Abstract
Background and Objectives
In multiple sclerosis (MS), immune cells invade the CNS and destroy myelin. Macrophages contribute to demyelination and myelin repair, and their role in each process depends on their ability to acquire specific phenotypes in response to external signals. In this article, we assess whether defects in MS patient macrophage responses may lead to increased inflammation or lack of neuroregenerative effects.
Methods
CD14+CD16− monocytes from patients with MS and healthy controls (HCs) were activated in vitro to obtain homeostatic-like, proinflammatory, and proregenerative macrophages. Macrophage activation profiles were assessed through RNA sequencing and metabolomics. Surface molecule expression of CD14, CD16, and HLA-DR and myelin phagocytic capacity were evaluated with flow cytometry. Macrophage supernatant capacity to influence oligodendrocyte precursor cell differentiation toward an astrocytic or oligodendroglia fate was also tested.
Results
We observed that MS patient monocytes ex vivo recapitulate their preferential activation toward the CD16+ phenotype, a subset of proinflammatory cells overrepresented in MS lesions. Functionally, MS patient macrophages display a decreased capacity to phagocytose human myelin and a deficit of processing myelin after ingestion. In addition, MS patient macrophage supernatant favors astrocytes over oligodendrocyte differentiation when compared with HC macrophage supernatant. Furthermore, even when exposed to homeostatic or proregenerative stimuli, MS patient macrophages uphold a proinflammatory transcriptomic profile with higher levels of cytokine/chemokine. Of interest, MS patient macrophages exhibit a distinct metabolic signature with a mitochondrial energy metabolism blockage. Transcriptomic data are further substantiated by metabolomics studies that reveal perturbations in the corresponding metabolic pathways.
Discussion
Our results show an intrinsic defect of MS patient macrophages, reminiscent of innate immune cell memory in MS, lifting macrophage importance in the disease and as potential therapeutic targets.
Introduction
Multiple sclerosis (MS) is an inflammatory disease in which peripheral immune cells infiltrate the CNS and destroy myelin, a neuroprotective and conduction-enhancing substance. The infiltrating cells consist of lymphocytes and monocytes which, together with microglia (the tissue-resident macrophages of the CNS), induce and maintain neuroinflammation. Infiltrating monocytes differentiate into macrophages and along with microglial cells play an important role in the disease.1 They contribute to myelin destruction through perpetuation of the inflammatory environment, recruitment of leukocytes, antigen presentation, and damage to neural cells through toxic effector mechanisms.2-4
Furthermore, macrophages play a role in an endogenous repair process termed “remyelination.” Remyelination capacity varies greatly between patients, and a high capacity is correlated with positive outcomes.5,6 In animal models, depletion of macrophages leads to reduced proliferation and differentiation of oligodendrocyte precursor cells (OPCs), resulting in fewer myelin-producing oligodendrocytes.7,8
Macrophages respond to cues in the environment that dictate different activation states, which are characterized by distinct transcriptomic and metabolic changes.9 The activation states are typically described with terms such as “proinflammatory” (typically induced with interferon [IFN] γ and lipopolysaccharide [LPS] in vitro) and “anti-inflammatory” (typically induced with interleukin (IL)-4 in vitro). While macrophages in an organism often display features of different activation states, simultaneously in vitro activation of macrophages with proinflammatory or anti-inflammatory stimuli has highlighted the extensive macrophage response inducing this functional diversity.10,11
Proinflammatory macrophages demonstrate an increased production of proinflammatory cytokines and cytotoxic molecules, indicating their potentially detrimental role in MS. In the case of effects on remyelination, in vivo and in vitro results show that proinflammatory macrophages promote OPC proliferation while anti-inflammatory macrophages promote OPC differentiation.7 The same study showed that, in vivo, the early stages of successful remyelination includes a switch in the macrophage population from a majority of proinflammatory cells to proregenerative macrophages. Thus, pathologic activation of macrophages seems as a potential culprit in both destruction and lack of repair.
In the CNS of a patient with MS, loss of homeostatic marker purinergic receptor P2Y12 (P2RY12) is seen in both lesions and normal-appearing white matter.12 Proinflammatory macrophage markers are abundant in active lesions and slowly expanding lesion rims,12,13 both of which show active demyelination. However, it is not known to what extent this perturbed state is driven by a pathologic environment or by intrinsic features of infiltrating macrophages. Emerging evidence points to intrinsic features of infiltrating macrophages in MS. For instance, patients with MS show an increase in CD16+ monocyte subpopulations in blood14 and/or CSF,15 implying differential activation of peripheral monocytes. Furthermore, recent study has provided insights into a dysregulation of bone marrow hematopoietic stem cell differentiation, resulting in an augmented production of myeloid cells, which could contribute to the accumulation of proinflammatory macrophages in MS.16 Intrinsic defects in MS monocytes were also reported with a decreased capacity for myelin phagocytosis compared with healthy control monocytes.17,18 All these clues suggest intrinsic defects in MS macrophages, but understanding the underlying mechanisms would help establish whether the innate immune system could be targeted in novel treatment strategies.
Using freshly isolated monocytes from blood samples and differentiating them in vitro, we test the macrophage's ability to induce proinflammatory or proregenerative gene expression in an artificial and controlled environment. Our original approach explores the interplay between disease status and essential aspects of macrophage activation at the functional and molecular levels.
Methods
Participants
A total of 47 patients with MS and 46 healthy controls (HCs) were included in the study. Patients were recruited from multicase families as part of a project studying the MS patient phenotype. 6 patients were single cases while the remaining 30 were from 15 sibling pairs. All patients met MS diagnostic criteria, and individuals with other inflammatory or neurologic disorders were excluded. Clinical evaluation, including Expanded Disability Status Scale testing and documentation of treatment history, was conducted on the same day as blood sampling.
Standard Protocol Approvals, Registrations, and Patient Consents
Approval was obtained from the French Ethics Committee and the Ministry of Research (NCT03369106), and written informed consent was obtained from all participants.
Macrophage Culture and Activation
Blood was sampled in acid citrate dextrose tubes. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Paque Plus (GE Healthcare Life Sciences) and by centrifugation (2200 rpm, 20 minutes) (detailed PBMC isolation and culture given in eMethods).
Human Myelin Extraction
Human myelin was extracted from normal-appearing white matter of a postmortem MS patient brain tissue (detailed methods given in eMethods).
Phagocytosis Assay and Flow Cytometry
Macrophage phagocytic capacity was evaluated through flow cytometry. After 24 hours of activation, macrophages in 24-well plates were exposed to labeled human myelin (25 µg/mL) in RPMI + 10% FBS for 1 hour at 37°C. Negative controls were included. After incubation, cells were washed with PBS, detached using Trypsin-EDTA, and washed again by centrifugation. Labeling was performed with anti-HLA-DR-PB and anti-CD16-FITC (Duraclone precoated tubes or separate antibodies, all from Beckman-Coulter) and anti-CD14-PEvio770 (Miltenyi) at room temperature in darkness for 30 minutes (precoated tubes) or 45 minutes at 4°C (liquid antibodies) in PBS + 5% FBS. After washing, cells were analyzed with MACSQuant (Miltenyi). Results were analyzed with FlowLogic software (Inivai Technologies). Cells were gated for size and granulosity (FSC/SCC) and singlets (FSC-A/FSC-H).
Multiway Generalized Canonical Correlation Analysis
Flow cytometric data were analyzed using Multiway Regularized Generalized Canonical Correlation Analysis (MGCCA). MGCCA was used to explore the complex relationships between flow cytometric data measured at 3 different states of activation and the response variable (HC vs MS) (detailed analysis methods given by eMethods).
RNA Sequencing and Differential Expression (DE) Analysis
Macrophage samples were processed 24 hours after activation for RNA extraction using the Nucleospin RNA extraction kit. Transcriptome sequencing was performed on 28 patients and 11 HCs using a Truseq stranded mRNA kit on a NextSeq 500 sequencer. Raw data quality was assessed with FastQC and trimmed with Fastp. Reads were aligned to the hg38 genome using Star v2.5.3a, and gene abundances were quantified with RSEM 1.2.28 (detailed analysis methods given in eMethods).
Weighted Gene Co-expression Network Analysis
Weighted gene co-expression network analysis (WGCNA)19 was performed on batch-corrected data to identify co-expression modules, considering overrepresented KEGG pathways and GO terms (detailed analysis methods given in eMethods).
Metabolomics Analysis
Metabolomics analysis was conducted on GM-CSF–exposed macrophages from 29 patients (16 treated, 13 untreated) and 12 healthy controls (HCs). Cell preparation mirrored that of RNASeq samples until lysis, where cells were detached with 0.25% Trypsin-EDTA, then processed, and stored. Samples were homogenized in 0.1% formic acid and methanol, then sonicated, and centrifuged before LC-MS analysis. LC-MS experiments were performed using a HILIC phase chromatographic column coupled to a Q-Exactive mass spectrometer (detailed analysis methods given in eMethods).
Macrophage-Conditioned Medium Effect on OPC Differentiation
Macrophage-conditioned media were collected 24 hours after activation, conserved at −80°C, and thawed just before use. OPC cell line CG420 was plated in 96-well plates (4 × 103 cells per well) coated with polylysine in the N1-B104 medium. After 1 day of culture, the medium was removed and replaced by human macrophage–conditioned media. Cells were fixed using 2% PFA and cold methanol after 72 hours.
Immunocytochemistry was performed using O4 (Home made Hybridome) and GFAP (Dako Z0334) antibody to evaluate oligodendroglial cells or astrocyte quantity. Images were taken using a plate scanner CellInsight NXT (ThermoFisher) and converted to TIFF for quantification when needed. To assess CG4 differentiation, the proportion of O4+ and GFAP+ cells on the total number of cells was evaluated using ImageJ software.
Other Statistical Analyses
Other statistical analyses are given in eMethods.
Data Availability
The omics data used in this article are accessible on GEO at accession number GSE269706.
Results
Generation of Monocyte-Derived Macrophages
Monocytes were isolated from PBMCs of a total of 47 patients with MS and 46 age-matched HCs (Table and eTable 1) using CD14-specific magnetic separation. After isolation, patient samples showed a slightly reduced proportion of CD14+CD16− cells compared with HC, but this was not explained by an increase in another specific population (eFigure 1D). Monocytes were then differentiated in vitro, and we refer to these samples as MGM-CSF, MIFNγ+LPS, and MIL4, respectively.
Table.
Characteristics of Continuous Variables of Study Participants, Given as Mean ± SD
| Age (y) | Female sex | Disease duration (y) | % MS-treated | % RRMS | EDSS | |
| HC | 39 ± 10 | 23/44 | ||||
| MS | 40 ± 12 | 35/44 | 12 ± 8 | 26/44 | 33/36 | 2.3 ± 1,5 |
Abbreviations: CIS = clinically isolated syndrome; MSSS = multiple sclerosis severity score; RRMS = relapsing-remitting MS; SP = secondary progressive.
MS Macrophages Differ From HC in Expression of Cell Surface Markers and in Function
Having started off with a primarily CD14+CD16− monocyte population, we examined how expression of CD14 and CD16, as well as HLA-DR, differed between HC and MS macrophages after each set of activation stimuli. We also studied their myelin-phagocytosing capacity. Using Cy5-labeled human myelin, we evaluated samples from 34 patients and 34 HCs for phagocytic and high-phagocytic cells and CD14 and CD16 positivity.
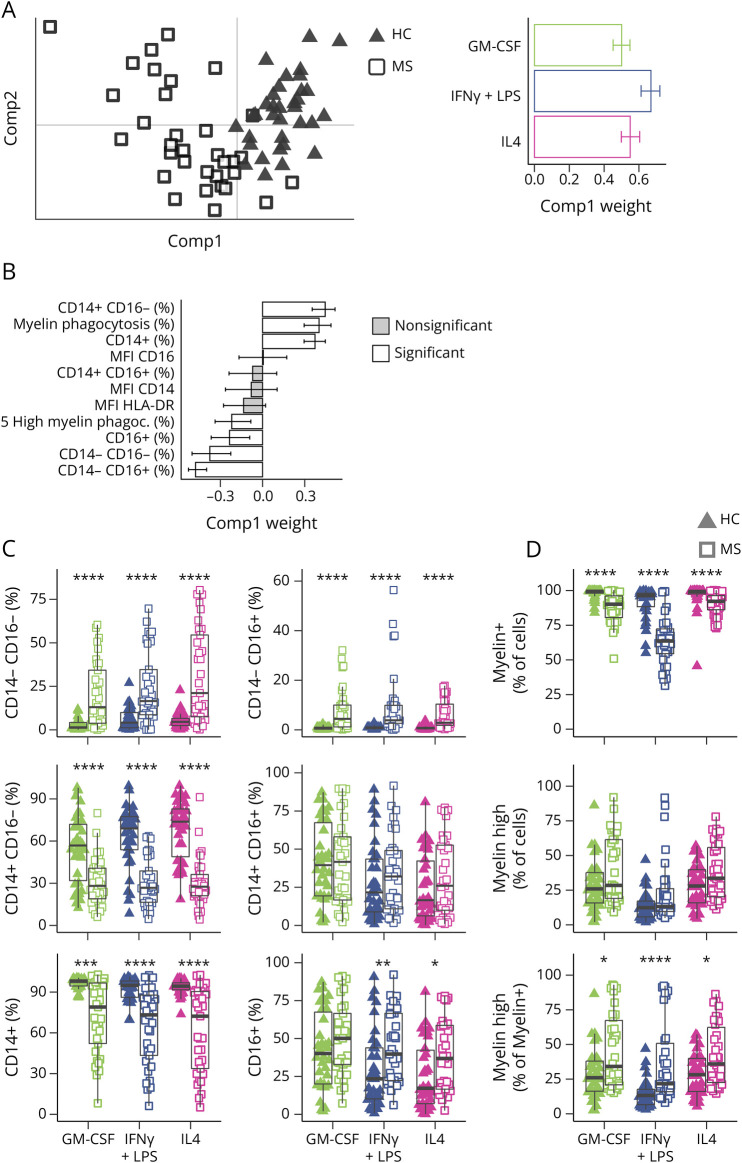
To efficiently identify the key differences among these multiple parameters, MGCCA was performed with disease status (HC or MS) as the target feature. The 3 activation states were considered by constructing a 3-order tensor (individuals×biological markers×activation states) (in eMethods). The first component successfully separated 88% of patients from controls using information from all 3 activation states (Figure 1A). This difference in the first component was not confounded by sex, age, or family relations between individuals (eFigure 2, A–C). The major variables contributing to the first component indicated higher percentages of CD14−/CD16+, CD14−/CD16−, CD16+, and highly phagocytosing cells in MS and higher percentages of CD14+, phagocytosing cells, and CD14+/CD16− cells in HCs (Figure 1B).
Figure 1. MS Patient Macrophages Exhibit Altered Levels of Cell Surface Marker and Functionalities Compared With HC Macrophages.
(A) PCA with the first 2 components of MGCCA targeting the differences between MS (squares) and HC (triangles) across all activation states and variables. The right panel represents the weights of each activation state to the first component of A. White bars represent variables that significantly contribute to the scores of samples. (B) Weights of each flow cytometry variable to the first component of A. White bars represent variables that significantly contribute to the scores of samples. (C) Distribution of CD14/CD16 population proportions in MS and HC for each activation state. (D) Myelin phagocytosis capacity of HC and MS patient macrophages. Data are shown for myelin positivity, high myelin positivity, and high myelin positivity as a percentage of myelin-positive cells. *p < 0.05, **p < 0.01, ***p < 0.001, and p < 0.0001 in the Mann-Whitney U test between HC and MS for each activation state. Error bars indicate 95% CI as calculated by bootstrapping (C and D). For box plots, values are grouped by activation state and disease (HC: triangles; MS: squares; GM-CSF: green; IFNγ+LPS: blue; IL4: pink). HC = healthy controls (n = 34); MS = patients with MS (n = 34).
There were fewer CD14+ cells in MS samples in all 3 conditions (Figure 1C), and a higher percentage of CD16+ cells in MS was most notable in MIFNγ+LPS and MIL4, although the CD14−/CD16+ population was more present in MGMCSF as well (Figure 1C). There were no differences in MFI of CD14, CD16, or HLA-DR (eFigure 3A).
The lower percentage of myelin-phagocytosing cells in MS samples was evident in all 3 activation states (Figure 1D). However, myelin-positive cells from MS samples were more likely to be gated as highly positive for myelin (Figure 1D, lower panel). Thus, while MS macrophages were less likely to phagocytose myelin, the actively phagocytosing cells were seemingly ingesting more myelin and/or failing to process it after ingestion.
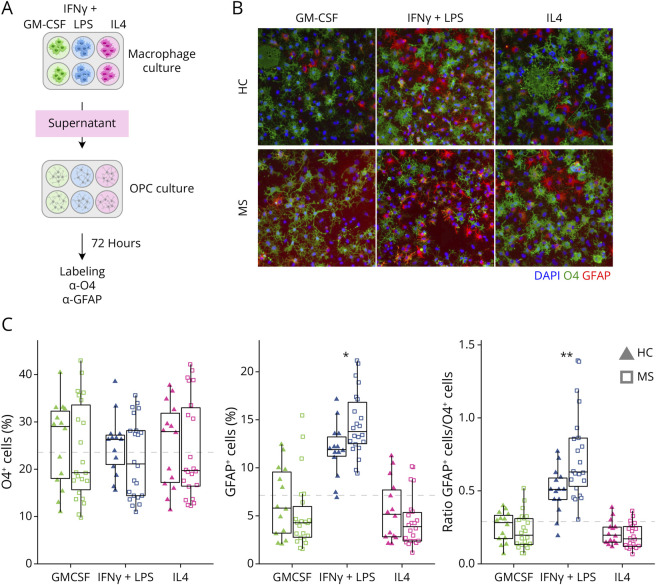
In addition, we tested the effect of macrophage-conditioned media on OPC differentiation (Figure 2A). Conditioned media of MS MIFNγ+LPS were more potent to drive cells toward an astrocytic lineage compared with HC MIFNγ+LPS–conditioned media (Figure 2B, quantification in Figure 2C), suggesting an exacerbated proinflammatory phenotype of MS MIFNγ+LPS macrophages.
Figure 2. MS Patient Macrophage-Conditioned Media Induce More Astrocytic Differentiation of OPC Than HC Macrophage-Conditioned Media.
(A) Experimental overview for the OPC differentiation assay: HC and MS macrophage-conditioned media were collected after 24 hours of treatment with GM-CSF, IFNγ+LPS, or IL4 and added to cell cultures from the CG4 rat OPC line. (B) Cell fate toward an OL or astrocytic lineage was evaluated using O4 (green, h) and GFAP (red, h) antibodies, respectively. (C) Distribution of percentages of O4+ and GFAP+ cells and the ratio of the 2, grouped by disease group and activation states. The dashed gray line corresponds to the level of O4, GFAP, and the ratio of GFAP/O4 in CG4 cells in absence of macrophage-conditioned media. *p < 0.05, **p < 0.01, ***p < 0.001, and p < 0.0001 in the Mann-Whitney U test between HC and MS for each activation state. For box plots, values are grouped by activation state and disease (HC: triangles; MS: squares; GM-CSF: green; IFNγ+LPS: blue; IL4: pink). HC = healthy controls (n = 14); MS = patients with MS (n = 2).
Next, we examined the differences between activation states for each individual to approximate their responses to stimuli. For each variable analyzed above, we calculated the difference in log2-transformed values between MGMCSF and MIFNγ+LPS or MIL4 samples. Repeating the MGCCA (eFigure 3B), we again saw a shift between MS and HC in the first component (eFigure 3C) and this difference was present in both activation stimuli (eFigure 3D). This difference in the first component was not confounded by age, treatment, or family relations between individuals (eFigure 2, A–C). A slight difference could be seen between men and women, with male MS and HC samples showing less separation than female samples, although the number of men was too small to draw reliable conclusions from this (eFigure 2, D and E). We noted that there was a reduction of CD16+ cells in HC MIFNγ+LPS and MIL4 compared with MGMCSF, but not in the MS equivalents (eFigure 3, E and F). Instead, there was a reduction of CD14+ cells in MS not seen in HCs (eFigure 3, E and F). Changes in CD14, CD16, and HLA-DR MFI values were equivalent between MS and HC samples (eFigure 3 G). We also observed a more extensive decrease in phagocytic capacity after proinflammatory stimulation in MS compared with HC (eFigure 3H). This suggests a more responsive phenotype to proinflammatory stimuli in MS macrophages with a higher antiphagocytic response. The effect of macrophage-conditioned media on OPCs did not show significant differences in relative changes, but a trend of increased response to proinflammatory stimuli in the promotion of astrocyte lineage differentiation could be seen (eFigure 3I).
Transcriptomic Profiles of MS Patient Macrophages Differ From Those of HC Macrophages
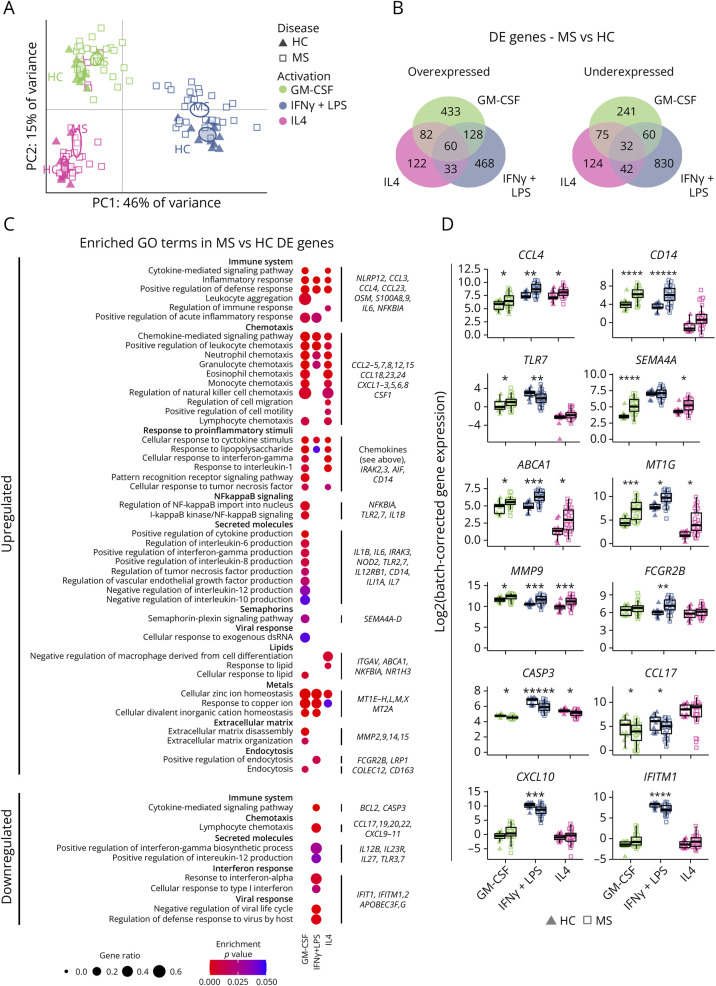
Transcriptomic profiles were analyzed through RNASeq in samples generated as above from 28 patients with MS and 11 HCs. The efficacy of our activation stimuli in HC macrophages was confirmed by the expression of known activated genes (eFigure 4). Principal component analysis (PCA) of all samples showed that while the major sources of variance were the activation stimuli, MS and HC samples were significantly different in all activation states (no overlap in 95% confidence intervals calculated by bootstrapping) along principal components 1 and 2 (61% of variance explained) (Figure 3A, HC: black triangles, MS: white squares). Disease status contributed to intersample variation, but to a lesser degree than activation stimuli. The major differences in transcriptomic profiles were independent of patient sex, age, treatment, and sibling effects (eFigure 5).
Figure 3. MS Macrophages Show a More Proinflammatory Transcriptomic Profile Compared With That of HC.
(A) PCA with each observation representing 1 sample from 1 individual (HC: triangles; MS: squares; GM-CSF: green; IFNγ+LPS: blue; IL4: pink). Ellipses indicate 95% CI of the group mean based on bootstrapping. (B) Venn diagrams showing the number of differentially overexpressed and underexpressed genes (absolute log2(fold-change) >0.5, q < 0.05) between HC and MS samples in each activation state. (C) A selection of GO terms, sorted by broader functionality, with dots indicating a significant overrepresentation in a given set of DE genes (adjusted p < 0.05). The proportion of genes from the term and the p value are indicated by the size and color of the dots, respectively. Examples of DE genes from at least 1 list of genes are provided on the right. (D) Sample distribution for a subset of genes highlighted in D. Expression is given in log2 and grouped by activation state and disease as in C. *q < 0.05, **q < 0.01, ***q < 0.001, ****q < 0.0001, and *****q < 0.00001 in limma DE analysis. GO = gene ontology; HC = healthy controls (n = 11); MS = patients with MS (n = 28).
MS Patient Macrophages Show a Globally Proinflammatory Transcriptomic Profile
We next performed DE analyses comparing MS and HC samples in each activation state independently (log2 fold-change ≥0.5, q < 0.05). In MGM-CSF, MIFNγ+LPS, and MIL4, respectively, 703, 689, and 297 genes were overexpressed in MS, whereas 408, 964, and 273 genes were underexpressed (Figure 3B). Several genes were differentially expressed in more than 1 activation state (Figure 3B).
GO term analysis revealed overrepresentation of inflammatory pathways in all activation states (Figure 3C). Multiple chemokine-coding genes, including CCL4 (Figure 3D), were increased in multiple activation states, suggesting an increased capacity to attract other proinflammatory immune cells. Conversely, fewer chemokines such as CCL17 and CCL22, known for their anti-inflammatory function, were underexpressed (Figure 3D). Proinflammatory genes such as CD14 and TLR7 were upregulated in 1 or more states, along with metallothionein genes (MT1G) (Figure 3D). Differences were also seen in apoptosis-related genes such as CASP3 (Figure 3D), downregulated most notably in MIFNγ+LPS.
In MGM-CSF, the DE genes were consistently indicative of a proinflammatory profile, with alterations of genes related to cytokine secretion, semaphorins (SEMA4A, Figure 3D), and extracellular matrix disassembly (MMP9, Figure 3D). Different genes related to endocytosis (FCGR2B, Figure 3D) were overexpressed in MS MGM-CSF and MIFNγ+LPS.
MIL4 showed a profile similar to that of MGM-CSF, albeit with fewer significant genes and GO terms. The only terms that were specific to MIL4 concerned response to lipid, with an upregulation of genes such as ABCA1 (Figure 3D). However, the overexpression of these genes was not specific to MIL4. Rather, the smaller total number of significant genes permitted the lipid-response genes to emerge as significantly overrepresented.
On the contrary, MIFNγ+LPS showed a state-specific underexpression of a subset of inflammatory genes, including chemokines such as CXCL9, CXCL10, and CXCL11 (CXCL10, Figure 3D), known to recruit Th1 cells, pattern recognition receptors such as TLR3 and TLR7 (TLR7, Figure 3D), and interferon response genes of the IFIT and IFITM families (IFITM1, Figure 3D).
Overall, while MIFNγ+LPS showed the broadest dysregulation of genes in the number of significant DE genes (Figure 3B), the differences in MGM-CSF seemed more cohesively indicative of a proinflammatory state (Figure 3C). The combination of both overexpression (Figure 3C) and underexpression (Figure 3C) of proinflammatory genes in MIFNγ+LPS implies a mixed phenotype with a partially incorrect response to proinflammatory stimuli.
The major differences in transcriptomic profiles were independent of patient sex, age, treatment, and sibling effects (eFigure 5).
Patients With MS Exhibit an Altered Response to Proinflammatory Stimuli Compared With HC Macrophages
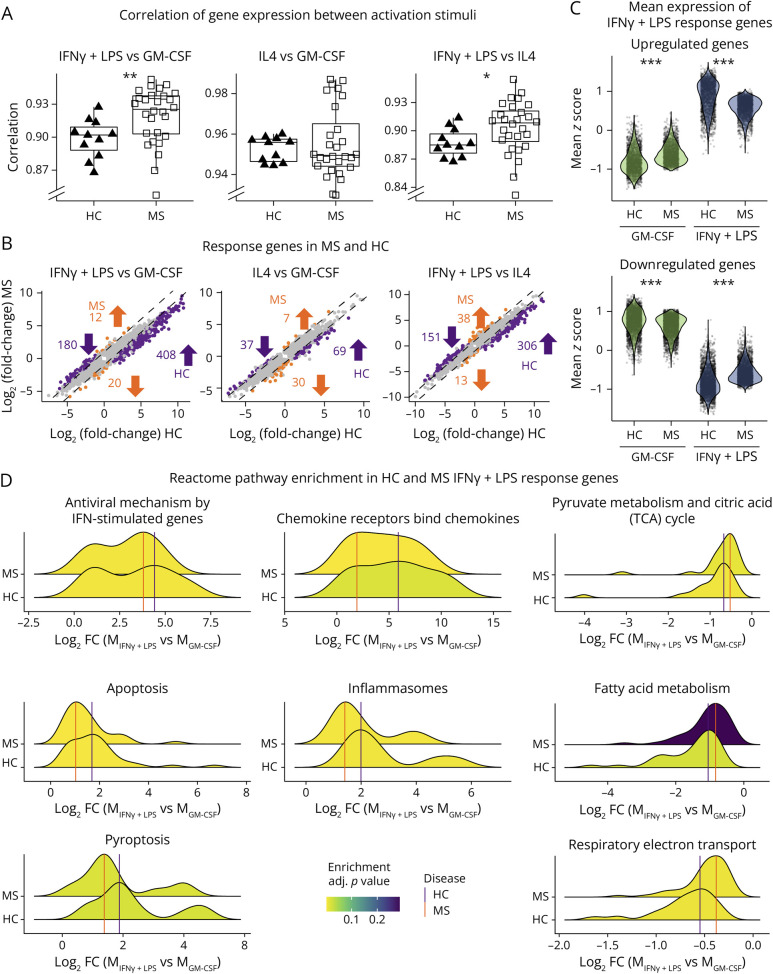
We observed a weakened functional response to proinflammatory stimuli in MS cells, suggesting a disturbed transcriptomic response. Pearson correlation coefficients of gene expression between pairs of stimuli were lower in MS samples compared with HC in both the MGM-CSF vs MIFNγ+LPS and MIFNγ+LPS vs MIL4 comparisons (p < 0.01 and p < 0.05, respectively, Mann-Whitney U test) (Figure 4A).
Figure 4. MS Macrophages Present a Limited Amplitude of Response When Stimulated With IFNγ+LPS.
(A) Boxplots of the Pearson correlation coefficient between gene expression in 2 activation states (IFNγ+LPS vs GM-CSF, IL4 vs GM-CSF, and IFNγ+LPS vs IL4), calculated for each individual (HC: triangles; MS: squares). (B) Scatterplots showing log2(fold-change) between 2 states in HC (x-axis) and MS (y-axis) for each gene that is DE between the 2 states in MS and/or HC. Genes with a difference in mean and median log2(fold-change) greater than 0.5 between HC and MS are highlighted in orange (larger absolute fold change in HC) and purple (larger absolute fold change in MS). Values represent numbers of genes specific to each group and direction of regulation. (C) Distributions of mean expression z-scores of IFNγ+LPS response genes, defined as upregulated (top panel) or downregulated (bottom panel) DEGs between MIFNγ+LPS and MGM-CSF in HCs. Each dot represents one gene for each group (activation stimuli and patient status). (D) Ridgeplots display the distribution of log2(fold-change) of differentially expressed genes between MIFNγ+LPS vs MGM-CSF in HCs (upper densities) and patients with MS (lower densities), divided into enriched reactome pathways. HC = healthy controls (n = 11); MS = patients with MS (n = 28). *p < 0.05, **p < 0.01, and ***p < 0.00001 in the Mann-Whitney U test (A) or Wilcoxon signed-rank test (C).
DE analyses revealed a higher number of genes specific to HC comparisons than in MS in each pair of stimuli, particularly noticeable between MGM-CSF and MIFNγ+LPS (Figure 4B). Indeed, 588 genes showed stronger response in HC than in MS, compared with 32 genes in MS than in HC. The weaker response was not simply a saturation of proinflammatory gene expression because the upregulated genes defined in HC conditions showed an average expression that was both higher in MS MGM-CSF and lower in MS MIFNγ+LPS and vice versa for downregulated genes (Figure 4C). Response genes for each group were enriched with genes from inflammation and metabolism-related reactome pathways, such as inflammasomes and respiratory electron transport, indicating a weaker response in patients with MS (Figure 4D).
Coexpressed Genes Involved in Migration and Metabolism Are Dysregulated in MS Macrophages
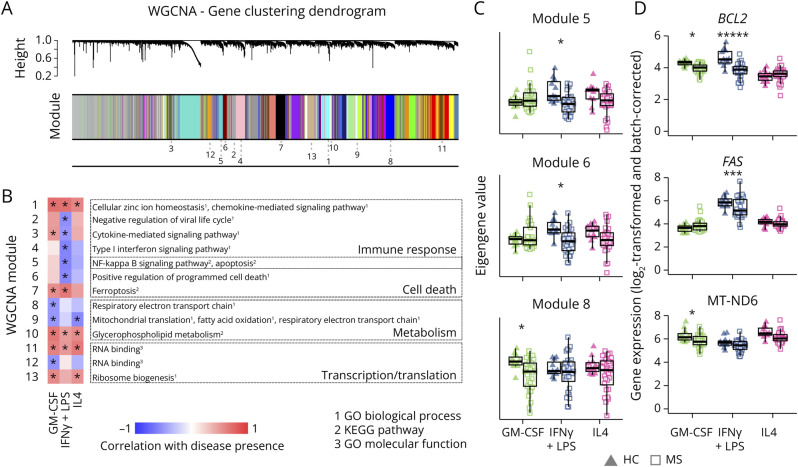
Next, we performed a WGCNA. To avoid that the differences between activation states mask the effect of the disease, we constructed the network by using the maximum correlation for each gene pair across activation states (Figure 5A, eFigure 6). After identifying modules, we analyzed their genes for overrepresented GO terms and KEGG pathways (eFigure 6B). The eigengene value for each module was calculated, and its correlation with disease status was assessed (eFigure 6, B and C). Modules significantly correlated with disease (FDR-adjusted asymptotic p < 0.05) in at least 1 activation state and overrepresented GO term or KEGG pathway are depicted in Figure 5B. 1 module, strongly correlated with disease, contained chemokine and metal homeostasis genes, echoing DE analysis differences.
Figure 5. Modules of Coexpressed Genes Involved in Inflammatory and Metabolic Pathways Show Altered Expression in MS Macrophages Compared With HC Macrophages.
(A) Clustering of genes in the multidataset WGCNA, with module assignment indicated as follows. The multidataset network was produced by comparing correlations between genes for each state individually and basing clustering on the maximum correlation for each gene pair. (B) Heatmap of correlation between presence of disease and eigengene values of each module and set of stimuli. Only modules with significant correlation and functional annotation terms are shown. Functional annotations were given by Enrichr from databases GO biological process, GO molecular function, and KEGG pathways (adjusted p < 0.05). (C) Eigengene values in 3 modules, for each sample and module, grouped according to disease (HC: triangles; MS: squares) and activation states (GM-CSF: green; IFNγ+LPS: blue; IL4: pink). (D) Expression of examples of genes from each module shown in C (grouped and visualized as in C, HC = healthy controls [n = 11]; MS = patients with MS [n = 28]). *p < 0.05. Correlations were calculated with FDR-corrected default WGCNA functions (B and C), and differential expression was tested with limma D.
Six modules were negatively correlated with disease in MIFNγ+LPS, including interferon-response genes and genes involved in cell death (modules 5 and 6, Figure 5C). However, some genes within these modules promote survival, such as BCL2 and FAS (Figure 5D).
Two modules with several genes involved in the respiratory electron transport chain were negatively correlated with disease in MGM-CSF (modules 8 and 9, Figure 5C and eFigure 6C). Although individual gene dysregulation was modest, the collective decreased expression suggests a global reduction of oxidative metabolism, a key feature of anti-inflammatory cells.21
MS Homeostatic-Like Macrophages Present Dysregulated Mitochondrial Energy Metabolism
A metabolomic analysis was performed using liquid chromatography mass spectrometry (LC-MS) on 39 samples (13 HC, 26 MS). Because the transcriptomic changes related to metabolism were mostly noted in MGM-CSF, we focus on this activation state. 4 samples were excluded from analysis after being identified as outliers using robust PCA. Separating between treated (n = 13) and untreated (n = 9) patients, we noted significant differences in several metabolites between untreated patients and HCs as well as between treated and untreated patients while no differences were seen between HCs and treated patients (Figure 6 and 7).
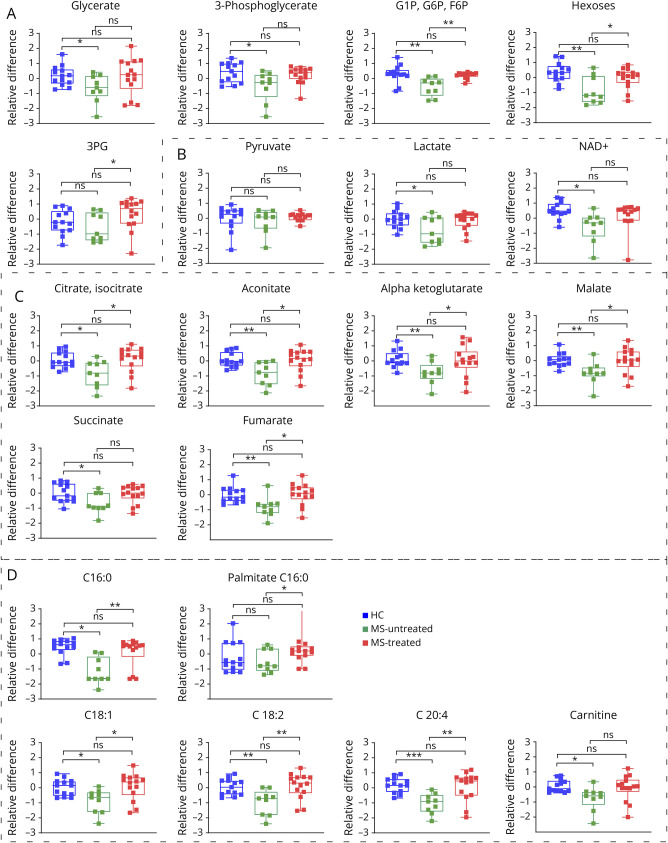
Figure 6. Boxplots of Abundance of Metabolites in HCs and MS.
HC = healthy controls (blue triangle); MS = patients with MS (untreated: green squares, treated: red squares); G1P = glucose-1-phosphate; G6P = glucose-6-phosphate; 3 PG = 3-phosphoglyceric acid. Boxes highlight metabolites implicated, respectively, in (A) glycolysis, (C) TCA cycle, and (D) fatty acid oxidation. *p < 0.05, **p < 0.01, and ***p < 0.001 in the Mann-Whitney U test.
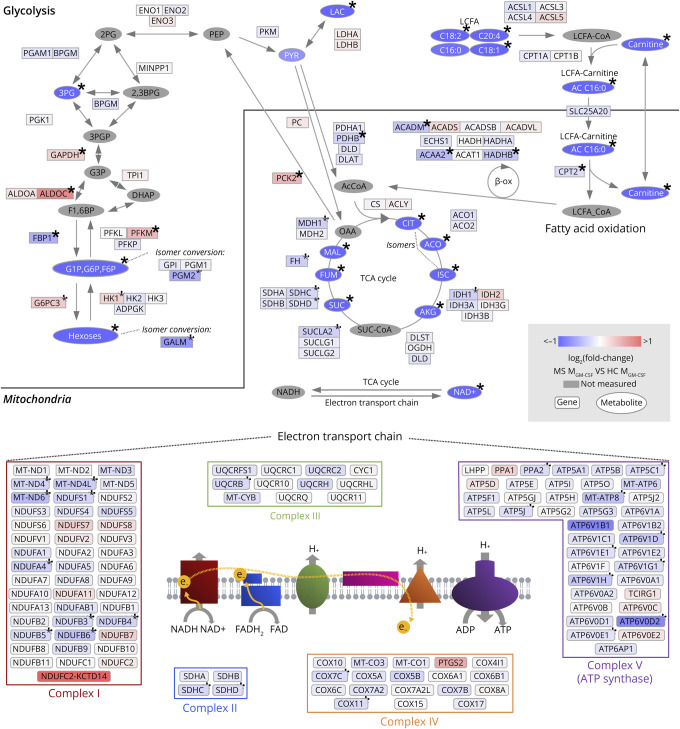
Figure 7. MS Homeostatic-Like Macrophages Show Several Signs of Alteration in Both the TCA Cycle and Respiratory Chain at the Gene Expression and Metabolic Levels.
Genes and metabolites included in the KEGG pathways, glycolysis, fatty acid degradation, TCA cycle, and oxidative phosphorylation, are organized according to pathways. Each element is colored to indicate fold change in MS MGM-CSF relative to HC (Δlog2(FC), overexpression in red, underexpression in blue). Genes involved in oxidative phosphorylation are organized into the complexes in which their gene products take part. Genes that were not expressed in at least 1 sample were excluded from the figure. *Significantly altered genes (q < 0.05) in differential expression analysis (genes) or one-way ANOVA (metabolites). HCs n = 12 (metabolites) or 11 (genes); patients with MS n = 13 (metabolites) or 29 (genes). 2,3BPG = 2,3-bisphosphoglyceric acid; 2 PG = 2-phosphoglyceric acid; 3 PG = 3-phosphoglyceric acid; 3PGP = 3-phospho-d-glyceroyl phosphate; AcCoA = acetyl coenzyme A; ACO = aconitate; AKG = alpha-ketoglutarate; CIT = citrate; DHAP = dihydroxyacetone phosphate; F1,6BP = fructose-1,6-bisphosphate; FUM = fumarate; G1P = glucose-1-phosphate; G3P = 3-phosphoglycerate; G6P = glucose-6-phosphate; HC = healthy controls; ISC = isocitrate; LAC = lactate; LCFA = long-chain fatty acid; MAL = malate; MS = patients with MS; OAA = oxaloacetate; OXSUC = oxalosuccinate; PEP = phosphoenolpyruvic acid; PYR = pyruvate; SUC = succinate; SUC-CoA = succinyl coenzyme A; β-ox = beta-oxidation.
There was a global downregulation of the included metabolites in patients with MS (Figure 6), including significantly lower levels of intermediates of glycolysis (Figure 6A) and the tricarboxylic acid (TCA) cycle (Figure 6C), multiple fatty acids (Figure 6D), and NAD+ (Figure 6B), which is generated by the mitochondrial electron transport chain. This suggests a downregulation of major energy metabolic pathways.
We combined the metabolomic data from HC and untreated MS patient samples with the transcriptomic data to visualize 4 main pathways of energy metabolism: fatty acid oxidation, the TCA cycle, oxidative phosphorylation, and glycolysis (Figure 7). The reduced levels of fatty acids were accompanied by a downregulation of genes encoding for 4 major enzymes in beta-oxidation (CPT2, HADHB, ACADM, and ACAA2). Similarly, the reduction of TCA cycle intermediates was consistent with lower expression of genes encoding for TCA enzymes (IDH1, SUCLA2, SDHC, SDHD, and MDH1). Finally, the lower abundance of NAD+ was accompanied by a global downregulation of genes necessary for each of the 5 mitochondrial complexes involved in oxidative phosphorylation (Figure 7).
We also found significantly decreased levels of hexoses, glucose-1-phosphate/glucose-6-phosphate, 3-phosphoglyceric acid, and, importantly, lactate (Figure 6A and 7), underlining the utilization of all cellular energetic resources. Conversely, 4 key genes (HX1, PFKM, ALDOC, and GAPDH) involved in glycolysis and 2 regulators of gluconeogenesis (PCK2 and G6PC3) were significantly overexpressed in patients with MS (Figure 7), suggesting the possible activation of compensatory mechanisms.
Strikingly, key metabolites related to glycolysis, fatty acid oxidation, and the TCA cycle in treated patients were comparable with HC (Figure 6), highlighting the beneficial effect of treatments on this cellular energy deficiency.
Discussion
Expression of CD14 and CD16 showed significant differences between patients with MS and HCs with a larger proportion of CD16+cells. This finding is consistent with the presence of CD16+ monocytes in active MS lesions, participating in blood-brain barrier breakdown and T-cell invasion of the CNS.15
MS macrophages exhibit reduced capacity to phagocytose and process myelin debris, as seen previously in MS monocytes.17,22 Our study provides evidence that this defect persists even in proregenerative conditions and is exacerbated with proinflammatory stimuli. This exacerbated response to proinflammatory stimuli is further strengthened by the preferential differentiation of CG4 cells to an astrocytic lineage in response to MS MIFNγ+LPS–conditioned media. Because myelin clearance and OPC differentiation toward oligodendrocytes are crucial for remyelination, our findings suggest that MS macrophages inherently lack the ability to properly orchestrate this repair process.7,23
The differential expression of cell surface markers was mirrored by multiple transcriptomic changes. MS macrophages overexpressed several cytokines at the gene and protein levels, including CCL5, IL6, and IL8 (eFigure 7), promoting a proinflammatory environment and immune cell infiltration into the CNS. Furthermore, genes encoding for ligands of CCR2 and CCR5 receptors (CCLs 2–5, 7, 8, and 13) were significantly overexpressed in at least 1 activation state. This implicates macrophages in the previously described increased percentage of CD4+CCR2+CCR5+ cells in the CSF in MS during relapse.24 These CD4+CCR2+CCR5+ cells in turn produced high levels of proinflammatory cytokines and were reactive to myelin basic protein (MBP).24 In addition, CCL2 and CCL5 induce stronger in vitro migratory capacities in monocytes from patients with MS than from HCs,25 implicating MS macrophages in increased recruitment of infiltrating monocytes. Of interest, MS patient macrophages underexpressed CCL17 that is known to attract Th2 and regulatory T cells, subtypes of anti-inflammatory lymphocytes.26 In total, these transcriptomic data suggest that infiltration of disease-associated cell types could be exaggerated by macrophage defects observed in MS conditions.
Another family of genes that was overexpressed in MS macrophages of all activation states was MTs, which are cysteine-rich proteins capable of binding metals. They are important for copper and zinc homeostasis, protection against oxidative stress, and sequestration of heavy metals.27,28 Several roles for MT in immune regulation have been proposed (Subramanian Vignesh and Deepe 2017), and MT overexpression has been described in MS CNS.29 Beyond immune regulation, MT expression could be related to the reduced concentration of zinc observed in MS30 through sequestration. Because zinc binds to myelin proteins such as MBP and seems important for myelin structure and/or function,31,32 its reduction may be of direct importance in the degeneration of myelin.
Overall, we see that macrophages derived from peripheral monocytes of patients with MS present a phenotype that is both reflective of known characteristics in MS lesions and indicates a role of macrophages in important pathologic events.
The activation as proinflammatory and anti-inflammatory macrophages relies on specific metabolic profiles, with increased anaerobic glycolysis and oxidative metabolism, respectively.21 We observed that MS patient macrophages, in absence of proinflammatory signals (MGM-CSF), exhibited an increased expression of glycolytic genes when compared with HC macrophages. However, we could not confirm these results at the metabolite level because we were unable to detect several glycolysis intermediates. Perivascular macrophages with high glycolytic capacity have been identified in MS animal models and showed increased transmigratory functions,33 involving this metabolic switch in immune cell infiltration in MS.
We found a global reduction of electron transport chain gene expression in MS patient macrophages, which was also described in MS postmortem tissue34 and on a protein level in MS patient lymphocytes.35 Reactive oxygen species (ROS) are believed to be a main instigator of oligodendrocyte alterations and neurotoxicity,36 and ROS production is linked to mitochondrial energy metabolism.37 The reduced mitochondrial metabolism seen in MS macrophages could thus reflect a destructive phenotype contributing to neural damage.
In addition to the changes in the electron transport chain, we saw disturbances in the TCA cycle and fatty acid metabolism, indicating overall reduction of oxidative metabolism in MS. Oxidative metabolism is both a hallmark of and necessary for the anti-inflammatory state in macrophages.38,39 The observed imbalance of oxidative metabolism may thus be a result and/or a cause of the pro-inflammatory MS phenotype even in the absence of pro-inflammatory signals.
A few metabolic treatment strategies have been proposed to reduce degeneration in neural cells in progressive MS.40 It is, however, recognized that many metabolic therapeutics could act on macrophages as well40 and more specifically that a metabolic switch toward oxidative phosphorylation is essential for promotion of a proregenerative states. One of the most common first-line treatments in MS, dimethyl fumarate, is believed to function through metabolic alterations,40 although it is unclear which cell types are primarily implicated in this correction. It is important to note that key metabolites related to glycolysis, fatty acid oxidation, and the TCA cycle in treated patients with MS displayed similar levels to HCs, which suggests some beneficial effects of the treatment on innate immune cells.
Our results importantly highlight that macrophages in patients with MS show a predisposition toward a proinflammatory phenotype, suggesting an intrinsic defect that may exacerbate MS lesions. Similar features are seen in other inflammatory diseases such as atherosclerosis and systemic lupus erythematous, where monocytes/macrophages exhibit increased proinflammatory cytokine expression,41,42 metabolic changes,42,43 and overrepresentation of proinflammatory CD14+CD16+ cells.42,44
This predisposition to a proinflammatory phenotype known as trained innate immunity43 was first identified after microbial infections. Increased proinflammatory cytokine production and altered metabolism are both hallmarks of this state.45 Although monocytes are short-lived, trained immunity is epigenetically programmed in the bone marrow, reflecting previous antigen exposure.46 Trained innate immunity is hypothesized to play a role in inflammatory diseases, contributing to disease initiation, maintenance, or aggravation.47 Our results support the role of innate immune memory in MS, although the initial trigger remains unknown. Of interest, the infectious etiology of MS has been considered for a long time, and numerous studies confirm a strong correlation between Epstein-Barr virus infection and the occurrence of MS.48 In addition to infectious triggers, trained innate immunity has also been described in the context of dietary factors such as a Western diet,49 a finding potentially relevant to MS considering the correlation between disease incidence and adolescence obesity.50
In conclusion, our data imply a complex perturbed macrophage response in MS, reminiscent of innate immune memory. We propose that the predisposition to a proinflammatory state and the response to activating stimuli must be considered in an MS-specific context when predicting how the innate immune system can be targeted in treatment.
Acknowledgment
The authors are grateful to all the patients and the volunteers who participated to this study, together with the members of the Zujovic/Nait Oumesmar team for their support. The authors are indebted to the Bouvet-Labruyère family and the OCIRP foundations for their constant support to MS research at the ICM. The authors acknowledge Justine Guegan, Dr Michel Mallat, and Dr. Jaime de Juan-Sanz for their critical reviews of the work. The authors thank the cell culture (CELIS), sequencing (i-GENSEQ), biostatistics facilities (DAC) but also the fundraising, scientific affairs and the administrative department of the Paris Brain Institute. The authors also thank Sarah Taieb Tamacha from the Paris Brain Institute-CIC for her efficient management of MS patient recruitment of the MS-BIOPROGRESS cohort and for supervision of blood sampling from patients with MS.
Glossary
- DE
differential expression/differentially expressed
- EDSS
Expanded Disease Severity Scale
- FBS
fetal bovine serum
- GM-CSF
granulocyte macrophage colony-stimulating factor
- GO
gene ontology
- HC
healthy control
- IFN
interferon
- IL
interleukin
- LPS
lipopolysaccharide
- MGCCA
multiway regularized canonical correlation analysis
- MFI
mean fluorescence intensity
- MS
multiple sclerosis
- MSSS
multiple sclerosis severity score
- MT
metallothionein
- OPC
oligodendrocyte precursor cell
- PBMC
peripheral blood mononuclear cell
- PCA
principal component analysis
- ROS
reactive oxygen species
- TCA
tricarboxylic acid
- Th
T helper
- WGCNA
weighted gene co-expression network analysis
Appendix. Authors
| Name | Location | Contribution |
| Jennifer Fransson, PhD | Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital Pitié Salpétrière Univ. Hosp., DMU Neuroscience 6 | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; analysis or interpretation of data |
| Corinne Bachelin, PhD | Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital Pitié Salpétrière Univ. Hosp., DMU Neuroscience 6 | Major role in the acquisition of data |
| Farid Ichou, PhD | Inst. of Cardiometabolism and Nutrition, Sorbonne-universités-Upmc 06, INSERM, CNRS | Major role in the acquisition of data; analysis or interpretation of data |
| Léna Guillot-Noël, MSc | Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital Pitié Salpétrière Univ. Hosp., DMU Neuroscience 6 | Major role in the acquisition of data |
| Maharajah Ponnaiah, PhD | Inst. of Cardiometabolism and Nutrition, Sorbonne-universités-Upmc 06, INSERM, CNRS | Analysis or interpretation of data |
| Arnaud Gloaguen, PhD | Laboratoire des Signaux et Systèmes (L2S), CNRS-CentraleSupélec, Université Paris-Saclay | Analysis or interpretation of data |
| Elisabeth Maillart, MD | Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital Pitié Salpétrière Univ. Hosp., DMU Neuroscience 6 | Major role in the acquisition of data |
| Bruno Stankoff, MD, PhD | Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital St. Antoine-HUEP | Major role in the acquisition of data |
| Arthur Tenenhaus, PhD | Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital Pitié Salpétrière Univ. Hosp., DMU Neuroscience 6; Laboratoire des Signaux et Systèmes (L2S), CNRS-CentraleSupélec, Université Paris-Saclay | Drafting/revision of the manuscript for content, including medical writing for content; analysis or interpretation of data |
| Bertrand Fontaine, MD, PhD | INSERM, SU, AP-HP, Centre de recherche en Myologie-UMR974 and Service of Neuro-Myology, Institute of Myology, University hospital Pitié-Salpêtriere | Study concept or design |
| Fanny Mochel, MD, PhD | Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital Pitié Salpétrière Univ. Hosp., DMU Neuroscience 6 | Drafting/revision of the manuscript for content, including medical writing for content; analysis or interpretation of data |
| Celine Louapre, MD, PhD | Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital Pitié Salpétrière Univ. Hosp., DMU Neuroscience 6 | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data |
| Violetta Zujovic, PhD | Sorbonne Université, Institut du Cerveau - Paris Brain Institute - ICM, Inserm, CNRS, APHP, Hôpital Pitié Salpétrière Univ. Hosp., DMU Neuroscience 6 | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data |
Study Funding
This work was supported by the OCIRP foundation, Bouvet Labruyère Prize, SANOFI Innovation Awards program (iAwards) 2018 and the programs “Investissements d'Avenir” ANR-10-IAIHU-06, “Translational Research Infrastructure for Biotherapies in Neurosciences” ANR-11-INBS-0011-NeurATRIS and “Idex Sorbonne Université dans le cadre du soutien de l'Etat aux programmes Investissements d'Avenir.”
Disclosure
The authors do not have financial interest in relation to the paper but report some grants and personal fees from private companies: B. Stankoff reports grants and personal fees for lectures from ROCHE, SANOFI-GENZYME, and MERCK-SERONO and personal fees for lectures from NOVARTIS, BIOGEN and TEVA, all outside the submitted work. E. Maillart reports grants and personal fees from Biogen, Novartis, and Roche and personal fees from Merck-Serono, Teva, Sanofi-Genzyme, and Ad Scientiam outside of the submitted work. C. Louapre has received consulting or travel fees from Biogen, Novartis, Roche, Sanofi, Teva and Merck Serono and research grant from Biogen, none related to the present work. V. Zujovic received SANOFI Innovation Awards program (iAwards) 2018, related to this work. Go to Neurology.org/NN for full disclosures.
References
- 1.Wang J, Wang J, Wang J, Yang B, Weng Q, He Q. Targeting microglia and macrophages: a potential treatment strategy for multiple sclerosis. Front Pharmacol. 2019;10:286. doi: 10.3389/fphar.2019.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith KJ, Lassmann H. The role of nitric oxide in multiple sclerosis. Lancet Neurol. 2002;1(4):232-241. doi: 10.1016/s1474-4422(02)00102-3 [DOI] [PubMed] [Google Scholar]
- 3.Anthony DC, Miller KM, Fearn S, et al. Matrix metalloproteinase expression in an experimentally-induced DTH model of multiple sclerosis in the rat CNS. J Neuroimmunol. 1998;87(1-2):62-72. doi: 10.1016/s0165-5728(98)00046-0 [DOI] [PubMed] [Google Scholar]
- 4.Prineas JW, Lee S. Microglia subtypes in acute, subacute, and chronic multiple sclerosis. J Neuropathol Exp Neurol. 2023;82(8):674-694. doi: 10.1093/jnen/nlad046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patrikios P, Stadelmann C, Kutzelnigg A, et al. Remyelination is extensive in a subset of multiple sclerosis patients. Brain. 2006;129(Pt 12):3165-3172. doi: 10.1093/brain/awl217 [DOI] [PubMed] [Google Scholar]
- 6.Bodini B, Veronese M, García-Lorenzo D, et al. Dynamic imaging of individual remyelination profiles in multiple sclerosis. Ann Neurol. 2016;79(5):726-738. doi: 10.1002/ana.24620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miron VE, Boyd A, Zhao JW, et al. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat Neurosci. 2013;16(9):1211-1218. doi: 10.1038/nn.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotter MR, Zhao C, van Rooijen N, Franklin RJM. Macrophage-depletion induced impairment of experimental CNS remyelination is associated with a reduced oligodendrocyte progenitor cell response and altered growth factor expression. Neurobiol Dis. 2005;18(1):166-175. doi: 10.1016/j.nbd.2004.09.019 [DOI] [PubMed] [Google Scholar]
- 9.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958-969. doi: 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol. 2016;17(1):26-33. doi: 10.1038/ni.3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piccolo V, Curina A, Genua M, et al. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat Immunol. 2017;18(5):530-540. doi: 10.1038/ni.3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zrzavy T, Hametner S, Wimmer I, Butovsky O, Weiner HL, Lassmann H. Loss of 'homeostatic' microglia and patterns of their activation in active multiple sclerosis. Brain. 2017;140(7):1900-1913. doi: 10.1093/brain/awx113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jäckle K, Zeis T, Schaeren-Wiemers N, et al. Molecular signature of slowly expanding lesions in progressive multiple sclerosis. Brain. 2020;143(7):2073-2088. doi: 10.1093/brain/awaa158 [DOI] [PubMed] [Google Scholar]
- 14.Gjelstrup MC, Stilund M, Petersen T, Møller HJ, Petersen EL, Christensen T. Subsets of activated monocytes and markers of inflammation in incipient and progressed multiple sclerosis. Immunol Cell Biol. 2018;96(2):160-174. doi: 10.1111/imcb.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waschbisch A, Schröder S, Schraudner D, et al. Pivotal role for CD16+ monocytes in immune surveillance of the central nervous system. J Immunol. 2016;196(4):1558-1567. doi: 10.4049/jimmunol.1501960 [DOI] [PubMed] [Google Scholar]
- 16.Shi KB, Li H, Chang T, et al. , . Bone marrow hematopoiesis drives multiple sclerosis progression. Cell. 2022;185(13):2234-2247.e17. doi: 10.1016/j.cell.2022.05.020 [DOI] [PubMed] [Google Scholar]
- 17.Natrajan MS, Komori M, Kosa P, et al. Pioglitazone regulates myelin phagocytosis and multiple sclerosis monocytes. Ann Clin Translational Neurol. 2015;2(12):1071-1084. doi: 10.1002/acn3.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Healy LM, Perron G, Won SY, et al. MerTK is a functional regulator of myelin phagocytosis by human myeloid cells. J Immunol. 2016;196(8):3375-3384. doi: 10.4049/jimmunol.1502562 [DOI] [PubMed] [Google Scholar]
- 19.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis JC, Magal E, Muir D, Manthorpe M, Varon S. CG-4, a new bipotential glial-cell line from rat-brain, is capable of differentiating invitro into either mature oligodendrocytes or type-2 astrocytes. J Neurosci Res. 1992;31(1):193-204. doi: 10.1002/jnr.490310125 [DOI] [PubMed] [Google Scholar]
- 21.Kelly B, O'Neill LAJ. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015;25(7):771-784. doi: 10.1038/cr.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natrajan MS, de la Fuente AG, Crawford AH, et al. Retinoid X receptor activation reverses age-related deficiencies in myelin debris phagocytosis and remyelination. Brain. 2015;138(Pt 12):3581-3597. doi: 10.1093/brain/awv289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotter MR, Li WW, Zhao C, Franklin RJM. Myelin impairs CNS remyelination by inhibiting oligodendrocyte precursor cell differentiation. J Neurosci. 2006;26(1):328-332. doi: 10.1523/jneurosci.2615-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato W, Tomita A, Ichikawa D, et al. CCR2+CCR5+ T cells produce matrix metalloproteinase-9 and osteopontin in the Pathogenesis of multiple sclerosis. J Immunol. 2012;189(10):5057-5065. doi: 10.4049/jimmunol.1202026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer HJ, Finck TLK, Pellkofer HL, Reichardt HM, Luhder F. Glucocorticoid therapy of multiple sclerosis patients induces anti-inflammatory polarization and increased chemotaxis of monocytes. Front Immunol. 2019;10:1200. doi: 10.3389/fimmu.2019.01200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshie O, Matsushima K. CCR4 and its ligands: from bench to bedside. Int Immunol. 2015;27(1):11-20. doi: 10.1093/intimm/dxu079 [DOI] [PubMed] [Google Scholar]
- 27.Mezzaroba L, Alfieri DF, Colado Simão AN, Vissoci Reiche EM. The role of zinc, copper, manganese and iron in neurodegenerative diseases. Neurotoxicology. 2019;74:230-241. doi: 10.1016/j.neuro.2019.07.007 [DOI] [PubMed] [Google Scholar]
- 28.Ruttkay-Nedecky B, Nejdl L, Gumulec J, et al. The role of metallothionein in oxidative stress. Int J Mol Sci. 2013;14(3):6044-6066. doi: 10.3390/ijms14036044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penkowa M, Espejo C, Ortega-Aznar A, Hidalgo J, Montalban X, Martínez Cáceres EM. Metallothionein expression in the central nervous system of multiple sclerosis patients. Cell Mol Life Sci. 2003;60(6):1258-1266. doi: 10.1007/s00018-003-3021-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bredholt M, Frederiksen JL. Zinc in multiple sclerosis: a systematic review and meta-analysis. ASN Neuro. 2016;8(3):1759091416651511. doi: 10.1177/1759091416651511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Earl C, Chantry A, Mohammad N, Glynn P. Zinc ions stabilise the association of basic protein with brain myelin membranes. J Neurochem. 1988;51(3):718-724. doi: 10.1111/j.1471-4159.1988.tb01803.x [DOI] [PubMed] [Google Scholar]
- 32.Tsang D, Tsang YS, Ho WKK, Wong RNS. Myelin basic protein is a zinc-binding protein in brain: possible role in myelin compaction. Neurochem Res. 1997;22(7):811-819. doi: 10.1023/a:1022031825923 [DOI] [PubMed] [Google Scholar]
- 33.Kaushik DK, Bhattacharya A, Mirzaei R, et al. Enhanced glycolytic metabolism supports transmigration of brain-infiltrating macrophages in multiple sclerosis. J Clin Invest. 2019;129(8):3277-3292. doi: 10.1172/jci124012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandit A, Vadnal J, Houston S, Freeman E, McDonough J. Impaired regulation of electron transport chain subunit genes by nuclear respiratory factor 2 in multiple sclerosis. J Neurol Sci. 2009;279(1-2):14-20. doi: 10.1016/j.jns.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 35.Gonzalo H, Nogueras L, Gil-Sánchez A, et al. Impairment of mitochondrial redox status in peripheral lymphocytes of multiple sclerosis patients. Front Neurosci. 2019;13:938. doi: 10.3389/fnins.2019.00938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quijano C, Trujillo M, Castro L, Trostchansky A. Interplay between oxidant species and energy metabolism. Redox Biol. 2016;8:28-42. doi: 10.1016/j.redox.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang SCC, Everts B, Ivanova Y, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15(9):846-855. doi: 10.1038/ni.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vats D, Mukundan L, Odegaard JI, et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4(1):13-24. doi: 10.1016/j.cmet.2006.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heidker RM, Emerson MR, LeVine SM. Metabolic pathways as possible therapeutic targets for progressive multiple sclerosis. Neural Regen Res. 2017;12(8):1262-1267. doi: 10.4103/1673-5374.213542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peruzzotti-Jametti L, Pluchino S. Targeting mitochondrial metabolism in Neuroinflammation: towards a Therapy for progressive multiple sclerosis. Trends Mol Med. 2018;24(10):838-855. doi: 10.1016/j.molmed.2018.07.007 [DOI] [PubMed] [Google Scholar]
- 41.Bekkering S, van den Munckhof I, Nielen T, et al. Innate immune cell activation and epigenetic remodeling in symptomatic and asymptomatic atherosclerosis in humans in vivo. Atherosclerosis. 2016;254:228-236. doi: 10.1016/j.atherosclerosis.2016.10.019 [DOI] [PubMed] [Google Scholar]
- 42.Mukherjee R, Kanti Barman P, Kumar Thatoi P, Tripathy R, Kumar Das B, Ravindran B. Non-classical monocytes display inflammatory features: validation in sepsis and systemic lupus erythematous. Scientific Rep. 2015;5:13886. doi: 10.1038/srep13886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Netea MG, Quintin J, van der Meer JWM. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011;9(5):355-361. doi: 10.1016/j.chom.2011.04.006 [DOI] [PubMed] [Google Scholar]
- 44.Rogacev KS, Cremers B, Zawada AM, et al. CD14++CD16+ monocytes independently predict cardiovascular events: a cohort study of 951 patients referred for elective coronary angiography. J Am Coll Cardiol. 2012;60(16):1512-1520. doi: 10.1016/j.jacc.2012.07.019 [DOI] [PubMed] [Google Scholar]
- 45.Netea MG, Domínguez-Andrés J, Barreiro LB, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol. 2020;20(6):375-388. doi: 10.1038/s41577-020-0285-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaufmann E, Sanz J, Dunn JL, et al. , . BCG Educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 2018;172(1-2):176-190.e19. doi: 10.1016/j.cell.2017.12.031 [DOI] [PubMed] [Google Scholar]
- 47.Arts RJW, Joosten LAB, Netea MG. The potential role of trained immunity in autoimmune and autoinflammatory disorders. Front Immunol. 2018;9:298. doi: 10.3389/fimmu.2018.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Läderach F, Münz C. Epstein barr virus Exploits Genetic Susceptibility to increase multiple sclerosis Risk. Microorganisms. 2021;9(11):2191. doi: 10.3390/microorganisms9112191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christ A, Günther P, Lauterbach MAR, et al. Western diet triggers NLRP3-Dependent innate immune reprogramming. Cell. 2018;172(1-2):162-175.e14. doi: 10.1016/j.cell.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harroud A, Mitchell RE, Richardson TG, et al. Childhood obesity and multiple sclerosis: a Mendelian randomization study. Mult Scler. 2021;27(14):2150-2158. doi: 10.1177/13524585211001781 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The omics data used in this article are accessible on GEO at accession number GSE269706.