Abstract
Diarrhoeal disease caused by Cryptosporidium is a major cause of morbidity and mortality in young and malnourished children from low- and middle-income countries, with no vaccine or effective treatment. Here we describe the discovery of EDI048, a Cryptosporidium PI(4)K inhibitor, designed to be active at the infection site in the gastrointestinal tract and undergo rapid metabolism in the liver. By using mutational analysis and crystal structure, we show that EDI048 binds to highly conserved amino acid residues in the ATP-binding site. EDI048 is orally efficacious in an immunocompromised mouse model despite negligible circulating concentrations, thus demonstrating that gastrointestinal exposure is necessary and sufficient for efficacy. In neonatal calves, a clinical model of cryptosporidiosis, EDI048 treatment resulted in rapid resolution of diarrhoea and significant reduction in faecal oocyst shedding. Safety and pharmacological studies demonstrated predictable metabolism and low systemic exposure of EDI048, providing a substantial safety margin required for a paediatric indication. EDI048 is a promising clinical candidate for the treatment of life-threatening paediatric cryptosporidiosis.
Subject terms: Drug discovery and development, Pharmacodynamics
EDI048 is a gastrointestinal-targeted Cryptosporidium PI(4)K inhibitor that undergoes a predictable metabolism and limits systemic exposure without compromising its anti-parasitic activity.
Main
Diarrhoea remains a leading driver of paediatric morbidity and mortality in low- and middle-income countries despite progress achieved through preventative interventions focused on clean water, hygiene and immunization against rotavirus1. Although Cryptosporidium has long been recognized as a major pathogen in people living with HIV/AIDS2, recent multicountry epidemiologic studies have demonstrated cryptosporidiosis to be one of the most important causes of moderate-to-severe diarrhoeal disease in young children up to the age of 24 months3–6. Cryptosporidiosis is caused by Cryptosporidium spp. and is an independent predictor of mortality7,8. While cryptosporidiosis is usually a self-limiting infection in otherwise healthy adults and older children, there are substantial unmet medical needs in younger children in sub-Saharan Africa and South Asia where an estimated 3–5 million Cryptosporidium-attributable cases result in 60,000–200,000 deaths annually8. Asymptomatic, prolonged or recurring Cryptosporidium infection in young children is also associated with long-term sequelae including linear growth faltering and cognitive deficits9,10. Other highly vulnerable populations with Cryptosporidium infection include malnourished children and immunocompromised patients with HIV/AIDS11,12. Even in high-income countries, disease outbreaks are common, with more than half of waterborne infections in the United States caused by Cryptosporidium13. Such outbreaks are likely to be aggravated by flooding and human migration due to climate change14. Currently, there is no vaccine against cryptosporidiosis and treatment options are limited15. Nitazoxanide, a static parasitic agent, is the only FDA-approved therapy for cryptosporidiosis, but it is not approved for children less than 1 year old, has limited efficacy in malnourished children and is not effective in immunocompromised patients16–18. Hence, there is an urgent need for safe and effective chemotherapeutics to treat paediatric cryptosporidiosis in low-resource settings19.
In humans, cryptosporidiosis is predominantly caused by two species, Cryptosporidium hominis and C. parvum. Cryptosporidium is an apicomplexan parasite transmitted through the faecal–oral route by highly infectious tough-walled oocysts that are resistant to chlorine disinfection and can survive for 2–6 months in damp environments. After ingestion, oocysts release sporozoites that infect intestinal epithelial cells, forming intracellular but extracytoplasmic parasitophorous vacuoles20. Within the vacuole, the parasite undergoes multiple rounds of its asexual life cycle followed by sexual reproduction, eventually leading to the formation of millions of infectious oocysts that cause re-infection, or are shed in the faeces. Prolonged oocyst shedding is common in a paediatric clinical population with cryptosporidiosis, and a longer duration of Cryptosporidium shedding has been associated with growth faltering in infants21,22. Cryptosporidiosis is predominantly a gastrointestinal (GI) infection. Although extra-GI tract Cryptosporidium infections have been reported23, their implication in paediatric diarrhoea leading to mortality and morbidity is yet to be established, and thus we focused on treating the enteric infection.
Previously, we have validated Cryptosporidium PI(4)K (phosphatidylinositol-4-OH kinase) as a promising molecular target to treat cryptosporidiosis24; however, the earlier candidate KDU731 could not be developed due to safety considerations. Cryptosporidiosis predominantly affects young vulnerable children who live in areas with limited healthcare infrastructure in low- and middle-income countries, hence ease of use, low cost of goods and an outstanding safety profile are critical for paediatric cryptosporidiosis drug development. Safety considerations for treating paediatric diseases are also critical due to the relative immaturity of organ systems and rapid postnatal growth and development in infants. Here we describe the discovery of an oral GI-targeted Cryptosporidium PI(4)K inhibitor, EDI048, a parasiticidal agent to treat paediatric enteric cryptosporidiosis. EDI048 is a soft drug engineered to undergo a predictable metabolism and limit systemic exposure without compromising its anti-parasitic activity in the GI tract. Extensive preclinical safety and pharmacological evaluation demonstrated that EDI048 has a large safety margin, and the candidate is currently in phase I clinical trials25. The discovery efforts described in this paper provide a comprehensive framework to pursue GI-targeted drug delivery strategies for cryptosporidiosis and other GI indications.
Results
Systemic exposure alone is not sufficient for efficacy
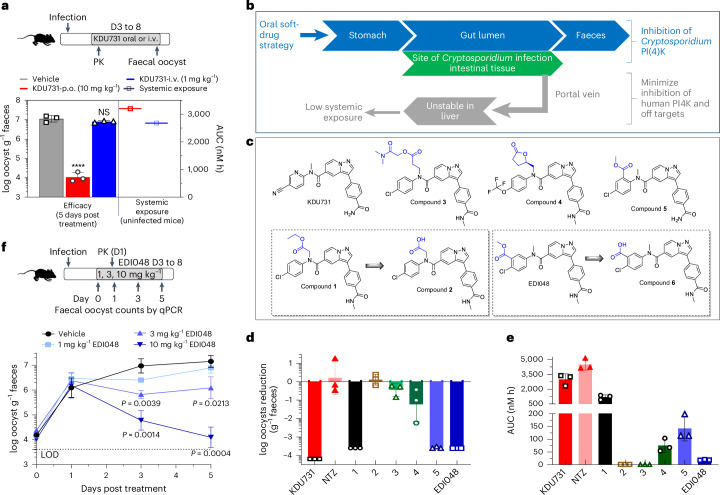
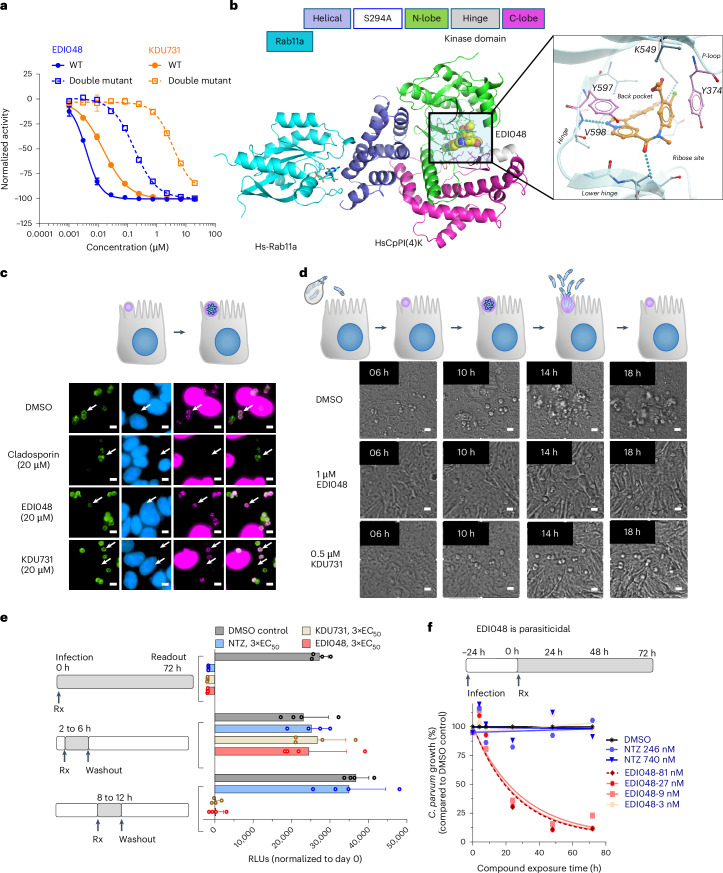
The primary site for Cryptosporidium infection is the enterocytes in the small intestine where the parasite resides outside of the cytoplasm in a parasitophorous vacuole facing the lumen directly underneath the enterocyte’s apical membrane. The relevance of systemic drug exposure for in vivo efficacy in cryptosporidiosis remains contentious and poorly understood as several compounds with high26,27 or limited27,28 systemic exposure show good efficacy in preclinical models. We previously identified the Cryptosporidium PI(4)K inhibitor KDU731, which is a poorly soluble, highly permeable and metabolically stable compound with promising anti-cryptosporidial activity24. Oral treatment with KDU731 at 10 mg kg−1 for 5 days in an immunocompromised (IFN-γ KO) mouse model resulted in an ~3 log reduction in faecal oocyst shedding with systemic exposure (total area under the curve (AUC)last = 3,200 nM h). To understand whether systemic exposure drives in vivo efficacy, we compared the efficacy of KDU731 oral treatment with intravenous administration at similar exposure. In contrast to the oral treatment, the intravenous injection had negligible effects on the parasite load (Fig. 1a). This result shows that systemic exposure alone is not sufficient for in vivo efficacy and suggests that direct GI exposure is necessary for anti-Cryptosporidium activity.
Fig. 1. Oral soft drug strategy to engineer GI stability and limit systemic exposure to treat cryptosporidiosis.
a, Schematic representation of C57BL/6 IFN-γ knockout mouse efficacy model. Infected animals were treated with vehicle (grey) or KDU731 orally (p.o.) at 10 mg kg−1 (red) or intravenously (i.v.) at 1 mg kg−1 (blue) with 3 mice per group. Parasite shedding per gram faeces was measured by qPCR; data are mean ± s.d. of 3 replicates. Statistical analysis was done using one-way analysis of variance (ANOVA). NSP = 0.5797; ****P < 0.0001. KDU731 systemic exposure in mice was measured in uninfected mice (mean data from n = 3 mice for oral and n = 1 for i.v. dosing are shown). b, Overview of an oral soft drug strategy to maximize intestinal exposure and minimize systemic exposure to limit off-target toxicity. c, Chemical structures of compounds described in this report to summarize structure–activity relationship for soft drug strategy by introducing metabolically labile spots shown in blue. Two soft drug candidates 1 and EDI048 along with their respective primary metabolites are shown in dashed boxes. d,e, In vivo efficacy (d) and systemic exposure (e) of the various analogues. Experimental design is as described in a. All animals were orally treated once daily with vehicle or 10 mg kg−1 for all compounds except nitazoxanide (NTZ) (100 mg kg−1) for 5 days. log10 oocyst reduction per gram faeces compared to vehicle control (d) and AUC (e) are mean ± s.d. of 3 replicates. AUC is the area under the curve from time 0 to the last timepoint measured from the mouse efficacy studies. The PK parameters are for active metabolite Tizoxanide following NTZ dosing at 100 mg kg−1. f, In vivo dose–response study with EDI048. On day 3 post infection, mice were treated once daily with vehicle (black) or indicated doses of EDI048 for 5 days, 3 mice per group. The PK was measured on day 1 post treatment and PK parameters are shown in Extended Data Table 2. Mean ± s.d. of oocysts shed per gram faeces (n = 3) are shown. Dashed line indicates the qPCR assay LOD. Statistical analyses were performed comparing untreated versus EDI048-treated groups (n = 3 each) on respective days using multiple unpaired parametric t-test (Holm–Šídák approach).
Rational design of intestinal targeting PI(4)K inhibitor
Given our results showing that systemic exposure is not sufficient for cryptosporidiosis therapeutic activity, we endeavoured to design PI(4)K inhibitors with limited or no systemic exposure. Limiting drug exposure to the site of therapeutic action is an attractive strategy to enhance safety margin29. One approach is the introduction of a structural metabolic liability (for example, a metabolic soft spot) into the compound leading to rapid metabolism and elimination after it had its intended therapeutic effect at the desired site; this is referred to as a soft-drug strategy. Ester functionalities are the most frequent moiety incorporated in the design of soft drugs due to their high catalytic efficiency, broad substrate specificity and rapid conversion to acids mediated by esterases, proteases and cytochrome P450s30. The carboxylic acid metabolite is potentially actively eliminated from the body via organic anion transporters, reducing systemic exposure. The desirable profile of a GI-targeted oral PI(4)K inhibitor to treat enteric cryptosporidiosis includes (1) potent Cryptosporidium PI(4)K inhibition, (2) high selectivity against the human orthologue (HsPI(4)K), (3) adequate solubility and permeability in enterocytes to reach the intracellular parasite, (4) metabolic stability in the GI lumen and (5) rapid clearance post absorption with efficient first-pass metabolism (Fig. 1b). To minimize the systemic exposure of Cryptosporidium PI(4)K inhibitors, we synthesized a few hundred soft-drug analogues of pyrazolopyridines by incorporating metabolically labile spots31. A few representative candidates are presented here to illustrate the optimization of an oral soft-drug candidate to treat enteric cryptosporidiosis (Fig. 1c).
The challenge in developing an oral Cryptosporidium PI(4)K selective GI-targeted drug is to strike a balance between reducing systemic exposure through increased metabolism while maintaining necessary solubility and permeability for effective anti-Cryptosporidium activity. Ethyl glycinate 1 and its primary carboxylic acid metabolite 2 were equally active on the CpPI(4)K enzyme (half maximal inhibitory concentration (IC50) = 2–7 nM). Acid 2 was inactive in the Cryptosporidium cellular assay (half maximal effective concentration (EC50) > 20 µM), probably due to low permeability, and was not efficacious in vivo (Table 1 and Fig. 1d). While 1 was highly efficacious in the mouse model and showed higher metabolism in vitro in hepatocytes, it was only moderately metabolized in vivo and still had systemic exposure similar to KDU731 (Fig. 1d,e and Table 1). Extending the ester into propionate 3 increased hepatic and plasma clearance but also decreased stability in the GI tract and in vitro assay media, which rendered the compound inactive in vitro and in vivo (Table 1 and Fig. 1d). Introduction of a cyclic ester lactone 4 also yielded increased in vitro clearance, which resulted in negligible systemic exposure (Fig. 1e) but unfortunately poor in vivo efficacy despite in vitro anti-Cryptosporidium activity ~2× lower than KDU731 (EC50 = 0.35 µM) (Fig. 1d), potentially suggesting that for a soft-drug candidate we may need higher in vitro potency. m-Chlorobenzyol ester 5 exhibited an excellent on-target activity and in vitro ADME profile but had poor selectivity over HsPI(4)K (Table 1 and Fig. 1d,e).
Table 1.
Anti-Cryptosporidium activity, in vitro and in vivo parameters of soft drug and other compounds
| Compound names | Anti-Cryptosporidium activity | Selectivity | In vitro PK | In vitro stability (M/H) | In vivo PKa | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cp CPE EC50 (µM) | Ch CPE EC50 (µM) | CpPI(4)K IC50 (µM) | HsPI(4)K IC50 (µM) | HepG2 CC50 (µM) | Solubility pH 6.8/FaSSIF (mM) | Permeability (×10−6 cm s−1) A–B/B–A/ratio | IS9 (T1/2, min) | Hepatocytes ER (%) (min) | Plasma (T1/2, min) | Cmax (nM) | AUCb (nM h) | |
| KDU731 | 0.156 ± 0.028 | 0.207 ± 0.002 | 0.017 ± 0.008 | 1.935 ± 1.082 | 59.982 ± 0.000 | 0.019/0.048 | 1.95/15.49/7.93 | >240/– | 18/60 (>175/57.9) | 85.2/>120 | 659 | 2,982 |
| NTZ | 18.837 ± 0.903 | >20 ± 0.000 | >10 ± 0.000 | >30 ± 0.000 | 34.900 ± 0.000 | – | – | – | – | – | 2,080c | 4,493c |
| 1 | 0.071 ± 0.016 | 0.066 ± 0.000 | 0.007 ± 0.002 | 1.141 ± 0.296 | 14.747 ± 0.000 | 0.031/0.086 | 7.89/7.4/0.94 | 120/120 | 74/– (13.4/–) | 13.6/>120 | 293 (1,656d) | 1,184 (3,775d) |
| 2 | >20 ± 0.000 | >20 ± 0.000 | 0.002 ± 0.000 | 0.135 ± 0.033 | >100 ± 0.000 | 0.708/1.0 | 0.89/1.65/1.85 | >240/– | 52/– (35.8/–) | >120/>120 | 50 | –e |
| 3 | >20 ± 0.000 | >20 ± 0.000 | 0.005 ± 0.000 | 1.034 ± 0.485 | 38.228 ± 0.000 | 0.011/0.355 | – | 9.2/45 | 93/– (<2.8/–) | – | <LLOQf | <LLOQf |
| 4 | 0.349 ± 0.181 | 0.503 ± 0.053 | 0.018 ± 0.011 | 1.380 ± 0.130 | 32.249 ± 0.000 | 0.037/– | – | 96/215 | –/95 (–/4.8) | <5/>120 | 31.7 | 75 |
| 5 | 0.039 ± 0.008 | 0.032 ± 0.001 | 0.005 ± 0.002 | 0.203 ± 0.047 | >100 ± 0.000 | 0.110/0.147 | 2.55/15.65/6.15 | 60/106 | 91/– (4/–) | >120/>120 | 32.7 | 142 |
| EDI048 | 0.052 ± 0.013 | 0.050 ± 0.001 | 0.004 ± 0.002 | 1.032 ± 0.470 | 28.083 ± 0.000 | 0.054/0.198 | 7.92/12.86/1.62 | 45/235 | 93/97 (2.8/<3) | 15.3/>120 | 8.4 (33.4g) | 19 (73.4g) |
| 6 | >20 ± 0.000 | ND | 0.031 ± 0.024 | 0.484 ± 0.119 | >100 ± 0.000 | >1/– | 0.56/1.04/1.86 | >240/– | 18/33 (>175/>175) | – | – | – |
| 7 | >20 ± 0.000 | ND | 0.076 ± 0.000 | 11.25 ± 0.00 | >100 ± 0.000 | – | – | – | – | – | – | – |
Data shown are mean ± s.e.m., n = 3 biological replicates for most of the in vitro data. M/H, mouse/human; ND, not determined; LLOQ, lower limit of quantification; Cp, C. parvum; Ch, C. hominis; FaSSIF, fasted state simulated intestinal fluid; A–B, apical to basolateral; ratio, B–A/A–B; ER, extraction ratio.
aIn vivo PK from the mouse efficacy studies with 10 mg kg−1 oral dosing except Nitazoxanide dosed at 100 mg kg−1.
bAUC is area under the curve from time 0 to the last timepoint measured.
cPK parameters are for active metabolite Tizoxanide following Nitazoxanide dosing at 100 mg kg−1.
dCompound 2 systemic concentrations following Compound 1 dosing.
eNot calculable (except at Cmax concentration, all other timepoints were below limit of quantification, 10.8 nM for Compound 2).
fAll concentrations were below limit of quantification, 8.9 nM for Compound 3.
gCompound 6 systemic concentration following EDI048 dosing.
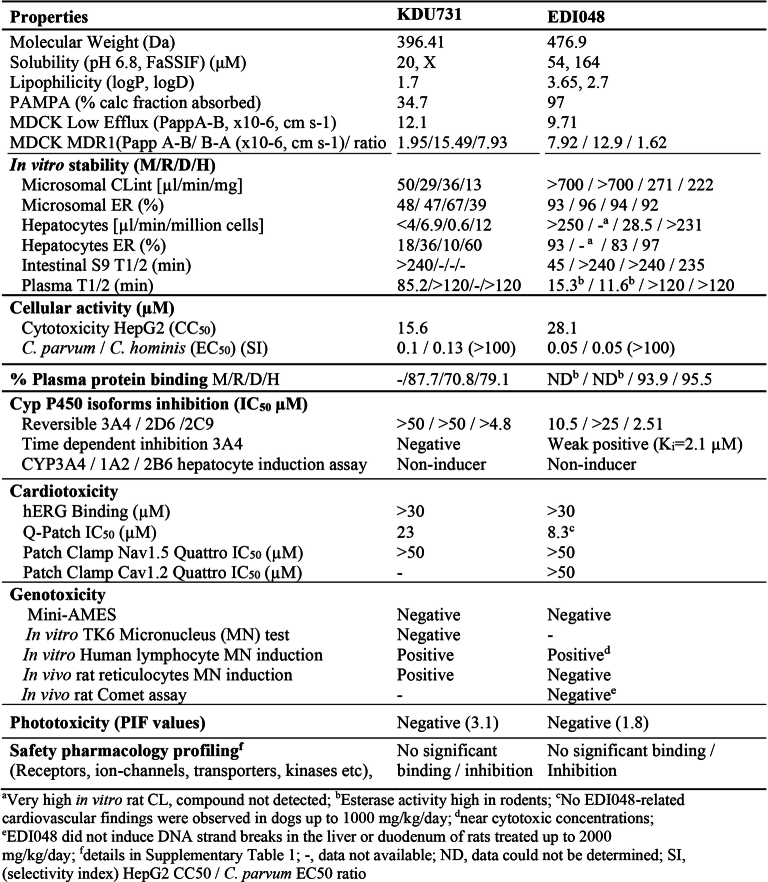
EDI048, with an o-chlorobenzoyl ester, demonstrated in vitro anti-Cryptosporidium activity, selectivity (>300-fold with HsPI(4)K), desirable human intestinal stability (T1/2 = 235 min) and high predicted extraction by the liver (human = 97%) (Table 1). As anticipated, the ester hydrolysis of EDI048 led to the formation of an inactive carboxylic acid metabolite compound 6 (C. parvum EC50 > 20 µM). Compound 6 has a low permeability and no notable off-target liability risks (Table 1, Extended Data Table 1 and Supplementary Table 1). Following oral administration, EDI048 was as efficacious as KDU731 (>3 log reduction in faecal oocyst shedding), despite systemic exposure near the detection limit (AUC = 20.4 nM h) in contrast to KDU731 (AUC = 3,200 nM h) (Extended Data Table 2). Nitazoxanide at 100 mg kg−1 was not efficacious in the immunocompromised mouse model, consistent with previous reports (Fig. 1d)18,24. EDI048 demonstrated dose-dependent efficacy with a 0.3, 1.1 and 3.1 log reduction in oocyst shedding at 1, 3 and 10 mg kg−1 doses, respectively. At the two lower doses, no detectable EDI048 systemic exposure was observed and at 10 mg kg−1, the observed maximum serum EDI048 concentration (Cmax) was 8.4 nM, notably below the in vitro potency (Fig. 1f and Extended Data Table 2). These data unambiguously confirm that GI exposure is necessary and sufficient for in vivo efficacy. Overall, EDI048 is a potent CpPI(4)K inhibitor (IC50 = 5.2 nM) with anti-parasitic activity against both C. parvum and C. hominis, is active on Cryptosporidium in the gastrointestinal epithelium in the mouse model and once absorbed is rapidly metabolized to an inactive carboxylic acid metabolite.
Extended Data Table 1.
Physicochemical properties and safety profiling data for EDI048 and KDU731
Extended Data Table 2.
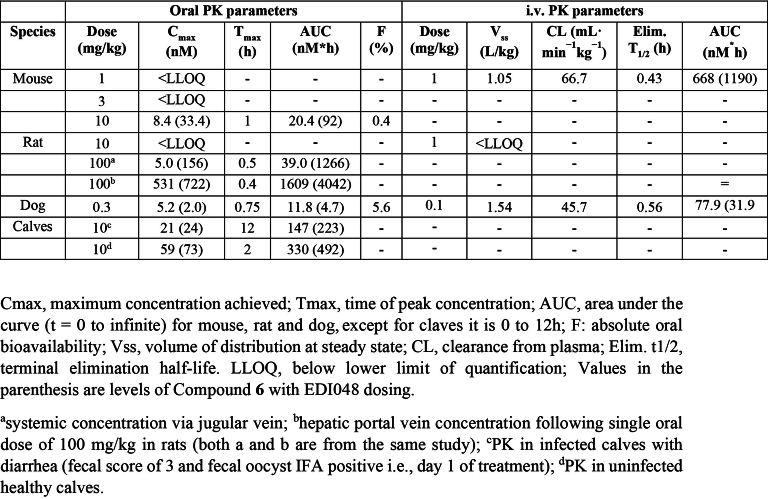
In vivo pharmacokinetic analysis of EDI048 in mouse, rats, dogs and neonatal calves
EDI048 undergoes rapid in vitro and in vivo metabolism
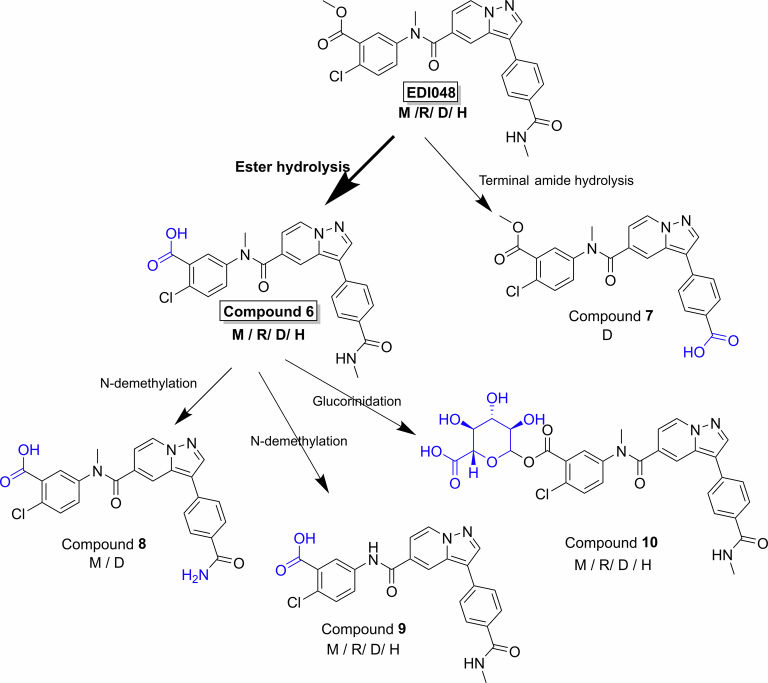
Understanding the metabolism and pharmacokinetics (PK) of EDI048 is critical, as both the parent drug and its metabolites play important roles in efficacy, safety and drug–drug interactions. EDI048 is a low-solubility, moderately permeable ester-containing soft drug which is designed to undergo rapid metabolism. The stability of EDI048 was assessed in selected preclinical species (mouse, rat, dog) and humans in vitro using microsomes, hepatocytes, intestinal S9 (IS9) fraction and plasma (Extended Data Table 1). EDI048 is stable in dog and human plasma (T1/2 > 240 min) but has a short half-life in rodent plasma (T1/2 = 12–15 min) potentially due to higher esterase activity in rodents compared with dogs and humans32. EDI048 displayed a desirable high extraction ratio in liver, both in microsomes and hepatocytes across all species. Metabolite identification studies in primary hepatocytes revealed that carboxylic acid metabolite 6 was the primary metabolite in all tested species (Extended Data Fig. 1). N-Demethylation (compounds 8 and 9) and glucuronidation (compound 10) were also observed in vitro, and no unique human-specific metabolites were observed in primary hepatocytes. In dogs, terminal amide hydrolysis of EDI048 led to the formation of inactive acid 7 (Table 1 and Extended Data Fig. 1).
Extended Data Fig. 1. Schematic representation of cross species metabolism for EDI048 in primary hepatocytes.
EDI048 was metabolized via ester hydrolysis to Compound 6 in all species, with amide hydrolysis to Compound 7 observed in dogs. Further, Compound 6 undergoes N-demethylation to Compound 8 or 9, and/or glucuronidation to Compound 10. No unique human specific metabolites were observed using hepatocytes in vitro.
Further in vivo pharmacokinetics studies confirmed the rapid metabolism of EDI048 across all tested preclinical species, with high systemic clearance and short terminal elimination half-life (Extended Data Table 2). In rodents, EDI048 was rapidly metabolized potentially due to high levels of esterase activity in tissues and blood, while in dogs the major site of metabolism was in the liver, resulting in measurable systemic exposure. The oral bioavailability of EDI048 was low in all species (0.4–5.6%) due to a high first-pass effect, and substantial levels of acid 6 were observed (Extended Data Table 2). To understand the factors contributing to low systemic exposure in vivo and to assess relative local enterocytes concentration, a pharmacokinetic study with EDI048 oral dosing was conducted in portal vein cannulated rats. EDI048 concentration in the portal vein (AUC = 1,609 nM h) was ~40-fold higher than systemic levels (AUC = 39 nM h), suggesting that EDI048 undergoes high first-pass metabolism in the liver, and supporting that EDI048 concentrations at the site of infection were higher than plasma exposure (Extended Data Table 2). These metabolism and pharmacokinetic studies confirmed that EDI048 is a soft drug in all tested preclinical species and had no human-specific metabolites, warranting further safety and efficacy profiling.
Interaction of EDI048 in the ATP-binding site of the Cryptosporidium PI(4)K
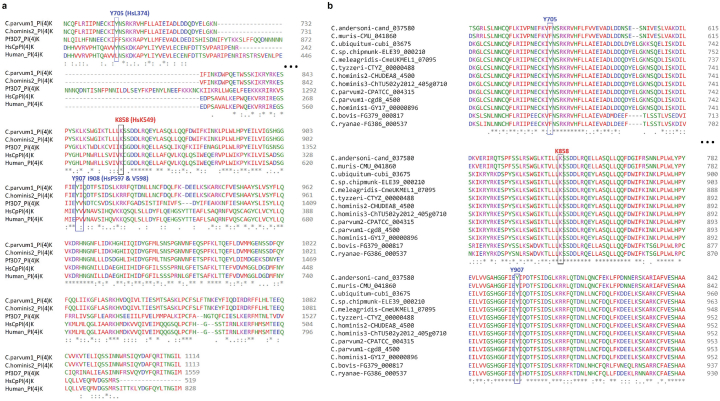
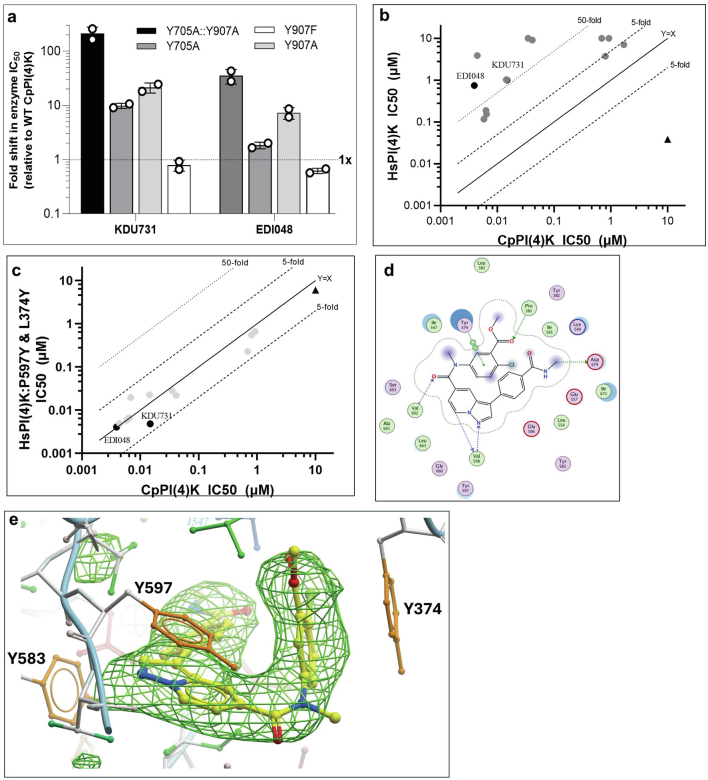
Phosphatidylinositol-4-kinase is a lipid kinase that plays a central role in cell signalling and membrane trafficking. This class of enzymes are highly conserved in eukaryotes, particularly in the ATP-binding pocket33. EDI048 is an ATP-competitive inhibitor of Cryptosporidium PI(4)K, similar to KDU731 (ref. 24). The essentiality of PI(4)K enzymes makes achieving high selectivity over the human lipid kinome a key design principle. EDI048 has a 300-fold selectivity window for Cryptosporidium PI(4)K over HsPI(4)K, and we sought to understand the molecular basis of selectivity (Table 1). Despite multiple attempts, we were unable to solve the Cryptosporidium PI(4)K crystal structure to gain insights into the ligand binding. To rationalize the selectivity and understand the key residues that contribute to the specificity of EDI048, we built a CpPI(4)K homology model on the basis of the HsPI(4)K structure34 and identified the non-conserved residues in the ligand-binding pocket of CpPI(4)K. Two key human residues, proline (P597) on the upper hinge and leucine (L374) in the P-loop, were deemed to provide the greatest differences in the ATP-binding pocket. Both are tyrosine residues (Y907 and Y705) in the Cryptosporidium enzyme (Extended Data Fig. 2a). To evaluate the importance of these residues for EDI048 binding, we expressed and purified a double mutant CpPI(4)K::Y705A:Y907A. EDI048 and KDU731 are 35–200× less potent against the CpPI(4)K::Y705A:Y907A mutant compared with wild-type CpPI(4)K (Fig. 2a and Extended Data Fig. 3a). Conversely, a double mutant of human PI(4)K, that is, HsPI(4)K (P597Y and L374Y) shifted the sensitivity of several PI(4)K inhibitors to more like CpPI(4)K (Extended Data Fig. 3b,c). Hence, human PI(4)K double mutant HsPI(4)K::P597Y:L374Y is referred to as HsCpPI(4)K chimaera. These results suggest that Y705 and Y907 have a profound effect on EDI048 binding and confer selectivity for the parasite enzyme.
Extended Data Fig. 2. Multiple sequence alignment of Cryptosporidium PI(4)K homologues highlighting key residues interacting with EDI048.
a, Multiple sequence alignment of CpPI(4)K, ChPI(4)K, PfPI(4)K and HsPI(4)K, highlighting key residues interacting with EDI048 along with catalytic lysine (K858). The critical residues identified to be interacting with EDI048 (Cryptosporidium Y705, Y907 and I908) are highlighted in blue box. b, Multiple sequence alignment of PI(4)K from various Cryptosporidium species. CpPI(4)K sequences were obtained from CryptoDB are labeled with their species name followed by a hyphen and the CryptoDB gene identifier. Highlighted are the two key tyrosine residues (Y705 and Y907) that interact with EDI048 and the conserved Lysine (K858) in C. parvum cgd8_4500.
Fig. 2. Structural insights into the ligand binding pocket of EDI048 and its mechanism of anti-Cryptosporidium activity in the enterocyte infection model.
a, Concentration–response curves for CpPI(4)K wild type (solid lines) and CpPI(4)K::Y705A:Y907A double mutant (dashed lines) with EDI048 (blue) or KDU731 (orange). Data are mean ± s.d. of at least 2 replicates. b, X-ray co-crystal structure of HsCpPI(4)K-HsRab11 complex with EDI048, highlighting the hinge H-bond between V598-NH and the pyrazolopyridine core (C atoms, orange; N atoms, blue), Pi-stacking of Y597 (lavender) with the pyarzolopyridine core (C atoms, orange), Pi-stacking between Y374 (lavender) with the chlorophenyl moiety of EDI048 (C atoms, orange), and H-bonding between K549 and carbonyl of the phenyl-amide moiety. c, EDI048 does not affect DNA synthesis. Cryptosporidium-infected cells were treated with compounds at 3 h post infection and 10 µM of EdU was added at 9 h post infection and 2 h later washed, fixed, stained and imaged with incorporated EdU representing replicating parasites (magenta), nuclei (blue) and parasitophorous vacuoles (green). White arrows indicate representative Cryptosporidium parasites. Scale bar, 5 µm. d, EDI048 inhibits formation of functional merozoites and blocks egress. Cryptosporidium-infected cells were treated with compounds at 3 h post infection followed by time-lapse imaging (also see Extended Data Fig. 4c and Supplementary Videos 1, 2 and 3 for time-lapse videos). Scale bar, 5 µm. Images in c and d are representative of 2 independent experiments, each with 2 technical replicates. e, Effect and reversibility of CpPI(4)K inhibition on early asexual stages of Cryptosporidium growth. Left: cells infected with nanoluciferase-expressing Cryptosporidium were treated with compounds as indicated in blocks of 4 h post infection, followed by drug washout, and then allowed to continue growing until 72 h post infection. Right: RLU normalized to time 0 are plotted, and data are mean ± s.d. of 4 technical replicates and representative of 2 biological replicates. NTZ, nitazoxanide. f, Time-kill curve with EDI048 compared to nitazoxanide. Cells were infected with nanoluciferase-expressing parasites and at 24 h post infection treated with indicated concentrations of EDI048 or NTZ (also see Supplementary Fig. 1 for complete dose–response data). Data are mean ± s.d. of at least 2 replicates.
Extended Data Fig. 3. Highly conserved tyrosine residues in Cryptosporidium PI(4)K (Y705 and Y907) enable inhibition of Cryptosporidium over human PI(4)K.
a, Characterization of single and double mutations in CpPI(4)K at Y705A and Y907A. Fold shift in IC50 of KDU731 and EDI048 against the CpPI(4)K single and double mutants over wild-type enzyme are plotted, the data is the mean of two independent experiments. (b, c), Introducing two-point mutations P597Y & L374Y (corresponding to Y907 and Y705 in CpPI(4)K) in the HsPI(4)K that is, HsCpPI(4)K significantly shifted the IC50s of CpPI(4)K inhibitor analogs (solid circles) to be much more like that of CpPI(4)K. HsPI(4)K specific inhibitor shown in solid triangle. Data are mean of at least 2 replicates. Schematic diagrams of protein-EDI048 interaction plot using Molecular Operating Environment (MOE) (d) and the simulated annealing OMIT mFo-DFc electron density map for EDI048 (e) are shown.
To gain structural insights, X-ray crystal structures of the HsCpPI(4)K chimaera with HsRab11a has been solved with EDI048 at 3.0 Å resolution (Fig. 2b and Extended Data Table 3; PDB 8VOF). As shown in the ligand-interaction plot and omit density maps (Extended Data Fig. 3d,e), EDI048 makes four primary interactions in the ATP-binding pocket: (1) π-stacking between Y597 and the pyrazolopyridine core in the hinge region, (2) π-stacking between Y374 and the chlorophenyl moiety in the P-loop, (3) H-bonding to the hinge between the V598-NH and the pyrazolopyridine moiety and (4) H-bonding between the catalytic K549 and the carbonyl of the secondary amide. Both π-stacking interactions were conspicuously missing in HsPI(4)K, thus rationalizing the selectivity of EDI048. Sequence analysis of Cryptosporidium PI(4)K in all clinical isolates reported in CryptoDB revealed that the residues critical for ligand-binding were highly conserved, suggesting that EDI048 will probably be effective against a diverse range of clinical isolates (Extended Data Fig. 2b). Thus, biochemical, mutational and structural studies with this innovative chimaeric approach have provided the molecular basis for EDI048 specificity and selectivity.
Extended Data Table 3.
Data collection and refinement statistics for cocrystal structure in presence of HsCpPI(4)K/HsRab11a with EDI048 (PDB ID: 8VOF)
Cryptosporidium PI(4)K inhibition leads to parasiticidal activity
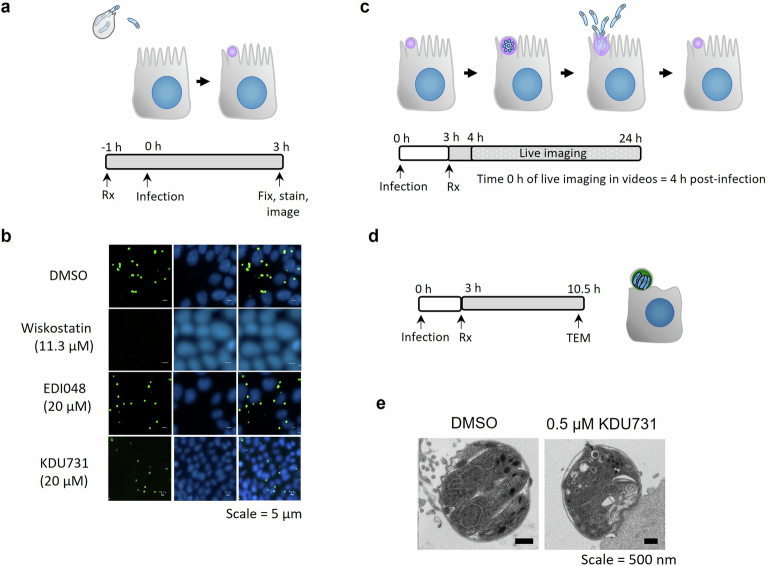
C. parvum oocysts release infectious sporozoites that invade enterocytes, forming trophozoites that grow for 6–8 h, followed by DNA replication and formation of functional merozoites. Merozoites egress from the infected cell by 10–14 h post infection, causing host cell death and re-infecting neighbouring host cells20,35. We used microscopy to probe the effect of PI(4)K inhibition on the growth and development of different asexual life stages of the parasite. EDI048 treatment does not affect sporozoite invasion, growth of trophozoites or new DNA synthesis (Fig. 2c and Extended Data Fig. 4a). Live imaging of C. parvum-infected HCT-8 cells demonstrated that upon EDI048 treatment, parasites were arrested at the meront stage, preventing egress (Fig. 2d, Extended Data Fig. 4c and Supplementary Videos 1–3), in contrast to DMSO-treated infected cells that burst open to egress functional merozoites ~10–12 h post infection. Transmission electron microscopy (TEM) studies also confirmed a lack of formation of distinct merozoite membranes with CpPI(4)K inhibition (Extended Data Fig. 4c). This is in line with the earlier observation in Plasmodium and Cryptosporidium36,37, suggesting that inhibition of parasite PI(4)K by EDI048 blocks membrane biogenesis, preventing the formation of functional merozoites.
Extended Data Fig. 4. EDI048 does not affect C. parvum sporozoites invasion, parasitophorous vacuole formation or growth but arrests C. parvum at meront stage.
a, Cartoon of life stages investigated in the invasion assay along with schematic of the assay. HCT-8 cells were pre-treated with 2x concentration of compounds for 1 hour (h). Primed C. parvum Iowa strain oocysts were then added and allowed to excyst and invade in presence of 1x compound concentration. At 3 h post-infection parasites that did not invade were washed off and then cells stained for parasitophorous vacuoles using FITC conjuated Vicia villosa lectin (green) and nuclei (blue). The active control wiskostatin (11.3 µM), an inhibitor of N-WASp and its activation by cdc42, prevented sporozoite invasion and formation of trophozoites, whereas 20 µM of EDI048 and KDU731 were inactive (b). c, Graphical representation of stages visualized by time-lapse microscopy and schematic representation of the experimental methodology. HCT-8 cells were infected by oocysts for 3 hours (h) and then washed and treated with 1 µM EDI048 or 0.5 µM KDU731. Time-lapse microscopy images were taken every 20 minutes from 4 h post-infection, that is, 1 h after compound addition (time 0 h in the videos) up to 24 h post-infection and are shown in Supplementary Videos 1 (DMSO), 2 (EDI048), and 3 (KDU731). d, Experimental overview to determine the effect of KDU731 on meronts by transmission electron microscopy (TEM). HCT-8 cells were infected with C. parvum oocysts induced for excystation and 0.5 µM of KDU731 was added at 3 h post-infection and parasite morphology analyzed by TEM at 10.5 h post-infection (e). Scale bars: 5 µm (b and Supplementary Videos 1 to 3) and 500 nm (e). Images and videos are representative of at least 2 independent experiments.
Sensitivity and reversibility of inhibition by EDI048 at different stages of the Cryptosporidium life cycle was further investigated using washout experiments. EDI048 does not affect parasite growth with a short 4 h exposure (2–6 h post infection) in the initial trophozoite development stage, suggesting that Cryptosporidium PI(4)K is not critical at this stage or its effect is recoverable. However, a short exposure to EDI048 during the merozoite maturation stage (8–12 h post infection) prevented parasite recovery, indicating that its effect on the parasite is irreversible, leading to cidal activity (Fig. 2e). In contrast, nitazoxanide is not cidal regardless of the stage of drug exposure. The parasiticidal activity of EDI048 is a highly desired property for cryptosporidiosis treatment in malnourished and immunocompromised patients. Kill-kinetics studies using nanoluciferase-expressing C. parvum show that, upon exposure to EDI048 at 24 h post infection, the parasites are rapidly eliminated. In contrast, nitazoxanide is static (Fig. 2f and Supplementary Fig. 1) and does not clear infection38. In vitro parasite elimination by EDI048 was exponential with a half-life of ~15 h, and the maximum rate of parasite elimination (>99% parasite reduction) was achieved at ≥27 nM (Fig. 2f and Supplementary Fig. 1). Consequently, 27 nM could be used as a target enterocyte concentration for achieving maximal parasiticidal activity.
Efficacy in a neonatal calf clinical model of cryptosporidiosis
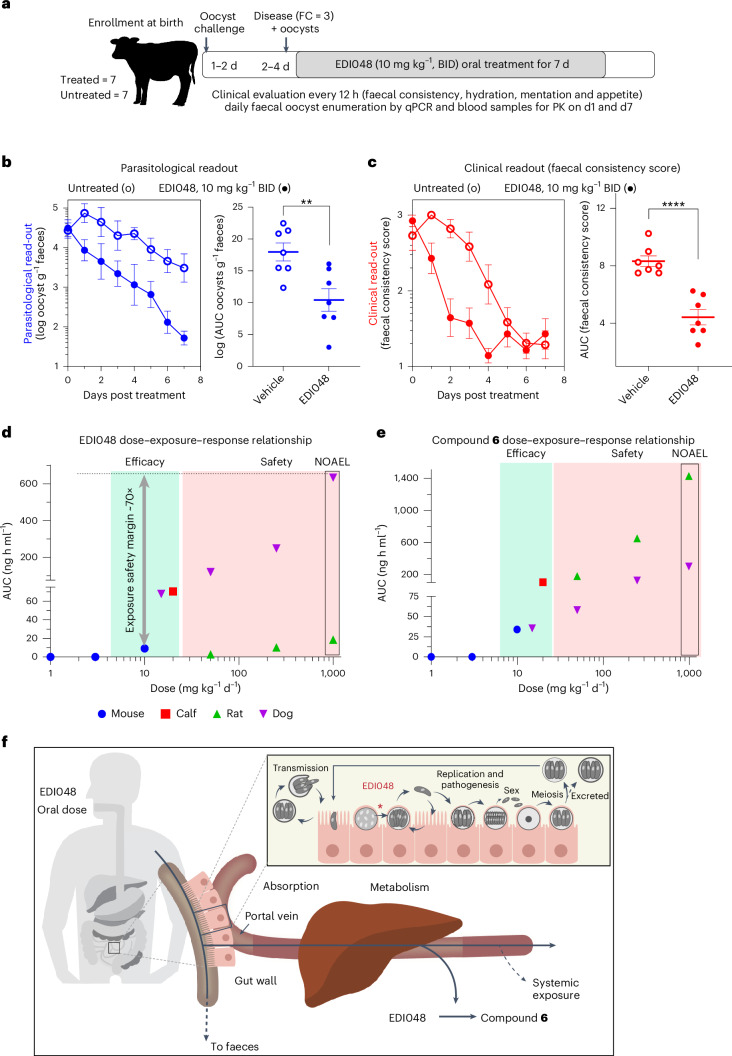
The pharmacokinetic–pharmacodynamic relationship of drugs acting in the GI tract can be complex due to the presence of watery diarrhoea and variability of intestinal transit time. The jejuno-caecal transit time is known to be reduced in cryptosporidiosis patients with diarrhoea relative to infected patients without watery diarrhoea39. Newborn calves are naturally susceptible to C. parvum infection and, unlike mice, neonatal calves develop profuse watery diarrhoeal symptoms similar to human patients. To understand the impact of Cryptosporidium infection and watery diarrhoea on the pharmacokinetics and therapeutic efficacy of EDI048, we evaluated EDI048 in a neonatal calf clinical model. Fourteen newborn calves were orally challenged with 5 × 107 oocysts and half of them received 10 mg kg−1 of EDI048 every 12 h for 7 days upon development of diarrhoea and confirmed oocyst shedding (Fig. 3a). Clinical symptoms were evaluated every 12 h (Supplementary Table 2), and quantitative faecal oocyst shedding was determined every 24 h using qPCR. The plasma pharmacokinetic profile of EDI048 and compound 6 were determined in uninfected calves and compared with infected calves with diarrhoea on day 1.
Fig. 3. Therapeutic efficacy in clinical neonatal calf model of cryptosporidiosis and in vivo safety of EDI048.
a, Schematic representation of a cryptosporidiosis calf model monitoring parasitological and clinical efficacy of EDI048. Calves were orally challenged with 5 × 107 C. parvum oocysts within 48 h of birth. Clinical parameters were assessed every 12 h and parasites shed in faeces were quantified daily by qPCR. Upon onset of diarrhea and detection of oocysts in faeces, calves were treated twice daily with 10 mg kg−1 (body weight) of EDI048 for 7 days. b,c, Compared with vehicle control (open circles), EDI048 (filled circles)-treated calves shed significantly fewer oocysts in faeces (b) and had improved clinical scores of diarrhea (faecal consistency scores) (c). Parasitological and clinical time-course data shown are mean ± s.e.m. of 7 calves tested per group. For AUC analysis, each circle represents an individual calf over 7 days and lines are mean ± s.e.m. of n = 7 calves per group. Statistical analyses were performed using two-tailed unpaired parametric t-tests; **P = 0.0062, ****P < 0.0001. d,e, Dose–exposure–response relationship for EDI048 (d) and compound 6 (e). Mouse exposure at 1, 3 and 10 mg kg−1 d−1 (blue, AUClast), calf exposure at 10 mg kg−1 twice a day (BID) (red, AUC0–12), rat exposures at 50, 250 and 1,000 mg kg−1 d−1 (green, AUC0–8) and dog exposures at 15, 50, 250 and 1,000 mg kg−1 d−1 (purple, AUC0–8) are shown. Overall, efficacy exposures in green and safety exposures in pink zone. 1,000 mg kg−1 d−1 is the NOAEL dose in both rat and dog toxicity study. Exposure multiples for EDI048 is 70× comparing between 1,000 mg kg−1 d−1 NOAEL exposure (day 14) and 10 mg kg−1 d−1 mouse efficacious exposure. f, Schematic representation of Cryptosporidium life cycle and site of action of EDI048 in the GI tract. Orally delivered EDI048 is absorbed by the intestinal cells at the site of infection. Herein, EDI048 demonstrates parasiticidal activity on the intracellular Cryptosporidium parasites by inhibiting membrane biogenesis. The absorbed EDI048 is metabolized in the liver, limiting systemic exposure and thus increasing safety margins.
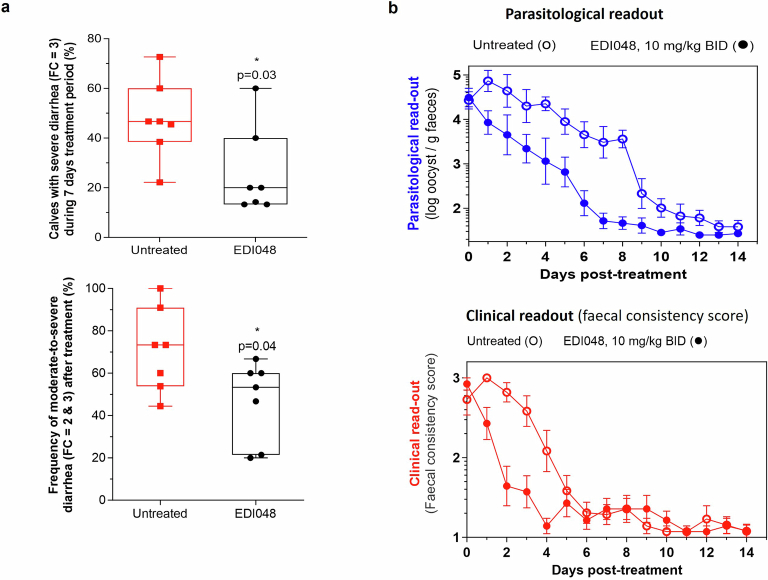
All calves tolerated the full 7 day course of EDI048 treatment without compound-related abnormalities. Treated calves shed significantly fewer oocysts (P = 0.0062) and had improved faecal consistency (P < 0.0001) when compared with vehicle-treated calves (Fig. 3b,c). Moreover, EDI048-treated calves started showing signs of resolution of diarrhea by 48 h post treatment (Fig. 3c). EDI048-treated calves suffered fewer days of severe diarrhea (P = 0.03) and moderate-to-severe diarrhea (P = 0.04) than vehicle-treated calves (Extended Data Fig. 5a). No recrudescence of infection was observed up to 7 days following cessation of treatment (Extended Data Fig. 5b). In vitro PK showed that EDI048 is stable in calf intestinal S9 (T1/2 = 175 min) and unstable in liver S9 fraction (T1/2 = 8.3 min). In vivo PK analysis revealed that EDI048 is substantially metabolized to compound 6 in calves, with levels of compound 6 being higher than that of EDI048 (Extended Data Table 2). As intended, despite limited systemic exposure, oral treatment with EDI048 resulted in potent reduction of faecal oocyst shedding and rapid resolution of clinical illness in young calves, suggesting that EDI048 may be suitable for clinical development.
Extended Data Fig. 5. EDI048 improves clinical diarrhea symptoms in experimentally challenged calves.
a, EDI048 reduces overall frequency of severe diarrhea and moderate-to-severe diarrhea. There were 7 calves per group, and each square (untreated) and circle (EDI048 treated) represents frequency of diarrhea for an individual calf over 7 days. p values were determined using unpaired, two-tailed t test. Data shown as a ‘box and whiskers’ plot; the box extends from the 25th to 75th percentiles, and whiskers with minimum to maximum showing all data points, and the center is median. b. Microbiological and clinical end-points measured 7 days post-cessation of drug treatment, details as described in Fig. 3b,c.
Safety profile of EDI048 enabled human clinical trial
Safety is paramount for any novel cryptosporidiosis clinical candidate considering that the patient population is mostly young, highly vulnerable children living in places with limited resources for providing adequate medical supervision. To assess the intrinsic cytotoxic activity of EDI048, we measured the half maximal cytotoxic concentration (CC50) in vitro with hepatic and intestinal cell lines in comparison with its anti-parasitic activity. EDI048 has >500-fold selectivity for cellular activity against C. parvum and C. hominis over cytotoxicity against a HepG2 cell line (Table 1). Further, EDI048 and its metabolites, compounds 6 and 7, were profiled in a broad range of pharmacological safety assays, including human recombinant receptors and relevant proteases and kinases (Supplementary Table 1). EDI048 displayed no notable liability for most targets, except for a modest activity (IC50 = 445 nM) against ATM (Ataxia-telangiectasia mutated kinase) with a >100-fold selectivity over CpPI(4)K inhibition. EDI048 had no relevant cardiotoxicity, mutagenicity, genotoxicity or phototoxicity (Extended Data Table 1). Children with cryptosporidiosis in endemic countries frequently suffer from comorbidities including HIV, tuberculosis and other infections; anti-cryptosporidial drugs are likely to be used along with other medications, and the risk of drug–drug interactions need to be minimized. EDI048 has minimal inhibitory effect against the major cytochrome P450 enzymes and did not induce human pregnane X receptor (Extended Data Table 1). In addition, limited systemic exposure of EDI048 offers opportunities to minimize in vivo off-target systemic effects and reduces the risks of drug–drug interactions40.
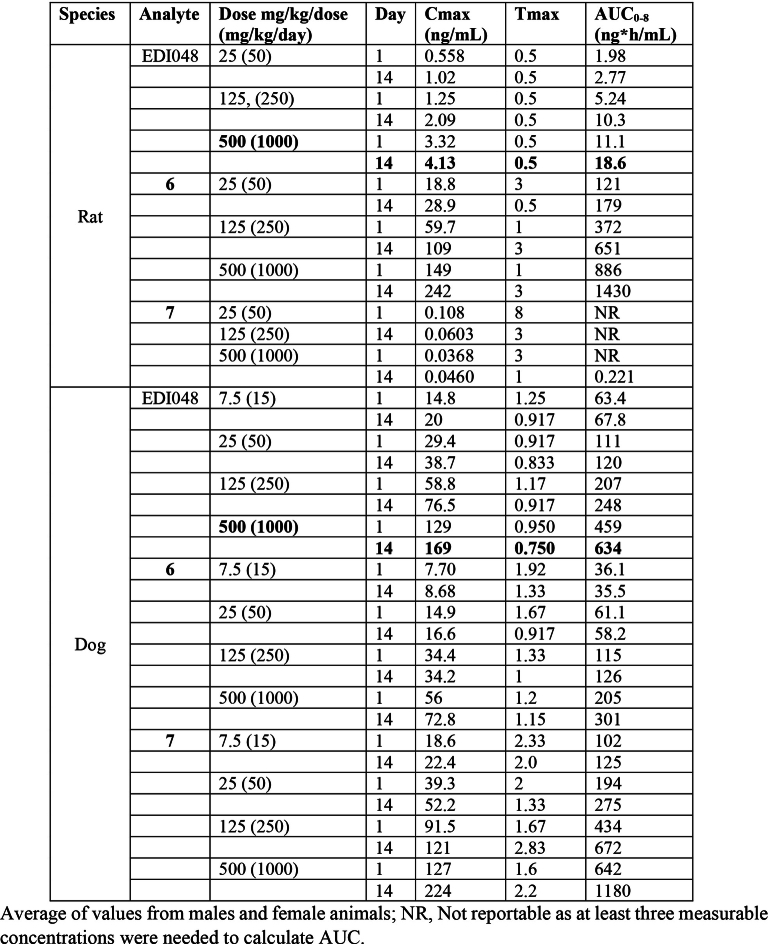
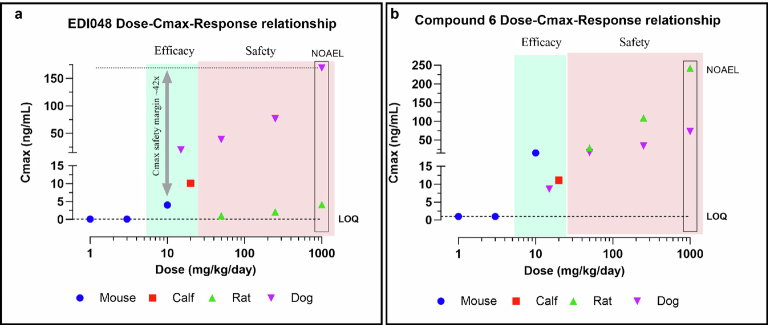
To assess EDI048 in vivo safety and enable clinical development, we performed 14 day repeat oral toxicity studies in rats and dogs in compliance with good laboratory practice. A high dose of 1,000 mg kg−1 d−1 EDI048 was selected for toxicity studies on the basis of favourable results of the pilot toxicity studies and regulatory guidance41. EDI048 was well tolerated in both species with no adverse toxicological findings identified, and the No Observed Adverse Effect Level (NOAEL) for both species was the highest dose studied (1,000 mg kg−1 d−1). Also, no EDI048-related cardiovascular findings were observed in dogs up to 1,000 mg kg−1 d−1. EDI048 was absorbed quickly (Tmax of 0.5 h in rats to ≤1.25 h in dogs). The peak plasma concentration (Cmax) and exposure (AUC0–8h) generally increased with the dose, albeit underproportionally, and no accumulation was observed upon repeated dosing. EDI048 was rapidly metabolized to acid 6 in rats, and acids 6 and 7 in dogs in line with in vitro observations (Fig. 3d,e and Extended Data Table 4). EDI048 metabolism was remarkably higher in rats, leading to barely detectable systemic levels (AUC0–8h = 18.6 ng h ml−1 at 1,000 mg kg−1 d−1 dose), and 75× higher level of compound 6 was observed (Fig. 3d,e and Extended Data Table 4). In dogs, at the NOAEL dose, the exposure (AUC0–8h) and Cmax values for EDI048 were 634 ng h ml−1 and 169 ng ml−1, respectively (Extended Data Table 4). Detection of EDI048 in dogs at NOAEL dose enabled the calculation of safety margins with respect to systemic exposure measured at the efficacious dose in mice. We have observed 70-fold (AUC0–8h) and 42-fold (Cmax) safety margins compared with the efficacious dose of 10 mg kg−1 in an immunocompromised mouse model (Fig. 3d and Extended Data Fig. 6a). Collectively, these data demonstrate that EDI048 has a safety and pharmacological profile suitable for a new drug candidate for the treatment of cryptosporidiosis, and EDI048 is currently being evaluated in a phase 1 clinical trial25.
Extended Data Table 4.
Toxicokinetic analysis of EDI048 and its metabolites from a 2 weeks GLP rat and dog toxicology studies
Extended Data Fig. 6. Dose Cmax response relationship for EDI048 and Compound 6.
Dose and Cmax relationship for EDI048 (a) and Compound 6 (b) in mouse Cmax at 1, 3 and 10 mg/kg/day (Blue). Calf Cmax at 10 mg/kg BID (Red), Rat Cmax at 50, 250 and 1000 mg/kg/day (Green) and dog Cmax at 15, 50, 250 and 1000 mg/kg/day (Purple). Overall, efficacy Cmax in green zone and safety Cmax in pink zone. 1000 mg/kg/day is the NOAEL dose in both rat and dog tox study. Cmax multiples for EDI048 is 42× comparing between 1000 mg/kg/day NOAEL Cmax (Day 14) and 10 mg/kg/day mouse efficacious Cmax.
Discussion
Cryptosporidiosis causes major diarrhoeal morbidity and mortality in vulnerable young and malnourished children in developing countries. EDI048 is a CpPI(4)K inhibitor with potent anti-parasitic activity against C. parvum and C. hominis, the two predominant species in patients in low- and middle-income countries. Biochemical activity with wild-type versus mutant CpPI(4)K enzyme and a high-resolution co-crystal structure of HsCpPI(4)K chimaera revealed that EDI048 makes Cryptosporidium-specific π-stacking interactions with two tyrosine residues in the ATP-binding pocket, providing desirable selectivity against HsPI(4)K. Amino acid residues critical for EDI048 binding are highly conserved across Cryptosporidium spp. of clinical relevance including C. hominis, C. parvum, C. meleagridis and so on42, suggesting that EDI048 is likely to be effective against diverse clinical isolates. In apicomplexan parasites, PI(4)K is involved in Rab11a‐mediated vesicular trafficking, which is essential for inner membrane complex formation43. The inhibition of PI(4)K blocks well-orchestrated cytokinesis leading to parasiticidal activity. Cytokinesis is not only critical during the asexual stages which are captured in the in vitro HCT-8 infection system, but also during sexual gametogenesis and sporogenesis. Hence, inhibition of PI(4)K is likely to be detrimental to multiple stages of the Cryptosporidium life cycle in vivo. Taken together, EDI048 will not only inhibit parasite replication, preventing further damage to the host intestinal villi leading to resolution of diarrhea, but will also significantly decrease faecal oocyst shedding and reduce disease transmission.
EDI048 is designed as an oral soft drug to be stable in the GI tract, exert its parasiticidal activity in the enterocytes and once absorbed, undergo predictable metabolism limiting systemic exposure (Fig. 3f). EDI048 is efficacious in an immunocompromised mouse model of cryptosporidiosis with barely detectable systemic exposure. In addition, in a neonatal calf cryptosporidiosis model, EDI048 reduced parasite faecal oocyst shedding and resulted in significant improvement of diarrhoeal symptoms. Furthermore, no recrudescence in infection was observed even after the cessation of EDI048 treatment (Extended Data Fig. 5b). Unlike the CpMetRS inhibitor44, resistance did not emerge during treatment with EDI048. Therefore, based on the limited data available, the perceived risk of resistance to EDI048 appears to be relatively lower. Nonetheless, this risk should be further assessed during EDI048 clinical development. These results unambiguously demonstrate that GI exposure is necessary and sufficient for in vivo efficacy for a direct anti-parasitic agent. One of the perceived limitations of the oral soft-drug approach is that EDI048 may not be efficacious against extra-GI infections such as respiratory cryptosporidiosis23,45, and may have limited efficacy if the parasite cycles between extra-GI and GI sites. The impact of Cryptosporidium respiratory infections on the global burden of paediatric diarrhea and its relevance for relapse or re-infection is yet to be established. Nevertheless, enteric cryptosporidiosis causes a life-threatening diarrhea in young children, and Cryptosporidium infection is strongly associated with long-term growth stunting and developmental defects19. The development of safe and effective oral therapeutics to treat enteric cryptosporidiosis in these vulnerable patient populations is of vital importance. Our results suggest that engineering a GI-targeted parasiticidal candidate, such as EDI048, may provide an opportunity to treat intracellular enteric Cryptosporidium infections with a favourable and large safety margin, potentially opening the opportunity for empirical treatment in outbreak settings16. Similar soft-drug approaches could be generalized to the treatment of other GI indications including additional enteric infections, CFTR protein-mediated secretory diarrhea, inflammatory-bowel disease, pouchitis, colon cancer and other diseases associated with transporters and receptors expressed in intestinal epithelia46. The major advantages of GI-targeted drugs include minimal systemic exposure, lowering the risk for systemic toxicity effects, thereby substantially increasing tolerability, and reducing potential drug–drug interaction liabilities40,47.
Taken together, our results show that EDI048 is a selective Cryptosporidium PI(4)K inhibitor with potent parasiticidal activity against Cryptosporidium species and a favourable in vivo preclinical safety profile. Currently, a first-in-human healthy volunteer clinical phase 1 study to evaluate the safety, tolerability and pharmacokinetics of EDI048 is in progress25. EDI048, if proven safe and effective in clinical trials, could be the first therapeutic designed to treat life-threatening paediatric cryptosporidiosis.
Methods
Compounds
KDU731 was synthesized as described earlier24. KDU731 was found to induce micronuclei in vitro in human peripheral blood lymphocytes and also in in vivo micronucleus assay in rat peripheral blood. The genotoxicity liability of KDU731 is mitigated by substituting cyanopyridine with chlorobenzene moiety while retaining potency and selectivity. All the soft-drug analogues described in this report were made with chlorobenzene/trifluoro-oxy-benzene substitution. Synthesis of EDI048, its metabolites and all other soft-drug candidates described in chemical synthesis along with analytical data is described below and also in a related patent31. Chemical structures were drawn using ChemDraw Professional v.22.0.0.22 (PerkinElmer).
KDU731 3–(4-carbamoylphenyl)-N-(5-cyanopyridin-2-yl)-N-methylpyrazolo [1,5-a]pyridine-5-carboxamide
KDU731 is a yellow-colour solid. 1H NMR (400 MHz, DMSO-d6): δ 8.87 (dd, J = 2.3, 0.8 Hz, 1H), 8.72 (dd, J = 7.2, 0.9 Hz, 1H), 8.53 (s, 1H), 8.26 (dd, J = 8.6, 2.3 Hz, 1H), 8.01 (s, 1H), 7.99–7.94 (m, 3H), 7.66–7.59 (m, 3H), 7.37 (s, 1H), 6.86 (dd, J = 7.2, 1.8 Hz, 1H), 3.54 (s, 3H); ESILC/MS: m/z 398 [M + H]+. J is the coupling constant and is a measure of the interaction between spin-coupled protons.
Compound 1 (ethyl N-(4-chlorophenyl)-N-(3-(4-(methylcarbamoyl)phenyl)pyrazolo[1,5-a]pyridine-5-carbonyl)glycinate)
Ethyl (4-chlorophenyl)glycinate (1.3 g, 5.9 mmol) was added to a solution of 3-bromopyrazolo[1,5-a]pyridine-5-carbonyl chloride (1.4 g, 5.4 mmol) in dichloromethane (DCM) (5 ml), followed by N,N-diisopropyl-N-ethylamine (1.9 ml, 10.8 mmol). The mixture was stirred overnight and concentrated under vacuum onto silica gel, followed by purification on additional silica gel with a 0% to 100% ethyl acetate (EtOAc) in heptane ramped to isolate ethyl N-(3-bromopyrazolo[1,5-a]pyridine-5-carbonyl)-N-(4-chlorophenyl)glycinate (1.0 g, 2.29 mmol, 42.4% yield) as a solid. This solid was dissolved in 1,4-dioxane (10 ml) and water (2.0 ml) in a microwave vial. N-Methyl-4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)benzamide (492 mg, 2.75 mmol), K2HPO4 (1.5 g, 6.9 mmol) and PdCl2(dppf)·CH2Cl2 (340 mg, 0.46 mmol) were added and the solution degassed with N2. The mixture was heated to 100 °C for 10 min, then filtered, concentrated and purified by supercritical fluid chromatography (SFC) to yield the title compound as a solid (27.6 mg, 2.4% yield). LC–MS (m/z): 491.3 [M + 1]+, retention time (RT) = 0.79 min. 1H NMR (500 MHz, DMSO-d6): δ 8.69 (d, J = 7.2 Hz, 1H), 8.47 (d, J = 13.6 Hz, 2H), 7.92 (d, J = 4.4 Hz, 2H), 7.74 (s, 1H), 7.50–7.42 (m, 4H), 7.37 (t, J = 4.9 Hz, 2H), 6.87–6.82 (m, 1H), 4.64 (s, 2H), 4.16 (q, J = 7.1 Hz, 2H), 2.82 (d, J = 4.5 Hz, 3H), 1.21 (t, J = 7.1 Hz, 3H).
Compound 2 (N-(4-chlorophenyl)-N-(3-(4-(methylcarbamoyl)phenyl)pyrazolo[1,5-a]pyridine-5-carbonyl)glycine)
Ethyl N-(4-chlorophenyl)-N-(3-(4-(methylcarbamoyl)phenyl)pyrazolo[1,5-a]pyridine-5-carbonyl)glycinate (250 mg, 0.509 mmol) and LiOH (12 mg, 0.509 mmol) were dissolved in 3 ml 1:1:1 methanol (MeOH):1,4-dioxane:water and stirred overnight. Product was precipitated with 6 N hydrochloric acid (HCl), and the solid was filtered and washed with water. The material was purified by SFC to yield the title compound as a solid (80 mg, 33.6%). LC–MS (m/z): 463.1 [M + 1]+, RT = 1.389 min. 1H NMR (400 MHz, DMSO-d6): δ 8.67 (dd, J = 7.2, 0.9 Hz, 1H), 8.46 (d, J = 6.6 Hz, 2H), 7.95–7.86 (m, 2H), 7.72 (d, J = 1.6 Hz, 1H), 7.49–7.40 (m, 4H), 7.39–7.30 (m, 2H), 6.83 (dd, J = 7.2, 1.8 Hz, 1H), 4.54 (s, 2H), 2.81 (d, J = 4.5 Hz, 3H).
Compound 3 (2-(dimethylamino)-2-oxoethyl 3-(N-(4-chlorophenyl)-3-(4-(methylcarbamoyl)phenyl)pyrazolo[1,5-a]pyridine-5-carboxamido)propanoate)
3-(N-(4-Chlorophenyl)-3-(4-(methylcarbamoyl)phenyl)pyrazolo[1,5-a]pyridine-5-carboxamido)propanoic acid (100 mg, 0.21 mmol) was added to a solution of 2-bromo-N,N-dimethylacetamide (30 mg) in dimethylformamide (2 ml). K2CO3 (87 mg) was added and the mixture stirred for 12 h. The mixture was cooled to 0 °C and poured into ice water. The aqueous layer was extracted with EtOAc and the combined organic layers dried over Na2SO4, filtered and concentrated. The material was purified by reverse phase HPLC (Zorbax C18, H2O/CAN, 18 ml min−1) to provide the title compound as a solid (25 mg, 21%). LC–MS (m/z): 561.75 [M + 1]+, RT = 1.42 min. 1H NMR (400 MHz, DMSO-d6): δ 8.65 (d, J = 7.2 Hz, 1H), 8.50–8.45 (m, 2H), 7.96–7.89 (m, 2H), 7.81 (s, 1H), 7.52 (d, J = 8.0 Hz, 2H), 7.45 (s, 4H), 6.88 (dd, J = 7.3, 1.8 Hz, 1H), 4.75 (s, 2H), 4.12 (t, J = 7.3 Hz, 2H), 2.90 (s, 3H), 2.83 (d, J = 4.4 Hz, 3H), 2.77 (s, 3H), 2.74 (t, J = 7.4 Hz, 2H).
Compound 4 ((R)-3-(4-(methylcarbamoyl)phenyl)-N-((5-oxotetrahydrofuran-2-yl)methyl)-N-(4-(trifluoromethoxy)phenyl)pyrazolo[1,5-a]pyridine-5-carboxamide)
To a suspension of (R)-3-bromo-N-((5-oxotetrahydrofuran-2-yl)methyl)-N-(4-(trifluoromethoxy)phenyl)pyrazolo[1,5-a]pyridine-5-carboxamide (65 mg, 0.13 mmol) in tetrahydrofuran (THF) (2 ml) was added (4-(methylcarbamoyl)phenyl)boronic acid (58 mg), triethylamine (Et3N) (55 µl) and H2O (1 ml), and the mixture was purged with N2. PdCI2(dtbpf) (8.5 mg) was added. The mixture was stirred at 100 °C for 2 h, then filtered, concentrated and subjected to SFC (2-ethylpyridine column, CO2/MeOH, 80 ml min−1, 2.18 min) to provide the title compound (27 mg, 37%) as a solid. LC–MS (m/z): 553.3 [M + 1]+, RT = 0.97 min. 1H NMR (500 MHz, DMSO-d6): δ 8.66 (d, J = 7.2 Hz, 1H), 8.46 (d, J = 5.9 Hz, 2H), 7.94–7.89 (m, 2H), 7.76 (s, 1H), 7.55–7.47 (m, 4H), 7.38 (d, J = 8.4 Hz, 2H), 6.86 (dd, J = 7.2, 1.8 Hz, 1H), 4.83–4.77 (m, 1H), 4.19 (dd, J = 14.6, 3.9 Hz, 1H), 4.11 (dd, J = 14.6, 8.0 Hz, 1H), 2.82 (d, J = 4.4 Hz, 1H), 2.56–2.49 (m, 2H), 2.34–2.24 (m, 1H), 1.94 (dq, J = 12.7, 9.1 Hz, 1H).
Compound 5 (methyl 2-(3-(4-carbamoylphenyl)-N-methylpyrazolo[1,5-a]pyridine-5-carboxamido)-5-chlorobenzoate)
Methyl 2-(3-bromo-N-methylpyrazolo(1,5-a)pyridine-5-carboxamido)-5-chlorobenzoate (0.79 g, 1.9 mmol), (4-carbamoylphenyl)boronic acid (0.46 g), PdCI2(dtbpf) (0.27 g) and K3PO4 (1.2 g) were taken up in dioxane (15 ml) and water (3.1 ml) in a microwave vial. The vial was purged with N2 for 10 min and then heated in a microwave at 100 °C for 10 min. The mixture was filtered, concentrated and purified by HPLC (amino column C3 20–25, CO2/MeOH, 80 ml min−1) to yield the title compound (650 mg, 1.4 mmol, 75% yield) as a yellow solid. LC–MS (m/z): 463.2 [M + 1]+, RT = 0.76 min. 1H NMR (400 MHz, chloroform-d): δ 8.69 (d, J = 1.8 Hz, 1H), 8.51 (d, J = 7.2 Hz, 1H), 8.27 (s, 1H), 8.12 (d, J = 8.2 Hz, 2H), 8.02 (s, 1H), 7.84 15 (dd, J = 8.6, 2.1 Hz, 1H), 7.56 (d, J = 8.2 Hz, 2H), 7.38 (s, 1H), 6.78 (d, J = 7.2 Hz, 1H), 3.96 (s, 3H), 3.64 (s, 3H).
EDI048 (methyl 2-chloro-5-(N-methyl-3-(4-(methylcarbamoyl)phenyl)pyrazolo[1,5-a]pyridine-5-carboxamido)benzoate)
To a suspension of methyl 5-(3-bromo-N-methylpyrazolo[1,5-a]pyridine-5-carboxamido)-2-chlorobenzoate (14.4 g, 34.1 mmol) in THF (360 ml) was added (4-(methylcarbamoyl)phenyl)boronic acid (8.6 g), Et3N (14.3 ml) and H2O (67 ml). The mixture was degassed and purged with N2 three times. PdCI2(dtbpf) (222 mg) was added. The mixture was stirred at 53 °C for 3 h, diluted with EtOAc and water, and then filtered. The layers were separated, the organic layer dried over Na2SO4, filtered and some of the volatiles removed. The resulting slurry was filtered to collect the solids. The solids were dissolved in ethanol and EtOAc, stirred with Pd-scavenging resin, filtered and fully concentrated. The solid was dissolved in hot EtOAc (150 ml), slowly cooled and collected to provide the title compound (10 g, 21 mmol, 62% yield) as a yellow solid. LC–MS (m/z): 477.0 [M + H]+, RT = 0.75 min (LC–MS source data provided). 1H NMR (400 MHz, chloroform-d): δ 8.33–8.25 (m, 1H), 8.06 (s, 1H), 7.80–7.75 (m, 2H), 7.71 (d, J = 2.80 Hz, 1H), 7.52 (d, J = 15 0.80 Hz, 1H), 7.32 (d, J = 8.80 Hz, 1H), 7.24–7.17 (m, 3H), 7.04 (dd, J = 2.80, 8.40 Hz, 1H), 6.83 (dd, J = 2.00, 7.20 Hz, 1H), 6.39 (br s, 1H), 3.92–3.82 (m, 3H), 3.51–3.40 (m, 3H), 3.00 (d, J = 5.20 Hz, 3H).
Compound 6 (2-chloro-5-(N-methyl-3-(4-(methylcarbamoyl)phenyl)pyrazolo[1,5-a]pyridine-5-carboxamido)benzoic acid)
t-Butyl 2-chloro-5-(N-methyl-3-(4-(methylcarbamoyl)phenyl)pyrazolo[1,5-a]pyridine-5-carboxamido)benzoate (130 mg, 0.25 mmol) was taken up in DCM (5.0 ml). The solution was cooled to 0 °C and trifluoroacetic acid (5.0 ml) was added. The mixture was stirred at r.t. for 6 h and the volatiles were removed under vacuum. The resulting solid was washed with diethyl ether and purified by HPLC (Kinetex EVO, 150 mm × 21.2 mm, 20 ml min−1; A = 0.1% trifluoroacetic acid in water, B = acetonitrile, 20–30% B over 2 min, 30–44% B over 7 min) to provide the title compound (25 mg, 0.055 mmol, 22%) as a solid. LC–MS (m/z): 463.15 [M + H]+, RT = 0.63 min. 1H NMR (400 MHz, DMSO-d6): δ 8.68 (d, J = 7.2 Hz, 1H), 8.52–8.40 (m, 2H), 7.95–7.82 (m, 4H), 7.57 (d, J = 8.0 Hz, 2H), 7.49 (d, J = 3.2 Hz, 2H), 6.87 (d, J = 7.2 Hz, 1H), 3.42 (s, 3H), 2.81 (d, J = 4.4 Hz, 3H).
Compound 7 (4-(5-((4-chloro-3-(methoxycarbonyl)phenyl)(methyl)carbamoyl)pyrazolo[1,5-a]pyridin-3-yl)benzoic acid)
(4-(tert-Butoxycarbonyl)phenyl)boronic acid (74 mg, 0.33 mmol) was dissolved in THF (3 ml), H2O (560 µl) and triethylamine (100 µl). Methyl 5-(3-bromo-N-methylpyrazolo[1,5-a]pyridine-5-carboxamido)-2-chlorobenzoate (100 mg, 0.24 mmol) was added and the solution purged with N2. PdCl2(dtbpf) (7.7 mg) was added. The mixture was stirred at 72 °C for 1 h, when the reaction was judged complete by LC–MS. The mixture was cooled to r.t. and diluted with EtOAc and water. The organic layer was washed with brine, dried with Na2SO4, concentrated and purified by flash chromatography (0–100% EA/Hep) to give methyl 5-(3-(4-(tert-butoxycarbonyl)phenyl)-N-methylpyrazolo[1,5-a]pyridine-5-carboxamido)-2-chlorobenzoate (112 mg, 0.215 mmol, 91% yield). This solid was diluted in DCM, and 4.0 M HCl in dioxane (538 µl) was added. Reaction was completed by LC–MS in 2 h. The volatiles were removed under vacuum to dryness and purified by preparative HPLC. The material was isolated and the resulting solid was collected and dried under air to yield the title compound (86 mg, 0.18 mmol, 84% yield). LC–MS (m/z): 464.3 [M + 1]+, RT = 0.82 min. 1H NMR (500 MHz, DMSO-d6): δ 12.93 (s, 1H), 8.72 (d, J = 7.1 Hz, 1H), 8.50 (s, 1H), 8.00 (d, J = 8.0 Hz, 2H), 7.96 (d, J = 2.6 Hz, 1H), 7.84 (s, 1H), 7.59–7.51 (m, 3H), 7.48 (d, J = 8.6 Hz, 1H), 6.95 (d, J = 7.2 Hz, 1H), 3.84 (s, 3H), 3.43 (s, 3H).
HsPI(4)K specific inhibitor (MI14)
The HsPI(4)K specific inhibitor (MI14) used in this study for enzymatic assays was synthesized as described earlier48.
Cell and parasites
Human ileocaecal colorectal adenocarcinoma cells (HCT-8) were purchased from American Type Culture Collection (ATCC) (CCL-34). Cells were cultured at 37 °C with 5% CO2 in a humidified tissue culture incubator using complete media consisting of RMPI-1640 medium (Gibco, A1049101) supplemented with either 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich, 12306C), 120 U ml−1 penicillin and 120 µg ml−1 streptomycin (Gibco, 15140-122). For cytopathic effect (CPE) assays, RPMI media were supplemented with 10% heat-inactivated horse serum (Thermo Fisher, 26050), 1× MEM nonessential amino acids (Thermo Fisher, 11140), 10 mM HEPES (Thermo Fisher, 15630), 100 U ml−1 penicillin and 100 U ml−1 streptomycin at 37 °C in a humidified incubator with 5% CO2. HCT-8 cells were only used for experiments between passage number P7 and P30. Cells were counted using a ChemoMetec NucleoCounter with NucleoView software v.1.2.0.0. C. parvum Iowa isolate oocysts were purchased from Bunch Grass Farm (Deary, Idaho) and the C. hominis TU502 isolate purchased from Dr Saul Tzipori (Tufts University Cummings School of Veterinary Medicine, North Grafton, Massachusetts). C. parvum Iowa nanoluciferase-expressing oocysts were a kind gift from D. Boris Striepen from the University of Pennsylvania49 and routinely passaged in interferon gamma knockout (IFNγ KO) or NOD SCID gamma (NSG) mouse models50.
In vitro activity against Cryptosporidium species
The inhibition of the asexual life cycle in the standard (48 h) assays was determined using the CPE assay51. Oocysts were artificially excysted using 10 mM HCl in 1X Hank’s balanced salt solution (HBSS) for 10 min at 37 °C using a shaker at 1,000 r.p.m., followed by exposure to pre-warmed, pre-gassed 2 mM sodium taurocholate in parasite infection medium (PIM, 1:1 formulation of Leibovitz’s L-15 medium and UltraCULTURE medium) for 5 min at 37 °C. HCT-8 cells were infected in a T-175 flask with oocysts triggered for excystation at a multiplicity of infection (MOI) of 3 for C. parvum and MOI of 4 for C. hominis. At 3 h post infection, the infected HCT-8 cells were dissociated via TrypLE (Gibco, 12604021) treatment for 30 min at 37 °C in 5% CO2. Cryptosporidium-infected cells were then pelleted, resuspended in HCT-8 culture media and seeded at a cell density of 2.75 × 104 cells per well in 30 µl per well. For seeding, we used a MultiDrop reagent dispenser (Thermo Fisher) and standard tube dispensing cassette at a high speed setting, onto a black polystyrene 384-well plate (Greiner, 781091). The 384-well plate was previously dry spotted with 60 nl per well of compounds (diluted in DMSO) in a ten-point dose response with 3-fold compound dilution or with controls (DMSO or KDU731 at a final concentration of 2 µM) using an Echo Acoustic liquid handler (BioTek Instruments). Assay plates were incubated at 37 °C in a humidified incubator with 5% CO2 for 48 h, and cells were then lysed with 20 µl r.t. CellTitre-Glo 2.0 (Promega, G9241) and incubated for 30 min at r.t. in the dark. After 30 min, luminescence was measured using the luminometer (CLARIOstar with software v.5.40 R2, BMG Labtech) at 0.1 s per well and 3,500 gain. Raw data files were exported and results were expressed as percent stimulation, where 100% stimulation was equal to the median of the active control wells and 0% stimulation was equal to the median of the DMSO-treated negative control wells. Cell viability curves were analysed using DAVID Helios software v.3.01.00.360 (ref. 52).
Cell-based mode of action, washout and time-kill assays
The invasion and DNA replication assay using fluorescence microscopy, live imaging and transmission electron microscopy were performed as described earlier35. Washout and time-kill assays were adapted from previous studies35,53 and performed using nanoluciferase-expressing parasites. For all assays, oocysts were primed for excystation by treatment with 10 mM HCl in water for 10 min at 37 °C, followed by exposure to 2 mM sodium taurocholate in Dulbecco’s phosphate-buffered saline with calcium and magnesium for 10 min at 16 °C. Primed oocysts were pelleted at 14,000 g for 4.5 min at 4 °C and resuspended in warm complete media for infection. Complete media consisted of RMPI-1640 medium (Gibco, A1049101) supplemented with either 10% heat-inactivated FBS (Sigma-Aldrich, 12306C), 120 U ml−1 penicillin and 120 µg ml−1 streptomycin (Gibco, 15140-122).
Host cell invasion was assayed by allowing C. parvum to invade host cell monolayers in the presence of compound and enumerating parasites and host cells after just 3 h (that is, before completion of a parasite division cycle). Wiskostatin, a known inhibitor of Neural Wiskott–Aldrich Syndrome protein, was used as an active control as host cell actin remodelling is required for Cryptosporidium invasion and parasitophorous vacuole formation35. HCT-8 cells were seeded at 25 µl per well in 384-well black polystyrene assay plates (Corning, 353962) such that the wells were 95–100% confluent at the time of infection. At 1 h before infection, DMSO control, wiskostatin and test compounds were added at twice the indicated concentrations incubated at 37 °C in a humidified 5% CO2 incubator for 1 h. At 1 h post compound addition, 50,000 primed oocysts at 25 µl per well were used to infect each well such that the final concentration of compounds was the same as the indicated concentrations. Assay plates were then incubated for 3 h in a humidified incubator at 37 °C under 5% CO2. For all the following steps, 1X phosphate-buffered saline (PBS) was used (prepared by diluting 10X PBS (Thermo Fisher, 70011-044) 10-fold with distilled water). At 3 h post infection, assay plates were washed 3 times with 1X PBS with 0.1% Tween 20, fixed with 4% paraformaldehyde (PFA) in 1X PBS for 15 min at r.t. and permeabilized with 0.25% Triton X-100 in 1X PBS for 10 min at 37 °C. Following permeabilization of the monolayer, HCT-8 cells were washed 3 times with 1X PBS with 0.1% Tween 20 and blocked with 4% bovine serum albumin (BSA) in 1X PBS for 2 h at 37 °C or 4 °C overnight. Parasitophorous vacuoles were stained with 1.33 µg ml−1 of fluorescein-labelled Vicia villosa lectin (VVL) (Vector laboratories, FL-1231) diluted in 1% BSA in 1X PBS for 1 h at 37 °C, followed by nuclei staining with Hoechst 33258 (AnaSpec, AS-83219) at a final concentration of 0.09 mM diluted in water for 15 min at 37 °C. Assay plates were then washed 5 times in 1X PBS with 0.1% Tween 20, and thereafter were ready for imaging. A Nikon Eclipse Ti2 epifluorescence microscope with a motorized stage and a Nikon DS-Qi2 camera with a wide field of view was used for imaging. NIS-Elements Advanced Research (AR) software v.5.02.01 (Nikon), incorporating the Nikon Perfect Focus System, was used to automatically focus and collect images from each well with a CFI Plan Apo Lambda ×20, NA 0.75 objective. For image analysis, NIS-Elements AR Analysis software v.5.20.02 and ImageJ v.1.53t were also used.
The DNA synthesis assay measures replication of the parasites inside intestinal cells. HCT-8 cells were seeded in 384-well polystyrene glass-bottomed plates (Cellvis, P384-1.5-N) pre-treated with 20 µl per well of freshly dissolved 40 µg ml−1 fibronectin in 1X PBS (prepared as described under the invasion assay method above) for 2 h at r.t. or at 4 °C overnight. Cells were seeded at 25 µl per well at a range of concentrations between 5,000 to 25,000 cells per well such that the cells were 90–100% confluent at the time of infection. Growth media were removed and cells were infected with 5 × 104 oocysts artificially triggered for excystation as described above. Assay plates were incubated for 3 h at 37 °C in 5% CO2 before the addition of different concentrations of EDI048 or controls. DMSO was used as a neutral control, and the lysyl-rRNA synthetase inhibitor cladosporin26 was used as an active control. Assay plates were incubated at 37 °C in 5% CO2 until ~9 h post infection. At ~9 h post infection, 10 µM 5-ethynyl-2’deoxyuridine (EdU) was added to all wells and incubated at 37 °C in 5% CO2 for 2 h. At 11 h post infection, assay plates were washed twice with 1X PBS, fixed with 4% PFA in 1X PBS for 15 min at r.t. and left in 2% PFA in 1X PBS fixative at 4 °C overnight. In the morning, fixative was removed and wells were washed twice with 3% BSA in 1X PBS. Cells were then permeabilized with 0.5% Triton X-100 for 20 min at r.t. and washed again twice with 3% BSA in 1X PBS. The Click-ITTM EdU Alexa Fluor 647 imaging kit was then used for EdU staining (Invitrogen, 10340). Briefly, this involved making a Click-iT reaction cocktail and incubating for 30 min at r.t., protected from light. Post-EdU staining, wells were again washed with 3% BSA in 1X PBS, and parasitophorous vacuole and nuclei were stained as described above. Wells were then washed 3 times with 1X PBS and imaged. Imaging was as described above for the invasion assay, except that a CFI S Plan Fluor ELWD ×40, NA 0.6 objective was used.
Live imaging of C. parvum was performed to assay the life stage effect of PI(4)K inhibitors. Fibronectin-coated glass-bottomed 96-well plates were infected with 150,000 primed oocysts per well. Assay plates were incubated for 3 h (h) at 37 °C under 5% CO2. At 3 h post infection, wells were washed 3 times with 2 ml of warm complete media and then incubated with 0.5 µM KDU731 (approximate C. parvum CPE assay EC90) or DMSO. Thereafter, live microscopy on the Nikon Eclipse Ti2 epifluorescence microscope was set up within the live humidified chamber at 37 °C under 5% CO2. Cells were imaged at 20 min intervals with a CFI S Plan Fluor ELWD 40XC, NA 0.6 objective. All images were strung together to make a video from 6 h to 21 h post infection.
TEM was performed to visualize the effect of compounds on meronts. HCT-8 cells were grown to >90% confluence in polystyrene 12-well cell culture plates (Corning, cCLS3513), and complete media were removed before infection. HCT-8 cells were infected with 100,000 primed oocysts per well. At 3 h post infection, wells were washed 6 times with 2 ml of warm complete media and then incubated with 0.5 µM (approximate C. parvum CPE assay EC90) KDU731 or DMSO in complete media for 10.5 h at 37 °C under 5% CO2. At 10.5 h post infection, Cryptosporidium-infected cells were de-adhered via 0.25% Trypsin–2.21 mM EDTA treatment for 5 min at 37 °C. Infected cells were pelleted at 150 g for 5 min and resuspended in 1 ml half-strength Karnovsky’s fixative: 1% PFA, 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2). Infected cells were fixed for 1 h at 4 °C and then pelleted as before. Fixed cells were thereafter washed 3 times in total with 0.1 M sodium cacodylate buffer wash reagent for 5 min at r.t. The washed and fixed samples were shipped to the University of Vermont Microscopy Imaging Centre for further treatment and generation of TEM images. Briefly, this involved crosslinking and dehydration of samples, followed by semi-thin sectioning (60–80 nm) and mounting onto a mesh copper grid for imaging with a JEM 1400 transmission electron microscope (JEOL).
C. parvum Iowa isolate oocysts expressing nanoluciferase (Nluc) under control of the enolase promoter were a kind gift from Dr Boris Striepen from the University of Pennsylvania. C. parvum was passaged in mice following published methods50. Briefly, NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice were purchased from The Jackson Laboratory and orally infected with 104 Nluc-expressing oocysts, and fresh faeces were collected within 2 h of excretion for several weeks post infection. Luminescence readings were taken for a fraction of the faeces to quantify the Nluc signal using the Nano-Glo Luciferase Assay System kit (Promega, N1130) and the CLARIOstar microplate reader with software v.5.40 R2 (BMG LABTECH). Oocysts were purified from faeces by homogenization, filtration and centrifugation, followed by sucrose and caesium chloride gradients. Purified oocysts were confirmed for expression of nanoluciferase by phase contrast microscopy and by quantifying luminescence reading, and were confirmed for viability by microscopic evaluation of excystation. Purified oocysts were stored at 4 °C and used within 2 months from the date of shedding.
Time-kill assays were used to assess whether EDI048 is parasiticidal or parasitistatic for C. parvum. HCT-8 cells were seeded in 384-well white-bottomed plates (Greiner, 781207) such that the wells were ~90% confluent at the time of infection. HCT-8 cells were then infected with oocysts primed for excystation immediately after resuspending oocysts in warm complete media. Oocysts (400 per well) were added and ~24 h after infection, compounds were added at indicated concentrations ranging from 20 µM to 1 nM. Time of compound addition (~24 h post infection) was considered time 0. At timepoints 0, 4, 8, 24, 48 and 72 h, luminescence (relative luminescence units, RLU) was measured using the Nano-Glo Luciferase Assay System kit (Promega, N1130) following manufacturer instructions and readings taken using the CLARIOstar microplate reader with software v.5.40 R2. For each timepoint, luminescence from each well was normalized to the average luminescence of the DMSO control wells to calculate percent (%) RLU relative to DMSO control. The RLU values as well as the DMSO control-normalized % RLU for each timepoint and each concentration of EDI048 and nitazoxanide were plotted using GraphPad Prism v.8.1.2.
Washout assays were performed to evaluate the effect of compounds on parasite growth stages. HCT-8 cells were seeded in 384-well white-bottomed plates (Greiner, 781207), and upon achieving ~90% confluence were infected with 400 per well of total Nluc-expressing oocysts primed for excystation immediately after resuspending oocysts in warm complete media. Compounds were added and washed at the indicated concentrations and timepoints. For each wash, media were removed and cells were washed at least two times with 100 µl per well of complete media. RLU was measured at indicated times using the Nano-Glo Luciferase Assay System kit (Promega, N1130) following manufacturer instructions and readings taken using the CLARIOstar microplate reader with software v.5.40 R2. The RLU values taken soon after infection were considered day 0 RLU and were used to normalize RLU values at each timepoint. Data were plotted using GraphPad Prism v.8.1.2.
Cytotoxicity assay
Cytotoxicity against HepG2 ATCC CRL-10741 was determined as previously described24. Briefly, cells were seeded into 384-well plates at 400 cells per well, incubated at 37 °C for 24 h and exposed to 3-fold serially diluted compounds for 96 h. Cell viability was monitored using Cell Counting Kit-8.
Expression and purification of CpPI(4)K and HsPI(4)K recombinant proteins for enzyme assays and structural studies
Baculovirus cloning, expression and purification of the full-length mutant C. parvum PI(4)K constructs were performed as previously described for wild-type enzyme24. Briefly, the full-length coding sequence of C. parvum PI(4)K (cgd8_4500) with the mutations described (Y705A, Y907A or both) was codon optimized for baculovirus expression, synthesized and cloned into pFastBac-HTb (Invitrogen) in frame with an amino (N)-terminal polyhistidine tag using the BamHI and HindIII restriction sites. Primers for mutagenesis were designed using NEBaseChanger (https://nebasechanger.neb.com/) from New England BioLabs and were ordered from Elim Biopharm. Mutagenesis was performed using the Q5 Mutagenesis kit (New England BioLabs, E0552S). Recombinant pFastBac-HTb-C. parvum PI(4)K bacmid clones were generated by site-specific transposition in Escherichia coli DH10Bac (Thermo Fisher). Bacmid isolation, transfection and selection of the recombinant viruses were performed according to manufacturer protocol (Bac-to-Bac System, Thermo Fisher, 10359-016). SF9 cells, cultured in ESF921 protein-free medium (Expression Systems, 96-001-01), were transfected with liposomes and recombinant baculovirusat 4:1 ratio and incubated at 27 °C for 4–7 days. For protein expression, Sf21 cells were infected with 3%, Passage2, virus at a cell density of 1.5 × 106 cells per ml. Cells were collected at 48 h post infection by centrifugation at 1,000 × g for 30 min at 4 °C and resuspended in cell lysis buffer (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1 mM dithiothreitol (DTT), 20 mM imidazole, 0.01% Triton X-100 and 1X complete protease inhibitor cocktail without ethylenediamine tetra-acetic acid (EDTA, Roche)). The cell suspension was lysed by sonication and the clarified supernatant was loaded onto a 1 ml HisTrap affinity column (GE Healthcare) pre-equilibrated with buffer A (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1 mM DTT, 20 mM imidazole and 1X complete protease inhibitor cocktail without EDTA). The column was washed with buffer A containing 45 mM imidazole and the bound protein was eluted with buffer A with 90 mM imidazole. Fractions containing mutant C. parvum PI(4)K were identified by size-exclusion chromatography, pooled, concentrated using Amicon Ultra-15, purified by a gel-filtration column (Hi-Load 26/60 Superdex 200, GE Healthcare) and equilibrated with 20 mM Tris pH 7.5, 300 mM NaCl, 1 mM DTT and 1X protease inhibitor cocktail without EDTA. Aliquots were flash frozen in liquid nitrogen and immediately stored at −80 °C.
Full-length wild-type Human PI(4)Kb isoform 2 was codon optimized for expression in Spodoptera frugiperda. The gene was split into two gene tiles with overlap and cloned into a linearized pFastBac1 vector at BamH1 and Xho1 sites.
Truncated HsPI(4)K proteins for X-ray crystallography and enzyme assays were all based on the construct described earlier54 in which HsPI(4)K isoform 2 (1–801) was truncated at the N (1–121) and C (785–801) termini, deleted internally at 249–287 and 408–507 and mutated at S294A. The sequence was codon optimized for expression in E. coli and cloned into pET24 at Nco1 and BamH1 sites with an N-terminal hexa-His tag followed by a TEV protease sequence. For the enzyme activity assays, a variant of the HsPI(4)K X-ray crystallography construct that retains C-terminal residues 785–801 was cloned into pET28a containing an N-terminal hexa-His, MBP, TEV sequence. It has been previously shown that the C termini of HsPI(4)K are necessary for the enzyme activity34,54. The HsCpPI(4)K chimaeric construct included the site-directed mutations, L374Y and P597Y. Both these tyrosine residues are highly conserved across all apicomplexan PI(4)K enzymes and are expected to have a profound effect on the binding pocket surface and allow Pi-stacking with aromatic scaffolds seen in the structures of other PIKK members55. For probing enzyme activity of the HsCpPI(4)K chimaera, the dynamic C-terminal region (C-tail, residues 785–801) of HsPI(4)K was retained as described above.
All four plasmids (HsPI(4)K and HsCpPI(4)K for both X-ray crystallography and enzyme assays) were transformed into the BL21 Star expression strain, grown in 2YT media (+antibiotic) in an incubator/shaker at 37 °C and 250 r.p.m., induced with isopropyl β-d-1-thiogalactopyranoside at 1 optical density (OD), then incubated overnight at 18 °C. Cells were collected by centrifugation (15 min at 5,000 × g) and cell pellets stored at −70 °C until purification. All purification buffers had 20 mM Tris pH 8.0, 150 mM NaCl, 5% glycerol and 1 mM Tris(2-carboxyethyl)phosphine in common, and all column fractions were analysed by gel electrophoresis and intact mass MS before pooling. Cells were lysed with a microfluidizer in buffer containing a commercial protease inhibitor cocktail and universal nucleases. HsPI(4)K proteins for X-ray crystallography were purified by a 3-step protocol: (1) HisTrap (5 ml) capture and 0–250 mM imidazole elution, (2) simultaneous overnight TEV cleavage and imidazole dialysis of the pooled PI(4)K fractions, then passage over a second HisTrap column to capture His-tagged TEV and impurities, followed by (3) final purification of XRC protein in a SEC column (Superdex 200). The pooled fractions were concentrated by centrifugation to ~10 mg ml−1 and flash frozen into liquid nitrogen for storage. Proteins for enzyme activity were purified by a variation of this 3-step protocol in that a no-cleavage step was performed due to the stabilizing effect of the MBP solubility tag. Assay proteins were purified by HisTrap capture and elution, followed by two rounds of SEC chromatography. E. coli expressed HsPI(4)K and HsCpPI(4)K enzymes showed comparable biochemical activity. Recombinant HsRab11 protein used for XRC studies was expressed and purified as described earlier34.
For PI(4)K enzyme assays, all solutions were prepared in deionized water. Triton X-100 (X100-500ML) and manganese(II) chloride tetrahydrate (M3634-100G) were purchased from Sigma-Aldrich. Tris-HCl solutions were prepared in-house as 1 M stocks with appropriate pH. Octylglucoside (OG) powder was used to prepare a 3% solution in 10 mM Tris pH 7.0. OG was used to solubilize 10 mg bovine liver l-α-phosphatidylinositol (PI; Avanti Polar Lipids, 84002P) to a final concentration of 30 mM PI. After addition of 3% OG into the glass vial, it was sealed with parafilm and allowed to sit on ice for 30 min. The solution was then vortexed until all PI was fully solubilized. ADP-Glo reagent, kinase detection reagent, Ultra Pure ATP and adenosine diphosphate (ADP) were purchased from Promega as an ADP-Glo kit (V9101). Luminescence measurements were carried out in white 384-well polypropylene plates (Greiner, 781207). Compound-mediated inhibition of PI(4)K variants was assessed in a total reaction volume of 10 μl. First, 5 µl of PI(4)K enzyme at twice the final concentration (6 nM enzyme for full-length enzymes or 10 nM for truncated constructs) in 10 mM Tris-HCl pH 7.5, 1 mM DTT, 0.05% Triton X-100 and 5 mM MnCl2 was dispensed into assay-ready plates dry spotted with compound (dissolved in DMSO) via Echo liquid handler (Labcyte). The enzyme was allowed to incubate with compound for 5 min at r.t. The reaction was started with the addition of substrate at twice the final concentration (20 μM PI, 6 μM ATP, 10 mM Tris-HCl pH 7.5, 1 mM DTT, 0.05% Triton X-100, 5 mM MnCl2). The final ATP concentration of 3 μM is below the ATP KM. No notable differences in KM were observed for mutant PI(4)K constructs relative to their corresponding wild-type enzymes. The reaction was allowed to proceed for 50 min at r.t. Subsequent steps to quench and detect were followed as described in the ADP-Glo kit manual. The reaction was quenched by the addition of 10 μl of ADP-Glo reagent and allowed to incubate at r.t. for at least 40 min. After this incubation, 20 μl of kinase detection reagent was added and allowed to incubate at r.t. for 30 min, after which the plate was read using a BMG Labtech CLARIOstar plate reader (with software v.5.40 R2) using a gain of 3,600, 0.2 s measurement interval and focal height of 10 mm. IC50 values were determined using in-plate duplicate 10-point 3-fold serial dilutions of each compound covering a concentration range of 20 μM to 1 nM. The neutral control (NC; DMSO) and the active control (AC; 10 μM KDU731) were present in columns 1, 12, 13 and 24 of each plate. The active control was also present in dose response on each plate. Data were converted to % activity, with 0% representing no inhibition (comparable to DMSO) and −100% representing full inhibition (comparable to the AC) based on the equation −100 × ((x − NC) / (AC − NC)), where x is the measured value. DAVID Helios software v.3.01.00.360 (ref. 52) was used to further analyse the data. The normalized values were plotted against compound concentration, and the IC50 was calculated using four-parameter nonlinear regression. The reported IC50 values were calculated as µM concentrations.
Crystallization and structure determination of HsCpPI(4)K/HsRab11a with EDI048
Crystallization
The protein used for X-ray crystallography was HsPI(4)KIIIβ with truncated dynamic regions34 and containing two mutations (P597Y and L374Y) in the ATP-binding pocket to mimic Cryptosporidium enzymes ligand-binding pocket. This truncated HsPI(4)KIIIβ::P597Y: L374Y enzyme is referred to as ‘HsCpPI(4)K’. HsCpPI(4)K/Rab11a complex was crystallized by vapour diffusion methods by incubating 10 mg ml−1 HsCpPI(4)K protein with 12 mg ml−1 HsRab11a in an ~1:1 molar ratio for 30 min at a final concentration of ~5–6 mg ml−1. Hanging crystallization drops were set up containing 1 µl complex and 1 µl well solution in 15-well crystallization plates. The well solution was composed of 500 mM ammonium sulfate, 88 mM sodium citrate, 5% glycerol and 875 mM lithium sulfate. The plates were then incubated for crystallization at 18–25 °C for 2–3 days. Crystals were collected for compound soaking in cryo-soak solutions. For soaking, equal volumes of 100% glycerol and 1 M HEPES pH 7.5 were mixed with sufficient mass of EDI048 to result in a final concentration of 100 mM. Protein crystallization mixture (20 µl) was dispensed and mixed with 5 µl of the compound solution to make a cryo-soaking solution. Cryo-soaking solution (10 µl) was dispensed into sitting drop wells in a 24-well plate, and HsCpPI(4)K/HsRab11a crystals were applied for soaking for ~24 h. Soaked crystals were collected and flash frozen in liquid nitrogen using a cryoprotectant composed of 25% ethylene glycol and 75% cryo-soak.
Data collection
A full data set (useable data to 2.9 Å) was collected at APS on the MCAT beamline. The data collection was performed at 100 K and wavelength = 1.0000 Å. The crystal used was of the space group P212121, with average unit cell lengths of a = 48.57 Å, b = 105.22 Å, c = 186.41 Å; α = β = γ = 90°. The raw data were integrated, indexed and scaled using the Autoproc suite from Global Phasing.
Refinement
Phaser (Phenix Consortium) as implemented in the CCP4 Suite of programs was used for the molecular replacement. An in-house structure of Hs-Cp-PI(4)K/HsRab11a was used as the search model. Refinement and manual rebuilding were performed in iterative cycles using Phenix Refine (Phenix Consortium) and Coot software v.0.9.8.92 (ccp4)56. Partial fitting of the protein was performed before refinement of the inhibitor. Idealized atomic coordinates for the inhibitor were generated with Molecular Operating Environment (MOE) v.2022.02 (Chemical Computing Group) and fit into an Fo−Fc difference electron density map contoured at the 3σ level using Coot. Structural parameters for EDI048 were generated by grade/mogul (Global Phasing, CCDC). The inhibitor was then refined along with the rest of the structure until the refinement converged. Solvent molecules were located and automatically assigned by Coot and Phenix.refine. Solvent that had good geometries (that is, trajectory and distances consistent with potential hydrogen bonds) were modelled and checked for consistent 2Fo−Fc difference density contoured at the 1σ level in subsequent cycles. Ramachandran statistics used were: Outliers: 0%; Allowed: 4%; Favoured: 96%. Relevant data and refinement statistics are presented in Extended Data Table 3. The PDB (Protein Data Bank) accession code for the HsCpPI(4)K/HsRab11a-EDI048 co-crystal structure is 8VOF. A ligand-interaction plot was generated using MOE v.2022.02 (Chemical Computing Group). A simulated annealing omit map (mFo−DFc) was calculated as additional confirmation of the ligand-binding mode (Phenix Consortium).
Solubility and permeability assays
Solubility was measured using a high-throughput equilibrium solubility assay employing a miniaturized shake-flask approach and streamlined HPLC analysis as described earlier57. In vitro permeability parallel artificial membrane assays were carried out using standard protocol58. Passive permeability along with P-glycoprotein-mediated efflux was measured in MDR-transfected MDCK cells as described earlier59.
In vitro liver microsomes and hepatocytes metabolic stability assessment
The metabolic stability in liver microsomes and hepatocytes was determined using the compound depletion approach and quantified by LC–MS/MS. The assay measures the rate and extent of metabolism as determined by the disappearance of the parent compound, which allows the determination of in vitro half-life (t1/2), intrinsic clearance (Clint), prediction of metabolic clearance (CL) and hepatic extraction ratio in various species60,61.
In vitro stability in intestinal S9 fraction, in vitro plasma protein binding and plasma stability
Stability assays in plasma and intestinal S9 fraction were performed as previously described62. Briefly, the metabolic reaction was initiated by addition of compounds to the matrices. Incubation was conducted in a shaking water bath at 37 °C and timepoint measurements were sequentially taken. Samples were quenched with ice-cold acetonitrile and analysed by LC–MS/MS. The t1/2 was calculated from the rate of compound depletion. In vitro plasma protein binding was determined using the rapid equilibrium dialysis method63.
Cytochrome P450 analysis
EDI048 was subjected to CYP450 inhibition analysis using 3 different isoforms following standard procedure64 and assessed for time-dependent inhibition using CYP3A4 (ref. 65). CYP induction assay was carried out using PXR reporter gene assay. A PXR binding-based hepatoma cell assay was employed to measure CYP3A4 transactivation. This assay uses a human hepatoma cell line DPX-2 overexpressing the human PXR and a CYP3A4 promoter-luciferase reporter gene66,67.
Cardiotoxicity and phototoxicity
Cardiotoxicity risk was measured as previously described67. Briefly, for hERG binding assay, concentration–response curves were typically generated as 6-point dose response from 0.37 to 30 µM using hERG-expressing HEK293 cells; for hNav1.5 and hCav1.2 patch clamp assays, as 7-point dose response from 0.1 to 50 µM in HEK293 cells expressing respective ion channels. The effect of the compound on hERG potassium channel current was evaluated (4-point dose response of 1–30 µM) at near-physiological temperature in stably transfected mammalian cells that express the hERG gene. Phototoxicity assay was performed following the OECD Guideline for Testing of Chemicals 432: ‘in vitro 3T3 NRU phototoxicity test’68. Briefly, in vitro photo safety evaluation was conducted in 3T3 fibroblast cells (seeded into 96-well microtitre plates treated with a range of concentration of compound) using neutral red uptake assay, tested up to the limit of solubility in HBSS in accordance with current regulatory guidelines.
Mini-ames, TK6 micronucleus assay and in vitro safety profiling assays
Mini-ames genotoxicity risk was measured as previously described69. Briefly, mutagenic potential of the compound was evaluated by its effects on one or more histidine-auxotrophic Salmonella typhimurium strains TA98 and TA100 in the absence or presence of rat liver metabolizing S9 system. The in vitro micronucleus test in TK6 cells was performed as previously described70. All assays for binding to proteins known to bear potential safety liabilities in humans were high-throughput competitive binding assays using specific radiolabelled ligands67.
In vitro human lymphocyte (HuLy) micronucleus assay
In vitro HuLy micronucleus assay was performed using pooled blood (two donors), in accordance with Labcorp standard operating procedures, the United Kingdom Good Laboratory Practice Monitoring Authority, Good Laboratory Practice Regulations 1999 and in compliance with the OECD guidelines71. The clastogenic and aneugenic potential of EDI048 was evaluated by analysing the frequency of micronuclei formation in cultured human peripheral blood lymphocytes treated in the absence and presence of a rat liver metabolizing system (S9). Briefly, 50–500 µg ml−1 of EDI048 was incubated for either 3 h or 24 h with 21 h or 24 h recovery period, respectively, with and without S9. Cells were centrifuged and resuspended in a minimal amount of fresh fixative, and slide analysis was carried out using fluorescence microscopy.
In vivo rat micronucleus and Comet assays
On days 3 and 15, blood samples for peripheral blood micronucleus assay were collected in randomized order from 2 weeks rat toxicology study animals and analysed by flow cytometer as described in ref. 72. Dosing details are described in ‘Rat toxicology study’. For in vivo Comet assay with EDI048, groups of 6 male Wistar rats were dosed at 200, 600 and 2,000 mg kg−1 d−1 for 2 days. Liver and duodenum tissue were sampled at ~Tmax following the second dose, and Comet assays were performed as described previously73.
In vitro identification of metabolites in hepatocytes
The metabolic disposition of EDI048 was assessed following in vitro incubation with mouse, rat, dog and human hepatocytes after 2 h of incubation. Briefly, 10 µM concentration of test compound was added directly to wells containing hepatocytes. Time zero samples were immediately quenched with an equal volume of acetonitrile and mixed well. Two-hour samples were incubated at 37 °C in a humidified CO2 incubator (5% CO2 and 90% relative humidity) for 2 h on an orbital shaker at 400 r.p.m. Following this, hepatocyte incubations were quenched with acetonitrile, centrifuged (~6,000 g for 10 min at ~20 °C) and the supernatant analysed using LC–UV/MS with accurate mass detection.
In vivo pharmacokinetic analysis
In vivo pharmacokinetic studies were conducted using non-randomized C57BL/6 male mice (n = 3, 8–10 weeks old), Wister male Sprague Dawley rats (n = 3, 8–10 weeks old) and male Beagle dogs (n = 3, 12–14 weeks old). Neonatal calf pharmacokinetic studies were performed as part of the efficacy study on day 1 of treatment. All procedures involving animals were reviewed and approved by the respective Institutional Animal Care And Use Committees (IACUC). Mice PK was performed according to the IACUC regulations of Charles River Laboratories (No. 2100230). Mice were housed in Innovive disposable micro-isolator cages with bedding. Rat PK was performed according to the IACUC regulations of Novartis Institute for BioMedical Research (No. 2100231). The jugular vein catheter/portal vein catheter rats were housed individually in Allentown reusable cages. Dog PK was performed according to the IACUC regulations of Charles River Laboratories (No. 2100232). All animals were group housed in pens with contact bedding. Calf PK was carried out in compliance with the Virginia Tech Institutional Animal Care and Use Committee (No. 00271) and housing conditions are elaborated in ‘Neonatal calf efficacy study’. No statistical methods were used to predetermine sample size. Sample size was determined on the basis of the minimum number of animals required for good data distribution and statistics. Although animals were selected randomly for each group, animal dosing was not blinded, as compounds need to be formulated in vehicle at different concentrations. However, for the sample collection and analysis, analysts were blinded or followed an unbiased approach using an automated platform. EDI048 was formulated in a suspension formulation for oral dosing (methylcellulose:Tween 80:water at 0.5:0.1:99.4 w/w for mouse, rats and dogs; methylcellulose:Tween 80:water at 0.5:0.5:99 w/w for calves) and solution formulation for intravenous dosing (NMP:4% BSA in PBS 10/90 v/v for mouse; NMP:PEG200 10/90 v/v for rats and dogs).
The blood samples for pharmacokinetic studies were collected between 0 and 24 h post dose. Blood samples were collected into 2 ml tubes containing 6 mg Na2EDTA and 3 mg sodium fluoride (BD Hemogard) and kept on ice until centrifugation. These samples were centrifuged within 30 min of collection for 10 min in a refrigerated centrifuge (set to maintain at 4 °C) at ~2,000 g. The resultant plasma was collected into fresh tubes placed on ice until transferred to a freezer, set to maintain −60 to −80 °C. The concentrations of compounds at various timepoints were determined by high-performance liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS). Plasma samples from pharmacokinetic studies were extracted with acetonitrile:methanol (50:50) containing CHIR073911 as internal standard, using a 4:1 extractant:plasma ratio. Analyte quantitation was performed by LC–MS/MS. Liquid chromatography was performed using a Waters ultra-performance liquid chromatography (UPLC) system with the Waters C18 SB column (2.1 × 50 mm) at an oven temperature of 50 °C, coupled with an API6500 triple quadruple mass spectrometer (Sciex Applied Biosystems). Instrument control and data acquisition were performed using Applied Biosystems software Analyst v.1.6.2. Pharmacokinetic parameters were determined by non-compartmental analysis using Phoenix WinNonLin v.8.3 (Certara).
Rat toxicology study
The rat toxicology study including analyses was conducted at designated test sites in compliance with the principles of Good Laboratory Practice Standards. All procedures involving animals were reviewed and approved by the IACUC of Covance Laboratories (No. 1970604). EDI048 was formulated in a suspension formulation (methylcellulose:Tween 80:water at 0.5:0.5:99 w/w) and orally administered twice daily to 10 male and 10 female Wistar rats at a daily oral dose of 50, 250 or 1,000 mg kg−1 body weight d−1 for 2 weeks. The number of animals in the toxicity protocols is considered to be the minimum necessary for statistical, regulatory and scientific reasons. For rat toxicology studies, 10 animals per sex per group was considered the minimum number that would account for the expected variability among these animals. Rats were obtained from Envigo and subjected to 5 days of quarantine and acclimatization before the study began. Rats were ~9–10 weeks at initiation of dosing and were pair or triple housed, by group and sex, in solid bottom cages with cellulose-based contact bedding. An enrichment device as well as a hut, tunnel or house were provided in each cage at all times. All animals were subjected to daily clinical observation, and body weight and food consumption were determined appropriately for all animals enrolled in the study. Clinical laboratory evaluations (haematology and clinical chemistry) were performed at the scheduled necropsy on day 15. Organs were examined for gross pathology and weighed before fixation and preparation for histology. Samples from organs and tissues prepared from animals assigned to control and high-dose groups were examined microscopically. Specifically, the adrenal glands, aorta (thoracic), bone marrow (femur, including joint, sternal), brain, caecum, colon, duodenum, epididymides, oesophagus, eyes, harderian glands, heart, ileum, jejunum, kidneys, lacrimal glands, liver, lymph nodes (mandibular, mesenteric), mammary gland, nerve (optic, sciatic), ovaries, pancreas, Peyer’s patches, pituitary, prostate, rectum, salivary glands (mandibular, parotid, sublingual), seminal vesicles, skeletal muscle, skin, spinal cord, spleen, stomach, testes, thymus, thyroid (with parathyroid), tongue, trachea, urinary bladder, uterus, cervix, vagina and gross lesions were examined for histopathological changes. Blood samples were collected on days 1 and 14 predose and at ~0.5, 1, 3 and 8 h post the first daily dose. These samples were processed to generate plasma, and concentrations were analysed for EDI048, and compounds 6 and 7 as described above.
Dog toxicology study
The dog toxicology study including analyses was conducted at designated test sites in compliance with the principles of Good Laboratory Practice Standards. All procedures involving animals were reviewed and approved by the IACUC of Covance Laboratories (No. 1970605). EDI048 was formulated in a suspension formulation (methylcellulose:Tween 80:water at 0.5:0.5:99) and orally administered twice daily to 3 male and 3 female beagle dogs at a daily oral dose of 15, 50, 150 or 1,000 mg kg−1 body weight d−1 for 2 weeks. The number of animals in the toxicity protocols is considered to be the minimum necessary for statistical, regulatory and scientific reasons. For dog toxicology studies, 3 animals per sex per group was considered the minimum number to account for the expected variability among these animals. Dogs were obtained from Marshall BioResources and subjected to at least 1 week of quarantine and acclimatization before the study began. Dogs were prepubertal/pubertal (~10–11 months old at dose initiation) and were housed in pairs or triplets, by group and sex, in elevated stainless-steel cages. All animals were subjected to daily clinical observation and body weight and food consumption were determined appropriately for all animals enrolled in the study. Clinical laboratory evaluations (haematology and clinical chemistry) were performed on day 7 and at the scheduled necropsy on day 15. Organs were examined for gross pathology and weighed before fixation and preparation for histology. All tissue samples described above in the rat toxicology section were collected and examined microscopically. Blood samples were collected from all animals on days 1 and 14 predose and at ~0.5, 1, 3 and 8 h post the first daily dose. These samples were processed to generate plasma, and concentrations were analysed for EDI048, and compounds 6 and 7 as described above.
Mouse efficacy study
All mouse studies described in this section were reviewed and approved by the IACUC of the Novartis Institute for Biomedical Research (animal use protocol no. 2017-055). Female C57BL/6 IFN-γ-knockout mice (B6.129S7-Ifngtm1Ts/J, Jackson Laboratories) aged 6–8 weeks were selected randomly for each group (n = 3) and infected with 10,000 C. parvum oocysts (Iowa isolate purified from experimentally challenged neonatal calves obtained from Bunch Grass Farms; oocysts used in these experiments were within 3–4 months from shedding). Sample size was determined on the basis of the minimum number of animals, technical and biological replicates required for good data distribution and statistics. For all these studies, n = 3 animals per dose group was used. EDI048 was formulated in 0.5% w/v methylcellulose and 0.5% w/v polysorbate in water and administered to mice for 5 days by oral gavage on day 3 post infection. Control mice were given only vehicle. Animal dosing was not blinded, as compounds need to be formulated in vehicle at different concentrations; however, for the sample collection and analysis, analysts were blinded or followed an unbiased approach using an automated platform. Faecal samples were collected daily from days 3 to 8 post infection. Measurements were performed on faecal material collected from cage-wide (pooled) collections. To measure oocyst load by qPCR, DNA was isolated from 50–100 mg of pooled faecal samples using the Quick-DNA faecal/soil microbe prep kit (Zymo Research). Samples were shaken for 20 min (25 °C, 1,450 r.p.m.) on an Eppendorf ThermoMixer F1.5 and subjected to 4–5 rounds of freeze–thaw cycles using liquid nitrogen. qPCR was performed using sample DNA extracted from faeces of infected mice along with DNA standards prepared by spiking varying numbers of oocysts into uninfected faeces. PCR primers JVAF and JAVR and probe 5’FAM labelled JVAR74 were used with the following cycling parameters: denaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 30 s. Each 10 μl PCR reaction contained 5 μl of SSoAdvanced universal probes supermix (Bio-Rad), 0.075 μM of each primer, 0.125 μM probe and 2 μl DNA. A standard calibration curve was prepared to estimate the number of oocysts per gram faeces. Oocyst dilutions (100 µl) ranging from 1 × 102 to 1 × 106 were added to each tube containing ~50 mg of faecal samples. DNA isolation and qPCR were performed as described above. PBS (100 µl) was used as a negative control. A calibration curve was freshly prepared and analysed with every set of study samples. Based on the observed cycle threshold (Ct) values corresponding to the lowest oocyst concentration, the limit of detection ranged between 80 and 280 oocysts per 50 g faeces (corresponding Ct values were 35 and above). To determine EDI048 and compound 6 concentrations, blood samples were collected on day 1 at various timepoints between 0 and 24 h post dose, processed and analysed as described above.
Neonatal calf efficacy study
Calf enrollment
All calves used in this study were cared for in compliance with the Virginia Tech IACUC. Sample size was calculated assuming that 85% of treated calves stop shedding oocysts by the end of the study observation period as compared with 15% of control calves. Assuming a type I error risk of 5% and a type II error risk of 80%, 7 calves were needed in each group of the Cryptosporidium-infected group. Fifteen Holstein–Friesian breed bull and heifer calves (Bos taurus taurus) were purchased from a local commercial dairy and enrolled into the study at birth. Study personnel attended the births to ensure that calves were delivered aseptically and that exposure to pathogens was limited. All calves enrolled were randomized to treatment with EDI048 (n = 7), positive infection control (n = 7) and negative infection control (n = 1) at birth. The perineum of the dam was thoroughly cleaned with povidone–iodine scrub and calves were delivered onto single-use plastic sheets to prevent exposure to environmental pathogens. Calves with abnormal physical examination findings and those weighing less than 30 kg at birth were excluded. Enrolled calves received 4 l ≥50 g IgG/L Land O’Lakes colostrum replacer (Purina Mills) and a 3 ml subcutaneous injection of vitamin E and selenium (BoSe, Merck). Calves were then transported from the commercial dairy farm to Cornell University (College of Veterinary Medicine, Ithaca, New York) in a dedicated trailer bedded with sterile straw.
Calf management and oocyst challenge
At Cornell University, calves were housed in individual box stalls in a Biosafety Level 2 facility. Shatter-proof mirrors were provided for enrichment. Within the first 48 h of birth, blood samples were collected and evaluated for adequate passive transfer of colostral immunity. Calves were offered a commercial 20% protein/20% fat non-medicated milk replacer (Land O’Lakes) every 12 h via nipple bucket. At each feeding, calves were fed an average of 7.6 g dry matter kg−1 birth weight for the duration of the study. Water was provided ad libitum. All calves randomized to the EDI048 treatment or positive control group were experimentally challenged within the first 48 h of life with 5 × 107 C. parvum oocysts (Iowa II strain, Bunch Grass Farm) through the rigid portion of an oroesophageal feeding tube. Oocysts were within 1 month of isolation, cleaned in 0.6% sodium hypochlorite for 1 min and then washed four times with PBS. The negative control calf was sham challenged to maintain blinding of study personnel, and study personnel were blinded during drug dosing, clinical scoring and qPCR analysis of faecal samples. At the end of the study, calves treated with the experimental compound were humanely euthanized using American Veterinary Medical Association approved methods to prevent accidental introduction into the food chain, following federal regulations. Calves that did not receive experimental compound were offered for adoption.
Administration of EDI048 and biological sampling
To facilitate collection of blood for plasma pharmacokinetic analysis, a long-term intravenous catheter (MILACATH, MILA) was aseptically placed in the jugular vein of each calf within the first 48–72 h of life. Calves were sedated with 0.1 ml intravenous xylazine (20 mg ml−1) (Akorn Animal Health). Sedation was reversed with 0.1 ml intramuscular atipamezole (5 mg ml−1) (Zoetis). After oral oocyst challenge, a faecal sample was collected directly from the rectum of each calf every 24 h. A complete physical examination was performed every 12 h, and clinical data including appetite, mentation, faecal consistency and hydration status were recorded. Clinical data were evaluated on a scale of 1 (normal) to 3 (severe) in accordance with previously described methods75,76.
EDI048 was prepared as 5 mg ml−1 suspension formulation in 0.5% w/v methylcellulose and 0.5% w/v Tween 80 in water, and EDI048 treatment was initiated when a calf began shedding oocysts in the stool and had a faecal consistency score of 3. Calves were induced to suckle and EDI048 was then given orally via an oral dosing syringe. Calves were treated every 12 h for 7 days at a dose of 10 mg kg−1 birth weight at least 2 h after feeding. Pharmacokinetic sampling was conducted on day 1 of treatment. Blood was drawn before and at multiple timepoints (between 0 and 12 h) after EDI048 administration. To determine the concentration of EDI048 and QPL621, blood samples were processed and analysed as described above. A faecal sample was collected at 1 h and 12 h post administration. On day 3 of life, a faecal sample was tested for Escherichia coli K99 and on day 7 for Salmonella, rotavirus and coronavirus.
Faecal oocyst enumeration
Oocysts counts were interpolated by qPCR at the Cornell Animal Health Diagnostic Center using serial dilutions of commercially purified C. parvum oocysts (Waterborne). Total nucleic acid was extracted from supernatants of 200 mg of faecal sample, oocyst suspension or negative control homogenized in 400 µl of PBS using a magnetic-bead-based automated procedure (AM1840, Applied Biosytems). An exogenous control (MS2 phage) was added to the lysis buffer to control for PCR inhibition77. qPCR for Cryptosporidium spp. 18S rRNA was performed on an Applied Biosystems 7500-FAST platform using commercial master mix (ToughMix, Quantabio) and oligonucleotides as previously described78. This count was standardized by the faecal dry weight percentage. A 5–10 g portion of each original faecal sample was dried at 108 °C for a minimum of 24 h (Squaroid Vaccuum Oven, Labline) and weighed75.
Data analysis
log normalized oocyst shedding per gram of faecal dry matter data over time was determined to have a normal distribution by Shapiro–Wilk, Anderson–Darline, D’Agostino and Pearson, and Kolmogorov–Smirnov tests, and were analysed using Student’s t-test (two-tailed, paired). LOD to detect parasites in the faecal samples by qPCR assay was determined to be 50 oocysts per gram faeces. Any samples that were undetected or below the lower limit of LOD that is, LOD/2 were represented as 25 oocysts per gram faeces. The clinical scores, that is, severe diarrhea (score = 3), moderate-to-severe dehydration (scores 2 or 3), mentation (scores 2 or 3) and appetite (scores of 2 or 3) were calculated for each calf, and statistical differences between vehicle control and EDI048-treated calves were analysed using the Student’s t-test (unpaired, two-tailed). Data were analysed using GraphPad Prism v.9.1.2 and v.9.5.1. In this study, infected calves showed diarrhoeal symptoms as measured by faecal consistency scores, the primary symptom for cryptosporidiosis. However, the neonatal calves did not develop severe impact on mentation, dehydration or appetite scores. Thus, the clinical effect of EDI048 compared to untreated calves was evaluated on faecal consistency scores only (Fig. 3c and Extended Data Fig. 5). The calf efficacy source data used for the analysis shown in Fig. 3 and Extended Data Fig. 5 are included as Microsoft Excel files (v.2405) in the Supplementary Information.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Fig. 1, and Tables 1 and 2.
Time-lapse microscopy images were taken every 20 min from 4 h post infection, that is, 1 h after compound addition (time 0 h in the videos) up to 24 h post infection and are shown in Supplementary Videos 1 (DMSO), 2 (EDI048) and 3 (KDU731).
Time-lapse microscopy images were taken every 20 min from 4 h post infection, that is, 1 h after compound addition (time 0 h in the videos) up to 24 h post infection and are shown in Supplementary Videos 1 (DMSO), 2 (EDI048) and 3 (KDU731).
Time-lapse microscopy images were taken every 20 min from 4 h post infection, that is, 1 h after compound addition (time 0 h in the videos) up to 24 h post infection and are shown in Supplementary Videos 1 (DMSO), 2 (EDI048) and 3 (KDU731).
HsCpPI(4)K X-ray crystal structure PDB validation report.
EDI048 LC–MS data.
Source data
Source data (efficacy and PK raw data).
Source data (concentration–response, reversibility and time-kill curve raw data).
Source data (parasitological readout, clinical (faecal consistency score) readout, dose–exposure response raw data).
Source data (fold shift in IC50 raw data).
Source data (parasitological readout and clinical (faecal consistency score) readout raw data).
Source data (dose–Cmax response raw data).
Source data (raw RLU data for parasitistatic/parasiticidal assay and one-phase exponential decay curve fit).
Acknowledgements
We thank B. Striepen for the generous gift of nanoluciferase-expressing C. parvum; D. V. Nydam for support in conducting the calf model; the staff at the University of Vermont’s microscopy core facility for help with electron microscopy; J. Burke for insights on human crystallography constructs; other Novartis colleagues for support and scientific discussion; M. Saldiva and B. Thomas for help with graphics, T. Krucker and G. Feng for partnership and portfolio management; the members of the Cryptosporidium Drug Accelerator consortium for helpful discussions. This work was funded by Novartis BioMedical Research, and in part by the Wellcome Trust (Project No. 219639/Z/19/Z) to T.T.D. and the Bill and Melinda Gates Foundation to J.A.Z. (Project No. OPP1117400).
Extended data
Author contributions
U.H.M., S.B.L., R.S.J. and T.T.D. conceived and designed the study. U.H.M., N.A. and T.T.D. wrote grant applications. J.M.Y., M.T. and P.L. performed compound synthesis. A.T.C., K.C., J.E.G., U.H.M. and R.S.J. performed and analysed in vitro potency assays. J.M.Y., R.S.J., J.E.G., U.H.M., C.S. and S.B.L. analysed the structure–activity relationship. L.C.W., K.C., A.T.C., U.H.M. and R.S.J. performed and analysed cell-based assays. C.L., J.S., L.C.W. and J.C. performed protein expression and purification. D.P., L.C.W., A.T.C. and J.E.G. executed enzyme assays. M.K., M.M. and C.C. performed protein crystallography experiments. L.X. analysed in vitro stability data. Y.-B.C. analysed in vivo bioanalytical data. U.K. and J.L. optimized formulation. I.H. and S.B.L analysed in vitro and in vivo pharmacokinetics data. A.T.C. and J.-R.G. analysed in vitro and in vivo toxicology results. K.H.D. executed the mouse efficacy model. K.H.D., S.B.L. and C.S.O. analysed in vivo mouse efficacy data. J.A.Z. and S.K.C. conducted the calf efficacy study. S.K.C. performed calf stool analysis. J.A.Z. wrote the grant application for calf studies. J.A.Z., U.H.M., S.B.L. and R.S.J. analysed calf data. U.H.M., N.A. and T.T.D. managed project priorities and resources. U.H.M., S.B.L., R.S.J. and T.T.D. wrote the manuscript with contributions from all co-authors.
Peer review
Peer review information
Nature Microbiology thanks Roberta O’Connor, Wesley Van Voorhis, Stephen Ward and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The atomic model of X-ray crystallography data has been deposited in the Protein Data Bank with the accession code PDB 8VOF. All compounds and reagents can be obtained through a materials transfer agreement from Novartis by contacting the corresponding authors, and a response may be expected in a few weeks. Source data are provided with this paper.
Competing interests
Multiple authors (U.H.M., S.B.L., R.S.J., A.T.C., J.M.Y., J.E.G., M.K., I.H., J.-R.G., J.C., U.K., M.T., P.L., K.H.D., K.C., D.P., M.M., C.L., C.C., J.S., L.X., Y.-B.C., J.L., C.S.O., N.A., C.S. and T.T.D.) are employees of Novartis and some of them have shares in the company. Compounds described in this report have been patented by Novartis, with J.M.Y., M.T. and P.L. listed as authors (US Patent WO/2022/079616). The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ujjini H. Manjunatha, Suresh B. Lakshminarayana, Rajiv S. Jumani.
Contributor Information
Ujjini H. Manjunatha, Email: manjunatha.ujjini@novartis.com
Thierry T. Diagana, Email: thierry.diagana@novartis.com
Extended data
is available for this paper at 10.1038/s41564-024-01810-x.
Supplementary information
The online version contains supplementary material available at 10.1038/s41564-024-01810-x.
References
- 1.GBD 2016 Diarrhoeal Disease Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis.18, 1211–1228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meisel, J. L., Perera, D. R., Meligro, C. & Rubin, C. E. Overwhelming watery diarrhea associated with a Cryptosporidium in an immunosuppressed patient. Gastroenterology70, 1156–1160 (1976). [PubMed] [Google Scholar]
- 3.Hossain, M. J. et al. Clinical and epidemiologic features of Cryptosporidium-associated diarrheal disease among young children living in Sub-Saharan Africa: the Vaccine Impact on Diarrhea in Africa (VIDA) study. Clin. Infect. Dis.76, S97–S105 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotloff, K. L. et al. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter Study (GEMS). Lancet Glob. Health7, e568–e584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotloff, K. L. et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet382, 209–222 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Platts-Mills, J. A. et al. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob. Health3, e564–e575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil, I. A. et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob. Health6, e758–e768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sow, S. O. et al. The burden of Cryptosporidium diarrheal disease among children <24 months of age in moderate/high mortality regions of Sub-Saharan Africa and South Asia, utilizing data from the Global Enteric Multicenter Study (GEMS). PLoS Negl. Trop. Dis.10, e0004729 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnee, A. E. et al. Identification of etiology-specific diarrhea associated with linear growth faltering in Bangladeshi infants. Am. J. Epidemiol.187, 2210–2218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerrant, D. I. et al. Association of early childhood diarrhea and cryptosporidiosis with impaired physical fitness and cognitive function four-seven years later in a poor urban community in northeast Brazil. Am. J. Trop. Med. Hyg.61, 707–713 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Mondal, D., Haque, R., Sack, R. B., Kirkpatrick, B. D. & Petri, W. A. Attribution of malnutrition to cause-specific diarrheal illness: evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh. Am. J. Trop. Med. Hyg.80, 824–826 (2009). [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, H. C. et al. High levels of CXCL10 are produced by intestinal epithelial cells in AIDS patients with active cryptosporidiosis but not after reconstitution of immunity. Infect. Immun.75, 481–487 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Painter, J. E. et al. Cryptosporidiosis surveillance—United States, 2011–2012. MMWR Suppl.64, 1–14 (2015). [PubMed] [Google Scholar]
- 14.Mora, C. et al. Over half of known human pathogenic diseases can be aggravated by climate change. Nat. Clim. Change12, 869–875 (2022). [DOI] [PMC free article] [PubMed]
- 15.Checkley, W. et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis.15, 85–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashigbie, P. G. et al. Use-case scenarios for an anti-Cryptosporidium therapeutic. PLoS Negl. Trop. Dis.15, e0009057 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amadi, B. et al. Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomised controlled trial. Lancet360, 1375–1380 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Amadi, B. M. et al. High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: a randomised controlled trial. BMC Infect. Dis.2, 195 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, I. H. et al. Safe and effective treatments are needed for cryptosporidiosis, a truly neglected tropical disease. BMJ Glob. Health8, e012540 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.English, E. D., Guerin, A., Tandel, J. & Striepen, B. Live imaging of the Cryptosporidium parvum life cycle reveals direct development of male and female gametes from type I meronts. PLoS Biol.20, e3001604 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korpe, P. et al. Prospective cohort study of Cryptosporidium infection and shedding in infants and their households. Clin. Infect. Dis.76, 2178–2186 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansen, O. H. et al. Oocyst shedding dynamics in children with cryptosporidiosis: a prospective clinical case series in Ethiopia. Microbiol. Spectr.10, e0274121 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iroh Tam, P. Y., Chisala, M., Nyangulu, W., Thole, H. & Nyirenda, J. Respiratory cryptosporidiosis in Malawian children with diarrheal disease. PLoS Negl. Trop. Dis.15, e0009643 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manjunatha, U. H. et al. A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature546, 376–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.First in Human Study to Assess the Safety, Tolerability and Pharmacokinetics of EDI048 in Healthy Volunteers (ClinicalTrials.gov, 2023); https://ClinicalTrials.gov/show/NCT05275855
- 26.Baragana, B. et al. Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis. Proc. Natl Acad. Sci. USA116, 7015–7020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinayak, S. et al. Bicyclic azetidines kill the diarrheal pathogen Cryptosporidium in mice by inhibiting parasite phenylalanyl-tRNA synthetase. Sci. Transl. Med.12, eaba8412 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnold, S. L. M. et al. Necessity of bumped kinase inhibitor gastrointestinal exposure in treating Cryptosporidium infection. J. Infect. Dis.216, 55–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchwald, P. Soft drugs: design principles, success stories, and future perspectives. Expert Opin. Drug Metab. Toxicol.16, 645–650 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Di, L. The impact of carboxylesterases in drug metabolism and pharmacokinetics. Curr. Drug Metab.20, 91–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young, J. M., Turner, M. R. & Lu, P. Compounds and compositions for the treatment of cryptosporidiosis. US patent 12,018,024 B2 (assigned to Novartis AG) (2022).
- 32.Bahar, F. G., Ohura, K., Ogihara, T. & Imai, T. Species difference of esterase expression and hydrolase activity in plasma. J. Pharm. Sci.101, 3979–3988 (2012). [DOI] [PubMed] [Google Scholar]
- 33.McPhail, J. A. & Burke, J. E. Drugging the phosphoinositide 3-kinase (PI3K) and phosphatidylinositol 4-kinase (PI4K) family of enzymes for treatment of cancer, immune disorders, and viral/parasitic infections. Adv. Exp. Med. Biol.1274, 203–222 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Burke, J. E. et al. Structures of PI4KIIIbeta complexes show simultaneous recruitment of Rab11 and its effectors. Science344, 1035–1038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jumani, R. S. et al. A suite of phenotypic assays to ensure pipeline diversity when prioritizing drug-like Cryptosporidium growth inhibitors. Nat. Commun.10, 1862 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Funkhouser-Jones, L. J., Ravindran, S. & Sibley, L. D. Defining stage-specific activity of potent new inhibitors of Cryptosporidium parvum growth in vitro. mBio11, e00052-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNamara, C. W. et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature504, 248–253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jumani, R. S. et al. A novel piperazine-based drug lead for cryptosporidiosis from the Medicines for Malaria Venture Open-access Malaria Box. Antimicrob. Agents Chemother.62, e01505-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharpstone, D. et al. Small intestinal transit, absorption, and permeability in patients with AIDS with and without diarrhoea. Gut45, 70–76 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charmot, D. Non-systemic drugs: a critical review. Curr. Pharm. Des.18, 1434–1445 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guidance for Industry M3(R2) Nonclinical Safety Studies for the Conduct of Human Clinical Trials and Marketing Authorization for Pharmaceuticals (US Food and Drug Administration, 2010). [PubMed]
- 42.Xiao, L. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol.124, 80–89 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Harding, C. R. & Meissner, M. The inner membrane complex through development of Toxoplasma gondii and Plasmodium. Cell. Microbiol.16, 632–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasan, M. M. et al. Spontaneous selection of Cryptosporidium drug resistance in a calf model of infection. Antimicrob. Agents Chemother.65, e00023-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mor, S. M. et al. Expectoration of Cryptosporidium parasites in sputum of human immunodeficiency virus-positive and -negative adults. Am. J. Trop. Med. Hyg.98, 1086–1090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorel, R., Wong, A. R. & Crawford, J. J. Trust your gut: strategies and tactics for intestinally restricted drugs. ACS Med. Chem. Lett.14, 233–243 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerard, L., Garey, K. W. & DuPont, H. L. Rifaximin: a nonabsorbable rifamycin antibiotic for use in nonsystemic gastrointestinal infections. Expert Rev. Anti Infect. Ther.3, 201–211 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Mejdrova, I. et al. Rational design of novel highly potent and selective phosphatidylinositol 4-kinase IIIbeta (PI4KB) inhibitors as broad-spectrum antiviral agents and tools for chemical biology. J. Med. Chem.60, 100–118 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Vinayak, S. et al. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature523, 477–480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pawlowic, M. C., Vinayak, S., Sateriale, A., Brooks, C. F. & Striepen, B. Generating and maintaining transgenic Cryptosporidium parvum parasites. Curr. Protoc. Microbiol.46, 20B.2.1–20B.2.32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chao, A. T. et al. Development of a cytopathic effect-based phenotypic screening assay against Cryptosporidium. ACS Infect. Dis.4, 635–645 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Gubler, H. et al. Helios: history and anatomy of a successful in-house enterprise high-throughput screening and profiling data analysis system. SLAS Discov.23, 474–488 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Love, M. S. et al. A high-throughput phenotypic screen identifies clofazimine as a potential treatment for cryptosporidiosis. PLoS Negl. Trop. Dis.11, e0005373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fowler, M. L. et al. Using hydrogen deuterium exchange mass spectrometry to engineer optimized constructs for crystallization of protein complexes: case study of PI4KIIIbeta with Rab11. Protein Sci.25, 826–839 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu, Y. et al. Rationally designed PI3Kalpha mutants to mimic ATR and their use to understand binding specificity of ATR inhibitors. J. Mol. Biol.429, 1684–1704 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Emsley, P. & Debreczeni, J. E. The use of molecular graphics in structure-based drug design. Methods Mol. Biol.841, 143–159 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Zhou, L., Yang, L., Tilton, S. & Wang, J. Development of a high throughput equilibrium solubility assay using miniaturized shake-flask method in early drug discovery. J. Pharm. Sci.96, 3052–3071 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Faller, B. Artificial membrane assays to assess permeability. Curr. Drug Metab.9, 886–892 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Irvine, J. D. et al. MDCK (Madin–Darby canine kidney) cells: a tool for membrane permeability screening. J. Pharm. Sci.88, 28–33 (1999). [DOI] [PubMed] [Google Scholar]
- 60.Lau, Y. Y. et al. The use of in vitro metabolic stability for rapid selection of compounds in early discovery based on their expected hepatic extraction ratios. Pharm. Res.19, 1606–1610 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Obach, R. S. Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: an examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab. Dispos.27, 1350–1359 (1999). [PubMed] [Google Scholar]
- 62.Di, L., Kerns, E. H., Hong, Y. & Chen, H. Development and application of high throughput plasma stability assay for drug discovery. Int. J. Pharm.297, 110–119 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Waters, N. J., Jones, R., Williams, G. & Sohal, B. Validation of a rapid equilibrium dialysis approach for the measurement of plasma protein binding. J. Pharm. Sci.97, 4586–4595 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Bell, L. et al. Evaluation of fluorescence- and mass spectrometry-based CYP inhibition assays for use in drug discovery. J. Biomol. Screen.13, 343–353 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Zimmerlin, A., Trunzer, M. & Faller, B. CYP3A time-dependent inhibition risk assessment validated with 400 reference drugs. Drug Metab. Dispos.39, 1039–1046 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Wang, J., Urban, L. & Bojanic, D. Maximising use of in vitro ADMET tools to predict in vivo bioavailability and safety. Expert Opin. Drug Metab. Toxicol.3, 641–665 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Faller, B. et al. High-throughput in vitro profiling assays: lessons learnt from experiences at Novartis. Expert Opin. Drug Metab. Toxicol.2, 823–833 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Test No. 432: In Vitro 3T3 NRU Phototoxicity Test (OECD, 2019).
- 69.Ames, B. N., Lee, F. D. & Durston, W. E. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc. Natl Acad. Sci. USA70, 782–786 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lukamowicz, M., Woodward, K., Kirsch-Volders, M., Suter, W. & Elhajouji, A. A flow cytometry based in vitro micronucleus assay in TK6 cells–validation using early stage pharmaceutical development compounds. Environ. Mol. Mutagen.52, 363–372 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Test No. 487: In Vitro Mammalian Cell Micronucleus Test (OECD, 2016).
- 72.Cammerer, Z., Elhajouji, A. & Suter, W. In vivo micronucleus test with flow cytometry after acute and chronic exposures of rats to chemicals. Mutat. Res.626, 26–33 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Test No. 489: In Vivo Mammalian Alkaline Comet Assay (OECD, 2016).
- 74.Jothikumar, N., da Silva, A. J., Moura, I., Qvarnstrom, Y. & Hill, V. R. Detection and differentiation of Cryptosporidium hominis and Cryptosporidium parvum by dual TaqMan assays. J. Med. Microbiol.57, 1099–1105 (2008). [DOI] [PubMed] [Google Scholar]
- 75.Bellosa, M. L. et al. A comparison of fecal percent dry matter and number of Cryptosporidium parvum oocysts shed to observational fecal consistency scoring in dairy calves. J. Parasitol.97, 349–351 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Zambriski, J. A. et al. Cryptosporidium parvum: determination of ID50 and the dose–response relationship in experimentally challenged dairy calves. Vet. Parasitol.197, 104–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dreier, J., Stormer, M. & Kleesiek, K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J. Clin. Microbiol.43, 4551–4557 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Operario, D. J., Bristol, L. S., Liotta, J., Nydam, D. V. & Houpt, E. R. Correlation between diarrhea severity and oocyst count via quantitative PCR or fluorescence microscopy in experimental cryptosporidiosis in calves. Am. J. Trop. Med. Hyg.92, 45–49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1, and Tables 1 and 2.
Time-lapse microscopy images were taken every 20 min from 4 h post infection, that is, 1 h after compound addition (time 0 h in the videos) up to 24 h post infection and are shown in Supplementary Videos 1 (DMSO), 2 (EDI048) and 3 (KDU731).
Time-lapse microscopy images were taken every 20 min from 4 h post infection, that is, 1 h after compound addition (time 0 h in the videos) up to 24 h post infection and are shown in Supplementary Videos 1 (DMSO), 2 (EDI048) and 3 (KDU731).
Time-lapse microscopy images were taken every 20 min from 4 h post infection, that is, 1 h after compound addition (time 0 h in the videos) up to 24 h post infection and are shown in Supplementary Videos 1 (DMSO), 2 (EDI048) and 3 (KDU731).
HsCpPI(4)K X-ray crystal structure PDB validation report.
EDI048 LC–MS data.
Source data (efficacy and PK raw data).
Source data (concentration–response, reversibility and time-kill curve raw data).
Source data (parasitological readout, clinical (faecal consistency score) readout, dose–exposure response raw data).
Source data (fold shift in IC50 raw data).
Source data (parasitological readout and clinical (faecal consistency score) readout raw data).
Source data (dose–Cmax response raw data).
Source data (raw RLU data for parasitistatic/parasiticidal assay and one-phase exponential decay curve fit).
Data Availability Statement
The atomic model of X-ray crystallography data has been deposited in the Protein Data Bank with the accession code PDB 8VOF. All compounds and reagents can be obtained through a materials transfer agreement from Novartis by contacting the corresponding authors, and a response may be expected in a few weeks. Source data are provided with this paper.