Abstract
Tumor heterogeneity and tumor evolution contribute to cancer treatment failure. To understand how selective pressures drive heterogeneous tumor evolution, it would be useful to image multiple important components and tumor subclones in vivo. We propose a supercontinuum-tailoring two-photon microscope (SCT-TPM) and realize simultaneous observation of nine fluorophores with a single light beam, breaking through the ‘color barrier’ of intravital two-photon fluorescence imaging. It achieves excitation multiplexing only by modulating the phase of fiber supercontinuum (SC), allowing to capture rapid events of multiple targets with maintaining precise spatial alignment. We employ SCT-TPM to visualize the spatiotemporal dynamics of heterogeneous tumor evolution under host immune surveillance, particularly the behaviors and interactions of six tumor subclones, immune cells and vascular network, and thus infer the trajectories of tumor progression and clonal competition. SCT-TPM opens up the possibility of tumor lineage tracking and mechanism exploration in living biological systems.
Subject terms: Optical imaging, Multiphoton microscopy, Time-lapse imaging, Tumour heterogeneity, Cancer imaging
Studying tumour heterogeneity and tumour evolution remains a challenging task. Here, the authors propose a supercontinuum-tailoring two-photon microscope to enable the imaging of multiple fluorophores with a single light beam. They use this tool to monitor the dynamics of heterogeneous tumour evolution.
Introduction
Cancer ecosystems are complex and consist of multiple tumor subclones with distinct genetic and phenotypic characteristics. The heterogeneous tumors continuously evolve and adapt to selective pressures from the tumor microenvironment (TME), such as immune surveillance and therapeutic interventions, resulting in cells with stronger resistant or metastatic capacity winning the clonal competition and leading to therapeutic failure1–3. Recent advances in single-cell and genomic techniques have enabled the dissection of the degree and spatial distribution of tumor heterogeneity with greater precision4–6. However, our understanding of the temporal course of heterogeneous tumor evolution and associated biological events has lagged, mainly due to the difficulties in performing multi-target longitudinal observations in vivo.
To investigate how multiple elements such as heterogeneous tumor cells, immune cells, vascular network, and stromal components interact to shape TME and regulate tumor development and cell fate7–10, new multicolor imaging techniques are needed to visualize dynamic biological processes in real time. Techniques such as flow cytometry11, immunofluorescence imaging12, and Stimulated Raman Scattering13,14 perform well for multi-target observations of cells and molecules, but they are not suitable for imaging in live biological systems. Two-photon microscopy has long been the predominant intravital imaging technique. But conventional two-photon microscopy can only image 3-4 targets simultaneously, which is known as the ‘color barrier’13. Two strategies have been developed to overcome the ‘color barrier’: detection multiplexing and excitation multiplexing. Spectral imaging is a prime example of detection multiplexing, which enables multicolor imaging by using multiple detection bands spanning the visible spectrum15,16. However, due to limited temporal resolution and signal-to-noise ratio, spectral imaging is unsuitable for dynamic signals or long-term observations in highly scattered tissues. Excitation multiplexing involves using multiple laser lines or non-linear processes such as wavelength mixing to optimally excite more fluorophores with different emission spectra simultaneously17,18. However, the combined beam power typically increases the risk of phototoxicity and bleaching, and optical systems with multiple laser lines are challenging to align. Supercontinuum has brought about novel excitation sources that can excite nearly all common fluorophores by using a single light beam, effectively lowering illumination and reducing collimation difficulties19–21. However, current supercontinuum imaging techniques typically utilize the broad spectra for excitation multiplexing, and only multiplex at the extremities of the supercontinuum spectrum. This leads to a limited number of fluorescence colors that can simultaneously observed, typically 3-4 colors, and the optimal excitation of fluorophores has not yet been achieved. In addition, the supercontinuum light source also suffers from overlapping fluorophore excitation or emission spectra, as well as limitations in signal intensity, temporal and spatial resolution, and system stability performance. To date, existing supercontinuum methods have not been able to achieve imaging with more than 4 colors in a live mammalian model. Thus, the current challenge in intravital fluorescence microscopy is to effectively and robustly excite and distinguish more fluorophores while maintaining low damage and photobleaching, thus enabling multi-target longitudinal observations of dynamic biological events.
Here, we propose supercontinuum-tailoring two-photon microscopy (SCT-TPM), which employs both excitation and detection multiplexing to realize intravital multicolor imaging. In the excitation multiplexing part, spectra, and phase patterns of the SC are tailored to produce expected two-photon absorption processes for multiple fluorophores. The SC was generated by combining a conventional fs laser with a highly nonlinear photonic crystal fiber (PCF). By tailoring the spectral phases of SC, two-photon absorption processes at a given transition frequency occur for all photon pairs spanning the SC. Therefore, two-photon absorptions for different transition frequencies can be controlled by modulating the SC phase patterns. In the detection multiplexing part, the fluorescence signals from multiple fluorophores are split by three channels and the NMF unmixing algorithm. SCT-TPM enabled us to image nine fluorophores simultaneously and capture rapid events of multiple targets while maintaining precise spatial alignment, demonstrating high spatiotemporal resolution, high excitation efficiency, high robustness, and low photodamage. Using SCT-TPM, we visualized the spatiotemporal dynamics of heterogeneous tumor evolution under host immune surveillance. We monitored the number, spatial distribution, behavior, and interactions of eight targets in TME, including immune cells, vascular network, and six tumor subclones, and monitored the long-term course of tumor evolution for up to 10 days. According to the motility features of immune cells and six tumor subclones, it was possible to infer the trajectories of tumor progression and clonal competition, for example, after several rounds of host immune attack, one of the six subclones might derive into highly metastatic or immune-resistant lineages. To the best of our knowledge, this is the first intravital imaging of six tumor subclones in the context of the host immune microenvironment. It reveals the prevailing tumor heterogeneity and tumor evolutionary routines and provides an opportunity to customize therapeutic strategies for specific tumor progression stages or for specific subclones. SCT-TPM expands the multicolor capabilities of intravital imaging and, in addition to tumor heterogeneity, will be a useful visualization tool for biomedical research.
Results
Excitation multiplexing by SC-tailoring
Excitation multiplexing is achieved by tailoring the spectra and phases of SC. The SCT-TPM system is illustrated in Fig. 1a and Supplementary Fig. 1. 100-fs, 800 nm pulses (Fig. 1b, left) were pumped into PCF to generate a SC ranging from 700 to 900 nm (Fig. 1b, right), which covered most of the two-photon absorption peak of existing fluorophores. The chirped pulses in SC were compensated into near transform-limited (TL) pulses by a pulse shaper to improve two-photon excitation efficiency (Fig. 1c and Supplementary Fig. 2). Moreover, phase modulations of the SC by the pulse shaper can control nonlinear processes for fluorophores that have a different peak wavelength of two-photon absorption spectra (Fig. 1f, g).
Fig. 1. Schematic illustrating how SCT-TPM achieves multicolor two-photon imaging.
a Principle of the SCT-TPM. SCT-TPM applied both excitation multiplexing and detection multiplexing. The 100-fs, 800 nm laser pulse is pumped into the PCF to generate an SC ranging from 700 to 900 nm by the dispersion and nonlinear effects of PCF. The 4 f reflective pulse shaper was programmed to compensate dispersion of the entire optical system to make the output SC beam near transform-limited, and then the designed phase patterns were applied to achieve SC tailoring. Different dichroic mirrors and optical filters were used in the detection path to realize detection multiplexing. Fluorescence signals were collected by photomultiplier tube (PMT). PCF, photonic crystal fiber; SLM, spatial light modulator; PSF, point spread function. b 100-fs, 800 nm pump pulse (left), and SC spectrum (right). c Phase modulation by SLM. d Measured PSF of identical 200 nm fluorescent bead during SC-tailoring. e Three blue fluorescent proteins, EBFP2.0, TagBFP, and mCerulean, to verify the discernibility of SCT-TPM. Two independent repeated experiments with similar results. f Two-photon absorption spectra (upper) and fluorescence emission spectra (down) of three blue fluorescent proteins. g The detailed process of supercontinuum tailoring. Two-photon absorption at different desired wavelengths occurs preferentially in different phase patterns. Phase pattern 1 corresponds to 770 nm (first row), Phase pattern 2 corresponds to 810 nm (second row), and Phase pattern 3 corresponds to 850 nm (third row). EBFP2.0, TagBFP, and mCerulean were optimally excited by phase patterns 1, 2, and 3, respectively. Individual fluorescent proteins could be discriminated against after unmixing. Purple arrows, EBFP2.0-expressing cells; blue arrows, TagBFP-expressing cells; cyan arrows, mCerulean-expressing cells (not marked all cells with arrows). Scale bars: 100 μm. Source data are provided as a Source Data file.
Phase modulation of SC is the key to realizing excitation multiplexing by SC-tailoring. We designed three-phase patterns for transition frequencies of 770, 810, and 850 nm, respectively (Fig. 1g and Supplementary Fig. 3). To accurately realize the transition frequency both at the ends and in the middle of the SC, phase patterns were designed with hybrid phase functions (Supplementary Note 1). Through numerical simulations, the sinusoidal phase (Fig. 1g, green line for 810 nm) and binary phase with genetic algorithm (Fig. 1g, blue line for 770 nm and red line for 850 nm) were determined as three-phase patterns. A pulse shaper with the spatial light modulator (SLM) was used to load the phase patterns onto the SC. By modulating the phase of individual frequency components (Fig. 1c, 1g first column), the quantum interference effect between them was controlled to excite nonlinear processes for different fluorophores (Fig. 1g, second column). Second harmonic generation (SHG) from a BBO crystal was measured to verify the SC-tailoring (Fig. 1g, third column). The peaks of SHG with different phase patterns corresponding to 385, 405, and 425 nm, indicate that the SC has been tailored as desired. Sequential switching of the three phases represents one imaging cycle. Because the response time of switching phase pattern is limited only by SLM, SCT-TPM can complete an imaging cycle within 3.9 s to capture rapid events in live cells.
To characterize the point spread function (PSF) of SCT-TPM, we captured 3D images of 200 nm yellow-green fluorescent beads with different phase patterns (Fig. 1d). The PSFs showed that the lateral and axial resolution could reach 0.57 μm and 2.65 μm, respectively (Supplementary Fig. 4). When switching the phases of SC, the excitation volumes of SCT-TPM overlaid without spatial shift or distortion, ensuring the strong robustness of imaging during SC tailoring.
To demonstrate the concept of excitation multiplexing in SCT-TPM, we imaged mixed cultures of three blue-color B16 cells labeled with EBFP2.0, TagBFP, and mCerulean, respectively (Fig. 1e) using only one PMT (Ch B). EBFP2.0, TagBFP, and mCerulean exhibit similar emission spectra (Fig. 1f, bottom) but different two-photon absorption spectra (Fig. 1f, upper). After an imaging cycle, raw (Fig. 1g, fifth column) and clear images after unmixing (Fig. 1g, sixth column) of the three fluorophores were obtained. The excitation specificity of each fluorophore was calculated by using its measured SHG and two-photon absorption spectra, to provide guidance for designing multicolor labeling strategies (Fig. 1g, fourth column and Supplementary Fig. 3). As long as these three fluorophores detected in the same channel correspond to different optimal phase patterns, they can be identified in the subsequent unmixing process.
Detection multiplexing and nine-color imaging of live cells
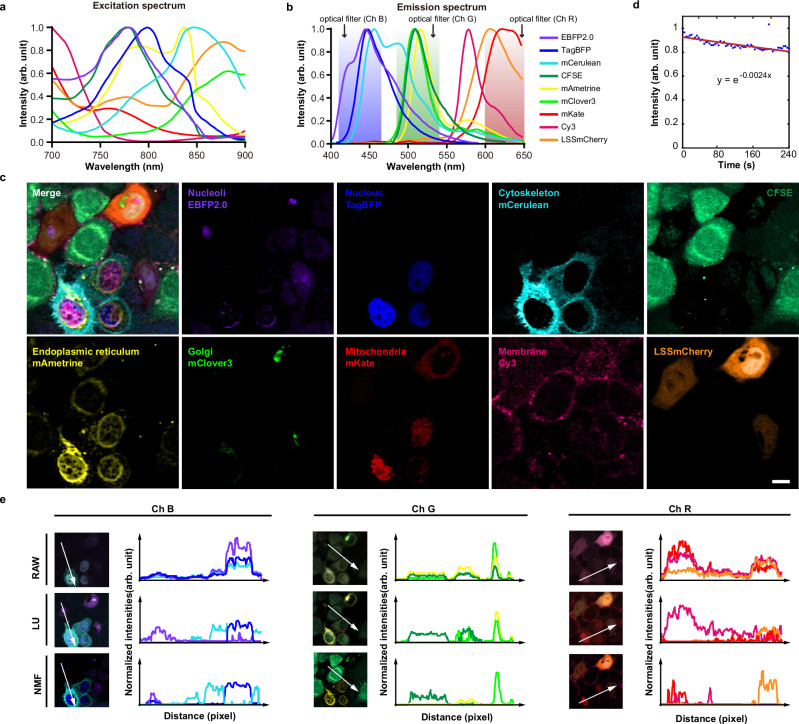
The multicolor capacity could be further augmented with detection multiplexing. Based on the above experiments, we added two more channels for detection multiplexing. The central wavelengths of the three channels were 447, 520, and 629 nm, and the bandwidth of the filters in front of the detectors was ~ 50–60 nm. Nine fluorophores, including seven fluorescent proteins- EBFP2.0, TagBFP, mCerulean, mAmetrine, mClover3, mKate, and LSSmCherry, and two fluorescent dyes- CFSE and Cy3, were selected to label different structures of HeLa cells. The labeling strategy is illustrated in Supplementary Fig. 5 and Supplementary Table 1. Two to five organelles or subcellular structures in each cell were labeled with different fluorophores. Although all fluorophores were highly overlapping in both two-photon absorption and emission spectra, nine images of each fluorophore were obtained after NMF unmixing in one imaging cycle (Fig. 2a–c and Supplementary Figs. 5–8).
Fig. 2. SCT-TPM imaging of live HeLa cells labeled with nine fluorophores.
a Two-photon excitation spectra, and (b) Fluorescence emission spectra of nine fluorophores. c NMF unmixed pseudo-color images of live HeLa cells labeled with nine fluorophores. Two independent repeated experiments with similar results. d Signal intensity curves of continuous 60-frames in 240 s. The average photobleaching constant was determined to be 0.0024. Blue dots, average of nine fluorescence signals; solid red line, exponential fitting. e Intensity profiles for comparison of raw images, linear unmixing (LU), and NMF unmixing. The x-axis denoted the distance along the arrows drawn in the left images; the y-axis denoted the intensity value of three fluorophores collected by a channel. Three panels, Ch B, G, and R, denoted three channels with filters 447/60, 520/60, and 629/53, respectively. Colors in (a) and (e) match rendering in (b). Scale bars: 10 μm. Source data are provided as a Source Data file.
One of the key advantages of SCT-TPM is its low photobleaching and photodamage. Within 240 s, we performed continuous 60-cycle imaging of live HeLa cells to measure the degree of photobleaching. Referring to the method of Wei et al., the average photobleaching coefficient of nine fluorophores was calculated to be only 0.0024 (Fig. 2d)13. Two factors contributed to the low photobleaching: firstly, a single beam was utilized as the excitation light source, resulting in only 15 mW of laser power behind the objective lens. Secondly, the rapid switching speed of phase modulation also shortened the exposure time of the laser on living samples. These factors permitted SCT-TPM to minimize the optical damage to the sample during the imaging process, making it ideal for long-term imaging of live cells or organisms.
Another advantage of SCT-TPM is that it shows multicolor capacity in live cells or complex biological environments. To unmix high crosstalk images for intravital imaging, we developed the NMF unmixing algorithm based on sparsity constraints22,23. This algorithm does not require a priori information needed for regular linear unmixing. Only an identity matrix was needed for initialization to predict the spectra of fluorophores, and then the iteration with a sparse constraint can reduce the crosstalk between images (Supplementary Note 2). Figure 2e showed intensity profiles of different channels with images before unmixing (raw images), after linear unmixing24, and after NMF unmixing. Less overlap between the profiles after NMF unmixing indicated a more effective extraction of fluorophores. In addition, Root-mean-square error (RMSE), spectral angular distance (SAD), and spectral information divergence (SID) were calculated to evaluate the quality of NMF unmixing quantitatively. All parameters were smaller than the expected standard for unmixing hyperspectral images25. The two-dimensional correlation coefficients15 between all images were also calculated before and after unmixing, demonstrating that the crosstalk between the unmixed images was significantly reduced (Supplementary Tables 2–4 and Supplementary Note 3). Combined with the NMF unmixing algorithm, SCT-TPM exhibited exceptional multicolor imaging capabilities, allowing for simultaneous visualization of nine targets within live cells with subcellular resolution. This was confirmed by the precise correspondence between the subcellular localization of each fluorophore and the predetermined labeling strategy (Fig. 2c, Supplementary Fig. 5, and Supplementary Table 1).
Validation of multicolor time-lapse imaging capability and monitoring of cell proliferation heterogeneity
We demonstrated the multicolor time-lapse imaging capability of SCT-TPM by monitoring organelle dynamics after pro-proliferative signal stimulation. Seven-color B16 cells were treated with epidermal growth factor (EGF, an endogenous cytokine that promotes cell proliferation) for 4 h before imaging (Fig. 3a and Supplementary Table 1). Time-lapse imaging demonstrated that multiple organelles responded to EGF stimulation, preparing energy, materials, forces, and information for cell proliferation26–29. SCT-TPM captured a mitotic tumor cell undergoing chromosome segregation after EGF stimulation (Fig. 3b and Supplementary Movie 1). Active movements of mitochondria, Golgi apparatus, and actin filaments were also observed (Fig. 3c–f and Supplementary Movie 2–4). Note that SCT-TPM enables the tracking of the rapid movements of mitochondria (migrating approximately 6.24 μm along the actin axis in 156 s) with sufficient temporal resolution (3.9 s per cycle) (Fig. 3d). Secondly, the PSF of SCT-TPM is spatially stable and allows monitoring the oscillation of fine actin filaments without spatial misalignment (Fig. 3f). Third, SCT-TPM has low phototoxicity and photobleaching, which minimizes negative impacts on cell viability and fluorescence signal intensity even after up to 80 imaging cycles. Thus short-wavelength fluorescent proteins EBFP2.0 and TagBFP can be used for SCT-TPM imaging, whereas they were previously challenging for time-lapse imaging due to their susceptibility to photobleaching30.
Fig. 3. Time-lapse imaging of live B16 cells in response to pro-proliferative signals.
a Dynamics of organelles after the treatment of 40 ng/ml EGF for 4 h: b chromosome separation, c mitochondria fusing and fissioning, d a mitochondrion moving along the actin axis, e activated Golgi apparatus and f the oscillating actin filaments. Subcellular structures of B16 cells were labeled by different fluorescent proteins, respectively. Scale bars: 5 μm. g Mitotic behavior after the treatment of EGF for 8 h: h interphase, i prophase, j prometaphase, k metaphase, l anaphase, and (m) telophase. n–q Tumor cells undergoing mitosis: n EBFP2.0-B16 cell and mKate-B16 cell, o mKate-B16 cell, p EBFP2.0-B16 cell and (q) TagBFP-B16 cell. r The relative numbers of mitotic cells after being treated with EGF or not. A two-tailed unpaired t test was performed, n = 6, p = 0.0008, 95% CI = [14.98–41.72], t = 4.724. Inset: the percentage of six subclones in all mitotic cells. s Cell growth rates of different B16 subclones detected by CCK-8. Ordinary one-way ANOVA analysis and Tukey’s multiple comparison tests were performed; n = 7, p = 0.0002, 0.0033, 95% CI = [− 0.3519 to − 0.09068], [− 0.3065 to − 0.04527], F = 6.106. Data in (r, s) were presented as mean ± SEM. t Mitotic arrest and cell apoptosis after the treatment of 40 ng/ml EGF for 8 h + 50 μM Paclitaxel for 2 h: u chromosome doubling, v abnormal binucleated tumor cell, w cell lysis, x release of extracellular vesicle, y intercellular adhesion, and (z) intercellular phagocytosis. Scale bars: 20 μm. Source data are provided as a Source Data file.
Furthermore, SCT-TPM was capable of monitoring the proliferation process of heterogeneous tumor cells (Fig. 3g and Supplementary Movie 5). Mixed cultures of six subclones of B16 cells (EBFP2.0-B16, TagBFP-B16, mCerulean-B16, mAmetrine-B16, mKate-B16 and LSSmCherry-B16, abbreviated as EBFP-B16, TagB-B16, Ceru-B16, mAme-B16, Kate-B16 and LSSm-B16 when necessary, respectively, Supplementary Fig. 9) were treated with EGF for 8 h and then stained with Hoechst 33342 before imaging. SCT-TPM captured the delicate cell cycle dynamics of different tumor subclones after EGF stimulation (Fig. 3h–m) and recorded the mitotic time courses of different subclones (Fig. 3n–q). A newly proliferated EBFP2.0-B16 cell took 22 min to exit mitosis and enter interphase (chromosomes decondensation, white arrowhead in Fig. 3n). During the same period, a mKate-B16 cell progressed from interphase to prometaphase (cell rounding and chromatin condensation, white arrow in Fig. 3n). Another pair of mKate-B16 cells transitioned from anaphase to telophase within 16 min (Fig. 3o). Particularly, an EBFP2.0-B16 cell (Fig. 3p) quickly progressed from metaphase to telophase in only 10 min, whereas a TagBFP-B16 cell (Fig. 3q) took 26 min to complete the same process. Therefore, SCT-TPM revealed differences in the mitotic velocities of tumor cells, indicating temporal heterogeneity in cell proliferation. SCT-TPM also allowed us to visualize the pro-proliferative effect of EGF and the clonal heterogeneity of cell proliferation. A total of 164 mitotic cells were observed in independently repeated imaging experiments (control group: n = 6 replicates; EGF group: n = 6 replicates). Compared to the control group, the number of mitotic cells increased two-fold after EGF stimulation (Fig. 3r). Of these mitotic cells, the percentage of mKate-B16 cells was the highest, whereas the percentage of TagBFP-B16 cells was the lowest (inset box in Fig. 3r). The differences in proliferation behaviors of the six subclones were also confirmed by CCK-8 assay, with EBFP2.0-B16 and mKate-B16 growing faster than TagBFP-B16 (Fig. 3s).
Cell proliferation induced by EGF could be completely counteracted by Paclitaxel (an anti-microtubule drug that inhibits mitosis) (Fig. 3t and Supplementary Movie 6). Tumor cells were able to accomplish chromosome doubling but could not divide successfully (Fig. 3u) and instead formed binucleated or multinucleated cells (Fig. 3v). These abnormal cells would eventually undergo apoptosis, accomplished by cell lysis (Fig. 3w) and release of extracellular vesicles (Fig. 3x). More interestingly, cell-cell interactions between different subclones were observed. Paclitaxel induced abnormal adhesion between adjacent cells labeled with different colors (Fig. 3y) and phagocytosis of vesicles released from apoptotic cells by another tumor cell (Fig. 3z). These suggest that tumor cells respond cooperatively to drug stimulation through intercellular communication and substance exchange, a phenomenon that has received less attention in previous studies.
Intravital 5D imaging of dynamic biological events during tumor evolution
In addition to the proliferation heterogeneity of tumor cells themselves, the continuous tumor evolution is regulated by many dynamical biological events occurring in the TME, such as the behavior and interactions of heterogeneous tumor cells, immune cells, vascular networks, and stromal components. Therefore, intravital 5D imaging (three-dimensional spatial structure, time, and color) is needed to monitor the spatiotemporal dynamic processes of heterogeneous tumor evolution. We established an eight-color-labeled murine melanoma model for intravital imaging by inoculating six subclones of B16 cells into the skin-fold window of EGFP mouse and labeling the arteries with AlexaFluorTM 633 hydrazide (Supplementary Figs. 9–11). Where the mixed inoculation of six subclones of B16 cells labeled with different fluorescent proteins was designed to somewhat mimic tumor heterogeneity in the natural microenvironment and to monitor the possible interactions between different tumor cells.
Intravital 5D imaging was performed on days 3–8 post-inoculation. Eight targets in live mice were simultaneously observed for up to 2.5 h, and the imaging was repeated for 2-3 days per mouse. Based on the observed morphological features and motility behaviors, we could determine that the majority of the EGFP-positive cells infiltrating into the tumor were host-derived immune cells. This was confirmed by immunofluorescence staining experiments with tumor tissue sections, wherein more than 90% of the EGFP-positive cells in the tumor area were stained by the primary antibody anti-CD45-AF647 (CD45, a leukocyte common antigen that is a molecular marker of immune cells) on days 5 to 15 post-inoculation (Supplementary Figs. 12, 13 and Supplementary Method 3). The remaining 10% of the EGFP-positive signals were probably mainly from neovascularization, which could be inferred from their morphology. Time-lapse imaging showed that infiltration and patrolling of host-derived immune cells (the moving EGFP-positive cells) in the tumor area had already occurred on Day 3-4 post-inoculation (Fig. 4a left, Supplementary Figs. 14–16 and Supplementary Movie 7). In addition, we observed several immune cells besieging a single tumor cell (Fig. 4b), as well as the release of tumor extracellular vesicles (Fig. 4c). The tumor antigens carried by these vesicles might trigger a stronger adaptive immune response. On Days 4–6, tumor cell apoptosis (Fig. 4d) and sparse tumor cell density were observed (Fig. 4a middle, Supplementary Figs. 17–19 and Supplementary Movie 8). However, on Days 7 and 8, the density of tumor cells rebounded significantly (Fig. 4a right, Supplementary Figs. 20, 21, and Supplementary Movie 9). Some tumor cells that survived host immune surveillance were observed undergoing mitosis (Fig. 4e) or moving (Fig. 4f), exhibiting enhanced proliferative and migratory activity. These more aggressive tumor cells, along with the disorganized neovascular networks (Fig. 4h, i compared to Fig. 4g), would eventually drive the tumor into a more malignant stage. Therefore, intravital 5D imaging provided insight into the number, spatial distribution, behavior, and interactions of immune cells, vascular networks, and six tumor subclones during tumor evolution.
Fig. 4. Intravital 5D imaging of dynamics of eight targets in TME.
a Images of TME in different stages of tumor evolution. Seven imaging sets were recorded (Mouse #1 on Day 3 abbreviated as Day3_M1, the remaining six sets is Day4_M1, Day4_M2, Day5_M2, Day6_M2, Day7_M3 and Day8_M3). b Immune cells were besieging a mCerulean-B16 cell. c The release of extracellular vesicles. d Cell apoptosis and lysis. e Cell mitosis. f Cell migration. g–i Disorganized neovascular network appeared on Day 7 (h, i) compared to that on Day 3 (g). The magenta channel was extracted in (g–i) for clearer demonstration. Scale bars: 20 μm. j Movement trajectories of immune cells. k The time-dependent curves of the motility coefficient (M). l Chemotactic index of the three groups. One-way ANOVA, n = 23, 30, 19 cells, p < 0.0001, = 0.0190, < 0.0001, 95% CI = [0.1276 to 0.2500], [− 0.1479 to − 0.01098], [− 0.3330 to − 0.2035], F = 55.87. j–l showed results for Day 3-M1, and data for the other six sets were detailed in Supplementary Figs. 15–21. m The proportion of different groups changed during tumor progression. All the moving immune cells observed in the seven imaging sets were counted in (m). n Frequency of interaction between different tumor subclones and immune cells. One-way ANOVA, n = 7, p = 0.0333, 0.0155, 0.0072, 0.0011, 95% CI = [− 0.4124 to − 0.01131], [0.03137–0.4324], [0.05060–0.4517], [− 0.4970 to− 0.09593], F = 5.782. o The movement speed of different subclones. One-way ANOVA, n = 7, p = 0.0171, 0.0163, 0.0462, 0.0152, 95% CI = [− 0.4236 to − 0.02848], [− 0.4249 to − 0.02975], [0.002194–0.3973], [0.03152–0.4266], F = 3.71. p The movement length of different subclones. One-way ANOVA, n = 7, p = 0.0248, 0.0415, 0.0001, < 0.0001, 95% CI = [− 0.4324 to − 0.01975], [− 0.4180 to − 0.005362], [0.1525–0.5652], [0.1540–0.5666], F = 7.513. q The movement displacement of different subclones. One-way ANOVA, n = 7, p = 0.0199, 0.0466, 0.0394, 95% CI = [− 0.4616 to − 0.02713], [0.002134–0.4366], [0.007228– 0.4417], F = 3.088. All data is presented as mean ± SEM. Source data are provided as a Source Data file.
We also found that the motility behavior of immune cell populations changed with tumor progression. Immune cells (the moving EGFP-positive cells) were classified into three groups based on their movement trajectories (Fig. 4j) and motility coefficients (Fig. 4k): Brownian walking cells, directed migrating cells, and constrained moving cells, which showed different mean velocity, arrest coefficient and chemotactic index (Fig. 4i and Supplementary Figs. 15–21). It had been reported that different motility behaviors suggested that the immune cells were performing different tasks. Brownian walking cells might be responsible for random patrolling to monitor possible pathogens, directed migrating cells might be recruited toward the diseased region, while constrained moving cells might be surrounding invaders and collecting antigenic information31–34. Quantitative analysis revealed that from Day 3 to Day 4, the proportion of directed migrating cells increased; from Day 4 to Day 6, the proportion of directed migrating cells decreased, and the sum of the other two increased; from Day 7 to Day 8, the proportion of constrained moving cells decreased and the proportion of Brownian walking cells rebounded (Fig. 4m). Thus, it is possible to visualize the time course of tumor immunosurveillance based on the motility behavior of EGFP-positive cell populations, without labeling and distinguishing complex immune cell surface markers (Supplementary Fig. 14). Longitudinal monitoring of immune cells in TME with SCT-TPM was expected to be a new potential strategy for assessing tumor progression.
The dynamic behavior of the six B16 subclones during tumor evolution was further analyzed. It was found that LSSmCherry-B16 and mCerulean-B16 interacted with EGFP-positive cells most frequently, while it was the opposite for mAmetrine-B16 and mKate-B16 (Fig. 4n). Furthermore, tumor cells displayed different motility, with mAmetrine-B16 exhibited the highest movement speed (Fig. 4o), longest movement path (Fig. 4p) and largest displacement (Fig. 4q). Due to the differences in growth rate (Fig. 3r, s), migration ability (Fig. 4o–q), and frequency of interactions with host-derived immune cells (Fig. 4n), subclones originating from the same parental cell line may undergo distinct selective evolutionary trajectories, resulting in tumor heterogeneity.
Longitudinal monitoring of tumor evolution under host immune surveillance
To investigate the trajectories and outcomes of heterogeneous tumor evolution regulated by these dynamic biological events, we further employed SCT-TPM to longitudinally monitor tumor progression in live mice from a more macroscopic perspective (millimeter-scale). With the help of blood vessels to localize the range and depth of intravital imaging, we performed large-field-of-view imaging of the entire tumor area in each mouse for 8–10 days (Fig. 5a and Supplementary Figs. 22, 23).
Fig. 5. Large-field multicolor imaging of tumor evolution under host immune surveillance.
a Large-field multicolor images of the TME in Mouse #A acquired from Day 3 to Day 10 after inoculation (Images acquired from the other two mice (Mouse #B and #C) were provided in Supplementary Figs. 22, 23). White dashed line: tumor boundary; green dashed line: venous vessels; red dashed line: arterial vessels; orange dashed line: region near the blood vessel. Scale bars: 100 μm. b Based on the vascular localization, the tumor boundary contours of Days 3, 5, 7, and 9 were overlaid. c The relative number of tumor cells. Ordinary one-way ANOVA analysis and Tukey’s multiple comparison tests were performed, n = 3, p = 0.0487, 0.0464, 95% CI = [− 1.482 to -0.003025], [− 1.488 to − 0.008539], F = 2.769. d The relative density of host-derived EGFP cells infiltrating into the tumor area. One-way ANOVA, n = 3, p = 0.0476, 0.0122, 95% CI = [− 1.676 to − 0.006448], [0.1775–1.847], F = 3.551. e The relative number changes of different subclones during Days 3–10. Ordinary one-way ANOVA and Tukey’s multiple comparison tests were performed to compare the relative proportions of each subclone per day, n = 3, *: p < 0.05. For Day 5, TagB-B16 vs. LSSm-B16: p = 0.0484, 95% CI = [− 29.23 to − 0.08267], F = 2.96; for Day 8, TagB-B16 vs. mAme-B16: p = 0.0396, 95% CI = [− 50.92 to − 1.026], F = 2.887; for Day 9, TagB-B16 vs. mAme-B16: p = 0.0385, 95% CI = [− 33.21 to− 0.7522], Ceru-B16 vs. mAme-B16: p = 0.0291, 95% CI = [− 34.01 to − 1.555], mAme-B16 vs. Kate-B16: p = 0.0354, 95% CI = [0.9915 to 33.45], F = 3.914. All data is presented as mean ± SEM. Source data are provided as a Source Data file.
Longitudinal imaging revealed that the tumor area and its boundary varied daily, showing a fluctuating trend of initial expansion, subsequent retraction, and re-expansion (Fig. 5a–c). During the initial stage of tumor development, the boundary expansion was attributable to the rapid proliferation of tumor cells (Day 3-6 in Fig. 5a–c). At the same time, host immunity was activated, which increased the number of EGFP-positive cells infiltrating the tumor area (mainly recruited immune cells, as evidenced by Supplementary Figs. 12, 13). This resulted in the retraction of the tumor boundary on day 7 (Day 7 in Fig. 5a–c). Subsequently, tumor cells that escaped from the first round of host immune killing survived and induced a new proliferation peak, leading to the outward expansion of the tumor boundary once again (Day 8, 9 in Fig. 5a–c). The second proliferation peak might also active stronger immune effects to inhibit tumor growth (Day 10 in Fig. 5a–c). We speculated that after several more rounds of tumor-immune interactions, an immune-tolerant microenvironment would eventually develop, leading to an irreversible malignant evolution of the tumor. Furthermore, the fluctuation of EGFP-positive cell density lagged that of tumor cell numbers (time of first peaking: Day 5 vs. Day 8 in Fig. 5c, d), indicating a time delay between immune infiltration and tumor expansion. This was because immune cells needed time for recruitment, delaying their arrival at the tumor site. It should also be noted that the number and speed of tumor cell proliferation, EGFP-positive cell infiltration, and tumor cell apoptosis were higher in the vicinity of blood vessels compared to areas distant from blood vessels, revealing the dual function of blood vessels in delivering nutrients to support tumor growth and delivering immune cells to kill tumor cells (Fig. 5a, circled area marked by the orange dashed line). Thus, SCT-TPM visualized the temporal and spatial heterogeneity of tumor evolution in the natural immune microenvironment.
The proportions of the six subclones showed different fluctuations (Fig. 5e). For example, the relative proportion of mAmetrine-B16 cells increased most significantly over time (yellow line in Fig. 5e), which was attributed to their lower host immune response and higher migration capacity (Fig. 4n-q). TagBFP-B16 cells showed the lowest proportion duo to their slower proliferation rates (blue line in Figs. 5e, 3r, s). Similarly, changes in the proportions of the other four subclones were co-determined by their proliferation rate, motility, and susceptibility to host immunity (Figs. 3r, s, 4n–q). Predictably, the TagBFP-B16 subclone might gradually lose its competitiveness during tumor evolution, while the mAmetrine-B16 subclone was more likely to differentiate into highly metastatic lineages or immune-tolerant lineages. This initial prediction demonstrated the ability of longitudinal multicolor imaging for tumor lineage analysis. Nevertheless, it is important to point out that future studies with longer periods of validation experiments on more animals are needed to fully characterize the entire dynamic process of clonal competition in heterogeneous tumors.
Discussion
In this paper, we present a multicolor two-photon imaging system, SCT-TPM, which provides insights into the heterogeneous tumor evolution by visualizing the spatiotemporal dynamics of multiple tumor subclones in the context of their TME. By using a broad spectrum as the excitation source, performing multiple nonlinear processes simultaneously through phase modulation, and ensuring that all spectral components are focused at the same spatial location, SCT-TPM exhibits three advantages compared to other intravital imaging methods: (1) more fluorophores can be used simultaneously so that up to nine targets can be visualized in live animals; (2) rapid dynamic response of more targets can be monitored in the range of seconds (3.9 s) with precise spatial alignment; and (3) relatively low photodamage enables longitudinal observation in living animals over several consecutive days.
In addition, SCT-TPM offers great flexibility for other potential application scenarios. The SC window can be altered by changing the PCF, and the central wavelength of two-photon excitation can be easily changed by designing various hybrid phase patterns so that different fluorophores can be flexibly selected for different applications. Furthermore, compared to conventional linear unmixing, our NMF unmixing algorithm performs much better to cope with spectral drift caused by variations in acidity, oxygen content, etc. in living organisms12. The objective function of NMF can also be redefined depending on the application requirements, to be compatible with different multicolor visualization schemes. Expanding the imaging colors of SCT-TPM should also be considered in the future. By customizing more and higher quality fluorescent probes to optimize in vivo multicolor labeling strategies, it will be possible to multiplex more colors. It would be advantageous to expand the excitation multiplexing window by generating a wider supercontinuum, such as 300 nm, or to expand the detection multiplexing by increasing the number of PMT. It is crucial to carefully adjust the power of the excitation spectrum, integration time of detection, and other imaging parameters in order to maintain optimal image signal-to-noise ratio, imaging depth, and temporal and spatial resolution. Once these technical issues are addressed, it becomes feasible to expand color capabilities in SCT-TPM. Based on current conditions and experimental requirements, 9-color imaging is considered the optimal outcome at this stage.
With SCT-TPM, we have performed multiparametric (8 targets), multidimensional (5D data), long-term (up to 10 days), and multiscale (micro-perspective: subcellular and single-cell behavior; macro-perspective: tumor boundary expansion) intravital imaging of TME. First, simultaneous observation of six color-labeled tumor subclones not only clearly delineates cell boundaries in dense tumor tissue for single-cell distinction but also facilitates tracking the behaviors of different subclones, which is promising for monitoring the evolutionary direction of heterogeneous tumors and the process of tumor lineage competition35. Second, multi-subclone tracking can also be used to distinguish tumor cells with different functions or different genotypes, such as distinguishing between high- and low-metastatic cells, cancer stem cells and non-stem cells, cells harboring driver mutations or not, and cells with high- or low-expression of target genes, to visualize molecular mechanisms of tumor development in the same in vivo conditions36. Last, visualizing the spatiotemporal dynamics of TME will help to intuitively determine the stage and subtype of tumor progression and to evaluate tumor response to therapy. Behavioral features of multiple factors are also expected to serve as potential biomarkers to guide tumor biology studies. Incorporating multiple targets into a comprehensive assessment of tumor progression and therapeutic response is probably more valuable than the traditional evaluation of a single biomarker.
Certainly, our study has some limitations. The supercontinuum spectrum causes axial lengthening of the PSF, limiting the axial resolution of 3D imaging to about 2 μm. Secondly, the excitation multiplexing of SCT-TPM requires the use of some large Stokes-shifted fluorophores, which may be slightly less stable than normal fluorophores. However, it is expected that the system can be improved by incorporating devices or modules to eliminate axial chromatic aberration. Using large Stokes-shifted fluorophores to label targets with more stable properties can optimize the combination labeling strategy. Third, we preliminarily demonstrate how SCT-TPM can be used to visualize the dynamics of heterogeneous tumor evolution. However, more observational data and further biological mechanism studies are needed to fully understand tumor development and progression.
Overall, SCT-TPM is expected to be used in various fields of tumor research, including tumor heterogeneity (temporal and spatial heterogeneity, intra-tumor heterogeneity, and extra-tumor heterogeneity), tumor molecular mechanisms, tumor immunity, and tumor pharmacology. SCT-TPM will facilitate tumor studies from discrete static snapshots to continuous dynamic monitoring, and from local single-target observation to systematic multi-target research. It is foreseeable that SCT-TPM can also be used in a variety of biomedical studies beyond tumor biology. For example, when combined with multi-color brainbow technology37, model animal, and nano-drug delivery system, SCT-TPM is capable of observing brain structure and function, embryonic stem cell differentiation, drug metabolism, and efficacy37,38. SCT-TPM is also directly compatible with the detection of multicolor immunofluorescence or multicolor-labeled fresh tissue sections, which will hopefully be used in clinical pathology studies and in vitro diagnostics. In conclusion, SCT-TPM expands the capability of multi-target observation in intravital imaging and will be a useful visualization tool for biomedical research.
Methods
Ethics statement
All animal studies were approved by the Hubei Provincial Animal Care and Use Committee and followed the experimental guidelines of the Animal Experimentation Ethics Committee of Huazhong University of Science and Technology (S719).
Optical setup for SCT-TPM
The SCT-TPM was set up on an isolator optical table (Supplementary Fig. 1). The femtosecond laser (Mai Tai BB, Spectra-Physics) beam first passed through a Faraday isolator (IO-5-NIR-LP, Thorlabs) and then was expanded to 6 mm to fully fill the back aperture of coupling objective (UplanSApo, NA 0.75, 20 ×, Olympus). The expanded laser beam was focused onto a PCF (NL-1050-NEG-01, NKT photonics) to produce SC39. The SC covered the wavelength range of 700–900 nm (Fig. 1b) with an output (input) power of 400 mW (680 mW), and the SC stability during imaging was monitored by using a feedback control loop. The SC beam collimated by an objective (L Plan, NA 0.6, 40 ×, Daheng Optics) was sent into a lab-built reflective 4 f pulse shaper, which contained a grating (53004BK02-340R, Richarson grating), a flipping square mirror, a concave mirror (CM750-200-P01, Thorlabs), and a one-dimensional 640-pixel SLM (SLM-640-P-VN, Cambridge Research & Instrumentation). Multiphoton intrapulse interference phase scan (MIIPS)40 was used to measure and compensate for phase distortion of the SC so that the fiber SC reaches near-TL pulse. Then the designed phase patterns were applied for SC-tailoring. The shaped pulses from the reflective 4 f pulse shaper were raster scanned by a galvanometer mirror pair (6215H, Cambridge Technology). The scanned beam was relayed by a scan lens (AC254-050-B-ML, Thorlabs) and a tube lens (AC508-300-B-ML, Thorlabs) to the rear focal plane of the microscope objective (XLPLN25XWMP2, NA 1.05, 25 ×, Olympus). The microscope objective was mounted on an objective scanning system (PD72Z4CAQ, Physik Instrumente) for volume imaging. The power of the objective focal point was 15 mW, and the typical field-of-view of 0.25 × 0.25 mm2. Two-photon excitation fluorescence signals were spectrally separated from the excitation pulses by long-pass dichroic mirrors (FF665-Di02, Semrock). The fluorescence signals were split and purified by dichroic mirrors (FF495-Di03, FF565-Di01, Semrock) and bandpass filters (FF02-447/60 or 475/50, FF01-525/45 or 520/60, FF01-629/53, Semrock), and finally collected by three PMT (H7422-40, Hamamatsu). A manual translation stage (M561D-XYZ, Newport) was used to rapidly adjust the imaging focal plane, and a two-dimensional motorized precision stage (H117E1, Prior Scientific) was used for mosaic-mode imaging. Pixel dwelling time ranged from 5–12 μs depending on different applications (detailed parameters for all SCT-TPM imaging were summarized in Supplementary Table 5). A custom LabView program was used for hardware control and image acquisition.
Phase control by pulse shaper
As mentioned above, SC-tailoring was based on multiphoton intrapulse interference (MII) theory (Fig.1g). A multiphoton transition process simultaneously involved several photons (e.g., 2 photons for two-photon). The final transition probability depended on the interference of all possible frequency combinations, and this interference could be controlled by modulating the phase of the pulses. To illustrate the role of the spectral phase in a two-photon transition process, considering the SHG generation, the SHG power spectrum was written:
| 1 |
where and were the spectral amplitude and phase of the femtosecond laser pulse, respectively. Equation 1 reflected the fact that two-photon transitions occur for all pairs of photons between frequencies ω + Ω and ω – Ω.
Phase design for SC-tailoring
Since the participation of frequency components symmetric about the transition frequencies was required in the MII effect, and the transition frequencies at which the two-photon absorption process is enhanced will be expanded into bands, they could not be set so far apart as to close to the two ends of the SC. To achieve a pronounced excitation multiplexing, we kept the FWHM at about 40 nm and finally set the transition frequencies at 770 nm, 810 nm, and 850 nm, respectively (Supplementary Note 1).
The sinusoidal function is the most commonly chosen in phase modulation41,42, which can easily change the transition frequency by changing parameters such as amplitude, period, or horizontal shift of the function (Supplementary Figs. 24, 25 and Supplementary Movies 10, 11). However, due to the periodicity and symmetry of the sinusoidal function, sub-transition frequencies are easily generated when the transition frequency is set at the two ends of SC, and thus the binarization function is used at both ends of the SC. It modulated phase by setting the spectral phase to 0 or π, with which the TL pulse could be maintained according to Eq. 1. The specific distribution of 0 and π is determined by the genetic algorithm (GA)43 through continuous iteration. When the transition frequency is located at the two ends of SC, it is more difficult to form pairs of frequencies, and the total number of photon pairs that can participate in two-photon absorption will be less. Therefore, when determining the binarized phase patterns, the bandwidth of the transition frequencies for two-photon absorption processes is appropriately increased, and the last three phase patterns are shown in Fig. 1g. Meanwhile, the SC-tailoring of the hybrid phase mask was tested by BBO crystal with the SHG central wavelength was 385, 405, and 425 nm, respectively (Fig. 1g, third column), which meant that the two-photon excitation energy was concentrated at the central wavelength of 770, 810, and 850 nm, respectively. The hybrid phase mask was applied in all imaging sessions.
Calculation of excitation specificity and selection of fluorophore
Assuming that the fluorophore i was uniformly distributed at a certain point in the sample and the concentration of fluorophore was saturated, without considering the PSF of the excitation light at the focal point, the objective could completely eliminate the chromatic aberration, according to two-photon excitation probability44 the particles in the fluorescent molecule due to two-photon excitation in a unit time could be described as
| 2 |
where I was the light intensity of excitation light, squared because of absorption of two photons, and denoted the two-photon absorption cross-section of i. The light source used in this system was an SC spectrum, which was a superposition of light with different frequency components, then Eq. 2 could be written:
| 3 |
where was the two-photon excitation spectrum of i, and was the spectrum of the excitation light after objective. Assuming that there was no excited emission and self-bleaching in an excitation window, the average signal of fluorescence that could be collected by the channel be expressed as
| 4 |
denoted the fluorescence quantum yield, and denoted the fluorescence collection efficiency of the channel, which could be constants at the same imaging condition. substituting the formula derived in the previous section here
| 5 |
We could directly see the factors influencing fluorescent intensity in Eq. 5. Since the SC contains many frequency components when it was used as an excitation source, the fluorescence intensity produced by the excited fluorophore could be regarded as the sum of each frequency. The fluoresce intensity of excited fluorophore was closely related to both the fluorophore’s two-photon excitation spectra and the light source’s spectra (Supplementary Fig. 3). Obviously when modulating the phase of the SC, the fluorescence emitted by the fluorophore would be a function of its two-photon absorption spectrum.
To simulate the nonlinear effects, we used SHG spectra mentioned in the last session as the excitation source to calculate the fluorescence intensity of different fluorophores under different phase patterns. This was the index for us to choose fluorophores to carry out multi-labeling. The value measured by spectrometer was the average value, then Eq. 4 transformed to:
| 6 |
where was related to the shape of the excitation light pulse, here we considered the excitation light as a Gaussian shape, g = 0.664. When we loaded phase1-phase3 to excite samples in sequence, we could estimate the fluorescence intensity of each fluorophore under three phase patterns with their reference two-photon absorption spectra by Eq. 6. We wrote this, and then define the excitation specificity of fluorophores under each phase pattern as
| 7 |
The maximum value of specificity was 1, under the phase pattern best suited to excite a particular fluorophore. When the emission spectra were similar, the fluorophores could be used simultaneously in SCT-TPM as long as their specificity values were maximized under different phase patterns (Supplementary Fig. 3d). We calculated excitation specificity of various fluorophores, and finally use it as the criteria for selecting fluorophores.
NMF unmixing
Because the fluorescence energies emitted by the fluorophores during multicolor imaging were insufficient to trigger nonlinear effects, it can be assumed that the signal acquired in each channel is a linear combination of all fluorophores, and the weights correspond to the contribution of each fluorophore in the sample:
| 8 |
Where, were images acquired from m channels, were the n fluorophores in the sample, denoted the ratio of the jth fluorophore’s fluorescence collected in the ith channel to its total energy, could be referred to the fingerprint of fluorophore j, determined by the channel settings of SCT-TPM and the fluorophores.
For NMF unmixing, we performed non-negative matrix factorization to unmix bleed-through signals. The preprocessed multi-channel data cube was flattened to an image matrix and normalized it. directly factorizes the image matrix22. Corresponding to Eq. 8, W was the abundance matrix, which represented the decomposed fluorophore, and H was the feature matrix, which was the fingerprint of different fluorophores. The elements in W and H were restricted to be nonnegative during matrix decomposition. Using optimization theory through an iterative approach to find W and H, and using a great likelihood estimated to accurately represent the original image.
The objective function of matrix decomposition was set to , where denoted the Euclidean distance between the image matrix V and the product of W and H obtained from the decomposition, indicating the residual of the decomposition, and was the penalty term in the objective function, reflecting the sparsity constraint for the abundance matrix in the decomposition process, aiming to reduce the crosstalk between the unmixed images. , to limit the value of sparseness (H), the optimization strategy of projected gradient descent was used during the iteration. spH was a constant that denoted the target of sparseness (H), which could be set among [0, 1] according to the fingerprints of the fluorophores. An identity matrix could be used as H0, which could effectively speed up the convergence of the objective function and also eliminate the randomness of the unmixed results. The extraction of fluorophores was completed by recovering the feature matrix into an image after the matrix MATLAB (MATLAB R2022b, MathWorks, USA) codes (Supplementary Software 1).
For stitching, z-stack, and time-lapse imaging, NMF unmixing would be conducted in each single slice at first. However, because the labels in each slice were different, the unmixing precision would be slightly different at different slices, to uniform image data, we chose the feature matrix H of the slice with the minimized SAD, SID, and RMSE as the fingerprint, and then perform liner unmixing with pinv function in MATLAB to calculate the endmember matrix W at the volume in the entire acquisition time.
For linear unmixing in 9-color HeLa cells, image matrix V and fingerprint H0 was input to lsqnonneg function in MATLAB, and the endmember matrix W could be acquired by pixels directly. The fingerprints were acquired by single label cells as shown in Supplementary Figs. 6–8.
Images processing
Pre-processing processes such as coordination of pixel shift and subtraction of scaled images from other channels were performed before NMF unmixing. Denoising with a Gaussian filter, contrast enhancement, pseudo-color and composited with maximum intensity projection were performed on images after NMF unmixing. Mosaic stitching was performed after unmixing and linear fusion was used at the image edges. All processing was conducted using custom-written MATLAB codes.
Tracking and analyzing cell movement behavior
The positions and movement trajectories of moving EGFP-positive cells and tumor cells were tracked by the ‘spot’ function in Imaris (Imaris x64 9.0.1, Oxford Instrument, UK). The tracking speed, tracking length, and tracking displacement length data were extracted to characterize the movement of tumor cells. The frequency of interaction between tumor cells and immune cells was characterized by their distance. An immune cell is considered to be interacting with a tumor cell if the distance between the two cells is greater than the sum of their radii and less than the sum of their diameters.
The movement trajectories of immune cells that moved within the observation volume throughout the imaging period were extracted for analysis. The trajectories of all immune cells were normalized to the origin, and the cell trajectory plot was obtained by overlaying all trajectories. The velocity of immune cell movement was calculated based on displacement, which , and denoted the time interval between two frames. The mean velocity was obtained by averaging the instantaneous velocities of the cell . The arrest coefficient was defined as the ratio of time that the instantaneous velocity of the cell was below 1.5 μm/min, with larger values indicating longer stagnation times and vice versa. The chemotactic index was defined as the ratio of the displacement to the total length of the cell’s path, with larger values indicating the larger the range involved in cell movement and the more obvious the tendency to move in a straight line, and vice versa. The mean displacement (m.d.) of the cell indicated the average of the displacements. The mean square displacement (m.s.d.) was the average of the squared displacements. The motility coefficient M was similar to the diffusion coefficient in Brownian motion, and in the quantitative analysis of three-dimensional space, . m.d., m.s.d., and M reflected the speed and tendency of cell movement, and the larger the parameter value, the faster the cell moved, and the more obvious the tendency was31,32.
The M-t linear regression was used to classify moving immune cells into three groups. M was constant in Brownian walking (slope = 0), increased with time in directed migrating (slope > 0), and decreased with time in constrained motility (slope < 0)33. Then, the mean velocity, arrest coefficient, chemotactic index, mean displacement, mean square displacement, and instantaneous velocity were statistically analyzed.
Preparation of transient transfected cells
For HeLa cell imaging, fusion proteins that linked different fluorescent proteins to various intracellular proteins were expressed by transient transfection. The fluorescent protein EBFP2.0 fused to fibrillarin was expressed in the nucleolus; TagBFP fused to H2B was expressed throughout the nucleus; mCerulean fused to Lifeact was expressed in the cytoskeleton; mAmetrine fused to Calreticulin, and KDEL was expressed in the endoplasmic reticulum (ER); mClover3 fused to SiT-15 was expressed in the Golgi apparatus; mKate fused to pyruvate dehydrogenase was expressed in mitochondria; LSSmCherry was expressed in the whole cell. Details of the plasmid construction and cell transfection processes are described in Supplementary Method 1, Supplementary Fig. 5, and Supplementary Table 1. For B16 cells imaging in Fig. 3a, similar to the fluorescent HeLa cells described above, different subcellular structures of B16 cells were labeled with seven fluorescent proteins by transient transfection (Fig. 3a, Supplementary Method 1, and Supplementary Table 1). Seven-color B16 cells were mixed and cultured overnight in a confocal imaging dish. The culture medium was replaced with fresh medium containing 40 ng/ml EGF four hours prior to imaging.
HeLa cells and B16 cells were stored in-house by the laboratory. HeLa and B16 cell lines were authenticated by STR Multi-amplification Kit and tested negative for mycoplasma contamination. HeLa cells were cultured in the recommended medium supplemented (89% DMEM medium, 10% FBS, and 1% penicillin-streptomycin, Gibco) overnight (at 37 °C in a 5% CO2 incubator). B16 cells were cultured in the recommended medium supplemented (containing 89% RPMI 1640 medium, 10% FBS, and 1% penicillin-streptomycin, Gibco).
Preparation of monoclonal fluorescent cells
Six fluorescent proteins (EBFP2.0, TagBFP, mCerulean, mAmetrine, mKate, and LSSmCherry) were chosen to construct six kinds of plasmids. Each plasmid contains one fluorescent protein, with a puromycin resistance gene sequence inserted as the selecting marker for stable transfection. Details of stable transfection and monoclonal fluorescent cell screening are described in Supplementary Method 2 and Supplementary Fig. 9. The obtained monoclonal cell lines would be used for time-lapse imaging of live cells and intravital imaging of live mice.
For time-lapse imaging of B16 cells in Fig. 3g, mix-cultured multicolor B16 cells were treated with 40 ng/mL EGF for eight hours. For time-lapse imaging of live cells in Fig.3t, mix-cultured multicolor B16 cells were treated with 40 ng/mL EGF for eight hours prior to imaging. Two hours before imaging, 50 μM Paclitaxel was added to the dish. The nuclei of tumor cells were stained with Hoechst 33342 prior to imaging. For the cell growth rate assay in Fig. 3s, each of the six B16 subclones was seeded into 96-well plates at a density of 5000 per well. After 48 h incubation, cells were treated with 10% CCK-8 (Zoman Bio, China) diluted in a normal culture medium and incubated for another 1.5 h. The absorbance was measured spectrophotometrically at 450 nm.
Multiple labeling of TME in live mice
C57BL/6-Tg (CAG-EGFP)/J female mice (9–12 weeks), which expressed EGFP throughout the entire body, excluding erythrocytes and hair, were prepared for establishing intravital TME imaging models34,45. All mice were housed in a specific pathogen-free barrier facility (room temperature of 20–26 °C; relative humidity: 40%–70%; alternating light/dark time: 12/12 h) at the Animal Center of Wuhan National Laboratory for Optoelectronics. For intravital imaging, a skin-fold window chamber (APJ Trading Co., Inc., Ventura, CA) was implanted into the back of the mouse. 1 × 106 tumor cells (mixture of six subclones) resuspended in 25 μL PBS were injected between the fascia and dermis of the rear skin. The entire surgical process was conducted under sterile conditions to avoid infection. To relieve pain associated with surgery and inflammation, the mice receive Tolfedine via i.p. injection (16.25 mg/kg−1, Vétoquinol, Québec, Canada) immediately and within 24 h after implantation. The mice were allowed to recover for 24 h, and then anesthetized using isoflurane (1% in oxygen) and maintained at 37 °C for imaging. The time of each imaging was kept to less than 3 h to avoid complications of prolonged anesthesia. To label the elastin fibers of the arterial vessels, 100 µL of 0.25 mM Alexa FluorTM 633 hydrazide (A30634, Life Technologies) in saline46 was injected into the EGFP mouse via the tail vein 3 h before imaging, and AF633 could maintain a stable labeling for 3–5 days. The full experimental procedure can be found in Supplementary Fig. 10. Tumor volume in all mice did not exceed the maximum ethically permissible tumor burden (maximum tumor diameter not exceeding 15 mm).
All multicolor fluorescent labeling strategies involved in this paper were summarized in Supplementary Table 6.
Statistics and reproducibility
Unless otherwise noted, data used in statistical analyses were presented as mean ± SEM. The Kolmogorov-Smirnov normality test was used to determine whether data values were normally distributed. When values followed Gaussian distribution, the unpaired two-sided t test or ordinary one-way ANOVA analysis with Tukey’s multiple comparison tests was performed to determine statistical significance. When values were not normally distributed, a non-parametric test (Kruskal-Wallis test and Dunn’s multiple comparisons test for more than two groups) was used instead. All data were first normalized and then statistically analyzed by GraphPad Prism 9 (GraphPad Software, Inc., La Jolla, CA). The sample size and number of replications of the experiment have been described in the legend of each figure. No statistical method was used to predetermine the sample size. No data were excluded from the analyses. The experiments were not randomized, and the Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by the National Natural Science Foundation of China No.62475063 (L.Fu), Collaborative Innovation Center Fund XTCX2022JKB12 (L.Fu), National Key Research and Development Program of China No.2022YFC2404401 (L.Fu, Q.Liu, X.Gao), and Open Project Program of Wuhan National Laboratory for Optoelectronics No. 2023WNLOKF013 (L.Fu, X.Gao). The authors thank the Optical Bioimaging Core Facility of WNLO-HUST for the support in data acquisition.
Author contributions
L.Fu designed the study. X.Gao, X.Huang, and Z.Chen designed the experiments. X.Gao, X.Huang, Z.Chen, Y.Zhou, L.Yang, and Z.Hou finished the experiments and analyzed the results. S.Qi, Z.Zhang, and J.Yang helped in the experimental design and provided cell and mouse samples. Q.Luo, Z.Zhang, Q.Liu, S.Qi, and Z.Liu provided supervision and support review. X.Gao, X.Huang, Z.Chen, L.Fu, and Q.Luo wrote and edited the paper.
Peer review
Peer review information
Nature Communications thanks Brad Amos, Wei Min, and Frank Winkler for their contribution to the peer review of this work. A peer review file is available.
Data availability
Data generated in the study are included in the manuscript and its Supplementary Information. Source data are provided in this paper.
Code availability
The unmixing code written in MATLAB is contained in Supplementary Software along with a demo image and instruction (Supplementary Software 1).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiujuan Gao, Xinyuan Huang, Zhongyun Chen.
Contributor Information
Qingming Luo, Email: qluo@hainanu.edu.cn.
Ling Fu, Email: lfu@mail.hust.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53697-1.
References
- 1.Vitale, I., Shema, E., Loi, S. & Galluzzi, L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med.27, 212–224 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Nguyen, P. H. et al. Intratumoural immune heterogeneity as a hallmark of tumour evolution and progression in hepatocellular carcinoma. Nat. Commun.12, 227 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutierrez, C. et al. Multifunctional barcoding with ClonMapper enables high-resolution study of clonal dynamics during tumor evolution and treatment. Nat. Cancer2, 758–772 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathur, R. et al. Glioblastoma evolution and heterogeneity from a 3D whole-tumor perspective. Cell187, 446–463 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tavernari, D. et al. Nongenetic evolution drives lung adenocarcinoma spatial heterogeneity and progression. Cancer Discov.11, 1490–1507 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Ma, L. et al. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol.75, 1397–1408 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell168, 613–628 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Binnewies, M. et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med.24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei, X. et al. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol. Cancer20, 7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turley, S. J., Cremasco, V. & Astarita, J. L. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol.15, 669–682 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Brandi, J., Wiethe, C., Riehn, M. & Jacobs, T. OMIP-93: A 41-color high parameter panel to characterize various co-inhibitory molecules and their ligands in the lymphoid and myeloid compartment in mice. Cytom. Part A103, 624–630 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Seo, J. et al. PICASSO allows ultra-multiplexed fluorescence imaging of spatially overlapping proteins without reference spectra measurements. Nat. Commun.13, 2475 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei, L. et al. Super-multiplex vibrational imaging. Nature544, 465–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi, L. et al. Highly-multiplexed volumetric mapping with Raman dye imaging and tissue clearing. Nat. Biotechnol.40, 364–373 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bares, A. J. et al. Hyperspectral multiphoton microscopy for in vivo visualization of multiple, spectrally overlapped fluorescent labels. Optica7, 1587–1601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cutrale, F. et al. Hyperspectral phasor analysis enables multiplexed 5D in vivo imaging. Nat. Methods14, 149–152 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Mahou, P. et al. Multicolor two-photon tissue imaging by wavelength mixing. Nat. Methods9, 815–818 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Rakhymzhan, A. et al. Synergistic strategy for multicolor two-photon microscopy: Application to the analysis of germinal center reactions in. Vivo. Sci. Rep.7, 7101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudley, J. M., Genty, G. & Coen, S. Supercontinuum generation in photonic crystal fiber. Rev. Mod. Phy.78, 1135–1184 (2006). [Google Scholar]
- 20.Tu, H. & Boppart, S. A. Coherent fiber supercontinuum for biophotonics. Laser Photon. Rev.7, 628–645 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McConnell, G. Confocal laser scanning fluorescence microscopy with a visible continuum source. Opt. Express12, 2844–2850 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Lee, D. D. & Seung, H. S. Learning the parts of objects by non-negative matrix factorization. Nature401, 788–791 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Hoyer, P. O. Modeling receptive fields with non-negative sparse coding. Neurocomputing52-54, 547–552 (2003). [Google Scholar]
- 24.Zimmermann, T., Marrison, J., Hogg, K. & O’Toole, P. in Confocal Microscopy: Methods and Protocols (ed Stephen W. Paddock) 129–148 (Springer New York, 2014).
- 25.Keshava, N. & Mustard, J. F. Spectral unmixing. IEEE Signal Process. Mag.19, 44–57 (2002). [Google Scholar]
- 26.Vyas, S., Zaganjor, E. & Haigis, M. C. Mitochondria and cancer. Cell166, 555–566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fletcher, D. A. & Mullins, R. D. Cell mechanics and the cytoskeleton. Nature463, 485–492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lappalainen, P., Kotila, T., Jégou, A. & Romet-Lemonne, G. Biochemical and mechanical regulation of actin dynamics. Nat. Rev. Mol. Cell Biol.23, 836–852 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Tang, D. & Wang, Y. Cell cycle regulation of Golgi membrane dynamics. Trends Cell Biol.23, 296–304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valm, A. M. et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature546, 162–167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matheu, M. P., Cahalan, M. D. & Parker, I. Immunoimaging: studying immune system dynamics using two-photon microscopy. Cold Spring Harb. Protoc.2011, pdb.top99 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Ruocco, M. G. et al. Suppressing T cell motility induced by anti-CTLA-4 monotherapy improves antitumor effects. J. Clin. Invest.122, 3718–3730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cahalan, M. D. & Parker, I. Choreography of cell motility and interaction dynamics imaged by two-photon microscopy in lymphoid organs. Ann. Rev. Immunol.26, 585–626 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi, S. et al. Long-term intravital imaging of the multicolor-coded tumor microenvironment during combination immunotherapy. ELife5, 10.7554/elife.14756 (2016). [DOI] [PMC free article] [PubMed]
- 35.Rios, A. C. et al. Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell35, 618–632 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Laughney, A. M. et al. Single-cell pharmacokinetic imaging reveals a therapeutic strategy to overcome drug resistance to the microtubule inhibitor eribulin. Sci. Transl. Med.6, 261ra152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature450, 56–62 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Momoh, J. et al. Intravital microscopy for real-time monitoring of drug delivery and nanobiological processes. Adv. Drug Deliv. Rev.189, 114528 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Liang, X. B. & Fu, L. Enhanced self-phase modulation enables a 700-900 nm linear compressible continuum for multicolor two-photon microscopy. IEEE J. Sel. Top. Quantum Electron. 20, 10.1109/JSTQE.2013.2253449 (2014).
- 40.Lozovoy, V. V., Pastirk, I. & Dantus, M. Multiphoton intrapulse interference.IV.Ultrashort laser pulse spectral phase characterization and compensation. Opt. Lett.29, 775–777 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Pastirk, I., Cruz, J. M. D., Walowicz, K. A., Lozovoy, V. V. & Dantus, M. Selective two-photon microscopy with shaped femtosecond pulses. Opt. Express11, 1695–1701 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Meshulach, D. & Silberberg, Y. Coherent quantum control of two-photon transitions by a femtosecond laser pulse. Nature396, 239–242 (1998). [Google Scholar]
- 43.Isobe, K. et al. Multifarious control of two-photon excitation of multiple fluorophores achieved by phase modulation of ultra-broadband laser pulses. Opt. Express17, 13737–13746 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Xu, C. & Webb, W. W. Measurement of two-photon excitation cross sections of molecular fluorophores with data from 690 to 1050 nm. J. Opt. Am. B13, 481–491 (1996). [Google Scholar]
- 45.Zhou, Z. et al. Adaptive optical microscopy via virtual-imaging-assisted wavefront sensing for high-resolution tissue imaging. PhotonX3, 3 (2022). [Google Scholar]
- 46.Shen, Z., Lu, Z., Chhatbar, P. Y., O’Herron, P. & Kara, P. An artery-specific fluorescent dye for studying neurovascular coupling. Nat. Methods9, 273–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Data generated in the study are included in the manuscript and its Supplementary Information. Source data are provided in this paper.
The unmixing code written in MATLAB is contained in Supplementary Software along with a demo image and instruction (Supplementary Software 1).