Abstract
Cartilage rarely heals spontaneously once damaged. Osteoarthritis (OA) is the most common degenerative joint disease among the elderly; however, effective treatment for OA is currently lacking. Autologous chondrocyte implantation (ACI), an innovative regenerative technology involving the implantation of healthy chondrocytes, may restore damaged lesions. Chondrocytes for ACI may potentially be induced from differentiated somatic cells using retrovirus (RV)-mediated transduction of three reprogramming factors (SOX9, KLF4, and c-MYC). However, the efficiency of the current induction system needs to be improved and the safety issues arising from the genomic integration of the vector DNA have to be addressed. To solve these problems, we used an RNA vector, termed the replication-defective and persistent Sendai virus vector (SeVdp), to express reprogramming factors for chondrocyte induction. Our results showed that the SeVdp-based vector induced chondrocytes more efficiently than the RV vector, probably because of robust and rapid expression of the transgenes, without any apparent integration of the SeVdp vector. The induced chondrocytes formed cartilage-like tissues when injected subcutaneously into mice. Thus, the SeVdp-based system for inducing chondrocytes may act as a foundation for developing safer and more effective treatments for damaged cartilage.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-77508-1.
Keywords: Chondrocyte, Direct reprogramming, Sendai virus vector, Osteoarthritis
Subject terms: Transdifferentiation, Reprogramming, Gene delivery
Introduction
Osteoarthritis (OA) is a degenerative joint disease that damages the articular hyaline cartilage, predominantly in the elderly, and causes chronic pain that limits mobility in patients with OA. Joint cartilage plays a crucial role in limb movements by absorbing impacts and lubricating the surfaces of joint bones. Owing to its poor vascularity, the capacity of joint cartilage to regenerate when damaged or injured is limited. Although various therapies are available to manage pain and improve mobility, an effective cure for OA is currently lacking. Autologous chondrocyte implantation (ACI) is an innovative technology for repairing cartilage lesions using chondrocytes that have been isolated from patients and expanded ex vivo1,2. However, ACI has not been widely used in treating OA, partially because of the limited availability of healthy cartilage for treating patients with OA. Therefore, an alternative source for obtaining chondrocytes in large amounts needs to be developed for enhancing the applicability of ACI in treating OA.
Chondrocyte generation by direct reprogramming is a promising method for obtaining sufficient numbers of chondrocytes for ACI. Chondrocytes can be directly induced from differentiated somatic cells by transducing three reprogramming factors (SOX9, KLF4, and c-MYC)3,4. The method depends on the forced expression of the reprogramming factors, KLF4 and c-MYC, together with the chondrocyte-specific transcription factor SOX9, generating chondrocytes directly from somatic cells, such as dermal fibroblasts, without going through the iPSC stage. This reprogramming system enables the generation of large numbers of chondrocytes from cells readily available in patient tissues. Despite these potential advantages, improvements in the efficiency of chondrocyte induction and safety of gene transduction are required before applying direct reprogramming of chondrocytes to ACI.
In a previous study, we performed alanine scanning mutagenesis of SOX9 and identified a SOX9 variant (SOX9H131A/K398A) with potent chondrogenic activity5. The use of SOX9H131A/K398A improves the efficiency of chondrocyte induction compared with the original wild-type SOX9. Nevertheless, the induction efficiency remains low, possibly because of the heterogeneous expression of reprogramming factors from retroviral (RV) vectors. Moreover, the use of RV vectors involves genomic integration6,7, which ensures long-term expression of transduced genes, but also entails a potential risk of accumulating undesired mutations and inappropriate activation of genes, which may eventually lead to malignancy after transplantation8. For instance, tumor formation in transplanted iPSCs, possibly caused by multiple RV integrations,9 and leukemia development after RV-based gene therapy for X-linked severe combined immunodeficiency have been reported 10. Thus, gene transduction without genomic integration is imperative for the clinical application of chondrocytes generated via direct reprogramming.
RNA viruses that do not integrate into the host genome, including negative-sense single-stranded RNA viruses, provide a safe platform for vectors to transduce exogenous genes into cells. Sendai virus (SeV)-based vectors are attractive options because of their broad cell tropism and low pathogenicity in human cells. SeV vectors have been used for transcription factor-mediated reprogramming to generate iPS cells11 or cardiomyocyte-like cells12 free of genomic integration. One of the Sev-based vectors, the replication-defective and persistent SeV vector (SeVdp), originally developed from the non-cytopathic and persistent SeV Cl.151 strain11,13, remains stable in cells at a non-permissive temperature (38oC) without chromosomal integration. SeVdp vectors not only have excellent transduction efficiency and low cytopathic effects but also express multiple genes stably at high levels from a single vector.
In this study, we used the SeVdp vector to develop a reprogramming system that directly induces chondrocytes from mouse embryonic fibroblasts (MEFs). This SeVdp-based vector expressed reprogramming factors (SOX9H131A/K398A, KLF4, and c-MYC) more rapidly and at higher levels than the RV vectors, and induced chondrocytes more efficiently. Genomic polymerase chain reaction (PCR) analysis did not detect integration of the SeVdp vector in the genome of the induced chondrocytes. Cartilage-like tissues were formed in vivo when SeV-induced chondrocytes were subcutaneously transplanted into mice. Thus, the SeVdp-based vector for direct reprogramming will boost further improvements in the development of a safe and efficient chondrocyte induction system.
Results
Sendai virus-based vector directs reprogramming of fibroblasts to chondrocytes
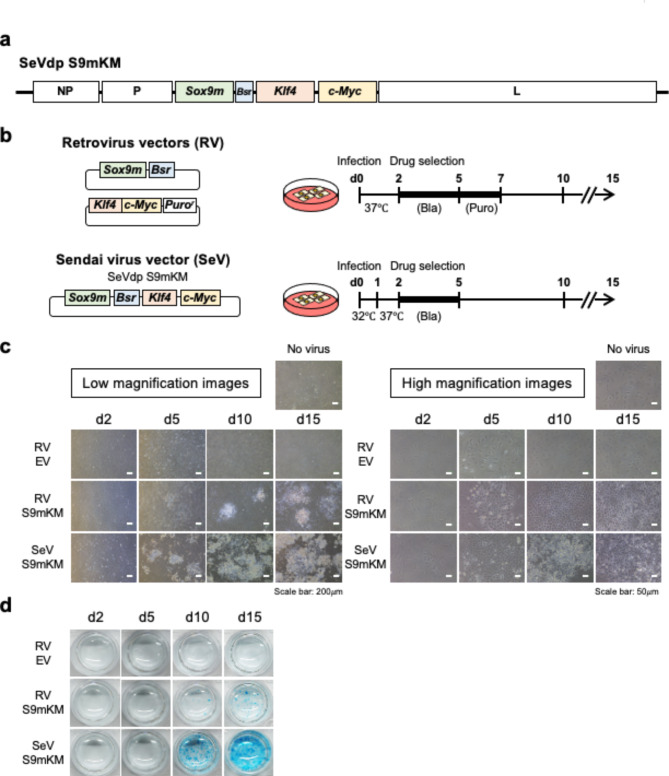
To prepare an SeV vector that expresses the reprogramming factors, SOX9, KLF4, and c-MYC, to induce chondrocytes, we inserted cognate genes and the blasticidin resistance gene (Bsr) into an SeVdp vector11 (Fig. 1a). The SeVdp vector, originally developed from the non-cytopathic strain Cl.151, is capable of stable and long-term expression of multiple transgenes with minimal cytopathic effects11. Moreover, instead of wild-type SOX9, we used SOX9H131A/K398A (SOX9m), which has a mutated DNA-binding domain and SUMOylation site, to improve the efficiency of chondrocyte induction5 (Supplementary Fig. S1). The SeV-based vector was used to infect MEFs (Fig. 1b) and uninfected cells were removed via blasticidin treatment before chondrocyte induction. The SeV vector also induced chondrocytes in mouse dermal fibroblasts (MDFs) and adipose tissue-derived mesenchymal stem cells, which are readily available for regenerative medicine (Supplementary Fig. S2). For comparison, chondrocyte induction from MEFs using RV-based vectors was performed as described previously5 (Fig. 1b). When MEFs were infected with RV vectors, polygonal or round chondrocytes appeared in clusters on day 5 and expanded further on day 10 (Fig. 1c, RV S9mKM). No such morphological changes were observed in cells infected with the empty RV vector (Fig. 1c, RV EV). In contrast, MEFs infected with the SeV vector generated similar chondrocyte clusters in greater numbers than those infected with the RV vector (Fig. 1c, SeV S9mKM). The cell clusters observed in SeV- or RV vector-infected cells were positively stained with Alcian blue, with more intense staining observed in SeVdp-infected cells, indicating that SeV vector-induced chondrocytes secreted extracellular matrix more abundantly (Fig. 1d). These data indicated that the SeV vector induced chondrocytes more efficiently than the RV vectors used in our previous study.
Fig. 1.
Sendai-virus-based direct reprogramming system for chondrocytes. (a) Structure of the Sendai virus (SeV) vector for inducing chondrocytes. Complementary DNAs encoding SOX9H131A/K398A (SOX9m), blasticidin-resistant gene (Bsr), KLF4, and c-MYC were inserted into the replication-deficient persistent SeVdp vector as shown. (b) Structures of the retrovirus vectors and Sendai virus vector used in this study. Experimental schemes for inducing chondrocytes using the retorivirus vectors or the Sendai virus vector are shown. (c) Morphological changes of virus-infected MEFs 2, 5, 10, and 15 d after infection with the empty retrovirus vector (RV EV), retrovirus vectors with the direct reprogramming factors (RV S9mKM), or Sendai virus vector with the direct reprogramming factors (SeV S9mKM). (d) Alcian blue staining of the differentiated chondrocytes.
SeV-based reprogramming vector induces chondrocytes rapidly and efficiently
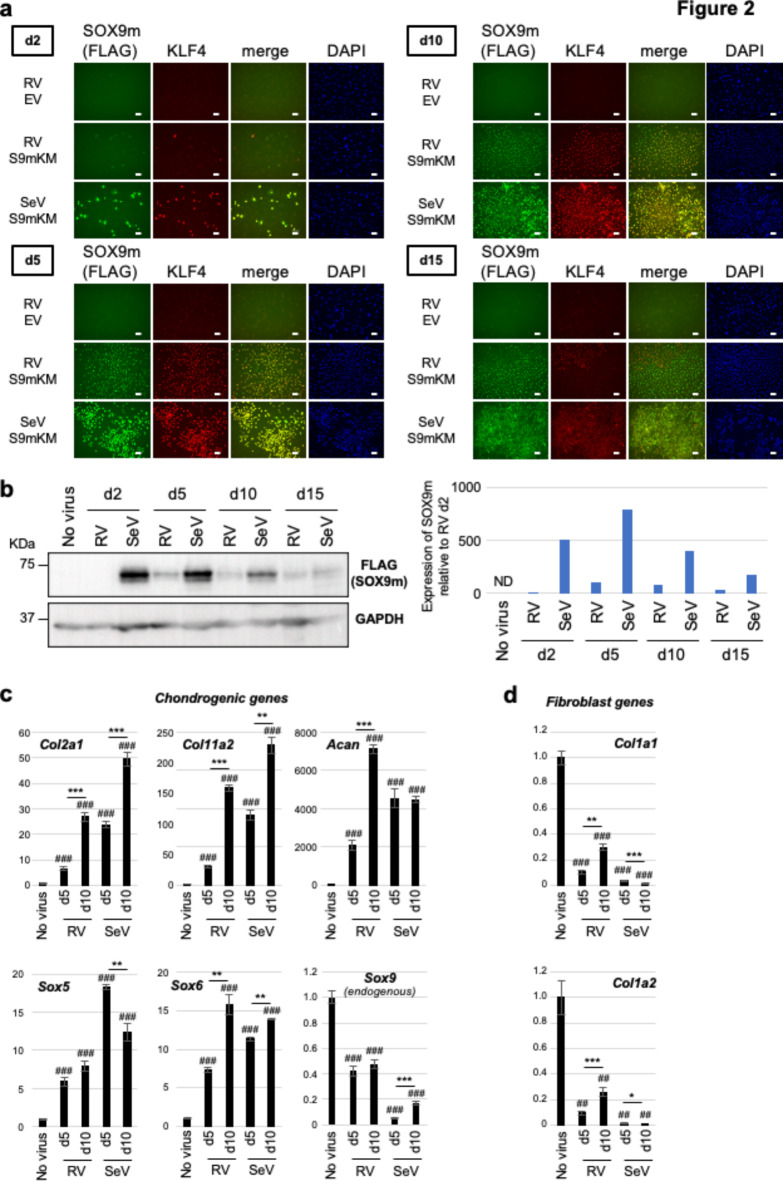
To quantitatively assess the molecular mechanism via which the SeV vector efficiently induced chondrocytes, we analyzed the levels of reprogramming factors expressed from each vector during chondrocyte induction. We first performed immunofluorescence staining of RV- and SeV-infected cells using antibodies against the FLAG tag (FLAG-tagged SOX9m), KLF4, and c-MYC. These reprogramming factors were expressed at low levels in a small fraction of RV-infected cells on day 2 but showed higher expression levels in a large number of cells on days 5 and 10 (Fig. 2a and Supplementary Fig. S3a, RV S9mKM). As compared to RV-infected cells, SeV-infected cells expressed SOX9m, KLF4, and c-MYC on day 2 at a higher level in a larger fraction of cells, and high expression levels of the reprogramming factors were maintained on days 5 and 10 (Fig. 2a and Supplementary Fig. S3a, SeV S9mKM). Importantly, SOX9m and KLF4 expression was more homogenous in SeV-infected cells than in RV-infected cells (Fig. 2a, merge). Western blot analysis also showed that the level of SOX9m expression from the SeV vector was high on days 2 and 5 and decreased gradually after day 5. In contrast, SOX9m expression from the RV vector was detectable on day 5, but was lower than that from the SeV vector and decreased slightly on days 10 and 15 (Fig. 2b). Similar expression patterns were observed for KLF4 (Supplementary Fig. S3b). These data suggest that stronger and more rapid expression of reprogramming factors by the SeV vector than by the RV vectors may be critical for the efficient induction of chondrocytes.
Fig. 2.
Rapid and strong expression of the direct reprogramming factors by the Sendai virus vector. (a) Immunofluorescence staining of the retrovirus (RV)- or Sendai virus (SeV)-infected cells using an anti-FLAG tag (Sox9H131A/K398A) antibody and an anti-KLF4 antibody. (b) Western blotting analysis of Sox9H131A/K398A expression in RV- or SeV-infected cells. Quantified data were normalized to GAPDH levels and are shown in the graph on the right. (c) Expression levels of the chondrogenic genes in RV- or SeV-infected cells. n = 3. **p < 0.01; ***p < 0.001; # represents a significant difference versus uninfected cells (No virus) (###p < 0.001). (d) Expression levels of the fibroblast genes in RV- or SeV-infected cells. n = 3. *p < 0.05; **p < 0.01; ***p < 0.001; # represents a significant difference versus uninfected cells (##p < 0.01; ###p < 0.001).
Consistent with the strong and rapid expression of SOX9m, KLF4, and c-MYC (Fig. 2a, b, and Supplementary Fig. S3), SeV infection upregulated the expression of chondrogenic genes (Col2a1, Col11a2, Acan, Sox5, and Sox6) on day 5 to levels comparable with those upregulated in RV-infected cells on day 10 (Fig. 2c). The expression of endogenous Sox9 correlated inversely with the levels of vector-derived SOX9m (Fig. 2b, c). In addition, fibroblast-related genes (Col1a1 and Col1a2) were significantly repressed 5 days after viral infection in both RV- and SeV-infected cells; however, these fibroblast genes were derepressed on day 10 in RV-infected cells, possibly indicating the dedifferentiation of chondrocytes in 2D culture conditions14. In contrast, the derepression of fibroblast genes was not observed in SeV-infected cells (Fig. 2d). Together, these results suggest that the strong and rapid expression of reprogramming factors from the SeV vector enables efficient induction of chondrocytes and may prevent dedifferentiation of the induced chondrocytes.
SeV-based reprogramming vector induces chondrocytes in three-dimensional (3D) pellet culture
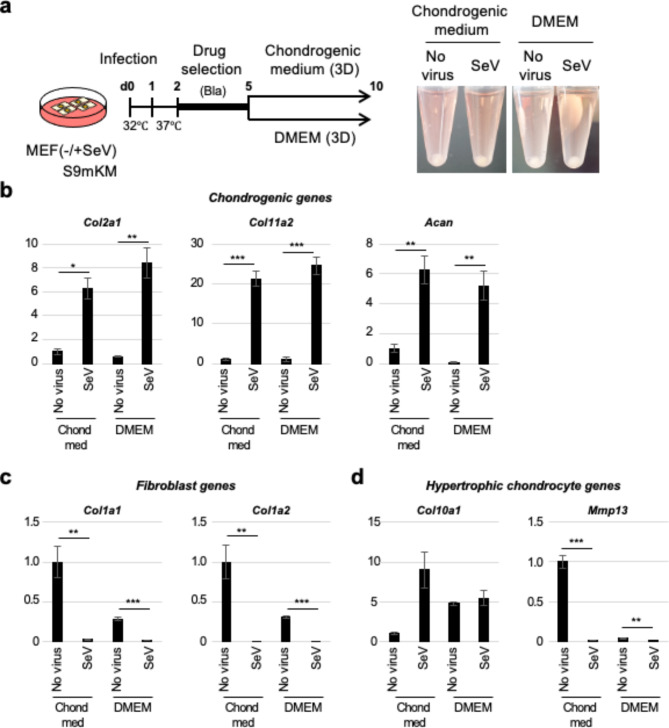
We then tested SeV-mediated chondrocyte induction in 3D culture, a more physiologically relevant culture system that allows extensive cell-cell interactions that mimic the native in vivo environment of cartilage. SeV-infected cells were selected with blasticidin and then cultured as a pellet, either in standard cell culture medium (Dulbecco’s modified Eagle medium [DMEM] plus 10% fetal bovine serum) or chondrogenic medium that included TGFβ and GDF5 (Fig. 3a). Regardless of the medium used (Fig. 3b), the SeV-infected cells expressed chondrogenic genes (Col2a1, Col11a2, and Acan) at significantly higher levels than the uninfected control cells, and repressed fibroblast gene expression (Col1a1 and Col1a2) to almost undetectable levels (Fig. 3c). In addition, there was no significant increase in the expression of the hypertrophic chondrocyte gene, Col10a1, in SeV-infected cells (Fig. 3d). Moreover, the expression of Mmp13 was significantly downregulated in SeV-infected cells (Fig. 3d). These data show that the 3D pellet culture of the SeV vector-infected cells efficiently induce chondrocytes and may minimize the requirement for growth factors, such as TGFβ and GDF5.
Fig. 3.
Three-dimensional pellet culture of SeV-induced chondrocytes. (a) Experimental scheme for chondrocyte induction from SeV-infected cells in three-dimensional (3D) pellet culture. The right panels show macroscopic observations of the SeV-infected cells on day 10. (b) Expression of chondrogenic genes in the 3D pellet cultured cells. n = 2 ~ 4. *p < 0.05; **p < 0.01; ***p < 0.001 (c) Expression of fibroblast genes in the 3D pellet cultured cells. n = 3 ~ 4. **p < 0.01; ***p < 0.001 (d) Expression of the hypertrophic chondrocyte genes in the 3D pellet cultured cells. n = 2 ~ 4. **p < 0.01; ***p < 0.001.
SeV-based reprogramming vector does not integrate into the genome of induced chondrocytes
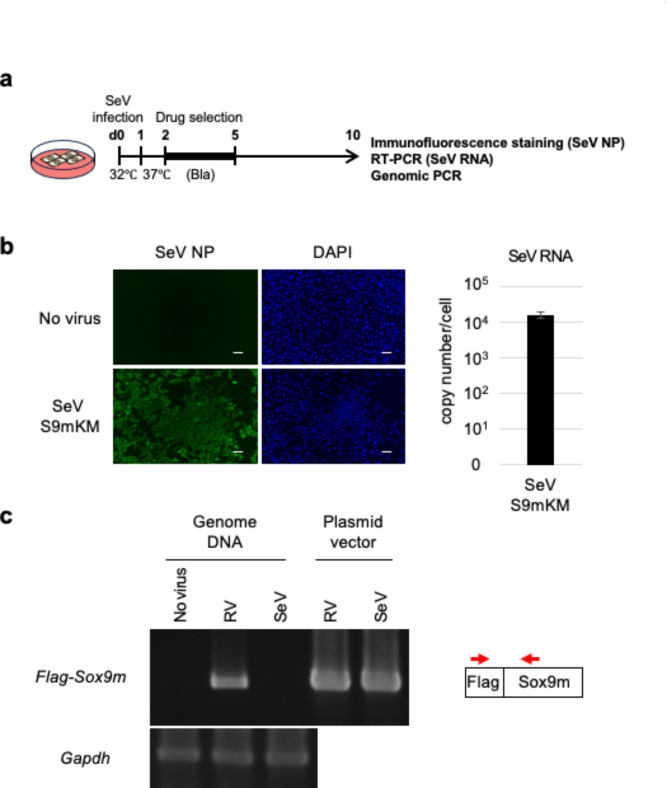
RV-based vectors are integrated into the host genome and may cause insertional mutations or malignant transformation of host cells, posing a potentially serious risk to cell-based therapies. In contrast, the SeV vector genome is a negative-sense single-stranded RNA that resides in the cytoplasm and replicates via an RNA intermediate (positive-stranded anti-genomic RNA). Theoretically, because of this unique replication mechanism, SeVs and their derived vectors are not believed to integrate into the host genome. To confirm that the SeV vector-derived sequence was not present in the genome of the induced chondrocytes, we performed PCR analysis of the genomic DNA of the induced chondrocytes (Fig. 4a). As shown in Fig. 4b, most SeV-infected cells expressed high levels of the viral NP protein, and the SeV RNA was detected at the expected level (~ 4 × 104 copies per cell)13. Genomic PCR using a primer set specific for the virus-specific FLAG-tagged Sox9 detected PCR products of the expected size in the RV-infected cells but not in the SeV-infected or uninfected cells. As a control, the cellular gene encoding glyceraldehyde 3-phosphate dehydrogenase (Gapdh), was detected in the genomes of both RV- and SeV-infected cells, and the RV and SeV vector plasmids generated PCR products of the same size (Fig. 4c). These data indicated that the SeV vector-derived sequence was not integrated into the genome of the induced chondrocytes.
Fig. 4.
No integration of the SeV vector in induced chondrocytes. (a) Experimental scheme for chondrocyte induction and genomic PCR. (b) Detection of the SeV NP protein using immunofluorescence staining (left panel) and SeV RNA using RT-qPCR (right panel) in SeV-infected cells. Scale bar: 100 μm. (c) PCR analysis of the virus vector-derived sequence (FLAG-tagged Sox9m) in genomic DNA prepared from noninfected, RV-infected, or SeV-infected cells. The position of the primers used for genomic PCR is shown in red arrows.
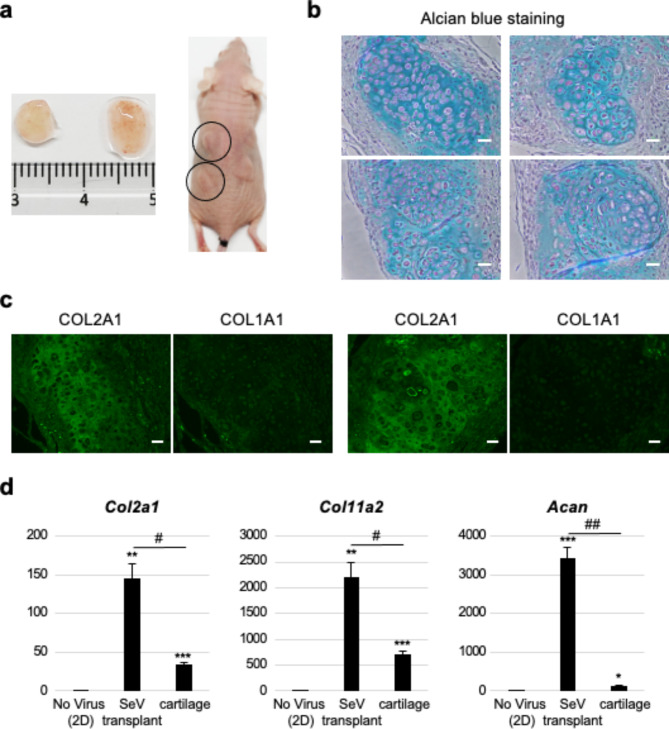
Chondrocytes induced by the SeV-based reprogramming vector form cartilage-like tissuesin vivo
To validate the ability of the SeV-induced chondrocytes to form cartilage in vivo, we injected SeV-infected cells into the subcutaneous space on the back of immunodeficient mice. Three weeks after the injection, the mice were euthanized and the transplants were isolated (Fig. 5a). Alcian blue staining of the tissue sections revealed chondrocytes in the lacunae surrounded by abundant extracellular matrix, which was consistent with the histological features observed in the cartilage (Fig. 5b). Notably, some areas of the cartilaginous tissue were populated with an array of cells that formed a zone-like structure reminiscent of chondrocytes undergoing endochondral ossification (Fig. 5b, right lower panel). As expected, the transplants showed significant induction of chondrogenic genes (Fig. 5d) and type II collagen, but not type I collagen (Fig. 5c). The expression levels of chondrogenic genes were higher than those in the mouse cartilage (Fig. 5d), probably because of the strong transcriptional induction by SeV-derived reprogramming factors. These results demonstrated that SeV-induced chondrocytes retained their ability to form cartilage-like tissues in vivo
Fig. 5.
In vivo cartilage formation from SeV-induced chondrocytes. (a) Formation of cartilaginous tissues from the transplanted SeV-induced chondrocytes in mice. (b) Alcian blue staining of the tissue sections. Scale bar: 25 μm (c) Expression of chondrogenic genes in the cartilaginous tissues generated from SeV-induced chondrocytes. n = 2. **p < 0.01; ***p < 0.001.
Discussion
In this study, we developed an SeV vector that directly induces chondrocytes from MEFs by simultaneously expressing three reprogramming factors (SOX9H131A/K398A, KLF4, and c-MYC) and showed that the newly developed SeV vector was superior to our previous RV vector in inducing chondrocytes. First, the SeV-based vector induced chondrocytes more efficiently than the RV vectors (Figs. 1 and 2), which is consistent with the high efficiency of gene delivery and robust expression of reprogramming factors by SeV-based vectors reported for reprogramming somatic cells into iPSCs11 and fibroblasts directly into cardiomyocytes11,12. The high expression levels of reprogramming factors from SeV vectors are possibly due to their high copy number (104–105 copies per cell)13 compared to only ~ 10 copies per cell of RVs15. Second, the SeV vector achieved more immediate expression of reprogramming factors than our previous RV vectors. This is presumably because SeVs possess an RNA-dependent RNA polymerase that instantaneously initiates transcription and replication in the cytoplasm16. In contrast, RVs undergo reverse transcription and integration into the host genome7, where viral gene expression typically occurs after host cell division17, which is entirely dependent on the cellular transcriptional machinery. Third, the SeV vector showed a relatively uniform expression of transgenes11, which ensured the production of a more homogenous population of induced chondrocytes. SeV vectors incorporate multiple exogenous genes into a single vector, from which the genes are expressed in a relatively constant stoichiometry. In addition, the levels of transgene expression among the infected cells varied to a lesser extent than those expressed by the RV vectors, for which the expression varied widely with the integrated genomic sites. Overall, the newly developed SeV vector enables the efficient induction of a more homogenous population of chondrocytes in a shorter period of cell culture and is therefore better suited for the scalable production of chondrocytes.
The presence of integrated transgenes in the genome is an obvious cause of concern when transcription factor-induced chondrocytes are used in regenerative medicine, largely because of their potential to form tumors after transplantation. This is particularly true when the reprogramming system requires c-Myc and Klf4, which act as oncogenes18,19. Indeed, chondrocytes induced by RV vector-directed reprogramming have been reported to generate tumors after transplantation3. Considering that SeV is a negative-sense single-stranded RNA virus (NSRV), it is generally believed that SeV does not integrate into the host genome. However, some reports have suggested that RNA viruses that do not generate DNA intermediates for replication can accidentally integrate into the host genome20–22. As shown in Fig. 4c, we did not observe integration of the current SeV vector into the genome of induced chondrocytes, at least during the period required for cell culture, indicating that the current SeV vector poses little concern for integration into the host genome.
Despite these substantial advantages of SeV vectors over RV vectors, certain limitations must be overcome to further improve the transcription factor-mediated induction of chondrocytes. As shown in Fig. 5b, some chondrocytes appeared enlarged, possibly indicating hypertrophic changes due to differentiation via the endochondral ossification pathway. Articular chondrocytes in osteoarthritic lesions often display a hypertrophic phenotype that is believed to play a causative role in OA progression. Although gene expression analyses indicated that SeV-induced chondrocytes did not display any evident gene expression signature of hypertrophic chondrocytes, they may easily respond to external cues to become hypertrophic. If these chondrocytes are implanted into an osteoarthritic lesion, they can exacerbate rather than ameliorate OA. To prevent this untoward hypertrophic differentiation, an SeV vector carrying a dominant negative form of a transcription factor (such as Runx or Osterix) that plays crucial roles in chondrocyte hypertrophy and subsequent differentiation into osteocytes may be helpful23,24.
Another limitation of current SeV vectors is their immunogenicity. Although the SeV vector used in this study lacks envelope-related genes and largely escapes the innate immune response, it still elicits an acquired immune response25 that could eliminate transplanted cells in vivo. For instance, in immunodeficient mice, transplanted SeV-infected cells remain intact for at least 60 days; however, they disappear after only a few days in immunocompetent mice, even when immunosuppressants are administered26. Therefore, removal of the SeV vector from induced chondrocytes before implantation may be beneficial for the long-term preservation of implanted cells. Possible methods for removing SeV vectors from reprogrammed cells include the use of a vector backbone derived from temperature-sensitive mutations27, treatment of reprogrammed cells with an siRNA targeting the SeV L gene11, inclusion of a target sequence of cell-type-specific miRNAs,28 or installation of a Csy4 endoribonuclease-based system in the vector29,30. The incorporation of these technologies into the current vector may lead to the development of an SeV vector that generates chondrocytes free of the SeV vector initially used to induce them.
In summary, the SeV vector-based direct reprogramming system reported in this study not only represents a considerable advancement in generating induced chondrocytes for cell therapy but also provides a solid foundation for further improvements in the scalable production of vector-free chondrocytes.
Methods
Preparation of viral vectors
The recombinant DNA experiments performed in this study were approved by the Recombinant DNA Experiment Committee of the University of Tsukuba (approval numbers: 190121, 210260, and 220144). Complementary DNAs encoding human SOX9H131A/K398A, KLF4, c-MYC, and blasticidin resistance genes were inserted into the SeVdp vector11 (Fig. 1a). Using the SeVdp vector and expression vectors for SeV proteins, a virus-containing culture medium was prepared as described previously11, except that NIH3T3 cells were used to determine the titer. The retroviral vectors were prepared as described previously5.
Induction of chondrocytes using the SeV-based direct reprogramming system
MEFs were prepared from C57BL/6J mouse embryos as described previously31. MEFs were cultured at 37℃ in an atmosphere of 5% O2 and 5% CO2 in DMEM containing 10% (v/v) fetal bovine serum, 100 units/mL penicillin, and 100 µg/mL streptomycin (hereafter referred to as standard medium). The MEFs were seeded in a 24-well plate at a density of 4 × 104 cells/well and were infected with SeV on the next day at a multiplicity of infection (MOI) of approximately 1.0 for 24 h at 32℃ in an atmosphere of 5% CO2. The virus-infected cells were then cultured at 37℃ in an atmosphere of 5% O2 and 5% CO2 for 24 h. The uninfected cells were removed by culturing in standard medium containing 8 µg/mL blasticidin for 3 d. The surviving cells were further cultured at 37℃ in an atmosphere of 5% O2 and 5% CO2 in standard medium without blasticidin, and the medium was changed every 2 d. The chondrogenic medium was DMEM containing 1% (v/v) fetal bovine serum, 10 ng/mL GDF5 (Biolegend, San Diego, CA, USA; 779506), 10 ng/mL TGF-β1 (Biolegend, 580702), 50 µg/mL L-ascorbic acid 2-phosphate (Sigma, St Louis, MO, USA; A8960), 0.1 µM dexamethasone (Nacalai Tesque, Inc., Tokyo, Japan; 11107-64), 1 mM sodium pyruvate (Nacalai Tesque, Inc.; 06977-34), and ITS (Thermo Fisher Scientific Inc., Waltham, MA, USA; 41400045). Phase-contrast images of the cells were acquired using a Nikon ECLIPS TS100 microscope.
Preparation of stromal vascular fraction (SVF) cells derived from inguinal white adipose tissue (iWAT) and mouse dermal fibroblasts (MDFs)
iWAT was isolated from a 5-month-old female mouse and was minced. The minced iWAT was dispersed using collagenase (Roche, ~ 1.5 units/g tissue) and dispase II (Roche, ~ 2.4 units/g tissue) in phosphate buffered saline (PBS) containing 10 mM CaCl2 (~ 1 ml/g tissue) at 37℃ for 1 h. The dispersed cells were centrifuged at 700 × g for 10 min, and the pellet was isolated as SVF cells. The SVF cells suspended in standard medium were filtered through a 70 μm strainer and cultured at 37℃ in an atmosphere of 20% O2 and 5% CO2. To isolate MDFs, the hair around the chest of the same female mouse was shaved, and the exposed skin was removed and minced. The skin tissue was dispersed using collagenase and dispase II, and the dispersed cells were filtered and cultured as described for the SVF cells. Chondrocytes were induced using the same method as used for MEFs, except that the cells were cultured in the presence of 20% O2 instead of 5% O2.
Alcian blue staining
The induced chondrocytes were washed with PBS and fixed in methanol at room temperature for 2 min. After removing methanol, the cells were washed using 0.1 M HCl and stained with Alcian blue staining solution (pH 2.5, Nacalai Tesque, Inc.) at room temperature for approximately 60 min. The cells were washed with 0.1 M HCl.
Immunofluorescence staining
The virus-infected cells were washed with PBS and fixed using 10% formalin (v/v) at room temperature for 10 min. Then, the cells were washed with PBS, treated with 50 mM NH4Cl at room temperature for 10 min, treated with 0.1% (w/v) Triton X-100 at room temperature for 5 min, and washed with PBS. The cells were then incubated with the primary antibody in 0.1% (w/v) saponin/PBS at room temperature for 60 min. The cells were washed with PBS and incubated with fluorescent-labeled secondary antibodies in 0.1% (w/v) saponin/PBS at room temperature for 60 min. After washing with 0.1% (w/v) saponin/PBS, the cells were treated with one drop of VECTASHIELD containing DAPI (Nacalai Tesque, Inc.). Fluorescent images were acquired using an All-in-One Fluorescence Microscope BZ-710 (Keyence corp.). The antibodies and dilutions used for immunofluorescence staining are listed in Supplementary Table S1.
Immunoblotting
MEFs were seeded in 60 mm plate at a density of 4 × 105 cells/plate and infected with RVs or SeV. The virus-infected cells were collected 2, 5, 10, and 15 days after virus infection and suspended in radioimmunoprecipitation assay buffer containing protease inhibitor cocktail, 1 µM MG132, and 0.5 mM phenylmethylsulfonyl fluoride (approximately 5 × 105 cells/50 µL). After brief sonication using Bioruptor II (30 s ON-30 s OFF, two cycles, power: high; BM Equipment Co., Ltd., Tokyo, Japan), the insoluble fraction was removed via centrifugation at 20,000 × g at 4℃ for 5 min. The cell extracts (approximately 28 µg protein each) were used for immunoblotting with an anti-DYKDDDDK (Flag-SOX9) antibody (1:1,500 dilution). Chemiluminescent images were acquired using a Fusion FX7. EDGE (M&S Instruments Inc.) and quantification were performed using the Evolution Capt software, and the original images of the blots are shown in Supplementary Fig. S1.
Reverse transcription and quantitative PCR (RT-qPCR)
Total RNA was extracted from the induced chondrocytes and RT-qPCR was performed as described previously5. The primer sets used for qPCR are listed in Supplementary Table S2. P-values were calculated using the Student’s t-test. To quantify the SeV genomic RNA, total RNA was purified from virus-infected cells and used for RT-qPCR. The amount of SeV genomic RNA was calculated based on a standard curve constructed using the plasmid DNA vector used for the SeV preparation.
Genomic PCR
Chondrocytes were induced by RV or SeV in a 12-well plate (approximately 105 cells/well) for 10 d and lysed using 100 µL of lysis buffer (50 mM Tris-HCl, pH 7.5, 1% SDS, 20 mM EDTA, 0.1 M NaCl) containing 50 µg/mL RNase A at 37℃ for 15 min. Proteinase K (2 µg) was added and the cells were incubated at 55℃ for 5 h. Genomic DNA was extracted using 100 µL of phenol/chloroform and precipitated by adding 100 µL of 2-propanol. After centrifugation at 15,500 × g at 4℃ for 7 min, the pellet was rinsed with 70% ethanol. The pellet was dissolved in 10 µL of TE (pH 8.0). For genomic PCR, 10 ng of genomic DNA or 1 ng of the plasmid vector was used as the template. To detect the virus vector-derived sequence (Flag-tagged Sox9m-coding gene), PCR was performed using KOD-plus-Neo (TOYOBO) with a two-step cycle (94℃, 2 min > [98℃, 10 s; 68℃, 30 s] × 35 cycles > 4℃). To detect Gapdh, PCR was performed using a three-step cycle (94℃, 2 min > [98℃, 10 s; 58℃, 30 s; 68℃, 20 s] × 35 cycles > 4℃). The primers used for PCR are listed in Supplementary Table S3. The PCR products were analyzed by electrophoresing on a 1% agarose gel. Original images of the gels are shown in Supplementary Fig. S2.
Transplantation of SeV-induced chondrocytes in mice
The animal experiments performed in this study were approved by the Animal Experimental Committee of the University of Tsukuba (approval numbers: 22–488 and 23–312). All experiments were performed in accordance with the relevant guidelines, regulations, and ARRIVE guidelines. MEFs were infected with SeV at an MOI approximately 1.0 in 100 mm plates, and uninfected cells were removed by treating with blasticidin (8 µg/mL) for 3 d. The virus-infected cells were cultured without blasticidin for 1 d and transplanted subcutaneously into the backs of immunodeficient mice (BALB/cAJc1-nu/nu, 6 weeks old, female). The mice were euthanized via cervical dislocation under isoflurane anesthesia 3 weeks after transplantation and the grafts were isolated. Paraffin sections were prepared from the grafts and stained using Alcian Blue. Immunofluorescence staining of paraffin-embedded sections was performed according to standard protocols using anti-COL2A1 or anti-COL1A1 antibody (Santa Cruz Biotechnology). Total RNA was extracted from the grafts and gene expression was analyzed using RT-qPCR.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the other laboratory members for their advice and helpful discussions. This work was supported by grants from JSPS KAKENHI grant numbers JP19K07343 and JP22K06878 (AF), JP19H03203 and JP23K27100 (KN), JP17H04036 and JP21H02678 (KH), and AMED JP22ym0126803 (AF). The funders had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation. We thank Editage for editing and reviewing the manuscript for English language.
Author contributions
J.Z., A.F., and Y.S. performed the experiments and analyzed the data. M.S. and K.N. provided experimental resources. A.F. managed the project and wrote the first manuscript draft. K.H. supervised the study and edited the manuscript. All the authors have reviewed and approved the final manuscript.
Data availability
The data underlying this article are available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Koji Hisatake, Email: kojihisa@md.tsukuba.ac.jp.
Aya Fukuda, Email: fukudaa@md.tsukuba.ac.jp.
References
- 1.Brittberg, M. et al. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N. Eng. J. Med.331, 889–895 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Peterson, L., Brittberg, M., Kiviranta, I., Åkerlund, E. L. & Lindahl, A. Autologous chondrocyte transplantation. Am. J. Sports Med.30, 2–12 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Hiramatsu, K. et al. Generation of hyaline cartilaginous tissue from mouse adult dermal fibroblast culture by defined factors. J. Clin. Investig.121, 640–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Outani, H. et al. Direct induction of chondrogenic cells from human dermal fibroblast culture by defined factors. PLoS ONE8, e77365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekiguchi, Y., Fukuda, A., Nishimura, K. & Hisatake, K. Engineering critical residues of SOX9 discovers a variant with potent capacity to induce chondrocytes. Stem Cells41, 1157–1170 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Kurian, K. M., Watson, C. J. & Wyllie, A. H. Retroviral vectors. J. Clin. Pathol. Mol. Pathol.53 (2000). [DOI] [PMC free article] [PubMed]
- 7.Chameettachal, A., Mustafa, F. & Rizvi, T. A. Understanding retroviral life cycle and its genomic RNA packaging. J. Mol. Biol.435, 167924 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Li, Z. et al. Murine leukemia induced by retroviral gene marking. Science1979(296), 497–497 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Aoi, T. et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science321, 699–702 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina, S. et al. LMO2 -associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science302, 415–419 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Nishimura, K. et al. Development of defective and persistent Sendai virus vector: A unique gene delivery/expression system ideal for cell reprogramming. J. Biol. Chem.286, 4760–4771 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyamoto, K. et al. Direct in vivo reprogramming with Sendai virus vectors improves cardiac function after myocardial infarction. Cell Stem Cell22, 91-103.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Nishimura, K. et al. Persistent and stable gene expression by a cytoplasmic RNA replicon based on a noncytopathic variant sendai virus. J. Biol. Chem.282, 27383–27391 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Benya, P. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell30, 215–224 (1982). [DOI] [PubMed] [Google Scholar]
- 15.Kustikova, O. S. et al. Dose finding with retroviral vectors: Correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood102, 3934–3937 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Bloyet, L.-M. The nucleocapsid of paramyxoviruses: structure and function of an encapsidated template. Viruses13, 2465 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salas-Briceno, K., Zhao, W. & Ross, S. R. Murine leukemia virus infection of non-dividing dendritic cells is dependent on nucleoporins. PLoS Pathog.20, e1011640 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhanasekaran, R. et al. The MYC oncogene—The grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol.19, 23–36 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, Z., He, J. & Xie, K. KLF4 transcription factor in tumorigenesis. Cell Death Discov.9, 118 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horie, M. et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature463, 84–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horie, M. & Tomonaga, K. Non-retroviral fossils in vertebrate genomes. Viruses3, 1836–1848 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, L. et al. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. USA118, 34 (2021). [DOI] [PMC free article] [PubMed]
- 23.Enomoto, H. et al. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J. Biol. Chem.275, 8695–8702 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Oh, J.-H., Park, S.-Y., de Crombrugghe, B. & Kim, J.-E. Chondrocyte-specific ablation of Osterix leads to impaired endochondral ossification. Biochem. Biophys. Res. Commun.418, 634–640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami, Y. et al. Newly-developed Sendai virus vector for retinal gene transfer: reduction of innate immune response via deletion of all envelope-related genes. J. Gene Med.10, 165–176 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Yamaki, Y. et al. Utilization of a novel Sendai virus vector in ex vivo gene therapy for hemophilia A. Int. J. Hematol.113, 493–499 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Ban, H. et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. USA108, 14234–14239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura, K. et al. Simple and effective generation of transgene-free induced pluripotent stem cells using an auto-erasable Sendai virus vector responding to microRNA-302. Stem Cell Res.23, 13–19 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Haurwitz, R. E., Jinek, M., Wiedenheft, B., Zhou, K. & Doudna, J. A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science329, 1355–1358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishimoto, T. et al. An engineered ligand-responsive Csy4 endoribonuclease controls transgene expression from Sendai virus vectors. J. Biol. Eng.18, 9 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu, L., Lai, W., Stumpo, D. & Blackshear, P. Mouse embryonic fibroblast cell culture and stimulation. Bio. Protoc.6 (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available from the corresponding author on reasonable request.