Abstract
Sialic acids are commonly found on the terminal ends of biologically important carbohydrates, including intestinal mucin O-linked glycans. Pathogens such as Clostridium perfringens, the causative agent of necrotic enteritis in poultry and humans, have the ability to degrade host mucins and colonize the mucus layer, which involves removal of the terminal sialic acid by carbohydrate-active enzymes (CAZymes). Here, we present the structural and biochemical characterization of the GH33 catalytic domains of the three sialidases of C. perfringens and probe their substrate specificity. The catalytically active domains, which we refer to as NanHGH33, NanJGH33, and NanIGH33, displayed differential activity on various naturally occurring forms of sialic acid. We report the X-ray crystal structures of these domains in complex with relevant sialic acid variants revealing the molecular basis of how each catalytic domain accommodates different sialic acids. NanHGH33 displays a distinct preference for α-2,3-linked sialic acid, but can process α-2,6-linked sialic acid. NanJGH33 and NanIGH33 both exhibit the ability to process α-2,3- and α-2,6-linked sialic acid without any significant apparent preference. All three enzymes were sensitive to generic and commercially available sialidase inhibitors, which impeded sialidase activity in cultures as well as the growth of C. perfringens on sialylated glycans. The knowledge gained in these studies can be applied to in vivo models for C. perfringens growth and metabolism of mucin O-glycans, with a view toward future mitigation of bacterial colonization and infection of intestinal tissues.
Keywords: sialidase, glycoside hydrolase, neuraminic acid, Clostridium perfringens, carbohydrate-processing, inhibitor, X-ray crystal structure
Sialic acids are commonly found on the terminal ends of biologically important carbohydrates (1). They are abundant in the mucin glycans of humans and animals, mammalian milk oligosaccharides, bacterial surfaces, and the extracellular matrices of marine biofilms (2, 3, 4). Within the gastrointestinal (GI) tract, the microbial catabolism of sialic acid, which is typically harvested from host mucin glycans, can confer advantages to select groups of commensal bacteria commonly overrepresented during microbiome dysbiosis, such as Escherichia coli (5) and Ruminococcus gnavus (6). This process can influence the virulence of bacterial pathogens and contribute to inflammation during disease (7, 8, 9, 10, 11). Clostridium perfringens, an opportunistic bacterial pathogen that commonly inhabits animal and human GI tracts, is one of the earliest known microbes to produce sialidase(s), also referred to as neuraminidases, that are capable of processing sialylated glycoconjugates [for example see (12)]. Abundant genomic evidence now indicates that the genomes of all C. perfringens strains contain at least one sialidase encoding gene with many strains containing up to three. The sialidases have shown properties consistent with contributing to adherence to and colonization of the mammalian GI mucosa (13, 14, 15, 16). The ability of poultry-adapted strains of C. perfringens to degrade host mucin through sialic acid metabolism is also predicted to have a major role in virulence, specifically in poultry necrotic enteritis (NE) (17). NE results in billion-dollar losses annually to the poultry industry (18), particularly following the phasing out of prophylactic antimicrobial use in the livestock industry. Thus, further understanding the metabolism of sialic acid by C. perfringens may aid efforts to develop alternative strategies for managing the disease (19).
Recent work has demonstrated that when C. perfringens CP4, a poultry NE-causing strain, is grown in the presence of mucins a statistically significant decrease in sialylated and sulfated mucin O-glycans occurs and corresponds with increases in several neutral glycan species (17). Upon quantification of the two forms of sialic acids N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc), a specific and significant decrease in Neu5Ac was seen, suggesting it was being preferentially consumed by the pathogen. The large repertoire of carbohydrate-active enzymes (CAZymes) within the genome of C. perfringens strains positions this bacterium to dismantle host mucin O-glycans effectively and efficiently. It has been postulated that the removal of terminal sialic acids from mucin glycans is the rate-limiting step in the total erosion of protective glycans (20). Continuing detailed characterization of the enzymatic machinery C. perfringens used for the hydrolysis of sialic acids from mucin glycans is critical to understanding mucin degradation and metabolism by this bacterium.
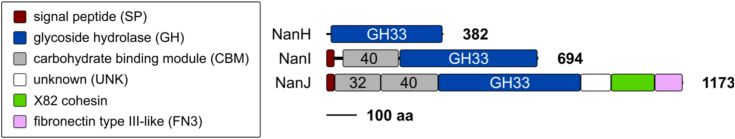
The genomes of most C. perfringens strains contain three separate genes encoding highly conserved GH33 CAZymes targeting sialic acid glycosidic linkages: the nonsecreted 43 kDa NanH, and the two secreted 77 kDa NanI and 129 kDa NanJ (20). NanH consists solely of the catalytic GH33 domain. NanI possess an additional N-terminal family 40 carbohydrate binding module (CBM). NanJ is highly multimodular comprising a central GH33 domain flanked on the N terminus by family 32 and 40 CBMs and on the C-terminal side by an unknown domain, X82 cohesin, and fibronectin type III-like domains (Fig. 1) (21, 22). CBM40 family members have been shown to bind sialic acid (23, 24), whereas polyspecific family 32 CBMs have demonstrated binding to other mucin-related glycans including galactose, lactose (25, 26), and N-acetyllactosamine (27). The multimodularity of C. perfringens CAZymes with CBMs, cohesin domains, and fibronectin type III-like domains has been proposed to promote binding and degradation of the complex glycans within the mucus layer (20). The X-ray crystallographic protein structure of the NanI catalytic domain was solved previously, as well as complexes with a known inhibitor and a catalytic intermediate. The enzyme adopts an overall β-propeller fold with an active site pocket for exo-sialidase activity (28) consistent with other bacterial (23, 29, 30, 31, 32), viral (33), and human (34) exo-sialidases. NanI displays the evolutionary retained elements that are consistent in the active site of sialidases (28, 35). The carboxylate group of a bound sialic acid interacts with the tri-arginine cluster (Arg266, Arg555, and Arg615 in NanI). A hydrophobic pocket (Phe347, Phe353, Phe460, Thr345 Ile327) accommodates the N-acetyl group of the sialic acid. A conserved aspartic acid (Asp291) functions as an acid/base catalyst (28). To date, structural information for the catalytic domains of NanH and NanJ is absent, while detailed functional comparisons of the three enzymes is limited. On the basis of sialidase activity in the extracellular supernatants of isogenic C. perfringens (strain CN3718) sialidase mutants, the three enzymes were reported to differ in their preference for sialic acid linkages (36). NanH was reported to be preferentially active on α-2,8 linkages (α-2,8 > α-2,3 > α-2,6), NanI on α-2,3 linkages (α-2,3 > α-2,6 > α-2,8), and NanJ on α-2,6-linkages (α-2,6 > α-2,8 > α-2,3). However, more detailed analyses of the isolated enzymes are needed to confirm their substrate preferences and to establish structure-function relationships conferring their preferences.
Figure 1.
Characterization of substrate scope of Clostridium perfringens sialidases. The full-length sialidases of C. perfringens are displayed, with domain boundaries based on published data, or predicted by dbCAN (53) and InterProScan (54). Schematics are to scale, with polypeptide chain length indicated. Adapted from Low et al. (2021) (20). Note that this study examined recombinant constructs consisting of the catalytic GH33 domain only of each.
Given the expansion of GH33 encoding genes in C. perfringens, we hypothesized that the different versions of the enzymes may have different properties relevant to the processing of complex mucin O-glycan structures when deployed for nutrient harvesting. Here, we investigate more detailed structure-function relationships for the isolated GH33 catalytic domains of the three sialidases of C. perfringens. We refer to these constructs as NanHGH33, NanIGH33, and NanJGH33. We found that these enzymes display distinct abilities to process variably modified forms of terminal sialic acids and differential specificities for unique stereochemical linkages of sialic acids. The data presented here have informed in vivo models for C. perfringens growth and metabolism of mucin O-glycans. We demonstrate the potential/proof of concept for inhibition of the breakdown of these glycans through treatment with a sialidase inhibitor. Together these data present a basis for future development of mitigation strategies via small molecule and substrate mimetics to treat C. perfringens-causative disease.
Results
Activities of the three C. perfringens GH33 enzymes
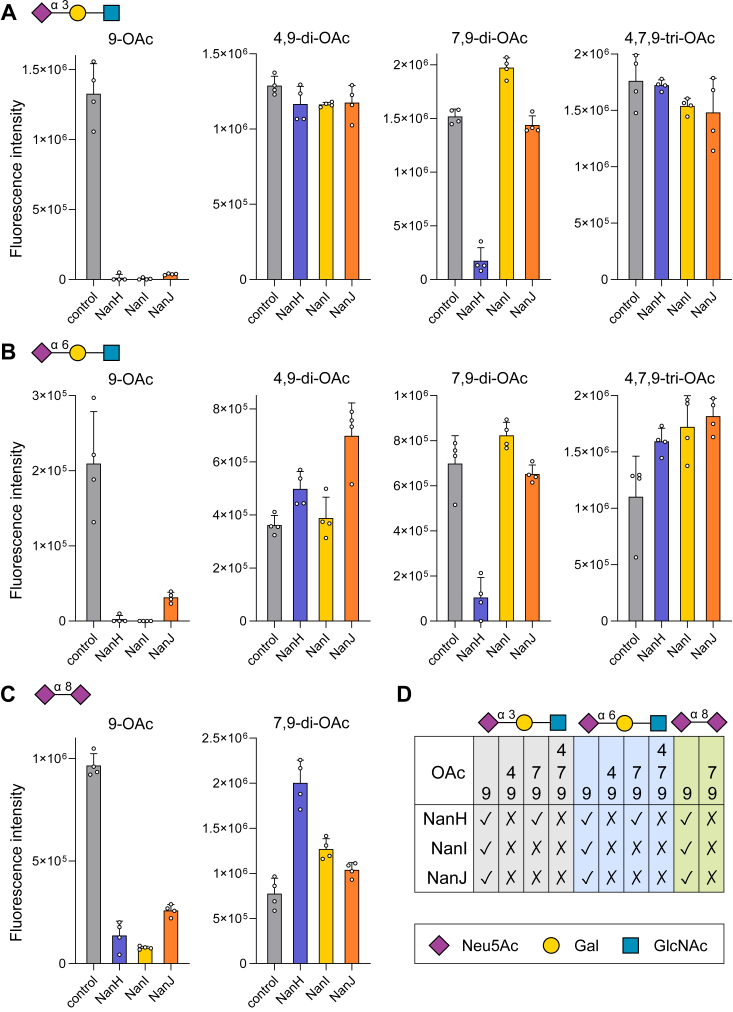
The isolated GH33 domains of NanH, NanI, and NanJ (referred to as NanHGH33, NanIGH33, and NanJGH33) were obtained by expression of the gene fragments encoding them in E. coli and subsequent protein purification by immobilized metal affinity chromatography. To provide initial insight into the substrate selectivities of the GH33 enzymes we used a previously described microarray method (37) to probe accommodation of sialic acid acetylation patterns in the context of how the sialic acid is linked to a preceding sugar moiety. The array of acetylated sialosides was treated with NanHGH33, NanIGH33, or NanJGH33, followed by using bovine coronavirus hemagglutinin-esterase (BCoV HE) to probe the result of the sialidase treatment. Loss of BCoV HE binding after sialidase treatment indicates removal of the sialic acid. All three sialidases were found to be active on 9-O-Ac forms of α-2,3-sialyllactose, α-2,6-sialyllactose, and α-2,8-disialic acid (Fig. 2). The presence of a 4,9-O-Ac abrogated activity of all the enzymes. Only NanHGH33 was active on 3′- and 6′-sialyllactosamines with a 7,9-di-O-Ac Neu5Ac. None of the enzymes had activity on the 4,7,9-tri-O-Ac Neu5Ac sialosides. This suggests that a 4-O-Ac modification is inhibitory to all the enzymes while a 7-O-Ac modification is inhibitory to NanIGH33 and NanJGH33.
Figure 2.
Probing the substrate tolerance of Clostridium perfringens sialidases using the acetylated sialosides microarray. The acetylated sialosides array was treated with NanHGH33, NanIGH33, and NanJGH33 (20 μg/ml in pH 7.4 PBS buffer) at 37 °C for 1 h. Bovine coronavirus hemagglutinin-esterase (BCoV HE) was then used to probe the result of the sialidase treatment. Loss of binding signal after sialidase treatment suggests cleavage of the sialic acid. Microarray data for the untreated control and the NanHGH33, NanIGH33, and NanJGH33 treated slides are shown for (A) acetylated 2,3-sialosides, (B) acetylated 2,6-sialosides, and (C) acetylated 2,8-sialosides. Data are shown as quadruplicate values, with the error bar representing standard deviation of the mean. D, summary of substrate tolerance of the sialidase on the acetylated sialosides.
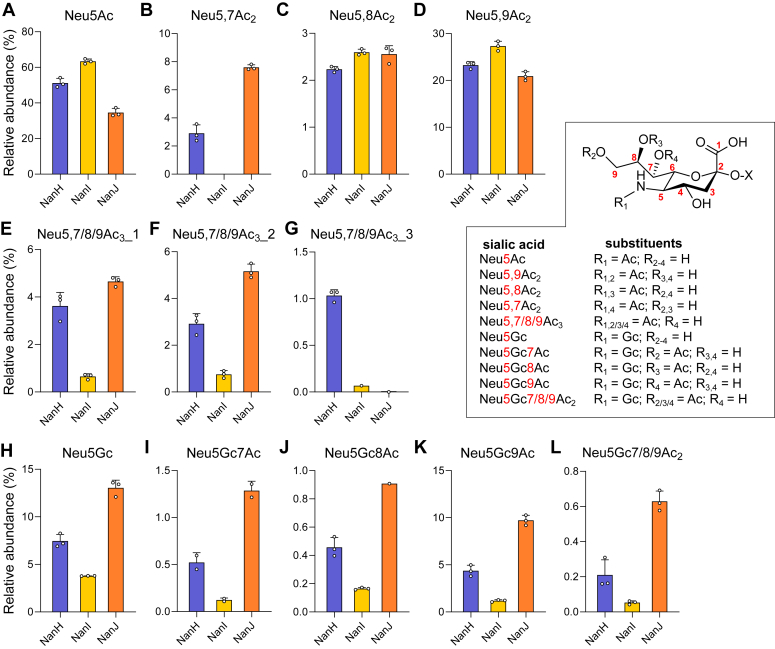
We further investigated the ability NanHGH33, NanIGH33, and NanJGH33 to hydrolyze sialic acid variants by incubating bovine submaxillary mucin (BSM), a well-characterized source of O-acetylated sialic acids (38), with the enzymes and analyzing the released sialic acids by high-performance liquid chromatography coupled to mass spectrometry (Fig. 3). Neu5Ac was the most abundant sialic acid released. Diacetylated versions with additional 7-O-Ac, 8-O-Ac, and 9-O-Ac modifications, and triacetylated versions were also detected (Fig. 3, A–G). However, in the latter cases, the specific positions of the acetyl groups could not be resolved but were assumed to be the three possibilities involving pairs of 7-O-Ac, 8-O-Ac, and 9-O-Ac modifications in addition to the 5-N-Ac. NanIGH33 was unable to release Neu5,7Ac2 indicating clearly that the 7-O-Ac group inhibits the hydrolysis these terminal sialic acid variants. NanJGH33 was able to release this sialic acid variant, but was inactive on Neu5,7,9Ac3 in the arrays, suggesting that accommodation of the 7-O-Ac group by NanJGH33 may be context dependent. Indeed, one of the tri-acetylated variants was resistant to both NanIGH33 and NanJGH33, like the Neu5,7,9Ac3 on the arrays, and we thus propose what we have annotated as Neu5,7/8/9Ac3_3 is Neu5,7,9Ac3. In general, NanIGH33 displayed comparatively poor ability to release triacetylated sialic acid variants from BSM, whereas NanJGH33 was active on the two versions that are assumed to be Neu5,8,9Ac3 and Neu5,7,8Ac3, and NanHGH33 was active on all of them.
Figure 3.
Assessing the tolerance of C. perfringens sialidases against O-acetylated sialic acids.A–L, release of O-acetylated sialic acid species indicated in the panels by each enzyme. Bovine submaxillary mucin (BSM) was treated with NanIGH33, NanJGH33, and NanHGH33 for 5.5 h after which liberated sialic acids were derivatized and analyzed by HPLC-MS. Activities of each sialidase were normalized based on their rate of hydrolyzing 4MU-Neu5Ac. Three replicates were assessed per sialidase, with error bars representing the standard deviation of the mean. In all instances, MS detector responses for each unique sialic acid were normalized to the total detectible sialic acid pool in each sample. Except for Neu5Ac, Neu5Gc, and Neu5,9Ac2, all assignments are putative albeit Neu5,7Ac2, Neu5,8Ac2 (and their Neu5Gc congeners) are consistent with their previously reported retention times and fragmentation patterns. In E-G, triacetylated substrates at four positions (5 and 7/8/9) were not confidently identified and are thus listed as Neu5,7/8/9_1, _2, and _3. Based on microarray data (Fig. 2), we propose what we have annotated as Neu5,7/8/9Ac3_3 is Neu5,7,9Ac3. Neu5Ac, N-acetylneuraminic acid; Neu5Gc, N-glycolylneuraminic acid; HPLC-MS, high-performance liquid chromatography coupled to mass spectrometry.
All of the enzymes displayed some capacity to release from BSM Neu5Gc and acetylated variants thereof, though we were unable to assign absolute structures to these sugars and they are thus inferred (Fig. 3, H–L). Based on the relative abundances of the released Neu5Gc variants, and comparison across the three enzymes, it appears that NanIGH33 is the least able at releasing Neu5Gc, whereas NanJGH33 may be marginally better than NanHGH33.
Overall, these results indicate that the acetylation of sialic acid plays a strong role in determining whether this terminal residue is a substrate for the C. perfringens sialidases, with NanHGH33 being the most tolerant, followed by NanJGH33, and finally NanIGH33 being the least tolerant of acetyl groups in particular positions and combinations. To probe the structural features underlying this selectivity, we used X-ray crystallography to determine the structures of the three enzymes in complex with various ligands.
Structural analysis of the C. perfringens sialidases
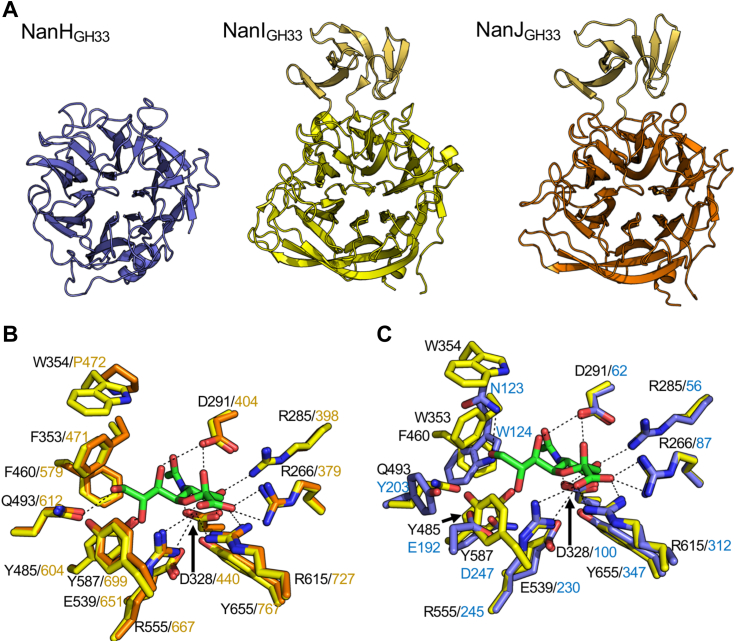
A comparison of the amino acid sequence of the three sialidases indicates that NanIGH33 and NanJGH33 share 60% amino acid sequence identity, compared with a relatively low 29% and 27% amino acid sequence identity with NanHGH33, respectively. We solved the unliganded structures of NanHGH33 and NanJGH33 and compared them with the previously determined structure of NanIGH33 (Protein Data Bank (PDB) ID 2VK5 (28) (Fig. 4). The structures of NanHGH33 and NanJGH33 display the characteristic 6-bladed β-propeller fold seen in exo-acting sialidase GH33 domains (39). The structures of NanJGH33 and NanIGH33 were very similar with an RMSD of 1.1 Å and with both proteins containing the same β-sheet domain insertion within the catalytic domain (between residues 477–546 in NanJGH33 and 361–427 in NanIGH33). The inserted domain is not present in NanHGH33 and the RMSD of NanHGH33 with the other two structures is 1.7 Å. The inserted domain is not within the proximity of the catalytic site of the sialidases, and therefore likely does not directly impact the activity of the sialidase.
Figure 4.
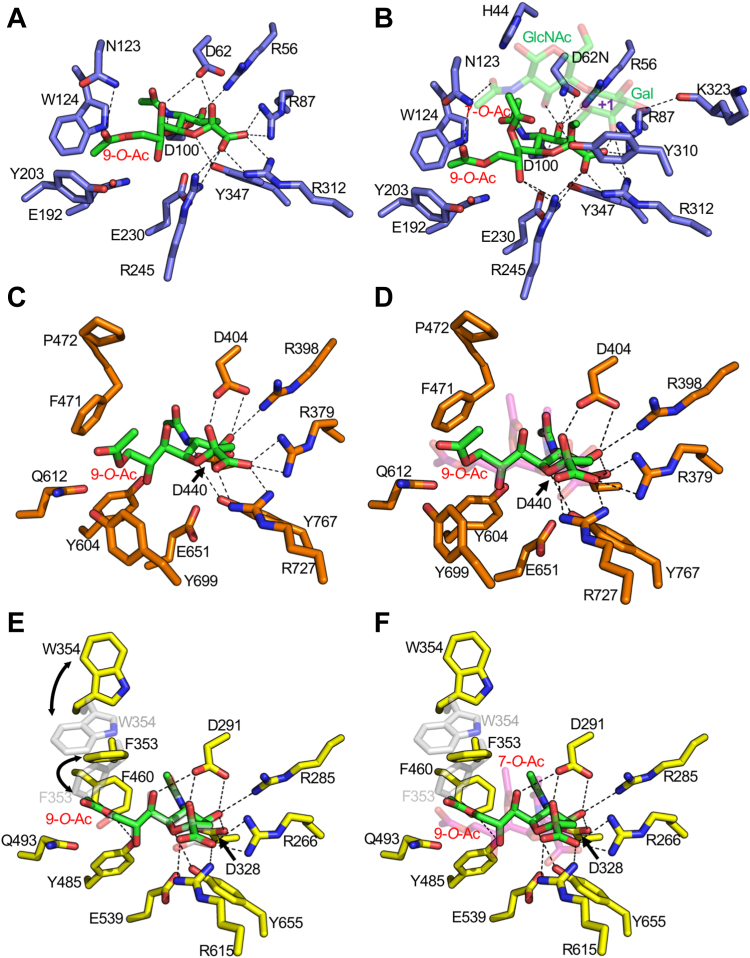
Structures of NanI, NanJGH33, and NanHGH33. A, a comparison of the catalytic domains of NanJGH33 (orange), NanHGH33 (blue), and NanI (yellow, PDB code: 2VK5). The insertion loop in NanJGH33 and NanI is highlighted in light orange/yellow. B, Neu5Ac complexed in the −1 binding site of NanJGH33 and NanI (PDB code: 2BF6). C, Neu5Ac complexed in the −1 binding site of NanHGH33 and NanI (PDB code: 2BF6). The distance cutoff for H-bonds, shown as dashed black lines, was chosen as 3.3 Å. Neu5Ac, N-acetylneuraminic acid; PDB, Protein Data Bank.
The structures of NanHGH33 and NanJGH33 in complex with the Neu5Ac (Fig. S1) compared to the existing structure of NanI in complex with Neu5Ac (PDB ID 6BF6) revealed the properties of the −1 subsites in these enzymes. The amino acid side chains forming the active site pockets of NanJGH33 and NanIGH33 were identical, though a notable position in a second shell of active site residues showed a tryptophan (W354 in NanIGH33) versus proline (P472 in NanJGH33) difference (Fig. 4B). Similarly, the amino acid sidechains in the active sites of NanHGH33 and NanIGH33 that interact with the Neu5Ac ring structure, but not the glycerol substituent, were also identical (Fig. 4C). Further, this specific region of the active site is highly conserved across all three enzymes and prevents recognition of 4-O-acetylated sialic acid variants via an arginine residue (R285 in NanIGH33, R398 in NanJGH33, and R56 in NanHGH33) that blocks accommodation of extensions on C4. NanHGH33, however, has a considerably different organization in the active site region that interacts with the glycerol moiety, suggesting this may contribute to the ability of this enzyme to recognize additional acetylated variants of sialic acid.
We explored this hypothesis by determining the structure of NanHGH33 with bound Neu5,9Ac2 and an inactive D62N mutant in complex with a trisaccharide comprising N-acetyllactosamine terminating in α-2-6-linked Neu5,7,9Ac3 (referred to as SAc3LacNAc) (Fig. 5, A and B, Fig. S1). Neither structure showed any significant perturbation of active site residues relative to either the unliganded or Neu5Ac complex. The 9-O-Ac group of both ligands project into a preformed pocket formed largely by N123, W124, and Y203. The 7-O-Ac group of SAc3LacNAc also projects up into a preexisting space in the active site. This reveals how these modifications are accommodated by the active site of NanHGH33, which is notable as they both involve the residues in the active site region of NanHGH33 that are not conserved in NanIGH33 and NanJGH33. In NanIGH33 and NanJGH33, the 9-O-Ac binding site is apparently blocked off by phenylalanine residues (F353 and F471 in NanIGH33 and NanJGH33, respectively) (Fig. 5, C–F). In NanHGH33, the 7-O-Ac group of SAc3LacNAc nestles against N123, which is spatially occupied in NanIGH33 and NanJGH33 by the larger phenylalanine residue that also blocks the 9-O-Ac pocket.
Figure 5.
Structures of NanIGH33, NanJGH33, and NanHGH33in complex with sialic acid variants.A, NanHGH33 in complex with Neu5,9Ac. B, NanHGH33 D62N in complex with SAc3LacNAc. The N-acetyllactosamine (LacNAc) moiety is shown as transparent to focus on the Neu5,7,9Ac residue in the −1 subsite. The location of the +1 subsite is labeled in purple. C, NanJGH33 in complex with Neu5,9Ac. D, NanJGH33 in complex with Neu5,9Ac overlapped with the Neu5,7,9Ac residue from SAc3LacNAc in the NanHGH33 D62N complex; Neu5,7,9Ac is shown as transparent magenta sticks. E, NanIGH33 in complex with Neu5,9Ac (yellow and green) overlapped with the NanI Neu5Ac complex (transparent gray, PDB ID 2BF6). Arrows highlight the conformational differences of specific sidechains. F, NanIGH33 in complex with Neu5,9Ac (yellow and green) with conformationally flexible sidechains in the NanI Neu5Ac complex (transparent gray, PDB ID 2BF6) overlapped with the Neu5,7,9Ac residue from SAc3LacNAc in the NanHGH33 D62N complex; Neu5,7,9Ac is shown as transparent magenta sticks. The distance cutoff for H-bonds, shown as dashed black lines, was chosen as 3.3 Å. Neu5Ac, N-acetylneuraminic acid; PDB, Protein Data Bank.
The positions of the active site residues in the NanJGH33 Neu5,9Ac2 complex remained essentially identical to those in the NanJGH33 Neu5Ac complex, continuing to suggest a relatively rigid active site (Fig 5C, Fig. S1). The 9-O-Ac group was accommodated by this group being orientated up and away from the active site, rather than continuing the trajectory of the glycerol chain into the a 9-O-Ac binding pocket as in NanHGH33 (Fig. 5D). With knowledge of how the 7-O-Ac group of SAc3LacNAc is accommodated by NanHGH33, there is nothing to suggest in the NanJGH33 Neu5Ac complex that NanJGH33 would be unable to recognize a 7-O-Ac modification in very similar conformation. Indeed, NanJGH33 was able to release Neu5,7Ac2 from BSM. The presence of the 9-O-Ac group, however, and how it is positioned in the NanJGH33 active site appears to change the context of other acetyl groups. The conformation of the 9-O-Ac group in the NanJGH33 complex would be anticipated to clash with the conformation of a 7-O-Ac group if positioned in a manner that would allow accommodation in the NanJGH33 active site. We propose this as a plausible structural explanation for why NanJGH33 was inactive on Neu5,7,9Ac3 on the arrays but is active on the individual Neu5,7Ac2 and Neu5,9Ac2 variants.
In contrast to NanHGH33 and NanJGH33 binding Neu5,9Ac2, the binding of this monosaccharide to NanIGH33 caused a structural rearrangement in the active site. The side chain of W354 swivels approximately 180o, which in turn allows the sidechain of F353 to move ∼90o clockwise and occupy the vacated space (Fig. 5E, Fig. S1). This opens up the 9-O-Ac binding pocket that was otherwise blocked by F353 allowing Neu5,9Ac2 to be bound by NanIGH33 in a conformation similar to that observed in the NanHGH33 complex. This structural rearrangement is not possible in NanJGH33 due to the presence of P472, which is locked in place and prevents significant movement of the F471 sidechain, thus requiring the structural rearrangement of the Neu5,9Ac2 instead. The inability of NanIGH33 to recognize 7-O-Ac modified sialic acid in any capacity is likely due to the F353/W354 combination (Fig. 5E). When the 9-O-Ac pocket is unoccupied it appears that W354 blocks the region that would otherwise accommodate a 7-O-Ac (this position is the smaller P472 residue in NanJGH33). Though W354 moves when binding a ligand with a 9-O-Ac, the rotation of F353 would cause its sidechain to block 7-O-Ac binding.
We did not have simple ligands or substrates with 8-O-Ac modifications and thus we were unable to directly probe the structural basis of how this group is accommodated by the three enzymes. The structures we obtained, however, revealed conserved water in a pocket beneath O8 of the glycerol moiety (Fig. 6). This space is likely sufficient in all of the enzymes to accommodate the acetyl group of an 8-O-Ac modification.
Figure 6.
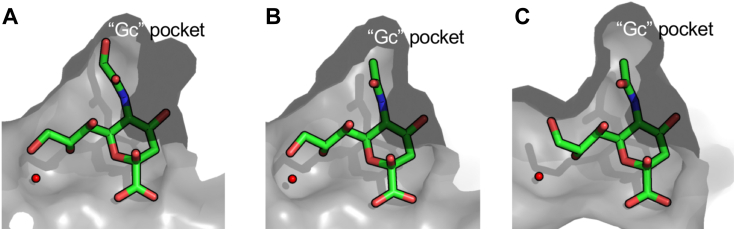
NanIGH33in complex with Neu5Gc reveals a “Gc” binding pocket.A, solvent accessible surface of the crystal structure of NanIGH33 in complex with Neu5Gc. B, solvent accessible surface of the −1 subsite in the crystal structure of NanJGH33 Neu5Ac complex and (C) the crystal structure of NanHGH33 Neu5Ac complex. The pocket accommodating the glycerol group of Neu5Gc is highlighted in each panel. The red sphere in each panel is the conserved water molecule that occupies a putative 8-O-Ac binding pocket. Neu5Ac, N-acetylneuraminic acid; Neu5Gc, N-glycolylneuraminic acid.
Generally, the dominant form of sialic acid in O-glycans is Neu5Ac, and is thus the most likely substrate of the C. perfringens sialidases. However, Neu5Gc (and its alternately acetylated species) is a naturally occurring variant of sialic acid that is present in BSM and was released to some extent by all of the enzymes, though low abundance made it difficult to confidently assign the structures of the acetylated variants. We were able to generate a complex of NanIGH33 with Neu5Gc, which highlighted a “Gc pocket” that can accommodate the additional O11 hydroxyl group of Neu5Gc (Fig. 6A). This pocket also appears to be present in NanJGH33 and NanHGH33, which is consistent with their ability to release this monosaccharide (Fig. 6, B and C).
Linkage specificity of the C. perfringens sialidases
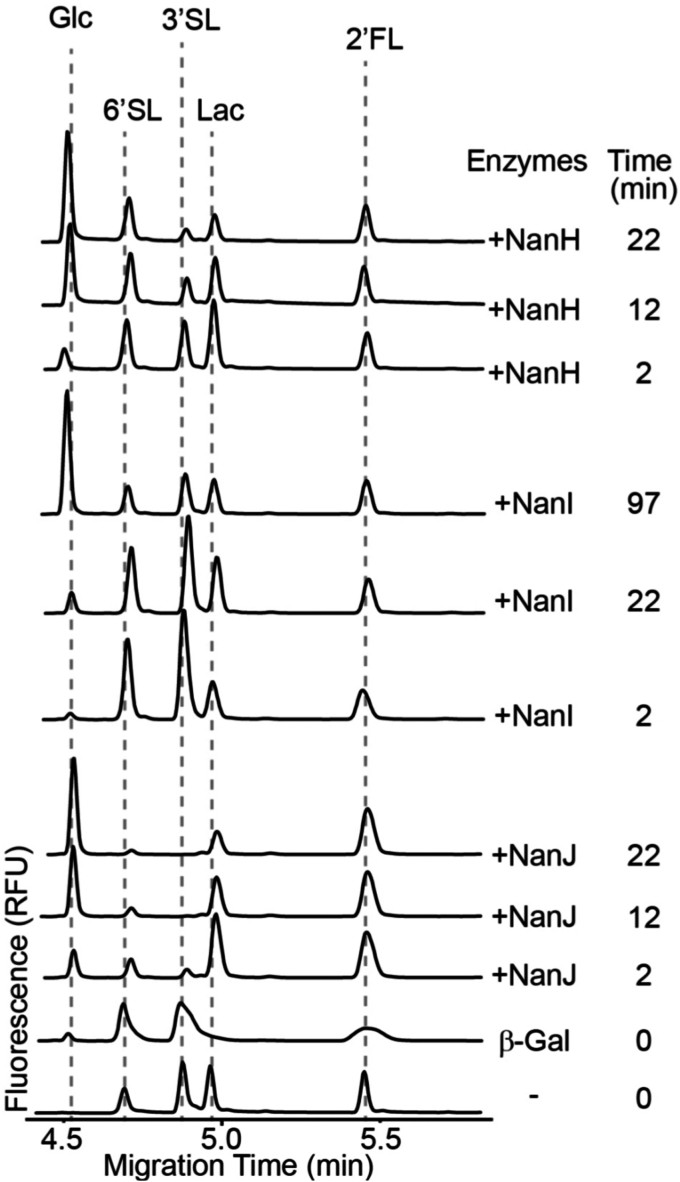
While the selectivity of the three C. perfringens sialidases has been hinted at previously, we sought to take a more quantitative in vitro approach to examining the selectivity of the enzymes for the most common terminal sialic acid linkages. We used capillary electrophoresis with laser induced flourescence for detection (CE-LIF) to monitor 3′-sialyllactose (3′-SL) and 6′-sialyllactose (6′-SL) hydrolysis in a competition assay with equal amounts of each substrate over relatively short time periods (Fig. 7). Reactions included a comparatively high concentration of β-galactosidase to remove the lactose product and prevent the potent trans-sialidase activity of the enzymes. When provided with both substrates at the same time, NanHGH33 hydrolyzed 3′-SL but did not show any significant capacity to hydrolyze 6′-SL (Fig. 7). However, in the absence of a competing α-2,3-linked sialic acid substrate NanHGH33 was clearly able to hydrolyze α-2,6-linked sialic acid in the microarray analysis (Fig. 2B). NanIGH33 showed roughly equal rates of 3′-SL and 6′-SL depletion, revealing little to no selectivity (Fig. 7). NanJGH33 processed both substrates very rapidly, again suggesting limited selectivity for α-2,3- versus α-2,6-linked sialic acid (Fig. 7) when both substrates are provided simultaneously.
Figure 7.
CE-LIF-based substrate reduction assays reveal activity against α2,3/6-linked sialosides for NanHGH33, NanIGH33, and NanJGH33. Mixtures of fluorogenically labeled 3’/6′-SL contain lactose (Lac) which was removed from the assay mixture by pretreatment with β-galactosidase to yield labeled glucose (Glc). Neuraminidases were added and immediately analyzed by CE-LIF, the first time point reflecting the 2 min capillary preconditioning prior to sample injection. CE-LIF, capillary electrophoresis with laser induced flourescence.
Inhibition of the C. perfringens sialidases
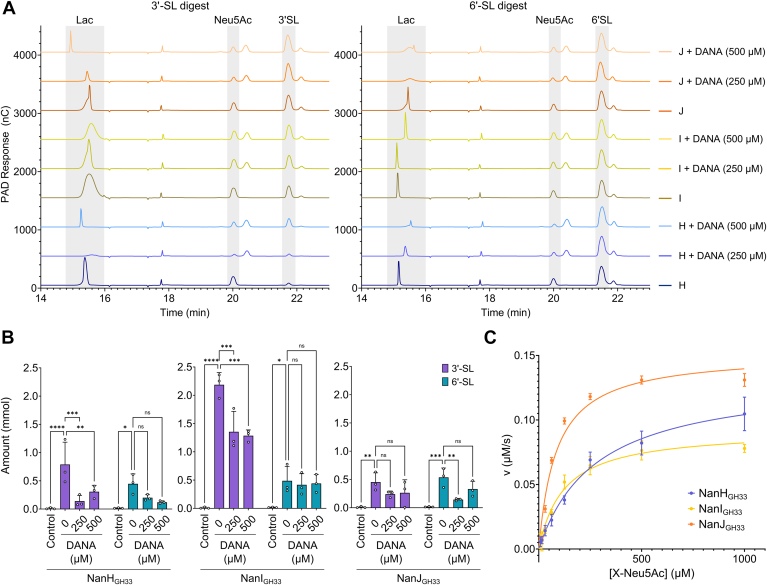
There exists numerous sialidase inhibitors with previously demonstrated activity against human (40) and viral (33, 41, 42) neuraminidases. The inhibition of C. perfringens sialidases has been previously investigated, however, limited by the use of cell culture supernatants (36) or unspecified enzyme (43). Here, we assessed known neuraminidase-active compounds, including inhibitors of human and influenza neuraminidases, for their potential inhibition of the three purified C. perfringens GH33 enzymes: N-acetyl-2,3-dehydro-2-deoxyneuraminic acid (DANA), (3S, 4S, 5R, 6R)-6-(acetylamino)-4,5-dihydroxy-3-piperidinecarboxylic acid (siastatin B), (1S,2S,3R,4R)-3-[(1S)-1-(acetylamino)-2-ethylbutyl]-4-[(aminoiminomethyl)amino]-2-hydroxycyclopentanecarboxylic acid (peramivir), 4-guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid (zanamivir), and (3R,4R,5S)-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylic acid ethyl ester (oseltamivir). Initially, DANA was used to investigate the potential for sialidase inhibitors to protect against hydrolysis of sialyllactose substrates (Fig. 8, A and B). Indeed, DANA was found to inhibit the activity of the three GH33 sialidases and reduced the amount of lactose released by NanHGH33, NanIGH33, and NanJGH33 from 3′- and 6′-SL. To better quantify inhibition, we used the chromogenic substrate X-Neu5Ac, beginning with determination of the kinetic parameters for all three sialidases at each of the enzymes pH optimum (Fig. S2). The resulting data were fit to the standard Michaelis-Menten model (Fig. 8C and Fig. S3). NanJGH33 was found to be the most kinetically efficient at hydrolysis of X-Neu5Ac, with approximately 2-fold and 3.6-fold the kcat/Km of NanIGH33 and NanHGH33, respectively (Table 1). Enzyme inhibition kinetics of the five inhibitors tested revealed that zanamivir and oseltamivir did not display any detectable inhibition of the three GH33 domains at the concentrations tested (up to 1 mM) (Table 1). Peramivir was found to inhibit NanJGH33 weakly with a Ki of 104.0 ± 28.4 μM but it did not inhibit NanHGH33 or NanIGH33. DANA and siastatin B were found to inhibit all three GH33s. DANA was equally potent against NanIGH33 and NanJGH33 with Kis of 3.1 ± 0.5 μM and 3.2 ± 0.3 μM, respectively; it inhibited NanHGH33 to a lesser extent with a Ki of 33.9 ± 4.4 μM. Siastatin B was a less potent inhibitor of DANA, with higher Ki values against NanHGH33, NanIGH33, and NanJGH33, of 125.0 ± 16.2 μM, 10.7 ± 2.0 μM, and 13.3 ± 1.6 μM, respectively. Both DANA and siastatin B were slightly weaker inhibitors of NanHGH33 than NanIGH33 and NanJGH33. All inhibition data were consistent with a competitive mode of inhibition, as one would predict for sialic acid derivatives.
Figure 8.
Enzymatic hydrolysis of sialyllactose and X-Neu5Ac substrates.A, enzymatic digests of 3′-sialyllactose and 6′-sialyllactose substrates by the sialidases were analyzed by HPAEC-PAD in triplicate, with one replicate trace shown. B, released Lac, as analyzed by HPAEC-PAD, was quantified using Chromeleon software (Thermo Fisher Scientific) and the known concentration series of standards as outlined. Amounts of Lac disaccharide are shown as averages of triplicate HPAEC-PAD injections, and error bars represent the standard deviation of the mean. Statistical significance is indicated by asterisks (∗ = p < 0.05, ∗∗ = p < 0.005, ∗∗∗ = p < 0.0005, ∗∗∗∗ = p < 0.0001), as calculated using a 2-way ANOVA. C, the initial rates of X-Neu5Ac substrate hydrolysis by the GH33 domains were obtained and fit to the Michaelis-Menten equation. Data were obtained in triplicate, and error bars represent the standard error of the mean. Neu5Ac, N-acetylneuraminic acid; X-Neu5Ac, 5-bromo-4-chloro-3-indolyl α-D-N-acetylneuraminic acid; HPAEC-PAD, high-performance anion-exchange chromatography with pulsed amperometric detection.
Table 1.
Kinetic parameters for uninhibited and inhibited hydrolysis of X-Neu5Ac
| Kinetic constant | NanH | NanI | NanJ |
|---|---|---|---|
| Vmax (μM/s) | 0.13 ± 0.01 | 0.09 ± 0.01 | 0.13 ± 0.01 |
| Km (μM) | 280.0 ± 62.7 | 123.0 ± 25.4 | 87.0 ± 10.7 |
| kcat (1/s) | 13.3 ± 1.2 | 9.2 ± 0.6 | 15.3 ± 0.6 |
| kcat/Km | 0.05 ± 0.01 | 0.08 ± 0.02 | 0.18 ± 0.02 |
| Ki (μM) | |||
| DANA | 33.9 ± 4.4 | 3.1 ± 0.5 | 3.2 ± 0.3 |
| Siastatin B | 125.0 ± 16.2 | 10.7 ± 2.0 | 13.3 ± 1.6 |
| Peramivir | ND | ND | 104.0 ± 28.4 |
| Zanamivir | ND | ND | ND |
| Oseltamivir | ND | ND | ND |
Standard error is shown. ND indicates inhibition too weak to be detected or quantified.
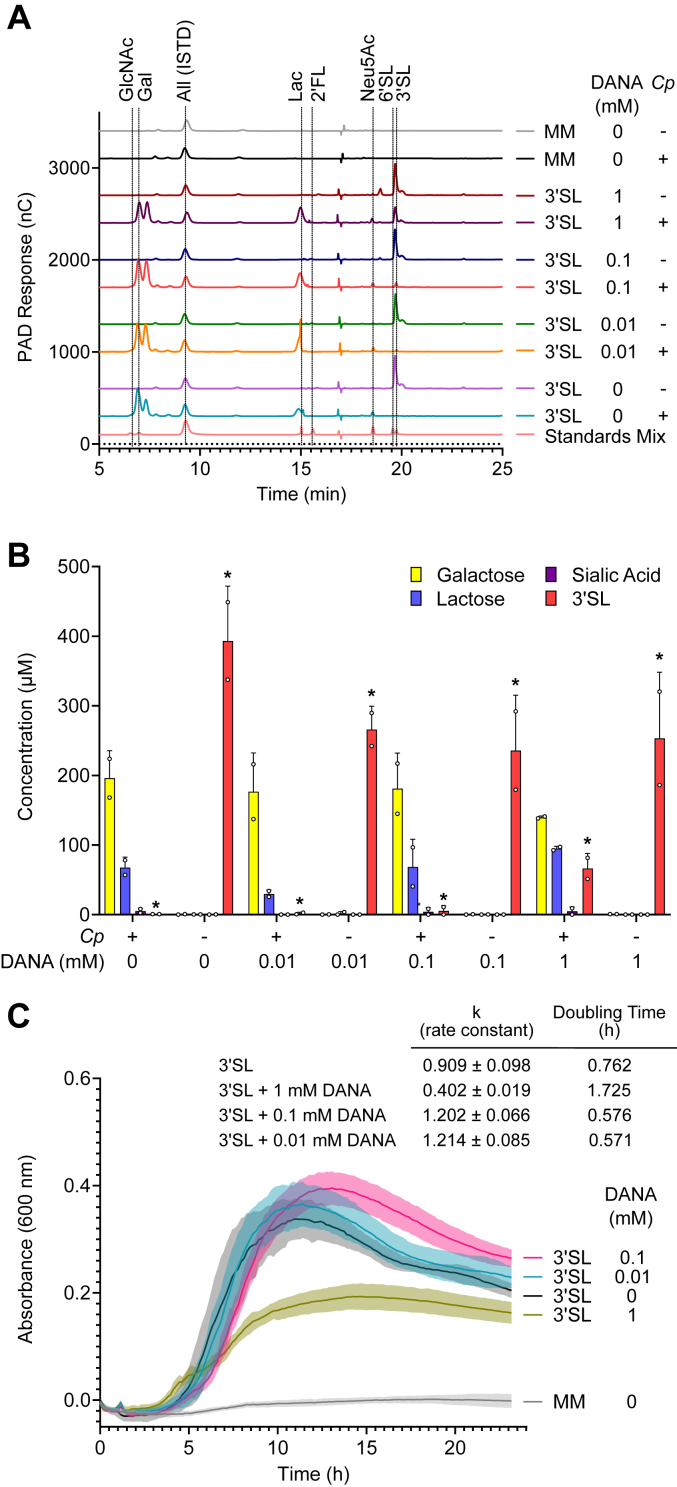
To investigate if sialidase inhibitors would demonstrate potential in treatment of C. perfringens colonization and infection we used a C. perfringens growth assay and DANA as a model inhibitor, as it was found to effectively inhibit the three GH33 domains of NanH, NanI, and NanJ. DANA was found to protect against C. perfringens consumption of 3′-SL, as assessed by high-performance anion-exchange chromatography-pulsed amperometric (HPAEC-PAD) quantification (Fig. 9, A and B). Furthermore, DANA was also found to reduce the growth of C. perfringens in the presence of 3′SL (Fig. 9C) but not with glucose as the sole carbohydrate source (data not shown). However, a high concentration of DANA (1 mM) was required to demonstrate a clear effect. This may be due to the relatively low potency of DANA against the sialidases in the direct enzyme assays as well as the complexity of potential cellular interplay of all three sialidases at once.
Figure 9.
Detection of carbohydrates remaining in Clostridium perfringens culture supernatants.A, representative HPAEC-PAD traces of filtered cell-free supernatants from 24 h cultures or negative controls containing 0.5% 3′-sialyllactose (3′-SL) and indicated concentrations of DANA. A dilution series of a standards mix of known milk oligosaccharides and monosaccharides was run in parallel with the samples to allow for quantification with an allose internal standard (ISTD) to account for variation in instrument injection volumes. Peaks from standards in the mix are indicated. B, quantification of known carbohydrates from C. perfringens supernatants. Error bars show the standard deviation of the mean (SD; n = 2 biological replicates) and asterisks indicate significant (p < 0.05) differences relative to the leftmost treatment group (+C. perfringens, - DANA) using a 2-way ANOVA. C, Clostridium perfringens growth curves in the presence of increasing concentrations of DANA neuraminidase inhibitor using 3′-SL as a substrate, with optical density monitored by absorbance quantitation at 600 nm. Averages of six growth curves are shown, with areas representing the standard deviation (SD). MM indicates minimalized medium with no carbohydrate. Bacterial growth kinetics were calculated based on exponential growth (GraphPad Prism), and rate constants and doubling times are listed for each growth condition. DANA, N-acetyl-2,3-dehydro-2-deoxyneuraminic acid; HPAEC-PAD, high-performance anion-exchange chromatography with pulsed amperometric detection.
Discussion
The genomes of most C. perfringens strains encode three GH33 isozymes, suggesting the potential for diversified, rather than redundant, function. The observation that these isozymes only share 30 to 60% amino acid sequence identity and different modular architectures further supports this concept. Here, we have specifically probed this idea through structural and functional analysis of the GH33 domains from these enzymes. As a cohort of enzymes they possess broad specificity and provide the machinery to process multiple different variants of terminal sialic acid attached via different linkages.
NanJGH33 was more efficient than NanIGH33 at hydrolyzing Neu5Gc and could accommodate 7-O-Ac modification, but the latter only when 9-O-acetylation was absent. NanIGH33 was most stringent being entirely unable to accommodate 7-O-Ac modification and was comparatively poor at hydrolyzing Neu5Gc variants. Neither enzyme could hydrolyze sialic acid with 4-O-Ac modifications. These two enzymes, which are predicted to be extracellular, are therefore somewhat redundant in their apparent specificity. NanIGH33 also proved to be the less efficient enzyme on a synthetic chromogenic substrate.
NanHGH33 proved to be the enzyme with the broadest activity with respect to accommodations of sialic acid variants, being able to process all common variants of sialic acid, except those with 4-O-Ac modifications, and did so with little obvious preference. The broad selectivity of NanHGH33 for these variants in comparison to NanIGH33 and NanJGH33 appears to come from differences in the region responsible for recognition of 9-O-Ac and 7-O-Ac modifications. The NanHGH33 active site is preformed and suited to accommodate these modifications without any obvious conformational changes to the protein or substrate, unlike the requirements of NanIGH33 and NanJGH33.
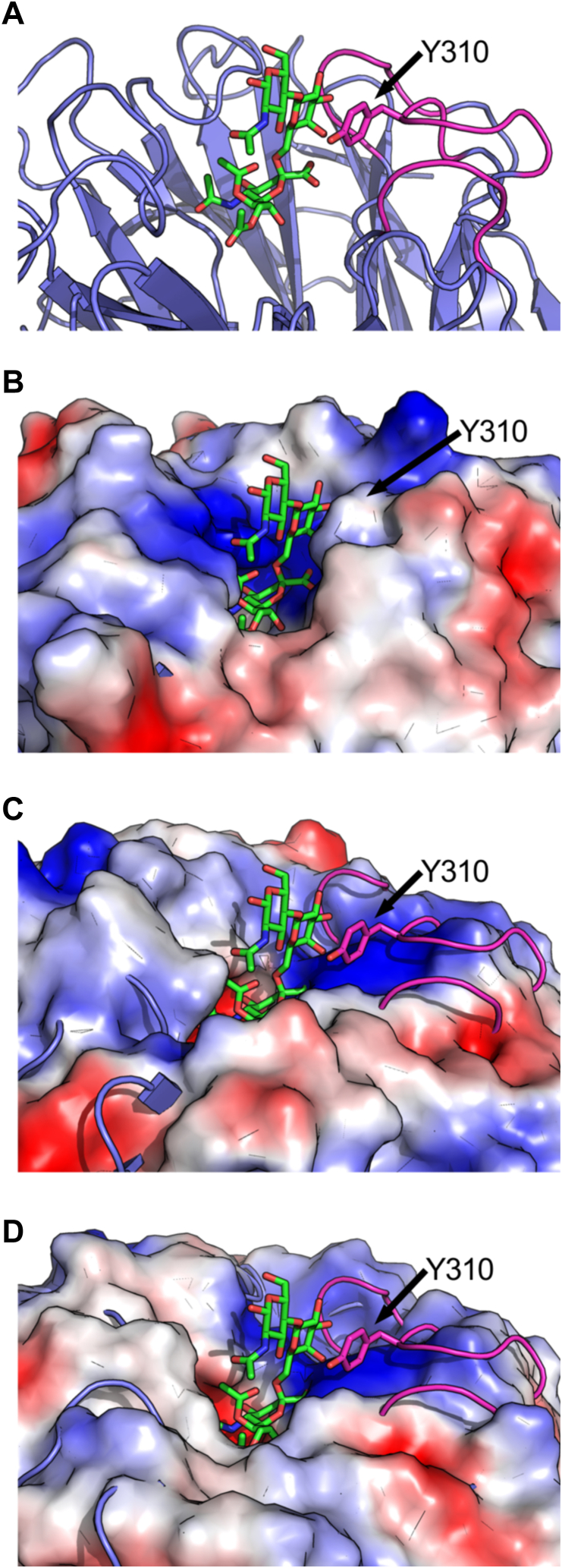
The most notable difference in NanH is in the loop structures surrounding the active site. The catalytic site of GH33 sialidases is in the cavity formed at the center of the 6-blades of the β-propellor fold. The loops of the propellers form a rim around the cavity. In NanHGH33 loops on 3-blades forming the rim of one side of the cavity are extended, thus deepening the active site (Fig. 10, A and B). The center blade of the three has a solvent exposed tyrosine residue, Y310, that projects into the active site. In the SAc3LacNAc Michaelis complex this tyrosine packs against the galactose to which the sialic acid is α-2,6-linked, providing additional interactions in a +1 subsite. In contrast, these loops are relatively minimal in NanIGH33 and NanJGH33, creating a somewhat shallow sialic acid binding site (Fig. 10, C and D). This lack of a +1 binding site in either NanIGH33 or NanJGH33 supports a general absence of selectivity for α-2,3- or α-2,6- linkages. In the case of the NanHGH33 SAc3LacNAc, however, the O4 of the galactose residue in the +1 subsite is oriented toward the aromatic ring of Y310, possibly making this less optimal interaction. Presumably, a similar interaction involving Y310 occurs when NanHGH33 binds α-2,3-linked sialoglycosides but in an as yet unknown but more favorable relative orientation of the galactose in the +1 subsite. Other GH33 sialidases have a +1 subsite with an aromatic amino acid sidechain, like NanHGH33 [e.g. TcTs from Trypanosoma cruzi (44) and NanC from Streptococcus pneumoniae (31)], whereas others have shallow active site like NanIGH33 and NanJGH33 [e.g. PG0352 from Porphyromonas gingivalis (45)]. However, current data are insufficient to provide a clear link between the presence of a +1 subsite and linkage or aglycon specificity, thus further work is required to provide an explanation.
Figure 10.
NanH has an additional +1 subsite.A, secondary structure of NanHGH33 (blue) in complex with SAc3LacNAc (green sticks) showing the three extended loops surrounding the active site (pink). Y310 that packs against the galactose in the +1 subsite is shown as sticks. B, surface of NanHGH33 showing the packing of the extended glycan against the extended surface of Y310 and the loops. C, NanIGH33 (shown as a surface) overlapped with NanHGH33 in complex with SAc3LacNAc (as shown in panel A). D, NanJGH33 (shown as a surface) overlapped with NanHGH33 in complex with SAc3LacNAc (as shown in panel A). Surfaces are shown as color ramped from red to white to blue as by their predicted negative to neutral to positive charge.
Overall, consistent with our hypothesis, the three C. perfringens sialidases do display some diversification of their substrate selectivity; however, there are more redundancies than differences, and it does not adequately justify the expansion of these enzymes in this bacterium. The most profound difference in the enzymes regards their likely modes of deployment. NanI and NanJ are both modular enzymes (Fig. 1), including modules characterized as having carbohydrate binding activity (22). However, NanI is predicted to be fully secreted while NanJ has a cohesin module that participates in the formation of CAZyme complexes and an FN3 domain that is postulated to mediate attachment to the bacterial cell-surface (21, 46). Thus, these extracellular enzymes likely have different modes of interacting with the host. In contrast, NanH is predicted to be localized to the cytoplasm of the bacterium (i.e. it lacks any obvious export signals), suggesting it does not interact directly with the host. The biological role of NanH is more likely in processing glycans that are imported with their terminal sialic acids intact. This could be the case considering that NanI and NanJ are less efficient (or unable) to process certain acetylated variants. NanH may play the role of “clean up” after sialylated glycans that are recalcitrant to NanI and NanJ are released extracellularly by other glycoside hydrolases and imported into the bacterium. This would rely on one or more hypothetical endo-acting glycanases that could hydrolyze and thereby release sialylated glycans. There is precedent for such endo-acting enzymes in other O-glycan metabolizing bacteria but they remain unidentified in C. perfringens (47). We suggest that the context of cellular environment is more important to the biological role of the three enzymes than the nuances in their substrate selectivities.
Regardless of cellular environment and locale in the host, sialidase inhibition is a well-sought therapeutic target for the treatment of viral and bacterial pathogens (48). The neuraminidase inhibitor DANA served as a scaffold to develop compounds for influenza viral sialidases, such as oseltamivir, zanamivir, and peramivir (41, 42, 49) and continual development is necessary to counteract the rapid mutation rate of the viral enzymes. These influenza-directed compounds have also been tested against the sialidases of the bacterial pathogen S. pneumoniae, with limited effect (50). While no commercial inhibitors were found to be significantly effective against the C. perfringens sialidases in this study (Ki values < 1.0 μM), the inhibition of bacterial growth by DANA demonstrated that interfering with the ability of this pathogen to remove terminal sialic acids from glycans is a viable strategy. Indeed, further modifications of DANA at the C9 or C5 positions have been shown to increase selectivity for Vibrio cholera over human sialidases (51), which lends further support to this approach. The structural determinants of the activity of the three C. perfringens sialidases elucidated herein will enable the design of compounds that can maximize interactions within the active site clefts for the development of more potent, specific inhibitors in the treatment of C. perfringens causative disease.
Experimental procedures
Gene synthesis and recombinant protein expression in E. coli
C. perfringens CP4 nucleotide sequences were taken from NCBI (52), analyzed by dbCAN2 (53) and InterProScan (54), and modeled using Phyre2 (55). For nanI and nanJ genes, Phyre2 models were manually curated with PyMOL (Schrödinger, Inc; https://pymol.org/), and GH33 domain amino acid sequence boundaries were chosen based on these analyses; NanI256-707, NanJ192-639, referred to as NanIGH33 and NanJGH33, respectively. The full-length nucleotide sequence for the nanH gene was used for all downstream studies; the nanH gene encodes for a GH33 domain without any ancillary domains, but for consistency was referred to as NanHGH33. All GH33 nucleotide sequences were codon optimized for expression in E. coli and gene synthesized including flanking NdeI and XhoI restriction sites at the 5′ and 3′ ends, respectively, and a stop codon at the 3′ end (BioBasic Inc). Genes were ordered synthesized into the pET28a vector for recombinant protein expression with an N-terminal His6-tag for downstream protein purification. A D62N mutation in the NanHGH33 construct was introduced via the quick-change method to create a catalytically inactivated version of the enzyme for crystallography.
Protein production for crystallization
The pET28a plasmids containing the gene fragments encoding NanHGH33, NanHGH33D62N, NanIGH33, and NanJGH33 were transformed into E. coli BL21 (DE3∗) and grown in 2× YT media supplemented with 50 μg mL-1 kanamycin at 37 °C. Once the cultures reached an A600 of 0.6 to 0.8, production of recombinant protein was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside followed by continued incubation at 16 °C. The cells were harvested by centrifugation at 6000 rpm for 15 min at 4 °C. The collected cell pellet was resuspended in sucrose solution (25% sucrose, 20 mM Tris–HCl pH 8.0, and 10 mg mL-1 lysozyme), before completing the lysis with a solution containing 1% deoxycholate, 0.1% Triton X-100, 500 mM NaCl, 0.2 μg/ml DNase in 20 mM Tris pH 8.0, and 2 mM MgCl2.
The produced recombinant protein was isolated by immobilized metal affinity chromatography (STREAMLINE Chelating beads, GE HealthCare). Fractions that contained the protein of interest were determined by SDS-PAGE and pooled. This was concentrated using a stirred ultrafiltration unit with a 10 kDa membrane (Millipore Sigma) before buffer exchanging by dialysis into 20 mM Tris–HCl pH 8.0 with 100 mM NaCl (the buffer used for NanH also included 1 mM dithiothreitol). The concentrated proteins were further purified by size exclusion chromatography (Sephadex S-200) equilibrated in 20 mM Tris–HCl pH 8.0, with 100 mM NaCl buffer. Fractions containing purified protein were concentrated again in a stirred ultrafiltration cell to 20 mg mL-1.
Crystallization and optimization
NanHGH33, NanHGH33D62N, and NanJGH33 proteins (20 mg mL-1) were crystallized by sitting drop vapor diffusion method at 18 °C. The crystallization conditions for NanHGH33 and NanJGH33 were screened by mixing a 1:1 ratio of purified protein with a range of crystallization conditions found in the MCSG 1 to 4 (Anatrace) and Index (Hampton Research) crystallization screens. NanHGH33 and the mutant crystallized in the condition 0.1 M Hepes:NaOH pH 7.5, and 20% PEG 8000. NanJGH33 crystals were obtained in 0.1 M Hepes, pH 7.5, 2.0 M ammonium sulfate. NanIGH33 (15 g L-1) was crystallized following the methods described by Newstead et al. (28), using the hanging drop vapor diffusion method, with a 1:1 ratio of purified protein to crystallization solution (20% PEG 3350, 0.2 M KNO3 pH 7.0). To obtain the NanIGH33 complexes with ligand, the protein was cocrystallized with 15 mM of either Neu5,9Ac or Neu5Gc. The substrate and inhibitor complexes of the NanHGH33 and NanJGH33 (both WT and catalytic mutants) were obtained by soaking the crystal in 10 mM of the respective monosaccharide ligands for 30 min. The NanHGH33D62N complex with SAc3LacNAc was obtained by adding compound in solid form to a drop containing a crystal and soaking for ∼30 min. All crystals were cryoprotected using the crystallization condition supplemented with 20% v/v ethylene glycol prior to cooling directly in an N2 stream at 100 K.
Data collection, structure solution, and refinement
Diffraction data were collected on an instrument comprising a Pilatus 200K 2D detector coupled with a MicroMax-007HF X-ray generator, a VariMax-HF ArcSec Confocal Optical System, and an Oxford Cryostream 800. Data sets were processed using HKL2000 (Table S1). Using the coordinates of apo-NanIGH33 (PDB code 2VK6) and Phaser (56) (https://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software), the structures of NanHGH33, NanIGH33, and NanJGH33 were determined by molecular replacement. Initial refinement of these structures was carried out using REFMAC (57) (https://www2.mrc-lmb.cam.ac.uk/groups/murshudov/content/refmac/refmac.html) before structural corrections and the addition of waters were manually done in COOT (58) (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/). This model was further improved using Phenix.Refine (59). All the data sets were monitored by flagging 5% of all observation as “free” (60). Validation of the model was performed using MolProbity (61). Final refinement and model statistics are given in Table S1.
Protein production for enzyme assays
Protein construct vectors were transformed into E. coli BL21 Tuner (DE3) cells (Novagen). Cells were grown in LB Miller broth containing 50 μg mL-1 kanamycin at 37 °C to an A600 of 0.6 to 0.8, when protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.5 mM. The cell culture was incubated at 37 °C for 4 h prior to being harvested by centrifugation at 6500g for 20 min at 4 °C. Cell pellets were stored at −20 °C until needed.
The cell pellet from 1 L of bacterial culture was thawed and resuspended in 50 ml of lysis buffer (20 mM Tris pH 8.0, 500 mM NaCl, 0.1 mg mL-1 lysozyme). Cells were homogenized by sonication for 2 min of 1 s intervals of sonic pulses at an intensity amplitude of 30 (Fisherbrand Model 705 Sonic Dismembrator and probe; Thermo Fisher Scientific). Cellular debris was removed by centrifugation at 17,500g for 45 min at 4 °C. The filtrate was loaded onto a HisTrap FF column (Ni Sepharose 6 Fast Flow; Cytiva) and purified by immobilized metal affinity chromatography; recombinant protein was eluted by an increasing gradient 0 to 500 mM imidazole in 20 mM Tris pH 8.0 and 500 mM NaCl. Protein was concentrated by centrifugation with 30 kDa cutoff Amicon Ultra centrifugal concentrators (Millipore Sigma). His6-tagged protein was further purified using a HiLoad 16/60 Superdex 200 prep-grade size exclusion column (GE HealthCare) in 20 mM Tris pH 8, 500 mM NaCl, 2% glycerol (the buffer used for NanHGH33 also included 1 mM DTT). Pure protein fractions were pooled and concentrated. Protein purification was monitored throughout by SDS-PAGE. All recombinant proteins were used freshly prepared for downstream enzyme activity assays.
Activity against O-acetylated sialic acids: BSM assay
Neuraminidase specificity on different sialic acid types was determined using BSM as a model substrate based on a published assay (62). Briefly, 250 μg BSM fortified with 0.5 μg 13C3-Neu5Ac as an internal standard (ISTD) was dissolved in 50 mM sodium acetate buffer, pH 4.5 and combined with 0.1 mU of neuraminidase based on units predetermined by activity on 4-methylumbelliferone-Neu5Ac. Reactions were incubated at 37 °C for 5.5 h and then stopped by removal of enzyme using 10 kDa molecular weight cutoff spin columns centrifuged at 14,000g for 10 min. After this, samples were labeled with 400 μl 24 mM N, N-dimethylbenzylamine in 2M acetic acid at 60 °C for 1 h. Immediately following this, a reverse phase extraction was performed to remove excess N, N-dimethylbenzylamine using 500 mg C18 columns preconditioned with 2 ml 80% acetonitrile/0.1% trifluoroacetic acid, followed by 8 ml of ultrapure water. Samples were then applied to columns, washed with 4 ml ultrapure water, and then eluted in 1.2 ml of 50% acetonitrile. Elutions were dried on the speedvac in the dark, after which they were resuspended in 50 μl of 30% methanol and transferred to vials for ultra high-performance liquid chromatography coupled to mass spectrometry analysis using a Phenomenex Kinetex Biphenyl 100 mm × 2.1 mm column with H2O and methanol mobile phases and an Agilent Technologies 6530 QTOF mass spectrometer operating in positive ion mode with an Agilent Jet Stream electrospray ionization source as previously published (63, 64). MS peak areas were quantitated relative to the ISTD.
Microarray printing and screening procedure
The synthesis of the acetylated sialosides was as previously reported (37). All the biotinylated compounds were printed on streptavidin-coated glass slides (SuperStreptavidin Microarray Substrate Slides, Arrayit Inc) using a Scienion sciFLEXARRAYER S3 noncontact microarray equipped with a Scienion PDC80 nozzle (Scienion Inc). Individual compounds were dissolved in PBS buffer (10 mM, pH 7.4) at a concentration of 100 μM and were printed in replicates of six with spot volume ∼ 400 pL, at 20 °C and 50% humidity. Each slide has 21 subarrays in a 3 × 8 layout. The slides were stored at 4 °C after printing and were blocked with TSM binding buffer (20 mM Tris·Cl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, and 2 mM MgCl2, 0.05% Tween-20, and 1% bovine serum albumin) for 1 h at 4 °C prior to use. The slides were incubated with different sialidases, including NanH, NanI, and NanJ (20 μg/ml in pH 7.4 PBS buffer) at 37 °C for 1 h. Each slide was then washed with TSM washing buffer, TSM buffer, water, and spun dry. The human-Fc tagged BCoV HE was mixed with goat anti-human IgG antibody (Alexa Fluor 647 conjugated, Jackson ImmunoResearch 109–605–008) in a 1:1 ratio with a final concentration of 3 μg/ml in TSM binding buffer and incubated at 4 °C for 1 h. The glass slide was incubated with the premixed solution at 4 °C for 1 h, washed with TSM washing buffer, TSM buffer, water, and spun dry. The washing steps could be carried out with the cassette attached to protect the unused blocks if necessary. The slides were scanned using a GenePix 4000B microarray scanner (Molecular Devices) at the appropriate excitation wavelength with a resolution of 10 μm. Various gains and PMT values were used in the scanning to ensure that all the signals were within the liner range of the scanner’s detector and there was no saturation of signals. The image was analyzed using GenePix Pro 7 software (version 7.2.29.2, Molecular Devices; https://support.moleculardevices.com/s/article/GenePix-Pro-7-Microarray-Acquisition-Analysis-Software-Download-Page). The data were analyzed with an Excel macro (65) to provide the results. The highest and the lowest value of the total fluorescence intensity of the six replicates spots were removed, and the four values in the middle were used to provide the mean value and standard deviation.
Enzymatic cleavage of oligosaccharides and HPAEC-PAD analysis
Purified GH33 enzymes were screened for activity on 3′-sialyllactose (Biosynth), 6′-sialyllactose (Biosynth), and disialyllactose (Biosynth). Reactions (250 μl) contained 10 nM enzyme, 1 mg mL-1 substrate, and 0 or 500 μM DANA in 20 mM phosphate buffer at the optimal pH for each enzyme (NanHGH33 at 7.0, NanIGH33 at 6.5, and NanJGH33 at 6.8). Reactions were incubated at 37 °C for 60 min, after which the reactions were treated at 95 °C for 10 min to denature the enzyme and terminate the reaction. Samples were then shortly centrifuged at 8000g to pellet denatured protein from the product. HPAEC was performed with a Dionex ICS-3000 chromatography system (Thermo Fisher Scientific) equipped with an autosampler as well as a PAD detector. Taken from the soluble fraction of the enzyme digests, the samples were diluted one-fifth and fortified with 100 μM allose as an ISTD and 10 μl were injected onto an analytical (3 × 150 mm) CarboPac PA20 column (Thermo Fisher Scientific) with a CarboPac PA20 guard column (Thermo Fisher Scientific). Samples were eluted at a 0.4 ml min-1 flow rate with a background of sodium hydroxide (NaOH; buffer A) and a gradient of sodium acetate (NaOAc; buffer B) (Table S2). The elution was monitored with a PAD detector (standard quadratic waveform). For quantification, a mixture of monosaccharide and oligosaccharide standards were used in a known concentration series with the following concentrations for a 1× solution, and added 100 μM allose ISTD: 100 μM fucose (Fuc), 100 μM glucose (Glc), 100 μM N-acetylgalactosamine (GalNAc), 100 μM lactose (Lac), 100 μM 2′-fucosyllactose (2′-FL), 500 μM Neu5Ac, 100 μM 6′-sialyllactose (6′-SL), 100 μM 3′-sialyllactose (3′-SL). Note that higher amounts of Neu5Ac standard were required to produce an approximately equivalent PAD response to those of other saccharide standards with these instrument conditions (i.e. absent a postcolumn addition of NaOH). Quantification of monooligosaccharides and oligosaccharides was performed in Chromeleon software (version 6.80, Thermo Fisher Scientific; https://www.thermofisher.com/order/catalog/product/CHROMELEON6) from triplicate HPAEC-PAD injections.
Substrate reduction assays using CE-LIF
The relative specificities of each neuraminidase for differing sialic acid linkages were screened using 3′-SL and 6′-SL in CE-based assays (66). 8-Aminopyrene-1,3,6-trisulfonate -labeled 3′- and 6′-SL were produced via reductive amination and subsequently excess fluorogenic reagents and unlabeled oligosaccharides were removed using tetrabutylammonium bromide-assisted liquid-liquid extraction (67). Each assay was performed directly in CE-LIF vials which contained both 3′-SL and 6′-SL and similarly prepared 2′-fucosyllactose as an internal standard. Reactions were carried out in freshly prepared 50 mM sodium acetate buffer, pH 4.5, at a temperature of 37 °C. To eliminate the trans-sialidase activity of each neuraminidase 1.2 U of β-galactosidase (Megazyme) were added to each reaction. Total reaction volumes were 50 μl for each enzyme. Samples were hydrodynamically injected every 10 min and separated by CE-LIF exactly as previously described with the exception that fused silica capillaries (DigiKey) were used (67). Note that the first recorded time point (after addition of neuraminidase) was 2 min, reflecting the 2 min capillary rinse before sample injection.
Enzymatic hydrolysis of the chromogenic substrate X-Neu5Ac, and catalysis and inhibition kinetics
pH optima for the three GH33 domains were assayed using the chromogenic substrate 5-bromo-4-chloro-3-indolyl α-D-N-acetylneuraminic acid (X-Neu5Ac) (Sigma-Aldrich). Hydrolysis was measured spectrophotometrically at 630 nm with 10 nM enzyme and 500 μM X-Neu5Ac in 50 mM citrate-phosphate buffer from pH 3 to 8. Assays were carried out at 25 °C in triplicate in 96-well microtitre plates and measured using a Synergy HT plate reader (BioTek). Data analysis was performed in Prism 9 (GraphPad; https://www.graphpad.com/features). Product equivalents were calculated based on the A630 of the total cleavage of 100 μM X-Neu5Ac by NanHGH33, and thereby calculating a conversion factor.
Enzyme kinetics assays were performed with concentrations of X-Neu5Ac ranging from 7.8 μM to 1 mM and inhibitor concentrations from 0 to 100 μM in 20 mM phosphate buffer at the optimal pH for each enzyme (NanHGH33 at 7.0, NanIGH33 at 6.5, and NanJGH33 at 6.8). Inhibitors were sourced from Sigma-Aldrich: DANA, siastatin B, peramivir, oseltamivir, and zanamivir. The rate of hydrolysis was quantified using A620 at 25 °C. Enzyme and inhibitor were incubated for 5 min prior to reaction initiation by the addition of the X-Neu5Ac substrate. Triplicate readings were obtained using a SpectraMax iD3 plate reader (Molecular Devices), and a product conversion factor was calculated as for the pH optima assays but measured at 620 nm. The initial rates were determined by fitting a straight line to the linear portion of the progress curves. The resulting rate data were fitted to Michaelis-Menten curves, and Ki, Km, and Vmax values for the reactions were obtained from simultaneously fitting the uninhibited and inhibited data to the full mixed enzyme inhibition equation in Prism 9 (GraphPad) (Fig. S3). For those inhibitors that appeared to follow competitive inhibition kinetics, the data were then fit to the competitive inhibition equation for determination of Ki.
Growth and inhibition of C. perfringens on sialylated substrates
C. perfringens CP1 was pregrown in Columbia broth for 16 h from a glycerol stock. The following day, the bacteria were subcultured into 5 ml Columbia and grown for an additional 2 h, after which were pelleted by centrifugation at 5000g for 10 min and resuspended in a previously established Clostridium perfingens minimalized medium (17). Cultures were adjusted to inoculate at A600 of 0.1 in 200 μl minimalized medium supplemented with filter-sterilized 0.5% 3′SL or negative control of filter-sterilized ultrapure water, and in the absence or presence of DANA at 0.01 mM, 0.1 mM, or 1 mM concentrations. Experiments were conducted in six technical triplicates in 96-well plates in an anaerobic chamber at 37 °C for 24 h, with absorbance recorded at 600 nm using a Stratus plate reader (Cerillo), after which culture supernatants were collected, pooled between technical replicates, and filter-sterilized with a 0.2 μm membrane. A 1/10 dilution of the supernatant was then analyzed by HPAEC-PAD using a Dionex ICS-3000 instrument with a CarboPac PA20 column using the same elution method as above (Table S1). The runs also included 50 μg/ml standards of the oligosaccharides used to assess growth or their monosaccharide and disaccharide constituents to annotate metabolites. Bacterial growth kinetics were calculated using GraphPad Prism for exponential growth with log (population).
Data availability
The accession codes for the crystal structures are available on the PDB as follows: NanHGH33 (apo: 8UB5; Neu5Ac complex: 8UL7; Neu59Ac complex: 8ULE; SAc3LacNAc complex: 8UM0), NanJGH33 (apo: 8URL; Neu5Ac complex: 8UVV; Neu5,9Ac complex: 9C20) and NanIGH33 (Neu5,9Ac: 8U2A; Neu5Gc: 8U5O).
Supporting information
This article contains supporting information.
Conflict of interest
A. B. Boraston is an Editorial Board Member/Editor-in-Chief/Associate Editor/Guest Editor for JBC and was not involved in the editorial review or the decision to publish this article. The other authors declare they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
B. J. M., K. E. L., J. D. W. I., L. K., P. D., L. L., J. M. G., L. K., and W. F. Z. investigation; B. J. M., K. E. L., L. L., W. F. Z., D. W. A., and A. B. B. visualization; B. J. M., K. E. L., and A. B. B. writing–original draft; B. J. M., W. F. Z., and A. B. B. validation; K. E. L., W. F. Z., D. W. A., and A. B. B. writing–review and editing; G. D. I., G.-J. B., W. F. Z., D. W. A., and A. B. B. supervision; G. D. I. resources; W. F. Z., D. W. A., and A. B. B. project administration; W. F. Z., D. W. A., and A. B. B. funding acquisition; W. F. Z., D. W. A., and A. B. B. conceptualization; A. B. B. formal analysis.
Funding and additional information
This work was supported by funding from the Canadian Glycomics Network (Project ID-04) and Alberta Agriculture and Forestry/Results Driven Agricultural Research (RDAR) (Proposal 2019H001R). J. M. G. was supported by an NSERC - PDF fellowship. W. F. Z. received infrastructure support from the Canadian Foundation for Innovation (John Evans Leaders Fund) and the BC Knowledge Development Fund.
Reviewed by members of the JBC Editorial Board. Edited by Chris Whitfield
Contributor Information
Wesley F. Zandberg, Email: wesley.zandberg@ubc.ca.
D. Wade Abbott, Email: wade.abbott@agr.gc.ca.
Alisdair B. Boraston, Email: boraston@uvic.ca.
Supporting information
References
- 1.Varki A., Schauer R. 9th Ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2009. Sialic Acids, Chapter 14; pp. 1–19. [PubMed] [Google Scholar]
- 2.Severi E., Hood D.W., Thomas G.H. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 3.Hobbs M., Jahan M., Ghorashi S.A., Wang B. Current Perspective of Sialylated Milk Oligosaccharides in Mammalian Milk: Implications for Brain and Gut Health of Newborns. Foods. 2021;10:473. doi: 10.3390/foods10020473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Graaff D.R., Felz S., Neu T.R., Pronk M., van Loosdrecht M.C.M., Lin Y. Sialic acids in the extracellular polymeric substances of seawater-adapted aerobic granular sludge. Water Res. 2019;155:343–351. doi: 10.1016/j.watres.2019.02.040. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y.L., Chassard C., Hausmann M., von Itzstein M., Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat. Commun. 2015;6:8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell A., Brunt J., Crost E., Vaux L., Nepravishta R., Owen C.D., et al. Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut. Nat. Microbiol. 2019;4:2393–2404. doi: 10.1038/s41564-019-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jennings M.P., Day C.J., Atack J.M. How bacteria utilize sialic acid during interactions with the host: snip, snatch, dispatch, match and attach. Microbiology (Reading) 2022;168:001157. doi: 10.1099/mic.0.001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juge N., Tailford L., Owen C.D. Sialidases from gut bacteria: a mini-review. Biochem. Soc. Trans. 2016;44:166–175. doi: 10.1042/BST20150226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almagro-Moreno S., Boyd E.F. Sialic acid catabolism confers a competitive advantage to pathogenic vibrio cholerae in the mouse intestine. Infect Immun. 2009;77:3807–3816. doi: 10.1128/IAI.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahya H.F., Andrew P.W., Yesilkaya H. Deacetylation of sialic acid by esterases potentiates pneumococcal neuraminidase activity for mucin utilization, colonization and virulence. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis A.L., Lewis W.G. Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell Microbiol. 2012;14:1174–1182. doi: 10.1111/j.1462-5822.2012.01807.x. [DOI] [PubMed] [Google Scholar]
- 12.Burton R.M. The action of neuraminidase from Clostridium perfringens on gangliosides. J. Neurochem. 1963;10:503–512. doi: 10.1111/j.1471-4159.1963.tb09853.x. [DOI] [PubMed] [Google Scholar]
- 13.Li J., Navarro M.A., Uzal F.A., McClane B.A. NanI sialidase contributes to the growth and adherence of Clostridium perfringens type F Strain F4969 Presence Adherent Mucus. Infect Immun. 2021;89 doi: 10.1128/IAI.00256-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., McClane B.A. Contributions of NanI sialidase to Caco-2 cell adherence by Clostridium perfringens type A and C strains causing human intestinal disease. Infect Immun. 2014;82:4620–4630. doi: 10.1128/IAI.02322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navarro M.A., Li J., McClane B.A., Morrell E., Beingesser J., Uzal F.A. NanI sialidase is an important contributor to Clostridium perfringens type F strain F4969 intestinal colonization in mice. Infect Immun. 2018;86 doi: 10.1128/IAI.00462-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J., Sayeed S., Robertson S., Chen J., McClane B.A. Sialidases affect the host cell adherence and epsilon toxin-induced cytotoxicity of Clostridium perfringens type D strain CN3718. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacMillan J.L., Vicaretti S.D., Noyovitz B., Xing X., Low K.E., Inglis G.D., et al. Structural analysis of broiler chicken small intestinal mucin O-glycan modification by Clostridium perfringens. Poult. Sci. 2019;98:5074–5088. doi: 10.3382/ps/pez297. [DOI] [PubMed] [Google Scholar]
- 18.Zahoor I., Ghayas A., Basheer A. Genetics and genomics of susceptibility and immune response to necrotic enteritis in chicken. A. Rev. Mol. Biol. Rep. 2018;45:31–37. doi: 10.1007/s11033-017-4138-8. [DOI] [PubMed] [Google Scholar]
- 19.M'Sadeq S.A., Wu S., Swick R.A., Choct M. Towards the control of necrotic enteritis in broiler chickens with in-feed antibiotics phasing-out worldwide. Anim. Nutr. 2015;1:1–11. doi: 10.1016/j.aninu.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low K.E., Smith S.P., Abbott D.W., Boraston A.B. The glycoconjugate-degrading enzymes of Clostridium perfringens: tailored catalysts for breaching the intestinal mucus barrier. Glycobiology. 2021;31:681–690. doi: 10.1093/glycob/cwaa050. [DOI] [PubMed] [Google Scholar]
- 21.Adams J.J., Gregg K., Bayer E.A., Boraston A.B., Smith S.P. Structural basis of clostridium perfringens toxin complex formation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12194–12199. doi: 10.1073/pnas.0803154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boraston A.B., Ficko-Blean E., Healey M. Carbohydrate recognition by a large sialidase toxin from Clostridium perfringens. Biochemistry. 2007;46:11352–11360. doi: 10.1021/bi701317g. [DOI] [PubMed] [Google Scholar]
- 23.Crennell S., Garman E., Laver G., Vimr E., Taylor G. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure. 1994;2:535–544. doi: 10.1016/s0969-2126(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 24.Moustafa I., Connaris H., Taylor M., Zaitsev V., Wilson J.C., Kiefel M.J., et al. Sialic acid recognition by vibrio cholerae neuraminidase. J. Biol. Chem. 2004;279:40819–40826. doi: 10.1074/jbc.M404965200. [DOI] [PubMed] [Google Scholar]
- 25.Grondin J.M., Chitayat S., Ficko-Blean E., Houliston S., Arrowsmith C.H., Boraston A.B., et al. An unusual mode of galactose recognition by a family 32 carbohydrate-binding module. J. Mol. Biol. 2014;426:869–880. doi: 10.1016/j.jmb.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 26.Newstead S.L., Watson J.N., Bennet A.J., Taylor G. Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. Acta. Crystallogr. D. Biol. Crystallogr. 2005;61:1483–1491. doi: 10.1107/S0907444905026132. [DOI] [PubMed] [Google Scholar]
- 27.Ficko-Blean E., Boraston A.B. The interaction of a carbohydrate-binding module from a clostridium perfringens n-acetyl-beta-hexosaminidase with its carbohydrate receptor. J. Biol. Chem. 2006;281:37748–37757. doi: 10.1074/jbc.M606126200. [DOI] [PubMed] [Google Scholar]
- 28.Newstead S.L., Potter J.A., Wilson J.C., Xu G., Chien C.H., Watts A.G., et al. The structure of Clostridium perfringens NanI sialidase and its catalytic intermediates. J. Biol. Chem. 2008;283:9080–9088. doi: 10.1074/jbc.M710247200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crennell S.J., Garman E.F., Laver W.G., Vimr E.R., Taylor G.L. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc. Natl. Acad. Sci. U. S. A. 1993;90:9852–9856. doi: 10.1073/pnas.90.21.9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaskell A., Crennell S., Taylor G. The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure. 1995;3:1197–1205. doi: 10.1016/s0969-2126(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 31.Owen C.D., Lukacik P., Potter J.A., Sleator O., Taylor G.L., Walsh M.A. Streptococcus pneumoniae nanc: structural insights into the specificity and mechanism of a sialidase that produces a sialidase inhibitor. J. Biol. Chem. 2015;290:27736–27748. doi: 10.1074/jbc.M115.673632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park K.H., Kim M.G., Ahn H.J., Lee D.H., Kim J.H., Kim Y.W., et al. Structural and biochemical characterization of the broad substrate specificity of bacteroides thetaiotaomicron commensal sialidase. Biochim. Biophys. Acta. 2013;1834:1510–1519. doi: 10.1016/j.bbapap.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 33.Russell R.J., Haire L.F., Stevens D.J., Collins P.J., Lin Y.P., Blackburn G.M., et al. The structure of h5n1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 34.Chavas L.M., Tringali C., Fusi P., Venerando B., Tettamanti G., Kato R., et al. Crystal structure of the human cytosolic sialidase Neu2. Evidence for the dynamic nature of substrate recognition. J. Biol. Chem. 2005;280:469–475. doi: 10.1074/jbc.M411506200. [DOI] [PubMed] [Google Scholar]
- 35.Taylor G. Sialidases: structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- 36.Li J., McClane B.A. The sialidases of clostridium perfringens type d strain cn3718 differ in their properties and sensitivities to inhibitors. Appl. Environ. Microbiol. 2014;80:1701–1709. doi: 10.1128/AEM.03440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., Lang Y., Liu L., Bunyatov M.I., Sarmiento A.I., de Groot R.J., et al. Synthetic o-acetylated sialosides facilitate functional receptor identification for human respiratory viruses. Nat. Chem. 2021;13:496–503. doi: 10.1038/s41557-021-00655-9. [DOI] [PubMed] [Google Scholar]
- 38.Klein A., Diaz S., Ferreira I., Lamblin G., Roussel P., Manzi A.E. New sialic acids from biological sources identified by a comprehensive and sensitive approach: liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) of SIA quinoxalinones. Glycobiology. 1997;7:421–432. doi: 10.1093/glycob/7.3.421. [DOI] [PubMed] [Google Scholar]
- 39.Chuzel L., Ganatra M.B., Rapp E., Henrissat B., Taron C.H. Functional metagenomics identifies an exosialidase with an inverting catalytic mechanism that defines a new glycoside hydrolase family (GH156) J. Biol. Chem. 2018;293:18138–18150. doi: 10.1074/jbc.RA118.003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo T., Heon-Roberts R., Zou C., Zheng R.B., Pshezhetsky A.V., Cairo C.W. Selective inhibitors of human neuraminidase 1 (neu1) J. Med. Chem. 2018;61:11261–11279. doi: 10.1021/acs.jmedchem.8b01411. [DOI] [PubMed] [Google Scholar]
- 41.Kim C.U., Lew W., Williams M.A., Liu H., Zhang L., Swaminathan S., et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 1997;119:681–690. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 42.von Itzstein M., Wu W.Y., Kok G.B., Pegg M.S., Dyason J.C., Jin B., et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 43.Holzer C.T., von Itzstein M., Jin B., Pegg M.S., Stewart W.P., Wu W.Y. Inhibition of sialidases from viral, bacterial and mammalian sources by analogues of 2-deoxy-2,3-didehydro-n-acetylneuraminic acid modified at the c-4 position. Glycoconj J. 1993;10:40–44. doi: 10.1007/BF00731185. [DOI] [PubMed] [Google Scholar]
- 44.Amaya M.F., Watts A.G., Damager I., Wehenkel A., Nguyen T., Buschiazzo A., et al. Structural insights into the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Structure. 2004;12:775–784. doi: 10.1016/j.str.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 45.Clark N.D., Pham C., Kurniyati K., Sze C.W., Coleman L., Fu Q., et al. Functional and structural analyses reveal that a dual domain sialidase protects bacteria from complement killing through desialylation of complement factors. PLoS Pathog. 2023;19 doi: 10.1371/journal.ppat.1011674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ficko-Blean E., Gregg K.J., Adams J.J., Hehemann J.H., Czjzek M., Smith S.P., et al. Portrait of an enzyme, a complete structural analysis of a multimodular beta-N-acetylglucosaminidase from Clostridium perfringens. J. Biol. Chem. 2009;284:9876–9884. doi: 10.1074/jbc.M808954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crouch L.I., Liberato M.V., Urbanowicz P.A., Basle A., Lamb C.A., Stewart C.J., et al. Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown. Nat. Commun. 2020;11:4017. doi: 10.1038/s41467-020-17847-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowles W.H.D., Gloster T.M. Sialidase and sialyltransferase inhibitors: targeting pathogenicity and disease. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.705133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babu Y.S., Chand P., Bantia S., Kotian P., Dehghani A., El-Kattan Y., et al. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based Drug design. J. Med. Chem. 2000;43:3482–3486. doi: 10.1021/jm0002679. [DOI] [PubMed] [Google Scholar]
- 50.Walther E., Xu Z., Richter M., Kirchmair J., Grienke U., Rollinger J.M., et al. Dual acting neuraminidase inhibitors open new opportunities to disrupt the lethal synergism between Streptococcus pneumoniae and influenza. Virus Front. Microbiol. 2016;7:357. doi: 10.3389/fmicb.2016.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khedri Z., Li Y., Cao H., Qu J., Yu H., Muthana M.M., et al. Synthesis of selective inhibitors against V. cholerae sialidase and human cytosolic sialidase NEU2 Org. Biomol. Chem. 2012;10:6112–6120. doi: 10.1039/c2ob25335f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022;50:D20–D26. doi: 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H., Yohe T., Huang L., Entwistle S., Wu P., Yang Z., et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., et al. Interproscan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the ccp4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DiMaio F., Echols N., Headd J.J., Terwilliger T.C., Adams P.D., Baker D. Improved low-resolution crystallographic refinement with phenix and rosetta. Nat. Methods. 2013;10:1102–1104. doi: 10.1038/nmeth.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brunger A.T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 61.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., et al. Molprobity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hunter C.D., Porter E.M., Cairo C.W. Human neuraminidases have reduced activity towards modified sialic acids on glycoproteins. Carbohydr. Res. 2020;497 doi: 10.1016/j.carres.2020.108139. [DOI] [PubMed] [Google Scholar]
- 63.Gray T.E., Narayana K., Garner A.M., Bakker S.A., Yoo R.K.H., Fischer-Tlustos A.J., et al. Analysis of the biosynthetic flux in bovine milk oligosaccharides reveals competition between sulfated and sialylated species and the existence of glucuronic acid-containing analogues. Food Chem. 2021;361 doi: 10.1016/j.foodchem.2021.130143. [DOI] [PubMed] [Google Scholar]
- 64.Wylie A.D., Zandberg W.F. Quantitation of sialic acids in infant formulas by liquid chromatography-mass spectrometry: an assessment of different protein sources and discovery of new analogues. J. Agric. Food Chem. 2018;66:8114–8123. doi: 10.1021/acs.jafc.8b01042. [DOI] [PubMed] [Google Scholar]
- 65.Liu L. Zenodo; Athens, GA: 2017. Enthalpyliu/carbohydrate-microarray-processing: Carbohydrate Microarray Processing v1.0 Ed. [Google Scholar]
- 66.Gregg K.J., Zandberg W.F., Hehemann J.H., Whitworth G.E., Deng L., Vocadlo D.J., et al. Analysis of a new family of widely distributed metal-independent alpha-mannosidases provides unique insight into the processing of n-linked glycans. J. Biol. Chem. 2011;286:15586–15596. doi: 10.1074/jbc.M111.223172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kinoshita M., Nakatani Y., Yamada K., Yamamoto S., Suzuki S. A rapid and facile preparation of APTS-labeled N-glycans by combination of ion pair-assisted extraction and HILIC-SPE for routine glycan analysis. J. Pharm. Biomed. Anal. 2021;195 doi: 10.1016/j.jpba.2020.113875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession codes for the crystal structures are available on the PDB as follows: NanHGH33 (apo: 8UB5; Neu5Ac complex: 8UL7; Neu59Ac complex: 8ULE; SAc3LacNAc complex: 8UM0), NanJGH33 (apo: 8URL; Neu5Ac complex: 8UVV; Neu5,9Ac complex: 9C20) and NanIGH33 (Neu5,9Ac: 8U2A; Neu5Gc: 8U5O).