Abstract
Background
Mango is a tropical fruit with high economic value. The selection of suitable dwarf mango varieties is an important aspect of mango breeding. However, the mechanisms that regulate mango dwarfing remain unclear.
Results
In this study, we compared the transcriptomes and metabolomes of mango varieties Guiqi (a dwarfed variety) and Jinhuang (an arborized variety). A total of 4,954 differentially expressed genes and 317 differentially abundant metabolites were identified between the two varieties, revealing the molecular mechanism of the gibberellin 3β-hydroxylase gene GA3ox in regulating dwarfing traits in mangoes using joint transcriptome and metabolome analyses. The results showed that differentially expressed genes were enriched in the diterpenoid biosynthesis pathway and that differentially abundant metabolites were annotated to their upstream pathway, the terpenoid backbone biosynthesis. A gene regulation network based on these two pathways was constructed, indicating the upregulation of the GA3ox gene and the accumulation of gibberellin in dwarfed mangoes. We then transferred the GA3ox gene to tobacco plants following the application of gibberellin, and the morphology and height of the transgenic tobacco plants largely recovered the phenotype.
Conclusions
These results demonstrated that GA3ox plays a role in the regulation of dwarf traits. Our study provides an important theoretical basis for studying the regulatory mechanisms underlying mango dwarfism to facilitate mango breeding.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05700-6.
Keywords: Mango, Gibberellin 3β-hydroxylase, Dwarf plants, Alpha-linolenic acid metabolism, MAPK signaling pathway, Linoleic acid metabolism
Background
Mango (Mangifera indica L.) is a tropical fruit belonging to the family Anacardiaceae. Mango is known as the king of fruits because of its fresh fragrance and unique flavor. Mango has high nutritional value and is rich in various nutrients, such as proteins, fats, minerals, sugars, and nutrients that lower cholesterol and prevent cardiovascular diseases [1]. Mangoes are healthy tropical fruits that quench the thirst [2]. China is a major mango-producing country [3]. Mango plants can grow to more than ten meters tall, making the selection of suitable dwarf mango varieties an important aspect of mango breeding. Mangoes can be dwarfed through appropriate pruning, hormone control, and other cultivation techniques. However, the regulatory mechanisms that underlie mango dwarfing remain largely unknown.
Studies have shown that plant height is closely related to the synthesis of plant hormones, such as gibberellins (GAs). Gibberellin is present throughout the plant life cycle [4, 5] and regulate plant morphogenesis, promote flower bud differentiation, and induce the synthesis of long fibers and tubular molecules [6]. Reduced levels and activities of endogenous gibberellins in plants have been shown to affect plant height [7]. Mutations in the structure or altered expression levels of key enzyme genes in the gibberellin biosynthesis signaling pathway can affect plant height [8, 9]. Gibberellin 3β-hydroxylase (GA3ox) is a key enzyme that regulates gibberellin synthesis in plants [10]. Mutations in GA3ox or changes in its expression levels can alter the amount of active gibberellic acid (GA3) synthesis in plants, thereby affecting plant traits such as height. The GA3ox gene controlling dwarf architecture was identified in watermelon (Citrullus lanatus L.). Moreover, the functional loss of GA3ox leads to GA3 reduction and a consequent dwarfism phenotype [10]. GA3ox has been reported to contribute to regulation of the dwarf phenotype through Lacd1 in sponge gourds (Luffa acutangular) [11]. Gibberellin 3β-hydroxylase performs key functions in gibberellin biosynthesis and promotes root elongation in Arabidopsis [12]. Compared with average height phenotype, GA3ox was expressed significantly differently in GA-sensitive maize dwarfism (Zea mays L.). This suggests that GA3ox may be involved in the regulation of dwarfing traits [13]. Gibberellin 3β-hydroxylase is also involved in the modulation of GA metabolism, which regulates cell elongation in rice (Oryza sativa) [14]. However, whether GA and GA3ox influence mango dwarfing remains unknown.
In this study, we compared the transcriptomes and metabolomes of the leaves of dwarfed (Guiqi; Variety A) and the arborized (Jinhuang; Variety Q) varieties of mango. The dwarfed variety is the largest cultivated mango variety among domestic self-breeding varieties, with remarkable market application value. Its dwarf habit, disease-, and insect-resistant characteristics make it suitable for intensive cultivation in large-scale production orchards. Guiqi mangoes have moderate tree vigor with an open stance and an oval-shaped canopy. These serve as ideal characteristics for dwarf-oriented breeding, with a phenotype where the main stem and tree height are approximately 0.2 and 2.0 m, respectively. The arborized variety exhibited strong growth potential with an open, round-headed canopy and long, upright, and sturdy branches. Guiqi mangoes form a distinct contrast to the widely cultivated cypress-type Jinhuang mangoes. By comparing the transcriptomes and metabolomes of these two varieties, we found that the gene regulation network was directed toward GA biosynthesis and GA3ox. Then, the GA3ox function was studied by transferring it to tobacco plants. Our study aimed to identify the key genes involved in the regulatory mechanisms of mango dwarfism and provide potential gene resources for mango dwarfism breeding. In the present study, we demonstrated for the first time that GA3ox is involved in the regulation of dwarfism in mangoes. These results provided a theoretical basis for constructing a mango dwarfing gene regulatory network. This provides a solution for studying the mechanisms of mango dwarfing.
Results
Morphological characteristics
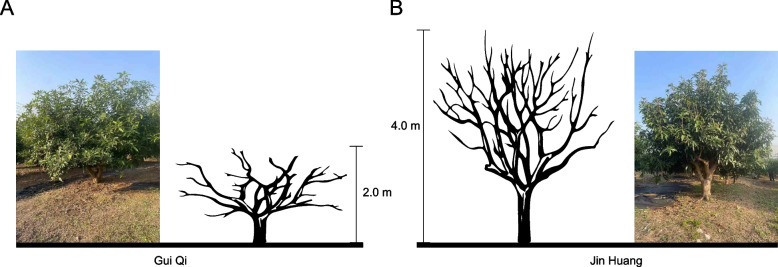
Variety A exhibited an expansive tree structure with an oval-shaped crown and moderately short stature (2 ± 0.16 m on average) (Fig. 1A). The inflorescence was sizable, forming a conical shape, with an average single fruit weight of 271 ± 59 g (Fig. 1A). In contrast, Variety Q reached a height of 4 ± 0.23 m and featured larger fruits, with individual weights ranging from 1 to 2.5 kg. It had a strong tendency to bear fruit, resulting in high yield (Fig. 1B). Leaves of Variety A were narrow and elongated, whereas those of Variety Q were large and thick (Fig. 1). The branches of Variety A were upright with small branching angles, whereas those of Variety Q had dispersed branches with large branching angles (Fig. 1).
Fig. 1.
Comparison of two mango varieties. Photos of the dwarf mango variety Guiqi (A) and the tall mango variety Jinhuang (B)
Analysis of transcriptome between arborized and dwarfed mango
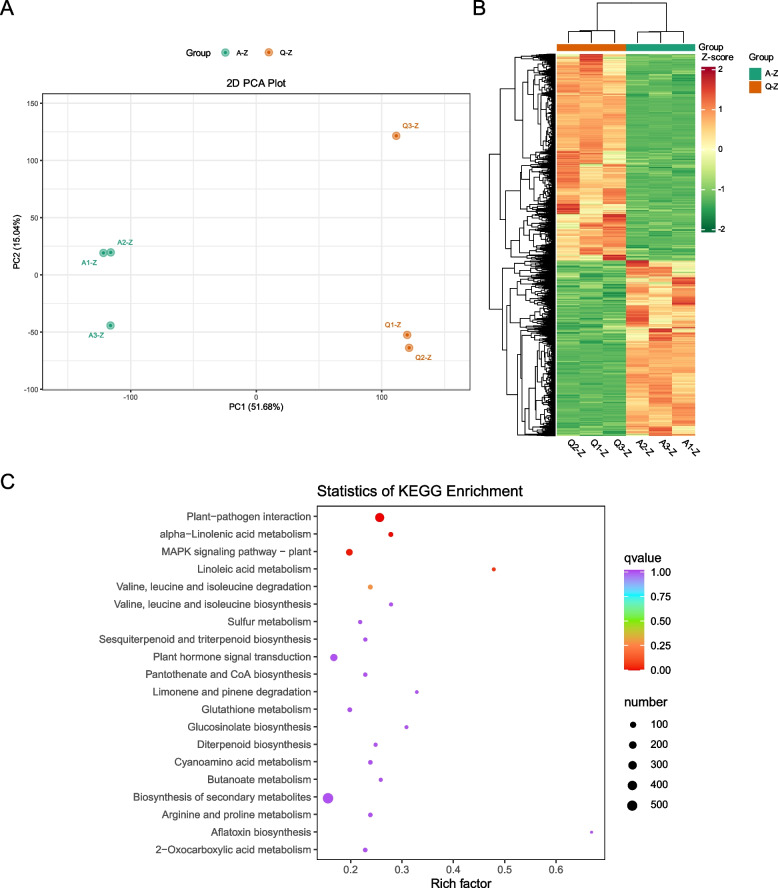
Transcriptome sequencing generated an average of 46,855,952 reads per sample with 45,907,199 clean reads per sample (Table S2). The proportion of uniquely mapped reads ranged from 81.63 to 83.61% (Table S2). Principal component analysis (PCA) results indicated that samples within each group clustered together and samples between groups exhibited considerable separation, suggesting good consistency within the groups and significant differences between the two varieties (Fig. 2A).
Fig. 2.
Transcriptome data analysis: (A) PCA scatter plot displaying separation of sample groups, B heatmap of differentially expressed genes (DEGs), and C bubble chart showing KEGG enrichment of DEGs, where the size of the points represents the number of DEGs within each pathway
A total of 4,954 differentially expressed genes (DEGs) between the two varieties were identified (Table S3). Among these, the number of upregulated DEGs in Variety Q was slightly higher than that of the downregulated DEGs when compared to Variety A (Fig. 2B). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed that the DEGs were primarily enriched in pathways related to plant-pathogen interactions; alpha-linolenic acid metabolism; mitogen-activated protein kinase (MAPK) signaling; linoleic acid metabolism; valine, leucine, and isoleucine degradation; arginine and proline metabolism; cyanoamino acid metabolism; 2-oxocarboxylic acid metabolism; glucosinolate biosynthesis; biosynthesis of secondary metabolites; valine, leucine and isoleucine biosynthesis; butanoate metabolism; pantothenate and CoA biosynthesis; diterpenoid biosynthesis, sesquiterpenoid and triterpenoid biosynthesis; Limonene and pinene degradation; and plant hormone signal transduction (Fig. 2C).
Analysis of metabolome between arborized and dwarfed mango varieties
The metabolome analysis identified 1,004 differentially available metabolites (DAMs) (Table S4). Distinct clustering of samples from Varieties A and Q was revealed through PCA, as well as quality control samples with substantial inter-group separation, indicating good intra-group sample reproducibility (Fig. 3A). The identified metabolites were predominantly phenolic acids, flavonoids, lipids, amino acids and their derivatives, organic acids, and alkaloids (Fig. 3B).
Fig. 3.
Metabolomic data analysis: (A) PCA scatter plot, B heatmap of abundance for all metabolites identified in the two varieties, C heatmap of metabolite abundance for differential metabolites in the two varieties, and (D) KEGG pathway enrichment analysis for the differential metabolites in (C)
Compared to Variety Q, Variety A showed upregulation of 171 and downregulation of 146 DAMs (Table S5). Among these, a substantial number belonged to the phenolic acid and flavonoid categories (Fig. 3C). The DAMs were predominantly enriched in pathways such as C5-branched dibasic acid metabolism; tyrosine metabolism; alanine, aspartate, and glutamate metabolism; and plant hormone signal transduction (Fig. 3D).
Detection of gibberellin between arborized and dwarfed mango varieties
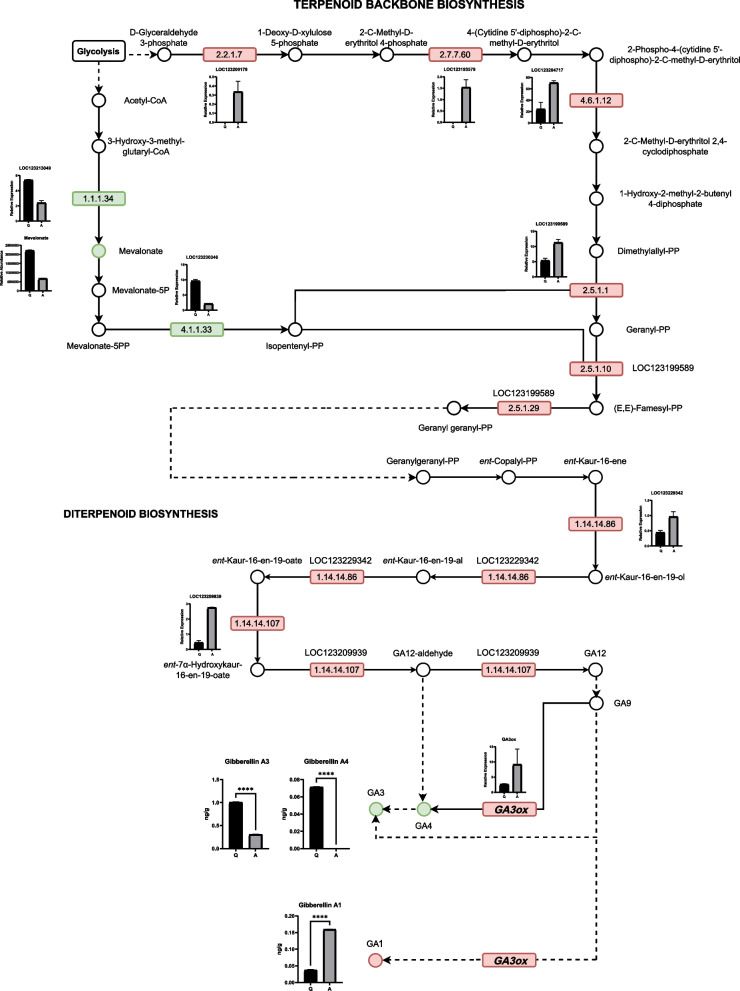
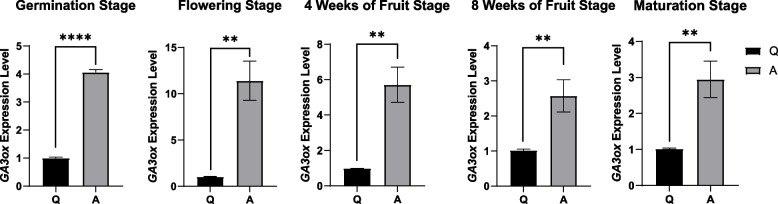
The transcriptomic results showed the enrichment of DEGs in the diterpenoid biosynthesis pathway, and the DAMs in the metabolome were annotated to their upstream pathway, terpenoid backbone biosynthesis. Therefore, we constructed a gene regulatory network that included the DEGs and DAMs in these two pathways. Significant changes were observed in the terpenoid backbone biosynthesis pathway in Variety A, with the downregulation of LOC123213049 (3-hydroxy-3-methylglutaryl-coenzyme A reductase 1-like) and LOC123230346 (diphosphomevalonate decarboxylase MVD2, peroxisomal-like) genes, as well as mevalonate (Fig. 4). However, most of the other related genes were up-regulated, including LOC123200179 (probable 1-deoxy-D-xylulose-5-phosphate synthase 2, chloroplastic), LOC123193579 (uncharacterized), LOC123204717 (2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, chloroplastic-like), LOC123199589 (geranylgeranyl pyrophosphate synthase, chloroplastic-like), and in the diterpenoid biosynthesis pathway, LOC123229342 (ent-kaurene oxidase, chloroplastic-like), LOC123209939 (ent-kaurenoic acid oxidase 1-like), and LOC123207275 (GA3ox) (Fig. 4). Additionally, the levels of GA3 and GA4 were significantly downregulated, whereas those of GA1 were significantly upregulated (Fig. 4). The expression levels of GA3ox were confirmed by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The results indicated that the expression level of the GA3ox gene in Variety A was significantly higher than that in Variety Q at all five growth stages (p < 0.05), including the germination, flowering, and maturation stages and at four and eight weeks of fruit development (Fig. 5), indicating the potential importance of GA3ox upregulation in dwarf mangoes.
Fig. 4.
Schematic representation of the pathways involved in gibberellin synthesis which is associated with dwarfism, including terpenoid backbone biosynthesis and diterpenoid biosynthesis. The squares and circles represent genes and metabolites, respectively, and the inset bar plots represent the expression/abundance of the corresponding genes and metabolites. Q indicates the Jinhuang mango variety, and A indicates the Guiqi mango variety
Fig. 5.
qRT-PCR detection of GA3ox gene expression at the germination stage, flowering stage, four weeks of fruit development, eight weeks of fruit development, and maturation stage, respectively. Q indicates the Jinhuang mango variety, and A indicates the Guiqi mango variety. Note. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001
Expression of GA3ox in tobacco
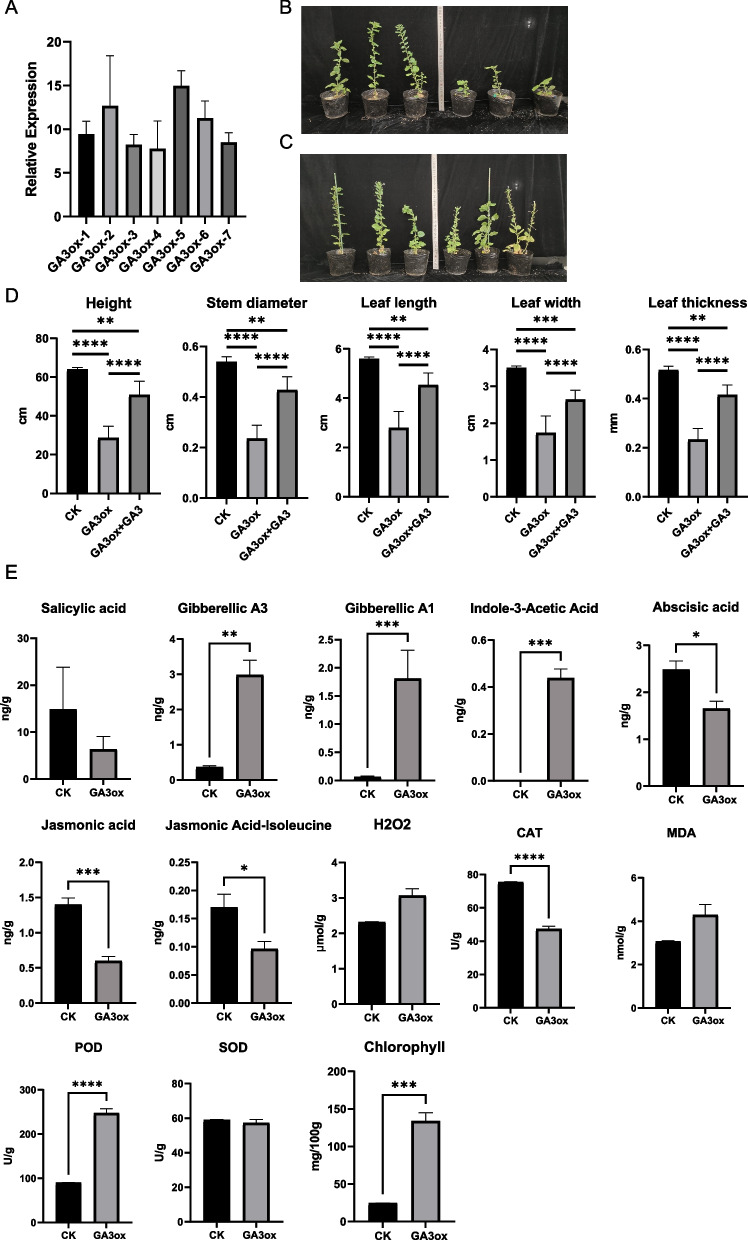
To further investigate the functionality of GA3ox, the gene was introduced into N. benthamiana and the transformation was validated using qPCR (Fig. 6A). Both wild-type and transgenic tobacco plants flowered; however, only the transgenic tobacco plants exhibited a pronounced dwarf phenotype (Fig. 6B). Following the application of GA3, the morphology and height of the transgenic tobacco plants largely recovered to the wild phenotype (Fig. 6C). Significant differences were observed among wild-type, transgenic, and transgenic tobacco after GA3 application in terms of plant height, stem diameter, leaf length, leaf width, and leaf thickness (p < 0.05) (Fig. 6D).
Fig. 6.
Expression of GA3ox in tobacco. A qPCR validation of positive transformation of GA3ox. B Photos of wild-type and GA3ox transgenic tobacco with the three biological replicates of wild-type on the left and those of the transgenic on the right. C Photos of wild-type and GA3ox transgenic tobacco applied with GA3, with the three biological replicates of wild-type on the left and those of the transgenic on the right. D The comparison of plant height, stem diameter, leaf length, leaf width, and leaf thickness between the wild-type, transgenic, and transgenic tobacco subjected to gibberellic acid (GA3) application. E The levels of salicylic acid, gibberellic A3, gibberellic A1, indole-3-acetic acid, abscisic acid, jasmonic acid, jasmonic acid-isoleucine, H2O2, catalase (CAT) activity, malondialdehyde (MDA) content, peroxidase (POD) activity, superoxide dismutase (SOD) activity, and chlorophyll in wild type and transgenic tobacco. Note. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001
To further investigate the molecular basis of the dwarf phenotype induced by GA3ox in transgenic tobacco, we measured the levels of plant hormones involved in the regulation of growth and development, including GA1, GA3, indole-3-acetic acid (auxin), jasmonic acid, jasmonoyl isoleucine, and abscisic acid. Compared to that in the wild type, the levels of GA1 and GA3 were significantly upregulated in transgenic tobacco, confirming that the expression of the GA3ox gene led to increased gibberellin levels (p < 0.05) (Fig. 6E). Correspondingly, the auxin level also exhibited upregulation (p < 0.05) (Fig. 6E), suggesting a potential positive regulatory relationship between auxin and gibberellins. Conversely, the levels of jasmonic acid, jasmonoyl isoleucine, and abscisic acid were significantly downregulated (p < 0.05) (Fig. 6E), implying a potential antagonistic interaction with gibberellins. Plant hormones play a crucial role in regulating antioxidant enzyme activity, thereby influencing plant stress tolerance. The results showed that peroxidase (POD) activity was significantly upregulated in transgenic tobacco plants, whereas catalase (CAT) activity was significantly downregulated (p < 0.05) (Fig. 6E).
Discussion
Plant height is a crucial agronomic trait for crop growth and high-yield cultivation. Several studies have been conducted to understand the molecular and genetic basis of plant height in model plants and crops [15]. However, the regulatory mechanisms underlying dwarfing traits in mango remain unelucidated. In the present study, transcriptome analysis of arborized and dwarfed mangoes showed that 4,954 DEGs were enriched in the alpha-linolenic acid metabolism and MAPK signaling pathways. Alpha-linolenic acid is a precursor of jasmonic acid [16], a phytohormone that regulates plant growth. Several studies have shown that jasmonates are involved in many plant expansion events such as primary root growth, leaf senescence, and reproductive development [17, 18]. Our results indicated that alpha − linolenic acid is involved in the regulation of the dwarfing phenotype in mangoes through the synthesis of jasmonic acid. Consistent with our results, alpha-linolenic acid metabolism is related to peanut dwarf mutants and may be a key factor in the dwarf phenotype [19]. Additionally, alpha-linolenic acid metabolism has been reported to play a crucial role in maize dwarf habit [20].
Melatonin (MT) also regulates plant growth. Alpha-linolenic acid metabolism is related to the composition and metabolism of MT in cotton, and participates in the regulation of dwarfing traits [21]. MAPK mutations cause dwarfing phenotypes in Arabidopsis [22] and tobacco [23]. Compared to traditional bread fruit (Artocarpus altilis), DEGs of the dwarf phenotype after grafting were enriched in the MAPK signaling pathway [24]. Researchers have found that the MAPK signaling pathway participates in the regulation of dwarfing in bananas [25]. However, previous studies have focused only on pulp color regulation in mangoes [26], and the regulatory mechanism of the mango dwarfing phenotype via the MAPK signaling pathway remains unelucidated. Mitogen-activated protein kinase cascades were elucidated as unified signaling modules downstream of receptor-like proteins in plant growth [27]. Our research indicates that the MAPK signaling pathway is involved in the regulation of dwarf traits in mangoes. These results could be due to the participation of the MAPK signaling pathway in polar transport, biosynthesis, and signaling of auxin, which can regulate plant growth [28].
Gibberellins are a major determinant of plant height [29, 30]. We focused on the upstream pathways of GA synthesis by analyzing the metabolomes of arborized and dwarfed mangoes. Gibberellic acid is a plant hormone with a tetracyclic diterpenoid structure that is involved in various important developmental processes, including plant growth, seed germination, stem elongation, and leaf expansion [31]. Gibberellins promote plant growth via cell expansion and division [32]. OsNac120 regulates plant growth via GA signaling in rice [33]. Strigolactone–gibbberellin crosstalk mediated by a distant silencer fine-tunes plant height in upland cotton [34]. Genome-wide analysis of the GA oxidase family members in four Purnus species showed that GAs play a dominant role in plant height [35]. This gene module regulates vegetative growth via GA biosynthesis in tomato [36]. Regulation of GA3 was applied to grape (Vitis vinifera L.) berry size [37]. Mutations in GA metabolism and signaling pathways have led to the development of semi-dwarf varieties [38]. Beneficial semi-dwarfism is conferred in wheat and rice by the mutant reduced height-1 (Rht-1) and semi-dwarf1 (sd1) alleles [39]. The sd1 allele reduces the abundance of bioactive GA [40], whereas Rht-B1b and Rht-D1b proteins resist GA-stimulated destruction [41]. In 2023, researchers discovered that GA synthesis inhibitors could stably repress wheat growth, leading to a semi-dwarf phenotype [42]. In chrysanthemum (Chrysanthemum morifolium), plant height was regulated by modulating GA biosynthesis [43]. The dwarf-type wintersweet (Chimonanthus praecox) can be recovered by exogenous GA application. Further studies have shown that GA synthesis-related genes regulate dwarfing mechanisms in C. praecox [44]. Transcriptome analysis has shown that GA functions as a regulator of the maize dwarfing phenotype [45]. Gibberellic acid promotes the degradation of DELLA proteins, which are negative regulators [46]. CRISPR/Cas9 genome editing in tomatoes creates a gibberellin-responsive dominant dwarf DELLA allele [47]. Gibberellins include the functionally active molecules GA1, GA3, GA4, and GA7 [4]. In the present study, we identified DEGs in the GA synthesis signaling pathway in both arborized and dwarfed mangoes. The levels of gibberellins, including GA3 and GA4, were also significantly downregulated. In agreement with our findings, higher GA4 content was also found in watermelon dwarf plants belonging to the phylum Magnoliaceae [10]. These results indicate that GA synthesis is closely related to the development of dwarf mango plants. These results suggest that various feedback, feed-forward, and cross-regulatory loops are involved in the maintenance of GA homeostasis. GA has been reported to block flavonoid biosynthesis [48], leading to the dwarfing phenotype in plants [49–51]. Regulation of the dwarfing phenotype may be related to these mechanisms.
Jasmonic acid is a lipid-derived plant hormone essential for plant development and an important endogenous signal that regulates the plant dwarf phenotype [52]. In the present study, jasmonic acid and GA were found to be jointly involved in the regulation of mango dwarfism (Fig. 6E). This is consistent with previous results for peaches (Prunus persica) [53]. In maize, jasmonic acid and GAs exhibit interactive relationships that lead to plant dwarfism [54]. Antioxidant enzyme activity plays a crucial role in stress tolerance in tobacco [55]. This result is consistent with the findings of the present study. Peroxidase regulates growth and development in rice [56]. Peroxidase genes are associated with cell wall remodeling, which can regulate wheat height decline [57]. In the present study, CAT activity was significantly downregulated (p < 0.05) in transgenic tobacco plants. This may lead to increased levels of reactive oxygen species [58]. Reactive oxygen species promote pollen and seed development by triggering programmed cell death and tapetal cell degradation, resulting in the dwarf phenotype in plants [59].
Gibberellin 3β-hydroxylase (GA3ox) is important for the production of biologically active GAs [8]. In monocots, mutations in GA3ox, such as Dwarf1 (D1) in maize [60] and OsGA3ox1 and OsGA3ox2 (Dwarf18 or D18) in rice, result in dwarfism [61]. OsGA3ox genes regulate rice plant height by synthesizing diverse active GAs [62]. Moreover, some GA3ox genes have been characterized as growth regulators in cucurbit crops, such as watermelon [63], cucumber [64], and pumpkin [65]. Mutations in GA3ox that cause dwarf phenotypes have been identified in Arabidopsis [66] and Medicago sativa [67]. In a previous study, GA3ox was cloned into mangoes. Compared with arborized mangoes, GA3ox-transfected mangoes showed a dwarf phenotype-biased expression pattern [68]. In this study, to further investigate its function, GA3ox was transferred to tobacco. The transgenic plants showed a distinct dwarfing phenotype. Significant differences in plant height, stem diameter, leaf length, leaf width, and leaf thickness were observed among the wild-type, transgenic, and transgenic tobacco plants after GA3 application. Translocated GA3ox catalyzes the final step of bioactive GA synthesis, which converts GA20 to GA1 and GA5 to GA3 [60], leading to the upregulation of GA1 and GA3 levels. Furthermore, auxin was significantly upregulated in the transgenic tobacco plants. Auxin overaccumulation is insensitive to gravitropic stimuli and results in dwarfism in plants [69]. Consistent with our findings, auxin overexpression leads to the dwarf phenotype in Arabidopsis [66], rice [70], apples [71], and maize [72]. In contrast, stunted plant growth led to markedly down-regulated levels of jasmonic acid, jasmonic acid-isoleucine and abscisic acid in transgenic tobacco.
Conclusions
Selection of dwarf mango varieties can improve mango quality and increase economic benefits. Regulation of dwarfism in mango varieties is closely related to hormones and genes. In the present study, we used transcriptomics and metabolomics to reveal the key differences between arborized and dwarf mango varieties. Transcriptome analysis identified 4,954 DEGs in the two varieties. KEGG pathway enrichment analysis revealed that the DEGs were primarily enriched in alpha-linolenic acid metabolism, the plant MAPK signaling pathway, and linoleic acid metabolism. In total, 1,004 DAMs were characterized using metabolomic analysis. Next, we focused on pathways upstream of GA synthesis. GA3ox is crucial for the biological synthesis of GA and was introduced into N. tabacum resulting in a dwarf tobacco phenotype. Our study revealed that GA3ox regulates dwarfing traits in mangoes, which may contribute to the elucidation of the molecular mechanisms of dwarfing regulation.
Materials and methods
Plant material and sampling
Guiqi (dwarfed) and Jinhuang (arborized) mango varieties were selected as the study species and labeled Varieties A and Q, respectively. Leaves from three individuals of each variety were collected for transcriptomic and metabolomic analyses. Samples were collected from the Mango Germplasm Repository at the Subtropical Crops Research Institute of Guangxi Zhuang Autonomous Region. Tender leaves, freshly sprouted and free from diseases and pests, were collected from each variety after four weeks of fruit development, with three biological replicates per variety. The collected samples were immediately frozen in liquid nitrogen and stored at -80 °C for subsequent analyses, including transcriptomic profiling, metabolomic assessments, and GA quantification. Leaves were collected during the germination, flowering, and maturation stages, and after four and eight weeks of fruit development. These samples were specifically designated for qRT-PCR analysis of the GA3ox gene (Gene Bank Number: PP378951).
RNA extraction and RNA-seq analysis
The RNAprep Pure Plant Plus kit (Tiangen Biotech, Shanghai, China) was used for RNA extraction, and a total amount of 1 μg RNA per sample was used to purify the mRNA using poly-T oligo attached magnetic beads. Sequencing libraries were generated using the NEBNext® Ultra™ II RNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s recommendations. RNA degradation and contamination were assessed using 1% agarose gel electrophoresis. RNA purity was verified using a NanoPhotometer spectrophotometer (NanoPhotometer, Implen Inc., Westlake Village, CA, USA). The RNA concentration was quantified with the Qubit® RNA Assay Kit on the Qubit®2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). The RNA integrity was evaluated using a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA) and an RNA Nano 6000 Assay Kit (Agilent Technologies).
Transcriptome analysis
Raw data were subjected to quality control using fastp v0.19.3 to obtain clean reads [73]. Clean reads were aligned with the reference genome (NCBI accession number: GCF_011075055.1) using HISAT v2.1.0 [74]. Gene expression was quantified using featureCounts v1.6.2 [75]. Differential expression analysis between varieties was conducted using DESeq2 v1.22.1 [76]. The Benjamini & Hochberg method was employed to adjust p-values, with a threshold of corrected p < 0.05 and |Log2FC|≥ 1 considered as significant differential expression.
Sample preparation and LC–MS
The leaves were freeze-dried using a vacuum freeze dryer (Scientz-100F; Ningbo, China) and ground in a mixer mill (Retsch MM 400; Haan, Germany). Approximately 50 mg of the freeze-dried powder was mixed with 1.2 mL of 70% methanol solution and vortexed for 30 s every 30 min for six vortexing cycles. After centrifugation at 12,000 rpm for 3 min, the supernatant was filtered through a SCAA-104 (0.22 μm pore size; ANPEL, Shanghai, China) and subjected to metabolite profiling using a UPLC-ESI–MS/MS system (UPLC, Shimadzu Nexera X2, Tokyo, Japan; MS, Applied Biosystems 6500 Q TRAP, Carlsbad, CA, USA). The conditions were as follows: Agilent SB-C18 column (1.8 µm, 2.1 mm * 100 mm) for UPLC; mobile phase consisting of solvent A (pure water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). Sample measurements were performed using a gradient program, starting with 95% A and 5% B. Within 9 min, the linear gradient was adjusted to 5% A and 95% B and maintained for 1 min. Subsequently, within 1.1 min, the composition was reverted to 95% A and 5.0% B and maintained for 2.9 min. The flow rate was set at 0.35 mL/min, column temperature was set at 40 °C, and the injection volume was 2 μL. The eluent was alternately connected to an ESI-triple quadrupole-linear ion trap (QTRAP)-MS. The ESI source operation parameters were as follows: source temperature 500 °C; ion spray voltage (IS) in positive mode 5500 V/negative mode -4500 V; ion source gas I (GSI), gas II (GSII), and curtain gas (CUR) were set at 50, 60, and 25 psi, respectively; collision-activated dissociation (CAD) was set to high [77].
Metabolome analysis
Principal component analysis was performed using PrComp (v3.5.1) [78]. Differential metabolites were identified based on variable importance in projection (VIP ≥ 1) and absolute Log2FC (|Log2FC|≥ 1.0) criteria. VIP values were obtained from the OPLS-DA results using the R package MetaboAnalystR (v1.0.1) [79]. Prior to OPLS-DA, the data underwent log transformation (log2) and mean centering. To prevent overfitting, a permutation test was conducted using 200 permutations [80].
qRT-PCR detection of GA3ox gene
As described previously, qRT-PCR was performed to quantify the expression of the GA3ox gene [81]. Purified RNA was reverse transcribed to cDNA using All-in-One™ First-Strand cDNA Synthesis Kit (GeneCopoeia, Guangzhou, China), and 1 μg total RNA was included in each reverse transcriptase reaction. The 2−ΔΔCT method was employed for the analysis of qRT-PCR data to determine relative quantification [82]. Gene expression levels were normalized to the geometric mean of β-actin (ACTB). Three biological replicates were used for each variety at each stage. The primers used are listed in Supplementary Table S1.
Detection of gibberellin content of mango
Gibberellins A1, A3, and A4 were detected using targeted metabolomics. Approximately 100 mg of each plant sample was extracted in 1 ml of ice-cold 50% aqueous acetonitrile (v/v). The samples were sonicated for 3 min at 4 °C and subsequently extracted using a benchtop laboratory rotator (IKA®-Werke GmbH & Co. KG, Staufen, Germany) for 30 min at 4 °C. After centrifugation at 12, 000 rpm for 10 min, the supernatant was collected and purified using C18 reverse-phase polymer-based solid-phase extraction (RP-SPE) cartridges. A UPLC-Orbitrap-MS system (UPLC, Vanquish, Thermo Fisher Scientific, Waltham, MA, UDSA; MS, QE) was used to analyze the sample extracts. The analytical conditions were as follows: UPLC column, Waters ACQUITY UPLC HSS T3 (1.8 μm, 2.1 mm*50 mm); column temperature, 40 °C; flow rate, 0.3 mL/min; injection volume, 2 μL; solvent system, water (0.1% acetic acid): acetonitrile (0.1% acetic acid); gradient program at 0 min, 1.0 min, 5.0 min, 7.0 min, 7.1 min, and 9.0 min, at 90:10 V/V, 90:10 V/V, 10:90 V/V, 10:90 V/V, 90:10 V/V, and 90:10 V/V, respectively. High-resolution mass spectrometry data were recorded on a Q-Exactive hybrid Q-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a heated ESI source. The ESI source parameters were set as follows: spray voltage, -2.8 kV; sheath gas pressure, 40 arb; auxiliary gas pressure, 10 arb; sweep gas pressure, 0 arb; capillary temperature, 320 °C; and auxiliary gas heater temperature, 350 °C. Data were acquired on the Q-Exactive using Xcalibur 4.1 (Thermo Fisher Scientific) and processed using TraceFinder™ 4.1 Clinical (Thermo Fisher Scientific).
Transgenic expression of GA3ox in tobacco
Tobacco (Nicotiana benthamiana) seeds were provided by Jiangsu Sanshu Biotechnology Co. (Nantong, China). Seeds were sown on MS medium and left to grow for approximately five weeks before subsequent genetic transformation. The full-length coding sequence of the GA3ox gene from tobacco was amplified by PCR using two gene-specific primers: GA3ox Kpnl F (atttggagaggacagggtaccATGTCTTCAATTCCTGAAACTTCCA) and GA3ox Xbal R (tgcctgcaggtcgactctagaCTAAGCCACTTCTGCATTACTATCATC). The GA3ox gene was cloned into the pCAMBIA1301S vector to construct the recombinant plasmids. The resulting plasmid vectors were used to transform Agrobacterium tumefaciens strain LBA4404. Subsequently, A. tumefaciens was used to introduce the GA3ox expression cassette into the young leaves of sterile tobacco seedlings. The young leaves were exposed to inoculation for 20 min with agitation every 2–3 min, and cultivated under weak light at 22 °C for 3 d. The calli obtained were sub-cultured on MS medium supplemented with BA (1 mg/L), NAA (0.2 mg/L), Cb (500 mg/L), and Hgy (15 mg/L) for shoot differentiation, and the regenerated shoots were transferred to rooting medium. Positive transformation was verified by qPCR.
Following transformation, wild-type and transgenic GA3ox tobacco plants were subjected to GA3 spray treatment at a concentration of 100 mg/L (applied every 3 d for a continuous period of 30 d). Simultaneously, an equal volume of clean water was used for spray treatment in the control group (applied every 3 d for a continuous period of 30 d). On day 50 after spray application, the plant height, stem diameter, leaf length, leaf width, and leaf thickness of both wild-type and transgenic GA3ox tobacco plants, as well as those treated with GA3, were measured using a ruler for comparative analysis. Three biological replicates were used for each treatment group.
Physiological measurement of transgenic tobacco
Plant hormones, including salicylic acid, GA3, GA1, auxin, abscisic acid, jasmonic acid, and jasmonic acid isoleucine, were detected using targeted metabolomics [83]. Levels of H2O2, malondialdehyde, and chlorophyll were measured using previously published methods [84]. Mango pulp tissues (3 g) were homogenized with 10% trichloroacetic acid (15 mL) and centrifuged at 15,000 × g for 20 min. A mixture of 1 ml of supernatant and 0.5% 2-thiobarbituric acid of 3 mL was made, followed by warming for 20 min at 95 ℃, and immediate cooling in a cold water bath. Absorbance was spectrophotometrically measured at 532 nm (UV 1600 PC, Shimadzu) after centrifugation at 3000 × g for 10 min, along with the subtraction of nonspecific absorbance values at 600 nm. The estimation of the quantity of malondialdehyde was calculated using the equation: (μM/gFW) = [6.45 (OD532 − OD600) − 0.56 OD450] × 5 mL /0.25 g.
Catalase, POD, and superoxide dismutase activities were measured as previously described [85]. A total of 0.1 M sodium phosphate buffer (pH 7.0) of 2.5 mL was used to draw mango pulp tissues (1 g) for 10 min at 4 ℃, with extracting liquid being centrifuged for 15 min at 12,000 × g. The enzymatic activities of CAT, POD, and superoxide dismutase were determined by collecting the supernatant and recording the absorbance at 240 nm for 3 min at 25 ℃, at 470 nm for 3 min at 25 ℃, and at 560 nm, respectively.
Statistical analysis
Comparison of GA3ox expression between the two mango varieties, plant height, stem diameter, leaf length, leaf width, and leaf thickness, and changes in physiological indicators between the two control and transgenic tobacco plants were performed using Student’s t-test.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- DEG
Differentially Expressed Gene
- DAM
Differentially Abundant Metabolite
- GA3ox
Gibberellin 3β-hydroxylase
- GA
Gibberellic Acid
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
Mitogen-activated Protein Kinase
- PCA
Principal Component Analysis
- QC
Quality Control
- POD
Peroxidase
- CAT
Catalase
- MT
Melatonin
- Rht-1
Reduced height-1
- Sd1
Semi-dwarf1
- qRT-PCR
Quantitative Reverse Transcription Polymerase Chain Reaction
- MDA
Malondialdehyde
- SOD
Superoxide Dismutase
Authors’ Contributions
Y.Z., X.P., G.H. and S.W. designed the experiments and wrote the manuscript; X.P.,M.L. and J.Z. performed the materials harvest and measurement; Y.Z. and Y.T. extracted RNA and prepared the sequencing libraries; Y.Z. prepared the samples for LC-MS; M.L and Y.Z detected the expression of GA3ox in tobacco; Y.Z., X.P. and J.Z analyzed the data. The authors read and approved the final manuscript.
Funding
This research was funded through Local Scientific and Technology Development Guided by Central Government (Grant No. Guike ZY21195011); Guangxi Natural Science Foundation (Grant No. 2021GXNSFAA196043); Guangxi Science and Technology Major Program (Grant No. GuikeAA22068098) and Basic Scientific and Research Project of Guangxi Academy of Agricultural Sciences (Grant No. Guinongke 2024YP127).
Data availability
The datasets generated and analysed during the current study are available in the NCBI SRA repository (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA1026664).
Declarations
Ethics approval and consent to participate
The study complies with relevant institutional, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu Zhang and Xinhua Pang contributed equally to this work.
Contributor Information
Yu Zhang, Email: gxrzszy0624@163.com.
Guodi Huang, Email: Huangguodi2010@126.com.
Shaolong Wei, Email: nngt51578@126.com.
References
- 1.Lebaka VR, Wee YJ, Ye W, Korivi M. Nutritional composition and bioactive compounds in three different parts of mango fruit. Int J Environ Res Public Health. 2021;18(2):741. 10.3390/ijerph18020741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirza B, Croley CR, Ahmad M, Pumarol J, Das N, Sethi G, Bishayee A. Mango (Mangifera indica L.): a magnificent plant with cancer preventive and anticancer therapeutic potential. Crit Rev Food Sci Nutr. 2021;61(13):2125–51. 10.1080/10408398.2020.1771678. [DOI] [PubMed]
- 3.Liang Q, Song K, Lu M, Dai T, Yang J, Wan J, Li L, Chen J, Zhan R, Wang S. Transcriptome and metabolome analyses reveal the involvement of multiple pathways in flowering intensity in mango. Front Plant Sci. 2022;13:933923. 10.3389/fpls.2022.933923. [DOI] [PMC free article] [PubMed]
- 4.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–51. 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed]
- 5.Achard P, Genschik P. Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot. 2009;60(4):1085–92. 10.1093/jxb/ern301. [DOI] [PubMed]
- 6.Plackett AR, Thomas SG, Wilson ZA, Hedden P. Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 2011;16(10):568–78. 10.1016/j.tplants.2011.06.007. [DOI] [PubMed]
- 7.Huang J, Tang D, Shen Y, Qin B, Hong L, You A, Li M, Wang X, Yu H, Gu M, Cheng Z. Activation. of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J Genet Genomics. 2010;37(1):23–36. 10.1016/S1673-8527(09)60022-9. [DOI] [PubMed]
- 8.Suo H, Ma Q, Ye K, Yang C, Tang Y, Hao J, Zhang ZJ, Chen M, Feng Y, Nian H. Overexpression of AtDREB1A causes a severe dwarf phenotype by decreasing endogenous gibberellin levels in soybean [Glycine max (L.) Merr]. PLoS One. 2012;7(9):e45568. 10.1371/journal.pone.0045568. [DOI] [PMC free article] [PubMed]
- 9.Xu Q, Krishnan S, Merewitz E, Xu J, Huang B. Gibberellin-regulation and genetic variations in leaf elongation for tall fescue in association with differential gene expression controlling cell expansion. Sci Rep. 2016;6:30258. 10.1038/srep30258. [DOI] [PMC free article] [PubMed]
- 10.Sun Y, Zhang H, Fan M, He Y, Guo P. A mutation in the intron splice acceptor site of a GA3ox gene. confers dwarf architecture in watermelon (Citrullus lanatus L.). Sci Rep. 2020;10(1):14915. 10.1038/s41598-020-71861-7. [DOI] [PMC free article] [PubMed]
- 11.Zhao G, Luo C, Luo J, Li J, Gong H, Zheng X, Liu X, Guo J, Zhou L, Wu H. A mutation in LacDWARF1 results in a GA-deficient dwarf phenotype in sponge gourd (Luffa acutangula). Theor Appl Genet. 2021;134(10):3443–57. 10.1007/s00122-021-03938-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker R, Fernandez Garcia MN, Powers SJ, Vaughan S, Bennett MJ, Phillips AL, Thomas SG, Hedden P. Mapping sites of gibberellin biosynthesis in the Arabidopsis root tip. New Phytol. 2021;229(3):1521–34. 10.1111/nph.16967. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Wang Y, Zhao J, Huang J, Shi Y, Deng D. Unveiling gibberellin-responsive coding and. long noncoding RNAs in maize. Plant Mol Biol. 2018;98(4–5):427–438. 10.1007/s11103-018-0788-8. [DOI] [PubMed]
- 14.Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C. Brassinosteroid regulates. cell. elongation by modulating gibberellin metabolism in rice. Plant Cell. 2014;26(11):4376–93. 10.1105/tpc.114.132092. [DOI] [PMC free article] [PubMed]
- 15.Henchiri H, Rayapuram N, Alhoraibi HM, Caïus J, Paysant-Le Roux C, Citerne S, Hirt H, Colcombet J, Sturbois B, Bigeard J. Integrated multi-omics and genetic analyses reveal molecular determinants underlying Arabidopsis snap33 mutant phenotype. Plant J. 2024;118(4):1016–35. 10.1111/tpj.16647. [DOI] [PubMed] [Google Scholar]
- 16.Ghorbel M, Brini F, Sharma A, Landi M. Role of jasmonic acid in plants: the molecular point of view. Plant Cell Rep. 2021;40(8):1471–94. 10.1007/s00299-021-02687-4. [DOI] [PubMed]
- 17.Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot. 2013;111(6):1021–58. 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed]
- 18.Kim J, Chang C, Tucker ML. To grow old: regulatory role of ethylene and jasmonic acid in senescence. Front Plant Sci. 2015;29(6):20. 10.3389/fpls.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo F, Hou L, Ma C, Li G, Lin R, Zhao Y, Wang X. Comparative transcriptome analysis of the peanut semi-dwarf mutant 1 reveals regulatory mechanism involved in plant height. Gene. 2021;791:145722. 10.1016/j.gene.2021.145722. [DOI] [PubMed]
- 20.Liu J, Zhang L, Huang L, Yang T, Ma J, Yu T, Zhu W, Zhang Z, Tang J. Uncovering the gene regulatory network of maize hybrid ZD309 under heat stress by transcriptomic and metabolomic analysis. Plants (Basel). 2022;11(5):677. 10.3390/plants11050677. [DOI] [PMC free article] [PubMed]
- 21.Li L, Yan X, Li J, Wu X, Wang X. Metabolome and transcriptome association analysis revealed key factors involved in melatonin mediated cadmium-stress tolerance in cotton. Front Plant Sci. 2022;13:995205. 10.3389/fpls.2022.995205. [DOI] [PMC free article] [PubMed]
- 22.Takagi M, Hamano K, Takagi H, Morimoto T, Akimitsu K, Terauchi R, Shirasu K, Ichimura K. Disruption of the MAMP-Induced MEKK1-MKK1/MKK2-MPK4 Pathway Activates the TNL Immune Receptor SMN1/RPS6. Plant Cell Physiol. 2019;60(4):778–87. 10.1093/pcp/pcy243. [DOI] [PubMed] [Google Scholar]
- 23.Jin H, Axtell MJ, Dahlbeck D, Ekwenna O, Zhang S, Staskawicz B, Baker B. NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev Cell. 2002;3(2):291–7. 10.1016/s1534-5807(02)00205-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, Underhill SJR. Differential transcription pathways associated with rootstock-induced dwarfing in breadfruit (Artocarpus altilis) scions. BMC Plant Biol. 2021;21(1):261. 10.1186/s12870-021-03013-6. [DOI] [PMC free article] [PubMed]
- 25.Qin L, Tian D, Guo C, Wei L, He Z, Zhou W, Huang Q, Li B, Li C, Jiang M. Discovery of gene regulation mechanisms associated with uniconazole-induced cold tolerance in banana using integrated transcriptome and metabolome analysis. BMC Plant Biol. 2024;24(1):342. 10.1186/s12870-024-05027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Qin Y, Xie Z, Wang P, Zhao Z, Huang X, Chen Q, Huang Z, Chen Y, Gao A. Combined. transcriptome and metabolome analysis reveal that the white and yellow mango pulp colors are associated with carotenoid and flavonoid accumulation, and phytohormone signaling. Genomics. 2023;115(5):110675. 10.1016/j.ygeno.2023.110675. [DOI] [PubMed]
- 27.Zhang M, Su J, Zhang Y, Xu J, Zhang S. Conveying endogenous and exogenous signals: MAPK. cascades in plant growth and defense. Curr Opin Plant Biol. 2018;45(Pt A):1–10. 10.1016/j.pbi.2018.04.012. [DOI] [PubMed]
- 28.Lee JS, Wang S, Sritubtim S, Chen JG, Ellis BE. Arabidopsis mitogen-activated protein kinase. MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J. 2009; 57(6):975–85. 10.1111/j.1365-313X.2008.03741.x. [DOI] [PubMed]
- 29.Komorisono M, Ueguchi-Tanaka M, Aichi I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M, Sazuka T. Analysis of the rice mutant dwarf and gladius leaf 1. Aberrant katanin-mediated microtubule organization causes up-regulation of gibberellin biosynthetic genes independently of gibberellin signaling. Plant Physiol. 2005;138(4):1982–93. 10.1104/pp.105.062968. [DOI] [PMC free article] [PubMed]
- 30.Shani E, Hedden P, Sun TP. Highlights in gibberellin research: A tale of the dwarf. and the slender. Plant Physiol. 2024;195(1):111–34. 10.1093/plphys/kiae044. [DOI] [PMC free article] [PubMed]
- 31.Ueguchi-Tanaka M. Gibberellin metabolism and signaling. Biosci Biotechnol Biochem. 2023;87(10):1093–101. 10.1093/bbb/zbad090. [DOI] [PubMed] [Google Scholar]
- 32.Hernández-García J, Briones-Moreno A, Blázquez MA. Origin and evolution of gibberellin. signaling and metabolism in plants. Semin Cell Dev Biol. 2021;109:46–54. 10.1016/j.semcdb.2020.04.009. [DOI] [PubMed]
- 33.Xie Z, Jin L, Sun Y, Zhan C, Tang S, Qin T, Liu N, Huang J. OsNAC120 balances plant growth and drought tolerance by integrating GA and ABA signaling in rice. Plant Commun. 2024;5(3):100782. 10.1016/j.xplc.2023.100782. [DOI] [PMC free article] [PubMed]
- 34.Tian Z, Chen B, Li H, Pei X, Sun Y, Sun G, Pan Z, Dai P, Gao X, Geng X, Peng Z, Jia Y, Hu D, Wang L, Pang B, Zhang A, Du X, He S. The strigolactone-gibberellin crosstalk mediated by a distant silencer fine-tunes plant height in upland cotton. Mol Plant. 2024;S1674–2052(24):00262–4. 10.1016/j.molp.2024.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Zhang J, Guo X, Qiu L, Chen K, Wang J, Cheng T, Zhang Q, Zheng T. Genome-Wide Analysis of the Gibberellin-Oxidases Family Members in Four PrunusSpecies and a Functional Analysis of PmGA2ox8 in Plant Height. Int J Mol Sci. 2024;25(16):8697. 10.3390/ijms25168697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Y, Fan Y, Huang L, Lu H, Tan B, Ramirez C, Xia C, Niu X, Chen S, Gao M, Zhang C, Liu Y, Xiao F. The SINA1-BSD1 Module Regulates Vegetative Growth Involving Gibberellin Biosynthesis in Tomato. Adv Sci (Weinh). 2024:e2400995. 10.1002/advs.202400995. [DOI] [PMC free article] [PubMed]
- 37.Li WF, Zhou Q, Ma ZH, Zuo CW, Chu MY, Mao J, Chen BH. Regulatory. mechanism of GA3 application on grape (Vitis vinifera L.) berry size. Plant Physiol Biochem. 2024;210:108543. 10.1016/j.plaphy.2024.108543. [DOI] [PubMed]
- 38.Boss PK, Thomas MR. Association of dwarfism and floral induction with a grape ‘green revolution’. mutation. Nature. 2002; 416(6883):847–50. 10.1038/416847a. [DOI] [PubMed]
- 39.Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416(6882):701–2. 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 40.Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y, Harberd NP, Fu X. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature. 2018;560(7720):595–600. 10.1038/s41586-018-0415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van De Velde K, Thomas SG, Heyse F, Kaspar R, Van Der Straeten D, Rohde A. N-terminal. truncated RHT-1 proteins generated by translational reinitiation cause semi-dwarfing of wheat Green Revolution alleles. Mol Plant.2021;14(4):679–87. 10.1016/j.molp.2021.01.002. [DOI] [PubMed]
- 42.Song L, Liu J, Cao B, Liu B, Zhang X, Chen Z, Dong C, Liu X, Zhang Z, Wang W, Chai L, Liu J, Zhu J, Cui S, He F, Peng H, Hu Z, Su Z, Guo W, Xin M, Yao Y, Yan Y, Song Y, Bai G, Sun Q, Ni Z. Reducing brassinosteroid signalling enhances grain yield in semi-dwarf wheat. Nature. 2023;617(7959):118–24. 10.1038/s41586-023-06023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Ding L, Song A, Li S, Liu J, Zhao W, Jia D, Guan Y, Zhao K, Chen S, Jiang J, Chen F. DWARF AND ROBUST PLANT regulates plant height via modulating gibberellin biosynthesis in chrysanthemum. Plant Physiol. 2022;190(4):2484–500. 10.1093/plphys/kiac437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu T, Liu B, Liu N, Xu J, Song X, Li S, Sui S. Gibberellin-related genes regulate dwarfing. mechanism in wintersweet. Front Plant Sci. 2022; 13:1010896. 10.3389/fpls.2022.1010896. [DOI] [PMC free article] [PubMed]
- 45.Wang Y, Wang X, Deng D, Wang Y. Maize transcriptomic repertoires respond to gibberellin. stimulation. Mol Biol Rep. 2019; 46(4):4409–21. 10.1007/s11033-019-04896-3. [DOI] [PubMed]
- 46.Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is. regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14(1):57–70. 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed]
- 47.Tomlinson L, Yang Y, Emenecker R, Smoker M, Taylor J, Perkins S, Smith J, MacLean D, Olszewski NE, Jones JDG. Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnol J. 2019;17(1):132–40. 10.1111/pbi.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun H, Cui H, Zhang J, Kang J, Wang Z, Li M, Yi F, Yang Q, Long R. Gibberellins Inhibit Flavonoid. Biosynthesis and Promote Nitrogen Metabolism in Medicago truncatula. Int J Mol Sci. 2021;22(17):9291. 10.3390/ijms22179291. [DOI] [PMC free article] [PubMed]
- 49.Zang X, Liu J, Zhao J, Liu J, Ren J, Li L, Li X, Yang D. Uncovering mechanisms governing stem. growth in peanut (Arachis hypogaea L.) with varying plant heights through integrated transcriptome and metabolomics analyses. J Plant Physiol. 2023;287:154052. 10.1016/j.jplph.2023.154052. [DOI] [PubMed]
- 50.Zhao X, Sun XF, Zhao LL, Huang LJ, Wang PC. Morphological, transcriptomic and metabolomic. analyses of Sophora davidii mutants for plant height. BMC Plant Biol. 2022;22(1):144. 10.1186/s12870-022-03503-1. [DOI] [PMC free article] [PubMed]
- 51.Qin L, Li C, Guo C, Wei L, Tian D, Li B, Wei D, Zhou W, Long S, He Z, Huang S, Wei S. Integrated. metabolomic and transcriptomic analyses of regulatory mechanisms associated with uniconazole-induced dwarfism in banana. BMC Plant Biol. 2022; 22(1):614. 10.1186/s12870-022-04005-w. [DOI] [PMC free article] [PubMed]
- 52.Ren Y, Zhang S, Xu T, Kang X. Morphological, Transcriptome, and Hormone Analysis of. Dwarfism in Tetraploids of Populus alba × P. glandulosa. Int J Mol Sci. 2022;23(17):9762. 10.3390/ijms23179762. [DOI] [PMC free article] [PubMed]
- 53.Lloret A, Quesada-Traver C, Conejero A, Arbona V, Gómez-Mena C, Petri C, Sánchez-Navarro JA, Zuriaga E, Leida C, Badenes ML, Ríos G. Regulatory circuits involving bud dormancy factor PpeDAM6. Hortic Res. 2021;8(1):261. 10.1038/s41438-021-00706-9. [DOI] [PMC free article] [PubMed]
- 54.Best N, Dilkes B. Genetic evidence that brassinosteroids suppress pistils in the maize. tassel independent of the jasmonic acid pathway. Plant Direct. 2023;7(7):e501. 10.1002/pld3.501. [DOI] [PMC free article] [PubMed]
- 55.Huang Y, Li X, Duan Z, Li J, Jiang Y, Cheng S, Xue T, Zhao F, Sheng W, Duan Y. Ultra-low concentration of chlorine dioxide regulates stress-caused premature leaf senescence in tobacco by modulating auxin, ethylene, and chlorophyll biosynthesis. Plant Physiol Biochem. 2022;186:31–9. 10.1016/j.plaphy.2022.06.029. [DOI] [PubMed]
- 56.Zhang H, He Y, Tan X, Xie K, Li L, Hong G, Li J, Cheng Y, Yan F, Chen J, Sun Z. The. Dual Effect of the Brassinosteroid Pathway on Rice Black-Streaked Dwarf Virus Infection by Modulating the Peroxidase-Mediated Oxidative Burst and Plant Defense. Mol Plant Microbe Interact. 2019;32(6):685–96. 10.1094/MPMI-10-18-0285-R. [DOI] [PubMed]
- 57.Borrill P, Mago R, Xu T, Ford B, Williams SJ, Derkx A, Bovill WD, Hyles J, Bhatt D, Xia. X, MacMillan C, White R, Buss W, Molnár I, Walkowiak S, Olsen OA, Doležel J, Pozniak CJ, Spielmeyer W. An autoactive NB-LRRgene causes Rht13 dwarfism in wheat. Proc Natl Acad Sci U S A. 2022;119(48):e2209875119. 10.1073/pnas.2209875119. [DOI] [PMC free article] [PubMed]
- 58.Habib S, Lwin YY, Li N. Down-Regulation of SlGRAS10in Tomato Confers Abiotic Stress. Tolerance Genes (Basel). 2021;12(5):623. 10.3390/genes12050623. [DOI] [PMC free article] [PubMed]
- 59.Yan MY, Xie DL, Cao JJ, Xia XJ, Shi K, Zhou YH, Zhou J, Foyer CH, Yu JQ. Brassinosteroid-mediated reactive oxygen species are essential for tapetum degradation and pollen fertility in tomato. Plant J. 2020;102(5):931–47. 10.1111/tpj.14672. [DOI] [PubMed]
- 60.Chen Y, Hou M, Liu L, Wu S, Shen Y, Ishiyama K, Kobayashi M, McCarty DR, Tan BC. The maize DWARF1 encodes a gibberellin 3-oxidase and is dual localized to the nucleus and cytosol. Plant Physiol. 2014;166(4):2028–39. 10.1104/pp.114.247486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. Cloning and. functional analysis of two gibberellin 3 beta-hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci U S A. 2001; 98(15):8909–14. 10.1073/pnas.141239398. [DOI] [PMC free article] [PubMed]
- 62.Hao XH, Hu S, Zhao D, Tian LF, Xie ZJ, Wu S, Hu WL, Lei H, Li. DP. OsGA3ox genes regulate rice fertility and plant height by synthesizing diverse active GA. Yi Chuan. 2023;45(9):845–55. 10.16288/j.yczz.23-058. [DOI] [PubMed]
- 63.Kang HG, Jun SH, Kim J, Kawaide H, Kamiya Y, An G. Cloning of gibberellin 3 beta-hydroxylase. cDNA and analysis of endogenous gibberellins in the developing seeds in watermelon. Plant Cell Physiol. 2002; 43(2):152–8. 10.1093/pcp/pcf016. [DOI] [PubMed]
- 64.Pimenta Lange MJ, Liebrandt A, Arnold L, Chmielewska SM, Felsberger A, Freier E, Heuer M, Zur D, Lange T. Functional characterization of gibberellin oxidases from cucumber. Cucumis sativus L Phytochemistry. 2013;90:62–9. 10.1016/j.phytochem.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 65.Pimenta Lange MJ, Knop N, Lange T. Stamen-derived bioactive gibberellin is essential for male. flower development of Cucurbita maxima L. J Exp Bot. 2012;63(7):2681–91. 10.1093/jxb/err448. [DOI] [PMC free article] [PubMed]
- 66.Sato A, Yamamoto KT. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant. 2008;133(2):397–405. 10.1111/j.1399-3054.2008.01055.x. [DOI] [PubMed] [Google Scholar]
- 67.Dalmadi A, Kaló P, Jakab J, Saskoi A, Petrovics T, Deák G, Kiss GB. Dwarf plants of diploid Medicago sativa carry a mutation in the gibberellin 3-beta-hydroxylase gene. Plant Cell Rep. 2008;27(8):1271–9. 10.1007/s00299-008-0546-5. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Huang GD, Rong T, Zhang J, Zheng J, Zhao Y, Tang YJ, Zhao ZC. Cloning, Subcellular Localization and Expression analysis of GA3ox Gibberellin Oxidase Gene in Mango. Molecular Plant Breeding. 2023;21(04):1111–6. 10.13271/j.mpb.021.001111.
- 69.Nakamura A, Umemura I, Gomi K, Hasegawa Y, Kitano H, Sazuka T, Matsuoka M. Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 2006;46(2):297–306. 10.1111/j.1365-313X.2006.02693.x. [DOI] [PubMed] [Google Scholar]
- 70.Song Y, Xu ZF. Ectopic overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) gene OsIAA4 in rice induces morphological changes and reduces responsiveness to Auxin. Int J Mol Sci. 2013;14(7):13645–56. 10.3390/ijms140713645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao D, Wang Y, Feng C, Wei Y, Peng X, Guo X, Guo X, Zhai Z, Li J, Shen X, Li T. Overexpression of MsGH3.5 inhibits shoot and root development through the auxin and cytokinin pathways in apple plants. Plant J. 2020;103(1):166–83. 10.1111/tpj.14717. [DOI] [PubMed]
- 72.Li Z, Zhang X, Zhao Y, Li Y, Zhang G, Peng Z, Zhang J. Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol J. 2018;16(1):86–99. 10.1111/pbi.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory. requirements. Nat Methods. 2015;12(4):357–60. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed]
- 75.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for. assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. 10.1093/bioinformatics/btt656. [DOI] [PubMed]
- 76.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for. RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed]
- 77.Glauser G, Grund B, Gassner AL, Menin L, Henry H, Bromirski M, Schütz F, McMullen. J, Rochat B. Validation of the Mass-Extraction-Window for Quantitative Methods Using Liquid Chromatography High Resolution Mass Spectrometry. Anal Chem. 2016; 88(6):3264–71. 10.1021/acs.analchem.5b04689. [DOI] [PubMed]
- 78.Wang Q, Cai WJ, Yu L, Ding J, Feng YQ. Comprehensive Profiling of Phytohormones in. Honey by Sequential Liquid-Liquid Extraction Coupled with Liquid Chromatography-Mass Spectrometry. J Agric Food Chem. 2017;65(3):575–85. 10.1021/acs.jafc.6b04234. [DOI] [PubMed]
- 79.Chong J, Yamamoto M, Xia J. MetaboAnalystR 2.0: From Raw Spectra to Biological. Insights. Metabolites. 2019;9(3):57. 10.3390/metabo9030057. [DOI] [PMC free article] [PubMed]
- 80.Balcke GU, Handrick V, Bergau N, Fichtner M, Henning A, Stellmach H, Tissier A, Hause. B, Frolov A. An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods. 2012;8(1):47. 10.1186/1746-4811-8-47. [DOI] [PMC free article] [PubMed]
- 81.Willems E, Hu TT, Soler Vasco L, Buyse J, Decuypere E, Arckens L, Everaert N. Embryonic protein undernutrition by albumen removal programs the hepatic amino acid and glucose metabolism during the perinatal period in an avian model. PLoS ONE. 2014;9(4):e94902. 10.1371/journal.pone.0094902. [DOI] [PMC free article] [PubMed]
- 82.Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2ˆ(-delta delta CT) method for. quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013;3(3):71–85. [PMC free article] [PubMed]
- 83.Albaseer SS, Rao RN, Swamy YV, Mukkanti K. An overview of sample preparation and. extraction of synthetic pyrethroids from water, sediment and soil. J Chromatogr A. 2010;1217(35):5537–54. 10.1016/j.chroma.2010.06.058. [DOI] [PubMed]
- 84.Gao, J. Experimental Instruction In Plant Physiology (Higher Education Press, 2006).
- 85.Li Li, Changbao Li, Jian Sun, Ming Xin, Ping Yi, Xuemei He, Jinfeng Sheng, Zhugui Zhou, Dongning Ling, Fengjin Zheng, Jiemin Li, Guoming Liu, Zhichun Li, Yayuan Tang, Ying Yang, Jie Tang. Synergistic effects of ultraviolet light irradiation and high-oxygen. modified atmosphere packaging on physiological quality, microbial growth and lignification metabolism of fresh-cut carrots. Postharvest Biology and Technology. 2021;173,111365. 10.1016/j.postharvbio.2020.111365.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available in the NCBI SRA repository (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA1026664).