Abstract
Previous studies in patients with mature B-cell lymphomas (MBCL) have shown that pathogenic TP53 aberrations are associated with inferior chemotherapeutic efficacy and survival outcomes. In solid malignancies, p53 immunohistochemistry is commonly used as a surrogate marker to assess TP53 mutations, but this correlation is not yet well-established in lymphomas. This study evaluated the accuracy of p53 immunohistochemistry as a surrogate marker for TP53 mutational analysis in a large real-world patient cohort of 354 MBCL patients within routine diagnostic practice. For each case, p53 IHC was assigned to one of three categories: wild type (staining 1–50% of tumor cells with variable nuclear staining), abnormal complete absence or abnormal overexpression (strong and diffuse staining > 50% of tumor cells). Pathogenic variants of TP53 were identified with a targeted next generation sequencing (tNGS) panel. Wild type p53 expression was observed in 267 cases (75.4%), complete absence in twenty cases (5.7%) and the overexpression pattern in 67 cases (18.9%). tNGS identified a pathogenic TP53 mutation in 102 patients (29%). The overall accuracy of p53 IHC was 84.5% (95% CI 80.3–88.1), with a robust specificity of 92.1% (95% CI 88.0- 95.1), but a low sensitivity of 65.7% (95% CI 55.7–74.8). These results suggest that the performance of p53 IHC is insufficient as a surrogate marker for TP53 mutations in our real-world routine diagnostic workup of MBCL patients. By using p53 immunohistochemistry alone, there is a significant risk a TP53 mutation will be missed, resulting in misevaluation of a high-risk patient. Therefore, molecular analysis is recommended in all MBCL patients, especially for further development of risk-directed therapies based on TP53 mutation status.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00428-023-03676-6.
Keywords: B-cell lymphoma, Molecular diagnostics, Targeted therapy, Immunohistochemistry, Hematopathology
Introduction
Lymphomas encompass more than 80 different types of malignancies, each distinguished by unique clinical, morphological, immunohistochemical, molecular, and cytogenetic characteristics [1]. The p53 protein encoded by the TP53 gene is an important tumor suppressor that mediates cell-cycle arrest, DNA repair, transcription, signalling, metabolism, apoptosis, and autophagy [2, 3]. In human cancers, nonsynonymous or missense mutations in TP53, often accompanied by loss of heterozygosity, is the most common mechanism leading to altered p53 sequence and structure. This is consistent with the so called ‘two-hit’ hypothesis; inactivation of both copies of a tumor suppressor gene are required for dysfunction, often resulting in gain-of-function (pro-oncogenic) or loss-of-function (decreased tumor suppression) [4]. The overall prevalence of TP53 mutations in cancer is around 50%, reaching 100% in some carcinomas such as high-grade ovarian cancer [4–6]. In contrast, homozygous deletions of TP53 are exceedingly rare (0.2%) [7]. TP53 mutations resulting in p53 dysfunction are less frequent in lymphoid malignancies than in other types of cancer [2, 8]. In mature B-cell lymphomas (MBCL) the frequency varies across different subtypes, ranging from approximately 10% in chronic lymphocytic leukemia (CLL) to 25% in diffuse large B-cell lymphoma not otherwise specified (DLBCL, NOS) and Burkitt lymphoma (BL). Several studies in MBCL patients have shown that pathogenic TP53 aberrations are associated with inferior therapeutic efficacy and survival outcomes [3, 9–11]. TP53 mutational status has a central role in the current treatment algorithms for both CLL and mantle cell lymphoma (MCL). In TP53-aberrant CLL/MCL patients, therapy aims to block the B-cell receptor pathway with Bruton tyrosine kinase (BTK) inhibitors or B-cell lymphoma-2 (bcl2) inhibitors, which act independently of the TP53 pathway as opposed to treatment with cytotoxic agents [9, 10].
DLBCL, NOS is the most prevalent subtype of MBCL and characterized by a poor prognosis, aggressive disease course, and significant genetic heterogeneity. The standard first-line therapy for DLBCL, NOS is the ‘one-size-fits-all’ immunochemotherapy regimen R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone). While this treatment can cure a proportion of patients, approximately 40% of patients develop disease recurrence and require additional therapy [12]. While TP53 is at present only used as a therapy stratifier in MCL and CLL, there are multiple studies that have shown that pathogenic TP53 aberrations are associated with inferior therapeutic efficacy and survival outcomes in other mature B-cell lymphomas as well, most notably in (D)LBCL[3, 11]. In DLBCL, NOS but also in other lymphomas, there is an urgent need for further development of targeted and risk-directed therapies that improve clinical outcomes by selecting the most optimal treatment for each patient based on intrinsic tumor factors. TP53 mutational status has the potential to provide important prognostic and predictive information if reliable assessment can be achieved in routine clinical practice.
Regardless of the effect of TP53 mutational status on the clinical course, targeted next generation sequencing (tNGS) is currently not standard practice in the diagnostic workup of lymphomas in most centers. In solid tumors, p53 immunohistochemistry (IHC) is an accurate and frequently used surrogate marker to assess TP53 mutations. P53 IHC is widely available, interpretable by a pathologist, faster and relatively inexpensive compared to tNGS. Both overexpression or complete absence of p53 expression may indicate a pathogenic TP53 mutation or deletion [13–17]. Gain-of-function TP53 aberrations accompanied by loss of the tumor-suppressive function typically arise from missense mutations or in-frame deletions, disrupting MDM2 mediated ubiquitin degradation of p53, causing it to accumulate in the tumor cells leading to overexpression. Loss-of-function is usually caused by nonsense mutations, splicing mutations, or frameshifts, resulting in a premature stop gain and therefore no translated protein and IHC expression [18, 19]. Given the clinical impact of pathogenic TP53 mutations, p53 IHC may be a valuable marker in MBCL diagnostics.
Since in solid tumors p53 IHC is an accurate and frequently used surrogate marker to assess TP53 mutations, p53 IHC is also frequently used for lymphomas, while in this entity the performance of p53 IHC has currently not been definitively established. Studies in this area are lacking, particularly in indolent B-cell lymphomas (IBCL) and have primarily centred on (D)LBCL and MCL, rather than the broader MBCL population, thus not reflecting real-life everyday diagnostics [2, 3, 20]. With the implementation of tNGS in our center since 2017 for routine daily diagnostic procedures for MBCLs, we were in the unique setting to directly compare p53 IHC with TP53 mutational analyses for many years. Therefore, in this retrospective study the diagnostic accuracy of p53 IHC as a surrogate marker for TP53 mutational tNGS analysis was evaluated in a large real-world cohort of various MBCL subtypes.
Methods
Patient selection
For this retrospective study 354 patients diagnosed with BCL between 2017–2022 were selected. The cases were diagnosed according to the revised fourth edition WHO classification (2016) and included (diffuse) large B-cell lymphoma ((D)LBCL), mantle cell lymphoma (MCL) and indolent B-cell lymphoma (IBCL). The study was conducted in accordance with the Dutch Code for Proper Secondary Use of Human Tissue, the local institutional board requirements, and the revised Declaration of Helsinki (2008). Approval with a waiver of consent was obtained from the LUMC's medical ethics committee (B16.048).
Immunohistochemistry and in situ hybridization
A representative formalin-fixed paraffin-embedded (FFPE) tumor block was selected for each case, and three µm thin tissue sections were prepared. Tumor cell percentage was at least 20%, and for most cases more than 60%. The slides were stained for p53 antibody expression using the Dako Auto Stainer Link 48 (Dako Omnis, Monoclonal Mouse Anti Human, p53 clone DO-7, Glostrup, Denmark), according to standard procedures and as previously reported by our institute for endometrial carcinoma [13]. The p53 staining patterns were classified as wild type or abnormal, with cut-offs used in our daily diagnostics and as previously published in lymphoma patients[3, 21]. Wild type expression was defined as p53 expression in 1–50% of tumor nuclei with variable nuclear staining intensity (including weak, moderate, and strong). Abnormal p53 expression was defined as strong diffuse positive p53 expression in > 50% of the tumor nuclei (abnormal overexpression) or complete absence of p53 staining with a positive internal and/or external control (abnormal null mutant). Representative examples of the staining categories are shown in Fig. 1. All p53 IHC slides were independently scored by two hematopathologists (LMH and PMJ) who were blinded to the clinical and molecular data. All discordant cases were discussed at a consensus meeting attended by both pathologists where a definitive category was assigned. As described before, according to standard diagnostic lymphoma workup for LBCL, MYC, and when positive, BCL2, and BCL6 rearrangements were analysed with fluorescence in situ hybridization (FISH), using break-apart probes [22]. The Epstein-Barr virus (EBV) status was determined using EBV-encoded RNA in situ hybridization (ISH).
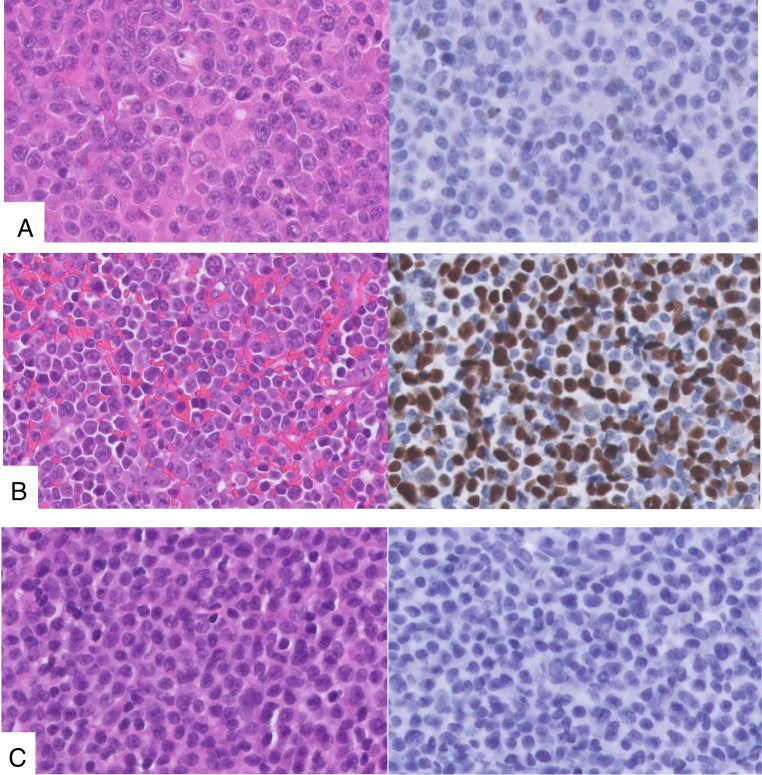
Fig. 1.
Representative microscopy of p53 immunohistochemistry staining patterns. A Diffuse large B-cell lymphoma, not otherwise specified (hematoxylin & Eosin × 80) with P53 wild type pattern, × 80. B Diffuse large B-cell lymphoma, not otherwise specified (hematoxylin & Eosin × 80) with P53 abnormal overexpression pattern, × 80. C Mantle cell lymphoma (hematoxylin & Eosin × 80) with P53 abnormal deletion pattern, × 80
Targeted next-generation sequencing
DNA alterations in TP53 were investigated using two tNGS panels. Only 10% (n = 36) of the samples were sequenced using amplicons depicted in online resource 1a, while the majority (n = 318, 90%) were sequenced using amplicons depicted in online resource 1b. Libraries prepared with the tNGS panels were sequenced using the Ion GeneStudio™ S5 System and the sequenced reads were mapped against the human reference genome (GRCh37/hg19) using iontorrent aligner (TMAP) and variant caller (TVC) using default parameters for somatic variant calling. Only variants with a predefined minimum coverage of 100 reads and variant allele frequency (VAF) of 0.10 (10%) were considered. Subsequently interpretation of the variants was done using Franklin genoox and GenomeNexus [23, 24], aggregating data from all public databases (including Cosmic, Clinvar and TP53 databases) and classified into class 1 (benign), class 2 (likely benign), class 3 (unknown significance), class 4 (likely pathogenic), or class 5 (pathogenic) [25]. Variants classified as class 4 and 5 were selected for further analysis.
Statistical analysis
The statistical analysis was conducted with RStudio (version 4.2.1). The inter-rater reliability was assessed using Cohen's kappa coefficient. Unpaired t-tests, Chi-square tests, and one-way ANOVA were used to determine statistically significant differences between the TP53 wild type and mutated groups. The diagnostic accuracy of p53 IHC was evaluated by calculating its accuracy, sensitivity, specificity, area under the curve (AUC), positive predictive value (PPV), negative predictive value (NPV), receiver operating characteristic curve (ROC) and area under the curve (AUC) compared to TP53 tNGS analysis. A two-sided p-value < 0.05 was considered statistically significant.
Results
Patient characteristics
The cohort included various MBCL and comprised a (D)LBCL group consisting of Burkitt lymphoma (n = 4), DLBCL, NOS (n = 167), primary cutaneous DLBCL, leg type (PC DLBCL, LT, n = 13), high grade B-cell lymphoma (HGBL, n = 11), primary central nervous system lymphoma (PCNSL, n = 17) and primary mediastinal B-cell lymphoma (PMBL, n = 3). Furthermore, mantle cell lymphoma (MCL, n = 18) and a heterogeneous group of IBCL (n = 76), including NOS (n = 4), follicular lymphoma (FL, n = 52), hairy cell leukemia/lymphoma (HCL, n = 2), lymphoplasmacytic lymphoma (LPL, n = 12), mucosa-associated lymphoid tissue (MALT, n = 8), nodal marginal zone lymphoma (NMZL, n = 17), chronic lymphocytic leukemia (CLL, n = 13) and primary cutaneous follicle center lymphoma (PCFCL, n = 3). All baseline characteristics are depicted in Table 1. The mean age was 63 years (19–95) with a slight male predominance (59%). Analysis was primarily performed on tissue of time at primary diagnosis (n = 252, 71%) and 29% (n = 102) in relapsed or secondary acquired tissue. In the HGBL group, nine patients were classified as double hit genotype (for n = 7 patients translocations MYC + /BCL2 + /BCL6- and 2 cases with translocations MYC + /BCL6 + /BCL2-) and two cases were triple hit genotype.
Table 1.
Patient and tumor characteristics
| Baseline characteristics | ||||
|---|---|---|---|---|
| Total | TP53 mutated | TP53 wild type | p-value | |
|
Patients (%) Age y, mean (range) |
354 63 (19–95) |
102 (29) 62 (21–95) |
254 (71) 64 (19–89) |
0.24 |
| Gender | 0.21 | |||
| Female (%) | 145 (41) | 36 (25) | 109 (75) | |
| Male (%) | 209 (59) | 66 (32) | 143 (68) | |
| WHO classification, 2016 (%) | ||||
| (D)LBCL (%) | 215 (61) | 65 (30) | 150 (70) | |
| Burkitt | 4 (1) | 4 (100) | 0 | |
| DLBCL, NOS | 167 (47) | 52 (31) | 115 (69) | |
| PCDLBCL LT | 13 (4) | 2 (15) | 11 (85) | |
| HGBL | 11 (3) | 5 (45) | 6 (55) | |
| PCNSL | 17 (5) | 2 (12) | 15 (88) | |
| PMBL | 3 (1) | 0 | 3 (100) | |
| MCL | 28 (8) | 16 (57) | 12 (43) | |
| IBCL (%) | 111 (31) | 21 (19) | 90 (81) | |
| NOS | 4 (1) | 1 (5) | 3 (95) | |
| FL | 52 (47) | 9 (45) | 43 (55) | |
| HCL | 2 (2) | 1 (50) | 1 (50) | |
| LPL | 12 (11) | 1 (8) | 11 (92) | |
| MALT | 8 (7) | 1 (13) | 7 (87) | |
| NMZL | 17 (15) | 1 (6) | 16 (94) | |
| CLL | 13 (12) | 6 (46) | 7 (54) | |
| PCFCL | 3 (3) | 1 (33) | 2 (67) | |
| Tissue | 0.11 | |||
| Primary | 252 (71) | 66 (26) | 186 (84) | |
| Secondary | 102 (29) | 36 (35) | 66 (65) | |
WHO, World Health Organization; (D)LBCL, (diffuse) large B-cell lymphoma; DLBCL, NOS, diffuse large B-cell lymphoma, not otherwise specified; PC DLBCL LT, primary cutaneous diffuse large B-cell lymphoma leg type; HGBL, high grade B-cell lymphoma; PCNSL, primary central nervous system lymphoma; PMBL, primary mediastinal B-cell lymphoma; MCL, mantle cell lymphoma; IBCL, indolent B-cell lymphoma; NOS, not otherwise specified; FL, follicular lymphoma; HCL, hairy cell lymphoma; LPL, lymphoplasmacytic lymphoma; MALT, mucosa-associated lymphoid tissue; NMZL, nodal marginal zone lymphoma; CLL, chronic lymphocytic lymphoma; PCFCL, primary cutaneous follicle centre lymphoma
P53 IHC
P53 IHC staining quality was sufficient for interpreting the staining patterns in all cases. In the p53 IHC assessment, the two hematopathologists were discordant in sixteen cases, most often between the categories wild type and overexpression (n = 11) vs wild type and complete absence (n = 5). Consensus was achieved through shared evaluation in all cases. The resulting Cohen’s kappa correlation coefficient was 0.88, indicating a strong level of agreement. Wild type p53 expression was observed in 267 cases (75.4%), while an abnormal complete absence pattern was observed in twenty cases (5.7%), and an overexpression pattern was observed in 67 cases (18.9%). All our overexpression cases showed > 50% of tumor nuclei with high-intensity expression of p53 IHC.
TP53 assessment by tNGS
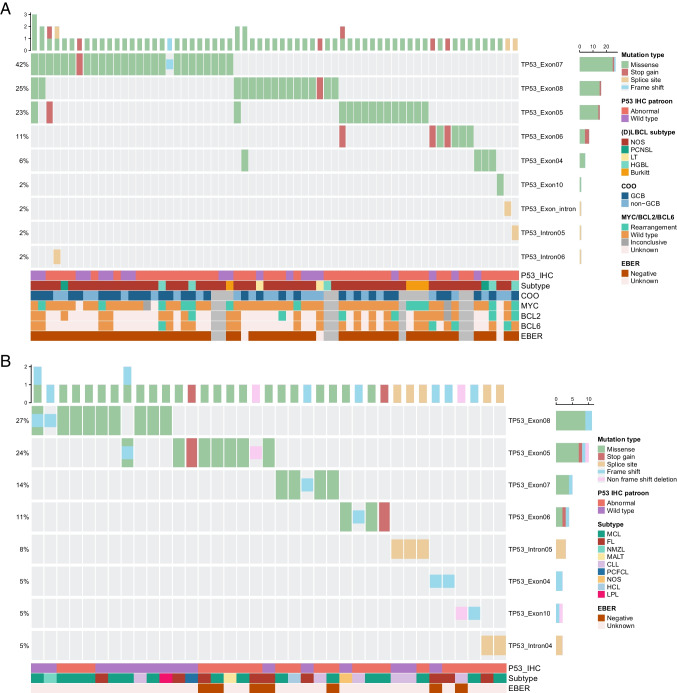
Out of the 354 patients, 102 (29%) had a TP53 mutation, while 252 (71%) had a wild type status. No significant differences were observed between the mutated and wild type groups in baseline characteristics (Table 1). TP53 mutations were most commonly observed in (D)LBCL (n = 65), followed by the IBCL group (n = 21), and MCL (n = 16). Overall, most patients had one pathogenic variation in TP53 (n = 91), nine patients had two different pathogenic mutations and one patient had four different mutations. Using the current tNGS approach, there were no TP53 gene deletions detected in this cohort. In the TP53 mutated (D)LBCL group, the most frequent type of mutation observed was a missense mutation (87%), followed by nonsense (8%), splice site (4%) and frame shift mutations (1%). Exon 7 (39%), exon 8 (21%), exon 5 (20%), and exon 6 (9%) were the most common locations of mutation. In MCL (n = 16, 16%), missense mutations (69%) were the most frequent, followed by frameshift (12%), splice site (12%), and nonsense mutations (6%). Exon 8 (44%) and exon 5 (19%) were the most affected locations. The mutation type distribution in IBCL was slightly different, with missense (50%), frameshift (29%), and splice site mutations (13%) being the most frequent types observed. The least frequent types were non-frameshift deletion (8%) and nonsense (4%) mutations. Exon 5 (33%) and exon 8 (17%) were the most affected locations in IBCL. The exon location did not differ significantly between the three groups (p = 0.122). However, the type of mutation was significantly different between LBCL, MCL and IBCL (p = 0.001). Details are presented in a TP53 mutation oncoplot (Fig. 2).
Fig. 2.
Tumor and molecular characteristics of all TP53 mutated cases. No pattern with discordant p53 wild type IHC could be identified. A: (D)LBCL. B: MCL and IBCL. IHC, immunohistochemistry; COO, cell of origin; EBER, Epstein-Barr virus encoded ribonucleic acid; (D)LBCL, diffuse large B-cell lymphoma; NOS, not otherwise specified; PCNSL, primary central nervous system lymphoma; LT, leg type; HGBL, high-grade B-cell lymphoma; GCB, germinal center type; MCL, mantle cell lymphoma; FL, follicular lymphoma; NMZL, nodal marginal zone lymphoma; MALT, mucosa-associated lymphoid tissue; CLL, chronic lymphocytic lymphoma; PCFCL, primary cutaneous follicle centre lymphoma; NOS, not otherwise specified; HCL, hairy cell lymphoma; LPL, lymphoplasmacytic lymphoma
Diagnostic performance of p53 IHC
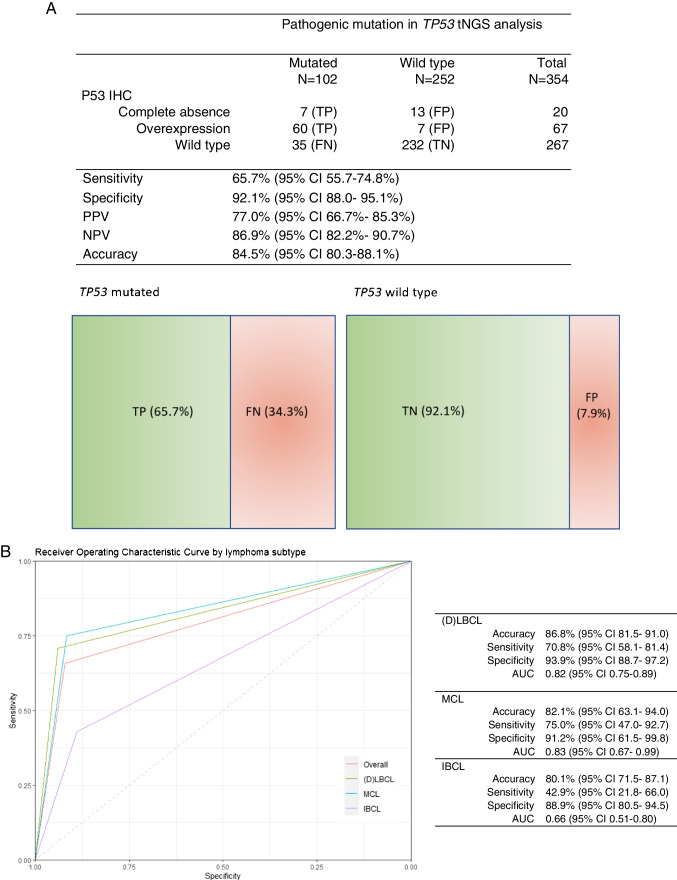
Within the TP53 mutated group (n = 102, 29%), concordant results with the immunohistochemical assay were found in 67 cases (65.7%). An overexpression pattern was observed in 60 cases (58.9%) of which 54 were missense mutations (90%), one frame shift (2%) and one non frame shift deletion (2%). Two cases had both a missense and nonsense mutation (3%), one case a missense and splice site (2%) and one case a missense and frame shift (2%). A complete absence pattern was seen in seven cases (6.9%), of which six had a truncating mutation. Three cases were splice site mutations (43%), two frame shifts (29%) and one nonsense (14%). One of these cases had a missense mutation (14%). The remaining 35 discordant cases (34.3%) had a false negative p53 wild type staining pattern, despite a confirmed TP53 mutation. 20 cases in this discordant group had missense mutations (57%), five a nonsense mutation (14%), four a frame shift (11%), four splice sites (11%) and one a frame shift deletion (3%). One case displayed both a missense and frame shift (3%). In the TP53 wild type group (n = 252, 71%), a concordant wild type p53 IHC pattern was observed in 232 cases (92.1%). In contrast, thirteen cases (5.2%) showed a false positive complete absence staining pattern and seven cases (2.8%) an overexpression pattern, despite the absence of TP53 mutation. Overall, concordant results between the p53 IHC and tNGS were observed in 299 of 354 cases, resulting in an accuracy of 84.5% (95% CI 80.3–88.1), with a corresponding sensitivity of 65.7% (95% CI 55.7–74.8) and specificity of 92.1% (95% CI 88.0- 95.1). The PPV was 77.0% (95% CI 66.7- 85.3), while the NPV was 86.9% (95% CI 82.2- 90.7), as illustrated in Fig. 3a.
Fig. 3.
A Correlation p53 immunohistochemistry pattern and TP53 mutational analysis, tNGS, targeted next generation sequencing; IHC, immunohistochemistry; TP, true positive; FP, false positive; FN, false negative; TN, true negative; CI, confidence interval. B Diagnostic performance of p53 immunohistochemistry by lymphoma subtype with corresponding ROC curves, IHC, immunohistochemistry; (D)LBCL, diffuse large B-cell lymphoma; MCL, mantle cell lymphoma; IBCL, indolent B-cell lymphoma; AUC, area under the curve
Additional analyses on cases excluding all decalcified samples (n = 313) demonstrated similar overall performance rates. Overall accuracy was 86.0% (95% CI 81.6–74.8) with a corresponding sensitivity of 64.5% (95% CI 53.9–74.2) and specificity of 95.0% (95% CI 91.2–97.5).
Diagnostic performance per subtype
The diagnostic performance rates per subtype for (D)LBCL, MCL and IBCL were respectively overall accuracy 86.8 (95% CI 81.5- 91.0), 82.1% (95% CI 63.1- 94.0), and 80.1% (95% CI 71.5- 87.1). The sensitivity was 70.8 (95% CI 58.1- 81.4), 75.0% (95% CI 47.0- 92.7), and 42.9% (95% CI 21.8- 66.0). Lastly the specificity was 93.9% (95% CI 88.7- 97.2), 91.2% (95% CI 61.5- 99.8) and 88.9% (95% CI 80.5- 94.5). IBCL demonstrated a similar accuracy and specificity to (D)LBCL and MCL, however the sensitivity of IBCL was lower. In Fig. 3b the ROC curves of LBCL, MCL and IBCL are plotted with the corresponding AUC.
Discordant cases
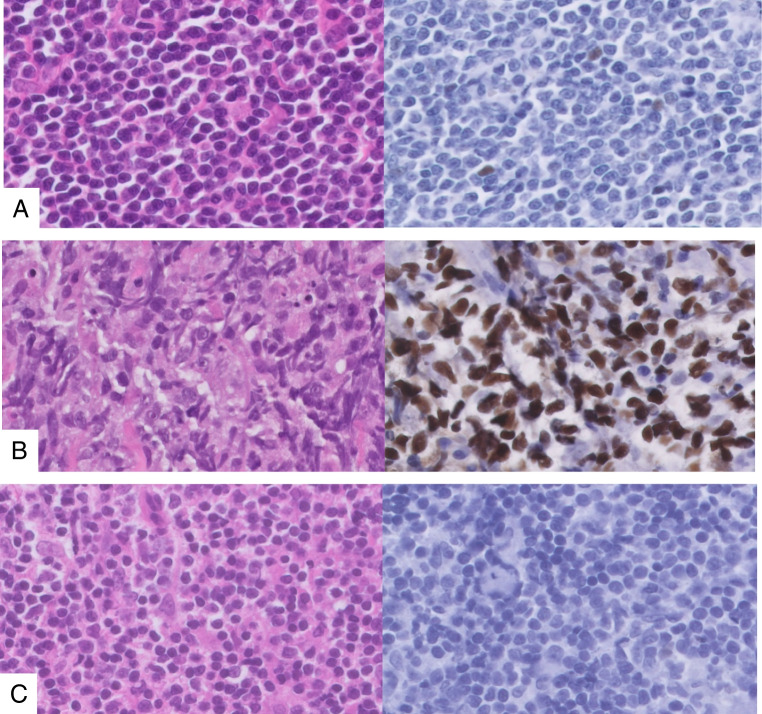
A total of 55 cases (16%), 20 FP and 35 FN, including (D)LBCL (n = 28, 51%), MCL (n = 5, 9%), and IBCL (n = 22, 40%), demonstrated discordant results between p53 IHC and TP53 mutational analysis, as indicated in online resource 2. For these cases. needle biopsies (n = 39, 71%) and excisions (n = 16, 29%) were both present, and the tumor percentage was > 50%. Several TP53 pathogenic variants, sequenced with tNGS panel A or B, were found on various exons, and the variant allele frequency (VAF) was usually > 20%. No significant differences between the false negative and false positive IHC group could be identified in mutation type (p = 0.11), exon location (p = 0.80) or lymphoma subtype (p = 0.10) (Fig. 2 and online resource 3). Upon further analysis of the tNGS data, one LPL case manifested a class IV (likely pathogenic) TP53 variant, but with an extremely low VAF of 1.6%. Similarly, one other LPL case had a class V (pathogenic) TP53 variant with a low VAF of 2.4%. For the entire TP53 wild type group, no gains or losses of TP53 could be found. Representative examples of discordant cases are shown in Fig. 4.
Fig. 4.
Representative p53 immunohistochemistry of discordant cases, A Mantle cell lymphoma (hematoxylin & Eosin × 80) with wild type p53 expression pattern and TP53 missense mutation, × 80, B Diffuse large B-cell lymphoma, not otherwise specified (hematoxylin & Eosin × 80) with abnormal p53 overexpression pattern and no confirmed TP53 mutation, × 80, C Follicular lymphoma (hematoxylin & Eosin × 80) with abnormal p53 complete absence pattern and no confirmed TP53 mutation, × 80
Discussion
This study assessed the reliability of p53 IHC as a surrogate marker for TP53 mutations in a large cohort of MBCL patients. For many years, the implementation of tNGS for routine daily diagnostic procedures for MBCLs placed us in the unique setting to directly compare p53 IHC with TP53 mutation analyses. Our results showed an overall accuracy of 84.5% for p53 IHC, with a corresponding sensitivity of 65.7% and specificity of 92.1%. Although the specificity was high, the sensitivity was comparatively low, indicating a significant risk of missing TP53 mutations when using p53 IHC alone, potentially leading to the misevaluation of high-risk patients. Hence, these results indicate that p53 IHC is inadequate as a surrogate marker for TP53 mutations in the diagnostic workup of MBCL patients. The performance of p53 IHC varied across different subtypes of MBCLs. Specifically, our data suggest that p53 IHC had the least sensitivity for identifying TP53 mutations in patients with IBCL. This highlights the heterogeneity of MBCLs and the importance of selecting appropriate diagnostic techniques. Therefore, molecular analysis is advised instead of p53 immunohistochemistry in all patients with MBCL, particularly for further advancements of risk-directed therapies based on TP53 mutation status.
Numerous studies in solid tumors have demonstrated the efficacy of p53 IHC as a surrogate marker for TP53 mutations, with overall accuracies and sensitivities exceeding 90% [13–15]. Although our high specificity rates are comparable to previous findings in solid tumors, this study shows a lower overall accuracy and sensitivity of p53 IHC in MBCL [13–17]. Studies evaluating the accuracy of p53 IHC in comparison to TP53 mutation status in lymphomas are scarce and previous studies have primarily centred on (D)LBCL and MCL, rather than the broader MBCL population. While the sensitivity of the IHC was the lowest performance rate in our study, these other studies have found even lower overall sensitivity rates. For instance, in a DLBCL cohort, Xu-Monette et al. observed a sensitivity of 48.9% and a specificity of 94.9%, using the same p53 expression cutoff of > 50% (3). Zenz et al. reported a sensitivity of 57.1% and specificity of 97.5% in (D)LBCL [26]. In DLBCL, NOS Peroja et al. reported a sensitivity of 55.6% and specificity of 90.8% with corresponding positive and negative predictive values of 31.3% and 96.4%, respectively [21]. In MCL, Rodrigues et al. reported a sensitivity of 82% and specificity of 100%, which is slightly higher than our findings of 75% and 91%, respectively [27]. In summary, while p53 IHC may serve as an effective surrogate marker for TP53 mutations in solid tumors, its efficacy in MBCL is less reliable. The reasons for the lower sensitivity of MBCL compared to solid tumors are not yet fully understood. Possible explanations could include technical processing of the tissue or the intrinsic tumor factors. Our study and previous findings highlight the need for alternative or complementary techniques to accurately identify TP53 mutations and stratify high-risk patients in lymphomas.
In this study, TP53 mutations were detected in 102 (29%) patients. The prevalence rates of TP53 mutations in CLL, MCL, and (D)LBCL have been reported to range from approximately 10% to 30%, which aligns with our findings and those of other studies [2, 4, 8]. Similar to our findings, previous studies on (D)LBCL, MCL and IBCL have shown that nonsynonymous or missense mutations in the TP53 gene are the most frequent, often accounting for over 80% of mutations, followed by nonsense mutations [3, 28–31]. Consistent with other studies, our study found that TP53 mutations were most often located in exons 5 through 8 in (D)LBCL and MCL. In IBCL, TP53 mutations are relatively uncommon, ranging from 10 to 20%, and are often associated with poor prognosis and/or transformation [32]. There is limited data on specific exon locations in IBCL, but overall and similar to (D)LBCL, exons 5 through 8 are frequently affected, without a clear subtype-specific pattern [33–35]. Notably, TP53 mutations in exon 5–8 are found in 94.2% of all tumors, as reported in the IARC database [4].
A major strength of this study is the inclusion of a large patient cohort with a wide variety of MBCL subtypes, including IBCL. This is a retrospective study including 354 patients diagnosed with BCL between 2017–2022. At our center, the implementation of molecular analysis with our in-house developed B-cell tNGS panel in the diagnostic workup of lymphoma patients has expanded greatly over the years. In the beginning (2017–2019), the panel was mainly used for (D)LBCL patients, which explains why some subtypes (e.g., MCL, CLL and FL) are underrepresented in this study. In the subsequent period (2019–2022), tNGS has been applied to all our B-cell lymphoma patients.
Another strength is that both IHC and tNGS were executed and interpreted in a single laboratory, following our standard diagnostic routine, reducing potential technical errors or inter-variabilities between independent hospitals. Incorrect processing, interlaboratory differences, and decalcified tumor tissue can all negatively impact the performance of p53 IHC and result in false negative and false positive staining patterns. Therefore, for interpretation of the IHC it is best if one protocol is used, as performed in our study. A total of 55 cases were discordant between p53 IHC and TP53 mutational analysis, 20 FP and 35 FN. Of the thirteen cases with complete absence of p53 expression without a mutation, eight were decalcified bone (marrow) biopsies, of which most were LPL (n = 7) and one DLBCL (online resource 2). The suboptimal performance of the IHC in these cases could be attributed to antigen loss during the decalcification process, underscoring a critical limitation of the p53 IHC in routine diagnostic procedures. However, this explanation is not certain, and additional analyses excluding all decalcified samples demonstrated similar performance rates overall and per lymphoma subtype.
There were seven cases with an overexpression pattern, while no TP53 mutation was detected. No additional mutational analysis was performed to confirm the wild type TP53 status. DNA and tNGS analysis of all discordant cases were of good quality. Details of all discordant cases are depicted in online resource 3. The tNGS panel used in this study is particularly suitable and widely accepted for the identification of pathogenic mutations, so the likelihood that additional mutational analysis would detect a pathogenic TP53 mutation is very low. With this NGS panel Copy Number Variant (CNV) analysis was not possible, therefore an undetected TP53 deletion could be the underlying cause of some of the false positive cases. However, in the case of a TP53 deletion without an accompanying mutation, the function of the p53 protein is typically preserved with an accompanying wild type IHC expression pattern. In addition, homozygous TP53 deletions are exceedingly rare.
Our data suggests that wild type cases with strong staining in < 50% of tumor cells cannot be attributed to the relative high number of false negative wild type cases and are also present in the true negative category. Strong staining in tumor cells < 50% would be classified as wild type, however we encountered this expression pattern only once in the false negative category (n = 35), and eight times in the true negative category (n = 232). The other 258 cases had an unambiguous wild type staining pattern with p53 expression in 10–20% of the tumor cells with weak to moderate staining intensity.
Of the seven cases with a complete absence p53 IHC pattern, six had an expected truncating mutation. There was one case with a missense mutation and an unexpected null mutant IHC pattern, without a direct causal explanation.
In recent years, molecular research has gained considerable importance not only in routine diagnostics for lymphoma subtype classification, but also in prognosis and thus patient management. There is a significant research focus on stratifying lymphoma patients who are at high risk of treatment failure with conventional chemotherapy for targeted treatment strategies based on mutational profiles, with TP53 serving as a critical genetic marker. As mentioned earlier, integrating routine lymphoma diagnostics with prospective clinical data is crucial to comprehensively understand the real-world impact of risk-based profiling and make important advancements in this field [12]. Currently, p53 IHC is a widely available marker that is implemented in most diagnostic pathology departments as part of the routine diagnostic workup. Although p53 IHC is a convenient and well-interpretable marker that is faster and less expensive than tNGS, this study demonstrates that the sensitivity of the IHC alone is insufficient and may result in the misclassification of a high-risk patient. Secondly, despite the finding that a true overexpression pattern of p53 IHC is strongly indicative of a TP53 mutation (specificity 92%), it is not certain. The false-positive rate remains substantial, especially when considering an important treatment decision based on TP53 mutational status. Therefore, tNGS confirmation over p53 IHC should be strongly considered in MBCL. In cases where additional molecular analysis is not possible or available, p53 IHC alone could be used with caution considering the limitations that were shown here.
In conclusion, this study shows that p53 IHC is an insufficient surrogate marker for TP53 mutational status in MBCL patients in an unique real-world daily diagnostic setting. By using p53 IHC alone there is a significant risk a TP53 mutation will be missed, resulting in misclassification of a high-risk patient. Therefore, molecular analysis is recommended over p53 IHC in all patients with MBCL, especially for further development of risk-directed therapies based on TP53 mutation status.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Lieneke Steeghs and Demi van Egmond of the Department of Pathology of the LUMC for their technical assistance.
Author contribution
P.M.J., L.M.H., A.H.G.C. and A.D assessed the pathology. R.A.L, T.W., D.R., R.E., and J.S.P.V. designed and optimized the (lymfV1) tNGS sequencing panel. D.R., F.A.G and R.E. analyzed and interpreted the molecular data. L.M.H., P.M.J. and J.S.P.V analyzed and interpreted the data and wrote the manuscript. R.A.L.G., F.A.G., T.N., A.D., T.J.A.D., A.S.S. and H.V. performed review and revision of the paper. All authors read and approved the final paper.
Funding
The authors did not receive support from any organization for the submitted work.
Arjan Diepstra: research funding Takeda.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with the Dutch Code for Proper Secondary Use of Human Tissue, the local institutional board requirements, and the revised Declaration of Helsinki (2008). Approval with a waiver of consent was obtained from the LUMC's medical ethics committee (B16.048).
Conflicts of interest
The authors do not declare any conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Patty M. Jansen and Joost S. P. Vermaat Contributed equally to this work.
References
- 1.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R et al (2016) The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127(20):2375–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu-Monette ZY, Medeiros LJ, Li Y, Orlowski RZ, Andreeff M, Bueso-Ramos CE et al (2012) Dysfunction of the TP53 tumor suppressor gene in lymphoid malignancies. Blood 119(16):3668–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu-Monette ZY, Wu L, Visco C, Tai YC, Tzankov A, Liu WM et al (2012) Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood 120(19):3986–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P et al (2007) Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 28(6):622–629 [DOI] [PubMed] [Google Scholar]
- 5.Vazquez A, Bond EE, Levine AJ, Bond GL (2008) The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov 7(12):979–987 [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Chen C, Xu Z, Scuoppo C, Rillahan CD, Gao J et al (2016) Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531(7595):471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng J, Demeulemeester J, Wedge DC, Vollan HKM, Pitt JJ, Russnes HG et al (2017) Pan-cancer analysis of homozygous deletions in primary tumours uncovers rare tumour suppressors. Nat Commun 8(1):1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M et al (2019) Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep 28(5):1370–84.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallek M, Al-Sawaf O (2021) Chronic lymphocytic leukemia: 2022 update on diagnostic and therapeutic procedures. Am J Hematol 96(12):1679–1705 [DOI] [PubMed] [Google Scholar]
- 10.Malcikova J, Pavlova S, Kunt Vonkova B, Radova L, Plevova K, Kotaskova J et al (2021) Low-burden TP53 mutations in CLL: clinical impact and clonal evolution within the context of different treatment options. Blood 138(25):2670–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiappella A, Diop F, Agostinelli C, Novo M, Nassi L, Evangelista A et al (2022) Prognostic impact of TP53 mutation in newly diagnosed diffuse large B-cell lymphoma patients treated in the FIL-DLCL04 trial. Br J Haematol 196(5):1184–1193 [DOI] [PubMed] [Google Scholar]
- 12.Vermaat JS, Pals ST, Younes A, Dreyling M, Federico M, Aurer I et al (2015) Precision medicine in diffuse large B-cell lymphoma: hitting the target. Haematologica 100(8):989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vermij L, Léon-Castillo A, Singh N, Powell ME, Edmondson RJ, Genestie C et al (2022) p53 immunohistochemistry in endometrial cancer: clinical and molecular correlates in the PORTEC-3 trial. Mod Pathol 35(10):1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raffone A, Travaglino A, Cerbone M, De Luca C, Russo D, Di Maio A et al (2020) Diagnostic accuracy of p53 immunohistochemistry as surrogate of TP53 sequencing in endometrial cancer. Pathol Res Pract 216(8):153025 [DOI] [PubMed] [Google Scholar]
- 15.Li J, Wang J, Su D, Nie X, Liu Y, Teng L et al (2021) p53 Immunohistochemistry Patterns Are Surrogate Biomarkers for TP53 Mutations in Gastrointestinal Neuroendocrine Neoplasms. Gastroenterol Res Pract 2021:2510195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung YN, Kim D, Kim J (2022) p53 immunostaining pattern is a useful surrogate marker for TP53 gene mutations. Diagn Pathol 17(1):92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köbel M, Piskorz AM, Lee S, Lui S, LePage C, Marass F et al (2016) Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J Pathol Clin Res 2(4):247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M et al (2015) Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525(7568):206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivlin N, Brosh R, Oren M, Rotter V (2011) Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2(4):466–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Young KH, Medeiros LJ (2018) Diffuse large B-cell lymphoma. Pathology 50(1):74–87 [DOI] [PubMed] [Google Scholar]
- 21.Peroja P, Pedersen M, Mantere T, Nørgaard P, Peltonen J, Haapasaari KM et al (2018) Mutation of TP53, translocation analysis and immunohistochemical expression of MYC, BCL-2 and BCL-6 in patients with DLBCL treated with R-CHOP. Sci Rep 8(1):14814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Groen RAL, van Eijk R, Böhringer S, van Wezel T, Raghoo R, Ruano D et al (2021) Frequent mutated B2M, EZH2, IRF8, and TNFRSF14 in primary bone diffuse large B-cell lymphoma reflect a GCB phenotype. Blood Adv 5(19):3760–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bruijn I, Li X, Sumer SO, Gross B, Sheridan R, Ochoa A et al (2022) Genome Nexus: A Comprehensive Resource for the Annotation and Interpretation of Genomic Variants in Cancer. JCO Clin Cancer Inform 6:e2100144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franklin by Genoox [Available from: https://franklin.genoox.com/clinical-db/home.
- 25.Thompson BA, Spurdle AB, Plazzer JP, Greenblatt MS, Akagi K, Al-Mulla F et al (2014) Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet 46(2):107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zenz T, Kreuz M, Fuge M, Klapper W, Horn H, Staiger AM et al (2017) TP53 mutation and survival in aggressive B cell lymphoma. Int J Cancer 141(7):1381–1388 [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues JM, Hassan M, Freiburghaus C, Eskelund CW, Geisler C, Räty R et al (2020) p53 is associated with high-risk and pinpoints TP53 missense mutations in mantle cell lymphoma. Br J Haematol 191(5):796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young KH, Leroy K, Møller MB, Colleoni GW, Sánchez-Beato M, Kerbauy FR et al (2008) Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood 112(8):3088–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eskelund CW, Dahl C, Hansen JW, Westman M, Kolstad A, Pedersen LB et al (2017) TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood 130(17):1903–1910 [DOI] [PubMed] [Google Scholar]
- 30.Halldórsdóttir AM, Lundin A, Murray F, Mansouri L, Knuutila S, Sundström C et al (2011) Impact of TP53 mutation and 17p deletion in mantle cell lymphoma. Leukemia 25(12):1904–1908 [DOI] [PubMed] [Google Scholar]
- 31.Greiner TC, Dasgupta C, Ho VV, Weisenburger DD, Smith LM, Lynch JC et al (2006) Mutation and genomic deletion status of ataxia telangiectasia mutated (ATM) and p53 confer specific gene expression profiles in mantle cell lymphoma. Proc Natl Acad Sci U S A 103(7):2352–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krug U, Ganser A, Koeffler HP (2002) Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene 21(21):3475–3495 [DOI] [PubMed] [Google Scholar]
- 33.O’Shea D, O’Riain C, Taylor C, Waters R, Carlotti E, Macdougall F et al (2008) The presence of TP53 mutation at diagnosis of follicular lymphoma identifies a high-risk group of patients with shortened time to disease progression and poorer overall survival. Blood 112(8):3126–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koduru PR, Raju K, Vadmal V, Menezes G, Shah S, Susin M et al (1997) Correlation between mutation in P53, p53 expression, cytogenetics, histologic type, and survival in patients with B-cell non-Hodgkin’s lymphoma. Blood 90(10):4078–4091 [PubMed] [Google Scholar]
- 35.Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E et al (2011) Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol 29(16):2223–2229 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.