Abstract
We have generated transgenic mice carrying wild-type and mutant forms of the apolipoprotein (apo)A-I/apoCIII gene cluster. Mutations were introduced either in one or in three SP1 binding sites of the apoCIII enhancer. In mice carrying the wild-type transgene, major sites of apoA-I mRNA synthesis were liver and intestine and minor sites were kidney and, to a lesser extent, other tissues. The major site of chloramphenicol acetyl transferase (CAT) activity (used as a reporter for the apoCIII gene) was liver and minor sites intestine and kidney. A mutation in one SP1 binding site reduced the expression of the apoA-I gene to ~23 and 19% in the liver and intestine, respectively, as compared to the control wild-type. The hepatic expression of the CAT gene was not affected whereas the intestinal expression was nearly abolished. Mutations in three SP1 binding sites reduced the hepatic and intestinal expression of the apoA-I and CAT genes to 14 and 4%, respectively, as compared to the wild-type control, and abolished CAT expression in all tissues. The findings suggest that the SP1 sites of the apoCIII enhancer are required for the expression of the apoCIII gene and also contribute significantly to the hepatic and intestinal expression of the apoA-I gene in vivo.
INTRODUCTION
Previous studies using deletion, nucleotide substitution and footprinting analysis of the apolipoprotein (apo)A-I promoter, as well as chloramphenicol acetyl transferase (CAT) assays, localized the promoter elements required for hepatic transcription of the apoA-I gene in cell cultures downstream of nucleotide –255. This region was insufficient to drive intestinal transcription in cell cultures (1).
Similar analysis of the apoCIII promoter in cell cultures localized the promoter elements required for hepatic and intestinal transcription downstream of nucleotide –890 (2,3). This region contains four proximal and six distal regulatory elements designated A to J. The regulatory element B contains a hormone response element (HRE) which is recognized by various members of the orphan- and ligand-dependent nuclear receptor family (4–6) and elements C and D bind the CCAAT enhancer box binding protein (C/EBP). The distal regulatory region contains two HREs on elements G and I4 as well as three binding sites for the ubiquitous transcription factor SP1 (7) on elements F, H and I. Only the HRE of element I4 is recognized by hepatocyte nuclear factor-4 (HNF-4) (4,7).
Initial in vitro (1,6–9) and subsequent in vivo experiments (10–13) showed that the distal regulatory region –590/–800 of the apoCIII promoter is a common enhancer of the apoA-I/apoCIII/apoA-IV gene cluster. The enhancer could increase the activity of proximal apoCIII promoter segments in HepG2 and CaCo-2 cells 7–10-fold (2). The enhancer could also increase the activity of the proximal (–255/–5) apoA-I promoter in HepG2 cells 5–13-fold and the activity of the –1500/–5 apoA-I promoter in CaCo-2 cells in the presence of HNF-4 7-fold (1).
Previous mutagenesis studies have established that mutations in SP1 sites of the apoCIII enhancer diminished the activity of the apoCIII promoter and enhancer cluster in cells of hepatic (HepG2) or intestinal (CaCo-2) origin (7). The most severe inactivation of the apoCIII promoter/enhancer cluster was caused by mutation in the SP1 binding site on element H or by a deletion which eliminated two of the SP1 sites on elements H and I (7). Similarly, the activity of the apoA-I promoter/apoCIII enhancer cluster was reduced by 30–60% in HepG2 cell cultures by point mutations in the SP1 binding sites (1). These findings indicate the importance of the SP1 binding sites of the apoCIII enhancer for the overall activity of the apoA-I promoter/enhancer cluster.
To validate that the in vitro data on apoA-I and apoCIII gene regulation we have generated transgenic mice containing the wild-type (WT) and mutant regulatory regions of the apoA-I/apoCIII gene cluster. Mutations were designed to destroy either one or all three of the SP1 binding sites of the apoCIII enhancer. The transgenic mouse lines were used to decipher the role of SP1, which binds to the apoCIII enhancer, on the transcriptional regulation of the apoCIII and the closely linked apoA-I gene in vivo.
Our findings suggest that the SP1 binding sites of the apoCIII enhancer are essential for the overall expression of the apoCIII gene. The SP1 binding sites are also crucial for intestinal expression and contribute greatly to the hepatic expression of the apoA-I gene. It appears that multiple SP1 molecules that are bound to the apoCIII enhancer synergize with HNF-4 bound to this enhancer, as well as to the proximal promoters, to activate the hepatic and intestinal transcription of the two linked genes.
MATERIALS AND METHODS
Materials
Reagents were purchased from the following sources. The Klenow fragment of the DNA polymerase I restriction enzymes, T4 ligase, T4 polynucleotide kinase and Vent polymerase from New England Biolabs (Beverly, MA). [γ-32P]ATP (5000 Ci/mmol), 14C chloramphenicol, [α-32P]dCTP and [α-32P]dGTP (3000 Ci/mmol) from New England Nuclear (Boston, MA). Bactotryptone and bacto-yeast extracts from Difco (Detroit, MI). Acrylamide, sodium dodecyl sulfate (SDS), urea and Tris from International Biotechnologies, Inc. (Rochester, NY). Bacterial XL-1 Blue cells from Stratagene (La Jolla, CA). Custom-made oligonucleotides and plasmid pSPORT-1 from Gibco BRL (Rockville, MD). S1 nuclease from Boehringer Mannheim (Indianapolis, IN). Proteinase K, spermine, phenylmethylsulfonyl fluoride (PMSF) and ammonium acetate from Sigma (St Louis, MO).
Methods
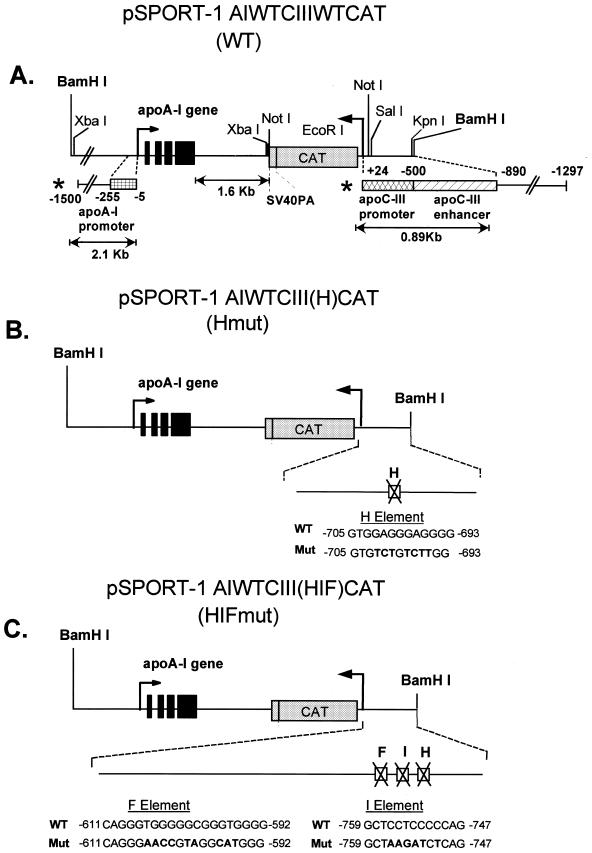
Plasmid constructions. To generate the constructs containing the apoA-I/apoCIII gene cluster and to introduce mutations in the SP1 sites of the enhancer, initially the human –890/+24 apoCIII promoter was amplified using the pUC-19 CIII promoter plasmid described previously (14) as a template and the CIII 890K and CIII 24E oligonucleotides (Table 1) as primers. The primers introduced KpnI and EcoR1 sites at the 5′ and 3′ ends of the –890/+24 apoCIII promoter/enhancer region. Following digestion with KpnI and EcoRI, the amplified fragment was cloned into the corresponding sites of the pSPORT-1 plasmid (Gibco BRL). The EcoRI site of this plasmid was subsequently blunted by EcoRI digestion and filling of the 5′ protruding end using the Klenow fragment of the DNA polymerase I to generate the modified pSPORT-1 CIII WT plasmid. To introduce the apoA-I gene and its regulatory sequences into the pSPORT-1 CIII WT, a 5.5 kb genomic sequence (15) containing the entire apoA-I gene and a 2.1 kb promoter segment was excised from the pUC19 A-I CIII plasmid by XbaI digestion and cloned into the XbaI site of the modified pSPORT-1 CIII WT plasmid to generate the pSPORT-1 –890/+24 CIII WT A-I WT plasmid. This plasmid, which contains the apoA-I and apoCIII genomic sequences, was digested with NotI and ligated with the CAT cDNA flanked by NotI sites to generate the pSPORT-1 CIII WT CAT A-I WT plasmid (Fig. 1A). The WT gene cluster where the apoCIII gene was substituted by the CAT cDNA was excised by BamHI digestion and the mouse lines carrying this transgene were designated WT (Fig. 1A). The CAT cDNA sequence utilized above was obtained by PCR amplification of the CAT cDNA template (2) in a way that introduced NotI sites at the 5′ and 3′ ends as well as a unique EcoRI site that allows identification of the transgenic mice.
Table 1. Oligonucleotide sequence of primers used in PCR amplifications.
| Name | Sequence | Location of Sequence |
|---|---|---|
| CIII 890K |
ATAGGTACCGTTCCTCCCAGTTGCTCC |
CIII –910/–893 antisense containing a KpnI site |
| CIII 24E |
AAAGAATTCCAGCTGCCTCTAGGGATG |
CIII +27/+10 antisense containing an EcoRI site |
| PN2 |
AAAGAATTCCTGCCTCTAGGGATGAACTGAGCA |
CIII +24/+1 sense containing an EcoRI site |
| PBN1 |
ATAGGTACCGGATCCGTTCTTCCCAGTTGCTCCCACAGC |
CIII –890/-868 sense containing KpnI and BamHI sites |
| CIIIIM1-5 |
–756 AAG ATC TCA GGG ATG TTA TCA GTG GGT CCA –727 |
CIII –756/–727 sense mutagenic |
| CIIIIM1-3 |
–739 TAA CAT CCC TGA GAT CTT AGC TGG TCT CAT GTC TGG GCC –777 |
CIII –739/–777 antisense mutagenic |
| CIIIHM-5 |
–702 TCT GTC TTG GCA AAG GCC TCG GGC T –678 |
CIII –702/–678 sense mutagenic |
| CIIIHM-3 |
–689 CCT TTG CCA AGA CAG ACA CCA GGC TCC CTA TTT TG –721 |
CIII –689/–721 antisense mutagenic |
| CIIIFM3-5 |
–606 AAC CGT AGG CAT GGG GGC TGC TGG GTG AGC AG C –574 |
CIII –606/–574 antisense mutagenic |
| CIIIFM3-3 | -586 GCA GCC CCC ATG CCT ACG GTT CCC TGT GTG CCC CAC CGC CGC –627 | CIII –586/–627 sense mutagenic |
Sequences of the apoCIII promoter region are described by Ogami et al. (2). Bold letters indicate mutations in the original sequence.
Figure 1.
pSPORT-1 plasmid derivatives containing WT (A) and mutated apoCIII enhancer either in one SP1 binding site (B) or three SP1 binding sites (C). The names of the resulting transgenic lines harboring these transgenes are indicated in parentheses. The WT and mutant sequences of the SP1 sites of the apoCIII enhancer are shown in (B) and (C), respectively. Nucleotide substitutions in the mutated sequences are depicted by bold characters. The regions of the apoCIII promoter and enhancer or the apoA-I promoter and enhancer utilized in in vitro experiments (shown in Fig. 6A and B) are indicated by asterisks.
The mutations in the SP1 sites of elements H and the triple mutation on elements H, I and F of the apoCIII enhancer were generated by amplification and mutagenesis of the –890/+24 apoCIII promoter region. In a typical amplification/mutagenesis reaction, the 5′ external primer and the 3′ mutagenic primer were used to amplify the upstream region of interest and the 5′ mutagenic primer and 3′ external primer were used to amplify the downstream region of interest. Following amplification, an aliquot of 5 µl containing the two amplification products were mixed and amplified using the 5′ and 3′ external primers. For the introduction of a single mutation in the SP1 site of the apoCIII enhancer, we used the oligonucleotides PN2 and PBN1 as 5′ and 3′ external primers and the oligonucleotides CIII HM-5 and CIII HM-3 as 5′ and 3′ mutagenic primers respectively (Table 1), and the pSPORT-1 CIII WT (–890/+24) plasmid as a template. The amplified mutant –890/+24 apoCIII promoter region was excised via KpnI–EcoRI digestion and cloned into the corresponding sites of pSPORT-1 plasmid to generate the pSPORT-1 CIII H plasmid.
This intermediate plasmid was used as a template for two rounds of further mutagenesis of the enhancer region using the PN2 and PBN1 oligonucleotides as external primers, and CIII IM1-5 and CIII IM1-3 as mutagenic primers in the first round and CIII FM3-3 and CIII FM3-5 in the second round of amplification (Table 1). The mutated –890/+24 apoCIII promoter/enhancer region containing mutations in three SP1 sites in elements H, I and F of the enhancer were digested with KpnI and EcoRI and cloned into the corresponding sites of the pSPORT-1 plasmids. These clonings generated the pSPORT-1 CIII HIF plasmid derivative. The EcoRI site of the mutated apoCIII enhancer plasmids was subsequently eliminated as described for the WT construct, and the modified derivatives were digested with SalI and BamHI to provide the mutated apoCIII regulatory sequences. The introduction of the mutations in the SP1 sites of the apoCIII enhancer were confirmed by DNA sequencing.
To generate the transgenic constructs containing the WT apoA-I and apoCIII regulatory sequences carrying the single or the triple mutations in the apoCIII enhancer, a 6.89 kb fragment encompassing the 2.1 kb apoA-I promoter region and the entire apoA-I gene and CAT gene was obtained by BamHI and SalI digestion of the pSPORT-1 A-I WT CIII WT CAT plasmid (Fig. 1B). The 0.89 kb mutated –890/+24 apoCIII promoter/enhancer region and the 6.89 kb apoA-I gene and CAT gene sequences obtained by BamHI and SalI digestion as described above were introduced by triple ligation into the BamHI site of the pSPORT-1 plasmid to generate the two mutated plasmids pSPORT-1AI WT CIII (H) CAT and the pSPORT-1A-I WT CIII (HIF) CAT respectively (Fig. 1B and C). The mutated gene cluster was excised by BamHI digestion and the mouse lines carrying the transgene mutated at the H site or the HIF sites were designated Hmut and HIFmut respectively (Fig. 1B and C).
Generation and characterization of the transgenic mice. Heterozygous transgenic mice used in these studies were generated using standard transgenic mice methodologies (16,17). To obtain fertilized oocytes, C57BL-6 female embryo donor mice at 6–8 weeks of age were injected with pregnant mare’s serum followed by injection of human chorionic gonadotropins 48 h later. The female embryo donors were mated with C57/BL-6 males, and on the following day the fertilized oocytes were recovered in M2 medium (17). DNA of the construct of interest was microinjected into the embryo’s pronuclei at a concentration of 2 µg/ml in injection buffer (10 mM Tris, pH 7.4, EDTA 0.1 mM). Embryos that survived microinjection were maintained for 30 min in M2 medium (17) and then implanted into the oviduct of pseudopregnant female mice (Swiss Webster) mated previously with vasectomized male mice (Swiss Webster). Thirty embryos were implanted in each female pseudopregnant mouse. Recipient mice progressed through gestation, and ~70% of them gave birth to transgenic mice. Typically, ~20–30% of the littermates were identified as transgenic founders (Fo) by Southern blotting of DNA isolated from tail biopsies of 4-week-old mice. Identification was confirmed by Southern blotting of mouse genomic DNA following EcoRI digestion and hybridization with the 978 bp apoA-I probe. This probe contained 540 bp of exon 4 and 438 bp of the intergenic sequence between the apoA-I and apoCIII genes. This analysis detected an 8.2 kb band in the transgenic mice that corresponds to microinjected transgene. F1 progeny of transgenic mice were obtained through breeding of the Fo founders. Two to three transgenic mouse lines were generated per construct to overcome positional effects and ensure that the pattern of expression is characteristic of a specific construct.
Lipid, lipoprotein and apoA-I profile of transgenic mice. Levels of total serum cholesterol and serum TG were determined using commercially available enzymatic kits (Boehringer Mannheim).
RNA isolation, northern blotting and S1 nuclease analyses. Total cellular RNA was isolated from liver and other tissues using the guanidine isothiocyanate method, Qiagen RNA/DNA kit and purified using a medium-size column. RNA was eluted with high salt buffer, precipitated by isopropanol and dissolved in RNase-free water. Equal quantities of RNA (10 µg) were separated by electrophoresis in 1.0% agarose–formaldehyde gels. Following transfer to a HyBond N+ nylon membrane (Amersham Pharmacia Biotech), the RNA was cross-linked to the filter by UV irradiation (Stratalinker, Stratagene) at 0.12 J/cm2 for 1 min. The apoA-I probe for hybridization is 438 bp in length. This probe contains 290 bp of exon 4 of human apoA-I and 148 bp of the intergenic sequence between the apoA-I and apoCIII genes and does not cross-hybridize with the mouse apoA-I mRNA. The mouse 28S rRNA probe was obtained from Ambion (Austin, TX). This oligonucleotide probe contains 43 nucleotides of highly conserved sequences among higher eukaryotes. The 28S rRNA probe was labeled by 5′ end labeling using the T4 polynucleotide kinase method. The apoA-I mRNA signal of each sample was normalized by dividing with the 28S rRNA signal. The apoA-I probe was prepared as described (2). Hybridization reactions contained 1–2 × 106 c.p.m. 32P-labeled DNA per ml buffer. Unhybridized probe was removed by washing at 65°C with 2× SSC, 0.1% SDS, followed by 15–30 min washes with 1× SSC and then with 0.5× SSC, as needed. Quantitation of X-ray film was performed by a PhosphoImager (Molecular Dynamics) using the ImageQuant program. The relative levels (%) of apoA-I mRNA represent the ratio of apoA-I to 28S ribosomal RNA signal. S1 nuclease mapping (18) was also used to detect minor sites of apoA-I mRNA synthesis. The apoA-I signal was compared with the β-actin mRNA signal. The actin probe (ATCC) represented a 240 bp AvaI–BamHI restriction fragment of the mouse β-actin gene and contained 100 bp of exon I and 140 bp of the 5′ end of the gene (19). A 978 bp apoA-I probe described above has been used for the detection of the apoA-I mRNA by the S1 nuclease analysis. A specific 540 bp apoA-I mRNA signal and a 100 bp mouse β-actin signal was obtained using this analysis.
CAT assays. For the CAT assay, F1 mice were sacrificed at 2 months of age. Tissues were collected and used immediately, or frozen and stored at –80°C. CAT assays were performed as described (20). Briefly, the tissue was homogenized in a polytron homogenizer on ice using 5 µl/mg tissue of extraction buffer containing 15 mM Tris–Cl pH 8.0, 1 mM dithiothreitol (DTT), 0.4 mM PMSF, 50 mM KCl, 15 mM NaCl, 2 mM EDTA and 0.15 mM spermine. The supernatant of the homogenate was heated at 65°C for 8 min, centrifuged in a microcentrifuge for 3 min. The supernatant was used for CAT and protein determination assays. The specific activity was calculated as pmol product/mg protein/min.
The concentration of soluble protein was determined by the Bio-Rad protein assay. The percentage of chloramphenicol converted to acetylated forms was determined either by densitometric scanning of autoradiograms or by scraping individual spots from the thin-layer chromatography (TLC) and counting in a scintillation counter. CAT activities were expressed as pmol of acetyl chloramphenicol generated per min/mg of protein, after subtracting the background for each tissue from control mice that do not express the CAT gene.
RESULTS
Generation and comparison of transgenic mice harboring the WT apoA-I/apoCIII gene cluster with mice harboring a cluster mutated in the SP1 site(s) of the apoCIII enhancer
We have generated and studied comparatively three sets of mouse lines expressing the constructs shown in Figure 1A–C. The top construct, designated WT, contains the 2.1 kb of the 5′ regulatory sequence, the entire coding sequence of apoA-I gene as well as a 1.81 kb segment containing the intragenic sequence and a 1.6 kb segment containing the CAT gene in front of the –890/+24 regulatory sequence of the apoCIII gene. In this construct, the apoCIII gene was replaced by the CAT cDNA sequence (Fig. 1A). The middle construct, designated Hmut, contains mutations in one SP1 binding site (on element H) of the apoCIII enhancer (Fig. 1B). The third construct, designated HIFMut, contains mutations in all three SP1 sites (on elements H, I and F) of the apoCIII enhancer (Fig. 1C). Transgenes containing the WT and the two mutant apoA-I/CIII promoter constructs were excised from the corresponding pSPORT-1 constructs by BamHI digestion. The purified DNA fragments were dissolved in 10 mM Tris–HCl pH 7.4, 0.1 mM EDTA, at a concentration of 2 ng/µl, and were microinjected into fertilized eggs from C57BL/6J females mated with males of the same strain. The names of the transgenes are shown in parentheses in Figure 1A–C. These names are used throughout the text.
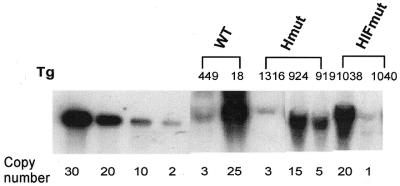
Transgenic founders (Fo) were identified by Southern blotting analysis using as probe the 978 bp apoA-I fragment described in Materials and Methods. The number of transgene copies incorporated into the genome of each transgenic founder (Fo) was determined by Southern blotting and by comparison of the intensity of the bands formed when increasing amounts of the transgene were diluted in non-transgenic DNA (Fig. 2). The two transgenic lines shown in Figure 2 and line N:6 (not shown) which carry the WT transgene contain 3, 25 and 5 copies of the transgene, respectively. The transgenic lines carrying the Hmut transgene (which was altered in one SP1 site) of the apoCIII enhancer contain 3, 5 or 15 copies of the transgene, respectively. The transgenic lines carrying the HIFmut transgene (which was altered in three SP1 sites) of the apoCIII enhancer contain 1 or 20 copies of the transgene, respectively (Fig. 2).
Figure 2.
Gene copy number of mouse lines expressing WT and mutated apoCIII enhancer constructs. The names of transgenic mouse lines analyzed are described in Figure 1A–C. Results are summarized in Tables 3 and 4.
Lipid profiles of mice expressing the apoA-I and apoCIII genes under the control of the WT apoA-I/apoCIII promoter and enhancer or mice carrying mutations in the apoCIII enhancer
The lipid levels of mice expressing the human apoA-I gene were determined as explained in the Materials and Methods (Table 2). The triglyceride levels were within the normal range. The total cholesterol and high density lipoprotein (HDL) levels of the transgenic mice expressing the WT construct were, in general, slightly elevated relative to the lines expressing the mutant transgenes (Table 2).
Table 2. Lipid profiles in control and transgenic micea.
| Constructs | Mouse line | Total cholesterol (mg/dI) | Triglyceride (mg/dI) |
|---|---|---|---|
| C57BL/6 | 73 | 68 | |
| WT | #449 | 163 | 78 |
| #18 | 141 | 66 | |
| Hmut | #919 | 112 | 79 |
| #924 | 124 | 69 | |
| #1316 | 118 | 71 | |
| HIFmut | #1040 | 97 | 60 |
| #1038 | 112 | 75 |
aMouse plasmas were collected by pooling the blood from two to three mice of each line in plastic tubes containing EDTA to achieve a final concentration of 5 mM EDTA.
Comparison of the expression of the CAT gene under the control of the WT apoA-I promoter and either WT or mutated apoCIII enhancer
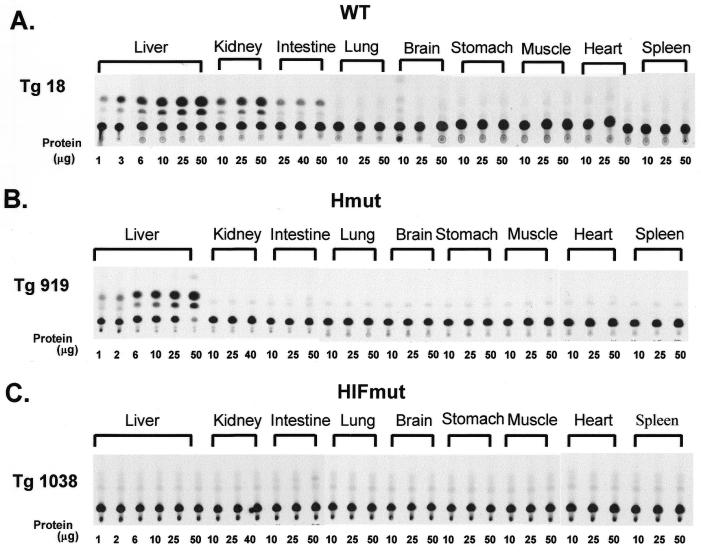
The activity of the WT or mutated apoCIII promoter/enhancer was measured by the activity of the reporter CAT gene as described in the Materials and Methods. The mouse lines carrying the WT promoter/enhancer construct displayed high levels of CAT activity in the liver, low levels of expression in the intestine and kidney and no expression in the lung, spleen, brain, stomach, muscle and heart (Fig. 3A; Table 3). The level of CAT activity in the intestine and kidney in mice expressing the WT construct was ~5 and 3%, respectively (Table 3), of that observed in the liver. With the exception of the ectopic expression in the kidney, the observed pattern of expression mimics the expression of the apoCIII gene in fetal human tissues and rat and rabbit tissues (21–23).
Figure 3.
CAT assays of transgenic mouse tissues expressing constructs containing the WT and mutated apoCIII enhancer segments shown in Figure 1A–C. (A) Mice expressing the CAT gene under the control of the WT apoA-I promoter/WT apoCIII promoter and enhancer. (B) Mice expressing the CAT genes under the control of the WT apoA-I promoter/mutant apoCIII enhancer (in one SP1 binding site on element H). (C) Mice expressing the CAT gene under the control of WT apoA-I promoter/mutant apoCIII enhancer (in all three SP1 binding sites on elements H, I and F). The names of the transgenic lines are described in Figure 1A–C. CAT assays were performed in triplicate as described in Materials and Methods.
Table 3. Comparison of CAT-specific activity of mice carrying the WT regulatory sequences of the human apoA-I and apoCIII gene and those carrying mutations in the SP1 sites of the apoCIII enhancer.
| Construct | Mouse line | Copy number | na | CAT activity (pmol/mg protein/min) | |||
|---|---|---|---|---|---|---|---|
| Liver | Intestine | Kidney | Other tissues | ||||
| WT | 18 | (25) | 4 | 7105 ± 1651 | 516 ± 109 | 183.3 ± 31 | – |
| 449 | (3) | 3 | 5193 ± 1121 | 211 ± 29 | 243 ± 50 | – | |
| 6 | (5) | 3 | 9151.2 ± 41 | 320 ± 2.9 | 130 ± 2.8 | – | |
| Hmut | 919 | (5) | 3 | 6427 ± 43.7 | – | – | – |
| 924 | (15) | 3 | 9038 ± 717.1 | 8.2 ± 4.5 | 25.4 ± 2.9 | – | |
| 1316 | (3) | 3 | 10225 ± 212 | – | – | – | |
| HIFmut | 1038 | (20) | 3 | – | – | – | – |
| 1040 | (1) | 3 | – | – | – | – | |
aNumber of mice analyzed.
A mutation in one SP1 site on element H of the apoCIII enhancer eliminated the intestinal expression in two out of the three mouse lines studied but did not significantly affect the hepatic expression of the apoCIII gene (Fig. 3B; Table 3). This finding indicates the existence of different mechanisms of transcriptional activation of the apoCIII gene in the liver and the intestine. Mutations in all three SP1 sites of the enhancer abolished the expression of the reporter CAT gene in all tissues. The findings suggest that the apoCIII enhancer is required for the overall transcription of the apoCIII gene and that the SP1 sites are essential for the activity of the apoCIII promoter/enhancer cluster.
Comparison of the expression of the apoA-I gene under the control of the WT apoA-I promoter and either WT or mutated apoCIII enhancer
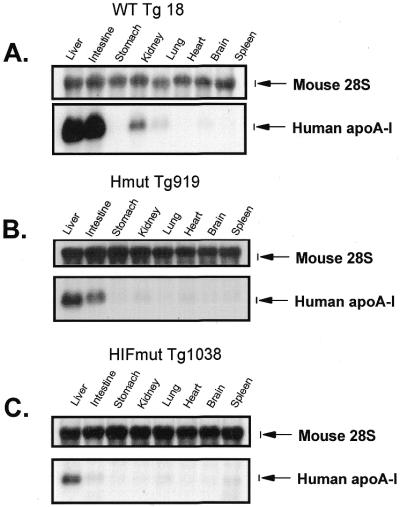
The expression of the apoA-I gene was determined by northern blotting as described in the Materials and Methods. The analysis of the mouse lines showed that the WT apoA-I promoter/WT apoCIII enhancer directs high levels of expression in the liver and intestine, moderate levels of expression in the kidney and low levels of expression in the lung. S1 nuclease mapping also detected very low levels of expression in the stomach, kidney, heart, spleen and muscle and no expression in the brain (Fig. 4; Table 4). This pattern of expression mimics the expression of the apoA-I gene in fetal human tissues (21).
Figure 4.
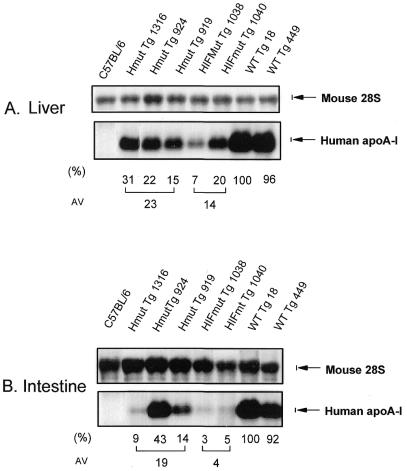
Northern blot analysis of apoA-I mRNA obtained from tissues of transgenic mice expressing constructs containing the WT and mutated apoA-I and apoCIII enhancer constructs shown in Figure 1A–C. (A) Mice expressing the apoA-I gene under the control of the WT apoA-I promoter/WT apoCIII promoter and enhancer. (B) Mice expressing the apoA-I gene under the control of the WT apoA-I promoter/mutant apoCIII enhancer in one SP1 site on element H. (C) Mice expressing the apoA-I gene under the control of WT apoA-I promoter mutant/apoCIII enhancer in three SP1 sites on elements H, I and F. The names of the transgenic lines are described in Figure 1A–C. Northern blotting was performed as described in the Materials and Methods. In (A–C) the relative apoA-I mRNA levels in different tissues of specific mouse lines are illustrated. Comparative levels of apoA-I mRNA in liver and intestine of different mouse lines are shown in Figure 5A and B.
Table 4. Comparisons of the levels of expression of the human apoA-I gene in transgenic mice carrying the WT apoA-I and apoCIII regulatory regions with mice carrying mutations in the SP1 sites of the apoCIII enhancer.
| Construct | Mouse line | Copy number | n | Relative mRNA expression | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver | Intestine | Stomach | Kidney | Lung | Heart | Brain | Spleen | ||||
| WT | 18 | (25) | 4 | ++++ | +++ | ± | ++ | + | ± | – | ± |
| 449 | (3) | 3 | ++++ | +++ | + | ++ | + | ± | – | ± | |
| 6 | (5) | 3 | ++++ | +++ | + | ++ | + | ± | – | ± | |
| Hmut | 919 | (5) | 3 | ++ | ++ | – | ± | ± | ± | – | ± |
| 924 | (15) | 3 | ++ | ++ | – | – | ± | ± | – | ± | |
| 1316 | (3) | 3 | ++ | + | – | – | ± | ± | – | ± | |
| HIFmut | 1038 | (20) | 3 | ++ | ± | – | ± | ± | ± | – | ± |
| 1040 | (1) | 3 | ++ | ± | – | ± | ± | ± | – | ± | |
++++, The highest expression 100%; +++, high expression 60–100%; ++, intermediate expression 15–30%; +, low expression 5–10%; ±, very low expression <1–2%; n, number of mice analyzed.
Mutations in one or three SP1 sites of the apoCIII enhancer did not alter qualitatively the expression of the apoA-I gene in the major tissues (Fig. 4B and C). However, quantitation of the relative levels of expression showed that the steady-state apoA-I mRNA levels in the liver and intestine of mice carrying mutations in one SP1 site on element H of the apoCIII enhancer were reduced by 77 and 81%, respectively, as compared to the expression in mice carrying the WT transgene (Fig. 5A and B). Similar quantitation showed that the relative levels of apoA-I mRNA in the liver and intestine in mice carrying three SP1 mutations on element HIF were reduced by 86 and 96% in the liver and intestine, respectively, as compared to the apoA-I mRNA levels of mice carrying the WT transgene (Fig. 5A and B).
Figure 5.
Northern blotting analysis of 10 µg of total RNA obtained from liver (A) and intestine (B) of transgenic mice expressing constructs containing the WT and mutated apoCIII enhancer shown in Figure 1A–C. The nylon membrane was hybridized simultaneously with the apoA-I mouse 28S RNA. The names of the transgenic lines are given in Figure 1A–C. The ratio of apoA-I to 28S signal of samples obtained from mice carrying the WT apoA-I WT apoCIII construct was arbitrarily set to 100%. The ratios of apoA-I/28S bands provide a measure of their abundance relative to those of the WT control.
The findings indicate that the hepatic activity of the apoA-I promoter is reduced to ~14% of its original strength when the apoCIII enhancer is nearly inactivated by elimination of its SP1 binding sites, indicating that SP1 serves an important function for the overall apoCIII enhancer activity. Comparison of the in vivo and in vitro data of apoA-I and apoCIII gene regulation are shown in Figure 6A and B.
Figure 6.
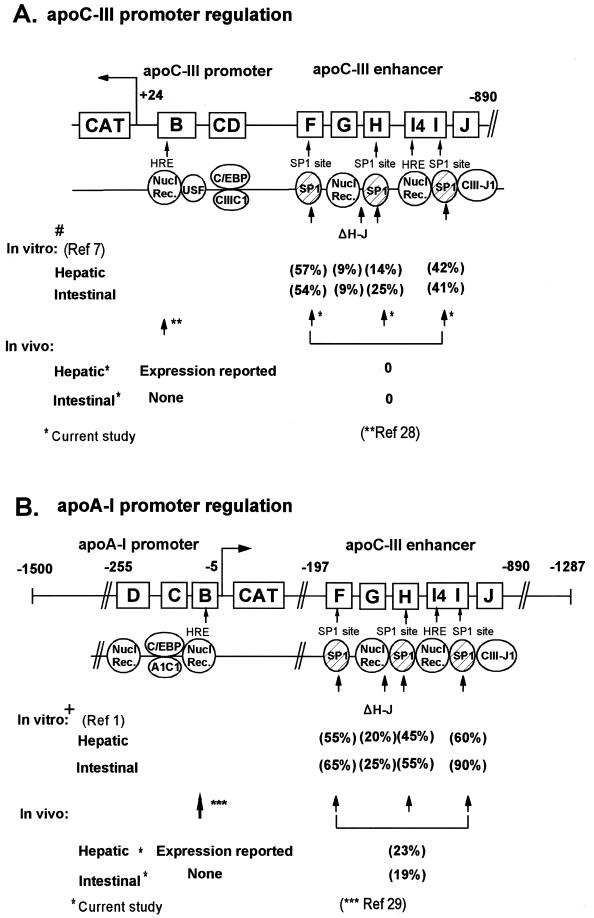
(A) Schematic representation of the WT and mutated –890/+24 apoCIII promoter/enhancer, the –255/–5 apoA-I promoter –197/–1287 apoCIII enhancer regions showing the binding specificity of the regulatory sites in vitro. Numbers in parentheses indicate the percentage activity of the promoter/enhancer cluster when it is mutated such that the indicated factors do not bind to the mutated site (1,7). Factors are symbolized by ovals. In (A) # indicates the elements and factors contributing to the hepatic and intestinal activity of the apoCIII promoter/enhancer cluster in cell culture (7). In (B) + indicates that the proximal apoA-I promoter required for hepatic expression in cell cultures extends to nucleotide –255 (2) and, for the intestinal expression, to nucleotide –1500. Intestinal expression in cell cultures required the presence of HNF4 (1). Single asterisks refer to the present transgenic studies. The triple asterisks refer to expression studies in transgenic mice carrying 256 bp of the apoA-I promoter (29). The double asterisks refer to expression studies in transgenic mice carrying 200 bp of the apoCIII promoter (28).
DISCUSSION
Background
Previous studies have established that there is a linkage (23) and a common regulatory mechanism of the apoA-I/apoCIII/apoA-IV gene cluster (1,4,7,8,24). A common feature of this gene cluster is that all three genes contain HREs in their proximal promoters (1,5,7,25–27). In addition, the distal regulatory region of the apoCIII promoter acts as a common enhancer for the three closely linked genes (1,7,8). The enhancer contains two HREs and three SP1 binding sites (5,7). Mutagenesis analysis showed that alterations that prevent the binding of SP1 to the enhancer diminished the promoter/enhancer activity in cell cultures (1,7,8). Other transcription factors bind to the proximal promoters of the apoA-I and apoCIII genes and may affect their overall activity and tissue specificity (2,27).
The apoCIII enhancer confers correct tissue-specific expression of the apoCIII gene in vivo. The SP1 sites of the apoCIII enhancer are required for the expression of the apoCIII gene in vivo
In the current study, regulatory elements representing the –890/+24 apoCIII promoter directed expression of the apoCIII gene in a pattern similar to that observed in fetal human tissues and rat and rabbit tissues (21–23). In these species the apoCIII is expressed predominantly in the liver and to a lesser extent in the intestine (21–23). The only difference in the present study was that in mice carrying the WT transgene there was a low level of ectopic expression in the kidney. It is possible that the construct utilized lacks a silencer which may be involved in the repression of the expression of the apoCIII gene in the kidney. The current study using transgenic mice also attempted to elucidate the importance of the SP1 sites of the enhancer on the overall activity and tissue specificity of the apoA-I and apoCIII promoters. To address this question we generated two sets of transgenic mice with mutations either only in element H or in elements H, I and F, which abolish the binding of SP1 to these sites. The mutation in one SP1 site on element H did not affect the hepatic expression, but abolished the intestinal expression in two out of three mouse lines. The findings suggest that the apoCIII enhancer is important for intestinal transcription and that the mechanism of hepatic and intestinal transcription of the apoCIII gene may require different combinations of transcription factors in vivo. The in vitro data had shown previously that mutation in the SP1 binding site of element H reduced the hepatic and intestinal activity of the apoCIII promoter/enhancer cluster by 86 and 75%, respectively (7). A deletion that eliminated two of the SP1 binding sites on elements H and I reduced the activity of the promoter/enhancer cluster by 91% (7) (Fig. 6A). These in vitro data are in agreement with the findings of the present study, which indicate that the presence of the SP1 sites of the apoCIII enhancer is required for the overall activity of the apoCIII promoter/enhancer in vivo. When all three SP1 sites are mutated the expression of the CAT gene (used as a reporter for the apoCIII gene) is then abolished in all tissues.
A previous report showed that the hepatic expression of apoCIII gene in transgenic mice could be achieved by a construct containing 200 bp of the apoCIII promoter (28). It is possible that the changes introduced on the apoCIII enhancer in the present study have a negative effect on the residual activity of the proximal apoCIII promoter. Another possibility is that the integration of the apoCIII gene construct in the previous study might have occurred in the vicinity of SP1 sites of another regulatory sequence and, in turn, this might have activated the proximal apoCIII promoter (28).
The apoCIII enhancer confers tissue-specific expression of the apoA-I gene. The SP1 sites of the ApoCIII enhancer contribute to the hepatic and intestinal expression of this gene
In the current study, regulatory elements representing the –2100/+5 apoA-I promoter directed expression of the apoA-I gene in a pattern similar to that observed in fetal human tissues, where apoA-I is expressed abundantly in both the liver and intestine (21). These findings are consistent with previous findings showing that the apoCIII enhancer could direct the intestinal expression of the apoA-I gene, but did not restrict gene expression to the villus cells (10). Previous studies had shown that the activity of the apoA-I promoter/apoCIII enhancer cluster was reduced by 30–60% in HepG2 cell cultures by point mutations in individual SP1 binding sites (Fig. 6B), indicating that these sites may be important for the overall activity of the apoA-I promoter (1,7). Other studies have also suggested that DNA sequence present in the intergenic region between the apoCIII and apoA-IV gene (11) which contain the apoCIII enhancer (16) are required for the correct intestinal expression of the apoA-IV gene which is located 5 kb downstream of the apoCIII gene (24).
A previous transgenic study established that the proximal apoA-I promoter extending to nucleotide –256 was sufficient to direct hepatic expression of the apoA-I gene in vivo and in vitro (12,29), suggesting that this region contains at least some of the necessary elements for the hepatic expression of the apoA-I gene. The current study established that mutations in all three SP1 binding sites of the apoCIII enhancer reduced the hepatic expression by 86% as compared to the expression in mice carrying the WT transgene and nearly abolished the intestinal expression. Furthermore, a mutation in a single SP1 site on element H reduced both the hepatic and intestinal expression to ~20% of the WT control. The fact that the apoA-I transgene is expressed at lower levels when the apoCIII enhancer is mutated in either one or three SP1 sites may explain the decrease in cholesterol levels of the transgenic mice as compared to the non-transgenic littermates (Table 2). It is known that the majority of cholesterol in the transgenic mice is found in HDL and the HDL levels correlate with plasma apoA-I levels. Thus, the observed difference in total cholesterol levels of the non-transgenic, WT and mutant transgenic mice may be accounted for by the different levels of expression of the human apoA-I transgene.
Potential mechanisms of transcriptional enhancement in vivo
The present study shows that the apoA-I promoter alone can contribute ~14% to the hepatic transcriptional activity when the apoCIII enhancer is inactivated by mutations in all three SP1 sites. When both the promoter and the enhancer are functional, the activity of the apoA-I promoter/apoCIII enhancer cluster increases 7-fold to 100%. It appears that in these tissues, promotion of transcription mandates synergy between the factor bound to proximal promoter and the apoCIII enhancer. Preliminary findings by us indicate that this synergy involves protein–protein interactions between SP1 bound to the enhancer and the orphan nuclear receptor HNF-4, which binds both to the proximal promoter and the apoCIII enhancer (30). Our findings suggest that enhancement of the hepatic transcription of the apoA-I gene may be achieved by recruitment of the proteins of the basal transcription complex by factors which bind both to the proximal apoA-I promoter and to the apoCIII enhancer. In this regard, SP1 directly or indirectly may play an essential role in the observed transcriptional enhancement. A similar mechanism may also apply for the closely linked apoCIII and apoA-IV genes. The transcriptional mechanism described for the liver may somehow be different in the intestine and the kidney.
Numerous studies have established that increases in plasma apoA-I and HDL concentrations are associated with protection from cardiovascular disease (31–34). In addition, alteration in apoCIII levels has been shown to affect the catabolism of triglyceride-rich lipoproteins (28,35–38). Thus, the in vivo transcriptional regulatory mechanisms which emerge from this and similar studies may provide rational approaches for correcting low plasma HDL levels and reducing the plasma levels of atherogenic triglyceride-rich lipoprotein particles in humans.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Markella Zanni for editorial corrections and comments and Anne Plunkett for preparing the manuscript. This work was supported by grants from the National Institutes of Health (HL33952).
REFERENCES
- 1.Kardassis D., Tzameli,I., Hadzopoulou-Cladaras,M., Talianidis,I. and Zannis,V.I. (1997) Distal apolipoprotein C-III regulatory elements F to J act as a general modular enhancer for proximal promoters that contain hormone response elements. Synergism between hepatic nuclear factor-4 molecules bound to the proximal promoter and distal enhancer sites. Arterioscler. Thromb. Vasc. Biol., 17, 222–232. [DOI] [PubMed] [Google Scholar]
- 2.Ogami K., Hadzopoulou-Cladaras,M., Cladaras,C. and Zannis,V.I. (1990) Promoter elements and factors required for hepatic and intestinal transcription of the human ApoCIII gene. J. Biol. Chem., 265, 9808–9815. [PubMed] [Google Scholar]
- 3.Reue K., Left,T. and Breslow,J.L. (1988) Human apolipoprotein CIII gene expression is regulated by positive and negative cis-acting elements and tissue-specific protein factors. J. Biol. Chem., 263, 6857–6864. [PubMed] [Google Scholar]
- 4.Lavrentiadou S.N., Hadzopoulou-Cladaras,M., Kardassis,D. and Zannis,V.I. (1999) Binding specificity and modulation of the human ApoCIII promoter activity by heterodimers of ligand-dependent nuclear receptors. Biochemistry, 38, 964–975. [DOI] [PubMed] [Google Scholar]
- 5.Ladias J.A.A., Hadzopoulou-Cladaras,M., Kardassis,D., Cardot,P., Cheng,J., Zannis,V.I. and Cladaras,C. (1992) Transcriptional regulation of human apolipoprotein genes apoB, apoCIII and apoAII by members of the steroid hormone receptor superfamily HNF-4, ARP-1, EAR-2 and EAR-3. J. Biol. Chem., 267, 15849–15860. [PubMed] [Google Scholar]
- 6.Mietus-Snyder M., Sladek,F.M. Ginsburg,G.S., Kuo,C.F., Ladias,J.A.A., Darnell,J.E.,Jr and Karathanasis,S.K. (1992) Antagonism between apolipoprotein AI regulatory protein 1, Ear3/COUP-TF and hepatocyte nuclear factor 4 modulates apolipoprotein CIII gene expression in liver and intestinal cells. Mol. Cell. Biol., 12, 1708–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talianidis I., Tambakaki,A., Toursounova,J. and Zannis,V.I. (1995) Complex interactions between SP1 bound to multiple distal regulatory sites and HNF-4 bound to the proximal promoter lead to transcriptional activation of liver-specific human APOCIII gene. Biochemistry, 34, 10298–1030. [DOI] [PubMed] [Google Scholar]
- 8.Ktistaki E., Lacorte,J.-M., Katrakili,N., Zannis,V.I. and Talianidis,I. (1994) Transcriptional regulation of the apolipoprotein A-IV gene involves synergism between a proximal orphan receptor response element and a distant enhancer located in the upstream promoter region of the apolipoprotein C-III gene. Nucleic Acids Res., 22, 4689–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginsburg G.S., Ozer,J. and Karathanasis,S.K. (1995) Intestinal apolipoprotein AI gene transcription is regulated by multiple distinct DNA elements and is synergistically activated by the orphan nuclear receptor, hepatocyte nuclear factor 4. J. Clin. Invest., 96, 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisaha J.G., Simons,T.C., Gordon,J.L. and Breslow,J.L. (1995) Characterization of an enhancer element in the human apolipoprotein C-III gene that regulates human apolipoprotein A-I gene expression in the intestinal epithelium. J. Biol. Chem., 270, 19979–19988. [DOI] [PubMed] [Google Scholar]
- 11.Lauer S.J., Simonet,W.S., Bucay,N., de Silva,H.V. and Taylor,J.M. (1991) Tissue-specific expression of the human apolipoprotein A-IV gene in transgenic mice. Circulation, 84, 17. [Google Scholar]
- 12.Walsh A., Azrolan,N., Wang,K., Marcigliano,A., O’Connel,A. and Breslow,J.L. (1993) Intestinal expression of the human apoA-I gene in transgenic mice is controlled by a DNA region 3′ to the gene in the promoter of the adjacent convergently transcribed apoC-III gene. J. Lipid Res., 34, 617–623. [PubMed] [Google Scholar]
- 13.Kan H.Y., Gerorgopoulos,S. and Zannis,I. (2000) A hormone response element in the human apolipoprotein CIII (ApoCIII) enhancer is essential for intestinal expression of the apoA-I and apoCIII gene and contributes to the hepatic expression of the two linked genes in transgenic mice. J. Biol. Chem., 275, 30423–30431. [DOI] [PubMed] [Google Scholar]
- 14.Roghani A. and Zannis,V.I. (1988) Alterations of the glutamine residues of human apolipoprotein AI propeptide by in vitro mutagenesis. Characterization of the normal and mutant protein forms. Biochemistry, 27, 7428–7435. [DOI] [PubMed] [Google Scholar]
- 15.Karathanasis S.K., Zannis,V.I. and Breslow,J.L. (1983) Isolation and characterization of the human apolipoprotein A-I gene. Proc. Natl Acad. Sci. USA, 80, 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Beyec J., Chauffeton,V., Kan,H.-Y., Janvier,P.-L., Cywiner,C., Chatelet,F.-P., Kalopissis,A.D., Zannis,V., Chambaz,J., Pinçon-Raymond,M. and Cardot,P. (1999) The –700/–310 fragment of the apolipoprotein A-IV gene combined with the –890/–500 apolipoprotein C-III enhancer is sufficient to direct a pattern of gene expression similar to that for the endogenous apolipoprotein A-IV gene. J. Biol. Chem., 274, 4954–4961. [DOI] [PubMed] [Google Scholar]
- 17.Hogan B. (1986) Manipulating the Mouse Embryo. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Berk A.J. and Sharp,P.A. (1977) Ultraviolet mapping of the adenovirus 2 early promoters. Cell, 12, 721–732. [DOI] [PubMed] [Google Scholar]
- 19.Alonso S., Minty,A., Bourlet,Y. and Buckingham,M. (1986) Comparison of three actin-coding sequences in the mouse; evolutionary relationships between the actin genes of warm-blooded vertebrates. J. Mol. Evol., 23, 11–22. [DOI] [PubMed] [Google Scholar]
- 20.Pothier F., Quellet,M., Julien,J.P. and Guerin,S.L. (1992) An improved CAT assay for promoter analysis in either transgenic mice or tissue culture cells. DNA Cell Biol., 11, 83–90. [DOI] [PubMed] [Google Scholar]
- 21.Zannis V.I., Cole,F.S., Jackson,C.L., Kurnit,D.M. and Karathanasis,S.K. (1985) Distribution of apolipoprotein A-I, C-II, C-III and E mRNA in fetal human tissues. Time-dependent induction of apolipoprotein E mRNA by cultures of human monocyte-macrophages. Biochemistry, 24, 4450–4455. [DOI] [PubMed] [Google Scholar]
- 22.Lenich C., Brecher,P., Makrides,S., Chobanian,A. and Zannis,V.I. (1988) Apolipoprotein gene expression in the rabbit: abundance, size and distribution of apolipoprotein mRNA species in different tissues. J. Lipid Res., 29, 755–764. [PubMed] [Google Scholar]
- 23.Haddad I.A., Ordovas,J.M., Fitzpatrick,T. and Karathanasis,S.K. (1986) Linkage, evolution and expression of the rat apolipoprotein A-I, C-III and A-IV genes. J. Biol. Chem., 28, 13268–13277. [PubMed] [Google Scholar]
- 24.Karathanasis S.K. (1985) Apolipoprotein multigene family: tandem organization of human apolipoprotein AI, CIII and AIV genes. Proc. Natl Acad. Sci. USA, 82, 6374–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rottman J.N., Widom,R.L., Nadal-Ginard,B., Mahdavi,V. and Karathanasis,S.K. (1991) A retinoic acid-responsive element in the apolipoprotein AI gene distinguishes between two different retinoic acid response pathways. Mol. Cell. Biol., 11, 3814–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzameli I. and Zannis,V.I. (1996) Binding specificity and modulation of the apoA-I promoter activity by homo- and heterodimers of nuclear receptors. J. Biol. Chem., 271, 8402–8415. [DOI] [PubMed] [Google Scholar]
- 27.Papazafiri P., Ogami,K., Ramji,D.P., Nicosia,A., Monaci,P., Cladaras,C. and Zannis,V.I. (1991) Promoter elements and factors involved in hepatic transcription of the human ApoA-I gene positive and negative regulators bind to overlapping sites. J. Biol. Chem., 266, 5790–5797. [PubMed] [Google Scholar]
- 28.deSilva H.V., Lauer,S.J., Wang,J., Simonet,W.S., Weisgraber,K.H., Mahley,R.W. and Taylor,J.M. (1994) Overexpression of human apolipoprotein C-III in transgenic mice results in an accumulation of apolipoprotein B48 remnants that is corrected by excess apolipoprotein E.E. J. Biol. Chem., 269, 2324–2335. [PubMed] [Google Scholar]
- 29.Walsh A., Ito,Y. and Breslow,J.L. (1989) High levels of human apolipoprotein A-I in transgenic mice result in increased plasma levels of small high density lipoprotein (HDL) particles comparable to human HDL3. J. Biol. Chem., 264, 6488–6494. [PubMed] [Google Scholar]
- 30.Kardassis D., Falvey,E., Tsantili,P., Hadzopoulou-Cladaras,M. and Zannis,V.I. (2000) Direct physical interactions between SP1 and HNF-4 mediates synergistic transactivation of the apolipoprotein CIII promoter. Circulation, 102, 294. [DOI] [PubMed] [Google Scholar]
- 31.Castelli W.P., Doyle,J.T., Gordon,T., Hames,C.H., Hjortland,M.C., Hulley,S.B., Kagan,A. and Zukel,W.J. (1977) HDL cholesterol and other lipids in coronary heart disease. The cooperative lipoprotein phenotyping study. Circulation, 55, 767–772. [DOI] [PubMed] [Google Scholar]
- 32.Heiss G. and Tyroler,H. (1982) Quantification (NIH Publication no. 83-1266). US Department of Health and Human Services, National Institutes of Health, Bethesda, MD, pp. 7–24.
- 33.Glueck C.J., Gartside,P., Fallat,R.W., Sielski,J. and Steiner,P.M. (1976) Longevity syndromes: familial hypoβ and familial hyperα lipoproteinemia. J. Lab. Clin. Med., 88, 941–957. [PubMed] [Google Scholar]
- 34.Rubin E.M., Krauss,R.M., Spangler,E.A., Verstuyft,J.G. and Clift,S.M. (1991) Inhibition of early atherogenesis in transgenic mice by human apolipoprotein AI. Nature, 353, 265–269. [DOI] [PubMed] [Google Scholar]
- 35.Brown W.V. and Baginsky,M.L. (1972) Inhibition of lipoprotein lipase by an apoprotein of human very low density lipoprotein. Biochem. Biophys. Res. Commun., 46, 375–382. [DOI] [PubMed] [Google Scholar]
- 36.Windler E., Chao,Y. and Havel,R.J. (1980) Determinants of hepatic uptake of triglyceride-rich lipoproteins and their remnants in the rat. J. Biol. Chem., 255, 5475–5480. [PubMed] [Google Scholar]
- 37.Quarfordt S.H., Michalopoulos,G. and Schirmer,B. (1982) The effect of human C apolipoproteins on the in vitro hepatic metabolism of triglyceride emulsions in the rat. J. Biol. Chem., 257, 14642–14647. [PubMed] [Google Scholar]
- 38.Ito Y., Azrolan,N., O’Connel,A., Walsh,A. and Breslow,J.L. (1990) Hypertriglyceridemia as a result of human apo CIII gene expression in transgenic mice. Science, 249, 790–793. [DOI] [PubMed] [Google Scholar]