Abstract
Regulation of protein expression can be achieved through destruction of proteins by the 26S proteasome. Cellular processes that are regulated by proteolysis include cell cycle progression, stress responses and differentiation. Several nucleotide excision repair proteins in yeast and humans, such as Rad23, Rad4 and XPB, have been shown to co-purify with Cim3 and Cim5, AAA ATPases of the 19S proteasome regulatory subunit. However, it has not been determined if nucleotide excision repair is regulated through protein destruction. We measured nucleotide excision repair in yeast mutants that are defective in proteasome function and found that the repair of the transcribed and non-transcribed strands of an RNA polymerase II-transcribed reporter gene was increased in the absence of proteasome function. Additionally, overexpression of the Rad4 repair protein, which is bound to the repair/proteolytic factor Rad23, conferred higher rates of nucleotide excision repair. Based on our data we suggest that a protein (or proteins) involved in nucleotide excision repair or in regulation of repair is degraded by the 26S proteasome. We propose that decreased proteasome function enables increased DNA repair, due to the transient accumulation of a specific repair factor, perhaps Rad4.
INTRODUCTION
Nucleotide excision repair is a pathway, conserved from yeast to humans, that removes many bulky chemical adducts and UV-induced photoproducts from DNA in a relatively error-free manner. This pathway is evolutionarily conserved and defects in this process in humans are associated with increased incidence of cancer. Identification and cloning of genes involved in nucleotide excision repair, as well as expression of their products, has enabled the reconstitution of repair in vitro with purified proteins (1,2). Nucleotide excision repair is a multistep process by which DNA damage is recognized and incisions are made such that the damage is removed as part of an oligonucleotide. The resulting gap is filled in by DNA polymerases δ and ɛ, and the 3′ end of the newly synthesized DNA is covalently attached to the flanking parental DNA by DNA ligase. The entire process in eukaryotes requires approximately 30 different proteins (3).
The expression of several proteins involved in nucleotide excision repair is induced following exposure to DNA damage (4–7), presumably to hasten removal of DNA adducts. In addition, levels of some of these proteins increase during the meiotic cell cycle (4,5,8,9). However, protein levels do not fluctuate during the mitotic cell cycle in the absence of DNA damage (8,9). Although it is not known if nucleotide excision repair is regulated by the ubiquitin-mediated protein degradation pathway, several DNA repair proteins have been linked to the proteolytic pathway (10,11).
The ubiquitin-mediated protein degradation pathway is involved in many cellular processes, such as cell cycle progression, stress response and cell differentiation (12). Like the nucleotide excision repair pathway, ubiquitin-mediated protein degradation is conserved from yeast to humans. Ubiquitin is activated by ubiquitin-activating enzyme (Uba/E1) and is transferred to ubiquitin-conjugating enzymes (Ubc/E2) and ubiquitin protein ligases (E3) through a series of transesterification reactions. Ubiquitin is ultimately attached to lysine residues in target proteins through the formation of isopeptide bonds. Additional specificity of the ubiquitin system resides in ubiquitin protein ligases (E3). Once the first ubiquitin is attached to a protein, ubiquitin chain assembly factors (E4) may promote the assembly of multiubiquitin chains, which target the protein to the 26S proteasome (13,14).
The 26S proteasome consists of two different subunits—the 19S regulatory subunit and the 20S catalytic core subunit (12). The 20S subunit is a barrel-shaped protein complex with the catalytic sites on the interior surface. Narrow ports at each end may regulate access to the interior. Astride each end of the 20S core is a 19S regulatory subunit. Proteins are delivered to the 19S subunit to be unfolded before being translocated into the interior of the 20S catalytic core for degradation.
The 19S regulatory particle consists of at least 17 subunits, which can bind substrate-linked multiubiquitin chains, unfold the tethered proteins and translocate the polypeptide chains into the 20S catalytic core. A key feature of the 19S subunit is the presence of six homologous AAA ATPases that are thought to unfold substrate proteins in an ATP-dependent manner. Conditional yeast mutants exist for some of these AAA ATPases such as Cim3 (Sug1/Rpt6) and Cim5 (Rpt1/Yta3). They appear to display some substrate specificity, indicating that they are not redundant. The subcellular distribution of the proteasome is a topic of much interest, and several lines of inquiry suggest that it is associated with the nuclear envelope in Saccharomyces cerevisiae (15,16). It is not clear if the intact 26S proteasome is necessary for all proteasomal function or whether the 19S regulatory subunit can function on its own independently of the 20S catalytic core.
Several lines of evidence suggest a role for ubiquitylation and protein degradation in the cellular response to DNA damage. Yeast Rad6/Ubc2 is a ubiquitin-conjugating enzyme (E2) involved in translesion synthesis following exposure to DNA-damaging agents, telomeric silencing and sporulation (5,17). Another ubiquitin-conjugating enzyme implicated in DNA repair is Ubc13, which interacts with the newly identified E4 protein Mms2 (14). Together these proteins assemble novel K63-linked polyubiquitin chains for signaling in DNA repair. Mutant rad6, ubc13 and mms2 strains display increased UV sensitivity compared to wild-type strains. It is not known if substrates that are ubiquitylated by Rad6 or Ubc13/Mms2 eventually get degraded or recycled.
We speculate that DNA repair proteins might be degraded following completion of repair in a proteasome-dependent manner. This may constitute an important regulatory step to prevent incision of DNA structures that naturally occur during the cellular transactions involving DNA. We tested this hypothesis by measuring transcription-coupled repair and overall genomic nucleotide excision repair in the presence and absence of proteasome function. We took advantage of conditional proteasome mutants that degrade proteins at the permissive temperature, but are significantly impaired at the non-permissive temperature. The conditional alleles are in genes encoding the AAA ATPases Cim3 (Sug1/Rpt6) and Cim5 (Yta3/Rpt1). We found that repair is increased following UV irradiation at the non-permissive temperature in the proteasome mutant relative to the parent strain. These results indicate that the accumulation of DNA repair factors in the proteasome mutant may enable faster repair. The Rad23 protein has been shown to provide a link between nucleotide excision repair and the ubiquitin-mediated protein degradation pathway. Recent studies showed that Rad23 is a negative regulator of proteolysis (18). Genetic studies have suggested that Rad23 performs a regulatory role in DNA repair (19). The Rad4 nucleotide excision repair protein is always present in a complex with Rad23 (20–22), and is essential for repair. We therefore investigated if Rad4 might be regulated by proteolysis. We overexpressed Rad4 in a repair-proficient strain and observed increased rates of nucleotide excision repair, similar to those observed in the conditional proteasome mutants at the non-permissive temperature.
MATERIALS AND METHODS
Yeast strains, media, plasmids and probes
The yeast strains used in this study (Table 1) were manipulated using standard techniques and grown in media prepared as described by Adams et al. (23). YPD is 1% yeast extract, 2% peptone and 2% glucose (Gibco BRL). Synthetic Complete (SC) is 0.67% bacto-yeast nitrogen base without amino acids, 2% glucose and 0.2% drop-out mix. SC minus leucine (SC-leu) is SC missing the leucine from the drop-out mix. Where specified, 100 µm cupric sulfate was added to the media. Plasmid pKS212 is a Bluescript vector (Stratagene) into which the internal 1.0 kb EcoRI–XhoI fragment from RPB2 was inserted (24). Plasmid pKS212 was linearized by cleaving with XhoI or EcoRI and incubated with [α-32P]CTP (Amersham Corp.), rNTPs and T7 RNA polymerase or T3 RNA polymerase (Boehringer Mannheim), respectively, under conditions recommended by the manufacturer to generate radioactive RNA probes to detect the transcribed or the non-transcribed strand, respectively, of RPB2.
Table 1. Yeast strains used in this study.
| CMY394 |
MATa ura3-52 leu2Δ1 his3Δ-200 trp1Δ1 lys2-801 ade2-101 |
| CMY762 |
MATa cim3-1 ura3-52 leu2Δ1 his3Δ-200 |
| CMY791 |
MATa cim5-1 ura3-52 leu2Δ1 his3Δ-200 |
| LCY652 | MATa RAD4-HA ura3-52 leu2Δ1 his3Δ-200 trp1Δ1 lys2-801 ade2-101 |
Strain LCY652 was constructed by transforming CMY394 with two plasmids, pCS28 and pCS31. Two plasmids, containing the 5′ and 3′ portions of the RAD4 gene, were necessary since expression of intact Rad4 is toxic to Escherichia coli (25). Plasmid pCS28 contains 750 bp of the RAD4 gene encoding the N-terminal portion of Rad4 protein with a C-terminal HA-tag. Plasmid pCS31 contains 1.66 kb of the RAD4 gene encoding the C-terminal portion of the Rad4 protein with a C-terminal HA-tag. There is a 150 bp overlap among the RAD4 sequences on the two plasmids. The plasmid pCS28 was digested with BglII and pCS31 was digested with EcoRI and HindIII. Yeast was co-transformed with the digested plasmids to generate RAD4-HA. Recombination between the two plasmids in yeast generates a plasmid encoding full-length Rad4 protein with a HA-tag at the C-terminus (26).
Growth and UV irradiation of yeast cells
The growth and irradiation of strains and purification of DNA was basically as described previously (27). Strains were grown in the specified media at the temperatures stated in the figure legends. Exponentially growing cultures were collected at 4°C by centrifugation at 1740 g for 4 min and resuspended in cold phosphate-buffered saline at 1 × 107 cells/ml. The cell suspension was distributed as a thin layer and subjected to gentle shaking during irradiation with 60 J/m2 of predominantly 254 nm UV light using an American Ultraviolet Co. germicidal lamp. Following irradiation the cells were collected by centrifugation and either spheroplasted and lysed immediately or returned to their pre-irradiation media and temperature for the specified repair times, and then spheroplasted and lysed. All manipulations were performed under yellow light to preclude photoreactivation.
Isolation and restriction of yeast DNA
Cells were digested for 30 min at 37°C with 2 mg/ml Zymolyase 100T (ICN Biochemicals) to generate spheroplasts (27). After digestion, spheroplasts were collected at 4°C by centrifugation at 1125 g for 3 min and resuspended in 0.2 ml of cold Zymolyase buffer lacking Zymolyase (0.25 M EDTA, 1 M sorbitol and 20 mM 2-mercaptoethanol). Spheroplasts were then diluted with the gradual addition of 2.8 ml of cold 0.05 M Tris–HCl (pH 8.0), 0.05 M EDTA and were lysed by the addition of 0.2 ml of cold 20% Sarkosyl (28). The mixture was incubated on ice for 10 min. Cellular debris and Sarkosyl were precipitated by the addition of 0.64 ml of 5 M potassium acetate. Mixtures were incubated at 4°C overnight and then centrifuged at 5000 r.p.m. in a Sorvall HL-6000 rotor at 4°C for 25 min. Supernatants containing chromosomal DNA were transferred to fresh tubes and further purified by phenol/chloroform extraction and ethanol precipitation (29). The samples were then treated with RNase A (final concentration, 50 µg/ml) and the DNA was digested to completion with PvuI and PvuII restriction enzymes. Restricted DNA was then ethanol precipitated and resuspended in TE (10 mM Tris–HCl pH 8.0, 1 mM EDTA pH 8.0).
Repair analysis
The incidence of cyclobutane pyrimidine dimers (CPDs) in each strand of the PvuI–PvuII restriction fragment of the RPB2 gene was determined by previously developed methods (30,31). In brief, equal amounts of purified, restricted DNA was either mock treated (10 mM Tris–HCl pH 7.5, 0.1 M NaCl, 10 mM EDTA, 1 mg/ml BSA) or treated with the enzyme T4 endonuclease V in mock buffer for 20 min at 37°C. T4 endonuclease V is a CPD-specific DNA glycosylase/AP lyase which produces a single-stranded break specifically at each CPD. DNA was denatured, electrophoresed through 0.5% alkaline agarose gels to separate full-length restriction fragments from the digestion products of T4 endonuclease V, transferred to Hybond N+ (Amersham) membranes and hybridized with strand-specific 32P-labeled RNA probes generated by in vitro transcription of linearized pKS212. The membranes were then deprobed and hybridized with the complementary strand-specific probe. The strand-specific probe to be used first was determined randomly. Autoradiograms were generated and signal intensities determined using a Hewlett-Packard Scanjet II and the application NIH Image 1.62. The Poisson expression was applied to calculate the number of CPDs per fragment from the ratio of intensities of bands from T4 endonuclease V treated and mock-treated DNA samples.
RESULTS
Proficient transcription-coupled repair in conditional proteasome mutants at the permissive temperature
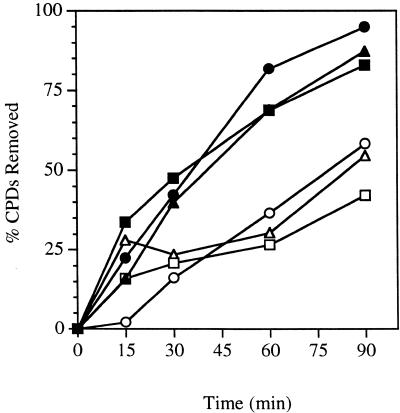
We examined the repair capability of yeast strains harboring conditional mutations in the 19S regulatory subunit of the 26S proteasome. We measured the removal of UV-induced CPDs from either strand of the reporter gene RPB2 in heat-sensitive cim3-1 and cim5-1 mutants and the parent strain at the permissive temperature. An autoradiogram from one such experiment is shown in Figure 1. In the assay used, successful DNA repair is reflected by the restoration of the full-length DNA restriction fragment, following T4 endonuclease-treatment. Dimer frequencies were calculated from the measured incidences of CPDs per restriction fragment by densitometric scanning of autoradiograms such as the ones shown in Figure 1. Figure 2 shows a graphical representation of the repair in cim3-1, cim5-1 and the parent strain. Repair of the transcribed strand of RPB2 is rapid in the cim3-1 and cim5-1 mutants and the parent strain, reaching 45–50% in the first 30 min following UV irradiation. Consistent with previous studies of other repair-proficient strains, repair of the non-transcribed strand of RPB2 is slower than that of the transcribed strand in cim3-1, cim5-1 and the parent strain. Approximately 20–30% of CPDs were removed from the non-transcribed strand during the first 30 min following UV irradiation.
Figure 1.
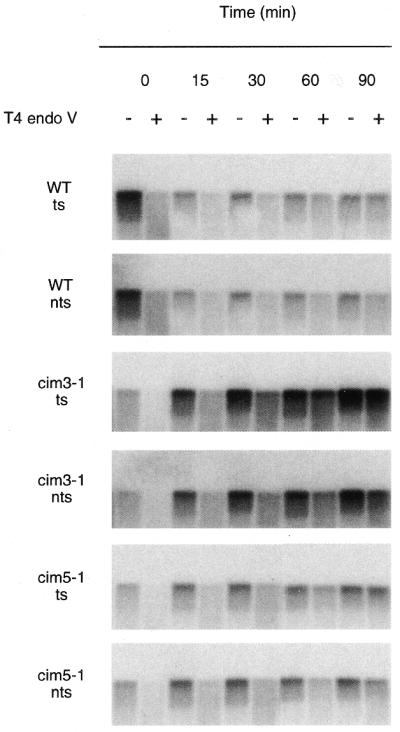
Proficient repair in conditional proteasome mutants at the permissive temperature. Autoradiograms illustrating removal of CPDs from each of the strands of the RPB2 gene in the wild-type parent strain (CMY394), a heat-sensitive cim3-1 mutant (CMY762) and a heat-sensitive cim5-1 mutant (CMY791) at the permissive temperature. Exponentially growing cultures in YPD media at 24°C were UV-irradiated and then incubated for the times indicated. DNA was purified from the cells as described in Materials and Methods. DNA was transferred to Hybond N+ membrane and was hybridized with an RNA probe specific for one strand of the RPB2 gene and an autoradiogram was generated. The probe was stripped off and the immobilized DNA was then hybridized with a probe specific for the complementary strand of RPB2. The autoradiograms show the 5.3 kb PvuI–PvuII restriction fragment of RPB2. Transcribed strand, ts; non-transcribed strand, nts.
Figure 2.
Time course for removal of CPDs from the RPB2 gene in conditional proteasome mutants at the permissive temperature. Exponentially growing cultures in YPD at 24°C were UV-irradiated with 60 J/m2 and then incubated in YPD at 24°C for the times indicated. Repair was determined from the measured incidences of CPDs in each strand of the PvuI–PvuII restriction fragment of the RPB2 gene. Transcribed strand, wild-type (filled squares); non-transcribed strand, wild-type (open squares); transcribed strand, cim3-1 (filled triangles); non-transcribed strand, cim3-1 (open triangles); transcribed strand, cim5-1 (filled circles); non-transcribed strand, cim5-1 (open circles).
Faster repair in proteasome mutants
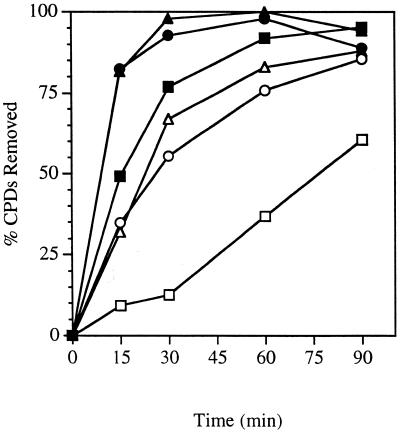
We next examined the repair capability of conditional proteasome mutants and the parent strain at the non-permissive temperature. At the non-permissive temperature, proteolysis of a number of substrate proteins is deficient (32). We measured the removal of CPDs from either strand of RPB2 in heat-sensitive cim3-1 and cim5-1 mutants and the parent strain at the non-permissive temperature. An autoradiogram from one representative experiment is shown in Figure 3. Repair of the transcribed strand of RPB2 was very rapid in all three strains. Remarkably, repair of the transcribed strand was significantly faster in the cim3-1 and cim5-1 mutants than in the parent strain. Repair of the non-transcribed strand of RPB2 was slower than that of the transcribed strand in all three strains. However, repair of the non-transcribed strand in cim3-1 and cim5-1 was noticeably faster than in the parent strain. Figure 4 shows a graphical representation of the repair in cim3-1, cim5-1 and the parent strain at the non-permissive temperature. More than 90% of the CPDs were removed from the transcribed strand in the cim3-1 and cim5-1 mutants during the first 30 min following UV irradiation. In contrast, the parent strain removed 75–80% of the CPDs from the transcribed strand during the first 30 min following UV irradiation, suggesting that repair of the transcribed strand of RPB2 may be faster in the cim3-1 and cim5-1 mutants than in the parent strain.
Figure 3.
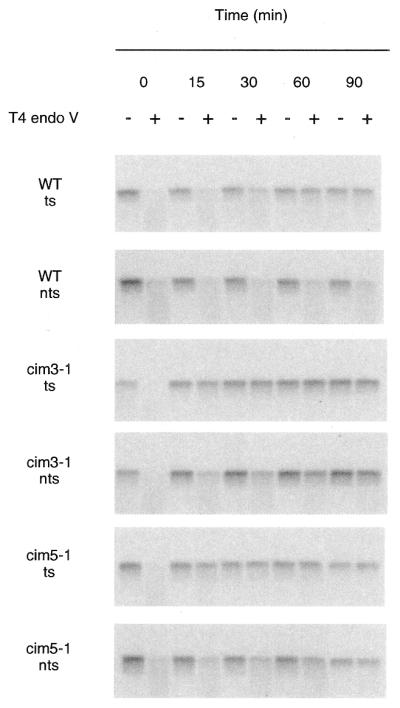
Enhanced repair in conditional proteasome mutants at the non-permissive temperature. Autoradiograms illustrating removal of CPDs from each of the strands of the RPB2 gene in the wild-type parent strain (CMY394), a heat-sensitive cim3-1 mutant (CMY762), and a heat-sensitive cim5-1 mutant (CMY791) at the non-permissive temperature. Exponentially growing cultures in YPD media at 24°C were shifted to 37°C for 2 h, then UV-irradiated and incubated in YPD at 37°C for the times indicated. DNA was purified from the cells and analyzed as described in Figure 1. DNA was transferred to Hybond N+ membrane and hybridized with strand-specific RNA probes and autoradiograms were generated. Transcribed strand, ts; non-transcribed strand, nts.
Figure 4.
Time course for removal of CPDs from the RPB2 gene in conditional proteasome mutants at the non-permissive temperature. Exponentially growing cultures in YPD at 24°C were shifted to 37°C for 2 h, then UV-irradiated and incubated in YPD at 37°C for the times indicated. Repair was determined from the measured incidences of CPDs in each strand of the PvuI–PvuII restriction fragment of the RPB2 gene. Transcribed strand, wild-type (filled squares); non-transcribed strand, wild-type (open squares); transcribed strand, cim3-1 (filled triangles); non-transcribed strand, cim3-1 (open triangles); transcribed strand, cim5-1 (filled circles); non-transcribed strand, cim5-1 (open circles).
In contrast to the repair observed for the non-transcribed strand at the permissive temperature, we observed striking differences in the repair of the non-transcribed strand in cim3-1 and the parent strain at the non-permissive temperature. As can be seen in Figures 3 and 4, repair of the non-transcribed strand in the cim3-1 mutant reached almost 67% during the first 30 min following UV irradiation. During this same time period, <15% of the CPDs were removed from the non-transcribed strand of RPB2 in the parent strain. Similarly, repair of the non-transcribed strand was significantly faster in the cim5-1 mutant than in the parent strain (Fig. 4). Thus, in the absence of either Cim3 or Cim5 function, repair is increased for both the transcribed and non-transcribed strands of RPB2.
Overexpression of Rad4 results in faster repair
We wished to determine whether the stability of one of the proteins involved in nucleotide excision repair was regulated by 26S-mediated degradation. One likely candidate is Rad4 (26), which forms a high-affinity interaction with Rad23. Rad23 proteins from yeast and humans interact with catalytically active 26S proteasome. The Rad23 protein is a novel inhibitor of multiubiquitin chain formation (18). Consequently, we considered the possibility that Rad23 might protect Rad4 from degradation. We examined the repair capability of a yeast strain overexpressing Rad4 following UV irradiation.
We measured the removal of CPDs from each strand of RPB2 in a strain overexpressing a HA-tagged Rad4 under the control of the inducible CUP1 promoter. Expression from the CUP1 promoter was induced by the addition of copper to the medium (100 µM CuSO4). An autoradiogram from one such experiment is shown in Figure 5. Repair of the transcribed strand was rapid in the strain overexpressing Rad4 and the parent strain, reaching >60% at 30 min post-irradiation. Repair of the non-transcribed strand was ~55% after 30 min, much faster than repair of the non-transcribed strand of the parent strain (25–30% of CPDs were removed by 30 min) (Fig. 6). The repair observed upon overexpression of Rad4 in the parent strain resembles the repair observed in the conditional proteasome mutants at the non-permissive temperature, suggesting that Rad4 might be a target for degradation.
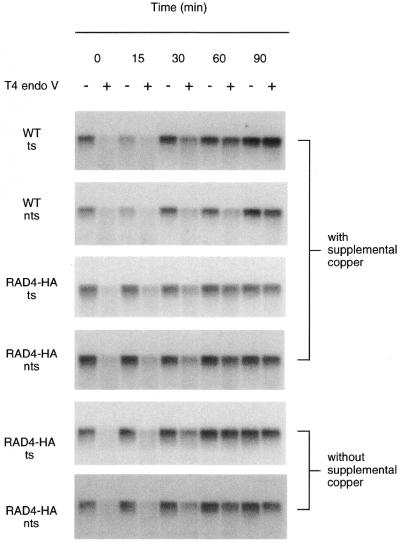
Figure 5.
Overexpression of Rad4 increases global genomic repair. Autoradiograms illustrating removal of CPDs from the RPB2 gene in the parent strain (CMY394) and in the strain overexpressing a HA-tagged Rad4 under control of the CUP1 promoter (LCY652). Exponentially growing cultures at 30°C in media (SC for CMY394, SC-leu for LCY652) supplemented or not with 100 µM CuSO4 were UV-irradiated and then incubated for the times indicated. DNA purified from the cells was analyzed as described in Figure 1. DNA was transferred to Hybond N+ membrane and hybridized with stand-specific RNA probes and autoradiograms were generated. Transcribed strand, ts; non-transcribed strand, nts.
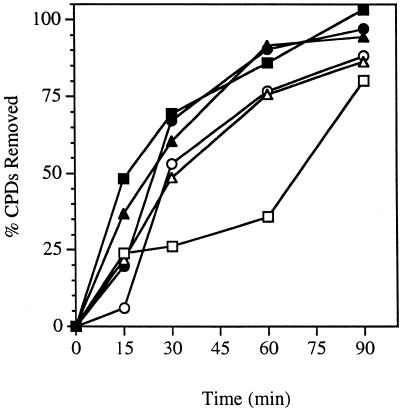
Figure 6.
Time course for removal of CPDs from the RPB2 gene in the parent strain and in the strain overexpressing a HA-tagged Rad4. Exponentially growing cultures at 30°C in media (SC for CMY394, SC-leu for LCY652) supplemented or not with 100 µM CuSO4 were UV-irradiated with 60 J/m2 and then incubated in media (SC for CMY394, SC-leu for LCY652) supplemented or not with 100 µM CuSO4 at 30°C for the times indicated. Repair was determined from the measured incidences of CPDs in each strand of the PvuI–PvuII restriction fragment of the RPB2 gene. Transcribed strand, wild-type with copper (filled squares); non-transcribed strand, wild-type with copper (open squares); transcribed strand, RAD4-HA with copper (filled circles); non-transcribed strand, RAD4-HA with copper (open circles); transcribed strand, RAD4-HA without copper (filled triangles); non-transcribed strand, RAD4-HA without copper (open triangles).
Experiments were repeated for the Rad4-overexpressing strain without addition of excess copper to the medium. An autoradiogram from one such experiment is shown in Figure 5. Repair in the absence of supplemental copper was similar to the repair observed in the presence of supplemental copper. Therefore, sufficient divalent cations are already present in the medium to induce expression from the CUP1 promoter as reported previously (33–35). Repair of the transcribed and non-transcribed strands of RPB2 is rapid in the Rad4-overexpressing strain (Fig. 6). Approximately 70% of the CPDs are removed from the transcribed strand in the first 30 min following UV irradiation. In the absence of supplemental copper, repair of the non-transcribed strand in the strain overexpressing Rad4 is >50% CPDs removed after 30 min following UV irradiation (Fig. 6).
DISCUSSION
We have demonstrated that conditional mutations in the 19S regulatory subunit of the 26S proteasome result in increased nucleotide excision repair in vivo. We measured nucleotide excision repair in yeast conditional mutants (cim3-1 and cim5-1), under permissive and non-permissive conditions and found that repair of both the transcribed strand and the non-transcribed strand of an RNA polymerase II-transcribed reporter gene was increased in the absence of proteasome function. Our data lead us to speculate that a protein (or proteins) involved in nucleotide excision repair or regulation of nucleotide excision repair might be degraded by the 26S proteasome. In the absence of proteasome function the repair protein (or proteins) is presumed to accumulate and enhance repair throughout the genome. One likely candidate is Rad4 which, when overexpressed, increased efficiency of overall genomic repair. These studies demonstrate that the 26S proteasome regulates a specific DNA repair process.
We found that Rad4, a crucial factor for nucleotide excision repair, is probably negatively regulated through proteolysis. Overexpression of Rad4 resulted in increased nucleotide excision repair rates, similar to the increased rates of repair observed for the conditional proteasome mutants at the non-permissive temperature. The increase in repair rates upon overexpression of Rad4 may mean that degradation of another factor does not normally limit nucleotide excision repair. The absence of a functional proteasome is not by itself sufficient for increasing the rate of repair because a functional proteasome is present in the Rad4 overexpressing strain, which also displays increased repair rates. Thus, accumulation of Rad4 appears to be necessary for rapid overall genomic repair. The accumulation of Rad4 might be achieved by modulating proteasome function.
We observed that repair of the transcribed strands of the reporter gene was not as fast in the strain overexpressing Rad4 as repair of the transcribed strands in the proteasomal mutants at the non-permissive temperature. Repair in the wild-type strain in the presence of 100 µM CuSO4 is very similar to the repair in the wild-type strain in the absence of copper (compare Figs 4 and 6). Thus, it is unlikely that addition of copper to the medium is inhibiting nucleotide excision repair. It is likely that another protein is stabilized in the proteasomal mutants at the non-permissive temperature that enables faster repair of the transcribed strands of class II genes. A possible candidate protein that is stabilized is Ssl2(Rad25), as the human homolog XPB has already been shown to interact with human Cim3 homolog, hSug1 (36).
XPB, a subunit of human transcription initiation/repair factor IIH (TFIIH), interacts with hSUG1, the human homolog of Cim3 (36). No evidence for ubiquitylation of XPB or association of XPB with the 26S proteasome was reported. However, ubiquitylated XPB could be degraded rapidly such that it would be undetectable by western analysis with anti-ubiquitin antibodies. Furthermore, the purification of Sug1–HA complexes in the absence of ATP would dissociate the 26S proteasome into its constituent 19S and 20S particles (37,38). Microinjection of mRNA encoding hSUG1 (under the control of the strong CMV promoter) into fibroblast nuclei led to a dramatic decrease in transcription, suggesting that hSUG1 can inhibit transcription by binding to TFIIH via XPB. Weeda et al. speculated that the proteasome may be required to activate proteins by processing the inactive precursors, i.e., acting as a molecular chaperone to properly fold proteins and/or assemble the repair proteins into complexes with other proteins involved in repair (36). Their model suggests that defects in proteasome function could cause defects in repair protein folding and complex assembly, resulting in defective repair. However, their results are inconsistent with our findings which show that repair is increased in cells with defective proteasome function and are also inconsistent with the model of the 26S proteasome activating a factor by folding or assembling complexes.
Our results are at odds with a recently published report describing a role for the 19S regulatory subunit in nucleotide excision repair. It was proposed that the 19S regulatory subunit may function independently from the catalytic 20S subunit in transcription and nucleotide excision repair. Russell et al. inhibited 19S regulatory function by two different methods and found that nucleotide excision repair was impaired. Using an in vitro nucleotide excision repair assay, Russell et al. were able to reduce nucleotide excision repair by 45% by immunodepleting the proteasome subunit (Cim3) (22). It was not shown whether the residual repair was due to inefficient depletion of Cim3. They also observed that the 20S particle was dispensable for nucleotide excision repair. Their conclusion was that the proteolytic activity of the proteasome was not necessary for nucleotide excision repair.
A potential explanation for the discrepancies between our results and those of Russell et al. may reside in the different assays used. Russell et al. used extracts from yeast cells, added purified components and examined DNA incision and repair synthesis. However, post-incision steps are not easily examined by this method. Furthermore, a potential role for the proteasome in recycling and dismantling repair complexes was not considered. Perhaps most importantly, the study by Russell et al. did not consider the possibility that the proteasome may play a negative role in nucleotide excision repair.
The expression of several proteins involved in nucleotide excision repair is induced following exposure to DNA damage, presumably to hasten removal of DNA adducts. In addition, levels of many of these proteins also increase during the meiotic cell cycle. However, protein levels do not fluctuate during the mitotic cell cycle in the absence of DNA damage. Once the induced proteins are no longer needed, the proteins are presumably degraded and a basal level of protein abundance is restored.
Schauber et al. showed that Rad23 and Rad4 interact, and are associated with the 26S proteasome (26). Rad23 has been shown to interact with other proteins involved in nucleotide excision repair, in addition to Rad4 (20–22). Schauber et al. proposed a role for the proteasome in the disassembling and recycling of repair proteins in a Rad23-dependent manner. Recent studies suggest that Rad23 may prevent premature degradation of specific proteins, but may then escort proteins to the 26S proteasome for destruction. Alternatively, Rad23 may have an independent role as a molecular chaperone responsible for the assembly of the nucleotide excision repair complex (11). Both suggestions are consistent with the DNA repair defect of rad23Δ mutants, which display severely reduced levels of nucleotide excision repair (39) (data not shown).
We speculate that following induction of repair proteins and completion of DNA repair in normal cells, the repair proteins are degraded in a proteasome-dependent manner to prevent them from incising DNA structures that arise during other cellular transactions involving DNA. Following exposure to UV radiation, RAD4 mRNA levels increase several fold. We overexpressed Rad4 in wild-type cells and observed increased repair in both strands of the RPB2 gene, suggesting that Rad4 may be normally degraded in the absence of DNA damage, but is transiently stabilized in the presence of lesions to promote repair. This result also indicates that Rad4 is in limiting amounts in undamaged cells. The stabilization of Rad4 in the proteasomal mutants would permit increased rates of transcription-coupled repair and nucleotide excision repair. Future experiments will examine the levels of native Rad4 following UV irradiation and determine if Rad4 abundance is reduced to pre-irradiation levels by the time repair is completed.
We suggest that, following formation of DNA damage, there are conflicting forces within cells—one is to efficiently remove DNA damage and the other is to keep overall levels of repair proteins low to prevent gratuitous cleavage of undamaged DNA. These conflicting forces may be addressed by enabling faster repair of essential sequences as is seen in transcription-coupled repair. Repair of the non-transcribed sequences is proficient in wild-type strains, but slower than the rapid repair observed for non-transcribed sequences in the proteasomal mutants at the non-permissive temperature and the transcribed strands in wild-type cells. A balance is achieved between sufficient repair of non-transcribing sequences to enable survival following UV irradiation and excessive cleavage of undamaged DNA by repair proteins resulting in genomic instability.
REFERENCES
- 1.Huang J.-C., Svoboda,D.L., Reardon,J.T. and Sancar,A. (1992) Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl Acad. Sci. USA, 89, 3664–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzder S.N., Habraken,Y., Sung,P., Prakash,L. and Prakash,S. (1995) Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A and transcription factor TFIIH. J. Biol. Chem., 270, 12973–12976. [DOI] [PubMed] [Google Scholar]
- 3.Aboussekhra A., Biggerstaff,M., Shivji,M.K., Vilpo,J.A., Moncollin,V., Podust,V.N., Protic,M., Hubscher,U., Egly,J.M. and Wood,R.D. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell, 80, 859–868. [DOI] [PubMed] [Google Scholar]
- 4.Robinson G.W., Nicolet,C.M., Kalainov,D. and Friedberg,E.C. (1986) A yeast excision-repair gene is inducible by DNA damaging agents. Proc. Natl Acad. Sci. USA, 83, 1842–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madura K., Prakash,S. and Prakash,L. (1990) Expression of the Saccharomyces cerevisiae DNA repair gene RAD6 that encodes a ubiquitin conjugating enzyme, increases in response to DNA damage and in meiosis but remains constant during the mitotic cell cycle. Nucleic Acids Res., 18, 771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones J.S. and Prakash,L. (1991) Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD18 increase in UV irradiated cells and during meiosis but not during the mitotic cell cycle. Nucleic Acids Res., 19, 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoeijmakers J.H.J. (1993) Nucleotide excision repair I: from E.coli to yeast. Trends Genet., 9, 173–177. [DOI] [PubMed] [Google Scholar]
- 8.Madura K. and Prakash,S. (1990) The Saccharomyces cerevisiae DNA repair gene RAD2 is regulated in meiosis but not during the mitotic cell cycle. Mol. Cell. Biol., 10, 3256–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones J.S., Prakash,L. and Prakash,S. (1990) Regulated expression of the Saccharomyces cerevisiae DNA repair gene RAD7 in response to DNA damage and during sporulation. Nucleic Acids Res., 18, 3281–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jentsch S., McGrath,J.P. and Varshavsky,A. (1987) The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature, 329, 131–134. [DOI] [PubMed] [Google Scholar]
- 11.Watkins J.F., Sung,P., Prakash,L. and Prakash,S. (1993) The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol., 13, 7757–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varshavsky A. (1997) The ubiquitin system. Trends Biochem. Sci., 22, 383–387. [DOI] [PubMed] [Google Scholar]
- 13.Koegl M., Hoppe,T., Schlenker,S., Ulrich,H.D., Mayer,T.U. and Jentsch,S. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell, 96, 635–644. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann R.M. and Pickart,C.M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell, 96, 645–653. [DOI] [PubMed] [Google Scholar]
- 15.Enenkel C., Lehmann,A. and Kloetzel,P.M. (1998) Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J., 17, 6144–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enenkel C., Lehmann,A. and Kloetzel,P.M. (1999) GFP-labelling of 26S proteasomes in living yeast: insight into proteasomal functions at the nuclear envelope/rough ER. Mol. Biol. Rep., 26, 131–135. [DOI] [PubMed]
- 17.Dohmen R.J., Madura,K., Bartel,B. and Varshavsky,A. (1991) The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl Acad. Sci. USA, 88, 7351–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortolan T.G., Tongaonkar,P., Lambertson,D., Chen,L. and Madura,K. (2000) The Rad23 DNA repair protein is a negative regulator of multiubiquitin chain assembly. Nature Cell Biol., 2, 601–608. [DOI] [PubMed] [Google Scholar]
- 19.Miller R.D., Prakash,L. and Prakash,S. (1982) Defective excision of pyrimidine dimers and interstrand DNA crosslinks in rad7 and rad23 mutants of Saccharomyces cerevisiae. Mol. Gen. Genet., 188, 235–239. [DOI] [PubMed] [Google Scholar]
- 20.Guzder S.N., Bailly,V., Sung,P., Prakash,L. and Prakash,S. (1995) Yeast DNA repair protein RAD23 promotes complex formation between transcription factor TFIIH and DNA damage recognition factor RAD14. J. Biol. Chem., 270, 8385–8388. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z., Wei,S., Reed,S.H., Wu,X., Svejstrup,J.Q., Feaver,W.J., Kornberg,R.D. and Friedberg,E.C. (1997) The RAD7, RAD16 and RAD23 genes of Saccharomyces cerevisiae: requirement for transcription-independent nucleotide excision repair in vitro and interactions between the gene products. Mol. Cell. Biol., 17, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell S.J., Reed,S.H., Huang,W., Friedberg,E.C. and Johnston,S.A. (1999) The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol. Cell, 3, 687–695. [DOI] [PubMed] [Google Scholar]
- 23.Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1998) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, 1997 Edn. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 24.Sweder K.S. and Hanawalt,P.C. (1992) Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc. Natl Acad. Sci. USA, 89, 10696–10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleer R., Nicolet,C.M., Pure,G.A. and Friedberg,E.C. (1987) RAD4 gene of Saccharomyces cerevisiae: molecular cloning and partial characterization of a gene that is inactivated in Escherichia coli. Mol. Cell. Biol., 7, 1180–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schauber C., Chen,L., Tongaonkar,P., Vega,I., Lambertson,D., Potts,W. and Madura,K. (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature, 391, 715–718. [DOI] [PubMed] [Google Scholar]
- 27.Sweder K.S. and Hanawalt,P.C. (1994) The COOH terminus of suppressor of stem loop (SSL2/Rad25) in yeast is essential for overall genomic excision repair and transcription-coupled repair. J. Biol. Chem., 269, 1852–1857. [PubMed] [Google Scholar]
- 28.Nasmyth K.A. (1982) The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell, 30, 567–578. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) In Nolan,C. (ed.), Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Bohr V.A., Smith,C.A., Okumoto,D.S. and Hanawalt,P.C. (1985) DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell, 40, 359–369. [DOI] [PubMed] [Google Scholar]
- 31.Mellon I., Spivak,G. and Hanawalt,P.C. (1987) Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell, 51, 241–249. [DOI] [PubMed] [Google Scholar]
- 32.Ghislain M., Udvardy,A. and Mann,C. (1993) S.cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature, 366, 358–362. [DOI] [PubMed] [Google Scholar]
- 33.Gorman J.A., Clark,P.E., Lee,M.C., Debouck,C. and Rosenberg,M. (1986) Regulation of the yeast metallothionein gene. Gene, 48, 13–22. [DOI] [PubMed] [Google Scholar]
- 34.Szczypka M.S. and Thiele,D.J. (1989) A cysteine-rich nuclear protein activates yeast metallothionein gene transcription. Mol. Cell. Biol., 9, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen L.T., Howard,W.R., Strain,J.J., Winge,D.R. and Culotta,V.C. (1996) Enhanced effectiveness of copper ion buffering by CUP1 metallothionein compared with CRS5 metallothionein in Saccharomyces cerevisiae. J. Biol. Chem., 271, 18514–18519. [DOI] [PubMed] [Google Scholar]
- 36.Weeda G., Rossignol,M., Fraser,R.A., Winkler,G.S., Vermeulen,W., van’t Veer,L.J., Ma,L., Hoeijmakers,J.H.J. and Egly,J.-M. (1997) The XPB subunit of repair/transcription factor TFIIH directly interacts with SUG1, a subunit of the 26S proteasome and putative transcription factor. Nucleic Acids Res., 25, 2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glickman M.H., Rubin,D.M., Fried,V.A. and Finley,D. (1998) The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol., 18, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verma R., Chen,S., Feldman,R., Schieltz,D., Yates,J., Dohmen,J. and Deshaies,R.J. (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell, 11, 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller J.P. and Smerdon,M.J. (1996) Rad23 is required for transcription-coupled repair and efficient overall repair in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2361–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]