Abstract
RNA-binding proteins (RBPs) regulate translation and plasticity which are required for memory. RBP dysfunction has been linked to a range of neurological disorders where cognitive impairments are a key symptom. However, of the 2,000 RBPs in the human genome, many are uncharacterized with regards to neurological phenotypes. To address this, we used the model organism C. elegans to assess the role of 20 conserved RBPs in memory. We identified eight previously uncharacterized memory regulators, three of which are in the C. elegans Y-Box (CEY) RBP family. Of these, we determined that cey-1 is the closest ortholog to the mammalian Y-Box (YBX) RBPs. We found that CEY-1 is both necessary in the nervous system for memory ability and sufficient to promote memory. Leveraging human datasets, we found both copy number variation losses and single nucleotide variants in YBX1 and YBX3 in individuals with neurological symptoms. We identified one predicted deleterious YBX3 variant of unknown significance, p.Asn127Tyr, in two individuals with neurological symptoms. Introducing this variant into endogenous cey-1 locus caused memory deficits in the worm. We further generated two humanized worm lines expressing human YBX3 or YBX1 at the cey-1 locus to test evolutionary conservation of YBXs in memory and the potential functional significance of the p.Asn127Tyr variant. Both YBX1/3 can functionally replace cey-1, and introduction of p.Asn127Tyr into the humanized YBX3 locus caused memory deficits. Our study highlights the worm as a model to reveal memory regulators and identifies YBX dysfunction as a potential new source of rare neurological disease.
Author summary
Memory relies on RNA translation, often controlled by RNA-binding proteins (RBPs). RBP dysfunction has been linked to memory issues in many neurological diseases. Here, we take advantage of the model organism Caenorhabditis elegans to determine the role of 20 RBPs in memory. We identify eight new memory regulators, including members of the CEY RBP family, which are closely related to the human YBX RBPs. We found that CEY-1 is required for memory in C. elegans and enhances memory when overexpressed. Taking advantage of human genome sequencing data, we found three individuals with the same genetic variant in YBX3, two of which have neurological symptoms including intellectual disability. Introducing this same variant into the CEY-1 protein caused memory deficits, suggesting the variant may cause dysfunction in vivo. To confirm this in the context of the mammalian genes, we made C. elegans that express the YBX3 variant protein instead of CEY-1 to model the functional consequences of the human YBX3 variant. Again, we observed memory deficits. Our study shows the utility of C. elegans in discovering new memory-regulating RBPs and highlights a potentially conserved role for the CEY/YBX proteins in memory. Moreover, our findings suggest that dysfunction in the YBX RBPs may contribute to human neurological disease, beckoning future human and mammalian studies.
Introduction
Careful regulation of RNA localization, translation, and stability is essential for the proper functioning of all tissues but is especially important in the nervous system. Neurons are composed of subcellular compartments, specifically somatic, axonal, and dendritic regions, each of which possesses a specialized and functionally distinct proteome. Moreover, these subcellular compartments must respond dynamically, and often uniquely, to external stimuli. A prominent example of this is local mRNA translation in response to plasticity-inducing stimuli, which is necessary for learning and memory [1,2]. In this instance, both the pre- and post-synaptic compartment can undergo local protein synthesis in response to the same plasticity-inducing stimulus [3,4], yet mRNAs translated in response to that stimulus are compartment-specific, likely due to distinct mRNA regulators.
RNA-binding proteins (RBPs) are involved in each step of RNA regulation in neurons, controlling target RNA splicing, polyadenylation, localization, translation, and stability [5]. As such, their proper function is essential for neuronal plasticity and its associated behaviors–especially learning and memory. The importance of understanding the role of RBPs in the nervous system is underscored by a growing body of evidence that their dysfunction results in neurological disease, including disorders ranging from neurodevelopmental disorders to neurodegenerative disease [6–8]. RBP-associated disorders often include symptoms of behavioral abnormalities and cognitive impairments, including Fragile X Messenger Ribonucleoprotein (FMR1) in Fragile X syndrome involving cognitive impairment, PUMILIO1 (PUM1) associated developmental disability, ataxia, and seizures (PADDAS), Quaking (QKI) in schizophrenia, and both Heterogeneous Ribonucleoprotein K (HNRNPK) in Au-Kline syndrome and SON in Zhu-Tokita-Takenouchi-Kim (ZTTK) syndrome both involving intellectual disability [9–13]. In fact, RBPs outweigh all other protein types in their prevalence of predicted deleterious variation in Mendelian disorders, especially those that are diseases of the nervous system [7]. However, despite their importance, many RBPs are unexamined in the nervous system. Barriers remain to systematic characterization of RBPs, as such studies remain relatively slow in mammals and the human genome encodes nearly 2000 RBPs, which would require extensive time and effort to comprehensively study [14,15]. Moreover, studying the tissue-specific function of RBPs is even more difficult as it requires challenging and costly genetic approaches. However, these issues can be circumvented in simpler model organisms like the nematode worm Caenorhabditis elegans that allow for low-cost, rapid screening of molecules in a tissue-specific manner.
C. elegans have a rich history of use in high-throughput approaches and led to the discovery of many new regulators of neurological phenotypes, including molecularly conserved behaviors [16–18]. Indeed, the use of the worm resulted in the discovery of many conserved regulators of disease, including those involving cognitive deficits [19–24]. The C. elegans genome encodes almost 900 RBPs, over 80% of which have mammalian orthologs [25], and can be studied using thousands of publicly available mutants, including those that allow for neuron-specific RNAi for rapid tissue-specific genetic screening [26,27]. Importantly, C. elegans offers the advantage of using these genetic tools to investigate the role of RBPs in memory and cognition, as they form molecularly conserved associative memories, including short-term (STM), intermediate-term (ITM), and long-term memories (LTM) [20,28–32]. Given its wealth of molecular tools, ease of genetic screening, and molecularly conserved memory, C. elegans is an excellent model system to define the functions of neuronal RBPs in learning and memory in vivo.
Here, we have used the worm to perform a neuron-specific, targeted screen to discover conserved RBPs that regulate associative memory. Altogether, we find that 80% of 20 screened RBPs are memory regulators, eight of which are novel memory molecules. Importantly, our approach reveals a family of RBPs that are practically uncharacterized in their nervous system function and are essential for associative memory ability in the worm: the C. elegans Y-Box (CEY) RNA binding protein family.
Of this family, we determine that CEY-1 is most closely related to mammalian YBXs and requires further study to understand the potentially conserved role of YBXs in the nervous system. We found that CEY-1 acts specifically in the nervous system to promote memory. We discovered that large copy number losses including the genomic loci of YBX genes, as well as other known neurodevelopment disorder associated genes, may be associated with neurological symptoms such as intellectual disability. Furthermore, we similarly describe one rare heterozygous variant in the human YBX3 gene identified in two unrelated individuals with neurological symptoms. Importantly, introduction of the conserved YBX3 variant into either endogenous cey-1 locus or animals where the cey-1 locus has been humanized results in memory deficits in the worm, supporting the idea that the YBX3 variant we identified may contribute to human disease.
Altogether, we uncovered new associative memory regulators in a targeted screen of 20 conserved neuronal RBPs. We describe in detail a novel associative memory regulator, CEY-1/YBX, and highlight C. elegans as a discovery organism for associative memory regulators and potential disease-causing RBPs.
Results
Targeted screen identifies novel RBPs that regulate learning and memory
The C. elegans genome encodes a total of 887 predicted RBPs. Of those, 60% are conserved with mammals [33,34]; thus, the worm is an excellent system to rapidly screen RBPs for conserved roles in memory. We prioritized candidate RBPs based on the following criteria to increase the likelihood of identifying memory regulators: 1) nervous system expression based upon transcriptomic studies, 2) evolutionary conservation with mammals, and 3) detection in synaptic regions across species, as RBPs are known to regulate plasticity at the synapse. By cross-referencing adult-only, neuron-specific transcriptomic datasets [24,35] as well as orthology tools including Wormcat and OrthoList2 [36,37], we identified 550 neuronally expressed RBPs that are orthologs of known mammalian RBPs. After filtering this list based upon detection in C. elegans and mammalian synapses [35,38–42], 313 RBPs met screening criteria. Of those, we proceeded with the top 20 most synaptically-enriched RBPs based on our previous work identifying the adult C. elegans synaptic transcriptome [38,43] for behavioral study.
We assessed the roles of these 20 RBPs in memory by performing adult-only RNAi in neuronally RNAi-sensitized animals (punc119::sid-1) [27] and measuring the effects of knockdown on well-established assays of positive olfactory associative memory [29,30]. In these assays, animals are trained to form a positive association with the neutral odorant butanone and food, and by varying the number of food-butanone pairings can elicit short- and intermediate-term (STM and ITM) and long-term memories (LTM). The molecular requirements for these memories, including glutamate receptors (STM, ITM, LTM), calcium-dependent signaling cascades (STM, ITM, LTM), protein synthesis (ITM, forgetting, and LTM), and CREB-dependent transcription (LTM), are shared with mammals [20,28–30] and other C. elegans associative behavioral assays such as aversive olfactory learning and memory, memory for touch habituation, gustatory memory, imprinted olfactory memory, and aversive pathogen memories, to name a few [22,32,44–50].
In our screen, we assayed the following behaviors after positive olfactory training: learning (0 min after training), protein synthesis-independent STM (30 min after training), and translation-dependent ITM (1 hr after training) (Fig 1A) [29,30]. Effects of RBP knockdown on transcription and translation-dependent LTM were assessed in RNAi-sensitized animals with an additional egl-30 gain of function mutation (egl-30(js126), punc119::sid-1)) that increases Gαq signaling and confers the ability to form a LTM that is molecularly identical to wild-type animals with only one round of training, making them amenable to high-throughput screening (Fig 1B) [51].
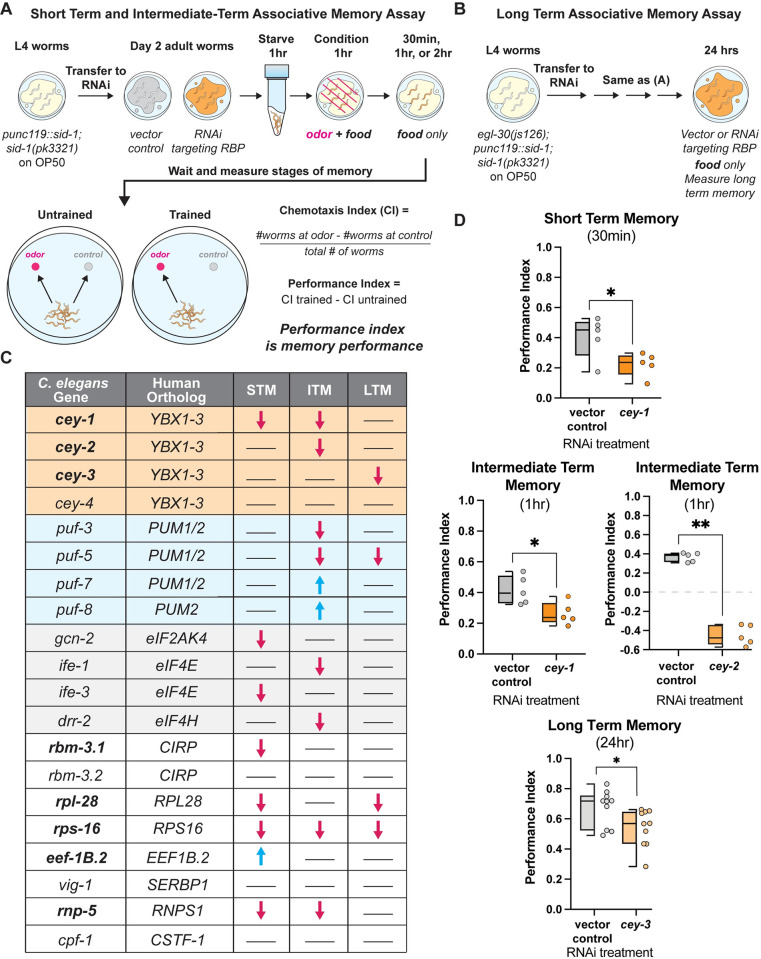
Fig 1. Targeted screen of 20 conserved neuronal RBPs reveals eight novel associative memory regulators.
(A) Short-term and intermediate-term associative memory (STM and ITM) assay workflow using adult-only RNAi knockdown. Worms that are neuronally sensitive to RNAi, LC108(usIS69[myo-2p::mCherry + unc119p::sid-1]), were starved for 1 hour, conditioned for 1 hour, and tested immediately for 1x learning (0 min) via chemotaxis assays, or transferred onto holding plates with OP50 for 0.5, 1, or 2 hours after conditioning, then tested for STM and ITM changes. Memory performance is calculated by comparing the trained and untrained chemotaxis indices. (B) The long-term associative memory (LTM) assay workflow is identical to that for STM/ITM except worms have an additional egl-30(js126) mutation that allows for LTM formation after just 1x training. Worms are tested immediately for 1x learning (0 min) via chemotaxis assays or transferred onto holding plates with RNAi for 16–24 hours after conditioning, then tested for LTM changes. (C) Results of our targeted screen of 20 RBPs reveals eight novel associative memory regulators. All RBPs screened are shown and include the C. elegans gene, its mammalian protein ortholog, and any STM, ITM, or LTM changes. Blue arrows indicate enhanced memory while pink arrows indicate decreased memory; lines indicate memory did not change. Novel memory regulators not reported in existing literature are bolded. For full screen results, see S4 Fig, and for significance quantification see S3 Fig. (D) Each member of the CEY RBP family plays a novel, unique role in memory. Adult-only RNAi knockdown of cey-1 in neuronally-sensitized worms causes STM and ITM deficits, cey-2 knockdown causes ITM deficits, and cey-3 knockdown causes LTM deficits. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. Mann-Whitney test comparing ranks. n = 10 per RNAi treatment. *p<0.05, **p<0.01.
Of the 20 RBPs tested, 80% regulate at least one type of memory (Fig 1C). Two classes of proteins known to be required for memory across species had multiple members represented in our screen candidates: the Pumilio and FBF RBP family (Pumilio1/2 orthologs puf-3, puf-5, puf-7, and puf-8) as well as four proteins involved in translation initiation (eIF4/2α orthologs gcn-2, ife-1, ife-3, and drr-2). Pumilios have been linked to both LTM and working memory in Drosophila melanogaster and mice [52,53], and we have previously described their importance in C. elegans memory [38,43]. Similarly, eukaryotic translation initiation machinery eIF4E and eIF2α are required for memory formation, STM, and LTM in mice [54,55]. However, to the best of our knowledge the requirement for this molecular machinery in olfactory associative memory was not previously demonstrated in the worm. Validating these two classes of known memory regulators strengthens the likelihood that novel molecules identified in our screen have a conserved function in higher organisms. Indeed, we found that 40% of memory regulating RBPs in our screen were not previously associated with memory phenotypes, and are therefore novel, bolded in Fig 1C. Importantly, there were no significant differences in naïve chemotaxis to a chemoattractive concentration (0.1%) of butanone (S2 Fig) for any RBP condition and additionally, no learning deficits were detected after knockdown of any RBP in our screen (S1 Fig). Together, these results suggest the deficits or enhancements we identified in our screen are specific to memory and are not due to broad behavioral or neurological disruption. For all behavior results plotted quantitatively, see S3 Fig, and for raw data for behavior timepoints where RNAi significantly altered memory, see S4 Fig.
Identification of CEY family as novel memory regulators
Strikingly, all members of the C. elegans Y-Box (CEY) RBP family (cey-1-4), orthologs of the mammalian Y-box binding (YBX) family, met our screening criteria despite being uncharacterized in learning and memory behavior. The CEY RBPs regulate fertility in the worm [56,57], and mammalian YBX proteins are translational regulators involved in cancer metastasis and cell cycle progression [58–61]. Here we found that three of the four CEY proteins screened regulate memory, with each CEY involved in its own molecularly distinct form of memory (Fig 1D); cey-1 is required for STM and ITM, cey-2 is required for ITM, cey-3 is required for LTM, and cey-4 knockdown had no detectable effect on behavior. To our knowledge, this is the first documented behavioral phenotype regulated by CEY RBPs.
As there are no reported neuronal phenotypes associated with the CEY RBPs, we further examined their expression pattern based on previous transcriptomic profiling datasets. Single-cell RNA-seq data generated from neurons isolated from L4 larval animals revealed broad expression of cey-1 and cey-4, while cey-2 is only detected in eight neurons (S5 Fig) [62,63]. cey-3 is not detected in any neurons at L4 but is detected in adult neuron-specific datasets [24,40], highlighting the importance of compiling multiple transcriptomic datasets to choose our screen candidates.
Similar to their C. elegans orthologs, the mammalian YBX proteins are relatively understudied with regards to behavioral phenotypes or their function in the adult nervous system, though YBX1 is reported to be involved in neural stem cell development [64]. Similar to cey-1, YBX1 is broadly expressed throughout the nervous systems of humans, adult macaques, and rats [65]. In these mammals, YBX1 is expressed in memory-regulating regions including the hippocampus, parahippocampal cortex, and dentate gyrus [65]. YBX3 is also broadly expressed, including in memory-regulating regions, while YBX2 is thought to be restricted to non-neuronal tissues [66,67]. Importantly, YBX1 is reported to bind to plasticity-associated mRNAs, including GluR2 mRNA and CaM1 mRNA, in an activity-dependent manner, making the YBXs promising candidates as novel essential memory molecules in mammals [68]. Moreover, both YBX1 and YBX3 proteins are detected in mouse hippocampal post-synaptic densities, suggesting that they have the potential to regulate local translation in a brain region critical for memory [69]. In non-neuronal tissues, CEYs and YBX proteins appear functionally conserved; both worm and mammalian proteins regulate polysome formation [56,70,71], suggesting that we can study conserved functions of this class of RBP in the worm.
To strengthen the translatability of our findings by focusing on conserved functions, we decided to study the C. elegans ortholog(s) that are the most closely related to the YBX proteins [60,61,64,72,73]. To this end, we performed phylogeny and homology analysis using MegaX and DIOPT [74,75] and found that cey-1 is the primary C. elegans YBX ortholog, closer in similarity to the YBXs than other CEY family members (Fig 2A and 2B). For example, the linker regions (NC9 and CC13) and Cold-Shock Domain (CSD) of CEY-1 are extremely similar to those of the YBX proteins (approximately 97% similar) (Fig 2C) [76,77]. At both the N- and C-termini, the CEY-1/YBX proteins are moderately conserved (approximately 60%). These are within low complexity regions, otherwise known as intrinsically disordered regions, responsible for protein-protein binding and stress granule formation [71,78].
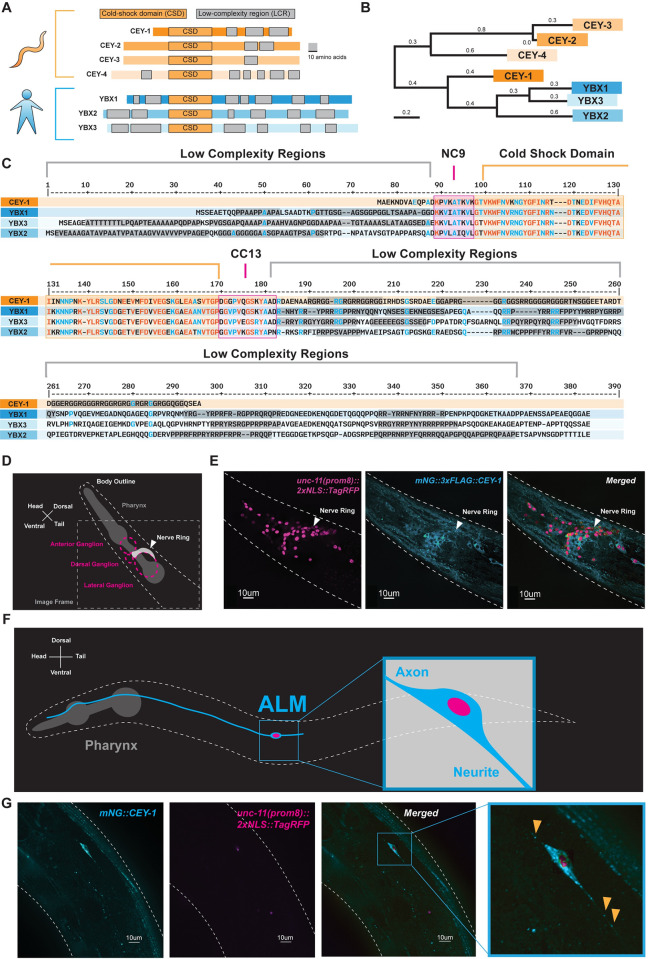
Fig 2. CEY-1 is the closest ortholog to the human Y-box binding proteins.
(A) Diagram of the conserved protein domains of the C. elegans CEY RBPs and human YBX RBPs. C. elegans have four CEY RNA-binding proteins, CEY-1, CEY-2, CEY-3, and CEY-4, that are orthologs of the three mammalian Y-box binding proteins, YBX1, YBX2, and YBX3. All CEY/YBX RNA-binding proteins have a conserved cold-shock domain (CSD) that mediates RNA binding as well as one or more low complexity regions (LCRs) thought to be important for protein-protein interactions. Grey scale bar is 10 amino acids. (B) Phylogenetic tree generated using MegaX reveals that CEY-1 is more similar to the human YBX proteins than the other CEY RNA-binding proteins. (C) Protein sequence alignment of the CEY and human YBX binding proteins made with MultiAlin showing organization of protein domains. Highly conserved residues are depicted in red and blue. The cold-shock domains are highlighted in orange, the NC9 and CC13 linkers are highlighted in pink, and the low complexity regions are highlighted in grey. Domains were annotated according to SMART. (D) Diagram of microscopy images shown in (E). C. elegans head is labeled including the location of the pharynx and main neuronal ganglia/nerve rings. (E) CEY-1 is primarily localized to the cytoplasm at baseline conditions. Representative image of Day 2 adult worms with RFP-labeled neuronal nuclei (unc-11(prom8)::2xNLS::TagRFP) pseudocolored magenta and an endogenously mNeonGreen-tagged CEY-1 (mNG::CEY-1) pseudocolored cyan. (F) Diagram of microscopy images shown in (G). ALM sensory neuron has a stereotyped, easily identifiable anterior axonal projection and posterior neurite. (G) CEY-1 is localized to neurite and axon of the ALM sensory neuron at baseline conditions. Representative image of Day 2 adult worms with RFP-labeled neuronal nuclei (unc-11(prom8)::2xNLS::TagRFP) pseudocolored magenta and an endogenously mNeonGreen-tagged CEY-1 (mNG::CEY-1) pseudocolored cyan. Orange arrows indicate CEY-1 puncta.
Based on gene expression data, the expression pattern of cey-1 and the mammalian YBX proteins is similarly broad across the nervous system [62,65–67,79–81]. Though previous studies using promoter-fusion approaches corroborate the transcriptomic evidence that cey-1 is broadly expressed in the nervous system [56], we sought to verify that the protein is indeed expressed in neurons. We crossed the previously published cey-1::GFP promoter-reporter system [56] with a marker of neuronal nuclei (unc-11(prom8)::2xNLS::TagRFP) and validated their reported neuronal expression (S6 Fig). We confirmed these findings by verifying the neuronal localization of endogenous CEY-1 by crossing animals expressing an endogenously mNeonGreen-tagged CEY-1 (mNG::CEY-1) with unc-11(prom8)::2xNLS::TagRFP expressing animals (Fig 2D and 2E). We also examined if CEY-1 could potentially be localized to synaptic regions. As the protein is expressed in multiple neurons and tissues (Fig 2D and 2E) which interferes with visualization of specific synapses in the nerve ring, we examined if we could detect CEY-1 in the projections of the ALM neuron. The ALM neuron is isolated in the midbody and has an easily detectable and stereotyped anterior axonal projection and posterior neurite (Fig 2F and 2G). Though the posterior neurite of the ALM does not form any known synapses and may not behave in the same manner as a true dendrite [82,83], imaging the ALM would allow us to determine if CEY-1 was present in any neuronal projection. Endogenous CEY-1 was observed in both the neurite and axon of the ALM (Fig 2G), suggesting that this RBP could have the potential to localize to synaptic regions.
CEY-1/YBX acts in neurons to regulate memory
To determine if cey-1 acts in the nervous system to regulate learning and memory, we next tested if nervous-system specific cey-1 knockdown causes memory deficits by performing adult-only, neuron-specific knockdown of cey-1 using worms only expressing sid-1 in the nervous system (punc119::sid-1; sid-1(pk3321)). Contrary to the mutants used in our initial screen, which can have off-target effects in other tissues, these worms lack sid-1 in all tissues other than the nervous system, leading to RNAi specifically in the nervous system [27]. We confirmed that RNAi treatment in these animals resulted in reduced expression of CEY-1, by generating neuronally RNAi-sensitized mNG::CEY-1 animals (S7 Fig). Our behavioral findings using this strain replicated the initial results of our screen: cey-1 knockdown decreases both STM and ITM but not learning (Fig 3A). Importantly, we detected no motility or butanone chemotaxis deficits, although there are mild sensory deficits for isoamyl alcohol and nonanol (S7 Fig).
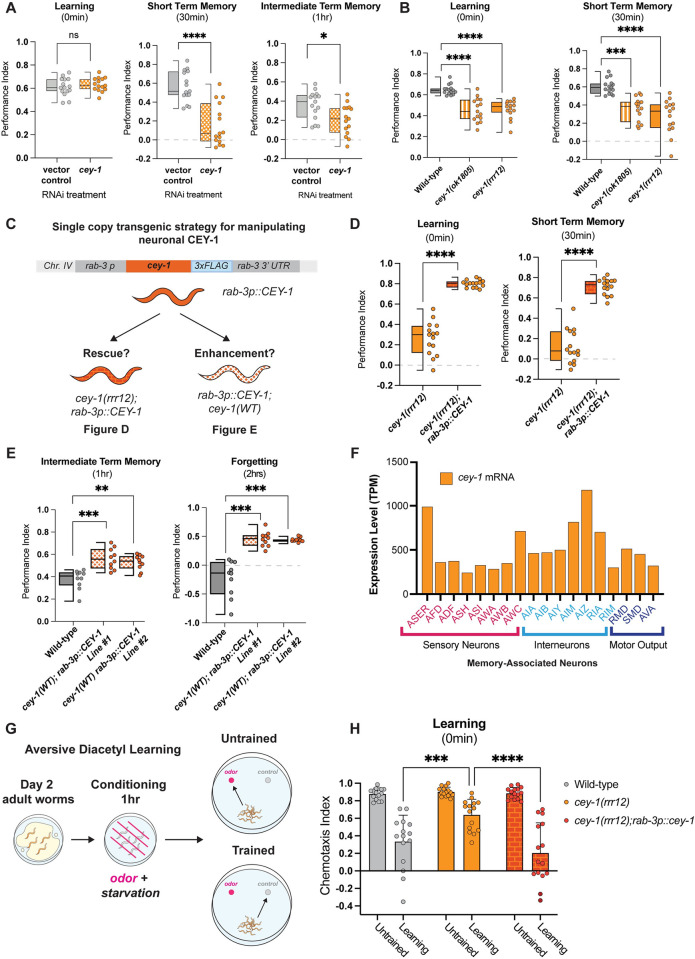
Fig 3. CEY-1 promotes learning and memory in the C. elegans nervous system.
(A) Adult-only, neuron-specific knockdown of cey-1 in punc119::sid-1; sid-1(pk3321) animals causes STM and ITM deficits. Box and whisker plot as in Fig 1. n = 15 per RNAi treatment. *p<0.05, ****p<0.0001. ns, not significant (p>0.05). (B) Loss-of-function mutants cey-1(rrr12) and cey-1(ok1805) have severe learning and memory deficits. Box and whisker plot as in Fig 1. n = 15 per genotype. ***p<0.001, ****p<0.0001. (C) Diagram of strategy to determine whether neuronal cey-1 is sufficient to rescue and enhance memory. Worms that express a single copy of cey-1 only in the nervous system (rab-3p::cey-1::rab-3 3’UTR; cey-1(rrr12)) were tested for phenotypic rescue in (D) while worms that have a single extra copy of cey-1 only in the nervous system (rab-3p::cey-1::rab-3 3’UTR; cey-1(+)) were tested in (E). (D) Single copy expression of cey-1 in the nervous system (rab-3p::cey-1::rab-3 3’UTR; cey-1(rrr12)) rescues loss-of-function learning and memory loss. Box and whisker plot as in Fig 1. n = 15 per genotype. ****p<0.0001. (E) Single copy overexpression of cey-1 in the nervous system is sufficient to enhance memory (rab-3p::cey-1::rab-3 3’UTR; cey-1(+)). Box and whisker plot as in Fig 1. n = 15 per genotype. **p<0.01,***p<0.001. (F) Single-cell RNA-seq data from L4 hermaphrodites show that cey-1 is expressed in every previously described memory-associated neuron, including those involved in aversive olfactory learning. (G) Workflow for aversive olfactory learning assays. Adult worms are starved for 1 hour in the presence of 100% diacetyl and tested immediately for 1x learning (0 min) via chemotaxis assays to 1% diacetyl. Learning performance is calculated by comparing the trained and untrained chemotaxis indices. (H) CEY-1 is required for learning in aversive paradigms, and behavioral defects in cey-1(rrr12) loss-of-function mutants are restored by nervous system-specific CEY-1 rescue. Box and whisker plot as in Fig 1. n = 15. ***p<0.001,****p<0.0001.
However, given recent evidence that the punc119::sid-1; sid-1(pk3321) worms we used for may have leaky cey-1 knockdown upon RNAi treatment in non-neuronal tissues such as the intestine [84], we pivoted to other genetic approaches to study cey-1 in the nervous system. We examined the behavior of animals bearing two different loss-of-function cey-1 alleles. Both animals exhibited severe deficits in learning and, as a result, inability to form a memory (Fig 3B). We ensured that both mutant alleles exhibited normal naïve chemotaxis to 10% butanone (S8 Fig), and further confirmed that the cey-1(rrr12) animals have no thrashing, motility, or butanone sensing deficits driving these results, as we used them for subsequent experiments (S8 Fig). Together, these results suggest the deficits we observed are learning and memory-specific and that cey-1 is necessary for associative behaviors. We believe that by knocking down cey-1 during adulthood (Fig 3A) we are circumventing loss of cey-1 during neuronal development, which may be causing the more severe effects on learning seen in cey-1 mutants (Fig 3B).
Elevated neuronal CEY-1 is sufficient to enhance memory
We next asked if restoring cey-1 function in nervous system was sufficient to rescue learning and memory deficits observed upon loss of cey-1. We generated cey-1(rrr12) loss of function animals that also express a single-copy knock-in transgene driving expression of cey-1 specifically in the nervous system (rab-3p::cey-1::rab-3 3’UTR; cey-1(rrr12)) (Fig 3C). We found that neuron-specific cey-1 rescue is sufficient to restore the learning and memory deficits seen in cey-1(rrr12) mutants back to wild-type levels (Figs 3D and S8). Importantly, neither the cey-1 loss of function mutants nor the neuronal cey-1 rescue worms have deficits in butanone sensing, thrashing, or motility, suggesting that phenotypes recorded are memory-specific (S8 Fig).
Thus far, our data suggested that cey-1 acts in the nervous system to promote memory. To test this hypothesis, we determined whether increasing cey-1 expression in the nervous system would have beneficial effects on behavior. A single round of food-butanone training results in memory that persists for no longer than two hours in wild-type animals, and we asked if additional neuronal cey-1 increased the duration of this memory, as has been observed with other memory promoting genetic manipulations [28,29,43]. Worms with (rab-3p::cey-1::rab-3 3’UTR; cey-1(+)) showed enhanced memory (Fig 3E), as their memory persists longer than the average two hours, consistent with previous memory enhancers [28,29,43]. Collectively, our results show that cey-1 is sufficient to increase memory when expressed only in the nervous system, suggesting that cey-1 is a memory enhancer.
CEY-1 is required for multiple associative behaviors
Despite its broad expression in the nervous system, we wanted to determine if cey-1 was expressed in neurons involved in multiple types of learning and memory, and thus could be required for other behaviors. Because the C. elegans nervous system is invariant [82,85], the identities of specific neurons involved in a variety of associative behaviors are known. Based upon previously generated L4 single-cell RNA-seq data, we find that cey-1 is indeed detected in all canonical learning and memory-associated neurons, including those associated with gustatory plasticity, thermotaxis, and olfactory memory (Fig 3F) [51,86–92]. Therefore, we decided to test whether cey-1(rrr12) loss-of-function mutants also exhibit impaired learning in an aversive olfactory associative memory paradigm. In this assay, worms are starved in the presence of 100% diacetyl that is normally chemoattractive, forming a negative association (Fig 3G). Learning after aversive diacetyl conditioning requires neurons [32] distinct from those known to be involved in our appetitive memory paradigms [20,28,51]. We found that CEY-1 is also required for learning in this aversive paradigm, and the learning defects in cey-1 loss-of-function mutants are restored by nervous system-specific CEY-1 rescue (Fig 3H). These results suggest that CEY-1 may be a broad regulator of the molecular mechanisms required for learning and memory across multiple neuronal circuits and paradigms.
Copy number variant deletions in YBX1 and YBX3 may be associated with neurological symptoms, including intellectual disability
Human genomic datasets are increasingly linking RBP dysfunction to neurodevelopmental and neurological disorders, including those characterized by intellectual disability and cognitive dysfunction [6–8,93]. Therefore, we used copy number variation (CNV) datasets to determine if loss of the YBXs may potentially be associated with neurological dysfunction in humans. We used an open-access database, DECIPHER, that reports CNVs from 50,000 individuals [94]. We focused specifically on CNVs in YBX1 and YBX3, as YBX2 is not expressed in the adult human brain [65,67].
We found that many individuals with CNV deletions that include either YBX1 or YBX3, as well as other genes have neurological symptoms (Fig 4A). The CNV deletions were variable in size, ranging from 1.59 Mb to 24.59 Mb. The most common symptom reported in individuals with CNV deletions including the YBXs is intellectual disability, as 37.5% of individuals with CNV deletions containing YBX1 and 80% of individuals with CNV deletions in YBX3 have intellectual disability (Fig 4B). Other common symptoms for individuals with YBX1-containing CNV deletions include epicanthus, microcephaly, and seizures, while individuals with YBX3-containing CNV deletions often exhibit strabismus and scoliosis. However, as is the case with the majority of CNVs, all YBX-containing CNV deletions included other genes that may be contributing to these individuals’ phenotypes. In the case of YBX1, the minimal overlap between CNV losses affecting the YBX1 loci is 1.08 Mb containing 34 genes, including two loci that are predicted to be loss of function intolerant: Forkhead Box J3 (FOXJ3) [95] and Solute Carrier Family 2 Member 1 (SLC2A1) [96]. SLC2A1 is associated with autosomal dominant Dystonia 9 (MIM#601042) [96,97], GLUT1 deficiency syndrome 1 (MIM#606777) [98,99], GLUT1 deficiency syndrome 2 (MIM#612126) [100,101], Stomatin-deficient cryohydrocytosis with neurologic defects (MIM#608885) [102,103], and susceptibility to idiopathic generalized epilepsy 12 (MIM#614847) [104,105]. In the case of YBX3, the minimal overlap between CNV deletions affecting the YBX3 loci is 3.24 Mb containing 68 genes, including three loci that are predicted to be loss of function intolerant: ETS Variant Transcription Factor 6 (ETV6) [97], Low Density Lipoprotein Receptor-related Protein 6 (LRP6) [98], and Dual-Specificity Phosphatase 16 (DUSP16) [99]. However, unlike the loci surrounding YBX1, none of the loss of function intolerant loci around YBX3 are known to cause monogenic neurological disease. Summary statistics for CNV DECIPHER data for YBX1 can be found in S9 Fig and for YBX3 in S10 Fig. All individuals with CNV deletions in YBX1 or YBX3 as well as their DECIPHER phenotypic data can be found in S1 Table.
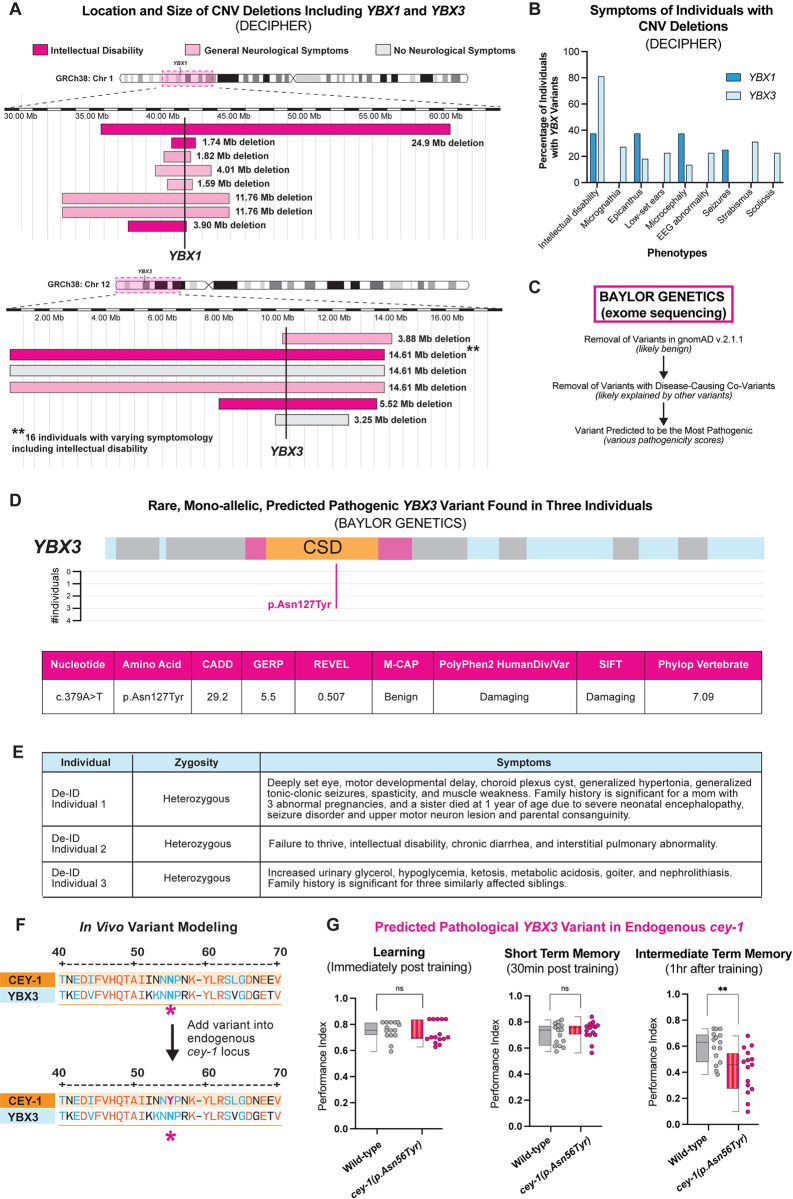
Fig 4. Copy number variants in YBX1 and YBX3 found in individuals with neurological symptoms and in vivo modeling of a predicted pathogenic single nucleotide variant in YBX3.
(A) Location, size, and neurological symptoms associated with copy number variant (CNV) deletions reported in DECIPHER. The exact location of YBX1 and YBX3 are shown with a bold vertical line. The area around the gene affected by the CNV deletions is highlighted with a pink box on their respective chromosome ideograms at the top of the figure. CNVs where the individual had neurological symptoms are in light pink, and CNVs where the individual had associated intellectual disability are in dark pink. (B) Intellectual disability is the most often associated symptom of patient CNV deletions containing the YBX1 or YBX3 loci based on DECIPHER. (C) Workflow to examine single nucleotide variants (SNVs) that may be disease-linked using the postnatal exome sequencing database from Baylor Genetics. Several strategies were used to remove likely non-pathogenic variants, including removing cases with known co-variants and removal of variants found in the general population (gnomAD v2.1.1). (D) Location and pathogenicity score of a rare heterozygous SNV in YBX3 found in three de-identified individuals, two with indicated neurological symptoms. The variant we identified, p.Asn127Tyr, had the highest predicted pathogenicity using seven different metrics (see text). (E) Table of p.Asn127Tyr cases from Baylor Genetics de-identified exome sequencing dataset. Two of the three individuals have neurological symptomology reported, such as intellectual disability and seizures. (F) Location of p.Asn127Tyr variant shows conserved residues between CEY-1 and YBX3 and location of variant when introduced into the endogenous CEY-1 protein. (G) Introduction of p.Asn127Tyr into the endogenous cey-1 locus (p.Asn56Tyr) causes ITM deficits. Box and whisker plot as in Fig 1. n = 15 per genotype. **p<0.01, ns, not significant (p>0.05).
Taken together, these data suggest that loss of function CNVs including the YBX1 and YBX3 loci may be associated with neurological dysfunction; however, the confounding variable of multi-gene deletions prevents any direct association. As a result, we decided to model the rare YBX3 c.379A>T (p.Asn127Tyr) variant for biological function in vivo.
Introducing a predicted deleterious variant in YBX3 into the endogenous cey-1 locus causes memory deficits in vivo
The minimal overlap regions for CNV deletions containing YBX1 and YBX3 include loci beyond the gene of interest that are associated with monogenic neurological disorders and other diseases in humans. Therefore, single-nucleotide variants (SNVs) in the genes of interest may better link deleterious variants in single genes directly to neurological disorders [10,12,100–102] compared to CNVs with multi-gene deletions.
We received a deidentified dataset of YBX variants found through exome and genome sequencing completed at Baylor Genetics [103] to identify potentially deleterious SNVs in the YBXs. To determine variants of interest, we curated rare, heterozygous variants in postnatal individuals, as these are the most likely to be deleterious [104,105]. We first removed SNVs that were most likely not contributing to early developmental brain disorders by eliminating variants found in gnomAD v2.1.1 as well as cases where the subject’s symptomology was explained by other variants in the genome (Fig 4C). We then used seven different pathogenicity metrics to determine which variants were the most likely to be deleterious, including CADD [106,107], GERP [108], REVEL [109], M-CAP [110], PolyPhen 2 [111], SIFT [112], and Phylop Vertebrate [113]. The variant with the highest overall pathogenicity scores was in YBX3: c.379A>T resulting in the amino acid change p.Asn127Tyr. The affected amino acid is in the Cold-Shock Domain of the YBX3 protein and, significantly, this variant is not reported in gnomAD v.2.1.1 nor gnomAD v4, suggesting it is rare (Fig 4D). We found this variant in three different individuals, two of which present with neurological symptoms such as intellectual disability and seizures (Fig 4E).
The p.Asn127 residue is highly conserved across species, including between C. elegans CEY-1 and human YBX3 (Fig 4F). Therefore, we decided to test the potential significance of this variant in vivo in C. elegans by introducing it into the endogenous cey-1 locus and examining its effect on memory. We used CRISPR to introduce the same mutation into the endogenous cey-1 gene at the same conserved residue as in YBX3: p.Asn56Tyr (cey-1 p.Asn56Tyr). We then tested for memory deficits at learning, STM, and ITM. Remarkably, introduction of the p.Asn56Tyr variant into cey-1 caused ITM deficits, suggesting the variant may have consequences on human neurological function (Fig 4G). Importantly, CEY-1 p.Asn56Tyr worms had no butanone sensing deficits or motility impairments (S11 Fig), suggesting that in this case, neurological dysfunction is memory-specific. We performed qRT-PCR and verified that while the cey-1(rrr12) allele is a true knockout that undergoes nonsense mediated decay [56], our p.Asn56Tyr variant has no significant effect on levels of cey-1 mRNA, suggesting this SNV affects protein function rather than gene expression levels (S11 Fig). By using the worm to model one predicted damaging variant in YBX3, our findings suggest that YBX3 may be of interest for human disease associations. Therefore, we decided to take our rare variant modeling a step further by using humanized worm strains expressing YBX1/3.
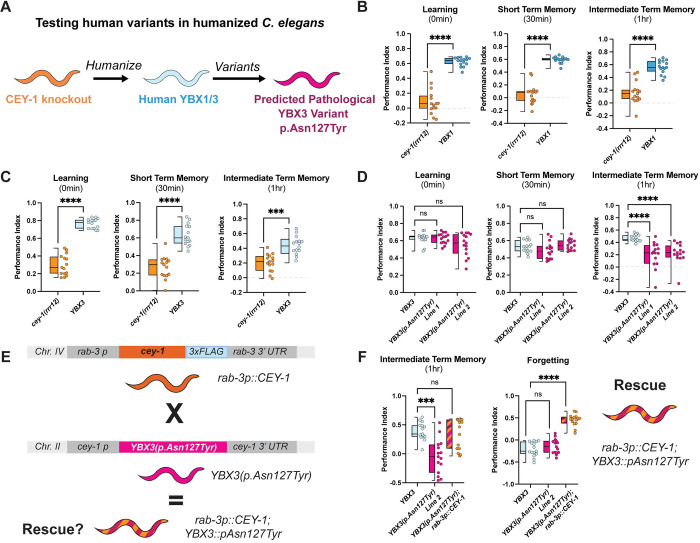
YBX1 and YBX3 functionally replace cey-1 in a humanized model and introduction of a predicated deleterious variant into YBX3 causes memory deficits in vivo
Recent studies have highlighted the utility of humanized worm lines in phenotypic modeling of rare human gene variants in vivo [114–117]. We decided to use this CRISPR-based approach to determine whether the predicted damaging variant YBX3 p.Asn127Tyr, which we found in YBX3 in two human individuals with neurological symptoms and one individual without neurological symptoms, affects YBX3 function in the context of memory. We created two humanized worm lines expressing either human YBX1 or YBX3 at the endogenous cey-1 locus, thereby replacing cey-1 (Fig 5A). These human genes are expressed similarly to endogenous cey-1 at the mRNA level, as measured by qPCR (S12 Fig). In both cases, the human orthologs were able to functionally replace cey-1: learning, STM, and ITM were comparable to that of wild-type animals (Figs 5B, 5C and S12). Intriguingly, worms expressing YBX3 displayed behavior that was nearly indistinguishable from wild-type animals, exhibiting normal forgetting at two hours post-training, while worms expressing YBX1 still maintained a modest memory two hours post-training, suggesting potential differences in regulation during active forgetting (S12 Fig). Given YBX3 functionally replaces cey-1 in memory, we introduced the rare predicted deleterious variant p.Asn127Tyr into the humanized YBX3 locus to determine if the variant would affect memory. Like when we mutated the endogenous cey-1 gene (S11 Fig), this variant had no significant effect on YBX3 mRNA levels (S12 Fig). When we examined their behavior, worms expressing YBX3(p.Asn127Tyr) have severe intermediate term memory deficits (Fig 5D), mirroring our findings with the equivalent variant inserted into the endogenous cey-1 locus (Fig 4G). None of the humanized strains had impaired butanone sensing or motility, suggesting these deficits are memory-specific (S12 Fig). Taken together, our findings suggest a high degree of functional conservation between cey-1 and YBX3.
Fig 5. Human YBX1 and YBX3 functionally replace cey-1 in memory in vivo and insertion of a rare SNV into YBX3 disrupts ITM.
(A) Diagram of workflow for humanizing C. elegans to express either human YBX1 or human YBX3, then generating YBX3 c.379A>T (p.Asn127Tyr). (B) Insertion of human YBX1 into the endogenous cey-1 locus rescues learning and memory deficits seen in cey-1 knockout animals (cey-1(rrr12)) back to wild-type levels (also see S12 Fig). (C) Insertion of human YBX3 into the endogenous cey-1 locus rescues learning and memory deficits seen in cey-1 knockout animals (cey-1(rrr12)) back to wild-type levels (also see S12 Fig). (D) Introduction of c.379A>T(p.Asn127Tyr) into the humanized YBX3 locus causes ITM deficits. Box and whisker plot as in Fig 1. n = 15 per genotype. ****p<0.0001, ns, not significant (p>0.05). (E) Diagram of strategy to determine whether neuronal cey-1 is sufficient to rescue and enhance memory in worms expressing YBX3(p.Asn127Tyr). Worms that express a single copy of cey-1 only in the nervous system (rab-3p::cey-1::rab-3 3’UTR; cey-1(rrr12)) were crossed to those expressing patient variant YBX3(p.Asn127Tyr) and subsequently tested for phenotypic rescue in (F). (F) Neuronal expression of CEY-1 rescues intermediate term memory deficits observed in YBX3(p.Asn127Tyr) animals. n = 15, ***p<0.001, ****p<0.0001, ns, not significant (p>0.05).
We next attempted to determine the effect of the p.Asn127Tyr mutation on YBX3 function. Based upon Alphafold2 [118] predictions, the Asn127 residue is part of a long, disordered loop that connects two β-sheets of the YBX3 cold shock domain (S13 Fig). Since this residue is not buried within the organized part of the protein, it is not obvious how this mutation could affect protein structure. We turned to two prediction models, AlphaMissense [119] and Functional Characterization via Evolutionary Scale Models (FunC-ESMs, [120]), that predict the effect of mutations on protein function. Both models predicted that p.Asn127Tyr would be detrimental to protein function, with an AlphaMissense score of 0.998 on a scale of 0.0–1.0, which is ‘likely pathogenic’, and FunC-ESMs predicting a ‘Total Loss’. Together, these prediction tools suggest that this residue is important for function.
Because the prediction models suggest that YBX3(p.Asn127Tyr) may produce a protein with reduced function, we tested whether or not we could rescue the behavioral phenotypes of worms expressing YBX3(p.Asn127Tyr) with a wild-type copy of CEY-1. We generated YBX3(p.Asn127Tyr);rab-3p::cey-1::rab-3 3’UTR animals (Fig 5E) and found that introduction of a functional CEY-1 in the nervous system rescued the ITM deficits of YBX3(p.Asn127Tyr) animals (Fig 5F). Moreover, the YBX3(p.Asn127Tyr) variant did not appear to suppress the beneficial effects of an extra copy of wild-type CEY-1 on forgetting behaviors (Fig 5F). Together, these results suggest that YBX3(p.Asn127Tyr) is not dominant negative. As YBX3 has a probability of loss-of-function intolerance (pLI) score of 0, this means the gene is tolerant of the functional loss of one copy of the gene. Therefore, our results suggest that YBX3(pAsn127Tyr) may confer a neomorphic effect.
Discussion
C. elegans as a discovery platform for novel associative memory regulators
We performed a targeted screen of 20 synaptically-enriched RBPs examining their roles in learning and memory and identified eight novel associative memory regulators, each regulating memory differently. While we focused on the CEY RBP family in this work, five other RBPs remain to be investigated. We found that rbm-3.1 is required for STM, rnp-5 is required for STM and ITM, rpl-28 is required for STM and LTM, rps-16 is required for STM, ITM, and LTM, and eef-1B.2 suppresses STM. It is worth noting that the LTM-regulating RBPs were discovered using egl-30(gf) mutants with enhanced LTM that are extremely amenable to screening [51].Though previous work suggests that this memory requires the same molecules and neurons as LTM in wild-type animals [51], these results should be verified in wild-type animals prior to further study. rbm-3.1, ortholog of CIRP, is reported to mediate alcohol-induced spatial memory deficits in mice [121] but has never been associated with memory independently of alcohol use prior to this study. In addition, eef-1B.2 and rnp-5 are associated with human neurological disease. Pathogenic variants in EEF1B.2 cause autosomal recessive intellectual disability [122], while rnp-5 ortholog RNPS1 is a risk factor for both intellectual disability and autism [123,124]. To our knowledge, neither rps-16 nor rpl-28 have been studied in the nervous system of any organism.
Here, we demonstrated novel roles for eight RBPs in memory using C. elegans. Our results suggest that these RBPs play a conserved role in memory that remains understudied, and future studies should combine worm behavior with human genetic data to examine these and other uncharacterized genes. Taken together, our study highlights the utility of C. elegans as a high-throughput screening tool for novel associative memory regulators, including those that regulate different, molecularly distinct forms of memory.
Three of the four CEY RBPs are involved in memory
We identified three members of the CEY RBP family, orthologs of the mammalian YBX RBPs, as novel memory molecules. We found that each CEY RBP has its own distinct role in memory: cey-1 regulates STM/ITM, cey-2 regulates ITM, and cey-3 regulates LTM. Of note, cey-4 does not appear to be required for memory ability but is expressed throughout the adult nervous system [62,80,81], and therefore is likely involved in other neuronal functions. The unique role of each CEY in memory is not explained by their expression patterns, as cey-1 and cey-4 are both broadly expressed in the nervous system while cey-2 and cey-3 are found in very few neurons [62,80,81]. While evidence from the germline suggests the CEY RBPs are promiscuous in binding and are not affiliated with specific mRNA subsets [56], it is unclear whether there is more mRNA target specificity in the nervous system. Future studies examining how each CEY RBP regulates the translational landscape of neurons are warranted to determine the independent mechanisms by which the CEYs proteins regulate memory.
CEY-1 is a novel neuronal memory-promoting RBP
Our study of CEY-1, the closest C. elegans ortholog to the mammalian YBX proteins, revealed that it acts in the nervous system to promote memory, as neuron-specific rescue of CEY-1 ameliorates behavioral defects cause by the loss of cey-1 in multiple associative learning and memory paradigms, and an additional copy of CEY-1 only in the nervous system improves memory performance. Future investigations are needed to unveil the molecular mechanisms by which CEY-1 regulates memory, and whether or not it acts specifically at synapses, but based on known CEY/YBX biology and the fact that it may be generally required for learning and memory, we hypothesize that CEY-1 controls transcription and/or translation of mRNAs involved in memory.
YBX1 is reported to bind to plasticity-associated mRNAs in an activity-dependent manner in vitro [68], and both CEY-1/YBX proteins regulate polysome formation and translation [56,70,71]. Furthermore, in non-neuronal tissues, YBX1 is reported to interact with known regulators of plasticity-dependent translation, specifically mTOR, AKT and translation initiation machinery [60,70,125]; therefore, it is likely that CEY-1/YBX is involved in these processes as well. Unlike other RBPs linked to plasticity-dependent translation, such as FMRP [126], YBX proteins are relatively non-specific with regards to target mRNAs [71,127]. It is unclear if a subset of molecules required for memory are regulated by YBX, or if it is involved in controlling broad machinery involved in plasticity and memory such as general protein synthesis. Future work will be necessary to determine the exact mechanisms by which CEY-1 promotes memory.
YBX3 p.Asn127Tyr decreases intermediate term memory in a humanized C. elegans model
Given the lack of experimental evidence linking the mammalian YBX proteins to memory, we queried human variant datasets to explore potential associations between YBX dysfunction and neurological symptoms. We focused on YBX1 and YBX3 because they are expressed in the adult nervous system and found that several large CNVs involving either the YBX1 or YBX3 loci, as well as one rare missense YBX3 variant, are present in individuals with neurological symptoms including intellectual disability. However, the CNV analysis is limited by other disease-associated genes in the minimal overlap regions involving YBX1 and YBX3 and the SNV analysis is limited by the sample size and availability of de-identified clinical findings.
To assess the biological significance of rare, mono-allelic SNVs in either YBX1 or YBX3, we conducted in vivo modeling of the variant with the highest pathogenicity scores. The YBX3 p.Asn127Tyr variant was identified in three individuals, two of whom exhibit neurological symptoms and one individual without neurological symptoms. The YBX3 c.379A>T (p.Asn127Tyr) variant is located within the YBX3 CSD at a highly conserved site [76,77,127]. Introduction of the corresponding p.Asn56Tyr variant into the endogenous cey-1 gene caused ITM deficits. We then decided to perform the same variant study in the context of human YBX3. Both YBX1 and YBX3 are able to functionally replace cey-1, as substitutions using either gene at the cey-1 locus does not impair learning and memory. Introducing the YBX3 c.379A>T (p.Asn127Tyr) variant into the humanized C. elegans line caused similar ITM deficits as the introduction of the variant into endogenous cey-1. Overall, these results suggest three key findings: one, that cey-1 and YBX3 are functionally conserved, two, that these data implicate both YBX1 and YBX3 in memory for the first time, and three, that YBX3 c.379A>T (p.Asn127Tyr) may be of interest for future studies exploring the role of YBX3 in human disease.
We believe that ITM deficits born from the introduction of the p.Asn127Tyr SNV may stem from dysregulation of mRNA translation by CEY-1/YBX3. The CSD, responsible for RNA binding, often lacks a highly specific affinity for a particular RNA motif [77,127]. As a result, proteins with CSDs are considered master regulators of the mRNA landscape. Though it is unclear how this mutation could affect CSD function and mRNA regulation based on the location of the amino acid, the p.Asn127Tyr mutation does add a bulky residue that could cause steric hindrance. A few possibilities for how this mutation might affect protein function, and therefore ability to properly regulate translation, include 1) disrupting interactions with other proteins, 2) affecting 3D conformation such that it impairs protein function, or 3) altering protein stability due to misfolding. Though the mechanism is currently unknown, our data indicate that rare YBX3 variants may be biologically significant and potentially affect neurologic function.
Foundations for mammalian in vivo studies of YBX proteins in plasticity and neurological disease
Our finding of functional conservation between C. elegans cey-1 and human YBX1 and YBX3 with regards to memory underscores the importance of future mammalian studies to determine the role of the YBXs in the nervous system. To date, studying the function of specific YBX proteins in mammalian models is challenging due to the potential for YBXs to genetically compensate for one another and the low survival rate of double knockouts [64]. Here, we report that either depleting CEY-1/YBX levels specifically in adulthood or introducing the rare YBX3 c.379A>T (p.Asn127Tyr) variant detected in three human individuals disrupts memory. Each of these manipulations is less likely to be lethal or induce genetic compensation than complete loss during development. Future studies employing similar approaches in mammalian models will be extremely valuable. Moreover, future work will need to be considerate of loss of function mechanisms of variants. YBX1 is predicted to be loss of function intolerant (pLI = 1, gnomAD v4.0), suggesting haploinsufficient variants may be deleterious. Conversely, YBX3 is predicted to be loss of function tolerant (pLI = 0, gnomAD v4.0). Therefore, it is predicted that rare variants in YBX3 that cause dysfunctions may be dominant-negative or result in gain-of-function. Interestingly, our behavioral data suggest that YBX3p.Asn127Tyr may be a neomorphic change manifesting as a reduction of function in our assays, as defects can be rescued with a wild-type copy of the protein. It is possible that other factors influence the penetrance of this mutation, which agrees with the finding that not all individuals with this variant exhibit neurological symptoms. Future identification of rare variants in the YBXs using comprehensive exome or genomic human datasets and modeling variants in vivo in mammals will enhance our understanding of mechanisms by which YBX dysfunction may play a role in human disease.
Taken together, our results uncover the CEY-1/YBX RBPs as novel associative memory regulators and potentially new contributors to rare neurological disease. Our current findings are limited to the functional testing of a single variant in YBX3. However, further examination of this protein class will provide new insights into the molecular mechanisms of memory and deepen our understanding of how disruption of RBPs contributes to disorders of the nervous system.
Materials and methods
C. elegans maintenance
All strains were maintained at 20°C on 10cm plates made from standard nematode growth medium (NGM: 3g/L NaCl, 2.5g/L of Bacto-peptone, 17g/L Bacto-agar in milliQ water) or high nematode growth medium (HGM: 3g/L NaCl, 20g/L of Bacto-peptone, 30g/L Bacto-agar in milliQ water). After autoclaving and allowing molten agar to cool slightly, we added 1mL/L cholesterol (5mg/mL in ethanol), 1mL/L 1M CaCl2, 1mL/L 1M MgSO4, and 25mL/L 1M potassium phosphate buffer (pH 6.0) [128]. Experiments were performed using NGM plates seeded with OP50 E. coli as the food source for ad libitum feeding [128].
Hypochlorite population synchronization was performed by collecting eggs from gravid hermaphrodites via exposure to an alkaline-bleach solution (85mL water, 15mL sodium hypochlorite, 5mL 5M NaOH), followed by repeated washing of collected eggs in 1mL of M9 buffer (6g/L Na2HPO4, 3g/L KH2PO4, 5g/L NaCl and 1mL/L 1M MgSO4 in milliQ water [128]).
Strains
Wild-type: (N2 Bristol)
Mutants: NM1380(egl-30(js126)), LC108(uIs69[punc-119::sid-1; pmyo-2::mCherry]), TU3401(usIS69[myo-2p::mCherry + unc119p::sid-1]; sid-1(pk3321)), RAF5(cey-1(rrr12)), VC1310(cey-1(ok1805)), GLW51(cey-1(utx43[mNG::3xFLAG::cey-1]) II), OH13605(otIs619[unc-11(prom8)::2xNLS::TagRFP] X) were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN).
Nuclear GFP reporter for cey-1 #1087(cey-1p::PEST::GFPH2B::cey-1u; unc-119 was described previously [56] and generously gifted by Rafal Ciosk.
Strains made in collaboration with InVivo Biosystems include: COP2655(knuSi967[(rab-3p::CEY-1::3xFLAG::rab-3u, unc-119(+))] IV; unc-119(ed3) III) and COP2656(knuSi968[(rab-3p::CEY-1::3xFLAG::rab-3u, unc-119(+))] IV; unc-119(ed3) III) are two lines with the same genotype, COP2689(cey-1(knu1233 [N56Y])); COP2721((knu1251[YBX3]) II); COP2763((knu1275[YBX1]) II); COP2774((knu1286[p.N127Y]) II) and COP2775((knu1287[p.N127Y]) II) are two lines with the same genotype.
The following strains were generated by crosses: RNA1(egl-30(js126); uIs69[(pCFJ90)myo-2p::mCherry + unc-119p::sid-1]) was made by crossing NM1380(egl-30(js126)) with LC108(uIs69[(pCFJ90)myo-2p::mCherry + unc-119p::sid-1]). RNA20(cey-1(utx43[mNG::3xFLAG::cey-1]) II; otIs619[unc-11(prom8)::2xNLS::TagRFP] X) was made by crossing OH13605(otIs619[unc-11(prom8)::2xNLS::TagRFP] X) with GLW51(cey-1(utx43[mNG::3xFLAG::cey-1]) II). RNA26(cey-1p::PEST::GFPH2B::cey-1u; unc-119(+); otIs619[unc-11(prom8)::2xNLS::TagRFP] X) was made by crossing OH13605(otIs619[unc-11(prom8)::2xNLS::TagRFP] X) with cey-1 #1087(cey-1p::PEST::GFPH2B::cey-1u; unc-119(+)). RNA40(cey-1(rrr12); knuSi968[(rab-3p::CEY-1::3xFLAG::rab-3u, unc-119(+)] IV; unc-119(ed3) III) was made by crossing RAF5(cey-1(rrr12)) with COP2656(knuSi968[(rab-3p::CEY-1::3xFLAG::rab-3u, unc-119(+))] IV; unc-119(ed3) III). RNA44 (cey-1(utx43[mNG::3xFLAG::cey-1]) II; usIS69[myo-2p::mCherry + unc119p::sid-1]; sid-1(pk3321)) was made by crossing GLW51(cey-1(utx43[mNG::3xFLAG::cey-1]) II) with TU3401(usIS69[myo-2p::mCherry + unc119p::sid-1]; sid-1(pk3321)). RNA47(knu1286[p.N127Y]) II; knuSi968[(rab-3p::CEY-1::3xFLAG::rab-3u, unc-119(+))] IV; unc-119(ed3) III) was made by crossing COP2774((knu1286[p.N127Y]) II) with COP2656(knuSi968[(rab-3p::CEY-1::3xFLAG::rab-3u, unc-119(+))] IV; unc-119(ed3) III).
RNAi treatment
For adult-only RNAi, worms that are neuronally sensitive to RNAi, RNA1(egl-30(js126); uIs69[(pCFJ90)myo-2p::mCherry + unc-119p::sid-1])) and LC108 (uIs69[(pCFJ90)myo-2p::mCherry + unc-119p::sid-1]), or allow RNAi only in the nervous system, TU3401(usIS69[myo-2p::mCherry + unc119p::sid-1]; sid-1(pk3321)), were synchronized by bleaching. Then, at L4, worms were transferred from plates seeded with OP50 to plates seeded with RNAi (HT115 E. coli) for ad libitum feeding and allowed to grow for two days. For all RNAi experiments, standard NGM molten agar was supplemented with 1mL/L IPTG (isopropyl β-d-1-thiogalactopyranoside) and 1mL/L 100mg/mL carbenicillin.
Short-term and intermediate-term positive olfactory associative memory assays
Wild-type, mutant, or RNAi-treated worms were trained and tested for short- and intermediate-term memory changes as previously described [28]. Briefly, synchronized Day 2 adult worms were washed off plates with M9 buffer. Worms were then allowed to settle by gravity and washed twice more with M9 buffer to remove any bacteria. After washing, the worms were starved for one hour in M9 buffer. For 1x food-butanone pairing, hereby called conditioning, starved worms were transferred to 10cm NGM conditioning plates seeded with OP50 E. coli bacteria and with a total of 16μL of 10% butanone (Sigma Aldrich) diluted in ethanol streaked on the lid in a ‘#’ shape for one hour. After conditioning, the trained population of worms were tested for chemotaxis to 10% butanone and to an ethanol control using standard, previously described chemotaxis conditions [87]. Different stages of memory were tested by measuring chemotaxis of different subpopulations of worms at different timepoints at molecularly distinct stages of memory [29,30]. These stages are immediately after training (0min, learning) or after being transferred to 10cm NGM plates with fresh OP50 for 30 minutes (short-term associative memory), 1 hour (intermediate-term associative memory), or 2 hours (forgetting).
Chemotaxis indices for each timepoint were calculated as
Chemotaxis index = (#wormsbutanone-#wormsethanol)/(total#worms).
Performance index is the change in the chemotaxis index after training relative to the untrained chemotaxis index, or
Performance index = Chemotaxis indextrained—Chemotaxis indexuntrained
Long-term positive olfactory associative memory assays
As previously published, egl-30(js126) animals form a long-term memory after just one round of training [51]. For LTM screening, we administered RNAi to adult, neuronally RNAi sensitive egl-30(js126);punc119::sid-1 worms, performed S/ITM training (1 CS-US pairing), and measured learning immediately after training and long-term memory 16–20 hours post-training. Chemotaxis index and performance index were calculated in the same manner as short term and intermediate term associative memory assays.
Aversive associative memory assays
As previously published [32], worms were tested for learning changes in response to aversive diacetyl training. Briefly, synchronized Day 2 adult worms were washed off plates with M9 buffer. Worms were then allowed to settle by gravity and washed twice more with M9 buffer to remove any bacteria. After washing, the worms were conditioned for one hour by starving on 10cm NGMs with no bacteria with 16uL of 100% diacetyl (Sigma Aldrich) streaked on the lid in a ‘#’ shape. Immediately after conditioning, the trained population of worms were tested for chemotaxis to 1% diacetyl and to an ethanol control using standard, previously described chemotaxis conditions [87].
Baseline chemosensation assays
Chemotaxis, or chemosensation experiments, were performed based on previously published assays [87]. In brief, assays were performed on unseeded 10cm NGMs. Two marks were made on the back of the plate on opposite sides of the plate, approximately 0.5cm from the edge. 1μL of sodium azide (Thermo Fisher) was placed on both spots and allowed to dry before adding 1μL of test odorant diluted in ethanol on one side and ethanol on the other. Odorants included 0.1% and 10% butanone (vol/vol), 0.1% nonanol (2-nonanone)(vol/vol), 1% isoamyl alcohol (vol/vol), 1% benzaldehyde (vol/vol), 10% pyrazine (weight/vol), and 1% diacetyl (vol/vol) (all from Sigma Aldrich). Worms were washed off their plates and subsequently washed three times with M9 buffer, then placed near the bottom center of the plate, equidistant between the two marks, and allowed to chemotax for an hour. Chemotaxis indices for each timepoint were calculated as
Chemotaxis index = (#wormsodorant—#wormsethanol)/(total#worms).
% Origin motility assays
We measured motility as previously published [20] by finding the average percentage of worms remaining at the origin of the plate after a naïve (untrained) chemotaxis assay using an attractive concentration of butanone (0.1% vol/vol).
Solid media motility assays
Worms were synchronized by hypochlorite treatment. 15–20 well-fed Day 2 adult worms on NGM plates seeded with OP50 E. coli were recorded for 60 second increments per group using a Nikon DS-Fi3 camera and NIS Elements Imaging Software. Videos were analyzed using wrMTrck software [129]. Data were thresholded such that only tracks of 15 seconds or longer were analyzed. Motility is reported as Average Speed (μm/sec) or Total distance in μm per track.
Thrashing assays
Trashing assays were performed by first filling an unseeded 35mm NGM agar plate with 1mL of M9 buffer solution. Next, 10–15 well-fed Day 2 adult worms were transferred to an unseeded NGM agar plate to remove any bacteria stuck to the worms. All worms were then placed into the buffer for 1 minute to acclimate to the new environment. Thrashing behavior was recorded in 30 second increments per group of worms using a Nikon DS-Fi3 camera and NIS Elements Imaging Software. The number of thrashes per worm were quantified by manually counting the number of body bends; one body bend movement of the worm is defined by swinging its head and tail to the same side to form a C shape and then back to the initial position. This protocol was adapted from Nawa and Matsuoka 2012 [130].
Confocal microscopy
Worms were paralyzed with fresh 4% levamisole diluted in M9 buffer and imaged for up to 30 minutes once put onto the slide to preserve protein dynamics. In all experiments, imaging of Day 2 adult worms was performed on a Nikon Ti2E inverted microscope system with a W1 spinning disk confocal unit at 100x magnification. For general CEY-1::mNG visualization, we used an excitation wavelength of 488nm for GFP and 561nm for RFP, used a pixel size of 0.11um/px, and z stacks with a z-step of 0.6μm. For quantification of CEY-1::mNG fluorescence after RNAi treatment, worms were imaged at 100X magnification, an excitation wavelength of 488nm, and z stacks with a z-step of 0.2 μm. Images were processed in Nikon NIS Elements software. The same settings for laser power and detector gain were used for all genotypes. For all images, red fluorescence was pseudo colored magenta and green fluorescence were pseudo colored cyan for colorblindness inclusivity. To quantify CEY-1::mNG fluorescence after RNAi knockdown, ImageJ [131] was used to quantify total fluorescence over a 100 μm square centered over the nerve ring after background subtraction.
Variant identification and pathogenicity analysis
We received a deidentified dataset of human YBX gene variants identified through clinical exome and genome sequencing completed at Baylor Genetics [103] and removed any variants where symptomology listed could be explained by other genetic alterations found in the individual’s genome. Next, we removed any variants found in gnomAD v.2.1.1, even if only one case was reported [132], as these variants would not likely cause severe disease if found in the general population. We then selected variants for potential in vivo modeling based on the presence of neurological symptoms, such as seizures, microcephaly, autism, and intellectual disability. Variants of interest were then assessed using multiple pathogenicity metrics to determine the final variant for in vivo testing: specifically, M-CAP [110], SIFT [112], PolyPhen 2 [111], GERP [108], CADD [106,107], REVEL [109], and Phylop Vertebrate [113]. All metrics were found and visualized using the UCSC genome browser [133]. We selected the variant with the highest overall predicted pathogenicity across all score metrics as opposed to any specific numerical cutoffs. Pathogenicity of the p.Asn127Tyr variant on YBX3 protein function was predicted by using the YBX3 UniProt ID (P16989) as input for the following publicly available pipelines: AlphaMissense [119] and FunC-ESMs [120].
Phylogenetic analysis
To analyze the evolutionary history and determine the similarity of RNA binding proteins between phyla, we used maximum likelihood (ML) analysis with the JTT matrix-based model [134]. We obtained protein sequences of the C. elegans CEY proteins as well as the human YBX proteins from UniProt [135] then used these sequences to generate a tree with the highest log likelihood score (-4517.57) using the Neighbor-Join and BioNJ algorithms to calculate pairwise distances from the JTT model. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site and a 0.20 scale bar indicating the relative distance between nodes. All analyses were performed using MEGA X [74,136].
Orthology analysis
Drosophila RNAi Screening Center Integration Ortholog Prediction Tool (DIOPT) is an online bioinformatics resource designed to facilitate the identification of orthologous genes across various species [75].
RNA isolation, cDNA synthesis and qRT-PCR
Worms of a particular genotype were crushed in liquid nitrogen and added to Trizol (Thermo Fisher Scientific). RNA was isolated per manufacturer’s instructions, followed by DNase treatment (Qiagen). cDNA was synthesized with an oligo dT primer and Superscript III reverse transcriptase enzyme (Thermo Fisher Scientific). cDNA was mixed with buffers, primers, SYBR green, and hot start Taq polymerase in a master mix prepared by a manufacturer (Thermo Fisher Scientific). Using a Quant Studio 7 Pro Dx Real-Time PCR System (Thermo Fisher Scientific), PCR reactions were run followed by a dissociation reaction to determine specificity of the amplified product. The amount of gene expression was quantified using the ΔΔCt method using pmp-3 as a reference gene. Primer sets were as follows:
cey-1 For: 5’-GGATCCAAGTATGCTGCCGA -3’
cey-1 Rev: 5’- CCATCTGTGTCACGAGCAGT -3’
pmp-3 For: 5’- AGTTCCGGTTGGATTGGTCC -3’
pmp-3 Rev: 5’- CCAGCACGATAGAAGGCGAT-3’
YBX3 WT For: 5’- CCACCGTAACCCAACCTACC -3’
YBX3 WT Rev: 5’- CTTGGCCTCCTTTCCGTCTT -3’
YBX3 p.Asn127Tyr For: 5’- CGTCGTTACCGTCGTGGATA -3’
YBX3 p.Asn127Tyr Rev: 5’- TGGATACGGTTTGGGTGTGG -3’YBX1 For: 5’- CTACCGTCGTTACCCACGTC -3’
YBX1 Rev: 5’- ATCAGCTCCCTCCATGACCT -3’
Statistical analysis
Statistical data is reported in the main text, figures, and tables as noted. Significance threshold of p < 0.05 was used. The symbols *, **, ***, and **** refer to p < 0.05, 0.01, 0.001, and 0.0001, respectively. For the comparison of performance indices between two behavior conditions (e.g. vector control vs cey-1 RNAi), a Mann-Whitney test comparing ranks was used because it does not assume normality. For comparison of performance indices between three or more groups (e.g. wild-type vs two different cey-1 loss-of-function mutants), one-way analysis of variances followed by Bonferroni post hoc tests for multiple comparisons were performed. For comparison of motility one-way analysis of variances followed by Games-Howell’s multiple comparisons tests (due to larger Ns) were performed. Two-way ANOVAs were used for evaluating effects between genotype (Wild-type, cey-1(rrr12), YBX1, YBX3) and timepoint (0hr, 0.5hr, 1hr, 2hr) on performance indices with a significant interaction between factors (p<0.0001) prompting Bonferroni post-hoc analyses to determine differences between individual groups. All experiments were repeated on separate days with separate populations to confirm reproducibility of results. Sample size n represents the number of chemotaxis assays performed for behavior, with each assay containing approximately 50–150 worms each.
Statistical analysis software
All statistics and code were run in GraphPad Prism 10, using standard toolboxes.
Supporting information
Boxes signify 3+ RBPs in the same protein family or class (blue box include PUF RBPs, orange box includes CEY RBPs, grey box includes translation initiation machinery). Box and whisker plots are shown for the learning timepoints from the STM/ITM assays for each RBP screened. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. ns, not significant (p>0.05).
(TIF)
All RBPs in the screen are shown grouped by experiment. Bar represents mean. Whiskers mark the standard error of the mean. n = 10 per RNAi treatment. ns, not significant.
(TIF)
All RBPs are shown where pink circles are decreased memory and blue circles are increased memory. The size of circle represents the p value from a combined n ≥ 5–10 per RNAi treatment.
(TIF)
All RBPs with significantly altered memory are shown divided by memory timepoint. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. n ≥ 5–10 per RNAi treatment. *p<0.05,**p<0.01, ***p<0.001, ****p<0.0001.
(TIF)
While cey-1 and cey-4 have broad expression, cey-2 is only located in eight neurons at L4.
(TIF)
(A) Neuron-specific and/or single-cell RNA-seq data compiled from five different publications suggest cey-1 mRNA is expressed in the adult nervous system [24, 35, 38, 43, 80, 137, 138]. (B) Diagram of microscopy images shown in (C). C. elegans head is labeled including the location of the pharynx and main neuronal ganglia/nerve rings. (C) A transcriptional cey-1 reporter suggests the gene is broadly expressed in the neurons in the head at baseline conditions. Representative image of Day 2 adult worms with RFP-labeled neuronal nuclei (unc-11(prom8)::2xNLS::TagRFP)) pseudocolored magenta and a nuclear GFPH2B cey-1 promoter fusion (cey1p::GFPH2B) pseudocolored cyan show colocalization.
(TIF)
(A) Naïve battery of both negative and positive odors reveals deficits in both nonanol and isoamyl alcohol chemotaxis, but not in benzaldehyde, pyrazine, or diacetyl chemotaxis at attractive concentrations. (B) Baseline chemosensation for butanone at neutral concentration of 10% is unaffected by neuron-specific knockdown of cey-1. n = 15 per RNAi treatment. (C) Baseline chemosensation for butanone at an attractive concentration of 0.1% is unaffected by neuron-specific knockdown of cey-1. n = 15 per RNAi treatment. (D) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by neuron-specific loss of cey-1. n = 15 per RNAi treatment. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. **p<0.01, ****p<0.0001. ns, not significant (p>0.05). (E) Representative images of adult-only, neuron-specific knockdown RNAi treatment (vector or cey-1 RNAi) in neuronally RNAi-sensitized mNG::CEY-1 animals. (F) Quantification of total fluorescent intensity. n = 26 for each RNAi condition. *p<0.05.
(TIF)
(A) Naïve chemotaxis towards 10% butanone prior to behavioral conditioning does not significantly differ between wild-type animals, cey-1(ok1805), and cey-1(rrr12) mutants. n = 15 per genotype. (B) Baseline chemosensation for butanone at an attractive concentration of 0.1% is unaffected by whole-body loss of cey-1. n = 15 per genotype. (C) Baseline chemosensation for butanone at an attractive concentration of 0.1% is unaffected by neuron-specific rescue of cey-1. n = 15 per genotype. (D) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by whole-body loss of cey-1. n = 15 per genotype. (E) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by neuron-specific rescue of cey-1. n = 15 per genotype. (F) Day 2 adult speed measured in μm/sec by Wrmtrck. n = 20–30 worms per genotype, Tracks were thresholded at 15 seconds duration, with multiple tracks per worm possible. ns, not significant. (G) Day 2 average track distance (in μm) traveled as measured by Wrmtrck. n = 20–30 worms per genotype, Tracks were thresholded at 15 seconds duration, with multiple tracks per worm possible. ns, not significant. Of note, for both F and G, wild-type controls for this figure are the same as S11 Fig because experiments were performed the same day each time; data was put into two separate graphs for organization. (H) Day 2 thrashing comparing number of body bends per 30 seconds of wild-type, cey-1(rrr12) knockout worms, and nervous system specific rescue worms (cey-1(rrr12);rab-3p::CEY-1) shows that cey-1 mutants do not have thrashing deficits, and indeed exhibit more body bends per minute. n = 50 for each genotype. (I) Associative memory curve comparing wild-type, cey-1(rrr12) knockout worms, and nervous system specific rescue worms (cey-1(rrr12);rab-3p::CEY-1) shows that while knockouts have no associative learning or memory, neuron-specific rescue of cey-1 allows for learning and memory equivalent to wild-type levels. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. ns, not significant (p>0.05).
(TIF)
(A) Percentages of individuals reported with gain or loss CNVs that include YBX1. (B) Percentages of mechanisms of inheritance of gain and loss CNVs that include YBX1. (C) Size of CNV gain and losses that include YBX1. (D) Predictive scores for YBX1 from gnomAD v.2.11 [132] suggest that YBX1 is intolerant to loss of function variants and is haploinsufficient. (E) Phenotypes of patients specifically with deletion/loss CNVs including YBX1 include epicanthus, delayed speech and development, intellectual disability, and other neurological features.
(TIF)
(A) Percentages of individuals reported with gain or loss CNVs that include YBX3. (B) Percentages of mechanisms of inheritance of gain and loss CNVs that include YBX3. (C) Size of CNV gain and losses that include YBX3. (D) Predictive scores for YBX3 from gnomAD v.2.11 [132] suggest that YBX3 is tolerant to loss of function variants and is haplosufficient, suggesting variants may instead be deleterious by being dominant negative or gain-of-function. (E) Phenotypes of patients specifically with deletion/loss CNVs including YBX3, primarily intellectual disability, low-set ears, micrognathia, and other neurological features.
(TIF)
(A) Baseline chemosensation for butanone at an attractive concentration of 0. 1% is unaffected by the p.Asn56Tyr variant in cey-1. n = 15 per genotype. (B) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by p.Asn56Tyr variant in cey-1. n = 15 per genotype. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. ns, not significant (p>0.05). (C) qRT-PCR of cey-1 mRNA levels in Day 2 adults shows that while cey-1 knockouts (cey-1(rrr12)) worms undergo nonsense mediated decay [56] resulting in reduced cey-1 mRNA levels, introduction of the p.Asn56Tyr variant has no significant effect on cey-1 expression. n = 6 per genotype. **p<0.01. (D) Day 2 thrashing comparing number of body bends per 30 seconds of wild-type, neuron-specific CEY-1 overexpression (cey-1(WT);rab-3p::CEY-1), and variant worms with p.Asn56Tyr in endogenous CEY-1 (cey-1(p.Asn127Tyr) shows that cey-1 mutants do not have thrashing deficits, and indeed exhibit more body bends per minute. n = 50 for each genotype. Of note, wild-type controls for this figure are the same as S8 Fig because experiments were performed the same day each time; data was put into two separate graphs for organization. (E) Day 2 adult speed measured in μm/sec by Wrmtrck. n = 20–30 worms per genotype, Tracks were thresholded at 15 seconds duration, with multiple tracks per worm possible. ns, not significant. (F) Day 2 average track distance (in μm) traveled as measured by Wrmtrck. n = 20–30 worms per genotype, Tracks were thresholded at 15 seconds duration, with multiple tracks per worm possible. ns, not significant. Of note, for both E and F, wild-type controls for this figure are the same as S8 Fig because experiments were performed the same day each time; data was put into two separate graphs for organization.
(TIF)
(A) Associative memory curve comparing wild-type, cey-1 knockouts, and humanized lines expressing either YBX1 or YBX3 at the endogenous cey-1 locus. While worms expressing YBX3 have normal memory performance, those expressing YBX1 appear to have extended memory, as they have a strong association for butanone even two hours post-training. (B) Baseline chemosensation for butanone at an attractive concentration of 0.1% is normal in worms expressing YBX1. n = 15 per genotype. (C) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by YBX1. n = 15 per genotype. (D) Baseline chemosensation is normal in worms expressing YBX3 at the cey-1 locus. n = 15 per genotype. (E) Motility is unaffected by YBX3. n = 15 per genotype. (F) Baseline chemosensation is unaffected by expressing YBX3(p.Asn127Tyr) at the cey-1 locus. n = 15 per genotype. (G) Motility is unaffected by YBX3(p.Asn127Tyr). n = 15 per genotype. (H) qRT-PCR of cey-1 and YBX mRNA levels in Day 2 adults shows that levels of YBX1, YBX3, and YBX3(pAsn127Tyr) are not significantly reduced compared to wild-type cey-1 mRNA levels and therefore, introduction of the p.Asn127Tyr variant has no significant effect on expression. n = 5 per genotype. ns, not significant.
(TIF)
(A) Forward-facing view of the protein with the β sheet that binds RNA facing the viewer. (B) Top-down view of the YBX3 protein depicts the location of the Asn127 residue.
(TIF)
(XLSX)
(XLSX)
Acknowledgments
We thank the Ciosk lab for generously sharing the cey-1 #1087 strain; the CGC for strains; Ben Jussila and In Vivo Biosystems for strains and helpful discussion; Peter Boag for helpful discussion; and members of the Arey lab, particularly Katie L Brandel and Catherine Stuart for their detailed feedback on the manuscript. We thank Drs. Jill Rosenfeld and Hongzheng Dai from Baylor Genetics for confirming the presence and relevance of the YBX3 p.Asn127Tyr variant. We thank Dr. Katka Cermakova from Baylor College of Medicine, and Dr. Amelie Stein from University of Copenhagen for discussion of potential effects of the p.Asn127Tyr variant on protein function. This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from https://deciphergenomics.org/about/stats and via email from contact@deciphergenomics.org. DECIPHER is hosted by EMBL-EBI.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by a NIH Director’s New Innovator Award (DP2NS132372), support from the Whitehall Foundation (https://www.whitehall.org/), and Glenn Foundation for Medical Research and AFAR Grant for Junior Faculty (https://www.afar.org/) to RNA. ANH is supported by and F31 NRSA F31NS129312-01 and Baylor Research Advocates for Student Scientists https://give.bcm.edu/get-involved/brass/). HTC’s research effort is partially supported by The Robert and Janice McNair Foundation (https://mcnairfoundation.org/), Burroughs Wellcome Fund (https://www.bwfund.org/), Cain Pediatric Neurology Research Foundation Laboratories (https://www.bcm.edu/departments/pediatrics/research/cain-foundation-laboratories), Anne and Bob Graham, and the Child Neurology Foundation and Society. (https://www.childneurologyfoundation.org/about-cnf/). KBA is supported by a NSF GRFP (DGE-349937), EWP is supported by an NRSA (F31GM151846), EJL is funded by an NRSA (F31AG081095), and ESG is funded by BCM PREP (R25GM069234). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, et al. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5 [DOI] [PubMed] [Google Scholar]
- 3.Hafner A-S, Donlin-Asp PG, Leitch B, Herzog E, Schuman EM. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science. 2019:364. doi: 10.1126/science.aau3644 [DOI] [PubMed] [Google Scholar]
- 4.Akins MR, Berk-Rauch HE, Fallon JR. Presynaptic translation: stepping out of the postsynaptic shadow. Front Neural Circuits. 2009;3:17. doi: 10.3389/neuro.04.017.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schieweck R, Ninkovic J, Kiebler MA. RNA-binding proteins balance brain function in health and disease. Physiol Rev. 2021;101:1309–1370. doi: 10.1152/physrev.00047.2019 [DOI] [PubMed] [Google Scholar]
- 6.Doxakis E. RNA binding proteins: a common denominator of neuronal function and dysfunction. Neurosci Bull. 2014;30:610–626. doi: 10.1007/s12264-014-1443-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebauer F, Schwarzl T, Valcárcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22:185–198. doi: 10.1038/s41576-020-00302-y [DOI] [PubMed] [Google Scholar]
- 8.Prashad S, Gopal PP. RNA-binding proteins in neurological development and disease. RNA Biol. 2021;18: 972–987. doi: 10.1080/15476286.2020.1809186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauriat TL, Shiue L, Haroutunian V, Verbitsky M, Ares M, Ospina L, et al. Developmental expression profile of quaking, a candidate gene for schizophrenia, and its target genes in human prefrontal cortex and hippocampus shows regional specificity. J Neurosci Res. 2008;86:785–796. doi: 10.1002/jnr.21534 [DOI] [PubMed] [Google Scholar]
- 10.Au PYB, Goedhart C, Ferguson M, Breckpot J, Devriendt K, Wierenga K, et al. Phenotypic spectrum of Au-Kline syndrome: a report of six new cases and review of the literature. Eur J Hum Genet. 2018;26:1272–1281. doi: 10.1038/s41431-018-0187-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingemans AJM, Truijen KMG, Kim J-H, Alaçam Z, Faivre L, Collins KM, et al. Establishing the phenotypic spectrum of ZTTK syndrome by analysis of 52 individuals with variants in SON. Eur J Hum Genet. 2022;30:271–281. doi: 10.1038/s41431-021-00960-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quartier A, Poquet H, Gilbert-Dussardier B, Rossi M, Casteleyn A-S, Portes V, et al. Intragenic FMR1 disease-causing variants: a significant mutational mechanism leading to Fragile-X syndrome. Eur J Hum Genet. 2017;25:423–431. doi: 10.1038/ejhg.2016.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gennarino VA, Palmer EE, McDonell LM, Wang L, Adamski CJ, Koire A, et al. A Mild PUM1 Mutation Is Associated with Adult-Onset Ataxia, whereas Haploinsufficiency Causes Developmental Delay and Seizures. Cell. 2018;172:924–936.e11. doi: 10.1016/j.cell.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130 [DOI] [PubMed] [Google Scholar]
- 15.Olshansky SJ, Goldman DP, Zheng Y, Rowe JW. Aging in America in the twenty-first century: demographic forecasts from the MacArthur Foundation Research Network on an Aging Society. Milbank Q. 2009;87:842–862. doi: 10.1111/j.1468-0009.2009.00581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim KW, Tang NH, Piggott CA, Andrusiak MG, Park S, Zhu M, et al. Expanded genetic screening in Caenorhabditis elegans identifies new regulators and an inhibitory role for NAD+ in axon regeneration. eLife. 2018:7. doi: 10.7554/eLife.39756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poole RJ, Bashllari E, Cochella L, Flowers EB, Hobert O. A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002109. doi: 10.1371/journal.pgen.1002109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doitsidou M, Flames N, Topalidou I, Abe N, Felton T, Remesal L, et al. A combinatorial regulatory signature controls terminal differentiation of the dopaminergic nervous system in C. elegans. Genes Dev. 2013;27:1391–1405. doi: 10.1101/gad.217224.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDiarmid TA, Belmadani M, Liang J, Meili F, Mathews EA, Mullen GP, et al. Systematic phenomics analysis of autism-associated genes reveals parallel networks underlying reversible impairments in habituation. Proc Natl Acad Sci USA. 2020;117:656–667. doi: 10.1073/pnas.1912049116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakhina V, Arey RN, Kaletsky R, Kauffman A, Stein G, Keyes W, et al. Genome-wide functional analysis of CREB/long-term memory-dependent transcription reveals distinct basal and memory gene expression programs. Neuron. 2015;85: 330–345. doi: 10.1016/j.neuron.2014.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sohrabi S, Mor DE, Kaletsky R, Keyes W, Murphy CT. High-throughput behavioral screen in C. elegans reveals Parkinson’s disease drug candidates. Commun Biol. 2021;4: 203. doi: 10.1038/s42003-021-01731-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vukojevic V, Gschwind L, Vogler C, Demougin P, de Quervain DJ-F, Papassotiropoulos A, et al. A role for α-adducin (ADD-1) in nematode and human memory. EMBO J. 2012;31: 1453–1466. doi: 10.1038/emboj.2012.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenyves BG, Arnold A, Gharat VG, Haab C, Tishinov K, Peter F, et al. Dual Role of an mps-2/KCNE-Dependent Pathway in Long-Term Memory and Age-Dependent Memory Decline. Curr Biol. 2021;31: 527–539.e7. doi: 10.1016/j.cub.2020.10.069 [DOI] [PubMed] [Google Scholar]
- 24.Yao V, Kaletsky R, Keyes W, Mor DE, Wong AK, Sohrabi S, et al. An integrative tissue-network approach to identify and test human disease genes. Nat Biotechnol. 2018. doi: 10.1038/nbt.4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharifnia P, Jin Y. Regulatory roles of RNA binding proteins in the nervous system of C. elegans. Front Mol Neurosci. 2014;7: 100. doi: 10.3389/fnmol.2014.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30: 313–321. doi: 10.1016/s1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- 27.Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7: 554–559. doi: 10.1038/nmeth.1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010;8: e1000372. doi: 10.1371/journal.pbio.1000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein GM, Murphy CT. C. elegans positive olfactory associative memory is a molecularly conserved behavioral paradigm. Neurobiol Learn Mem. 2014;115: 86–94. doi: 10.1016/j.nlm.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kauffman A, Parsons L, Stein G, Wills A, Kaletsky R, Murphy C. C. elegans positive butanone learning, short-term, and long-term associative memory assays. J Vis Exp. 2011. doi: 10.3791/2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rankin CH, Beck CD, Chiba CM. Caenorhabditis elegans: a new model system for the study of learning and memory. Behav Brain Res. 1990;37: 89–92. doi: 10.1016/0166-4328(90)90074-o [DOI] [PubMed] [Google Scholar]
- 32.Stetak A, Hörndli F, Maricq AV, van den Heuvel S, Hajnal A. Neuron-specific regulation of associative learning and memory by MAGI-1 in C. elegans. PLoS ONE. 2009;4: e6019. doi: 10.1371/journal.pone.0006019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30: 1427–1464. doi: 10.1093/nar/30.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matia-González AM, Laing EE, Gerber AP. Conserved mRNA-binding proteomes in eukaryotic organisms. Nat Struct Mol Biol. 2015;22: 1027–1033. doi: 10.1038/nsmb.3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaletsky R, Yao V, Williams A, Runnels AM, Tadych A, Zhou S, et al. Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 2018;14: e1007559. doi: 10.1371/journal.pgen.1007559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holdorf AD, Higgins DP, Hart AC, Boag PR, Pazour GJ, Walhout AJM, et al. WormCat: An Online Tool for Annotation and Visualization of Caenorhabditis elegans Genome-Scale Data. Genetics. 2020;214: 279–294. doi: 10.1534/genetics.119.302919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim W, Underwood RS, Greenwald I, Shaye DD. OrthoList 2: A New Comparative Genomic Analysis of Human and Caenorhabditis elegans Genes. Genetics. 2018;210: 445–461. doi: 10.1534/genetics.118.301307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arey RN, Kaletsky R, Murphy CT. Nervous system-wide profiling of presynaptic mRNAs reveals regulators of associative memory. Sci Rep. 2019;9: 20314. doi: 10.1038/s41598-019-56908-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostroff LE, Santini E, Sears R, Deane Z, Kanadia RN, LeDoux JE, et al. Axon TRAP reveals learning-associated alterations in cortical axonal mRNAs in the lateral amgydala. eLife. 2019;8. doi: 10.7554/eLife.51607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaletsky R, Lakhina V, Arey R, Williams A, Landis J, Ashraf J, et al. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature. 2016;529: 92–96. doi: 10.1038/nature16483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nijssen J, Aguila J, Hoogstraaten R, Kee N, Hedlund E. Axon-Seq Decodes the Motor Axon Transcriptome and Its Modulation in Response to ALS. Stem Cell Reports. 2018;11: 1565–1578. doi: 10.1016/j.stemcr.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74: 453–466. doi: 10.1016/j.neuron.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arey R, Kaletsky R, Murphy CT. Profiling of presynaptic mRNAs reveals a role for axonal PUMILIOs in associative memory formation. BioRxiv. 2019. doi: 10.1101/733428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freytag V, Probst S, Hadziselimovic N, Boglari C, Hauser Y, Peter F, et al. Genome-Wide Temporal Expression Profiling in Caenorhabditis elegans Identifies a Core Gene Set Related to Long-Term Memory. J Neurosci. 2017;37: 6661–6672. doi: 10.1523/JNEUROSCI.3298-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau HL, Timbers TA, Mahmoud R, Rankin CH. Genetic dissection of memory for associative and non-associative learning in Caenorhabditis elegans. Genes Brain Behav. 2013;12: 210–223. doi: 10.1111/j.1601-183X.2012.00863.x [DOI] [PubMed] [Google Scholar]
- 46.Timbers TA, Rankin CH. Tap withdrawal circuit interneurons require CREB for long-term habituation in Caenorhabditis elegans. Behav Neurosci. 2011;125: 560–566. doi: 10.1037/a0024370 [DOI] [PubMed] [Google Scholar]
- 47.Rose JK, Kaun KR, Chen SH, Rankin CH. GLR-1, a non-NMDA glutamate receptor homolog, is critical for long-term memory in Caenorhabditis elegans. J Neurosci. 2003;23: 9595–9599. doi: 10.1523/JNEUROSCI.23-29-09595.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin X, Pokala N, Bargmann CI. Distinct circuits for the formation and retrieval of an imprinted olfactory memory. Cell. 2016;164: 632–643. doi: 10.1016/j.cell.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim JP, Fehlauer H, Das A, Saro G, Glauser DA, Brunet A, et al. Loss of CaMKI Function Disrupts Salt Aversive Learning in C. elegans. J Neurosci. 2018;38: 6114–6129. doi: 10.1523/JNEUROSCI.1611-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H, Wu T, Canales XG, Wu M, Choi M-K, Duan F, et al. Forgetting generates a novel state that is reactivatable. Sci Adv. 2022;8: eabi9071. doi: 10.1126/sciadv.abi9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arey RN, Stein GM, Kaletsky R, Kauffman A, Murphy CT. Activation of gαq signaling enhances memory consolidation and slows cognitive decline. Neuron. 2018;98: 562–574.e5. doi: 10.1016/j.neuron.2018.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dubnau J, Chiang A-S, Grady L, Barditch J, Gossweiler S, McNeil J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13: 286–296. doi: 10.1016/s0960-9822(03)00064-2 [DOI] [PubMed] [Google Scholar]
- 53.Dong H, Zhu M, Meng L, Ding Y, Yang D, Zhang S, et al. Pumilio2 regulates synaptic plasticity via translational repression of synaptic receptors in mice. Oncotarget. 2018;9: 32134–32148. doi: 10.18632/oncotarget.24345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129: 195–206. doi: 10.1016/j.cell.2007.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gindina S, Botsford B, Cowansage K, LeDoux J, Klann E, Hoeffer C, et al. Upregulation of eIF4E, but not other translation initiation factors, in dendritic spines during memory formation. J Comp Neurol. 2021;529: 3112–3126. doi: 10.1002/cne.25158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnold A, Rahman MM, Lee MC, Muehlhaeusser S, Katic I, Gaidatzis D, et al. Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes. Nucleic Acids Res. 2014;42: 13353–13369. doi: 10.1093/nar/gku1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calculli G, Lee HJ, Shen K, Pham U, Herholz M, Trifunovic A, et al. Systemic regulation of mitochondria by germline proteostasis prevents protein aggregation in the soma of C. elegans. Sci Adv. 2021;7. doi: 10.1126/sciadv.abg3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Budkina K, El Hage K, Clément M-J, Desforges B, Bouhss A, Joshi V, et al. YB-1 unwinds mRNA secondary structures in vitro and negatively regulates stress granule assembly in HeLa cells. Nucleic Acids Res. 2021;49: 10061–10081. doi: 10.1093/nar/gkab748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon E, Todorova K, Wang J, Horos R, Lee KK, Neel VA, et al. The RNA-binding protein YBX1 regulates epidermal progenitors at a posttranscriptional level. Nat Commun. 2018;9: 1734. doi: 10.1038/s41467-018-04092-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J-Z, Zhu H, You P, Liu H, Wang W-K, Fan X, et al. Upregulated YB-1 protein promotes glioblastoma growth through a YB-1/CCT4/mLST8/mTOR pathway. J Clin Invest. 2022;132. doi: 10.1172/JCI146536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehta S, McKinney C, Algie M, Verma CS, Kannan S, Harfoot R, et al. Dephosphorylation of YB-1 is Required for Nuclear Localisation During G2 Phase of the Cell Cycle. Cancers (Basel). 2020;12. doi: 10.3390/cancers12020315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor SR, Santpere G, Reilly M, Glenwinkel L, Poff A, McWhirter R, et al. Expression profiling of the mature C. elegans nervous system by single-cell RNA-Sequencing. BioRxiv. 2019. doi: 10.1101/737577 [DOI] [Google Scholar]
- 63.Liska D, Wolfe Z, Norris A. VISTA: Visualizing the Spatial Transcriptome of the C. elegans Nervous System. BioRxiv. 2023. doi: 10.1101/2023.04.28.538711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans MK, Matsui Y, Xu B, Willis C, Loome J, Milburn L, et al. Ybx1 fine-tunes PRC2 activities to control embryonic brain development. Nat Commun. 2020;11: 4060. doi: 10.1038/s41467-020-17878-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Unkrüer B, Pekcec A, Fuest C, Wehmeyer A, Balda MS, Horn A, et al. Cellular localization of Y-box binding protein 1 in brain tissue of rats, macaques, and humans. BMC Neurosci. 2009;10: 28. doi: 10.1186/1471-2202-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Q, Ding S-L, Li Y, Royall J, Feng D, Lesnar P, et al. The allen mouse brain common coordinate framework: A 3D reference atlas. Cell. 2020;181: 936–953.e20. doi: 10.1016/j.cell.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. The human cell atlas. eLife. 2017;6. doi: 10.7554/eLife.27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka T, Ohashi S, Funakoshi T, Kobayashi S. YB-1 binds to GluR2 mRNA and CaM1 mRNA in the brain and regulates their translational levels in an activity-dependent manner. Cell Mol Neurobiol. 2010;30: 1089–1100. doi: 10.1007/s10571-010-9541-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Distler U, Schmeisser MJ, Pelosi A, Reim D, Kuharev J, Weiczner R, et al. In-depth protein profiling of the postsynaptic density from mouse hippocampus using data-independent acquisition proteomics. Proteomics. 2014;14: 2607–2613. doi: 10.1002/pmic.201300520 [DOI] [PubMed] [Google Scholar]
- 70.Nekrasov MP, Ivshina MP, Chernov KG, Kovrigina EA, Evdokimova VM, Thomas AAM, et al. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J Biol Chem. 2003;278: 13936–13943. doi: 10.1074/jbc.M209145200 [DOI] [PubMed] [Google Scholar]
- 71.Skabkin MA, Kiselyova OI, Chernov KG, Sorokin AV, Dubrovin EV, Yaminsky IV, et al. Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res. 2004;32: 5621–5635. doi: 10.1093/nar/gkh889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kretov DA. Role of Y-Box Binding Proteins in Ontogenesis. Biochemistry Mosc. 2022;87: S71–S4. doi: 10.1134/S0006297922140061 [DOI] [PubMed] [Google Scholar]
- 73.Fotovati A, Abu-Ali S, Wang P-S, Deleyrolle LP, Lee C, Triscott J, et al. YB-1 bridges neural stem cells and brain tumor-initiating cells via its roles in differentiation and cell growth. Cancer Res. 2011;71: 5569–5578. doi: 10.1158/0008-5472.CAN-10-2805 [DOI] [PubMed] [Google Scholar]
- 74.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35: 1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12: 357. doi: 10.1186/1471-2105-12-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang X-J, Zhu H, Mu S-R, Wei W-J, Yuan X, Wang M, et al. Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA. J Biol Chem. 2019;294: 10998–11010. doi: 10.1074/jbc.RA119.007545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Budkina KS, Zlobin NE, Kononova SV, Ovchinnikov LP, Babakov AV. Cold Shock Domain Proteins: Structure and Interaction with Nucleic Acids. Biochemistry Mosc. 2020;85: S1–S19. doi: 10.1134/S0006297920140011 [DOI] [PubMed] [Google Scholar]
- 78.Kretov DA, Curmi PA, Hamon L, Abrakhi S, Desforges B, Ovchinnikov LP, et al. mRNA and DNA selection via protein multimerization: YB-1 as a case study. Nucleic Acids Res. 2015;43: 9457–9473. doi: 10.1093/nar/gkv822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eliseeva IA, Sogorina EM, Smolin EA, Kulakovskiy IV, Lyabin DN. Diverse Regulation of YB-1 and YB-3 Abundance in Mammals. Biochemistry Mosc. 2022;87: S48–S167. doi: 10.1134/S000629792214005X [DOI] [PubMed] [Google Scholar]
- 80.Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, et al. Molecular topography of an entire nervous system. Cell. 2021;184: 4329–4347.e23. doi: 10.1016/j.cell.2021.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hammarlund M, Hobert O, Miller DM, Sestan N. The cengen project: the complete gene expression map of an entire nervous system. Neuron. 2018;99: 430–433. doi: 10.1016/j.neuron.2018.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314: 1–340. doi: 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- 83.Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82: 358–370. doi: 10.1016/0012-1606(81)90459-0 [DOI] [PubMed] [Google Scholar]
- 84.Gahlot S, Singh J. Caenorhabditis elegans neuronal RNAi does not render other tissues refractory to RNAi. Proc Natl Acad Sci USA. 2024;121: e2401096121. doi: 10.1073/pnas.2401096121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, et al. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature. 2019;571: 63–71. doi: 10.1038/s41586-019-1352-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishida Y, Sugi T, Nonomura M, Mori I. Identification of the AFD neuron as the site of action of the CREB protein in Caenorhabditis elegans thermotaxis. EMBO Rep. 2011;12: 855–862. doi: 10.1038/embor.2011.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74: 515–527. doi: 10.1016/0092-8674(93)80053-h [DOI] [PubMed] [Google Scholar]
- 88.Chou JH, Bargmann CI, Sengupta P. The Caenorhabditis elegans odr-2 gene encodes a novel Ly-6-related protein required for olfaction. Genetics. 2001;157: 211–224. doi: 10.1093/genetics/157.1.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi JI, Lee HK, Kim HS, Park SY, Lee TY, Yoon K-H, et al. Odor-dependent temporal dynamics in Caenorhabitis elegans adaptation and aversive learning behavior. PeerJ. 2018;6: e4956. doi: 10.7717/peerj.4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoshida K, Hirotsu T, Tagawa T, Oda S, Wakabayashi T, Iino Y, et al. Odour concentration-dependent olfactory preference change in C. elegans. Nat Commun. 2012;3: 739. doi: 10.1038/ncomms1750 [DOI] [PubMed] [Google Scholar]
- 91.Luo L, Gabel CV, Ha H-I, Zhang Y, Samuel ADT. Olfactory behavior of swimming C. elegans analyzed by measuring motile responses to temporal variations of odorants. J Neurophysiol. 2008;99: 2617–2625. doi: 10.1152/jn.00053.2008 [DOI] [PubMed] [Google Scholar]
- 92.Kunitomo H, Sato H, Iwata R, Satoh Y, Ohno H, Yamada K, et al. Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt concentration chemotaxis in Caenorhabditis elegans. Nat Commun. 2013;4: 2210. doi: 10.1038/ncomms3210 [DOI] [PubMed] [Google Scholar]
- 93.Nussbacher JK, Tabet R, Yeo GW, Lagier-Tourenne C. Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron. 2019;102: 294–320. doi: 10.1016/j.neuron.2019.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84: 524–533. doi: 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Online Mendelian Inheritance in Man, O. MIM 616035. John Hopkins University; 2019 Jun.
- 96.Online Mendelian Inheritance in Man, O. MIM 138140. John Hopkins University; 2023 Dec.
- 97.Online Mendelian Inheritance in Man, O. MIM 600618. John Hopkins University; 2021 Jan.
- 98.Online Mendelian Inheritance in Man, O. MIM 603507. John Hopkins University; 2022 Dec.
- 99.Online Mendelian Inheritance in Man, O. MIM 607175. John Hopkins University; 2009 Dec.
- 100.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80: 155–165. doi: 10.1016/0092-8674(95)90460-3 [DOI] [PubMed] [Google Scholar]
- 101.Wang X, You B, Yin F, Chen C, He H, Liu F, et al. A presumed missense variant in the U2AF2 gene causes exon skipping in neurodevelopmental diseases. J Hum Genet. 2023;68: 375–382. doi: 10.1038/s10038-023-01128-2 [DOI] [PubMed] [Google Scholar]
- 102.Molitor L, Bacher S, Burczyk S, Niessing D. The molecular function of PURA and its implications in neurological diseases. Front Genet. 2021;12: 638217. doi: 10.3389/fgene.2021.638217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312: 1870–1879. doi: 10.1001/jama.2014.14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mani A. Pathogenicity of de novo rare variants: challenges and opportunities. Circ Cardiovasc Genet. 2017;10. doi: 10.1161/CIRCGENETICS.117.002013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Järvelä I, Määttä T, Acharya A, Leppälä J, Jhangiani SN, Arvio M, et al. Exome sequencing reveals predominantly de novo variants in disorders with intellectual disability (ID) in the founder population of Finland. Hum Genet. 2021;140: 1011–1029. doi: 10.1007/s00439-021-02268-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46: 310–315. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47: D886–D894. doi: 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010;6: e1001025. doi: 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99: 877–885. doi: 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jagadeesh KA, Wenger AM, Berger MJ, Guturu H, Stenson PD, Cooper DN, et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016;48: 1581–1586. doi: 10.1038/ng.3703 [DOI] [PubMed] [Google Scholar]
- 111.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7: 248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31: 3812–3814. doi: 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20: 110–121. doi: 10.1101/gr.097857.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lins J, Brock TJ, Hopkins CE, Hart AC. Generation of a C. elegans tdp-1 null allele and humanized TARDBP containing human disease-variants. MicroPubl Biol. 2023;2023. doi: 10.17912/micropub.biology.000693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Illiano P, Lanzo A, Leo D, Paglione M, Zampi G, Gainetdinov RR, et al. A Caenorhabditis elegans model to study dopamine transporter deficiency syndrome. Eur J Neurosci. 2017;45: 207–214. doi: 10.1111/ejn.13366 [DOI] [PubMed] [Google Scholar]
- 116.Zhu B, Mak JCH, Morris AP, Marson AG, Barclay JW, Sills GJ, et al. Functional analysis of epilepsy-associated variants in STXBP1/Munc18-1 using humanized Caenorhabditis elegans. Epilepsia. 2020;61: 810–821. doi: 10.1111/epi.16464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCormick K, Brock T, Wood M, Guo L, McBride K, Kim C, et al. A Gene Replacement Humanization Platform for Rapid Functional Testing of Clinical Variants in Epilepsy-associated STXBP1. BioRxiv. 2021. doi: 10.1101/2021.08.13.453827 [DOI] [Google Scholar]
- 118.Varadi M, Bertoni D, Magana P, Paramval U, Pidruchna I, Radhakrishnan M, et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2024;52: D368–D375. doi: 10.1093/nar/gkad1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cheng J, Novati G, Pan J, Bycroft C, Žemgulytė A, Applebaum T, et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science. 2023;381: eadg7492. doi: 10.1126/science.adg7492 [DOI] [PubMed] [Google Scholar]
- 120.Cagiada M, Jonsson N, Lindorff-Larsen K. Decoding molecular mechanisms for loss of function variants in the human proteome. BioRxiv. 2024. doi: 10.1101/2024.05.21.595203 [DOI] [Google Scholar]
- 121.Jacob A, Ma Y, Nasiri E, Ochani M, Carrion J, Peng S, et al. Extracellular cold inducible RNA-binding protein mediates binge alcohol-induced brain hypoactivity and impaired cognition in mice. Mol Med. 2019;25: 24. doi: 10.1186/s10020-019-0092-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gong P, Liu J, Jiao X, Niu Y, Wang J, Wang X, et al. Novel biallelic loss of EEF1B2 function links to autosomal recessive intellectual disability. Hum Mutat. 2022;43: 299–304. doi: 10.1002/humu.24329 [DOI] [PubMed] [Google Scholar]
- 123.Nguyen LS, Kim H-G, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22: 1816–1825. doi: 10.1093/hmg/ddt035 [DOI] [PubMed] [Google Scholar]
- 124.Gonatopoulos-Pournatzis T, Wu M, Braunschweig U, Roth J, Han H, Best AJ, et al. Genome-wide CRISPR-Cas9 Interrogation of Splicing Networks Reveals a Mechanism for Recognition of Autism-Misregulated Neuronal Microexons. Mol Cell. 2018;72: 510–524.e12. doi: 10.1016/j.molcel.2018.10.008 [DOI] [PubMed] [Google Scholar]
- 125.Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 synthesis is regulated by mTOR signaling pathway. PLoS ONE. 2012;7: e52527. doi: 10.1371/journal.pone.0052527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146: 247–261. doi: 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kleene KC. Y-box proteins combine versatile cold shock domains and arginine-rich motifs (ARMs) for pleiotropic functions in RNA biology. Biochem J. 2018;475: 2769–2784. doi: 10.1042/BCJ20170956 [DOI] [PubMed] [Google Scholar]
- 128.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nussbaum-Krammer CI, Neto MF, Brielmann RM, Pedersen JS, Morimoto RI. Investigating the spreading and toxicity of prion-like proteins using the metazoan model organism C. elegans. J Vis Exp. 2015; 52321. doi: 10.3791/52321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nawa M, Matsuoka M. The Method of the Body Bending Assay Using Caenorhabditis elegans. Bio Protoc. 2012;2. doi: 10.21769/BioProtoc.253 [DOI] [Google Scholar]
- 131.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9: 676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen S, Francioli LC, Goodrich JK, Collins RL, Kanai M, Wang Q, et al. A genome-wide mutational constraint map quantified from variation in 76,156 human genomes. BioRxiv. 2022. doi: 10.1101/2022.03.20.485034 [DOI] [Google Scholar]
- 133.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12: 996–1006. doi: 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8: 275–282. doi: 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- 135.Consortium UniProt. Uniprot: the universal protein knowledgebase in 2023. Nucleic Acids Res. 2023;51: D523–D531. doi: 10.1093/nar/gkac1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stecher G, Tamura K, Kumar S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol. 2020;37: 1237–1239. doi: 10.1093/molbev/msz312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang X, Jiang Q, Song Y, He Z, Zhang H, Song M, Zhang X, Dai Y, Karalay O, Dieterich C, et al. Ageing induces tissue-specific transcriptomic changes in Caenorhabditis elegans. EMBO J. 2022. 41, e109633. doi: 10.15252/embj.2021109633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ange J., Weng Y, Stevenson ME, Kaletsky R, Moore RS, Zhou S, and Murphy CT. Adult Single-nucleus Neuronal Transcriptomes of Insulin Signaling Mutants Reveal Regulators of Behavior and Learning. BioRxiv. 2024. doi: 10.1101/2024.02.07.579364 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Boxes signify 3+ RBPs in the same protein family or class (blue box include PUF RBPs, orange box includes CEY RBPs, grey box includes translation initiation machinery). Box and whisker plots are shown for the learning timepoints from the STM/ITM assays for each RBP screened. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. ns, not significant (p>0.05).
(TIF)
All RBPs in the screen are shown grouped by experiment. Bar represents mean. Whiskers mark the standard error of the mean. n = 10 per RNAi treatment. ns, not significant.
(TIF)
All RBPs are shown where pink circles are decreased memory and blue circles are increased memory. The size of circle represents the p value from a combined n ≥ 5–10 per RNAi treatment.
(TIF)
All RBPs with significantly altered memory are shown divided by memory timepoint. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. n ≥ 5–10 per RNAi treatment. *p<0.05,**p<0.01, ***p<0.001, ****p<0.0001.
(TIF)
While cey-1 and cey-4 have broad expression, cey-2 is only located in eight neurons at L4.
(TIF)
(A) Neuron-specific and/or single-cell RNA-seq data compiled from five different publications suggest cey-1 mRNA is expressed in the adult nervous system [24, 35, 38, 43, 80, 137, 138]. (B) Diagram of microscopy images shown in (C). C. elegans head is labeled including the location of the pharynx and main neuronal ganglia/nerve rings. (C) A transcriptional cey-1 reporter suggests the gene is broadly expressed in the neurons in the head at baseline conditions. Representative image of Day 2 adult worms with RFP-labeled neuronal nuclei (unc-11(prom8)::2xNLS::TagRFP)) pseudocolored magenta and a nuclear GFPH2B cey-1 promoter fusion (cey1p::GFPH2B) pseudocolored cyan show colocalization.
(TIF)
(A) Naïve battery of both negative and positive odors reveals deficits in both nonanol and isoamyl alcohol chemotaxis, but not in benzaldehyde, pyrazine, or diacetyl chemotaxis at attractive concentrations. (B) Baseline chemosensation for butanone at neutral concentration of 10% is unaffected by neuron-specific knockdown of cey-1. n = 15 per RNAi treatment. (C) Baseline chemosensation for butanone at an attractive concentration of 0.1% is unaffected by neuron-specific knockdown of cey-1. n = 15 per RNAi treatment. (D) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by neuron-specific loss of cey-1. n = 15 per RNAi treatment. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. **p<0.01, ****p<0.0001. ns, not significant (p>0.05). (E) Representative images of adult-only, neuron-specific knockdown RNAi treatment (vector or cey-1 RNAi) in neuronally RNAi-sensitized mNG::CEY-1 animals. (F) Quantification of total fluorescent intensity. n = 26 for each RNAi condition. *p<0.05.
(TIF)
(A) Naïve chemotaxis towards 10% butanone prior to behavioral conditioning does not significantly differ between wild-type animals, cey-1(ok1805), and cey-1(rrr12) mutants. n = 15 per genotype. (B) Baseline chemosensation for butanone at an attractive concentration of 0.1% is unaffected by whole-body loss of cey-1. n = 15 per genotype. (C) Baseline chemosensation for butanone at an attractive concentration of 0.1% is unaffected by neuron-specific rescue of cey-1. n = 15 per genotype. (D) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by whole-body loss of cey-1. n = 15 per genotype. (E) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by neuron-specific rescue of cey-1. n = 15 per genotype. (F) Day 2 adult speed measured in μm/sec by Wrmtrck. n = 20–30 worms per genotype, Tracks were thresholded at 15 seconds duration, with multiple tracks per worm possible. ns, not significant. (G) Day 2 average track distance (in μm) traveled as measured by Wrmtrck. n = 20–30 worms per genotype, Tracks were thresholded at 15 seconds duration, with multiple tracks per worm possible. ns, not significant. Of note, for both F and G, wild-type controls for this figure are the same as S11 Fig because experiments were performed the same day each time; data was put into two separate graphs for organization. (H) Day 2 thrashing comparing number of body bends per 30 seconds of wild-type, cey-1(rrr12) knockout worms, and nervous system specific rescue worms (cey-1(rrr12);rab-3p::CEY-1) shows that cey-1 mutants do not have thrashing deficits, and indeed exhibit more body bends per minute. n = 50 for each genotype. (I) Associative memory curve comparing wild-type, cey-1(rrr12) knockout worms, and nervous system specific rescue worms (cey-1(rrr12);rab-3p::CEY-1) shows that while knockouts have no associative learning or memory, neuron-specific rescue of cey-1 allows for learning and memory equivalent to wild-type levels. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. ns, not significant (p>0.05).
(TIF)
(A) Percentages of individuals reported with gain or loss CNVs that include YBX1. (B) Percentages of mechanisms of inheritance of gain and loss CNVs that include YBX1. (C) Size of CNV gain and losses that include YBX1. (D) Predictive scores for YBX1 from gnomAD v.2.11 [132] suggest that YBX1 is intolerant to loss of function variants and is haploinsufficient. (E) Phenotypes of patients specifically with deletion/loss CNVs including YBX1 include epicanthus, delayed speech and development, intellectual disability, and other neurological features.
(TIF)
(A) Percentages of individuals reported with gain or loss CNVs that include YBX3. (B) Percentages of mechanisms of inheritance of gain and loss CNVs that include YBX3. (C) Size of CNV gain and losses that include YBX3. (D) Predictive scores for YBX3 from gnomAD v.2.11 [132] suggest that YBX3 is tolerant to loss of function variants and is haplosufficient, suggesting variants may instead be deleterious by being dominant negative or gain-of-function. (E) Phenotypes of patients specifically with deletion/loss CNVs including YBX3, primarily intellectual disability, low-set ears, micrognathia, and other neurological features.
(TIF)
(A) Baseline chemosensation for butanone at an attractive concentration of 0. 1% is unaffected by the p.Asn56Tyr variant in cey-1. n = 15 per genotype. (B) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by p.Asn56Tyr variant in cey-1. n = 15 per genotype. Box and whisker plot: the center line denotes the median value (50th percentile) while the box contains the 25th to 75th percentiles. Whiskers mark the 5th and 95th percentiles. ns, not significant (p>0.05). (C) qRT-PCR of cey-1 mRNA levels in Day 2 adults shows that while cey-1 knockouts (cey-1(rrr12)) worms undergo nonsense mediated decay [56] resulting in reduced cey-1 mRNA levels, introduction of the p.Asn56Tyr variant has no significant effect on cey-1 expression. n = 6 per genotype. **p<0.01. (D) Day 2 thrashing comparing number of body bends per 30 seconds of wild-type, neuron-specific CEY-1 overexpression (cey-1(WT);rab-3p::CEY-1), and variant worms with p.Asn56Tyr in endogenous CEY-1 (cey-1(p.Asn127Tyr) shows that cey-1 mutants do not have thrashing deficits, and indeed exhibit more body bends per minute. n = 50 for each genotype. Of note, wild-type controls for this figure are the same as S8 Fig because experiments were performed the same day each time; data was put into two separate graphs for organization. (E) Day 2 adult speed measured in μm/sec by Wrmtrck. n = 20–30 worms per genotype, Tracks were thresholded at 15 seconds duration, with multiple tracks per worm possible. ns, not significant. (F) Day 2 average track distance (in μm) traveled as measured by Wrmtrck. n = 20–30 worms per genotype, Tracks were thresholded at 15 seconds duration, with multiple tracks per worm possible. ns, not significant. Of note, for both E and F, wild-type controls for this figure are the same as S8 Fig because experiments were performed the same day each time; data was put into two separate graphs for organization.
(TIF)
(A) Associative memory curve comparing wild-type, cey-1 knockouts, and humanized lines expressing either YBX1 or YBX3 at the endogenous cey-1 locus. While worms expressing YBX3 have normal memory performance, those expressing YBX1 appear to have extended memory, as they have a strong association for butanone even two hours post-training. (B) Baseline chemosensation for butanone at an attractive concentration of 0.1% is normal in worms expressing YBX1. n = 15 per genotype. (C) Motility, measured as proportion of worms at the origin of a chemotaxis plate, is unaffected by YBX1. n = 15 per genotype. (D) Baseline chemosensation is normal in worms expressing YBX3 at the cey-1 locus. n = 15 per genotype. (E) Motility is unaffected by YBX3. n = 15 per genotype. (F) Baseline chemosensation is unaffected by expressing YBX3(p.Asn127Tyr) at the cey-1 locus. n = 15 per genotype. (G) Motility is unaffected by YBX3(p.Asn127Tyr). n = 15 per genotype. (H) qRT-PCR of cey-1 and YBX mRNA levels in Day 2 adults shows that levels of YBX1, YBX3, and YBX3(pAsn127Tyr) are not significantly reduced compared to wild-type cey-1 mRNA levels and therefore, introduction of the p.Asn127Tyr variant has no significant effect on expression. n = 5 per genotype. ns, not significant.
(TIF)
(A) Forward-facing view of the protein with the β sheet that binds RNA facing the viewer. (B) Top-down view of the YBX3 protein depicts the location of the Asn127 residue.
(TIF)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.