Abstract
The zinc transporter Slc30a1 plays an essential role in maintaining cellular zinc homeostasis. Despite this, its functional role in macrophages remains largely unknown. Here, we examine the function of Slc30a1 in host defense using mice models infected with an attenuated stain of Salmonella enterica Typhimurium and primary macrophages infected with the attenuated Salmonella. Bulk transcriptome sequencing in primary macrophages identifies Slc30a1 as a candidate in response to Salmonella infection. Whole-mount immunofluorescence and confocal microscopy imaging of primary macrophage and spleen from Salmonella-infected Slc30a1flag-EGFP mice demonstrate Slc30a1 expression is increased in infected macrophages with localization at the plasma membrane and in the cytosol. Lyz2-Cre-driven Slc30a1 conditional knockout mice (Slc30a1fl/fl;Lyz2-Cre) exhibit increased susceptibility to Salmonella infection compared to control littermates. We demonstrate that Slc30a1-deficient macrophages are defective in intracellular killing, which correlated with reduced activation of nuclear factor kappa B and reduction in nitric oxide (NO) production. Notably, the model exhibits intracellular zinc accumulation, demonstrating that Slc30a1 is required for zinc export. We thus conclude that zinc export enables the efficient NO-mediated antibacterial activity of macrophages to control invading Salmonella.
Research organism: Mouse
Introduction
Salmonella, the gram-negative bacterium that causes Salmonellosis, is a well-characterized and relatively common foodborne disease. According to the 2015 Global Burden of Disease Study, Salmonella infection is one of the leading causes of diarrhea-related death, particularly among children under 5 years of age (Troeger et al., 2017). Severe cases of Salmonella septicemia are associated with a 25% mortality rate (Feasey et al., 2012). Based on epidemiological reports, zinc deficiency has been linked to the incidence of Salmonella infection (Troeger et al., 2017). As an essential micronutrient, zinc is involved in a wide range of fundamental biological processes, including modulating the immune system’s response to invading pathogens (Wessels et al., 2021). However, whether zinc plays a protective role in response to Salmonella infection remains unclear.
In mammalian cells, zinc homeostasis is tightly regulated by two families of zinc transporters, namely solute carrier family 30 (Slc30a, also known as ZnTs) and solute carrier family 39 (Slc39a, also known as ZIPs), which export and import cellular zinc, respectively. Zinc imbalance caused by the dysfunction of zinc transporters contributes to impaired hematopoiesis and macrophage survival (He et al., 2023; Gao et al., 2017). Slc30a1 (ZnT1) is ubiquitously expressed in mammalian cells and is localized primarily at the plasma membrane (Nishito and Kambe, 2019; Golan et al., 2015; Guo et al., 2010), where it enables the export of cytoplasmic zinc to the extracellular space (Shusterman et al., 2017; Kambe et al., 2015; Kimura and Kambe, 2016). In mice, loss of Slc30a1 causes early embryonic death (Andrews et al., 2004), suggesting that Slc30a1 is essential for early development and survival. Recent data provide evidence that Slc30a1 expression is induced by a wide range of pathogens, including bacteria (Botella et al., 2011; Stocks et al., 2021), fungi (Rossi et al., 2021), and viruses (Moskovskich et al., 2019).
Macrophages serve as the front line in the innate immune system by detecting and eliminating foreign pathogens. When Salmonella invades tissues, the bacteria are rapidly engulfed and killed by resident macrophages. This rapid engulfment and killing of Salmonella by macrophages result in bacterial clearance, cytokine production, and the recruitment of polymorphonuclear phagocytes (Mastroeni et al., 2009). Despite evidence suggesting a link between zinc regulators and macrophage function (Gao et al., 2018), whether and how Slc30a1 regulates macrophage function during infection remains poorly understood.
Here, we perform transcriptome sequencing analysis to identify Slc30a1 as an important host factor in Salmonella-infected primary macrophages. We then used Slc30a1flag-EGFP reporter mice to assess subcellular localization and cell-specific expression of Slc30a1 at the protein level. To study the role of Slc30a1 in macrophages, we generate Slc30a1fl/fl;Lyz2-Cre (Slc30a1 cKO) mice and induce Salmonella infection. Lastly, we used primary macrophages differentiated from the bone marrow of Slc30a1 cKO mice to directly investigate the killing capacity and the antimicrobial response of macrophages to Salmonella infection. Our findings provide interesting new insights into the role of Slc30a1 in manipulating cellular zinc homeostasis to support an antimicrobial response in macrophages in response to infectious diseases.
Results
Slc30a1 expression is induced in mouse macrophages upon Salmonella infection
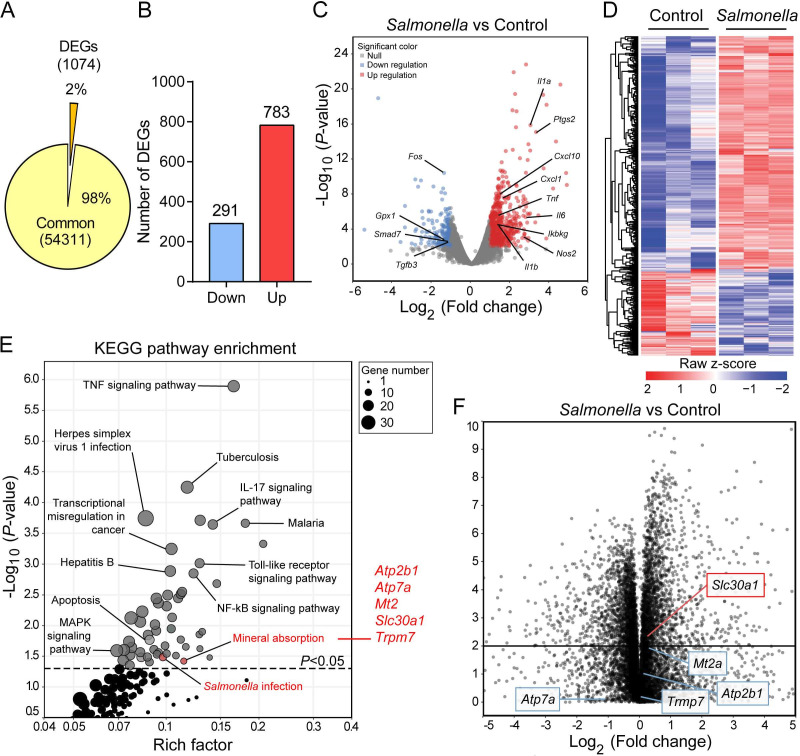
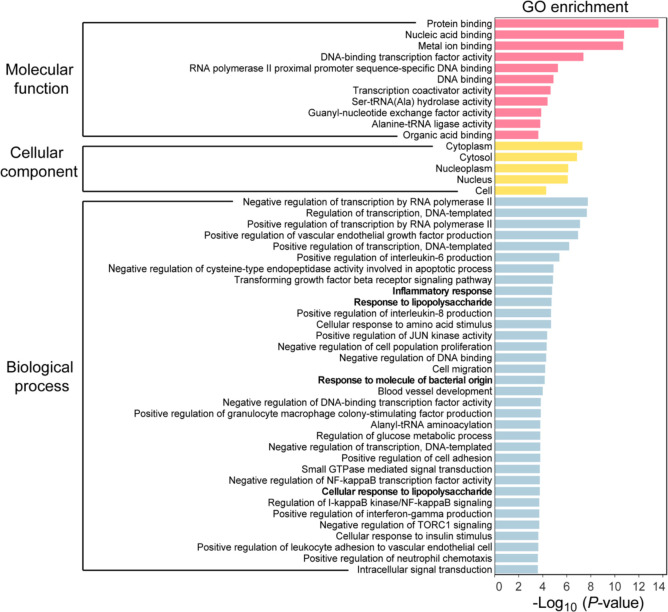
We performed RNA sequencing (RNA-seq) on bone marrow-derived macrophages (BMDMs) that were isolated from wild-type C57BL/6 mice and then exposed to an attenuated stain of Salmonella enterica Typhimurium with a multiplicity of infection (MOI) of 1 for 2 hr, the approximate time that has been closely correlated with the initial replication of Salmonella in macrophages (Helaine et al., 2010). Our analysis revealed a total of 1074 differentially expressed genes (DEGs), defined as an absolute log2 fold change >1 and an adjusted p value of <0.05 compared to control-treated BMDMs (Figure 1A, B). Among these 1074 genes, significantly downregulated genes were enriched in anti-inflammatory pathways, including Gpx1, Smad7, and Tgfb3, and significantly upregulated genes were enriched in infectious and inflammatory pathways, including Tnf, Il6, Il1β, Ptgs2, and Nos2, suggesting a potent antimicrobial response in infected macrophages (Figure 1C); as shown in a heatmap, many genes were upregulated following Salmonella infection (Figure 1D). Gene ontology (GO) analysis of the DEGs revealed that the top 50 GO terms generally contributed to proinflammatory signaling, inflammatory cytokines, and responses to molecules of bacterial origin (Figure 1—figure supplement 1 and Supplementary file 1), while Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of these DEGs yielded 54 pathways (p < 0.05) involved largely in infectious diseases (Figure 1E and Supplementary file 2).
Figure 1. Slc30a1 expression is upregulated in Salmonella-infected macrophages.
(A) Pie chart summarizing the percentage of unchanged genes and differentially expressed genes (DEGs) in wild-type (WT) bone marrow-derived macrophages (BMDMs) 2 h after Salmonella infection (multiplicity of infection [MOI] = 1) (n = 3) using RNA sequencing (RNA-seq) analysis. (B) Bar graph displaying the number of DEGs including up- and downregulated genes identified by p < 0.05 and an absolute log2 fold change >1; shown at the right is the total number of down- and upregulated genes. (C) Volcano plot displaying the fold change in gene expression and corresponding p values for Salmonella-infected cells versus control (uninfected) cells. (D) Heatmap showing the pattern of DEGs in BMDMs measured between Salmonella-infected cells and control cells. (E) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathways (p < 0.05) of DEGs between Salmonella-infected cells and control cells. (F) Schematic diagram (left) illustrating the use of RNA-seq analysis on human monocyte-derived macrophages (MDMs) infected with Salmonella for 4 hr and volcano plot (right) of DEGs in Salmonella-infected MDMs based on a previously published dataset (GSE67427); the Atp2b1, Atp7a, Mt2, Slc30a1, and Trpm7 genes are indicated.
Figure 1—figure supplement 1. Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs) in bone marrow-derived macrophages (BMDMs) with or without Salmonella infection.
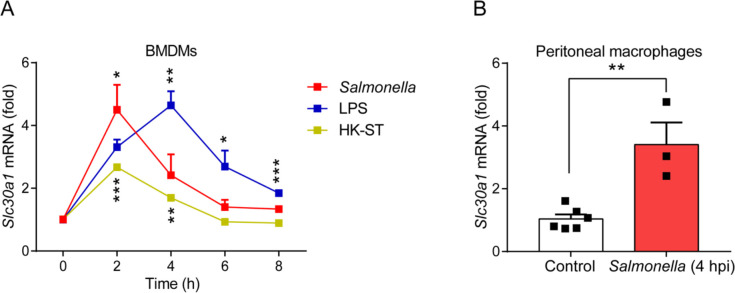
Figure 1—figure supplement 2. The gene expression of Slc30a1 in macrophages is induced by Salmonella infection.
Using KEGG pathway analysis, we identified mineral absorption as a predominant pathway, with upregulation of the Atp2b1, Atp7a, Mt2, Slc30a1, and Trpm7 genes. To narrow down the list of genes, we analyzed publicly available bulk RNA-seq data (GSE67427) obtained from human monocyte-derived macrophages that were infected for 4 hr with Salmonella typhimurium and found that the DEGs in our mouse RNA-seq dataset were also differentially regulated in human macrophages (Figure 1F). Importantly, Slc30a1 was ranked as the most significantly upregulated gene and was therefore selected for further study. To validate the RNA-seq data, we then used RT-qPCR(Real-time quantitative polymerase chain reaction) to measure Slc30a1 mRNA levels in BMDMs at various time points following exposure to attenuated Salmonella, lipopolysaccharide (LPS), or heat-killed Salmonella typhimurium (HK-ST). We found that Slc30a1 expression peaked at 2 and 4 hr in the Salmonella- and LPS-treated cells, respectively, while HK-ST produced a significantly smaller response that also peaked at 2 hr (Figure 1—figure supplement 2A). To validate this in vitro finding in vivo, we gave C57BL/6 mice an intraperitoneal (i.p.) injection of a sublethal dose of attenuated Salmonella (1 × 105 CFU per mouse); at 4 hr post-infection (4 hpi), Slc30a1 expression was significantly higher in peritoneal macrophages compared to uninfected mice (Figure 1—figure supplement 2B). Together, these data show that Slc30a1 is upregulated in macrophages during Salmonella infection.
Macrophages in Salmonella-infected Slc30a1 reporter mice have increased levels of Slc30a1 protein
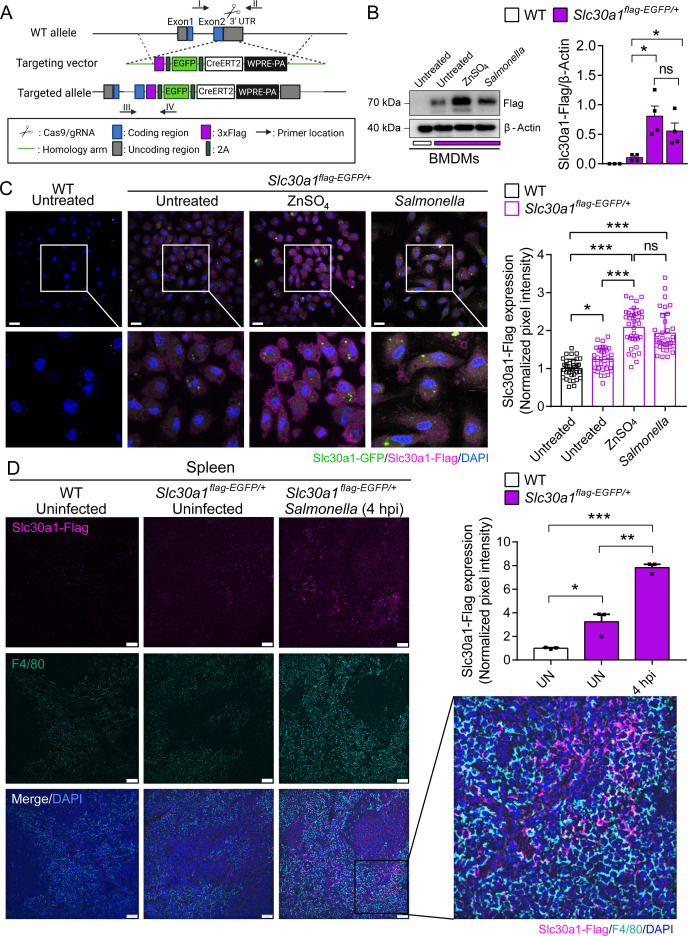
To detect the levels of Slc30a1 protein in macrophages following Salmonella infection, we generated a Slc30a1 reporter mouse line expressing 3xFlag-2A-EGFP-2A-CreERT2-Wpre-pA under the control of the Slc30a1 promoter (Figure 2A) and studied heterozygous offspring mice (Slc30a1flag-EGFP/+). BMDMs were then isolated from Slc30a1flag-EGFP/+ mice and either infected with attenuated Salmonella (MOI = 1) or treated with ZnSO4 (40 µM) for 4 hr, followed by western blot analysis using an anti-flag antibody. Compared to untreated WT and untreated Slc30a1flag-EGFP/+ BMDMs, both the Salmonella-infected and ZnSO4-treated Slc30a1flag-EGFP/+ BMDMs had significantly higher levels of the reporter protein (Figure 2—source data 1). Given that Slc30a1 localizes primarily to the plasma membrane (Nishito and Kambe, 2019; Golan et al., 2015; Guo et al., 2010), we examined the localization of Slc30a1 in Salmonella-infected and ZnSO4-treated cells using confocal microscopy. As expected, we found that ZnSO4 increased the levels of the Slc30a1-flag reporter, primarily in the plasma membrane (Figure 2C). Interestingly, however, the Slc30a1-flag reporter was present both in the cytosol and in the plasma membrane in Salmonella-infected cells, suggesting that Slc30a1 may contribute to both zinc efflux and intracellular zinc movement in response to Salmonella infection.
Figure 2. Characterization of a Slc30a1 reporter mouse with or without Salmonella infection.
(A) Strategy for generating an Slc30a1 reporter mouse line expressing 3xFlag-EGFP under the control of the endogenous Slc30a1 promotor (Slc30a1flag-EGFP/+). (B) Western blot analysis of Slc30a1-flag in bone marrow-derived macrophages (BMDMs) isolated from Slc30a1flag-EGFP/+mice and exposed to ZnSO4 treatment (40 µM) or Salmonella (multiplicity of infection [MOI] = 1) for 4 hr. (C) Confocal fluorescence images of BMDMs treated as shown in B and immunostained using an anti-flag antibody (magenta); the nuclei were counterstained with DAPI(4′,6-diamidino-2-phenylindole) (blue), and GFP(Green fluorescent protein) was visualized directly based on green fluorescence. Scale bars, 20 µm. Shown at the right is the summary of the normalized pixel intensity of Slc30a1-flag expression in BMDMs in each group. (D) Confocal fluorescence images of spleen samples obtained from a Salmonella-infected mouse at 4 hpi; also shown are samples of uninfected WT and Slc30a1flag-EGFP/+ mice. The samples were stained with anti-flag (magenta) and anti-F4/80 (cyan), and the nuclei were counterstained with DAPI (blue). Scale bars, 50 µm. Shown at the right is the summary of the normalized pixel intensity of Slc30a1-flag expression in the spleen in each group. Data are presented as mean ± SEM(Standard Error of the Mean). p values were determined using two-tailed unpaired Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, and ns, not significant.
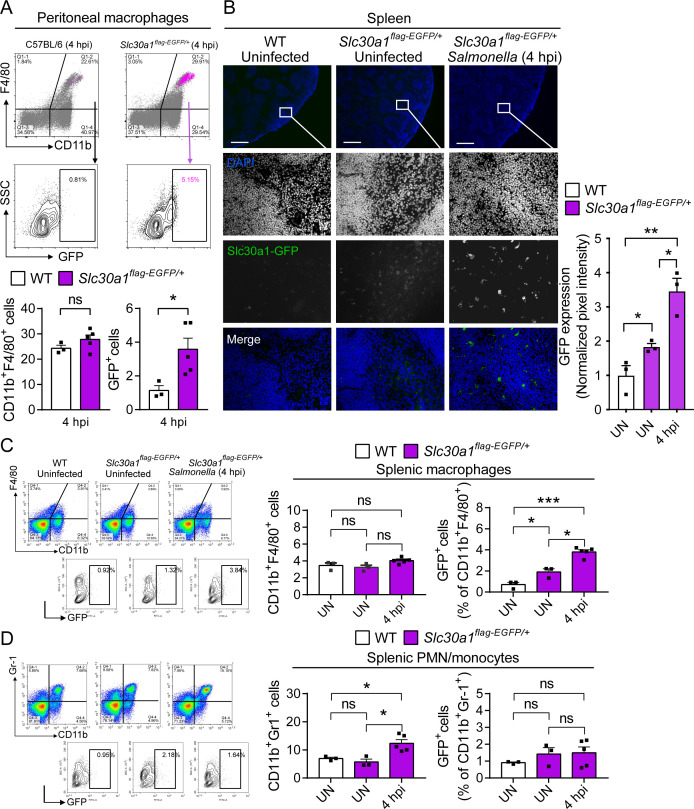
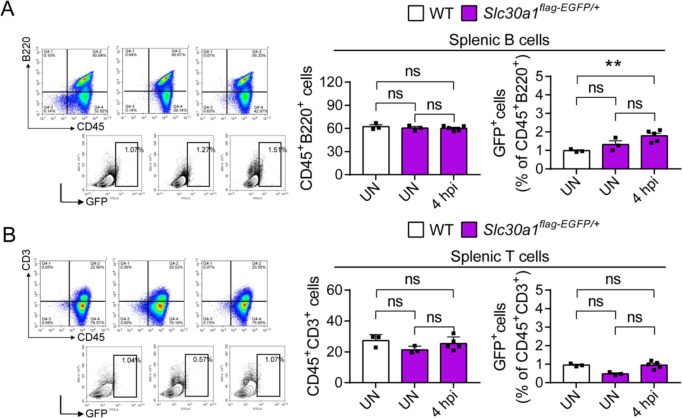
To examine the in vivo expression of Slc30a1, we infected Slc30a1flag-EGFP/+ mice with an i.p. injection of attenuated Salmonella (1 × 105 CFU per mouse). Given that the spleen is the largest secondary lymphoid organ (Lewis et al., 2019) and is a major source of bacterial burden during systemic infection (Carreno et al., 2021), we dissected the spleen 4 hr after infection and examined Slc30a1-flag expression in splenic macrophages. Immunostaining for Slc30a1-flag and the macrophage biomarker F4/80 revealed that Slc30a1 is expressed at high levels, specifically in splenic F4/80‒positive cells at the area of red pulp in infected mice, but not in uninfected mice (Figure 2D). Moreover, Salmonella-induced expression of Slc30a1 was confirmed by measuring high GFP expression in CD11b+F4/80+ peritoneal macrophages (Figure 3A) and splenic macrophages (Figure 3B, C), while there was no GFP expression in other splenic immune cells (Figure 3—figure supplement 1).
Figure 3. Induction of Slc30a1 expression in macrophages of a Slc30a1 reporter mouse upon Salmonella infection.
(A) FACS(Fluorescence-activated cell sorting) plots of GFP expression in CD11b+F4/80+ macrophages isolated from the peritoneal cavity of Salmonella-infected WT mice and Salmonella-infected Slc30a1flag-EGFP/+ mice at 4 hpi (n = 3–5 mice/group). (B) Fluorescence microscopy images of spleen samples obtained from a Salmonella-infected mouse at 4 hpi; also shown are samples of uninfected WT and Slc30a1flag-EGFP/+ mice. The green region indicated the Slc30a1-GFP expression, and the blue region indicated the nuclei staining by DAPI. Scale bar, 500 μm. FACS plots of GFP expression in CD11b+F4/80+ splenic macrophages (C) and CD11b+Gr-1+ splenic PMN/monocyte (D) isolated from the spleens of uninfected WT mice, uninfected Slc30a1flag-EGFP/+ mice, and Salmonella-infected Slc30a1flag-EGFP/+ mice at 4 hpi (n = 3–5 mice/group). Shown at the right is the summary of the percentage of CD11b+F4/80+ splenic macrophages and CD11b+Gr-1+ splenic PMN/monocyte in each group. Data in this figure are represented as mean ± SEM. p values were determined using two-tailed unpaired Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, and ns, not significant.
Figure 3—figure supplement 1. Proportion of splenic white blood cells in Slc30a1flag-EGFP/+ after Salmonella infection.
Lyz2-Cre-driven Slc30a1 conditional knockout mice have increased susceptibility to Salmonella infection
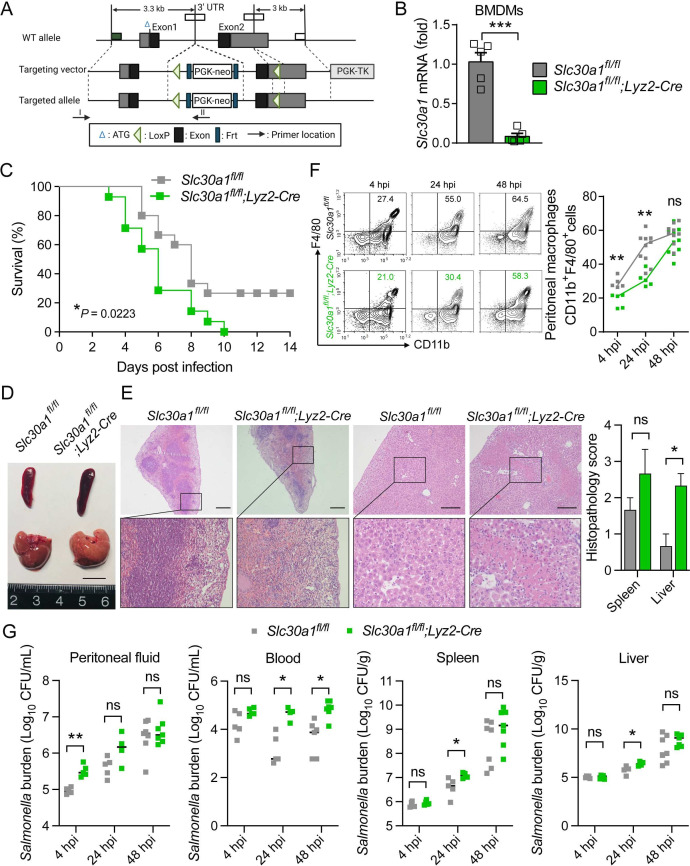
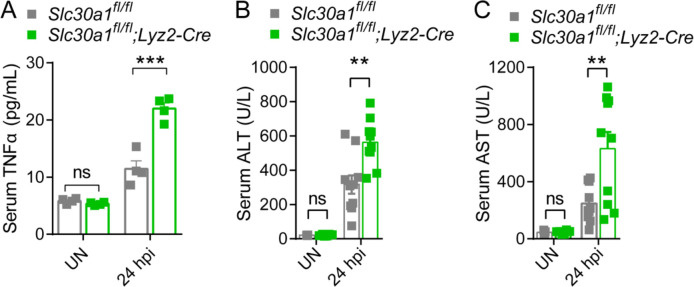
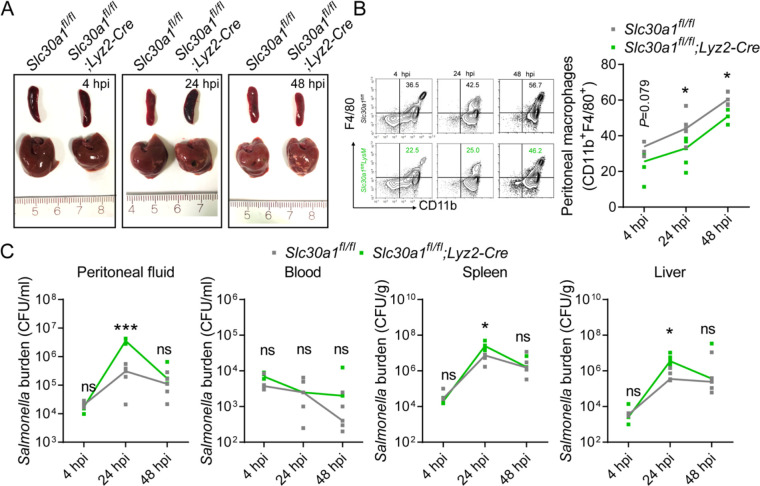
To examine the role of Slc30a1 in macrophages with respect to protecting against Salmonella infection, we generated the conditional knockout mice with a loss-of-function allele of Slc30a1 in myeloid cells including macrophages by crossing mice carrying a floxed Slc30a1 allele (Slc30a1fl/fl) with Lyz2-Cre recombinase mice (Figure 4A); we then used the homozygous floxed mice with heterozygous Cre (Slc30a1fl/fl;Lyz2-Cre or Slc30a1 cKO(Conditional knockout)), with homozygous floxed littermates lacking Cre (Slc30a1fl/fl) serving as a control group. Loss of Slc30a1 expression was confirmed in Slc30a1 cKO BMDMs, with a more than 95% reduction in Slc30a1 mRNA levels compared to Slc30a1fl/fl cells (Figure 4B). Next, we injected attenuated Salmonella (1 × 105 CFU per mouse) into male Slc30a1 cKO and Slc30a1fl/fl littermates; we then monitored survival for 2 weeks and sacrificed a subgroup of mice 4–48 hpi. We found that the Salmonella-infected Slc30a1 cKO mice had significantly lower survival compared to Salmonella-infected Slc30a1fl/fl mice (Figure 4C). At 24 hpi, we observed severe tissue damage in the spleen (i.e., deformation of white pulp tissue) and liver (i.e., necrotic tissue) of Slc30a1 cKO mice when compared with Slc30a1fl/fl mice (Figure 4D, E), as well as higher levels of serum TNFα, alanine aminotransferase (ALT), and aspartate aminotransferase (AST) compared to Slc30a1fl/fl mice (Figure 4—figure supplement 1). Interestingly, we also found that the Salmonella-infected Slc30a1 cKO mice contained a smaller proportion of CD11b+F4/80+ peritoneal macrophages compared to Salmonella-infected Slc30a1fl/fl mice (Figure 4F). In addition, we found significantly fewer neutrophils and monocytes in blood samples obtained at 24 hpi from infected Slc30a1 cKO mice compared to controls without interruption of other blood parameters (Supplementary file 3), possibly due to the reported expression of Lyz2-Cre in neutrophils (Clausen et al., 1999).
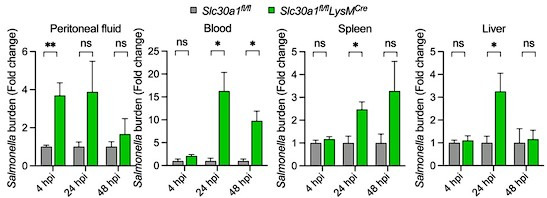
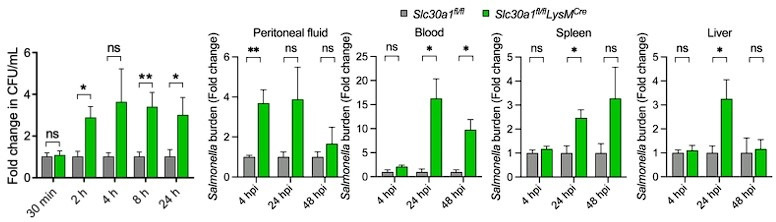
Figure 4. Lyz2-Cre-driven Slc30a1 conditional knockout mice are highly susceptible to Salmonella infection.
(A) Strategy for generating floxed Slc30a1 mice. Crossing this mouse with a heterozygous Lyz2-Cre mouse produces Slc30a1fl/fl;Lyz2-Cre and Slc30a1fl/fl littermates. (B) RT-qPCR analysis of relative Slc30a1 mRNA levels in bone marrow-derived macrophages (BMDMs) isolated from Slc30a1fl/fl;Lyz2-Cre and Slc30a1fl/fl mice (n = 5). (C) Kaplan–Meier survival curve of Salmonella-infected mice (n = 14–15 mice/group). (D, E) Gross and hematoxylin and eosin (H&E)-stained images of spleen and liver obtained from Salmonella-infected mice at 24 hpi. Scale bars, 1 cm, 5 mm and 10 mm, respectively. (F) FACS plots (left) and summary (right) of CD11b+F4/80+ peritoneal macrophages obtained from Salmonella-infected mice at 4, 24, and 48 hpi (n = 5–10 mice/group). (G) Summary of Salmonella CFUs measured in the peritoneal fluid, blood, spleen, and liver of Salmonella-infected mice at 4, 24, and 48 hpi (n = 3–5 mice/group). Data in this figure are represented as mean ± SEM. p values of survival in D were determined using Log-rank test, in B and F–K using two-tailed unpaired Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, and ns, not significant.
Figure 4—figure supplement 1. Elevated levels of proinflammatory cytokine and injury markers in the serum of Slc30a1fl/fl;Lyz2-Cre mice.
Figure 4—figure supplement 2. Salmonella infection leads to significantly decreased macrophages and increased tissue bacterial burden in Slc30a1fl/fl;Lyz2-Cre mice.
We also quantified Salmonella burden at 4, 24, and 48 hpi and found significantly higher numbers of bacterial cells in the peritoneal cavity, blood, spleen, and liver of infected Slc30a1 cKO mice compared to control mice, particularly at 24 hpi (Figure 4G), indicating severe systemic infection. Furthermore, we performed in vivo infection studies in a separate group of mice using a non-lethal dose of attenuated Salmonella (1 × 104 CFU per mouse) and found the same pattern of tissue damage (Figure 4—figure supplement 2A), peritoneal macrophage recruitment (Figure 4—figure supplement 2B), and bacterial burden (Figure 4—figure supplement 2C). Together, these in vivo data provide compelling evidence that Slc30a1 in macrophages may play an important protective role against Salmonella infection.
Loss of Slc30a1 in macrophages reduces bactericidal capacity by reducing iNOS(Inducible nitric oxide synthase) and nitric oxide production
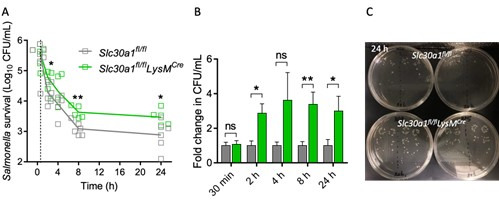
Killing intracellular microbes is a key biological function of macrophages (Flannagan et al., 2009). To study whether Slc30a1 mediates this function in macrophages, we isolated BMDMs from Slc30a1 cKO and Slc30a1fl/fl mice and exposed the cells to attenuated Salmonella (MOI = 10) in vitro for 24 hr. We found that Slc30a1 cKO BMDMs contained a significantly higher intracellular bacterial load compared to control cells at both 8 and 24 hr (Figure 5A). Moreover, examining the cells at high magnification using transmission electron microscopy revealed increased numbers of bacterial cells in the phagosomes of Slc30a1 cKO BMDMs at 24 hr compared to control cells (Figure 5B).
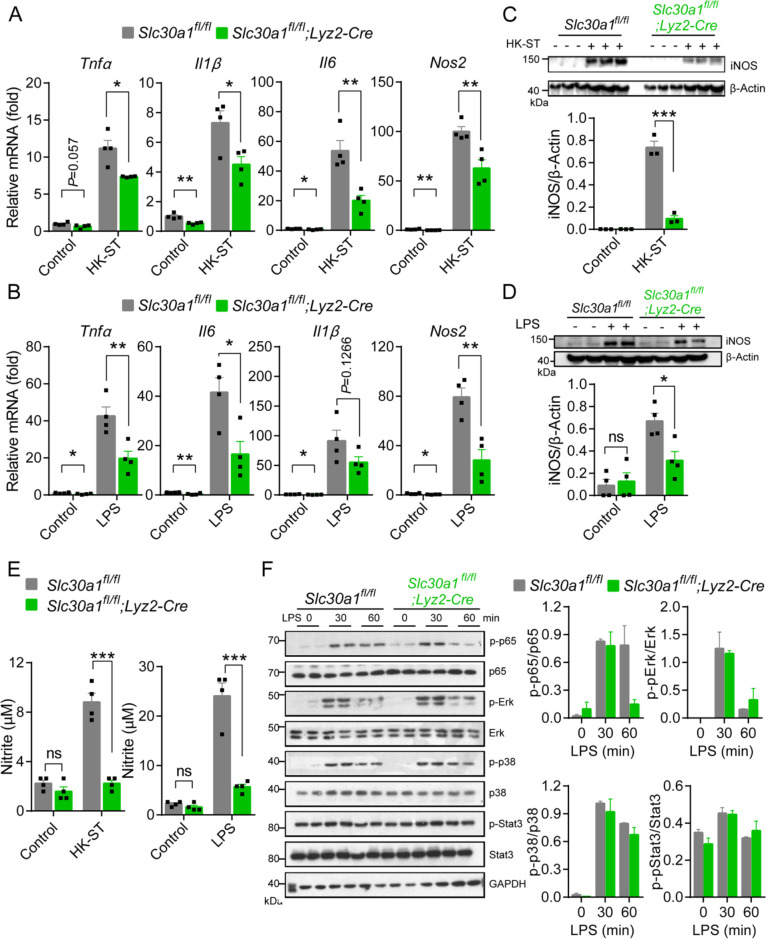
Figure 5. Slc30a1 is required for iNOS/nitric oxide (NO) production and intracellular pathogen-killing capacity of macrophages in response to Salmonella infection.
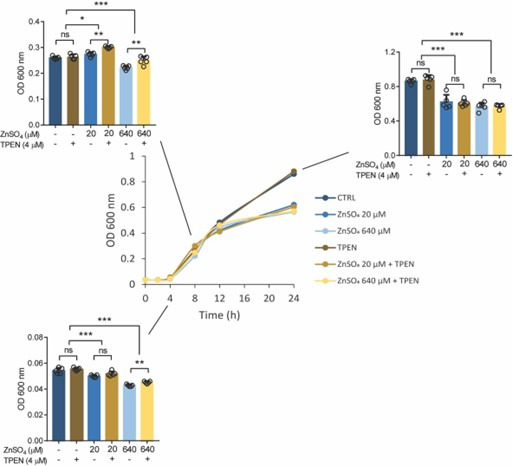
(A) Time course of the intracellular pathogen-killing capacity of Salmonella-infected Slc30a1fl/fl;Lyz2-Cre and Slc30a1fl/fl bone marrow-derived macrophages (BMDMs) measured in colony-forming units per ml (n = 5). (B) Transmission electron microscopy images of Salmonella-infected BMDMs at 24 hpi. Red arrows indicate bacterial-containing phagosomes, and the insets show magnified images of bacterial engulfment. Scale bars, 5 µm. (C–E) RT-qPCR analysis of mRNAs encoding inflammatory cytokines (Tnfα, Il1β, and Il6), chemokines (Ccr2, Ccl2, Cxcl1, and Cxcl10) and macrophage biomarkers (Nos2 and Arg1) in BMDMs measured at the indicated times after Salmonella infection (multiplicity of infection [MOI] = 1) (n = 5). (F) Western blot analysis and summary of iNOS protein in Slc30a1fl/fl;Lyz2-Cre and Slc30a1fl/fl BMDMs either untreated or stimulated with Salmonella for 24 hr (n = 3). (G) Summary of nitrite concentration measured in the cell culture supernatant of BMDMs either untreated or stimulated with Salmonella for 24 hr (n = 5). (H) Western blot analysis and summary of p-p65 and p65 measured at the indicated times in Salmonella-infected BMDMs (n = 3). (I) RT-qPCR analysis of mRNA levels of genes involved in nuclear factor kappa B (NF-κB) signaling (p65, MyD88, Ikkα, and Ikkβ) measured in BMDMs 60 min after Salmonella infection (n = 5). (J) Western blot analysis and summary of p-p65 and p65 measured in uninfected and Salmonella-infected BMDMs either with or without N,N,N′,N′-tetrakis-(2-pyridyl-methyl)-ethylenediamine (TPEN) (4 µM) for 60 min (n = 4). (K) RT-qPCR analysis of p65, MyD88, Ikkα, and Ikkβ mRNA in uninfected and Salmonella-infected BMDMs either with or without TPEN for 60 min (n = 6). Data in this figure are represented as mean ± SEM. p values were determined using two-tailed unpaired Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, and ns, not significant.
Figure 5—figure supplement 1. Loss of Slc30a1 in the murine macrophages reduces iNOS and nitric oxide (NO) production by inhibiting the nuclear factor kappa B (NF-κB) pathway.
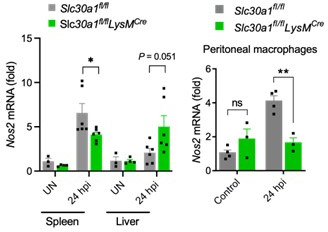
Infected macrophages typically exhibit a classically activated (i.e., M1-like) phenotype. During Salmonella infection, these activated macrophages produce several proinflammatory cytokines and chemokines such as IL-1β(Interleukin-1 beta), TNF, and CCL2(C-C motif chemokine ligand 2) in order to clear the pathogen via mechanisms that include cell death, the recruitment of polymorphonuclear phagocytes, and oxidative processes (Brennan and Cookson, 2000; Pham et al., 2020; Depaolo et al., 2005). We therefore infected BMDMs with attenuated Salmonella (MOI = 1) and measured the mRNA levels of genes expressing key proinflammatory cytokines (Tnfα, Il1β, and Il6; Figure 5C), chemokines (Ccl2, Cxcl1, and Cxcl10; Figure 5D), and macrophage polarization‒related factors (Arg1 and Nos2; Figure 5E). We found significantly reduced expression of proinflammatory cytokine- and chemokine-related genes in infected Slc30a1 cKO BMDMs compared to control cells. In particular, we found extremely low levels of Nos2 mRNA in infected Slc30a1 cKO BMDMs; Nos2 encodes iNOS, a hallmark of classically activated macrophages that contributes to the host defense system by producing NO. Downregulation of the Nos2 gene was supported by the low levels of iNOS (Figure 5F—source data 1) and nitrite concentrations (Figure 5G) in Salmonella-infected BMDMs. In addition, we found reduced expression of Nos2 and a number of proinflammatory cytokine-related genes in Slc30a1 cKO BMDMs exposed to either HK-ST or LPS compared to control cells (Figure 5—figure supplement 1A, B). Western blot analysis of iNOS protein levels confirmed that both HK-ST and LPS (Figure 5—figure supplement 1C, D, Figure 5—source data 2) downregulate Nos2 in Slc30a1 cKO BMDMs compared to control cells. Moreover, we measured significantly lower levels of nitrite (NO2−), a product of NO metabolism, in the cell culture supernatant of Slc30a1 cKO BMDMs 24 hr after HK-ST and LPS (Figure 5—figure supplement 1E) compared to control cells.
A variety of cellular signaling pathways have been reported to regulate Nos2 expression in macrophages in response to pathogens, particularly the NF-κB (nuclear factor kappa B) pathway (Xie et al., 1994), the MAPK (mitogen-activated protein kinase) pathway (Chan and Riches, 2001), and the transcription factor STAT3 (signal transducer and activator of transcription 3) (Ahuja et al., 2020). We therefore measured the phosphorylated (i.e., activated) state of the principal proteins in these pathways—namely, phosphorylated p65 (in the NF-κB pathway), Erk and p38 (in the MAPK pathway), and Stat3—in Slc30a1 cKO and Slc30a1fl/fl BMDMs 30 and 60 min after LPS stimulation. Western blot analysis revealed that p-p65, p-Erk, p-p38, and p-Stat3 levels were raised within 30 min in both Slc30a1 cKO and Slc30a1fl/fl BMDMs; however, p-p65 levels were significantly lower in Slc30a1 cKO BMDMs at 60 min (Figure 5—figure supplement 1F, Figure 5—source data 2). Similarly, Salmonella infection (MOI = 1) caused lower levels of p-p65 protein in Slc30a1 cKO BMDMs at 60 min compared to infected Slc30a1fl/fl cells (Figure 5H—source data 1), as well as reduced mRNA levels of several genes upstream of the NF-κB pathway, including p65, MyD88, Ikkα, and Ikkβ (Figure 5I). Notably, these Salmonella-induced changes in the expression of these key NF-κB signaling molecules were prevented by treating cells with the membrane-permeable zinc chelator TPEN (N, N,N′,N′-tetrakis-(2-pyridyl-methyl)-ethylenediamine (TPEN)) (Figure 5J—source data 1, Figure 5K). These results suggest that Slc30a1 in macrophages protects against Salmonella infection by regulating intracellular zinc.
Altered zinc distribution in Salmonella-infected Lyz2-Cre-driven Slc30a1 knockout mice
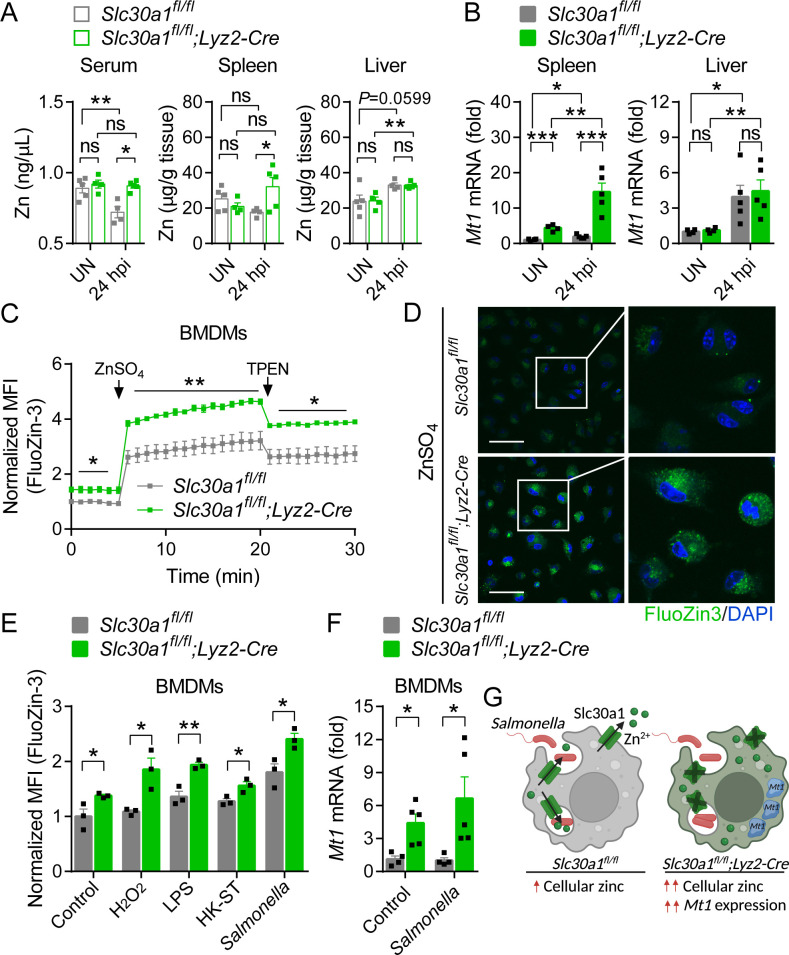
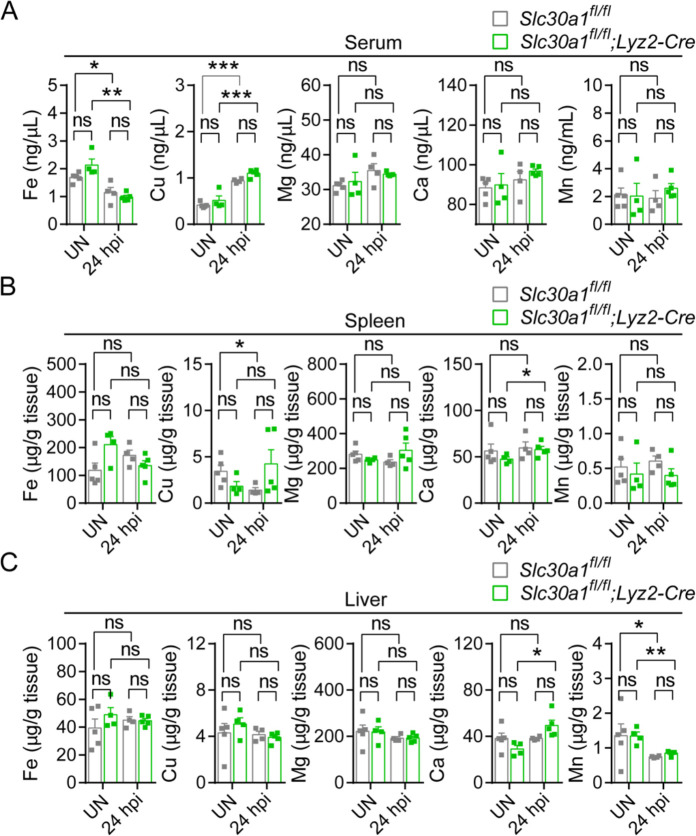
Next, we examined whether Slc30a1 in macrophages regulates systemic zinc homeostasis by measuring zinc concentration in the serum, spleen, and liver of Slc30a1 cKO and Slc30a1fl/fl mice infected with attenuated Salmonella (1 × 105 CFU per mouse) for 24 hr using inductively coupled plasma mass spectrometry (ICP-MS); we also measured other minerals, including iron, copper, magnesium, calcium, and manganese (Figure 6—figure supplement 1A–C). We found that compared to Salmonella-infected control mice, Salmonella-infected Slc30a1 cKO mice had significantly higher levels of zinc in the serum and spleen but no difference in the liver (Figure 6A), consistent with the previous finding that during systemic infection serum zinc is redistributed to the liver in order to increase monocyte maturation, boost the immune system, and restrict the delivery of nutrient metals to pathogens (Alker and Haase, 2018). In addition, elevated serum zinc may impair the function of immune cells, leading to increased susceptibility to Salmonella infection. Moreover, the spleen serves to filter the blood filter and is a hub for monocytes (Swirski et al., 2009); thus, the high zinc content in the spleen may be due to circulating high zinc‒containing macrophages, a notion supported by our finding of increased mRNA levels of Mt1, which encodes the principal splenic zinc reservoir protein metallothionein 1, in the spleen of Salmonella-infected Slc30a1 cKO mice, but not in the liver (Figure 6B).
Figure 6. Loss of Slc30a1 in macrophages causes intracellular zinc accumulation.
(A) Summary of zinc (Zn) content measured in the serum, spleen, and liver of uninfected (UN) and Salmonella-infected mice at 24 hpi (n = 4–5 mice/group). (B) RT-qPCR analysis of Mt1 mRNA in the spleen and liver of the indicated mice. (C) Time course of normalized FluoZin-3 mean fluorescence intensity (MFI) measured in bone marrow-derived macrophages (BMDMs); where indicated, ZnSO4 (100 µM) and N, N,N′,N′-tetrakis-(2-pyridyl-methyl)-ethylenediamine (TPEN) (4 µM) were applied to the cells. (D) Confocal fluorescence images of BMDMs stained with FluoZin-3 (green) after treatment with ZnSO4 for 15 min; the nuclei were counterstained with DAPI (blue). Scale bars, 50 µm. (E) Summary of normalized FluoZin-3 MFI measured in BMDMs 30 min after application of H2O2 (1 mM), lipopolysaccharide (LPS; 1 µg/ml), heat-killed Salmonella typhimurium (HK-ST) (multiplicity of infection [MOI] = 100), or Salmonella (MOI = 10) (n = 3). (F) RT-qPCR analysis of Mt1 mRNA in uninfected and Salmonella-infected BMDMs (n = 5). (G) Model showing the predicted effects of the loss of Slc30a1 on cellular zinc trafficking and intracellular zinc accumulation in BMDMs in response to Salmonella infection. Data in this figure are represented as mean ± SEM. p values were determined using two-tailed unpaired Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, and ns, not significant.
Figure 6—figure supplement 1. Except for Zn, Lyz2-Cre-mediated genetic deletion of Slc30a1 in mice does not affect the content of other trace minerals.
Figure 6—figure supplement 2. Slc30a1 deficiency increase sensitivity to zinc stress.
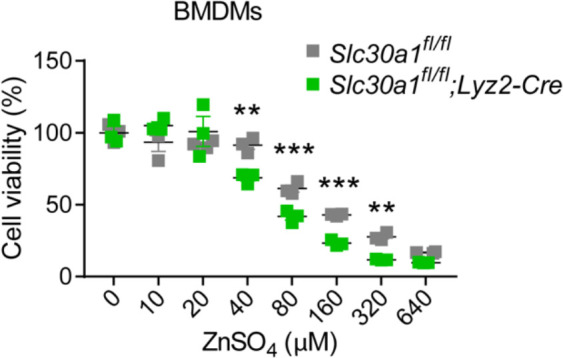
Next, we used the fluorescent zinc probe FluoZin-3 to quantify the accumulation of intracellular zinc in BMDMs isolated from Slc30a1 cKO and control mice following ZnSO4 treatment. Consistent with the well-documented role of Slc30a1 in zinc resistance (Palmiter, 2004), we found that Slc30a1 cKO BMDMs were significantly more sensitive to ZnSO4-induced toxicity compared to control cells (Figure 6—figure supplement 2), and ZnSO4 treatment (100 µM) caused a significantly higher FluoZin-3 signal in Slc30a1 cKO BMDMs compared to control cells (Figure 6C, D).
Previous studies showed that cytosolic-free zinc increases in macrophages following exposure to LPS, Escherichia coli, and Salmonella (Haase et al., 2008; Brieger et al., 2013, Wu et al., 2017). We, therefore, measured intracellular free zinc in Salmonella-infected Slc30a1 cKO and control BMDMs. We found that 30 min after Salmonella infection (MOI = 1), FluoZin-3 fluorescence was significantly higher in Slc30a1 cKO cells; similar results were obtained when the cells were exposed to H2O2 (1 mM), LPS (1 µg/ml), or HK-ST (MOI = 100) (Figure 6E). In addition, we measured higher levels of Mt1 mRNA in Slc30a1 cKO BMDMs compared to Slc30a1fl/fl cells, even in uninfected cells (Figure 6F). Based on these results, we hypothesize that the loss of Slc30a1 leads to an abnormal increase in intracellular zinc during Salmonella infection and increases the expression of Mt1, possibly to compensate for the excessive intracellular zinc by increasing Mt1-mediated zinc storage (Figure 6G).
Discussion
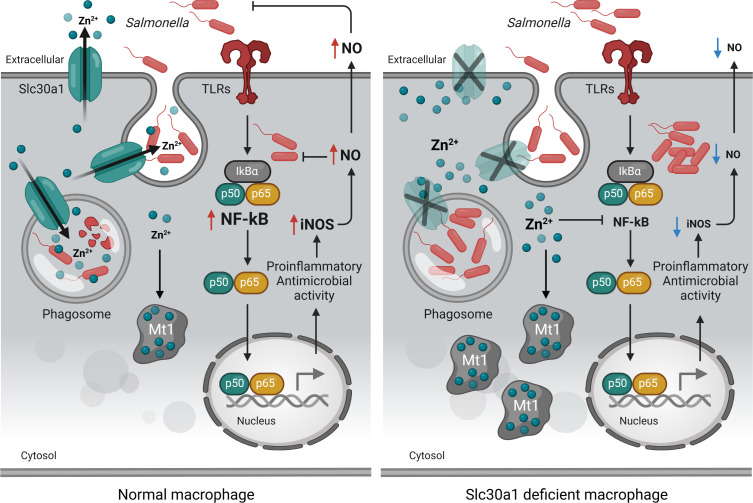
Here, we provide evidence that Slc30a1 expression in macrophages is required for host resistance against pathogens, as illustrated schematically in Figure 7. Based on this model, Slc30a1-deficient macrophages have increased Salmonella burden due to reduced iNOS and NO formation via reduced NF-κB signaling. This effect is due primarily to the inhibitory effects of high intracellular zinc, reflected by the compensatory upregulation of the zinc-binding protein Mt1.
Figure 7. Putative protective function of Slc30a1 in macrophages during Salmonella infection.
Under normal conditions, Slc30a1 is upregulated in response to Salmonella infection, generating a short-term decrease in cytosolic zinc concentration and increasing zinc toxicity in Salmonella-containing phagosomes. Loss of Slc30a1 leads to an accumulation of intracellular zinc, thereby upregulating Mt1 overexpression and reducing iNOS and nitric oxide (NO) production via reduced nuclear factor kappa B (NF-κB) signaling, reducing the cell’s bacterial clearance capacity.
The cellular zinc exporter Slc30a1 is a key regulator of cellular zinc homeostasis (Kambe et al., 2015; Liuzzi and Cousins, 2004; Lichten and Cousins, 2009). Previous in vitro studies have shown that Slc30a1 in macrophages mediates the intracellular killing of invading Mycobacterium tuberculosis and E. coli by increasing zinc levels within phagosomes, thereby promoting bacterial clearance (Botella et al., 2011; Stocks et al., 2021). Here, we provide the first in vivo evidence that Slc30a1 in macrophages plays a key role in host protection against Salmonella infection. We show that mice lacking Slc30a1 in macrophages are highly susceptible to Salmonella infection.
The killing capacity of macrophages has been studied extensively, and it mediates the host defense by ingesting and then destroying invading microbes (Flannagan et al., 2009). Using BMDMs isolated from our Lyz2-Cre-driven Slc30a1 conditional knockout mice, we show that these macrophages have reduced pathogen-killing capacity via reduced NF-κB signaling, decreasing iNOS and NO production. Interestingly, we found that treating these cells with the zinc chelator TPEN improves NF-κB signaling, suggesting that changes in cellular zinc levels likely contribute to their reduced antimicrobial function. Wu et al. previously reported that zinc can suppress iNOS/NO production by inhibiting NF-κB in Salmonella-infected macrophages (Wu et al., 2017). Moreover, recent studies have shown that zinc can reduce NF-κB activation in macrophages (Haase et al., 2008; Brieger et al., 2013; Wu et al., 2017; Jeon et al., 2000). With respect to the underlying mechanism, studies have shown that zinc inhibits the NF-κB pathway by reducing the activity of recombinant IKKα and IKKβ by blocking IκBα phosphorylation and degradation (Jeon et al., 2000; Zhou et al., 2004; Liu et al., 2013). Consistent with this notion, our finding of the role of Slc30a1 in regulating the macrophage response to Salmonella may help explain how the genetic deletion of Slc30a1, specifically in this professional phagocyte leads to the observed disease outcome in our mouse model.
An interesting finding from our in vivo experiments is that our Lyz2-Cre-driven Slc30a1 conditional knockout mice fail to redistribute zinc levels in the serum and spleen following Salmonella infection. In general, serum zinc is rapidly taken up by the liver as part of the host defense process during systemic infection (Alker and Haase, 2018). In mice, defects in this process—for example, due to high zinc supplementation—have negative consequences for neutrophil extracellular traps (Kuźmicka et al., 2020). In infants, consuming a full-fat diet of powdered cow’s milk containing excessive zinc content can lead to a decreased phagocytic capacity of monocytes (Schlesinger et al., 1993). In adults, excessive consumption of zinc supplements can impair both the migration and bactericidal capacity of polymorphonuclear leukocytes (Chandra, 1984). This is consistent with our finding that a high zinc serum level in our Lyz2-Cre-driven Slc30a1 conditional knockout mice affects the percentage of neutrophils and monocytes and the migration of macrophages, thereby reducing bacterial clearance and increasing their susceptibility to Salmonella infection. Furthermore, nutrient-dense conditions such as excessive serum zinc can be beneficial to the growth of Salmonella. Previous studies reported that a high-affinity Zn2+ uptake system ZnuABC of Salmonella protects the bacterial cells from nitrosative stress in macrophages (Fitzsimmons et al., 2018). Zinc rich also induces peptide deformylase, an essential protein contributing to Salmonella maturation that can be inhibited by NO (Singhal and Fang, 2020).
Our measurements of intracellular zinc levels revealed high zinc concentrations in Slc30a1-deficient macrophages. We also detected an upregulation of Mt1, possibly indicating a cellular response to intracellular zinc saturation. Mt1 is a low molecular weight cysteine-rich protein with a high affinity for divalent metals that is essential for buffering surplus zinc to maintain zinc homeostasis (Krężel and Maret, 2017). Smith et al. showed that the level of Mt1 expression was positively correlated with intracellular free zinc concentration (Smith et al., 2008). In this respect, Mt1 overexpression may serve as zinc storage for reducing the side effects of excessive cytosolic-free zinc due to Slc30a1 deletion.
Although we found that genetically deleting Slc30a1 in macrophages reduces the killing capacity by suppressing iNOS and nitrite production through zinc-medicated NF-κB activation, the underlying mechanism that connects NO inactivation and excessive intracellular zinc remains uncertain. In this regard, additional experiments will be needed to investigate whether the antimicrobial function of Slc30a1-deficient macrophages is restricted to the defect of NO using selective inhibitors of iNOS. Furthermore, although we focused our study on BMDMs, macrophages may respond differently to infection in the context of the full immune system in vivo. Thus, generating a mouse line in which both Slc30a1 and Nos2 are knocked out in macrophages (Slc30a1fl/flNos2fl/fl;Lyz2-Cre) may provide insight into underlying mechanisms behind failed bacterial clearance that affects disease outcome.
In conclusion, we report that Slc30a1 of macrophages plays a protective role against Salmonella infection by regulating iNOS and NO activities, which are essential for bacterial clearance through NF-κB signaling. These findings underscore the notion that cellular zinc regulated by zinc transporter Slc30a1 is important for host defenses by maintaining an intracellular killing capacity of macrophages. However, the precise mechanism of how Slc30a1 drives NO-mediated antibacterial activity remains misty, and further studies are needed to explore this aspect.
Materials and methods
Mouse strains
Slc30a1flag-EGFP/+ mice were generated by Shanghai Biomodel Organism Science & Technology Development Co Ltd. The donor vector contains the 5′ homologous arm, 3xFlag-2A-EGFP-2A-CreERT2-Wpre-pA, and the 3′ homologous arm. In the 3′ UTR of the Slc30a1 locus, the 3xFlag-2A-EGFP-2A-CreERT2-Wpre-pA cassette was inserted downstream of exon 2. Slc30a1 floxed mice were generated by Shanghai Biomodel Organism Science & Technology Development Co Ltd. The floxed Slc30a1 allele contains a single loxP site upstream of exon 2 and a single loxP site neo cassette downstream of exon 2. Heterozygous Slc30a1 floxed mice (Slc30a1fl/+) were backcrossed to the C57BL/6 background for more than 5 generations and then crossed with Lyz2-Cre-expressing mice (The Jackson Laboratory, 004781) to generate Lyz2-Cre-driven Slc30a1 conditional knockout (Slc30a1 cKO) mice. All mice were genotyped using genomic PCR. Cre-negative floxed mice (Slc30a1fl/fl) were used as the control group. Under standard conditions, an 8-week-old male Slc30a1 cKO presents with no obvious phenotype (data not shown). Wild-type C75BL/6 mice were purchased from Shanghai SLRC Laboratory Animal Co, Ltd. Eight-week-old male mice were used in this study. All animals were fed a standard chow diet with access to drinking water and were housed at constant temperature (23°C) under a 12:12 hr light/dark cycle in specific pathogen-free conditions. All in vivo experiments were conducted in accordance with the National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee at Zhejiang University.
DNA isolation and genotyping
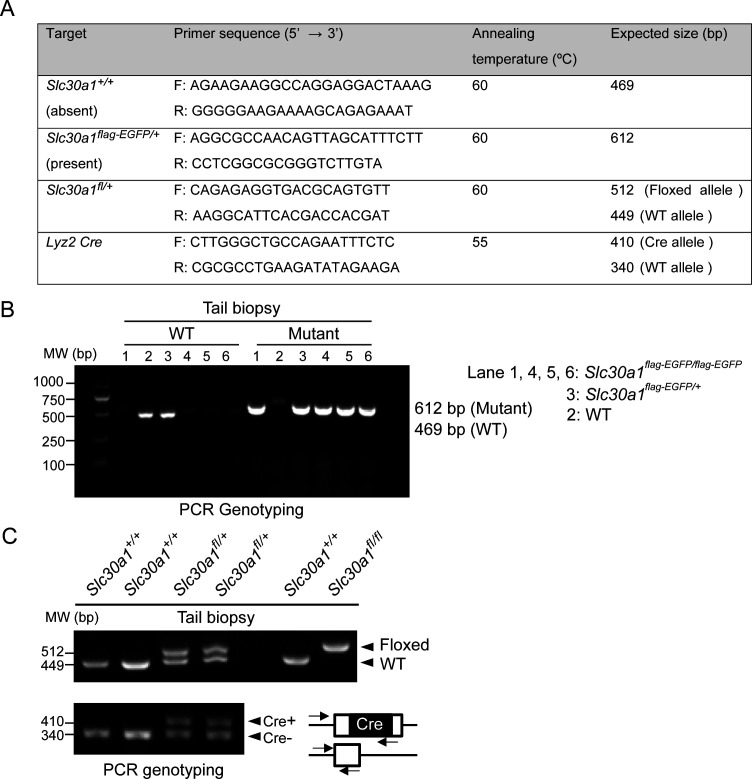
Genomic DNA of mouse tissue biopsies was extracted using the Tissue & Blood DNA Extraction kit-250prep (Zhejiang Easy-Do Biotech Co, Ltd, #DR0301250) in accordance with the manufacturer’s instructions. DNA samples were quantified using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). PCR amplification was conducted using 50 ng DNA per reaction, 2xTaq PCR StarMix (GenStar, #A012-101), and specific primers (Figure 8A) in a T100 Thermal Cycler (Bio-Rad, #10878655). Finally, the PCR products were separated by agarose gel electrophoresis to determine the genotypes (Figure 8B, C, Figure 8—source data 1).
Figure 8. PCR-based genotyping.
(A) List of primers used for routine genotyping. PCR was performed using genomic DNA from mouse tail biopsies. (B) Genotyping of Slc30a1-3xflag-EGFP (Slc30a1flag-EGFP/+) mouse line. The PCR fragment length for the wild-type (WT) Slc30a1 allele is 612 and 469 bp for the mutant allele, respectively. (C) The conditional knockout (Slc30a1fl/fl;Lyz2-Cre) mice were recognized by genomic PCR rendering one band of flox/flox allele (512 bp) with two bands of Lyz2-Cre heterozygous (410 and 340 bp). Each lane represents one individual mice.
Bacterial preparation
The experiments were performed with attenuated S. enterica subsp. enterica serovar Typhimurium strain MeganVac1 (Accession: CP112994.1). The bacterial stain was grown in tryptic soy broth medium (Solarbio Life Science, #T8880) overnight at 37°C under sterile conditions. Bacterial inoculum was compared to a 0.5 McFarland turbidity standard (approximately 1 × 108 CFU/ml) and adjusted to 105 colony-forming units per ml (CFU/ml) with sterile phosphate-buffered saline (PBS). The bacterial count in the original suspension was verified using a plate counting method. HK-ST was prepared by incubating the bacterial inoculum in a 60°C water bath for 2 hr, and a bacterial viable count was performed to confirm ≥99.99% reduction in viability.
Primary cell culture
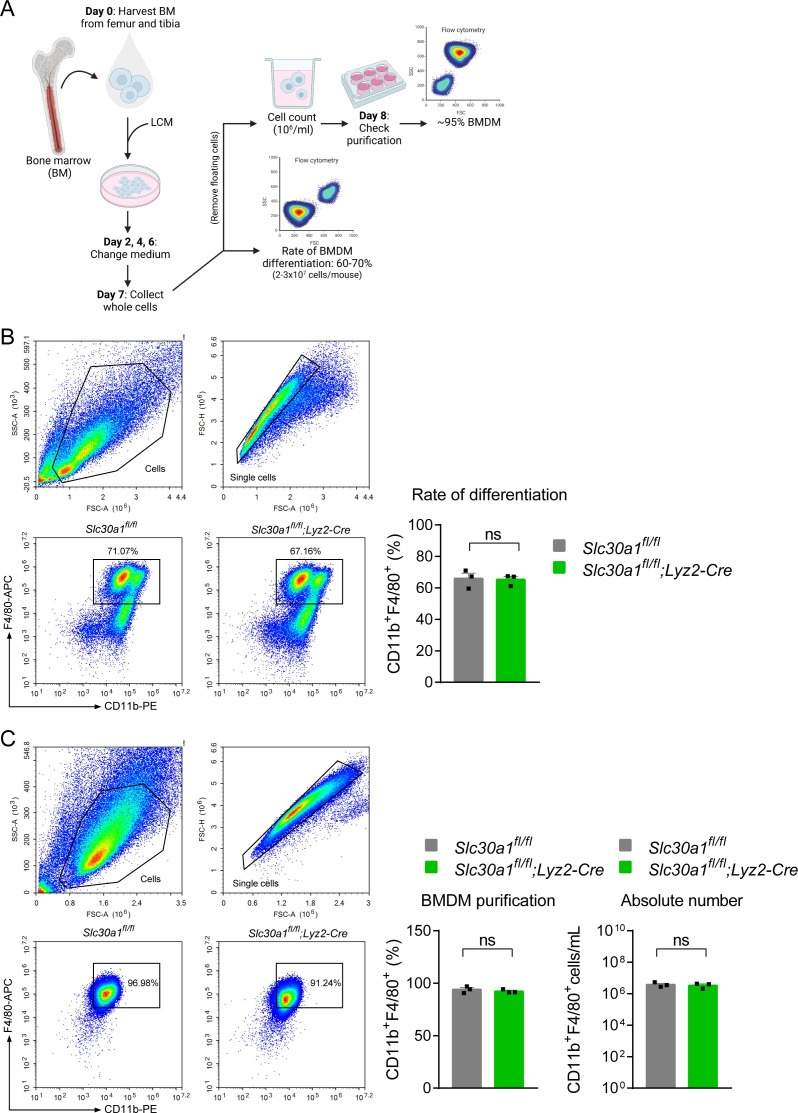
BMDMs were differentiated from bone marrow cells obtained from 8-week-old male mice (Figure 9). In brief, bone marrow cells from the femur and tibia were isolated by flushing with sterile PBS (Corning, #21-040-CV) and filtered through a 40-μm nylon membrane filter (Falcon, #350). The resulting cell suspension was centrifuged at 300 × g for 5 min, and the cell pellet was resuspended in RPMI 1640 (Corning, #21-040-CVR) containing 1% (vol/vol) penicillin–streptomycin (HyClone, #SV30010), 20% (vol/vol) fetal bovine serum (FBS) (Gibco, #10270-106), and 30% L929 conditioned medium. BMDMs were then differentiated for 7 days under 37°C in humidified air containing 5% CO2, refreshing the differentiation medium every 2 days. These BMDMs were then transferred to RPMI 1640 supplemented with 1% penicillin–streptomycin and 10% FBS at 37°C and used the next day. We found no difference in BDMD differentiation between the genotypes used in this study.
Figure 9. Lyz2-Cre-mediated genetic deletion of Slc30a1 in mice does not affect the differentiation of bone marrow-derived macrophages (BMDMs).
(A) Schematic illustration depicting the strategy used to measure the BMDM differentiation procedure. Bone marrow (BM) cells were harvested from the femur and tibia and cultured in L929 cell-conditioned medium (LCM) for 7 days. (B) Flow cytometry analysis and summary of the percent BMDM differentiation (CD11b+F4/80+) from BM samples obtained from the indicated mice measured at day 7 (n = 3). (C) Percent BMDM purification and absolute number of BMDMs measured after removing floating cells and measuring cell concentration. Data in this figure are represented as mean ± SEM. p values were determined using two-tailed unpaired Student’s t-test. ns, not significant.
RNA-seq and data analysis
BMDMs from biological triplicates of C75BL/6 mice were infected with Salmonella for 2 hr, and total RNA was extracted for RNA-seq. RNA-seq analysis was performed by LC Sciences (Hangzhou, China). GO and KEGG enrichment analyses were performed using the DAVID database. Heatmaps were generated using the OmicStudio tool (https://www.omicstudio.cn/tool). Volcano plots and bar plots of the GO and KEGG pathways were generated using the Tableau (2019.3. Ink) software desktop system.
Immunofluorescence
BMDMs grown on glass coverslips in 6-well plates were exposed to ZnSO4 (40 µM) or infected with Salmonella (MOI = 1) for 4 hr. The cells were then fixed in 4% paraformaldehyde (PFA) for 15 min, permeabilized in 0.2% Triton X-100 (Sigma, #T9284) for 10 min, and then blocked in 5% (wt/vol) BSA(Bovine serum albumin) (Sigma, #A6003) for 30 min at room temperature. The fixed cells were then incubated in monoclonal anti-FLAG M2-Peroxidase (HRP) antibody (Sigma, #A8592) overnight at 4°C, followed by fluorophore-conjugated secondary antibody, Alexa Fluor 647 secondary antibody (Thermo Fisher Scientific, #A32728) for 1 hr at room temperature in the dark. The nuclei were stained with 3 µM DAPI (4′,6-diamidino-2-phenylindole; BioLegend, #422801) for 5 min; after gently rinsing three times with PBS, the glass coverslips were mounted cell-side down on clean plain glass slides with 80% glycerol (Sigma, #G5516) and imaged using a CSU-W1 confocal microscope (Olympus).
Salmonella infection in vivo
Eight-week-old male mice were injected intraperitoneally (i.p.) with 200 μl PBS containing Salmonella at a final dose of 1 × 105 CFU per mouse or sterile PBS as uninfected groups. At the indicated time points, the mice were euthanized, and peritoneal cavity cells were subsequently collected to determine the peritoneal macrophage population. Blood samples were obtained via cardiac puncture, and the spleen and liver were dissected. The organs were either immediately placed on ice in sterile PBS to count bacterial burden or immersed in 4% PFA for later histological analysis. A portion of the blood was kept in serum collection tubes and allowed to be clotted for 2 hr at room temperature; clear serum was then obtained by centrifugation at 1000 rpm for 10 min. For survival experiments, after infection with Salmonella, the mice were monitored daily for 2 weeks.
Bacterial burden in tissues
Approximately 100–200 mg of liver and spleen samples were homogenized in sterile PBS. A 100 µl of tissue homogenate, peritoneal cavity fluid, and blood were serially diluted 10-fold in PBS and plated on TSA(Tryptic soy agar) agar (Solarbio life science, #T8650). The plates were incubated at 37°C overnight, and viable bacteria colonies were counted as CFU/ml or CFU per gram of tissue (CFU/g).
Serum ALT and AST measurements
The levels of serum AST and ALT from mice infected with or without Salmonella for 24 hr were measured using an alanine aminotransferase assay kit and aspartate aminotransferase assay kit (ShenSuoYouFu) according to the manufacturer’s protocol.
ELISA
Cytokine levels of TNFα were quantified in serum obtained from mice infected with and without Salmonella for 24 hr by the mouse/rat TNF-A Valukine ELISA kit (R&D Systems, #VAL609) following the product protocol.
Histopathology
Spleen and liver samples were immersed in 4% PFA solution for 24 hr and then embedded in paraffin. The tissue samples were then sectioned and stained with hematoxylin and eosin using standard protocols. The sections were examined using bright-field microscopy (Nikon Eclipse Ni-U). Three adjacent sections of each sample were quantified using ImageJ software (National Institutes of Health).
Flow cytometry
Cell suspensions (1 × 105 cells/ml) were immunostained using specific anti-mouse antibodies in the dark at 4°C for 30 min. The following fluorochrome-conjugated antibodies were used in this study (all from BioLegend): CD11b-Pacific Blue (clone M1/70, #101224), CD11b-PE (clone M1/70, #1101208), F4/80-APC (clone BM8, #123116), Gr-1-APC (clone RB6-8C5, #108412), CD45-APC/Cyanine7 (clone 30-F11, #103116), CD45R/B220-Pacific Blue (clone RA3-6B2, #103227), and CD3-PerCP/Cyanine 5.5 (clone 17A2, #100218). All antibodies were used at a dilution of 1:200. Live versus dead cells were identified based on DAPI staining. Cell sorting was performed using the NovoCyte flow cytometer (ACEA Biosciences Inc).
Trace mineral analysis using ICP-MS
Metal content was measured using ICP-MS. Tissue samples (200 mg) were digested in 4 ml of EMSURE ISO nitric acid 65% solution (Merck, #1.00456.2508) using a MARS 6 microwave extraction system. In brief, the samples were heated from room temperature to 200°C in a microwave oven (1000 W, 2,450 MHz) for up to 20 min. The microwave power was maintained for an additional 30 min, followed by a cooling-down period of 15 min. The samples were then completely dried at 110°C for 3 hr. The digested samples were diluted to a final volume of 5 ml in high-purity deionized water obtained using a Milli-Q Integral 10 purification system (EMD Millipore, #C205110). The resulting samples were subjected to metal analysis using an Agilent 7700x ICP-MS equipped with an Agilent ASX 520 auto-sampler.
Gene expression analysis using RT-qPCR
Total RNA was extracted from the samples using TransZol Up (TransGen Biotech, #ET111-01), and 1 μg of total RNA was used as a template to synthesize cDNA using the HiScript II 1st Strand cDNA Synthesis Kit (Yeasen Biotech, #11123ES60) in a ProFlex PCR System (Life technologies). Each RT-qPCR reaction consisted of 5 μl of SYBR Green PCR Master Mix (Bimake, #B21202), 2 μl of forward and reverse primers (1 µM), 0.2 µl of cDNA, and sufficient RNase-free water to yield a final volume of 10 μl. RT-qPCR was performed in duplicate for each sample using the LightCycler 480 Real-Time PCR System (F. Hoffmann-La Roche, Ltd) with the following conditions: initial denaturation at 95°C for 3 min, 40 cycles of amplification at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, followed by denaturation at 95°C for 30 s, and annealing-extension at 40°C for 30 s. Data were analyzed using the 2−ΔCt method (Pfaffl, 2001) to calculate the relative target gene expression normalized to the endogenous housekeeping gene Gapdh (glyceraldehyde-3-phosphate dehydrogenase). The oligonucleotides used in this study are provided in Supplementary file 4.
Bacterial killing assay
BMDMs were plated at 2.5 × 105 cells/well in a 12-well plate and allowed to adhere overnight. The cells were then infected with Salmonella (MOI = 10) for 30 min by centrifuging the plate at 1000 rpm for 10 min and incubating for an additional 20 min. The plate was then rinsed twice with PBS containing 100 μg/ml gentamicin (Sigma, #345814) to remove the remaining extracellular bacteria. The cells were then incubated in a fresh medium containing 10 μg/ml gentamicin to prevent the growth of extracellular bacteria. Where indicated, infected cells were lysed with 0.1% Triton X-100, and serial dilutions of the cell lysates were plated on TSA agar, followed by incubation at 37°C overnight. The number of colonies appearing on the agar plate was counted in order to quantify the surviving bacteria. Intracellular killing of macrophages was verified after 24 hr infection using transmission electron microscopy. BMDMs were harvested and subjected to routine processing, post-fixed, embedded, sectioned, and mounted at the Center for Cryo-Electron Microscopy (CCEM), Zhejiang University. Finally, thin sections (~70 nm) were examined in an FEI Tecnai 10 (100 kV) transmission electron microscope.
NO measurement
Nitric oxide (NO) was determined through the formation of nitrite (NO2−), the primary, stable metabolite of NO. Extracellular nitrite released from BMDMs (1 × 105 cells/well) stimulated with either HK-ST (MOI = 10) or 100 ng/ml LPS (Sigma, #L7895) was measured using a colorimetric assay based on the Griess reaction (Green et al., 1982). At the indicated time points, cell culture supernatants were collected and placed into a fresh 96-well plate. The supernatants were then mixed with freshly prepared Griess solution consisting of 0.1% N-1-naphthyl ethylenediamine dihydrochloride (Sigma, #N9125) and 1% sulfanilamide (Sigma, #V900220) in 5% phosphoric acid (Sigma, #P5811) at a ratio of 1:1. The plate was incubated at room temperature for 10 min in the dark. Absorbance at 540 nm was then measured using a microplate reader (Biotek Eon, Gen5), and nitrite production was calculated using a NaNO2 calibration curve.
Cell viability
Cell viability of BMDMs under high zinc conditions was measured using the plate-based colorimetric tetrazolium salt assay. First, BMDMs were plated in 96-well plates at 2 × 104 cells/well. The next day, twofold serial dilutions of ZnSO4 (Sigma, #Z0251) were added to the culture media, and the cells were incubated at 37°C for 24 hr. Next, the cell culture medium was carefully removed and replaced with 100 µl of fresh medium containing 5 mg/ml thiazolyl blue tetrazolium blue (Sigma, #M2128). The cells were incubated for an additional 4 hr, and then the cell culture media was discarded. Blue crystals catalyzed by the mitochondrial enzyme succinate dehydrogenase were solubilized in 100% DMSO(Dimethyl sulfoxide), and the intensity was measured colorimetrically at 570 nm.
Intracellular zinc measurement
A cell-based microplate assay was used to measure intracellular free zinc ions (Zn2+) using the cell-permeable fluorescent probe FluoZin-3 AM (Invitrogen, #F24195). BMDMs were plated into 96-well clear-bottom black polystyrene microplates (Corning, #3603) at 1 × 105 cells/well and allowed to attach onto the bottom surface overnight. The cells were then incubated in culture media containing 2 μM FluoZin-3 AM for 30 min in the dark. After washing twice with PBS, the cells were incubated in fresh PBS for an additional 10 min to allow de-esterification. To detect cellular zinc accumulation, 100 μM ZnSO4 (Sigma, #Z0251) and/or 4 μM TPEN (Sigma, #P4413) was added to the cells, and FluoZin-3 fluorescence was measured at 1-min intervals for 30 min at 37°C using a SynergyMx M5 fluorescence microplate reader (Molecular Devices) with 485 nm excitation and 535 nm emission (Brieger et al., 2013). The levels of intracellular zinc in response to pathogens and H2O2 were measured after 30 min stimulation. Images of FluoZin-3 fluorescence were acquired using a Nikon A1R confocal microscopy and analyzed using NIS-Elements Viewer imaging software, version 4.50.
Protein quantification using western blot analysis
Proteins from BMDMs were extracted using RIPA buffer (Solarbio, #R0020) containing Pierce protease inhibitor (Thermo Fisher Scientific, #A32965) and phosphatase inhibitor (Roche, #04906845001). Total protein content was quantified using the Bradford dye-binding method (Sigma, #B6916). A total of 50 μg of cellular proteins were mixed with 5× loading buffer and boiled for 5 min. Equal amounts of prepared proteins were electrophoresed in 10% SDS–PAGE(sodium dodecyl sulfate-polyacrylamide gel electrophoresis) gel at 100 V. After gel electrophoresis, separated proteins were transferred onto polyvinylidene difluoride membrane (Bio-Rad, #1620177) at 300 mA for 90 min using transfer buffer containing 25 mM Tris, 192 mM glycine, and 20% methanol. The membranes were then blocked in 5% (wt/vol) skim milk in Tris-buffered saline containing 0.05% Tween-20 (TBST) for 90 min at room temperature. The membranes were then washed three times with TBST for 10 min each and then probed using the following primary antibodies overnight at 4°C (1:1000 dilution): iNOS antibody (#39898S), NF-κB p65 (D14E12) XP rabbit mAb (#8242S), or phosphor-NF-κB p65 (Ser536) (93H1) rabbit mAb (#3033) (all from Cell Signaling Technology). A monoclonal anti-FLAG M2-Peroxidase (HRP) antibody was used to detect the flag-tagged fusion protein. The following day, the membranes were washed with TBST and incubated with a secondary antibody, HRP-conjugated goat anti-rabbit IgG (H+L) (ABclonal, #AS014) diluted in 5% milk/TBST (1:2000) at room temperature for 2 hr. The membranes were then washed in TBST three times for 10 min each to remove excessive antibodies and developed using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, #32106). The bands were quantified using the ChemiDoc Touch Imaging System (Bio-Rad).
Statistical analysis
Statistical significance between the two groups was determined using a two-tailed, unpaired Student t-test (for comparing two groups). Data are presented as mean ± SEM. For comparison of survival curves, a Log-rank (Mantel–Cox) test was performed. The sample size for each statistical analysis is provided in the figure legends. Data were analyzed and generated using GraphPad Prism version 7.04 (GraphPad Software Inc, La Jolla, CA, USA). For all studies, p values below 0.05 were considered to be statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32330047, 31930057 to FW and 31970689 to JM).
Funding Statement
The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Contributor Information
Junxia Min, Email: junxiamin@zju.edu.cn.
Fudi Wang, Email: fwang@zju.edu.cn.
Sophie Helaine, Harvard Medical School, United States.
Dominique Soldati-Favre, University of Geneva, Switzerland.
Funding Information
This paper was supported by the following grants:
National Natural Science Foundation of China 32330047 to Fudi Wang.
National Natural Science Foundation of China 31930057 to Fudi Wang.
National Natural Science Foundation of China 31970689 to Junxia Min.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing - original draft.
Data curation, Investigation, Methodology.
Data curation, Investigation, Methodology.
Data curation, Investigation, Methodology.
Conceptualization, Resources, Supervision, Funding acquisition, Validation, Visualization, Project administration, Writing - review and editing.
Conceptualization, Resources, Supervision, Funding acquisition, Validation, Visualization, Project administration, Writing - review and editing.
Ethics
All in vivo experiments were conducted in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal experiments were approved by the Institutional Animal Care and Use Committee, Zhejiang University. Committee protocol #AIRB-2023-1479 at the institution.
Additional files
Data availability
The RNA-seq datasets used to generate figure 1A–F have been deposited in NCBI Gene Expression Omnibus ID GSE239543 and GSE67427, respectively.
The following dataset was generated:
Na-Phatthalung P, Wang F, Min J. 2024. RNA-seq data from mouse BMDMs infected with Salmonella enterica serovar Typhimurium. NCBI Gene Expression Omnibus. GSE239543
The following previously published dataset was used:
Blischak JD, Tailleux L, Mitrano A, Barreiro LB, Gilad Y. 2015. Mycobacterial infection induces a specific human innate immune response. NCBI Gene Expression Omnibus. GSE67427
References
- Ahuja A, Kim E, Sung GH, Cho JY. STAT3 differentially regulates tlr4-mediated inflammatory responses in early or late phases. International Journal of Molecular Sciences. 2020;21:7675. doi: 10.3390/ijms21207675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alker W, Haase H. Zinc and Sepsis. Nutrients. 2018;10:976. doi: 10.3390/nu10080976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews GK, Wang H, Dey SK, Palmiter RD. Mouse zinc transporter 1 gene provides an essential function during early embryonic development. Genesis. 2004;40:74–81. doi: 10.1002/gene.20067. [DOI] [PubMed] [Google Scholar]
- Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charrière GM, Waddell SJ, Foti M, Lugo-Villarino G, Gao Q, Maridonneau-Parini I, Butcher PD, Castagnoli PR, Gicquel B, de Chastellier C, Neyrolles O. Mycobacterial p1-type atpases mediate resistance to zinc poisoning in human macrophages. Cell Host & Microbe. 2011;10:248–259. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan MA, Cookson BT. Salmonella induces macrophage death by caspase-1-dependent necrosis. Molecular Microbiology. 2000;38:31–40. doi: 10.1046/j.1365-2958.2000.02103.x. [DOI] [PubMed] [Google Scholar]
- Brieger A, Rink L, Haase H. Differential regulation of TLR-dependent MyD88 and TRIF signaling pathways by free zinc ions. Journal of Immunology. 2013;191:1808–1817. doi: 10.4049/jimmunol.1301261. [DOI] [PubMed] [Google Scholar]
- Carreno D, Wanford JJ, Jasiunaite Z, Hames RG, Chung WY, Dennison AR, Straatman K, Martinez-Pomares L, Pareek M, Orihuela CJ, Restrepo MI, Lim WS, Andrew PW, Moxon ER, Oggioni MR. Splenic macrophages as the source of bacteraemia during pneumococcal pneumonia. EBioMedicine. 2021;72:103601. doi: 10.1016/j.ebiom.2021.103601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan ED, Riches DW. IFN-gamma + LPS induction of iNOS is modulated by ERK, JNK/SAPK, and p38(mapk) in a mouse macrophage cell line. American Journal of Physiology. Cell Physiology. 2001;280:C441–C450. doi: 10.1152/ajpcell.2001.280.3.C441. [DOI] [PubMed] [Google Scholar]
- Chandra RK. Excessive intake of zinc impairs immune responses. JAMA. 1984;252:1443–1446. doi: 10.1001/jama.1984.03350110043027. [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Research. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Depaolo RW, Lathan R, Rollins BJ, Karpus WJ. The chemokine CCL2 is required for control of murine gastric Salmonella enterica infection. Infection and Immunity. 2005;73:6514–6522. doi: 10.1128/IAI.73.10.6514-6522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons L, Liu L, Porwollik S, Chakraborty S, Desai P, Tapscott T, Henard C, McClelland M, Vazquez-Torres A. Zinc-dependent substrate-level phosphorylation powers Salmonella growth under nitrosative stress of the innate host response. PLOS Pathogens. 2018;14:e1007388. doi: 10.1371/journal.ppat.1007388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan RS, Cosío G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nature Reviews. Microbiology. 2009;7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhao L, Wang H, Xie E, Wang X, Wu Q, Yu Y, He X, Ji H, Rink L, Min J, Wang F. Metal transporter Slc39a10 regulates susceptibility to inflammatory stimuli by controlling macrophage survival. PNAS. 2017;114:12940–12945. doi: 10.1073/pnas.1708018114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Dai W, Zhao L, Min J, Wang F. The role of zinc and zinc homeostasis in macrophage function. Journal of Immunology Research. 2018;2018:6872621. doi: 10.1155/2018/6872621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan Y, Berman B, Assaraf YG. Heterodimerization, altered subcellular localization, and function of multiple zinc transporters in viable cells using bimolecular fluorescence complementation. The Journal of Biological Chemistry. 2015;290:9050–9063. doi: 10.1074/jbc.M114.617332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Analytical Biochemistry. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- Guo L, Lichten LA, Ryu MS, Liuzzi JP, Wang F, Cousins RJ. STAT5-glucocorticoid receptor interaction and MTF-1 regulate the expression of ZnT2 (Slc30a2) in pancreatic acinar cells. PNAS. 2010;107:2818–2823. doi: 10.1073/pnas.0914941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase H, Ober-Blöbaum JL, Engelhardt G, Hebel S, Heit A, Heine H, Rink L. Zinc signals are essential for lipopolysaccharide-induced signal transduction in monocytes. Journal of Immunology. 2008;181:6491–6502. doi: 10.4049/jimmunol.181.9.6491. [DOI] [PubMed] [Google Scholar]
- He X, Ge C, Xia J, Xia Z, Zhao L, Huang S, Wang R, Pan J, Cheng T, Xu PF, Wang F, Min J. The zinc transporter slc39a10 plays an essential role in embryonic hematopoiesis. Advanced Science. 2023;10:e2205345. doi: 10.1002/advs.202205345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S, Thompson JA, Watson KG, Liu M, Boyle C, Holden DW. Dynamics of intracellular bacterial replication at the single cell level. PNAS. 2010;107:3746–3751. doi: 10.1073/pnas.1000041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon KI, Jeong JY, Jue DM. Thiol-reactive metal compounds inhibit NF-kappa B activation by blocking I kappa B kinase. Journal of Immunology. 2000;164:5981–5989. doi: 10.4049/jimmunol.164.11.5981. [DOI] [PubMed] [Google Scholar]
- Kambe T, Tsuji T, Hashimoto A, Itsumura N. The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiological Reviews. 2015;95:749–784. doi: 10.1152/physrev.00035.2014. [DOI] [PubMed] [Google Scholar]
- Kimura T, Kambe T. The functions of metallothionein and zip and znt transporters: an overview and perspective. International Journal of Molecular Sciences. 2016;17:336. doi: 10.3390/ijms17030336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krężel A, Maret W. The functions of metamorphic metallothioneins in zinc and copper metabolism. International Journal of Molecular Sciences. 2017;18:1237. doi: 10.3390/ijms18061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuźmicka W, Manda-Handzlik A, Cieloch A, Mroczek A, Demkow U, Wachowska M, Ciepiela O. Zinc supplementation modulates nets release and neutrophils’ degranulation. Nutrients. 2020;13:51. doi: 10.3390/nu13010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM, Williams A, Eisenbarth SC. Structure and function of the immune system in the spleen. Science Immunology. 2019;4:eaau6085. doi: 10.1126/sciimmunol.aau6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten LA, Cousins RJ. Mammalian zinc transporters: nutritional and physiologic regulation. Annual Review of Nutrition. 2009;29:153–176. doi: 10.1146/annurev-nutr-033009-083312. [DOI] [PubMed] [Google Scholar]
- Liu MJ, Bao S, Gálvez-Peralta M, Pyle CJ, Rudawsky AC, Pavlovicz RE, Killilea DW, Li C, Nebert DW, Wewers MD, Knoell DL. ZIP8 regulates host defense through zinc-mediated inhibition of NF-κB. Cell Reports. 2013;3:386–400. doi: 10.1016/j.celrep.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi JP, Cousins RJ. Mammalian zinc transporters. Annual Review of Nutrition. 2004;24:151–172. doi: 10.1146/annurev.nutr.24.012003.132402. [DOI] [PubMed] [Google Scholar]
- Mastroeni P, Grant A, Restif O, Maskell D. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nature Reviews. Microbiology. 2009;7:73–80. doi: 10.1038/nrmicro2034. [DOI] [PubMed] [Google Scholar]
- Moskovskich A, Goldmann U, Kartnig F, Lindinger S, Konecka J, Fiume G, Girardi E, Superti-Furga G. The transporters SLC35A1 and SLC30A1 play opposite roles in cell survival upon VSV virus infection. Scientific Reports. 2019;9:10471. doi: 10.1038/s41598-019-46952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishito Y, Kambe T. Zinc transporter 1 (ZNT1) expression on the cell surface is elaborately controlled by cellular zinc levels. The Journal of Biological Chemistry. 2019;294:15686–15697. doi: 10.1074/jbc.RA119.010227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD. Protection against zinc toxicity by metallothionein and zinc transporter 1. PNAS. 2004;101:4918–4923. doi: 10.1073/pnas.0401022101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham THM, Brewer SM, Thurston T, Massis LM, Honeycutt J, Lugo K, Jacobson AR, Vilches-Moure JG, Hamblin M, Helaine S, Monack DM. Salmonella-driven polarization of granuloma macrophages antagonizes tnf-mediated pathogen restriction during persistent infection. Cell Host & Microbe. 2020;27:54–67. doi: 10.1016/j.chom.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DCP, Figueroa JAL, Buesing WR, Candor K, Blancett LT, Evans HM, Lenchitz R, Crowther BL, III, Elsegeiny W, Williamson PR, Rupp J, Deepe GS., Jr A metabolic inhibitor arms macrophages to kill intracellular fungal pathogens by manipulating zinc homeostasis. Journal of Clinical Investigation. 2021;131:34237029. doi: 10.1172/JCI147268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger L, Arevalo M, Arredondo S, Lönnerdal B, Stekel A. Zinc supplementation impairs monocyte function. Acta Paediatrica. 1993;82:734–738. doi: 10.1111/j.1651-2227.1993.tb12548.x. [DOI] [PubMed] [Google Scholar]
- Shusterman E, Beharier O, Levy S, Zarivach R, Etzion Y, Campbell CR, Lee IH, Dinudom A, Cook DI, Peretz A, Katz A, Gitler D, Moran A. Zinc transport and the inhibition of the L-type calcium channel are two separable functions of ZnT-1. Metallomics. 2017;9:228–238. doi: 10.1039/c6mt00296j. [DOI] [PubMed] [Google Scholar]
- Singhal A, Fang FC. Direct and indirect inhibition of Salmonella peptide deformylase by nitric oxide. mBio. 2020;11:e01383-20. doi: 10.1128/mBio.01383-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PJ, Wiltshire M, Furon E, Beattie JH, Errington RJ. Impact of overexpression of metallothionein-1 on cell cycle progression and zinc toxicity. American Journal of Physiology. Cell Physiology. 2008;295:C1399–C1408. doi: 10.1152/ajpcell.00342.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks CJ, von Pein JB, Curson JEB, Rae J, Phan M-D, Foo D, Bokil NJ, Kambe T, Peters KM, Parton RG, Schembri MA, Kapetanovic R, Sweet MJ. Frontline Science: LPS-inducible SLC30A1 drives human macrophage-mediated zinc toxicity against intracellular Escherichia coli. Journal of Leukocyte Biology. 2021;109:287–297. doi: 10.1002/JLB.2HI0420-160R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, Figueiredo JL, Kohler RH, Chudnovskiy A, Waterman P, Aikawa E, Mempel TR, Libby P, Weissleder R, Pittet MJ. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–616. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troeger C, Forouzanfar M, Rao PC, Khalil I, Brown A, Reiner RC, Jr, Fullman N, Thompson RL, Abajobir A, Ahmed M, Alemayohu MA, Alvis-Guzman N, Amare AT, Antonio CA, Asayesh H, Avokpaho E, Awasthi A, Bacha U, Barac A, Betsue BD, Beyene AS, Boneya DJ, Malta DC, Dandona L, Dandona R, Dubey M, Eshrati B, Fitchett JRA, Gebrehiwot TT, Hailu GB, Horino M, Hotez PJ, Jibat T, Jonas JB, Kasaeian A, Kissoon N, Kotloff K, Koyanagi A, Kumar GA, Rai RK, Lal A, El Razek HMA, Mengistie MA, Moe C, Patton G, Platts-Mills JA, Qorbani M, Ram U, Roba HS, Sanabria J, Sartorius B, Sawhney M, Shigematsu M, Sreeramareddy C, Swaminathan S, Tedla BA, Jagiellonian RT-M, Ukwaja K, Werdecker A, Widdowson M-A, Yonemoto N, El Sayed Zaki M, Lim SS, Naghavi M, Vos T, Hay SI, Murray CJL, Mokdad AH. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. The Lancet Infectious Diseases. 2017;17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels I, Fischer HJ, Rink L. Dietary and physiological effects of zinc on the immune system. Annual Review of Nutrition. 2021;41:133–175. doi: 10.1146/annurev-nutr-122019-120635. [DOI] [PubMed] [Google Scholar]
- Wu A, Tymoszuk P, Haschka D, Heeke S, Dichtl S, Petzer V, Seifert M, Hilbe R, Sopper S, Talasz H, Bumann D, Lass-Flörl C, Theurl I, Zhang K, Weiss G. Salmonella utilizes zinc to subvert antimicrobial host defense of macrophages via modulation of nf-κb signaling. Infection and Immunity. 2017;85:e00418-17. doi: 10.1128/IAI.00418-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie QW, Kashiwabara Y, Nathan C. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. The Journal of Biological Chemistry. 1994;269:4705–4708. [PubMed] [Google Scholar]
- Zhou Z, Wang L, Song Z, Saari JT, McClain CJ, Kang YJ. Abrogation of nuclear factor-kappaB activation is involved in zinc inhibition of lipopolysaccharide-induced tumor necrosis factor-alpha production and liver injury. The American Journal of Pathology. 2004;164:1547–1556. doi: 10.1016/s0002-9440(10)63713-3. [DOI] [PMC free article] [PubMed] [Google Scholar]