binding to wild-type (WT) CD320 peptide (blue), anti-CD320 binding to a mutant CD320 peptide with the autoepitope replaced with a stretch of 10 alanines (green), or isotype control antibody binding wild-type CD320 peptide (black). (D) Sanger sequencing of the heavy-chain constant region compared with the immunoglobulin heavy constant gamma 1 (IGHG1) consensus sequence. (E) Dose response of anti-CD320 on holotranscobalamin uptake (n = 3, one-way ANOVA with Tukey’s multiplicity correction, means ± SE). (F) Cytotoxicity in untreated HEK293T cells (brown) or HEK293T cells treated with isotype control antibody (black) or anti-CD320 (blue) (n = 3, one-way ANOVA, means ± SE). (G) Schematic of the in vitro model of the human BBB. iPSCs were differentiated into endothelial cells and plated on a semi-permeable transwell membrane, and B12 transport was measured in the presence of anti-CD320 or isotype control by serial sampling of holotranscobalamin in the apical and basolateral media. (H) Ratio of basolateral to apical holotranscobalamin concentration (measured by ELISA) in isotype control (black)– or anti-CD320 (blue)– treated wells over 48 hours. A higher ratio represents greater B12 transport (n = 3, two-way ANOVA, means ± SD). N.S., not significant.
We then interrogated the autoantibody’s effect on B12 transport using an in vitro model of the human BBB (Fig. 3G). Human induced pluripotent stem cells (hiPSCs) were differentiated into endothelial cells using a previously established protocol and plated onto semiper-meable transwell membranes (22). Barrier integrity and tight junction formation were confirmed using trans-endothelial electrical resistance (TEER; fig. S4B) (23). Upon formation of a high TEER barrier, we replaced the basolateral space with B12-depleted medium and replaced the apical space with the same medium supplemented with holotranscobalamin and isotype control antibody or anti-CD320. Serial measurement of the holotranscobalamin concentration in apical and basolateral media enabled assessment of B12 transport dynamics. Compared with the isotype control, anti-CD320 impaired transport of holotranscobalamin across the barrier (Fig. 3H).
Identification of additional cases
We retrospectively analyzed PhIP-seq data from 254 individuals enrolled in the same neuroinflammatory disease study as the index patient. We identified seven additional cases harboring a CSF anti-CD320 autoantibody targeting the same epitope (cases 2 to 8; Fig. 4A and fig. S5, A to C). These seven individuals were clinically diverse, with a wide range of autoantibody titers (Fig. 4B and fig. S5D). CSF from all seven cases functionally impaired holotranscobalamin uptake (Fig. 4C). Compared with antibody-negative healthy controls, B12 was low and MMA was elevated in the CSF of four of the seven cases (Fig. 4D and fig. S5E). Holotranscobalamin was low in the CSF of all seven cases (Fig. 4E) (24). In contrast, serum B12 was normal in the four individuals with paired blood samples available (table S3). Case 3 had T2 hyperintensities in the cerebellar peduncles and dorsal brainstem resembling the brain MRI of the index patient (Fig. 4F). Case 6 developed progressive ataxia, cognitive decline, and white matter disease in the setting of a persistently elevated anti-CD320 titer on longitudinal CSF sampling (fig. S5F). Case 8 presented with sub-acute combined degeneration of the dorsal columns and lateral corticospinal white matter tracts, a classic manifestation of traditional B12 deficiency (Fig. 4G and table S4). He partially recovered on B cell depletion therapy and high-dose oral B12 supplementation, and his MRI abnormalities resolved (Fig. 4G).
Fig. 4. Identification of additional cases with CD320 autoantibodies.
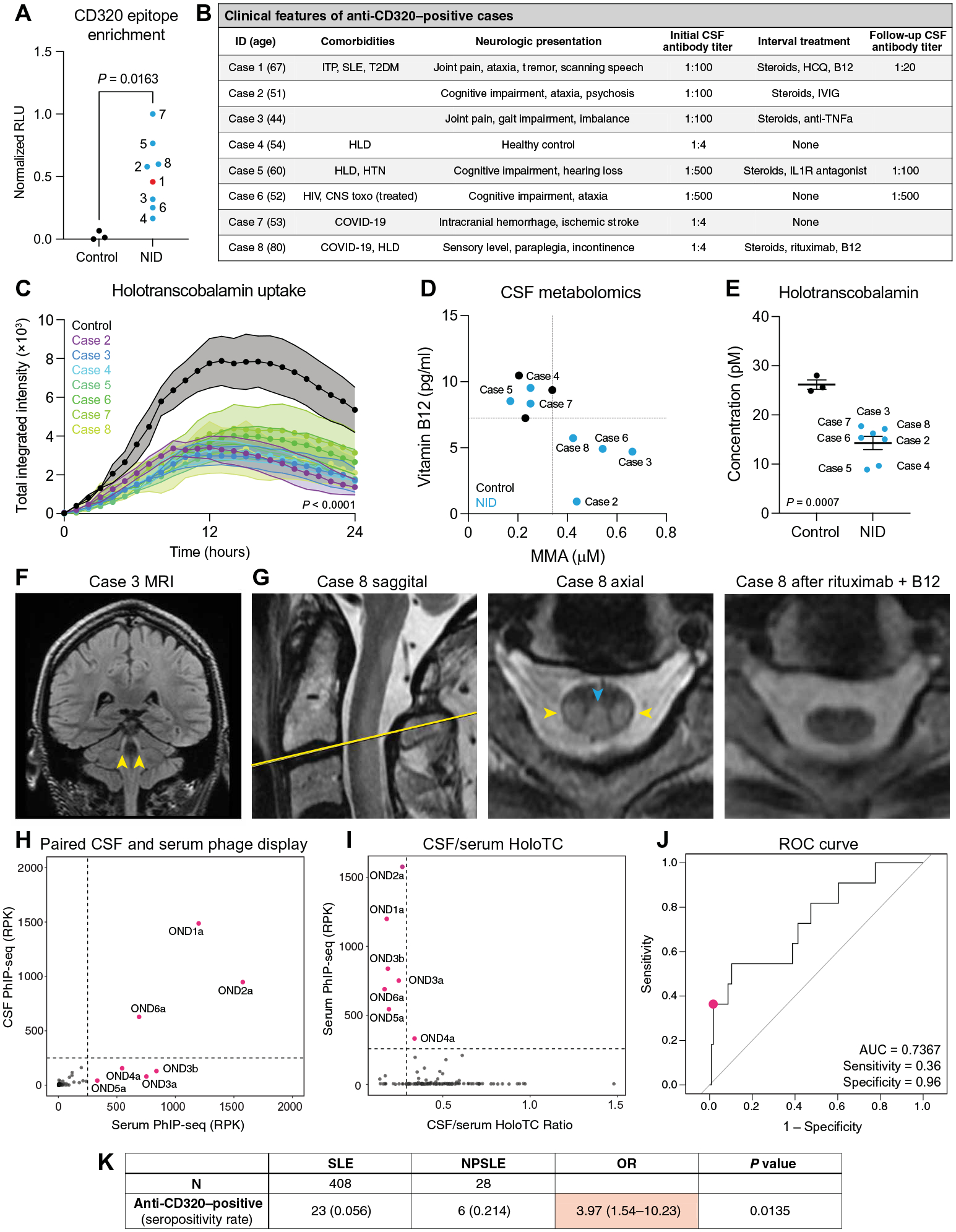
(A) Enrichment of the CD320 target peptide by CSF immunoglobulin from three healthy controls (black) and eight individuals enrolled in a neuroinflammatory disease study (NID; blue), including the index patient (red), using a split-luciferase binding assay (two-sided t test). (B) Clinical characteristics of individuals harboring CSF anti-CD320 autoantibodies. Autoantibody titer was determined by cell-based assay. HLD, hyperlipidemia; HTN, hypertension. (C) Holotranscobalamin uptake by cells treated with CSF from the seven additional cases compared with treatment with healthy control CSF (n = 3, one-way ANOVA with Tukey’s multiplicity correction, means ± SE). (D) Total vitamin B12 (y axis) and MMA (x axis) concentrations in CSF from the seven cases enrolled in a neuroinflammatory disease study (blue dots) and three healthy controls (black dots). Dotted lines show the lowest CSF vitamin B12 or highest MMA level among healthy controls. (E) Holotranscobalamin concentrations in CSF from healthy controls and cases in the neuroinflammatory disease cohort (two-sided t test; means ± SE). (F) Coronal T2-weighted FLAIR MRI of case 3. Yellow arrows indicate symmetric T2-FLAIR hyperintensities involving the bilateral cerebellar peduncles. (G) Sagittal and axial cuts of the T2-weighted C-spine MRI for case 8 before (left two images) and axial cut after (right image) treatment with rituximab and B12 supplementation. Blue arrow indicates T2 hyperintensity in the dorsal columns. Yellow arrows indicate T2 hyperintensity in the lateral corticospinal tracts. (H) CD320 enrichment of paired serum and CSF samples from 132 patients with MS or other neurologic diseases. Enrichment determined by PhIP-seq, where RPK represents depth-normalized sequencing reads. Labeled samples exhibit CD320 enrichment equal to or greater than 250 RPK (dotted line). (I) CSF/serum holotranscobalamin (HoloTC) ratio versus serum anti-CD320 enrichment. The CSF/serum holotranscobalamin threshold (0.2) was chosen on the basis of previously reported reference ranges in each biofluid (4, 49). (J) Receiver operator characteristic (ROC) curve showing the performance of serum anti-CD320 seropositivity for predicting a low CSF/serum holotranscobalamin level. RPK threshold was chosen to stratify seropositive versus seronegative and is not a proxy for autoantibody titer. (K) Contingency analysis of anti-CD320 seropositivity in non-neurologic SLE and NPSLE. Fisher’s exact test, 95% confidence interval. RLU, relative light unit; AUC, area under the curve.
Because of the presence of anti-CD320 in the CSF of a healthy control participant (case 4) and in a noninflammatory control participant (case 7) of the neuroinflammatory disease study, we screened sera from a cohort of 84 additional healthy controls and identified anti-CD320 antibodies targeting the same epitope in five individuals, yielding a seropositivity rate of ~6% (fig. S6A). To determine the clinical relevance of this finding, we leveraged 132 paired serum and CSF samples from a cohort of patients with multiple sclerosis (MS; n = 123) or other neurologic diseases (n = 9). Anti-CD320 autoantibodies were detected in 5.7% of this cohort, concordant with the seropositivity rate in healthy controls (Fig. 4H). High-titer CSF samples impaired holotranscobalamin uptake (fig. S6B). Detection of anti-CD320 in the serum was 36% sensitive and 96% specific for a low CSF to serum holotranscobalamin ratio (Fig. 4, I and J), and holotranscobalamin accumulated in the sera of seropositive individuals (fig. S6C). Furthermore, anti-CD320 seropositivity predicted elevated CSF MMA, a metabolic marker of B12 deficiency, with a 78% positive predictive value (fig. S6, D and E).
SLE is a multi-organ autoimmune disease that occasionally attacks the nervous system, leading to neuropsychiatric SLE (NPSLE) (25). NPSLE is clinically heterogeneous and difficult to diagnose because of the lack of a validated biomarker (26–28). Because the index patient was eventually diagnosed with SLE, we wondered whether anti-CD320 is enriched in other patients with systemic autoimmune disease. We compared anti-CD320 seropositivity in 408 patients with non-neurologic SLE and 28 patients with NPSLE, including the index patient. The seropositivity rate in non-neurologic SLE was 5.6%, concordant with the seropositivity rate in healthy controls (fig. S6F). In contrast, anti-CD320 was nearly four times as prevalent in NPSLE [21.4%, odds ratio (OR) = 3.97; Fig. 4K]. Five of the six patients with seropositive NPSLE presented with cognitive symptoms, and three of the six presented with myelopathy (table S5). These findings suggested that comorbid anti-CD320 may contribute to neurologic deficits in a subset of patients with NPSLE.
Whole-genome CRISPR screen uncovers an alternative pathway for B12 uptake
Despite the presence of anti-CD320 in the serum, seropositive individuals displayed none of the hematologic manifestations of vitamin B12 deficiency (table S3). Therefore, we hypothesized that alternative B12 uptake pathways exist in blood cells to compensate for systemic impairment of CD320. To identify this alternative receptor, we performed a genome-wide CRISPR interference (CRISPRi) screen using the granulocyte-lineage leukemia cell line, K562, which has previously been shown to internalize holotranscobalamin (Fig. 5A) (29). K562 cells expressing nuclease-dead Cas9 fused to the transcriptional repressor, Krüppel associated box (KRAB), were infected with a library of about 200,000 single guide RNAs (sgRNAs) targeting transcription start sites of all protein coding genes and about 10,000 control sgRNAs (30, 31). We incubated this pool of single knockdown cells with holotranscobalamin conjugated to a pH-sensitive fluorescent dye. This allowed us to readout B12 uptake and lysosomal trafficking via a red fluorescent signal. We sorted cells with high and low holotranscobalamin uptake by fluorescence-activated cell sorting (fig. S7A). Next, we sequenced genomic DNA from each population, compared the sgRNA distribution between populations, and estimated the effect size and P value for each gene knockdown using the Cas9 high-throughput maximum likelihood estimator (casTLE) (32). This screen identified 68 hits with a false discovery rate of less than 0.1%, including CD320 (Fig. 5B and table S6).
Fig. 5. LDLR is an alternative B12 uptake receptor.
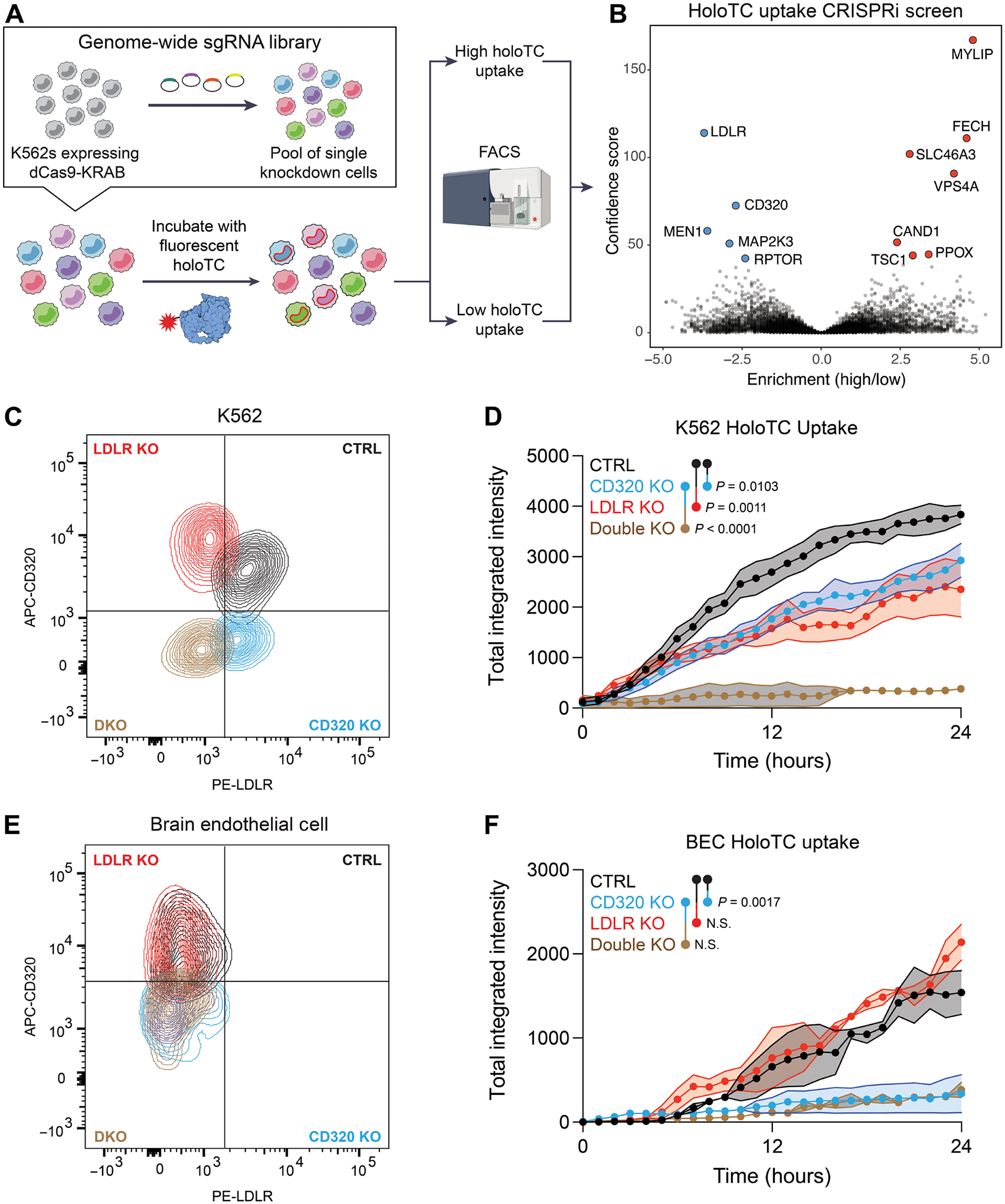
(A) Schematic of a CRISPRi screen for genetic modifiers of holotranscobalamin uptake. K562 cells expressing dCas9-KRAB were infected with a library of sgRNAs targeting the transcriptional start sites of all protein coding genes. This pool of single knockdown cells was then incubated with holotranscobalamin conjugated to a pH-sensitive fluorescent dye. Cells with high (top 5% of red fluorescent signal) and low (bottom 5%) holoTC uptake were separated by FACS and then sequenced and analyzed using casTLE. Screen was performed in technical duplicate. (B) Volcano plot of the genes which when repressed impair (blue) or promote (red) holoTC uptake. The top 12 hits are labeled, meeting an adjusted P value of <0.00001 (Benjamini-Hochberg). (C) Flow cytometry confirmation of control (black), CD320 KO (blue), LDLR KO (red), and double KO (brown) K562 cells. (D) Holotranscobalamin uptake by control (black), CD320 KO (blue), LDLR KO (red), or double KO (brown) K562 cells (n = 3, one-way ANOVA with Tukey’s multiplicity correction, means ± SE). (E) Flow cytometry confirmation of control (black), CD320 KO (blue), LDLR KO (red), and double KO (brown) primary human brain endothelial cells. (F) Holotranscobalamin uptake by control (black), CD320 KO (blue), LDLR KO (red), or double KO (brown) primary brain endothelial cells (n = 2, one-way ANOVA with Tukey’s multiplicity correction, means ± SE). APC, allophycocyanin; PE, phycoerythrin.
The top knockdown that impaired B12 uptake was the LDLR, a promiscuous cell-surface receptor whose primary function is cellular uptake of cholesterol-rich lipids (33). Conversely, the top knockdown that promoted B12 uptake was myosin regulatory light chain interacting protein, an E3 ubiquitin-protein ligase that mediates proteasomal degradation of LDLR (34). Whereas CD320 is relatively enriched at the BBB, LDLR is relatively depleted (fig. S7B). Furthermore, CD320 binds holotranscobalamin via two LDLR-A domains (15). Together, these findings nominated LDLR as a candidate for noncanonical B12 uptake outside the CNS.
To validate this hit, we knocked out LDLR alone or in combination with CD320 in K562 cells, primary brain endothelial cells, and HEK293T cells. Compared with the control, LDLR KO or CD320 KO in K562 cells resulted in impaired holotranscobalamin uptake (Fig. 5, C and D). However, combined KO of both LDLR and CD320 was necessary to eliminate uptake in this hematopoietic cell line. Similar results were observed in HEK293T cells (fig. S7, C and D). In contrast, knocking out CD320 alone was sufficient to eliminate holotranscobalamin uptake in brain endothelial cells, which do not appreciably express LDLR at baseline (Fig. 5, E and F). These findings suggested that LDLR contributes to holotranscobalamin internalization in hematopoietic cells, providing an alternate pathway for B12 uptake when CD320 is impaired.
DISCUSSION
We identified an autoantibody targeting the transcobalamin receptor in a patient with an unusual neurologic presentation. Anti-CD320 impaired uptake and transport of B12 in vitro and was associated with CNS-specific B12 deficiency in vivo. We retrospectively found autoantibodies reactive to the same epitope of CD320 in seven other patients with unexplained neurologic symptoms, 6% of healthy controls, and 21.4% of patients with NPSLE. Although CD320 was necessary for B12 uptake in brain endothelial cells, a CRISPR screen uncovered LDLR as an alternative uptake pathway in a hematopoietic cell line. These findings suggest a model in which anti-CD320 impairs transport of B12 across the BBB, leading to ABCD with varied neurologic manifestations but sparing peripheral manifestations of B12 deficiency (fig. S8A).
The neurologic spectrum of traditional systemic B12 deficiency is wide, ranging from peripheral neuropathy to encephalomyelopathy. Before the onset of these symptoms, individuals progress from replete to deficient through a state of inadequacy known as subclinical B12 deficiency (35). We detected anti-CD320 in a wide range of cases, including healthy controls, patients with other neurologic conditions (MS and NPSLE), and a patient with classic signs of B12 deficiency despite a normal serum B12 concentration (case 8). This heterogeneity may reflect differences in anti-CD320 antibody avidity, the degree and duration of CSF B12 deficiency, or host factors conferring differential susceptibility to disease (fig. S8B). Although not directly addressed in the present study, the specificity and positive predictive value of anti-CD320 in the blood for detecting early markers of B12 deficiency in the CSF led us to speculate that seropositive healthy controls may be at risk for ABCD. In contrast to neuronal surface autoantibodies in autoimmune encephalitides [such as N-methyl-d-aspartate (NMDA) receptor antibody or leucine-rich glioma-inactivated 1 antibody] that are directly neuromodulatory (36–38), CD320 autoantibodies may exert a second-order effect on brain function by first decreasing CSF concentrations of B12 that then lead to insidious neurologic sequelae. This study suggests that the measurement of vitamin B12 metabolites in CSF should be considered for patients with unexplained neurologic deficits who are anti-CD320 seropositive.
The clinical relevance of anti-CD320 in healthy controls remains uncertain. Unique autoantibody repertoires have previously been measured in healthy individuals (39–42). Many of these autoantibodies are likely nonpathogenic and represent an immunologic fingerprint of past exposures. However, some of these antibodies are speculated to cause subclinical disease, to predispose individuals to disease, or even to protect individuals from disease (43). Deciphering where anti-CD320 lies on this spectrum will require larger cohorts and longitudinal sampling of the general population.
All cases shared autoantibodies targeting a common epitope in the extracellular domain of the transcobalamin receptor (Pro183-Thr197). An autoantibody roughly mapping to a similar region (Thr169-Tyr229) has also been described in patients with cutaneous arteritis (44). Future studies will be necessary to determine whether neurologic and dermatologic manifestations of anti-CD320 autoantibodies coexist.
Several patients improved after immunosuppression (cases 1, 2, 3, and 8), and two of these patients (cases 1 and 8) received concurrent high-dose systemic B12 supplementation. It is unclear whether immunosuppression mediated improvement in these cases via dampening of alternative neuroinflammatory mechanisms or via mitigation of anti-CD320 autoantibody production. The increased B12 concentration in posttreatment CSF from case 1 suggests that high serum B12 concentrations may suffice to overcome BBB transport defects caused by anti-CD320.
The identification of tissue-specific B12 uptake pathways provides a possible explanation for isolated neuropsychiatric manifestations of systemic B12 deficiency (11). CD320 autoantibodies may be over-represented in these patients, impairing the transport of B12 into the brain, whereas LDLR is still able to transport B12 into blood cells.
This study has several limitations. First, without prospective recruitment and empiric B12-targeted treatment of seropositive patients, it is not possible to determine whether anti-CD320 autoantibodies primarily drive disease pathogenesis, exacerbate underlying pathology (“second hit”), or passively spectate. Second, anti-CD320 sensitivity and specificity calculations were based on measurements in a cohort of patients with other neurologic diseases (primarily MS), which may not accurately reflect the clinical relevance of the autoantibody in healthy individuals. Future studies will be necessary to evaluate whether anti-CD320 autoantibodies modulate clinical outcomes in the general population and in other neurologic diseases such as MS and NPSLE. Last, this study is entirely reliant on human tissues for the characterization of patient-derived autoantibodies and alternative B12 uptake mechanisms. Although mouse and human B12 biology diverge in certain key aspects, animal models will be important to evaluate the pathogenicity of anti-CD320 and the role of LDLR in B12 metabolism in vivo (45).
These findings parallel cerebral folate deficiency syndrome, in which autoantibodies targeting folate receptors at the choroid plexus are associated with low 5-methyltetrahydrofolate levels in the CSF despite normal levels in the blood (46). Correction of central vitamin B12 deficiency may provide clinical benefit with minimal risk. Future studies will be necessary to determine the therapeutic effect of high-dose vitamin B12 supplementation and/or immunomodulatory therapy.
MATERIALS AND METHODS
Study design
The aims of this study were to (i) identify and characterize novel autoantibodies in the index patient, (ii) screen for this autoantibody in additional controls and patients, and (iii) dissect tissue-specific B12 uptake mechanisms. Sample sizes, when not limited by human sample availability, were chosen on the basis of similar studies in the literature. All experiments included negative controls and, when available, validated positive controls. No data were excluded, all outliers are shown as individual data points, and experimental end points were predetermined at the onset of the study. None of the experiments required randomization, and all microscopy-based quantifications were performed by blinded observers. Experiments were performed in technical and biological replicate (exact number specified in figure legends) and, when possible, validated with orthogonal techniques.
Patient enrollment and data collection
Patients were enrolled in a research study to detect novel autoantibodies in suspected neuroinflammatory disease [University of California, San Francisco (UCSF) IRB# 13–12236]. CSF samples were frozen and stored at −80°C before aliquoting as undiluted CSF or diluted 1:1 in antibody sample buffer [final concentration: 20% glycerol, 20 mM Hepes, and 0.02% sodium azide in phosphate-buffered saline (PBS)]. Healthy volunteers aged 18 to 65 were enrolled in a research study (UCSF IRB# 22–36117) for the collection of serum samples. Individuals were excluded if they were currently pregnant, were currently receiving immunosuppressive medications, had received chemotherapy in the past 5 years, had been diagnosed with a bleeding disorder, or had received intravenous immunoglobulin in the last 30 days. Paired serum and CSF were collected from a cohort of patients with MS or other neurologic diseases (IRB# 14–15278). Patients with NPSLE were enrolled under the aforementioned study protocol (UCSF IRB# 13–12236). NPSLE was defined according to a modification of the 2001 American College of Rheumatology criteria. Specifically, patients were classified as NPSLE if they (i) carried a clinical diagnosis of SLE and (ii) had one or more of the following CNS manifestations of disease not otherwise explained by an alternative etiology: aseptic meningitis, cerebrovascular disease (not otherwise explained by anti-phospholipid antibody syndrome), demyelinating syndrome, movement disorder, myelopathy (not otherwise explained by anti-AQP4), seizure disorder, acute confusional state, cognitive dysfunction (moderate or severe), severe acute depression, and psychosis. Patients with non-neurologic SLE were enrolled in the California Lupus Epidemiology Study.
Programmable phage display
We adapted a previously published protocol for phage immunoprecipitation sequencing (PhIP-seq) (13). A library containing 731,724 49–amino acid peptides with 25–amino acid overlaps was cloned into T7 bacteriophage. Patient CSF was incubated with 1010 plaque-forming units of the phage library, antibodies were enriched with protein A/G magnetic beads, and antibody-bound phage was amplified in Escherichia coli before a second round of immunoprecipitation. Enriched phage lysates were adaptor-ligated and barcoded before pair-end sequencing on an Illumina NovaSeq to a depth of 2 million reads per sample. Reads were trimmed, aligned at the amino acid level using RAPSearch, and normalized to sequencing depth to generate reads per 100,000 bases (RPK) for each sample. Enriched peptides were identified by calculating the fold change of normalized counts between samples immunoprecipitated with CSF or magnetic beads only. The Benjamini-Hochberg method was used to correct for multiplicity and calculate false discovery rates. A step-by-step protocol is available at www.protocols.io/view/scaled-moderate-throughput-multichannel-phip-proto-8epv5zp6dv1b/v1.
Cell-based overexpression assay
HEK293T cells [American Type Culture Collection (ATCC)] were transfected with a FLAG-tagged CD320 construct packaged within the pCMV6 entry plasmid (OriGene, RC200073) or a FLAG-tagged MOG construct (GenScript OHu14941) using Lipofectamine 3000 according to the manufacturer’s protocol. After 24 hours, cells were fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature (RT), blocked and permeabilized, and stained with patient CSF and rabbit anti-FLAG [1:800; Cell Signaling Technology (CST), 14793] at 4°C overnight. Secondary staining was conducted with Alexa Fluor–conjugated antibodies at RT for 1 hour. Fluorescent images were acquired on a Zeiss LSM780 confocal laser scanning microscope.
Western blot
A recombinant CD320 protein containing a C-terminal GST tag and the putative epitope (Abnova, catalog no. H00051293-P01) and a recombinant truncated CD320 protein containing an N-terminal His-tag (Novus Biologicals, NBP189348PEP) were loaded into the wells of a 4 to 12% Criterion XT Bis-Tris PAGE Gel (Bio-Rad, #3450117) at 1.5 μg per well. After separation for 1 hour at 180 V, the proteins were transferred onto Immun-Blot Low Fluorescence polyvinylidine fluo-ride (PVDF) membranes (Bio-Rad, #1620264) using ice-cold transfer buffer containing 15% methanol and 85% 1x tris/glycine transfer buffer (Bio-Rad, #1610734) at 100 V for 1 hour on ice. After protein transfer, the PVDF membranes were incubated in Intercept PBS Blocking Buffer (LiCor, 927–70001) for 30 min at RT. The membranes were then incubated in CSF and plasma-diluted in Intercept PBS Blocking Buffer at dilutions of 1:10 and 1:100, respectively, at 4°C overnight. Membranes were rinsed five times in 1X tris-buffered saline (TBS) containing 0.1% Tween 20 (TBS-T) and stained with goat–anti-human IRDye 800CW (LiCor, 926–32232) at 1:10,000 at RT for 1 hour. After five more rinses using 1X TBS-T, the membranes were imaged on a LI-COR Odyssey.
CRISPR-Cas9 KO
Primary human brain endothelial cells (Cell Systems) were cultured in complete classic medium in T75 flasks coated with attachment factor to 50% confluency. Ribonucleoprotein complexes consisting of purified Cas9 and a pool of three chemically modified sgRNAs targeting CD320 (GGCGUCACUCACCUGCGGCC, CGGCCUCCAGGCCUAGUCCG, and GGAUGGCGCAGGUUGGAGCG), LDLR (GCCGUGGGCUCUGUCAAGCU, GGUCGCUCUGGACACGGAGG, and UCACCUGCAGAUCAUUCUCU), or both CD320 and LDLR were transfected into cells using the Lipofectamine CRISPRMAX Transfection Reagent according to the manufacturer’s protocol (Synthego). After 72 hours, single-transfected cells were clone-sorted into a 96-well plate using a BD FACSAria III. Successful KO was confirmed by flow cytometry and immunofluorescence using a commercial anti-CD320 antibody (5 μg/mL; R&D Systems, AF1557) and anti-LDLR antibody (1:100; BD, 565653). The same lipofection protocol was used for generation of single KOs in HEK293T cells. For K562 cells (ATCC), ribonucleoprotein complexes were transfected by nucleofection using the Lonza SF Cell Line 4D-Nucleofector X Kit S (V4XC-2032) according to the manufacturer’s protocol using the FF-120 program.
Holotranscobalamin uptake assay
Recombinant human transcobalamin II (R&D Systems, 7895-TC-050) was conjugated to a pH-sensitive fluorescent dye (pHrodo Red iFL NHS ester) at a dye:protein molar ratio of 4:1. Free dye was removed using a 40-kDa MWCO Zeba Spin Desalting Column. pHrodo-conjugated transcobalamin was incubated with a 3X molar ratio of cyanocobalamin at RT for 1 hour to form pHrodo-holotranscobalamin. Wild-type and CD320 KO HEK293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) to 80% confluency before plating in a 96-well tissue culture–treated plate at 20,000 cells per well 1 day before the assay. Cells were washed with PBS and treated with 10% healthy control CSF, patient CSF, or anti-CD320 (1 μg/ml; R&D Systems, AF1557) in serum-free DMEM for 30 min at 37°C. A total of 100 ng of pHrodo-holotranscobalamin was added to each well, and cellular uptake was monitored every hour for 24 hours using a live-cell time-lapse fluorescence microscope (Incucyte SX5). The average total integrated orange intensity across technical duplicates was used to compare uptake dynamics between conditions.
Affinity purification
For total IgG affinity purification, 10 μl of Pierce Protein A/G magnetic beads per reaction was washed on a magnetic stand three times with DMEM. A total of 10 μl of CSF was diluted in 90 μl of DMEM, and 10 μl of washed beads was added. The mixture was incubated for 1 hour at RT with mixing. After incubation, the mixture was placed on a ring magnet, the supernatant was saved, and the beads were washed three times with 0.1% Tween 20 in PBS. After the last wash, beads were resuspended in 100 μl of 0.1 M glycine (pH 2.0) and incubated for 10 min at RT to elute IgG off beads. The mixture was placed on a magnetic stand, and the eluant was collected. A total of 10 μl of 1 M tris was added to neutralize the eluant. Buffer was exchanged with DMEM using a 40-kDa MWCO Zeba column.
For antigen-specific IgG affinity purification, an N-terminal Biotin-Ahx–tagged peptide (EGDATTMGPPVTLESVTSLRNATTMGPPVTLESVPSVGNATSSSAGDQ) corresponding to the target epitope of anti-CD320 was used as an affinity reagent (GenScript). CSF was diluted 1:10 in DMEM, and 100 μg/ml of peptide was added and incubated 1 hour at RT with mixing. The mixture was then incubated with Pierce Streptavidin magnetic beads for 1 hour with mixing. From there, washing and elution were carried out identically to the total IgG affinity purification protocol.
Enzyme-linked immunosorbent assay
Vitamin B12 concentrations in serum and CSF were measured with a commercial competitive enzyme-linked immunosorbent (ELISA) (Novus Biologicals, NBP2–60196) according to the manufacturer’s protocol with two adaptations. First, precoated wells were blocked overnight with sample buffer at 4°C. Second, CSF samples were diluted 1:10 before addition to blocked wells, and serum samples were diluted 1:100. Prior studies have demonstrated the stability of vitamin B12 with repeated freeze-thaw cycles (47). Holotranscobalamin concentration in serum and CSF was measured by a commercial ELISA assay (Tecan, AX53101) according to the manufacturer’s protocol. Total IgG concentration was measured using a commercial ELISA assay (Invitrogen, BMS2091TEN) using a dilution of 1:2500 for CSF samples.
Colocalization immunofluorescence
Primary human brain endothelial cells were cultured in complete classic medium (Cell Systems, ACBRI 376) at 20,000 cells per well in a 12-well removable chamber slide (Ibidi) coated with attachment factor (Cell Systems). On day 2, the medium was replaced with complete serum-free medium (Cell Systems) containing 10% CSF. After 5 hours of treatment, AF647-conjugated wheat germ agglutinin (1:200; Vector Laboratories) was added to wells for 5 min at RT to label the surface glycocalyx, and cells were subsequently washed and fixed with 4% PFA for 10 min at RT. Cells were permeabilized and blocked with 0.3% Triton X-100, 0.1% Tween 20, and 3% normal donkey serum in PBS for 30 min at RT and then stained overnight at 4°C with goat anti-CD320 (1:200; R&D Systems, AF1557) and rabbit anti–lysosomal-associated membrane protein 1 (1:100; CST, D2D1). Primary stain was washed out three times, and then cells were stained with secondary antibodies (1:1000), followed by mounting with Prolong Gold. Slides were imaged on a Zeiss LSM780 at ×63. Colocalization analysis was performed using Coloc 2 (ImageJ) with identical threshold parameters across conditions.
Gas chromatography mass spectrometry
MMA was measured by targeted gas chromatography–tandem mass spectrometry (GC-MS/MS) at Bevital AS (Norway) using a previously published protocol (48). Homocysteine measurements were below the limit of detection.
Holotranscobalamin binding assay
HEK293T cells were incubated with goat polyclonal anti-CD320 (R&D Systems, AF1557) at varying concentrations on ice for 30 min. Recombinant human transcobalamin II was conjugated to a fluorescent dye (Alexa Fluor 647 NHS ester) at a dye:protein molar ratio of 10:1. Free dye was removed using a 40-kDa MWCO Zeba Spin Desalting Column. Alexa Fluor 647–conjugated transcobalamin was incubated with a 3× molar ratio of cyanocobalamin at RT for 1 hour to form AF647-holotranscobalamin. A total of 100 ng of AF647-holotranscobalamin was added to pretreated cells and incubated on ice for 30 min. Cells were washed with PBS and analyzed for fluorescent signal on a BD LSRFortessa flow cytometer.
Antibody internalization assay
Goat polyclonal anti-CD320 (R&D Systems, AF1557) and an isotype control antibody were conjugated to a pH-sensitive fluorescent dye (pHrodo Red iFL NHS ester) at a dye:protein molar ratio of 10:1. Free dye was removed using a 40-kDa MWCO Zeba Spin Desalting Column. Wild-type and CD320 KO HEK293T cells were grown in DMEM containing 10% FBS to 80% confluency. Cells were washed with PBS and treated with pHrodo-conjugated anti-CD320 or isotype control antibody (1 μg/ml), and cellular uptake was monitored every hour for 24 hours using a live-cell time-lapse fluorescence microscope (Incucyte SX5). The average total integrated orange intensity across technical duplicates was used to compare uptake dynamics between conditions.
Split-luciferase binding assay
Anti-CD320 autoantibodies were detected in healthy control sera using a HiBit bioluminescent protein detection platform. A construct containing the nucleotide sequence corresponding to the previously identified CD320 autoantibody epitope and an 11–amino acid HiBit tag was used to in vitro transcribe a labeled peptide. The labeled peptide was subsequently column-purified, immunoprecipitated with patient serum on Sephadex protein A/G beads, and co-incubated with a complementary LgBit peptide in the presence of luciferase substrate. Bioluminescence was measured using a Promega GloMAX plate reader.
Beacon optofluidic antibody isolation
In preparation for Beacon screening, two cryopreserved PBMC donor samples were thawed using Opto Memory B Discovery PBMC Thawing Kits (catalog no. 750-08254, Bruker Cellular Analysis), then subjected to enrichment of memory B cells using the Switched Memory B Cell Isolation Kit, Human (catalog no. 130-093-617, Miltenyi Biotec), and lastly activated for 5 days in culture using feeder cell-free medium and protocols included in the Opto Memory B Discovery Sample Prep Kit, Human (catalog no. 750-08323, Bruker Cellular Analysis). After 5 days of incubation, activated human B cells were loaded onto the Beacon system, automatically isolating tens of thousands of single-activated B cells into NanoPen chambers on four OptoSelect 20k chips (catalog no. 750-00019, Bruker Cellular Analysis).
After loading, activated human B cells were screened on-chip using a multiplexed fluorescent assay for secreted IgG identification and determination of antibody specificity. In-channel capture assay reagents were prepared using 6.25% anti-human IgG polystyrene particles (catalog no. HUP5-60-5, Spherotech), AF647 anti-human IgG (3 μg/ml; catalog no. 109-606-170, Jackson Immunoresearch), and of a soluble tetramer of biotinylated CD320 target peptide conjugated to BB515 streptavidin (1.5 μg/ml; catalog no. 564453, BD Bio-sciences). The CD320:streptavidin tetramer was prepared through three stepwise additions of BB515 streptavidin to 10 μg of biotin-CD320 at a 4:1 molar ratio over 30 min. Cell Analysis Suite (CAS) v3.1.1.134 software used machine learning to automatically score assay images and identify NanoPen chambers that contained B cells secreting CD320 peptide–specific antibodies. Users manually verified assay results using Image Analyzer v3.1.1.134 software before performing targeted cell export from the chips.
Single antigen-specific B cells were recovered into 96-well plates followed by off-chip cDNA synthesis and amplification using the Opto B Discovery cDNA Synthesis Kit (cat. no. 750–02030, Bruker Cellular Analysis). Paired antibody heavy/light chain sequences were amplified from the cDNA of recovered cells using the Opto B Discovery Sanger Prep Kit, Human (750-02041, Bruker Cellular Analysis), which includes a primer for conventional Sanger sequencing.
Biolayer interferometry
SAX 2.0 streptavidin biosensors (Sartorius) were loaded with 50 nM biotinylated CD320 peptide in Octet Kinetic Buffer containing 0.02% Tween 20 and 0.2% bovine serum albumin on an Octet RED384. Loading was terminated in the linear phase once a response of at least 0.5 nm was achieved. Loaded probes were then equilibrated in kinetic buffer before association/dissociation with 50 nM anti-CD320. Curves were normalized to reference wells, aligned to baseline, and processed using Savitzky-Golay filtering.
Cytotoxicity assay
HEK293T cells (20,000) were plated in each well of a 96-well plate in DMEM + penicillin G/streptomycin sulfate + 10% FBS. One day after plating, the medium was aspirated and replaced with serum-free DMEM and increasing concentrations of anti-CD320 or isotype control antibody (Bio X Cell, BE0297) and near-infrared (NIR) Incucyte Cytotox reagent (1:4000). Images were acquired every hour for 24 hours to assess antibody-dependent cytotoxicity.
iPSC-derived BBB model
Extended endothelial cell culture method (EECM) brain microvascular endothelial cell (BMEC)–like cells were differentiated as previously described with minor modifications (21). Briefly, hiPSCs [WiCell, iPS(IMR90)-4] were passaged at least twice but no more than 10 times after thawing and differentiated into endothelial progenitor cells (EPCs); CD31+ cells were purified by magnetic assisted cell sorting using an EasySep FITC Positive Selection Kit II (STEMCELL Technologies, #17682) and a fluorescein isothiocyanate (FITC)–conjugated human anti-CD31 antibody (Miltenyi Biotec, #130-117-390). At the end of the 8-day protocol, these purified EPCs were grown in human endothelial serum-free medium (Thermo Fisher Scientific, 11111–044) supplemented with B27 (Thermo Fisher Scientific, 17504044) and recombinant fibroblast growth factor 2 (Tocris, 233-FB-500) and selectively passaged by Accutase (500 ml; Sigma-Aldrich, #A6964) between three and five times to yield purified EECM-BMEC–like cells, at which time they were seeded onto collagen/fibronectin-coated Transwell inserts (Corning, #3401) at a density of 1 × 105 cells per apical chamber at day 0. Cells were maintained in this differentiation medium for 4 days with media changes every 2 days. TEER was measured every 24 hours from days 1 to 6 using an epithelial volt/ohm meter (EVOM3, WPI).
At day 4 after seeding, B12-deficient medium was prepared by incubating endothelial cell medium with anti-B12–conjugated magnetic beads (Molecular Depot, B2010667) at a dilution of 1:100 for 1 hour at 4°C with constant rotation. Medium was then placed on a magnetic stand, and supernatant devoid of magnetic beads was collected. Both apical and basolateral compartments were replaced with B12-depleted media on day 4. Isotype control antibody (Bio X Cell, BE0297) or anti-CD320 was added to the apical compartment of transwells at a concentration of 5 μg/ml in technical triplicate. Wells were pretreated with antibodies for 30 min at 37° before addition of 1 μg/ml of recombinant transcobalamin (R&D Systems, 7895-TC-050) precomplexed to 3× molar excess of vitamin B12. Medium from apical and basolateral compartments was sampled at 0, 12, 24, and 48 hours, concomitant with TEER measurements. Holotranscobalamin concentration was measured by ELISA (Tecan, AX53101) according to the manufacturer’s protocol, prediluting the sample 1:500 in H2O before 1:1 dilution in pretreatment (final dilution of 1:1000). The ratio of basolateral to apical holotranscobalamin was calculated and normalized to t = 0 hours.
Whole-genome CRISPRi screen
The 10 sgRNA-per-gene CRISPRi repression library (30) was synthesized, cloned, and infected into dCas9-KRAB K562 cells as previously described (31). Briefly, ~300 million K562 cells stably expressing EF1alpha-dCas9-KRAB were infected with the 10 guide per gene sgRNA library at a multiplicity of infection < 1. Infected cells underwent puromycin selection (1 μg/ml) for 5 days, after which point puromycin was removed and cells were resuspended in normal growth medium without puromycin. After selection, sgRNA infection was confirmed by flow cytometry, which indicated that >90% of cells expressed the mCherry reporter. Sufficient sgRNA library representation was confirmed by deep sequencing after selection. Cells were cultured and maintained at 1000× coverage for 1 week. Staining reagent was prepared by conjugating human transcobalamin II to a pH-sensitive fluorescent dye (pHrodo Deep Red iFL NHS ester) at a dye:protein molar ratio of 4:1. Free dye was removed using a 40 kDa MWCO Zeba Spin Desalting Column. pHrodo-conjugated transcobalamin was incubated with a 3× molar ratio of cyanocobalamin at RT for 1 hour to form pHrodo-holotranscobalamin. This complex (100 μg) was incubated with 300 million library cells per replicate (2×) in 150-ml medium at 37°C for 4 hours before sorting. Cells were then washed and prepared for fluorescence-activated cell sorting (FACS). Cells that internalized the top 5% (15 million cells) and bottom 5% (15 million cells) of phrodo-holoTC were sorted for downstream processing. Genomic DNA was extracted for all populations separately using a QIAGEN Blood Midi Kit. Deep sequencing of sgRNA sequences on an Illumina HiSeq was used to monitor library composition. Guide composition was analyzed and compared with the plasmid library and between conditions using casTLE (https://github.com/elifesciences-publications/dmorgens-castle) (34). Briefly, casTLE compares each set of gene-targeting guides with the negative controls, comprising nontargeting and nongenic (“safe-targeting”) sgRNAs. The enrichment of individual guides was calculated as the log ratio between high and low populations, and gene-level effects were calculated from 10 guides targeting each gene. P values were then calculated by permutating the targeting guides.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (v10), R, or Python. Two-sided Student’s t test was used for two group comparisons, one-way analysis of variance (ANOVA) with Tukey’s multiple hypothesis correction was used for three or more group comparisons, two-way ANOVA with Šidák multiple hypothesis correction was used for iPSC-BBB treatment × time point comparisons, and Fisher’s exact test was used for contingency analysis. Uncertainty is expressed as means ± 2 SD, mean ± 2 SE, or 95% confidence intervals. Proteome-wide phage display data were analyzed using edgeR with multiplicity correction using the Benjamini-Hochberg method. Genome-wide CRISPR screen data were analyzed using casTLE.
Supplementary Material
Acknowledgments:
We thank J. Chan for valuable experimental advice; M. Zia, A. Khazaei, S. Sidhu, and S. Vanasupa for sample processing; P. Chen and C. Gerungan for equipment and reagents; the ORIGINS/EPIC research team for patient recruitment and sample collection; and the research participants and their families for participation in this study.
Funding:
This work was supported by the NIMH (R01MH122471 to M.R.W., J.L.D., and S.J.P.), NINDS (U01NS120836, to M.R.W., S.J.P., J.M.G., A.J.G., and A.N. and R35NS111644 to S.L.H.), NCCDPHP (U01DP006701 to J.Y.), Department of Defense (W81XWH-21-1-0979 to J.Z., M.R.W., and S.J.P.), HDFCCC Laboratory for Cell Analysis Shared Resource Facility (NIH P30CA082103), National Multiple Sclerosis Society, Valhalla Foundation, and the Westridge Foundation (M.R.W. and A.J.G.).
Competing interests:
J.V.P., J.L.D., S.J.P., and M.R.W. are coinventors on a patent application related to this work (PCT/US2024/018105, “Compositions and methods related to transcobalamin receptor autoantibodies”). M.R.W. receives unrelated research grant funding from Roche/Genentech and Novartis; received speaking honoraria from Genentech, Takeda, WebMD, and Novartis; and is a founder and paid consultant for Delve Bio Inc. J.L.D. is a founder and paid consultant for Delve Bio Inc. and a paid consultant for the Public Health Company and Allen & Co. M.R.W. and J.L.D. receive licensing fees from CDI Laboratories. C.M.B. is a physician consultant for the Neuroimmune Foundation. J.M.G. receives research support from Hoffman LaRoche and Vigil Neurosciences for clinical trials and is a paid consultant for Arialys and Ventyx Bio. S.L.H. currently serves on the scientific advisory board of Accure, Alector, and Annexon; has previously consulted for BD, Moderna, NGM Bio, and Pheno Therapeutics; previously served on the board of directors of Neurona; and has received travel reimbursement and writing support from F. Hoffmann-La Roche and Novartis AG for anti-CD20 therapy–related meetings and presentations. A.J.G. is an Associate Editor for JAMA Neurology and received advisory board fees from Pipeline Therapeutics outside the submitted work. J.Y. has received grant support from Gilead, Aurinia, and BMS Foundation and performed consulting for Astra Zeneca, Pfizer, and UBC. The other authors declare that they have no competing interests.
Footnotes
Supplementary Materials
This PDF file includes:
Other Supplementary Material for this manuscript includes the following:
Data and materials availability:
All data associated with this study are present in the paper or the Supplementary Materials, and raw sequencing data have been deposited in Dryad (50).
REFERENCES AND NOTES
- 1.Green R, Allen LH, Bjørke-Monsen A-L, Brito A, Guéant J-L, Miller JW, Molloy AM, Nexo E, Stabler S, Toh B-H, Ueland PM, Yajnik C, Vitamin B12 deficiency. Nat. Rev. Dis. Primers 3, 17040 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Stabler SP, Vitamin B12 deficiency. N. Engl. J. Med 368, 149–160 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Hooshmand B, Appold F, Fissler P, Perneczky R, Otto M, Tumani H, Kivipelto M, von Arnim CAF, Markers of vitamin B12 status in relation to cerebrospinal fluid biomarkers of Alzheimer’s disease and cognitive performance. Ann. Neurol 94, 223–231 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Christine CW, Auinger P, Saleh N, Tian M, Bottiglieri T, Arning E, Tran NK, Ueland PM, Green R, Parkinson study group-DATATOP investigators, relationship of cerebrospinal fluid vitamin B12 status markers with Parkinson’s disease progression. Mov. Disord 35, 1466–1471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oki R, Izumi Y, Fujita K, Miyamoto R, Nodera H, Sato Y, Sakaguchi S, Nokihara H, Kanai K, Tsunemi T, Hattori N, Hatanaka Y, Sonoo M, Atsuta N, Sobue G, Shimizu T, Shibuya K, Ikeda K, Kano O, Nishinaka K, Kojima Y, Oda M, Komai K, Kikuchi H, Kohara N, Urushitani M, Nakayama Y, Ito H, Nagai M, Nishiyama K, Kuzume D, Shimohama S, Shimohata T, Abe K, Ishihara T, Onodera O, Isose S, Araki N, Morita M, Noda K, Toda T, Maruyama H, Furuya H, Teramukai S, Kagimura T, Noma K, Yanagawa H, Kuwabara S, Kaji R, Efficacy and safety of ultrahigh-dose methylcobalamin in early-stage amyotrophic lateral sclerosis. JAMA Neurol 79, 575–583 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen MJ, Rasmussen MR, Andersen CBF, Nexø E, Moestrup SK, Vitamin B12 transport from food to the body’s cells—a sophisticated, multistep pathway. Nat. Rev. Gastroenterol. Hepatol 9, 345–354 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Lai S-C, Nakayama Y, Sequeira JM, Wlodarczyk BJ, Cabrera RM, Finnell RH, Bottiglieri T, Quadros EV, The transcobalamin receptor knockout mouse: A model for vitamin B12 deficiency in the central nervous system. FASEB J 27, 2468–2475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora K, Sequeira JM, Hernández AI, Alarcon JM, Quadros EV, Behavioral alterations are associated with vitamin B12 deficiency in the transcobalamin receptor/CD320 KO mouse. PLOS ONE 12, e0177156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernàndez-Roig S, Lai S-C, Murphy MM, Fernandez-Ballart J, Quadros EV, Vitamin B12 deficiency in the brain leads to DNA hypomethylation in the TCblR/CD320 knockout mouse. Nutr. Metab 9, 41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quadros EV, Lai S-C, Nakayama Y, Sequeira JM, Hannibal L, Wang S, Jacobsen DW, Fedosov S, Wright E, Gallagher RC, Anastasio N, Watkins D, Rosenblatt DS, Positive newborn screen for methylmalonic aciduria identifies the first mutation in TCblR/CD320, the gene for cellular uptake of transcobalamin-bound vitamin B(12). Hum. Mutat 31, 924–929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindenbaum J, Healton EB, Savage DG, Brust JC, Garrett TJ, Podell ER, Marcell PD, Stabler SP, Allen RH, Neuropsychiatric disorders caused by cobalamin deficiency in the absence of anemia or macrocytosis. N. Engl. J. Med 318, 1720–1728 (1988). [DOI] [PubMed] [Google Scholar]
- 12.Larman HB, Zhao Z, Laserson U, Li MZ, Ciccia A, Gakidis MAM, Church GM, Kesari S, Leproust EM, Solimini NL, Elledge SJ, Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol 29, 535–541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandel-Brehm C, Dubey D, Kryzer TJ, O’Donovan BD, Tran B, Vazquez SE, Sample HA, Zorn KC, Khan LM, Bledsoe IO, McKeon A, Pleasure SJ, Lennon VA, DeRisi JL, Wilson MR, Pittock SJ, Kelch-like protein 11 antibodies in seminoma-associated paraneoplastic encephalitis. N. Engl. J. Med 381, 47–54 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun B, Ramberger M, O’Connor KC, Bashford-Rogers RJM, Irani SR, The B cell immunobiology that underlies CNS autoantibody-mediated diseases. Nat. Rev. Neurol 16, 481–492 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang W, Nakayama Y, Sequeira JM, Quadros EV, Mapping the functional domains of TCblR/CD320, the receptor for cellular uptake of transcobalamin-bound cobalamin. FASEB J 27, 2988–2994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alam A, Woo J-S, Schmitz J, Prinz B, Root K, Chen F, Bloch JS, Zenobi R, Locher KP, Structural basis of transcobalamin recognition by human CD320 receptor. Nat. Commun 7, 12100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Null N, Jones RC, Karkanias J, Krasnow MA, Pisco AO, Quake SR, Salzman J, Yosef N, Bulthaup B, Brown P, Harper W, Hemenez M, Ponnusamy R, Salehi A, Sanagavarapu BA, Spallino E, Aaron KA, Concepcion W, Gardner JM, Kelly B, Neidlinger N, Wang Z, Crasta S, Kolluru S, Morri M, Pisco AO, Tan SY, Travaglini KJ, Xu C, Alcántara-Hernández M, Almanzar N, Antony J, Beyersdorf B, Burhan D, Calcuttawala K, Carter MM, Chan CKF, Chang CA, Chang S, Colville A, Crasta S, Culver RN, Cvijović I, D’Amato G, Ezran C, Galdos FX, Gillich A, Goodyer WR, Hang Y, Hayashi A, Houshdaran S, Huang X, Irwin JC, Jang S, Juanico JV, Kershner AM, Kim S, Kiss B, Kolluru S, Kong W, Kumar ME, Kuo AH, Leylek R, Li B, Loeb GB, Lu W-J, Mantri S, Markovic M, McAlpine PL, de Morree A, Morri M, Mrouj K, Mukherjee S, Muser T, Neuhöfer P, Nguyen TD, Perez K, Phansalkar R, Pisco AO, Puluca N, Qi Z, Rao P, Raquer-McKay H, Schaum N, Scott B, Seddighzadeh B, Segal J, Sen S, Sikandar S, Spencer SP, Steffes LC, Subramaniam VR, Swarup A, Swift M, Travaglini KJ, Van Treuren W, Trimm E, Veizades S, Vijayakumar S, Vo KC, Vorperian SK, Wang W, Weinstein HNW, Winkler J, Wu TTH, Xie J, Yung AR, Zhang Y, Detweiler AM, Mekonen H, Neff NF, Sit RV, Tan M, Yan J, Bean GR, Charu V, Forgó E, Martin BA, Ozawa MG, Silva O, Tan SY, Toland A, Vemuri VNP, Afik S, Awayan K, Botvinnik OB, Byrne A, Chen M, Dehghannasiri R, Detweiler AM, Gayoso A, Granados AA, Li Q, Mahmoudabadi G, McGeever A, de Morree A, Olivieri JE, Park M, Pisco AO, Ravikumar N, Salzman J, Stanley G, Swift M, Tan M, Tan W, Tarashansky AJ, Vanheusden R, Vorperian SK, Wang P, Wang S, Xing G, Xu C, Yosef N, Alcántara-Hernández M, Antony J, Chan CKF, Chang CA, Colville A, Crasta S, Culver R, Dethlefsen L, Ezran C, Gillich A, Hang Y, Ho P-Y, Irwin JC, Jang S, Kershner AM, Kong W, Kumar ME, Kuo AH, Leylek R, Liu S, Loeb GB, Lu W-J, Maltzman JS, Metzger RJ, de Morree A, Neuhöfer P, Perez K, Phansalkar R, Qi Z, Rao P, Raquer-McKay H, Sasagawa K, Scott B, Sinha R, Song H, Spencer SP, Swarup A, Swift M, Travaglini KJ, Trimm E, Veizades S, Vijayakumar S, Wang B, Wang W, Winkler J, Xie J, Yung AR, Artandi SE, Beachy PA, Clarke MF, Giudice LC, Huang FW, Huang KC, Idoyaga J, Kim SK, Krasnow M, Kuo CS, Nguyen P, Quake SR, Rando TA, Red-Horse K, Reiter J, Relman DA, Sonnenburg JL, Wang B, Wu A, Wu SM, Wyss-Coray T, The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paik DT, Tian L, Williams IM, Rhee S, Zhang H, Liu C, Mishra R, Wu SM, Red-Horse K, Wu JC, Single-cell RNA sequencing unveils unique transcriptomic signatures of organ-specific endothelial cells. Circulation 142, 1848–1862 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quadros EV, Nakayama Y, Sequeira JM, The protein and the gene encoding the receptor for the cellular uptake of transcobalamin-bound cobalamin. Blood 113, 186–192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson CA, Vitamin B12 levels in the serum and cerebrospinal fluid in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 27, 174–177 (1964). [PMC free article] [PubMed] [Google Scholar]
- 21.Stabler SP, Allen RH, Barrett RE, Savage DG, Lindenbaum J, Cerebrospinal fluid methylmalonic acid levels in normal subjects and patients with cobalamin deficiency. Neurology 41, 1627–1632 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Nishihara H, Gastfriend BD, Kasap P, Palecek SP, Shusta EV, Engelhardt B, Differentiation of human pluripotent stem cells to brain microvascular endothelial cell-like cells suitable to study immune cell interactions. STAR Protoc 2, 100563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishihara H, Perriot S, Gastfriend BD, Steinfort M, Cibien C, Soldati S, Matsuo K, Guimbal S, Mathias A, Palecek SP, Shusta EV, Pasquier RD, Engelhardt B, Intrinsic blood-brain barrier dysfunction contributes to multiple sclerosis pathogenesis. Brain 145, 4334–4348 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nexo E, Hoffmann-Lücke E, Holotranscobalamin, a marker of vitamin B-12 status: Analytical aspects and clinical utility. Am. J. Clin. Nutr 94, 359S–365S (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanly JG, Diagnosis and management of neuropsychiatric SLE. Nat. Rev. Rheumatol 10, 338–347 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Jeltsch-David H, Muller S, Neuropsychiatric systemic lupus erythematosus: Pathogenesis and biomarkers. Nat. Rev. Neurol 10, 579–596 (2014). [DOI] [PubMed] [Google Scholar]
- 27.DeGiorgio LA, Konstantinov KN, Lee SC, Hardin JA, Volpe BT, Diamond B, A subset of lupus anti-DNA antibodies cross-reacts with the NR2 glutamate receptor in systemic lupus erythematosus. Nat. Med 7, 1189–1193 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Varley JA, Andersson M, Grant E, Berretta A, Zandi MS, Bondet V, Duffy D, Hunt D, Piehl F, Waters P, Irani SR, Absence of neuronal autoantibodies in neuropsychiatric systemic lupus erythematosus. Ann. Neurol 88, 1244–1250 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Gick GG, Arora K, Sequeira JM, Nakayama Y, Lai S-C, Quadros EV, Cellular uptake of vitamin B12: Role and fate of TCblR/CD320, the transcobalamin receptor. Exp. Cell Res 396, 112256 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, Weissman JS, Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, e19760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgens DW, Deans RM, Li A, Bassik MC, Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol 34, 634–636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgens DW, Wainberg M, Boyle EA, Ursu O, Araya CL, Tsui CK, Haney MS, Hess GT, Han K, Jeng EE, Li A, Snyder MP, Greenleaf WJ, Kundaje A, Bassik MC, Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat. Commun 8, 15178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilleron J, Zeigerer A, Endosomal trafficking in metabolic homeostasis and diseases. Nat. Rev. Endocrinol 19, 28–45 (2023). [DOI] [PubMed] [Google Scholar]
- 34.Zelcer N, Hong C, Boyadjian R, Tontonoz P, LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 325, 100–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carmel R, Subclinical cobalamin deficiency. Curr. Opin. Gastroenterol 28, 151–158 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Hughes EG, Peng X, Gleichman AJ, Lai M, Zhou L, Tsou R, Parsons TD, Lynch DR, Dalmau J, Balice-Gordon RJ, Cellular and synaptic mechanisms of anti-NMDA receptor encephalitis. J. Neurosci 30, 5866–5875 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikasova L, De Rossi P, Bouchet D, Georges F, Rogemond V, Didelot A, Meissirel C, Honnorat J, Groc L, Disrupted surface cross-talk between NMDA and Ephrin-B2 receptors in anti-NMDA encephalitis. Brain 135, 1606–1621 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Ramberger M, Berretta A, Tan JMM, Sun B, Michael S, Yeo T, Theorell J, Bashford-Rogers R, Paneva S, O’Dowd V, Dedi N, Topia S, Griffin R, Ramirez-Franco J, El Far O, Baulac S, Leite MI, Sen A, Jeans A, McMillan D, Marshall D, Anthony D, Lightwood D, Waters P, Irani SR, Distinctive binding properties of human monoclonal LGI1 autoantibodies determine pathogenic mechanisms. Brain 143, 1731–1745 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larman HB, Laserson U, Querol L, Verhaeghen K, Solimini NL, Xu GJ, Klarenbeek PL, Church GM, Hafler DA, Plenge RM, Nigrovic PA, De Jager PL, Weets I, Martens GA, O’Connor KC, Elledge SJ, PhIP-seq characterization of autoantibodies from patients with multiple sclerosis, type 1 diabetes and rheumatoid arthritis. J. Autoimmun 43, 1–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shome M, Chung Y, Chavan R, Park JG, Qiu J, LaBaer J, Serum autoantibodyome reveals that healthy individuals share common autoantibodies. Cell Rep 39, 110873 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreu-Sánchez S, Bourgonje AR, Vogl T, Kurilshikov A, Leviatan S, Ruiz-Moreno AJ, Hu S, Sinha T, Vich Vila A, Klompus S, Kalka IN, de Leeuw K, Arends S, Jonkers I, Withoff S, Brouwer E, Weinberger A, Wijmenga C, Segal E, Weersma RK, Fu J, Zhernakova A, Phage display sequencing reveals that genetic, environmental, and intrinsic factors influence variation of human antibody epitope repertoire. Immunity 56, 1376–1392.e8 (2023). [DOI] [PubMed] [Google Scholar]
- 42.Bodansky A, Yu DJ, Rallistan A, Kalaycioglu M, Boonyaratanakornkit J, Green DJ, Gauthier J, Turtle CJ, Zorn K, O’Donovan B, Mandel-Brehm C, Asaki J, Kortbawi H, Kung AF, Rackaityte E, Wang C-Y, Saxena A, de Dios K, Masi G, Nowak RJ, O’Connor KC, Li H, Diaz VE, Casaletto KB, Gontrum EQ, Chan B, Kramer JH, Wilson MR, Utz PJ, Hill JA, Jackson SW, Anderson MS, DeRisi JL, Unveiling the autoreactome: Proteome-wide immunological fingerprints reveal the promise of plasma cell depleting therapy. medRxiv 23300188 [Preprint] (2023); 10.1101/2023.12.19.23300188. [DOI] [Google Scholar]
- 43.Jaycox JR, Dai Y, Ring AM, Decoding the autoantibody reactome. Science 383, 705–707 (2024). [DOI] [PubMed] [Google Scholar]
- 44.Matsuda KM, Kotani H, Yamaguchi K, Okumura T, Fukuda E, Kono M, Hisamoto T, Kawanabe R, Norimatsu Y, Kuzumi A, Fukayama M, Fukasawa T, Ebata S, Yoshizaki-Ogawa A, Okamura T, Shoda H, Fujio K, Goshima N, Sato S, Yoshizaki A, Significance of anti-transcobalamin receptor antibodies in cutaneous arteritis revealed by proteome-wide autoantibody screening. J. Autoimmun 135, 102995 (2023). [DOI] [PubMed] [Google Scholar]
- 45.Hygum K, Lildballe DL, Greibe EH, Morkbak AL, Poulsen SS, Sorensen BS, Petersen TE, Nexo E, Mouse transcobalamin has features resembling both human transcobalamin and haptocorrin. PLOS ONE 6, e20638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramaekers VT, Rothenberg SP, Sequeira JM, Opladen T, Blau N, Quadros EV, Selhub J, Autoantibodies to folate receptors in the cerebral folate deficiency syndrome. N. Engl. J. Med 352, 1985–1991 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Jee P, Fernandez L, Perkins SL, Brooks SPJ, Effect of storage and repeated freeze/thaw on (S) vitamin B12. Clin. Biochem 47, 344 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Windelberg A, Arseth O, Kvalheim G, Ueland PM, Automated assay for the determination of methylmalonic acid, total homocysteine, and related amino acids in human serum or plasma by means of methylchloroformate derivatization and gas chromatography-mass spectrometry. Clin. Chem 51, 2103–2109 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Hvas A-M, Nexo E, Holotranscobalamin—A first choice assay for diagnosing early vitamin B12 deficiency? J. Intern. Med 257, 289–298 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Pluvinage J, Transcobalamin receptor antibodies in autoimmune vitamin B12 central deficiency, Dryad (2024); 10.5061/dryad.6djh9w18j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials, and raw sequencing data have been deposited in Dryad (50).


