Abstract
The prevalence of resistance to harmful human pathogens is steadily rising, emphasizing the urgent need to identify novel antimicrobial compounds. For this purpose, plants stand out as a significant source of bioactives worthy of exploration. Among these, alkaloids, a vast and structurally diverse category of plant secondary metabolites, have emerged as a foundation for crucial antibacterial medications such as metronidazole and the quinolones. In the current work, the crude methanol leaf extract of Psammogeton biternatum Edgew collected from District Bannu, Pakistan, was subjected to TLC (indirect) bioautography and X-ray crystallography for the isolation of potential antibacterial agents. From the crude extract, a novel quinoline alkaloid called quinoline dione ((3R,3aS,5aR)-3,5a,9-trimethyl-3a,4,5,5a-tetrahydro-2H-isoxazolo[2,3-a] quinoline-2,8(3H)-dione (C14H17NO3)) was isolated. The crystal information (M = 247.296 g/mol) is as follows: orthorhombic, P212121, a = 7.7339(14) Å, b = 10.7254(19) Å, c = 15.730(2) Å, V = 1304.8(4) Å3, Z = 4, T = 296 K, μ(Mo Kα) = 0.088 mm–1, ρ calc = 1.259 g/cm3, 13928 reflections measured (5.86° ≤ 2Θ ≤ 51.98°), 2478 unique (Rint = 0.1613, Rσ = 0.1335). The final R1 was 0.1098 (I ≥ 2u(I)), and wR2 was 0.2183. The antibacterial activity for both crude extract of leaves and quinoline dione was determined by a well diffusion method. The quinoline dione alkaloid demonstrated excellent inhibition zones against methicillin-resistant Staphylococcus aureus (18 mm), Bacillus subtills (17 mm), Escherichia coli (20 mm), and Pseudomonas aeruginosa (23 mm) compared to the crude extract. The antibiofilm potential was recorded against Pseudomonas aeruginosa by the 96-well microtiter plate method. A dose-dependent biofilm inhibition response was recorded, which increased with the increase in concentration. Moreover, quinoline dione showed a greater antibiofilm effect as compared to the crude extract, which may be linked to the presence of a particular active functional group positioned on the compound isolated in its pure form. Through in silico studies, i.e., molecular docking, quinoline dione shows strong binding energies with the LasR transcriptional regulator (6MVN) at −9.3 and LasR transcriptional activator (3IX4) at −9.2 kcal/mol, as well as moderate affinities with other targets such as AHL synthase LasI (PDB ID 1RO5) and OprM channel (PDB ID 3D5K), indicating its potential as a quorum sensing inhibitor. Thus, the antibacterial and antibiofilm potential of quinoline dione was confirmed.
Introduction
Medicinal plants have been used for years and long before the global use of antibiotics to effectively treat different human ailments, and a number of plants have been tested for their potential antimicrobial activity.1 Plant-based natural products act as a rich source of bioactive compounds, which are characterized by their chemical and structural diversity and offer numerous opportunities for the discovery of novel drugs.2,3 Plants contain active metabolites including alkaloids, terpenoids, flavonoids, and phenolic derivatives, which serve as direct precursors in the pharmaceutical industry. It is presumed that plant extracts display diverse target sites and will show activity against drug-resistant microbial pathogens.4
It is now an established fact that the presence of antimicrobial resistance (AMR) poses a high risk to the general health of both humans and animals.5 According to estimation, the annual death toll at the present time is about 700 000 people, which is mainly due to antibiotic resistance; if no efforts are taken to stop the rising AMR, by 2050, approximately 10 million people are at high risk.6 Biofilm formation is one of the important strategies of resistance used by bacteria, which makes them more resistant to antibiotics as compared to their planktonic counterparts.7
A biofilm is a complex matrix where free-floating (planktonic) bacterial cells fixed together in a gel-like structure called extracellular polymeric substances that consist of proteins, polysaccharides, nucleic acids, and other organic elements.8 Biofilms enable bacteria to tightly bind to biotic and abiotic surfaces and develop resistance to host defenses and antibacterial agents.9 The transformation of bacteria into biofilms form may add up to 1000-fold more resistance to commonly used antibacterial agents compared to bacteria in a planktonic mode.10 It is estimated that about 60% microbial-induced infections occur due to biofilms, and in humans about two-thirds of bacterial infections are the result of biofilms.11
The screening of nontargeted and effect-directed techniques has presented novel opportunities to streamline the drug discovery process, reducing both time and financial costs. It helps to find active antimicrobial compounds irrespective of their complexity and ensures a series of systematic processes including characterization, activity-based identification, and isolation of compounds.12 Quinoline derivatives, particularly alkaloids such as 3,3-dibenzyl-4-hydroxy-3,4-dihydro-1H-quinolin-2-ones, 6,7-sustituted 5,8-quinolinequinone, and others are noteworthy due to their diverse medicinal properties and renowned for their potent antimicrobial and other biological activities.2,13 In addition, some hydrazone derivatives possessing a quinoline moiety are renowned for their potent antimicrobial and other biological properties.14
While exploring natural products, medicinal plants are an important prime source. There are approximately 1572 genera and nearly 6000 wild plant species, predominantly concentrated in the Himalaya, Hindukush, and Karakorum regions. Among these, around 600 species have been meticulously catalogued for their medicinal attributes. Consequently, there is a pressing need for comprehensive research to harness the potential of these resources, as only a limited number of medicinal plants in Pakistan have been investigated for their potential antimicrobial activities.4,15
The present targeted plant is Psammogeton biternatum Edgew. (P. biternatum), a common herb belonging to the genus Psammogeton, which consists of 6–8 species distributed across Pakistan, Afghanistan, Iran, and India. It is locally known as izbotk and is traditionally used for the treatment of malaria, cough, typhoid, and chest problems.16 Despite its potential medicinal properties, this species remains largely unexplored, both pharmacologically and phytochemically. This highlights the need for a comprehensive study to uncover the potential bioactive compounds within this plant species and address the existing void in the scientific literature.
The antibacterial and antibiofilm potential of P. biternatum leaf extract was investigated. Furthermore, a novel quinoline dione compound was isolated (Figure 1) using a TLC-based autobiography technique, identified using X-ray crystallography, and evaluated for its antibacterial potential both in vitro and using an in silico approach. This study not only emphasizes the importance of medicinal plants in modern drug discovery but also highlights the methodologies that can discover new bioactive molecules with significant medical potential.
Figure 1.
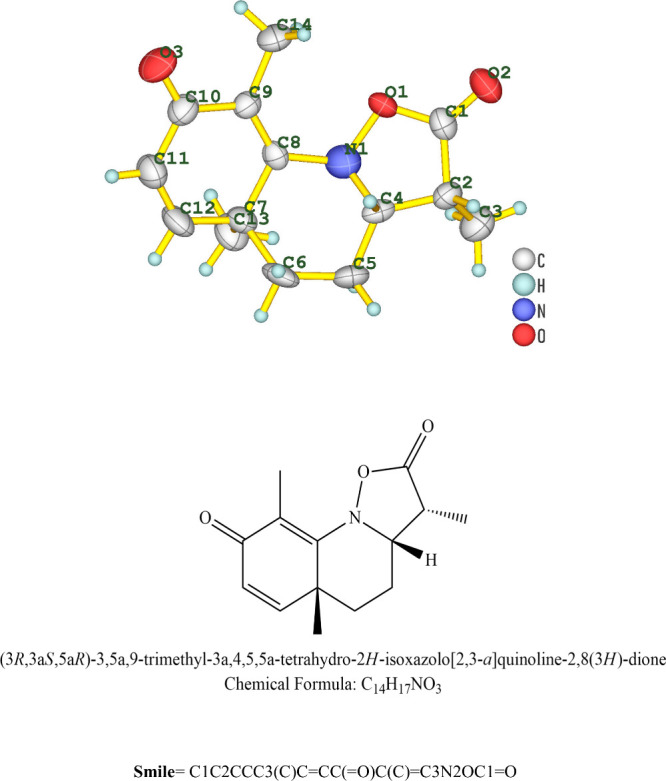
Structure and chemical formula of quinoline dione.
Materials and Methods
Plant Collection and Identification
The study was performed in the phytochemistry laboratory of the Department of Botany, Hazara University Mansehra, Pakistan. P. biternatum Edgew. was collected from different parts of District Bannu and identified at the department of Botany, Hazara University, and a dried specimen (voucher number VN52292) was deposited in the university herbarium.
Plant Extract Preparation
The leaves of the selected plant were collected and dried under shade for 2 weeks, and the dried plant was then chopped and ground to a coarse powder form. The powder (350 g) was then soaked in methanol (1 L) for 3 days with constant vigorous daily stirring. The extract was then filtered through Whatman (41) filter paper, and the filtrate was concentrated under reduced pressure to avoid thermal decomposition on rotary evaporator at 43 °C. The process was repeated three times to get the complete crude extract, which was then stored in the refrigerator for future activities.17
Media and Bacterial Strains
The bacterial strains used were methicillin-resistant Staphylococcus aureus (MRSA), Bacillus subtills (Gram-positive), and Pseudomonas aeruginosa (PAO1) and Escherichia coli (Gram-negative). The turbidity standard of 0.5 McFarland, which is equal to 1.5 × 108 colony forming units (CFU)/mL, was maintained on nutrient agar (NA) at 4 °C. The bacterial strains were cultured in nutrient broth (NB) media and incubated at 37 °C for 24 h.18
TLC Bioautography
To isolate the plant bioactive compound, the plant extract was dissolved in methanol and loaded on precoated silica gel TLC plates.19 Four TLC plates (Merk, silica gel 60 F254, 20 × 20 cm) were cut in equal width (20 mm) and length (70 mm), and 5 μL (5 mg/mL) of the crude extract was loaded onto TLC plates on the horizontal application line and eluted using an ethyl acetate/acetone (3:1) mobile solvent system. The developed plates were dried using forced air drying to remove solvent traces on the plates. After drying, all the spots on plates were seen under visible (white), short UV (254 nm), and long UV (366 nm) lights. The developed plates were then placed aseptically onto the respective bacterial strains swabbed onto nutrient agar Petri plates and incubated overnight at 37 ± 2 °C. The movement of the extract separating spot was expressed by its retention factor:
where the distance travelled by the solute is the distance travelled from the point of application to the center of the spot.
Purification of Quinoline Dione and X-ray Crystallography
After confirming that the active zones are around 0.6 and 0.9, preparative TLC was employed for the isolation of the active compounds using the same solvent system of ethyl acetate/acetone (3:1). After repetitive development of preparative TLC, the silica along with the active region was scraped from the TLC plate based on its Rf value, i.e., 0.6 or 0.9. The scraped region was then soaked in the same solvent system for 4–5 min and subsequently washed with methanol to remove impurities. Upon filtering the silica and solvent evaporation, pure white crystals (11 mg) were obtained from the 0.6 region and oily substances were obtained from the 0.9 region. Only the crystals from the 0.6 region were then subjected to compound identification using a Bruker APEX-II CCD diffractometer at the Scientific and Technological Research Application and Research Center, Sinop University, Turkey. The crystal was maintained at 296 K during data collection. X-ray beams were directed at the crystals, and the X-rays were diffracted by the crystal’s atoms in multiple directions. The angle and intensity of the diffracted beams were used to create a 3D electron density map. The structure was solved using the Olex2 software with charge flipping and refined with olex2.refine.20,21
Refinement Model
The constraints (0) and restraint numbers (26) were used in the refinement process using OLex2-1.5.20 The rest of the details used were as follows: twinned data refinement scales, 2(4)–1(4); fixed Uiso at 1.2 times of all C(H) groups and all C(H,H) groups; fixed Uiso at 1.5 times of all C(H,H,H) groups. Refinement of riding coordinates: ternary CH groups, C2(H2), C4(H4); secondary CH2 groups, C5(H5a,H5b), C6(H6a,H6b); aromatic/amide H groups, C11(H11), C12(H12); idealized Me groups refined as rotating group, C3(H3a,H3b,H3c), C13(H13a,H13b,H13c), C14(H14a,H14b,H14c).20
Agar Well Diffusion Assay
The antibacterial potential of P. biternatum crude leaves extract was evaluated by the agar well diffusion method. Nutrient agar plates (NAP) were prepared by pouring 75 mL of nutrient agar into a Petri plate and incubating the sample overnight at 37 °C. After incubation, sterile cotton swabs were dipped in the bacterial suspension of each inoculum (0.5 McFarland) and gently rubbed on the surface of NAP to evenly spread the bacteria. Then, wells (6 mm) were made with the help of a cork borer in the seeded agar plates, and 100 μL of extract was transferred into each well. The plates were then incubated at 37 °C for 24 h. The bacterial inhibition was noted by recording the diameter of inhibition zone (IZ) in millimeter (mm). Since the crude extract was dissolved in dimethyl sulfoxide (DMSO), pure DMSO (100%) was added in one of the well as a negative control and antibiotics (tetracycline) were used as a positive control, and each assay was performed in triplicate.22
Biofilm Inhibition Assay
A modified biofilm formation assay was carried out using a 96-well microtiter plate method. For biofilm formation, P. aeruginosa was grown overnight and diluted (1:100) into nutrient broth media. For the antibiofilm assay, M63 medium supplemented with 0.2% glucose, 0.4% arginine, 1 mM MgSO4·7H2O, and salts was used. 90 μL of M63 medium and extract was pipetted into the 96-well microtiter plate; twofold dilution was done, and 10 μL of the normalized bacterial suspension was added. The plates were then kept in the incubator for 24 h at 37 °C. After incubation, the plate was washed with distilled water to discard any planktonic cells. After drying, 125 μL of 0.1% crystal violet dye was used as staining adherent and added into each well. To resolubilize the adherent biofilm cells, 130 μL of 30% acetic acid was added into each well, and the plate was then analyzed using a microplate absorbance reader at 550 nm for biofilm inhibition. M63 medium without inoculum served as a negative control, while M63 medium plus inoculum served as a positive control. All assays were done in triplicate.23
In Silico Antibacterial Evaluation
Preparation of Receptors and Ligands for Redocking and Docking
The identified compound was converted into PDB formation using the PDB Web site. The protein/targets LasR transcriptional activator (PDB ID 3IX4), AHL synthase LasI (PDB ID 1RO5), OprM channel (PDB ID 3D5K), and LasR transcriptional regulator (PDB ID 6MVN) were prepared by removing water molecules, and heteroatoms and missing hydrogen were added, followed by the addition of Kollman charges and Ganister charges using Dock prep of ChimeraX.24 Among the targeted proteins, 3IX4 and 6MVN were used for redocking, as these proteins were experimentally cocrystallized with their inhibitors.25 The ligands were extracted from their complex form using Discovery Studio 2024, and the respective chains were separated for redocking purposes while following the same docking procedure. For docking with other ligands, the grid box was set as center (x = 5.0759, y = 11.3238, z = 2.158, spacing of 0.375 Å) with a size of x = y = z = 25 for 3IX4 (chain A); center (x = 41.1649, y = −10.3846, z = −13.6178) with a size of x = 50.322239399, y = 45.9206489944, and z = 46.2245800018 for 1RO5; center (x = −21.0983, y = −2.115, z = −46.8884) with a size of x = 73.5098215103, y = 76.8415097427, and z = 139.539057007 for 3D5K; and center (x = 6.9568, y = −13.9221, z = −38.4412) with a size of x = y = z = 25 for 6MVN (chain A). Exhaustiveness was equal to 8. The same parameters were recorded from the configuration file, ligand, and protein were converted to pdbqt file format, and molecular docking was carried out using the Python Prescription 0.8 interface with the prescribed codes of autodock vina 1.2.5 win.26 The ligand molecules were downloaded from the PubChem Web site, while the quinoline dione was prepared using Discovery Studio 2024.
Results and Discussion
TLC Bioautography
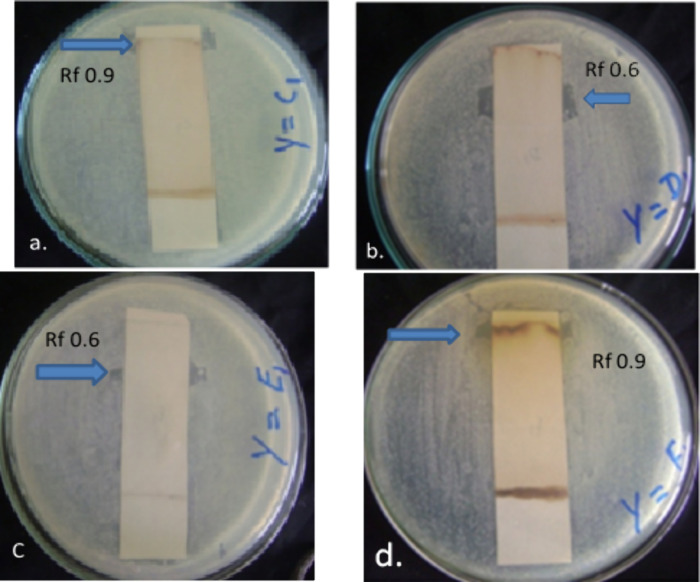
P. biternatum Edgew leaf extract was subjected to TLC and contact bioautography (indirect) assays to identify the active antibacterial agent against MRSA (C1), Pseudomonas aeruginosa (D1), Escherichia coli (E1), and Bacillus subtilis (F1), respectively. TLC was initially performed as a qualitative method to document the extract constituents, and contact bioautography was used to determine the antibacterial activity of the compounds separated by TLC. More than about 25% of conventional medicines have been being derived from active plant metabolites directly or indirectly.27 The bacterial inhibition was shown by representing active bands (ABs) with their respective Rf values (0.9 and 0.6) on the TLC chromatogram as shown in Figure 2a–d and Table 1. A strong antibacterial effect was observed against Pseudomonas aeruginosa and E. coli on AB with Rf values of 0.6 each as revealed by the clear inhibition zone formation (Figure 2b and c). The other AB was observed against MRSA and Bacillus subtilis each with an Rf value of 0.9 as shown in Figure 2a and d, respectively. A potent antibacterial potential has been recorded for a number of herbal plants against different drug-resistant bacterial strains, which provides a valuable source for the isolation of antimicrobial agents.28 This property of medicinal plants can help greatly if exploited properly to eradicate the emergence of multidrug-resistant bacterial strains.29
Figure 2.
Antibacterial activity of P. biternatum against tested bacterial strains: (a) MRSA, (b) Pseudomonas aeruginosa, (c) Escherichia coli, (d) Bacillus subtilis.
Table 1. Active Bands with Rf values against tested strains.
| bacterial strains | active band/Rf values |
|---|---|
| MRSA (C1) | 0.9 |
| P. aeruginosa (D1) | 0.6 |
| E. coli (E1) | 0.6 |
| B. subtilis (F1) | 0.9 |
Crystal Structure Determination of Quinoline Dione
After subjecting the white crystals to X-ray crystallography, the compound was identified as an alkaloid quinoline dione. The crystallographic data for C14H17NO3 (M = 247.296 g/mol) are as follows: the crystal system is orthorhombic, the space group is P212121 (no. 19), a = 7.7339(14) Å, b = 10.7254(19) Å, c = 15.730(2) Å, V = 1304.8(4) Å3, Z = 4, T = N/A K, μ(Mo Kα) = 0.088 mm–1, ρcalc = 1.259 g/cm3, 13928 reflections measured (5.86° ≤ 2Θ ≤ 51.98°), and 2478 unique (Rint = 0.1613, Rσ = 0.1335), which were used in all calculations. The final R1 was 0.1098 (I ≥ 2u(I)), and wR2 was 0.2183 (all data). The crystallographic data are given in Table 2, while the structure is given in Figure 1. The structure has been deposited at the Cambridge Crystallographic Data Centre, CCDC no. 2368888.
Table 2. Crystal Data and Structure Refinement for Quinoline Dione.
| empirical formula | C14H17NO3 |
|---|---|
| formula weight | 247.296 |
| temperature (K) | 296 |
| crystal system | orthorhombic |
| space group | P212121 |
| a (Å) | 7.7339(14) |
| b (Å) | 10.7254(19) |
| c (Å) | 15.730(2) |
| α (°) | 90 |
| β (°) | 90 |
| γ (°) | 90 |
| volume (Å3) | 1304.8(4) |
| Z | 4 |
| ρcalc (g/cm3) | 1.259 |
| μ (mm–1) | 0.088 |
| F(000) | 528.4 |
| crystal size (mm3) | 0.06 × 0.03 × 0.02 |
| radiation | Mo Kα (λ = 0.71073) |
| 2θ range for data collection (°) | 5.86 to 51.98 |
| index ranges | –9 ≤ h ≤ 9, −13 ≤ k ≤ 13, −18 ≤ 1 ≤ 19 |
| reflections collected | 13928 |
| independent reflections | 2478 [Rint = 0.1613, Rσ = 0.1335] |
| data/restraints/parameters | 2478/0/167 |
| goodness-of-fit on F2 | 1.158 |
| final R indexes [I ≥ 2σ (I)] | R1 = 0.1098, wR2 = 0.1811 |
| final R indexes [all data] | R1 = 0.1961, wR2 = 0.2183 |
| largest diff. peak/hole (e/Å3) | 0.80/–0.75 |
| Flack parameter | 0.3(18) |
As given in Table 2, the orthorhombic space group P212121 is frequently encountered in organic and pharmaceutical compounds, indicating a common structural motif that contributes to stability and functionality. Similar crystallographic characteristics are observed in various alkaloids and synthetic organic molecules used in medicinal chemistry.30 The unit cell dimensions and volume are crucial for comparing structural similarities with compounds of related structures. These parameters are consistent with the dimensions found in the literature for molecules of similar size and composition, suggesting typical packing arrangements and intermolecular interactions.31 The calculated density and low absorption coefficient are in line with those of organic compounds, which generally have lower densities compared to inorganic materials. This consistency supports the validity of the structural data and the refinement process. The R indexes (R1 and wR2) and goodness-of-fit values are essential for evaluating the quality of the crystal structure. The values obtained are comparable to those reported in other studies involving complex organic molecules, demonstrating that the refinement is within acceptable limits and that the structure is reliable. The literature often cites similar R values for well-refined structures, affirming the accuracy of our model.32
The Flack parameter is particularly relevant for determining the absolute configuration of chiral molecules. A parameter close to 0.3, as seen, is often reported in the literature for partially resolved noncentrosymmetric structures. This is significant for understanding the stereochemistry and potential biological activity of the compound.33 The size of the crystal and the range of data collection (2θ range) are typical for high-quality crystallographic analysis, as seen in numerous studies. The chosen crystal size ensures sufficient diffraction without excessive absorption, aligning with standard practices in crystallography literature.34
As given in Table 3, the atomic coordinates (x, y, z) specify the positions of the atoms within the unit cell in fractional coordinates. Each value represents the atom’s location as a fraction of the unit cell dimensions along the respective axis. These coordinates are crucial for defining the geometry and bonding environment of the crystal structure. Equivalent isotropic displacement parameter (Ueq) values provide information about the thermal vibrations of the atoms. Higher Ueq values indicate greater atomic displacement, suggesting increased thermal motion or possible disorder at that atomic site. Atoms like C3 (Ueq = 65) and N1 (Ueq = 68.8) show relatively higher displacement parameters, which could be due to thermal motion or less well-defined positions. The variation in Ueq indicates differences in thermal motion among the carbon atoms, which can be influenced by their bonding environment and positional disorder. The positions and displacement parameters of the aromatic carbons (e.g., C8, C9, C10, C11, and C12) are particularly relevant for understanding the stability and planarity of the aromatic ring system in the structure. The higher Ueq value of 68.8(17) of N1 is important for evaluating the reliability of the nitrogen position and its involvement in any hydrogen bonding or coordination. O1, O2, and O3 generally exhibit higher Ueq values compared to most carbon atoms, indicating substantial thermal motion at these sites, which is typical for oxygen atoms due to their involvement in hydrogen bonding or lone pair interactions. The variability in Ueq values aligns with the typical thermal motion observed in small-molecule crystallography, as reported in studies of comparable compounds.35
Table 3. Fractional Atomic Coordinates (x, y, z × 104), and Equivalent Isotropic Displacement Parameter Ueq (Å2 × 103) of Quinoline Dione.
| atom | x | y | z | Ueq |
|---|---|---|---|---|
| C1 | 8727(8) | 11727(5) | 10237(4) | 46.5(15) |
| C2 | 9371(7) | 11990(5) | 9350(4) | 44.6(15) |
| C3 | 8508(9) | 13152(6) | 9003(4) | 65(2) |
| C4 | 8933(7) | 10769(5) | 8923(3) | 39.5(14) |
| C5 | 8659(9) | 10680(6) | 7970(4) | 55.7(18) |
| C6 | 8274(9) | 9309(6) | 7765(4) | 57.0(18) |
| C7 | 6749(9) | 8757(5) | 8288(4) | 45.6(16) |
| C8 | 6971(7) | 9001(4) | 9244(3) | 36.5(13) |
| C9 | 6923(7) | 8105(5) | 9824(4) | 38.4(13) |
| C10 | 6855(8) | 6782(6) | 9575(4) | 50.2(17) |
| C11 | 6754(10) | 6519(6) | 8669(4) | 57.9(18) |
| C12 | 6693(9) | 7404(6) | 8090(4) | 59.2(19) |
| C13 | 5006(9) | 9312(6) | 7999(4) | 65(2) |
| C14 | 6904(9) | 8308(5) | 10780(3) | 59.4(19) |
| N1 | 7274(8) | 10379(5) | 9371(4) | 68.8(17) |
| O1 | 7597(5) | 10768(3) | 10248(2) | 44.8(10) |
| O2 | 9059(6) | 12283(4) | 10881(3) | 64.6(14) |
| O3 | 6904(7) | 5944(4) | 10093(3) | 73.9(14) |
As given in Table 4, the Uij values for carbon atoms (C1–C14) exhibit varying degrees of anisotropy. For instance, C3 has relatively high U11 (82) and U33 (62), indicating significant thermal motion along these directions. C6 shows pronounced anisotropy with U11 (85) and U22 (65) compared to U33 (21), suggesting that it is more displaced along the a- and b-axes. The U12, U13, and U23 values are generally smaller, indicating less cross-axis displacement, which is typical for stable molecular structures. The nitrogen atom (N1) has high Uij values, particularly U11 (93), indicating substantial anisotropic thermal motion. This could be due to the nitrogen’s involvement in potential intermolecular interactions or lone pair effects. O1, O2, and O3 generally have higher Uij values compared to carbon atoms. O3, in particular, has significant anisotropy with U11 (105) and U33 (68), reflecting substantial thermal motion and possible involvement in hydrogen bonding. The cross-axis displacement parameters (U12, U13, amd U23) for oxygen atoms are generally more noticeable due to their higher electron density and nonbonding interactions, showing them to be more mobile, i.e., have greater thermal vibrations. The anisotropic displacement parameters reveal the dynamic nature of the crystal structure. The greater thermal motion is important for the understanding of the stability and reactivity of the compound. This analysis provides insight into the molecular dynamics within the crystal lattice, contributing to the broader understanding of the compound’s behavior under crystallographic conditions.
Table 4. Anisotropic Displacement Parameters (Å2 × 103) for Quinoline Dionea.
| atom | U11 | U22 | U33 | U12 | U13 | U23 |
|---|---|---|---|---|---|---|
| C1 | 48(4) | 45(4) | 46(4) | –5(3) | –5(3) | –9(3) |
| C2 | 45(3) | 48(4) | 41(4) | –11(3) | 4(3) | 5(3) |
| C3 | 82(5) | 51(4) | 62(5) | –6(4) | 5(4) | 8(3) |
| C4 | 36(3) | 48(3) | 34(3) | 1(3) | 4(3) | 13(3) |
| C5 | 71(5) | 63(4) | 32(4) | –9(4) | 14(3) | 9(3) |
| C6 | 85(5) | 65(4) | 21(3) | 5(4) | –2(3) | –6(3) |
| C7 | 58(4) | 48(4) | 31(3) | –2(3) | –4(3) | –2(3) |
| C8 | 38(3) | 34(3) | 38(3) | 3(3) | 1(3) | –8(3) |
| C9 | 39(3) | 38(3) | 38(3) | –2(3) | 0(3) | 4(3) |
| C10 | 55(4) | 43(4) | 52(5) | –4(3) | –1(3) | 7(3) |
| C11 | 79(5) | 44(4) | 51(4) | –2(3) | –5(4) | –8(3) |
| C12 | 85(5) | 57(4) | 36(4) | –2(4) | –13(4) | –12(3) |
| C13 | 73(5) | 65(4) | 58(5) | –5(4) | –24(4) | 0(4) |
| C14 | 81(5) | 55(4) | 42(4) | –8(4) | 2(4) | 14(3) |
| N1 | 93(5) | 60(4) | 53(4) | –7(3) | 1(4) | 3(3) |
| O1 | 60(2) | 48(2) | 27(2) | –6(2) | 4.0(18) | –8.0(19) |
| O2 | 87(3) | 59(3) | 48(3) | –7(2) | –7(2) | –14(2) |
| O3 | 105(4) | 49(3) | 68(3) | –3(3) | 8(3) | 18(3) |
The anisotropic displacement factor exponent takes the form: π2[h2a × 2U11 + 2hka × b × U12 + . . .] where Uij are the elements of the anisotropic displacement tensor and h, k, and l are the Miller indices.
As given inTable 5, the C–C bond lengths in quinoline dione vary from 1.316(8) Å (C11–C12) to 1.556(9) Å (C6–C7). The shorter bond lengths (e.g., C8–C9 at 1.327(7) Å and C11–C12 at 1.316(8) Å) are consistent with typical aromatic or double-bond character, indicating a partial double-bond nature within the quinoline ring system. Longer C–C bonds, such as C6–C7 at 1.556(9) Å, suggest single-bond character and potential flexibility in these parts of the molecule, which could be due to less steric hindrance or electronic effects. The C=O bond lengths, such as C1–O2 at 1.203(7) Å and C10–O3 at 1.214(6) Å, are indicative of carbonyl groups, consistent with the typical double-bond character in such functional groups. The C–O single-bond lengths, such as C1–O1 at 1.350(6) Å and N1–O1 at 1.462(6) Å, reflect the single-bond nature with partial double-bond character due to resonance or conjugation effects.
Table 5. Bond Lengths for Quinoline Dione.
| atom | atom | length (Å) | atom | atom | length (Å) |
|---|---|---|---|---|---|
| C1 | C2 | 1.509(8) | C7 | C12 | 1.485(8) |
| C1 | O1 | 1.350(6) | C7 | C13 | 1.542(8) |
| C1 | O2 | 1.203(7) | C8 | C9 | 1.327(7) |
| C2 | C3 | 1.516(8) | C8 | N1 | 1.509(7) |
| C2 | C4 | 1.510(8) | C9 | C10 | 1.472(8) |
| C4 | C5 | 1.517(8) | C9 | C14 | 1.520(8) |
| C4 | N1 | 1.523(8) | C10 | C11 | 1.455(9) |
| C5 | C6 | 1.535(9) | C10 | O3 | 1.214(6) |
| C6 | C7 | 1.556(9) | C11 | C12 | 1.316(8) |
| C7 | C8 | 1.535(8) | N1 | O1 | 1.462(6) |
The C–N bond lengths, such as C4–N1 at 1.523(8) Å and C8–N1 at 1.509(7) Å, are typical for single bonds in amide or aromatic systems. These values are consistent with the literature reports for similar compounds, indicating standard bonding interactions without significant strain or distortion. The aromatic carbon–carbon bonds, such as C8–C9 at 1.327(7) Å and C11–C12 at 1.316(8) Å, are shorter than typical single bonds, reflecting the delocalized π-electron system in the quinoline ring that contributes to the stability and planarity of the aromatic ring. Bond lengths observed in quinoline dione are in agreement with those reported for other quinoline derivatives and related aromatic compounds, e.g., the C=O bond lengths around 1.20 Å and C–N bond lengths around 1.51–1.52 Å are typical for such structures.36 The information about the bond lengths in quinoline dione gives insights into the electronic structure and potential reactivity that align with established inclinations in crystallography, supporting the validity of the structural model and providing a basis for further analysis and application.37
As given in Table 6, the carbon–oxygen–carbon angles (C1) are as follows: O1–C1–C2, 111.6(5)°; O2–C1–C2, 128.0(5)°; and O2–C1–O1, 120.4(6)°. These angles around C1 reflect a trigonal planar geometry typical of carbonyl-containing structures. The bond angle of O2–C1–C2 (128.0°) proposes a significant electron-withdrawing effect from the carbonyl group, leading to a broader angle. Carbon–carbon–carbon angles (C2, C4, C7, C9, C10) are as follows: C3–C2–C1, 110.0(5)°; C4–C2–C1, 100.1(4)°; C5–C4–C2, 121.6(5)°; N1–C4–C2, 102.8(5)°; N1–C4–C5, 108.8(5)°; C6–C5–C4, 107.1(5)°; C7–C6–C5, 113.6(5)°; and C12–C7–C6, 106.5(5)°. These angles show typical variations seen in substituted aromatic and heteroaromatic systems. The bond angles imitate both electronic and steric effects, representing the general planar structure of the aromatic system.
Table 6. Bond Angles of Quinoline Dione.
| atom | atom | atom | angle (°) | atom | atom | atom | angle (°) |
|---|---|---|---|---|---|---|---|
| O1 | C1 | C2 | 111.6(5) | C13 | C7 | C12 | 106.9(5) |
| O2 | C1 | C2 | 128.0(5) | C9 | C8 | C7 | 123.1(5) |
| O2 | C1 | O1 | 120.4(6) | N1 | C8 | C7 | 108.3(4) |
| C3 | C2 | C1 | 110.0(5) | N1 | C8 | C9 | 128.5(5) |
| C4 | C2 | C1 | 100.1(4) | C10 | C9 | C8 | 121.1(5) |
| C4 | C2 | C3 | 117.1(5) | C14 | C9 | C8 | 125.3(5) |
| C5 | C4 | C2 | 121.6(5) | C14 | C9 | C10 | 113.6(5) |
| N1 | C4 | C2 | 102.8(5) | C11 | C10 | C9 | 116.7(5) |
| N1 | C4 | C5 | 108.8(5) | O3 | C10 | C9 | 122.2(5) |
| C6 | C5 | C4 | 107.1(5) | O3 | C10 | C11 | 121.0(5) |
| C7 | C6 | C5 | 113.6(5) | C12 | C11 | C10 | 122.6(6) |
| C8 | C7 | C6 | 111.6(5) | C11 | C12 | C7 | 124.0(6) |
| C12 | C7 | C6 | 106.5(5) | C8 | N1 | C4 | 109.8(5) |
| C12 | C7 | C8 | 112.0(5) | O1 | N1 | C4 | 102.4(5) |
| C13 | C7 | C6 | 111.1(5) | O1 | N1 | C8 | 115.5(4) |
| C13 | C7 | C8 | 108.7(5) | N1 | O1 | C1 | 108.4(4) |
Nitrogen–carbon–nitrogen/carbon–oxygen–nitrogen angles are as follows: N1–C8–C7, 108.3(4)°; N1–C8–C9, 128.5(5)°; C8–N1–C4, 109.8(5)°; O1–N1–C4, 102.4(5)°; O1–N1–C8, 115.5(4)°; and N1–O1–C1, 108.4(4)°. The bond angles around N1 reflect the tetrahedral geometry typical of sp3 hybridized nitrogen.
Aromatic ring angles (C8–C9–C10, C9–C14–C8, etc.) are as follows: C9–C8–C7, 123.1(5)°; C10–C9–C8, 121.1(5)°; C11–C10–C9, 116.7(5)°; O3–C10–C9, 122.2(5)°; C12–C11–C10, 122.6(6)°; and C11–C12–C7, 124.0(6)°. The angles within the aromatic ring are close to 120°, characteristic of sp2 hybridized carbons in a benzene-like ring structure. Minor deviations are due to the presence of substituents.
The bond angles are consistent with those reported for similar aromatic- and quinoline-based structures. For instance, bond angles around sp2 hybridized carbons in aromatic systems typically range from 118° to 123°, and angles around sp3 hybridized carbons and nitrogens range from 105° to 115°, aligning well with our data. By comparing these bond angles with established data in the literature, we can conclude that the molecular geometry of quinoline dione is typical for such compounds, reinforcing the reliability of our structural data and its relevance in understanding the compound’s chemical behavior.38
The details about torsion angle are given in Table 7. The C1–C2–C4–C5 angle of 155.9(4)° indicates a near-planar arrangement with a slight deviation, which is typical in conjugated systems to reduce steric strain. The smaller C1–C2–C4–N1 angle of 34.0(5)° suggests a nonplanar conformation involving the nitrogen, which could be due to steric interactions or lone pair repulsion affecting the dihedral angle. The C1–O1–N1–C4 and C1–O1–N1–C8 angles of 26.6(4)° and 145.8(4)°, respectively, reflect the orientation of the O1–N1 bond relative to the C1 and C4/C8 atoms. The larger angle indicates a significant deviation from planarity, likely due to the spatial arrangement of the oxygen and nitrogen atoms. The C2–C4–C5–C6 angle of 179.0(6)° is nearly 180°, indicating a trans configuration, which is common in extended conjugated systems to minimize steric interactions. The negative C2–C4–N1–C8 and C2–C4–N1–O1 angles of −161.0(4)° and −37.8(4)°, respectively, suggest different orientations of the N1–C8 and N1–O1 bonds relative to the C2–C4 axis, reflecting the molecule’s flexibility. The C4–C5–C6-C7 and C4–N1–C8–C7 angles of 54.2(5)° and −62.1(5)°, respectively, indicate significant deviations from planarity, showing the influence of the substituents on the ring conformation. The torsion angles of C5–C6–C7–C8 and C5–C6–C7–C12 of −50.8(6)° and −173.3(5)°, respectively, reflect the spatial orientation of the substituents on the ring, with one nearly trans and the other significantly twisted. The C6–C7–C8–C9 and C6–C7–C8–N1 angles of −126.2(5)° and 52.6(5)°, respectively, show substantial nonplanarity, indicating a distorted ring conformation due to the substituents. The near-zero C7–C8–C9–C10 angle of 7.3(7)° suggests a nearly planar configuration, while the larger negative C7–C8–C9–C14 angle of −171.7(6)° indicates a trans configuration. The C7–C8–N1–O1 and C7–C12–C11–C10 angles of −177.2(5)° and 1.1(9)°, respectively, show that the N1–O1 bond is almost anti to the C7–C8 bond, while the C12–C11 bond is nearly planar. The C8–C9–C10–C11 and C8–C9–C10–O3 angles of −3.0(6)° and 176.0(6)°, respectively, indicate a near-planar configuration for C8–C9–C10–C11 and a trans configuration for C8–C9–C10–O3, respectively. The C9–C10–C11–C12 angle of −1.2(7)° shows a nearly planar conformation, suggesting minimal steric hindrance at this part of the molecule.
Table 7. Torsion Angles for Quinoline Dione.
| A | B | C | D | angle (°) | A | B | C | D | angle (°) |
|---|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C4 | C5 | 155.9(4) | C5 | C6 | C7 | C13 | 70.7(6) |
| C1 | C2 | C4 | N1 | 34.0(5) | C6 | C7 | C8 | C9 | –126.2(5) |
| C1 | O1 | N1 | C4 | 26.6(4) | C6 | C7 | C8 | N1 | 52.6(5) |
| C1 | O1 | N1 | C8 | 145.8(4) | C6 | C7 | C12 | C11 | 124.9(6) |
| C2 | C4 | C5 | C6 | 179.0(6) | C7 | C8 | C9 | C10 | 7.3(7) |
| C2 | C4 | N1 | C8 | –161.0(4) | C7 | C8 | C9 | C14 | –171.7(6) |
| C2 | C4 | N1 | O1 | –37.8(4) | C7 | C8 | N1 | O1 | –177.2(5) |
| C4 | C5 | C6 | C7 | 54.2(5) | C7 | C12 | C11 | C10 | 1.1(9) |
| C4 | N1 | C8 | C7 | –62.1(5) | C8 | C9 | C10 | C11 | –3.0(6) |
| C4 | N1 | C8 | C9 | 116.7(5) | C8 | C9 | C10 | O3 | 176.0(6) |
| C5 | C6 | C7 | C8 | –50.8(6) | C9 | C10 | C11 | C12 | –1.2(7) |
| C5 | C6 | C7 | C12 | –173.3(5) |
The torsion angles observed in quinoline dione are consistent with those reported for similar heterocyclic and aromatic compounds. Literature on quinoline derivatives often reports similar conformational flexibility and deviations from planarity due to steric and electronic effects.37 Understanding the torsion angles is crucial for predicting the compound’s reactivity, interaction with other molecules, and overall stability. The observed torsion angles highlight the balance between electronic effects, steric hindrance, and the molecule’s inherent flexibility.
Hydrogen coordinates (x, y, z) are given in fractional values relative to the unit cell dimensions see Table 8. These positions help define the precise geometry of the molecule and confirm the hydrogen placement on the carbon framework. Isotropic Ueq values for hydrogen atoms range from 47.4(17) to 98(3), indicating the extent of thermal motion. Higher Ueq values suggest greater mobility or positional disorder of the hydrogen atoms. Specific hydrogen atom analysis for H2 is as follows: coordinates, x = 10628(7), y = 12105(5), z = 9358(4); Ueq = 53.5(18). Positioned on C2, H2 has moderate thermal motion, which is typical for a hydrogen atom in an aromatic ring.
Table 8. H Atom Coordinates x, y, and z (Å × 104) and Isotropic Displacement Parameters (Å2 × 103) of Quinoline Dione.
| atom | x | y | z | Ueq |
|---|---|---|---|---|
| H2 | 10628(7) | 12105(5) | 9358(4) | 53.5(18) |
| H3a | 8790(40) | 13852(9) | 9357(16) | 98(3) |
| H3b | 8910(40) | 13300(20) | 8435(11) | 98(3) |
| H3c | 7277(10) | 13037(16) | 9000(20) | 98(3) |
| H4 | 9832(7) | 10166(5) | 9077(3) | 47.4(17) |
| H5a | 7698(9) | 11204(6) | 7798(4) | 67(2) |
| H5b | 9688(9) | 10954(6) | 7670(4) | 67(2) |
| H6a | 8006(9) | 9237(6) | 7165(4) | 68(2) |
| H6b | 9304(9) | 8819(6) | 7874(4) | 68(2) |
| H11 | 6733(10) | 5692(6) | 8492(4) | 69(2) |
| H12 | 6610(9) | 7167(6) | 7523(4) | 71(2) |
| H13a | 5050(20) | 10205(6) | 8040(30) | 98(3) |
| H13b | 4780(30) | 9070(30) | 7422(10) | 98(3) |
| H13c | 4098(12) | 9010(30) | 8360(18) | 98(3) |
| H14a | 8063(11) | 8450(40) | 10977(5) | 89(3) |
| H14b | 6200(40) | 9020(20) | 10912(4) | 89(3) |
| H14c | 6440(50) | 7583(16) | 11055(4) | 89(3) |
Coordinates for the methyl group hydrogens (H3a, H3b, H3c, H13a, H13b, H13c, H14a, H14b, H14c) vary significantly within each methyl group, reflecting the three-dimensional rotation. Ueq is generally higher (98(3) for H3a, H3b, and H3c; 89(3) for H14a, H14b, and H14c). These values indicate substantial thermal motion due to free rotation around the C–C bond. Coordinates for other aliphatic hydrogens (H5a, H5b, H6a, and H6b) are distributed around the parent carbon atoms. Ueq values of 67(2) and 68(2) reflect the flexible nature of the aliphatic chain, with moderate thermal motion. Coordinates for aromatic hydrogens (H4, H11, and H12) were positioned according to the aromatic ring structure. Ueq = 47.4(17) for H4, with higher values for H11 (69(2)) and H12 (71(2)). H11 and H12 exhibit higher thermal motion, possibly due to their positions at the ends of the aromatic ring, where the ring might experience more flexibility. The observed Ueq values and coordinates are consistent with typical hydrogen atom behavior in organic compounds. In the literature, hydrogen atoms bonded to sp2 carbons generally show lower Ueq values compared to those bonded to sp3 carbons or in methyl groups, where free rotation is possible. Similar studies report comparable Ueq ranges and positional variability for hydrogen atoms in organic molecules, indicating that our data align well with established trends. The hydrogen atom coordinates and isotropic displacement parameters provide a comprehensive understanding of the molecular geometry of quinoline dione. The values align with expected behavior based on the molecular structure and its environment, supporting the reliability and accuracy of the crystallographic analysis.37,39
The radial distribution function, g(r), describes the probability of finding a particle at a distance r from a reference particle in a system. For a crystal, it reflects the periodic arrangement of atoms or molecules.40 The base for calculating the pair distribution function, g(r), from the particle positions is given by the formula
where ρ is the number density, N is the total number of particles, ri and rj are the positions of particles i and j, respectively, and δ is the delta function.
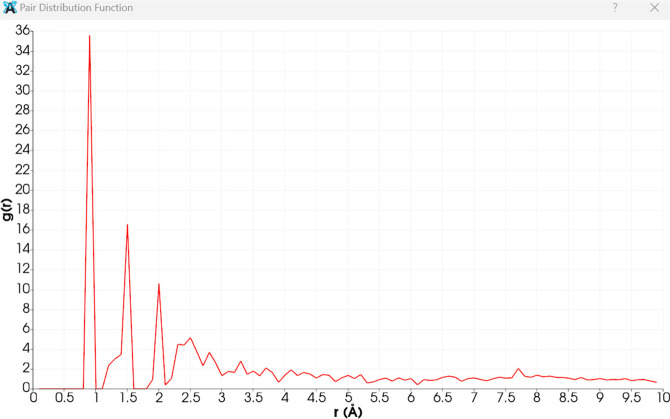
Figure 3has 3 peaks. The first and most intense peak around 1 Å is typical for nearest-neighbor distances, often corresponding to bonds between light atoms like hydrogen–carbon or carbon–carbon in organic molecules. The second peak (around 1.5 Å) suggests the next-nearest neighbor distances, which might be indicative of other bonded pairs, such as carbon–nitrogen or carbon–oxygen bonds. The third peak (around 2 Å) could correspond to more distant neighbors, potentially indicating secondary interactions or longer bond distances. The peaks beyond 2 Å, although less intense, indicate further atomic correlations in the structure. These might be due to more extended molecular frameworks or secondary structural features. Decay of peaks is a gradual decrease in peak intensity with increasing r, indicating that the probability of finding atoms decreases with distance, as expected. This reflects the diminishing influence of the reference atom over longer distances. The sharp and well-defined peaks suggest a well-ordered structure typical of crystalline materials. The heights and positions of these peaks provide insight into the atomic arrangements and bonding distances within the structure. For quinoline dione, these peaks likely correspond to specific bond lengths and spatial arrangements discussed in the tables of bond lengths and angles. The first peak likely corresponds to typical C–H or C–C bonds, while subsequent peaks relate to longer bonds or intermolecular interactions. The PDF plot is consistent with the crystallographic data provided for quinoline dione. The observed peaks align with the expected bond distances and atomic arrangements in the molecule, reaffirming the structural information obtained from X-ray crystallography. This analysis helps us to understand the local atomic environment and the overall structural order within the material.
Figure 3.
Pair distribution function of the quinoline dione crystal (g(r) is plotted against the radial distance r in angstroms).
Structural identification of quinoline derivatives through X-ray crystallography is essential for explaining their biological activities. Previous research, such as the study by Zhang et al.36 on the crystal structure of 5-chloro-2-(quinolin-8-yl)isoindoline-1,3-dione and that by Sawada et al.41 on the crystal structure of 2,4-dimethyl-4,4a-dihydro-1H-[1,3,5]-triazino-[1,2-a]-quinoline-1,3,6(2H)-trione and corresponding dehydrated compound, has provided valued insights into the conformational properties and binding affinities of these compounds. These findings are crucial for detailed structural characterization in the context of drug discovery and development. In our study, the structural analysis of quinoline dione revealed a triclinic crystal system with parameters that closely resemble those reported for similar compounds. Specifically, the unit cell dimensions and space group symmetry are comparable, suggesting a similar mode of molecular packing and potential interaction sites. This similarity indicates that quinoline dione, like the compound studied by Zhang et al., exhibits significant hydrogen bonding and π–π stacking interactions. These interactions are critical to the stability and functionality of the molecule. Moreover, the structural characteristics of quinoline dione, such as its planar conformation and extensive hydrogen bonding network, suggest a strong potential for interactions with biological targets such as in antimicrobial applications, where such interactions can play a pivotal role in the compound’s efficacy. The detailed comparison of these structural features highlights the potential of quinoline dione as a bioactive agent and aligns it with other well-studied quinoline derivatives.36,41
Antibacterial Activity
The results obtained from agar well diffusion assay for both crude and isolated compounds indicated a sharp increase in antibacterial potency, as shown in Table 9. Moreover, the crude extract was more active against Gram-positive bacteria compared to the compound isolated, which showed a high degree of activity against Gram-negative bacteria. It is reported that the compound isolated in its pure form is more active than its crude extract, which may be attributed to the presence of specific active functional group they possessed. In our study, the activity may be associated with quinoline moiety that is present in a number of alkaloids and is linked with its antimicrobial potential.42
Table 9. Antibacterial Potency (mm) of Crude Leaf Extract and Quinoline Dione (5 mg/mL)a.
| bacteria | crude extract | tetracycline | quinoline dione |
|---|---|---|---|
| MRSA (C1) | 12 ± 0.13 | 26 ± 012 | 18 ± 0.14 |
| P. aeruginosa (D1) | 13 ± 0.25 | 25 ± 0.17 | 23 ± 0.11 |
| E. coli (E1) | 12 ± 0.12 | 24 ± 0.15 | 20 ± 0.15 |
| B. subtilis (F1) | 15 ± 0.25 | 22 ± 0.11 | 17 ± 0.20 |
Data are expressed as mean ± standard error (n = 3).
Antibiofilm Activity
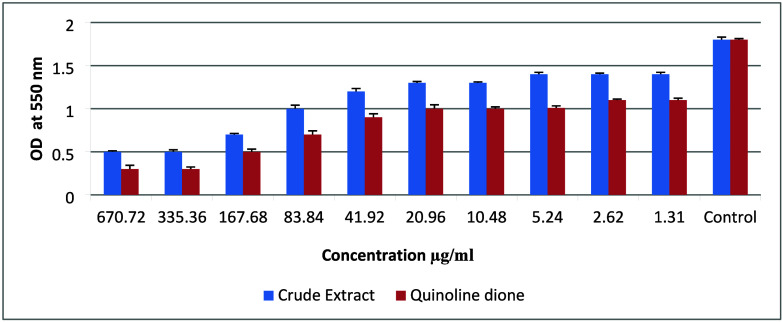
The inhibition of biofilm formation was studied in P. aeruginosa (PAO1) as it is a model organism for biofilm-related studies. In the assay, dose-dependent biofilm inhibition was recorded for both the crude extract and quinoline dione, and an increase in antibiofilm activity was also observed for the isolated compound compared to the crude extract as shown in Figure 4. This property of quinoline dione can play an important role in stopping the initial attachment of P. aeruginosa to form the biofilm and quorum sensing in preformed biofilms, as it can potentially contain an active group responsible for disrupting the biofilm-forming structure.43 Previous studies have highlighted the antibacterial properties of quinoline dione derivatives and demonstrated the efficacy of the isolated chemical skeleton.44 Plants extracts or derived compounds show promising results in pursuit of eradicating infections caused by bacterial biofilms as they contain ingredients that can disrupt the biofilm structure far more easily than commercially available antibiotics.45 This report suggests the potential for synthesizing and screening quinoline dione compounds for other pharmacological activities beyond their established antibacterial effects.
Figure 4.
Comparison of leaf extract and quinoline dione against biofilm inhibition of P. aeruginosa. Data are expressed as mean ± standard error (n = 3).
In Silico Studies
Redocking and Validation
To validate our docking protocol, we performed a redocking procedure using the crystal structure of chain A from the 6MNV protein. The cocrystallized ligand, N-(3-oxo-decanoyl)-homoserine lactone, was extracted and used as a standard to assess whether our predicted docking poses matched the experimental data. The minimum binding energy for the redocked 6MNV chain A and its ligand was determined to be −8.3 kcal/mol. The poses of the cocrystallized ligand and the docked ligand were analyzed and compared using Discovery 2024, yielding coordinates of 5.092684, −18.250263, and −35.991053 for the cocrystallized ligand and 5.543474, −18.751526, and −36.429895 for the docked ligand, respectively. The RMSD between the cocrystallized and docked poses was calculated to be 1.158 Å using ChimeraX.
Similarly, the redocking procedure was performed for the protein–ligand complex with PDB ID 3IX4. The binding energy for the cocrystallized ligand (TX1) was calculated to be −14 kcal/mol for chain A with an RMSD of 1.382 Å between the experimental and predicted poses. For both redocking experiments, the RMSD values were found to be lesser than 2 Å, supporting that our docking procedure is reliable.46 The similarity in coordinates after visualizing the ligand binding interactions for both experimental and predicted poses further confirmed the accuracy of our software for showing the same interactions.
Docking
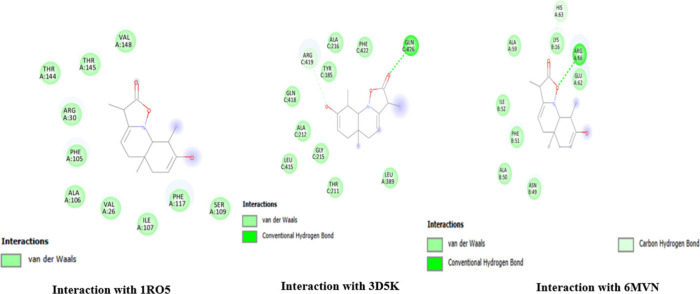
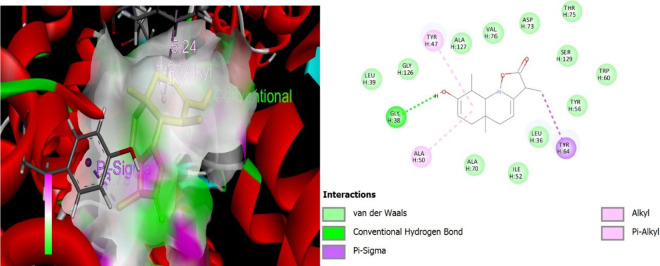
During the in silico evaluation, promising results were obtained against LasR receptors with the lowest binding energies, as shown in Table 10, while the interactive sites can be seen in Figures 5–7. In P. aeruginosa, OprM is known for its role in multidrug resistance, and ligands that could potentially interact with OprM might include small molecules or compounds that have the potential to inhibit the efflux pump, thereby reducing the antibiotic resistance in P. aeruginosa. Understanding the role of LasR and quorum sensing in P. aeruginosa is important for studying the pathogenicity of the bacterium and developing potential therapeutic strategies. Disrupting quorum sensing mechanisms may interfere with bacterial virulence and could be a target for controlling bacterial infections.47 Our in silico studies also confirm the antibacterial action against LasR with the lowest binding energy of −9.8 kcal/mol, LasI with −6.8 kcal/mol, and −6.9 kcal/mol binding energy against OprM and LasR (transcriptional regulator), thus possessing a possible potential to be a new antibacterial agent.
Table 10. Binding Energies of All of the Ligands with the Protein Targets Using Molecular Docking Analysis.
| sample (PubChem ID) | PDB ID 3IX4 chain Aa (kcal/mol) | PDB ID 1RO5b(kcal/mol) | PDB ID 3D5Kc (kcal/mol) | PDB ID 6MVN chain Ad (kcal/mol) |
|---|---|---|---|---|
| ciprofloxacin (2764) | –10.4 | –6.9 | –8 | –8.5 |
| amoxicillin (33613) | –8.7 | –5.9 | –6.8 | –7.8 |
| dodecyl sulfate (8778) | –6.7 | –5.7 | –4.9 | –6.8 |
| quinoline dione | –9.2 | –5.9 | –7.4 | –9.3 |
| tetracycline (54675776) | –5.7 | –6.7 | –7.8 | –5.5 |
LasR, a transcriptional activator.
AHL synthase LasI.
OprM channel.
LasR transcriptional regulator.
Figure 5.
Visualization of ciprofloxacin–Protein (PDB ID 3IX4) interactions. The diagram illustrates the interactions between a ligand and surrounding amino acid residues in a protein. Different types of interactions are highlighted using distinct color codes: van der Waals interactions are represented in light green, carbon hydrogen bonds are colored light cyan, π–π stacking interactions are indicated in pink, alkyl interactions are marked in light purple, and π–alkyl interactions are displayed in light pink. Key interacting amino acid residues include ILE C:52, LEU C:36, TYR C:64, TYR C:56, TRP C:60, ASP C:73, ALA C:127, VAL C:76, CYS C:79, LEU C:125, SER C:129, THR C:75, and GLY C:38. Each residue is labeled with its name and chain identifier to provide clear insights into the specific interactions occurring between the ligand and the protein.
Figure 7.
Interaction site of the ligand (quinoline dione) with receptors. The figure illustrates the interactions between quinoline dione and different protein targets as identified by their PDB IDs: 1RO5, 3D5K, and 6MVN. Interaction with 1RO5: quinoline dione interacts primarily through van der Waals forces. Key residues involved include THR A:144, THR A:145, ARG A:30, PHE A:105, ALA A:106, VAL A:26, ILE A:107, and VAL A:148. Interaction with 3D5K: The interaction involves both van der Waals forces and conventional hydrogen bonds. Notable residues participating in the interaction include ALA C:216, PHE C:422, GLN C:426 (hydrogen bond), GLN C:418, ALA C:212, TYR C:185, and LEU A:389. Interaction with 6MVN: This interaction is characterized by van der Waals forces, conventional hydrogen bonds, and carbon hydrogen bonds. Significant residues include HIS A:63, LYS B:16 (hydrogen bond), ARG A:66 (hydrogen bond), GLU A:62 (carbon hydrogen bond), PHE B:51, and ALA B:50.
Figure 6.
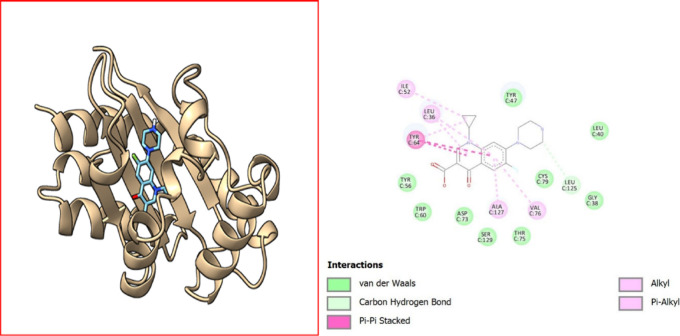
Visualization of quinoline dione–protein (PDB ID 3IX4) interactions. Molecular interaction diagram of a quinoline dione with surrounding amino acid residues in a protein. The interactions depicted include van der Waals forces (green circles), a conventional hydrogen bond (green dotted line) with GLY H:38, π–σ interactions (purple dashed lines) with TYR H:47 and TYR H:64, alkyl interactions (pink circles) with ALA H:50, and π–alkyl interactions (pink circles with purple shade) with TYR H:47. The ligand is shown at the center, surrounded by the amino acid residues identified by their three-letter codes and sequence positions (e.g., GLY H:126, LEU H:39). This diagram highlights the key binding interactions between the ligand and the protein.
The Table 10 provides binding affinities (in kcal/mol) for various compounds interacting with different protein targets. The lower (more negative) the value, the stronger the binding energy of the compound to the protein. Ciprofloxacin shows the highest binding energy with the LasR transcriptional activator (3IX4) with a binding energy of −10.4 kcal/mol. This suggests a strong interaction, potentially inhibiting the quorum sensing mechanism in Pseudomonas aeruginosa. It shows moderate binding affinities with other targets. Amoxicillin shows moderate binding affinities across all targets with the highest binding energy observed for the LasR transcriptional activator at −9.3 kcal/mol. It is generally known for its action against cell wall synthesis rather than interaction with these protein targets. Dodecyl sulfate shows relatively weak binding affinities with all targets except for the LasR transcriptional regulator (6MVN), where it shows a slightly better binding energy (−6.8 kcal/mol). Quinoline dione shows a strong binding energy with the LasR transcriptional regulator (6MVN) at −9.3 kcal/mol, indicating its potential as a quorum sensing inhibitor. It shows moderate affinity for other targets. Tetracycline shows consistent and relatively strong binding affinities across all targets with the highest binding energy observed for the OprM channel (3D5K) at −7.8 kcal/mol. This suggests its potential broad-spectrum effectiveness.
Conclusion
Th antibacterial and antibiofilm efficacy of Psammogeton biternatum Edgew was explored, which led to the identification of a novel quinoline alkaloid, quinoline dione, using X-ray crystallography. The crude methanol extract exhibited significant antibacterial activity against various strains, particularly against Gram-positive bacteria. However, the isolated quinoline dione demonstrated superior antibacterial efficacy against Gram-negative bacteria, notably MRSA, B. subtilis, E. coli, and P. aeruginosa.
The antibiofilm activity, assessed using the 96-well microtiter plate method, revealed a dose-dependent inhibition of P. aeruginosa biofilm formation, with quinoline dione showing markedly higher efficacy compared with the crude extract. This enhanced activity is likely attributed to the specific active functional groups present in the pure isolated compound.
Furthermore, in silico studies confirmed the strong binding energy of quinoline dione with the LasR transcriptional regulator, suggesting its potential role as a quorum sensing inhibitor. The molecular docking studies also indicated moderate affinities with other targets, such as LasI and OprM, supporting its potential as a multifunctional antimicrobial agent. The isolated compound holds potential for further progress and testing in both in vitro and in vivo models to establish its efficacy and safety. Future investigations should focus on its synthesis and pharmacokinetic and pharmacodynamic studies to explore its therapeutic potential in clinical settings.
Acknowledgments
The authors extend their appreciation to University Higher Education Fund for funding this research work under the Research Support Program for Central Laboratories at King Khalid University through project number CL/PRI/A/2.
The authors declare no competing financial interest.
References
- Romha G.; Admasu B.; Hiwot Gebrekidan T.; Aleme H.; Gebru G. Antibacterial activities of five medicinal plants in Ethiopia against some human and animal pathogens. Evidence-Based Complementary Altern. Med. 2018, 2018, 2950758. 10.1155/2018/2950758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sha’alan N. H. Antimicrobial activity and spectral, magnetic and thermal studies of some transition metal complexes of a Schiff base hydrazone containing a quinoline moiety. Molecules 2007, 12 (5), 1080–1091. 10.3390/12051080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H.; Ma Q.; Ye L.; Piao G. The traditional medicine and modern medicine from natural products. Molecules 2016, 21 (5), 559. 10.3390/molecules21050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A.; Chandraker S.; Patel V.; Ramteke P. Antibacterial activity of medicinal plants against pathogens causing complicated urinary tract infections. Indian journal of pharmaceutical sciences 2009, 71 (2), 136. 10.4103/0250-474X.54279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrence R.; Ramakrishnan A.; Sundaramoorthy N.; Shyam A.; Mohan V.; Subbarao H.; Ulaganathan V.; Raman T.; Solomon A.; Nagarajan S. Norfloxacin salts of carboxylic acids curtail planktonic and biofilm mode of growth in ESKAPE pathogens. Journal of applied microbiology 2018, 124 (2), 408–422. 10.1111/jam.13651. [DOI] [PubMed] [Google Scholar]
- Yelin I.; Kishony R. Antibiotic resistance. Cell 2018, 172 (5), 1136–1136. 10.1016/j.cell.2018.02.018. [DOI] [PubMed] [Google Scholar]
- De La Fuente-Núñez C.; Korolik V.; Bains M.; Nguyen U.; Breidenstein E. B.; Horsman S.; Lewenza S.; Burrows L.; Hancock R. E. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56 (5), 2696–2704. 10.1128/AAC.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazargani M. M.; Rohloff J. Antibiofilm activity of essential oils and plant extracts against Staphylococcus aureus and Escherichia coli biofilms. Food control 2016, 61, 156–164. 10.1016/j.foodcont.2015.09.036. [DOI] [Google Scholar]
- Jamal M.; Ahmad W.; Andleeb S.; Jalil F.; Imran M.; Nawaz M. A.; Hussain T.; Ali M.; Rafiq M.; Kamil M. A. Bacterial biofilm and associated infections. Journal of the chinese medical association 2018, 81 (1), 7–11. 10.1016/j.jcma.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Cepas V.; López Y.; Muñoz E.; Rolo D.; Ardanuy C.; Martí S.; Xercavins M.; Horcajada J. P.; Bosch J.; Soto S. M. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microbial Drug Resistance 2019, 25 (1), 72–79. 10.1089/mdr.2018.0027. [DOI] [PubMed] [Google Scholar]
- de la Fuente-Núñez C.; Cardoso M. H.; de Souza Cândido E.; Franco O. L.; Hancock R. E.W. Synthetic antibiofilm peptides. Biochim. Biophys. Acta 2016, 1858 (5), 1061–1069. 10.1016/j.bbamem.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvinen P.; Nybond S.; Marcourt L.; Ferreira Queiroz E.; Wolfender J.-L.; Mettälä A.; Karp M.; Vuorela H.; Vuorela P.; Hatakka A.; et al. Cell-based bioreporter assay coupled to HPLC micro-fractionation in the evaluation of antimicrobial properties of the basidiomycete fungus Pycnoporus cinnabarinus. Pharm. Biol. 2016, 54 (6), 1108–1115. 10.3109/13880209.2015.1103754. [DOI] [PMC free article] [PubMed] [Google Scholar]; Móricz Á. M.; Ott P. G.; Habe T. T.; Darcsi A.; Boszormenyi A.; Alberti Á.; Krüzselyi D. n.; Csontos P.; Béni S.; Morlock G. E. Effect-directed discovery of bioactive compounds followed by highly targeted characterization, isolation and identification, exemplarily shown for Solidago virgaurea. Analytical chemistry 2016, 88 (16), 8202–8209. 10.1021/acs.analchem.6b02007. [DOI] [PubMed] [Google Scholar]
- Gemma S.; Savini L.; Altarelli M.; Tripaldi P.; Chiasserini L.; Coccone S. S.; Kumar V.; Camodeca C.; Campiani G.; Novellino E.; et al. Development of antitubercular compounds based on a 4-quinolylhydrazone scaffold. Further structure-activity relationship studies. Bioorg. Med. Chem. 2009, 17 (16), 6063–6072. 10.1016/j.bmc.2009.06.051. [DOI] [PubMed] [Google Scholar]; Bispo M. d. L.; de Alcantara C. C.; de Moraes M. O.; do ÓPessoa C.; Rodrigues F. A.; Kaiser C. R.; Wardell S. M.; Wardell J. L.; de Souza M. V. A new and potent class of quinoline derivatives against cancer. Monatshefte für Chemie-Chemical Monthly 2015, 146, 2041–2052. 10.1007/s00706-015-1570-0. [DOI] [Google Scholar]; Selvi G.; Priyadarshini G. S. Antimicrobial activities of the synthesised Fe (II), Co (II), Ni (II) & Cu (II) Quinoline Schiff Base Complexes. Adv. Appl. Res. 2013, 5 (2), 161–165. [Google Scholar]
- Duval A. R.; Carvalho P. H.; Soares M. C.; Gouvêa D. P.; Siqueira G. M.; Lund R. G.; Cunico W. 7-Chloroquinolin-4-yl arylhydrazone derivatives: synthesis and antifungal activity. Scientific World Journal 2011, 11, 1489–1495. 10.1100/tsw.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen Y.-L.; Chen I.-L.; Lu C.-M.; Tzeng C.-C.; Tsao L.-T.; Wang J.-P. Synthesis and anti-inflammatory evaluation of 4-anilinofuro [2, 3-b] quinoline and 4-phenoxyfuro [2, 3-b] quinoline derivatives. Part 3. Bioorganic & medicinal chemistry 2004, 12 (2), 387–392. 10.1016/j.bmc.2003.10.051. [DOI] [PubMed] [Google Scholar]; Charris J.; Lobo G.; Camacho J.; Ferrer R.; Barazarte A.; Dominguez J.; Gamboa N.; Rodrigues J.; Angel J. Synthesis and antimalarial activity of (E) 2-(2’-chloro-3′-quinolinylmethylidene)-5, 7-dimethoxyindanones. Letters in Drug Design & Discovery 2007, 4 (1), 49–54. 10.2174/157018007778992865. [DOI] [Google Scholar]
- Abbasi A. M.; Khan M.; Ahmad M.; Zafar M.; Jahan S.; Sultana S. Ethnopharmacological application of medicinal plants to cure skin diseases and in folk cosmetics among the tribal communities of North-West Frontier Province, Pakistan. Journal of ethnopharmacology 2010, 128 (2), 322–335. 10.1016/j.jep.2010.01.052. [DOI] [PubMed] [Google Scholar]
- Tareen R. B.; Bibi T.; Khan M. A.; Ahmad M.; Zafar M. Indigenous knowledge of folk medicine by the women of Kalat and Khuzdar regions of Balochistan, Pakistan. Pak J. Bot. 2010, 42 (3), 1465–1485. [Google Scholar]; Manzoor M.; Durrani M.; Jabeen R.; Irfan S.; Ayub F.; Bibi S. An Ethno botanical Study for the Treatment of Cancer and Malaria Used by the People of Quetta City. Int. J. Basic Appl. Sci. 2012, 1 (2), 137–149. 10.14419/ijbas.v1i2.72. [DOI] [Google Scholar]
- Khan I.; Khan U.; Khan K.; Nawaz M. A.; Khan N. A.; Ali F. In vitro anti-pseudomonal potential of Juglans regia and Otostegia limbata leaves extract against planktonic and biofilm form of Pseudomonas aeruginosa. Pak. J. Bot. 2018, 50 (2), 827–833. [Google Scholar]
- Khan I.; Khan U.; Khan W.; Subhan M.; Nawaz M. A.; Pervez S.; Khan K.; Jan A. K.; Ahmad S. Antibacterial and antibiofilm potential of leaves extracts of Mirabilis jalapa L. and Ajuga bracteosa wall. against Pseudomonas aeruginosa. Pure Appl. biol. 2017, 6 (2), 605–613. 10.19045/bspab.2017.60062. [DOI] [Google Scholar]
- Khurram M.; Khan M. A.; Hameed A.; Abbas N.; Qayum A.; Inayat H. Antibacterial activities of Dodonaea viscosa using contact bioautography technique. Molecules 2009, 14 (3), 1332–1341. 10.3390/molecules14031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolomanov O. V.; Bourhis L. J.; Gildea R. J.; Howard J. A.; Puschmann H. OLEX2: a complete structure solution, refinement and analysis program. Journal of applied crystallography 2009, 42 (2), 339–341. 10.1107/S0021889808042726. [DOI] [Google Scholar]
- Bourhis L. J.; Dolomanov O. V.; Gildea R. J.; Howard J. A.; Puschmann H. The anatomy of a comprehensive constrained, restrained refinement program for the modern computing environment-Olex2 dissected. Acta Crystallographica Section A: Foundations and Advances 2015, 71 (1), 59–75. 10.1107/S2053273314022207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabbir U.; Mumtaz S.; Khan I.; Yameen M. A.; Riaz M.; Khan U.; Ilyas N.; Nawaz I.; Ahmad R.; Khan W. Effect of honey use with Seriphidium chitralense podlech on growth and biofilm formation of Pseudomonas aeruginosa. Kuwait Journal of Science 2023, 50 (4), 703–708. 10.1016/j.kjs.2023.05.017. [DOI] [Google Scholar]
- O’Toole G. A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, (47), e2437 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng E. C.; Goddard T. D.; Pettersen E. F.; Couch G. S.; Pearson Z. J.; Morris J. H.; Ferrin T. E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32 (11), e4792 10.1002/pro.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y.; Nair S. K. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chemistry & biology 2009, 16 (9), 961–970. 10.1016/j.chembiol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; McCready A. R.; Paczkowski J. E.; Henke B. R.; Bassler B. L. Structural determinants driving homoserine lactone ligand selection in the Pseudomonas aeruginosa LasR quorum-sensing receptor. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (1), 245–254. 10.1073/pnas.1817239116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry 2010, 31 (2), 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A. The impact of natural products upon modern drug discovery. Curr. Opin. Chem. Biol. 2008, 12 (3), 306–317. 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Vaou N.; Stavropoulou E.; Voidarou C.; Tsigalou C.; Bezirtzoglou E. Towards advances in medicinal plant antimicrobial activity: A review study on challenges and future perspectives. Microorganisms 2021, 9 (10), 2041. 10.3390/microorganisms9102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney M. G. Natural products as a source for novel antibiotics. Trends in pharmacological sciences 2016, 37 (8), 689–701. 10.1016/j.tips.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Su Y.; Shi D.; Xiong B.; Xu Y.; Hu Q.; Huang H.; Yang J.; Yu C. Solid-State Forms of Koumine Hydrochloride: Phase Transformations and the Crystal Structure and Properties of the Stable Form. ACS omega 2022, 7 (34), 29692–29701. 10.1021/acsomega.2c02175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham F.; Crowder C.; Reid J.; Fawcett T.; Faber J. X-ray powder diffraction analysis of imipenem monohydrate. Powder Diffraction 2012, 27 (1), 20–24. 10.1017/S0885715612000048. [DOI] [Google Scholar]
- Khalid M.; Ullah M. A.; Adeel M.; Khan M. U.; Tahir M. N.; Braga A. A. C. Synthesis, crystal structure analysis, spectral IR, UV-Vis, NMR assessments, electronic and nonlinear optical properties of potent quinoline based derivatives: Interplay of experimental and DFT study. J. Saudi Chem. Soc. 2019, 23 (5), 546–560. 10.1016/j.jscs.2018.09.006. [DOI] [Google Scholar]
- Karle J. M.; Karle I. L. Crystal structure of (−)-mefloquine hydrochloride reveals consistency of configuration with biological activity. Antimicrob. Agents Chemother. 2002, 46 (5), 1529–1534. 10.1128/AAC.46.5.1529-1534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabel V.; Svorc L.; Sivy J.; Marchalin S.; Safar P. Structural characterization and crystal packing of the isoquinoline derivative. European Journal of Chemistry 2018, 9 (3), 189–193. 10.5155/eurjchem.9.3.189-193.1758. [DOI] [Google Scholar]
- Dunitz J. D.X-ray Analysis and the Structure of Organic Molecules; VCH, 1995. [Google Scholar]
- Zhang W.; Zhang Y.; Feng S.-H.; Wang J.-J. The crystal structure of 5-chloro-2-(quinolin-8-yl) isoindoline-1, 3-dione, C17H9ClN2O2. Zeitschrift für Kristallographie-New Crystal Structures 2021, 236 (6), 1155–1156. 10.1515/ncrs-2021-0256. [DOI] [Google Scholar]
- Allen F. H.; Bruno I. J. Bond lengths in organic and metal-organic compounds revisited: X—H bond lengths from neutron diffraction data. Acta Crystallographica Section B: Structural Science 2010, 66 (3), 380–386. 10.1107/S0108768110012048. [DOI] [PubMed] [Google Scholar]
- Scharfer C.; Schulz-Gasch T.; Ehrlich H.-C.; Guba W.; Rarey M.; Stahl M. Torsion angle preferences in druglike chemical space: a comprehensive guide. J. Med. Chem. 2013, 56 (5), 2016–2028. 10.1021/jm3016816. [DOI] [PubMed] [Google Scholar]
- Müller P.; Herbst-Irmer R.; Spek A.; Schneider T.; Sawaya M.. Crystal Structure Refinement: A Crystallographer’s Guide to SHELXL; Oxford University Press, 2006. [Google Scholar]
- Baranyai A.Physical Chemistry of Bulk Water. In Encyclopedia of Interfacial Chemistry, Wandelt K., Ed.; Elsevier, 2018; pp 227–231. [Google Scholar]
- Sawada M.; Tanaka T.; Takai Y.; Furukawa Y.; Hanafusa T.; Misumi S. X-ray crystal structures of 2, 4-dimethyl-4, 4a-dihydro-1H-(1, 3, 5)-triazino (1, 2-a) quinoline-1, 3, 6 (2H, 5H)-trione and its dehydrogenated derivative. Bond length and EI mass fragmentation. Bull. Chem. Soc. Jpn. 1986, 59 (3), 949–951. 10.1246/bcsj.59.949. [DOI] [Google Scholar]
- Kiser T. H.; Obritsch M. D.; Jung R.; McLaren R.; Fish D. N. Efflux pump contribution to multidrug resistance in clinical isolates of Pseudomonas aeruginosa. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy 2010, 30 (7), 632–638. 10.1592/phco.30.7.632. [DOI] [PubMed] [Google Scholar]
- Kadela-Tomanek M.; Bębenek E.; Chrobak E.; Boryczka S. 5, 8-Quinolinedione scaffold as a promising moiety of bioactive agents. Molecules 2019, 24 (22), 4115. 10.3390/molecules24224115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh N. H.; Len V. T.; Hang B. T. Y.; Hoa L. T. Synthesis and Reactions of a New Quinone Quinoline 7-(Carboxymethoxy)-3-sulfoquinoline-5, 6-dione. Journal of Heterocyclic Chemistry 2019, 56 (3), 1048–1054. 10.1002/jhet.3490. [DOI] [Google Scholar]
- Kostić M.; Ivanov M.; Stojković D.; Ćirić A.; Soković M. Antibacterial and antibiofilm activity of selected polyphenolic compounds: An in vitro study on Staphylococcus aureus. Lekovite sirovine 2020, 40, 57–61. 10.5937/leksir2040057K. [DOI] [Google Scholar]; Kim H.-S.; Park H.-D. Ginger extract inhibits biofilm formation by Pseudomonas aeruginosa PA14. PloS one 2013, 8 (9), e76106 10.1371/journal.pone.0076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A. N. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. Journal of medicinal chemistry 2003, 46 (4), 499–511. 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- Fagerlind M. G.; Rice S. A.; Nilsson P.; Harlén M.; James S.; Charlton T.; Kjelleberg S. The role of regulators in the expression of quorum-sensing signals in Pseudomonas aeruginosa. Journal of molecular microbiology and biotechnology 2004, 6 (2), 88–100. 10.1159/000076739. [DOI] [PubMed] [Google Scholar]