Abstract
Current genome-wide association studies (GWAS) for kidney function lack ancestral diversity, limiting the applicability to broader populations. The East-Asian population is especially under-represented, despite having the highest global burden of end-stage kidney disease. We conducted a meta-analysis of multiple GWASs (n = 244,952) on estimated glomerular filtration rate and a replication dataset (n = 27,058) from Taiwan and Japan. This study identified 111 lead SNPs in 97 genomic risk loci. Functional enrichment analyses revealed that variants associated with F12 gene and a missense mutation in ABCG2 may contribute to chronic kidney disease (CKD) through influencing inflammation, coagulation, and urate metabolism pathways. In independent cohorts from Taiwan (n = 25,345) and the United Kingdom (n = 260,245), polygenic risk scores (PRSs) for CKD significantly stratified the risk of CKD (p < 0.0001). Further research is required to evaluate the clinical effectiveness of PRSCKD in the early prevention of kidney disease.
Subject terms: End-stage renal disease, Genome-wide association studies
Here the authors present a large genetic study in East Asians that identifies 97 genetic regions linked to kidney function. These findings aim at better understanding chronic kidney disease in diverse populations.
Introduction
No cure has yet been developed for chronic kidney disease (CKD), which affects more than 700 million people worldwide and thus places a substantial socioeconomic burden on the global economy and public health systems1. Sodium–glucose cotransporter-2 (SGLT2) inhibitors are the only oral drugs approved by the U.S. Food and Drug Administration to slow the progression of CKD. This therapeutic effect was an unexpected discovery2–4. Accordingly, an effective drug discovery platform for CKD should be established5. Regardless of which type of therapy is examined, such as cell therapy or antisense oligonucleotides6, identifying the genetic variants associated with CKD is essential for accelerating drug development efforts7.
More than 250 genetic loci associated with kidney functional markers, including the estimated glomerular filtration rate (eGFR) and the urine albumin-to-creatinine ratio, are highly replicated in large-scale population-based cohorts, such as those from the Million Veteran Program, Biobank Japan (BBJ) Project, and UK Biobank (UKB)8–10. However, undetermined pathogenic pathways, a dominant distribution in noncoding regions, and extensive linkage disequilibrium (LD) across common variants in these kidney function-related loci have prevented researchers from elucidating the functionality and pinpointing the casual variants8,11. In addition, the relatively limited ancestral and ethnic diversity of the individuals analyzed in existing large-scale genome-wide association studies (GWASs), which predominantly focus on Caucasian populations, may lead to significant limitations in regional practice and healthcare policy development for the Asian population12. Therefore, GWASs involving diverse populations with a range of kidney functional markers are urgently required for clinical and therapeutic translation.
Taiwan has the highest prevalence (3679 per million population) and incidence (823 per million population) of end-stage kidney disease (ESKD) worldwide, making it particularly suitable for CKD-related GWAS13,14. In a population-based family study that involved data obtained from Taiwan’s National Health Insurance Research Database, a high kidney disease heritability of 31.1% was observed, indicating that further examination of genetic inheritance is warranted15. In this study, to determine the effect of genetic heritability on common kidney functional markers in regions with a high prevalence of ESKD, we conducted a systematic analysis of GWAS findings in populations from the Taiwan Biobank (TWB), the BBJ, and a large hospital-based cohort from China Medical University Hospital (CMUH) in Taiwan.
Results
Discovery of genetic associations with eGFR through a meta-analysis of GWASs involving the BBJ and TWB
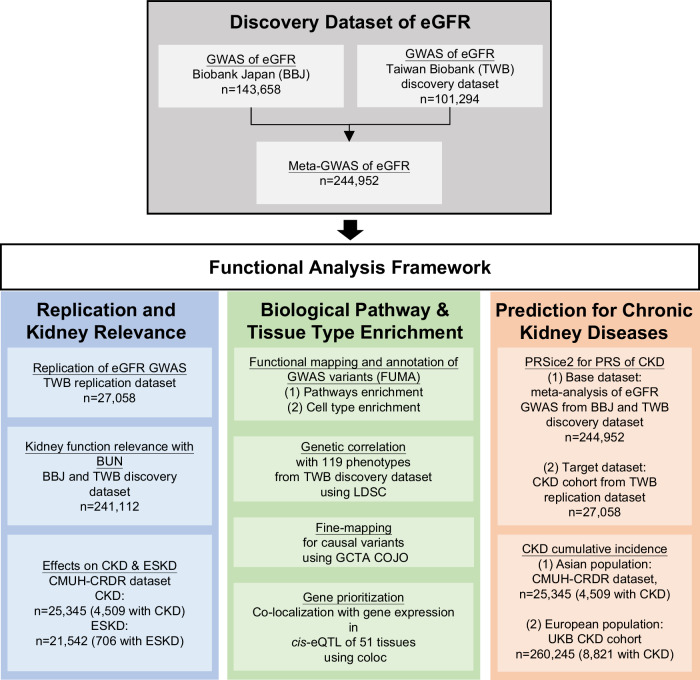
The present study design is depicted in Fig. 1. We conducted a fixed-effects inverse variance-weighted meta-analysis of GWASs involving two large biobanks containing samples from individuals of East Asian ancestry, namely the BBJ (n = 143,658) and TWB (n = 101,294). The mean eGFRs (standard deviations [SDs]) of the BBJ, TWB, and pooled populations were 79.93 ( ± 15.42), 101.95 ( ± 14.75), and 89.04 ( ± 15.14) mL/min/1.73 m2, respectively, and their corresponding mean ages (SDs) were 62.9 ( ± 13.2), 50.4 ( ± 10.9), and 57.7 ( ± 12.2) years, respectively (Supplementary Data 1). The genotypes used in the two GWASs were imputed from the East Asian reference panels of the 1000 Genomes Project Phase 316. As shown in Fig. 2 and Supplementary Data 2, a meta-analysis of these GWASs revealed 5790 genome-wide significant (P < 5 × 10−8) single-nucleotide polymorphisms (SNPs). In these genome-wide significant SNPs, 2140 were unreported in the previous eGFR GWASs10,17. A total of 238 independently significant SNPs (LD r2 < 0.6) were represented by 111 lead SNPs (LD r2 < 0.1) located in 97 genomic risk loci, of which 26 loci were unreported in other GWASs (Supplementary Data 3). Genomic risk loci were defined as nonoverlapping on the basis of a window size of ±250 kb around LD block of lead SNPs, and these loci were merged into a single locus if the distance between them was shorter than 250 kb10,17. The previously unreported genome-wide significant SNP with the lowest P value (P = 5.1 × 10−45) was rs754331108, which was located in the intronic region of CASP9. The minor alleles across genome-wide significant SNPs both increased and decreased the eGFR, with lower-frequency alleles exhibiting stronger effects (Supplementary Fig. 1a). The effects of genome-wide significant SNPs on the eGFR were largely homogeneous (median I2 = 0) across the two biobank populations (Supplementary Fig. 1b). According to meta-GWAS summary statistics, the estimated genetic heritability (h2) of the eGFR was 10.9%, and the LD score regression intercept was 1.05, indicating that the effects of bias due to population stratification and cryptic relatedness were negligible.
Fig. 1. Flowchart of the study design.
A meta-analysis of an eGFR GWAS was conducted using the BBJ and TWB. Replication was performed using an independent TWB-derived replication dataset. The relevance of the eGFR to kidney function was validated through the associations of the eGFR with BUN, CKD, and ESKD in the TWB and BBJ. Pathways and tissue types were enriched through the FUMA platform. Genetic correlation analysis of 119 traits was conducted using LD score regression. Fine mapping of causal variants was performed using GCTA-COJO, and gene prioritization with tissue-specific cis-eQTLs was conducted using the R package “coloc.” The PRS for CKD was derived using PRSice-2 and tested using patient data obtained from a Taiwanese hospital cohort (CMUH-CRDR CKD) and community-based data obtained from a UK cohort (UKB CKD). BBJ Biobank Japan, BUN blood urea nitrogen, cis-eQTL cis-expression quantitative trait locus, CKD chronic kidney disease, CMUH-CRDR Clinical Research Data Repository of China Medical University Hospital, eGFR estimated glomerular filtration rate, ESKD end-stage kidney disease, FUMA Functional Mapping and Annotation of Genome-Wide Association Studies, GCTA-COJO genome-wide complex trait analysis–conditional and joint analysis, GWAS genome-wide association study, LDSC linkage disequilibrium score regression, PRS polygenic risk score, SNP single-nucleotide polymorphism, TWB Taiwan Biobank, UKB UK Biobank.
Fig. 2. A circular Manhattan plot from a meta-analysis of eGFR-derived GWASs (TWB, n = 101,294; BBJ, n = 143,658; total n = 244,952).
The green band corresponds to –log10(P) for association with eGFR in the meta-analysis by chromosomal position. The blue band corresponds to –log10(P) for association with eGFR in the TWB-derived discovery dataset by chromosomal position. The orange band corresponds to –log10(P) for association with eGFR in the BBJ dataset by chromosomal position. The solid red line indicates genome-wide significance (P = 5 × 10–8). Genes labeled in black indicate SNPs exclusively identified in the meta-analysis, whereas genes labeled in blue indicate SNPs identified in the meta-analysis and additionally detected in the TWB or BBJ or both. A total of 5790 SNPs had P values of <5 × 10−8, of which 4732 had a consistent effect direction. The lowest P value was observed for rs62435145 near UNCX on chromosome 7 (P = 5.23 × 10−67 Supplementary Data 2). All statistical tests employed two-sided P values. BBJ Biobank Japan, eGFR estimated glomerular filtration rate, GWAS genome-wide association study, SNP single-nucleotide polymorphism, TWB Taiwan Biobank.
Replication of eGFR-associated SNPs in an independent TWB dataset
We evaluated the replication of eGFR-associated SNPs, defined as genome-wide significant SNPs, in an independent replication dataset that comprised data from 27,058 individuals in the TWB. Of the 5790 eGFR-associated SNPs identified during the discovery stage, 5342 were available in the replication dataset (Supplementary Data 4). These SNPs had a consistent effect direction in the TWB-based replication dataset, with 3899 of them having a P value of <0.05. The effect estimates strongly correlated with those from the discovery stage, exhibiting almost complete consistency in directionality (Pearson’s r = 0.97; P < 0.0001; Fig. 3). Of the 3899 replicated eGFR-associated SNPs, 387 from 10 independent genomic risk loci were genome-wide significant in the TWB-based replication dataset. These SNPs were located close to genes including ABCG2, BCAS3, CSP9, CCDC158, DCDC1, DNAJC16, LRP2, MRTFA, MUC1, NRG4, SCARNA21B, SHROOM3, SLC34A1, STBD1, THBS3, and TRIM46 (Supplementary Data 4).
Fig. 3. Replication of eGFR-associated SNPs in an independent replication dataset derived from the TWB (n = 27,058).

Data regarding 5342 out of 5790 eGFR-associated SNPs were available in the TWB-derived replication dataset. Of these eGFR-associated SNPs, 3899 were replicated in the TWB-derived replication dataset (two-sided P < 0.05, consistent effect direction), plotted as blue dots (“Yes” indicated by blue dots, with a consistent effect, two-sided P < 0.05; “Inconclusive” indicated by gray dots, two-sided P ≥ 0.05). The blue line represents the best fit of the blue dots. Pearson’s r is 0.97 (two-sided P < 0.0001). The data present the effect estimates, and error bars correspond to 95% CIs. Further details are provided in Supplementary Data 4. BBJ Biobank Japan, eGFR estimated glomerular filtration rate, SNP single-nucleotide polymorphism, TWB Taiwan Biobank.
Correlations of eGFR loci with blood urea nitrogen and CKD
We examined the correlation between eGFR-associated SNPs and blood urea nitrogen (BUN) to determine whether the identified SNPs were directly related to kidney function rather than creatinine metabolism10. Urea nitrogen is a harmful waste product that may accumulate in the blood if kidney function is impaired18. Our BUN-related meta-analysis of GWASs involved BBJ and TWB datasets (n = 241,112). A total of 2365 replicated eGFR-associated SNPs were associated with BUN (P < 0.05; Supplementary Fig. 2a and Supplementary Data 5). The effect sizes of 2064 replicated eGFR-associated SNPs from 31 independent genomic risk loci associated with eGFR and BUN were strongly and inversely correlated (defined as kidney-relevant); this finding was consistent with the established understanding of kidney pathophysiology (Pearson’s r = −0.86; P < 0.0001).
To determine whether the replicated eGFR-associated SNPs altered the risk of kidney diseases (e.g., CKD or ESKD), we conducted a logistic regression of a validation longitudinal cohort from the Clinical Research Data Repository of CMUH (CMUH-CRDR), which contains data regarding CKD follow-up status (CKD, n = 4509; control, n = 20,836). In addition, we examined the effect of replicated eGFR-associated SNPs on CKD; the results indicated that the effect direction of 397 replicated eGFR-associated SNPs on the eGFR was negatively correlated with the relevant effect direction of CKD (P < 0.05; maximum, median, and minimum odds ratios [ORs] = 1.02, 1.01, and 0.98, corresponding 95% confidence intervals [CIs] = 1.01–1.03, 1.00–1.01, 0.97–1.00, respectively; Pearson’s r = −0.94, P < 0.0001; Supplementary Fig. 2b and Supplementary Data 6). These SNPs included 160 from 14 independent genomic risk loci likely related to kidney function, which were located close to the following genes: ASCC3, DCDC1, F12, FAM47E, FBXO22, HCRTR2, KNG1, LRP2, RAI14, NRG4, PAX8, PDILT, SIM1, STC1, TINAG, UBE2Q2, and WDR72.
Data from the CMUH-CRDR (ESKD, n = 706; control, n = 20,836) were employed to clinically validate the effect of replicated eGFR-associated SNPs on the risk of ESKD. A total of 162 replicated eGFR-associated SNPs exhibited negative correlations with the effect directions of the eGFR and ESKD (P < 0.05; maximum, median, and minimum ORs= 1.01, 1.00, and 0.99, corresponding 95% CIs = 1.01–1.02, 0.99–1.00, 0.98–1.00, respectively; Pearson’s r = −0.96, P < 0.0001; Supplementary Fig. 2c and Supplementary Data 7), including 69 likely kidney-related SNPs from 7 genomic risk loci close to the following genes: KNG1, F12, HLA-DQA1, TINAG, RSBN1L, and TMEM60.
Genetic correlations of the eGFR and BUN with other phenotypes
We explored the genome-wide genetic correlations (rg) of eGFR and BUN with 71 quantitative and 48 binary phenotypes from a TWB discovery dataset to understand their shared genetic basis (Supplementary Data 8). Totals of 13 and 2 statistically significant genetic correlations were identified for eGFR and BUN, respectively (P < 4.2 × 10−4 = 0.05/119; Supplementary Data 8). With the exception of serum creatinine (S-Cre), the strongest genetic correlations observed between the eGFR and phenotypes were those with BUN (rg = −0.30, P = 1.13 × 10−8), urinary albumin (rg = 0.25, P = 5.92 × 10−7), uric acid (rg = −0.24, P = 7.15 × 10−10), and muscle mass (rg = −0.14, P = 1.67 × 10−7; Supplementary Fig. 3a). For BUN, the strongest genetic correlation observed was that with S-Cre (rg = 0.28, P = 4.06 × 10−8; Supplementary Fig. 3b). Genetic correlation analysis revealed that muscle mass was correlated with eGFR but not with BUN. These results indicated that the eGFR-associated SNPs reflected the regulatory roles of renal excretion and muscle generation in S-Cre levels.
Functional enrichment and pathway enrichment analyses
To determine whether the eGFR-associated SNPs were mechanistically linked to kidney function, we conducted serial enrichment analyses to characterize tissue-specific gene expression, regulatory annotations, and pathway dynamics.
Multimarker Analysis of GenoMic Annotation (MAGMA) software was used to prioritize genes for gene sets, pathways, and cell types based on the results of a meta-analysis of eGFR GWASs. We identified significantly enriched tissues and cell types with the strongest enrichment of eGFR-associated SNPs observed in the kidney medulla (P = 1.01 × 10−6) and kidney cortex (P = 1.26 × 10−6) tissues in Genotype-Tissue Expression (GTEx) version 8 (54 tissue types; Fig. 4 and Supplementary Data 9). Pathway enrichment analysis revealed nine significant canonical pathways, including several pathways relevant to kidney function, such as urate metabolism (Bonferroni-corrected P = 2.0 × 10−4) and abacavir transmembrane transport (Bonferroni-corrected P = 5.0 × 10−4; Table 1 and Supplementary Data 10). Enrichment analysis of BUN-associated SNPs in specific tissues and cell types revealed a similar expression pattern to that of eGFR-associated SNPs, including in kidney medulla and kidney cortex tissues (Supplementary Data 9); this finding supports the use of BUN for prioritizing loci that are highly likely to be associated with kidney function.
Fig. 4. Tissue-specific analysis of eGFR GWASs.
Functional analysis of an eGFR-derived GWAS was conducted using GTEx version 8 (54 tissue types) in MAGMA. Kidney medulla and kidney cortex tissues had P values of <0.05 (above the dashed line). All statistical tests employed two-sided P values. Further details are provided in Supplementary Data 9. eGFR estimated glomerular filtration rate, GTEx Genotype-Tissue Expression, GWAS genome-wide association study, MAGMA Multimarker Analysis of GenoMic Annotation.
Table 1.
Gene set enrichment analysis of GWASs for the eGFR in MAGMA
| Database | Category | Gene set | No. of genes | Beta | Standard deviation of beta | Standard error of beta | P value | Bonferroni-corrected P value |
|---|---|---|---|---|---|---|---|---|
| Gene Ontology | Biological process | Urate metabolic process | 10 | 1.92 | 0.04 | 0.34 | 1.29 × 10−8 | 0.0002 |
| Reactome | Curated gene set | Abacavir transmembrane transport | 5 | 2.68 | 0.04 | 0.49 | 3.03 × 10−8 | 0.0005 |
| Gene Ontology | Biological process | Cellular response to endogenous stimulus | 1263 | 0.16 | 0.04 | 0.03 | 1.51 × 10−7 | 0.0023 |
| Gene Ontology | Biological process | Enzyme-linked receptor protein signaling pathway | 947 | 0.18 | 0.04 | 0.04 | 2.90 × 10−7 | 0.0045 |
| Gene Ontology | Biological process | Negative regulation of RNA biosynthetic process | 1104 | 0.15 | 0.04 | 0.03 | 6.11 × 10−7 | 0.0095 |
| Gene Ontology | Biological process | Response to endogenous stimulus | 1501 | 0.14 | 0.04 | 0.03 | 6.25 × 10−7 | 0.0097 |
| Gene Ontology | Biological process | Contact inhibition | 8 | 1.50 | 0.03 | 0.32 | 1.05 × 10−6 | 0.0163 |
| Gene Ontology | Molecular function | Regulatory region nucleic acid binding | 849 | 0.17 | 0.04 | 0.04 | 1.12 × 10−6 | 0.0174 |
| Gene Ontology | Cellular compartment | Activin receptor complex | 7 | 2.44 | 0.05 | 0.52 | 1.40 × 10−6 | 0.0217 |
eGFR estimated glomerular filtration rate, GWAS genome-wide association study, MAGMA Multimarker Analysis of GenoMic Annotation.
All statistical tests employed two-sided P values. Bonferroni correction was applied for multiple testing adjustment of P values.
Stratified LD score regression was used to estimate the contributions of cell-type-specific functional genomic elements and tissue-specific gene expression to heritability through GWAS summary statistics pertaining to the eGFR and BUN. Cell-type-specific functional genomic elements were sourced from the Roadmap Epigenomics database19, and tissue-specific gene expression data were derived from the GTEx20 and Franke Lab databases21. In the eGFR GWAS, fetal kidney tissues were regarded as the enriched tissues for functional genomic elements, and kidney (A05.810.453.kidney), pancreas, and kidney cortex tissues were regarded as the most significant tissues for tissue-specific gene expression, followed by fetal kidney tissues (Supplementary Data 11). In the BUN GWAS, fetal kidney tissues were the enriched tissues for functional genomic elements, and kidney cortex tissues were the enriched tissues for tissue-specific gene expression (Supplementary Data 11). These findings indicated that kidney-specific epigenomic elements and gene expression contributed to the heritability of the eGFR and BUN.
Statistical fine mapping of causal variants from eGFR GWAS
To identify the causal variants among the eGFR-associated SNPs, a stepwise conditional analysis was conducted using genome-wide complex trait analysis–conditional and joint analysis (GCTA-COJO) with an in-sample LD reference. Subsequently, statistical fine mapping of eGFR loci was conducted through summary statistics-based conditional analyses for 238 independent significant SNPs mapped to 97 genomic risk loci. We found a credible set with a cumulative posterior probability (PP) of more than 99% and a median set size of 52 SNPs for independent significant SNPs (Supplementary Data 12).
The potential causal variants within the small credible set were evaluated to determine their functional impact and regulatory potential. Missense SNPs with a cumulative PP of over 99% or mapping to a small credible set (n < 5) are particularly important, as they suggest a direct involvement of the affected gene. As shown in Fig. 5a and Table 2, five missense SNPs were identified. Among these missense SNPs, the rs17730281 missense SNP in WDR72 had a combined annotation-dependent depletion (CADD) score of >15, which supported its potential deleterious effect (Table 2). To determine the regulatory potential of SNPs from small credible sets within the kidney, we associated these SNPs with DNase I hypersensitivity sites identified from the Roadmap Epigenomics database, which includes multiple kidney cell types. We subsequently prioritized 23 eGFR-associated SNPs that were mapped to one of these epigenomic annotations with a credible set size of <5 and a PP of >95%, indicating their potential as causal regulatory variants (Supplementary Data 13). The rs9895661 SNP close to the BCAS3 locus had a PP of >95% and a CADD score of 17.47, which suggested its regulatory potential for gene expression in kidney tissues (Fig. 5b).
Fig. 5. Fine mapping of credible sets of exonic and regulatory SNPs.
a Fine mapping of exonic SNPs. The triangles represent exonic SNPs, and their sizes correspond to their CADD scores. The red triangles indicate exonic SNPs with a credible set size of <5 or a PP of >99%. b Fine mapping of regulatory SNPs. Each color corresponds to a unique tissue type, as indicated by Roadmap Epigenomics data. The labels indicate credible set sizes of ≤10 and PPs of >95%. All statistical tests employed two-sided P values. Further details are provided in Supplementary Data 13. CADD combined annotation-dependent depletion, PP posterior probability, SNP single-nucleotide polymorphism.
Table 2.
Missense SNPs with a small credible set size (n < 5) or a PP of 50%
| rsID | Credible set size | PP | Gene | Exonic function | CADD | Regulatory function | Summary |
|---|---|---|---|---|---|---|---|
| rs76872124 | 5 | 0.63 | TRIM46 | p.Thr474Ile | 18 | Roadmap Others | TRIM46 is a gene that encodes a protein belonging to the tripartite motif family. This gene is associated with interferon-gamma signaling and cytokine signaling in the immune system96. It is also associated with inflammatory bowel disease97. The SNPs in TRIM46 are associated with measurements of uric acid98 and BUN97. |
| rs140449886 | 3 | 0.25 | GON4L | p.Asp1522Gly | 0.777 | None | GON4L is a gene that encodes a protein with transcriptional repressor activity. This activity is presumably mediated by the formation of a complex with YY1, SIN3A, and HDAC199. GON4L is required for B-cell lymphopoiesis100. It is also associated with chronic obstructive pulmonary disease. The SNPs in GON4L are associated with measurements of BUN10. |
| rs2304456 | 1 | 1 | KNG1 | p.Ile197Met | 3.084 | None | KNG1 is a gene that encodes kininogen 1—a protein that serves as an inhibitor of thiol proteases. KNG1 is associated with several diseases, including high-molecular-weight kininogen deficiency101 and type 6 hereditary angioedema102. The SNPs in KNG1 are associated with measurements of creatinine103 and BUN17. |
| rs2231137 | 1 | 1 | ABCG2 | p.Val12Met | 4.287 | None | ABCG2 is a gene that encodes ATP-binding cassette subfamily G member 2—a protein that belongs to the ATP-binding cassette transporter family. ABCG2 is associated with several diseases, including hyperuricemia104 and gout105. The SNPs in ABCG2 are associated with measurements of uric acid9, hyperuricemia106, gout107, and creatinine97. |
| rs17730281 | 1 | 0.99 | WDR72 | p.Leu819Pne | 25.7 | None | WDR72 is a gene that encodes WD repeat domain 72—a protein that is presumably involved in the localization of the calcium transporter SLC24A4 to the ameloblast cell membrane108. WDR72 is associated with several diseases, including CKD10 and hypomaturation amelogenesis imperfecta109,110. The SNPs in WDR72 are associated with measurements of the estimated glomerular filtration rate111, creatinine103, and urinary pH112. |
CADD combined annotation-dependent depletion, BUN blood urea nitrogen, SNP single-nucleotide polymorphism, PP posterior probability.
Statistical colocalization for causal gene prioritization
Colocalization analyses of 111 eGFR-associated lead SNPs were conducted using cis-expression quantitative trait locus (cis-eQTL) data and eGFR GWASs within ±100 kb regions of the lead SNPs. Specifically, cis-eQTLs across 51 tissues were sourced from GTEx version 8 and the Human Kidney eQTL Atlas, which includes data regarding tissues from renal glomerular and tubulointerstitial compartments11. A PP of >80% for colocalization was observed in 287 genes encompassing multiple tissue types, including 43 genes in at least one kidney tissue type (Supplementary Data 14). Changes in the expression of these 43 genes in the kidney may be linked to the eGFR, as several genes have been reported10. For instance, UMOD was identified as a casual gene for CKD22, with higher UMOD gene expression associated with a lower eGFR (Supplementary Fig. 4). We observed that the rs77924615 SNP was associated with high UMOD gene expression and a low eGFR in both glomerular and tubulointerstitial compartments (Supplementary Fig. 4), while Wuttke et al. observed the rs77924615 SNP only in tubulointerstitial compartments10. Overall, our findings underscore the key roles of several kidney function-related genes—including UNCX, TBX2, and SHROOM3—and are consistent with those of related studies10,23,24.
According to our colocalization findings, FGF5 and F12 are potential effector genes that influence an individual’s eGFR. Notably, the rs16998073 SNP associated with FGF5 gene expression was detected in both renal glomerular and tubulointerstitial compartments (Supplementary Fig. 4). FGF5, which encodes fibroblast growth factor 5, has been associated with blood pressure, coronary artery disease, and kidney function10. In the present study, the rs16998073 SNP associated with F12 gene expression was exclusively identified in renal tubulointerstitial compartments (Supplementary Fig. 4). F12, which encodes coagulation factor XII protein, plays a role in the renin–angiotensin–aldosterone system, which may be linked to kidney function25. Our analyses revealed that eGFR-associated SNPs demonstrate colocalization with eQTL in kidney tissues. Among these SNPs, some also colocalized with gene expressions in other tissue types and showed consistent effect directions, similar to those observed as in kidney tissue, with the exceptions of CEP89 and PAX8 (Supplementary Fig. 4).
Cumulative incidence of CKD throughout an individual’s life, stratified by polygenic risk scores
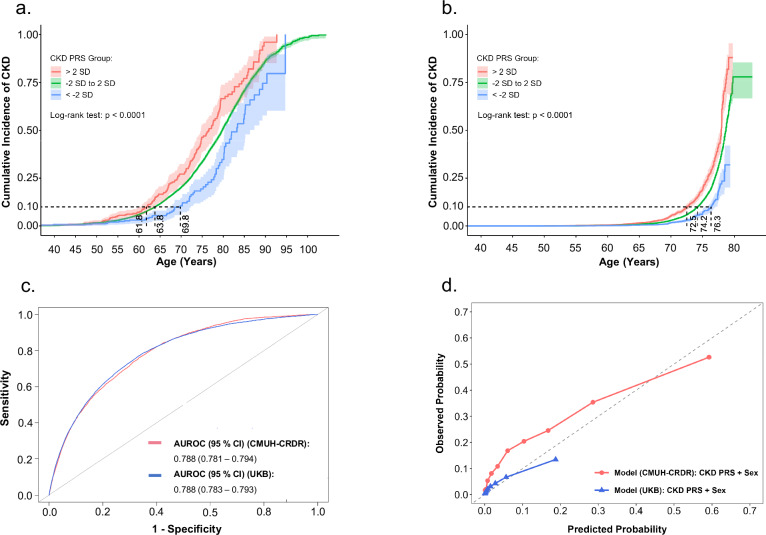
To examine the relevance of our findings regarding genetic susceptibility to CKD, we constructed a polygenic risk score (PRS) for CKD (PRSCKD) by using GWAS-derived summary statistics for the eGFR. An optimal PRS model was obtained through a meta-analysis of eGFR-related GWAS summary statistics, with BBJ- and TWB-derived discovery data used as the base datasets and with TWB-derived replication data regarding CKD used as the target dataset (with a cut-off P value of 0.048). Information regarding the onset of CKD is particularly useful for validating the longitudinal predictive performance of PRSs derived from eGFR-associated SNPs. We observed a significantly higher cumulative incidence of CKD in patients with a PRSCKD two SDs above the mean compared with those with a PRSCKD two SDs below the mean in an external Taiwanese dataset. This difference in cumulative incidence remained constant throughout the lives of the participants, particularly from age 50 to age 80 (Fig. 6a). An analysis of the adjusted hazard ratio (HR) of CKD revealed a dose–response pattern between patients with a PRSCKD two SDs above the mean and those with a PRSCKD within two SDs of the mean, with the corresponding adjusted HRs for patients with a PRSCKD two SDs below the mean being 2.3 (95% CI = 1.7–3.0; P < 0.0001) and 1.6 (95% CI = 1.3–2.1; P < 0.0001) (Supplementary Fig. 5), respectively. We also examined the time to reach a 10% cumulative incidence of CKD in the three PRSCKD groups. This threshold is close to the global CKD prevalence of 9.1%1. The time to reach a 10th percentile cumulative incidence of CKD in the three PRSCKD groups (above, within, and below two SDs) were 61.8, 63.8, and 69.8 years after birth, respectively. Notably, the PRSCKD effectively differentiated between high- and low-risk groups in both East Asians and White British populations, as demonstrated in the UKB (Fig. 6b). The area under the receiver-operating characteristic curve (AUROC) for PRSCKD was consistent across the Taiwanese and White British populations, namely 0.788 for both (95% CIs = 0.781–0.794 and 0.783–0.793, respectively; Fig. 6c). In addition, the calibration curve for our PRSCKD model indicated that the predicted probability was highly consistent with the observed probability, particularly between 0.4 and 0.5, indicating the model’s accuracy (Fig. 6d).
Fig. 6. Cumulative incidence of CKD based on PRS stratification with a Taiwanese dataset obtained from the CMUH-CRDR and a White British dataset obtained from the UKB.
The high-PRS group exhibited a higher cumulative incidence of CKD than did the low-PRS group across age in the a external Taiwanese dataset (CMUH-CRDR, n = 25,345, P = 2.06 × 10−7) and b White British dataset (UKB, n = 260,245, P = 2.60 × 10−29). The data present the PRS and error bars correspond to 95% CIs. All statistical tests employed two-sided P values. The dashed line represents a CKD cumulative incidence of 10%, which is an estimate of the global prevalence of CKD. c The AUROC of the CKD PRS model is 0.788 in both the CMUH-CRDR and UKB datasets. d The calibration curve of our PRSCKD model indicates that the predicted probability was closely aligned with the observed probability when the predicted probability ranged between 0.4 and 0.5 in the CMUH-CRDR dataset and between 0.0 and 0.2 in the UKB dataset. AUROC area under the receiver-operating characteristic curve, CI confidence interval, CKD chronic kidney disease, CMUH-CRDR Clinical Research Data Repository of China Medical University Hospital, PRS polygenic risk score, SD standard deviation, UKB UK Biobank.
Discussion
This study represents a large GWAS examining the eGFR of East Asian populations from Taiwan and Japan, which are among the regions with the highest prevalence of ESKD worldwide. A total of 26 unreported genomic risk loci closely linked to kidney function were identified. We discovered that individuals whose PRSCKD was within the highest two SDs of the PRSCKD distribution were more likely than others to develop CKD and also developed CKD approximately 8 years earlier than did those whose PRSCKD was within the lowest two SDs. When PRSCKD was combined with age and sex, it demonstrated good discriminative and calibrating performance in forecasting CKD development, achieving an AUROC of 0.788. Functional enrichment analysis revealed that the identified biological pathways were primarily associated with urate metabolic process. This finding aligns with the fine-mapping result, which identified a missense mutation in ABCG2 as a potential causal variant.
A major gap between research regarding translation of genetic nephrology and real-world practice manifests as the underrepresentation of certain ancestry groups, particularly Asian populations26,27. The present study sought to address this gap and expand the current level of understanding regarding the long-term epidemiology and susceptibility patterns of CKD in Taiwan28. The escalating prevalence of ESKD in Taiwan imposes a substantial socioeconomic burden, yet its primary causes remain unidentified29. Over the preceding three decades, extensive efforts have been made to determine the factors underlying Taiwan’s high incidence of CKD. Some researchers have suggested the involvement of Chinese herbal medicine30 or environmental exposures, such as arsenic31. Others have speculated that Taiwan’s universal healthcare system may have resulted in an increase in CKD diagnoses32. However, these theories lack causal evidence. Conversely, the present findings uniquely demonstrate a genetic predisposition to early CKD onset in the Taiwanese population, thereby corroborating the results of a population-based familial aggregation study that revealed a moderate heritability rate of 31.1% for the phenotypic variance of ESKD15.
After applying the highest PRSCKD threshold of two SDs, we observed an earlier onset of CKD, by an average of 8 years, among individuals in their mid-50s who were at the highest genetic risk of CKD. Although the predictive performance of PRSCKD was modest in the White British dataset, it effectively differentiated between groups with high and low risk of developing CKD as defined by PRSCKD. These findings underscore the importance of shifting toward early prevention of CKD through genetic counseling. Our proposed, PRS-based risk stratification has the potential to provide an opportunity for the early implementation of CKD preventive care strategies, such as intensive prediabetes management and rigorous blood pressure monitoring and optimization. Nevertheless, a more comprehensive pragmatic trial is required to assess the real-world effectiveness of PRSCKD in preventing CKD or slowing its progression.
From the enrichment analysis, we identified three eGFR-associated genes that were physiologically relevant, which may serve as potential targets for CKD drug development33. Compared to prior findings mainly derived from Caucasian and African American populations, this study focused on East Asian populations and revealed that UMOD but not APOL1 was significantly associated with the eGFR10,34. The UMOD variant was discovered in a 2009 GWAS that employed the S-Cre-based eGFR as a phenotype35. Extensive research has underscored the promising prognostic value of UMOD for CKD in general populations of various ethnicities22. A recent discovered variant of UMOD, namely p.Thr62Pro, has been found to have a moderate effect on the risk of CKD in a large proportion of individuals with European ancestry, with endoplasmic reticulum homeostasis and maturation being potential pathogenic pathways36. A large Mendelian Randomization (MR) study with a sample size of 567,460 provided evidence further supporting a causal relationship between UMOD and the risk of CKD37. These findings highlight the potential for drug discovery by exploring whether modulating UMOD expression could be a viable strategy for CKD prevention.
The F12 gene was consistently identified in all our replication and validation analyses (Supplementary Data 15). F12 gene encodes the plasma coagulation factor XIIa, which can subsequently cleave plasma pre-kallikrein to kallikrein, thereby initiating the kallikrein-kinin system (KKS)38. In a subset of twin and sibling participants recruited from southern California, F12 was found to involve the renin–angiotensin system (RAS) pathway and potentially regulate blood pressure25. Association of the F12 polymorphism and serum osteopontin (OPN) levels was identified in the German Chronic Kidney Disease (GCKD) study39. OPN is a phosphorylated glycoprotein encoded by SPP1; it is predominantly synthesized in kidney tissue and has been associated with kidney fibrosis in animal experiments and in the German Chronic Kidney Disease (GCKD) study39,40. In addition, the association of the F12 gene with CKD may be attributable to dysregulated blood pressure and chronic inflammation resulting from the activation of the RAS and KKS, respectively25,39. A recent study from China has identified F12 as a druggable target through extensive MR and colocalization analysis, using data from the CKDGen Consortium and cell-type-dependent eQTL data from kidney tubular and glomerular samples41. Continued research efforts are required to elucidate the potential beneficial role of targeting F12 in patients across a range of kidney disease spectrums, with a focus on replication studies and large clinical trials.
Among the three index genome-wide-significantly SNP associated with OPN, rs10011284 was mapped to an intergenic region between SPP1 and MEPE, and MEPE was found to be involved in bone mineralization, phosphate homeostasis, and bone turnover39,42,43. The variant rs10011284, also linked to gout, may be influenced by ABCG2, which is located near SPP1 and MEPE. This proximity suggests a potential pathogenic role for ABCG2 in the development of CKD39. Although the ABCG2 gene is well known for its connection with serum uric acid levels and gout, as demonstrated in previous GWASs44–46, mechanistic studies47,48, and human research49–51, its independent role in CKD development remains to be clarified. The ABCG2 protein, also known as the breast cancer resistance protein, is a multidrug transporter and a high-capacity urate exporter47,52,53. In a human study, reduced ABCG2 function was associated with a rapid decline in the age-dependent eGFR among individuals with serum uric acid levels exceeding 6 mg/dL. This finding underlines the potential role of ABCG2 in eGFR decline, especially given the high prevalence of hyperuricemia in East Asia54. These insights underscore the importance of further exploring the feasibility of integrating ABCG2 function quantification into routine practice to enhance CKD care and disease prevention.
We identified three of five causal missense SNPs in the present study differ from those reported in previous CKD genetic studies8,10,54. The genes associated with these missense SNPs, such as TRIM46 and GON4L, have been linked to tubular fibrosis and BUN levels, respectively10,55. Our study highlights the genetic diversity in CKD risk between populations of European ancestry and East Asian ancestry. For instance, rs4715491 in the FAM83B gene and rs4148155 in the ABCG2 gene were found to be associated with eGFR exclusively in Asian populations. The genetic variants contributing to CKD risk across different ancestries warrant cautious interpretation and limited generalizability from regional evidence, potentially arising from the following reasons: population-specific variations such as founder effects, changes in allele frequency due to genetic drift and local selection12, and interactions between genetic backgrounds and environmental exposure56. Therefore, the null association between rs4148155 and eGFR decline in European populations may be underestimated, as the allele frequency of rs4148155 in the ABCG2 gene is only 0.104 in European populations, compared to 0.319 in East Asian populations57. The variance in rs4148155 allele frequency indicates the necessity of larger European sample sizes for achieving statistical significance. In addition, the effect of rs4148155 in ABCG2 may be amplified by the purine-rich diet prevalent in Taiwan, which is known to exacerbate gout and hyperuricemia—both of which are risk factors for CKD. This concern underscores the importance of utilizing regional genetic data to develop feasible PRSs for disease prevention and early CKD screening in specific populations. Future studies should investigate how the distinct genetic risk patterns defined by PRSCKD interact with environmental exposures and contribute to geographical disparities in CKD epidemiology worldwide.
This study had several methodological strengths, including a large sample size comprising relatively homogenous East Asian populations; the use of standardized, high-quality genotype imputation and phenotype identification; and a comprehensive bioinformatic analysis of eGFR-associated genes and pathways. In addition, the potential causal association in this study was suggested by the validation of PRSCKD through longitudinal prediction with the phenotype of CKD onset in an independent dataset. This study also had some limitations. First, our findings may not be generalizable to people with non-Asian ancestry. In addition, the assumption of fixed-allele effects across multiple geographical areas in Asia may not hold given the potential for geographical differences in environment–gene interaction58. Despite this inherent limitation, we observed similar PRSCKD model performance in both the independent Taiwanese and White British populations. Second, the proposed PRS for risk of CKD was derived from the GWAS for kidney function estimated by eGFR based on S-Cre rather than CKD, the responsible phenotype. While this approach has been debated in a previous study59, it may not be feasible to perform GWAS directly for CKD as CKD itself stands for a highly heterogenous disease spectrum from arbitrary definitions to etiologies of primary or secondary kidney degeneration. Third, the possibility of an underestimation of potential eGFR-associated SNPs and their genetic impact on CKD development cannot be excluded, despite our study having a large sample size among those conducted on East Asian populations to date9,60,61.
In this GWAS of eGFR using large data exclusively from an East Asian population, we identified 2140 previously unreported genome-wide significant SNPs. One of the prioritized genes was, F12, has been potentially linked to CKD through RAS and KKS dysregulation and supported by a recent GWAS-based drug repurposing study41. The robust association between PRSCKD and the cumulative incidence of CKD across different ancestries sets the stage for further research to verify its clinical effectiveness in early CKD prevention.
Methods
Biobank data source
BBJ
The BBJ is a hospital-based registry of DNA samples, serum samples, and clinical information collected from approximately 200,000 patients with one or more of a collection of 47 common diseases (e.g., cancers, neurological diseases, cardiovascular diseases, and infectious diseases) identified by physicians at 66 hospitals affiliated with 12 medical institutions between 2003 and 200762. Informed consent was obtained from all participants in writing, and the ethics committees of the RIKEN Center for Integrative Medical Sciences and the Institute of Medical Sciences, at the University of Tokyo approved the study. The RIKEN Center made available the GWAS summary statistic for public download at the Japanese Encyclopedia of Genetic Associations by Riken [http://jenger.riken.jp/en/result] without requiring data application. The BBJ comprised 143,658 available eGFR and 139,817 available BUN from patient population with a mean age (SD) of 62.9 ( ± 13.2) years, a mean eGFR (SD) of 79.93 ( ± 15.42) mL/min/1.73 m2, and a mean (SD) BUN level of 15.44 ( ± 4.77) mg/dL9; 54.9% of this group were male.
TWB
The TWB is an ongoing project that contains data and samples from a national prospective cohort of the Taiwanese population; its goal is to longitudinally collect a wide range of phenotypic measurements and genomic data from Han Chinese individuals aged 20–70 years without cancer history63. The TWB includes two customized arrays, namely TWBv1 and TWBv2, which are specifically designed for the Taiwanese population (see “Genotyping and imputation”). In the present study, in addition to demographic and phenotypic data, we obtained TWBv2 genotyping data for 101,294 individuals for our discovery dataset. The TWB-based discovery dataset included individuals with a mean age (SD) of 50.4 ( ± 10.9) years, a mean eGFR (SD) of 101.95 ( ± 14.75) mL/min/1.73 m2, and a mean BUN level (SD) of 13.07 ( ± 3.90) mg/dL.; 45.6% of the individuals were male. We also obtained TWBv1 genotyping data for 27,058 individuals for our replication dataset. The TWB-based replication dataset included individuals with a mean age (SD) of 49.26 ( ± 11.10) years, a mean eGFR (SD) of 101.83 ( ± 15.22) mL/min/1.73 m2, and a mean BUN level (SD) of 13.26 (4.00) mg/dL; 49.9% of the individuals were male. The data utilization and research conduct were approved by the Ethics Committees of Academia Sinica, the TWB, and CMUH25.
UKB
The UKB is a large prospective database of biological samples obtained from ~500,000 individuals aged 40–69 years with extensive phenotypic data (http://www.ukbiobank.ac.uk)64. Genotyping was performed using the Affymetrix UK BiLEVE Axiom array on an initial set of 50,000 participants; the Affymetrix UKB Axiom array was then used on the remaining set of participants. In the UKB, 91.7%, 1.9%, 0.8%, and 5.6% of the included data are from European (White) individuals, Asian individuals, Black individuals, and individuals of other ethnicities, respectively. In this study, we used data for 260,245 White British participants as validation data to confirm the predictive performance of PRSCKD. The validation cohort had a mean age (SD) of 54.98 ( ± 18.20) years and 8821 cases of CKD, and 45.7% of them were male.
CMUH-CRDR
We used independent data from the CMUH-CRDR as clinical validation data. Between 2003 and 2020, the CMUH-CRDR documented the electronic medical records data of 3,077,895 patients, including demographic (e.g., age, self-report sex) and administrative data, diagnostic data, surgical data, laboratory measurements, and mortality data, from the Health and Welfare Data Science Center of the Ministry of Health and Welfare65. CMUH also conducted its Precision Medicine Project to gather genetic data using the TWBv2 genotyping array66. The CMUH’s Precision Medicine Project consecutively enrolled outpatient participants since 201867. A total of 25,345 patients with eGFR measurements were identified from the combined CMUH-CRDR (clinical data) and CMUH’s Precision Medicine Project (genotyping data) cohort. This cohort had a mean age (SD) of 56.72 ( ± 15.73) years, 4509 cases of CKD, and 706 cases of ESKD; 44.3% of the cohort was male. All the participants provided written informed consent. The study protocol was approved by the Big Data Center and the Research Ethics Committee and Institutional Review Board (REC/IRB) of CMUH (IRB No. CMUH105-REC3-068, CMUH110-REC2-145, CMUH111-REC3-138, and CMUH112-REC2-036).
Phenotype definition
The primary phenotype of concern was the eGFR, which is based on S-Cre measurements. S-Cre and BUN levels were measured using the Jaffe rate method and enzymatic conductivity rate method, respectively, on a Beckman UniCel DxC 800 immunoassay system (Beckman Coulter, Brea, CA, USA). S-Cre values were calibrated using an isotope dilution mass spectrometry reference method68. The Chronic Kidney Disease Epidemiology Collaboration equation was employed to determine eGFR values69. Before GWAS analysis, BUN and eGFR values were normalized using rank-based inverse normal transformation (RINT)70. In addition, the predictive performance of the proposed PRS was validated using the CKD phenotype, which was differentially defined in the three study populations. In the TWB validation cohort, CKD was identified by a baseline eGFR of <60 mL/min/1.73 m2, whereas in the CMUH-CRDR validation cohort, it was identified by the presence of at least two outpatient eGFR measurements of <60 mL/min/1.73 m2, with a minimum interval of 90 days between measurements. In the sensitivity analysis of the UKB cohort, CKD was defined in accordance with the International Classification of Diseases (ICD) codes for CKD (ICD-10 codes N18.3, N18.4, N18.5, and N18.6). CKD controls were defined as individuals with no CKD and a baseline eGFR of >90 mL/min/1.73 m2. In the CMUH-CRDR cohort, progression to ESKD was defined as the initiation of long-term renal replacement therapy (peritoneal dialysis, hemodialysis, or kidney transplantation) identified from certificates of catastrophic illness issued by the National Health Insurance Administration, Ministry of Health and Welfare, Taiwan. ESKD controls were individuals with no ESKD and a baseline eGFR of >90 mL/min/1.73 m2.
Genotyping and imputation
Genotyping was conducted using two custom SNP arrays based on the Taiwanese population, namely TWBv1 and TWBv263,71,72. The TWBv1 array was based on the Thermo Fisher Axiom Genome-Wide CHB Array, with customized content containing ~650,000 markers on the GRCh37 coordinates, aimed at capturing functional variants. The TWBv2 array covers rare coding risk alleles based on whole genomic sequence data obtained from 946 TWB samples. It also contains data on ~690,000 markers aligned to the GRCh38 reference build.
For imputation, low-quality samples and variants were filtered out, including those with an SNP genotype call rate of <95%, an individual call rate of <95%, a minor allele frequency (MAF) of <1%, and a Hardy–Weinberg equilibrium (HWE) P value of <1 × 10−4. The 1000 Genome Project Phase 3 was used as the reference to exclude samples obtained from populations of non-East Asian ethnicity, as determined through principal component analysis (PCA). The genotyping data were converted from GRCh38 to GRCh37 using LiftOver73. Then, we pre-phased genotyping data using SHAPEIT274 and imputed it using IMPUTE275, which generated 81,698,455 imputed SNPs. We further filtered out imputed SNPs with an INFO score of lower than 0.7, resulting in the retention of 18,609,316 imputed SNPs. All quality control procedures were conducted using PLINK v2.076.
GWAS
We conducted a GWAS by using a machine-learning method, called REGENIE77. Before REGENIE, both raw and imputed genotyping data were subjected to quality control checks at an SNP genotype call rate of <99%, a MAF of <1%, an HWE P value of <1 × 10−15, an individual call rate of <98%, and a heterozygosity rate greater than ±3 SDs. In addition, PCA was conducted using the reference population of the 1000 Genomes Project16. In the first step of REGENIE, we calculated the genetic correlation of the samples and then used this information as covariates in the second step of REGENIE. In the second step, we applied a linear regression test for quantitative traits, assuming an additive genetic model, and conducted a Firth logistic regression test for binary traits. To adjust for potential confounding effects, we employed age, sex, and the first six principal components (PCs) as covariates in the GWAS of the eGFR. The circular Manhattan plot was generated using the R package “circlize” in R software v.4.3.1 (R Foundation for Statistical Computing, Vienna, Austria)78.
Meta-analysis of GWASs
A fixed-effects inverse variance-weighted meta-analysis was conducted using METAL software to increase the accuracy of effect estimates and their standard errors79. Genomic control (GC) correction was applied when the GC factor λGC exceeded 1. This meta-analysis yielded 5790 SNPs for the eGFR and 3600 SNPs for BUN, with a genome-wide significance level of 5 × 10−8. Heterogeneity among studies was evaluated using the I2 statistic. We used LD with an r² threshold of 0.6 to identify independently significant SNPs, and an LD r² threshold of 0.1 to identify lead SNPs from among these independently significant SNPs. Genomic risk loci were defined as regions with a window size of ±250 kb centered on independently significant SNPs. In addition, the genomic risk loci were merged into a single locus if the distance between them was shorter than 250 kb80.
Genetic heritability
We examined the genetic heritability of the eGFR by using GWAS summary statistics obtained from our meta-analysis, LD score regression was used to estimate genetic heritability81. LD scores were estimated from the East Asians panel of the 1000 Genomes Project Phase 3 by using an r2 estimator with 1 cM windows. SNPs with a MAF of less than 1% and loci with significantly large effect sizes or long-range LD were excluded from all regressions81. This approach enabled us to avoid potential bias from confounding factors, such as cryptic relatedness or population stratification. Each SNP’s contribution was evaluated by examining the correlation between test statistics and LD16.
Replication in an independent TWB dataset
A replication procedure was conducted to validate the findings of the original GWAS meta-analysis through an independent cohort. The TWB dataset, which comprises data regarding 27,705 individuals with available eGFR and TWBv1 genotyping data, was used as a replication dataset. The quality control steps applied for the replication dataset were the same as those described in the subsection of this paper titled “GWAS.” To assess the association between SNPs and RINT (eGFR) in the replication cohort, a linear regression model was used in REGENIE. A SNP was considered replicated if it had the same effect direction as the corresponding SNP in the original GWAS and a P value of <0.05 in the replication dataset.
Associations of eGFR-associated SNPs to BUN level, CKD, and ESKD status
To investigate the eGFR-associated SNPs with other markers of kidney function, we conducted a meta-analysis of BUN levels by using data from the BBJ- and TWB-based discovery datasets. We then examined the associations between eGFR-associated SNPs and BUN levels. SNPs were considered relevant to kidney function if they had an opposite effect direction on BUN levels relative to the eGFR and a P value of <0.05. Logistic regression modeling was used to evaluate the association between these eGFR-associated SNPs and the risk of developing CKD or ESKD in the CMUH-CRDR cohort. We searched for an opposite effect direction between eGFR-associated SNPs and CKD or ESKD as well as a P value of <0.05.
Genetic correlations of eGFR and BUN with other complex traits and diseases
To examine the genetic correlations between complex quantitative traits and diseases, a GWAS-based analysis of 119 phenotypes was conducted using the TWB-based discovery dataset. Genetic correlations were estimated using LD score regression81, which involved two steps. First, the LD scores of each SNP in two GWAS summary statistics were calculated using predetermined East Asian population-based LD scores from the 1000 Genomes Project. Second, cross-trait LD score regression was conducted, which involved regressing the product of the LD scores of SNPs in the two GWAS summary statistics against the observed correlation in effect sizes between traits.
Gene prioritization, gene set enrichment, and tissue enrichment analysis
Functional analysis was conducted for genes, gene sets, and tissue enrichment by using MAGMA software version 1.0882, which was integrated into the Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA) platform version 1.5.280. Data regarding East Asian populations from the 1000 Genomes Project were used for LD calculation. Gene prioritization was performed using the mean P value of SNPs located within 1 kb upstream or downstream of genes. Gene set enrichment analysis was conducted on curated gene sets and Gene Ontology (GO) terms obtained from MSigDB version 7.083. For all tested gene sets, the P values utilized in the MAGMA-based gene set analysis were adjusted using Bonferroni correction. A total of 10,678 gene sets were used; these sets were divided into 4761 curated gene sets and 5917 GO terms. Tissue enrichment analysis was conducted using the GTXv8 database20.
Cell-type-specific enrichment through stratified LD score regression
Stratified LD score regression was used to identify tissues and cell types relevant to eGFR and BUN GWAS meta-analysis results. Heritability was partitioned from GWAS summary statistics to sets of cell-type-specific regulatory elements and gene expression84,85. The precalculated LD score for East Asians was obtained from the LD score regression resource website (https://alkesgroup.broadinstitute.org/LDSCORE/). The regulatory annotation datasets for cell-type-specific analysis contained 220 cell types featuring four histone markers, namely H3K4me1, H3K4me3, H3K9ac, and H3K27ac86–88. The gene expression datasets for cell-type-specific analysis were from GTEx and Franke Lab. GTEx contains RNA sequencing data of 53 human cell types, while Franke Lab includes array data of 152 human and mouse cell types21. The significance threshold was set at a false discovery rate of less than 5%.
Fine mapping and credible set identification in a meta-analysis
Genomic risk loci containing functional SNPs were subjected to statistical fine mapping using GCTA-COJO-Slct, a stepwise model selection procedure for identifying independent SNPs89. Approximate conditional analyses were then performed to determine the conditional effect sizes of all remaining independent SNPs in a locus using the GCTA-COJO-Cond algorithm89. For each SNP within a locus, the approximate Bayes factors (ABFs) derived from the effect estimate on the eGFR and its standard error of conditional estimates were used to compute the posterior probability (PP) of the SNP being responsible for the association signal (potential causal variant). To calculate the ABF for each SNP, the R package “gtx” version 2.1.6 (https://github.com/tobyjohnson/gtx) was employed, applying Wakefield’s formula90. The 99% credible sets were calculated by summing the PP-ranked SNPs until the cumulative PP exceeded 99%, thereby representing the credible set of SNPs that included the SNP responsible for the association with eGFR. For the deleterious scoring of functional SNPs in genomic risk loci, we utilized the FUMA platform to integrate annotations from CADD (version 1.4)80,91.
Colocalization analysis of eGFR-associated SNPs with cis-eQTLs
Colocalization analysis was conducted through a Bayesian test to examine the question of whether two traits can share a causal variant92. We examined the correlation between the eGFR and gene expression by evaluating the colocalization between eGFR-associated SNPs and cis-eQTLs from the Human Kidney eQTL Atlas for tubules and glomeruli11 and by using GTEx version 8 for 49 tissues20. Both cis-eQTLs and GWAS-derived effect alleles were harmonized and identified within ±100 kb of each GWAS-derived lead SNP for colocalization analysis. We used the R package “coloc” (version 5) with default settings to identify loci with a PP of >80%. In cases where the harmonized effect alleles had the same effect directions in both the eQTL and GWAS data, the aligned allelic effect direction was defined as positive.
PRS for CKD
To determine whether our findings can support the genetic susceptibility of CKD, we established a PRS for CKD on the basis of GWAS summary statistics for the eGFR. Clumping and thresholding were used to calculate the PRS93,94. Briefly, PRSice-2 software was used to calculate the PRS and to evaluate the most appropriate PRS model with the highest R2 value95. The base dataset used in PRSice-2 was derived from the meta-analysis summary statistics of eGFR GWASs involving the BBJ- and TWB-based discovery datasets. The target dataset in PRSice-2 consisted of independent samples from the TWB replication dataset with CKD status available, defined as a baseline eGFR of <60 mL/min/1.73 m2. These samples underwent the same quality control procedure as that described in the GWAS section. The weight of the proposed PRS model was determined from the beta coefficient of the eGFR GWAS summary statistic of the base dataset. After SNPs were clumped with an LD r2 of 0.1 and a window size of ±250 kb, a P value threshold was established to select independent significant SNPs for inclusion in the PRS. The PRS model was established through adjustment for age, sex, and the first six PCs. Since the PRS model adopts beta coefficients from eGFR GWAS, a higher PRS indicates higher eGFR and consequently low CKD risk. To better interpret the role of the PRS in CKD risk predictions, we multiplied the PRS by −1 as the operative value of PRSCKD.
Predictive performance of the PRS for CKD
The predictive performance of the PRS for CKD development was evaluated in two independent datasets by using a cumulative incidence curve, which was used to represent the probability of a CKD event over time. After we examined the predictive performance of the PRS by using the CMUH-CRDR dataset (n = 25,345), we externally verified this performance by using the UKB dataset (n = 260,245).
The Kaplan–Meier method was used to generate cumulative incidence curves, and a log-rank test was conducted to determine the differences between the curves of different PRSCKD subgroups, categorized as <−2 SDs, −2 to 2 SDs, and >2 SDs of the mean PRSCKD. The index date was set as each patient’s date of birth, and follow-up was conducted until the first CKD diagnosis was established, until the patient died, until the patient was lost to follow-up, or until the administrative censor date. In the CMUH-CRDR validation cohort, each patient’s date of death was verified by the National Death Registry of the Ministry of Health and Welfare of Taiwan. In the UKB cohort, the date of death was verified by NHS Digital for patients in England and Wales and by the NHS Central Register (part of the National Records of Scotland) for patients in Scotland. The administrative censor dates were December 31, 2021, for the CMUH-CRDR validation cohort and December 31, 2016, for the UKB cohort. To examine the level of CKD risk associated with PRSCKD, a competing risk analysis with deaths considered as censoring events was conducted using cause-specific Cox proportional hazards modeling, with age used as the time scale. The discriminative performance of the Cox proportional hazards model that incorporated PRSCKD and sex data was evaluated using the AUROC. In addition, we plotted the observed versus predicted risk probability to evaluate the calibration of the Cox proportional hazards model.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors thank the staff of the iHi Platform at the Big Data Center of CMUH for their assistance with data exploration, statistical analysis, and manuscript preparation. The authors thank the Health and Welfare Data Science Center, Ministry of Health Welfare, and the Health Data Science Center, CMUH, for providing administrative, technical, and funding support. This study was supported by the National Science and Technology Council of Taiwan (grant no. 112-2321-B-468-001 to C.-C. Kuo, 111-2320-B-039-052-MY3 to C.-C Kuo, 113-2634-F039-001 to C.-C. Kuo), Academia Sinica, Taiwan (grant no. AS-HLGC-111-04 to Y.-T. Lin), and CMUH (grant nos. DMR-112-119 to H.-L. Chen, and DMR-112-188 to C.-C. Kuo, DMR-113-177 to C.-C. Kuo).
Author contributions
Manuscript writing: H.-L. Chen, H.-Y. Chiang, David R. Chang, C.-F. Cheng, and C.-C. Kuo. Study design: H.-L. Chen, C.-F. Cheng, and C.-C. Kuo. Management of an individual contributing study: H.-L. Chen, C.-F. Cheng, Y.-T. Lin, and C.-C. Lin. Statistical analysis: H.-L. Chen, C.-F. Cheng, Y.-T. Lin, C.-C. Lin, P.-T. Yu, and C.-F. Hung. Bioinformatics: H.-L. Chen, C.-F. Cheng, Charles C.-N. Wang, T.-P. Lu, C.-Y. Lee, A. Chattopadhyay, and C.-H. Lin. Interpretation of the results: H.-L. Chen, David R. Chang, C.-F. Cheng, A. Tin, and C.-C Kuo. Critical review of the manuscript: H.-L. Chen, H.-Y. Chiang, Charles C.-N. Wang, T.-P. Lu, H.-C. Yeh, I.-W. Ting, H.-K. Tsai, E.-Y. Chuang, F.-J. Tsai, A. Tin, and C.-C. Kuo. Subject recruitment: F.-J. Tsai, H.-Y. Chiang, Y.-T. Lin, C.-C. Lin, and C.-C. Kuo.
Peer review
Peer review information
Nature Communications thanks Matthias Wuttke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The GWAS summary data supporting our findings are available on Figshare (10.6084/m9.figshare.24356587), and the statistical data are available in the Supplementary Data. The data from Taiwan Biobank (TWB; Application No. TWBR11111-02) and UK Biobank (UKB; Application No. 81803) were obtained through approved applications and they are publicly available to approved researchers for health-related research (TWB: https://www.biobank.org.tw/english.php; UKB: https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access). The GWAS summary data from Biobank Japan (BBJ) are publicly available without permission at http://jenger.riken.jp/en/result. The individual-level raw data from CMUH are unavailable because they contain information that may compromise participant privacy.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fuu-Jen Tsai, Chin-Chi Kuo.
Contributor Information
Fuu-Jen Tsai, Email: 000704@tool.caaumed.org.tw.
Chin-Chi Kuo, Email: chinchik@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53516-7.
References
- 1.Collaboration GBDCKD. Global, regional, and national burden of chronic kidney disease, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet395, 709–733 (2020). [DOI] [PMC free article] [PubMed]
- 2.Chertow, G. M. et al. Effects of dapagliflozin in stage 4 chronic kidney disease. J. Am. Soc. Nephrol.32, 2352–2361 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heerspink, H. J. L. et al. Dapagliflozin in patients with chronic kidney disease. New Engl. J. Med383, 1436–1446 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Jafar, T. H. FDA approval of dapagliflozin for chronic kidney disease: a remarkable achievement? Lancet398, 283–284 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Savage, N. Tapping into the drug discovery potential of AI. Biopharm. Deal. B37–B39 https://www.nature.com/articles/d43747-021-00045-7 (2021).
- 6.Aghajan, M. et al. Antisense oligonucleotide treatment ameliorates IFN-γ-induced proteinuria in APOL1-transgenic mice. JCI Insight4, e126124 (2019). [DOI] [PMC free article] [PubMed]
- 7.Hubaud, A. & Singh, A. P. Genetics in drug discovery. Trends Genet.37, 603–605 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Tin, A. & Kottgen, A. Genome-wide association studies of CKD and related traits. Clin. J. Am. Soc. Nephrol.15, 1643–1656 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanai, M. et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet.50, 390–400 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Wuttke, M. et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet.51, 957–972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheng, X. et al. Mapping the genetic architecture of human traits to cell types in the kidney identifies mechanisms of disease and potential treatments. Nat. Genet.53, 1322–1333 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirugo, G., Williams, S. M. & Tishkoff, S. A. The missing diversity in human genetic studies. Cell177, 1080 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.United States Renal Data System. 2020 USRDS Annual Data Report: Epidemiology of kidney disease in the United States. (National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, 2020).
- 14.Bello, A. K. et al. ISN–Global Kidney Health Atlas: a report by the International Society of Nephrology: an assessment of global kidney health care status focussing on capacity, availability, accessibility, affordability and outcomes of kidney disease. https://www.theisn.org/wp-content/uploads/media/ISN%20Atlas_2023%20Digital.pdf (International Society of Nephrology, Brussels, 2023).
- 15.Wu, H. H. et al. Family aggregation and heritability of ESRD in Taiwan: a population-based study. Am. J. Kidney Dis.70, 619–626 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Genomes, ProjectC. et al. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanzick, K. J. et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat. Commun.12, 4350 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki, M. et al. Blood urea nitrogen is independently associated with renal outcomes in Japanese patients with stage 3-5 chronic kidney disease: a prospective observational study. BMC Nephrol.20, 115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein, B. E. et al. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol.28, 1045–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium, G. T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fehrmann, R. S. et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet.47, 115–125 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Devuyst, O. & Pattaro, C. The UMOD locus: insights into the pathogenesis and prognosis of kidney disease. J. Am. Soc. Nephrol.29, 713–726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao, J. et al. An early prediction model for chronic kidney disease. Sci. Rep.12, 2765 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kispert, A. T-Box genes in the kidney and urinary tract. Curr. Top. Dev. Biol.122, 245–278 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Biswas, N. et al. Polymorphisms at the F12 and KLKB1 loci have significant trait association with activation of the renin-angiotensin system. BMC Med. Genet.17, 21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köttgen, A. et al. Genetics in chronic kidney disease: conclusions from a Kidney Disease: Improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int.101, 1126–1141 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet.51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bikbov, B. et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet395, 709–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu, C. C., Hsu, Y. H., Wu, M. S. & Hwang, S. J. Achievements and challenges in chronic kidney disease care in Taiwan. J. Formos. Med. Assoc.121, S3–s4 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Wu, F. L. et al. Does Chinese herb nephropathy account for the high incidence of end-stage renal disease in Taiwan? Nephron Clin. Pr.120, c215–c222 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Hsu, L. I. et al. Arsenic exposure from drinking water and the incidence of CKD in low to moderate exposed areas of Taiwan: a 14-year prospective study. Am. J. Kidney Dis.70, 787–797 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Yang, W. C. & Hwang, S. J. Incidence, prevalence and mortality trends of dialysis end-stage renal disease in Taiwan from 1990 to 2001: the impact of national health insurance. Nephrol. Dial. Transpl.23, 3977–3982 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Reay, W. R. & Cairns, M. J. Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet.22, 658–671 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Friedman, D. J. & Pollak, M. R. APOL1 nephropathy: from genetics to clinical applications. Clin. J. Am. Soc. Nephrol.16, 294–303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Köttgen, A. et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet.41, 712–717 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olinger, E. et al. An intermediate-effect size variant in UMOD confers risk for chronic kidney disease. Proc. Natl. Acad. Sci. USA119, e2114734119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponte, B. et al. Mendelian randomization to assess causality between uromodulin, blood pressure and chronic kidney disease. Kidney Int.100, 1282–1291 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Kalinin, D. V. Factor XII(a) inhibitors: a review of the patent literature. Expert Opin. Ther. Pat.31, 1155–1176 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Cheng, Y. et al. Genetics of osteopontin in patients with chronic kidney disease: the German Chronic Kidney Disease study. PLoS Genet.18, e1010139 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irita, J. et al. Osteopontin deficiency protects against aldosterone-induced inflammation, oxidative stress, and interstitial fibrosis in the kidney. Am. J. Physiol. Ren. Physiol.301, F833–F844 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Chen, X. et al. Drug repurposing opportunities for chronic kidney disease. iScience27, 109953 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marks, J., Churchill, L. J., Debnam, E. S. & Unwin, R. J. Matrix extracellular phosphoglycoprotein inhibits phosphate transport. J. Am. Soc. Nephrol.19, 2313–2320 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowe, P. S. The chicken or the egg: PHEX, FGF23 and SIBLINGs unscrambled. Cell Biochem Funct.30, 355–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dehghan, A. et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet372, 1953–1961 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamatani, Y. et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet.42, 210–215 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Tin, A. et al. Target genes, variants, tissues and transcriptional pathways influencing human serum urate levels. Nat. Genet.51, 1459–1474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takada, T. et al. ABCG2 dysfunction increases serum uric acid by decreased intestinal urate excretion. Nucleosides Nucleotides Nucleic Acids33, 275–281 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Woodward, O. M. et al. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc. Natl. Acad. Sci. USA106, 10338–10342 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kannangara, D. R. W. et al. Hyperuricaemia: contributions of urate transporter ABCG2 and the fractional renal clearance of urate. Ann. Rheum. Dis.75, 1363–1366 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Matsuo, H. et al. ABCG2 dysfunction causes hyperuricemia due to both renal urate underexcretion and renal urate overload. Sci. Rep.4, 3755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhatnagar, V. et al. Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: potential role of remote sensing and signaling. Clin. Kidney J.9, 444–453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ejendal, K. F. & Hrycyna, C. A. Multidrug resistance and cancer: the role of the human ABC transporter ABCG2. Curr. Protein Pept. Sci.3, 503–511 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Kukal, S. et al. Multidrug efflux transporter ABCG2: expression and regulation. Cell Mol. Life Sci.78, 6887–6939 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohashi, Y. et al. Urate transporter ABCG2 function and asymptomatic hyperuricemia: a retrospective cohort study of CKD progression. Am. J. Kidney Dis.81, 134–144.e131 (2023). [DOI] [PubMed] [Google Scholar]
- 55.Liao, L. et al. TRIM46 upregulates Wnt/beta-catenin signaling by inhibiting Axin1 to mediate hypoxia-induced epithelial-mesenchymal transition in HK2 cells. Mol. Cell Biochem.477, 2829–2839 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Tremblay, J. & Hamet, P. Environmental and genetic contributions to diabetes. Metabolism100S, 153952 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozlitina, J., Xing, C., Pertsemlidis, A. & Schucany, W. R. Power of genetic association studies with fixed and random genotype frequencies. Ann. Hum. Genet.74, 429–438 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Khan, A. et al. Genome-wide polygenic score to predict chronic kidney disease across ancestries. Nat. Med.28, 1412–1420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen, Y. C. et al. Genome-wide association study for eGFR in a Taiwanese population. Clin. J. Am. Soc. Nephrol.17, 1598–1608 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee, D. J. et al. Genome-wide association study and fine-mapping on Korean biobank to discover renal trait-associated variants. Kidney Res Clin. Pr.43, 299–312 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirata, M. et al. Cross-sectional analysis of BioBank Japan clinical data: a large cohort of 200,000 patients with 47 common diseases. J. Epidemiol.27, S9–S21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng, Y. A. et al. Taiwan Biobank: a rich biomedical research database of the Taiwanese population. Cell Genom.2, 100197 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.King, E. K. et al. Prediction of non-responsiveness to pre-dialysis care program in patients with chronic kidney disease: a retrospective cohort analysis. Sci. Rep.11, 13938 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu, T. Y. et al. Comparison of multiple imputation algorithms and verification using whole-genome sequencing in the CMUH genetic biobank. Biomedicine11, 57–65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun, T. H. et al. Utility of polygenic scores across diverse diseases in a hospital cohort for predictive modeling. Nat. Commun.15, 3168 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siekmann, L. Determination of creatinine in human serum by isotope dilution-mass spectrometry. Definitive methods in clinical chemistry, IV. J. Clin. Chem. Clin. Biochem.23, 137–144 (1985). [PubMed] [Google Scholar]
- 69.Levey, A. S. et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med.150, 604–612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCaw, Z. R., Lane, J. M., Saxena, R., Redline, S. & Lin, X. Operating characteristics of the rank-based inverse normal transformation for quantitative trait analysis in genome-wide association studies. Biometrics76, 1262–1272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen, C. H. et al. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum. Mol. Genet.25, 5321–5331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei, C. Y. et al. Genetic profiles of 103,106 individuals in the Taiwan Biobank provide insights into the health and history of Han Chinese. NPJ Genom. Med.6, 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hinrichs, A. S. et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res.34, D590–D598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Connell, J. et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet.10, e1004234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet.5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet.53, 1097–1103 (2021). [DOI] [PubMed] [Google Scholar]
- 78.Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun.8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet.47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol.11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet.50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet.47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet.45, 124–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet.44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wakefield, J. Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol.33, 79–86 (2009). [DOI] [PubMed] [Google Scholar]
- 91.Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet.46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet.10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prive, F., Vilhjalmsson, B. J., Aschard, H. & Blum, M. G. B. Making the most of clumping and thresholding for polygenic scores. Am. J. Hum. Genet.105, 1213–1221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Novembre, J. et al. Addressing the challenges of polygenic scores in human genetic research. Am. J. Hum. Genet.109, 2095–2100 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience8, giz082 (2019). [DOI] [PMC free article] [PubMed]
- 96.Zhu, W., Deng, Y. & Zhou, X. Multiple membrane transporters and some immune regulatory genes are major genetic factors to gout. Open Rheumatol. J.12, 94–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee, C. J. et al. Phenome-wide analysis of Taiwan Biobank reveals novel glycemia-related loci and genetic risks for diabetes. Commun. Biol.5, 1175 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boocock, J. et al. Genomic dissection of 43 serum urate-associated loci provides multiple insights into molecular mechanisms of urate control. Hum. Mol. Genet.29, 923–943 (2020). [DOI] [PubMed] [Google Scholar]
- 99.Lu, P. et al. The developmental regulator protein Gon4l associates with protein YY1, co-repressor Sin3a, and histone deacetylase 1 and mediates transcriptional repression. J. Biol. Chem.286, 18311–18319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu, P. et al. The Justy mutation identifies Gon4-like as a gene that is essential for B lymphopoiesis. J. Exp. Med.207, 1359–1367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Adenaeuer, A. et al. Severe high-molecular-weight kininogen deficiency: clinical characteristics, deficiency-causing KNG1 variants, and estimated prevalence. J. Thromb. Haemost.21, 237–254 (2023). [DOI] [PubMed] [Google Scholar]
- 102.Santacroce, R., D’Andrea, G., Maffione, A. B., Margaglione, M. & d’Apolito, M. The genetics of hereditary angioedema: a review. J. Clin. Med. 10, 2023 (2021). [DOI] [PMC free article] [PubMed]
- 103.Sinnott-Armstrong, N. et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet.53, 185–194 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lin, C. T. et al. The ABCG2 rs2231142 polymorphism and the risk of nephrolithiasis: a case-control study from the Taiwan biobank. Front. Endocrinol.14, 1074012 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garcia-Nieto, V. M. et al. Gout associated with reduced renal excretion of uric acid. Renal tubular disorder that nephrologists do not treat. Nefrologia42, 273–279 (2022). [DOI] [PubMed] [Google Scholar]
- 106.Chen, C. J. et al. ABCG2 contributes to the development of gout and hyperuricemia in a genome-wide association study. Sci. Rep.8, 3137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakayama, A. et al. Subtype-specific gout susceptibility loci and enrichment of selection pressure on ABCG2 and ALDH2 identified by subtype genome-wide meta-analyses of clinically defined gout patients. Ann. Rheum. Dis.79, 657–665 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Katsura, K. et al. WDR72 regulates vesicle trafficking in ameloblasts. Sci. Rep.12, 2820 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang, H. et al. WDR72 mutations associated with amelogenesis imperfecta and acidosis. J. Dent. Res.98, 541–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kuechler, A. et al. A novel homozygous WDR72 mutation in two siblings with amelogenesis imperfecta and mild short stature. Mol. Syndromol.3, 223–229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morris, A. P. et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat. Commun.10, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Benonisdottir, S. et al. Sequence variants associating with urinary biomarkers. Hum. Mol. Genet.28, 1199–1211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The GWAS summary data supporting our findings are available on Figshare (10.6084/m9.figshare.24356587), and the statistical data are available in the Supplementary Data. The data from Taiwan Biobank (TWB; Application No. TWBR11111-02) and UK Biobank (UKB; Application No. 81803) were obtained through approved applications and they are publicly available to approved researchers for health-related research (TWB: https://www.biobank.org.tw/english.php; UKB: https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access). The GWAS summary data from Biobank Japan (BBJ) are publicly available without permission at http://jenger.riken.jp/en/result. The individual-level raw data from CMUH are unavailable because they contain information that may compromise participant privacy.