Abstract
Background
Asthma poses a significant global health challenge, characterized by high rates of morbidity and mortality. Despite available treatments, many severe asthma patients remain poorly managed, highlighting the need for novel therapeutic strategies. This study aims to identify potential drug targets for asthma by examining the influence of circulating plasma proteins on asthma risk.
Methods
This study employs summary-data-based Mendelian randomization (MR) and two-sample MR methods to investigate the association between 2940 plasma proteins from the UK Biobank study and asthma. The analysis includes discovery (FinnGen cohort) and replication (GERA cohort) phases, with Bayesian colocalization used to validate the relationships between proteins and asthma. Furthermore, protein–protein interaction and druggability assessments were conducted on high-evidence strength protein biomarkers, and candidate drug prediction and molecular docking were performed for proteins without targeted drugs. Given the complexity of asthma pathogenesis, the study also explores the relationships between plasma proteins and asthma-related endpoints (e.g., obesity-related asthma, infection-related asthma, childhood asthma) to identify potential therapeutic targets for different subtypes.
Results
In the discovery cohort, 75 plasma proteins were associated with asthma, including IL1RAP, IL1RL1, IL6, CXCL5, and CXCL8. Additionally, 6 proteins (IL4R, LTB, CASP8, MAX, PCDH12, and SCLY) were validated through co-localization analysis and validation cohort. The assessment of drug targetability revealed potential drug targets for IL4R, CASP8, and SCLY, while candidate drugs were predicted for LTB and MAX proteins. MAX exhibited strong binding affinity with multiple small molecules indicating a highly stable interaction and significant druggability potential. Analysis of the 75 proteins with 9 asthma-related endpoints highlighted promising targets such as DOK2, ITGAM, CA1, BTN2A1, and GZMB.
Conclusion
These findings elucidate the link between asthma, its related endpoints, and plasma proteins, advancing our understanding of molecular pathogenesis and treatment strategies. The discovery of potential therapeutic targets offers new insights into asthma drug target research.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05782-8.
Keywords: Asthma, Drug target, Mendelian randomization, Plasma proteome
Background
Asthma is a chronic respiratory disease that affects millions of people around the world, which is one of the major global public health problems, with high morbidity and mortality rates and a heavy economic burden [1–3]. Despite the availability of several therapeutic options, a subset of patients with severe asthma remains refractory to conventional treatments [4, 5]. In addition to traditional pharmacological methods, there have been efforts to investigate the potential of natural products and traditional Chinese medicine for treating asthma and other diseases [6–9]. However, the clinical applications of these approaches are limited [10]. In recent years, the introduction of biologics has provided new therapeutic options for asthma management. By specifically targeting the inflammatory pathways involved in the disease’s pathogenesis, biological therapies have demonstrated effectiveness in controlling exacerbations, reducing side effects, and improving the quality of life for selected patients [11]. For instance, real-world evaluations for children, adolescents, and young adults show that monoclonal antibodies like Omalizumab significantly enhance asthma control, nearly eliminating seasonal acute attacks and reducing dependence on additional medications [12].
However, targeted therapies for asthma face significant challenges. Currently, FDA-approved biologics primarily target downstream pathways of type 2 (T2) inflammation, with many of these drugs mainly benefiting adult patients [11]. Moreover, there is a lack of effective treatment for non-eosinophilic asthma [13]. Furthermore, the mechanisms underlying neutrophilic inflammation associated with non-T2 pathways, as well as their relationship with interleukin (IL) 17 (IL-17) and IL-8 in asthma, are not yet well understood, leading to the unsuccessful development of relevant biologics [14]. Therefore, there is an urgent need to identify new potential therapeutic targets for asthma.
In recent years, with the development of sequencing technologies, genome-wide association studies (GWAS) have identified genetic variants associated with plasma protein levels, known as protein quantitative trait locus (pQTLs) [15]. These findings provide an opportunity to use Mendelian randomization (MR) to study the causal impact of potential drug targets on the phenome of human diseases, including asthma. To minimize confounding factors and examine potential causal associations between risk factors (such as plasma proteins) and disease outcomes (such as asthma), MR uses genetic variants as instrumental variables to infer causality [16, 17]. Additionally, colocalization methods can help identify potential functional variants and regulatory elements, further revealing the associations between genomic variation and phenotype [18].
In this study, we analyzed 2940 plasma proteins from the UK Biobank (UKBB) alongside asthma GWAS data from the FinnGen cohort. MR and colocalization analysis were employed, with validation using the GERA cohort, to identify potential therapeutic targets. Considering the complexity of asthma pathogenesis, the study also explored the association between proteins and different asthma subtypes, aiming to provide new insights for drug target development in asthma treatment.
Methods
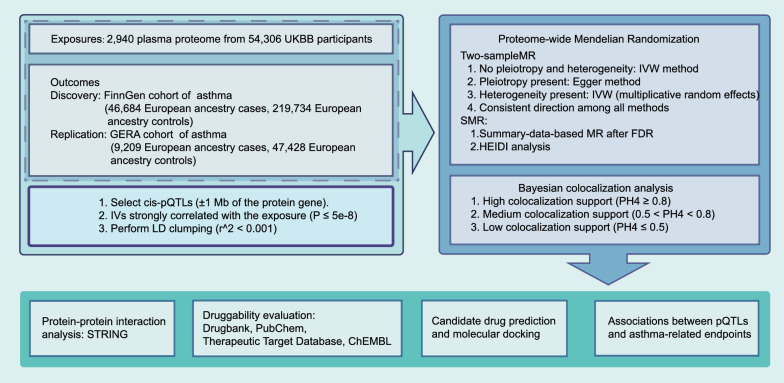
As illustrated in Fig. 1, the study design aims to investigate the association between protein pQTLs and asthma. The study establishes causal relationships by using summary-data-based MR (SMR) and two-sample MR methods, including discovery and replication phases. Bayesian colocalization is used to validate the relationships between protein biomarkers and asthma. Subsequently, to investigate the interactions of the identified protein biomarkers and assess whether the target proteins have the potential to serve as drug targets, protein–protein interaction (PPI) analysis, and druggability evaluations will be conducted. Lastly, to precisely identify potential therapeutic targets for different subtypes, we will analyze the relationships between pQTLs and asthma-related endpoints using the previously described methods, following the identification of relevant proteins associated with asthma.
Fig. 1.
Study design illustrating the investigation of the association between protein pQTLs (protein quantitative trait locus) and asthma
Study population and data resource
The protein data utilized in this study were sourced from participants enrolled in the UKBB. Employing the antibody-based Olink Explore 3072 PEA [19], protein profiling was performed on plasma samples collected from 54,306 UKBB participants. This comprehensive analysis encompassed 2,923 unique proteins. The discovery and validation of GWAS summary data about asthma were derived from the independent FinnGen cohort (46,684 cases, 219,734 controls) [20] and the GERA cohort (9,209 cases, 47,428 controls) [21]. Additionally, GWAS results for asthma-related endpoints were sourced from FinnGen. Importantly, there is no overlap between these cohorts. It is significant to note that all GWAS participants were of European ancestry and provided informed consent, with the study receiving ethical approval from the relevant authorities. Detailed information regarding these datasets can be found in Table S1.
Proteome-wide MR analysis
This study utilized the "TwoSampleMR" package for MR analysis, as with all MR analyses, there are three core assumptions, including relevance, independence, and exclusion restriction [22]. The selection of instrumental variables followed these steps: (1) Selection of cis-pQTLs, where the SNP is located within 1 Mb of the transcription region of the protein-coding gene. (2) Selection of SNPs associated with protein (P ≤ 5 × 10−8). (3) LD clumping was performed to determine independent pQTLs for each protein (r2 < 0.001), using reference data from the 1000 Genomes Project of European ancestry.
A variety of analytical methods were employed in this study. The inverse-variance weighted (IVW) method was used for proteins with multiple instruments to estimate MR effects. For proteins with only one instrument, the Wald ratio method was used to calculate the log odds change in risk per standard deviation increment of circulating protein levels. Additionally, other methods such as simple mode, weighted mode, weighted median, MR-Egger, and IVW multiplicative random effects were applied. The MR-Egger method was used to address horizontal pleiotropy when indicated by the intercept [19]. In the presence of heterogeneity, IVW multiplicative random effects were employed for evaluation. Results were deemed valid if they showed consistent effect directions across all methods.
Furthermore, SMR analysis was used as a complementary method to explore the causal relationship between proteins and asthma. Unlike traditional MR, the SMR and HEIDI methods utilize summary-level data from GWAS and pQTLs studies to test whether the protein and phenotype are correlated due to shared causal variants [23]. To combine the findings from MR and illustrate the causality of the results, positive results from either two-sample MR or SMR were considered indicative of causal effects. SMR software (SMR v1.3.1) was used for SMR and HEIDI tests. The false discovery rate (FDR) based on the Benjamini–Hochberg method with a threshold of α = 0.05 was used for multiple testing corrections [24].
Bayesian colocalization analysis
To assess the concordance of two associated signals with a shared causal variant, Bayesian colocalization analysis was conducted using coloc.abf and coloc.susie functions from the "coloc" package [25, 26]. This approach allows the assessment of hypotheses for single and multiple causal variables. We tested five hypotheses, with particular emphasis on the fourth hypothesis (PH4) which suggests a shared causal variant for both the protein and the GWAS (asthma). Default parameters were used for the analysis. High support for colocalization was defined as PH4 ≥ 0.8, medium support as 0.5 < PH4 < 0.8, and low support as PH4 ≤ 0.5 [27].
PPI analysis
After identifying numerous causally related proteins through MR, we conducted a comprehensive PPI analysis and functional enrichment analysis to explore the direct (physical) and indirect (functional) interactions between the identified proteins and verify their functional relevance in the pathogenesis of asthma. Bioinformatics tools and databases, such as STRING [28] (https://string-db.org/), were used to systematically construct and analyze the PPI networks of the relevant proteins. To ensure the reliability of the interactions, we applied a stringent threshold, setting the combined PPI score above 0.7 to prioritize high-confidence protein interactions.
Druggability evaluation
Several databases, including DrugBank [29], PubChem [30], Therapeutic Target Database [31], and ChEMBL [32], were queried to assess the current status of existing therapies targeting the identified proteins with co-localization evidence. The drugs were categorized into three groups: approved, investigational/experimental, and not found. Detailed information regarding drug names and their developmental stages targeting the identified proteins was compiled for comprehensive analysis.
Candidate drug prediction and molecular docking
After evaluating the current status of drug development for the target proteins, the study used the Drug Signatures Database (DSigDB) [33] to predict potential drug candidates for the identified proteins lacking targeted therapies. Subsequently, molecular docking analyzed binding affinity and interaction patterns between potential drug candidates and therapeutic targets, aiming to identify ligands with high binding affinities and favorable interaction patterns for potential therapeutic candidates and pharmacological targets [34]. Autodock 4.2.6 was employed for this analysis. Drug structural data were obtained from PubChem, and protein structural information was retrieved from the Protein Data Bank [35]. To further validate the robustness of this method, we conducted docking experiments involving classical asthma protein targets, including the beta-2 adrenergic receptor, glucocorticoid receptor, adenosine receptor A1, and muscarinic acetylcholine receptor M1, along with their corresponding existing therapeutic drugs, such as formoterol, hydrocortisone, theophylline, and methacholine to compare the binding energies between our predicted drugs and their targets.
Results
Associations between plasma proteins and asthma
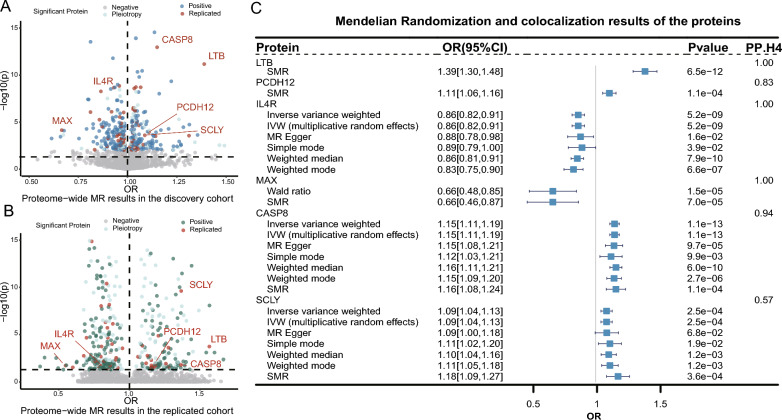
This study systematically investigated the causal relationship between 2940 proteins and asthma risk using MR analysis. In the discovery cohort (FinnGen cohort), MR analysis covered 1997 proteins after excluding plasma proteins lacking genetic instruments. Two-sample MR and SMR were employed for the analysis. All instrumental variables had F-values exceeding 30, indicating their reliability and effectiveness in capturing genetic associations. After FDR correction, it was found that 301 proteins were genetically associated with asthma, including interleukin-1 receptor accessory protein (IL1RAP), interleukin-1 receptor-like 1 (IL1RL1), IL6, C-X-C motif chemokine 5 (CXCL5), and C-X-C motif chemokine (CXCL8). Detailed results are provided in Fig. 2A and Table S2, offering a comprehensive understanding of the identified relationships.
Fig. 2.
A A volcano plot illustrating the proteome-wide Mendelian randomization (MR) results in the asthma discovery cohort (FinnGen cohort). The x-axis represents the odds ratio of MR results, while the y-axis represents the -log10P of MR results. Gray points represent non-significant results (P > 0.05, after FDR correction), light green points indicate pleiotropic loci (P < 0.05, after FDR correction, but with pleiotropy), blue points indicate significant loci (P < 0.05, after FDR correction), and red points indicate proteins validated in the validation cohort and after colocalization validation. B A Volcano plot illustrating the proteome-wide Mendelian randomization (MR) results in the asthma validation cohort (GERA cohort). The x-axis represents the odds ratio of MR results, while the y-axis represents the -log10P of MR results. Gray points represent non-significant results (P > 0.05, after FDR correction), light green points indicate pleiotropic loci (P < 0.05, after FDR correction, but with pleiotropy), green points indicate significant loci (P < 0.05, after FDR correction), and red points indicate proteins that are significant in both the discovery and validation cohorts and after colocalization validation. C The forest plots for 6 proteins in Mendelian randomization analysis of asthma. Each forest plot represents the effect size of different proteins under each method. The horizontal line represents the 95% confidence interval, reflecting the credibility range of the effect size. The point represents the effect size of the gene (odds ratio). PP.H4 represents a certain confidence level, possibly the posterior probability of hypothesis 4, indicating the probability that a locus simultaneously affects two features
To ensure the robustness and validity of these findings, validation was conducted using the GERA cohort, as outlined in Table S3. In this validation cohort, a broad assessment was conducted on 1797 proteins after excluding pQTLs lacking instrumental variables, with 392 proteins demonstrating significance. Among the proteins identified in the discovery cohort, 75 were successfully validated, indicating their reproducibility and reliability (P < 0.05). The results are shown in Fig. 2B. Subsequently, co-localization analysis was performed for these 75 validated proteins, revealing the relationships of shared causal variations among the 6 proteins associated with asthma. The effects of these 6 proteins are detailed in Fig. 2C. Specifically, we found 5 proteins with high-level co-localization support, namely interleukin-4 receptor subunit alpha (IL4R), lymphotoxin-beta (LTB), caspase 8 (CASP8), protein max (MAX), and protocadherin 12 (PCDH12). The sum of the single effects (SuSiE) method also provided strong evidence of colocalization for the associations of IL4R and MAX. Additionally, the selenocysteine lyase (SCLY) protein displayed medium-level co-localization support, while the remaining 69 proteins all showed low-level evidence of co-localization support. Further details can be found in Table S4 and Table S5.
PPI evidence
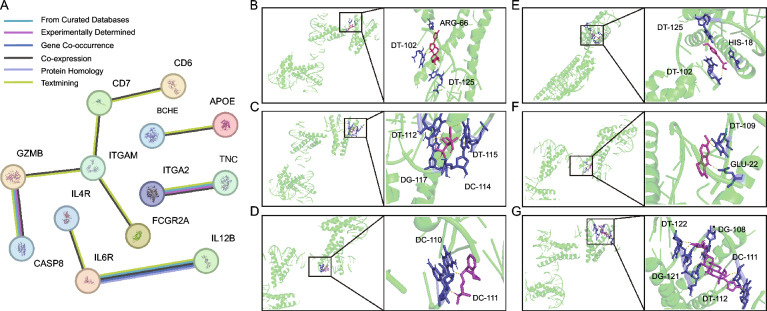
The PPI network revealed high-confidence interactions among the 75 proteins associated with asthma, including co-expression and protein homology, as illustrated in Fig. 3A, these proteins are involved in multiple shared biological functions. KEGG pathway enrichment analysis further demonstrated that these proteins are significantly enriched in pathways such as hematopoietic cell lineage, phagosome, complement, and coagulation cascades. This suggests that they play crucial roles in immune cell development and function, inflammatory responses, blood coagulation, and cellular phagocytosis. Please refer to Table S6 for specific results.
Fig. 3.
A The network of protein–protein interactions (PPI) results. A stringent threshold was set, with the comprehensive PPI score above 0.7. The network nodes represent proteins, while the edges indicate protein–protein associations. These associations are specific and meaningful, as the proteins collectively contribute to shared functions. B–G Docking results of MAX protein with small molecules. B MAX docking with digitoxigenin, C MAX docking with zoledronic acid D. MAX docking with deptropine E. MAX docked with methyl 4-methoxycinnamate, F MAX docking with harmaline, G. MAX docked with ouabain
Druggability evaluation
We then conducted a drug database search for six proteins with moderate to high co-localization support from the shared co-localization analysis with asthma, aiming to evaluate their therapeutic development status. Our search revealed that IL4R has two targeted drugs for asthma. Dupilumab, a monoclonal antibody, is already on the market for treating asthma in adolescents and adults. Additionally, AER001, an IL4/13 receptor antagonist used to treat severe asthma and eczema, is currently in phase 2 studies. Targeting the drug of CASP8 is primarily used for cancer treatment while targeting the drug of SCLY is mainly utilized for various supplements and vitamin supplements. However, no targeted drugs were found for PCDH12, LTB, and MAX in the searched drug database. Please refer to Table S7 for specific results.
Candidate drug prediction
The DSigDB database was used in this study to predict potentially effective intervention drugs for the proteins PCDH12, LTB, and MAX, for which no targeted drugs were found. The top 10 potential chemical compounds were identified based on adjusted P values, as shown in Table S8. Due to the unresolved macromolecular structure of LTB, Autodock was employed to determine the binding sites and interactions between the 6 drug candidates and the proteins encoded by MAX. Subsequently, we generated the corresponding binding energies for each interaction, as detailed in Table S9 and Fig. 3B-G. Notably, the MAX-digitoxigenin interaction exhibited the lowest binding energy of − 8.24 kcal/mol, reflecting a highly stable affinity.
To verify the stability of MAX and its predicted small molecule drugs, we docked classical asthma targets along with their corresponding small molecule drugs and calculated their binding energies. For instance, formoterol binds to the beta-2 adrenergic receptor with a binding energy of − 5.54 kcal/mol, while hydrocortisone interacts with annexin A1 at − 6.54 kcal/mol and the glucocorticoid receptor, demonstrating a strong affinity of − 11.94 kcal/mol, among others. These results indicate similar strong binding affinities, consistent with the binding energies for other medications predicted by MAX (ranging from − 4.2 kcal/mol to − 8.24 kcal/mol). Detailed results can be found in Table S10 and Supplementary Figure.
Associations between plasma proteins and asthma-related endpoints
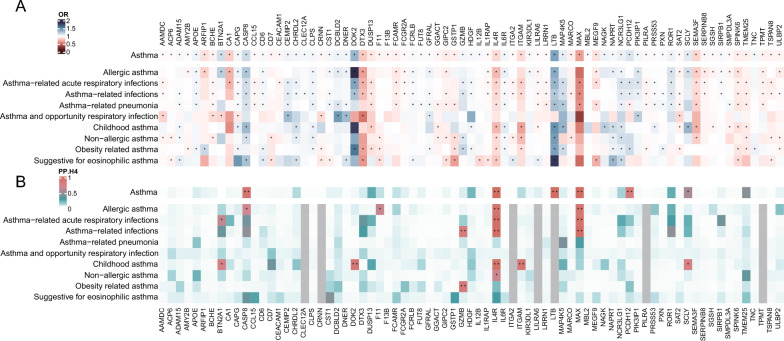
We conducted a series of detailed analyses on asthma-related endpoints following the identification of key asthma-associated proteins. Figure 4A summarizes the MR analysis results of 75 plasma proteins associated with asthma-related endpoints as detailed in Table S11. The study found a general consistency between the direction of association of these proteins with asthma and their association with asthma-related endpoints.
Fig. 4.
A Heatmap of Mendelian randomization results of 75 proteins with asthma and asthma-related endpoints. The color mapping represents the odds ratio, and asterisks indicate significant results. B Heatmap of colocalization results of 75 proteins with asthma and asthma-related endpoints. The color mapping represents the PH4, with one asterisk indicating medium colocalization support (0.8 > PH4 > 0.5), and two asterisks indicating high colocalization support (PH4 ≥ 0.8)
Further co-localization analysis, as depicted in Fig. 4B, showed that IL4R and MAX are moderate to strongly associated with most asthma-related endpoints, including allergic asthma, asthma-related acute respiratory infections, and asthma-related infections, as detailed in Table S12 and Table S13. In contrast, proteins such as docking protein 2 (DOK2), integrin alpha-M (ITGAM), and carbonic anhydrase 1 (CA1) exhibit moderate to strong co-localization evidence with childhood asthma in different methods, which differs from asthma. Additionally, granzyme B (GZMB) shows high co-localization evidence with obesity-related asthma. We found robust evidence that the following target proteins are associated with infection-related asthma: butyrophilin subfamily 2 member A1 (BTN2A1), and GZMB.
Discussion
In this study, proteome-wide MR and colocalization analysis were conducted using large-scale pQTLs data to investigate the roles of 2,940 circulating proteins in asthma. The results were replicated in an independent cohort to ensure robustness, identifying 75 asthma-associated proteins, including IL1RAP, IL1RL1, and IL6. Colocalization analysis further validated the associations between asthma and 6 proteins: IL4R, LTB, CASP8, MAX, PCDH12, and SCLY. Moreover, the relationships among these proteins were explored through protein–protein interactions, which revealed key functional connections. For proteins lacking targeted drugs such as LTB and MAX, potentially effective intervention drugs were predicted, and molecular docking. The results indicate that the binding energies of MAX and the predicted drugs are similar to those of classical asthma targets and their corresponding small molecule drugs, demonstrating significant potential for drug development. Additionally, MR and colocalization analysis results for these 75 proteins with 9 asthma-related endpoints were demonstrated, identifying proteins with strong evidence, including DOK2, ITGAM, CA1, BTN2A1, and GZMB.
The pathogenesis of asthma involves several aspects: airway inflammation (high T2 and low T2), airway hyperresponsiveness, and airway remodeling [36–39]. Our findings identified key proteins, including IL1RAP, IL1RL1, IL6, CXCL5, and CXCL8, which align with numerous previous studies [40–44]. Although these proteins were not replicated or colocalized in our study, their discovery remains significant for asthma drug target development. IL1RAP plays a critical role in high T2 inflammation, forming a complex with IL-33 and ST2 that activates T2 cytokine release, facilitates B-cell differentiation into IgE-producing cells, and promotes eosinophil proliferation, thereby establishing a T2 inflammatory environment in the lungs or other tissues [45]. Regarding low T2 inflammation, CXCL8 drives neutrophilic airway inflammation, likely a primary pathway for non-Th2 asthma inflammation. Inhibiting CXCL8/IL-8 may aid in treating virus-induced asthma exacerbations [46]. Furthermore, mouse studies have shown that a monoclonal antibody targeting IL-6 effectively treats airway inflammation and remodeling in severe asthma patients [44].
Additionally, we identified and validated associations between IL4R and MAX with asthma, along with several asthma-related endpoints, including allergic asthma, asthma-related acute respiratory infections, and asthma-associated infections. IL-4 has been extensively studied and is considered the primary switch for Th2 cells, driving these cells to produce other pro-allergic cytokines such as IL-5 and IL-13 [47, 48]. Therefore, interrupting the cascade reaction initiated by IL-4 is a therapeutic approach for asthma. Monoclonal antibodies targeting IL4R, such as Dupilumab, have been approved for asthma treatment [49, 50], confirming the therapeutic potential of this pathway in our study. MAX is a helix-loop-helix protein with limited pathway-related studies, but our molecular docking identified its strong druggability. Moreover, tissue-specific analysis shows high expression of MAX in the lungs [51], perhaps indicating its complex association with asthma pathogenesis, warranting further exploration. Furthermore, some strong evidence for potential targeted proteins was identified. A study revealed that LTBR expressed in airway smooth muscle cells can activate sustained signaling pathways involved in smooth muscle remodeling, lung function impairment, and antigen-induced airway hyper-responsiveness [52]. Further research is needed to elucidate the molecular actions of LTB due to its unresolved structure. In addition, CASP8 was found to be associated with severe asthma, including severe T2-high and non-T2-high asthma phenotypes [53]. It may be a potential target for severe asthma treatment. Studies further indicate differences in gender distribution, exacerbating factors, associated complications, severity, and disease progression mechanisms among distinct asthma phenotypes [54, 55]. Therefore, results regarding protein associations with various asthma subtypes can guide the development of specific targeted therapies. Our results indicate a strong colocalization signal between GZMB and obesity-related asthma. Genetic deletion of granzyme B in mice can alleviate the severity of contact dermatitis and improve impaired wound healing associated with calorie and diabetes-related injuries, demonstrating developmental potential for GZMB in obesity-related asthma. Additionally, our analysis revealed an association between childhood asthma and the proteins DOK2, ITGAM, and CA1. ITAM integrin ITGAM/ITGB2 is associated with various adhesive interactions of monocytes, macrophages, and granulocytes, mainly manifested in skin and mucosal involvement [56]. This finding may support the view that childhood asthma is caused by impaired skin and other epithelial surface barrier functions [57, 58].
However, our study has some limitations. Despite excluding biases caused by cascade imbalance through colocalization analysis, MR analysis is susceptible to unmeasured factors and pleiotropy [59, 60]. Additionally, the whole-genome association study data for exposures and outcomes we used are from European populations. Although limiting the study population can reduce population structure bias, it also limits the generalizability of our results to other populations. Furthermore, this study primarily focused on circulating proteins in plasma, potentially overlooking the effects of alternative treatments. Despite these limitations, our study provides a reliable analysis of large-scale plasma proteins associated with asthma. Future research could further explore the exact roles of these proteins in asthma pathogenesis and the expression differences in different asthma subtypes. Moreover, the feasibility of these proteins as potential drug targets and more experimental validation could be explored. Integrating other omics data (such as transcriptomics and metabolomics) and clinical data could provide a more comprehensive understanding of asthma pathogenesis and personalized treatment strategies. Ultimately, we hope these efforts may contribute to the development of personalized asthma treatment and precision medicine.
Conclusions
These findings illuminate the connection between asthma, its related endpoints, and plasma proteins, advancing our understanding of the molecular pathogenesis and treatment strategies for asthma. This provides unique insights into exploring potential drug targets for asthma and offers ideas for future research on personalized treatment approaches and precision medicine.
Supplementary Information
Supplementary Material 1. TableS1. Source information for genome-wide association studies. TableS2. Proteome-wide Mendelian randomization results in the asthma discovery cohort. TableS3. Proteome-wide Mendelian randomization results in the asthma replicated cohort. TableS4. Colocalization results in the asthma discovery cohort. TableS5. Sussie results in the asthma discovery cohort. TableS6. KEGG pathways involving the 75 proteins associated with asthma. TableS7. Identification of druggable targets. TableS8. Candidate drug predicted using DSigDB. TableS9. Docking results of MAX protein with small molecules. TableS10. Docking results of traditional targets and their small molecule therapeutic drugs. TableS11. Mendelian randomization results in the asthma-related endpoints. TableS12. Colocalization results in the asthma-related endpoints. TableS13. Susie method results in asthma-related endpoints
Supplementary Material 2. Supplementary Figure: Docking results of traditional targets and their small molecule therapeutic drugs
Acknowledgements
We are grateful for the efforts of consortia mentioned in Table S1 in providing high-quality public GWAS data available for download.
Abbreviations
- T2
type 2
- IgE
Immunoglobulin E
- IL
Interleukin
- GWAS
Genome-wide association studies
- pQTLs
Quantitative trait locus
- MR
Mendelian randomization
- UKBB
UK Biobank
- PPI
Protein–protein interaction
- IVW
Inverse-variance weighted
- FDR
False discovery rate
- PH4
fourth hypothesis
- DSigDB
Drug Signatures Database
- IL1RAP
Interleukin-1 receptor accessory protein
- IL1RL1
Interleukin-1 receptor-like 1
- CXCL5
C-X-C motif chemokine 5
- CXCL8
C-X-C motif chemokine 8
- IL4R
Interleukin-4 receptor subunit alpha
- LTB
Lymphotoxin-beta
- CASP8
Caspase 8
- MAX
Protein max
- PCDH12
Protocadherin 12
- SCLY
Selenocysteine lyase
- SuSiE
Sum of the single effects
- DOK2
Docking protein 2
- ITGAM
Integrin alpha-M
- CA1
Carbonic anhydrase 1
- GZMB
Granzyme B
- BTN2A1
Butyrophilin subfamily 2 member A1
Author contributions
Y. J., Y.W., and J.G. wrote the manuscript and analyzed the data. Z.W., X.W. and X.Y. performed the research. Y.Z. and H.Y. designed the research.
Funding
This study was supported by the Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-040A), the Tianjin Municipal Health Commission Key Discipline Special Fund (TJWJ2022XK038) and the National Natural Science Foundation of China (grant number: 72104179).
Availability of data and materials
The GWAS summary data for plasma proteins and asthma analyzed in this study, including findings from UKBB, FinnGen, and GERA, are included in Table S1.
Declarations
Ethics approval and consent to participate
The UKBB has ethical approval from the North West Multi-Centre Research Ethics Committee (MREC), which covers the UK, and all participants provided written informed consent. This project received ethical approval from the Institutional Human Research Ethics Committee, University of Queensland.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yuhan Jiang, Yifan Wang and Ju Guo share equal contribution.
Contributor Information
Hongxi Yang, Email: yanghongxi@tmu.edu.cn.
Yingxue Zou, Email: zouyingxue2015@tju.edu.cn.
References
- 1.Backman H, Räisänen P, Hedman L, Stridsman C, Andersson M, Lindberg A, et al. Increased prevalence of allergic asthma from 1996 to 2006 and further to 2016—results from three population surveys. Clin Exp Allergy. 2017;47(11):1426–35. [DOI] [PubMed] [Google Scholar]
- 2.Cloutier MM, Dixon AE, Krishnan JA, Lemanske RF, Pace W, Schatz M. Managing asthma in adolescents and adults: 2020 asthma guideline update from the National Asthma Education and Prevention Program. JAMA. 2020;324(22):2301–17. [DOI] [PubMed] [Google Scholar]
- 3.The Economic Impact That Asthma Has on the Economy and Families. https://cms.illinois.gov/benefits/stateemployee/bewell/financialwellness/economic-impact-of-asthma.html. Accessed 8 May 2024.
- 4.De Keyser HH, Szefler S. Asthma attacks in children are always preceded by poor asthma control: myth or maxim? Breathe (Sheff). 2020;16(3): 200169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morjaria JB, Polosa R. Recommendation for optimal management of severe refractory asthma. J Asthma Allergy. 2010;26(3):43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iğdır University Vocational School of Technical Sciences, Iğdır, Türkiye, Karadağ M. USE OF Prunus armeniaca L. SEED OIL AND PULP IN HEALTH AND COSMETIC PRODUCTS. ABES. 2024;9(Special Issue):105–10.
- 7.Huntley A, Ernst E. Herbal medicines for asthma: a systematic review. Thorax. 2000;55(11):925–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miryusifova K. The saffron effects on the dynamics of experimental epilepsy. ABES. 2024;9(1):196–202. [Google Scholar]
- 9.Gashimova U, Guliyeva R, Javadova K, Ibishova A, Panakhova E. Histological examination of retinal function and the effects of curcuma longa on memory correction in experimental olfactory bulbectomy rat models. Adv Biol. 2024.
- 10.Zhou B, Liu H, Jia X. The role and mechanisms of traditional chinese medicine for airway inflammation and remodeling in asthma: overview and progress. Front Pharmacol. 2022;13: 917256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGregor MC, Krings JG, Nair P, Castro M. Role of biologics in asthma. Am J Respir Crit Care Med. 2019;199(4):433–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busse WW, Morgan WJ, Gergen PJ, Mitchell HE, Gern JE, Liu AH, et al. Randomized trial of omalizumab (anti-IgE) for asthma in inner-city children. N Engl J Med. 2011;364(11):1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteban-Gorgojo I, Antolín-Amérigo D, Domínguez-Ortega J, Quirce S. Non-eosinophilic asthma: current perspectives. J Asthma Allergy. 2018;29(11):267–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinks TSC, Levine SJ, Brusselle GG. Treatment options in type-2 low asthma. Eur Respir J. 2021;57(1):2000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49(12):1752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013;42(4):1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta V, Walia GK, Sachdeva MP. ‘Mendelian randomization’: an approach for exploring causal relations in epidemiology. Public Health. 2017;145:113–9. [DOI] [PubMed] [Google Scholar]
- 18.Kanduri C, Bock C, Gundersen S, Hovig E, Sandve GK. Colocalization analyses of genomic elements: approaches, recommendations and challenges. Bioinformatics. 2019;35(9):1615–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun BB, Chiou J, Traylor M, Benner C, Hsu YH, Richardson TG, et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023;622(7982):329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guindo-Martínez M, Amela R, Bonàs-Guarch S, Puiggròs M, Salvoro C, Miguel-Escalada I, et al. The impact of non-additive genetic associations on age-related complex diseases. Nat Commun. 2021;12(1):2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;30(7): e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Zhao J, Jiang F, Wang L, Xiao Q, Han F, et al. Identification of novel protein biomarkers and drug targets for colorectal cancer by integrating human plasma proteome with genome. Genome Med. 2023;19(15):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol). 1995;57(1):289–300. [Google Scholar]
- 25.Giambartolomei C, Zhenli Liu J, Zhang W, Hauberg M, Shi H, Boocock J, et al. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018;34(15):2538–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 2021;17(9): e1009440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan S, Xu F, Li X, Chen J, Zheng J, Mantzoros CS, et al. Plasma proteins and onset of type 2 diabetes and diabetic complications: proteome-wide Mendelian randomization and colocalization analyses. Cell Rep Med. 2023;4(9): 101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest | Nucleic Acids Research | Oxford Academic. https://academic.oup.com/nar/article/51/D1/D638/6825349. Accessed 23 Sep 2024. [DOI] [PMC free article] [PubMed]
- 29.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(4):D1202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu F, Han B, Kumar P, Liu X, Ma X, Wei X, et al. Update of TTD: therapeutic target database. Nucleic Acids Res. 2010;38(Database issue):D787–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1100–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo M, Shin J, Kim J, Ryall KA, Lee K, Lee S, et al. DSigDB: drug signatures database for gene set analysis. Bioinformatics. 2015;31(18):3069–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11(5):905–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varricchi G, et al. Biologics and airway remodeling in severe asthma. Allergy. 2022. 10.1111/all.15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalfaoui L, Pabelick CM. Airway smooth muscle in contractility and remodeling of asthma: potential drug target mechanisms. Expert Opin Ther Targets. 2023;27(1):19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell RJ, Brightling C. Pathogenesis of asthma: implications for precision medicine. Clin Sci. 2017;131(14):1723–35. [DOI] [PubMed] [Google Scholar]
- 39.Habib N, Pasha MA, Tang DD. Current understanding of asthma pathogenesis and biomarkers. Cells. 2022;11(17):2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clifford RL, Patel JK, John AE, Tatler AL, Mazengarb L, Brightling CE, et al. CXCL8 histone H3 acetylation is dysfunctional in airway smooth muscle in asthma: regulation by BET. Am J Physiol Lung Cell Mol Physiol. 2015;308(9):L962-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badi YE, Salcman B, Taylor A, Rana B, Kermani NZ, Riley JH, et al. IL1RAP expression and the enrichment of IL-33 activation signatures in severe neutrophilic asthma. Allergy. 2023;78(1):156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon ED, Palandra J, Wesolowska-Andersen A, Ringel L, Rios CL, Lachowicz-Scroggins ME, et al. IL1RL1 asthma risk variants regulate airway type 2 inflammation. JCI Insight. 2016;1(14): e87871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Platelet Response to Allergens, CXCL10, and CXCL5 in the Context of Asthma - PMC. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9936497/. Accessed 5 May 2024. [DOI] [PMC free article] [PubMed]
- 44.Rincon M, Irvin CG. Role of IL-6 in asthma and other inflammatory pulmonary diseases. Int J Biol Sci. 2012;8(9):1281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu M, Zheng X, Huang J, Hu X. Association of IL33, IL1RL1, IL1RAP polymorphisms and asthma in chinese han children. Front Cell Dev Biol. 2021;15(9): 759542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pizzichini MM, Pizzichini E, Efthimiadis A, Chauhan AJ, Johnston SL, Hussack P, et al. Asthma and natural colds. Inflammatory indices in induced sputum: a feasibility study. Am J Respir Crit Care Med. 1998;158(4):1178–84. [DOI] [PubMed] [Google Scholar]
- 47.Nur Husna SM, Md Shukri N, Mohd Ashari NS, Wong KK. IL-4/IL-13 axis as therapeutic targets in allergic rhinitis and asthma. PeerJ. 2022;30(10): e13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bao K, Reinhardt RL. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine. 2015;75(1):25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricciardolo FLM, Bertolini F, Carriero V. The role of dupilumab in severe asthma. Biomedicines. 2021;9(9):1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castro M, et al. Dupilumab efficacy and safety in moderate-to-severe uncontrolled asthma. New Engl J Med. 2018. 10.1056/NEJMoa1804092. [DOI] [PubMed] [Google Scholar]
- 51.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251(4998):1211–7. [DOI] [PubMed] [Google Scholar]
- 52.Miki H, Kiosses WB, Manresa MC, Gupta RK, Sethi GS, Herro R, et al. Lymphotoxin beta receptor signaling directly controls airway smooth muscle deregulation and asthmatic lung dysfunction. J Allergy Clin Immunol. 2023;151(4):976-990.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qi X, Gurung P, Malireddi RKS, Karmaus PWF, Sharma D, Vogel P, et al. Critical role of caspase-8-mediated IL-1 signaling in promoting Th2 responses during asthma pathogenesis. Mucosal Immunol. 2017;10(1):128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pividori M, Schoettler N, Nicolae DL, Ober C, Im HK. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. Lancet Respir Med. 2019;7(6):509–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jenkins CR, Boulet LP, Lavoie KL, Raherison-Semjen C, Singh D. Personalized treatment of asthma: the importance of sex and gender differences. J Allergy Clin Immunol Pract. 2022;10(4):963–71. [DOI] [PubMed] [Google Scholar]
- 56.Herb M, Gluschko A, Schramm M. LC3-associated phagocytosis initiated by integrin ITGAM-ITGB2/Mac-1 enhances immunity to Listeria monocytogenes. Autophagy. 2018;14(8):1462–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Epithelial cell dysfunction, a major driver of asthma development - PMC. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7496351/. Accessed 5 May 2024. [DOI] [PMC free article] [PubMed]
- 58.Smits HH, van der Vlugt LE, von Mutius E, Hiemstra PS. Childhood allergies and asthma: new insights on environmental exposures and local immunity at the lung barrier. Curr Opin Immunol. 2016;1(42):41–7. [DOI] [PubMed] [Google Scholar]
- 59.Zhu X, Li X, Xu R, Wang T. An iterative approach to detect pleiotropy and perform Mendelian Randomization analysis using GWAS summary statistics. Bioinformatics. 2020;37(10):1390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spiller W, Slichter D, Bowden J, Davey SG. Detecting and correcting for bias in Mendelian randomization analyses using gene-by-environment interactions. Int J Epidemiol. 2019;48(3):702–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1. TableS1. Source information for genome-wide association studies. TableS2. Proteome-wide Mendelian randomization results in the asthma discovery cohort. TableS3. Proteome-wide Mendelian randomization results in the asthma replicated cohort. TableS4. Colocalization results in the asthma discovery cohort. TableS5. Sussie results in the asthma discovery cohort. TableS6. KEGG pathways involving the 75 proteins associated with asthma. TableS7. Identification of druggable targets. TableS8. Candidate drug predicted using DSigDB. TableS9. Docking results of MAX protein with small molecules. TableS10. Docking results of traditional targets and their small molecule therapeutic drugs. TableS11. Mendelian randomization results in the asthma-related endpoints. TableS12. Colocalization results in the asthma-related endpoints. TableS13. Susie method results in asthma-related endpoints
Supplementary Material 2. Supplementary Figure: Docking results of traditional targets and their small molecule therapeutic drugs
Data Availability Statement
The GWAS summary data for plasma proteins and asthma analyzed in this study, including findings from UKBB, FinnGen, and GERA, are included in Table S1.