Abstract
Cancer cells undergo metabolic reprogramming that is intricately linked to malignancy. Protein acylations are especially responsive to metabolic changes, influencing signal transduction pathways and fostering cell proliferation. However, as a novel type of acylations, the involvement of malonylation in cancer remains poorly understood. In this study, we observed a significant reduction in malonyl-CoA levels in hepatocellular carcinoma (HCC), which correlated with a global decrease in malonylation. Subsequent nuclear malonylome analysis unveiled nucleolin (NCL) malonylation, which was notably enhanced in HCC biopsies. we demonstrated that NCL undergoes malonylation at lysine residues 124 and 398. This modification triggers the translocation of NCL from the nucleolus to nucleoplasm and cytoplasm, binding to AKT mRNA, and promoting AKT translation in HCC. Silencing AKT expression markedly attenuated HCC cell proliferation driven by NCL malonylation. These findings collectively highlight nuclear signaling in modulating AKT expression, suggesting NCL malonylation as a novel mechanism through which cancer cells drive cell proliferation.
Keywords: malonylation(Kmal), malonyl-CoA, nucleolin, hepatocellular carcinoma, cell proliferation, Akt
Protein post-translational modification (PTM) is one of the core mechanisms for organisms to diversify their functional proteome and fine-tune the dynamic signal networks in response to intracellular or environmental cues. As a group of newly defined PTMs, protein acylations compete with metabolic flux for CoA species as the substrate of modification. These processes enable cells to sense metabolic changes, via the alterations of protein conformation, localization, degradation, and overall functions, finally achieving metabolic homeostasis and reprogramming cell fate (1, 2). Hitherto, with acetylation (Kace) being comprehensively characterized, functions of most non-acetyl acylations remain to be uncovered.
Lysine malonylation (Kmal) is an evolutionarily conserved modification, in which malonyl-CoA provides malonyl moiety to the lysine residue of targeted proteins and changes the charge on lysine from +1 to −1 (2). This change could disrupt preexisting interactions between an unmodified lysine and a negative charge-carrying molecule or protein, thus impacting cellular interactome and rewiring cell fate. Malonyl-CoA is the metabolic switch of fatty acid metabolism. On one hand, it is a potent endogenous inhibitor of CPT1α, the critical molecule responsible for mitochondrial entrance and oxidation of long-chain fatty acids. On the other hand, malonyl-CoA could be converted from acetyl-CoA which is catalyzed by acetyl-CoA carboxylase 1 (ACC) as the first rate-limiting step of de novo lipogenesis. Like acetylation and other short-chain acylations (3), protein malonylation can be initiated through both enzymatic and non-enzymatic mechanisms (4). This feature suggests that the regional concentration of malonyl-CoA may serve as a trigger to alter the function of modified proteins and subsequently control cell fate. Therefore, the precise characterization of malonyl-CoA level is important for predicting acylation tendency under physiological or pathological conditions.
Since 2011, more than 3000 proteins have been identified as Kmal targets in E. coli, mice, and humans, exhibiting diverse subcellular distributions in mitochondria, cytosol, or the nucleus (5). While a large number of proteins undergo malonylation, the molecular function of Kmal in cell-fate decisions remains poorly understood. Sirt5, an NAD+ dependent lysine deacylase, is responsible for the removal of malonyl-, glutaryl-, and succinyl-group from lysine (6, 7). Proteomic analyses using Sirt5 knockout mice have revealed that increased Kmal is associated with impaired mitochondrial function, fatty acid oxidation and glycolysis by targeting multiple enzymes in the corresponding metabolic pathways (8, 9). Recent studies focusing on specific genes, such as mTOR (10), GAPDH (11), mDia2 (12), and TKT1 (13), have suggested that Kmal may have impacts on angiogenesis, inflammation, cytoskeleton organization and DNA damage responses. Despite being a widespread PTM, the connection between Kmal and cancer development, especially in cancer types tightly linked to lipid metabolism deregulation, such as hepatocellular carcinoma (HCC) (14), has yet to be established.
Liver is a metabolic active organ, which is extremely efficient in fatty acid oxidation upon glucose shortage. However, in the presence of excessive nutrient supply, liver becomes a major site for de novo lipogenesis. As fatty liver is one of the leading causes of HCC worldwide (15), we therefore asked whether HCC-associated lipidomic reprogramming contributes to cancer progression through malonylation. In this study, we quantified lipid components and acyl-CoA profiles in HCC tissues. We surprisingly identified malonyl-CoA as one of the most different CoA species between tumor and normal tissues. Further nuclear malonylome analysis identified HCC-associated malonylation of NCL, which facilitates NCL binding to AKT mRNA and promoting AKT translation, thus driving cell proliferation and tumor growth in HCC. Our findings highlight a great potential of malonylation as an oncogenic signal by reinforcing NCL-AKT axis in HCC.
Results
Profiles of lipid components and acyl-CoA species of human HCC tissues
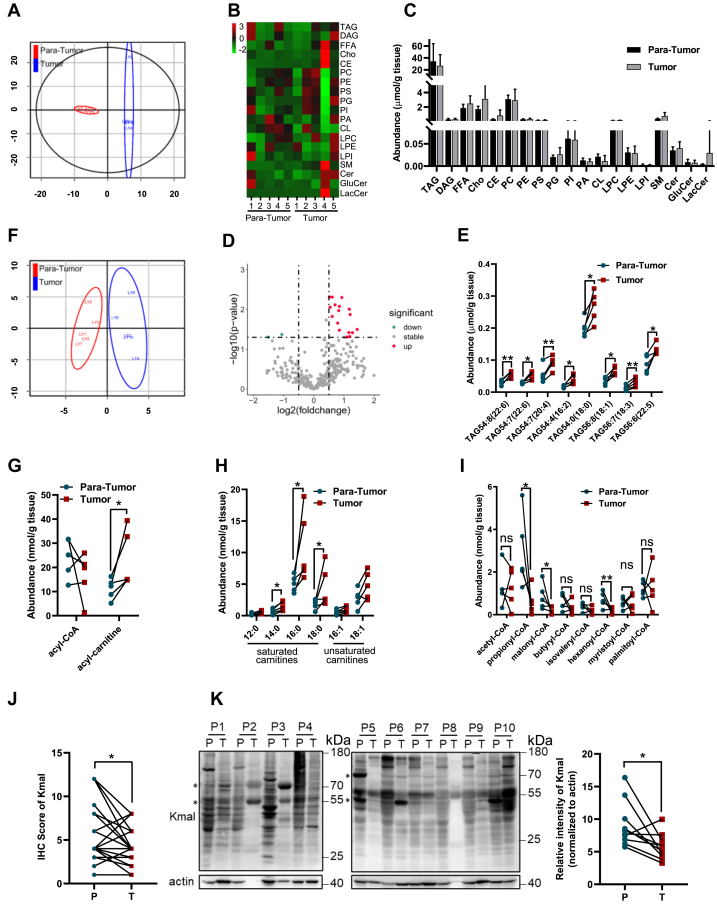
To investigate changes in lipid metabolism in HCC, we collected tumor biopsies and adjacent normal liver tissues (para-HCC) from five patients and conducted a lipidomic analysis. Systematic changes in lipidomic profiles across different tissues were evaluated by the partial least squares discriminant analysis (PLS-DA). In total, we identified 317 lipids, including 165 phospholipids and 135 neutral lipids (Fig. S1A). Orthogonal partial least square discriminant analysis (OPLS-DA) revealed significant differences in lipid profiles between HCC and para-HCC tissues (Fig. 1A), although the total levels of individual lipid components did not exhibit significant changes, likely due to the comparatively smaller sample size (Fig. 1, B and C). Among the lipids with notable differences, phosphatidylcholine (36:5) and diacylglycerol-36:4 (18:2/18:2) were slightly reduced in HCC, while some sphingomyelin and triacylglycerols (TAGs) such as TAG-54:8 (22:6), TAG-54:7 (22:6), TAG-54:7 (20:4), TAG-54:4 (16:2), TAG-54:0 (18:0), TAG-56:8 (18:1), TAG-56:7 (18:3) and TAG-56:6 (22:5), were increased in HCC compared to para-HCC tissue (Fig. 1D and Table S1). This suggests an augmented potential for lipid storage in HCC cells (Fig. 1E).
Figure 1.
HCC tissues revealed lower malonyl-CoA level and reduced global malonylation.A, orthogonal projections to latent structures-discrimination analysis (OPLS-DA) score plots of total lipids derived from the ultra-performance liquid chromatography mass spectrometry (LC/MS) spectra of tumor and para-cancer samples from patients diagnosed with HCC. OPLS-DA was performed in R 4.0.5 using ropls package. B, systematic lipidomic changes occurring in different tissues. Result shown as heatmap. Pseudocolors indicated positive (red) or negative (green) correlation values. The scale represents the correlation values from 3 to −2. C, abundances of the total level of each lipid species in HCC and para-HCC tissues. D, volcano plot displaying fold change of lipids between HCC and para-HCC tissues. (p < 0.05, absolute log2 fold change > 0.5 or < −0.5, unpaired t test). Red dots: metabolites increased in tumor tissues; green dots: metabolites increased in normal liver tissues; gray dots: metabolites with no significant difference between the two groups. E, abundances of TAGs showing significant difference between HCC and para-HCC tissues. F, OPLS-DA score plots of acyl-CoAs and acyl-carnitines derived from the LC/MS spectra of HCC and para-HCC tissues. OPLS-DA was performed in R 4.0.5 using ropls package. G, abundances of total acyl-CoA and acyl-carnitine between HCC and para-HCC tissues. H, intensity of acyl-carnitines in HCC and para-HCC tissues. I, abundances of major acyl-CoA species showing significant difference between HCC and para-HCC tissues. J, statistical analysis of IHC scores. IHC analysis was carried out with pan-Kmal antibody on a tissue microarray containing 42 HCC samples as well as the corresponding para-HCC samples. ∗p < 0.05, n = 42. K, immunoblotting showing global Kmal pattern in HCC and para-HCC tissues. Actin was used as a loading control. Asterisks indicate those bands with increased signal intensity in HCC samples. Controls are para-cancer tissue samples from patients. Right panel, statistical analysis of immunoblotting result, in which the intensity of Kmal-recognized signals were normalized by actin expression. ∗p < 0.05, n = 10.
Acyl-CoAs are essential intermediate metabolites for fatty acid oxidation. Long-chain fatty acids need to transfer the acyl group from acyl-CoA to carnitine through CPT1α for the entrance into mitochondria. Using an integrated analytical platform covering short-, medium-, and long-chain fatty acyl-CoAs and their corresponding acylcarnitines (16), we identified 31 acyl-CoAs and 6 acyl-carnitines (Fig. S1B). While the OPLS-DA results revealed global changes in the sum of CoAs and carnitines between HCC and para-HCC tissues (Fig. 1F), only total acyl-carnitines were significantly increased in HCC tissues (Fig. 1G). Of note, increased carnitine species were primarily derived from saturated long-chain fatty acids (14:0, 16:0, 18:0), while unsaturated fatty acyl-carnitines remained unchanged (Fig. 1H), indicating a great potential of HCC cells to predominantly break down saturated fatty acids. Additionally, short-chain fatty acyl-CoAs gradually reduced along with the increase of carbons in aliphatic tails (2–6 carbons), and medium-chain fatty acyl-CoAs (7–12 carbons) remained unchanged but at very low levels, while long-chain fatty acyl-CoAs displayed an increasing pattern with the extension of aliphatic tails from 13 to 18 carbons, regardless of HCC or para-HCC tissues (Fig. S2). Among different acyl-CoAs, short-chain acyl-CoAs in particular propionyl-CoA (C3:0), malonyl-CoA and hexanoyl-CoA (C6:0), were significantly reduced in HCC tissues compared to para-HCC tissues (Fig. 1I). Instead, acetyl-CoA, myristoyl-CoA and palmitoyl-CoA, which had been well characterized in protein acylations, remained unchanged.
Malonyl-CoA is the precursor of Kmal. In line with previous reports (9), protein malonylation is highly abundant in the liver (Fig. S3, A and B). We next explored whether the reduced malonyl-CoA affected Kmal levels in HCC. Initially, we examined Kmal levels using a pan anti-malonyllysine antibody through immunohistochemistry staining on a tissue chip comprising samples from 42 HCC patients. The results indicated that Kmal levels in para-HCC tissues were significantly higher than those in HCC tissues (Figs. S3C and 1J). Subsequently, tumor tissues and para-HCC tissues from ten patients with primary HCC were collected for global Kmal level analysis via immunoblotting. Consistently, the overall Kmal levels were decreased in HCC samples, although some specific bands showed increased intensity in tumor samples (Fig. 1K). Collectively, our results suggest that both malonyl-CoA and global Kmal levels are significantly reduced in HCC. Given that malonyl-CoA can transfer the malonyl group to lysine through a non-enzymatic mechanism (4), it is reasonable that the reduced hepatic Kmal accompanies the decreases in malonyl-CoA. Intriguingly, those HCC-specific increased Kmal targets may be more indicative and indicate certain signals that are selectively enhanced by cancer cells.
Nucleolin is malonylated at K124 and K398 in hepatocarcinoma cells
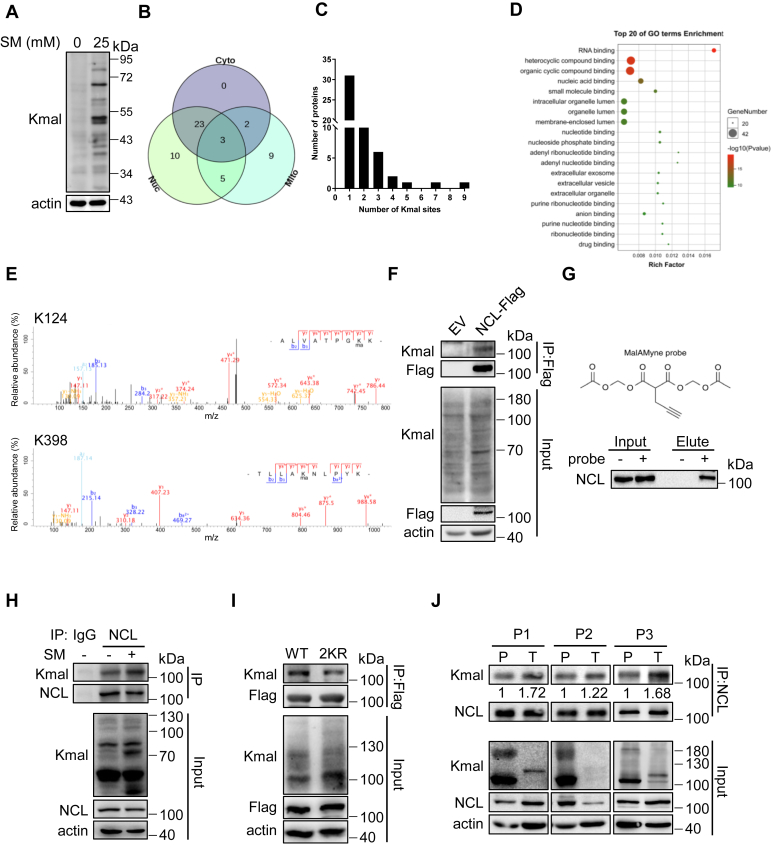
As nucleus is the major cellular compartment of genetic information and the core hub to regulate cell activities, it is important to identify HCC-associated nuclear Kmal targets. HepG2 cells were pre-treated with sodium malonate (SM) to maximize the presentation of the Kmal spectrum (Fig. 2A). Then, Kmal-modified peptides from the nuclear fraction were enriched with an anti-Kmal pan-antibody, and followed by further analysis using the liquid chromatography-tandem mass spectrometry (LC-MS/MS). Through this approach, we identified 99 Kmal sites on 52 proteins, of which 41 were known to have nuclear localization (Fig. 2B). Compared with the other two analyses performed with human-derived samples (10, 11), about 50% (20 out of 41, Table S2) of the nuclear targets from our analysis were unique while the others overlapped with previous studies. Among the identified proteins, 31 (59.6%) had a single Kmal site, and 21 (40.4%) had two or more Kmal sites (Fig. 2C). Gene Ontology (GO) analysis revealed an enrichment of nucleotide-binding proteins in Kmal-modified proteins, with RNA-binding proteins being prominently featured (Fig. 2D).
Figure 2.
Nucleolin is malonylated in HCC cells at lysine 124 and 398.A, immunoblotting of Kmal from HepG2 cells treated with 25 mM SM for 72 h. B, Venn diagram displaying the number of Kmal proteins and their subcellular distributions as determined by the COMPARTMENTS database. Cyto indicates cytoplasmic proteins; Mito indicates mitochondrial proteins; Nuc indicates nuclear proteins. C, histogram of the number of Kmal sites per protein. D, results from DAVID gene ontology (GO) analysis showing malonylated proteins categorized by molecular function for visualization. Top 20 pathways were shown. E, the lysine-124 and -398 residues of NCL were identified to be Kmal targeting sites by mass spectrometry. F, immunoprecipitation of Flag from HepG2 cells with stable expression of empty vector (EV) or NCL-Flag. G, HepG2 cells were incubated with MalAM-yne (200 μM) or vehicle control for 2 h, followed by click chemistry to label the probe with biotin. Then, immunoprecipitation was performed using streptavidin beads. Samples were blotted for NCL via immunoblotting. H, immunoprecipitation showing Kmal of endogenous NCL in HepG2 cells with or without sodium malonate treatment (25 mM, 72 h). I, immunoprecipitation showing Kmal of ectopic NCL in HepG2 cells with stable expression of NCL-Flag (WT) or NCL2KR-Flag (2 KR). J, immunoprecipitation showing Kmal of endogenous NCL in HCC and para-HCC biopsy of volunteered patients. Kmal of NCL in each tumor tissue was normalized by the modification level in corresponding paracancerous tissue.
Nucleolin (NCL) is one of the most abundant non-ribosomal proteins of the nucleolus and plays important roles in regulating cell proliferation through its involvement in ribosome biogenesis, mRNA translation, and chromatin remodeling (17). Nuclear malonylome analysis identified two malonylated lysine residues in NCL (K124 and K398), which are located within the N-terminal acidic domain and the central RNA-binding domain (RBD), respectively (Fig. 2E). Immunoprecipitation of ectopically expressed NCL-Flag confirmed the malonylation of overexpressed NCL (Fig. 2F). We next employed the malonylation chemical probe (MalAM-yne) to specifically label malonylated lysines (Bao et al., 2013). Consistently, endogenous NCL was successfully labeled and enriched using the biotin-streptavidin system (Fig. 2G). Furthermore, immunoprecipitation of endogenous NCL from HepG2 cells demonstrated that NCL could indeed be malonylated, and malonate treatment enhanced NCL malonylation (Fig. 2H). Additionally, site-directed mutagenesis of K124 and K398 to arginines remarkably reduced the Kmal level of NCL (Fig. 2I), but not levels of acetylation (Kace) and succinylation (Ksucc) (Fig. S4, A–D), further validating the MASS data that K124 and K398 sites are primarily subject to malonylation modifications in HCC cells. To investigate whether Kmal of NCL is related to HCC in vivo, we collected biopsies for tissue immunoprecipitation. Interestingly, we found that HCC tissues displayed an increased NCL Kmal level compared to para-HCC tissues in all three tested patients (Fig. 2J). Collectively, our findings demonstrate that NCL is malonylated at lysine 124 and 398, and NCL Kmal is significantly increased in HCC.
NCL malonylation promotes cell proliferation in HCC cells
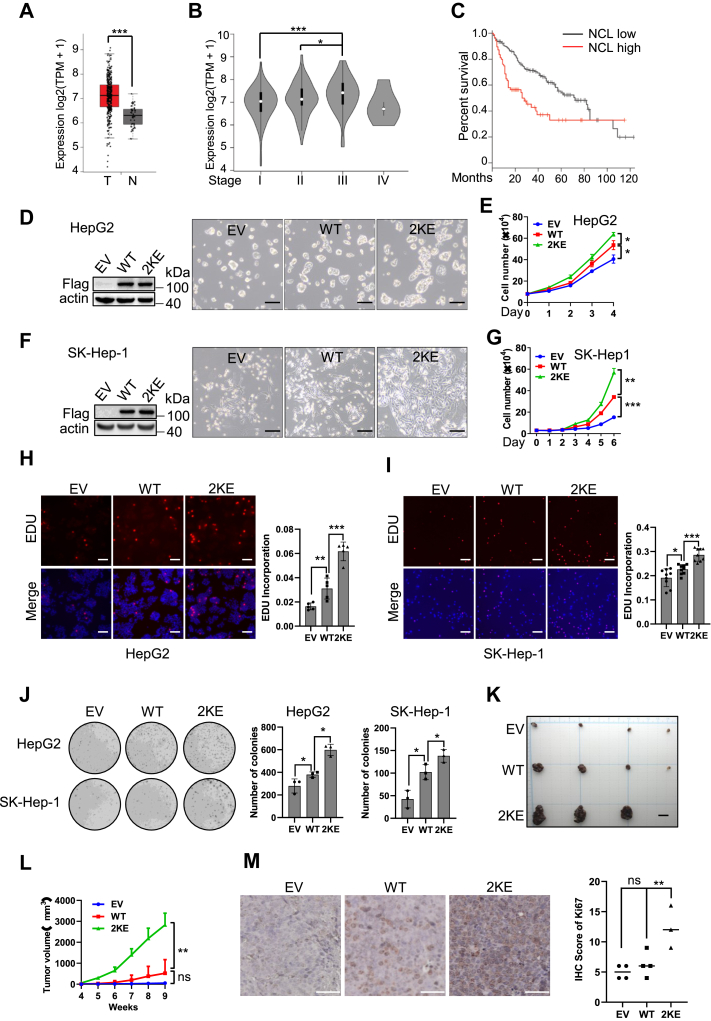
High levels of NCL expression have been related to poor prognosis in many cancer types, including breast cancer, gastric cancer, leukemia and glioblastoma (Fig. S5) (18). In patients with HCC [TCGA database, analysed by GEPIA (19)], the hepatic expression of NCL mRNA is significantly increased compared with normal tissue, and is positively correlated with tumor progression stage (Fig. 3, A and B). Consistently, the higher level of NCL indicates poorer overall survival (OS) (Fig. 3C), suggesting an oncogenic function of NCL in HCC. Mutation of the lysine to a glutamic acid (E) can replace the positive charge with negatively charged carboxylic acid, and has been commonly deployed to mimic lysine malonylation (9, 10). To explore the function of NCL malonylation, we generated stable HCC cells in which both K124 and K398 of NCL were mutated to glutamic acid (NCL2KE). Compared with empty vector, ectopic expression of wild-type NCL (NCLWT) significantly promoted HCC cell proliferation, which was further enhanced by NCL2KE mutant in HepG2 cells (Fig. 3, D and E) and SK-Hep1 cells (Fig. 4, F and G). Moreover, EdU incorporation (Fig. 3, H and I) and colony formation assays (Fig. 3J) further supported that NCL2KE promoted HCC cell proliferation in vitro. We next investigated the importance of NCL malonylation for tumor growth using a xenograft model. HepG2 cells with stable expression of Mock, NCLWT and NCL2KE were subcutaneously injected into nude mice. We found that NCL2KE expression robustly accelerated tumor growth compared to the mock and NCLWT group (Fig. 3, K and L). Histological analysis showed that NCL2KE-associated tumor growth was accompanied by enhanced Ki-67 expression (Fig. 3, M and N), further indicating that the NCL malonylation is a potential driver of HCC cell proliferation.
Figure 3.
NCL malonylation promotes HCC cell proliferation in vitro and in vivo.A, comparative analysis of NCL mRNA expression between liver tumor specimens and the normal tissues was performed. The mRNA data was derived from the cancer genome atlas (TCGA) database and analyzed by GEPIA2. n(T) = 369, n(N) = 160. B, comparative analysis of NCL expression between different stages of liver cancer. Stage I, n = 168; stage II, n = 84; stage III, n = 82; stage IV, n = 6. C, Kaplan-Meier survival curve of patients with low (n = 91) or high (n = 91) level expression of NCL. (p = 0.00046, Log-rank test). The quartile cutpoint is used to separate the low (below the 25th percentile) and high (above the 75th percentile) groups. D–I, HepG2 cells and SK-Hep1 cells were infected with lentivirus containing EV, WT NCL (NCLWT), or malonylation mimicking mutant (NCL2KE), followed by selection with puromycin. Polyclonal stable cells were used for cell proliferation assay. Representative pictures showing the confluence (24 h after seeding) of HepG2 (D) and SK-Hep1 cells (F); scale bar indicating 200 μm. E, cell growth curve for HepG2 cells within 96 h. G, cell growth curve for SK-Hep1 cells within 144 h. (H) HepG2 and (I) SK-Hep-1 stable cells were stained with EDU (red) and Hoechst (blue). Representative pictures were shown on the left panel. Scale bar indicates 100 μm. Right panel showing the bar graph in which EDU signal was quantified and normalized by Hoechst signal intensity. J, colony formation ability of HepG2 and SK-Hep-1 cells with stable expression of the indicated ectopic protein. Left panel: representative pictures; right panel showing the quantification of colonies. K–M, HepG2 cells with stable expression of the indicated ectopic protein were subcutaneously injected into nude mice to establish HCC xenograft model. Nine weeks later, mice were sacrificed. K, image of xenograft tumors. Scale bar indicates 10 mm. L, tumor volumes were measured and recorded every week. M, Ki-67 staining in xenograft tumors. Left panel: representative images of Ki-67 staining, scale bar indicating 50 μm; right panel: quantification of Ki-67 staining. ∗p < 0.05,∗∗∗p < 0.001,∗∗∗∗p < 0.0001, n = 3. Error bars represent ± SD.
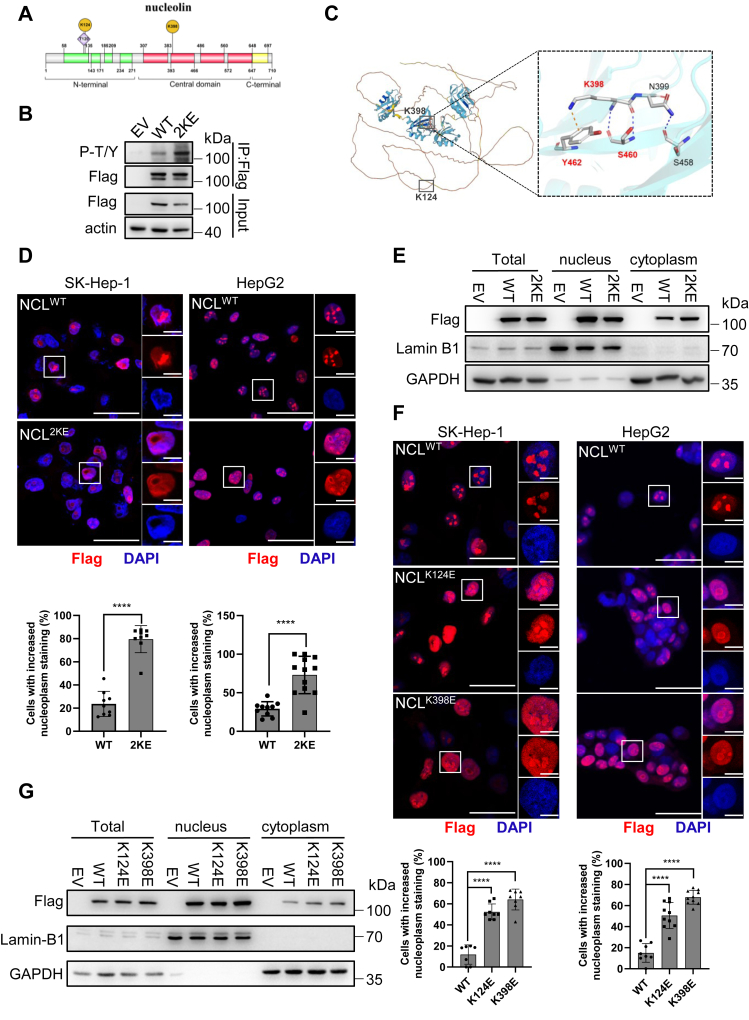
Figure 4.
Lysine malonylation fine-tunes NCL phosphorylation and promotes NCL release from nucleolus.A, schematic representation of functional domains of human nucleolin protein. B, immunoprecipitation analysis of NCL phosphorylation in HepG2 cells with the pan-phospho antibody recognizing p-tyrosine and p-threonine (p-T/Y). C, AlphaFold structure predicted with the full-length sequence of human NCL. Color represents model confidence, with yellow indicating very low confidence and blue indicating very high confidence. The location of K124 and K398 were indicated with a black box, and the Zoom-in box showed the predicted binding pocket of K398 based on molecular docking simulation. D, the subcellular distribution of NCLWT and NCL2KE in SK-Hep1 and HepG2 cells. Flag-tagged protein was labeled with Alexa594 (red). DAPI was included to visualize nuclei. Upper panel: representative pictures of the indicated cells with zoom-in nuclei; lower panel: the proportion of cells with nucleoplasmic localization of NCLWT and NCL2KE cells. Over 200 cells were counted in for each cell type. Scale bar indicates 50 μm (full size) and 10 μm (insets). E, subcellular fractionation showing the distribution of NCLWT and NCL2KE in HepG2 cells. Lamin B1 was included as a marker of nuclear fraction, and GAPDH was included as a marker of cytosolic fraction. F, the subcellular distribution of NCLK124E and NCLK398E in SK-Hep1 and HepG2 cells. Scale bar indicates 50 μm (full size) and 10 μm (insets). G, subcellular fractionation showing the distribution of NCLK124E and NCLK398E in HepG2 cells. Error bars represent ± SD. ∗p < 0.05, ∗∗p < 0.01.
Lysine malonylation fine-tunes NCL phosphorylation and subcellular localization
As a multifunctional protein, NCL is ubiquitously distributed in nucleoli, nucleoplasm, cytoplasm and membrane. PTMs in particular phosphorylation play an important role for NCL to redistribute in different cellular compartments and achieve multiple functions (17). Lysine 124 is located in the N-terminal domain which contains multiple TPXK motifs and is heavily phosphorylated, while lysine 398 resides in the third RNA binding domain of the central region (Fig. 4A). This prompted us to test whether the two lysine malonylation affect NCL phosphorylation and subcellular distributions. Interestingly, the pan-phospho-threonine/tyrosine (p-T/Y) antibody specifically recognized a band at NCL molecular weight, and the signal was markedly increased in cells with NC2KE mutant (Fig. 4B), indicating that NCL malonylation increased its phosphorylation either at threonine or tyrosine residues. We next used alphafold to predict the structure of NCL. While lysine 124 falls into an unpredicted region with very low confident, lysine 398 displayed great potential to react with tyrosine 462 and serine 460 (Fig. 4C), two possible phosphorylation sites. We then performed immunofluorescent staining to determine the subcellular localization of malonylated NCL. As shown in Figure 4D, NCLWT predominantly localized in the nucleolus in both HepG2 and SK-Hep1 cells, while the NC2KE mutant exhibited considerable nucleoplasmic localization. Furthermore, subcellular fractionation showed increased NC2KE accumulated in cytoplasm fraction compared to NCLWT (Fig. 4E), suggesting NCL Kmal promotes its translocation from nucleolus into nucleoplasm and cytoplasm. This observation was further evidenced by immunofluorescence and subcellular fractionation of each Kmal-mimicking mutant, K124 E and K398 E, which revealed increased expression in both nucleoplasm and cytoplasm (Fig. 4, F and G).
Lysine malonylation promotes NCL binding to AKT mRNA and facilitates AKT translation, thus enhancing cell proliferation
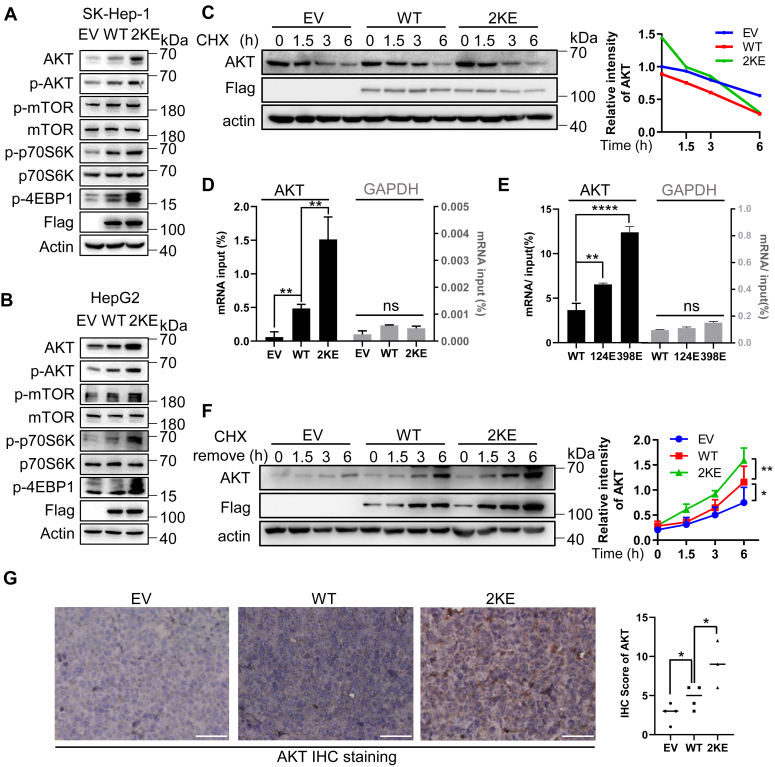
Upon phosphorylation, nucleolus NCL translocates to cytoplasm where it binds to target mRNAs, including those of AKT, p53 and Bcl-2, regulating their translation process (18, 20). As AKT is a master regulator of cell proliferation, we first examined AKT expression. Overexpression of NCL increased AKT expression, with the NCL2KE mutant further enhancing this effect. Importantly, elevated AKT expression promoted the proliferative AKT-mTOR cascade, indicated by enhanced phosphorylation of AKT, mTOR, p70S6K and 4EBP1 in NCL2KE HCC cells (Fig. 5, A and B). Malonyl-CoA and Orlistat, both inducers of global malonylation, increased AKT expression only in the presence of NCL in HepG2 cells (Fig. S6A), further substantiating that NCL malonylation facilitates AKT expression. To elucidate the mechanism by which NCL2KE regulates AKT protein expression, we analyzed AKT transcription, degradation, and translation. Neither NCLWT or NCL2KE affected AKT mRNA levels (Fig. S6B). We then used cycloheximide (CHX) to block the cellular translation machinery and tested the stability of AKT protein. The results showed that the half-life of AKT in SK-Hep1 cells with stable expression of NCLWT and NCL2KE were comparable (Fig. 5C). Next, we performed RNA-immunoprecipitation (RNA-IP) to assess whether the Kmal of NCL influences its binding affinity to AKT mRNA. Compared with NCLWT, an equal amount of NCL2KE bound to significantly more AKT mRNA (Fig. 5D). Consistent findings were observed in the NCLK124 E and NCLK398 E (Fig. 5E). GAPDH mRNA served as a negative control to verify RNA-IP specificity. In line with the increased NCL-AKT mRNA interaction, the CHX-removal assay confirmed that NCL malonylation enhanced de novo synthesis of AKT protein (Fig. 5F). Consistently, histological analysis of xenografts showed significantly increased AKT expression in NCL2KE tumors compared to empty vector control and NCLWT (Fig. 5G).
Figure 5.
Malonylated NCL binds to AKT mRNA and facilitates AKT translation.A–B, protein expression of the PI3K/AKT/mTOR pathway in SK-Hep1(A) and HepG2(B) stable cells. Total protein of each cell line was processed by immunoblotting using antibodies against AKT, p-AKT, mTOR, p-mTOR, P70S6K1, p-P70S6K1, p-4E-BP1. Protein loading was normalized with actin. C, half-life of AKT protein in EV, NCLWT, and NCL2KE stable SK-Hep1 cells. The indicated SK-Hep1 cells were treated with 100 μg/ml cycloheximide (CHX) for the time-course as indicated. Total cell lysates were collected for immunoblotting of AKT, Flag, and actin. D–E, RNA-IP showing the binding affinity of indicated ectopic NCL to AKT mRNA. Cell lysates from EV, NCLWT, and NCL2KE (D) or NCLWT, NCLK124E, and NCLK398E (E) stable SK-Hep-1 cells were immunoprecipitated with anti-Flag antibody. The co-precipitated AKT or GAPDH (included as a negative control) mRNA was quantified using RT-qPCR. Results (the mean ± SD, n = 3) were presented as percentages of IP signal/input signal (% input). F, EV, NCLWT, and NCL2KE stable SK-Hep-1 cells were pretreated with 100 μg/ml cycloheximide for 12 h. Cycloheximide was then washed out, and cells were incubated for the time course as indicated. AKT synthesis levels were detected by immunoblotting. Right panel: AKT expression was quantified by ImageJ, with each point representing the mean ± SD. G, histological analysis of AKT expression in HCC xenografts. HepG2 xenograft mice with indicated expression of NCLWT and NCL2KE were established as described above. Left panel: representative images of AKT IHC staining; right penal: AKT staining was quantified and the scores were presented in the bar graph. Scale bar indicates 50 μm. Each point represents the mean ± SD, ns: not significant, ∗p < 0.01, ∗∗p < 0.01, ∗∗∗p < 0.001.
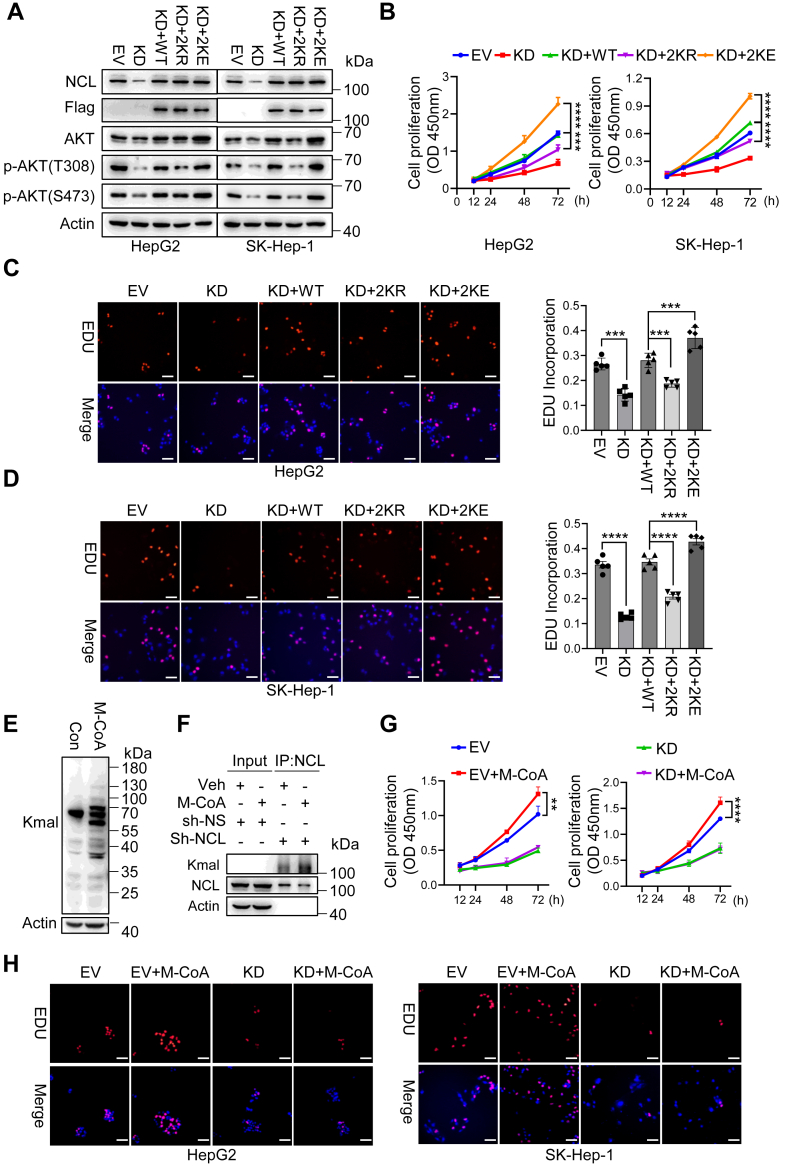
To determine whether enhanced AKT expression driven by NCL malonylation is associated to HCC cell proliferation, we constructed stable NCL knockdown cells and assessed AKT activity, and cell proliferative potential. Silencing NCL remarkably reduced AKT expression, which was restored by re-introducing NCLWT, and further increased by NCL2KE. More importantly, NCL2kE cells exhibited the highest levels of phosphorylated AKT at both threonine 308 and serine 473, indicating superior activation compared to other cells. In contrast, overexpression of malonylation-deficient NCL2KR in NCL knockdown cells failed to restore AKT activity (Fig. 6A). Consistent with these findings, while NCL knockdown substantially restricted cell proliferation, NCLWT cells restored cell proliferative capacity, NCL2KE significantly enhanced cell proliferation beyond that observed in NCLWT and NCL2KR (Fig. 6B). This observation was further validated through EDU incorporation assay (Fig. 6, C and D). To further explore the biological relevance of NCL malonylation in HCC proliferation, we treated HepG2 cells with malonyl-CoA, which induced both total malonylation and NCL malonylation (Fig. 6, E and F). As malonyl-CoA effectively induced AKT expression and phosphorylation (Fig. S6A), and enhanced cell proliferation, knockdown of NCL expression significantly compromised malonyl-CoA-induced AKT expression and cell proliferation (Figs. 6, G and H, and S7). Collectively, these findings suggest that malonylated NCL has a higher affinity for AKT mRNA, thereby accelerating AKT translation and promoting HCC cell proliferation through enhanced AKT-mTOR signaling.
Figure 6.
NCL malonylation enhances AKT expression and mediates cell proliferation.A–D, HepG2 and SK-Hep-1 cells were infected with lentivirus containing sh-NS (EV), sh-NCL (KD), sh-NCL and NCLWT (KD + WT), sh-NCL and NCL2KR (KD+2 KR), and sh-NCL and NCL2KE (KD+2 KE). Polyclonal stable cells were used for (A) immunoblotting, (B) cell viability assay, and (C–D) EDU incorporation assay. Scale bar indicates 100 μm. E, HepG2 cells were treated with 10 μM Malonyl-CoA for 24 h. Total cell lysates were immunoblotted with pan-Kmal antibody. F, HepG2 stable cells with sh-NS or sh-NCL were treated with 10 μM Malonyl-CoA for 24 h. Total cell lysates were proceeded to immunoprecipitation with anti-NCL antibody, followed by the immunoblotting with pan-Kmal and NCL antibodies. G, cell viability of HepG2 and SK-Hep-1 cells treated with Malonyl-CoA (10 μM) on cell proliferation in the presence and absence of NCL. H, representative images showing the EDU incorporation in HepG2 and SK-Hep-1 cells treated with vehicle or Malonyl-CoA (10 μM) for 72 h, in the presence or absence of sh-NCL. Scale bar indicates 100 μm. Each point represents the mean ± SD, ns indicates not significant, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, n = 6. Error bars represent ± SD.
Silencing AKT expression compromises NCL malonylation-associated HCC cell proliferation
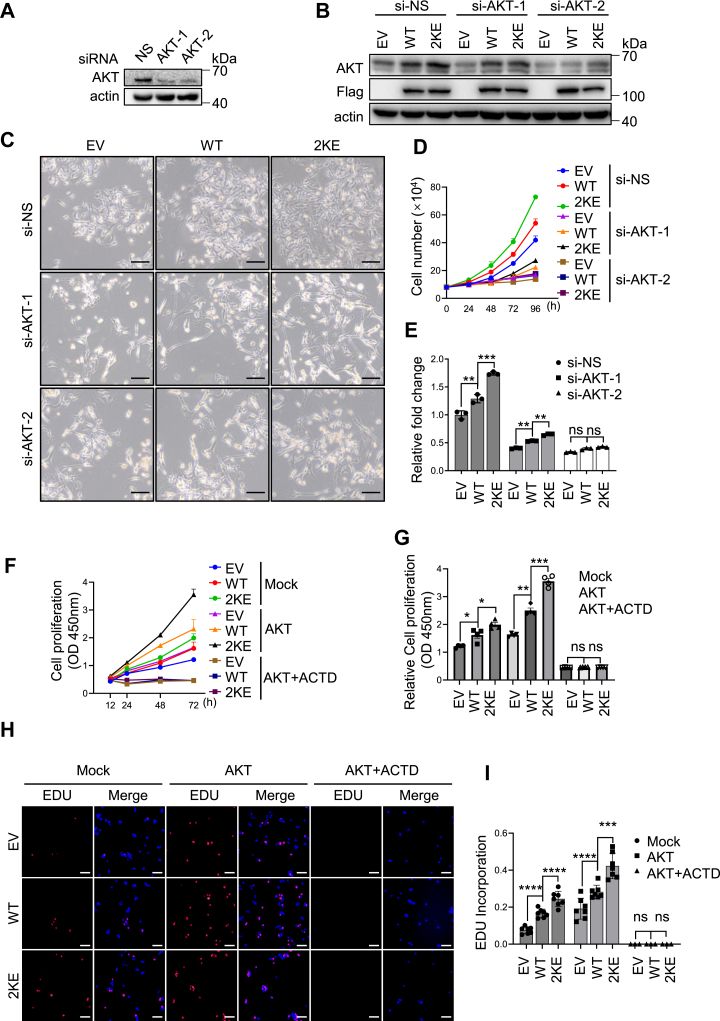
We next investigated whether AKT is essential for the growth promotion observed in HCC cells carrying the NCL Kmal-mimicking mutant. SK-Hep1 cells stably expressing either NCLWT or NCL2KE were transfected with AKT-targeting siRNAs to achieve efficient AKT knockdown (Fig. 7, A and B). With partial silencing of AKT using siAKT-1, the augmentation of cell proliferation by NCL2KE was slightly reduced compared to non-silencing siRNA (si-NS). Notably, the more intensive knockdown of AKT by siAKT-2 treatment almost completely abrogated the growth promotion induced by NCL2KE, indicating that AKT is indispensable for the malonylated NCL to boost cell proliferation (Fig. 7, C–E). Furthermore, we overexpressed AKT in AKT-low HepG2 cells (Fig. S8). AKT overexpression significantly increased cell proliferation in HepG2 cells, particularly in the NCL2kE cell line, compared to NCLWT controls. Notably, blocking mRNA synthesis by actinomycin D eliminated NCL malonylation-associated proliferative tendency even in the presence of ectopic AKT, implying that the effect of NCL2KE on AKT-mediated cell proliferation requires the presence of mRNA (Fig. 7, F and G). This observation was further corroborated by EDU incorporation assay (Fig. 7, H and I). Taken together, our findings suggest that NCL malonylation promotes HCC cell proliferation by facilitating AKT translation.
Figure 7.
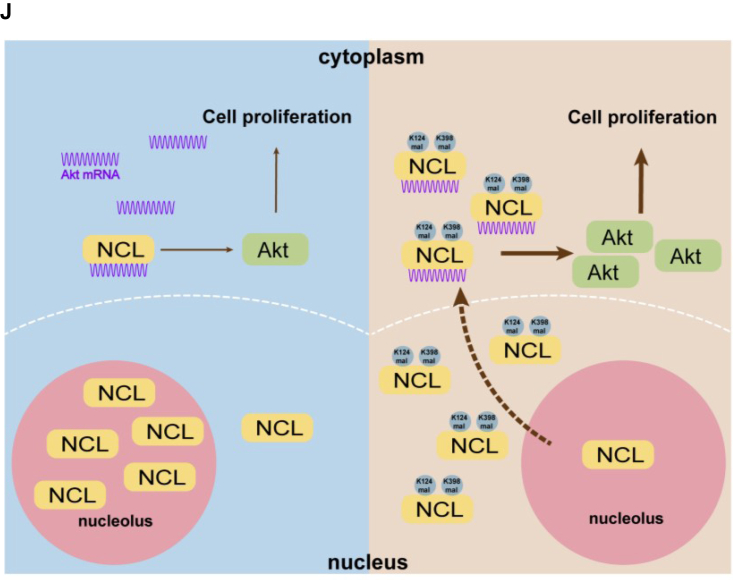
AKT is required for the enhanced cell proliferation driven by NCL malonylation.A, immunoblotting of AKT expression to validate the knockdown efficiency of AKT-specific siRNAs. SK-Hep1 cells were transfected with control siRNA (siNS), siAKT-1, and siAKT-2 for 36 h, and total cell lysates were analyzed by immunoblotting. B–E, EV, NCLWT, and NCL2KE stable SK-Hep1 cells were transfected with control or AKT-specific siRNA for 36 h. Cells were then collected for immunoblotting or subculture for further analysis. B, total cell lysates were analyzed by immunoblotting with AKT, Flag, and actin antibodies. C, representative pictures of EV, NCLWT, and NCL2KE stable SK-Hep1 cells after indicated siRNAs transfection. Scale bar indicates 200 μm. D, cell growth curve of SK-Hep1 cells with indicated treatment. E, statistical analyses of cell numbers 96 h after subculture. Bar graph showed the mean ± SD. ns: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n = 3. F, cell viability of HepG2 cells with indicated treatments. EV, WT, and 2 KE stable HepG2 cells were infected with mock or AKT expressing lentivirus, followed by the treatment of 200 nM actinomycin D (ACTD) or vehicle. CCK-8 assay was performed to evaluate the proliferation. G, statistical analyses of cell viability 72 h after subculture. H, EDU incorporation in HepG2 cells with indicated treatments. EV, WT, and 2 KE stable HepG2 cells were infected with mock or AKT expressing lentivirus, followed by the treatment of 200 nM actinomycin D (ACTD) or vehicle. EDU (red) and Hoechst (blue) was used to stain cells. Representative pictures were shown on the left panel. Scale bar indicates 100 μm. I, statistical analyses of EDU incorporation. EDU signal was quantified and normalized by Hoechst signal intensity. ∗∗∗p < 0.001,∗∗∗∗p < 0.0001, n = 6. Error bars represent ± SD. J, schematic model showing NCL(Kmal)–AKT axis in regulating cell proliferation in HCC.
Discussion
Metabolic reprogramming is one of the emerging hallmarks of cancer development. Cancer cells hijack metabolic networks to support cell proliferation, adapt to metastatic cascade, and escape from immune surveillance (21, 22, 23). Despite extensive research in metabolic control by genomes, the retrograde regulation of signal transduction by metabolites remains unclear. Protein acylation represents a major mechanism for cells to sense metabolic cues and bridge metabolome with functional proteomics (1). Cancer is a metabolic disease, yet the systematic identification and functional characterization of cancer-specific non-acetyl acylations is still a poorly understood area. We here analyzed lipid and CoA profiling in HCC and identified malonyl-CoA as one of the major different acyl-CoAs between tumor and normal liver tissues. Through a further nuclear malonylome analysis, we found that NCL is highly malonylated in HCC. Mechanistically, malonylated nucleolin translocates from nucleolus to nucleoplasm and cytoplasm, binding to AKT mRNA and facilitating AKT translation. Consequently, the NCL(Kmal)-AKT axis promotes cell proliferation in HCC cells. Our findings together suggest malonylation as a novel mechanism by which cancer cells can drive cell proliferation through NCL (Fig. 7J).
In recent years, several new types of short-chain fatty acylations have been identified, including malonylation, propionylation, butyrylation, succinylation, glutarylation, lactylation and some others (7, 24, 25). As proteomic analysis revealed thousands of targeting proteins for each of these acylations, our knowledge to most of them is still limited to the involved gene signatures and enriched cellular events. Therefore, it is important to thoroughly define the influence of novel acylations on specific genes, in particular those critical for physiology or pathology. Kmal, as a potential communication process between lipid metabolism and protein function, only a few targets have thus far been carefully validated and functionally characterized. Sensing the accumulation of malonyl-CoA, Kmal of mTORC1 was reported to inhibit cell proliferation in human endothelial cells and suppress angiogenesis (10). In the immune system, Kmal of GAPDH led to its dissociation from TNF-α mRNA and thus promotes TNF-α translation in macrophages in the presence of lipopolysaccharide (LPS) (11). While Kmal of hepatic acetyl-CoA carboxylase 1(ACC1) was essential to maintain its stability and subsequently promote hepatosteatosis (26), mDia2 malonylation drove nuclear translocation of actin and thus promote HCC metastasis (12). Moreover, TKT1 malonylation resulted in its inactivation and thus compromised pentose phosphate pathway, leading to increased vulnerability of colorectal cancer cells to DNA damage (13).
To elucidate whether Kmal is critically involved in HCC progression, we systematically investigated the lipidome and malonylome of HCC. Probably due to a smaller sample size, our lipidomic analysis revealed comparatively limited information. We found that some sphingomyelin, TAGs (Tabel S1) and saturated long-chain fatty acyl-carnitines are up-regulated in HCC tissue, which is consistent with the previous reports (27, 28). As a major membrane lipid, an increase in sphingomyelin could reduce membrane fluidity and increase its rigidity, potentially resulting in the loss of intercellular communication and contact inhibition, and consequently leading to uncontrolled cell proliferation. On the other hand, cellular TAG levels are controlled by FAO and lipogenesis. In highly proliferative HCC cells, increased de novo lipogenesis provides building blocks for the biosynthesis of DNA and membrane, leading to the accumulation of TAGs. Otherwise, we are the first to compare acyl-CoAs in cancer versus normal tissue. Interestingly, hepatic short-chain acyl-CoA, medium-chain acyl-CoA, and long-chain acyl-CoA exhibited specific but different patterns along with the extension of aliphatic tails, respectively (Fig. S2). More importantly, our results highlighted malonyl-CoA and propionyl-CoA, instead of acetyl-CoA, as the major different hits between HCC and para-HCC tissues, implying that protein Kmal and Kprop (propionylation) could be intensively rewired during HCC development.
Nuclear contains the whole genome and is responsible for ribosome biogenesis, DNA replication and gene expression. Over the past decade, the vast majority of effort has focused on the epigenetic modification of nuclear proteins, including histones, transcription factors and transcriptional cofactors (29, 30, 31). Similar to other lysine-acylations, Kmal is also capable of modifying histone, indicating a possible entry of malonyl-CoA into nucleus (32, 33). Therefore, it is intriguing to further characterize non-histone nuclear targets of Kmal and uncover their molecular functions in tumorigenesis. To avoid the potential submergence of trace nuclear signals in the massive information from whole cell lysate, we directly applied the nuclear fraction of HepG2 cells to mass spectrometry for proteomic analysis. About 50% (Table S2) of the nuclear targets from our analysis are unique while the others have been identified by previous studies (10, 11). Among them, we validated NCL, the most abundant non-ribosomal protein in nucleolus, as one of the HCC-associated substrates of malonylation. Interestingly, Kmal of NCL increased in HCC tissue compared to the corresponding para-HCC tissue, which somehow contradicts the reduced malonyl-CoA (Fig. 1I) and decreased global Kmal level (Fig. 1, J and K) in HCC tissues. Based on the observations from other lysine acylations (3, 4), malonyl-CoA may transfer the malonyl moiety to the target lysine through both enzymatic and non-enzymatic mechanisms. However, the biochemistry of Kmal was way underestimated. Currently, Sirt5 is the only characterized “eraser” of Kmal, but it simultaneously catalyzes the de-acylation of lysine acetylation, succinylation and glutarylation (6, 7). This reality makes it difficult to clean and clearly define the function of Kmal by manipulating Sirt5. On the other hand, the mammalian malonyltransferase as the “writer” of Kmal has not been clearly defined so far, except for one work regarding the involvement of known acetyltransfease in Kmal process (34). In our case, the controversy between the overall reduced Kmal in HCC and the NCL-specific increase of Kmal could be attributed to the crosstalk between NCL and a potential malonyltransferase, which specifically regulate NCL Kmal in HCC. The potential presence of an HCC-associated malonyltransferase could be partially evidenced by the pan-Kmal antibody immunoblot result (Fig. 1K), in which several bands were remarkably increased in HCC tissues regardless of the reduced malonyl-CoA level. Therefore, to further elucidate the function of Kmal in cancer development, future work should be focused on the discovery of mammalian malonyltransferase(s) in the cancer context.
NCL has been found in nucleoli, nucleoplasm, cytoplasm and cell membrane. In cancer cells, nucleoplasmic NCL coordinates with RNA polymerase II (Pol II) to regulate the transcription of several oncogenes, thus preventing cells from apoptosis. Moreover, cytoplasmic NCL has been reported to directly bind to the UTR region of Bcl-2, AKT and p53, and regulate either mRNA stability or the translation capacity (35, 36). In our study, we found that Kmal at lysine-124 and lysine-398 of NCL increased its phosphorylation and caused its translocation from nucleolus to nucleoplasm and cytosol, yet the molecular detail is still elusive. NCL has three conserved functional domains: the N-terminal acidic domain that is important for protein-protein interaction and is highly phosphorylated by Casein Kinase 2 (CK2) and cyclin-dependent kinase 1 (CDK1), the central region containing four RBDs, and the C-terminal domain bearing several methylation sites (Fig. 4A) (37). PTMs play crucial roles in determining the subcellular distribution of NCL (38). CDK1 phosphorylates human NCL on threonine residues within a TPXKK repeat, including threonine 59, 76, 84, 92, 99, 106, 121, 129, and 220 (38). Coincidently, T121 and K124 fall into the same TPXKK motif, indicating a possible allosteric regulation between the two modifications. Using the TG-3 antibody which was specifically immunoreacted with phosphorylated NCL (p-NCL) catalyzed by CDK, Alex et al., found that non-phosphorylated NCL was restricted in nucleolus while phosphorylated NCL diffused in nucleoplasm (39). In the meanwhile, the predicted structure with alphafold strongly suggested that K398 may react with tyrosine-462 and serine-460, and its neighbor residue glutaric acid-399 may react with serine-458, which together indicate some new unknown phosphorylation sites of NCL (Fig. 4B). Therefore, it is plausible that malonylation at K124 and K398 expel NCL from nucleolus by fine-tuning its phosphorylation. This hypothesis needs to be further investigated in the future.
Akt is a key component of a wide range of growth signals and plays a crucial role in regulating cell proliferation, survival, migration and metabolism (40). As one of the leading oncogenes, the regulatory network of Akt, particularly its phosphorylation and downstream targets has been comprehensively investigated. However, the translational control of Akt remains elusive. Recently, CITED2 has been reported to recruit PRMT5 and p300 to nucleolin (NCL) in the nucleoplasm, leading to the methylation and acetylation of NCL. The modified NCL then translocates to cytoplasm where it directly binds to the mRNA of AKT, facilitating AKT translation (20). We here identified malonylation as a novel PTM of NCL, which fine-tunes NCL phosphorylation and subcellular distribution, thereby boosting cell proliferation by driving AKT translation in HCC cells. Although not validated, other proteomic studies have identified acetylation and succinylation at K124, and acetylation and ubiquitination at K398 (41, 42). It remains unclear whether the newly discovered Kmal could compete or coordinate with other PTMs at the same lysine residues, thus regulating cancer progression. Our results demonstrated that the levels of acetylation and succinylation of NCL in NCL2KR mutant showed no significant decreases compared to NCLWT, whereas the malonylation level exhibited a substantial decrease (Fig. 2I). This finding suggests that the K124 and K398 sites are primarily subject to Kmal modifications, but not Kace and Ksucc in HCC cells. Additionally, unlike other PTMs on NCL, malonylation could be responsive to the cellular level of de novo lipogenesis, and thus bridges metabolic status with cell proliferation. Given that the biosynthesis and metabolic flux of malonyl-CoA are highly dynamic in cytoplasm in response to metabolic alterations, our findings highlight lysine malonylation as a novel signal exchanging mechanism between nuclear and cytoplasm, which may be critical for eukaryotic cells to sense metabolic cues and determine to “go” or “stop” in term of cell proliferation.
Experimental procedures
Cell culture
Human liver cancer cell lines (HepG2,SK-HEP-1) and 293T cell line were purchased from the Shanghai Institute of Biological Sciences, Chinese Academy of Sciences. HepG2,SK-HEP-1 and 293T were cultured in Dulbecco’s Modified Eagle Medium(DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, at 37 °C in 5% CO2 incubator.
Clinical samples
Hepatocarcinomas and adjacent normal liver tissues were obtained from the Affiliated Hospital of Xuzhou Medical University. This research was approved by the Ethical Review Committee of this hospital. The study in this work abided by the Declaration of Helsinki principles. The specimens were collected and stored in liquid nitrogen immediately after the surgery. Total protein was extracted and subjected to WB analysis.
Constructs and transfection
Human NCL was cloned into pCDH-CMV-MCS-EF1-Flag and was used as the template for site-directed mutagenesis of lysine 124 and lysine 398 to glutamate. The following primers were used for mutagenesis PCR reactions:
NCL(K124R)F GGTAGCAACTCCTGGTAGGAAGGGTGCTGCCATC; NCL(K124R)R GATGGCAGCACCCTTCCTACCAGGAGTTGCTACC; NCL(K398R)F GAACACTTTTGGCTAGAAATCTCCCTTACAAAG; NCL(K398R)R CTTTGTAAGGGAGATTTCTAGCCAAAAGTGTTC;
NCL(K124E)F GGTAGCAACTCCTGGTGAGAAGGGTGCTGCCATC; NCL(K124E)R GATGGCAGCACCCTTCTCACCAGGAGTTGCTACC; NCL(K398E)F GAACACTTTTGGCTGAAAATCTCCCTTACAAAG; NCL(K398E)R CTTTGTAAGGGAGATTTTCAGCCAAAAGTGTTC.
For the construction of sh-NCL, we targeted 3′UTR region of human NCL at CCTTGGAAATCCGTCTAGTTA, which was cloned into pLKO.1 lentivirus vector.
Plasmids were transfected into 293T cells to package lentivirus. HCC cells were infected with the obtained lentivirus and selected with 2 μg/ml puromycin for polyclonal stable cells. For knocking down of Akt expression, Akt siRNAs were purchased from RIBOPharm Co., Ltd. The following stealth siRNA duplex was used to knockdown human Akt: siAkt-1-sense-CCUCA CAGCCCUGAAGUACUCUUTC; siAkt-1-antisense-GAAAGAGUACUUC AGGGCUGUGAGG; siAkt-2-sense- ACAAAAACGUCUUUCCAUCUG; siAkt-2-antisense-GAUGGAAAGACGUUUUUGUGC.
RNA isolation and qPCR analysis
Total RNA was isolated with Trizol reagent (9109) and served as the template for reverse-transcription to cDNA using a PrimeScript reverse transcription (RT) reagent kit (R223-01) according to the manufacturer’s instructions. qPCR was performed with 2xChamQ SYBR Qpcr Master (Low ROX Premixed) on Applied Biosystems 7500 Real-Time PCR System (Bio-Rad Laboratories). Human gapdh was included as an internal control. The primers for the target genes were as follows: Akt-forward: AGCGACGTGGCTATTGTGAAG, Akt-reverse: GCCATCATTCTTGAGG AGGAAGT; gapdh-forward: CTCCATCCTGGCCTCGCTGT; gapdh-reverse: GCTGTCACCTTCACCGTTCC.
RNA-immunoprecipitation
RNA-IP was performed as previously described (43). In brief, 2 × 107 SK-Hep-1 stable cells were collected, washed with ice-cold PBS twice and lysed in 2 ml NP-40 Buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl,0.5% NP-40,PMSF and RNase inhibitor were add freshly). The cell lysates were precleaned with protein G Sepharose beads and then incubated with the indicated antibodies or IgG control on a rotator at 4 °C overnight. The antibody-RNA complexes were collected. The immunoprecipitated RNA was eluted and extracted for real-time PCR analysis.
MalAMyne labelling and Cu(I)-catalyzed click chemistry
The MalAMyne chemical probe was synthesized as described previously (44). Briefly, cells were incubated with 200 μM MalAMyne or DMSO for 1 h. Total cell lysates were applied to biotin conjugation through a Cu(I)-catalysed click chemistry reaction, followed by streptavidin affinity enrichment. Elutes was analyzed by immunoblotting with the indicated antibody.
Immunoblotting and Co-immunoprecipitation analysis
Immunoblotting was performed as described previously (45). Cells were lysed with NP-40 buffer (50 mM Tris-HCL PH 7.5, 150 mM NaCl, 50 mM NaF, 0.5%NP-40, add PMSF and DTT freshly). Total cell lysates were separated by SDS-PAGE gel and analyzed by immunoblotting. β-actin was used as internal control. Kmal pan-antibody was diluted in 5% bovine serum albumin (BSA) for incubation. Other antibodies were diluted in 5% non-fat milk for further incubation. Membrane was incubated with anti-NCL antibody (Affinity Biosciences: DF12542, Proteintech: 10556-1-AP), anti-Akt antibody (Zen BioScience: 310021, Cell Signaling: 4691S), anti-malonyllysine antibody (PTM-Biolab: PTM-901; Cell Signaling: 14942S), anti-acetyllysine antibody (PTM-Biolab: PTM-102), anti-succinyllysine antibody (PTM-Biolab: PTM-401), anti-Flag antibody (Sigma: F1804), anti-Phospho-Threonine/Tyrosine antibody (Cell Signaling: 9381S), anti-Sirt5 antibody (Proteintech: 15122-1-AP), anti-Sirt5 antibody (Proteintech: 15122-1-AP), anti-mTOR antibody (Cell Signaling: 2972S), anti-P-mTOR antibody (Cell Signaling: 5536S), anti-p70 S6 Kinase antibody (Cell Signaling: 9202S); anti-P-p70 S6 Kinase antibody (Cell Signaling: 9208S), anti-P-4E-BP1 antibody (Cell Signaling: 2855S), anti-P-Akt (S473) antibody (Cell Signaling: 4060S), anti-P-Akt (T308) antibody (Cell Signaling: 13038S), and anti-β-actin antibody (Proteintech: 66009-1-1g).
Co-immunoprecipitation was modified from the protocol described previously (46). HepG2 cells were lysed using 0.1% NP-40 buffer (50 mM Tris-HCL PH 7.5, 150 mM NaCl, 50 mM NaF, 0.1%NP-40, add fresh PMSF when used). Total cell lysates were precleaned with CL-4B to remove non-specific binding. One microgram of indicated antibodies were added to cell lysates containing 1 mg protein, and the mixture was incubated at 4 °C overnight. IgG antibody was purchased from Beyotime (A7016). Protein A/G (B23202, Bimake) magnetic beads were used to precipitate protein complex through incubation at 4 °C for 2 h. Samples were washed three times with 0.1% NP-40 buffer, followed by elution with SDS Loading buffer.
Immunofluorescence
HepG2 and SK-Hep1 cells were fixed in polyformaldehyde at room temperature for 30 min, followed by permeabilization with 0.2% Triton-X100 on ice for 10 min. Cells were then washed and blocked with PBS-BSA buffer, followed by incubation with the anti-Flag antibody (F1804,sigma) at 4 °C overnight. Alexa Fluor 594-conjugated goat anti-human IgG Fc (VA023, VICMED) were used as secondary antibody. DAPI (D9542, Sigma) was used for the staining of nucleus. Fluorescent signals were detected using a Zeiss LSM-880 scanning confocal microscope.
HCC xenografts
Six weeks old female BALB/c nude mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The mice were randomly grouped and subcutaneously injected with the indicated HepG2 cells (1 × 107 cells in 100 μl serum-free medium containing 0.25 v/v Matrigel) in the right flank. The tumor volumes were measured once a week and calculated as length × width2 × 0.5. Nine weeks later, mice were sacrificed and the tumors were carefully removed and imaged. Then the tumors were fixed with 4% paraformaldehyde overnight for further Immunohistochemistry analysis. All of the animal experimental procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals through the Laboratory Animal Ethics Committee of Xuzhou Medical University.
Immunohistochemistry analysis
The liver tissue microarray slide was purchased from rom Shanghai Xinchao Company. Microarray slide or xenograft tumor sections were dewaxed at 65 °C overnight in xylene. Then the hydrated sample was incubated in peroxidase inhibitor for 10 min at room temperature. The sample was next put into boiled CBS and heated for another 15 min. After the sample was cooling to room temperature, 5% bovine serum albumin (BSA) was applied for blocking for an hour, followed by incubation with antibody at 4 °C overnight (K-mal 1:150 dilution,14942S, CST; Ki-67 1:100 dilution, 12202S, CST; AKT 1:100 dilution, 4691S, CST). The sample was then incubated with dedicated enzyme-labeled goat anti-rabbit IgG complex (Rabbit two-step detection kit, PV-9001,ZSGB-BIO) at room temperature for 30 min. Afterwards, the sample was stained with DAB (DAB color development kit, ZLI-9018,ZSGB-BIO) and pictures were taken with a Nikon ECLIPSE microscope.
5-Ethynyl-20-deoxyuridine assay
5000 cells were seeded into 96-well plates, ensuring that the cells remained in the logarithmic growth phase throughout the experiment. Added 100 μl of medium containing 50 μM EDU (C10310, RiboBio) to each cell well. After 2 h of incubation, cells were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.5% TritonX-100 for 20 min. Next, cells were stained with 100 μl Apollo dye solution for 30 min. Finally, cells were stained with Hoechst 33,342 for 30 min.
Subcellular fractionation
5 × 106 cells were collected for separation. One-sixth were reserved as Input, and remaining cells were resuspended in 1 ml of TM2 buffer (10 mM Tris-HCl PH = 7.4, 4 mM MgCl2, PMSF added freshly). Samples were incubated at room temperature for 1 min, and then incubated on ice for 5 min 333 μl of 2 %-TritonX were added in samples for a final concentration of 0.5%. Samples were perforated on ice for 10 min, and then centrifuged at 15,000 rpm at 4 °C for 10 min. The supernatant was aspirated into another 1.5 ml EP tube as the cytoplasmic fraction; the precipitate was the nuclear fraction. The remaining steps were the same as those for immunoblotting.
Metabolomics analysis
Metabolomics analysis was performed at LipidALL Technologies Co. Ltd. For Lipid extraction: Lipid were extracted from tissues using a modified version of the Bligh and Dyer’s method as described previously (47). Briefly, tissues were homogenized in 750 μl of chloroform:methanol 1:2 (v/v) with 10% deionized water on a bead ruptor, following by incubation at 4 °C at 1500 rpm for 30 min. At the end of the incubation, 350 μl of deionized water and 250 μl of chloroform were added. The samples were then centrifuged and the lower organic phase containing lipids was extracted into a clean tube. Lipid extraction was carried out twice and the lipid extracts were pooled into a single tube and dried in the SpeedVac under OH mode. Samples were stored at −80 °C until further analysis.
For lipid analyses by normal phase LC/MS, polar lipids were analyzed using an Exion UPLC system coupled with a triple quadrupole/ion trap mass spectrometer (QTRAP 6500 Plus; SCIEX) as described previously (48). Separation of individual lipid classes of polar lipids by normal phase (NP)-HPLC was carried out using a Phenomenex Luna 3μ-silica column (internal diameter 150 × 2.0 mm) with the following conditions: mobile phase A (chloroform: methanol:ammonium hydroxide, 89.5:10:0.5) and mobile phase B (chloroform:methanol:ammonium hydroxide:water, 55:39:0.5:5.5). MRM transitions were set up for comparative analysis of various polar lipids. Individual lipid species were quantified by referencing to spiked internal standards. PC-14:0/14:0, PE-14:0/14:0, PS34:1-d31, PA-17:0/17:0, PG-14:0/14:0, Cer d18:1/17:0, SM d18:1/12:0, GluCer d18:one-eighth:0, LacCer d18:one-eighth:0, Sph d17:1 were obtained from Avanti Polar Lipids. Dioctanoyl phosphatidylinositol (PI) (16:0-PI) was obtained from Echelon Biosciences, Inc. Gb3-C17:0 was obtained from Matreya LCC and GM3 d18:1/17:0 was synthesized in-house.
For lipid analyses by reverse-phase LC/MS, glycerol lipids (diacylglycerides, DAG; triacylglycerides, TAG) were analyzed using a modified version of reverse phase HPLC/ESI/MS/MS described previously (49). Briefly, separation of lipids aforementioned was carried out on a Phenomenex Kinetex 2.6μm-C18 column (i.d. 4.6 × 100 mm) using an isocratic mobile phase chloroform:methanol:0.1 M ammonium acetate (100:100:4) at a flow rate of 160 μl/min for 22 min. Using neutral loss-based MS/MS techniques, the levels of TAG were calculated as relative contents to the spiked d5-TAG 42:0, d5-TAG 48:0 and d5-TAG 54:0 internal standards (CDN isotopes), while DAG species were quantified using d5-DAG (16:0/16:0) and d5-DAG (18:1/18:1) as internal standards (Avanti Polar Lipids).
Free cholesterols and total cholesteryl esters were analyzed using HPLC/APCI/MS/MS as previously described with corresponding d6-Cho and d6-CE-18:1 (CDN isotopes) as internal standards (50).
Mass spectrometry for proteomic analysis
Mass spectrometry was performed at Jingjie PTM Biolabs Inc. In brief, tissue sample were grinded by liquid nitrogen into cell powder and then transferred to a 5-ml centrifuge tube. After that, four volumes of lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail) was added to the cell powder, followed by sonication three times on ice using a high intensity ultrasonic processor (Scientz). The remaining debris was removed by centrifugation at 12,000 g at 4 °C for 10 min. Finally, the supernatant was collected and the protein concentration was determined with BCA kit according to the manufacturer’s instructions.
For trypsin digestion, the protein solution was reduced with 5 mM dithiothreitol for 30 min at 56 °C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. The protein sample was then diluted by adding 100 mM NH4HCO3 to urea concentration less than 2M. Finally, trypsin was added at 1:50 trypsin-to-protein mass ratio for the first digestion overnight and 1:100 trypsin-to-protein mass ratio for a second 4 hour-digestion.
To enrich Kmal modified peptides, tryptic peptides dissolved in NETN buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, 0.5% NP-40, pH 8.0) were incubated with pre-washed antibody beads (PTM-901, PTM Biolabs) at 4 °C overnight with gentle shaking. Then the beads were washed four times with NETN buffer and twice with H2O. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid. Finally, the eluted fractions were combined and vacuum-dried. For LC-MS/MS analysis, the resulting peptides were desalted with C18 ZipTips (Millipore) according to the manufacturer’s instructions.
For LC-MS/MS analysis, the tryptic peptides were dissolved in 0.1% formic acid (solvent A), directly loaded onto a home-made reversed-phase analytical column (15-cm length, 75 μm i.d.). The gradient was comprised of an increase from 6% to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, 23% to 35% in 8 min and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 400 nl/min on an EASY-nLC 1000 UPLC system. The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350 to 1800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were then selected for MS/MS using NCE setting as 28 and the fragments were detected in the Orbitrap at a resolution of 17,500. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans with 15.0s dynamic exclusion. Automatic gain control (AGC) was set at 5E4.
The resulting MS/MS data were processed using Maxquant search engine (v.1.5.2.8). Tandem mass spectra were searched against Kmal database concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to 4 missing cleavages. The mass tolerance for precursor ions was set as 20 ppm in First search and 5 ppm in Main search, and the mass tolerance for fragment ions was set as 0.02 Da. FDR was adjusted to < 1% and minimum score for modified peptides was set > 40.
Cell proliferation and colony formation assays
Cell proliferation assays were performed by cell counting and a Cell counting Kit 8 (CCK-8) (C0038, Beyotime). 500 cells were seeded into 6-well plates for the colony formation assay. After 14 days of incubation, the plates were fixed with methanol for 20 min and stained with 0.2% crystal violet for 20 min.
NCL structure prediction
Human full-length NCL protein sequence was applied to AlphaFold for structure predication (51). The predicted structure was pulled out from the website (https://alphafold.ebi.ac.uk/entry/P19338). The predicted binding pocket around K398 was visualized and recolored with PyMOL (http://www.pymol.org).
Statistical analysis
GraphPad Prism 8.0 (GraphPad) was used for all of the statistical analyses. Data samples were compared using a one-tailed Student’s t test unless specified, and a p < 0.05 was considered significant.
Data availability
The data generated in this study are within the article or in Supporting Information files. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD048727.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
L. S., H. M., and T. L. writing–original draft; L. S., H. M., T. L., Y. T., Y. S., G. S., L. Z., S. G., and Y. L. methodology; L. S., H. M., T. L., Q. Z., M. X., Z. Z., Y. Q., H. C., X. Z., K. C., S. G., and Y. L. investigation; L. S., H. M., T. L., and S. G. formal analysis; L. S., H. M., T. L., Q. Z., M. X., Z. Z., Y. Q., H. C., X. Z., K. C., and Y. L. data curation; Y. T., Y. S., B. Z., J. D., G. S., L. Z., S. G., and Y. L. writing–review and editing; J. Z. supervision; J. Z. and Y. L. resources; J. Z. and Y. L. project administration; Y. L. validation; Y. L. funding acquisition; Y. L. conceptualization.
Funding and additional information
This work was supported by the National Natural Science Foundation of China (Grant Nos. 82373073, 92054106, and 81972723), and the Jiangsu Distinguished Professorship Program to Y. L., and the Scientific Research Foundation of Xuzhou Medical University (RC20552204) to K. C.
Reviewed by members of the JBC Editorial Board. Edited by Mike Shipston
Contributor Information
Junnian Zheng, Email: jnzheng@xzhmu.edu.cn.
Shutao Guo, Email: stguo@nankai.edu.cn.
Yong Liu, Email: liuymito@xzhmu.edu.cn.
Supporting information
References
- 1.Trub A.G., Hirschey M.D. Reactive acyl-CoA species modify proteins and induce carbon stress. Trends Biochem. Sci. 2018;43:369–379. doi: 10.1016/j.tibs.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirschey M.D., Zhao Y. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell Proteomics. 2015;14:2308–2315. doi: 10.1074/mcp.R114.046664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner G.R., Bhatt D.P., O'Connell T.M., Thompson J.W., Dubois L.G., Backos D.S., et al. A class of reactive acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metab. 2017;25:823–837.e8. doi: 10.1016/j.cmet.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulkarni R.A., Worth A.J., Zengeya T.T., Shrimp J.H., Garlick J.M., Roberts A.M., et al. Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem. Biol. 2017;24:231–242. doi: 10.1016/j.chembiol.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu H., Zhou J., Lin S., Deng W., Zhang Y., Xue Y. PLMD: an updated data resource of protein lysine modifications. J. Genet. Genomics. 2017;44:243–250. doi: 10.1016/j.jgg.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Du J., Zhou Y., Su X., Yu J.J., Khan S., Jiang H., et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M., et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.012658. M111 012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colak G., Pougovkina O., Dai L., Tan M., Te Brinke H., Huang H., et al. Proteomic and biochemical studies of lysine malonylation suggest its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Mol. Cell Proteomics. 2015;14:3056–3071. doi: 10.1074/mcp.M115.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishida Y., Rardin M.J., Carrico C., He W., Sahu A.K., Gut P., et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell. 2015;59:321–332. doi: 10.1016/j.molcel.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruning U., Morales-Rodriguez F., Kalucka J., Goveia J., Taverna F., Queiroz K.C.S., et al. Impairment of angiogenesis by fatty acid synthase inhibition involves mTOR malonylation. Cell Metab. 2018;28:866–880.e15. doi: 10.1016/j.cmet.2018.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galvan-Pena S., Carroll R.G., Newman C., Hinchy E.C., Palsson-McDermott E., Robinson E.K., et al. Malonylation of GAPDH is an inflammatory signal in macrophages. Nat. Commun. 2019;10:338. doi: 10.1038/s41467-018-08187-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Q., Wu D., Zhao J., Yan Z., Chen L., Guo S., et al. TFAM loss induces nuclear actin assembly upon mDia2 malonylation to promote liver cancer metastasis. EMBO J. 2022;41 doi: 10.15252/embj.2021110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H.L., Chen Y., Wang Y.Q., Tao E.W., Tan J., Liu Q.Q., et al. Sirtuin5 protects colorectal cancer from DNA damage by keeping nucleotide availability. Nat. Commun. 2022;13:6121. doi: 10.1038/s41467-022-33903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul B., Lewinska M., Andersen J.B. Lipid alterations in chronic liver disease and liver cancer. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang D.Q., El-Serag H.B., Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021;18:223–238. doi: 10.1038/s41575-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam SM Z.T., Li J., Zhang S.H., Chua G.H., Li B.W., Shui G. A robust, integrated platform for comprehensive analyses of acyl-coenzyme as and acyl-carnitines revealed chain length-dependent disparity in fatty acyl metabolic fates across Drosophila development. Sci. Bull. 2020;65:1840–1848. doi: 10.1016/j.scib.2020.07.023. [DOI] [PubMed] [Google Scholar]
- 17.Jia W., Yao Z., Zhao J., Guan Q., Gao L. New perspectives of physiological and pathological functions of nucleolin (NCL) Life Sci. 2017;186:1–10. doi: 10.1016/j.lfs.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 18.Berger C.M., Gaume X., Bouvet P. The roles of nucleolin subcellular localization in cancer. Biochimie. 2015;113:78–85. doi: 10.1016/j.biochi.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 19.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin S.H., Lee G.Y., Lee M., Kang J., Shin H.W., Chun Y.S., et al. Aberrant expression of CITED2 promotes prostate cancer metastasis by activating the nucleolin-AKT pathway. Nat. Commun. 2018;9:4113. doi: 10.1038/s41467-018-06606-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J., Thompson C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019;20:436–450. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leone R.D., Powell J.D. Metabolism of immune cells in cancer. Nat. Rev. Cancer. 2020;20:516–531. doi: 10.1038/s41568-020-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergers G., Fendt S.M. The metabolism of cancer cells during metastasis. Nat. Rev. Cancer. 2021;21:162–180. doi: 10.1038/s41568-020-00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S.C., et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan M., Peng C., Anderson K.A., Chhoy P., Xie Z., Dai L., et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao H., Cai Q., Guo W., Su Q., Qin H., Wang T., et al. Malonylation of Acetyl-CoA carboxylase 1 promotes hepatic steatosis and is attenuated by ketogenic diet in NAFLD. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112319. [DOI] [PubMed] [Google Scholar]
- 27.Li Z., Guan M., Lin Y., Cui X., Zhang Y., Zhao Z., et al. Aberrant lipid metabolism in hepatocellular carcinoma revealed by liver lipidomics. Int. J. Mol. Sci. 2017;18:2550. doi: 10.3390/ijms18122550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu X., Zhang X., Zhang Y., Zhang K., Zhan C., Shi X., et al. Metabolic profiling analysis upon acylcarnitines in tissues of hepatocellular carcinoma revealed the inhibited carnitine shuttle system caused by the downregulated carnitine palmitoyltransferase 2. Mol. Carcinog. 2019;58:749–759. doi: 10.1002/mc.22967. [DOI] [PubMed] [Google Scholar]
- 29.Narita T., Weinert B.T., Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019;20:156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Z., Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biol. 2019;20:245. doi: 10.1186/s13059-019-1870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabari B.R., Zhang D., Allis C.D., Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017;18:90–101. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishiguro T., Tanabe K., Kobayashi Y., Mizumoto S., Kanai M., Kawashima S.A. Malonylation of histone H2A at lysine 119 inhibits Bub1-dependent H2A phosphorylation and chromosomal localization of shugoshin proteins. Sci. Rep. 2018;8:7671. doi: 10.1038/s41598-018-26114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amamoto Y., Aoi Y., Nagashima N., Suto H., Yoshidome D., Arimura Y., et al. Synthetic posttranslational modifications: chemical catalyst-driven regioselective histone acylation of native chromatin. J. Am. Chem. Soc. 2017;139:7568–7576. doi: 10.1021/jacs.7b02138. [DOI] [PubMed] [Google Scholar]
- 34.Zhang R., Bons J., Scheidemantle G., Liu X., Bielska O., Carrico C., et al. Histone malonylation is regulated by SIRT5 and KAT2A. iScience. 2023;26 doi: 10.1016/j.isci.2023.106193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otake Y., Soundararajan S., Sengupta T.K., Kio E.A., Smith J.C., Pineda-Roman M., et al. Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood. 2007;109:3069–3075. doi: 10.1182/blood-2006-08-043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takagi M., Absalon M.J., McLure K.G., Kastan M.B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 37.Mongelard F., Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Rong Cong S.D., Bouvet P. The multiple properties and functions of nucleolin. Nucleolus. 2011;9:185–212. [Google Scholar]
- 39.Dranovsky A., Vincent I., Gregori L., Schwarzman A., Colflesh D., Enghild J., et al. Cdc2 phosphorylation of nucleolin demarcates mitotic stages and Alzheimer's disease pathology. Neurobiol. Aging. 2001;22:517–528. doi: 10.1016/s0197-4580(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 40.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinert B.T., Schölz C., Wagner S.A., Iesmantavicius V., Su D., Daniel J.A., et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 42.Wu Q., Cheng Z., Zhu J., Xu W., Peng X., Chen C., et al. Suberoylanilide hydroxamic acid treatment reveals crosstalks among proteome, ubiquitylome and acetylome in non-small cell lung cancer A549 cell line. Sci. Rep. 2015;5:9520. doi: 10.1038/srep09520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di J., Zhao G., Wang H., Wu Y., Zhao Z., Zhu B., et al. A p53/CPEB2 negative feedback loop regulates renal cancer cell proliferation and migration. J. Genet. Genomics. 2021;48:606–617. doi: 10.1016/j.jgg.2021.05.011. [DOI] [PubMed] [Google Scholar]
- 44.Bao X., Zhao Q., Yang T., Fung Y.M., Li X.D. A chemical probe for lysine malonylation. Angew. Chem. Int. Ed. Engl. 2013;52:4883–4886. doi: 10.1002/anie.201300252. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y., He Y., Jin A., Tikunov A.P., Zhou L., Tollini L.A., et al. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2414–E2422. doi: 10.1073/pnas.1315605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y., Clegg H.V., Leslie P.L., Di J., Tollini L.A., He Y., et al. CHCHD2 inhibits apoptosis by interacting with Bcl-x L to regulate Bax activation. Cell Death Differ. 2014;22:1035–1046. doi: 10.1038/cdd.2014.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam S.M., Chua G.H., Li X.J., Su B., Shui G. Biological relevance of fatty acyl heterogeneity to the neural membrane dynamics of rhesus macaques during normative aging. Oncotarget. 2016;7:55970–55989. doi: 10.18632/oncotarget.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam S.M., Tong L., Duan X., Petznick A., Wenk M.R., Shui G. Extensive characterization of human tear fluid collected using different techniques unravels the presence of novel lipid amphiphiles. J. Lipid Res. 2014;55:289–298. doi: 10.1194/jlr.M044826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shui G., Guan X.L., Low C.P., Chua G.H., Goh J.S., Yang H., et al. Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry: application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Mol. Biosyst. 2010;6:1008–1017. doi: 10.1039/b913353d. [DOI] [PubMed] [Google Scholar]
- 50.Shui G., Cheong W.F., Jappar I.A., Hoi A., Xue Y., Fernandis A.Z., et al. Derivatization-independent cholesterol analysis in crude lipid extracts by liquid chromatography/mass spectrometry: applications to a rabbit model for atherosclerosis. J. Chromatogr. A. 2011;1218:4357–4365. doi: 10.1016/j.chroma.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 51.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are within the article or in Supporting Information files. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD048727.