Abstract
Online Mendelian Inheritance in Animals (OMIA) is a freely available curated knowledgebase that contains information and facilitates research on inherited traits and diseases in animals. For the past 29 years, OMIA has been used by animal geneticists, breeders, and veterinarians worldwide as a definitive source of information. Recent increases in curation capacity and funding for software engineering support have resulted in software upgrades and commencement of several initiatives, which include the enhancement of variant information and links to human data resources, and the introduction of ontology-based breed information and categories. We provide an overview of current information and recent enhancements to OMIA and discuss how we are expanding the integration of OMIA into other resources and databases via the use of ontologies and the adaptation of tools used in human genetics.
Keywords: Mendelian inheritance, Likely causal variants, Phene, Web database, Ontology
Introduction
OMIA (Nicholas et al. 2024) is a freely available, curated, online knowledgebase that provides users with up-to-date summary information on the known functional variants in vertebrate animals, together with background information on known inherited disorders and other inherited traits. OMIA is modelled on and reciprocally hyperlinked to Online Mendelian Inheritance in Man (OMIM, Online Mendelian Inheritance in Man, 2024), and provides further links to PubMed and Gene records at the National Center for Biotechnology Information (NCBI, Sayers et al. 2024), to the European Bioinformatics Institute (EBI)’s Ensembl (Martin et al. 2023) and recently introduced links to the Mondo disease ontology (Mondo, Vasilevsky et al. 2022) and the Vertebrate Breed Ontology (VBO, Mullen et al. 2024).
At the beginning of 2021, the central OMIA team increased from one to two curators, and bequest funding from 2021 onwards has enabled regular software engineering support. In this paper we provide an overview of OMIA data and a summary of recent software updates, major enhancements to likely causal variant tables and OMIA-OMIM hyperlinks, introduction of links to Mondo and VBO, and the launch of Pioneers of Mendelian Inheritance in Animals (PMIA). We also provide an update on current initiatives that focus on the use of ontologies to expand the interoperability of OMIA with other resources such as the Anstee Hub for Inherited Diseases in Animals (AHIDA, Tammen et al. 2021).
History of OMIA
The origin and development of OMIA have been described by Nicholas (2021). Briefly, one of us (FN) began a compendium of mainly Mendelian phenes in animals (called Mendelian Inheritance in Animals, MIA) way back in 1980, in the pre-internet age. The ‘O’ was added when MIA became available on the internet on 26th May 1995. Details of the developments over the decades, and the people involved, are recorded in the Acknowledgements tab (https://omia.org/acknowledgements/) of the OMIA home page.
Until roughly five years ago, curation was predominately conducted single-handedly by one curator (FN), and limited funding restricted access to urgently needed software upgrades and modifications. The funding situation began to improve in 2017 when the Sydney Informatics Hub (SIH) started to provide University-funded bioinformatic/software-engineering resources for OMIA enhancement projects. From 2021 a bequest to the Sydney School of Veterinary Science has provided funding for the creation and continual support of AHIDA, a portal for reporting and surveillance of inherited diseases in animals in Australia. Because OMIA is a vital resource for AHIDA, some of the AHIDA funding has been allocated to SIH for the enhancement and continual maintenance of OMIA.
Since August 2010, the OMIA database and website have been using Django software, a high-level Python web framework. In July 2021, software packages were upgraded from Python 2 to Python 3 (van Rossum and Drake 2009) and from Django 1.9 to 3.2, and in 2024 to Django 4.2 (Django Software Foundation 2023), and automated tests were added with the aim of future-proofing OMIA. Site code was optimised to improve response times. Furthermore, search options were refined to increase the number of fields that are searched in a ‘quick’ search to improve user experience.
Scope of OMIA
OMIA focuses on traits and diseases (‘phenes’) with confirmed or suspected Mendelian modes of inheritance. In addition, several phenes with unknown or complex modes of inheritance and phenes caused by somatic variants, genetically engineered modifications or genome editing are also included. While most OMIA entries are for the major domesticated animal species, more than 500 (mainly vertebrate) animal species have entries in OMIA. Information about humans and model organisms such as mouse, rat, zebrafish, and western clawed frog (xenopus) are not included, as they have dedicated species-specific resources.
As a means of highlighting and summarising advances in Mendelian inheritance in animals, OMIA also has a tab that lists Landmark papers (reporting major advances), Reviews (summarising current knowledge) and (genomic) Resource papers (reporting species maps and genome assemblies), all listed in chronological order, to provide an historical perspective.
OMIA is a globally used knowledgebase. Google Analytics user data for the past year identified 131,000 sessions by 54,000 users from 116 countries. In June 2024, OMIA included information on 2,533 phenes across 540 species, contained 5,133 phene-species entries and included a total of 32,767 references. Core statistics for key species are summarised in Table 1.
Table 1.
Summary of OMIA phene information (accessed June 21 2024). For an up-to-date list with hyperlinks to individual entries see the summary table at the OMIA homepage: https://omia.org/home/
| Traits (phenes) | Dog | Cattlea | Cat | Pig | Sheep | Horse | Chicken | Goat | All species |
|---|---|---|---|---|---|---|---|---|---|
| All traits: disease and non-disease | 948 | 686 | 436 | 391 | 322 | 295 | 259 | 126 | 5133 |
| All Mendelian traits: disease and non-disease | 426 | 308 | 144 | 141 | 127 | 63 | 138 | 26 | 1936 |
| • with at least one known likely causal variant | 348 | 208 | 113 | 68 | 61 | 48 | 57 | 17 | 1196 |
| • with references to genetically engineered or modified animals | 5 | 15 | 2 | 98 | 12 | 0 | 11 | 6 | 237 |
| Mendelian diseases | 387 | 270 | 116 | 111 | 91 | 48 | 98 | 12 | 1403 |
| • with at least one known likely causal variant | 322 | 189 | 93 | 53 | 43 | 37 | 34 | 8 | 875 |
a taurine cattle only, additional entries exist for indicine cattle
Recent enhancements: text mining
Ever since PubMed started in 1997, OMIA has been conducting daily searches of all the new papers that have been entered into PubMed in the previous 24 h. Over the years, the simple keyword-based searches were improved, providing an effective but rather labour-intensive means of identifying new papers relevant to OMIA. During 2022, for example, the daily automated PubMed literature search resulted in 17,653 hits, of which 719 papers (on average 2 papers/day) were identified to be added to OMIA. In other words, the number of false positives was still depressingly high. Realising that the advent of text-mining tools should provide less labour-intensive strategies, several were tried when they became available in the early 2020s. By far the most successful turned out to be a machine-learning model developed using Microsoft’s PubMedBERT (Gu et al. 2021), which was adapted for OMIA in February 2023, and has been in daily use ever since. A total of 2,811 relevant papers were added to OMIA in the first 482 days of using the tool (average of 5.8 papers/day).
Recent enhancements: standardised nomenclature and ontologies in OMIA
Integration of phenotype category, breed and disease ontologies
Three major current projects relate to the introduction of ontologies (Table 2) for phenotype category, breed and disease definition into OMIA. Ontologies are controlled vocabularies that represent knowledge both by their meaning and their relationship to each other and provide unique numerical identifiers to enable advanced computational analysis. We aim to harmonize breed and disease definitions in OMIA in a computer-accessible format, thus enabling integration with other global online resources and integration with the submission portal of AHIDA (Tammen et al. 2021), which was launched in Australia in 2023.
Table 2.
Phenotype, breed and disease ontologies used in OMIA
| Ontology | ID | Description | Reference |
|---|---|---|---|
| Mammalian Phene Ontology | MP | An ontology of pre-coordinated phenotype terms, definitions and synonyms that can be used to describe mammalian phenotypes. It organizes phenotype terms into major biological system headers. | Smith and Eppig 2009 |
| Mondo disease ontology | Mondo | A semi-automatically constructed ontology that merges multiple disease resources to yield a coherent merged ontology. | Vasilevsky et al. 2022 |
| Vertebrate Breed Ontology | VBO | An ontology created to serve as a single computable resource for vertebrate breed names. | Mullen et al. 2024 |
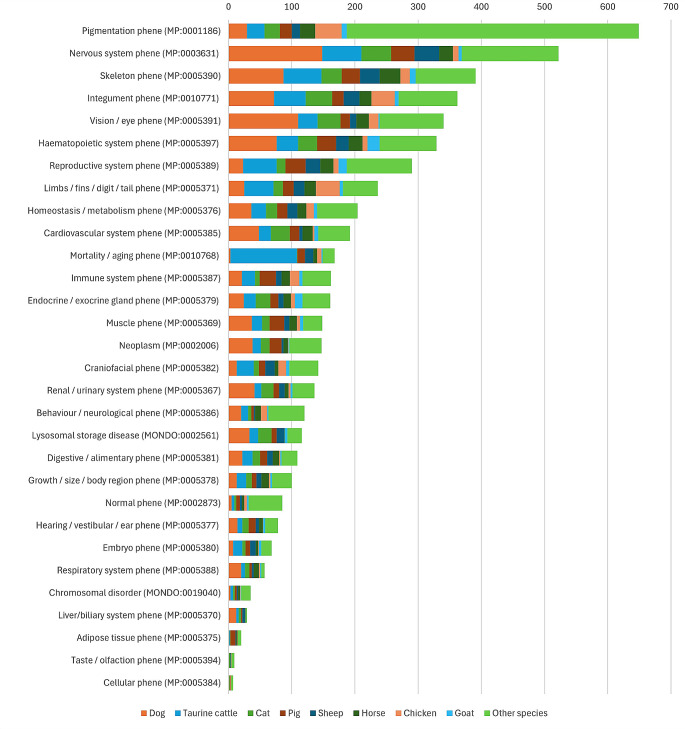
OMIA previously included a home-grown list of 20 ‘phene categories’ that could be used in ‘Advanced Search’ and in the ‘Browse’ page to create category-specific phene lists, but many OMIA phenes did not have a phene category specified. To allow for comprehensive categorisation and for easy identification of phenes based on body system, OMIA’s 20 phene categories have been replaced with 28 major biological system headers from the Mammalian Phenotype (MP) Ontology (Smith and Eppig 2009) and two headers from Mondo (Vasilevsky et al. 2022) (Fig. 1). To facilitate this, the ‘category’ field has been included in phene-species pages and each phene has been linked to at least one category by a curator (IT). Entries for the category ‘Chromosomal disorder (MONDO:0019040)’ are low as chromosomal abnormalities were previously listed in Online Cytogenetics of Animals (OCOA), hosted by the University of Sydney. This resource is no longer maintained, and we aim to transfer information to OMIA in the future.
Fig. 1.
Summary of OMIA phene category information for key species (accessed 21 June 2024). Corresponding Mammalian Phenotype Ontology (MP) and Mondo identifiers are included for each OMIA category. For an up-to-date list with hyperlinks to individual entries see the OMIA ‘Browse’ page: https://omia.org/browse/
Recognising the need to replace OMIA’s home-grown breed list with a computable comprehensive list of standardised breed names, the OMIA team instigated the creation of the Vertebrate Breed Ontology (VBO, Mullen et al. 2024) in a project led by the Monarch Initiative, in collaboration with colleagues from Iowa State University and with FAO colleagues responsible for the Domestic Animal Diversity Information System (DAD-IS). Breed information in OMIA phene-species entries and in variant tables has been replaced with links to VBO, and OMIA advanced searches allow searches by VBO breed name. Breed information is currently not entered for all phene-species or variants and is likely to be incomplete for some entries, as this update requires review of more than 5000 phene-species entries and the information is continuously changing. Currently 1297 (25%) of OMIA’s 5133 phene-species entries list at least one breed. We aim to prioritise adding this information to Mendelian phenes with known variants and variants. Of 1198 such phenes, 796 (66%) include breed information and 1194 (75%) of the 1590 likely causal variants in OMIA have at least one breed listed. It is noted that not all entries are expected to have breed information (e.g. non-domesticated species, entries relating to genetically engineered or modified animals). Recent publications reporting genotyping of large cohorts of animals for known likely causal variants (e.g. Anderson et al. 2022; Donner et al. 2023; Durward-Akhurst et al. 2024) can be useful to update breed information for known variants and this process is greatly facilitated if OMIA phene and OMIA variant and VBO breed identifiers are included.
Finally, we are working towards integrating OMIA information into Mondo (Vasilevsky et al. 2022), a global disease ontology that aims to harmonise disease definitions across the world. So far, a new field has been created in OMIA to enable addition of hyperlinks to Mondo when an OMIA phene name is a close match to a human disease term in Mondo. Currently 221 of such links have been included in OMIA. Mondo also lists terms for non-human animal diseases and to date has used OMIA as a source for 971 inherited disease terms for non-human animals. However, the non-human animal information in Mondo is still under development.
Standardised nomenclature for likely causal variants in OMIA variant tables
The introduction of variant tables in OMIA was reported by Tammen and Nicholas (2018), who indicated that the ultimate aim was to provide an European Variation Archive (EVA) rs ID (European Variation Archive 2024) for all variants, to reduce the need to standardise and update variant information in OMIA. In 2018, OMIA listed 926 likely causal variants of which 43% had a genomic position and only 7% were listed with an EVA rs ID (Tammen and Nicholas 2018). We recognised that EVA does not accommodate all types of variants, very few authors of OMIA-relevant papers submit variant information to EVA, and new EVA IDs are allocated infrequently.
Consequently, with the help of many colleagues (see OMIA’s acknowledgement page for details: https://omia.org/acknowledgements/) we reviewed and standardised historic variant information in OMIA and applied Human Genome Variation Society (HGVS) recommendations (den Dunnen et al. 2016), with the aim to report genomic, cDNA and protein location coordinates based on a recent reference genome assembly where possible. In October 2021, variants listed in OMIA that were lacking an EVA ID but had standardised location information were submitted to EVA using a new OMIA pipeline for export of variant information in VCF for submission to EVA. Currently 436 (27%) of the 1590 likely causal variants in OMIA list an rs ID and 1241 (78%) of the variants list genomic coordinates in a reference genome. For key domesticated species (dog, cattle, cat, horse and donkey) more than 90% of the variants list coordinates in HGVS format (Table 3).
Table 3.
Summary of OMIA variant information (accessed 21 June 2024). For an up-to-date list, see https://omia.org/results/?search_type=advanced&result_type=variant&singlelocus=yes&characterised=yes
| OMIA variants | Dog | Cattlea | Cat | Pig | Sheep | Horse | Chicken | Goat | All species |
|---|---|---|---|---|---|---|---|---|---|
| All known likely causal variants for all Mendelian traits: disease and non-disease | 528 | 274 | 195 | 67 | 88 | 107 | 72 | 21 | 1590 |
| • All known likely causal variants for Mendelian diseases | 483 (91%) | 247 (90%) | 147 (75%) | 50 (75%) | 49 (56%) | 89 (83%) | 39 (54%) | 11 (52%) | 1193 (75%) |
| • Variants with g. coordinates | 477 (90%) | 244 (89%) | 184 (94%) | 41 (61%) | 80 (91%) | 101 (94%) | 45 (63%) | 12 (57%) | 1241 (78%) |
| • Variants with EVA rs ID | 59 (11%) | 185 (68%) | 44 (23%) | 30 (45%) | 23 (26%) | 66 (62%) | 18 (25%) | 8 (38%) | 435 (27%) |
a taurine cattle only, additional entries exist for indicine cattle
Since June 2024, OMIA has been implementing the addition of NCBI-based identifiers for genomic, transcript and protein IDs to the coordinates for each variant. This process has now been completed for horse and donkey variants and is in progress for other species. However, such information is lengthy (e.g. the coordinates for the variant causing hyperkalemic periodic paralysis (HYPP) in horses are listed as NC_009154.3:g.15474228C>G; NM_001081761.1:c.4248C>G; NP_001075230.1:p.(F1416L)) and these will change when a new reference genome becomes available.
To provide unique, unchanging and easy-to-use identifiers for all likely causal variants in animals that can be used widely (e.g. by genotyping service providers to ensure greater transparency in relation to DNA testing), OMIA numerical variant identifiers (OMIAvariant ID) have been presented since 2021 in the first column of all OMIA variant tables. Pleasingly, several review papers have started to include OMIAvariant IDs in their tables (e.g. Anderson et al. 2022; Donner et al. 2023; Meadows et al. 2023). Information about likely causal variants is summarised in Table 3.
Review of OMIA-OMIM hyperlinks
Since 1997, OMIA has been reciprocally hyperlinked to OMIM. Links to OMIM are created by OMIA curators when new phenes are entered into OMIA. In the past, this focused on adding OMIM phenotype identifiers (IDs), while OMIM gene IDs were rarely added. OMIM automatically downloads OMIA phene IDs that have an OMIM ID link once a week and updates OMIM accordingly. In OMIA, separate fields for ‘OMIM phene’ and ‘OMIM gene’ hyperlinks were recently created, and we reviewed OMIA-OMIM hyperlinks for all OMIA phenes for which a likely causal variant has been identified in at least one species. OMIM links were confirmed, deleted, or added. This review resulted in confirmation of 607 OMIM links, addition of 683 OMIM links and deletion of 46 OMIM links. Most of the added OMIM links were OMIM gene IDs (n = 493), as these were in the past not routinely added to OMIA. OMIA currently lists 2769 models of human traits based on links to OMIM and 2228 OMIM entries have a link to OMIA. However, a large list of OMIA phenes without known likely causal variants have not yet been reviewed, as it is more speculative to identify homology between human and animal phenes if the likely causative gene has not yet been identified. The revision of OMIA-OMIM hyperlinks and the above-mentioned links to Mondo will facilitate comparative-medicine-related research approaches.
Recent enhancements: PMIA
In 2022, Pioneers of Mendelian Inheritance in Animals (PMIA) was added to OMIA, accessible from the home page. This project comprises a series of commentaries on papers that illustrate the early discoveries of Mendelian inheritance in animals. PMIA was first announced on Mendel Day (8 March) in 2022, and launched as part of OMIA on the 8th of July 2022, two weeks before the bicentenary of Mendel’s birth on the 22nd. Currently PMIA includes detailed commentaries by FN on 15 papers that illustrate the early discoveries of Mendelian inheritance in animals. Included are the first report of Mendelian inheritance in non-rodent animals, namely the inheritance of five trait pairs in chickens (including some drawings by Bateson) and the first reports in cattle and goats (all in 1902); followed by the first reports in guinea pigs, rabbits and fish (1903); cats (1904); and sheep (1905).
Future enhancements under development
Development of ontology-based clinical synopses in OMIA
Due to the large number of known single gene diseases in humans (OMIM currently lists 6,474 single gene diseases and traits) and the increasing number of known single gene diseases in non-human animals, it can be very challenging for medical clinicians and veterinarians to diagnose rare inherited diseases. In human medicine the term “diagnostic odyssey” is used to highlight that patients frequently undergo unnecessary tests and procedures and experience lengthy delays in getting a correct diagnosis and access to effective care. Tools have been developed to assist medical professionals to more efficiently identify lists of differentials for suspected inherited diseases based on clinical presentation of their patients to shorten this diagnostic odyssey. One of these tools is Phenomizer (Köhler et al. 2009, 2019), which, in essence, matches a set of clinical signs coded as human phene ontology (HPO) concepts with documented inherited phenes in compendia such as Online Mendelian Inheritance in Man (OMIM) to provide a list of ranked differentials. To enable such automated matching OMIM includes a clinical synopsis in addition to free text inforamtion for each disease. The clinical synopsis is a list of ontology-based terms that describe the disease.
To enable the development of a similar tool for veterinarians, we are aiming to include ontology-based clinical synopses to OMIA and are currently implementing the use of PhenoTagger (Luo et al. 2021) to facilitate this process. This tool uses text-mining algorithms and machine learning to extract concepts from unstructured text, such as exists in current OMIA curation fields for clinical signs and pathological findings and published papers, which are then compared with a dictionary of concepts embodied in phene ontologies such as the Human Phenotype Ontology (HPO). In other words, the unstructured text can be turned into short lists of computerised, standardised words in ontologies, depicting particular phenes. We are currently training PhenoTagger using the cross-species phenotype ontology upheno2 (Matentzoglu et al. 2019) and have created curation fields that will in the near future show clinical synopsis for OMIA entries. So, by combining PhenoTagger with Phenomizer, each trained on and/or adapted to non-human and non-model-animal information such as exists in OMIA, it should be possible to use these tools to greatly assist the diagnosis of isolated occurrences of “novel” clinical cases of inherited diseases in the veterinary context. Then, via the long-established reciprocal links between OMIM and OMIA (described above), combined with the existing links between OMIM and model-animal resources such as the Mouse Phenome Database (Bogue et al. 2023), it should be possible to greatly strengthen the potential of comparative biology to be applied in the diagnosis of inherited diseases across all vertebrates.
Some “novel” cases will be readily shown to be new cases of inherited diseases that have already been reported, and for which a DNA test may already exist. Others will have been reported in the same animal species but with no likely causal variant yet discovered, in which case the “novel” case, combined with relevant samples from relatives, can be added to the samples already available for research into the causal variant. Others will have been reported in other vertebrate species, in one or more of which a likely causal variant has also been reported, in which case a functional candidate causal gene will have been revealed, greatly enhancing the chance of discovering a likely causal variant in the species in which the “novel” case occurred.
Evidence-based classification of the predicted pathogenicity of likely causal variants
While the OMIA list of likely causal variants is as up-to-date as possible, it is realised that there is substantial variation in the extent to which variants in that list are truly causal. In general, variants are added to the OMIA list if they are claimed to be likely causal in a peer-reviewed paper. The danger is that breeders and genotyping providers will interpret the presence of a variant in the OMIA list as a licence to use that variant as a diagnostic direct DNA test. To address this danger, a warning sign has been included at the top of all OMIA lists of likely causal variants: “Inclusion of a variant in this table does not automatically mean that it should be used for DNA testing”.
This same problem was long-ago addressed for human likely causal variants by human geneticists. Indeed, in 2015 a set of guidelines was developed by the American College of Medical Genetics and Genomics (ACMG), the Association for Molecular Pathology (AMP) and the College of American Pathologists (CAP). Developed in the context of inherited disease, these guidelines provide evidence-based criteria for classifying likely causal variants on the basis of all available evidence (not just the initial publication) as ‘pathogenic’, ‘likely pathogenic’, ‘uncertain significance’, ‘likely benign’, and ‘benign’ (Richards et al. 2015). In the ensuing years, these five categories have become the global standard for classifying human likely causal variants. The criteria are under continual review, now under the umbrella of the Gene Curation Coalition (GenCC; DiStefano et al. 2022). A web-based tool, the Variant Curation Interface (VCI; Preston et al. 2022) has been developed to facilitate the classification of variants.
At its 2023 conference, the International Society for Animal Genetics (ISAG) created a Standing Committee for Animal Genetic Testing Standardisation. One of its four purposes is to enable the above global standards for human variants to be adapted and applied to non-human and non-model animal species. To this end, an ISAG Variant Pathogenicity Work Group is about to commence work. Its task will be to work in conjunction with GenCC to allocate each likely causal variant in the OMIA list to one of the above five categories, which can then be clearly indicated in the OMIA variant tables. This will be a great and significant step forward.
Another important development concerning OMIA likely causal variants is a recently published paper from a human genetics team in Toronto, Canada (Haque et al. 2024). Led by Gregory Costain, this group showed how a knowledge of OMIA variants can substantially enhance the effectiveness of pathogenicity classification of human likely causal variants. Thus, human genetics can benefit from knowledge of homologous non-human non-model animal variants; and vice versa.
The obvious way forward is to exploit every opportunity to provide seamless joint interrogation of all available data from all vertebrate species.
As Haque et al. (2024) conclude. “The degree to which the exponential growth of human and veterinary genomic datasets can be integrated and harnessed to improve variant interpretation across both contexts warrants additional study.”
Conclusion
Our vision for the future is that in addition to summarising information about inherited conditions in animals, OMIA becomes a global repository for standardised information (including pathogenicity classification) on likely causal variants for inherited diseases and other traits. We envisage that OMIA and the Anstee Hub for Inherited Diseases in Animals (AHIDA) will become joint resources for semi-automated diagnosis and for enhancement of research effectiveness in discovering new DNA diagnostics, for rare or emerging inherited conditions in animals. A key challenge is the workload of curating and enhancing OMIA which is currently done by a small team of academics with limited funding support. It is inevitable that omissions or errors occur, and we encourage anyone to send an email to omia.admin@sydney.edu.au to suggest additions or changes.
In human medicine, phene-level tools such as PhenoTagger and Phenomizer are now being combined with (or incorporated into) genomic-level tools such as Exomiser (Cipriani et al. 2020) and Seqr (Pais et al. 2022) to further enhance the chances of discovering a likely causal variant in “novel” cases.
The potential of adapting these same combinations/incorporations of phene and genomic tools so they can be applied jointly to all vertebrate species is currently being investigated.
The overall aim is to enable clinicians and geneticists across all vertebrate species to be able to utilise the same set of tools, so as to be able to exploit to the greatest extent possible the full potential of comparative biology to the benefit of all vertebrate species.
Acknowledgements
The Acknowledgements tab on the OMIA home page (https://omia.org/acknowledgements/) provides a detailed account of the contributions of the many people who have contributed to the development of OMIA since its inception. We are particularly grateful to Stephanie Mei Ying Shields, Xinwei Luo and Tim White for their assistance towards implementing PhenoTagger in OMIA, and to Sony Jufri and Peter Thomson for implementing and evaluating a machine-learning model for text-mining. Recent software engineering support has been possible due to support of the Sydney Informatics Hub, a core research facility of the University of Sydney, and funding from the Ronald Bruce Anstee Bequest to the Sydney School of Veterinary Science.
Author contributions
OMIA is co-authored by curator IT and founder FN with regular curation contributions from TL and software engineering support from MM IT and FN wrote the main manuscript text. All authors reviewed the manuscript.
Funding
IT and FN benefit from funding for software engineering support of OMIA (provided by MM and other colleagues within the Sydney Informatics Hub, The University of Sydney) via the Ronald Bruce Anstee Bequest to the Sydney School of Veterinary Science for the establishment and operation of the Anstee Hub for Inherited Disease of Animals (AHIDA).
Open Access funding enabled and organized by CAUL and its Member Institutions
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Anderson H, Davison S, Lytle KM, Honkanen L, Freyer J, Mathlin J, Kyöstilä K, Inman L, Louviere A, Chodroff Foran R, Forman OP, Lohi H, Donner J (2022) Genetic epidemiology of blood type, disease and trait variants, and genome-wide genetic diversity in over 11,000 domestic cats. PLoS Genet 18:e1009804. 10.1371/journal.pgen.1009804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue MA, Ball RL, Walton DO, Dunn MH, Kolishovski G, Berger A, Lamoureux A, Grubb SC, Gerring M, Kim M, Liang H, Emerson J, Stearns T, He H, Mukherjee G, Bluis J, Davis S, Desai S, Sundberg B, Kadakkuzha B, Kunde-Ramamoorthy G, Philip VM, Chesler EJ (2023) Mouse phenome database: curated data repository with interactive multi-population and multi-trait analyses. Mamm Genome 34:509–519. 10.1007/s00335-023-10014-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani V, Pontikos N, Arno G, Sergouniotis PI, Lenassi E, Thawong P, Danis D, Michaelides M, Webster AR, Moore AT, Robinson PN, Jacobsen JOB, Smedley D (2020) An improved phenotype-driven tool for rare mendelian variant prioritization: Benchmarking exomiser on real patient whole-exome data. Genes (Basel) 11:460. 10.3390/genes11040460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, Roux AF, Smith T, Antonarakis SE, Taschner PE (2016) HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat 37:564–569. 10.1002/humu.22981 [DOI] [PubMed] [Google Scholar]
- DiStefano MT, Goehringer S, Babb L, Alkuraya FS, Amberger J, Amin M, Austin-Tse C, Balzotti M, Berg JS, Birney E, Bocchini C, Bruford EA, Coffey AJ, Collins H, Cunningham F, Daugherty LC, Einhorn Y, Firth HV, Fitzpatrick DR, Foulger RE, Goldstein J, Hamosh A, Hurles MR, Leigh SE, Leong IUS, Maddirevula S, Martin CL, McDonagh EM, Olry A, Puzriakova A, Radtke K, Ramos EM, Rath A, Riggs ER, Roberts AM, Rodwell C, Snow C, Stark Z, Tahiliani J, Tweedie S, Ware JS, Weller P, Williams E, Wright CF, Yates TM, Rehm HL (2022) The Gene Curation Coalition: a global effort to harmonize gene-disease evidence resources. Genet Med 24:1732–1742. 10.1016/j.gim.2022.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Django Software Foundation (2023) Django (Version 4.2) [Computer Software], Available via: https://www.djangoproject.com/. Accessed 18 Jun 2024
- Donner J, Freyer J, Davison S, Anderson H, Blades M, Honkanen L, Inman L, Brookhart-Knox CA, Louviere A, Forman OP, Chodroff Foran R (2023) Genetic prevalence and clinical relevance of canine mendelian disease variants in over one million dogs. PLoS Genet 19:e1010651. 10.1371/journal.pgen.1010651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durward-Akhurst SA, Marlowe JL, Schaefer RJ, Springer K, Grantham B, Carey WK, Bellone RR, Mickelson JR, McCue ME (2024) Predicted genetic burden and frequency of phenotype-associated variants in the horse. Sci Rep 14:8396. 10.1038/s41598-024-57872-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Variation Archive (EVA) (2024) Available via: https://sciencegateways.org/resources/europeanvariationarchiveeva. Accessed 18 Jun 2024
- Gu Y, Tinn R, Cheng H, Lucas M, Usuyama N, Liu X, Naumann T, Gao J, Poon H (2021) Domain-specific language model pretraining for biomedical natural language processing. ACM Trans Comput Healthc 3:1–23. 10.1145/3458754 [Google Scholar]
- Haque B, Guirguis G, Curtis M, Mohsin H, Walker S, Morrow MM, Costain G (2024) A comparative medical genomics approach may facilitate the interpretation of rare missense variation. J Med Genet 20. 10.1136/jmg-2023-109760. :jmg-2023-109760 [DOI] [PMC free article] [PubMed]
- Köhler S, Schulz MH, Krawitz P, Bauer S, Dölken S, Ott CE, Mundlos C, Horn D, Mundlos S, Robinson PN (2009) Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet 85:457–464. 10.1016/j.ajhg.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Øien NC, Buske OJ, Groza T, Jacobsen JOB, McNamara C, Vasilevsky N, Carmody LC, Gourdine JP, Gargano M, McMurry JA, Danis D, Mungall CJ, Smedley D, Haendel M, Robinson PN (2019) Encoding clinical data with the human phenotype ontology for computational differential diagnostics. Curr Protoc Hum Genet 103:e92. 10.1002/cphg.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Yan S, Lai PT, Veltri D, Oler A, Xirasagar S, Ghosh R, Similuk M, Robinson PN, Lu Z (2021) PhenoTagger: a hybrid method for phenotype concept recognition using human phenotype ontology. Bioinformatics 27:1884–1890. 10.1093/bioinformatics/btab019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FJ, Amode MR, Aneja A, Austine-Orimoloye O, Azov AG, Barnes I, Becker A, Bennett R, Berry A, Bhai J, Bhurji SK, Bignell A, Boddu S, Branco Lins PR, Brooks L, Ramaraju SB, Charkhchi M, Cockburn A, Da Rin Fiorretto L, Davidson C, Dodiya K, Donaldson S, El Houdaigui B, El Naboulsi T, Fatima R, Giron CG, Genez T, Ghattaoraya GS, Martinez JG, Guijarro C, Hardy M, Hollis Z, Hourlier T, Hunt T, Kay M, Kaykala V, Le T, Lemos D, Marques-Coelho D, Marugán JC, Merino GA, Mirabueno LP, Mushtaq A, Hossain SN, Ogeh DN, Sakthivel MP, Parker A, Perry M, Piližota I, Prosovetskaia I, Pérez-Silva JG, Salam AIA, Saraiva-Agostinho N, Schuilenburg H, Sheppard D, Sinha S, Sipos B, Stark W, Steed E, Sukumaran R, Sumathipala D, Suner MM, Surapaneni L, Sutinen K, Szpak M, Tricomi FF, Urbina-Gómez D, Veidenberg A, Walsh TA, Walts B, Wass E, Willhoft N, Allen J, Alvarez-Jarreta J, Chakiachvili M, Flint B, Giorgetti S, Haggerty L, Ilsley GR, Loveland JE, Moore B, Mudge JM, Tate J, Thybert D, Trevanion SJ, Winterbottom A, Frankish A, Hunt SE, Ruffier M, Cunningham F, Dyer S, Finn RD, Howe KL, Harrison PW, Yates AD, Flicek P (2023) Ensembl 2023. Nucleic Acids Res 51:D933–D941. 10.1093/nar/gkac958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matentzoglu N, Osumi-Sutherland D, Balhoff JP, Bello S, Bradford Y, Cardmody L, Grove C, Harris MA, Harris N, Köhler S, McMurry J, Mungall C, Munoz-Torres M, Pilgrim C, Robb S, Robinson PN, Segerdell E, Vasilevsky N, Haendel M (2019) uPheno 2: Framework for standardised representation of phenotypes across species. Available via: 10.7490/f1000research.1116540.1. Accessed 18 Jun 2024
- Meadows JRS, Kidd JM, Wang GD, Parker HG, Schall PZ, Bianchi M, Christmas MJ, Bougiouri K, Buckley RM, Hitte C, Nguyen AK, Wang C, Jagannathan V, Niskanen JE, Frantz LAF, Arumilli M, Hundi S, Lindblad-Toh K, Ginja C, Agustina KK, André C, Boyko AR, Davis BW, Drögemüller M, Feng XY, Gkagkavouzis K, Iliopoulos G, Harris AC, Hytönen MK, Kalthoff DC, Liu YH, Lymberakis P, Poulakakis N, Pires AE, Racimo F, Ramos-Almodovar F, Savolainen P, Venetsani S, Tammen I, Triantafyllidis A, vonHoldt B, Wayne RK, Larson G, Nicholas FW, Lohi H, Leeb T, Zhang YP, Ostrander EA (2023) Genome sequencing of 2000 canids by the Dog10K consortium advances the understanding of demography, genome function and architecture. Genome Biol 24:187. 10.1186/s13059-023-03023-7. Erratum in: Genome Biol. (2023) 24:255. https://doi.org/10.1186/s13059-023-03101-w
- Mullen KR, Tammen I, Matentzoglu NA, Mather M, Mungall CJ, Haendel MA, Nicholas FW, Toro S, the Vertebrate Breed Ontology Consortium (2024) The Vertebrate Breed Ontology: Towards effective breed data standardization. arXiv:2406.02623. 10.48550/arXiv.2406.02623
- Nicholas FW (2021) Online mendelian inheritance in animals (OMIA): a record of advances in animal genetics, freely available on the internet for 25 years (. Anim Genet 52:3–9. 10.1111/age.13010 [DOI] [PubMed] [Google Scholar]
- Nicholas FW, Tammen I, Sydney Informatics Hub (2024) Online Mendelian Inheritance in Animals (OMIA) [dataset]. Available via: https://omia.org/. 10.25910/2AMR-PV70. Accessed 18 Jun 2024
- Online Mendelian Inheritance in Man, OMIM® McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). Available via: https://omim.org/. Accessed 18 Jun 2024
- Pais LS, Snow H, Weisburd B, Zhang S, Baxter SM, DiTroia S, O’Heir E, England E, Chao KR, Lemire G, Osei-Owusu I, VanNoy GE, Wilson M, Nguyen K, Arachchi H, Phu W, Solomonson M, Mano S, O’Leary M, Lovgren A, Babb L, Austin-Tse CA, Rehm HL, MacArthur DG, O’Donnell-Luria A (2022) Seqr: a web-based analysis and collaboration tool for rare disease genomics. Hum Mutat 43:698–707. 10.1002/humu.24366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CG, Wright MW, Madhavrao R, Harrison SM, Goldstein JL, Luo X, Wand H, Wulf B, Cheung G, Mandell ME, Tong H, Cheng S, Iacocca MA, Pineda AL, Popejoy AB, Dalton K, Zhen J, Dwight SS, Babb L, DiStefano M, O’Daniel JM, Lee K, Riggs ER, Zastrow DB, Mester JL, Ritter DI, Patel RY, Subramanian SL, Milosavljevic A, Berg JS, Rehm HL, Plon SE, Cherry JM, Bustamante CD, Costa HA, Clinical Genome Resource (ClinGen) (2022) ClinGen Variant Curation Interface: a variant classification platform for the application of evidence criteria from ACMG/AMP guidelines. Genome Med. 2022 14:6. 10.1186/s13073-021-01004-8 [DOI] [PMC free article] [PubMed]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Beck J, Bolton EE, Brister JR, Chan J, Comeau DC, Connor R, DiCuccio M, Farrell CM, Feldgarden M, Fine AM, Funk K, Hatcher E, Hoeppner M, Kane M, Kannan S, Katz KS, Kelly C, Klimke W, Kim S, Kimchi A, Landrum M, Lathrop S, Lu Z, Malheiro A, Marchler-Bauer A, Murphy TD, Phan L, Prasad AB, Pujar S, Sawyer A, Schmieder E, Schneider VA, Schoch CL, Sharma S, Thibaud-Nissen F, Trawick BW, Venkatapathi T, Wang J, Pruitt KD, Sherry ST (2024) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 52:D33–D43. 10.1093/nar/gkad1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Eppig JT (2009) The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip Rev Syst Biol Med 1:390–399. 10.1002/wsbm.44 [DOI] [PMC free article] [PubMed]
- Tammen I, Nicholas FW (2018) Online Mendelian Inheritance in Animals (OMIA): inclusion of a hyperlinked table of likely causal variants for inherited disorders and traits. In: Proceedings of the 11th World Congress on Genetics Applied to Livestock Production (WCGALP 2018), p. 402, Massey University, Auckland
- Tammen I, Wade C, Waud B, Gimeno M, Velie BD, O’Rourke B, Nicholas FW (2021) The Anstee Hub for Inherited Diseases of Animals (AHIDA) – development of a new online platform for surveillance, reporting and control of inherited diseases in animals. Proc Assoc Advmt Anim Breed Genet 24: 386–389. Available via: http://www.aaabg.org/aaabghome/AAABG24papers/96Tammen24386.pdf. Accessed 18 Jun 2024
- Van Rossum G, Drake FL (2009) Python 3 reference Manual. CreateSpace, Scotts Valley, CA [Google Scholar]
- Vasilevsky NA, Matentzoglu NA, Toro S, Flack JE, Hegde H, Unni DR, Alyea GF, Amberger JS, Babb L, Balhoff JP, Bingaman TI, Burns GA, Buske OJ, Callahan TJ, Carmody LC, Cordo PC, Chan LE, Chang GS, Christiaens SL, Dumontier M, Failla LE, Flowers MJ, Garrett HA, Goldstein JL, Gration D, Groza T, Hanauer M, Harris NL, Hilton JA, Himmelstein DS, Hoyt CT, Kane MS, Köhler S, Lagorce D, Lai A, Larralde M, Lock A, López Santiago I, Maglott DR, Malheiro AJ, Meldal BHM, Munoz-Torres MC, Nelson TH, Nicholas FW, Ochoa D, Olson DP, Oprea TI, Osumi-Sutherland D, Parkinson H, Pendlington ZM, Rath A, Rehm HL, Remennik L, Riggs ER, Roncaglia P, Ross JE, Shadbolt MF, Shefchek KA, Similuk MN, Sioutos N, Smedley D, Sparks R, Stefancsik R, Stephan R, Storm AL, Stupp D, Stupp GS, Sundaramurthi JC, Tammen I, Tay D, Thaxton CL, Valasek E, Valls-Margarit J, Wagner AH, Welter D, Whetzel PL, Whiteman LL, Wood V, Xu CH, Zankl A, Zhang XA, Chute CG, Robinson PN, Mungall CJ, Hamosh A, Haendel MA (2022) Mondo: Unifying diseases for the world; by the world. medRxiv 2022041322273750. 10.1101/2022.04.13.22273750
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.