Abstract
Mt. Erebus, Antarctica, is the southernmost active volcano in the world and harbors diverse geothermally unique ecosystems, including “Subglacial” and “Exposed” features, surrounded by a vast desert of ice and snow. Previous studies, while limited in scope, have highlighted the unique and potentially endemic biota of Mt. Erebus. Here, we provide an amplicon-based biodiversity study across all domains of life and all types of geothermal features, with physicochemical and biological data from 48 samples (39 Exposed and 9 Subglacial) collected through various field seasons. We found potentially high taxonomic novelty among prokaryotes and fungi, supporting past hypotheses of high endemism due to the distinctive and isolated environment; in particular, the large number of taxonomically divergent fungal sequences was surprising. We found that different site types had unique physicochemistry and biota; Exposed sites were warmer than Subglacial (median: 40°C versus 10°C for Exposed and Subglacial, respectively) and tended to have more photosynthetic organisms (Cyanobacteria and Chlorophyta). Subglacial sites had more Actinobacteriota, correlated with greater concentrations of Ca and Mg present. Our results also suggest potential human impacts on these remote, highly significant sites, finding evidence for fungal taxa normally associated with wood decay. In this study, we provide a blueprint for future work aimed at better understanding the novel biota of Mt. Erebus.
Keywords: Antarctica, biodiversity, endemism, environmental microbiology, human impacts, thermophiles
Mt. Erebus, Antarctica, harbors the southernmost active geothermal sites in the world, which contain highly taxonomically novel organisms across several unique site types, each with distinct biota.
Introduction
Mt. Erebus, located on Ross Island, Antarctica, stands as the southernmost active volcano on Earth and the sole known active volcano harboring a permanent phonolite lava lake (Kyle 1990, Kelly et al. 2008, Sims et al. 2021). The sustained volcanic activity of Mt. Erebus gives rise to diverse geothermal features, including ice caves, fumaroles, and exposed warm soils, supporting a surprisingly rich diversity of life (Soo et al. 2009, Herbold et al. 2014a, Tebo et al. 2015, Fraser et al. 2018, Noell et al. 2022). These habitats possess unique characteristics. First, they are profoundly influenced by local volcanic activity, with CO2-rich fumarolic gases traversing highly altered soils, depositing minerals and freely available water, thereby lowering soil pH (Ugolini and Starkey 1966, Soo et al. 2009, Ilanko et al. 2019, Hill et al. 2022). Second, these isolated geothermal hot soils serve as oases of heat, reductant, and liquid water amid a vastly different, extremely cold, and dry environment. The closest similar geothermal sites, Mt. Melbourne and Mt. Rittman, are 359 km and 456 km away to the North, respectively (Herbold et al. 2014b).
The interplay of hot soils and steam with cold air results in a variety of unique geothermal features such as ice towers, ice hummocks, ice caves, and bare hot soils. Generally, the geothermal areas are categorized as “Exposed” (rarely covered by ice) or “Subglacial” (covered by a ceiling of ice for most of the year). Many pockets of ice-free soil can be clustered into “Hot” sites, such as Western Crater and Tramway Ridge, the latter of which is designated an Antarctic Specially Protected Area (ASPA) (Fig. 1). The unique nature and diversity of geothermal soils and associated features on Mt. Erebus, combined with its isolation, are predicted to result in a wide range of novel and potentially endemic biota (Herbold et al. 2014a) and are also predicted to serve as refugia for life during glacial cycles (Fraser et al. 2014). Previous biodiversity studies on Mt. Erebus have been constrained in scope, often having a narrow sampling scale and/or relying on culture (for prokaryotes/fungi) or direct enumeration-based (for nonfungal eukaryotes) methods (Janetschek 1963, Ugolini and Starkey 1966, Broady 1984, Hudson and Daniel 1988, Hudson et al. 1989, Skotnicki et al. 2001), both of which are known to underestimate the true diversity present across all domains of life (Staley and Konopka 1985).
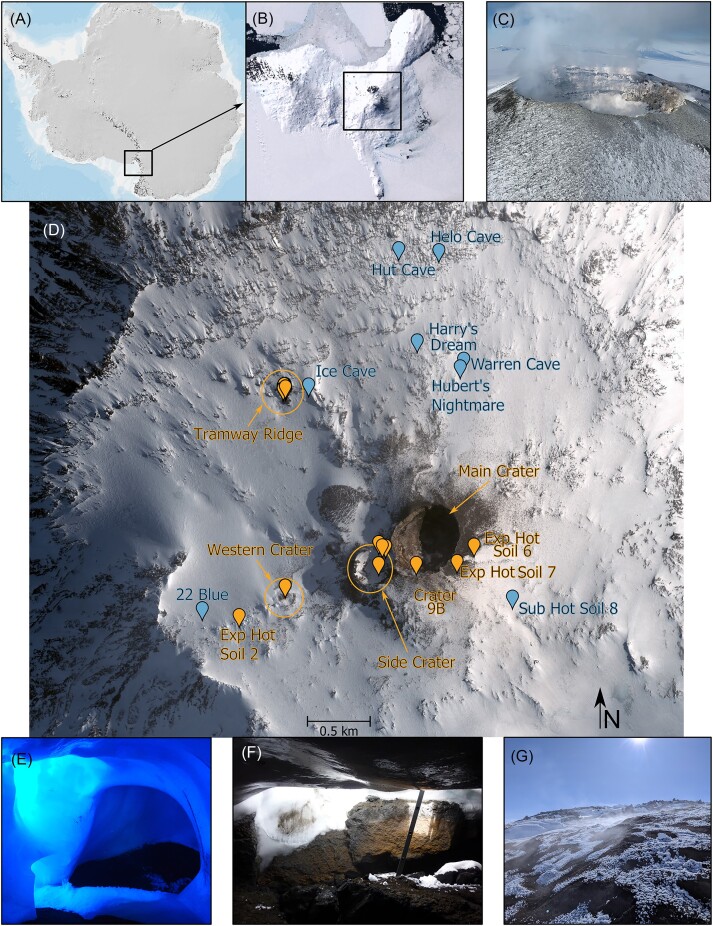
Figure 1.
Overview of sampling sites on Mt. Erebus. (A) Map of Antarctica; area enlarged in (B) is shown in the box. (B) Ross Island, Victoria Land, Antarctica. The location of Mt. Erebus is denoted in the box. (A and B) are from the Antarctic Digital Database Map Viewer https://www.add.scar.org/, Open Source. (C) Main Crater of Mt. Erebus, harboring a permanent lava lake. (D) Satellite image of Mt. Erebus, showing sampling locations. Sites where more than two samples were collected are indicated with circles. The markings denote either Exposed or Subglacial sites. Satellite image was purchased from DigitalGlobe Incorporated, Longmont, CO, USA (2019). (E–G) Representative images of selected sampling sites. (E) Entrance to 22 Blue Cave, as well as the sampling site (mostly dark). (F) Entrance to Helo Cave. (G) Geothermally heated soil at Tramway Ridge. Images in are courtesy of Ian McDonald, Craig Cary, Stephen Noell, and Matthew Stott.
For prokaryotes, culture-independent (genetic-based) studies on Mt. Erebus have been described at only two Exposed hot soil sites (Tramway Ridge and Western Crater) and three Subglacial sites (Hubert’s Nightmare, Harry’s Dream, and Warren Cave). At the Exposed Tramway Ridge and Western Crater sites, distinct communities of primarily chemoautotrophic prokaryotes driven by differences in soil pH were described (Noell et al. 2022). In particular, past studies of Tramway Ridge features have revealed mainly cosmopolitan thermophilic prokaryotes at the surface, including many culturable heterotrophs (Ugolini and Starkey 1966, Hudson and Daniel 1988, Hudson et al. 1989, Soo et al. 2009, Herbold et al. 2014a, Vickers et al. 2016). However, within a few centimeters of the surface, the community shifts to one dominated by a relatively small number of uncultured novel taxa (Herbold et al. 2014a). At Subglacial sites, autotrophic CO and CO2 fixation has been identified as a major trait of the prokaryotes present (Tebo et al. 2015, Yabe et al. 2022). Culture-independent studies of fungi have been limited to Subglacial sites but revealed diverse Basidiomycota and Ascomycota (Connell and Staudigel 2013, Fraser et al. 2018) and abundant Malassezia, which is often human-associated but is also widespread in environmental samples (Amend 2014, Steinbach et al. 2023) and a known contaminant in polymerase chain reaction (PCR) reagents (Czurda et al. 2016). Culture-based fungal studies have produced only a few thermotolerant (Gostinčar et al. 2023) fungi from Tramway Ridge (Ugolini and Starkey 1966, Greenfield 1982). For nonfungal eukaryotes, only one culture-independent study has been conducted on Mt. Erebus, finding evidence for the presence of arthropods, oligochaetes, and nematodes at a few Exposed and Subglacial sites (Fraser et al. 2018). Past observational studies have noted mosses and epiphytic green algae, with reports of soil fauna confined to limited sightings of amoebae and rotifers inhabiting the moss beds at Tramway Ridge below 40°C (Janetschek 1963, Broady 1984, Bargagli et al. 2004). Despite this past work on Mt. Erebus, we are not aware of any single survey using consistently applied culture-independent methodologies across all three domains of life.
In this study, we conducted an in-depth survey of the Mt. Erebus biota across the three domains of life, encompassing almost all described geothermal sites with the goal of better understanding the microbial ecology of this unique ecosystem. We proposed three hypotheses: (1) that the two different site categories, Subglacial and Exposed, would have different biological communities (in terms of community composition and potential functions) given the known differences in physicochemical factors, and that Tramway Ridge would differ from other Exposed samples in physicochemistry and biological community, based on past results (Noell et al. 2022); (2) that Erebus hosts a large number of taxonomically novel species; (3) and that Subglacial samples would have the highest signature of potential human impact. We based this final hypothesis on a past study that identified fungal markers for human activity at Subglacial sites (Connell and Staudigel 2013), likely due to their use in the past as refuges of warmth for explorers and scientists on Mt. Erebus. To test these hypotheses, we employed 16S rRNA gene, 18S rRNA gene, and ITS region amplicon sequencing, alongside a diverse range of physicochemical measurements. This study represents the first comprehensive, culture-independent survey of life at geothermal features on Mt. Erebus.
Materials and methods
Sample site descriptions and soil collection
We collected 48 soil samples from 30 different sites near the summit of Mt. Erebus at elevations from 3170 to 3710 m. These areas were generally divided into “Exposed” or “Subglacial” categories. An overview of sample sites is seen in Fig. 1 and Fig. S1 and details given in Table S1.
Exposed hot soils on Mt. Erebus are areas where the soil is thermally heated, to the extent that snow and ice do not accumulate. There are four main sites where Exposed hot soils are found: Tramway Ridge, Western Crater, Side Crater, and around the Main Crater. Tramway Ridge, protected under ASPA 175 since 2002, lies at an altitude of 3350 m and is ~1.5 km northwest from the Main Crater. We obtained a specialized permit from the New Zealand Ministry of Foreign Affairs and Trade, the governing body overseeing access by NZ scientists to ASPAs, to enter this ASPA and conduct sampling. Tramway Ridge has numerous, actively steaming fumaroles, with the highest measured surface temperatures on Mt. Erebus (65°C) (Hudson and Daniel 1988, Soo et al. 2009). Steep temperature and chemical gradients occur around these fumarole openings due to the perennially cold air temperatures (less than −20°C), with extensive moss beds and cyanobacterial mats at suitable ranges (generally 10°C–45°C) within the gradients (Hudson and Daniel 1988, Soo et al. 2009, Herbold et al. 2014b). Approximately 1 km west from the Main crater lies Western Crater, a shallow depression that is highly exposed to maritime winds. Unlike Tramway Ridge, there are few actively steaming fumaroles at Western Crater and no visible moss or cyanobacterial mats, with soil temperatures only reaching 50°C, and most hot soil spots are at least partially covered by ice hummocks (small transient snow enclosures; see Fig. S1K for an example) (Noell et al. 2022). Side Crater, located immediately adjacent to Main Crater, is (currently) volcanically inactive; the bottom is covered with ice, but several areas of hot soil and actively steaming fumaroles are present, primarily on the steep eastern side of the crater (Panter and Winter 2008). The active Main Crater, located at the summit of Mt. Erebus, is 250 m wide and 120 m deep, and harbors a persistent lava lake that produces regular strombolian eruptions (Kelly et al. 2008, Sims et al. 2021). The walls of the Main Crater are inaccessible to sampling, but numerous hot soil sites exist around the rim of the Main Crater with high soil temperatures (>60°C). In addition to these primary sites, small patches of Exposed hot soil can be found at various places around the summit (see Fig. 1).
Subglacial hot soils on Mt. Erebus are mainly found in ice caves distributed around the summit (Tebo et al. 2015, Ilanko et al. 2019). These caves are formed when gas emissions from the crust melt the ice covering the surface; the water refreezes, creating a roof of ice (Ilanko et al. 2019). Many of the ice caves are interconnected and thought to constitute a “cave system” rather than individual features (Giggenbach 1976). Although the soils in ice caves are generally similar in chemical characteristics, the ice caves vary in size, temperature, and light penetration (Giggenbach 1976, Tebo et al. 2015). Reported soil temperatures from ice caves are so far generally lower than at Exposed sites (Tebo et al. 2015). Subglacial hot soils other than ice caves can be found scattered around the summit, usually in the form of small ice hummocks (often <0.5 m in diameter). While ice caves are quite persistent, ice hummocks are ephemeral structures since the ice often collapses between seasons.
The 48 soil samples were collected during the field seasons (southern summer) of 2006–2007, 2009–2010, 2010–2011, 2011–2012, and 2019–2020 from various sites around Erebus. Before collecting the samples, soil temperatures were measured 0.5 cm below the soil surface using a CheckTempt1 thermometer (Hanna Instruments, Rhode Island, USA). Prior to collection of soil, the top mat-like layer (if present) was removed and either discarded or used as a separate sample (“Mat” samples) due to its distinct appearance from the underlying soil. Then, the topsoil (0–2 cm or 0–4 cm) below the mat was transferred into sterile centrifuge tubes using sterile spatulas; samples that are exceptions list the sample depth in the sample name (e.g. 1–4 cm). All collected samples were kept frozen once collected and at −80°C in the laboratory prior to processing.
Geochemistry
Elemental composition of each soil sample was measured using inductively coupled plasma–mass spectrometry (ICP–MS). Dried samples were ground using a Mixer Mill MM 400 (Retch, Haan, Germany) at a frequency of 27.5 s−1 for 1 min and 50 s. 1 g of each sample was weighed into a 50-ml falcon tube, and acid digested using 10 ml of 2 M HCl and 4 ml of 5.33 M HNO3 with reflux at 80°C for 30 min, or until no bubbles were observed upon gentle agitation. The digested contents were rinsed into a volumetric flask and diluted with Milli-Q water to a final volume of 100 ml and left overnight. To remove soil particles, and reduce residual chloride concentration, samples were filtered using 0.45 µm membrane filter into 50 ml volumetric flasks and made up to 50 ml with Milli-Q water. 10 ml of each sample was then tipped into a 15-ml falcon tube and 200 µl of concentrated (16 M) HNO3 was added. Elemental composition was measured using an Agilent 8900 ICP-MS with a triple quadrupole (Agilent Technologies, Santa Clara, CA, USA) at the University of Waikato ICP-MS facility. The following elements were analysed: Na, Mg, K, Ca, Sr, Ba, P, S, Se, Al, Tl, Pb, B, As, V, Cr, Mn, Fe, Co, Ni, Cu, Zn, Cd, and U. The results from ICP–MS were obtained from different instrument calibrations causing some variation in the isotopes of the elements measured and upper detection limits. Since Al, K, and Na reached their detection limits (5000 ppm, 10 000 ppm, and 10 000 ppm, respectively) in some runs but not others, these elements were removed from some of the statistical analyses. Experimental and calibration controls were included in every run to allow for the identification of background elemental composition.
pH and conductivity were measured from a suspension of soil and Milli-Q water with a 1:2.5 w/w ratio. pH was measured using a HI2213 Basic pH/ORP/°C Meter/3-point calibration (Hanna Instruments) and adjusted for temperature. Conductivity was measured using a Thermo Scientific Orion 4-Star Benchtop pH/Conductivity Meter (Thermo Fisher Scientific, Massachusetts, USA).
Total water content in soil samples was estimated gravimetrically by drying 3 g of each soil sample in a drying oven at 105°C. When the weight of a sample was unchanged over a 24 h period, the initial and final weight difference was converted into the water content percentage. The resulting dried samples were then prepared for total carbon and nitrogen measurements via grinding, as for ICP–MS analysis. Total carbon and nitrogen were measured from 1 g of each sample on a Vario EL Cube (Elementar, Langeselbold, Germany).
DNA extraction, 16S rRNA gene PCR, sequencing, and quality control
A modification of the CTAB protocol developed previously (Herbold et al. 2014a) was used to perform triplicate DNA extractions on 0.7–0.9 g of soil. Additional extractions were performed for samples with low DNA yields. DNA from replicate extractions was pooled prior to analysis. The quality and concentration of the extracted DNA was checked using a Nanodrop DN-1000 spectrophotometer (Nanodrop Inc., Delaware, USA) and a Qubit 2.0 Fluorometer (Thermo Fisher Scientific).
16S rRNA amplicons were generated by triplicate 25 µl PCR reactions, using Ion Torrent Earth Microbiome fusion primers based on the Earth Microbiome Project 16S rRNA gene primers; the version used was the most recent version of these primer sets: 515YF (5′CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXXXXXXXXXGATGTGYCAGCMGCCGCGGTAA3′) and 926R (5′CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGATCCGYCAATTYMTTTRAGTTT3′) (Apprill et al. 2015, Parada et al. 2016, Caporaso et al. 2018). PCR was conducted on DNA from each sample using a specific forward primer containing a unique barcode. Each reaction contained 6 mM MgCl2, 0.24 mM dNTPs, 1.2x PCR buffer, 1.6 × 10−5 mg µl−1 BSA, 0.024 U µl−1 Taq polymerase (Thermo Fisher Scientific), 0.2 mM of each primer, and 1–3 ng of sample DNA. PCR reactions were run on an Applied Biosystems ProFlex PCR System (Thermo Fisher Scientific) and the cycling conditions were: 94°C for 3 min followed by 30 cycles of 94°C for 45 s, 50°C for 1 min, and 72°C for 1.5 min with a final extension of 72°C for 10 min.
Triplicate PCR reactions were pooled, and 5 µl of each was visualized on 1% agarose gel. The Invitrogen SequalPrep Normalization kit (Thermo Fisher Scientific) was used to purify and normalize 25 µl of each amplicon, 2 µl of each normalized amplicon was pooled to make up the sequence library. The library was quantified using Qubit 2.0 Fluorometer (Thermofisher) and treated with Ion PGMTM Template IA 500 Kit (Thermo Fisher Scientific) before preparation for sequencing with using Ion PGMTM Hi-QTM View Sequencing Kit (Thermo Fisher Scientific). The library was added to an Ion 318TM Chip Kit v2 BC (Thermo Fisher Scientific) and sequenced on an Ion Torrent PGM (Thermo Fisher Scientific).
Resulting 16S rRNA gene sequences were filtered for quality and trimmed to generate amplicon sequence variants (ASVs) using the DADA2 v3.14 pipeline (see https://benjjneb.github.io/dada2/tutorial.html) with the modifications recommended for Ion Torrent data (e.g. HOMOPOLYMER_GAP_PENALTY = –1, BAND_SIZE = 32); this pipeline includes built-in chimera removal (Callahan et al. 2016). The ASVs were assigned taxonomy using the SILVA database nr. 99 v138, including the species level assignment (addSpecies command) (McLaren 2020). Alternative taxonomic assignment was conducted using the DADA2 formatted GTDB release 214.1 (v4.4) (Alishum et al. 2023). Throughout the manuscript, we have used the taxonomic names and ranks given in SILVA for consistency but also cite alternative taxonomic names where appropriate. Sequences assigned to eukaryotes, mitochondria, and chloroplast were removed. The R-packages Decipher (Wright 2016) and phangorn (Schliep 2011) were used to generate an unrooted phylogenetic tree by the neighbor-joining method. See Table S8 for number of reads at all stages in processing.
Samples taken from the different site categories had similar mean read counts (Fig. S2A and B). However, two samples stood out; WC–10 and Crater 1 Mat had notably lower read counts (2462 reads) and higher read counts (105 524), respectively, compared to other samples. Leaving these samples out from the dataset did not affect the calculated distributions in a principal coordinates analysis (PCoA) analysis (Fig. S2C and D). Lastly, rarefaction curves with the species richness were plotted (Fig. S2E). The rarefaction curves all reached plateau, including WC-10 (Fig. S2F). This indicates that the majority of the richness present in the samples has been covered, despite the low number of reads in the WC-10 sample.
18S rRNA gene and ITS region PCR, sequencing, and quality control
Select samples (17) were chosen for ITS region (used for fungal classification) and 18S rRNA gene (used for nonfungal eukaryote classification) sequencing based on the type of geothermal feature to ensure representation from a variety of environments. DNA from these samples (also used for 16S rRNA gene sequencing) was sent to Omega Biosciences (Norcross, GA, USA) for amplification using the Earth Microbiome Project 18S rRNA gene or ITS region primers without blocking sequences (the most updated versions of the EMP 18S and ITS primers were used); for 18S rRNA gene, the primers used were 1391f (5′GTACACACCGCCCGTC3′) and EukBr (5′TGATCCTTCTGCAGGTTCACCTAC3′); for ITS region, the primers used were IT1f (5′CTTGGTCATTTAGAGGAAGTAA3′) and ITS2 (5′GCTGCGTTCTTCATCGATGC3′) (Caporaso et al. 2018). They then underwent paired-end sequencing on an Illumina MiSeq v3 with 300 bp reads.
The resulting ITS region and 18S rRNA gene sequences were adaptor trimmed using Cutadapt (Martin 2011) and quality checked using FastQC (Andrews). Resulting trimmed sequences were processed using the DADA2 pipeline with default parameters for the respective workflows (for ITS, see https://benjjneb.github.io/dada2/ITS_workflow.html; for 18S, see https://benjjneb.github.io/dada2/tutorial.html), except in the filterAndTrim step for both workflows, the following change was made: maxEE = c(3,5). For ITS sequences, taxonomy was assigned using the UNITE database v9 with singletons set as RefS (Abarenkov et al. 2022). For 18S rRNA gene sequences, taxonomy was assigned using the SILVA v132 18S database formatted for DADA2 (Morien and Parfrey 2018). For the 18S data set, all ASVs assigned to fungal groups were removed, while all ASVs assigned to nonfungal groups were removed from the ITS data set (see Table S8 for number of reads at all stages in processing).
After generating the ITS ASVs with DADA2, we found that a large number of fungal sequences were poorly classified. We therefore conducted a phylogenetic analysis to identify putative clades of related ASVs and their taxonomic identities. We compiled a database consisting of (1) All ASV sequences, (2) The highest similarity sequence in the UNITE-SH database (Abarenkov et al. 2022) where that match had a % identity >90% over >75% of the length, (3) Where there was no match in the UNITE-SH database, the highest similarity sequence in the UNITE+INSD database where that match had a % identity >90% over >75% of the length, and (4) additional sequences from the NCBI nucleotide database in an iterative adaptive heuristic search strategy, attempting to improve the phylogenetic analysis and resolve identifications wherever possible. ASV and added sequences were aligned using default options in MAFFT (Katoh and Standley 2013); only references that were ITS1-5.8S-ITS2 were used to improve placement. Sequences were not trimmed. A nucleotide substitution model was determined using standard options in Jmodeltest (invgamma option) (Darriba et al. 2012) and a phylogeny was then constructed using MrBayes (Ronquist et al. 2012) using default options.
From the phylogeny, high-level phylogenetic clades were identified based on >50% support. We named each clade on the basis of the lowest-numbered ASV present within that clade. As a measure of phylogenetic novelty, we also recorded the number of matches in the database where there were fewer than 5 at >90% identity and >75% of the ASV sequence length, and at >80% identity and >75% of the ASV sequence length. Lastly, for those clades or subclades where no ASVs matched sequences in UNITE databases, we searched the representative ASV sequences against the full NCBI nucleotide database (https://blast.ncbi.nlm.nih.gov/). Using the full NCBI database allowed inclusion of unidentified environmental sequences in the analysis, and also served as a check for any nonfungal sequences remaining in the data. Final taxonomic classification of ASVs in clades was based on the taxonomic classifications of reference sequences that fit into these phylogenetic clades.
Statistical analyses
Data analysis was conducted using R v. 4.2.2 and all plots were visualized using ggplot2 v. 3.4.0 (Wickham 2016) with beautification of figures in Inkscape (https://www.inkscape.org/). In general, we began our statistical analyses by comparing the two site categories of “Exposed” and “Subglacial;” given the already observed differences in communities between Tramway Ridge and Western Crater, we also chose to break out the Exposed category into the subcategories “Tramway Ridge” and “Exp. Hot Soil,” the latter referring to all Exposed sites other than Tramway Ridge. Snippets of code are provided below where appropriate; complete code used for analyses is provided on Github (see the section “Data availability”).
Site similarity was visualized using PCoA (based on Euclidean distances of scaled variables using the functions cmdscale, scale, and vegdist). For testing physicochemical factors for significant differences between sites, outliers were first removed using the interquartile range method (Smiti 2020). Next, physicochemical parameters that were returned as significantly different between Subglacial and Exposed categories based on significant P-values (Benjamini–Hochberg adjusted) from a two-sided t-test were selected for further analysis (number of tests: 27). For this smaller list of parameters (n = 12), significant differences between Site Subcategories were detected using a Krustal–Wallis test followed by Dunn’s test within ggstatsplot v. 0.12.3 (Patil 2021), using the command ggbetweenstats and P-value adjustment (number of tests performed: 12).
Alpha diversity was measured with phyloseq v.1.40.0 using the Shannon index (McMurdie and Holmes 2013). Unconstrained beta diversity of the microbial community structures was investigated by center log-ratio transforming sequence abundances on a Unifrac distance matrix using microbiome v. 1.18.0 (Lahti and Shetty 2017) and phyloseq, respectively. Different ways of categorizing sample sites were tested for statistical differences in the microbial community composition by using vegan to calculate a permutational analysis of variance (PERMANOVA) followed by an analysis of multivariate homogeneity (PERDISP) to verify that the dispersion of the tested samples did not differ significantly. The differential abundance test (trans_diff) within the microeco package was used to find specific taxa (at the phylum level) whose abundances differed significantly between either Site category or subcategory, using the random forest method (method = “rf”) and Benjamini–Hochberg correction. We used the R package indicspecies v. 1.7.14 to calculate Indicator Values for ASVs, with the command multipatt (Cáceres and Legendre 2009); only ASVs with strong indicator values (above 0.85) were included.
Environmental factors correlating with microbial community composition were identified using mantel tests within microeco, using Spearman’s correlation and Bray’s distance matrix and Benjamini–Hochberg corrected P-values. The environmental factors that correlated significantly (P < .05, Benjamini–Hochberg correction) with microbial community structure were plotted using a distance-based redundancy analysis (dbRDA) within microeco (plot_ordination). Autocorrelations between environmental measures were investigated using the function “cor.test” from the package stats with Spearman’s test, with Benjamini–Hochberg correction. To identify correlations between specific taxa and environmental factors, we used an RDA at the phylum level within microeco. Correlations between taxa and environmental factors were calculated within microeco using a Spearman’s correlation within the $cal_cor function with a Benjamini–Hochberg correction across all data (p_adjust_type = “All). The resulting heatmap was plotted using $plot_cor within microeco.
To predict functional profiles for the prokaryotic ASVs, we used a functional prediction program (FAPROTAX) (Louca et al. 2016) that uses taxonomic closeness to cultured strains with known functional traits in microeco. The FAPROTAX database currently contains over 7600 functional annotations covering over 4600 taxa. Assignment to function is based on matches between taxonomy of ASVs in our samples with taxonomy in the database, not on sequence similarity. Correlations between predicted functional profiles and environmental factors were calculated with the $cal_cor using Spearman’s test with a Benjamini–Hochberg correction across all data (p_adjust_type = “All”).
To examine co-occurrence patterns between prokaryotes and eukaryotes, we constructed a co-occurrence network. First, to reduce compositional bias, we normalized each separate dataset (16S, 18S, and ITS) using center-log-ratio method (CLR) within the microbiome package v. 1.18.0. The data sets were then combined and ASVs were trimmed to only retain ASVs that had at least a CLR of 4.5 in at least three samples, resulting in 228 ASVs (Table S9). Then, using NetCoMi v. 1.1.0 (Peschel et al. 2021), a co-occurrence network was constructed, using the netConstruct command with Pearson as the calculation method, and “t-test” as the sparsification method with multiple testing adjustment using the adaptive Benjamini–Hochberg method.
Clustering was performed using the fast and greedy option under the command netAnalyze, and singletons were removed prior to plotting.
Results
The physicochemistry of Mt. Erebus Subglacial and Exposed soils differ
Despite all samples sites being within 3 km of each other, the physicochemical conditions of geothermal features across Mt. Erebus varied greatly. Given past results indicating that Tramway Ridge microbial communities are distinct, we chose to split the category of Exposed samples into the subcategories of Tramway Ridge and other Exposed samples (from here, “Exp. Hot Soils”). This decision was confirmed by the results of a PcoA, which showed that Subglacial samples tended to cluster separately from Tramway Ridge samples, which were more scattered (Fig. S3). Other Exp. Hot Soil samples either clustered more with Tramway Ridge samples or Subglacial samples, with the latter being characterized by elevated Ca, Mg, and Sr concentrations. The Exp. Hot Soil samples that clustered with Subglacial samples had similar concentrations of Ca, Mg, and Sr as Subglacial samples (Fig. S3). Thus, we continued to analyse the physicochemical data with Tramway Ridge samples split from Exp. Hot Soils samples.
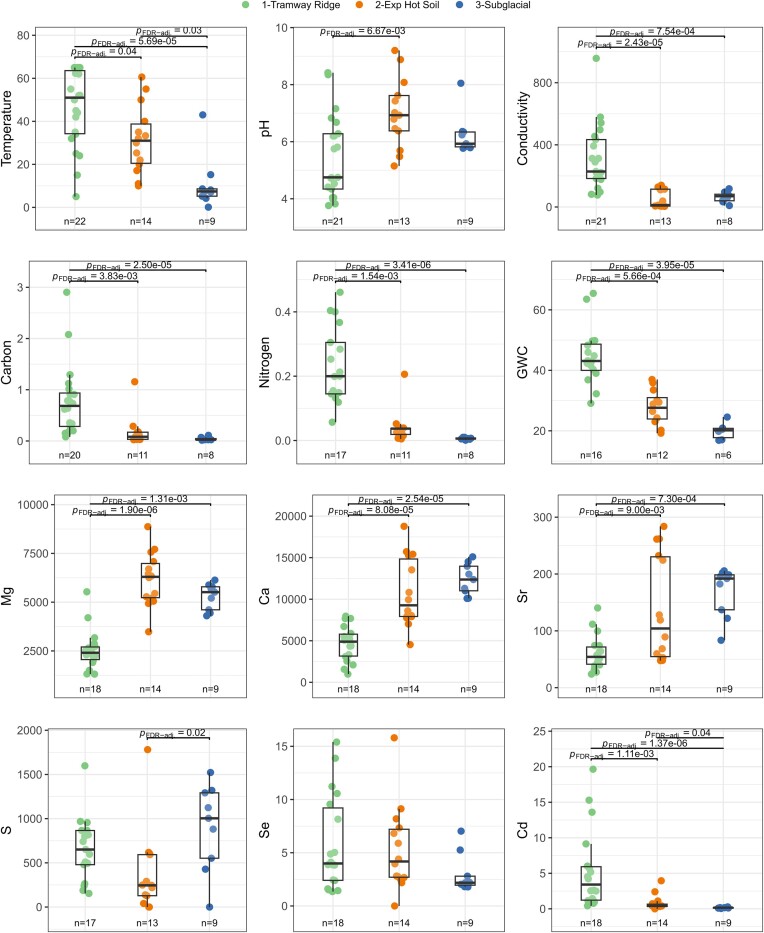
The three site subcategories were different in many physicochemical parameters (Dunn’s test, adjusted P-values < .05, Benjamini–Hochberg correction; Fig. 2). Exp. Hot Soils and Tramway Ridge samples were distinguishable from Subglacial samples by having higher temperatures (medians: 51°C, 31°C, and 7.4°C for Tramway Ridge, Exp. Hot Soils, and Subglacial, respectively) (Fig. 2). Most other parameters (conductivity, total carbon, total nitrogen, water content, and Cd) were significantly higher at Tramway Ridge compared to all other site subcategories (Fig. 2). Notably, Tramway Ridge had significantly lower median values for Mg (2406 ppb, 6299 ppb, 5514 ppb for Tramway Ridge, Exp. Hot Soils, and Subglacial, respectively), Ca (4894 ppb, 9269 ppb, and 12 358 ppb for Tramway Ridge, Exp. Hot Soils, and Subglacial, respectively), and Sr (54.2 ppb, 104.2 ppb, and 192.0 ppb for Tramway Ridge, Exp. Hot Soils, and Subglacial, respectively) compared to other site subcategories. Tramway Ridge samples also tended to be more acidic than other samples, although this difference was only significant for Exp. Hot Soils compared to Tramway Ridge (medians: 4.75 and 6.93 for Tramway Ridge and Exp. Hot Soils, respectively).
Figure 2.
The range of select measured physicochemical parameters within Tramway Ridge, Exposed Hot Soils, or Subglacial from Mt. Erebus geothermal soils. Only parameters that were significantly (two-sided t-test, Benjamini–Hochberg adjusted P-value < .05) different between Subglacial and all Exposed samples are shown. P-values given in the figure (adjusted via a Benjamini–Hochberg correction) are from a Dunn’s test, following a significant result from a Krustal–Wallis test. Only significant P-values are shown. GWC = gravitational water content. Number of Dunn’s tests performed: 12. Data presented has outliers removed, as specified in the section “Materials and methods”.
Initial analysis of biological communities reveals large proportions of unclassified microorganisms
To assess the diversity of microorganisms inhabiting these sites, we determined the microbial community composition via amplicon sequencing of the 16S rRNA gene (prokaryotes), ITS region (fungi), and 18S rRNA gene (nonfungal eukaryotes). We observed 2734 prokaryotic ASVs, 214 fungal ASVs, and 512 nonfungal eukaryote ASVs (see Tables S2, S3, and S4 for ASV taxonomies and abundance profiles). Of note, a substantial proportion of ASVs in all three communities appeared to be novel with 59.4% and 86% of prokaryotic and nonfungal eukaryotic taxa unable to be assigned to any genera in the SILVA database nr. 99 v138, respectively (5.3% and 6% of ASVs could not be assigned to a prokaryotic and nonfungal eukaryotic phylum). When we attempted taxonomic assignment for prokaryotes using GTDB release 214.1, we found similar proportions (46% were unassigned at the genus level, 15.8% were unassigned at the phylum level).
For the fungal community, we found many ASVs that were taxonomically unclassified and poorly resolved phylogenetically, which led to a refinement of phylogenetic methodology. This analysis identified 87 lineages (>50% support on phylogenetic tree branch) of fungi within a large polytomy (lineages with <50% support were defined as polytomies; it was expected that the basal phylogeny would not be resolved given the limitations of the ITS region; Fig. S4). 46 lineages were comprised of a single ASV, 17 lineages formed clades of two ASVs, and the remainder contained up to 14 ASVs. Many single ASV lineages are likely biologically relevant, given that 14 of the 46 single ASV clades had relative abundances greater than 1% in at least one sample, with three having relative abundances above 40% in at least one sample (Table S3). Based on BLASTn sequences that overlapped 75% or more of the query length, 45 lineages could be tentatively identified based on >90% similarity to a known reference sequence in the UNITE database and six additional lineages matched a sequence in the wider UNITE+INSD database. An additional 10 lineages matched at least one sequence in the NCBI database, with six of these only matching a single sequence at >90% identity. In total, 20 lineages had no matching sequences in any database at >80% identity (Table S5). While most of these lineages had only a single ASV, one contained five ASVs and another contained three.
Differential abundances of several taxonomic groups distinguish site types
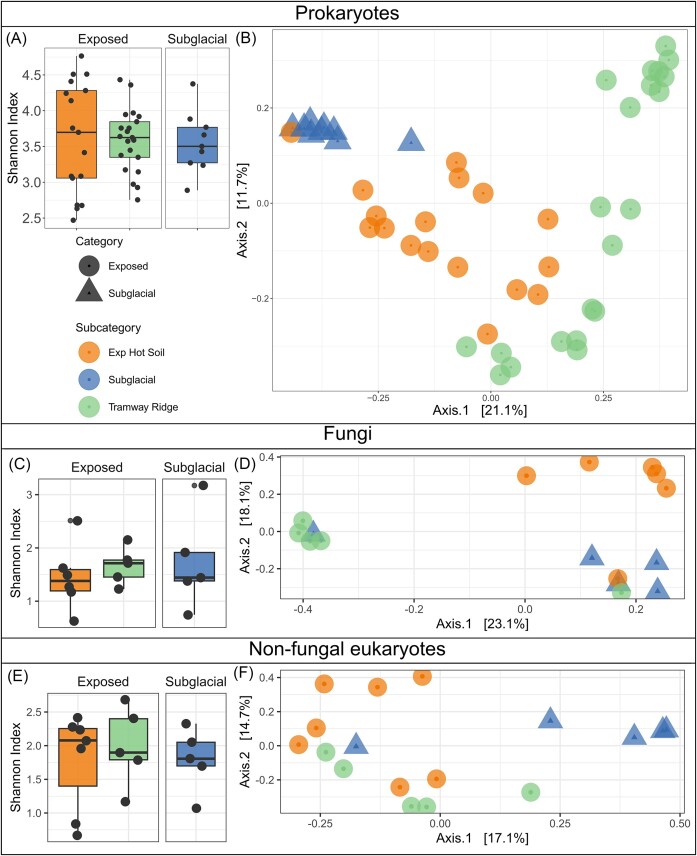
Unlike in the physicochemical results, we found that mean community alpha diversity (measured via the Shannon index) was similar across site categories (Exposed and Subglacial), although there was substantial variation within site categories for the prokaryotes and nonfungal eukaryotes (Fig. 3A, C, and E). For prokaryotes, Shannon diversity had mean values of 3.5–3.75; for nonfungal eukaryotes, this range was 1.6–2.2 and for fungi was 1.7–2.1.
Figure 3.
Biodiversity analysis of prokaryotic (16S rRNA gene), fungal (ITS) and nonfungal eukaryote (18S rRNA gene) datasets across Mt. Erebus Exposed and Subglacial geothermal sites. (A, C, and E) Alpha diversity measured with the Shannon index. (B, D, and E) Beta diversity measured with a PCoA using Unifrac distances.
However, in our beta diversity analysis (Fig. 3) using PCoA, we found that, among prokaryotic, fungal, and nonfungal eukaryotic data sets, Subglacial and Exposed samples generally clustered separately from each other, although to different extents depending on the data set (Fig. 3B, D, and F). This trend was confirmed with significant PERMANOVA test results (P = .001). Furthermore, a PERMDISP2 test between categories was nonsignificant, indicating that the significant differences between categories (from the PERMANOVA test) is not due to a significant difference in spread or dispersion between categories for both the fungal and nonfungal eukaryotic data sets. Exposed hot soil samples were different both from Tramway Ridge samples (P = .009 and .024 for fungal and nonfungal eukaryotes, respectively) and from Subglacial samples (P = .01 and .016 for fungal and nonfungal eukaryotes, respectively), but Tramway Ridge and Subglacial samples were not significantly different (P = .12 and 0.06 for fungal and nonfungal eukaryotes, respectively). Due to the different dispersions (or variance; PERMDISP2 test) of the samples, a reliable PERMANOVA test for differences in prokaryotic communities between sample categories or subcategories was not feasible. In the prokaryotic data set, it was especially notable that one Exposed hot soil sample (Crater 6 low) clustered with the Subglacial samples in both prokaryotic and physicochemical data (Fig. 3A). We did not observe a clustering of samples based on the year in which they were sampled (Fig. S5).
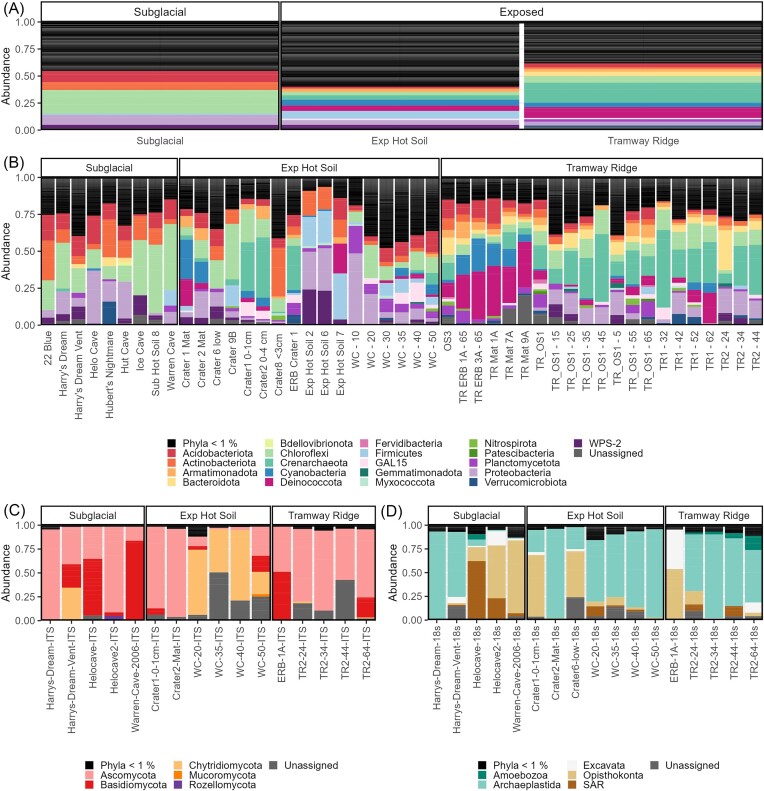
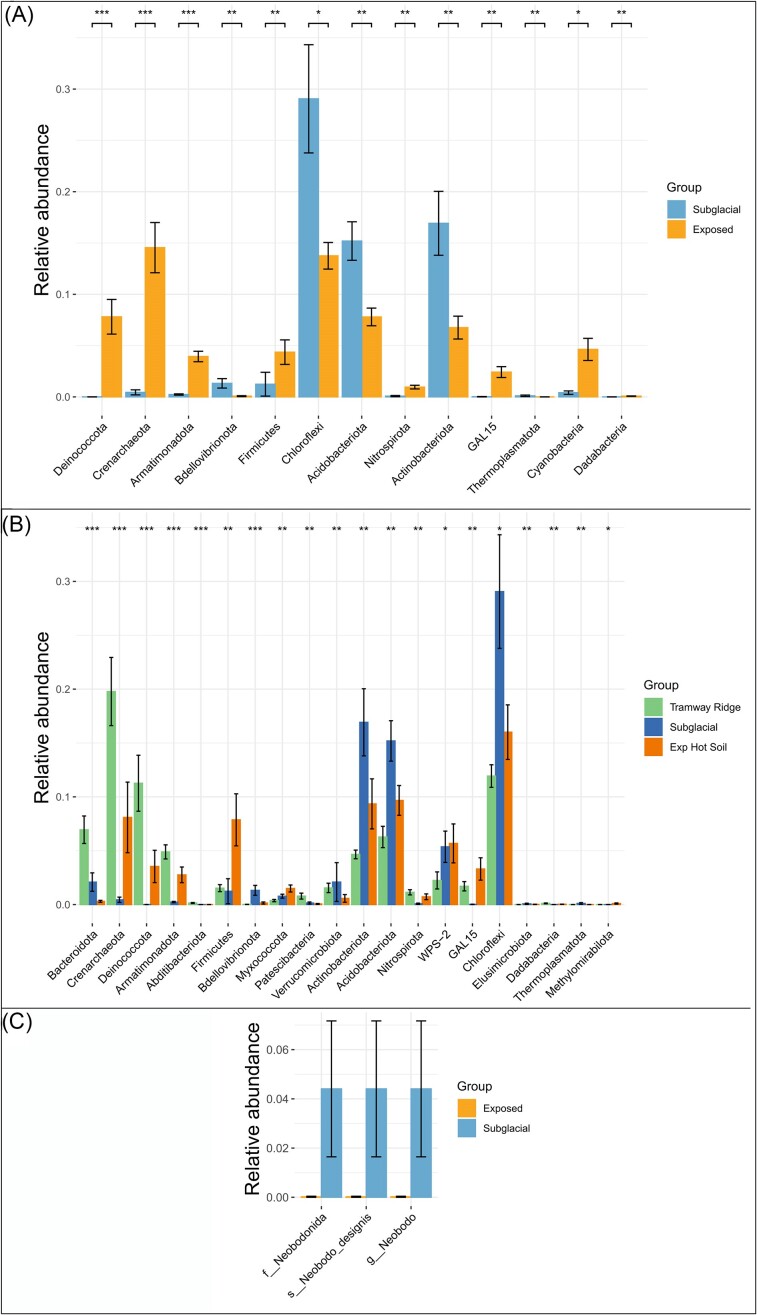
To identify the taxonomic groups responsible for the differential clustering of the site categories, we used a differential abundance test within the microeco package in R and an indicator species analysis. In the prokaryotic data, our differential abundance test identified several phyla that had higher relative abundance averages in Exposed samples. Here, we report taxonomic names as assigned by SILVA but provide alternative taxonomic names (from Oren and Garrity 2021) in parentheses. Phyla of note with differential abundances include: Crenarchaeota (especially class Nitrososphaeria; now Thermoproteota), Deinococcota, Armatimonadota, Cyanobacteria (now Cyanobacteriota), Nitrospirota, Firmicutes (now Bacillota), and GAL15 (adjusted P < .05, Benjamini–Hochberg correction) (Figs 4A, B, and 5A). Conversely, the average relative abundances of Actinobacteriota (mainly classes Actinobacteria and Thermoleophilia; now Actinomycetota), Chloroflexi (now Chloroflexota; especially classes AD3–now candidate phylum Ca. Dormibacteriota—and Ktedonobacteria), Acidobacteria (now Acidobacteriota), and Alphaproteobacteria were greater in Subglacial sites (adjusted P < .05, Benjamini–Hochberg correction) (Fig. 5A, Fig. S4A). Exposed hot soils differed from Tramway Ridge by having significantly higher relative abundances of Firmicutes and Actinobacteriota, although the latter had a high variance in relative abundance at Exposed hot soils (from <1% up to 32.9% in Crater 8 < 3 cm) (Fig. 5B). Our indicator species analysis at the ASV level confirmed these broad trends, with a few nuances (Table S6). Of particular note was the finding that the genus Fischerella is an indicator for Tramway Ridge, while Leptolyngbya is an indicator more broadly for Exposed sites; also, we also found four indicator species for Tramway Ridge that were from the phylum Bacteroidota, which was not identified in the differential abundance analysis (Table S6).
Figure 4.
Microbial community composition shown as relative abundances of prokaryotic (A and B) and eukaryotic phyla (C and D). The average relative abundance of prokaryotic phyla in Subglacial, Exposed hot soils, and Tramway Ridge samples are seen in A, while B shows the relative abundances for each sample individually. Similarly, C and D shows the relative abundances of fungal and other eukaryotic phyla in individual samples.
Figure 5.
Results of a differential abundance test. (A and B) Prokaryotic phyla that significantly differed in relative abundances between (A) Subglacial versus Exposed samples or (B) all three subcategories. (C) Taxa at any rank that were significantly different between Exposed and Subglacial sites in the nonfungal eukaryotic data set. For all, significant difference in abundance was determined using the random forest method within the microeco package. The significance stars correspond to the following Benjamini–Hochberg adjusted P-values: * = .05, ** = .01, *** = .001, and **** = .0001.
For the fungi, we found that the three site subcategories were generally distinguishable by the dominance of certain phyla (Fig. 4C). For Subglacial samples, ASVs from Ascomycota and Basidiomycota were predominant in most samples. Samples from Western Crater were distinguishable by very high abundances of ASVs from Chytridiomycota, while Tramway Ridge samples were almost completely dominated by ASVs from Ascomycota (relative abundance >50% in all samples). At taxonomic levels below phylum, conclusions were challenging to draw due to the large number of ASVs that could not be assigned even to a class (Fig. S6B). However, when examining the distribution patterns of different clades (generated from our phylogenetic approach), we found that the most highly divergent clades (that could not be reliably assigned at phylum or class level; e.g. clades 1, 3, 4, 5, and 42) were found in Exposed samples (e.g. clade 1, predominantly found at Western Crater, is a highly divergent member of Chytridiomycota). On the other hand, clades that were most abundant in Subglacial samples were from better characterized groups (e.g. clade 32–Leotiomycetes, clade 14–Agaricomycetes, and so on) (Fig. S7). In our differential abundance analysis, there were no fungal taxonomic groups identified as significantly different between sample categories. However, our indicator species analysis identified four ASVs from Ascomycota as indicators for Tramway Ridge, two of which were from the Family Herpotrichiellaceae (Table S6).
At a broad level, the nonfungal eukaryotic data was dominated by ASVs from the Archaeplastida group (Chloroplastida), apart from three Subglacial samples (two Helo cave samples, one Warren Cave sample). These three Subglacial samples were almost completely lacking in Archaeplastida ASVs, instead being dominated by ASVs from the SAR group and nonfungal Opisthokonta. The Chloroplastida ASVs detected were mainly from green algal groups (Pseudochlorella, Coccomyxa, Coenocystis, and so on) or moss chloroplasts. Nonphotosynthetic eukaryotes detected included alveolates (mainly ciliates), amoeboids (e.g. the cercazoan Gymnophrys, and amoebozoan Tubulinea, Vermamoeba, and Flamella), and other protists (e.g. the Excavata Neobodo). In our differential abundance analysis, only one taxonomic group was identified as significantly different between the three site categories: the species Neobodo designis, which was present only at Subglacial sites at 4% relative abundance (Fig. 5C). Again, our indicator species analysis identified a number of interesting species; of note were four Chloroplastida ASVs (two each at Tramway Ridge and Exposed Hot Soils), ASVs from the genera Monodus and Didinium in Exposed Hot Soils samples (yellow–green unicellular algal genus and carnivorous ciliate, respectively), N. designis at Subglacial samples, and ASVs from the genera Tubulinea and Gymnophrys (amoeba and predatory amoeboid Cercazoa, respectively) (Table S6).
Network analysis
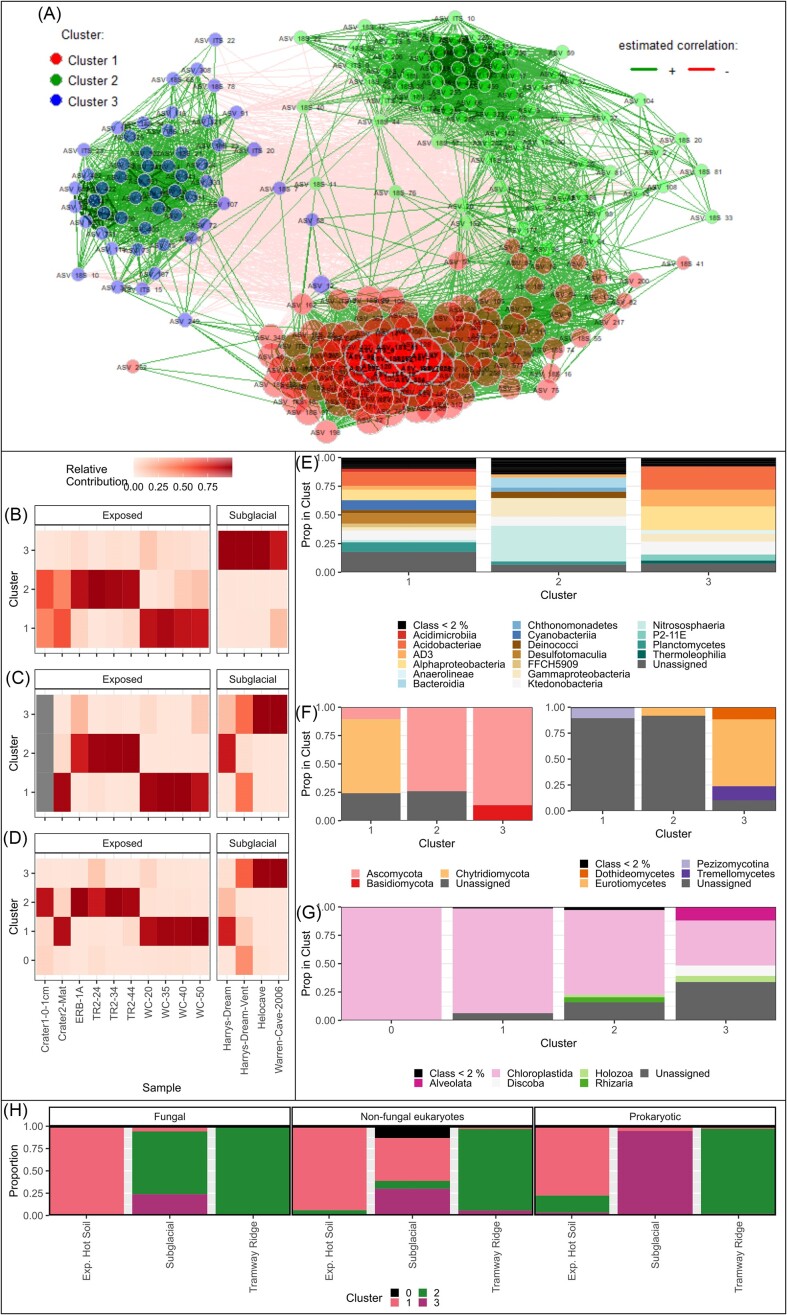
We used a co-occurrence network to investigate whether co-occurrence patterns across all domains of life also point toward partitioning of Erebus biota into the three site subcategories, Subglacial, Tramway Ridge, and Exposed Hot Soils. Based on our other results, we expected that ASVs from each of the three site subcategories would show greater interconnectedness within subcategory than to other subcategories, resulting in clustering of ASVs based on site subcategory. After trimming to high abundance ASVs and transforming the counts, we obtained a co-occurrence network with four clusters (based on the optimization algorithm), one of which only contained one ASV (Fig. 6A). The modularity of the network was 0.43, indicating medium connectivity; 85.6% of the connections were positive correlations.
Figure 6.
Co-occurrence network to investigate abundance patterns in the abundant eukaryotes, nonfungal eukaryotes, and prokaryotes. (A) Co-occurrence network using Pearson correlations, where each dot is an ASV, colored by cluster; ASVs within clusters are more connected to each other than to ASVs outside of the cluster. (B–D) Heatmaps showing the relative abundance of each cluster at each site for (B) prokaryotes, (C) fungi, and (D) nonfungal eukaryotes. (E–G) Bar graphs showing what proportion of each cluster belongs to what class for (E) prokaryotes, (F) fungi, or (G) nonfungal eukaryotes. For (F), proportion of cluster belonging to fungal phyla is shown on the left and class on the right, given the large proportion of fungal ASVs that were unassigned at the class level. (H) Bar plot showing the contribution of ASVs from each cluster to the total number of reads in each site subcategory for each type of organism.
Based on the total abundance of reads from each cluster, we found support for our hypothesis of partitioning based on site subcategory, with each of the clusters generally corresponding to one of three site subcategories: Exp. Hot Soils (Cluster 1), Tramway Ridge (Cluster 2), or Subglacial (Cluster 3) (Fig. 6B–D, H). In general, ASVs from one of the three clusters comprised over 80% of the reads in any given subcategory. However, this was not the case for the fungal and the nonfungal eukaryotes, where ASVs from clusters other than Cluster 3 had strong contributions to Subglacial communities (Fig. 6C, D, and H).
We also observed that Clusters 1 and 2 were more connected to each other than to Cluster 3 (162 positive connections between 1 and 2, versus 39 and 8 for Clusters 3 and 1 and Clusters 3 and 2, respectively), indicating more similarity in co-occurrence patterns within Exposed sites than between Exposed and Subglacial sites (Fig. 6A). The composition of the clusters generally matched our community composition analyses (Fig. 4); in particular, Cluster 3 had few photosynthetic organisms (Cyanobacteria or Chloroplastida in the 16S or 18S rRNA gene surveys, respectively), Clusters 1 and 2 had much larger proportions of novel fungi at the class level than Cluster 3, and Cluster 2 prokaryotes were enriched in the classes Nitrososphaeria and Deinococcus (Fig. 6E–G).
Does microbial community composition correlate with environmental factors?
We next looked at what physicochemical factors might be influencing the different microbial communities observed at the three site types using a Mantel test and a db-RDA. However, we first observed that several factors were strongly correlated with each other across all site types, especially Mg, Ca, and Sr, as well as total carbon and nitrogen (Table S7). As autocorrelation can strongly impact the results of Mantel tests, we chose to remove Sr from this analysis as it is not known to have any strong relationship with biotic communities. We then removed either Mg or Ca, or total carbon or nitrogen, and reran the Mantel test, with the finding that the correlation coefficients and adjusted P-values for all factors remained almost identical between all tests, indicating that our findings are robust to autocorrelation (Table 1).
Table 1.
Physicochemical factors that correlate with prokaryotic communities determined via a Mantel test (Spearman correlation on Bray–Curtis distance matrix between biotic communities) to identify. Only factors with significant adjusted P-values are shown. Test was rerun twice with autocorrelated factors Sr and one of Mg or Ca removed. Adjusted P-values are the result of a Benjamini–Hochberg adjustment for multiple comparisons; number of tests performed: 27 prior to removing any factors, 25 after removing Sr and Ca or Mg, and 26 after removing total carbon or nitrogen.
| Variables | Correlation coefficient | P-value | Adjusted P-value |
|---|---|---|---|
| Temperature | 0.350658773 | .002 | .0108 |
| pH | 0.198262798 | .004 | .0135 |
| Conductivity | 0.208247108 | .003 | .011571429 |
| Carbon | 0.212146947 | .005 | .015 |
| Nitrogen | 0.273662297 | .003 | .011571429 |
| GWC | 0.332289989 | .001 | .00675 |
| Mg | 0.411655232 | .001 | .00675 |
| Ca | 0.464576911 | .001 | .00675 |
| Sr | 0.41058035 | .001 | .00675 |
| Rerun without Sr and Mg | |||
| Temperature | 0.350658773 | .001 | .008333333 |
| pH | 0.198262798 | .006 | .025 |
| Conductivity | 0.208247108 | .005 | .025 |
| Carbon | 0.212146947 | .008 | .028571429 |
| Nitrogen | 0.273662297 | .002 | .0125 |
| GWC | 0.332289989 | .001 | .008333333 |
| Ca | 0.464576911 | .001 | .008333333 |
| Rerun without Sr and Ca | |||
| Temperature | 0.350658773 | .001 | .008333333 |
| pH | 0.198262798 | .008 | .028571429 |
| Conductivity | 0.208247108 | .007 | .028571429 |
| Carbon | 0.212146947 | .006 | .028571429 |
| Nitrogen | 0.273662297 | .002 | .0125 |
| GWC | 0.332289989 | .001 | .008333333 |
| Mg | 0.411655232 | .001 | .008333333 |
| Rerun without total nitrogen | |||
| Temperature | 0.350659 | .001 | .0052 |
| pH | 0.198263 | .004 | .017333 |
| Conductivity | 0.208247 | .005 | .018571 |
| Carbon | 0.212147 | .008 | .026 |
| GWC | 0.33229 | .001 | .0052 |
| Mg | 0.411655 | .001 | .0052 |
| Ca | 0.464577 | .001 | .0052 |
| Sr | 0.41058 | .001 | .0052 |
| Rerun without total carbon | |||
| Temperature | 0.350659 | .001 | .0052 |
| pH | 0.198263 | .005 | .018571 |
| Conductivity | 0.208247 | .006 | .0195 |
| Nitrogen | 0.273662 | .002 | .008667 |
| GWC | 0.33229 | .001 | .0052 |
| Mg | 0.411655 | .001 | .0052 |
| Ca | 0.464577 | .001 | .0052 |
| Sr | 0.41058 | .001 | .0052 |
GWC: gravitational water content.
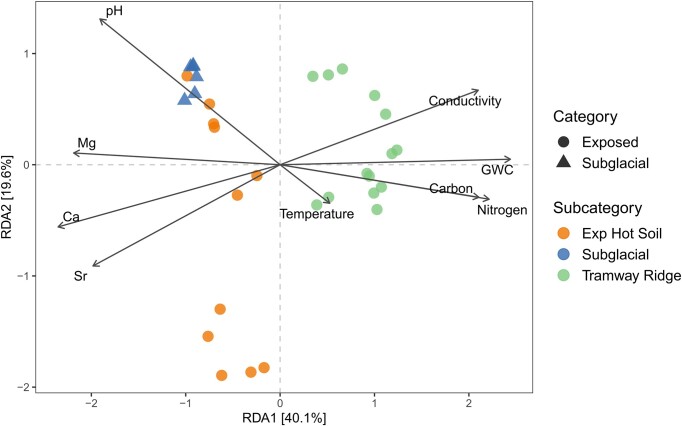
For prokaryotes, analyses revealed that the measured physicochemical factors responsible for the separation of Tramway Ridge from Subglacial and Exposed hot soil samples along the first axis were elevated temperature, nitrogen, carbon, gravimetric water content, and conductivity in Tramway Ridge samples (Fig. 7). Subglacial samples and some Exposed hot soil samples were characterized by elevated concentrations of Ca and Mg. An RDA analysis was conducted to explore the contribution of different phyla in explaining the grouping patterns observed in Fig. 7 (Fig. S8A). The top 10 taxa that explained the most variation in the axes were included as explanatory variables, increasing the percentage of variation explained by the axes (40.1%–60.7% for the first axis). The phyla Deinococcota, Bacteroidota, and Crenarchaeota were found to be associated with samples from Tramway Ridge. Conversely, Actinobacteriota were associated with other Exposed hot soils and Subglacial soils. The dispersion of Exposed hot soil and Subglacial soil samples along the second axis was mainly explained by several phyla: Proteobacteria (now Pseudomonadota), WPS-2, and Firmicutes were associated with multiple Subglacial samples as well as Western Crater samples. Acidobacteria, Planctomycetota, and Chloroflexi were associated with two Subglacial samples and several Exposed hot soil samples. Furthermore, several phyla that contributed to the variation observed in the RDA analysis were also correlated with physicochemical factors (Spearman; P < .05, Benjamini–Hochberg adjustment) (Fig. S8B). This provides additional support to the hypothesis that these physicochemical factors play an important role in shaping the distinct taxonomic compositions of the different environments, although it is equally possible that the differences in physicochemical factors may be a result of biotic influences.
Figure 7.
dbRDA of environmental factors that correlated with the prokaryotic communities across different Exposed and Subglacial geothermal sites on Mt. Erebus. Environmental factors shown had a significant (P < .05) correlation with the communities based on a Mantel test after adjusting for multiple comparisons using a Benjamini–Hochberg adjustment (see Table 1).
For both the fungal and nonfungal eukaryotic communities, it was more difficult to infer correlations between physicochemical parameters and community compositions due to the small number of samples from each sample category. Indeed, we did not find any physicochemical parameters that were correlated with community compositions using a Mantel test (P < .05, Benjamini–Hochberg correction).
We used a functional prediction program (FAPROTAX) to investigate correlations between predicted functions and environmental drivers of prokaryote biodiversity, determined above (Fig. S9). Results and discussion of this analysis can be found in Supplemental Text 1.
Potential human impacts on Mt. Erebus biota
We examined our data set for potential markers of human contamination, specifically to identify if any sites are more impacted than others. Previously, Malasezzia yeast species were identified in Warren Cave in 2013 at high abundance (37% relative abundance), with their presence attributed to human contamination (Connell and Staudigel 2013), although this attribution has been called into question lately (Amend 2014, Czurda et al. 2016, Steinbach et al. 2023). We did observe Malasezzia species in our data set, with lesser abundance (<15% relative abundance) at Helo Cave and one sample each from Western Crater and Tramway Ridge. We found none at Warren Cave, however. We also observed fungi highly related to known wood decay fungi in our data set: clade 26 (100% ID, full length match to Gymnopilus penetrans) and clade 63 (highly similar match to Polyporales, white rot fungi), found at Helo Cave, Western Crater 20°C, and Western Crater 50°C. Helo Cave is located within 50 m of the wooden Lower Erebus Hut, while Western Crater is much further removed from the closest hut site. ASVs from both clades were present only in very low abundance (<2% relative abundance), except for ASV 155, 16% at ERB-1A. We did not find evidence for any known human-related bacteria (e.g. Staphylococcus, Bacteroidales, or Escherichia coli) in our prokaryotic data set. We did observe several 18S ASVs that were identified as human DNA (ASVs 186 and 265). These ASVs were at low abundance in only a few samples (0.8% relative abundance at Helo Cave, 0.1% relative abundance at Western Crater 20°C).
Discussion
Our investigation of the diversity of biota inhabiting geothermal ice-free soils on Mt. Erebus using culture-independent methods provides the broadest survey to-date of life in Earth’s most remote geothermal site. Our findings provide evidence that the many environments supported by thermal features on Mt. Erebus have provided unique habitats for diverse biotic communities. Additionally, our biological data suggest high levels of taxonomically novel microorganisms inhabiting thermal features on Mt. Erebus. Finally, we show limited evidence of human contamination signatures on Mt. Erebus.
Hypothesis 1: different habitats on Erebus support distinct biota.
We found support for our hypothesis that there are distinct biological communities, functions, and correlated physicochemical parameters at Subglacial, Tramway Ridge, and Exposed sites (Figs 2–6, Fig. S8). We base this on the separate clustering of the three subcategories in the physicochemical data (Fig. S3), beta diversity plots (Fig. 3), and the co-occurrence network (Fig. 6), as well as the significant PERMANOVA results and the distinct indicator species identified for each subcategory. A previous study also found that Tramway Ridge and Western Crater harbored distinct microbial communities (Noell et al. 2022), but here we extend those results to show that Tramway Ridge has, for the most part, distinct biological communities compared to all other Exposed sites and all Subglacial sites. The exceptions to these trends seem to be mainly driven by unique chemistry (as will be discussed below for Actinobacteriota) or higher light levels (as will be discussed below for certain Subglacial sites). Below, we discuss the distinguishing physicochemical and biological features of each site subcategory.
Distinguishing features of subglacial sites: Actinobacteriota, elevated Ca/Mg/Sr, and eukaryotic bacteriovores
For our prokaryotic data set, one of our more surprising findings was the strong correlation between high concentrations of calcium (Ca), magnesium (Mg), and strontium (Sr) in Subglacial samples and the presence of Actinobacteriota (Fig. S8). Ca exhibited the greatest correlation coefficient with the structure of microbial communities (Table 1), 2.3 times greater than pH. Indeed, the clustering of Exposed sample Crater-6-low (from Side Crater) with other Subglacial samples, despite its high temperature (60.6°C), seems to be driven more by mineral content than temperature (Fig. 2). This sample had high abundances of Actinobacteriota (Fig. 3B) and similar amounts of Mg, Ca, and Sr as Subglacial samples (Fig. S2). Because Ca, Mg, and Sr are colinear variables, it is impossible to conclude which variable drives the Actinobacteriota abundance. However, past research has found that Ca has a significant impact on the growth of many Actinobacteriota species (Fang et al. 2017, Gonzalez-Pimentel et al. 2018, Araujo et al. 2020, Buresova‐Faitova et al. 2022). It is unlikely that sporulation, known to be influenced by Ca (Fang et al. 2017), is a factor, given that most Actinobacteriota in our data set are not known to be spore-forming (e.g. genera Conexibacter, Mycobacterium, and Crossiella; Monciardini et al. 2003, Parte et al. 2012). Ca is also known to be involved in antibiotic resistance in some Actinobacteriota (Bekker et al. 2008) and in the production of Ca-dependent antibiotics (Nakaew et al. 2009, Quintana et al. 2013, Riquelme et al. 2017, Paun et al. 2021, Gonzalez-Pimentel et al. 2022). It could be that these antibiotics provide a competitive advantage, but this hypothesis remains to be tested. Alternatively, it might be that the Actinobacteriota in Subglacial samples increase the levels of Ca in the soil due to the biomineralization of dissolved Ca ions, a hypothesis that was proposed for some cave Actinobacteriota (Cuezva et al. 2012, Lange-Enyedi et al. 2024). It is therefore not clear whether high Ca concentrations are beneficial for actinobacterial growth or purely a result of their metabolic activity.
For our fungal data set, few meaningful patterns were observed in the Subglacial samples. However, in the nonfungal eukaryote dataset, Subglacial samples were characterized by the presence of N. designis, flagellates that are bacteriovores generally found in aquatic environments, but have also been found in soil (von der Heyden and Cavalier-Smith 2005). Their absence from Exposed sites could either indicate a preference for cooler temperatures or that they can find greater concentrations of bacterial prey at Subglacial sites. Similarly, ciliate ASVs (also heterotrophic eukaryotes that generally feed on bacteria) were only present at Subglacial samples (Fig. S6), indicating similar constraints on distribution as Neobodo.
Distinguishing features of exposed sites: influence of sunlight and higher temperatures
The most apparent difference between Exposed and Subglacial sites is the variation in available sunlight. This disparity is evident in the presence of photosynthetic Cyanobacteria, algae (order Chlorophyta and genus Monodus), and fungi from the class Eurotiomycetes, which is primarily a lichenized class, at only Exposed sites and Harry’s Dream, the cave in our data set with the highest light penetration (Tebo et al. 2015) (Figs 4A and 6, Table S6). Many of the cyanobacterial ASVs identified belong to the families Nostocaceae and Leptolyngbyaceae. These Cyanobacteria have been previously found at Tramway Ridge and are renowned for being cosmopolitan and thermophilic (Soo et al. 2014, Alcorta et al. 2019, Noell et al. 2022, Strunecký et al. 2023). The most common algae at Exposed samples in our data set have been observed previously on Mt. Erebus, especially Chlorella (Broady 1984). Similar to past descriptions of these species, we found them mainly in Harry’s Dream samples and did not find them in samples with temperatures above 35°C. On the other hand, Coccomyxa were primarily found in Exposed samples, especially at the Tramway Ridge 44°C sample, indicating a higher temperature tolerance than Chlorella. Our finding of putatively lichenized Eurotiomycetes differs from previous fungi-based results in Antarctica which identified Lecanoromycetes, another primarily lichenized fungal class (Coleine et al. 2018, Biagioli et al. 2023). In our 18S and 16S rRNA gene data sets, we identified two common photosynthetic partners of lichenized fungi: the green alga Trebouxia and the cyanobacterium Nostoc. Both were primarily found at Western Crater, indicating that the Eurotiomycetes species on Mt. Erebus are either not lichenized or use unusual photosynthetic partners.
We observed clear differences in the types of fungi observed at different Exposed sites; however, the ecological implications of these patterns is unclear given their phylogenetic novelty. We did observe that fungi predominant at Western Crater were mostly from clade 1, a highly divergent group of Chytridiomycota that showed similarity to fungi found in biological soil crusts in nearby Victoria Land (Canini et al. 2020). Chytridiomycota are best known for parasitic infections and degrading complex organic matter (Rosenblum et al. 2010); their presence at Western Crater, which has extremely low organic content, is intriguing.
Finally, our nonfungal eukaryotic data set identified amoebae primarily in Exposed samples (Table S6), although the most abundant amoeba ASV in our data set was a Vermamoeba species that was found in Helo Cave at 5% relative abundance. This particular species is known to be widespread and thermotolerant (Delafont et al. 2018). Amoebae have previously been found on Mt. Erebus at Exposed sites (Broady et al. 1987) and viable amoeba cysts have been found in Arctic permafrost before (Shmakova et al. 2016), including similar groups to those found in some of our samples (especially Harry’s Dream).
Distinguishing features of Tramway Ridge sites: novel Nitrososphaeria
Overall, our expanded dataset of microbial ecosystems on Mt. Erebus further confirms that Tramway Ridge is an outlier across all other prokaryotic communities on Mt. Erebus, as has also been previously reported (Noell et al. 2022) (Fig. 2). The prokaryotic communities characteristic of Tramway Ridge have been discussed previously (Soo et al. 2009, Herbold et al. 2014a, Noell et al. 2022), with our extended sample set confirming these previous findings. Of particular note is confirmation that Tramway Ridge has characteristically high abundances of the former archaeal phylum Thaumarchaeota, (Herbold et al. 2014a), in particular ASV_1, which we classified to the order SCGC AB-179-E04 (now belonging to the phylum Thermoproteota; Oren and Garrity 2021, Rinke et al. 2021) within the class Nitrososphaeria. This class is common in marine habitats (Kerou et al. 2016), with members known for being autotrophic ammonia oxidizers. Some deep-branching, thermophilic members of the Nitrososphaeria have been shown to lack the genes needed for ammonia oxidation and instead are likely heterotrophic (Beam et al. 2014, Ren et al. 2019, Kato et al. 2021, Luo et al. 2024). Recent metagenomic work has shown that the organism represented by ASV_1 also lacks ammonia oxidation genes (Herbold et al. 2024).
We also note the identification of two fungi from the family Herpotrichiellaceae as indicator species for Tramway Ridge. Species from this family are generally rock-dwelling fungi, many of which are known to be opportunistic human pathogens (Thitla et al. 2023).
Hypothesis 2: Large numbers of unexpected fungi and taxonomically novel prokaryotes.
Past studies have hypothesized that the biota of Mt. Erebus is enriched in taxonomically novel and potentially endemic organisms due to the unique geothermal features, geochemistry, and isolation of these sites. Three lines of evidence from our data support this hypothesis. First, a large proportion of fungal sequences in our data set had either no significant similarity (20 lineages) or a very limited numbers of matches (three lineages) to any DNA sequence in any database. It is likely that these sequences represent highly novel groups of fungi, given the clear patterns we observed in abundance profiles of these novel clades at different sites (Fig. S7), the paucity of thermophilic fungi at low-organic sites in previous studies (de Oliveira et al. 2015, Coleine et al. 2022), and our finding that the novelty of clades increased at Exposed sites, where one would expect larger accumulations of dispersed spores and thus, less novel fungi. Indeed, a recent study of fungal diversity in biological soil crusts in nearby Victoria Land, Antarctica, also found a large proportion of putatively unknown fungal species (Canini et al. 2020). Second, we found a comparatively high (5.3% for Silva and 15.8% for GTDB) proportion of the prokaryotic community was unassigned at the phylum level. This number is dependent on the database used and when the database was accessed. Nonetheless, in general in soils, the number of unassigned sequences at the phylum level is generally <1% (Delgado-Baquerizo 2019), while in hot springs, this number can be around 3% (Podar et al. 2020). Finally, only a small proportion of the prokaryotic community across all site categories (25% of ASVs, comprising 21.8% of reads) had a predicted functional profile using the FAPROTAX dataset, due to the large number of ASVs that were unassignable at high taxonomic levels in our dataset, a number on the low end of previously published results (Van Rossum et al. 2020, Sansupa et al. 2021). Taxonomic novelty is generally considered a marker for endemicity, but this hypothesis remains to be fully tested.
The detection in our data set of fungal species at high temperature sites is surprising since, although fungi are known to be capable of survival in many different types of extreme environment (e.g. extreme cold, dry, radiation, and so on), truly thermophilic fungi are rare (Coleine et al. 2022). Thermophilic fungi are generally thought to be mesophiles that broadened their temperature range to take advantage of high nutrient situations (e.g. compost piles, bird nests, and so on) (de Oliveira et al. 2015). However, we note that our results are only indicative of presence, not activity. This is important given that a previous study comparing the taxonomy of active vs. present fungi at Antarctic sites found a stark difference between the two communities (Cox et al. 2019). Additionally, a study examining growth tolerances of fungi isolated from sites within Victoria Land, Antarctica, including geothermally heated soils from Mt. Melbourne, only found one fungal isolate that showed any growth at temperatures above 40°C (Zucconi et al. 1996). Thus, our results should be treated as preliminary.
These results highlight the need for further studies, such as metagenomics, sequencing of full-length ribosomal sequences (for fungi), growth/activity measurements to identify active members of the fungal community, and/or culture-based studies, to truly understand the taxonomy and functions of the novel fungi/prokaryotes present. More diversity studies of fungi in Antarctica are particularly needed due to their current underrepresentation in databases. Indeed, a recent study reported the successful cultivation of a member of the WPS-2 phylum (now Ca. Eremiobacterota) from Warren Cave on the Mt. Erebus summit that is a novel aerobic anoxygenic photoheterotrophic bacterium (Yabe et al. 2022).
Hypothesis 3: Potential human influence on sites is limited to putative wood decay fungi
Although a past study identified large amounts of potentially human-associated fungal contaminants in Subglacial Erebus samples, the results from our data set on human impacts are more mixed. In fact, the presence of Malasezzia in some of our samples and in previously published Warren Cave samples does not necessarily stem from human activity on site, given their discovery in a broad range of habitats around the world (Amend 2014) and their propensity to contaminate PCR reagents (Czurda et al. 2016). We were particularly interested to find fungi closely related to known wood decay fungi in our data set, given the almost complete lack of woody plants in Antarctica (Peña-Méndez et al. 2005, Pautler et al. 2013). The high identity matches (100% ID in one-third cases) suggest these species could have been introduced with construction material used to build structures on Erebus (e.g. Lower Erebus hut, located within 50 m of Helo Cave, where these fungi were identified), or with wooden equipment used at the sites (e.g. wooden ladders have been used in the past to descend into caves like Helo Cave). Alternatively, these taxa could have broader ecological niches than previously recognized. We also note with concern the presence of 192 ASVs from Animalia across all sites, although half of these have less than 10 reads in any sample. This highlights the need for a follow-up study using qPCR with specific primers and more stringent contamination protocols during sampling and sample processing, to eliminate the possibility of human contamination of samples during processing. We also note that the absence of other signatures of human contamination in our data set (e.g. human-associated bacteria) does not mean that those signatures are not present.
Conclusion
Our survey of the biota inhabiting Mt. Erebus has given insights into the broad diversity of organisms inhabiting the southernmost active geothermal site in the world. Ice-free features on Mt. Erebus ranging from ice caves to steaming hot soils all harbor an impressive array of taxa, from novel Nitrososphaeria, to a wealth of novel fungi, to a range of photosynthetic microorganisms, with distinct communities at Subglacial sites, Exposed sites, and Tramway Ridge in particular. We hope this study will serve as a springboard for further, targeted, and in-depth studies of these unique and potentially endemic organisms, in addition to providing a rich starting point for comparison studies with other Antarctic geothermal sites. Additionally, we hope the community will continue to consider how to preserve these precious sites, as well as adhering to the rules that are already in place around access.
Supplementary Material
Acknowledgements
We especially recognize the logistical support from Antarctica New Zealand for fieldwork on Mt. Erebus and support in the field from Jon Tyler. We thank Drs Alexis Marshall and Mafalda S. Baptista for support with quality control of sequencing data and scripts for ASV generation. We dedicate this paper to our wonderful colleague and dear friend, S. Craig Cary, who passed away in February 2024. He was a pioneering and inspiring researcher in the study of microbial life in extreme environments. He spent 17 seasons in Antarctica studying the ecology of microbial communities in polar deserts, meltwater ponds, and high temperature soils in Antarctic geothermal areas. This latter work is one of the most significant projects he was involved in at the time of his death. His passion for and enthusiasm about scientific research remains one of his enduring legacies. He will be missed by all of us.
Contributor Information
Trine Bertram Rasmussen, Thermophile Research Unit, Te Aka Mātuatua – School of Science, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand; International Centre for Terrestrial Antarctic Research, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand.
Stephen E Noell, Thermophile Research Unit, Te Aka Mātuatua – School of Science, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand; International Centre for Terrestrial Antarctic Research, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand.
Craig W Herbold, Te Kura Pūtaiao Koiora – School of Biological Sciences, Te Whare Wānanga o Waitaha – University of Canterbury, Christchurch, Aotearoa 8041, New Zealand.
Ian A Dickie, Te Kura Pūtaiao Koiora – School of Biological Sciences, Te Whare Wānanga o Waitaha – University of Canterbury, Christchurch, Aotearoa 8041, New Zealand.
Roanna Richards-Babbage, Thermophile Research Unit, Te Aka Mātuatua – School of Science, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand; International Centre for Terrestrial Antarctic Research, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand.
Matthew B Stott, Te Kura Pūtaiao Koiora – School of Biological Sciences, Te Whare Wānanga o Waitaha – University of Canterbury, Christchurch, Aotearoa 8041, New Zealand.
S Craig Cary, Thermophile Research Unit, Te Aka Mātuatua – School of Science, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand; International Centre for Terrestrial Antarctic Research, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand.
Ian R McDonald, Thermophile Research Unit, Te Aka Mātuatua – School of Science, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand; International Centre for Terrestrial Antarctic Research, Te Whare Wānanga o Waikato – University of Waikato, Hamilton, Aotearoa 3216, New Zealand.
Author contributions
Trine Bertram Rasmussen (Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing), Stephen E. Noell (Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing), Craig W. Herbold (Conceptualization, Data curation, Investigation, Visualization, Writing – review & editing), Ian A. Dickie (Formal analysis, Investigation, Methodology, Software, Writing – review & editing), Roanna Richards-Babbage (Conceptualization, Data curation, Methodology, Resources, Writing – review & editing), Matthew B. Stott (Conceptualization, Data curation, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing), S. Craig Cary (Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing), and Ian R. McDonald (Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing)
Conflict of interest
None declared.
Funding
This work was supported by the Royal Society of New Zealand (Marsden Grant 18-UOW-028 to S.C., M.S., and I.M.).
Data Availability
All R scripts used to analyse the data are available on Github at https://github.com/ThermophileResearchUnit/Erebus_survey_manuscript. All sequence data is available in DDBJ/EMBL/GenBank. 16S rRNA gene sequence data has been deposited as accession KIGZ00000000 version KIGZ01000000, except for samples TR1-*, TR2-*, and WC-* which have been deposited as accession KIGX00000000 version, KIGX01000000. 18S rRNA gene sequence data have been deposited as SUB14470052. ITS sequence data have been deposited as SUB14470177.
References
- Abarenkov K, Zirk A, Piirmann T et al. UNITE general FASTA release for fungi. UNITE Community, 2022. 10.15156/BIO/2483911. [DOI] [Google Scholar]
- Alcorta J, Vergara-Barros P, Antonaru LA et al. Fischerella thermalis: a model organism to study thermophilic diazotrophy, photosynthesis and multicellularity in cyanobacteria. Extremophiles. 2019;23:635–47. [DOI] [PubMed] [Google Scholar]
- Alishum A, Paul G, Claus C. DADA2 formatted 16S rRNA gene sequences for both bacteria & archaea. Zenodo, 2023. 10.5281/zenodo.10403693. [DOI] [Google Scholar]
- Amend A. From dandruff to deep-sea vents: Malassezia-like fungi are ecologically hyper-diverse. PLoS Pathog. 2014;10:e1004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC. Babraham Bioinformatics. [Google Scholar]
- Apprill A, McNally S, Parsons R et al. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015;75:129–37. [Google Scholar]
- Araujo R, Gupta VVSR, Reith F et al. Biogeography and emerging significance of actinobacteria in Australia and Northern Antarctica soils. Soil Biol Biochem. 2020;146:107805. [Google Scholar]
- Bargagli R, Skotnicki ML, Marri L et al. New record of moss and thermophilic bacteria species and physico-chemical properties of geothermal soils on the northwest slope of Mt. Melbourne (Antarctica). Polar Biol. 2004;27:423–31. [Google Scholar]
- Beam JP, Jay ZJ, Kozubal MA et al. Niche specialization of novel thaumarchaeota to oxic and hypoxic acidic geothermal springs of Yellowstone National Park. ISME J. 2014;8:938–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker OB, Elizarov SM, Alekseeva MT et al. Ca2+-dependent modulation of antibiotic resistance in Streptomyces lividans 66 and Streptomyces coelicolor A3(2). Microbiology. 2008;77:559–67. [PubMed] [Google Scholar]
- Biagioli F, Coleine C, Buzzini P et al. Positive fungal interactions are key drivers in Antarctic endolithic microcosms at the boundaries for life sustainability. FEMS Microbiol Ecol. 2023;99:1–8. [DOI] [PubMed] [Google Scholar]
- Broady PA. Taxonomic and ecological investigations of algae on steam-warmed soil on Mt Erebus, Ross Island, Antarctica. Phycologia. 1984;23:257–71. [Google Scholar]
- Broady P, Given D, Greenfield L et al. The biota and environment of fumaroles on Mt Melbourne, northern Victoria Land. Polar Biol. 1987;7:97–113. [Google Scholar]
- Buresova-Faitova A, Kopecky J, Sagova-Mareckova M et al. Comparison of actinobacteria communities from human-impacted and pristine karst caves. Microbiologyopen. 2022;11. 10.1002/mbo3.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres MD, Legendre P. Associations between species and groups of sites: indices and statistical inference. Ecology. 2009;90:3566–74. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canini F, Geml J, D’Acqui LP et al. Exchangeable cations and pH drive diversity and functionality of fungal communities in biological soil crusts from coastal sites of Victoria Land. Fungal Ecol. 2020;45:100923. [Google Scholar]
- Caporaso JG, Ackermann G, Apprill A et al. Earth microbiome project: EMP 16S illumina amplicon protocol. Protocols.io. 2018, 1–7. [Google Scholar]
- Coleine C, Stajich JE, Selbmann L. Fungi are key players in extreme ecosystems. Trends Ecol Evol. 2022;37:517–28. [DOI] [PubMed] [Google Scholar]
- Coleine C, Stajich JE, Zucconi L et al. Antarctic cryptoendolithic fungal communities are highly adapted and dominated by lecanoromycetes and dothideomycetes. Front Microbiol. 2018;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell L, Staudigel H. Fungal diversity in a dark oligotrophic volcanic ecosystem (DOVE) on Mount Erebus, Antarctica. Biology. 2013;2:798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox F, Newsham KK, Robinson CH. Endemic and cosmopolitan fungal taxa exhibit differential abundances in total and active communities of Antarctic soils. Environ Microbiol. 2019;21:1586–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuezva S, Fernandez-Cortes A, Porca E et al. The biogeochemical role of actinobacteria in Altamira Cave. FEMS Microbiol Ecol. 2012;81:281–90. [DOI] [PubMed] [Google Scholar]
- Czurda S, Smelik S, Preuner-Stix S et al. Occurrence of fungal DNA contamination in PCR reagents: approaches to control and decontamination. J Clin Microbiol. 2016;54:148–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R et al. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delafont V, Rodier M-H, Maisonneuve E et al. Vermamoeba vermiformis: a free-living amoeba of interest. Microb Ecol. 2018;76:991–1001. [DOI] [PubMed] [Google Scholar]
- Delgado-Baquerizo M. Obscure soil microbes and where to find them. ISME J. 2019;13:2120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira TB, Gomes E, Rodrigues A. Thermophilic fungi in the new age of fungal taxonomy. Extremophiles. 2015;19:31–7. [DOI] [PubMed] [Google Scholar]
- Fang B-Z, Salam N, Han M-X et al. Insights on the effects of heat pretreatment, pH, and calcium salts on isolation of rare actinobacteria from karstic caves. Front Microbiol. 2017;8. 10.3389/fmicb.2017.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CI, Connell L, Lee CK et al. Evidence of plant and animal communities at exposed and subglacial (cave) geothermal sites in Antarctica. Polar Biol. 2018;41:417–21. [Google Scholar]
- Fraser CI, Terauds A, Smellie J et al. Geothermal activity helps life survive glacial cycles. Proc Natl Acad Sci USA. 2014;111:5634–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giggenbach WF. Geothermal ice caves on Mt Erebus, Ross Island, Antarctica. NZ J Geol Geophys. 1976;19:365–72. [Google Scholar]
- Gonzalez-Pimentel JL, Dominguez-Moñino I, Jurado V et al. The rare Actinobacterium crossiella sp. is a potential source of new bioactive compounds with activity against bacteria and fungi. Microorganisms. 2022;10:1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pimentel JL, Miller AZ, Jurado V et al. Yellow coloured mats from lava tubes of La Palma (Canary Islands, Spain) are dominated by metabolically active. Sci Rep. 2018;8:1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostinčar C, Stajich JE, Gunde-Cimerman N. Extremophilic and extremotolerant fungi. Curr Biol. 2023;33:R752–6. [DOI] [PubMed] [Google Scholar]
- Greenfield L. Thermophilic fungi and actinomycetes from Mt Erebus and a fungus pathogenic to Bryurn antarcticum at Cape Bird. NZ. New Zealand Antarctic Record. Wellington: DSIR Antarctic Division, 1982, 10–1. [Google Scholar]
- Herbold CW, Lee CK, McDonald IR et al. Evidence of global-scale aeolian dispersal and endemism in isolated geothermal microbial communities of Antarctica. Nat Commun. 2014a;5. 10.1038/ncomms4875. [DOI] [PubMed] [Google Scholar]
- Herbold CW, Mcdonald IR, Cary SC. Microbial ecology of geothermal habitats in Antarctica. In: Cowan D (ed.), Antarctic Terrestrial Microbiology: Physical and Biological Properties of Antarctic Soils. Berlin, Heidelberg: Springer, 2014b, 181–216. [Google Scholar]
- Herbold CW, Noell SE, Lee CK et al. Nutritional niches of endemic, facultatively anaerobic heterotrophs from an isolated Antarctic terrestrial hydrothermal refugium elucidated through metagenomics. 2024. PREPRINT (Version 1) available at Research Square. 10.21203/rs.3.rs-4805162/v1. [DOI]
- Hill GJ, Wannamaker PE, Maris V et al. Trans-crustal structural control of CO2-rich extensional magmatic systems revealed at Mount Erebus Antarctica. Nat Commun. 2022;13:2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JA, Daniel RM. Enumeration of thermophilic heterotrophs in geothermally heated soils from Mount Erebus, Ross Island, Antarctica. Appl Environ Microbiol. 1988;54:622–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JA, Daniel RM, Morgan HW. Acidophilic and thermophilic Bacillus strains from geothermally heated Antarctic soil. FEMS Microbiol Lett. 1989;60:279–82. [Google Scholar]
- Ilanko T, Fischer TP, Kyle P et al. Modification of fumarolic gases by the ice-covered edifice of Erebus volcano. J Volcanol Geotherm Res. 2019;381:119–39. [Google Scholar]
- Janetschek H. On the terrestrial fauna of the Ross-Sea area, Antartica. Pac Insects. 1963;5:305–11. [Google Scholar]
- Kato S, Ohnishi M, Nagamori M et al. Conexivisphaera calida gen. nov., sp. nov., a thermophilic sulfur- and iron-reducing archaeon, and proposal of Conexivisphaeraceae fam. nov., Conexivisphaerales ord. nov., and Conexivisphaeria class. nov. in the phylum Thaumarchaeota. Int J Syst Evol Microbiol. 2021;71. 10.1099/ijsem.0.004595. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PJ, Kyle PR, Dunbar NW et al. Geochemistry and mineralogy of the phonolite lava lake, Erebus volcano, Antarctica: 1972–2004 and comparison with older lavas. J Volcanol Geotherm Res. 2008;177:589–605. [Google Scholar]
- Kerou M, Eloy Alves RJ, Schleper C. Nitrososphaeria. Bergey’s Manual of Systematics of Archaea and Bacteria. Hoboken, NJ: Wiley, 2016, 1–8. [Google Scholar]
- Kyle PR. A. McMurdo Volcanic Group western Ross Embayment. In: Volcanoes of the Antarctic Plate and Southern Oceans. Antarctic Research Series. Vol. 48, Washington, DC: American Geophysical Union, 1990, 19–139. [Google Scholar]
- Lahti L, Shetty S. Tools for microbiome analysis in R. GitHub, 2017. [Google Scholar]
- Lange-Enyedi NT, Németh P, Borsodi AK et al. Calcium carbonate precipitating extremophilic bacteria in an Alpine ice cave. Sci Rep. 2024;14:2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353:1272–7. [DOI] [PubMed] [Google Scholar]
- Luo Z-H, Li Q, Xie Y-G et al. Temperature, pH, and oxygen availability contributed to the functional differentiation of ancient Nitrososphaeria. ISME J. 2024;18. 10.1093/ismejo/wrad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011;17:10. [Google Scholar]
- McLaren MR. Silva SSU taxonomic training data formatted for DADA2 (Silva version 138) (Version 2). Zenodo, 2020. 10.5281/zenodo.3986799. [DOI] [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monciardini P, Cavaletti L, Schumann P et al. Conexibacter woesei gen. nov., sp. nov., a novel representative of a deep evolutionary line of descent within the class Actinobacteria. Int J Syst Evol Microbiol. 2003;53:569–76. [DOI] [PubMed] [Google Scholar]
- Morien E, Parfrey LW. SILVA v128 and v132 dada2 formatted 18 s “train sets”. Zenodo, 2018. 10.5281/zenodo.1447330. [DOI] [Google Scholar]
- Nakaew N, Pathom-aree W, Lumyong S. Generic diversity of rare actinomycetes from Thai cave soils and their possible use as new bioactive compounds. Actinomycetologica. 2009;23:21–6. [Google Scholar]
- Noell SE, Baptista MS, Smith E et al. Unique geothermal chemistry shapes microbial communities on Mt. Erebus, Antarctica. Front Microbiol. 2022;13. 10.3389/fmicb.2022.836943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A, Garrity GM. Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol. 2021;71. 10.1099/ijsem.0.005056. [DOI] [PubMed] [Google Scholar]
- Panter KS, Winter B. Geology of the Side Crater of the Erebus volcano. J Volcanol Geotherm Res. 2008;177:578–88. [Google Scholar]
- Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14. [DOI] [PubMed] [Google Scholar]
- Parte A, Whitman WB, Goodfellow M et al. et al. (eds.), The Actinobacteria, Part B. Bergey’s Manual of Systematic Bacteriology. New York, NY: Springer, 2012, 1379–83. [Google Scholar]
- Patil I. Visualizations with statistical details: the “ggstatsplot” approach. JOSS. 2021;6:3167. [Google Scholar]
- Paun VI, Lavin P, Chifiriuc MC et al. First report on antibiotic resistance and antimicrobial activity of bacterial isolates from 13,000-year old cave ice core. Sci Rep. 2021;11:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautler BG, Dubnick A, Sharp MJ et al. Comparison of cryoconite organic matter composition from Arctic and Antarctic glaciers at the molecular-level. Geochim Cosmochim Acta. 2013;104:1–18. [Google Scholar]
- Peña-Méndez EM, Gajdošová D, Novotná K et al. Mass spectrometry of humic substances of different origin including those from Antarctica. Talanta. 2005;67:880–90. [DOI] [PubMed] [Google Scholar]
- Peschel S, Müller CL, von Mutius E et al. NetCoMi: network construction and comparison for microbiome data in R. Brief Bioinform. 2021;22. 10.1093/bib/bbaa290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar PT, Yang Z, Björnsdóttir SH et al. Comparative analysis of microbial diversity across temperature gradients in hot springs from Yellowstone and Iceland. Front Microbiol. 2020;11. 10.3389/fmicb.2020.01625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana ET, Badillo RF, Maldonado LA. Characterisation of the first actinobacterial group isolated from a Mexican extremophile environment. Antonie Van Leeuwenhoek. 2013;104:63–70. [DOI] [PubMed] [Google Scholar]
- Ren M, Feng X, Huang Y et al. Phylogenomics suggests oxygen availability as a driving force in Thaumarchaeota evolution. ISME J. 2019;13:2150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Chuvochina M, Mussig AJ et al. A standardized archaeal taxonomy for the Genome Taxonomy Database. Nat Microbiol. 2021;6:946–59. [DOI] [PubMed] [Google Scholar]
- Riquelme C, de L Enes Dapkevicius M, Miller AZ et al. Biotechnological potential of actinobacteria from Canadian and Azorean volcanic caves. Appl Microbiol Biotechnol. 2017;101:843–57. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum EB, Voyles J, Poorten TJ et al. The deadly chytrid fungus: a story of an emerging pathogen. PLoS Pathog. 2010;6:e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansupa C, Wahdan SFM, Hossen S et al. Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil bacteria?. Appl Sci. 2021;11:688. [Google Scholar]
- Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakova L, Bondarenko N, Smirnov A. Viable species of flamella (Amoebozoa: variosea) isolated from ancient Arctic permafrost sediments. Protist. 2016;167:13–30. [DOI] [PubMed] [Google Scholar]
- Sims KWW, Aster RC, Gaetani G et al. Chapter 7.2 Mount Erebus. Memoirs. 2021;55:695–739. [Google Scholar]
- Skotnicki ML, Selkirk PM, Broady P et al. Dispersal of the moss Campylopus pyriformis on geothermal ground near the summits of Mount Erebus and Mount Melbourne, Victoria Land, Antarctica. Antartic Sci. 2001;13:280–5. [Google Scholar]
- Smiti A. A critical overview of outlier detection methods. Comput Sci Rev. 2020;38:100306. [Google Scholar]
- Soo RM, Skennerton CT, Sekiguchi Y et al. An expanded genomic representation of the phylum Cyanobacteria. Genome Biol Evol. 2014;6:1031–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo RM, Wood SA, Grzymski JJ et al. Microbial biodiversity of thermophilic communities in hot mineral soils of Tramway Ridge, Mount Erebus, Antarctica. Environ Microbiol. 2009;11:715–28. [DOI] [PubMed] [Google Scholar]
- Staley JT, Konopka A. Measurement of in situ activtities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–46. [DOI] [PubMed] [Google Scholar]
- Steinbach RM, El Baidouri F, Mitchison-Field LMY et al. Malassezia is widespread and has undescribed diversity in the marine environment. Fungal Ecol. 2023;65:101273. [Google Scholar]
- Strunecký O, Ivanova AP, Mareš J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. J Phycol. 2023;59:12–51. [DOI] [PubMed] [Google Scholar]
- Tebo BM, Davis RE, Anitori RP et al. Microbial communities in dark oligotrophic volcanic ice cave ecosystems of Mt Erebus, Antarctica. Front Microbiol. 2015;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitla T, Kumla J, Hongsanan S et al. Exploring diversity rock-inhabiting fungi from northern Thailand: a new genus and three new species belonged to the family Herpotrichiellaceae. Front Cell Infect Microbiol. 2023;13. 10.3389/fcimb.2023.1252482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini FC, Starkey RL. Soils and micro-organisms from Mount Erebus, Antarctica. Nature. 1966;211:440–1. [Google Scholar]
- Van Rossum T, Ferretti P, Maistrenko OM et al. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol. 2020;18:491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers CJ, Herbold CW, Cary SC et al. Insights into the metabolism of the high temperature microbial community of Tramway Ridge, Mount Erebus, Antarctica. Antarct Sci. 2016;28:241–9. [Google Scholar]
- von der Heyden S, Cavalier-Smith T. Culturing and environmental DNA sequencing uncover hidden kinetoplastid biodiversity and a major marine clade within ancestrally freshwater Neobodo designis. Int J Syst Evol Microbiol. 2005;55:2605–21. [DOI] [PubMed] [Google Scholar]
- Wickham H. Ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer, 2016. [Google Scholar]
- Wright ES. Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 2016;8:352–9. [Google Scholar]
- Yabe S, Muto K, Abe K et al. Vulcanimicrobium alpinus gen. nov. sp. nov., the first cultivated representative of the candidate phylum “Eremiobacterota”, is a metabolically versatile aerobic anoxygenic phototroph. ISME Commun. 2022;2:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucconi L, Pagano S, Fenice M et al. Growth temperature preferences of fungal strains from Victoria Land, Antarctica. Polar Biol. 1996;16:53–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All R scripts used to analyse the data are available on Github at https://github.com/ThermophileResearchUnit/Erebus_survey_manuscript. All sequence data is available in DDBJ/EMBL/GenBank. 16S rRNA gene sequence data has been deposited as accession KIGZ00000000 version KIGZ01000000, except for samples TR1-*, TR2-*, and WC-* which have been deposited as accession KIGX00000000 version, KIGX01000000. 18S rRNA gene sequence data have been deposited as SUB14470052. ITS sequence data have been deposited as SUB14470177.