Abstract
Inspired by the success of large language models (LLMs), we develop a long-context generative model for genomes. Our multiscale transformer model, megaDNA, is pre-trained on unannotated bacteriophage genomes with nucleotide-level tokenization. We demonstrate the foundational capabilities of our model including the prediction of essential genes, genetic variant effects, regulatory element activity and taxonomy of unannotated sequences. Furthermore, it generates de novo sequences up to 96 K base pairs, which contain potential regulatory elements and annotated proteins with phage-related functions.
Subject terms: Machine learning, Computational models, Software
MegaDNA, a long-context genomic language model, generates DNA sequences up to 96 K base pairs with annotated proteins and potential regulatory elements. It predicts essential genes, genetic variant effects, and regulatory element activity.
Introduction
Large pre-trained language models have drastically transformed the natural language processing (NLP) field1,2. Drawing on the similarity of natural language and genome sequences, genomic language models have been developed3. These models are trained on large-scale genomic datasets, and they effectively predict regulatory elements, uncover co-regulation patterns in proteins, and identify the genome-wide variant effects4–8. However, it remains an open question whether language models can be tailored to generate genome-scale sequences with functional elements while retaining the capacity to decipher the intricate relationships within DNA sequences.
Most of the current genomic language models use masked language modeling like bidirectional encoder representations from transformers (BERT)1. This approach is not ideal for tasks that require generating new content. In addition, they face technical constraints such as short context size and the aggregation of sequences in k-mer tokenization. These limitations hinder their ability to learn from genome-scale data with the level of resolution needed for designing functional elements.
In this work, we introduce megaDNA, a long-context language model that demonstrates foundational capacities in understanding and generating genomic sequences. Our model draws inspiration from the generative pre-trained transformers (GPT) model2, which is renowned for its proficiency in generating long and coherent texts. We utilize a multiscale transformer structure, developed by Yu et al.9, which enables us to train the model on unannotated whole bacteriophage genomes at the single-nucleotide level in a self-supervised manner. Without further fine-tuning, our model can predict gene essentiality across the phage genome in a zero-shot manner. The sequence embeddings computed by our model can be directly applied to predict the functional properties of both regulatory elements and proteins. Moreover, the trained model generates sequences up to 96 K base pairs (bp), sharing a similar genomic structure with natural bacteriophage genomes. We find functional promoters and ribosome binding sites (RBS) in the 5′ untranslated regions (5′UTR) of the predicted genes. The proteins from the generated sequences are predicted to be structurally plausible. Our model is available from GitHub.
Results
megaDNA allows zero-shot prediction of gene essentiality
To construct the training dataset, we collected bacteriophage genomes with high confidence from three sources including the NCBI GenBank, the metagenomic gut virus (MGV) catalog10, and the gut phage database (GPD)11 (Supplementary Fig. 1). After data cleaning, we constructed a dataset of 99.7 K bacteriophage genomes to pre-train our model (Methods), and the training data was nucleotide-level tokenized, where each nucleotide is treated as a separate token.
Traditionally, transformer-based language models only process a few thousand tokens of context because the computational cost of the self-attention mechanism scales quadratically with sequence length. This context window is not sufficient to model nucleotide-level tokenized phage genomes. To overcome this problem, we employed a multi-scale transformer structure9 to model the long-range context information. This architecture consists of three decoder-only transformer layers with multi-head attention, and each layer captures sequence information at different resolutions: the local layer processes embeddings of tokenized sequences within a 16 bp window. Its output serves as the input to the middle layer, which has a context window of 1024 bp. Finally, the global layer utilizes information from the middle layer to model sequence dependencies across the whole input context (96 K bp).
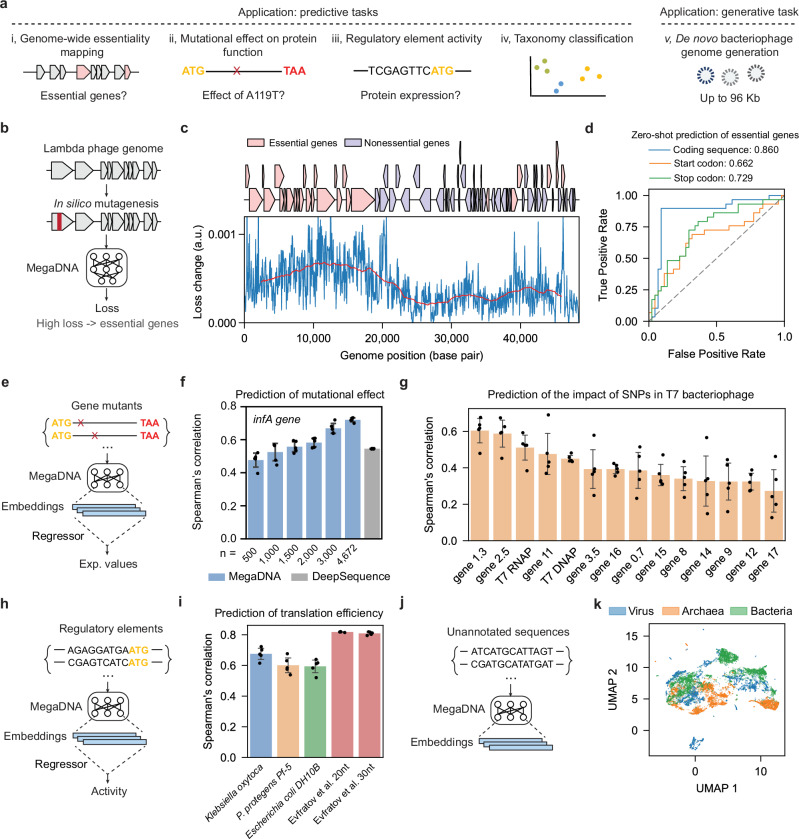
We hypothesize that our pretrained language model captures the structural patterns of bacteriophage genomes in our training dataset, allowing the model’s loss to approximate the fitness of genome sequences. To test this hypothesis, we conducted in silico mutagenesis analysis to predict essential genes in the lambda phage genome12 (Fig. 1b). Without any supervised training, we found mutations within the coding sequences of essential genes result in higher losses than non-essential genes (Fig. 1c). Consequently, changes in model loss can be used as a zero-shot predictor of essential genes (AUROC: 0.86, Fig. 1d). Similarly, mutations in the start and stop codons of essential genes lead to higher model losses than non-essential genes (Fig. 1d, Supplementary Fig. 2). We further analyzed the similarity of sequences in the training dataset to the lambda phage genome (Supplementary Fig. 3). We found that 847 training sequences aligned with the lambda phage genome through BLAST analysis13. Among these sequences, 50 show a sequence identity above 0.4, and 8 have an identity exceeding 0.9. This finding indicates that our model’s performance benefits from a broad spectral of related references in the training dataset, predominantly consisting of low-similarity sequences and supplemented by a small proportion of highly similar ones. In addition, about 34% of the phage genomes have more representations in the training sequences than the lambda phage (Supplementary Fig. 3), suggesting that the megaDNA model could be potentially utilized to study essential genes within these genomes.
Fig. 1. Foundational capacities of our language model.
a Overview of the model applications. b In silico mutagenesis analysis to identify essential genes in the bacteriophage genome. c Model loss variation across the lambda phage genome in the mutagenesis analysis. Upper, essential, and non-essential genes in the genome. Lower: changes in model loss for 50 bp non-overlapping windows across the genome (blue). The step size is 50 bp, and moving averages of model loss across 5000 bp windows are denoted in red. d Zero-shot prediction of essential genes by calculating the effects of mutations in the gene coding region (blue), start codon (orange) and stop codon (green). Area under the ROC curve (AUROC) scores are reported. e Prediction of mutational effects on protein functions using model embeddings. f Prediction of mutational effects for the deep mutational scanning experiment of the infA gene. Spearman correlation coefficients of the predicted and reported fitness from fivefold cross-validation tests are reported (Blue: megaDNA, gray: DeepSequence). n is the number of training samples. g Prediction of the impacts of Single Nucleotide Polymorphisms (SNPs) in the T7 bacteriophage genome. Spearman correlation of the predicted and reported fitness from fivefold cross-validation tests is reported. h Prediction of regulatory element activity using model embeddings. i Prediction of translation efficiencies for non-model organisms and high-throughput gene expression libraries. For K. oxytoca, P. protegens, and E. coli DH10B, we evaluated the model performance on endogenous genes. Fivefold cross-validation tests were used for all calculations. j Classifying taxonomies of unannotated sequences using model embeddings. k UMAP visualization of model embeddings for sequences from bacteriophages, bacteria, and archaea (model middle layer, sample size: n = 5000 per group). For f, g, and i, data are presented as mean values ± SEM from fivefold cross-validation tests (n = 5 folds). Source data are provided as a Source Data file.
megaDNA learns the functional properties of proteins and regulatory elements
Sequence embeddings are high-dimensional representations of the model input that capture rich contextual information. These embeddings can be used to make predictions about the quantitative property related to the original input. Since our megaDNA model takes DNA sequences as the input, we could harness the model’s learned representations for a wide range of predictive tasks. We first evaluated our model’s ability to predict the effects of sequence mutations on protein functions using a deep mutational scanning (DMS) dataset for the E. coli essential gene infA14 (Fig. 1e). This dataset includes all possible single codon mutations for infA and the corresponding mutational effects measured as fitness values through a growth competitive assay. To model protein fitness, mutated gene coding sequences were used as inputs. Then a linear regression model was trained on sequence embeddings derived from the internal activities of neurons within the megaDNA model to predict the mutational effects. The standard deviation of the prediction error is 0.16, which is much smaller than the full dynamic range of the protein fitness measurement (Supplementary Fig. 4). Our model’s prediction performance closely matched the state-of-the-art model DeepSequence15 (Fig. 1f), including for a protein not existing in the training dataset (Supplementary Fig. 5). Moreover, our model successfully predicted the impact of SNPs across the T7 bacteriophage genome16 (Fig. 1g). In this case, the mutated gene sequences were used as model input and the sequence embeddings were utilized to train regression models that predict SNP impacts, quantified as rates of mutability16. It is worth noting that this dataset covers only about 15% of all possible SNPs (Supplementary Fig. 6), which is substantially smaller than the DMS dataset. Despite this constraint, the Spearman correlation coefficient between predicted and measured impact is 0.71 for gene 1.3 and 0.54 for T7 RNAP (Supplementary Fig. 4). For genes with lower prediction accuracy, such as gene 17, our model still identifies mutations with significant fitness effects.
Expression of phage genes relies on the protein synthesis machinery in host bacteria cells. We investigated the potential of the model embeddings to predict regulatory element activity in bacteria genomes (Fig. 1h). The 5′UTR sequences were used as model input to derive embeddings, which were then used to predict their translation efficiencies via linear regression. We leveraged the paired RNA-seq and ribosome profiling datasets to quantify the translation efficiency of 5′UTR across three bacteria genomes17,18. The translation efficiency was defined as the ratio of the normalized ribosome density and the RNA level for the genes. Our model effectively predicted the translation efficiencies of 5′UTR in both model and non-model organisms, including K. Oxytoca, P. Protegens, and E. coli (Fig. 1i). In addition to the endogenous regulatory elements, we also benchmarked the predictive performance of our approach on the high-throughput measurement of the translational activity of a random 5’UTR library in E. coli19 (Fig. 1i). The Spearman correlation coefficients range from 0.62 to 0.82 for these datasets, illustrating our model’s capacity to capture translation-related sequence features. We also found that the model’s performance is robust to the training sample size (Supplementary Fig. 7).
Lastly, we extended our model to identify the taxonomy of unannotated sequences at the domain level (Fig. 1j). We collected unannotated sequences from bacteriophage, bacteria, and archaea genomes, which were used as model input to calculate their embeddings. Then these embeddings were mapped into a low-dimensional space, where we observed clear separations among different domains (Fig. 1k). By training logistic regression models based on sequence embeddings and the domain labels, we achieved a high classification accuracy in the cross-validation tests (average AUROC of 0.98, Supplementary Fig. 8). This high level of accuracy was consistent across different layers of our model (Supplementary Fig. 8). The prediction performance of the embeddings from the local and global layer was slightly lower compared to the middle layer. This difference may result from the local layer’s short context window and the global layer’s limited resolution for local sequence features. In contrast, the middle layer’s context window provides an optimal balance of length and resolution, enabling effective distinction of sequences from different domains. We further weighted the model predictions on the test sequences based on their similarity to the training datasets. A weight of 0 excludes test sequences that have at least one matched training sequence, and a weight of 1 includes all test sequences in the AUROC calculation (methods). Our results indicate that completely ruling out similar test sequences only results in a reduction of AUROC by 0.03, 0.01, and 0.02 for different model layers (Supplementary Fig. 9). Since the training data doesn’t contain genome sequences of bacteria or archaea, these results demonstrate the broad applicability of our model.
megaDNA generates de novo genomic sequences
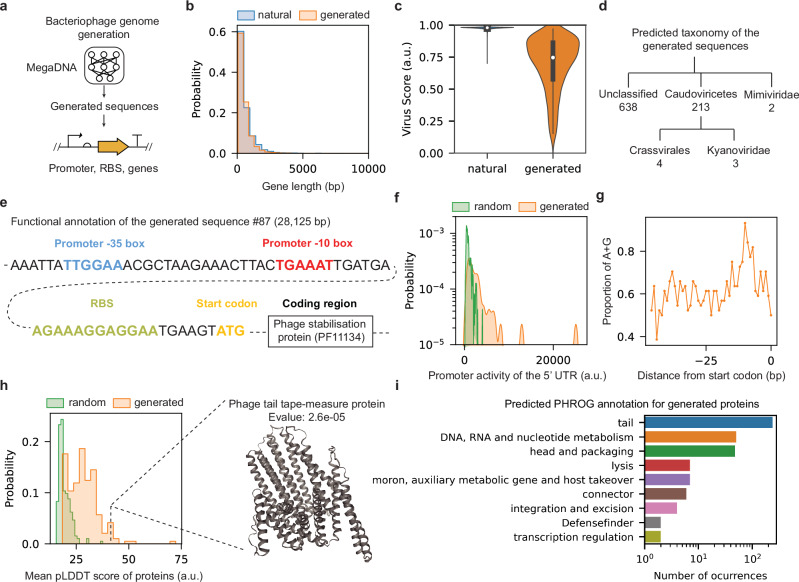
Our approach enables de novo generation of the genome sequence of bacteriophage (Fig. 2a). We generated a total of 1024 sequences longer than 1 K bp and applied geNomad20 for functional annotation of the generated sequences. The average sequence length is 43 K bp, and the average number of predicted genes per sequence is 67, which is similar to the training dataset (mean length: 48 K bp, average number of predicted genes: 68). The gene length distribution of the generated sequences is close to that of the training dataset (Fig. 2b, average gene length: 558 bp vs 646 bp), while the gene number distribution shows wider spread (Supplementary Fig. 10). In addition, the gene densities of the generated and training sequences are close with each other (Supplementary Fig. 10). The median virus score of all generated sequences is 0.75 and the maximum score is 0.97. These scores are lower than those of natural bacteriophage genomes, which have a median value of 0.98 (Fig. 2c). 228 out of all generated sequences (22%) are predicted to be Caudoviricetes by geNomad (Fig. 2d). As a comparison, 98% of the genomes in the training dataset were classified as Caudoviricetes. We further analyzed the generated sequences using vContact221 and found that all generated sequences form singletons, indicating a high diversity among them.
Fig. 2. Language model generates sequences with functional genomic structures.
a The workflow schematic. b Comparison of gene length distributions between randomly sampled subsets of predicted genes in generated sequences and training dataset (sample size: n = 2000). Two-sided Kolmogorov–Smirnov test: p value = 0.15. c Comparison of the predicted virus scores for generated sequences (sample size: n = 1024) and the training dataset (sample size: n = 99,429). Median virus scores are indicated by white dots. Black bars denote the interquartile ranges (25th to 75th percentiles). d Predicted taxonomy for the generated sequences predicted as viral. Only taxonomies with >1 sequence are shown. geNomad20 was used to produce results in (c, d). e Functional annotation of a selected sequence fragment (generated sequence #87). f Predicted promoter activity for all the 5′UTRs in the generated sequence #87 (sample size: n = 44), along with the promoter activity of the random sequences with the same length. Promoter activities were calculated using the Promoter Calculator22. Two-sided Kolmogorov–Smirnov test: p value = 6.3 × 10−8. g Proportions of adenine (A) and guanine (G) nucleotides preceding the start codon of all the predicted genes in the generated sequence #87. h Mean predicted pLDDT scores for the ESMFold predicted structures23. We focused on proteins with geNomad markers from generated sequences (sample size: n = 343; median value: 28) against random peptide sequences of the same lengths (sample size: n = 343; median value: 18). A sample generated protein is shown on the right. Two-sided Kolmogorov–Smirnov test: p value = 6.7 × 10−42. i Top 10 predicted functions of proteins derived from the generated sequences, as identified by phold24. Source data are provided as a Source Data file.
We then examined the 5′UTR of the annotated genes in the generated sequences to determine if they contain potential regulatory elements such as promoters and RBS to initiate transcription and translation. We chose the generated sequence #87 for further analysis due to its high predicted virus score (0.96) and its relatively small size (28 K bp). Using a machine learning tool (Promoter Calculator)22, we identified the −35 box and −10 box of the promoter within the 5’UTR of the predicted phage stabilization protein. Notably, their sequences are close to the established consensus motifs: TTGACA and TATAAT (Fig. 2e). Prior to the start codon of the same gene, we observed a region enriched in adenine (A) and guanine (G) nucleotides, indicative of a functional ribosome binding site (Fig. 2e). Analyzing all 5’UTR of the predicted genes from this sequence, we found a significantly higher promoter activity compared to random sequences of the same length (Fig. 2f). Intriguingly, the proportion of A and G nucleotides peaked around 10 bp upstream of the start codon, aligning closely to the optimal position for an RBS to drive translation initiation (Fig. 2g). In another sequence example (#212), we observed similar promoter activities and RBS characteristics, and we found a consensus −10 sequence TATAAT that located upstream of the N-terminus of the major capsid protein (Supplementary Fig. 11). This trend of A/G enrichment and high promoter activity within the 5′UTRs is also consistent across all the generated sequences (Supplementary Figs. 12 and 13). We found a low correlation between the promoter activities and virus scores, which may suggest that the model learns the two features independently (Supplementary Fig. 12). In short, our generated sequences harbor potential regulatory sequences that could enable the expression of the predicted genes.
In the generated sequences, 343 annotated genes were predicted to match geNomad markers. These genes share very little homology with the training dataset (Supplementary Table 1). The query genes and their matches do not belong to the same gene family. We employed ESMFold23 to predict their structures and calculated the average predicted Local Distance Difference Test (pLDDT) scores. This score reflects the confidence of ESMFold in the predicted structures. The median pLDDT score for these proteins is higher than that of random peptide sequences (28 vs 18, Fig. 2h). We further randomly sampled 10 K annotated genes from all the generated sequences and found high pLDDT scores for them (median value of 36, Supplementary Fig. 14). These results may suggest inherent similarities between the generated sequences and sequences used in training ESMFold. We further annotated gene functions using phold24. In brief, phold leverages a protein language model25 to derive structural information from protein sequences. This information is compared against a structural database via Foldseek26 to obtain PHROG annotations27. Although the generated sequences do not include a coherent set of genes necessary for the full phage life cycle, our analysis reveals several large protein families associated with phage-related functions, such as head and packaging, and nucleotide metabolism (Fig. 2i). Among these, several proteins were predicted to have DNA-binding activity, and the predicted structure also resembles the canonical helix-turn-helix (HTH) domain in this protein family (Supplementary Fig. 15). Moreover, the predicted structure of a generated protein aligns with the experimental structure in the PDB database, further demonstrating the model’s ability to capture biologically relevant features (Supplementary Fig. 15).
Discussion
In this work, we present a long-context generative model for genomic sequences, which effectively learns the language of gene coding and regulatory sequences via a single step of self-supervised training on unannotated whole genomes. We demonstrated that the model loss can be utilized for unsupervised prediction of essential genes in phage genomes, and the information-rich sequence embeddings enable the prediction of genetic variant effects and regulatory element activities across a wide range of organisms. The generated sequences match the length of natural bacteriophage genomes and display functional genomic architectures. It is worth noting that these sequences have not been optimized at the codon or gene level to allow for efficient protein expression in bacteria. In addition, the training dataset only covers a limited subset of the global phage diversity, which may impact the model’s ability to generalize to phage taxa not included in the training data. A recent study by Ratcliff et al. also shows that the generated sequences are compositionally more similar to certain bacteriophage families than the others28. Lastly, our pretrained model still lacks the ability to generate the complete set of genes covering all the necessary functions of the phage life cycle. Despite these limitations, we envision that our generative genomic model represents the first step towards the de novo design of the whole functional genome, paving the way for advancements in medicine, agriculture, and environmental science. Improvements of generative language models have been driven by the scaling-up of training dataset and model size, along with techniques for model fine-tuning and alignment with human input. Such approaches are likely to further improve the performance of the megaDNA model. This field also faces ongoing challenges in ethical considerations, biosafety, and regulatory frameworks, which are critical for the responsible advancement of generative modeling in synthetic biology29.
Methods
Model training
Our training dataset was curated from three sources. Firstly, we downloaded all the complete virus genomes from NCBI GenBank (as of February 2023) and retained only those with “phage” in the organism’s name. Secondly, the phage genomes from MGV (version 1.0) were downloaded, and we only included genomes with a completeness score larger than 95% and classified under the order Caudovirales. Our third source was GPD (https://www.sanger.ac.uk/data/gut-phage-database/), and we kept all the genomes with a completeness score above 0.95. Following the initial collection, we undertook an additional round of filtering. We used geNomad (version 1.6.1, default parameters with “—relaxed” flag) to predict the taxonomy of these genomes and then deleted all the genomes whose predicted host is not a unicellular organism. All genomes smaller than 96 K bp were then collected to construct the final training dataset.
Our megaDNA model utilized a three-layer transformer structure9. Each layer had a depth of 8 and progressively larger dimensions (local: 196, middle: 256, global: 512). The context lengths for the three layers are 16, 64, and 128. The model contains 145 M parameters in total. We assigned numerical tokens (1, 2, 3, and 4) to the nucleotides A, T, C, and G, respectively. For model training, we used a batch size of 1 and set the learning rate at 0.0002. The learning rate was progressively increased during the initial 50,000 steps as part of a warmup schedule. We used the Adam optimizer and applied gradient clipping with a norm of 0.5 to prevent gradient explosion.
In silico mutagenesis of phage genomes
Lambda phage genome sequence and annotations were downloaded from NCBI (Accession number: NC_001416.1). Essential genes were identified according to Piya et al.12. We conducted in silico mutagenesis using a 50 bp sliding window across the genome, and each nucleotide was randomly mutated to A, T, C, or G with equal probability. The impact of mutations was assessed by computing the model loss, which is further compared with their original counterparts. For both essential genes and non-essential genes, we calculated the mean model loss for all the windows within the gene coding region. Mann–Whitney U test was used to evaluate statistical differences between these two groups (scipy.stats.mannwhitneyu). Furthermore, mutations targeting the start and stop codons of all coding genes were simulated. We also generated a control set comprising an equivalent number of 3-nt mutations randomly distributed across the genome. The effect of these mutations on model loss was analyzed to infer their impact on fitness. The changes in model loss were used as a predictor of gene essentiality, and we computed the receiver operating characteristic (ROC) curve and the area under the ROC curve (AUROC) based on model predictions and experimental results12. We used the BLAST analysis to identify training sequences similar to the lambda phage genome (version 2.8.1+, default parameters and max_target_seqs set to 30,000).
Prediction of mutational effects on protein function
Sequence embeddings are high-dimensional representations of the model input produced by the language models. The assumption is that these embeddings capture complex patterns and relationships within the data, which has been supported by previous studies in the field of protein language models30,31. Technically speaking, model embeddings are vectors corresponding to the activities of a series of neurons within our model. We used the DNA sequences of the mutated genes as the model input and embeddings from three different layers of the model were extracted (dimensional = 196, 256, 512). These embeddings were concatenated to form a 964-dimensional vector representing each gene coding sequence. We then used a ridge regression model to predict the fitness value of the mutated sequence based on its embedding (sklearn.linear_regression.RidgeCV). We used the 5-fold cross validation to evaluate the prediction performance of our model. In each fold, one-fifth of the data was held out as test data while the remaining data were used as training data. The predictive performance of this model was then evaluated on the test dataset. The infA gene dataset was obtained from Kelsic et al.14. For the T7 bacteriophage dataset16, the genome sequence and annotations were downloaded from NCBI (Accession number: V01146.1). The model performance was evaluated for each gene in the same manner.
Prediction of translation efficiency
We assessed the translation efficiency (TE) of genes in Klebsiella oxytoca, Pseudomonas protegenes Pf-5, and Escherichia coli DH10B by calculating the ratio of average ribosome density (RD) to mRNA expression. The ribosome density of each gene was calculated by averaging ribosome occupancies over the length of the gene. The mRNA expression in FPKM (fragments per kilobase of transcript per million mapped reads) of each gene was calculated by normalizing the read counts to the gene length and the total number of mapped reads. The ribosome profiling and RNA-seq datasets of K. oxytoca and P. protegenes Pf-5 were obtained from the Sequence Read Archive with the accession code PRJNA57976717. The E. coli DH10B datasets were obtained from the NCBI GEO database with accession number GSE15266418. We used the DNA sequences spanning from −160 to 160 relative to the start codon as the input to our model. Model embeddings were extracted from three layers (dim = 196, 256, 512) and concatenated together to form a 964-dim vector for each input sequence. To mitigate the influence of lowly expressed genes on TE calculations, we focused on the top 25% expressed genes in Klebsiella oxytoca and Escherichia coli DH10B, and the top 20% expressed genes in P. protegenes Pf-5. We used 5-fold cross validation to evaluate the performance of our model. In each fold, a ridge regression model was trained on the input and TE values in the training dataset with default parameters (sklearn.linear_regression.RidgeCV). The trained model was then used to predict TE values in the test dataset. For the Evfratov et al. dataset19, 20 nt and 30 nt 5′UTR sequences were used as the input. The model performance was evaluated as previously described.
Classification taxonomy of unannotated sequences
We collected complete genome sequences of bacteria, archaea, and bacteriophage from NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes, n = 5000 each) via the wget command in Linux. The data can also be accessed from https://ftp.ncbi.nlm.nih.gov/genomes/genbank/. We randomly sampled 10 K bp sequence fragments from genomes longer than 10 K bp. For a total of 15,000 sequences, sequence embeddings were generated using our model across three layers (dim = 196, 256, 512). For embedding visualization, we used Uniform Manifold Approximation and Projection (UMAP)32 as implemented in the Python package umap-learn. To classify these sequences at the domain level, we employed a logistic regression model to evaluate the predictive performance of the sequence embeddings across multiple classes (sklearn.linear_model.LogisticRegression). The model’s performance was assessed using a 5-fold stratified cross-validation test. This method ensures that each fold is a good representative of the whole by maintaining the same proportion of samples for each class as in the complete dataset. Within each fold, the model was trained on a subset of the data and then used to predict probabilities on the test subset. The results for each domain were reported using AUROC (sklearn.metrics.roc_auc_score).
To further control for the sequence similarity between the test and training datasets, we introduced a weighted scheme in our model evaluation. For each fold, sequences in the test dataset that have at least one match in the training dataset (identified through the BLAST analysis with default parameters and max_target_seqs set to 30,000) are assigned a weight (w) ranging from 0 to 1. A weight of 0 removes these sequences from the AUROC calculation completely, while a weight of 1 includes all test sequences.
Model inference
We generated sequences from the trained model using a predefined set of parameters. Specifically, we adjusted the temperature to 0.95 to ensure a balance between variety and coherence in the sequences and kept the filter threshold at 0.0 to avoid limiting the range of token probabilities. For model training and inference, we utilized Nvidia’s A100 GPU (40GB) and 3090 Ti GPU (24GB) and used the PyTorch version 2.1.1 software package.
Analysis of the generated sequence
geNomad20 was used to annotate generated sequences with default parameters and the “—relaxed” flag (version 1.6.1). The 100 base pair regions preceding the start codon of each predicted gene were designated as the 5′UTR. We employed the Promoter Calculator (v1.0)22 to find the promoters in these regions. Only the promoter with the highest predicted activity in the forward direction was annotated. For protein structure prediction, we used the ESMFold model v123. The chunk size of the model was set to 64 for proteins longer than 700 amino acids (AA) and 128 for shorter proteins. We limited our structure calculation to proteins less than 1000 AA in length. Function prediction for these proteins within each PHROGs category was carried out using pharokka33 (version 1.7.1) and phold24 (version 0.1.4) with default parameters, as available on GitHub (https://github.com/gbouras13/pharokka, https://github.com/gbouras13/phold). Genome similarities of the generated sequences were analyzed using vContact221 with default parameters.
Statistics and reproducibility
No statistical method was used to predetermine sample size; No data were excluded from the analyses; The experiments were not randomized; The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We are grateful to Y. Yan, E. Heppenheimer, and H. Lee for their helpful discussions and to all the reviewers for their constructive feedback. B.S. acknowledges the open-source efforts of P. Wang.
Author contributions
B.S. conceived the research project. B.S. and J.Y. implemented the model. B.S. carried out model training, performed the computational and statistical analysis, and wrote the paper. All the authors discussed the results and contributed to the revision of the paper.
Peer review
Peer review information
Nature Communications thanks Jeremy Ratcliff and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The bacteriophage genomes were downloaded from public databases including: NCBI GenBank (https://ftp.ncbi.nlm.nih.gov/genomes/genbank/), n = 16,609; MGV, n = 53,032; GPD (https://www.sanger.ac.uk/data/gut-phage-database/), n = 30,032. Details about the training dataset including the accession codes of genome sequences are available from GitHub. Source data are provided in this paper.
Code availability
Our trained model and model inference codes are available from GitHub.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53759-4.
References
- 1.Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. Bert: pre-training of deep bidirectional transformers for language understanding. Preprint at arXiv10.48550/arXiv.1810.04805 (2018).
- 2.Brown, T. et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst.33, 1877–1901 (2020). [Google Scholar]
- 3.Benegas, G., Ye, C., Albors, C., Li, J. C. & Song, Y. S. Genomic language models: opportunities and challenges. Preprint at arXiv10.48550/arXiv.2407.11435 (2024).
- 4.Ji, Y., Zhou, Z., Liu, H. & Davuluri, R. V. DNABERT: pre-trained Bidirectional Encoder Representations from transformers model for DNA-language in genome. Bioinformatics37, 2112–2120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalla-Torre, H. et al. The nucleotide transformer: building and evaluating robust foundation models for human genomics. bioRxiv10.1101/2023.01.11.523679 (2023).
- 6.Benegas, G., Batra, S. S. & Song, Y. S. DNA language models are powerful predictors of genome-wide variant effects. Proc. Natl Acad. Sci. USA120, e2311219120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang, Y., Cornman, A. L., Kellogg, E. H., Ovchinnikov, S. & Girguis, P. R. Genomic language model predicts protein co-regulation and function. bioRxiv2023, 2024 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen, E. et al. HyenaDNA: Long-range genomic sequence modeling at single nucleotide resolution. Adv. Neural Inf. Process Syst. 36, (2024).
- 9.Yu, L. et al. Megabyte: Predicting million-byte sequences with multiscale transformers. Adv. Neural Inf. Process Syst. 36, 78808–78823 (2023).
- 10.Nayfach, S. et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol.6, 960–970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camarillo-Guerrero, L. F., Almeida, A., Rangel-Pineros, G., Finn, R. D. & Lawley, T. D. Massive expansion of human gut bacteriophage diversity. Cell184, 1098–1109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piya, D. et al. Systematic and scalable genome-wide essentiality mapping to identify nonessential genes in phages. PLoS Biol.21, e3002416 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGinnis, S. & Madden, T. L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res.32, W20–W25 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelsic, E. D. et al. RNA structural determinants of optimal codons revealed by MAGE-Seq. Cell Syst.3, 563–571.e6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riesselman, A. J., Ingraham, J. B. & Marks, D. S. Deep generative models of genetic variation capture the effects of mutations. Nat. Methods15, 816–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robins, W. P., Faruque, S. M. & Mekalanos, J. J. Coupling mutagenesis and parallel deep sequencing to probe essential residues in a genome or gene. Proc. Natl Acad. Sci. USA110, E848–E857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu, M.-H. et al. Control of nitrogen fixation in bacteria that associate with cereals. Nat. Microbiol.5, 314–330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espah Borujeni, A., Zhang, J., Doosthosseini, H., Nielsen, A. A. K. & Voigt, C. A. Genetic circuit characterization by inferring RNA polymerase movement and ribosome usage. Nat. Commun.11, 5001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evfratov, S. A. et al. Application of sorting and next generation sequencing to study 5΄-UTR influence on translation efficiency in Escherichia coli. Nucleic Acids Res.45, 3487–3502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camargo, A.P., Roux, S., Schulz, F. et al. Identification of mobile genetic elements with geNomad. Nat. Biotechnol.42, 1303–1312 (2024). [DOI] [PMC free article] [PubMed]
- 21.Bin Jang, H. et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol.37, 632–639 (2019). [DOI] [PubMed] [Google Scholar]
- 22.LaFleur, T. L., Hossain, A. & Salis, H. M. Automated model-predictive design of synthetic promoters to control transcriptional profiles in bacteria. Nat. Commun.13, 5159 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023). [DOI] [PubMed] [Google Scholar]
- 24.G. Bouras. Phage Annotation using Protein Structures. https://github.com/gbouras13/phold
- 25.Heinzinger, M. et al. Bilingual language model for protein sequence and structure. bioRxiv10.1101/2023.07.23.550085 (2024).
- 26.van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol.42, 243–246 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terzian, P. et al. PHROG: families of prokaryotic virus proteins clustered using remote homology. NAR Genom. Bioinform3, lqab067 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratcliff, J. Transformer model generated bacteriophage genomes are compositionally distinct from natural sequences. NAR Genom. Bioinform.6, lqae129 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker, D. & Church, G. Protein design meets biosecurity. Science 383, 349 (2024). [DOI] [PubMed] [Google Scholar]
- 30.Marquet, C. et al. Embeddings from protein language models predict conservation and variant effects. Hum. Genet.141, 1629–1647 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villegas-Morcillo, A., Gomez, A. M. & Sanchez, V. An analysis of protein language model embeddings for fold prediction. Brief. Bioinform23, bbac142 (2022). [DOI] [PubMed] [Google Scholar]
- 32.McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: uniform manifold approximation and projection. J. Open Source Softw.3, 861 (2018).
- 33.Bouras, G. et al. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics39, btac776 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bacteriophage genomes were downloaded from public databases including: NCBI GenBank (https://ftp.ncbi.nlm.nih.gov/genomes/genbank/), n = 16,609; MGV, n = 53,032; GPD (https://www.sanger.ac.uk/data/gut-phage-database/), n = 30,032. Details about the training dataset including the accession codes of genome sequences are available from GitHub. Source data are provided in this paper.
Our trained model and model inference codes are available from GitHub.