Abstract
Artemisinin-based combination therapy (ACT) is the mainstay of effective treatment of Plasmodium falciparum malaria. However, the long-term utility of ACTs is imperiled by widespread partial artemisinin resistance in Southeast Asia and its recent emergence in parts of East Africa. This underscores the need to identify chemotypes with new modes of action (MoAs) to circumvent resistance to ACTs. In this study, we characterized the asexual blood stage antiplasmodial activity and resistance mechanisms of LDT-623, a 4-aminoquinoline (4-AQ). We also detected LDT-623 activity against multiple stages (liver schizonts, stage IV-V gametocytes, and ookinetes) of Plasmodium’s life cycle, a feature unlike other 4-AQs such as chloroquine (CQ) and piperaquine (PPQ). Using heme fractionation profiling and drug uptake studies in PfCRT-containing proteoliposomes, we observed inhibition of hemozoin formation and PfCRT-mediated transport, which constitute characteristic features of 4-AQs’ MoA. We also found minimal cross-resistance to LDT-623 in a panel of mutant pfcrt or pfmdr1 lines, but not the PfCRT F145I mutant that is highly resistant to PPQ resistance yet is very unfit. No P. falciparum parasites were recovered in an in vitro resistance selection study, suggesting a high barrier for resistance to emerge. Finally, a competitive growth assay comprising >50 barcoded parasite lines with mutated resistance mediators or major drug targets found no evidence of cross-resistance. Our findings support further exploration of this promising 4-AQ.
Author summary
The emergence of artemisinin-resistant malaria parasites, first in Southeast Asia and more recently East Africa, highlights the urgency to identify and optimize chemotypes with antimalarial activity that are not compromised by known resistance mechanisms in clinical settings. Using a computer-aided drug discovery approach, we previously identified a 4-AQ (LDT-611) that was active against P. falciparum asexual blood-stage (ABS) parasites. Herein, we report the multistage antimalarial activity of a structural analog, a side-chain-modified 4-AQ (LDT-623). This analog shares 4-AQ characteristics, such as PfCRT-mediated transport, as well as inhibition of hemozoin formation, that occurs in the parasite digestive vacuole. Cross-resistance profiling showed that this compound was not compromised by known mutations in the digestive vacuole transporters PfCRT or PfMDR1 or by other gene products that govern resistance to antimalarials under development. Efforts to evolve resistance to LDT-623 in vitro yielded parasite populations lacking noticeable phenotypic changes. In conclusion, we report a 4-AQ whose MoA is likely to differ from licensed antimalarials within this chemical class, paving the way for further studies on this chemotype as a therapeutic avenue to treat multidrug-resistant malaria.
1. Introduction
Malaria remains a major global health concern, with the World Health Organization (WHO) reporting 249 million cases and 608,000 related deaths across 85 malaria-endemic countries and areas in 2022 [1]. Of the human-infecting Plasmodium species, P. falciparum is the most devastating due to its impact on mortality and morbidity, especially in young children and pregnant women. Currently, artemisinin (ART)-based combination therapies (ACTs) constitute the frontline antimalarial treatment policy [1]. ACTs comprise a rapid-acting yet short-lived ART derivative that eliminates most of the initial parasite load, partnered with a longer-lasting molecule with a distinct MoA to eliminate surviving parasites. The efficacy of ACTs has, however, been compromised by the emergence of mutant k13 parasites in Southeast Asia [2–4] and parts of East Africa [5–7]. Parasites with specific k13 mutations exhibit partial resistance to artemisinin, characterized by delayed clearance time upon treatment with an ART derivative or an ACT [8–10] and higher levels of ring-stage parasite survival in vitro [11]. This threat to the long-term therapeutic utility of ACTs calls for urgent efforts to identify and develop new molecules, preferably with divergent MoAs, no cross-resistance to existing molecules, and pan-activity across multiple stages of the Plasmodium lifecycle.
In the human host, the P. falciparum lifecycle involves the invasion of hepatocytes by sporozoites injected by a feeding female Anopheles mosquito. Liver-stage parasites mature and are released into the bloodstream as merozoites that invade red blood cells (RBCs) to initiate the pathogenic asexual blood stage (ABS) cycle. This cycle lasts ~48 hours and is characterized by differentiation of the merozoites into ring forms then into trophozoites and ultimately schizonts, from which more merozoites egress and reinvade other RBCs. Some merozoites can also differentiate into sexual gametocyte forms that once mature can be transmitted to a feeding mosquito.
ABS parasites digest host hemoglobin (Hb) in their acidic digestive vacuole (DV), leading to the release of essential amino acids for intracellular growth and free heme as a by-product. In its labile form, free heme is toxic due to its ability to induce free radical formation. Parasites counter this toxicity by converting heme into inert hemozoin (Hz) crystals [12]. Extensive evidence shows that 4-AQ drugs such as chloroquine (CQ) and piperaquine (PPQ) exert their antiplasmodial activity by accumulating within the DV and binding to heme monomers and dimers, effectively interfering with this detoxification pathway [13]. This antiplasmodial activity is predicated on the structurally unique pharmacophore of these compounds that comprises the 4-amino-7-chloroquinoline nucleus for heme binding and inhibition of Hz formation as well as a tertiary amino group in the side chain for vacuolar drug accumulation [14,15]. However, point mutations in the P. falciparum chloroquine resistance transporter (PfCRT) [16], an oligopeptide and drug transporter located on the DV membrane [17], have been identified as the main driver of resistance to CQ, PPQ, and some related quinolines [18]. Polymorphic PfCRT variants carry 4 to 10 mutations that can differentially modify parasite susceptibility to CQ and PPQ [19,20]. However, several 4-AQs maintain similar activity against both CQ-sensitive and CQ-resistant parasites, suggesting that they are not recognized and exported by mutant PfCRT [21]. This has informed the introduction of more versatile and potent 4-AQs like amodiaquine, pyronaridine, and ferroquine in clinical use as effective alternatives to address resistance [22].
Herein, we identify the multistage antiplasmodial activity of LDT-623. This compound is a side-chain-modified 4-AQ structurally analogous to LDT-611 that was previously identified in a screening campaign on a library of ~120,000 natural products and derivatives for potential antiplasmodial activity [23]. To evaluate the activity of LDT-623 against other stages of the P. falciparum lifecycle, we tested its gametocytocidal potential and ability to inhibit other exoerythrocytic forms of the parasite. Furthermore, we investigated its MoA and possible cross-resistance with known drug targets. Our results open up the possibility of further exploring this compound as a potential antimalarial.
2. Material and methods
2.1. Compound preparation
Screening compounds were purchased from providers Chembridge, MedChemExpress, InterBioScreen, ChemDiv Inc, and BLD Pharmatech Ltd, through the MolPort website (www.molport.com) and were further dissolved in DMSO in 10 mM stock solutions. Standard antimalarial drugs were purchased from Sigma-Aldrich and prepared as 10 mM stocks.
2.2. Virtual screening workflow for the identification of LDT-611 structural analog molecules
Based on the premise that structurally similar compounds may possess similar biological activity, we carried out a virtual screening of the MolPort library of ~7.5 million compounds to select molecules with relevant structural similarity to LDT-611. We employed the “Find similar” function that utilizes the Tanimoto coefficient, a quantitative measure of chemical scaffold similarity, to filter molecules with structural similarity. A similarity cutoff of 78% (Tc = 0.78) was defined based on a visual inspection of analog compounds, to keep those that were 4-AQ and resembled LDT-611’s structure. Further steps removed double entries and molecules whose antimalarial activity had been previously reported on PubChem. A final filter removed acridines from the dataset. Finally, the selected analogs were purchased and evaluated in vitro.
2.3. P. falciparum in vitro culture and ABS assays
P. falciparum ABS parasites were cultured at 3% hematocrit in human O+ RBCs in RPMI-1640 media supplemented with 25 mM HEPES, 50 mg/L hypoxanthine, 2 mM L-glutamine, 0.21% sodium bicarbonate, 0.5% (w/v) AlbuMAXII (Invitrogen) and 10 μg/mL gentamycin. Culture flasks were kept in modular incubator chambers (Billups-Rothenberg) in a 5% O2, 5% CO2, and 90% N2 atmosphere at 37°C. Synchronized ring-stage parasites were obtained by two consecutive rounds of treatment with 5% D-sorbitol in 48-hour intervals [24]. ABS parasite survival assays were performed to define half-maximal compound concentrations that inhibit 50% or 90% parasite growth (IC50 and IC90). This was done by exposing cultures at 1% hematocrit and 0.3% parasitemia in 96-well plates in the presence of drugs in a two-fold 10 or 12-point serial dilution in duplicate along with drug-free controls. After a 72-hour incubation, parasite survival was assessed by flow cytometry on an iQue flow cytometer (Sartorius) using SYBR Green and MitoTracker Deep Red FM (Life Technologies) as nuclear stain and vital dye, respectively. Parasite survival was determined as a percentage relative to drug-free control. IC50 values were interpolated from log concentration versus response curves in Prism 9 (GraphPad Prism Software Inc.).
2.4. Cytotoxicity assays
Human hepatoma (HepG2) and fibroblast-like monkey kidney cells (COS-7) were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% heat-inactivated fetal bovine serum and gentamycin (40 mg/L) at 37°C in a 5% CO2 atmosphere. Cytotoxicity of test compounds was measured by metabolic reduction of mitochondrial enzymes using the MTT assay (3-[4,5-dimethyl-thiazol-2-yl]-2,5-diphenyltetrazolium chloride) [25]. Cells were seeded at 105 cells/well in 96-well plates and incubated for 72 hours in the presence of a 2-fold 12-point serial dilution of drugs along with drug-free controls. After adding 15 μL of MTT (5 mg/mL) to the wells, plates were centrifuged at 317g for 5 minutes and the pellet was resuspended in isopropanol. Absorbance was measured at 570 nm (CLARIOStar, Labtech BMG). Cell viability was determined by formazan production, and was expressed as a percentage of untreated control wells.
2.5. P. falciparum gametocyte production and growth inhibition assay
P. falciparum NF54-Pfs16-GFP-Luc (MRA-1217), a highly gametocyte-producing strain, was cultured following standard procedures [26]. NF54-Luc ABS parasites were sorbitol-synchronized as previously described [24] and cultured to reach 8–10% parasitemia as mature trophozoites prior to starting gametogenesis induction via nutrient deficit stress [27]. Briefly, stress was induced by replacing only 40% of the culture medium and returning cultures to a 37°C incubation. Cultures were treated with 50 mM N-acetylglucosamine for 10 days to remove ABS parasites. Mature gametocytes (stages IV-V) were isolated using magnetic columns and incubated at 2% gametocytemia and 1.5% hematocrit in 96-well plates containing the test compounds for 48 hours at 37°C, following standard P. falciparum culturing conditions. Methylene blue was used as a gametocytocidal control alongside drug-free wells. Luciferase activity was assessed by adding 40 μL of luciferin substrate (Promega) to 40 μL of parasite lysates and detecting bioluminescence at an integration constant of 10s with the BioTek Cytation 5 Cell Imaging Multimode Reader [28].
2.6. In vitro inhibition of P. berghei ookinete formation
To measure the inhibition of gamete conversion into ookinetes, a mutant luciferase-expressing P. berghei line (nLuc) was intraperitoneally injected into BALB/c mice [29]. After 3–4 days, gametocyte-carrying blood samples were collected. In vitro compound testing was carried out by exposing 4 μL of infected blood mixed with 80 μL of culture medium (RPMI-1640 supplemented with 25 mM HEPES, 50 mg/L hypoxanthine, and 1% penicillin/streptomycin/neomycin at pH 8.3) to a range of drug concentrations for 24 hours at 21°C. Following the incubation, 20 μL of nLuc substrate (Nano-Glo Luciferase Assay System, Promega) was added to each well, and luminescence was measured in a luminometer. Increased light emission correlated with nLuc production and subsequent luciferase activity due to in vitro fertilization and ookinete conversion, as luciferase expression in this parasite line is regulated by the CTRP promoter that is ookinete-specific [30]. Conversion inhibition was calculated as the percentage of luciferase activity of drug-exposed wells relative to untreated ones, from which IC50 values were interpolated.
2.7. In vitro inhibition of liver stage infection
In vitro inhibition of Plasmodium liver stage infection was assessed using bioluminescence data from Huh-7 cells infected with a firefly-luciferase-expressing P. berghei line, PbGFP-Luccon [31], in the presence of various compound concentrations. Huh-7 cells, a human hepatoma cell line, were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, 1% nonessential amino acids, 1% glutamine, 1% penicillin/streptomycin and 10 mM HEPES at pH 7 at 37°C under 5% CO2. One day before drug treatment and infection, 104 Huh-7 cells/well were seeded in 96-well plates. The medium was then replaced by an infection medium (culture medium supplemented with 50 μg/mL gentamycin and 0.8 μg/mL amphotericin B) containing desired concentrations of test compounds. Fresh sporozoites recovered from salivary glands of infected female Anopheles stephensi mosquitoes were added to seeded Huh-7 cells at a 1:1 proportion, followed by centrifugation at 1700g for 5 minutes and incubation for 46 hours at 37°C under 5% CO2. Compound toxicity against Huh-7 cells was assessed by measuring cell confluency using the Alamar Blue assay (Invitrogen). IC50 values were calculated using nonlinear regression analysis (GraphPad Prism version 9).
2.8. PfCRT transport measurement assays
Purified PfCRT variants were reconstituted in preformed liposomes made of E. coli total lipids:cholesteryl hemisuccinate 97:3 (w/w) at a protein-to-lipid ratio of 1:150 (w/w). The lumen of the proteoliposomes was composed of 100 mM KPi, pH 7.5, and 2 mM β-mercaptoethanol. Uptake of [3H]CQ (1 Ci/mmol) or [3H]PPQ (1 Ci/mmol) was performed by diluting PfCRT-containing proteoliposomes (30 ng of PfCRTper reaction) in an uptake buffer containing 100 mM Tris/MES, pH 5.5, in the presence or absence of 1 μM of the test compounds. In addition, 1 μM valinomycin was added to the reaction to generate a K+ diffusion potential-driven membrane potential (ΔΨ). Reactions were stopped after 30 seconds by the addition of ice-cold 100 mM KPi, pH 6.0, and 100 mM LiCl, and filtered through 0.45-μm nitrocellulose filters (Millipore). Filters were dried and incubated in a scintillation cocktail, and the radioactivity captured on the filters was counted in a Hidex SL300 scintillation counter. The efficiency of detection was calculated with a standard curve of known concentrations of each radiolabeled compound, and this was used to transform decays per minute (dpm) into pmol. The nonspecific interaction of each compound with nitrocellulose filters was determined by measuring mock uptake in the absence of liposomes or proteoliposomes. These values (determined for each experiment) were used to calculate background uptake in liposomes or proteoliposomes. Drug-specific uptake was determined by subtracting the time-dependent accumulation of the tested compounds in control liposomes (lacking PfCRT) from that measured in PfCRT-containing proteoliposomes [20].
Solid Supported Membrane (SSM)-based electrophysiological measurements were performed using SURFE2R N1 technology (Nanion Technologies, Inc.) according to published protocols [32,33]. Briefly, sensors were coated with 1-octadecanethiol (Sigma-Aldrich), rinsed, and 1,2-diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, Inc) was added. Immediately, the sensor was filled with 50 μL non-activating buffer (10 mM Tris-MES, pH 5.5) and PfCRT-containing proteoliposomes (PLs). To generate a K+ diffusion potential-driven ΔΨ, valinomycin was included at a final concentration of 80 nM [34,35]. The activating solution (10 mM Tris-MES, pH 5.5 containing either 10 μM CQ, PPQ, LDT-623, pyrimethamine, or atovaquone) was applied using a single-solution exchange protocol (activating buffer incubation for 1 second). Each measurement was repeated at least twice employing at least three different sensors. Peak currents for PfCRT were corrected by subtracting the peak currents recorded with control liposomes (lacking this transporter). To acquire the PfCRT-elicited charge (Coulomb) movement associated with drug transport, the area under the curve (current as a function of time) was analyzed with GraphPad Prism version 10.
2.9. Cellular heme fractionation assay
A heme fractionation assay was used to measure the inhibition of Hz formation in cultured ABS P. falciparum 3D7 parasites as previously described [36]. Briefly, ring-stage 3D7 parasites were treated with consecutive D-sorbitol and Percoll solutions to obtain tightly synchronized 3 hours post-invasion young rings that were incubated with test drugs (0.5–3.0 × IC50) alongside a vehicle control in standard culturing conditions (see above). Late trophozoites were harvested after 32 hours of incubation by lysing RBCs with 0.05% saponin followed by multiple washes with 1 × PBS (pH 7.5) to remove any residual traces of hemoglobin. The parasite pellet was resuspended in PBS and the total number of trophozoites isolated was quantified using flow cytometry. Contents of the trophozoite pellet were released by hypotonic lysis and sonication. After centrifugation, treatments with HEPES buffer (pH 7.4), sodium dodecyl sulfate, pyridine, and NaOH, allowed us to separate the fractions corresponding to the different heme species of interest. A microplate reader was employed to measure the UV-visible spectrum of each heme fraction as a Fe(III)heme-pyridine complex. Fractions of undigested hemoglobin, “free” heme, and Hz were quantified using a heme standard curve based on the mass of each heme-Fe species per mature trophozoite (fg/cell), obtained by dividing the total amount of each heme species by the corresponding number of parasites in that fraction as determined by flow cytometry. Statistical comparisons and analyses for trends employed Students’ t-tests (GraphPad Prism version 9).
2.10. Resistance selection experiments
Resistance to LDT-623 was assessed using single-step in vitro resistance selection protocols [37]. Duplicate flasks with 1 × 109 P. falciparum Dd2-Pol δ parasites were exposed to 3 × IC50 under two different drug regimens: (1) removal of drug pressure as soon as live parasites cleared, and (2) drug pressure kept constant throughout the experiment. Dd2-Pol δ is a hypermutable line with two point mutations in DNA polymerase δ that compromise its proofreading activity [38] Cultures were monitored daily by thin blood smears until live parasites were no longer detectable. Drug-containing media was changed daily until parasites cleared, and every other day thereafter. Cultures were passaged once weekly by reducing the volume by 25% and replacing 1/3 of RBCs. Recrudescence monitoring was carried out twice a week by thin blood smears for 60 days. In the event of recrudescence, parasite susceptibility to LDT-623 was tested as described above.
2.11. Resistome growth assay
The resistome growth assay employs a parasite pool consisting of 51 barcoded lines including both Dd2 and 3D7 genetic backgrounds, with wild-type lines for both (S3 Table). Most lines used were derived from in vitro resistance selection experiments, with others having been CRISPR edited. All lines were barcoded via CRISPR editing of a 101bp barcode cassette into the non-essential pfpare locus (PF3D7_0709700) [39]. Parasites were cultured in RPMI-1640 medium supplemented with HEPES (5.94 g/L), NaHCO3 (2.1 g/L), glutamax (1×), hypoxanthine (50 mg/L), gentamycin (25 μg/L), and AlbumaxII (5 g/L) at a 50% hematocrit. Parasite growth was monitored over 14 days by flow cytometry (Cytoflex, Beckman Coulter) by staining with 1× SYBR Green and 0.2 μM MitoTracker Deep Red. Parasitemia adjustments were made to maintain a healthy range of <5% parasitemia. Parasite cultures were harvested at day 14 and lysed by saponin (0.05%), with pellets washed twice with PBS. Barcodes were PCR amplified using 5 μL of parasite pellet and sequenced by Illumina amplicon sequencing [40].
3. Results
3.1. Identification and activity profiling of LDT-611 and its analogs
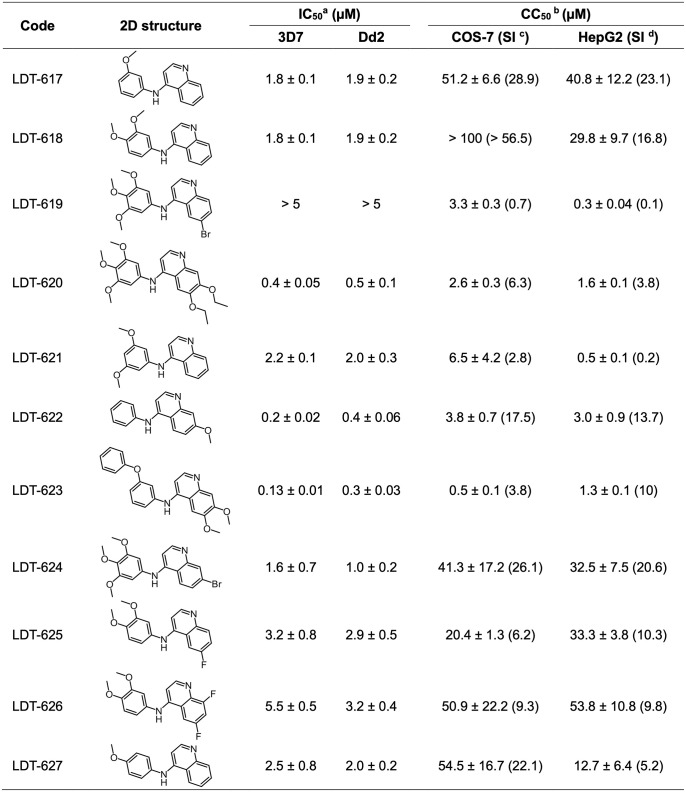
In a previous study, we identified several biodiversity-derived compounds with in vitro activity against P. falciparum ABS parasites in vitro [23]. To further explore the antimalarial potential of these hits, we chose LDT-611 (S1A Fig) due to the combination of its sub-micromolar activity against ABS parasites, selectivity index > 10, and relatively low structural similarity to clinical antimalarials. LDT-611 inhibited the growth of CQ-sensitive (3D7) and CQ- and mefloquine-resistant (W2) parasite strains with IC50 values of 760 and 820 nM, respectively (S1B Fig). Its equipotency against wild-type and mutant PfCRT isoforms suggests that this compound can circumvent CQ resistance. To identify a more potent compound, we used the Tanimoto coefficient as part of an optimization pipeline that yielded 11 available analogs (S1C Fig). Tanimoto coefficients (S1D Fig) showed that at least 78% of their structures were shared (Tc = 0.78), with the potent antimalarial 4-AQ nucleus remaining intact. These 11 analogs were then evaluated for their activity against P. falciparum 3D7 and Dd2 ABS parasites, along with cytotoxicity assays with COS-7 and HepG2 mammalian cell lines (Fig 1).
Fig 1. In vitro activity of LDT-611 analogs against P. falciparum asexual blood stages and mammalian cell lines.
a IC50: Mean half-maximal inhibitory concentration and their respective standard errors, assayed against P. falciparum 3D7 and Dd2 ABS parasites (N = 3); b CC50: Mean half-maximal cytotoxic concentration against mammalian cell lines (N = 3); c SI: Selectivity index was calculated by dividing the CC50 (against COS-7) by the IC50 (against 3D7); d SI: Selectivity index was calculated by dividing the CC50 (against HepG2) by the IC50 (against 3D7). Data are represented by mean ± standard deviation.
3.2. LDT-623, the most potent analog, exhibits multistage antiplasmodial activity
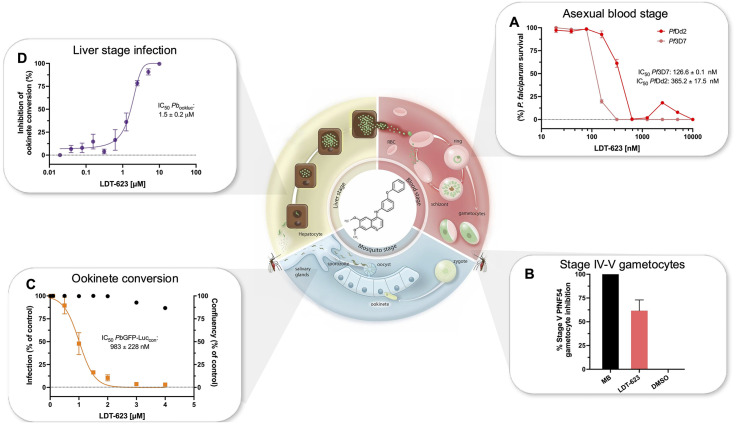
LDT-623 was the most potent of the analogs tested against ABS parasites, with IC50 values of 127 and 365 nM for 3D7 and Dd2 ABS parasites, respectively) (Fig 2A). This compound was therefore selected for further experimental evaluation throughout the Plasmodium lifecycle (S1 Table).
Fig 2. Antiplasmodial activity of LDT-623 throughout the Plasmodium lifecycle.
(A) LDT-623 in vitro activity against ABS P. falciparum CQ-sensitive 3D7 and CQ-resistant Dd2 strains. Mean IC50: 126.6 and 365.2 nM, respectively. Parasite survival was calculated as a percentage of untreated control. Data represent mean ± SEM (N = 2–3 independent assays, with technical duplicates). (B) Percent activity of LDT-623 against late-stage P. falciparum NF54 gametocytes at 5 μM. Gametocyte viability was assessed via bioluminescence. Data represent mean ± SEM (N = 3). MB: methylene blue; DMSO: dimethyl sulfoxide. (C) Dose-response curve for LDT-623 concentration versus inhibition of P. berghei ookinete formation compared to vehicle-treated control. IC50: 1.5 μM. Data represent mean ± SEM (N = 7). (D) Pre-erythrocytic activity of LDT-623. IC50: 983 nM. Dose-response curve represents hepatic stage infection as a percentage of drug-free control assessed by luminescence 48h after infection of Huh-7 cells with P. berghei sporozoites, while black dots express host cell confluency. Data represent mean ± SEM (N = 2).
The potential transmission-blocking activity of LDT-623 was assessed by exposing NF54 stage IV-V gametocytes to 5 μM of test compounds. LDT-623 inhibited 62% of mature gametocyte viability compared to untreated control (Fig 2B), an unusual activity for 4-AQs [41]. Gametocytocidal activity was assessed alongside methylene blue, a positive control that showed 100% activity at the same 5 μM concentration. We also employed a P. berghei model to evaluate LDT-623’s inhibition of ookinete formation in vitro, using a conversion assay that evaluates the inhibition of gamete fertilization into a zygote and subsequent conversion into an ookinete. LDT-623 yielded an IC50 of 1.5 μM (Fig 2C). For reference, CQ inhibited 56.7% of zygote conversion into ookinetes at 10 μM (S2 Fig).
We also observed pre-erythrocytic activity of LDT-623 using an in vitro firefly luciferase-expressing P. berghei line by measuring inhibition of hepatocyte infection based on luminescence intensity [31]. In agreement with its IC50 of 983 nM (Fig 2D), LDT-623 showed 52.1% inhibition of hepatic infection at 1 μM. The positive control atovaquone almost completely ablated infection at 10 nM, consistent with its known liver stage activity [42]. Of note, we observed minimal to no dose-dependent toxicity towards Huh-7 host cells, as evidenced by over 86% cell viability at the maximum concentration of 4 μM LDT-623.
3.3. LDT-623 inhibits intracellular hemozoin formation
The 4-AQ nucleus, contained by antimalarial drugs such as CQ and PPQ, interferes with the heme detoxification pathway inside the parasite DV through complexation with heme, leading to inhibition of Hz formation [43,44]
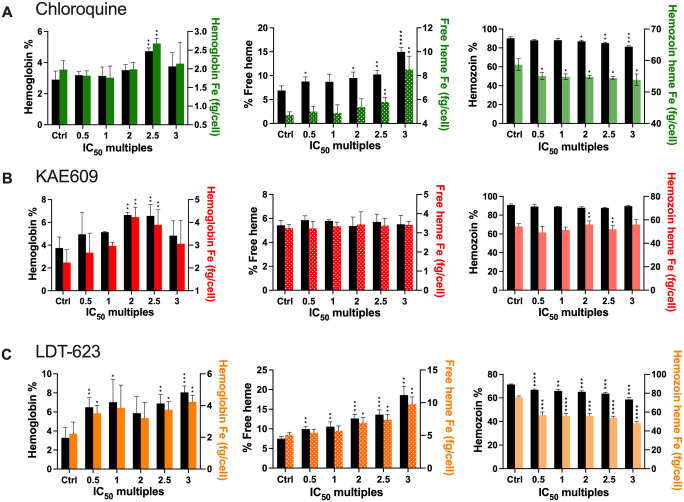
Heme fractionation profiling of LDT-623 revealed a statistically significant increase in “free” heme in a concentration-dependent manner compared to untreated control (Fig 3A and S2 Table). Concurrently, we also observed significant concentration-dependent decreases in the levels of Hz. Moreover, an augmented percentage of undigested Hb was observed starting at 0.5 × IC50 of LDT-623. CQ, used as a positive control, also exhibited a significant increase in “free” heme starting at 0.5 × IC50 while Hz levels only decreased significantly from 2 × IC50. Interestingly, the proportion of “free” heme present as undigested Hb within the DV after incubation was significant at 2.5 × IC50 of CQ only (Fig 3B). As a negative control, parasites were treated with KAE609 (cipargamin), a PfATP4 inhibitor, which did not affect Hz formation [45]. Besides a subtle yet significant increment in Hb levels observed under 2 and 2.5 × IC50, no striking perturbations were noted for any of the heme species investigated under KAE609 treatment (Fig 3C). Altogether, these results demonstrate that LDT-623 interferes with the vital Hz formation pathway, presumably due to its quinoline core.
Fig 3. LDT-623 interferes with the hemozoin formation pathway within the digestive vacuole.
(A) Parasites treated with LDT-623 display a more pronounced heme fractionation profile than CQ. (B) Heme fractionation profile of CQ-treated P. falciparum 3D7 parasites showing an increase in free heme and a decrease in Hz, as determined 32 h post-drug exposure. (C) KAE609 treatment did not interfere with heme or Hz accumulation. Percent levels of heme species (hemoglobin, free heme, or hemozoin) are represented by the black bars on the left y-axis, and absolute heme amounts (hemoglobin Fe, free heme Fe, or hemozoin heme Fe) determined from a heme standard curve and measured in femtograms per cell are represented in different colors (LDT-623 in red, CQ in orange, and KAE609 in green) on the right y-axis. Statistical comparisons of the drug-treated lines with their untreated controls were performed using two-tailed Student’s tests (with Welch’s correction). *p < 0.05; **p < 0.01; ***p < 0.001; ****p <0.0001.
3.4. LDT-623 impacts PfCRT variant-specific drug efflux
PPQ and CQ resistance-conferring mutations in PfCRT allow positively charged drugs and other solutes to efflux into the cytosol [20,46]. Consequently, CQ- or PPQ-resistant parasites accumulate lower amounts of drugs within their DVs and prevent inhibition of heme detoxification. This contrasts with drug-sensitive parasites, where the increased intravacuolar concentrations of these drugs inhibit heme detoxification and result in subsequent parasite death.
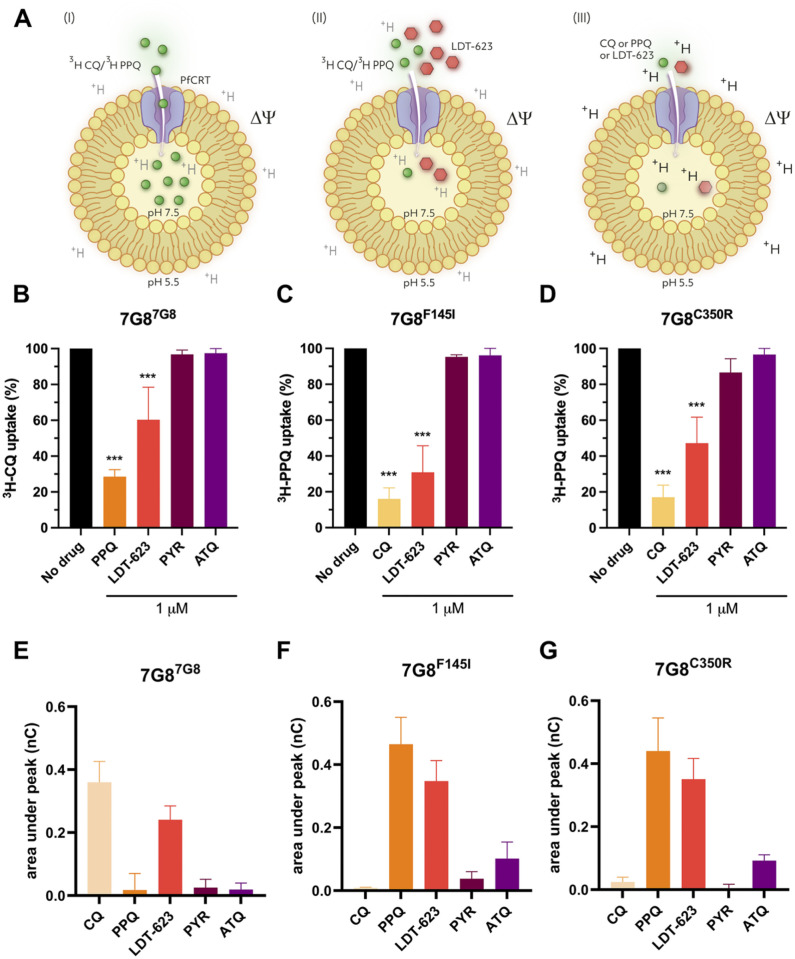
Using PfCRT-containing proteoliposomes as a surrogate for drug transport, we employed an assay that mimics an “inside-out” parasite DV. This assay employed the CQ-resistant, PPQ-sensitive 7G8 PfCRT isoform, as well as two variants (7G8F145I and 7G8C350R) that include an additional mutation that is observed in Southeast Asia and South America, respectively, and that mediate PPQ resistance and a reversion to CQ sensitivity [19,47]. Assays were designed to evaluate the impact of LDT-623 on PfCRT-mediated drug uptake in these 7G8 isoforms. The uptake of [3H]CQ or [3H]PPQ was measured in PfCRT-containing proteoliposomes using an internally directed pH gradient and ΔΨ (Fig 4A) and was normalized to that of control liposomes lacking PfCRT [20]. Drug-specific transport (here measured by [3H]-labeled drug accumulation) can be differentially mediated by distinct PfCRT isoforms, in agreement with varying resistance phenotypes as seen with the PfCRT isoforms used: 7G87G8 (CQ-resistant, PPQ-sensitive), 7G8F145I (CQ-sensitive, PPQ-resistant) and 7G8C350R (CQ-sensitive, PPQ-resistant).
Fig 4. Uptake of the 4-AQs CQ, PPQ, and LDT-623 in PfCRT-containing proteoliposomes.
(A) Schematic representation of the drug uptake assay highlighting the “inside-out” digestive vacuole aspect of PfCRT-containing proteoliposomes. Uptake of radiolabeled CQ or PPQ in the absence (I) or presence (II) of LDT-623 using the accumulation of radiolabeled CQ or PPQ as read-out or measuring 4-AQ transport-associated current fluxes (III) with electrophysiology. Uptake of 100 nM [3H]CQ (B) or [3H]PPQ (C-D) measured for 30 seconds in proteoliposomes containing the indicated PfCRT variants normalized to control liposomes. Data represent mean ± SEM of N = 3 independent experiments with n = 7 technical replicates each. (E-G) Transport of 10 μM CQ, PPQ, LDT-623, PYR, or ATQ by proteoliposomes containing indicated PfCRT variants using the SURFE2R N1-based SSM electrophysiological measurements. Data represent PfCRT isoform-specific currents (corrected for the non-specific signal measured in control liposomes lacking PfCRT) and are the mean ± SEM of N = 3 independent experiments with n = 6–9 technical replicates each.
In the CQ-resistant 7G8 PfCRT isoform, the drug-free control showed that [3H]CQ uptake was maximal, as expected, based on its resistance phenotype (S3 Table). The addition of 1 μM LDT-623, however, reduced the uptake of [3H]CQ (to an average of 60.2% relative to [3H]CQ without another drug) (Fig 4B). As for the PPQ-resistant 7G8F145I and 7G8C350R isoforms, the addition of [3H]CQ also yielded a reduced percentage of [3H]PPQ uptake (30.% and 47.2%, respectively, relative to [3H]PPQ alone) (Fig 4C and 4D). No competitive inhibition of [3H]CQ or [3H]PPQ transport was observed with pyrimethamine (PYR) or atovaquone (ATQ), which have entirely distinct MoAs (inhibition of folate synthesis and the mitochondrial electron transport chain, respectively). Of note, [3H]CQ uptake by the PfCRT 7G8 isoform and [3H]PPQ uptake by the 7G8F145I and 7G8C350R isoforms were inhibited by the addition of the reciprocal agents PPQ and CQ (see Fig 4B–4D), to levels slightly greater than those observed with LDT-623. We interpret these data as evidence that PPQ and CQ can antagonize each other’s transport via appropriately mutated PfCRT variant isoforms. Earlier studies have shown that PPQ and CQ can bind drug-resistant and -sensitive PfCRT isoforms with similar Kd values, with the difference being that the resistant isoforms acquire transport properties [20].
Since both [3H]CQ and [3H]PPQ uptake are reduced in the presence of LDT-623 in the PfCRT isoforms tested herein, it can be speculated that LDT-623 competes with CQ and PPQ for transport by the distinct PfCRT isoforms. This notion is supported by the robustness of variant-specific 4-AQ interactions with PfCRT and by the minimal impact of ATQ- or PYR–cytochrome bc1 or dihydrofolate reductase inhibitors, respectively [48,49], on the uptake of [3H]CQ or [3H]PPQ (Fig 4B–4D). To provide further experimental evidence for the interaction of LDT-623 with PfCRT, we established a robust platform to screen electrogenic events in the same PfCRT-containing proteoliposome-based model system using solid-supported membrane (SSM) electrophysiology [33,50]. This system enables the detection of ion fluxes associated with 4-AQ, e.g., CQ, PPQ, or LDT-623, transport. Our data (Fig 4E–4G) reveal that the addition of the three tested 4-AQs to the distinct PfCRT isoforms–as validated with parallel [3H]CQ and [3H]PPQ uptake studies (Fig 4B–4D)–elicited currents. We conclude that these currents, reflective of the change of the membrane potential in the PfCRT-containing proteoliposomes, are the result of 4-AQ transport-associated flux of protons (H+), according to an H+/4-AQ co-transport (symport) mechanism as we recently demonstrated [20].
3.5. Profiling of LDT-623 in pfcrt and pfmdr1-edited P. falciparum lines illustrate variant-mediated cross-resistance profiles
To correlate drug uptake results with parasite susceptibility phenotypes, we used pfcrt-modified isogenic parasite lines previously generated [19,20,51–53] via zinc-finger nuclease editing (Table 1 and Fig 5A). We hypothesized that distinct PfCRT isoforms could display different binding transport activity patterns for 4-AQs, thereby modulating parasite susceptibility to LDT-623.
Table 1. Panel of pfcrt-edited P. falciparum lines used in this study.
| Parasite line | Origin | Phenotype | PfCRT amino acid at listed positions | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CQ | PPQ | 72 | 74 | 75 | 76 | 93 | 97 | 145 | 218 | 220 | 271 | 326 | 343 | 350 | 353 | 356 | 371 | ||
| Dd2Dd2 | SE Asia | R | S | C | I | E | T | T | H | F | I | S | E | S | M | C | G | T | I |
| Dd2T93S | Cambodia | R | R | C | I | E | T | S | H | F | I | S | E | S | M | C | G | T | I |
| Dd2H97Y | Cambodia | R | R | C | I | E | T | T | Y | F | I | S | E | S | M | C | G | T | I |
| Dd2F145I | Cambodia | S a | R | C | I | E | T | T | H | I | I | S | E | S | M | C | G | T | I |
| Dd2I218F | Cambodia | S | R | C | I | E | T | T | H | F | F | S | E | S | M | C | G | T | I |
| Dd2M343L | Cambodia | R | R | C | I | E | T | T | H | F | I | S | E | S | L | C | G | T | I |
| Dd2G353V | Cambodia | S | R | C | I | E | T | T | H | F | I | S | E | S | M | C | V | T | I |
| Dd23D7 | Africa | S | S | C | M | N | K | T | H | F | I | A | Q | N | M | C | G | I | R |
| 7G87G8 | S. America, W. Pacific | R | S | S | M | N | T | T | H | F | I | S | Q | D | M | C | G | L | R |
| 7G8F145I | Experimental | S | R | S | M | N | T | T | H | I | I | S | Q | D | M | C | G | L | R |
| 7G8C350R | French Guiana | S | R | S | M | N | T | T | H | F | I | S | Q | D | M | R | G | L | R |
a Sensitized to CQ compared to the parental line but not fully sensitive. pfcrt editing by customized zinc-finger nucleases was confirmed using Sanger sequencing. Differences from the 3D7 wild-type allele are shown in gray shading for both Dd2 and 7G8. Bold indicates mutations identified from piperaquine-resistant field isolates. S: sensitive, R: resistant. Table adapted from [19,20,51].
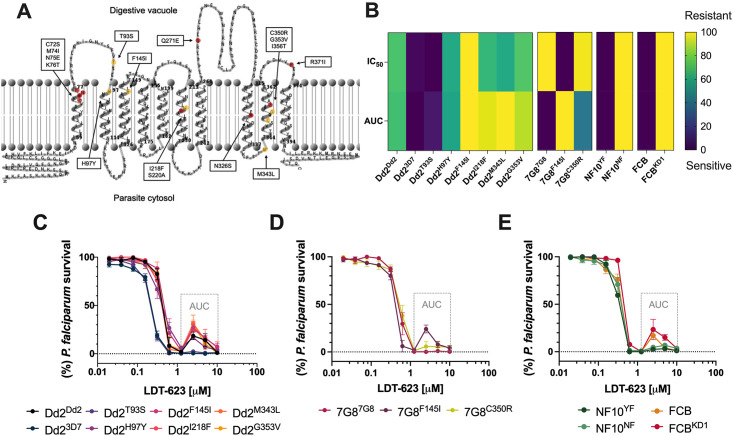
Fig 5. LDT-623 dose–response curves illustrate distinct variant PfCRT-mediated resistance profiles.
(A) Mapping of CQ (red) and PPQ (yellow) resistance-conferring PfCRT amino acid substitutions on the 2D structure of PfCRT3D7 using TMRPres2D [59]. (B) Cross-resistance heatmap of IC50 and AUC values for LDT-623 across pfcrt and pfmdr1-edited P. falciparum lines (N = 2–4). Data were normalized across parameters and scaled into a 0–100 interval for heatmap coloring. (C-D) Dose-response curves for pfcrt-edited clones. Data show mean ± SEM percent parasite survival (relative to drug-free controls) as a function of LDT-623 concentration (N = 2–4). (D) Dose-response curves for pfmdr1-edited clones. Data show mean ± SEM percent parasite survival (relative to drug-free controls) as a function of LDT-623 concentration (N = 3).
PfMDR1, another DV-localized transporter, is a major determinant of pleiotropic antimalarial resistance in the field. Studies have shown that changes in both copy number variations and amino acids at position 86 in pfmdr1 significantly impact parasite susceptibility to several antimalarial drugs [54,55]. Because there appears to be an interplay between specific pfcrt and pfmdr1 alleles [56,57], we also explored the role of genetically manipulated pfmdr1 on the susceptibility of P. falciparum to LDT-623. We used the NF10YF clone (progeny of a genetic cross between 7G8, from Brazil, and GB4 parasites, from Gabon) that harbors a single copy of the GB4 pfmdr1 allele encoding the N86Y/Y184F (YF) haplotype, and the NF10NF clone, genetically engineered to express the N86/Y184F (NF) haplotype [55]. Additionally, we assessed the impact of pfmdr1 amplification on the activity of LDT-623 using the parental line FCB and FCBKD1 that express two or one copies of pfmdr1, respectively [54].
Most of the pfcrt-edited P. falciparum lines assessed did not show substantial rightward shifts in their survival curves (Fig 5C–5E). For comparison, previously published CQ and PPQ susceptibility data for the parasite strains tested were listed alongside LDT-623 results (S4 and S5 Tables). Even though initial LDT-623 concentrations similarly killed different parasite lines, specific sets of mutations differentially impacted parasite survival under higher drug concentrations, resulting in incomplete killing. This incomplete killing phenotype, characterized by biphasic dose-response curves and associated with PPQ resistance [19,58], reveals atypical profiles for typical 72 h dose-response assays, complicating the IC90 determination. In this way, AUC was calculated within the region of the survival curve limited by drug concentrations from 1.25 to 10 μM, encompassing a range of biphasic responses. Lower, yet not statistically significant, AUC for Dd23D7 and Dd2T93S (p-value: 0.1, Mann-Whitney U test) suggest an increase in susceptibility to LDT-623 compared to the isogenic Dd2Dd2 (Fig 5B). 7G8 clones 7G8F145I and 7G8C350R, linked to PPQ resistance, were the closest to statistical significance in the AUC analysis compared to the isogenic 7G87G8 (p-value: 0.06; Mann-Whitney U test), suggesting a trend towards diminished susceptibility to LDT-623 (Fig 5B). Moreover, the PfCRT F145I mutation seems to play a role in resistance to high concentrations of LDT-623 by protecting parasites via an incomplete killing phenotype, regardless of the parasite’s genetic background.
Regarding pfmdr1 genetic changes, the N-terminal N86 mutation in NF10NF did not show any impact on the activity of LDT-623 compared to its parent NF10YF at the IC50 level (1.1-fold increase, p-value: 0.7, Mann-Whitney U-test). While analyzing pfmdr1 copies, the knockdown clone FCBKD1 showed a 1.2-fold increase in IC50 compared to FCB (Fig 5B), a behavior like that previously observed for CQ [54]. However, this increase was not statistically significant (p-value: 0.1, Mann-Whitney U-test). Finally, the knockdown FCBKD1 displayed a more pronounced biphasic curve compared to its FCB parent but no statistical significance was achieved between their AUC values (p-value: 0.4; Mann-Whitney U test). Overall, even though there appears to be a tendency of slight resistance regarding a few of the lines assayed, no striking differences in parasite susceptibility via AUC analysis were observed. None of the pfcrt or pfmdr1 relevant field mutations engineered into these parasite lines yielded significant cross-resistance.
3.6. LDT-623 is refractory to resistance selection in vitro
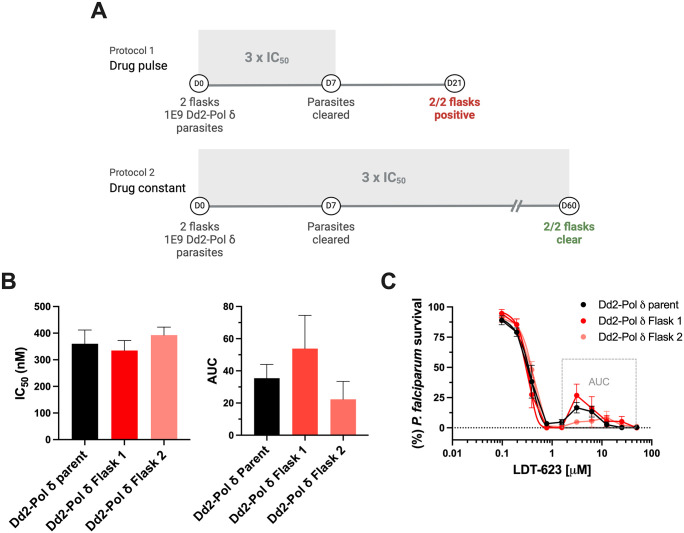
In a bid to obtain additional insights into the potential molecular targets or pathways that drive the multistage antiplasmodial activity of LDT-623, we carried out two parallel resistance selection protocols to generate drug-resistant parasites (Fig 6A).
Fig 6. LDT-623 does not readily select for resistance in vitro.
(A) Selection of resistance to LDT-623 using the Dd2-Polδ line. (A) Parasites were cultured in the presence of 1 μM LDT-623 (~3 × IC50). Parasite inoculum was set at 1 × 109 in duplicate flasks. Shading indicated the presence of drug pressure in the protocol. (B) LDT-623 IC50 and AUC values for parental and recrudescent Dd2-Polδ lines under LDT-623 pressure. (C) Dose-response curves of LDT-623 for parental line (Dd2-Polδ) versus drug-pressured lines (Dd2-Polδ Flask 1 and Dd2-Polδ Flask 2). Arrow represents the 3 × IC50 concentration used for resistance selection. Data represent mean ± SEM (N = 5). Statistical significance was determined by Mann-Whitney U tests, *p<0.05.
No parasite recrudescence was observed in the set-up where constant pressure was applied throughout the 60 days of selection. In the set-up in which drug pressure was removed after day 7 following parasite clearance, recrudescence was observed in both flasks on day 21. However, drug response assays showed no difference in IC50 between the Dd2-Polδ parent culture and the recrudescent bulk parasites from either flask (Fig 6B). Given the biphasic shape of the parasite survival curves, an AUC analysis was performed. While the mean AUC for the Dd2-Polδ parent culture was 35.4, parasites recovered in Flasks 1 and 2 had values of 53.8 and 22.3, respectively (p-values: 0.7 and 0.3, Mann-Whitney U test; Fig 6B), revealing no significant difference between the curves (Fig 6C and S6 Table). With no LDT-623-resistant parasites recovered under constant drug pressure, the log minimum inoculum of resistance (MIR) calculated for LDT-623 is higher than 9 (i.e. >109 infected RBCs), providing evidence for a robust barrier for resistance.
3.7. Failure to recover parasites in the resistome assay indicates no cross-resistance of LDT-623 with major targets and resistance mechanisms
Cross-resistance profiling of compounds against parasite lines with known resistance mutations is another approach to gain insight into the potential resistance liabilities of a compound. Leveraging drug-resistant lines generated predominantly by the MalDA consortium using in vitro evolution or genetic engineering has aided the composition of a library of >50 parasites that, altogether, cover a broad range of currently known resistance mechanisms as well as notable targets advancing in the contemporary antimalarial drug discovery pipeline [60–62]. CRISPR-based barcode insertion allowed these representative parasite lines to be pooled and co-cultured, with their relative abundances quantified by barcode sequencing (S7 Table) [40]. Using a multiplexed competitive growth assay, we subjected this pool of barcoded parasites to LDT-623 pressure, as a means to identify potential cross-resistance targets.
Selective pressure of 3 × IC50 LDT-623 for Dd2 (~790 nM) was applied to the pool of barcoded parasites and maintained constant throughout the assay. At the same time, cumulative growth profiles were recorded for LDT-623, KDU691 – a known PI4-kinase inhibitor [63], and an untreated control (S3 Fig). After 14 days of incubation, no growth recovery was observed for the pool of barcoded parasites treated with LDT-623 and thus no barcodes could be detected by PCR. In contrast, treatment of the pool with the PI4-kinase inhibitor KDU691 yielded recrudescence after 9 days and enrichment of barcode profiles of PI4-kinase mutant parasites (S3B Fig). Failure of these parasite lines to grow under LDT-623 pressure is a strong indicator of no cross-resistance with major targets and resistance mechanisms evaluated, such as ABC transporter I (ABCI3), P-type ATPase (ATP4), cyclic amine resistance locus (CARL), and translation elongation factor 2 (eEF2) [64], among others [62] (S7 Table).
4. Discussion and conclusions
Even though 4-AQs have been relatively well-explored, the history of drug development endorses their adoption as partner drugs in antimalarial therapy, as exemplified by amodiaquine, PPQ, and pyronaridine. Based on a previously characterized antiplasmodial molecule [23], a similarity screening coupled with in vitro assays identified LDT-623, a 4-AQ with nanomolar ABS activity. Previously described as an ABS inhibitor in high throughput screening endeavors [65], we aimed to explore this molecule’s potential by demonstrating its hepatic schizonticidal and transmission-blocking activity in P. falciparum mature gametocytes and P. berghei ookinetes. This moderate yet unusual multistage activity that distinguishes LDT-623 from the typical 4-AQ phenotype encouraged us to investigate it further.
Using a cellular heme fractionation assay, we demonstrated that LDT-623 interferes with the heme detoxification pathway within the DV in a concentration-dependent manner. This can be linked to its MoA. Numerous point mutations in pfcrt have been proven to mediate CQ [18] and PPQ resistance [66] to varying extents. These mutation-driven phenotypes support parasite evasion from the inhibition of Hz formation within the DV: outward drug transport is significantly greater in PfCRT-mutated forms, leading to a lesser extent of drug accumulation in its site of action, thus rendering the parasite resistant. To further dissect the contribution of these complimentary mechanisms to the MoA of LDT-623, we resorted to isoforms of PfCRT-reconstituted proteoliposomes [20]. Measurement of the active uptake of radiolabeled CQ and PPQ showed that LDT-623 partially interferes with drug transport by PfCRT, suggesting that LDT-623 can compete with the other 4-AQs for their interaction with the same functional site. Indeed, electrophysiological measurements support this notion, as LDT-623 elicits the same magnitude of currents as CQ and PPQ. These currents are considered to reflect PfCRT-mediated H+-coupled 4-AQ symport, according to which the transport of 4-AQ compounds is associated with the flux of H+ in the same direction [20].
Functionally, differential drug transport affinities between these PfCRT variants might mirror distinct drug sensitivity profiles. To this end, we put together a panel of pfcrt-edited P. falciparum lines. To evaluate the potential cross-resistance of LDT-623 with globally relevant pfcrt mutations, we selected not only those haplotypes mutated at positions 72–76 from Southeast Asia/Asia (CVIET) and South America (SVMNT) that drive CQ resistance [52] but also PPQ-resistance-conferring mutations such as H97Y, F145I, M343L, or G353V, observed in Southeast Asia [19], and the C350R mutation that has been recently shown to impact over 2/3 of parasite populations analyzed in the Guianas [67–69]. Most Dd2 lines assayed, except for Dd23D7 (wild-type PfCRT) and Dd2T93S, show a biphasic response at high LDT-623 concentrations. This same phenotype is not observed in the 7G87G8 line with the South American SVMNT PfCRT isoform. We believe that CVIET PfCRT could be the major driver of these bimodal dose-response curves. Overall, we hypothesize that a gain of compound transport by mutant pfcrt might be the underlying reason that LDT-623 loses some activity in a range of higher concentrations, as also seen by increased PPQ transport with the highly PPQ-resistant Dd2F145 and Dd2T93S parasite lines [20,52]. Although LDT-623 activity hints towards diminished potency against F145I, data is not a proxy for potential clinical settings, as this mutation is highly unfit in the Dd2 background [19] and no reports have shown its emergence in 7G8 parasites.
Because there appears to be an interplay between particular pfcrt and pfmdr1 alleles [56,57], we explored the impact of pfmdr1—a marker of resistance to lumefantrine and mefloquine (partner drugs in ACTs), on activity of LDT-623. The FCB line, besides its CVIET PfCRT haplotype, also harbors the pfmdr1 N86Y mutation (cognate to NF10YF, yet in a distinct genetic background). Interestingly, although NF10 are also CVIET-bearing parasites, they seem to preserve the sigmoidal curve shape—as opposed to the biphasic response from FCB parasites. Revisiting the trend observed for Dd2 regarding the bimodal curve believed to be driven by CVIET, in this case, we believe that the incomplete killing for both FCB and FCBKD1 might be due to either a strain-specific behavior or even to possible specific interactions between these pfcrt and pfmdr1 alleles. A few of the mutant PfCRT and PfMDR1 haplotypes evaluated in this study also play a role in the parasite susceptibility to other antimalarial drugs, as seen with increased lumefantrine susceptibility and sensitivity to ART derivatives [70–72]. Additionally, increased pfmdr1 copy numbers are commonly observed in Southeast Asia [54,72]. These amplified pfmdr1 copies, fortunately, do not usually extend to African parasite populations, where the fitness costs potentially do not support their expansion in highly endemic areas [73]. Given that we found no pre-existing cross-resistance with pfcrt- or pfmdr1-mutated parasites, the results brought by the present study are timely within the context of considering 4-AQs as effective partner drugs. Our finding that inhibition of Hz formation is part of the MoA of LDT-623, along with the absence of the heme detoxification pathway in both liver and insect parasite stages [13] implies a multifaceted MoA, whose basis requires further investigation. Exploring mechanisms of antimalarial compounds beyond ABS is notoriously difficult given the lower parasite loads available in parasite hepatic and sexual stages compared to ABS. Similarly, insect stages are of extremely low density in the mosquito midgut.
We then resorted to hypermutable Dd2-Pol δ ABS parasites that can display an increased mutation rate of up to 28-fold under drug pressure [38], hence expanding the genetic diversity available for the parasite populations during the selective pressure and increasing our chances of identifying a potential separate MoA of LDT-623. Even though this parasite line does not represent a direct proxy for field outcomes, it is a reliable tool to gain further insights into a drug’s MoA. This strategy has been successful in generating mutants for antimalarial compounds dubbed “irresistible” [38,74], including ones that have previously failed to select for mutations in 3D7 and Dd2 parasites. The failure of LDT-623 to select resistant parasites in vitro suggests that this event might be unlikely to occur in the field. Among numerous potential reasons for this compound not to select for resistance, the bimodal phenotype for LDT-623 recrudescent parasites may reveal a subset of parasites surviving high drug concentrations. This biphasic response could be linked to concentration-dependent activation of drug efflux mechanisms [16,75,76] or even a polypharmacological trait translating into multiple MoAs of this compound [75], which corroborates the multistage antiplasmodial activity observed for LDT-623. Moreover, considering the absence of evidence of lumefantrine and pyronaridine resistance in P. falciparum strains to date and the fact that amodiaquine remains widely effective [77], the study is timely when contemplating 4-AQs as effective partner drugs. With ferroquine undergoing phase 2 trials as a first-line partner drug [72,78–82], including in combination with OZ439, revisiting 4-AQs is appealing. In light of this, a novel 4-AQ antimalarial, such as LDT-623, showing no cross-resistance to these relevant genetic changes, may constitute a valuable tool for further drug optimization efforts.
Finally, we explored parasite modulators of LDT-623 susceptibility and gain insight into its MoA using a fitness competition assay employed to identify cross-resistant barcoded parasite lines able to outgrow drug pressure [40]. Strikingly, none of the parasite strains assayed were retrieved. It is also important to mention that compounds like LDT-623, which not only seem to possess a multifaceted MoA but also leverage host-specific processes (such as the heme detoxification pathway) are compelling tools that have the potential to make it more challenging for resistance to evolve [83,84]. Altogether, by leveraging LDT-623’s ability to bypass drug efflux mechanisms in mutant PfCRT and PfMDR1, two major resistance determinants in ACT partner drugs, our data encourage revisiting quinolines as components of malaria treatment strategies such as triple ACTs, as demonstrated for other antimalarial combinations [19,51].
Supporting information
(A) Structure of compound LDT-611. (B) In vitro growth inhibition of chloroquine-sensitive and resistant P. falciparum strains for LDT-611 and chloroquine, an antimalarial standard (data are represented as mean ± SD, N = 3). (C) Virtual screening workflow for the identification of LDT-611 structural analog molecules. (D) Radial plot showing structural similarity between LDT-611 and its structural analogs selected with Tanimoto coefficient > 0.78.
(PNG)
Luciferase activity (light emission in relative light units) is expressed as means ± SEM and is directly proportional to gamete fertilization and ookinete conversion. (N = 2–3).
(PNG)
(A) Cumulative growth profiles for LDT-623 (4-AQ), a known PI4-kinase inhibitor KDU691, and an untreated control. Drug pressure of 3 x IC50 was maintained constant for 14 days and the pool exhibited no growth recovery for LDT-623, whereas recrudescence was observed after 7 days with KDU691. (B) Barcode profiles (days 0 and 14) for the no-drug control and the recovered parasites treated with KDU691. Cultures treated with KDU691 showed enrichment of PI4-kinase mutant parasites. Cultures were run in triplicate.
(JPG)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Strain background (3D7 or Dd2), gene name and ID, mutation, and input % are indicated. Growth of parasite pool is shown in S3 Fig.
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo - FAPESP (2019/02171-3 and 2021/13809-9 to LTF, 2017/18611-7 to FTMC, 2021/06769-0 to DYB, and 2018/24878-9 to JC), Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (441038/2020-4 and 440373/2022-0 to CHA, and 162117/2018-3 to LCSA), and the National Institutes of Health – NIH (R01 AI124678 and R37 AI050234 to DAF, R01 AI147628 to MQ, DAF). CHA was also supported by Fundação de Amparo à Pesquisa do Estado de Goiás (202010267000272). DYB acknowledges funding from Instituto Serrapilheira (G-1709-16618). MP is funded by “la Caixa” (HR21-848) and European Union Horizon Europe programme (101080744). PS was supported by the Swedish Research Council - Vetenskapsrådet (2021-03667) and The Swedish Foundation for International Cooperation in Research and Higher Education - STINT (BR2018-8017). MCSL gratefully acknowledges funding from the Bill and Melinda Gates Foundation (OPP1054480). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. World Malaria Report 2023. 2023.
- 2.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;57: 742–68. doi: 10.1038/nature12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Pluijm RW, Imwong M, Chau NH, Hoa NT, Thuy-Nhien NT, Thanh NV, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis. 2019;19: 952–61. doi: 10.1016/S1473-3099(19)30391-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imwong M, Dhorda M, Myo Tun K, Thu AM, Phyo AP, Proux S, et al. Molecular epidemiology of resistance to antimalarial drugs in the Greater Mekong subregion: an observational study. Lancet Infect Dis. 2020;20: 1470–80. doi: 10.1016/S1473-3099(20)30228-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad MD, Asua V, Garg S, Giesbrecht D, Niaré K, Smith S, et al. Evolution of partial resistance to artemisinins in malaria parasites in Uganda. N Engl J Med. 2023;389: 722–32. doi: 10.1056/NEJMoa2211803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fola AA, Feleke SM, Mohammed H, Brhane BG, Hennelly CM, Assefa A, et al. Plasmodium falciparum resistant to artemisinin and diagnostics have emerged in Ethiopia. Nat Microbiol. 2023. 8:10. 2023;8: 1911–9. doi: 10.1038/s41564-023-01461-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mihreteab S, Platon L, Berhane A, Stokes BH, Warsame M, Campagne P, et al. Increasing prevalence of artemisinin-resistant HRP2-negative malaria in Eritrea. N Engl J Med. 2023;389: 1191–202. doi: 10.1056/NEJMoa2210956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. The Lancet. 2012;379: 1960–1966. doi: 10.1016/S0140-6736(12)60484-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flegg JA, Guerin PJ, White NJ, Stepniewska K. Standardizing the measurement of parasite clearance in falciparum malaria: the parasite clearance estimator. Malar J. 2011;10: 339. doi: 10.1186/1475-2875-10-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dondorp AM, François N, Poravuth Y, Debashish D, Phae PA, Joel T, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361: 455–67. doi: 10.1056/NEJMoa0808859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis. 2013; 13: 1043–9. doi: 10.1016/S1473-3099(13)70252-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Guha M, Choubey V, Maity P, Bandyopadhyay U. Antimalarial drugs inhibiting hemozoin (β-hematin) formation: A mechanistic update. Life Sci. 2007;80: 813–28. doi: 10.1016/j.lfs.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 13.de Villiers KA, Egan TJ. Heme Detoxification in the malaria parasite: a target for antimalarial drug development. Acc Chem Res. 2021;54: 2649–59. doi: 10.1021/acs.accounts.1c00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaschula CH, Egan TJ, Hunter R, Basilico N, Parapini S, Taramelli D, et al. Structure-activity relationships in 4-aminoquinoline antiplasmodials. The role of the group at the 7-position. J Med Chem. 2002;45: 3531–9. doi: 10.1021/jm020858u [DOI] [PubMed] [Google Scholar]
- 15.Egan TJ, Hunter R, Kaschula CH, Marques HM, Misplon A, Walden J. Structure-function relationships in aminoquinolines: Effect of amino and chloro groups on quinoline-hematin complex formation, inhibition of β- hematin formation, and antiplasmodial activity. J Med Chem. 2000;43: 283–91. doi: 10.1021/jm990437l [DOI] [PubMed] [Google Scholar]
- 16.Wicht KJ, Mok S, Fidock DA. Molecular mechanisms of drug resistance in Plasmodium falciparum malaria. Annu Rev Microbiol. 2020;74: 431–54. doi: 10.1146/annurev-micro-020518-115546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez CP, Manson EDT, Cubel SM, Mandel L, Weidt SK, Barrett MP, et al. The knock-down of the chloroquine resistance transporter PfCRT is linked to oligopeptide handling in Plasmodium falciparum. Microbiol Spectr. 2022;10: e0110122. doi: 10.1128/spectrum.01101-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fidock D, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6: 861–71. doi: 10.1016/s1097-2765(05)00077-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, Kümpornsin K, et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun 2018;9: 3314. doi: 10.1038/s41467-018-05652-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Tan YZ, Wicht KJ, Erramilli SK, Dhingra SK, Okombo J, et al. Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature. 2019;576: 315–20. doi: 10.1038/s41586-019-1795-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hrycyna CA, Summers RL, Lehane AM, Pires MM, Namanja H, Bohn K, et al. Quinine dimers are potent inhibitors of the Plasmodium falciparum chloroquine resistance transporter and are active against quinoline-resistant P. falciparum. ACS Chem Biol. 2014;9: 722–30. doi: 10.1021/cb4008953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravindar L, Hasbullah SA, Rakesh KP, Hassan NI. Recent developments in antimalarial activities of 4-aminoquinoline derivatives. Eur J Med Chem. 2023;256: 115458. doi: 10.1016/j.ejmech.2023.115458 [DOI] [PubMed] [Google Scholar]
- 23.Ferreira LT, Borba JVB, Moreira-Filho JT, Rimoldi A, Andrade CH, Costa FTM. QSAR-based virtual screening of natural products database for identification of potent antimalarial hits. Biomolecules. 2021; 11: 459. doi: 10.3390/biom11030459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65: 418–20. . [PubMed] [Google Scholar]
- 25.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65: 55–63. doi: 10.1016/0022-1759(83)90303-4 . [DOI] [PubMed] [Google Scholar]
- 26.Vaughan AM, Mikolajczak SA, Camargo N, Lakshmanan V, Kennedy M, Lindner SE, et al. A transgenic Plasmodium falciparum NF54 strain that expresses GFP-luciferase throughout the parasite life cycle. Mol Biochem Parasitol. 2012;186: 143–7. doi: 10.1016/j.molbiopara.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 27.Fivelman QL, McRobert L, Sharp S, Taylor CJ, Saeed M, Swales CA, et al. Improved synchronous production of Plasmodium falciparum gametocytes in vitro. Mol Biochem Parasitol. 2007;154: 119–23. doi: 10.1016/j.molbiopara.2007.04.008 [DOI] [PubMed] [Google Scholar]
- 28.Swann J, Corey V, Scherer CA, Kato N, Comer E, Maetani M, et al. High-throughput luciferase-based assay for the discovery of therapeutics that prevent malaria. ACS Infect Dis. 2016;2: 281–93. doi: 10.1021/acsinfecdis.5b00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calit J, Drobescu I, Gaitán XA, Borges MH, Ramos MS, Eastman RT, et al. Screening the Pathogen Box against Plasmodium sexual stages using a new nanoluciferase based transgenic line of P. berghei identifies transmission-blocking compounds. Antimicrob Agents Chemother. 2018;62: e1053–18. doi: 10.1128/AAC.01053-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuda M, Sawai T, Chinzei Y. Structure and expression of an adhesive protein–like molecule of mosquito invasive-stage malarial parasite. J Exp Med. 1999;189: 1947–52. doi: 10.1084/jem.189.12.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ploemen IHJ, Prudêncio M, Douradinha BG, Ramesar J, Fonager J, Van Gemert GJ, et al. Visualization and quantitative analysis of the rodent malaria liver stage by real-time imaging. PLoS One. 2009;4: 1–12. doi: 10.1371/journal.pone.0007881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bazzone A, Barthmes M. Functional characterization of SLC transporters using solid supported membranes. In: Postis VLG, Goldman A, editors. Biophysics of membrane proteins: methods and protocols. New York, NY: Springer US; 2020. pp. 73–103. [DOI] [PubMed] [Google Scholar]
- 33.Weng J, Zhou X, Wiriyasermkul P, Ren Z, Chen K, Gil-Iturbe E, et al. Insight into the mechanism of H+-coupled nucleobase transport. Proc Natl Acad Sci USA. 2023;120: e2302799120. doi: 10.1073/pnas.2302799120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szabo G, Eisenman G, Laprade R, Ciani SM, Krasne S. Experimentally observed effects of carriers on the electrical properties of bilayer membranes—equilibrium domain. With a contribution on the molecular basis of ion selectivity. Membranes. 1973;2: 179–328. . [PubMed] [Google Scholar]
- 35.Jung H, Tebbe S, Schmid R, Jung K. Unidirectional reconstitution and characterization of purified Na+/proline transporter of Escherichia coli. Biochemistry. 1998;37: 11083–8. doi: 10.1021/bi980684b [DOI] [PubMed] [Google Scholar]
- 36.Combrinck JM, Fong KY, Gibhard L, Smith PJ, Wright DW, Egan TJ. Optimization of a multi-well colorimetric assay to determine haem species in Plasmodium falciparum in the presence of anti-malarials. Malar J. 2015;14: 253. doi: 10.1186/s12936-015-0729-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng CL, Fidock DA. Plasmodium falciparum in vitro drug resistance selections and gene editing. Methods Mol Biol. 2019;2013: 123–40. doi: 10.1007/978-1-4939-9550-9_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kümpornsin K, Kochakarn T, Yeo T, Okombo J, Luth MR, Hoshizaki J, et al. Generation of a mutator parasite to drive resistome discovery in Plasmodium falciparum. Nat Commun. 2023;14: 3059. doi: 10.1038/s41467-023-38774-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Istvan ES, Mallari JP, Corey VC, Dharia N V., Marshall GR, Winzeler EA, et al. Esterase mutation is a mechanism of resistance to antimalarial compounds. Nat Commun. 2017;8: 14240. doi: 10.1038/ncomms14240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrasquilla M, Drammeh NF, Rawat M, Sanderson T, Zenonos Z, Rayner JC, et al. Barcoding genetically distinct Plasmodium falciparum strains for comparative assessment of fitness and antimalarial drug resistance. mBio. 2022;13. doi: 10.1128/MBIO.00937-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plouffe DM, Wree M, Du AY, Meister S, Li F, Patra K, et al. High-throughput assay and discovery of small molecules that interrupt malaria transmission. Cell Host Microbe. 2016;19: 114–26. doi: 10.1016/j.chom.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouzón-Arnáiz I, Avalos-Padilla Y, Biosca A, Caño-Prades O, Román-Álamo L, Valle J, et al. The protein aggregation inhibitor YAT2150 has potent antimalarial activity in Plasmodium falciparum in vitro cultures. BMC Biol. 2022;20: 1–27. doi: 10.1186/s12915-022-01374-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fong KY, Wright DW. Hemozoin and antimalarial drug discovery. Future Med Chem. 2013;5: 1437–50. doi: 10.4155/fmc.13.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan DJ. Quinolines block every step of malaria heme crystal growth. Proc Natl Acad Sci USA. 2017;114: 7483–5. doi: 10.1073/pnas.1708153114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spillman NJ, Allen RJW, McNamara CW, Yeung BKS, Winzeler EA, Diagana TT, et al. Na+ regulation in the malaria parasite Plasmodium falciparum involves the cation ATPase PfATP4 and is a target of the spiroindolone antimalarials. Cell Host Microbe. 2013;13: 227–37. doi: 10.1016/j.chom.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riegel B, Roepe PD. Altered drug transport by Plasmodium falciparum chloroquine resistance transporter isoforms harboring mutations associated with piperaquine resistance. Biochemistry. 2020;59: 2484–93. doi: 10.1021/acs.biochem.0c00247 [DOI] [PubMed] [Google Scholar]
- 47.Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, et al. Adaptive evolution of malaria parasites in French Guiana: Reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA. 2015;112: 11672–7. doi: 10.1073/pnas.1507142112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Birth D, Kao WC, Hunte C. Structural analysis of atovaquone-inhibited cytochrome bc1 complex reveals the molecular basis of antimalarial drug action. Nat Commun. 2014;5: 4029. doi: 10.1038/ncomms5029 [DOI] [PubMed] [Google Scholar]
- 49.Walter RD. Altered dihydrofolate reductase in pyrimethamine-resistant Plasmodium falciparum. Mol Biochem Parasitol. 1986;19: 61–6. doi: 10.1016/0166-6851(86)90066-6 [DOI] [PubMed] [Google Scholar]
- 50.Assur Sanghai Z, Liu Q, Clarke OB, Belcher-Dufrisne M, Wiriyasermkul P, Giese MH, et al. Structure-based analysis of CysZ-mediated cellular uptake of sulfate. eLife. 2018;7: e27829. doi: 10.7554/eLife.27829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Small-Saunders JL, Hagenah LM, Wicht KJ, Dhingra SK, Deni I, Kim J, et al. Evidence for the early emergence of piperaquine-resistant Plasmodium falciparum malaria and modeling strategies to mitigate resistance. PLoS Pathog. 2022;18: e1010278. doi: 10.1371/journal.ppat.1010278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dhingra SK, Small-Saunders JL, Ménard D, Fidock DA. Plasmodium falciparum resistance to piperaquine driven by PfCRT. Lancet Infect Dis. 2019;19: 1168–9. doi: 10.1016/S1473-3099(19)30543-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dhingra SK, Gabryszewski SJ, Small-Saunders JL, Yeo T, Henrich PP, Mok S, et al. Global spread of mutant PfCRT and its pleiotropic impact on Plasmodium falciparum multidrug resistance and fitness. mBio. 2019;10: e02731–18. doi: 10.1128/mBio.02731-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sidhu ABS, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194: 528–35. doi: 10.1086/507115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, Uhlemann AC, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun. 2016;7: 11553. doi: 10.1038/ncomms11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Happi CT, Gbotosho GO, Folarin OA, Sowunmi A, Bolaji OM, Fateye BA, et al. Linkage disequilibrium between two distinct loci in chromosomes 5 and 7 of Plasmodium falciparum and in vivo chloroquine resistance in Southwest Nigeria. Parasitol Res. 2006;100: 141–8. doi: 10.1007/s00436-006-0246-4 [DOI] [PubMed] [Google Scholar]
- 57.Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106: 18883–9. doi: 10.1073/pnas.0911317106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bopp S, Magistrado P, Wong W, Schaffner SF, Mukherjee A, Lim P, et al. Plasmepsin II–III copy number accounts for bimodal piperaquine resistance among Cambodian Plasmodium falciparum. Nat Commun. 2018. 9: 1769. doi: 10.1038/s41467-018-04104-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spyropoulos IC, Liakopoulos TD, Bagos PG, Hamodrakas SJ. TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics. 2004;20: 3258–60. doi: 10.1093/bioinformatics/bth358 [DOI] [PubMed] [Google Scholar]
- 60.Corey VC, Lukens AK, Istvan ES, Lee MCS, Franco V, Magistrado P, et al. A broad analysis of resistance development in the malaria parasite. Nat Commun. 2016;7: 11901. doi: 10.1038/ncomms11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cowell AN, Istvan ES, Lukens AK, Gomez-Lorenzo MG, Vanaerschot M, Sakata-Kato T, et al. Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science 2018;359: 191–9. doi: 10.1126/science.aan4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forte B, Ottilie S, Plater A, Campo B, Dechering KJ, Gamo FJ, et al. Prioritization of molecular targets for antimalarial drug discovery. ACS Infect Dis. 2021;7: 2764–76. doi: 10.1021/acsinfecdis.1c00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McNamara CW, Lee MC, Lim CS, Lim SH, Roland J, Simon O, et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature. 2013;504: 248–53. doi: 10.1038/nature12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baragaña B, Hallyburton I, Lee MCS, Norcross NR, Grimaldi R, Otto TD, et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature. 2015;522: 315–20. doi: 10.1038/nature14451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gamo FJ, Sanz LM, Vidal J, De Cozar C, Alvarez E, Lavandera JL, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465: 305–10. doi: 10.1038/nature09107 [DOI] [PubMed] [Google Scholar]
- 66.Mathieu LC, Cox H, Early AM, Mok S, Lazrek Y, Paquet JC, et al. Local emergence in Amazonia of Plasmodium falciparum K13 C580Y mutants associated with in vitro artemisinin resistance. eLife. 2020;9: 1–21. doi: 10.7554/eLife.51015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Florimond C, de Laval F, Early AM, Sauthier S, Lazrek Y, Pelleau S, et al. Impact of piperaquine resistance in Plasmodium falciparum on malaria treatment effectiveness in The Guianas: a descriptive epidemiological study. Lancet Infect Dis. 2024;24: 161–71. doi: 10.1016/S1473-3099(23)00502-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parobek CM, Parr JB, Brazeau NF, Lon C, Chaorattanakawee S, Gosi P, et al. Partner-drug resistance and population substructuring of artemisinin-resistant Plasmodium falciparum in Cambodia. Genome Biol Evol. 2017;9: 1673–86. doi: 10.1093/gbe/evx126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leang R, Taylor WRJ, Bouth DM, Song L, Tarning J, Char MC, et al. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother. 2015;59: 4719–26. doi: 10.1128/AAC.00835-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sisowath C, Petersen I, Veiga MI, Mårtensson A, Premji Z, Björkman A, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009;199: 750–7. doi: 10.1086/596738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conrad MD, LeClair N, Arinaitwe E, Wanzira H, Kakuru A, Bigira V, et al. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis. 2014;210: 344–53. doi: 10.1093/infdis/jiu141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Venkatesan M, Gadalla NB, Stepniewska K, Dahal P, Nsanzabana C, Moriera C, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am J Trop Med Hyg. 2014;91: 833–43. doi: 10.4269/ajtmh.14-0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheeseman IH, Miller B, Tan JC, Tan A, Nair S, Nkhoma SC, et al. Population structure shapes copy number variation in malaria parasites. Mol Biol Evol. 2016;33: 603–20. doi: 10.1093/molbev/msv282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoo E, Schulze CJ, Stokes BH, Onguka O, Yeo T, Mok S, et al. The antimalarial natural product Salinipostin A identifies essential α/β serine hydrolases involved in lipid metabolism in P. falciparum parasites. Cell Chem Biol. 2020;27: 143–57. doi: 10.1016/j.chembiol.2020.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le Manach C, Paquet T, Wicht K, Nchinda AT, Brunschwig C, Njoroge M, et al. Antimalarial lead-optimization studies on a 2,6-imidazopyridine series within a constrained chemical space to circumvent atypical dose-response curves against multidrug-resistant parasite strains. J Med Chem. 2018;61: 9371–85. doi: 10.1021/acs.jmedchem.8b01333 [DOI] [PubMed] [Google Scholar]
- 76.Patel V, Booker M, Kramer M, Ross L, Celatka CA, Kennedy LM, et al. Identification and characterization of small molecule inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. J Biol Chem. 2008;283: 35078–85. doi: 10.1074/jbc.M804990200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blasco B, Leroy D, Fidock DA. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med. 2017;23: 917–28. doi: 10.1038/nm.4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCarthy JS, Rückle T, Djeriou E, Cantalloube C, Ter-Minassian D, Baker M, et al. A Phase II pilot trial to evaluate safety and efficacy of ferroquine against early Plasmodium falciparum in an induced blood-stage malaria infection study. Malar J. 2016;15: 469. doi: 10.1186/s12936-016-1511-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Held J, Supan C, Salazar CLO, Tinto H, Bonkian LN, Nahum A, et al. Ferroquine and artesunate in African adults and children with Plasmodium falciparum malaria: a phase 2, multicentre, randomised, double-blind, dose-ranging, non-inferiority study. Lancet Infect Dis. 2015;15: 1409–19. doi: 10.1016/S1473-3099(15)00079-1 [DOI] [PubMed] [Google Scholar]
- 80.Wells TN, H van Huijsduijnen R. Ferroquine: welcome to the next generation of antimalarials. Lancet Infect Dis. 2015;15: 1365–6. doi: 10.1016/S1473-3099(15)00148-6 [DOI] [PubMed] [Google Scholar]
- 81.Gansane A, Lingani M, Yeka A, Nahum A, Bouyou-Akotet M, Mombo-Ngoma G, et al. Randomized, open-label, phase 2a study to evaluate the contribution of artefenomel to the clinical and parasiticidal activity of artefenomel plus ferroquine in African patients with uncomplicated Plasmodium falciparum malaria. Malar J. 2023;22: 1–16. doi: 10.1186/s12936-022-04420-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beshir K, Sutherland CJ, Merinopoulos I, Durrani N, Leslie T, Rowland M, et al. Amodiaquine resistance in Plasmodium falciparum malaria in Afghanistan is associated with the pfcrt SVMNT allele at codons 72 to 76. Antimicrob Agents Chemother. 2010;54: 3714–6. doi: 10.1128/AAC.00358-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kelly JX, Smilkstein MJ, Brun R, Wittlin S, Cooper RA, Lane KD, et al. Discovery of dual function acridones as a new antimalarial chemotype. Nature. 2009;459: 270–3. doi: 10.1038/nature07937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gunsaru B, Burgess SJ, Morrill W, Kelly JX, Shomloo S, Smilkstein MJ, et al. Simplified reversed chloroquines to overcome malaria resistance to quinoline-based drugs. Antimicrob Agents Chemother. 2017;61: e01913–16. doi: 10.1128/AAC.01913-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Structure of compound LDT-611. (B) In vitro growth inhibition of chloroquine-sensitive and resistant P. falciparum strains for LDT-611 and chloroquine, an antimalarial standard (data are represented as mean ± SD, N = 3). (C) Virtual screening workflow for the identification of LDT-611 structural analog molecules. (D) Radial plot showing structural similarity between LDT-611 and its structural analogs selected with Tanimoto coefficient > 0.78.
(PNG)
Luciferase activity (light emission in relative light units) is expressed as means ± SEM and is directly proportional to gamete fertilization and ookinete conversion. (N = 2–3).
(PNG)
(A) Cumulative growth profiles for LDT-623 (4-AQ), a known PI4-kinase inhibitor KDU691, and an untreated control. Drug pressure of 3 x IC50 was maintained constant for 14 days and the pool exhibited no growth recovery for LDT-623, whereas recrudescence was observed after 7 days with KDU691. (B) Barcode profiles (days 0 and 14) for the no-drug control and the recovered parasites treated with KDU691. Cultures treated with KDU691 showed enrichment of PI4-kinase mutant parasites. Cultures were run in triplicate.
(JPG)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Strain background (3D7 or Dd2), gene name and ID, mutation, and input % are indicated. Growth of parasite pool is shown in S3 Fig.
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.