Abstract
Biomedical image segmentation is essential in clinical practices, offering critical insights for accurate diagnosis and strategic treatment approaches. Nowadays, self-attention-based networks have achieved competitive performance in both natural language processing and computer vision, but the computational cost has reduced their popularity in practical applications. The recent study of Convolutional Neural Network (CNN) explores linear functions within modified CNN layer demonstrating pure CNN-based networks can still achieve competitive results against Vision Transformer (ViT) in biomedical image segmentation, with fewer parameters. The modified CNN, i.e., Depthwise CNN, however, leaves the features cleaved off in the channel dimension and prevents the extraction of features in the perspective of channel interaction. To effectively further explore the feature learning power of modified CNN with biomedical image segmentation, we design a lightweight multi-convolutional multi-scale convolutional network block (MSConvNeXt) for U-shape symmetrical network. Specifically, a network block consisting of a depthwise CNN, a deformable CNN, and a dilated CNN is proposed to capture semantic feature information effectively while with low computational cost. Furthermore, channel shuffling operation is proposed to dynamically promote an efficient feature fusion among the feature maps. The network block hereby is properly deployed within U-shape symmetrical encoder-decoder style network, named TriConvUNeXt. The proposed network is validated on a public benchmark dataset with a comprehensive evaluation in both computational cost and segmentation performance against 13 baseline methods. Specifically, TriConvUNeXt achieves 1% higher than UNet and TransUNet in Dice-Coefficient while 81% and 97% lower in computational cost. The implementation of TriConvUNeXt is made publicly accessible via https://github.com/ziyangwang007/TriConvUNeXt.
Keywords: Biomedical image segmentation, Convolutional Neural Network, Image semantic segmentation, Depthwise convolution
Introduction
Biomedical imaging has long been a cornerstone in the field of healthcare, offering clinicians a non-invasive window into the inner workings of the human body. The ability to visualize and interpret these images, especially in delineating specific anatomical structures, can often dictate the course of patient diagnosis and treatment. As such, the segmentation of biomedical images, which involves the partitioning of an image into meaningful regions, has been the subject of intense research focus [1–6]. In recent years, deep learning has shown promising performance in image processing [7–10], ushering in networks capable of unprecedented accuracy and efficiency. Early efforts saw the rise of Convolutional Neural Network (CNN) with architectures such as Fully Convolutional Network (FCN) leading the way [3]. UNet architecture then set a new benchmark, enhancing the quality of segmentation with its unique encoder-decoder design with skip connection [1]. More recently, the self-attention mechanisms from transformer have been explored in computer vision, introducing the potential for even greater detail recognition and spatial understanding in biomedical images [11–14]. This continual evolution underscores the rapid advancements in the field, each innovation bringing us closer to more precise and efficient biomedical image segmentation.
In the study of biomedical image segmentation, UNet stands as the most popular network which has been widely explored and utilized [1]. The symmetrical encoder-decoder design adeptly balances the acquisition of both high-level and low-level features, cementing its reputation as a preferred network for numerous improved version. Paving the way for 3D imaging, both 3D UNet [20] and V-Net [21] advanced the original UNet to accommodate volumetric data, thus widening the ambit to encompass 3D biomedical imaging. Taking a different trajectory, PSPNet harnessed the concept of pyramid scene parsing, enabling nuanced region-based contextual captures that underscored its competence in a spectrum of segmentation tasks [22]. As an enhancement to the classic UNet, MultiResUNet incorporated multiple residual connections into skip connections [23], while LinkNet redefined the encoder-decoder topology for expedited inference [5, 24]. UNet has also been a fertile ground for innovations, birthing variants like UNet++ [25] and UNet3+ [26]. Leveraging advanced techniques—ranging from novel loss designs and attention mechanisms to residual learning and dense connections—these iterations represent the evolution and adaptability of the UNet in biomedical image segmentation [27]. The nnU-Net, with its meticulous architecture tweaks, emerged as a benchmark in adaptability across an array of datasets [28].
Motivated by the success of self-attention in natural language processing [17], recent studies have witnessed the transformer with segmentation networks [11–13, 29–33]. Specifically, TransUNet introduced ViT to be utilized into bottleneck of UNet to model global range dependencies [34], and SwinUNet further synthesize the prowess of shift window-based ViT with the UNet design for biomedical image segmentation [13, 35]. The computational cost of ViT-based network, however, is normally significantly higher than CNN-based network.
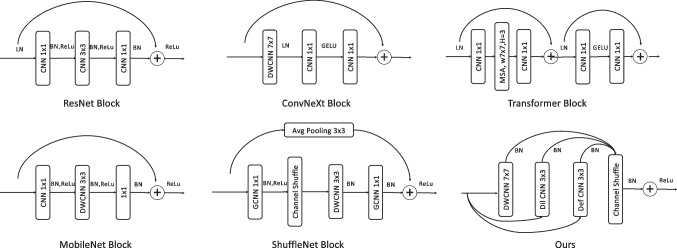
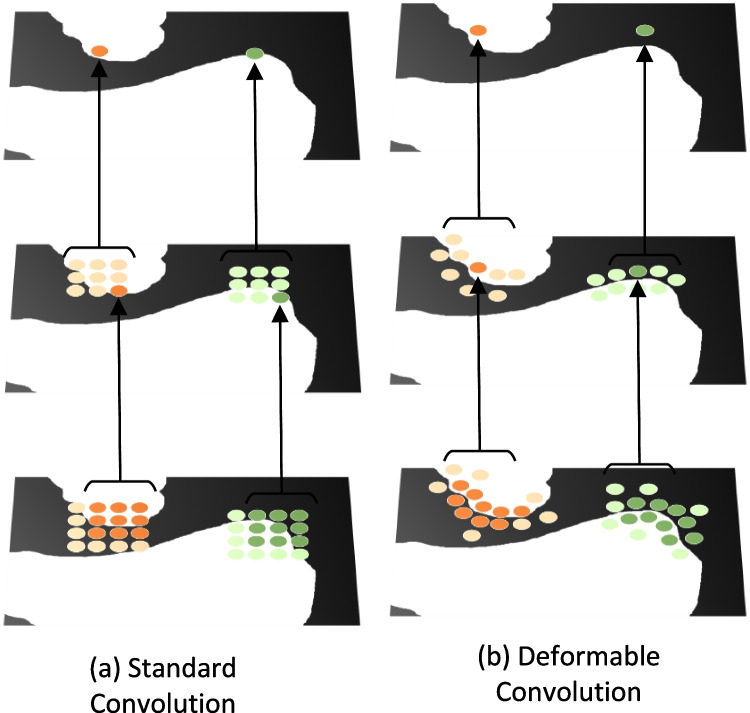
Outside the realm of segmentation-specific architectures, various generalized CNN have contributed foundational principles, and some of example advanced network blocks is briefly sketched in Fig. 1. The neural network architecture engineering such as ResNet have marked their significance by introducing the innovative concept of residual connections, which allows gradients to flow freely in deep networks, avoiding the vanishing gradient problem [15]. InceptionNet, also known as GoogLeNet, employs a multi-scale processing approach with its inception blocks, containing filters of varying sizes that operate concurrently, capturing diverse feature scales [38]. ShuffleNet, on the other hand, is renowned for its unique channel shuffle operation, enhancing the efficiency of information flow across channels, while MobileNet stands out with its depthwise separable convolutions, decomposing standard convolutions into more efficient operations suitable for mobile applications [18, 19]. DenseNet’s distinctiveness comes from its dense connections, fostering feature reuse and improved gradient flow [39]. The transformer architectures, initially tailor-made for natural language processing, employ self-attention mechanisms and have found their way into the vision domain [17, 40–43]. ConvNeXt employs modified convolutions, balancing computational cost with network capacity which demonstrates superior performance against Transformer [16]. The ConvNeXt, however, leverage only depthwise CNN, could be further explored for feature information extraction. Beyond these architectures, specialized techniques like depthwise CNN, graph CNN, dilated CNN, and channel shuffle have expanded the capabilities of traditional CNN [6, 19, 44–47], and some of typical CNN is illustrated in Fig. 2. The provided Fig. 1 offers a visual summation of these architectural blocks, including ResNet [15], ConvNeXt [16], Transformer [11], MobileNet [18], ShuffleNet [19], and the custom block, emphasizing their unique structures and functionalities in the realm of deep learning.
Fig. 1.
The illustration of ResNet [15], ConvNeXt [16], Transformer [17], MobileNet [18], ShuffleNet [19], and our proposed MSConvNeXt network blocks
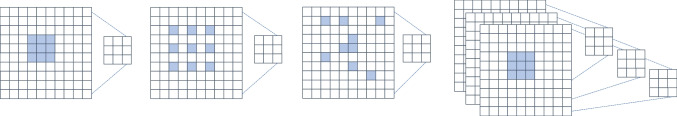
Fig. 2.
The illustration of conventional CNN, dilated CNN [36], deformable CNN [37], and depthwise CNN [18]
Following the recent successes in network architecture engineering and the above concern of computational cost, we hereby introduce TriConvUNeXt which is with triple modified CNN-based Multi-Scale Convolutional Network Blocks (MSConvNeXt). The contribution can be considered five-fold:
Integration of the dilated CNN within the network block that adeptly captures multi-scale contextual information without adding significant computational overhead.
Deployment of depthwise CNN within the network block, which enhances efficiency by applying a single filter per input channel, thus substantially reducing the number of parameters and associated computations.
Incorporation of deformable CNN within the network block, empowering TriConvUNeXt to capture expansive spatial information, further refining feature extraction.
The integration of channel shuffling strategies with three modified CNN resulting in MSConvNeXt, devised to promote a richer cross-talk between feature channels, resulting in more diverse and detailed feature representations.
The proper design with MSConvNeXt to come up with a lightweight yet robust U-shape encoder-decoder network, named TriConvUNeXt, which outperforms existing CNN- and ViT-based baseline methods.
Approach
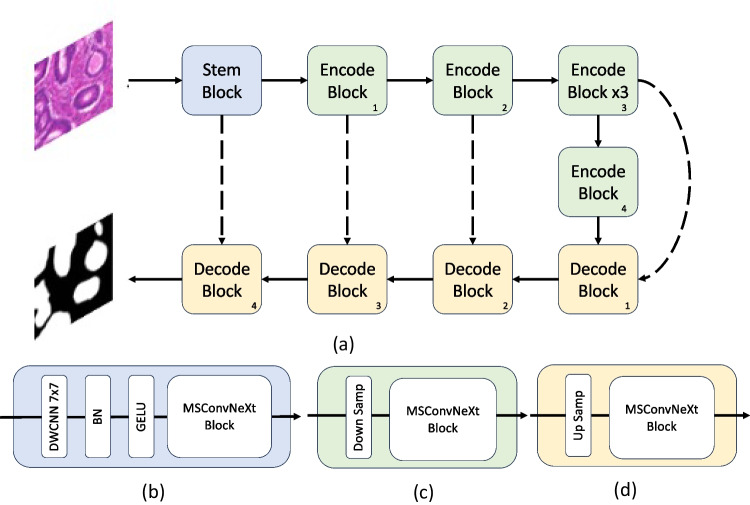
The framework for the proposed TriConvUNeXt is illustrated in Fig. 3. The architecture adopts a U-shape design, characterized by channel contraction and channel expansion pathways, termed as encoder i and decoder i, respectively, where represents the encoder and decoder at various levels, and both of the encoders and decoders possess identical structures (seen in Fig. 3c, d). To counteract the potential loss of surface-level information, skip connections are utilized between the encoders and decoders, augmenting the data flow transfer, which is with black dash line . Upon inputting an image into the network, it initially traverses a stem block (seen in Fig. 3b). The stem block solely modifies the channel without altering the shape of input feature. Subsequently, the feature courses through successive encoder modules, each reducing the feature size. This successive reduction permits the network to grasp features across expansive receptive fields. Notably, drawing inspiration from ConvUNext’s design [48], we incorporated three iterations in the third encoder stage as shown in . This intricate design bolsters the networks’ capability to distill surface-level features. Following the encoders, the feature streams into a series of cascaded decoders. Herein, bilinear interpolation progressively enlarges the feature’s shape while simultaneously reducing the channel count. This enlarged feature map, teeming with the segmentation target’s rich textural features, empowers the network to adeptly extract the feature’s nuanced details within the feature domain. In the final stage, a convolution tailgates the decoder to align with the number of classes of segmentation inference.
Fig. 3.
The illustration of the proposed network with corresponding network blocks. a The architecture of TriConvUNeXt, b the stem network block, c the encoder block with downsampling, and d the decoder block with upsampling
Redesign CNN-Based Network Blocks
The ViT has marked significant strides in biomedical imaging, as exemplified by networks such as TransUNet [34] and SwinUnet [35]. While the prevalent hypothesis attributes this success to the critical role of self-attention [11, 17], the work in ConvNeXt [16] offers a contrasting perspective, demonstrating that, by emulating the transformer architecture, incorporating rudimentary convolutional structures can yield competencies on par with more intricate designs.
Drawing from ConvNeXt [16], the self-attention module was supplanted by a depthwise CNN, modeled in line with the transformer. Additionally, the multi-layer perception (MLP) of transformer was substituted with an inverted bottleneck. The resultant architecture manifested exceptional performance metrics. However, a discernible limitation was observed: the utilization of a large-kernel depthwise, while advantageous for capturing extensive feature range within individual channels, overlooked inter-channel feature correlations. The inverted bottleneck module, despite its ability to amalgamate channel information, does not adequately address this limitation.
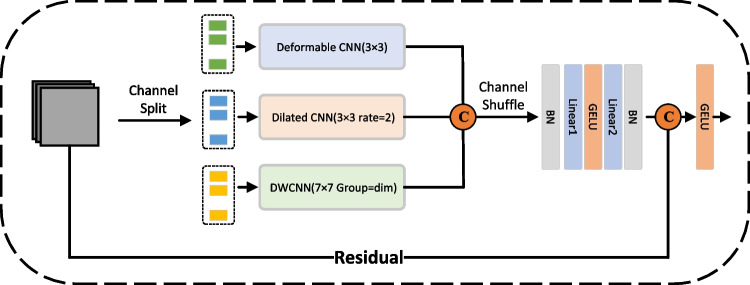
To rectify this, we segregated the semantic feature information into three channel-based clusters, each undergoing distinctive convolutional operations to fortify network robustness, named MSConvNeXt. The emergent features from these groups are then concatenated along the channel dimension. In a subsequent step, a channel shuffling operation is employed, ensuring diverse channel groupings for subsequent iterations. This design not only accommodates inter-channel target correlations but also judiciously minimizes the network’s parameter count. The details of proposed MSConvNeXt are illustrated in Fig. 4.
Fig. 4.
The illustration of the proposed MSConvNeXt block, which consists of deformable CNN, dilated CNN, and depthwise CNN (DWCNN)
Towards Lightweight UNet
The MSConvNeXt block incorporates three distinct convolution types in parallel, each tailored for specific feature extraction purposes. At the beginning of the feature inflow into the MSConvNeXt block, 25%, 25%, and 50% of the input features are randomly selected in the channel dimension and fed to the deformed CNN, dilated CNN, and depthwise CNN, respectively. A shuffled-channel process is then deployed to enhance the relevance of the semantic information in the channel dimension, enabling that all CNN to fully extract the feature information motivated by ShuffleNet [19]. In the last step of each MSConvNeXt block sequential GELU activation [49] and batch normalization [50] as well as linear are applied. Details of each modified CNN used in the MSConvNeXt blocks are given below:
-
(i)Deformable CNN: Given the irregular boundary characteristics of biomedical images, deformable convolution is employed to capture intricate boundary details. Unlike standard convolutions, deformable convolutions introduce random offsets in their kernel, enhancing adaptability for irregular structures (seen in Fig. 5). The standard convolution is defined as:
where Y, X, W denote output feature map, input feature map, and convolutional kernel weight, : a position in the convolution kernel, : a position in the output feature map, : sum operation on all positions in the convolution kernel.Incorporating deformable convolution introduces an offset , resulting in:The key distinction lies in the altered sampling position of X. Despite its adaptability, the computational complexity of deformable convolution can be high, particularly with larger kernels. Therefore, we set the kernel size for deformable convolutional operation with .
-
(ii)Dilated CNN: The dilated CNN introduces a dilation rate to the conventional convolution process without additional computational cost [36]. By manipulating the dilation rate, the receptive field (RP) of convolution kernels expands, enhancing both local and global context comprehension. An example comparison of receptive field between conventional CNN with dilated CNN is briefly sketched in Fig. 2a, b. The RP of a convolutional operation can be calculated as:
where k is the kernel size of the standard convolutional operations, and r is the dilation rate [51]. The parametric quantities par1 are where denotes the number of channels of the input tensor and the output tensor.When the same size of the receptive field is realized with standard convolution, the size of the convolution kernel employed is . At this point, the number of parameters par2 required for standard convolution is:
As can be seen, we can use a smaller number of parameters while increasing the receptive field to obtain more valuable global features. -
(iii)
Depthwise CNN: Depthwise separable convolution processes each input feature map channel individually, subsequently merging outputs via pointwise convolution [18, 52]. Relative to traditional convolution, depthwise drastically diminishes computational requirements. In aligning with the long-range feature extraction prowess of the transformer, we maintain a larger depthwise kernel in MSConvNeXt, sizing it at 7 with a padding of 3, mirroring ConvNeXt [16].
To juxtapose the parameter count between depthwise and standard convolution, consider:
For an input feature map of dimensions , with M output channels and a convolution kernel, the standard convolution parameters are:The parameters of depthwise convolution :The comparative ratio is:Depthwise CNN offers a significant reduction in convolutional parameters, optimizing efficiency without compromising performance.
Fig. 5.
The direct comparison of a the standard convolutional operation and b the deformable convolutional operation
Experiments and Results
Dataset
The Glas dataset, originating from Gland Segmentation in Colon Histology Images Challenge Contest (GlaS) from MICCAI Challenge 2015 [53]. It includes 165 images obtained from 16 H &E-stained histological sections of colorectal adenocarcinoma at stage T3 or T4. Each section originates from a unique patient and was processed separately in the lab, leading to significant variations across subjects in terms of stain distribution and the structure of the tissue. The conversion of these histological sections into digital whole-slide images (WSIs) was performed using a Zeiss MIRAX MIDI Slide Scanner, achieving a pixel resolution of 0.465 µm. The dataset has been splited into 50% for training, and the rest 50% is used to test the network with randomly selection, ensuring no overlap between each sets.
Implementation Details
We implemented the proposed method using Python with Pytorch library [54]. The experiments were conducted on a single Nvidia GeForce RTX 4090 GPU, which offered robust computational power and facilitated faster network convergence. The entire runtime, accounting for data transfer, network training, and inference, averaged around 0.5 h.
For the training process, we employed an optimizer of Adam [55], with an initial learning rate set to 0.0015. The learning rate was subject to a decay policy, reducing it by a factor of 1e-4 every 1 epochs. A batch size of 16 was found to strike a good balance between computational efficiency and network stability.
Loss
The loss function used to train our network was Dice-Coefficient-based and Cross-Entropy-based loss, denoted as Dice and CE, which has proven effective for segmentation tasks, ensuring the network’s focus on both local and global features. The CE loss can be illustrated as
where is the actual label value and is the probability value predicted by the network. log is the natural logarithm. The mathematical representation of the Dice loss is as follows:
where is the predicted value and i represents the index of each pixel. is the true value. Smooth is a very small constant (e.g., 1e-5) that prevents the denominator from being 0 and improves the stability of the calculation.
Finally, the total of loss in the experiment can be illustrated as
where is a loss weight and . It adjusts the relative importance of and . The is set to 0.5 in the experiment.
Baseline Methods
We conducted a direct comparison of TriConvUNeXt against a series of established baseline methods from the literature, including FPN [56], PAN [57], PSPNets [22], DeepLabV3 [58], UNet [1], ResUNet [59], UNext [60], TransUNet [34], SwinUnet [35], ConvUNeXT [48], MRNet-Seg [61], DSCNet [62], and TransAttUnet [63].
Evaluation Metrics
The evaluation metrics utilized in our experiments are reported as percentages (%) includes Dice-Coefficient (Dice), accuracy (Acc), precision (Pre), sensitivity (Sen), and specificity (Spe). In this context, higher values for these metrics signify better network performance. In the following formulas, TP, FP, TN, and FN represent true positives, false positives, true negatives, and false negatives, respectively. These metrics provide a comprehensive assessment of the network’s performance across various aspects. In the context of segmentation or classification:
True positives (TP): Correctly predicted positive cases.
False positives (FP): Incorrectly predicted positive cases (actually negative).
True negatives (TN): Correctly predicted negative cases.
False negatives (FN): Incorrectly predicted negative cases (actually positive).
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
Additionally, the computational cost of our proposed method with all baseline methods, as quantified by the number of network parameters (Par), underscores its efficiency, and practical applicability is also reported.
Qualitative Results
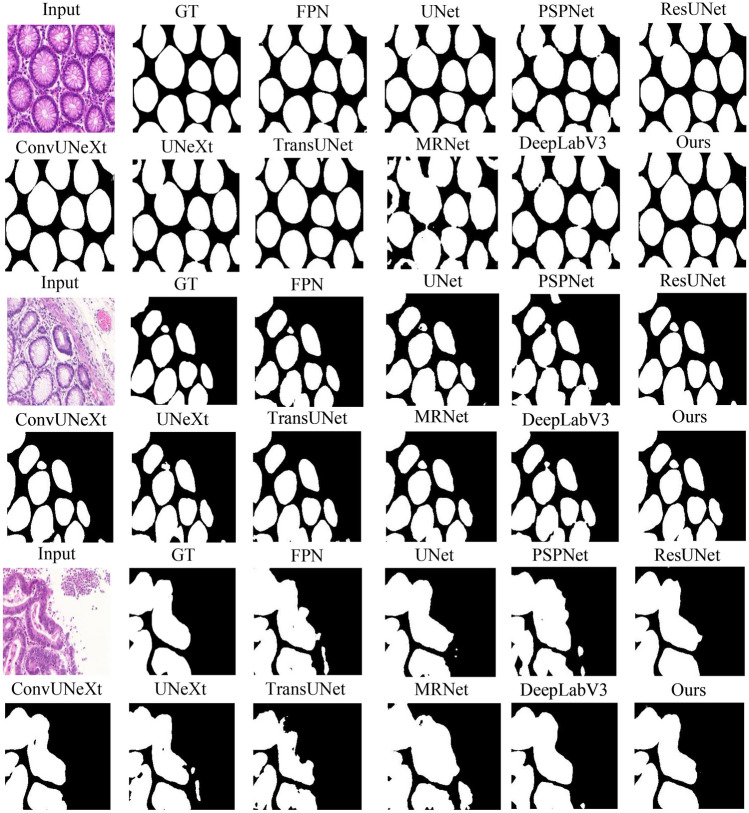
Figure 6 demonstrates example raw images, corresponding ground truth (GT), and the segmentation predictions by TriConvUNeXt as well as all baseline methods. These visual results provide a clear demonstration of the efficacy of TriConvUNeXt in capturing intricate details and patterns in the images. It is worth mentioning that SwinUNet shows the worst performance compared to other networks [35]. We speculate this is due to the dataset size limitation, which makes it difficult for self-attention to obtain valuable long-range features, and the self-attention “breaks down” in shallow networks.
Fig. 6.
The example raw images, corresponding ground truth, and segmentation inferences
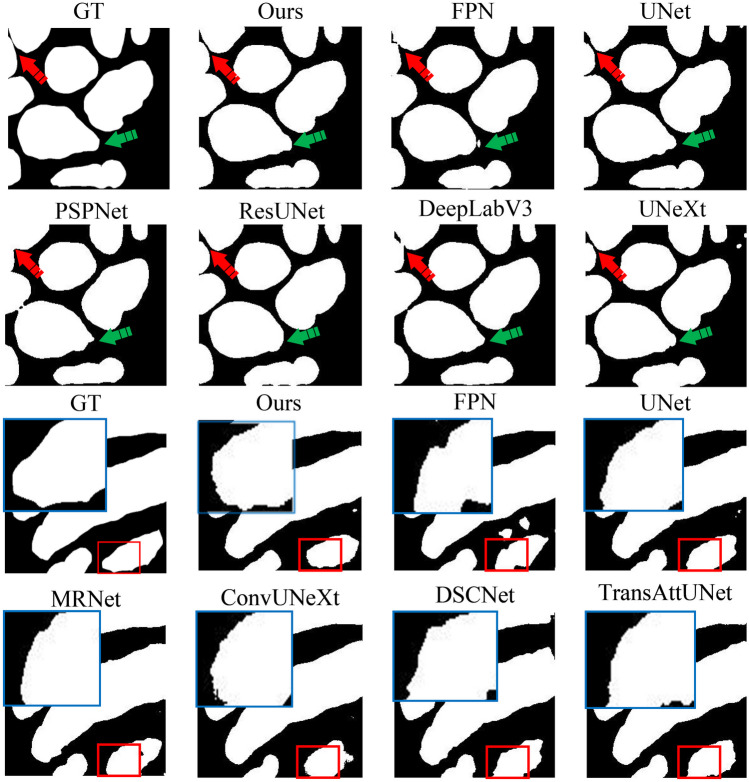
Figure 7 illustrates the detailed comparisons of the segmentation result boundaries produced by each network. Significant boundary differences are highlighted with red arrows, and the blue arrows indicate where two cells are distinctly segmented, whereas other networks merge two cells into a single ROI. The boundary of the ROI by TriConvUNeXt is also emphasized with a larger zoomed-in blue box, demonstrating smoother transitions. TriConvUNeXt achieves greater robustness in complex segmentation tasks compared to other baseline networks.
Fig. 7.
The example ground truth and corresponding segmentation inferences
Quantitative Results
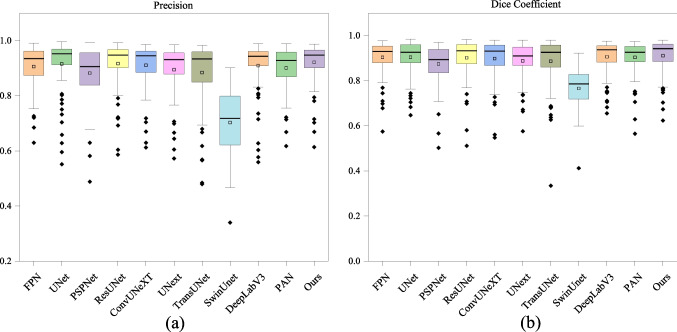
Our quantitative analysis provides a direct comparison of TriConvUNeXt against other existing baseline methods in Table 1. A comprehensive evaluation of segmentation performance and computational cost is reported. A deeper dive into the performance of each image on test set is visualized in Fig. 8, which presents via box plots for the distribution of Dice, specificity, and precision metrics. These plots effectively depict that the TriConvUNeXt is more likely to predict with more TP and TN but less FP and FN pixels. In additional, our proposed TriConvUNeXt is with only 3.3M trainable parameters in total, which is significantly lower than current popular networks such as TransUNet [34], SwinUNet [35], MRNet-Seg [61], ResUNet [59], DeepLab [58], and TransAttUNet [63].
Table 1.
The experimental results of TriConvUNeXt against other baseline methods on test set
| Network | Par | Dice | Acc | Pre | Sen | Spe |
|---|---|---|---|---|---|---|
| UNet [1] | 17.3M | 90.98 | 90.95 | 92.23 | 89.92 | 91.87 |
| FPN [56] | 4.2M | 90.88 | 90.74 | 91.34 | 90.54 | 90.56 |
| PAN [57] | 2.4M | 90.75 | 90.56 | 90.65 | 90.96 | 89.80 |
| PSPNet [22] | 0.9M | 88.30 | 88.17 | 88.98 | 87.73 | 88.27 |
| DeepLabV3 [58] | 12.7M | 91.19 | 91.04 | 91.32 | 91.13 | 90.64 |
| ResUNet [59] | 32.5M | 90.45 | 90.44 | 91.69 | 89.43 | 91.19 |
| UNext [60] | 1.5M | 89.28 | 89.20 | 90.03 | 88.65 | 89.40 |
| TransUNet [34] | 108.1M | 89.81 | 89.53 | 89.37 | 90.32 | 88.52 |
| SwinUnet [35] | 41.3M | 77.37 | 74.40 | 70.51 | 85.90 | 62.32 |
| ConvUNeXT [48] | 3.5M | 90.31 | 90.19 | 90.91 | 89.78 | 90.40 |
| MRNet-Seg [61] | 36.2M | 88.85 | 88.40 | 87.24 | 90.63 | 85.68 |
| DSCNet [62] | 2.1M | 88.13 | 87.63 | 86.46 | 89.90 | 84.88 |
| TransAttUnet [63] | 26.0M | 91.03 | 90.83 | 90.80 | 91.32 | 90.26 |
| TriConvUNeXt | 3.3M | 91.41 | 91.32 | 92.41 | 90.57 | 91.80 |
The best performance is highlighted with bold
Fig. 8.
Box plots representing the distribution of Dice and precision on the test set
Ablation Study
To validate the individual contribution of each component in our proposed network block, we conducted an exhaustive ablation study. The results of this study are summarized in Table 2. Starting with a basic version within only a single modified CNN been deployed, we incrementally incorporated deformable convolutions, dilated convolutions, and depthwise convolutions (denoted as DWCNN in Table 2). Each of these components plays a critical role (with ) in improving the segmentation performance, as evident from the consistent improvement in the Dice, Acc, Pre, Sen, and Spe.
Table 2.
The ablation study of proposed MSConvNeXt block within the TriConvUNeXt network
| Deformable | Dilated | Depthwise | Dice | Acc | Pre | Sen | Spe |
|---|---|---|---|---|---|---|---|
| 0.8153 | 0.8054 | 0.7906 | 0.8426 | 0.7667 | |||
| 0.8935 | 0.8889 | 0.8730 | 0.9158 | 0.8586 | |||
| 0.9020 | 0.9018 | 0.9077 | 0.8981 | 0.9036 | |||
| 0.9027 | 0.9019 | 0.9077 | 0.9003 | 0.9013 | |||
| 0.8976 | 0.8958 | 0.8937 | 0.9024 | 0.8871 | |||
| 0.9100 | 0.9092 | 0.9171 | 0.9040 | 0.9119 | |||
| 0.9141 | 0.9132 | 0.9241 | 0.9057 | 0.9180 |
The best performance is highlighted with bold
A few key observations from our ablation study include the following:
The combination of deformable and dilated convolutions resulted in superior performance in terms of Dice and Acc compared to using them solely.
Depthwise convolution brought a significant boost in precision without significantly affecting the network’s specificity.
The full configuration, which includes all three components—deformable, dilated, and depthwise CNN—yielded the best overall results, outperforming all other combinations.
These findings affirm that our proposed network block, built upon a synergy of deformable, dilated, and depthwise convolutions, is both innovative and valuable. The inclusion of channel shuffle further harmonizes the interplay of these components, ensuring a lightweight yet robust performance for biomedical image segmentation.
Advantages and Limitations
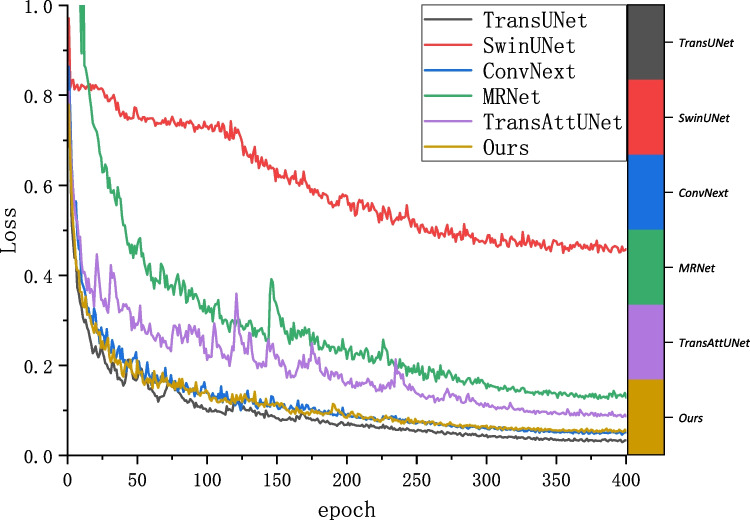
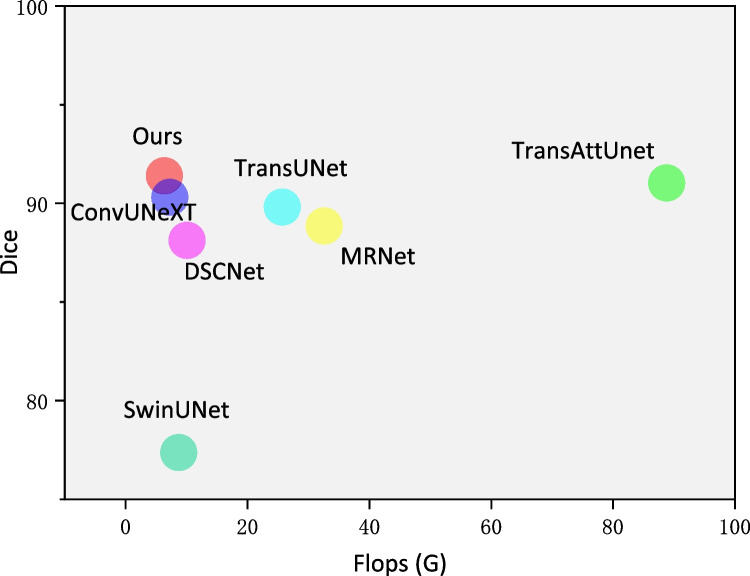
The segmentation robust and the computational complexity comparison are analyzed to better represent the advantages of the proposed TriConvUNeXt. Figure 9 illustrates the training history of the loss convergence of TriConvUNeXt as well as other networks. Although TriConvUNeXt is with a slightly higher segmentation loss compared with TransUNet [34], TriConvUNeXt demonstrates better segmentation performance on the test set, which proves that our proposed lightweight network is able to avoid the occurrence of overfitting. In addition, TriConvUNeXt outperforms the transformer-based network with significantly less computational resources, which is detailed illustrated in Fig. 10 demonstrating that the TriConvUNeXt with lowest FLOPs accompanied by the highest segmentation performance compared to other networks.
Fig. 9.
The training history of segmentation loss according to training epoch
Fig. 10.
Analysis between computational cost and segmentation performance of TriConvUNeXt against baseline networks
Conclusion
In the rapidly evolving realm of biomedical image segmentation, the quest for efficiency without compromising accuracy is paramount. Our work, centered on the design and implementation of TriConvUNeXt, stands as a testament to this endeavor. By ingeniously amalgamating dilated CNN, depthwise CNN, and deformable CNN, we introduced a distinctive MSConvNeXt network block that adeptly captures intricate image features. The incorporation of channel shuffling further enhances the interplay between feature channels, ensuring richer representations. A publicly available dataset is utilized for evaluation of the proposed method and other 13 state-of-the-art methods. It is worth noting that our proposed U-shaped network, TriConvUNeXt, achieves competitive performance against ViT-based UNet and other modified UNets with only 3.3M trainable parameters, which is over 30 lower than TransUNet.
In conclusion, TriConvUNeXt not only sets a new benchmark in terms of performance metrics but also underscores the potential of innovative design choices in advancing the state-of-the-art. As biomedical imaging continues to be a linchpin in modern healthcare, computationally efficient networks with promising performance pave the way for real-world clinical settings. In the future, we aim to further study convolutional operations, especially when tackling regions of interest with tubular structures, which are challenging but common in biomedical image segmentation tasks.
Declarations
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ronneberger, O., al: U-net: Convolutional networks for biomedical image segmentation. In: MICCAI (2015)
- 2.Wang, Z., Voiculescu, I.: Dealing with unreliable annotations: a noise-robust network for semantic segmentation through a transformer-improved encoder and convolution decoder. Applied Sciences 13(13), 7966 (2023) [Google Scholar]
- 3.Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015) [DOI] [PubMed]
- 4.Wang, Z., Voiculescu, I.: Weakly supervised medical image segmentation through dense combinations of dense pseudo-labels. In: MICCAI Workshop on Data Engineering in Medical Imaging, pp. 1–10 (2023). Springer
- 5.Chaurasia, A., Culurciello, E.: Linknet: Exploiting encoder representations for efficient semantic segmentation. In: 2017 IEEE Visual Communications and Image Processing (VCIP), pp. 1–4 (2017). IEEE
- 6.Zhang, Z., Li, S., Wang, Z., Lu, Y.: A novel and efficient tumor detection framework for pancreatic cancer via ct images. In: 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pp. 1160–1164 (2020). IEEE [DOI] [PubMed]
- 7.Wang, Z., Dong, N., Voiculescu, I.: Computationally-efficient vision transformer for medical image semantic segmentation via dual pseudo-label supervision. In: 2022 IEEE International Conference on Image Processing (ICIP), pp. 1961–1965 (2022). IEEE
- 8.Sun, S., Ren, W., Wang, T., Cao, X.: Rethinking image restoration for object detection. Advances in Neural Information Processing Systems 35, 4461–4474 (2022)
- 9.Wang, Y., Jin, X., Castro, C.: Accelerating the characterization of dynamic dna origami devices with deep neural networks. Scientific Reports 13(1), 15196 (2023) [DOI] [PMC free article] [PubMed]
- 10.Sun, S., Ren, W., Li, J., Zhang, K., Liang, M., Cao, X.: Event-aware video deraining via multi-patch progressive learning. IEEE Transactions on Image Processing (2023) [DOI] [PubMed]
- 11.Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., et al.: An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
- 12.Zhang, D., Zhou, F.: Self-supervised image denoising for real-world images with context-aware transformer. IEEE Access 11, 14340–14349 (2023)
- 13.Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin, S., Guo, B.: Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10012–10022 (2021)
- 14.Wang, Z., Su, M., Zheng, J.-Q., Liu, Y.: Densely connected swin-unet for multiscale information aggregation in medical image segmentation. In: 2023 IEEE International Conference on Image Processing (ICIP), pp. 940–944 (2023). IEEE
- 15.He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
- 16.Liu, Z., Mao, H., Wu, C.-Y., Feichtenhofer, C., Darrell, T., Xie, S.: A convnet for the 2020s. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 11976–11986 (2022)
- 17.Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł., Polosukhin, I.: Attention is all you need. Advances in neural information processing systems 30 (2017)
- 18.Howard, A.G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., Weyand, T., Andreetto, M., Adam, H.: Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861 (2017)
- 19.Zhang, X., Zhou, X., Lin, M., Sun, J.: Shufflenet: An extremely efficient convolutional neural network for mobile devices. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 6848–6856 (2018)
- 20.Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3d u-net: learning dense volumetric segmentation from sparse annotation. In: MICCAI (2016). Springer
- 21.Milletari, F., Navab, N., Ahmadi, S.-A.: V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation (2016)
- 22.Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J.: Pyramid scene parsing network. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2881–2890 (2017)
- 23.Ibtehaz, N., Rahman, M.S.: Multiresunet: Rethinking the u-net architecture for multimodal biomedical image segmentation. Neural networks 121, 74–87 (2020) [DOI] [PubMed]
- 24.Wang, Z., Voiculescu, I.: Triple-view feature learning for medical image segmentation. In: MICCAI Workshop on Resource-Efficient Medical Image Analysis, pp. 42–54 (2022). Springer
- 25.Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: Unet++: A nested u-net architecture for medical image segmentation. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4, pp. 3–11 (2018). Springer [DOI] [PMC free article] [PubMed]
- 26.Huang, H., Lin, L., Tong, R., Hu, H., Zhang, Q., Iwamoto, Y., Han, X., Chen, Y.-W., Wu, J.: Unet 3+: A full-scale connected unet for medical image segmentation. In: ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1055–1059 (2020). IEEE
- 27.Oktay, O., Schlemper, J., Folgoc, L.L., Lee, M., Heinrich, M., Misawa, K., Mori, K., McDonagh, S., Hammerla, N.Y., Kainz, B., et al.: Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 (2018)
- 28.Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods (2021) [DOI] [PubMed]
- 29.Zhou, F., Fu, Z., Zhang, D.: High dynamic range imaging with context-aware transformer. In: 2023 International Joint Conference on Neural Networks (IJCNN), pp. 1–8 (2023). IEEE
- 30.Wang, Z., Zhang, H., Liu, Y.: Weakly-supervised self-ensembling vision transformer for mri cardiac segmentation. In: 2023 IEEE Conference on Artificial Intelligence (CAI), pp. 101–102 (2023). IEEE
- 31.Wang, Z., Yang, C.: Mixsegnet: Fusing multiple mixed-supervisory signals with multiple views of networks for mixed-supervised medical image segmentation. Engineering Applications of Artificial Intelligence 133, 108059 (2024)
- 32.Wang, Z., Li, T., Zheng, J.-Q., Huang, B.: When cnn meet with vit: Towards semi-supervised learning for multi-class medical image semantic segmentation. In: European Conference on Computer Vision, pp. 424–441 (2022). Springer
- 33.Wang, Z., Zhao, C., Ni, Z.: Adversarial vision transformer for medical image semantic segmentation with limited annotations. British Machine Vision Conference (2022)
- 34.Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., Lu, L., Yuille, A.L., Zhou, Y.: Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021)
- 35.Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., Wang, M.: Swin-unet: Unet-like pure transformer for medical image segmentation. In: European Conference on Computer Vision, pp. 205–218 (2022). Springer
- 36.Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE transactions on pattern analysis and machine intelligence 40(4), 834–848 (2017) [DOI] [PubMed]
- 37.Dai, J., Qi, H., Xiong, Y., Li, Y., Zhang, G., Hu, H., Wei, Y.: Deformable convolutional networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 764–773 (2017)
- 38.Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., Vanhoucke, V., Rabinovich, A.: Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015)
- 39.Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
- 40.Lyu, W., Zheng, S., Ling, H., Chen, C.: Backdoor attacks against transformers with attention enhancement. In: ICLR 2023 Workshop on Backdoor Attacks and Defenses in Machine Learning (2023)
- 41.Lyu, W., Dong, X., Wong, R., Zheng, S., Abell-Hart, K., Wang, F., Chen, C.: A multimodal transformer: Fusing clinical notes with structured ehr data for interpretable in-hospital mortality prediction. In: AMIA Annual Symposium Proceedings, vol. 2022, p. 719 (2022). American Medical Informatics Association [PMC free article] [PubMed]
- 42.Liu, Q., Deng, H., Lian, C., Chen, X., Xiao, D., Ma, L., Chen, X., Kuang, T., Gateno, J., Yap, P.-T., et al: Skullengine: a multi-stage cnn framework for collaborative cbct image segmentation and landmark detection. In: Machine Learning in Medical Imaging: 12th International Workshop, MLMI 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, September 27, 2021, Proceedings 12, pp. 606–614 (2021). Springer [DOI] [PMC free article] [PubMed]
- 43.Wang, Z., Ma, C.: Dual-contrastive dual-consistency dual-transformer: A semi-supervised approach to medical image segmentation. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 870–879 (2023)
- 44.Tan, M., Le, Q.: Efficientnet: Rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, pp. 6105–6114 (2019). PMLR
- 45.Zhang, S., Tong, H., Xu, J., Maciejewski, R.: Graph convolutional networks: a comprehensive review. Computational Social Networks 6(1), 1–23 (2019) [DOI] [PMC free article] [PubMed]
- 46.Yu, F., Koltun, V.: Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:1511.07122 (2015)
- 47.Chen, X., Wu, J., Lyu, W., Zou, Y., Thung, K.-H., Liu, S., Wu, Y., Ahmad, S., Yap, P.-T.: Brain tissue segmentation across the human lifespan via supervised contrastive learning. arXiv preprint arXiv:2301.01369 (2023)
- 48.Han, Z., Jian, M., Wang, G.-G.: Convunext: An efficient convolution neural network for medical image segmentation. Knowledge-Based Systems 253, 109512 (2022)
- 49.Hendrycks, D., Gimpel, K.: Gaussian error linear units (gelus). arXiv preprint arXiv:1606.08415 (2016)
- 50.Ioffe, S., Szegedy, C.: Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: International Conference on Machine Learning, pp. 448–456 (2015). pmlr
- 51.Wang, Z., Voiculescu, I.: Quadruple augmented pyramid network for multi-class covid-19 segmentation via ct. In: 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pp. 2956–2959 (2021). IEEE [DOI] [PubMed]
- 52.Chollet, F.: Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1251–1258 (2017)
- 53.Sirinukunwattana, K., Pluim, J.P., Chen, H., Qi, X., Heng, P.-A., Guo, Y.B., Wang, L.Y., Matuszewski, B.J., Bruni, E., Sanchez, U., et al: Gland segmentation in colon histology images: The glas challenge contest. Medical image analysis 35, 489–502 (2017) [DOI] [PubMed]
- 54.Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., Killeen, T., Lin, Z., Gimelshein, N., Antiga, L., et al.: Pytorch: An imperative style, high-performance deep learning library. Advances in neural information processing systems 32 (2019)
- 55.Kingma, D.P., Ba, J.A., Adam, J.: A method for stochastic optimization. arxiv 2014. arXiv preprint arXiv:1412.6980106 (2020)
- 56.Kirillov, A., He, K., Girshick, R., Dollár, P.: A unified architecture for instance and semantic segmentation. In: CVPR (2017)
- 57.Li, H., Xiong, P., An, J., Wang, L.: Pyramid attention network for semantic segmentation. arXiv preprint arXiv:1805.10180 (2018)
- 58.Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 801–818 (2018)
- 59.Wang, Z., Zhang, Z., Voiculescu, I.: Rar-u-net: a residual encoder to attention decoder by residual connections framework for spine segmentation under noisy labels. In: 2021 IEEE International Conference on Image Processing (ICIP), pp. 21–25 (2021). IEEE
- 60.Valanarasu, J.M.J., Patel, V.M.: Unext: Mlp-based rapid medical image segmentation network. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 23–33 (2022). Springer
- 61.Lin, H.-Y., Liu, H.-W., et al: Multitask deep learning for segmentation and lumbosacral spine inspection. IEEE Transactions on Instrumentation and Measurement 71, 1–10 (2022)
- 62.Qi, Y., He, Y., Qi, X., Zhang, Y., Yang, G.: Dynamic snake convolution based on topological geometric constraints for tubular structure segmentation. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 6070–6079 (2023)
- 63.Chen, B., Liu, Y., Zhang, Z., Lu, G., Kong, A.W.K.: Transattunet: Multi-level attention-guided u-net with transformer for medical image segmentation. IEEE Transactions on Emerging Topics in Computational Intelligence (2023)