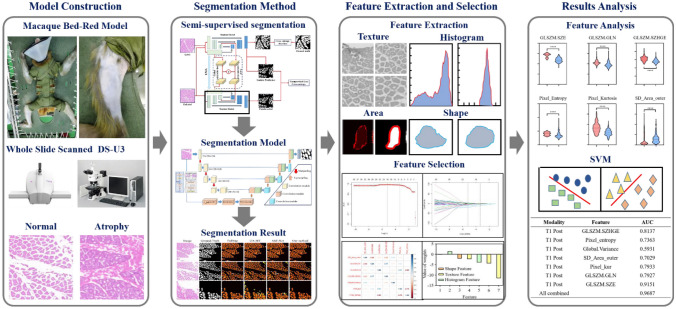
Abstract
Radiomics features have been widely used as novel biomarkers in the diagnosis of various diseases, but whether radiomics features derived from hematoxylin and eosin (H&E) images can evaluate muscle atrophy has not been studied. Therefore, this study aims to establish a new biomarker based on H&E images using radiomics methods to quantitatively analyze H&E images, which is crucial for improving the accuracy of muscle atrophy assessment. Firstly, a weightless muscle atrophy model was established by laying macaques in bed, and H&E images of the shank muscle fibers of the control and bed rest (BR) macaques were collected. Muscle fibers were accurately segmented by designing a semi-supervised segmentation framework based on contrastive learning. Then, 77 radiomics features were extracted from the segmented muscle fibers, and a stable subset of features was selected through the LASSO method. Finally, the correlation between radiomics features and muscle atrophy was analyzed using a support vector machine (SVM) classifier. The semi-supervised segmentation results show that the proposed method had an average Spearman’s and intra-class correlation coefficient (ICC) of 88% and 86% compared to manually extracted features, respectively. Radiomics analysis showed that the AUC of the muscle atrophy evaluation model based on H&E images was 96.87%. For individual features, GLSZM_SZE outperformed other features in terms of AUC (91.5%) and ACC (84.4%). In summary, the feature extraction based on the semi-supervised segmentation method is feasible and reliable for subsequent radiomics research. Texture features have greater advantages in evaluating muscle atrophy compared to other features. This study provides important biomarkers for accurate diagnosis of muscle atrophy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-024-01122-w.
Keywords: Radiomics, H&E images, Muscle atrophy, Biomarkers semi-supervised segmentation
Introduction
Long term in orbit flight can easily lead to the occurrence of weightless muscle atrophy, which greatly hinders the development of the aerospace industry. According to previous research reports, during space flights, the volume of the gastrocnemius and quadriceps muscles decreased by 6.3% and 6.0%, respectively, and this reduction in muscle volume was caused by an imbalance in protein metabolism in muscle tissue [1–3]. Therefore, accurate diagnosis of weightless muscle atrophy and studying its biological mechanisms are important guarantees for successfully completing space missions. At present, muscle imaging and muscle biopsy are the two most important diagnostic methods in clinical research, but muscle imaging cannot directly reflect the biological mechanisms and determining factors of muscle atrophy, making it difficult to serve as a diagnostic standard for muscle atrophy [4]. On the contrary, muscle biopsy can understand the essence of muscle atrophy from the level of protein and muscle fiber structure and can directly and truly reflect the exact mechanism of muscle atrophy, thus becoming the recognized gold standard for the diagnosis of muscle atrophy at present [5, 6]. In previous studies, clinicians often focused more on the relationship between morphometric parameters such as cross-sectional area (CSA), perimeter, and diameter of muscle fibers in H&E images and muscle atrophy when evaluating muscle atrophy. For example, with regard to the addressing muscle atrophy caused by Duchenne muscular dystrophy (DMD), Lau et al. evaluated muscle atrophy by measuring the CSA, diameter, and central nucleated fibers (CNF) of muscle fibers [7]. Emily et al. investigated the effects of 14 days of bed rest (BR) on the content of skeletal muscle satellite cells and fiber type atrophy in middle-aged individuals by measuring the CSA of various muscle fiber types [8]. However, the changes in these parameters may not be unique to muscle atrophy and may be influenced by the patient’s physical activity, dietary intake, and specific muscles, which make it difficult to accurately evaluate muscle atrophy [9]. Therefore, it is urgent to explore a new biomarker based on H&E images to improve the accuracy of muscle atrophy diagnosis.
Previous studies have shown that H&E images contain rich fine structural information of cells and tissues, which helps to better understand the biological mechanisms of diseases [10]. Therefore, a significant amount of research is devoted to analyzing the relationship between the quantitative results of fine cellular structure and the biological mechanism of disease. Researchers have found that texture features based on gray-level symbiosis extracted from multiple spatial scales are closely related to disease biology such as cervical cells, breast cancer, and osteosarcoma [11–13]. Based on these premises in our previous research on muscle atrophy, we found that the loss of muscle-derived proteins and increased protein hydrolysis in muscle fibers can have a certain impact on the density of normal muscle tissue, and this impact may be reflected in changes in potential features of H&E images [14, 15]. Therefore, quantitative analysis of derived features from muscle fiber H&E images may provide a new method for accurately evaluating muscle atrophy. Based on this assumption, it is crucial to effectively and quickly obtain H&E image-derived features, and radiomics provides a highly feasible analysis tool for this purpose. Radiomics can extract a large number of quantitative features reflecting morphological, intensity, and texture attributes from medical images through high-throughput calculations, exploring the relationship between medical images and potential visual features. Many studies have demonstrated the ability of radiomics features as new biomarkers for evaluating diseases [16, 17]. For example, Granata et al. demonstrated the effectiveness of radiomics features obtained from computed tomography (CT) as biomarkers in lung adenocarcinoma patients who benefited from immunotherapy in 2021 [18]. Guerrisi et al. explored a biomarker for survival prediction and response imaging in patients with metastatic melanoma treated with PD-1 inhibitor nivolumab using CT texture features [19]. In addition, radiomics has recently gained extensive exploration in analyzing the relationship between features derived from H&E images and the biological mechanisms of diseases, indicating the ability of radiomics features as biomarkers in evaluating disease pathology [20]. However, whether radiomics features derived from H&E images can reflect the state of muscle atrophy has not been studied yet. Therefore, based on this goal, exploring and establishing the quantitative relationship between radiomics features derived from H&E images and muscle atrophy will provide a new approach for accurately diagnosing patients’ muscle atrophy status.
However, there are some limitations to analyzing H&E images by radiomics, among them is ROI segmentation. At present, manual delineation of image ROI regions and their feature extraction still has certain potential value, especially for models that are easier to interpret and the ability to learn on finite datasets. However, this manual delineation method poses certain challenges in solving issues such as the large number of muscle fibers and low-contrast H&E images. Therefore, it is very useful to study a segmentation method that learns a large amount of unlabeled data given a small amount of labeled data. With the introduction of deep learning (DL), semi-supervised segmentation methods have emerged [21] and were successfully applied in nuclear segmentation of pathological images, demonstrating more replicable and effective radiomics features [22–24]. Based on the above analysis, we assume that the combination of semi-supervised segmentation and radiomics can extract more valuable information from H&E images, thereby better evaluating and classifying the state of muscle fibers in muscle atrophy. To our knowledge, there are currently no complete reports on the combination of semi-supervised segmentation of muscle fibers based on H&E images and radiomics research.
The aim of this study is to develop a method to evaluate the relationship between the state of muscle fibers during muscle atrophy and radiomics features derived from H&E images and to explore a potential biomarker that can evaluate muscle atrophy. To address the time-consuming and labor-intensive issue of manually labeling muscle fibers, this study proposes a semi-supervised segmentation framework based on contrastive learning and explores the feasibility of this method in radiomics analysis. The segmentation model is mainly composed of a transformer and CNN, which uses a dual encoder branch, instead of the traditional single encoder branch structure to extract rich local features and global contextual information. Through enhancing the intra-class compactness and inter-class separability of local and global information of muscle fibers, valuable information is extracted from unlabeled data. The whole workflow is shown in Fig. 1.
Fig. 1.
Overall workflow of the radiomics framework
Related Work
Evaluation of Muscle Atrophy Based on Pathological Images
Skeletal muscle is one of the largest organs in the human body. However, long-term exposure to microgravity can lead to severe decline of the skeletal muscle system, which is mainly manifested as the reduction in muscle volume, the decrease in muscle fiber CSA, and the change of muscle fiber types. Previous research results have shown that neuromuscular activity decreases by 35–40% after a space flight, and the average leg extensor muscle strength of astronauts decreases by 20% after 28 days of flight [25–27]. To reduce these risks, maintaining astronaut strength through early diagnosis and treatment of muscle atrophy is crucial for achieving long-term space travel and exploration.
At present, muscle histological examination is a routine method for studying muscle atrophy. In clinical practice, H&E staining and immunofluorescence staining are commonly used to examine muscle histology and muscle fiber types, respectively. H&E staining can be used for morphological analysis of muscle fibers, such as muscle fiber diameter, CSA, and regenerated muscle fibers. By calculating the CSA, perimeter, and diameter of each muscle fiber in the H&E image, the degree of muscle atrophy and changes can be evaluated [28, 29]. Compared to the currently used imaging systems, including CT, magnetic resonance imaging (MRI), ultrasound, and electromyography, histological analysis methods are very effective tools for determining the morphological indicators of muscle health status. Quantitative analysis of muscle tissue morphometry can provide insights into the dynamic adaptation of skeletal muscle fibers during muscle atrophy [30]. However, evaluating muscle atrophy by calculating parameters such as CSA has certain limitations, as these indicators are influenced by the patient’s physical activity, dietary intake, and specific muscles, making it difficult to effectively evaluate muscle atrophy [9]. Therefore, it is necessary to search for a new potential biomarker for evaluating muscle atrophy through H&E images. In this study, an analysis framework for muscle atrophy based on radiomics was established using H&E images obtained from the shank muscle tissue of macaques as the analysis object.
A Semi-supervised Medical Image Segmentation Method Using DL
In recent years, semi-supervised learning has attracted increasing attention in the field of medical imaging. Compared with fully supervised methods, semi-supervised methods utilize a large amount of available unlabeled data to supplement a small number of labeled datasets in order to address the drawbacks of manual annotation work being time-consuming and labor-intensive, as well as the scarcity of labeled data [21]. Traditional semi-supervised segmentation methods usually use artificially designed shallow features, which have limited representation ability, especially for medical images with severe noise interference and low contrast, and cannot achieve ideal segmentation results. Compared with the above segmentation methods, the semi-supervised method based on DL utilizes representation learning to learn data-driven features, which can provide accurate segmentation results [31, 32]. In this study, in order to better utilize unlabeled data, we adopted consistency learning and contrastive learning to improve the segmentation performance of the network. For consistency learning, a mean teacher (MT) model was proposed in 2017, which updates the teacher’s parameters through weight averaging and performs consistency learning under different data perturbations [33]. Subsequently, on the basis of MT, more and more studies have improved it based on different medical image characteristics and tasks and used different consistency strategies to improve the prediction accuracy of unlabeled data. For example, Yu et al. proposed a new uncertainty aware semi-supervised framework that effectively utilizes unlabeled data by encouraging consistent predictions of the same input under different perturbations [34]. However, the learning ability of the model is subject to error supervision from unlabeled data, neglecting the relationship between labeled and unlabeled data, resulting in inaccurate prediction results. Lei et al. proposed a dynamic convolutional-based adversarial self-ensembling network (ASE-Net) to address this issue. Firstly, adversarial consistency training was used to obtain a prior relationship between labeled and unlabeled data, and then, a dynamic convolution-based bidirectional attention component (DyBAC) was used to improve the feature representation ability of ASE-Net [35]. However, the network models lack the utilization of the feature structure of the entire dataset, ignoring the similarities or differences between different features, which can lead to unstable training and reduced segmentation performance. To address this issue, Lou et al. proposed a semi-supervised segmentation network based on contrastive learning with minimum–maximum similarity (MMS), which evaluates the consistency of unlabeled predictions through pixel contrast loss between positive and negative pairs [36]. In order to make it easier for the deep model to learn the representation of target distribution, Wu et al. added a patch level contrastive learning module on the basis of pixel level contrastive learning, strengthening the compactness between features and better segmenting of the nucleus in tissue pathology images [37]. Based on the above research foundation, we propose a semi-supervised segmentation network based on local–global feature contrastive learning tasks to learn better general features from muscle fibers in unlabeled H&E images in this study.
Radiomics
Radiomics is a method of extracting quantitative image features from medical images, which improves decision support for tumor diagnosis in a low-cost and non-invasive manner. The features extracted by radiomics include the intensity, shape, texture, and wavelet information of the target area, which can reveal the diagnosis, prediction, and prognosis of cancer patients through their correlation with objective response criteria such as survival or treatment response [38, 39]. Although radiomics is often applied to tumor differentiation and grading in medical images, it has recently expanded to predict pathological tissue images. Chen et al. established a machine learning (ML) algorithm based on pathological data to construct and cross-verify the automatic diagnosis and prognosis model, solving the problem that traditional histopathology conducted by pathological experts through the naked eye cannot accurately diagnose bladder cancer [40]. Kim et al. utilized tissue feature engineering technology to extract texture features based on first-order statistics (FOS) from staining channels and classified tumors based on important features [41]. Based on the above research reports, radiomics models have shown potential in quantitative characterization of pathological tissues, providing new ideas for evaluating muscle atrophy. To our knowledge, there have been no published studies on whether a radiomics model based on H&E pathological images can be used to evaluate muscle atrophy. In this study, we utilized radiomics techniques to identify quantitative imaging features related to changes in muscle fibers after muscle atrophy and explore their ability to evaluate novel biomarkers for muscle atrophy.
Materials and Method
Animal Model Establishment
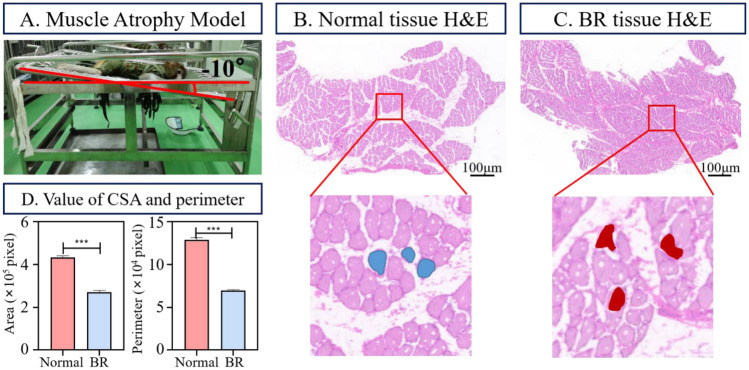
This study investigated muscle atrophy caused by weightlessness in a simulated space environment by establishing a BR macaque model (Fig. 2A). We collaborated with the Chinese Astronaut Training Animal Experiment Center and used six normal macaques as the experimental subjects, with an average age of 6 years and all males. They were randomly divided into control groups (n = 3) and BR groups (n = 3); the BR group was subjected to a − 10° continuous 42-day BR to model a space weightlessness environment and establish a weightless muscle atrophy model [42]. The CSA and perimeter changes of each experimental group on day 42 are shown in Fig. 2D. All animal experiments were approved by Institutional Animal Care and Use Committee of China Astronaut Research and Training Center (ACC-IACUC-2019–2002).
Fig. 2.
Example of H&E muscle fiber images. A The process of establishing a weightlessness model for macaques. B The H&E image of normal shank muscle tissue, with the blue area indicating normal muscle fibers. C H&E image of the atrophic shank muscle tissue, with the red area indicating the atrophic muscle fibers. D On the 42nd day, changes in CSA and perimeter of normal and BR macaque muscle fibers
Semi-supervised Segmentation Method
Overview
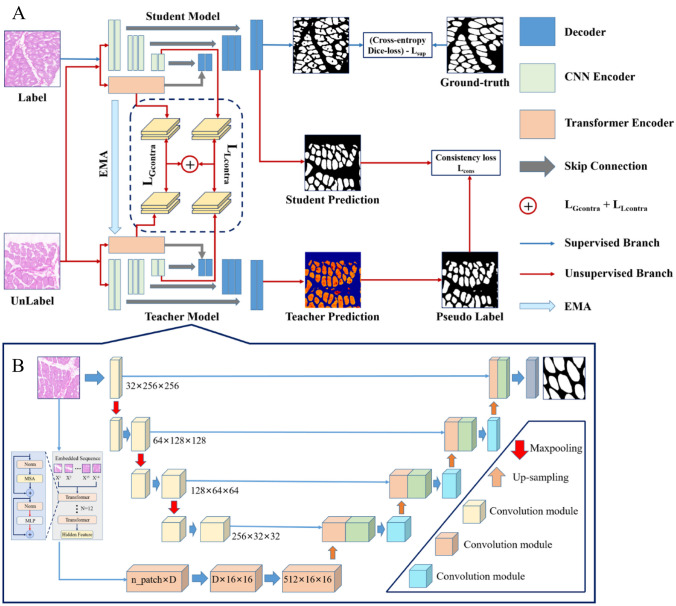
The proposed semi-supervised segmentation method is based on the MT framework, and the specific structure is shown in Fig. 3A. The student and teacher models share the same architecture, consisting of a segmentation network and a projector. In the segmentation network, a dual encoder segmentation network composed of a U-net framework–based transformer and CNN is used to capture both local features and global contextual information of muscle fibers. In the supervised branch, the labeled data are utilized by calculating the loss (cross-entropy + Dice loss) between the predicted results of the network and the ground truth (Fig. 3, blue arrows). In unsupervised branches, the global features extracted from the transformer encoder branch and the local features extracted from the CNN encoder branch are compared and learned through the output features of the projector, and the prediction results output by the student model and the pseudo labels output by the teacher model are uniformly regularized. This can drive the network to extract useful information from unlabeled data. Figure 4 shows the visual process of the pseudo-label evolution of the teacher model output, the training updates every ten epochs, the contour of muscle fibers gradually approaches ground truth, and non-muscle fiber areas gradually decrease. In the tenth round, most of the muscle fibers have been accurately predicted.
Fig. 3.
Overall architecture. A The semi-supervised muscle fiber segmentation framework updates the student network model through the weighted sum of , , , and , while the teacher model accelerates the update by performing EMA based on the weight of the student model. Blue arrows represent supervised branches, while red arrows represent unsupervised branches. B The main body segmentation network architecture consists of a transformer and a CNN for the input branch of the network
Fig. 4.
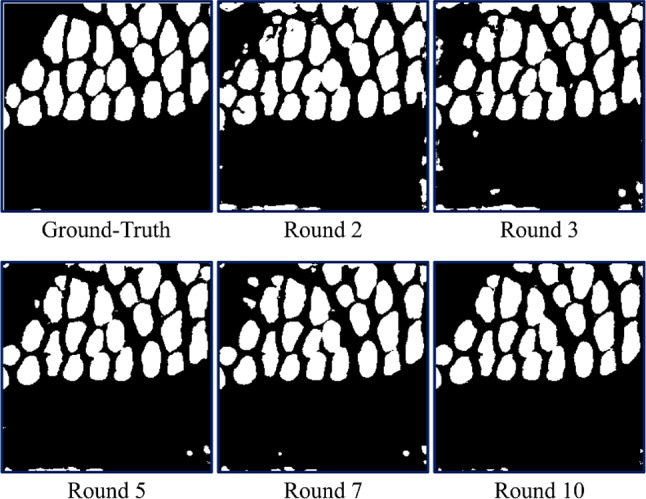
Example of pseudo-labels
The weighted sum formula for the total loss of the entire semi-supervised segmentation model is as follows:
| 1 |
| 2 |
where and represent the contrastive learning loss of global contextual information and the contrastive learning loss of local features, respectively, is an unsupervised consistency loss based on cross-entropy, and is the weight factor used to balance the impact of each loss on the model. We set as a slope weighting coefficient, as shown in formula (2), where is the maximum epoch for the current training, ep is the current training epoch, , , and . In this study, the training parameters of teacher model were updated by setting the exponential moving average (EMA) of student model weights.
Local–Global Feature Contrastive Learning
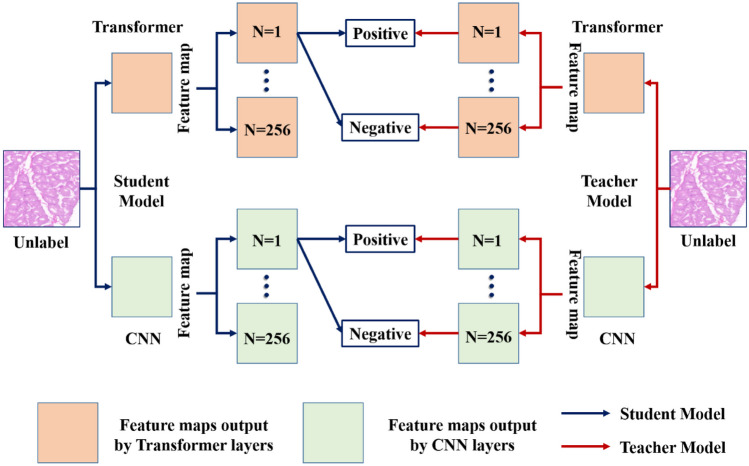
In this work, due to the irregular shape changes and low contrast between muscle fibers and non-muscle fibers in H&E images, it is difficult to accurately distinguish the differences between muscle fibers and background pixels. Therefore, we designed a dual encoder input branch on the major segmentation framework, which integrates the global context features extracted by the transformer encoder on the basis of local features extracted by traditional convolutional neural network (CNN) encoders (as shown in Fig. 3B). We transferred the same image to the student and teacher models separately and projected the features output by the dual encoder onto the low-dimensional feature map. We sampled positive/negative pairs in the global and local features of the muscle fibers. The function of projectors is to retain key contextual information in extracted features, which has been proven to be beneficial for contrastive learning [43]. Considering the feature of muscle fiber H&E images, our local–global feature contrastive learning module, as shown in Fig. 5, is developed based on the idea of contrastive learning from the original feature map. A feature map will be selected from the student model, and its corresponding position will be selected from the teacher model to sample its corresponding feature. Between these two patches, features with the same position form patch level positive pairs, while opposite positions form negative pairs. The purpose is to pull in the distance between positive samples and push away irrelevant samples. Finally, we will calculate losses separately on the global–local features, aiming to provide the model with a deeper understanding of the interaction between local and global contextual information in H&E muscle fiber images.
Fig. 5.
Proposed local–global feature contrastive learning module for semi-supervised muscle fiber segmentation
Radiomics Feature Extraction
We extracted radiomics features of muscle fibers in H&E images, including normal and atrophic muscle fibers. We extracted three sets of radiomics features to describe the ROI of muscle fiber regions: (1) shape features, (2) intensity features, and (3) texture features. In radiology, shape features denote representation information of physical features of muscle fibers, such as CSA, perimeter, diameter, and Fourier descriptors, which include the roundness, scattering, slenderness, and eccentricity of muscle fibers. The grayscale intensity feature is based on the differences in signal intensity histograms and distributions within muscle fibers. Texture features refer to the spatial distribution and grayscale variation patterns of pixels in an image, which extract quantitative texture features from gray-level co-occurrence matrix features (GLCM), gray-level size zone matrix features (GLSZM), gray-level run length matrix features (GLRLM), and neighborhood gray-tone difference matrix features (NGTDM). All radiomics feature extraction and analyses were completed in the MATLAB toolkit [44]. In this study, a total of 77 radiomics features were extracted, including 24 shape features, 11 intensity features, and 42 texture features.
Radiomics Feature Selection
In order to improve the evaluation effect of muscle atrophy, it is necessary to screen out features related to muscle atrophy from a large amount of feature data. Firstly, normalize the radiomics features into radiomics labels, which can eliminate the unit limitation of each feature data. Then, in order to solve the problem of overfitting caused by excessive features, we use the binary discrete LASSO method to reduce the dimensionality of the feature space to construct a logistic regression model [45]. LASSO is a contraction method that restricts model coefficients by adding L1 regularization, which limits as many model coefficients as possible to 0. This allows for feature selection, reducing unselected feature coefficients to 0. The objective function of LASSO regression is shown in formula (3):
| 3 |
| 4 |
| 5 |
where y is the dependent variable, X is the independent variable, β is the vector of regression coefficients to be estimated, and λ is the hyperparameter controlling the strength of regularization. The value of is minimized by satisfying , where t ≥ 0 is the contraction parameter. For the LASSO filtering method, we use the “glmnet” package in R software to complete the feature selection and use a tenfold cross-validation method to select the most suitable λ. We use the remaining non-zero variables to construct a radiomics model.
Establishment and Evaluation of Radiomics Models
In this study, we first divided the data into a training set and a test set in a 4:1 ratio. Then, a muscle atrophy evaluation model was established based on the selected radiomics features and SVM. The SVM classifier constructs a hyperplane that provides the best separation boundary, which maximally separates objects in high-dimensional space. SVM is widely used because it can effectively avoid overfitting and has stable classification performance [46]. We analyzed each feature and all feature sets using an SVM classifier and established a model. In order to obtain a stable radiomics model, we used a tenfold cross-validation method to optimize the hyperparameters on the training set. Finally, the performance of the radiomics model was evaluated by the mean area under curve (AUC) and mean accuracy (ACC) in the training and validation sets.
Statistical Analysis
We used Matlab2019a (MathWorks, Natick, USA) and R software version 4.2.1 for the statistical analysis. We used Matlab2019a to extract features and calculate AUC of muscle fibers in H&E images. We used R software package “glmnet” for LASSO feature selection. We used a bilateral double sample t-test or Mann–Whitney test to evaluate the inter-group differences between normal and atrophic muscle fibers in each feature, depending on whether they are normally distributed. If the p-value is less than 0.05, the results are considered statistically significant. Spearman’s correlation coefficient and intra-class correlation coefficient (ICC) were used to evaluate the impact of segmentation on radiomics features, where ICC evaluates the consistency between automatic segmentation and manual segmentation features [47].
Experiments and Results
Dataset
Macaque Dataset
This study used H&E images of the calves of six macaques as experimental data. We prepared glass slides according to standard procedures, including slicing and fixed frozen sections on a freezing microtome (Cryotome E, Thermo), followed by cutting 3-µm-thick sections and staining them with H&E, where eosin renders the cytoplasm red. We placed tissue slices under a pathological scanner (Pannoramic MIDI, 3DHISTECH, Hungary) to capture images at a magnification of × 20. A total of six muscle tissue H&E images were obtained, with the size of 1920 × 1017 for each H&E image (Fig. 2B, C). The muscle fiber labeling in all six H&E whole-slice images was performed by two experienced pathologists (over 2 years of work experience) on the Matlab Image Labeler App. In order to adapt to the training of semi-supervised segmentation models, this study used the overlap method to crop 6 H&E images and ground truth into 540 small patches of size 256 × 256, with an overlap step size of 64. In this study, 540 images were allocated in an 8:2 ratio, with a training set of 432 and a testing set of 108. We randomly selected 25% of the images in the training set as labeled data, leaving 324 images as unlabeled data. The 108 images included in this test set all have manually labeled ground truth.
MoNuSeg Dataset
This dataset contains a total of 44 H&E images, of which 2163 annotated nuclei from 30 H&E images were used for training and 7223 annotated nuclei from 14 H&E images were used for testing [48]. The size of each image is 1000 × 1000. These images come from 18 different centers and 7 different organs. In order to increase the amount of data, we use four data augmentation techniques to alleviate overfitting: (1) random rotation on [0, 10], (2) random rotation on [− 10, 0], (3) mirroring, and (4) Gaussian blur.
Implementation Details of Semi-supervised Segmentation
For the training of the H&E image muscle fiber dataset, we used the Adaptive Moment Estimation optimizer (Adam) with an initial learning rate of 0.01 and an input image size of 256 × 256, with a training epoch of 100, and we set the batch size for supervised and unsupervised training to 16. In order to achieve better initialization of the segmentation network and reduce the impact of deviating from pseudo labels, the unsupervised branch only starts training from the sixth epoch. Poly scheduling is used to reduce the learning rate in the training process, . Due to the limited amount of imaging data, we conducted five data augmentation techniques, including (1) random rotation within the range of [− 10, 10], (2) mirroring, (3) translation, (4) Gaussian blur, and (5) image scaling.
Comparison with State-of-the-Art Methods
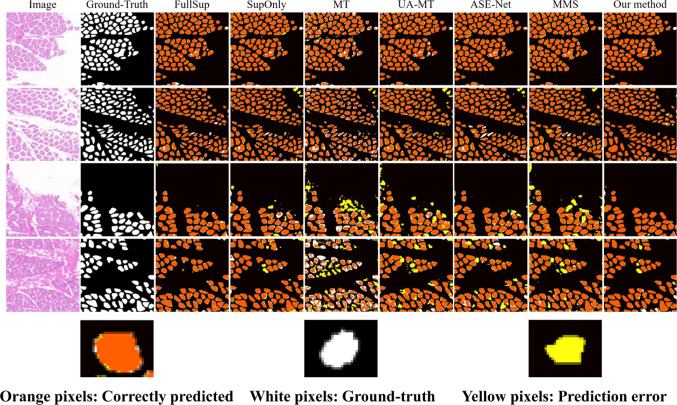
We compared the proposed method with the following semi-supervised semantic segmentation methods: MT [33], UA-MT [34], ASE-Net [35], and MMS [36]. We used the same segmentation network and experimental environment to implement four state-of-the-art semi-supervised segmentation methods to ensure fairness. To demonstrate the effectiveness and accuracy of our method, five evaluation metrics, Dice similarity coefficient (DSC), intersection-over-union (IoU), precision, recall, and F1-score, were calculated on the H&E muscle fiber image dataset and compared with existing methods. Among them, DSC and IoU are the main indicators for evaluating the accuracy of medical image segmentation. From Table 1, it can be seen that the proposed method outperforms only by 9.57%, 10.19%, 5.73%, and 1.95% under 1/16, 1/8, and 1/4 partition protocols, respectively. Overall, the proposed method outperforms other semi-supervised segmentation methods, especially for 1/4 labeled data, with a DSC exceeding 2.11% of MMS. In addition, we visualized and compared FullSup, SupOnly, and four semi-supervised segmentation methods in Fig. 6. From it, it can be observed that the proposed method can better predict the contours and details of muscle fibers, and there are fewer false predictions (yellow pixels).
Table 1.
Performance comparison with state-of-the-art semi-supervised segmentation methods on the H&E image muscle fiber dataset
| Label | Method | DSC (%) | IoU (%) | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|---|---|---|
| 1/16 (27) | SupOnly | 64.88 | 50.48 | 78.46 | 58.52 | 67.04 |
| MT [29] | 51.16 | 37.05 | 53.29 | 56.08 | 54.64 | |
| UA-MT [30] | 68.56 | 57.39 | 72.15 | 69.49 | 70.79 | |
| ASE-Net [31] | 71.46 | 58.51 | 77.63 | 72.38 | 74.91 | |
| MMS [32] | 72.88 | 59.87 | 78.94 | 66.68 | 72.29 | |
| Our method | 74.45 | 61.68 | 79.81 | 72.12 | 75.77 | |
| 1/8 (54) | SupOnly | 73.28 | 60.32 | 78.68 | 71.97 | 75.18 |
| MT [29] | 66.43 | 51.67 | 71.12 | 67.36 | 69.19 | |
| UA-MT [30] | 79.82 | 68.65 | 86.85 | 77.24 | 81.76 | |
| ASE-Net [31] | 82.98 | 73.15 | 82.69 | 83.71 | 83.19 | |
| MMS [32] | 82.63 | 70.09 | 87.65 | 78.27 | 82.69 | |
| Our method | 83.47 | 73.75 | 85.59 | 83.23 | 84.39 | |
| 1/4 (108) | SupOnly | 83.53 | 73.58 | 84.35 | 83.97 | 84.16 |
| MT [29] | 73.98 | 61.22 | 82.42 | 70.15 | 75.79 | |
| UA-MT [30] | 84.56 | 75.09 | 91.18 | 81.36 | 85.99 | |
| ASE-Net [31] | 86.82 | 78.31 | 91.51 | 94.46 | 93.07 | |
| MMS [32] | 87.15 | 76.62 | 90.47 | 83.39 | 86.79 | |
| Our method | 89.26 | 82.13 | 91.54 | 88.69 | 89.77 | |
| 100% | FullSup | 90.14 | 83.27 | 90.25 | 90.35 | 90.29 |
Values in bold indicate the best experimental results
Fig. 6.
Visualization comparison of segmentation results with different advanced methods. Full leverage labeled images (FullSup) represent training with 100% labeled data, while only leverage labeled images (SupOnly) represent training with only 1/4 labeled data. These two results represent the upper and lower bounds of the semi-supervised segmentation method. Other methods are trained using 1/4 labeled data and 3/4 unlabeled data. Orange pixels represent the correctly segmented area, white pixels represent ground truth, and yellow pixels represent incorrectly segmented area
In radiomics analysis, the segmentation method has a significant impact on the repeatability and reliability of radiomics features. We extracted 77 radiomics features from segmented and manually delineated regions, respectively. The reliability of radiomics parameters evaluated by Spearman’s correlation coefficients is shown in Supplementary Fig. 1, where radiomics features exhibit a high correlation between automatic segmentation and manual segmentation, with an average Spearman’s correlation coefficient of 0.88 (95% CI, 0.85–0.91). The repeatability of radiomics features evaluated by ICC has an average value of 0.86 (95% CI, 0.83–0.89). The Spearman’s correlation coefficients and ICC for all 77 features are presented in Supplementary Table 1.
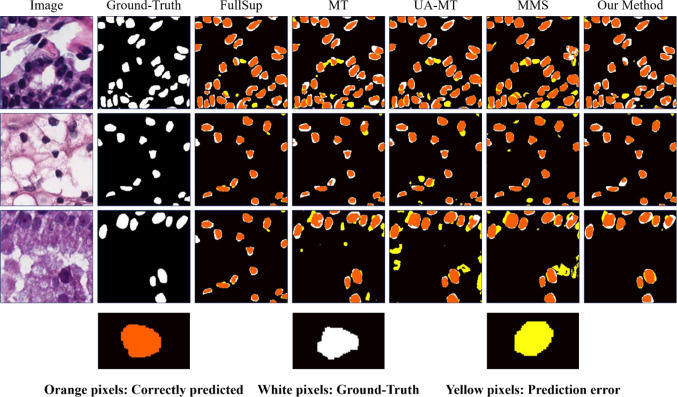
Due to the limited amount of data in muscle fiber H&E images, this may affect the generalization ability of our segmentation method. Therefore, we conducted experiments on the MoNuSeg dataset using the proposed semi-supervised segmentation model. The quantitative results are shown in Table 2, and it is clear that our method outperforms other state-of-the-art semi-supervised segmentation models under different numbers of labeled images. Especially in the 1/4 labeled image quantity, our method improved by 2.59% and 4.14% on DSC and IoU, respectively, compared to MMS. Figure 7 shows the visualization results of segmentation. Compared with other methods, our method can accurately segment cell regions and has a significantly lower false-positive rate (yellow pixels).
Table 2.
Performance comparison with state-of-the-art semi-supervised segmentation methods on the MoNuSeg dataset
| Label | Method | DSC (%) | IoU (%) | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|---|---|---|
| 1/16 | MT [29] | 71.03 | 55.68 | 69.34 | 76.24 | 72.71 |
| UA-MT [30] | 72.08 | 57.20 | 67.82 | 81.11 | 73.87 | |
| MMS [32] | 72.64 | 56.71 | 64.84 | 83.78 | 73.10 | |
| Our method | 74.04 | 59.12 | 70.92 | 78.52 | 74.53 | |
| 1/8 | MT [29] | 72.52 | 57.48 | 71.31 | 76.95 | 73.77 |
| UA-MT [30] | 74.48 | 59.90 | 69.19 | 83.91 | 75.84 | |
| MMS [32] | 75.35 | 60.25 | 72.19 | 80.10 | 75.94 | |
| Our method | 76.06 | 61.73 | 73.14 | 80.25 | 76.53 | |
| 1/4 | MT [29] | 74.32 | 59.48 | 72.41 | 78.00 | 75.10 |
| UA-MT [30] | 76.18 | 61.80 | 75.98 | 78.69 | 77.31 | |
| MMS [32] | 76.33 | 61.30 | 74.00 | 78.23 | 76.06 | |
| Our method | 78.92 | 65.44 | 77.80 | 80.90 | 79.32 | |
| 100% | FullSup | 79.27 | 65.90 | 75.06 | 84.54 | 79.52 |
Values in bold indicate the best experimental results
Fig. 7.
Visualization comparison of segmentation results with different advanced methods in nuclei segmentation. Orange pixels represent the correctly segmented area, white pixels represent ground truth, and yellow pixels represent incorrectly segmented area
Ablation Studies
This ablation experiment mainly explores the performance of different components in muscle fiber segmentation in H&E images. As shown in Table 3, we conducted ablation experiments on the proposed semi-supervised segmentation method. In this experiment, only 1/4 of the training data were labeled for the following ablation experiments.
Table 3.
Comparison of the quantitative results of ablation experiments for each module based on the training results of 1/4 labeled data as a benchmark
| Method | Loss | Evaluation index | |||||
|---|---|---|---|---|---|---|---|
| DSC (%) | IoU (%) | F1-score (%) | |||||
| Method 1 | √ | √ | 86.58 | 77.85 | 87.25 | ||
| Method 2 | √ | √ | √ | 87.65 | 79.19 | 88.93 | |
| Method 3 | √ | √ | √ | 87.96 | 79.84 | 88.52 | |
| Method 4 | √ | √ | √ | 87.99 | 79.87 | 88.54 | |
| Method 5 | √ | √ | √ | 88.07 | 80.14 | 88.69 | |
| Our method | √ | √ | √ | √ | 89.25 | 82.13 | 89.77 |
Values in bold indicate the best experimental results
Ablation Studies for and
In this study, the features of the teacher and student models in the same position are formed as positive pairs, while those in different positions are negative pairs. In order to discuss the performance of contrastive learning, in model 3 and model 5, it can be seen that the segmentation performance decreased by 1.29% and 1.18%, respectively, by calculating only the global feature information and the local feature information. In model 1, we did not use contrastive learning loss, but the result showed a significant decrease in DSC, from 89.25 to 86.58%, proving that contrastive learning can effectively extract feature information from unlabeled data, and integrating local and global information can better learn the diversity of muscle fiber features.
Ablation Studies for and
In order to ensure better convergence of the teacher and student models, this study used cross-entropy loss as the consistency loss to perform consistency regularization on the predicted results of the teacher and student models. From model 2 in Table 3, it can be seen that adding consistency loss increased DSC by 1.6%. Due to the fact that the number of positive samples is smaller than the number of negative samples in some H&E images after cropping, it is challenging to develop accurate semi-supervised segmentation algorithms. To address this imbalance issue, we introduce Dice loss into the supervised branch and combine it with cross-entropy loss to improve the performance of the training model. From model 4 and our method, it can be seen that adding Dice loss improves both DSC and IoU.
Radiomics Feature Selection and Analysis
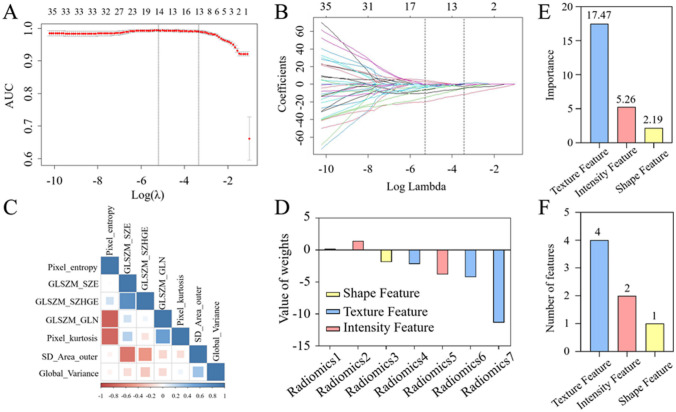
In order to determine the predictive factors related to muscle atrophy, 77 features were selected from H&E images, of which 64 features had an ICC ≥ 0.75. All 64 radiomics features were included in the LASSO logistic regression algorithm, and the dependent variable for normal muscle fibers was set to 0, while the dependent variable for atrophic muscle fibers was set to 1. As shown in Fig. 8, the features with non-zero coefficients were selected using the one-time standard deviation λ determined in Fig. 8A. Fig. 8B shows the distribution of LASSO coefficients for 64 radiomics features. Based on the LASSO regression model and muscle state, 7 potential predictive factors were selected from 64 features in the training set after removing features with zero regression coefficients, including 4 texture features, 2 intensity features, and 1 shape feature. Correlation analysis shows that the correlation coefficients between the radiomics features of the optimal feature subset obtained from the Spearman correlation coefficient are all less than 0.85 (Fig. 8C). Table 4 shows the relationship between the selected seven radiomics features and muscle atrophy status. “ − ” indicates a negative correlation between radiomics features and muscle atrophy status, while “ + ” indicates a positive correlation. Fig. 8D shows the coefficients of the seven selected radiomics features and the importance of their impact on the dependent variable. From Fig. 8D, it can be seen that for a single radiomics feature, the absolute coefficient value of GLSZM_SZE for texture features is the highest, at 11.36. For single class radiomics feature groups, the absolute coefficient value and quantity of texture feature groups are the highest, at 17.48 (Fig. 8E, F). Therefore, texture features have a greater impact on distinguishing between normal and atrophic muscle fibers.
Fig. 8.
Radiomics feature selection based on the LASSO method. A The penalty term in the LASSO model through a tenfold cross-validation based on the minimum criterion λ. The y-axis represents binomial deviation. The lower x-axis represents the logarithm (λ). The upper x-axis represents the average number of predictive factors. The red dots indicate average deviance values for each λ where different models have different deviance. B LASSO coefficient spectrum of 77 radiomics features. C The correlation between radiomics features related to muscle status, and the depth of their color indicates the degree of correlation between different features. D The coefficients of seven radiomics features filtered out through the LASSO algorithm. We selected the minimum standard based on tenfold cross-validation (λ = 0.0353956, the 1 − SE criterion). E The total LASSO coefficients showing the radiomics features of each class are presented. F The number of selected radiomics features in each class
Table 4.
Definition, name, class, and weight value of seven selected radiomics features
| Definition | Name of the feature | Class | Value of weights |
|---|---|---|---|
| Radiomics 1 | GLSZM_SZHGE | Texture feature | − 0.01118295 |
| Radiomics 2 | Pixel_Entropy | Intensity feature | 1.42789398 |
| Radiomics 3 | Global.Variance | Texture feature | − 1.87595125 |
| Radiomics 4 | SD_Area_outer | Shape feature | − 2.19355039 |
| Radiomics 5 | Pixel_ kurtosis | Intensity feature | − 3.82902944 |
| Radiomics 6 | GLSZM_GLN | Texture feature | − 4.22294183 |
| Radiomics 7 | GLSZM_SZE | Texture feature | − 11.36601396 |
Analyzing Muscle Status Based on Individual Radiomics Features
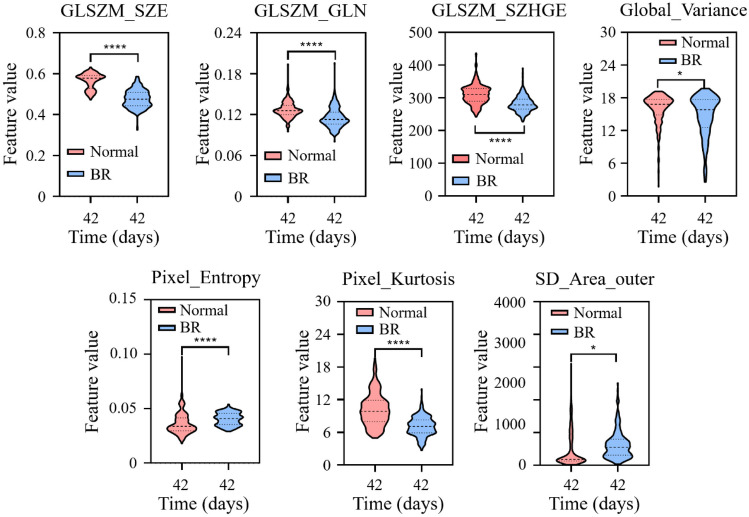
In order to further analyze the p-values and differences of radiomics features one to seven between normal and atrophic muscle fibers, calculate the p-value of individual radiomics features for two muscle states using the t-test statistical analysis method. As shown in Fig. 9, the seven radiomics features showed extremely significant differences in normal and atrophic muscle fibers. Figure 9 shows that as the muscle fiber status transitions from normal to atrophy, only Global_Variance and SD_Area_outer show a significant increase. From Fig. 9, it can be seen that the GLSZM_SZE, GLSZM_GLN, and GLSZM_SZHGE features are significantly reduced, and all have extremely significant differences. However, the Global_Variance value (15.97) of atrophic muscle fibers is higher than that of normal muscle fibers (14.74), and the difference is not as significant as the first three texture features. Pixel_Entropy and Pixel_ kurtosis in Fig. 9 represent entropy and kurtosis in intensity features, with the most significant decrease in kurtosis compared to entropy, decreasing from 10.19 to 7.11, respectively. Through the selection of seven features, it was found that the changes in muscle fibers are also reflected in their shape, as shown in Fig. 9. Compared with the texture and intensity features, the standard deviation of the circumscribed area from normal muscle fibers to atrophic muscle fibers increased by 11.2%, indicating that after muscle fiber atrophy, the shape changed significantly and became irregular.
Fig. 9.
The trend of changes in the seven selected radiomics features in normal and atrophy fibers
Performance Differences of Individual Feature Models Compared to Combined Models
We randomly selected 80% of H&E imaging data as the training set and the remaining 20% as the test set. We constructed an SVM prediction model using the features selected in Table 4 and evaluated it through AUC and ACC. As shown in Table 5, the feature combination composed of seven radiomics features achieved good classification performance in distinguishing normal and atrophic muscle fibers, with an AUC and ACC of 0.9687 and 0.8841 in the test set, respectively. To evaluate the performance of individual radiomics features, we constructed SVM classification models for each feature and compared them with feature combinations. The results in Table 5 indicate that GLSZM_SZE is the single radiomics feature with the highest AUC value in the test queue (0.9151), although it is lower than the overall model obtained by combining seven features (AUC = 0.9687). This demonstrates the importance of GLSZM_SZE in evaluating changes in muscle fibers during muscle atrophy. In addition, a comparison was made between the predictive performance of models based solely on CSA features that were more clinically focused and the radiomics features screened in this study. The results showed that the AUC of any single radiomics feature was higher than that based on CSA features (Table 5), demonstrating that radiomics features can better distinguish between normal and atrophic muscle fibers.
Table 5.
Summary of radiomics features related to muscle atrophy and performance based on individual features and feature combinations in the test queue
| Modality | Feature type | Feature | AUC | ACC |
|---|---|---|---|---|
| Individual feature | Muscle morphometric parameters | CSA | 0.5454 | 0.6147 |
| Radiomics 1 | Texture feature | GLSZM_SZHGE | 0.8137 | 0.7248 |
| Radiomics 2 | Intensity feature | Pixel_Entropy | 0.7363 | 0.6697 |
| Radiomics 3 | Texture feature | Global_Variance | 0.5931 | 0.4862 |
| Radiomics 4 | Shape feature | SD_Area_outer | 0.7029 | 0.5910 |
| Radiomics 5 | Intensity feature | Pixel_Kurtosis | 0.7933 | 0.7773 |
| Radiomics 6 | Texture feature | GLSZM_GLN | 0.7927 | 0.7193 |
| Radiomics 7 | Texture feature | GLSZM_SZE | 0.9151 | 0.8440 |
| All combined | 0.9687 | 0.8841 |
Values in bold indicate the best experimental results
The Stability of the GLSZM_SZE Feature
In order to further analyze whether texture features and GLSZM_SZE features are affected by image contrast [49], this resulted in excellent performance of this feature in evaluating muscle atrophy. We extracted radiomics features from three different contrast H&E images and analyzed them. As shown in Fig. 10, the LASSO coefficients of the texture feature group are the highest in low-contrast and high-contrast images at 13.01 and 13.46, respectively. In the texture feature group, the maximum LASSO coefficients of the GLSZM feature group are 11.78 and 12.61, respectively. This proves that the GLSZM feature group has a greater advantage in evaluating changes in normal and atrophic muscle fibers. In addition, GLSZM_SZE, as the feature with the highest coefficient in the GLSZM_SZE feature group, still contributes the most in evaluating muscle atrophy in three different contrast images, indicating that GLSZM_SZE features are not affected by image contrast.
Fig. 10.
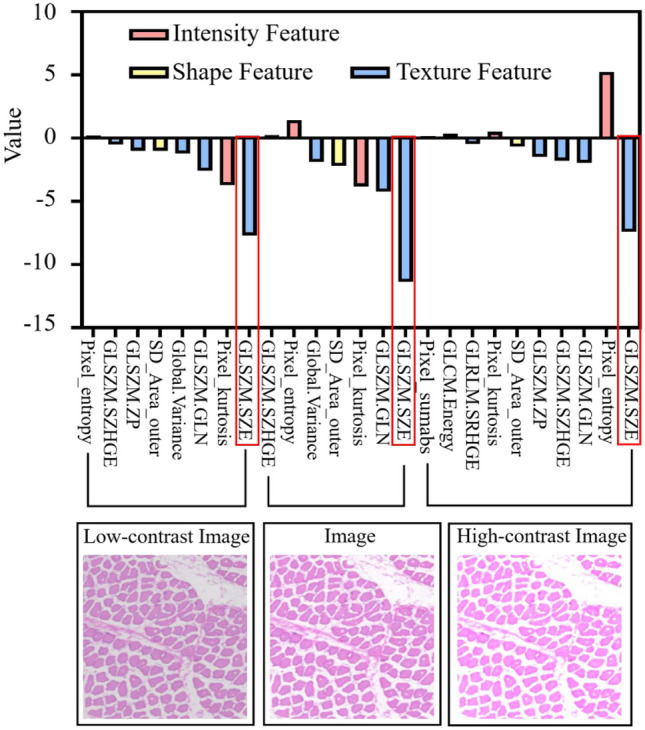
The importance of features under different H&E image contrasts. The x-axis represents the name of the radiomics feature, and the y-axis represents the coefficient of the radiomics feature. All experimental groups used the LASSO method for feature selection, with eight radiomics features selected from low-contrast images and ten radiomics features selected from high-contrast images
Discussion
Long-term space flight and microgravity environments can lead to a decrease in mass and strength of skeletal muscles. The most direct reason for this is the reduced contraction of muscle fibers, which will affect the efficiency of astronauts’ work and their ability to adapt to gravity when returning to Earth. Accurately evaluating weightless muscle atrophy is of great significance for the early diagnosis and treatment of muscle atrophy. At present, in the field of muscle disease and rehabilitation, researchers usually use pathological biopsy to evaluate quantifiable indicators (CSA, perimeter) of muscle fibers. However, these indicators are influenced by patients’ physical activity, dietary intake, and specific muscles, which have a certain impact on the accurate assessment of weightless muscle atrophy [9]. Hence, we attempted to explore a potential biomarker for evaluating muscle atrophy based on H&E images using radiomics methods. The main work includes the following two points. Firstly, a semi-supervised segmentation model based on contrastive learning is proposed to solve the problem of manual labeling difficulty due to the large number of muscle fibers. Then, we analyzed the radiomics features of normal and atrophic muscle fibers and demonstrate that radiomics features derived from H&E images have predictive value for evaluating muscle atrophy.
ROI segmentation is the first step in the radiomics workflow. When manually performing target segmentation, the data have a high degree of variability. However, hundreds or even thousands of muscle fibers need to be analyzed in the study of muscle fibers in H&E images, making manual segmentation very time-consuming. With the introduction and continuous improvement of DL, DL models have achieved significant success in segmenting cells in histopathological images [37]. However, in order to achieve good segmentation performance, a large amount of annotated data are required. Such labeled data is quite time-consuming in the collection process and even requires multiple labeling of the target area. Therefore, how to train deep models with only a few labeled data has become a hot topic of concern for researchers. Currently, semi-supervised segmentation models have become a more promising learning method, with the advantage of requiring only a few pixel level annotated data and being able to learn from a large amount of unlabeled data [33, 34]. Based on the above insights, we propose a semi-supervised segmentation algorithm based on contrastive learning to demonstrate its effectiveness on our own H&E muscle fiber image dataset. As shown in Table 1, the proposed method is more accurate in segmenting muscle fiber regions compared to other state-of-the-art methods. In addition, we use the segmentation results obtained from fully supervised training as the upper limit of semi-supervised segmentation performance and include different amounts of unlabeled data in the training. From Table 3, it can be seen that the 1/4 labeled data is closer to fully supervised segmentation performance, which can prove that the proposed method can achieve fully supervised performance by only using a small amount of labeled training data. Although encoder and decoder structures have been widely used in medical image segmentation, a single encoder captures global features while obtaining local features, making it difficult to accurately distinguish the differences between muscle fibers and the background, especially in challenging situations such as low contrast between muscle fibers and background noise, and irregular muscle fiber shapes [50]. In order to achieve better feature extraction capability of the model, we adopted a dual encoder input branch composed of a transformer and CNN in the feature extraction module to encourage the network to capture both global contextual information and local features of muscle fibers simultaneously. In addition, we use the contrast loss of global and local features to enhance pixel level intra-class compactness and inter-class separability, in order to extract important feature information from unlabeled data. In recent years, radiomics features have been proven to be closely related to muscle biological characteristics and disease assessment. However, segmentation methods have a significant impact on the repeatability and reliability of radiomics features. Therefore, in this study, the average Spearman’s correlation coefficient and ICC between semi-supervised segmentation results and manual segmentation results were 0.88 and 0.86, respectively. This proves that the features extracted by the semi-supervised segmentation method have high repeatability and reliability and can replace manual segmentation methods.
Based on the above segmentation results, another purpose of this study is to demonstrate the significant value of radiomics features derived from H&E images in evaluating muscle atrophy. Based on the LASSO algorithm, seven radiomics features were selected, including four texture features, two intensity features, and one shape feature. Among the four texture features, three are GLSZM and one is a global texture feature. The two intensity features are kurtosis and entropy, and the one shape feature is the standard deviation of the circumscribed circle area. The kurtosis feature highlights the intensity changes in the muscle fiber region, while the entropy value reflects the texture changes of the muscle fibers, which is similar to the texture feature. On the other hand, GLSZM accounts for the most in texture features, representing the number of areas with a grayscale value of i and a connected area of j in the image, reflecting the fineness of the image texture [51]. As shown in Fig. 9, it can be seen that normal muscle fibers and atrophic muscle fibers exhibit significant strength differences, and changes in pixel strength can reflect the differences between texture features of GLSZM. Based on this observation, it can be concluded that GLSZM-derived features are potentially effective representations of muscle fiber H&E images to evaluate muscle atrophy. Previous studies have shown that the elimination of anti-gravity loading may lead to a decrease in protein synthesis [52] and a decrease in the expression of MyHC-I [53], which may be reflected in the texture refinement of H&E images. These features are related to the changes in the types of proteins inside muscle fibers. The more proteins there are, the higher the expression level of their proteins, indicating that the texture information inside muscle fibers is more abundant. The small zone emphasis (SZE) in GLSZM features refers to the proportion of the small- and medium-sized area of the image in the whole connected area, which precisely reflects this feature [54]. Figure 9A shows the significant difference between normal and atrophic muscle fibers in GLSZM_SZE. The GLSZM_SZE value of normal muscle fibers is 0.1 higher than that of atrophic muscle fibers, indicating that the larger the GLSZM_SZE value, the smaller the regions, and the finer the image texture. This proves that the features of GLSZM_SZE may be related to changes in certain key proteins in muscle atrophy. The results in Table 5 also confirm that GLSZM_SZE performs best in evaluating muscle atrophy.
To further validate the performance of radiomics features in evaluating muscle atrophy, this study used SVM models to test the performance of individual radiomics features and the performance of combined features. Previous studies have shown that pathologists are more focused on morphometric indicators such as CSA and the perimeter of muscle fibers [27, 28]. After 42 days of BR in macaques, there was a significant decrease in the CSA and perimeter of muscle fibers (Fig. 2D). However, in this study, the texture feature group had a 20.37% higher prediction accuracy than the muscle morphology measurement group in the performance comparison of feature categories (Table 6), indicating that texture features have a significant advantage in evaluating muscle atrophy. For the evaluation performance of individual features, the performance of the established muscle classification model is higher than that of CSA. As shown in Table 5, particularly GLSZM_SZE and GLSZM_SZHGE are 36.97% and 26.83% higher than CSA feature in AUC indicators. This indicates that during the process of muscle atrophy, the fine-grained texture of muscle fibers can better reflect changes in muscle atrophy than CSA. In order to evaluate the importance of GLSZM_SZE without the influence of H&E image contrast, we used three types of contrast images for validation, extracting radiomics features and using the LASSO algorithm to filter features. As shown in Fig. 10, although the number of features included in low-contrast and high-contrast images has changed compared to the original image, GLSZM_SZE remains the same among all features and has the greatest impact. This indicates that the importance of GLSZM_SZE features in evaluating muscle atrophy is not affected by image contrast and can be considered as a potential biomarker for evaluating muscle atrophy in the future. Based on the above analysis, this study can conclude that the radiomics features of H&E images have potential value in evaluating the state of muscle fibers in muscle atrophy, and the precision of texture features may help further explain the changes in some proteins of muscle fibers during the atrophy process.
Table 6.
Evaluate performance based on each feature class in the test queue
| Modality | Feature class | Feature | AUC | ACC |
|---|---|---|---|---|
| Feature subset | Muscle morphometric parameters |
CSA Perimeter Diameter |
0.7174 | 0.6697 |
| Feature subset | Intensity feature |
Pixel_Entropy Pixel_Kurtosis |
0.8050 | 0.7706 |
| Feature subset | Texture feature |
GLSZM_SZHGE Global_Variance GLSZM_GLN GLSZM_SZE |
0.9460 | 0.8734 |
| All combined | 0.9687 | 0.8841 |
Values in bold indicate the best experimental results
Our research still has the following limitations. Firstly, due to the difficulty in establishing a BR model for macaques in this study, six macaques were used as experimental subjects, with three as the control group and three as the BR group. However, a small sample size can have a certain impact on the training of segmentation models. For example, when taking patches, if the overlap coefficient is too high, it can cause too many duplicate images, resulting in poor reliability of the results. On the contrary, if the overlap coefficient is equal to 0, it will cause a decrease in data volume, which is not conducive to improving segmentation results. In future work, the amount of data will increase, which may be beneficial for improving the accuracy of segmentation and evaluation of muscle atrophy. Secondly, this study only focuses on the evaluation of radiomics features and does not take into account the changes in molecular level indicators in muscle atrophy. Therefore, more molecular indicators need to be collected in future work, which not only helps to better explain the significance of radiomics features but may also construct more accurate evaluation models.
Conclusion
This study explores a radiomics feature based on H&E images as a potential biomarker for evaluating muscle atrophy. By establishing a semi-supervised automatic segmentation method based on contrastive learning, the inherent limitations of limited training data annotations are addressed. The experimental results show that the semi-supervised automatic segmentation method has high accuracy in muscle fiber H&E images, and the extracted features have high repeatability and reliability. We performed a radiomics analysis using features extracted from automatically segmented muscle fiber regions to evaluate muscle atrophy and find a potential biomarker. The experimental results indicate that the fineness of the texture can better reflect the difference between normal and atrophic muscle fibers. For a single feature, GLSZM_SZE may be a potential biomarker for effectively evaluating weightless muscle atrophy, with the best performance in AUC and ACC, with values of 0.915 and 0.844, respectively. In summary, the predictive model established in this study and the explored biomarkers provide a powerful auxiliary tool for the early diagnosis and treatment of weightless muscle atrophy in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
All authors contributed to the study conception and design. Conceptualization, methodology, software, validation, formal analysis, investigation, writing—original draft preparation, and writing—review and editing were performed by Getao Du. Data acquisition, funding acquisition, and H&E slice preparation were performed by Peng Zhang, Xu Zhou, and Jiajie Jia. The establishment of animal model was performed by Guanghan Kan. Resources was performed by Jianzhong Guo. Writing—review and editing and funding acquisition were performed by Xiaoping Chen. Conceptualization, resources, and supervision were performed by Jimin Liang. Conceptualization, resources, writing review and editing, supervision, project administration, and funding acquisition were performed by Yonghua Zhan.
Funding
This work was supported, in part, by the National Natural Science Foundation of China (32171173), the National Key Laboratory of Space Medicine Fundamentals and Application (No. SMFA20A02), the Space Medical Experiment Project of China Manned Space Program (HYZHXM01017), and the Natural Science Basic Research Program of Shaanxi (Program No. 2024JC-ZDXM-47).
Data Availability
All data in this study can be obtained by contacting the corresponding authors upon reasonable request.
Declarations
Ethics Approval
The animal and the experimental protocol used in the paper were reviewed and ethically approved by the Institutional Animal Care and Use Committee of China Astronaut Research and Training Center (ACC-IACUC-2019–002).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaoping Chen, Email: xpchen2009@163.com.
Jimin Liang, Email: jiminliang@gmail.com.
Yonghua Zhan, Email: yhzhan@xidian.edu.cn.
References
- 1.Guo N, Fan XY, Wu YT, Li ZL, Liu SJ, Wang LJ, Yao J, Li YH, Effect of Constraint Loading on the Lower Limb Muscle Forces in Weightless Treadmill Exercise. JOURNAL OF HEALTHCARE ENGINEERING 2018: 8487308, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne MWC, Williams DR, Trudel G, Space flight rehabilitation. AMERICAN JOURNAL OF PHYSICAL MEDICINE & REHABILITATION 86: 583-591, 2007. [DOI] [PubMed] [Google Scholar]
- 3.Kubat GB, Bouhamida E, Ulger O, Turkel I, Pedriali G, Ramaccini D, Ekinci O, Ozerklig B, Atalay O, Patergnani S, Sahin BN, Morciano G, Tuncer M, Tremoli E, Pinton P, Mitochondrial dysfunction and skeletal muscle atrophy: Causes, mechanisms, and treatment strategies. MITOCHONDRION 72: 33-58, 2023. [DOI] [PubMed] [Google Scholar]
- 4.Gouzi F, Maury J, Molinari N, Pomiès P, Mercier J, Préfaut C, Hayot M, Reference values for vastus lateralis fiber size and type in healthy subjects over 40 years old: a systematic review and metaanalysis. JOURNAL OF APPLIED PHYSIOLOGY 115: 346-354, 2013. [DOI] [PubMed] [Google Scholar]
- 5.Guttsches AK, Rehmann R, Schreiner A, Rohm M, Forsting J, Froeling M, Tegenthoff M, Vorgerd M, Schlaffke L, Quantitative Muscle-MRI Correlates with Histopathology in Skeletal Muscle Biopsies. Journal of Neuromuscular Diseases 8: 669-678, 2021. [DOI] [PubMed] [Google Scholar]
- 6.Baguet A, Everaert I, Hespel P, Petrovic M, Achten E, Derave W, A New Method for Non-Invasive Estimation of Human Muscle Fiber Type Composition. Plos One 6: e21956, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau YS, Xu L, Gao YD, Han RZ, Automated muscle histopathology analysis using CellProfiler. SKELETAL MUSCLE 8: 32, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arentson-Lantz EJ, English KL, Paddon-Jones D, Fry CS, Fourteen days of bed rest induces a decline in satellite cell content and robust atrophy of skeletal muscle fibers in middle-aged adults. JOURNAL OF APPLIED PHYSIOLOGY 120: 965-975, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumitru A, Radu BM, Radu M, Cretoiu SM, Muscle Changes During Atrophy. ADVANCES IN EXPERIMENTAL MEDICINE AND BIOLOGY 1088: 73-92, 2018. [DOI] [PubMed] [Google Scholar]
- 10.Chan JKC, The Wonderful Colors of the Hematoxylin-Eosin Stain in Diagnostic Surgical Pathology. INTERNATIONAL JOURNAL OF SURGICAL PATHOLOGY 22: 12-32, 2014. [DOI] [PubMed] [Google Scholar]
- 11.Ming X, Oei RW, Zhai RP, Kong FF, Du CR, Hu CS, Hu WG, Zhang Z, Ying HM, Wang JZ, MRI-based radiomics signature is a quantitative prognostic biomarker for nasopharyngeal carcinoma. SCIENTIFIC REPORTS 9: 10412, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chong GO, Park SH, Park NJY, Bae BK, Lee YH, Jeong SY, Kim JC, Park JY, Ando Y, Han HS, Predicting Tumor Budding Status in Cervical Cancer Using MRI Radiomics: Linking Imaging Biomarkers to Histologic Characteristics. CANCERS 13: 5140, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma MM, Gan LY, Liu YH, Jiang Y, Xin L, Liu Y, Qin NS, Cheng YJ, Liu Q, Xu L, Zhang YF, Wang XP, Zhang XD, Ye JM, Wang XY, Radiomics features based on automatic segmented MRI images: Prognostic biomarkers for triple-negative breast cancer treated with neoadjuvant chemotherapy. EUROPEAN JOURNAL OF RADIOLOGY 146: 110095, 2022. [DOI] [PubMed] [Google Scholar]
- 14.Zhang P, Chen XP, Fan M, Signaling mechanisms involved in disuse muscle atrophy. MEDICAL HYPOTHESES 69: 310-321, 2007. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, Zhang P, Liu HJ, Fan M, Chen XP, Proteomic analysis of mouse soleus muscles affected by hindlimb unloading and reloading. Muscle Nerve 52: 803-811, 2015. [DOI] [PubMed] [Google Scholar]
- 16.Mayerhoefer ME, Materka A, Langs G, Häggström I, Szczypinski P, Gibbs P, Cook G, Introduction to Radiomics. JOURNAL OF NUCLEAR MEDICINE 61: 488-495, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bortolotto C, Lancia A, Stelitano C, Montesano M, Merizzoli E, Agustoni F, Stella G, Preda L, Filippi AR, Radiomics features as predictive and prognostic biomarkers in NSCLC. EXPERT REVIEW OF ANTICANCER THERAPY 21: 257-266, 2021. [DOI] [PubMed] [Google Scholar]
- 18.Granata V, Fusco R, Costa M, Picone C, Cozzi D, Moroni C, La Casella GV, Montanino A, Monti R, Mazzoni F, Grassi R, Malagnino VR, Cappabianca S, Grassi R, Miele V, Petrillo A, Preliminary Report on Computed Tomography Radiomics Features as Biomarkers to Immunotherapy Selection in Lung Adenocarcinoma Patients. CANCERS 13: 3992, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerrisi A, Russillo M, Loi E, Ganeshan B, Ungania S, Desiderio F, Bruzzaniti V, Falcone I, Renna D, Ferraresi V, Caterino M, Solivetti FM, Cognetti F, Morrone A, Exploring CT Texture Parameters as Predictive and Response Imaging Biomarkers of Survival in Patients With Metastatic Melanoma Treated With PD-1 Inhibitor Nivolumab: A Pilot Study Using a Delta-Radiomics Approach. FRONTIERS IN ONCOLOGY 11: 704607, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brancato V, Cavaliere C, Garbino N, Isgrò F, Salvatore M, Aiello M, The relationship between radiomics and pathomics in Glioblastoma patients: Preliminary results from a cross-scale association study. FRONTIERS IN ONCOLOGY 12: 1005805, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke ZH, Wang DY, Yan Q, Ren J, Lau RWH, Dual student: Breaking the limits of the teacher in semi-supervised learning [C]// Proceedings of the 2019 IEEE/CVF INTERNATIONAL CONFERENCE ON COMPUTER VISION. 2019: 6727–6735.
- 22.Su H, Yin ZZ, Huh SG, Kanade T, Zhu J, Interactive Cell Segmentation Based on Active and Semi-Supervised Learning. IEEE TRANSACTIONS ON MEDICAL IMAGING 35: 762-777, 2016. [DOI] [PubMed] [Google Scholar]
- 23.Kucharski D, Kleczek P, Jaworek-Korjakowska J, Dyduch G, Gorgon M, Semi-Supervised Nests of Melanocytes Segmentation Method Using Convolutional Autoencoders. SENSORS 20: 1546, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar N, Uppala P, Duddu K, Sreedhar H, Varma V, Guzman G, Walsh M, Sethi A, Hyperspectral Tissue Image Segmentation Using Semi-Supervised NMF and Hierarchical Clustering. IEEE TRANSACTIONS ON MEDICAL IMAGING 38: 1304-1313, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandenburgh H, Chromiak J, Shansky J, Del Tatto M, Lemaire J, Space travel directly induces skeletal muscle atrophy. FASEB JOURNAL 13: 1031-1038, 1999. [DOI] [PubMed] [Google Scholar]
- 26.Ventadour S, Attaix D, Mechanisms of skeletal muscle atrophy. CURRENT OPINION IN RHEUMATOLOGY 18: 631-635, 2006. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimitsu K, Shiba N, Matsuse H, Takano Y, Matsugaki T, Inada T, Tagawa Y, Nagata K, Development of a Training Method for Weightless Environment Using Both Electrical Stimulation and Voluntary Muscle Contraction. TOHOKU JOURNAL OF EXPERIMENTAL MEDICINE 220: 83-93, 2010. [DOI] [PubMed] [Google Scholar]
- 28.Viggars MR, Wen Y, Peterson C, Jarvis JC, Automated cross-sectional analysis of trained, severely atrophied, and recovering rat skeletal muscles using MyoVision 2.0. JOURNAL OF APPLIED PHYSIOLOGY 132: 593–610, 2022. [DOI] [PubMed]
- 29.Salaun E, Lefeuvre-Orfila L, Cavey T, Martin B, Turlin B, Ropert M, Loreal O, Derbré F, Myriocin prevents muscle ceramide accumulation but not muscle fiber atrophy during short-term mechanical unloading. JOURNAL OF APPLIED PHYSIOLOGY 120: 178-187, 2016. [DOI] [PubMed] [Google Scholar]
- 30.Retting A, Haase T, Pletnyov A, Kohl B, Ertel W, von Kleist M, Sunkara V, SLCV-a supervised learning-computer vision combined strategy for automated muscle fibre detection in cross-sectional images. Peerj 7: e7053, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu XN, Sanford T, Turkbey B, Xu S, Wood BJ, Yan PK, Shadow-Consistent Semi-Supervised Learning for Prostate Ultrasound Segmentation. IEEE TRANSACTIONS ON MEDICAL IMAGING 41: 1331-1345, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farooq MU, Ullah Z, Gwak J, Residual attention based uncertainty-guided mean teacher model for semi-supervised breast masses segmentation in 2D ultrasonography. COMPUTERIZED MEDICAL IMAGING AND GRAPHICS 104: 102173, 2023. [DOI] [PubMed] [Google Scholar]
- 33.Tarvainen A, Valpola H, Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results[C]// Neural Information Processing Systems (nips). 2017: 1195–1204.
- 34.Yu LQ, Wang SJ, Li XM, Fu CW, Heng PA, Uncertainty-Aware Self-ensembling Model for Semi-supervised 3D Left Atrium Segmentation[C]// Proceedings of the 22nd International Conference on Medical Image Computing and Computer-Assisted Intervention. 2019: 605–613.
- 35.Lei T, Zhang D, Du XG, Wang X, Wan Y, Nandi AK, Semi-Supervised Medical Image Segmentation Using Adversarial Consistency Learning and Dynamic Convolution Network. IEEE TRANSACTIONS ON MEDICAL IMAGING 42: 1265-1277, 2023. [DOI] [PubMed] [Google Scholar]
- 36.Lou A, Tawfik K, Yao X, Liu ZT, Noble J, Min-Max Similarity: A Contrastive Semi-Supervised Deep Learning Network for Surgical Tools Segmentation. IEEE TRANSACTIONS ON MEDICAL IMAGING 42: 2832-2841, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu HS, Wang ZZ, Song YY, Yang L, Qin J, Cross-patch Dense Contrastive Learning for Semi-supervised Segmentation of Cellular Nuclei in Histopathologic Images[C]// Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2022: 11656–11665.
- 38.Gillies RJ, Schabath MB, Radiomics Improves Cancer Screening and Early Detection. CANCER EPIDEMIOLOGY BIOMARKERS & PREVENTION 29: 2556-2567, 2020. [DOI] [PubMed] [Google Scholar]
- 39.Lu C, Shiradkar R, Liu ZY, Integrating pathomics with radiomics and genomics for cancer prognosis: A brief review. CHINESE JOURNAL OF CANCER RESEARCH 33: 563-573, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen WT, Gong MC, Zhou DS, Zhang LJ, Kong J, Jiang F, Feng SX, Yuan RQ, CT-based deep learning radiomics signature for the preoperative prediction of the muscle-invasive status of bladder cancer. FRONTIERS IN ONCOLOGY 12: 1019749, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim CH, Bhattacharjee S, Prakash D, Kang S, Cho NH, Kim HC, Choi HK, Artificial Intelligence Techniques for Prostate Cancer Detection through Dual-Channel Tissue Feature Engineering. CANCERS 13: 1524, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Pang XS, Li SM, Li WJ, Chen XP, Zhang P, ER Stress is Activated and Involved in Disuse-Induced Muscle Atrophy. FRONTIERS IN BIOSCIENCE-LANDMARK 28: 136, 2023. [DOI] [PubMed] [Google Scholar]
- 43.Wu YW, Zeng DW, Wang ZP, Shi YY, Hu JT, Distributed contrastive learning for medical image segmentation. MEDICAL IMAGE ANALYSIS 81: 102564, 2022. [DOI] [PubMed] [Google Scholar]
- 44.Bagher-Ebadian H, Chetty IJ, Technical Note: ROdiomiX: A validated software for radiomics analysis of medical images in radiation oncology. MEDICAL PHYSICS 48: 354-365, 2021. [DOI] [PubMed] [Google Scholar]
- 45.Kong CL, Zhao ZW, Chen WY, Lv XL, Shu GF, Ye MQ, Song JJ, Ying XH, Weng QY, Weng W, Fang SJ, Chen MJ, Tu JF, Ji JS, Prediction of tumor response via a pretreatment MRI radiomics-based nomogram in HCC treated with TACE, EUROPEAN RADIOLOGY 31: 7500-7511, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang XH, Wan Q, Chen HJ, Li YF, Li XC, Classification of pulmonary lesion based on multiparametric MRI: utility of radiomics and comparison of machine learning methods. EUROPEAN RADIOLOGY 30: 4595-4605, 2020. [DOI] [PubMed] [Google Scholar]
- 47.Yunus MM, Sabarudin A, Karim MKA, Nohuddin PNE, Zainal IA, Shamsul MSM, Yusof AKM, Reproducibility and Repeatability of Coronary Computed Tomography Angiography (CCTA) Image Segmentation in Detecting Atherosclerosis: A Radiomics Study. DIAGNOSTICS 12: 2007, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar N, Verma R, Anand D, Zhou YN, Onder OF, Tsougenis E, et al, A Multi-Organ Nucleus Segmentation Challenge. IEEE TRANSACTIONS ON MEDICAL IMAGING 39: 1380-1391, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Radzi SFM, Karim MKA, Saripan MI, Abd Rahman MA, Osman NH, Dalah EZ, Noor NM, Impact of Image Contrast Enhancement on Stability of Radiomics Feature Quantification on a 2D Mammogram Radiograph. IEEE ACCESS 8: 127720-127731, 2020. [Google Scholar]
- 50.Strange H, Scott I, Zwiggelaar R, Myofibre segmentation in H&E stained adult skeletal muscle images using coherence-enhancing diffusion filtering. BMC MEDICAL IMAGING 14: 38, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bogowicz M, Riesterer O, Stark LS, Studer G, Unkelbach J, Guckenberger M, Tanadini-Lang S, Comparison of PET and CT radiomics for prediction of local tumor control in head and neck squamous cell carcinoma. ACTA ONCOLOGICA 56: 1531-1536, 2017. [DOI] [PubMed] [Google Scholar]
- 52.Schakman O, Gilson H, Kalista S, Thissen JP, Mechanisms of Muscle Atrophy Induced by Glucocorticoids. HORMONE RESEARCH 72: 36-41, 2009. [DOI] [PubMed] [Google Scholar]
- 53.Sharlo CA, Lomonosova YN, Turtikova OV, Mitrofanova OV, Kalamkarov GR, Bugrova AE, Shevchenko TF, Shenkman BS, The Role of GSK-3β Phosphorylation in the Regulation of Slow Myosin Expression in Soleus Muscle during Functional Unloading. BIOLOGICHESKIE MEMBRANY 34: 164-171, 2018. [Google Scholar]
- 54.Bourbonne V, Fournier G, Vallières M, Lucia F, Doucet L, Tissot V, Cuvelier G, Hue S, Du HL, Perdriel L, Bertrand N, Staroz F, Visvikis D, Pradier O, Hatt M, Schick U, External Validation of an MRI-Derived Radiomics Model to Predict Biochemical Recurrence after Surgery for High-Risk Prostate Cancer. CANCERS 12: 814, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this study can be obtained by contacting the corresponding authors upon reasonable request.