Abstract
Background
Hepatitis B virus (HBV) infection is a persistent global public health problem, and curing for chronic hepatitis B (CHB) through the application of existing antiviral drugs is beset by numerous challenges. The viral protein HBx is a critical regulatory factor in the life cycle of HBV. Targeting HBx is a promising possibility for the development of novel therapeutic strategies.
Methods
The Nano-Glo® HiBiT Lysis Detection System was used to screen the herbal monomer compound library for compounds that inhibit HBx expression. Western blotting was used to examine proteins expression. Southern blotting or Northern blotting were used to detect HBV DNA or HBV RNA. ELISA was performed to detect the HBsAg level. The effect of asiatic acid on HBV in vivo was investigated by using recombinant cccDNA mouse model.
Results
Asiatic acid, an extract of Centella asiatica, significantly reduced the HBx level. Mechanistic studies demonstrated that asiatic acid may promote the degradation of HBx in an autophagy pathway-dependent manner. Subsequently, asiatic acid was found to reduce the amount of HBx bound to covalently closed circular DNA (cccDNA) microchromosomes, and repressive chromatin modifications then occurred, ultimately inhibiting cccDNA transcriptional activity. Moreover, in HBV-infected cells and a mouse model of persistent HBV infection, asiatic acid exhibited potent anti-HBV activity, as evidenced by decreased levels of HBV RNAs, HBV DNA and HBsAg.
Conclusions
Asiatic acid was identified as a compound that targets HBx, revealing its potential for application as an anti-HBV agent.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12985-024-02535-3.
Keywords: Asiatic acid, HBx, Hepatitis B virus
Introduction
Hepatitis B virus (HBV) infection can lead to numerous serious health complications, including hepatitis, cirrhosis, and hepatocellular carcinoma [1, 2]. It is estimated that 257.5 million people worldwide were infected with HBV in 2022, with a global prevalence of HBV of 3.2% [3]. Current clinical treatments including interferon (IFN) and nucleoside analogs (NAs), can effectively inhibit HBV replication, but cannot completely clear HBV [4]. The key obstacle is that cccDNA is difficult to eradicate. It is stable, with a long half-life, and can be replenished through new cycles of infection and through intracellular amplification pathways. As the template for the transcription of all HBV RNAs, cccDNA persists in the nucleus in liver cells as minichromosomes [5]. The transcription of HBV cccDNA is regulated by multiple factors, including transcription factors, viral proteins, and epigenetic modifications. Therefore, targeting viral proteins or host factors may reduce its transcriptional activity [6].
The viral protein HBx is a multifunctional regulatory factor that plays a crucial role in regulating cccDNA transcriptional activity both in vivo and in vitro [7]. HBx, promotes the binding of histone acetyltransferases (HATs) to cccDNA while reducing the interaction of histone deacetylases (HDACs) with cccDNA, resulting in increased acetylation of cccDNA-bound histones and increased cccDNA transcriptional activity [8–10]. In the absence of HBx, transcriptional repressors such as SETDB1, PRMT1, and Spindlin1 bind to cccDNA, leading to increases in cccDNA-bound histone H3 lysine 9 di- and trimethylation (H3K9me2/3) and a decrease in H3K4me3, therefore causing a decrease in cccDNA transcriptional activity [11–13]. The deletion of HBx also contributes to the recruitment of the heterochromatin protein HP1 to cccDNA, which leads to the formation of a more compact chromatin structure and impedes transcription [13]. Considering the critical role of HBx in cccDNA transcription, therapeutic strategies targeting HBx may contribute to controlling chronic HBV infection.
Traditional Chinese medicines (TCMs), as alternative and complementary medicines, have been widely used in clinical practice in China. They are easily accessible, easily absorbed and metabolized by the human body, and have low cytotoxicity [14]. Epigallocatechin-3-gallate (EGCG) has been demonstrated to inhibit HBV replication by increasing lysosomal acidification and inhibiting HBV-induced incomplete autophagy [15]. In addition, Sophora flavescens polysaccharides were found to markedly inhibit the secretion of HBsAg in HepG2.2.15 cells [16]. Furthermore, saikosaponin C (SSc) can inhibit the synthesis of HBV pregenomic RNA (pgRNA) by attenuating the expression of HNF1α and HNF4α, thereby exerting anti-HBV effects [17]. Studies have demonstrated that TCMs and the related active ingredients have considerable potential for the treatment of HBV. Therefore, we screened hepatoprotective formulas and traditional Chinese herbs from the Chinese Pharmacopoeia and Compendium of Materia Medica, and constructed a compound library containing 715 types of Chinese herbal monomers. Preliminary findings using the Nano-Glo® HiBiT Lysis Detection System indicated that asiatic acid (AA) could reduce the HBx protein level. Asiatic acid is a pentacyclic triterpenoid isolated from the traditional Chinese medicinal herb Centella asiatica. Evidence suggests that asiatic acid can alleviate liver fibrosis, mitigate liver damage, and exhibit significant hepatoprotective effects [18]. However, the role of asiatic acid in HBV has not been reported.
In this study, we found that asiatic acid may promote the degradation of HBx through the autophagy–lysosomal pathway, which in turn reduces the binding of HBx to cccDNA microchromosomes, leading to a decrease in the level of an activating histone modification (H3K4me3) and increases in the levels of inhibitory modifications (H3K9me3 and H3K27me3) on cccDNA-bound histones, and ultimately affecting cccDNA transcriptional activity. Our study provides new insight into the development of novel antiviral strategies for inhibiting cccDNA transcriptional activity.
Materials and methods
Cell culture and antibodies
HepG2-NTCP and HepAD38 cells were kindly donated by Prof. Ningshao Xia (Xiamen University, China), Primary human hepatocytes (PHHs) were obtained from Liver Biotechnology (China), and Huh-7 cells were obtained from Health Science Research Resource Blank. HepG2-NTCP and Huh-7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin. HepAD38 cells were cultured in DMEM medium containing 10% foetal bovine serum, 100 U/mL penicillin and 100 µg/mL streptomycin, with the addition of 400 µg/mL G418 (Merck) to the medium. PHHs were cultured in Williams E medium supplemented with 5 µg/mL transferrin, 10 ng/mL epidermal growth factor (EGF), 3 µg/mL insulin, 2 mM L-glutamine, 18 µg/mL hydrocortisone, 40 ng/mL dexamethasone, 5 ng/mL sodium selenite, 2% dimethyl sulfoxide (DMSO), 100 U/mL penicillin, and 100 µg/mL streptomycin. All cell lines were cultured in a 37ºC cell culture incubator with a 5% CO2 concentration.
Rabbit anti-HBsAg (NB100-62652) was purchased from Novus (USA). Rabbit anti-LC3B (3868 S), Mouse anti-SQSTM1/p62 (88588 S), Rabbit anti-Flag (#14793S) and Rabbit anti-HA (#3724) were purchased from Cell Signaling Technology (USA). Rabbit anti-HBx (RD981038100) was purchased from BioVendor (USA). Rabbit anti-β-actin monoclonal antibody (sc-1616-R) was obtained from Santa Cruz Biotechnology (USA). Rabbit anti-H3K9me3 polyclonal antibody (17–625), rabbit anti-H3K27me3 polyclonal antibody (17–622), mouse anti-H3K4me3 monoclonal antibody (17–678) were obtained from Merck Millpore (Germany).
Nano-Glo® HiBiT lysis assay
To screen for small molecules targeting HBx, we used the Nano-Glo® HiBiT Lytic system (N3030, Promega, Wisconsin, USA) as described in detail previously [19]. In brief, after transfection of HiBiT-HBx plasmid into cells, the cells were treated with different herbal monomer compounds for 2 days. After washing the cells with PBS, 200 µL of Nano-Glo® HiBiT Lytic Buffer was added to each well to lyse the cells. The lysate was transferred to a new 1.5 mL Ep tube. After centrifugation, 10 µL of Nano-Glo® HiBiT Lytic Substrate was added to 50 µL of clarified lysate to measure luciferase activity. The measured luminescence signal was proportional to the amount of HiBiT-tagged HBx in the cell lysate.
MTT assay
(3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was used to test cell viability. Cells were seeded into 96-well plates, and drug concentrations from 0 µM to 200 µM were added to the plates when the cell density reached 80%. After 72 h of incubation, 5 mg/mL MTT was added to each well, and the plates were incubated for 4 h in the dark. Then, 100 µL of DMSO was added to each well to dissolve the crystals, and a microplate reader was used to measure the half-maximal cytotoxic concentration (CC50).
Alamar blue assay
Various cell lines were seeded in twelve-well culture plates at appropriate densities. The cells were treated with different concentrations of asiatic acid. After the indicated time, 1× Alamar Blue was added to the cells, which were then incubated at 37 °C for 4 h. Absorbance values were detected using an enzyme labeling instrument with the excitation and emission wavelengths set to 560 nm and 590 nm, respectively.
Viruses and infection
HBV virus was collected from the culture supernatant of HepAD38 cells and concentrated with 5% PEG8000. Virus diluted with infection medium was mixed with 5% PEG8000 and added to the well plates. The following day, the medium was discarded and the cells were washed twice with PBS for further experiments.
RNA extraction and realtime PCR
TRNzol Reagent (Tiangen, China) was used to extract RNA. After removing genomic DNA (gDNA), the RNA was reverse transcribed to complementary DNA (cDNA) by using the FastKing RT Kit (KR116-02, Tiangen, China). Then, cDNA was amplified using Fast Start Universal SYBR Green Master (Roche, Switzerland). The fluorescence signals were analyzed by the 2−ΔΔCt method to determine the relative mRNA expression levels. β-actin mRNA was used as an internal reference. The specific primers used are listed in Table S1.
Western blotting
Western blotting was used to analyze proteins expression. First, proteins were extracted from cells by using RIPA with protease inhibitor. Then, the BCA protein assay kit was employed to determine the protein concentration, and the protein content in the lysate was calculated based on a standard curve. Subsequently, 30 µg extracted proteins were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to the polyvinylidene fluoride (PVDF) membranes. Next, the membranes were blocked with 5% non-fat dry milk for 2 h and incubated overnight at 4 °C with diluted primary antibodies. The membranes were then washed and incubated with secondary antibodies at room temperature for 2 h. Finally, the proteins were visualized using enhanced chemiluminescence (ECL) reagents, with β-actin serving as a reference control.
Cycloheximide (CHX) chase assay
HBV-infected HepG2-NTCP cells were treated with 20 µM asiatic acid and then incubated with 10 µg/mL CHX for 1, 2, 3 h. Then, the cells were collected for protein extraction. The level of HBx was measured by Western blotting.
Immunofluorescence staining
Cells were seeded on slides and treated according to the requirements of specific experiments. The cells were fixed with 4% paraformaldehyde for 20 min, permeabilized at room temperature with 0.3% Triton X-100, and blocked with 5% BSA for 1 h. The cells were then incubated with anti-LC3B antibodies for 1 h, followed by staining with fluorochrome conjugated secondary antibodies. The nuclei were counterstained using 4′,6-diamidino-2-phenylindole (DAPI), and images were captured using a confocal laser scanning microscope (Leica DMi8, Germany).
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed following the manufacturer’s protocol (17-10086, Merck Millipore, Darmstadt, Germany). Cells cultured in 10-cm dishes were harvested by scraping in cold PBS and centrifuged at 800 × g and 4 °C for 5 min. Cell lysis buffer was used to resuspend the cells. After centrifugation, the nuclei were fixed with 1% formaldehyde at room temperature for 10 min to cross-link DNA with proteins. Then, the pellets were resuspended in nuclear lysis buffer and DNA was fragmented by sonication. The supernatants were diluted with dilution buffer at a 1:10 ratio and 1% of the dilution was used as the input. The remaining diluted chromatin was added with the antibody targeting the protein and 20 µL of protein A/G magnetic beads (17-10086, Merck Millipore, Germany) at 4 °C overnight. On the following day, the supernatant was removed and the beads were washed sequentially with 500 µL each of Low Salt Immune Complex Wash Buffer, High Salt Immune Complex Wash Buffer, LiCl Immune Complex Wash Buffer, and TE Buffer for 5 min each time. The beads were resuspended using 100 µL of ChIP Elution Buffer containing Proteinase K and incubated at 62 °C for 2 h, followed by further incubation at 95 °C for 10 min. Immunoprecipitated chromatin was extracted and purified by using phenolchloroform, isopropanol and ethanol, and analyzed by Taq-man probe PCR. The specific HBV cccDNA primer and probe used are listed in Table S1.
HBV core DNA extraction and quantitation
Cells were treated with the lysate (10 mM Tris-HCl, 1 mM EDTA, 0.5% NP-40, 2% sucrose, pH 8.0) and incubated for 15 min at 37 °C. After centrifugation, 10 mM MgCl2 and 40 U/mL DNase were added to the supernatant of the collected lysate, followed by incubating for 4 h at 37 °C. The HBV core capsids were precipitated with 5% PEG8000. Then, the precipitate was incubated with proteinase K at 45 °C overnight. Finally, HBV core DNA was extracted with phenol-chloroform. Fast Start Universal SYBR Green Master Mix was used for quantitative analysis with the pCH9/3091 plasmid as standard. The specific HBV core DNA primers used are listed in Table S1.
Hirt cccDNA extraction and detection
Cells were lysed in 500 µL cell lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 150 mM NaCl, 1% SDS) at 37 °C for 20 min. Then the lysate solution was transferred to a new tube, 125 µL KCl (2.5 M) was added to the cell lysate, and the solution was incubated overnight at 4 °C. The next day, after centrifugation at 4℃ and 12,000 × g for 30 min, the supernatant was collected. Then 500 µL of phenolchloroform was added with repeated mixing by oscillation. After centrifugation at 16,000 × g for 3 min, the supernatant was collected. An equal volume of isopropanol (approximately 500 µL) and 1 µL of glycogen were added to the supernatant. After centrifugation at 16,000 × g for 30 min at 4 °C, the supernatant was discarded and added 1 mL of 75% ethanol to wash the precipitate. Then, the DNA precipitation was dried at 37 °C in a metal bath, and 20 µL ddH2O was added to dissolve the precipitate overnight. HBV cccDNA expression levels were detected by Taq-man probe PCR. The specific HBVcccDNA primers used are listed in Table S1.
Southern blotting/northern blotting
The DIG-High Prime DNA Labeling and Detection Starter Kit or DIG Northern Starter Kit was used to analyze HBV DNA or total HBV RNA, respectively. DNA and RNA samples were separated on a 1% agarose gel. DNA or RNA in the gel was transferred to a nylon membrane by siphoning. Then DNA or RNA was immobilized on the membrane by using UV cross-linking, and hybridized with labeled HBV DNA or RNA probes overnight at 42 °C (DNA) or 68 °C (RNA). The membrane was washed with washing buffer to remove unbound probes and was then incubated with anti-digoxin antibody. Finally, X-ray film was used to visualize the results.
Enzyme-linked immunosorbent assay (ELISA)
Commercial enzyme linked immunosorbent assay kit (KHB, China) was used to measure the HBsAg in supernatant. Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) concentrations in mouse serum were measured using a commercially enzyme assay (Wanleibio, China) according to the manufacturer’s protocol.
Mouse model
A precursor cccDNA (prcccDNA)-based model of persistent HBV infection was used to validate the inhibitory effect of asiatic acid on HBV. The 4 µg prcccDNA plasmid and 4 µg Cre recombinase plasmid were injected into mice by hydrodynamic injection. After one week, all the mice were divided into the following four groups: vehicle group, ETV group (0.02 mg/kg), AA (30 mg/kg) and AA + ETV, with six mice per group. The mice were treated with asiatic acid or saline every two days, and serum was collected every four days. After 24 days, the mice were sacrificed to harvest liver tissue. All animal experiments were approved by the Animal Ethics Committee of Chongqing Medical University.
Immunohistochemistry
Liver tissues from the mouse model were used to prepare paraffin sections. After dewaxing, tissue antigens were retrieved by microwaving in sodium citrate buffer (10 mmol/L, pH 6.0). Then, the sections were permeated with 0.5% Triton X-100 (Sigma, USA) and the endogenous peroxidase was sequestered. And goat serum was added for blocking. Next, the specimens were incubated with primary antibody (anti-HBs working solution, ZM-0122, Zhongshan Jinqiao Biological Technology) overnight at 4 °C and were then incubated with a secondary antibody. Finally, the sections were stained with DAB and counterstained with hematoxylin.
Statistical analysis
The data analyses and related statistical graphs in this study were performed using GraphPad Prism 8.0 software. The significance of the results was analyzed using the Mann-Whitney U test. Each experiment was conducted at least three times, and the results were expressed as the mean ± standard deviation. Differences were considered statistically significant when P < 0.05.
Results
Asiatic acid reduced HBx expression
To identify herbal monomers that target the HBx protein, a library of Chinese medicinal monomers was constructed and screened by the process shown in the flow chart (Fig. 1A). HiBiT-HBx was previously constructed and applied to screen for candidate molecules targeting HBx via the HiBiT lytic detection system [19]. In the first round of screening, the HiBiT system was used to identify compounds that depressed the level of HiBiT-HBx. Eight compounds, namely, isoscoparin, 6’-O-β-D-glucosylgentiopicroside, caffeic acid, pregnenolone, b-D-Glucopyranoside, forsythoside A, asiatic acid and forsythoside E, decreased HiBiT-HBx level by at least 50% of the baseline value (Fig. 1B). To determine whether these eight compounds also inhibit HBV transcription, HBV-infected HepG2-NTCP cells were incubated with each compound. The intracellular levels of cccDNA transcription products, including total HBV RNAs and HBV 3.5-kb RNA, were measured. Three of the compounds de-creased the intracellular level of HBV RNAs. Interestingly, asiatic acid (AA) treatment diminished the intracellular levels of HBV RNAs in a dose-dependent manner (Fig. 1C-D). Therefore, asiatic acid was selected for further study.
Fig. 1.
Screening process for compounds targeting HBx. (A) Schematic depiction of screening small-molecule compounds targeting HBx; (B) The inhibitory effect of 715 natural compounds on HBx; (C,D) HepG2-NTCP cells were inoculated with 1000 vge of HBV particles, subsequently subjected to treatment with 8 candidate compounds at concentrations of 10 and 30 µM. Total HBV RNAs and HBV 3.5-kb RNA were quantified by real-time PCR. The β-actin mRNA level was used as an internal control; (E,F) HepG2-NTCP cells transfected with 3xFlag and 2xHA-tagged HBx plasmids were treated with asiatic acid at different concentrations (10, 20 and 30 µM) or ETV (100 nM). The level of the corresponding protein was examined by western blotting analysis. The mRNA levels of HBx were quantified by real-time PCR using specific primers. The β-actin mRNA level was used as an internal control. Representative data are from at least three independent experiments. Data are shown as mean ± SD, *P < 0.05; **P < 0.01
To validate the effect of asiatic acid on the HBx level, the cytotoxicity of asiatic acid in several cell lines was tested by using MTT assay and Alamar blue assay. The CC50 of asiatic acid in all cell lines was greater than 50 µM (Figure S1A). The Alamar blue experiment showed that asiatic acid had no effect on HepG2-NTCP cell activity at concentrations of less than 30 µM (Figure S1B). Therefore, 10, 20 and 30 µM asiatic acid were used to treat HepG2-NTCP cells transfected with 3×Flag- and 2×HA-tagged HBx plasmids. Western blot assay demonstrated that asiatic acid dose-dependently repressed the HBx protein level (Fig. 1E). The validation on Huh-7 cells got similar result (Figure S1C). Meanwhile, the realtime PCR results proved that asiatic acid treatment did not cause a significant change in the 3×Flag- or 2×HA-tagged HBx mRNA level (Fig. 1F). Thus, asiatic acid did not affect HBx mRNA level to bring down its protein level. In addition, asiatic acid did not affect the levels of other viral proteins, including HBs, HBc and HBp (Figure S1D). The above results suggested that asiatic acid specifically promoted the degradation of the HBx protein.
Asiatic acid may promote HBx degradation through the autophagy-lysosomal pathway
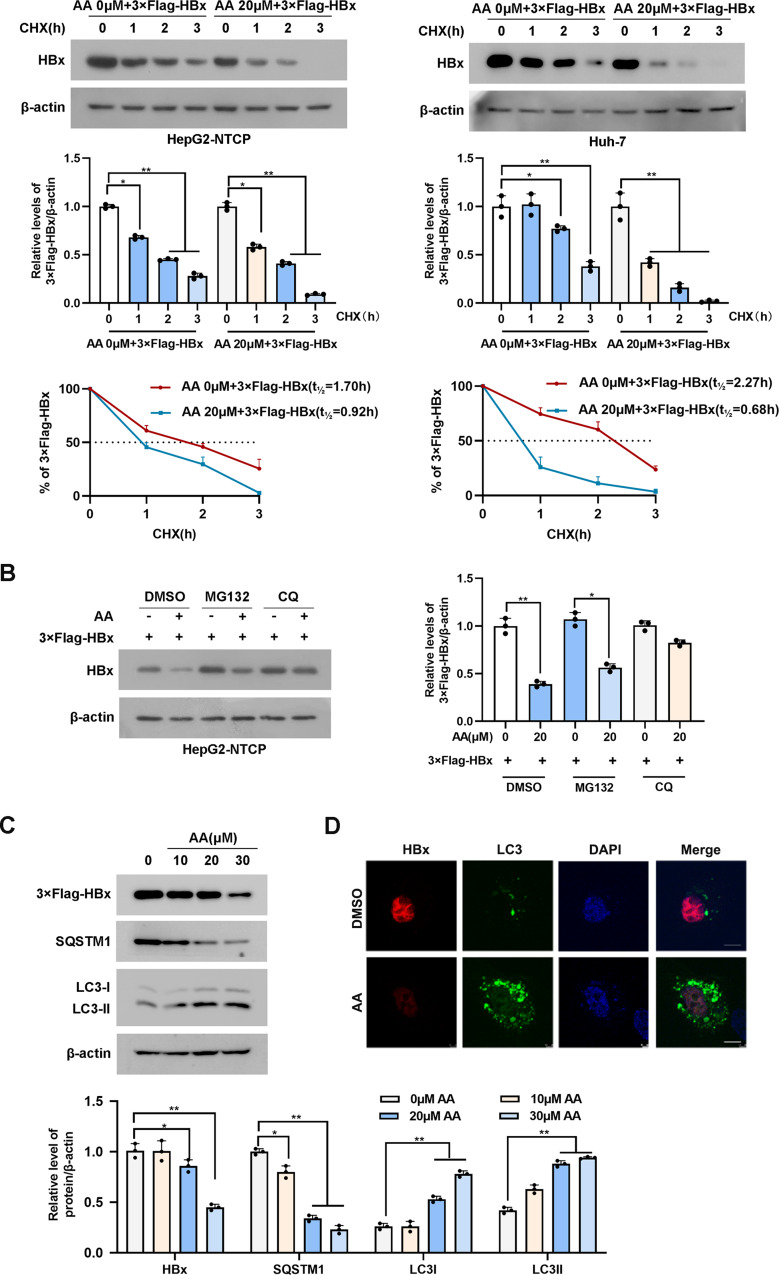
To elucidate the molecular mechanism by which asiatic acid reduces the HBx protein level, CHX Chase Assay was used to detect changes in the half-life of the HBx protein. The degradation of the HBx protein in both HepG2-NTCP and Huh-7 cells treated with asiatic acid was notably accelerated (Fig. 2A). There are two main pathways for protein degradation: the ubiquitin-proteasome pathway and the autophagy-lysosomal pathway. HepG2-NTCP cells were incubated with the proteasome inhibitor MG132 or the autophagy pathway inhibitor chloroquine (CQ). Western blot assay demonstrated that when HepG2-NTCP were exposed to MG132, the HBx protein level still sharply decreased in response to asiatic acid treatment. In contrast, in the presence of CQ, the HBx protein level was less affected by asiatic acid (Fig. 2B). To minimize the undesired consequences of chloroquine’s off-target effects, HepG2-NTCP cells were first transfected with either HBx or siATG5, a key regulatory gene instrumental in autophagosome assembly. Following this, the cells were treated with 20 µM of asiatic acid. Consistent with our expectations, the silencing of ATG5 significantly attenuated the suppressive effect of asiatic acid on HBx protein levels (Figure S2). In addition, asiatic acid treatment increased the expression of LC3-II (a marker of autophagosome formation and maturation) and markably decreased the expression of SQSTM1 (a cargo for autophagic degradation) (Fig. 2C). Immunofluorescence assay further confirmed that asiatic acid increased the number of LC3 puncta and inhibited HBx expression (Fig. 2D). These results showed that asiatic acid may promote HBx degradation through the autophagy pathway.
Fig. 2.
Asiatic acid decreased HBx stability. (A) HepG2-NTCP or Huh-7 cells transfected with 3xFlag-HBx plasmids were exposed to 20 µM asiatic acid, and then subjected to 10 µg/ml cycloheximide at the indicated times. HBx was examined by western blotting analysis using anti-Flag antibody; (B) HepG2-NTCP cells transfected 3×Flag-HBx plasmid were exposed to 20 µM asiatic acid, and then subjected to ubiquitin-proteasome inhibitor MG132 (10 µM) or autophagy pathway inhibitor CQ (50 µM) treatment for 8 h before the cells were harvested. The levels of the proteins were examined by western blotting analysis, and β-actin was used as the loading control; (C) SQSTM1, LC3-I, LC3-II (autophagy-related markers) were detected by western blotting assay. (D) Huh-7 cells were transfected with GFP-LC3 and subjected to 20 µM asiatic acid, the expression level of HBx and LC3 puncta were visualized by immunofluorescent staining. Scale bar = 10 μm. Representative data are from at least three independent experiments. Data are shown as mean ± SD, *P < 0.05; **P < 0.01
Asiatic acid reduced HBx level and then regulated HBV cccDNA minichromosome through epigenetic modifications
HBx can epigenetically modify cccDNA and thereby activate cccDNA transcription. Given the regulatory effect of HBx on cccDNA transcription, we investigated the effect of asiatic acid on cccDNA transcriptional activity. Asiatic acid reduced the level of the endogenous HBx protein in HBV-infected HepG2-NTCP cells (Figure S3A). Moreover, the level of HBV cccDNA did not significantly change, but the levels of total HBV RNAs and HBV 3.5 kb RNA were markedly decreased. (Fig. 3A-B, Figure S3B). More importantly, we calculated the total HBV RNAs/cccDNA and HBV 3.5-kb RNA/cccDNA ratios and found that asiatic acid inhibited the transcriptional activity of cccDNA (Fig. 3C). HBx can bind to cccDNA and regulate the levels of various histone modifications on cccDNA microchromosomes. Therefore, the ChIP assay was performed in HBV-infected HepG2-NTCP cells. The results indicated that asiatic acid treatment decreased the amount of HBx bound to cccDNA microchromosomes (Fig. 3D). Furthermore, asiatic acid treatment attenuated the enrichment of a transcriptional activation marker, H3K4me3, but notably increased the enrichment of two transcriptional repressive markers, H3K9me3 and H3K27me3, on cccDNA microchromosomes (Fig. 3E-G). Moreover, it was observed that chloroquine treatment could attenuate the effect of asiatic acid on HBV transcription and HBx binding to cccDNA as well as cccDNA minichromosome epigenetic modification. These results reveal that the degradation of HBx mediated by the autophagy pathway leads to suppressive epigenetic regulation of cccDNA minichromosome.
Fig. 3.
Asiatic acid affected cccDNA epigenetic modifications. (A-C) HepG2-NTCP cells were incubated with asiatic acid (20µM), following with or without CQ (50µM). The levels of total HBV RNAs and HBV 3.5-kb RNA were detected by real-time PCR using specific primers. The β-actin mRNA level was used as an internal control. The ratios of total HBV RNAs/cccDNA and HBV 3.5-kb RNA/cccDNA were calculated as indicators of cccDNA transcriptional activity; (D) The level of HBx associated with cccDNA in HBV-infected HepG2‐NTCP cells was examined by ChIP-qPCR assay. Cross-linked chromatin was immunoprecipitated with specific anti-HBx or anti-IgG antibody, followed by PCR quantification of HBV cccDNA using specific primers, with the host genes GAPDH and MYH6 used as controls; (E-G) The levels of H3K9me3, H3K27me3 and H3K4me3 associated with cccDNA, GAPDH or MYH6 promoter were analysed by ChlP assay with anti-H3K9me3, anti-H3K27me3, anti-H3K4me3 and anti-IgG, respectively. Representative data are from at least three independent experiments. Data are shown as mean ± SD, *P < 0.05; **P < 0.01
Evaluation of the antiviral effect of asiatic acid in HBV-infected cellular model
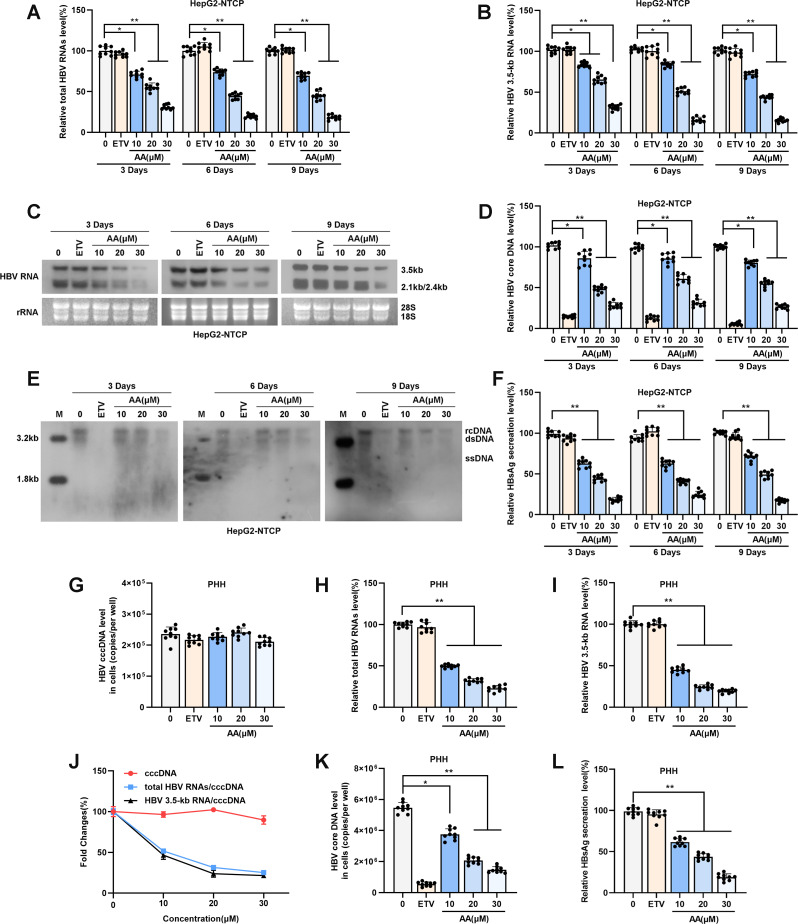
To further evaluate the antiviral effect of asiatic acid in vitro, HBV-infected HepG2-NTCP cells were incubated with 10 µM, 20 µM and 30 µM asiatic acid for 3, 6, or 9 days, after which the cells and culture supernatants were collected. Realtime PCR and Northern blot assay verified that asiatic acid treatment decreased the intracellular HBV RNAs level (Fig. 4A-C). Asiatic acid treatment also decreased the amount of HBV core DNA detected by real-time PCR and Southern blotting (Fig. 4D-E). In addition, asiatic acid treatment led to a significant decrease in the HBsAg level (Fig. 4F). Notably, asiatic acid exerted continuous antiviral activity in a concentration-dependent manner (Fig. 4A-F). Moreover, in primary hepatocytes (PHHs), asiatic acid treatment did not affect the level of HBV cccDNA, but suppressed the levels of HBV RNAs, HBV core DNA and HBsAg, also suggesting the effect of asiatic acid on inhibiting HBV cccDNA transcriptional activity (Fig. 4G-L). Together, these data suggest that asiatic acid exerts antiviral effects on HBV-infected HepG2-NTCP cells and PHHs.
Fig. 4.
Asiatic acid inhibited HBV replication and transcription. (A-B) HBV-infected HepG2-NTCP cells were treated with asiatic acid(10, 20 and 30 µM) or ETV for 3, 6, 9 days. Total HBV RNAs and HBV 3.5-kb RNA were detected by real-time PCR; (C) HBV 3.5-kb RNA and 2.4/2.1-kb RNA were examined by Northern blotting, ribosomal RNAs (28 S and 18 S) were served as a loading control; (D,E) The level of HBV core DNA was analyzed by real-time PCR and Southern blot; (F) ELISA was performed to detect the HBsAg level in supernatant of HBV-infected HepG2-NTCP cells; (G-J) PHHs were treated with asiatic acid(10, 20, 30µM) or ETV for 4 days. HBV cccDNA was detected by Taq-man probe PCR. Total HBV RNAs and HBV 3.5-kb RNA were quantified by real-time PCR. The ratios of total HBV RNAs/cccDNA and HBV 3.5-kb RNA/cccDNA were calculated as indicators of cccDNA transcription activity; (K,L) The level of HBV core DNA was analyzed by real-time PCR analysis and HBsAg in supernatant of HBV-infected HepG2-NTCP cells was subjected to ELISA. Representative data are from at least three independent experiments. Data are shown as mean ± SD, *P < 0.05; **P < 0.01
Validate the antiviral effect of asiatic acid in the HBV recombinant-cccDNA mouse model
We further explored the antiviral effects of asiatic acid in vivo. First, the appropriate concentration of asiatic acid was selected based on existing reports to test for hepatotoxicity in normal C57BL/6 mice [20]. C57BL/6 mice were treated with 15 mg/kg, 30 mg/kg, or 50 mg/kg asiatic acid by intragastric administration. Serum ALT and AST concentrations were measured by ELISA every 4 days. This preliminary evaluation demonstrated that asiatic acid did not exhibit hepatotoxicity at doses of 15 mg/kg and 30 mg/kg (Figure S4A-B). Moreover, as shown by H&E staining of mouse liver tissues, asiatic acid did not cause significant liver damage (Figure S4C).
Then, the recombinant-cccDNA mouse model was chosen for subsequent experiments [21]. First, we injected the prcccDNA and Cre recombinase plasmids through hydrodynamic tail vein injection. The mice were administered 30 mg/kg asiatic acid, and 0.02 mg/kg ETV as a control every 2 days (Fig. 5A). Serum was collected every 4 days, and the body weight was monitored. Compared with mice in the vehicle group, mice in the asiatic acid group exhibited markedly lower serum levels of HBsAg, HBeAg and HBV DNA without significant changes in body weight (Fig. 5B-E). In addition, after 24 days of asiatic acid treatment, all the mice were sacrificed, and the livers were harvested. It was observed that asiatic acid prominently reduced the levels of HBV RNA and HBV DNA in liver tissues, without affecting the level of cccDNA (Fig. 5F-H, Figure S4D-E). Besides, HBsAg was decreased under asiatic acid treatment, as evidenced by immunohistochemistry (Fig. 5I). In conclusion, these results confirmed that asiatic acid can also exert antiviral effects in vivo.
Fig. 5.
Asiatic acid played an antiviral role in vivo. (A) Schematic depiction of experiments in HBV recombinant-cccDNA mouse model; (B) The mouse body weight was recorded by an electronic scale during the whole administration period; (C) The level of HBV DNA in the serum was analyzed by real-time PCR; (D,E) The levels of HBsAg and HBeAg in the serum were detected by ELISA; (F,G) The levels of total HBV RNAs, HBV 3.5-kb RNA in liver tissues were analyzed by real-time PCR; (H) The level of HBV DNA in liver tissues was analyzed by real-time PCR; (I) HBsAg in liver tissues were analyzed by immunohistochemistry staining. Scale bar = 100 μm. Representative data are from at least three independent experiments. Data are shown as mean ± SD, *P < 0.05; **P < 0.01
Discussion
During HBV transcription and replication, the viral protein HBx, a transcriptional coactivator of cccDNA, can initiate and maintain cccDNA transcription [6]. Thus, HBx is an important target for the development of new antiviral drugs. Currently, drugs targeting HBx include mainly FXR agonists and HBx-DDB1 complex inhibitors. Vonafexor (EYP001), a second-generation nonbile acid farnesol X receptor (FXR) agonist, inhibits HBV pgRNA transcription and HBV DNA replication in a HBx-dependent manner by modulating FXR receptor activity [22]. MLN4924, a NEDD8 activating enzyme inhibitor, and nitazoxanide (NTZ), a thiazolide anti-infective drug, have been shown to interfere with the interaction between HBx and DDB1, thereby inhibiting HBV transcription [23, 24]. In addition, the NQO1 inhibitor dicoumarol has been shown to significantly promote the degradation of HBx via a ubiquitin-independent pathway, which in turn inhibits the transcription of HBV cccDNA [19]. Although these compounds are effective at inhibiting the expression or function of HBx, they have some shortcomings, which may limit their application. To develop novel anti-HBV strategies, we utilized the Nano-Glo® HiBiT Lysis Detection System to screen our constructed compound library for herbal monomer compounds capable of inhibiting the expression of HBx. We found that asiatic acid effectively reduced the level of HBx. Moreover, we employed various models to further validate the inhibitory effect of asiatic acid on the HBx protein level in a concentration gradient-dependent manner, and the results suggested that it has an anti-HBV effect.
Asiatic acid, a known constituent of the herb Centella asiatica, has anti-oxidant, anti-apoptotic and anti-inflammatory properties [25]. In addition to its hepatoprotective properties, asiatic acid also has considerable potential for assisting in the treatment of malignant tumors, cardiovascular diseases, and neurological disorders. It can reverse liver fibrosis by inactivating the TGF-β/Smad pathway and rebalance Smad3/Smad7 signaling to suppress the progression of melanoma and lung carcinoma [26, 27]. Moreover, asiatic acid can decrease cardiac hypertrophy induced by pressure overload or angiotensin II through the activation of the AMPKα signaling pathway [28]. Furthermore, asiatic acid has potential anti-enterovirus 71 (EV71) and anti-HIV effects [29, 30]. In this study, we found that asiatic acid reduced the level of HBx protein, and then decreased the level of HBx bound on cccDNA, leading to a marked decrease in H3K4me3 and increases in H3K9me3 and H3K27me3 on cccDNA, thereby inhibiting cccDNA transcriptional activity. Additionally, asiatic acid can significantly decrease the levels of virological markers of HBV, including HBV DNA, HBV RNAs and HBsAg in vitro and in vivo without obvious cytotoxicity, demonstrating the value of asiatic acid in anti-HBV. Unfortunately, we did not find HBV cccDNA can be eradicated by the asiatic acid. Since asiatic acid can reduce the intracellular HBV core DNA level, the long-term effect of asiatic acid may potentially reduce the replenishment of the cccDNA pool. However, this inference remains to be further investigated. In addition, to increase the application prospects of asiatic acid as an antiviral drug, the antiviral effect of asiatic acid should also be verified in more HBV infection models such as human liver chimeric uPA/SCID mice. The pharmacokinetics of asiatic acid also needs to be evaluated. Moreover, as a candidate antiviral compound, structural optimization of asiatic acid to reduce cytotoxicity is also a further step.
The major pathways for protein degradation in eukaryotic cells are the ubiquitin-proteasome pathway and the autophagy pathway. In this study, we found that asiatic acid can activate the autophagy pathway and that inhibiting autophagy can weaken the inhibitory effect of asiatic acid on the HBx protein level, revealing that the autophagy pathway may be involved in HBx protein degradation mediated by asiatic acid. Consistent with our findings, the activation of the autophagy system by asiatic acid ameliorates diabetic nephropathy fibrosis, ischemic myocardial injury, and acute hepatic injury [31, 32]. According to the choice of substrates for degradation, autophagy can be categorized into non-selective and selective autophagy. Selective autophagy recognizes specific cytoplasmic “cargoes” by autophagy receptors, which then interact with LC3 in the autophagic membrane to transfer the “cargoes” to autophagosomes and send them to lysosomes for degradation [33, 34]. This research reveals that asiatic acid specifically enhances the degradation of HBx protein, without significantly altering the levels of other viral proteins. This finding implies that asiatic acid might regulate the expression of specific autophagy receptors or interfere with their binding to HBx protein, ultimately facilitating the targeted degradation of HBx protein via the selective autophagy pathway. However, the molecular mechanism by which asiatic acid regulates the autophagy pathway to degrade HBx needs to be further elucidated.
Conclusion
Based on our constructed library of herbal monomer compounds and our screen of this library with the Nano-Glo® HiBiT Lysis Detection System, we revealed that asiatic acid reduced the HBx level. Asiatic acid may destabilize HBx via the autophagy pathway. In HBV-infected cells and mouse models, asiatic acid showed significant antiviral activity, especially through epigenetic modification mediated suppression of cccDNA transcription.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- AA
Asiatic acid
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- CC50
50% cytotoxicity concentrations
- cccDNA
Covalently closed circular DNA
- cDNA
Complementary DNA
- CHB
Chronic hepatitis B
- CHX
Cycloheximide
- CQ
Chloroquine
- DEPC
Diethyl pyrocarbonate
- DMEM
Dulbecco’s modified eagle medium
- DMSO
Dimethyl sulfoxide
- EGCG
Epigallocatechin-3-gallate
- EDTA
Ethylene diamine tetraacetic acid
- ELISA
Enzyme linked immunosorbent assay
- ETV
Entecavir
- FBS
Fetal bovine serum
- FXR
Farnesol X receptor
- HDACs
Histone deacetylases
- HATs
Histone acetyltransferases
- HBV
Hepatitis B virus
- HBeAg
Hepatitis B e antigen
- HBx
HBV X protein
- IFN
Interferon
- MOPS
4-Popanesulfonyl morpholine
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NAs
Nucleos(t)ide analogs
- NTCP
Sodium taurocholate cotransporting polypeptide
- NTZ
Nitazoxanide
- PBS
Phosphate buffer solution
- PCR
Polymerase chain reaction
- PEG8000
Polyethylene glycol 8000
- pgRNA
Pregenomic RNA
- PHH
Primary human hepatocytes
- PVDF
Poly(1,1-difluoroethylene)
- SDS
Sodium dodecyl sulfate
- SSc
Saikosaponin C
- TBS
Tris buffered saline
- TCM
Traditional Chinese medicine
Author contributions
ZY and HBY designed the study. RRL, CDW, ZY, NNT and ZHL performed the experiments and data analysis. KXX, ZZZ, SYH, JHR and FL gave resources. RRL and CDW wrote the original manuscript. All the authors reviewed the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (82472262, 82302507), the Chongqing Natural Science Foundation (CSTB2022NSCQ-MSX0864, CSTB2023NSCQ-BHX0170, cstc2021jcyj-bshX0179, CSTB2023NSCQ-MSX0480), Science and Technology Research Project of Chongqing Municipal Education Commission (KJQN202100429, KJQN202300483), Future Medical Youth Innovation Team of Chongqing Medical University (W0040).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
The animal study was reviewed and approved by the Laboratory Animal Center of Chongqing Medical University.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ranran Li and Chunduo Wang contributed equally to this work.
Contributor Information
Nana Tao, Email: taona929@163.com.
Zhihong Li, Email: zhihongli@cqmu.edu.cn.
Zhen Yang, Email: yangzhen547@163.com.
Haibo Yu, Email: 306842@hospital.cqmu.edu.cn.
References
- 1.Lanini S, Ustianowski A, Pisapia R, Zumla A, Ippolito G. Viral Hepatitis: etiology, Epidemiology, Transmission, Diagnostics, Treatment, and Prevention. Infect Dis Clin North Am. 2019;33:1045–62. [DOI] [PubMed] [Google Scholar]
- 2.Tang LSY, Covert E, Wilson E, Kottilil S, Chronic Hepatitis B, Infection. Rev JAMA. 2018;319:1802–13. [DOI] [PubMed] [Google Scholar]
- 3.Polaris Observatory Collaborators. Global prevalence, cascade of care, and prophylaxis coverage of hepatitis B in 2022: a modelling study. Lancet Gastroenterol Hepatol. 2023;8:879–907. [DOI] [PubMed] [Google Scholar]
- 4.Lin C-L, Yang H-C, Kao J-H. Hepatitis B virus: new therapeutic perspectives. Liver Int. 2016;36(Suppl 1):85–92. [DOI] [PubMed] [Google Scholar]
- 5.Wu TT, Coates L, Aldrich CE, Summers J, Mason WS. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. 1990;175:255–61. [DOI] [PubMed] [Google Scholar]
- 6.Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, et al. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol. 2011;55:996–1003. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q, Yan L, Xu B, Wang X, Sun X, Han N, et al. Screening of the HBx transactivation domain interacting proteins and the function of interactor Pin1 in HBV replication. Sci Rep. 2021;11:14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A. 2009;106:19975–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poziello A, Nebbioso A, Stunnenberg HG, Martens JHA, Carafa V, Altucci L. Recent insights into histone Acetyltransferase-1: biological function and involvement in pathogenesis. Epigenetics. 2021;16:838–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molecular insights into Spindlin. 1-HBx interplay and its impact on HBV transcription from cccDNA minichromosome - PubMed [Internet]. [cited 2024 May 27]. https://pubmed.ncbi.nlm.nih.gov/37537164/ [DOI] [PMC free article] [PubMed]
- 11.Benhenda S, Ducroux A, Rivière L, Sobhian B, Ward MD, Dion S, et al. Methyltransferase PRMT1 is a binding partner of HBx and a negative regulator of hepatitis B virus transcription. J Virol. 2013;87:4360–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ducroux A, Benhenda S, Rivière L, Semmes OJ, Benkirane M, Neuveut C. The Tudor domain protein Spindlin1 is involved in intrinsic antiviral defense against incoming hepatitis B virus and herpes simplex virus type 1. PLoS Pathog. 2014;10:e1004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivière L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel M-L, et al. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol. 2015;63:1093–102. [DOI] [PubMed] [Google Scholar]
- 14.Xia J, Inagaki Y, Song P, Sawakami T, Kokudo N, Hasegawa K, et al. Advance in studies on traditional Chinese medicines to treat infection with the hepatitis B virus and hepatitis C virus. Biosci Trends. 2016;10:327–36. [DOI] [PubMed] [Google Scholar]
- 15.Zhong L, Hu J, Shu W, Gao B, Xiong S. Epigallocatechin-3-gallate opposes HBV-induced incomplete autophagy by enhancing lysosomal acidification, which is unfavorable for HBV replication. Cell Death Dis. 2015;6:e1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, Zhou Z, He L, Ma H, Qu W, Yin J, et al. Hepatoprotective and inhibiting HBV effects of polysaccharides from roots of Sophora flavescens. Int J Biol Macromol. 2018;108:744–52. [DOI] [PubMed] [Google Scholar]
- 17.Pan Y, Ke Z, Ye H, Sun L, Ding X, Shen Y, et al. Saikosaponin C exerts anti-HBV effects by attenuating HNF1α and HNF4α expression to suppress HBV pgRNA synthesis. Inflamm Res. 2019;68:1025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan S-L, Yang H-T, Lee Y-J, Lin C-C, Chang M-H, Yin M-C. Asiatic acid ameliorates hepatic lipid accumulation and insulin resistance in mice consuming a high-fat diet. J Agric Food Chem. 2014;62:4625–31. [DOI] [PubMed] [Google Scholar]
- 19.Cheng S-T, Hu J-L, Ren J-H, Yu H-B, Zhong S, Wai Wong VK, et al. Dicoumarol, an NQO1 inhibitor, blocks cccDNA transcription by promoting degradation of HBx. J Hepatol. 2021;74:522–34. [DOI] [PubMed] [Google Scholar]
- 20.Qiu F, Yuan Y, Luo W, Gong Y-S, Zhang Z-M, Liu Z-M, et al. Asiatic acid alleviates ischemic myocardial injury in mice by modulating mitophagy- and glycophagy-based energy metabolism. Acta Pharmacol Sin. 2022;43:1395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi Z, Li G, Hu H, Yang C, Zhang X, Leng Q, et al. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice. J Virol. 2014;88:8045–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erken R, Andre P, Roy E, Kootstra N, Barzic N, Girma H, et al. Farnesoid X receptor agonist for the treatment of chronic hepatitis B: a safety study. J Viral Hepat. 2021;28:1690–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie M, Guo H, Lou G, Yao J, Liu Y, Sun Y, et al. Neddylation inhibitor MLN4924 has anti-HBV activity via modulating the ERK-HNF1α-C/EBPα-HNF4α axis. J Cell Mol Med. 2021;25:840–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inhibition of HBV Transcription From. cccDNA With Nitazoxanide by Targeting the HBx-DDB1 Interaction - PubMed [Internet]. [cited 2024 May 23]. https://pubmed.ncbi.nlm.nih.gov/30704981/ [DOI] [PMC free article] [PubMed]
- 25.Islam MT, Ali ES, Uddin SJ, Khan IN, Shill MC, de Castro E, Sousa JM, et al. Anti-cancer effects of Asiatic Acid, a Triterpene from Centilla Asiatica L: a review. Anticancer Agents Med Chem. 2020;20:536–47. [DOI] [PubMed] [Google Scholar]
- 26.Asiatic acid isolated from Centella asiatica inhibits TGF. -β1-induced collagen expression in human keloid fibroblasts via PPAR-γ activation - PubMed [Internet]. [cited 2024 May 23]. https://pubmed.ncbi.nlm.nih.gov/24250248/ [DOI] [PMC free article] [PubMed]
- 27.Lian G-Y, Wang Q-M, Tang PM-K, Zhou S, Huang X-R, Lan H-Y. Combination of Asiatic Acid and Naringenin modulates NK Cell Anti-cancer immunity by Rebalancing Smad3/Smad7 signaling. Mol Ther. 2018;26:2255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Z-G, Dai J, Wei W-Y, Zhang W-B, Xu S-C, Liao H-H, et al. Asiatic Acid protects against Cardiac Hypertrophy through activating AMPKα Signalling Pathway. Int J Biol Sci. 2016;12:861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao C, Xu J, Zhang Y, Zhao L, Feng B. Inhibition of human enterovirus 71 replication by pentacyclic triterpenes and their novel synthetic derivatives. Chem Pharm Bull (Tokyo). 2014;62:764–71. [DOI] [PubMed] [Google Scholar]
- 30.Jaisi A, Prema null, Madla S, Lee Y-E, Septama A, Morita H. Investigation of HIV-1 viral protein R inhibitory activities of Twelve Thai Medicinal Plants and their commercially available major constituents. Chem Biodivers. 2021;18:e2100540. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X-X, Liu Y, Xu S-S, Yang R, Jiang C-H, Zhu L-P, et al. Asiatic acid from Cyclocarya paliurus regulates the autophagy-lysosome system via directly inhibiting TGF-β type I receptor and ameliorates diabetic nephropathy fibrosis. Food Funct. 2022;13:5536–46. [DOI] [PubMed] [Google Scholar]
- 32.Pang X, Qiao Q, Vonglorkham S, Feng Z, Pang L, Chen S, et al. Asiatic acid ameliorates acute hepatic injury by reducing endoplasmic reticulum stress and triggering hepatocyte autophagy. Biomed Pharmacother. 2020;129:110375. [DOI] [PubMed] [Google Scholar]
- 33.Yao W, Li Y, Chen Y, Chen Y, Xie Y, Ye M, et al. Atg1-mediated Atg11 phosphorylation is required for selective autophagy by regulating its association with receptor proteins. Autophagy. 2023;19:180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aman Y, Schmauck-Medina T, Hansen M, Morimoto RI, Simon AK, Bjedov I, et al. Autophagy in healthy aging and disease. Nat Aging. 2021;1:634–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.