Abstract
Precision medicine research benefits from machine learning in the creation of robust models adapted to the processing of patient data. This applies both to pathology identification in images, i.e., annotation or segmentation, and to computer-aided diagnostic for classification or prediction. It comes with the strong need to exploit and visualize large volumes of images and associated medical data. The work carried out in this paper follows on from a main case study piloted in a cancer center. It proposes an analysis pipeline for patients with osteosarcoma through segmentation, feature extraction and application of a deep learning model to predict response to treatment. The main aim of the AWESOMME project is to leverage this work and implement the pipeline on an easy-to-access, secure web platform. The proposed WEB application is based on a three-component architecture: a data server, a heavy computation and authentication server and a medical imaging web-framework with a user interface. These existing components have been enhanced to meet the needs of security and traceability for the continuous production of expert data. It innovates by covering all steps of medical imaging processing (visualization and segmentation, feature extraction and aided diagnostic) and enables the test and use of machine learning models. The infrastructure is operational, deployed in internal production and is currently being installed in the hospital environment. The extension of the case study and user feedback enabled us to fine-tune functionalities and proved that AWESOMME is a modular solution capable to analyze medical data and share research algorithms with in-house clinicians.
Keywords: OHIF, Interactive image segmentation, Girder, Web-viewer, Radiomics, Deep learning, Medical imaging
Introduction
Precision medicine (PM) seeks to offer patients a treatment adapted to the characteristics of their disease. In other words, it can be described as the process that enables the identification and classification of individuals into subgroups whose responses to a specific treatment will differ for the same disease [1]. PM is becoming more and more widespread, and can be applied to various diseases, from the prediction of treatment response for different types of cancers [2], especially in radiology [3, 4], to the prevention and management of diabetes [5]. PM has an increasing need of large well-annotated volumes of data in order to allow the development of sufficiently complex, robust, relevant and generalizable models, in particular in statistical learning and deep learning.
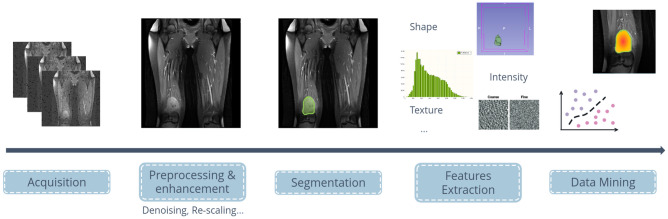
Medical imaging processing involves several stages. They are described in Fig. 1. Insights from experts help to ensure the quality of the analysis and of the generated models. The first stage in the process is data acquisition and gathering. A number of problems can be encountered at this first stage, particularly in the case of multicentric studies (data collection from several hospitals), where the different MR systems and acquisition parameters have a major impact on the quality and appearance of the image, even on associated metadata [6]. Pre-processing algorithms or techniques can be applied to acquired data to remove noise, re-scaling data or enhance contrast at stage two. The third stage is visualization, enabling clinicians to clearly identify the pathology through a shared taxonomy [7]. At this stage, the clinician can choose to annotate the anomalies detected, for example, by delimiting them as for tumor segmentation. The fourth part of the process, as shown in Fig. 1, is optional. It corresponds to features extraction; this can be the application of more or less sophisticated processing to extract and exploit hidden information as radiomics [8]. Data mining is the last step of the image processing and exploits the data previously acquired (raw or obtained during segmentation and/or features extraction phases). Usually associated with machine learning or deep learning, this phase produces models to discriminate pattern in the data (classification or prediction to treatment).
Fig. 1.
Illustration of the processing stages of medical image analysis: from acquisition to data mining for computer-aided diagnosis
All these steps and the interpretation of the result depend on the quality of execution of the different tasks and the quality of the process evaluation [9]. Clinician’s involvement is of major importance to obtain good quality labels. Image annotation and quantitative analysis of large mass of data are however time-consuming tasks, especially with non-ergonomic tools, and require to take in hand the adapted tools.
Related Works
A wide range of tools and functionality in medical image processing exists. In the present work, the focus was made on open-source tools, summarized in Table 1.
Table 1.
Overview of the previous comparable tools mentioned by analysis stage. SEG means segmentation, FEATS for feature extraction and DIAG for diagnosis. Extendable refers to the tool’s ability to be enhanced by third-party plugins
| Name | Type | Actions in analysis | Extendable |
|---|---|---|---|
| ITK-Snap | Desktop | SEG (manual) | No |
| MITK | Desktop | SEG (manual+auto) | Yes |
| XNAT | Hybrid | SEG (auto) | Yes |
| IBEX | Desktop | SEG + FEATS | Yes |
| 3DSlicer | Desktop | SEG (manual+auto) | Yes |
| ISB-CGC | Web | SEG + FEATS | Partially |
| MonaiLabel Server | Web | SEG (manual+auto) | Yes |
| CIRCUS | Web | SEG + DIAG | No |
| ePAD | Web | SEG + FEATS | Yes |
| Studierfenster | Web | SEG + FEATS | Partially |
Many applications currently use locally installed desktop software. Among the most widespread software, ITK-Snap [10] and MITK [11] both propose segmentation tools for medical imaging. More recently, the latter offers fully automated segmentation tools, but is still limited to the Linux operating system. XNAT [12], an advanced data management and segmentation platform, offers a semi-automatic tool to interpolate segmentation between two pre-segmented slices. Much less widespread, IBEX [13] has features for editing regions of interest for radiomics extraction. It has been tested as a proof of concept by researchers with a variety of skills sets [14]. One of the most comprehensive and complex solutions available is 3DSlicer [15]. Its broad functionalities allow to manage most of the phase of the analysis and can be enhanced by plugins.
While these tools serve many clinician needs, they also have significant limitations. Indeed, special authorizations are required to install such software for secure reasons in hospital environment, and it might be complicated to install on institutional computers due to access restriction. The clinician may then be reduced to a single point of access to the software, which is generally not accessible remotely, limiting its use in everyday clinical routine [16].
In parallel, another type of platform has emerged: web viewers. Some of them focus on a single application case [17, 18], while others seek to have a more global range of functions. The simplest ones propose a platform dedicated to visualization only [19, 20]. Others, more accomplished, are interested in the following steps of the processing chain. Starting with annotation and segmentation, notably with the Open Health Imaging Foundation (OHIF) platform [16] which is a zero-footprint web architecture offering a few simple tools. This platform has the advantage of being able to be enriched, and has been widely used as a building block in other projects. It was, for example, used in the Crowds Cure Cancer project1 for the collaborative annotation of cancer patient data cohorts or again in the ISB-CGC initiative [21] for the analysis of cancer-related data too. MonaiLabel Server [22], which adds a computational server to OHIF, offers more complete and advanced functionalities for segmentation with semi and fully automatic models. This solution can also be plugged into 3D Slicer, making it a hybrid approach for both local and web-based use. The Clinical Infrastructure for Radiologic Computation of United Solutions (CIRCUS) [23] provides a web-based solution for computer-assisted detection and diagnosis. It also focuses on models for annotation. It was successfully deployed in two hospitals. Next, the ePAD [24] and Studierfenster [25] platforms are designed for the fourth processing stage. The former includes bio-markers visualization. The latter offers the possibility to use machine learning models and to extract radiomics for several cases.
From all the software mentioned, it appears that none of them allow to perform every single step of the processing stages in one ergonomic interface. Moreover, regulation and protection of personal data, especially in the context of medical imaging is increasing. The overall objective of this project is to provide a secure platform to facilitate transfer between research and clinical application with a web-based software solution for image processing. The AWESOMME platform, presented in this paper, innovates by offering visualization, annotation and feature extraction tools and links data to computer-aided diagnostic models (classification, prediction...) generated during data mining projects. This platform aims to enhance available tools in place, without loss of previous works and in a distant future replace those tools.
The following section presents the different datasets used in this project, the components of the infrastructure to develop and the associated tools to implement different stages modules. Each chosen module focuses on a specific concept or need. The “Results” section describes usage and implementation of plugins and modules to create this software. The generic aspect and efficiency of the platform are tested in this “Results” section. The action perimeter, its limitations and its comparisons with other platforms are discussed in the fourth and last section of this paper.
Materials and Methods
AWESOMME was developed as part of a cross-disciplinary research project. It was born from a collaboration with the Léon Bérard Cancer Centre (CLB) in Lyon (France) looking for a tool to centralize the use of tools created during an anterior project. The use case was completed by examples of data collected in two other datasets.
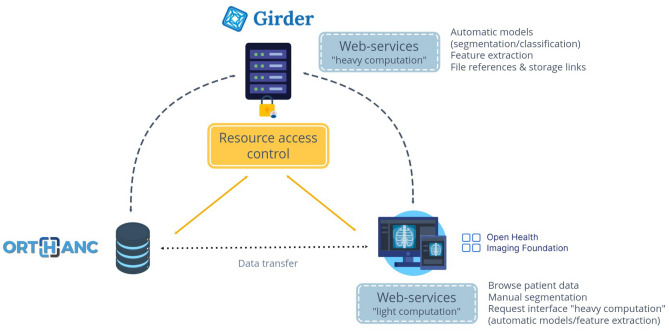
This platform is based on a three-component architecture (Fig. 2). It relies on a data storage system using the international standard format for storing medical data Digital Imaging and Communication in Medicine (DICOM) [23], a computational server also used as a storage for other data format (such as radiomic files) and a web interface used as a single access point for all processing steps. This type of architecture is the most common existing solutions but had to be improved to meet the requirement of this project.
Fig. 2.
Overview of the architecture of the AWESOMME Platform. Three main components are visible, for each new plugins and communication protocol for user traceability were developed
Those requirements can be articulated around four notions, security and user authentication, segmentation (manual and automatic), feature extraction and classification for computer-aided diagnosis.
Vocabulary specific for each component is used in the following sections; Table 2 proposes descriptions for ambiguous terms.
Table 2.
Definitions of technical terms
| Vocabulary | Definition |
|---|---|
| Model | Machine learning term, algorithm trained on data to realize a specific action (segmentation or classification) |
| Module | Refers to the plugin created for each stage of the processing chain |
| Mode | Specific term used in OHIF. Designs a specific viewer for a dedicated application |
| Plugin | An extension to an existing element, implements new functionalities and installed directly on the base element |
Datasets
Use Case Study
Led by the CLB, an Osteosarcoma cohort [26] was set up with 177 patients from 3 multi-centers (site gathering data from several sub-centers in a region). The data were collected anonymized after institutional review board approval, from the two external centers and extracted from the CLB’s Picture Archiving Communication System (PACS) [27] in the standard DICOM format. The entire dataset was segmented by the source provider, and the segmentations obtained were transmitted and integrated into the database in The Neuroimaging Informatics Technology Initiative (NIfTI) format.
Models
This cohort has been exploited in two previous research works. Each produced a machine learning model, one for 2D automatic segmentation (collaboration with Altran-Capgemini during an internship) and the other for outcome prediction classification based on the patient radiomic signature. The prediction model used ReliefF technique to analyze redundancy and reduce the number of features, and train a SVM classifier. Details about the training and the validation can be found in the article [3]. These models were to be integrated on the platform for inference and not for performance evaluation.
Other Datasets
To complete the main study use case, two other independent datasets were introduced inside the platform. The first one is a subset of the LIDC-IDRI dataset [34]. A lung tumors segmentation model has been trained on it. The architecture of the network is a 3D iUnet [35] to cope with the large GPU memory such large images require. The network was first trained on patches centered on the tumors, then random patches all over the images were used.
The second subset is extracted from a single-center ancillary study of patients in acute respiratory distress syndrome (ARDS). The study was approved by their institutional ethics committee (CSE HCL20 [36]). The model was trained with 316 CT volumes from 97 patients (yielding a Dice similarity coefficient of 0.972 on the training set). The lung segmentation model is a deep 3D convolutional neural network using a modified version of the 3D U-net architecture, optimized with the Adam optimizer through a Dice-based loss function [37].
Models
Two models produced during these works were integrated in the platform: the lung volume segmentation model and the lungs tumors segmentation model for inference, only as a proof of concept and not to assess the validity of the models.
Architecture
The infrastructure was deployed on the CPU on a dedicated virtual machine with a 10Tb.
Image Data Server: Orthanc
Orthanc Server is an open-source, small footprint lightweighted vendor neutral archive that can act as a PACS. It comes with its own database engine based and assures DICOM interoperability using the DICOMTK Toolkit. This makes Orthanc Server a robust DICOM store. It is expendable by internal plugins, some existing plugins are already developed and can be modified. Since Orthanc Server provides a full programmatic access to its core features through a REST API [28], higher-level applications can also be built on top of it by driving the REST API.
This service can be reached at: https://covid.creatis.insa-lyon.fr/awesomme-orthanc/.
Web-Services Server: Girder
Girder is a free open-source platform [29]. It is developed by Kitware and allows transparent data management functionalities, as storage and serve data from back-end storage engine (such as MongoDB databases). It is divided in a server (Python) client (JavaScript) architecture, the web interface act as the client and interacts with the server part thanks to a single and expandable RESTful web API, that can be used in other applications to interact with Girder. Girder provides a flexible architecture that allows users to extend its functionality through plugins. These plugins enable developers to add custom features, tools, and integration to Girder without modifying its core codebase. Plugins are implemented as Python packages, and this mechanism can be used to add new custom routes and endpoints. It provides also a hooks system that can be used to extend or modify native Girder behaviors and events.
Girder also provides authentication and user management methods, and most importantly it enables access control to its resources through an authorization system. With the customization of routes, Girder was extended with new plugins to act as a centralized authentication system and to modify Girder behavior to be a computational web-services launcher (to extract radiomic and start docker image applications).
One can access the Girder interface at: https://covid.creatis.insa-lyon.fr/awesomme-girder/.
Web Viewer: OHIF
The OHIF viewer is an open-source, zero-footprint web-based viewer [16]. OHIF has been widely adopted by the developer community as evidenced by the large number of projects based on it. It can connect to Image Archives using the DICOMweb standard web service. It is based on Cornerstone to decode and render DICOM images.
It includes a few interactive tools, such as windowing or leveling, and offers measurement and limited manual segmentation tools. Still, OHIF natively lacks semi-automatic of fully segmentation tools that will allow clinicians and researchers to quickly annotate large amounts of data. It lacks the functionality to save corrections made on existing segmentation. Furthermore, OHIF does not offer feature extraction or machine learning classification models.
Version 2.0 of OHIF introduces a side panel system for a unique expendable viewer, into which items can easily be added. The latest version benefits from a more complex architecture, as it also uses a system of modes, allowing access to viewers that do not have the same tools or the same analysis objectives. This project was based on the version 3.3, the improvements were initially implemented in version 2.0 of OHIF and then re-integrated into version 3.3 using both mode and panel extension.
The web viewer service is available at: https://covid.creatis.insa-lyon.fr/awesomme-ohif/.
Modules Methods
Authentication
The three components described above have their own authentication system. The challenge is to offer a single system for the entire infrastructure.
Orthanc can simply contain identification pairs in its configuration file, or rely on plugins to link up with KeyCloack, for example. The Girder authentication system is comprehensive, secure and autonomous. It can be connected remotely via API routes. Girder’s system was found to be the most robust and expandable. Thus, the next step is to connect the Orthanc and OHIF components to the Girder authentication system. Orthanc’s plugins (Python Plugin and Orthanc-Explorer-2) were used to connect Orthanc to the Girder API Routes and simple requests were used in OHIF.
Segmentation
The segmentation module addresses several needs.
Firstly, in the osteosarcoma case study, segmentation files were already available. They were done manually or with a semi-automatic tool with corrections. It corresponds to a long and painstaking process done by a clinician. It was particularly important to preserve the work already done and re-integrate it into the platform. For that purpose, an import module was needed in the infrastructure. Our case study contained images and segmentations in The Neuroimaging Informatics Technology Initiative (Nifti) format but the data server Orthanc only supports DICOM format. Several conversion tools exist and the following list is not an exhaustive: dcm-js (javascript), pydicom (python).
Secondly, the developed module integrates different aspects of manual segmentation. An important requirement was to propose tools familiar to the clinicians to create detailed annotation on the image. To this end, AWESOMME strongly relies on tools proposed by OHIF with the Cornerstone suite.
Finally, pre-trained segmentation models were made available on the platform. To integrate those models easily, Docker was used in order to wrap the development environment of the model. That way, all needed modules and weights of models were put inside a Docker image that is called by a script. As OHIF is the entry point for the clinicians, it sends a request of execution of a specific model and its inputs. The instruction is transmitted to the Girder server, which can as a coordinator then launch the execution commands via Docker using python-on-whales library in Girder’s python side.
Feature Extraction
The feature extraction module focuses on radiomic extraction since this was the initial subject of our case study. The implementation of the extraction and storage were done on the Girder Server as for segmentation using API Routes.
As the use of radiomic features is expanding rapidly, a few concerns have emerged such as lack of reproducibility. Two initiatives were taken into account to answer those challenges. Concerning the extraction, the PyRadiomics [30] library was used to respect the Image Biomarker Standardisation Initiative (IBSI) [31].
Radiomic features and additional information storage are critical to share clinical and omics data in oncology research. As the results, the methodology to conserve information of extraction followed the recommendations of the French GrOup inter-SIRIC sur le paRtage et l’Intégration des donnéeS clinico-biologiques en cancérologie initiative (OSIRIS) [32].
Classification
The classification module is based on the same methodology as the previous two modules. In the same way as for the automatic segmenting models, each classification model used for prognosis or diagnosis must also be integrated into a docker system. The results files must be accessible via the platform in a text file, or a file readable by the clinicians. In the main use case, the inputs for the predictive model were a single radiomic file in the CSV format.
Results
This section describes the main implementations made to improve and upgrade the 3 components to address the specific needs mentioned earlier and to offer diverse functionalities with a section dedicated to deployment phase results.
Several plugins were developed to meet different needs and were divided into two main categories: security including user management and data protection, and data analysis.
Deployment Phase Results
A first deployment phase was carried out in our laboratory. AWESOMME was deployed on a server and could be available for registration.
A public account has been set up for demonstrations. The aim of this launch was to obtain initial feedback on the platform before deploying a corrected, robust and extended version inside the hospital.
A demonstration of the platform and the different functionalities described in this article is available at: https://covid.creatis.insa-lyon.fr/awesomme-demo/.
Security: User Management and Data Protection
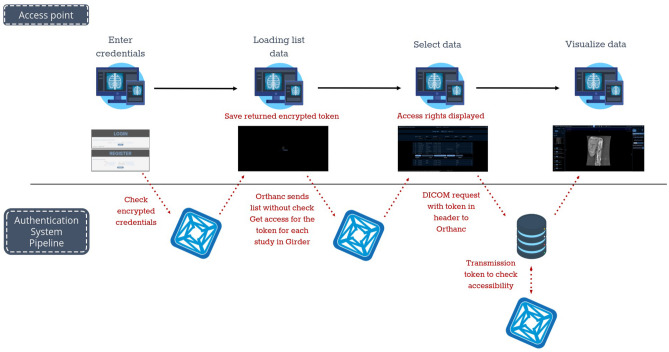
The precise workflow for user verification and access control is presented in Fig. 3.
Fig. 3.
Check-in workflow to access data
User Management
Link Girder Credentials to Other Components
The first plugin created auth_ohif enabled two links: Orthanc-Girder and OHIF-Girder. It retrieved encrypted user credentials and sent an encrypted token back to OHIF, which stores it and then interacts with Girder for processing. This plugin is called when a function with an encrypted token is used (either from OHIF or Orthanc). Each time a data is accessed by OHIF modules, the token is verified, for example, to check if a user has access to the data (accessing the link between user groups and Orthanc data saved in the resources_ohif plugin described below).
Orthanc-Girder link was built on already existing plugins: Orthanc-Explorer-2 and Orthanc-Python. The original plugins, OrthancExplorer2 used Keycloack authorization, if both installed, provided an authentication mechanism through a modern and user-friendly interface. Meanwhile, OrthancExplorer2 was enhanced to restrict access only to those registered in the Girder database and without needing Keycloack authorization plugin. This development ensured a well-controlled access to a unique and secure system, facilitating its management while also suppressing all unsecured URLs provided by Orthanc’s default UI. This advancement strengthened the confidentiality and sharing of medical data.
OHIF-Girder link required a new user interface on OHIF side to enter credentials. This was added and simply used the API routes with the encrypted credentials to save the encrypted token in as a state in an authentication React service.
Advanced Management
The advanced_management plugin was created to ease clinicians’ registration. New API Routes were created for administrators enabling them to generate random codes in the Girder interface (or in OHIF). The codes were divided in admin codes and member codes, meaning that an administrator can decide to open the data collection to other user with administrator rights. New users registering on the OHIF platform can then enter such a code. The code used determined the user rights and automatically granted them to the user.
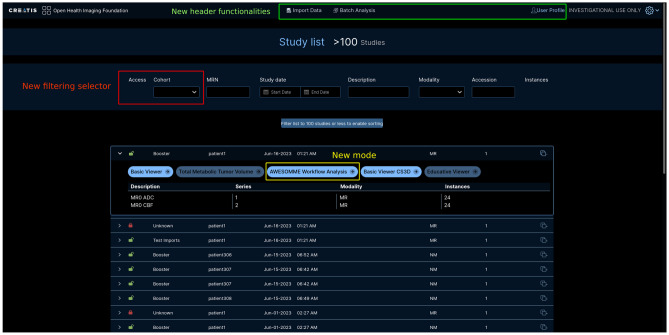
A new user interface was implemented in OHIF to enable the use of the plugin by clinicians directly on the web-viewer. This functionality is available on the page under the user profile top-right button (Fig. 4).
Fig. 4.
New functionalities on the page. Two modification categories are displayed: the new filtering selector shown in red and new buttons in the header part to access new functionalities forms shown in green at the top
Actions are made available depending on the current user access rights. An admin could add another user in a group of data or generate codes to share to several other users. A non-admin user in a group could request membership by providing such a code.
Data Protection
Data Collections
The resources_ohif plugin was implemented to create two new data models, series and derived objects from series (SEG, RTStruct, SR...). These models kept track of which cohort(s) each data file (DICOM images on the Orthanc Server or radiomics/diagnostics on the Girder Server) belongs to.
It enabled filtering, granting or prohibiting access to specific data for a group of user via the auth_ohif plugin. The cohort filter is called from OHIF using new API Routes.
Corresponding developments were then been implemented in OHIF to render access rights saved in Girder, as highlighted in the middle of Fig. 4. New filtering criteria allowed to focus on a specific group of all the data available on the server and to reduce the number of patient data displayed in the list. In addition, name in which the data is included and a visual indicator for permission access were added for the sake of readability for each data item.
Traceability
The traceability plugin provided an interface for tracking the activities of each user on the platform. It was made only accessible for administrators. A list of action of interest was compiled, including both internal Girder actions and new actions created by the developed plugins.
For every action of this list, an event is generated, and these events are then monitored by the traceability plugin. For some actions, events were already triggered. For new developed actions, event triggers were added in the code, but native Girder code which did not include events, the plugin listened to API Route request end notification. Every event triggered the apparition of a new log in the journal stored in the Girder’s database. A log entry includes the name of the action, parameters, the user who triggered it and the date. As part of the actions listed, creation of a cohort’s access code or request for automatic segmentation can be named.
This plugin did not need a counterpart in OHIF since it was only created for administrator and developers of AWESOMME and it was not relevant to access those information in the medical viewer.
Data Import Module
This OHIF module was created to upload data on the Orthanc server from local data or from a temporary connection to a PACS. The user can choose the data group to which the import is added, a user can only add data to a group for which they are an administrator.
From the PACS, the transfer of data for a specific patient to the Orthanc Server of AWESOMME went through an anonymization script. A form should be filled to get only very specific data from the PACS and not huge data collection. The correspondence between the patient on the PACS and the patient on the platform is transmitted only to the user so that only the user keeps this information.
Local import functionality was achieved to support either DICOM or NiFti format. As data is saved in the Orthanc Server via the DICOM API Routes, NiFti were to be converted to DICOM format. The conversion was made directly in OHIF thanks to JavaScript modules. However, the converted DICOM did not contain automatically pertinent information as Patient ID or modality of the image etc... So, the user must provide the NiFti file with a specific file name to complete information on the DICOM after conversion. Nifti files could also be paired to an Excel file with additional metadata fields to inject in the DICOM file.
Data Analysis Implementations
This part presents the new features implemented on the web-viewer. Most of the modifications were first made on OHIF version 2.0, then transposed and completed on version 3.3. The figures below show the final platform, i.e., the enhanced version 3.3. In OHIF version V2, all modifications were made in the only viewer available, so all data had access to the same interface and functions. In OHIF version V3, the mode system was leveraged to offer a new mode with the new functionalities without affecting the existing mode. OHIF enables access to the different modes in the patient expanded rows of the home view. The internal functions from OHIF were used to add a new mode which appeared automatically next to the others. This mode access was restrained with specific mode function that verifies if the mode is valid for each patient. In this paper case, the mode was made valid for all data collection.
Data analysis included segmentation or delineation (manual or automatic), feature extraction and computer-aided diagnosis (as classification). These actions should be performed by expert in the user interface, but OHIF could not support intensive calculation thus a plugin was created for the most computationally intensive function (such as running a segmentation model).
Batch Analysis Module
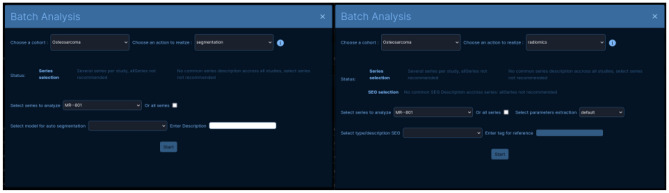
Some steps of the processing chain could be made by batch. Batch analysis, made available directly on the home page, was limited to one cohort at a time.
Currently, the batch analysis system is enabled for fully autonomous processes such as automatic segmentation and radiomics extraction. Figure 5 shows pieces of information displayed and the form to be filled in by the user. Data belonging to a group were not necessarily homogeneous, i.e., the relevant acquisition for each patient may differ. They were then identified by their acquisition type (MR/CT...) and number or by their description for segmentation files. The indications at the top of the form were added to guide the user through the choice of inputs for analysis. The “Enter Description” and “Tag for reference” fields, for automatic segmentation and for feature extraction respectively, were built to identify outputs.
Fig. 5.
Batch analysis forms for segmentation (left) and radiomics extraction (right)
The batch analysis module relies on the same implementations as described below, the request can be transmitted as a list instead.
Segmentation
Manual Tools and New Functions
This part was proper to the OHIF interface, as it included only light computation functions.
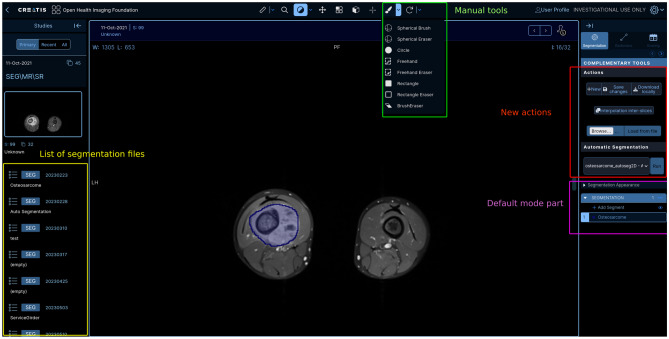
Whether in version v2 or v3, OHIF offers default tools, using an external Cornerstone library, including specifically manual segmentation and annotation tools. However, one of the default tool in OHIF version 2.0 did not have its equivalent in the version 3.3: the Freehand ROI Tool, because of the switch between the CornerstoneTools in the version 2.0 to Cornerstone3D library in the latest version. This tool provides greater precision compared to the rest of the ROI tools. Thus it was manually added to the list. The complete list of tools available is displayed in Fig. 6 at the top.
Fig. 6.
Enhanced OHIF: shows the global view with data visualization and tools with the new segmentation panel
Then a custom segmentation panel, in which new functionalities are implemented in the top part of the panel, was created with the default mode segmentation panel as a base. This default panel was activated when segmentation was loaded into the viewer.
The default panel already authorized a few actions as renaming segments but for other actions, such as creating a new segment in a segmentation (which corresponds to the file and segments to the delimited content — one segmentation should be able to have one or several delimited zones), there was an existence but no user interface was available to use them. The first enhancement made was enabling those interactions in the default segmentation panel. Figure 6 presents the new functionalities created to improve the clinicians’ experience on the platform.
Then, an import functionality has been implemented directly in the home page. A similar function was designed in the panel to make for segmentation import. It was conceived to import previous masks (DICOM of NiFti) made on local tools. From NiFti, conversion relies on the same method as for the home view import data functionality. Though, a specific file name was not required as the converted file is linked to the current displayed series. A few check-ups were made on the size of the data and the spacing. The user was asked to enter information to complete the metadata of the segmentation file as the description. It was necessary for a better tracking of actions to describe the segment and indicate its nature (automatic or manual) as well as the person or model who created it.
Also, an interpolation tool was created: it is particularly useful to reduce the cost of the manual segmentation task by clinicians as it offered the possibility to interpolate a segment data between only two segmented slices ensuring shapes coherence.
Automatic Segmentation
Trained automatic segmentation models were also made available in the panel (middle part selector). Since it required more computer resources, they were accessible via a request system to the heavy computation server.
The aimodules_ohif plugin in Girder allowed the retrieval of requests and instructions sent by the web viewer for the launch of automatic models or the launch of radiomics extraction (also thanks to new API Routes). Appropriate data are retrieved via a communication link to Orthanc before launching the appropriate processing and sending back the result to the viewer. For the first step of analysis, three automatic segmentation models have been added.
Osteosarcoma: The original study case model which is used to delineate bone tumor
Lungs: A model to delineate abnormalities in the lungs
Lungs: A model to delineate volume of the lungs
Models are linked to different application cases, using the cohort implemented. Thus, an automatic segmentation model can only be called up on data belonging to a specific group (information retained by the previous plugin). The Girder API Route system did not allow to return file, so only the pixel array in NIfTI format was sent back to OHIF. Conversion back to DICOM was achieved in the OHIF part using the library dcmjs.
A preliminary analysis about time cost of inference of a model was made on the Osteosarcoma segmentation model. Table 3 presents two criteria investigated to have an impact on the total time of the model. In this case, the number of slices had a significant impact on the model temporal cost but for the communication temporal cost, total file size was the most critical point. Transfer of data from Orthanc to Girder and conversion temporal cost are not significant.
Table 3.
Temporal cost of automatic model request depending on some data characteristics
| Sample info | Time per phased | Ratio phase over total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Criteria | Slicesa | Resolutionb | Sizec | Modeld | Data handlinge | Communicationf | Total | Model | Data handling | Communication |
| Sample 1 | 16 | 512*512 | 8.6 | 11.75 | 0.46 | 6.79 | 19 | 0.63 | 0.02 | 0.36 |
| Sample 2 | 60 | 352*352 | 15.1 | 22.18 | 0.96 | 11.8 | 34 | 0.65 | 0.03 | 0.29 |
| Sample 3 | 26 | 1024*1024 | 54.6 | 13 | 2.21 | 42.79 | 58 | 0.22 | 0.04 | 0.55 |
Values in bold are there to clearly identify problematic values in processing time
aNumber of slices or instances contained in the series sample
bResolution is expressed in pixels
cSize refers to the total of MB for all instances combined
dTemporal cost are in seconds
eModel temporal cost designates the time during which the model is running on the data to produce the result
fData handling includes data download from Orthanc to Girder’s local folder, data conversion and results upload to Girder
gCommunication describe the response of the request sending data back to OHIF from Girder
As models could be part of very different application cases (here osteosarcoma or lungs segmentation), they should not be called on every data. A filter was directly built in this panel to ensure that all models were retrieved but only models linked to the collection of the current data were available in the selector. Information as time cost or confidence was added. If necessary, metadata should be entered in the form. Only segmentation with a unique segment is fully supported (loading, saving, import) in the current version.
Feature Extraction
On Girder’s side, feature extraction was implemented in the aimodules_ohif plugin as well. For radiomics, the instructions should follow the same strategy: the request should contain the information about the series and segmentation to retrieve from the data server before launching extraction. Pre-saved parameters files are saved in Girder but the request could also contain new sets of parameters in the JSON format. In order to comply with the OSIRIS initiative, those parameters and the version of the module used to extract radiomics were saved in the same file as the radiomics in Girder, allowing future communication with OSIRIS server.
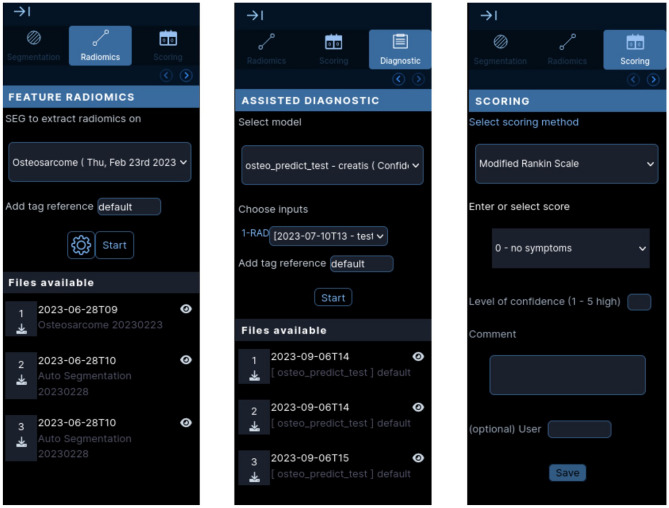
Figure 7 (left) shows how the panel is organized in two parts on OHIF’s side. Top part of the panel is to start the extraction, containing segmentation input, tag reference and parameter window. The bottom part lists the radiomic files downloable for the series. After each launch and except if an error occurred (signaled by a notification), a file should be added to the list without further action from the user.
Fig. 7.
Enhanced OHIF: (left) only shows the radiomics panel, it is accessible the same way as the segmentation panel on the side on the window. (middle) shows the side panel for launching diagnostic. (right) Presentation of the scoring panel. One can select a type of score, and then the chosen score. Here it uses a scale of pre-defined values but for other cases a text box can display. A physician can enter the confidence in their scoring and add a comment before saving it to the Girder server
Computer-Aided Classification and Prognostic
As the two previous actions, automatic computer-aided classification for prognostic was also implemented in the same Girder plugin. For the moment, one model to predict patient treatment response for Osteosarcoma was integrated. This model required a radiomic file as an input. that should be passed as an argument to the API request. The reference to this file is part of the request parameters and then mounted from the Girder data storage to the docker volume.
The automatic models from data mining were implemented in a third panel. It was built as a two-part panel, the first part for launching the automatic models and the second part for the results files. Number and type of inputs were stored with the model information in Girder and were transmitted to OHIF to create a custom and generic input form for each model. Figure 7 (middle) presents the form for the osteosarcoma treatment response prediction model.
Other Panel
An emerging need was to be able to score or evaluate data. This involved either scoring a piece of data according to a specific criterion, or giving a confidence index to a segmentation, an image or a value. This scoring module was built for the Modified Rankin Scale. It was designed with generalization in mind. Figure 7 (right) shows how a user can select the type and enter the score either from pre-defined choices or a text box. The scores are then recorded and associated with the data (primary or derived) for subsequent statistical analysis or evaluation.
Discussion
The medical viewer AWESOMME has been designed to help clinicians and radiologists to easily create research cohorts of data including metadata and annotation in a setting designed to fit in with their daily hospital routine. It was created with security of access and traceability in mind, while promoting ease of access and use. Meaning, the platform should be accessible on any machine in the hospital network and should include familiar analysis tools. Three different actions were implemented in the platform, segmentation, feature extraction and classification for computer-aided diagnosis.
To answer the first requirement, the decision was made to develop the solution as web-based platform. Desktop software solutions such as original Slicer 3D require independent multi-installations for multi-machine use. A single installation, accessible on all Internet-connected machines in the hospital environment, ensures the same controlled, secure and homogeneous environment across all hospital machines.
The architecture generally encountered in the implementation of a medical web viewer is a set of at least three components: a data server, a computational server and a web interface. This is the architecture on which both the MonaiLabel solution and the XNAT software are based. XNAT uses the web viewer as a plugin rather than the main component of its solution while our solution uses it as the main interface for the user.
The choice of data server is a keypoint question. The component chosen for the web interface, OHIF, is based on a DICOMweb communication protocol. In our hospital case, PACS uses a similar system, so the platform connection to the hospital database should be straightforward. However, this raises a few questions. Uploading research data to PACS from the platform, particularly DICOM segmentation files, is currently prohibited to avoid overloading clinical PACS. That’s why the choice was made in favor of the Orthanc data server, which can also be enhanced. On the other hand, it was necessary to maintain a link to PACS for ongoing data import. This has justified the need of a PACS import module. OHIF used as the main interface was chosen as it is an ergonomic, web-based viewer. From literature and developments made during this project, OHIF has appeared as an easily expendable and ergonomic tool. In spite of that, these two components (Orthanc and OHIF) do not allow the user to run heavy calculations such as machine learning models. The third component in the architecture was used for this purpose. Girder was chosen because it is a modular component that also serves as a data repository for non-DICOM files. Girder does not use the protocol required for OHIF justifying the user of the Orthanc data server. Girder’s native features as users and groups management have been used to offer a strong cohort filter for data. It relies on matching data on the data server Orthanc and a list of references in Girder. References should be manually added on Girder’s interface or using the requests. In our case, the requests are sent by OHIF which have the import module for users. It meets security requirement by restricting access to data regarding user’s right.
In the AWESOMME infrastructure, computational services and management of user rights are associated in one web-services server. This coupling implies that computing resources are directly linked to the deployment environment. In our case, AWESOMME is offered on a server with dedicated GPU resources. These resources are more than sufficient for the proof of concept. On the other hand, in the event of an increase in data use and computational demands, the computational part could easily be deported to a possible computational cluster. Unfortunately, having several components also brings the disadvantage to have more parts to maintain, in different programming languages. The evolution from OHIF version 2 to version 3.3 is a perfect example of the difficulties that can be encountered: changes to sub-modules imply the deletion/modification of functionalities in the interface.
Another major limitation of this architecture is also the regular exchange of information between the different components. It appears from preliminary results of a time cost analysis that the characteristics of the data (number of slices and resolution) have an impact on the time it takes to complete the request. Two aspects are taken into consideration: the time of the model and the time during the communication of the results between Girder and OHIF. The first is part of the optimization of the model, but the latter should be decreased as much as possible as it is strongly dependent on the quality of the network connection. In the study case of this article, emergency and fast response is not a key point but it can be in other circumstances. Limitations surrounding temporal costs should be the subject of a specific and more complete analysis to fully comprehend their impact on machine learning models’ uses during clinical routine. The possibilities for optimizing models do not fall within the scope of this manuscript, although the use of GPUs for inference could perhaps improve model calculation times. We are currently studying strategies for optimizing data exchanges but we have yet not concluded. For example, by avoiding going through the results of the query and directly uploading the results from Girder by transmitting only the information from the segmentation series to OHIF so that it can reload this data specifically.
To the best of our knowledge, no other web-based platform included a complete workflow of analysis: segmentation, feature extraction and diagnostic support. MonaiLabel offers a comprehensive solution for segmentation including functionalities for continuous learning. Both MonaiLabel and XNAT solutions rely on the component OHIF that the platform AWESOMME uses as well since OHIF already offers manual tools for segmentation. The segmentation panel for these solutions does not satisfy all the functionalities required, notably the import of previous segmentation done on a desktop software or feature extraction. The recent version of the plugin XNAT-OHIF based its current interface for the segmentation panel on MonaiLabel and does not include feature extraction. One of the key points of this project was to be able to re-inject segmentation made on desktop software. As manual segmentation is very time-consuming, it was important that the platform was able to continue to use previously performed segmentation, thus saving clinicians the trouble of having to do it all over again. This functionality is not implemented in MonaiLabel and not available in the web-viewer access point of XNAT (but in the data management interface). AWESOMME enables it, for both DICOM (SEG) and NiFti. Other formats are widely used for segmentation, as DICOM RTStruct and NRRD for example, import function for these formats should be made available shortly.
Integration of the main use case model was completed by the addition of other models extending possible case studies. It was the opportunity to analyze the process of automatic model integration into the platform. Although the Docker system is quite useful to connect easily Girder to new model, models themselves can be harder to handle. As they are developed by different researchers in different study cases, inputs are not homogeneous from one model to another. The types of the inputs can change, from DICOM to NiFti for image input format and models can even be based on different type of data (radiomic) or image for classification. It highlights the need of specific guidelines for future models, all pre-processing and post-processing should be included in the docker system. Data could be provided in a specific format, initiatives as Brain Imaging Data Structure (BIDS) [33] are example of data structures that could be replicated in AWESOMME. New automatic model should rely on the same data structure or change it directly in pre-processing steps in their docker image. The same strategy should apply to outputs. Integration of automatic models was successful as model trained for research were used on new incoming data. As previously mentioned, it is important to notice that analysis of the automatic models results is outside of the scope of this project and should be further evaluated in a follow-up study.
Regarding feature extraction, AWESOMME offers radiomics extraction based on a specific python module. The extraction of radiomics in the platform environment is intended to be reproducible as the choice of the module was made to ensure the compliance with international initiatives on radiomics guidelines. It could be relevant to add more compliant modules for radiomic extraction. The platform could also be used in other cases of computer-aided diagnosis, with the extraction of non-radiomics features. It has been designed to integrate the addition of other feature extraction approaches with minimal effort.
The laboratory instance is already successfully deployed and the hospital instance is currently being deployed. The operating system available in the laboratory is different from that in the hospital, so a test phase was first necessary to check that all the components were working properly. The final ongoing step is the provision of a dedicated machine from the hospital.
Initial user’s feedback from within the laboratory and from the radiologists was promising. The possibility of adding new data and functionalities specific to research projects was mentioned. For example, the import module has been created at the request of clinicians and colleagues. The platform has then demonstrated to be modular and adaptable to different study cases. Teams which provided data samples are eager to use the platform, naming the scoring project and the lungs volume segmentation. It leads to new functionality development enabling, for example, visualization of scores or adding new computer-aided diagnosis model. A new need also arose to enable inter-expert evaluation, especially in the case of multi-center cohorts. It means that analysis done by an expert (either scoring, annotation etc...) could be compared to another expert inputs.
Conclusion
The purpose of this platform is to provide a direct link between machine learning algorithms and clinical research and diagnosis for the analysis of large volumes of patient data while ensuring protection and traceability of sensitive data. As several tools already exist for specific applications or actions, this solution aims to centralize all processing steps to visualize and produce expert data more efficiency.
Based on a three-component architecture, enhancements of the basic elements have been proposed allowing the implementation of secure and generic use of AWESOMME. Import of data on the platform has been facilitated with anonymization and upload functionalities in a data collection system. Users access and data restriction were implemented through a filter on these cohorts. Regarding the actions available, automatic models for both segmentation and classification (computer-aided diagnosis) were wrapped independently in Docker images callable from one on the major architecture component. Feature extraction was developed to comply with existing initiative and ensure repeatability.
Thanks to the authentication and user access control, patient data and information are secured and user actions are tracked down at any time. Different types of data and application have been considered to demonstrate scalability and generalization. AWESOMME’s simplified multi-expert management opens up the possibility of creating cohorts enriched with annotations and inter-expert variability at low cost. The platform AWESOMME has then the capacity to provide a new approach for multi-sources data processing (images, radiomics etc...) helping to better understand patient outcomes classification and prediction thanks to machine learning research through a clinician-friendly interface.
Author Contributions
All authors contributed to the study conception. Tiphaine Diot and Frederic Cervenansky contribute to the design of AWESOMME. Data collection and analysis were performed by Amine Bouhamama and Benjamin Leporq. The first draft of the manuscript was written by Tiphaine Diot and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the CNRS via the INS2I single call. This work was performed within the framework of the LABEX PRIMES (ANR-11-LABX-0063) of Université de Lyon, within the program "Investissements d'Avenir" operated by the French National Research Agency (ANR).
Data Availability
The datasets used during and/or analyzed during the current study are not available; they belong to the producer/host center: CLB. The data used in this study adhere to the tenets of the Declaration of Helsinki.
Declarations
Ethics Approval
This is a technical study. The CLB Research Ethics Committee has confirmed that no ethical approval is required.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent to Publication
The authors affirm that human research participants provided informed consent for publication of the images in Fig. 6.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.König, I.R., Fuchs, O., Hansen, G., al.: What is precision medicine? Eur. [R]espir. [J]. 50 (2017) 10.1183/13993003.00391-2017
- 2.Hingorani, A.D., Windt, D.A., Riley, R.D.e.a.: Prognosis research strategy (progress) 4: stratified medicine research. BMJ 346 (2013) 10.1136/bmj.e5793 [DOI] [PMC free article] [PubMed]
- 3.Bouhamama, A.: Can Radiomic Predict Response to Neoadjuvant Chemotherapy of Osteosarcomas? European Congress of Radiology, (2019). 10.26044/ECR2019/C-0930
- 4.Sun, R., Lerousseau, M., Henry, T., Carré, A.e.a.: Intelligence artificielle en radiothérapie : radiomique, pathomique, et prédiction de la survie et de la réponse aux traitements. Cancer [R]ad. 25(6-7), 630–637 (2021) 10.1016/j.canrad.2021.06.027 [DOI] [PubMed]
- 5.Xie, F., Chan, J.C., Ma, R.C.: Precision medicine in diabetes prevention, classification and management. J. of [D]ia. [I]nv. 9(5), 998–1015 (2018) 10.1111/jdi.12830 [DOI] [PMC free article] [PubMed]
- 6.Bleker, J., Kwee, T.C., Yakar, D.: Quality of multicenter studies using mri radiomics for diagnosing clinically significant prostate cancer: A systematic review. Life 12(7) (2022) 10.3390/life12070946 [DOI] [PMC free article] [PubMed]
- 7.Zhang, Z., Xie, Y., Xing, F., McGough, M., Yang, L.: Mdnet: A semantically and visually interpretable medical image diagnosis network. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3549–3557 (2017)
- 8.Lambin, P., Rios-Velazquez, E., Leijenaar, R.e.a.: Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. [C]ancer 48, 441–6 (2012) 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed]
- 9.Haomin, C., Caalina, G., Chien-Ming, H., Mathias, U.: Explainable medical imaging ai needs human-centered design: guidelines and evidence from a systematic review. npj [D]ig. [M]ed. 5 (2022) 10.1038/s41746-022-00699-2 [DOI] [PMC free article] [PubMed]
- 10.Yushkevich, P.A., Piven, J., Hazlett, H.C., Gimpel Smith, R.e.a.: User-guided 3d active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage 31(3), 1116–1128 (2006) 10.1016/j.neuroimage.2006.01.015 [DOI] [PubMed]
- 11.Goch, C.J., Metzger, J., Nolden, M.: Abstract: Medical research data management using MITK and XNAT. In: Informatik Aktuell, pp. 305–305 (2017). 10.1007/978-3-662-54345-0_68
- 12.Doran, S., Sa’d, M.A., Petts, J., Darcy, J.e.a.: Integrating the OHIF viewer into XNAT: Achievements, challenges and prospects for quantitative imaging studies. Tomo. 8(1), 497–512 (2022) 10.3390/tomography8010040 [DOI] [PMC free article] [PubMed]
- 13.Zhang, L., Fried, D.V., Fave, X.J., Hunter, L.A., Yang, J., Court, L.E.: An open infrastructure software platform to facilitate collaborative work in radiomics. Med. [P]hy. 42(3), 1341–1353 (2015) 10.1118/1.4908210 [DOI] [PMC free article] [PubMed]
- 14.Korte, J.C., Cardenas, C., Hardcastle, N.e.a.: Radiomics feature stability of open-source software evaluated on apparent diffusion coefficient maps in head and neck cancer. Scientific Reports 11(17633) (2021) 10.1038/s41598-021-96600-4 [DOI] [PMC free article] [PubMed]
- 15.Fedorov, A., Beichel, R., Kalpathy-Cramer, J., Finet, J.e.a.: 3d slicer as an image computing platform for the quantitative imaging network. Magn. [R]es. [I]mag. 30(9), 1323–1341 (2012) 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed]
- 16.Ziegler, E., Urban, T., Brown, D., Petts, J., Pieper, S.D.e.a. : Open health imaging foundation viewer: An extensible open-source framework for building web-based imaging applications to support cancer research. JCO [C]lin. [C]an. [I]nf. (4), 336–345 (2020). 10.1200/cci.19.00131 [DOI] [PMC free article] [PubMed]
- 17.Han, S., Shin, J., Jung, H., Ryu, J.e.a.: ADAS-viewer: web-based application for integrative analysis of multi-omics data in alzheimer’s disease. npj [S]yst. [B]iol. [A]ppl. 7(1) (2021) 10.1038/s41540-021-00177-7 [DOI] [PMC free article] [PubMed]
- 18.Keshavan, A., Datta, E., McDonough, I.M.e.a.: Mindcontrol: A web application for brain segmentation quality control. NeuroImage 170, 365–372 (2018) 10.1016/j.neuroimage.2017.03.055 [DOI] [PubMed]
- 19.Lajara, N., Espinosa-Aranda, J.L., Deniz, O., Bueno, G.: Optimum web viewer application for DICOM whole slide image visualization in anatomical pathology. Comp. [M]eth. and [P]rog. [B]iomed. 179, 104983 (2019) 10.1016/j.cmpb.2019.104983 [DOI] [PubMed]
- 20.Gustafson, C., Bug, W.J., Nissanov, J. BMC Bioinformatics : a client-server system for browsing 3d biomedical image data sets. BMC Bioinformatics 8(1) (2007). 10.1186/1471-2105-8-40 [DOI] [PMC free article] [PubMed]
- 21.Reynolds, S.M., Miller, M., Lee, P.e.a.: The ISB cancer genomics cloud: A flexible cloud-based platform for cancer genomics research. Cancer [R]es. 77(21), 7–10 (2017) 10.1158/0008-5472.can-17-0617 [DOI] [PMC free article] [PubMed]
- 22.Diaz-Pinto, A., Alle, S., Ihsani, A., Asad, M.e.a.: Monai label: A framework for ai-assisted interactive labeling of 3d medical images (2022) arXiv:2203.12362 [DOI] [PubMed]
- 23.Nomura, Y., Miki, S., Hayashi, N.e.a.: Novel platform for development, training, and validation of computer-assisted detection/diagnosis software. Int. J. of [C]omp. [A]ss. Rad. [S]ur. 15, 661–672 (2020) 10.1007/s11548-020-02132-z [DOI] [PMC free article] [PubMed]
- 24.Rubin, D.L., Akdogan, M.U., Altindag, C., Alkim, E.: ePAD: An image annotation and analysis platform for quantitative imaging. Tomography 5(1), 170–183 (2019) 10.18383/j.tom.2018.00055 [DOI] [PMC free article] [PubMed]
- 25.Egger, J., Wild, D., Weber, M., Ramirez Bedoya, C.A.e.a.: Studierfenster: an open science cloud-based medical imaging analysis platform. J. [D]ig. [I]ma. 35(2), 340–355 (2022) 10.1007/s10278-021-00574-8 [DOI] [PMC free article] [PubMed]
- 26.Bouhamama, A., Leporq, B., Khaled, W., Nemeth, A.e.a.: Prediction of histologic neoadjuvant chemotherapy response in osteosarcoma using pretherapeutic mri radiomics. Rad. [I]ma. [C]an. 4(5) (2022) 10.1148/rycan.210107 [DOI] [PMC free article] [PubMed]
- 27.Bick, U., Lenzen, H.: PACS: the silent revolution. Eur. [R]ad. 9(6), 1152–1160 (1999) 10.1007/s003300050811 [DOI] [PubMed]
- 28.Richardson, L., S., R.: Restful Web Services, (2007)
- 29.Grauer, M., Rose, L., Choudhury, R.: Understanding the resonant platform. Kitware (2016)
- 30.Griethuysen, J.J.M., Fedorov, A., Parmar, C.e.a.: Computational radiomics system to decode the radiographic phenotype. Cancer [R]es. 77(21), 104–107 (2017) 10.1158/0008-5472.can-17-0339 [DOI] [PMC free article] [PubMed]
- 31.Zwanenburg, A., Vallières, M., Abdalah, M., Aerts, H.e.a.: The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295(2), 328–338 (2020) 10.1148/radiol.2020191145 [DOI] [PMC free article] [PubMed]
- 32.Guérin, J., Laizet, Y., Le Texier, V., Chanas, L.e.a.: OSIRIS: A minimum data set for data sharing and interoperability in oncology. JCO [C]lin. Cancer [I]nf. (5), 256–265 (2021) 10.1200/cci.20.00094 [DOI] [PMC free article] [PubMed]
- 33.Gorgolewski, K., Auer, T., Calhoun, V.e.a.: The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci/ Data 3(160044) (2016) 10.1038/sdata.2016.44 [DOI] [PMC free article] [PubMed]
- 34.Samuel G., Armato Geoffrey, McLennan Luc, Bidaut Michael F., McNitt‐Gray Charles R., Meyer Anthony P., Reeves Binsheng, Zhao Denise R., Aberle Claudia I., Henschke Eric A., Hoffman Ella A., Kazerooni Heber, MacMahon Edwin J. R., van Beek David, Yankelevitz Alberto M., Biancardi Peyton H., Bland Matthew S., Brown Roger M., Engelmann Gary E., Laderach Daniel, Max Richard C., Pais David P.‐Y., Qing Rachael Y., Roberts Amanda R., Smith Adam, Starkey Poonam, Batra Philip, Caligiuri Ali, Farooqi Gregory W., Gladish C. Matilda, Jude Reginald F., Munden Iva, Petkovska Leslie E., Quint Lawrence H., Schwartz Baskaran, Sundaram Lori E., Dodd Charles, Fenimore David, Gur Nicholas, Petrick John, Freymann Justin, Kirby Brian, Hughes Alessi, Vande Casteele Sangeeta, Gupte Maha, Sallam Michael D., Heath Michael H., Kuhn Ekta, Dharaiya Richard, Burns David S., Fryd Marcos, Salganicoff Vikram, Anand Uri, Shreter Stephen, Vastagh Barbara Y., Croft Laurence P., Clarke (2011) The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans Medical Physics 38(2) 915-931. 10.1118/1.3528204 [DOI] [PMC free article] [PubMed]
- 35.Etmann C., K.R., -B., S.: iUNets: Learnable Invertible Up- and Downsam- pling for Large-Scale Inverse Problems, vol. 11 (2020)
- 36.Jean-Christophe, Richard Florian, Sigaud Maxime, Gaillet Maciej, Orkisz Sam, Bayat Emmanuel, Roux Touria, Ahaouari Eduardo, Davila Loic, Boussel Gilbert, Ferretti Hodane, Yonis Mehdi, Mezidi William, Danjou Alwin, Bazzani Francois, Dhelft Laure, Folliet Mehdi, Girard Matteo, Pozzi Nicolas, Terzi Laurent, Bitker (2022) Response to PEEP in COVID-19 ARDS patients with and without extracorporeal membrane oxygenation. A multicenter case–control computed tomography study Abstract Critical Care 26(1). 10.1186/s13054-022-04076-z [DOI] [PMC free article] [PubMed]
- 37.Ludmilla, Penarrubia Aude, Verstraete Maciej, Orkisz Eduardo, Davila Loic, Boussel Hodane, Yonis Mehdi, Mezidi Francois, Dhelft William, Danjou Alwin, Bazzani Florian, Sigaud Sam, Bayat Nicolas, Terzi Mehdi, Girard Laurent, Bitker Emmanuel, Roux Jean-Christophe, Richard (2023) Precision of CT-derived alveolar recruitment assessed by human observers and a machine learning algorithm in moderate and severe ARDS Abstract Intensive Care Medicine Experimental 11(1). 10.1186/s40635-023-00495-6 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during and/or analyzed during the current study are not available; they belong to the producer/host center: CLB. The data used in this study adhere to the tenets of the Declaration of Helsinki.