Abstract
Colorectal cancer (CRC) stands out as one of the most prevalent global cancers. The accurate localization of colorectal polyps in endoscopy images is pivotal for timely detection and removal, contributing significantly to CRC prevention. The manual analysis of images generated by gastrointestinal screening technologies poses a tedious task for doctors. Therefore, computer vision-assisted cancer detection could serve as an efficient tool for polyp segmentation. Numerous efforts have been dedicated to automating polyp localization, with the majority of studies relying on convolutional neural networks (CNNs) to learn features from polyp images. Despite their success in polyp segmentation tasks, CNNs exhibit significant limitations in precisely determining polyp location and shape due to their sole reliance on learning local features from images. While gastrointestinal images manifest significant variation in their features, encompassing both high- and low-level ones, a framework that combines the ability to learn both features of polyps is desired. This paper introduces UViT-Seg, a framework designed for polyp segmentation in gastrointestinal images. Operating on an encoder-decoder architecture, UViT-Seg employs two distinct feature extraction methods. A vision transformer in the encoder section captures long-range semantic information, while a CNN module, integrating squeeze-excitation and dual attention mechanisms, captures low-level features, focusing on critical image regions. Experimental evaluations conducted on five public datasets, including CVC clinic, ColonDB, Kvasir-SEG, ETIS LaribDB, and Kvasir Capsule-SEG, demonstrate UViT-Seg’s effectiveness in polyp localization. To confirm its generalization performance, the model is tested on datasets not used in training. Benchmarking against common segmentation methods and state-of-the-art polyp segmentation approaches, the proposed model yields promising results. For instance, it achieves a mean Dice coefficient of 0.915 and a mean intersection over union of 0.902 on the CVC Colon dataset. Furthermore, UViT-Seg has the advantage of being efficient, requiring fewer computational resources for both training and testing. This feature positions it as an optimal choice for real-world deployment scenarios.
Keywords: Colorectal cancer, Polyp localization, CNN, Vision transformer, Attention mechanism
Introduction
Colorectal cancer ranks among the most frequently diagnosed malignancies and stands as a primary contributor to cancer-related fatalities among both men and women worldwide [1]. Recent statistics from the USA place colorectal cancer as the third major factor contributing to cancer-related mortality, estimating 53,020 new cases and 52,550 associated deaths. Additionally, other digestive system cancers, such as esophageal, stomach, and small intestine cancers, significantly contribute to the overall mortality rate related to cancer [2]. Gastrointestinal (GI) diseases such as polyps, ulcers, and bleeding pose significant challenges for doctors in detecting and localizing inside the small bowel. The detection of polyps, specifically, presents a notably difficult challenge due to their various sizes, shapes, and diverse morphological features. However, numerous initiatives have been undertaken to enhance diagnostic methods and technological advancements aimed at improving the accuracy and efficiency of polyp detection. Colonoscopy stands as one of the primary tools employed for screening to prevent CRC cancer. The procedure involves examining the large intestine with a flexible tube inserted through the rectum or mouth. This tube is equipped with a small camera that enables physicians to visualize the interior of the colon [3]. During this process, specialists search for polyps and determine whether cancer is present. In general, polyps fall into two main categories: non-neoplastic and neoplastic polyps. Non-neoplastic polyps typically do not develop into cancer, while neoplastic polyps have the potential to progress into cancer [1]. Colorectal polyps are categorized into three different sizes: diminutive (5 mm), small (6 to 9 mm), and advanced or extensive (10 mm) [4]. While colonoscopy is effective at detecting and removing polyps, it does have several limitations. This invasive procedure involves inserting a flexible tube called a colonoscope into the rectum and colon, which can lead to discomfort and may require sedation. Additionally, colonoscopy may miss some polyps, especially smaller ones or those located in hard-to-reach areas [5]. However, examining the small bowel within the GI tract presents significant challenges due to its intricate structure and considerable length of approximately 6 ms. Hence, wireless capsule endoscopy (WCE) has emerged as an innovative diagnostic tool for the GI tract. Utilizing a compact 11 mm capsule that is ingested, WCE navigates through the human digestive system, equipped with a small integrated camera to capture images of the GI tract. Remarkably, it can generate approximately 55,000 images in a single examination, all of which are transmitted to external devices for comprehensive analysis [6]. The key advantage of WCE over colonoscopy lies in its non-invasive nature, eliminating the need for staff preparation. Furthermore, the non-invasive nature of WCE allows individuals to continue their daily activities during the examination, offering added comfort [7, 8]. Despite these tools enhancing the GI tract examination process, both colonoscopy and WCE data require manual analysis and treatment by doctors. Interpreting a large volume of images is a labor-intensive and time-consuming task for doctors. Therefore, there is a pressing demand for the development of a medical support system to assist doctors in making swift, cost-effective, and dependable decisions.
Recent developments in computer-aided detection (CAD) utilizing artificial intelligence (AI) systems have led to significant advancements in the analysis of GI medical images [9, 10]. Machine learning (ML) and deep learning (DL) methods are the major contributors to the GI malignancies analysis. These techniques have shown great promise in improving the accuracy and efficiency of diagnosis of gastrointestinal conditions, helping healthcare professionals to make more informed decisions, and improving patient care. Handcrafted methods, such as Local Binary Pattern (LBP), Scale-Invariant Feature Transform (SIFT), and Histogram of Oriented Gradients, among others, have been employed to characterize images from both colonoscopy and WCE images [11–13]. Various machine learning algorithms, including Support Vector Machines (SVM), Random Forest (RF), K-Nearest Neighbors (KNN), Decision Trees (DT), and others, have been used as classifiers to identify abnormal region features extracted by classical descriptors [14–16]. However, handcrafted approaches are constrained in their ability to generalize, as they are typically customized for specific tasks or datasets. The process of selecting suitable WCE and colonoscopy features can also be demanding in terms of time and labor.
In contrast, deep learning-based CNNs have demonstrated promising performance in learning features from GI images [17–19]. The superiority of CNNs over handcrafted methods lies in their ability to automatically extract relevant features. Various CNN-based encoder-decoder architectures, such as U-Net [20], SegNet [21], and others, have been widely employed for effectively localizing small polyps with impressive performance. However, CNNs come with two significant drawbacks: they demand a substantial amount of training data, and they may struggle to effectively account for global information and spatial context. These limitations can significantly impact model precision, especially in terms of detection and segmentation, as most polyp datasets contain a combination of global and local features, both of which are crucial for accurately identifying the regions that contain polyps. On the other hand, Vision Transformers (ViTs) have emerged as the front-runners in many computer vision tasks in recent years, and their performance surpasses that of CNNs on the ImageNet dataset. Transformers have proven highly effective due to their architecture built on the self-attention mechanism (SEA), which is used to capture and identify long-term dependencies and global context by considering the relationships between all regions [22]. Harnessing the capabilities of CNNs for acquiring local features and leveraging the potential of ViTs for capturing global context can offer an effective approach to enhance polyp segmentation.
Many efforts have been already made in the literature to improve and generalize the polyp segmentation. Achieving precise identification of small polyps continues to be a formidable task, given the numerous hurdles present in polyp datasets. These obstacles include low contrast, intricate backgrounds, and substantial variations in both shape and texture. As shown in Fig. 1, it can be challenging to visually distinguish polyps from the healthy or normal tissues surrounding them because they may appear very similar in terms of color and texture. Therefore, there is an urgent need to develop decision-support systems to assist doctors in interpreting the polyp regions in GI images. This paper presents a deep learning framework based on an encoder-decoder architecture for polyp segmentation. The encoder is built upon the vision transformer, which employs a self-attention mechanism to capture long-term dependencies and learn global information. Meanwhile, the decoder harnesses the power of CNNs combined with the squeeze and excitation (SE) and dual attention mechanism (DAM) to extract local patterns and identify the most important regions in the images. The main contributions of this study can be highlighted as follows:
A deep learning approach based on the transformer encoder and the CNN decoder is proposed for small polyps segmentation.
The encoder is constructed using the vision transformer, which is applied to extract global patterns and spatial context from colonoscopy and WCE images.
The decoder part exhibits the potential of CNNs in learning invariant local features.
An efficient dual attention-based residual block is employed to capture intricate features and enhance the model’s ability to focus on relevant information for both spatial and channel details.
The squeeze and excitation block is strategically employed at different stages in the decoder section to enhance the model’s performance in learning features, allowing the model to dynamically emphasize important information.
The proposed approach undergoes training and validation on five public datasets, with each dataset used for training while the others are utilized for testing, and vice versa.
The model performance demonstrates its ability to identify polyps regions with respectable precision, outperforming the state-of-the-art methods for polyps segmentation.
The model can be used in the real-world scenario for polyp segmentation due to its ability to generalize and locate polyps on unseen datasets.
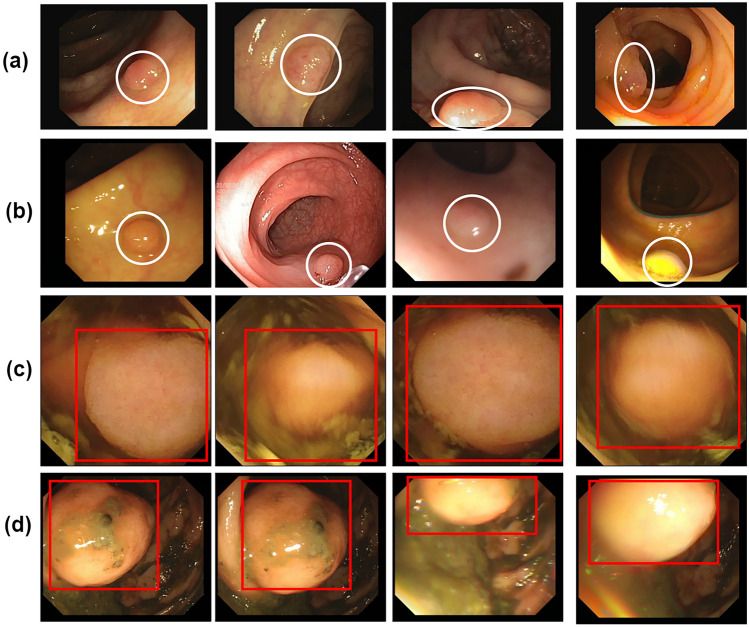
Fig. 1.
Some examples of polyps: the first two rows a, b display small polyps, while the last two rows c, d feature large polyps
The remainder of the paper is structured as follows: Section “Related Works” presents a summary of various related studies. Section “Methodology” describes the proposed model for abnormality detection in WCE images. Section “Results and Discussion” covers the datasets, experimental results, and analysis. Finally, the study concludes in “Conclusion” section.
Related Works
GI disease segmentation and localization is an active area for researchers, and significant efforts have been undertaken to develop a support decision system for polyp segmentation [23–25]. Polyps, in particular, are among the most common diseases targeted for early detection and precise segmentation, considering the challenges doctors face in identifying and locating them. Various methods were proposed in the past to detect and localize polyp regions in GI images. The first methods proposed were traditional techniques based on handcrafted descriptors to obtain low-level features such as texture and geometric features. Iwahori et al. [12] present an automated method for polyp detection, employing the Hessian filter in combination with Histogram of Oriented Gradients (HOG). The Hessian filter is used to extract potential polyp regions, followed by extracting HOG features from these regions. The selection of these features is done in an adaptive manner using Random Forests and RealAdaBoost. Finally, they combine the various polyp regions using the K-Means++ method. Their findings indicate that Random Forests achieve high sensitivity and specificity when compared to RealAdaBoost. Jain et al. [26] have used features based on fractal dimensions from RGBL channel images to distinguish between normal and abnormal conditions within the gastrointestinal tract. They present results for classification on two distinct datasets: the KID dataset and a private dataset. In their experiments, they employed various machine learning classifiers, including KNN, SVM, RF, and Naive Bayesian networks. In their study, Sanchez-Gonzalez et al. [27] extracted features like shape, color, and curvature of edges and their regions from images within the CVC dataset. They then utilized various classifiers, such as Naive Bayes, MultiBoost AB, and AdaBoost, to selectively choose these features. This approach allowed them to identify the boundaries of the polyp-affected area, and subsequent segmentation was carried out by connecting these identified edges. However, the variability of features within polyp datasets poses a limitation on the performance of handcrafted methods in identifying polyp regions and accurately locating them.
Deep learning-based convolution neural networks have demonstrated their effectiveness in polyp region analysis due to their ability to learn features automatically [28, 29]. Guo et al. [30] adapted the FCN network for polyp segmentation by transforming it into a Dilated ResFCN, in which they incorporated residual connections, employed high-kernel-sized dilated convolutions, and integrated fusion sub-networks. Their work made use of the Endoscopic Vision GIANA Polyp Segmentation Challenge dataset, including CVC-ColonDB [31] and CVC-ClinicDB [32]. In their study [33], the authors presented a framework named PolypSegNet. Their approach presents a deep learning architecture based on an encoder-decoder. It incorporates deep dilated inception (DDI) blocks to capture the features of various receptive areas. In addition, a deep fusion skip module (DFSM) efficiently links the encoder and decoder layers, enabling the passage of contextual information. Additionally, a deep reconstruction module (DRM) optimizes multi-scale decoded feature maps for improved reconstruction. Jha et al. [34] introduce the ResUNet++ approach, a neural network for semantic segmentation. It integrates residual and squeeze blocks, attention blocks, and atrous spatial pyramidal pooling (ASPP). Their model demonstrates efficiency even when dealing with a small image dataset. In their study, Ta et al. [35] introduced Boundary-Aware Attention blocks within a boundary enhancement module. This module leverages the encoder features and the partial decoder output to produce the polyp mask. Qadir et al. [36] utilized Mask R-CNN as a feature extractor to perform polyp area segmentation in colonoscopy images, employing various CNN architectures, including ResNet50, ResNet101, and InceptionResNetV2. This approach achieved acceptable results on the CVC-ColonDB dataset. In [37], the authors introduced a deep convolutional neural network designed for the detection of anomalies in WCE images. Initially, they trained an effective attention-based CNN to categorize images into one of four classes: polyp, vascular, inflammatory, or normal. Subsequently, an amalgamation of Grad-CAM++ and a custom SegNet was employed to perform the segmentation of anomalous regions within abnormal images. In their study, Lafraxo et al. [38] introduced an end-to-end architecture named AttResU-Net, which combines the attention mechanism and residual units within the U-Net framework. This integration enhanced the performance of polyp and bleeding segmentation, providing improved segmentation results. On the other hand, transformers have had a remarkable influence on GI abnormalities analysis, even though they were originally designed for natural language processing tasks [39]. Dosovitskiy et al. [22] proposed the first visual transformer based on the self-attention mechanism. The work presented in [40] utilizes a transformer neural network with spatial pooling configuration to tackle binary and multi-classification challenges within WCE images. Through the application of pooling, the spatial design of the vision transformer exhibits enhanced performance, as evidenced by notable improvements in the small-scale capsule endoscopy dataset [41]. The authors in [42] introduced an approach that relies on vision transformers to identify gastrointestinal diseases from WCE images. They conducted a comparative examination, where they compared the performance of the transformer-based approach to a pre-trained CNN model, specifically DenseNet201. Their experiments demonstrate that the vision transformer outperformed DenseNet201 across a range of quantitative performance assessment metrics. Chen et al. [43] introduced the TransUnet framework, which employs a transformer-based network featuring a hybrid ViT encoder alongside an upsampled CNN decoder. This hybrid ViT integrates both CNN and transformer elements. The results achieved by TransUnet underscore the effectiveness of using a transformer in the encoder for polyp segmentation. The authors in [44] have introduced a deep learning framework known as SR-AttNet, specially designed for segmenting colorectal polyps. This technique employs an encoder-decoder architecture that combines both non-expanded and expanded filtering to capture information from both nearby and distant regions while also understanding the depth of the image. To enhance the model, they have included four-fold skip connections between each spatial encoder-decoder and implemented a Feature-to-Mask pipeline to manage expanded and non-expanded features for the ultimate prediction. Furthermore, their approach incorporates an attention mechanism named SR-Attention, which draws inspiration from the Stretch-Relax concept. This attention system creates spatial features with significant variations, which are utilized to generate valuable attention masks for feature selection.
Methodology
In this section, we will describe the proposed approach for localizing existing GI tract polyps, which utilizes a 2D segmentation architecture inspired by the Unet-based encoder-decoder model. The encoder employs a vision transformer to capture global features, and the decoder uses multiple convolution block-based attention mechanisms to extract local features from images. Here’s an overview of our system.
We build an encoder-based vision transformer to obtain higher-level features.
A decoder-based CNN is used to extract features from the decoder at different scales through multiple skip connections.
Squeeze and excitation attention is applied at various points within the decoder to enhance its focus on the most crucial image regions.
To prevent vanishing gradients in the decoder, we employ a residual block with a custom attention mechanism directly from the encoder to maintain essential features.
The specific details of each component of the proposed system are provided in the following subsections, while the overall diagram illustrating the methodology for localizing polyps within GI images is presented in Fig. 2.
Fig. 2.
The overall structure of the proposed system for polyp localization in Colonoscopy and WCE images
Data Preparation
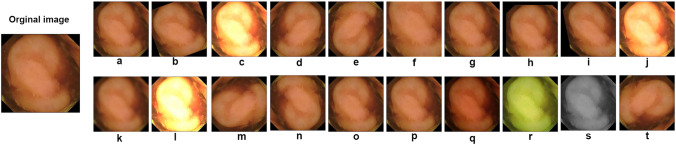
In the proposed system, we conducted experiments using five datasets: CVC clinicDB [32], CVC colonDB [31], kvasir-SEG [45], ETIS-Larib PolypDB [46], and Kvasir Capsule-SEG [47]. Each dataset exhibits unique feature distributions and often necessitates various augmentations due to a limited number of images. To address this challenge, we applied data augmentation techniques to each dataset. Common augmentation methods, including flipping, brightness, cropping, rotation, Gaussian noise, and zooming, were applied to all datasets except the Kvasir Capsule-SEG dataset, which comprises only 55 polyp images. For this particular dataset, we executed 20 augmentation operations on each image, resulting in a total of 1,100 synthetic images with distinct features. Figure 3 showcases examples of image augmentation for the CVC clinic, CVC colon, Kvasir-SEG, and Etis Larib datasets, while Fig. 4 provides the twenty augmentation strategies applied to the Kvasir Capsule-SEG dataset.
Fig. 3.
Some data augmentation applied to CVC Clinic, CVC Colon, Kvasir-SEG, and Etis larib datasets
Fig. 4.
Representation of the twenty augmentation techniques applied to the Kvasir Capsule-SEG dataset: a Zoom (0.7); b Rotate (20); c Brightness (2.0); d Vertical flip; e Horizontal flip; f Random Crop (256, 256); g Random Noise (0.6); h Translate (20, 20); i Shear (0.2); j Contrast (1.5); k Blur (5); l Sharpen (1.5); m Rotate 90 (1); n Rotate 90 (2); o Scale (0.8); p Scale (1.2); q Gamma Correction (1.5); r Color Shift (20); s Grayscale (1); t Rotated 90° counter-clockwise
U-Net Overview
U-Net, originally introduced by Ronneberger et al. [20] for biomedical image segmentation, has gained widespread popularity as a go-to architecture for various image segmentation tasks. This architecture is rooted in the foundation of fully convolutional networks [48]. The U-Net model comprises two key components: the encoder and decoder pathways. The encoder’s role is to extract deep features using a series of convolution layers with 3x3 filters, followed by ReLU activation and MaxPooling layers. Conversely, the decoder is responsible for creating the output segmentation map, employing operations such as upsampling, convolution, ReLU activation, and MaxPooling layers. Notably, a crucial aspect of U-Net is the integration of skip connections, linking each stage of the encoder to its corresponding stage in the decoder. These skip connections play a vital role in providing essential features, ultimately enhancing the model’s capability to produce increasingly accurate segmentation results.
The Vision Transformer Encoder
Drawing inspiration from the U-net description provided earlier, we integrated a Vision Transformer [22] into the encoder section of our proposed system. The Vision Transformer stands as the first visual transformer designed specifically for image classification. Its architecture is rooted in the self-attention mechanism found in the transformer encoder, which was originally used for natural language processing tasks. In the proposed system, we developed the vision transformer from scratch and incorporated it as the encoder component of the proposed method for polyp segmentation. This integration aimed to capture overarching dependencies present in WCE and colonoscopy images.
The ViT encoder model is designed to process input images with dimensions (H, W, C), where H, W, and C denote the image’s height, width, and number of channels, respectively. This image is then divided into N flattened patches, denoted as with dimensions , where each patch has a size of (P, P). The total number of patches, N, is calculated as N = . After converting these patches into a single vector X with dimensions , a dense layer is applied to transform X into a new vector E of size D, known as the Embedding E[N, D]. The class embedding is not added as the ViT is implemented for the polyp segmentation task. To produce the final vector Z, a 1D positional embedding is introduced to the linearly embedded image patches E, which is crucial for preserving the sequence order in transformers. Subsequently, the Z vector obtained in the previous step is fed into the transformer encoder block to generate the context vector. The transformer encoder consists of multiple layers of Multi-Head Self Attention (MHSA) and MLP blocks. Layer normalization is applied before each block, and skip connections are present after each block. The MLP module comprises two fully connected layers, employing Gaussian Error Linear Units (GELU) as the activation function for non-linearity. The skip connections, , where i ranges from 1 to 4, are employed to pass features to the decoder part at various scales. These skip connections play a crucial role in facilitating the fusion of high-level semantic information from the encoder with the fine-grained details from the decoder.
CNN Decoder-Based Attention
The decoder employed in the proposed approach shares similarities with the decoder used in the U-Net architecture. It consists of a series of convolutional layer blocks from scratch and serves the purpose of resampling the feature set obtained from the encoder. Much like the U-Net network, where different feature resolutions are fused with the decoder, we incorporate a set of skip connections denoted as , ranging from 1 to 4. These connections enable us to extract a sequence of features from the transformer encoder, each with a size of , subsequently reshaped to . For each resolution , the embedding features undergo a transformation back into the input space. This transformation involves a block that includes 3x3 convolutional layers, followed by the application of a normalization layer. The output features from the stacked transformers experience a resolution increase through a deconvolution layer. Additionally, the output features from the final transformer go through a similar resolution enhancement step, accomplished by applying a deconvolution layer and concatenating the results with the output from the previous transformer. These fused features then pass through a convolutional layer and are upsampled via a deconvolution layer. This entire process is duplicated for all successive layers, extending up to the initial input resolution. Finally, the outcome is processed through a convolutional layer featuring a softmax activation function, which generates the output mask prediction.
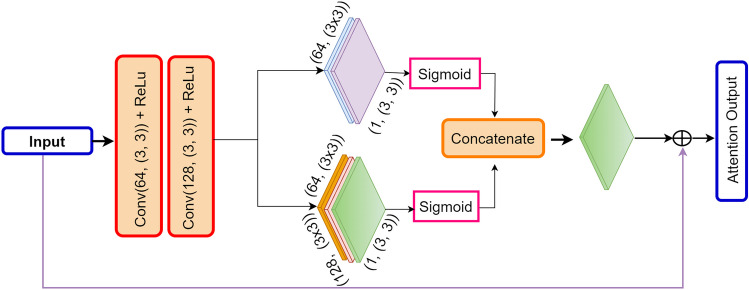
To enhance the decoder’s ability to emphasize essential image regions, we integrated a squeeze and excitation attention mechanism at various points in the decoder architecture. As depicted in Fig. 5, the SE mechanism is a two-step process. Initially, it condenses feature maps into a single channel through global average pooling, averaging values across spatial dimensions. Subsequently, a compact neural network module is employed to acquire a collection of channel-specific weights. This module takes the condensed feature map as input and applies non-linear transformations to generate the weight set.
Fig. 5.
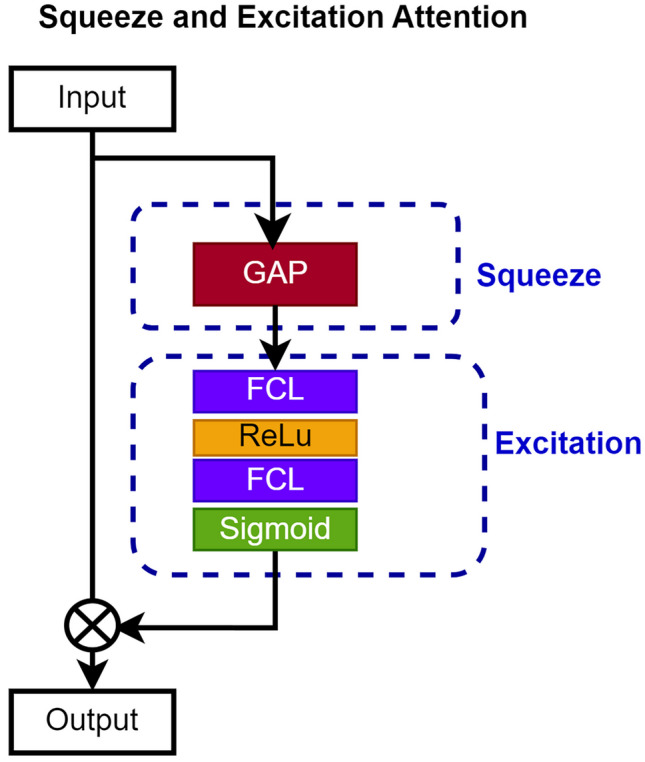
A schematic representation illustrating the attention mechanism of squeeze and excitation
Furthermore, as shown in Figs. 6 and 7, we incorporate a dual attention mechanism within a residual block to ensure that the valuable features acquired from the last transformer are retained. This serves a twofold purpose: preventing the vanishing gradient problem and facilitating the transmission of the most critical features to the final convolutional layer in the decoder network. The DAM employs a streamlined and efficient process that combines both pixel-wise and channel-wise attention mechanisms. Illustrated in Fig. 7, the pixel-wise attention mechanism consists of two consecutive convolutional layers, one of 3x3 with 28 filters and the other with a single filter. On the other hand, the channel attention mechanism encompasses three convolutional layers, each of 3x3 with 128, 64, and 1 filter, respectively. The output of the DAM is integrated with the input features of the residual block, which corresponds to the output of the last transformer in the encoder section. The DAM features are obtained following the equations described in Eqs. 1, 2, 3, 4, and 5. The indices (i, j) represent the location in the feature map matrix, while m, n represent the spatial location in the convolution kernel, and is a pixel value in the input feature map W at location . , , are the weights of the convolution operation at location (m, n), and b represent a bias term. This strategic step plays a crucial role, particularly given the prevalence of small-sized polyps. Leveraging a residual block-based attention mechanism empowers the model to effectively preserve both high- and low-level features, contributing to the overall success of the proposed system.
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
Fig. 6.
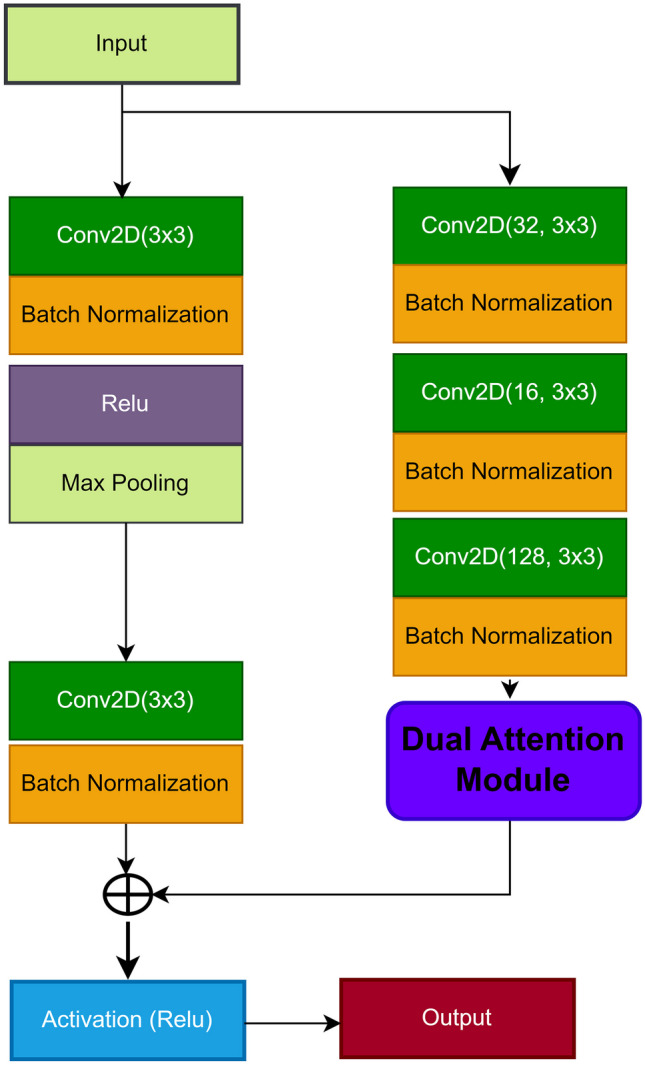
A visual representation of the residual block incorporating the dual attention mechanism
Fig. 7.
The structure of the dual attention mechanism used within the residual block
Results and Discussion
This section provides insight into the datasets used for polyp segmentation, implementation details, and the various experiments conducted. Additionally, it presents the quantitative and qualitative results of the proposed model, comparing them with the latest state-of-the-art methods for polyp localization in the GI tract.
Datasets and Implementation Detail
The proposed system leverages five datasets in its experiments: CVC clinicDB [32], CVC colonDB [31], kvasir-SEG [45], ETIS-Larib PolypDB [46], and Kvasir Capsule-SEG [47]. All of these datasets are publicly available, with each dataset characterized by its distinct feature distribution. Table 1 shows some details about each dataset. As outlined in “Data Preparation” section, we apply data augmentation techniques to the training set to generate additional synthetic polyp images. Offline data augmentation strategies have been utilized for this purpose. To reduce computational requirements, all images are resized to , and each dataset is split in a ratio of 80:20 for training and testing.
Table 1.
The number of images containing polyps in each dataset before and after data augmentation techniques
| Dataset | Before augmentation | After augmentation | Image size |
|---|---|---|---|
| CVC-ClinicDB | 612 | 2712 | 384 × 288 |
| Kvasir-SEG | 1000 | 4800 | 626 × 546 |
| CVC-Colon | 300 | 1344 | 574 × 500 |
| ETIS-Larib | 196 | 1020 | 1256 × 966 |
| Kvasir Capsule-SEG | 55 | 1100 | 336 × 336 |
The proposed system undergoes training and assessment across all datasets, utilizing each dataset for training purposes and the remaining ones for testing. Implementation is carried out using Keras with the latest TensorFlow version as the backend, operating on an Intel i7 processor with 64 GB RAM and a 24 GB NVIDIA GeForce RTX 3090 GPU. The training process involves processing RGB images through 100 epochs, employing the Adam optimizer with a learning rate of 0.001, and integrating three loss functions: categorical cross-entropy (BCE), dice loss (Dice), and BCE+Dice. Additionally, an early stopping mechanism is in place, resulting in the reduction of the learning rate by a factor of 0.01 when validation loss stagnates after 15 epochs to mitigate overfitting issue. The whole hyper-parameters of the proposed encoder-decoder system are listed in Table 2. To evaluate the performance of the proposed approach, we have utilized common metrics for the segmentation task, including mean Dice coefficient (mDc), mean Intersection over Union (mIoU), Precision (P), Recall (R), and -score. The following equations present the mathematical formulas for each metric, along with the loss functions considered in this study. The terms TP, TN, FP, and FN represent True Positive, True Negative, False Positive, and False Negative, respectively. Here, y and indicate the true and predicted outputs, respectively.
| 6 |
| 7 |
| 8 |
| 9 |
| 10 |
| 11 |
| 12 |
| 13 |
Table 2.
Hyper-parameters used in the training of the proposed encoder-decoder for polyp localization
| Hyper-parameters | Values |
|---|---|
| Image size | |
| Patch size | |
| Projection dimension (k) | 64 |
| Number of heads | 8 |
| Transformer units | [128, 64] |
| Transformer layers | 13 |
| Batch size | 16 |
| Optimizer | Adam |
| Learning rate | 0.001 |
| Weight decay rate | 0.0001 |
| Number of epochs | 100 |
Performance Evaluation and Analysis
Quantitative Results
The proposed system features an architecture similar to the Unet model, leveraging ConvNets layers in both the encoder and decoder to extract and resample features. The proposed approach combines the potential of convolutional operations for extracting local features with the transformative power of transformers for learning global information from images. The encoder employs the ViT to capture high-level features and significant regions, utilizing a multi-head attention mechanism. Simultaneously, feature resampling and distribution are achieved through an attention-based convolution block. We initiate our analysis with an experiment of three loss functions as defined in Eqs. 11, 12, and 13. The purpose of employing these loss functions is to quantify the disparity between the predicted pixel masks generated by the proposed model and the true pixel masks. In essence, a loss function serves as a measure of the error between the predicted values and the actual values. By evaluating different loss functions, each encapsulating distinct aspects of the prediction error, we aim to discern which one is more suitable for our specific task. As depicted in Table 3, the combined loss (BCE + Dice) consistently outperforms across almost all metrics for all datasets, with the exception of recall and score for the CVC clinic dataset. Notably, the BCE loss function achieves the highest recall and score specifically on the CVC clinic dataset, while implementing the same loss function yields optimal results for Dice coefficient and score on the Etis Larib dataset. When training incorporates Dice as the loss function, the model excels in achieving the highest recall on both the CVC colon and Etis Larib datasets. Furthermore, the effectiveness of the combined loss in the optimization process is underscored, demonstrating effective results in all metrics, with only slight differences in some cases. The experiment substantiates its capability to minimize errors between true pixel masks and predicted ones. As a result, we have chosen it as the primary loss function for the remainder of our experiments.
Table 3.
The comparison results of using three different loss functions in the training of the proposed model
| Datasets | Loss | P | R | mIoU | mDc | |
|---|---|---|---|---|---|---|
| CVC Clinic | BCE | 0.8989 | 0.9106 | 0.8956 | 0.9046 | 0.9082 |
| Dice | 0.9006 | 0.9049 | 0.8917 | 0.9025 | 0.9039 | |
| BCE+Dice | 0.9038 | 0.9081 | 0.8981 | 0.9059 | 0.9072 | |
| CVC colon | BCE | 0.9042 | 0.9180 | 0.9016 | 0.9107 | 0.9150 |
| Dice | 0.9047 | 0.9182 | 0.9020 | 0.9109 | 0.9151 | |
| BCE+Dice | 0.9072 | 0.9178 | 0.9042 | 0.9121 | 0.9154 | |
| Kvasir-SEG | BCE | 0.7916 | 0.8135 | 0.7724 | 0.8001 | 0.8073 |
| Dice | 0.8001 | 0.8177 | 0.7850 | 0.8064 | 0.8104 | |
| BCE+Dice | 0.8144 | 0.8211 | 0.8012 | 0.8230 | 0.8352 | |
| Etis Larib | BCE | 0.9155 | 0.9101 | 0.9046 | 0.9175 | 0.9140 |
| Dice | 0.9161 | 0.9174 | 0.9025 | 0.9016 | 0.9051 | |
| BCE+Dice | 0.9196 | 0.9145 | 0.9131 | 0.9118 | 0.9134 | |
| Kvasir Capsule-SEG | BCE | 0.4829 | 0.5278 | 0.4661 | 0.5018 | 0.5161 |
| Dice | 0.4919 | 0.5301 | 0.4703 | 0.5089 | 0.5133 | |
| BCE+Dice | 0.5009 | 0.5418 | 0.4912 | 0.5122 | 0.5372 |
To assess the model’s performance and the contribution of each element in the proposed system, we employed an experiment known as the ablation study to discern the impact of each component on the proposed UViT-Seg for polyp localization performance. The experiment involves evaluating the proposed system by removing and adding the squeeze attention mechanism and the residual-based dual attention block. Table 4 displays the obtained results of this experiment, indicating that the proposed approach achieves the best results by combining all its components. On the CVC Clinic dataset, the proposed model demonstrates nuanced results when considering individual elements. Employing the attention mechanism alone and excluding the residual block in the decoder yields a high precision of 89%. In contrast, the mIoU, mDc, and show elevated values when the residual block is used in isolation, and SE attention is omitted. For the Kvasir-SEG and CVC Colon datasets, the exclusive use of the residual block within the proposed model showcases its superiority over relying on attention alone. The mDc and demonstrate high values of 89.23% and 89.98%, respectively, on the Etis Larib dataset when SE attention is employed. Meanwhile, the mIoU, precision, and recall achieve their second peak results with values of 89.66%, 88.67%, and 89.26%, respectively, through the exclusive integration of the residual block. In the last dataset used in this experiment, Kvasir Capsule-SEG, precision and mDc showcase strong second-tier performance when the system is constructed solely with the residual block. In contrast, the mIoU, recall, and exhibit high-level performance through the exclusive incorporation of attention. The lowest results are obtained with the exclusion of both SE attention and the residual block. However, the results of ablation experiments conclusively demonstrate that incorporating both of these elements into the proposed system leads to the optimal model performance for localizing polyps in GI images. An additional experiment is conducted on five datasets, involving the evaluation of the model with and without the proposed dual attention mechanism within the residual block. The aim of this experiment is to assess the impact of incorporating the dual attention mechanism within the residual block on the model’s performance. The results from this experiment, showcased in Table 5, vividly demonstrate the potential of the dual attention mechanism within the residual block. Integrating DAM into the residual block yields superior results, achieving high performance across all datasets and metrics, with the exception of the CVC Colon dataset. In this particular case, the version without DAM within the residual block exhibits a notable recall of 91.82%. Nevertheless, the overall experiment solidly validates the effectiveness of the DAM module. It plays a pivotal role in aiding the residual block to capture the most crucial regions, derived from resampled features processed by the last deconvolution layer positioned after the skip connection S4.
Table 4.
The outcomes of the ablation experiments conducted on the proposed model across the five datasets
| Datasets | SE mechanism | Resdual block | P | R | mIoU | mDc | |
|---|---|---|---|---|---|---|---|
| CVC Clinic | 0.8648 | 0.8724 | 0.8537 | 0.8684 | 0.8607 | ||
| ✓ | 0.8900 | 0.8959 | 0.8715 | 0.8959 | 0.8909 | ||
| ✓ | 0.8828 | 0.8976 | 0.8868 | 0.8900 | 0.8985 | ||
| ✓ | ✓ | 0.9038 | 0.9081 | 0.8981 | 0.9059 | 0.9072 | |
| Kvasir-SEG | 0.7647 | 0.7993 | 0.7417 | 0.7836 | 0.7973 | ||
| ✓ | 0.7874 | 0.7583 | 0.7148 | 0.7651 | 0.7595 | ||
| ✓ | 0.7837 | 0.8026 | 0.7731 | 0.8005 | 0.8127 | ||
| ✓ | ✓ | 0.8144 | 0.8211 | 0.8012 | 0.8230 | 0.8352 | |
| CVC Colon | 0.8817 | 0.8833 | 0.8606 | 0.8790 | 0.8871 | ||
| ✓ | 0.8847 | 0.8943 | 0.8765 | 0.8877 | 0.8916 | ||
| ✓ | 0.8872 | 0.8907 | 0.8787 | 0.8975 | 0.8991 | ||
| ✓ | ✓ | 0.9072 | 0.9178 | 0.9042 | 0.9121 | 0.9154 | |
| Etis Larib | 0.8857 | 0.8739 | 0.8891 | 0.8847 | 0.8742 | ||
| ✓ | 0.8901 | 0.8954 | 0.8891 | 0.8923 | 0.8998 | ||
| ✓ | 0.8966 | 0.8867 | 0.8926 | 0.8861 | 0.8964 | ||
| ✓ | ✓ | 0.9196 | 0.9145 | 0.9131 | 0.9118 | 0.9134 | |
| Kvasir Capsule-SEG | 0.4878 | 0.4710 | 0.4313 | 0.4736 | 0.4803 | ||
| ✓ | 0.4419 | 0.5090 | 0.4651 | 0.4826 | 0.5132 | ||
| ✓ | 0.4928 | 0.4999 | 0.4528 | 0.4939 | 0.4966 | ||
| ✓ | ✓ | 0.5009 | 0.5418 | 0.4912 | 0.5122 | 0.5372 |
Table 5.
Effect of Dual Attention mechanism within the residual blocks on the proposed model performance, where the bold values indicate the superior results
| Residual Block | P | R | mIoU | mDc | |
|---|---|---|---|---|---|
| CVC Clinic | |||||
| With DAM | 0.9038 | 0.9081 | 0.8981 | 0.9059 | 0.9072 |
| Without DAM | 0.8868 | 0.8906 | 0.8835 | 0.8935 | 0.8977 |
| CVC Colon | |||||
| With DAM | 0.9072 | 0.9178 | 0.9042 | 0.9121 | 0.9154 |
| Without DAM | 0.8996 | 0.9182 | 0.8969 | 0.8980 | 0.9038 |
| Kvasir-SEG | |||||
| With DAM | 0.8144 | 0.8211 | 0.8012 | 0.8230 | 0.8352 |
| Without DAM | 0.7731 | 0.8243 | 0.7648 | 0.7957 | 0.8111 |
| Etis Larib | |||||
| With DAM | 0.9196 | 0.9145 | 0.9131 | 0.9118 | 0.9134 |
| Without DAM | 0.8903 | 0.9091 | 0.8984 | 0.9046 | 0.9073 |
| Kvasir Capsule-SEG | |||||
| With DAM | 0.5009 | 0.5418 | 0.4912 | 0.5122 | 0.5372 |
| Without DAM | 0.4692 | 0.5154 | 0.4469 | 0.4893 | 0.5038 |
In order to assess the generalization performance of the trained model, a comprehensive set of experiments was conducted on the five datasets, each employed exclusively for testing the model trained on another. This thorough approach aimed to evaluate how well the model performs across the used datasets and whether it can effectively generalize its learned patterns. Table 6 presents the comprehensive results of this experiment, revealing insightful trends and variations in performance metrics. The model exhibits robust segmentation capabilities when trained on one dataset and tested on others, excluding the Kvasir Capsule-SEG dataset. Particularly, training on the CVC Clinic dataset demonstrates commendable performance across CVC Colon, Kvasir-SEG, and Etis Larib datasets during testing, highlighting its versatility. Similarly, the model trained on CVC Colon showcases reciprocal effectiveness, producing noteworthy results on CVC Clinic, Kvasir-SEG, and Etis Larib datasets. Training on Kvasir-SEG and Etis Larib datasets also demonstrates acceptable generalization performance. In the broader context of the Kvasir-SEG as training dataset scenario, the segmentation model exhibits commendable generalization across multiple datasets, showcasing robust performance metrics. Notably, when applied to the CVC Clinic dataset, the model achieves impressive precision, recall, IoU, score, and dice coefficient values of 87.27%, 90.51%, 86.43%, 88.76%, and 89.77%, respectively. This trend continues across other datasets, as evidenced by substantial results on the CVC Colon dataset, where precision, recall, mIoU, score, and mDc reach 87.70%, 91.09%, 86.73%, 89.15%, and 90.23%, demonstrating the model’s consistent effectiveness. Furthermore, the Etis Larib dataset also showcases the model’s proficiency, recording values of 91.98%, 89.85%, 87.77%, 90.67%, and 90.13% for precision, recall, IoU, score, and mDc. These findings underscore the model’s versatility in adapting to diverse datasets and highlight its ability to maintain high segmentation performance across varying data characteristics. However, an exception arises with the Kvasir Capsule-SEG dataset during both training and testing, attributing lower results to its limited size (55 original images) and potential impact from augmentation strategies. Furthermore, the presence of large polyps in the original images of Kvasir Capsule-SEG, unlike the predominantly small polyps in other datasets, may contribute to this divergence in performance.
Table 6.
The generalization performance of the proposed UViT-Seg, where each dataset is used for training and the others for evaluation
| Evaluation Data/Metrics | P | R | mIoU | mDc | |
|---|---|---|---|---|---|
| CVC clinic: Train | |||||
| CVC Colon | 0.8692 | 0.9042 | 0.8542 | 0.8837 | 0.8951 |
| Kvasir-SEG | 0.7374 | 0.8015 | 0.7107 | 0.7653 | 0.7844 |
| Etis Larib | 0.8915 | 0.8877 | 0.8486 | 0.8888 | 0.8879 |
| Kvasir Capsule-SEG | 0.3624 | 0.5405 | 0.3566 | 0.4250 | 0.4817 |
| CVC colon: Train | |||||
| CVC Clinic | 0.8617 | 0.9106 | 0.8584 | 0.8843 | 0.8996 |
| Kvasir-SEG | 0.7271 | 0.8314 | 0.7255 | 0.7744 | 0.8050 |
| Etis Larib | 0.8967 | 0.9372 | 0.8931 | 0.9159 | 0.9284 |
| Kvasir Capsule-SEG | 0.4003 | 0.5502 | 0.3987 | 0.4507 | 0.4980 |
| Kvasir-SEG: Train | |||||
| CVC clinic | 0.8727 | 0.9051 | 0.8643 | 0.8876 | 0.8977 |
| CVC Colon | 0.8770 | 0.9109 | 0.8673 | 0.8915 | 0.9023 |
| Etis Larib | 0.9198 | 0.8985 | 0.8777 | 0.9067 | 0.9013 |
| Kvasir Capsule-SEG | 0.4395 | 0.5488 | 0.4368 | 0.4814 | 0.5164 |
| Etis Larib: Train | |||||
| CVC clinic | 0.8679 | 0.8365 | 0.7944 | 0.8475 | 0.8399 |
| CVC Colon | 0.8642 | 0.9044 | 0.8492 | 0.8810 | 0.8940 |
| Kvasir-SEG | 0.7444 | 0.8043 | 0.7180 | 0.7691 | 0.7880 |
| Kvasir Capsule-SEG | 0.3528 | 0.5461 | 0.3485 | 0.4205 | 0.4822 |
| Kvasir Capsule-SEG: Train | |||||
| CVC Clinic | 0.8776 | 0.7990 | 0.7681 | 0.8324 | 0.8112 |
| CVC Colon | 0.8900 | 0.7798 | 0.7572 | 0.8290 | 0.7983 |
| Kvasir-SEG | 0.7865 | 0.6895 | 0.6504 | 0.7261 | 0.7020 |
| Etis Larib | 0.9166 | 0.7445 | 0.7291 | 0.8137 | 0.7695 |
To evaluate the efficacy of our model, we performed a thorough comparative analysis, assessing its performance in conjunction with six cutting-edge methods designed for polyp segmentation, namely UNet [20], UNet++ [49], ResUNet [50], MultiResUNet [51], and AttentionUNet [52]. The configuration of the training for each of these five architectures closely mirrored the parameters used in training the proposed system across all five datasets. The results of this extensive evaluation are presented in Table 7, offering a comprehensive comparative overview between the proposed model and the aforementioned systems. Examination of Table 7 reveals a substantial variation among comparable models. Both the proposed system and the Multi-ResNet model exhibit effectiveness across the majority of metrics used for evaluation. In most cases, the proposed approach outshines other models, except in instances where Multi-Resnet and AttentionUNet demonstrate superior performance. Turning attention to the CVC clinic, the proposed model attains the highest recall, score, and mDc with impressive results of 90.81%, 90.59%, and 90.72%, respectively. Meanwhile, Multi-ResNet surpasses all these systems in terms of precision and mIoU, boasting values of 90.48% and 90.19%, respectively. The identical scenario unfolds in the context of the Kvasir-SEG dataset, where the proposed approach distinguishes itself among all models. Multi-ResNet emerges as the second-best performer across all metrics, except for recall, where Unet attains the highest value of 82.75%. This trend persists in the Kvasir-Capsule-SEG dataset, with the Unet model surpassing all others in terms of recall by achieving a superior value of 54.39%. The sole exception we observed in this experiment pertains to the CVC colon dataset, where a discernible variation in performance among the evaluated models was noted. Both Unet++ and ResUnet exhibited an equally high recall of 92.10%, while the AttentionUnet secured the position of the second-best model with a recall of 91.97%. Notably, the proposed approach surpassed other models in terms of precision, intersection over union, , and mDc, achieving peak values of 90.72%, 90.42%, 90.21%, and 91.54%, respectively. The AttentionUNet model exhibits the second-best performance across three key metrics that are precision, recall, and mIoU, attaining impressive scores of 92.95%, 93.96%, and 90.81%, respectively, on the Etis Larib dataset. In this scenario, both the proposed model and the Multi-ResNet model emerge as effective, each showcasing its superiority in distinct metrics. Nevertheless, the results obtained from both the evaluated systems and the proposed approach exhibit varying patterns, with, in most instances, noteworthy levels of performance achieved. The case of Kvasir Capsule-SEG is somewhat distinctive, owing to several challenges associated with the images, including a small dataset size, and all images portraying polyps in their larger dimensions. Moreover, the applied data augmentation strategies may exert a substantial influence on the models’ performance. In addition, we conducted a thorough examination of these models’ generalization performance by employing a 5-fold validation split on subsets of each dataset. The primary objective of this analysis is to assess how the models respond to variations in polyp features across different datasets. The findings, detailed in Table 8, reveal notable disparities in the outcomes. Notably, the proposed approach demonstrates superior performance on the Etis Larib dataset, achieving an mDc of 91% and an mIoU of 89%. Conversely, across other datasets, our model exhibits significant variability in segmentation accuracy. This variability may stem from the utilization of 5-fold validation as the primary training mode. Notably, results obtained via holdout training mode surpass those from k-fold validation, suggesting that the latter’s efficacy might be attributed to the limited image samples in each dataset, thereby limiting the advantages of cross-validation.
Table 7.
Comparative analysis of UViT-Seg with common medical imaging segmentation methods. Values in bold font indicate the best results, while blue-colored values represent the second-best results
| Models/Metrics | P | R | mIoU | mDc | |
|---|---|---|---|---|---|
| CVC Clinic | |||||
| Unet [20] | 0.8080 | 0.8120 | 0.7961 | 0.8048 | 0.7991 |
| Unet ++ [49] | 0.7882 | 0.8011 | 0.7456 | 0.7773 | 0.7869 |
| ResUnet [50] | 0.8408 | 0.8640 | 0.8408 | 0.8746 | 0.8574 |
| MultiResUnet [51] | 0.9048 | 0.8711 | 0.9019 | 0.8879 | 0.8798 |
| AttentionUnet [52] | 0.8214 | 0.8349 | 0.8125 | 0.7630 | 0.7841 |
| The proposed Model | 0.9038 | 0.9081 | 0.8981 | 0.9059 | 0.9072 |
| CVC Colon | |||||
| Unet | 0.8976 | 0.9181 | 0.8952 | 0.9071 | 0.9135 |
| Unet ++ | 0.8594 | 0.9210 | 0.8594 | 0.8868 | 0.9062 |
| ResUnet | 0.8594 | 0.9210 | 0.8594 | 0.8868 | 0.9062 |
| MultiResUnet | 0.9014 | 0.9192 | 0.8999 | 0.9098 | 0.9153 |
| AttentionUnet | 0.8917 | 0.9197 | 0.8907 | 0.9043 | 0.9131 |
| The proposed Model | 0.9072 | 0.9178 | 0.9042 | 0.9121 | 0.9154 |
| Kvasir-SEG | |||||
| Unet | 0.7867 | 0.8275 | 0.7810 | 0.8048 | 0.8173 |
| Unet ++ | 0.7163 | 0.8335 | 0.7162 | 0.7678 | 0.8038 |
| ResUnet | 0.7721 | 0.8201 | 0.7598 | 0.7934 | 0.8083 |
| MultiResUnet | 0.8042 | 0.8264 | 0.7971 | 0.8144 | 0.8210 |
| AttentionUnet | 0.7990 | 0.8210 | 0.7868 | 0.8088 | 0.8155 |
| The proposed Model | 0.8144 | 0.8211 | 0.8012 | 0.8230 | 0.8352 |
| Etis Larib | |||||
| Unet | 0.9268 | 0.9391 | 0.9048 | 0.9028 | 0.9065 |
| Unet ++ | 0.8893 | 0.9411 | 0.8893 | 0.8936 | 0.8997 |
| ResUnet | 0.9173 | 0.9370 | 0.8934 | 0.9067 | 0.9028 |
| MultiResUnet | 0.9310 | 0.9401 | 0.9000 | 0.9102 | 0.9118 |
| AttentionUnet | 0.9295 | 0.9396 | 0.9081 | 0.9045 | 0.9075 |
| The proposed Model | 0.9196 | 0.9145 | 0.9131 | 0.9118 | 0.9134 |
| Kvasir Capsule-SEG | |||||
| Unet | 0.4805 | 0.5439 | 0.4748 | 0.5088 | 0.5284 |
| Unet ++ | 0.4657 | 0.4266 | 0.3678 | 0.4364 | 0.4283 |
| ResUnet | 0.4602 | 0.5310 | 0.4465 | 0.4918 | 0.5142 |
| MultiResUnet | 0.5010 | 0.5356 | 0.4866 | 0.5159 | 0.5268 |
| AttentionUnet | 0.4637 | 0.5243 | 0.4438 | 0.4898 | 0.5090 |
| The proposed Model | 0.5009 | 0.5418 | 0.4912 | 0.5122 | 0.5372 |
Table 8.
Segmentation performance of the five common medical imaging methods and UViT-Seg with 5-fold validation
| Datasets | CVC | CVC | Kvasir | Etis | Kvasir | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clinic | Colon | SEG | Larib | Capsule-SEG | ||||||
| Models | mDc | mIoU | mDc | mIoU | Dc | mIoU | mDc | mIoU | mDc | mIoU |
| [20] | 0.890 | 0.869 | 0.870 | 0.851 | 0.801 | 0.774 | 0.903 | 0.890 | 0.450 | 0.388 |
| [49] | 0.874 | 0.840 | 0.863 | 0.820 | 0.765 | 0.713 | 0.885 | 0.850 | 0.425 | 0.357 |
| [50] | 0.877 | 0.846 | 0.832 | 0.800 | 0.756 | 0.701 | 0.900 | 0.881 | 0.436 | 0.371 |
| [51] | 0.896 | 0.901 | 0.893 | 0.872 | 0.805 | 0.782 | 0.891 | 0.864 | 0.474 | 0.422 |
| [52] | 0.889 | 0.867 | 0.876 | 0.844 | 0.797 | 0.768 | 0.908 | 0.881 | 0.430 | 0.365 |
| UViT-Seg | 0.886 | 0.860 | 0.876 | 0.859 | 0.727 | 0.655 | 0.914 | 0.890 | 0.411 | 0.336 |
Moreover, to evaluate if the proposed UViT-Seg performs well on the five datasets for polyp localization, we conducted another statistical experiment using the Wilcoxon rank-sum test (WRST). The WRST is particularly suitable for this analysis due to its ability to handle non-parametric data and its robustness against outliers, making it well-suited for comparing performance metrics across the datasets considered in this study. The comparison included the proposed model and the models mentioned in Table. As previously mentioned, both the proposed UViT-Seg and other models were trained using identical settings, with mDc and mIou recorded for each run. The results of the WRST are listed in Table 9. In this experiment, we notate the method pairs as follows: A (Unet, UViT-Seg), B (AttentionUnet, UViT-Seg), C (Mult-ResUnet, UViT-Seg), D (ResUnet, UViT-Seg), and E (Unet++, UViT-Seg). It can be seen that the p-value (P-v) is less than 0.05 for all the pairs on the CVC-Colon and Kvasir-SEG datasets. This suggests that there is statistical significance in the results obtained, leading to the rejection of the null hypothesis. In the case of CVC clinic and ETIS Larib, the P-v is greater than 0.05 for the pairs A, B, and C. This implies that the differences in performance between the methods within these pairs are not statistically significant. In the case of the Kvasir Capsule-SEG dataset, the P-v is greater than 0.05 for all the pairs except E. This suggests that it was not possible to determine the statistical significance of the results obtained from the proposed UViT-Seg.
Table 9.
Statistical analysis of the proposed UViT-Seg against common state-of-the-art methods using Wilcoxon rank-sum tests. The values highlighted in bold font show the p-value 0.05
| Dataset | CVC | CVC | Kvasir | Etis | Kvasir | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clinic | Colon | SEG | Larib | Capsule-SEG | ||||||
| P-v | P-v | P-v | P-v | P-v | P-v | P-v | P-v | P-v | P-v | |
| (Dc) | (IoU) | (Dc) | (IoU) | (Dc) | (IoU) | (Dc) | (IoU) | (Dc) | (IoU) | |
| A | 0.009 | 0.073 | 0.007 | 0.007 | 0.001 | 0.004 | 0.007 | 0.081 | 0.070 | 0.078 |
| B | 0.051 | 0.032 | 0.009 | 0.009 | 0.002 | 0.006 | 0.006 | 0.004 | 0.084 | 0.076 |
| C | 0.061 | 0.048 | 0.009 | 0.009 | 0.004 | 0.004 | 0.042 | 0.063 | 0.075 | 0.068 |
| D | 0.018 | 0.005 | 0.004 | 0.043 | 0.017 | 0.033 | 0.009 | 0.008 | 0.073 | 0.076 |
| E | 0.003 | 0.041 | 0.005 | 0.011 | 0.032 | 0.037 | 0.046 | 0.030 | 0.022 | 0.001 |
In addition, we compare the performance of the proposed model with five state-of-the-art models for polyp segmentation: SFA [53], PraNet [54], HarDNet-MSEG [55], Focus U-Net [56], and SR-AttNet [44]. Table 10 presents the evaluation results on four datasets. The proposed model excels across three datasets, achieving an mDc score of 0.915 and an mIoU of 0.902 on the CVC Colon dataset, as well as an mDc of 0.913 and an mIoU of 0.913 on the CVC Clinic dataset. In the case of the CVC Clinic dataset, the proposed model attains a superior mIoU of 0.898, while the Focus U-Net outperforms all evaluated methods in terms of dice score, reaching the highest result of 0.938. In the Kvasir-SEG case, the work named HarDNet-MSEG demonstrated the best performance with a dice score of 0.912 and an mIoU of 0.857, surpassing other approaches and showcasing an improvement of 9.22% in mDc and 6.99% in mIoU. The proposed model demonstrates a notable improvement of 4.21% and 12.19% in mDc and mIoU, respectively. Additionally, it exhibits gains of 9.73% and 20.63% in mDc and mIoU on the CVC Colon and Etis Larib datasets, outperforming the Focus U-net and securing the second-best segmentation performance on both datasets. However, the proposed model for polyp segmentation consistently demonstrates effective results compared to other evaluated approaches in the state of the art and it demonstrates its ability to identify accurately polyps region in GI images.
Table 10.
Results of the comparative study of UViT-Seg with recent polyp segmentation approaches
| CVC ColonDB | CVC Clinic | Kvasir-SEG | Etis LaribDB | |||||
|---|---|---|---|---|---|---|---|---|
| Models | mDc | mIoU | mDc | mIoU | mDc | mIoU | mDc | mIoU |
| SFA [53] | 0.469 | 0.347 | 0.700 | 0.607 | 0.723 | 0.611 | 0.297 | 0.217 |
| PraNet [54] | 0.709 | 0.640 | 0.899 | 0.849 | 0.898 | 0.840 | 0.628 | 0.567 |
| HarDNet-MSEG [55] | 0.731 | 0.660 | 0.932 | 0.882 | 0.912 | 0.857 | 0.677 | 0.613 |
| Focus U-Net [56] | 0.878 | 0.804 | 0.938 | 0.889 | 0.910 | 0.853 | 0.832 | 0.757 |
| SR-AttNet [44] | 0.665 | 0.539 | 0.786 | 0.693 | 0.871 | 0.806 | 0.476 | 0.355 |
| UViT-Seg | 0.915 | 0.902 | 0.907 | 0.898 | 0.835 | 0.801 | 0.913 | 0.9131 |
The complexity of the model can impact its deployment in real-world scenarios, as clinical services require not only high accuracy but also efficiency and interpretability. In the context of medical image segmentation, such as the tasks considered in our study, deploying a highly complex model may introduce challenges in practical implementation within clinical settings. Table 11 shows the complexity analysis between the proposed approach and other polyp segmentation methods. The proposed UViT-Seg stands out compared to other methods, as it demands fewer computational resources and significantly reduces training time.
Table 11.
Trainable parameters of the proposed approach and other approaches
| Models | Trainable Parameters (M) |
|---|---|
| Unet | 31.04 |
| Unet++ | 9.04 |
| ResUnet | 8.22 |
| Multi-ResUnet | 7.24 |
| Attention Unet | 8.13 |
| ParaNet | 32.55 |
| HarDNet-MSEG | 33.34 |
| SR-AttNet | 31.1 |
| UViT-Seg | 2.24 |
Qualitative Results
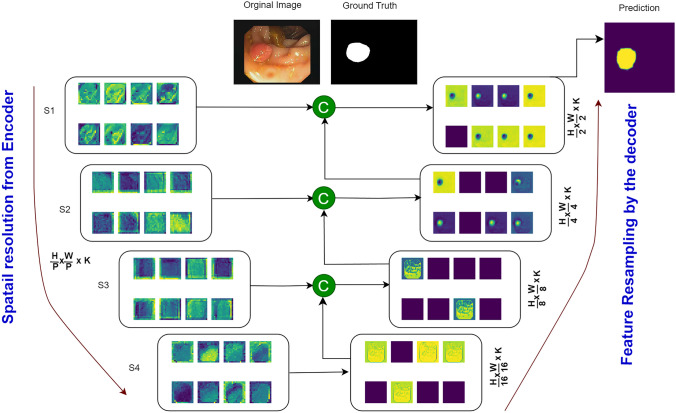
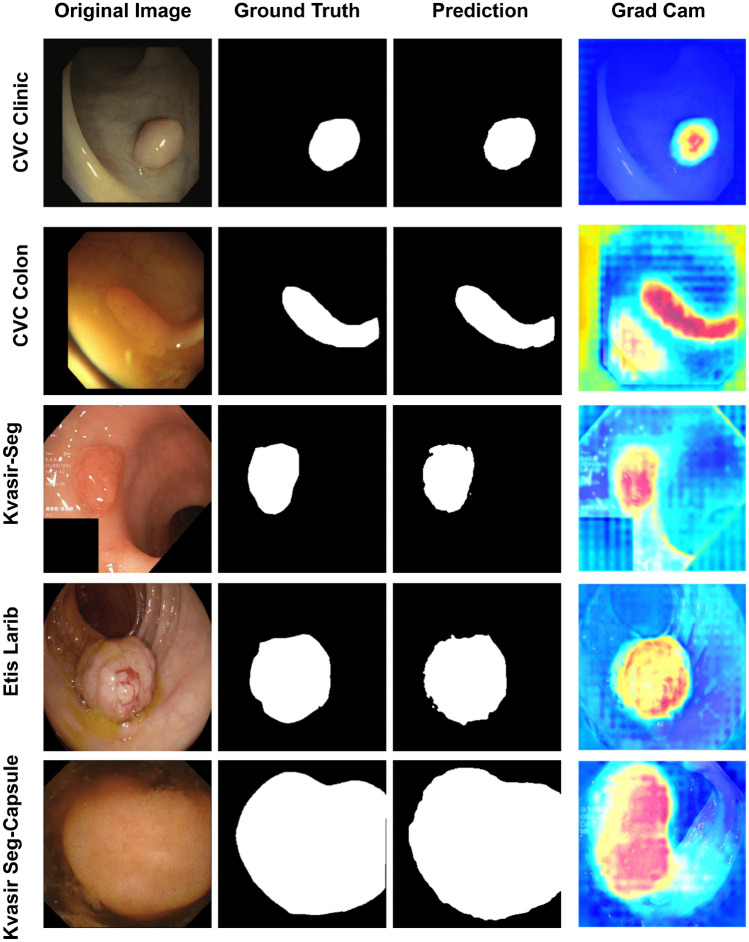
In this section, we delve into the visualization performance of the proposed UViT-Seg for polyp localization. Our exploration begins with visualizing the features at each stage of both the encoder and decoder. Figure 8 provides a visual representation of the features obtained at each skip connection , where i . The features within each set of maintain a consistent resolution of and are fed into the decoder to facilitate feature reconstruction at various stages. The output of each undergoes resampling via a deconvolution layer and subsequently passes through a set of convolution layers, enhancing feature capture. The generated features are then fused with the next output of . Examining Fig. 8, we observe that the feature concatenation process enables the model to systematically capture essential features at each stage. This sequential capture of features proves crucial in obtaining interesting insights at different levels, aiding the model in effectively localizing the polyp region well before generating the final prediction from the last layer. After a detailed examination of features at each stage and a thorough understanding of the model’s interpretation, especially its emphasis on intricate details, we present a visual representation of the model’s performance on five distinct datasets across different train-test splits in Fig. 9. This figure showcases the original image, ground truth (GT), predicted mask, and Grad-CAM visualization. The datasets considered in our study exhibit significant variations in polyp features, including differences in texture, shape, and size (ranging from small to large polyps). The model exhibits commendable proficiency in identifying the majority of polyps within each test dataset, irrespective of their size. This robust performance suggests the model’s adaptability to diverse polyp characteristics, making it a versatile tool for consistent polyp detection across a spectrum of conditions. Moreover, the Grad-CAM analysis provides additional insights into the model’s decision-making process, highlighting its capability to focus on regions containing polyps. This visualization not only validates the model’s predictions but also aids in understanding the features that contribute most to its accuracy. The successful identification of key regions through Grad-CAM emphasizes the model’s potential for precise localization of polyps within GI images.
Fig. 8.
Visualizing predicted features across various resolutions using the proposed UViT-Seg, The tested image form CVC clinic dataset
Fig. 9.
The visual representation of the predicted masks generated by UViT-Seg alongside highlighted regions identified with Grad CAM
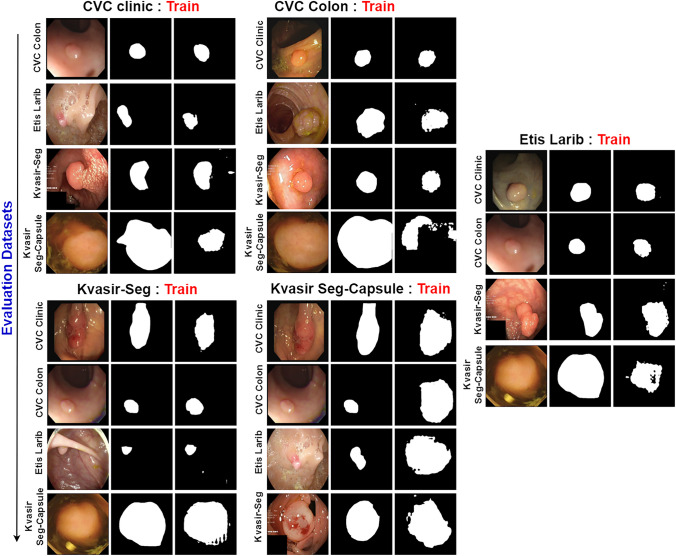
The primary challenge faced by every intelligent system lies in its performance on new data with a distinct feature distribution. In addressing this challenge, we present an additional visual representation of the proposed UViT-Seg to validate its generalization ability. The experiment entails training the model on one dataset and subsequently evaluating its performance on other datasets. Our objective is to assess the model’s adaptability to diverse conditions of polyps, encompassing variations in size, shape, and a range of other features. Specifically, the datasets used in this work each display their own unique features. For example, the Kvasir Capsule-SEG dataset includes images captured using WCE technology under specific conditions, where most of the polyps are large in size. In contrast, other datasets were captured with digestive tubes, resulting in entirely different features, particularly in their representation of small-sized polyps. However, Fig. 10 presents the visualization results of this experiment and the generalization capabilities of the model on datasets that display small and median polyps (CVC Clinic, CVC colon, Etis Larib and Kvasir-SEG). Even with their significant variation in features, the model demonstrates a remarkable ability to identify and localize polyp regions in the majority of cases. The visual analysis reveals that the model, trained on one dataset, successfully extends its performance to datasets with diverse small and median polyps, indicating a promising level of generalization. The case of the Kvasir Capsule-SEG dataset is unique, as mentioned earlier. It comprises only 55 original polyps and their corresponding masks, and all these polyps are large in size. These specific conditions might influence the model’s generalization performance. As illustrated in Fig. 10, when this dataset is used for testing, the predicted polyps in most cases appear small or medium-sized, except for the case of training the model with Kvasir-SEG, where the predicted polyps maintain their larger shape. Similarly, when employed for training, the predicted polyps are consistently displayed in their larger size. This discrepancy raises considerations about how the dataset’s characteristics may affect the model’s ability to generalize across different polyp sizes. Despite the substantial challenges posed by Kvasir Capsule-SEG, UViT-Seg demonstrates the ability to predict the polyp region in certain cases. In others, it adeptly identifies polyp regions with variations in size. Conversely, when dealing with other datasets, the model proves its ability to generalize, showcasing its adaptability, a crucial attribute for real-world scenarios where polyps can exhibit considerable diversity in terms of morphology and size. In addition, we have conducted a comparative analysis of the proposed model against several common medical image segmentation methods, including Unet, Unet++, ResUnet, AttentionUnet, and Mult-ResUnet. The aim is to assess the performance of the proposed model in comparison to other established medical segmentation approaches. Figure 11 provides a visual representation of the compared methods, revealing that the proposed model consistently achieves outstanding performance in accurately locating polyps across all evaluated datasets. AttentionUnet also exhibits the ability to locate polyp regions with slight variations in shape. However, Unet and Multi-ResUnet show some confusion, particularly in the case of CVC Clinic and CVC Colon datasets. Notably, Unet++ and ResUnet models encounter challenges and exhibit significant difficulties in effectively segmenting polyps, with the exception of cases such as Etis Larib and Kvasir Capsule-SEG, where the ResUnet model successfully identifies the polyps region.
Fig. 10.
Visualizing the generalization performance of UViT-Seg across all datasets, where each dataset is used for training, while others are employed for evaluation
Fig. 11.
Visualization of the performance of UViT-Seg and other common medical image segmentation methods
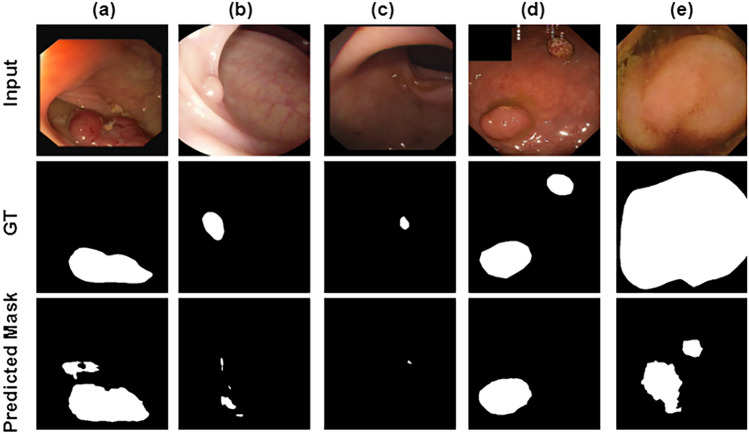
However, while the proposed UViT-Seg model demonstrates its effectiveness in polyp segmentation within GI images, it also exhibits certain limitations. In Fig. 12, we present a visual representation of the failure cases associated with the proposed UViT-Seg. In our experiments, we identified three main reasons for failed predictions. Firstly, in numerous cases, the normal region displays very similar features compared to the area containing a polyp, posing significant challenges for the model in distinguishing between them. Secondly, the proposed method may struggle to accurately locate polyps situated in deep regions with unclear boundaries. Lastly, in some instances, the normal region in certain images shares features with polyp regions in others, potentially impacting the predictive performance of UViT-Seg. To address these limitations, ongoing efforts are essential. The incorporation of additional data enhancement techniques holds promise for improving the model’s ability to distinguish between features. Moreover, preparing a more extensive dataset, specifically focusing on diverse polyps, particularly the larger ones, may alleviate the challenges associated with failed predictions in the Kvasir Capsule-SEG and enhance the generalization process.
Fig. 12.
The visual representation of the failure cases associated with the proposed UViT-Seg. Examples images from each dataset
Conclusion
Polyp segmentation presents a significant challenge within the medical field, primarily due to various complexities associated with polyp features such as color, texture, shape, and other factors related to GI screening technologies. Recent advancements in deep learning models have showcased remarkable performance owing to their innate ability to extract features effectively. The majority of these models adopt an encoder-decoder architecture employing convolutional layers, which may not be sufficiently robust in learning the diverse global representations of polyp regions in GI images. In this study, we introduce UViT-Seg, a framework designed for automated polyp segmentation in both colonoscopy and WCE images. Unlike the majority of CNN-based methods for medical image segmentation, especially in polyps, the distinctive feature of UViT-Seg lies in its ability to extract feature representations from images. This is achieved by deploying a vision transformer in the encoder, capturing high-level features to gain a deeper understanding of the polyp region. Simultaneously, the decoder module captures low-level features using a set of deconvolution and convolution blocks based on the squeeze and excitation mechanism. Additionally, the dual attention mechanism, integrated with the residual block, enhances the model’s capability to retain essential features from input GI images. Experimental results are derived from five distinct datasets used for training and testing, each with a unique feature distribution and diverse polyp conditions. The results confirm UViT-Seg capability to precisely locate polyps within images. Furthermore, UViT-Seg utilizes one dataset for training and the remaining four for evaluation, validating its ability to generalize and adapt to new variations in features within unseen data. Performance comparisons against five common medical segmentation methods demonstrate UViT-Seg effectiveness. Moreover, when compared to several recent approaches for polyp segmentation, the proposed UViT-Seg exhibits optimal results, standing out for its complexity efficiency. It requires fewer computational resources for both training and testing, making it more suitable for real-world deployment scenarios.
Nevertheless, while the UViT-Seg model yields effective results, it still encounters certain limitations. One challenge lies in its ability to distinguish between normal regions and those containing polyps, as the feature distributions in these regions are very similar. Additionally, it struggles to accurately locate polyps in deep regions, particularly when the boundaries are unclear. Furthermore, due to the utilization of five distinct datasets, each with its unique characteristics, the model faces difficulties in generalizing its findings, especially when dealing with the localization of larger polyps. By addressing these challenges and implementing enhancements to the model in future work, our aim is to achieve more accurate localization and identification of polyp regions in WCE and colonoscopy images. This approach is particularly crucial for polyps with unclear boundaries and larger sizes. Furthermore, these improvements will advance the development of a comprehensive system for polyp detection, making it more capable of adapting to the diverse feature distributions present in polyp regions in GI images.
Author Contributions
Y.O., A.G., Z.K., M.E., L.K., and A.F.E. wrote the main manuscript text. All authors reviewed the manuscript.
Funding
This work was supported by the Ministry of National Education by Vocational Training; in part by the Higher Education and Scientific Research through the Ministry of Industry, Trade, and Green and Digital Economy; in part by the Digital Development Agency (ADD); and in part by the National Center for Scientific and Technical Research (CNRST) under Project ALKHAWARIZMI/2020/20.
Data Availability
All the datasets used in this study are indicated and cited correctly in the paper.
Declarations
Ethics Approval
Institutional Review Board approval was obtained.
Consent to Participate
All study participants provided written informed consent.
Consent for Publication
The manuscript contains no identifiable individual data or images that would require consent to publish from any participant.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel, R.L., Miller, K.D., Wagle, N.S., Jemal, A.: Cancer statistics, 2023. CA: a cancer journal for clinicians 73(1), 17–48 (2023) [DOI] [PubMed]
- 2.Siegel, R.L., Wagle, N.S., Cercek, A., Smith, R.A., Jemal, A.: Colorectal cancer statistics, 2023. CA: a cancer journal for clinicians 73(3), 233–254 (2023) [DOI] [PubMed]
- 3.Hazewinkel, Y., Dekker, E.: Colonoscopy: basic principles and novel techniques. Nature reviews Gastroenterology & hepatology 8(10), 554–564 (2011) [DOI] [PubMed] [Google Scholar]
- 4.Holzheimer, R.G., Mannick, J.A.: Surgical treatment: evidence-based and problem-oriented (2001) [PubMed]
- 5.Tranquillini, C.V., Bernardo, W.M., Brunaldi, V.O., MOURA, E.T.d., Marques, S.B., MOURA, E.G.H.d.: Best polypectomy technique for small and diminutive colorectal polyps: A systematic review and meta-analysis. Arquivos de gastroenterologia 55, 358–368 (2018) [DOI] [PubMed]
- 6.Costamagna, G., Shah, S.K., Riccioni, M.E., Foschia, F., Mutignani, M., Perri, V., Vecchioli, A., Brizi, M.G., Picciocchi, A., Marano, P.: A prospective trial comparing small bowel radiographs and video capsule endoscopy for suspected small bowel disease. Gastroenterology 123(4), 999–1005 (2002) [DOI] [PubMed] [Google Scholar]
- 7.Iddan, G., Meron, G., Glukhovsky, A., Swain, P.: Wireless capsule endoscopy. Nature 405(6785), 417–417 (2000) [DOI] [PubMed] [Google Scholar]
- 8.Omori, T., Hara, T., Sakasai, S., Kambayashi, H., Murasugi, S., Ito, A., Nakamura, S., Tokushige, K.: Does the pillcam sb3 capsule endoscopy system improve image reading efficiency irrespective of experience? a pilot study. Endoscopy international open 6(06), 669–675 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jha, D., Ali, S., Tomar, N.K., Johansen, H.D., Johansen, D., Rittscher, J., Riegler, M.A., Halvorsen, P.: Real-time polyp detection, localization and segmentation in colonoscopy using deep learning. Ieee Access 9, 40496–40510 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urban, G., Tripathi, P., Alkayali, T., Mittal, M., Jalali, F., Karnes, W., Baldi, P.: Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 155(4), 1069–1078 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross, S., Stehle, T., Behrens, A., Auer, R., Aach, T., Winograd, R., Trautwein, C., Tischendorf, J.: A comparison of blood vessel features and local binary patterns for colorectal polyp classification. In: Medical Imaging 2009: Computer-Aided Diagnosis, vol. 7260, pp. 758–765 (2009). SPIE
- 12.Iwahori, Y., Hattori, A., Adachi, Y., Bhuyan, M.K., Woodham, R.J., Kasugai, K.: Automatic detection of polyp using hessian filter and hog features. Procedia computer science 60, 730–739 (2015) [Google Scholar]
- 13.Amber, A., Iwahori, Y., Bhuyan, M.K., Woodham, R.J., Kasugai, K.: Feature point based polyp tracking in endoscopic videos. In: 2015 3rd International Conference on Applied Computing and Information Technology/2nd International Conference on Computational Science and Intelligence, pp. 299–304 (2015). IEEE
- 14.Sasmal, P., Bhuyan, M.K., Iwahori, Y., Kasugai, K.: Colonoscopic polyp classification using local shape and texture features. IEEE Access 9, 92629–92639 (2021) [Google Scholar]
- 15.Pogorelov, K., Ostroukhova, O., Jeppsson, M., Espeland, H., Griwodz, C., de Lange, T., Johansen, D., Riegler, M., Halvorsen, P.: Deep learning and hand-crafted feature based approaches for polyp detection in medical videos. In: 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS), pp. 381–386 (2018). IEEE
- 16.Hmoud Al-Adhaileh, M., Mohammed Senan, E., Alsaade, W., Aldhyani, T.H.H., Alsharif, N., Abdullah Alqarni, A., Uddin, M.I., Alzahrani, M.Y., Alzain, E.D., Jadhav, M.E.: Deep learning algorithms for detection and classification of gastrointestinal diseases. Complexity 2021, 1–12 (2021)
- 17.Goel, N., Kaur, S., Gunjan, D., Mahapatra, S.: Dilated cnn for abnormality detection in wireless capsule endoscopy images. Soft Computing, 1–17 (2022)
- 18.Jain, S., Seal, A., Ojha, A.: A convolutional neural network with meta-feature learning for wireless capsule endoscopy image classification. Journal of Medical and Biological Engineering 43(4), 475–494 (2023) [Google Scholar]
- 19.Jia, X., Meng, M.Q.-H.: A deep convolutional neural network for bleeding detection in wireless capsule endoscopy images. In: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 639–642 (2016). IEEE [DOI] [PubMed]
- 20.Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18, pp. 234–241 (2015). Springer
- 21.Badrinarayanan, V., Kendall, A., Cipolla, R.: Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE transactions on pattern analysis and machine intelligence 39(12), 2481–2495 (2017) [DOI] [PubMed] [Google Scholar]
- 22.Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., et al.: An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
- 23.Jia, X., Meng, M.Q.-H.: Gastrointestinal bleeding detection in wireless capsule endoscopy images using handcrafted and cnn features. In: 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 3154–3157 (2017). IEEE [DOI] [PubMed]
- 24.Yuan, Y., Li, B., Meng, M.Q.-H.: Bleeding frame and region detection in the wireless capsule endoscopy video. IEEE journal of biomedical and health informatics 20(2), 624–630 (2015) [DOI] [PubMed] [Google Scholar]
- 25.Yuan, Y., Wang, J., Li, B., Meng, M.Q.-H.: Saliency based ulcer detection for wireless capsule endoscopy diagnosis. IEEE transactions on medical imaging 34(10), 2046–2057 (2015) [DOI] [PubMed] [Google Scholar]
- 26.Jain, S., Seal, A., Ojha, A., Krejcar, O., Bureš, J., Tachecí, I., Yazidi, A.: Detection of abnormality in wireless capsule endoscopy images using fractal features. Computers in biology and medicine 127, 104094 (2020) [DOI] [PubMed] [Google Scholar]
- 27.Sánchez-González, A., García-Zapirain, B., Sierra-Sosa, D., Elmaghraby, A.: Automatized colon polyp segmentation via contour region analysis. Computers in biology and medicine 100, 152–164 (2018) [DOI] [PubMed] [Google Scholar]
- 28.Jia, X., Xing, X., Yuan, Y., Xing, L., Meng, M.Q.-H.: Wireless capsule endoscopy: A new tool for cancer screening in the colon with deep-learning-based polyp recognition. Proceedings of the IEEE 108(1), 178–197 (2019) [Google Scholar]
- 29.Shin, Y., Balasingham, I.: Comparison of hand-craft feature based svm and cnn based deep learning framework for automatic polyp classification. In: 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 3277–3280 (2017). IEEE [DOI] [PubMed]
- 30.Guo, Y., Bernal, J., J. Matuszewski, B.: Polyp segmentation with fully convolutional deep neural networks–extended evaluation study. Journal of Imaging 6(7), 69 (2020) [DOI] [PMC free article] [PubMed]
- 31.Tajbakhsh, N., Gurudu, S.R., Liang, J.: Automated polyp detection in colonoscopy videos using shape and context information. IEEE transactions on medical imaging 35(2), 630–644 (2015) [DOI] [PubMed] [Google Scholar]
- 32.Bernal, J., Sánchez, F.J., Fernández-Esparrach, G., Gil, D., Rodríguez, C., Vilariño, F.: Wm-dova maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Computerized medical imaging and graphics 43, 99–111 (2015) [DOI] [PubMed]
- 33.Mahmud, T., Paul, B., Fattah, S.A.: Polypsegnet: A modified encoder-decoder architecture for automated polyp segmentation from colonoscopy images. Computers in Biology and Medicine 128, 104119 (2021) [DOI] [PubMed] [Google Scholar]
- 34.Jha, D., Smedsrud, P.H., Riegler, M.A., Johansen, D., De Lange, T., Halvorsen, P., Johansen, H.D.: Resunet++: An advanced architecture for medical image segmentation. In: 2019 IEEE International Symposium on Multimedia (ISM), pp. 225–2255 (2019). IEEE
- 35.Ta, N., Chen, H., Lyu, Y., Wu, T.: Ble-net: boundary learning and enhancement network for polyp segmentation. Multimedia Systems 29(5), 3041–3054 (2023) [Google Scholar]
- 36.Qadir, H.A., Shin, Y., Solhusvik, J., Bergsland, J., Aabakken, L., Balasingham, I.: Polyp detection and segmentation using mask r-cnn: Does a deeper feature extractor cnn always perform better? In: 2019 13th International Symposium on Medical Information and Communication Technology (ISMICT), pp. 1–6 (2019). IEEE
- 37.Jain, S., Seal, A., Ojha, A., Yazidi, A., Bures, J., Tacheci, I., Krejcar, O.: A deep cnn model for anomaly detection and localization in wireless capsule endoscopy images. Computers in Biology and Medicine 137, 104789 (2021) [DOI] [PubMed] [Google Scholar]
- 38.Lafraxo, S., Souaidi, M., El Ansari, M., Koutti, L.: Semantic segmentation of digestive abnormalities from wce images by using attresu-net architecture. Life 13(3), 719 (2023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oukdach, Y., Kerkaou, Z., El Ansari, M., Koutti, L., Fouad El Ouafdi, A., De Lange, T.: Vitca-net: a framework for disease detection in video capsule endoscopy images using a vision transformer and convolutional neural network with a specific attention mechanism. Multimedia Tools and Applications, 1–20 (2024)
- 40.Bai, L., Wang, L., Chen, T., Zhao, Y., Ren, H.: Transformer-based disease identification for small-scale imbalanced capsule endoscopy dataset. Electronics 11(17), 2747 (2022) [Google Scholar]
- 41.Oukdach, Y., Kerkaou, Z., Ansari, M.E., Koutti, L., Ouafdi, A.F.E.: Conv-vit: Feature fusion-based detection of gastrointestinal abnormalities using cnn and vit in wce images. In: 2023 10th International Conference on Wireless Networks and Mobile Communications (WINCOM), pp. 1–6 (2023). 10.1109/WINCOM59760.2023.10322944
- 42.Hosain, A.S., Islam, M., Mehedi, M.H.K., Kabir, I.E., Khan, Z.T.: Gastrointestinal disorder detection with a transformer based approach. In: 2022 IEEE 13th Annual Information Technology, Electronics and Mobile Communication Conference (IEMCON), pp. 0280–0285 (2022). IEEE
- 43.Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., Lu, L., Yuille, A.L., Zhou, Y.: Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021)
- 44.Alam, M.J., Fattah, S.A.: Sr-attnet: An interpretable stretch–relax attention based deep neural network for polyp segmentation in colonoscopy images. Computers in Biology and Medicine 160, 106945 (2023) [DOI] [PubMed] [Google Scholar]
- 45.Jha, D., Smedsrud, P.H., Riegler, M.A., Halvorsen, P., de Lange, T., Johansen, D., Johansen, H.D.: Kvasir-seg: A segmented polyp dataset. In: MultiMedia Modeling: 26th International Conference, MMM 2020, Daejeon, South Korea, January 5–8, 2020, Proceedings, Part II 26, pp. 451–462 (2020). Springer
- 46.Silva, J., Histace, A., Romain, O., Dray, X., Granado, B.: Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. International journal of computer assisted radiology and surgery 9, 283–293 (2014) [DOI] [PubMed] [Google Scholar]
- 47.Jha, D., Tomar, N.K., Ali, S., Riegler, M.A., Johansen, H.D., Johansen, D., de Lange, T., Halvorsen, P.: Nanonet: Real-time polyp segmentation in video capsule endoscopy and colonoscopy. In: Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), pp. 37–43 (2021)
- 48.LeCun, Y., Bottou, L., Bengio, Y., Haffner, P.: Gradient-based learning applied to document recognition. Proceedings of the IEEE 86(11), 2278–2324 (1998) [Google Scholar]
- 49.Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: Unet++: A nested u-net architecture for medical image segmentation. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4, pp. 3–11 (2018). Springer [DOI] [PMC free article] [PubMed]
- 50.Xiao, X., Lian, S., Luo, Z., Li, S.: Weighted res-unet for high-quality retina vessel segmentation. In: 2018 9th International Conference on Information Technology in Medicine and Education (ITME), pp. 327–331 (2018). IEEE
- 51.Ibtehaz, N., Rahman, M.S.: Multiresunet: Rethinking the u-net architecture for multimodal biomedical image segmentation. Neural networks 121, 74–87 (2020) [DOI] [PubMed] [Google Scholar]
- 52.Oktay, O., Schlemper, J., Folgoc, L.L., Lee, M., Heinrich, M., Misawa, K., Mori, K., McDonagh, S., Hammerla, N.Y., Kainz, B., et al.: Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 (2018)
- 53.Fang, Y., Chen, C., Yuan, Y., Tong, K.-y.: Selective feature aggregation network with area-boundary constraints for polyp segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part I 22, pp. 302–310 (2019). Springer
- 54.Fan, D.-P., Ji, G.-P., Zhou, T., Chen, G., Fu, H., Shen, J., Shao, L.: Pranet: Parallel reverse attention network for polyp segmentation. In: International Conference on Medical Image Computing and Computer-assisted Intervention, pp. 263–273 (2020). Springer
- 55.Huang, C.-H., Wu, H.-Y., Lin, Y.-L.: Hardnet-mseg: A simple encoder-decoder polyp segmentation neural network that achieves over 0.9 mean dice and 86 fps. arXiv preprint arXiv:2101.07172 (2021)
- 56.Yeung, M., Sala, E., Schönlieb, C.-B., Rundo, L.: Focus u-net: A novel dual attention-gated cnn for polyp segmentation during colonoscopy. Computers in biology and medicine 137, 104815 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the datasets used in this study are indicated and cited correctly in the paper.