Abstract
The translation of pre‐clinical anti‐cancer therapies to regulatory approval has been promising, but slower than hoped. While innovative and effective treatments continue to achieve or seek approval, setbacks are often attributed to a lack of efficacy, failure to achieve clinical endpoints, and dose‐limiting toxicities. Successful efforts have been characterized by the development of therapeutics designed to specifically deliver optimal and effective dosing to tumour cells while minimizing off‐target toxicity. Much effort has been devoted to the rational design and application of synthetic nanoparticles to serve as targeted therapeutic delivery vehicles. Several challenges to the successful application of this modality as delivery vehicles include the induction of a protracted immune response that results in their rapid systemic clearance, manufacturing cost, lack of stability, and their biocompatibility. Extracellular vesicles (EVs) are a heterogeneous class of endogenous biologically produced lipid bilayer nanoparticles that mediate intercellular communication by carrying bioactive macromolecules capable of modifying cellular phenotypes to local and distant cells. By genetic, chemical, or metabolic methods, extracellular vesicles (EVs) can be engineered to display targeting moieties on their surface while transporting specific cargo to modulate pathological processes following uptake by target cell populations. This review will survey the types of EVs, their composition and cargoes, strategies employed to increase their targeting, uptake, and cargo release, and their potential as targeted anti‐cancer therapeutic delivery vehicles.
Keywords: cancer, communication, engineering, extracellular vesicles, uptake
1. INTRODUCTION
It is essential for cells to communicate effectively with one another, and with their microenvironment to sustain proper tissue structure and function during development and to maintain homeostasis (Lowenstein, 1990; Plotnikov et al., 2017). Altered communication between cells and their microenvironment is pivotal in the development, progression, and metastasis of many tumours (Dominiak et al., 2020; Ebrahim et al., 2024; Zefferino et al., 2021). Cells communicate with neighbouring cells and their environment by direct interactions mediated by cell adhesion transmembrane proteins, including integrins (Pang et al., 2023), cadherins (Yulia et al., 2018), selectins (D'Arcy & Kiel, 2021), and immunoglobulin superfamily members (Brummendorf & Lemmon, 2001). Cells also communicate with neighbouring and distant cells by secreting diverse classes of soluble biomolecules, such as cytokines (Altan‐Bonnet & Mukherjee, 2019), growth factors, chemokines (Singh & Gray, 2021), ion channels (Jackson, 2022), neurotransmitters (Heasley, 2001), and hormones (Padmanabhan & Cardoso, 2020). A novel mechanism that has emerged as critical for intercellular communication is the biogenesis and export of membrane‐bound extracellular vesicles (EVs) and amembranous extracellular particles (EPs) that serve as active carriers of biologically important material (Dominiak et al., 2020; Ebrahim et al., 2024; Zefferino et al., 2021). EVs have generated intense interest for their potential to serve as powerful therapeutic tools for the targeted delivery of anti‐cancer agents.
1.1. The diversity of EVs
EVs comprise a diverse collection of particles, including apoptotic bodies, cancer‐derived large oncosomes, microvesicles, and exosomes (Figure 1). EVs are distinguished by their mode of biogenesis, unique cargo composition, and role in cellular communication (Théry et al., 2018; Welsh et al., 2024). When classified by mode of biogenesis, EVs are divided into exosomes that arise via inward budding from endosomal membranes, and ectosomes that bud outward from the plasma membrane. Apoptotic bodies (∼0.8–5 µM), arising as byproducts from programmed cell death, participate in immune responses and microenvironmental homeostasis (Kakarla et al., 2020). The makeup of their cargo and their interactions with immune cells play a critical role in controlling the immune response in the tumour microenvironment (TME). Cancer‐derived large oncosomes (∼1–10 µM) are membrane‐bound EVs generated by cancer cells that contain a variety of biomolecules, including proteins, nucleic acids, lipids, organelles, and smaller EVs (Choi et al., 2017; Choi et al., 2019; Meehan et al., 2016). Cancer‐derived large oncosomes can facilitate intercellular communication and provide a means for cancer cells to manipulate their microenvironment, promoting tumour growth, metastasis, and immune evasion. Microvesicles (0.1–100 µM), are shed directly from the cell membrane and can influence cancer cell survival and angiogenesis (Skog et al., 2008). These membrane‐bound vesicles play a crucial role in advancing tumour development by aiding in the creation of new blood vessels to sustain the expanding tumour. Exosomes (50–120 nm), small membrane‐bound vesicles of endocytic origin, contribute to tumour progression and immune evasion (Johnson et al., 2023). The biogenesis of exosomes is a complex process that involves the inward budding of vesicles in the multivesicular body (MVB). Their small size allows for the efficient intercellular transport of bioactive molecules, shaping the communication network within the TME.
FIGURE 1.
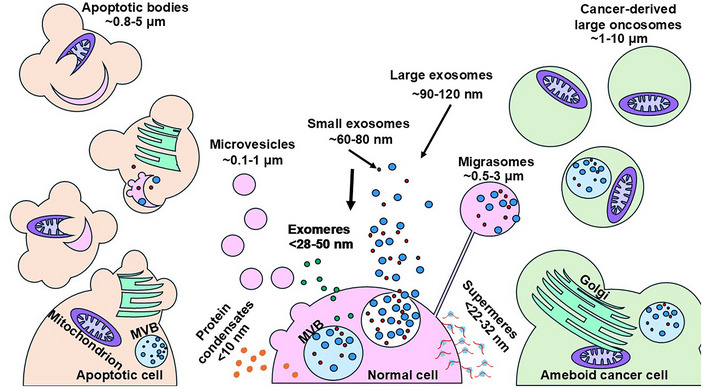
Diversity of extracellular nanoparticles. EVs are a heterogeneous assortment of lipid bilayer nanoparticles comprised of particles classified based on several criteria. Small and large exosomes are generated by inward budding of endosomal membranes. The membrane encloses an assortment of bioactive macromolecules, including DNA, coding, and non‐coding RNAs, proteins, lipids, and metabolites. Ectosomes (microvesicles, exomeres, supermeres, apoptotic bodies, and cancer‐derived large oncosomes) result from budding from the plasma membrane. EPs are amembranous particles comprised of cell‐free proteins (histones), nucleic acids (dsDNA, mRNA, miRNA), and lipids (HDL, IDL, LDL) and can induce a TLR‐mediated inflammatory response. EP, extracellular particles; EV, extracellular vesicles; TLR, toll‐like receptor.
Within the realm of EPs, a diverse array of structures has been identified, encompassing exomeres, supermeres, migrasomes, and protein condensates (Figure 1). Exomeres (< 28–50 nm), one of the more recently identified EPs, are smaller in size and have been implicated in various physiological and pathological processes such as immune modulation, cancer progression, and tissue generation. Although originally dismissed as artefacts of ultracentrifugation, exomeres have been detected by asymmetric field flow fractionation (Zhang et al., 2018). Although exosomes and exomeres contain some of the same lipids and proteins, there are distinct differences. Exomeres, for example, are enriched in the Argonaute (Ago 1–3) proteins and in several enzymes involved in various metabolic pathways (Jeppesen et al., 2023). Supermeres (22–32 nm), another class of EPs, other extracellular condensates, dynamic biomolecular assemblies, and larger subpopulations of EVs and EPs collectively shape the complex language of intercellular communication within the TME (Zhang et al., 2022). Migrasomes (∼0.5–3 µm), recently identified as a distinct type of extracellular vesicle, are involved in cell migration and cancer metastasis (Jiang et al., 2023; Zhang et al., 2022). These vesicles are formed at retraction fibres of the cellular migration‐associated protrusions and contain various proteins and lipids that aid in cell movement and invasion into surrounding tissues (Jiang et al., 2023). Additionally, protein condensates (< 10 nm) are formed through liquid‐liquid phase separation, and play a significant role in gene expression, protein translation, and signal transduction, thereby contributing to the broad range of cellular processes and interactions (Jiang et al., 2020).
1.2. The diversity of EV cargo
The intense interest in the study of EVs is motivated primarily by two factors: their potential to serve as a non‐invasive source of diagnostic and prognostic surrogate biomarkers for many diseases including cancer (Liu et al., 2021; Sunkara et al., 2016; Xu et al., 2020; Zhou et al., 2020) and as paracrine signalling mediators capable of modifying target cell phenotypes (Zocco et al., 2014; Zou, Li et al., 2023; Zou, Lei et al., 2023). The copious literature on exploiting EVs as a source of biomarkers for cancer and other diseases has been extensively reviewed elsewhere (Liu et al., 2021, Sunkara et al., 2016; Xu et al., 2020; Zhou et al., 2020). This review will examine the characteristics of EVs that render them attractive as potential therapeutic tools, particularly as delivery vehicles for anti‐cancer therapies. EVs can carry a complex array of bioactive macromolecules as depicted in Figure 2. EV cargo molecules are comprised of proteins, lipids, metabolites, and nucleic acids, including DNA and most coding and non‐coding RNA species (Colombo et al., 2014; Jeppesen et al.; 2019, Lee et al., 2024). EV cargo composition varies based on the genes expressed in the cell of origin when EVs are being packaged and released. Changes in the physiologic state of the cell therefore alter EV cargo composition (Haraszti et al., 2016; Tancini et al., 2019; van Niel et al., 2018; Zhang et al., 2023). EV functional properties are conferred by macromolecules displayed on the outer surface of their lipid membranes or carried within the vesicle lumen (Colombo et al., 2014; Jeppesen et al., 2019; Haraszti et al., 2016; Mause & Weber, 2010; Lee et al., 2024; Tancini et al., 2019; Zhang et al., 2023). Since EV cargoes can modify cellular processes impacting tumour progression, immune modulation, and microenvironmental dynamics, EVs have tremendous potential to serve as attractive therapeutic tools for cancer and other diseases (Batrakova & Kim, 2015; Ou et al., 2021; van Niel et al., 2018; Wiklander et al., 2015; Zhu et al., 2017). This potential is enhanced by the ability to engineer EVs to increase their cell‐specific targeting and their biocompatibility as evidenced by the low levels of toxicity and immunogenicity induced by prolonged dosing in vivo (Zhu et al., 2017).
FIGURE 2.
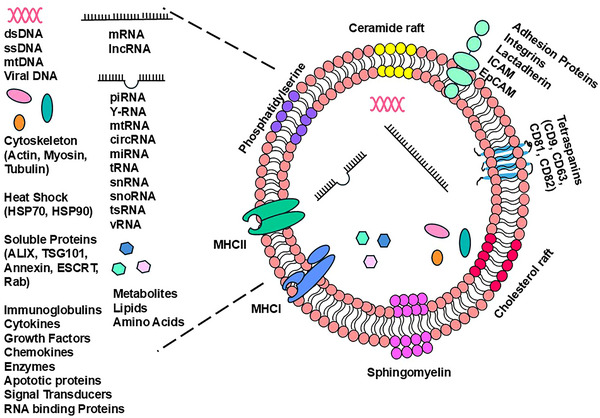
Diversity of exosome cargo and components. Exosomes are bound by a bilayer membrane containing various lipids including ceramides, sphingomyelin, and cholesterol. Peripheral and integral proteins are present in the membrane including tetraspanins, receptors, and major histocompatibility complex proteins. Exosomal cargo is diverse and includes nucleic acids, soluble and cytoskeletal proteins, lipids, metabolites, and amino acids.
Understanding the mechanisms regulating the cellular uptake and cargo unloading of diverse endogenous EV subtypes is pivotal for exploiting them as novel treatment modalities and devising engineering strategies to maximize their potential. The following comprehensive exploration offers insights into the intricate dynamics of EV uptake and cargo release in cancer delving further into the complex landscape to foster a deeper understanding of the molecular and cellular interactions modulated by EVs that govern disease progression.
1.3. Shared properties of EVs and viruses
EVs, particularly exosomes, share many properties with enveloped viruses, such as Influenza type A, HIV, vaccinia, and SARS‐CoV‐2 (Badierah et al., 2021; Metzner & Zaruba, 2021; Nolte‐’t Hoen et al., 2016). These viral particles could be considered as a type of pathogenic exosome (Badierah et al., 2021; Gould et al., 2003; Metzner & Zaruba, 2021; Nolte‐’t Hoen et al., 2016). Viral particles enter cells and deliver cargo (the viral genome and proteins) that coopt normal cellular functions to replicate new viral particles whereas EVs enter target cells to deliver cargo (nucleic acids, proteins, lipids) that modify cellular phenotypes. Remarkably, exosomes produced by virus‐infected cells can contain the entire viral genome and capsid proteins, thus facilitating viral spread to non‐infected cells (Boson et al., 2021; Skehel & Wiley, 1995, Locker et al., 2013; Weiss & Göttlinger, 2011). The source of the membrane that envelops viral particles and EVs depends on their source. Some enveloped viruses, such as HIV‐1 and Influenza type A, derive their membranes by budding from lipid raft regions enriched in virally encoded envelope proteins, resembling the budding of the ectosomal microparticles, microvesicles, and apoptotic bodies (Skehel & Wiley, 1995; Weiss & Göttlinger, 2011). Other enveloped viruses, such as vaccinia viruses and SARS‐CoV‐2, bud from endoplasmic reticulum and trans‐Golgi, whereas exosomes are generated by inward budding from late endosomes and multivesicular bodies (Boson et al., 2021; Locker et al., 2013). Viral membranes contain glycoproteins that bind to specific target cell membrane receptors and mediate the fusion of viral and plasma membranes (Mebatsion et al., 1996; Yamauchi & Helenius, 2013). Membrane fusion has also been proposed as an EV entry mechanism (Bonsergent & Lavieu, 2019; Hemler, 2003; Yao et al., 2018).
Viral vectors are employed in gene therapy approaches to take advantage of the naturally evolved ability of viruses to enter target cells and deliver nucleic acid cargo. The effectiveness of these vectors is diminished by their ability to trigger an immune response and often by the induction of insertional mutagenesis (Thomas et al., 2003). Since EVs demonstrate lower toxicity and immunogenicity (Zhu et al., 2017) and do not induce insertional mutations, they offer an attractive alternative to viral vectors as a system to deliver therapeutic bioactive cargo to target cells.
1.4. Mechanisms of EV uptake
1.4.1. Non‐endocytic mechanisms
Direct receptor engagement
The most direct route for EVs to modify target cell phenotypes is first by direct interaction between proteins in the outer leaflet of the EV lipid bilayer with surface receptors on target cell plasma membranes. For example, EV‐associated Distal‐like canonical Notch ligand 4 (Dll4) inhibits Notch signalling in target endothelial cells (Sheldon et al., 2010). It has also been reported that exosomes isolated from mature CD8+ dendritic cells carry intercellular adhesion molecule 1 (ICAM1) on their surface membrane and can bind to lymphocyte function‐associated antigen 1 (LFA‐1) on the surface of activated T cells (Segura et al., 2007) and antigen‐presenting cells (Nolte‐’t Hoen et al., 2009) inducing their proliferation. Multiple studies have demonstrated that exosomal major histocompatibility complex (MHC)‐peptide complexes can activate target T cells by directly binding to the T cell receptor (Admyre et al., 2006; Muntasell et al., 2007; Nolte‐’t Hoen et al., 2009; Raposo et al., 1996).
Membrane fusion
EVs interacting with proteins on the plasma membrane outer leaflet can undergo fusion of the EV bilayer membrane to the target cell plasma membrane. This allows for the direct release of EV cargo into the cytoplasm and bypasses the need for endosomal sorting and cargo escape. This membrane fusion involves the two outer membrane leaflets forming a hemifusion due to their proximity (Chernomordik et al., 1987; Chernomordik & Kozlov, 2008; Jahn et al., 2003). Expansion of this hemifusion region allows the opening of a fusion pore, permitting the two hydrophobic leaflets to mix and form one structure. Membrane fusion is exploited by many viruses to gain entry of viral cores into target cells. Membrane fusion requires the action of several protein families such as Ras‐associated binding (Rab) proteins, soluble N‐ethylmaleimide‐sensitive factor attachment protein receptors (SNARE), and Sec1/Mammalian homolog of Caenorhabditis elegans uncoordinated gene 18 (Munc‐18) related proteins (Jahn & Scheller, 2006; Han et al., 2017; Südhof & Rothman, 2009).
1.4.2. Endocytic mechanisms
While the direct engagement of EVs with target cell membrane proteins and receptors is a key component of EV‐mediated cell‐to‐cell communication, most EVs exert their effects after being endocytosed by target cells. The major types of endocytosis, as depicted in Figure 3, encompass clathrin‐ and dynamin‐dependent endocytosis, caveolin‐ and dynamin‐dependent endocytosis, lipid raft‐dependent endocytosis, macropinocytosis, and phagocytosis, as well as other minor pathways (Colombo et al., 2014; Gurung et al., 2021; Joshi et al., 2020). EV endocytosis is an energy‐dependent process that can occur as rapidly as 15 min after dosing (Christianson et al., 2013; Montecalvo et al., 2012; Tian et al., 2014; Xu et al., 2022) and requires a functional actin cytoskeletal network (Atay et al., 2011; Escrevente et al., 2011; Feng et al., 2010; Fitzner et al., 2011; Grimmer et al., 2002; Gurung et al., 2021; Hao et al., 2007; Joshi et al., 2020; Montecalvo et al., 2012; Morelli et al., 2004; Obregon et al., 2009; Ridley, 2006; Svensson et al., 2013; Swanson, 2008; Tian et al., 2014; Xu et al., 2022). Heparan sulphate proteoglycans (HSPG) of both the glypican and syndecan families were found on the surface of exosomes and were required for their efficient uptake by endocytosis (Christianson et al., 2013).
FIGURE 3.
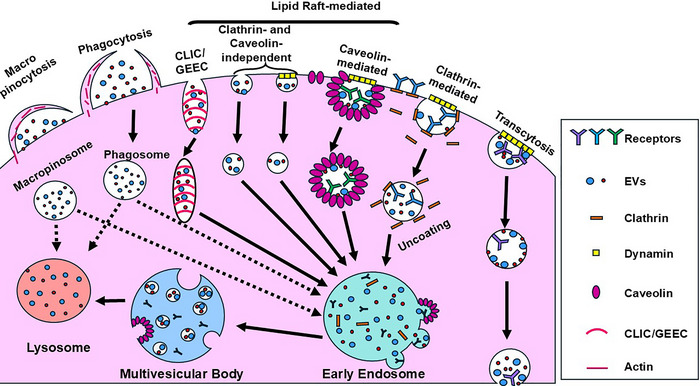
Overview of endocytosis pathways. Cells employ various mechanisms to internalize particles. Macropinocytosis involves the engulfment and import of large particles whereas phagocytosis refers to the import of smaller particles. Both processes require actin‐mediated extension of the plasma membrane to engulf the particles. Clathrin‐mediated endocytosis is a complex process requiring the formation of clathrin‐coated pits that trap extracellular particles and mediate their internalization and intracellular transport within the cell. Caveolin‐dependent endocytosis occurs via the association of particles with the caveolin‐1 protein, the major component of caveolae. Dynamin is required for both clathrin‐mediated and caveolin‐mediated endocytosis. There are other clathrin‐ and caveolin‐mediated endocytosis pathways, some requiring dynamin as well as dynamin‐independent pathways such as the CLIC/GEEC. During transcytosis, EVs endocytosed by clathrin‐ or caveolin‐mediated endocytosis, or by macropinocytosis are transported intracellularly from one luminal surface to another and released. It occurs in central nervous system endothelial cells and allows EVs to cross the blood‐brain barrier. Following uptake by the cell by any of these pathways, the internalized cargo is shuttled intracellularly (solid and dotted arrows), eventually fusing with the early endosome or the lysosome. (Dotted arrows indicate evidence of alternative destinations.). CLIC, clathrin‐independent carrier; GEEC, glycosylphosphatidylinositol‐anchored protein‐enriched endosomal compartments.
Clathrin‐dependent endocytosis
Clathrin‐dependent endocytosis of EVs involves the interaction of EV membrane ligand proteins with areas of the outer leaflet of the target cell plasma membrane characterized by clusters of the molecular scaffold protein clathrin (Kirchhausen, 2000, Kirchhausen et al., 2014). The clathrin molecules are arranged in a cage‐like lattice in 60–120 nm structures called clathrin‐coated pits. Dynamin‐2 mediates the inward membrane deformation to form a vesicular bud that subsequently pinches off to form clathrin‐coated vesicles. Clathrin then uncoats from the vesicles and is recycled to the plasma membrane and the vesicles fuse to the endosome for sorting back to the Golgi apparatus or the plasma membrane. Vesicles and proteins to be degraded are sorted by members of the endosomal sorting complex required for transport (ESCRT) protein family and Rab proteins into intraluminal vesicles that bud off from the endosomal membrane which transforms the late endosome into the MVB (Gurung et al., 2021; Joshi et al., 2020; Kirchhausen, 2000; Kirchhausen et al., 2014; Ridley, 2006; Xu et al., 2022). Non‐degraded EVs can fuse to the endosomal membrane, releasing their cargo into the cytoplasm.
Caveolin‐dependent endocytosis
The best characterized clathrin‐independent endocytosis pathway that forms 50–80 nm invaginations on cellular plasma membranes is the caveolin‐dependent endocytosis pathway (D'Alessio, 2023; Dalton et al., 2023; Kiss & Botos, 2009; Simón et al., 2020). Caveolae are enriched in sphingolipids and cholesterol that are associated with caveolin‐1, an integral membrane protein (D'Alessio, 2023, Pelkmans et al., 2002). The molecular cascade that triggers caveolae formation begins with the assembly of a hetero‐oligomeric complex composed of caveolin‐1 and caveolin‐2 in the endoplasmic reticulum. This complex moves to the Golgi complex where it binds to cholesterol followed by shuttling from the Golgi to the plasma membrane (D'Alessio, 2023; Dalton et al., 2023; Kiss & Botos, 2009; Simón et al., 2020). After the accumulation and fusion of multiple caveolae to the plasma membrane, other proteins such as cavins, dynamin, the Eps15 homology domain‐containing 2 (EHD2) ATPase, and protein kinase C and casein kinase II interacting protein 2 (PACSIN2) are recruited to stabilize the caveolae structure (D'Alessio, 2023; Dalton et al., 2023; Kiss & Botos, 2009; Simón et al., 2020). Interactions between EV membrane ligands and plasma membrane receptors mediate the association between EVs and the caveolae. Tyrosine phosphorylation of caveolins triggers caveolin internalization while dynamin mediates membrane deformation and pinching off caveolin‐lined vesicles that travel along the actin cytoskeleton to fuse with the endosomal compartment (D'Alessio, 2023, Pelkmans et al., 2002).
Lipid raft‐dependent endocytosis
Lipid raft‐dependent endocytosis encompasses an array of clathrin‐independent, cholesterol‐sensitive endocytosis pathways. As with caveolin‐dependent endocytosis, lipid raft domains are membrane protein microdomains enriched in sphingolipids and cholesterols as well as in proteins bound to glycosylphosphatidylinositol anchor protons (GPI‐APs) that are associated with the outer leaflet of the plasma membrane and in transmembrane proteins (Dermine et al., 2001; Lajoie & Nabi, 2007; Li et al., 2016; Sezgin et al., 2017). The GPI‐APs and transmembrane proteins can interact with EV‐associated membrane proteins. Lipid raft‐dependent endocytosis can be further subdivided based on whether dynamin‐mediated pinching to form the mature vesicles is required and are classified by the specific cellular membrane proteins such as adenosine diphosphate‐ribosylation factor 6 (ARF6), flotillin, Ras homolog A (RhoA), or GTPase regulator associated with focal adhesion kinase‐1 (GRAF1) that are present (Dermine et al., 2001; Lajoie & Nabi, 2007; Li et al., 2016; Sezgin et al., 2017).
Macropinocytosis
Macropinocytosis is initiated by the actin‐ and microtubule‐dependent formation of membrane ruffles that fold over and fuse, forming large (> 250 nm) intracellular vacuoles called macropinosomes (Ahram et al., 2000; Dermine et al., 2001; Grimmer et al., 2002; Ridley, 2006; Stephens et al. 2002; Swanson, 2008). Phosphatidylinositol‐3 kinase can stimulate membrane ruffle formation (Hoeller et al., 2013). This process is independent of membrane receptor engagement and results in the specific engulfment of extracellular material. Macropinocytosis is mediated by small non‐Rho GTPases such as Cell division cycle 42 (Cdc42), Ras‐related C3 botulinum toxin substrate 1 (Rac1), and Rac2 that promote Actin‐related protein 2/3 (Arp2/3)‐dependent branched actin polymerization (Ahram et al., 2000; Dermine et al., 2001). The Rab5 and Rab34 GTPases are required for the fusion of macropinosomes to the early endosome whereas Rab7 mediates fusion to the late endosomes (Harrison et al., 2003; Kasmapour et al., 2012; Kasmapour et al., 2013).
Phagocytosis
Phagocytosis results in the engulfment and ingestion of extracellular material ranging from exosomes to large particles such as apoptotic bodies. The cascade of phagocytic events is initiated by membrane receptor engagement that triggers the actin‐dependent formation of membrane pseudopods that engulf the particle and form phagosomes (Ridley, 2006; Stephens et al., 2002; Swanson, 2008). Phosphatidylinositol‐3 kinase stimulates membrane ruffle formation, which occurs independent of membrane receptor engagement and results in the engulfment of extracellular material (Hoeller et al., 2013). The phagosomes then fuse with the early endosomes, then shuttle to late endosomes and eventually lysosomes. Phagocytosis, as the name implies, commonly occurs in dedicated phagocytic cells such as dendritic cells, neutrophils, monocytes, and macrophages; however, the process has been described in numerous non‐phagocytic cells (Ridley, 2006; Stephens et al., 2002; Swanson, 2008).
Transcytosis
A specialized, tightly regulated form of transport by which biomolecules and larger complexes, including EVs, can cross the blood–brain barrier has been termed transcytosis (Bora et al., 2022; Erickson et al., 2023; Morad et al., 2019; Pandit et al., 2020). In this process, macromolecules and/or EVs are endocytosed by central nervous system endothelial cells, shuttled through the cytoplasm, and released into the brain. Transcytosis can be mediated by clathrin‐, caveolae‐, and lipid raft‐dependent endocytosis.
1.5. EV uptake in normal and cancer cells
For EVs to mediate intercellular communication in cancer and other diseases, and to serve as therapeutic cargo delivery vehicles, they must deliver their cargo to the cytoplasm of the target cells. Determining the cell type‐specific EV uptake pathways involved in this process is a major focus of investigation. Identifying the specific endocytic pathway responsible for EV uptake in distinct cell types is usually accomplished by treating cells with chemical inhibitors of each pathway or by siRNA directed against specific genes that mediate the pathways and comparing changes in uptake amongst treated and control cells. Complete inhibition of EV uptake following treatment with one of these inhibitors is rare and has been interpreted as an indication that several uptake pathways are operating simultaneously. The potential for an inhibitor to interfere with more than one pathway must also be considered when attempting to attribute EV uptake to a specific pathway.
Research into EV uptake by target cells is motivated by the desire to better understand the factors that can be modified or engineered to maximize the uptake of therapeutic vesicles by the target cell while minimizing off‐target delivery to bystander cells. Table 1 summarizes the pathways employed for EV uptake by twenty‐one normal and cancer cell types in seven cell classes from twenty‐six studies (Cheung et al., 2016; Cooper et al., 2018; Eguchi et al., 2019; Escrevente et al., 2011; Feng et al., 2010; Fitzner et al., 2011; Horibe et al., 2018; Li,Wang et al., 2020; Li, Pinilla‐Macua et al., 2020; Li et al., 2021; Montecalvo et al., 2012; Morishita et al., 2021; Nanbo et al., 2013; Sagar et al., 2016; Skelton et al., 2023; Svensson et al., 2013; Temchura et al., 2008; Tian et al., 2014; Tu et al., 2021; Verdera et al., 2017; Wang et al., 2020; Wei et al., 2017; Yoon et al., 2020; Zhang et al., 2022; Zheng et al., 2019). A direct comparison of pathway utilization in malignant and normal cells derived from the same tissue was not possible from the existing data, but some general trends emerged.
TABLE 1.
EV endocytic pathways by cell type.
| Cell class | Cell type | Pathway | Reference | |||||
|---|---|---|---|---|---|---|---|---|
| CA | CL | LR | MF | MP | PC | |||
| Cancer | Multiple myeloma | X | X | X | Tian et al., 2014, Tu et al., 2021, Zheng et al., 2019 | |||
| Neuroblastoma | X | X | X | Li et al., 2020 | ||||
| Ovarian | X | X | X | Escrevente et al., 2011, Verdera et al., 2017 | ||||
| Epithelial | Hepatocytes | X | X | X | Li et al., 2021 | |||
| Cancer | Colon | X | X | Horibe et al., 2018 | ||||
| Mesenchymal | Dermal fibroblasts | X | X | Cooper et al., 2018 | ||||
| Neural | Neural stem cells | X | X | Zhang et al., 2022 | ||||
| Cancer | Glioblastoma | X | X | Tian et al., 2014, Svensson et al., 2013 | ||||
| Non‐small Cell lung | X | X | Wei et al., 2017 | |||||
| Endothelial | Uterine MVEC | X | X | Li et al., 2020 | ||||
| Mesenchymal | Adipocytes | X | X | Sagar et al., 2016 | ||||
|
Placental Fibroblasts |
X | X | Li et al., 2020 | |||||
| Cancer | Osteosarcoma | X | Skelton et al., 2023 | |||||
| Mesenchymal |
Vascular smooth Muscle |
X | Wang et al., 2020 | |||||
| Epithelial | Gastric | X | X | Yoon et al., 2020, Nanbo et al., 2013 | ||||
| Immune |
Mouse Macrophage‐like |
X | X | Morishita et al., 2021 | ||||
| Immune | Dendritic | X | X | X | Montecalvo et al., 2012, Temchura et al., 2008, Morishita et al., 2021 | |||
| Immune | Psoriatic T cell | X | Cheung et al., 2016 | |||||
| Mesenchymal | Cardiomyocytes | X | Eguchi et al., 2019 | |||||
| Neural | Microglia | X | Fitzner et al., 2011 | |||||
| Immune | Phagocytes | X | Feng et al., 2010 |
Abbreviations: CA, caveolin‐dependent; CL, clathrin‐dependent; EV, extracellular vesicles; LR, Lipid Raft‐dependent (caveolin‐ and clathrin independent); MF, membrane fusion; MP, macropinocytosis; PC, phagocytosis. MVEC, microvascular endothelial cells.
The most common endocytosis pathways utilized across all cell types were caveolin‐dependent, clathrin‐dependent, and macropinocytosis. The most reported pathway was caveolin‐dependent endocytosis with 14 of the 21 cell types utilizing this pathway. Twelve cell types employed clathrin‐dependent endocytosis, 11 cell types used micropinocytosis, 2 cell types used lipid‐raft dependent endocytosis, and dendritic cells and phagocytic cells utilized membrane fusion and phagocytosis, respectively.
One of the most striking observations from pooling existing reports is that 15 of the 21 cell types employed multiple endocytosis pathways for EV uptake, with 5 cell types utilizing 3 pathways and 10 cell types using two pathways. Amongst the cells utilizing 3 pathways, 4 cell types (multiple myeloma cells, neuroblastoma cells, ovarian cancer cells, and hepatocytes) used caveolin‐dependent endocytosis, clathrin‐dependent endocytosis, and macropinocytosis, while dendritic cells employed clathrin‐dependent endocytosis, membrane fusion, and macropinocytosis. Six cell types used a single endocytosis pathway. Both osteosarcoma cells and vascular smooth muscle cells employed only caveolin‐dependent endocytosis, while psoriatic T‐cells and cardiomyocytes used only clathrin‐dependent endocytosis. In contrast, microglia utilized macropinocytosis, and phagocytic cells employed phagocytosis exclusively.
Intriguingly, all seven cancer cell lines and four of the five mesenchymal cell lines utilized caveolin‐dependent endocytosis. Caveolin‐1 expression is upregulated in many cancers in vivo and cancer cell lines including lung, breast, liver, kidney, and colon (Burgermeister et al., 2008). Caveolin‐1 can act either as a tumour suppressor or an oncogene depending on the tumour type or stage (Gupta et al., 2014). Caveolin‐1 facilitates tumorigenesis by facilitating anchorage‐independent growth metastasis, and drug resistance while inhibiting apoptosis (Goetz et al., 2008; Núñez‐Wehinger et al., 2014). It is an important regulator of transforming growth factor‐β (TGF‐β). TGF‐β is critical in the induction of epithelial‐to‐mesenchymal transition (EMT) which is often activated during cancer progression (Kannan et al., 2014), and may be a major regulator of caveolin‐mediated EV uptake in these cells.
1.6. EV cargo release
The final step of EV‐mediated cell‐to‐cell communication is mediated by the modification of the target cell phenotype by the EV cargo. This can be accomplished either by direct engagement of target cell membrane receptors, described previously, or by releasing EV contents into the cytoplasm following EV uptake into the cell. Direct evidence of EV cargo release is minimal due to the challenges of efficiently labelling and following the fate of limited quantities of EV cargo but instead has been inferred to have occurred based on the modification of cell phenotypes following the introduction of miRNA or protein cargoes (Chen et al., 2018; Flemming et al., 2020; Ohno et al., 2013; Overmiller et al., 2017; Usman et al., 2018; Valadi et al., 2007; Yang et al., 2017; Zhang et al., 2014). Although direct fusion of EVs is one potential mechanism to deliver EV cargo into the cytoplasm, endocytosis is the dominant mechanism of EV uptake (Ahram et al., 2000; Atay et al., 2011; D'Alessio, 2023; Dalton et al., 2023; Dermine et al., 2001; Escrevente et al., 2011; Feng et al., 2010; Fitzner et al., 2011; Grimmer et al., 2002; Gurung et al., 2021; Hao et al., 2007; Hoeller et al., 2013; Joshi et al., 2020; Kirchhausen et al., 2014; Kirchhausen, 2000; Kiss & Botos, 2009; Lajoie & Nabi, 2007; Li et al., 2016; Morelli et al., 2004; Obregon et al., 2009; Pelkmans et al., 2002; Ridley, 2006; Sezgin et al., 2017; Simón et al., 2020; Stephens et al., 2002; Svensson et al., 2013; Swanson, 2008; Stephens et al., 2002; Xu et al., 2022). Therefore, some mechanism that permits endosomal escape is needed to allow cargo access to host cytoplasmic components, such as the RNA‐induced silencing complex. Several mechanisms for endosomal escape have been proposed including endosomal lysis, endosomal membrane permeabilization, and fusion of the EV and endosomal membranes (Heusermann et al., 2016; Stewart et al., 2016). Recently, methods employing the use of dual fluorescent and luminescent reporter constructs have permitted closer and more quantitative analysis of EV uptake and cargo unloading (Hung & Leonard, 2016; Lai et al., 2015).
A recent elegant study used innovative molecular tools and a combination of light and electron microscopy (EM) to trace EV cargo unloading following EV uptake (Joshi et al., 2020). EVs were loaded with GFP fused to the C‐terminus of the CD63 tetraspanin protein expressed in EV‐producing cells. These EVs displayed the GFP moiety on the cytoplasmic EV surface. An HEK293 cell line was engineered to express an anti‐GFP fluobody that recognizes and binds to the cargo. This allowed tracking the location and fate of the cargo following EV uptake and cargo release. Following endocytic EV uptake, a cytoplasmic punctate staining pattern was detected by light microscopy. Electron microscopy localized the EVs to the endosomal compartment. Transducing these cells with a construct encoding an Azumi green‐tagged galectin‐3 protein that can bind to β‐galactosides in the luminal endosomal membrane revealed the absence of endosomal permeabilization. Generating EVs containing an N‐terminal GFP‐CD63 cargo protein where the GFP moiety faces the EV lumen allowed for detecting released EV cargo since the GFP would be inaccessible to the anti‐GFP fluobody in intact EVs. Using this approach, EV cargo release was localized to MVB/late endosomes and lysosomes (Joshi et al., 2020).
Since the ability of viruses to fuse with endosomes is regulated by cholesterol and endosomal pH, target cells in another study were treated either with Bafilomycin A, an inhibitor of endosomal acidification (Pinilla‐Macua et al., 1998), or with U18666A, a cholesterol transport inhibitor (Elgner et al., 2016). Treatment of cells with these inhibitors blocked cargo release, indicating that the fusion of the EV membrane to the endosomal membrane was the mechanism used for cargo release in these cells.
1.7. EVs as therapeutics
1.7.1. Nanoparticles and targeted anticancer therapies
Anticancer drugs are, by their very nature, extremely toxic mostly due to off‐target effects (Basak et al., 2021; Kirk, 2012). The same can be said of many anticancer genetic therapies (Kohn et al., 2023; Mullard, 2021). It is therefore vital to design methods capable of delivering the highest possible effective dose of these agents thereby minimizing systemic drug‐ or treatment‐associated toxicity. Such methods are designed to achieve four primary goals (Mills & Needham, 1999). First, the desired effective dose of the agent must be retained and delivered by the vehicle. Second, the delivery system must evade the immune response to maximize its presence in the circulation and tumour site. Third, once arriving at the tumour, the system must specifically interact with the target cells to minimize off‐target effects and toxicity. Finally, the drug or biological agent must be released at the tumour site while retaining its antitumor function.
Traditional drug delivery pathways such as oral, inhalation, parenteral, and intravascular administration rely on systemic circulatory dispersal of ‘naked’ drugs or biological agents such as nucleic acids or proteins. Since the 1990s, synthetic nanoparticles (NPs) have been touted as ideal drug delivery systems that can be engineered to solve the barriers associated with effective targeted delivery of therapeutic agents (Hoffman, 2008). These barriers include the insolubility of many drugs in the aqueous circulatory system environment (ten Tije et al., 2003), the vulnerability of drugs or molecular agents to chemical or enzymatic degradation (Erikson et al., 2008), the rapid clearance of therapeutic agents from the circulation by macrophages and other immune system components (Krishna & Nadler, 2016), non‐specific uptake by cells, and toxicity (Lin et al., 2019; Li et al., 2022).
NPs encompass a diverse collection of particles ranging in size from ∼20 nm to 1 µM composed of biodegradable polymers, gold nanoparticles, silica nanoparticles, dendrimers, iron‐oxide nanoparticles, liposomes, or carbon nanotubes (Briolay et al., 2021; Eftekhari et al., 2023; Merisko‐Liversidge & Liversidge, 2008; Sousa de Almeida et al., 2021). Synthetic NPs can facilitate high‐capacity loading and delivery of hydrophobic water‐insoluble drugs by carrying nanosuspensions of pure drug particles in a stabilized solution of surfactants, increasing drug bioavailability while reducing solvent‐associated toxicity (Zakir et al., 2011; Zhao et al., 2018; Zu et al., 2014). In addition, encapsulating drugs and biological agents in NPs protects them from degradation. Conversely, the NPs themselves can be modified to degrade, enabling controlled drug or biological agent release (Kamaly et al., 2016; Perrigue et al., 2021; Vuong et al., 2022). The targeting of synthetic NPs to specific cell populations can be accomplished by conjugating or coating them with ligands or antibodies that specifically bind to receptors or other proteins overexpressed on the cell subpopulation to be targeted (Gao et al., 2019; Kutova et al., 2019; Yoo et al., 2019).
Following their intravenous (IV) administration, synthetic NPs encounter another obstacle due to their rapid clearance from the circulatory system by phagocytic cells, primarily macrophages, leukocytes, dendritic cells, and platelets. Interactions with these immune cells are facilitated by the acquisition of a protein corona composed of plasma proteins adsorbed to the NP surface (Bashiri et al., 2023; Kopac, 2021; Meng et al., 2022; Rampado et al., 2020). An inner ‘hard’ corona is bound directly to the NP surface whereas the outer ‘soft’ corona associates with the ‘hard’ corona through weak protein–protein interactions’ (Bashiri et al., 2023, Kopac, 2021, Meng et al., 2022; Rampado et al., 2020). NP size, shape, and molecular composition affect protein corona makeup as does the solution temperature, pH, plasma protein size, and protein concentration. The major components of the synthetic nanoparticle protein corona are comprised of anti‐thrombin III, complement C3, complement factor H, factor V, fibronectin, and immunoglobulin G (Zhang et al., 2020). Intriguingly, variations in the composition of the NP protein corona can serve as diagnostic and pharmacodynamic biomarkers (Kamaly et al., 2022; Meng et al., 2022).
IV administration of NPs loaded with antitumor drugs and biological agents disperses them throughout the entire body. Unlike normal tissues, the vasculature in a solid tumour is irregular and heterogeneous, with leaky vessels and poor lymphatic drainage (Konerding et al., 1999; Warren et al., 1978). This allows NPs that are too large for excretion by the kidney yet small enough to escape entrapment by the reticuloendothelial system to accumulate at the tumour site, a phenomenon known as the enhanced permeability and retention (EPR) effect (Kobayashi & Brechbiel, 2005; Maeda et al., 2016; Matsumura & Maeda, 1986). The magnitude of this effect can vary significantly; however, leakage is slow and often achieves only a 2‐fold increase in NP delivery, which is typically insufficient to allow accumulation of the agent to effective therapeutic levels (Prabhakar et al., 2013).
One strategy to subvert immune cell‐mediated clearance of NPs and extend their circulation lifetime is the grafting of poly(ethylene glycol), or PEGylation, to the NP surface (Hamilton et al., 2002; Harris & Chess, 2003). This can extend the circulatory persistence of NPs from minutes to hours. Treatment with PEGylated NPs, however, can result in the appearance of anti‐PEG IgG and IgM antibodies that activate the complement system followed by rapid synthetic NP clearance and trigger hypersensitivity reactions (Estapé Senti et al., 2022; Kozma et al., 2020). More recently, cloaking synthetic NPs with natural or engineered cell membranes has emerged as a promising alternative to PEGylation to aid in evading immune cell‐mediated NP clearance and promoting cell‐specific targeting (Alyami et al., 2020; Jiang et al., 2019; Jiang et al., 2020; Li et al., 2018; Nie et al., 2020; Xuan et al., 2018; Zhang et al., 2019).
Synthetic NPs can address, at least to some extent, the barriers to effective targeted therapies. However, despite their promise to increase anticancer drug delivery and the efficacy of drugs and biological agents, significant concerns persist. These include their potential to provoke a protracted immune response, the costs of their manufacture, lack of stability, and their compatibility with human tissue. The shedding of vesicles containing the transferrin receptor and other membrane‐associated proteins during the maturation of cultured reticulocytes into erythrocytes is the first reported description of what came to be called exosomes (Johnstone et al., 1987; Pan et al., 1985). These reports led to the hypothesis that these shedded vesicles or exosomes served to rid the cells of unnecessary proteins (Johnstone, 1992). It was the observation that these vesicles could be internalized by other cells that led to the realization that EVs function as more than a passive cellular waste disposal system but carry biomolecular cargo between cells. This potential for EVs to serve as therapeutic vehicles has been the focus of subsequent intensive research.
Since the intercellular communication abilities of EVs have evolved in the very organisms in which they would be employed, their intrinsic properties offer multiple advantages over their synthetic NP counterparts as drug delivery vehicles. Unlike synthetic NPs, EVs are cell‐derived, biocompatible, less toxic, and are assembled and loaded by endogenous cellular processes (Jiang et al., 2019; Jiang et al., 2020; Mendt et al., 2018; Schindler et al., 2019; Sun et al., 2016; Walker et al., 2019). Since they are naturally recycled as part of the endosomal system, EVs are biodegradable (de Gassart et al., 2004; O'Brien et al., 2022) and, because EVs are isolated from benign or autologous sources that do not express exogenous proteins, they are less likely to be cleared by phagocytic cells or to trigger an immune response compared to NPs (Mendt et al., 2018; Sun et al., 2016; Zhu et al., 2017). Importantly, unlike viral‐based gene therapy approaches, EVs are not mutagenic and cannot replicate (Elsharkasy et al., 2020; Liu et al., 2022). Finally, EVs possess the ability to cross biological barriers such as the endosomal and plasma membranes as well as the blood–brain barrier, which renders them a valuable resource for the treatment of neurologic disorders including cancer (Elliott & He, 2021; Elsharkasy et al., 2020; Kawikova & Askenase, 2015; Kawikova & Askenase, 2015; Liu et al., 2022; Moyano et al., 2016; Zheng et al., 2019). Exosomes may also be able to bypass P‐glycoprotein efflux‐mediated multi‐drug resistance (MDR) in cancer cells (Kim et al., 2016).
EVs, like synthetic NPs, also accumulate a protein corona when exposed to human serum or plasma and the proteins present may be influenced by an individual's health status (Dietz et al., 2023; Palviainen et al., 2020; Tóth et al., 2021). Exposing monocyte‐derived dendritic cells to EVs isolated from cultured THP1 cells that were exposed plasma isolated from healthy donors or from donors with rheumatoid arthritis demonstrated that both healthy and RA‐derived protein coronas efficiently induced TNF‐α production and dendritic cell maturation, suggesting the possibility of a tonic proinflammatory effect of the protein corona (Tóth et al., 2021). The EVs isolated from patients with systemic lupus erythematosus display elevated levels of complement C3d‐opsonized immune complexes and decreased levels of C3b and C3ib (Winberg et al., 2017). While this reduced EV phagocytosis, it also prolonged EV transit time in the circulation leading to prolonged inflammatory conditions.
The highly variable composition of the protein corona surrounding synthetic NPs and cell‐derived EVs is influenced by various factors including the particle size, composition, and surface features, and also reflects the individual demographic characteristics and health status of the source of the plasma or other biological fluids. A subsequent study demonstrated that EVs can accumulate this corona in vitro and that its components are derived from proteins present in the culture as well as the cytoplasm of the producer cells (Liam‐Or et al., 2024). Albumin is the major protein found in the EV protein corona (Dietz et al., 2023; Liam‐Or et al., 2024), along with a core of 17 other proteins including integrins, lipoproteins ApoA1 and ApoB, and complement proteins (Singh et al., 2020). Some of the functions attributable to the EV protein corona are increased EV uptake (Dietz et al., 2023; Liam‐Or et al., 2024), alterations in immune responses, angiogenesis, regeneration (Gomes et al., 2022; Wolf et al., 2022), and EV in vivo distribution (Liam‐Or et al., 2024).
It is important to acknowledge that despite the advantages of EVs as targeted therapeutic vehicles, several drawbacks exist. Rational engineering of EVs is a major research focus to address three of the major weaknesses of EV‐mediated therapies: targeting EVs to specific cell subpopulations, augmenting target cell uptake of therapeutic EVs, and maximizing EV cargo loading/release (Esmaeili et al., 2022; Rhim et al., 2023). Another challenge to the successful deployment of EV‐based therapies lies in their heterogeneity. Multiple EV subpopulations that differ in size, cell of origin, biogenesis mechanism, and composition have been described and this heterogeneity may result in differences in cell‐specific targeting, mode of uptake by target cells, and the phenotypic changes they mediate (Colombo et al., 2013; Kim et al., 2019; Sharma et al., 2020). Methods to isolate desired EV subpopulations consistently and efficiently need to be developed. Therapeutic EVs must be free from contaminating viruses, and bacteria. They must be isolated from serum‐free culture media to prevent contamination by serum EVs before they can be deemed suitable as human disease therapies (Burnouf et al., 2019).
1.8. Tumour microenvironment influence on EV properties and function
Several crucial factors need to be carefully considered before undertaking EV engineering for use as targeted therapeutics. To successfully study the effects of EV‐mediated signalling in cancer and to develop appropriately engineered therapeutic EVs, efforts should be focused on developing clinically relevant in vitro models that replicate the pathophysiology of the relevant TME as closely as possible. The TME is comprised of cancer cells with a heterogeneous population of tissue‐specific stromal and immune cells that communicate with each other and with the extracellular matrix (ECM) in which they are embedded (Li et al., 2024; Nishida‐Aoki & Ochiya, 2024; Wang et al., 2017). EV‐mediated signalling has been implicated in multiple aspects of tumour initiation and progression including tumour cell proliferation, immunosuppression, angiogenesis, invasion, metastasis, and the development of multidrug resistance (Kanada et al., 2016; Kalluri, 2016; Minciacchi et al., 2015; Tkach & Théry, 2016).
Although animal models, usually rodent‐based, can address some shortcomings of 2D cell culture systems in replicating the TME in vitro, they entail significant investments of time and cost with no guarantee that the results will be replicated in human trials (Doncheva et al., 2021; Shanks et al., 2009). The development of patient‐derived xenograft models allows for the retention of patient genomic features of different cancer stages and subtypes and permits the assessment of responses to various treatments. These models may not accurately mimic the native TME since the xenograft cells are often implanted at non‐physiologic sites and the issues of cost and time compared to in vitro systems remain (Jung, 2014; Lee et al., 2019).
To address deficiencies of 2D cell culture systems and to provide alternatives to animal models, much effort is now being focused on the development of 3D cell culture models including organoids (Saito et al., 2019; Yin et al., 2016), spheroids (Nunes et al., 2019; Zoetemelk et al., 2019), microfluidics (Jiang et al., 2024; Kühlbach et al., 2018), hydrogels (Castellote‐Borrell et al., 2023; Lupu et al., 2023), 3D scaffolds (Górnicki et al., 2024; Kuriakose et al., 2019), and 3D bioprinting (Langer et al., 2019; Zhang et al., 2024) that more accurately mimic the TME. These approaches permit the co‐culture of multiple cell types and more closely replicate the cell‐cell and cell‐matrix interactions that occur in the native TME. As a result, they produce EVs with different compositions and cargo than those that were isolated from 2D cultures (Brown & Wilson, 2004; Wilson & Hay, 2011; Zhang et al., 2024). Importantly, the 3D conformations achieved by these techniques can duplicate the hypoxic and low pH found in the native TME (Bister et al., 2020; Han et al., 2023, Yan et al.; 2023). Hypoxic and low pH culture conditions stimulate EV biogenesis and release, and target cell uptake of EVs (Liu et al., 2020; Ural et al., 2021; Zhang et al., 2017; Zhu et al., 2018). The composition, cargo, and downstream functional effects of EVs isolated from cells cultured in hypoxia or low pH conditions are altered in a cell type‐specific way compared with EVs produced by cells cultured at neutral pH or normoxic conditions (Parolini et al., 2009).
The choice of the cell source from which the EVs will be isolated profoundly affects their innate properties. EV homing capacity to target cells, EV membrane, and cargo composition, and their therapeutic effects in target cells are all affected by the phenotypic characteristics of the EV‐producing cells (Chiou et al., 2018; Jiang et al., 2022; Lázaro‐Ibáñez et al., 2017). It is also important to consider the effect of artificial 2D monolayer cell culture systems on EV biogenesis, secretion, and composition. Cells grown in monolayer display altered morphology, gene expression, mRNA splicing, differential cell motility, cytokine, growth factor signalling, integrin, and chemokine expression compared with cells grown in 3D culture systems (Baker & Chen, 2012; Fitzgerald et al., 2018; Hoarau‐Véchot et al., 2018; Hurwitz et al., 2016; Sung et al., 2015; Sung et al., 2020; Yamada & Cukierman, 2007). Many of these altered cellular processes affect vesicle biogenesis and cargo sorting (Tauro et al., 2013; Zanetti‐Domingues et al., 2020), generating a population of EVs that do not accurately represent the composition and cargo of the EVs produced by these cells in vivo nor will they produce the same effects in target cells.
1.9. Engineering EVs to serve as therapeutics
1.9.1. Maximizing EV targeting
As discussed previously, despite the ability to efficiently synthesize lipid nanoparticles as drug delivery vehicles, their utility is limited by several considerations, such as their rapid accumulation in the liver following systemic administration (Di et al., 2022); instability due to several factors such as pH, temperature, and lyophilization (Young et al., 2022); and the difficulty of achieving high drug loading (Yoon et al., 2013). The area of most intense research has focused on strategies to improve cell‐specific or tissue‐specific ligand‐mediated targeting of synthetic nanoparticles using insertion of targeting ligands into preformed lipid particles and direct conjugation of ligands to lipids (Lee et al., 2023). Alternatively, methodologies are being evaluated to actively target without ligands by modification of nanoparticle components such as cationic lipids, cholesterol, phospholipids, or polyethylene glycol (Lee et al., 2023).
The major factors limiting the potential of EVs as therapeutic agents involve their ability to effectively target specific cell populations, the efficiency of EV uptake by the targeted cells, and the ability of the unloaded EV cargo to avoid lysosomal degradation (Gurung et al., 2021; Joshi et al., 2020; Xu et al., 2022). EV targeting and uptake are not completely independent since the first step in the uptake process involves an interaction between the EV and plasma membranes. Although numerous methods to improve EV targeting have been described, they all employ a similar underlying strategy. This entails engineering the display of a molecule on the outer surface of the EV membrane that interacts with a molecule that occurs at a higher density on the target cell plasma membrane than it does on non‐target cells. It is crucial to carefully select the molecules involved in this interaction to maximize the specificity of the interaction (Briolay et al., 2021; Eftekhari et al., 2023; Sousa de Almeida et al., 2021).
Many proteins, receptors, lipids, or other molecules have been reported to be differentially expressed on the plasma membranes of a specific tumour cell subpopulation. Although this heterogeneity has been known for decades, with the advent of novel techniques such as spatial transcriptomics, mass spectrometry, and artificial intelligence (AI)‐mediated deep learning for novel biomarker discovery correlated with histologic features, it is evident that the selection of optimal molecules for targeted therapeutics is not a simple matter of identifying overexpressed antigens (Bae, 2009; Capone et al., 1984; Li & Hall, 2010; Wen et al., 1995; Williams et al., 2024). At a minimum, the optimal target molecules selected for EV engineering methods should be present at high density on the target cell membranes but at very low density on non‐target cell membranes. This will maximize specific EV targeting while minimizing off‐target effects (Bae, 2009; Capone et al., 1984; Li & Hall, 2010; Wen et al., 1995; Williams et al., 2024).
Some commonly used methods that can achieve EV cell‐specific targeting are depicted in Figure 4. EV engineering can be accomplished either by manipulating the EV biogenesis and assembly process in the producing cells (Figure 4a‐c) or by direct modification of the EVs after their isolation (Figure 4d‐4f). The most reported methodology involves the modification of EV membrane proteins to display a moiety that can bind to a molecule in the target cell plasma membrane (Figure 4a). A diverse array of molecules can be used as targeting moieties including proteins, peptides, antibodies, single‐chain variable fragments (scFv), and aptamers.
FIGURE 4.
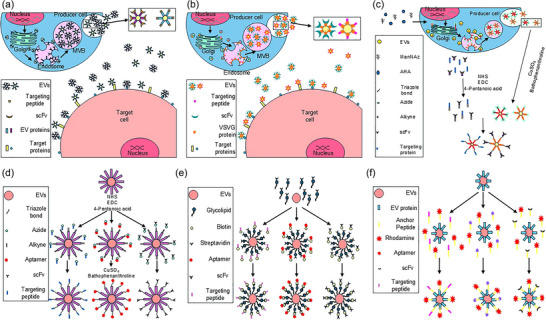
Exosome engineering strategies. Two generalized approaches are employed to engineer exosomes to improve cell‐specific targeting. The first approach (a–c) relies on modifying the exosome biogenesis and assembly process in producing cells. The second approach (d–f) involves direct modification of isolated exosomes. Producing cell modification: (a) Fusing EV membrane proteins with cell type‐specific binding moieties; (b) Insertion of chimeric VSVG/cell type‐specific targeting moieties into exosome membrane; (c) Metabolic reprogramming of producing cells. Culturing producer cells in growth media supplemented with modified amino acid analogues (e.g., ADA) and/or azide‐containing saccharides introduces active sites for bioconjugation to target cell‐specific moieties via azide‐involved biorthogonal reactions, that is, click chemistry. Direct exosome modification: (d) Introduction of alkyl groups to exosome membrane proteins via modification by NHS, EDC, and 4‐pentanoic acid allows for bioconjugation to azide‐containing cell type‐specific cargo moieties mediated by CuSO4 and bathophenanthroline; (e) Binding biotinylated cell type‐specific targeting moieties to streptavidin‐conjugated glycolipids that insert into the lipid bilayer exosome envelope; (f) Insertion of targeting moiety/anchoring peptides that recognize and bind to exosomal membrane proteins. AHA, L‐azidohomoalanine; CuSO4, cupric sulphate; EDC,5‐ethynl‐2‐deoxycytidine; EV, extracellular vesicles; ManNAz, tetraacetylated N‐acetylazido‐D mannosamine; MVB, multivesicular body; NHS, N‐hydroxysuccinimide; scFv, single chain variable fragments.
Numerous tumour‐targeting peptides have been described that bind to proteins overexpressed on specific cancer cells. The K237 peptide specifically binds the VEGFR2 receptor on breast tumours (Yu et al., 2010), whereas the IL4RPep1 peptide targets the interleukin 4 receptor (IL4R) on lung, breast, and colon tumours (Chi et al., 2015; Guruprasath et al., 2017; Jeon et al., 2013; Kim et al., 2012; Namgung et al., 2014). The LyP‐1 peptide targets the mitochondrial p32 protein in melanoma tumours (Karmali et al., 2009). Glioblastoma and melanoma tumours overexpressing integrin αvβ3 can be targeted using the iRD peptide (Chen et al., 2017; Su et al., 2013; Yu et al., 2013), whereas the RGD4C peptide that binds to the same integrin has been used to target melanoma, glioblastoma, colon, and ovarian tumours (Chen et al., 2022; Echigo et al., 2022; Kato et al., 2022). Finally, CD206 which is overexpressed on breast tumour cells can be targeted by the mUNO peptide (Figueir, 2021).
Exosome membranes are characterized by a high density of proteins found in the MVB from which the vesicles were generated. These include proteins such as the endosomal sorting complex required for transport (ESCRT) protein family involved in membrane transport, MVB biogenesis proteins such as the tumour susceptibility gene 101 (TSG101) and ALG‐2‐interacting protein X (ALIX), and members of the tetraspanin protein family (Escola et al., 1998; Kalra et al., 2016). Some tetraspanins (CD9, CD63, CD81, and CD151) are present in most EV subpopulations whereas others (CD37, CD53, and CD82) are more restricted in their distribution (Maecker et al., 1997; Wright et al., 2004). The levels of EV‐associated CD9 are decreased in many solid tumours and the EVs produced by these tumours, whereas CD151 levels are increased (Brzozowski et al., 2018).
The first reported example of a targeting peptide approach genetically modified EVs to express a fusion of the lysosomal‐associated membrane protein 2B (LAMP2B) with the rabies virus glycoprotein peptide that enabled targeting of the EVs to oligodendrocytes, microglia, and neurons (Alvarez‐Erviti et al., 2011). Fusion of LAMP2B to a targeting moiety is one of the most frequently reported strategies, with fusions to CD63 and EV‐associated tetraspanins also commonly used (Bellavia et al., 2017; Kanuma et al., 2017; Tian et al., 2014). The inclusion of glycosylation motifs to the outward‐facing portion of the protein can enhance the display of these fusion proteins by protecting them from acid‐mediated endosomal proteolysis (Hung & Leonard, 2015). Even more complex fusion transmembrane proteins designed to display a targeting moiety on the outer EV membrane fused to specific cargo proteins or nucleic acids on the inner EV membrane have been reported (Liu et al., 2024; Zheng et al., 2023). A recent study that assessed the surface and luminal display capabilities of 115 EV‐associated transmembrane proteins and 129 EV‐associated non‐transmembrane proteins by a bioluminescence assay found 24 conserved proteins present in EVs isolated from five different cell types (Zheng et al., 2023).
Another common modification strategy transfects EV producer cells with a construct encoding the targeting moiety fused to a viral fusion protein such as the vesicular stomatitis virus glycoprotein (Figure 4b). VSV‐G, one of five structural proteins encoded in the single‐stranded RNA genome of the vesicular stomatitis virus (VSV), is a class III fusion protein found in the viral envelope whose function is to promote the fusion of the viral and cellular membranes (Backovic & Jardetzky, 2009; Sun et al., 2008). VSV‐G‐based lentiviral vectors have been developed as oncolytic therapies and vaccine vectors (Diaz et al., 2007; Ebert et al., 2003; Lichty, Power et al., 2004; Lichty, Stojdl et al., 2004; Obuchi et al., 2003; Roberts et al., 1999).
Employing the VSV‐G protein in EV engineering permits the incorporation of targeting moieties and protein or nucleic acid cargoes into the fusion protein, and facilitates EV uptake by direct fusion to the target cell plasma membrane (Meyer et al., 2017; Ovchinnikova et al., 2021). Cells engineered to express VSV‐G produce significantly higher levels of EV release (Rolls et al., 1994). The fusogenic activity of the VSV‐G protein is induced by low pH, which may render it particularly efficacious for targeting EVs to solid tumours (Yao et al., 2003).
A major limitation of VSV‐G approaches for EV engineering is that this viral glycoprotein triggers a robust primary immune response, producing strongly neutralizing antibodies and complement‐mediated inactivation (DePolo et al., 2000; Kelley et al., 1972). The effectiveness of these anti‐VSV‐G antibodies prevents repeated administrations of VSV‐G‐containing EVs. This limitation has been overcome for vaccine vectors by employing viral pseudotypes (Rose et al., 2000) and by incorporating sequences for complement regulatory proteins (Schauber‐Plewa et al., 2005); however, the limitation remains for VSV‐G‐based EV therapeutics.
Click chemistry, a class of simple, efficient, and high‐yield cycloaddition chemical reactions used to join two molecular entities was first described in 2001 (Evans, 2007; Kolb et al., 2001). This type of reaction has been developed for targeting and isolating specific molecules in complex biological environments to generate products that are physiologically stable and non‐toxic (Prescher & Bertozzi, 2005, Prescher et al., 2004; Sletten & Bertozzi, 2009). These biorthogonal reactions have since become a powerful tool for studying and modifying various molecules in living organisms, without generating toxic byproducts and with little reactivity to endogenous functional groups (Scinto et al., 2021). Click chemistry‐based engineering of EVs can be used for the modification of EV‐producing cells (Figure 4c) or directly on purified EVs (Figure 4d) to attach targeting moieties to EV membrane proteins (Smyth et al., 2014; Wang et al., 2015). The latter option will be essential for modifying patient EVs for treatments employing autologous EVs (Murphy et al., 2019).
It is also possible to employ modified glycolipids to attach surface‐displayed targeting moieties to EV membranes. A modular synthesis pathway termed ‘membrane cloaking’ has been described that modifies PEGylated 1,2‐Dimyristoyl‐sn‐glycero‐phosphoethanolamine (DMPE‐PEG), a water‐soluble derivative of phosphatidylethanolamine, by conjugating it to streptavidin (Figure 4e). Incubation of streptavidin‐DMPE‐PEG and biotinylated targeting peptides or antibodies results in the insertion of the modified lipid conjugated to the targeting moiety into the EV membrane (Antes et al., 2018). Another direct EV modification technique (Figure 4f) incubates EVs with targeting peptides, scFv, antibodies, or aptamers conjugated to membrane anchor peptides (Gao et al., 2018).
1.9.2. Maximizing EV uptake and cargo release
Although contact between the EV and plasma membranes is necessary for EV uptake, this interaction is not necessarily sufficient to ensure the uptake of these EVs into the targeted cells. The various endogenous endocytic pathways employed by which native EVs are taken up by cells have already been described. The result of EV uptake via these pathways usually results in the EVs entering the endosome. The capacity for native EVs and their cargo to affect the target cell phenotype depends on their ability to exit the endosomal/lysosomal degradation pathway. Although it is not known definitively what percentage of EVs and their cargo is degraded, it is believed that it is substantial. Thus, methods allowing EVs and their cargo to either bypass the endosomal/lysosomal pathway and escape the endosome with its function intact are being developed. Two such approaches are depicted in Figure 5.
FIGURE 5.
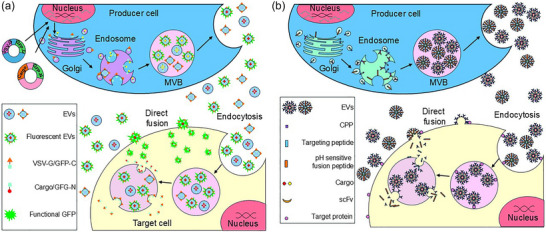
Strategies for increasing EV uptake and cargo release. (a) Simultaneously increasing EV uptake and active loading of desired cargo into engineered exosomes utilizing split GFP complementation. A plasmid encoding a fusogenic protein or peptide and a portion of GFP is cotransfected with a second plasmid encoding the remaining GFP sequence linked to the desired cargo protein or nucleic acid. (b) Improving EV uptake by target cells employing either fusogenic proteins, usually virally derived proteins such as VSV‐G, or short CPPs. The fusion of two lipid bilayer membranes is generally accomplished by forming a hemistable pore that is gradually expanded, allowing for the transfer of contents from the vesicle fused to the cells. CPPs are short hydrophobic peptides, often modified from viral fusogenic sequences joined by a linker sequence with a cell‐specific targeting sequence. CPP, cell‐penetrating peptides; EV, extracellular vesicles; VSV‐G, vesicular stomatitis virus protein G.
The first technique involves modifying target cells to express the VSV‐G viral fusion protein (Figure 5A). An elegant study describes a procedure to generate two‐component fluorescent ‘gectosomes’ carrying specific cargo proteins employing a GFP complementation strategy (Zhang et al., 2020). This method employs a soluble engineered two‐part self‐assembling green fluorescent protein (GFP) system (Cabantous et al., 2005). GFP1‐10 encodes the first ten β‐strands while GFP‐11 encodes the 16 amino acid (aa) eleventh β‐strand. When separated, the fragments exhibit minimal fluorescence; however, they become highly fluorescent following association and self‐assembly. Transfecting EV‐producing cells with an expression construct encoding the VSV‐G sequence fused with the GFP1‐10 sequence and a second construct encoding a specific target protein fused with the GFP11 sequence generates ‘gectosomes’ by budding from the plasma membrane. Those ectosomes containing only the VSV‐G/GFP1‐10 protein or the cargo/GFP11 will be non‐fluorescent whereas ectosomes containing the self‐assembled VSV‐G/GFP1‐11/cargo protein will be fluorescent.
‘Gectosome’ production allows the flow cytometry‐mediated purification of the fluorescent particles, theoretically reducing the concentration required to achieve a therapeutic effect and permitting tracking of EV uptake by the target cells. Although an elegant method, the previously described immune response to the viral VSV‐G protein limits the possibility for the repeated administration of VSV‐G‐based EVs and seriously limits their potential usefulness. It should be possible to modify the procedure to fuse only the VSV‐G fusion peptide to the GFP1‐10 sequence, thereby minimizing any immune response (Zhang et al., 2020).
As a result of the immunogenicity of viral fusogenic proteins, attempts have been made to identify human endogenous fusion proteins. One such protein, syncytin‐1, has been found to enhance EV entry into cells with higher efficiency than VSV‐G (Bui et al., 2023). Syncytin‐1 is expressed from the envelope sequence of an ancestral retroviral integration into the human genome (Blond et al., 2000; Tolosa et al., 2012). The protein is highly fusogenic and immunomodulatory and participates in the fusion of syncytiotrophoblast cells in the human placenta. Syncytin‐1 fusogenic activity requires interaction with the human sodium‐dependent neutral amino acid transporter type 1 (hASCT‐1) or hASCRT‐2 genes which are human type D mammalian retrovirus receptors. The full‐length protein consists of a surface subunit containing a signal peptide and a transmembrane subunit containing a fusion peptide, an immunosuppressive domain, a transmembrane domain, and a cytoplasmic domain (Cheynet et al., 2005; Grandi & Tramontano, 2018; Tolosa et al., 2015).
Despite the finding that syncytin‐1 enhances EV release by producer cells and mediates EV membrane fusion to target cells and cargo cytoplasmic release (Bui et al., 2023), caution is warranted. Elevated syncytin‐1 has been reported to be associated with multiple sclerosis, schizophrenia, and bipolar disorder (Antony et al., 2004; Chignola et al., 2019; Liu et al., 2018; Perron et al., 2008; Wang et al., 2018). Syncytin‐1 has also been reported to promote tumorigenesis, proliferation, and metastasis, in several cancers including leukaemia, breast, endometrial, urothelial cell, and hepatocellular cancers (Chignola et al., 2019; Liu et al., 2018; Sun et al., 2017; Yu et al., 2014; Zhou et al., 2021). Future research into the use of syncytin‐1 could focus on using only the syncytin‐1 fusion peptide rather than the full‐length protein to avoid potential oncogenic or neural pathological effects. Human endogenous retroviral integrations comprise ∼8% of the human genome making it likely that there are numerous other human fusogens with the potential to increase EV fusion‐mediated uptake by target cells without the risk of inducing pathogenic effects (Lander et al., 2001).
A second EV engineering technique to increase EV uptake and cargo release relies on pH‐sensitive fusogenic peptides (Figure 5B). Cell‐penetrating peptides (CPPs), as their name suggests, have the capacity to cross cellular membranes (Angelova et al., 2013; Jafari et al., 2015; Azmi et al., 2018; Varnamkhasti et al., 2020). The first CPPs, penetratin and TAT, were derived from the Antennapedia homeodomain protein and the human immunodeficiency virus Trans‐Activator of Transcription (HIV‐TAT protein) (Prochiantz, 2000). CPPs initially attracted attention for their high transduction efficiency in directly transporting attached drugs or biomolecules into cells (Guidotti et al., 2017; Kang et al., 2017). The attachment of these molecules may be covalent, but more commonly the attachment is mediated through electrostatic interactions (Bolhassani et al., 2017). With the realization of the potential of EVs to serve as therapeutic drug delivery vehicles, the use of CPPs to facilitate EV uptake is an area of active investigation (Derakhshankhah & Jafari, 2018; Kim et al., 2018; Morimoto et al., 2024; Noguchi et al., 2021; Varnamkhasti et al., 2020). Most CPPs are cationic and are often enriched in arginine, but there are also amphipathic and hydrophobic CPPs (, Hade et al., 2023; Nakase et al., 2017). A 30‐amino acid amphiphilic synthetic peptide GALA forms a random coil at neutral pH but assumes an amphipathic α‐helix at acidic pH, allowing it to mediate endosomal escape (Li et al., 2004). This peptide has now been employed for maximizing EV cargo escape from endosomal/lysosomal degradation (Morishita et al., 2017; Nakase & Futaki, 2015).
The ability of natural and synthetic drug delivery systems such as EVs and synthetic lipid NPs to transfer and release chemotherapeutic and biomolecular cargo in target cells while reducing off‐target toxicity has focused interest on strategies for lysosomal‐mediated drug activation (Sun et al., 2023). The efficient intracellular release of therapeutic cargo is a critical step to maximize therapeutic efficacy (Ahmad et al., 2019; Fu et al., 2020). The benefit of the strategic exploitation of lysosomal characteristics to ensure the proper release of therapeutic agents has been demonstrated for the treatment of multiple diseases including cardiovascular diseases (Bonam et al., 2019; Kong et al., 2022), lysosomal storage disease (Fernández‐Pereira et al., 2021), autoimmune diseases (Schrezenmeier & Dörner, 2020), and cancer (Biswas & Torchilin, 2014; Wang et al., 2021). An array of pH‐sensitive moieties and other strategies to facilitate EV cargo endolysosomal escape are being adapted to enhance the intracellular release of synthetic lipid nanoparticle therapeutic cargo (Sun et al., 2023). The research will continue to focus on how best to exploit the low lysosomal pH that ranges from 3.5 to 5.5 to engineer pH‐sensitive nanovesicles employing design elements such as protonable amino groups, acid‐decomposable calcium carbonate, acid‐degradable bonds (e.g., boronic esters, acetal, benzoic‐imine bonds, hydrazone), endosomolytic peptides, drug‐intercalated DNA structures to maximize either cargo release into the endolysosomal compartment or the cytoplasm by causing disruption of the endolysosome.
1.10. Importance of EV isolation techniques
The selection of the most efficacious method for EV isolation is of paramount importance for research applications and the use of EVs in therapeutic applications. A major factor in determining the proper technique is the source from which the EVs must be isolated. Each EV source can introduce different potential contaminants, thus requiring modification of the isolation protocol to obtain the purest sample possible. EVs isolated from bodily fluids like serum, plasma, urine, or saliva will require the elimination of contaminating microbes, lipids, lipoproteins, nucleic acids, and protein aggregates (Brinkmann et al., 2004; György et al., 2011; Phillipson & Kubes, 2011; Vickers et al., 2011; Williams & Mackman, 2011). EVs isolated from cell culture media can be contaminated by a variety of substances. The addition of fetal bovine serum (FBS) or other substances such as culture‐derived recombinant cytokines or growth factors to the culture media will introduce FBS‐derived EVs at significant concentrations (Jeppesen et al., 2014; Théry et al., 2006). Contaminating particles have also been detected in non‐supplemented culture media (Jeppesen et al., 2014; Szatanek et al., 2015). Therefore, extended ultracentrifugation of all media components may be warranted to prevent contamination EV characterization of cargo and composition for biomarker analysis or skewing of phenotypic changes induced in target cells.
Numerous methods for EV isolation and purification have been described with sequential ultracentrifugation being one of the most common techniques (Sunkara et al., 2016; Théry et al., 2006; Witwer et al., 2013). The subpopulation of EVs to be isolated will determine the parameters of g‐force, temperature, and duration employed. Apoptotic bodies and cancer‐derived large oncosomes require a minimum of 2000 × g at 4°C for 20 min (Liu et al., 2018) or 10,000 × g at 4°C for 30 min (Minciacchi et al., 2015), respectively for their isolation, whereas microvesicles require 20,000 x g at 4°C for 90 min (Tutuianu et al., 2024). Exosomes can be isolated at 100,000 × g at 4°C for 16 h (Flemming et al., 2020; Overmiller et al., 2017) whereas migrasomes, exomeres, and supermeres can be isolated at 150,000 × g at 4°C for 4 h (Ma et al., 2015), 167,000 x g or 267,000 × g at 4°C for 16 h (Zhang et al., 2021), respectively, and it is common that these four small EV subpopulations are subjected to additional purification by sucrose or iodixanol gradients (Cantin et al., 2008; Dettenhofer & Yu, 1999; Keller et al., 2011; Poliakov et al., 2009; Théry et al., 2006), immunoaffinity purification (Filipović et al., 2022; Nakai et al., 2016), or size exclusion chromatography (Böing et al., 2014; Taylor et al., 2002).
A recent study has reported a new technique that employs direct imaging flow cytometry on EVs labelled with the fluorescent lipophilic dialkyl carbocyanine Dil (1,1′–dioctadecyl‐3,3,3′,3′‐tetramethyl indocarbocyanine perchlorate) dye (Viveiros et al., 2022). This method permits the direct imaging and characterization of EVs with minimal sample manipulation. This yields an analysis of an EV population without processing‐associated EV loss. These advanced imaging techniques will enable the analysis of EV biogenesis, release, biodistribution, and their role in disease pathogenesis (Verweij et al., 2021). Finally, asymmetric‐field‐flow fractionation can be used to detect, measure, and isolate discrete, highly purified EV subpopulations without extensive ultracentrifugation (Wiedmer et al., 2023; Zhang et al., 2018).
1.11. EV production by tumours
The role of EVs in the pathogenesis of cancer is extensive and ever‐growing (Liao et al., 2023). EVs produced by tumour, stromal, and immune cells promote tumorigenesis, immune evasion, drug resistance, angiogenesis, and the development of premetastatic niches in solid cancers including breast (Dioufa et al., 2017; Piombino et al., 2021; Yáñez‐Mó et al., 2015), pancreatic (Costa‐Silva et al., 2015; Jin et al., 2020; Ray, 2015; Stefanius et al., 2019), lung (Chen et al., 2017; Lobb et al., 2017; Pritchard et al., 2020; Wang et al., 2016), and melanoma (Andrade et al., 2019, Hoshino et al., 2015; Logozzi et al., 2009, Lattmann & Levesque, 2022; Pfeffer et al., 2015, Pompili et al., 2022). EVs are released at elevated levels from cancer cells and exosomal miRNA can be detected in solid tumour cells (Logozzi et al., 2009; Pfeffer et al., 2015), suggesting EV contents may serve as a form of intercellular communication between neighbouring tumour cells. Cancer‐derived EVs also possess the ability to set up distant environments more permissive to tumour metastasis. Increased release of tumour‐promoting EVs by tumour cells may contribute to therapy resistance (Andrade et al., 2019). Expression of drug efflux pumps, the capture of monoclonal antibodies administered for cancer therapy, or the transfer of nucleic acids, lipids, or proteins on EVs that confer physiologic changes in recipient cells to promote the resistant phenotype has been demonstrated in various tumour types (Pompili et al., 2022). EV production by melanoma, breast, pancreatic, and lung cancer cells increases after chemotherapy exposure, and this elevated production is accompanied by increased chemoresistance (Lattmann & Levesque, 2022; Logozzi et al., 2009; Nannan et al., 2020; Shechter et al., 2020). Moreover, tumour‐derived EVs can skew macrophages toward an anti‐inflammatory M2 phenotype and foster alterations in stromal and tumour cell biology that promote tumour growth (Andrade et al., 2019).
1.12. EVs as cancer therapeutics
In addition to their ability to influence disease development and progression, EVs also offer promising therapeutic targets for improving patient outcomes. Several strategies for targeting EV as cancer therapeutics are reviewed here, including manipulating the tumour microenvironment via EV, engineering EV for targeted, minimally toxic drug delivery, using EV as cancer vaccines, and modulating tumour‐derived EV production.
1.12.1. EV‐induced antitumor immunity
The tumour microenvironment is commonly characterized as anti‐inflammatory and tumour‐promoting. Tumor‐derived EVs may contribute to this immunosuppressive microenvironment. In one study, EVs released by tumour cells in vitro and in vivo enhance the suppression of cytotoxic T cells by myeloid‐derived suppressor cells, thereby stimulating tumour growth (Chalmin et al., 2010). When EV release was inhibited by amiloride, enhanced killing of tumour cells by cyclophosphamide was observed (Chalmin et al., 2010). Thus, targeting tumour‐derived EV production or using EV‐based immunotherapeutic approaches to overcome mechanisms of immunosuppression in the tumour microenvironment offers potential interventions for improving anti‐tumour immunity.
Based on our current understanding, the most direct approaches involve targeting the maturation or activation of immune cells toward a more pro‐inflammatory phenotype. For example, combined administration of engineered EVs loaded with either the TGF‐β receptor 1 kinase inhibitor (SD‐208) or the Toll‐like receptor (TLR) 7/8 agonist, resiquimod, promoted the release of IL‐6 and tumour necrosis factor (TNF)‐alpha by dendritic cells (DCs) and macrophages (Lee et al., 2022). These proinflammatory cytokines induced the transformation of macrophages from an anti‐inflammatory to a proinflammatory phenotype which markedly restrained prostate and melanoma tumour cell growth (Lee et al., 2022). EVs have also been engineered to promote the expression of FAP (fibroblast activation protein) in DCs leading to their maturation, which improved anti‐melanoma immunity (Hu et al., 2021). Modification of the glycosylation patterns on EVs may be another strategy for promoting DC maturation and activation of T cells. High mannose glycans are a natural ligand of ICAM‐3 grabbing non‐integrin (DC‐SIGN) that is expressed on DCs. Engagement of this receptor results in cargo internalization and antigen processing for presentation on MHC. Overexpression of these glycans on malignant melanoma‐derived apoptotic EVs led to increased CD4/CD8 T cell activation (Morishita et al., 2017). The ability of EVs to carry many conjugated peptides on their surface may allow for the development of antibody‐based cancer therapies that include the coordination of multiple components of the tumour microenvironment to enhance tumour lysis. In one proof‐of‐concept study, the expression of α‐EGFR/α‐CD3 antibodies on EV enhanced the killing of EGFR+ breast cancer cells in vitro (Cheng et al., 2018).
1.12.2. EVs as cancer vaccines
EVs display high safety profiles, are easy to store and preserve, can be mass‐produced, and show enhanced specificity of the immune response, making their use in vaccines fairly practical (Xu et al., 2020). Cancer vaccines can have as their therapeutic goal either cancer prophylaxis or treatment. EVs can be designed to function either to deliver therapeutic agents or to function as a vaccine to prevent disease. A popular strategy for cancer vaccine design invokes tumour neoantigens to promote antigen‐presenting cell (APC)‐T‐cell interactions, triggering an anti‐tumour adaptive immune response and immunologic memory (Liu et al., 2022). In several preclinical lung cancer models, EV‐based vaccination demonstrated favourable results. Most such strategies engineered EVs to contain immunostimulatory molecules or pro‐inflammatory cytokines to promote anti‐tumour immunity. In one study, lung tumour cell derived EVs modified to express CD40L led to a robust proliferation and activation of cytotoxic T‐cells and slowed tumour progression (Wang et al., 2014). In another, antigenic similarity between embryonic and tumour cells allowed EVs from GM‐CSF modified murine embryonic stem cells to diminish the implantation and expansion of lung cancer lines in vivo (Yaddanapudi et al., 2019). A challenge to EV‐based vaccination is ensuring that infusions reach lymphatic tissues to induce their desired effects; however, EV modifications such as the addition of high mannose glycans can trigger activation, phagocytosis and trafficking of APC to areas of immune surveillance (Choi et al., 2019).
While Phase I and II clinical trials of DC and ascites‐derived EV vaccines for cancer failed to achieve primary endpoints or evoke antigen‐specific T cell responses, they did demonstrate the ability to invoke innate natural killer (NK) immunity (Besse et al., 2015), and it is generally believed that EV vaccines remain promising, particularly when used in combination with other in cancer immunotherapeutic approaches, such as immune checkpoint inhibitors, traditional chemotherapies, and oncolytic viruses to enhance the efficacy of each (, Garofalo et al., 2018; Lv et al., 2021; Ren et al., 2022). One relatively recent vaccination technique combines EV‐ and nanoparticle‐based technologies. In this study, microfluidic sonication was used to envelop poly(lactic‐co‐glycolic acid) (PLGA) nanoparticles in EV membranes, with the goal of exploiting the membrane fusion properties and innate immune evasion of proteins expressed on EV membranes (Liu et al., 2019). The same system also delivered CD80/86 with an anti‐PD‐1 antibody to reverse tumour‐mediated immune suppression and mediate remodelling of exhausted T cells in lung and colon carcinoma cell models (Liu et al., 2022).
Another innovative anti‐cancer vaccine strategy attempts to harness the antibacterial immune response to prevent cancer. The inadequate activation of cancer‐specific T‐cells and an immunosuppressive TME are the primary hindrances of anti‐cancer immune responses (Dersh et al., 2021; Rabinovich et al., 2007). The primary mechanism that underlies the efficacy of PD‐1/PD‐L1 targeted immunotherapy is its ability to induce the selective proliferation and reinvigoration of exhausted cancer‐antigen‐responsive T‐cells (Im et al., 2016). However, 80%–90% of tumours do not respond well to antiPD‐1/antiPD‐L1 therapies due to insufficient T‐cell activation or the ineffective recruitment of activated T‐cells to the tumour (Galon & Bruni, 2019).
In the 19th century, William Coley published reports of his comprehensive studies on the induction of tumour regression following injection of Coley's toxins, mixtures of extracts of heat‐inactivated bacteria (Carlson et al., 2020; Coley, 1991). Re‐evaluation of these studies provide the rationale for employing EVs derived from the outer bacterial membrane of Gram‐positive and Gram‐negative bacteria to induce potent and persistent immune responses, mediated by interferon‐gamma (IFN‐γ) and C‐X‐C motif chemokine ligand 10 (CXCL10), that successfully eliminated tumours without significant toxicity (Kim et al., 2015). EVs from Gram‐negative bacteria that were engineered to either display the PD‐1 ectodomain on the EV outer membrane (Li et al., 2020) or that contained a DNA plasmid encoding PD‐1 (Pan et al., 2022) were able to abrogate the inhibition of the PD‐1/PD‐L1 system on T‐cell effector functions. Intraperitoneal injection of these outer bacterial membrane‐derived EVs that contain copious amounts of pathogen‐associated molecular patterns (PAMPs) was found to induce trained immunity by the activation of inflammasome‐mediated innate immune signalling and IL‐1β secretion (Liu et al., 2024). This primes the immune response, generating antigen‐presenting cell progenitors and increasing the anti‐cancer response following tumour antigen‐based vaccination.
Synthetic bacterial vesicles (SBVs), engineered by enzymatic treatment of outer Escherichia coli membranes followed by sonication to form vesicles to minimize their ability to trigger a toxic systemic innate immune proinflammatory reaction were found to induce activation of mouse bone marrow‐derived dendritic cells (Park et al., 2021). Coimmunization of mice with SBVs and EVs isolated from melanoma cancer cells induced a Th‐1 mediated immune response that was enhanced by an anti‐PD‐1 inhibitor and resulted in tumour regression in a melanoma xenograft model.
The large‐scale production of bacterial EVs, a necessary step to render this treatment strategy feasible as a novel therapeutic agent, has recently been reported (Won et al., 2023). The mass‐produced E. coli‐derived outer membrane vesicles were able to induce complete tumour regression in syngeneic xenograft models in C57BL/6 and BALB/c mice by mediating the increased infiltration of cancer antigen‐specific CD8+ T‐cells with high expression levels of Tumor cell factor 1 (TCF1) and PD‐1. These studies highlight the enormous potential of bacteria‐derived EVs to serve as potent novel tools for the induction of anti‐cancer immunity.
1.12.3. EVs as targeted therapeutic delivery vehicles
One of the most exciting uses of EVs in cancer therapy is the targeted and encapsulated delivery of traditionally toxic and poorly tolerated chemotherapies. Various designs for tumour‐specific delivery of therapeutic EVs with reduced off‐target toxicity have been proposed. Both nucleic acid aptamer and peptide‐based targeting molecules on EV surfaces have shown enhanced delivery and tumour‐specific killing of traditional chemotherapeutics (Hosseini et al., 2022; Lin et al., 2019). Although targeting with specific binding epitopes on EV surfaces seems ideal, studies have shown that simply encapsulating drugs within EVs enhances efficacy and reduces off‐target effects, suggesting enhanced uptake of EV‐encapsulated therapeutics by tumour cells relative to healthy tissue (Bagheri et al., 2020; Li et al., 2020). The ability of EVs to be loaded with multiple therapeutic molecules allows for multiplexed approaches to treatment, such as addressing drug resistance as well as drug delivery at once (Liang et al., 2020).
1.13. Targeting tumour‐derived EV production and the premetastatic niche
As mentioned above, EV production is often upregulated in cancers and tumour‐derived EV exerts both local and distant pro‐tumour effects. Tumour‐derived EV tropism for various organs may help set up premetastatic niches and partially explain patterns of metastasis. Understanding these patterns may identify targets for cancer therapy. An intriguing recent study employed a multiparametric analysis that combined metabolomic, tissue, and spatial single‐cell transcriptomics of pre‐metastatic liver biopsies to classify the risk of liver metastasis in patients who had received pancreatic cancer resections (Bojmar et al., 2024). They were able to classify patients into early or late liver metastasis, extrahepatic metastasis, and disease‐free survivor groups and to identify prognostic indicators for each group. A machine‐based learning model was developed that could predict patient outcomes before surgery with 78% accuracy. The characterization of EVs from these patients could potentially increase the accuracy of this prognostic model.
It has been reported that integrin expression on EVs from melanoma, breast, and pancreatic tumour cell lines defines organ tropism of metastasis (Hoshino et al., 2015; Lattmann & Levesque, 2022). The expression of α6β4 and α6β1 integrins on EVs has been associated with lung metastasis, while the expression of integrin αvβ5 on EVs was linked to liver metastasis (Hoshino et al., 2015). EVs expressing these integrins can redirect the metastasis of tumour cells to organs to which they would not normally metastasize (Hoshino et al., 2015). Melanoma also commonly metastasizes to the brain. While the precise mechanism of how EVs cross the blood‐brain barrier remains poorly defined, CD46 has been described as the major receptor for the uptake of EV derived from a brain metastatic melanoma cell line (Sk‐Mel‐28) by human blood–brain barrier endothelial cells (hCMEC/D3) cells (Kuroda et al., 2019). Reducing EV production by the tumour could reduce the risk of metastasis, and characterizing integrin expression could identify sites most likely to harbour metastatic disease or identify pathways to deter progression from local to more advanced disease.
To create distant premetastatic niches, tumour‐derived EVs promote neovascularization and lymphatic remodelling. The pathways involved in this process could potentially be exploited to trigger a desirable therapeutic effect or counter an undesired effect of EVs. For example, urokinase plasminogen activator receptor (uPAR, CD87) is expressed at low levels in healthy tissue, but its expression increases on malignancy and correlates with invasion and metastasis (Zhai et al., 2022). uPAR expression on melanoma EVs promotes angiogenesis and helps to establish the blood supply of the premetastatic niche by inducing VE‐cadherin, EGFR, uPAR and ERK1/2 signalling in endothelial cells (Biagioni et al., 2021; Lattmann & Levesque, 2022). EVs expressing elevated levels of uPAR have been isolated from highly metastatic pancreatic tumour cells (Mu et al., 2013). Similarly, melanoma EV expression of cellular adhesion molecules, such as VCAM‐1, targets them for uptake into lymphatic endothelial cells, altering gene expression and proliferation of recipient cells (Lattmann & Levesque, 2022; Leary et al., 2022). A key driver of melanoma EV lymphatic endothelial cell targeting, lymphangiogenesis, and tumour cell adhesion is NGFR expression (García‐Silva et al., 2021; Lattmann & Levesque, 2022). In the B16 model, NGFR+ EVs promote lymphangiogenesis via induction of the ERK/NFKB pathway and tumour cell adhesion via ICAM‐1. Ablation of NGRF signalling decreased nodal metastasis and extended patient survival (García‐Silva et al., 2021; Lattmann & Levesque, 2022). These receptors may provide targets for countering the tumour‐permissive properties of melanoma‐derived EVs. Compared to melanoma and other tumours, considerably less is understood about EV production in non‐melanoma skin cancer (NMSC). We recently demonstrated that desmoglein 2 (Dsg2) expression modulates cutaneous squamous cell carcinoma (cSCC)‐associated EV production and cargo sorting of cytokines, including IL‐8, which has chemotactic and angiogenic properties (Flemming et al., 2020; Overmiller et al., 2017). The ability of Dsg2 to increase EV release from cSCC and alter cytokine properties could be manipulated in the design of EV‐based therapeutics.
A more global approach to decreasing the ability of tumour‐derived EVs to set up pro‐tumour metastatic niches could be downregulating EV production by tumour cells. Rab proteins are an essential part of vesicular and membrane trafficking that help regulate the abundance and subcellular localization of proteins within cells and cellular signalling (Bhuin & Roy, 2014). Over 70 Rab family members have been defined as contributing to vesicle formation, mobility, and membrane fusion in normal and tumour cells. Specific Rab protein impairments can manifest distinct disease phenotypes. Characterization of the unique Rab proteins contributing to exosome packaging and production by tumour cells could lead to therapeutic targets for the downregulation of EV production by tumour cells, thus downregulating their ability to foster metastasis. Rab27a knockdown in highly metastatic melanoma cells led to a reduction in exosome production, primary tumour size, and metastases (Peinado et al., 2012). One challenge to this approach is the large number of Rab proteins and the potential for redundancy in cellular functions or off‐target effects.
2. CONCLUSIONS
This review has highlighted the tremendous potential of EVs to serve as therapeutic tools in human diseases, particularly cancer, while also recognizing the significant challenges to be overcome if they are to be successfully translated into viable targeted therapeutic delivery vehicles. Their advantages include their low immunogenicity, low toxicity, ease of engineering, and ability to protect drugs and bioactive molecules from degradation. Current research is focused on developing strategies to improve the targeting specificity, uptake by target cells, and escape from lysosomal degradation. We presented examples of the effectiveness of strategies to increase EV targeting, uptake by target cells, and cargo release in various solid tumours including melanoma, breast, lung, and pancreatic cancers. This review has endeavoured to present an overview of the complex pathways involved in EV uptake and how these pathways may be exploited to maximize EV therapeutic utility. This comprehensive survey of the EV literature aims to provide insights into the current understanding of EV‐mediated intercellular communication and bring focus to their potential to serve as a novel and powerful addition to the therapeutic arsenal available to treat cancer.
AUTHOR CONTRIBUTIONS
Stephanie R. Jackson Cullison: Conceptualization (supporting); investigation (supporting); writing—original draft (supporting); writing—review & editing (supporting). Joseph P. Flemming: Conceptualization (supporting); investigation (supporting); writing—original draft (supporting); writing—review & editing (supporting). Kubra Karagoz: Writing—original draft (supporting); writing—review & editing (supporting). Peter J. Wermuth: Conceptualization (lead); investigation (lead); writing—original draft (lead); writing—review & editing (lead). Mỹ G. Mahoney: Conceptualization (lead); funding acquisition (lead); project administration (lead); resources (lead); supervision (lead); writing—original draft (lead); writing—review & editing (lead).
CONFLICT OF INTEREST STATEMENT
The authors state no conflict of interest.
ACKNOWLEDGEMENTS
We sincerely thank Ana Pivovarnik for her assistance with the references. This study was supported by a grant from the National Institutes of Health National Institute of Arthritis and Musculoskeletal and Skin Diseases R01‐AR074314 to M.G.M.
Jackson Cullison, S. R , Flemming, J. P. , Karagoz, K. , Wermuth, P. J. , & Mahoney, M. G. (2024). Mechanisms of Extracellular Vesicle Uptake and Implications for the Design of Cancer Therapeutics. Journal of Extracellular Biology, 3, e70017. 10.1002/jex2.70017
Stephanie R. Jackson Cullison and Joseph P. Flemming are co‐first authors.
Peter J. Wermuth and Mỹ G. Mahoney are co‐last authors.
REFERENCES
- Admyre, C. , Johansson, S. M. , Paulie, S. , & Gabrielsson, S. (2006). Direct exosome stimulation of peripheral human T cells detected by ELISPOT. European Journal of Immunology, 36, 1772–1781. [DOI] [PubMed] [Google Scholar]
- Ahmad, A. , Khan, J. M. , & Haque, S. (2019). Strategies in the design of endosomolytic agents for facilitating endosomal escape in nanoparticles. Biochimie, 160, 61–75. [DOI] [PubMed] [Google Scholar]
- Ahram, M. , Sameni, M. , Qiu, R. G. , Linebaugh, B. , Kirn, D. , & Sloane, B. F. (2000). Rac1‐induced endocytosis is associated with intracellular proteolysis during migration through a three‐dimensional matrix. Experimental Cell Research, 260, 292–303. [DOI] [PubMed] [Google Scholar]
- Altan‐Bonnet, G. , & Mukherjee, R. (2019). Cytokine‐mediated communication: A quantitative appraisal of immune complexity. Nature Reviews, Immunology, 19, 205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez‐Erviti, L. , Seow, Y. , Yin, H. , Betts, C. , Lakhal, S. , & Wood, M. J. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotechnology, 29, 341–345. [DOI] [PubMed] [Google Scholar]
- Alyami, M. Z. , Alsaiari, S. K. , Li, Y. , Qutub, S. S. , Aleisa, F. A. , Sougrat, R. , Merzaban, J. S. , & Khashab, N. M. (2020). Cell‐type‐specific CRISPR/Cas9 delivery by biomimetic metal organic frameworks. Journal of American Chemical Society, 142, 1715–1720. [DOI] [PubMed] [Google Scholar]
- Andrade, L. N. S. , Otake, A. H. , Cardim, S. G. B. , da Silva, F. I. , Ikoma Sakamoto, M. M. , Furuya, T. K. , Uno, M. , Pasini, F. S. , & Chammas, R. (2019). Extracellular vesicles shedding promotes melanoma growth in response to chemotherapy. Scientific Reports, 9, 14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova, A. , Angelov, B. , Drechsler, M. , & Lesieur, S. (2013). Neurotrophin delivery using nanotechnology. Drug Discovery Today, 18, 1263–1271. [DOI] [PubMed] [Google Scholar]
- Antes, T. J. , Middleton, R. C. , Luther, K. M. , Ijichi, T. , Peck, K. A. , Liu, W. J. , Valle, J. , Echavez, A. K. , & Marbán, E. (2018). Targeting extracellular vesicles to injured tissue using membrane cloaking and surface display. Journal of Nanobiotechnology, 16, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony, J. M. , van Marle, G. , Opii, W. , Butterfield, D. A. , Mallet, F. , Yong, V. W. , Wallace, J. L. , Deacon, R. M. , Warren, K. , & Power, C. (2004). Human endogenous retrovirus glycoprotein‐mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nature Neuroscience, 7, 1088–1095. [DOI] [PubMed] [Google Scholar]
- Atay, S. , Gercel‐Taylor, C. , & Taylor, D. D. (2011). Human trophoblast‐derived exosomal fibronectin induces pro‐inflammatory IL‐1β production by macrophages. American Journal of Reproductive Immunology, 66, 259–269. [DOI] [PubMed] [Google Scholar]
- Azmi, I. D. M. , Østergaard, J. , Stürup, S. , Gammelgaard, B. , Urtti, A. , Moghimi, S. M. , & Yaghmur, A. (2018). Cisplatin encapsulation generates morphologically different multicompartments in the internal nanostructures of nonlamellar liquid‐crystalline self‐assemblies. Langmuir : The Acs Journal Of Surfaces and Colloids, 34, 6570–6581. [DOI] [PubMed] [Google Scholar]
- Backovic, M. , & Jardetzky, T. S. (2009). Class III viral membrane fusion proteins. Current Opinion in Structural Biology, 19, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badierah, R. A. , Uversky, V. N. , & Redwan, E. M. (2021). Dancing with Trojan horses: An interplay between the extracellular vesicles and viruses. Journal of Biomolecular Structure & Dynamics, 39, 3034–3060. [DOI] [PubMed] [Google Scholar]
- Bae, Y. H. (2009). Drug targeting and tumor heterogeneity. Journal of Controlled Release: Official Journal of the Controlled Release Society, 133, 2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri, E. , Abnous, K. , Farzad, S. A. , Taghdisi, S. M. , Ramezani, M. , & Alibolandi, M. (2020). Targeted doxorubicin‐loaded mesenchymal stem cells‐derived exosomes as a versatile platform for fighting against colorectal cancer. Life Sciences, 261, 118369. [DOI] [PubMed] [Google Scholar]
- Baker, B. M. , & Chen, C. S. (2012). Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. Journal of Cell Science, 125(Pt 13), 3015–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak, D. , Arrighi, S. , Darwiche, Y. , & Deb, S. (2021). Comparison of anticancer drug toxicities: paradigm shift in adverse effect profile. Life, 12, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashiri, G. , Padilla, M. S. , Swingle, K. L. , Shepherd, S. J. , Mitchell, M. J. , & Wang, K. (2023). Nanoparticle protein corona: from structure and function to therapeutic targeting. Lab on a Chip, 23, 1432–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batrakova, E. V. , & Kim, M. S. (2015). Using exosomes, naturally‐equipped nanocarriers, for drug delivery. Journal of Controlled Release: Official Journal of the Controlled Release Society, 219, 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia, D. , Raimondo, S. , Calabrese, G. , Forte, S. , Cristaldi, M. , Patinella, A. , Memeo, L. , Manno, M. , Raccosta, S. , Diana, P. , Cirrincione, G. , Giavaresi, G. , Monteleone, F. , Fontana, S. , De Leo, G. , & Alessandro, R. (2017). Interleukin 3‐ receptor targeted exosomes inhibit in vitro and in vivo chronic myelogenous leukemia cell growth. Theranostics, 7, 1333–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse, B. , Charrier, M. , Lapierre, V. , Dansin, E. , Lantz, O. , Planchard, D. , Le Chevalier, T. , Livartoski, A. , Barlesi, F. , Laplanche, A. , Ploix, S. , Vimond, N. , Peguillet, I. , Théry, C. , Lacroix, L. , Zoernig, I. , Dhodapkar, K. , Dhodapkar, M. , Viaud, S. , Soria, J. C., … Chaput, N. (2015). Dendritic cell‐derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology, 5, e1071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuin, T. , & Roy, J. K. (2014). Rab proteins: the key regulators of intracellular vesicle transport. Experimental Cell Research, 328, 1–19. [DOI] [PubMed] [Google Scholar]
- Biagioni, A. , Laurenzana, A. , Menicacci, B. , Peppicelli, S. , Andreucci, E. , Bianchini, F. , Guasti, D. , Paoli, P. , Serratì, S. , Mocali, A. , Calorini, L. , Del Rosso, M. , Fibbi, G. , Chillà, A. , & Margheri, F. (2021). uPAR‐expressing melanoma exosomes promote angiogenesis by VE‐Cadherin, EGFR and uPAR overexpression and rise of ERK1,2 signaling in endothelial cells. Cellular and Molecular Life Sciences : CMLS, 78, 3057–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bister, N. , Pistono, C. , Huremagic, B. , Jolkkonen, J. , Giugno, R. , & Malm, T. (2020). Hypoxia and extracellular vesicles: a review on methods, vesicular cargo and functions. Journal of Extracellular Vesicles, 10, e12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, S. , & Torchilin, V. P. (2014). Nanopreparations for organelle‐specific delivery in cancer. Advanced Drug Delivery Review, 66, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond, J. L. , Lavillette, D. , Cheynet, V. , Bouton, O. , Oriol, G. , Chapel‐Fernandes, S. , Mandrand, B. , Mallet, F. , & Cosset, F. L. (2000). An envelope glycoprotein of the human endogenous retrovirus HERV‐W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. Journal of Virology, 74, 3321–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böing, A. N. , van der Pol, E. , Grootemaat, A. E. , Coumans, F. A. , Sturk, A. , & Nieuwland, R. (2014). Single‐step isolation of extracellular vesicles by size‐exclusion chromatography. Journal of Extracellular Vesicles, 3, 10.3402/jev.v3.23430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojmar, L. , Zambirinis, C. P. , Hernandez, J. M. , Chakraborty, J. , Shaashua, L. , Kim, J. , Johnson, K. E. , Hanna, S. , Askan, G. , Burman, J. , Ravichandran, H. , Zheng, J. , Jolissaint, J. S. , Srouji, R. , Song, Y. , Choubey, A. , Kim, H. S. , Cioffi, M. , van Beek, E. , … Lyden, D. (2024). Multi‐parametric atlas of the pre‐metastatic liver for prediction of metastatic outcome in early‐stage pancreatic cancer. Nature Medicine, 30, 2170–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhassani, A. , Jafarzade, B. S. , & Mardani, G. (2017). In vitro and in vivo delivery of therapeutic proteins using cell penetrating peptides. Peptides, 87, 50–63. [DOI] [PubMed] [Google Scholar]
- Bonam, S. R. , Wang, F. , & Muller, S. (2019). Lysosomes as a therapeutic target. Nature Review Drug Discovery, 18, 923–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsergent, E. , & Lavieu, G. (2019). Content release of extracellular vesicles in a cell‐free extract. FEBS Letters, 593, 1983–1992. [DOI] [PubMed] [Google Scholar]
- Bora, K. , Wang, Z. , Yemanyi, F. , Maurya, M. , Blomfield, A. K. , Tomita, Y. , & Chen, J. (2022). Endothelial cell transcytosis assay as an in vitro model to evaluate inner blood‐retinal barrier permeability. Journal of Visualized Experiments : JoVE, 184, e64076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boson, B. , Legros, V. , Zhou, B. , Siret, E. , Mathieu, C. , Cosset, F. L. , Lavillette, D. , & Denolly, S. (2021). The SARS‐CoV‐2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus‐like particles. The Journal of Biological Chemistry, 296, 100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann, V. , Reichard, U. , Goosmann, C. , Fauler, B. , Uhlemann, Y. , Weiss, D. S. , Weinrauch, Y. , & Zychlinsky, A. (2004). Neutrophil extracellular traps kill bacteria. Science, 303, 1532–1535. [DOI] [PubMed] [Google Scholar]
- Briolay, T. , Petithomme, T. , Fouet, M. , Nguyen‐Pham, N. , Blanquart, C. , & Boisgerault, N. (2021). Delivery of cancer therapies by synthetic and bio‐inspired nanovectors. Molecular Cancer, 20, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. M. , & Wilson, W. R. (2004). Exploiting tumour hypoxia in cancer treatment. Nature Reviews. Cancer, 4, 437–447. [DOI] [PubMed] [Google Scholar]
- Brummendorf, T. , & Lemmon, V. (2001). Immunoglobulin superfamily receptors: cis‐interactions, intercellular adaptors, and alternative splicing regulate adhesion. Current Opinion in Cell Biology, 13, 611–618. [DOI] [PubMed] [Google Scholar]
- Brzozowski, J. S. , Bond, D. R. , Jankowski, H. , Goldie, B. J. , Burchell, R. , Naudin, C. , Smith, N. D. , Scarlett, C. J. , Larsen, M. R. , Dun, M. D. , Skelding, K. A. , & Weidenhofer, J. (2018). Extracellular vesicles with altered tetraspanin CD9 and CD151 levels confer increased prostate cell motility and invasion. Scientific Reports, 8, 8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui, S. , Dancourt, J. , & Lavieu, G. (2023). Virus‐free method to control and enhance extracellular vesicle cargo loading and delivery. ACS Applied Bio Materials, 6, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgermeister, E. , Liscovitch, M. , Röcken, C. , Schmid, R. M. , & Ebert, M. P. (2008). Caveats of caveolin‐1 in cancer progression. Cancer Letters, 268, 187–201. [DOI] [PubMed] [Google Scholar]
- Burnouf, T. , Agrahari, V. , & Agrahari, V. (2019). extracellular vesicles as nanomedicine: hopes and hurdles in clinical translation. International Journal of Nanomedicine, 14, 8847–8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabantous, S. , Terwilliger, T. C. , & Waldo, G. S. (2005). Protein tagging and detection with engineered self‐assembling fragments of green fluorescent protein. Nature Biotechnology, 23, 102–107. [DOI] [PubMed] [Google Scholar]
- Cantin, R. , Diou, J. , Bélanger, D. , Tremblay, A. M. , & Gilbert, C. (2008). Discrimination between exosomes and HIV‐1: purification of both vesicles from cell‐free supernatants. Journal of Immunological Methods, 338, 21–30. [DOI] [PubMed] [Google Scholar]
- Capone, P. M. , Papsidero, L. D. , & Chu, T. M. (1984). Relationship between antigen density and immunotherapeutic response elicited by monoclonal antibodies against solid tumors. Journal of the National Cancer Institute, 72, 673–677. [PubMed] [Google Scholar]
- Carlson, R. D. , Flickinger, J. C. Jr. , & Snook, A. E. (2020). Talkin' toxins: From Coley's to modern cancer immunotherapy. Toxins, 12, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellote‐Borrell, M. , Merlina, F. , Rodríguez, A. R. , & Guasch, J. (2023). Biohybrid hydrogels for tumoroid culture. Advanced Biology, 7, e2300118. [DOI] [PubMed] [Google Scholar]
- Chalmin, F. , Ladoire, S. , Mignot, G. , Vincent, J. , Bruchard, M. , Remy‐Martin, J. P. , Boireau, W. , Rouleau, A. , Simon, B. , Lanneau, D. , De Thonel, A. , Multhoff, G. , Hamman, A. , Martin, F. , Chauffert, B. , Solary, E. , Zitvogel, L. , Garrido, C. , Ryffel, B. , Borg, C., … Ghiringhelli, F. (2010). Membrane‐associated Hsp72 from tumor‐derived exosomes mediates STAT3‐dependent immunosuppressive function of mouse and human myeloid‐derived suppressor cells. The Journal of Clinical Investigation, 120, 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , Song, X. , Gong, T. , Fu, Y. , Yang, L. , Zhang, Z. , & Gong, T. (2017). nRGD modified lycobetaine and octreotide combination delivery system to overcome multiple barriers and enhance anti‐glioma efficacy. Colloids and Surfaces. B, Biointerfaces, 156, 330–339. [DOI] [PubMed] [Google Scholar]
- Chen, W. , Jiang, J. , Xia, W. , & Huang, J. (2017). Tumor‐related exosomes contribute to tumor‐promoting microenvironment: an immunological perspective. Journal of Immunology Research, 1073947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Jia, L. , Zhu, G. , Wang, W. , Geng, M. , Lu, H. , Zhang, Y. , Zhou, M. , Zhang, F. , & Cheng, X. (2022). Sortase A‐mediated cyclization of novel polycyclic RGD peptides for ανβ3 integrin targeting. Bioorganic & Medicinal Chemistry Letters, 73, 128888. [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Wang, H. , Xia, Y. , Yan, F. , & Lu, Y. (2018). Therapeutic potential of mesenchymal cell‐derived miRNA‐150‐5p‐expressing exosomes in rheumatoid arthritis mediated by the modulation of MMP14 and VEGF. Journal of Immunology, 201, 2472–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Q. , Shi, X. , Han, M. , Smbatyan, G. , Lenz, H. J. , & Zhang, Y. (2018). Reprogramming exosomes as nanoscale controllers of cellular immunity. Journal of the American Chemical Society, 140, 16413–16417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik, L. V. , & Kozlov, M. M. (2008). Mechanics of membrane fusion. Nature Structural & Molecular Biology, 15, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik, L. V. , Melikyan, G. B. , & Chizmadzhev, Y. A. (1987). Biomembrane fusion: a new concept derived from model studies using two interacting planar lipid bilayers. Biochimica et Biophysica Acta, 906, 309–352. [DOI] [PubMed] [Google Scholar]
- Cheung, K. L. , Jarrett, R. , Subramaniam, S. , Salimi, M. , Gutowska‐Owsiak, D. , Chen, Y. L. , Hardman, C. , Xue, L. , Cerundolo, V. , & Ogg, G. (2016). Psoriatic T cells recognize neolipid antigens generated by mast cell phospholipase delivered by exosomes and presented by CD1a. The Journal of Experimental Medicine, 213, 2399–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheynet, V. , Ruggieri, A. , Oriol, G. , Blond, J. L. , Boson, B. , Vachot, L. , Verrier, B. , Cosset, F. L. , & Mallet, F. (2005). Synthesis, assembly, and processing of the Env ERVWE1/syncytin human endogenous retroviral envelope. Journal of Virology, 79, 5585–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, L. , Na, M. H. , Jung, H. K. , Vadevoo, S. M. , Kim, C. W. , Padmanaban, G. , Park, T. I. , Park, J. Y. , Hwang, I. , Park, K. U. , Liang, F. , Lu, M. , Park, J. , Kim, I. S. , & Lee, B. H. (2015). Enhanced delivery of liposomes to lung tumor through targeting interleukin‐4 receptor on both tumor cells and tumor endothelial cells. Journal of Controlled Release : Official Journal of The Controlled Release Society, 209, 327–336. [DOI] [PubMed] [Google Scholar]
- Chignola, R. , Sega, M. , Molesini, B. , Baruzzi, A. , Stella, S. , & Milotti, E. (2019). Collective radioresistance of T47D breast carcinoma cells is mediated by a Syncytin‐1 homologous protein. PLoS One, 14, e0206713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, N. T. , Kageyama, R. , & Ansel, K. M. (2018). Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Reports, 25, 3356–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, D. , Lee, T. H. , Spinelli, C. , Chennakrishnaiah, S. , D'Asti, E. , & Rak, J. (2017). Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation. Seminars in Cell & Developmental Biology, 67, 11–22. [DOI] [PubMed] [Google Scholar]
- Choi, D. , Spinelli, C. , Montermini, L. , & Rak, J. (2019). Oncogenic regulation of extracellular vesicle proteome and heterogeneity. Proteomics, 19, e1800169. [DOI] [PubMed] [Google Scholar]
- Choi, E. S. , Song, J. , Kang, Y. Y. , & Mok, H. (2019). Mannose‐modified serum exosomes for the elevated uptake to murine dendritic cells and lymphatic accumulation. Macromolecular Bioscience, 19, e1900042. [DOI] [PubMed] [Google Scholar]
- Christianson, H. C. , Svensson, K. J. , van Kuppevelt, T. H. , Li, J. P. , & Belting, M. (2013). Cancer cell exosomes depend on cell‐surface heparan sulfate proteoglycans for their internalization and functional activity. Proceedings of the National Academy of Sciences of the United States of America, 110, 17380–17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley, W. B. (1991). The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. Clinical Orthopaedics and Related Research, 262, 3–11. [PubMed] [Google Scholar]
- Colombo, M. , Moita, C. , van Niel, G. , Kowal, J. , Vigneron, J. , Benaroch, P. , Manel, N. , Moita, L. F. , Théry, C. , & Raposo, G. (2013). Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. Journal of Cell Science, 126(Pt 24), 5553–5565. [DOI] [PubMed] [Google Scholar]
- Colombo, M. , Raposo, G. , & Théry, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual Review of Cell and Developmental Biology, 30, 255–289. [DOI] [PubMed] [Google Scholar]
- Cooper, D. R. , Wang, C. , Patel, R. , Trujillo, A. , Patel, N. A. , Prather, J. , Gould, L. J. , & Wu, M. H. (2018). Human adipose‐derived stem cell conditioned media and exosomes containing malat1 promote human dermal fibroblast migration and ischemic wound healing. Advances in Wound Care, 7, 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa‐Silva, B. , Aiello, N. M. , Ocean, A. J. , Singh, S. , Zhang, H. , Thakur, B. K. , Becker, A. , Hoshino, A. , Mark, M. T. , Molina, H. , Xiang, J. , Zhang, T. , Theilen, T. M. , García‐Santos, G. , Williams, C. , Ararso, Y. , Huang, Y. , Rodrigues, G. , Shen, T. L. , … Lyden, D. (2015). Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nature Cell Biology, 17, 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio, A. (2023). Unraveling the cave: A seventy‐year journey into the caveolar network, cellular signaling, and human disease. Cells, 12, 2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton, C. M. , Schlegel, C. , & Hunter, C. J. (2023). Caveolin‐1: A review of intracellular functions, tissue‐specific roles, and epithelial tight junction regulation. Biology, 12, 1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcy, C. , & Kiel, C. (2021). Cell adhesion molecules in normal skin and melanoma. Biomolecules, 11, 1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gassart, A. , Géminard, C. , Hoekstra, D. , & Vidal, M. (2004). Exosome secretion: the art of reutilizing nonrecycled proteins?. Traffic, 5, 896–903. [DOI] [PubMed] [Google Scholar]
- DePolo, N. J. , Reed, J. D. , Sheridan, P. L. , Townsend, K. , Sauter, S. L. , Jolly, D. J. , & Dubensky, T. W. Jr. (2000). VSV‐G pseudotyped lentiviral vector particles produced in human cells are inactivated by human serum. Molecular Therapy, 2, 218–222. [DOI] [PubMed] [Google Scholar]
- Derakhshankhah, H. , & Jafari, S. (2018). Cell penetrating peptides: A concise review with emphasis on biomedical applications. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 108, 1090–1096. [DOI] [PubMed] [Google Scholar]
- Dermine, J. F. , Duclos, S. , Garin, J. , St‐Louis, F. , Rea, S. , Parton, R. G. , & Desjardins, M. (2001). Flotillin‐1‐enriched lipid raft domains accumulate on maturing phagosomes. The Journal of Biological Chemistry, 276, 18507–18512. [DOI] [PubMed] [Google Scholar]
- Dersh, D. , Hollý, J. , & Yewdell, J. W. (2021). A few good peptides: MHC class I‐based cancer immunosurveillance and immunoevasion. Nature Reviews. Immunology, 21, 116–128. [DOI] [PubMed] [Google Scholar]
- Dettenhofer, M. , & Yu, X. F. (1999). Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. Journal of Virology, 73, 1460–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di, J. , Du, Z. , Wu, K. , Jin, S. , Wang, X. , Li, T. , & Xu, Y. (2022). Biodistribution and non‐linear gene expression of mRNA LNPs affected by delivery route and particle size. Pharmaceutical Research, 39, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, R. M. , Galivo, F. , Kottke, T. , Wongthida, P. , Qiao, J. , Thompson, J. , Valdes, M. , Barber, G. , & Vile, R. G. (2007). Oncolytic immunovirotherapy for melanoma using vesicular stomatitis virus. Cancer Research, 67, 2840–2848. [DOI] [PubMed] [Google Scholar]
- Dietz, L. , Oberländer, J. , Mateos‐Maroto, A. , Schunke, J. , Fichter, M. , Krämer‐Albers, E. M. , Landfester, K. , & Mailänder, V. (2023). Uptake of extracellular vesicles into immune cells is enhanced by the protein corona. Journal of Extracellular Vesicles, 12(12), e12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioufa, N. , Clark, A. M. , Ma, B. , Beckwitt, C. H. , & Wells, A. (2017). Bi‐directional exosome‐driven intercommunication between the hepatic niche and cancer cells. Molecular Cancer, 16, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominiak, A. , Chełstowska, B. , Olejarz, W. , & Nowicka, G. (2020). Communication in the cancer microenvironment as a target for therapeutic interventions. Cancers, 12, 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheva, N. T. , Palasca, O. , Yarani, R. , Litman, T. , Anthon, C. , Groenen, M. A. M. , Stadler, P. F. , Pociot, F. , Jensen, L. J. , & Gorodkin, J. (2021). Human pathways in animal models: possibilities and limitations. Nucleic Acids Research, 49, 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert, O. , Shinozaki, K. , Huang, T. G. , Savontaus, M. J. , García‐Sastre, A. , & Woo, S. L. (2003). Oncolytic vesicular stomatitis virus for treatment of orthotopic hepatocellular carcinoma in immune‐competent rats. Cancer Research, 63, 3605–3611. [PubMed] [Google Scholar]
- Ebrahim, T. , Ebrahim, A. S. , & Kandouz, M. (2024). Diversity of intercellular communication modes: A cancer biology perspective. Cells, 13, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echigo, H. , Mishiro, K. , Munekane, M. , Fuchigami, T. , Kitamura, Y. , Kinuya, S. , & Ogawa, K. (2022). Development and evaluation of a theranostic probe with RGD peptide introduced platinum complex to enable tumor‐specific accumulation. Bioorganic & Medicinal Chemistry, 70, 116919. [DOI] [PubMed] [Google Scholar]
- Eftekhari, A. , Kryschi, C. , Pamies, D. , Gulec, S. , Ahmadian, E. , Janas, D. , Davaran, S. , & Khalilov, R. (2023). Natural and synthetic nanovectors for cancer therapy. Nanotheranostics, 7, 236–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi, S. , Takefuji, M. , Sakaguchi, T. , Ishihama, S. , Mori, Y. , Tsuda, T. , Takikawa, T. , Yoshida, T. , Ohashi, K. , Shimizu, Y. , Hayashida, R. , Kondo, K. , Bando, Y. K. , Ouchi, N. , & Murohara, T. (2019). Cardiomyocytes capture stem cell‐derived, anti‐apoptotic microRNA‐214 via clathrin‐mediated endocytosis in acute myocardial infarction. The Journal of Biological Chemistry, 294, 11665–11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgner, F. , Ren, H. , Medvedev, R. , Ploen, D. , Himmelsbach, K. , Boller, K. , & Hildt, E. (2016). The intracellular cholesterol transport inhibitor U18666A inhibits the exosome‐dependent release of mature hepatitis C virus. Journal of Virology, 90, 11181–11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, R. O. , & He, M. (2021). Unlocking the power of exosomes for crossing biological barriers in drug delivery. Pharmaceutics, 13, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsharkasy, O. M. , Nordin, J. Z. , Hagey, D. W. , de Jong, O. G. , Schiffelers, R. M. , Andaloussi, S. E. , & Vader, P. (2020). Extracellular vesicles as drug delivery systems: Why and how?. Advanced Drug Delivery Reviews, 159, 332–343. [DOI] [PubMed] [Google Scholar]
- Erickson, M. A. , Shulyatnikova, T. , Banks, W. A. , & Hayden, M. R. (2023). Ultrastructural remodeling of the blood‐brain barrier and neurovascular unit by lipopolysaccharide‐induced neuroinflammation. International Journal Of Molecular Sciences, 24, 1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erikson, A. , Tufto, I. , Bjønnum, A. B. , Bruland, Ø. S. , & Davies, C. (2008). The impact of enzymatic degradation on the uptake of differently sized therapeutic molecules. Anticancer Research, 28(6A), 3557–3566. [PubMed] [Google Scholar]
- Escola, J. M. , Kleijmeer, M. J. , Stoorvogel, W. , Griffith, J. M. , Yoshie, O. , & Geuze, H. J. (1998). Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B‐lymphocytes. The Journal of Biological Chemistry, 273, 20121–20127. [DOI] [PubMed] [Google Scholar]
- Escrevente, C. , Keller, S. , Altevogt, P. , & Costa, J. (2011). Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer, 11, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili, A. , Alini, M. , Baghaban Eslaminejad, M. , & Hosseini, S. (2022). Engineering strategies for customizing extracellular vesicle uptake in a therapeutic context. Stem Cell Research &Therapy, 13, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estapé Senti, M. , de Jongh, C. A. , Dijkxhoorn, K. , Verhoef, J. J. F. , Szebeni, J. , Storm, G. , Hack, C. E. , Schiffelers, R. M. , Fens, M. H. , & Boross, P. (2022). Anti‐PEG antibodies compromise the integrity of PEGylated lipid‐based nanoparticles via complement. Journal of Controlled Release : Official Journal of the Controlled Release Society, 341, 475–486. [DOI] [PubMed] [Google Scholar]
- Evans, R. A. (2007). The rise of azide‐alkyne 1,3‐dipolar ‘click’ cycloaddition and its application to polymer science and surface modification. Australian Journal of Chemistry, 60, 384–395. [Google Scholar]
- Feng, D. , Zhao, W. L. , Ye, Y. Y. , Bai, X. C. , Liu, R. Q. , Chang, L. F. , Zhou, Q. , & Sui, S. F. (2010). Cellular internalization of exosomes occurs through phagocytosis. Traffic, 11, 675–687. [DOI] [PubMed] [Google Scholar]
- Fernández‐Pereira, C. , San Millán‐Tejado, B. , Gallardo‐Gómez, M. , Pérez‐Márquez, T. , Alves‐Villar, M. , Melcón‐Crespo, C. , Fernández‐Martín, J. , & Ortolano, S. (2021). Therapeutic approaches in lysosomal storage diseases. Biomolecules, 11, 1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo, P. , Lepland, A. , Scodeller, P. , Fontana, F. , Torrieri, G. , Tiboni, M. , Shahbazi, M. A. , Casettari, L. , Kostiainen, M. A. , Hirvonen, J. , Teesalu, T. , & Santos, H. A. (2021). Peptide‐guided resiquimod‐loaded lignin nanoparticles convert tumor‐associated macrophages from M2 to M1 phenotype for enhanced chemotherapy. Acta Biomater rialia, 133, 231–243. [DOI] [PubMed] [Google Scholar]
- Filipović, L. , Spasojević, M. , Prodanović, R. , Korać, A. , Matijaševic, S. , Brajušković, G. , de Marco, A. , & Popović, M. (2022). Affinity‐based isolation of extracellular vesicles by means of single‐domain antibodies bound to macroporous methacrylate‐based copolymer. New Biotechnology, 69, 36–48. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, W. , Freeman, M. L. , Lederman, M. M. , Vasilieva, E. , Romero, R. , & Margolis, L. (2018). A system of cytokines encapsulated in extracellular vesicles. Scientific Reports, 8, 8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzner, D. , Schnaars, M. , van Rossum, D. , Krishnamoorthy, G. , Dibaj, P. , Bakhti, M. , Regen, T. , Hanisch, U. K. , & Simons, M. (2011). Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. Journal of Cell Science, 124, 447–458. [DOI] [PubMed] [Google Scholar]
- Flemming, J. P. , Hill, B. L. , Haque, M. W. , Raad, J. , Bonder, C. S. , Harshyne, L. A. , Rodeck, U. , Luginbuhl, A. , Wahl, J. K. 3rd , Tsai, K. Y. , Wermuth, P. J. , Overmiller, A. M. , & Mahoney, M. G. (2020). miRNA‐ and cytokine‐associated extracellular vesicles mediate squamous cell carcinomas. Journal of Extracellular Vesicles, 9, 1790159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X. , Shi, Y. , Qi, T. , Qiu, S. , Huang, Y. , Zhao, X. , Sun, Q. , & Lin, G. (2020). Precise design strategies of nanomedicine for improving cancer therapeutic efficacy using subcellular targeting. Signal Transductional and Targeted Therapy, 5, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon, J. , & Bruni, D. (2019). Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nature Reviews. Drug Discovery, 18, 197–218. [DOI] [PubMed] [Google Scholar]
- Gao, S. , Yang, D. , Fang, Y. , Lin, X. , Jin, X. , Wang, Q. , Wang, X. , Ke, L. , & Shi, K. (2019). Engineering nanoparticles for targeted remodeling of the tumor microenvironment to improve cancer immunotherapy. Theranostics, 9, 126–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. , Ran, N. , Dong, X. , Zuo, B. , Yang, R. , Zhou, Q. , Moulton, H. M. , Seow, Y. , & Yin, H. (2018). Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Science Translational Medicine, 10, eaat0195. [DOI] [PubMed] [Google Scholar]
- García‐Silva, S. , Benito‐Martín, A. , Nogués, L. , Hernández‐Barranco, A. , Mazariegos, M. S. , Santos, V. , Hergueta‐Redondo, M. , Ximénez‐Embún, P. , Kataru, R. P. , Lopez, A. A. , Merino, C. , Sánchez‐Redondo, S. , Graña‐Castro, O. , Matei, I. , Nicolás‐Avila, J. Á. , Torres‐Ruiz, R. , Rodríguez‐Perales, S. , Martínez, L. , Pérez‐Martínez, M. , … Peinado, H. (2021). Melanoma‐derived small extracellular vesicles induce lymphangiogenesis and metastasis through an NGFR‐dependent mechanism. Nature Cancer, 2, 1387–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo, M. , Saari, H. , Somersalo, P. , Crescenti, D. , Kuryk, L. , Aksela, L. , Capasso, C. , Madetoja, M. , Koskinen, K. , Oksanen, T. , Mäkitie, A. , Jalasvuori, M. , Cerullo, V. , Ciana, P. , & Yliperttula, M. (2018). Antitumor effect of oncolytic virus and paclitaxel encapsulated in extracellular vesicles for lung cancer treatment. Journal of Controlled Release : Official Journal of the Controlled Release Society, 283, 223–234. [DOI] [PubMed] [Google Scholar]
- Goetz, J. G. , Lajoie, P. , Wiseman, S. M. , & Nabi, I. R. (2008). Caveolin‐1 in tumor progression: the good, the bad and the ugly. Cancer Metastasis Reviews, 27, 715–735. [DOI] [PubMed] [Google Scholar]
- Gomes, F. G. , Andrade, A. C. , Wolf, M. , Hochmann, S. , Krisch, L. , Maeding, N. , Regl, C. , Poupardin, R. , Ebner‐Peking, P. , Huber, C. G. , Meisner‐Kober, N. , Schallmoser, K. , & Strunk, D. (2022). Synergy of human platelet‐derived extracellular vesicles with secretome proteins promotes regenerative functions. Biomedicines, 10, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górnicki, T. , Lambrinow, J. , Golkar‐Narenji, A. , Data, K. , Domagała, D. , Niebora, J. , Farzaneh, M. , Mozdziak, P. , Zabel, M. , Antosik, P. , Bukowska, D. , Ratajczak, K. , Podhorska‐Okołów, M. , Dzięgiel, P. , & Kempisty, B. (2024). Biomimetic scaffolds‐a novel approach to three dimensional cell culture techniques for potential implementation in tissue engineering. Nanomaterial, 14, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S. J. , Booth, A. M. , & Hildreth, J. E. (2003). The Trojan exosome hypothesis. Proceedings of the National Academy of Sciences of the United States of America, 100, 10592–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, N. , & Tramontano, E. (2018). HERV envelope proteins: Physiological role and pathogenic potential in cancer and autoimmunity. Frontiers in Microbiology, 9, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer, S. , van Deurs, B. , & Sandvig, K. (2002). Membrane ruffling and macropinocytosis in A431 cells require cholesterol. Journal of Cell Science, 115, 2953–2962. [DOI] [PubMed] [Google Scholar]
- Guidotti, G. , Brambilla, L. , & Rossi, D. (2017). Cell‐penetrating peptides: From basic research to clinics. Trends in Pharmacological Sciences, 38, 406–424. [DOI] [PubMed] [Google Scholar]
- Gupta, R. , Toufaily, C. , & Annabi, B. (2014). Caveolin and cavin family members: Dual roles in cancer. Biochimie, 107, 188–202. [DOI] [PubMed] [Google Scholar]
- Gurung, S. , Perocheau, D. , Touramanidou, L. , & Baruteau, J. (2021). The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Communication and Signaling : CCS, 19, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasath, P. , Kim, J. , Gunassekaran, G. R. , Chi, L. , Kim, S. , Park, R. W. , Kim, S. H. , Baek, M. C. , Bae, S. M. , Kim, S. Y. , Kim, D. K. , Park, I. K. , Kim, W. J. , & Lee, B. (2017). Interleukin‐4 receptor‐targeted delivery of Bcl‐xL siRNA sensitizes tumors to chemotherapy and inhibits tumor growth. Biomaterials, 142, 101–111. [DOI] [PubMed] [Google Scholar]
- György, B. , Módos, K. , Pállinger, E. , Pálóczi, K. , Pásztói, M. , Misják, P. , Deli, M. A. , Sipos, A. , Szalai, A. , Voszka, I. , Polgár, A. , Tóth, K. , Csete, M. , Nagy, G. , Gay, S. , Falus, A. , Kittel, A. , & Buzás, E. I. (2011). Detection and isolation of cell‐derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood, 117, e39–48. [DOI] [PubMed] [Google Scholar]
- Hade, M. D. , Suire, C. N. , & Suo, Z. (2023). Effective peptide‐based platform for efficient exosomal loading and cellular delivery of a microRNA. ACS Applied Materials & Interfaces, 15, 3851–3866. [DOI] [PubMed] [Google Scholar]
- Hamilton, A. , Biganzoli, L. , Coleman, R. , Mauriac, L. , Hennebert, P. , Awada, A. , Nooij, M. , Beex, L. , Piccart, M. , v an Hoorebeeck, I. , Bruning, P. , & de Valeriola, D. (2002). EORTC 10968: a phase I clinical and pharmacokinetic study of polyethylene glycol liposomal doxorubicin (Caelyx, Doxil) at a 6‐week interval in patients with metastatic breast cancer. European Organization for Research and Treatment of Cancer. Annals of Oncology : Official Journal of the European Society for Medical Oncology, 13, 910–918. [DOI] [PubMed] [Google Scholar]
- Han, J. , Pluhackova, K. , & Böckmann, R. A. (2017). The multifaceted role of SNARE proteins in membrane fusion. Frontiers in Physiology, 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M. , Zhang, Z. , Liu, Z. , Liu, Y. , Zhao, H. , Wang, B. , Zhang, C. , Shang, H. , Li, Y. , Wang, S. , & Xin, T. (2023). Three‐dimensional‐cultured MSC‐derived exosome with hydrogel for cerebral ischemia repair. Biomaterial Advances, 149, 213396. [DOI] [PubMed] [Google Scholar]
- Hao, S. , Bai, O. , Li, F. , Yuan, J. , Laferte, S. , & Xiang, J. (2007). Mature dendritic cells pulsed with exosomes stimulate efficient cytotoxic T‐lymphocyte responses and antitumour immunity. Immunology, 120, 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraszti, R. A. , Didiot, M. C. , Sapp, E. , Leszyk, J. , Shaffer, S. A. , Rockwell, H. E. , Gao, F. , Narain, N. R. , DiFiglia, M. , Kiebish, M. A. , Aronin, N. , & Khvorova, A. (2016). High‐resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. Journal of Extracellular Vesicles, 5, 32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, J. M. , & Chess, R. B. (2003). Effect of pegylation on pharmaceuticals. Nature Reviews. Drug Discovery, 2, 214–221. [DOI] [PubMed] [Google Scholar]
- Harrison, R. E. , Bucci, C. , Vieira, O. V. , Schroer, T. A. , & Grinstein, S. (2003). Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Molecular and Cellular Biology, 23, 6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasley, L. E. (2001). Autocrine and paracrine signaling through neuropeptide receptors in human cancer. Oncogene, 20, 1563–1569. [DOI] [PubMed] [Google Scholar]
- Hemler, M. E. (2003). Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annual Review of Cell and Developmental Biology, 19, 397–422. [DOI] [PubMed] [Google Scholar]
- Heusermann, W. , Hean, J. , Trojer, D. , Steib, E. , von Bueren, S. , Graff‐Meyer, A. , Genoud, C. , Martin, K. , Pizzato, N. , Voshol, J. , Morrissey, D. V. , Andaloussi, S. E. , Wood, M. J. , & Meisner‐Kober, N. C. (2016). Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. The Journal of Cell Biology, 213, 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau‐Véchot, J. , Rafii, A. , Touboul, C. , & Pasquier, J. (2018). Halfway between 2D and animal models: are 3D cultures the ideal tool to study cancer‐microenvironment interactions?. International Journal Of Molecular Sciences, 19, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller, O. , Bolourani, P. , Clark, J. , Stephens, L. R. , Hawkins, P. T. , Weiner, O. D. , Weeks, G. , & Kay, R. R. (2013). Two distinct functions for PI3‐kinases in macropinocytosis. Journal of Cell Science, 126(Pt 18), 4296–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, A. S. (2008). The origins and evolution of “controlled” drug delivery systems. Journal of Controlled Release : Official Journal of the Controlled Release Society, 132, 153–163. [DOI] [PubMed] [Google Scholar]
- Horibe, S. , Tanahashi, T. , Kawauchi, S. , Murakami, Y. , & Rikitake, Y. (2018). Mechanism of recipient cell‐dependent differences in exosome uptake. BMC Cancer, 18, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino, A. , Costa‐Silva, B. , Shen, T. L. , Rodrigues, G. , Hashimoto, A. , Tesic Mark, M. , Molina, H. , Kohsaka, S. , Di Giannatale, A. , Ceder, S. , Singh, S. , Williams, C. , Soplop, N. , Uryu, K. , Pharmer, L. , King, T. , Bojmar, L. , Davies, A. E. , Ararso, Y. , Zhang, T., … Lyden, D. (2015). Tumour exosome integrins determine organotropic metastasis. Nature, 527, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini, N. F. , Amini, R. , Ramezani, M. , Saidijam, M. , Hashemi, S. M. , & Najafi, R. (2022). AS1411 aptamer‐functionalized exosomes in the targeted delivery of doxorubicin in fighting colorectal cancer. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 155, 113690. [DOI] [PubMed] [Google Scholar]
- Hu, S. , Ma, J. , Su, C. , Chen, Y. , Shu, Y. , Qi, Z. , Zhang, B. , Shi, G. , Zhang, Y. , Zhang, Y. , Huang, A. , Kuang, Y. , & Cheng, P. (2021). Engineered exosome‐like nanovesicles suppress tumor growth by reprogramming tumor microenvironment and promoting tumor ferroptosis. Acta Biomater ialia, 135, 567–581. [DOI] [PubMed] [Google Scholar]
- Hung, M. E. , & Leonard, J. N. (2015). Stabilization of exosome‐targeting peptides via engineered glycosylation. Journal of Biological Chemistry, 290, 8166–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, M. E. , & Leonard, J. N. (2016). A platform for actively loading cargo RNA to elucidate limiting steps in EV‐mediated delivery. Journal of Extracellular Vesicles, 5, 31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz, S. N. , Conlon, M. M. , Rider, M. A. , Brownstein, N. C. , & Meckes, D. G. Jr. (2016). Nanoparticle analysis sheds budding insights into genetic drivers of extracellular vesicle biogenesis. Journal of Extracellular Vesicles, 5, 31295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im, S. J. , Hashimoto, M. , Gerner, M. Y. , Lee, J. , Kissick, H. T. , Burger, M. C. , Shan, Q. , Hale, J. S. , Lee, J. , Nasti, T. H. , Sharpe, A. H. , Freeman, G. J. , Germain, R. N. , Nakaya, H. I. , Xue, H. H. , & Ahmed, R. (2016). Defining CD8+ T cells that provide the proliferative burst after PD‐1 therapy. Nature, 537, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, W. F. (2022). Endothelial ion channels and cell‐cell communication in the microcirculation. Frontiers in Physiology, 13, 805149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari, S. , Maleki Dizaj, S. , & Adibkia, K. (2015). Cell‐penetrating peptides and their analogues as novel nanocarriers for drug delivery. Bioimpacts: BI, 5, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, R. , Lang, T. , & Südhof, T. C. (2003). Membrane fusion. Cell, 112, 519–533. [DOI] [PubMed] [Google Scholar]
- Jahn, R. , & Scheller, R. H. (2006). SNAREs–Engines for membrane fusion. Nature Reviews. Molecular Cell Biology, 7, 631–643. [DOI] [PubMed] [Google Scholar]
- Jeon, J. O. , Kim, S. , Choi, E. , Shin, K. , Cha, K. , So, I. S. , Kim, S. J. , Jun, E. , Kim, D. , Ahn, H. J. , Lee, B. H. , Lee, S. H. , & Kim, I. S. (2013). Designed nanocage displaying ligand‐specific Peptide bunches for high affinity and biological activity. ACS Nano, 7, 7462–7471. [DOI] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Q. , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177, 428–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Hvam, M. L. , Primdahl‐Bengtson, B. , Boysen, A. T. , Whitehead, B. , Dyrskjøt, L. , Orntoft, T. F. , Howard, K. A. , & Ostenfeld, M. S. (2014). Comparative analysis of discrete exosome fractions obtained by differential centrifugation. Journal of Extracellular Vesicles, 3, 25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Zhang, Q. , Franklin, J. L. , & Coffey, R. J. (2023). Extracellular vesicles and nanoparticles: emerging complexities. Trends in Cell Biology, 33, 667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H. , Zhao, H. , Zhang, M. , He, Y. , Li, X. , Xu, Y. , & Liu, X. (2022). Hypoxia induced changes of exosome cargo and subsequent biological effects. Frontiers in Immunology, 13, 824188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. , Guo, K. , Chen, Y. , & Xiang, N. (2024). Droplet microfluidics for current cancer research: from single‐cell analysis to 3D cell culture. ACS Biomaterials Science & Engineering, 10, 1335–1354. [DOI] [PubMed] [Google Scholar]
- Jiang, Q. , Liu, Y. , Guo, R. , Yao, X. , Sung, S. , Pang, Z. , & Yang, W. (2019). Erythrocyte‐cancer hybrid membrane‐camouflaged melanin nanoparticles for enhancing photothermal therapy efficacy in tumors. Biomaterials, 192, 292–308. [DOI] [PubMed] [Google Scholar]
- Jiang, S. , Fagman, J. B. , Chen, C. , Alberti, S. , & Liu, B. (2020). Protein phase separation and its role in tumorigenesis. Elife, 9, e60264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. , Krishnan, N. , Zhou, J. , Chekuri, S. , Wei, X. , Kroll, A. V. , Yu, C. L. , Duan, Y. , Gao, W. , Fang, R. H. , & Zhang, L. (2020). Engineered cell‐membrane‐coated nanoparticles directly present tumor antigens to promote anticancer immunity. Advanced Materials, 32, e2001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. , Liu, X. , Ye, J. , Ma, Y. , Mao, J. , Feng, D. , & Wang, X. (2023). Migrasomes, a new mode of intercellular communication. Cell communication and signaling : CCS, 21, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, G. , Hong, W. , Guo, Y. , Bai, Y. , & Chen, B. (2020). Molecular mechanism of pancreatic stellate cells activation in chronic pancreatitis and pancreatic cancer. Journal of Cancer, 11, 1505–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, V. , Vasu, S. , Kumar, U. S. , & Kumar, M. (2023). Surface‐engineered extracellular vesicles in cancer immunotherapy. Cancers, 15, 2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone, R. M. (1992). The Jeanne Manery‐Fisher Memorial Lecture 1991. Maturation of reticulocytes: formation of exosomes as a mechanism for shedding membrane proteins. Biochemistry and Cell Biology = Biochimie et Biologie Cellulaire, 70, 179–190. [DOI] [PubMed] [Google Scholar]
- Johnstone, R. M. , Adam, M. , Hammond, J. R. , Orr, L. , & Turbide, C. (1987). Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). Journal of Biological Chemistry, 262, 9412–9420. [PubMed] [Google Scholar]
- Joshi, B. S. , de Beer, M. A. , Giepmans, B. N. G. , & Zuhorn, I. S. (2020). Endocytosis of extracellular vesicles and release of their cargo from endosomes. ACS Nano, 14, 4444–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J. (2014). Human tumor xenograft models for preclinical assessment of anticancer drug development. Toxicological research, 30, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarla, R. , Hur, J. , Kim, Y. J. , Kim, J. , & Chwae, Y.‐J. (2020). Apoptotic cell‐derived exosomes: Messages from dying cells. Experimental & Molecular Medicine, 52, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri, R. (2016). The biology and function of exosomes in cancer. Journal of Clinical Investigation, 126, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra, H. , Drummen, G. P. , & Mathivanan, S. (2016). Focus on extracellular vesicles: introducing the next small big thing. International Journal of Molecular Sciences, 17, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaly, N. , Farokhzad, O. C. , & Corbo, C. (2022). Nanoparticle protein corona evolution: from biological impact to biomarker discover. Nanoscale, 14, 1606–1620. [DOI] [PubMed] [Google Scholar]
- Kamaly, N. , Yameen, B. , Wu, J. , & Farokhzad, O. C. (2016). Degradable controlled‐release polymers and polymeric nanoparticles: Mechanisms of controlling drug release. Chemical Reviews, 116, 2602–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanada, M. , Bachmann, M. H. , & Contag, C. H. (2016). Signaling by extracellular vesicles advances cancer hallmarks. Trends in Cancer, 2, 84–94. [DOI] [PubMed] [Google Scholar]
- Kang, J. H. , Jung, M. Y. , Yin, X. , Andrianifahanana, M. , Hernandez, D. M. , & Leof, E. B. (2017). Cell‐penetrating peptides selectively targeting SMAD3 inhibit profibrotic TGF‐β signaling. The Journal of Clinical Investigation, 127, 2541–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan, A. , Krishnan, A. , Ali, M. , Subramaniam, S. , Halagowder, D. , & Sivasithamparam, N. D. (2014). Caveolin‐1 promotes gastric cancer progression by up‐regulating epithelial to mesenchymal transition by crosstalk of signalling mechanisms under hypoxic condition. European Journal of Cancer, 50, 204–215. [DOI] [PubMed] [Google Scholar]
- Kanuma, T. , Yamamoto, T. , Kobiyama, K. , Moriishi, E. , Masuta, Y. , Kusakabe, T. , Ozasa, K. , Kuroda, E. , Jounai, N. , & Ishii, K. J. (2017). CD63‐mediated antigen delivery into extracellular vesicles via DNA vaccination results in robust CD8+ T Cell responses. Journal of Immunology, 198, 4707–4715. [DOI] [PubMed] [Google Scholar]
- Karmali, P. P. , Kotamraju, V. R. , Kastantin, M. , Black, M. , Missirlis, D. , Tirrell, M. , & Ruoslahti, E. (2009). Targeting of albumin‐embedded paclitaxel nanoparticles to tumors. Nanomedicine : Nanotechnology, Biology, and Medicine, 5, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasmapour, B. , Cai, L. , & Gutierrez, M. G. (2013). Spatial distribution of phagolysosomes is independent of the regulation of lysosome position by Rab34. The International Journal Of Biochemistry & Cell Biology, 45, 2057–2065. [DOI] [PubMed] [Google Scholar]
- Kasmapour, B. , Gronow, A. , Bleck, C. K. , Hong, W. , & Gutierrez, M. G. (2012). Size‐dependent mechanism of cargo sorting during lysosome‐phagosome fusion is controlled by Rab34. Proceedings of the National Academy of Sciences of the United States of America, 109, 20485–20490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, N. , Sato, T. , Fuchigami, Y. , Suga, T. , Geng, L. , Tsurumaru, M. , Hagimori, M. , Mukai, H. , & Kawakami, S. (2022). Synthesis and evaluation of a novel adapter lipid derivative for preparation of cyclic peptide‐modified PEGylated liposomes: application of cyclic RGD peptide European Journal of Pharmaceutical Sciences : Official Journal of the European Federation for Pharmaceutical Sciences, 176, 106239. [DOI] [PubMed] [Google Scholar]
- Kawikova, I. , & Askenase, P. W. (2015). Diagnostic and therapeutic potentials of exosomes in CNS diseases. Brain Research, 1617, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, S. , Ridinger, J. , Rupp, A. K. , Janssen, J. W. , & Altevogt, P. (2011). Body fluid derived exosomes as a novel template for clinical diagnostics. Journal of Translational Medicine, 9, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, J. M. , Emerson, S. U. , & Wagner, R. R. (1972). The glycoprotein of vesicular stomatitis virus is the antigen that gives rise to and reacts with neutralizing antibody. Journal of Virology, 10, 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. , Kitamatsu, M. , & Ohtsuki, T. (2018). Enhanced intracellular peptide delivery by multivalent cell‐penetrating peptide with bioreducible linkage. Bioorganic & Medicinal Chemistry Letters, 28, 378–381. [DOI] [PubMed] [Google Scholar]
- Kim, J. H. , Bae, S. M. , Na, M. H. , Shin, H. , Yang, Y. J. , Min, K. H. , Choi, K. Y. , Kim, K. , Park, R. W. , Kwon, I. C. , Lee, B. H. , Hoffman, A. S. , & Kim, I. S. (2012). Facilitated intracellular delivery of peptide‐guided nanoparticles in tumor tissues. Journal of Controlled Release : Official Journal of the Controlled Release Society, 157, 493–499. [DOI] [PubMed] [Google Scholar]
- Kim, J. H. , Lee, J. , Park, J. , & Gho, Y. S. (2015). Gram‐negative and Gram‐positive bacterial extracellular vesicles. Seminars in Cell & Developmental Biology, 40, 97–104. [DOI] [PubMed] [Google Scholar]
- Kim, M. S. , Haney, M. J. , Zhao, Y. , Mahajan, V. , Deygen, I. , Klyachko, N. L. , Inskoe, E. , Piroyan, A. , Sokolsky, M. , Okolie, O. , Hingtgen, S. D. , Kabanov, A. V. , & Batrakova, E. V. (2016). Development of exosome‐encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine, 12, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. Y. , Khanal, D. , Kalionis, B. , & Chrzanowski, W. (2019). High‐fidelity probing of the structure and heterogeneity of extracellular vesicles by resonance‐enhanced atomic force microscopy infrared spectroscopy. Nature Protocols, 14, 576–593. [DOI] [PubMed] [Google Scholar]
- Kirchhausen, T. (2000). Clathrin. Annual Review of Biochemistry, 69, 699–727. [DOI] [PubMed] [Google Scholar]
- Kirchhausen, T. , Owen, D. , & Harrison, S. C. (2014). Molecular structure, function, and dynamics of clathrin‐mediated membrane traffic. Cold Spring Harbor Perspectives in Biology, 6, a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, R. (2012). Targeted therapies: the toxic reality of new drugs. Nature Reviews. Clinical Oncology, 9, 488. [DOI] [PubMed] [Google Scholar]
- Kiss, A. L. , & Botos, E. (2009). Endocytosis via caveolae: alternative pathway with distinct cellular compartments to avoid lysosomal degradation?. Journal of Cellular and Molecular Medicine, 13, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, H. , & Brechbiel, M. W. (2005). Nano‐sized MRI contrast agents with dendrimer cores. Advanced Drug Delivery Reviews, 57(15), 2271–2286. [DOI] [PubMed] [Google Scholar]
- Kohn, D. B. , Chen, Y. Y. , & Spencer, M. J. (2023). Successes and challenges in clinical gene therapy. Gene Therapy, 30, 738–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb, H. C. , Finn, M. G. , & Sharpless, K. B. (2001). Click chemistry: diverse chemical function from a few good reactions. Angewandte Chemie, 40, 2004–2021. [DOI] [PubMed] [Google Scholar]
- Konerding, M. A. , Malkusch, W. , Klapthor, B. , van Ackern, C. , Fait, E. , Hill, S. A. , Parkins, C. , Chaplin, D. J. , Presta, M. , & Denekamp, J. (1999). Evidence for characteristic vascular patterns in solid tumours: quantitative studies using corrosion casts. British Journal of Cancer, 80, 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, P. , Cui, Z. Y. , Huang, X. F. , Zhang, D. D. , Guo, R. J. , & Han, M. (2022). Inflammation and atherosclerosis: Signaling pathways and therapeutic intervention. Signal Transductional and Targeted Therapy, 7, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopac, T. (2021). Protein corona, understanding the nanoparticle‐protein interactions and future perspectives: A critical review. International Journal of Biological Macromolecules, 169, 290–301. [DOI] [PubMed] [Google Scholar]
- Kozma, G. T. , Shimizu, T. , Ishida, T. , & Szebeni, J. (2020). Anti‐PEG antibodies: Properties, formation, testing and role in adverse immune reactions to PEGylated nano‐biopharmaceuticals. Advanced Drug Delivery Reviews, 154–155, 163–175. [DOI] [PubMed] [Google Scholar]
- Krishna, M. , & Nadler, S. G. (2016). Immunogenicity to biotherapeutics—the role of anti‐drug immune complexes. Frontiers in Immunology, 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbach, C. , da Luz, S. , Baganz, F. , Hass, V. C. , & Mueller, M. M. (2018). A microfluidic system for the investigation of tumor cell extravasation. Bioengineering, 5, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose, A. E. , Hu, W. , Nguyen, K. T. , & Menon, J. U. (2019). Scaffold‐based lung tumor culture on porous PLGA microparticle substrates. PLoS One, 14, e0217640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, H. , Tachikawa, M. , Yagi, Y. , Umetsu, M. , Nurdin, A. , Miyauchi, E. , Watanabe, M. , Uchida, Y. , & Terasaki, T. (2019). Cluster of differentiation 46 is the major receptor in human blood‐brain barrier endothelial cells for uptake of exosomes derived from brain‐metastatic melanoma cells (SK‐Mel‐28) Molecular Pharmaceutics, 16, 292–304. [DOI] [PubMed] [Google Scholar]
- Kutova, O. M. , Guryev, E. L. , Sokolova, E. A. , Alzeibak, R. , & Balalaeva, I. V. (2019). Targeted delivery to tumors: Multidirectional strategies to improve treatment efficiency. Cancers, 11, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C. P. , Kim, E. Y. , Badr, C. E. , Weissleder, R. , Mempel, T. R. , Tannous, B. A. , & Breakefield, X. O. (2015). Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nature Communications, 6, 7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie, P. , & Nabi, I. R. (2007). Regulation of raft‐dependent endocytosis. Journal of Cellular and Molecular Medicine, 11, 644–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S. , Linton, L. M. , Birren, B. , Nusbaum, C. , Zody, M. C. , Baldwin, J. , Devon, K. , Dewar, K. , Doyle, M. , FitzHugh, W. , Funke, R. , Gage, D. , Harris, K. , Heaford, A. , Howland, J. , Kann, L. , Lehoczky, J. , LeVine, R. , McEwan, P. , McKernan, K., … International Human Genome Sequencing Consortium . (2001). Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Langer, E. M. , Allen‐Petersen, B. L. , King, S. M. , Kendsersky, N. D. , Turnidge, M. A. , Kuziel, G. M. , Riggers, R. , Samatham, R. , Amery, T. S. , Jacques, S. L. , Sheppard, B. C. , Korkola, J. E. , Muschler, J. L. , Thibault, G. , Chang, Y. H. , Gray, J. W. , Presnell, S. C. , Nguyen, D. G. , & Sears, R. C. (2019). Modeling tumor phenotypes in vitro with three‐dimensional bioprinting. Cell Reports, 26, 608–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattmann, E. , & Levesque, M. P. (2022). The role of extracellular vesicles in melanoma progression. Cancers, 14, 3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro‐Ibáñez, E. , Lunavat, T. R. , Jang, S. C. , Escobedo‐Lucea, C. , Oliver‐De La Cruz, J. , Siljander, P. , Lötvall, J. , & Yliperttula, M. (2017). Distinct prostate cancer‐related mRNA cargo in extracellular vesicle subsets from prostate cell lines. BMC Cancer, 17, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary, N. , Walser, S. , He, Y. , Cousin, N. , Pereira, P. , Gallo, A. , Collado‐Diaz, V. , Halin, C. , Garcia‐Silva, S. , Peinado, H. , & Dieterich, L. C. (2022). Melanoma‐derived extracellular vesicles mediate lymphatic remodelling and impair tumour immunity in draining lymph nodes. Journal of Extracellular Vesicles, 11, e12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. Y. , Amirthalingam, S. , Lee, C. , Rajendran, A. K. , Ahn, Y. H. , & Hwang, N. S. (2023). Strategies for targeted gene delivery using lipid nanoparticles and cell‐derived nanovesicles. Nanoscale Advances, 5, 3834–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Jo, D. H. , Kim, J. H. , Cho, C. S. , Han, J. E. , Kim, Y. , Park, H. , Yoo, S. H. , Yu, Y. S. , Moon, H. E. , Park, H. R. , Kim, D. G. , Kim, J. H. , & Paek, S. H. (2019). Development of a patient‐derived xenograft model of glioblastoma via intravitreal injection in mice. Experimental & Molecular Medicine, 51, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. H. , Song, J. , Kim, I. G. , You, G. , Kim, H. , Ahn, J. H. , & Mok, H. (2022). Exosome‐mediated delivery of transforming growth factor‐β receptor 1 kinase inhibitors and toll‐like receptor 7/8 agonists for combination therapy of tumors. Acta Biomater ialia, 141, 354–363. [DOI] [PubMed] [Google Scholar]
- Lee, Y. J. , Shin, K. J. , & Chae, Y. C. (2024). Regulation of cargo selection in exosome biogenesis and its biomedical applications in cancer. Experimental & Molecular Medicine, 56, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Pinilla‐Macua, I. , Ouyang, Y. , Sadovsky, E. , Kajiwara, K. , Sorkin, A. , & Sadovsky, Y. (2020). Internalization of trophoblastic small extracellular vesicles and detection of their miRNA cargo in P‐bodies. Journal of Extracellular Vesicles, 9, 1812261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Zhen, X. , Lyu, Y. , Jiang, Y. , Huang, J. , & Pu, K. (2018). Cell membrane coated semiconducting polymer nanoparticles for enhanced multimodal cancer phototheranostics. ACS Nano, 12, 8520–8530. [DOI] [PubMed] [Google Scholar]
- Li, M. , Mei, S. , Yang, Y. , Shen, Y. , & Chen, L. (2022). Strategies to mitigate the on‐ and off‐target toxicities of recombinant immunotoxins: An antibody engineering perspective. Antibody Therapeutics, 5, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Nicol, F. , & Szoka, F. C. Jr. (2004). GALA: a designed synthetic pH‐responsive amphipathic peptide with applications in drug and gene delivery. Drug Delivery Reviews, 56, 967–985. [DOI] [PubMed] [Google Scholar]
- Li, W. , Zhu, J. , Li, J. , Jiang, Y. , Sun, J. , Xu, Y. , Pan, H. , Zhou, Y. , & Zhu, J. (2024). Research advances of tissue‐derived extracellular vesicles in cancers. Journal of Cancer Research and Clinical Oncology, 150, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Chen, R. , Kemper, S. , & Brigstock, D. R. (2021). Structural and functional characterization of fibronectin in extracellular vesicles from hepatocytes. Frontiers in Cell and Developmental Biology, 9, 640667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Gao, L. , Tan, X. , Li, F. , Zhao, M. , & Peng, S. (2016). Lipid rafts‐mediated endocytosis and physiology‐based cell membrane traffic models of doxorubicin liposomes. Biochimica et Biophysica Acta, 1858, 1801–1811. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Wang, J. , Chen, S. , Wu, P. , Xu, S. , Wang, C. , Shi, H. , & Bihl, J. (2020). miR‐137 boosts the neuroprotective effect of endothelial progenitor cell‐derived exosomes in oxyhemoglobin‐treated SH‐SY5Y cells partially via COX2/PGE2 pathway. Stem Cell Research & Therapy, 11, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zhao, R. , Cheng, K. , Zhang, K. , Wang, Y. , Zhang, Y. , Li, Y. , Liu, G. , Xu, J. , Xu, J. , Anderson, G. J. , Shi, J. , Ren, L. , Zhao, X. , & Nie, G. (2020). Bacterial outer membrane vesicles presenting programmed death 1 for improved cancer immunotherapy via immune activation and checkpoint inhibition. ACS Nano, 14, 16698–16711. [DOI] [PubMed] [Google Scholar]
- Li, Y. J. , Wu, J. Y. , Wang, J. M. , Hu, X. B. , Cai, J. X. , & Xiang, D. X. (2020). Gemcitabine loaded autologous exosomes for effective and safe chemotherapy of pancreatic cancer. Acta Biomaterialia, 101, 519–530. [DOI] [PubMed] [Google Scholar]
- Li, Y. M. , & Hall, W. A. (2010). Targeted toxins in brain tumor therapy. Toxins, 2, 2645–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liam‐Or, R. , Faruqu, F. N. , Walters, A. , Han, S. , Xu, L. , Wang, J. T. , Oberlaender, J. , Sanchez‐Fueyo, A. , Lombardi, G. , Dazzi, F. , Mailaender, V. , & Al‐Jamal, K. T. (2024). Cellular uptake and in vivo distribution of mesenchymal‐stem‐cell‐derived extracellular vesicles are protein corona dependent. Nature Nanotechnology, 19, 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, G. , Zhu, Y. , Ali, D. J. , Tian, T. , Xu, H. , Si, K. , Sun, B. , Chen, B. , & Xiao, Z. (2020). Engineered exosomes for targeted co‐delivery of miR‐21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. Journal of Nanobiotechnology, 18, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. , Yi, Q. , He, J. , Huang, D. , Xiong, J. , Sun, W. , & Sun, W. (2023). Extracellular vesicles in tumorigenesis, metastasis, chemotherapy resistance and intercellular communication in osteosarcoma. Bioengineered, 14, 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichty, B. D. , Power, A. T. , Stojdl, D. F. , & Bell, J. C. (2004). Vesicular stomatitis virus: re‐inventing the bullet. Trends in Molecular Medicine, 10, 210–216. [DOI] [PubMed] [Google Scholar]
- Lichty, B. D. , Stojdl, D. F. , Taylor, R. A. , Miller, L. , Frenkel, I. , Atkins, H. , & Bell, J. C. (2004). Vesicular stomatitis virus: a potential therapeutic virus for the treatment of hematologic malignancy. Human Gene Therapy, 15, 821–831. [DOI] [PubMed] [Google Scholar]
- Lin, A. , Giuliano, C. J. , Palladino, A. , John, K. M. , Abramowicz, C. , Yuan, M. L. , Sausville, E. L. , Lukow, D. A. , Liu, L. , Chait, A. R. , Galluzzo, Z. C. , Tucker, C. , & Sheltzer, J. M. (2019). Off‐target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Science Translational Medicine, 11, eaaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Q. , Qu, M. , Zhou, B. , Patra, H. K. , Sun, Z. , Luo, Q. , Yang, W. , Wu, Y. , Zhang, Y. , Li, L. , Deng, L. , Wang, L. , Gong, T. , He, Q. , Zhang, L. , Sun, X. , & Zhang, Z. (2019). Exosome‐like nanoplatform modified with targeting ligand improves anti‐cancer and anti‐inflammation effects of imperialine. Journal of Controlled Release : Official Journal of the Controlled Release Society, 311–312, 104–116. [DOI] [PubMed] [Google Scholar]
- Liu, C. , Liu, X. , Xiang, X. , Pang, X. , Chen, S. , Zhang, Y. , Ren, E. , Zhang, L. , Liu, X. , Lv, P. , Wang, X. , Luo, W. , Xia, N. , Chen, X. , & Liu, G. (2022). A nanovaccine for antigen self‐presentation and immunosuppression reversal as a personalized cancer immunotherapy strategy. Nature Nanotechnology, 17, 531–540. [DOI] [PubMed] [Google Scholar]
- Liu, C. , Xu, J. , Wen, F. , Yang, F. , Li, X. , Geng, D. , Li, L. , Chen, J. , & Zheng, J. (2018). Upregulation of syncytin‐1 promotes invasion and metastasis by activating epithelial‐mesenchymal transition‐related pathway in endometrial carcinoma. OncoTargets and Therapy, 12, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Zhang, W. , Li, Y. , Chang, J. , Tian, F. , Zhao, F. , Ma, Y. , & Sun, J. (2019). Microfluidic sonication to assemble exosome membrane‐coated nanoparticles for immune evasion‐mediated targeting. Nano Letters, 19, 7836–7844. [DOI] [PubMed] [Google Scholar]
- Liu, D. , Kou, X. , Chen, C. , Liu, S. , Liu, Y. , Yu, W. , Yu, T. , Yang, R. , Wang, R. , Zhou, Y. , & Shi, S. (2018). Circulating apoptotic bodies maintain mesenchymal stem cell homeostasis and ameliorate osteopenia via transferring multiple cellular factors. Cell Research, 28, 918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G. , Ma, N. , Cheng, K. , Feng, Q. , Ma, X. , Yue, Y. , Li, Y. , Zhang, T. , Gao, X. , Liang, J. , Zhang, L. , Wang, X. , Ren, Z. , Fu, Y. X. , Zhao, X. , & Nie, G. (2024). Bacteria‐derived nanovesicles enhance tumour vaccination by trained immunity. Nature Nanotechnology, 19, 387–398. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Fu, M. , Wang, M. , Wan, D. , Wei, Y. , & Wei, X. (2022). Cancer vaccines as promising immuno‐therapeutics: platforms and current progress. Journal of Hematology & Oncology, 15, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M. , Zhang, Y. , He, J. , Liu, W. , Li, Z. , Zhang, Y. , Gu, A. , Zhao, M. , Liu, M. , & Liu, X. (2024). Fusion with ARRDC1 or CD63: a strategy to enhance p53 loading into extracellular vesicles for tumor suppression. Biomolecules, 14, 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Wu, X. , Chandra, S. , Lyon, C. , Ning, B. , Jiang, L. , Fan, J. , & Hu, T. Y. (2022). Extracellular vesicles: Emerging tools as therapeutic agent carriers. Acta Pharmaceutica Sinica. B, 12, 3822–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. Y. , Liao, Y. , Hosseinifard, H. , Imani, S. , & Wen, Q. L. (2021). Diagnostic role of extracellular vesicles in cancer: A comprehensive systematic review and meta‐analysis. Frontiers in Cell and Developmental Biology, 9, 705791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Li, L. , Rong, Y. , Qian, D. , Chen, J. , Zhou, Z. , Luo, Y. , Jiang, D. , Cheng, L. , Zhao, S. , Kong, F. , Wang, J. , Zhou, Z. , Xu, T. , Gong, F. , Huang, Y. , Gu, C. , Zhao, X. , Bai, J. , Wang, F., … Cai, W. (2020). Hypoxic mesenchymal stem cell‐derived exosomes promote bone fracture healing by the transfer of miR‐126. Acta Biomater ialia, 103, 196–212. [DOI] [PubMed] [Google Scholar]
- Lobb, R. J. , van Amerongen, R. , Wiegmans, A. , Ham, S. , Larsen, J. E. , & Möller, A. (2017). Exosomes derived from mesenchymal non‐small cell lung cancer cells promote chemoresistance. International Journal of Cancer, 141, 614–620. [DOI] [PubMed] [Google Scholar]
- Krijnse Locker, J. , Chlanda, P. , Sachsenheimer, T. , & Brügger, B. (2013). Poxvirus membrane biogenesis: rupture not disruption. Cellular Microbiology, 15, 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logozzi, M. , De Milito, A. , Lugini, L. , Borghi, M. , Calabrò, L. , Spada, M. , Perdicchio, M. , Marino, M. L. , Federici, C. , Iessi, E. , Brambilla, D. , Venturi, G. , Lozupone, F. , Santinami, M. , Huber, V. , Maio, M. , Rivoltini, L. , & Fais, S. (2009). High levels of exosomes expressing CD63 and caveolin‐1 in plasma of melanoma patients. PLoS One, 4, e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein, W. R. (1990). Cell‐to‐cell communication and the control of growth. The American Review of Respiratory Disease, 142, S48–S53. [DOI] [PubMed] [Google Scholar]
- Lupu, A. , Gradinaru, L. M. , Gradinaru, V. R. , & Bercea, M. (2023). Diversity of bioinspired hydrogels: from structure to applications. Gels, 9, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, P. , Chen, X. , Fu, S. , Ren, E. , Liu, C. , Liu, X. , Jiang, L. , Zeng, Y. , Wang, X. , & Liu, G. (2021). Surface engineering of oncolytic adenovirus for a combination of immune checkpoint blockade and virotherapy. Biomaterials Science, 9, 7392–7401. [DOI] [PubMed] [Google Scholar]
- Ma, L. , Li, Y. , Peng, J. , Wu, D. , Zhao, X. , Cui, Y. , Chen, L. , Yan, X. , Du, Y. , & Yu, L. (2015). Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Research, 25, 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker, H. T. , Todd, S. C. , & Levy, S. (1997). The tetraspanin superfamily: Molecular facilitators. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology, 11, 428–442. [PubMed] [Google Scholar]
- Maeda, H. , Tsukigawa, K. , & Fang, J. (2016). A retrospective 30 years after discovery of the enhanced permeability and retention effect of solid tumors: Next‐generation chemotherapeutics and photodynamic therapy—problems, solutions, and prospects. Microcirculation, 23, 173–182. [DOI] [PubMed] [Google Scholar]
- Matsumura, Y. , & Maeda, H. (1986). A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Research, 46, 6387–6392. [PubMed] [Google Scholar]
- Mause, S. F. , & Weber, C. (2010). Microparticles: protagonists of a novel communication network for intercellular information exchange. Circulation Research, 107, 1047–1057. [DOI] [PubMed] [Google Scholar]
- Mebatsion, T. , Konig, M. , & Conzelmann, K. K. (1996). Budding of rabies virus particles in the absence of the spike glycoprotein. Cell, 84, 941–951. [DOI] [PubMed] [Google Scholar]
- Meehan, B. , Rak, J. , & Di Vizio, D. (2016). Oncosomes—Large and small: What are they, where they came from?. Journal of Extracellular Vesicles, 5, 33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendt, M. , Kamerkar, S. , Sugimoto, H. , McAndrews, K. M. , Wu, C. C. , Gagea, M. , Yang, S. , Blanko, E. V. R. , Peng, Q. , Ma, X. , Marszalek, J. R. , Maitra, A. , Yee, C. , Rezvani, K. , Shpall, E. , LeBleu, V. S. , & Kalluri, R. (2018). Generation and testing of clinical‐grade exosomes for pancreatic cancer. JCI Insight, 3, e99263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Y. , Chen, J. , Liu, Y. , Zhu, Y. , Wong, Y. K. , Lyu, H. , Shi, Q. , Xia, F. , Gu, L. , Zhang, X. , Gao, P. , Tang, H. , Guo, Q. , Qiu, C. , Xu, C. , He, X. , Zhang, J. , & Wang, J. (2022). A highly efficient protein corona‐based proteomic analysis strategy for the discovery of pharmacodynamic biomarkers. Journal of Pharmaceutical Analysis, 12, 879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merisko‐Liversidge, E. M. , & Liversidge, G. G. (2008). Drug nanoparticles: Formulating poorly water‐soluble compounds. Toxicologic Pathology, 36, 43–48. [DOI] [PubMed] [Google Scholar]
- Metzner, C. , & Zaruba, M. (2021). On the relationship of viral particles and extracellular vesicles: implications for viral vector technology. Viruses, 13, 1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, C. , Losacco, J. , Stickney, Z. , Li, L. , Marriott, G. , & Lu, B. (2017). Pseudotyping exosomes for enhanced protein delivery in mammalian cells. International Journal of Nanomedicine, 12, 3153–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, J. K. , & Needham, D. (1999). Targeted drug delivery. Expert Opinion on Therapeutic Patents, 9, 1499–1513. [Google Scholar]
- Minciacchi, V. R. , Freeman, M. R. , & Di Vizio, D. (2015). Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Seminars in Cell & Devolopmental Biology, 40, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minciacchi, V. R. , You, S. , Spinelli, C. , Morley, S. , Zandian, M. , Aspuria, P. J. , Cavallini, L. , Ciardiello, C. , Reis Sobreiro, M. , Morello, M. , Kharmate, G. , Jang, S. C. , Kim, D. K. , Hosseini‐Beheshti, E. , Tomlinson Guns, E. , Gleave, M. , Gho, Y. S. , Mathivanan, S. , Yang, W. , Freeman, M. R., … Di Vizio, D. (2015). Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor‐derived extracellular vesicles. Oncotarget, 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecalvo, A. , Larregina, A. T. , Shufesky, W. J. , Stolz, D. B. , Sullivan, M. L. , Karlsson, J. M. , Baty, C. J. , Gibson, G. A. , Erdos, G. , Wang, Z. , Milosevic, J. , Tkacheva, O. A. , Divito, S. J. , Jordan, R. , Lyons‐Weiler, J. , Watkins, S. C. , & Morelli, A. E. (2012). Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood, 119, 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morad, G. , Carman, C. V. , Hagedorn, E. J. , Perlin, J. R. , Zon, L. I. , Mustafaoglu, N. , Park, T. E. , Ingber, D. E. , Daisy, C. C. , & Moses, M. A. (2019). Tumor‐derived extracellular vesicles breach the intact blood‐brain barrier via transcytosis. ACS Nano, 13, 13853–13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli, A. E. , Larregina, A. T. , Shufesky, W. J. , Sullivan, M. L. , Stolz, D. B. , Papworth, G. D. , Zahorchak, A. F. , Logar, A. J. , Wang, Z. , Watkins, S. C. Jr. , Falo, L. D , & Thomson, A. W. (2004). Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood, 104, 3257–3266. [DOI] [PubMed] [Google Scholar]
- Morimoto, K. , Ishitobi, J. , Noguchi, K. , Kira, R. , Kitayama, Y. , Goto, Y. , Fujiwara, D. , Michigami, M. , Harada, A. , Takatani‐Nakase, T. , Fujii, I. , Futaki, S. , Kanada, M. , & Nakase, I. (2024). Extracellular microvesicles modified with arginine‐rich peptides for active macropinocytosis induction and delivery of therapeutic molecules. CS Applied Materials & Interfaces, 16, 17069–17079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita, M. , Horita, M. , Higuchi, A. , Marui, M. , Katsumi, H. , & Yamamoto, A. (2021). Characterizing different probiotic‐derived extracellular vesicles as a novel adjuvant for immunotherapy. Molecular Pharmaceutics, 18, 1080–1092. [DOI] [PubMed] [Google Scholar]
- Morishita, M. , Takahashi, Y. , Nishikawa, M. , Ariizumi, R. , & Takakura, Y. (2017). Enhanced class I tumor antigen presentation via cytosolic delivery of exosomal cargos by tumor‐cell‐derived exosomes displaying a pH‐sensitive fusogenic peptide. Molecular Pharmaceutics, 14, 4079–4086. [DOI] [PubMed] [Google Scholar]
- Moyano, A. L. , Li, G. , Boullerne, A. I. , Feinstein, D. L. , Hartman, E. , Skias, D. , Balavanov, R. , van Breemen, R. B. , Bongarzone, E. R. , Månsson, J. E. , & Givogri, M. I. (2016). Sulfatides in extracellular vesicles isolated from plasma of multiple sclerosis patients. Journal of Neuroscience Research, 94, 1579–1587. [DOI] [PubMed] [Google Scholar]
- Mu, W. , Rana, S. , & Zöller, M. (2013). Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia, 15, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard, A. (2021). Gene therapy community grapples with toxicity issues, as pipeline matures. Nature Reviews Drug Discovery, 20, 804–805. [DOI] [PubMed] [Google Scholar]
- Muntasell, A. , Berger, A. C. , & Roche, P. A. (2007). T cell‐induced secretion of MHC class II‐peptide complexes on B cell exosomes. The EMBO Journal, 26, 4263–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, D. E. , de Jong, O. G. , Brouwer, M. , Wood, M. J. , Lavieu, G. , Schiffelers, R. M. , & Vader, P. (2019). Extracellular vesicle‐based therapeutics: natural versus engineered targeting and trafficking. Experimental & Molecular Medicine, 51, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, W. , Yoshida, T. , Diez, D. , Miyatake, Y. , Nishibu, T. , Imawaka, N. , Naruse, K. , Sadamura, Y. , & Hanayama, R. (2016). A novel affinity‐based method for the isolation of highly purified extracellular vesicles. Scientific Reports, 6, 33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase, I. , & Futaki, S. (2015). Combined treatment with a pH‐sensitive fusogenic peptide and cationic lipids achieves enhanced cytosolic delivery of exosomes. Scientific Reports, 5, 10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase, I. , Noguchi, K. , Aoki, A. , Takatani‐Nakase, T. , Fujii, I. , & Futaki, S. (2017). Arginine‐rich cell‐penetrating peptide‐modified extracellular vesicles for active macropinocytosis induction and efficient intracellular delivery. Scientific Reports, 7, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgung, R. , Mi Lee, Y. , Kim, J. , Jang, Y. , Lee, B. H. , Kim, I. S. , Sokkar, P. , Rhee, Y. M. , Hoffman, A. S. , & Kim, W. J. (2014). Poly‐cyclodextrin and poly‐paclitaxel nano‐assembly for anticancer therapy. Nature Communications, 5, 3702. [DOI] [PubMed] [Google Scholar]
- Nanbo, A. , Kawanishi, E. , Yoshida, R. , & Yoshiyama, H. (2013). Exosomes derived from Epstein‐Barr virus‐infected cells are internalized via caveola‐dependent endocytosis and promote phenotypic modulation in target cells. Journal of Virology, 87, 10334–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannan, L. , Oudart, J. B. , Monboisse, J. C. , Ramont, L. , Brassart‐Pasco, S. , & Brassart, B. (2020). Extracellular vesicle‐dependent cross‐talk in cancer‐focus on pancreatic cancer. Frontiers in Oncology, 10, 1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, D. , Dai, Z. , Li, J. , Yang, Y. , Xi, Z. , Wang, J. , Zhang, W. , Qian, K. , Guo, S. , Zhu, C. , Wang, R. , Li, Y. , Yu, M. , Zhang, X. , Shi, X. , & Gan, Y. (2020). Cancer‐cell‐membrane‐coated nanoparticles with a yolk‐shell structure augment cancer chemotherapy. Nano Letters, 20, 936–946. [DOI] [PubMed] [Google Scholar]
- Nishida‐Aoki, N. , & Ochiya, T. (2024). Impacts of tissue context on extracellular vesicles‐mediated cancer‐host cell communications. Cancer Science, 115(6), 1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, K. , Obuki, M. , Sumi, H. , Klußmann, M. , Morimoto, K. , Nakai, S. , Hashimoto, T. , Fujiwara, D. , Fujii, I. , Yuba, E. , Takatani‐Nakase, T. , Neundorf, I. , & Nakase, I. (2021). Macropinocytosis‐inducible extracellular vesicles modified with antimicrobial protein CAP18‐derived cell‐penetrating peptides for efficient intracellular delivery. Molecular Pharmaceutics, 18, 3290–3301. [DOI] [PubMed] [Google Scholar]
- Nolte‐’t Hoen, E. , Cremer, T. , Gallo, R. C. , & Margolis, L. B. (2016). Extracellular vesicles and viruses: Are they close relatives?. Proceedings of the National Academy of Sciences of the United States of America, 113, 9155–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte‐’t Hoen, E. N. , Buschow, S. I. , Anderton, S. M. , Stoorvogel, W. , & Wauben, M. H. (2009). Activated T cells recruit exosomes secreted by dendritic cells via LFA‐1. Blood, 113, 1977–1981. [DOI] [PubMed] [Google Scholar]
- Nunes, A. S. , Barros, A. S. , Costa, E. C. , Moreira, A. F. , & Correia, I. J. (2019). 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnology and Bioengineering, 116, 206–226. [DOI] [PubMed] [Google Scholar]
- Núñez‐Wehinger, S. , Ortiz, R. J. , Díaz, N. , Díaz, J. , Lobos‐González, L. , & Quest, A. F. (2014). Caveolin‐1 in cell migration and metastasis. Current Molecular Medicine, 14, 255–274. [DOI] [PubMed] [Google Scholar]
- Obregon, C. , Rothen‐Rutishauser, B. , Gerber, P. , Gehr, P. , & Nicod, L. P. (2009). Active uptake of dendritic cell‐derived exovesicles by epithelial cells induces the release of inflammatory mediators through a TNF‐alpha‐mediated pathway. The American Journal of Pathology, 175, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, K. , Ughetto, S. , Mahjoum, S. , Nair, A. V. , & Breakefield, X. O. (2022). Uptake, functionality, and re‐release of extracellular vesicle‐encapsulated cargo. Cell Reports, 39, 110651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuchi, M. , Fernandez, M. , & Barber, G. N. (2003). Development of recombinant vesicular stomatitis viruses that exploit defects in host defense to augment specific oncolytic activity. Journal of Virology, 77(16), 8843–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. , Takanashi, M. , Sudo, K. , Ueda, S. , Ishikawa, A. , Matsuyama, N. , Fujita, K. , Mizutani, T. , Ohgi, T. , Ochiya, T. , Gotoh, N. , & Kuroda, M. (2013). Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Molecular Therapy : The Journal of the American Society of Gene Therapy, 21, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, Y. H. , Zou, S. , Goh, W. J. , Wang, J. W. , Wacker, M. , Czarny, B. , & Pastorin, G. (2021). Cell‐derived nanovesicles as exosome‐mimetics for drug delivery purposes: Uses and recommendations. Methods in Molecular Biology, 2211, 147–170. [DOI] [PubMed] [Google Scholar]
- Ovchinnikova, L. A. , Filimonova, I. N. , Zakharova, M. Y. , Balabashin, D. S. , Aliev, T. K. , Lomakin, Y. A. , & Gabibov, A. G. (2021). Targeting extracellular vesicles to dendritic cells and macrophages. Acta Naturae, 13, 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmiller, A. M. , Pierluissi, J. A. , Wermuth, P. J. , Sauma, S. , Martinez‐Outschoorn, U. , Tuluc, M. , Luginbuhl, A. , Curry, J. , Harshyne, L. A. , Wahl, J. K. 3rd , South, A. P. , & Mahoney, M. G. (2017). Desmoglein 2 modulates extracellular vesicle release from squamous cell carcinoma keratinocytes. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology, 31, 3412–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, V. , & Cardoso, R. C. (2020). Neuroendocrine, autocrine, and paracrine control of follicle‐stimulating hormone secretion. Molecular and Cellular Endocrinology, 500, 110632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palviainen, M. , Saraswat, M. , Varga, Z. , Kitka, D. , Neuvonen, M. , Puhka, M. , Joenväärä, S. , Renkonen, R. , Nieuwland, R. , Takatalo, M. , & Siljander, P. R. M. (2020). Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo‐Implications for biomarker discovery. PLoS One, 15, e0236439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, B. T. , Teng, K. , Wu, C. , Adam, M. , & Johnstone, R. M. (1985). Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. Journal of Cell Biology, 101, 942–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J. , Li, X. , Shao, B. , Xu, F. , Huang, X. , Guo, X. , & Zhou, S. (2022). Self‐blockade of PD‐L1 with bacteria‐derived outer‐membrane vesicle for enhanced cancer immunotherapy. Advanced Materials, 34, e2106307. [DOI] [PubMed] [Google Scholar]
- Pandit, R. , Koh, W. K. , Sullivan, R. K. P. , Palliyaguru, T. , Parton, R. G. , & Götz, J. (2020). Role for caveolin‐mediated transcytosis in facilitating transport of large cargoes into the brain via ultrasound. Journal of Controlled Release : Official Journal of the Controlled Release Society, 327, 667–675. [DOI] [PubMed] [Google Scholar]
- Pang, X. , He, X. , Qiu, Z. , Zhang, H. , Xie, R. , Liu, Z. , Gu, Y. , Zhao, N. , Xiang, Q. , & Cui, Y. (2023). Targeting integrin pathways: Mechanisms and advances in therapy. Signal Transduction and Targeted Therapy, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K. S. , Svennerholm, K. , Crescitelli, R. , Lässer, C. , Gribonika, I. , & Lötvall, J. (2021). Synthetic bacterial vesicles combined with tumour extracellular vesicles as cancer immunotherapy. Journal of Extracellular Vesicles, 10, e12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolini, I. , Federici, C. , Raggi, C. , Lugini, L. , Palleschi, S. , De Milito, A. , Coscia, C. , Iessi, E. , Logozzi, M. , Molinari, A. , Colone, M. , Tatti, M. , Sargiacomo, M. , & Fais, S. (2009). Microenvironmental pH is a key factor for exosome traffic in tumor cells. The Journal of Biological Chemistry, 284, 34211–34222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado, H. , Alečković, M. , Lavotshkin, S. , Matei, I. , Costa‐Silva, B. , Moreno‐Bueno, G. , Hergueta‐Redondo, M. , Williams, C. , García‐Santos, G. , Ghajar, C. , Nitadori‐Hoshino, A. , Hoffman, C. , Badal, K. , Garcia, B. A. , Callahan, M. K. , Yuan, J. , Martins, V. R. , Skog, J. , Kaplan, R. N. , … Lyden, D. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro‐metastatic phenotype through MET. Nature Medicine, 18, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans, L. , Püntener, D. , & Helenius, A. (2002). Local actin polymerization and dynamin recruitment in SV40‐induced internalization of caveolae. Science, 296, 535–539. [DOI] [PubMed] [Google Scholar]
- Perrigue, P. M. , Murray, R. A. , Mielcarek, A. , Henschke, A. , & Moya, S. E. (2021). Degradation of drug delivery nanocarriers and payload release: A review of physical methods for tracing nanocarrier biological fate. Pharmaceutics, 13, 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, H. , Mekaoui, L. , Bernard, C. , Veas, F. , Stefas, I. , & Leboyer, M. (2008). Endogenous retrovirus type W GAG and envelope protein antigenemia in serum of schizophrenic patients. Biological Psychiatry, 64, 1019–1023. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S. R. , Grossmann, K. F. , Cassidy, P. B. , Yang, C. H. , Fan, M. , Kopelovich, L. , Leachman, S. A. , & Pfeffer, L. M. (2015). Detection of exosomal miRNAs in the plasma of melanoma patients. Journal of Clinical Medicine, 4, 2012–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillipson, M. , & Kubes, P. (2011). The neutrophil in vascular inflammation. Nature Medicine, 17, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinilla‐Macua, I. , Bayer, N. , Schober, D. , Prchla, E. , Murphy, R. F. , Blaas, D. , & Fuchs, R. (1998). Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. Journal of Virology, 72, 9645–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piombino, C. , Mastrolia, I. , Omarini, C. , Candini, O. , Dominici, M. , Piacentini, F. , & Toss, A. (2021). The role of exosomes in breast cancer diagnosis. Biomedicines, 9, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov, E. Y. , Silachev, D. N. , Popov, V. A. , Zorova, L. D. , Pevzner, I. B. , Zorov, S. D. , Jankauskas, S. S. , Babenko, V. A. , Sukhikh, G. T. , & Zorov, D. B. (2017). Intercellular signalling cross‐talk: To kill, to heal and to rejuvenate. Heart, Lung & Circulation, 26, 648–659. [DOI] [PubMed] [Google Scholar]
- Poliakov, A. , Spilman, M. , Dokland, T. , Amling, C. L. , & Mobley, J. A. (2009). Structural heterogeneity and protein composition of exosome‐like vesicles (prostasomes) in human semen. The Prostate, 69, 159–167. [DOI] [PubMed] [Google Scholar]
- Pompili, S. , Vetuschi, A. , Sferra, R. , & Cappariello, A. (2022). Extracellular vesicles and resistance to anticancer drugs: A tumor skeleton key for unhinging chemotherapies. Frontiers in Oncology, 12, 933675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar, U. , Maeda, H. , Jain, R. K. , Sevick‐Muraca, E. M. , Zamboni, W. , Farokhzad, O. C. , Barry, S. T. , Gabizon, A. , Grodzinski, P. , & Blakey, D. C. (2013). Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Cancer Research, 73, 2412–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher, J. A. , & Bertozzi, C. R. (2005). Chemistry in living systems. Nature Chemical Biology, 1, 13–21. [DOI] [PubMed] [Google Scholar]
- Prescher, J. A. , Dube, D. H. , & Bertozzi, C. R. (2004). Chemical remodelling of cell surfaces in living animals. Nature, 430, 873–877. [DOI] [PubMed] [Google Scholar]
- Pritchard, A. , Tousif, S. , Wang, Y. , Hough, K. , Khan, S. , Strenkowski, J. , Chacko, B. K. , Darley‐Usmar, V. M. , & Deshane, J. S. (2020). Lung tumor cell‐derived exosomes promote M2 macrophage polarization. Cells, 9, 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochiantz, A. (2000). Messenger proteins: Homeoproteins, TAT and others. Current Opinion in Cell Biology, 12, 400–406. [DOI] [PubMed] [Google Scholar]
- Rabinovich, G. A. , Gabrilovich, D. , & Sotomayor, E. M. (2007). Immunosuppressive strategies that are mediated by tumor cells. Annual Review of Immunology, 25, 267–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampado, R. , Crotti, S. , Caliceti, P. , Pucciarelli, S. , & Agostini, M. (2020). Recent advances in understanding the protein corona of nanoparticles and in the formulation of “Stealthy” nanomaterials. Frontiers in Bioengineering and Biotechnology, 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo, G. , Nijman, H. W. , Stoorvogel, W. , Liejendekker, R. , Harding, C. V. , Melief, C. J. , & Geuze, H. J. (1996). B lymphocytes secrete antigen‐presenting vesicles. The Journal of Experimental Medicine, 183, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, K. (2015). Pancreatic cancer: Pancreatic cancer exosomes prime the liver for metastasis. Nature Reviews. Gastroenterology & Hepatology, 12, 371. [DOI] [PubMed] [Google Scholar]
- Ren, Y. , Miao, J. M. , Wang, Y. Y. , Fan, Z. , Kong, X. B. , Yang, L. , & Cheng, G. (2022). Oncolytic viruses combined with immune checkpoint therapy for colorectal cancer is a promising treatment option. Frontiers in Immunology, 13, 961796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim, W. K. , Kim, J. Y. , Lee, S. Y. , Cha, S. G. , Park, J. M. , Park, H. J. , Park, C. G. , & Han, D. K. (2023). Recent advances in extracellular vesicle engineering and its applications to regenerative medicine. Biomaterial Research, 27, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A. J. (2006). Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends in Cell Biology, 16, 522–529. [DOI] [PubMed] [Google Scholar]
- Roberts, A. , Buonocore, L. , Price, R. , Forman, J. , & Rose, J. K. (1999). Attenuated vesicular stomatitis viruses as vaccine vectors. Journal of Virology, 73, 3723–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls, M. M. , Webster, P. , Balba, N. H. , & Rose, J. K. (1994). Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self‐replicating RNA. Cell, 79, 497–506. [DOI] [PubMed] [Google Scholar]
- Rose, N. F. , Roberts, A. , Buonocore, L. , & Rose, J. K. (2000). Glycoprotein exchange vectors based on vesicular stomatitis virus allow effective boosting and generation of neutralizing antibodies to a primary isolate of human immunodeficiency virus type 1. Journal of Virology, 74, 10903–10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar, G. , Sah, R. P. , Javeed, N. , Dutta, S. K. , Smyrk, T. C. , Lau, J. S. , Giorgadze, N. , Tchkonia, T. , Kirkland, J. L. , Chari, S. T. , & Mukhopadhyay, D. (2016). Pathogenesis of pancreatic cancer exosome‐induced lipolysis in adipose tissue. Gut, 65, 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y. , Muramatsu, T. , Kanai, Y. , Ojima, H. , Sukeda, A. , Hiraoka, N. , Arai, E. , Sugiyama, Y. , Matsuzaki, J. , Uchida, R. , Yoshikawa, N. , Furukawa, R. , & Saito, H. (2019). Establishment of patient‐derived organoids and drug screening for biliary tract carcinoma. Cell Reports, 27, 1265–1276. [DOI] [PubMed] [Google Scholar]
- Schauber‐Plewa, C. , Simmons, A. , Tuerk, M. J. , Pacheco, C. D. , & Veres, G. (2005). Complement regulatory proteins are incorporated into lentiviral vectors and protect particles against complement inactivation. Gene Therapy, 12, 238–245. [DOI] [PubMed] [Google Scholar]
- Schindler, C. , Collinson, A. , Matthews, C. , Pointon, A. , Jenkinson, L. , Minter, R. R. , Vaughan, T. J. , & Tigue, N. J. (2019). Exosomal delivery of doxorubicin enables rapid cell entry and enhanced in vitro potency. PLoS One, 14, e0214545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrezenmeier, E. , & Dörner, T. (2020). Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nature Reviews. Rheumatology, 16, 155–166. [DOI] [PubMed] [Google Scholar]
- Scinto, S. L. , Bilodeau, D. A. , Hincapie, R. , Lee, W. , Nguyen, S. S. , Xu, M. , Am Ende, C. W. , Finn, M. G. , Lang, K. , Lin, Q. , Pezacki, J. P. , Prescher, J. A. , Robillard, M. S. , & Fox, J. M. (2021). Bioorthogonal chemistry. Nature Reviews. Methods Primers, 1, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura, E. , Guérin, C. , Hogg, N. , Amigorena, S. , & Théry, C. (2007). CD8+ dendritic cells use LFA‐1 to capture MHC‐peptide complexes from exosomes in vivo. Journal of Immunology, 179, 1489–1496. [DOI] [PubMed] [Google Scholar]
- Sezgin, E. , Levental, I. , Mayor, S. , & Eggeling, C. (2017). The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nature Reviews. Molecular Cell Biology, 18, 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks, N. , Greek, R. , & Greek, J. (2009). Are animal models predictive for humans?. Philosophy, Ethics, and Humanities in Medicine, 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S. , LeClaire, M. , Wohlschlegel, J. , & Gimzewski, J. (2020). Impact of isolation methods on the biophysical heterogeneity of single extracellular vesicles. Science Reports, 10, 13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter, D. , Harel, M. , Mukherjee, A. , Sagredo, L. M. , Loven, D. , Prinz, E. , Avraham, S. , Orian‐Rousseau, V. , Geiger, T. , Shaked, Y. , & Wolfenson, H. (2020). Breast cancer‐derived microparticles reduce cancer cell adhesion, an effect augmented by chemotherapy. Cells, 9, 2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, H. , Heikamp, E. , Turley, H. , Dragovic, R. , Thomas, P. , Oon, C. E. , Leek, R. , Edelmann, M. , Kessler, B. , Sainson, R. C. , Sargent, I. , Li, J. L. , & Harris, A. L. (2010). New mechanism for Notch signaling to endothelium at a distance by Delta‐like 4 incorporation into exosomes. Blood, 116, 2385–2394. [DOI] [PubMed] [Google Scholar]
- Simón, L. , Campos, A. , Leyton, L. , & Quest, A. F. G. (2020). Caveolin‐1 function at the plasma membrane and in intracellular compartments in cancer. Cancer Metastasis Review, 39, 435–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. J. , & Gray, J. W. (2021). Chemokine signaling in cancer‐stroma communications. Journal of Cell Communication and Signaling, 15, 361–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. , Szigyártó, I. C. , Ricci, M. , Zsila, F. , Juhász, T. , Mihály, J. , Bősze, S. , Bulyáki, É. , Kardos, J. , Kitka, D. , Varga, Z. , & Beke‐Somfai, T. (2020). Membrane active peptides remove surface adsorbed protein corona from extracellular vesicles of red blood cells. Frontiers in Chemistry, 8, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel, J. J. , & Wiley, D. C. (1995). Influenza viruses and cell membranes. American Journal of Respiratory and Critical Care Medicine, 152, S13–S15. [DOI] [PubMed] [Google Scholar]
- Skelton, A. M. , Cohen, D. J. , Boyan, B. D. , & Schwartz, Z. (2023). Osteoblast‐derived matrixvesicles exhibit exosomal traits and a unique subset of microRNA: Their caveolae‐dependent endocytosis results in reduced osteogenic differentiation. International Journal Of Molecular Sciences, 24, 12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog, J. , Würdinger, T. , van Rijn, S. , Meijer, D. H. , Gainche, L. , Sena‐Esteves, M. , Curry, W. T. Jr. , Carter, B. S. , Krichevsky, A. M. , & Breakefield, X. O. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology, 10, 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten, E. M. , & Bertozzi, C. R. (2009). Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angewandte Chemie, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, T. , Petrova, K. , Payton, N. M. , Persaud, I. , Redzic, J. S. , Graner, M. W. , Smith‐Jones, P. , & Anchordoquy, T. J. (2014). Surface functionalization of exosomes using click chemistry. Bioconjugate Chemistry, 25, 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa de Almeida, M. , Susnik, E. , Drasler, B. , Taladriz‐Blanco, P. , Petri‐Fink, A. , & Rothen‐Rutishauser, B. (2021). Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine. Chemical Society Reviews, 50, 5397–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanius, K. , Servage, K. , de Souza Santos, M. , Gray, H. F. , Toombs, J. E. , Chimalapati, S. , Kim, M. S. , Malladi, V. S. , Brekken, R. , & Orth, K. (2019). Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. Elife, 8, e40226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, L. , Ellson, C. , & Hawkins, P. (2002). Roles of PI3Ks in leukocyte chemotaxis and phagocytosis. Current Opinion in Cell Biology, 14, 203–213. [DOI] [PubMed] [Google Scholar]
- Stewart, M. P. , Lorenz, A. , Dahlman, J. , & Sahay, G. (2016). Challenges in carrier‐mediated intracellular delivery: moving beyond endosomal barriers. Wiley Interdisciplinary Reviews. Nanomedicine and Nanobiotechnology, 8, 465–478. [DOI] [PubMed] [Google Scholar]
- Su, S. , Wang, H. , Liu, X. , Wu, Y. , & Nie, G. (2013). iRGD‐coupled responsive fluorescent nanogel for targeted drug delivery. Biomaterials, 34, 3523–3533. [DOI] [PubMed] [Google Scholar]
- Südhof, T. C. , & Rothman, J. E. (2009). Membrane fusion: grappling with SNARE and SM proteins. Science, 323, 474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Xu, R. , Sun, X. , Duan, Y. , Han, Y. , Zhao, Y. , Qian, H. , Zhu, W. , & Xu, W. (2016). Safety evaluation of exosomes derived from human umbilical cord mesenchymal stromal cell. Cytotherapy, 18, 413–422. [DOI] [PubMed] [Google Scholar]
- Sun, X. , Belouzard, S. , & Whittaker, G. R. (2008). Molecular architecture of the bipartite fusion loops of vesicular stomatitis virus glycoprotein G, a class III viral fusion protein. Journal of Biological Chemistry, 283, 6418–6427. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Sha, Y. , Cui, G. , Meng, F. , & Zhong, Z. (2023). Lysosomal‐mediated drug release and activation for cancer therapy and immunotherapy. Advanced Drug Delivery Reviews, 192, 114624. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Zhu, H. , Song, J. , Jiang, Y. , Ouyang, H. , Dong, T. , Tao, R. , Fan, X. , & Zhang, G. (2017). Expression of leukocytic syncytin‐1 in B‐cell acute lymphoblastic leukemia and acute myeloid leukemia patients. Clinical Laboratory, 63, 1567–1574. [DOI] [PubMed] [Google Scholar]
- Sung, B. H. , Ketova, T. , Hoshino, D. , Zijlstra, A. , & Weaver, A. M. (2015). Directional cell movement through tissues is controlled by exosome secretion. Nature Communications, 6, 7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, B. H. , von Lersner, A. , Guerrero, J. , Krystofiak, E. S. , Inman, D. , Pelletier, R. , Zijlstra, A. , Ponik, S. M. , & Weaver, A. M. (2020). A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nature Communications, 11, 2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkara, V. , Woo, H. K. , & Cho, Y. K. (2016). Emerging techniques in the isolation and characterization of extracellular vesicles and their roles in cancer diagnostics and prognostics. The Analyst, 141, 371–381. [DOI] [PubMed] [Google Scholar]
- Svensson, K. J. , Christianson, H. C. , Wittrup, A. , Bourseau‐Guilmain, E. , Lindqvist, E. , Svensson, L. M. , Mörgelin, M. , & Belting, M. (2013). Exosome uptake depends on ERK1/2‐heat shock protein 27 signaling and lipid Raft‐mediated endocytosis negatively regulated by caveolin‐1. The Journal of Biological Chemistry, 288, 17713–17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, J. A. (2008). Shaping cups into phagosomes and macropinosomes. Nature Reviews. Molecular Cell Biology, 9, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatanek, R. , Baran, J. , Siedlar, M. , & Baj‐Krzyworzeka, M. (2015). Isolation of extracellular vesicles: Determining the correct approach (Review). International Journal Of Molecular Medicine, 36, 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancini, B. , Buratta, S. , Sagini, K. , Costanzi, E. , Delo, F. , Urbanelli, L. , & Emiliani, C. (2019). Insight into the role of extracellular vesicles in lysosomal storage disorders. Genes, 10, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauro, B. J. , Greening, D. W. , Mathias, R. A. , Mathivanan, S. , Ji, H. , & Simpson, R. J. (2013). Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell‐derived organoids. Molecular & Cellular Proteomics : MCP, 12, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D. D. , Lyons, K. S. , & Gerçel‐Taylor, C. (2002). Shed membrane fragment‐associated markers for endometrial and ovarian cancers. Gynecologic Oncology, 84, 443–448. [DOI] [PubMed] [Google Scholar]
- Temchura, V. V. , Tenbusch, M. , Nchinda, G. , Nabi, G. , Tippler, B. , Zelenyuk, M. , Wildner, O. , Uberla, K. , & Kuate, S. (2008). Enhancement of immunostimulatory properties of exosomal vaccines by incorporation of fusion‐competent G protein of vesicular stomatitis virus. Vaccine, 26, 3662–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Tije, A. J. , Verweij, J. , Loos, W. J. , & Sparreboom, A. (2003). Pharmacological effects of formulation vehicles: Implications for cancer chemotherapy. Clinical Pharmacokinetics, 42, 665–685. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Amigorena, S. , Raposo, G. , & Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology, Chapter 3, 3, 22. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , Beckham, C. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C. E. , Ehrhardt, A. , & Kay, M. A. (2003). Progress and problems with the use of viral vectors for gene therapy. Nature Reviews. Genetics, 4, 346–358. [DOI] [PubMed] [Google Scholar]
- Tian, T. , Zhu, Y. L. , Zhou, Y. Y. , Liang, G. F. , Wang, Y. Y. , Hu, F. H. , & Xiao, Z. D. (2014). Exosome uptake through clathrin‐mediated endocytosis and macropinocytosis and mediating miR‐21 delivery. The Journal of Biological Chemistry, 289, 22258–22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y. , Li, S. , Song, J. , Ji, T. , Zhu, M. , Anderson, G. J. , Wei, J. , & Nie, G. (2014). A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials, 35, 2383–2390. [DOI] [PubMed] [Google Scholar]
- Tkach, M. , & Théry, C. (2016). Communication by extracellular vesicles: Where we are and where we need to go. Cell, 164, 1126–1232. [DOI] [PubMed] [Google Scholar]
- Tolosa, J. M. , Parsons, K. S. , Hansbro, P. M. , Smith, R. , & Wark, P. A. (2015). The placental protein syncytin‐1 impairs antiviral responses and exaggerates inflammatory responses to influenza. PLoS One, 10, e0118629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolosa, J. M. , Schjenken, J. E. , Clifton, V. L. , Vargas, A. , Barbeau, B. , Lowry, P. , Maiti, K. , & Smith, R. (2012). The endogenous retroviral envelope protein syncytin‐1 inhibits LPS/PHA‐stimulated cytokine responses in human blood and is sorted into placental exosomes. Placenta, 33, 933–941. [DOI] [PubMed] [Google Scholar]
- Tóth, E. Á. , Turiák, L. , Visnovitz, T. , Cserép, C. , Mázló, A. , Sódar, B. W. , Försönits, A. I. , Petővári, G. , Sebestyén, A. , Komlósi, Z. , Drahos, L. , Kittel, Á. , Nagy, G. , Bácsi, A. , Dénes, Á. , Gho, Y. S. , Szabó‐Taylor, K. É. , & Buzás, E. I. (2021). Formation of a protein corona on the surface of extracellular vesicles in blood plasma. Journal of Extracellular Vesicles, 10, e12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, C. , Du, Z. , Zhang, H. , Feng, Y. , Qi, Y. , Zheng, Y. , Liu, J. , & Wang, J. (2021). Endocytic pathway inhibition attenuates extracellular vesicle‐induced reduction of chemosensitivity to bortezomib in multiple myeloma cells. Theranostics, 11, 2364–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutuianu, A. , Anene, C. A. , Shelton, M. , Speirs, V. , Whitelaw, D. C. , Thorpe, J. , Roberts, W. , & Boyne, J. R. (2024). Platelet‐derived microvesicles isolated from type‐2 diabetes mellitus patients harbour an altered miRNA signature and drive MDA‐MB‐231 triple‐negative breast cancer cell invasion. PLoS One, 19, e0304870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ural, E. E. , Toomajian, V. , Hoque Apu, E. , Veletic, M. , Balasingham, I. , Ashammakhi, N. , Kanada, M. , & Contag, C. H. (2021). Visualizing extracellular vesicles and their function in 3D tumor microenvironment models. International Journal of Molecular Sciences, 22, 4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman, W. M. , Pham, T. C. , Kwok, Y. Y. , Vu, L. T. , Ma, V. , Peng, B. , Chan, Y. S. , Wei, L. , Chin, S. M. , Azad, A. , He, A. B. , Leung, A. Y. H. , Yang, M. , Shyh‐Chang, N. , Cho, W. C. , Shi, J. , & Le, M. T. N. (2018). Efficient RNA drug delivery using red blood cell extracellular vesicles. Nature Communications, 9, 2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekström, K. , Bossios, A. , Sjöstrand, M. , Lee, J. J. , & Lötvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- van Niel, G. , D'Angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews. Molecular Cell Biology, 19, 213–228. [DOI] [PubMed] [Google Scholar]
- Varnamkhasti, B. S. , Jafari, S. , Taghavi, F. , Alaei, L. , Izadi, Z. , Lotfabadi, A. , Dehghanian, M. , Jaymand, M. , Derakhshankhah, H. , & Saboury, A. A. (2020). Cell‐penetrating peptides: As a promising theranostics strategy to circumvent the blood‐brain barrier for CNS diseases. Current Drug Delivery, 17, 375–386. [DOI] [PubMed] [Google Scholar]
- Costa Verdera, H. , Gitz‐Francois, J. J. , Schiffelers, R. M. , & Vader, P. (2017). Cellular uptake of extracellular vesicles is mediated by clathrin‐independent endocytosis and macropinocytosis. Journal of Controlled Release : Official Journal of the Controlled Release Society, 266, 100–108. [DOI] [PubMed] [Google Scholar]
- Verweij, F. J. , Balaj, L. , Boulanger, C. M. , Carter, D. R. F. , Compeer, E. B. , & D'Angelo, G. (2021). The power of imaging to understand extracellular vesicle biology in vivo. Nature Methods, 18, 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, K. C. , Palmisano, B. T. , Shoucri, B. M. , Shamburek, R. D. , & Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high‐density lipoproteins. Nature Cell Biology, 13, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros, A. , Kadam, V. , Monyror, J. , Morales, L. C. , Pink, D. , Rieger, A. M. , Sipione, S. , & Posse de Chaves, E. (2022). In‐cell labeling coupled to direct analysis of extracellular vesicles in the conditioned medium to study extracellular vesicles secretion with minimum sample processing and particle loss. Cells, 11, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong, M. D. L. , Haouas, M. , Ural, M. S. , Desmaële, D. , Martineau‐Corcos, C. , & Gref, R. (2022). Degradation of polymer‐drug conjugate nanoparticles based on lactic and itaconic acid. International Journal of Molecular Science, 23, 14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, S. , Busatto, S. , Pham, A. , Tian, M. , Suh, A. , Carson, K. , Quintero, A. , Lafrence, M. , Malik, H. , Santana, M. X. , & Wolfram, J. (2019). Extracellular vesicle‐based drug delivery systems for cancer treatment. Theranostics, 9, 8001–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Li, J. , Cheng, C. , & Liu, S. (2020). Angiotensin‐converting enzyme 2 augments the effects of endothelial progenitor cells‐exosomes on vascular smooth muscle cell phenotype transition. Cell and Tissue Research, 382, 509–518. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Li, Y. , & Nie, G. (2021). Multifunctional biomolecule nanostructures for cancer therapy. Nature Review Materials, 6, 766–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Wang, L. , Lin, Z. , Tao, L. , & Chen, M. (2014). More efficient induction of antitumor T cell immunity by exosomes from CD40L gene‐modified lung tumor cells. Molecular Medicine Reports, 9, 125–131. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Altinoglu, S. , Takeda, Y. S. , & Xu, Q. (2015). Integrating protein engineering and bioorthogonal click conjugation for extracellular vesicle modulation and intracellular delivery. PLoS One, 10(11), e0141860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Zhao, J. , Zhang, L. , Wei, F. , Lian, Y. , Wu, Y. , Gong, Z. , Zhang, S. , Zhou, J. , Cao, K. , Li, X. , Xiong, W. , Li, G. , Zeng, Z. , & Guo, C. (2017). Role of tumor microenvironment in tumorigenesis. Journal of Cancer, 8, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Liu, Z. , Wang, P. , Li, S. , Zeng, J. , Tu, X. , Yan, Q. , Xiao, Z. , Pan, M. , & Zhu, F. (2018). Syncytin‐1, an endogenous retroviral protein, triggers the activation of CRP via TLR3 signal cascade in glial cells. Brain Behavior and Immunity, 67, 324–334. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Yi, J. , Chen, X. , Zhang, Y. , Xu, M. , & Yang, Z. (2016). The regulation of cancer cell migration by lung cancer cell‐derived exosomes through TGF‐β and IL‐10. Oncology Letters, 11, 1527–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, B. A. , Shubik, P. , Wilson, R. , Garcia, H. , & Feldman, R. (1978). The microcirculation in two transplantable melanomas of the hamster. II. Scanning electron microscopy. Cancer Letters, 4, 117–124. [DOI] [PubMed] [Google Scholar]
- Wei, F. , Ma, C. , Zhou, T. , Dong, X. , Luo, Q. , Geng, L. , Ding, L. , Zhang, Y. , Zhang, L. , Li, N. , Li, Y. , & Liu, Y. (2017). Exosomes derived from gemcitabine‐resistant cells transfer malignant phenotypic traits via delivery of miRNA‐222‐3p. Molecular Cancer, 16, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E. R. , & Göttlinger, H. (2011). The role of cellular factors in promoting HIV budding. Journal of Molecular Biology, 410, 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh, J. A. , Goberdhan, D. C. I. , O'Driscoll, L. , Buzas, E. I. , Blenkiron, C. , Bussolati, B. , Cai, H. , Di Vizio, D. , Driedonks, T. A. P. , Erdbrügger, U. , Falcon‐Perez, J. M. , Fu, Q. L. , Hill, A. F. , Lenassi, M. , Lim, S. K. , Mahoney, M. G. , Mohanty, S. , Möller, A. , Nieuwland, R. , Ochiya, T., … Witwer, K. W. (2024). Minimal information for studies of extracellular vesicles (MISEV2023): From basic advanced approaches. Journal of Extracellular Vesicles, 13, e12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, D. Y. , Hall, W. A. , Conrad, J. , Godal, A. , Flørenes, V. A. , & Fodstad, O. (1995). In vitro and in vivo variation in transferrin receptor expression on a human medulloblastoma cell line. Neurosurgery, 36, 1158–1163. [DOI] [PubMed] [Google Scholar]
- Wiedmer, S. K. , Multia, E. , Liangsupree, T. , & Riekkola, M. L. (2023). Automated on‐line isolation and fractionation method for subpopulations of extracellular vesicles. Methods in Molecular Biology, 2668, 99–108. [DOI] [PubMed] [Google Scholar]
- Wiklander, O. P. , Nordin, J. Z. , O'Loughlin, A. , Gustafsson, Y. , Corso, G. , Mäger, I. , Vader, P. , Lee, Y. , Sork, H. , Seow, Y. , Heldring, N. , Alvarez‐Erviti, L. , Smith, C. I. , Le Blanc, K. , Macchiarini, P. , Jungebluth, P. , Wood, M. J. , & Andaloussi, S. E. (2015). Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. Journal of Extracellular Vesicles, 4, 26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, H. L. , Frei, A. L. , Koessler, T. , Berger, M. D. , Dawson, H. , Michielin, O. , & Zlobec, I. (2024). The current landscape of spatial biomarkers for prediction of response to immune checkpoint inhibition. NPJ Precision Oncology, 8, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. C. , & Mackman, N. (2011). MPs or ICs? Blood, 117(4), 1101–1102. [DOI] [PubMed] [Google Scholar]
- Wilson, W. R. , & Hay, M. P. (2011). Targeting hypoxia in cancer therapy. Nature Reviews. Cancer, 11, 393–410. [DOI] [PubMed] [Google Scholar]
- Winberg, L. K. , Nielsen, C. H. , & Jacobsen, S. (2017). Surface complement C3 fragments and cellular binding of microparticles in patients with SLE. Lupus Science & Medicine, 4, e000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Buzás, E. I. , Bemis, L. T. , Bora, A. , Lässer, C. , Lötvall, J. , Nolte‐'t Hoen, E. N. , Piper, M. G. , Sivaraman, S. , Skog, J. , Théry, C. , Wauben, M. H. , & Hochberg, F. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles, 27, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, M. , Poupardin, R. W. , Ebner‐Peking, P. , Andrade, A. C. , Blöchl, C. , Obermayer, A. , Gomes, F. G. , Vari, B. , Maeding, N. , Eminger, E. , Binder, H. M. , Raninger, A. M. , Hochmann, S. , Brachtl, G. , Spittler, A. , Heuser, T. , Ofir, R. , Huber, C. G. , Aberman, Z. , … Strunk, D. (2022). A functional corona around extracellular vesicles enhances angiogenesis, skin regeneration and immunomodulation. Journal of Extracellular Vesicles, 11, e12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won, S. , Lee, C. , Bae, S. , Lee, J. , Choi, D. , Kim, M. G. , Song, S. , Lee, J. , Kim, E. , Shin, H. , Basukala, A. , Lee, T. R. , Lee, D. S. , & Gho, Y. S. (2023). Mass‐produced gram‐negative bacterial outer membrane vesicles activate cancer antigen‐specific stem‐like CD8+ T cells which enables an effective combination immunotherapy with anti‐PD‐1. Journal of Extracellular Vesicles, 12, e12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, M. D. , Moseley, G. W. , & van Spriel, A. B. (2004). Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue Antigens, 64, 533–542. [DOI] [PubMed] [Google Scholar]
- Xu, K. , Liu, Q. , Wu, K. , Liu, L. , Zhao, M. , Yang, H. , Wang, X. , & Wang, W. (2020). Extracellular vesicles as potential biomarkers and therapeutic approaches in autoimmune diseases. Journal of Translational Medicine, 18, 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M. , Ji, J. , Jin, D. , Wu, Y. , Wu, T. , Lin, R. , Zhu, S. , Jiang, F. , Ji, Y. , Bao, B. , Li, M. , Xu, W. , & Xiao, M. (2022). The biogenesis and secretion of exosomes and multivesicular bodies (MVBs): Intercellular shuttles and implications in human diseases. Genes & Diseases, 10, 1894–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Zeng, S. , Gong, Z. , & Yan, Y. (2020). Exosome‐based immunotherapy: a promising approach for cancer treatment. Molecular Cancer, 19, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan, M. , Shao, J. , Zhao, J. , Li, Q. , Dai, L. , & Li, J. (2018). Magnetic mesoporous silica nanoparticles cloaked by red blood cell membranes: applications in cancer therapy. Angewandte Chemie, 57, 6049–6053. [DOI] [PubMed] [Google Scholar]
- Yaddanapudi, K. , Meng, S. , Whitt, A. G. , Al Rayyan, N. , Richie, J. , Tu, A. , Eaton, J. W. , & Li, C. (2019). Exosomes from GM‐CSF expressing embryonic stem cells are an effective prophylactic vaccine for cancer prevention. Oncoimmunology, 8, 1561119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K. M. , & Cukierman, E. (2007). Modeling tissue morphogenesis and cancer in 3D. Cell, 130, 601–610. [DOI] [PubMed] [Google Scholar]
- Yamauchi, Y. , & Helenius, A. (2013). Virus entry at a glance. Journal of Cell Science, 126, 1289–1295. [DOI] [PubMed] [Google Scholar]
- Yan, Z. , Yin, H. , Wu, J. , Tian, G. , Li, M. , Liao, Z. , He, S. , Deng, H. , Ning, C. , Ding, Z. , Yuan, X. , Sui, X. , Chen, M. , Liu, S. , & Guo, Q. (2023). Engineering exosomes by three‐dimensional porous scaffold culture of human umbilical cord mesenchymal stem cells promote osteochondral repair. Materials Today. Bio, 19, 100549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R. , Andreu, Z. , Zavec, A. B. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Cordeiro‐da Silva, A. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. , … De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Zhang, X. , Chen, X. , Wang, L. , & Yang, G. (2017). Exosome mediated delivery of miR‐124 promotes neurogenesis after ischemia. Molecular Therapy. Nucleic Acids, 7, 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y. , Ghosh, K. , Epand, R. F. , Epand, R. M. , & Ghosh, H. P. (2003). Membrane fusion activity of vesicular stomatitis virus glycoprotein G is induced by low pH but not by heat or denaturant. Virology, 310, 319–332. [DOI] [PubMed] [Google Scholar]
- Yao, Z. , Qiao, Y. , Li, X. , Chen, J. , Ding, J. , Bai, L. , Shen, F. , Shi, B. , Liu, J. , Peng, L. , Li, J. , & Yuan, Z. (2018). Exosomes exploit the virus entry machinery and pathway to transmit alpha interferon‐induced antiviral activity. Journal of Virology, 92, e01578–016718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, X. , Mead, B. E. , Safaee, H. , Langer, R. , Karp, J. M. , & Levy, O. (2016). Engineering stem cell organoids. Cell Stem Cell, 18, 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, J. , Park, C. , Yi, G. , Lee, D. , & Koo, H. (2019). Active targeting strategies using biological ligands for nanoparticle drug delivery systems. Cancers, 11, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, G. , Park, J. W. , & Yoon, I. S. (2013). Solid lipid nanoparticles (SLNs) and nanostructured lipid carriers (NLCs): Recent advances in drug delivery. Journal of Pharmaceutical Investigation, 43, 353–362. [Google Scholar]
- Yoon, J. H. , Ashktorab, H. , Smoot, D. T. , Nam, S. W. , Hur, H. , & Park, W. S. (2020). Uptake and tumor‐suppressive pathways of exosome‐associated GKN1 protein in gastric epithelial cells. Gastric Cancer : Official Journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association, 23, 848–862. [DOI] [PubMed] [Google Scholar]
- Young, R. E. , Hofbauer, S. I. , & Riley, R. S. (2022). Overcoming the challenge of long‐term storage of mRNA‐lipid nanoparticle vaccines. Molecular Therapy : The Journal of the American Society of Gene Therapy, 30, 1792–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D. H. , Lu, Q. , Xie, J. , Fang, C. , & Chen, H. Z. (2010). Peptide‐conjugated biodegradable nanoparticles as a carrier to target paclitaxel to tumor neovasculature. Biomaterials, 31, 2278–2292. [DOI] [PubMed] [Google Scholar]
- Yu, H. , Liu, T. , Zhao, Z. , Chen, Y. , Zeng, J. , Liu, S. , & Zhu, F. (2014). Mutations in 3'‐long terminal repeat of HERV‐W family in chromosome 7 upregulate syncytin‐1 expression in urothelial cell carcinoma of the bladder through interacting with c‐Myb. Oncogene, 33, 3947–3958. [DOI] [PubMed] [Google Scholar]
- Yu, K. F. , Zhang, W. Q. , Luo, L. M. , Song, P. , Li, D. , Du, R. , Ren, W. , Huang, D. , Lu, W. L. , Zhang, X. , & Zhang, Q. (2013). The antitumor activity of a doxorubicin loaded, iRGD‐modified sterically‐stabilized liposome on B16‐F10 melanoma cells: In vitro and in vivo evaluation. International Journal Of Nanomedicine, 8, 2473–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yulia, M. , Kusters, D. H. M. , & Nusrat, A. (2018). Cadherins: cellular adhesive molecules serving as signalling mediators. The Journal of Physiology, 596, 3883–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakir, F. , Sharma, H. , Kaur, K. , Malik, B. , Vaidya, B. , Goyal, A. K. , & Vyas, S. P. (2011). Nanocrystallization of poorly water soluble drugs for parenteral administration. Jorunal of Biomedical Nanotechnology, 7, 127–129. [DOI] [PubMed] [Google Scholar]
- Zanetti‐Domingues, L. C. , Bonner, S. E. , lyer, R. S. , Martin‐Fernandez, M. L. , & Huber, V. (2020). Cooperation and interplay between EGFR signalling and extracellular vesicle biogenesis in cancer. Cells, 9, 2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zefferino, R. , Piccol, C. , Di Gioia, S. , Capitanio, N. , & Conese, M. (2021). How cells communicate with each other in the tumor microenvironment: Suggestions to design novel therapeutic strategies in cancer disease. International Journal of Molecular Sciences, 22, 2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, B. T. , Tian, H. , Sun, J. , Zou, J. B. , Zhang, X. F. , Cheng, J. X. , Shi, Y. J. , Fan, Y. , & Guo, D. Y. (2022). Urokinase‐type plasminogen activator receptor (uPAR) as a therapeutic target in cancer. Journal of Translational Medicine, 20, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Freitas, D. , Kim, H. S. , Fabijanic, K. , Li, Z. , Chen, H. , Mark, M. T. , Molina, H. , Martin, A. B. , Bojmar, L. , Fang, J. , Rampersaud, S. , Hoshino, A. , Matei, I. , Kenific, C. M. , Nakajima, M. , Mutvei, A. P. , Sansone, P. , Buehring, W. , … Lyden, D. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field‐flow fractionation. Nature Cell Biology, 20, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Meng, X. , Yang, Z. , Cao, Y. , Cheng, Y. , Wang, D. , Lu, H. , Shi, Z. , Dong, H. , & Zhang, X. (2019). Cancer cell membrane camouflaged nanoprobe for catalytic ratiometric photoacoustic imaging of microRNA in living mice. Advanced Materials, 31, e1807888. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Li, C. , Huang, R. , Teng, H. , Zhang, Y. , Zhou, M. , Liu, X. , Fan, B. , Luo, H. , He, A. , Zhao, A. , Lu, M. , Chopp, M. , & Zhang, Z. G. (2022). Cerebral endothelial cell derived small extracellular vesicles improve cognitive function in aged diabetic rats. Frontiers in Aging Neuroscience, 14, 926485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Jeppesen, D. K. , Higginbotham, J. N. , Graves‐Deal, R. , Trinh, V. Q. , Ramirez, M. A. , Sohn, Y. , Neininger, A. C. , Taneja, N. , McKinley, E. T. , Niitsu, H. , Cao, Z. , Evans, R. , Glass, S. E. , Ray, K. C. , Fissell, W. H. , Hill, S. , Rose, K. L. , Huh, W. J. , Washington, M. K., … Coffey, R. J. (2021). Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nature Cell Biology, 23, 1240–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Zhou, X. , Yao, Q. , Liu, Y. , Zhang, H. , & Dong, Z. (2017). HIF‐1‐mediated production of exosomes during hypoxia is protective in renal tubular cells. American Journal of Physiology. Renal Physiology, 313, F906–F913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Xu, Q. , Zi, Z. , Liu, Z. , Wan, C. , Crisman, L. , Shen, J. , & Liu, X. (2020). Programmable extracellular vesicles for macromolecule delivery and genome modifications. Developmental Cell, 55, 784–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Li, L. , Yu, J. , Zhu, D. , Zhang, Y. , Li, X. , Gu, H. , Zhang, C. Y. , & Zen, K. (2014). Microvesicle‐mediated delivery of transforming growth factor β1 siRNA for the suppression of tumor growth in mice. Biomaterials, 35, 4390–4400. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Liang, F. , Zhang, D. , Qi, S. , & Liu, Y. (2023). Metabolites as extracellular vesicle cargo in health, cancer, pleural effusion, and cardiovascular diseases: An emerging field of study to diagnostic and therapeutic purposes. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 157, 114046. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Wu, J. L. Y. , Lazarovits, J. , & Chan, W. C. W. (2020). An analysis of the binding function and structural organization of the protein corona. Jouernal of the American Chemical Society, 142, 8827–8836. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, M. , Xie, Z. , Ding, Y. , Huang, J. , Yao, J. , Lv, Y. , & Zuo, J. (2022). Research progress and direction of novel organelle‐migrasomes. Cancers, 15, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Chen, X. , Gao, S. , Fang, X. , & Ren, S. (2024). 3D bioprinted tumor model: a prompt and convenient platform for overcoming immunotherapy resistance by recapitulating the tumor microenvironment. Cell Oncology, 47, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Liu, J. , Guan, L. , Zhang, Y. , Dong, P. , Li, J. , Liang, X. , & Komiyama, M. (2018). Fabrication of aqueous nanodispersion from natural DNA and chitosan as eminent carriers for water‐insoluble bioactives International Jorunal of Biological Macromolecules, 118(Pt A), 263–270. [DOI] [PubMed] [Google Scholar]
- Zheng, M. , Huang, M. , Ma, X. , Chen, H. , & Gao, X. (2019). Harnessing exosomes for the development of brain drug delivery systems. Bioconjugate Chemistry, 30, 994–1005. [DOI] [PubMed] [Google Scholar]
- Zheng, W. , Rädler, J. , Sork, H. , Niu, Z. , Roudi, S. , Bost, J. P. , Görgens, A. , Zhao, Y. , Mamand, D. R. , Liang, X. , Wiklander, O. P. B. , Lehto, T. , Gupta, D. , Nordin, J. Z. , & El Andaloussi, S. (2023). Identification of scaffold proteins for improved endogenous engineering of extracellular vesicles. Nature Communications, 14, 4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. , Tu, C. , Zhang, J. , & Wang, J. (2019). Inhibition of multiple myeloma derived exosomes uptake suppresses the functional response in bone marrow stromal cell. International Journal Of Oncology, 54, 1061–1070. [DOI] [PubMed] [Google Scholar]
- Zhou, B. , Xu, K. , Zheng, X. , Chen, T. , Wang, J. , Song, Y. , Shao, Y. , & Zheng, S. (2020). Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduction and Targeted Therapy, 5, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Liu, L. , Liu, Y. , Zhou, P. , Yan, Q. , Yu, H. , Chen, X. , & Zhu, F. (2021). Implication of human endogenous retrovirus W family envelope in hepatocellular carcinoma promotes MEK/ERK‐mediated metastatic invasiveness and doxorubicin resistance. Cell Death Discovery, 7(1), 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. P. , Tian, T. , Wang, J. Y. , He, J. N. , Chen, T. , Pan, M. , Xu, L. , Zhang, H. X. , Qiu, X. T. , Li, C. C. , Wang, K. K. , Shen, H. , Zhang, G. G. , & Bai, Y. P. (2018). Hypoxia‐elicited mesenchymal stem cell‐derived exosomes facilitates cardiac repair through miR‐125b‐mediated prevention of cell death in myocardial infarction. Theranostics, 8, 6163–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Badawi, M. , Pomeroy, S. , Sutaria, D. S. , Xie, Z. , Baek, A. , Jiang, J. , Elgamal, O. A. , Mo, X. , Perle, K. , Chalmers, J. , Schmittgen, T. D. , & Phelps, M. A. (2017). Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. Journal of Extracellular Vesicles, 6, 1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocco, D. , Ferruzzi, P. , Cappello, F. , Kuo, W. P. , & Fais, S. (2014). Extracellular vesicles as shuttles of tumor biomarkers and anti‐tumor drugs. Frontiers in Oncology, 4, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetemelk, M. , Rausch, M. , Colin, D. J. , Dormond, O. , & Nowak‐Sliwinska, P. (2019). Short‐term 3D culture systems of various complexity for treatment optimization of colorectal carcinoma. Scientific Reports, 9, 7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, X. , Lei, Q. , Luo, X. , Yin, J. , Chen, S. , Hao, C. , Shiyu, L. , & Ma, D. (2023). Advances in biological functions and applications of apoptotic vesicles. Cell Communication and Signaling : CCS, 21, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Z. , Li, H. , Xu, G. , Hu, Y. , Zhang, W. , & Tian, K. (2023). Current knowledge and future perspectives of exosomes as nanocarriers in diagnosis and treatment of diseases. International Journal of Nanomedicine, 18, 4751–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu, Y. , Wu, W. , Zhao, X. , Li, Y. , Wang, W. , Zhong, C. , Zhang, Y. , & Zhao, X. (2014). Enhancement of solubility, antioxidant ability and bioavailability of taxifolin nanoparticles by liquid antisolvent precipitation technique. International Journal of Pharmaceutics, 471, 366–376. [DOI] [PubMed] [Google Scholar]


