Abstract
Peroxisomes are organelles that are central to lipid metabolism and chemical detoxification. Despite advances in our understanding of peroxisome biogenesis, the mechanisms maintaining peroxisomal membrane proteins remain to be fully elucidated. We show here that mammalian FAF2/UBXD8, a membrane-associated cofactor of p97/VCP, maintains peroxisomal homeostasis by modulating the turnover of peroxisomal membrane proteins such as PMP70. In FAF2-deficient cells, PMP70 accumulation recruits the autophagy adaptor OPTN (Optineurin) to peroxisomes and promotes their autophagic clearance (pexophagy). Pexophagy is also induced by p97/VCP inhibition. FAF2 functions together with p97/VCP to negatively regulate pexophagy rather than as a factor for peroxisome biogenesis. Our results strongly suggest that p97/VCPFAF2-mediated extraction of ubiquitylated peroxisomal membrane proteins (e.g., PMP70) prevents peroxisomes from inducing nonessential autophagy under steady state conditions. These findings provide insight into molecular mechanisms underlying the regulation of peroxisomal integrity by p97/VCP and its associated cofactors.
Subject terms: Pexophagy, Ubiquitylation, Membrane proteins
The mechanisms maintaining peroxisomal membrane proteins remain to be fully elucidated. Here, the authors report that p97/VCP and FAF2/UBXD8 modulate the turnover of peroxisomal membrane proteins to prevent autophagic degradation of peroxisomes.
Introduction
Peroxisomes, which are crucial for lipid peroxidation, are maintained through the counter actions of biogenesis and degradation. While much of our understanding of peroxisome biogenesis is based on peroxisome deficiency disorders1, the detailed molecular mechanisms underlying peroxisome degradation remain enigmatic. Recent studies, however, suggest that ubiquitylation is important for autophagic peroxisomal degradation (pexophagy) (Fig. 1a)2–4.
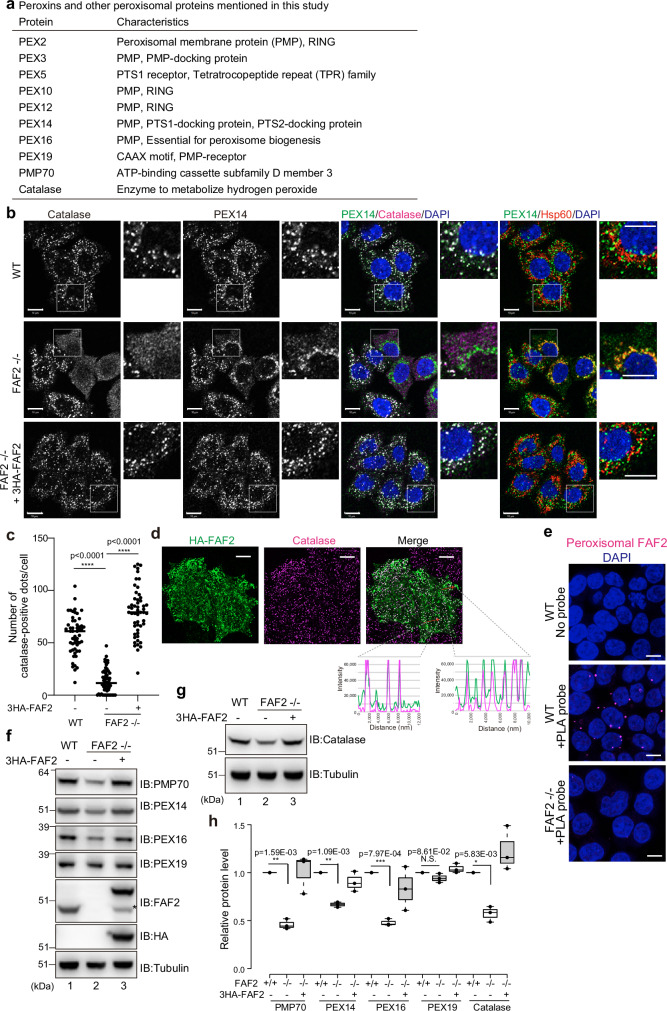
Fig. 1. FAF2 deletion reduces peroxisomal abundance.
a Characteristics of the peroxisomal proteins mentioned in this study. b Immunocytochemical images of WT, FAF2-/-, and FAF2-/- HCT116 cells stably expressing 3HA-FAF2 (+3HA-FAF2). Cell nuclei were stained with DAPI. PEX14 and catalase colocalize in WT and FAF2-/- HCT116 cells stably expressing 3HA-FAF2. Peroxisome-deficient cells (i.e., reduced number of catalase-positive dots) were observed in FAF2-/- HCT116 cells. In some FAF2-/- cells, catalase was cytosolic, and PEX14 colocalized with Hsp60 (mitochondrial marker). Higher magnification images of the boxed regions are shown. Scale bars, 10 µm. c. The number of catalase-positive peroxisomes per cell. Peroxisome abundance is significantly reduced in FAF2-/- cells. Dots represent individual data points from three independent experiments. Total number of cells from three independent experiments; n = 53 cells (17, 17, 19 cells/experiments; WT), n = 60 cells (20 cells/experiments; FAF2-/-), and n = 52 cells (17, 17, 18 cells/experiments; FAF2-/- cells + 3HA-FAF2). Bar, median. Statistical significance was calculated using one-way ANOVA; ****p < 0.0001. d Representative images of HA-FAF2 colocalized with peroxisomal catalase. HeLa cells transiently expressing HA-FAF2 were immunostained with anti-HA and anti-catalase antibodies. A line scan (red arrows in the merged panel) shows the colocalization of HA-FAF2 (green line) and catalase (magenta line). Scale bars, 10 µm. e Endogenous FAF2 localized to PMP70-positive peroxisomes. Localization was determined using an in situ proximity ligation assay. WT cells and FAF2-/- cells were reacted with anti-FAF2 and anti-PMP70 antibodies, followed by oligonucleotide-conjugated secondary antibodies (PLA probes). Red dots indicate FAF2-PMP70 interactions. Cell nuclei were stained with DAPI. Scale bars, 10 µm. f, g PMP70, PEX14, PEX16, and catalase are reduced in FAF2-/- cells. WT, FAF2-/-, and FAF2-/- HCT116 cells stably expressing 3HA-FAF2 were immunoblotted with the indicated antibodies. The asterisk indicates an exogenous FAF2 cleavage product that lacks the N-terminal 3HA-tag. h Quantitative analysis of peroxisomal protein levels for cells in f and g. Protein levels were normalized to WT HCT116 cells, which were set to 1. Dots represent individual data points from three independent experiments. Statistical significance was calculated using a one-tailed Welch’s t-test; *p < 0.05, **p < 0.01, ***p < 0.001; N.S. not significant. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Source data are provided as a Source Data file.
Ubiquitylation of organelles by ubiquitin ligase (E3s) accelerates their degradation, whereas deubiquitylase (DUB) activity prevents degradation. For example, Parkin-mediated ubiquitylation induces mitochondrial degradation via autophagy (mitophagy), whereas DUB USP30 counteracts mitophagy5–10. In concert with DUB activities, the ubiquitin content of organelles is reduced by extracting ubiquitylated membrane proteins via p97/VCP, an AAA+ ATPase ortholog of the yeast Cdc48 that is essential for many processes11. Ubiquitylation triggers p97/VCP-mediated extraction of mitofusin 2 (Mfn2) from the outer mitochondrial membrane and targets it for proteasomal degradation12,13. p97/VCP similarly extracts MITOL from damaged mitochondrial membranes following Parkin-mediated ubiquitylation, but with the extracted MITOL targeted to peroxisomes14. p97/VCP activity requires accessory proteins, and its substrate specificity is cofactor dependent13,15–17. The p97/VCP cofactor FAF2/UBXD8 (hereafter referred to as FAF2) localizes to multiple organelles such as mitochondria, the endoplasmic reticulum (ER), and lipid droplets16,18–20. FAF2 has multiple domains, including UBA, HP, and a ubiquitin-regulatory X (UBX) domain, and associates with organellar membranes via hairpin (HP) structure20,21. The yeast ortholog of FAF2, Ubx2, mediates interactions between Cdc48, ubiquitin ligases, and their substrates in response to ER-associated protein degradation (ERAD)21–24. FAF2 also removes precursor proteins arrested within the translocator channel in the outer mitochondrial membrane25. A link between FAF2 and peroxisomes was suggested by the role that peroxins PEX19 and PEX3 play in the post-translational insertion of FAF2 into ER subdomains20. In addition, it has been recently reported that USP30 regulates pexophagy to maintain peroxisomal abundance26,27.
In this study, we report that FAF2, in conjunction with p97/VCP, prevents nonessential autophagic peroxisomal degradation by extracting ubiquitylated proteins such as PMP70 from peroxisomes. In FAF2-deficient cells, OPTN recruitment to peroxisomes promoted pexophagy, and functional inhibition of USP30 accelerated peroxisomal degradation. These results indicate that under steady-state conditions, FAF2-mediated extraction of ubiquitylated proteins from peroxisomal membranes governs ubiquitin-dependent peroxisomal degradation and that OPTN modulates pexophagy. Our study uncovered that FAF2 recruits p97/VCP to peroxisomes and maintains peroxisomal abundance by contributing to the efficient removal of peroxisomal membrane proteins.
Results
Genetic loss of FAF2 causes a significant reduction of peroxisomal abundance
p97/VCP-mediated extraction of MITOL from the mitochondrial membrane is ubiquitylation dependent14. To identify a p97/VCP cofactor for the MITOL extraction, we knocked down five of the ~30 known UBX-containing proteins—NSFL1C/p47, UFD1, NPLOC4, Rep8/UBXD6, and FAF2 (Supplementary Fig. 1a–d). FAF2 knockdown impaired MITOL extraction (Supplementary Fig. 1e), whereas knockdown of the other cofactors had no effects (Supplementary Fig. 1e). Since ubiquitylated MITOL translocates from mitochondria to peroxisomes, we examined peroxisomal abundance in the absence of FAF2. FAF2-/- cell lines, generated via a CRISPR/Cas9 gene editing (Supplementary Fig. 2a), had fewer peroxisomes (Fig. 1b, c), suggesting that the reduction in peroxisomes impairs MITOL translocation.
We next focused on the role of FAF2 in the homeostatic regulation of peroxisomes. In wild-type (WT) HCT116 cells, catalase, a peroxisomal matrix protein, colocalized with the membrane protein PEX14, but not the mitochondrial Hsp60 (Fig. 1b). Conversely, in some FAF2-/- cells, the catalase was cytosolic and PEX14 colocalized with Hsp60, suggesting that FAF2 deletion drives peroxisomal loss (Fig. 1b). Although peroxisomal abundance varied in the FAF2-/- cells, which were expanded from a single clone, the number of peroxisomes per cell was significantly lower than that in WT cells (Fig. 1c). The expression of exogenous 3HA-tagged FAF2 (3HA-FAF2) restored peroxisomal abundance in FAF2-/- cells (Fig. 1b, c), indicating FAF2 regulates the peroxisomal loss. Although FAF2 localizes in multiple organelles16,18–20, its peroxisomal localization is yet to be fully demonstrated.
Shifting cells to anabolic conditions by the addition of oleic acid promotes lipid droplet accumulation. Repeating this, we found that a portion of HA-tagged FAF2 (HA-FAF2) colocalized to lipid droplets (Supplementary Fig. 2b), which were largely oleic acid dependent. HA-FAF2 also colocalized with catalase (Fig. 1d), suggesting that a portion of FAF2 localizes at peroxisomes in HeLa cells. FAF2 likewise partially colocalized with the peroxisome marker PMP70 in WT cells, but not in FAF2-/- cells (Supplementary Fig. 2c). While non-specific nuclei staining was observed with the anti-FAF2 antibody, additional non-peroxisomal FAF2-staining may reflect FAF2 localization on other organelles16,28,29. To confirm the peroxisomal localization of endogenous FAF2, we utilized an in situ proximity ligation assay (PLA assay) (Fig. 1e). Rabbit anti-FAF2 and mouse anti-PMP70 antibodies were used as primary antibodies, and oligonucleotide-conjugated antibodies (PLA probes) were used as the secondary antibody. After incubating with a DNA polymerase and fluorescence-labeled oligonucleotides, the presence of PLA dots would indicate that FAF2 and PMP70 are in close proximity (<40 nm). Red discrete spots were observed in WT cells in a PLA probe-dependent manner but not in FAF2-/- cells (Fig. 1e), indicating that endogenous FAF2 localizes on peroxisomes. Further, FAF2-/- cells had reduced levels of the peroxisomal proteins PMP70, PEX14, PEX16, and catalase, which returned to WT levels with 3HA-FAF2 expression (Fig. 1f–h). FAF2 loss had little to no effect on PEX19, most likely because PEX19 localizes mostly in the cytosolic (Fig. 1f–h). These results indicate that FAF2 localizes to peroxisomes and FAF2 loss promotes peroxisome loss and a reduction in peroxisomal proteins.
Pexophagy is accelerated in FAF2-/- cells
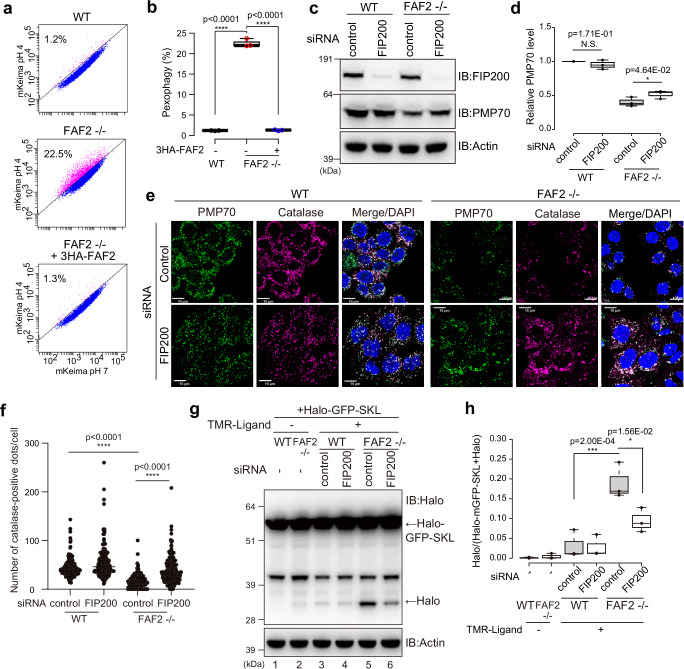
Two possibilities for peroxisomal loss in FAF2-/- cells are: (1) FAF2 depletion facilitates peroxisomal degradation, or (2) the deletion impedes peroxisomal biogenesis. To examine the former possibility, we established a flow cytometry method to monitor and quantitatively assess the pexophagy flux using a mKeima (Fig. 2a), a fluorescent protein with a pH-dependent excitation spectrum30. mKeima was targeted to the peroxisomal matrix by conjugating the SKL peroxisomal targeting signal to its C-terminus. If peroxisomes are delivered to lysosomes, the acidic environment of the lysosome will cause the excitation wavelength of mKeima-SKL to shift from 405-nm (neutral pH) to 561-nm. mKeima-SKL was stably expressed in WT or FAF2-/- cells (Supplementary Fig. 3a). Since PEX3 overexpression induces pexophagy31, WT cells expressing PEX3-YFP were used as a positive control. Pexophagy was observed in 15.0% of the PEX3-YFP-overexpressing cells (Supplementary Fig. 3b), but was 1.2% in WT cells overexpressing YFP alone (Fig. 2a). The pexophagy flux in FAF2-/- cells was ~20 times higher (22.5%) than that in WT cells (Fig. 2a, b) and was completely blocked by the lysosomal inhibitor bafilomycin A1 (Baf.A1) (Supplementary Fig. 3c,d). Exogenous expression of 3HA-FAF2 returned the flux to WT levels (1.3%), indicating that the observed acceleration in pexophagy was caused by the depletion of a FAF2 protein (Fig. 2a, b). To determine if peroxisome loss depends on macroautophagy, we knocked down an essential autophagy subunit (FIP200). PMP70 levels in FAF2-/- cells were slightly elevated relative to control siRNA-treated cells following FIP200 knockdown (Fig. 2c, d). The number of catalase-positive peroxisomes was elevated in FAF2-/- cells treated with FIP200 siRNA (Fig. 2e, f). To monitor pexophagy flux in FAF2-/- cells, we also used HaloTag (Halo)-based reporter assay32. Unlike ligand-free Halo, which is susceptible to lysosomal degradation, ligand-bound Halo is resistant. Consequently, when the SKL signal is used to target Halo-mGFP to the peroxisomal matrix, the ligand-dependent Halo band on immunoblots can be used as an indicator of pexophagy. Immunoblots of FAF2-/- cells stably expressing Halo-mGFP-SKL (peroxisome-targeted Halo-mGFP) had a prominent Halo band (Fig. 2g lane 5 and 2h), whereas the band in both FIP200 knockdown FAF2-/- cells (Fig. 2g lane 6) and WT cells (Fig. 2g lanes 3 and 4) was less pronounced. These results strongly suggest that the loss of peroxisomes in FAF2-/- cells is due to accelerated pexophagy. To further examine this, we treated FAF2-/- cells with Torin1, an mTOR inhibitor that induces autophagy by inhibiting the phosphorylation of ULK1 and Atg1333,34. While Torin1 treatment had no effect on the number of catalase-positive peroxisomes in WT cells or FAF2-/- cells expressing 3HA-FAF2, a significant reduction was observed in FAF2-/- cells (Supplementary Fig. 4a, b). Similar results were observed in HaloTag assay (Supplementary Fig. 4c, d). In the presence of the TMR-ligand, the intensity of the Halo band in FAF2-/- cells (Supplementary Fig. 4c lane 5) was more pronounced than in WT cells (Supplementary Fig. 4c lane 2). Torin1 treatment also enhanced the Halo bands, in WT and FAF2-/- cells (Supplementary Fig. 4c, d), and the Torin1-induced acceleration of pexophagy in FAF2-/- cells were reversed by expressing 3HA-FAF2 (Supplementary Fig. 4c). Similar results were observed using a Halo-mGFP was fused to the peroxisomal membrane protein PMP34 (Supplementary Fig. 4e, f). It has been reported that Torin1 induces the ubiquitylation of PMP70 in WT cells35, explaining why pexophagy is accelerated in FAF2-/- cells (Supplementary Fig. 4). These results indicate that FAF2 normally governs nonessential pexophagy and that when FAF2 deletion promotes the autophagic elimination of peroxisomes.
Fig. 2. Loss of FAF2 accelerates pexophagy.
a Representative FACS data (mKeima-SKL 561/488 ratio) with the percentage of pexophagy-positive cells indicated. b Quantitative analysis of the pexophagy flux for cells from a. Each circle represents an individual data point from three independent experiments. Statistical significance was calculated using one-way ANOVA; ****p < 0.0001. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. c PMP70 levels increased following the FIP200 knockdown. WT and FAF2-/- cells were transfected with control or FIP200 siRNAs and then immunoblotted with the indicated antibodies. d Quantitative analysis of PMP70 levels in cells from c. PMP70 levels were normalized to WT cells, which were set to 1. Dots represent individual data points from three independent experiments. Statistical significance was calculated using a one-tailed Welch’s t-test; *p < 0.05; N.S.—not significant. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. e FIP200 knockdown reversed the peroxisomal loss observed in FAF2-/- cells. WT and FAF2-/- HCT116 cells were treated with control or PMP70 siRNAs and then immunostained with anti-PMP70 and anti-catalase antibodies. Cell nuclei were stained with DAPI. Peroxisomal abundance increased following FIP200 knockdown in FAF2-/- cells. Scale bars, 10 µm. f Quantitative analysis of per cell peroxisome abundance for cells in e. The number of catalase-positive peroxisomes was plotted. Dots represent individual data points from three independent experiments. n = 257 cells (86, 92, 79 cells/experiments; WT + sicontrol), n = 198 cells (58, 75, 65 cells/experiments; WT + si FIP200), n = 209 cells (67, 62, 80 cells/experiments; FAF2-/- + sicontrol), and n = 212 cells (77, 68, 67 cells/experiments; FAF2-/- + siFIP200). Bars, median. Statistical significance was calculated using one-way ANOVA; ****p < 0.0001. g The pexophagy flux was evaluated using a HaloTag (Halo)-based reporter processing assay. The Halo fragment was more prominent in FAF2-/- cells than WT cells, indicating that pexophagy is accelerated in FAF2-/- cells. FIP200 knockdown reduced pexophagy in FAF2-/- cells. h Quantitative analysis of the pexophagy flux in cells from g. Halo band intensity was normalized to the sum of the Halo-mGFP-SKL and Halo band intensities. The normalized Halo intensities are shown as box plots with dots indicating individual data points from three independent experiments. Statistical significance was calculated using one-way ANOVA; *p < 0.05; ***p < 0.001. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Source data are provided as a Source Data file.
If FAF2 is involved in peroxisome biogenesis like PEX19, peroxisome may also disappear in FAF2-/- cells. To test this, we compared the levels of peroxisomal proteins in FAF2-/- cells and PEX19-/- cells. PEX19 is essential for peroxisomal biogenesis3,36–38 so that PEX19-/- cells exhibited peroxisomal loss14. Using previously generated PEX19-/- cells, PMP70 and PEX16 were nearly undetectable, whereas PEX14 was not affected (Supplementary Fig. 5a, b). The observed equivalency of PEX14 in WT and PEX19-/- cells is likely because PEX14 is targeted to mitochondria when peroxisomes are absent and it thus escapes proteasomal degeneration. Although PMP70 and PEX16 were reduced in FAF2-/- cells (Fig. 1f, h), it was not to the same extent as in the PEX19-/- cells, suggesting that the mechanism underlying peroxisomal loss may differ between the two cell lines.
To further exclude any potential role that FAF2 might have in de novo peroxisome biogenesis, we made a FAF2/FIP200 and PEX19/FIP200 double KO HCT116 cells (Supplementary Fig. 5c). If FAF2 loss accelerates pexophagy, then peroxisome levels in the FAF2/FIP200 double KO should be restored since FIP200 is essential for autophagy. However, if FAF2 functions in peroxisome biogenesis, then peroxisomes should remain sparse. We assessed peroxisome abundance by immunostaining for PMP70 and catalase. As shown in Supplementary Fig. 5d, the reduced number of peroxisomes in FAF2-/- cells was recovered in FAF2/FIP200 double KO cells (Supplementary Fig. 5e). Peroxisome loss in PEX19-/- cells, however, was not recovered with FIP200 knockout (Supplementary Fig. 5d, e). Furthermore, Baf.A1 treatment significantly increased peroxisomal abundance as well as PMP70 protein level in FAF2-/- cells but not in PEX19-/- cells (Supplementary Fig. 6a–d). Taken together, these results demonstrate that FAF2 is involved in pexophagy rather than peroxisome biogenesis.
USP30 knockdown enhances pexophagy in FAF2-deficient cells
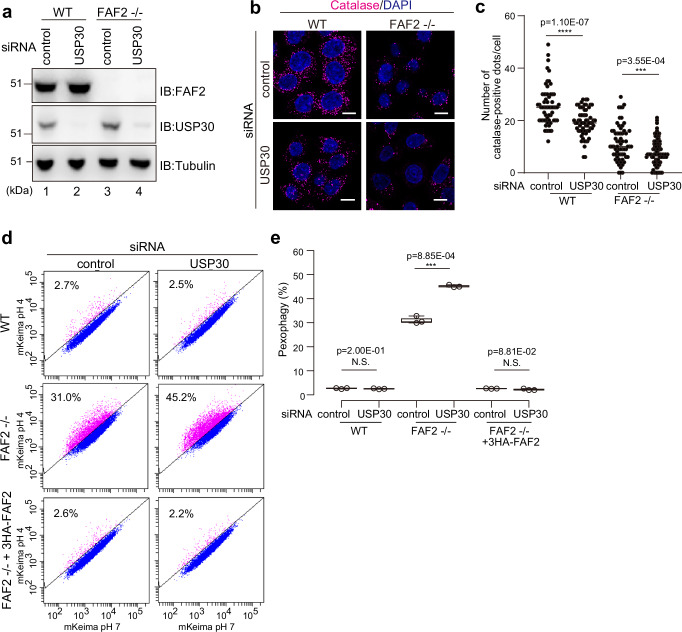
Because USP30 functions in peroxisome degradation26,27, we examined if USP30 is also involved in pexophagy in FAF2-/- cells. After knocking down USP30 in WT cells with siRNA (Fig. 3a), we found a slight reduction in peroxisome abundance (Fig. 3b, c). However, mKeima-based flow cytometry indicated that the USP30 knockdown effects on pexophagy were indistinguishable from WT cells treated with the control siRNA (Fig. 3d, e). In sharp contrast, pexophagy, as determined by immunostaining (Fig. 3b, c) and mKeima flow cytometry (Fig. 3d, e), was enhanced in FAF2-/- cells following USP30 knockdown. These results suggest that USP30 plays a critical role in the progression of pexophagy when FAF2 is depleted.
Fig. 3. USP30 depletion promotes pexophagy in FAF2-/- cells.
a WT and FAF2-/- HCT116 cells were treated with control or USP30 siRNAs and then immunoblotted with the indicated antibodies. b Immunocytochemical images of WT and FAF2-/- HCT116 cells stained with an anti-catalase antibody. Cells were transfected with control and USP30 siRNAs. Cell nuclei were stained with DAPI. Scale bars, 10 µm. c Quantitative analysis of per cell peroxisome abundance for cells in b. The number of catalase-positive peroxisomes was plotted. Dots represent individual data points from three independent experiments. n = 61 cells (19, 24, 18 cells/experiments; WT + sicontrol), n = 63 cells (25, 20, 18 cells/experiments; WT + siUSP30), n = 64 cells (17, 20, 27 cells/experiments; FAF2-/- cells + sicontrol), and n = 88 cells (47, 21, 20 cells/experiments; FAF2-/- + siUSP30). Bars, median. Statistical significance was calculated using a one-tailed Welch’s t-test; ***p < 0.001; ****p <0.0001. d USP30 knockdown promotes pexophagy in FAF2-/- cells. WT, FAF2-/-, or FAF2-/- cells stably expressing 3HA-FAF2 were transfected with control or USP30 siRNAs. Cells were analyzed by FACS 48 h post-siRNA transfection. e Quantitative analysis of the pexophagy flux in cells from d. Each circle represents an individual data point from three independent experiments. Statistical significance was calculated using a one-tailed Welch’s t-test; ***p < 0.001; N.S.—not significant. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Source data are provided as a Source Data file.
PMP70 is a genuine substrate of FAF2 for extraction from peroxisomal membrane
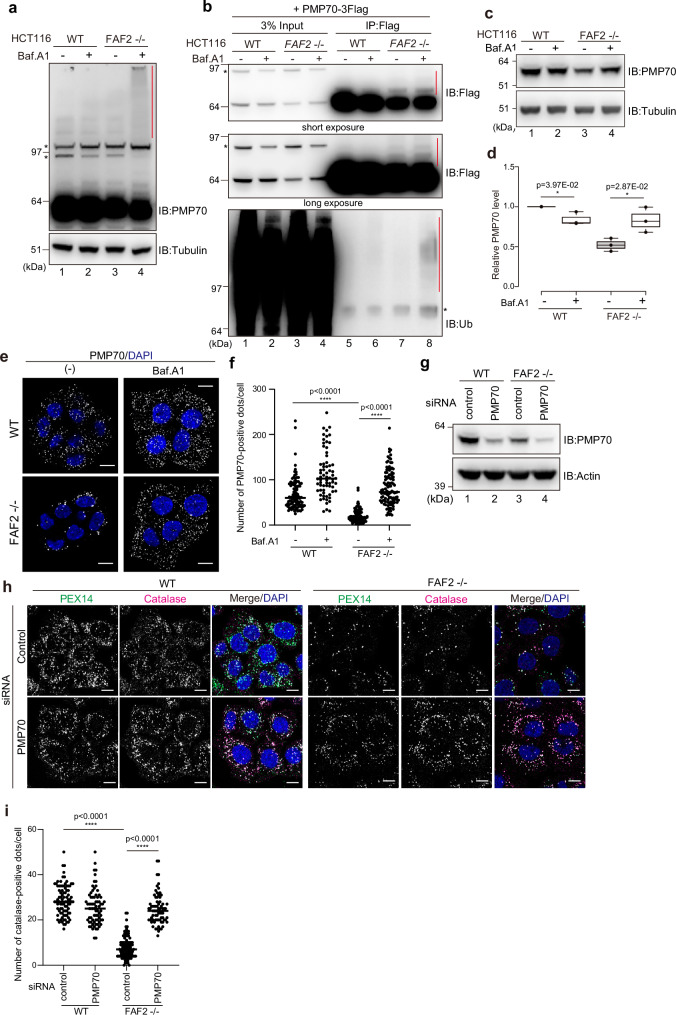
The yeast ortholog of FAF2, Ubx2, recognizes ubiquitylated substrates on target membranes24. If FAF2 negatively regulates pexophagy by extracting ubiquitylated peroxisomal proteins under steady-state conditions, then ubiquitylated proteins should accumulate when autophagy is inhibited in FAF2-/- cells. To test this possibility, we examined the levels of 3Flag-PEX14, 3Flag-PEX16, and endogenous PMP70 in FAF2-/- cells after 24 h with Baf.A1. While 3Flag-PEX16 and 3Flag-PEX14 were largely unaffected by Baf.A1 (Supplementary Fig. 7a, b), smeared, high molecular weight signals were detected for PMP70 (Fig. 4a lane 4). By immunoprecipitating PMP70-3Flag and then immunoblotting with an anti-ubiquitin antibody, we confirmed that the smeared signals corresponded to poly-ubiquitylated forms of PMP70 (Fig. 4b). 3Flag-PEX16 is ubiquitylated in FAF2-/- cells under steady state conditions (Supplementary Fig. 7c), and 3Flag-PEX14 ubiquitylation in WT and FAF2-/- cells was comparable both before and after Baf.A1 treatment (Supplementary Fig. 7d). While PMP70 and PEX5 were ubiquitylated by amino acid starvation4, ubiquitylated PEX5S did not accumulate in FAF2-/- cells even when treated with Baf.A1 (Supplementary Fig. 7e, f), suggesting that PEX5S ubiquitylation is not responsible for pexophagy induced by FAF2 loss.
Fig. 4. Extraction of excess PMP70 by the p97/VCP-FAF2 complex prevents pexophagy.
a WT or FAF2-/- HCT116 cells were treated with or without Baf.A1 for 24 h and immunoblotted with the indicated antibodies. The red vertical line denotes a smear in the PMP70 signal. The asterisks indicate cross-reactive bands. n = 3 assays. b PMP70 ubiquitylation in FAF2-/- cells. WT or FAF2-/- cells stably expressing PMP70-3Flag were treated with or without Baf.A1 for 24 h and then immunoprecipitated with anti-Flag antibodies. The samples were immunoblotted with anti-Flag and anti-ubiquitin antibodies. The red vertical lines denote ubiquitylation. The asterisks indicate cross-reactive bands. n = 2 assays. c PMP70 level in FAF2-/- cells increases in response to Baf.A1 treatment. The cells were treated with or without Baf.A1 for 24 h and then immunoblotted with the indicated antibodies. d Quantitative analysis of PMP70 levels in cells from c. PMP70 levels were normalized to WT cells, which were set to 1. Dots indicate individual data points from three independent experiments. Statistical significance was calculated using a one-tailed Welch’s t-test; *p < 0.05. The center lines correspond to the medians and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. e Peroxisomal abundance in FAF2-/- cells is recovered following Baf.A1 treatment. At 24 h post-Baf.A1 treatment, the cells were immunostained with anti-PMP70 antibodies. Cell nuclei were stained with DAPI. Scale bars, 10 µm. f Quantitative analysis of the per cell number of PMP70-positive peroxisomes for cells in e. The number of PMP70-positive peroxisomes per cell was plotted. Dots represent individual data points from three independent experiments. n = 103 cells (35, 35, 33 cells/experiments; WT), n = 68 cells (22, 22, 24 cells/experiments; WT + Baf.A1), n = 91 cells (32, 30, 29 cells/experiments; FAF2-/-), and n = 110 cells (37, 37, 36 cells/experiments; FAF2-/- + Baf.A1). Bars, median. Statistical significance was calculated using one-way ANOVA; ****p < 0.0001. g WT and FAF2-/- cells were treated with control or PMP70 siRNAs and then immunoblotted with the indicated antibodies. h PMP70 knockdown reverted the peroxisomal loss observed in FAF2-/- cells. WT and FAF2-/- cells were treated with control or PMP70 siRNAs and then immunostained with anti-PEX14 and anti-catalase antibodies. Cell nuclei were stained with DAPI. Scale bars, 10 µm. i Quantitative analysis of the per cell number of catalase-positive peroxisomes per cells in h. The dots indicate individual data points from three independent experiments. n = 86 cells (35, 26, 25 cells/experiments; WT + sicontrol), n = 73 cells (23, 26, 24 cells/experiments; WT + siPMP70, n = 100 cells (33, 34, 33 cells/experiments; FAF2-/- + sicontrol), and n = 73 cells (25, 25, 23 cells/experiments; FAF2-/- cells + siPMP70). Bars, median. Statistical significance was calculated using one-way ANOVA; ****p < 0.0001. Source data are provided as a Source Data file.
Because Baf.A1treatment increased both PMP70 protein levels (Fig. 4c, d) and the number of PMP70-positive peroxisomes in FAF2-/- cells (Fig. 4e, f), we turned our focus to PMP70. If PMP70 is a substrate of FAF2 and its extraction from peroxisomal membranes prevents pexophagy, then a reduction of PMP70 levels might rescue peroxisomal loss. PMP70 was knocked down in FAF2-/- cells using siRNA (Fig. 4g), and the abundance of catalase-positive (and PEX14-positive) peroxisomes was determined. Peroxisome abundance in FAF2-/- cells significantly increased following PMP70 knockdown (Fig. 4h, i). FAF2-/- cells expressing siRNA-resistant PMP70-3Flag were likewise assayed (Supplementary Fig. 8a). As expected, endogenous PMP70 was knocked down, while siRNA-resistant PMP70-3Flag was unaffected (Supplementary Fig. 8a). Under these conditions, the peroxisomal number increased in FAF2-/- cells following PMP70 siRNA treatment, whereas no change was observed in FAF2-/- cells expressing siRNA-resistant PMP70-3Flag (Supplementary Fig. 8b and 8c). PEX16 was also tested as a FAF2 substrate. Baf.A1 treatment had no effect on 3Flag-PEX16 ubiquitylation in FAF2-/- cells (Supplementary Fig. 7c), but PEX16 knockdown increased peroxisome abundance (Supplementary Fig. 9a–c). These results suggest that PMP70, as well as PEX16, are the FAF2 substrates that are extracted from peroxisomal membranes.
Interactions of FAF2 with other cofactors and PMP70
To gain insights into the proteins that interact with FAF2, HCT116 cells stably expressing 3HA-FAF2 were treated with a DSP crosslinker, and 3HA-FAF2 was immunoprecipitated with an anti-HA antibody. In addition to p97/VCP, endogenous PMP70, PEX16, and USP30 were co-immunoprecipitated (Fig. 5a). This is consistent with our earlier demonstration that PMP70 and PEX16 are FAF2 substrates (Fig. 4h, i and Supplementary Fig. 9) and that USP30 cooperatively regulates pexophagy with FAF2 (Fig. 3). In contrast, 3HA-FAF2 did not interact with the peroxisomal membrane protein PEX26 (Fig. 5b). The p97/VCP dominant-negative QQ (E305Q/E578Q) mutant, which has impaired ATP hydrolysis, tightly bound endogenous FAF2 (Fig. 5c). The stronger interactions between FAF2 and the p97/VCP QQ mutant than with p97/VCP WT suggest that ATP hydrolysis impairment stabilizes p97/VCP-FAF2 binding (Fig. 5c). FAF2 also interacts with NPLOC4 and UFD1 (Fig. 5d). These results indicate that, under basal conditions, p97/VCPFAF2 complex is associated with NPLOC4-UFD1 and that it interacts with PMP70 and PEX16 on peroxisomal membranes.
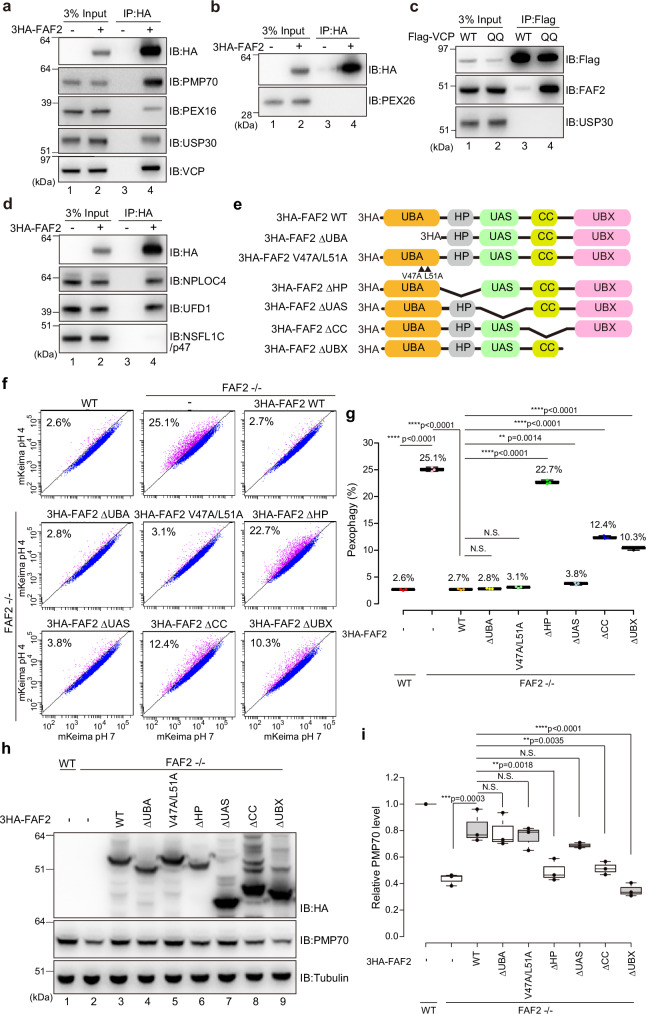
Fig. 5. FAF2 acts as a negative regulator of basal pexophagy.
a, b HCT116 cells stably expressing 3HA-FAF2 were treated with the chemical crosslinker DSP and then co-immunoprecipitated with anti-HA agarose beads. The samples were immunoblotted with the indicated antibodies. 3HA-FAF2 interacts with PMP70, PEX16, USP30, and p97/VCP, but not PEX26. n = 3 assays. c p97/VCP interacts with FAF2 but not USP30. HeLa cells transiently expressing Flag-tagged p97/VCP WT or the E305Q/E578Q mutant (QQ) were treated with DSP and then co-immunoprecipitated with anti-Flag beads. The samples were immunoblotted with the indicated antibodies. n = 2 assays. d 3HA-FAF2 forms a complex with NPLOC4 and UFD1 but not NSFL1C/p47. HCT116 cells stably expressing 3HA-FAF2 were treated with DSP and then immunoprecipitated with anti-HA agarose beads. The samples were immunoblotted with the indicated antibodies. n = 2 assays. e Schematic diagram of the FAF2 constructs used in this study. UBA, ubiquitin-associated domain; HP, hairpin; CC, coiled-coil domain; UBX, ubiquitin-regulatory X domain. To disrupt interactions with ubiquitin, V47A and L51A substitution were introduced into the UBA domain. deletion; Δ. f FACS-based analysis of the pexophagy flux. Representative FACS data (mKeima-SKL 561/488 ratio) for WT, FAF2-/-, or FAF2-/- cells stably expressing the indicated 3HA-FAF2 mutants with the percentage of pexophagy-positive cells indicated. FAF2 ΔUAS, FAF2 ΔUBA, and FAF2 V47A/L51A restored pexophagy in FAF2-/- cells. The effect of pexophagy suppression by FAF2 ΔHP, ΔCC, and ΔUBX was limited, indicating that the FAF2 UBA and UAS domains are dispensable for pexophagy suppression. deletion; Δ. g Quantitative analysis of the pexophagy flux in f. Dots represent individual data points from three independent experiments. Statistical significance was calculated using one-way ANOVA; **p < 0.01; **** p < 0.0001; N.S.—not significant. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. deletion; Δ. h Total cell lysates prepared from WT, FAF2-/-, and FAF2-/- HCT116 cells expressing 3HA-FAF2 WT or the indicated mutants were immunoblotted with the indicated antibodies. deletion; Δ. i Quantitative analysis of PMP70 levels in cells from h. PMP70 levels were normalized to WT HCT116 cells, which were set to 1. Dots indicate individual data points from three independent experiments. Statistical significance was calculated using one-way ANOVA; **p < 0.001; ***p < 0.001; ****p < 0.0001; N.S. not significant. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. deletion; Δ. Source data are provided as a Source Data file.
Functional analysis of FAF2 domains in pexophagy
We next sought to determine which FAF2 domain is crucial for preventing pexophagy. To test if the FAF2 UBA domain interacts with ubiquitin, we performed a GST-pull down assay (Supplementary Fig. 10) and in-cell experiments (Supplementary Fig. 11). Since K48- and K63-linked ubiquitin chains are the most abundant chain types, they were incubated in vitro with purified, GST-tagged FAF2 UBA domain (GST-FAF2 UBA). An immunoblot of the glutathione agarose-bound proteins showed that GST-FAF2 UBA interacted with both K48- and K63-linked ubiquitin chains whereas GST alone did not (Supplementary Fig. 10a). For NSFL1C/p47 (another p97/VCP cofactor), the alpha-3 hydrophobic side chains of L38 and Y42 contact multiple ubiquitin side chains39 (Supplementary Fig. 10b and 10c). Ala substitution of the corresponding amino acids in the FAF2 UBA domain (i.e. V47 and L51) inhibited ubiquitin interactions (Supplementary Fig. 10a). This suggests that FAF2 UBA domain-ubiquitin interactions are not specific for the K48- and K63-linked ubiquitin chains. Since L-leucyl-L-leucine methyl ester (LLOMe) induces ubiquitylation of lysosomes40,41, we next examined if FAF2 is recruited to ubiquitin-coated lysosomes following LLOMe treatment. Conversion of LLOMe into a membranolytic form (Leu-Leu)n-OMe (n > 3) by a lysosomal thiol protease dipeptidyl peptidase I causes lysosomal rupture and ubiquitylation. We confirmed that Flag-ubiquitin accumulates on lysosomes after treatment with LLOMe for 1.5 h (Supplementary Fig. 11a) but found that full-length FAF2 (WT) did not translocate to lysosomes (Supplementary Fig. 11a). To overcome this, we made a series of FAF2 domain deletion mutants (Fig. 5e). Deletion of HP domain enables FAF2 to translocate to lysosomes, indicating that lysosomal translocation of FAF2 is impeded by HP domain (Supplementary Fig. 11a). The FAF2 ΔUBA/ΔHP double mutant, however, was unable to translocate to lysosomes (Supplementary Fig. 11b). These results suggest that the HP domain is crucial for FAF2 membrane anchoring in its native organelles and that the UBA domain recognizes ubiquitin.
In addition to those domains, FAF2 also has a UAS/Thioredoxin-like domain20,42 and a coiled-coil domain (CC)20. These domains were subsequently deleted (Fig. 5e), and each of the deletion constructs were tested along with V47A/L51A mutant for pexophagy flux in FAF2-/- cells. The pexophagy flux in FAF2-/- cells expressing 3HA-FAF2 ΔUBA (2.8%), V47A/L51A (3.1%), or ΔUAS (3.8%) was comparable to that in FAF2-/- cells expressing 3HA-FAF2 WT (2.7%) (Fig. 5f, g). In contrast, 3HA-FAF2 ΔHP (22.7%), ΔCC (12.4%), and ΔUBX (10.3%) inhibited pexophagy recovery in FAF2-/- cells (Fig. 5f, g), even though the three mutants were sufficiently expressed (Fig. 5h). PMP70 levels were significantly reduced in FAF2-/- cells expressing 3HA-FAF2 ΔHP, ΔCC, or ΔUBX compared to FAF2-/- cells expressing 3HA-FAF2 WT (Fig. 5h, i). In contrast, the levels of PMP70 in the 3HA-FAF2 ΔUBA, V47A/L51A, and ΔUAS cells were comparable to FAF2-/- cells with 3HA-FAF2 WT (Fig. 5h, i), which is consistent with the flow cytometry data (Fig. 5f, g). We thus concluded that the FAF2 HP domain is essential and that the FAF2 UBA domain is dispensable for pexophagy prevention, even though it can interact with ubiquitin (see “Discussion”).
OPTN is required for pexophagy in FAF2-/- cells
In FAF2-/- cells, ubiquitylated peroxisomal membrane proteins such as PMP70 accumulate (Fig. 4a, b), and peroxisomes are degraded via autophagy (Fig. 2). Both ubiquitin and LC3B partially colocalized to a few remaining peroxisomes in FAF2-/- cells that had been labeled with GFP-SKL (Fig. 6a). LC3B recruitment to GFP-SKL positive peroxisomes was specific to FAF2 deletion (60.4%) since LC3B recruitment was negligible in WT cells (4.8%) or in FAF2-/- cells expressing 3HA-FAF2 (22.0%) (Fig. 6b). Similar results were obtained by colocalization with LC3B and PMP70 (Supplementary Fig. 12a), which suggest that a portion of ubiquitylated peroxisomes in FAF2-/- cells are sequestrated in autophagosomes.
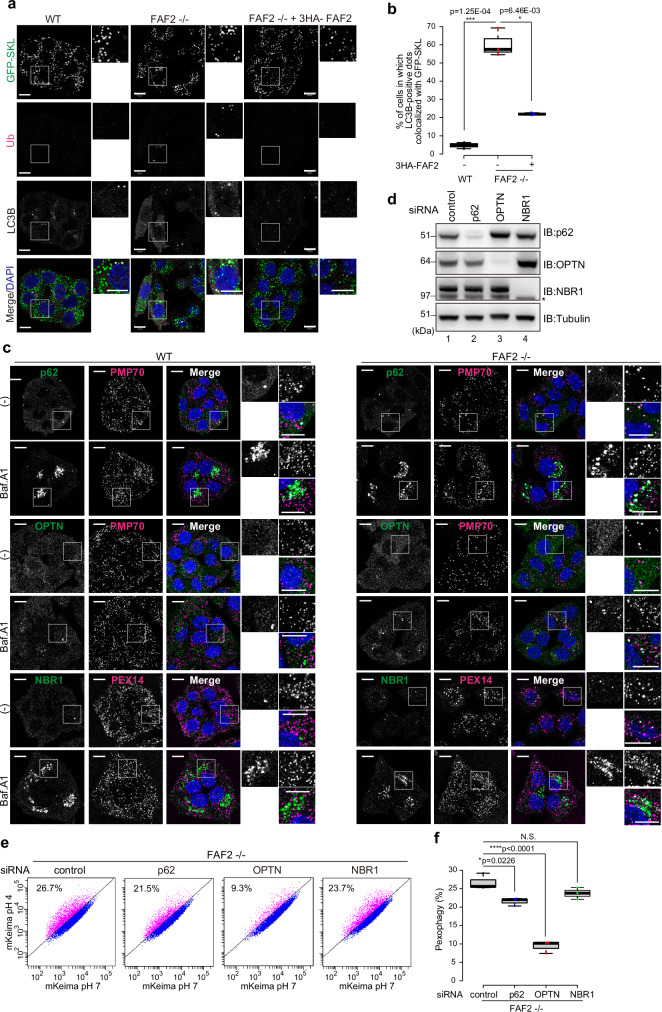
Fig. 6. OPTN is required for pexophagy in FAF2-deficient cells.
a Ubiquitin (Ub) and LC3B accumulate on GFP-SKL-positive peroxisomes in FAF2-/- cells. HCT116 cells stably expressing GFP-SKL were immunostained with anti-ubiquitin and anti-LC3B antibodies. Cell nuclei were stained with DAPI. Higher magnification images of the boxed regions are shown to the right. Scale bars, 10 µm. b Quantitative analysis of cells in a. Cells in which at least one GFP-SKL signal was also positive for LC3B was counted as a positive cell. Dots represent individual data points from three independent experiments. n = 227 cells (68, 79, 80 cells/experiments; WT), n = 167 cells (55, 65, 47 cells/experiments; FAF2-/-), and n = 214 cells (66, 84, 64 cells/experiments; FAF2-/- + 3HA-FAF2). Statistical significance was calculated using one-way ANOVA; *p < 0.05, ***p < 0.001. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. c WT or FAF2-/- HCT116 cells were treated with or without Baf.A1 for 24 h and then immunostained with the indicated antibodies. FAF2-/- cells that did not have complete peroxisomal loss were used to generate representative images of the cell line. Higher magnification images of the boxed regions are shown to the right. (-) indicates untreated. Scale bars, 10 µm. n = 2 assays. d HCT116 cells were transfected with control, p62, OPTN, and NBR1 siRNAs. The cells were immunoblotted with the indicated antibodies. The asterisk indicates cross-reactive bands. n = 2 assays. e FACS-based analysis of the pexophagy flux. Representative FACS data (mKeima-SKL 561/488) for the siRNA-transfected FAF2-/- cells are shown with the percentage of pexophagy-positive cells indicated. f Quantitative analysis of the pexophagy flux for cells in e. Dots represent individual data points from three independent experiments. Statistical significance was calculated using one-way ANOVA; *p < 0.05, ****p < 0.0001; N.S.- not significant. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Source data are provided as a Source Data file.
During ubiquitin-dependent selective autophagy, assembly of the autophagy machinery is facilitated by the recruitment of autophagy adaptors to target organelles43–46. Mammalians have five adaptors (NDP52/CALCOCO2, OPTN, TAX1BP1, p62/SQSTM1, and NBR1), several of which are important in pexophagy2,36,47. For pexophagy induced by PEX3-overexpression31, NBR1 is required, and p62 participates in peroxisomal clustering. NBR1 is also involved in PEX2-mediated pexophagy during starvation4. To clarify which adaptors are essential for pexophagy induced by FAF2 deletion, we examined the localization of each adaptor (Fig. 6c and Supplementary Fig. 12b). Under basal conditions, all five adaptors were cytosolic with a portion of p62 also localizing peroxisomes in WT and FAF2-/- cells (Fig. 6c). FAF2-/- cells that did not have complete peroxisomal loss were used to generate representative images of the cell line (Fig. 6c). Baf.A1 treatment caused most of the autophagy adaptors to accumulate, the lone exception was OPTN. FAF2-/- cells also appeared to have p62 and NBR1 signal aggregates that contained PMP70- or PEX14-labeled peroxisomes (Fig. 6c). Interestingly, some of the OPTN dot-like structures that formed following Baf.A1 treatment clearly colocalized with PMP70 in FAF2-/- cells (Fig. 6c). Neither NDP52 nor TAX1BP1 was observed on or around peroxisomes (Supplementary Fig. 12b). In addition to p62 and NBR1, immunofluorescence data suggest that OPTN may function in pexophagy. To further clarify whether p62, NBR1, and OPTN contribute to pexophagy, FAF2-/- cells stably expressing mKeima-SKL were analyzed by flow cytometry after siRNA-mediated knockdown of each of the adaptors (Fig. 6d–f). While knockdown of p62 or NBR1 slightly repressed pexophagy, the pexophagy flux in the siNBR1-treated cells was not statistically significant (Fig. 6e, f). Similarly, TAX1BP1 and NDP52 knockdown (Supplementary Fig. 12c) had little to no effect on the pexophagy flux (Supplementary Fig. 12d, e). In sharp contrast, significant prevention of pexophagy was observed following OPTN knockdown in FAF2-/- cells (Fig. 6e, f). We next sought to determine if PMP70 knockdown reduced OPTN recruitment to peroxisomes. After Baf.A1 treatment, the percentage of FAF2-/- cells with peroxisomal OPTN was significantly reduced in PMP70 knockdown cells as compared to the corresponding control (reduced from 55.4 to 15.1 %) (Supplementary Fig. 13a, b). These results strongly suggest that pexophagy requires PMP70-mediated recruitment of OPTN and that OPTN mainly recognizes ubiquitylated PMP70.
p97/VCPFAF2 complex prevents peroxisome from entering an autophagic pathway
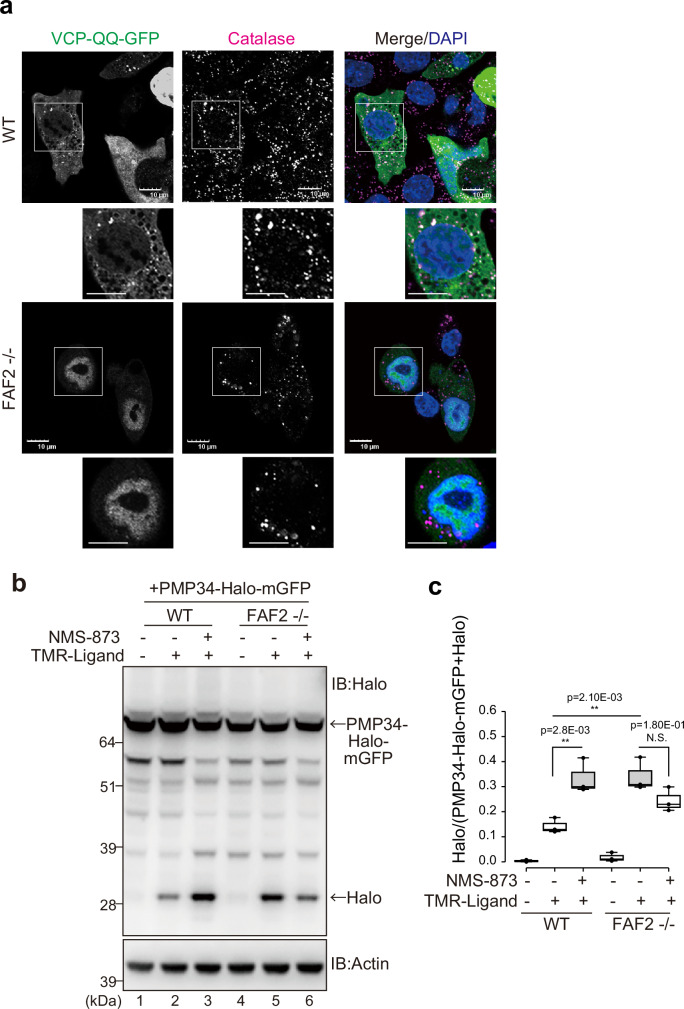
Since FAF2 is a p97/VCP cofactor and membrane protein extraction requires energy, it is possible that the ATPase activity of p97/VCP may be important. Since ATP-hydrolysis deficient p97/VCP stably binds FAF2 (Fig. 5c), we examined the peroxisomal localization of p97/VCP QQ (E305Q/E578Q)-GFP. In the presence of FAF2 (WT cells), p97/VCP-QQ-GFP partially localized to peroxisomes after Baf.A1 treatment (Fig. 7a) but did not in FAF2-/- cells (Fig. 7a). We next sought to determine if pexophagy was induced when p97/VCP-mediated protein extraction was inhibited. Cells were treated with the p97/VCP inhibitor NMS-873, and a HaloTag assay that incorporated PMP34-Halo-mGFP was used. The Halo band in WT cells was enhanced by NMS-873 treatment (Fig. 7b), suggesting that p97/VCP inhibition accelerates pexophagy. The effect was not observed in FAF2-/- cells (Fig. 7b and 7c). We thus concluded that ubiquitylated peroxisomal membrane proteins (e.g., PMP70) cannot be extracted by p97/VCP in cells with dysfunctional FAF2 and that subsequent accumulation of these proteins in peroxisomes induces pexophagy.
Fig. 7. p97/VCP-FAF2 prevents autophagic degradation of peroxisomes.
a p97/VCP is localized to peroxisomes in WT cells. WT or FAF2-/- cells were transfected with VCP-QQ-GFP and then immunostained with an anti-catalase antibody. Scale bars, 10 µm. b Pexophagy was induced following p97/VCP inhibition. WT or FAF2-/- cells stably expressing PMP34-Halo-mGFP were treated with NMS-873 (p97/VCP inhibitor). Total cell lysates were subjected to SDS-PAGE. c Quantitative analysis of the pexophagy flux for cells in b. Dots represent individual data points from three independent experiments. Statistical significance was calculated using a one-way ANOVA; **p < 0.005; N.S.- not significant. The center lines correspond to the medians, and the box limits indicate the 25th and 75th percentiles. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Source data are provided as a Source Data file.
Discussion
Peroxisomes are involved in various oxidative reactions associated with lipid metabolism and are susceptible to damage by exposure to peroxides, consequently peroxisomal quality control is pivotal. One aspect of this quality control involves the monitoring of peroxisomal matrix proteins by proteases in the peroxisome matrix, such as PsLon/Lonp2 and Tysnd148,49. While little is known about how peroxisomal membrane proteins are maintained, their half-life in cultured cells ranges from 2–6 h (PEX3) to 36 h (PEX16)50. In contrast, the half-life of peroxisomes is approximately 2–3 days. This suggests the involvement of specific molecular machinery that functions in the extraction and removal of damaged or old membrane proteins from peroxisomal membranes. Selective degradation of impaired peroxisomes via the pexophagy pathway is critical for maintaining peroxisomal integrity. As with mammalian mitophagy, ubiquitin plays a key role in pexophagy2,4,31,51. The amount of ubiquitin on the surface of an organelle is determined by a balance between E3-mediated conjugation and removal by DUB and p97/VCP. Although the ubiquitylation of organelles by E3 has been extensively studied (e.g., Parkin E3 activity in mitophagy), the counter-reaction of how DUB and p97/VCP remove ubiquitylated proteins to downregulate organellophagy is poorly understood.
Mammalian FAF2 and its yeast ortholog Ubx2 are p97/VCP cofactors linking ubiquitylated proteins to p97/VCP via a UBA domain (ubiquitin-binding domain) and a UBX domain (p97/VCP-binding domain). The fundamental function of the p97/VCP-cofactor complex is to unfold and extract ubiquitylated proteins from a membrane or a multiprotein complex. Yeast Ubx2 is crucial for the removal of precursor proteins arrested within the translocator channel of the outer mitochondrial membrane25. In this study, we found that the level of three peroxisomal membrane proteins (PMP70, PEX14, PEX16) and a matrix protein (catalase) are reduced in FAF2-/- cells and that peroxisome degradation via pexophagy is significantly accelerated. We surmised that FAF2 usually prevents pexophagy by removing ubiquitylated membrane proteins (e.g., PEX16 and PMP70) on peroxisomes. Since the knockdown of PMP70 increased peroxisome abundance in FAF2-/- cells, we further concluded that PMP70 is a genuine FAF2 substrate that is extracted from peroxisomal membranes and that the accumulation of ubiquitylated PMP70 or PMP70 interacting proteins triggers pexophagy (Fig. 4h, i). The role that other ubiquitylated peroxisomal proteins may have in modulating pexophagy remains to be determined.
USP30 knockdown in FAF2-/- cells facilitated pexophagy, suggesting that USP30 participates in FAF2-mediated peroxisomal homeostasis. USP30 counteracts Parkin-mediated mitophagy by removing ubiquitin from mitochondrial proteins that had been ubiquitylated by Parkin and counteracts Parkin-mediated mitophagy5–7. USP30 also removes ubiquitin from import substrates and components of the mitochondrial translocator, which improves translocation efficiency10,52. Furthermore, there are reports that USP30 also functions in pexophagy26,27. This study demonstrates the significant role that USP30 plays in the basal pexophagy of FAF2-/- cells. Interestingly, dysfunction of both USP30 and FAF2 had an additive effect that accelerated pexophagy, whereas dysfunction of either protein individually resulted in different phenotypes (Fig. 3). If the target proteins for FAF2 and USP30 are identical, the ubiquitylated proteins that accumulate in USP30-knockdown cells would be extracted by p97/VCP-FAF2 complex, while the ubiquitylated proteins that accumulate in FAF2-/- cells would be deubiquitylated by USP30. Consequently, the accelerated pexophagy phenotype would not be observed following disruption of only FAF2 or only USP30. While pexophagy was not as evident in cells with USP30 dysfunction, FAF2-depletion alone was sufficient to induce obvious pexophagy. The molecular basis for this discrepancy might be attributable to the specific types of polyubiquitin chains recognized by USP30. USP30 has a unique ubiquitin recognition mechanism that is specific for K6- and K11-linked ubiquitin chains7–9. If the ubiquitin chains that accumulate due to FAF2 dysfunction are primarily K48-linked or K63-linked, the narrow substrate specificity of USP30 could impede deubiquitylation and it would be unable to counteract the FAF2 dysfunction. However, if the p97/VCP-FAF2 complex recognizes K6- and K11-linked ubiquitin chains, the ubiquitylated proteins that accumulate in USP30-knockdown cells would be extracted by the p97/VCP-FAF2 complex and the phenotype derived from USP30 dysfunction would be concealed. Consequently, the differential selectivity in ubiquitin chain recognition between USP30 and p97/VCP-FAF2 complex may explain the distinct phenotypes observed with USP30-knockdown and in FAF2-/- cells while also accounting for the additive phenotype when both genes are simultaneously disrupted.
It is thought that the p97/VCP cofactor recognizes ubiquitylated substrates through its UBA domain. The FAF2 UBA domain, however, was dispensable to preventing pexophagy, whereas the HP domain was essential (Fig. 5f–i). In addition, FAF2 binds PMP70 in WT HCT116 cells (Fig. 5a), although the majority of PMP70 is not ubiquitylated. We thus considered that ubiquitylation might not be necessary for FAF2 substrate recognition. However, ubiquitylated PEX16 and PMP70 accumulated in FAF2-/- cells (Fig. 4a, b and Supplementary Fig. 7c), USP30 knockdown enhanced pexophagy (Fig. 3d, e), and poly-ubiquitin interactions with the FAF2 UBA domain were observed in vitro (Supplementary Fig. 10a). These results suggest that the extraction of substrates and promotion of pexophagy in FAF2-/- cells is ubiquitylation dependent. It is possible that FAF2 binds substrates like PMP70 irrespective of ubiquitylation (Fig. 5a) while also concomitantly interacting with the p97/VCP complex to recruit ubiquitin-recognition factors such as NPLOC4 and UFD1 (Fig. 5d).
Although FAF2 ΔUBA rescued pexophagy (Fig. 5f, g), FAF2 can also interact with additional cofactors, such as UFD1 and NPLOC4 (Fig. 5d). This suggests that poly-ubiquitin chains conjugated to PMP70 may be recognized by UFD1-NPLOC4, which is associated with the p97/VCP-FAF2 complex. Since FAF2 recruits the UFD1-NPLOC4- p97/VCP complex to peroxisomes, FAF2 may enhance the recognition of ubiquitylated PMP70 by the complex. UBX proteins are important for the efficient unfolding of ubiquitylated proteins by the UFD1-NPLOC4-p97/VCP complex53. We thus speculate that FAF2 functions as a hub for the recruiting of the complex to peroxisomes. This hypothetical mechanism thus provides a reasonable explanation for the non-essential role of the UBA domain in FAF2 prevention of pexophagy while still allowing for the importance of ubiquitylation in promoting pexophagy in FAF2-/- cells. Regardless, the relationship between FAF2 and ubiquitylation is more complex than expected. Since the deletion of the FAF2 UBX domain dramatically reduced interactions with p97/VCP (Supplementary Fig. 13c), we speculated that it would impede pexophagy from progressing. However, FAF2 ΔUBX partially recovered pexophagy (Fig. 5f, g). This could indicate that FAF2 ΔUBX tightly binds USP30, the effect of which would be a reduction in peroxisomal poly-ubiquitin chains due to USP30-mediated deubiquitylation.
Our study shows that pexophagy is induced in FAF2-/- cells by the accumulation of ubiquitylated proteins, such as PMP70, on peroxisomes and the subsequent recruitment of autophagy adaptors and LC3B. Adaptor proteins that recognize ubiquitin are critical for ubiquitylation-induced organellophagy54,55. Among the mammalian autophagy adaptors, NBR1 and p62 are known to be involved in pexophagy31,47. Importantly, our study shows that OPTN is also crucial for the pexophagy induced by FAF2 dysfunction (Fig. 6). This is reminiscent of the critical role that OPTN plays in Parkin- and ubiquitin-mediated mitophagy. To promote efficient mitophagy, OPTN links ubiquitylated mitochondria to LC3 family proteins through an LIR motif and to ATG9A vesicles via a leucine zipper domain56. We surmise that OPTN similarly assists in the de novo synthesis of autophagosomal membranes near ubiquitylated peroxisome in FAF2-/- cells. Although Deosaran et al reported that ubiquitylated PEX5 is required for the recruitment of NBR1 to peroxisomes during pexophagy47, PEX5S is not highly ubiquitylated in FAF2-/- cells (Supplementary Fig. 7e, f). The differences between OPTN and NBR1 in their contribution to pexophagy may be derived from differences in their respective substrates and, thus, the ubiquitylation pattern on the target peroxisomes.
In conclusion, FAF2 maintains peroxisomal integrity by preventing pexophagy, and the loss of FAF2 accelerates peroxisomal clearance. While pexophagy is inducible by PEX3-overexpression, reactive oxygen species, and amino acid starvation2,4,31, we revealed pexophagy under steady-state conditions and found that peroxisomal abundance is maintained by the p97/VCP-FAF2 complex, which antagonizes constitutive pexophagy. Even in the absence of pexophagy stimulation, such as PEX3-overproduction, cells are continuously at risk for accelerated pexophagy and the concomitant removal of peroxisomes. FAF2 maintains peroxisome abundance by preventing such accelerated pexophagy. Our study thus provides insights into the regulation of peroxisome integrity via ubiquitylation-mediated downregulation.
Methods
Plasmids and antibodies
To construct plasmids for transient expression of PEX3-YFP, the PEX3 coding sequence was amplified from pcDNAZeo/RnPEX3-HA2 and inserted into the BamHI/EcoRI sites of pEYFP-N1 (Clontech). For stable expression of mKeima-SKL, the coding region was inserted into the BamHI/EcoRI sites of the pMXs-Puro retroviral expression vector (CELL BIOLABS, INC.). To make pMXs-Puro/3HA-FAF2, 3HA-PEX5S, and 3Flag-PEX14, the respective coding regions were inserted into the BamHI/EcoRI site of pMXs-Puro/3HA-TEV or pMXs-Puro/3Flag-TEV as previously reported56. For stable expression of PMP70-3Flag, the PMP70 coding sequence was amplified from pcDNA-Zeo/MmPMP70-HA3 and inserted into the BamHI/EcoRI sites of pBABE-Puro-TEV-3Flag56. For stable expression of 3Flag-PEX16, the PEX16 coding sequence was amplified from pcDNA-Zeo/HA3-HsPEX16 and inserted into the BamHI/EcoRI sites of pMXs-Puro-3Flag-TEV56. pCMV2/Flag-p97/VCP was described previously57, and the E305Q/E578Q (p97QQ) plasmid was shared by Dr. Kakizuka (Kyoto University). pcDNAZeo/RnPEX3-HA2, pcDNA-Zeo/MmPMP70-HA3, and pcDNA-Zeo/HA3-HsPEX16 were kindly shared by Dr. Fujiki. For stable expression of Halo-mGFP-SKL, SKL coding sequence was inserted into pMRX-IBU-HaloTag7-mGFP by primer-based PCR. To construct PMP34-Halo-mGFP, PMP34 coding sequence was amplified by PCR and inserted into the HindIII site of pMRX-IBU-HaloTag7-mGFP. The pMRX-IBU-HaloTag7-mGFP plasmid was shared by Dr. Yamamoto (Nippon Medical School). For the siRNA-resistant PMP70-3Flag plasmid, we mutated PMP70 from CCTCTTATCTCTCTGGTT to CCCTTGATAAGCCTCGTG by primer-based PCR. For stable expression of PEX19-P2A-GFP-SKL, the PEX19 coding region was inserted into the BamHI site of the pMXs-Puro/P2A, and then the PEX19-P2A region was amplified by PCR and subsequently inserted into the BamHI site of the pMXspuro-GFP-SKL. pMXs-Puro/3Flag-OPTN and pMXs-Puro/P2A were shared by Dr. Yamano. Antibodies used in this study are listed in Supplementary Fig. 14.
Cell culture, transfections, and reagents
HeLa cells were cultured at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing 1× nonessential amino acids (Gibco), 1× sodium pyruvate (Gibco), 1× penicillin–streptomycin–glutamine (Gibco), and 10% fetal bovine serum. HCT116 cells were cultured in McCoy’s 5 A medium (Gibco) supplemented with 1× nonessential amino acids (Gibco), 1× GlutaMAX (Gibco), and 10% fetal bovine serum. HCT116 or HeLa cells stably expressing 3HA-FAF2, PMP70-3Flag, 3HA-PEX5S, 3Flag-PEX14, 3Flag-PEX16, 3Flag-MITOL/HA-parkin, 3Flag-OPTN, mKeima-SKL, GFP-SKL, Halo-mGFP-SKL, and PMP34-Halo-mGFP were established by recombinant retrovirus infection. Virus particles were produced in HEK293T cells by co-transfection with Gag-Pol, VSV-G, and the retrovirus plasmids using Lipofectamine LTX Reagent (Thermo Fisher Scientific)58. After 12 h, the transfection medium was replaced with fresh medium, and the cells were further cultivated for 24 h. Collected viral supernatants were then added to either HCT116 cells or HeLa cells with 8 µg/mL polybrene (SIGMA). Plasmid transfections were performed using Fugene6 Reagent (Roche) according to the manufacturer’s protocol.
MG132 (peptide Institute, Cat# 3175-v) was used at 10 µM to inhibit proteasome activity, and NMS-873 (Cat# SML1128, SIGMA) was used at 10 µM to inhibit p97/VCP activity. For autophagy inhibition and the induction of lysosomal rupture, Baf.A1 (Cat# BVT-0205-M001, AdipoGen) and LLOMe (Cat# L7393-500MG, SIGMA) were used at 100 nM and 0.3 mg/mL, respectively. Torin1 (Cat# 14379S, Cell Signaling) was used at 1 µM to promote autophagy. BSA (Cat# 8806-1 G, SIGMA) and oleic acid [Cat# 4954-1GM, Calbiochem (Millipore)] were used at 200 µM to observe lipid droplets.
RNA interference
Non-targeting control siRNA was shared by Dr. Kimura (Shizuoka University). For siRNA knockdown of USP30, OPTN, NDP52, TAX1BP1, FIP200/RB1CC1, and PEX16, Silencer Select Pre-designed siRNAs (IDs s39400, s19719, s19996, s16986, and s18994) and Stealth siRNA (HSS145149) were purchased from Thermo Fisher Scientific. siGENOME siRNA (D-010230-02-0005, D-010522-01-0005, and M-009909-01-0005) and custom siRNAs (LP_23815) for knockdown of p62, NBR1, NSFL1C/p47, and PMP70 were purchased from Dharmacon. Genesolution siRNAs (SI04323347 and SI04151343) for knockdown of Rep8/UBXD6 and FAF2 were purchased from QIAGEN.
The siRNA oligonucleotide sequences are as follows: control, 5’-CGUUAAUCGCGUAUAAUACGCGUAT-3’; USP30, 5’- CCAGAGUCCUGUUCGAUUUtt-3’; OPTN, 5’-GGAGACUGUUGGAAGCGAAtt-3’; NDP52, 5’-CCUUCAUGUGGGUUACUUUtt-3’; TAX1BP1, 5’-CUGAUACACUGGAACACGAtt-3’; FIP200/RB1CC1, 5’- GCCUAGAACAACUAACGAATT-3’; PEX16 5’-GGAUCCUACGGAAGGAGCUUCGGAA-3’; p62, 5’-GAUCUGCGAUGGCUGCAAU-3’; NBR1, 5’-GGAGUGGAUUUACCAGUUA-3’; NSFL1C/p47, 5’-CCAGCAUGUUGUACGGAAA-3’,5’-CCACUUGUUCCCACGAGAA-3’, 5’-UGACUACUUUCCCGAACAA-3’, and 5’-GGAGAGACCAGUAAACCGA-3’; Rep8/UBXD6, 5’-CTGGATGACGAGAATTGGGTA-3’; FAF2, 5’-CTCGTCTGTGGTTGAGTTTAT-3’; ABCD3/PMP70, 5’-GAGUAAGGCUCACUAAAUA-3’, 5’-CCAAAUACCUUGCCACUGU-3’, 5’-CCUCUUAUCUCUCUGGUUA-3’, and 5’-UCAAUAAGCUAGAUACGAA-3’. siRNAs for NPLOC4 and UFD1 knockdown were used as previously reported59. siRNAs were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. Transfection media was replaced with fresh media after 24 h, and the cells were grown for an additional 24 h.
CRISPR/Cas9-based generation of PEX19 and FAF2 knock-out cell lines
PEX19-/- HCT116 cells were established as previously reported14. FAF2-/- HCT116 cells were established by CRISPR/Cas9-based genome editing with an antibiotic-selection strategy. The gRNA target sequence (5’- GGATCAGTGTCGCCATACCTTGG-3’) was designed using an online CRISPR design tool (CRISPRdirect) to a region in exon 2 of FAF2. Two DNA oligonucleotides, hFAF2-ex2-2-CRISPR-F (5’-TGTATGAGACCAC GGATCAGTGTCGCCATACCT-3’) and hFAF2-ex2-2-CRISPR-R (5’-AAAC AGG TAT GGC GAC ACT GAT CC GTGGTCTCA−3’) were annealed and introduced into a linearized pEF1-hspCas9-H1-gRNA vector (Cas9 SmartNuclease TM; System Biosciences, LLC) according to the manufacturer’s protocol. The DNA fragment was verified by DNA sequencing. A 247-bp region of the 5’ and 3’ homology arms of FAF2 exon 2, which lacks the gRNA target sequence but has a BamHI site in the middle (total 500 bp), was synthesized and cloned into pUC57-Amp (GENEWIZ) to make pUC57-Amp/FAF2-ex2-donor. The neomycin-resistant (NeoR) and the hygromycin-resistant genes (HygroR) were extracted by BamHI digestion from pBSK/NeoR and pBSK/HygroR56 and inserted into the BamHI site of pUC-Amp/FAF2-ex2-donor. The resultant NeoR and HygroR donor plasmids containing the FAF2 exon 2 homology arm were then transfected into HCT116 cells along with the gRNA plasmid using FuGENE6 (Promega). Cells were grown in McCoy’s 5 A media containing 700 µg/mL G418 (Cat# G8168; SIGMA) and 100 µg/mL hygromycin B (Cat# 10687-10, Invitrogen). Single clones were isolated, and genomic DNA was prepared accordingly. FAF2-/- single clones were screened by PCR using hFAF2-ex2-2- check-F2 (5’-TTTGCTTAGCCCACAGGTTGAA-3’) and hFAF2-ex2-2-check-R2 (5’-GGAAAAACATACGGTTTTGACTCT-3’) primers to verify NeoR and HygroR insertion. Absence of the FAF2 protein was confirmed by immunoblotting with an anti-FAF2 antibody.
To establish FIP200 knockout HCT116 cells and PEX19/FIP200, FAF2/FIP200 double knockout HCT116 cells, the gRNA target sequence (5’- CAAGATTGCTATTCAACACC-3’) was designed using online CRISPR design tool (SYNTHEGO) to a region in exon 4 of FIP200. The oligonucleotide pair was annealed and introduced into WT HCT116, PEX19-/-, and FAF2-/- cell lines. Puromycin-resistant cells were seeded into 96-well plates, single clones were analyzed by immunoblotting, and genomic DNA was sequenced to confirm FAF2 knockout.
Immunoprecipitation
HCT116 cells grown in a 6-cm dish were solubilized with TNE-N+ buffer [20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and protease-inhibitor cocktail complete EDTA-free (Roche) in the presence or absence of 1 mM N-ethylmaleimide (NEM)]. After centrifugation, the supernatant was incubated with anti-DDDDK-tag mAb agarose beads (Cat # 3329, MBL) or anti-HA-tag mAb agarose beads (Cat # A2095, SIGMA) at 4 °C. The agarose was washed three times with TNE-N+ buffer, and proteins were extracted by adding 6× SDS-PAGE sample buffer [0.28 M Tris-HCl (pH 6.8), 30% glycerol, 0.5 M DTT, 10% SDS, and 0.05% bromophenol blue]. To stabilize protein-protein interactions via chemical crosslinking, cells were treated with 0.5 mM dithiobis(succinimidyl propionate) (DSP, Thermo Fisher Scientific) at room temperature for 30 min. The crosslinking reaction was quenched with 100 mM Tris-HCl, pH 7.5. After washing with TBS, the cells were lysed with TNE-N+ lysis buffer supplemented with protease-inhibitor cocktail complete EDTA-free (Roche). After centrifugation, the collected supernatant was incubated with equilibrated agarose beads.
Immunoblotting
HeLa and HCT116 cells were solubilized with TNE-N+ lysis buffer. After removing insoluble debris by centrifugation, supernatants were collected to obtain total cell lysates. The total protein concentration of the lysates was determined using a BCA Protein Assay Kit (Pierce). SDS-PAGE sample buffer was added to the lysates, and the samples were boiled at 98 °C for 5 min or incubated at room temperature for 1 h. Proteins were separated on 4-12% Bis-Tris SDS-PAGE gels (NuPAGE, Invitrogen) in MOPS buffer. Proteins were transferred to PVDF membranes (Merck), blocked with 1 or 5% skim milk/TBST, and incubated with primary antibodies. Membranes were then incubated with horseradish-peroxidase-conjugated goat anti-mouse, goat anti-rabbit, or donkey anti-rat (Cat # 315-035-048, # 111-035-144, and # 712-035-153; Jackson ImmunoResearch Inc.) secondary antibodies. Images were obtained using an ImageQuant LAS 4000 (GE Healthcare) or Fusion SOLO S system (VILBER). Band intensities were quantified by ImageJ.
HaloTag (Halo)-based reporter processing assay
A HaloTag (Halo)-based reporter processing assay32 was used to monitor pexophagy flux. Cells were incubated for 20 min with 100 nM TMR-ligand (Cat# G8251, Promega), and then washed twice with PBS. Cell lysates were collected after 18 h and subjected to SDS-PAGE.
Immunocytochemistry
HCT116 cells were fixed with 4% paraformaldehyde (Wako), permeabilized with 0.15% Triton X-100 (MP Biomedicals), 0.5% Triton X-100, or 0.15% saponin (SIGMA), and then incubated first with primary antibodies followed by 1:2000 secondary antibodies [Alexa Fluor 488-, 568-, or 647- conjugated goat or donkey anti-mouse, anti-rabbit, or anti-goat IgG antibody (Invitrogen)]. To stain the nucleus, cells were incubated with 0.3 µg/mL DAPI (Thermo Fisher Scientific) in PBS for 5 min. BODIPY 493/503 (Cat# D3922, Thermo Fisher Scientific) was used at 1 µM and was incubated with the secondary antibodies.
Microscopy images were acquired at room temperature on a laser-scanning microscope with a Plan Apochromat 63x/1.4 Oil objective (LSM710 or LSM780; Carl Zeiss) or with a PlanApo N 60x/1.4 oil objective lens (FV3000; Olympus). Image sizes were adjusted using Photoshop (Adobe). The number of peroxisomes per cell was determined using PMP70- or catalase-positive dots per cell in randomly selected cells.
FACS-based pexophagy assay
HCT116 cells transiently or stably expressing YFP and mKeima-SKL in a 6-well plate were resuspended in a sorting buffer (phosphate buffer with 2.5% FBS). Analysis was performed with FACSDiva on a BD LSRFortessa X-20 cell sorter. mKeima was measured using dual-excitation ratiometric pH measurements at 405-nm (pH 7) and 561-nm (pH 4) with 610/20-nm emission filters. For each sample, 10,000 mKeima-SKL positive cells were collected. The percentage of pexophagy-positive cells were plotted as box-plots with dots indicating individual data points, the center lines the medians, and the box limits the 25th and 75th percentiles as determined using R. The box-plot whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles.
In situ proximity ligation assay (PLA)
Cells were fixed with 4 % PFA and permeabilized with 0.15 % TritonX-100. After incubation with Blocking One P (Nakarai), cells were reacted with primary antibodies that consisted of rabbit anti-FAF2 and mouse anti-PMP70 antibodies. Secondary antibodies were oligonucleotide-conjugated antibodies (PLA probes). Following the inclusion of a DNA polymerase and fluorescence-labeled oligonucleotides, PLA dot signals would only be generated if the probes were in close proximity (<40 nm). The PLA probes (Cat# Duo92001-100RXN and Duo92005-100RXN), ligase, and polymerase (Cat# Duo92008-100RXN) were purchased from SIGMA (Duolink).
Bacterial expression of FAF2 UBA recombinant proteins
The coding sequence for the human FAF2 UBA domain was subcloned into pGEX-6p-1. E. coli BL21 (DE3) codon plus + RIL competent cells (Cat# 230245, Agilent) were transformed with the resultant plasmids (pGEX6p-1/FAF2 UBA or pGEX6p-1/FAF2 UBA V47A/L51A) and GST-tagged proteins were expressed with 0.1 mM IPTG at 18 °C overnight. GST-FAF2 UBA proteins were mixed with glutathione-sepharose 4B beads (GE Healthcare) in buffer A [50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 1 mM DTT] and eluted with 20 mM glutathione. The elution buffer was then exchanged with buffer A using a PD miditrap G-25 (GE Healthcare).
Preparation of polyubiquitin chains
Bovine ubiquitin was purchased from SIGMA (Cat# Ub253-25MG). Long unanchored K48- and K63-linked polyubiquitin chains were prepared using UBE2K/E2-25K and UBE2N/UBC13-UBE2V2/MMS2 as described previously60,61.
GST-FAF2 UBA pulldown of free-ubiquitin chains
Beads with immobilized FAF2 UBA proteins were incubated for 1 h at room temperature with polyubiquitin chains in buffer A containing 0.1% Triton X-100. After three washes with the same buffer, the proteins were eluted with SDS sample buffer.
Statistical analysis
Statistical analyses were performed using data obtained from two or more biologically independent experiments. Welch’s t-test was used for comparisons between two groups and ANOVA was used for multiple comparisons using GraphPad Prism.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Dr. Yoko Kimura (Shizuoka University) and Dr. Akira Kakizuka (Kyoto University) for the p97/VCP plasmid. We also thank Dr. Hikaru Tsuchiya for the E2 recombinant proteins (E2-25K, Ubc13/UBE2N, and MMS2/UBE2V2), and are grateful to Dr. Kanji Okumoto (Kyushu University) and Dr. Kosuke Tanegashima (Tokyo Metropolitan Institute of Medical Science) for valuable comments and technical support. We thank Dr. Daisuke Morito (Showa University) for the lipid droplet-staining protocol. This work was supported by JSPS KAKENHI Grant JP19H05712, AMED CREST Grant JP23gm1410004, Takeda Science Foundation, Nanken-Kyoten (2023-kokunai), and Joint Usage and Joint Research Programs of the Institute of Advanced Medical Sciences of Tokushima University (to N.M.); by JSPS KAKENHI Grant Numbers JP26000014, 19H00997, and the Takeda Science Foundation (to K.T.); by JSPS KAKENHI Grant Numbers JP15K19037, JP18K14708, JP21K06161, TMDU priority research areas grant, and MRI research grant FY2023 (to F.K.); by MEXT/JSPS KAKENHI Grant Numbers JP18H05500, JP16K18545, and JP18K06237 (to K.Y.); and by MEXT/JSPS KAKENHI Grant Numbers JP26116007, JP15K21743, and JP17H03675, and by Meiji (to Y.F.). During the revision of our manuscript, Dr. Keiji Tanaka passed away. We would like to express our sincere condolences.
Author contributions
Conceptualization: F.K., K.Y., N.M., K.T.; methodology: F.K., K.Y.; investigation: F.K., T.H.; writing—original draft: F.K.; review and editing: F.K., K.Y., Y.F., H.K., N.M., K.T.; funding acquisition: F.K., K.Y., Y.F., N.M., K.T.; and supervision: K.Y., N.M., K.T.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided in this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased: Keiji Tanaka.
Contributor Information
Fumika Koyano, Email: koyano-fm.biom@tmd.ac.jp.
Noriyuki Matsuda, Email: nr-matsuda.biom@tmd.ac.jp.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53558-x.
References
- 1.Fujiki, Y., Okumoto, K., Mukai, S., Honsho, M. & Tamura, S. Peroxisome biogenesis in mammalian cells. Front. Physiol.5, 307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang, J. et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol.17, 1259–1269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujiki, Y. et al. Recent insights into peroxisome biogenesis and associated diseases. J. Cell Sci.133, jcs236943 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Sargent, G. et al. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J. Cell Biol.214, 677–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bingol, B. et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature510, 370–375 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Liang, J. R. et al. USP30 deubiquitylates mitochondrial Parkin substrates and restricts apoptotic cell death. EMBO Rep.16, 618–627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham, C. N. et al. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol.17, 160–169 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Gersch, M. et al. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat. Struct. Mol. Biol.24, 920–930 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato, Y. et al. Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30. Nat. Struct. Mol. Biol.24, 911–919 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Ordureau, A. et al. Global landscape and dynamics of parkin and USP30-dependent ubiquitylomes in ineurons during mitophagic signaling. Mol. Cell77, 1124–1142.e1110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stach, L. & Freemont, P. S. The AAA+ ATPase p97, a cellular multitool. Biochem. J.474, 2953–2976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka, A. et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol.191, 1367–1380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura, Y. et al. Different dynamic movements of wild-type and pathogenic VCPs and their cofactors to damaged mitochondria in a Parkin-mediated mitochondrial quality control system. Genes Cells18, 1131–1143 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Koyano, F. et al. Parkin-mediated ubiquitylation redistributes MITOL/March5 from mitochondria to peroxisomes. EMBO Rep.20, e47728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araki, K. & Nagata, K. Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol.3, a007526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller, B., Klemm, E. J., Spooner, E., Claessen, J. H. & Ploegh, H. L. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl Acad. Sci. USA105, 12325–12330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Boom, J. & Meyer, H. VCP/p97-mediated unfolding as a principle in protein homeostasis and signaling. Mol. Cell69, 182–194 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Raman, M. et al. Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nat. Cell Biol.17, 1356–1369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olzmann, J. A., Richter, C. M. & Kopito, R. R. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl Acad. Sci. USA110, 1345–1350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrul, B. & Kopito, R. R. Peroxin-dependent targeting of a lipid-droplet-destined membrane protein to ER subdomains. Nat. Cell Biol.18, 740–751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, C. W. & Lee, S. C. The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J. Cell Sci.125, 2930–2939 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Neuber, O., Jarosch, E., Volkwein, C., Walter, J. & Sommer, T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat. Cell Biol.7, 993–998 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Schuberth, C. & Buchberger, A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat. Cell Biol.7, 999–1006 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Schuberth, C., Richly, H., Rumpf, S. & Buchberger, A. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep.5, 818–824 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mårtensson, C. U. et al. Mitochondrial protein translocation-associated degradation. Nature569, 679–683 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Marcassa, E. et al. Dual role of USP30 in controlling basal pexophagy and mitophagy. EMBO Rep.19, e45595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riccio, V. et al. Deubiquitinating enzyme USP30 maintains basal peroxisome abundance by regulating pexophagy. J. Cell Biol.218, 798–807 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng, J., Cao, Y., Yang, J. & Jiang, H. UBXD8 mediates mitochondria-associated degradation to restrain apoptosis and mitophagy. EMBO Rep.23, e54859 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganji, R. et al. The p97-UBXD8 complex regulates ER-Mitochondria contact sites by altering membrane lipid saturation and composition. Nat. Commun.14, 638 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katayama, H., Kogure, T., Mizushima, N., Yoshimori, T. & Miyawaki, A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol.18, 1042–1052 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Yamashita, S.-I., Abe, K., Tatemichi, Y. & Fujiki, Y. The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy10, 1549–1564 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yim, W. W., Yamamoto, H. & Mizushima, N. A pulse-chasable reporter processing assay for mammalian autophagic flux with HaloTag. Elife11. 10.7554/eLife.78923 (2022) [DOI] [PMC free article] [PubMed]
- 33.Hosokawa, N. et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell20, 1981–1991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirozh, O. et al. Nucleolar stress caused by arginine-rich peptides triggers a ribosomopathy and accelerates aging in mice. Mol. Cell84, 1527–1540.e1527 (2024). [DOI] [PubMed] [Google Scholar]
- 35.Zheng, J., Chen, X., Liu, Q., Zhong, G. & Zhuang, M. Ubiquitin ligase MARCH5 localizes to peroxisomes to regulate pexophagy. J. Cell Biol. 221. 10.1083/jcb.202103156 (2022) [DOI] [PMC free article] [PubMed]
- 36.Honsho, M., Yamashita, S. & Fujiki, Y. Peroxisome homeostasis: mechanisms of division and selective degradation of peroxisomes in mammals. Biochim. Biophys. Acta1863, 984–991 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Liu, Y., Yagita, Y. & Fujiki, Y. Assembly of peroxisomal membrane proteins via the direct Pex19p-Pex3p pathway. Traffic17, 433–455 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Matsuzono, Y. et al. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc. Natl Acad. Sci. USA96, 2116–2121 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan, X. et al. Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97. EMBO J.23, 1463–1473 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maejima, I. et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J.32, 2336–2347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okatsu, K. et al. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J. Cell Biol.209, 111–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, H. et al. UAS domain of Ubxd8 and FAF1 polymerizes upon interaction with long-chain unsaturated fatty acids. J. Lipid Res.54, 2144–2152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansen, T. & Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy7, 279–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Stolz, A., Ernst, A. & Dikic, I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol.16, 495–501 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Zaffagnini, G. & Martens, S. Mechanisms of selective autophagy. J. Mol. Biol.428, 1714–1724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deosaran, E. et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci.126, 939–952 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Okumoto, K., Kametani, Y. & Fujiki, Y. Two proteases, trypsin domain-containing 1 (Tysnd1) and peroxisomal lon protease (PsLon), cooperatively regulate fatty acid β-oxidation in peroxisomal matrix. J. Biol. Chem.286, 44367–44379 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita, A. et al. Depletion of LONP2 unmasks differential requirements for peroxisomal function between cell types and in cholesterol metabolism. Biol. Direct18, 60 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huybrechts, S. J. et al. Peroxisome dynamics in cultured mammalian cells. Traffic10, 1722–1733 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Kim, P. K., Hailey, D. W., Mullen, R. T. & Lippincott-Schwartz, J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl Acad. Sci. USA105, 20567–20574 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phu, L. et al. Dynamic regulation of mitochondrial import by the ubiquitin system. Mol. Cell77, 1107–1123 e1110 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Fujisawa, R., Polo Rivera, C. & Labib, K. P. M. Multiple UBX proteins reduce the ubiquitin threshold of the mammalian p97-UFD1-NPL4 unfoldase. Elife11. 10.7554/eLife.76763 (2022) [DOI] [PMC free article] [PubMed]
- 54.Heo, J. M., Ordureau, A., Paulo, J. A., Rinehart, J. & Harper, J. W. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell60, 7–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lazarou, M. et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature524, 309–314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamano, K. et al. Critical role of mitochondrial ubiquitination and the OPTN-ATG9A axis in mitophagy. J. Cell Biol.219, e201912144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noguchi, M. et al. ATPase activity of p97/valosin-containing protein is regulated by oxidative modification of the evolutionally conserved cysteine 522 residue in Walker A motif. J. Biol. Chem.280, 41332–41341 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Yamano, K. et al. Endosomal Rab cycles regulate Parkin-mediated mitophagy. Elife7, e31326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manno, A., Noguchi, M., Fukushi, J., Motohashi, Y. & Kakizuka, A. Enhanced ATPase activities as a primary defect of mutant valosin-containing proteins that cause inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Genes Cells15, 911–922 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J.25, 4877–4887 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato, Y. et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature455, 358–362 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided in this paper.