Abstract
Japanese macaques are able to learn how to use rakes to take food after only a few weeks of training. Since tool‐use training induced rapid morphological changes in some restricted brain areas, this system will be a good model for studying the neural basis of plasticity in human brains. To examine the mechanisms of tool‐use associated brain expansion on the molecular and cellular level, here, we performed comprehensive analysis of gene expressions with microarray. We identified various transcripts showing differential expression between trained and untrained monkeys in the region around the lateral and intraparietal sulci. Among candidates, we focused on genes related to synapse formation and function. Using quantitative reverse transcription–polymerase chain reaction and histochemical analysis, we confirmed at least three genes ( ADAM19, SPON2, and WIF1) with statistically different expression levels in neurons and glial cells. Comparative analysis revealed that tool use‐associated genes were more obviously expressed in macaque monkeys than marmosets or mice. Thus, our findings suggest that cognitive tasks induce structural changes in the neocortex via gene expression, and that learning‐associated genes innately differ with relation to learning ability.
Keywords: gene expression, macaque, microarray, parietal cortex, tool use
The macaque brain induced plastic changes by tool use training. In this study, by microarray screening, we identified three genes as tool use acquisition‐associated genes.
Introduction
The neocortex is an indispensable region for higher cognitive function. Humans in particular have a greatly expanded neocortex with multiple complex functional areas. In addition, the neocortex is not static, but rather is a highly dynamic structure that can change depending upon the environment. In fact, it has been demonstrated that learning complex cognitive tasks increases brain size, even in adults. For example, grey and white matter in specific cortical areas increased after subjects learned how to juggle (Draganski et al. 2004; Driemeyer et al. 2008; Kühn et al. 2014), and orchestra musicians have an expanded Broca's area (Sluming et al. 2002). Prolonged structural changes of the brain are caused by various factors, for example, neurogenesis, gliogenesis, axogenesis, synaptogenesis, and angiogenesis. Although imaging studies have revealed gross morphological changes in the neocortex, it remains unknown what mechanisms induce these changes. Because of obvious ethical reasons, human studies are limited to non‐invasive methods. Thus, a good animal model is needed for studying the neural mechanisms of cortical changes.
We have previously established a model system to analyze neural mechanisms of cortical plasticity. Japanese macaques (Macaca fuscata) are able to learn how to use rakes to retrieve food placed beyond the reach of their hands with only a few weeks of training (lriki et al. 1996; Iriki & Taoka 2012). We have found that this tool use training induces morphological and electrophysiological changes in the intraparietal sulcus (IPS) of the macaque brain (lriki et al. 1996; Maravita & Iriki 2004; Hihara et al. 2006; Iriki & Taoka 2012). Moreover, global analysis with voxel‐based morphometry (VBM) revealed that tool use training induces bilateral grey matter expansion in the lateral sulcus (LS) (including secondary somatosensory (SII) area) as well as the IPS (Quallo et al. 2009). SII is highly interconnected with the primary somatosensory area (SI), prefrontal cortex, and posterior insula (Iwamura 1998), and is essential for tactile object recognition and tactile sensorimotor learning, and is associated with retrieval of food and establishment of body schema (Ridley & Ettlinger 1976; Murray & Mishkin 1984; Caselli 1993; Binkofski et al. 1999a,b; Burton et al. 1999; Maravita & Iriki 2004; Taoka et al. 2013). Thus, we suspected that both IPS and SII were involved in tool use and its learning process, and that the IPS and SII areas were dynamically changed by tool‐use training. To explore the molecular basis of cortical plasticity, here, we performed comprehensive gene expression analysis using a cDNA microarray to identify the gene expression changes associated with tool use training in these areas.
Materials and methods
Ethics
Research protocols were approved by the Animal Care and Use Committee of RIKEN and conform to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Tool use training
Macaque monkeys were provided by the National BioResource Project ‘Japanese Monkeys’. Eight male and two female Japanese macaques (Macaca fuscata) were used for this study (Table 1). Tool use training was performed as previously described (Quallo et al. 2009). After a week of habituation to the training room and experimenters, we trained one female and four male monkeys as experimental animals, and one female and four male monkeys as control animals. Monkeys in the experimental group were trained to acquire food rewards (small pieces of sweet potato or apple) by rakes (Fig. 1A). During the training period, each monkey performed almost 250 trials in the training session and 50 trials in the timed test sessions each day, whereas each monkey in the control group performed almost 300 trials acquiring food with both hands. Control monkeys and monkeys before training showed no preference for hand use. Experimental monkeys learned how to use rakes over 1–2 weeks of training (Fig. 1B). Although we have previously found that tool use training immediately induced gene expression of neurotrophic factors and receptors just after training (Ishibashi et al. 2002a,b), our focus in this study was on the molecular mechanisms of prolonged structural changes rather than immediate plastic changes. Therefore, we avoided detection of immediate early genes by collecting samples from monkeys more than 24 h after the last training session and compared gene expression levels between experimental and control animals. All monkeys were trained to use rakes with their right hands. In this study, we analyzed the left hemisphere for all animals.
Table 1.
Monkeys we used in this study
| ID | age | Weight (kg) | Microarray | qRT–PCR |
|---|---|---|---|---|
| Control | ||||
| 339M | 5 years 5 months old | 9.0 | LS, IPS | |
| 340F | 4 years 6 months old | 5.3 | LS | LS |
| 347M | 5 years 4 months old | 7.3 | LS, IPS | LS, IPS |
| 354M | 5 years 10 months old | 9.2 | LS, IPS | LS, IPS |
| 358M | 3 years 9 month old | 8.6 | IPS | LS, IPS |
| Tool use | ||||
| 334M | 5 years 7 months old | 8.6 | LS, IPS | LS, IPS |
| 335F | 3 years 10 months old | 6.3 | LS | LS |
| 343M | 6 years | 8.6 | IPS | LS, IPS |
| 348M | 5 years 1 month old | 7.0 | LS, IPS | LS, IPS |
| 365M | 3 years 11 months old | 6.3 | LS, IPS | |
F, female; IPS, intraparietal sulcus; LS, lateral sulcus; M, male; qRT–PCR, quantitative reverse transcription–polymerase chain reaction.
Figure 1.
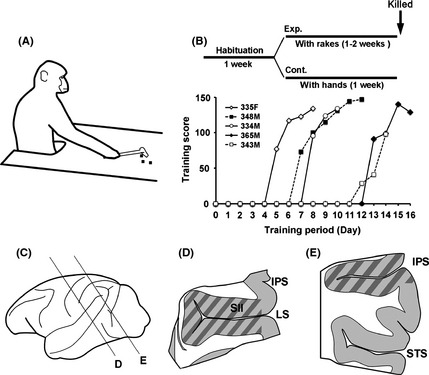
Tool use training of Rhesus macaque monkeys (A, B) and collected brain areas for gene expression analysis (C–E). Schematic representation of tool use training (A). Experimental procedure and learning curve of tool use training (B). Horizontal axis indicates learning periods. Vertical axis indicates the score of the tool use test. The score of 50 timed trial tests was calculated as follows: monkeys retrieved pieces of food on the first attempt (score 3), second attempt (score 2), multiple attempts (score 1) or more than five attempts or failed (score 0). Dissection and sample preparation of the macaque brain (C–E). Examples of dissected area from the lateral sulcus (LS) and the intraparietal sulcus (IPS) region (marked area) (D, E). RNAs were purified from the anterior region of the surrounding area of LS including SII and insula (Is), and from the posterior region of the surrounding area of the IPS. We collected both superior and inferior LS and IPS regions, because it was difficult to dissect half of these areas.
Sample preparation of macaques for gene expression analysis
Perfusion and preparation of frozen sections of macaque monkeys were performed as previously described (Matsunaga et al. 2015b). To prepare RNAs for microarray analysis or quantitative reverse transcription–polymerase chain reaction (qRT–PCR), we cut some serial sections at a 50 μm thickness (1 mm interval) and dissected brain slices with scalpels in a cold chamber. We collected tissues from the lateral sulcus (LS), including the secondary somatosensory area (SII) and insula (Fig. 1D, marked area), and from the posterior intraparietal sulcus (IPS; Fig. 1E, marked area). We purified RNAs with a RNeasy Lipid Tissue Mini Kit (Qiagen, Valencia, CA, USA). The quality of RNAs was checked by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Microarray analysis
After probes were labeled with a GeneChip 3′IVT Expression Kit (Affymetrix, Santa Clara, CA, USA), comprehensive gene expression analyses were performed with a GeneChip Rhesus Macaque Genome Array (Affymetrix). After all expression data were analyzed with GeneChip Operating software (Affymetrix) to generate CHP data with the MAS5 statistical algorithm, comparative gene expression analyses between each sample were performed with GeneSpring GX software (Agilent Technologies). We used three experimental animals and three control animals for microarray analysis of the LS and IPS regions. Probe intensity was normalized by the 75th percentile shift, and probes with one or more values in the 20–100th percentile range of raw data were selected. Transcripts with a fold change ≥1.4 and P value < 0.05 (Mann–Whitney U‐test) were identified as differentially expressed. We calculated P‐value without correction for multiple testing since it was too stringent to identify candidate genes for the experiments on GeneSpring GX software. Alternatively, we first screened candidate genes by microarray, and then performed the second screening by checking each gene expression level with qRT–PCR method. The microarray data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (the GEO Series accession number is GSE63657).
Sample preparation of marmosets and mice
Sample preparation of frozen sections of marmosets and mice were performed as previously described (Matsunaga et al. 2015a). We used three embryonic marmosets of either sex, two male and one female neonatal marmosets, and one male and two female adult marmosets. We used three E14 of either sex, three P14 of either sex, and three P56 mice of either sex.
qRT–PCR
We designed primers using Primer Express Software v3.0 (Life Technologies, Carlsbad, CA, USA). To prevent contamination by genomic DNA, we used a pair of primers that amplified specific intron‐spanning sequences. We purified RNA with the RNeasy Lipid Tissue Mini Kit (Qiagen) or the RecoverAll Total Nucleic Acid Isolation Kit for formalin‐fixed, paraffin‐embedded tissue (FFPE; Ambion, Austin, TX, USA). We generated cDNA with Superscript III polymerase (Life Technologies) and random primers (Life Technologies). Real‐time PCR was performed in triplicate with Power SYBR(R) Green PCR Master Mix (Life Technologies) and measured with a 7900HT Real Time PCR System (Life Technologies). We normalized with GAPDH since this is the gene that is the most coincident with the results of the microarray. Primers we used are summarized in Tables 2 and S1. Statistical analysis (Mann–Whitney U‐test) was done using RStudio version 0.98.932. (RStudio, Boston, MA, USA).
Table 2.
Primers used for quantitative reverse transcription–polymerase chain reaction (RT–PCR) and cDNA isolation
| qPCR primer | Forward primer | Reverse primer | Position | Length | ||
|---|---|---|---|---|---|---|
| GAPDH | GCACCGTGAAGGCTGAGAAC | AGGGATCTCGCTCCTGGAA | NM_001195426 | 233–307 | ORF | 75 |
| cjGAPDH | GGCGTGAACCATGAGAAGTATG | GGTGCAGGAGGCATTGCT | XM_002759682 | 471–530 | ORF | 60 |
| mGAPDH | CTCGTCCCGTAGACAAAATGG | TGACCAGGCGCCCAATA | GU214026 | 55–120 | ORF | 66 |
| ADAM19 | CTCCAAGCCGGCCAATT | CCTGGAGAAGTCCTGGGAAAG | XM_001105246 | 2580–2638 | ORF | 59 |
| cjADAM19 | CTTAGTTGGAGGCGCAAGCT | GAAGGACATGCCCGTGATTAA | XM_002744097 | 972–1043 | ORF | 72 |
| mADAM19 | GGGCCCTTCAGTTTACACATCA | TGTAGATCTTCCCGTTTCATTCTG | NM_009616 | 690–753 | ORF | 64 |
| SPON2 | GCCAAATACAGCATCACCTTCA | CGTACTGGTTCTTCCTCCACATG | XM_001092523 | 165–312 | ORF | 148 |
| cjSPON2 | TGTGGACAGCGCGTCAGT | GGACGACCACAGGGAAACTTC | XM_008993730 | 1003–1063 | ORF | 61 |
| mSPON2 | CCAGCCGAGGCAACGA | CCAGCGGTGTCTCTGGAACT | NM_133903 | 1067–1120 | ORF | 54 |
| WIF1 | TCCATGGAGTGAACTGTGACAAA | AACAGGTCCCTCCGTTAAAGC | NM_001260671 | 911–973 | ORF | 63 |
| cjWIF1 | GAAAGACGCATCTGCGAGTGT | GGTGTGCAAAGGGCTTTCTC | XM_002752707 | 964–1031 | ORF | 68 |
| mWIF1 | GAGAGCAGTGTGAACTCAGCAAA | CTTTTACCAATGCATTTACCTCCAT | NM_011915 | 1148–1214 | ORF | 67 |
| KLK15 | GGAGCCCCCAGTCACAAGT | AGATAATGCTGATGTTGGCACAAT | XM_001116190 | 464–526 | ORF | 63 |
| In situ probe | Forward primer | Reverse primer | Position | Length | ||
|---|---|---|---|---|---|---|
| ADAM19 | TGCTGCAATGCCTCTAATTG | ACCACAGGACCCACACTCTC | XM_001105246 | 1400–2187 | ORF | 788 |
| cjADAM19 | cDNA for macaque WIF1 | 95.4% homology to macaque ADAM19 | ||||
| mADAM19 | cDNA (Carninci et al., 2005) | AK147549 | 5′UTR+ORF+3′UTR | 6217 | ||
| SPON2 | CTGGACCTGTACCCCTACGA | CCTGGACAATGAAGGACGAT | XM_001092523 | 588–1381 | ORF+3′UTR | 794 |
| cjSPON2 | CTGGACCTGTACCCCTACGAa | CCTGGACAATGAAGGACGATa | XM_008993730 | 754–1551 | ORF+3′UTR | 798 |
| mSPON2 | CTTGTCTCAAGCCCCTTCTG | ACCACAGGGAAACCTCACAG | NM_133903 | 114–1141 | 5′UTR+ORF | 1028 |
| WIF1 | CTCCCTGGATAAAGGCATCA | TTAAGTGAAGGCGTGTGCTG | NM_001260671 | 321–1091 | ORF | 771 |
| cjWIF1 | cDNA for macaque WIF1 | 97.3% homology to macaque WIF1 | ||||
| mWIF1 | cDNA (Carninci et al., 2005) | AK077698 | 5′UTR+ORF+3′UTR | 2246 | ||
The primer was designed based on macaque cDNA (XM_001092523). Grey colour columns, macaque DNAs; cj, marmoset DNA; m, mouse DNA.
Histochemical analysis
Brain tissue from female marmosets or macaques was collected, RNA was purified, and cDNA was generated as described above to isolate cDNA fragment by PCR for probe preparation of in situ hybridization. Primers used for cDNA isolation were described (Table 2). Mouse ADAM19, SPON2 and WIF1 probes included all exons for marmoset and macaque probes. Marmoset and macaque SPON2 probes were located to the same region (because we isolated the gene by the same primers), and the same ADAM19 and WIF1 probe were used for marmoset and macaque sections.
In situ hybridization and immunohistochemical analyses were performed as previously described (Matsunaga et al. 2014). Antibodies used were anti‐NeuN mouse monoclonal (1:200; EMD Millipore, Billerica, MA, USA), anti‐rabbit polyclonal Tbr2 (1:200; Abcam, Cambridge, MA, USA), anti‐rabbit polyclonal Oligodendrocyte Specific Protein (1:100: ab7474; Abcam), and Cy3‐conjugated anti‐mouse or Rabbit IgG antibody (1:400; Jackson ImmunoResearch, West Grove, PA, USA). All images were captured using a NanoZoomer 2.0 slide scanner (Hamamatsu Photonics, Hamamatsu, Japan) or an ORCA‐Flash2.8 digital camera (Hamamatsu Photonics) under a BX‐50 microscope (Olympus, Tokyo, Japan) or a DP80 digital camera under a BX‐53 microscope (Olympus).
Results
Tool use training of macaque monkeys
One week after habituation to the training room and experimenters, monkeys were trained to use rakes to acquire food placed out of the animals' reach (Fig. 1A). Five monkeys were trained and although all monkeys learned how to use the rakes, their learning speeds differed (Fig. 1B). One or 2 days after the final trial test, we transcardially perfused the monkeys and obtained brain tissue samples.
Screening of tool use‐associated gene expression in the LS region
To screen candidate genes involved in plastic change of the LS region induced by tool use training, we performed comprehensive gene expression analysis with a GeneChip Rhesus macaque genome array (Affymetrix). We performed microarray screening with three experimental monkeys (fast‐learners, 334M, 335F, 348M in Fig. 1B) and three control monkeys for the LS. The monkeys we used were summarized in Table 1.
In the LS region, we identified 179 transcripts showing higher expression levels in the experimental group, and 148 transcripts showing higher expression levels in the control group. Among these candidates, we focused on genes involved in neural circuit formation and function for further screening.
To verify the results of the microarray, we next performed qRT–PCR analysis of candidate genes (primers used are shown in Table S1). We used five trained and five control monkeys for this analysis. We validated 34 candidate gene expression profiles and confirmed three genes (Table S1), A Disintegrin And Metalloprotease 19 (ADAM19), Kallikrein‐15 (KLK15), and Spondin‐2 (SPON2), with significantly differential expression between the experimental and control groups (Table 3).
Table 3.
Gene expression levels by microarray and quantitative reverse transcription–polymerase chain reaction (qRT–PCR) analysis
| Microarray | qRT–PCR | |||
|---|---|---|---|---|
| Fold change | P < 0.05 | Fold change | P < 0.05 | |
| LS region | ||||
| ADAM19 | 1.46 | a | 1.32 | a |
| KLK15 | 2.05 | a | 2.3 | a |
| SPON2 | 1.49 | a | 1.74 | a |
| IPS region | ||||
| KLK15 | 2.02 | a | 2.89 | a |
| WIF1 | 1.4 | a | 1.48 | a |
P < 0.05.Fold change is the ratio of gene expression in experimental animals compared to control animals. IPS, intraparietal sulcus; LS, lateral sulcus.
Screening of tool use‐associated gene expression in the IPS region
Next, we analyzed gene expression changes in the IPS region. By microarray analysis with three experimental monkeys and three control monkeys (Table 1), we identified 199 transcripts as more highly expressed in the experimental group, and 143 transcripts as more highly expressed in the control group in the IPS region.
Since we did not have IPS tissue derived from one monkey, we performed qRT–PCR with samples from four experimental animals and four control animals (Table 1), and checked 21 candidate genes (Table S1). We found two genes, KLK15 and Wnt inhibitory Factor‐1 (WIF1), with significantly different expression levels between the experimental and control groups (Table 3).
Candidate gene expressions in primary motor cortex
To explore whether tool use training‐associated induction/reduction was due to neural activity of hand use or complex cognitive learning, we next examined candidate gene expression in the primary motor area (M1). We collected tissues from the region corresponding to the hand field and performed qRT–PCR analysis (n = 5, each group). Although expression of candidate genes was significantly higher in the trained animals than control animals in the LS and IPS regions, there were no significant differences in M1, with the exception of KLK15 (Fig. 2A–D; expression differences in the LS and IPS region were larger than the M1 region.) Thus, it appears that changes in three tool use‐associated genes (ADAM19, SPON2, and WIF1) were due to complex cognitive learning rather than simple forelimb movement.
Figure 2.
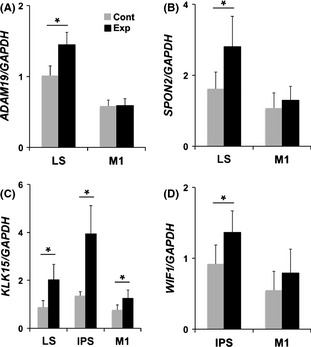
Quantitative analysis of tool use associate candidates by quantitative reverse transcription–polymerase chain reaction (qRT–PCR) (A–D). qRT–PCR was performed with five control and five experimental animals for the lateral sulcus (LS) and M1, and four experimental and four control animals for the intraparietal sulcus (IPS) region. Note that SPON2, ADAM19, and WIF1 in the LS and IPS showed higher expression in experimental animals than control animals, although no significant differences were seen in M1. *P < 0.05.
Histological analysis of tool use‐associated genes
Next, we performed in situ hybridization analysis to explore the expression patterns of the three identified tool use‐associated genes ADAM19, SPON2, and WIF1 in the macaque neocortex.
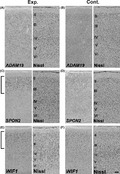
ADAM19‐expressing cells were dispersed broadly in the superior LS (upper bank of the LS) within layers II–VI (Fig. 3A,B). In contrast, SPON2‐expressing cells in the superior LS and WIF1‐expressing cells in the inferior IPS (lower bank of the IPS) were largely located in the upper layers of the cortex (Fig. 3C–F), which are highly evolved in the primate.
Figure 3.
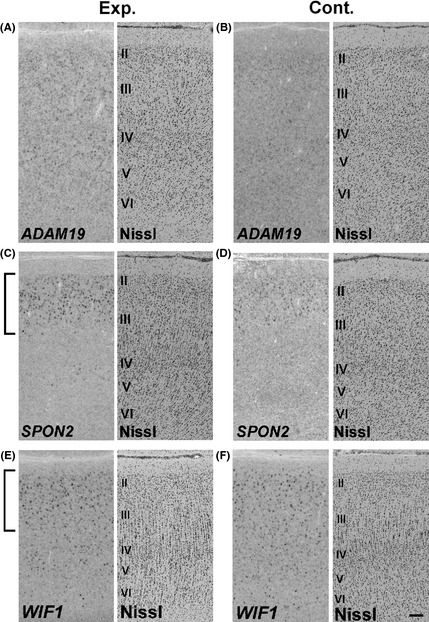
In situ hybridization of adult macaque superior lateral sulcus (LS) (A–D) and inferior intraparietal sulcus (IPS) regions (E, F) for ADAM19 (A, B), SPON2 (C, D), and WIF1 (E, F) of experimental (A, C, E) or control neocortex (B, D, F). Note that many SPON2‐ or WIF1‐expressing cells were located in the upper layers, which are particularly evolved in the primate brain. Expressions of all these genes were seen in the control monkeys, although their expression levels looked different between control and experimental monkeys. Scale bar is 100 μm.
Co‐staining with marker proteins revealed that ADAM19‐ or SPON2‐positive cells were NeuN‐positive (Fig. 4A–F), indicating that the tool use‐associate gene expression changes occurred in neurons. On the other hand, WIF1‐positive cells were not NeuN‐positive (Fig. 4G–I), but rather oligodendrocyte‐specific protein (OSP)‐positive (Fig. 4J–L), suggesting that neuronal activity by tool use indirectly modulated gene expression in glial cells.
Figure 4.
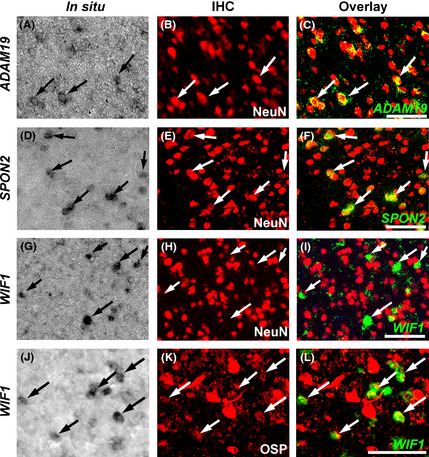
In situ hybridization for ADAM19, SPON2 and WIF1 (A, D, G, J), subsequent co‐immunostaining with cell type specific markers (B, E, H, K), and overlay images of in situ hybridization (green, pseudo‐colour) and IHC (red) (C, F, I, L) of sections derived from experimental monkeys. In situ hybridization of adult macaque superior lateral sulcus (LS) (A–F) and inferior intraparietal sulcus (IPS) regions (G–L) for ADAM19 (A–C), SPON2 (D–F), and WIF1 (G–L) with immunostaining for the neuronal marker NeuN (A–I) or oligodendrocyte marker OSP (J–L). ADAM19 and SPON2‐expressing cells were NeuN‐positive (A–F), and WIF1‐expressing cells were NeuN‐negative (G–I) but OSP‐positive (J–L). IHC, immunohistochemistry; OSP, oligodendrocyte specific protein. Scale bar is 100 μm.
Primate‐specific neocortical expression of SPON2 and WIF1
Next, we analyzed tool use‐associated gene expression in mice (Mus musculus; rodents) and marmosets (Callithrix jacchus; new world monkey) to verify whether these gene expression profiles are specific to macaques or broadly seen in other mammalian neocortices.
ADAM19 expression was seen in mouse, marmoset (Fig. 5A–E), and macaque neocortices (Fig. 4B), and was seen at comparable levels in all three species. Developmental changes of ADAM19 expression were similar between the rodent (mouse) and primate (marmoset) (Fig. 5D,E). Thus, ADAM19 expression does not seem to be primate‐specific, although its expression was changed by tool use training.
Figure 5.
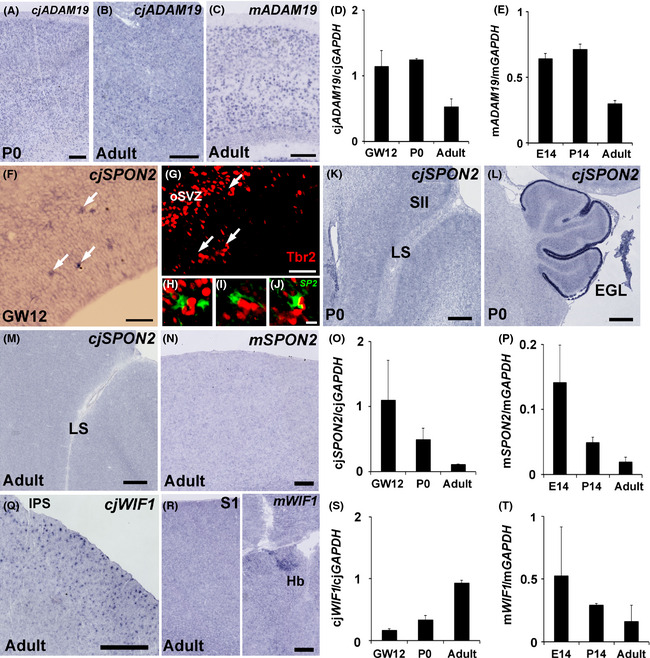
ADAM19, SPON2 and WIF1 expressions in the mouse and marmoset neocortex. Similar ADAM19 expression levels among the macaque, marmoset and mouse (A–E). In situ hybridization for ADAM19 in marmoset (A, B) and mouse (C) somatosensory cortex. Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) analysis of ADAM19 expression in developing marmoset (D) and mouse (E). Differential SPON2 expression between the macaque and the other species (F–P). In situ hybridization for SPON2 in the marmoset (F, K, L, M) and mouse (N). In situ hybridization for SPON2 (F) and subsequent immunostaining with a Tbr2 antibody (G) in GW12 marmoset neocortex. Overlay image of SPON2 (green, pseudo‐colour) and Tbr2 (red) staining (H, I, J). H, I, and J are magnified images of the areas indicated by arrows (F). In situ hybridization for SPON2 of the lateral sulcus (LS) area (K) and cerebellum (L) in P0 marmoset, and the LS area in adult marmoset (M) and adult mouse somatosensory cortex (N). qRT–PCR analysis of SPON2 expression in developing marmoset (O) and mouse (P). Note that SPON2 expression was reduced during development and disappeared by the adult stage for both species, in contrast to macaques. Differential WIF1 expression between marmoset and mouse neocortex (Q–T). In situ hybridization for WIF1 in marmoset (Q) and mouse caudal parietal cortex (R, left) and habenula (R, right). Note that in situ hybridization staining for WIF1 was seen in the habenula but no staining was seen in the neocortex. qRT–PCR analysis of WIF1 expression in developing marmoset (S) and mouse (T). qRT–PCR was performed with three animals at each stage. For qRT–PCR experiments, tissues were derived from the whole brain for E14 mouse embryos, the parietal cortex for P14 and adult mice, the whole neocortex for GW12 marmoset embryos, and the parietal cortex for P0 and adult marmosets. Scale bars are 500 μm (K, L, Q), 200 μm (A–C), 100 μm (N, R), 50 μm (F, G), and 10 μm (J).
In contrast to ADAM19, the two other tool use‐related genes SPON2 and WIF1 showed differential expression among different mammalian neocortices (Figs 3C,E, 5F–T). As previously reported (Feinstein et al. 1999), no clear SPON2 expression was seen in adult mouse neocortex (Fig. 5N). In marmosets, SPON2 expression was seen in the cortical area of the GW12 embryo (Fig. 5F). SPON2‐posisitive cells were located adjacent to Tbr2‐positive neuronal progenitor cells in the subventricular zone (Fig. 5G–J), suggesting the SPON2 functions as a secreted protein related to neurogenesis. Its expression in the neocortex was reduced across development. At the neonatal stage, its expression was not detected anywhere in the neocortex, although it was seen in the external granule cell layer (Fig. 5K,L), and this expression disappeared by the adult stage (Fig. 5M, data not shown). The reduction of SPON2 expression in the neocortex was also confirmed by qRT–PCR (Fig. 5O,P). In contrast to mice and marmosets, SPON2 expression was clearly seen in the upper layer of the macaque neocortex even without tool use training (Fig. 3D). On the other hand, WIF1 was expressed in both macaque and marmoset neocortices (Figs 3F, 5Q,S), though mouse neocortex showed no clear WIF1 expression in the adult brain (Fig. 5R,T). Thus, tool use‐associated gene expression was innately diverse among different mammalian neocortices even without tool use training (Fig. 6A).
Figure 6.
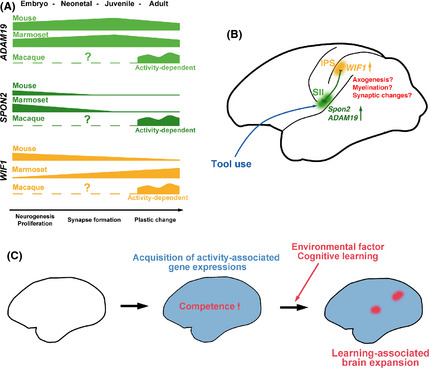
Summary of temporal change of gene expressions among mouse, marmoset and macaque neocortex (A) and hypothetical model of tool use associated changes (B) and brain expansion of primates (C). Tool use induces gene expressions (e.g. SPON2 and ADAM19) in efferent neurons of the SII area. Their gene products function in the intraparietal sulcus (IPS) and induce cortical changes. In turn, this makes subsequent gene expression changes (e.g. WIF1) in the neurons or oligodendrocytes of the IPS area. Reciprocal interaction of these gene products may cause plastic changes such as axogenesis, myelination or synaptic changes (B). Acquisition of activity‐dependent gene expressions might have accelerated brain expansion by both innate and behavior‐dependent changes simultaneously acquired (C).
Discussion
In this study, we performed tool use training and comprehensive gene expression analysis in Japanese macaques, and found more than 300 candidates in the LS and IPS regions of the parietal cortex, and ultimately identified three genes, ADAM19, SPON2 and WIF1, as tool use‐associated genes.
ADAM19 is a member of the ‘A Disintegrin And Metalloprotease’ (ADAM) family that modulates various signaling pathways by cleaving and shedding membrane‐bound growth factors or their receptors (Alfandari et al. 2009; Reiss & Saftig 2009). ADAM19 is involved in cardiovascular morphogenesis and neural and muscle development by cleavage of HB‐EGF, cadherin, or neuregulin‐1 (Shirakabe et al. 2001; Zhou et al. 2004; Komatsu et al. 2007; Neuner et al. 2008; Fukazawa et al. 2013; Schiffmacher et al. 2014).
SPON2 (also known as M‐spondin, Mindin) is a secreted protein containing thrombospondin‐type 1 repeats (TSR), originally identified as a protein structurally similar to F‐spondin (Higashijima et al. 1997; Umemiya et al. 1997), functions as an integrin ligand (Li et al. 2009), and induces neurite outgrowth in cultured hippocampal neurons (Feinstein et al. 1999).
WIF1 was originally identified in expression sequence tags derived from human retina (Hsieh et al. 1999). WIF1 encodes a secreted protein that functions as an inhibitor of Wnt signaling via direct binding to Wnt family proteins (Hsieh et al. 1999). Increasing evidence suggests that Wnt signaling controls synapse formation and function at pre‐ and postsynaptic sites (Rosso & Inestrosa 2013). Although WIF1 function in adult neocortex still remains unknown, it has been shown that WIF1 inhibits BDNF‐induced spine formation by antagonizing Wnt signaling in cultured cortical neurons derived from neonatal mouse cortex (Hiester et al. 2013).
SPON2 and WIF1 encode secreted proteins that control synapse formation and function (Feinstein et al. 1999; Hiester et al. 2013) and the gene product of ADAM19 is a metalloprotease cleaving synaptic molecule (Shirakabe et al. 2001), suggesting that all of these genes are involved in brain plasticity. Particularly, SPON2 and WIF1 were strongly expressed in the upper layers of the neocortex, where cortico‐cortical connections are particularly evolved in the primate brain. Thus, SPON2 and WIF1 are strong candidates for controlling tool use‐related cortical changes.
Co‐immunohistochemical analysis revealed that SPON2 was expressed in neurons, whereas WIF1 was expressed in oligodendrocytes. Tool use learning may induce prolonged changes in neuronal activity or gene expression directly or indirectly in neurons and glial cells. Both SPON2 and WIF1 are secreted proteins; therefore, neuron‐glia interactions may play an important role in grey and white matter expansion by tool use training, for example, through axogenesis, synaptogenesis, or myelination. Since SII neurons project to the surrounding area of the IPS (Cipolloni & Pandya 1999; Borra et al. 2008), interaction of SPON2‐expressing neurons and WIF1‐expressing oligodendrocytes may exist in the IPS region (Fig. 6B).
Comparative gene expression analysis in the adult neocortex among mouse, marmosets, and macaques revealed that strong SPON2 expression was macaque‐specific, strong WIF1 expression was primate‐specific, and ADAM19 expression was conserved across mice, marmosets, and macaques. SPON2 and WIF1 expression differed among species even without tool use training, suggesting it has innately differential expression among mammals. Thus, it appears that the capability of gene expression changes in response to tool use training is determined innately, that tool use competence differs among different mammalian/primate species, and that such competence is acquired by primate‐specific gene expressions. An acquisition of innate gene expressions responding to environmental factors may accelerate neocortical expansion in the primate brain via stimulations by various environmental factors (Iriki & Taoka 2012) (Fig. 6C).
Interestingly, SPON2 was expressed in embryonic and neonatal marmosets, but its expression disappeared during marmoset development. SPON2 and WIF1 expression was seen in embryonic mouse neocortex, but the expression of both genes disappeared during postnatal mouse development. Although we could not acquire tissue from embryonic or neonatal macaques, it is expected that marmoset and mouse neocortices lose the expression of one/both of these genes during postnatal development, whereas expression of both genes is maintained into adulthood in the macaque neocortex. Prolonged expression of these plasticity‐related genes may allow macaques to undergo higher levels of cognitive plasticity, such as in tool use training. Interestingly, although both macaques and marmosets are capable of learning to use rakes with their hands and show expansion of the parietal cortex with tool use, macaques learn tool use in a much shorter time period than marmosets (Quallo et al. 2009; Yamazaki et al. 2011; Iriki et al. 2014). Gene expression manipulation studies using viral vectors will reveal whether such expression differences are causally related to this cognitive diversity.
In this study, although only a few genes were identified as tool use‐associated genes, many unidentified genes may be involved in plastic changes during the learning process. We may have failed to detect a large number of differential gene expressions for a number of reasons. First, because the monkeys we used are not genetically controlled (in contrast to the mice), a large number of genetic changes may be masked by individual genetic differences. Second, we collected brain tissues from all layers of the cortex; however, if target genes are only seen in a subset of layers (as in the case of SPON2 and WIF1), it would be difficult to detect a subtle expression difference. Finally, the issue of the timing of sample collection is critical. We purposefully collected tissues at 1 or 2 days after tool use training to avoid contamination of immediate early gene expression because we focused on long‐term gene expression changes associated with structural plasticity in the neocortex rather than immediate gene changes induced by tool use training. We may have missed changes in gene expression that were transient but that are required for long‐term changes in neocortical plasticity and that wouldn't be present at the time point we harvested tissues. Further modifying the research strategy and technical improvements will be necessary for finding more tool use‐associated gene candidates, and will reveal the entire picture of molecular changes by tool use training.
Supporting information
Table S1. Primer sets used for 2nd screening by qRT‐PCR.
Acknowledgements
We thank Mr Keisuke Fukumoto (RIKEN BSI Research Resources Center: RRC) for technical assistance with microarray experiment, RIKEN RRC for DNA sequence analysis and the FANTOM Consortium Core Group for mouse cDNA clone, Drs Akihiro Kawasaki, Chihiro Yokoyama (RIKEN Center for Life Science Technologies) and Dr. Kimie Niimi (RIKEN RRC) for marmoset caesarian section, and Dr. Shigeyoshi Itohara for lab facility. The monkeys used in this study were provided by the National BioResource Project ‘Japanese Monkeys’ of the Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was supported by JSPS KAKENHI Grant number 25750403, and RIKEN Incentive Research Projects (to E.M.), the JSPS through the “Funding Program for World‐Leading Innovative R&D on Science and Technology (FIRST Program),” initiated by the Council for Science and Technology Policy (CSTP) and the Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) by the MEXT of Japan (to A.I.). The authors declare no competing financial interests.
References
- Alfandari, D. , McCusker, C. & Cousin, H. 2009. ADAM function in embryogenesis. Semin. Cell Dev. Biol. 20, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkofski, F. , Buccino, G. , Posse, S. , Seitz, R. J. , Rizzolatti, G. & Freund, H. 1999a. A fronto‐parietal circuit for object manipulation in man: evidence from an fMRI‐study. Eur. J. Neurosci. 11, 3276–3286. [DOI] [PubMed] [Google Scholar]
- Binkofski, F. , Buccino, G. , Stephan, K. M. , Rizzolatti, G. , Seitz, R. J. & Freund, H. J. 1999b. A parieto‐premotor network for object manipulation: evidence from neuroimaging. Exp. Brain Res. 128, 210–213. [DOI] [PubMed] [Google Scholar]
- Borra, E. , Belmalih, A. , Calzavara, R. , Gerbella, M. , Murata, A. , Rozzi, S. & Luppino, G. 2008. Cortical connections of the macaque anterior intraparietal (AIP) area. Cereb. Cortex 18, 1094–1111. [DOI] [PubMed] [Google Scholar]
- Burton, H. , Abend, N. S. , MacLeod, A. M. , Sinclair, R. J. , Snyder, A. Z. & Raichle, M. E. 1999. Tactile attention tasks enhance activation in somatosensory regions of parietal ortex: a positron emission tomography study. Cereb. Cortex 9, 662–674. [DOI] [PubMed] [Google Scholar]
- Carninci, P. , Kasukawa, T. , Katayama, S. , Gough, J. , Frith, M. C. , Maeda, N. , Oyama, R. , Ravasi, T. , Lenhard, B. , Wells, C. , Kodzius, R. , Shimokawa, K. , Bajic, V. B. , Brenner, S. E. , Batalov, S. , Forrest, A. R. , Zavolan, M. , Davis, M. J. , Wilming, L. G. , Aidinis, V. , Allen, J. E. , Ambesi‐Impiombato, A. , Apweiler, R. , Aturaliya, R. N. , Bailey, T. L. , Bansal, M. , Baxter, L. , Beisel, K. W. , Bersano, T. , Bono, H. , Chalk, A. M. , Chiu, K. P. , Choudhary, V. , Christoffels, A. , Clutterbuck, D. R. , Crowe, M. L. , Dalla, E. , Dalrymple, B. P. , de Bono, B. , Della Gatta, G. , di Bernardo, D. , Down, T. , Engstrom, P. , Fagiolini, M. , Faulkner, G. , Fletcher, C. F. , Fukushima, T. , Furuno, M. , Futaki, S. , Gariboldi, M. , Georgii‐Hemming, P. , Gingeras, T. R. , Gojobori, T. , Green, R. E. , Gustincich, S. , Harbers, M. , Hayashi, Y. , Hensch, T. K. , Hirokawa, N. , Hill, D. , Huminiecki, L. , Iacono, M. , Ikeo, K. , Iwama, A. , Ishikawa, T. , Jakt, M. , Kanapin, A. , Katoh, M. , Kawasawa, Y. , Kelso, J. , Kitamura, H. , Kitano, H. , Kollias, G. , Krishnan, S. P. , Kruger, A. , Kummerfeld, S. K. , Kurochkin, I. V. , Lareau, L. F. , Lazarevic, D. , Lipovich, L. , Liu, J. , Liuni, S. , McWilliam, S. , Madan Babu, M. , Madera, M. , Marchionni, L. , Matsuda, H. , Matsuzawa, S. , Miki, H. , Mignone, F. , Miyake, S. , Morris, K. , Mottagui‐Tabar, S. , Mulder, N. , Nakano, N. , Nakauchi, H. , Ng, P. , Nilsson, R. , Nishiguchi, S. , Nishikawa, S. , Nori, F. , Ohara, O. , Okazaki, Y. , Orlando, V. , Pang, K. C. , Pavan, W. J. , Pavesi, G. , Pesole, G. , Petrovsky, N. , Piazza, S. , Reed, J. , Reid, J. F. , Ring, B. Z. , Ringwald, M. , Rost, B. , Ruan, Y. , Salzberg, S. L. , Sandelin, A. , Schneider, C. , Schönbach, C. , Sekiguchi, K. , Semple, C. A. , Seno, S. , Sessa, L. , Sheng, Y. , Shibata, Y. , Shimada, H. , Shimada, K. , Silva, D. , Sinclair, B. , Sperling, S. , Stupka, E. , Sugiura, K. , Sultana, R. , Takenaka, Y. , Taki, K. , Tammoja, K. , Tan, S. L. , Tang, S. , Taylor, M. S. , Tegner, J. , Teichmann, S. A. , Ueda, H. R. , van Nimwegen, E. , Verardo, R. , Wei, C. L. , Yagi, K. , Yamanishi, H. , Zabarovsky, E. , Zhu, S. , Zimmer, A. , Hide, W. , Bult, C. , Grimmond, S. M. , Teasdale, R. D. , Liu, E. T. , Brusic, V. , Quackenbush, J. , Wahlestedt, C. , Mattick, J. S. , Hume, D. A. , Kai, C. , Sasaki, D. , Tomaru, Y. , Fukuda, S. , Kanamori‐Katayama, M. , Suzuki, M. , Aoki, J. , Arakawa, T. , Iida, J. , Imamura, K. , Itoh, M. , Kato, T. , Kawaji, H. , Kawagashira, N. , Kawashima, T. , Kojima, M. , Kondo, S. , Konno, H. , Nakano, K. , Ninomiya, N. , Nishio, T. , Okada, M. , Plessy, C. , Shibata, K. , Shiraki, T. , Suzuki, S. , Tagami, M. , Waki, K. , Watahiki, A. , Okamura‐Oho, Y. , Suzuki, H. , Kawai, J. & Hayashizaki, Y. ; FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) . 2005. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. [DOI] [PubMed] [Google Scholar]
- Caselli, R. J. 1993. Ventrolateral and dorsomedial somatosensory association cortex damage produces distinct somesthetic syndromes in humans. Neurology 43, 762–771. [DOI] [PubMed] [Google Scholar]
- Cipolloni, P. B. & Pandya, D. N. 1999. Cortical connections of the frontoparietal opercular areas in the rhesus monkey. J. Comp. Neurol. 403, 431–458. [PubMed] [Google Scholar]
- Draganski, B. , Gaser, C. , Busch, V. , Schuierer, G. , Bogdahn, U. & May, A. 2004. Changes in grey matter induced by training Newly honed juggling skills show up as a transient feature on a brain‐imaging scan. Nature 427, 311–312. [DOI] [PubMed] [Google Scholar]
- Driemeyer, J. , Boyke, J. , Gaser, C. , Bu, C. & May, A. 2008. Changes in gray matter induced by learning — revisited. PLoS ONE 3, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein, Y. , Borrell, V. , Garcia, C. , Burstyn‐cohen, T. , Tzarfaty, V. , Frumkin, A. , Nose, A. , Okamoto, H. , Higashijima, S. , Soriano, E. & Klar, A. 1999. F‐spondin and mindin: two structurally and functionally related genes expressed in the hippocampus that promote outgrowth of embryonic hippocampal neurons. Development 3648, 3637–3648. [DOI] [PubMed] [Google Scholar]
- Fukazawa, T. , Matsumoto, M. , Imura, T. , Khalesi, E. , Kajiume, T. , Kawahara, Y. , Tanimoto, K. & Yuge, L. 2013. Electrical stimulation accelerates neuromuscular junction formation through ADAM19/neuregulin/ErbB signaling in vitro. Neurosci. Lett. 545, 29–34. [DOI] [PubMed] [Google Scholar]
- Hiester, B. G. , Galati, D. F. , Salinas, P. C. & Jones, K. R. 2013. Neurotrophin and Wnt signaling cooperatively regulate dendritic spine formation. Mol. Cell Neurosci. 56, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima, S. , Nose, A. , Eguchi, G. , Hotta, Y. & Okamoto, H. 1997. Mindin/F‐spondin family: novel ECM proteins expressed in the zebrafish embryonic axis. Dev. Biol. 192, 211–227. [DOI] [PubMed] [Google Scholar]
- Hihara, S. , Notoya, T. , Tanaka, M. , Ichinose, S. , Ojima, H. , Obayashi, S. , Fujii, N. & Iriki, A. 2006. Extension of corticocortical afferents into the anterior bank of the intraparietal sulcus by tool‐use training in adult monkeys. Neuropsychologia 44, 2636–2646. [DOI] [PubMed] [Google Scholar]
- Hsieh, J. C. , Kodjabachian, L. , Rebbert, M. L. , Rattner, A. , Smallwood, P. M. , Samos, C. H. , Nusse, R. , Dawid, I. B. & Nathans, J. 1999. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 398, 431–436. [DOI] [PubMed] [Google Scholar]
- Iriki, A. , Tanaka, M. & Iwamura, Y. 1996. Coding of modified body schema during tool use by macaque postcentral neurones. NeuroReport 7, 2325–2330. [DOI] [PubMed] [Google Scholar]
- Iriki, A. & Taoka, M. 2012. Triadic (ecological, neural, cognitive) niche construction: a scenario of human brain evolution extrapolating tool use and language from the control of reaching actions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriki, A. , Yamazaki, Y. , Hikishima, K. , Saiki, M. , Inada, M. , Sasaki, E. , Lemon, R. , Price, C. & Okano, H. 2014. Brain structural changes through long‐term learning of tool use supported by sustained motivation for tool use in adult non‐human primates. Soc. Neurosci. Abst. 93.05/RR50. [Google Scholar]
- Ishibashi, H. , Hihara, S. , Takahashi, M. , Heike, T. , Yokota, T. & Iriki, A. 2002a. Tool‐use learning selectively induces expression of brain‐derived neurotrophic factor, its receptor trkB, and neurotrophin 3 in the intraparietal multisensorycortex of monkeys. Brain Res. Cogn. Brain Res. 14, 3–9. [DOI] [PubMed] [Google Scholar]
- Ishibashi, H. , Hihara, S. , Takahashi, M. , Heike, T. , Yokota, T. & Iriki, A. 2002b. Tool‐use learning induces BDNF expression in a selective portion of monkey anterior parietal cortex. Brain Res. Mol. Brain Res. 102, 110–112. [DOI] [PubMed] [Google Scholar]
- Iwamura, Y. 1998. Hierarchical somatosensory processing. Curr. Opin. Neurobiol. 8, 522–528. [DOI] [PubMed] [Google Scholar]
- Komatsu, K. , Wakatsuki, S. , Yamada, S. , Yamamura, K. , Miyazaki, J. & Sehara‐Fujisawa, A. 2007. Meltrin beta expressed in cardiac neural crest cells is required for ventricular septum formation of the heart. Dev. Biol. 303, 82–92. [DOI] [PubMed] [Google Scholar]
- Kühn, S. , Gleich, T. , Lorenz, R. C. , Lindenberger, U. & Gallinat, J. 2014. Playing Super Mario induces structural brain plasticity: gray matter changes resulting from training with a commercial video game. Mol. Psychiatry 19, 265–271. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Cao, C. , Jia, W. , Yu, L. , Mo, M. , Wang, Q. , Huang, Y. , Lim, J.‐M. , Ishihara, M. , Wells, L. , Azadi, P. , Robinson, H. , He, Y.‐W. , Zhang, L. & Mariuzza, R. A. 2009. Structure of the F‐spondin domain of mindin, an integrin ligand and pattern recognition molecule. EMBO J. 28, 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maravita, A. & Iriki, A. 2004. Tools for the body (schema). Trends Cogn. Sci. 8, 79–86. [DOI] [PubMed] [Google Scholar]
- Matsunaga, E. , Nambu, S. , Oka, M. & Iriki, A. 2015a. Comparative analysis of developmentally regulated expressions of Gadd45a, Gadd45b, and Gadd45 g in the mouse and marmoset cerebral cortex. Neuroscience 284, 566–580. [DOI] [PubMed] [Google Scholar]
- Matsunaga, E. , Nambu, S. , Oka, M. & Iriki, A. 2014. Complementary and dynamic type II cadherin expression associated with development of the primate visual system. Dev. Growth Differ. 56, 535–543. [DOI] [PubMed] [Google Scholar]
- Matsunaga, E. , Nambu, S. , Oka, M. , Tanaka, M. , Taoka, M. & Iriki, A. 2015b. Periostin, a neurite outgrowth‐promoting factor, is expressed at high levels in the primate cerebral cortex. Dev. Growth Differ. 57, 200–208. [DOI] [PubMed] [Google Scholar]
- Murray, E. A. & Mishkin, M. 1984. Relative contributions of SII and area 5 to tactile discrimination in monkeys. Behav. Brain Res. 11, 67–83. [DOI] [PubMed] [Google Scholar]
- Neuner, R. , Cousin, H. , McCusker, C. , Coyne, M. & Alfandari, D. 2008. Xenopus ADAM19 is involved in neural, neural crest and muscle development. Mech. Dev. 126, 240–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quallo, M. M. , Price, C. J. , Ueno, K. , Asamizuya, T. , Cheng, K. , Lemon, R. N. & Iriki, A. 2009. Gray and white matter changes associated with tool‐use learning in macaque monkeys. Proc. Natl Acad. Sci. USA 106, 18379–18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss, K. & Saftig, P. 2009. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin. Cell Dev. Biol. 20, 126–137. [DOI] [PubMed] [Google Scholar]
- Ridley, R. M. & Ettlinger, G. 1976. Impaired tactile learning and retention after removals of the second somatic sensory projection cortex (SII) in the monkey. Brain Res. 109, 656–660. [DOI] [PubMed] [Google Scholar]
- Rosso, S. B. & Inestrosa, N. C. 2013. WNT signaling in neuronal maturation and synaptogenesis. Front. Cell. Neurosci. 7, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmacher, A. T. , Padmanabhan, R. , Jhingory, S. & Taneyhill, L. A. 2014. Cadherin‐6B is proteolytically processed during epithelial‐to‐mesenchymal transitions of the cranial neural crest. Mol. Biol. Cell 25, 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakabe, K. , Wakatsuki, S. , Kurisaki, T. & Fujisawa‐Sehara, A. 2001. Roles of Meltrin beta /ADAM19 in the processing of neuregulin. J. Biol. Chem. 276, 9352–9358. [DOI] [PubMed] [Google Scholar]
- Sluming, V. , Barrick, T. , Howard, M. , Cezayirli, E. , Mayes, A. & Roberts, N. 2002. Voxel‐based morphometry reveals increased gray matter density in broca's area in male symphony orchestra musicians. NeuroImage 17, 1613–1622. [DOI] [PubMed] [Google Scholar]
- Taoka, M. , Tanaka, M. , Hihara, S. , Ojima, H. & Iriki, A. 2013. Neural response to movement of the hand and mouth in the secondary somatosensory cortex of Japanese monkeys during a simple feeding task. Somatosens. Mot. Res. 30, 140–152. [DOI] [PubMed] [Google Scholar]
- Umemiya, T. , Takeichi, M. & Nose, A. 1997. M‐spondin, a novel ECM protein highly homologous to vertebrate F‐spondin, is localized at the muscle attachment sites in the Drosophila embryo. Dev. Biol. 186, 165–176. [DOI] [PubMed] [Google Scholar]
- Yamazaki, Y. , Echigo, C. , Saiki, M. , Inada, M. , Watanabe, S. & Iriki, A. 2011. Tool‐use learning by common marmosets (Callithrix jacchus). Exp. Brain Res. 213, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Weskamp, G. , Chesneau, V. , Vortkamp, A. , Horiuchi, K. , Hahn, R. , Wilkes, D. , Fisher, P. , Baron, R. , Manova, K. , Basson, C. T. , Blobel, C. P. , Sahin, U. , Chiusaroli, R. & Hempstead, B. 2004. Essential role for ADAM19 in cardiovascular morphogenesis. Mol. Cell. Biol. 24, 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sets used for 2nd screening by qRT‐PCR.


