Summary
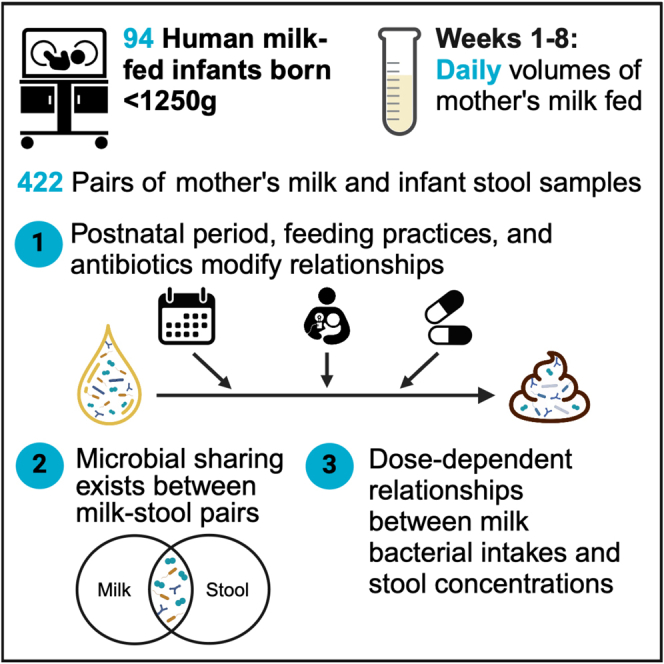
Mother’s milk contains diverse bacterial communities, although their impact on microbial colonization in very-low-birth-weight (VLBW, <1,500 g) infants remains unknown. Here, we examine relationships between the microbiota in preterm mother’s milk and the VLBW infant gut across initial hospitalization (n = 94 mother-infant dyads, 422 milk-stool pairs). Shared zero-radius operational taxonomic units (zOTUs) between milk-stool pairs account for ∼30%–40% of zOTUs in the VLBW infant’s gut. We show dose-response relationships between intakes of several genera from milk and their concentrations in the infant’s gut. These relationships and those related to microbial sharing change temporally and are modified by in-hospital feeding practices (especially direct breastfeeding) and maternal-infant antibiotic use. Correlations also exist between milk and stool microbial consortia, suggesting that multiple milk microbes may influence overall gut communities together. These results highlight that the mother’s milk microbiota may shape the gut colonization of VLBW infants by delivering specific bacteria and through intricate microbial interactions.
Keywords: preterm infant, very-low-birth-weight infant, microbiome, microbiota, mother’s milk, human milk, donor milk, nutrient fortification, direct breastfeeding, antibiotics
Graphical abstract
Highlights
-
•
Microbes shared between milk and stool make up 35% of bacteria in VLBW infant guts
-
•
Direct breastfeeding increases the likelihood of microbial sharing
-
•
Daily microbial intakes from milk show dose-response relationships in infant stool
-
•
Relationships are modified by postnatal period, feeding practices, and antibiotics
Shama and Asbury et al. demonstrate that, despite clinical interventions that perturb microbiomes, mother’s milk microbes and their dose are associated with gut microbial development in very-low-birth-weight (VLBW) infants. This highlights the potential for mother’s milk microbes to colonize the VLBW infant gut, especially with direct breastfeeding and limited antibiotics.
Introduction
Compared to healthy term-born infants, preterm very-low-birth-weight (VLBW, <1,500 g) infants disproportionately develop aberrant gut bacterial communities, characterized by low microbial diversity and an increased abundance of potentially pathogenic bacteria.1,2 Their perturbed microbial succession patterns have been associated with morbidities including necrotizing enterocolitis and late-onset sepsis.3,4 Encouragingly, mother’s milk feeding reduces the risk of these serious morbidities and improves neurodevelopment, making it the recommended source of nutrition for VLBW infants.5,6,7,8,9 In addition to its nutritive components, mother’s milk contains a myriad of bioactive components including microorganisms, human milk oligosaccharides (HMOs), and bioactive proteins that can favorably modify the gut microbial development of infants.
More recently, studies have demonstrated in healthy term mother-infant dyads that the complex community of bacteria in mother’s milk serve as pioneer colonizers in an infant’s gut.10,11,12,13,14,15,16 However, these findings may not be transferrable to mother-VLBW infant dyads, since they undergo distinct peri- and postnatal conditions that affect both the composition of mother’s milk and gut microbial colonization.17,18,19,20 VLBW infants and their mothers, for example, are frequently prescribed antibiotics prophylactically or for suspected or confirmed infections, and their use is associated with an altered microbiota in both preterm mother’s milk and the VLBW infant gut.1,19,21 VLBW infants are also born with immature swallowing and sucking reflexes, limiting their ability to feed at the breast or receive bottle feeds and resulting in mother’s milk that is often pumped, frozen, thawed, and delivered enterally through a feeding tube.22,23 Further, VLBW infants frequently require supplemental pasteurized donor human milk (PDHM), which no longer contains live bacterial cells due to the pasteurization process.24 Finally, both mother’s milk and PDHM are then nutrient enriched using multi-nutrient fortifiers to meet the elevated nutritional requirements of VLBW infants.25 While these nutritional practices are usually necessary, they do modify the nutritive and non-nutritive components (e.g., bacteria) delivered to the infant through mother’s milk, thereby impacting the infant’s gut bacterial environment.20,26,27,28 Given these distinct postnatal experiences, it is important to assess relationships between the microbiota in mother’s milk and VLBW infant’s gut independently from studies previously conducted in healthy term mother-infant dyads.
To our knowledge, no study has yet examined associations between the bacterial communities present in mother’s milk and the gut microbial colonization of VLBW infants. Furthermore, no study has accounted for the unique antibiotic and feeding practices among VLBW infant cohorts when assessing these relationships. Therefore, we aimed to investigate the relationships between the microbiota in preterm mother’s milk and the recipient VLBW infant gut and to establish whether these relationships are modified by postnatal period, in-hospital feeding practices (e.g., feeding type, fortifiers, feeding at the breast), and maternal-infant antibiotic exposures.
Results
Cohort characteristics
Ninety-four mother-infant dyads from the OptiMoM fortifier trial (NCT02137473) were included (Figure S1). The median (Q1, Q3) birth weight and gestational age of infants were 850 (730, 1047) grams and 27.4 (25.7, 29.1) weeks, respectively (Table S1). Approximately, 63.8% of infants were born by Cesarean section, and 92.6% were administered antibiotics for a median of 6 (3, 15) days. Over half (59.8%) of mothers received antibiotics either prenatally (within the two weeks prior to birth) or postnatally for a median of 1 (0, 4) day. Sixty-three (67.0%) infants were fed predominantly mother’s milk (i.e., ≥90% of enteral feeds) while the remaining 31 (33.0%) infants received mixed feeds with supplemental PDHM. Approximately half (54.3%) of infants were randomized to receive a human milk-based fortifier (HMBF), and probiotics were not part of clinical care for infants during this study. Seventy-nine (84.0%) infants were put to the breast (nutritive or non-nutritive sucking) before discharge, and feeding at the breast began at a median postmenstrual age of 34 (33, 35) weeks. We have previously characterized separately the microbiota of mother’s milk and infant stool samples collected from this cohort.19,20 In the present study, 422 milk-stool samples were paired to examine longitudinal relationships between the microbiota in mother’s milk and VLBW infant gut. Mother-infant dyads had a median of 5 (3, 6) milk-stool pairs collected during the first 8 postnatal weeks (Figure S2).
Microbial diversity of paired milk-stool samples
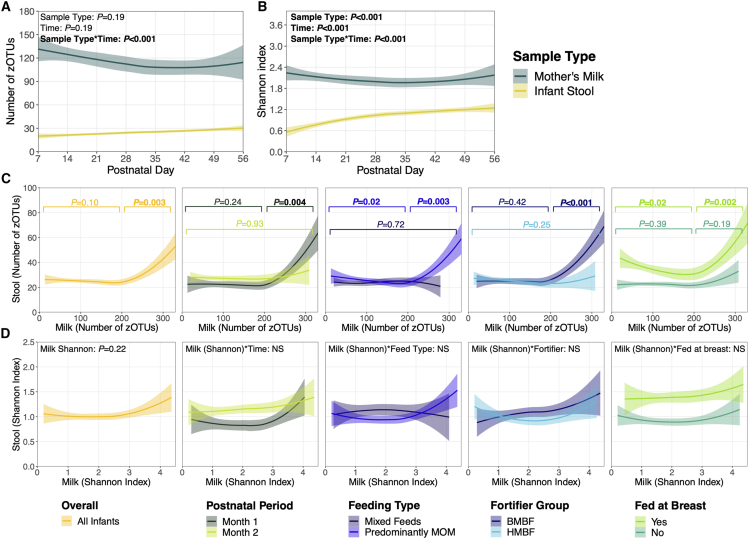
When comparing the overall alpha diversity between mother’s milk and infant stools, the number of zero-radius operational taxonomic units (zOTUs) and microbial diversity (by Shannon index) gradually decreased in mother’s milk over time but increased in infant stools (p < 0.001; Figures 1A and 1B). Notably, alpha diversity was higher in mother’s milk compared to infant stool samples. This may be attributed to the routine administration of antibiotics to infants in the early days postnatally and their limited microbial exposures during initial hospitalization. When examining the alpha diversity between paired milk-stool samples, a non-linear relationship was observed with the number of zOTUs. Specifically, no associations were identified between the number of zOTUs in mother’s milk and infant stools, unless milk samples contained >200 zOTUs, in which case a positive linear relationship was observed (p = 0.003; Figure 1C). Non-linear positive relationships were only present during the first (versus second) postnatal month (p = 0.004), with predominant mother’s milk (vs. mixed) feeding (p = 0.003), with bovine (versus human) milk-based fortification (p < 0.001), and after infants began feeding at the breast (p = 0.002). No relationships were observed between the microbial diversity (Shannon index) of paired milk-stool samples (Figure 1D). Of note, overall alpha diversity measures in mother’s milk and the infant stools independently increased once infants began feeding at the breast (Figure S3).
Figure 1.
Microbial alpha diversity of paired milk-stool samples
(A) Number of zOTUs and (B) Shannon index over time, stratified by sample type (n = 422 stools, n = 334 mother’s milk). Solid lines represent the mean alpha diversity over time, and shaded areas represent the 95% confidence interval. p values are from linear mixed-effects models adjusted for sample type, postnatal week, DNA extraction batch, and an interaction term between sample type and postnatal week. (C) Relationships between the number of zOTUs in paired milk-stool samples were examined for the entire cohort (in yellow) and by postnatal period and feeding variables of interest. Given the non-linear relationships observed (milk cut-point: zOTUs ≤ 200 or > 200), the number of zOTUs in paired milk-stool samples was investigated separately for each segment. p values (colored according to models stratified by each feeding variable of interest) are from unadjusted linear mixed-effects models due to sample size constraints within each segment. (D) Relationships with Shannon index in paired milk-stool samples were assessed using linear mixed-effects models adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest. Interaction terms were removed from final models if they were not statistically significant (p > 0.05; denoted with NS). Abbreviations: zOTUs, zero-radius operational taxonomic units; MOM, mother’s milk; HMBF, human milk-based fortifier; BMBF, bovine milk-based fortifier.
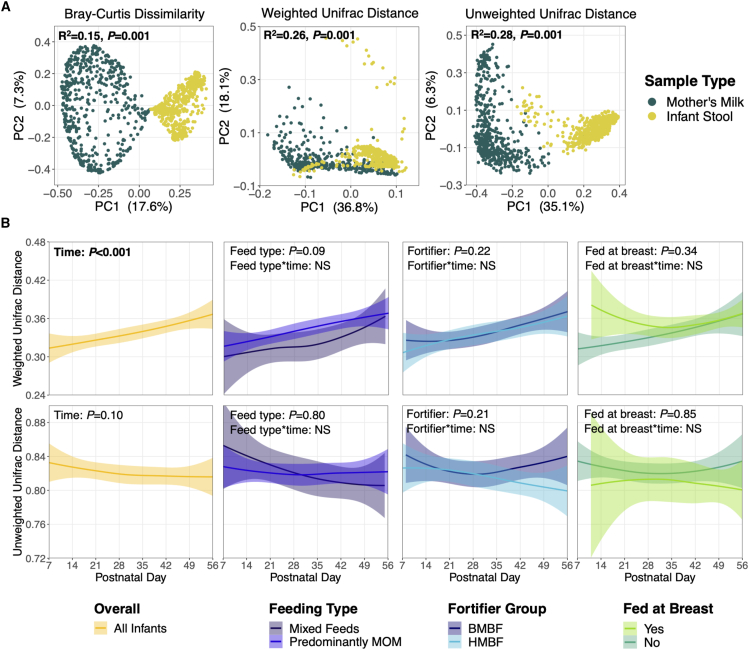
When comparing the overall beta diversity between mother’s milk and infant stool samples, distinct microbial community structures were observed (Bray-Curtis, R2 = 0.15, p = 0.001; weighted UniFrac, R2 = 0.26, p = 0.001; unweighted UniFrac, R2 = 0.28, p = 0.001; Figure 2A). Weighted UniFrac distances between paired milk-stools increased by postnatal week (p < 0.001), while unweighted UniFrac distances remained unchanged (p = 0.10; Figure 2B), suggesting that the microbiota in paired milk-stool samples became more dissimilar across time and that this divergence was driven by changes in bacterial abundances rather than the presence or absence of specific taxa. These observed relationships were not dependent on feeding type, fortifier type, or feeding at the breast.
Figure 2.
Microbial beta diversity of paired milk and stool samples
(A) Principal coordinate analysis plots of beta diversity metrics. R2 and p values are from adonis models adjusted for postnatal week, DNA extraction batch, and participant identification. (B) UniFrac distances between paired milk-stool samples were assessed over time for the entire cohort (in yellow) and by feeding variables of interest. Solid lines represent the mean distance between paired samples, while shaded areas represent 95% confidence intervals. p values are from linear mixed-effects models adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest. Interaction terms were tested between the feeding variables and postnatal week but were removed from final models if they were not statistically significant (p > 0.05; denoted with NS). Abbreviations: PC, principal component; MOM, mother’s milk; HMBF, human milk-based fortifier; BMBF, bovine milk-based fortifier.
Microbial taxa shared between paired milk-stool samples
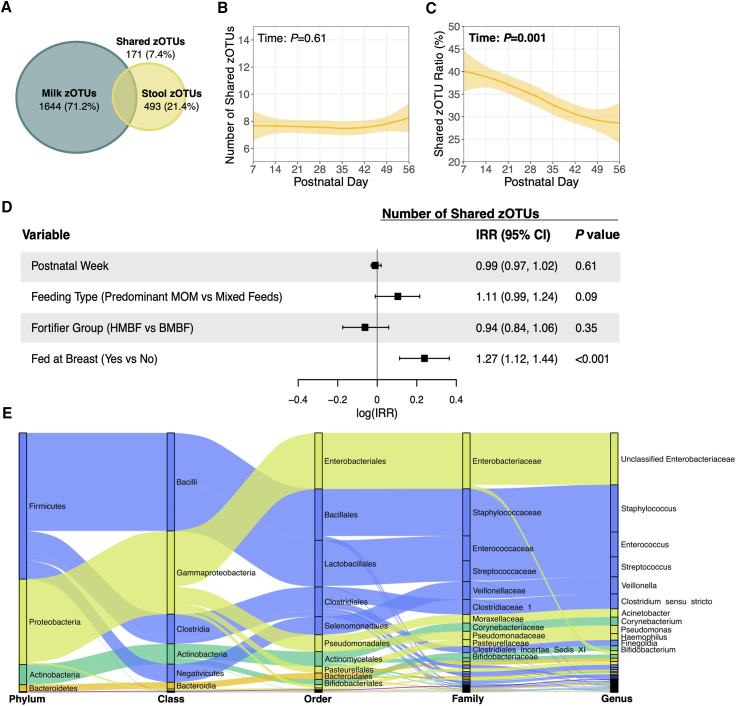
A total of 2,308 zOTUs were identified in mother’s milk and infant stool samples; of these, only 171 zOTUs (7.4%) were shared between paired milk-stool samples (Figure 3A). On average, 7 zOTUs (95% confidence interval [CI]: 7, 8) were shared within a given milk-stool pair, and this number did not change over time (Figure 3B). Shared zOTUs represented 43.1% (95% CI: 41.0, 45.2) of all zOTUs present in the infant gut during postnatal week 1, and this decreased to 28.7% (95% CI: 27.5, 29.9) by week 8 (p = 0.001; Figure 3C). Of note, the average shared zOTU ratio on a per mother-infant dyad basis was 34.4% (95% CI: 32.2, 36.5), which was consistent with the average shared zOTU ratio calculated across all milk-stool pairs (33.9%, 95% CI: 32.6, 35.4) (Table S2). We also examined the consistency of zOTU sharing in milk-stool pairs, where a milk sample was paired with two stool samples from the same infant (n = 26) (Table S3). The number of zOTUs shared between the first and second milk-stool pairs was largely consistent, and several zOTUs shared in one pair were also shared in the other.
Figure 3.
Shared microbial taxa between paired milk-stool samples
(A) Venn diagram showing the total number of zOTUs unique to milk and stool, and the total number of zOTUs that were shared in paired milk-stool samples. (B) Number of zOTUs shared between paired milk-stool samples over time. Solid lines represent the mean number of shared zOTUs over time, while the shaded areas represent 95% confidence intervals. p values are from linear mixed-effects models adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest. (C) Shared zOTU ratio was calculated as the number of shared zOTUs between paired milk-stool samples divided by the total number of zOTUs in each corresponding stool sample. This model was adjusted as described in (B). (D) Relationships between postnatal week, feeding variables of interest, and the likelihood of sharing a greater number of zOTUs between milk-stool pairings were assessed using an adjusted repeated measures Poisson regression model as described in (B). (E) Alluvial diagram depicting the taxonomy of shared zOTUs between paired milk-stool samples. Node sizes represent the number of milk-stool pairings that shared a zOTU mapping back to the specified taxa. For visual clarity, only taxa that were shared in approximately 10% of milk-stool pairings are listed. Abbreviations: zOTUs, zero-radius operational taxonomic units; IRR, incidence rate ratio; MOM, mother’s milk; HMBF, human milk-based fortifier; BMBF, bovine milk-based fortifier.
Feeding infants at the breast increased the likelihood of sharing a greater number of zOTUs (incidence rate ratio: 1.27, 95% CI: 1.12, 1.44; p < 0.001), whereas postnatal period, feeding type, and fortifier type did not change the likelihood of microbial zOTU sharing (Figure 3D). Shared zOTUs (n = 171) mapped to 60 different genera (Table S2); 12 genera were shared in ≥10% of milk-stools pairs (Figure 3E) and, therefore, used in subsequent analyses as “commonly shared genera.” These included unclassified Enterobacteriaceae, Staphylococcus, Enterococcus, Streptococcus, Veillonella, Clostridium sensu stricto, Acinetobacter, Corynebacterium, Pseudomonas, Haemophilus, Finegoldia, and Bifidobacterium (Table S2).
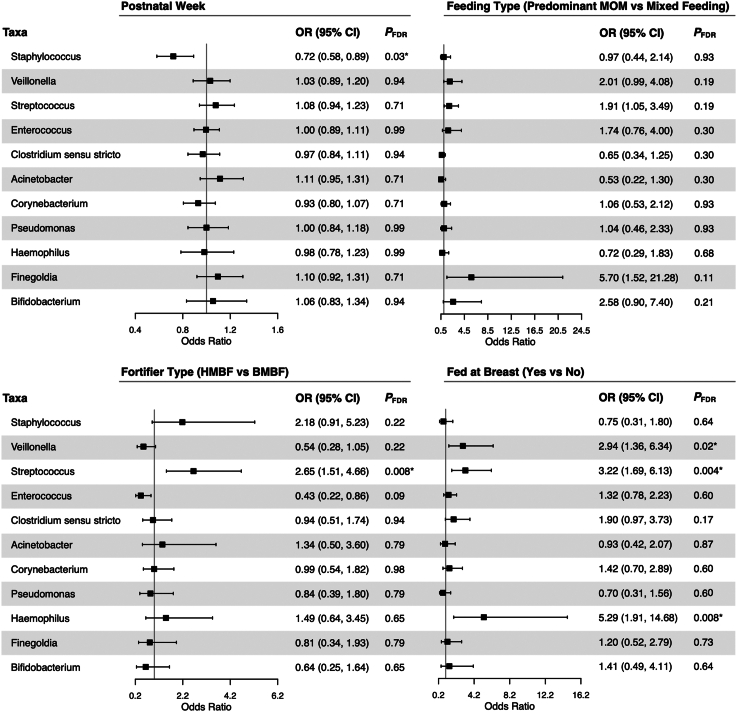
When assessing how postnatal period and in-hospital feeding practices were associated with zOTU sharing of the most commonly shared genera, we observed that the likelihood of sharing a zOTU mapping to Staphylococcus decreased with each additional postnatal week (odds ratio [OR] 0.72; 95% CI: 0.58, 0.89; pFDR = 0.03; Figure 4). Additionally, fortifier type was associated with an increased likelihood of having a shared zOTU mapping to Streptococcus, such that paired milk-stools collected from HMBF-fed (vs. bovine milk-based fortifier [BMBF]-fed) infants were more likely to share Streptococcus zOTUs (OR 2.65; 95% CI: 1.51, 4.66; pFDR = 0.008). Feeding an infant at the breast also increased the likelihood of sharing zOTUs mapping to Veillonella (OR 2.94; 95% CI: 1.36, 6.34; pFDR = 0.02), Streptococcus (OR 3.22; 95% CI: 1.69, 6.13; pFDR = 0.004), and Haemophilus (OR 5.29; 95% CI: 1.91, 14.68; pFDR = 0.008). Similarly, when we conducted a hierarchical clustering analysis to determine patterns of zOTU sharing (e.g., whether certain zOTUs are shared together), it was confirmed that once infants started feeding at the breast, they were more likely to have zOTU sharing patterns characterized by several genera including Veillonella, Streptococcus, Haemophilus, and Clostridium sensu stricto (Figure S4). Of note, independent of milk-stool sharing, the concentrations of these same 4 bacterial genera in the infant stool increased once infants began feeding at the breast (Figure S3).
Figure 4.
Likelihood of zOTU sharing in paired milk-stool samples depending on postnatal period and feeding practices
Repeated measures logistic regressions were used to assess how postnatal period and feeding practices influence the likelihood of sharing a zOTU within the 12 most commonly shared genera. Separate models were run for each genus, with the outcome being whether a paired milk-stool sample had a shared zOTU mapping to the specified genera (yes/no). Models were adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest. p values were adjusted using a Benjamini-Hochberg false discovery rate to account for multiple comparisons. Results for unclassified Enterobacteriaceae are not reported since almost all milk-stool pairs had a shared zOTU mapping back to this family, leading to model convergence issues. Abbreviations: OR, odds ratio; CI, confidence interval; MOM, mother’s milk; HMBF, human milk-based fortifier; BMBF, bovine milk-based fortifier.
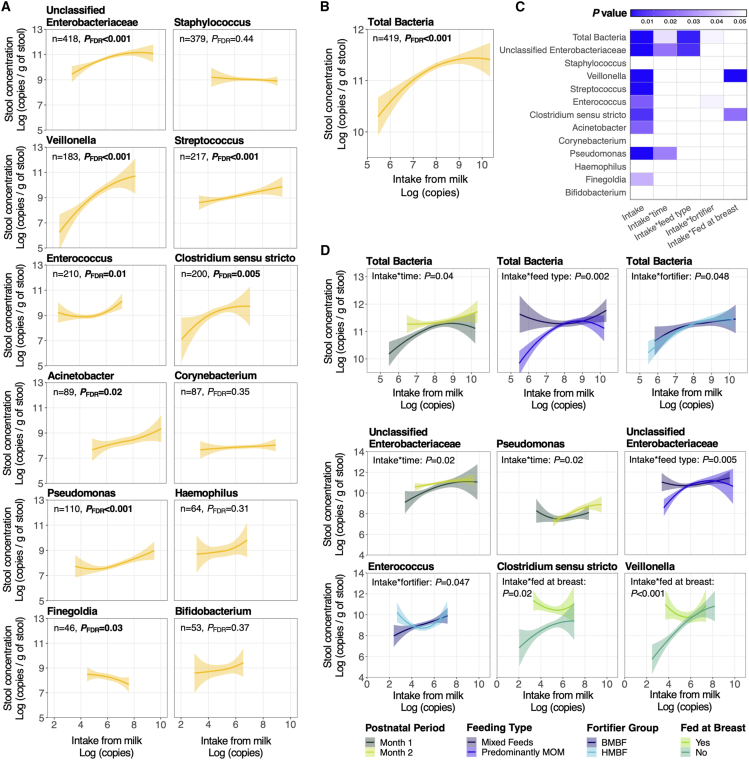
Dose-response relationships between bacterial intakes from mother’s milk and microbial concentrations in infant stools
Figure S5 illustrates both the bacterial concentration in mother’s milk and the bacterial intakes of VLBW infants from mother’s milk across initial hospitalization. Bacterial intakes were calculated from the bacterial concentrations in mother’s milk and the daily volume of mother’s milk fed (n = 3,508 days of bacterial intakes calculated across all infants). Total bacterial intakes from mother’s milk increased over time (p < 0.001), as did milk intakes of Staphylococcus, Pseudomonas, Acinetobacter, unclassified Enterobacteriaceae, and Finegoldia (p < 0.001). Intakes decreased for Clostridium sensu stricto and Haemophilus (p < 0.001) over time and remained unchanged for Corynebacterium, Streptococcus, Veillonella, Bifidobacterium, and Enterococcus.
We then investigated whether linear relationships exist between the dose of genera from mother’s milk and their concentrations in the infant’s gut. For these analyses, we included only milk-stool pairs where the given genus was present in both the mother’s milk and infant stool sample. From a clinical perspective, this was intended to identify potential genera with dose-response relationships that could be targeted for future studies and clinical applications. Cumulative bacterial intakes 3 days before each paired stool were positively associated with total bacterial concentrations in infant stools (p < 0.001; Figure 5B). For 7 of the 12 genera examined, positive associations were observed between 3-day bacterial intakes from mother’s milk and subsequent bacterial concentrations in infant stools (p = 0.03– < 0.001, Figure 5A); these positive associations were strongest for Veillonella (p < 0.002), Clostridium sensu stricto (p = 0.002), and Pseudomonas (p < 0.001). A negative association was observed between 3-day intakes of Finegoldia and its concentration in infant stools, while no associations were observed between 3-day intakes of Staphylococcus, Corynebacterium, Haemophilus, or Bifidobacterium and their respective concentrations in infant stools.
Figure 5.
Dose-response relationships between bacterial intakes from mother’s milk and their concentrations in infant stools
(A) Relationships between cumulative 3-day milk bacterial intakes of commonly shared taxa and their concentrations in corresponding infant stools were assessed using unadjusted linear mixed-effects models. Sample pairs were included in these models if both milk and stool contained the respective taxa, and p values were adjusted using a Benjamini-Hochberg false discovery rate. Solid lines represent the mean and shaded areas represent the 95% confidence interval. (B) Relationship between the cumulative 3-day total bacterial intake from mother’s milk and the total bacterial concentration in stool, as described in (A). (C) Summary of all p values from unadjusted linear mixed-effects models in (A) and (B) in addition to models stratified by postnatal period and feeding variables of interest. p values from the main effects were FDR-adjusted to account for multiple comparisons. (D) Statistically significant relationships between cumulative 3-day intakes of total bacteria and commonly shared taxa with corresponding concentrations in infant stools, stratified by postnatal period and feeding variables of interest. Abbreviations: NS, non-significant; FDR, false discovery rate; MOM, mother’s milk; HMBF, human milk-based fortifier; BMBF, bovine milk-based fortifier.
We then tested whether relationships between 3-day milk bacterial intakes and their concentrations in infant stools were modified by postnatal period and in-hospital feeding practices (Figures 5C, 5D, and Table S4). During postnatal month 1 (versus 2), stronger positive relationships were observed between 3-day bacterial intakes and concentrations in infant stools of total bacteria (p = 0.04) and unclassified Enterobacteriaceae (p = 0.02). In contrast, a positive relationship was observed between the 3-day milk intake of Pseudomonas and concentrations in infant stools, but only during postnatal month 2 (p = 0.02). Infants fed predominantly mother’s milk (versus mixed feeding with PDHM) showed stronger positive associations between 3-day bacterial intakes and concentrations in stools for total bacteria (p = 0.002) and unclassified Enterobacteriaceae (p = 0.005). Fortifier type also modified relationships between 3-day milk bacterial intakes and their concentrations in infant stools, such that a positive association was observed between the 3-day milk bacterial intakes of Enterococcus and their concentrations in infant stools, but only in BMBF-fed infants (p = 0.047). Lastly, prior to initiating direct breastfeeding, the 3-day milk intake of Clostridium sensu stricto (p = 0.02) and Veillonella (p < 0.001) was positively associated with their respective concentrations in infant stools; however, once infants started feeding at the breast, these relationships were no longer observed.
Importantly, some milk-stool pairs contained the 12 commonly shared genera in only the milk sample and not in the paired stool. We therefore aimed to establish whether postnatal period and feeding practices were associated with the odds of observing a given genera in both milk-stool samples rather than mother’s milk alone. The likelihood of Staphylococcus being present in both milk-stool samples (versus mother’s milk alone) decreased with each additional postnatal week (OR: 0.73, 95% CI: 0.59, 0.89; pFDR = 0.02; Figure S6). Feeding infants at the breast also increased the likelihood of observing Veillonella (OR: 3.35, 95% CI: 1.62, 6.91; pFDR = 0.006) and Haemophilus (OR: 4.13, 95% CI: 2.03, 8.42; pFDR < 0.001) in both the mother’s milk and stool samples paired.
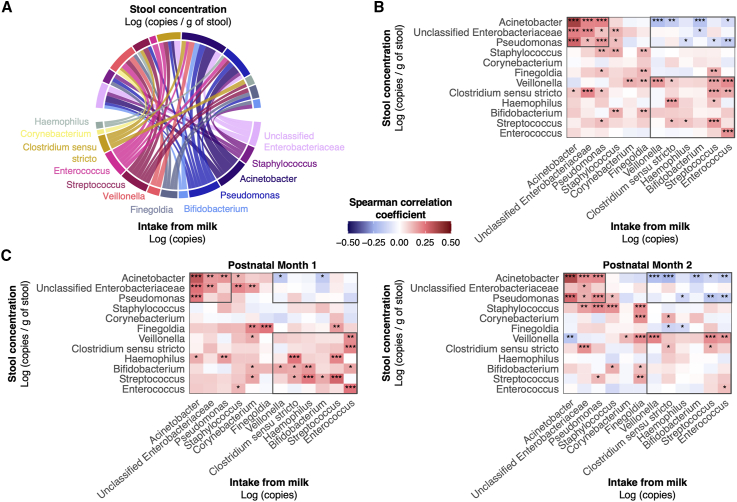
Microbial interactions exist between mother’s milk bacterial intakes and microbial concentrations in infant stools
To investigate the complex relationships among different mother’s milk bacterial genera (intake) and subsequent concentrations of various genera (concentration) in the infant’s gut, correlations between the 12 most commonly shared genera in milk and infant stools were assessed simultaneously. Forty-five of 144 potential correlations were statistically significant, showing varying strengths of correlation (Figure 6A). Apart from Corynebacterium, Clostridium sensu stricto, Haemophilus, and Bifidobacterium, the 3-day intake of a given bacterial genus was positively correlated with the concentration of the same genus in the paired infant stool sample (p < 0.05, Figure 6B). The intake of Acinetobacter, unclassified Enterobacteriaceae, and Pseudomonas clustered together into a positive correlation matrix (Spearman ρ = 0.12 to 0.37; p < 0.05), such that higher intakes of any of these genera present in mother’s milk were positively associated with each other’s stool concentrations. Importantly, the concentrations of these 3 genera in infant stools were negatively correlated with higher milk intakes of other bacterial genera, including Veillonella, Bifidobacterium, and Enterococcus (Spearman ρ = −0.20 to −0.10; p < 0.05).
Figure 6.
Correlations between milk bacterial intakes and concentrations in infant stools
(A) Chord diagram displaying statistically significant relationships between cumulative 3-day milk bacterial intakes of commonly shared genera and their concentrations infant stools. The width of the linkage is proportional to the strength of the correlation. (B) Heatmap displaying spearman rank correlations between 3-day milk bacterial intakes and concentrations in infant stools for the entire cohort. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (C) Heatmaps showing the spearman rank correlations as described in (A) stratified by postnatal period. Black borders are used to highlight key findings across heatmaps.
To determine whether these correlations were dependent on postnatal period, stratified correlation matrices were constructed (Figure 6C). For example, in month 1 samples, many genera associated with gut microbial maturity in preterm cohorts (e.g., Bifidobacterium, Streptococcus, and Veillonella) were positively correlated with each other across milk intakes and stool concentrations (e.g., Veillonella intake from mother’s milk was positively correlated with Bifidobacterium concentrations in the infant stool). In contrast, fewer positive correlations were observed between these “microbial maturity” taxa during month 2 but, instead, their intakes from mother’s milk showed negative correlations with stool concentrations of Gammaproteobacteria (e.g., Pseudomonas and Acinetobacter), which are associated with diseases in preterm infants (e.g., Streptococcus intake from mother’s milk was negatively correlated with Pseudomonas concentrations in the infant stool). Many of these same positive correlations (similar to month 1) were observed with predominant mother’s milk and BMBF feeding, whereas many of the negative correlations (similar to month 2) were observed with mixed feeding (Figure S7). Furthermore, the strength of these positive and negative correlations increased after infants started feeding at the breast.
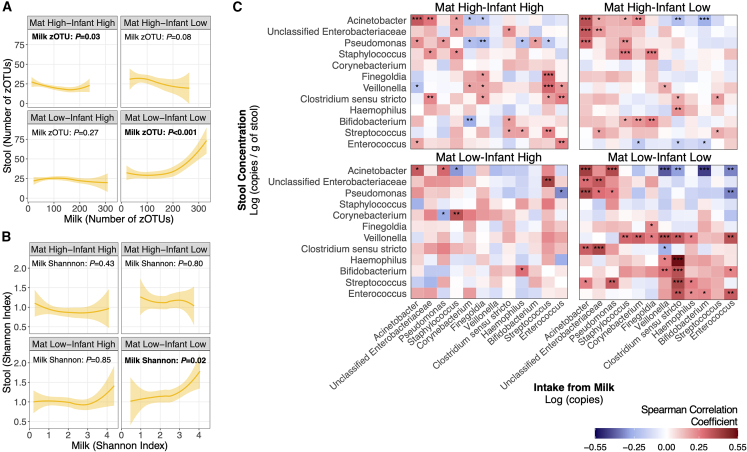
Maternal and infant antibiotic exposures modify relationships between the microbiota present in mother’s milk and infant stools
To examine whether antibiotic exposure, in either the mother or their infant, modified relationships between the microbiota in mother’s milk and the VLBW infant gut, mother-infant dyads were divided into 4 antibiotic groups (maternal high-infant high, maternal high-infant low, maternal low-infant high, and maternal low-infant low). High antibiotic exposure was defined as >1 day for mothers (prenatally or postnatally) and >3 days for infants. These cutoffs were selected to differentiate between the routine prophylactic administration of antibiotics prescribed to this population (e.g., to mother’s during Cesarean section delivery or to VLBW infants to reduce the risk of early-onset sepsis) and amounts used to treat suspected or confirmed infections.29,30 Relationships with alpha diversity of paired milk-stools were modified by antibiotic exposure in mothers and their infants (Figures 7A and 7B). Specifically, positive associations between the number of zOTUs (p < 0.001) and Shannon index (p = 0.02) in mother’s milk and infant stools were only observed in mother-infant dyads with low antibiotic exposure (maternal low-infant low group). Correlations between 3-day intakes of the 12 most commonly shared bacterial genera in mother’s milk and their concentrations in infant stools were also modified by antibiotic group. Similar positive and negative correlations with “microbial maturity” taxa and Gammaproteobacteria were observed, as we have previously described, and were strongest in the maternal low-infant low group compared to the other antibiotic treatment groups (p < 0.05).
Figure 7.
Relationships between the microbiotas in mother’s milk and infant stools according to maternal-infant antibiotic exposure
Relationships with (A) the number of zOTUs and (B) Shannon index in paired milk-stool samples stratified by antibiotic group: maternal low-infant low (n = 22 mother-infant dyads, n = 80 paired milk-stool samples), maternal low-infant high (n = 34 mother-infant dyads, n = 166 paired milk-stool samples), maternal high-infant low (n = 11 mother-infant dyads, n = 52 paired milk-stool samples), and maternal high-infant high (n = 27 mother-infant dyads, n = 124 paired milk-stool samples). High antibiotic use was defined as >1 day for mothers and >3 days for infants. Solid lines represent the mean alpha diversity, and shaded areas represent the 95% confidence interval. p values are from unadjusted linear mixed-effects models. (C) Heatmaps showing spearman rank correlations between cumulative 3-day milk bacterial intakes of commonly shared genera and their concentrations in infant stools stratified by antibiotic group. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Discussion
Our findings show that, despite the many challenges associated with preterm birth, which disrupt gut colonization, numerous relationships exist between the microbiota in mother’s milk and the VLBW infant gut. Specifically, the intakes of microbes from mother’s milk were positively associated with increased microbial diversity and with bacterial concentrations in the infant’s gut. These associations were modified by postnatal period (first versus second month), and by in-hospital feeding practices including feeding type (predominantly mother’s milk versus mixed feeding with PDHM), fortifier type (BMBFs versus HMBFs), and whether direct breastfeeding had been initiated. We also showed that microbes commonly associated with microbial maturity had positive correlations between mother’s milk and infant stools, while these same microbes in mother’s milk had negative correlations with Gammaproteobacteria in the infant gut. These relationships and others observed between the microbiota in milk-stool pairs were also significantly modified by prolonged antibiotic exposure in either the mother or the infant.
Dose-dependent positive relationships were observed between mother’s milk intakes of total bacteria and taxa such as unclassified Enterobacteriaceae, Veillonella, Streptococcus, Enterococcus, and Clostridium sensu stricto and their concentrations in the VLBW infant gut. To the best of our knowledge, no other study has examined relationships between daily bacterial intakes from mother’s milk and bacterial concentrations present in a preterm or term infant’s gut. However, previous cross-sectional work using relative abundance data shows similar patterns to our findings.11,31 Specifically, positive correlations were observed between the relative abundances of select bacterial genera (e.g., Streptococcus) in mother’s milk and infant stools. In our study, we also showed that milk intakes of Veillonella, Clostridium sensu stricto, Haemophilus, Bifidobacterium, Streptococcus, and Enterococcus were positively correlated with each other in the infant gut during postnatal month 1, and negatively associated with different Gammaproteobacteria (e.g., Pseudomonas and Acinetobacter) during month 2. Previous studies that have examined inter-genera correlations between bacteria in mother’s milk and an infant’s gut (term and moderate to late preterm) show a limited number of significant correlations11,31; however, this may be due to the cross-sectional and compositional (i.e., relative abundance) nature of their data, which can mask underlying community dynamics.32 It is possible that the positive correlations we observed during postnatal month 1 are attributed to having available ecological niches in the infant’s gut early after birth; however, over time, as these niches become more occupied, interactions between bacterial intakes from mother’s milk and resident microbes in the infant’s gut may become more competitive, as reflected by the negative correlations we observed during postnatal month 2. Future appropriately sized studies are now needed to understand whether these observed relationships impact the growth and health outcomes of VLBW infants.
It is important to note that probiotics were not given to either mothers or infants during the course of this study; however, using daily bacterial intakes from mother’s milk (expressed as log copies), we estimated that VLBW infants consume ∼0.033 × 109 colony-forming unit (CFU) of bacteria daily from mother’s milk, which is considerably lower than concentrations of probiotics currently used in North American neonatal intensive care units (NICUs) (0.1–2 × 109 CFU) (Figure S5B). Future work is needed to understand whether probiotics (and their current dose formulations) modify the relationships observed between the microbiota in mother’s milk and the VLBW infant gut.
Previous studies in term-born infants have shown that mother’s milk contributes up to 40% of the bacteria present in the infant’s gut.33,34,35 Despite their perturbed postnatal environments, we similarly observed that shared bacteria, identified between milk-stool pairs, accounted for 30%–40% of the bacteria in the VLBW infant’s gut across initial hospitalization. Our findings are also in line with a previous study (n = 34 term mother-infant dyads, Denmark), which showed that the absolute number of bacteria shared between milk-stool pairs remained unchanged over time but that the contribution of milk microbes decreased as the infant’s gut diversified.36 Across hospitalization, we observed a decreased likelihood of sharing zOTUs mapping to Staphylococcus. No other study, to the best of our knowledge, has examined how microbial sharing between milk-stool pairs changes across time; however, it is biologically plausible to observe a decrease in microbial sharing of Staphylococcus since its abundance diminishes over time in the milk microbiota of mothers delivering preterm.19,37
To our knowledge, no study to date has used high-throughput sequencing methods to examine whether a shared microbiota exists between mother’s milk and the VLBW infant’s gut. However, several taxa identified in our study as commonly shared, including Veillonella, Streptococcus, Staphylococcus, Haemophilus, Clostridium sensu stricto, and Bifidobacterium, were also reported as shared in previous term-born cohorts.11,36,38,39 The abundance of these shared bacterial genera may be particularly important for short- and long-term health outcomes in VLBW infants. For example, an increased abundance of Veillonella in the gut of infants has previously been associated with a reduced risk of developing asthma later in life, and with appropriate growth in extremely preterm infants.40,41 Bifidobacterium and Veillonella can also convert HMOs and lactate, respectively, into short-chain fatty acids (SCFAs), offering several benefits to the VLBW infant.42,43,44 Lastly, Clostridium may be a key bacterial taxon since higher abundances in the gut of preterm infants have been associated with improved intestinal barrier function and decreased permeability.45 Select Clostridia species may also contribute to the development and tolerance of the immune system by promoting the differentiation and expansion of regulatory T cells and by producing SCFA.46
Feeding type modulated several relationships observed between the microbiota in mother’s milk and the VLBW infant gut. Infants fed predominantly mother’s milk (≥90%) showed stronger positive associations and a greater number of correlations between the milk intake of bacteria and their concentrations in the infant’s gut, compared to infants receiving mixed feeds with PDHM. The more pronounced associations observed with predominant mother’s milk feeding likely reflect higher doses of viable bacteria and other milk bioactive proteins (e.g., lactoferrin) delivered with greater volumes of mother’s milk versus PDHM. It is unclear whether components and fragments of dead bacterial cells in PDHM differentially impact the gut microenvironment and microbial development of VLBW infants.47 Importantly, in our cohort, mother’s milk made up a high percentage of infant feeds (on average, 85.1% of feeds), even in the mixed feeding group (57.8% of feeds), which may have limited our ability to detect all significant relationships modified by feeding type.
Fortifier type also modified several observed associations, such that relationships between the milk and infant gut microbiota for BMBF-fed infants more closely resembled those observed with predominant mother’s milk feeding. In contrast, relationships observed for HMBF-fed infants more closely resembled those observed with mixed feedings. For example, a positive association between the microbial diversity of paired milk-stool samples was only observed for BMBF-fed infants and with predominant mother’s milk feeding. It is possible that these relationships were modified by fortifier group since HMBFs displace 40% of the mother’s milk at the caloric strength (28 kcal/oz) commonly fed in our study, whereas BMBFs only minimally displaced the volume of mother’s milk fed.20 Alternatively, the composition of each fortifier may differentially promote the proliferation of bacteria in mother’s milk and the infant’s gut and modify the overall gut environment.20 In particular, BMBFs are currently devoid of HMOs, while HMBFs, produced from large pools of donated human milk, contain a range of HMO structures produced by both secretor and non-secretor donors.48,49 Given that HMO concentrations differ in the milk of secretor versus non-secretor mothers, we hypothesize that the diverse range of HMO structures, provided from HMBFs, may influence the microbial development of VLBW infants and consequently the relationships observed between the microbiota in mother’s milk and the infant’s gut.50
We identified direct breastfeeding as a key modifier of microbial sharing between mother’s milk and the infant gut. Not only did the number of zOTUs shared between mother’s milk and stools increase after infants were fed directly at the breast but a consistent pattern of enhanced sharing of Veillonella, Streptococcus, Haemophilus, and Clostridium sensu stricto was also identified. No other study to date has examined whether direct breastfeeding modifies microbial sharing of taxa between milk-stool pairs; however, our results mirror those previously reported in full-term and moderate-late preterm infants, showing greater microbial diversity and abundance of Streptococcus and Haemophilus in mother’s milk samples collected after initiating direct breastfeeding.37,51 It is possible that multiple mechanisms underlie these relationships. First, given the “backwash” that occurs during direct breastfeeding, bacteria can be transferred to mother’s milk from the infant’s oral cavity or nasopharyngeal tract (such as Streptococcus and Haemophilus) and subsequently delivered from mother’s milk to lower segments of the infant’s gut.52 Future studies exploring the unique microbial contributions to mother’s milk, from various maternal (mother’s skin) and infant sources (infant’s mouth), and their subsequent impact on the VLBW infant’s gut microbial development will be informative. Second, we suspect that, by feeding infants at the breast, anaerobic bacteria (e.g., Veillonella, Clostridium sensu stricto) are directly translocated to the infant without prolonged exposure to air that occurs after mother’s milk is pumped. Similarly, as reported in a recent study, the provision of fresh versus frozen mother’s milk likely improves the viability of these microbes and thereby increases their proliferation within the preterm infant gut.53 Lastly, nutritive direct breastfeeding provides milk without the use of enteral feeding tubes, which are initially used to deliver feeds to VLBW infants. While necessary, enteral feeding tubes are known to harbor bacterial biofilms and strip human milk feeds of fat via adhesion to plastic tubing.28,54 However, it is unknown whether these tubes also impact the amount and composition of mother’s milk microbiota delivered to the infant.
Antibiotic exposure significantly modified relationships between the microbiota present in mother’s milk and in the VLBW infant gut. Specifically, the observed positive association between the alpha diversity in mother’s milk and infant stools no longer existed when mothers or their infants were exposed to high antibiotic use. Further, the presence and strength of correlations between bacterial intakes from mother’s milk and subsequent bacterial concentrations in the infant’s gut were also modified. It is likely that these relationships are dependent on both maternal and infant antibiotic exposures, since the former is associated with changes in the milk microbiota19 and delivering low-dose antibiotics through milk,55,56,57 while the latter alters the infant’s gut environment making it less favorable for certain susceptible milk bacteria to thrive.58 We emphasize that these findings should not discourage the use of mother’s milk during antibiotic exposure; mother’s milk is a complex biological fluid encompassing multiple vital antimicrobial and immunologic components for the developing infant.59 Instead, our findings highlight that discussions around antibiotic stewardship are needed for both mothers and their preterm infants, since exposure in either group may carry potential short- and long-term consequences for the developing infant.
Strengths of this analysis include the use of normalized abundance data32 and detailed daily feeding data, which allowed us to calculate both the intakes of genera from mother’s milk and their concentrations in the infant’s stool. Furthermore, milk-stool samples were paired for each mother-infant dyad weekly, ensuring that any temporal trends in the mother’s milk and infant stool microbiota could be accounted for.
Limitations of the study
Importantly, this work is not without its limitations. First, 16S rRNA gene sequencing is unable to distinguish between viable and dead bacteria in mother’s milk and their differential impact on the infant’s gut microbiota. Furthermore, this sequencing method limits the ability to examine relationships at the strain level, which is needed to confirm vertical transmission of bacterial strains from mother’s milk to the VLBW infant gut. Metagenomic sequencing would identify transferred bacterial strains and their functional potential; however its feasibility for studying the preterm mother’s milk microbiota remains hindered. Namely, the predominance of human DNA, which overshadows microbial DNA, and the ethical challenges of collecting sufficient volumes of milk from the limited amounts typically produced by mothers of preterm infants present significant barriers. Finally, due to their widespread and heterogeneous use, we were also unable to examine how different types and combinations of antibiotics, administered to mothers and their infants, modified relationships between the mother’s milk and infant gut microbiota. This warrants future investigation as different classes of antibiotics are likely to impact bacterial taxa differently.19
In conclusion, results from our longitudinal cohort study show that the microbiota in mother’s milk is associated with the developing gut microbiota of VLBW infants, often in a dose-dependent manner and modified by postnatal period, in-hospital feeding practices, and maternal-infant antibiotic exposure. These findings highlight that mother’s milk holds promise for being a potential vehicle to alter the gut microbiota of VLBW infants, which may in turn improve both their short- and long-term health. We anticipate that this work can be leveraged to encourage direct breastfeeding in the NICU and improve antibiotic stewardship among VLBW mother-infant dyads, given the widespread modifying effect these factors had on microbial sharing between mother’s milk and the infant gut. In combination with future research, findings from the present study may serve as a basis to formulate future microbial products for VLBW infants (e.g., probiotics or potentially postbiotics) that provide appropriate doses of microbes, which work synergistically to favorably shape and potentially rehabilitate the gut environment of vulnerable VLBW infants.
Resource availability
Lead contact
Further requests for information should be directed to and will be fulfilled by the lead contact, Deborah L. O’Connor (deborah.oconnor@utoronto.ca).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Raw sequences for this study are available at the NCBI Sequence Read Archive under the BioProject accession numbers PRJNA607284 and PRJNA723326 (http://www.ncbi.nlm.nih.gov/sra).
-
•
This study used pre-existing software without generating new custom code or algorithms.
-
•
Requests for additional information required to reanalyze the data should be directed to and will be fulfilled by the lead contact.
Consortia
Members of the OptiMoM Feeding Group are as follows: Susanne Aufreiter, Joanna Benec, Alan Daneman, Jane Francis, Carleigh Jenkins, Michael Jory, Iryna Muzyka, Aneta Plaga, Celina Purugganan, and Brock Williams (The Hospital for Sick Children); Edmond Kelly, Kirsten Kotsopoulos, Adel Mohamed, Prakesh Shah, and Karen Weishuhn (Sinai Health); Anwar Asady, Ann Bayliss, and Sandra Gabriele (Trillium Health Partners); Shirley Sit and Sue Ekserci (Humber River Hospital); Mahmud AlMadani (Lakeridge Health); Navneet Sharma (Markham Stouffville Hospital); Shaheen Doctor (North York General Hospital); Debbie Stone (Rogers Hixon Ontario Human Milk Bank); Peter Azzopardi and Karen Chang (Scarborough Health Network); David Gryn (Mackenzie Health); Jelena Popovic (Michael Garron Hospital); Douglas Campbell and Debby Arts-Rodas (Unity Health Toronto); Sandra Payne and Charmaine van Schaik (Southlake Regional Health Centre); and Ilona Burkot, Judy Gibson-Stoliar, and Simone Vaz (William Osler Health System).
Acknowledgments
We gratefully acknowledge the families for their participation, the diet technicians for preparing study feeds, Michael Jory for assisting with data management, and Victoria Forte, Maimuna Gias, and Jong Yup Sa for assisting with DNA extractions. The graphical abstract was created with BioRender.com. The OptiMoM Fortifier Study is supported by grants from the Canadian Institutes of Health Research (CIHR) (FHG129919; FDN143233). S.S. received graduate stipends from CIHR (FRN#181488), a clinician-scientist training program award from The Hospital for Sick Children, and the National Institutes of Health (R01HD111018). M.R.A. received postdoctoral fellowships from the Killam Trusts, the Molly Towell Perinatal Research Foundation, and L’Oreal Canada-UNESCO. A.S. holds funding from the Ontario Ministry of Economic Development and Innovation (project 13440), Genome Canada and the Ontario Genomics Institute (OGI-149), and CIHR (ECD-144627). E.M.C. received funding from the Lawson Family Chair in Microbiome Nutrition Research at the University of Toronto. All funders had no involvement in the study design, analysis, interpretation, or manuscript preparation.
Author contributions
S.S., M.R.A., N.B., and D.L.O. designed the study; N.B., C.T., S.U., and D.L.O. oversaw the trial and the collection of samples and data; S.S., M.R.A., A.T., E.M.C., A.G., and A. Kothari were involved in conducting qPCR; J.K.C. and P.W.W. performed the sequencing and bioinformatics; S.S., M.R.A., and A. Kiss were involved in data analyses, statistics, and figure generation; S.S., M.R.A., J.B., E.M.C., P.M.S., A.S., C.T., S.U., and D.L.O. provided interpretations of the data; S.S. wrote the initial draft of the manuscript; all authors approved the final submission; D.L.O. has primary responsibility for the final manuscript.
Declaration of interests
E.M.C. acknowledges research support from Lallemand Health Solutions and Ocean Spray and consultant fees, speaker, and/or travel support from Danone, Nestlé, and Lallemand Health Solutions. P.M.S. is a stockholder and an advisory board member for Antibe Therapeutics Inc. A.S. is a co-founder of MedBiome, a clinical microbiomics company.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Mother’s milk (1 mL) | (O’Connor et al., 2018)59 | Clinicaltrials.gov, NCT02137473 |
| Infant stool samples | (O’Connor et al., 2018)59 | Clinicaltrials.gov, NCT02137473 |
| Critical commercial assays | ||
| NucleoSpin Food DNA Isolation Kit | Macherey-Nagel | Cat#740945.50 |
| DNeasy PowerSoil Kit | Qiagen | Cat#12855-100 |
| KAPA2G Robust HotStart Ready Mix | KAPA Biosystems | Cat#KK5702 |
| 1.8X Ampure XP Magnetic Beads | Agencourt | Cat#A63881 |
| MiSeq Reagent Kit V2 (150 bp x 2) | Illumina | Cat#MS-102-2002 |
| TaqMan™ Gene Expression Master Mix | Thermo Fisher Scientific | Cat#4444964 |
| Custom TaqMan™ Gene Expression Assay | Thermo Fisher Scientific | Cat#4332078 |
| Deposited data | ||
| Milk raw sequencing data | (Asbury et al., 2020)19 | 16S rRNA gene sequence data (NCBI) BioProject: PRJNA607284 |
| Stool raw sequencing data | (Asbury et al., 2022)20 | 16S rRNA sequence data (NCBI) BioProject: PRJNA723326 |
| Oligonucleotides | ||
| 16S rRNA Forward Primer for V4 region 515F: GTGCCAGCMGCCGCGGTAA | (Caporaso et al., 2012)60 | N/A |
| 16S rRNA Reverse Primer for V4 region 806R: GGACTACHVGGGTWTCTAAT | (Caporaso et al., 2012)60 | N/A |
| 16S rRNA Forward Primer for all bacteria: CG GTGAATACGTTCCCGG | (Furet et al., 2009)61 | N/A |
| 16S rRNA Reverse Primer for all bacteria: TA CGGCTACCTTGTTACGACTT | (Furet et al., 2009)61 | N/A |
| 16S rRNA Probe for all bacteria: 50-CTTGTAC ACACCGCCCGTC-30 | (Furet et al., 2009)61 | N/A |
| Software and algorithms | ||
| USEARCH version 11.0.667 | (Edgar 2010, 2013, 2016)62,63,64 | http://www.drive5.com/usearch/ |
| VSEARCH version 2.10.4 | (Rognes et al., 2016)65 | https://github.com/torognes/vsearch |
| QIIME1 version 1.9.1 | (Caporaso et al., 2010)66 | http://qiime.org |
| FastTree version 2.1.11 | (Price et al., 2009)67 | http://www.microbesonline.org/fasttree/ |
| R version 4.3.1 | (R Core Team, 2017)68 | https://www.r-project.org |
| Phyloseq version 1.44.0 | (McMurdie et al., 2013)69 | https://joey711.github.io/phyloseq/ |
| Decontam version 1.13.0 | (Davis et al., 2018)70 | https://benjjneb.github.io/decontam/ |
| SAS University Edition | SAS Institute Inc | N/A |
| Vegan version 2.6.4 | (Oksanen et al., 2022)71 | https://github.com/vegandevs/vegan |
| ggplot2 version 3.4.3 | (Wickham et al., 2016)72 | https://ggplot2.tidyverse.org |
| forestplot version 3.1.3 | (Gordon et al., 2023)73 | https://rdrr.io/cran/forestplot/ |
| ggalluvial version 0.12.5 | (Brunson et al., 2018)74 | https://corybrunson.github.io/ggalluvial/ |
| pheatmap version 1.0.12 | (Kolde et al., 2018)75 | https://github.com/raivokolde/pheatmap |
| circlize version 0.4.15 | (Gu et al., 2014)76 | https://jokergoo.github.io/circlize_book/book/ |
| Other | ||
| ZymoBIOMICS Mock Community Standard | Zymo Research | Cat#D6300 |
| ZymoBIOMICS Mock Community DNA Standard | Zymo Research | Cat#D6306 |
Experimental model and study participant details
Study participants and design
Mother-infant dyads from the OptiMoM Fortifier Study (Optimizing Mothers’ Milk for Preterm Infants; NCT02137473), are included in this analysis.77 The primary objective of this multi-center, triple-blind, randomized clinical trial was to determine whether an HMBF, compared with a BMBF, improved feeding tolerance in infants born <1250g. Examination of the impact of fortifier type, as randomized, on the gut microbiota was pre-planned and these findings have been published as was the characterization of the mother’s milk microbiota.19,20 In the present paper, we determined whether the mother’s milk microbiota was associated with the gut microbial colonization of VLBW infants. Infants were eligible to participate in the original trial if they were born <1250g and if parents consented to using supplemental PDHM whenever there were insufficient mother’s milk volumes to meet the infant’s nutritional needs. Exclusion criteria for the trial included having a congenital or chromosomal anomaly affecting growth, enrollment in another study affecting nutritional management, receiving formula or a BMBF prior to study day 1 (first day of fortifier use), enteral feeds not anticipated to begin within 14 days of birth, or if the infant would likely be transferred to an NICU without available research ethics approval. Infants were recruited from two tertiary NICUs in Toronto, Canada between August 2014 and November 2015. The trial continued even if infants were transferred to any of the 16 participating level II NICUs and lasted until postnatal day 84, hospital discharge, or until the infant received two full oral feeds (breast or bottle) over three consecutive days. During the trial, mother’s milk was always fed first, followed by PDHM (Holder method 62.5°C for 30 min) if volumes were insufficient. Nutrient fortification began at ≥100 mL/kg/d and full enteral feeds were considered achieved at 160 mL/kg/d. Participating NICUs routinely conducted skin-to-skin care, however at the time of the trial, probiotics were not yet integrated into standard practice and therefore were not administered to infants. The study was approved by research ethics boards at each participating hospital and informed consent was obtained from parents of participating infants.
Clinical data and sample collection
Details of data and sample collection can be found elsewhere.19,20,77 Briefly, maternal and infant characteristics, including demographics, morbidity, and antibiotic use, were collected prospectively from medical charts. Maternal antibiotic use was collected daily for 2 weeks before birth and continued daily for both mothers and their infants from delivery until infant stool sample collection was complete at 8 postnatal weeks or hospital discharge. Collection of mother’s milk samples for our research is intended to reflect clinical practice. As part of routine care, mothers were educated on hygienic practices to express their milk and to clean their pumping devices. Generally, they express their milk by pump, but hand expression is also encouraged, and milk is stored at −20°C until use. At the time of this study, all milk preparation rooms used a “first in” “first out” inventory system. All mother’s milk fed to preterm infants (unfortified or fortified) was prepared in designated milk preparation rooms and packaged into feeding syringes under a laminar flow hood by trained staff. Milk was thawed and prepared once per day for feeding the infant over the subsequent 24-h and an aliquot of this pooled milk was collected for research purposes and stored at −80°C.19 For ethical reasons, samples were collected only if milk remained after enteral feed preparation. At the unit level, syringes were administered via a feeding tube and bedside nurses recorded the volume of enteral feeds consumed in the infant’s medical record. Members of our study team checked the medical record daily to ensure all feeds were recorded and to collect the volumes consumed for our study database.19 Infant stool samples were collected weekly from birth to 8 postnatal weeks from infant diapers into sterile containers and stored at −20°C.20 Within 24 h of collection, stool samples were transported to the Hospital for Sick Children on ice and stored at −80°C.
Method details
Pairing mother’s milk and infant stool samples within mother-infant dyads across hospitalization
During the 8 postnatal weeks of weekly mother’s milk and infant stool collections, infant stools were paired with the nearest milk sample collected within a +/− 7-day range; however, preference was given to milk samples collected in the 7-days prior to each stool. Importantly, mother’s milk samples were paired according to the actual date they were fed to the infant, and not the day of expression. Mother’s milk samples could have been paired with two stool samples from the same infant (n = 26 cases) provided they fell within the +/− 7-day range in order to maximize the number of milk-stool pairs in our analyses; we treated each milk-stool pair as a distinct data point with a unique pairing ID, which was retained for multivariable models. Similarly, for mothers of twins (n = 6 mothers) or triplets (n = 3 mothers), milk samples were paired with stools collected from each participating infant. In cases where no mother’s milk sample was collected within the +/− 7-day range, no pairing was made and the stool sample was removed from our analyses.
DNA extraction, PCR amplification, and 16S rRNA gene sequencing
DNA extraction and sequencing procedures have previously been described for the mother’s milk and infant stool samples included in this analysis.19,20 Briefly, DNA extraction was conducted for mother’s milk and infant stool samples using the NucleoSpin Food DNA Isolation Kit and the DNeasy PowerSoil Kit, respectively.60 The V4 hypervariable region of the 16S rRNA gene was then amplified and sequenced using forward (515F) and reverse (806R) primers on a MiSeq platform (Illumina, San Diego, CA, USA).61 Negative (DNA elution buffer and template-free, respectively) and positive (ZymoBIOMICS Mock Community Standards) controls were included for each DNA extraction batch and sequencing run.
Quantitative PCR for total bacteria
Total bacterial density in the infant stool samples was previously determined by quantitative PCR (qPCR)20; the same protocol has now been applied to the mother’s milk samples in this present study. Briefly, 1 μL of extracted DNA (10 ng/uL), 5 μL of 2X TaqMan Gene Expression Master Mix (Applied Biosystems, Thermo Fisher Scientific, Foster City, CA, USA), 0.5 μL of custom TaqMan assay targeting the 16S rRNA gene,62 and 3.5 μL of nuclease-free water were combined to perform each qPCR reaction (10 μL per reaction). A 7900HT Fast Real-Time PCR System (Applied Biosystems) run with default thermal settings was then used to run each reaction in triplicate. The total bacterial concentration of samples was then determined using a pGEM T-Easy-plasmid based standard curve. Calculations accounted for the amount of DNA used for qPCR (ng) and the total amount of DNA extracted per gram of stool (ng/g) or per mL of milk (ng/mL). Each qPCR run contained a template-free negative control and a custom bacterial community as a positive control. Total bacterial concentrations for milk and stool samples are expressed as 16S copy numbers/mL of milk or 16S copy numbers/g of stool, respectively.
Categorizing in-hospital feeding variables
Feeding type was categorized as predominantly mother’s milk fed (≥90% mother’s milk fed during hospitalization) and mixed feeding with PDHM (0–89% mother’s milk fed during hospitalization). These categories aimed to differentiate between infants requiring PDHM temporarily as a bridge during the first postnatal week until sufficient volumes of their mother’s milk became available, and those who continued to require PDHM as a supplement throughout hospitalization. Fortifier type was categorized as either BMBF or HMBF based on the initial trial randomization. One infant received the incorrect fortifier as randomized and was therefore excluded from these analyses. Feeding at the breast (yes/no) was determined using the first postnatal day infants were put to the breast (this included both nutritive and non-nutritive sucking). Paired milk-stool samples collected after this first postnatal day were categorized as a “yes” to indicate that direct breastfeeding was initiated.
Quantification and statistical analysis
Pre-processing of microbial data
Sequencing data were checked for quality, trimmed, and filtered using USEARCH (version 11.0.667) and VSEARCH (version 2.10.4) through the UNOISE pipeline, as previously described.19,20,63,64,65,78 Sequencing were de-replicated using VSEARCH, filtered to remove singleton reads, and UNOISE3, accessed through USEARCH, was used to denoise and remove chimeras. Retained sequences were then used to generate zOTUs through exact sequence variant clustering. Of note, zOTUs are synonymous with amplicon sequence variants.79 Taxonomy was assigned using the USEARCH SINTAX algorithm and the Ribosomal Database Project database version 16 and a minimum cut-off of 0.8 was applied.67 zOTU FASTA sequences were then aligned using QIIME1 and those that did not align were removed. FastTree was used to build a phylogenetic tree of the aligned sequences.66,68
Data analyses were performed in R (version 4.3.1)70 and pre-processing steps were conducted separately for mother’s milk and infant stool samples to avoid the unintentional removal of mother’s milk samples with lower read counts. First, contaminant zOTUs, identified in DNA extraction and PCR amplification negative controls, were removed using decontam (version 1.13.0).69 This step was conducted separately for milk and stool samples since DNA extraction and sequencing were performed separately for each sample type. The isContaminant function in decontam was used with parameters method = ”prevalence” and neg = ”isNeg” to identify and remove potential contaminant zOTUs specific to each sample type. Phyloseq (version 1.44.0) was then used to remove singleton and doubleton zOTUs as well as those mapping to Cyanobacteria/Chloroplast. Data for mother’s milk and infant stools were rarified to 3,500 and 10,000 reads per sample, respectively, based on minimum sequence coverage; samples with read counts below these thresholds were removed.72 Microbial data was then merged between mother’s milk and infant stool samples for all downstream analyses. Data visualization was conducted using the packages ggplot2 (version 3.4.3),74 ggalluvial (version 0.12.5),75 pheatmap (version 1.0.12),73 forestplot (version 3.1.3),76 and circlize (version 0.4.15).80
Calculating normalized taxa abundances
To overcome limitations of compositional data (relative abundances), normalized taxa abundances were calculated for both mother’s milk and infant stool samples. The predicted 16S rRNA gene copy numbers of zOTUs were obtained from USEARCH and the UNBIAS database and used to normalize zOTU counts.81 Copy number-corrected zOTU counts were then converted to relative abundances and multiplied by the total bacterial concentration of milk and infant stool samples obtained from qPCR. This provided the normalized abundance of taxa in milk and stool samples, expressed as 16S rRNA gene copy numbers per mL of milk or per gram of stool, respectively. Normalized abundances were then used for all analyses in our study.
Calculating bacterial intakes
Daily bacterial intakes from mother’s milk were calculated by multiplying the total bacterial density (16S rRNA gene copies/mL of milk) and normalized abundance of individual taxa in milk samples with the daily volume of mother’s milk fed (mL of milk) to infants. When matching the microbial data from milk samples to the infant’s daily feeding data, the collection date for each mother’s milk sample was considered the first day it was fed. Microbial data from a given milk sample were used in the calculation of intakes until a subsequent milk sample was collected from the same mother. Furthermore, microbial data from a given milk sample was only used for a maximum of 7 days following its collection, to account for the known temporal changes that occur in preterm mother’s milk.19 Collectively, this approach provided the daily intake of total bacteria and commonly shared genera expressed in the units, 16S rRNA gene copies.
To examine relationships between bacterial intakes from mother’s milk and their concentrations in infant stool samples, cumulative mother’s milk volumes fed over the 3-days prior to each stool collection were used in the calculation of bacterial intakes. This was done to account for the varied gastrointestinal transit time in VLBW infants.82,83 Therefore, the term “3-day intake from milk” is used to represent the cumulative intake of total bacteria and commonly shared taxa over this specified period.
Statistical analyses
Statistical analyses were performed in R (4.3.1) and SAS University Edition (SAS Institute, Cary, NC, USA). Non-parametric models were run when outcome measures were not normally distributed (based on Shapiro-Wilk tests and histograms), and all models accounted for repeated observations. Prior to building models, collinearity between independent variables was also examined and a variance inflation factor >2.5 was used as the cut-off to identify multicollinearity; none were identified. Multivariable models (linear, Poisson, logistic) were used across all analyses, and stratified analyses were only conducted under two circumstances. First, when interaction terms in our adjusted multivariable models were statistically significant, stratified analyses were then performed to further explore and visualize these relationships. Interaction terms that were not statistically significant were removed from multivariable models to improve precision. Second, we used stratified analyses to examine whether Spearman correlations, between milk bacterial intakes and their concentrations in infant stools, differed by postnatal period or in-hospital feeding practices.
Alpha and beta diversity analyses
To test whether the alpha diversity (number of zOTUs, Shannon index) observed in mother’s milk differs from infant stool over time, linear mixed-effects models were built and adjusted for DNA extraction batch, postnatal week, sample type, and an interaction term between postnatal week and sample type (Figure 1). To then assess whether relationships exist with alpha-diversity between paired milk (independent variable) and stool samples (dependent variable), linear mixed-effects models were again used and adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum (<1,000 and 1,000–1,249g), and our feeding variables of interest (fortifier group, feeding type, and feeding at the breast). These covariates were selected a priori based on previous work identifying them as key drivers of the VLBW infant gut microbiota, or to include unique in-hospital feeding variables specific to VLBW infants that have not been previously explored (i.e., feeding at the breast). We did not include delivery mode as a covariate in our multivariable models because, unlike in term infants, studies in preterm infants show limited associations between delivery mode and their gut microbiota.21,71 Interaction terms were tested between the alpha diversity in mother’s milk and postnatal week. Of note, our multivariable models included postnatal week as a covariate, however, when significant interactions emerged with this continuous variable, we further examined these relationships by converting postnatal week into a discrete variable. To tease apart and visualize these interactions, the initial 8-week hospitalization period was divided into the first and second postnatal month. Interaction terms were also tested between the alpha-diversity in mother’s milk and each feeding variable of interest.
When plotting the relationship between number of zOTUs in paired milk-stool samples, we observed a strong non-linear relationship that changed around 200 zOTUs in mother’s milk. In order to capture the non-linear nature of these data, an interaction term (number of zOTUs in milk and milk cut-point: zOTUs ≤200 or >200) was tested in our linear mixed-effects models, as described above. Results from this initial test confirmed that the relationship between the number of zOTUs in paired milk-stool samples was dependent on whether milk samples contained ≤200 or >200 zOTUs and, therefore, should be interpreted separately. To assess whether postnatal period and in-hospital feeding practices further modified this relationship, separate models were run to test 3-way interactions between the number of zOTUs in milk, the milk cut-point, and postnatal period or the feeding variable of interest (feeding type, fortifier type, or feeding at the breast); these models were adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum (<1,000 and 1,000–1,249g), and our feeding variables of interest (fortifier group, feeding type, and feeding at the breast). Significant 3-way interactions were further explored using stratified linear-mixed effects models that were left unadjusted due to sample size constraints after stratification.
To compare the overall microbial communities in mother’s milk and infant stools, adonis models stratified by participant identification and adjusted for postnatal week, sample type, and DNA extraction batch were built for beta-diversity measures (Bray-Curtis dissimilarity, weighted/unweighted Unifrac distance) using the vegan (version 2.6.4) R package (Figure 2).84 To then assess the overall microbial communities and their temporal trends in paired milk-stool samples, beta-diversity (weighted/unweighted Unifrac distances) distances between each milk-stool pair were calculated and tested using linear mixed-effects models adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest (fortifier group, feeding type, and feeding at the breast). Notably, we were unable to use Bray-Curtis dissimilarity to assess temporal trends between milk and infant stool bacterial communities since the sample pairs were deemed to be almost completely dissimilar (mean Bray-Curtis dissimilarity: 0.99998); this is likely due to this measure’s particular sensitivity to taxa abundance.85
Shared microbial taxa analyses
The number of shared zOTUs was determined between each paired milk and infant stool sample (Figure 3). Shared zOTU ratio was calculated as the number of shared zOTUs between a paired milk-stool sample divided by the total number of zOTUs in the respective stool sample. To assess the total number of shared zOTUs and shared zOTU ratio between paired milk-stool samples over time, linear mixed-effects models were used and adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest (fortifier group, feeding type, and feeding at the breast). A repeated measures Poisson regression model (adjusted using the same aforementioned covariates) was then used to examine whether the likelihood of sharing a greater number of zOTUs in milk-stool pairs differs depending on postnatal period and in-hospital feeding practices. We then assessed the taxonomy of shared zOTUs in paired milk-stool samples at the genus-level. Genera (with at least 1 shared zOTU) shared in at least 10% of milk-stool pairings were considered to be “commonly shared”. This resulted in 12 commonly shared genera that we tested in subsequent analyses.
To assess whether the odds of sharing commonly shared genera between paired milk-stool samples differs depending on postnatal period or feeding practices, repeated measures logistic regression models were used (Figure 4). Separate models were run for each genera, with the outcome being whether a paired milk-stool sample had a shared zOTU mapping to the specified genera (yes/no). These models were adjusted using the same aforementioned covariates and p values were corrected using a Benjamini-Hochberg false discovery rate (FDR) to account for multiple comparisons. Results for unclassified Enterobacteriaceae are not reported since almost all paired milk-stool samples had a shared zOTU mapping back to this taxon (99.5%), leading to issues with model convergence.
To analyze patterns of zOTU sharing at the genus-level, hierarchical clustering was performed and clusters were identified based on the within-cluster sum of squares (Figure S4). Paired milk-stool samples were clustered into 3 groups according to the number of unique zOTUs within all shared genera on a per sample basis. To explore differences between the 3 clusters, unadjusted repeated measures Poisson regression models were used to determine how the number of unique zOTUs within each individual commonly shared genera differed according to cluster assignment. Furthermore, to understand whether postnatal period or feeding practices were associated with the likelihood of belonging to a specific cluster, a multinomial logistic regression model with repeated measures was performed; this model was adjusted for postnatal week, feeding type, fortifier group, and feeding at the breast.
Bacterial intakes from mother’s milk and infant stool concentrations
To determine how bacterial concentrations in mother’s milk change over time, linear mixed-effects models were built and adjusted for postpartum week of expression, DNA extraction batch, and gestational age (Figure S5). Of note, although we adjusted for infant sex assigned at birth and birth weight stratum in other models, we were unable to include these variables in these models since some mothers of twins and triplets delivered infants with differing sexes and birth weights; instead, gestational age was used as a proxy for birth weight stratum. However, when examining changes in bacterial intakes from mother’s milk over time (i.e., on a per infant basis), we again used linear mixed-effects models but adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest (feeding type, fortifier group, and feeding at the breast).
To examine dose-response relationships between cumulative 3-day intakes of commonly shared bacteria from mother’s milk and their concentrations in infant stools, we only included sample pairs that contained the taxa of interest in both the milk and stool sample (versus milk-stool pairs that contained the taxa in either the milk or stool sample, or were absent in both samples) (Figure 5). These relationships were assessed using unadjusted linear-mixed effects models, due to sample size constraints. p values from these models were adjusted using a Benjamini-Hochberg FDR to account for multiple comparisons. Individual models were also built to test interactions between 3-day bacterial intakes from mother’s milk and postnatal period and feeding practices.
Given that some milk-stool pairs contained the 12 commonly shared genera in only the milk sample (not the stool), we aimed to establish whether postnatal period and feeding practices were associated with the odds of observing a given genera in both milk-stool samples rather than milk alone (Figure S6). For this, we used unadjusted, repeated measures logistic regressions and p values were FDR-corrected for multiple comparisons.
To examine whether inter-genera correlations existed amongst the 12 commonly shared genera, Spearman rank correlations were used to test microbial associations between 3-day bacterial intakes from mother’s milk (log copies) and their concentrations in infant stool samples (log copies/g stool) (Figure 6). These correlations were visualized using a chord diagram and heatmaps. Furthermore, correlations were tested for the overall cohort and then again by stratifying according to postnatal period and feeding practices.
Maternal and infant antibiotic exposure analyses
To explore the modulatory potential of antibiotic use, mother-infant dyads were divided into 4 antibiotic groups: maternal high-infant high, maternal high-infant low, maternal low-infant high, and maternal low-infant low. High antibiotic exposure was defined as >1 day for mothers (prenatally or postnatally) and >3 days for infants (Figure 7). To then assess whether relationships between alpha-diversity in paired milk (independent variable) and stool samples (dependent variable) were modified by antibiotic group, we used linear mixed-effects models; these models included an interaction term between alpha-diversity in the mother’s milk and antibiotic group, and were adjusted for antibiotic group, postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest (fortifier group, feeding type, and feeding at the breast). Significant interaction terms from these models confirmed that relationships between alpha-diversity in mother’s milk and infant stool samples were dependent on antibiotic group. To further explore and visualize these relationships, linear-mixed effects models stratified by antibiotic group were run. To then determine whether correlations between 3-day bacterial intakes from mother’s milk and their concentrations in infant stools were dependent on antibiotic group, we constructed stratified correlation matrices for each antibiotic group using heatmaps.
Additional resources
Data and samples for this study were collected as part of the OptiMoM Fortifier study which is registered at clinicaltrials.gov (accession number: NCT02137473).
Published: September 6, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101729.
Supplemental information
References
- 1.Aguilar-lopez M., Dinsmoor A.M., Ho T.T.B., Donovan S.M., Dinsmoor A.M., Ho T.T.B., Sharon M., Aguilar-lopez M. A systematic review of the factors influencing microbial colonization of the preterm infant gut preterm infant gut. Gut Microb. 2021;13:1–33. doi: 10.1080/19490976.2021.1884514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unger S., Stintzi A., Shah P., Mack D., O’Connor D.L. Gut microbiota of the very-low-birth-weight infant. Pediatr. Res. 2015;77:205–213. doi: 10.1038/pr.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pammi M., Cope J., Tarr P.I., Warner B.B., Morrow A.L., Mai V., Gregory K.E., Kroll J.S., McMurtry V., Ferris M.J., et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: A systematic review and meta-analysis. Microbiome. 2017;5 doi: 10.1186/s40168-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart C.J., Embleton N.D., Marrs E.C.L., Smith D.P., Fofanova T., Nelson A., Skeath T., Perry J.D., Petrosino J.F., Berrington J.E., Cummings S.P. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome. 2017;5:75. doi: 10.1186/s40168-017-0295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meek J.Y., Noble L., Section on Breastfeeding Policy Statement: Breastfeeding and the Use of Human Milk. Pediatrics. 2022;150 doi: 10.1542/peds.2022-057988. [DOI] [PubMed] [Google Scholar]
- 6.Horta B.L., Loret de Mola C., Victora C.G. Breastfeeding and intelligence: a systematic review and meta-analysis. Acta Paediatr. 2015;104:14–19. doi: 10.1111/apa.13139. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization . 2020. Protecting, Promoting and Supporting Breastfeeding: The Baby-Friendly Hospital Initiative for Small, Sick and Preterm Newborns. [Google Scholar]
- 8.Miller J., Tonkin E., Damarell R.A., McPhee A.J., Suganuma M., Suganuma H., Middleton P.F., Makrides M., Collins C.T. A Systematic Review and Meta-Analysis of Human Milk Feeding and Morbidity in Very Low Birth Weight Infants. Nutrients. 2018;10:707. doi: 10.3390/nu10060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lechner B.E., Vohr B.R. Neurodevelopmental Outcomes of Preterm Infants Fed Human Milk. Clin. Perinatol. 2017;44:69–83. doi: 10.1016/j.clp.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Asnicar F., Manara S., Zolfo M., Truong D.T., Scholz M., Armanini F., Ferretti P., Gorfer V., Pedrotti A., Tett A., Segata N. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. mSystems. 2017;2:1–13. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehr K., Moossavi S., Sbihi H., Boutin R.C.T., Bode L., Robertson B., Yonemitsu C., Field C.J., Becker A.B., Mandhane P.J., et al. Breastmilk Feeding Practices Are Associated with the Co-Occurrence of Bacteria in Mothers’ Milk and the Infant Gut: the CHILD Cohort Study. Cell Host Microbe. 2020;28:285–297.e4. doi: 10.1016/j.chom.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Pannaraj P.S., Li F., Cerini C., Bender J.M., Yang S., Rollie A., Adisetiyo H., Zabih S., Lincez P.J., Bittinger K., et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jost T., Lacroix C., Braegger C.P., Rochat F., Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014;16:2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 14.Duranti S., Lugli G.A., Mancabelli L., Armanini F., Turroni F., James K., Ferretti P., Gorfer V., Ferrario C., Milani C., et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5 doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milani C., Mancabelli L., Lugli G.A., Duranti S., Turroni F., Ferrario C., Mangifesta M., Viappiani A., Ferretti P., Gorfer V., et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 2015;81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards C.A., Van Loo-Bouwman C.A., Van Diepen J.A., Schoemaker M.H., Ozanne S.E., Venema K., Stanton C., Marinello V., Rueda R., Flourakis M., et al. A systematic review of breast milk microbiota composition and the evidence for transfer to and colonisation of the infant gut. Benef. Microbes. 2022;13:365–381. doi: 10.3920/BM2021.0098. [DOI] [PubMed] [Google Scholar]
- 17.Lau C. Breastfeeding Challenges and the Preterm Mother-Infant Dyad: A Conceptual Model. Breastfeed. Med. 2018;13:8–17. doi: 10.1089/bfm.2016.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thinkhamrop J., Hofmeyr G.J., Adetoro O., Lumbiganon P., Ota E. In: Cochrane Database of Systematic Reviews. Thinkhamrop J., editor. John Wiley & Sons, Ltd; 2015. Antibiotic prophylaxis during the second and third trimester to reduce adverse pregnancy outcomes and morbidity. [DOI] [Google Scholar]
- 19.Asbury M.R., Butcher J., Copeland J.K., Unger S., Bando N., Comelli E.M., Forte V., Kiss A., LeMay-Nedjelski L., Sherman P.M., et al. Mothers of Preterm Infants Have Individualized Breast Milk Microbiota that Changes Temporally Based on Maternal Characteristics. Cell Host Microbe. 2020;28:669–682.e4. doi: 10.1016/j.chom.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Asbury M.R., Shama S., Sa J.Y., Bando N., Butcher J., Comelli E.M., Copeland J.K., Forte V., Kiss A., Sherman P.M., et al. Human milk nutrient fortifiers alter the developing gastrointestinal microbiota of very-low-birth-weight infants. Cell Host Microbe. 2022;30:1328–1339.e5. doi: 10.1016/j.chom.2022.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Butcher J., Unger S., Li J., Bando N., Romain G., Francis J., Mottawea W., Mack D., Stintzi A., O’Connor D.L. Independent of Birth Mode or Gestational Age, Very-Low-Birth-Weight Infants Fed Their Mothers’ Milk Rapidly Develop Personalized Microbiotas Low in Bifidobacterium. J. Nutr. 2018;148:326–335. doi: 10.1093/jn/nxx071. [DOI] [PubMed] [Google Scholar]
- 22.Parker M.G., Stellwagen L.M., Noble L., Kim J.H., Poindexter B.B., Puopolo K.M., SECTION ON BREASTFEEDING, COMMITTEE ON NUTRITION, COMMITTEE ON FETUS AND NEWBORN Promoting human milk and breastfeeding for the very low birth weight infant. Pediatrics. 2021;148 doi: 10.1542/peds.2021-054272. [DOI] [PubMed] [Google Scholar]
- 23.Senterre T. Practice of enteral nutrition in very low birth weight and extremely low birth weight infants. World Rev. Nutr. Diet. 2014;110:201–214. doi: 10.1159/000358468. [DOI] [PubMed] [Google Scholar]
- 24.Quigley M., Embleton N.D., McGuire W. Formula versus donor breast milk for feeding preterm or low birth weight infants. Cochrane Database Syst. Rev. 2018;6 doi: 10.1002/14651858.CD002971.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown J.V., Lin L., Embleton N.D., Harding J.E., McGuire W. Multi-nutrient fortification of human milk for preterm infants. Cochrane Database Syst. Rev. 2020;6 doi: 10.1002/14651858.CD000343.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumbhare S.V., Jones W.D., Fast S., Bonner C., Jong G. ‘t, Van Domselaar G., Graham M., Narvey M., Azad M.B. Source of human milk (mother or donor) is more important than fortifier type (human or bovine) in shaping the preterm infant microbiome. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong X., Judge M., Xu W., Diallo A., Janton S., Brownell E.A., Maas K., Graf J. Influence of Feeding Type on Gut Microbiome Development in Hospitalized Preterm Infants. Nurs. Res. 2017;66:123–133. doi: 10.1097/NNR.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vongbhavit K., Salinero L.K., Kalanetra K.M., Masarweh C., Yu A., Taft D.H., Mills D.A., Underwood M.A. A comparison of bacterial colonization between nasogastric and orogastric enteral feeding tubes in infants in the neonatal intensive care unit. J. Perinatol. 2022;42:1446–1452. doi: 10.1038/s41372-022-01452-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puopolo K.M., Benitz W.E., Zaoutis T.E., Cummings J., Juul S., Hand I., Eichenwald E., Poindexter B., Stewart D.L., Aucott S.W., et al. Management of Neonates Born at ≤34 6/7 Weeks’ Gestation With Suspected or Proven Early-Onset Bacterial Sepsis. Pediatrics. 2018;142 doi: 10.1542/peds.2018-2896. [DOI] [Google Scholar]
- 30.World Health Organization . 2021. WHO Recommendation on Prophylactic Antibiotics for Women Undergoing Caesarean Section. [PubMed] [Google Scholar]
- 31.Ramani S., Stewart C.J., Laucirica D.R., Ajami N.J., Robertson B., Autran C.A., Shinge D., Rani S., Anandan S., Hu L., et al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat. Commun. 2018;9:5010–5012. doi: 10.1038/s41467-018-07476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao C., Coyte K.Z., Bainter W., Geha R.S., Martin C.R., Rakoff-nahoum S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 2021;591:633–638. doi: 10.1038/s41586-021-03241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pannaraj P.S., Li F., Cerini C., Bender J.M., Yang S., Rollie A., Adisetiyo H., Zabih S., Lincez P.J., Bittinger K., et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogaert D., van Beveren G.J., de Koff E.M., Lusarreta Parga P., Balcazar Lopez C.E., Koppensteiner L., Clerc M., Hasrat R., Arp K., Chu M.L.J.N., et al. Mother-to-infant microbiota transmission and infant microbiota development across multiple body sites. Cell Host Microbe. 2023;31:447–460.e6. doi: 10.1016/j.chom.2023.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Corona-Cervantes K., García-González I., Villalobos-Flores L.E., Hernández-Quiroz F., Piña-Escobedo A., Hoyo-Vadillo C., Rangel-Calvillo M.N., García-Mena J. Human milk microbiota associated with early colonization of the neonatal gut in Mexican newborns. PeerJ. 2020;2020 doi: 10.7717/peerj.9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laursen M.F., Pekmez C.T., Larsson M.W., Lind M.V., Yonemitsu C., Larnkjær A., Mølgaard C., Bode L., Dragsted L.O., Michaelsen K.F., et al. Maternal milk microbiota and oligosaccharides contribute to the infant gut microbiota assembly. ISME Commun. 2021;1 doi: 10.1038/s43705-021-00021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biagi E., Aceti A., Quercia S., Beghetti I., Rampelli S., Turroni S., Soverini M., Zambrini A.V., Faldella G., Candela M., et al. Microbial community dynamics in mother’s milk and infant’s mouth and gut in moderately preterm infants. Front. Microbiol. 2018;9:1–10. doi: 10.3389/fmicb.2018.02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corona-Cervantes K., García-González I., Villalobos-Flores L.E., Hernández-Quiroz F., Piña-Escobedo A., Hoyo-Vadillo C., Rangel-Calvillo M.N., García-Mena J. Human milk microbiota associated with early colonization of the neonatal gut in Mexican newborns. PeerJ. 2020;8 doi: 10.7717/peerj.9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogaert D., van Beveren G.J., de Koff E.M., Lusarreta Parga P., Balcazar Lopez C.E., Koppensteiner L., Clerc M., Hasrat R., Arp K., Chu M.L.J.N., et al. Mother-to-infant microbiota transmission and infant microbiota development across multiple body sites. Cell Host Microbe. 2023;31:447–460.e6. doi: 10.1016/j.chom.2023.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Younge N.E., Newgard C.B., Cotten C.M., Goldberg R.N., Muehlbauer M.J., Bain J.R., Stevens R.D., O’Connell T.M., Rawls J.F., Seed P.C., Ashley P.L. Disrupted Maturation of the Microbiota and Metabolome among Extremely Preterm Infants with Postnatal Growth Failure. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-44547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arrieta M.C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S., Kuzeljevic B., Gold M.J., Britton H.M., Lefebvre D.L., et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 42.Tsukuda N., Yahagi K., Hara T., Watanabe Y., Matsumoto H., Mori H., Higashi K., Tsuji H., Matsumoto S., Kurokawa K., Matsuki T. Key bacterial taxa and metabolic pathways affecting gut short-chain fatty acid profiles in early life. ISME J. 2021;15:2574–2590. doi: 10.1038/s41396-021-00937-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zoetendal E.G., Raes J., Van Den Bogert B., Arumugam M., Booijink C.C.G.M., Troost F.J., Bork P., Wels M., De Vos W.M., Kleerebezem M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 45.Ma B., Mccomb E., Gajer P., Yang H., Humphrys M., Okogbule-Wonodi A.C., Fasano A., Ravel J., Viscardi R.M. Microbial biomarkers of intestinal barrier maturation in preterm infants. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., Fukuda S., Saito T., Narushima S., Hase K., et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 47.Salminen S., Collado M.C., Endo A., Hill C., Lebeer S., Quigley E.M.M., Sanders M.E., Shamir R., Swann J.R., Szajewska H., Vinderola G. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021;18:649–667. doi: 10.1038/s41575-021-00440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prolacta Bioscience Technical Bulletin Human Milk Oligosaccharides. HMOs) 2019;22 https://www.prolacta.com/en/resource-library/human-milk-oligosaccharides-hmos-technical-bulletin/ [Google Scholar]
- 49.Bode L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masi A.C., Stewart C.J. Untangling human milk oligosaccharides and infant gut microbiome. iScience. 2022;25 doi: 10.1016/j.isci.2021.103542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kordy K., Gaufin T., Mwangi M., Li F., Cerini C., Lee D.J., Adisetiyo H., Woodward C., Pannaraj P.S., Tobin N.H., Aldrovandi G.M. Contributions to human breast milk microbiome and enteromammary transfer of Bifidobacterium breve. PLoS One. 2020;15:e0219633. doi: 10.1371/journal.pone.0219633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moossavi S., Azad M.B. Origins of human milk microbiota: new evidence and arising questions. Gut Microb. 2020;12 doi: 10.1080/19490976.2019.1667722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merter Ö.S., Altay N. Effect of Feeding Fresh or Frozen Breast Milk on the Gut Microbiota of Premature Infants: A Prospective Observational Study. Biol. Res. Nurs. 2024;26:78–90. doi: 10.1177/10998004231191728. [DOI] [PubMed] [Google Scholar]
- 54.Castro M., Asbury M., Shama S., Stone D., Yoon E.W., O’Connor D.L., Unger S. Energy and Fat Intake for Preterm Infants Fed Donor Milk Is Significantly Impacted by Enteral Feeding Method. JPEN - J. Parenter. Enter. Nutr. 2019;43:162–165. doi: 10.1002/jpen.1430. [DOI] [PubMed] [Google Scholar]
- 55.Thomas S.P., Denizer E., Zuffa S., Best B.M., Bode L., Chambers C.D., Dorrestein P.C., Liu G.Y., Momper J.D., Nizet V., et al. Transfer of antibiotics and their metabolites in human milk: Implications for infant health and microbiota. Pharmacotherapy. 2023;43:442–451. doi: 10.1002/phar.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Wattum J.J., Leferink T.M., Wilffert B., ter Horst P.G.J. Antibiotics and lactation: An overview of relative infant doses and a systematic assessment of clinical studies. Basic Clin. Pharmacol. Toxicol. 2019;124:5–17. doi: 10.1111/bcpt.13098. [DOI] [PubMed] [Google Scholar]
- 57.Patangia D.V., Ryan C.A., Dempsey E., Stanton C., Ross R.P. Vertical transfer of antibiotics and antibiotic resistant strains across the mother/baby axis. Trends Microbiol. 2022;30:47–56. doi: 10.1016/j.tim.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 58.Patel A.L., Mutlu E.A., Sun Y., Koenig L., Green S., Jakubowicz A., Mryan J., Engen P., Fogg L., Chen A.L., et al. Longitudinal survey of microbiota in hospitalized preterm very-low-birth-weight infants. J. Pediatr. Gastroenterol. Nutr. 2016;62:292–303. doi: 10.1097/MPG.0000000000000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brockway M.M., Daniel A.I., Reyes S.M., Gauglitz J.M., Granger M., McDermid J.M., Chan D., Refvik R., Sidhu K.K., Musse S., et al. Human Milk Bioactive Components and Child Growth and Body Composition in the First 2 Years: A Systematic Review. Adv. Nutr. 2024;15 doi: 10.1016/j.advnut.2023.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LeMay-Nedjelski L., Copeland J., Wang P.W., Butcher J., Unger S., Stintzi A., O’Connor D.L. In: Microbiome Analysis: Methods and Protocols. Beiko R.G., Hsiao W., Parkinson J., editors. Springer New York; 2018. Methods and Strategies to Examine the Human Breastmilk Microbiome; pp. 63–86. [DOI] [PubMed] [Google Scholar]
- 61.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Furet J.P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., Doré J., Corthier G. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 2009;68:351–362. doi: 10.1111/j.1574-6941.2009.00671.x. [DOI] [PubMed] [Google Scholar]
- 63.Edgar, R.C. UNOISE2: Improved Error-Correction for Illumina 16S and ITS Amplicon Sequencing. Preprint at bioRxiv. 10.1101/081257. [DOI]
- 64.Edgar R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 65.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 66.Price M.N., Dehal P.S., Arkin A.P. Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:1–14. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Core Team . 2017. R: A Language and Environment for Statistical Computing (R Foundation of Statistical Computing)https://www.r-project.org/ [Google Scholar]
- 71.Beck L.C., Masi A.C., Young G.R., Vatanen T., Lamb C.A., Smith R., Coxhead J., Butler A., Marsland B.J., Embleton N.D., et al. Strain-specific impacts of probiotics are a significant driver of gut microbiome development in very preterm infants. Nat. Microbiol. 2022;7:1525–1535. doi: 10.1038/s41564-022-01213-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McMurdie P.J., Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kolde R. Pheatmap: Pretty heatmaps. R package version 1.0.12. 2019 https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf [Google Scholar]
- 74.Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 75.Brunson J. ggalluvial: Alluvial Plots in “ggplot2”. 2023. https://corybrunson.github.io/ggalluvial
- 76.Gordon M., Lumley T. forestplot: Advanced Forest Plot Using “grid” Graphics. R package version 3.1.3. 2023 https://cran.r-project.org/web/packages/forestplot/forestplot.pdf [Google Scholar]
- 77.O’Connor D.L., Kiss A., Tomlinson C., Bando N., Bayliss A., Campbell D.M., Daneman A., Francis J., Kotsopoulos K., Shah P.S., et al. Nutrient enrichment of human milk with human and bovine milk–based fortifiers for infants born weighing <1250 g: a randomized clinical trial. Am. J. Clin. Nutr. 2018;108:108–116. doi: 10.1093/ajcn/nqy067. [DOI] [PubMed] [Google Scholar]
- 78.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016;4:1–22. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nearing J.T., Douglas G.M., Comeau A.M., Langille M.G.I. Denoising the Denoisers: An independent evaluation of microbiome sequence error- correction approaches. PeerJ. 2018;6:e5364. doi: 10.7717/peerj.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gu Z., Gu L., Eils R., Schlesner M., Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 81.Edgar, R.C. UNBIAS: An attempt to correct abundance bias in 16S sequencing, with limited success. Preprint at bioRxiv 10.1101/124149. [DOI]
- 82.McClure R.J., Newell S.J. Randomised controlled trial of trophic feeding and gut motility. Arch. Dis. Child. Fetal Neonatal Ed. 1999;80:F54–F58. doi: 10.1136/fn.80.1.F54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bodé S., Dreyer M., Greisen G. Gastric Emptying and Small Intestinal Transit Time in Preterm Infants: a Scintigraphic Method. J. Pediatr. Gastroenterol. Nutr. 2004;39:378–382. doi: 10.1002/j.1536-4801.2004.tb00870.x. [DOI] [PubMed] [Google Scholar]
- 84.Oksanen J., Simpson G.L., Blanchet F.G., Kindt R., Legendre P., Minchin P.R., O’Hara R.B., Solymos P., Stevens M.H.H., Szoecs E., et al. Vegan: community ecology package. R package version 2.6-4. 2001 doi: 10.32614/CRAN.package.vegan. [DOI] [Google Scholar]
- 85.Ricotta C., Podani J. On some properties of the Bray-Curtis dissimilarity and their ecological meaning. Ecol. Complex. 2017;31:201–205. doi: 10.1016/j.ecocom.2017.07.003. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw sequences for this study are available at the NCBI Sequence Read Archive under the BioProject accession numbers PRJNA607284 and PRJNA723326 (http://www.ncbi.nlm.nih.gov/sra).
-
•
This study used pre-existing software without generating new custom code or algorithms.
-
•
Requests for additional information required to reanalyze the data should be directed to and will be fulfilled by the lead contact.