Abstract
Spine fractures represent a critical health concern with far-reaching implications for patient care and clinical decision-making. Accurate segmentation of spine fractures from medical images is a crucial task due to its location, shape, type, and severity. Addressing these challenges often requires the use of advanced machine learning and deep learning techniques. In this research, a novel multi-scale feature fusion deep learning model is proposed for the automated spine fracture segmentation using Computed Tomography (CT) to these challenges. The proposed model consists of six modules; Feature Fusion Module (FFM), Squeeze and Excitation (SEM), Atrous Spatial Pyramid Pooling (ASPP), Residual Convolution Block Attention Module (RCBAM), Residual Border Refinement Attention Block (RBRAB), and Local Position Residual Attention Block (LPRAB). These modules are used to apply multi-scale feature fusion, spatial feature extraction, channel-wise feature improvement, segmentation border results border refinement, and positional focus on the region of interest. After that, a decoder network is used to predict the fractured spine. The experimental results show that the proposed approach achieves better accuracy results in solving the above challenges and also performs well compared to the existing segmentation methods.
Keywords: Feature fusion, Medical image analysis, CT, Spine fracture
Introduction
The spine is a crucial part of the human body, responsible for providing structural support and protecting the spinal cord. Spinal fractures occur from different causes, including trauma, osteoporosis, or pathological conditions [1]. The human spine, also known as the vertebral column or backbone, is a crucial component of the skeletal system, consisting of multiple vertebrae stacked on top of each other and categorized into regions [2]. It includes the cervical spine (C1-C7) in the neck, the thoracic spine (T1-T12) in the upper back, the lumbar spine (L1-L5) in the lower back, the sacrum at the base, and the coccyx at the very bottom. These segments are connected by intervertebral discs and form natural curves that provide stability and allow for a wide range of movements. The spine’s primary functions include supporting the body’s weight, protecting the spinal cord, facilitating motion, and maintaining posture.
Accurate segmentation of spinal fractures is challenging due to factors such as the complex anatomical structure of the spine, variations in fracture shapes and sizes, image noise, and partial occlusions [3]. Differentiating between normal and fractured vertebrae is difficult, especially when fractures are subtle or located near healthy tissue. Achieving high accuracy and robustness in spine fracture segmentation is crucial because of the limited availability of datasets. Spine fracture segmentation is a vital application of medical image analysis that leverages advanced technologies, including deep learning [4], to assist healthcare professionals in the accurate identification and treatment of spinal fractures, ultimately improving patient care and outcomes.
In this research, a novel deep learning model is proposed for the segmentation of spine fractures using CT images. The proposed model is an encoder-decoder-based segmentation network architecture. In the encoder part, a Feature Fusion Module (FFM) is proposed that aims to fully fuse all the hierarchical features and capture multi-scale features. Squeeze and Excitation (SEM) block is inserted to emphasize the channel-level features of the feature maps. Atrous Spatial Pyramid Pooling (ASPP) layer is then used to capture multi-scale features from the feature maps using different kernel sizes. After that, the Residual Convolution Block Attention Module (RCBAM) is used to improve the features on the spatial level that improve the segmentation accuracy. Residual Border Refinement Attention Block (RBRAB) is added to refine the segmentation results of the feature maps. The Local Position Residual Attention Block (LPRAB) is used to capture the local position information of the object within the dataset. After applying all these modules, the feature maps are passed to the decoder part which consists of an initial block, three bottleneck encoder blocks, and two decoder blocks, followed by a convolutional transpose layer, batch normalization, and a final convolutional layer to generate the predicted masks of the spine fractures.
The proposed model achieves higher segmentation results as compared to the previous deep learning models. The main contributions of the proposed approach are given below:
A novel deep learning model is proposed for the automated segmentation of spine fractures using CT images.
The Feature Fusion Module is proposed to fully fuse semantic information into low-level features and spatial resolution into high-level features and capture multi-scale features for generating robust discriminative feature representation.
An innovative RCBAM module combining residual block inside the CBAM block is proposed to improve the features on the spatial level that increases the segmentation results.
To refine the segmentation feature maps captured from the input dataset, the RBRAB module is also proposed which is a combination of residual blocks and attention module.
Local position information is extracted using an LPRAB module in which residual blocks are combined to extract deep features.
Related Work
The advantage of two deep learning models was proposed by Kim et al. [5] for automated lumbar spine segmentation. First, pose-driven learning is used to identify the lumbar vertebrae accurately. After that, each lumbar vertebrae was segmented with the help of M-Net. The final tuning segmentation was used with the level-set method and the previous segmentation results. The proposed model was evaluated using 160 lumbar x-ray images and obtained a dice score of 91.60%. Golla et al. [6] used U-Net for automated cervical spine fracture segmentation using a multi-loss function. A four-fold cross-validation is used to segment the spine along with two image reformation methods. The proposed approach achieved 87.2% cervical spine fractures.
The manual segmentation of spine fractures is a time-consuming process that can increase the treatment duration of the patient. To overcome this challenge, Park et al. [3] proposed an automated algorithm for spine fracture segmentation and differentiate between benign and malignant fractures. A convolutional neural network was trained on benign and malignant spine fracture datasets. After that, a radiomics model was constructed to compare the segmentation process between human experts and the proposed model. The model achieved better results from the human-expert segmentation with a dice score of 93%. Chan et al. [7] proposed a new system for spine segmentation and classification of compression fractures. The results from the proposed model achieved 97.8% classification accuracy.
Detecting spine fractures using deep learning faces a multitude of challenges, encompassing issues such as the scarcity and quality of labeled data, class imbalance, the potential for overfitting, and ethical concerns related to patient privacy and responsible AI usage. Integrating deep learning into clinical workflows, the need for substantial computational resources, vulnerability to adversarial attacks, and the requirement for continual learning to adapt to evolving data and medical practices further compound the complexities associated with this crucial medical imaging task. Addressing these challenges necessitates interdisciplinary collaboration, constant research, and innovative solutions to enhance the accuracy and practicality of spine fracture detection through deep learning.
In this research, a novel deep learning model is introduced for the segmentation of spine fractures in CT images. The model follows encoder-decoder architecture and incorporates several key components to enhance accuracy. The encoder features a Feature Fusion Module (FFM) for comprehensive hierarchical feature fusion and multi-scale feature capture. A Squeeze and Excitation (SEM) block emphasizes channel-level features, while an Atrous Spatial Pyramid Pooling (ASPP) layer captures multi-scale features. The model further improves spatial feature accuracy with a Residual Convolution Block Attention Module (RCBAM) and refines results with a Residual Border Refinement Attention Block (RBRAB). To capture local position information, a Local Position Residual Attention Block (LPRAB) is employed. The decoder includes several blocks and layers to generate predicted spine fracture masks. Notably, this proposed model outperforms previous deep learning approaches, achieving superior segmentation results.
Methodology
Artificial Intelligence (AI) plays a significant role in various medical imaging tasks, including spine fracture segmentation [8]. Segmentation refers to the process of identifying and delineating specific structures or regions of interest within medical images, such as X-rays, CT scans, or MRI scans. Machine learning and deep learning techniques, such as Convolutional Neural Networks (CNNs) [9], are trained to classify pixels or voxels in the medical images as either part of a fracture or not. Challenges in spine fracture segmentation include variations in fracture types, image quality, and anatomical differences among patients [10].
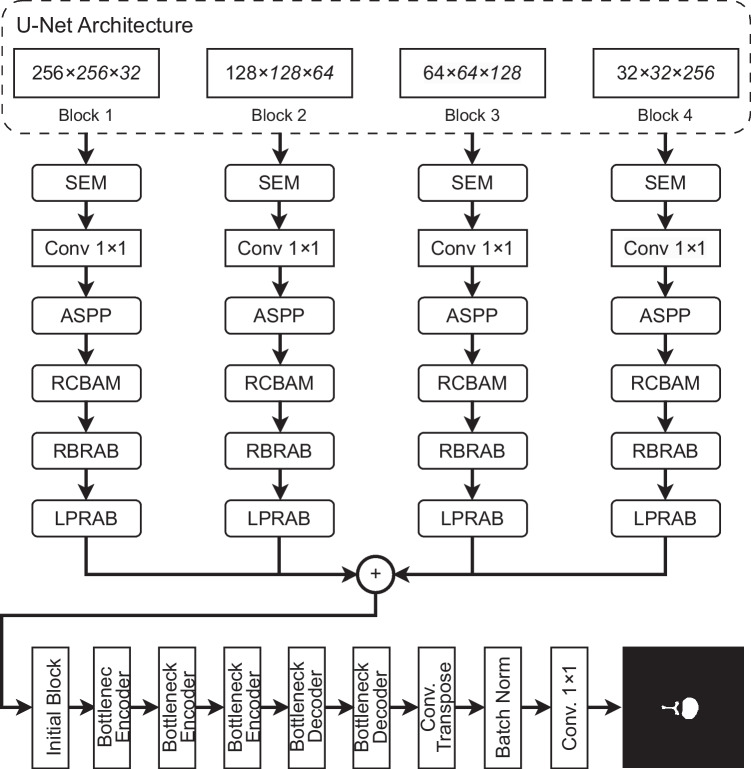
The proposed deep learning model is shown in Fig. 1, consists of two parts: (1) The segmentation network contains a Feature Fusion Module (FFM) that make each hierarchical feature to fully fused with other hierarchical features, Squeeze and Excitation Module (SEM) aims to capture and emphasize the most informative features from different channels within a convolutional layer, Atrous Spatial Pyramid Pooling (ASPP) used to capture multi-scale information from input images, Residual Convolution Block Attention Module (RCBAM) enhances the feature representation by selectively emphasizing informative channels and spatial regions, Residual Border Refinement Attention Block (RBRAB) is added to refine the segmentation results and improve the accuracy of object boundary delineation, and Local Position Residual Attention Block (LPRAB) is designed to capture and emphasize local positional information within the input dataset, enhancing a model’s ability to recognize patterns based on the spatial relationships and structures. (2) these features are passed to the second part which consists of an initial block, three bottleneck encoder blocks, two bottleneck decoder blocks, a transpose layer followed by batch normalization, and a fully connected layer to generate the final predicted mask. Each block of the proposed model is discussed below in detail and the working of the network is also explained.
Fig. 1.
The overview of the proposed deep learning model for spine fracture segmentation
Segmentation Encoder Blocks
Feature Fusion Module (FFM)
In image segmentation, both low-level and high-level features play crucial roles in achieving accurate and meaningful results. The combination of these two types of features helps a segmentation model understand and delineate objects or regions of interest within an image effectively improves the results [11–13]. Inspired by the discovery mentioned earlier, we introduce an innovative feature fusion technique in which each hierarchical feature is thoroughly integrated with the remaining three hierarchical features as shown in Fig. 1. The U-Net standard architecture is used for Feature Fusion Module. The output from blocks 2, 3, 4, and 5 of U-Net is taken to process full feature fusion.
Squeeze and Excitation Module (SEM)
Squeeze and Excitation Block (SEM) [14] allows class-specific feature enhancement, ensuring that each class channel receives the most relevant information. This helps the model focus on the distinctive characteristics of each class, leading to improved segmentation accuracy. The enhanced feature representation can help in accurately delineating object boundaries, which is essential in semantic segmentation. In the squeeze phase, the SE block reduces the spatial dimensions of the input feature map by applying Global Average Pooling (GAP) along the spatial dimensions. This operation calculates the average activation value for each channel independently, resulting in a single value for each channel. Following the squeeze phase, the SE block utilizes two fully connected (dense) layers to learn the channel-wise relationships within the feature map. These layers act as a small neural network that takes the channel-wise averages from the previous step and outputs two sets of parameters: weights for feature scaling (excitation) and biases. The basic architecture of SEM block is shown in Fig. 2. After that, a convolutional layer 1 × 1 is used to resize all the features into the same spatial dimension of .
Fig. 2.
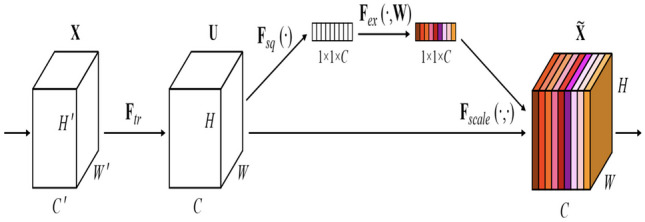
The Squeeze and Excitation block [14]
Atrous Spatial Pyramid Pooling (ASPP)
The Atrous Spatial Pyramid Pooling (ASPP) [15] is designed to capture multi-scale contextual information from an input feature map, allowing a network to make more accurate pixel-wise predictions, which is essential in semantic segmentation where each pixel is classified into a specific class. Its effectiveness in capturing contextual information has contributed to improved segmentation performance. After resizing all the feature maps, ASPP module extracts multi-scale contextual features from all the feature maps that enhance the results. Figure 3 shows the architecture of the ASPP module.
Fig. 3.
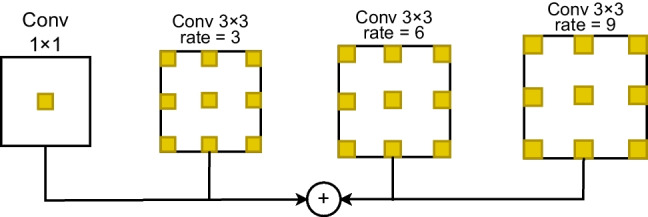
The Atrous Spatial Pyramid Pooling module
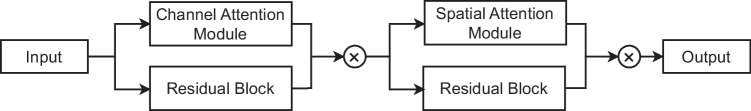
Residual Convolution Block Attention Module (RCBAM)
The Residual Convolution Block Attention Module (RCBAM) is used to enhance feature representation and improve the model’s performance. The architecture of the proposed RCBAM block is shown in Fig. 4. The input is given to the channel attention module to enhance feature representation by emphasizing informative channels within a feature map while suppressing less relevant ones. The residual block extracts more deep features from the input feature map. The output from the channel attention module is concatenated and this feature map is given to the spatial attention module and residual block. The spatial attention module is used to enhance feature representation by selectively emphasizing or de-emphasizing certain spatial regions within an input feature map. The output from the spatial attention module and the residual block are again concatenated to produce a refined feature map.
Fig. 4.
The proposed attention based residual convolution block
Residual Border Refinement Attention Block (RBRAB)
The Residual Border Refinement Attention Block (RBRAB) is used to refine the segmentation results and improve the accuracy of object boundary delineation. It combines aspects of residual connections, border refinement, and attention mechanisms to enhance feature representation and context-aware feature selection. The proposed architecture of the RBRAB is shown in Fig. 5. The RBRAB consists of a convolutional layer with 3 × 3 kernel size, a batch normalization layer, and a ReLU activation function. These three layers are repeated again and a self-attention module is added at the end of the block. Finally, the input feature map is concatenated with the output of the self-attention module and a refined segmentation mask is produced.
Fig. 5.
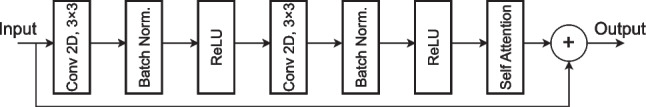
The attention based residual border refinement module
Local Position Residual Attention Block (LPRAB)
Local Position Residual Attention Block (LPRAB) is proposed to focus on the position of the object of interest and enhance the performance of the model. It takes an input feature map that contains pixel-wise information and focuses on capturing local positional information including the spatial coordinates of pixels and patterns within the image. A residual block is added which consists of two paths: the identity path and the residual path. The identity path simply passes the input feature map through, while the residual path learns a transformation that is added to the input. This facilitates the training of deep networks and helps in feature propagation. Figure 6 shows the proposed architecture of the LPRAB.
Fig. 6.
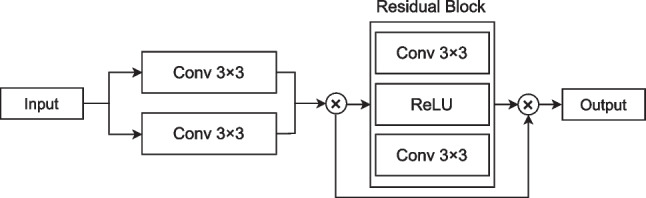
The proposed local position module with residual block
Segmentation Decoder Blocks
Initial Block
The initial block is a fundamental building block of the proposed decoder part. It starts with two successive convolutional layers with 3 × 3 kernels, followed by BatchNorm layers after each convolution. This configuration helps in feature extraction, normalization, and the stabilization of network training. The output of this initial block is a set of feature maps that serve as a foundation for subsequent layers in the neural network to extract higher-level features and make predictions for the given task. The layered architecture of the initial block is shown in Fig. 7.
Fig. 7.
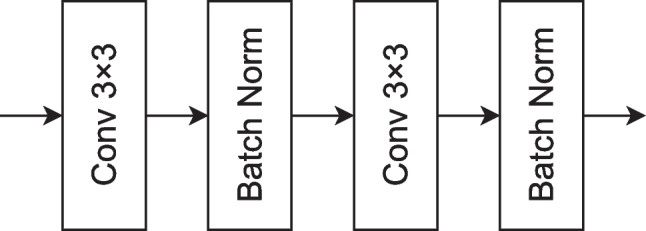
The initial block of the proposed model for fracture segmentation
Bottleneck Encoder
The bottleneck encoder block employs separable convolution layers, which consist of depthwise and pointwise convolutions, followed by Batch Normalization layers. This configuration is designed to efficiently capture basic features from the input data while ensuring stable and accelerated training of deep networks. The output of this initial block is a set of feature maps that serve as a foundation for subsequent layers in the neural network to extract higher-level features and make predictions for the given computer vision task. The architecture of the bottleneck encoder is shown in Fig. 8.
Fig. 8.
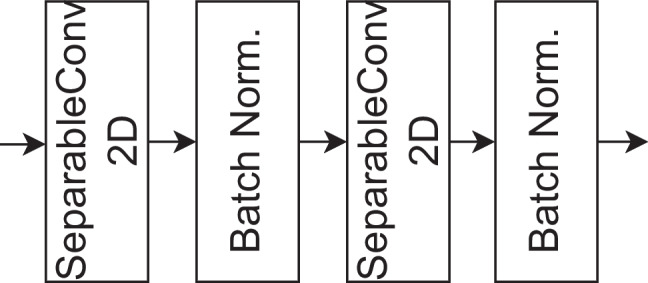
The bottleneck encoder of the proposed model for fracture segmentation
Bottleneck Decoder
The bottleneck decoder block consists of an Upsampling2D layer with a 3×3 kernel, followed by a convolutional layer with 1 × 1 kernel size and batch normalization. This block is employed to increase the spatial resolution of feature maps and refine feature representations. By employing the Upsampling2D layer, the spatial detail of the feature maps is expanded, followed by a 1 × 1 convolutional layer that performs channel-wise adjustments. Batch normalization ensures stable training. The bottleneck decoder part is shown in Fig. 9. After the decoder block, a convolutional transposed layer, followed by batch normalization and a final convolutional layer with 1 × 1 kernel size is used to predict the spine fractures.
Fig. 9.
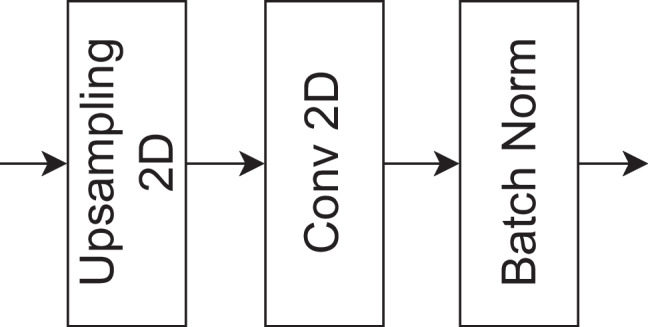
The bottleneck decoder of the proposed model for fracture segmentation
Model Training Parameters
After normalizing, cropping, and resampling the dataset, the subsequent stage involved training the model to autonomously extract spine fracture segments. The training dataset is divided into three parts in which 70% of the dataset is used for training, 10% dataset is used for validation, and 20% of the dataset is used for testing. The hyperparameter optimization is used to select the best model. The network is trained with 150 epochs using a learning rate of 0.001. The size of the batch is 16 and the network uses Adam [16] optimizer for training. Early stopping is used that will terminate the training process if there is no improvement of validation dataset after 5 epochs. The learning rate also decreases by multiplying a factor of 0.4 when there is no improvement.
Experimental Results
Dataset Definition
In this study, a comprehensive dataset of spine fractures has been assembled, comprising a total of 235 CT images obtained from multiple patients from a single center. The dataset represents a diverse range of spine fracture cases, reflecting the complexity and variations encountered in clinical practice. The study included CT spine images obtained from a single center located in Lahore, Pakistan. Inclusion criteria comprised adult patients (age 18 years) presenting with symptoms suggestive of spinal pathology who underwent CT imaging of the spine. Exclusion criteria included contraindications to CT, such as metallic implants or pregnancy. CT scans were performed using a 64-slice Siemens SOMATOM Definition scanner, with the following parameters: slice thickness of 1 mm, FOV of 250 mm. The study population consisted of 150 adult patients (75 males and 75 females) with a mean age of 50 years (range: 20–80 years). The majority of patients presented with back pain (60%), followed by trauma-related injuries (30%) and spinal deformities (10%).
Evaluation Metrics
Dice Coefficient Score (DSC)
The Dice Coefficient Score (DSC) is commonly used as an evaluation metric to assess the accuracy of the segmentation results. It measures the similarity between the predicted segmentation mask and the ground truth segmentation mask for an image. The DSC for segmentation is calculated using the following formula:
| 1 |
where
A represents the set of pixels in the predicted segmentation mask.
B represents the set of pixels in the ground truth segmentation mask.
|A| represents the total number of pixels in A.
|B| represents the total number of pixels in B.
represents the number of pixels that are common to both A and B.
Intersection over Union (IoU)
Intersection over Union (IoU), also known as the Jaccard index, is a widely used evaluation metric in computer vision and image processing, particularly for tasks like object detection and image segmentation. It measures the similarity between two sets, typically the predicted and ground truth regions in these tasks. IoU is used to assess the accuracy of the overlap between the predicted region and the true region. The IoU is calculated using the following formula:
| 2 |
where
A represents the set of pixels in the predicted region or mask.
B represents the set of pixels in the ground truth region or mask.
represents the number of pixels that are common to both A and B (the intersection).
represents the number of pixels that are in either A or B (the union).
Results and Discussion
Comprehensive Analysis
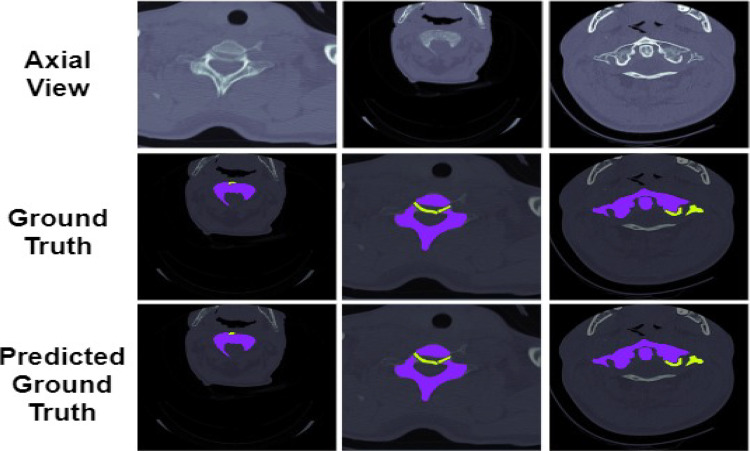
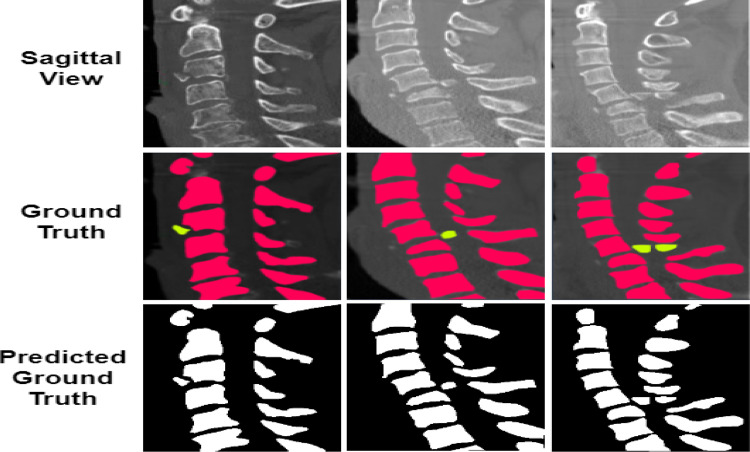
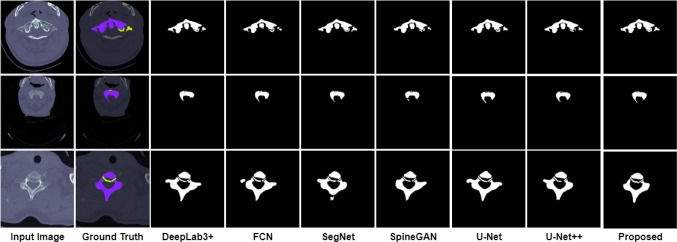
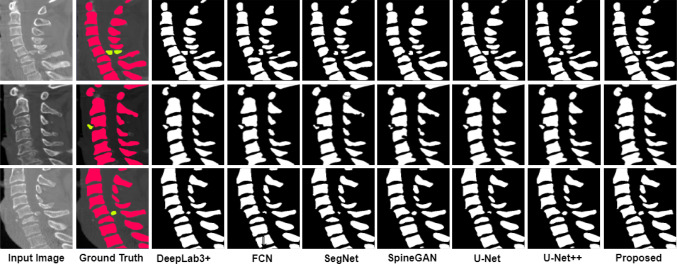
The comparison of the ground truth with the segmented maps predicted by the proposed model is shown in Figs. 10 and 11. From the figure, it can be seen that the segmentation of the spine fractures obtained by the proposed model is similar to the ground truth. The proposed method solves the problem of structural variabilities in the segmentation of spine fractures. The proposed model achieves DSC and IoU as 91.87% and 93.36% respectively. It is observed that the proposed model achieved higher results in quantitative analysis as compared to the other methods.
Fig. 10.
Comparison of the Axial segmentation masks predicted by the proposed model with the actual ground truth
Fig. 11.
Comparison of the Sagittal segmentation masks predicted by the proposed model with the actual ground truth
Ablation Study
Ablation experiments are performed to demonstrate the best results using the proposed deep learning model. The quantitative segmentation results are shown in Table 1. Multiple experiments are conducted and the results between the ablation experiments and the proposed model are significant.
Table 1.
Quantitative results of spine fracture segmentation with ablation study
| Method | FFM | SEM | ASPP | RCBAM | RBRAB | LPRAB | DSC | IoU |
|---|---|---|---|---|---|---|---|---|
| Proposed Model | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 91.87 | 93.36 |
| No FFM | ✓ | ✓ | ✓ | ✓ | ✓ | 86.18 | 87.39 | |
| No SEM | ✓ | ✓ | ✓ | ✓ | ✓ | 86.08 | 85.62 | |
| No ASPP | ✓ | ✓ | ✓ | ✓ | ✓ | 79.93 | 82.48 | |
| No RCBAM | ✓ | ✓ | ✓ | ✓ | ✓ | 88.69 | 89.01 | |
| No RBRAB | ✓ | ✓ | ✓ | ✓ | ✓ | 82.95 | 83.17 | |
| No LPRAB | ✓ | ✓ | ✓ | ✓ | ✓ | 87.23 | 86.93 |
FFM
First, the FFM was removed from the proposed model; it can be seen that the proposed model achieves DSC and IoU as 86.18%, 87.39%, decreased by 4.59%, and 4.73%. The FFM helps in achieving the multi-scale feature fusion information which increases the fracture segmentation results. Therefore, the proposed model with FFM in the encoder part can help generate more important features and provide better segmentation results.
SEM
From Table 1, the segmentation results without SEM module achieve DSC and IoU as 86.08% and 85.62%, decreased by 4.7% and 5.21% respectively. The SEM module plays an important role in the proposed model with a confidence of 95% due to its segmentation improvements. Therefore, it supports that the SEM module can enhance spatial inconsistencies and improve the fracture segmentation performance.
ASPP
The proposed model achieves DSC and IoU as 79.93% and 82.48%, decreased by 11.51% and 8.69% respectively. The ASPP block is a critical component in spine segmentation by enabling the neural network to capture and integrate multi-scale contextual information effectively. This helps the network make informed decisions about the location and boundaries of the spine structures in medical images, ultimately leading to more accurate and reliable segmentation results. Therefore, by adding an ASPP block, the multi-scale features can enhance the performance of the proposed model.
RCBAM
The absence of RCBAM block decreased the model’s results by 1.81% and 1.46% of DSC and IoU respectively by achieving 88.69% and 89.01%. An RCBAM block is designed to enhance the representational power of convolutional layers through attention mechanisms. When integrated with a residual block, which is a core building block of deep neural networks like ResNet, it results in improved feature extraction and representation learning. The enhanced residual block with CBAM is useful in capturing both fine-grained details and high-level semantics that improve the segmentation results.
RBRAB
The proposed model achieves 82.95% DSC and 83.17% IoU, decreased performance by 8.16% and 7.92% without using RBRAB. By adding another residual block, the network can iteratively refine the features, progressively emphasizing object boundaries and improving the accuracy of boundary segmentation.
LPRAB
By removing the LPRAB block from the proposed model, the results are 87.23% and 86.93% for DSC and IoU respectively, decreased by 3.43% and 3.76%. A Local Position Attention Module with a Residual Block combines the concept of capturing fine-grained spatial information within local regions of an image with the benefits of residual learning. It enhances the network’s ability to capture fine-grained spatial relationships and leverage residual learning for feature enhancement.
Comparison with Other Methods
To prove the superiority of the proposed model, the existing deep learning models such as U-Net [17], U-Net++ [18], SegNet [19], FCN [20], SpineGAN [21], and DeepLabv3+ [22] are compared with the proposed model. The segmentation masks obtained by different models and its actual masks are shown in Figs. 12 and 13. From the results, it can be seen that the segmentation masks generated by the proposed model are the most similar to the ground truth. Among all the selected methods, SpineGAN shows the least accuracy, followed by SegNet, U-Net, FCN, U-Net++, and DeepLabv3+. The results obtained by these six methods are given in Table 2 quantified using DSC, and IoU. The best results are obtained by DeepLabv3+ with 87.36% DSC and 88.15% IoU which are less than the proposed approach. The SpineGAN achieves the worst segmentation results with 78.31% DSC and 75.49% IoU. The proposed deep learning model performs well compared to the existing segmentation methods for spine fracture segmentation.
Fig. 12.
Axial Comparison of the segmentation masks predicted by the proposed model with the actual ground truth
Fig. 13.
Sagittal Comparison of the segmentation masks predicted by the proposed model with the actual ground truth
Table 2.
Quantitative segmentation results of the proposed model with existing methods
Conclusion
Segmenting spine fractures in medical images is a challenging task that requires advanced computer vision and image analysis techniques due to the heterogeneity of fractures, noise, and artifacts. In this paper, a novel encoder-decoder deep learning model is proposed for automated spine fracture segmentation using an CT dataset. First, the Feature Fusion Module is implemented by using a U-Net standard architecture. The output of different encoder blocks is extracted and given to the other modules such as SEM, ASPP, RCBAM, RBRAB, and LPRAB. After applying these modules, the refined feature maps are given to the decoder part for the final segmentation task which consists of an initial block, three encoder, and two decoder blocks. Finally, a transpose convolutional layer, batch normalization, and a convolutional layer are used to generate the segmented spine fracture. The proposed model achieves a DSC and IoU of 91.87% and 93.36% respectively. The proposed model also achieved better performance as compared to the state-of-the-art six deep learning models for spine fracture segmentation.
Author Contribution
Muhammad Usman Saeed and Wang Bin conceptualized the study, with contributions from Jinfang Sheng. Methodology, formal analysis, data curation, and writing original draft were prepared by Muhammad Usman Saeed. Methodology development, validation, supervision, and review original draft was done by Wang Bin. Muhammad Usman Saeed and Wang Bin validated the methodology. Formal analysis, investigation efforts, data curation, and supervision were conducted by Jinfang Sheng. Data curation, validation, and formal analysis were conducted by Hussain Mobarak Albarakati.
Declarations
Ethical Approval
Not applicable
Informed Consent
Not applicable
Consent for Publication
Not applicable
Competing Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Glessgen, C.G., Cyriac, J., Yang, S., Manneck, S., Wichtmann, H.M., Wasserthal, J., Kovacs, B.K., Harder, D.: Segment and slice: A two-step deep learning pipeline for opportunistic vertebral fracture detection in computed tomography. In: medRxiv (2022). https://api.semanticscholar.org/CorpusID:254066460
- 2.Healthline: Spine. https://www.healthline.com/human-body-maps/sternum (2023)
- 3.Park, T., Yoon, M.A., Cho, Y.C., Ham, S.J., Ko, Y., Kim, S., Jeong, H., Lee, J.: Automated segmentation of the fractured vertebrae on ct and its applicability in a radiomics model to predict fracture malignancy. Scientific Reports 12 (2022) [DOI] [PMC free article] [PubMed]
- 4.Zhang, Q., Du, Y., Wei, Z., Liu, H., Yang, X., Zhao, D.: Spine medical image segmentation based on deep learning. Journal of Healthcare Engineering 2021 (2021) [DOI] [PMC free article] [PubMed] [Retracted]
- 5.Kim, K.C., Cho, H.C., Jang, T.J., Choi, J.M., Seo, J.K.: Automatic detection and segmentation of lumbar vertebrae from x-ray images for compression fracture evaluation. Computer Methods and Programs in Biomedicine 200, 105833 (2021) 10.1016/j.cmpb.2020.105833 [DOI] [PubMed] [Google Scholar]
- 6.Golla, A.-K., Lorenz, C., Buerger, C., Lossau, T., Klinder, T., Mutze, S., Arndt, H., Spohn, F., Mittmann, M., Goelz, L.: Cervical spine fracture detection in computed tomography using convolutional neural networks. Physics in Medicine & Biology 68(11), 115010 (2023) 10.1088/1361-6560/acd48b [DOI] [PubMed] [Google Scholar]
- 7.Chan, Y.-K., Lin, C.-S., Lin, H.-J., Yip, K.-T.: Segmentation of spinal mri images and new compression fracture detection. Software Engineering, Artificial Intelligence, Networking and Parallel/Distributed Computing (2022) [Google Scholar]
- 8.Benzakour, A., Altsitzioglou, P., Lemée, J.-M., Ahmad, A.A., Mavrogenis, A.F., Benzakour, T.: Artificial intelligence in spine surgery. International Orthopaedics 47, 457–465 (2022) [DOI] [PubMed] [Google Scholar]
- 9.Small, J.E., Osler, P.M., Paul, A., Kunst, M.M.: Ct cervical spine fracture detection using a convolutional neural network. American Journal of Neuroradiology 42, 1341–1347 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunder, A., Chhabra, H.S., Aryal, A.: Geriatric spine fractures - demography, changing trends, challenges and special considerations: A narrative review. Journal of clinical orthopaedics and trauma 43, 102190 (2023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, X., Zhang, R., Yan, P.: Feature fusion encoder decoder network for automatic liver lesion segmentation. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 430–433 (2019)
- 12.Wu, H., Zhang, J., Huang, K.: Sparsemask: Differentiable connectivity learning for dense image prediction. 2019 IEEE/CVF International Conference on Computer Vision (ICCV), 6767–6776 (2019)
- 13.Zhang, Z., Zhang, X., Peng, C., Cheng, D., Sun, J.: Exfuse: Enhancing feature fusion for semantic segmentation. In: European Conference on Computer Vision (2018). https://api.semanticscholar.org/CorpusID:262349636
- 14.Hu, J., Shen, L., Albanie, S., Sun, G., Wu, E.: Squeeze-and-excitation networks. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 7132–7141 (2017)
- 15.Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K.P., Yuille, A.L.: Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Transactions on Pattern Analysis and Machine Intelligence 40, 834–848 (2016) [DOI] [PubMed] [Google Scholar]
- 16.Bock, S., Weiß, M.G.: A proof of local convergence for the adam optimizer. 2019 International Joint Conference on Neural Networks (IJCNN), 1–8 (2019)
- 17.Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. ArXiv abs/1505.04597 (2015)
- 18.Zhou, Z., Siddiquee, M.M.R., Tajbakhsh, N., Liang, J.: Unet++: A nested u-net architecture for medical image segmentation. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support : 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, held in conjunction with MICCAI 2018, Granada, Spain, S... 11045, 3–11 (2018) [DOI] [PMC free article] [PubMed]
- 19.Badrinarayanan, V., Kendall, A., Cipolla, R.: Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence 39, 2481–2495 (2015) [DOI] [PubMed] [Google Scholar]
- 20.Shelhamer, E., Long, J., Darrell, T.: Fully convolutional networks for semantic segmentation. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 3431–3440 (2014) [DOI] [PubMed]
- 21.Han, Z., Wei, B., Mercado, A., Leung, S., Li, S.: Spine-gan: Semantic segmentation of multiple spinal structures. Medical Image Analysis 50, 23–35 (2018) [DOI] [PubMed] [Google Scholar]
- 22.Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: European Conference on Computer Vision (2018). https://api.semanticscholar.org/CorpusID:3638670