Abstract
The generation of antibody-drug conjugates with optimal functionality depends on many parameters. These include binder epitope, antibody format, linker composition, conjugation site(s), drug-to-antibody ratio, and conjugation method. The production of matrices that cover all possible parameters is a major challenge in identifying optimal antibody-drug conjugates. To address this bottleneck, we adapted our Format Chain Exchange technology (FORCE), originally established for bispecific antibodies, toward the generation of binder-format-payload matrices (pair-FORCE). Antibody derivatives with exchange-enabled Fc-heterodimers are combined with payload-conjugated Fc donors, and subsequent chain-exchange transfers payloads to antibody derivatives in different formats. The resulting binder-format-conjugate matrices can be generated with cytotoxic payloads, dyes, haptens, and large molecules, resulting in versatile tools for ADC screening campaigns. We show the relevance of pair-FORCE for identifying optimal HER2-targeting antibody-drug conjugates. Analysis of this matrix reveals that the notion of format-defines-function applies not only to bispecific antibodies, but also to antibody-drug conjugates.
Subject terms: Biologics, Drug delivery
Designing optimal antibody-drug conjugates (ADCs) involves screening many complex parameters. Here, the authors present a payload-coupled chain-exchange technology for efficient ADC matrix production, demonstrating its power in designing ADCs targeting HER2.
Introduction
Antibody derivatives containing attached cytotoxic payloads—known as antibody-drug conjugates (ADCs) —are currently being developed as targeted cancer therapeutics. Currently, 13 ADCs have been approved by the FDA, and many more are in clinical trials1,2. For successful development, ADCs must retain preferentially uncompromised antigen binding and internalization, in addition to high cytotoxic payload potency3,4. Additionally, ADCs should be of a defined composition, because heterogeneous conjugation methods can lead to a mixture of over-conjugated and under-conjugated antibodies, which can result in reduced stability, poor pharmacokinetics (PK), decreased tumor penetration, increased systemic toxicity, and decreased efficacy5–8. Such challenges were faced during the development of first-and second-generation ADCs, many of which contain payloads heterogeneously conjugated via interchain disulfide reduction and cysteine labeling, or N-hydroxysuccinimide (NHS)-based conjugation onto surface-exposed lysine residues1,8–11. These conjugation approaches result in a stochastic distribution of attached payloads, thereby producing ADCs with variable conjugation sites and drug-to-antibody ratios (DAR)8,11,12. An additional concern with NHS conjugation approaches is a potential decrease in binding affinity due to conjugation of lysines located in complementarity-determining regions (CDRs) or in close proximity to CDRs13,14. While lysines are generally underrepresented in antibody CDRs, this is not the case for all antibodies, particularly those with lambda light chains15. In order to overcome issues caused by heterogeneous payload conjugation, third-generation ADCs often employ site-specific conjugation technologies8,16 (described in more detail below). In contrast to NHS conjugation approaches, conjugation of selectively reduced cysteine residues in the hinge region and the CH1/CL domains results in better homogeneity and control of conjugation sites, but can still result in a heterogeneous DAR1,16,17. This is a challenge for ADC discovery and screening approaches, as it can be challenging to ensure identical, or even similar DAR and conjugation sites between different antibodies with varying formats.
Issues associated with nonspecific payload coupling can be addressed by site-specific payload conjugation. Most of these approaches are based on the introduction of mutated residues that serve as targets for site-directed conjugation. Examples include THIOMABs, which carry additional exposed cysteine residues for maleimide conjugation18,19, and antibodies with unnatural amino acids introduced through stop-codon suppression20–23. Alternatively, antibodies can be engineered with recognition sequences that enable spontaneous or enzyme-based conjugation by means of inteins, SNAP, sortase, or transglutaminase9,24–28. These technologies address the generation of defined entities, but production and scaling is still complex and laborious in early project phases, since each conjugate must be produced and analyzed individually.
Site-specific conjugation can address the homogeneity of ADCs, but other challenges still remain. One major issue is that ADCs with desired functionalities are not simply the result of site-specific conjugation of a preferred cytotoxic compound onto a well-performing antibody. Instead, a wide variety of factors determine the successful development of ADCs with good efficacy and a sufficiently large therapeutic index. Binding modules need to be compatible with the conjugated cytotoxic payload in terms of target binding affinity and internalization kinetics. Binder affinity, valency, and format (the position of binders relative to the Fc domain) can influence tumor-cell selectivity, tumor penetration, and internalization kinetics, and are thus parameters that need to be optimized in ADC screening campaigns29–32. Linker composition and conjugation chemistry also play a role in ADC stability and efficacy33. Additionally, the conjugation positions must be compatible with binder functionality and can influence the stability, PK, and potency of the ADC34–36. The drug-to-antibody ratio (DAR), i.e., how many payloads are coupled per antibody, also affects ADC functionality and can modulate biophysical and PK properties6,7,9,37. Therefore, the generation, screening, and selection of optimal ADCs requires the combination of different binders, formats, and linker-payload modules, all while testing different conjugation positions with varying DAR. Selecting optimal ADCs thus requires the assessment of complex matrices that combine these parameters. Even when combining just a few variables for each parameter (binder, format, linker, payload, conjugation position, and DAR), this can still result in large matrices. The production of comprehensive ADC matrices to cover such a design space is tedious and is a major hurdle in early development.
The challenges listed here for ADCs are very similar to those described for the generation of bispecific antibodies (bsAbs). BsAbs are antibody derivatives that combine two different binders in one molecule, in contrast to standard ADCs, where the antibody contains one binder specificity paired with one type of cytotoxic payload1. In the development of bsAbs, multiple binding modules need to be compatible with each other, and numerous variables can influence the biological function, activity, and biophysical properties of the bsAb. These include binder format and position, linker lengths and flexibility, binder valency, and binding kinetics of the individual binders29,38–40. Therefore, the identification of desired bsAbs requires the generation and screening of comprehensive binder-format-linker-valency matrices, similar to the binder-payload-conjugation site-linker-DAR matrices required in ADC screening campaigns.
We have recently addressed the bottleneck of generating large binder-format bsAb matrices with a chain-exchange-based technology (FORCE)29. FORCE is an efficient, high-throughput-automation-compatible platform, and produces combinations of bispecific antibodies in different formats from monospecific input molecules in vitro. Precursor molecules are applied as half antibodies complemented with dummy chains, with both chains containing partially destabilized CH3 interfaces. Combining complementary precursors in a reducing environment triggers a chain-exchange reaction that generates bsAb matrices covering binder specificity, binder format, and binder valency as design variables29.
Here we show that the FORCE technology can be modified and expanded to cover large design spaces for ADCs. We expanded the technology from covering binder specificity, binder format, and binder valency as design variables to cover the payload, conjugation position, DAR, binder specificity, binder format, and binder valency as variables. We refer to this modified technology as payload-redirecting chain-exchange (pair-FORCE). Our newly developed pair-FORCE technology combines different binding modules with various payload-coupled, exchange-enabled Fc donor counterparts. We demonstrate that a variety of payload types are compatible with the pair-FORCE technology, enabling applications that are valuable for ADC screening campaigns as well as other related uses, such as the generation of tool reagents for epitope binning and imaging assays. This includes high-throughput conjugation of different binders with (1) cytotoxic linker-payload combinations to generate defined ADCs and identify molecules with optimal properties. (2) pH-sensitive fluorescent dyes to assess internalization efficacy, (3) fluorescent dyes/proteins and enzymes for screening of binding efficacy and epitope binning, and (4) site-specific biotinylation for generation of versatile assay reagents.
To demonstrate that pair-FORCE is a robust, rapid, and reliable platform for generating defined and comprehensive ADC matrices, we generated and analyzed a matrix of HER2-targeted ADCs covering the variables of binder specificity, binder format, binder valency, conjugation position, payload, and DAR. The analysis of this ADC matrix demonstrates significant differences between different ADC constructs in terms of internalization and cytotoxic activity, and highlights the importance of screening a comprehensive set of variables during ADC development. Similar to the development of bispecific antibodies, the notion of format-defines-function29,39 holds true for ADC activity. Pair-FORCE provides a robust, rapid, and reliable method to enable comprehensive screening of ADC parameters in a high-throughput manner. Given the importance of screening a large design space during ADC development, pair-FORCE has the potential to significantly improve ADC design cycles.
Results
Payload-redirecting chain-exchange (pair-FORCE)
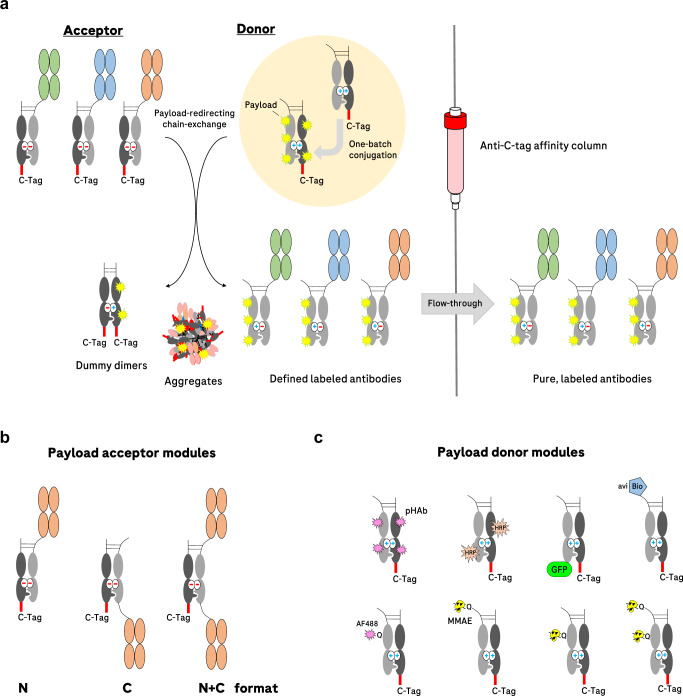
The principle of pair-FORCE is depicted in Fig. 1a. Different binder modules (depicted in green, blue, and orange) are exchange-enabled by carrying partially destabilized knob-into-hole CH3 interfaces with repulsive charges, as previously described29. These antibody derivatives with different Fab/Fv regions serve as acceptor entities for complementary Fc donor molecules. The payload-donor molecules are partially destabilized knob-into-hole Fc precursors that are conjugated with one or more payloads, either in a site-specific or nonspecific manner. The design, generation, and characterization of Fc-payload-donor molecules and binder-acceptor molecules is described in detail in Supplementary Fig. 1.
Fig. 1. Overview of the payload-redirecting chain-exchange concept (pair-FORCE).
a Different binders (in green, blue, and orange) have varying sequences in their Fab regions. On the upper right (highlighted in the yellow circle), a single conjugation reaction with an Fc donor molecule is depicted. The conjugated donor molecule is then applied to the exchange reaction with each of the different binders (acceptors) in the presence of the reducing agent TCEP, which reduces the disulfide bridges in the hinge region (shown as dashed lines). After the chain-exchange reaction, the mixture is applied to a C-tag affinity column, which captures dummy-dimers, aggregates, and non-reacted educt molecules. The products - labeled antibodies with a defined composition —are present in the flow-through with high purity. The hinge disulfides of the product antibodies reoxidize after C-tag purification. b Antibody-derived acceptor molecules can be produced in three different formats (N, C, and N + C) and can be combined with different payload-donor entities. c A variety of payload-donor modules can be produced through nonspecific conjugation or site-specific conjugation approaches (depicted here as either biotinylation of an Avi tag by the BirA enzyme44 or transglutaminase-mediated Q-tag conjugation26). After pairing with binder-containing acceptor modules through pair-FORCE, the labeled antibody derivatives can be utilized for a variety of approaches, including monitoring of internalization, immunoblotting, FACS, epitope binning, SPR assays, immunoprecipitation, and ADC cytotoxicity assays. pHAb pH-sensitive dye13, HRP horseradish peroxidase, GFP green fluorescent protein, avi recognition sequence for site-specific biotinylation44, AF488 Alexa Fluor 488, MMAE monomethyl auristatin E.
Mixing the acceptor and donor entities under mild reducing conditions for limited reduction of hinge disulfides (see Methods section and ref. 29) triggers an exchange reaction driven by repulsive charges in the CH3 interface of the precursor molecules. This reaction results in the formation of productive antibody derivatives containing the payload as well as dummy-dimers as by-products (Fig. 1a). Dummy-dimers, donor precursors, and acceptor precursors (but not the pair-FORCE products) harbor C-terminal C-tags and can be removed from the reaction mixture by adsorption on a C-tag affinity column. The flow-through contains the payload-coupled binders (pair-FORCE products). Because the exchange reaction transfers one defined conjugate from the Fc donor molecule to sets of many different binder-acceptors (Fig. 1b), conjugation-mediated variabilities between the resulting pair-FORCE products are excluded. All products contain identical payloads, conjugation positions, and DAR. Exchange-mediated, defined payload-attachment can be performed with various payloads coupled to Fc donor modules with different conjugation methods (Fig. 1c). This includes nonspecific conjugation to donor molecules (which are thereafter still converted into defined products), site-specific conjugation, or genetic fusion to donor modules, as described below.
Defined and comparable antibody-payload conjugates generated from NHS-conjugated donor modules
Conjugation of NHS-modified payloads onto surface-exposed lysine residues is a robust first- and second-generation workhorse technology for ADC generation41. This type of conjugation is considered random because the reaction leads to stochastic coupling of payloads onto numerous exposed lysine residues, resulting in heterogeneous DAR42,43. A heterogeneous DAR can lead to poor PK, instability of the conjugated payload, low tumor penetration, and decreased payload delivery8,11,16. Additionally, since lysine residues can be located in proximity to, or in the CDRs of select antibodies15, coupling payloads to these residues can influence antigen binding in some cases13,14. Nevertheless, it can sometimes be advantageous to use stochastic conjugation methods like NHS-lysine labeling in initial ADC screening campaigns, as these methods are simple and efficient. However, it can be challenging to generate sets of ADCs in which all molecules retain comparable DAR and conjugation positions. The pair-FORCE technology overcomes the issue of DAR variability between ADCs produced with nonspecific conjugation methods. Figure 2 shows how heterogeneously conjugated payloads on Fc donor molecules are converted into sets of labeled antibodies with identical DAR and unmodified binding regions. In this two-step method, the payload is first conjugated to the Fc donor molecule, rather than directly to the antibody of interest. In the second step, the payload-conjugated Fc donor then transfers a consistent quantity of payload onto the different antibody acceptor modules (Fig. 2a, b). This generates homogenous products with comparable DAR and purity.
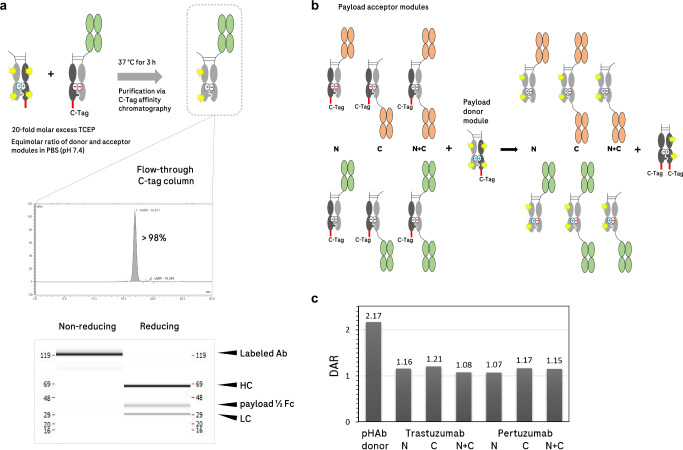
Fig. 2. Chain-exchange-mediated attachment of fluorescent dyes from NHS-conjugated donor modules with pair-FORCE.
a Upper panel: reactant molecules are mixed in equimolar concentrations in PBS under mild reducing conditions (20-fold molar excess TCEP). The mixture is incubated at 37 °C for 3 h while shaking. The reaction is then loaded onto a C-tag affinity column and the product is collected in the flow-through. Lower panel: The labeled product, a HER2 binder (Trastuzumab-derived) in the N-format with conjugated pHAb fluorescent dye molecules, has a purity of >98% as measured by analytical SEC. The capillary electrophoresis (CE-SDS) reveals defined bands of expected sizes under both non-reducing and reducing conditions. HC heavy chain, LC light chain, payload ½ Fc payload-conjugated Fc chain, Ab antibody. b A binder-format matrix of two different HER2 binders (derived from Trastuzumab and Pertuzumab) in three different formats (N, C, and N + C) was combined in a pair-FORCE reaction with an Fc donor molecule that was conjugated with pHAb (a pH-sensitive fluorophore) through NHS-amine labeling. c Analysis of the pair-FORCE products via mass spectrometry reveals a similar DAR (drug-to-antibody ratio) for all products, irrespective of the format. As expected, the pHAb-conjugated Fc donor molecule (Payload-donor module in 1c) has an approximately twofold greater DAR than the product molecules. Shown are results from one independent experiment (n = 1), see Methods section for details.
As an example of this approach, we combined two different HER2-targeting acceptor molecules (derived from Trastuzumab and Pertuzumab) with an Fc donor molecule that was labeled with a pH-sensitive dye (pHAb, Promega13) by nonspecific NHS conjugation on reactive amine groups. The chain-exchange reaction was performed in PBS with a 20-fold molar excess of TCEP (reducing agent) to reduce the hinge disulfides. After incubation for 3 h at 37 °C and orbital shaking at 300 rpm, the mixture was applied to a C-tag column. The flow-through contained the labeled product at a high purity level (>98% by analytical SEC), with the expected chain composition (Fig. 2a). In order to show that the exchange reaction is independent of the binder sequence and the format of the acceptor antibody, we produced a pair-FORCE matrix with the two different HER2 binders in three different formats, and evaluated the DAR by mass spectrometry (Fig. 2b). Figure 2c shows that all products contain a similar DAR after pair-FORCE. We also observed that the Fc donor precursor carried twice as many fluorophores than that observed in the products (Fig. 2c). This confirms the mode of the chain-exchange reaction, in which half of the donor molecule is transferred to the acceptor molecule, while the other half ends up in the dummy-dimer by-product. Pair-FORCE-mediated conversion of NHS-labeled conjugates into defined molecules with unaffected binding regions is not limited to low-molecular-weight payloads such as fluorescent dyes. The approach can also be utilized to attach enzymes such as horseradish peroxidase (HRP) conjugated via NHS chemistry to an Fc donor molecule (Supplementary Fig. 2).
Antibody-payload fusion proteins generated by chain-exchange
The pair-FORCE chain-exchange technology is not only restricted to transferring chemically conjugated entities. Payloads can also be attached in a site-specific manner to Fc donor molecules as fusion proteins (Fig. 1c). The Fc-payload-fusion protein can then be transferred to binder-acceptor modules, resulting in different antibody-fusion protein combinations. This application can attach defined fusion proteins to many different antibodies in a high-throughput manner. As an example to highlight this versatility, we produced Fc-payload-donor molecules harboring C-terminal fusions with enhanced GFP (EGFP). Such donor molecules can be expressed with similar yields compared to standard IgGs, and have favorable biophysical properties. Therefore, they can be produced as universal stock reagents for chain-exchange-mediated attachment to any exchange-enabled antibody derivative. The EGFP fusion serves as a payload transfer donor in the same manner as nonspecific- or site-specific Fc donor-payload conjugates. As an example, we used pair-FORCE to transfer EGFP to a HER2-binding acceptor module (Trastuzumab-derived) (Fig. 3). Such EGFP fusions can be generated in a simple and robust manner for many different binder combinations. They can be used for efficient epitope binning screens (Supplementary Fig. 3) and for monitoring the expression of cognate targets on the surface of tumor cells (Fig. 3). Thus, a C-terminal fusion protein on the Fc donor module does not interfere with chain-exchange and results in effective transfer of a fluorescent protein to binder-acceptor molecules.
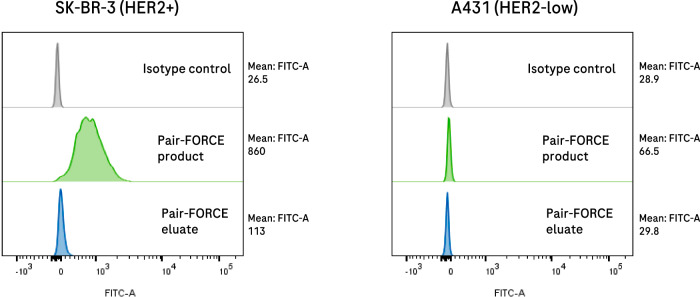
Fig. 3. Antibody-GFP fusion proteins can be generated by pair-FORCE.
EGFP can be transferred to antibody derivatives in different formats by pair-FORCE and can be applied to assess binding and the presence of target antigens by flow cytometry (see Methods section for flow cytometry details). A C-terminal EGFP molecule was transferred via pair-FORCE from a donor Fc-EGFP fusion to a HER2 binder-acceptor module in the N-format (derived from Trastuzumab). The pair-FORCE product (C-tag column flow-through) and pair-FORCE eluate (C-tag column eluate, contains by-products and unreacted Fc donor molecules) were incubated with HER2-positive SK-BR-3 cells or HER2-low A431 cells, and binding was determined by flow cytometry. EGFP fluorescence was monitored in the FITC channel. The successful transfer of EGFP to the Trastuzumab acceptor module was confirmed by the increased EGFP fluorescence on HER2-positive SK-BR-3 cells treated with the pair-FORCE product. Target-specific binding was retained after pair-FORCE, as only minimal fluorescence was detected on HER2-low A431 cells that were treated with the pair-FORCE product (right panel). FITC fluorescein isothiocyanate.
Payload transfer from Fc donor modules with site-specific conjugation
In addition to nonspecific payload conjugation and fusion protein generation, payloads can also be coupled to donor modules in a site-specific manner and can subsequently be transferred to sets of different binder-acceptor molecules in different formats. Examples of Fc donors that carry site-specific modifications introduced by enzyme-mediated coupling are shown in Fig. 1c. These include Fc donors that contain an Avi-tag, which enables site-specific biotinylation of binder-format combinations with pair-FORCE44. Such biotinylated antibodies can be immobilized onto SPR chips for investigation of binding kinetics, or used for immunoprecipitation through binding to streptavidin-coated beads. As an example, a biotinylated EGFR-binding antibody derivative generated by pair-FORCE was utilized to immunoprecipitate EGFR from a total cell lysate (Supplementary Fig. 4).
The enzymatic linkage between glutamine and lysine side chains by transglutaminase is a key approach we applied to conjugate Fc donor modules with payloads in a site-specific manner. To do so, we placed Q-tag sequences, which are specifically recognized by a unique microbial transglutaminase from Kutzneria albida (MTG)26, at different positions in the productive chain of the Fc donor molecules. The selected positions are located N-terminal of the hinge at amino acid 221, in the CH2 domain at amino acid 297, and the combination of both positions 297 + 221 (Fig. 1c). This MTG enzyme recognizes the Q-tag(s) on the Fc donor molecule and facilitates the formation of an isopeptide bond between a glutamine residue in the Q-tag and a lysine residue in the K-tag26. The simplest application of this MTG enzyme is the coupling of a payload harboring a K-tag to an acceptor molecule containing a Q-tag. For ADC generation and screening, we utilized a robust and versatile two-step procedure for payload coupling, similar to a previously described method28,45 (see Supplementary Figs. 1d, 5 for more information). Here, the MTG enzyme26 is used to conjugate a K-tag-containing linker-azide moiety onto a Q-tag in the Fc donor molecule. This results in an Fc donor precursor that can be coupled in a subsequent step to dibenzocyclooctyne (DBCO)-containing payloads via copper-free click chemistry28,46 (Supplementary Fig. 5a). The first reaction - the generation of azide-conjugated Fc donor modules using MTG—is efficient and quantitative, as measured by HIC (Supplementary Fig. 5b). The second reaction, which produces the desired payload-conjugated Fc donor molecule, is an azide-DBCO copper-free click reaction. Such reactions are robust and well established, and many DBCO-containing payloads for various applications are commercially available47–49.
One example is the attachment of Alexa Fluor 488-DBCO (AF488-DBCO) to an Fc donor that harbors an MTG-conjugated linker-azide moiety at amino acid position 297. The two-step conjugation method resulted in quantitative or near-quantitative conjugation of AF488 to various Fc donor molecules, as assessed by mass spectrometry (MS) (Table 1). Because the MTG-mediated site-directed payload conjugation targets only the productive chain of the Fc donor, DARs in the educts and the products of the exchange reaction are identical using this approach, as opposed to 50% transfer with a randomly conjugated donor (compare to Fig. 2b, c).
Table 1.
DARs of Fc donors and ADC products after two-step Fc donor conjugation and pair-FORCE
| Acceptor | Format | pHAb (NHS) | MMAE (Q221) | MMAE (Q297) | MMAE (Q221 + Q297) |
|---|---|---|---|---|---|
| Fc donor DAR | 2.2 | 0.6 | 0.8 | 1.3 | |
| Trastuzumab (HER2) | N | 1.2 | 0.7 | 0.8 | 1.6 |
| Trastuzumab (HER2) | C | 1.2 | 0.6 | 0.6 | 0.5* |
| Trastuzumab (HER2) | N + C | 1.1 | 0.7 | 0.8 | 1.3 |
| Pertuzumab (HER2) | N | 1.1 | 0.7 | 0.8 | 1.6 |
| Pertuzumab (HER2) | C | 1.2 | 0.6 | 0.7 | 1.4 |
| Pertuzumab (HER2) | N + C | 1.2 | 0.7 | 0.8 | 1.4 |
| Acceptor | Format | pHAb (NHS) | AF488 (Q297) | MMAE (Q297) |
|---|---|---|---|---|
| Fc donor DAR | 2.2 | 1.0 | 0.8 | |
| Cetuximab (EGFR) | N | 1.0 | 1.0 | 0.7 |
| Cetuximab (EGFR) | C | 1.1 | 1.0 | 0.8 |
| Cetuximab (EGFR) | N + C | 1.0 | 1.0 | 0.9 |
| Imgatuzumab (EGFR) | N | 1.1 | 0.9 | 0.8 |
| Imgatuzumab (EGFR) | C | 1.1 | 0.9 | 0.7 |
| Imgatuzumab (EGFR) | N + C | 1.1 | 1.0 | 0.8 |
Listed above are DAR values determined by mass spectrometry (MS) of different Fc donors or pair-FORCE products after either random conjugation of pHAb to the Fc donor (NHS chemistry), or after the two-step MTG-mediated azide-DBCO conjugation (AF488, MMAE). See the Methods section for details of the MS analysis. The DAR of AF488-conjugated HER2 antibody derivatives was not measured, but is expected to be similar to EGFR-binding antibody derivatives, as the same Fc donor was used. The DAR in Fc donor modules can be further increased by adding additional structurally compatible and accessible transglutaminase recognition sites. Exemplary intact MS spectra for MMAE-conjugated Fc donors and HER2-targeting MMAE ADCs are provided in Supplementary Fig. 6. *Strong adducts were observed for this molecule, which influenced the DAR determination (likely induced during MS measurement).
The two-step Fc donor preparation followed by chain-exchange-mediated payload transfer was also applied to generate a matrix of HER2 and EGFR-targeting ADCs conjugated with the antimitotic compound monomethyl auristatin E (MMAE). We tested different HER2 and EGFR binders in various formats, combined with different MMAE conjugation positions. In this case, the azide-DBCO reaction with commercial DBCO-MMAE reagents was only ~70% efficient, using the same azide-conjugated Fc donor precursors that were used for 100% effective AF488 conjugation. MS analyses revealed an MMAE DAR of ~0.7 for donors with one Q-tag (Q221 or Q297), and a DAR of ~1.4 for donors that harbor two Q-tags at positions Q221 + Q297 (Table 1 and Supplementary Fig. 6). The pair-FORCE reaction was very efficient in transferring the MMAE payload from the Fc donor to the antibody acceptors, with 100% transfer efficiency achieved for many formats and binders (compare the DAR of the Fc donors to the DAR of the pair-FORCE products in Table 1). Therefore, the DAR of most products was identical to the DAR of the Fc donor molecules, irrespective of the binder sequence or format. Thus, the compositions and DAR of pair-FORCE-derived products in the ADC binder-format-matrix are directly comparable with each other (with the sole exception of Trastuzumab in the C-format, Q221 + Q297, see Table 1 for details).
To confirm the robustness of the input molecules and resulting ADCs, we assessed the purity, thermal stability, and aggregation propensity of Fc donor molecules, HER2 binder-acceptor molecules, and HER2-MMAE ADCs. The results of these analyses (SEC and CE-SDS for molecule purity, nanoDSF and static light-scattering for thermal stability and aggregation propensity, respectively) indicate that all HER2-MMAE ADCs are of high purity with favorable thermal stability (Supplementary Figs. 7, 8). No aggregates were detected in the SEC analysis of most pair-FORCE generated HER2-MMAE ADCs, with a minority showing a maximum aggregation percentage of 0.5%. CE-SDS analysis showed correct chain composition and high levels of purity for all constructs (Supplementary Fig. 7). The majority of HER2-MMAE ADCs have Tm values above 63 °C and Tagg values above 69 °C (Supplementary Fig. 8). Importantly, HER2 ADC products generated by pair-FORCE show similar thermal stability attributes to their corresponding binder-acceptor modules, indicating that chain-exchange with an MMAE-conjugated Fc monomer does not impact thermal stability.
Fc donor molecules overall had reduced thermal stability compared to binder-acceptor molecules and pair-FORCE products, possibly due to the introduced Q-tag(s) (Supplementary Fig. 8). In addition, Fc donors conjugated with MMAE displayed slightly lower thermal stability compared to their unconjugated counterparts. However, since the conjugated Fc donor molecules are used as ADC-delivery reagents, and the resulting ADCs showed overall favorable thermal stability, we believe their thermal stability is sufficient for this application.
Chain exchange with MMAE-conjugated Fc donors should not affect the antibody binding regions. To confirm this, we investigated the binding kinetics of HER2-MMAE ADCs and HER2 binder-acceptor molecules using SPR (Supplementary Fig. 9). As expected, the binding kinetics of pair-FORCE-generated HER2-MMAE ADCs were nearly identical to those of the parental binder-acceptor antibodies (Supplementary Fig. 9). This demonstrates a major advantage of pair-FORCE, namely that the two-step site-specific conjugation and chain-exchange protocol does not interfere with the antibody binding regions.
Application example—HER2 binder-format-payload matrices for ADC screening
Parameters that influence the therapeutic applicability and efficacy of ADCs include (i) binding to tumor-cell surface antigens and accumulation on target-cell surfaces, (ii) internalization into target cells, (iii) metabolism of the ADC inside target cells, and (iv) cytotoxic activity combined with specificity towards target cells50. The desired activity is dependent on various molecule parameters, including the binder sequence and paratope, the targeted epitope, the binder format (which may affect functionality and internalization), the cytotoxic payload, the conjugation positions, and the DAR of the cytotoxic payload8,51. Combinations of these individual parameters result in complex matrices. To demonstrate that pair-FORCE can cover these aspects in an effective and comprehensive manner, we used this technology to transfer payloads to two different HER2-binding antibodies (Trastuzumab and Pertuzumab-derived binders) in the three different formats described in Fig. 1b (N, C, and N + C).
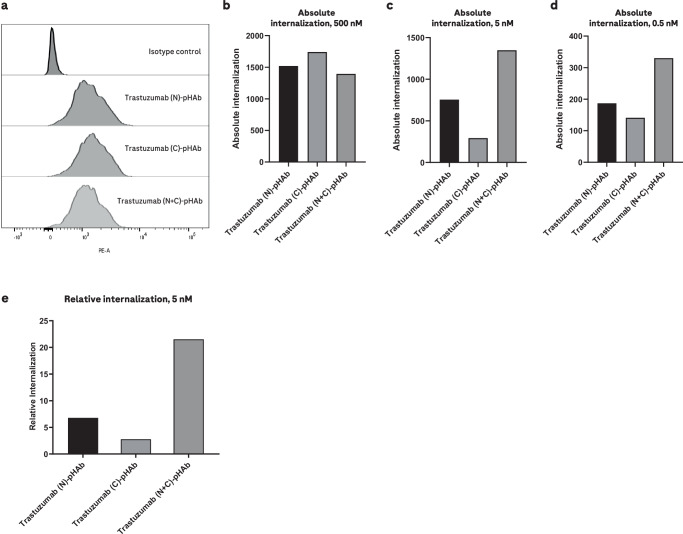
Target-cell binding of binder-format combinations was assessed with pair-FORCE-generated AF488-labeled molecules, generated using the two-step MTG conjugation and click chemistry approach (see above and Table 1), and internalization was monitored with molecules labeled with the pH-sensitive fluorescent dye pHAb (Fig. 4 and Table 1). Because the binder-format-AF488 and pHAb variants were generated by pair-FORCE, they contain very similar or identical DARs. This enables a direct comparison of binding as well as internalization efficacy of the different binder-format variants in the screening matrix. To determine the absolute internalization efficacy of Trastuzumab-derived HER2 binders in different formats and valencies, we incubated HER2-expressing SK-BR-3 cells with pair-FORCE-generated, pHAb-labeled HER2 antibodies at different concentrations, and measured pHAb fluorescence by flow cytometry (Fig. 4a–d). In parallel, we incubated the SK-BR-3 cells with pair-FORCE-generated, AF488-labeled HER2 antibodies and measured AF488 fluorescence by flow cytometry to determine the absolute binding. Using these two values, we calculated the relative internalization efficacy (the ratio of absolute internalization to absolute binding). The results of these analyses show that both absolute internalization (Fig. 4a–d) and relative internalization of HER2 binders (Fig. 4e) are clearly increased in the bivalent N + C format compared to the monovalent formats, confirming previous observations32. Monovalent HER2 binders require high concentrations to cause internalization, with monovalent binders in the N-terminal format showing better internalization than monovalent binders in the C-terminal format (Fig. 4e).
Fig. 4. Internalization of pair-FORCE-generated HER2 antibodies in different formats on HER2-expressing SK-BR-3 cells.
Internalization of Trastuzumab-derived HER2 binders in the N, C, and N + C format by HER2-expressing SK-BR-3 cells was assessed by flow cytometry using HER2 antibodies conjugated with the pH-sensitive dye pHAb. The conjugated HER2 antibodies were generated with pair-FORCE technology. The fluorescence of the pHAb dye is poor at neutral pH but increases substantially upon trafficking into the acidic environment of the endo-/lysosomal pathway13. For all panels, n = 1 independent replicates were performed. a Fluorescence histogram from flow cytometry experiments of SK-BR-3 cells treated with pHAb-conjugated HER2 binders at a saturating concentration of 500 nM. pHAb fluorescence was measured in the PE (phycoerythrin) channel. b The same experiment as (a), but absolute internalization is depicted as a bar graph. Absolute internalization is defined as the geometric mean of pHAb fluorescence in flow cytometry experiments. c Absolute internalization of pHAb-conjugated HER2 antibodies as in (b), but at a concentration of 5 nM. d Absolute internalization of pHAb-conjugated HER2 antibodies as in (b, c), but at a concentration of 0.5 nM. e Relative internalization of Trastuzumab-derived HER2 antibodies at a concentration of 5 nM. Relative internalization is calculated as the ratio of absolute internalization to absolute binding, which is derived from flow cytometry binding experiments with AF488-labeled HER2 antibodies generated by pair-FORCE (see Methods section for details). The absolute binding is defined as the geometric mean of AF488 fluorescence of SK-BR-3 cells treated with 200 nM of AF488-conjugated HER2 antibodies. N, C, and N + C refer to the binder formats in Fig. 1b. Source data for Fig. 4b–e are provided as a Source Data file.
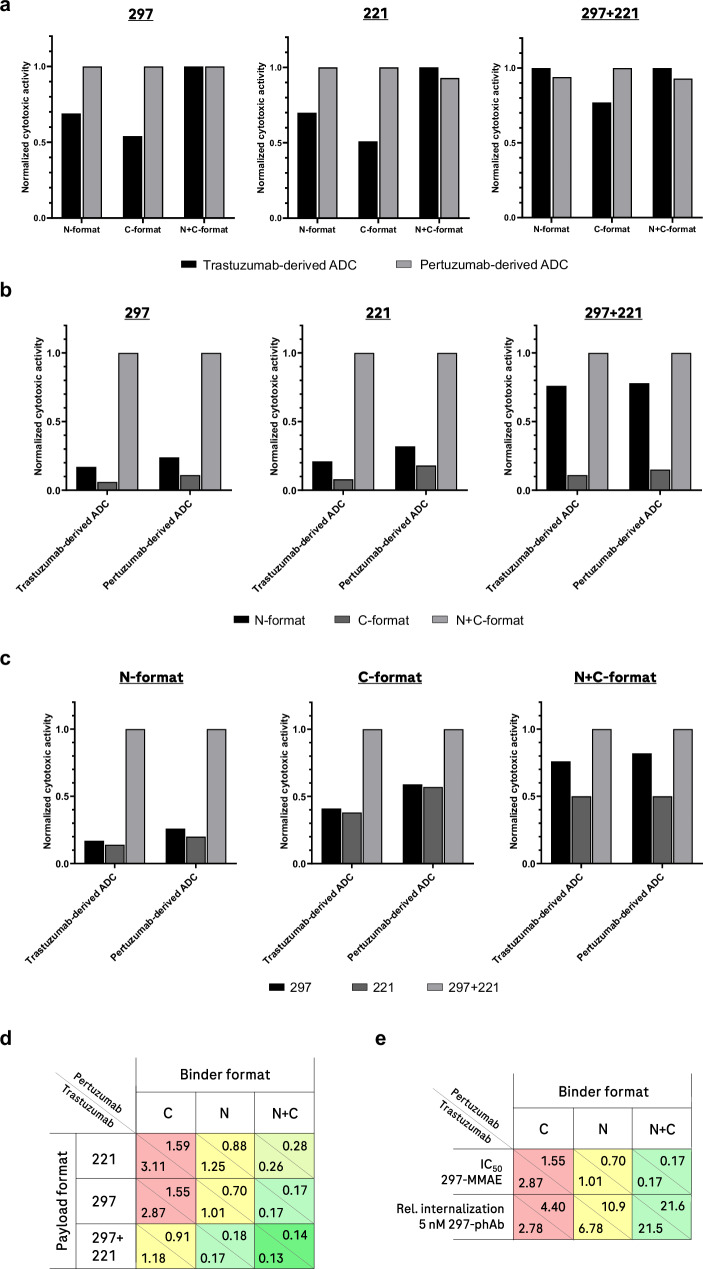
Next, the specificity and potency of HER2-MMAE ADCs in various binder-format-DAR combinations was assessed in cellular proliferation assays. Inhibition of cellular proliferation through bromodeoxyuridine (BrdU) incorporation was used as a readout for cytotoxicity (see Methods section). The results of these experiments show that the cytotoxicity of HER2-MMAE ADCs on HER2-expressing SK-BR-3 cells varies in potency depending on the HER2 binder, the format, and the DAR (Fig. 5). For further details, dose-response curves of HER2- and EGFR-MMAE ADCs, and controls, see Supplementary Fig. 10. Analyzing the effect of each parameter individually, we observed that the choice of HER2 binder (derived from Pertuzumab or Trastuzumab) has limited overall influence on potency (Fig. 5a). Both binders show the greatest (and very similar) activities when applied in bivalent formats, or with dual MMAE-occupancy (Q-tags at positions 297 + 221). However, in monovalent formats combined with one MMAE-conjugation site, Pertuzumab-derived binders performed slightly better than Trastuzumab-derived counterparts, underscoring the value of pair-FORCE in efficiently screening ADC parameters (Fig. 5a, d).
Fig. 5. Influence of individual and combined parameters on HER2-MMAE ADC activity.
In Fig. 5a–c, in order to define rules for each variable, the highest activity (i.e., the lowest IC50 value in cell proliferation/cytotoxicity assays) for a set of variables in each individual subgroup was set to 1.0, and the respective activities (IC50 values) were normalized relative to this value. This allows for the determination of the relative influence of the HER2 binder, format, and conjugation position/DAR on ADC activity. Relative values are comparable only within the same subgroup in each bar graph. Parameters that are compared in each panel are listed in the legend below the bar graphs. The values 297 and 221 refer to the MMAE conjugation site (Q-tag at positions 297 and 221 in the Fc donor productive chain). Individual data sets underlying this analysis are provided in Supplementary Fig. 10, based on triplicate experiments (n = 3). Source data for (5a–c) are provided as a Source Data file. a Comparison of the HER2 binders Trastuzumab vs Pertuzumab on HER2-MMAE ADC activity. b Influence of the binder format on HER2-MMAE ADC activity. c Influence of MMAE conjugation position and DAR on HER2-MMAE ADC activity. d Summary of the influence of binder, format, conjugation position, and DAR variables on HER2-MMAE ADC activity (indicated as IC50 values in nM, see dose-response curves in Supplementary Fig. 10). The relative activity of each construct is color-coded as a gradient ranging from red (least active), to yellow (medium activity), and finally to bright green (most active). e The binder format influences internalization, which correlates with activity. In the first row of the table, IC50 values from cell proliferation/cytotoxicity assays are indicated in nM, as in (5d). See Supplementary Fig. 10 for dose-response curves. In the second row, relative (rel.) internalization efficacies are indicated as in Fig. 4e. The ADC activity and relative internalization are color-coded as in (5d).
The format and valency of the binder entities had a much greater influence on ADC potency. Figure 5b shows that bivalent ADCs showed generally better potencies than monovalent formats, regardless of whether the binder entities were derived from Pertuzumab or Trastuzumab. Interestingly, in the monovalent setting, the N-format combined with dual MMAE conjugates (Q-tags at positions 297 + 221) retained similar (only slightly reduced) activity compared to bivalent ADCs. This was irrespective of the binder being derived from Pertuzumab or Trastuzumab. All other ADCs in monovalent binder formats showed inferior potencies to bivalent formats, with C-formats performing worse than N-formats.
Assessment of the influence of MMAE conjugation position and DAR (Fig. 5c) reveals that conjugation of MMAE on two sites (Q-tags at positions 297 + 221) generates ADCs with greater potencies compared to corresponding binder-format variants with only one MMAE conjugation site. This difference was observed with all formats and binders tested, with the largest difference between two conjugation sites and one conjugation site observed in the N-terminal format. Interestingly, smaller differences were observed in the N + C bivalent format, with Q-tag position 297 retaining similar (only slightly reduced) activity compared to Q-tag positions 297 + 221. This result demonstrates that increasing the binder valency can partially overcome the lower DAR from molecules with only one conjugation site. Monovalent ADCs with one conjugation site appeared to be slightly more active with MMAE conjugated to position 297 compared to MMAE conjugated to position 221, but the difference was minor. However, in the N + C bivalent format, position 297 was notably more active than position 221 (Fig. 5c). Figure 5d shows the effects of all parameters summarized in one matrix, revealing the combined effects of format, binder valency, conjugation position, and DAR for both HER2 binders. The format-dependent effect on internalization is a major factor for ADC efficacy, as shown in Fig. 5e. ADC activity was dependent on expression of the target antigen, as HER2- and EGFR-MMAE ADCs were inactive in all formats and combinations on low-HER2 and low-EGFR expressing MCF-7 cells (Supplementary Fig. 11). ADC activity also correlated with target-antigen density, as shown by the potency differences on cell lines with different levels of HER2 expression (Supplementary Fig. 12).
Discussion
The original FORCE technology29 generates matrices that cover binder specificity, binder format, and binder valency in order to identify optimal designs and compositions of bsAbs. We have demonstrated that the format-defines-function concept, which defines optimal bsAbs39, also applies to ADCs (Fig. 5d). Parameters that should be evaluated in ADC design campaigns include binder specificity and format, binder valency, type of cytotoxic payload, conjugation position, linker composition, and DAR. Thus, the identification of ADCs with optimal functionality requires the generation and screening of matrices that cover large design spaces. Therefore, we expanded and modified the chain-exchange-based, high-throughput-automation-compatible FORCE technology to enable the coverage of large design spaces for ADCs. We refer to this modified chain-exchange technology as pair-FORCE.
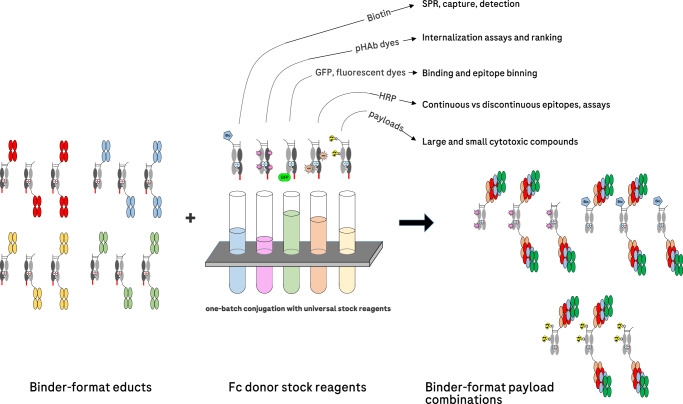
The advantage of pair-FORCE for generating ADC matrices compared to individual conjugations lies in the application of Fc donor stock reagents harboring payloads such as cytotoxic payloads, fluorescent dyes, or fusion proteins. These payloads can be conjugated to stock Fc donor molecules by different conjugation methods at different positions with varying DAR. These stock Fc donor reagents can then be used to transfer the conjugated productive chain to binder-acceptor educts in a defined manner (Fig. 1). The combination of one set of binder-acceptor educts with different Fc donor-payload stock reagents generates antibody-payload combinations to assess a variety of parameters important for ADC development (Fig. 6). The same chain-exchange technology used to generate ADCs with different payloads, conjugation sites, and DAR can be applied using different Fc donor stocks to cover the simple and robust generation of additional reagents valuable for ADC development. This includes the transfer of donors that harbor GFP or fluorophores to address binding efficacy, binding specificity, and internalization. Furthermore, biotinylated or enzyme-labeled donors generate versatile assay reagents (e.g., for investigating binding kinetics via SPR). The generation of comprehensive ADC matrices by robust automation-compatible exchange reactions using universal, exchange-enabled Fc-payload stock reagents enables the differentiation of ADC functionalities (Figs. 4, 5). In addition, the analysis of comprehensive matrices generated by pair-FORCE may provide rules of design for the development of ADCs for given targets and payloads.
Fig. 6. Combination of binder/format acceptor-educts with payload-conjugated Fc donor stock reagents generates binder-format-payload matrices for ADC screening and other related applications.
Pair-FORCE is a versatile technique that can be applied for numerous applications relating to ADC screening and development, as described above. The versatility is enabled by the robust chain-exchange reaction with payload-conjugated Fc donor stock reagents. SPR surface plasmon resonance, pHAb pH-sensitive dye13, GFP green fluorescent protein, HRP horseradish peroxidase.
The use of pre-conjugated Fc donor stock reagents is not only a simple and robust approach to generate ADC matrices; it also eliminates potential variabilities that may otherwise be introduced by individual conjugation reactions. For optimal evaluation of parameters influencing ADC function, it is crucial that ADCs in the matrix are well defined. Pair-FORCE allows for tunable control of all parameters, most importantly the site-specific payload conjugation of the Fc donor molecule, which allows for control of the DAR and conjugation positions. As next-generation ADCs are increasingly focusing on site-specific conjugation approaches to overcome challenges caused by heterogeneous conjugation methods (e.g., poor pharmacokinetic properties, low payload delivery, and a suboptimal therapeutic index)8,9,11,16, pair-FORCE presents a simple and modular site-specific conjugation approach to enable screening of important ADC parameters. An additional advantage of the site-specific conjugation method employed in pair-FORCE is that any effect on the integrity of binding regions in the resulting ADCs can be excluded, such as potential effects caused by nonspecific conjugation of lysine residues in select antibodies.
The demonstrated example of a HER2 ADC matrix generated by pair-FORCE highlights the value of this technique in screening for optimal ADCs and defining rules of design. We determined that for tumor cells expressing a high level of HER2, the optimal ADCs in terms of cytotoxic activity contained bivalent HER2 binders (Fig. 5d), which correlated with increased internalization compared to monovalent HER2 binders (Figs. 4e, 5e). The best-performing ADCs in cytotoxicity assays contained bivalent HER2 binders and two MMAE conjugation sites, corresponding to a DAR of ~1.4 (Fig. 5d and Table 1). Due to the granularity of the pair-FORCE matrix, we also could determine that monovalent HER2 binders in the N-format with two MMAE conjugation sites had a similar potency to bivalent HER2 binders with one MMAE conjugation site (Fig. 5d). This result demonstrates that internalization and DAR can be tuned to achieve similar cytotoxic potencies. This is important because it shows that varying one parameter can overcome the influence of another parameter on overall cytotoxic potency. Therefore, screening and optimization of parameters simultaneously using pair-FORCE is advantageous because it enables the selection of parameters that result in optimal potency, while allowing for variation in other parameters important for ADC development. Parameters including DAR, linker composition, and conjugation site have been shown to affect the hydrophobicity, stability, and aggregation propensity of ADCs, all of which can affect PK, tumor delivery, and the therapeutic index9. They are thus crucial parameters influencing the development of ADCs. For example, a greater DAR has been shown to lead to increased hydrophobicity, lower thermal stability, and correlates with less-favorable pharmacokinetic properties6,37,52. Pair-FORCE allows for tuning of these parameters in complex matrices of different binders with varying formats, all while keeping the DAR, conjugation sites, and linker composition nearly identical between the ADC constructs.
Designed as a robust screening technology, molecules generated by pair-FORCE do not always represent final ADC formats. However, despite the format difference to standard IgGs, bivalent pair-FORCE HER2-MMAE (N + C) constructs showed greater internalization and cytotoxicity than monovalent constructs, confirming previous observations about increased HER2 internalization with bivalent HER2 binders compared to monovalent binders32 (Fig. 5e and Supplementary Fig. 10). We tested two conjugation sites in this study, with a theoretical maximum DAR of 2. The DAR could be further increased by including additional Q-tags in the Fc donor molecule, or by using branched linkers with multiple-click reaction sites53,54. The pair-FORCE technology is also currently incompatible with certain conjugation approaches, for example maleimide alkylation of payloads on reduced hinge-cysteines. However, Fc donor modules could potentially be reduced and alkylated with payloads in the hinge region, while binder-acceptor modules could be engineered with mutated hinge regions lacking disulfide bonds. We have already demonstrated that chain-exchange of antibody derivatives is functional without the use of reducing agents, when hinge disulfides are removed55. Nevertheless, despite some final format limitations, pair-FORCE is suitable to rapidly screen for cytotoxic activity, binding, internalization, and biophysical properties between various antibody formats. The findings can thereafter be transferred to a final ADC format, where potential rules for design can be tested. Pair-FORCE is, therefore, a versatile and valuable technology with the potential to increase the throughput of ADC design campaigns and enable the establishment of rules of design for specific targets and cytotoxic payloads.
Methods
Antibody binder sequences and design of pair-FORCE educts
The following binder clones were used in this study to produce antibody derivatives for pair-FORCE. HER2: Trastuzumab, clone 4D5-856, Pertuzumab, clone rhuMAb 2C457. EGFR: Cetuximab, clone C22558, Imgatuzumab, clone GA-20159. Pair-FORCE educts were designed in a similar manner as already described for bsAb FORCE educts29, containing the destabilizing charge mutations E357K and K370E in the CH3 domains of the corresponding dummy chains. Heterodimerization of heavy chains was induced using knob-into-hole mutations in the CH3 domains60,61. The knob mutation used in this study was T366W, and the hole mutations were T366S, L368A, and Y407V. Similar to FORCE dummy chains, pair-FORCE dummy chains do not contain the engineered cysteine residues at positions 354/349 in the CH3 domains, which normally forms an extra disulfide bridge between the CH3 domains29,61. The pair-FORCE products do, however, contain the engineered disulfide bridge in the CH3 domain after chain exchange. The dummy chains were designed with a C-terminal C-tag composing the amino acids EPEA, which allows for selective binding of unreacted educts, dummy-dimers, and aggregates to a CaptureSelect™ C-tagXL affinity column (Thermo Fisher Scientific). For MTG-mediated conjugation, the Q-tag sequence YRYRQ was included in the Fc donor molecule in the positions stated in the text. In the linker-azide molecule, the K-tag sequence was RYESK26,45. For site-specific biotinylation of Fc donor constructs, the BirA recognition sequence GLNDIFEAQKIEWHE was included N-terminal of the hinge. The Lys residue in the recognition sequence is biotinylated by BirA44.
Expression and purification of antibody derivatives
Expression plasmids encoding the respective heavy and light chains of antibody derivatives in this study contained a CMV promoter and an IgG VH signal sequence, which leads to the secretion of the antibodies into the cell culture supernatant29. Recombinant antibody derivatives were expressed in transiently transfected HEK293 cells (Expi293F™, Thermo Fisher Scientific) according to the manufacturer’s instructions, and as previously described29. Expression yields for pair-FORCE Fc donor molecules and binder-acceptor molecules was comparable to standard IgGs, as previously described29. The supernatants were harvested by centrifugation at 3500×g for 45 min, followed by sterile filtration (0.22 µm filter). Antibody derivatives were purified from the supernatant using a HiTrap™ MabSelect SuRe protein A column (Cytiva, 11003494) followed by size exclusion chromatography using a HiLoad® 26/600 Superdex® 200 column (Cytiva, 28989335). Peak fractions were analyzed by reducing and non-reducing SDS-PAGE. Fractions containing the correct chain composition were pooled and concentrated using an Amicon Ultra-15 centrifugal filter with the appropriate molecular weight cut-off (Merck Millipore).
Analytical SEC and capillary SDS electrophoresis (CE-SDS)
To analyze the aggregation and monomeric purity of the purified antibody derivatives, an analytical SEC was performed. For analytical SEC, 40 µg of antibody derivatives were loaded onto a BioSuite 250, 5 µm HR SEC column (7.8 mm × 300 mm, Waters) connected to an UltiMate 3000 UHPLC (Thermo Fisher Scientific), with a running buffer of 200 mM KH2P04, 250 mM KCl at pH 6.2. The data were analyzed with Chromeleon CDS software (Thermo Fisher Scientific). In order to quantify purity and chain composition, as in Fig. 2a, non-reducing and reducing capillary SDS electrophoresis (CE-SDS) was performed. For this purpose, antibody derivatives were analyzed using the LabChip® GXII Touch™ HT Protein Characterization System (Perkin Elmer), according to the manufacturer’s instructions.
DAR determination of conjugated Fc donor molecules and pair-FORCE products by mass spectrometry
In order to calculate the DAR and to check the integrity of the conjugated Fc donor molecules and pair-FORCE products, mass spectrometry (MS) was performed. Before measurement, the samples were deglycosylated by adding N-glycosidase F (Roche Diagnostics, Penzberg, Germany). The deglycosylation was performed in 0.1 M sodium phosphate buffer at pH 7.1, at a ratio of 0.14 U/µg antibody. The reaction was incubated for 16 h at 37 °C, and samples were subsequently separated by reverse-phase chromatography. This was performed using a PLRP-S column (Agilent, Waldbronn, Germany) with mobile phase A containing 0.1% (v/v) formic acid in UPLC grade water, and mobile phase B containing acetonitrile (Fisher Chemical, Schwerte, Germany). The column temperature was 75 °C, and a gradient of 25% to 40% mobile phase B was used for separation. MS spectra were acquired using a MaXis Q-TOF instrument (Bruker Daltonics, Bremen, Germany) controlled by Compass 6.2 software. A total of 49 samples were analyzed, with one technical replicate each. One control sample (two technical replicates), consisting of a mixture of two different bispecific antibodies in different formats and one standard IgG, was run within the sequence to check LC separation and MS data quality. For data evaluation, in-house-developed software was used. The data annotation was performed using m/z spectra, and a deviation of a maximum of 75 ppm between theoretical and experimental mass was used to confirm the identity of the species. For DAR estimation, the average intensity ratio of each suitable charge state was used. Detailed MS acquisition settings were as follows: ESI Apollo source parameters: Capillary: 5000 V, Nebulizer: 1.6 Bar, Dry Gas 9 l/min, and 230 °C dry temp. Further details are provided in Table 2.
Table 2.
Additional mass spectrometry measurement parameters (see Methods section)
| MMAE-conjugated Fc donors | pair-FORCE products | |
|---|---|---|
| Transfer isCID | 0 eV | 110 eV |
| Collision Cell RF | 2200 Vpp | 4000 Vpp |
| Collision Transfer time/pre pulse storage | 120 µs/12.0 µs | 120 µs/15.0 µs |
| m/z range | 600–2000 | 1000–4000 |
Payload conjugation to exchange-enabled Fc donor molecules
Fc donor molecules were conjugated with payloads to generate Fc donor-payload entities for chain exchange reactions. Enhanced GFP (EGFP) was genetically fused to the C-terminus of the Fc donor with a Gly4Ser (G4S) linker. Biotin was conjugated in a site-directed manner via AviTag/BirA technology using the BirA Bulk Kit (Avidity LLC), as per the manufacturer’s instructions. Conjugation of reactive amines on Fc donor molecules with the NHS-coupled pH-sensitive fluorescent dye pHAb was performed using the Promega pHAb Amine Reactive Dye Kit (Promega, G9845), according to the manufacturer’s instructions (including the DAR calculation). Labeling of reactive amines with horseradish peroxidase (HRP) was performed using the EZ-Link® Activated Peroxidase Antibody Labeling Kit (Thermo Fisher Scientific, 31497), as per the manufacturer’s instructions. Site-directed MTG-mediated conjugation of K-tag containing azide-linker adapters to Q-tags on Fc donor molecules is described below.
Two-step, site-specific Fc donor conjugation with MMAE and fluorescent dyes
In order to conjugate MMAE and AF488 onto Fc donor molecules for ADC matrices, a two-step conjugation method was performed in a similar manner as previously described28,45. In the first step, a linker-azide moiety was conjugated onto a Q-tag or multiple Q-tags on the Fc donor molecule using a specialized MTG enzyme26. A tenfold molar excess of linker-azide was mixed with the antibody, along with the MTG enzyme at a 1:50 molar ratio of enzyme to antibody. The reaction mixture was incubated for 3 h at 37 °C and was terminated by adding excess ammonium sulfate. The linker-azide-conjugated Fc donor molecule was then purified by SEC. In the second step, the linker-azide-conjugated Fc donor was coupled with MMAE or AF488 using strain-promoted azide-alkyne cycloaddition (SPAAC) (Cu-free click chemistry reaction). For this purpose, the linker-azide-conjugated Fc donor was mixed with a fivefold molar excess of DBCO-(PEG)3-VC-PAB-MMAE (MedChem-Express, Cat. No. HY-111012) or DBCO-AF488 (Jena Bioscience, Cat. No. CLK-1278-1). The reaction was incubated overnight at 25°C, and the final conjugated Fc donor molecule was purified by SEC. Conjugation efficiency was analyzed by HIC after each step.
Hydrophobic interaction chromatography (HIC)
In order to assess the conjugation efficiency of the two-step, site-specific Fc donor conjugation, analytical HIC was performed. Conjugation of Fc donor educts with the azide-linker adapter and subsequent conjugation of DBCO-containing payloads with Cu-free click chemistry increases the hydrophobicity of the molecule, which can be analyzed by HIC. Briefly, 40 µg of each sample was applied to a TSKgel Butyl-NPR HPLC column (2.5 μm, 4.6 mm × 35 mm; Tosoh Bioscience, 0014947) using an UltiMate 3000 UHPLC (Thermo Fisher Scientific). Binding was performed in HIC buffer A (20 mM Na2HPO4, 1.5 M (NH4)2SO4, pH 7.0), with 5% of HIC buffer B (20 mM Na2HPO4, 25 % (v/v) isopropanol, pH 7). A gradient of 5–100% HIC buffer B was applied during the run.
Transfer of payloads from Fc donor-payload educts to binder-acceptor educts by chain exchange
The chain-exchange reaction was performed essentially as previously described29, with some minor modifications. Briefly, Fc donor-payload and binder-acceptor educts were combined in equimolar amounts at a concentration of 1 mg/ml in PBS, pH 7.4. In order to reduce the disulfide bridges in the hinge region, a 20-fold molar excess of TCEP containing 0.05% Tween 20 was added, which initiates the chain-exchange reaction. The mixture was incubated for 3 h at 37 °C while shaking at 300 rpm. The chain-exchange products were purified by application of the reaction mixture onto a CaptureSelect C-tagXL column (Thermo Fisher Scientific, 494307205), which binds to C-tag-containing aggregates, unreacted educts, and dummy-dimers. The flow-through contains the pair-FORCE product. Pair-FORCE products were incubated for 5 days at 4°C to allow for reoxidation of disulfide bridges. The products were further analyzed by analytical SEC and CE-SDS, as described above.
Cell culture
The cell lines SK-BR-3 (ATCC, HTB-30), MCF-7 (ATCC, HTB-22), A431 (ATCC, CRL-1555), SK-OV-3 (ATCC, HTB-77), MDA-MB-453 (ATCC, HTB-131), NCI-H1650 (ATCC, CRL-5883), and MDA-MB-468 (ATCC, HTB-132) were cultivated in RPMI-1640 media supplemented with 10% FCS and 2 mM glutamine. For A431 cultivation, the media was supplemented with 1 mM sodium pyruvate. The cell lines were cultured at 37 °C with 5% CO2 and 80% humidity. For sub-culturing, the cells were detached with Accutase (PAN Biotech) and were counted using a Vi-CELL XR cell counter (Beckman Coulter).
Immunoprecipitation and Western blot
In Supplementary Fig. 4, immunoprecipitation of EGFR from whole cell lysate was performed using a biotinylated EGFR-binding antibody derivative generated by pair-FORCE. A total of 3 × 106 A431 cells were resuspended on ice in 1 ml RIPA lysis and extraction buffer (Thermo Fisher Scientific, 89900). Cell debris was removed by centrifugation at 15,000×g for 15 min, and the supernatant was collected. Immunoprecipitation with the biotinylated C225 anti-EGFR antibody derivative was performed with Pierce™ Streptavidin Magnetic Beads (Thermo Fisher Scientific, 88816), according to the manufacturer’s instructions. Briefly, the biotinylated C225 antibody was incubated with the A431 lysate for 2 h at room temperature. The antibody-lysate mixture was added to streptavidin magnetic beads pre-washed with PBS, and the mixture was incubated for 1 h at room temperature with rotation. The beads were washed three times with PBS, and elution was performed with 50 µl of NuPAGE™ LDS Sample Buffer (Thermo Fisher Scientific, NP0007), with heating at 95 °C for 5 min. After the separation of the magnetic beads, 20 µl of eluate was loaded onto a 4–12% Bis-Tris gel (Invitrogen, NP0322). Blotting was performed using Trans-Blot Turbo Mini 0.2 µm PVDF Transfer Packs (Bio-Rad, 1704156) with the Trans-Blot Turbo Transfer System (Bio-Rad), according to the manufacturer’s instructions. The membrane was blocked for 30 min at room temperature in a blocking buffer (TBS with 5% skim milk and 0.05% Tween 20). The membrane was then incubated overnight in a blocking buffer containing an anti-EGFR primary antibody (Abcam, ab264540, 1:1000 dilution). On the following day, the membrane was washed three times with TBS-T (TBS with 0.05% Tween 20) and was incubated with the secondary antibody (Polyclonal Goat anti-Mouse Immunoglobulins/HRP; Agilent Dako, P044701-2, 1:1500 dilution) in blocking buffer for 1 h at RT. After three rounds of washing with TBS-T, chemiluminescence was detected using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, 34579) and the Gel Doc XR+ Gel Documentation System (Bio-Rad). For testing the functionality of the pair-FORCE-generated C225-HRP antibody derivative (Supplementary Fig. 2), the western blot procedure was performed in a similar manner. Briefly, A431 whole cell lysate was loaded onto the gel, and blotting was performed as above. C225-HRP was applied as the primary antibody at a concentration of 0.24 µg/ml in blocking buffer, and the detection protocol was performed as above, but with SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, 34095).
Flow cytometry: binding and internalization assays
In Supplementary Fig. 3, the binding of an EGFP-conjugated HER2 pair-FORCE product to target cells was assessed by flow cytometry. For this purpose, a total of 2 × 105 A431 cells were incubated with 200 nM of antibody derivatives in FACS buffer (PBS with 2% FCS) for 1 h on ice. Cells were then washed twice with cold PBS, and fluorescence was measured in the FITC channel of a FACS Canto II instrument (BD Biosciences). FlowJo software (BD Biosciences) was used for data analysis and visualization. Gating for single viable cells was performed for all flow cytometry analyses using standard forward-scatter and side-scatter analysis. For epitope binning assays (Supplementary Fig. 3), flow cytometry analyses were performed as above, with additional pre-incubation of A431 target cells with 200 nM of different anti-EGFR antibodies (P2X, mab806, and P1X), which bind to known epitopes on EGFR domains I, II, and III, respectively62,63. FITC intensity was measured using a FACS Canto II instrument as above. To assess internalization of HER2-targeting, pair-FORCE-generated molecules (Figs. 4, 5e), 0.75 × 105 SK-BR-3 cells were seeded out in flat-bottom 96-well plates and treated with various concentrations of HER2 binders conjugated with pHAb by pair-FORCE (500 nM, 50 nM, 5 nM, 0.5 nM, 0.05 nM, and 0 nM final antibody concentration), in a total volume of 200 µl. The cells were incubated for 24 h at 37 °C in a humidified 5% CO2 atmosphere. The cells were then detached with 100 µl Accutase (Pan Biotech, P10-21100), washed twice with PBS, and analyzed via flow cytometry in the PE channel as described above. In order to calculate the relative internalization efficacy of pair-FORCE-generated HER2 binders (defined as absolute internalization divided by absolute binding), binding studies were performed with pair-FORCE-generated HER2 binders labeled with AF488. Briefly, 2 × 105 SK-BR-3 cells were incubated with 200 nM of AF488-labeled HER2 binders in FACS buffer for 30 min on ice. The cells were subsequently washed twice with cold PBS, and fluorescence was measured by flow cytometry in the FITC channel, as described above. Data were collected for 20,000 cells per condition. Absolute internalization and absolute binding were defined as the geometric mean of fluorescence histograms from flow cytometry internalization (pHAb) and binding (AF488) experiments, respectively, and were calculated using FlowJo software. For visualization of bar graphs, GraphPad Prism 9 software was used (GraphPad Software, San Diego, CA, USA).
HER2 binding kinetics of pair-FORCE ADCs and HER2 binder-acceptor modules
For the assessment of the HER2 binding kinetics of HER2-targeting pair-FORCE ADCs and HER2 binder-acceptor modules, a Biacore 8 K+ (GE Healthcare) SPR system was used. First, an anti-human Fab capture ligand was immobilized onto a Series S CM4 (Cytiva 29104989) sensor chip via amine coupling, according to the manufacturer’s instructions (Human Fab Capture Kit, Cytiva, 28958325). Pair-FORCE antibodies were captured on the chip at a flow rate of 10 µl/min for 180 s, and capture ranged between 360–600 RU. An Fc-fused HER2 extracellular domain (ECD) was flowed as an analyte at a rate of 30 µl/min for 120 s, followed by dissociation for 600 s at the same flow rate. The analyte concentrations tested were 0, 5, 25, and 125 nM. Regeneration was performed using regeneration buffer (10 mM glycine, pH 2.0) for 60 s at a flow rate of 30 µl/min at the end of each SPR cycle. The equilibrium constant (KD) and kinetic rate constants were determined by fitting the data to a 1:1 Langmuir interaction model using Biacore™ Insight software (Cytiva).
Thermal stability measurements
Thermal stability measurements of MMAE-conjugated Fc donor molecules, HER2-targeting binder-acceptor molecules, and pair-FORCE-generated HER2-targeting MMAE ADCs were performed using an Uncle instrument (Unchained Labs). Briefly, 9 µl of each sample was loaded in duplicates into the capillaries of the Uncle Uni. As a standard, an in-house reference antibody was used at a concentration of 1 mg/ml. The Unis were loaded into the Uncle instrument and a thermal ramp from 30–90 °C was performed with a ramp rate of 0.1 °C/min. Uncle software was used to calculate the Tm of each sample using the first derivative of the barycentric mean (BCM) of intrinsic fluorescence intensity. The Tagg of each sample was calculated according to the standard Uncle algorithm using the intensity of scattered light at 266 nm.
Cell proliferation assays for assessment of ADC cytotoxicity
The cytotoxicity of pair-FORCE-generated ADCs was assessed using the bromodeoxyuridine (BrdU) colorimetric cell proliferation assay (Roche, Cat. No. 11647229001). Cells were seeded into 96-well flat-bottom tissue culture-treated microplates at a density of 7500 cells per well for MCF-7 and A431 cells, and 10,000 cells per well for SK-BR-3 cells (to account for the slower growth rate of SK-BR-3 cells). The cells were incubated for 24 h at 37 °C, 5% CO2, and 80% humidity, and were subsequently treated with serial dilutions of pair-FORCE-generated ADCs (500, 50, 5, 0.5, and 0.05 nM) for 72 h. After 72 h, the cells were incubated with BrdU for 3 h, fixed using FixDenat for 30 min, and incubated with Anti-BrdU POD for 90 min. The cells were washed with washing solution and the substrate solution was added and incubated for 3 min. Subsequently, cell proliferation was recorded by measuring absorbance at 370 nm using a plate reader (Tecan Group AG). The cytotoxic activity of ADCs was calculated from the BrdU assay, in which the measured signal represents DNA synthesis in viable cells. The percentage of viable cells depicted on the Y axis of cytotoxicity assays was calculated as the percentage of BrdU signal reduction relative to untreated cells. The IC50 value was determined by fitting the data with the equation y = 100/(1 + x/IC50) in GraphPad Prism 9, where x corresponds to the ADC concentration in nM, and y corresponds to the percentage of cell viability. Triplicates were performed for each data point.
Quantification of HER2 receptors on different cell lines
Quantification of HER2 receptors on different cell lines was performed using the QIFIKIT® (Agilent, K0078), according to the manufacturer’s instructions. Briefly, 100 µl of setup and calibration beads were washed three times with 1 ml of FACS buffer (PBS + 2% FCS). The beads were then resuspended in 98 µl of FACS buffer. A total of 3 × 105 cells (SK-OV-3, SK-BR-3, MDA-MB-453, NCI-H1650, MCF-7, MDA-MB-468) were incubated with 3.75 µg of anti-human CD340 (Clone 24D2, BioLegend) or isotype control (Mouse IgG1 clone MOPC-21, BioLegend) in FACS buffer for 45 min on ice. The cells were then washed three times with FACS buffer and resuspended in a total volume of 97 µl. Subsequently, 3 µl of FITC-conjugated anti-mouse antibody (QIFIKIT®) were added to both the cells and the beads, followed by another 45 min incubation on ice. After three additional washes, the cells and beads were resuspended in FACS buffer in a total volume of 125 µl (cells) and 200 µl (beads). Measurement and data acquisition of the beads and cells were performed according to the manufacturer’s instructions (QIFIKIT®), using a FACS Canto II instrument (BD Biosciences). Ten thousand cells were measured per condition. Generation of the calibration curve and HER2 quantification were carried out according to the manufacturer’s instructions (QIFIKIT®), using FlowJo software (BD Biosciences) and GraphPad Prism 9.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
V.V. and S. Dickopf were supported by the Roche Postdoctoral Fellowship (RPF) program.
Author contributions
V.V., S. Dickopf, T.S., S. Dengl, H.D., and U.B. devised the concept. V.V., S. Dickopf, N.S., R.-S.R., M.F., K.M., T.S., H.D., and B.N. designed, performed, and analyzed experiments. V.L. performed mass spectrometry characterization and analyzed the data. J.A.B. designed, performed, and analyzed the SPR experiments. V.M. designed, performed, and analyzed the thermal stability experiments. V.V., S. Dickopf, and U.B. wrote the manuscript. All coauthors reviewed and approved of the final manuscript.
Peer review
Peer review information
Nature Communications thanks Itai Benhar and Adrian Hugenmatter for their contribution to the peer review of this work. A peer review file is available.
Data availability
The ADC cytotoxicity data generated in this study and source data for all quantitative figures are provided in the Source Data file. All other data were available from the corresponding author(s) upon request. Source data are provided with this paper.
Competing interests
All coauthors are either employed by Roche or were previously employed by Roche at the time the work was conducted. Roche has an interest in, and patents/patent applications including V.V., S. Dickopf, K.M., H.D., S. Dengl, and U.B. as inventors that relate to FORCE technologies and antibody engineering.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Vedran Vasic, Steffen Dickopf.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53730-3.
References
- 1.Dumontet, C., Reichert, J. M., Senter, P. D., Lambert, J. M. & Beck, A. Antibody-drug conjugates come of age in oncology. Nat. Rev. Drug Discov.22, 641–661 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Liu, K. et al. A review of the clinical efficacy of FDA-approved antibody‒drug conjugates in human cancers. Mol. Cancer23, 62 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akkapeddi, P. et al. Construction of homogeneous antibody-drug conjugates using site-selective protein chemistry. Chem. Sci.7, 2954–2963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drago, J. Z., Modi, S. & Chandarlapaty, S. Unlocking the potential of antibody-drug conjugates for cancer therapy. Nat. Rev. Clin. Oncol.18, 327–344 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross, P. L. & Wolfe, J. L. Physical and chemical stability of antibody drug conjugates: current status. J. Pharm. Sci.105, 391–397 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Hamblett, K. J. et al. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin. Cancer Res.10, 7063–7070 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Anami, Y. et al. Homogeneity of antibody-drug conjugates critically impacts the therapeutic efficacy in brain tumors. Cell Rep.39, 110839 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riccardi, F., Dal Bo, M., Macor, P. & Toffoli, G. A comprehensive overview on antibody-drug conjugates: from the conceptualization to cancer therapy. Front. Pharm.14, 1274088 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck, A., Goetsch, L., Dumontet, C. & Corvaïa, N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat. Rev. Drug Discov.16, 315–337 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Sau, S., Alsaab, H. O., Kashaw, S. K., Tatiparti, K. & Iyer, A. K. Advances in antibody-drug conjugates: A new era of targeted cancer therapy. Drug Discov. Today22, 1547–1556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, Z., Li, S., Han, S., Shi, C. & Zhang, Y. Antibody drug conjugate: the “biological missile” for targeted cancer therapy. Signal Transduct. Target Ther.7, 93 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baah, S., Laws, M. & Rahman, K. M. Antibody-drug conjugates-a tutorial review. Molecules26, 2943 (2021). [DOI] [PMC free article] [PubMed]
- 13.Nath, N. et al. Homogeneous plate based antibody internalization assay using pH sensor fluorescent dye. J. Immunol. Methods431, 11–21 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Sadiki, A. et al. Site-specific conjugation of native antibody. Antib. Ther.3, 271–284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mejias-Gomez, O. et al. A window into the human immune system: comprehensive characterization of the complexity of antibody complementary-determining regions in functional antibodies. MAbs15, 2268255 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuchikama, K., Anami, Y., Ha, S. Y. Y. & Yamazaki, C. M. Exploring the next generation of antibody-drug conjugates. Nat. Rev. Clin. Oncol.21, 203–223 (2024). [DOI] [PubMed] [Google Scholar]
- 17.Nadkarni, D. V. Conjugations to endogenous cysteine residues. Methods Mol. Biol.2078, 37–49 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Junutula, J. R. et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat. Biotechnol.26, 925–932 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Pillow, T. H. et al. Site-specific trastuzumab maytansinoid antibody-drug conjugates with improved therapeutic activity through linker and antibody engineering. J. Med. Chem.57, 7890–7899 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Axup, J. Y. et al. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc. Natl Acad. Sci. USA109, 16101–16106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian, F. et al. A general approach to site-specific antibody drug conjugates. Proc. Natl Acad. Sci. USA111, 1766–1771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmerman, E. S. et al. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug. Chem.25, 351–361 (2014). [DOI] [PubMed] [Google Scholar]
- 23.VanBrunt, M. P. et al. Genetically encoded azide containing amino acid in mammalian cells enables site-specific antibody-drug conjugates using click cycloaddition chemistry. Bioconjug. Chem.26, 2249–2260 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Hussain, A. F. et al. Toward homogenous antibody drug conjugates using enzyme-based conjugation approaches. Pharmaceuticals14, 343 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Möhlmann, S., Bringmann, P., Greven, S. & Harrenga, A. Site-specific modification of ED-B-targeting antibody using intein-fusion technology. BMC Biotechnol.11, 76 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffen, W. et al. Discovery of a microbial transglutaminase enabling highly site-specific labeling of proteins. J. Biol. Chem.292, 15622–15635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider, H., Deweid, L., Avrutina, O. & Kolmar, H. Recent progress in transglutaminase-mediated assembly of antibody-drug conjugates. Anal. Biochem.595, 113615 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Dennler, P. et al. Transglutaminase-based chemo-enzymatic conjugation approach yields homogeneous antibody-drug conjugates. Bioconjug. Chem.25, 569–578 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Dengl, S. et al. Format chain exchange (FORCE) for high-throughput generation of bispecific antibodies in combinatorial binder-format matrices. Nat. Commun.11, 4974 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slaga, D. et al. Avidity-based binding to HER2 results in selective killing of HER2-overexpressing cells by anti-HER2/CD3. Sci. Transl. Med.10, eaat5775 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Zwaagstra, J. C. et al. Binding and functional profiling of antibody mutants guides selection of optimal candidates as antibody drug conjugates. PLoS ONE14, e0226593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos, M. K. et al. Valency of HER2 targeting antibodies influences tumor cell internalization and penetration. Mol. Cancer Ther.20, 1956–1965 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Su, D. & Zhang, D. Linker design impacts antibody-drug conjugate pharmacokinetics and efficacy via modulating the stability and payload release efficiency. Front. Pharmacol.12, 687926 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strop, P. et al. Location matters: site of conjugation modulates stability and pharmacokinetics of antibody drug conjugates. Chem. Biol.20, 161–167 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Kaempffe, A. et al. Effect of conjugation site and technique on the stability and pharmacokinetics of antibody-drug conjugates. J. Pharm. Sci.110, 3776–3785 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Shen, B. Q. et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat. Biotechnol.30, 184–189 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Sun, X. et al. Effects of drug-antibody ratio on pharmacokinetics, biodistribution, efficacy, and tolerability of antibody-maytansinoid conjugates. Bioconjug. Chem.28, 1371–1381 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Brinkmann, U. & Kontermann, R. E. The making of bispecific antibodies. MAbs9, 182–212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickopf, S., Georges, G. J. & Brinkmann, U. Format and geometries matter: structure-based design defines the functionality of bispecific antibodies. Comput. Struct. Biotechnol. J.18, 1221–1227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labrijn, A. F., Janmaat, M. L., Reichert, J. M. & Parren, P. Bispecific antibodies: a mechanistic review of the pipeline. Nat. Rev. Drug Discov.18, 585–608 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Kang, M. S., Kong, T. W. S., Khoo, J. Y. X. & Loh, T. P. Recent developments in chemical conjugation strategies targeting native amino acids in proteins and their applications in antibody-drug conjugates. Chem. Sci.12, 13613–13647 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang, H., D’Agostino, G. D., Cole, P. A. & Dempsey, D. R. Selective protein N-terminal labeling with N-hydroxysuccinimide esters. Methods Enzymol.639, 333–353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panowski, S., Bhakta, S., Raab, H., Polakis, P. & Junutula, J. R. Site-specific antibody drug conjugates for cancer therapy. MAbs6, 34–45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckett, D., Kovaleva, E. & Schatz, P. J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci.8, 921–929 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sela, T. et al. Diligent design enables antibody-ASO conjugates with optimal pharmacokinetic properties. Bioconjug. Chem.34, 2096–2111 (2023). [DOI] [PubMed] [Google Scholar]
- 46.Sletten, E. M. & Bertozzi, C. R. From mechanism to mouse: a tale of two bioorthogonal reactions. Acc. Chem. Res.44, 666–676 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim, E. & Koo, H. Biomedical applications of copper-free click chemistry: in vitro, in vivo, and ex vivo. Chem. Sci.10, 7835–7851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Debets, M. F. et al. Aza-dibenzocyclooctynes for fast and efficient enzyme PEGylation via copper-free (3+2) cycloaddition. Chem. Commun.46, 97–99 (2010). [DOI] [PubMed]
- 49.Kuzmin, A., Poloukhtine, A., Wolfert, M. A. & Popik, V. V. Surface functionalization using catalyst-free azide-alkyne cycloaddition. Bioconjug. Chem.21, 2076–2085 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Boghaert, E. R., Cox, M. C. & Vaidya, K. S. Pathophysiologic and pharmacologic considerations to improve the design and application of antibody-drug conjugates. Cancer Res.82, 1858–1869 (2022). [DOI] [PubMed] [Google Scholar]
- 51.Deonarain, M. P., Yahioglu, G., Stamati, I. & Marklew, J. Emerging formats for next-generation antibody drug conjugates. Expert Opin. Drug Discov.10, 463–481 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Beckley, N. S., Lazzareschi, K. P., Chih, H. W., Sharma, V. K. & Flores, H. L. Investigation into temperature-induced aggregation of an antibody drug conjugate. Bioconjug. Chem.24, 1674–1683 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Yamazaki, C. M. et al. Antibody-drug conjugates with dual payloads for combating breast tumor heterogeneity and drug resistance. Nat. Commun.12, 3528 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anami, Y. et al. Enzymatic conjugation using branched linkers for constructing homogeneous antibody-drug conjugates with high potency. Org. Biomol. Chem.15, 5635–5642 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Dickopf, S. et al. Prodrug-activating chain exchange (PACE) converts targeted prodrug derivatives to functional bi- or multispecific antibodies. Biol. Chem.403, 495–508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carter, P. et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc. Natl Acad. Sci. USA89, 4285–4289 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams, C. W. et al. Humanization of a recombinant monoclonal antibody to produce a therapeutic HER dimerization inhibitor, pertuzumab. Cancer Immunol. Immunother.55, 717–727 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldstein, N. I., Prewett, M., Zuklys, K., Rockwell, P. & Mendelsohn, J. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin. Cancer Res.1, 1311–1318 (1995). [PubMed] [Google Scholar]
- 59.Gerdes, C. A. et al. GA201 (RG7160): a novel, humanized, glycoengineered anti-EGFR antibody with enhanced ADCC and superior in vivo efficacy compared with cetuximab. Clin. Cancer Res.19, 1126–1138 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Ridgway, J. B., Presta, L. G. & Carter, P. Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng.9, 617–621 (1996). [DOI] [PubMed] [Google Scholar]
- 61.Merchant, A. M. et al. An efficient route to human bispecific IgG. Nat. Biotechnol.16, 677–681 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Kearns, J. D. et al. Enhanced targeting of the EGFR network with MM-151, an oligoclonal anti-EGFR antibody therapeutic. Mol. Cancer Ther.14, 1625–1636 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Johns, T. G. et al. Identification of the epitope for the epidermal growth factor receptor-specific monoclonal antibody 806 reveals that it preferentially recognizes an untethered form of the receptor. J. Biol. Chem.279, 30375–30384 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ADC cytotoxicity data generated in this study and source data for all quantitative figures are provided in the Source Data file. All other data were available from the corresponding author(s) upon request. Source data are provided with this paper.