Abstract
Background
We aimed to examine whether PTEN pathogenic variants (mutPTEN) were associated with overall survival (OS) in patients with advanced soft tissue sarcoma (STS) with the presence of one or more of the most common genomic alterations including p53, CDKN2A, RB1, and ATRX pathogenic variants.
Methods
This study included patients from Kaiser Permanente Northern California and Stanford Cancer Center with grade 2 or higher locally advanced and metastatic STS.
Results
A total of 174 patients had leiomyosarcoma (LMS), 136 had undifferentiated pleomorphic sarcoma (UPS), 78 had Liposarcoma (LPS), and 214 had other histology subtypes (Others). Among all patients with STS, OS was worse for those with mutPTEN versus wild-type PTEN (wtPTEN, adjusted HR [aHR] = 1.58 [95% CI, 1.11–2.23]), mutCDKN2A vs wtCDKN2A (aHR = 1.33 [95% CI .99–1.80]), and mutRB1 vs wtRB1 (aHR = 1.26 [95% CI 0.93–1.70[), while OS was similar for mutp53 vs wtp53 and mutATRX vs wtATRX. MutPTEN versus wtPTEN was consistently associated with worse OS in histologic subtypes including LMS and UPS and molecular subgroups.
Conclusion
MutPTEN vs wtPTEN was associated with worse OS in advanced STS. If confirmed, our findings could be helpful for prognostic stratification in clinical practice and for further understanding the molecular mechanisms of STS.
Introduction
Soft tissue sarcomas (STSs) are a group of relatively uncommon malignancies that are characterized by heterogeneity, with leiomyosarcoma (LMS), undifferentiated pleomorphic sarcoma (UPS), and Liposarcoma (LPS) the most common histologic subtypes [1]. Several dozen genomic alterations have been identified in advanced STSs including fusions, deletions, missense pathogenic variants, and others [2, 3]. Different histologic subtypes share some common genomic alterations but also harbor unique alterations. For example, MDM2 and CDK4 amplifications are universally present in dedifferentiated liposarcoma, while p53 (mutp53), CDKN2A (mutCDKN2A), RB1 (mutRB1), PTEN (mutPTEN) and ATRX (mutATRX) deletions or pathogenic variants are the most common alterations in LMS and UPS [2–4]. Other histologic subtypes, such as synovial sarcoma, are characterized by a unique fusion gene SS18-SSX [5, 6].
The treatment options for advanced STSs have evolved only modestly over the past three decades with very limited options still available [7–10]. Success with systemic therapy has been far from satisfactory for the most part [1, 11–13]. The recent advances with immunotherapy including immune checkpoint inhibitors and adoptive cell therapy have shown only modest efficacy [13–19]. The prognosis for patients with intermediate to high-grade STSs remains very poor. How the common genomic alterations affect the prognosis of advanced STSs has not been well studied. Previous studies have suggested that mutp53 is associated with poorer disease-free survival (DFS) and overall survival (OS) [20, 21]. Other studies have suggested that mutCDKN2A is associated with poorer OS [8, 22]. Little is known about the potential impact of mutPTEN, mutRB1 and mutATRX on the prognosis of STS. The current prognostic stratifications for STS remain dependent on clinical factors [1, 23, 24].
The role of mutPTEN in the prognosis of advanced STS remains unclear. In this study, we examined genomic pathogenic variants and OS among a combined cohort of 602 adult patients with advanced STS from Kaiser Permanente Northern California (KPNC) and Stanford Cancer Center using next-generation sequencing (NGS) data, with focus on the role of mutPTEN in the presence of co-pathogenic variants including mutp53, mutCDKN2A, mutRB1, and mutATRX.
Materials and methods
Study population
Our dataset included a total of 602 eligible patients (297 from KPNC and 305 from Stanford Cancer Center) with grade 2 or higher locally advanced (unresectable) or metastatic STS [25]. The KPNC cohort had NGS performed using StrataNGS (Ann Arbor, Michigan) from November 2017 to June 2022 and the Stanford cohort had NGS performed from February 2015 to April 2022 (see below). Patient data on demographics, Charlson comorbidity index (CCI), performance status (PS), and receipt of systemic therapy were obtained from the electronic medical record (Epic) and cancer registry database. CCI was based on the 12-month period prior to diagnosis of locally advanced or metastatic STS. This study was approved by the KPNC and Stanford Cancer Center institutional review boards with a waiver of consent.
NGS
StrataNGS is currently a 429-gene, pan-solid tumor, NGS assay for formalin-fixed paraffin-embedded (FFPE) tumor tissue, performed on co-isolated DNA and RNA [26]. ATRX was included in StrataNGS panel in August 2020. For the Stanford cohort, NGS was performed by FoundationOne (Foundation Medicine, Cambridge, MA) [27, 28], Tempus (Tempus Labs, Inc., Chicago, IL) [28, 29], and Altera™ (Natera, Inc., Austin, TX) [30]. All platforms evaluated the most common genomic alterations of interest, including p53, CDKN2A, RB1, PTEN and ATRX. MutCDKN2A includes CDKN2A deletion, pathogenic variants, CDK4 and CCND1 amplification. Approximately 60 genes with pathogenic variants were identified. The NGS assays were performed using three multiplexed PCR-based panels (two DNA and one RNA), simultaneously assessed single nucleotide variant, short indels, short structural variants and copy number variation, and all variant classes were analyzed and subjected to independent quality control metrics and bioinformatics pipelines for reporting following the American College of Medical Genetics and Genomics (ACMG) criteria.
Histology
We included only grade 2 or higher locally advanced (unresectable) or metastatic STS (either de novo or recurrent). We excluded gastrointestinal stromal tumor, benign histology, and grade 1 histology such as well-differentiated LPS, low-grade myxoid LPS, etc. Bone sarcoma was excluded from this analysis. We classified all patients into four histology subtypes to facilitate analysis: LMS, which included uterine LMS (uLMS) and extra-uterine LMS (extra-uLMS), UPS, LPS, and other histology subtypes (Others). LPS included pleomorphic LPS, grade 2 and 3 myxoid LPS and dedifferentiated LPS. UPS included unspecified high-grade sarcoma. The histologic subtypes of Others are shown in Data Supplement (Table A1).
Treatment
Treatment included the administration of chemotherapy, targeted therapeutics (such as pazopanib) and checkpoint inhibitors after a patient was diagnosed with locally advanced or metastatic disease, either alone or in a certain combination.
Definition of hotspot TP53 pathogenic variants
We previously defined mutp53 R175H, R248Q, R248W, R249S, R273H, R273L, and R282W as gain-of-function pathogenic variants based on our literature review [31]. The literature on these pathogenic variants were predominantly in carcinoma cell lines and animal models. There is a lack of studies of these pathogenic variants in sarcoma cell lines and animal models; therefore, the functions of these pathogenic variants in sarcoma remain unclear. For this reason, in our sarcoma studies, we have included all missense pathogenic variants involving six hotspot pathogenic variant codons (R175, G245, R248, R249, R173, and W282) as “hotspot pathogenic variants” instead of gain-of-function pathogenic variants. The rest of TP53 pathogenic variants were grouped as non-hotspot pathogenic variants. There were 33 patients with hotspot pathogenic variant.
Statistical analysis
OS was measured from the date of diagnosis of locally advanced or metastatic STS to the date of death or end of study follow-up (July 28, 2022 for KPNC cohort and December 27, 2022 for Stanford cohort), whichever came first. We used Pearson’s χ2 test to assess differences in distributions of demographic and clinical factors and in p53, CDKN2A, RB1, PTEN and ATRX pathogenic variants. We used the one-way ANOVA test to assess differences in continuous variables. We used the Kaplan–Meier plot (log-rank test) to perform unadjusted (univariate) OS analysis and to estimate the median OS. The number of patients at risk in the Kaplan–Meier OS curves accounted for delayed entry into the cohort at the time of receipt of NGS results (i.e., left-truncation, with median study entry of 7.7 months post-diagnosis) [32]. Cox proportional hazards regression models were used to estimate the adjusted hazard ratio (aHR) and 95% confidence intervals (CI) for the association between pathogenic variant subsets and OS, adjusted for covariates. Time since diagnosis of advanced STS was the time scale used in the regression models, allowing for delayed entry into the cohort [32]. Covariates included in our main regression models (and unless otherwise stated) were age (continuous), sex (male, female), ethnicity (Non-Hispanic White, Black, Asian, Hispanic, other/unknown), PS (0–1, 2–4), CCI (continuous), and treatment received (yes, no). We examined the effect of specific pathogenic variants in a model that included all five pathogenic variants simultaneously: p53 [yes, no], CDKN2A [yes, no], RB1 [yes, no], PTEN (yes, no), and ATRX (yes, no, unknown) pathogenic variants, as well as four different histologic subtypes including LMS (yes, no), UPS (yes, no), LPS (yes, no), and Others (yes, no). We conducted subgroup analyses for the effect of each specific pathogenic variant with or without other specified co-pathogenic variants. The statistical analysis was performed using SAS software version 9.4, R (R Core Team, 2020).
Result
Demographic and clinical characteristics of the cohort by histology and pathogenic variant status
Mutp53 was detected in 76.4% of patients with LMS, 59.6% with UPS, 15.4% with LPS and 32.7% with Others. The percent of patients with mutCDKN2A, mutRB1, mutPTEN or mutATRX also varied by histologic subtype (Table 1). Patients with mutPTEN had higher percentages of mutRB1, mutp53, and mutCDKN2A compared to patients with wtPTEN (Table 2). Patients with mutp53 versus wtp53 were generally older and more commonly female. In addition, the histology of their tumors was more commonly LMS, had fewer mutCDKN2A but more mutPTEN, mutRB1, and mutATRX (data supplement, Table A2). The demographic characteristics of patients with hotspot vs. non-hotspot mutp53 were not substantially different (Table A3). Patients with mutCDKN2A versus wild-type CDKN2A (wtCDKN2A) were generally older, with lower percentage of females, had a higher CCI, had more UPS or LMS, and fewer mutp53, mutRB1, mutPTEN or mutATRX (Data supplement, Table A4). Patients with mutRB1 versus wild-type RB1 (wtRB1) had a higher percentage of female patients and had a better CCI, and was essentially mutually exclusive with mutCDKN2. In contrast, tumors of patients with mutRB1 had a higher percentage of mutPTEN and mutATRX (Data supplement, Table A5). The histology of tumors of patients with mutPTEN versus wild-type PTEN (wtPTEN) was more likely to be LMS and had mutRB1 and mutp53 (Data supplement, Table A5). The histology of tumors of patients with mutATRX versus wild-type ATRX (wtATRX) were more commonly LMS, and had higher percentage of mutp53 and mutRB1 but lower percentage of mutCDKN2A (Data supplement, Table A6).
Table 1.
Demographics by histologic subtypes.
| LMS (n = 174) | UPS (n = 136) | LPS (n = 78) | Others (n = 214) | P value | ||
|---|---|---|---|---|---|---|
| Median age | 59 (23–88) | 63 (22–97) | 63 (35–90) | 57 (19–94) | <0.001 | |
| Female | 133 (76.4) | 72 (52.9) | 37 (47.4) | 108 (50.5) | <0.001 | |
| Race | Asian | 35 (20.1) | 22 (16.2) | 14 (17.9) | 44 (20.6) | 0.18 |
| Black | 11 (6.3) | 5 (3.7) | 2 (2.6) | 7 (3.3) | ||
| Hispanic | 26 (14.9) | 12 (8.8) | 16 (20.5) | 39 (18.2) | ||
| White | 99 (56.9) | 89 (65.4) | 44 (56.4) | 115 (53.7) | ||
| Others | 3 (1.7) | 8 (5.9) | 2 (2.6) | 9 (4.2) | ||
| PS | 0–1 | 155 (89.1) | 110 (80.9) | 63 (80.8) | 194 (90.7) | 0.01 |
| 2–4 | 10 (5.7) | 21 (15.4) | 12 (15.4) | 13 (6.1) | ||
| Unknown | 19(5.2) | 15(3.7) | 3 (3.8) | 7 (3.3) | ||
| CCI | 1 (0–9) | 2 (0–9) | 1 (0–7) | 1 (0–7) | <0.001 | |
| Treatment | Yes | 146 (83.9) | 103 (75.7) | 52 (66.7) | 173 (80.8) | 0.01 |
| No | 28 (16.1) | 33 (24.3) | 26 (33.3) | 41 (19.2) | ||
| TP53 | wt | 41 (23.6) | 55 (40.5) | 66 (76.9) | 144 (67.3) | <0.001 |
| mut | 133 (76.4) | 81 (59.5) | 12 (23.1) | 70 (32.7) | ||
| CDKN2A | wt | 166 (95.4) | 100 (73.5) | 20 (25.6) | 177 (82.7) | <0.001 |
| mut | 8 (4.6) | 36 (26.5) | 58 (74.4) | 37 (17.3) | ||
| RB1 | wt | 96 (55.2) | 109 (80.1) | 69 (88.5) | 197 (92.1) | <0.001 |
| mut | 78 (44.8) | 27 (19.9) | 9 (11.5) | 17 (7.9) | ||
| PTEN | wt | 142 (81.6) | 124 (91.2) | 74 (94.9) | 205 (95.8) | <0.001 |
| mut | 32 (18.4) | 12 (8.8) | 4 (5.1) | 9 (4.2) | ||
| ATRX | wt | 85 (48.9) | 96 (70.6) | 52 (66.7) | 147 (68.7) | <0.001 |
| mut | 34 (19.5) | 14 (10.3) | 5 (6.4) | 14 (6.5) | ||
| unknown | 55 (31.6) | 26 (19.1) | 21 (26.9) | 53 (24.8) | ||
The number inside the parenthesis represents the percent except for median age.
PS performance status, CCI Charlson comorbidity index, LMS leiomyosarcoma, UPS undifferentiated pleomorphic sarcoma, LPS liposarcoma.
Table 2.
Demographic and clinical characteristics of patients with PTEN pathogenic variant (mutPTEN) vs wild-type PTEN (wtPTEN).
| Wt (n = 545) | Mut (n = 57) | P | ||
|---|---|---|---|---|
| Median age | 60 (19–92) | 56 (35–81) | 0.56 | |
| Female | 315 (57.8) | 35 (61.4) | 0.61 | |
| Race | Asian | 106 (19.4) | 9 (15.8) | 0.06 |
| Black | 19 (3.5) | 6 (10.5) | ||
| Hispanic | 84 (15.4) | 9 (15.8) | ||
| White | 314 (57.5) | 33 (57.9) | ||
| Others | 22 (4.0) | 0 | ||
| PS | 0–1 | 474 (87.0) | 48 (84.2) | 0.71 |
| 2–4 | 49 (9.0) | 7 (12.3) | ||
| Unknown | 22 (4.0) | 2 (3.5) | ||
| CCI | 1 (0–9) | 2 (0–6) | 0.07 | |
| Treatment | Yes | 429 (78.7) | 45 (78.9) | 0.97 |
| No | 116 (21.3) | 12 (21.2) | ||
| Histology | LMS | 142 (26.1) | 32 (56.1) | <0.001 |
| UPS | 124 (22.8) | 12 (21.1) | ||
| LPS | 74 (13.6) | 4 (7.0) | ||
| Others | 205 (37.6) | 9 (15.8) | ||
| RB1 | wt | 440 (80.7) | 31 (54.4) | <0.001 |
| mut | 105 (19.3) | 26 (45.6) | ||
| TP53 | wt | 294 (53.9) | 12 (21.1) | <0.001 |
| mut | 251 (46.1) | 45 (78.9) | ||
| CDKN2A | wt | 410 (75.2) | 53 (93.0) | 0.002 |
| mut | 135 (24.8) | 4 (7.0) | ||
| ATRX | wt | 349 (64.0) | 31 (54.4) | 0.30 |
| mut | 58 (10.6) | 9 (15.8) | ||
| unknown | 138 (25.3) | 17 (29.8) | ||
The number inside the parenthesis represents the percent except for median age.
PS performance status, CCI Charlson comorbidity index, LMS leiomyosarcoma, UPS undifferentiated pleomorphic sarcoma, LPS liposarcoma.
MutPTEN vs wtPTEN was associated with a worse prognosis in the full cohort
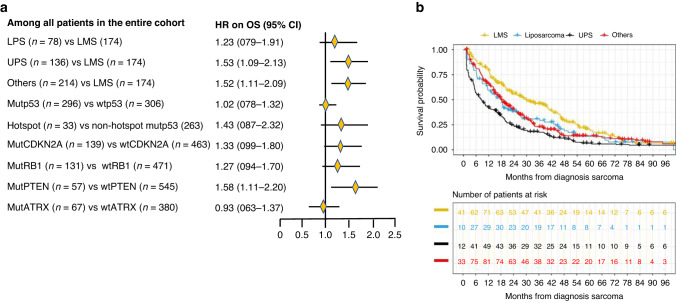
When examining single genes, OS appeared to be worse for patients with mutPTEN versus wtPTEN (aHR = 1.58 [95% CI, 1.11–2.23]), and for patients with mutCDKN2A versus wtCDKN2A (aHR = 1.33 [95% CI 0.99–1.80]), mutRB1 versus wtRB1 (aHR = 1.26 [95% CI 0.93–1.70]). In contrast, mutATRX versus wtATRX did not appear to be associated with worse OS (aHR = 0.93 [95% CI 0.63–1.37]) (Fig. 1a). While mutp53 versus wtp53 was also not associated with OS (aHR = 1.02 [95% CI 0.78–1.32]), hotspot versus non-hotspot mutp53 did appear to be associated with worse OS (aHR = 1.43 [95% CI 0.87–2.32]) (Fig. 1a).
Fig. 1.
a Forest plot of adjusted hazard ratios of OS associated with histologic subtypes and for wild-type vs pathogenic variant comparisons in single genes. aHR adjusted hazard ratio, OS overall survival, LMS leiomyosarcoma, UPS undifferentiated pleomorphic sarcoma, LPS liposarcoma, Others other histology subtypes than LMS, UPS and LPS, Mut pathogenic variant, Wt wild-type. b Kaplan–Meyer OS curves of four different histology subtypes. Histology subtypes include uLMS and extra-uLMS; UPS undifferentiated pleomorphic sarcoma, Liposarcoma (including dedifferentiated liposarcoma, pleomorphic liposarcoma, and grade 2 and 3 myxoid liposarcoma), Others (including all other grade 2 and 3 histology subtypes than LMS, UPS, or liposarcoma). The number of patients at risk accounted for left-truncation. Patients who were still alive by the data lock date were censored.
Compared to patients with LMS, OS appeared to be worse for patients with LPS (aHR = 1.22 [95% CI 0.78–1.91]), UPS (aHR = 1.53 [95% CI 1.11–2.14]) or with Other subtypes (aHR = 1.52 [95% CI 1.11–2.14]). (Fig. 1a). Median OS was 9.2 months for UPS, 15.4 months for LS, 14.4 for Other subtypes and 29.5 months for LMS, respectively (Fig. 1b).
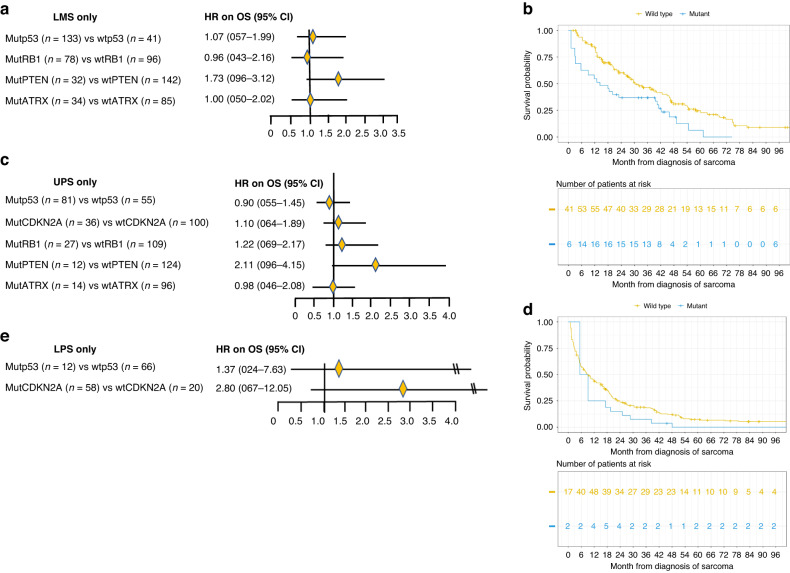
MutPTEN vs wtPTEN was associated with worse OS in LMS and UPS
LMS
Among patients with LMS, mutPTEN versus wtPTEN was associated with substantially worse OS (aHR = 1.73 [95% CI 0.96–3.12]) (Fig. 2a), with median OS of 14 versus 32 months (Fig. 2b), while mutp53 versus wtp53 (aHR = 1.07 [95% CI 0.57–1.99]), mutRB1 versus wtRB1 (aHR = 0.96 [95% CI 0.43–2.16]), mutATRX versus wtATRX (aHR = 1.00 [95% CI 0.50–2.02]) were not associated with substantial OS difference.
Fig. 2.
a Forest plot of hazard ratios of OS for patients with leiomyosarcoma (LMS). aHR adjusted hazard ratio, OS overall survival, uLMS uterine leiomyosarcoma, extra-uLMS extra-uterine leiomyosarcoma, Mut pathogenic variant, Wt wild-type. b Kaplan–Meir OS curves of LMS patients with mutPTEN versus with wtPTEN. The number of patients at risk accounted for left-truncation. Patients who were still alive by the data lock date were censored. c Forest plot of hazard ratios of OS for patients with undifferentiated pleomorphic sarcoma (UPS). aHR adjusted hazard ratio, OS overall survival, Mut pathogenic variant, Wt wild-type. d Kaplan–Meir OS curves of UPS patients with mutPTEN vs with wtPTEN. The number of patients at risk accounted for left-truncation. Patients who were still alive by the data lock date were censored. e Forest plot of adjusted hazard ratios of OS for patients with liposarcoma (LPS). aHR adjusted hazard ratio, OS overall survival, Mut pathogenic variant, Wt wild-type.
UPS
Among patients with UPS, OS with mutPTEN versus wtPTEN (HR = 2.11 [95% CI 0.96–4.15]) was substantially worse (Fig. 2c), with a median OS of 5.5 versus 9.0 months (Fig. 2d). However, the sample was small. OS with mutp53 versus wtp53 (aHR = 0.90 [95% CI 0.55–1.45]), OS with mutCDKN2A versus wtCDKN2A (aHR = 1.10 [95% CI 0.64–1.89]), mutRB1 versus wtRB1 (aHR = 1.22 [95% CI 0.69–2.17]), mutATRX versus wtATRX (HR = 0.98 [95% CI 0.46–2.08]) were not substantially different.
LPS
Among patients with LPS, OS with mutp53 versus wtp53 appears slightly worse (aHR = 1.37 [0.24–7.63]) but the sample size with mutp53 was small (n = 12) (Fig. 2e). OS with mutCDKN2A versus wtCDKN2A was worse (aHR = 2.80 [95% CI 0.67–12.05]) (Fig. 2e). The sample size with mutRB1, mutPTEN and mutATRX was too small to allow meaningful analysis.
Others
Among patients with Others, OS with mutPTEN versus wtPTEN (aHR = 1.50 [95% CI 0.68–3.31]) was worse, and OS with mutp53 versus wtp53 was also worse (aHR = 1.47 [95% CI 0.91–2.39]). OS with mutCDKN2A versus wtCDKN2A (aHR = 0.99 [95% CI 0.61–1.59]) and mutRB1 versus wtRB1 (aHR = 0.95 [95% CI 0.42–1.11]) were not substantially different, and OS with mutATRX versus wtATRX (aHR = 0.59 [95% CI 0.23–1.49]) was better but had small sample size (Data supplement, Fig. A1).
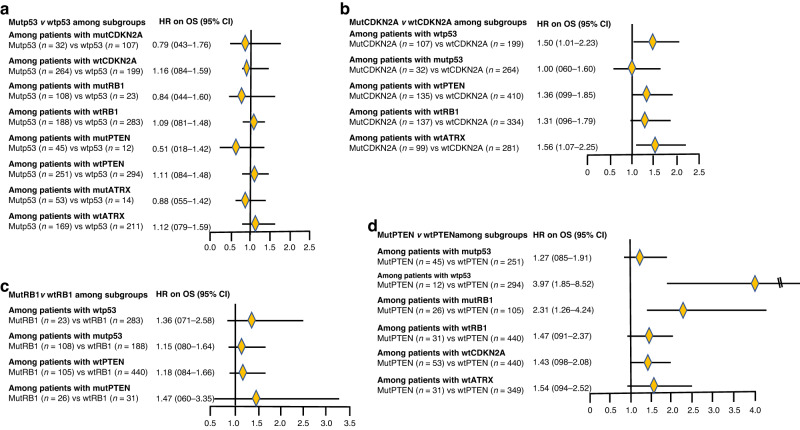
Association of mutp53 vs wtp53 with OS among subgroups based on co-pathogenic variants
Although many subgroups were small, there was a suggestion that mutp53 versus wtp53 was associated with modestly better OS among patients with mutCDKN2A (aHR = 0.79 [95% CI 0.43–1.76]) but not with wtCDKN2A (aHR = 1.16 [95% CI 0.84–1.59]), and among patients with mutRB1 (aHR = 0.84 [95% CI 0.44–1.60]) but not among patients with wtRB1 (HR = 1.09 [95% CI 0.81–1.48]), and among patients with mutPTEN (aHR = 0.51 [95% CI 0.18–1.42]) but not among patients with wtPTEN (aHR = 1.11 [95% CI 0.84–1.48]), and among patients with mutATRX (aHR = 0.88 p95% CI 0.55–1.42]) but not among patients with wtATRX (aHR = 1.12 [95% CI 0.79–1.59]) (Fig. 3a).
Fig. 3.
a Forest plot of adjusted hazard ratios of OS for patients with mutp53 versus wtp53 among subgroups. aHR adjusted hazard ratio, OS overall survival, Mut pathogenic variant, Wt wild-type. b Forest plot of adjusted hazard ratios of OS for patients with mutCDKN2A versus wtCDKN2A among subgroups. aHR adjusted hazard ratio, OS overall survival, Mut pathogenic variant, Wt wild-type. c Forest plot of adjusted hazard ratios of OS for patients with mutRB1 versus wtRB1 among subgroups. aHR adjusted hazard ratio, OS overall survival, Mut pathogenic variant, Wt wild-type. d Forest plot of adjusted hazard ratios of OS for patients with mutPTEN versus wtPTEN among subgroups. aHR adjusted hazard ratio, OS overall survival, Mut pathogenic variant, Wt wild-type.
Association of mutCDKN2A vs. wtCDKN2A with OS among subgroups based on co-pathogenic variants
MutCDKN2A versus wtCDKN2A was associated with substantially worse OS in among patients with wtp53 (aHR = 1.50 [95% CI 1.01–2.23]), among patients with wtPTEN (aHR = 1.36 [95% CI 0.99–1.85]), among patients with wtRB1 (aHR = 1.31 [95% CI 0.96–1.79), and among patients with wtATRX (aHR = 1.56 [95% CI 1.07–2.25]) (Fig. 3b). but not among patients with mutp53 (aHR = 1.00 [95% CI 0.60–1.60]) (Fig. 3b).
Association of mutRB1 vs wtRB1 with OS among subgroups based on co-pathogenic variants
MutRB1 versus wtRB1 was associated with modestly worse OS among patients with wtp53 (aHR = 1.36 [95% CI 0.71–2.58]) and among patients with mutPTEN (aHR = 1.47 [95% CI 0.60–3.35]), but slightly worse OS among patients with mutp53 (aHR = 1.15 [95% CI 0.80–1.64]), and among patients with wtPTEN (aHR = 1.18 [95% CI 0.84–1.66]) (Fig. 3c).
Association of mutPTEN vs wtPTEN with OS among subgroups based on co-pathogenic variants
MutPTEN versus wtPTEN was associated with slightly worse OS among patients with mutp53 (aHR = 1.27 [95% CI 0.85–1.91]) but substantially worse OS among patients with wtp53 (aHR = 3.97 [95% CI 1.85–8.52]) (Fig. 3d). OS with mutPTEN versus wtPTEN was substantially worse among patients with mutRB1 (aHR = 2.31 [95% CI 1.26–4.24]), and worse among patients with wtRB1 (aHR = 1.47 [95% CI 0.91–2.37]) and among patients with wtCDKN2A (aHR = 1.43 [95% CI 0.98–2.08]) and among patients with wtATRX (aHR = 1.54 [95% CI 0.94–2.52]) (Fig. 3d).
Discussion
In this study using the combined KPNC and Stanford cohort of 602 patients with advanced STS, we have shown that mutPTEN vs wtPTEN was associated with worse OS. We have also shown that histologically LMS had the best while UPS had the worst OS. It appeared that hotspot versus non-hotspot mutp53 but not mutp53 versus wtp53 was associated with worse OS; In addition, there was a suggestion that mutp53 versus wtp53 was associated with favorable OS among subgroups with mutCDKN2A, mutRB1, mutPTEN and mutATRX than among subgroups with wtCDKN2A, wtRB1, wtPTEN and wtATRX; Also, mutCDKN2A versus wtCDKN2A, mutRB1 versus wtRB1 and mutPTEN versus wtPTEN were associated with worse OS primarily among patients with wtp53 but not among patients with mutp53.
MutPTEN is a common pathogenic variant in advanced STS. To our knowledge, our study is the first to reveal that mutPTEN versus wtPTEN was associated with worse OS. MutPTEN versus wtPTEN was consistently associated with worse OS among the entire cohort, among histologic subtypes including LMS and UPS and among several molecular subgroups including patients with wtp53, wtCDKN2A, wtATRX, and mutRB1. This finding should be helpful for clinical practice in stratifying prognosis and potentially be helpful in subgroup stratification in clinical trials. Our findings are consistent with some of the previous reports on PTEN pathogenic variant and prognosis in other malignancies [33–35].
The worse OS associated with hotspot versus non-hotspot mutp53 appears to be consistent with our previous studies in metastatic colorectal cancer (CRC) and in advanced pancreatic ductal adenocarcinoma (PDAC) [31, 36]. However, the attenuation of the adverse OS effect of mutCDKN2A, mutRB1 and mutPTEN by mutp53 was unexpected and intriguing, though independently for others, mutp53 was associated with substantially worse OS. These results suggest that different advanced STS may be driven by driver pathogenic variant and co-pathogenic variants differently during the evolution of disease despite similar pathogenic variant profiles being present; some may adopt p53 pathogenic variant as a driver pathogenic variant and PTEN pathogenic variant as a co-pathogenic variant of passenger in nature, while some may adopt PTEN (or RB1, or CDKN2A or others) pathogenic variant as a driver pathogenic variant while p53 pathogenic variant as a subsequent pathogenic variant acting more of a passenger pathogenic variant, resulting in diverse biology with prognostic biases. Studies in STS on the association of mutp53 with prognosis have been limited. In the pooled analysis of MOSCATO and ProfiLER precision trials in sarcoma, mutp53 was found to be associated with worse DFS, but better response to anthracycline chemotherapy [20]. A previous study showed worse OS with mutp53 versus wtp53 in patients with metastatic sarcoma based on univariate analysis with a small sample size [21]. A small retrospective study with only 19 patients showed that mutp53 was predictive of response to pazopanib in patients with advanced STS.33 We speculate that certain TP53 pathogenic variants may present a vulnerability in STS for treatment targeting, similar to some of the well-characterized genomic lesions that have been targeted [37–41]. This notion appears consistent with the above-referenced studies showing that mutp53 was associated with a better response to doxorubicin and pazopanib [20, 33]. Certain chemical agents such as arsenic trioxide have been shown to be capable of restoring the conformation of multiple different p53 mutants into their original tumor suppressive function, indicating that mutant p53 proteins could be molecularly “differentiated” or “rescued” [42–45]. In our previous study in patients with metastatic CRC, mutp53 overall was not associated with prognosis, similar to our current finding in advanced STS; however, hotspot mutp53 was associated with worse OS than non-hotspot mutp53 in left-sided CRC (LCC) but not in right-sided CRC (RCC), while RCC was associated with worse OS than LCC only in patients with non-hotspot TP53. Our current results with advanced STS appear consistent with our previous findings in metastatic CRC as RCC and LCC possess distinct histologic characteristics [31]. The mechanisms of such a co-pathogenic variant-dependent OS differential would be interesting to investigate. This may be explored by functional studies and in prospective trials.
Our results showing that OS of mutCDKN2A versus wtCDKN2A and OS of mutPTEN versus wtPTEN were worse among patients whose STS retained wtp53 but not among patients whose tumor harbored mutp53 are also consistent with the results that mutp53 versus wtp53 was associated with better OS among patients with mutCDKN2A, mutRB1 and mutPTEN. This suggests that mutp53 could be functionally capable of overcoming the adverse effect by mutCDKN2A or mutPTEN in certain aspects. This would be quite intriguing if confirmed in additional studies as mutCDKN2A and mutPTEN would usually be considered cooperative with mutp53 to exacerbate cancer progression [30, 46–49], though better outcomes associated with mutp53 over wtp53 have been presented in some previous studies. In a study on patients with non-small cell lung cancer (NSCLC) bearing an STK11 pathogenic variant (mutSTK11), it was found that mutp53 conferred better OS over wtp53 [50]. In a separate study on patients with NSCLC bearing a mutSTK11, it was found that mutp53 was associated with better PFS and immunologically metabolic reprogramming compared to wtp53 [51]. Experimentally, a recent study showed that wtp53 could promote mitotic bypass and whole genome duplication under cyclin E-induced replicative stress, suggesting that wtp53 could exacerbate cancer progression under certain biological circumstances [52]. This finding brought about a novel concept on the roles of TP53 that warrants further investigations in animal and human studies.
Our study has several strengths. To our knowledge, our sample size is the largest compared to all the published STS genomic studies cohorts. In addition, our cohort includes patients from two large institutions with multi-specialty care centers. Furthermore, our cohort included the five most commonly mutated genes and the four most common histology subtypes in STS. We used Cox regression modeling to adjust for several patient and clinical variables. Our study also has limitations. First, it is a retrospective study and some patients did not have NGS performed at the time of diagnosis of advanced disease until months later. Nonetheless, we used appropriate statistical methods to address this issue [32]. Second, only approximately two-third of patients had known ATRX pathogenic variant status. Third, the sample size of some subgroups was small and analyses should be considered exploratory. Fourth, the results with Others could be more challenging to interpret as it is comprised of heterogeneous STS subtypes.
In summary, our study suggests that mutPTEN was associated with worse OS compared to wtPTEN and mutp53 appeared to attenuate the adverse OS effect by mutCDKN2A and mutPTEN co-pathogenic variants in advanced STS. If confirmed, our results provide new insight for further understanding the molecular mechanisms of STS and may improve prognostic stratifications in clinical practice and future clinical trial designs based on molecular subgroups and histologic subtypes.
Supplementary information
Acknowledgements
We thank M. van de Rijin, E. Moding and A. Kalbasi for helpful comments.
Author contributions
MP, KG, MYZ, CJ and LAH conceived the concept and designed the study and methodology. MP, CJ, MYZ, ZZ, JB, AS, NA, PT, AVS, and EC provided materials and performed data collection. MP, CJ, MYZ and ZZ performed data analysis. MP, MYZ, KG, WH, ST, and LAH wrote and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by The Permanente Medical Group and Stanford University and by Jiayan Foundation. No external funding was obtained for this study.
Data availability
Kaiser Permanente Northern California (KPNC) Institutional Review Board has not provided approval for StrataNGS data on individual patients used in this study to be placed in a public access repository. However, researchers can request access to use this study data by contacting the DOR Data Sharing Workgroup at DOR-DataSharingWorkgroup@kp.org.
Competing interests
MP reports advisory roles in Aadi Bioscience and Boehringer Ingelheim. The other authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Stanford University and Kaiser Permanente (protocol code 1710436–3 and date of approval April 3, 2023). Patient consent was waived by the Institutional Review Board.
Consent for publication
All authors have read and agreed to the published version of the manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Minggui Pan, Maggie Y. Zhou.
Supplementary information
The online version contains supplementary material available at 10.1038/s44276-023-00029-3.
References
- 1.Gamboa AC, Gronchi A, Cardona K. Soft-tissue sarcoma in adults: an update on the current state of histiotype-specific management in an era of personalized medicine. CA Cancer J Clin. 2020;70:200–29. 10.3322/caac.21605 [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–65.e28. 10.1016/j.cell.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nacev BA, Sanchez-Vega F, Smith SA, Antonesc CR, Rosenbarm E, Shi H, et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat Commun. 2022;13:3405. 10.1038/s41467-022-30453-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretina J, Taylor BS, Banerji S, Ramo AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–21. 10.1038/ng.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. 10.1016/j.cell.2013.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadoch C, Hargreaves DC, Hodges C, Elia L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seddon B, Strauss SJ, Whelan J, Leahy M, Woll P, Cowie F, et al. Gemcitabine and docetaxel versus doxorubicin as first-line treatment in previously untreated advanced unresectable or metastatic soft-tissue sarcomas (GeDDiS): a randomised controlled phase 3 trial. Lancet Oncol. 2017;18:1397–410. 10.1016/S1470-2045(17)30622-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan M, Trieu MK, Sidhu M, Yu J, Seto T, Ganjoo K. Fourteen-day gemcitabine-docetaxel chemotherapy is effective and safer compared to 21-day regimen in patients with advanced soft tissue and bone sarcoma. Cancers (Basel). 2021;13:1983. 10.3390/cancers13081983 [DOI] [PMC free article] [PubMed]
- 9.Demetri GD, von Mehren M, Jones RL, Hensley M, Schuietze SM, Staddon A, et al. Efficacy and safety of trabectedin or dacarbazine for metastatic liposarcoma or leiomyosarcoma after failure of conventional chemotherapy: results of a phase III randomized multicenter clinical trial. J Clin Oncol. 2016;34:786–93. 10.1200/JCO.2015.62.4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Graaf WT, Blay JY, Chawla SP, Kim B, Bui-Nguyen B, Casali P, et al. Pazopanib for metastatic soft-tissue sarcoma (PALETTE): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet. 2012;379:1879–86. 10.1016/S0140-6736(12)60651-5 [DOI] [PubMed] [Google Scholar]
- 11.Schoffski P, Chawla S, Maki RG, Italiano A, Gelderblom H, Choy E, et al. Eribulin versus dacarbazine in previously treated patients with advanced liposarcoma or leiomyosarcoma: a randomised, open-label, multicentre, phase 3 trial. Lancet. 2016;387:1629–37. 10.1016/S0140-6736(15)01283-0 [DOI] [PubMed] [Google Scholar]
- 12.Zhou M, Bui N, Bolleddu S, Lohman M, Becker HC, Ganjoo K. Nivolumab plus ipilimumab for soft tissue sarcoma: a single institution retrospective review. Immunotherapy. 2020;12:1303–12. 10.2217/imt-2020-0155 [DOI] [PubMed] [Google Scholar]
- 13.Wilky BA, Trucco MM, Subhawong TK, Fluoro V, Park W, Kwon D, et al. Axitinib plus pembrolizumab in patients with advanced sarcomas including alveolar soft-part sarcoma: a single-centre, single-arm, phase 2 trial. Lancet Oncol. 2019;20:837–48. 10.1016/S1470-2045(19)30153-6 [DOI] [PubMed] [Google Scholar]
- 14.Tawbi HA, Burgess M, Bolejack V, van Tine BA, Schetze M, Hu J, et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet Oncol. 2017;18:1493–501. 10.1016/S1470-2045(17)30624-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saerens M, Brusselaers N, Rottey S, Decruyenaere A, Creytens D, Lapeire L. Immune checkpoint inhibitors in treatment of soft-tissue sarcoma: a systematic review and meta-analysis. Eur J Cancer. 2021;152:165–82. 10.1016/j.ejca.2021.04.034 [DOI] [PubMed] [Google Scholar]
- 16.Italiano A, Bellera C, D’Angelo S. PD1/PD-L1 targeting in advanced soft-tissue sarcomas: a pooled analysis of phase II trials. J Hematol Oncol. 2020;13:55. 10.1186/s13045-020-00891-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Florou V, Rosenberg AE, Wieder E, Komanduri KV, Kolonias D, Uduman M, et al. Angiosarcoma patients treated with immune checkpoint inhibitors: a case series of seven patients from a single institution. J Immunother Cancer. 2019;7:213. 10.1186/s40425-019-0689-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravi V, Subramaniam A, Zheng J, Amini B, Trinh BA, Joseph J, et al. Clinical activity of checkpoint inhibitors in angiosarcoma: a retrospective cohort study. Cancer. 2022;128:3383–91. 10.1002/cncr.34370 [DOI] [PubMed] [Google Scholar]
- 19.Wagner MJ, Othus M, Patel SP, Ryan C, Sangal A, Powers, B, et al. Multicenter phase II trial (SWOG S1609, cohort 51) of ipilimumab and nivolumab in metastatic or unresectable angiosarcoma: a substudy of dual anti-CTLA-4 and anti-PD-1 blockade in rare tumors (DART). J Immunother Cancer. 2021;9:e002990. 10.1136/jitc-2021-002990 [DOI] [PMC free article] [PubMed]
- 20.Nassif EF, Auclin E, Bahleda R, Honore C, Mir O, Dumont S, et al. TP53 mutation as a prognostic and predictive marker in sarcoma: pooled analysis of MOSCATO and ProfiLER precision medicine trials. Cancers (Basel). 2021;13:3362. 10.3390/cancers13133362 [DOI] [PMC free article] [PubMed]
- 21.Ohnstad HO, Castro R, Sun J, Heintz K, Vassilev LT, Berkehagen B, et al. Correlation of TP53 and MDM2 genotypes with response to therapy in sarcoma. Cancer. 2013;119:1013–22. 10.1002/cncr.27837 [DOI] [PubMed] [Google Scholar]
- 22.Bui NQ, Przybyl J, Trabucco SE, Framptom G, Hastie T, van de Rijn M, et al. A clinico-genomic analysis of soft tissue sarcoma patients reveals CDKN2A deletion as a biomarker for poor prognosis. Clin Sarcoma Res. 2019;9:12. 10.1186/s13569-019-0122-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voss RK, Callegaro D, Chiang YJ, Fiore M, Miceli R, Keung EZ, et al. Sarculator is a good model to predict survival in resected extremity and trunk sarcomas in US patients. Ann Surg Oncol. 2022. 10.1245/s10434-022-11442-2 [DOI] [PubMed]
- 24.Pasquali S, Palmerini E, Quagliuolo V, Martin-Broto J, Lopez-Pousa A, Grignani G, et al. Neoadjuvant chemotherapy in high-risk soft tissue sarcomas: a Sarculator-based risk stratification analysis of the ISG-STS 1001 randomized trial. Cancer. 2022;128:85–93. 10.1002/cncr.33895 [DOI] [PubMed] [Google Scholar]
- 25.Pan M, Zhou MY, Jiang C, Zhang Z, Bui N, Bien J, et al. Sex-dependent prognosis of patients with advanced soft tissue sarcoma. Clin Cancer Res. 2023. 10.1158/1078-0432.CCR-23-1990 [DOI] [PMC free article] [PubMed]
- 26.Tomlins SA, Hovelson DH, Harms P, Drewery S, Falkner J, Fischer A, et al. Development and validation of StrataNGS, a multiplex PCR, semiconductor sequencing-based comprehensive genomic profiling test. J Mol Diagn. 2021;23:1515–33. 10.1016/j.jmoldx.2021.08.005 [DOI] [PubMed] [Google Scholar]
- 27.Takeda M, Takahama T, Sakai K, Shimizu S, Watanabe S, Kawakami H, et al. Clinical application of the FoundationOne CDx assay to therapeutic decision-making for patients with advanced solid tumors. Oncologist. 2021;26:e588–96. 10.1002/onco.13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng J, Johnson A, Shufean MA, Khale M, Yang D, Woodman S, et al. Operationalization of next-generation sequencing and decision support for precision oncology. JCO Clin Cancer Inform. 2019;3:1–12. 10.1200/CCI.19.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaubier N, Tell R, Lau D, Parsons J, Bush S, Perera J, et al. Clinical validation of the tempus xT next-generation targeted oncology sequencing assay. Oncotarget. 2019;10:2384–96. 10.18632/oncotarget.26797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White T, Szelinger S, LoBello J, King A, Aldrich J, Garinger N, et al. Analytic validation and clinical utilization of the comprehensive genomic profiling test, GEM ExTra((R)). Oncotarget. 2021;12:726–39. 10.18632/oncotarget.27945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan M, Jiang C, Tse P, Achacoso N, Alexeeff S, Solorzano AV, et al. TP53 gain-of-function and non-gain-of-function mutations are differentially associated with sidedness-dependent prognosis in metastatic colorectal cancer. J Clin Oncol. 2022;40:171–9. 10.1200/JCO.21.02014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernan MA, Sauer BC, Hernandez-Diaz S, Platt R, Shrier I. Specifying a target trial prevents immortal time bias and other self-inflicted injuries in observational analyses. J Clin Epidemiol. 2016;79:70–5. 10.1016/j.jclinepi.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vidotto T, Melo CM, Castelli E, Koti M, Dos Reis RB, Squire JA. Emerging role of PTEN loss in evasion of the immune response to tumours. Br J Cancer. 2020;122:1732–43. 10.1038/s41416-020-0834-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidotto T, Melo CM, Lautert-Dutra W, Chaves LP, Reis RB, Squire JA. Pan-cancer genomic analysis shows hemizygous PTEN loss tumors are associated with immune evasion and poor outcome. Sci Rep. 2023;13:5049. 10.1038/s41598-023-31759-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricci R, Maggiano N, Castri F, Rinelli A, Murazio M, Pacelli F, et al. Role of PTEN in gastrointestinal stromal tumor progression. Arch Pathol Lab Med. 2004;128:421–5. 10.5858/2004-128-421-ROPIGS [DOI] [PubMed] [Google Scholar]
- 36.Pan M, Jiang C, Zhang Z, Achacoso N, Alexeeff S, Solorzano AV, et al. TP53 gain-of-function and non-gain-of-function mutations are associated with differential prognosis in advanced pancreatic ductal adenocarcinoma. JCO Precis Oncol. 2023;7:e2200570. 10.1200/PO.22.00570 [DOI] [PubMed] [Google Scholar]
- 37.Lamm W, Natter C, Schur S, Kostler WJ, Reinthaller A, Krainer M, et al. Distinctive outcome in patients with non-uterine and uterine leiomyosarcoma. BMC Cancer. 2014;14:981. 10.1186/1471-2407-14-981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koehler K, Liebner D, Chen JL. TP53 mutational status is predictive of pazopanib response in advanced sarcomas. Ann Oncol. 2016;27:539–43. 10.1093/annonc/mdv598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan M, Ganjoo K, Karam A. Rapid response of a BRCA2/TP53/PTEN-deleted metastatic uterine leiomyosarcoma to olaparib: a case report. Perm J. 2021;25:20.251. 10.7812/TPP/20.251 [DOI] [PMC free article] [PubMed]
- 40.Turner NC, Huang X, Cristofanilli M. Palbociclib and fulvestrant in breast cancer. Reply. N Engl J Med. 2019;380:797. 10.1056/NEJMc1816595 [DOI] [PubMed] [Google Scholar]
- 41.Turner NC, Slamon DJ, Ro J, Bondarenko I, Im S, Masuda N, et al. Overall survival with palbociclib and fulvestrant in advanced breast cancer. N Engl J Med. 2018;379:1926–36. 10.1056/NEJMoa1810527 [DOI] [PubMed] [Google Scholar]
- 42.Sledge GW Jr., Frenzel M. Analysis of overall survival benefit of abemaciclib plus fulvestrant in hormone receptor-positive, ERBB2-negative breast cancer–reply. JAMA Oncol. 2020;6:1122–3. 10.1001/jamaoncol.2020.1518 [DOI] [PubMed] [Google Scholar]
- 43.Sledge GW Jr., Toi M, Neven P, Sohn J, Inoue K, Pivot X, et al. MONARCH 2: abemaciclib in combination with fulvestrant in women with HR+/HER2- advanced breast cancer who had progressed while receiving endocrine therapy. J Clin Oncol. 2017;35:2875–84. 10.1200/JCO.2017.73.7585 [DOI] [PubMed] [Google Scholar]
- 44.Chen S, Wu JL, Liang Y, Tang Y, Song H, Wu L, et al. Arsenic trioxide rescues structural p53 mutations through a cryptic allosteric site. Cancer Cell. 2021;39:225–39.e8. 10.1016/j.ccell.2020.11.013 [DOI] [PubMed] [Google Scholar]
- 45.Gummlich L. ATO stabilizes structural p53 mutants. Nat Rev Cancer. 2021;21:141. 10.1038/s41568-021-00337-1 [DOI] [PubMed] [Google Scholar]
- 46.Malarikova D, Berkova A, Obr A, Blahovkova B, Swaton M, Forstrova K, et al. Concurrent TP53 and CDKN2A gene aberrations in newly diagnosed mantle cell lymphoma correlate with chemoresistance and call for innovative upfront therapy. Cancers (Basel). 2020;12:2120. 10.3390/cancers12082120 [DOI] [PMC free article] [PubMed]
- 47.Reis GF, Pekmezci M, Hansen HM, Rice T, Marshall R, Molinano AM, et al. CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II-III) astrocytomas. J Neuropathol Exp Neurol. 2015;74:442–52. 10.1097/NEN.0000000000000188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng H, Ying H, Yan H, Kimmelman AC, Hillman DJ, Chen A, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–33. 10.1038/nature07443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng H, Ying H, Yan H, Kimmelman AC, Hillman DJ, Chen A, et al. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb Symp Quant Biol. 2008;73:427–37. 10.1101/sqb.2008.73.047 [DOI] [PubMed] [Google Scholar]
- 50.Naqash AR, Floudas CS, Maoz A, Xu J, Baca Y, Zeng J, et al. STK11/TP53 co-mutated non-small cell lung cancer (NSCLC) to display a unique tumor microenvironment (TME) and metabolic profile. J Clin Oncol. 2021;39:9087. 10.1200/JCO.2021.39.15_suppl.9087 [Google Scholar]
- 51.Bange E, Marmarelis ME, Hwang WT, Yang, Y, Thompson JC, Rosenbarm J, et al. Impact of KRAS and TP53 co-mutations on outcomes after first-line systemic therapy among patients with STK11-mutated advanced non-small-cell lung cancer. JCO Precis Oncol. 2019;3:PO.18.00326. 10.1200/PO.18.00326 [DOI] [PMC free article] [PubMed]
- 52.Zeng J, Hills SA, Ozono E, Diffley JFX. Cyclin E-induced replicative stress drives p53-dependent whole-genome duplication. Cell. 2023;186:528–42.e14. 10.1016/j.cell.2022.12.036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Kaiser Permanente Northern California (KPNC) Institutional Review Board has not provided approval for StrataNGS data on individual patients used in this study to be placed in a public access repository. However, researchers can request access to use this study data by contacting the DOR Data Sharing Workgroup at DOR-DataSharingWorkgroup@kp.org.