Abstract
Background
Members of the p24 (p24/gp25L/emp24/Erp) family of proteins have been shown to be critical components of the coated vesicles that are involved in the transportation of cargo molecules from the endoplasmic reticulum to the Golgi complex. The p24 proteins form hetero-oligomeric complexes and are believed to function as receptors for specific secretory cargo.
Results
Using sensitive sequence-profile analysis methods, we identified a novel β-strand-rich domain, the GOLD (Golgi dynamics) domain, in the p24 proteins and several other proteins with roles in Golgi dynamics and secretion. This domain is predicted to mediate diverse protein-protein interactions. Other than in the p24 proteins, the GOLD domain is always found combined with lipid- or membrane-association domains such as the pleckstrin homology (PH), Sec14p and FYVE domains.
Conclusions
The identification of the GOLD domain could aid in directed investigation of the role of the p24 proteins in the secretion process. The newly detected group of GOLD-domain proteins, which might simultaneously bind membranes and other proteins, point to the existence of a novel class of adaptors that could have a role in the assembly of membrane-associated complexes or in regulating assembly of cargo into membranous vesicles.
Background
The Golgi complex is the central secretory organelle of most eukaryotic cells and consists of membranous stacks called cisternae [1,2]. Secreted proteins, like all other proteins, are synthesized in the endoplasmic reticulum (ER) and are specifically packaged into vesicles that bud off from the ER in a GTP-dependent process [3,4]. These lipid vesicles are coated with the COPII coat protein-complex and are equipped with the ATP-dependent vesicle-fusion apparatus. They carry the secretory cargo to the cis surface of the Golgi complex, with which they fuse, delivering the cargo. A second type of vesicle, coated by the COPI coat-protein complex, is part of a retrograde pathway that buds off the Golgi membrane and returns proteins that are not targeted for secretion back to the endoplasm [3,4].
Studies on the secretory system in crown-group eukaryotes (plants, animals and fungi) have uncovered a family of proteins, the p24 (p24/gp25L/emp24/Erp) family, that have an important role in cargo selection and packaging into COPII-coated vesicles [5,6,7,8]. Additionally, they might also function in excluding secreted proteins from COPI-coated retrograde vesicles [9,10]. Members of the p24 family are type I membrane proteins, with a small carboxy-terminal cytoplasmic tail that interacts with the vesicle coat proteins and a globular lumenal region that probably interacts with the cargo [11,12]. They are abundantly distributed on the membranes of the vesicles budding off the ER and the cis Golgi membranes. The p24 proteins belong to at least four distinct subfamilies [8,12] and form hetero-oligomeric complexes that contain at least one member from each subfamily. This heteromerization of the p24 proteins has been shown to require a coiled-coil stretch at the extreme carboxyl terminus of their lumenal regions [10].
Improved understanding of the p24 family may throw light on evolution and function of the Golgi apparatus in eukaryotes. With this objective, we conducted a computational sequence analysis of the p24 proteins and show that they contain a conserved globular domain that is also present in several other Golgi and lipid-traffic proteins. We present evidence that this module is likely to serve as a common denominator in protein-protein interactions in several distinct contexts, such as in secretory vesicles and on the Golgi peripheral membrane. The proliferation of this superfamily appears to have been central to the diversification of the eukaryotic secretory apparatus.
Results and discussion
Identification of a conserved domain in p24 and other Golgi proteins
The bona fide p24 proteins contain a short carboxy-terminal tail that interacts with the COP-complex proteins through specific short peptide motifs. The amino-terminal region that faces the lumen is much larger and is predicted to form a compact globular unit. As this region of the protein is likely to contain a conserved globular domain that mediates other functional interactions of these proteins, we sought to investigate its complete diversity and potential evolutionary connections. We carried out a profile search of the Non-Redundant protein database (of the National Center for Biotechnology Information, NCBI) using the PSI-BLAST program [13], seeded with lumenal region of the Caenorhabditis elegans p24 family member K08E4.6 (the profile-inclusion threshold was set at 0.01 and the search iterated until convergence). This search readily detected the classical p24 family members that are found in six to nine copies in the proteomes of most organisms belonging to the eukaryotic crown group. In addition, this search retrieved several other proteins that do not belong to the p24 family with statistically significant expectation (E)-values (E < 0.001, see Figure 1 legend). These proteins include yeast Osh3p, a cytoplasmic oxysterol-binding protein, animal Sec14-like proteins that are involved in secretion, human GCP60 (also called PAP7, a peripheral-type benzodiazepine receptor-associated protein [14]), which interacts with the Golgi integral membrane protein Giantin, and several other uncharacterized eukaryotic proteins with different lipid-binding domains (Figure 1). Reciprocal searches initiated with this region from the newly detected proteins showed that they were more closely related to each other, but in subsequent iterations they recovered the classic p24 family members at significant E-values, suggesting that all these conserved regions define a novel superfamily of protein domains. Separate prediction of the secondary structure of this domain from the p24 family and the newly detected proteins, showed that the two groups had essentially the same core structural elements, further reinforcing their relationship. As this conserved domain is present in at least three distinct classes of proteins related to Golgi dynamics (animal Sec14 proteins, the p24 family and GCP60-like proteins), we name this conserved region the GOLD domain.
Figure 1.

A multiple alignment of the GOLD domain was constructed using T-Coffee [32] and realigning the sequences by parsing high-scoring pairs from PSI-BLAST search results. The PHD-secondary structure [15] is shown above the alignment, with E representing a β strand (upper-case letters indicate predictions with > 82% accuracy, and lower-case letters denote predictions with > 72% accuracy). A search with the lumenal region of Caenorhabditis elegans p24-family member K08E4.6 (region 20-191) recovers RALBP from Todarodes pacificus, the squid ortholog of SEC14L2 (E = 7 × 10-3, iteration 1). A reciprocal search with RALBP_Todarodes pacificus (region 189-343) recovers GCP60 (E = 2 × 10-10, iteration 1), FYCO1 (8 × 10-4, iteration 1), SPAC23H4.01c (10-3, iteration 2), KIAA0420 (10-3, iteration 2) and K08E4.6 (7 × 10-3, iteration 2). The 80% consensus shown below the alignment was derived using the following amino-acid classes: h, hydrophobic (ALICVMYFW, yellow shading); l, the aliphatic subset of the hydrophobic class (ALIVMC, yellow shading); a, aromatic (FHWY, yellow shading); s, small (ACDGNPSTV, green letters); u and p are the tiny subsets of the small class (u, GAS, green shading) and polar (p, CDEHKNQRST, blue letters). Y denotes a conserved tyrosine residue. The limits of the domains are indicated by the residue positions on each side. The numbers within the alignment indicate poorly conserved inserts that are not shown. The different families are shown on the right. A, p24 family; B, Osh3p family; C, CG9528 family (Sec14-like proteins with an amino-terminal PRELI/MSF1p' domain); D, Sec14-like proteins; E,GCP60 family; and F, FYCO1. The sequences are denoted by their gene name followed by the species abbreviation and GenBank Identifier. Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Top, Todarodes pacificus.
The presence of the GOLD domain at the extreme amino or carboxyl terminus of the Osh3p and animal Sec14 proteins, respectively, allowed us to establish accurate boundaries for it. The domain is typically between 90 and 150 amino acids long and, in the p24 family, it comprises almost the entire lumenal region, with the exception of an α-helical extension of approximately 50 amino acids that precedes the transmembrane segment. Most of the size difference observed in the GOLD-domain superfamily is traceable to a single large low-complexity insert that is seen in some versions of the domain. A secondary-structure prediction for the domain using the PHD [15] program reveals that it is likely to adopt a compact all-β-fold structure with six to seven strands. Most of the sequence conservation is centered on the hydrophobic cores that support these predicted strands. The predicted secondary-structure elements and the size of the conserved core of the domain suggests that it may form a β-sandwich fold with the strands arranged in two β sheets stacked on each other.
Experimental studies so far on diverse proteins containing GOLD domains point to a role for it in protein-protein interactions. A region of the GPC60 molecule that rather precisely encompasses the GOLD domain has been shown to bind to the cytoplasmic region of the Golgi membrane protein Giantin [16]. Cross-linking experiments have suggested that the p24 proteins interact directly with the cargo molecules that are present in the lumen of the COPII-coated vesicles and that they are, accordingly, cargo receptors [17]. However, yeast deletion mutants lacking all the p24 proteins grow similarly to wild type, although they show delays in translocation of a subset of cargo molecules such as invertase and Gas1p from the ER to the Golgi, and increased secretion of resident ER proteins [18]. Certain members of the p24 family from vertebrates have also been shown to bind to specific ligands such as the interleukin-1 receptor-like molecule T1/ST2 and might aid its proper expression on the cell surface [19]. These observations suggest that the p24 subset of the GOLD domains probably function as discriminators that selectively interact with particular proteins to influence their loading into vesicles. The GOLD domains show considerable variability in some of the loops that are predicted to extrude from the core β-sandwich-like structure (Figure 1). These loops might form exposed surfaces that provide the GOLD domains with the discriminatory capacity necessary for their interactions with diverse ligands.
The domain architecture and evolutionary history of GOLD-domain proteins: functional implications
With the exception of the p24 proteins, which have a simple architecture with the GOLD domain as their only globular domain, all other GOLD-domain proteins contain additional conserved globular domains (Figure 2). In these proteins, the GOLD domain co-occurs with lipid-, sterol- or fatty acid-binding domains such as PH [20,21], Sec14p [22], FYVE [23], oxysterol binding- and acyl CoA-binding domains, suggesting that these proteins may interact with membranes. The FYCO1 protein that combines a GOLD domain with a FYVE domain, also contains a RUN domain [24], an uncharacterized α-helical domain that may have a role in the interaction of various proteins with cytoskeletal filaments [24,25]. An orthologous group of proteins typified by human Sec14L1, which is conserved in all animals, has, in addition to the carboxy-terminal fusion of the Sec14p and GOLD domains, a previously unrecognized, conserved amino-terminal domain (Figures 2,3). This domain has so far been found only in eukaryotes, and occurs in stand-alone form in several proteins, including the human PRELI protein [26] and the yeast MSF1p' protein. The PRELI/MSF1p' domain is approximately 170 residues long and is predicted to assume a globular α + β fold with six β strands and four α helices (Figure 3). MSF1p' is proposed to be involved in mitochondrial protein sorting [27], suggesting that the PRELI/MSF1p' domain may also have a function associated with cellular membranes.
Figure 2.
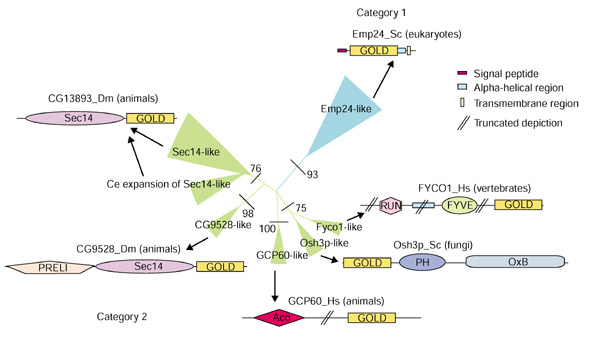
A phylogenetic tree of the GOLD-domain-containing proteins is shown along with the various architectures, drawn approximately to scale, and the phyletic distributions of individual architectural classes. The RELL bootstrap values for the major branches are shown at their base. The thickness of a given branch is approximately proportional to the number of proteins contained within it. PH, pleckstrin-homology domain; OxB, oxysterol-binding domain; Aco, acyl-CoA-binding domain; Sec14, domain found in Sec14 proteins; RUN (for RPIP8, UNC-14 and NESCA) and FYVE (for Fab1p, YOTB, Vac1p and EEA1).
Figure 3.
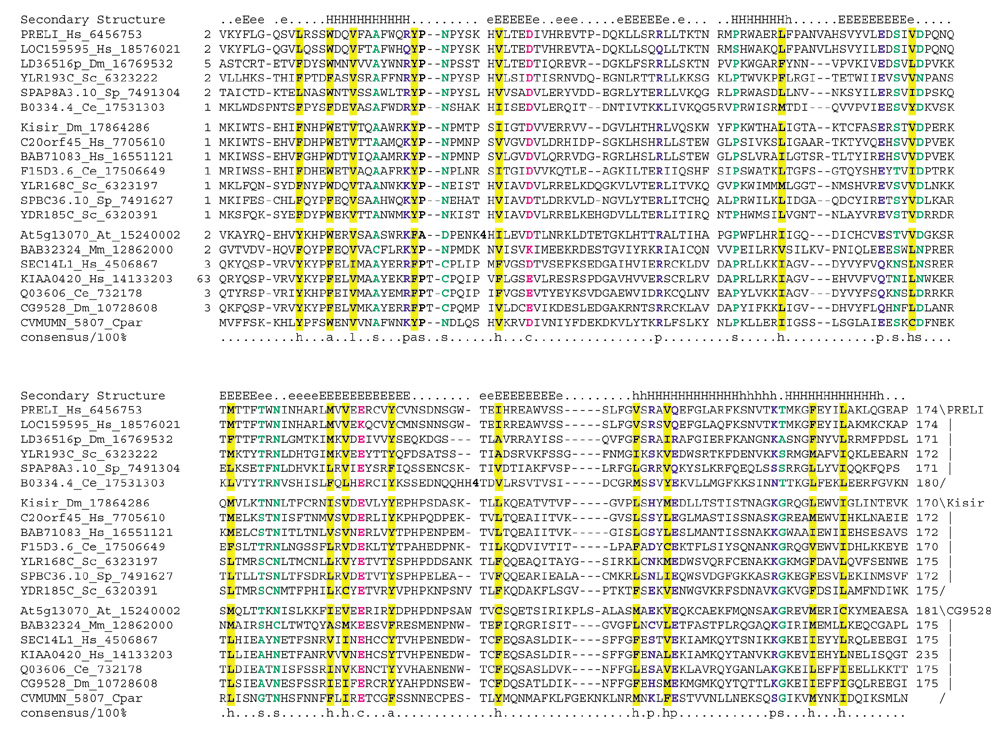
A multiple alignment of the PRELI/MSF1p' domain was constructed using T-Coffee [32] and realigning the sequences by parsing high-scoring pairs from PSI-BLAST search results. The PHD-secondary structure [15] is shown above the alignment with E representing a β strand (upper-case is for predictions with > 82% accuracy; lower-case denotes predictions with > 72% accuracy). The 100% consensus shown below the alignment was derived using the following amino-acid classes: h, hydrophobic (ALICVMYFW, yellow shading); l, the aliphatic subset of the hydrophobic class (ALIVMC, yellow shading); a, aromatic (FHWY, yellow shading); c, charged (DEHKR, pink letters); s, small (ACDGNPSTV, green letters) and p, polar (CDEHKNQRST, blue letters). The limits of the domains are indicated by the residue positions on each side (except for the unfinished genome of Cryptosporidium parvum). The numbers within the alignment are poorly conserved inserts that are not shown. The different families are shown on the right. The PRELI and Kisir subgroups contain stand-alone versions of the domain, whereas the CG9528 family comprises Sec14-like proteins with an amino-terminal PRELI and a carboxy-terminal GOLD domain. The sequences are denoted by their gene name followed by the species abbreviation and GenBank Identifier. At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Cpar, Cryptosporidium parvum; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe.
Thus, all GOLD-domain proteins can be divided into two architectural categories: the p24-like category, in which the GOLD domains project into the lumen, anchored in the membrane by the membrane-spanning helix (category 1); and proteins in which the GOLD domain occurs at the extreme amino or carboxyl terminus, with additional domains that are known to interact with lipid membranes (category 2) (Figure 2). GCP60, which is peripherally associated with the Golgi membrane, is one of the proteins in the second category that has been experimentally characterized. It has been shown that overexpression of a region of this protein encompassing the GOLD domain caused disassembly of the Golgi structure and abrogated protein transport from the ER to the Golgi [16].
These observations can be accommodated by two (not mutually exclusive) hypotheses regarding the functions of these proteins. The GOLD proteins belonging to the second architectural category could function as double-headed adaptors that interact with both a specific protein (via the GOLD domain) and different cellular lipid membranes. Thus, GCP60 and GOLD proteins with analogous architectures could help in the assembly of vesicular or Golgi-membrane-associated protein complexes by tethering specific proteins to the membranes, with the GOLD domain binding the protein targets and the lipid-binding protein to the membrane. Alternatively, at least some of the category-2 proteins could function as a previously unrecognized class of vesicular cargo-loading molecules that associate with the membrane via their lipid-binding domains and deliver their protein ligands via the GOLD domain. The observation that deletion mutants lacking all the p24 proteins still show normal trafficking of certain proteins such as carboxypeptidase Y, suggests that there are some protein-trafficking pathways that are unaffected by their absence. Thus, the GOLD-domain proteins of category 2 may have a specific role in regulating the secretion of molecules that are not affected by the p24 proteins. The hetero-oligomerization of the p24 proteins via the coiled-coil regions carboxy-terminal to the GOLD domain seems to help in generating combinatorial diversity for their interactions with multiple ligands. The presence of extensive coiled-coil segments in some of the category-2 GOLD-domain proteins, such as FYCO1, suggests that they might also form oligomers, like the p24 proteins.
Similarity-based clustering and phylogenetic analysis divides the GOLD domains into two primary divisions that precisely mirror the two categories established on the basis of domain architectures (Figure 2). This division was also supported by a synapomorphic (shared derived) feature in the form of two conserved cysteines, which is restricted to the p24 family (category-1 proteins). Likewise, the presence of a specific insert between strand 1 and 2 with a characteristic conserved tryptophan serves as a synapomorphic feature for category-2 GOLD domains (Figure 1). An analysis of the phyletic patterns suggests that the p24 family had already differentiated into at least four distinct subfamilies in the common ancestor of plants, animals and fungi. The detection of multiple members of the p24 family in the early branching eukaryotes such as Cryptosporidium parvum and kinetoplastids suggests that some of this diversification was probably already under way early in eukaryotic evolution. Within the eukaryotic crown group, we obtained evidence of specific instances of duplications and gene losses that are restricted to particular lineages. The most striking case is seen in Arabidopsis thaliana, which appears to have proliferated the Erv25 subfamily (five to six members), but lacks the Erp2p and Erp5p subfamilies. The second major family of GOLD domains (category 2) is so far only attested in the crown group. In fungi, this group is typified by Saccharomyces cerevisiae Osh3p, which combines an amino-terminal GOLD domain with PH and oxysterol-binding domains. The greatest architectural diversity of this group is seen in animals (Figure 2), suggesting that there was increased proliferation and domain shuffling among these proteins concomitant with the evolutionary emergence of the animals. This might correlate with the increased complexity of animal-specific secretory functions.
Conclusions
A novel β-strand-rich domain was identified in numerous eukaryotic proteins, including the p24 proteins, which appear to have a function related to the Golgi complex, secretion or protein sorting. These GOLD domains are predicted to be involved in specific protein-protein interactions. Other than the p24 proteins, GOLD domains are present in several proteins where they occur at the extreme termini and are combined with diverse membrane- or lipid-binding domains. These proteins are predicted to be double-headed adaptors that may help in the assembly of protein complexes on membranes or in the packaging of specific cargo molecules in membranous vesicles. The identification to the GOLD domain may help in a directed dissection of p24-family function and provide novel candidate molecules for experimental studies on secretion and sorting.
Materials and methods
The Non-Redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda) was searched using the BLASTP program [13]. Profile searches were conducted using the PSI-BLAST program with either a single sequence or an alignment used as the query, with a profile-inclusion expectation (E)-value threshold of 0.01, and were iterated until convergence [13,28]. Previously known conserved protein domains were detected using the corresponding PSI-BLAST-derived position-specific scoring matrices (PSSMs) [29]. The PSSMs were prepared by choosing one or more starting queries (seeds) for a set of most frequently encountered domains (see reference [28] for details) and run against the NR database until convergence with the -C option of PSI-BLAST to save the PSSM. We ensured that at convergence no false positives were included in the profiles. This profile database can be downloaded from [30] or used on the internet via the RPS-BLAST program [31]. All globular segments of proteins that did not map to domains with previously constructed PSSMs were searched individually using PSI-BLAST to detect any additional domains that may have been overlooked.
Multiple alignments were constructed using the T-Coffee program [32], followed by manual correction based on the PSI-BLAST results. Protein secondary structure was predicted using a multiple alignment as the input for the PHD program [15]. Signal peptides were predicted using the SIGNALP program [33,34] and the transmembrane regions were predicted using the TOPRED program [35]. Phylogenetic analysis was carried out using the maximum likelihood, neighbor-joining and least-squares methods [36,37]. Briefly, this process involved the construction of a least-squares tree using the FITCH program or a neighbor-joining tree using the NEIGHBOR program (both from the Phylip package) [38], followed by local rearrangement using the Protml program of the Molphy package [37] to arrive at the maximum likelihood (ML) tree. The statistical significance of various nodes of this ML tree was assessed using the relative estimate of logarithmic likelihood bootstrap (Protml RELL-BP) with 10,000 replicates.
Acknowledgments
Acknowledgements
We thank Eugene Koonin for providing useful comments on the manuscript.
References
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell New York and London: Garland Publishing: 1999.
- Hong W. Protein transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Sci. 1998;111:2831–2839. doi: 10.1242/jcs.111.19.2831. [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Nishimura N, Balch WE. Getting into the Golgi. Trends Cell Biol. 1998;8:21–25. doi: 10.1016/s0962-8924(97)01184-7. [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada I, Rindress D, Cameron PH, Ou WJ, Doherty JJ, 2nd, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJ. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- Schimmoller F, Singer-Kruger B, Schroder S, Kruger U, Barlowe C, Riezman H. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes MA, Craighead MW, Hoe MH, Lampen N, Geromanos S, Tempst P, Rothman JE. An integral membrane component of coatomer-coated transport vesicles defines a family of proteins involved in budding. Proc Natl Acad Sci USA. 1995;92:8011–8015. doi: 10.1073/pnas.92.17.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch M, Henthorn DC, Herrmann JM, Wilson R, Thomas DY, Bergeron JJ, Solari RC, Rowley A. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol Biol Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler K, Veit M, Stamnes MA, Rothman JE. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- Ciufo LF, Boyd A. Identification of a lumenal sequence specifying the assembly of Emp24p into p24 complexes in the yeast secretory pathway. J Biol Chem. 2000;275:8382–8388. doi: 10.1074/jbc.275.12.8382. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Herrmann JM, Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Dejgaard K, Fullekrug J, Dahan S, Fazel A, Paccaud JP, Thomas DY, Bergeron JJ, Nilsson T. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J Cell Biol. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Degenhardt B, Tobin D, Yao Zx ZX, Tasken K, Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor- and PKA (RIalpha)-associated protein. Mol Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yamamoto A, Yano A, Nakamura N, Ikehara Y. Identification and characterization of a novel Golgi protein, GCP60, that interacts with the integral membrane protein Giantin. J Biol Chem. 2001;276:45298–45306. doi: 10.1074/jbc.M108961200. [DOI] [PubMed] [Google Scholar]
- Muniz M, Nuoffer C, Hauri HP, Riezman H. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J Cell Biol. 2000;148:925–930. doi: 10.1083/jcb.148.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer S, Chen E, Duden R, Marzioch M, Rowley A, Hamamoto S, Merchant S, Schekman R. The p24 proteins are not essential for vesicular transport in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:4034–4039. doi: 10.1073/pnas.070044097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayle MA, Slack JL, Bonnert TP, Renshaw BR, Sonoda G, Taguchi T, Testa JR, Dower SK, Sims JE. Cloning of a putative ligand for the T1/ST2 receptor. J Biol Chem. 1996;271:5784–5789. doi: 10.1074/jbc.271.10.5784. [DOI] [PubMed] [Google Scholar]
- Haslam RJ, Koide HB, Hemmings BA. Pleckstrin domain homology. Nature. 1993;363:309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- Mayer BJ, Ren R, Clark KL, Baltimore D. A putative modular domain present in diverse signaling proteins. Cell. 1993;73:629–630. doi: 10.1016/0092-8674(93)90244-k. [DOI] [PubMed] [Google Scholar]
- Aravind L, Neuwald AF, Ponting CP. Sec14p-like domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Curr Biol. 1999;9:R195–R197. doi: 10.1016/s0960-9822(99)80127-4. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Aasland R. FYVE-finger proteins - effectors of an inositol lipid. J Cell Sci. 1999;112:4175–4183. doi: 10.1242/jcs.112.23.4175. [DOI] [PubMed] [Google Scholar]
- Callebaut I, de Gunzburg J, Goud B, Mornon JP. RUN domains: a new family of domains involved in Ras-like GTPase signaling. Trends Biochem Sci. 2001;26:79–83. doi: 10.1016/s0968-0004(00)01730-8. [DOI] [PubMed] [Google Scholar]
- Mari M, Macia E, Le Marchand-Brustel Y, Cormont M. Role of the FYVE finger and the RUN domain for the subcellular localization of Rabip4. J Biol Chem. 2001;276:42501–42508. doi: 10.1074/jbc.M104885200. [DOI] [PubMed] [Google Scholar]
- Guzman-Rojas L, Sims JC, Rangel R, Guret C, Sun Y, Alcocer JM, Martinez-Valdez H. PRELI, the human homologue of the avian px19, is expressed by germinal center B lymphocytes. Int Immunol. 2000;12:607–612. doi: 10.1093/intimm/12.5.607. [DOI] [PubMed] [Google Scholar]
- Nakai M, Takada T, Endo T. Cloning of the YAP19 gene encoding a putative yeast homolog of AP19, the mammalian small chain of the clathrin-assembly proteins. Biochim Biophys Acta. 1993;1174:282–284. doi: 10.1016/0167-4781(93)90198-m. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Wolf YI, Ponting CP, Koonin EV, Aravind L, Altschul SF. IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics. 1999;15:1000–1011. doi: 10.1093/bioinformatics/15.12.1000. [DOI] [PubMed] [Google Scholar]
- IMPALA ftp site ftp://ftp.ncbi.nih.gov/pub/impala/
- NCBI Conserved domain database search http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction: hydrophobicity analysis and the 'positive inside' rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Saitou N. On the maximum likelihood method in molecular phylogenetics. J Mol Evol. 1991;32:443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics. 1989;5:164–166. [Google Scholar]


