Abstract
Treatment of advanced prostate cancer is challenging due to a lack of effective therapies. Therefore, it is important to understand the molecular mechanisms underlying therapeutic resistance in prostate cancer and to identify promising drug targets offering significant clinical advantages. Given the pivotal role of dysregulated transcriptional programs in the therapeutic response, it is essential to prioritize translational efforts targeting cancer-associated transcription factors (TFs). The present study investigated whether chromatin accessibility was associated with therapeutic resistance in prostate cancer using Assay for Transposase-Accessible Chromatin with sequencing (ATAC-seq) data. The bioinformatics analysis identified differences in chromatin accessibility between the drug response (Remission) and drug resistance (Disease) groups. Additionally, a significant association was observed between chromatin accessibility, transcriptional output and TF activity. Among TFs, forkhead box protein M1 (FOXM1) was identified as a TF with high activity and expression in the Disease group. Notably, the results of the computational analysis were validated by FOXM1 knockdown experiments, which resulted in suppressed cell proliferation and enhanced therapeutic sensitivity in prostate cancer cells. The present findings demonstrated that chromatin accessibility and TF activity may be associated with therapeutic resistance in prostate cancer. Additionally, these results provide the basis for future investigations aimed at understanding the molecular mechanisms of drug resistance and developing novel therapeutic approaches for prostate cancer.
Keywords: Assay for Transposase-Accessible Chromatin with sequencing, bioinformatics, chromatin accessibility, therapeutic response, prostate cancer
Introduction
Androgen deprivation therapy remains an important therapy in the treatment of advanced prostate cancer; however, despite its initial effectiveness, therapeutic resistance often arises, thus resulting in the development of castration-resistant prostate cancer (CRPC) (1,2). CRPC has a limited response to current therapies, such as docetaxel with or without enzalutamide (MDV3100), and is therefore considered a clinical challenge (3–5). It is crucial to promptly identify new therapeutic or chemosensitizing targets for prostate cancer treatment.
Altered transcriptional programs confer survival advantages to cancer cells by enhancing their overall biological fitness across a range of different selective environments (6). Dysregulation of transcription factor (TF) expression drives carcinogenesis and determines the therapeutic responsiveness of cancer cells by modulating cancer hallmark signaling pathways (7–9). Consequently, translational efforts targeting cancer-associated TFs are crucial for advancing therapeutic strategies against various types of human cancer, including prostate cancer.
Assay for Transposase-Accessible Chromatin with sequencing (ATAC-seq) identifies accessible chromatin regions that are closely associated with gene expression in the genome, thus enabling the assessment of the gene regulatory landscape in human cancer (10,11). However, the relationship between chromatin accessibility and therapeutic response in prostate cancer remains poorly understood. The present study used ATAC-seq data from The Cancer Genome Atlas (TCGA) to examine the mechanisms underlying differential therapeutic responses in patients with prostate cancer. The results revealed the chromatin accessibility patterns associated with therapeutic resistance in prostate cancer, providing insights into strategies for overcoming therapeutic challenges.
Materials and methods
Data collection and preprocessing
Raw BAM ATAC-seq files of TCGA-prostate adenocarcinoma (PRAD) (n=26) were obtained via the NCI GDC Data Portal (https://portal.gdc.cancer.gov/). The peak calls for ATAC-seq profiles in prostate cancer were downloaded from TCGA Publication Page (https://gdc.cancer.gov/about-data/publications/ATACseq-AWG), as previously described (10). RNA-seq data and clinical information for TCGA-PRAD were retrieved from the cBioPortal for Cancer Genomics (https://www.cbioportal.org/study/summary?id=prad_tcga_pan_can_atlas_2018).
ATAC-seq peak clustering
DESeq2 (v1.34.0; http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) was employed to construct multifactorial models for ATAC-seq read counts in peaks. Subsequently, a variance stabilizing transformation was computed based on the DESeq2 model, and principal component analysis (PCA) was conducted to illustrate the distinction between the drug response (Remission) and drug resistance (Disease) groups (Fig. S1). The principal component (PC) 1 coefficient was set to 0 as the threshold to identify ambiguous samples in the PCA plot. A PC1 coefficient >0 indicates a positive correlation with the new PCA variable, whereas a coefficient <0 indicates a negative correlation. A total of 13 out of 20 complete response (CR)/partial response (PR) samples had a PC1 coefficient >0, whereas 7 CR samples had a PC1 coefficient <0 where progressive disease (PD)/stable disease (SD) samples dominated. These 7 CR samples cannot be clustered with the PD/SD group and were thus excluded from further analyses.
Differential peak accessibility
Using the countOverlaps function of the R packages GenomicAlignments (v1.30.0; http://bioconductor.org/packages/release/bioc/html/GenomicAlignments.html) and GenomicRanges (v1.46.1; http://bioconductor.org/packages/release/bioc/html/GenomicRanges.html), reads aligning to global peak regions were tallied. To mitigate the bias stemming from low-count peaks, 4,778 peaks with mean counts <50 across all samples were removed. DESeq2 (v1.34.0) was employed to evaluate differential peak accessibility. The significant peaks were displayed in a hierarchical clustering heatmap using the DESeq size-factor normalized read counts and the complete distance metric for clustering. The R packages ChiPseeker (v1.36.0; http://bioconductor.org/packages/release/bioc/html/ChIPseeker.html) and clusterProfiler (v4.8.1; https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) were used to visualize peak coverage across chromosomes and annotate peak regions. To analyze the relationship between chromatin accessibility and transcriptional output, log2 fold change was calculated by comparing the average peak calling for each gene from ATAC-seq data or mean expression of genes from the bulk RNA-seq data between Remission and Disease groups. Spearman correlation coefficient was used to measure the strength and direction of association. P<0.01 was considered to indicate a statistically significant difference.
De novo TF motif analysis
The Hypergeometric Optimization of Motif EnRichment (HOMER; v.4.11.1; http://homer.ucsd.edu/homer/motif/) utility findMotifsGenome.pl was employed to identify the top 10 TF motifs that were enriched in differentially accessible (DA) peaks. The analysis focused on 100 bp-wide regions around the DA peak summits, with hg19 serving as the reference genome. Additionally, custom background regions were generated, spanning a 150 bp-wide range around the peak summits. The top motifs were cross-referenced with the known motifs from the HOMER database and then manually curated to include only TFs that exhibited expression according to RNA-seq data, grouping similar motifs from TFs of the same family.
Association between TFs and therapeutic response groups
All accessible cis-regulatory elements (CREs) in the entire genome were analyzed using Cis-Regulatory Element Motif Activities (CREMA; http://crema.unibas.ch/crumara/). This tool can determine the intensity signals of each CRE, calculate TF motif activities, and identify the binding sites for hundreds of TFs within the CREs in each sample. The vector of mean TF activities was compared, and the association between TFs and therapeutic response groups was evaluated, using the Wilcoxon rank-sum test. Subsequently, the resulting P-values (one for each TF) underwent adjustment for multiple hypothesis testing using the false discovery rate (FDR) method. The analytical results were depicted as a scatterplot, with the x-axis representing the mean TF activity difference and the y-axis indicating FDR q-value. Significant TF motifs were selected based on an absolute mean TF activity difference >0.05 and an FDR q-value <0.05. The correlation of their activities across patients was calculated using Pearson correlation coefficient and was presented visually using a heatmap to assess the consistency of the significant TFs in both the Remission and Disease groups.
Pathway enrichment analysis
The Genomic Regions Enrichment of Annotations Tool (v1.26; http://great.stanford.edu/public/html/) was used to associate the sub-cluster of the DA peaks with genes. Pathway analysis was then performed to discover the functional significance of the DA peaks from the associated genes and to identify overrepresented pathways (12). PANTHER knowledgebase (http://www.pantherdb.org/) was used for the pathway analysis (hypergeometric test, adjusted P-value <0.05).
Cell culture and reagents
Prostate cancer cell lines PC-3 (cat. no. CRL-1435), LNCaP (clone FGC; cat. no. CRL-1740), DU145 (cat. no. HTB-81) and 22Rv1 (cat. no. CRL-2505) were purchased from the American Type Culture Collection. The normal prostate epithelial cell line PNT1A (cat. no. 95012614) was obtained from the European Collection of Authenticated Cell Cultures. Cells were cultured in RPMI1640 (PC-3, LNCaP, 22Rv1, PNT1A) or DMEM (DU145) containing 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 mg/ml). Cell culture reagents were obtained from Welgene, Inc. All cell lines tested negative for mycoplasma contamination using the Mycoplasma PCR Detection kit (Intron Biotechnology, Inc.). The cell lines were authenticated using short tandem repeat analysis. Cabazitaxel (cat. no. S3022), docetaxel (cat. no. S1148) and MDV3100 (cat. no. S1250) were purchased from Selleck Chemicals.
Small interfering RNA (siRNA) transfection
A siRNA against forkhead box (FOX)M1 (siFOXM1; sense, 5′-GCUCAUACCUGGUACCUAU-3′; antisense, 5′-AUAGGUACCAGGUAUGAGC-3′) (13) was obtained from Genolution, Inc. A control siRNA (ON-TARGETplus Non-targeting Pool; cat. no. D-001810-10), which was used as a negative control, was purchased from Horizon Discovery; Revvity, Inc. PC-3 (2.5×105 cells/well), LNCaP (2.5×105 cells/well), DU145 (2.5×105 cells/well), 22Rv1 (2.5×105 cells/well) and PNT1A (2.5×105 cells/well) cells were seeded into 6-well plates. Each cell line was transfected with 50 nM control siRNA or siFOXM1 for the indicated times at 37°C using Lipofectamine® RNAiMAX reagent (Invitrogen; Thermo Fisher Scientific, Inc.). For the MTT assay, cells were transfected with either siControl or siFOXM1 for 72 h. For the colony formation assay, cells were transfected with siControl or siFOXM1 once every 3 days for 9 days. To assess the effect of FOXM1 on drug response, cells were treated with each drug 24 h after transfection with siControl or siFOXM1. After 72 h of transfection, FOXM1 knockdown was confirmed by western blotting.
MTT assay
After 24 h of siFOXM1 transfection, PC-3, LNCaP, DU145, 22Rv1 and PNT1A cells were treated with 50 nM cabazitaxel or 0.5 nM docetaxel for 48 h, or 10 µM MDV3100 for 72 h at 37°C. The MTT assay (MilliporeSigma) was performed to assess cell viability, according to the manufacturer's instructions. The purple formazan was dissolved in DMSO and was quantitated by determining the absorbance at 570 nm on a BioTek SynergyMx microplate reader (BioTek; Agilent Technologies, Inc.).
Colony formation assay
PC-3 (3×103 cells/well), LNCaP (8×103 cells/well), DU145 (1×103 cells/well), 22Rv1 (3×103 cells/well) and PNT1A (3×103 cells/well) cells were plated into 6-well plates. Cells were transfected with 50 nM siControl or siFOXM1 once every 3 days for 9 days at 37°C. After the cells were fixed with 3.7% paraformaldehyde (MilliporeSigma) for 10 min at room temperature, they were stained with 0.5% crystal violet (MilliporeSigma) for 15 min at room temperature. Next, the number of colonies, defined as >50 cells/colony, was counted using ImageJ (1.8.0 172; National Institutes of Health).
Western blotting
The crude extracts from PC-3, LNCaP, DU145, 22Rv1 and PNT1A cells were prepared with RIPA buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS and 0.5 EDTA] including protease and phosphatase inhibitor cocktails (MilliporeSigma). The total protein concentration was quantified using the BCA assay kit (Thermo Fisher Scientific Inc.). Subsequently, the samples were separated by SDS-PAGE on 6% (FOXM1, 30 µg), 10% (β-tubulin, 10 µg) or 15% (caspase-3, 10 µg; cleaved caspase-3, 80 µg) gels and transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Inc.). The membranes were blocked with 5% skim milk in Tris-buffered saline-0.1% Tween 20 for 1 h at room temperature and were then incubated with anti-FOXM1 (1:1,000; cat. no. 5436S; Cell Signaling Technology, Inc.), anti-caspase-3 (1:1,000; cat. no. 9662S; Cell Signaling Technology, Inc.), anti-cleaved caspase-3 (1:1,000; cat. no. 9661S; Cell Signaling Technology, Inc.) and anti-β-tubulin (1:5,000; cat. no. T4026; MilliporeSigma) overnight at 4°C. β-tubulin was used as the loading control. After primary antibody incubation, the membranes were incubated with a goat anti-rabbit IgG-HRP antibody (1:5,000; cat. no. A120-101P; Bethyl Laboratories, Inc.) or a goat anti-mouse antibody (1:5,000, cat. no. A90-116P; Bethyl Laboratories, Inc.) for 1 h at room temperature. Subsequently, the signals were determined using Amersham ECL reagent (Cytiva). X-ray films were scanned on an EPSON scanner. The data are representative of at least three independent experiments.
Statistical analysis and data visualization
R version 4.1.1 (R Foundation for Statistical Computing) was used for all statistical analyses. The R package ggplot2 (v3.3.5; http://cran.r-project.org/web/packages/ggplot2/index.html) was used to create graphs, whereas the R package ComplexHeatmap (v2.10.0; http://www.bioconductor.org/packages/ComplexHeatmap/) was utilized to produce heatmaps. The P-values for multiple comparisons were adjusted using the Benjamini-Hochberg method. The Kaplan-Meier survival curve and log-rank test were employed to estimate overall survival (14–16). The samples were divided into FOXM1 high-expression (n=79) or low-expression (n=299) groups from TCGA transcriptomic data. To divide samples into high- and low-expression groups, the bifurcation point approach (17) was used because the cohort might have biased expression pattern of a specific gene, and the median or quantile is not the most natural cut-point for gene expression profiles and survival prediction. Subsequently, the Cox proportional hazards model was applied to calculate hazard ratios (HRs) and 95% confidence intervals. A one-way analysis of variance (ANOVA) was employed to compare means among the experimental groups, followed by Bonferroni's multiple comparisons test. A two-way ANOVA followed by Bonferroni's test was used for pairwise comparisons between siControl and siFOXM1, or siFOXM1 and its combinations with drugs (docetaxel, cabazitaxel, or MDV3100). P- or FDR q-values <0.05 were considered to indicate a statistically significant difference.
Results
Chromatin accessibility differences between the Remission and Disease groups
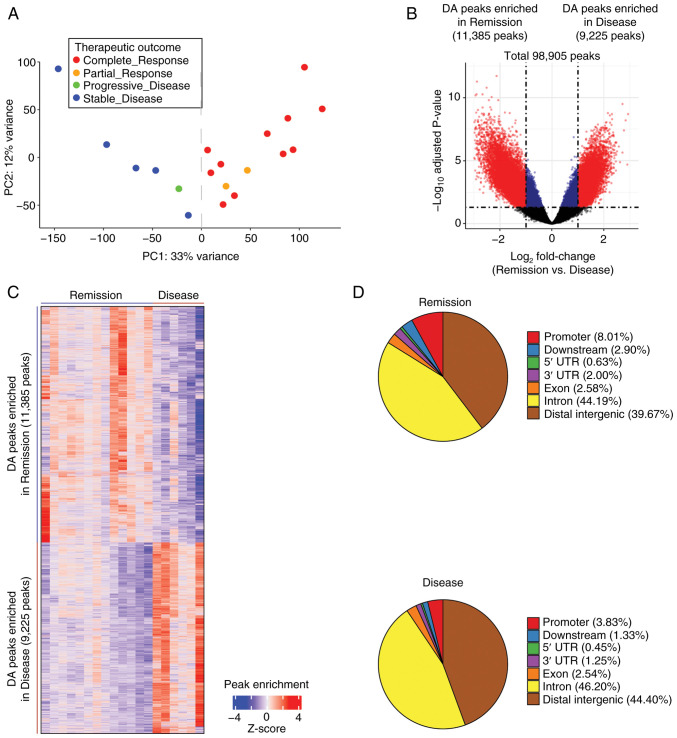
To examine the association between chromatin accessibility and therapeutic response in prostate cancer, ATAC-seq data consisting of 98,905 peaks from 26 TCGA-PRAD samples were analyzed (10) (Fig. S1). Therapeutic response was categorized into CR, PR, PD and SD based on response criteria (18). PCA of peak read count showed a clear separation between CR/PR samples (n=13) and PD/SD samples (n=6) when the 7 CR samples were excluded (Fig. S2A). In addition, it was confirmed that the 7 excluded CR samples exhibited a gene expression pattern more similar to PD/SD samples than to CR/PR samples (Fig. S2B). Excluding these samples allowed for more distinct CR/PR and PD/SD groups, despite the reduction in sample size. Henceforth, these two groups were designated as the Remission (n=13) and Disease (n=6) groups (Fig. 1A). The present study then investigated the chromatin accessibility differences between the Remission and Disease groups. A total of 20,610 DA peaks (absolute log2 fold change >1.0 and adjusted P<0.05) were identified, which accounted for 20.84% of the total ATAC-seq peaks (Fig. 1B and C). Among these, 11,385 peaks (55.24%) exhibited increased accessibility in the Remission group, whereas 9,225 peaks (44.76%) showed increased accessibility in the Disease group (Fig. 1B and C). The DA ATAC-seq peaks were distributed across various chromatin regions, including promoter, exon, intron and distal intergenic regions, for both the Remission and Disease groups (Fig. 1D). These results identified a distinct difference in chromatin accessibility between the two groups.
Figure 1.
Chromatin accessibility differences between the Remission and Disease groups. (A) Principal component analysis plot of the ATAC-seq signal showed distinct clusters between the complete response/partial response (n=13) and progressive disease/stable disease (n=6) groups, effectively reclassifying them as the Remission and Disease groups. (B) Volcano plot of ATAC-seq peaks exhibited discrete separation between the Remission and Disease groups. The adjusted P-values were obtained using DESeq2. The vertical dotted lines indicate absolute log2 fold change of 1.0, whereas the horizontal dotted line represents an adjusted P-value of 0.05. Among the total 98,905 peaks, significant peaks with log2 fold change >1.0 are highlighted in red, whereas those with log2 fold change <1.0 are depicted in blue. Notably, 20,610 significant DA peaks (highlighted in red) were respectively enriched in the Remission (n=11,385) and Disease (n=9,225) groups. (C) Hierarchical clustering of the 20,610 DA peaks in the Remission and Disease groups. The color indicator represents log2-transformed peak count data, which has been normalized by z-score row. (D) Pie charts of the distribution of DA ATAC-seq peaks (false discovery rate q-value <0.05) across various chromatin regions for the Remission and Disease groups. ATAC-seq, Assay for Transposase-Accessible Chromatin with sequencing.
Chromatin accessibility is associated with transcriptional output and TF activity between the Remission and Disease groups
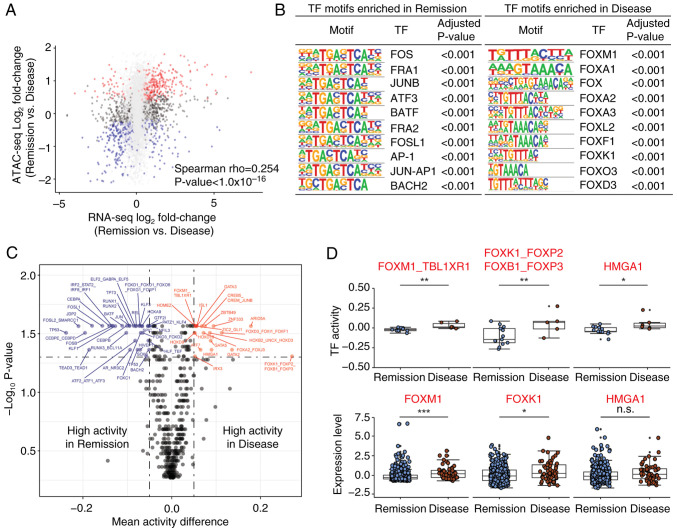
To explore the relationship between chromatin accessibility and transcriptional output, ATAC-seq data were compared with RNA-seq data. The chromatin accessibility of individual genes showed a moderate association with the corresponding gene expression between the Remission and Disease groups (Fig. 2A). Subsequently, to identify key TFs that drive the transcription program difference between the Remission and Disease groups, HOMER motif analysis of DA ATAC-seq peaks was performed. Several proto-oncogenes, such as FOS, FRA1 and JUNB, were enriched in the Remission group (Fig. 2B left), whereas FOX family genes, such as FOXM1, FOXA1 and FOXA2, were enriched in the Disease group (Fig. 2B right).
Figure 2.
TF activities are associated with therapeutic responses in prostate cancer. (A) Scatter plot of differential expression (x-axis) and differential accessibility (y-axis) between the Remission and Disease groups. The x-axis represents the log2 fold change of gene expression from the RNA-seq data, whereas the y-axis depicts the log2 fold change of the mean peaks associated with each gene from Assay for Transposase-Accessible Chromatin with sequencing data. Significant DA genes are highlighted in red (Remission) or blue (Disease). (B) TF binding motifs enriched in both the Remission and Disease groups were identified using Hypergeometric Optimization of Motif EnRichment, with the top 10 most enriched motifs selected. (C) Inferred TF motif activity differences between the Remission and Disease groups. The x-axis represents the mean TF activity differences, and the y-axis depicts the -log10 P-value. The vertical dotted lines indicate an absolute mean TF activity difference of 0.05, and the horizontal dotted line represents the false discovery rate q-value of 0.05 for significant TFs. (D) TFs with high TF activity (upper) and high mRNA expression (lower) specifically in the Disease group. *P<0.05, **P<0.01, ***P<0.005. FOX, forkhead box; n.s., not significant; TF, transcription factor.
Sample-specific TF binding motif activities were also inferred using CREMA, which enables the mapping of chromatin accessibility profiles to a lower-dimensional inferred TF activity. A total of 54 TF motif activities were significantly associated with therapeutic response, as determined by FDR q-value <0.05 and absolute mean activity difference >0.05 (Figs. 2C and S3A). TF activities from the same therapeutic response group (either the Remission or Disease) were also correlated across samples by Spearman correlation (Fig. S3B). Notably, the Wilcoxon rank-sum test showed that several TFs, including FOXM1, FOXK1 and HMGA1, consistently exhibited higher activity and mRNA expression in the Disease group compared with in the Remission group (Fig. 2D). Taken together, these results indicated that chromatin accessibility determines differential transcriptional output and TF activity between the two groups.
Experiments involving FOXM1 exemplify the usefulness of chromatin accessibility analysis for determining therapeutic response
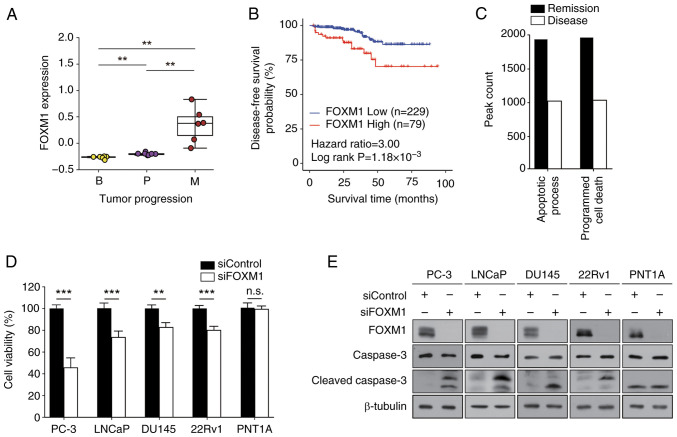
Notably, FOXM1 was frequently identified among the TFs highlighted in the present analyses (Fig. 2B and D). Therefore, to validate the results of computational analysis, TCGA-PRAD data were analyzed. The expression levels of FOXM1 were markedly upregulated in metastasis samples compared with those in benign or primary cancer samples (Fig. 3A), providing a potential underlying mechanism of elevated FOXM1 TF activity in metastasis (Fig. 2B and D). Kaplan-Meier analysis revealed that high expression levels of FOXM1 were associated with a worse prognosis in patients with prostate cancer (HR=3.0, P=1.18×10−3) (Fig. 3B). These results suggested that FOXM1 may serve a crucial role in determining therapeutic response and could be considered a useful prognostic marker for prostate cancer.
Figure 3.
Effects of FOXM1 on prostate cancer. (A) FOXM1 expression levels in patients with prostate cancer are represented in the box plots. The x-axis indicates three different stages of prostate cancer, and the y-axis depicts the normalized expression levels. (B) Disease-free survival curve for patients with prostate cancer based on the expression levels of FOXM1 from The Cancer Genome Atlas data. (C) Enrichment of PANTHER pathways of differential accessibilities in the Remission or Disease groups. The bar represents the number of peak region hits associated with the pathway. (D and E) PC-3, LNCaP, DU145, 22Rv1 and PNT1A cells were transfected with 50 nM siRNAs for 72 h prior to the MTT assay and western blot analysis. (D) Cell viability was expressed as a relative value compared with that of the siControl, which was set to 100%. Data are presented as the mean ± SEM (n=4). (E) Caspase-3 expression was assessed using anti-cleaved caspase-3 antibody. A representative image from three independent experiments is shown. **P<0.01, ***P<0.005. B, benign; FOX, forkhead box; M, metastatic; P, primary; si, small interfering.
To assess the role of FOXM1 in prostate cancer, pathway enrichment analysis was performed. The apoptotic process and programmed cell death pathways were distinct in the DA ATAC-seq peaks of Disease samples (Fig. 3C). To confirm the computational prediction results, MTT and colony formation assays were then performed. FOXM1 expression was knocked down and successful transfection was confirmed (Fig. 3E). FOXMI knockdown suppressed cell viability (Fig. 3D) and colony formation (Table I; Fig. S4) of prostate cancer cells (PC-3, LNCaP, DU145 and 22Rv1), but not of normal prostate epithelial cells (PNT1A). In addition, FOXM1 knockdown elevated the expression levels of cleaved caspase-3 in prostate cancer cells (Figs. 3E and S5). Taken together, these results indicated that FOXM1 may have a crucial role in regulating cell survival and proliferation in prostate cancer.
Table I.
Effect of FOXM1 knockdown on colony formation in prostate cancer cells.
| Number of colonies | ||
|---|---|---|
|
|
||
| Cell line | siControl | siFOXM1 |
| PC-3 | 310.70±6.49 | 10.67±1.45a |
| LNCaP | 332.00±20.42 | 12.33±3.48a |
| DU145 | 157.00±6.43 | 24.00±3.06a |
| 22Rv1 | 227.70±2.33 | 76.33±16.60a |
| PNT1A | 0 | 0 |
P<0.0005 vs. siControl. Data are presented as the mean ± SEM (n=3). FOXM1, forkhead box M1; si, small interfering.
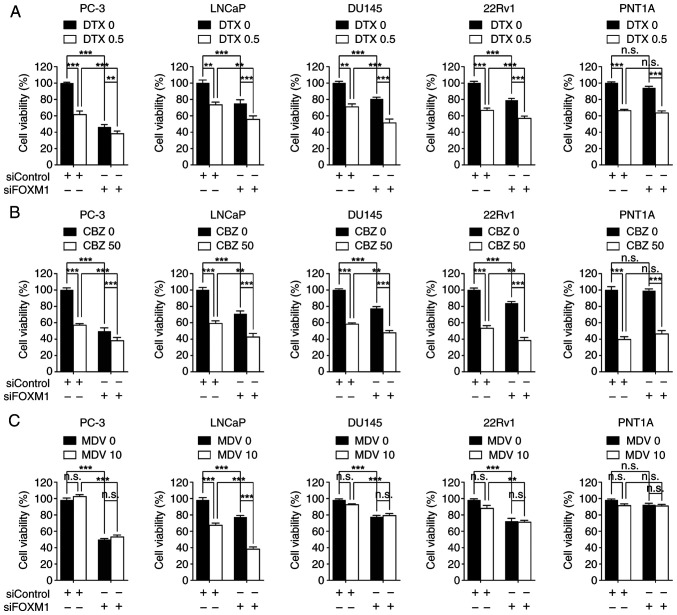
The present study also examined whether FOXM1 expression could influence the therapeutic response of prostate cancer cells to several therapeutic agents. Transfection with siFOXM1 increased the sensitivity of PC-3 [androgen receptor (AR)-negative], LNCaP [AR-positive, AR splice variant-7 (AR-V7)-negative], DU145 (AR-negative) and 22Rv1 (AR-positive, AR-V7-positive) cells to docetaxel or cabazitaxel compared with each drug alone (Fig. 4A and B). In addition, FOXM1 knockdown enhanced the therapeutic activity of the AR antagonist MDV3100 only in LNCaP cells (Fig. 4C). By contrast, siFOXM1 did not enhance chemotherapeutic effects in PNT1A cells (Fig. 4A-C). These results suggested that FOXM1 inhibition may be a potential strategy for improving the efficacy of nonselective chemotherapy or androgen blockade therapy. Notably, there was a weak, but significant, positive co-expressional correlation between FOXM1 and AR in TCGA transcriptomic data (Fig. S6), underscoring the clinical significance of the combined inhibition of FOXM1 and AR. These findings indicated that FOXM1 represents a promising therapeutic target for prostate cancer.
Figure 4.
FOXM1 knockdown increases therapeutic response in prostate cancer. Each cell line was transfected with 50 nM siRNAs for 24 h, and then cells were further treated with (A) 0.5 nM DTX, (B) 50 nM CBZ or (C) 10 µM MDV for 48 h prior to MTT assay. Cell viability was expressed as a relative value compared with that of the siControl, which was set to 100%. Data are presented as the mean ± SEM (n=4). **P<0.01, ***P<0.005. CBZ, cabazitaxel; DTX, docetaxel; FOX, forkhead box; MDV, MDV3100; n.s., not significant; si, small interfering.
Discussion
The present study described three main findings: i) Chromatin accessibility was associated with distinct therapeutic responses in prostate cancer; ii) chromatin accessibility was associated with distinct transcriptional output and TF activity in prostate cancer; and iii) FOXM1 exemplified the usefulness of chromatin accessibility analysis in assessing therapeutic response.
Balancing sample cluster integrity with sample size is a common challenge in bioinformatics, particularly when excluding outliers. The small sample size of 26 in the PRAD ATAC-seq study raised concerns. To address this, an initial PCA was conducted to identify 7 CR samples with gene expression patterns similar to PD/SD samples. These outliers likely represent technical or biological confounders, complicating the interpretation of their response status. Although excluding these 7 CR samples reduced the sample size, it improved the robustness of the findings, minimized data skew, and allowed for more accurate associations between molecular profiles and clinical outcomes. This step was critical for understanding chromatin accessibility differences underlying treatment response.
The prediction of therapeutic response remains a significant challenge in treating prostate cancer, with epigenetic changes serving a crucial role in its development (19). Chromatin accessibility profiling using ATAC-seq is an important tool for assessing epigenetic changes in cancer tissues (20); however, only a few studies have applied the ATAC-seq approach specifically to prostate cancer. Most research has analyzed the ATAC-seq data from prostate cancer cell lines (21), patient-derived xenografts (11) or murine organoids (22), rather than from clinical samples. While some studies have utilized ATAC-seq data from patient samples, they primarily focused on molecular classification, cell heterogeneity, or alterations in AR signaling of CRPC (23,24). By contrast, the present study integrated ATAC-seq data from primary human prostate cancer samples with individual patient therapeutic response data from TCGA. This approach revealed distinct chromatin accessibility signatures, regulatory pathways and TF activities linked to therapeutic response in prostate cancer. Notably, the present analysis uniquely identified FOXM1 as a modulator of therapeutic response; to the best of our knowledge, this has not been reported in other studies with ATAC-seq data. The present study provides a pioneering genome-wide analysis, revealing significant epigenetic differences between responsive and resistant cases, thus providing valuable insights into the mechanisms underlying therapeutic response in prostate cancer.
Chromatin profiling provides a valuable approach for classifying prostate cancer and discovering potential drug targets (11). In the present study, ATAC-seq data analysis was employed to identify distinct chromatin accessibility and specific TF activities associated with therapeutic response in prostate cancer. The analysis revealed that various FOX TF family genes were enriched in the Disease group. Dysregulation of FOX genes is frequently observed in human cancer (25), where they drive tumor development, progression and drug resistance (26,27). The present study demonstrated that FOXM1 was associated with poor clinical outcomes in patients with prostate cancer and served as a key TF governing transcriptional programs in therapeutic resistance in prostate cancer, which is consistent with previous findings that FOXM1 is functionally required not only for tumorigenesis, tumor proliferation, metastasis and invasion, but also serves as a poor prognostic marker and induces docetaxel resistance in CRPC (28).
Although the present study primarily focused on FOXM1, further investigation is needed to identify which other FOX isotypes may have a critical role in mediating therapeutic resistance. Additionally, future studies are required to investigate functional interaction among FOX family genes in therapeutic resistance and to assess the impact of their co-inhibition on cancer resistance. Moreover, the molecular mechanisms underlying how FOXM1 induces therapeutic resistance in prostate cancer are still unclear and require elucidation.
In conclusion, the present study demonstrated that chromatin accessibility may be associated with therapeutic response in prostate cancer. These findings provide valuable insights into the understanding of prostate cancer biology and could inform the development of therapeutic approaches for this disease.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This work was supported by a grant from the National Research Foundation of Korea grant funded by the Korea government (The Ministry of Science and ICT) (grant nos. 2018R1A4A1023822 and 2020R1A2C1102574), a grant from the Seoul National University Hospital Research Fund (grant no. 0320230140), and the Education and Research Encouragement Fund of Seoul National University Hospital.
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
SL, DYL, IS, JNC and JHJ designed the research and wrote the main manuscript text. SL analyzed the ATAC-seq data and made the figures. DYL and JNC designed and performed the validation experiment using prostate cancer cell lines. DYL, JNC and JHJ confirm the authenticity of all the raw data. IS contributed to data analysis and interpretation. All authors read and approved the final version of the manuscript, and consented to its publication.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chandrasekar T, Yang JC, Gao AC, Evans CP. Mechanisms of resistance in castration-resistant prostate cancer (CRPC) Transl Androl Urol. 2015;4:365–380. doi: 10.3978/j.issn.2223-4683.2015.05.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai C, Dehm SM, Sharifi N. Targeting the androgen signaling axis in prostate cancer. J Clin Oncol. 2023;41:4267–4278. doi: 10.1200/JCO.23.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park S, Kim YS, Kim DY, So I, Jeon JH. PI3K pathway in prostate cancer: All resistant roads lead to PI3K. Biochim Biophys Acta Rev Cancer. 2018;1870:198–206. doi: 10.1016/j.bbcan.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Cai M, Song XL, Li XA, Chen M, Guo J, Yang DH, Chen Z, Zhao SC. Current therapy and drug resistance in metastatic castration-resistant prostate cancer. Drug Resist Updat. 2023;68:100962. doi: 10.1016/j.drup.2023.100962. [DOI] [PubMed] [Google Scholar]
- 5.Gebrael G, Fortuna GG, Sayegh N, Swami U, Agarwal N. Advances in the treatment of metastatic prostate cancer. Trends Cancer. 2023;9:840–854. doi: 10.1016/j.trecan.2023.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Bradner JE, Hnisz D, Young RA. Transcriptional Addiction in Cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhagwat AS, Vakoc CR. Targeting transcription factors in cancer. Trends Cancer. 2015;1:53–65. doi: 10.1016/j.trecan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushweller JH. Targeting transcription factors in cancer-From undruggable to reality. Nat Rev Cancer. 2019;19:611–624. doi: 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiah JV, Johnson DE, Grandis JR. Transcription factors and cancer: Approaches to Targeting. Cancer J. 2023;29:38–46. doi: 10.1097/PPO.0000000000000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, Silva TC, Groeneveld C, Wong CK, Cho SW, et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362:eaav1898. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang F, Xu D, Wang S, Wong CK, Martinez-Fundichely A, Lee CJ, Cohen S, Park J, Hill CE, Eng K, et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science. 2022;376:eabe1505. doi: 10.1126/science.abe1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan L, Zhao M, Lu Y, Ning H, Yang S, Song Y, Chai W, Shi X. CENPE promotes lung adenocarcinoma proliferation and is directly regulated by FOXM1. Int J Oncol. 2019;55:257–266. doi: 10.3892/ijo.2019.4805. [DOI] [PubMed] [Google Scholar]
- 14.Park S, Lim JM, Chun JN, Lee S, Kim TM, Kim DW, Kim SY, Bae DJ, Bae SM, So I, et al. Altered expression of fucosylation pathway genes is associated with poor prognosis and tumor metastasis in non-small cell lung cancer. Int J Oncol. 2020;56:559–567. doi: 10.3892/ijo.2019.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee DY, Lee S, Kim YS, Park S, Bae SM, Cho EA, Park EJ, Park HH, Kim SY, So I, et al. Cyclosporin A inhibits prostate cancer growth through suppression of E2F8 transcription factor in a MELK-dependent manner. Oncol Rep. 2023;50:218. doi: 10.3892/or.2023.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, Park YR, Kim SH, Park EJ, Kang MJ, So I, Chun JN, Jeon JH. Geraniol suppresses prostate cancer growth through down-regulation of E2F8. Cancer Med. 2016;5:2899–2908. doi: 10.1002/cam4.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marco E, Karp RL, Guo G, Robson P, Hart AH, Trippa L, Yuan GC. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci USA. 2014;111:E5643–5650. doi: 10.1073/pnas.1408993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Conteduca V, Hess J, Yamada Y, Ku SY, Beltran H. Epigenetics in prostate cancer: Clinical implications. Transl Androl Urol. 2021;10:3104–3116. doi: 10.21037/tau-20-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandi FC, Modi H, Kampman L, Corces MR. Chromatin accessibility profiling by ATAC-seq. Nat Protoc. 2022;17:1518–1552. doi: 10.1038/s41596-022-00692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taavitsainen S, Engedal N, Cao S, Handle F, Erickson A, Prekovic S, Wetterskog D, Tolonen T, Vuorinen EM, Kiviaho A, et al. Single-cell ATAC and RNA sequencing reveal pre-existing and persistent cells associated with prostate cancer relapse. Nat Commun. 2021;12:5307. doi: 10.1038/s41467-021-25624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grbesa I, Augello MA, Liu D, McNally DR, Gaffney CD, Huang D, Lin K, Ivenitsky D, Goueli R, Robinson BD, et al. Reshaping of the androgen-driven chromatin landscape in normal prostate cells by early cancer drivers and effect on therapeutic sensitivity. Cell Rep. 2021;36:109625. doi: 10.1016/j.celrep.2021.109625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrestha R, Chesner LN, Zhang M, Zhou S, Foye A, Lundberg A, Weinstein AS, Sjöström M, Zhu X, Moreno-Rodriguez T, et al. An atlas of accessible chromatin in advanced prostate cancer reveals the epigenetic evolution during tumor progression. Cancer Res. 2024;84:3086–3100. doi: 10.1158/1538-7445.AM2024-LB196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bian X, Wang W, Abudurexiti M, Zhang X, Ma W, Shi G, Du L, Xu M, Wang X, Tan C, et al. Integration analysis of single-cell multi-omics reveals prostate cancer heterogeneity. Adv Sci (Weinh) 2024;11:e2305724. doi: 10.1002/advs.202305724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 27.Castaneda M, Hollander PD, Mani SA. Forkhead box transcription factors: Double-edged swords in cancer. Cancer Res. 2022;82:2057–2065. doi: 10.1158/0008-5472.CAN-21-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee DY, Chun JN, So I, Jeon JH. Oncogenic role of FOXM1 in human prostate cancer (Review) Oncol Rep. 2024;51:15. doi: 10.3892/or.2023.8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study may be requested from the corresponding author.