Abstract
Beyond the normal DNA transactions mediated by topoisomerase II, we have recently demonstrated that the cleavage activity of the two human topoisomerase II isoforms is several-fold stimulated if a ribonucleotide rather than a deoxyribonucleotide is present at the scissile phosphodiester in one strand of the substrate. Here we show that ribonucleotides exert a position-specific effect on topoisomerase II-mediated cleavage without altering the sequence specificity of the enzyme. Ribonucleotides located within the 4 bp cleavage stagger stimulate topoisomerase II-mediated cleavage, whereas ribonucleotides located outside the stagger in general have an inhibitory effect. Results obtained from competition experiments indicate that the position-specific effect of ribonucleotides on topoisomerase II activity is caused by altered substrate interaction. When cleavage is performed with substrates containing one ribonucleotide in both strands or several ribonucleotides in one strand the effect of the individual ribonucleotides on cleavage is not additive. Finally, although topoisomerase II recognizes substrates with longer stretches of ribonucleotides, an RNA/DNA hybrid where one strand is composed entirely of RNA is not cleaved by the enzyme. The positional effect of ribonucleotides on topoisomerase II-mediated cleavage shares many similarities to the positional effect exerted by either abasic sites or base mismatches, demonstrating a general influence of DNA imperfections on topoisomerase II activity.
INTRODUCTION
Eukaryotic topoisomerase II is a multifunctional nuclear enzyme with the main purpose of regulating the topological structure of DNA in response to various DNA metabolic processes (1). The dimeric enzyme performs the different DNA transactions by making a transient double-stranded break in the DNA backbone through which another DNA helix can be transferred before the break is religated (2,3). During the cleavage event topoisomerase II creates a 4 bp stagger with one subunit of the enzyme being covalently linked to each of the newly generated 5′-ends.
We have recently demonstrated that human topoisomerases IIα and IIβ, besides their activities on canonical DNA substrates, also cleave RNA-containing substrates, where they become covalently linked to a ribonucleotide rather than a deoxyribonucleotide (4). This observation has raised new perspectives for eukaryotic topoisomerase II and indicates a potential role of the enzyme in RNA modulation. Earlier studies with Escherichia coli topoisomerase III have shown that this enzyme can operate with similar activity on either DNA or RNA strands (5) and catalyze topological changes on circular RNA as well as DNA (6). Likewise, an endoribonuclease activity has recently been described for eukaryotic type I topoisomerases, including vaccinia virus and human topoisomerase I (7). The two enzymes recognize and cleave substrates composed partially of RNA and form covalent linkage to the ribonucleotide at the 3′-end of the cleavage site. The topoisomerase I–RNA intermediate in these cases decays rapidly through nucleophilic attack by the vicinal 2′-OH group of the ribose sugar, leading to formation of a 2′,3′-cyclic product and release of the enzyme from the complex (7). Taken together, the above findings indicate that DNA topoisomerases in general act as RNA processing enzymes or even as RNA topoisomerases.
To gain deeper insight into the mechanism of action and the cellular functions of eukaryotic topoisomerase II we have in the present study further investigated the influence of RNA or ribonucleotide-containing substrates on topoisomerase II activity. Our data demonstrate that ribonucleotides exert a position-specific effect on topoisomerase II-mediated cleavage and that the enzyme, besides cleaving substrates containing single ribonucleotides, also cleaves substrates with longer stretches of RNA. However, it is unable to cleave RNA/DNA hybrids and single-stranded RNA, disfavouring a role of topoisomerase II as an RNA topoisomerase.
MATERIALS AND METHODS
Oligonucleotides
All DNA and RNA oligonucleotides were synthesized on a Model 394 DNA synthesizer by DNA Technology Corp. and purified by preparative polyacrylamide gel electrophoresis (PAGE).
Preparation of cleavage substrates
The various cleavage substrates were prepared by hybridizing 10 pmol of the 25mer top strand to 10 pmol of the complementary 28mer bottom strand in 40 mM Tris–HCl, pH 7.5, 20 mM MgCl2 and 50 mM NaCl. The mixture was heated to 70°C for 2 min and allowed to cool slowly to room temperature. After hybridization, the top strand was 3′-end-labeled with [α 32P]dATP and Sequenase. For studies of topoisomerase II-mediated cleavage of the RNA/DNA hybrid the 25mer top strand was 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase before it was hybridized to the 28mer bottom DNA strand.
Purification of recombinant human topoisomerases IIα and IIβ
The recombinant human topoisomerase IIα and IIβ enzymes fused to a c-myc tag and a hexahistidine tail at the C-terminal end were overexpressed and purified from the protease-deficient yeast strain BJ201, carrying an expression vector containing either the topoisomerase IIα or IIβ cDNA under control of the yeast GAL1 promoter. The transformed cells were grown in rich YEP medium containing 2% glucose to a cell density of 5 × 107 cells/ml. The cells were then collected by centrifugation and resuspended in fresh YEP medium containing 2% galactose instead of glucose for inducible expression of topoisomerase II. After 16 h induction the cells were harvested and washed once in distilled water. The cell pellets were either flash frozen in liquid nitrogen for storage at –80°C or lysed immediately for enzyme purification. The enzymes were purified to near homogeneity as described previously (4). Briefly, the purification procedure utilized sequential chromatography of a cleared yeast lysate on a Ni2+–nitrilotriacetic acid matrix, a heparin–Sepharose column and a Source S column.
Topoisomerase II-mediated cleavage
A standard cleavage reaction was set up by incubating 1 pmol of topoisomerase II with 0.02 pmol of labelled substrate in 50 µl of 10 mM Tris–HCl, pH 7.0, 2.5 mM MgCl2, 2.5 mM CaCl2, 20 mM NaCl, 15 µg/ml bovine serum albumin and 0.1 mM EDTA (cleavage buffer) at 37°C for 10 min. SDS (1% final concentration) was then added to stop the reaction and the samples were analyzed by SDS–PAGE. Covalent complex formation was revealed by transfer of the radiolabelled oligonucleotide to the topoisomerase II polypeptide. Reaction products were visualized and quantified using a Molecular Dynamics PhosphorImager system. To determine the actual cleavage site of topoisomerase II on the various ribonucleotide-containing substrates, SDS-stopped samples were subjected to phenol extraction and the protein-linked cleavage complexes were recovered from the phenol/water interphase as described previously (8). The complexes were subsequently ethanol precipitated, proteinase K digested (1 mg/ml, 2 h at 37°C) and analyzed by electrophoresis on 12% denaturing polyacrylamide gels. To examine topoisomerase II-mediated cleavage of the 5′-end-labeled RNA/DNA hybrid, cleavage reactions were performed as described above, but in the presence of 40 U/reaction RNasin (Promega) to inhibit RNase reactions. The SDS-stopped samples were then ethanol precipitated and analyzed on a 12% denaturing polyacrylamide gel.
RESULTS
Ribonucleotides exert a position-specific effect on human topoisomerase II-mediated cleavage
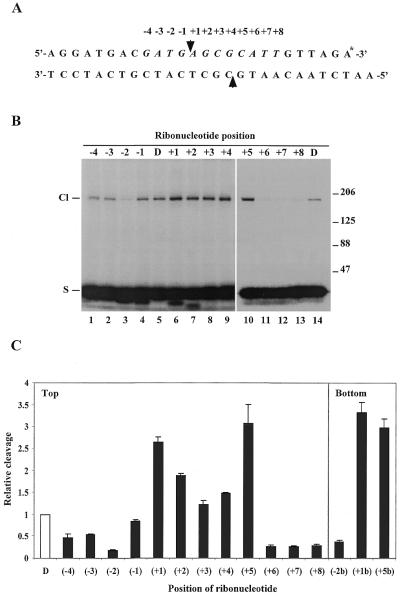
We have recently demonstrated that human topoisomerases IIα and IIβ have an increased cleavage activity on short DNA substrates containing a ribonucleotide rather than a deoxyribonucleotide at the scissile phosphodiester of one strand (4). The observation strongly indicates that topoisomerase II has the ability to operate on RNA or RNA-containing substrates. To gain further information on the interaction of topoisomerase II with different RNA-containing substrates, we have employed the same oligonucleotide-based system to investigate the position-specific effect of a single ribonucleotide on topoisomerase II-mediated cleavage. For this purpose a series of substrates were generated which were identical in sequence except that a single ribonucleotide substituted for the normal deoxyribonucleotide at different positions ranging from –4 to +8 relative to the topoisomerase II-mediated cleavage site in the top strand of a strong topoisomerase II recognition sequence (Fig. 1A). The 12 bp region encompassing positions –4 to +8 is likely to hold the sequence information determining the actual site of topoisomerase II-mediated cleavage (9). The ability of human topoisomerase IIα to cleave the various ribonucleotide-containing substrates was subsequently determined as described in Materials and Methods. Cleavage resulted in radioactive labelling of the enzyme due to its covalent linkage to the labelled top strand (Fig. 1B). The polypeptide bands visible in the autoradiogram migrated with an apparent mobility of 170 kDa, as predicted from the molecular weight of human topoisomerase IIα (10). The relative levels of cleavage were determined by PhosphorImager scanning of the gel and mean values from three independent experiments are presented in the histogram in Figure 1C. Evidently, the single ribonucleotides exerted a strong position-specific effect on topoisomerase IIα-mediated cleavage. Cleavage was stimulated when the substrates contained a ribonucleotide at positions +1 to +5, whereas it was inhibited when the ribonucleotide was located at positions –1 to –4 or +6 to +8. Similar results were obtained with human topoisomerase IIβ (data not shown). Thus, within the central 4 bp overhang generated by topoisomerase II-mediated cleavage ribonucleotide substitution enhanced the level of cleavage, whereas it was decreased if the ribonucleotide was incorporated either immediately upstream or downstream of the 4 bp stagger. The only exception was at position +5, where cleavage was stimulated rather than inhibited. When a ribonucleotide was incorporated at various positions in the bottom strand of the substrate and cleavage still measured on the labelled top strand a similar positional effect was observed (Fig. 1C). The results not only substantiate the position-specific effect of ribonucleotides on topoisomerase II-mediated strand scission, but also lend support to our previous finding that a single ribonucleotide equally influences cleavage of both strands, suggestive of a strong coordination between the two enzyme subunits during substrate recognition and cleavage (4).
Figure 1.
Position-specific effect of ribonucleotides on human topoisomerase II-mediated cleavage. (A) Schematic illustration of the topoisomerase II cleavage substrate. The arrowheads represent the points of topoisomerase II-mediated cleavage. A single ribonucleotide was incorporated into the top strand of the substrate at different positions ranging from –4 to +8 relative to the cleavage site as indicated above the sequence. The asterisk at the 3′-end of the top strand illustrates radioactive labelling. (B) Human topoisomerase IIα-mediated cleavage of the ribonucleotide-containing substrates. Cleavage was performed as described in Materials and Methods. The SDS-stopped samples were loaded directly on an 8% SDS–polyacrylamide gel and cleavage complex formation was visualized after autoradiography due to covalent linkage of the protein to the labelled top strand. The position of the ribonucleotide in the substrate is as indicated above the lanes. D represents the control DNA substrate. Cl and S indicate the positions of the protein-linked cleavage complex and the cleavage substrate, respectively. The numbers to the right of the gel illustrate the sizes of protein markers in kDa. (C) Schematic presentation of the positional effect of ribonucleotides on human topoisomerase IIα-mediated cleavage. Cleavage experiments were performed as described in (B) using substrates containing a single ribonucleotide located at different positions ranging from –4 to +8 in the top strand or at the –2, +1 or +5 position in the bottom strand, as indicated below the histogram. In all cases cleavage was measured on the top strand. Cleavage levels were determined after PhosphorImager scanning of the gels and are presented relative to the cleavage level obtained with the control DNA substrate. Data represent the averages of three independent experiments and standard deviations are indicated by error bars.
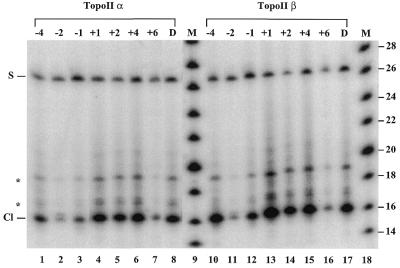
To investigate whether the position-specific effect of ribonucleotides on topoisomerase IIα- and IIβ-mediated cleavage reflects an altered sequence specificity of the enzymes, cleavage experiments were performed in which the protein-linked cleavage complexes were isolated from a phenol/water interphase and analyzed on a 12% denaturing polyacrylamide gel after proteinase K treatment (Fig. 2). Although cleavage levels varied significantly depending on the position of the ribonucleotide, both human topoisomerase IIα and IIβ gave rise to one prominent cleavage product with the various ribonucleotide-containing substrates. The cleavage products migrated to the same position as those obtained with the control DNA substrate, demonstrating that the site of topoisomerase II-mediated cleavage was not changed by the presence or the position of the ribonucleotide. Thus, human topoisomerase II does not cleave at the position of the ribonucleotide, rather, the presence of the ribonucleotide alters the interaction between the enzyme and its normal site of action.
Figure 2.
Determination of the sequence specificity of topoisomerase II on ribonucleotide-containing substrates. Topoisomerase II-mediated cleavage of substrates containing a ribonucleotide at different positions was performed as described in Materials and Methods. Cleavage products were isolated from a phenol/water interphase and analyzed by electrophoresis on a 12% denaturing polyacrylamide gel after proteinase K treatment. Lanes 1–8 and 10–17 show human topoisomerase IIα- and IIβ-mediated cleavage, respectively. Lanes 9 and 18 show DNA size markers (M) increasing in steps of two bases. The position of the ribonucleotide relative to the cleavage site is as indicated above the lanes. D represents the control DNA substrate. S indicates the cleavage substrate remaining in the interphase after phenol extraction. Cl indicates the cleavage product, for which migration is retarded by ∼1 base due to residual undigested protein. The asterisks indicate cleavage products having a longer protein fragment covalently linked due to partial proteinase K digestion (4,9).
The position-specific effect of ribonucleotides on topoisomerase II-mediated cleavage is caused by alterations in substrate interaction
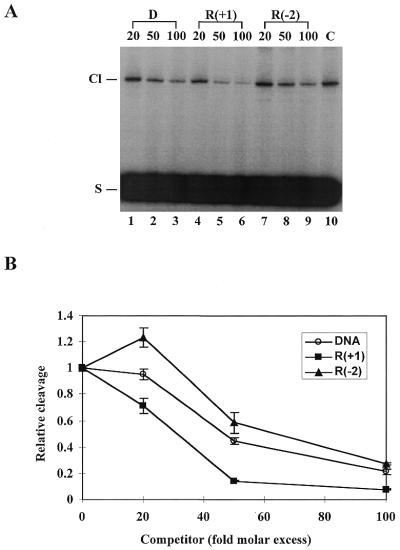
To further characterize the interaction of topoisomerase II with the various ribonucleotide-containing substrates, a competition experiment was performed, where topoisomerase II was incubated with the 3′-end-labelled control DNA substrate, either in the absence or presence of excess amounts of a cold competitor in a standard cleavage reaction. The cold competitor used in the experiment was the control DNA substrate or a ribonucleotide-containing substrate identical to the DNA control except that it had a ribonucleotide at position +1 or –2. These two positions were chosen since they give a high stimulatory or inhibitory effect on cleavage, respectively, upon insertion of a ribonucleotide. Each of the cold substrates competed with the 3′-end-labelled DNA substrate in a concentration-dependent manner, leading to reduced formation of the radiolabelled protein-linked cleavage complexes (Fig. 3A). The relative levels of labelled cleavage complexes obtained in the competition experiments were furthermore plotted as a function of the competitor concentration (Fig. 3B). Clearly, as compared to the DNA control, the substrate containing a ribonucleotide at the +1 position competed more efficiently for topoisomerase II, whereas the substrate with a ribonucleotide at the –2 position acted as a poor competitor. Thus, when a ribonucleotide-containing substrate is present together with a normal DNA substrate, the enzyme preferentially interacts with the ribonucleotide-containing substrate if the ribonucleotide is located at position +1, but prefers the DNA substrate if the ribonucleotide is located at position –2. The data therefore indicate that ribonucleotides exert their effect on topoisomerase II by affecting the ability of the enzyme to interact with the substrate, although the experimental set-up cannot exclude a direct influence on the cleavage reaction per se.
Figure 3.
Effect of ribonucleotides on the substrate preference of human topoisomerase II. (A) Competition experiment exploring the substrate preference of topoisomerase II. Cleavage experiments were performed by incubating human topoisomerase IIα with the 3′-end-labelled control DNA substrate in the absence and presence of excess amounts of cold competitor. Following incubation, the reactions were stopped with SDS. The samples were submitted to SDS–PAGE and cleavage complex formation was visualized by autoradiography. The cold competitor used in the experiment was either the control DNA substrate (lanes 1–3), the substrate containing a ribonucleotide at the +1 position (lanes 4–6) or the substrate containing a ribonucleotide at the –2 position (lanes 7–9) as indicated. The cold competitor was used in 20-, 50- or 100-fold molar excess relative to the labelled DNA substrate, as indicated above the lanes. C represents a control experiment performed in the absence of any competitor. Cl and S indicate the positions of the protein-linked cleavage complex and the cleavage substrate, respectively. (B) Graphic illustration of the competition experiment performed in (A). Cleavage levels were determined after PhosphorImager scanning of the gel and are presented relative to the cleavage level obtained in the absence of any competitor. Data represent the averages of three independent experiments and standard deviations are indicated by error bars.
The effect of ribonucleotides on topoisomerase II-mediated cleavage is not additive in nature
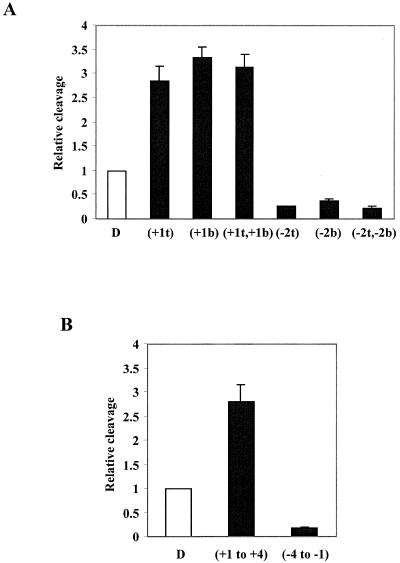
The dimeric topoisomerase II enzyme operates by creating double-strand breaks in the DNA backbone with the two enzyme subunits acting 4 bp apart on the two strands (11,12). We have demonstrated that the presence of a ribonucleotide in one of the strands in the topoisomerase II recognition sequence equally influences cleavage of both strands, suggestive of a strong coordination between the two enzyme subunits during the cleavage event (Fig. 1C; 4). We therefore addressed the question of whether co-existence of two ribonucleotides in the substrate, one in each strand, would exert an additive effect on topoisomerase II-mediated cleavage. For this purpose cleavage reactions were performed using substrates where ribonucleotides were inserted at the +1 (stimulatory) or –2 (inhibitory) positions in both strands. As seen from the histogram in Figure 4A, the substrates with two ribonucleotides gave approximately the same level of cleavage as the corresponding substrates with a ribonucleotide in only one of the strands, demonstrating that two ribonucleotides, when present one in each strand, cannot act in concert to further stimulate or inhibit cleavage. Rather, one ribonucleotide is sufficient to exert an optimal effect on the interaction between the dimeric enzyme and its substrate.
Figure 4.
Examination of the effect of multiple ribonucleotides on topoisomerase II-mediated cleavage. (A) Two ribonucleotides located one in each strand do not exert an additive effect on cleavage. Substrates harbouring a ribonucleotide in either one or both strands were incubated with human topoisomerase IIα as described in Materials and Methods and the SDS-stopped samples were analyzed on an 8% SDS–polyacrylamide gel. Cleavage levels were determined after PhosphorImager scanning of the gel and are schematically presented relative to the cleavage level obtained with the control DNA substrate (D). As indicated below the histogram, one set of substrates contained either a ribonucleotide at the +1 position of the top strand (+1t), of the bottom strand (+1b) or of both strands (+1t,+1b). The other set included substrates having a ribonucleotide at the –2 position of the top strand (–2t), of the bottom strand (–2b) or of both strands (–2t,–2b). Data represent the averages of three independent experiments and standard deviations are indicated by error bars. (B) Ribonucleotides present in the same strand do not exert an additive effect on cleavage. Cleavage experiments were performed as in (A). Cleavage levels were determined after PhosphorImager scanning of the gel and are schematically presented in the histogram relative to the cleavage level obtained with the control DNA substrate (D). The substrates used in the experiment contained a row of four consecutive ribonucleotides in the top strand either at positions –4 to –1 or +1 to +4 as indicated below the histogram. Data represent the averages of three independent experiments and standard deviations are indicated by error bars.
To further investigate the effect of ribonucleotides on topoisomerase II-mediated cleavage, experiments were performed to analyze the activity of topoisomerase II on DNA substrates containing longer stretches of RNA within the enzyme recognition sequence. Cleavage was thus performed using a substrate with a row of four consecutive ribonucleotides in the top strand, either in the cleavage stagger at positions +1 to +4 or upstream of the stagger at positions –4 to –1. As shown in the histogram in Figure 4B, both substrates were cleaved by human topoisomerase II, but the efficiencies of the cleavage reactions varied significantly. As compared to the control DNA substrate, cleavage was stimulated when the four ribonucleotides were present in the 4 bp stagger, but inhibited when the ribonucleotides were located upstream of the cleavage stagger, in accordance with our observations on single ribonucleotide substitutions. The level of cleavage stimulation or inhibition given by the four ribonucleotides was furthermore comparable to that obtained when a single ribonucleotide was located at the +1 or –2 position, respectively (compare Fig. 4A and B), demonstrating that ribonucleotides inserted in the same strand also exert a non-additive effect on topoisomerase II-mediated cleavage.
Human topoisomerase II does not operate on RNA/DNA hybrids
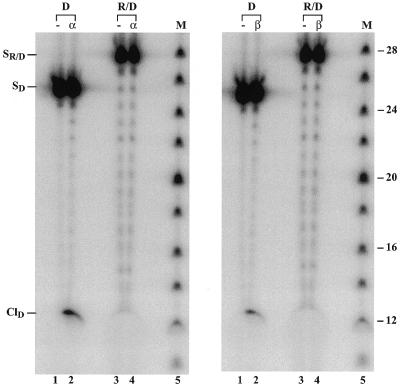
Based on the observed cleavage by topoisomerase II of DNA substrates with internal RNA stretches, we next investigated the ability of the enzyme to operate on an RNA/DNA hybrid. The hybrid used in this experiment contained a 5′-end-labelled top strand made up entirely of RNA, but was otherwise identical to the substrates used in the previous experiments. For comparison, a control experiment was included, where a normal DNA duplex was used as substrate, also labelled at the 5′-end of the top strand. As seen in Figure 5, both human topoisomerase IIα and IIβ cleaved the control DNA substrate, where cleavage resulted in a 12mer protein-free cleavage product. However, no cleavage occurred with the RNA/DNA hybrid, suggesting an inability of the human topoisomerase II isoforms to recognize and cleave substrates in which one strand is composed entirely of RNA around the topoisomerase II recognition site. Migration of the RNA strand in the gel was delayed as compared to that of the control DNA strand due to the presence of the 2′-OH group in the ribose residues.
Figure 5.
Topoisomerase II-mediated cleavage of an RNA/DNA hybrid. The RNA/DNA hybrid (R/D) or the control DNA substrate (D) labelled at the 5′-end of the top strand was incubated with human topoisomerase II as described in Materials and Methods. After ethanol precipitation the samples were submitted to electrophoresis on a 12% denaturing polyacrylamide gel and cleavage complex formation was visualized upon autoradiography. The left and right panels show human topoisomerase IIα- and IIβ-mediated cleavage, respectively. Lanes 1 and 2 (both panels) show experiments performed with the control DNA substrate in the absence and presence of topoisomerase II as indicated. Lanes 3 and 4 (both panels) represent cleavage reactions performed with the RNA/DNA hybrid in the absence and presence of topoisomerase II, as indicated. M represents DNA size markers increasing in steps of two bases. SR/D and SD indicate the positions of the hybrid and control DNA substrates having a labelled 25mer RNA and DNA strand, respectively. ClD represents the position of cleavage products obtained with the control DNA substrate. Low levels of substrate degradation are seen on the gel, which are most pronounced for the RNA strand due to the presence of residual RNase activity.
To further prove that lack of topoisomerase II-mediated cleavage of the RNA/DNA hybrid was not simply due to a failure in the sensitivity of our assay, cleavage reactions were performed in the presence of the anti-tumor drug mAMSA, which normally stimulates cleavage complex formation (13). Although mAMSA exhibited a substantial stimulatory effect on cleavage of the control DNA substrate, no cleavage could be detected of the RNA/DNA hybrid in the presence of the drug (data not shown), strongly indicating an intrinsic failure of topoisomerase II to interact with this substrate.
The results obtained with topoisomerase II share similarities to those obtained with eukaryotic topoisomerase I. The latter enzyme also cleaves DNA substrates with stretches of RNA (14), but does not cleave RNA/DNA hybrids (15). The inability of topoisomerase II to operate on the RNA/DNA hybrid disfavours the hypothesis that the enzyme has an intrinsic capability as an RNA topoisomerase. This is furthermore supported by the observation that topoisomerase II is unable to cleave a 25mer single-stranded RNA substrate (data not shown).
DISCUSSION
Since the discovery of topoisomerases in the 1970s (16), this class of enzymes has generally been considered as DNA modulators, which change the topological structure of DNA by cleaving one or both strands of the DNA duplex, subsequently transferring another DNA molecule through the transient gap (17). More recent discoveries on topoisomerases have, however, challenged this traditional definition and argue for the existence of an intrinsic RNA topoisomerase activity for at least some of these enzymes. Studies performed with E.coli topoisomerase III have demonstrated that this enzyme not only cleaves RNA, but also catalyzes topological changes in RNA substrates and therefore contains the expected capabilities of an RNA topoisomerase (6). Also, several eukaryotic type I topoisomerases have recently been shown to operate on different RNA-containing substrates, although no evidence has been found for a contribution of these enzymes as true RNA modulators (7,14,15).
Recently, we addressed the question of whether eukaryotic topoisomerase II acts on RNA-containing substrates and has activity as an RNA modulator. Our previous results showed that human topoisomerase II has increased cleavage activity if a deoxyribonucleotide is substituted by a ribonucleotide at one of the scissile phosphodiesters in the DNA duplex (4). In the present study we have demonstrated that ribonucleotides exert a position-specific effect on topoisomerase II-mediated cleavage. Ribonucleotides located inside the 4 bp cleavage stagger stimulate cleavage, whereas ribonucleotides located outside the stagger in general have an inhibitory effect. No matter the position of a ribonucleotide, it does not change the overall sequence specificity of the enzyme, but rather the ribonuclotide seems to influence the ability of the enzyme to interact with its normal recognition site. Although the enzyme cleaves substrates containing stretches of RNA, it is unable to cleave an RNA/DNA hybrid with one strand composed entirely of ribonucleotides or a single-stranded RNA molecule.
The fact that ribonucleotides located at different positions within the topoisomerase II recognition sequence can exert opposite effects on enzyme-mediated cleavage addresses the question of the mechanistic basis underlying the positional effect of ribonucleotides. The level of complex formation obtained during topoisomerase II-mediated cleavage reflects the equilibrium established between the forward cleavage reaction and the reverse ligation reaction, where a prerequisite to cleavage is binding of the enzyme to the substrate (3,18). We have previously demonstrated that the presence of a ribonucleotide at different positions in the substrate does not alter the rate of topoisomerase II-mediated ligation (4). In agreement with this, the results obtained from the competition experiments performed in the present study indicate that the ribonucleotide, according to its position, alters the preference of topoisomerase II for the specific substrate. This points to substrate binding as the step in the catalytic cycle which is influenced by ribonucleotide incorporation. Thus, depending on the position of the ribonucleotide, it might have a positive or negative effect on the direct contacts established between the enzyme and its substrate.
Intriguingly, the positional effect of ribonucleotides on topoisomerase II-mediated cleavage shows a remarkable similarity to the positional effect exerted on topoisomerase II by base mismatches and abasic sites. Like ribonucleotides, these DNA imperfections in general stimulate cleavage when they are located within the 4 bp stagger and inhibit cleavage when present outside this region (19–21). For ribonucleotides an exception to this rule is seen, when a ribonucleotide is located at position +5, since cleavage in this case is strongly stimulated. A base mismatch at this position may sometimes stimulate cleavage (19), whereas loss of a base here in general will inhibit cleavage (20,21). In support of the overall similarity in the positional effect given by the three different types of DNA imperfections, several studies have indicated that topoisomerase II primarily recognizes bases located outside the cleavage stagger, as sequence alterations here have a significant effect on topoisomerase II-mediated cleavage, whereas alterations occurring between the points of scission are easily tolerated (22,23).
It has been speculated that the stimulatory effect on topoisomerase II-mediated cleavage seen when either abasic sites or base mismatches are present in the 4 bp stagger results from a hampered ligation reaction. Thus, the destabilized duplex structure caused by the presence of base mismatches and abasic sites might decrease the probability of a proper ligation event due to ‘wobbling’ of the protein-linked DNA strand, whereby the level of cleavage complexes increases (19). However, in disfavour of this hypothesis, Osheroff and co-workers have demonstrated that, at least when abasic sites are present in the substrate, topoisomerase II-mediated ligation is unaffected or even increased (20,21), as we have recently shown for ribonucleotide substitutions (4). In further agreement with the data obtained on substrate preferences from the competition experiments presented in this study, they suggest that the structural changes caused by the abasic site in the DNA helix rather affect the ability of topoisomerase II to recognize and bind the substrate (21). Recently, the structural alterations caused in the DNA duplex by the presence of an abasic site have been determined by NMR spectroscopy. They involve loss of base stacking and production of a bend at the topoisomerase II cleavage site (24), which might severely influence the interaction between the enzyme and its substrate. The structural alterations caused by a base mismatch, an abasic site or a ribonucleotide substitution are thought to be very different in nature. The former two DNA lesions result from either base alteration or base loss, whereas a ribonucleotide when inserted into DNA represents a modification of the sugar ring. In addition, due to the increased stability of RNA/DNA hybrids (25,26), the presence of ribonucleotides in the substrate may locally strengthen hybridization between the two strands, whereas abasic sites and base mismatches might harm proper hybridization. Recently, another kind of DNA imperfection, cytosine arabinoside incorporation, has been found to affect topoisomerase II-mediated cleavage in a position-specific manner (27). Compared to DNA, cytosine arabinoside, like a ribonucleotide, has a modification in the sugar ring rather than in the base. The positional effect due to cytosine arabinoside shows similarities to that obtained with ribonucleotide substitutions, abasic sites and base mismatches, but deviates in the region upstream of the cleavage stagger, where insertion of cytosine arabinoside stimulates rather than inhibits cleavage. The fact that a ribonucleotide, which in DNA presents a modification resembling that due to cytosine arabinoside insertion, still exerts a positional effect more related to that obtained by base modifications demonstrates that a closer examination of the structural alterations occuring in the individual cases is required to understand the influence of the different DNA imperfections on topoisomerase II action.
In contrast to eukaryotic topoisomerase I, our previous studies have demonstrated that human topoisomerase II does not possess an endoribonuclease activity when the enzyme is covalently attached to a ribonucleotide (4). It has thus been attractive to speculate that topoisomerase II could provide the RNA topoisomerase activity for eukaryotic cells, as has been suggested for E.coli topoisomerase III in bacteria (5,6). However, our current data disfavour this hypothesis as the enzyme can cleave neither RNA/DNA hybrids nor single-stranded RNA. In addition, no activity of topoisomerase II on an RNA substrate forming an internal hairpin structure was observed (N.Osheroff and S.J.Froelich-Ammon, personal communication), further arguing against an action of topoisomerase II as an RNA topoisomerase. The enzyme may therefore be more likely to recognize ribonucleotides as a form of DNA damage leading to a distortion of the normal B-DNA helix. To this end, the fact that ribonucleotides affect topoisomerase II-mediated cleavage in a position-specific manner similar to other DNA lesions suggests that DNA imperfections trap topoisomerase II by a conserved mechanism. Whether enzyme trapping and subsequent cleavage at DNA imperfections may be linked to other as yet unknown functions of topoisomerase II awaits further experimentation. Nevertheless, the activities of topoisomerase II on different RNA-containing substrates and the apparent positional effects of ribonucleotides and other DNA imperfections on topoisomerase II-mediated cleavage not only add new perspectives to the already well-characterized enzyme, but also help to delineate mechanistic aspects of the enzyme catalyzed reactions.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Professor Neil Osheroff and Drs Lotte Bjergbæk, Morten K. Larsen, Ole F. Nielsen, and Thomas Ø. Tange for valuable discussions and to Kirsten Andersen for skillful technical assistance. This work was supported by the Danish Cancer Society (grant 97-100-32), the Danish Medical Research Council, the Danish Center for Molecular Geron-tology, the Danish Center for Respiratory Adaptation and the Thaysen Foundation.
REFERENCES
- 1.Wang J.C. (1996) Annu. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- 2.Vosberg H.P. (1985) Curr. Top. Microbiol. Immunol., 114, 19–102. [DOI] [PubMed] [Google Scholar]
- 3.Osheroff N., Zechiedrich,E.L. and Gale,K.C. (1991) Bioessays, 13, 269–273. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Knudsen,B.R., Bjergbaek,L., Westergaard,O. and Andersen,A.H. (1999) J. Biol. Chem., 274, 22839–22846. [DOI] [PubMed] [Google Scholar]
- 5.Di Gate R.J. and Marians,K.J. (1992) J. Biol. Chem., 267, 20532–20535. [PubMed] [Google Scholar]
- 6.Wang H., Di Gate,R.J. and Seeman,N.C. (1996) Proc. Natl Acad. Sci. USA, 93, 9477–9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekiguchi J. and Shuman,S. (1997) Mol. Cell, 1, 89–97. [DOI] [PubMed] [Google Scholar]
- 8.Andersen A.H., Sørensen,B.S., Christiansen,K., Svejstrup,J.Q., Lund,K. and Westergaard,O. (1991) J. Biol. Chem., 266, 9203–9210. [PubMed] [Google Scholar]
- 9.Lund K., Andersen,A.H., Christiansen,K., Svejstrup,J.Q. and Westergaard,O. (1990) J. Biol. Chem., 265, 13856–13863. [PubMed] [Google Scholar]
- 10.Tsai Pflugfelder M., Liu,L.F., Liu,A.A., Tewey,K.M., Whang Peng,J., Knutsen,T., Huebner,K., Croce,C.M. and Wang,J.C. (1988) Proc. Natl Acad. Sci. USA, 85, 7177–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L.F., Rowe,T.C., Yang,L., Tewey,K.M. and Chen,G.L. (1983) J. Biol. Chem., 258, 15365–15370. [PubMed] [Google Scholar]
- 12.Sander M. and Hsieh,T. (1983) J. Biol. Chem., 258, 8421–8428. [PubMed] [Google Scholar]
- 13.Robinson M.J. and Osheroff,N. (1990) Biochemistry, 29, 2511–2515. [DOI] [PubMed] [Google Scholar]
- 14.Sekiguchi J., Cheng,C. and Shuman,S. (1997) J. Biol. Chem., 272, 15721–15728. [DOI] [PubMed] [Google Scholar]
- 15.Shuman S. and Turner,J. (1993) J. Biol. Chem., 268, 18943–18950. [PubMed] [Google Scholar]
- 16.Wang J.C. (1971) J. Mol. Biol., 55, 523–533. [DOI] [PubMed] [Google Scholar]
- 17.Maxwell A. and Gellert,M. (1986) Adv. Protein Chem., 38, 69–107. [DOI] [PubMed] [Google Scholar]
- 18.Corbett A.H. and Osheroff,N. (1993) Chem. Res. Toxicol., 6, 585–597. [DOI] [PubMed] [Google Scholar]
- 19.Bigioni M., Zunino,F., Tinelli,S., Austin,C.A., Willmore,E. and Capranico,G. (1996) Biochemistry, 35, 153–159. [DOI] [PubMed] [Google Scholar]
- 20.Kingma P.S. and Osheroff,N. (1997) J. Biol. Chem., 272, 7488–7493. [DOI] [PubMed] [Google Scholar]
- 21.Kingma P.S. and Osheroff,N. (1997) J. Biol. Chem., 272, 1148–1155. [DOI] [PubMed] [Google Scholar]
- 22.Capranico G., Kohn,K.W. and Pommier,Y. (1990) Nucleic Acids Res., 18, 6611–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freudenreich C.H. and Kreuzer,K.N. (1993) EMBO J., 12, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cline S.D., Jones,W.R., Stone,M.P. and Osheroff,N. (1999) Biochemistry, 38, 15500–15507. [DOI] [PubMed] [Google Scholar]
- 25.Kotani H. and Kmiec,E.B. (1994) Mol. Cell. Biol., 14, 1949–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotani H. and Kmiec,E.B. (1994) Mol. Cell. Biol., 14, 6097–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cline S.D. and Osheroff,N. (1999) J. Biol. Chem., 274, 29740–29743. [DOI] [PubMed] [Google Scholar]