Abstract
Blimp-1 is a transcriptional repressor that is both required and sufficient to trigger terminal differentiation of B lymphocytes and monocyte/macrophages. Here we report the organization of the mouse Blimp-1 gene, an analysis of Blimp-1 homologs in different species, the characterization of Blimp-1 mRNA isoforms and initial studies on the transcription of Blimp-1. The murine Blimp-1 gene covers ~23 kb and contains eight exons. There are Blimp-1 homologs in species evolutionarily distant from mouse (Caenorhabditis elegans and Drosophila melanogaster) but no homolog was found in the unicellular yeast Saccharomyces cerevisiae. The three major Blimp-1 mRNA isoforms result from the use of different polyadenylation sites and do not encode different proteins. Run-on transcription analyses were used to show that the developmentally regulated expression of Blimp-1 mRNA in B cells is determined by transcription initiation. Multiple Blimp-1 transcription initiates sites were mapped near an initiator element and a region conferring basal promoter activity has been identified.
INTRODUCTION
Terminal differentiation, giving rise to post-mitotic cells performing dedicated functions, is a critical developmental process in multicellular organisms that begins during embryogenesis and continues throughout life. One well-studied example of terminal differentiation is the maturation of B lymphocytes into end-stage, antibody-secreting plasma cells. Although terminal differentiation of B cells involves complex changes in the expression of many genes (1–4), a single transcription factor, B lymphocyte-induced maturation protein-1 (Blimp-1), has been identified as a ‘master regulator’ of this process (5). Blimp-1 is a 98 kDa zinc finger-containing protein that functions as a transcriptional repressor (6–9). Expression of Blimp-1 is sufficient to trigger terminal differentiation of B-cell lymphoma line in vitro (5), and the developmental stage-specific expression of Blimp-1 in B cells in vivo (10) is consistent with its role as a master regulator of terminal B cell differentiation. Recently Blimp-1 has also been shown to be required for terminal differentiation of monocyte/macrophages (11), suggesting the possibility that it may play a role in terminal differentiation of many cell lineages. Consistent with this hypothesis, the Blimp-1 homologs in Xenopus (XBlimp1) and sea urchin (SpKrox1) are required during early embryo development of these two organisms (12,13).
Although the function of Blimp-1 has been the subject of many studies, virtually nothing is known about the structure of the Blimp-1 gene or regulation of Blimp-1 expression. Here we report the organization of mouse Blimp-1 gene, the characterization of the three most abundant Blimp-1 mRNA isoforms as well as a minor splice variant. We also show that Blimp-1 expression in terminally differentiated B cells is controlled by transcription initiation, and identify the transcription initiation sites as well as the basal promoter of the Blimp-1 gene.
MATERIALS AND METHODS
GenBank accession numbers
The accession numbers for mouse Blimp-1 gene are: AF305534 for the promoter and exon 1; AF305535 for exon 2 and 3; AF305536 for exon 4; AF305537 for exon 5; AF305538 for exon 6 and 7; and AF305539 for exon 8.
Genomic library screening
A genomic library from mouse (129 strain) in λfixII phage (Stratagene, La Jolla, CA) was screened with Blimp-1 cDNA using standard procedures (14,15). Three overlapping phage clones, φ21, φ25 and φ17 were obtained. The restriction fragments containing the Blimp-1 coding region were identified by hybridization and subjected to DNA sequencing.
Plasmids
For transient transfection experiments, Blimp-pGL3 was generated by cloning the 1.1 kb SacI–ScaI fragment from φ21, containing an ~900 bp region upstream of transcription initiation sites, into firefly luciferase reporter pGL3 (Promega, Madison, WI). The Renilla luciferase reporter pRL-tk was obtained from Promega. For nuclear run-on analysis, the following plasmids were used: pIg containing Cµ (16), pSK-Blimp-1 (5), pAlt-GAPDH (a gift from Dr M. Coutts) and pSV2-myc (17). For RNase protection assay, a 300 bp SacII fragment from the genomic phage, φ21, encompassing the putative transcriptional initiation sites was cloned into pBluescriptKS (Stratagene).
Cell lines and transient transfection
A plasmacytoma, P3X, and a pre-B cell line, 18-81, were maintained in RPMI-1640 supplemented with 10% fetal calf serum (FCS), 50 µM of β-ME and 10 µg/ml gentamicin. NIH 3T3 cells were maintained in DMEM supplemented with 10% FCS and 10 µg/ml gentamicin. A promonocytic cell line, U937, was maintained in RPMI-1640 supplemented with 10% FCS and 10 µg/ml gentamicin. P3X, 18-81 and U937 cells were transfected by electroporation using a Gene Pulser (Bio-Rad, Richmond, CA). Briefly, for P3X and 18-81 cells, 3 × 106 cells were pulsed in 300 µl of complete media at 960 µF and 240 V. The cells were allowed to recover for 16–18 h in complete media before harvesting. For U937 cells, 1 × 107 cells were pulsed in 300 µl of complete media at 960 µF and 300 V. The cells were recovered in complete media for 8 h before harvesting. NIH 3T3 cells were transfected by calcium phosphate–DNA complex (17). Briefly, 3 × 105 cells were incubated with DNA–calcium phosphate complex for 12–14 h. The media were changed and cells were harvested 24 h later. In all transfections, pRL-tk (Promega) encoding Renilla luciferase under control of HSV-tk promoter was co-transfected to correct for the transfection efficiency.
Luciferase assay and data analysis
After transient transfection, cells were harvested and lysed in passive lysis buffer (Promega). The levels of firefly and Renilla luciferase in each sample were assayed as described (17,18) and the light units were collected for 10 s with luminometer (Berthold Lumat LB9501). The relative light units from firefly luciferase were corrected for the transfection efficiency by the relative light units obtained from Renilla luciferase. All transfections were performed at least twice in triplicate with two to three preparations of DNA.
Northern analysis of P3X RNA
Total RNA was prepared from P3X by Trizol (Gibco, Rockville, MD). Total RNA (10–20 µg) was electrophoresed in a formaldehyde gel with 0.9% agarose (19) and transferred to a Hybond-N membrane (Amersham Pharmacia Biotech, Piscataway, NJ). The hybridization was performed in 50% formamide, 5× SSC, 5× Denhardt’s, 1% SDS and 100 µg/ml yeast total RNA at 45°C for DNA probes or 68°C for RNA probes or in 7% SDS, 10% PEG-8000, 1.5× SSPE and 100 µg/ml salmon sperm DNA at 68°C. Exon specific probes used were as follows. The exon 1 probe was a PCR product of φ21 DNA using primers derived from the 5′ untranslated (UT) region (5′-AGAGTAGTCAGTCGGTCGCTCA-3′ and 5′-GGAGAGAGTACAGGTGGGTCAG-3′). Exons 4 and 8 were generated as restriction fragments from phage φ25, which contains the respective exons. Exon 6 and 3′ UT probes were generated from cloned cDNA. Exon 6 was generated from the cDNA clone as a 300 bp AvrII–AflIII fragment. The probe for the 5′ most portion of the 3′ UT was a 0.8 kb BglII fragment, the probe for the middle portion of 3′ UT was a 1 kb BglII–XbaI fragment and the probe for the 3′ end of the 3′ UT was a 0.4 kb HincII–XhoI fragment.
Nuclear run-on
Nuclear run-on assays were performed according to standard protocols (20) with the following modification for the cell lysis step. One hundred million P3X and 18-81 cells were harvested and resuspended in TKM (50 mM Tris pH 7.5, 50 mM KCl, 15 mM MgCl2). Then an equal volume of TKM with 60% sucrose was added and cells were lysed by the addition of 10% NP-40 to a final concentration of 0.05%. The nuclei were collected and transcribed in the presence of 100 µCi UTP (800 Ci/mmol) for 20 min as described (20). The newly transcribed RNA was purified and hybridized to 5 µg of denatured cDNAs of Blimp-1, Cµ, GAPDH, c-myc and pUC19 immobilized on a nitrocellulose membrane. After the hybridization, the membrane was washed and treated with RNase as described (20). To determine the amount of newly transcribed RNA for each cDNA, the radioactivity was quantified by a PhosphoImager (Molecular Dynamics, Sunnyvale CA) and corrected for the uracil content in the cDNAs.
PCR for Blimp-1 isoform
Single-strand cDNA was synthesized from 20 µg of P3X total cellular RNA as described (21). The PCR reactions were performed with 1 µg of cDNA in 30 cycles at 94°C for 1 min, 55°C for 1 min and 72°C for 1 min. The PCR primers used were: 5′-GAAGAAACAGAATGGCAAGA-3′ (upstream primer), 5′-AGTTGCCCTTCAGGT-3′ ( downstream primer a in Fig. 3b), 5′-AAGACACTTTCAGACTGGT-3′ (downstream primer b in Fig. 3b), 5′-CCTAAGAAGCAACACGAA-3′ (downstream primer c in Fig. 3b).
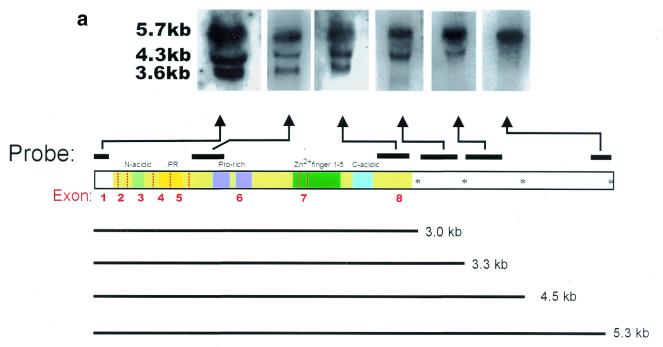
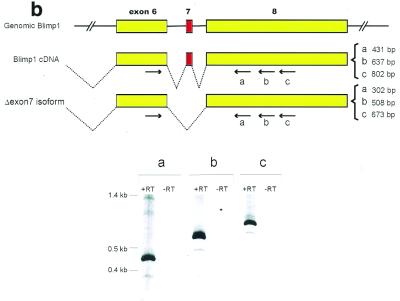
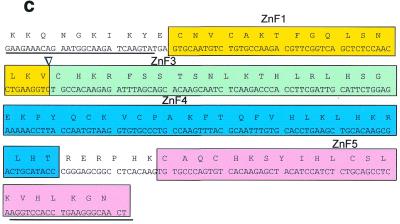
Figure 3.
Alternative polyadenylation and alternative splicing of Blimp-1 mRNA. (a) Northern blot analysis of Blimp-1 mRNAs. The diagram indicates portions of cDNA used as probes. Probes specific for the 5′ UT and the coding region were derived from either cDNA or genomic phage clones and are exon-specific. Probes derived from 3′ UT were derived from the cDNA. The asterisks in the diagram denote the putative poly(A) sites conserved between mouse and human Blimp-1 gene. The size of mRNA [without poly(A) tail] expected from the usage of each poly(A) site is indicated. (b) RT–PCR analysis of Blimp-1 mRNA. The diagram shows the possible alternative splicing involving exon 7 and the rationale for the PCR primers design. The expected sizes of the PCR products from the cloned cDNA and the potential Δexon7 isoform are indicated. The PCR products from each pair of primers were separated on 5% PAGE. +/– RT indicate the PCR products of cDNA synthesized in the presence or absence of AMV–RT respectively. (c) cDNA sequence and deduced amino acid sequence of the zinc finger domain of Δexon7 Blimp-1 isoform. The arrow indicates the exon splice junction.
Primer extension
Primer extension was performed with 50 µg of total cellular RNA as described (22) with the following modification. After the overnight hybridization of RNA with the primer, the primer extension reaction was performed using 4 U of Tth DNA polymerase (Boehringer Mannheim, Indianapolis, IN) in the Mn2+ buffer supplied by the manufacturer. The extension was carried out at 50°C for 10 min and then at 65°C for 10 min. The primer extension product was then electrophoresed on standard 8% PAGE with 7 M urea adjacent to a sequence ladder of the genomic Blimp-1 gene. The sequencing reaction of genomic Blimp-1 gene was generated with fmol sequencing kit from Promega. The sequence of primer used in both primer extension and sequencing reactions is 5′-CAGTGTCTGTGCGAGCGAGCGAGTGAGCGA-3′ and is specific for nucleotides 21–50 of cloned Blimp-1 cDNA.
RNase protection assay
RNase protection assay was carried out as described (23). Briefly, the antisense riboprobe was generated from genomic template encompassing the putative transcription initiation sites and hybridized to 20 µg of P3X total cellular RNA. After overnight hybridization, the reaction was treated with RNaseA/RNase T1 mixture and the RNase-resistant riboprobe was electrophoresed on standard 8% PAGE with 7 M urea. The RNA ladder was synthesized by T3 RNA polymerase transcription of PvuII-digested pKS and T7 RNA polymerase transcription of PvuII- or XhoI-digested pKS.
Gene analysis
A BLAST search (24,25) was performed on the whole GenBank database using the murine Blimp-1 amino acid sequence. The program used for the search was tblastn. Several of the species that matched Blimp-1 were translated from cDNA, however the Drosophila melanogaster and Caenorhabditis elegans amino acid sequences were derived from the genomic DNA. In an attempt to verify the accuracy of the predicted proteins, the genomic sequences from D.melanogaster and C.elegans were searched for splice sites using the Neural Network splice site predictor on the Berkeley Drosophila Genome Project page (http://www.fruitfly.org/seq_tools/splice.html) and the NetGene2 World Wide Web server which produces neural network predictions of splice sites in C.elegans (http://130.225.67.199/services/NetGene2/). Sites were used to construct the cDNA if they had a confidence interval score above 0.90 and resulted in cDNA sequence that was in-frame with the mouse Blimp-1 sequence. The resultant amino acid sequence was compared with the amino acid sequence predicted by the genome projects using the Clustal W program (26).
RESULTS
Organization of the murine Blimp-1 gene
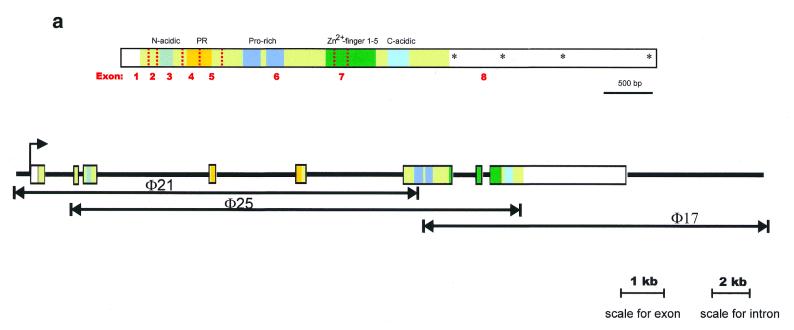
Three λ phage clones from a 129 mouse genomic phage library were isolated by virtue of their hybridization with a probe containing 5.3 kb Blimp-1 cDNA (Fig. 1a). Restriction mapping showed that these overlapping clones together span ~33 kb of genomic DNA. Restriction fragments encoding Blimp-1 mRNA were identified by hybridization to cDNA and sequenced to identify all exon–intron junctions (Fig. 1a). The Blimp-1 gene contains eight exons (Fig. 1a) spanning ~23 kb and all exon–intron junctions conform to the gt/ag rule. In general, all the identified functional domains (N-terminal acidic domain, PEST domain with proline-rich domain and C-terminal acidic domain) are encoded in separate exons with the exception of the PR domain (8) and the five C2H2-type zinc finger domains (Fig. 1a). The PR domain is encoded in exons 4 and 5 while the zinc finger domains are encoded in exons 6, 7 and 8. Of particular interest, zinc finger 1, one of two critical zinc fingers for Blimp-1’s ability to bind DNA (27), is encoded in exons 6 and 7.
Figure 1.
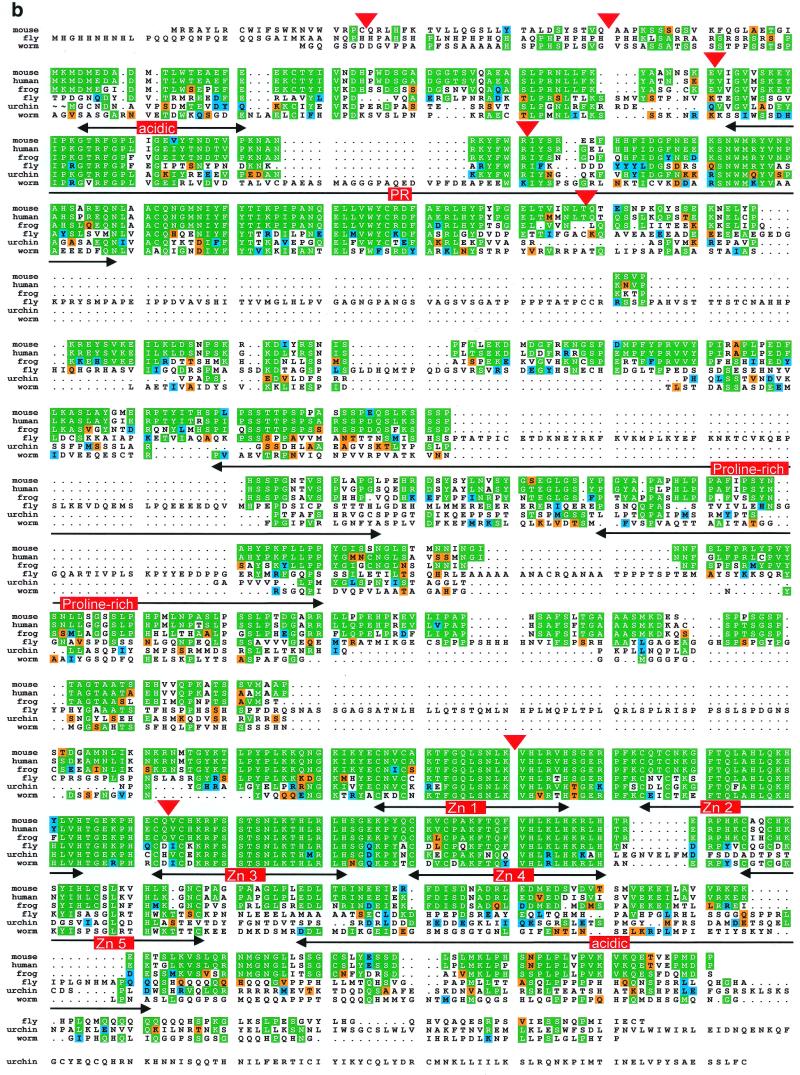
(Above) (a) Organization of the murine Blimp-1 gene. The top panel shows a diagram of Blimp-1 cDNA. The coding region is highlighted in light green, exon splice junctions are indicated by dotted red lines and the protein domains by colored blocks. The predicted poly(A) addition sites in the 3′ UT are indicated with asterisks. The bottom panel shows the location of each exon in the three overlapping phage clones, φ21, φ25 and φ17. (Opposite) (b) Comparison of amino acid sequences of Blimp-1 homologs from mouse (accession no. NP031574), human (accession no. NP001189), frog (accession no. AAF08791), fly (accession no. AAF50759), worm (accession no. T21336) and sea urchin (accession no. AAF30407). From the lineup, consensus sequence was derived (not shown). Amino acids identical to the derived consensus sequence are shown in green, amino acids with conservative change are shown in blue and semi-conservative changes in orange. The functional domains in Blimp-1 protein are indicated. Exon junctions in mouse Blimp-1 protein are indicated by red triangles.
Comparison between the murine and human Blimp-1 genes (GenBank accession no. AL358952 for human Blimp-1 genomic gene) shows that they have a very similar exon–intron organization (data not shown). The only exception is that the 5′ UT in human is encoded in only one exon while in the mouse it is encoded in two exons. Also, we could not find human sequence encoding the conserved histidines of zinc finger 1 in the human gene, because the sequence is still incomplete in this region.
When a BLAST search of the GenBank database was performed to identify Blimp-1 homologs in other species, nine sequences received a score that was significant. These sequences, from human, mouse, frog, worm, fly and sea urchin, were more related to the mouse Blimp-1 sequence than the mouse Blimp-1 sequence was to any other zinc finger or PR family protein. The sequences are 69% identical in the area of zinc fingers 1–4 and there is considerable homology in the PR domain and 5′ acidic region as shown in Figure 1b. Although there is little sequence homology in the proline-rich domain, the sequence from each species contains many prolines that are simply not aligned. The human (PRDI-BF1), Xenopus (Xblimp1) and sea urchin (SpKrox-1) cDNAs have been reported (7,12,13). However the worm and fly amino acid and cDNA sequences are predicted from genomic DNA determined by the C.elegans and Drosophila genome projects (28,29). We verified these predictions to the best of our ability using neural networks to predict splice sites. There was one major difference between the predicted proteins found with the BLAST search and the proteins that we constructed. In the C.elegans sequence there is an acceptor site used in the protein from the genome project that is not recognized by the neural network. The donor site is predicted and the splicing is necessary for the change of reading frame that allows the full protein to be produced. Additionally, there are several donor and acceptor sites that may or may not be used that would add or delete very small sequences that do not alter the match with mouse Blimp-1 significantly. However it is impossible to tell if these are included in the endogenous protein by computer predictions alone. Despite our attempts at predicting the amino acid sequence of the fly and worm Blimp-1, there are probably errors since there are stretches where these two sequences match neither each other nor the sequences from any other species. These non-homologous areas are unlikely to be due to evolutionary distance since the sea urchin Blimp-1 (SpKrox1) does not contain these unmatched gaps. Interestingly, the very 3′ end of sea urchin Blimp-1 contains a leucine-zipper that was not evolutionary conserved.
Blimp-1 has multiple transcription initiation sites and lacks a TATA element
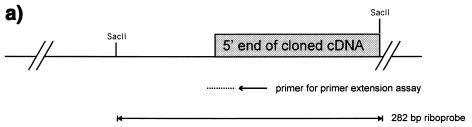
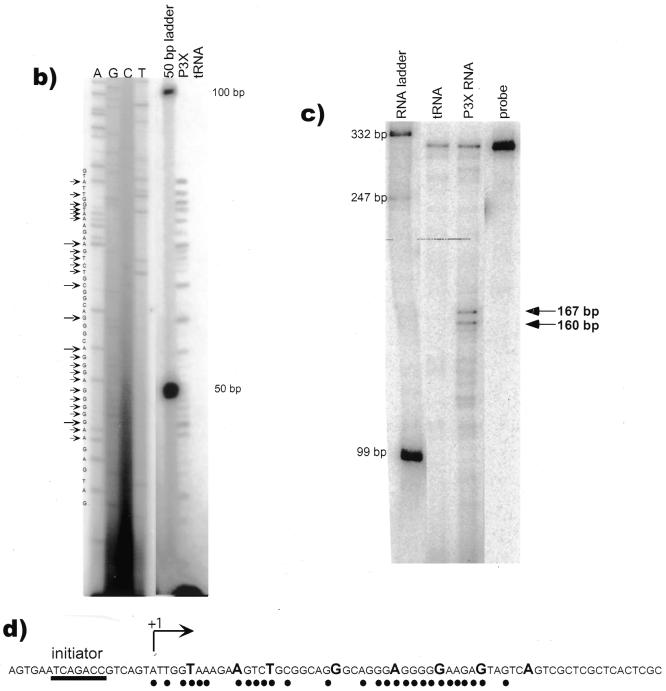
In order to map the transcription initiation sites of murine Blimp-1 gene, a primer extension assay was performed. A primer was chosen from the 5′ UT sequence of the cloned Blimp-1 cDNA (GenBank accession no. U08185). The results showed multiple extension products ranging from 40 to 80 bases (Fig. 2b), suggestive of multiple transcription initiation sites within a region of ~40 bp. However, some of the complexity could have been caused by the inability of the reverse transcriptase to transcribe completely in this GC-rich region. Furthermore, this assay indicates the length of 5′ UT mRNA from the primer but cannot show whether it is encoded by one or more exons. Therefore, a riboprobe protection assay was also performed to confirm the primer extension results and to map the location of transcription initiation sites in Blimp-1. A riboprobe from the genomic DNA encompassing the known 5′ UT and the putative initiation sites determined by primer extension was used (Fig. 2c). Multiple protected fragments were observed (Fig. 2c), in agreement with the primer extension results. The size of the two largest protected fragments, 167 and 160 bp ± 7 bp, is consistent with the size of the largest primer extension products (Fig. 2b). Taken together, these data show that Blimp-1 is transcribed from multiple transcription initiation sites located between 244 and 251 bp from the ATG start codon.
Figure 2.
Mapping transcription initiation site(s) on the Blimp-1 gene. (a) Rationale for the primer extension and RNase protection assays. The primer for primer extension is specific to nucleotides 21–50 of cloned cDNA. The riboprobe template was Blimp-1 genomic DNA encompassing the putative transcription initiation sites mapped by primer extension. (b) Primer extension assay. The first four lanes are a sequence ladder and the next lane size markers. The last two lanes show extension products derived from total RNA from plasmacytoma P3X or from control tRNA. The arrows indicate the putative transcription initiation sites on the Blimp-1 sequence. (c) RNase protection assay. The two major protected fragments are indicated by arrows. The sizes of RNA markers are indicated. (d) The sequence of transcription initiation sites. The sites mapped by primer extension assay are indicated by dots. The sites mapped by RNase protection assay are indicated by capital letters of the nucleotide sequence. The 5′-most transcription initiation site is assigned the +1 position. The initiator element likely to be responsible for Blimp-1 transcription initiation is underlined.
The occurrence of multiple transcription initiation sites is the hallmark of transcription from TATA-less promoters that usually contain initiator elements (30,31). We examined the DNA sequence in the vicinity of Blimp-1 transcriptional start sites and found no TATA sequence. However, there is an initiator element located 13 bp upstream to the most 5′ transcription initiation site. It is likely that this initiator is responsible for the transcription of Blimp-1 gene (Fig. 2d).
Three major Blimp-1 mRNA isoforms are generated by differential polyadenylation but a minor splice variant lacking exon 7 is also expressed
Knowing the exon–intron structure and the location of transcription initiation sites in the gene allowed us to characterize Blimp-1 mRNA isoforms. Blimp-1 mRNA is found in three major isoforms of ~5.7, 4.3 and 3.6 kb, determined by northern blot analysis (Fig. 3a) (11). The original cDNA clone (5) is 5.3 kb exclusive of poly(A), corresponding to the 5.7 kb mRNA isoform. Probes corresponding to all the protein-coding exons and to the 5′ UT hybridized to all three mRNA isoforms in northern blots of total RNA from P3X cells (Fig. 3a and data not shown) suggesting that the three isoforms encode the same protein. Analysis of the 2.5 kb 3′ UT portion of the cDNA sequence revealed several potential polyadenylation sites [poly(A) sites]. We therefore performed northern analyses using probes from different regions of the 3′ UT to determine whether the smaller isoforms were generated by the use of different poly(A) sites. A probe derived from the first 400 bp of the 3′ UT and a probe derived from the middle portion of the 3′ UT region detected the 5.7 and 4.3 kb isoforms but did not hybridize to the 3.6 kb isoform. A probe from the very 3′ end of the cDNA detected only the 5.7 kb isoform and did not hybridize to the 3.6 or 4.3 kb isoforms. Thus, the 3.6 kb isoform appears to result from polyadenylation at the poly(A) site located ~3.3 kb of the cDNA. The 4.3 kb isoform could result from usage of intermediate poly(A) site located at ~4.5 kb and the 5.7 kb isoform results from usage of the most 3′ poly(A) site. Thus, the three major Blimp-1 mRNA isoforms are generated by differential use of multiple poly(A) sites and all encode the same protein.
mRNAs encoding some zinc finger-containing proteins are expressed as alternatively spliced isoforms, differing in the presence of exons encoding specific zinc fingers (32–34). Since all of zinc finger 2 and parts of zinc fingers 1 and 3 are encoded by a small exon, exon 7, we examined whether an alternatively spliced isoform of Blimp-1 lacking exon 7 might be expressed. (Due to the small differences in size, such an isoform would not be revealed by northern analysis.) PCR primers for cDNA sequence 5′ of the region encoding zinc finger 1 and 3′ of the region encoding zinc finger 5 were used (Fig. 3b) in an RT–PCR reaction using RNA from plasmacytoma P3X. Each primer pair gave a product predicted for full-length Blimp-1 mRNA as well as another product ~120 bp smaller (Fig. 3b). The size of the smaller RT–PCR product is consistent with an alternatively spliced isoform lacking exon 7. When the smaller RT–PCR product was cloned and sequenced, it was indeed shown to be an alternatively spliced form lacking exon 7 (Fig. 3c).
RT–PCR (Fig. 3b) indicated that the isoform lacking exon 7 (Δexon7) constitutes <10% of total Blimp-1 mRNA in P3X cells. Riboprobe protection assays confirmed this proportion and showed that it did not vary when RNAs from various mouse tissues were examined (data not shown). The protein encoded by the Δexon7 isoform lacks the functional zinc finger 1 and 2 and is not predicted to bind DNA efficiently since Keller and Maniatis (27) showed that these two zinc fingers of Blimp-1 are critical for DNA binding activity. Indeed, the zinc finger domain synthesized in 293T cells from the Δexon7 cDNA did not bind to the Blimp-1 sequence from the c-myc promoter while binding of the zinc finger domain synthesized from the full-length cDNA was clearly evident (data not shown).
Developmental stage-specific expression of Blimp-1 in B cells is determined by transcription initiation
Blimp-1 protein and steady-state mRNA are present in plasmacytoma lines but are absent from cell lines representing earlier stages of B cell development (5,9). To determine if this B cell stage-specific expression is the result of differential transcriptional initiation, transcription run-on assays were performed. Nuclei were isolated from two B-lineage cell lines: P3X, a plasmacytoma that expresses Blimp-1, and 18-81, a pre-B line that does not express Blimp-1. The results showed no polymerase loading on the Blimp-1 gene in 18-81 nuclei, but significant polymerase loading on the Blimp-1 gene in P3X (Fig. 4). Transcription of Cµ, GAPDH and c-myc genes was also measured to provide positive controls for the assay. The amount of polymerase loading on Blimp-1 was 5-fold higher than that on c-myc, and ~7.5-fold lower than Cµ in P3X cells. These data show that developmental stage-specific expression of Blimp-1 in B cells is regulated by transcription initiation.
Figure 4.
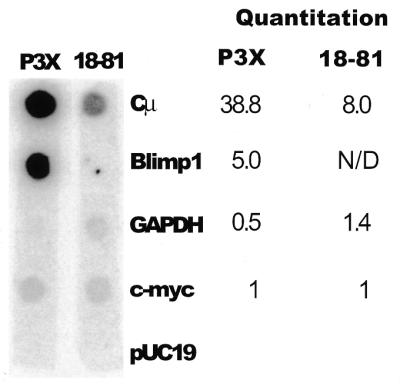
Polymerase loading on the Blimp-1 gene in B cell lines. Nuclear run-on assay showing the transcription of Cµ, Blimp-1, GAPDH and c-myc genes in nuclei from plasmacytoma (P3X) and pre-B (18-81) cells. pUC19 DNA was used as negative control. The amount of each transcript was quantified using a PhosphoImager and corrected for uridine content. The relative amount of each transcript, compared to c-myc, is shown.
The Blimp-1 basal promoter is contained within 900 bp 5′ of the transcription initiation sites
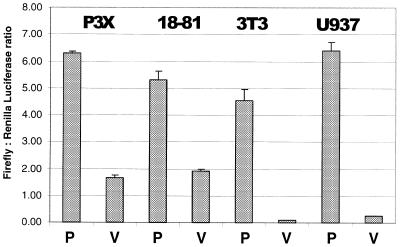
A transfection assay was used to search for the Blimp-1 promoter. A region from –918 to +204 bp relative to the most 5′ transcription initiation site of the Blimp-1 gene was engineered into a plasmid containing the firefly luciferase cDNA but lacking a TATA or initiator element. Upon transfection into P3X plasmacytoma cells, the plasmid containing Blimp-1 sequence, showed 3–4-fold more luciferase activity than the parent plasmid after correction for transfection efficiency (Fig. 5). Thus, the region containing ~900 bp 5′ of the initiation sites is sufficient to confer basal promoter activity. To determine if this region also contained elements capable of conferring B-cell developmental stage-specificity or tissue-specificity, these plasmids were also transfected into 18-81 preB cells, NIH 3T3 cells and human U937 promonocytic cells, which do not express endogenous Blimp-1. The Blimp-1 plasmid had similar promoter activity in all cell lines tested. Thus we conclude that additional regulatory elements, outside the 1100 bp present on our reporter, are likely to be required for tissue- and developmental stage-specific expression of Blimp-1.
Figure 5.
Identification of the Blimp-1 basal promoter. Transient transfection of a luciferase reporter dependent upon the Blimp-1 gene from –918 to +207 bp. (GenBank accession no. AF305534) in cell lines: P3X, 18-81, NIH 3T3 and U937. P denotes the reporter containing the promoter region; V denotes the parental reporter. The data were corrected for the transfection efficiency using Renilla luciferase and the mean ± standard deviation of triplicate data points is shown.
DISCUSSION
In this report, we analyzed the murine Blimp-1 gene and found that it contains eight exons, a transcriptional initiator and multiple transcription initiation sites. The organization of the human Blimp-1 gene is similar to that of the mouse. The three major Blimp-1 mRNA isoforms are generated by differential poly(A) site usage and a minor isoform, encoding a protein lacking zinc fingers 1 and 2, is generated by differential splicing. We showed that B-cell developmental stage-specific expression of Blimp-1 is controlled by transcription initiation. Functional analyses showed that a region 900 bp 5′ of the Blimp-1 transcription initiation sites is sufficient for basal, but not B-cell stage-specific, promoter activity.
Evolution of the Blimp-1 gene
Blimp-1 was originally identified in human (7) and murine (5) cells; subsequently Blimp-1 homologs were identified in Xenopus (12) and sea urchin (13). In this work we compared these Blimp-1 homologs as well as those in two more primitive eukaryotes, D.melanogaster and C.elegans (Fig. 1b) that have been identified by genome sequencing. However, Blimp-1 homologs were not found in the genomes of lower, unicellular eukaryotes, such as Saccharomyces cerevisiae, for which genome sequence is available. Thus, Blimp-1 appears to have arisen during early evolution of multicellular organisms, consistent with the suggestion of its role as an inducer for terminal differentiation.
In all of the Blimp-1 homologs, conservation of each known protein domain is evident, with the zinc finger domains having the highest degree of conservation. This domain conservation suggests that some/many biological functions of Blimp-1 are likely to be conserved between lower forms and mammals. Consistent with this idea is the finding of de Souza et al. (12) that mouse Blimp-1 can substitute XBlimp1 during frog embryogenesis. In mouse, Blimp-1 is important for terminal differentiation of B lymphoid and myeloid cells (9,11,35), is expressed and may play a role in other end-stage cells (11) and is required for development of the embryo (M.Davis, personal communication). Blimp-1 is a DNA-binding transcriptional repressor. One target of Blimp-1-dependent repression in hematopoietic cells is c-myc (9,11), a gene that plays key roles in cell growth and apoptosis (36,37) and which is repressed in terminally differentiated cells. However, we (J.Piskurich and K.Lin, unpublished data) and others (7) have also identified other genes that are targets of Blimp-1 repression in mice and human cells. It is interesting to note that although Xenopus (38) and Drosophila (39) have c-myc homologs, C.elegans does not. It will be particularly interesting to learn if Blimp-1 regulates cell proliferation or terminal differentiation in C.elegans, where, presumably, there are Blimp-1 targets not related to c-myc.
Blimp-1 mRNA isoforms
Although three major and one minor isoform of Blimp-1 mRNA have been identified, their significance is not readily apparent. The three major isoforms differ with respect to 3′ UT sequence but encode the same protein. The three major isoforms are found in human as well (11) where they are also likely to be the result of alternative polyadenylation. Although the 3′ UT region of human Blimp-1 cDNA has not been cloned, we could identify in the human Blimp-1 gene the same putative poly(A) addition sequences identified in the mouse 3′ UT region. Different 3′ UT sequence and/or poly(A) could affect mRNA stability or translatability. However, the proportion of these isoforms to one another is similar in most tissues of the adult mouse we have investigated (D.Chang, unpublished results). The data suggest, therefore, that these isoforms have neither functional nor regulatory significance.
The minor isoform lacking exon 7 encodes a protein that does not bind DNA. Theoretically this protein could interfere with the function of the full-length Blimp-1 either by forming a heterodimer with altered function or by sequestering Blimp-1-associating proteins required for function. A recent report shows that an aberrant splice product of the zinc finger protein Ikaros fails to bind DNA and blocks normal Ikaros function by dimerization (40). At present there is no evidence that Blimp-1 heterodimerizes, making this mechanism unlikely. However, Blimp-1 does require association with hGroucho (41) and HDAC (6) in order to function as a transcriptional repressor. If the small, non-DNA binding form is present at sufficient levels, it might sequester one or both of these proteins and block Blimp-1-dependent repression. Our analyses show that steady-state mRNA encoding the small form is usually 10-fold lower than that encoding full-length protein, making this mechanism unlikely unless the small protein is preferentially translated or has increased stability. When antisera become available to distinguish the two forms, it will be important to determine their relative abundance in different cells.
Regulation of Blimp-1 gene expression
In frogs, sea urchins and mice, Blimp-1 mRNA is expressed early in development (12,13). It is also expressed in various terminally differentiated cells of mouse and human and is induced upon terminal differentiation of both B lymphoid and myeloid cells. Thus, expression of Blimp-1 mRNA is subject to both lineage and developmental stage-specific regulation. The data presented here show that the developmental stage-specific expression of Blimp-1 mRNA in B cells is regulated at the level of transcription initiation. Transfection analyses showed that a 1.1 kb region containing the Blimp-1 transcription initiation sites, initiator element, and ~900 bp 5′ to the initiation sites has basal promoter activity. However, our studies did not reveal tissue or developmental stage specificity conferred by this region. More detailed analyses, possibly utilizing transgenic mice, may be necessary to unravel the transcriptional regulatory elements of the Blimp-1 gene. It is possible that regulation is mediated by transcriptional repressors that could not be identified by transient transfection. Furthermore, some or all of the transcriptional regulation may be mediated by elements such as enhancers or locus control regions, located distal to the promoter. The gene organization presented in this study provides the basis for a systematic search for such regulatory elements.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr M. Davis for providing the Blimp-1 cDNA, Dr M. Coutts for providing the GAPDH cDNA, Dr G. Siu for providing the genomic phage libraries and to members of the Calame laboratory for helpful discussions. This work was supported by RO1AI4576 to K.C. C.T. was supported by a Fellowship from the Leukemia and Lymphoma Society (no. 5295-98) and M.S. is supported by NIH Immunology Training Grant (5T32A107525).
DDBJ/EMBL/GenBank accession nos AF305534–AF305539
REFERENCES
- 1.Lamson G. and Koshland,M.E. (1984) Changes in J chain and mu chain RNA expression as a function of B cell differentiation. J. Exp. Med., 160, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krenacs L., Himmelmann,A.W., Quintanilla-Martinez,L., Fest,T., Riva,A., Wellmann,A., Bagdi,E., Kehrl,J.H., Jaffe,E.S. and Raffeld,M. (1998) Transcription factor B-cell-specific activator protein (BSAP) is differentially expressed in B cells and in subsets of B-cell lymphomas. Blood, 92, 1308–1316. [PubMed] [Google Scholar]
- 3.Oliver A.M., Martin,F. and Kearney,J.F. (1997) Mouse CD38 is down-regulated on germinal center B cells and mature plasma cells. J. Immunol., 158, 1108–1115. [PubMed] [Google Scholar]
- 4.Sanderson R.D., Lalor,P. and Bernfield,M. (1989) B lymphocytes express and lose syndecan at specific stages of differentiation. Cell Regul., 1, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner C.A. Jr, Mack,D.H. and Davis,M.M. (1994) Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell, 77, 297–306. [DOI] [PubMed] [Google Scholar]
- 6.Yu J., Angelin-Duclos,C., Greenwood,J., Liao,J. and Calame,K. (2000) Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol., 20, 2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keller A.D. and Maniatis,T. (1991) Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev., 5, 868–879. [DOI] [PubMed] [Google Scholar]
- 8.Huang S. (1994) Blimp-1 is the murine homolog of the human transcriptional repressor PRDI-BF1 [letter]. Cell, 78, 9. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y., Wong,K. and Calame,K. (1997) Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science, 276, 596–599. [DOI] [PubMed] [Google Scholar]
- 10.Angelin-Duclos C., Cattaretti,G., Lin,K.-I. and Calame,K.L. (2000) Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J. Immunol., 165, 5462–5471. [DOI] [PubMed] [Google Scholar]
- 11.Chang D., Angelin-Duclos,C. and Calame,K. (2000) BLIMP-1: trigger for differentiation of myeloid lineage. Nature Immunol., 1, 169. [DOI] [PubMed] [Google Scholar]
- 12.de Souza F.S., Gawantka,V., Gomez,A.P., Delius,H., Ang,S.L. and Niehrs,C. (1999) The zinc finger gene Xblimp1 controls anterior endomesodermal cell fate in Spemann’s organizer. EMBO J., 18, 6062–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Wikramanayake,A.H., Gonzalez-Rimbau,M., Vlahou,A., Flytzanis,C.N. and Klein,W.H. (1996) Very early and transient vegetal-plate expression of SpKrox1, a Kruppel/Krox gene from Stronglyocentrotus purpuratus. Mech. Dev., 60, 185–195. [DOI] [PubMed] [Google Scholar]
- 14.Quertermous T. (1989) In Ausubel,F.M. (ed.), Current Protocols in Molecular Biology. John Wiley & Son, Inc., New York, Vol. I, Unit 6.1.
- 15.Strauss W.M. (1989) In Ausubel,F.M. (ed.), Current Protocols in Molecular Biology. John Wiley & Son, New York, Vol. I, Unit 6.3.
- 16.Avitahl N. and Calame,K. (1996) A 125 bp region of the Ig VH1 promoter is sufficient to confer lymphocyte-specific expression in transgenic mice. Int. Immunol., 8, 1359–1366. [DOI] [PubMed] [Google Scholar]
- 17.Artandi S.E., Cooper,C., Shrivastava,A. and Calame,K. (1994) The basic helix-loop-helix-zipper domain of TFE3 mediates enhancer-promoter interaction. Mol. Cell. Biol., 14, 7704–7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews J.C., Hori,K. and Cormier,M.J. (1977) Substrate and substrate analogue binding properties of Renilla luciferase. Biochemistry, 16, 5217–5220. [DOI] [PubMed] [Google Scholar]
- 19.Brown T. (1989) In Ausubel,F.M. (ed.), Current Protocols in Molecular Biology. John Wiley & Son, Inc., New York, Vol. I, Unit 4.9.
- 20.Greenberg M.E. (1989) In Ausubel,F.M. (ed.), Current Protocols in Molecular Biology. John Wiley & Son, Inc., New York, Vol. I, Unit 4.10.
- 21.Klickstein L.B. (1989) In Ausubel,F.M. (ed.), Current Protocols in Molecular Biology. John Wiley & Son, Inc., New York, Vol. I, Unit 5.5.
- 22.Maniatis T., Fritsch,E.F. and Sambrook,J. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, NY.
- 23.Gilman M. (1989) In Ausubel,F.M. (ed.), Current Protocols in Molecular Biology. John Wiley & Son, Inc., New York, Vol. I, Unit 4.7.
- 24.Gish W. and States,D.J. (1993) Identification of protein coding regions by database similarity search. Nature Genet., 3, 266–272. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller A.D. and Maniatis,T. (1992) Only two of the five zinc fingers of the eukaryotic transcriptional repressor PRDI-BF1 are required for sequence-specific DNA binding. Mol. Cell. Biol., 12, 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The C.elegans sequencing consortium (1998) Genome sequence of the nematode C.elegans: a platform for investigating biology. [published errata appear in Science (1999) 283, 35 and (1999) 283, 2103 and (1999) 285, 1493]. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 29.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 30.Lo K. and Smale,S.T. (1996) Generality of a functional initiator consensus sequence. Gene, 182, 13–22. [DOI] [PubMed] [Google Scholar]
- 31.Javahery R., Khachi,A., Lo,K., Zenzie-Gregory,B. and Smale,S.T. (1994) DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol., 14, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Eyndhoven W.G., Gamper,C.J., Cho,E., Mackus,W.J. and Lederman,S. (1999) TRAF-3 mRNA splice-deletion variants encode isoforms that induce NF-kappaB activation. Mol. Immunol., 36, 647–658. [DOI] [PubMed] [Google Scholar]
- 33.Hahm K., Ernst,P., Lo,K., Kim,G.S., Turck,C. and Smale,S.T. (1994) The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol. Cell. Biol., 14, 7111–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabanillas A.M. and Darling,D.S. (1996) Alternative splicing gives rise to two isoforms of Zfhep, a zinc finger/homeodomain protein that binds T3-response elements. DNA Cell Biol., 15, 643–651. [DOI] [PubMed] [Google Scholar]
- 35.Schliephake D.E. and Schimpl,A. (1996) Blimp-1 overcomes the block in IgM secretion in lipopolysaccharide/anti-mu F(ab’)2-co-stimulated B lymphocytes. Eur. J. Immunol., 26, 268–271. [DOI] [PubMed] [Google Scholar]
- 36.Dang C.V. (1999) c-myc target genes involved in cell growth, apoptosis and metabolism. Mol. Cell. Biol., 19, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prendergast G.C. (1999) Mechanisms of apoptosis by c-myc. Oncogene, 18, 2967–2987. [DOI] [PubMed] [Google Scholar]
- 38.Principaud E. and Spohr,G. (1991) Xenopus laevis c-myc I and II genes: molecular structure and developmental expression. Nucleic Acids Res., 19, 3081–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallant P., Shiio,Y., Cheng,P.F., Parkhurst,S.M. and Eisenman,R.N. (1996) Myc and Max homologs in Drosophila. Science, 274, 1523–1527. [DOI] [PubMed] [Google Scholar]
- 40.Sun L., Goodman,P.A., Wood,C.M., Crotty,M.L., Sensel,M., Sather,H., Navara,C., Nachman,J., Steinherz,P.G., Gaynon,P.S. et al. (1999) Expression of aberrantly spliced oncogenic ikaros isoforms in childhood acute lymphoblastic leukemia [see comments]. J. Clin. Oncol., 17, 3753–3766. [DOI] [PubMed] [Google Scholar]
- 41.Ren B., Chee,K.J., Kim,T.H. and Maniatis,T. (1999) PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev., 13, 125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]