Abstract
Background
Plasmodium falciparum merozoite surface proteins 1 (PfMSP1) and 2 (PfMSP2) are potential candidates for malaria vaccine development. However, the genetic diversity of these genes in the global P. falciparum population presents a significant challenge in developing an effective vaccine. Hence, understanding the genetic diversity and evolutionary trends in the global P. falciparum population is crucial.
Methods
This study analyzed the genetic variations and evolutionary changes of pfmsp1 and pfmsp2 in P. falciparum isolates from the Central Highland and South-Central regions of Vietnam. DNASTAR and MEGA7 programs were utilized for analyses. The polymorphic nature of global pfmsp1 and pfmsp2 was also investigated.
Results
A total of 337 sequences of pfmsp1 and 289 sequences of pfmsp2 were obtained. The pfmsp1 and pfmsp2 from Vietnam revealed a higher degree of genetic homogeneity compared to those from other malaria-endemic countries. Remarkably, the allele diversity patterns of Vietnam pfmsp1 and pfmsp2 differed significantly from those of neighboring countries in the Greater Mekong Subregion. Declines in allele diversity and polymorphic patterns of Vietnam pfmsp1 and pfmsp2 were observed.
Conclusions
The Vietnam P. falciparum population might be genetically isolated from the parasite populations in other neighboring GMS countries, likely due to geographical barriers and distinct evolutionary pressures. Furthermore, bottleneck effects or selective sweeps may have contributed to the genetic homogeneity of Vietnam pfmsp1 and pfmsp2.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-024-10116-6.
Keywords: Plasmodium falciparum, Merozoite surface proteins, Genetic diversity, Vietnam
Introduction
Malaria, an acute febrile disease caused by infections with Plasmodium parasites, continues to be a significant global health challenge. According to the World Malaria Report 2023 from the World Health Organization (WHO), there were 249 million global malaria cases, making an increase of 5 million from the previous year, and an estimated 608,000 deaths were reported in 2022 [1]. Despite achievements in malaria control within the Greater Mekong Subregion (GMS) over recent decades, a surge in indigenous malaria cases from 90,082 to 170,527 was observed between 2021 and 2022. Specifically, the number of P. falciparum cases almost doubled during this period, with instances rising from 16,490 to 30,789 [1]. The rapid spread of antimalarial drug-resistant P. falciparum strains in the GMS poses a significant challenge to the region’s efforts to eliminate falciparum malaria [2, 3]. Although notable reductions in malaria morbidity and mortality have occurred in Vietnam over the last two decades [4], the disease remains endemic in the central and southern provinces, particularly in the Central Highland region where P. falciparum is the predominant species [5]. A total of 3,200 malaria cases were reported in Vietnam in 2019, among which 3,110 were P. falciparum cases [1]. The malaria incidences in Vietnam have decreased sharply since 2019, but the epidemic has continued with fluctuating annual cases thereafter. Understanding the genetic makeup and evolutionary trends of the P. falciparum population in certain endemic areas is crucial as it provides valuable insights to the epidemiological patterns and genetic characteristics of the parasites.
P. falciparum merozoite surface proteins (PfMSPs) are multigene family proteins that are expressed on the surfaces of merozoites and sporozoites of the parasite and play critical roles in the invasion of the parasite into host cells [6]. Over 10 distinct genes encoding PfMSPs have been identified in P. falciparum, among which pfmsp1 and pfmsp2 are the most intensively investigated due to their reliability as polymorphic markers. PfMSP1 is a transmembrane protein divided into 17 distinct blocks, with block II displaying the most significant polymorphic character [7, 8]. PfMSP1 is typically classified into three allelic variants, K1, MAD20, and RO33, based on the polymorphisms observed in block II [9, 10]. The K1 and MAD20 types contain different tripeptide repeat sequences, and the variations in the repeat sequences and different numbers of repeats generate genetic polymorphisms within each of these types. While, RO33 has quite different sequences lacking typical repeat sequences. PfMSP2 consists of a central repeat region (CRR) flanked by conserved N-terminal and C-terminal regions and is classified into two distinct variants, 3D7 and FC27, depending on the sequence variations in the CRR [11, 12]. In particular, high levels of size polymorphisms are identified in the two regions; R1 corresponding to the GSA-rich repeat units and R2 flanking the poly-threonine stretch. Due to these polymorphisms in pfmsp1 and pfmsp2, they are recognized as reliable polymorphic markers for studying genetic heterogenicity and evolutionary aspects of the P. falciparum population [13, 14].
Comprehensive studies analyzing the genetic nature of pfmsp genes in P. falciparum isolates from countries in the GMS have been conducted [15–19]. These studies indicated substantial genetic heterogeneities of pfmsp genes in the P. falciparum population in the GMS. The genetic characteristics of pfmsp genes in Vietnam P. falciparum have been partially characterized [20–22]. Similar to the pfmsp genes from other GMS countries, those in Vietnam P. falciparum also appear to exhibit considerable genetic diversity. However, these prior studies were limited by their reliance on simple typing instead of sequencing analysis and by being outdated, suggesting they may not accurately reflect the current genetic composition of the genes in Vietnam P. falciparum population. In this study, we explored the genetic diversities of pfmsp1 and pfmsp2 in Vietnam P. falciparum isolates collected in the Central Highland and South-Central regions of Vietnam between 2018 and 2022. We also comparatively analyzed the genetic differences among pfmsp1 and pfmsp2 populations from Vietnam and other malaria-endemic countries, including GMS countries.
Materials and methods
Parasite samples and study area
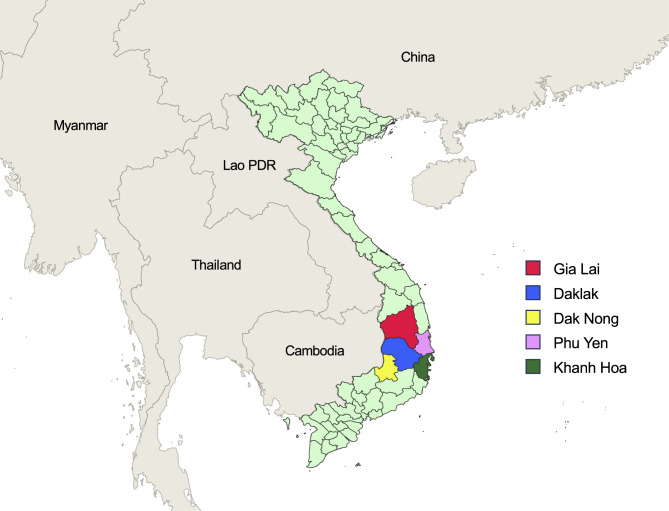
A total of 382 blood samples were collected from P. falciparum-infected individuals residing in three provinces in the Central Highlands (Dak Lak, Dak Nong, and Gia Lai) and two provinces in the South-Central regions (Phu Yen and Khanh Hoa), Vietnam, during 2018–2022 (Fig. 1). P. falciparum infection was diagnosed by microscopic analysis of thin and thick blood smears. The dried blood filter (DBF) was prepared from each patient by finger-prick method and P. falciparum infection was verified by a species-specific polymerase chain reaction (PCR) targeting the 18S ribosomal RNA (rRNA) gene as reported previously [5, 23]. The study protocol was reviewed and approved by the Ethics Committee of the Ministry of Health, Institute of Malariology, Parasitology and Entomology Quy Nhon, Vietnam (IRB Approval numbers: 386/VSR-LSDT, 45/VSR-NCDT, and 637/VSR-NCDT).
Fig. 1.
Map of blood collection areas. The blood samples used in this study were collected from P. falciparum-infected patients residing in five provinces of Vietnam from 2018 to 2022
Amplifications and sequence analyses of Vietnam pfmsp genes
Parasite DNA was isolated from each DBF using the QIAamp DNA Blood Kit (Qiagen, Hilden, Germany). Nested PCRs to amplify the pfmsp1 block II and pfmsp2 block III were performed using the primer sets and protocols described in previous studies [15, 17, 24]. Each PCR product was cloned into a T&A cloning vector (Real Biotech Corporation, Banqiao City, Taiwan) and transformed into Escherichia coli DH5α competent cells. The nucleotide sequences of the cloned gene were analyzed using the automated Sanger method with M13 forward and reverse primers. The nucleotide sequences of Vietnam pfmsp genes have been deposited in the GenBank with the accession numbers: pfmsp1 (OR425413–OR425789) and pfmsp2 (OR425790–OR426078).
Genetic diversity analysis
The nucleotide and inferred amino acid sequences of Vietnam pfmsp genes were analyzed using EditSeq and SeqMan in the DNASTAR package (DNASTAR, Madison, WI, USA). Sequences were analyzed based on the reference sequences: MAD20 (GenBank No.: X05624.2), K1 (GenBank No.: NC_004330.2), and RO33 (GenBank No.: M55001.1) for pfmsp1; 3D7 (GenBank No.: X53832) and FC27 (GenBank No.: J03828) for pfmsp2. The pfmsp sequences obtained in this study were also comparatively analyzed with the previously reported pfmsp sequences from Vietnam and other countries including the GMS countries (Supplemental File 1: Table S1).
Results
Genetic diversity of Vietnam pfmsp1 block II
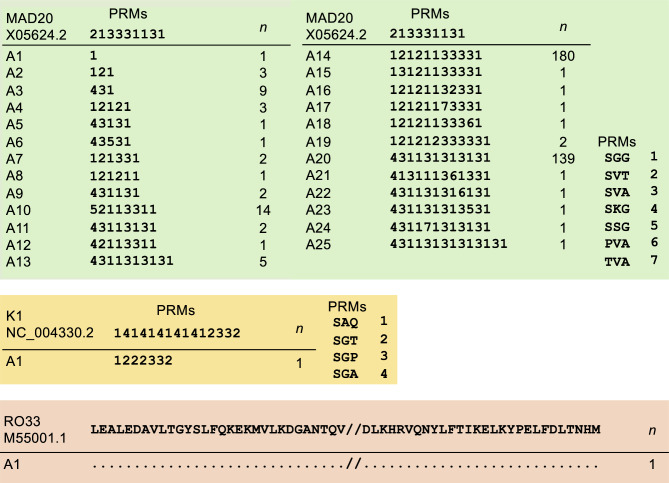
A total of 377 pfmsp1 block II sequences were successfully obtained from Vietnam P. falciparum, with MAD20 alleles being overwhelmingly predominant (375/377, 99.47%). Each of the K1 and RO33 alleles was identified only once, respectively (Fig. 2). Block II of the MAD20 alleles comprised various combinations and arrangements of seven distinct peptide repeat motifs (PRMs) of SGG, SVT, SVA, SKG, SSG, PVA, and TVA, yielding a total of 25 unique alleles of MAD20 (A1–A25). None of the allele shared the same PRM configuration with the reference sequence (X05624.2). The construction of each MAD20 allele varied, containing PRMs ranging from 1 to 14. The two alleles, A14 and A20, were the most prevalent, accounting for 180 and 139 sequences, respectively. Block II of the K1 allele also demonstrated genetic variation relative to the reference sequence (NC_004330.2) and contained only three PRMs: SAQ, SGT, and SGP. In contrast, block II of the RO33 allele was identical in sequence to the reference (M55001.1).
Fig. 2.
Polymorphic patterns in Vietnam pfmsp1 block II. A total of 27 different alleles were found in Vietnam pfmsp1 block II, including 25 alleles for MAD20 type, 1 allele for K1 type, and 1 allele for RO33 type. Vietnam MAD20 block II consisted of seven different PRMs, including SGG, SVT, SVA, SKG, SSG, PVA, and TVA. Meanwhile, Vietnam K1 block II included three distinct PRMs: SAQ, SGT, and SGP. Vietnam RO33 block II had sequences identical to the reference sequence. The total number of isolates for each allele is indicated
Genetic differences of pfmsp1 block II in the global population
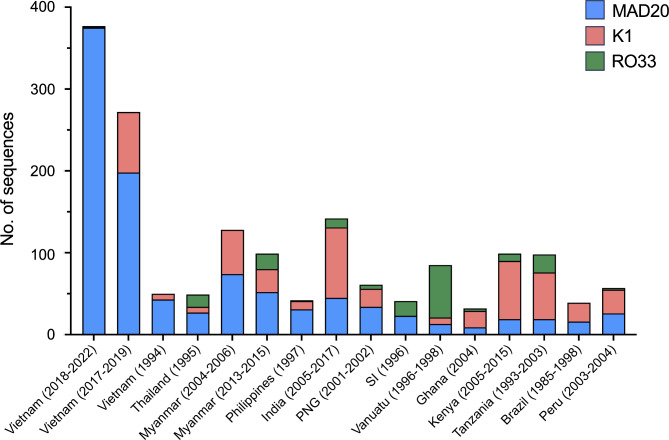
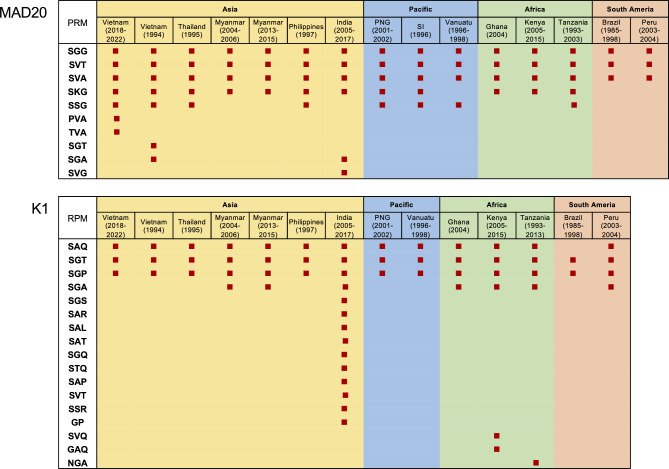
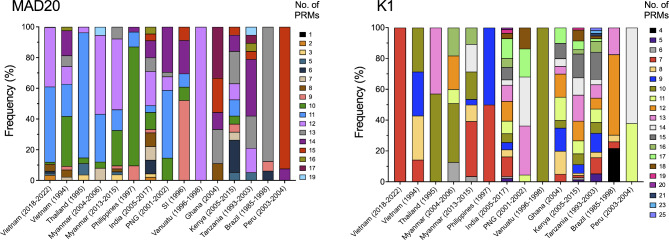
The allelic diversity of Vietnam pfmsp1 block II was compared with those of other countries and previously reported Vietnam pfmsp1 (Fig. 3). The MAD20 alleles were predominant across all Vietnam pfmsp1 populations, a trend similarly observed in pfmsp1 populations of GMS countries, including Thailand and Myanmar, as well as Pacific countries such as the Philippines, PNG, and SI. Conversely, K1 alleles were primarily found in pfmsp1 populations from India, Africa, and South American countries. Notably, the RO33 allele was predominant in the Vanuatu pfmsp1 population. The global pfmsp1 population displayed significant genetic heterogeneity in MAD20 and K1 alleles, attributed to various compositions and arrangements of PRMs. For MAD20 alleles, 10 different PRMs including SGG, SVT, SVA, SKG, SSG, PVA, TVA, SGT, SGA, and SVG were identified globally (Fig. 4). These PRMs were unequally distributed, with SGG, SVT, and SVA universally present across all populations. Additionally, SKG and SSG appeared in Asia, the Pacific, and Africa, but were absent in South American populations. The five PRMs, namely PVA, TVA, SGT, SGA, and SVG, were uniquely identified in either Vietnam or Indian populations, with PVA and TVA being novel PRMs first detected in the Vietnam MAD20 alleles analyzed in this study. A similar variation in PRM distribution was found in global K1 alleles (Fig. 4). A total of 17 distinct PRMs were detected in global K1 alleles. Among these, SGT and SGP were consistently observed across all populations, while SAG was present in K1 alleles of all countries analyzed except Brazil. SGA occurred in some countries across Asia, Africa, and South America, but not in the Pacific region. The remaining 13 PRMs showed unique occurrence in specific countries such as India, Kenya, or Tanzania. These PRMs contributed to various combinations and arrangements within global MAD20 and K1 alleles, resulting in significant size variations and genetic heterogeneity of pfmsp1 by country (Fig. 5). Worldwide, MAD20 alleles varied immensely, containing 1 to 19 PRMs, with prevalent sizes ranging from 9 to 15 PRMs in the global MAD20 population. In comparison, global K1 alleles exhibited even greater size diversity due to different compositions of PRM compositions, ranging from 4 to 25. Each country displayed variability in the number of PRMs present, with pronounce size variations of PRMs in both MAD20 and K1 alleles notably in pfmsp1 from India and African countries.
Fig. 3.
Allelic diversity of the global pfmsp1 block II. Different proportions of three pfmsp1 types including MAD20, K1, and RO33 were observed in the global pfmsp1 populations
Fig. 4.
PRM profiles of the global pfmsp1 block II. Differences in PRM types in the global MAD20 and K1 populations. MAD20 block II comprised 10 different PRMs, while K1 block II included 17 distinct PRMs
Fig. 5.
Size polymorphism patterns of the global pfmsp1 block II. Size differences in the global pfmsp1 block II were caused by varying numbers of PRMs. The global MAD20 block II exhibited 17 different sizes with PRM counts ranging from 1 to 19. The global K1 block II displayed 20 different size polymorphisms resulting from distinct PRMs counts that ranged from 4 to 25
Genetic diversity of Vietnam pfmsp2 block III
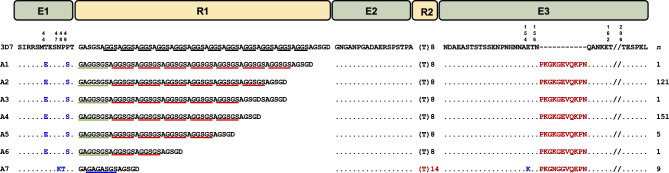
A total of 289 Vietnam pfmsp2 block III sequences were successfully obtained from the samples analyzed in this study. These sequences were categorized into 3D7 types. They displayed polymorphic characters, forming 7 distinct alleles (A1–A7) distinguished by sequence polymorphisms (Fig. 6). The A4 and A2 alleles were predominant, accounting for 52.2% (151/289) and 41.9% (121/289), respectively. In the E1 region, only four amino acid changes (T44E, N47K, P48T, and P49S) were observed, categorized into two groups of paired amino acid substitutions. Specifically, the T44E/P49S pair was present in 280 sequences (96.9%), while the N47K/P48T pair was found in 9 sequences (3.1%). Different numbers, types, and arrangements of PRMs were observed in the R1 region, contributing to the size polymorphisms of Vietnam pfmsp2 block III. GAGGSGSA, GGSGSA, and GAGASGSA served the fundamental units of PRMs. In the R2 region, all sequences exhibited poly-threonine (poly-T) signature characteristic of the 3D7 type. Alleles A1–A6 contained 8 threonines (T8), while A7 featured 14 threonines (T14). Compared to the 3D7 reference sequence, a notable characteristic in the E3 region of all Vietnam pfmsp2 was the insertion of 11 amino acids at position 156: PKGKGEVQKPN for A1–A6 alleles and PKGNGGVQEPN for the A7 allele. Additionally, an amino acid change of E154K was identified in the A7 allele.
Fig. 6.
Polymorphic patterns in Vietnam pfmsp2 block III. In Vietnam, seven distinct alleles of the 3D7 types (A1–A7) were identified in the pfmsp2 population
Genetic differences of pfmsp2 block III in the global population
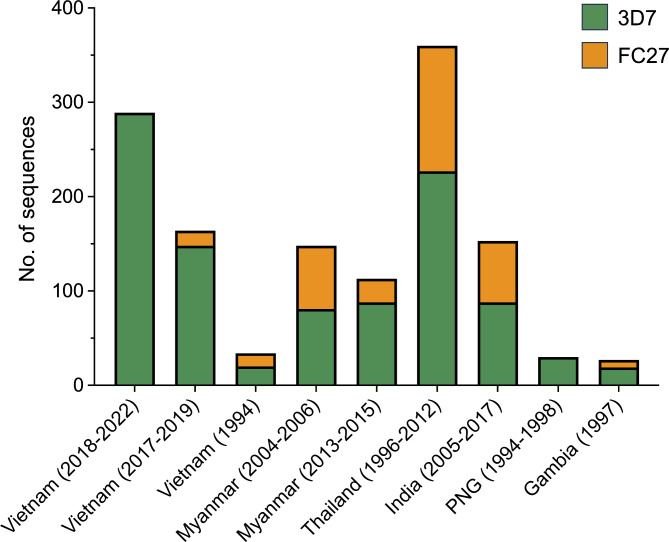
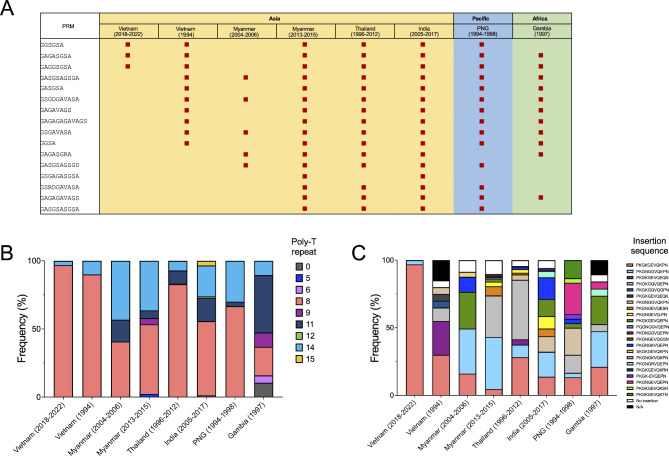
The distribution of alleles in the current Vietnam pfmsp2 was compared to pfmsp2 from Vietnam (1994 and 2017‒2019) and other countries including Myanmar, Thailand, India, PNG, and Gambia (Fig. 7). All populations exhibited the presence of both allelic types of 3D7 and FC27 in a substantial proportion except the current Vietnam and PNG. The previously reported Vietnam pfmsp2 (Vietnam 1994 and 2017‒2019) harbored both 3D7 and FC27, though they shared overlapping collection sites with the present study. Significant genetic diversity was observed in the global pfmsp2 block III. The regions of R1, R2, and E3 were the primary contributors to genetic diversity among and between pfmsp2 populations (Fig. 8). The R1 region exhibited exceptionally diverse polymorphisms, characterized by 16 different types of PRM with unequal arrangements and repetitions (Fig. 8A). Three Asian pfmsp2 populations, Myanmar (2013‒2015), Thailand, and India, possessed all PRM types. The PNG and Gambia populations also included 14 and 11 different PRM types, respectively. Notably, two Vietnam pfmsp2 populations exhibited distinct patterns. While the Vietnam pfmsp2 analyzed in this study displayed only 3 PRM types, Vietnam (1994) pfmsp2 comprised 10 different PRM types. In the R2 region, only two types of poly-T (T8 and T14) were identified in the Vietnam pfmsp2 population, whereas a variety of poly-T types were observed in the global population (Fig. 8B). The prevalence of poly-T types varied globally, with T8 and T14 being the dominant species. Intriguingly, T8 was markedly prevalent in the Vietnam pfmsp2 population. The E3 region of pfmsp2 also demonstrated substantial genetic diversity in the global population. A range of insertion types with diverse sequences was evident in the global pfmsp2 population (Fig. 8C). The PKGKGEVQKPN insertion appeared in all populations, albeit at varying frequencies by country. Additionally, pfmsp2 sequences lacking insertion were present in the global population, excluding PNG.
Fig. 7.
Allelic diversity of the global pfmsp2 block III. In global populations, different proportions of two pfmsp2 subtypes, namely 3D7 and FC27, were evident
Fig. 8.
Comparison of polymorphism patterns in the R1, R2, and E3 regions of the global pfmsp2 3D7 type. (A) Distinct PRM profiles in the R1 region. (B) Varied poly-T patterns in the R2 region. (C) Diverse insertion sequences in the E3 region. N/A, not available for analysis due to the sequence missing corresponding to the region
Discussion
PfMSPs are key contenders in the development of vaccines against falciparum malaria; however, the extensive genetic diversity of these genes within the global P. falciparum population poses significant challenges to vaccine development. In our study, we examined the genetic polymorphisms of two pfmsp genes, pfmsp1 and pfmsp2, in P. falciparum isolates from Vietnam and compared their genetic diversities with those from other countries, including those in the GMS.
It has been previously suggested that pfmsp1 block II and pfmsp2 block III exhibit high levels of polymorphism [13, 14]. In contrast, our analysis revealed a marked genetic uniformity in the Vietnam pfmsp1 and pfmsp2 genes, with the dominant presence of MAD20 alleles in pfmsp1 and 3D7 alleles in pfmsp2. This uniformity sets the Vietnamese isolates apart from their counterparts in neighboring GMS countries like Myanmar and Thailand. Notably, when compared with earlier analyses of the Vietnam pfmsp1 and pfmsp2 populations (2017‒2019) [22], a significant reduction in allelic variation was noted in recent populations, despite their shared geographical origins. No mixed infections of pfmsp1 and pfmsp2 were identified in the P. falciparum population analyzed in this study. However, in our previous study on the genetic makeup analysis of Vietnam pfmsp1, mixed infections with different alleles were detected [5], albeit at low frequencies. This discrepancy may be attributed to the preferred amplification and cloning of the major alleles in mixed infection samples, if present. Nevertheless, our study revealed that Vietnam pfmsp1 and pfmsp2 exhibited a higher degree of genetic homogeneity compared to those from other malaria-endemic countries. PRMs are significant factors in inducing genetic complexity in Vietnam pfmsp1 and pfmsp2. Common PRMs identified in other global pfmsp1 and pfmsp2 populations were also found in Vietnam pfmsp1 and pfmsp2 as major PRMs. However, two novel PRMs (PVA, and TVA) that not been reported in global pfmsp1 were also discovered in Vietnam pfmsp1, though their prevalence was low. The most notable features in Vietnam pfmsp2 included the low diversity of PRM types in the R1 region and the exceedingly high frequency of a single type of insertion (PKGKGEVQKPN, 96.9%) in the E3 region. Compared to previous Vietnam sequences (1994) that exhibited more than five different insertion in the E3 region, only two types of insertion sequence (PKGKGEVQKPN and PKGNGGVQEPN) were identified in the Vietnam pfmsp2 analyzed in this study. Malaria prevalence and transmission intensity, particularly P. falciparum, have been sharply declined in the endemic areas of Vietnam in recent few years [1, 4], The recent decrease of overall genetic diversity in Vietnam pfmsp1 and pfmsp2 populations could be attributed to declined transmission intensity in the country, limiting genetic exchange and recombination in the population.
Differences in genetic diversity of other vaccine candidates between Vietnam and other GMS countries have previously been reported. The pattern of restricted genetic diversity in Vietnam’s P. falciparum is not confined solely to MSP family genes but also extends to other genes. In Vietnam P. falciparum, pfama1 and pfeba-175 demonstrated notably lower genetic diversity and a unique genetic profile compared to other GMS countries, such as Myanmar and Thailand [25, 26]. This phenomenon was not exclusive to P. falciparum genes. P. vivax circumsporozoite surface protein (pvcsp) of Vietnam also exhibited a distinct allelic pattern relative to pvcsp from other GMS countries [27]. The genetic distinctiveness of Vietnam’s Plasmodium populations compared to those of other GMS countries could be attributed to geographic feature. The Truong Son Range (Annamite Range), stretching approximately 1,100 km across the north and south of western Vietnam, may act as a topographical barrier that impedes the transmission of parasites and mosquito vectors between western Vietnam and eastern Laos and Cambodia [26]. Foehn effect across the Range also renders dissimilar environmental conditions in the western and the eastern sides of the Range, resulting in different transmission settings that can influence transmission patterns and genetic dynamics of the parasites. Furthermore, this barrier could also restrict the migratory patterns of the human population, limiting transmission. Due to these environmental factors, the Plasmodium population in Central Vietnam could be undergoing unique evolutionary changes, independent from those in the neighboring GMS countries, resulting in the distinctive genetic lineage. However, this study still presents limitations. The samples from GMS countries were collected at varying time points, which challenges the distinction of precise genetic variations in contemporary P. falciparum populations. To better understand the genetic dynamics of Plasmodium species in this region, comprehensive examinations with larger-scale samples and more systematic methodologies are required, especially in Laos and Cambodia, which border Vietnam.
The results of this study also indicated a decrease the sequence diversity of Vietnam’s pfmsp1 and pfmsp2 compared to earlier periods, potentially due to population bottlenecks in the parasite population. The recent decrease in malaria cases in this endemic region, attributed to sustained control efforts, may lead to a bottleneck effect, significantly reducing genetic polymorphism [28]. Conversely, the reduction in polymorphism at these sites could result from a selective sweep, where newly developed directional selection favors certain alleles over others [29].
Conclusion
Vietnam pfmsp1 and pfmsp2 exhibit less genetic diversity than those from other malaria-endemic countries, including the GMS countries. Vietnam P. falciparum population is likely genetically isolated from the parasite populations in other neighboring GMS countries, possibly due to geographical barriers and unique evolutionary pressures. The bottleneck effect or selective sweep may contribute to the genetic homogeneity of Vietnam pfmsp1 and pfmsp2, which warrants further investigation. This study provides a comprehensive understanding of the genetic characteristics and population structure of the Vietnam P. falciparum population, but a more comprehensive analysis of the genetic diversity and evolutionary aspects of the population is also necessary.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the staff in the Tropical Diseases Clinical and Treatment ResearchDepartment, Institute of Malariology, Parasitology, and Entomology, Quy Nhon and the healthprofessionals for their contribution and technical support in field study. This research was supported by a National Research Foundation of Korea (NRF) grants from the Government of Korea (NRF-2023M3A9H506175721 and NRF-2024M3A9H5043141).
Abbreviations
- CRR
Central repeat region
- DBF
Dried blood filter
- GMS
Greater Mekong Subregion
- MSP
Merozoite surface protein
- P. falciparum
Plasmodium falciparum
- PfMSP1
P. falciparum merozoite surface protein 1
- PfMSP2
P. falciparum merozoite surface protein 2
- PCR
Polymerase chain reaction
- PRM
Peptide repeat motif
- poly-T
Poly-threonine
- rRNA
ribosomal RNA
- WHO
World Health Organization
Author contributions
TCV, HHQ, and BKN conceptualize the study. TCV, HGL, JMK, TMTN, and HHQ performed experiment or contributed in sample collection. TCV, HGL, JMK, HHQ, and BKN analyzed and interpreted the data. HHQ and BKN designed and supervised the experiments. TCV and BKN wrote the draft of the manuscript. HGL, JMK, TMTN, and HHQ reviewed the manuscript. All authors have agreed to the published version of the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by a National Research Foundation of Korea (NRF) grants from.
the Government of Korea (NRF-2023M3A9H506175721 and NRF-2024M3A9H5043141).
Data availability
Sequence data that support the findings of this study have been deposited in the GenBank with the accession numbers of pfmsp1 (OR425413–OR425789) and pfmsp2 (OR425790–OR426078).
Declarations
Ethics approval and consent to participate
The study protocol was reviewed and approved by the Ethics Committee of the Ministry of Health, Institute of Malariology, Parasitology and Entomology Quy Nhon, Vietnam (IRB Approval numbers: 386/VSR-LSDT, 45/VSR-NCDT, and 637/VSR-NCDT). Verbal informed consent was obtained from all the participants in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huynh Hong Quang, Email: huynhquangimpe@yahoo.com.
Byoung-Kuk Na, Email: bkna@gnu.ac.kr.
References
- 1.WHO. World Malaria Report. 2023. Word Malaria report Geneva: World Health Organization. 2023. Licence: CC. 2023.
- 2.Menard D, Dondorp A. <ArticleTitle Language=“En”>Antimalarial drug resistance: a threat to malaria elimination. Cold Spring Harbor Perspect Med. 2017;7:a025619. 10.1101/cshperspect.a025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lê HG, Naw H, Kang J-M, Võ TC, Myint MK, Htun ZT et al. Molecular Profiles of Multiple Antimalarial Drug Resistance Markers in Plasmodium falciparum and Plasmodium vivax in the Mandalay Region, Myanmar. Microorganisms. 2022;10:2021. [DOI] [PMC free article] [PubMed]
- 4.Goldlust SM, Thuan PD, Giang DDH, Thang ND, Thwaites GE, Farrar J, et al. The decline of malaria in Vietnam, 1991–2014. Malar J. 2018;17:226. 10.1186/s12936-018-2372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Võ TC, Lê HG, Kang JM, Naw H, Fan CK, Trinh NTM, et al. Molecular surveillance of malaria in the Central Highlands, Vietnam. Parasitol Int. 2021;83:102374. 10.1016/j.parint.2021.102374. [DOI] [PubMed] [Google Scholar]
- 6.Cowman AF, Baldi DL, Healer J, Mills KE, O’Donnell RA, Reed MB, et al. Functional analysis of proteins involved in Plasmodium falciparum merozoite invasion of red blood cells. FEBS Lett. 2000;476:84–8. [DOI] [PubMed] [Google Scholar]
- 7.Cooper JA. Merozoite surface antigen-I of Plasmodium. Parasitol Today. 1993;9:50–4. [DOI] [PubMed] [Google Scholar]
- 8.Miller LH, Roberts T, Shahabuddin M, McCutchan TF. Analysis of sequence diversity in the Plasmodium falciparum merozoite surface protein-1 (MSP-1). Mol Biochem Parasitol. 1993;59:1–14. [DOI] [PubMed] [Google Scholar]
- 9.Tanabe K, Mackay M, Goman M, Scaife JG. Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J Mol Biol. 1987;195:273–87. [DOI] [PubMed] [Google Scholar]
- 10.Ferreira MU, Kaneko O, Kimura M, Liu Q, Kawamoto F, Tanabe K. Allelic Diversity at the Merozoite Surface Protein-1 (MSP-1) Locus in Natural Plasmodium falciparum Populations: A Brief Overview. Mem Inst Oswaldo Cruz. 1998;93:631–8. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira MU, Hartl DL. Plasmodium falciparum: worldwide sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-2 (MSP-2). Exp Parasitol. 2007;115:32–40. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann EH, da Silveira LA, Tonhosolo R, Pereira FJ, Ribeiro WL, Tonon AP, et al. Geographical patterns of allelic diversity in the Plasmodium falciparum malaria-vaccine candidate, merozoite surface protein-2. Ann Trop Med Parasitol. 2001;95:117–32. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira MU, Ribeiro WL, Tonon AP, Kawamoto F, Rich SM. Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene. 2003;304:65–75. [DOI] [PubMed] [Google Scholar]
- 14.Weisman S, Wang L, Billman-Jacobe H, Nhan DH, Richie TL, Coppel RL. Antibody responses to infections with strains of Plasmodium falciparum expressing diverse forms of merozoite surface protein 2. Infect Immun. 2001;69:959–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang JM, Moon SU, Kim JY, Cho SH, Lin K, Sohn WM, et al. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum field isolates from Myanmar. Malar J. 2010;9:131. 10.1186/1475-2875-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thái TL, Jun H, Lee J, Kang J-M, Lê HG, Lin K, et al. Genetic diversity of merozoite surface protein-1 C-terminal 42 kDa of Plasmodium falciparum (PfMSP-142) may be greater than previously known in global isolates. Parasit Vectors. 2018;11:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lê HG, Kang JM, Jun H, Lee J, Thái TL, Myint MK, et al. Changing pattern of the genetic diversities of Plasmodium falciparum merozoite surface protein-1 and merozoite surface protein-2 in Myanmar isolates. Malar J. 2019;18:241. 10.1186/s12936-019-2879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanabe K, Mita T, Palacpac NMQ, Arisue N, Tougan T, Kawai S, et al. Within-population genetic diversity of Plasmodium falciparum vaccine candidate antigens reveals geographic distance from a Central sub-Saharan African origin. Vaccine. 2013;31:1334–9. 10.1016/j.vaccine.2012.12.039. [DOI] [PubMed] [Google Scholar]
- 19.Jongwutiwes S, Putaporntip C, Hughes AL. Bottleneck effects on vaccine-candidate antigen diversity of malaria parasites in Thailand. Vaccine. 2010;28:3112–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira MU, Liu Q, Zhou M, Kimura M, Kaneko O, Thien HV, et al. Stable patterns of allelic diversity at the Merozoite Surface Protein-1 locus of Plasmodium falciparum in clinical isolates from Southern Vietnam. J Eukaryot Microbiol. 1998;45:131–6. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann EHE, Ribolla PEM, Ferreira MU. Genetic relatedness of Plasmodium falciparum isolates and the origin of allelic diversity at the merozoite surface protein-1 (MSP-1) locus in Brazil and Vietnam. Malar J. 2003;2:24. 10.1186/1475-2875-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long BV, Allen G, Brauny M, Linh LTK, Pallerla SR, Huyen TTT, et al. Molecular surveillance and temporal monitoring of malaria parasites in focal Vietnamese provinces. Malar J. 2020;19:458. 10.1186/s12936-020-03561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang JM, Cho PY, Moe M, Lee J, Jun H, Lee HW, et al. Comparison of the diagnostic performance of microscopic examination with nested polymerase chain reaction for optimum malaria diagnosis in Upper Myanmar. Malar J. 2017;16:119. 10.1186/s12936-017-1765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aubouy A, Migot-Nabias F, Deloron P. Polymorphism in two merozoite surface proteins of Plasmodium falciparum isolates from Gabon. Malar J. 2003;2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang JM, Lê HG, Võ TC, Naw H, Yoo WG, Sohn WM, et al. Genetic polymorphism and natural selection of apical membrane antigen-1 in Plasmodium falciparum isolates from Vietnam. Genes. 2021;12:1903. 10.3390/genes12121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Võ TC, Lê HG, Kang J-M, Naw H, Yoo WG, Myint MK, et al. Genetic polymorphism and natural selection of the erythrocyte binding antigen 175 region II in Plasmodium falciparum populations from Myanmar and Vietnam. Sci Rep. 2023;13:20025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Võ TC, Trinh NTM, Lê HG, Kang J-M, Yoo WG, Quang HH, et al. Genetic Diversity of Circumsporozoite Surface Protein of Plasmodium vivax from the Central Highlands, Vietnam. Pathogens. 2022;11:1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. [DOI] [PubMed] [Google Scholar]
- 29.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 2007;89:391–403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the GenBank with the accession numbers of pfmsp1 (OR425413–OR425789) and pfmsp2 (OR425790–OR426078).