Abstract
Filoviruses are highly lethal in humans and nonhuman primates, likely due to potent antagonism of host interferon (IFN) responses early in infection. Filoviral protein VP35 is implicated as the major IFN induction antagonist, while Ebola virus (EBOV) VP24 or Marburg virus (MARV) VP40 are known to block downstream IFN signaling. Despite progress elucidating EBOV and MARV antagonist function, those for most other filoviruses, including Reston (RESTV), Sudan (SUDV), Taï Forest (TAFV), Bundibugyo (BDBV) and Ravn (RAVV) viruses, remain largely neglected. Thus, using standardized vectors and reporter assays, we characterized activities by each IFN antagonist from all known ebolavirus and marburgvirus species side-by-side. We uncover noncanonical suppression of IFN induction by ebolavirus VP24, differing potencies by MARV and RAVV proteins, and intriguingly, weaker antagonism by VP24 of RESTV. These underlying molecular explanations for differential virulence in humans could guide future investigations of more-neglected filoviruses as well as treatment and vaccine studies.
Keywords: Filoviruses, Ebolavirus, Marburgvirus, Neglected pathogens, Innate immune response, Viral immune evasion, Interferon pathway, Filoviral interferon antagonists, Virus-host interactions, Viral protein function
1. Introduction
Over the past fifty years, we have come to recognize many emerging viruses capable of spillover from their reservoir hosts and causing severe disease in non-adapted animals, including humans and non-human primates (NHPs) (Wong et al., 2014; Pappalardo et al., 2016; Messaoudi et al., 2015; Slenczka and Klenk, 2007; Choi et al., 2015). The family Filoviridae, negative-sense single-stranded RNA viruses consisting of genera Ebolavirus, Marburgvirus and recently-described Cuevavirus, include some of the most dangerous viruses known. Several family members can cause severe, often fatal illnesses in humans and NHPs during sporadic outbreaks, mostly in sub-Saharan Africa (Wong et al., 2014; Messaoudi et al., 2015; Slenczka and Klenk, 2007; Burk et al., 2016; Bukreyev et al., 2014). Filoviruses comprise seven known species, with eight viruses: marburgviruses Marburg virus (MARV) and Ravn virus (RAVV); ebolaviruses Ebola virus (EBOV), Sudan virus (SUDV), Reston virus (RESTV), Taï Forest virus (TAFV) and Bundibugyo virus (BDBV); and most recently, the cuevavirus Lloviu virus (LLOV) (Messaoudi et al., 2015; Slenczka and Klenk, 2007; Burk et al., 2016; Bukreyev et al., 2014; Kuhn et al., 2014). Human illnesses have been recorded for all of these filoviruses, with the notable exceptions of RESTV and LLOV. RESTV is frequently lethal in NHPs yet anecdotal evidence suggests it may be apathogenic in humans, while LLOV has not been isolated and so far only detected in dead bats (Messaoudi et al., 2015; Slenczka and Klenk, 2007; Burk et al., 2016).
Recent ecological breakthroughs have identified the cave-dwelling fruit bat, the Egyptian rousette (Rousettus aegyptiacus), as a natural reservoir of MARV and a likely source of virus transmission to humans (Towner et al., 2009, 2007; Amman et al., 2014, 2012). Natural and experimental evidence have shown that MARV replicates well in these bats with sustained viremia and oral shedding of virus, but no overt illness or mortality, suggesting MARV is well adapted to this particular reservoir host (Towner et al., 2009; Taylor et al., 2011; Jones et al., 2015; Amman et al., 2015). While it is still unknown precisely what factors mediate the variable pathogenicity seen among filoviruses in primates, or between reservoir and spillover hosts, public health interest to understand these ecological and molecular determinants has accelerated in recent years (Pappalardo et al., 2016; Messaoudi et al., 2015; Burk et al., 2016; Dunham et al., 2015).
The severe disease observed during human infections by filoviruses is thought to occur in part due to the ability of the virus to antagonize host innate immune responses (Wong et al., 2014; Messaoudi et al., 2015; Slenczka and Klenk, 2007; Cárdenas, 2010; Ramanan et al., 2011; Basler, 2015). This antagonism subverts proper and balanced expression of innate immune factors, and allows for virus replication to proceed and disseminate unchecked during critical early stages of infection (Wong et al., 2014; Messaoudi et al., 2015; Slenczka and Klenk, 2007; Cárdenas, 2010; Ramanan et al., 2011; Basler, 2015). This can dysregulate proinflammatory cytokines and clotting factors, promote CD4+ T lymphocyte apoptosis, cause dendritic cell inactivation and vascular defects, and impair or delay humoral immune responses, all of which can contribute to systemic illness and multisystem organ failure (Wong et al., 2014; Messaoudi et al., 2015; Slenczka and Klenk, 2007; Cárdenas, 2010; Ramanan et al., 2011; Basler, 2015; Bosio et al., 2003).
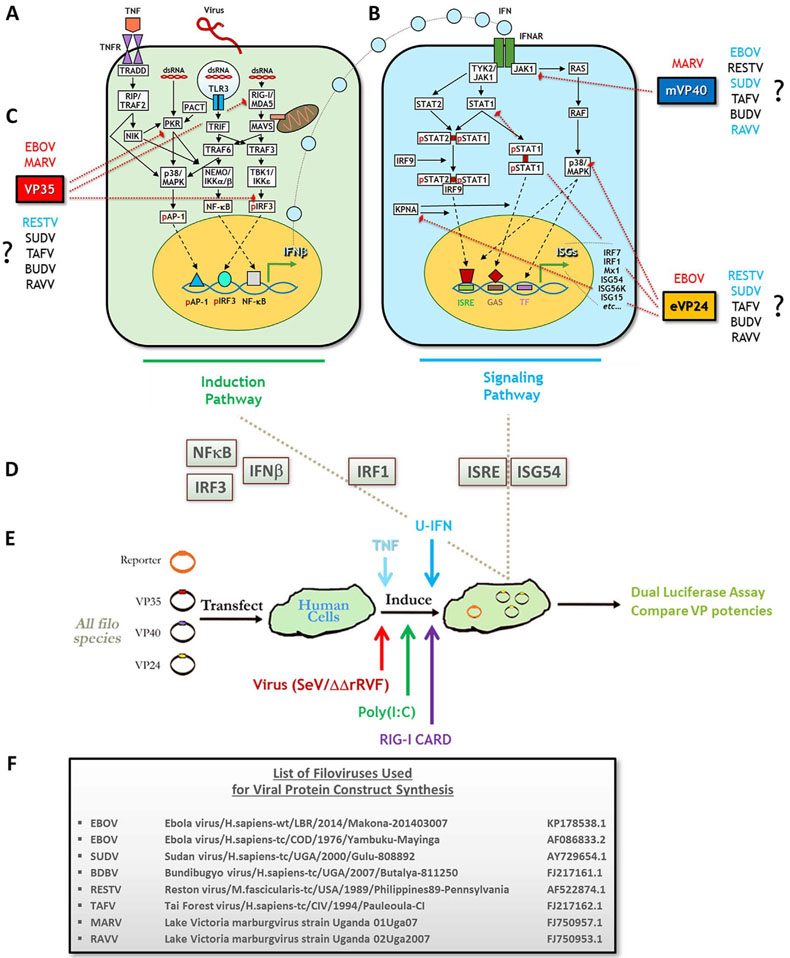
Innate immunity in virally-infected human cells is normally mediated by an interferon (IFN) antiviral response that consists of IFN induction and IFN signaling pathways, illustrated in Figures Fig. 1A and B (Wong et al., 2014; Cárdenas, 2010; Ramanan et al., 2011; Basler, 2015; Haller and Weber, 2009; Haller et al., 2006; Hoffmann et al., 2015). Briefly, viral double-stranded (ds)RNA is recognized by retinoic acid inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5), as well as protein kinase R (PKR) or toll-like receptors (TLRs) (Cárdenas, 2010; Ramanan et al., 2011; Basler, 2015; Haller and Weber, 2009; García et al., 2006; Kawai and Akira, 2006). These activate induction pathway intermediates, such as mitochondrial antiviral signaling protein (MAVS) and TNF receptor (TNFR) associated factor 6 (TRAF6), to recruit complexes including TANK-binding kinase 1 (TBK1) with inducible IκB kinase epsilon (IKKε), or IKKα/β with nuclear factor of kappa B (NF-κB) essential modifier (NEMO). The intermediates also initiate the mitogen-activated protein kinase (MAPK) cascade (Basler, 2015; Haller and Weber, 2009; García et al., 2006; Kawai and Akira, 2006; Halfmann et al., 2011). The culmination of these pathways lead to activation and nuclear translocation of the three main transcription factors necessary for IFN induction: interferon regulator factor 3 (IRF3), NF-κB and activating protein 1 (AP-1), which together transactivate the IFNβ promoter (Basler, 2015; Haller and Weber, 2009; Kawai and Akira, 2006; Halfmann et al., 2011; Yarilina and Ivashkiv, 2010). IFNs then trigger downstream signaling via IFNα receptor (IFNAR) binding and Janus kinase 1 (Jak1)/signal transducer and activator of transcription (STAT) pathway activation (Cárdenas, 2010; Ramanan et al., 2011; Basler, 2015; Haller and Weber, 2009; Hoffmann et al., 2015). Intermediate signaling allows the translocation factor karyopherin α to bind and shuttle STAT1 homo- or STAT1/2 heterodimers to the nucleus. STATs transactivate at interferon-stimulated gene (ISG) response elements (ISRE) or gamma-activated sequences (GAS) to induce numerous ISGs, such as IRF7, which mediate entry of the cell into an antiviral state that limits pathogen replication and dissemination (Cárdenas, 2010; Ramanan et al., 2011; Basler, 2015; Haller and Weber, 2009; Hoffmann et al., 2015; Reid et al., 2006, 2007).
Fig. 1.
Assessing a side-by-side panel of standardized filovirus antagonist constructs for inhibition of IFN induction or IFN signaling response pathways using response gene reporters and various inducers. The IFN antiviral response consists of upstream induction (left, in green) and downstream signaling (right, in blue) pathways. (A) First, infection by virus and intracellular release of viral dsRNA lead to activation of PRRs, including RIG-I/MDA5, TLR3 and PKR, to trigger diverse induction pathway intermediates, each with potential crosstalk. Intermediates phosphorylate and activate transcription factors AP-1, NF-κB and IRF3, and TNFR stimulation can further activate MAPK and NF-κB pathways. The three TFs translocate to the nucleus and transactivate the IFNβ promoter. (B) Next, the IFN signaling pathway is initiated when IFN is secreted and binds its receptor to trigger Jak1/Tyk. These factors activate STATs to homo- or heterodimerize, or initiate MAPK cascades via Ras/Raf. STAT heterodimers also associate with IRF9. Activated STATs then interact with translocation factor karyopherin α (KPNA), which shuttles them to the nucleus for transactivation at ISRE or GAS promoter elements, upregulating expression of dozens of ISGs. Activated MAPK intermediates such as p38 can enhance STAT transactivation or activate additional TFs, potentiating the response. ISG induction enters the cell into a full antiviral state. (C) Filoviral VP35 (red), VP40 (blue) and VP24 (orange) proteins antagonize IFN pathways. EBOV and MARV VP35 antagonize IFN induction by blocking PKR and RIG-I/MDA5 activities and preventing IRF3 phosphorylation; MARV (m)VP40 and EBOV(e)VP24 antagonize IFN signaling by inhibiting Jak1 activation (mVP40), or by inhibiting STAT1 activation, KPNA interaction with STAT1 or p38 phosphorylation (eVP24). While much is known about EBOV or MARV proteins, questions remain regarding the antagonists of the more neglected filoviruses (RESTV, SUDV, TAFV, BUDV and RAVV). Antagonists for each virus highlighted in blue have few descriptions in the literature, and those in black have no published studies. (D) Immune response reporters with activities/expression in either IFN induction (NF-κB, IRF3, IFNβ), IFN signaling (ISRE, ISG54) or both pathways (IRF1) were selected for the assays conducted as in (E) All VP constructs were singly transfected side-by-side into HEK293s with each of the described reporters in triplicate, induced by indicated agents and then harvested for luciferase activity (see Materials and Methods). (F) Each VP35, VP40 and VP24 WT or tagged construct was derived from the indicated representative isolates for all known ebolavirus and marburgvirus species. p=phosphorylated; SeV=Sendai virus; ΔΔrRVF=attenuated recombinant Rift Valley fever virus with NSs and NSm gene deletions; U-IFN=universal Type I IFN; Uga=Uganda (Pappalardo et al., 2016; Burk et al., 2016; Reid et al., 2007; Chakraborty et al., 2014; Zhang et al., 2012a; Kimberlin et al., 2010; Clifton et al., 2015; Valmas and Basler, 2011; Cong et al., 2015; Leung et al., 2010a; Feagins and Basler, 2014).
Filoviruses, in turn, have evolved potent strategies to inhibit and evade innate immune responses, primarily through the activities of three viral protein (VP) antagonists, VP35, VP24 and VP40. Most knowledge has come via studies focusing on these antagonists from two representative filoviruses, EBOV and MARV (Wong et al., 2014; Burk et al., 2016; Cárdenas, 2010; Ramanan et al., 2011; Basler, 2015). The three proteins antagonize different stages of induction or signaling pathways within the IFN response (Fig. 1C). VP35 of both EBOV and MARV binds to foreign dsRNA to suppress RIG-I-mediated virus recognition and subsequent IFN induction (Messaoudi et al., 2015; Slenczka and Klenk, 2007; Cárdenas, 2010; Ramanan et al., 2011, 2012; Ilinykh et al., 2015; Edwards et al., 2016; Bale et al., 2012; Yen et al., 2014). VP35 also binds to IKKε and TBK1 to inhibit phosphorylation of IRF3 and disrupt interactions with MAVS and IRF7 (Messaoudi et al., 2015; Ramanan et al., 2011; Basler, 2015; Edwards et al., 2016; Yen et al., 2014; Hartman et al., 2008a). Further, VP35 blocks activation of PKR and interaction between RIG-I and cellular PKR activator (PACT), preventing RIG-I stimulation (Messaoudi et al., 2015; Ramanan et al., 2011; Basler, 2015; Edwards et al., 2016; Hartman et al., 2008a; Kok et al., 2011; Schumann et al., 2009). Meanwhile, EBOV VP24 and MARV VP40 seem to have evolved independent strategies to antagonize downstream IFN signaling. VP24 binds to karyopherin α to prevent STAT1 translocation, and can also block p38 MAPK signaling; MARV VP40 prevents Jak1 activation (Ramanan et al., 2011; Halfmann et al., 2011; Reid et al., 2006; Zhang et al., 2012a, 2012b; Valmas and Basler, 2011; Mateo et al., 2010; Valmas et al., 2010).
Thus the current, canonical understanding of filoviral IFN antagonism has been that VP35 is the only protein to target IFN induction, while MARV VP40/EBOV VP24 target IFN signaling (Wong et al., 2014; Cárdenas, 2010; Ramanan et al., 2011; Basler, 2015). The interplay of these viral proteins to shut down host innate immune responses may be critical for subsequent viral replication and human disease (Kash et al., 2006). Indeed, our lab and others have shown that a single residue change in VP35 can ablate its antagonizing activity, and that infection by recombinant MARV or EBOV containing mutant forms of VP35 show robust IFN induction and concomitant, strongly attenuated viral growth in vitro, and for EBOV, a lack of severe disease in mice (Hartman et al., 2008a, 2006, 2008b; Albariño et al., 2015). To date, no IFN antagonistic function has been ascribed to VP40 of any ebolavirus (Ramanan et al., 2011; Basler, 2015; Valmas et al., 2010). Further, there remains little to no information in the literature describing the activities or efficiencies of the putative antagonist proteins for other filoviruses (Fig. 1C), which have considerable potential to cause human disease with varying degrees of severity (Burk et al., 2016; Feagins and Basler, 2015; Carroll et al., 2013; Towner et al., 2008; Kuhn et al., 2011). Recognizing this gap, we investigated side-by-side antagonist activities by VP35, VP24 and VP40 from representatives of all known filovirus isolates (Fig. 1F) in both IFN induction and signaling pathways. To this end, we used a standardized vector library and reporter-based system for single-assay measurement of protein potencies following induction by virus, dsRNA or cytokines in human cells (Fig. 1D and E). We report several new findings, including novel inhibitory roles for ebolavirus VP24 and marburgvirus VP40 proteins in IFN induction, moderate antagonistic activities by SUDV and TAFV VP40, differences between marburgvirus VP40 proteins, and an intriguing inefficiency by RESTV VP24 to inhibit IFN responses. We believe these findings could better uncover principles that govern filoviral antagonist function and host interactions, as well as allow for exploitation of these antagonists for the development of antiviral agents.
2. Materials and methods
2.1. Cell culture
Human embryonic kidney (HEK) 293 cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco), supplemented with 10% fetal bovine serum (FBS, HyClone), 1:100 penicillin/streptomycin (Gibco) and 1:100 GlutaMax (Gibco). Human HAP-1 WT or IFN receptor knockout (KO) cells (Horizon Discovery) were maintained in Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco), supplemented with 10% FBS and antibiotics. Cells were grown at 37 °C under 5% CO2 and split using 0.05% Trypsin-EDTA (Gibco). Cells were seeded prior to all experiments at 2.5–5×104/well in 96-well plates.
2.2. Vectors and antibodies
Newly-synthesized expression vectors were designed, based on human codon-optimized open reading frame (ORF) sequences of VP35, VP40 and VP24, for the following isolates from each filovirus species or variant (except LLOV, for which an isolate has yet to be recovered), as recorded in GenBank: Ebola virus 2014 Makona and 1976 Yambuku variants (EBOV, Ebola virus/H.sapiens-wt/LBR/2014/Makona-201403007 and Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Mayinga; GenBank accession numbers KP178538.1 and AF086833.2, respectively); Sudan virus (SUDV, Sudan virus/H.sapiens-tc/UGA/2000/Gulu-808892; AY729654.1); Bundibugyo virus (BDBV, Bundibugyo virus/H.sapiens-tc/UGA/2007/Butalya-811250; FJ217161.1); Taï Forest virus (TAFV, Tai Forest virus/H.sapiens-tc/CIV/1994/Pauleoula-CI; FJ217162.1); Reston virus (RESTV, Reston virus/M.fascicularis-tc/USA/1989/Philippines89-Pennsylvania; AF522874.1); Marburg virus (MARV, Lake Victoria marburgvirus strain Uganda 01Uga07; FJ750957.1); and Ravn virus (RAVV, Lake Victoria marburgvirus strain Uganda 02Uga2007; FJ750953.1). In total, 51 wild-type (WT) and FLAG-tagged VP expression vectors were constructed and used in our study. Each codon-optimized construct was generated by Genscript, Inc. (Piscataway, NJ) in pUC57 vector, and then was subcloned using EcoRI-XbaI restriction enzyme sites into pcDNA3.1(+) for expression of WT (untagged) proteins, or subcloned using KpnI-XbaI sites into pcDNA3.1(+)-N-DYK for expression of N-terminally FLAG-tagged proteins. EBOV VP24 from the Makona variant was mutagenized later in the study to produce VP24-W42A/K142A, which has been reported to have less-efficient binding and antagonism of karyopherin α (46). As a negative control, we used an empty pcDNA3.1(+) vector (Invitrogen). As a control of transfection and luciferase assay efficiencies, we also used a dual, constitutively-expressing green fluorescent protein (GFP) and luciferase vector (Cignal, Qiagen).
2.3. Transfections
Unless otherwise noted, antagonist activity experiments in human cells were performed adhering to the same overall protocol, in triplicate, via a standardized panel in 96-well plates (CellBIND, Corning). One plate allowed for simultaneous profiling of up to 29 of the WT or FLAG-tagged VP gene vectors in our library. For such assays, each VP construct was singly transfected at 80 ng/well of plasmid using TransIT-LT1 transfection reagent according to manufacturer’s recommendations (Mirus Bio, LLC) diluted into OptiMEM (Gibco), in conjunction with 10 ng/well of appropriate firefly luciferase-based, innate immune response reporter gene vector, and 10 ng of constitutive Renilla luciferase-based pRL-TK vector (Promega) used as a transfection efficiency control. IFN response reporters used in this study included: IFNβ expression promoter and IRF3 activity reporters (p125-Luc and p55-C1B-Luc, respectively, kind gifts of Takashi Fujita, Kyoto University); NF-κB activity reporter (Stratagene); IRF1 activity and ISRE promoter reporters (Cignal Reporter Assay Kits, Qiagen); and hamster (h)ISG54 promoter reporter (pISG54Lucter, a kind gift from Steven Goodbourn, University of London). Cignal reporters were transfected at 20 ng/well and without addition of pRL-TK, as this was already included within the kit. Except for FLAG-tagged VP24 assays (which used single transfection mix-derived replicates on the same plate that were then harvested for either reporter assays or western blot analysis, described below) and VP24 titration assays (which were conducted across two plates with 20–240 ng/well of VP vector), well positions for each construct were maintained across individual experiments. An equal amount of empty vector (EV) was transfected in dedicated wells for use as negative or positive controls of induction, or included in transfection mixes for VP24 titration assays to normalize amount of total DNA (up to 260 ng/well). Transfected cells were incubated at 37 °C for at least 18 h post-transfection (hpt) prior to induction. Assays shown are representative of multiple independent experiments. In a few experiments, select antagonists that were determined in preliminary data to share redundant activity to similar proteins (EBOV Mayinga VP24 and RAVV VP24) were replaced in our full panel with EBOV VP24-W42A/K142A.
2.4. Inductions
Transfected cells in 96-well VP antagonist panels above were induced the following day via 1) Sendai virus (SeV, Cantell strain, Charles River Laboratories) infection at either high MOI (~22.2), or increasing MOI between ~0.022 and ~22.2, diluted into fresh medium, unless otherwise noted; 2) by dsRNA mimic poly(I:C) at up to 2 μg/mL using Lipofectamine RNAiMax reagent (Thermo Fisher Scientific); 3) by transfection of RIG-I CARD domain vector (60 ng/well, a kind gift of Sonja Best, NIAID) using TransIT-LT1; 4) by universal Type I IFN (U-IFN, 500–1000 U/mL); 5) by tumor necrosis factor (TNF; 50 ng/mL); or 6) by an attenuated recombinant Rift Valley fever virus in which NSs and NSm IFN antagonists have been removed (rRVF-ΔNSs-ΔNSm, “ΔΔrRVF”), used at an MOI of ~2.2 (Bird et al., 2011). Untreated medium was used as an induction control. For SeV and ΔΔrRVF, cells were incubated for 1–2 h to allow for virus adsorption, and then exchanged for 100 μL fresh medium; for HAP-1 KO cells, an equal amount of medium was instead overlaid onto unremoved SeV adsorption medium. For poly(I:C), cells were exchanged with 90 μL fresh medium and then 10uL of transfection mixture diluted in OptiMEM was added to each well; RIG-I CARD transfection mixtures were added similarly to poly(I:C), except 100 μL fresh replacement media was used. For our qRT-PCR assays described below, we instead cotransfected RIG-I CARD vector simultaneously with reporter and antagonist vectors. Apparent cytopathic effects were not observed for any SeV assay in HEK293 cells and were modest with ΔΔrRVF or in HAP-1 cells. Cells were incubated at 4h to 24hpi and harvested using 1X passive lysis buffer (Promega) incubated on a rocker for at least 15 m, and then stored at −20 °C until use, or assayed immediately for relative luciferase activities.
2.5. Luciferase assays
Equal aliquots of induced cell lysates from 96-well plates were transferred to cognate wells in white bottom assay plates (Corning Costar), and firefly and Renilla luciferase activities assessed using the Dual Luciferase Assay Kit (Promega) for reporter gene and transfection control raw counts (relative light units, RLU), respectively, analyzed on a BioTek Synergy 4 microplate reader (Thermo). Relative firefly raw values were normalized to those of Renilla raw values across samples. Normalized values were then compared to EV negative control to obtain fold induction values, and then to EV positive control values to obtain percentage induction, set at 100% (fold change is also shown as “FC” above positive controls for each assay). For most assays, data was stratified into separated, relative activities for each of the three VPs across tested filoviruses.
2.6. Antibodies
For western blotting (see below), an anti-FLAG rabbit antibody (Sigma) was used at 1:2000 and 1:10000; and mouse anti-β-actin mAb (Genscript) was used at 1:2000 as an internal loading control. For neutralization assays (see below), a cocktail of IFNα, IFNβ and IFNα1 receptor (IFNAR1, PBL Biosciences) neutralizing antibodies (nAbs) were used at concentrations from 1000 U/mL, 2000 U/mL and 20 μg/mL, respectively, up to 20 μg/mL for each.
2.7. Western blotting
For each FLAG-tagged viral protein reporter assay, cells in single transfection mix-derived replicate wells on the same 96-well plate were lysed using up to 100 μL radioimmunoprecipitation assay (RIPA) buffer (Pierce), and then total lysates were centrifuged through QiaShredder columns (Qiagen) at maximum speed. Clarified lysates were transferred to fresh microcentrifuge tubes and stored at −20 °C, or equal aliquot amounts run immediately on a 4–12% NuPAGE Bis-Tris gel (Novex, Life Technologies) to separate proteins by gel electrophoresis. Protein gels were transferred onto nitrocellulose membranes using an iBlot apparatus (Invitrogen) and washed with deionized water and phosphate-buffered saline (PBS, Gibco) supplemented with 0.1% Tween-20 (PBST, Thermo). Blots were blocked for at least 45 m using 5% non-fat dry milk (BD Difco) in PBST, then washed and/or incubated with primary antibodies as indicated above overnight, or for 1h in the case of anti-FLAG used at 1:10000. Membranes were then washed, incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h and rewashed using a BenchPro 4100 Card Processing Station (Invitrogen). Finally, membranes were rinsed with water, and protein bands visualized using enhanced chemiluminescence (ECL, Pierce) on a ChemiDoc MP Imager (Bio-Rad).
2.8. Antibody neutralization assays
nAbs were added to reporter- and/or VP-transfected cells as cocktails in the concentrations described above either simultaneously along with SeV diluted into fresh medium, or as a cocktail diluted alone for pre-treatment of cells for at least 4 h; in either case, 50uL of medium was added per well. Following SeV adsorption, 50 μL per well of fresh medium was layered on top of conditioned medium; following pre-treatment of antibody, up to 50 μL per well of media containing SeV, or inducing cytokine at double working concentration, was layered on top.
2.9. RNA isolation and quantitative real-time RT-PCR
In a manner analogous to tagged vector-based reporter assays that were compared to western blot analysis, cells in single transfection mix-derived replicate wells on the same 96-well plate were lysed 24hpi, either by a high MOI of SeV (~22.2; induced 1dpt) or by simultaneous cotransfection/induction by RIG-I CARD vector along with antagonist and reporter vectors, for RNA extraction with 100 μL of 1x RNA Lysis/Binding Solution Concentrate (Thermo) followed by magnetic bead purification and TURBO DNase treatment using the MagMAX Total RNA Isolation kit according to the manufacturer’s recommendations (Thermo). Yields of purified RNA samples were verified by NanoDrop spectrophotometer (Thermo) and then interferon gene expression was assessed using human IFNβ and IFNλ gene-specific TaqMan-based primer-probes (Integrated DNA Technologies) as well as a primer-probe for RNase P (RPP30) as an internal control. 25 μL reaction mixes were produced using the SuperScript III One-Step RT-PCR system with Platinum Taq DNA polymerase and ROX dye according to the manufacturer’s recommendations (Thermo). Reactions were run on a Bio-Rad CFX96 Touch Real-Time PCR Detection System. Amplification data was corrected for fluorescence drift and baseline threshold, normalized to internal control, and fold change (“FC”) quantitated by ΔΔCt method in CFX Manager (Bio-Rad). As with reporter assays, data is compared relative to EV positive control induction values, set at 100%. Primer-probe sequences used were: IFNβ (Fwd: 5′-GCAA TTTTCAGTGTCAGAAGCTC-3′, Rev: 5′-TCCTGTCCTTGAGGCA GTATT-3′, Probe: 5′-TGTGGCAATTGAATGGGAGGCTT-3′); IFNλ (Fwd: 5′-GGAGCTAGCGAGCTTCAAGA-3′, Rev: 5′-ACTCCAGTTTTTC AGCTTGAGTG-3′, Probe: 5′-GCCAGGGACGCCTTGGAAGAG-3′); RPP30 (Fwd: 5′-AGATTTGGACCTGCGAGCG-3′, Rev: 5′-GAGCGG CTGTCTCCACAAGT-3′, Probe: 5′-TTCTGACCTGAAGGCTCTGCGC G-3′).
2.10. Statistical analyses
All statistics shown in figures was obtained by conducting a Student’s unpaired t-test for all values in which significance was indeterminate and that remained potentially significant at the highest MOI or protein concentration tested. Significant two-tailed p values were designated as follows: *=p < 0.05; **=p < 0.005; ***=p < 0.0005; ****=p < 0.0001. Standalone asterisks above each bar indicate t-tests that directly compare that antagonist activity value to the positive control induction value in each assay; asterisks above horizontal lines indicate t-tests that directly compare activity values between two antagonists. All statistics were calculated using QuickCalcs software (GraphPad).
3. Results
3.1. VP35 proteins of all ebolaviruses and RAVV are potent IFN induction and signaling antagonists
To conduct a side-by-side assessment of the potencies of the three known interferon (IFN) response antagonists, VP35, VP40 and VP24, in a single system, we generated a library of pcDNA3.1(+)-based vectors for expression of the three proteins from representative viral isolates for each filovirus species known to cause disease in humans or NHPs. These include reference isolates of EBOV from 2014 and 1976 outbreaks (Makona-201403007 and Yambuku-Mayinga; for clarity, these will be heretofore referred to as “EBOV Makona” and “EBOV Mayinga”), SUDV, RESTV, TAFV, BDBV, and related isolates of MARV and RAVV (both obtained from the same outbreak), as listed in Fig. 1F (Towner et al., 2009). LLOV was not included in this study as no isolate yet exists and there is no evidence of any human or NHP infection or disease. Because specific antibodies for probing many filoviral proteins are not available, vectors containing an N-terminally expressed FLAG tag were also generated, allowing assessment of antagonist expression using a single antibody (Burk et al., 2016). A total of 51 vectors that express WT or cognate tagged viral proteins were designed and used for the bulk of the analyses.
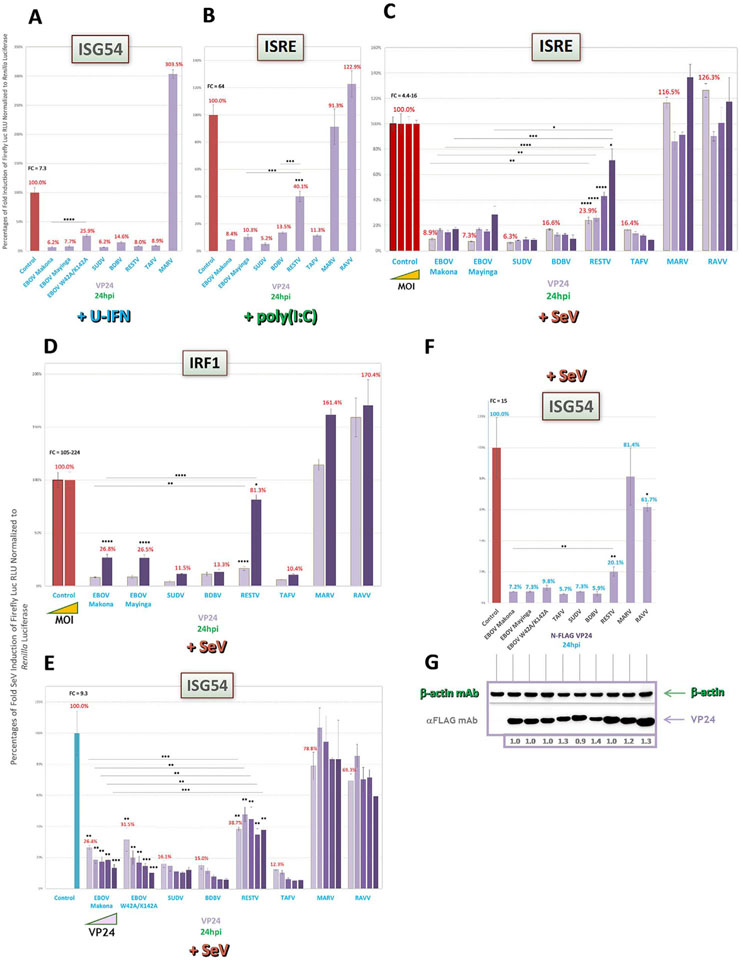
Along with our standardized library, we developed a standardized, 96-well plate reporter assay system for pan-filovirus assessment of IFN antagonist activities. In this format, all WT forms of antagonists in our library were transfected and induced in parallel, cotransfected with one of several luciferase-based reporters to measure responses during either upstream IFN induction (IRF3 and NF-κB activities, IFNβ promoter expression), downstream IFN signaling (ISRE and ISG54 promoter expression) or both (IRF1 activity, shown to play roles in both IFN induction and as well as ISG transactivation), as depicted in Fig. 1A–E (Yarilina and Ivashkiv, 2010; Hu et al., 2008; Kröger et al., 2002). Responses in the presence of filoviral proteins were compared to empty vector (EV) positive and negative controls to establish antagonist activity.
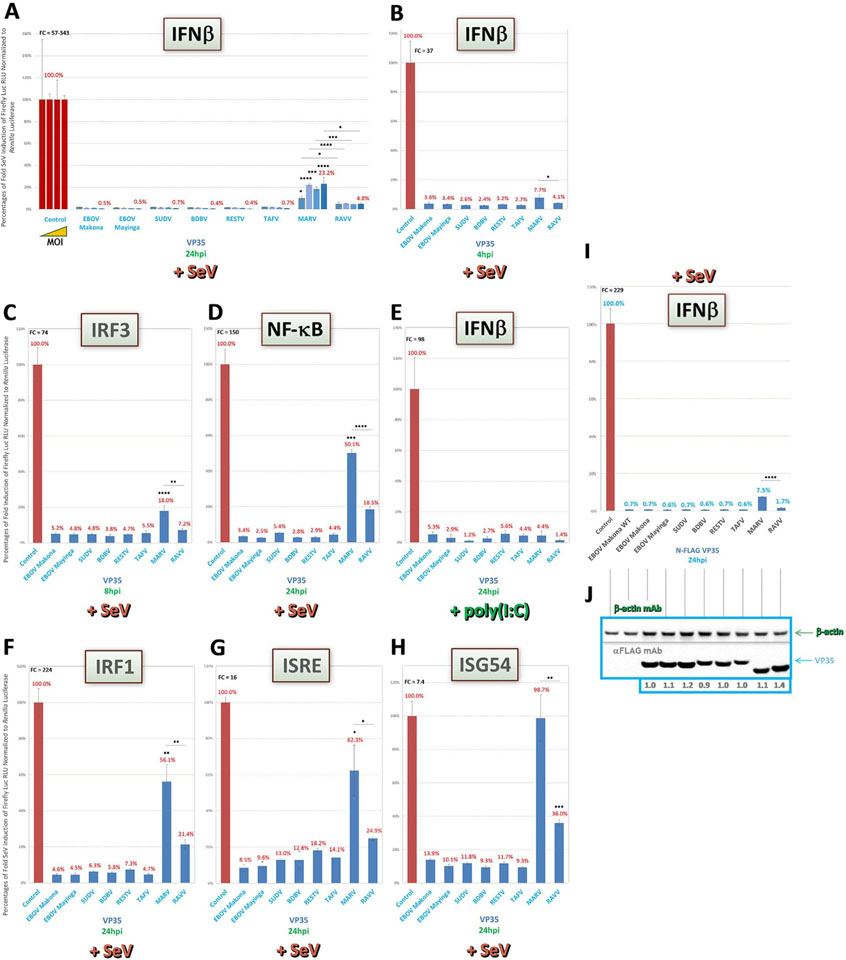
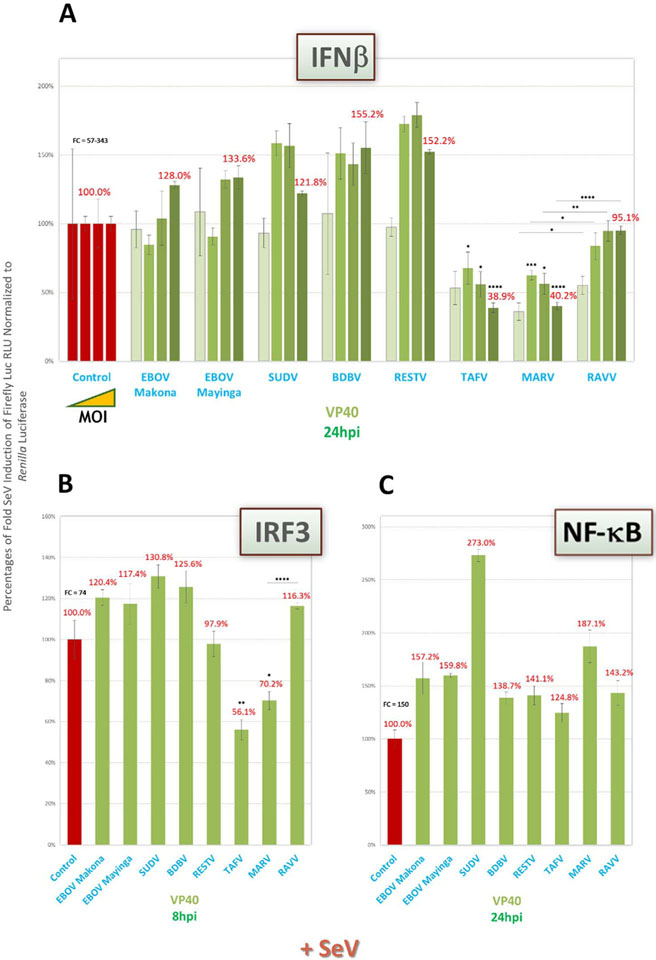
Prior studies with well-studied EBOV VP35 have shown nearlycomplete inhibition of IFNβ induction, while weaker antagonism was found for MARV VP35 (Dunham et al., 2015; Edwards et al., 2016; Albariño et al., 2015; Feagins and Basler, 2015; Basler et al., 2000). Thus, we first measured antagonism of IFNβ expression, predicting a similar pattern of activity for the other VP35 proteins. We found that all ebolavirus VP35 proteins showed equally potent, nearly-complete suppression of IFN reporter activity over a 1000-fold range of Sendai virus (SeV) infection from an MOI of ~0.022 (Fig. 2A). Indeed, very strong antagonism was already evident by 4hpi (Fig. 2B). MARV VP35, as expected, showed diminished IFN inhibition by comparison to ebolaviruses. Interestingly, RAVV VP35 showed approx. 3–5-fold more potent inhibitory activity than MARV VP35 across all SeV MOIs (Fig. 2A), even at the early induction timepoint (Fig. 2B).
Fig. 2.
VP35 of all ebolaviruses are the most potent IFN antagonist proteins on both induction and signaling pathways. Reporter assays, using HEK293 cells in 96-well plates containing our full panel of transfected VP constructs, were conducted as described in the Materials and Methods to assess pan-filovirus VP35 activities on upstream IFN induction or downstream IFN signaling pathways (reporters shown in shaded green boxes). (A) Tenfold increasing amounts of SeV, starting from an MOI of ~0.022, were used to induce cells cotransfected with IFNβ reporter vector, which were harvested 24hpi. (B) SeV-infected cells at a high MOI (~22.2) were harvested for IFNβ reporter assay at an early timepoint post-induction. (C-H) Cells cotransfected with indicated reporters for induction (C-E), signaling (G and H) or dual activities (for IRF1, which can transactivate both IFN and ISG genes, F) were induced by a high MOI of SeV, or by dsRNA mimic poly(I:C) for IFNβ (E), as indicated, and harvested 24hpi, except for the IRF1 reporter assay (F), which was harvested 8hpi. (I and J) Indicated untagged (EBOV Makona WT) or FLAG-tagged VP35- and IFNβ reporter-cotransfected cell lysate samples derived from the same experiment were harvested 24hpi by a high MOI of SeV from replicate wells on the same plate for reporter activity (I) or for western blot expression (J); western blots were probed as indicated using an anti-FLAG antibody, as well as anti-β-actin antibody for an internal loading control; densitometry values normalized to β-actin expression and set relative to tagged EBOV Makona VP35 are shown below each protein band. FC = fold change of positive induction relative to uninduced negative control. RLU = relative light units. *=p < 0.05; **=p < 0.005; ***=p < 0.0005; ****=p < 0.0001; asterisks above horizontal lines indicate direct comparison of activity values between two antagonists (Yarilina and Ivashkiv, 2010; Hu et al., 2008; Kröger et al., 2002).
We next tested VP35 potencies in cells transfected with activity reporters for IFN-transactivating proteins IRF3 or NF-κB. The VP35 antagonism profiles for both reporters were similar to that for IFNβ, with strong inhibition of SeV-induced IRF3 (Fig. 2C) or NF-κB (Fig. 2D) activity by all ebolavirus proteins, yet weaker activity by MARV VP35, and intermediate activity by RAVV VP35. Conversely, when we induced IFNβ using a surrogate for dsRNA, poly(I:C), a lesspotent, more MDA5-dependent inducer of the IFN pathway than SeV (Li et al., 2005; Gitlin et al., 2006; Kato et al., 2006; Reimer et al., 2008), the relative differences between marburgvirus and ebolavirus VP35 proteins disappeared (Fig. 2E).
We then assayed VP35 antagonistic activities on the downstream IFN signaling pathway, by using either the ISRE (Fig. 2G) or ISG54 promoter reporter (Fig. 2H). We also used our IRF1 dual induction/signaling reporter (Fig. 2F). In all cases, VP35 activity profiles in SeV-induced cells were analogous to those seen for IFNβ induction, with strong ebolavirus VP35, intermediate RAVV VP35 and weaker MARV VP35 antagonism.
Finally, since observed differences in VP35 activities between viruses could be due to inadequate expression of particular proteins in HEK293 cells, we validated these results using FLAG-tagged VP35 constructs transfected in two parallel sets of replicates and harvested for luciferase or western blot assays, respectively. All FLAG-tagged filovirus VP35 proteins were well expressed (Fig. 2J). We also found no impairment of tagged VP35 function compared to either untagged EBOV Makona VP35 directly, or to the cognate WT proteins from previous assays (Fig. 2A and B), as tagged proteins showed near-identical inhibitory effects on IFNβ reporter activity following SeV induction (Fig. 2I). Thus, these assays support that our previous observations for WT VP35 proteins were due to bona fide reporter gene antagonism and not to indirect effects at the level of protein expression.
Taken together, our data indicate that VP35 activity to block the IFN response, as has been observed for EBOV and MARV, are maintained across all filoviruses, but that RAVV VP35 has stronger-than-expected antagonism than previously reported for a marburgvirus.
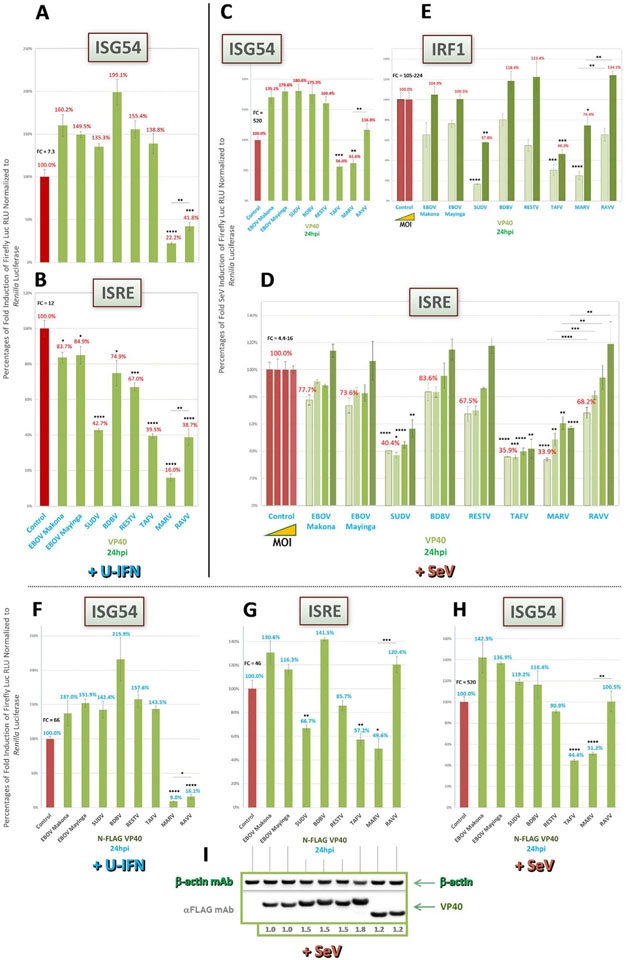
3.2. VP40 proteins of marburgviruses, SUDV and TAFV show conditional inhibitory activities on downstream IFN signaling
As MARV VP40 was originally identified to have antagonistic effects on Jak1 activation during downstream IFN signaling, we decided to first analyze pan-filovirus VP40 activities within our panel using the same two IFN signaling reporters, ISRE and ISG54, as well as IRF1 dual activity reporter, described above (Valmas and Basler, 2011). We anticipated that MARV VP40 would prevent signaling upon U-IFN treatment. Indeed, activation of both ISG54 (Fig. 3A) and ISRE (Fig. 3B) reporters were robustly inhibited by MARV VP40 under these conditions. Closely-related RAVV VP40, however, showed half the potency with both reporters after U-IFN induction (Fig. 3A and B). Meanwhile, as previous reports on EBOV VP40 activity during IFN signaling suggested (Ramanan et al., 2011; Basler, 2015; Valmas et al., 2010), every ebolavirus VP40 protein failed to inhibit U-IFN-induced ISG54 (Fig. 3A), and in fact even showed moderate reporter overstimulation, an effect seen in some assays by select proteins lacking inhibitory ability. These effects are likely due to mild background induction of IFN responses by ectopic filoviral proteins, which is observable only for those proteins that are then unable to counteract such responses. Most ebolavirus VP40 proteins also did not antagonize U-IFN-induced ISRE (Fig. 3B), but unexpectedly, we observed a moderate inhibition of ISRE by both SUDV and TAFV VP40 proteins, at levels similar to that of RAVV VP40 (Fig. 3B).
Fig. 3.
MARV and RAVV VP40 differentially block downstream IFN signaling in an inducer-specific manner while VP40 of SUDV and TAFV show moderate activities. Reporter assays as described in Fig. 2 were conducted to assess relative filovirus VP40 antagonism of Jak1 activation within the IFN signaling pathway, using the same signaling reporters. (A and B) Cells were induced 1 day post-transfection (dpt) by U-IFN for ISG54 and ISRE reporters, respectively. (C) Cells cotransfected with the ISG54 reporter were induced by a high MOI of SeV (~22.2). (D and E) Cells infected by tenfold increasing amounts of SeV, starting from an MOI of ~0.022, for ISRE expression (D), or low (~0.022) and high (~22.2) MOI of SeV for IRF1 activity (E), were harvested 24hpi. (F-I) Indicated FLAG-tagged VP40- and ISG54 (U-IFN, F; SeV, H) or ISRE (SeV, G) reporter-cotransfected cell lysate samples derived from the same experiment were harvested 24hpi by a high MOI of SeV from replicate wells on the same plate for reporter activity (F-H) or for western blot expression (I); western blots were probed as indicated using an anti-FLAG antibody, as well as anti-β-actin antibody for an internal loading control; densitometry values normalized to β-actin expression and set relative to tagged EBOV Makona VP40 are shown below each protein band. FC = fold change of positive induction relative to uninduced negative control. RLU = relative light units. *=p < 0.05; **=p < 0.005; ***=p < 0.0005; ****=p <0.0001; asterisks above horizontal lines indicate direct comparison of activity values between two antagonists (Valmas and Basler, 2011; Valmas et al., 2010).
Next, we looked at VP40 IFN signaling antagonism following infection by SeV, a more potent and broader inducer of the IFN pathway (Li et al., 2005; Peters et al., 2008; Zaritsky et al., 2015). In contrast to the strong activity by MARV or RAVV VP40 seen after U-IFN induction of ISG54 and ISRE (Fig. 3A and B), antagonism of these reporters was much weaker following SeV induction (Fig. 3C and D). Further, for ISRE induced by over a 1000-fold range of SeV from an MOI of ~0.022, we observed MOI-dependent effects on marburgvirus VP40 activity (Fig. 3D), with comparatively weaker retention of potencies for RAVV VP40 compared to MARV VP40 at each MOI. Conversely, both SeV-induced reporters showed mostly similar ebolavirus VP40 activity profiles (Fig. 3C and D) to those obtained by U-IFN (Fig. 3A and B). The exception was TAFV VP40, which, interestingly, showed moderate inhibition of SeV-induced ISG54 (Fig. 3C), an activity that was neither observed following U-IFN induction nor shared by SUDV VP40 (Fig. 3A). And unlike the reduced activity found for marburgvirus VP40, antagonism of SeV-induced ISRE by both SUDV and TAFV VP40 was maintained at equivalent levels (Fig. 3D) as compared to U-IFN induction (Fig. 3B), and their activities were mostly unaltered by SeV MOI (Fig. 3D). Further support for the activities of MARV, SUDV and TAFV VP40 proteins on the ISRE reporter could be found when we instead infected cells with an IFN antagonist-deficient recombinant Rift Valley fever virus (ΔΔrRVF). Antagonist activity profiles following induction by ΔΔrRVF (MOI of ~2.2, Supplemental Fig 1A) were highly similar to that obtained with SeV (Fig. 3D), indicating VP40 control of IFN responses are broader effects and are not specific only to SeV infection.
Meanwhile, when we tested IRF1 activity following SeV induction, we saw a VP40 activity profile at a low MOI (~0.022, Fig. 3E) that was analogous to that seen for U-IFN-induced ISRE (Fig. 3B), with more potent inhibitory activity by marburgvirus VP40 than that seen for SeV-induced ISRE and ISG54 (Fig. 3C and D), as well as stronger, MARV VP40-like levels of antagonism by TAFV and SUDV VP40. This effect on IRF1 activity was abrogated at a high MOI (~22.2, Fig. 3E), in which the potency profile returned to being similar to that seen for SeV-induced ISRE (Fig. 3D).
Finally, as we considered previously for VP35, differences in activities by VP40 proteins between viruses could be due to major inefficiencies in protein expression. Thus, we validated our data with FLAG-tagged VP40 constructs transfected in parallel sets of replicates and harvested for either luciferase or western blot assays. Indeed, as we had confirmed for tagged VP35 proteins, FLAG-tagged VP40 proteins were well expressed (Fig. 3I). The tagged proteins gave similar activity profiles for U-IFN-induced ISG54 (Fig. 3F), including strong inhibition by VP40 of marburgviruses, and for SeV-induced ISRE and ISG54 (Fig. 3G and H), including the moderate antagonism by SUDV and/or TAFV VP40, as compared to the cognate untagged proteins from previous assays (Fig. 3A, C and D), thereby corroborating our observations for WT VP40 activities and ruling out indirect protein expression-level effects.
These data suggest that SUDV and TAFV VP40 have a moderate IFN signaling-inhibitory function, an activity previously unreported for ebolaviruses. Meanwhile, VP40 of marburgviruses have filovirus- and inducer-specific potencies whereby MARV VP40, in agreement with prior studies (Valmas and Basler, 2011), is capable of suppressing IFN responses induced by high amounts of SeV or ΔΔrRVF, an activity that is not shared by RAVV VP40.
3.3. VP40 proteins of marburgviruses and TAFV show moderate inhibition of upstream IFN induction
Following our assessment of downstream IFN signaling, which was known to be inhibited by MARV VP40 (Valmas and Basler, 2011), we next decided to test filoviral VP40 proteins for any potentially unrecognized ability to antagonize the upstream IFN induction pathway. Indeed, as with our ISRE and ISG54 signaling reporters, IFNβ expression over a 1000-fold range of SeV infection from an MOI of ~0.022 (Fig. 4A) or IRF3 activity at high MOI (~22.2, Fig. 4B) were moderately antagonized by VP40 proteins of marburgviruses, but not of ebolaviruses, with the notable exception of TAFV. We also again observed more potent MARV VP40 than RAVV VP40 activity. While IRF3 reporter activity was modestly inhibited by these VP40 proteins, NF-κB activity was not suppressed by any filoviral VP40 (Fig. 4C).
Fig. 4.
VP40 of marburgviruses, as well as of TAFV, show moderate, noncanonical inhibition of upstream IFN induction. Reporter assays as described in Fig. 2 were conducted to assess relative filovirus VP40 antagonism of the upstream IFN induction pathway, using the same induction reporters. (A) Cells cotransfected with IFNβ reporter vector were induced 1dpt by tenfold increasing amounts of SeV during an experiment identical to that conducted for VP35 in Fig. 2A. (B and C) Cells were induced by a high MOI of SeV (~22.2) for IRF3 or NF-κB reporter assays and harvested at 8 h or 24hpi, respectively. FC = fold change of positive induction relative to uninduced negative control. RLU = relative light units. *=p < 0.05; **=p <0.005; ***=p < 0.0005; ****=p < 0.0001; asterisks above horizontal lines indicate direct comparison of activity values between two antagonists.
These results suggest that, in addition to its known IFN signaling antagonism, marburgvirus VP40 is also capable of moderate, previously-unreported inhibitory activity during upstream IFN induction; and that TAFV, but not SUDV, VP40 maintains a role in blocking IFN induction, as was shown for both ebolavirus proteins for IFN signaling. Our assays also suggest marburgvirus and TAFV VP40 may inhibit IFNβ expression through IRF3, but not through NF-κB, signaling.
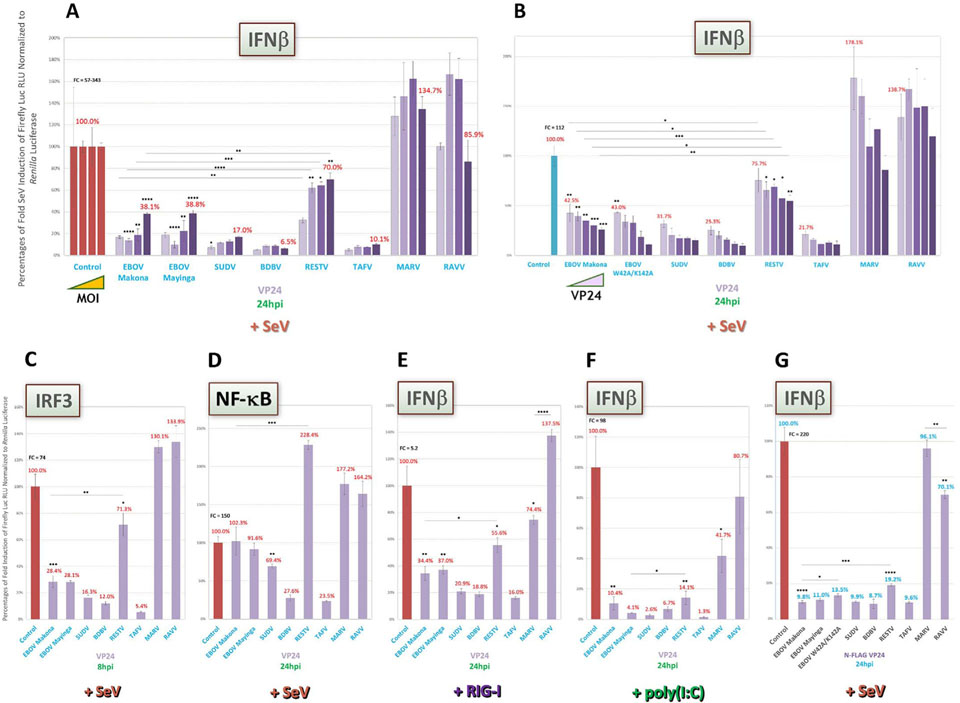
3.4. VP24 proteins of all ebolaviruses strongly inhibit downstream IFN signaling with the exception of RESTV, which shows weakened antagonism in an inducer-specific manner
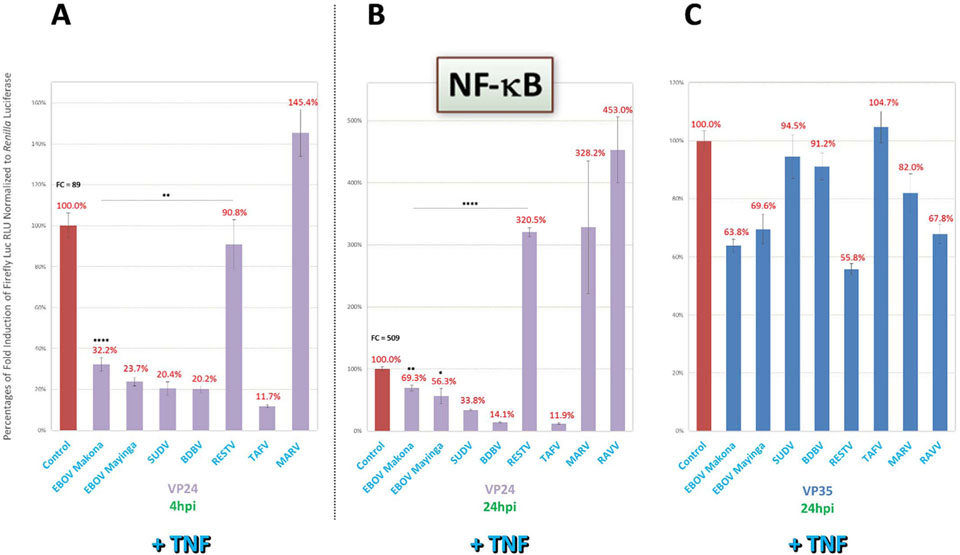
VP24 of EBOV has long been known to block downstream IFN signaling by binding to karyopherin α, which prevents STAT1 interaction and subsequent nuclear translocation (Reid et al., 2006, 2007; Zhang et al., 2012a; Mateo et al., 2010). Given this reported activity, we first decided to compare pan-filovirus VP24 activities within our panel using the ISRE and ISG54 IFN signaling reporters. As shown for ISRE-cotransfected cells induced by U-IFN (Fig. 5A), poly(I:C) (Fig. 5B) or SeV over a 1000-fold range from an MOI of ~0.022 (Fig. 5C), VP24 of ebolaviruses, but not of marburgviruses, strongly antagonized reporter activity for all tested conditions. Interestingly, RESTV VP24 displayed potent antagonism in U-IFN-induced cells, but unlike those of the other ebolaviruses, showed markedly weaker antagonism when cells were induced by poly(I:C) or higher levels of SeV. This impaired antagonism by RESTV VP24 relative to other ebolavirus proteins at increased SeV MOI (~22.2) was also found using our reporter for IRF1 dual activity (Fig. 5D). RESTV VP24 inhibition of ISG54 expression was similarly weak in a VP24-specific titration assay, while all other ebolavirus VP24 proteins had robust, dose-dependent increases in potency (Fig. 5E). So too was reduced RESTV VP24 activity obtained following ISRE induction by ΔΔrRVF (MOI of ~2.2, Suppl Fig. 1B), offering support that this inefficiency was not specific to SeV and poly(I:C) induction. Activity profiles for the other filoviral proteins following ΔΔrRVF induction (Suppl Fig. 1B) were also highly consistent to those seen for SeV-induced assays under similar conditions (Fig. 5B and C), indicative of broader, inherent abilities by ebolavirus VP24 proteins in controlling IFN responses. Among VP24 proteins across all assays, we observed strongest antagonism for SUDV, BDBV or TAFV (Fig. 5A–F; Suppl Fig. 1B).
Fig. 5.
VP24 of all ebolaviruses, except RESTV in an inducer-specific manner, strongly antagonize downstream IFN signaling. Reporter assays as described in Fig. 2 were conducted to assess pan-filovirus VP24 antagonism of downstream IFN signaling, using the same signaling reporters. (A–C) Cells cotransfected with ISRE reporter vector were induced 1dpt by U-IFN (A), by poly(I:C) (B) or by tenfold increasing amounts of SeV starting from an MOI of ~0.022 (C) and harvested 24hpi. (D) Low (~0.022) or high (~22.2) MOI of SeV were used to induce cells cotransfected with IRF1 dual activity reporter vector and harvested 24hpi. (E) Cells cotransfected with an increasing concentration of indicated filovirus VP24 vectors ranging from 20 to 240 ng/well, in a VP24-specific assay conducted across two plates, along with the ISG54 reporter were induced 1dpt by a high MOI of SeV (~22.2). (F and G) Indicated FLAG-tagged VP24- and ISG54 reporter-cotransfected cell lysate samples derived from the same experiment were harvested 24hpi by a high MOI of SeV from replicate wells on the same plate for reporter activity (F) or for western blot expression (G); western blots were probed as indicated using an anti-FLAG antibody, as well as anti-β-actin antibody for an internal loading control; densitometry values normalized to β-actin expression and set relative to tagged EBOV Makona VP24 are shown below each protein band. FC=fold change of positive induction relative to uninduced negative control. RLU = relative light units. *=p < 0.05; **=p < 0.005; ***=p < 0.0005; ****=p < 0.0001; asterisks above horizontal lines indicate direct comparison of activity values between two antagonists..
Further, as an added control for specificity of VP24 activity, we tested in a subset of our assays a mutated form of EBOV VP24 with alanine point mutations at W42 and K142, which have been reported to ablate IFN signaling antagonism due to weakened karyopherin α binding (Mateo et al., 2010). We noted marked differences between EBOV VP24 WT and this signaling antagonism-impaired EBOV VP24-W42A/K142A in U-IFN-induced cells cotransfected with the ISG54 reporter, as anticipated based on the available data (Fig. 5A) (Mateo et al., 2010); however, such reduced function by EBOV VP24-W42A/K142A was minimal in the context of SeV induction, conditions that have not been previously tested (Fig. 5E).
Finally, as we had validated for VP35 and VP40, we ensured that the observed differences in activities by VP24 proteins were not due to deficiencies in protein expression. We transfected FLAG-tagged VP24 constructs with two parallel sets of replicates and harvested cell lysates for luciferase or western blot assays. Like the other antagonists in our library, all tagged filovirus VP24 proteins were well expressed (Fig. 5G). FLAG-tagging had no deleterious effect on VP24 function, as all tagged VP24 proteins of ebolaviruses efficiently blocked SeV-induced ISG54 reporter activity with the exception of tagged RESTV VP24 (Fig. 5F), similarly to what we observed with WT RESTV VP24 (Fig. 5B–E). Meanwhile, tagged marburgvirus VP24 proteins showed no potency, whereas tagged EBOV VP24-W42A/K142A had minimally impaired activity compared to tagged EBOV VP24 WT, both of which had also been seen using the untagged forms of these proteins. These assays again confirmed that the observed effects for WT antagonists, in this case VP24, were due to actual inhibition of reporter activity rather than indirectly due to inefficient protein expression.
Together, our results indicate that VP24 of all ebolaviruses, but not marburgviruses, share robust suppression of IFN signaling except for VP24 of RESTV. Intriguingly, RESTV VP24 appears unable to efficiently antagonize IFN signaling elicited under most tested conditions. Conversely, these results indicate that residues putatively impairing VP24 antagonism of karyopherin α, such as those of EBOV VP24-W42A/K142A, may be less sufficient to unblock IFN signaling in certain inducing conditions than previously appreciated.
3.5. VP24 proteins of all ebolaviruses except RESTV have a novel activity to antagonize the IFN induction pathway
Similar to our investigation for VP40, we next decided to assess filoviral VP24 proteins for any previously unrecognized ability to inhibit upstream IFN induction. Unexpectedly, VP24 of all ebolaviruses except RESTV robustly suppressed SeV-induced IFNβ expression in either a SeV MOI-dependent (over a 1000-fold range from an MOI of ~0.022, Fig. 6A) or a VP24 dose-dependent (Fig. 6B) manner. In both experiments, VP24 of RESTV again showed minimal inhibitory activity, similar to that seen with downstream IFN signaling (Fig. 5).
Fig. 6.
VP24 of all ebolaviruses except RESTV show robust, noncanonical inhibition of upstream IFN induction under multiple conditions. Reporter assays as described in Fig. 2 were conducted to assess potential VP24 antagonism of upstream IFN induction, using the same induction reporters. (A) Cells cotransfected with IFNβ reporter vector were induced 1dpt by tenfold increasing amounts of SeV during the same experiment as that conducted for VP35 in Fig. 2A. (B) Cells cotransfected with an increasing concentration of indicated filovirus VP24 vectors ranging from 20 to 240 ng/well along with the IFNβ reporter across two plates were induced 1dpt by a high MOI of SeV (~22.2) during the same experiment as that conducted with the ISG54 reporter in Fig 5E. (C and D) Cells were induced 1dpt by a high MOI of SeV and harvested 8hpi or 24hpi to assess IRF3 (C) and NF-κB (D) reporter activities, respectively. (E and F) Cells were cotransfected with IFNβ reporter vector, induced 1dpt by human RIG-I CARD vector (E) or poly(I:C) (F) transfection and harvested 24hpi. (G) Activities of FLAG-tagged filovirus VP24 proteins in cells cotransfected with the IFNβ reporter and induced by a high amount of SeV 1dpt were harvested in an experiment analogous to that described in Fig. 5F. FC=fold change of positive induction relative to uninduced negative control. RLU = relative light units. *=p < 0.05; **=p < 0.005; ***=p < 0.0005; ****=p < 0.0001; asterisks above horizontal lines indicate direct comparison of activity values between two antagonists.
When VP24 proteins were studied using reporters of IRF3 (Fig. 6C) and NF-κB (Fig. 6D) activity following SeV infection, or following direct RIG-I CARD vector-based (Fig. 6E) or poly(I:C)-based (Fig. 6F) induction of IFNβ expression, antagonistic effects were similar, or in the case of NF-κB, appeared to be filovirus species specific. Consistent with our previous data, RESTV VP24 was always weaker compared to other ebolaviruses, even despite a relatively strong inhibition of poly(I:C)-based IFNβ induction, while TAFV and BDBV VP24 proteins were the most potent. Use of FLAG-tagged filoviral VP24 proteins, transfected at equal amounts to their untagged counterparts at well-expressed levels as previously shown (Fig. 5F), also strongly antagonized IFNβ expression after SeV induction (Fig. 6G), once again validating the activities obtained by the WT constructs. Despite more modest differences in this particular assay between tagged ebolavirus VP24 potencies, RESTV VP24 still showed overall weakest activity. Next, in an attempt to determine if the W42A/K142A mutations in EBOV VP24 showed different effects during IFN induction compared to IFN signaling, and thus suggest potentially separable functional regions within VP24 proteins that may be exploited by reverse genetics, we included both untagged and tagged EBOV VP24-W42A/K142A in a subset of induction assays. However, both forms of EBOV VP24-W42A/K142A showed similarly modest differences after SeV induction compared to EBOV VP24 WT (Fig. 6B and G) as we saw with IFN signaling reporters (Fig. 5E and F), making it difficult to assess the importance of putative karyopherin α-interacting residues for induction-specific targeting.
Finally, to further investigate if the apparent property of ebolavirus VP24 to inhibit upstream IFN induction was demonstrable by a different method of measurement, we conducted a side-by-side comparison of reporter assays with qRT-PCR-based IFN gene expression following SeV induction (MOI of ~22.2), analogously to those we conducted when assessing protein expression levels. Using a subset of VP24 proteins along with human TaqMan primer-probes for IFNβ and IFNλ, a Type III interferon gene known to be similarly induced and activate many of the same pathways as IFNβ (Hoffmann et al., 2015), we found that, when compared to the very strong antagonism seen by IFNβ reporter assay (Suppl Fig. 2A), tagged EBOV VP24 displayed a more modest, yet still significant inhibition of IFN gene expression (Suppl Fig. 2B and C), and in both assays, MARV VP24 was incapable of blocking IFN induction. TAFV and RESTV VP24 showed even weaker activity, with little distinction of activities between them (Suppl Fig. 2B and C). We reasoned that this much weaker observable antagonism following SeV induction was likely due to the activation of IFN responses in a substantial proportion of untransfected cells, leading to a high background accumulation of IFN in cell lysates that obscured the actual activity of respective VP24 proteins within transfected cells. To address this problem, we cotransfected RIG-I CARD vector along with VP24 proteins and reporter vector simultaneously, which we hypothesized would greatly increase the chances of viral protein transfection and response induction within the same cell population and thus better resolve VP24 antagonism of IFN expression. As anticipated, under these conditions we achieved highly correlative results between reporter (Suppl Fig. 2D) and qRT-PCR assays (Suppl Fig. 2E and F) for the same IFN gene targets, including potent antagonism by all ebolavirus proteins, but not MARV VP24, and weaker activity by RESTV VP24. Therefore, inhibition of endogenous IFN gene expression by ebolavirus VP24 as observed by qRT-PCR aligns well with activities previously measured in induction gene reporter-based assays and lends support that they were not a consequence of the luciferase reporter approach.
These data taken together suggest marked, noncanonical antagonism by ebolavirus VP24 proteins on upstream induction of both ectopic reporters and endogenous IFN genes, in some cases with variable potencies seen between viruses, but in which RESTV VP24, as was observed for IFN signaling, was consistently the weakest antagonist under all tested conditions.
3.6. No evidence of a role for positive feedback in the antagonistic effect on IFN induction by ebolavirus VP24
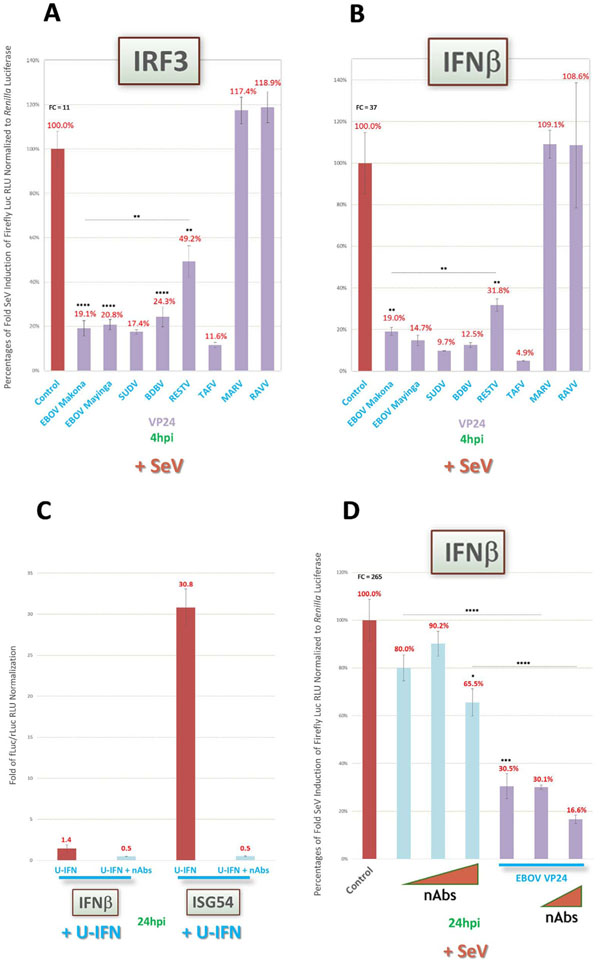
An open question in understanding the observed antagonistic activities by ebolavirus VP24 proteins on IFN induction is whether there is any indirect effect involving positive feedback (Feagins and Basler, 2015). In such a scenario, VP24 blockade of downstream IFN signaling would prevent IRF7 expression and the subsequent activation of a potential positive feedback loop that can further perpetuate IFNβ expression (Haller et al., 2006; Feagins and Basler, 2015; Sato et al., 1998). To rule out this possibility in our system, we first assessed our induction reporters at very early timepoints (4 h) post-SeV infection, near the limits of assay detection, when IRF7 induction and putative feedback would be unlikely to have a strong impact on results. We found that VP24 of ebolaviruses maintain similarly robust antagonism of both IRF3 activity (Fig. 7A) and IFNβ expression (Fig. 7B) as early as 4hpi. Indeed, VP24 inhibition was equally as strong, or even stronger, than that observed 24hpi as previously shown (Fig. 6A and B, D).
Fig. 7.
No evidence supporting novel ebolavirus VP24 inhibitory activity on upstream IFN induction pathway due to indirect IFN signaling antagonism via a putative feedback loop. Reporter assays as described in Fig. 2 were conducted to rule out a putative positive IFN signaling feedback loop that indirectly induces IFNβ and thus could confound interpretation of VP24 activity data. (A and B) Cells were induced 1dpt by a high MOI of SeV (~22.2) and harvested 4hpi to assess IRF3 activity (A) and IFNβ (B) reporter activities, respectively, for early response activation. (C) Cells transfected with IFNβ or ISG54 reporter vectors alone as indicated were induced by U-IFN in the presence or absence of a neutralizing antibody (nAb) cocktail targeting IFNα, IFNβ and IFNAR1, which was diluted in medium at final concentrations of 1000U, 2500U and 20 μg/mL, respectively. (D) Cells cotransfected with IFNβ reporter vectors, and EBOV Makona VP24 vector or EV control as indicated, were induced by a high MOI of SeV in the presence or absence of increasing concentrations of nAb cocktail up to 20 μg/mL for each antibody. Values for (C) are presented as fold change of reporter activity compared to uninduced replicates. FC = fold change of positive induction relative to uninduced negative control. fLuc/rLuc = firefly/Renilla luciferase. RLU = relative light units. *=p < 0.05; **=p < 0.005; ***=p < 0.0005; ****=p < 0.0001; asterisks above horizontal lines indicate direct comparison of activity values between two antagonists (Feagins and Basler, 2015).
For further support, we directly induced the IFN signaling pathway by U-IFN along with a cocktail of neutralizing antibodies (nAbs) targeting IFNα, IFNβ and the IFNα1 receptor (IFNAR1). We predicted that U-IFN would be unable to upregulate IFNβ, limiting a role for a putative loop, and would be blocked by the cocktail for appropriate ISG54 induction. As we show in Fig. 7C, U-IFN did not induce IFNβ expression regardless of the addition of nAbs; meanwhile, U-IFN strongly induced ISG54 reporter activity, which was completely abolished by the cocktail, thus validating the efficacy of our antibody cocktail and the apparent inability of active IFN signaling pathway components to successfully induce transactivation of the IFNβ promoter. Next, we used the same cocktail to test EBOV VP24 antagonism following SeV induction of IFNβ. We found that increasing amounts of nAbs minimally affected IFNβ reporter activity, and that no loss of EBOV VP24 potency to block IFNβ expression occurred upon nAb treatment (Fig. 7D).
Finally, we employed HAP-1 WT and IFN receptor knockout (IFNAR KO) cell lines to validate our results, as well as provide a cleaner and more direct comparison of VP24 activity in cells in which the IFN signaling pathway is completely abrogated. HAP-1 WT, but not KO, cells exhibited IFN signaling via ISG54 reporter activity as expected (Suppl Fig. 3A), while strong IRF1 reporter activity was not fully ablated in KO cells (Suppl Fig. 3B), likely due to the unmodulated role of IRF1 within the IFN induction pathway. As in HEK293 cells, we saw similarly robust antagonism of IRF1 activity in HAP-1 WT cells by EBOV VP35 and VP24 (Suppl Fig. 3C), and importantly, similar IRF1 inhibitory profiles for filoviral VP35 and VP24 WT proteins in the IFNAR KO (Suppl Fig. 3D and E).
Together, these data suggest that in our system, there is no substantial evidence for an indirect influence on VP24 antagonism by a potential positive feedback loop, and demonstrate that ebolavirus VP24 can indeed inhibit the upstream IFN induction pathway independently from any IFN signaling pathway component, thus appearing to represent a bona fide novel function of these antagonist proteins.
3.7. Ebolavirus VP24, but not VP35, can also inhibit IFN induction via early TNF-induced NF-κB signaling
As VP24 of all ebolaviruses except RESTV strongly inhibited IRF3 activity upon SeV induction (Fig. 6C), but showed variable inhibition of NF-κB activity in a species-specific manner, with weak antagonism by VP24 proteins of EBOV, SUDV or RESTV (Fig. 6D), we assessed NF-κB activity following direct TNF induction at early (4 h) or late (24 h) timepoints. NF-κB induction by TNF occurs through a pathway distinct from TLR and RIG-I signaling that may supersede the activation of induction intermediates such as MAVS or TRAF6 (Fig. 1A) (Yarilina and Ivashkiv, 2010; Palladino et al., 2003; Kumar et al., 2013). Under these conditions, we found that all ebolavirus VP24 proteins, except RESTV VP24, were capable of efficiently antagonizing NF-κB activity early after TNF induction (Fig. 8A), but that EBOV and SUDV proteins lost some degree of potency by 24hpi (Fig. 8B), as observed in prior SeV-based assays. Consistent with previous reports, we found that all filovirus VP35 proteins were rendered incapable of inhibiting TNF-induced NF-κB activity (Fig. 8 C), despite their strong antagonism when we previously had induced by SeV (Fig. 2D) (Chang et al., 2009).
Fig. 8.
VP24 of all ebolaviruses except RESTV also display strong inhibition of early TNF-induced NF-κB activity, and later, virus species-specific variable inhibition, despite prominent loss of VP35 activity. Reporter assays as described in Fig. 2 were conducted to further characterize putative species-specific VP24 antagonism on NF-κB activity. (A–C) Cells cotransfected with NF-κB activity reporter were directly induced 1dpt by TNF (50 ng/mL) and harvested 4hpi (A) or 24hpi (B) to assess VP24 inhibitory activity for early or late TNF receptor-based response activation, respectively, or 24hpi to assess VP35 inhibitory activity (C), as indicated. FC = fold change of positive induction relative to uninduced negative control. RLU = relative light units. *=p < 0.05; **=p < 0.005; ***=p < 0.0005; ****=p < 0.0001; asterisks above horizontal lines indicate direct comparison of activity values between two antagonists.
These results suggest that VP24 proteins of ebolaviruses, except of RESTV, are also capable of robustly inhibiting early NF-κB signaling when directly induced by TNF, an activity by which VP35 appears to be insufficient. This further suggests possible non-redundant functions of ebolavirus VP35 and VP24, which may together antagonize IFN induction under a variety of alternative and shifting host response conditions.
4. Discussion
In this report, we describe a standardized vector library and comprehensive, side-by-side activity profiling for VP35, VP40 and VP24 IFN antagonist proteins from representative isolates of all known filoviruses. Based on the results of our study, we reveal several previously-unrecognized filoviral antagonist activities. 1) All ebolavirus VP35 proteins universally shut down IFN induction and signaling, as has been previously shown for EBOV. Further, MARV VP35 is a comparatively less-efficient antagonist, as has also been shown. The novelty is that VP35 of RAVV, despite its close phylogenetic relationship to MARV, has activity more similar to that of ebolaviruses (Fig. 2A–D, F–I). 2) VP40 proteins of marburgviruses have inducerspecific activities in which potencies are robust when induced by U-IFN, but more modest when induced by SeV (Fig. 3A and B compared to C–E). There is also a filovirus-specific trend of clearly weaker antagonism by RAVV VP40 compared to MARV VP40 (Fig. 3). 3) SUDV and TAFV VP40 proteins display unexpected antagonistic activity on IFN signaling, which has not been seen before by any ebolavirus VP40 (Fig. 3B–E, G and H). 4) Both marburgvirus VP40 proteins, particularly MARV, as well as TAFV VP40 are capable of previously-unreported, modest inhibition of IFN induction via IRF3 but not via NF-κB signaling (Fig. 4). 5) While VP24 proteins of most ebolaviruses, but not of marburgviruses, show robust antagonism of IFN signaling, as was expected, VP24 of RESTV is conspicuously the only antagonist from any ebolavirus species incapable of similarly overcoming induction by higher amounts of SeV, by poly(I:C) or by ΔΔrRVF (Fig. 5B–F, Suppl Fig. 1), despite strong inhibitory activity upon induction by U-IFN (Fig. 5A). 6) Finally, we report the previously-unrecognized, noncanonical ability of all ebolavirus VP24 proteins except RESTV to directly and efficiently block upstream IFN induction via both IRF3 and early TNF-induced NF-κB signaling. We hypothesize that this inhibitory effect on IFN induction in HEK293 cells represents a bona fide novel VP24 function shared by most known ebolaviruses.
Much previous work has been done to elucidate the function of IFN antagonists in the past fifteen years, since the breakthrough 2000 study first showing EBOV VP35 blocked innate immune responses (Basler et al., 2000). However, these efforts have since been conducted across laboratories using multiple vector backbones (Dunham et al., 2015; Valmas and Basler, 2011; Ilinykh et al., 2015; Yen et al., 2014; Hartman et al., 2006; Basler et al., 2000, 2003); epitope tags (Reid et al., 2007; Valmas and Basler, 2011; Yen et al., 2014; Mateo et al., 2010); reporter systems (Reid et al., 2007; Ilinykh et al., 2015; Albariño et al., 2015; García-Dorival et al., 2014); in-house antibodies of differing efficacies and epitopes, with many antagonists lacking specific antibodies (Burk et al., 2016; Reid et al., 2007; Feagins and Basler, 2015); host cells (Reid et al., 2007; Valmas and Basler, 2011; Ilinykh et al., 2015; Yen et al., 2014; Feagins and Basler, 2015); and inducers (Dunham et al., 2015; Valmas and Basler, 2011; Ilinykh et al., 2015; Edwards et al., 2016; Yen et al., 2014; Feagins and Basler, 2015; Edwards and Basler, 2015). Therefore, our standardized viral protein library and reporter assay-based panel were developed to better understand pan-filovirus antagonism for all isolated species simultaneously. Our study also aimed to describe any functional differences that may have gone overlooked amid the variables associated with separate protein activity experiments. Finally, use of a luciferase reporter system to measure antagonist potencies has long been established to be robust and biologically relevant, with functional data correlating well with IFN responses obtained following filoviral infection (Basler, 2015; Ilinykh et al., 2015; Edwards et al., 2016; Hartman et al., 2008a; Kash et al., 2006; Albariño et al., 2015; Mateo et al., 2011).
Of the three types of filovirus IFN antagonist proteins we profiled, most information currently available in the literature describes EBOV and MARV VP35 activities (Reid et al., 2006; Valmas and Basler, 2011; Edwards et al., 2016; Bale et al., 2012; Basler et al., 2000, 2003). It was of little surprise that in our system, ebolavirus VP35 proteins were strong IFN induction and signaling antagonists (Fig. 2). However, the differences noted between MARV and RAVV were unexpected. It has been shown in previous reports by our lab and others that mutations within VP35, R312A (for EBOV) and analogous R301A (for MARV), specifically impede VP35 IFN antagonism without affecting virus replication or transcription, for which VP35 is essential (Bale et al., 2012; Ramanan et al., 2012; Hartman et al., 2008a, 2006, 2008b; Albariño et al., 2015; Zinzula and Tramontano, 2013). These mutations in recombinant viruses also attenuate viral growth due to robust cellular IFN responses, as well as in vivo virulence as observed in mouse models of filovirus infection (Hartman et al., 2008a, 2006). Despite clear impact by these mutations on EBOV and MARV virulence, it is intriguing that observed differences in VP35 WT potencies between ebolaviruses, MARV and RAVV do not also correlate to virulence of their respective virus. Indeed, while examples of human cases of RAVV are sparse, EBOV, MARV and RAVV seem to cause similarly lethal human disease, while RESTV, with equally potent VP35 activity, is apparently nonpathogenic in humans (Pappalardo et al., 2016; Messaoudi et al., 2015; Slenczka and Klenk, 2007; Burk et al., 2016; Kuhn et al., 2011). It remains unclear what factors govern such correlations between antagonist function and severity of disease, although we can speculate that certain physiological conditions may need to be met in order for filovirus-specific VP35 potencies to influence pathogenicity. Alternatively, specific interactions of each VP35 with host or virus proteins, or the antagonist’s functional mechanisms or kinetics, may play a more important role than overall differences in WT IFN antagonistic potencies between individual filoviruses. This may be the case for at least EBOV and MARV VP35, the latter of which recognizes dsRNA via alternative backbone coating and end-capping mechanisms (RAVV VP35 dsRNA binding remains untested) (Bale et al., 2012; Ramanan et al., 2012; Zinzula and Tramontano, 2013; Cárdenas et al., 2006; Leung et al., 2010b, 2009). This distinct recognition by MARV VP35, in turn, may favor inhibition of MDA5 and poly(I: C), hence explaining our observation that MARV VP35 shows equally strong poly(I:C)-induced IFNβ antagonism compared to other filoviruses (Fig. 2E) (Zinzula and Tramontano, 2013).
The reports that established marburgvirus VP40 as an antagonist of Jak1 activation assessed VP40 function by inducing cells with cytokine treatment (Valmas and Basler, 2011; Valmas et al., 2010). Our data agree with these results and show strong MARV VP40 activity in our U-IFN induction-based assays (Fig. 3A and B). However, using SeV infection to measure VP40 activity has not to our knowledge been previously reported. The unexpected reduction of marburgvirus VP40 antagonism following SeV induction was MOI dependent, as stronger inhibition of IRF1 activity and ISRE expression was seen at a lower SeV MOI (Fig. 3D and E), whereas both reporters, as well as ISG54, were only modestly antagonized at high MOI (Fig. 3C–E) compared to U-IFN (Fig. 3A and B). While SeV MOI-dependent effects were also found for SUDV and TAFV VP40, they appear to be promoter specific, as more potent antagonism was only observed at a lower MOI for IRF1 (Fig. 3E) but not ISRE (Fig. 3D). We also saw putative promoter specificity for SUDV VP40, which showed no antagonism of ISG54 regardless of inducer (Fig. 3A and C, F and H), as well as inducer specificity for TAFV VP40, which displayed MARV VP40-like levels of inhibition of ISG54 upon SeV infection (Fig. 3C) while having undetectable activity following U-IFN induction (Fig. 3A). The promoter specificity observed for the three IFN signaling reporters could be due to differences in respective sequence composition. The ISRE reporter is made up of tandem repeats of STAT1/2-responsive elements, while the ISG54 reporter contains an intact promoter region, including elements directly responsive to IRF3 (Basler et al., 2003); the IRF1 dual activity reporter is also responsive to IFNγ, which we did not assess.
Meanwhile, the observed inducer-specific effects for certain antagonist potencies, such as MARV VP35 (Fig. 2E) or RESTV VP24 (Fig. 5A), as well as the SeV MOI-dependent effects on others, such as EBOV and RESTV VP24 (Fig. 5C and D), or MARV and RAVV VP40 (Fig. 3D and E), are quite intriguing. We hypothesize two, not necessarily mutuallyexclusive, explanations for these effects. First, being the most powerful inducer, increasing amounts of SeV could simply trigger concomitantly stronger innate immune responses, which particular antagonists, such as RESTV VP24, cannot overcome due to less efficient activity. Second, SeV could produce a much broader and more dynamic range of IFN responses, inducing alternate states of differentially-expressed or -activated proteins that are dependent on SeV MOI (Zaritsky et al., 2015; Ye and Maniatis, 2011). For instance, SeV might activate larger, concentration-dependent subsets of Type I IFNs, as well as Type II and III IFNs (not assessed in the context of this study with the exception of IFNλ expression in Suppl Fig. 2, included since IFNλ is thought to be similarly induced and upregulate similar genes as Type I IFNs) that may affect viral protein function or alternatively act as a sort of immune pathway rheostat by which certain host proteins become subject to negative regulation at higher levels of induction (Hoffmann et al., 2015; Zaritsky et al., 2015; Ye and Maniatis, 2011). This could explain why in a few assays, some antagonists, such as TAFV and MARV VP40 (Fig. 4 A), counterintuitively appeared capable of modestly stronger inhibition at higher MOI, potentially the result of an altered immune repertoire that may favor the upregulation and/or activation of a different combination of host pathway intermediates and effectors as those induced at lower MOIs. The specific proteins upregulated within this high MOI subset may include those that have a reduced role in suppressing select antagonists, thus leading to stronger activity by those viral proteins. On the other hand, each of the other nonviral inducers likely triggers a more singular, focused response reliant on specific receptors or pathway intermediates, which also then alters respective antagonist activity profiles, like those we observed in assays induced by U-IFN (e.g. Fig. 3A and B, 5A), TNF (e.g. Fig. 8C) or poly(I:C) (e.g. Fig. 2E, 6F) (Kawai and Akira, 2006; Li et al., 2005; Zaritsky et al., 2015; Zinzula and Tramontano, 2013; Cárdenas et al., 2006; Ye and Maniatis, 2011; Harcourt et al., 1998). Recent work by Zaritsky et al. indeed suggests induction profile differences in monocytes dependent on MOI of SeV (Zaritsky et al., 2015), and the wide range of MOI used in our assays was designed to allow us to better discriminate which antagonists are potentially affected by either or both of our hypotheses, effects that could have gone overlooked using a single amount of virus. Finally, we rule out that C protein of SeV, a known STAT1 inhibitor (Zaritsky et al., 2015), indirectly interfered with our IFN signaling assays or could explain inducer specificity, at least in the context of IFN antagonism activity. This is supported by robust positive control inductions of IFN signaling reporters, corroborative antagonist activity profiles shared between signaling and induction reporters (e.g. Fig. 2D and G) as well as between SeV and other inducers, such as poly(I:C) (Fig. 5B) and ΔΔrRVF (an attenuated virus that lacks any functional IFN antagonist, Suppl Fig. 1), and the strong agreement of our activity data with those of comparable assays in prior studies that investigated overlapping filoviral proteins.
As with condition-specific inhibitory properties, it is similarly surprising to find MARV-like levels of antagonism by the ebolavirus VP40 proteins from SUDV (on IFN signaling) and TAFV (on induction and signaling), with TAFV VP40 being uniquely capable of inhibiting as many SeV-induced reporters as MARV VP40 (Fig. 3 and 4). To our knowledge, this is the first report of ebolavirus VP40 proteins being shown to have any IFN antagonizing activity. As SUDV, TAFV as well as BDBV VP35 and VP24 proteins showed equally strong or even stronger activities than respective EBOV proteins, our data suggest that, based on IFN antagonism, these viruses could be capable of comparable levels of virulence and corresponding human disease, although a relative lack of clinical cases makes their exact pathogenicity so far uncertain. It will be of interest for future studies to determine if the additional inhibitory effects observed for SUDV and TAFV VP40 proteins, but not for those of other ebolaviruses, influences viral pathogenesis, and what features of these two proteins are responsible for both their shared and disparate activities.
The fact that MARV VP40 has better potency than RAVV VP40 is also intriguing (Fig. 3 and 4). This effect was observed for all conditions tested and appears to be independent of SeV MOI. A prior study reported that MARV and RAVV VP40 proteins had similar inhibitory activities, however the authors’ use of different cell lines, assays and viral isolates may account for this discrepancy between our results and theirs (Valmas and Basler, 2011). As discussed earlier, the difference we found between MARV and RAVV VP35 potencies had been unexpected given the similar virulence of both viruses, at least based on the limited clinical data for RAVV (Burk et al., 2016). While speculative, it may be possible that RAVV divergence from MARV has shifted the antagonistic balance between VP40 and VP35, such that a less potent RAVV VP40 has evolved to compensate for stronger RAVV VP35 activity while maintaining overall similar levels of IFN response ablation compared to MARV. Future experiments to elucidate any consequences of this weaker RAVV VP40 activity, and possible functional compensation by RAVV VP35, on virus growth and pathogenesis could be informative.
Strong antagonism of IFN signaling by ebolavirus VP24 proteins was also anticipated under a variety of different conditions (Fig. 5), given the extensive literature describing EBOV VP24 preventing the interaction of karyopherin α with phosphorylated STAT1 (Basler, 2015;Reid et al., 2006, 2007; Ilinykh et al., 2015; Mateo et al., 2010, 2011; Zhang et al., 2012b; Feagins and Basler, 2015; García-Dorival et al., 2014). Nevertheless, the drastic reduction reported here in RESTV VP24 activity is unique among ebolaviruses (Fig. 5B–F). RESTV remains the only known ebolavirus species apparently incapable of causing severe human disease, while maintaining high pathogenicity in NHP species (Pappalardo et al., 2016; Burk et al., 2016; Kuhn et al., 2011). Few reports to date provide evidence of RESTV-specific attenuating factors between primate species (Pappalardo et al., 2016; Burk et al., 2016). Illuminating microarray analysis by Kash, et al., conducted just prior to identification of EBOV VP24 as an IFN signaling antagonist, compared transcriptomes of RESTV-infected cells to EBOV-infected cells, and showed stronger human immune responses upon RESTV infection (Kash et al., 2006). More importantly, their findings offered an early indication that these differences correlate with antagonistic effects on IFN signaling, as RESTV infection led to more robust levels of ISG expression and higher stimulation of additional ISGs upon IFN treatment compared to EBOV, the latter for which seems to imply that only VP24 should remain a viable antagonist (Kash et al., 2006). In a series of other studies, lethal adaptation of WT EBOV in mouse or guinea pig models, where it is normally nonpathogenic, was found to require point mutations that consistently occurred within VP24 or its 5′ noncoding region (Ramanan et al., 2011; Reid et al., 2007; Mateo et al., 2010, 2011).
Despite several RESTV-inclusive reports, only one previous paper has directly examined RESTV VP24 protein activity, which showed equivalent potency to EBOV VP24 upon U-IFN induction of human cells (Reid et al., 2007), conditions that gave us similar results and may thus have obscured the inefficiencies we saw with other inducers. While it is still unclear as to why RESTV VP24 lacks similar potency to other ebolaviruses, we speculate that certain amino acids conserved by other ebolaviruses, but not by RESTV, could impact VP24 function either alone or in combination. Indeed, Zhang, et al. suggested this may be the case following their examination of EBOV, SUDV and RESTV VP24 structures, which identified five potential sites where residue alterations in RESTV could weaken its affinity for karyopherin α binding (Zhang et al., 2012a). Similarly, a recent report by Pappalardo, et al. described all nonconserved residues of RESTV compared to the other ebolaviruses for each viral gene, and cited VP24 as a likely determinant of pathogenicity (Pappalardo et al., 2016). However, we cannot rule out that RESTV VP24 impairment may be species specific and unrelated to its nonconserved residues. Elucidation of any specific sites that might dictate RESTV VP24 inefficiency, as well as the possible impact such inefficiency has on RESTV growth and virulence, could be valuable to defining human-nonpathogenic determinants for development of better antiviral strategies and in understanding host-pathogen interactions in NHP models.
Finally, we report that ebolavirus VP24 proteins antagonize up-stream IFN induction pathways, largely independent of SeV MOI (Fig. 6–8, Suppl Fig. 2 and 3). We observed similar, albeit less potent, effects by MARV, RAVV and TAFV VP40 proteins (Fig. 4A and B). VP24 and VP40 proteins from some filoviruses are each capable of targeting downstream IFN signaling, but achieve this using distinct strategies. In this way, these two proteins, as well as others, may complement one another by providing redundant antagonistic functions necessary to ensure immune evasion of each filovirus in its respective ecological niche. From this perspective, the complex IFN response may be naturally sensitive to filovirus-mediated intervention whereby each virus species has evolved a different mechanism(s) to antagonize related critical steps in the pathway, exemplified by our data implicating both VP24 and VP40 from particular filoviruses as inhibitors of IFN induction.
This is not the first study to show noncanonical immunosuppressive activities by filoviral VP24 proteins. Halfmann, et al. identified that EBOV VP24 could prevent the IFNβ-induced phosphorylation of p38 (Halfmann et al., 2011). p38 is an important MAPK pathway intermediate that, among other activities, can upregulate ISG expression following JAK1 activation as well as PKR-mediated AP-1 transactivation (Fig. 1A and B) (García et al., 2006; Kawai and Akira, 2006; Halfmann et al., 2011). The authors described VP24 antagonism of p38 in the context of downstream IFN signaling, although evidence also suggests p38 plays an essential, cell-dependent role in upstream IFN induction upon viral infection (Halfmann et al., 2011; Mikkelsen et al., 2009). Meanwhile, the Basler lab recently showed that MARV VP24 can bind to Keap1, a repressor of IKKβ activity, which allows IKKβ to activate anti-apoptotic NF-κB signaling (Edwards and Basler, 2015).
Indeed, this is also not the first demonstration of filovirus VP24 inhibitory effects on IFNβ induction. Another recent study from the Basler lab showed strong antagonism of SeV-induced IFNβ expression by both EBOV and LLOV VP24 proteins (Feagins and Basler, 2015). However, the authors concluded that this effect was due to an indirect ability of VP24 IFN signaling inhibition to disrupt a putative positive feedback loop that might potentiate IFNβ expression through upregulation of RIG-I (Feagins and Basler, 2015). Attempts using our system to trigger such positive feedback by U-IFN on IFNβ reporter activity or to block induction by nAbs failed, as did nAb blockade of VP24 antagonism of IFNβ expression. Additional data using HAP-1 WT and IFNAR KO cells also showed that IFN induction-based IRF1 activity in an IFN signaling-deficient background was similarly inhibited by VP24 proteins (Suppl Fig. 3), while qRT-PCR-based assays of endogenous IFN gene expression displayed analogous inhibitory activity by ebolavirus but not MARV VP24 proteins in induced transfected cells (Suppl Fig. 2), a finding that corroborates our reporter-based assays.
All three lines of evidence support our hypothesis that ebolavirus VP24 proteins, except RESTV VP24, have a direct, novel function in the prevention of IFN induction. In fact, our current data, in agreement with the recent observation by the Basler lab and using a similar assay (Feagins and Basler, 2015), appears to be the only direct assessment of inhibitory activity by any filoviral VP24 protein on IFN induction. The canonical view that EBOV VP24 is only capable of blocking IFN signaling seems to have been established based on relatively limited evidence in the literature, much of which comes from two similarly-conducted immunofluorescence assays in Vero cells putatively showing a lack of ectopic GFP-IRF3 nuclear translocation in the presence of EBOV VP24 (Reid et al., 2006; Basler et al., 2003). We believe this, and in light of our current findings, leaves open an intriguing possibility for ebolavirus VP24 proteins to prevent IFN induction via a mechanism that may not be dependent on blocking IRF3 translocation.
A virus having more than one protein capable of antagonizing IFN induction could be advantageous to virus replication and evasion of innate immune responses under a variety of potentially changing conditions that may occur in an animal host. Filoviruses have apparently evolved multiple coordinated ways to counter the multilayered IFN response. This may provide one explanation for ebolavirus VP24 inhibition of early NF-κB activity (Fig. 8A). While VP24 was capable of efficient TNF-driven NF-κB antagonism, ebolavirus VP35 protein activity was debilitated under these conditions (Fig. 8C). This is supported by a previous report showing VP35 cannot block TNF or IκBα mRNA expression (Chang et al., 2009). NF-κB has been found to be among the earliest transactivators present at the IFNβ promoter (Balachandran and Beg, 2011; Malmgaard, 2004; Wang et al., 2010). TNF-induced NF-κB signaling is mediated by pathway intermediates TNF receptor type I-associated death domain protein (TRADD), receptor interacting protein (RIP) and NF-κB-inducing kinase (NIK), independently from RIG-I signaling mediated by MAVS and TRAF6 (Fig. 1A) (Yarilina and Ivashkiv, 2010; Balachandran and Beg, 2011; Wang et al., 2010; Hiscott et al., 2001; Aggarwal, 2003). Interestingly, TNF can stimulate the PKR pathway, which has been shown to activate both NF-κB and p38, as well as stimulate p38 directly (García et al., 2006; Halfmann et al., 2011; Yarilina and Ivashkiv, 2010; Palladino et al., 2003; Hiscott et al., 2001). As mentioned earlier, VP35 antagonizes PKR, and EBOV VP24 blocks p38 phosphorylation (Halfmann et al., 2011; Schumann et al., 2009). Thus, one can speculate that in certain conditions favoring TNF-mediated responses, and concomitant NF-κB activity that is now refractory to VP35 inhibition, ebolavirus VP24 provides nonredundant “backup” antagonism, via a putative interaction with an unknown intermediate (possibly within the TRADD/RIP/NIK pathway) to prevent critical early NF-κB signaling, as well as to potentially mitigate TNF-induced PKR and p38 activation.
In conclusion, our comparative analysis of filoviral IFN antagonist activities in HEK293 cells provide promising avenues for future investigations. Of particular interest will be a deeper understanding of the precise mechanisms that determine VP24- or VP40-mediated, noncanonical antagonistic activities on the IFN induction pathway, and conversely, the newly-recognized impairment of RESTV VP24. Finally, it will be of broader significance to put into context these novel functions by not only addressing how these proteins, working alongside VP35, coordinate exquisite inhibition of multiple aspects of host innate immune responses, but also how they separately and together influence the pathogenesis of each of their respective filoviruses.
Supplementary Material
Acknowledgments
The authors would like to thank Genscript, Inc. for their assistance in the synthesis of our filoviral antagonist library; Steven Goodbourn (University of London) for the hISG54 reporter vector; Sonja Best (NIAID) for the RIG-I CARD vector; Éric Bergeron, Laura McMullan, Florine Scholte, Lisa Guerrero, Marko Zivcec, Markus Kainulainen, Stephen Welch, Jessica Spengler and Jenna Kelly of VSPB for helpful advice, reporter plasmids or cell lines; and Christina Spiropoulou and Stuart Nichol for critical reading of the manuscript. This work was supported in part by a joint American Society for Microbiology/CDC Postdoctoral Fellowship in Infectious Disease and Public Health Microbiology. All research and conclusions described herein are solely those of the authors and do not necessarily represent the views of CDC.
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2016.11.015.
References
- Aggarwal BB, 2003. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol 3 (9), 745–756. [DOI] [PubMed] [Google Scholar]
- Albariño CG, Wiggleton Guerrero L., Spengler JR, Uebelhoer LS, Chakrabarti AK, Nichol ST, et al. , 2015. Recombinant Marburg viruses containing mutations in the IID region of VP35 prevent inhibition of host immune responses. Virology 476, 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, et al. , 2012. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 8 (10), e1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Nyakarahuka L, McElroy AK, Dodd KA, Sealy TK, Schuh AJ, et al. , 2014. Marburgvirus resurgence in Kitaka Mine bat population after extermination attempts, Uganda. Emerg. Infect. Dis 20 (10), 1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Jones ME, Sealy TK, Uebelhoer LS, Schuh AJ, Bird BH, et al. , 2015. Oral shedding of Marburg virus in experimentally infected Egyptian fruit bats (Rousettus aegyptiacus). J. Wildl. Dis 51 (1), 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Beg AA, 2011. Defining emerging roles for NF-kappaB in antivirus responses: revisiting the interferon-beta enhanceosome paradigm. PLoS Pathog. 7(10), e1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Julien JP, Bornholdt ZA, Kimberlin CR, Halfmann P, Zandonatti MA, et al. , 2012. Marburg virus VP35 can both fully coat the backbone and cap the ends of dsRNA for interferon antagonism. PLoS Pathog. 8 (9), e1002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, 2015. Innate immune evasion by filoviruses. Virology 479–480, 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Wang X, Mühlberger E, Volchkov V, Paragas J, Klenk HD, et al. ,2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97 (22), 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Mühlberger E, Bray M, et al. , 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol 77 (14), 7945–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird BH, Maartens LH, Campbell S, Erasmus BJ, Erickson BR, Dodd KA, et al. , 2011. Rift Valley fever virus vaccine lacking the NSs and NSm genes is safe, nonteratogenic, and confers protection from viremia, pyrexia, and abortion following challenge in adult and pregnant sheep. J. Virol 85 (24), 12901–12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio CM, Aman MJ, Grogan C, Hogan R, Ruthel G, Negley D, et al. , 2003. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J. Infect. Dis 188 (11), 1630–1638. [DOI] [PubMed] [Google Scholar]
- Bukreyev AA, Chandran K, Dolnik O, Dye JM, Ebihara H, Leroy EM, et al. ,2014. Discussions and decisions of the 2012–2014 international committee on taxonomy of viruses (ICTV) Filoviridae study group, January 2012–June 2013. Arch. Virol 159 (4), 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk R, Bollinger L, Johnson JC, Wada J, Radoshitzky SR, Palacios G, et al. ,2016. Neglected filoviruses. FEMS Microbiol. Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas WB, 2010. Evasion of the interferon-mediated antiviral response by filoviruses. Viruses 2 (1), 262–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas WB, Loo YM, Gale M Jr., Hartman AL, Kimberlin CR, Martinez-Sobrido L, et al. , 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol 80(11), 5168–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SA, Towner JS, Sealy TK, McMullan LK, Khristova ML, Burt FJ, et al. , 2013. Molecular evolution of viruses of the family Filoviridae based on 97 whole-genome sequences. J. Virol 87 (5), 2608–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Rao B, Asgeirsson B, Dandekar A, 2014. Correlating the ability of VP24 protein from Ebola and Marburg viruses to bind human karyopherin to their immune suppression mechanism and pathogenicity using computational methods. F1000Research. [Google Scholar]
- Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, et al. , 2009. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 5 (6), e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Jonsson-Schmunk K, Qiu X, Shedlock DJ, Strong J, Xu JX, et al. , 2015. A single dose respiratory recombinant adenovirus-based vaccine provides long-term protection for non-human primates from lethal Ebola infection. Mol. Pharm 12 (8), 2712–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton MC, Bruhn JF, Atkins K, Webb TL, Baydo RO, Raymond A, et al. , 2015. High-resolution crystal Structure of dimeric VP40 from Sudan ebolavirus. J. Infect. Dis 212 (Suppl. 2), S167–S171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Pei J, Grishin NV, 2015. Predictive and comparative analysis of Ebolavirus proteins. Cell Cycle 14 (17), 2785–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham EC, Banadyga L, Groseth A, Chiramel AI, Best SM, Ebihara H, et al. , 2015. Assessing the contribution of interferon antagonism to the virulence of West African Ebola viruses. Nat. Commun 6, 8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Basler CF, 2015. Marburg virus VP24 protein relieves suppression of the NF-kappaB pathway through interaction with Kelch-like ECH-associated protein1. J. Infect. Dis 212 (Suppl. 2), S154–S159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MR, Liu G, Mire CE, Sureshchandra S, Luthra P, Yen B, et al. , 2016. Differential regulation of interferon responses by Ebola and Marburg virus VP35 proteins. Cell Rep. 14 (7), 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagins AR, Basler CF, 2014. The VP40 protein of Marburg virus exhibits impaired budding and increased sensitivity to human tetherin following mouse adaptation. J. Virol 88 (24), 14440–14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagins AR, Basler CF, 2015. Lloviu virus VP24 and VP35 proteins function as innate immune antagonists in human and bat cells. Virology 485, 145–152. [DOI] [PubMed] [Google Scholar]
- García MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, et al. , 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. MMBR 70 (4), 1032–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Dorival I, Wu W, Dowall S, Armstrong S, Touzelet O, Wastling J, et al. , 2014. Elucidation of the Ebola virus VP24 cellular interactome and disruption of virus biology through targeted inhibition of host-cell protein function. J. Proteome Res. 13 (11), 5120–5135. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, et al. , 2006.Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 103 (22), 8459–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P, Neumann G, Kawaoka Y, 2011. The Ebolavirus VP24 protein blocks phosphorylation of p38 mitogen-activated protein kinase. J. Infect. Dis 204 (Suppl3), S953–S956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Weber F, 2009. The interferon response circuit in antiviral host defense.Verh. K. Acad. Geneeskd. Belg 71 (1–2), 73–86. [PubMed] [Google Scholar]
- Haller O, Kochs G, Weber F, 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344 (1), 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt BH, Sanchez A, Offermann MK, 1998. Ebola virus inhibits induction of genes by double-stranded RNA in endothelial cells. Virology 252 (1), 179–188. [DOI] [PubMed] [Google Scholar]
- Hartman AL, Dover JE, Towner JS, Nichol ST, 2006. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J. Virol 80 (13), 6430–6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Ling L, Nichol ST, Hibberd ML, 2008b. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J. Virol 82(11), 5348–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Bird BH, Towner JS, Antoniadou ZA, Zaki SR, Nichol ST, 2008a. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of ebola virus. J. Virol 82 (6), 2699–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J, Kwon H, Genin P, 2001. Hostile takeovers: viral appropriation of the NF- kappaB pathway. J. Clin. Investig 107 (2), 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HH, Schneider WM, Rice CM, 2015. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36 (3), 124–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Park-Min KH, Yarilina A, Ivashkiv LB, 2008. Regulation of STAT pathways and IRF1 during human dendritic cell maturation by TNF-alpha and PGE2. J. Leukoc. Biol 84 (5), 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilinykh PA, Lubaki NM, Widen SG, Renn LA, Theisen TC, Rabin RL, et al. , 2015. Different temporal effects of Ebola virus VP35 and VP24 proteins on global gene expression in human dendritic cells. J. Virol 89 (15), 7567–7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Schuh AJ, Amman BR, Sealy TK, Zaki SR, Nichol ST, et al. , 2015.Experimental inoculation of Egyptian Rousette bats (Rousettus aegyptiacus) with viruses of the Ebolavirus and Marburgvirus genera. Viruses 7 (7), 3420–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash JC, Mühlberger E, Carter V, Grosch M, Perwitasari O, Proll SC, et al. , 2006.Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J. Virol 80 (6), 3009–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. , 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441 (7089), 101–105. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S, 2006. TLR signaling. Cell Death Differ. 13 (5), 816–825. [DOI] [PubMed] [Google Scholar]
- Kimberlin CR, Bornholdt ZA, Li S, Woods VL Jr., MacRae IJ, Saphire EO,2010. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. USA 107 (1), 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok KH, Lui PY, Ng MH, Siu KL, Au SW, Jin DY, 2011. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe 9 (4), 299–309. [DOI] [PubMed] [Google Scholar]
- Kröger A, Köster M, Schroeder K, Hauser H, Mueller PP, 2002. Activities of IRF-1.J. Interferon Cytokine Res. 22 (1), 5–14. [DOI] [PubMed] [Google Scholar]
- Kuhn JH, Dodd LE, Wahl-Jensen V, Radoshitzky SR, Bavari S, Jahrling PB, 2011. Evaluation of perceived threat differences posed by filovirus variants. Biosecur. Bioterror. Biodef. Strateg. Pract. Sci 9 (4), 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Bào Y, Bavari S, Becker S, Bradfute S, Brauburger K, et al. , 2014. Virus nomenclature below the species level: a standardized nomenclature for filovirus strains and variants rescued from cDNA. Arch. Virol 159 (5), 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Abbas W, Herbein G, 2013. TNF and TNF receptor superfamily members in HIV infection: new cellular targets for therapy? Mediat. Inflamm, 484378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Ginder ND, Fulton DB, Nix J, Basler CF, Honzatko RB, et al. , 2009.Structure of the Ebola VP35 interferon inhibitory domain. Proc. Natl. Acad. Sci. USA 106 (2), 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Shabman RS, Farahbakhsh M, Prins KC, Borek DM, Wang T, et al. ,2010a. Structural and functional characterization of Reston Ebola virus VP35 interferon inhibitory domain. J. Mol. Biol 399 (3), 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, et al. , 2010b. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat. Struct. Mol. Biol 17 (2), 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Chen Z, Kato N, Gale M Jr., Lemon SM, 2005. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem 280 (17), 16739–16747. [DOI] [PubMed] [Google Scholar]
- Malmgaard L, 2004. Induction and regulation of IFNs during viral infections. J. Interferon Cytokine Res. 24 (8), 439–454. [DOI] [PubMed] [Google Scholar]
- Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE, 2010. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J. Virol 84(2), 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo M, Carbonnelle C, Reynard O, Kolesnikova L, Nemirov K, Page A, et al. , 2011. VP24 is a molecular determinant of Ebola virus virulence in guinea pigs. J. Infect. Dis 204 (Suppl 3), S1011–S1020. [DOI] [PubMed] [Google Scholar]
- Messaoudi I, Amarasinghe GK, Basler CF, 2015. Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat. Rev. Microbiol 13 (11), 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen SS, Jensen SB, Chiliveru S, Melchjorsen J, Julkunen I, Gaestel M, et al. , 2009. RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells: dependence on TRAF2 and TAK1. J. Biol. Chem 284 (16), 10774–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MA, Bahjat FR, Theodorakis EA, Moldawer LL, 2003. Anti-TNF-alpha therapies: the next generation. Nat. Rev. Drug Discov. 2 (9), 736–746. [DOI] [PubMed] [Google Scholar]
- Pappalardo M, Julia M, Howard MJ, Rossman JS, Michaelis M, Wass MN,2016. Conserved differences in protein sequence determine the human pathogenicity of Ebolaviruses. Sci. Rep 6, 23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K, Chattopadhyay S, Sen GC, 2008. IRF-3 activation by Sendai virus infection is required for cellular apoptosis and avoidance of persistence. J. Virol 82 (7), 3500–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P, Shabman RS, Brown CS, Amarasinghe GK, Basler CF, Leung DW,2011. Filoviral immune evasion mechanisms. Viruses 3 (9), 1634–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P, Edwards MR, Shabman RS, Leung DW, Endlich-Frazier AC, Borek DM, et al. , 2012. Structural basis for Marburg virus VP35-mediated immune evasion mechanisms. Proc. Natl. Acad. Sci. USA 109 (50), 20661–20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF, 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol 81 (24), 13469–13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, et al. ,2006. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol 80 (11), 5156–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer T, Brcic M, Schweizer M, Jungi TW, 2008. poly(I:C) and LPS induce distinct IRF3 and NF-kappaB signaling during type-I IFN and TNF responses in human macrophages. J. Leukoc. Biol 83 (5), 1249–1257. [DOI] [PubMed] [Google Scholar]
- Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N, 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441 (1), 106–110. [DOI] [PubMed] [Google Scholar]
- Schumann M, Gantke T, Mühlberger E, 2009. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J. Virol 83 (17), 8993–8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slenczka W, Klenk HD, 2007. Forty years of Marburg virus. J. Infect. Dis 196 (Suppl2), S131–S135. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Dittmar K, Ballinger MJ, Bruenn JA, 2011. Evolutionary maintenance of filovirus-like genes in bat genomes. BMC Evolut. Biol 11, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Pourrut X, Albariño CG, Nkogue CN, Bird BH, Grard G, et al. , 2007.Marburg virus infection detected in a common African bat. PLoS One 2 (8), e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Sealy TK, Khristova ML, Albariño CG, Conlan S, Reeder SA, et al. ,2008. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 4 (11), e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, et al. , 2009.Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 5 (7), e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmas C, Basler CF, 2011. Marburg virus VP40 antagonizes interferon signaling in a species-specific manner. J. Virol 85 (9), 4309–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmas C, Grosch MN, Schümann M, Olejnik J, Martinez O, Best SM, et al. , 2010. Marburg virus evades interferon responses by a mechanism distinct from ebola virus. PLoS Pathog. 6 (1), e1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Basagoudanavar SH, Wang X, Hopewell E, Albrecht R, García-Sastre A, et al. , 2010. NF-kappa B RelA subunit is crucial for early IFN-beta expression and resistance to RNA virus replication. J. Immunol 185 (3), 1720–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G, Kobinger GP, Qiu X, 2014. Characterization of host immune responses inEbola virus infections. Expert Rev. Clin. Immunol 10 (6), 781–790. [DOI] [PubMed] [Google Scholar]
- Yarilina A, Ivashkiv LB, 2010. Type I interferon: a new player in TNF signaling. Curr. Dir. Autoimmun 11, 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Maniatis T, 2011. Negative regulation of interferon-beta gene expression during acute and persistent virus infections. PloS One 6 (6), e20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen B, Mulder LC, Martinez O, Basler CF, 2014. Molecular basis for ebolavirusVP35 suppression of human dendritic cell maturation. J. Virol 88 (21), 12500–12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky LA, Bedsaul JR, Zoon KC, 2015. Virus multiplicity of infection affects type I interferon subtype induction profiles and interferon-stimulated genes. J. Virol 89(22), 11534–11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AP, Abelson DM, Bornholdt ZA, Liu T, Woods VL Jr., Saphire EO, 2012b. The ebolavirus VP24 interferon antagonist: know your enemy. Virulence 3(5), 440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AP, Bornholdt ZA, Liu T, Abelson DM, Lee DE, Li S, et al. , 2012a. The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PLoS Pathog. 8 (2), e1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula L, Tramontano E, 2013. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit. Antivir. Res 100 (3), 615–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.