Abstract
There have been several reports of skin manifestations in patients with coronavirus disease 2019 (COVID-19). However, it is unclear whether severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA can be detected on the skin surface, including the sebum, of these patients. In this study, SARS-CoV-2 RNA was detected using real-time reverse-transcription polymerase chain reaction (RT-PCR) assay of skin surface lipids (SSLs) collected using an oil-blotting film from the faces of hospitalized patients with COVID-19. Human transcriptome analysis was also performed using the same samples. In facial SSLs of patients with COVID-19, the RT-PCR positivity rate was 84.6% (11/13 samples) within 5 days and 30.4% (7/23 samples) by 6–10 days of symptom onset. In the transcriptome analysis, the most characteristic SSL-RNA profile was the upregulation of interferon-stimulated gene (ISG)-related genes, such as ISG15, IFITM1, and MX1. This study presents an alternative technique using SSLs for non-invasive SARS-CoV-2 RNA detection and simultaneous analysis of human molecular pathogenesis in patients with COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-77862-0.
Keywords: SARS-CoV-2, COVID-19, Skin, Sebum, Transcriptome, Skin surface lipid
Subject terms: Gene expression analysis; Isolation, separation and purification; Sequencing; Infection; Biomarkers
Introduction
Since the beginning of the coronavirus disease 2019 (COVID-19) pandemic, real-time reverse-transcription polymerase chain reaction (RT-PCR) of viral RNA isolated from nasopharyngeal swabs has been used for its clinical diagnosis. However, identification of the specimens carrying severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at detectable levels is still underway, and this can be critical in devising strategies to prevent SARS-CoV-2 transmission. It has been reported that SARS-CoV-2 can be detected in upper respiratory tract specimens, such as nasopharyngeal and oropharyngeal swabs, and in other specimens, such as saliva, sputum, bronchoalveolar fluid, feces, blood, and urine1,2. The collection of nasopharyngeal swab fluid is invasive, uncomfortable for patients, and poses a risk of infection to healthcare workers; therefore, SARS-CoV-2 nucleic acid detection using saliva specimens—which are as sensitive as nasopharyngeal swab fluid—is now widely used for diagnosis3. However, there are several limitations, such as collection of an insufficient amount of saliva samples from which SARS-CoV-2 nucleic acid cannot be detected4 and difficulty in collecting saliva samples from infants and children.
Cutaneous manifestations, such as morbilliform rash, urticaria, vesicles, pseudo-chilblains, and vaso-occlusive lesions, are observed in some patients with COVID-195. Additionally, SARS-CoV-2 RNA has been detected in skin biopsies of patients with COVID-196,7. Notably, the reports of SARS-CoV-2 RNA detection in sweat samples are controversial8–10. Moreover, it remains unclear whether SARS-CoV-2 can be detected on the skin surface.
We previously established a method using RNA isolated from skin surface lipids (SSLs) for biological analysis11. The method enables transcriptome analysis using RNA extracted from SSLs collected using an oil-blotting film; it can be used to detect pathological molecular changes in patients with atopic dermatitis and Parkinson’s disease12–14. It is a simple specimen collection method, and unlike blood or nasopharyngeal swabs, samples can be collected by nonmedical personnel, including the patients themselves.
In this study, we aimed to clarify whether SARS-CoV-2 RNA can be detected on the skin surface of patients with COVID-19. Facial SSLs of patients with COVID-19 were collected using an oil-blotting film, and RT-PCR and patient transcriptome analyses were performed on these samples. Here, we established an alternative, non-invasive method for the detection of SARS-CoV-2 and simultaneous analysis of the molecular pathogenesis of COVID-19 using an oil-blotting film.
Results
Patient demographic and clinical findings
Hospitalized patients with COVID-19 who were enrolled between April 2020 and April 2021 (n = 29) and healthy participants who underwent SSL sampling in 2019 (n = 18) were included in this study (Supplementary Table 1). COVID-19 occurrence in patients was confirmed by RT-PCR assay for detection of SARS-CoV-2 RNA from nasopharyngeal swabs at hospitals. The median age of patients with COVID-19 was 38 years (interquartile range: 28–53 years; range: 18–72 years), and 79.3% of the participants were male individuals. The first SSL sample was collected after a median of 6 days (interquartile range: 2.75–7 years; range: 1–10 days) after symptom onset, followed by a second SSL sample collection in some cases. Upon hospital admission, the severity of COVID-19 was classified as mild in 15 patients and moderate in 14 patients.
SARS-CoV-2 detection in SSLs obtained from patients with COVID-19
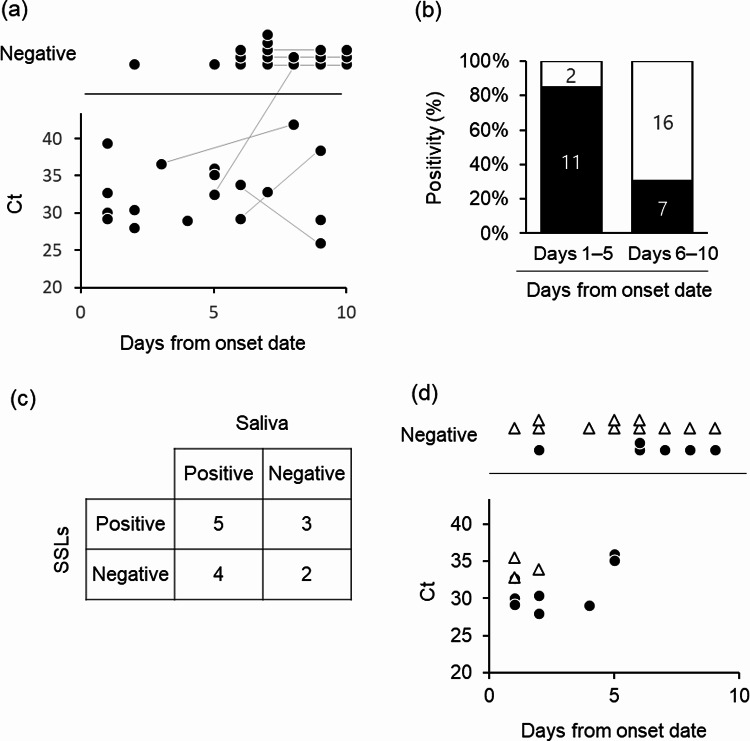
SSLs were collected from the entire face using an oil-blotting film before washing the face in the morning. SARS-CoV-2 was detected in 51.4% (19/37) SSL samples collected from the patients’ face within 10 days of symptom onset (one patient was asymptomatic) (Fig. 1a). The RT-PCR positivity rates of SSL samples within 5 and 6–10 days of onset were 84.6% (11/13) and 30.4% (7/23), respectively (Fig. 1b). Saliva contamination of the SSL samples was expected as they were obtained from the face; therefore, the results were compared with the RT-PCR results of saliva samples collected at the closest time point to the SSL samples. In 50% (7/14) of the samples, the results of both tests matched (five positive and three negative), whereas they did not in the remaining 50% of the samples (Fig. 1c, Supplementary Fig. 1, Supplementary Table 2). Interestingly, in a few patients, the saliva samples showed negative results but the SSL samples showed positive results (3/14). The expression of the salivary marker histatin 3 (HTN3)15 was not observed in five SSL samples that were positive for SARS-CoV-2 RNA and had transcriptome data. Furthermore, we tested SSL samples collected from the body (armpits, chest, and abdomen), where SARS-CoV-2 contamination from saliva via droplets was considered insignificant, and found that 3 out of 14 samples were positive. In particular, on the day after symptom onset, two of three samples were positive, and on the second day after symptom onset, one of three samples of SSLs on the body was positive (Fig. 1d). Differences in the amount of SSLs collected in each sample could affect the SARS-CoV-2-positivity rate; however, it was difficult to estimate the amount of SSLs in each sample because the quantity was low. Therefore, we compared human internal standard gene expression levels between SARS-CoV-2-positive and -negative samples using RT-PCR and found no significant differences in ACTB and GAPDH levels in the face and body (Fig. 2a, b, Supplementary Fig. 2a, b). The mean Ct value of human ACTB in samples obtained from the face and body was 13.9 and 17.7, respectively, suggesting that the amount of RNA collected was approximately 10 times lower in samples obtained from the body, which had fewer SSLs (Fig. 2c, Supplementary Fig. 2c). Finally, whole-genome sequencing of the SARS-CoV-2-positive SSL samples resulted in strain identification in 12 of the 17 samples (Table 1): 2 samples with B.1.1 and 4 samples with B.1.1.284 among the samples collected between April and July 2020; 3 samples with R.1 (EPI_ISL_12941049, EPI_ISL_12941050, EPI_ISL_18884079) and 3 samples with B.1.1.7 (EPI_ISL_12941051, EPI_ISL_12941052, EPI_ISL_18884080) among the samples collected between March and April 2021.
Fig. 1.
SARS-CoV-2 detection in SSLs from patients with COVID-19. (a) Scatter plots of Ct values obtained using the N2 primer set from facial SSL samples versus days from symptom onset. A total of 33 specimens from 24 patients were used in the analysis. Plots connected by lines indicate the same patient. (b) Positivity rates for SARS-CoV-2 in SSL samples collected on days 1–5 and 6–10 from symptom onset. The numbers in the bars indicate the number of individuals. (c) Comparison of SARS-CoV-2 detection results of RT-PCR in paired saliva and SSL samples (n = 14). (d) Scatterplot of Ct values obtained with the N2 primer set from the SSL samples versus days from symptom onset (n = 14) of face (●) and body (△).
Fig. 2.
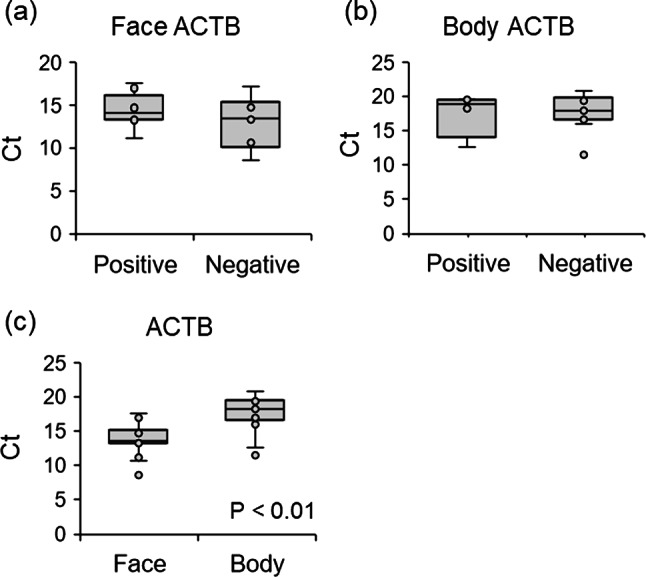
Influence of SSL-RNA content on SARS-CoV-2 detection. (a) Ct values of human ACTB for SARS-CoV-2-positive (n = 9) and -negative (n = 6) samples in facial SSLs. (b) Ct values of human ACTB for SARS-CoV-2-positive (n = 4) and -negative (n = 11) samples in SSLs of the body. (c) Ct values of human ACTB in SSL samples (n = 15) from the face and body of the same individual.
Table 1.
Determination of the lineage of SARS-CoV-2.
| PANGO Lineage | 2020/Apr–Jul | 2021/Mar–Apr |
|---|---|---|
| B.1.1 | 2 | 0 |
| B.1.1.284 | 4 | 0 |
| R.1 | 0 | 3 |
| B.1.1.7 | 0 | 3 |
| Failed | 2 | 3 |
Transcriptome analysis of SSL samples from patients with COVID-19
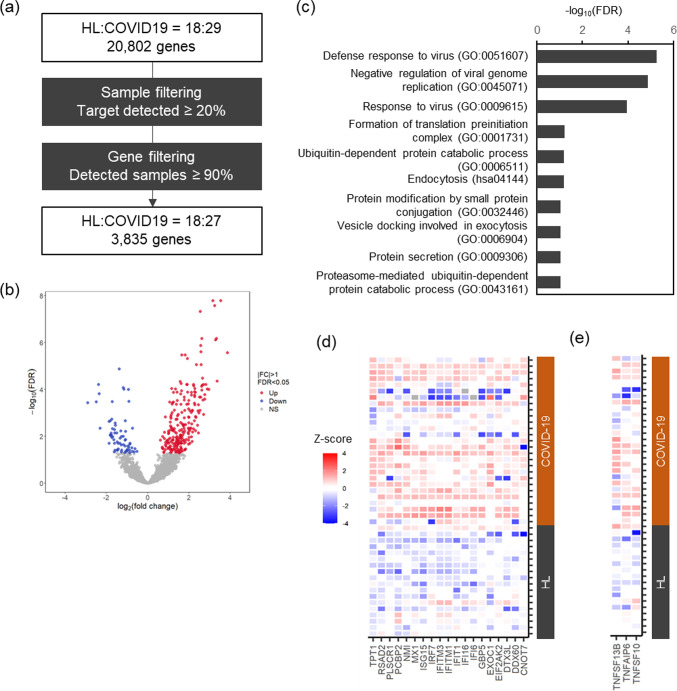
Previous studies have shown that it is possible to analyze the human transcriptome from SSL samples collected from the face using an oil-blotting film11. After preprocessing the sequence data, 45 samples (18 healthy participants and 27 patients with COVID-19) and 3,835 genes were analyzed (Fig. 3a). The false discovery rate (FDR) was set to 0.05, and a comparison of the two groups, healthy participants and patients with COVID-19, resulted in the identification of 302 differentially expressed genes (DEGs) (Fig. 3b, Supplementary Tables 3, 4). Analysis of 235 upregulated and 67 downregulated genes among the DEGs showed significant results in Gene Ontology (GO) for biological process categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The top three terms for the upregulated genes were defense response to the virus (GO:0051607), negative regulation of viral genome replication (GO:0045071), and response to the virus (GO:0009615) (Fig. 3c). These terms included a group of type I IFN-related genes that are essential for antiviral immunity, and the expression of genes such as ISG15, IRF7, IFITM1, IFITM3, IFI6, and MX1 was significantly upregulated in patients with COVID-19 (Fig. 3d). The expression of TNF pathway-related genes (TNFSF10, TNFAIP6, and TNFSF13) was significantly upregulated in patients with COVID-19 compared with that in healthy controls (Fig. 3e). No significant differences were observed for the expression of the inflammation-related genes IL1B and NF-kB (Supplementary Fig. 3). The expression of IL-6-induced genes, such as IL6R, SOCS3, and STAT3, was not included in the analysis because of their low expression levels. By contrast, analysis of downregulated genes revealed terms related to cytoplasmic translation (GO:0002181), translation (GO:0006412), and ribosomes (hsa03010), suggesting that potential alterations in protein synthesis occurred in response to this disease.
Fig. 3.
Transcriptome analysis of SSL samples from patients with COVID-19. (a) Data preprocessing. We selected 45 samples with a detected gene percentage greater than 20%. Normalization factors were calculated using DESeq2, and genes with non-zero read counts in more than 90% of samples (3,835 genes) were selected. (b) Volcano plot of gene expression changes in COVID-19. (c) Significant GO terms in biological processes and KEGG pathways for 235 genes upregulated in patients with COVID-19. (d) Heatmap of viral response-associated genes in the top three GO terms (GO:0051607, GO:0045071, and GO:0009615) extracted from the genes upregulated in patients with COVID-19. (e) Heatmap of TNF pathway-related genes upregulated in patients with COVID-19. Expression levels were converted to z-scores by genes. HL, healthy control.
Discussion
In this study, we attempted to detect SARS-CoV-2 and simultaneously analyze the human transcriptome using specimens obtained from a single oil-blotting film. To our knowledge, this is the first report of detection of SARS-CoV-2 RNA from the skin surface. The patient or nurse followed the instructions in the manual to collect SSLs from the patient’s face, and human mRNA—used as an indicator of proper sample collection—was detected in all 37 samples, suggesting that this oil-blotting film method is an easy and effective way for any individual to collect specimens. A key advantage of this technique is its simplicity and non-invasive nature, making it especially useful for patients for whom conventional sampling methods pose challenges, such as infants.
A positive rate of 84.6% was obtained with facial SSLs isolated within 5 days of symptom onset, indicating that the sensitivity of this technique for detecting SARS-CoV-2 RNA is relatively high in the early stages of symptom onset. Analysis of nasal swabs revealed that the SARS-CoV-2 RNA level peaked on day 4 of symptom onset16 and that the viral load decreased as time progressed17. Although the date of peak viral loads was not clear in this study because of the small sample size, the overall trend in SSL samples of a high positivity rate up to day 5 after symptom onset, followed by a decrease in the detection rate over time, was consistent with the results reported for nasal swabs.
There are two possibilities for the origin of SARS-CoV-2 nucleic acids detected in oil-blotting film samples: (i) persistence of droplets on the face and body and (ii) release of SARS-CoV-2 RNA on the skin via sebum or sweat. SSL collection was performed with new nitrile gloves; therefore, contamination from the hands was eliminated as much as possible; however, the possibility of contamination from the patient’s salivary droplets and nasal secretion on the face was not eliminated. Owing to the presence of RNase on the skin, it is unlikely that SARS-CoV-2 RNA in the droplets would remain undegraded. It has been found that fatty acids in sebum inhibit RNase activity11, and it is possible that SARS-CoV-2 RNA remains undegraded in environments where droplets evaporate quickly and sebum is abundant. Interestingly, in this study, there were some cases where SARS-CoV2 RNA was detected in SSL samples but not in saliva samples, and there were some cases where SARS-CoV2 RNA was detected even on bodies where droplet contamination was considered low. To our knowledge, there are no reports of SARS-CoV-2 detection in sebum. On the contrary, there is a report of SARS-CoV-2 RNA being detected in sweat10, as well as reports that SARS-CoV-2 RNA was not detected in sweat8,9. Considering the findings mentioned above, the possibility that SARS-CoV-2 RNA was released on the skin via sebum or sweat cannot be denied. The origin of SARS-CoV-2 RNA in the specimens collected by oil-blotting film in this study was difficult to determine, as there was insufficient evidence to reject the possibility of contamination from the patient’s salivary droplets and nasal secretion. However, this study provides the first knowledge of SARS-CoV-2 detection on the skin surface, and we propose this as a novel methodology for non-invasive SARS-CoV-2 RNA detection in patients with COVID-19.
In this study, human transcriptome analysis was performed using RNA obtained from the same oil-blotting film used for SARS-CoV-2 detection. The most characteristic SSL-RNA profile of patients with COVID-19 was the upregulation of interferon-stimulated (ISG)-related genes, such as ISG15, IFITM1, and MX1, which are typical genes involved in host immune response during SARS-CoV-2 infection18. This gene expression profile is similar to that of the blood transcriptome of patients with COVID-1919. Regarding the skin, the levels of IFN-related genes and inflammation-related molecules have been reported to be elevated in the biopsy gene expression profiles of skin lesions in patients with moderate-to-severe COVID-1920. In addition, the expression of BAFF (TNFSF13B) and TRAIL (TNFSF10), members of the TNF superfamily, was upregulated in areas with severe lung tissue damage21. It is difficult to discuss the association between the SSL-RNA gene profile and skin manifestations because the skin manifestations of patients with COVID-19 were not recorded in this study. The fact that the host response during acute respiratory infection was detectable in SSL-RNA, as well as in the skin and lung tissues, is interesting. The expression profile of SSL-RNA is presumed to reflect its expression primarily in the epidermis, hair follicles, and sebaceous glands11. ISG are also expressed in keratinocytes and cutaneous leukocytes, which are involved in immune responses in the skin22. It is important to note that it is unclear whether changes in the SSL-RNA gene profile are a reflection of changes in keratinocytes, the main cells of the skin, or whether they also reflect the changes in immune cells. Indeed, genes encoding macrophage receptor molecules (CD209 and CD163) and chemokines (CXCL2 and CCL2), which have been detected in skin punch biopsy20, were not detected in SSL-RNA profile owing to low expression levels. However, this methodology, which can noninvasively detect the biological response to acute infection without biopsy, will help us understand the pathogenesis of the disease.
A limitation of the present study is the small number of patients tested. For the comparative study of the positivity rates of saliva and SSLs, sampling was not conducted on the same day; therefore, sampling at the same time is necessary for a comprehensive discussion. It was not possible to determine the source of the SARS-CoV-2 RNA detected in SSLs. Additionally, the detection of SARS-CoV-2 nucleic acids using RT-PCR alone does not indicate that the presence of infectious virus on the skin and, therefore, this should be discussed with caution as a possible route of transmission. Finally, accurate negative controls need to be prepared and examined for possible future real-world diagnostic use of this method. If false-negative results are caused by cross-contamination between infected and healthy individuals (for example, patients with COVID-19 sneezing on healthy individuals), it may be necessary to reduce the false-negative rate, perhaps by waiting for a certain time after COVID-19 exposure before collecting sebum.
In conclusion, this study presents a method for the simultaneous detection of SARS-CoV-2 nucleic acids and host transcriptome analysis from SSLs collected using a single oil-blotting film. This method can be used as an alternative technique for convenient pathogen detection and understanding the molecular pathogenesis of acute respiratory infections.
Methods
Study design
This study was approved by the Medical Research Ethics Committee of the National Institute of Infectious Diseases (NIID; approval number: 1111) and the Ethical Committee of Kao Corporation (approval numbers: T293-200331 and T229-190615). This study was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all the participants or their legal representatives. The selection criteria were as follows: (i) patients aged 20 years or older, (ii) patients with confirmed SARS-CoV-2 infection who visited a collaborating medical institution (patients with COVID-19) or hospitalized patients with clinically suspected SARS-CoV-2 infection to be tested for infection, and (iii) patients who provided written informed consent themselves or through a legal representative. The exclusion criteria were as follows: (i) patients who were judged by their physicians to be inappropriate for inclusion and (ii) patients whose health condition would worsen by having their blood collected due to anemia or other reasons.
Patients from whom consent was obtained were enrolled as study participants, and information on age, sex, date of symptom onset, clinical symptoms related to COVID-19, and clinical examinations was obtained from their medical records. Severity was based on World Health Organization (WHO) categories. Specimens collected from hospitalized patients in Japan between April 2020 and April 2021 were used for analysis. In the case of healthy controls, data from a study conducted in 2019, in which SSLs were collected from healthy participants, were used. Our study examined both male and female participants, and similar findings have been reported for individuals of both sexes.
SSL collection and RNA preparation
SSLs were collected from the entire face (forehead, temples, cheeks, nose, and chin) or body (armpits, chest, and abdomen) of the participants before washing the face in the morning using an oil-blotting film (5.0 cm × 8.0 cm; 3 M Japan, Tokyo, Japan) by the patients or a nurse wearing a nitrile glove. The oil-blotting films with the SSLs were placed in RNase-free 25-mL tubes containing molecular sieves 13 × 1/8 (FUJIFILM Wako Pure Chemical Corporation, Tokyo, Japan) and stored at − 80 °C. After cutting the oil-blotting film into small pieces, QIAzol (1,425 µL; QIAGEN, Hilden, Germany) was added, and the samples were vortexed for 3 min; then, 1,300 µL of solution was collected in a tube. After adding 260 µL of chloroform, the tube was vortexed for 10 s and centrifuged at 12,000 × g for 15 min at 4 °C. The supernatant (aqueous layer; 750 µL) was collected and RNA was purified using the RNeasy Mini QIAcube Kit (QIAGEN) with DNase treatment, according to custom protocol. Briefly, 85% ethanol (750 µL) was added to samples (upper aqueous phase; 750 µL), which were then mixed well and transferred to a RNeasy mini column. After washing with RW1 buffer, DNase I treatment, re-washing with RW1 buffer, and washing with RPE buffer, the RNA was finally eluted by passing RNase-free water (50 µL) through the column twice to obtain RNA solution (100 µL). Subsequently, the purified RNA solution (100 µL) was ethanol precipitated with Ethachinmate (Nippon Gene, Tokyo, Japan) and redissolved in RNase-free water (10 µL).
SARS-CoV-2 detection
Of the 10 µL RNA obtained from the oil-blotting film, 1 µL was diluted five-fold with water and used for virus detection. Cycle threshold (Ct) values were measured using RT-PCR with the QuantiTect Probe RT-PCR kit (QIAGEN) and the NIID-N2 primer/probe set23 (Supplementary Table 5) targeting the SARS-CoV-2 nuclear protein (N) region on an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). The thermal cycling conditions were as follows: 50 °C for 30 min, 95 °C for 15 min, 45 cycles at 95 °C for 15 s, and 60 °C for 1 min.
Whole-genome sequencing
The sample RNA was reverse transcribed using the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific) or LunaScript RT SuperMix Kit (New England BioLabs, Ipswich, MA, USA), and DNA amplicons for amplifying the SARS-CoV-2 genomic region were obtained using multiplex PCR with ARTIC-N4 or N5 primers and Q5 Hot Start DNA polymerase (New England BioLabs)24. A library was prepared using the QIAseq FX DNA Library Kit (QIAGEN) and sequenced using the MiSeq System (Illumina, San Diego, CA, USA). The obtained reads were mapped to the SARS-CoV-2 reference genome MN908947.3. Whole-genome sequencing was performed by Takara Bio, Inc. or the National Institute of Infectious Diseases.
AmpliSeq transcriptome analysis
Transcriptome analysis was performed as previously described, with a few modifications11. Briefly, RNA solution (1.5 µL) was used for reverse transcription and sequencing library preparation using the Ion AmpliSeq Transcriptome Human Gene Expression Kit (Thermo Fisher Scientific, Waltham, MA, USA). In addition to the previously described methods, 250 and 100 ng of T4 Gene 32 Protein (New England Biolabs) were added to the reverse transcription reaction mix and first-round PCR mix, respectively. The library was then used for template preparation using the Ion 540 Chip Kit on an Ion Chef instrument (Thermo Fisher Scientific), and sequencing was performed on an Ion S5 XL System (Thermo Fisher Scientific). The read count data were analyzed using the R software. Samples in which the percentage of detected genes was less than 20% of the target 20,802 genes were excluded from the analysis as low-quality samples. Genes detected in more than 90% of the samples that met the criteria were normalized using the size factor in DESeq225 to extract genes with differential expression (FDR < 0.05, |Log2FC| > 1). Biological process and KEGG pathway analyses were performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID)26–28.
Internal standard gene expression analysis
Owing to the limited amount of SSL-RNA, the remaining samples from the AmpliSeq analysis were used for the evaluation of human internal standard genes using RT-PCR. In other words, pre-amplified amplicons were used as cDNA templates using an Ion AmpliSeq™ Transcriptome Human Gene Expression kit (Thermo Fisher Scientific). In accordance with the sequences included in the Ion AmpliSeq™ Transcriptome Human Gene Expression Core Panel provided by Thermo Fischer Scientific, the primers for RT-PCR were designed using Primer-BLAST (NCBI) and are shown in Supplementary Table 6. They were designed as nested primers, which were specified to amplify the region in the first PCR using the Ion AmpliSeq™ Transcriptome Human Gene Expression kit. Human ACTB and GAPDH were quantified as stably expressed genes in the SSL-RNA.
RT-PCR was performed with reaction mixtures comprising 0.75 µL of each forward and reverse primers of each gene (250 nM), 1.0 µL of template (1:10 dilution), and 2.5 µL of THUNDERBIRD Next SYBR qPCR Mix (TOYOBO, Osaka, Japan) using the QuantStudio™ 12 K Flex Real-time PCR System (Thermo Fisher Scientific). The thermal cycling conditions were as follows: 50℃ for 2 min, 95℃ for 10 min, 40 cycles at 95℃ for 15 s and at 60℃ for 1 min; specificity of the PCR products was evaluated using melting curve analysis.
Statistics
To assess the significance of the differences between the two groups, Welch’s t-test was used. We extracted DEGs between healthy controls and patients with COVID-19 using the likelihood ratio test with Benjamini–Hochberg’s FDR < 0.05 and |Log2FC| > 1 as threshold values.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the clinical staff at the Hospital of the Institute of Medical Science, University of Tokyo, National Center for Global Health and Medicine, and Tokyo Metropolitan Cancer and Infectious Diseases Center, Komagome Hospital. We also thank GISAID for sharing and comparing our data with data submitted globally.This work was supported by the Japan Agency for Medical Research and Development (AMED) [grant numbers: JP21fk0108104 and JP23fk0108637 (to T.S.)].
Author contributions
Conceptualization, T.S.; Methodology, T. Kuwano (T.K.1) and T.I.; Investigation, T.K.1, T. Kanno (T.K.2), M.T., Y.H., H.K., Y.U., M.I., and N.T.; Resources, M.K., H.N., T.T., N.Y., H.Y., S.K., Y.M., N.K.I., N.O., T. Kobayashi (T.K.3), K.F., M.T., and A.I.; Project Administration, A.K.T. and T.S.; Funding Acquisition, T.S.; Supervision, T.M., A.K.T., and T.S.; Writing – Original Draft, T.K.1; Writing –Review & Editing, T.K.1, T.I., T.M., A.K.T., and T.S.
Data availability
Source data are available as Supplementary Data Values file.
Competing interests
T. Kuwano (T.K.1), T. Kanno (T.K.2), M.T., N.T., T.I., T.M., A.K.T., and T.S. are the inventors of the patented method of SARS-CoV-2 detection in SSLs. The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tetsuya Kuwano, Email: kuwano.tetsuya@kao.com.
Tadaki Suzuki, Email: tksuzuki@niid.go.jp.
References
- 1.Wang, W. et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 323, 1843–1844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng, L. et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J. Med. Virol.92, 1676–1680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyllie, A. L. et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl. J. Med.383, 1283–1286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matic, N. et al. Practical challenges to the clinical implementation of saliva for SARS-CoV-2 detection. Eur. J. Clin. Microbiol. Infect. Dis.40, 447–450 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polly, S. & Fernandez, A. P. Common skin signs of COVID-19 in adults: An update. Cleve Clin. J. Med.89, 161–167 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Majumdar, R. et al. SARS-CoV-2 RNA detection in formalin-fixed paraffin-embedded (FFPE) tissue by droplet digital PCR (ddPCR). Clin. Chim. Acta532, 181–187 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marzano, A. V. et al. SARS-CoV-2 detection by digital polymerase chain reaction and immunohistochemistry in skin biopsies from 52 patients with different COVID-19-associated cutaneous phenotypes. Dermatology. 239, 584–591 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Fathizadeh, H. et al. Study presence of COVID-19 (SARS-CoV-2) in the sweat of patients infected with COVID-19. Microb. Pathog149, 104556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arslan, B., Bercin, S., Aydogan, S., Islamoglu, Y. & Dinc, B. SARS-CoV-2 is not found in the sweat of COVID-19 positive patients. Ir. J. Med. Sci.191, 27–29 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Recalcati, S., Tonolo, S., Meroni, E. & Fantini, F. SARS-CoV-2 in the sweat of COVID-19-positive patients: A possible route of transmission? J. Eur. Acad. Dermatol. Venereol.35, e865–e866 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue, T. et al. Non-invasive human skin transcriptome analysis using mRNA in skin surface lipids. Commun. Biol.5, 215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shima, K. et al. Non-invasive transcriptomic analysis using mRNAs in skin surface lipids obtained from children with mild-to-moderate atopic dermatitis. J. Eur. Acad. Dermatol. Venereol.36, 1477–1485 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto-Hanada, K. et al. mRNAs in skin surface lipids unveiled atopic dermatitis at 1 month. J. Eur. Acad. Dermatol. Venereol.37, 1385–1395 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Uehara, Y. et al. Non-invasive diagnostic tool for Parkinson’s disease by sebum RNA profile with machine learning. Sci. Rep.11, 18550 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song, M., Bai, H., Zhang, P., Zhou, X. & Ying, B. Promising applications of human-derived saliva biomarker testing in clinical diagnostics. Int. J. Oral Sci.15, 2 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frediani, J. K. et al. The new normal: Delayed peak SARS-CoV-2 viral loads relative to symptom onset and implications for COVID-19 testing programs. Clin. Infect. Dis.78, 301–307 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puhach, O., Meyer, B. & Eckerle, I. SARS-CoV-2 viral load and shedding kinetics. Nat. Rev. Microbiol.21, 147–161 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar, L., Liu, G. & Gack, M. U. ISG15: Its roles in SARS-CoV-2 and other viral infections. Trends Microbiol.31, 1262–1275 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadjadj, J. et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science369, 718–724 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domizio, J. D. et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature603, 145–151 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohue-Kitano, R. et al. Medium-chain fatty acids suppress lipotoxicity-induced hepatic fibrosis via the immunomodulating receptor GPR84. JCI Insight. 8, e165469 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg, E. et al. (ed, N.) Circadian control of interferon-sensitive gene expression in murine skin. Proc. Natl. Acad. Sci. U S A117 5761–5771 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirato, K. et al. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV-2019) in Japan. Jpn J. Infect. Dis.73, 304–307 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Itokawa, K., Sekizuka, T., Hashino, M., Tanaka, R. & Kuroda, M. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS One15, e0239403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc.4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Sherman, B. T. et al. A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res.50, W216–W221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are available as Supplementary Data Values file.