Abstract
Background
Common Variable Immunodeficiency (CVID) represents a heterogenic group of primary immunodeficiencies (PID) characterized by impaired antibody production and susceptibility to infections. Non-infectious complications, such as autoimmune diseases, lymphoproliferative disorders, and malignancies, now significantly impact prognosis. Moreover, both hematologic and solid organ malignancies are more frequently observed in CVID patients compared to other PIDs. The risk factors for carcinogenesis in CVID remain largely unknown.
Objective
This multicenter study aims to characterize the clinical profile of cancer in CVID patients in Spain and to identify independent risk factors associated with malignancy development, focusing on the role of immune dysregulation.
Methods
A nationwide, cross-sectional study was conducted from November 2019 to May 2022, involving 17 hospitals treating PID patients in Spain. Data were collected systematically on demographics, infectious and non-infectious comorbidities, immunological parameters, and treatment. Statistical analysis, including multivariate logistic regression, was performed to identify risk factors associated to malignancy.
Results
Of 250 CVID patients, 38 (15.26%) were diagnosed with cancer, predominantly non-Hodgkin lymphoma, gastric cancer, and lung adenocarcinoma. Cancer patients were significantly older (mean age 60.70 vs. 49.36 years, p<0.001) and had higher rates of immune dysregulation (81.58% vs. 59.7%, p=0.01). Immune dysregulation was an independent risk factor for cancer (OR 2.19, p=0.04), alongside previous immunosuppressant therapy (OR 2, p=0.031), higher IgM levels (OR 1.008 per SD, p=0.012), older age (OR 1.04, p<0.001), and lower CD4 cell counts at diagnosis (OR 0.997, p<0.001).
Conclusions
This study highlights the increased cancer risk in CVID patients, with immune dysregulation, prior immunosuppressant use, elevated IgM levels, and lower CD4 cell counts as conjointly associated. These findings underscore the need for vigilant cancer screening and tailored management strategies in CVID patients to improve outcomes. Future research should focus on elucidating the molecular mechanisms linking immune dysregulation and malignancy in CVID.
Keywords: common variable immunodeficiency, immune dysregulation, malignancy, cancer risk, immunosuppressants
1. Introduction
Common variable immunodeficiency (CVID) constitutes a heterogeneous group of primary immunodeficiency disorders (PID) with an estimated prevalence of 1:50,000 to 1:25,000 (1, 2). It is characterized by decreased levels of serum IgG, along with decreased IgM and/or IgA, after excluding secondary causes of hypogammaglobulinemia (1, 2). Historically, infectious diseases were the primary cause of morbidity and mortality in CVID patients until the introduction of immunoglobulin replacement therapy (IgRT) in the late 20th century. This treatment has significantly reduced infection-related complications, shifting the burden towards non-infectious complications such as autoimmune diseases, benign lymphoproliferative disorders, and cancer, which now have a larger impact on prognosis (3, 4). These immune dysregulation-related phenomena may affect up to 70% of patients, contributing to an 11-fold increased risk of death (5). Notably, IgRT does not seem to prevent or improve many of these conditions (5).
Moreover, both hematologic and solid organ malignancies are more frequently observed in CVID patients compared to other PIDs, and they are associated with poorer outcomes (3, 6). Various studies report a variable frequency of malignancy in CVID patients (3, 5–9), with the incidence of cancer around 10% (ranging from 1.5% to 20.7%). These malignancies typically occur during the 4th to 6th decades of life, with a risk 5-12 times higher than the general population (5, 10). The most frequently reported malignancies in CVID patients include non-Hodgkin lymphoma (NHL), gastric carcinoma, and leukemia (3, 5–7, 9).
The risk factors for carcinogenesis in CVID remain largely unknown. It is hypothesized that immune dysregulation may be associated with the development of neoplasia in this population (11–13). Several specific manifestations of this dysimmunity have been identified as potential risk factors for malignancy, such as the history of immune thrombocytopenic purpura with over a threefold increase in cancer risk (14), or the presence of arthritis, atrophic gastritis, or interstitial lung disease (ILD) (17).
Currently, there are no standardized protocols for cancer screening in CVID patients, nor are there validated tools to accurately assess the risk of neoplasia development in this population (17). The heterogeneity of CVID and its associated complications complicate the establishment of universal screening guidelines (7). Furthermore, the interplay between immune dysregulation and cancer development remains poorly understood, necessitating comprehensive research to elucidate these mechanisms.
The aim of this study was to characterize the clinical profile of cancer patients in CVID in Spain, and to identify the potential independent risk factors associated to its presence, exploring the impact of the immune dysregulation subpopulation and its potential role in malignancy.
2. Materials and methods
2.1. Study design, setting and population
A multicenter, cross-sectional, nationwide study of patients diagnosed with CVID was conducted in Spain from November 2019 to May 2022. Seventeen hospitals treating PIDs participated in establishing the GTEM-SEMI-CVID Registry, an initiative led by the Working Group of Rare Diseases of the Spanish Society of Internal Medicine (GTEM-SEMI). Patients aged 16 years and older with a confirmed diagnosis of CVID, according to the ESID working definitions (2), who were currently or had previously been under follow-up by the participating units, were eligible for inclusion. Patients with confirmed monogenic immunodeficiencies were excluded from eligibility.
2.2. Data collection and variables
The GTEM-SEMI-CVID-Registry systematically compiles comprehensive data on patients, including sociodemographics, epidemiology, genetics, comorbidities, imaging, laboratory results, treatments, and outcomes, as in Cabañero-Navalon et al. (7). Demographic information included sex, age at diagnosis, clinical onset, diagnostic delay, and follow-up duration. Genomic data, family history, and consanguinity were recorded. Infectious complications were registered, including major bacterial, opportunistic, and chronic infections. Non-infectious comorbidities included autoimmune cytopenias, organomegalies, systemic autoimmune disorders, and malignancies. Laboratory parameters such as immunoglobulin levels, lymphocyte subpopulations cell counts, and autoantibody presence were gathered, as well as the histopathological analyses of biopsied tissues. Therapeutic variables as IgRT and immunosuppressant use were noted. Further information on the GTEM-SEMI-CVID-Registry methodology can be found elsewhere (7).
2.3. Definitions
In this study examining variables associated to malignancy, cases were defined as patients who, at any point during their clinical follow-up, had a history of either hematologic and solid tumors, excluding basal cell and squamous cell skin carcinomas. Controls were patients without these conditions who mainly suffered infectious complications (iCVID).
Patients were screened for immune dysregulation (dCVID), defined by the presence of lymphadenopathy, immune cytopenias, non-infectious interstitial lung disease, splenomegaly, hepatomegaly, hepatic nodules, autoimmune organ-specific or systemic diseases, non-infectious enteropathy, and/or immune-mediated skin involvement.
2.4. Statistical analysis
The statistical analysis was performed using R software, version 4.3.0. The following information was considered as potentially associated to malignancy occurrence: sex, age, immunosuppression, immune dysregulation, IgG at diagnosis and last follow-up, IgM levels at diagnosis and last follow-up, IgA levels at diagnosis and last follow-up, CD4 cell count at diagnosis and last follow-up, CD8 cell count at diagnosis and last follow-up, and CD19 cell count at diagnosis and last follow-up. Median-based imputation was used to estimate missing values. Chi-square test was used to test whether the frequency of malignancy occurrence differed when patients were grouped based on gender, immunosuppression, and immune dysregulation. In addition, we aimed to explore the effect of immune dysregulation in this assessment. Therefore, for each continuous variable, a two-way ANOVA with iCVID and dCVID and malignancy was conducted. Effects or interactions with p-value below 0.05 were considered statistically significant. Partial-eta squared was used to measure the size of the effect of each main effect and interaction. Tukey post-hoc test (Bonferroni corrected when necessary) was applied when significant effects and interactions were found.
We used a multivariate logistic regression to study the likelihood of malignancy occurrence based on the different variables that showed significant Malignancy effect. Categorical variables showing significant relationship with malignancy were also included in the logistic regression. From the coefficients of the model, we calculated the Odds-ratio (OR), their confidence interval and the p-value associated to each coefficient (Wald test). We also studied the model’s performance by extracting the accuracy, sensibility, specificity, and the area under the receiver operating characteristic (ROC) curve (AUC). These performance parameters were extracted by using a leave-one-out cross-validation method. A nomogram was then created to represent the estimated probability of cancer at a given time for illustrative purposes.
2.5. Ethical statement
The development and protocol of the GTEM-SEMI-CVID Registry received independent approval from the Ethical Committees of all participating hospitals, each under their respective registry codes. The study was conducted in accordance with the Declaration of Helsinki and adhered to the STROBE guidelines. Anonymity and data confidentiality for all included patients were maintained in compliance with Spanish regulations governing observational studies.
3. Results
3.1. Study population
Out of a total of 250 patients included in the GTEM-SEMI-CVID Registry, 249 had been assessed for cancer prevalence. Among these, 38 (15.26%) were diagnosed with cancer during follow-up and were referred to as cases. Non-Hodgkin B lymphoma was the most frequent malignancy, occurring in 11 patients (4.41%). Gastric cancer and lung adenocarcinoma were reported in 5 (2.01%) and 3 (1.20%) patients, respectively. There were 2 cases of colorectal cancer, and 1 case each of breast and prostate cancer. Additionally, 3 patients had basal cell carcinoma, with 1 patient having both basal cell and squamous cell carcinoma of the skin. The remaining 7 cases included myeloma, splenic lymphoma, and cancers of the thyroid, kidney, uterus, and cervix.
All 13 patients with lymphoid malignancies showed immune dysregulation, with it being the first clinical symptom in 3 cases and malignancy in 2 at CVID debut. All five patients with gastric cancer also had immune dysregulation, and autoimmunity was the first symptom in two of these cases. Helicobacter pylori was present in 3, absent in 1, and never tested in the last. Atrophic gastritis was also present in those H. pylori positive patients.
Cases were significantly older than controls, with a mean age of 60.70 (SD 17.56) vs. 49.36 (17.88) years (p<0.001). There were no differences in sex distribution between the groups. No significant differences were seen in the diagnostic delay of CVID among groups. Immune dysregulation was present in 81.58% of cases compared to 59.7% of controls (p=0.01). Specifically, 50% of cancer patients had a history of cytopenia, compared to 30% of controls (p=0.02). Lymphadenopathy was more common in cases (57.9% vs. 30.8%, p=0.003), as was immune-mediated skin involvement (47.4% vs. 23.2%, p=0.003). There were no differences in the prevalence of splenomegaly, hepatomegaly, liver nodules, non-infectious interstitial lung disease, autoimmune diseases, or non-infectious enteropathy between the groups. Further details can be found in Table 1 .
Table 1.
Main characteristics of CVID patients with and without malignancy in the Spanish GTEM-SEMI-CVID cohort.
| Variable | Malignancy Mean (SD) – n (%) |
No malignancy Mean (SD) – n (%) |
P-value | |
|---|---|---|---|---|
| Age | 60.63 (17.56) | 49.36 (17.88) | 4x10-4* | |
| Sex | Male | 16 (42.11) | 105 (49.76) | 0.481 |
| Female | 22 (57.89) | 106 (50.24) | ||
| Immune dysregulation | 31 (81.58) | 126 (59.72= | 0.01* | |
| Cytopenias | 19 (50) | 64 (30.33) | 0.025* | |
| Lymphadenopathies | 22 (57.89) | 65 (30.81) | 0.003* | |
| Splenomegaly | 16 (42.11) | 65 (30.81) | 0.193 | |
| Hepatomegaly | 7 (18.42) | 39 (18.48) | 0.185 | |
| Liver nodules | 4 (10.53) | 12 (5.69) | 0.219 | |
| Lung disease | 26 (68-42) | 121 (57.35) | 0.282 | |
| Enteropathy | 14 (36.84) | 69 (32.70) | 0.71 | |
| Atrophic gastritis | 8 (21.05) | 24 (11.37) | 0.203 | |
| Autoimmune systemic disease | 8 (21.05) | 41 (19.43) | 0.853 | |
| Skin affectation | 17 (44.74) | 49 (23.22) | 0.002* | |
| Immunoglobulins | ||||
| IgG levels at diagnosis (mg/dL) | 391.56 (238.539 | 405.36 (215.47) | 0.475 | |
| Last follow-up IgG levels (mg/dL) | 850.07 (313.77) | 852.35 (235.26) | 0.982 | |
| IgM levels at diagnosis (mg/dL) | 52.32 (79.34) | 53.48 (90.78) | 0.720 | |
| Last follow-up IgM levels (mg/dL) | 72.31 (89.00) | 67.58 (224.77) | 0.164 | |
| IgA levels at diagnosis (mg/dL) | 39.09 (58.42) | 48.56 (81.43) | 0.976 | |
| Last follow-up IgA levels (mg/dL) | 122.29 (516.84) | 66.57 (166.60) | 0.810 | |
| Lymphocyte cell count | ||||
| CD4 cell count at diagnosis (cell/mm3) | 468.00 (243.41) | 715.15 (442.60) | 0.002* | |
| Last follow-up CD4 cell count (cell/mm3) | 596.83 (302.48) | 1,054.21 (4,228.46) | 0.596 | |
| CD8 cell count at diagnosis (cell/mm3) | 419.45 (216.18) | 537.04 (412.97) | 0.604 | |
| Last follow-up CD8 cell count (cell/mm3) | 478.33 (243.00) | 571.90 (429.06) | 0.604 | |
| CD19 cell count at diagnosis (cell/mm3) | 192.68 (144.50) | 237.26 (198.43) | 0.665 | |
| Last follow-up CD19 cell count (cell/mm3) | 188.76 (155.24) | 185.66 (191.21) | 0.282 | |
| Immunosuppressant therapy | 20 (52.63) | 74 (35.97) | 0.027* | |
| Corticosteroids | 17 (44.74) | 68 (32.23) | 0.134 | |
| Azathioprine | 7 (18.42) | 22 (10.43) | 0.168 | |
| Tacrolimus | 1 (2.63) | 6 (2.84) | 1 | |
| Rituximab | 8 (21.05) | 21 (9.95) | 0.056 | |
Mean and SD will be applied for quantitative variables, while N and percentage will be applied for qualitative variables.
* statistically significant.
In the Spanish GTEM-SEMI-CVID Registry, clearly-defined monogenic disorders under a CVID phenotype were excluded. However, analysis of genetic data revealed several variants of uncertain significance (VUS) across a range of genes associated with CVID. The most frequently observed VUS were in the TACI and NFKB1 genes, each identified in 7 patients. The CTLA4 gene was the next most frequently affected, with VUS identified in 5 patients, followed by MBL2, which showed mutations in 3 patients. Variants in IKAROS were noted in 2 patients. Additionally, single occurrences of VUS were observed in several genes, including BTK, NFKB2, LRBA, MLL2, PI3KCD, PI3KR1, PCLG2, PTPN2, RAG1, TFC3, and CD27.
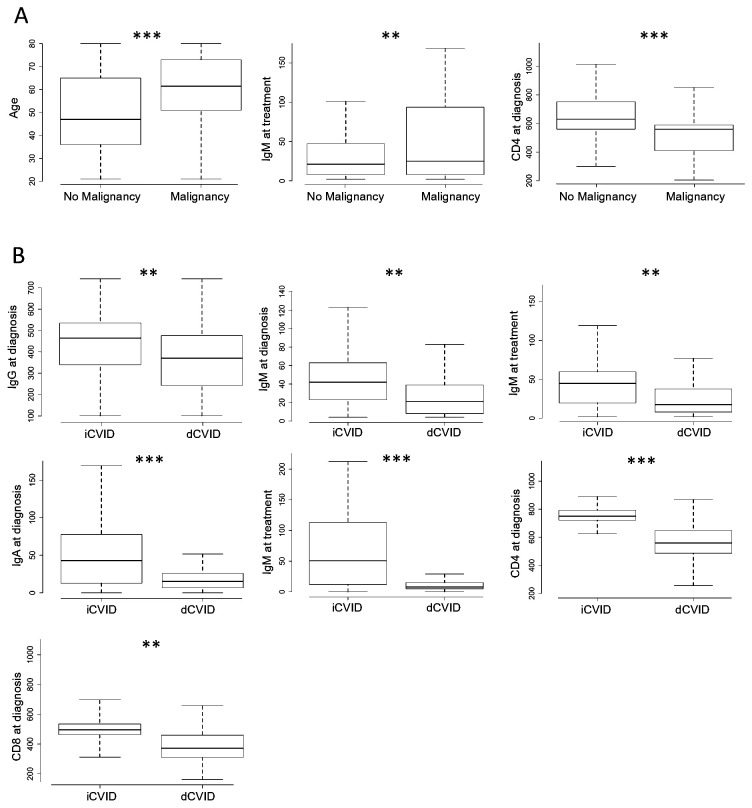
Cases had received more immunosuppressant therapy than controls during follow-up (55.6% vs. 35.7%, p=0.03). At diagnosis, CD4 cell counts were significantly lower in cases (468.01 cells/µL, SD 243.41) than in controls (715.16 cells/µL, SD 442) (p=0.023), although no differences were observed at the last follow-up. Total IgG, IgA, and IgM levels at diagnosis and during follow-up did not differ between the groups, nor did CD19 or CD8 cell counts. No significant differences were observed in the presence of antinuclear antibodies. However, antineutrophil cytoplasmic antibodies detection was significantly more frequently observed in cancer patients (3.57%) when compared to controls (2.6%) (p=0.023). Figure 1 displays box plots for those variables where significant main effects of immune dysregulation and malignancy were found (Tukey test).
Figure 1.
Box plot illustrating the results of the Tukey post-hoc test for variables with significant effects, showing (A) the impact of immune dysregulation and (B) the impact of malignancy. Statistically significant differences are marked as p < 0.05 and **p<0.01, ***p < 0.001.
Patients with immune dysregulation (dCVID) had significantly lower CD4 cell counts at diagnosis (643.57 cells/µL, SD 476.05) compared to patients without immune dysregulation (747.51 cells/µL, SD 306.49) (p=0.016), regardless of the presence of cancer. IgG levels at diagnosis were lower in dCVID patients (383.09 mg/dL, SD 230.97) compared to iCVID patients (440.79 mg/dL, SD 189.53) (p=0.04). IgA levels at diagnosis were also lower in dCVID patients (29.46 mg/dL, SD 44.35) compared to non-dCVID patients (79.30 mg/dL, SD 110.55) (p<0.001). This difference persisted at follow-up (45.72 mg/dL, SD 264 vs. 126.96 mg/dL, SD 235.92, p=0.007). There were no significant differences in IgM levels or CD19 and CD8 cell counts at diagnosis and last follow-up. These results were observed in the two-way ANOVA analysis, but no significant interaction was found for any variable, as seen in Table 2 .
Table 2.
Two-way ANOVA analysis assessing the effects of malignancy and immune dysregulation, as well as their interaction, on various clinical and immunological variables in CVID patients.
| Variable | Immune dysregulation effect | Malignancy effect | Malignancy x Immune dysregulation interaction |
|---|---|---|---|
| Age | F(1,245) = 2.4 | F(1,245) = 14.3***, ηp2 = 0.05 | F(1,245) = 1.4 |
| IgG levels at diagnosis (mg/dL) | F(1,245) = 10.3** ηp2 = 0.037 | F(1,245) = 0.2 | F(1,245) = 0.1 |
| Last follow-up IgG levels (mg/dL) | F(1,245) = 0 | F(1,245) =0 | F(1,245) = 0.9 |
| IgM levels at diagnosis (mg/dL) | F(1,245) = 8.2**, ηp2 = 0.037 | F(1,245) =0.11 | F(1,245) = 0.07 |
| Last follow-up IgM levels (mg/dL) | F(1,245) = 9.2**, ηp2 = 0.04 | F(1,245) =8.7**, ηp2 = 0.03 | F(1,245) = 0.07 |
| IgA levels at diagnosis (mg/dL) | F(1,245) = 30.6***, ηp2 = 0.1 | F(1,245) = 0.01 | F(1,245) = 0.01 |
| Last follow-up IgA levels (mg/dL) | F(1,245) = 61.3***, ηp2 = 0.2 | F(1,245) = 0.2 | F(1,245) = 0.1 |
| CD4 cell count at diagnosis (cell/mm3) | F(1,245) = 30.1***, ηp2 = 0.09 | F(1,245) = 7.4**, ηp2 = 0.02 | F(1,245) = 0.14 |
| Last follow-up CD4 cell count (cell/mm3) | F(1,245) = 0.3 | F(1,245) =0.06 | F(1,245) = 0.23 |
| CD8 cell count at diagnosis (cell/mm3) | F(1,245) = 6.3*, ηp2 = 0.02 | F(1,245) = 0.9 | F(1,245) = 0.001 |
| Last follow-up CD8 cell count (cell/mm3) | F(1,245) = 3.1 | F(1,245) = 0.4 | F(1,245) = 0.003 |
| CD19 cell count at diagnosis (cell/mm3) | F(1,245) = 0.07 | F(1,245) =0.8 | F(1,245) = 0.03 |
| Last follow up CD19 cell count (cell/mm3) | F(1,245) = 0.1 | F(1,245) =0.001 | F(1,245) = 0.1 |
The table presents the F-values and significance levels from a Two-Way ANOVA and highlighted in bold examining the effects of malignancy, immune dysregulation, and their interaction on various clinical and immunological variables in CVID patients. Significant results are marked with asterisks and highlighted in bold (*p < 0.05, **p < 0.01, ***p < 0.001), indicating a meaningful impact of the factor on the variable. Higher F-values represent a stronger effect. Additionally, partial eta squared (ηp²) is included to indicate the proportion of variance in the data explained by immune dysregulation or malignancy, with higher values representing greater effect sizes. For example, in IgA levels at diagnosis, ηp² = 0.1, suggesting that 10% of the variability in IgA levels is due to immune dysregulation.
3.2. Multivariable regression analysis
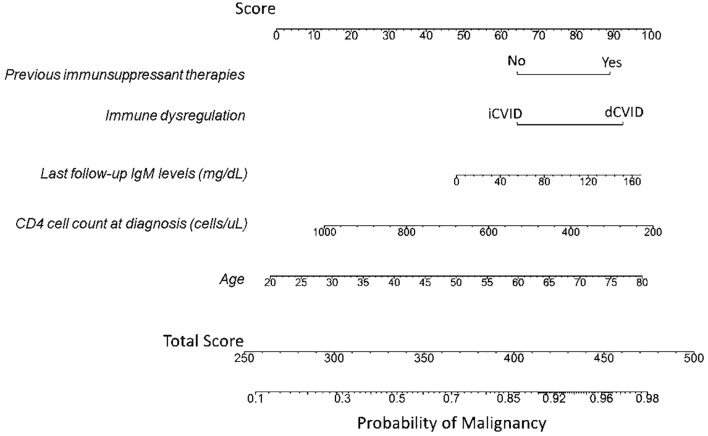
A multivariable logistic regression analysis was conducted to assess the contribution of various parameters to cancer occurrence in our cohort. Initially, all potential predictors for cancer, as outlined in the Methods section, were filtered based on VIF, data sparsity, and biological plausibility. The final model included the variables age, immunosuppressant therapy, immune dysregulation, IgM levels at last follow-up and CD4 cell count at diagnosis, which showed each a significant univariant relationship with malignancy occurrence and therefore were considered as independent associates of malignancy in our cohort. The logistic model was significant with respect of the null-model (Likelihood Ratio test, p<0.001). The ORs, the confidence interval of the ORs and the p-values obtained from the Wald test applied to each coefficient are reported in Table 3 . Immune dysregulation increased the odds of cancer by 2.19 times (p=0.04), and previous immunosuppressant therapies raised the odds by 2 (p=0.031). Increases in IgM levels in one standard deviation (203.3 mg/dL) raised the odds by 1.008 times (p=0.012). Furthermore, age and CD4 cell count at diagnosis were considered as risk factors with ORs of 1.04 (p<0.001) and 0.997 (p<0.001). Nomogram associated to this model is shown in Figure 2 .
Table 3.
Logistic regression analysis of the occurrence of Malignancy. Odds-ratio, p-value and the 95% confidence interval of each independent variable are shown.
| Variable | OR | CI 95% | P-value |
|---|---|---|---|
| Intercept | 0.13 | 0.02-0.66 | 0.014 |
| Immunosuppressant treatment | 2 | 1.06-3.79 | 0.031 |
| Immune dysregulation | 2.19 | 1.01-4.88 | 0.04 |
| Age | 1.04 | 1.02-1.06 | <0.001 |
| Last follow-up IgM levels | 1.008 | 1.001-1.01 | 0.012 |
| CD4 cell count at diagnosis | 0.997 | 0.995-0.998 | <0.001 |
Figure 2.
Nomogram illustrating the predicted probability of malignancy based on the predictive risk factors. The total score corresponds to the probability of malignancy occurring, shown on the bottom scale. To obtain the nomogram-predicted probability of malignancy, first locate the patient’s value for each of the five variables on their respective axes. Draw a vertical line from each variable value to the upper “Score” axis to determine the points attributed to each value. Sum the points for all variables, and then locate the total points on the “Total Score” axis. Finally, draw a vertical line from the total points to the “Probability of Malignancy” axis to determine the estimated probability of malignancy. Example 1: A 65-year-old patient with CVID presented with immune dysregulation manifested as immune thrombocytopenic purpura (ITP) requiring immunosuppressive therapy. The patient had an IgM level under IgRT of 125 mg/dL and a CD4 cell count at diagnosis of 350 cells/µL. For dCVID associated with immune dysregulation, plotting a vertical line to the “Score” axis yields approximately 92 points. Similarly, an age of 65 corresponds to 74 points, the use of immunosuppressants to 89 points, an IgM level of 125 mg/dL to 85 points, and a CD4 cell count of 350 cells/µL to about 90 points. The total score for this patient is 432 (92 + 74 + 89 + 85 + 90). Using this total score on the “Total Score” axis and drawing a vertical line to the “Probability of Malignancy” axis indicates an estimated malignancy risk of approximately 92%. Example 2: In contrast, a 25-year-old CVID patient without immune dysregulation, who has never received immunosuppressive therapy, has an IgM level under IgRT of 40 mg/dL and a CD4 cell count at diagnosis of 700 cells/µL. The absence of immune dysregulation corresponds to 62 points on the upper score line, an age of 25 to approximately 8 points, no history of immunosuppressive therapy to 63 points, an IgM level of 40 mg/dL to 60 points, and a CD4 cell count of 700 cells/µL to about 45 points. The total score for this patient is 238 (62 + 8 + 63 + 60 + 45), corresponding to a malignancy probability of less than 10%.
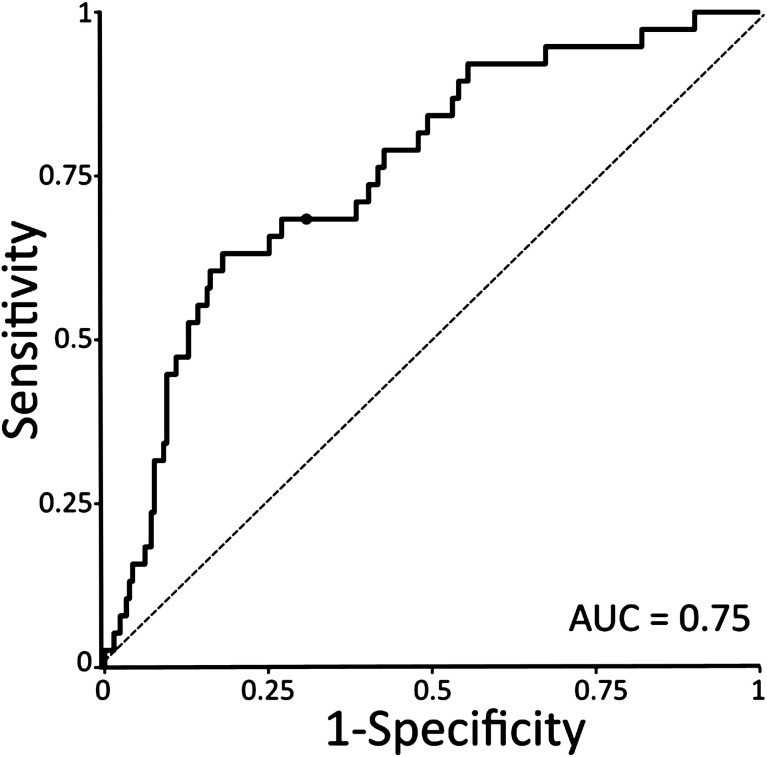
We performed a leave-one-out cross-validation to assess the predictive performance of this exploratory model. The model’s predictive performance, evaluated by the AUC, was 0.75 ( Figure 3 ). The model showed an overall accuracy of 0.69, with a sensitivity of 0.68 and a specificity of 0.69.
Figure 3.
Receiver Operating Characteristic (ROC) curve illustrating the performance of the logistic regression model in distinguishing between Malignancy and No malignancy. The diagonal dashed line represents the baseline performance of a random classifier. Dot point shows the optimal cut-off point based on the maximal specificity and sensibility, which corresponds to 0.5.
4. Discussion
In this study, we investigated the risk factors for cancer development in a cohort of 249 CVID patients, with particular emphasis on immune dysregulation. In the GTEM-SEMI-CVID Registry, older age, immune dysregulation, previous immunosuppressant therapies, higher IgM levels, and lower CD4 cell counts at diagnosis were independently associated to malignancy in CVID from a multivariable approach ( Figure 2 ).
Patients with CVID have a significantly increased risk of both hematological and solid malignancies, with a reported prevalence of approximately 10% (ranging from 1.5% to 20.7%). In our registry, the prevalence of cancer was 15.26%, one of the highest reported rates (5, 6, 8, 9, 15, 16), and significantly higher than the estimated global cancer prevalence of 1-2% in the general Spanish population aged 45-54. Consistent with prior studies, non-Hodgkin B-cell lymphoma was the most common malignancy, with a frequency of 4.4%, which is also higher than the global prevalence of non-Hodgkin lymphoma in the Spanish population for that same age range, estimated at 0.01-0.02%. This was followed by gastric cancer, lung tumors, and cutaneous malignancies.
Despite the evidence is very limited to date, some works suggest that immune dysregulation could be associated with the development of cancer both in pediatric (11) and adult CVID patients (12, 13). Several specific manifestations of this dysregulation have been identified as potential risk factors for malignancy. Namely, in a Czech study following 295 patients of which 22 developed cancer, history of Immune thrombocytopenic purpura (ITP) was established as a potential risk factor, with over 3 times higher risk of cancer development (14). Moreover, immune dysregulation manifesting as arthritis, atrophic gastritis, or interstitial lung disease (ILD) was associated with a cancer diagnosis in a German cohort of 219 patients, including 27 with cancer (17). However, the epidemiological profile of this population differs markedly from that of the Mediterranean, where arthritis is very uncommon (7).
Interestingly, our findings concur with some studies suggesting that elevated IgM is associated with an increased risk of malignancies, including lymphoma, in CVID patients. Several years ago, Resnick et al. (5) reported that higher IgM levels were associated with reduced survival in CVID, in the context of increased mortality rates associated with lymphoma. Indeed, in some subgroups of CVID patients, this IgM elevation has been suggested as a condition-specific marker of lymphoma (18). Other studies have reported this association also with lymphoproliferation and high risk of malignancies (19–21). The increase in this biomarker has also been linked to immune dysregulation, such as in interstitial lung disease and pulmonary B-cell hyperplasia (22).
The relationship between cancer and lower CD4 cell counts in CVID is not clearly established, as we have found in our cohort. However, some studies suggest that lower numbers and impaired function of CD4 T cells and natural killer cells are linked to a higher risk of malignancy in these patients (21, 23) by increasing inflammation, accelerating immunosenescence, and impairing immune surveillance (24). Importantly, an early decline in CD4 T cell counts may prolong the timeframe for neoplasia development, which could be particularly relevant for dCVID patients who present with lower CD4 T cell counts at diagnosis. Moreover, some reports of patients with CVID and several solid and hematologic neoplasia have highlighted very low CD4/CD8 ratios (25). These lower counts have also been more frequently found in a subgroup of CVID patients exhibiting increased autoimmunity, granulomas, splenomegaly, and expanded CD21low B cells (26, 27), which, in our cohort, was also independently associated to an increased risk of cancer. However, evidence could be more consistent regarding immune markers (17).
In this context, the association between immune dysregulation and higher IgM levels linked to persistent inflammation, a reduced CD4 T cell compartment, and the use of immunosuppressive therapies to manage these complications, synergistically contributes to the development of neoplasia. Although these treatments are necessary for controlling immune dysregulation, they have been shown to be independently associated to neoplasia in this cohort. Therefore, their use requires caution, coupled with a higher index of suspicion and more comprehensive screening measures.
Despite the association of iatrogenic immunosuppression to malignancy in other patient subgroups such as solid organ transplant recipients is well-studied, evidence is lacking in CVID patients and longitudinal data are strongly needed to guide management (28). Previous efforts have focused on infectious comorbidities in these both primarily and secondarily immunocompromised CVID individuals (7, 29). However, prospective analyses of neoplasia in these patients are especially important given the multifactorial interplay of genetics, immune dysregulation, and chronic infectious agents, including oncogenic microorganisms such as Epstein–Barr virus (EBV) (30) and H. pylori (7), which may be even more prevalent in this doubly immunocompromised population.
This nationwide, multicenter study systematically compiles extensive data on CVID patients, using advanced statistical methods to identify independent malignancy associates in a large CVID cohort under a robust temporal framework for data collection. However, it is not exempt from limitations. The retrospective cross-sectional nature of this work is an inherent bias as it limits the ability to establish causal relationships between predictors and malignancy development, and we can talk about potential associates that should be further explored in future prospective studies that can more rigorously test these hypotheses, both from clinical or basic and translational approaches. Furthermore, the study’s focus on Spain might introduce unique environmental and genetic factors affecting cancer prevalence and associated factors. The exclusion of patients with confirmed monogenic immunodeficiencies may reduce the generalizability of the findings to all CVID patients, especially considering some monogenic mutations could be linked to higher susceptibility to neoplasia, such as in the NFK-B signaling pathway implicated in many hallmarks for carcinogenesis (31). Additionally, the reliance on data from participating hospitals could introduce selection bias, as patients in specialized centers might have more severe disease manifestations. Moreover, the potential for missing data, despite the use of median-based or imputation, may affect the robustness of the statistical analyses. Finally, the retrospective nature of this study allows for the detection of associations but not causal relationships, necessitating prospective studies to establish causality.
5. Conclusion
This study identifies key potential predictors of malignancy in CVID patients, including older age, immune dysregulation, previous immunosuppressant therapies, elevated IgM levels, and lower CD4 cell counts at diagnosis. The findings underscore the heightened cancer risk in this population and highlight the necessity for vigilant monitoring and tailored screening protocols to improve patient outcomes, especially in those with increased inflammation and immune dysregulation. Further prospective research is needed to establish causality, enhance management strategies for CVID-associated malignancies, and deepen the molecular and genomic understanding of the underlying pathophysiology and common carcinogenic signaling pathways also implicated in CVID.
Acknowledgments
We would like to thank the Working Group of Rare Diseases of the Spanish Society of Internal Medicine (GTEM), the Spanish Society of Internal Medicine (SEMI), and the Valencian Community Society of Internal Medicine (SMICV) for their support. We also acknowledge the European Society for Immunodeficiencies (ESID) for their financial support to the first author. Additionally, we thank the Service of Data Science, Biostatistics, and Bioinformatics of the Health Research Institute La Fe for their assistance with the statistical analysis.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The article processing charge for publication has been funded by CSL Behring.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Health Research Institute La Fe, Valencia, Spain. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because Retrospective study with anonymous data.
Author contributions
MC: Conceptualization, Data curation, Methodology, Writing – original draft, Writing – review & editing, Funding acquisition, Investigation, Project administration. VG: Investigation, Methodology, Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Resources. HB: Conceptualization, Investigation, Writing – review & editing, Data curation. CB: Data curation, Writing – review & editing. LM: Data curation, Writing – review & editing. XS: Data curation, Writing – review & editing. JC: Data curation, Writing – review & editing. AR-M: Data curation, Writing – review & editing. FP: Data curation, Writing – review & editing. AB: Data curation, Writing – review & editing. NL: Data curation, Writing – review & editing. MT: Data curation, Writing – review & editing. AM: Data curation, Writing – review & editing. JN: Data curation, Writing – review & editing. NG: Data curation, Writing – review & editing. GC: Data curation, Writing – review & editing. RS: Data curation, Writing – review & editing. JB: Data curation, Writing – review & editing. AG: Data curation, Writing – review & editing. Jd: Data curation, Writing – review & editing. DL: Data curation, Writing – review & editing. AR: Data curation, Writing – review & editing. ACM: Data curation, Writing – review & editing. PM: Data curation, Writing – review & editing, Conceptualization, Investigation, Methodology, Project administration, Supervision, Writing – original draft.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Picard C, Bobby Gaspar H, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, et al. International union of immunological societies: 2017 primary immunodeficiency diseases committee report on inborn errors of immunity. J Clin Immunol. (2018) 38:96–128. doi: 10.1007/s10875-017-0464-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seidel MG, Kindle G, Gathmann B, Quinti I, Buckland M, van Montfrans J, et al. The european society for immunodeficiencies (ESID) registry working definitions for the clinical diagnosis of inborn errors of immunity. J Allergy Clin Immunol Pract. (2019) 7:1763–70. doi: 10.1016/j.jaip.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 3. Gathmann B, Mahlaoui N, CEREDIH. Gérard L, Oksenhendler E, Warnatz K, et al. Clinical picture and treatment of 2212 patients with common variable immunodeficiency. J Allergy Clin Immunol. (2014) 134:116–26. doi: 10.1016/j.jaci.2013.12.1077 [DOI] [PubMed] [Google Scholar]
- 4. Chapel H, Lucas M, Lee M, Bjorkander J, Webster D, Grimbacher B, et al. Common variable immunodeficiency disorders: division into distinct clinical phenotypes. Blood. (2008) 112:277–86. doi: 10.1182/blood-2007-11-124545 [DOI] [PubMed] [Google Scholar]
- 5. Resnick ES, Moshier EL, Godbold JH, Cunningham-Rundles C. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood. (2012) 119:1650–7. doi: 10.1182/blood-2011-09-377945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kiaee F, Azizi G, Rafiemanesh H, Zainaldain H, Sadaat Rizvi F, Alizadeh M, et al. Malignancy in common variable immunodeficiency: a systematic review and meta-analysis. Expert Rev Clin Immunol. (2019) 15:1105–13. doi: 10.1080/1744666X.2019.1658523 [DOI] [PubMed] [Google Scholar]
- 7. Cabañero-Navalon MD, Garcia-Bustos V, Nuñez-Beltran M, Císcar Fernández P, Mateu L, Solanich X, et al. Current clinical spectrum of common variable immunodeficiency in Spain: The multicentric nationwide GTEM-SEMI-CVID registry. Front Immunol. (2022) 13:1033666. doi: 10.3389/fimmu.2022.1033666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quinti I, Soresina A, Spadaro G, Martino S, Donnanno S, Agostini C, et al. Long-term follow-up and outcome of a large cohort of patients with common variable immunodeficiency. J Clin Immunol. (2007) 27:308–16. doi: 10.1007/s10875-007-9075-1 [DOI] [PubMed] [Google Scholar]
- 9. Ho HE, Cunningham-Rundles C. Non-infectious complications of common variable immunodeficiency: updated clinical spectrum, sequelae, and insights to pathogenesis. Front Immunol. (2020) 11:149. doi: 10.3389/fimmu.2020.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mellemkjaer L, Hammarstrom L, Andersen V, Yuen J, Heilmann C, Barington T, et al. Cancer risk among patients with IgA deficiency or common variable immunodeficiency and their relatives: a combined Danish and Swedish study. Clin Exp Immunol. (2002) 130:495–500. doi: 10.1046/j.1365-2249.2002.02004.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szczawińska-Popłonyk A, Ta Polska-Jóźwiak K, Schwartzmann E, Popłonyk N. Immune dysregulation in pediatric common variable immunodeficiency: implications for the diagnostic approach. Front Pediatr. (2022) 10:855200. doi: 10.3389/fped.2022.855200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wehr C, Houet L, Unger S, Kindle G, Goldacker S, Grimbacher B, et al. Altered spectrum of lymphoid neoplasms in a single-center cohort of common variable immunodeficiency with immune dysregulation. J Clin Immunol. (2021) 41:1250–65. doi: 10.1007/s10875-021-01016-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berbers RM, Drylewicz J, Ellerbroek PM, van Montfrans JM, Dalm VASH, van Hagen PM, et al. Targeted proteomics reveals inflammatory pathways that classify immune dysregulation in common variable immunodeficiency. J Clin Immunol. (2021) 41:362–73. doi: 10.1007/s10875-020-00908-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kralickova P, Milota T, Litzman J, Malkusova I, Jilek D, Petanova J, et al. CVID-Associated Tumors: Czech Nationwide Study Focused on Epidemiology, Immunology, and Genetic Background in a Cohort of Patients With CVID. Front Immunol. 2019. 9:3135. doi: 10.3389/fimmu.2018.03135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abolhassani H, Aghamohammadi A, Imanzadeh A, Mohammadinejad P, Sadeghi B, Rezaei N. Malignancy phenotype in common variable immunodeficiency. J Investig Allergol Clin Immunol. (2012) 22:133–4. [PubMed] [Google Scholar]
- 16. Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin Immunol. (1999) 92:34–48. doi: 10.1006/clim.1999.4725 [DOI] [PubMed] [Google Scholar]
- 17. Bruns L, Panagiota V, von Hardenberg S, Schmidt G, Adriawan IR, Sogka E, et al. Common variable immunodeficiency-associated cancers: the role of clinical phenotypes, immunological and genetic factors. Front Immunol. (2022) 13:742530. doi: 10.3389/fimmu.2022.742530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho HE, Cunningham-Rundles C. Seeking relevant biomarkers in common variable immunodeficiency. Front Immunol. (2022) 13:857050. doi: 10.3389/fimmu.2022.857050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gallo V, Cirillo E, Prencipe R, Lepore A, Del Vecchio L, Scalia G, et al. Clinical, immunological, and functional characterization of six patients with very high IgM levels. J Clin Med. (2020) 9:818. doi: 10.3390/jcm9030818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maglione PJ. Autoimmune and lymphoproliferative complications of common variable immunodeficiency. Curr Allergy Asthma Rep. (2016) 16:19. doi: 10.1007/s11882-016-0597-6 [DOI] [PubMed] [Google Scholar]
- 21. Gangemi S, Allegra A, Musolino C. Lymphoproliferative disease and cancer among patients with common variable immunodeficiency. Leuk Res. (2015) 39:389–96. doi: 10.1016/j.leukres.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 22. Maglione PJ, Gyimesi G, Cols M, Radigan L, Ko HM, Weinberger T, et al. BAFF-driven B cell hyperplasia underlies lung disease in common variable immunodeficiency. JCI Insight. (2019) 4:22728. doi: 10.1172/jci.insight.122728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shavit R, Maoz-Segal R, Frizinsky S, Haj-Yahia S, Offengenden I, Machnas-Mayan D, et al. Combined immunodeficiency (CVID and CD4 lymphopenia) is associated with a high risk of Malignancy among adults with primary immune deficiency. Clin Exp Immunol. (2021) 204:251–7. doi: 10.1111/cei.13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castilho JL, Bian A, Jenkins CA, Shepherd BE, Sigel K, Gill MJ, et al. CD4/CD8 ratio and cancer risk among adults with HIV. J Natl Cancer Inst. (2022) 114:854–62. doi: 10.1093/jnci/djac053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vorechovsky I, Litzman J, Lokaj J, Hausner P, Poch T. Common variable immunodeficiency and Malignancy: a report of two cases and possible explanation for the association. Cancer Immunol Immunother. (1990) 31:250–4. doi: 10.1007/BF01789177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salzer U, Warnatz K, Peter HH. Common variable immunodeficiency: an update. Arthritis Res Ther. (2012) 14:223. doi: 10.1186/ar4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Unger S, Seidl M, van Schouwenburg P, Rakhmanov M, Bulashevska A, Frede N, et al. The TH1 phenotype of follicular helper T cells indicates an IFN-γ-associated immune dysregulation in patients with CD21low common variable immunodeficiency. J Allergy Clin Immunol. (2018) 141:730–40. doi: 10.1016/j.jaci.2017.04.041 [DOI] [PubMed] [Google Scholar]
- 28. Wong GK, Huissoon AP. T-cell abnormalities in common variable immunodeficiency: the hidden defect. J Clin Pathol. (2016) 69:672–6. doi: 10.1136/jclinpath-2015-203351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gobert D, Bussel JB, Cunningham-Rundles C, Galicier L, Dechartres A, Berezne A, et al. Efficacy and safety of rituximab in common variable immunodeficiency-associated immune cytopenias: a retrospective multicentre study on 33 patients. Br J Haematol. (2011) 155:498–508. doi: 10.1111/j.1365-2141.2011.08880.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chua I, Quinti I, Grimbacher B. Lymphoma in common variable immunodeficiency: interplay between immune dysregulation, infection and genetics. Curr Opin Hematol. (2008) 15:368–74. doi: 10.1097/MOH.0b013e328302c7b6 [DOI] [PubMed] [Google Scholar]
- 31. Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr Opin Genet Dev. (2008) 18:19–26. doi: 10.1016/j.gde.2008.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.