Abstract
We describe a new species, Chrysospleniuminsularis J.E.Jang, K.H.Lee & H.Y.Gil, belonging to the family Saxifragaceae, from the southern islands of the Republic of Korea. Chrysospleniuminsularis is morphologically similar to C.japonicum (Maxim.) Makino but can be distinguished by fairly persistent bulbils, green to yellowish-green sepals, four stamens, and cylindrical papillose seeds. Chrysospleniuminsularis is also distinguished from C.alternifolium L., which is distributed in Europe, northern Russia, and the Caucasus, by the absence of stolons and green bracts. Phylogenetic analyses, based on one nuclear ribosomal (ITS) and two chloroplast (rbcL, matK) regions, confirmed that the new species was monophyletic and that C.insularis and C.alternifolium formed a sister relationship with robust support. Herein, we provide a detailed morphological description of C.insularis with its corresponding geographical distribution and comparison table and figures of related species.
Key words: Chrysosplenium , morphology, new species, phylogeny, taxonomy
Introduction
Chrysosplenium L. is a perennial herbaceous genus of the family Saxifragaceae, consisting of more than 70 species (Kim et al. 2019; Fu et al. 2020). Species of this genus are mainly distributed in temperate regions of the Northern Hemisphere, and their habitats are shady and humid areas in the mountains (Kim and Kim 2015; Kim et al. 2018; Zhao et al. 2022). Biogeographically, Chrysosplenium is known to have originated in East Asia, and several independent lineages have migrated from East Asia to the New World (Soltis et al. 2001; Deng et al. 2015; Liu et al. 2016).
The genus Chrysosplenium is distinguished from other genera in Saxifragaceae by its tetramerous flowers with petaloid sepals and four or eight stamens (Kim et al. 2018). However, species delimitation is often difficult in this genus because of extensive morphological variations owing to differences in growth periods and habitats (Qin et al. 2018; Kim et al. 2019; Choi et al. 2020). The genus is divided into two sections, Chrysospleniumsect.Alternifolia Franch. and C.sect.Oppositifolia Franch., based on the arrangement of the leaves (Franchet 1890). However, Hara (1957) proposed 17 series because of the high variability in the flower, capsule, and seed traits within each section. The infrageneric classification of Chrysosplenium species is based on several criteria, including leaf arrangement, seed surface, pedicel length, sterile branch position, capsule shape, stem surface, ovary position, stamen length, leaf surface, sepal length, and basal leaf size (Hara 1957; Fu et al. 2020). Several phylogenetic studies on Saxifragaceae genera, including Chrysosplenium, have been performed based on the chloroplast matK region, and their results have shown that C.sect.Oppositifolia and C.sect.Alternifolia are monophyletic (Nakazawa et al. 1997; Soltis et al. 2001; Deng et al. 2015). Recently, several new species have been described based on detailed and comprehensive morphological, molecular, and cytological studies (Liu et al. 2016; Kim et al. 2018, 2019; Wakabayashi et al. 2018; Fu et al. 2020, 2021).
Thirteen Chrysosplenium species belonging to seven series have been recognized in the Korean Peninsula to date (Nakazawa et al. 1997; Kim et al. 2019; Choi et al. 2020; Korea National Arboretum 2021). The following are these 13 species [Chrysospleniumser.Pilosa Maxim.: C.flaviflorum Ohwi, C.epigealum J.W.Han & S.H.Kang, C.ramosissimum Y.I.Kim & Y.D.Kim, C.valdepilosum (Ohwi) S.H.Kang & J.W.Han, C.aureobracteatum Y.I.Kim & Y.D.Kim, C.barbatum Nakai; C.ser.Oppositifolia Maxim.: C.ramosum Maxim.; C.ser.Nepalensia Maxim.: C.grayanum Maxim.; C.ser.Sinica Maxim.: C.sinicum Maxim.; C.ser.Macrostemon H. Hara: C.macrostemon Maxim. ex Franch. & Sav.; C.ser.Alternifolia Maxim.: C.japonicum (Maxim.) Makino, C.serreanum Hand.-Mazz.; C.ser.Flagellifera Maxim.: C.flagelliferum F.Schmidt], and among them, C.aureobracteatum, C.barbatum, C.epigealum, C.flaviflorum, and C.ramosissimum are endemic to Korea (Chung and Kim 1988; Korea National Arboretum 2021; Chung et al. 2023).
During a floristic survey in the southern part of Korea in March 2020, we found a new Chrysosplenium species that is restricted to the southern islands of Korea (Jeju-do and Gageo-do Islands). This species is readily distinguished from previously known Chrysosplenium species in Korea by its greenish-yellow to green bracteal leaves at flowering and a cylindrical papillose seed surface. This species is most similar to C.japonicum (Maxim.) Makino, which belongs to the C.ser.Alternifolia, and is distributed throughout Northeast Asia, including Southeast China, Japan, Korea, Russia (Manchuria), and Taiwan (Nakazawa et al. 1997; Pan and Ohba 2001; Hsu et al. 2011). The new species, however, is clearly distinguishable from C.japonicum by the form of bulbils, color of sepals, number of stamens, and surface of seeds. Based on thorough literature surveys, extensive field observations, detailed analysis of floral morphology and seed coat characteristics, we designated this new species as C.insularis J.E.Jang, K.H.Lee & H.Y.Gil. Here, we provide a detailed morphological description and phylogenetic position of C.insularis and its geographical distribution.
Materials and methods
Material collection
Field surveys were conducted from March 2020 to March 2023. Voucher specimens were deposited at the herbarium of the Korea National Arboretum (KH, http://www.nature.go.kr/kbi/plant/smpl/KBI_2001_030100.do). Materials preserved in 70% ethanol were used to observe and measure the floral parts. Morphological observations and measurements of the new species were conducted on live and dried specimens, including the materials preserved at KH. Quantitative characteristics were measured based on at least 30 samples. The terminology used for description and comparison was referenced from Choi et al. (2020), Pan and Ohba (2001), Lozina (1939), Wakabayashi (2001), Kim et al. (2018), Kim et al. (2019), Fu et al. (2020), Fu et al. (2021).
Microscopic observation
The seed morphology was observed under a stereomicroscope and a scanning electron microscope (SEM). The seeds were measured using a stereomicroscope (Carl Zeiss Microscopy GmbH, Stemi 508, Zeiss, Göttingen, Germany) with an Axiocam ERc 5s. Before SEM imaging, the seeds were dehydrated using 100% ethanol and sputter-coated with gold in a KIC-IA COXEM ion coater (COXEM Co., Ltd., Daejeon, Korea). SEM imaging was performed using a COXEM EM-30 PLUS+ table scanning electron microscope (COXEM) at 20 kV at the Seed Testing Laboratory of KH.
Phylogenetic analysis
Molecular phylogenetic analyses were conducted to confirm the phylogenetic position of the new putative species of Chrysosplenium. Sixteen accessions of four taxa, including the new and related species, were collected from seven localities in South Korea. Total DNA was extracted from silica gel-dried leaves using the DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA) in accordance with the manufacturer’s instructions. The nrDNA region (ITS) and two cpDNA regions (matK, rbcL) were subjected to polymerase chain reaction (PCR) (Choi et al. 2020) on a ProFlex 96-Well PCR System (Applied Biosystems, Foster City, CA, USA). The primers used and their sequences are listed in Table 1. Each reaction mixture contained AccuPower® PCR PreMix (Bioneer, Daejeon, South Korea), ca. 10 ng (1 μL) of genomic DNA, and 100 pM of primers in a total volume of 20 µL. The PCR conditions included an initial denaturation at 94 °C for 5 min, followed by 35 cycles of amplification at 94 °C for 1 min, 54 °C for 1 min, and 72 °C for 1 min, and a final extension at 72 °C for 7 min. The PCR products were visualized on 1% agarose gels and sequenced on an ABI 3730xl DNA analyzer using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). The sequences obtained were manually determined and aligned using MAFFT with Geneious Prime® 2022.1.1. (Biomatters Ltd., Auckland, NZ). The DNA sequences generated in this study have been deposited in GenBank and are indicated with an asterisk (*) in the voucher information in Table 2.
Table 1.
Primers used for phylogenetic analysis.
| Fragment | Primer | Sequence 5′ → 3′ | Reference |
|---|---|---|---|
| ITS | ITS1 | TCCGTAGGTGAACCTGCGG | White et al. (1990) |
| ITS4 | TCCTCCGCTTATTGATATGC | ||
| rbcL | rbcL_1F | ATGTCACCACAAACAGAAAC | Fay et al. (1998) |
| rbcL_724R | TCGCATGTACCTGCAGTAGC | ||
| matK | 3F_Kim_F | CGTACAGTACTTTTGTGTTTA | K.J.Kim, pers. comm. |
| 1R_Kim_R | ACCCAGTCCATCTGGAAATCT |
Table 2.
Voucher information and GenBank number of accessions used in this study (*newly generated sequences).
| Taxon | Locality | Voucher information | GenBank number | ||
|---|---|---|---|---|---|
| ITS | rbcL | matK | |||
| C.alternifolium | JAPAN: Shimane-ken | DG2019032310003 | OK315466 | OK315387 | OK315343 |
| C.aureobracteatum | KOREA: Gangwon-do, Mt. Gwangdeog | LeeJD et al. 17127-1 | MK989508 | MK989534 | MK989559 |
| KOREA: Gangwon-do, Mt. Gwangdeog | LeeJD et al. 17127-2 | MK989509 | MK989533 | MK989562 | |
| C.barbatum | KOREA: Jeollanam-do, Woldeung-myeon | LeeJD et al. 17008-1 | MK989505 | MK989538 | MK989560 |
| KOREA: Gyeongsangbuk-do, Mt. Danseok | LeeJD et al. 17020-1 | MK989506 | MK989536 | MK989564 | |
| KOREA: Gangwon-do, Mt. Gwangdeog | LeeJD et al. 17066 | MK989507 | MK989537 | MK989561 | |
| C.flagelliferum | KOREA: Gyeongsangbuk-do, Ulleung-gun, Gwanmobong | ESK21-267* | OR809214 | PP133187 | PP170153 |
| KOREA: Gyeongsangbuk-do, Ulleung-gun, Gwanmobong | ESK21-268* | OR809215 | PP133188 | PP170154 | |
| KOREA: Gyeongsangbuk-do, Ulleung-gun, Seonginbong | AP22-025* | OR809213 | PP133186 | PP170152 | |
| KOREA: Gyeoggi-do, Mt. Cheonma | LeeJD et al. 17014 | MK989499 | MK989530 | MK989585 | |
| KOREA: Gangwon-do, Mt. Cheongtae | LeeJD et al. 17052-1 | MK989500 | MK989529 | MK989583 | |
| KOREA: Gyeongsangbuk-do, Ulleung-do | LeeJD et al. 17122 | MK989501 | MK989531 | MK989584 | |
| C.flaviflorum | KOREA: Gangwon-do, Mt. Pokkye | ESK21-182* | OR809216 | PP133189 | PP170155 |
| KOREA: Gangwon-do, Mt. Pokkye | ESK21-183* | OR809217 | PP133190 | PP170156 | |
| KOREA: Gangwon-do, Mt. Pokkye | ESK21-184-1* | OR809218 | PP133191 | PP170157 | |
| KOREA: Gangwon-do, Mt. Pokkye | ESK21-184-2* | OR809219 | PP133192 | PP170158 | |
| KOREA: Chungcheongbuk-do, Mt. Gyemyeong | LeeJD et al. 17030 | MK989513 | MK989542 | MK989569 | |
| KOREA: Gyeongsangbuk-do, Mt. Cheonglyang | LeeJD et al. 17039 | MK989514 | MK989540 | MK989567 | |
| KOREA: Gangwon-do, Mt. Chiak | LeeJD et al. 17048 | MK989515 | MK989541 | MK989568 | |
| C.grayanum | JAPAN: Hokkaido, Sapporo, Mt. Maruyama | Nakamura 16401 | MK989524 | MK989554 | MK989574 |
| JAPAN: Hokkaido, Sapporo, Mt. Maruyama | Nakamura 16402 | MK989523 | MK989553 | MK989575 | |
| JAPAN: Hyogo prefecture, Sasayama | Lee JH & JS Shin s. n. | MK989525 | MK989551 | MK989576 | |
| KOREA: Jeollanam-do, Mt. Cheongtae | LeeJD et al. 17090-1 | MK989522 | MK989550 | MK989579 | |
| KOREA: Jeollanam-do, Mt. Cheongtae | LeeJD et al. 17090-2 | MK989520 | MK989555 | MK989578 | |
| KOREA: Jeollanam-do, Mt. Cheongtae | LeeJD et al. 17090-3 | MK989521 | MK989552 | MK989577 | |
| C.griffithii | CHINA | 13PXD035 | MH809138 | MN185317 | MN451058 |
| C.insularis | KOREA: Jeju-do, Seogwipo-si, Hogeun-dong | SOK-2022-175* | OR809225 | PP133198 | PP170164 |
| KOREA: Jeju-do, Seogwipo-si, Hogeun-dong | J.E.Jang et al. 230322* | OR809226 | PP133199 | PP170165 | |
| KOREA: Jeollanam-do, Gageodo | K.H.Lee 230514-1* | OR809227 | PP133200 | PP170166 | |
| KOREA: Jeollanam-do, Gageodo | K.H.Lee 230514-2* | OR809228 | PP133201 | PP170167 | |
| C.japonicum | KOREA: Gyeonggi-do, Mt. Cheonma | J.E.Jang 230325-1* | OR809220 | PP133193 | PP170159 |
| KOREA: Gyeonggi-do, Mt. Cheonma | J.E.Jang 230325-2* | OR809221 | PP133194 | PP170160 | |
| KOREA: Gangwon-do, Wonju-si | S.R.Lee et al. 230420-1* | OR809222 | PP133195 | PP170161 | |
| KOREA: Gangwon-do, Wonju-si | S.R.Lee et al. 230420-2* | OR809223 | PP133196 | PP170162 | |
| KOREA: Gangwon-do, Wonju-si | S.R.Lee et al. 230420-3* | OR809224 | PP133197 | PP170163 | |
| KOREA: Jeollabuk-do, Mt. Chaegye | LeeJD et al. 17022 | MK989502 | MK989548 | MK989586 | |
| KOREA: Chungcheongnam-do, Palbong-myeon | LeeJD et al. 17025-1 | MK989504 | MK989549 | MK989587 | |
| C.kamtschaticum | JAPAN: Hokkaido, Sapporo, Mt.Maruyama | Nakamura 16403 | MK989516 | MK989539 | MK989566 |
| C.ramosum | KOREA: Chungcheongbuk-do, Daegang-myeon Goseong | LeeJD et al. 17097-1 | MK989517 | MK989543 | MK989571 |
| KOREA: Gangwon-do, Mt. Taegi | LeeJD et al. 17147 | MK989518 | MK989545 | MK989573 | |
| KOREA: Gyeongsangbuk-do, Mt. Irwol | LeeJD et al. 17205-1 | MK989519 | MK989544 | MK989572 | |
| C.sinicum | KOREA: Jeju-do, Haean-dong | LeeJD et al. 17043 | MK989528 | MK989557 | MK989582 |
| KOREA: Gangwon-do, Mt. Cheongtae | LeeJD et al. 17051-1 | MK989526 | MK989556 | MK989580 | |
| KOREA: Chungcheongbuk-do, Gagok-myeon | LeeJD et al. 17086 | MK989527 | MK989558 | MK989581 | |
| C.valdepilosum | KOREA: Gangwon-do, Mt. Taegi | LeeJD et al. 17053-1 | MK989512 | MK989535 | MK989563 |
| KOREA: Jeollabuk-do, Mt. Jiri | LeeJD et al. 17057 | MK989510 | MK989532 | MK989565 | |
| Peltoboykiniatellimoides | JAPAN: Nagano, Kiso-Fukushima, | Okuyama 035251 | AB248847 | ||
| CHINA: Zhejiang, Suichang County | XXL170002-1 | MZ779205 | |||
We also included 32 accessions of 13 Chrysosplenium species deposited in GenBank and selected Peltoboykiniatellimoides (Maxim.) Hara as the outgroup (Soltis et al. 1996). A total of 47 accessions from 14 taxa were used for the phylogenetic analysis. Details of the voucher information and GenBank accession numbers of the species used in this study are provided in Table 2. Phylogenetic analyses were performed using the maximum likelihood (ML) method. For the ML analysis, the best-fit model was identified using ModelFinder in Phylosuite (Kalyaanamoorthy et al. 2017; Zhang et al. 2020). ML phylogenies were inferred using IQ-TREE (Nguyen et al. 2015) under the GTR+ F + R3 model in the ITS regions and the TIM+F+R2 model in the combined chloroplast regions (Minh et al. 2013).
Results and discussion
Taxonomic treatment
. Chrysosplenium insularis
J.E.Jang, K.H.Lee & H.Y.Gil sp. nov.
B5BC1FD3-D041-526D-8B66-D363DFB74D04
urn:lsid:ipni.org:names:77350706-1
Figure 1.
ChrysospleniuminsularisA habit B bulbils C stem D basal leaf E inflorescence, F bracteal leaves G flower H, I capsule J seed. Photographs by Ju Eun Jang and Kang-Hyup Lee.
Figure 4.
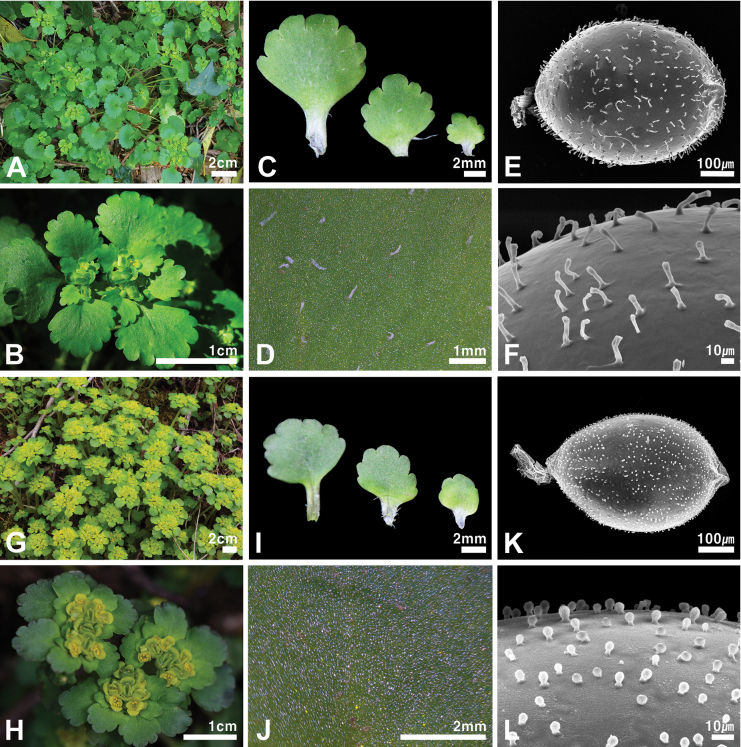
Comparative photographs of the habit (A, G), inflorescence (B, H), bracteal leaves (C, I), surface of bracteal leaves (D, J), and seed (E, F, K, L) of Chrysospleniuminsularis (A–F) and C.japonicum (G–L). Photographs by Ju Eun Jang and Kang-Hyup Lee.
Diagnosis.
Chrysospleniuminsularis differs from C.japonicum in having fairly persistent bulbils, green to yellowish-green sepals, four stamens, and cylindrical papillose seeds.
Type.
Korea • Jeju, Seogwipo-si, Hogeun-dong; 33.25084, 126.54434; elev. 58 m; 25 Mar 2020 [fl]; Kang-Hyup Lee JJ-200325-001 [holotype KH (Fig. 2); isotypes, 3 sheets, KH].
Figure 2.
Holotype of Chrysospleniuminsularis.
Description.
Perennial herbs, hermaphroditic, 5–15 cm tall. Bulbils present near stem base, fairly persistent, pink, turning darkish brown, pilose. Roots fibrous, white. Stems erect, cespitose, light green to green, sparsely hairy, without stolons. Basal leaves of flowering stems 1–6, opposite, simple, estipulate; petiole 3–9 cm long, glabrescent or sparsely hairy; blade reniform, 13–20 mm × 15–25 mm, apex rounded and often retuse, margins dentate to crenate, 13–17 teeth, base cordate, adaxially green, pilose, abaxially pale green, subglabrous. Cauline leaves of flowering stems 1–4, alternate, simple, estipulate; petiole 5–22 mm long, glabrescent or sparsely hairy; blade flabellate to reniform, 7–12 mm × 11–18 mm, apex retuse and often rounded or obtuse, margins dentate to crenate, 9–13 teeth, base cordate to broadly cuneate, adaxially green, pilose, abaxially pale green, subglabrous. Inflorescences terminal, 6–14 flowered cyme, surrounded by leaf-like bracts; peduncles 4.59–18.54 mm long; pedicels 0.5–1.5 mm long, sparsely pilose; bracteal leaves by inflorescence 3, petiole 0.2–4.7 mm long, glabrescent or sparsely hairy; blade subflabellate to orbicular, 2–18 × 2–14 mm, apex truncate and often retuse, margins dentate to crenate, 5–9 teeth, base broadly cuneate to subcordate, adaxially green, sparsely pilose to glabrescent, abaxially pale green, subglabrous. Flowers 4-merous, actinomorphic; sepals petaloid 4, free, erect to subspreading, ovate to broadly ovate, 1.2–2.1 × 1.5–3.1 mm, apex obtuse or rounded, yellowish green to green, glabrous; stamens 4; filaments narrow conical, 0.3–0.4 mm long; anther 0.2 mm long, yellow; pistil 2-carpellate, semi-inferior; ovary 1-locular; styles 2, free, erect, 0.2–0.3 mm long; stigma round; disc present. Capsules 2-lobed, horn shaped, lobes subequal, 2.8–3.7 × 3.8–5.2 mm long, green, glabrous, dehiscent along the adaxial suture. Seeds numerous, ovoid-ellipsoid, 0.7–0.9 × 0.5–0.6 mm, brown to dark brown, cylindrical papillose on smooth surfaces.
Phenology.
Flowering and fruiting from March to May.
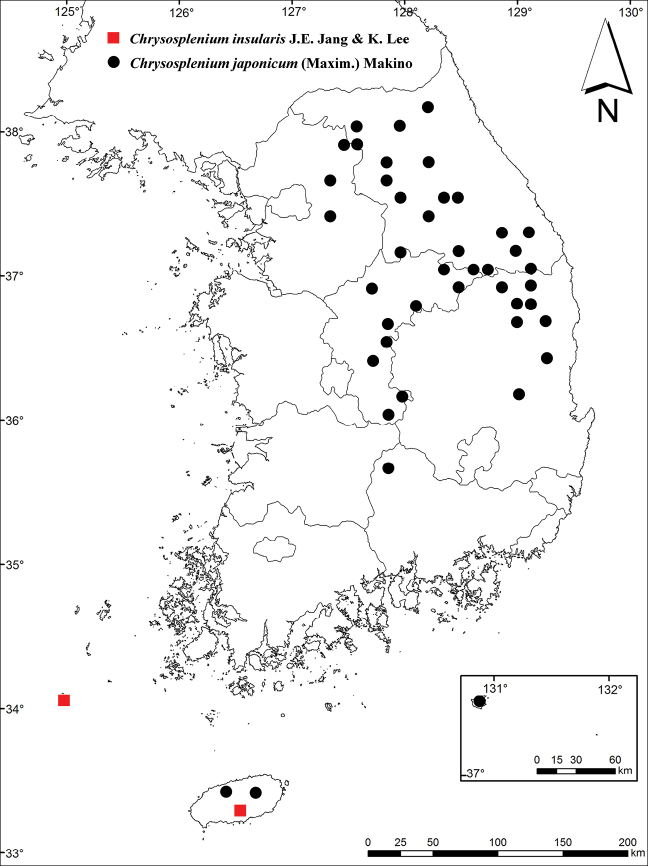
Distribution and habitat.
Southern coastal regions of Korea (Jeju-do and Gageo-do Islands). Forests, wet places in forests, shaded places on the riverside (Fig. 3).
Figure 3.
Distribution map of Chrysospleniuminsularis and C.japonicum in Korea (revised from Oh et al. 2016).
Etymology.
The specific epithet “insularis” refers to its distribution on islands.
Vernacular name.
Island golden saxifrage: Seom-gwaeng-i-nun (섬괭이눈).
Morphological assessment.
Among the species distributed in Korea, Chrysospleniuminsularis is morphologically similar to C.japonicum in terms of leaf arrangement, leaf margin, and bracteal leaf color. Despite these similarities, it is clearly differentiated by the form of bulbils [present, fairly persistent (Fig. 1B) vs. present], surface of bracteal leaves [adaxially sparsely pilose to glabrescent, abaxially subglabrous (Fig. 4D) vs. mainly glabrous (Fig. 4J)], color of sepals [green to yellowish green vs. yellowish green to yellow], number of stamens [4 vs. usually 8], and surface of seeds [cylindrical papillose (Fig. 4E, F) vs. papillose (Fig. 4K, L)]. Additionally, this new species is morphologically similar to C.alternifolium, which is distributed in northern Eurasia, but is distinguished by the following characteristics: stolon (absent vs. present), color of bracts (green vs. yellow), color of sepals (green to yellowish green vs. golden yellow), number of stamens (4 vs. 8), and surface of seeds [cylindrical papillose (Fig. 4E, K) vs. smooth (Fig. 4K, L)]. A comparison of the major characteristics of the new species with those of two closely related species, C.japonicum and C.alternifolium, is shown in Table 3.
Table 3.
Major characteristics of Chrysospleniuminsulalis and two closely related taxa (*: data from Lozina 1939; -: none known).
| Character | C.insulalis | C.japonicum | C.alternifolia* | |
|---|---|---|---|---|
| Bulbils | present, fairly persistent | present | - | |
| color | pink, turning to darkish brown | pink | - | |
| Stolon | absent | absent | present | |
| Bracteal leaves | color | green | yellowish green | yellow |
| surfaces | adaxially sparsely pilose to glabrescent, abaxially subglabrous | mainly glabrous | mainly glabrous | |
| Sepals | color | green to yellowish green | yellowish green to yellow | golden yellow |
| Stamens | number | 4 | usually 8 | 8 |
| Seeds | surfaces | cylindrical papillose | papillose | smooth |
| Fl. and fr. | Mar. to May | Apr. to Jun. | Apr. to Jul. | |
Phylogenetic analysis.
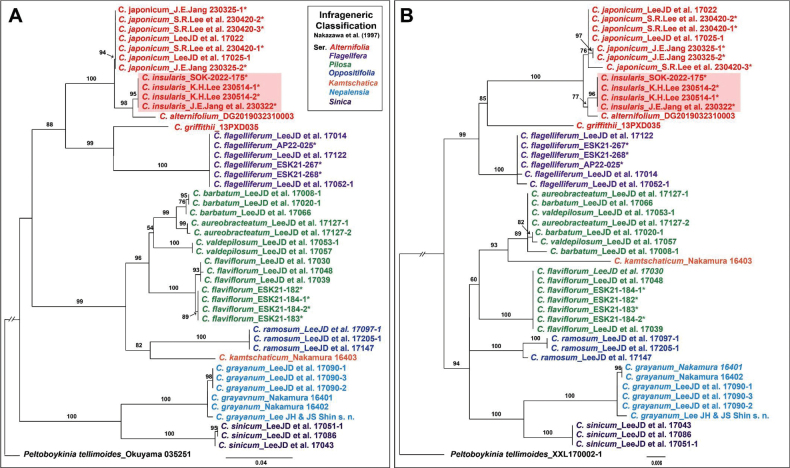
In total, 48 sequences of three regions (ITS, matK, and rbcL) were newly obtained from the 16 accessions of Chrysospleniuminsularis and the three related taxa. We also used 93 sequences from 32 accessions obtained from GenBank (12 species of Chrysosplenium and one Peltoboykiniatellimoides as an outgroup) for the phylogenetic analysis. The aligned matrix of the ITS region and combined chloroplast regions (matK and rbcL) contained 635 and 1407 characters, respectively. We found 242 variable sites and 193 parsimony-informative sites in the ITS regions, whereas 172 variable sites and 104 parsimony-informative sites were found in the combined chloroplast regions. The GC ratios were 46.2% and 37.4% for the ITS and combined chloroplast regions, respectively. The phylogenetic tree (Fig. 5) revealed a topology similar to that obtained in a previous study (Choi et al. 2020). The phylogenetic results showed some topological incongruence between the ITS and combined CP trees. In the ITS tree, the most basal clade (BS = 100%) included the monophyletic C.grayanum and C.sinicum and showed a sister relationship with other Chrysosplenium species. However, the CP tree was divided into two clades, with C.grayanum and C.sinicum sharing the most common ancestors with the C.ser.Pilosa, C.kamtschaticum, and C.ramosum (BS = 94%). The phylogenetic relationships among the three subclades were not fully resolved, and the C.ser.Pilosa. was not monophyletic, embedding C.kamtschaticum. Furthermore, the series Alternifolia was monophyletic in the CP tree but not in the ITS tree. Both trees strongly supported the monophyly of C.insularis (BS = 95% in ITS, BS = 96% in CP), and it shared the most common ancestor with C.alternifolium distributed in Japan (BS = 98% in ITS, BS = 77% in CP). The phylogenetic trees revealed that C.insularis formed an independent monophyletic clade from closely related taxa (i.e., C.japonicum and C.alternifolium), suggesting the newly recognized species of Chrysosplenium (Fig. 5).
Figure 5.
Phylogenetic tree of Chrysospleniuminsularis and related taxa based on ITS regions and combined CP regions (matK and rbcL) A ITS region B combined CP regions (matK and rbcL). The numbers above the branches are bootstrap values (BS > 50%) by the maximum likelyhood method. Newly generated sequences in this study are shown with an asterisk, and the new species are marked with a red box. The voucher information of all samples used in the analysis is indicated after the scientific names.
Additional specimens examined.
Chrysospleniuminsularis (Paratypes): Korea • Jeonnam, Sinan-gun, Heuksan-myeon, Gageodo-ri; 14 May 2023; K.H.Lee 230514-1 (KH). • Jeju, Seogwipo-si, Hogeun-dong; 28 Apr. 2020; PBK0118-001 (KH). • Jeju, Seogwipo-si, Hogeun-dong; 22 Mar. 2022; Hanon-220322-011 (KH) • Jeju, Seogwipo-si, Hogeun-dong; 22 Mar. 2023; J.E.Jang et al. 230322-1 (KH).
Chrysospleniumjaponicum: Korea • Gyeonggi, Gwangju-si, Chowol-eup, Mugap-ri, Mugapsan; 24 Apr. 2007; HNHM-A-158 • Gwangju-si, Toechon-myeon, Cheonjinam; 7 Apr. 2000; KNAH014041 • Gwangju-si, Toechon-myeon, Usan-ri, Aengjabong; 11 Apr. 2004; kjs040141 (KH) • Incheon-si, Ongjin-gun, Jawoldo Isl.; 8 Apr. 2009; NAPI-2009-1214 (KH) • Incheon-si, Ongjin-gun, Deokjeok-myeon, Mungap-ri, Gitdaebong; 9 Apr. 2014; Park140230 (KH) • Incheon-si, Ganghwa-gun, Ganghwado Isl.; 20 Apr. 2006; LeeGH6-35 (KH) • Gyeonggi, Namyangju-si, Onam-eup, Cheonmasan; 17 Apr. 2009; ParkSH90273 (KH) • Gyeonggi, Namyangju-si, Onam-eup, Palhyeon-ri; 25 May 2023; J.E.Jang 230325-1 (KH) • Gyeonggi, Namyangju-si, Joan-myeon, Ungilsan; 11 Apr. 2009; Y.M.Kang s.n. (KH) • Gyeonggi, Gwacheon-si, Makgye-dong, Cheonggyesan; 8 Apr. 2006; KHUS20110475 (KH) • Gangwon, Pyeongchang-gun, Yongpyeong-myeon, Jaesan-ri, Geumdangsan; 17 Apr. 2012; JSY120434 (KH) • Jeongseon-gun, Imgye-myeon; 23 Apr. 2011; 0307013 (KH) • Gangwon, Taebaek-si, Hasami-dong, Deokhangsan; 23 Apr. 2005; kjs050052 (KH) • Gangwon, Wonju-si, Panbu-myeon, Geumdae-ri; 20 Apr. 2023; S.R.Lee et al. 230420-1 (KH) • Chungbuk, Danyang-gun, Danyang-eup, Suchon-ri, Sobaeksan; 17 Apr. 2005; Sobaeksan-050417-070 (KH) • Chungbuk, Chungju-si, Sotae-myeon, Boktan-ri; 12 Apr. 2012; Namhan-548 (KH) • Gyeongbuk, Gunui-gun, Bugye-myeon, Dongsan-ri, Palgongsan; 22 Apr. 2006; CBU-070308 (KH) • Gyeongbuk, Bonghwa-gun, Myeongho-myeon, Bugok-ri, Cheongnyangsan; 27 Mar. 2006; CBU-070519 (KH) • Chungbuk, Cheongsong-gun, Hyeonseo-myeon, Bohyeonsan; 22 Apr. 2006; K.O.Yoo s.n. (KH) • Jeonbuk, Namwon-si, Ayeong-myeon, Gusang-ri, Bonghwasan; 1 May 2007; HNHM-A-283 (KH).
Key to the species of Chrysosplenium in South Korea modified from Choi et al. (2020)
| 1a | Cauline leaves alternate | 2 |
| 2a | Leaves heterophyllous; sterile branches developed; caluline and bracteal leaves 2–5 lobed | C.flagelliferum |
| 2b | Leaves isophyllous; sterile branch absent; cauline and bracteal leaves not lobed with 8–12 teeth | 3 |
| 3a | Sepals green; stamens 4 | C.insularis |
| 3b | Sepals yellowish green or golden yellow; stamens 8 | 4 |
| 4a | Stolons present; sepals golden yellow; seed surface smooth | C.alternifolium |
| 4b | Stolons absent; sepals yellowish green; seed surface papillose | C.japonicum |
| 1b | Cauline leaves opposite | 5 |
| 5a | Sepals green, spreading; capsules cup-shaped | C.ramosum |
| 5b | Sepals yellow, erect; capsules horn-shaped | 6 |
| 6a | Plants glabrous | 7 |
| 7a | Stamens 4 (-6); cylindrical papillae with roundish head at the tip on smooth seed | C.grayanum |
| 7b | Stamens 8; cylindrical papillae with truncate tip on scabrous seed surfaces | 8 |
| 8a | Sterile branches present; plant glabrous except petiole of sterile branches; stamens shorter than the sepals | C.sinicum |
| 8b | Sterile branches absent; plant glabrous; stamens longer than the sepals | C.macrostemon |
| 6b | Plants pubescent | 9 |
| 9a | Seeds without tubercules | 10 |
| 10a | Leaves of sterile branches congested at the distal end, with white variegated veins on the upper surface | C.flaviflorum |
| 10b | Leaves of sterile branches distantly arranged, with silvery dotted upper surface | C.epigealum |
| 9b | Seeds with tubercules | 11 |
| 11a | Seed tubercles arranged on inconspicuous longitudinal ridges | 12 |
| 12a | Sterile branches highly branched, ca. 30 cm long after fruiting; leaves of sterile branches with silvery dots, upper surface glabrous; bracteal leaves yellowish-green | C.ramosissimum |
| 12b | Sterile branches unbranched, less than 15 cm long after fruiting; leaves of sterile branches without silvery dots, upper surface pilose; bracteal leaves bright yellow | C.valdepilosum |
| 11b | Seed tubercles arranged on prominent longitudinal ridges | 13 |
| 13a | Leaves of sterile branches distantly arranged after fruiting; bracteal leaves golden yellow, greenish yellow at flowering | C.aureobracteatum |
| 13b | Leaves of sterile branches congested at the distal end after fruiting; bracteal leaves green at flowering | C.barbatum |
Supplementary Material
Acknowledgements
We greatly thank Seokmin Yoon and Jin Suk Kim for providing information about this new species and respect their passion for exploring nature.
Citation
Jang JE, Park BK, Lee K-H, Kim H-J, Gil H-Y (2024) Description and phylogenetic position of a new species, Chrysosplenium insularis J.E.Jang, K.H.Lee & H.Y.Gil (Saxifragaceae), from the southern islands of South Korea. PhytoKeys 248: 91–104. https://doi.org/10.3897/phytokeys.248.131291
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This study was financially supported by the research projects of the Korea National Arboretum [KNA 1-1-13, 14-1].
Author contributions
Conceptualization: KL. Resources: BKP. Visualization: KL, BKP, and JEJ. Supervision: HG. Writing the original draft: JEJ. Writing, review, and editing: JEJ, KL, HK, and HG.
Author ORCIDs
Ju Eun Jang https://orcid.org/0000-0002-6229-8477
Beom Kyun Park https://orcid.org/0000-0003-1894-3296
Kang-Hyup Lee https://orcid.org/0000-0002-7189-3235
Hyuk-Jin Kim https://orcid.org/0000-0002-3177-2914
Hee-Young Gil https://orcid.org/0000-0003-3714-0827
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Choi JE, In KH, Kim BS, Kim K, Kim JS, Kim YI, Lee BY, Lim CE. (2020) A new distribution record of Chrysospleniumgrayanum Maxim.(Saxifragaceae) in Korea: Evidence from morphological and molecular data. Journal of Species Research 9(1): 46–55. 10.12651/JSR.2020.9.1.046 [DOI] [Google Scholar]
- Chung YH, Kim YD. (1988) Monographic Study of Endemic Plants in Korea X. Taxonomy and Interspecific Relationship of the Genus Chrysosplenium. Korean Journal of Environmental Biology 6(2): 33–63.
- Chung GY, Jang H, Chang KS, Choi HJ, Son DC. (2023) A checklist of endemic plants on the Korean Peninsula II. Korean Journal of Plant Taxonomy 53(2): 79–101. 10.11110/kjpt.2023.53.2.79 [DOI] [Google Scholar]
- Deng JB, Drew BT, Mavrodiev EV, Gitzendanner MA, Soltis PS, Soltis DE. (2015) Phylogeny, divergence times, and historical biogeography of the angiosperm family Saxifragaceae. Molecular Phylogenetics and Evolution 83: 86–98. 10.1016/j.ympev.2014.11.011 [DOI] [PubMed] [Google Scholar]
- Fay MF, Bayer C, Alverson WS, de Bruijn AY, Chase MW. (1998) Plastid rbcL sequence data indicate a close affinity between Diegodendron and Bixa. Taxon 47(1): 43–50. 10.2307/1224017 [DOI] [Google Scholar]
- Franchet AR. (1890) Monographie du genere Chrysosplenium Tourn. Nouvelles Archives du Muséum d’Histoire Naturelle, Série 3 2: 87–114.
- Fu LF, Liao R, Lan DQ, Wen F, Liu H. (2020) A new species of Chrysosplenium (Saxifragaceae) from Shaanxi, north-western China. PhytoKeys 159: 127–135. 10.3897/phytokeys.159.56109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu LF, Yang TG, Lan DQ, Wen F, Liu H. (2021) Chrysospleniumsangzhiense (Saxifragaceae), a new species from Hunan, China. PhytoKeys 176: 21–32. 10.3897/phytokeys.176.62802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H. (1957) Synopsis of genus Chrysosplenium L. Journal of the Faculty of Science, University of Tokyo, Section III. Botany 7: 1–90. [Google Scholar]
- Hsu TC, Chung SW, Cheng YC. (2011) Chrysospleniumjaponicum (Saxifragaceae), Newly Recorded from Taiwan. Taiwania 56(4): 337–340. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature methods 14(6): 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Kim YD. (2015) Chrysospleniumaureobracteatum (Saxifragaceae), a new species from South Korea. Novon: A Journal for Botanical Nomenclature 23(4): 432–436. 10.3417/2013018 [DOI] [Google Scholar]
- Kim YI, Cho SH, Lee JH, Kang DH, Park JH, Kim YD. (2018) Chrysospleniumramosissimum Y.I.Kim & Y.D.Kim (Saxifragaceae), a new species from Korea. PhytoKeys 111: 1–10. 10.3897/phytokeys.111.27182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Shin JS, Lee S, Chen JH, Choi S, Park JH, Kim YD. (2019) A new species of Chrysosplenium (Saxifragaceae) from Northeastern China. PhytoKeys 135: 39–47. 10.3897/phytokeys.135.39036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korea National Arboretum (2021) Checklist of Vascular Plants in Korea (Native Plants). Korea National Arboretum, 1006 pp.
- Liu H, Luo J, Liu Q, Lan D, Qin R, Yu X. (2016) A new species of Chrysosplenium (Saxifragaceae) from Zhangjiajie, Hunan, central China. Phytotaxa 277(3): 287–292. 10.11646/phytotaxa.277.3.7 [DOI] [Google Scholar]
- Lozina AS. (1939) Chrysosplenium L. In: Komarov VL, Yuzepchuk SV. (Eds) Flora of the U.S.S.R, Vol. 9. Botanical Institute of Academy of Science, Leningrad, 156–158.
- Minh BQ, Nguyen MA, von Haeseler A. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30(5): 1188–1195. 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Wakabayashi M, Ono M, Murata J. (1997) Molecular phylogenetic analysis of Chrysosplenium (Saxifragaceae) in Japan. Journal of Plant Research 110(2): 265–274. 10.1007/BF02509315 [DOI] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh BU, Ko SC, Kang SH, Paik WK, Yoo KO, Im HT, Jang CG, Chung GY, Choi BH, Choi HJ, Lee YM, Shin CH, Choi K, Han JS, Park SH, Kim HJ, Chang KS, Yang JC, Jung SY, Lee CH, Oh SH, Jo DG. (2016) Distribution Maps of Vascular Plants in Korea. Korea National Arboretum, Pocheon, 216 pp. [Google Scholar]
- Pan JT, Ohba H. (2001) Chrysosplenium. In: Wu ZY, Raven PH. (Eds) Flora of China, Vol.8. Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis, 346–358.
- Qin R, Lan DQ, Huang W, Liu H. (2018) Research progress on chemical constituents and pharmacological activities of Chrysosplenium spp.(Saxifragaceae). Journal of South-Central University for Nationalities 37: 54–59. [Natural Science Edition] [Google Scholar]
- Soltis DE, Kuzoff RK, Conti E, Gornall R, Ferguson K. (1996) matK and rbcL gene sequence data indicate that Saxifraga (Saxifragaceae) is polyphyletic. American Journal of Botany 83(3): 371–382. 10.1002/j.1537-2197.1996.tb12717.x [DOI] [Google Scholar]
- Soltis DE, Nakazawa MT, Xiang QY, Kawano S, Murata J, Wakabayashi M, Jetter CH. (2001) Phylogenetic relationships and evolution in Chrysosplenium (Saxifragaceae) based on matK sequence data. American Journal of Botany 88(5): 883–893. 10.2307/2657040 [DOI] [PubMed] [Google Scholar]
- Wakabayashi M. (2001) Chrysosplenium L. In: Iwatsuki K, Boufford DE, Ohba H. (Eds) Flora of Japan, Vol.IIb. Kodansha Ltd., Tokyo, 58–70.
- Wakabayashi M, Takahashi H, Tomita S. (2018) Chrysospleniumsuzukaense (Saxifragaceae), a New Species from Yoro and Suzuka Mts., Central Honshu, Japan. Acta Phytotaxonomica et Geobotanica 69(1): 41–51. [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic Press, San Diego, California, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular ecology resources 20(1): 348–355. 10.1111/1755-0998.13096 [DOI] [PubMed] [Google Scholar]
- Zhao J, Qiu X, Zhao Y, Wu R, Wei P, Tao C, Wan L. (2022) A review of the genus Chrysosplenium as a traditional Tibetan medicine and its preparations. Journal of Ethnopharmacology 290: 115042. 10.1016/j.jep.2022.115042 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.