Abstract
Ubiquitination, a multifaceted post-translational modification, regulates protein function, degradation, and gene expression. The pivotal role of ubiquitination in the pathogenesis and progression of cancer, including colorectal, breast, and liver cancer, is well-established. Osteosarcoma, an aggressive bone tumor predominantly affecting adolescents, also exhibits dysregulation of the ubiquitination system, encompassing both ubiquitination and deubiquitination processes. This dysregulation is now recognized as a key driver of osteosarcoma development, progression, and chemoresistance. This review highlights recent progress in elucidating how ubiquitination modulates tumor behavior across signaling pathways. We then focus on the mechanisms by which ubiquitination influences osteosarcoma cell function. Finally, we discuss the potential for targeting the ubiquitin-proteasome system in osteosarcoma therapy. By unraveling the impact of ubiquitination on osteosarcoma cell physiology, we aim to facilitate the development of novel strategies for prognosis, staging, treatment, and overcoming chemoresistance.
Keywords: Ubiquitination, osteosarcoma, cancer development, therapeutic target
Introduction
Osteosarcoma (OS), a highly aggressive bone tumor, is characterized by the presence of malignant mesenchymal cells that form osteoid or immature bone tissue and exhibits a bimodal incidence that primarily affects children and adolescents in addition to individuals > 60 years of age1. Approximately 25% of patients have metastatic spread, most commonly to the lungs2. Despite advancements in treatment strategies, including extensive surgical resection and neoadjuvant/adjuvant chemotherapy, improving patient survival remains a significant challenge3,4. While targeted therapy offers promise due to selectivity and reduced toxicity, efficacy is hampered by rapid development of drug resistance in patients undergoing prolonged chemotherapy with relapse salvage rates hovering at approximately 30%5. Furthermore, the prognosis for patients with metastatic or recurrent osteosarcoma has had little improvement in recent decades6. This stagnation, coupled with limitations in early detection methods, presents a major obstacle to improving survival rates7.
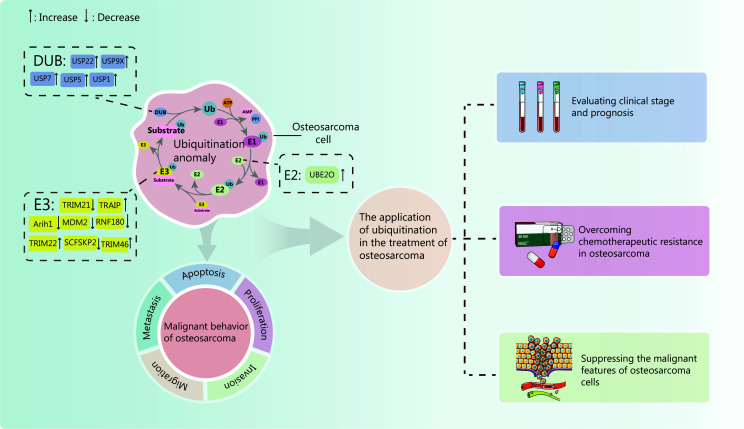
Ubiquitination, a ubiquitous post-translational modification, has a central role in orchestrating cellular processes8. Recent research has unveiled an intricate interplay between ubiquitination and deubiquitination in cancer development and progression9–12. This interplay holds promising clinical applications, including overcoming drug resistance in OS and providing novel targets for targeted therapy13–15. Therefore, understanding the role of ubiquitination in OS holds immense potential for improving both disease prognosis and therapeutic efficacy. This article examines the role of ubiquitination in regulating osteosarcoma proliferation, invasion, migration, and other malignant behaviors. Furthermore, the potential of ubiquitin-based research to create novel therapeutic strategies for OS is discussed. We posit that ubiquitination dysregulation is a primary driver of OS development and drug resistance. Consequently, targeting ubiquitination holds significant promise for future OS treatment. By comprehensively reviewing the involvement of ubiquitination in OS pathogenesis and the current state of research, this work identifies knowledge gaps and outlines potential avenues for future investigation. The findings provide a foundation for developing novel therapeutic strategies targeting the ubiquitination pathway in OS while deepening our understanding of the disease through a focused examination of the critical role of ubiquitination (Figure 1).
Figure 1.
Effect of ubiquitination imbalance on osteosarcoma cells and its clinical application. Aberrant expression of E1, E2, and E3 enzymes within the ubiquitin-proteasome system, as well as deubiquitinases, can disrupt ubiquitination and deubiquitination pathways, contributing to the malignant phenotypes of osteosarcoma cells, including proliferation, invasion, migration, and metastasis. By investigating the impact of these ubiquitin modification dysregulations on osteosarcoma, researchers have made significant strides in evaluating tumor prognosis, overcoming drug resistance, and inhibiting tumor progression. Together, these findings promote the development of osteosarcoma treatment, such as judging the prognosis of osteosarcoma, overcoming drug resistance, and inhibiting tumor occurrence. As illustrated in the figure, corresponding solid and dashed lines at the upstream and downstream of the signaling pathway indicate consistent activation or inhibition, respectively. The red line signifies a distinct process. Upon activation of the signaling pathway, expression or interaction of a downstream ubiquitin ligase is induced, subsequently influencing tumorigenesis. This effect is independent of the illustrated upstream signaling events.
Ubiquitination and osteosarcoma
Burden of OS
OS, the most common primary bone tumor, primarily affects adolescents and older individuals with pre-existing bone deformities16. OS typically arises in the ends of long bones, particularly the limbs17. Characteristically, this cancer involves the abnormal production of immature bone and osteoid by transformed osteoblasts, resulting in remarkable intratumoral heterogeneity in which different tissue types and behaviors co-exist18. This complexity underscores the challenge of managing this disease. Both environmental and genetic factors contribute to OS development, including age, exposure to ionizing radiation, and pre-existing bone conditions, such as osteoarthritis19. Patients typically present with localized swelling, pain, and limited joint movement. In approximately 15% of advanced cases there is radiographic evidence of metastasis, mainly to the lungs5. Imaging has a crucial role in diagnosis, revealing different patterns within the tumor, including osteogenic, osteolytic, or mixed forms. Notably, the triangular periosteal calcification at the tumor-bone interface, known as the Codman triangle, is a highly characteristic feature20,21.
The mainstay of OS treatment involves surgery, radiotherapy, and pre- or postoperative chemotherapy3. However, effective options remain limited for patients with advanced lung metastasis, resistance to chemotherapy, or drug intolerance. Furthermore, chemoresistance significantly hinders improvements in survival rates22. This grim reality highlights the urgent need for intensified research and clinical trials to overcome these limitations23,24.
Ubiquitin-proteasome system
Ubiquitination and deubiquitination dynamically regulate intracellular protein modification and gene expression. Dysregulation of these processes frequently drives tumorigenesis and progression by altering the expression of both modified and unmodified genes, leading to aberrant signaling and malignant behavior. The resulting altered gene expression profile offers potential as a cancer biomarker for prognostication and therapeutic targeting12,25.
Ubiquitination, a pivotal post-translational modification orchestrated by a coordinated enzymatic cascade, utilizes a 76-amino acid ubiquitin molecule13. This key modifier harbors a C-terminal tail with a conserved diglycine (GG) motif and seven lysine (Lys) residues, serving as attachment sites for additional ubiquitin moieties26. The cascade involves the following complex interplay of enzymes: E1 family members activate ubiquitin; E2 enzymes categorized by function accept ubiquitin; and E3 ligases (particularly HECT, RING, and RBR types) recognize the target protein and facilitate ubiquitin attachment to specific Lys residues27–29. This intricate collaboration orchestrates a vast array of cellular functions, highlighting the potential of ubiquitination as a therapeutic target. Notably, the hundreds of E3 ligases in eukaryotes exemplify the remarkable power and versatility of ubiquitination in regulating life30–32.
Deubiquitination, the counterpoint to ubiquitination, relies on deubiquitinating enzymes (DUBs) to release the attached ubiquitin molecule from its target protein, effectively reversing the functional impact33. DUBs are currently classified into seven main groups, each with unique structural and functional features: ubiquitin-specific protease (USP); ubiquitin C-terminal hydrolase (UCH); ovarian tumor protease (OTU); machado-josephin domain protease (MJD); Jab1/MPN+/Mov34 (JAMM) domain protease; monocyte chemoattractant protein inducible protein (MCPIP); and motifs interacting with ubiquitin-containing novel DUB family (MINDY)34.
Link between OS and ubiquitination
A growing body of research has highlighted dysregulation of the ubiquitin-proteasome system (UPS) in OS. Components of the UPS, including ubiquitin ligases and DUBs, are frequently aberrantly expressed in OS cells, which contributes to tumorigenesis, progression, and metastasis35,36. For example, several ubiquitin ligases have been shown to promote osteosarcoma cell growth and survival by targeting tumor suppressors for degradation. Conversely, DUBs stabilize oncoproteins, thereby driving tumor progression. Understanding the precise mechanisms by which ubiquitination contributes to OS pathogenesis is essential for developing novel therapeutic strategies. By elucidating the specific roles of ubiquitin-related enzymes in this cancer type, researchers can identify potential drug targets and biomarkers for patient stratification.
Subsequently, we will examine the current state of ubiquitination research across diverse tumor types. This will be followed by an in-depth exploration of the influence of ubiquitination on the malignant phenotype of OS. Finally, we will discuss the clinical implications and potential applications of targeting ubiquitination in OS treatment.
Ubiquitin modification of proteins in key signaling pathways in cancer
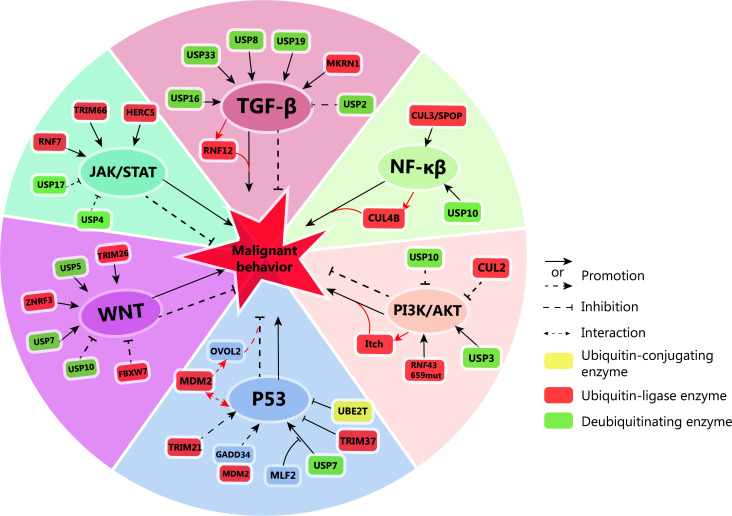
The intricate interplay between ubiquitination and deubiquitination tightly regulates diverse signaling pathways, demonstrably influencing tumorigenesis and cancer progression (Figure 2). This dynamic duo acts as a cellular switchboard, dictating the fate of proteins through targeted attachment and removal of ubiquitin molecules. In the following sections, we delve deeper into the specific effects of ubiquitination on tumor development, exploring how ubiquitination manipulates key signaling networks to promote cancer hallmarks.
Figure 2.
Ubiquitin-mediated regulation of key signaling pathways in tumor cells. Ubiquitin ligases and deubiquitinases have pivotal roles in modulating the activation or inhibition of TGF-β, NF-κβ, WNT, PI3K/AKT, JAK/STAT, and P53 signaling pathways in tumor cells. These pathways, in turn, influence the malignant behavior of tumors. Notably, the activation of some signaling pathways can also lead to the activation of downstream ubiquitin ligases, further impacting tumor progression. TGF-β: deubiquitinases ubiquitin-specific protease 16 (USP16), USP33, USP8, USP19, and the makorin ring finger protein 1 (MKRN1) activate the TGF-β signaling pathway. The activation of TGF-β further activates ring finger protein 12 (RNF12). These events collectively contribute to the malignant behavior of tumors. Conversely, USP2 inhibits TGF-β signaling, suppressing tumor growth. NF-κβ: USP10 and cullin-3/ speckle-type POZ protein (CUL3/SPOP) activate the NF-κβ signaling pathway, promoting tumor formation. NF-κβ activation also upregulates CUL4B, further driving tumor growth. WNT: USP7, USP5, and zinc and ring finger 3 (ZNRF3), tripartite motif containing 26 (TRIM26) activate the WNT pathway, promoting tumorigenesis. Conversely, USP10 and recombinant F-box and WD repeat domain containing protein 7 (FBXW7) inhibit tumorigenesis by suppressing the WNT pathway. PI3K/AKT: RNF43 and USP3 activate the PI3K/AKT pathway, promoting tumorigenesis. Conversely, CUL2 and USP10 inhibit PI3K/AKT, suppressing tumorigenesis. Activated PI3K/AKT can further activate ubiquitin ligase Itch, contributing to tumorigenesis. JAK/STAT: RNF7, TRIM66, and HERC5 activate the JAK/STAT pathway, promoting tumorigenesis. USP17 and USP4 inhibit the JAK/STAT pathway, suppressing tumorigenesis. p53: TRIM21 activates the p53 pathway, while growth arrest and DNA-damage-inducible gene 34 (GADD34) competitively binds to mousedouble minute 2 (MDM2) to promote p53 expression. These events collectively inhibit tumorigenesis. TRIM37 and ubiquitin-conjugating enzyme E2T (UBE2T) inhibit p53 expression, while myeloid leukemia factor 2 (MLF2) inhibits the binding of USP7 to p53, further promoting tumorigenesis. Additionally, p53 interacts with MDM2, which in turn promotes the expression of OVO-like zinc finger 2 (OVOL2) and inhibits tumorigenesis.
P53 signaling pathway
The p53 pathway, a critical tumor suppressor, is tightly regulated by ubiquitination and deubiquitination enzymes, offering promising opportunities for therapeutic intervention. Researchers have investigated the biological function of growth arrest and DNA-damage-inducible gene 34 (GADD34) by transfecting U2OS human OS cells. MDM2 was shown to mediate GADD34 ubiquitination. Interestingly, GADD34 acts as a ubiquitination competitor, reducing p53 ubiquitination and increasing p53 protein levels, thereby suppressing OS proliferation37. In breast and rectal cancers, TRIM21 acts as a gatekeeper, inhibiting mutant p53 accumulation and suppressing tumor growth38. Conversely, in hepatocellular carcinoma, TRIM37 promotes p53 ubiquitination and degradation, hindering cancer progression39. These contrasting examples highlight the context-dependent nature of ubiquitination in p53 regulation. Exploiting ubiquitination for targeted therapy presents exciting possibilities. In pancreatic cancer, UBE2T-mediated p53 degradation confers gemcitabine resistance, highlighting the potential of manipulating this pathway to overcome drug resistance40. Colorectal cancer (CRC) progression relies on myeloid leukemia factor 2 (MLF2), which disrupts the USP7-p53 deubiquitination complex, promoting tumorous growth41. Additionally, in breast cancer, p53 directly binds mousedouble minute 2 (MDM2), preventing ubiquitination of OVO-like zinc finger 2 (OVOL2), a transcriptional suppressor of metastasis. This interaction makes it possible to exploit the p53-MDM2-OVOL2 axis for therapeutic benefit42.
JAK/STAT signaling pathway
The JAK/STAT signaling pathway intricately collaborates with ubiquitination to influence tumor cell behavior. Destruction of the DUBs (usp17 and usp4) targeting PDGFR β/STAT3 in osteosarcoma will lead to the interruption of signal transduction and the uncontrolled proliferation of OS cells43. Silencing TRIM66 reduces Janus kinase2 (JAK2) activation and signal transducer and activator of transcription 3 (STAT3) phosphorylation in rectal cancer, leading to suppressed cell proliferation, migration, and invasion44. Conversely, under glucose starvation, protein tyrosine phosphatase mitochondrial 1 (PTPMT1)-mediated dephosphorylation of recombinant human eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1) safeguards it from HECT and RLD domain containing E3 ubiquitin protein ligase 5 (HERC5)-induced ubiquitination and degradation. Stabilized 4EBP1 promotes lung cancer cell apoptosis by competing with Jak and ERK for binding to STAT345. Interestingly, ring finger protein 7 (RNF7) activates STAT3 signaling by ubiquitinating recombinant suppressors of cytokine signaling (SOCS1) in renal cell carcinoma, thereby inhibiting apoptosis, promoting glycolysis, and reducing sensitivity to sunitinib46. These diverse examples highlight the multifaceted role of ubiquitination in modulating the JAK/STAT pathway in tumors.
PI3K/AKT signaling pathway
The PI3K/AKT pathway, a central hub controlling numerous cellular processes through the vast network of downstream effectors and intricate crosstalk with other signaling cascades, has a crucial role in cancer development and progression47. This multifaceted pathway has been extensively studied in various cancers, revealing the diverse and context-dependent influence. USP3 leads to activation of the PI3K/AKT signaling pathway in OS by binding to EPHA2, then reducing protein degradation. Overexpression of USP3 significantly increased OS cell proliferation, migration, and invasion48. A mutated form of RNF43 (RNF43 659mut) binds to p85 in rectal cancer, promoting ubiquitination and degradation, ultimately leading to enhanced PI3K signaling and increased tumor activity49. Conversely, USP10 deubiquitinates K63-linked ubiquitin chains on Akt in non-small cell lung cancer, weakening downstream signaling and inhibiting tumor cell proliferation50. USP10 further exerts a tumor suppressive role by stabilizing adenosine 5′monophosphate-activated protein kinase alpha (AMPKα) and phosphatase and tension homologue deleted from chromosome 10 (PTEN) in liver cancer, leading to mammalian target of rapamycin complex 1 (mTORC1) inhibition and reduced Akt phosphorylation51. Breast cancer presents another example of how ubiquitination modulates the PI3K/AKT pathway. Recombinant peptidyl prolyl cis/trans isomerase NIMA interacting protein 1 (Pin1), an isomerase activated by Akt phosphorylation, promotes the interaction between salt-induced protein kinase 1 (Sik1) and E3 ligase Itch, leading to Sik1 ubiquitination and degradation, ultimately contributing to tumorigenesis52. Interestingly, the N6-methyladenosine (m6A) modification (WEE2-AS1) stabilizes RPN2 protein by preventing Cul2-mediated K322 ubiquitination, thereby activating PI3K/AKT and promoting glioblastoma progression53.
NF-κβ signaling pathway
The NF-κβ signaling pathway has a pivotal role in tumor development and resistance to therapies. In osteosarcoma, the ubiquitin ligase, cullin 4B protein (CUL4B), orchestrated by NF-κβ subunits, promotes invasion by degrading the CDK inhibitor, p2154. The CUL3/SPOP E3 ligase promotes growth by targeting DRAK1 for degradation in paclitaxel-resistant cervical cancer cells, leading to enhanced NF-κβ activity55. USP10 deubiquitinates NLRP7 protein in CRC, stabilizing NLRP7 and prompting M2 macrophage polarization via NF-κβ-dependent secretion of monocyte chemoattractant protein-1 (MCP-1). This pro-tumorigenic cascade fuels CRC proliferation and metastasis56.
WNT signaling pathway
The WNT signaling pathway, which has an essential role in many cancers, presents promising therapeutic opportunities through modulation of ubiquitination and deubiquitination. USP7, a DUB, promotes migration by activating WNT signaling via EMT/β-catenin in OS57. For example, the development of proteolysis-targeting chimeras (PROTACs) targeting zinc and ring finger 3 (ZNRF3), a WNT-responsive ligase, leading to targeted degradation of transmembrane proteins and CRC regression in cell culture and animal models58. The interaction between prospero homeobox 1 (PROX1) and heterogeneous nuclear ribonucleoprotein H (hnRNPH) inhibits ubiquitination of breast cancer, promoting WNT pathway activation and tumor metastasis59. USP5 deubiquitinates and stabilizes β-catenin, thereby activating the WNT/β-catenin pathway and promoting stem cell characteristics in lung cancer60.
Conversely, Let-7b-5p acts as a tumor suppressor in colon cancer by blocking the ubiquitination-mediated degradation of adenomatous polyposis coli (APC) and NKD1, crucial molecules in the WNT pathway61. Similarly, the 185aa isoform of circFBXW7 regulates the WNT pathway by ubiquitinating and inhibiting β-catenin, effectively suppressing lung adenocarcinoma stem cell activity and reversing resistance to tyrosine kinase inhibitors62. Finally, the LKB1/AMPK pathway interacts with WNT/β-catenin, with USP10 acting as a central hub. By modulating metabolism and cell proliferation, the LKB1/AMPK axis inhibits tumor growth through USP10-mediated regulation63.
TGF-β signal pathway
The TGF-β signaling pathway presents a perplexing conundrum in cancer, acting as both a tumor suppressor and promoter depending on the specific cellular context64. This paradoxical role hinges on the delicate balance between ubiquitination and deubiquitination of key pathway components, highlighting the potential of targeting these enzymes for therapeutic intervention. USP8 promotes tumor progression and T cell depletion in breast cancer by deubiquitinating the TGF-β receptor, TβRII65. Targeting USP19 splicing or its enzymatic activity offers a promising avenue, as shown by the splicing regulator, herboxidiene, which suppresses cell migration by upregulating the anti-migratory USP19 isoform66. Dysregulation of deubiquitination can also drive cancer development, as exemplified in hepatocellular carcinoma (HCC) in which elevated recombinant diacylglycerol kinase gamma (DGKG) recruits USP16, leading to zinc finger E-box-binding homeobox 2 (ZEB2) deubiquitination and subsequent TGF-β1 upregulation, which promotes angiogenesis and regulatory T cell differentiation67. Similarly, makorin ring finger protein 1 gene (MKRN1) in colon cancer fosters TGF-β signaling and metastasis by ubiquitinating and degrading Smad nuclear interacting protein 1 (SNIP1)-β68. Adding another layer of complexity, Akt-mediated phosphorylation further intertwines ubiquitination with TGF-β signaling, as evident in RNF12, which promotes breast cancer metastasis upon Akt activation69. Deubiquitination enzymes also have crucial roles. USP33, for example, promotes pancreatic cancer malignancy through the TGFBR2/TGF-β pathway70, while inhibiting USP2 expression conversely activates TGF-β signaling and fuels glioblastoma development71.
Impact of ubiquitination on the malignant OS phenotype
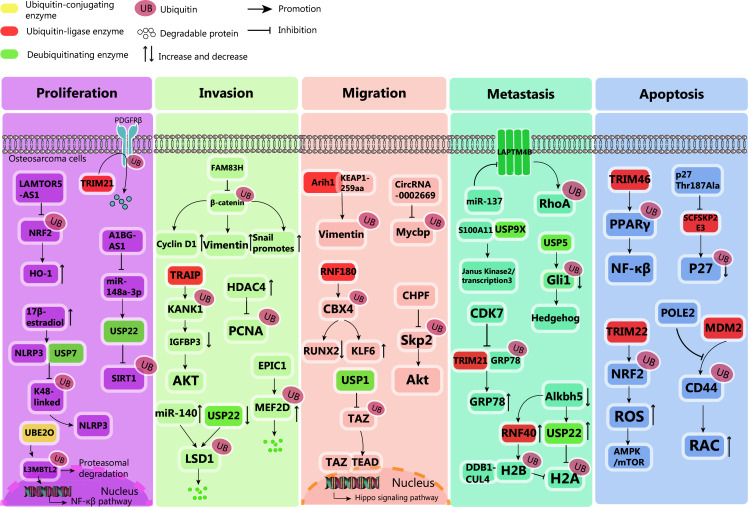
Ubiquitination, a process attaching a small protein tag to other proteins, has a central role in regulating various signaling pathways that profoundly influence the biological behavior of OS (Figure 3). The impact extends beyond mere cell proliferation, intricately modulating invasion, migration, and the threat of metastasis. This complex regulatory network presents a wealth of potential targets for therapeutic intervention, offering hope for improved outcomes in this challenging cancer.
Figure 3.
Dysregulated ubiquitination pathway drives osteosarcoma malignancy. Proliferation: Proliferation of osteosarcoma is regulated by intricate ubiquitination processes. UBE2O ubiquitinates L3MBTL2, bolstering NF-κβ expression and tumor growth. LAMTOR5-AS1 halts proliferation by blocking NRF2 ubiquitination, augmenting heme oxygenase-1 levels. TRIM21 reduces PDGFRβ via ubiquitination, inhibiting tumor expansion. A1BG-AS1 enhances proliferation by inhibiting miR-148a-3p and upregulating USP22, inhibiting SIRT1 ubiquitination. High E2 levels facilitate USP7-NLRP3 interaction, removing K48-linked ubiquitination, boosting NLRP3 inflammasome and proliferation. Invasion: Invasion is curbed by EPIC1-mediated MEF2D ubiquitination and degradation. TRAIP promotes Kank1 polyubiquitination, downregulating IGFBP3 and activating Akt, fostering invasion. FAM83H stabilizes β-catenin, upregulating cyclin D1, vimentin, and snail, promoting invasion. HDAC4 overexpression elevates PCNA while decreasing ubiquitinated PCNA, enhancing invasion. miR-140 or USP22 knockdown triggers LSD1 ubiquitination and degradation, inhibiting invasion. Migration: KEAP1-259aa interacts with ARIH1 to promote the proteasomal degradation of cytoplasmic vimentin, inhibiting osteosarcoma cell migration. CircRNA_0002669 promotes osteosarcoma cell migration by preventing MYCBP ubiquitination. USP1 interacts with TAZ, activating the Hippo signaling pathway and promoting cell migration. CHPF inhibits SKP2 ubiquitination and activates the Akt signaling pathway, promoting osteosarcoma migration. RNF180 ubiquitinates Cbx4, upregulating KLF6 and downregulating Runx2, inhibiting cell migration. Metastasis: S100A11 interacts with USP9X to stimulate the JAK2/STAT3 signaling pathway, promoting lung metastasis in osteosarcoma patients. USP5 deubiquitinates GLI1, activating the Hedgehog signaling pathway and promoting osteosarcoma metastasis. ALKBH5 increases the expression of USP22 and RNF40, inhibiting histone H2A single ubiquitination. RNF40, ubiquitinated on H2B, inhibits H2A ubiquitination in tumors, interacting with the DDB1-CUL4-based ubiquitin E3 ligase complex to promote osteosarcoma metastasis. CDK7 inhibits TRIM21-mediated GRP78 ubiquitination, and promoting osteosarcoma metastasis. miR-137 can inhibit osteosarcoma metastasis by targeting LAPTM4B to stabilize RhoA protein. Apoptosis: POLE2 enhances CD44 expression by inhibiting MDM2-mediated ubiquitination, subsequently activating the RAC signaling pathway to promote osteosarcoma cell apoptosis. TRIM22 inhibits osteosarcoma progression through Nrf2-mediated (ROS) imbalance. The p27 Thr187Ala knock-in mutation disrupts SCFSkp2 E3 ligase-mediated p27 ubiquitination, leading to p27 Thr187Ala accumulation and promoting osteosarcoma cell apoptosis. TRIM46 interacts with PPAR, ubiquitinates PPAR, activates the NF-κβ signaling pathway, and induces apoptosis.
Proliferation
Cell proliferation, the fundamental process driving primary tumor growth, is meticulously regulated within OS through a complex interplay of ubiquitination pathways. These pathways exert diverse and often opposing influences, underscoring the intricate regulatory network governing this critical biological process. Several ubiquitination axes have emerged as pivotal contributors. A negative correlation exists between lethal (3) malignant brain tumor-like protein 2 (L3MBTL2) and UBE2O within OS tissue. UBE2O ubiquitinates L3MBTL2, targeting L3MBTL2 for proteasomal degradation. The UBE2O/L3MBTL2 axis is indispensable for OS growth. Elevated UBE2O and diminished L3MBTL2 expression correlate with adverse clinical outcomes in OS patients. Pharmacologic inhibition of UBE2O using arsenic trioxide potentiates L3MBTL2-induced aggregates, consequently suppressing OS growth. Consequently, the UBE2O-L3MBTL2 axis is posited as a promising therapeutic target for OS72. High-throughput sequencing identified a novel long non-coding RNA (lncRNA) termed long non-coding RNA LAMTOR5 antisense RNA 1 (LAMTOR5-AS1). This lncRNA enhances nuclear factor erythroid 2-related factor 2 (NRF2) levels by impeding NRF2 ubiquitination and degradation, although the transcriptional capacity of NRF2 is compromised. Subsequently, elevated NRF2 upregulates the downstream gene, heme oxygenase-1 (HO-1). Moreover, NRF2 autoregulates expression by inducing LAMTOR5-AS1 transcription. LAMTOR5-AS1 significantly inhibits OS cell proliferation73. The E3 ubiquitin ligase, TRIM21, is characterized by a tripartite motif, facilitates platelet-derived growth factor receptor beta (PDGFRβ) ubiquitination within the U2OS OS cell line, thereby regulating basal PDGFRβ levels. siRNA-mediated depletion of TRIM21 attenuates PDGF-BB-induced PDGFRβ ubiquitination, promoting OS cell growth74.
Studies have revealed high expression of A1BG antisense RNA 1 (A1BG-AS1) and USP22 in addition to low miR-148a-3p levels in OS tissues and cells. Downregulation of A1BG-AS1 and USP22 or upregulation of miR-148a-3p inhibits the malignant behavior of OS cells. A1BG-AS1 functions as a miR-148a-3p sponge, while miR-148a-3p targets USP22, thereby suppressing its expression. Conversely, USP22 upregulation reverses the phenotypic inhibition of OS cells induced by A1BG-AS1 inhibition. Mechanistically, USP22 influences OS cell biology by deubiquitinating silent mating type information regulation 2 homolog-1 (SIRT1)75. In OS patients with high 17β-estradiol (E2) levels, E2 activates mTORC1, promoting USP7 stability. USP7 interacts with NLRP3 and removes K48-linked ubiquitination, enhancing the NLRP3 inflammasome pathway and promoting proliferation of OS cells76.
Invasion
OS invasion is meticulously regulated by ubiquitination-mediated protein degradation, with several key factors influencing this process. Notably, several proteins that suppress invasion are targeted for degradation. Researchers observed a significant increase in myocyte enhancer factor 2D (MEF2D) protein ubiquitination in EPIC1-overexpressing OS cells. Co-transfection of pCDNA-EPIC1 and pCDNA-MEF2D rescued the cell viability and invasion inhibition induced by EPIC1 overexpression. These findings indicated that EPIC1 suppresses OS cell survival and invasion by promoting MEF2D ubiquitination77. TRAIP, a key differentially expressed gene with prognostic significance in OS, was identified. TRAIP promotes Kank1 polyubiquitination and subsequent degradation in OS cells, downregulates insulin-like growth factor binding protein 3 (IGFBP3), and activates the Akt pathway, thereby fostering invasion. These results highlight the critical role of the TRAIP/Kank1/IGFBP3/Akt signaling axis in OS progression78. Family with sequence similarity 83 member H (FAM83H) inhibits β-catenin ubiquitination in OS, thereby stabilizing β-catenin. Consequently, increased expression of cyclin D1, vimentin, and snail promotes cellular invasion79. Overexpressed histone deacetylase 4 (HDAC4) increases PCNA protein levels without affecting PCNA mRNA levels, while decreasing ubiquitinated PCNA. HDAC4 promotes invasion, while HDAC4 knockout exhibits the opposite effect. Direct binding between HDAC4 and PCNA was confirmed80. In OS tissues and cells, miR-140 and p21 expression is decreased, while USP22 and lysine-specific demethylase 1 (LSD1) expression is increased. Overexpressing miR-140 or knocking down USP22 induces LSD1 ubiquitination and degradation, inhibiting OS cell invasion81.
Migration
Researchers discovered that Kelch-like ECH-associated protein 1-259aa (KEAP1-259aa) interacts with the E3 ubiquitin ligase, Arih1, to bind cytoplasmic vimentin, promoting proteasomal degradation and inhibiting OS cell migration82. CircRNA_0002669 directly binds to c-Myc binding protein (MYCBP), a positive regulator of c-Myc, preventing MYCBP ubiquitination and proteasomal degradation. Consequently, circRNA_0002669 promotes OS cell migration by safeguarding MYCBP from protein degradation and disrupting the miR-889-3p-mediated pathway83. USP1 directly interacts with the transcription co-activator, TAZ, in OS cell lines. Mechanistic analysis revealed that USP1 inhibition of the anti-OS effect is partially attributed to TAZ instability, reduced nuclear accumulation, and subsequent downregulation of Hippo signaling pathway genes. Similarly, the pharmacologic USP1 inhibitor, ML323, exerts analogous effects on the Hippo signaling pathway, inhibiting OS migration in vitro and in vivo84. Chondroitin polymerizing factor (CHPF), a glycosyltransferase strongly expressed in OS tissues and cells, promotes OS migration by counteracting s-phase kinase-associated protein 2 (SKP2) ubiquitination and activating the Akt signaling pathway85. The novel E3 ubiquitin ligase, RNF180, promotes chromobox homolog 4 (Cbx4) ubiquitination, upregulates Kruppel-like factor 6 (KLF6), and downregulates runt-related transcription factor 2 (Runx2) in OS cells, inhibiting cell migration86.
Metastasis
OS cell-derived extracellular vesicles (EVs) activate pulmonary interstitial macrophages by secreting the chemokine, CXCL2, and initiating an influx of granulocyte-macrophage colony-stimulating factor (GM-CSF)-mobilized dendritic cells (GM-DCs). Proteomic analysis of EVs revealed that S100A11, packaged within EVs, stimulates the Janus kinase 2/signal transducer and activator of transcription 3 signaling pathway in macrophages through interaction with USP9X. High expression of S100A11 or circulating GM-DCs is associated with lung metastasis and poor prognosis in OS patients87. USP5 promotes OS metastasis by activating the Hedgehog (Hh) signaling pathway, as demonstrated in cultured cells and animal models. Mechanistically, USP5 stabilizes and deubiquitinates GLI family zinc finger protein 1 (GLI1), a key mediator of the Hh pathway. The oncogenic effects of USP5 in OS are contingent on GLI1 stability88. Alkbh5-mediated m6A deficiency in OS leads to increased USP22 and RNF40 expression, inhibiting histone H2A mono-ubiquitination and inducing key oncogenes, thereby driving uncontrolled cell cycle progression, continuous replication, and DNA repair. RNF40, ubiquitinated on H2B, inhibits H2A ubiquitination in cancer by interacting with the DDB1-CUL4-based ubiquitin E3 ligase complex, affecting DDB1-CUL4-based ubiquitin E3 ligase complex stability and promoting OS metastasis89. The E3 ubiquitin ligase, TRIM21, binds and targets GRP78 for ubiquitination and degradation, while cyclin-dependent kinase 7 (CDK7) phosphorylates G protein-coupled receptor 78 (GRP78) at T69, inhibiting TRIM21 recruitment and stabilizing GRP78. Notably, THZ1, a specific CDK7 inhibitor, suppresses OS growth and metastasis. Combined treatment with CDK7 and GRP78 inhibitors exhibits a synergistic effect on OS growth and progression90. MiR-137 targets LAPTM4B, a lysosomal-associated protein transmembrane 4B. LAPTM4B stabilizes RhoA protein by inhibiting ubiquitin-mediated proteasomal degradation, regulating stress fiber organization and promoting OS cell metastasis91.
Apoptosis
Researchers discovered that DNA polymerase epsilon 2 (POLE2) enhances CD44 expression by inhibiting MDM2-mediated ubiquitination, subsequently activating the RAC signaling pathway to promote OS cell apoptosis92. TRIM22 interacts with Nrf2, accelerating degradation through Nrf2 ubiquitination. This process is dependent on TRIM22 E3 ligase activity and independent of Kelch-like ECH-associated protein 1 (Keap1), ultimately activating the ROS/AMPK/mTOR/autophagy signaling axis and inducing apoptosis. Mechanistically, TRIM22 inhibits OS progression via Nrf2-mediated intracellular reactive oxygen species (ROS) imbalance. Overexpressed TRIM22 significantly enhances ROS production, while inhibiting mitochondrial potential, thereby activating the AMPK/mTOR signaling cascade93. The p27 Thr187Ala knock-in mutation disrupts SCFSkp2 E3 ligase-mediated p27 ubiquitination and degradation, leading to p27 Thr187Ala accumulation and promoting OS cell apoptosis94. As an oncogene in OS, TRIM46 interacts with peroxisome proliferator-activated receptor α (PPAR), ubiquitinates PPAR, activates the NF-κβ signaling pathway, and induces apoptosis95.
Application of ubiquitination in the treatment of OS
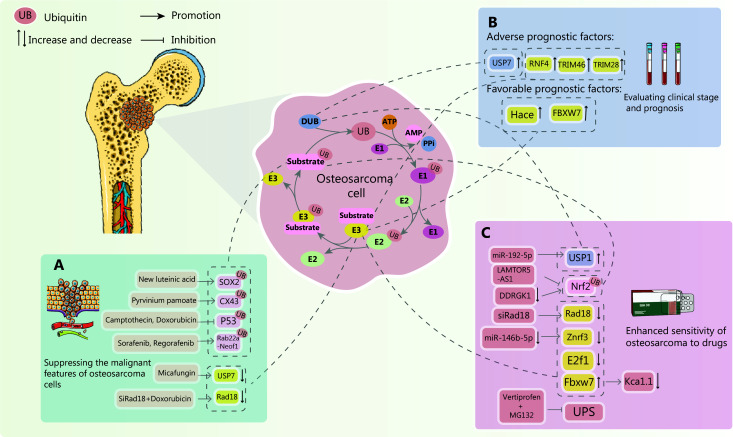
As our understanding of the intricate dance between ubiquitination and OS deepens, the potential for clinical diagnosis and treatment expands exponentially. Notably, tumor drug resistance remains a formidable foe in OS therapy, significantly impacting both treatment efficacy and patient outcomes. Overcoming this resistance, predicting OS progression with accuracy, and effectively curbing tumor growth and metastasis represent the holy grail in current treatment strategies. Exploiting ubiquitination pathways offers a glimmer of hope, a promising avenue to address these challenges and potentially revolutionize OS management (Figure 4).
Figure 4.
The potential of ubiquitination modulation in osteosarcoma treatment. In the context of osteosarcoma, targeting ubiquitination pathways has emerged as a promising therapeutic strategy. Recent research has yielded significant advancements in prognostic assessment, overcoming drug resistance, and inhibiting tumor development through ubiquitination-based interventions. (A) New Luteinic acid interacts with USP9X to directly target the ubiquitination and degradation of the key transcription factor, sex determining region Y box protein 2 (SOX2), significantly inhibiting osteosarcoma cell proliferation. Pyrvinium pamoate, a casein kinase 1 (CK1α) activator, promotes the ubiquitination and degradation of connexin 43 (CX43), hindering osteosarcoma metastasis. Sorafenib and remifentanil ubiquitinate and degrade RAB22A-NEOF1, blocking RAB22A-NEOF1-driven lung metastasis. Camptothecin or adriamycin treatment reduces p53 ubiquitination, stabilizing p53 protein levels, and inhibiting osteosarcoma cell proliferation. Micafungin exerts its anti-tumor effect on osteosarcoma by downregulating USP7. Adriamycin, encapsulated in nanoparticles and combined with siRNA targeting RAD18, significantly inhibits tumor growth and metastasis by reducing RAD18 expression. (B) Research has demonstrated that in osteosarcoma patients, elevated levels of ubiquitin ligases RNF4, TRIM46, TRIM28, and the deubiquitinase USP47 are associated with poor prognosis, whereas elevated levels of ubiquitin ligases HACE and FBXW7 indicate a favorable prognosis. (C) Mir-192-5p enhances the susceptibility of osteosarcoma cells to cisplatin by downregulating USP1 expression. Lamtor5-as1 increases chemosensitivity by inhibiting Nrf2 ubiquitination, leading to decreased Nrf2 transcriptional activity and disruption of the pro-survival loop in drug-resistant cells. Targeting DDRGK1 can reduce its expression and further promote Nrf2 ubiquitination, enhancing tumor drug sensitivity. RGD exosome-delivered SIRAD18 effectively silences the ubiquitin ligase RAD18, potentially enhancing anti-tumor drug activity. Inhibiting E2F1-mediated retinoic acid receptor α ubiquitination can improve drug efficacy in osteosarcoma treatment. Vertiprofen (VP), in combination with the proteasome inhibitor, MG132, simultaneously inhibits the ubiquitin-proteasome system (UPS) and enhances chemotherapy sensitivity by regulating autophagy and protein homeostasis. Inhibiting miR-146b-5p and downregulating the cell surface E3 ligase, ZNRF3, ultimately sensitizes osteosarcoma cells to chemotherapeutic drugs. The E3 ligase, FBXW7, downregulates the calcium-activated potassium channel, KCa1.1, overcoming osteosarcoma drug resistance.
Judging clinical staging and prognosis
RNF4 as a potential prognostic marker across various sarcomas, including OS. High levels of RNF4, but not the RNF4 partner, BMP6, correlate with significantly shorter survival times for patients96. Interestingly, elevated RNF4 alone predicts poorer outcomes, suggesting the independent prognostic value. Unlike RNF4, HACE1 appears to act as a tumor suppressor. Lower HACE1 expression is associated with both reduced survival rates and advanced disease stages97. Our research has shown significantly higher USP7 expression in advanced and metastatic OS compared to early-stage counterparts57. This positive correlation with tumor stage and metastasis strengthens the potential as a prognostic marker.
Fbxw7 serves as a guardian against OS. Lower expression of Fbxw7 in OS tissues is linked to worse outcomes, highlighting the potential as an independent prognostic marker98. TRIM46 leads to a contrasting outcome. An elevated expression of TRIM46 in OS suggests a pro-tumor role and patients with high TRIM46 levels exhibit significantly lower survival rates95. The PVT1/TRIM28 complex promotes tumorigenesis by enhancing the degradation of a tumor suppressor protein, ultimately activating mTOR signaling and promoting stem cell characteristics. Notably, high PVT1 expression is associated with a poor prognosis and decreased survival, underlining the potential of PVT1 as a therapeutic target99.
These diverse examples showcase the complex interplay of ubiquitination in regulating OS prognosis. Understanding these intricate mechanisms holds immense potential for developing novel prognostic tools and therapeutic strategies to improve patient outcomes.
Application in OS drug therapy
The notorious resistance of OS to chemotherapy poses a significant challenge in patient treatment. However, recent research offers promising avenues for overcoming this hurdle by targeting key pathways involved in drug resistance. DDRGK1, a protein stabilizing NRF2, emerges as a potential target. Disabling DDRGK1 destabilizes NRF2, unleashing a tide of ROS and enhancing sensitivity to drugs, such as doxorubicin and etoposide35. miR-192-5p, for example, bolsters OS cell susceptibility to cisplatin by targeting USP1 expression100. Rad18, implicated in adriamycin resistance, can be effectively silenced by chemically modified siRad18 loaded onto RGD exosomes, significantly enhancing the anti-tumor activity of adriamycin101.
LAMTOR5-AS1, an lncRNA, enhances chemotherapy sensitivity by boosting NRF2 activity and HO-1 expression, disrupting the pro-survival loop in resistant cells and leaving cells vulnerable to drug attack73. The calcium-activated potassium channel, KCa1.1, is linked to tumor progression and can be downregulated via Fbxw7, an E3 ligase, overcoming resistance to paclitaxel, adriamycin, and cisplatin102. Vertiprofen (VP), exhibiting dose-dependent cytotoxicity by inducing protein ubiquitination, can be combined with the proteasome inhibitor, MG132, to selectively target p53 for degradation, enhancing chemotherapy sensitivity by modulating autophagy and protein homeostasis103. miR-146b-5p, promoting tumor progression and chemoresistance, can be inhibited to upregulate MMP16 protein and downregulate Znrf3, a cell surface E3 ligase, ultimately sensitizing OS cells to chemotherapy drugs104. Inhibiting E2F1-mediated ubiquitination of retinoic acid receptorαcan amplify the efficacy of all-trans retinoic acid (ATRA) in OS treatment105.
Inhibition of malignant OS biological behavior
The aggressive growth and metastatic spread of OS are fueled by complex protein interactions, presenting promising targets for novel therapeutic interventions. New luteinic acid, a natural compound, interacts with USP9X, directly targeting the key transcription factor, Sox2, for ubiquitination and degradation, significantly inhibiting OS cell proliferation106. Pyrvinium pamoate, a CK1α activator, promotes CX43 phosphorylation at T437, triggering CX43 ubiquitination and degradation, thereby hindering OS metastasis107. Similarly, the kinase, PINK1, phosphorylates Rab22a-Neof1, marking it for ubiquitination and degradation by sorafenib and regorafenib, thus blocking Rab22a-Neof1-driven lung metastasis108. Proteins, like Gadd34, can act as “ubiquitination baits,” protecting other key proteins from degradation. Gadd34 upregulation following treatment with camptothecin or doxorubicin reduces p53 ubiquitination, thereby stabilizing p53 protein levels and inhibiting OS cell proliferation109.
Recent studies have demonstrated that micafungin effectively inhibits epithelial-mesenchymal transition (EMT) in OS cells by reducing the levels of ubiquitin-specific protease 7 (USP7), phospho-Akt, and phospho-GSK-3β. In vivo experiments using a xenograft tumor mouse model revealed significant tumor growth inhibition following daily intraperitoneal micafungin administration compared to the control group. These findings suggest that micafungin exerts its anti-tumor effects on OS by suppressing EMT via the USP7/Akt/GSK-3β pathway. USP7, a DUB, is a key oncogenic factor that stabilizes oncoproteins and inhibits tumor suppressors. Downregulation of USP7 by micafungin highlights USP7 potential as a therapeutic target. While preclinical studies have demonstrated promising anti-tumor effects of micafungin, rigorous human clinical trials are essential to evaluate safety and efficacy. Phase I trials will establish optimal dosage and identify potential adverse reactions, followed by phase II and III trials to assess therapeutic efficacy in OS patients. Given the complex nature of OS, combination therapies targeting USP7 in conjunction with chemotherapy or immunotherapy enhance anti-tumor responses and overcome drug resistance. Identifying predictive biomarkers of response to USP7 inhibitors is crucial for patient selection and treatment optimization110.
Another study demonstrated the potential of nanoparticle-based drug delivery for overcoming OS chemotherapy resistance. By encapsulating doxorubicin within nanoparticles and incorporating siRNA targeting Rad18, an E3 ubiquitin ligase implicated in doxorubicin resistance, the researchers achieved significant tumor growth inhibition and reduced metastasis. This approach also induced immunogenic cell death, suggesting a synergistic effect with immunotherapy. While these preclinical findings are encouraging, safety, efficacy, cost, and biomarker development must be addressed before clinical use. Rigorous clinical trials and innovative approaches to reduce production costs are essential to ensure widespread access to this promising therapy111.
Future research directions in clinical treatment
Ubiquitination holds significant potential for the treatment of OS. Addressing the challenges of drug specificity, resistance, and delivery is imperative for translating this potential into improved patient outcomes. Research focused on identifying and overcoming mechanisms of drug resistance is crucial for long-term therapeutic success. Additionally, identifying biomarkers predictive of patient response to ubiquitin-targeted therapy can personalize treatment strategies and enhance patient prognosis. The development of ubiquitination-related, OS-specific biomarkers is a promising future research direction. Common targets in OS treatment include VEGFR, MDM2, mTOR, and HDAC112. The development of specific inhibitors against MDM2 or other oncogenic E3 ligases can prevent p53 degradation and reactivate the tumor-suppressive function113. However, identifying highly selective inhibitors that avoid disrupting essential cellular processes remains a challenge. Conversely, drugs that activate specific DUBs can stabilize p53 and enhance anti-tumor effects in OS. Similar to E3 ligase inhibitors, achieving drug specificity remains an obstacle. The future holds promise for developing specific drugs targeting E3 ligases or DUBs that drive tumor growth or suppress its elimination. Additionally, combining these therapies with conventional approaches like chemotherapy could offer a multifaceted attack on the disease. Identifying patients most likely to benefit through ubiquitination-based biomarkers could further personalize treatment strategies. Targeted protein degradation (TPD) and other emerging technologies utilize engineered E3 ligase chimeras to target specific proteins for ubiquitination and degradation. This approach may eliminate oncogenic proteins lacking natural E3 ligase recognition motifs. While still in the preclinical stage, TPD is anticipated to target key proteins driving OS progression that are currently intractable to treatment114. Ensuring drug specificity to avoid unintended cellular disruptions, overcoming potential resistance mechanisms, and developing efficient delivery systems to tumor sites remain significant hurdles. Overall, by addressing these challenges, research on ubiquitination holds the key to unlocking novel and effective therapeutic strategies for OS.
Conclusions
The propensity of OS for distant metastases and resistance to targeted drugs remains a major obstacle, leading to poor patient outcomes. Deciphering the molecular mechanisms driving its development and progression is crucial for improving treatment efficacy and survival rates. Recent research has highlighted the pivotal role of ubiquitination, a post-translational protein modification, in this aggressive cancer. This review delves into the multifaceted impact of ubiquitination on OS cell biology and the potential for clinical application. Ubiquitin ligases and DUBs, the enzymes responsible for adding and removing ubiquitin tags, meticulously regulate key signaling pathways, such as Ras/PI3K/mTOR and NF-κβ. This intricate relationship impacts proteins within the WNT pathway, ultimately influencing cell proliferation, invasion, migration, and apoptosis. Furthermore, ubiquitination modulates the expression of genes, such as p53 and c-Myc, further shaping OS cell behavior.
The clinical spotlight shines brightly on the role of ubiquitination in OS resistance to chemotherapy. While targeted therapies offer high specificity and reduced side effects, drug resistance remains a formidable hurdle. Ubiquitination has a crucial role in regulating tumor drug sensitivity by influencing drug transport pathways, the apoptosis pathway, and target protein expression within the tumor microenvironment. Additionally, the levels of specific ubiquitin ligase and DUB expression hold promise as prognostic indicators for patients. Exciting recent advances in CRISPR/Cas9 library screening provide a powerful tool to identify genes essential for cancer cell survival, proliferation, migration, and drug resistance. This technology significantly bolsters research and development efforts aimed at overcoming resistance in OS targeted therapy, offering a beacon of hope for improved patient outcomes115.
Despite the promising potential of exploiting ubiquitination for targeted therapies, significant knowledge gaps remain that hinder progress. While downregulating TRIM7 shows promise in inhibiting metastasis and resensitizing tumors to chemotherapy, the exact role of abnormal m6A modifications in TRIM7 and the impact on OS progression and resistance are largely unexplored36. Similarly, the mechanisms by which TRIM22 suppresses tumor growth and metastasis in OS require further investigation93. Beyond TRIM7 and TRIM22, other intriguing targets exist. The identification of Bmi1, Ring1b, and USP22 as “cancer death” markers predicting poor prognosis highlights the potential of histone ubiquitination, but the intricate regulatory mechanisms in cancer remain unclear116. Additionally, the USP13/Mettl3 axis promoting OS progression through autophagy warrants further exploration, given the complex and multifaceted roles of autophagy in tumor biology117. Delving deeper into these knowledge gaps holds immense potential for unlocking novel therapeutic avenues. By elucidating the mechanisms underlying the observed effects of TRIM7, TRIM22, histone ubiquitination, and the USP13/Mettl3 axis, researchers can develop targeted therapies that effectively overcome drug resistance and improve clinical outcomes for OS patients. The rapid emergence of drug resistance during chemotherapy necessitates personalized treatment strategies. Tailoring future regimens based on individual patient bioindicators can minimize unnecessary drug exposure and maximize clinical benefit. In this context, leveraging ubiquitination modifications present an exciting avenue for enhancing the efficacy of existing targeted therapies while mitigating systemic toxicity.
While existing research has established that ubiquitination influences the malignant behavior of OS cells and is a critical factor in the disease, the precise identity and function of many ubiquitinated substrates remain unclear. Moreover, many studies have focused on a single OS subtype, disregarding disease heterogeneity. Consequently, the impact of ubiquitination across different cell subtypes remains largely unexplored. Furthermore, studies often rely solely on bioinformatics or basic in vitro experiments, hindering the comprehensive elucidation of the ubiquitin-mediated signaling network and its translation into clinical applications or drug development. Although progress has been made, most studies concentrate on a limited number of proteins, neglecting the broader ubiquitination landscape within OS cells. A comprehensive understanding of ubiquitination dynamics across different cellular compartments is still lacking. The complex interplay of ubiquitination with other post-translational modifications (e.g., methylation, glycosylation, and phosphorylation) complicates the isolation of its specific role in OS pathogenesis. Despite the identification of numerous ubiquitin-related proteins associated with OS, translating these findings into effective therapeutic targets remains a significant challenge. Additionally, the paucity of clinical data on ubiquitin-based therapies for OS patients hampers the development of treatment regimens. Addressing these knowledge gaps and pursuing focused research directions will be essential for advancing our understanding of OS and developing more effective treatment strategies.
Future studies should employ proteomic methods to identify novel ubiquitinated proteins in OS, providing fresh insights into disease mechanisms. However, research should extend beyond protein identification to encompass detailed investigations into the enzymatic activity, substrate specificity, and regulatory mechanisms of ubiquitin ligases and DUBs. Given the heterogeneity of OS and the complex intracellular environment, future research should encompass multiple cell subtypes and explore the interplay between ubiquitination and other post-translational modifications. By elucidating how these modifications synergistically regulate protein function in OS, groundbreaking discoveries may be achieved. In the clinical realm, developing specific inhibitors or activators of ubiquitin ligases or DUBs for OS treatment is essential. Furthermore, exploring the synergistic effects of targeting ubiquitination in combination with other therapies holds promise for improved patient outcomes. Ubiquitination represents a promising avenue for unraveling the complexities of OS, thereby enabling the development of novel therapeutic strategies and enhancing patient prognosis.
Acknowledgements
We thank The Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University, Southwest Medical University, Affiliated Hospital of Putian University, Affiliated Hospital of Guangdong Medical University, and Zhongda Hospital Affiliated to Southeast University for supporting this study.
Funding Statement
This work was funded by the Sichuan Provincial Central Leading Local Science and Technology Development Special Project (Grant No. 2023ZYD0072), the National Natural Science Foundation of China (Grant No. 82301785), and the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2019A1515111078).
Conflict of interest statement
No potential conflicts of interest are disclosed.
Author contributions
Conceived and designed the analysis: Huan Liu, Cheng Zhang, and Jianlin Shen.
Collected the data: Jianlin Shen, Yue Lai, Yanjiao Wu, and Xuan Lin.
Contributed data or analysis tools: Jianlin Shen, Yue Lai, and Xuan Lin.
Performed the analysis: Jianlin Shen and Yue Lai.
Wrote the paper: Jianlin Shen and Yue Lai.
References
- 1.Lv Y, Wu L, Jian H, Zhang C, Lou Y, Kang Y, et al. Identification and characterization of aging/senescence-induced genes in osteosarcoma and predicting clinical prognosis. Front Immunol. 2022;13:997765. doi: 10.3389/fimmu.2022.997765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nomura M, Rainusso N, Lee YC, Dawson B, Coarfa C, Han R, et al. Tegavivint and the β-Catenin/ALDH axis in chemotherapy-resistant and metastatic osteosarcoma. J Natl Cancer Inst. 2019;111:1216–27. doi: 10.1093/jnci/djz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. 2021;18:609–24. doi: 10.1038/s41571-021-00519-8. [DOI] [PubMed] [Google Scholar]
- 4.Chen C, Xie L, Ren T, Huang Y, Xu J, Guo W. Immunotherapy for osteosarcoma: fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. 2021;500:1–10. doi: 10.1016/j.canlet.2020.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Panez-Toro I, Muñoz-García J, Vargas-Franco JW, Renodon-Cornière A, Heymann MF, Lézot F, et al. Advances in osteosarcoma. Curr Osteoporos Rep. 2023;21:330–43. doi: 10.1007/s11914-023-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Zhang H, Liu J, Shang G. Targeted therapy for osteosarcoma: a review. J Cancer Res Clin Oncol. 2023;149:6785–97. doi: 10.1007/s00432-023-04614-4. [DOI] [PubMed] [Google Scholar]
- 7.Song J, Yuan X, Piao L, Wang J, Wang P, Zhuang M, et al. Cellular functions and molecular mechanisms of ubiquitination in osteosarcoma. Front Oncol. 2022;12:1072701. doi: 10.3389/fonc.2022.1072701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockram PE, Kist M, Prakash S, Chen SH, Wertz IE, Vucic D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021;28:591–605. doi: 10.1038/s41418-020-00708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampson C, Wang Q, Otkur W, Zhao H, Lu Y, Liu X, et al. The roles of E3 ubiquitin ligases in cancer progression and targeted therapy. Clin Transl Med. 2023;13:e1204. doi: 10.1002/ctm2.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang JT, Lee A, Kho C. Ubiquitin and ubiquitin-like proteins in cancer, neurodegenerative disorders, and heart diseases. Int J Mol Sci. 2022;23:5053. doi: 10.3390/ijms23095053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducker C, Shaw PE. Ubiquitin-mediated control of ETS transcription factors: roles in cancer and development. Int J Mol Sci. 2021;22:5119. doi: 10.3390/ijms22105119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewson G, Eichhorn PJA, Komander D. Deubiquitinases in cancer. Nat Rev Cancer. 2023;23:842–62. doi: 10.1038/s41568-023-00633-y. [DOI] [PubMed] [Google Scholar]
- 13.Dagar G, Kumar R, Yadav KK, Singh M, Pandita TK. Ubiquitination and deubiquitination: implications on cancer therapy. Biochim Biophys Acta Gene Regul Mech. 2023;1866:194979. doi: 10.1016/j.bbagrm.2023.194979. [DOI] [PubMed] [Google Scholar]
- 14.Meng Y, Sun H, Li Y, Zhao S, Su J, Zeng F, et al. Targeting ferroptosis by ubiquitin system enzymes: a potential therapeutic strategy in cancer. Int J Biol Sci. 2022;18:5475–88. doi: 10.7150/ijbs.73790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji Z, Shen J, Lan Y, Yi Q, Liu H. Targeting signaling pathways in osteosarcoma: mechanisms and clinical studies. MedComm (2020) 2023;4:e308. doi: 10.1002/mco2.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beird HC, Bielack SS, Flanagan AM, et al. Osteosarcoma. Nat Rev Dis Primers. 2022;8:77. doi: 10.1038/s41572-022-00409-y. [DOI] [PubMed] [Google Scholar]
- 17.Eaton BR, Schwarz R, Vatner R, Yeh B, Claude L, Indelicato DJ, et al. Osteosarcoma. Pediatr Blood Cancer. 2021;68:e28352. doi: 10.1002/pbc.28352. [DOI] [PubMed] [Google Scholar]
- 18.Pin F, Prideaux M, Bonewald LF, Bonetto A. Osteocytes and cancer. Curr Osteoporos Rep. 2021;19:616–25. doi: 10.1007/s11914-021-00712-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–35. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 20.Shanmugasundaram K, Nayak BK, Friedrichs WE, Kaushik D, Rodriguez R, Block K. NOX4 functions as a mitochondrial energetic sensor coupling cancer metabolic reprogramming to drug resistance. Nat Commun. 2017;8:997. doi: 10.1038/s41467-017-01106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salaroglio IC, Panada E, Moiso E, Buondonno I, Provero P, Rubinstein M, et al. PERK induces resistance to cell death elicited by endoplasmic reticulum stress and chemotherapy. Mol Cancer. 2017;16:91. doi: 10.1186/s12943-017-0657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwata S, Ishii T, Kawai A, Hiruma T, Yonemoto T, Kamoda H, et al. Prognostic factors in elderly osteosarcoma patients: a multi-institutional retrospective study of 86 cases. Ann Surg Oncol. 2014;21:263–8. doi: 10.1245/s10434-013-3210-4. [DOI] [PubMed] [Google Scholar]
- 23.Meltzer PS, Helman LJ. New horizons in the treatment of osteosarcoma. N Engl J Med. 2021;385:2066–76. doi: 10.1056/NEJMra2103423. [DOI] [PubMed] [Google Scholar]
- 24.Sheng G, Gao Y, Yang Y, Wu H. Osteosarcoma and metastasis. Front Oncol. 2021;11:780264. doi: 10.3389/fonc.2021.780264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang F, Nie L, Wei W. Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 2021;28:427–38. doi: 10.1038/s41418-020-00648-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han S, Wang R, Zhang Y, Li X, Gan Y, Gao F, et al. The role of ubiquitination and deubiquitination in tumor invasion and metastasis. Int J Biol Sci. 2022;18:2292–303. doi: 10.7150/ijbs.69411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barghout SH, Schimmer AD, Barker E. E1 Enzymes as therapeutic targets in cancer. Pharmacol Rev. 2020;73:1–56. doi: 10.1124/pharmrev.120.000053. [DOI] [PubMed] [Google Scholar]
- 28.Clague MJ, Heride C, Urbé S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25:417–26. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17:626–42. doi: 10.1038/nrm.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–57. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 31.Dikic I, Schulman BA. An expanded lexicon for the ubiquitin code. Nat Rev Mol Cell Biol. 2023;24:273–87. doi: 10.1038/s41580-022-00543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Bu F, Zhang W. The role of ubiquitination in regulating embryonic stem cell maintenance and cancer development. Int J Mol Sci. 2019;20:2667. doi: 10.3390/ijms20112667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antao AM, Tyagi A, Kim KS, Ramakrishna S. Advances in deubiquitinating enzyme inhibition and applications in cancer therapeutics. Cancers. 2020;12:1579. doi: 10.3390/cancers12061579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mennerich D, Kubaichuk K, Kietzmann T. DUBs, hypoxia, and cancer. Trends Cancer. 2019;5:632–53. doi: 10.1016/j.trecan.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Zhou T, Yang X, Cao X, Jin G, Zhang P, et al. DDRGK1 enhances osteosarcoma chemoresistance via inhibiting KEAP1-mediated NRF2 ubiquitination. Adv Sci (Weinh) 2023;10:e2204438. doi: 10.1002/advs.202204438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou C, Zhang Z, Zhu X, Qian G, Zhou Y, Sun Y, et al. N6-Methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. EBioMedicine. 2020;59:102955. doi: 10.1016/j.ebiom.2020.102955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomiyoshi G, Nakamura R, Shinmen N, Yoshida Y, Mine S, Machida T, et al. GADD34 activates p53 and may have utility as a marker of atherosclerosis. Front Med (Lausanne) 2023;10:1128921. doi: 10.3389/fmed.2023.1128921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Zhang C, Xu D, Chang CY, Wang J, Liu J, et al. The ubiquitin ligase TRIM21 regulates mutant p53 accumulation and gain of function in cancer. J Clin Invest. 2023;133:e164354. doi: 10.1172/JCI164354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge Y, Zhao R, Li B, Xiao B, Zhou L, Zuo S. Aerobic glycolysis and tumor progression of hepatocellular carcinoma are mediated by ubiquitin of P53 K48-linked regulated by TRIM37. Exp Cell Res. 2022;421:113377. doi: 10.1016/j.yexcr.2022.113377. [DOI] [PubMed] [Google Scholar]
- 40.Jiang X, Ma Y, Wang T, Zhou H, Wang K, Shi W, et al. Targeting UBE2T potentiates gemcitabine efficacy in pancreatic cancer by regulating pyrimidine metabolism and replication stress. Gastroenterology. 2023;164:1232–47. doi: 10.1053/j.gastro.2023.02.025. [DOI] [PubMed] [Google Scholar]
- 41.Fang D, Hu H, Zhao K, Xu A, Yu C, Zhu Y, et al. MLF2 negatively regulates P53 and promotes colorectal carcinogenesis. Adv Sci (Weinh) 2023;10:e2303336. doi: 10.1002/advs.202303336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Luo F, Luo S, Li L, Ren X, Lin J, et al. Transcriptional repression of aerobic glycolysis by OVOL2 in breast cancer. Adv Sci (Weinh) 2022;9:e2200705. doi: 10.1002/advs.202200705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarri N, Wang K, Tsioumpekou M, Castillejo-López C, Lennartsson J, Heldin CH, et al. Deubiquitinating enzymes USP4 and USP17 finetune the trafficking of PDGFRβ and affect PDGF-BB-induced STAT3 signalling. Cell Mol Life Sci. 2022;79:85. doi: 10.1007/s00018-022-04128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He T, Cui J, Wu Y, Sun X, Chen N. Knockdown of TRIM66 inhibits cell proliferation, migration and invasion in colorectal cancer through JAK2/STAT3 pathway. Life Sci. 2019;235:116799. doi: 10.1016/j.lfs.2019.116799. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Lei J, Zhang S, Wang X, Jin J, Liu Y, et al. 4EBP1 senses extracellular glucose deprivation and initiates cell death signaling in lung cancer. Cell Death Dis. 2022;13:1075. doi: 10.1038/s41419-022-05466-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao C, Zhang W, Hua M, Chen H, Yang B, Wang Y, et al. RNF7 inhibits apoptosis and sunitinib sensitivity and promotes glycolysis in renal cell carcinoma via the SOCS1/JAK/STAT3 feedback loop. Cell Mol Biol Lett. 2022;27:36. doi: 10.1186/s11658-022-00337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang N, Sun X, Li P, Liu X, Zhang X, Chen Q, et al. TRIM family contribute to tumorigenesis, cancer development, and drug resistance. Exp Hematol Oncol. 2022;11:75. doi: 10.1186/s40164-022-00322-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li A, Wang S, Nie J, Xiao S, Xie X, Zhang Y, et al. USP3 promotes osteosarcoma progression via deubiquitinating EPHA2 and activating the PI3K/AKT signaling pathway. Cell Death Dis. 2024;15:235. doi: 10.1038/s41419-024-06624-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang L, Ford-Roshon D, Russo M, O’Brien C, Xiong X, Gurjao C, et al. RNF43 G659fs is an oncogenic colorectal cancer mutation and sensitizes tumor cells to PI3K/mTOR inhibition. Nat Commun. 2022;13:3181. doi: 10.1038/s41467-022-30794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He Y, Jiang S, Mao C, Zheng H, Cao B, Zhang Z, et al. The deubiquitinase USP10 restores PTEN activity and inhibits non-small cell lung cancer cell proliferation. J Biol Chem. 2021;297:101088. doi: 10.1016/j.jbc.2021.101088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu C, Ning Z, Wang A, Chen D, Liu X, Xia T, et al. USP10 suppresses tumor progression by inhibiting mTOR activation in hepatocellular carcinoma. Cancer Lett. 2018;436:139–48. doi: 10.1016/j.canlet.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 52.Sun Z, Jiang Q, Gao B, Zhang X, Bu L, Wang L, et al. AKT blocks SIK1-mediated repression of STAT3 to promote breast tumorigenesis. Cancer Res. 2023;83:1264–79. doi: 10.1158/0008-5472.CAN-22-3407. [DOI] [PubMed] [Google Scholar]
- 53.Li B, Zhao R, Qiu W, Pan Z, Zhao S, Qi Y, et al. The N6-methyladenosine-mediated lncRNA WEE2-AS1 promotes glioblastoma progression by stabilizing RPN2. Theranostics. 2022;12:6363–79. doi: 10.7150/thno.74600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang C, Chen B, Jiang K, Lao L, Shen H, Chen Z. Activation of TNF-α/NF-κB axis enhances CRL4BDCAF11 E3 ligase activity and regulates cell cycle progression in human osteosarcoma cells. Mol Oncol. 2018;12:476–94. doi: 10.1002/1878-0261.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pang K, Lee J, Kim J, Park J, Park Y, Hong E, et al. Degradation of DRAK1 by CUL3/SPOP E3 Ubiquitin ligase promotes tumor growth of paclitaxel-resistant cervical cancer cells. Cell Death Dis. 2022;13:169. doi: 10.1038/s41419-022-04619-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, Qi ZP, He DL, Chen ZH, Liu JY, Wong MW, et al. NLRP7 deubiquitination by USP10 promotes tumor progression and tumor-associated macrophage polarization in colorectal cancer. J Exp Clin Cancer Res. 2021;40:126. doi: 10.1186/s13046-021-01920-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng Q, Li Z, Zhao X, Guo L, Yu C, Qin J, et al. Ubiquitin-specific protease 7 promotes osteosarcoma cell metastasis by inducing epithelial-mesenchymal transition. Oncol Rep. 2019;41:543–51. doi: 10.3892/or.2018.6835. [DOI] [PubMed] [Google Scholar]
- 58.Marei H, Tsai WTK, Kee YS, Ruiz K, He J, Cox C, et al. Antibody targeting of E3 ubiquitin ligases for receptor degradation. Nature. 2022;610:182–9. doi: 10.1038/s41586-022-05235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu L, Tian Q, Gao H, Wu K, Wang B, Ge G, et al. PROX1 promotes breast cancer invasion and metastasis through WNT/β-catenin pathway via interacting with hnRNPK. Int J Biol Sci. 2022;18:2032–46. doi: 10.7150/ijbs.68960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tung CH, Wu JE, Huang MF, Wang WL, Wu YY, Tsai YT, et al. Ubiquitin-specific peptidase 5 facilitates cancer stem cell-like properties in lung cancer by deubiquitinating β-catenin. Cancer Cell Int. 2023;23:207. doi: 10.1186/s12935-023-03059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dai Y, Liu J, Li X, Deng J, Zeng C, Lu W, et al. Let-7b-5p inhibits colon cancer progression by prohibiting APC ubiquitination degradation and the Wnt pathway by targeting NKD1. Cancer Sci. 2023;114:1882–97. doi: 10.1111/cas.15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li K, Peng ZY, Wang R, Li X, Du N, Liu DP, et al. Enhancement of TKI sensitivity in lung adenocarcinoma through m6A-dependent translational repression of Wnt signaling by circ-FBXW7. Mol Cancer. 2023;22:103. doi: 10.1186/s12943-023-01811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Liu J, Zheng S, Cao L, Li Y, Sheng R, et al. The deubiquitinase USP10 mediates crosstalk between the LKB1/AMPK axis and Wnt/β-catenin signaling in cancer. FEBS Lett. 2023;597:3061–71. doi: 10.1002/1873-3468.14763. [DOI] [PubMed] [Google Scholar]
- 64.Liu J, Jin J, Liang T, Feng XH. To Ub or not to Ub: a regulatory question in TGF-β signaling. Trends Biochem Sci. 2022;47:1059–72. doi: 10.1016/j.tibs.2022.06.001. [DOI] [PubMed] [Google Scholar]
- 65.Xie F, Zhou X, Li H, Su P, Liu S, Li R, et al. USP8 promotes cancer progression and extracellular vesicle-mediated CD8+ T cell exhaustion by deubiquitinating the TGF-β receptor TβRII. EMBO J. 2022;41:e108791. doi: 10.15252/embj.2021108791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, van Dinther M, Thorikay M, Gourabi BM, Kruithof BPT, Ten Dijke P. Opposing USP19 splice variants in TGF-β signaling and TGF-β-induced epithelial-mesenchymal transition of breast cancer cells. Cell Mol Life Sci. 2023;80:43. doi: 10.1007/s00018-022-04672-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, Xu J, Zhou S, Yao F, Zhang R, You W, et al. Endothelial DGKG promotes tumor angiogenesis and immune evasion in hepatocellular carcinoma. J Hepatol. 2024;80:82–98. doi: 10.1016/j.jhep.2023.10.006. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Li QS, Liu HL, Tang HT, Yang HL, Wu DQ, et al. MKRN1 promotes colorectal cancer metastasis by activating the TGF-β signalling pathway through SNIP1 protein degradation. J Exp Clin Cancer Res. 2023;42:219. doi: 10.1186/s13046-023-02788-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang Y, Liu S, Shan M, Hagenaars SC, Mesker WE, Cohen D, et al. RNF12 is regulated by AKT phosphorylation and promotes TGF-β driven breast cancer metastasis. Cell Death Dis. 2022;13:44. doi: 10.1038/s41419-021-04493-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu X, Xu J, Shen B, Xu J, Jiang J. USP33 promotes pancreatic cancer malignant phenotype through the regulation of TGFBR2/TGFβ signaling pathway. Cell Death Dis. 2023;14:362. doi: 10.1038/s41419-023-05871-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tu Y, Xu L, Xu J, Bao Z, Tian W, Ye Y, et al. Loss of deubiquitylase USP2 triggers development of glioblastoma via TGF-β signaling. Oncogene. 2022;41:2597–608. doi: 10.1038/s41388-022-02275-0. [DOI] [PubMed] [Google Scholar]
- 72.Zhong L, Wang J, Chen W, Lv D, Zhang R, Wang X, et al. Augmenting L3MBTL2-induced condensates suppresses tumor growth in osteosarcoma. Sci Adv. 2023;9:eadi0889. doi: 10.1126/sciadv.adi0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pu Y, Tan Y, Zang C, Zhao F, Cai C, Kong L, et al. LAMTOR5-AS1 regulates chemotherapy-induced oxidative stress by controlling the expression level and transcriptional activity of NRF2 in osteosarcoma cells. Cell Death Dis. 2021;12:1125. doi: 10.1038/s41419-021-04413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sarri N, Papadopoulos N, Lennartsson J, Heldin CH. The E3 ubiquitin ligase TRIM21 regulates basal levels of PDGFRβ. Int J Mol Sci. 2023;24:7782. doi: 10.3390/ijms24097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han X, Yin M, Gong C, Zhang C, Zhu G, Hu M, et al. A1BG-AS1 promotes the biological functions of osteosarcoma cells via regulating the microRNA-148a-3p/USP22 axis and stabilizing the expression of SIRT1 through deubiquitinase function. Expert Opin Ther Targets. 2023;27:1017–29. doi: 10.1080/14728222.2023.2263908. [DOI] [PubMed] [Google Scholar]
- 76.Yang Z, Yu W, Xu A, Liu B, Jin L, Tao H, et al. mTORC1 accelerates osteosarcoma progression via m6A-dependent stabilization of USP7 mRNA. Cell Death Discov. 2024;10:127. doi: 10.1038/s41420-024-01893-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao W, Zhang D, Qin P, Zhang J, Cui X, Gao J, et al. Long non-coding RNA EPIC1 inhibits viability and invasion of osteosarcoma cells by promoting MEF2D ubiquitylation. Int J Biol Macromol. 2019;128:566–73. doi: 10.1016/j.ijbiomac.2019.01.156. [DOI] [PubMed] [Google Scholar]
- 78.Li M, Wu W, Deng S, Shao Z, Jin X. TRAIP modulates the IGFBP3/AKT pathway to enhance the invasion and proliferation of osteosarcoma by promoting KANK1 degradation. Cell Death Dis. 2021;12:767. doi: 10.1038/s41419-021-04057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim KM, Hussein UK, Park SH, Kang MA, Moon YJ, Zhang Z, et al. FAM83H is involved in stabilization of β-catenin and progression of osteosarcomas. J Exp Clin Cancer Res. 2019;38:267. doi: 10.1186/s13046-019-1274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao K, Wang H, Fang Y, Wang Y, Wei L, Chen X, et al. Histone deacetylase 4 promotes osteosarcoma cell proliferation and invasion by regulating expression of proliferating cell nuclear antigen. Front Oncol. 2019;9:870. doi: 10.3389/fonc.2019.00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu W, Wang D, Liu L, Wang L, Yan M. miR-140 inhibits osteosarcoma progression by impairing USP22-mediated LSD1 stabilization and promoting p21 expression. Mol Ther Nucleic Acids. 2021;24:436–48. doi: 10.1016/j.omtn.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Liu Z, Zhong Z, Ji Y, Guo H, Wang W, et al. A tumor suppressor protein encoded by circKEAP1 inhibits osteosarcoma cell stemness and metastasis by promoting vimentin proteasome degradation and activating anti-tumor immunity. J Exp Clin Cancer Res. 2024;43:52. doi: 10.1186/s13046-024-02971-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Zhan Y, Liu Z, Guo H, Liu D, Chen C. Circ_0002669 promotes osteosarcoma tumorigenesis through directly binding to MYCBP and sponging miR-889-3p. Biol Direct. 2024;19:25. doi: 10.1186/s13062-024-00466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yuan P, Feng Z, Huang H, Wang G, Chen Z, Xu G, et al. USP1 inhibition suppresses the progression of osteosarcoma via destabilizing TAZ. Int J Biol Sci. 2022;18:3122–36. doi: 10.7150/ijbs.65428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen Y, Li J, Peng D, Liao L, Chen X, Zhong W, et al. Chondroitin polymerizing factor (CHPF) promotes cell proliferation and tumor growth in human osteosarcoma by inhibiting SKP2’s ubiquitination while activating the AKT pathway. Genes Dis. 2023;10:2125–36. doi: 10.1016/j.gendis.2022.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Q, Liu N, Xu T, Song K. RING finger gene 180 inhibits osteosarcoma progression through regulating chromobox homolog 4 ubiquitination. Cell Cycle. 2023;22:1246–58. doi: 10.1080/15384101.2023.2205201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deng C, Xu Y, Chen H, Zhu X, Huang L, Chen Z, et al. Extracellular-vesicle-packaged S100A11 from osteosarcoma cells mediates lung premetastatic niche formation by recruiting gMDSCs. Cell Rep. 2024;43:113751. doi: 10.1016/j.celrep.2024.113751. [DOI] [PubMed] [Google Scholar]
- 88.Wu Q, Liu R, Yang Y, Peng J, Huang J, Li Z, et al. USP5 promotes tumorigenesis by activating Hedgehog/Gli1 signaling pathway in osteosarcoma. Am J Cancer Res. 2024;14:1204–16. doi: 10.62347/JMFF8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yadav P, Subbarayalu P, Medina D, Nirzhor S, Timilsina S, Rajamanickam S, et al. M6A RNA methylation regulates histone ubiquitination to support cancer growth and progression. Cancer Res. 2022;82:1872–89. doi: 10.1158/0008-5472.CAN-21-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang T, Li J, Yang M, Ma X, Wang Z, Ma X, et al. CDK7/GRP78 signaling axis contributes to tumor growth and metastasis in osteosarcoma. Oncogene. 2022;41:4524–36. doi: 10.1038/s41388-022-02446-z. [DOI] [PubMed] [Google Scholar]
- 91.Yan R, Liu D, Wang J, Liu M, Guo H, Bai J, et al. miR-137-LAPTM4B regulates cytoskeleton organization and cancer metastasis via the RhoA-LIMK-Cofilin pathway in osteosarcoma. Oncogenesis. 2023;12:25. doi: 10.1038/s41389-023-00471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang B, Hu H, Wang X, Shao Z, Shi D, Wu F, et al. POLE2 promotes osteosarcoma progression by enhancing the stability of CD44. Cell Death Discov. 2024;10:177. doi: 10.1038/s41420-024-01875-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu W, Zhao Y, Wang G, Feng S, Ge X, Ye W, et al. TRIM22 inhibits osteosarcoma progression through destabilizing NRF2 and thus activation of ROS/AMPK/mTOR/autophagy signaling. Redox Biol. 2022;53:102344. doi: 10.1016/j.redox.2022.102344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang J, Aldahamsheh O, Ferrena A, Borjihan H, Singla A, Yaguare S, et al. The interaction of SKP2 with p27 enhances the progression and stemness of osteosarcoma. Ann N Y Acad Sci. 2021;1490:90–104. doi: 10.1111/nyas.14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang W, Cai X, Xu T, Liu K, Yang D, Fan L, et al. Tripartite motif-containing 46 promotes viability and inhibits apoptosis of osteosarcoma cells by activating NF-B signaling through ubiquitination of PPAR. Oncol Res. 2020;28:409–21. doi: 10.3727/096504020X15868639303417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Novak R, Ahmad YA, Timaner M, Bitman-Lotan E, Oknin-Vaisman A, Horwitz R, et al. RNF4~RGMb~BMP6 axis required for osteogenic differentiation and cancer cell survival. Cell Death Dis. 2022;13:820. doi: 10.1038/s41419-022-05262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.El-Naggar AM, Clarkson PW, Negri GL, Turgu B, Zhang F, Anglesio MS, et al. HACE1 is a potential tumor suppressor in osteosarcoma. Cell Death Dis. 2019;10:21. doi: 10.1038/s41419-018-1276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Z, Xiao J, Hu K, Wang G, Li M, Zhang J, et al. FBXW7 acts as an independent prognostic marker and inhibits tumor growth in human osteosarcoma. Int J Mol Sci. 2015;16:2294–306. doi: 10.3390/ijms16022294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsang SV, Rainusso N, Liu M, Nomura M, Patel TD, Nakahata K, et al. LncRNA PVT-1 promotes osteosarcoma cancer stem-like properties through direct interaction with TRIM28 and TSC2 ubiquitination. Oncogene. 2022;41:5373–84. doi: 10.1038/s41388-022-02538-w. [DOI] [PubMed] [Google Scholar]
- 100.Zhou S, Xiong M, Dai G, Yu L, Zhang Z, Chen J, et al. MicroRNA-192-5p suppresses the initiation and progression of osteosarcoma by targeting USP1. Oncol Lett. 2018;15:6947–56. doi: 10.3892/ol.2018.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Du M, Gu J, Liu C, Liu N, Yu Z, Zhou C, et al. Genome-wide CRISPR screen identified Rad18 as a determinant of doxorubicin sensitivity in osteosarcoma. J Exp Clin Cancer Res. 2022;41:154. doi: 10.1186/s13046-022-02344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ohya S, Kajikuri J, Endo K, Kito H, Elboray EE, Suzuki T, et al. Ca2+-activated K+ channel KCa1.1 as a therapeutic target to overcome chemoresistance in three-dimensional sarcoma spheroid models. Cancer Sci. 2021;112:3769–83. doi: 10.1111/cas.15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saini H, Sharma H, Mukherjee S, Chowdhury S, Chowdhury R. Verteporfin disrupts multiple steps of autophagy and regulates p53 to sensitize osteosarcoma cells. Cancer Cell Int. 2021;21:52. doi: 10.1186/s12935-020-01720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu E, Zhao J, Ma J, Wang C, Zhang C, Jiang H, et al. miR-146b-5p promotes invasion and metastasis contributing to chemoresistance in osteosarcoma by targeting zinc and ring finger 3. Oncol Rep. 2016;35:275–83. doi: 10.3892/or.2015.4393. [DOI] [PubMed] [Google Scholar]
- 105.Zhang L, Zhou Q, Zhang N, Li W, Ying M, Ding W, et al. E2F1 impairs all-trans retinoic acid-induced osteogenic differentiation of osteosarcoma via promoting ubiquitination-mediated degradation of RARα. Cell Cycle. 2014;13:1277–87. doi: 10.4161/cc.28190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen X, Zhang X, Cai H, Yang W, Lei H, Xu H, et al. Targeting USP9x/SOX2 axis contributes to the anti-osteosarcoma effect of neogambogic acid. Cancer Lett. 2020;469:277–86. doi: 10.1016/j.canlet.2019.10.015. [DOI] [PubMed] [Google Scholar]
- 107.Wang X, Qin G, Liang X, Wang W, Wang Z, Liao D, et al. Targeting the CK1α/CBX4 axis for metastasis in osteosarcoma. Nat Commun. 2020;11:1141. doi: 10.1038/s41467-020-14870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zeng C, Zhong L, Liu W, Zhang Y, Yu X, Wang X, et al. Targeting the lysosomal degradation of Rab22a-NeoF1 fusion protein for osteosarcoma lung metastasis. Adv Sci (Weinh) 2022;10:e2205483. doi: 10.1002/advs.202205483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tomiyoshi G, Nakamura R, Shinmen N, Yoshida Y, Mine S, Machida T, et al. GADD34 activates p53 and may have utility as a marker of atherosclerosis. Front Med (Lausanne) 2023;10:1128921. doi: 10.3389/fmed.2023.1128921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang QL, Wang L, Li QY, Li HY, Lin L, Wei D, et al. Micafungin exerts antitumor effect on breast cancer and osteosarcoma through preventing EMT in tumor cells in an USP7/AKT/GSK-3β pathway-dependent manner. Naunyn Schmiedebergs Arch Pharmacol. 2024;397:4447–59. doi: 10.1007/s00210-023-02903-w. [DOI] [PubMed] [Google Scholar]
- 111.Cao J, Zhu C, Cao Z, Ke X. CPPs-modified chitosan as permeability-enhancing chemotherapeutic combined with gene therapy nanosystem by thermosensitive hydrogel for the treatment of osteosarcoma. Int J Biol Macromol. 2024;267(Pt 2):130915. doi: 10.1016/j.ijbiomac.2024.130915. [DOI] [PubMed] [Google Scholar]
- 112.Hu Z, Wen S, Huo Z, Wang Q, Zhao J, Wang Z, et al. Current status and prospects of targeted therapy for osteosarcoma. Cells. 2022;11:3507. doi: 10.3390/cells11213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nirala BK, Yamamichi T, Yustein JT. Deciphering the signaling mechanisms of osteosarcoma tumorigenesis. Int J Mol Sci. 2023;24:11367. doi: 10.3390/ijms241411367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wells JA, Kumru K. Extracellular targeted protein degradation: an emerging modality for drug discovery. Nat Rev Drug Discov. 2024;23:126–40. doi: 10.1038/s41573-023-00833-z. [DOI] [PubMed] [Google Scholar]
- 115.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–21. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang C, Meng Y, Zhao J, Ma J, Zhao Y, Gao R, et al. Deubiquitinase USP13 regulates glycolytic reprogramming and progression in osteosarcoma by stabilizing METTL3/m6A/ATG5 axis. Int J Biol Sci. 2023;19:2289–303. doi: 10.7150/ijbs.82081. [DOI] [PMC free article] [PubMed] [Google Scholar]