Abstract
The TGFβ receptor inhibitor galunisertib showed promising efficacy in patients with pancreatic ductal adenocarcinoma (PDAC) in the phase 2 H9H-MC-JBAJ study. Identifying biomarkers for this treatment remains essential. Baseline plasma levels of chemokine CCL3 were integrated with clinical outcomes in PDAC patients treated with galunisertib plus gemcitabine (n = 104) or placebo plus gemcitabine (n = 52). High CCL3 was a poor prognostic factor in the placebo group (mOS 3.6 vs. 10.1 months; p < 0.01) but a positive predictor for galunisertib (mOS 9.2 vs. 3.6 months; p < 0.01). Mechanistically, tumor-derived CCL3 activates Tgfβ signaling in macrophages, inducing their M2 phenotype and Lif secretion, sustaining a mesenchymal/basal-like ecotype. TGFβ inhibition redirects macrophage polarization to M1, reducing Lif and shifting PDAC cells to a more epithelial/classical phenotype, improving gemcitabine sensitivity. This study supports exploring TGFβ-targeting agents in PDAC with a mesenchymal/basal-like ecotype driven by high CCL3 levels.
Subject terms: Predictive markers, Targeted therapies, Pancreatic cancer
Introduction
In 2020, approximately 496,000 individuals worldwide were diagnosed with pancreatic ductal adenocarcinoma (PDAC), positioning it as the seventh leading cause of cancer-related deaths, with an estimated 466,000 deaths1. Across all stages, the 5-year relative survival rate remains alarmingly low, standing at a mere 13%2. In the absence of significant advancements in early detection and treatment strategies, PDAC is projected to ascend to the rank of the second most prevalent cause of cancer-related mortality by 2040 in Western countries3. This poor prognosis for PDAC patients can be largely attributed to the limited effectiveness of currently approved chemotherapeutic treatments. Additionally, molecularly targeted therapies and immune checkpoint inhibitors, often developed for this disease in unselected populations without consideration of predictive biomarkers, have proven ineffective4. In this regard, the implementation of the different classifications proposed for molecular subtypes of PDAC5–7 is also facing a slow and difficult clinical development8. The reductionistic scenario that depicted two broad consensus subtypes—classical and basal-like—is instead, much more complex and dynamic. Most tumors contain a substantial fraction of cells co-expressing classical and basal-like markers, establishing a continuum of these two phenotypes, probably in response to gradients of cytokines secreted by tumor and stromal cells acting in a paracrine manner within different spatially confined microenvironments, or ecotypes9. The process through which tumor epithelial cells lose their cell polarity and cell-cell adhesion, and gain invasive, metastatic properties, becoming more resistant to treatment, is defined Epithelial-to-Mesenchymal Transition (EMT)10.
The largest efforts of integrated genomic analysis in understanding the molecular pathology of PDAC confirmed Transforming Growth Factor β (TGFβ) as one of the most recurrently mutated signal transduction pathways in PDAC, making the inhibition of this signaling a promising experimental therapeutic approach for the treatment of patients affected by this disease7. We have contributed to demonstrate the inhibition of the TGFβ signaling as a potential strategy in preclinical models of PDAC11,12. These results led us to investigate clinically the inhibition of TGFβ signaling in combination with chemo-13–16 or immunotherapeutic agents17 as a novel treatment strategy for PDAC patients. In the initial phase 2b/ randomized phase 2 H9H-MC-JBAJ study in patients with newly diagnosed unresectable PDAC, we measured a positive signal for improvement of the overall survival (OS) for the combination of the TGFβ receptor inhibitor galunisertib plus gemcitabine versus gemcitabine monotherapy13. Besides the overall clinical endpoints, the randomized design of this study allowed a wide range of translational analyses on biological samples, leading to the identification of those biological characteristics potentially useful as biomarkers for patients’ selection. In particular, we identified the chemokine (C-C motif) ligand 3 (CCL3), also known as macrophage inflammatory protein 1- α (MIP1-α), as a significant negative prognostic marker in the population of patients receiving single-agent gemcitabine in the control group. On the contrary, galunisertib dramatically reverted this chemoresistance, and CCL3 was the most significant positive predictive marker for the combination of this TGFβ inhibitor plus chemotherapy14.
Tumor-associated macrophages (TAMs) are among the major components of the PDAC microenvironment, contribute to tumor growth and limit the activity of conventional chemotherapeutic agents by sustaining an inflammatory milieu that acts as a protective niche for cancer cells18. TAMs mainly originate from circulating monocytes that can be recruited to the tumor microenvironment in response to diverse attractant chemokines. In response to different signals, TAMs may be functionally programmed towards two forms, namely M1 or ‘classic’, leading to antitumor responses and cytotoxicity, or M2 or ‘alternative’, sustaining tumor promotion, treatment resistance, and suppressing effective adaptive immune responses19. CCL3 plays an important role in the process by which TAMs influence tumor development through the binding to the cell surface G-protein-coupled receptors CCR1 and CCR520–22.
In this study, we dissected the molecular and cellular mechanisms between tumor cells and TAMs responsible for the exceptional response to the inhibition of TGFβ signaling pathway of PDAC characterized by this prognostically negative ecotype sustained by elevated levels of CCL3.
Methods
Study population
This study included patients enrolled in part 2 of the H9H-MC-JBAJ study clinical trial. H9H-MC-JBAJ was a multinational, 2-part study of oral galunisertib in combination with gemcitabine. The first part was a nonrandomized, open-label, multicenter dose-escalation phase. The second part was a a 2:1 randomized, double-blind, 2-arm study of galunisertib in combination with gemcitabine versus gemcitabine plus placebo. Patients were included in the study if they met all of the following criteria: 1. Histological or cytological diagnosis of PDAC that is locally advanced (Stage II, III) or metastatic (Stage IV) and not amenable to resection with curative intent. Patients with previous radical surgery for PDAC were eligible after progression was documented. If they received adjuvant chemotherapy or chemoradiotherapy with gemcitabine, they would have been enrolled if the treatment was completed 3 months before or longer; 2. Have a measurable disease or non-measurable disease, defined according to RECIST 1.1; 3. Males or females at least 18 years of age; 4. Have adequate organ function, including: (hematologic) absolute neutrophil count (ANC) ≥ 1.5 × 109/L, platelets ≥100 × 109/L, and hemoglobin ≥9 g/dL; (hepatic) bilirubin ≤1.5 times upper limits of normal (ULN), alkaline phosphatase (ALP), aspartate aminotransferase (AST), alanine aminotransferase (ALT) ≤ 2.5 times ULN; (renal) serum creatinine within normal limits, ≤1.5 times ULN. 5. Have a performance status of ≤2 on the Eastern Cooperative Oncology Group (ECOG) scale; 6. Patients with moderate or severe cardiac disease were excluded. 7. Patients with endocrine pancreatic tumors or ampullary cancer were excluded from the study13. Research involving human research participants, material, and data have been performed in accordance with the Declaration of Helsinki.
Cell cultures
RAW 264.7 cells were purchased from ATCC. Pancreatic cancer cells were kindly provided by Dr. Paola Cappello (University of Turin, Italy) and Prof. Vincenzo Bronte (Istituto Oncologico Veneto, Padova). Cells were isolated by tumors developed in LSL-KrasG12D/+; p53R172H/+; PdxCretg/+ (KPC) or LSL-KrasG12D/+; PdxCretg/+ (KC) mice. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Euroclone, Milan, Italy) containing 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (PS) solution (Gibco) and 1% Glutamine (glu) (Gibco), and maintained in humidified atmosphere at 37 °C in a 5% CO2 incubator. All cells were regularly tested by PCR to exclude Mycoplasma contamination.
Bone-marrow derived macrophages (BMDM) were isolated by flushing femurs of adult C57BL6/J mice and culturing the resulting cells in RPMI containing 10% FBS, 1% PS and 1% glu supplemented with 100 ng/mL recombinant m-CSF (Miltenyi Biotec) for 7 days, and then polarized for 48 h with IFN-γ (150 ng/mL) or IL-4 (100 ng/mL)/IL-13 (10 ng/mL) all from Miltenyi Biotec.
Co-culture experiments were performed using a hydrophobic silicon barrier in ø100 mm Petri dishes. 1 × 106 RAW 264.7 cells and 6 × 105 PDAC cells were plated in each half of the dish and let to attach overnight. The day after, the barrier was cut and cells allowed to communicate for 48 h in low serum conditions (DMEM 1% FBS) in absence or presence of treatments.
Compounds and treatments
Recombinant Ccl3 (rCcl3) (Cat. No.450-MA) and recombinant Lif (rLif) (Cat. No.8878-LF) were purchased from R&D System and used for 72 h at 50 ng/mL (Ccl3) or 100 ng/mL (Lif). Gemcitabine (Cat. No.S1149), maraviroc (Cat. No. S2003), EC330 (Cat. No. S0472), and galunisertib (Cat. No. S2230) were purchased from Selleckchem. For in vitro experiments, all compounds were dissolved in 100% dimethyl sulfoxide (DMSO) (Applichem) and used at the following concentrations: 0.78 nM–200 nM gemcitabine, 5 µM galunisertib, 5 μM maraviroc or 1 µM EC330. For in vivo treatments, galunisertib was resuspended in hydroxyethylcellulose (HEC) (0.25%) and administered via oral gavage at concentration of 100 mg/kg twice/day. Gemcitabine was resuspended in saline solution and administered intraperitoneally at concentration of 75 mg/Kg once/week.
Quantification of the effect of treatments
To evaluate the response of PDAC cell lines to the combination of gemcitabine plus galunisertib, 1 × 103 cells were seeded in 96-well plates and treated for 72 h with DMSO or increasing concentrations of gemcitabine, in presence or absence of a fixed dose of galunisertib. Crystal violet was used to measure cell viability using a plate reader (Victor X4, PerkinElmer). Mean EC50 values and 95% CIs were calculated using GraphPad Prism software (GraphPad Software Inc).
Plasmids and lentiviral production
Lentiviruses for gene knockdown were produced in HEK-293T cells by co-transfecting lentiviral vector, dR8.74 packaging plasmid (Addgene #22036) and pMD2.G envelope plasmid (Addgene #12259). shRNA vectors used were: pLKO.1-puro (scramble, LV-c; Addgene #8453), pLKO.1-puro-shCcl3.39 and pLKO.1-puro-shCcl3.40 targeting the coding region (CDS) of murine Ccl3 (targeting sequence 5′-CGCCAATTCATCGTTGACTAT-3′ and 5′-CTCTGTACCATGACACTCTGC-3′, respectively). When unspecified, gene silencing was performed by co-infection of PDAC cells with both Ccl3-targeting lentiviruses. Lentiviruses for gene overexpression were produced in HEK-293T by co- transfection of pLenti-GIII-CMV vector (Cat. No. 49645064, Applied Biological Materials Inc.) or pLenti-GIII-CMV-Ccl3 (Cat. No. 15367064, Applied Biological Materials Inc.), dR8.74 packaging plasmid (Addgene #22036) and pMD2.G envelope plasmid (Addgene #12259).
In vitro migration assay
For migration assay, 4 × 105 PDAC cells were seeded on the bottom of a plate using Culture2well silicone inserts (IBIDI) and let to adhere overnight in complete medium until they reached a confluence of ~95%. The inserts were then removed, and medium was replaced by serum-free DMEM. 4 × 105 RAW 264.7 cells were then seeded in transwell inserts with a pore diameter of 8μm that were placed on the upper side of the plate, and media were allowed to communicate in presence of 500 μg/ml Mitomycin C (Sigma-Aldrich). After 30 h migrating RAW 264.7 cells were stained with Diff Quick (MediCult Italia S.p.A) and counted. Cell-free scratches of PDAC cells were imaged at the indicated time points after insert removal until complete wound closure using a Zeiss ApoTome.2 microscope with 4X objective lens. The measure of the scratched area was performed with ImageJ software, and the migration rate for each well was calculated as mean of the relative covered area compared to T0 area for each well.
RNA extraction and real-time qPCR
RNA extraction was performed using PureLink RNA Mini Kit (Invitrogen) and quantified by using Nanodrop 8000. After DNase I treatment (Roche Diagnostics), 1 μg of RNA was reverse transcribed with High-Capacity RNA-to-cDNA™ Kit (Applied Biosystems). Quantitative real-time PCR was carried out at 60 °C using PowerUp SYBR Green Master Mix (Applied Biosystems) on a QuantStudio3 (Applied Biosystems). Primers were designed by using Primer3. Primer sequences are reported in Supplementary Table 2.
Chromatin immunoprecipitation (ChIP)
2 × 106 RAW264.7 were fixed with 1% formaldehyde for 30 min, followed by addition of 125 mM glycine for 5 min to stop fixation. Cells were lysed in Farnham lysis buffer (5 mM PIPES pH 8, 85 mM KCl, 0.5% NP-40) supplemented with phosphatase and protease inhibitors for 15 min. Nuclei were then collected by centrifugation at 800 × g for 10 min and then lysed in nuclear lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8) supplemented with phosphatase and protease inhibitors for 30 min. Chromatin was sonicated with an ultrasonic bath to an average size of 200–600 bp, diluted with Dilution Buffer (10 mM Tris-HCl pH 8, 2 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% SDS) and incubated overnight with 20 μl dynabeads protein G and 3 μg of mouse anti Stat3 (124H6) (#9139, Cell Signaling) or normal mouse IgG (sc-2025, Santa Cruz Biotechnology). Immunocomplexes were washed with increasing salt concentrations, DNA eluted at 65 °C with 1% SDS and recovered with PureLink RNA mini kit (Cat. 12183018 A, Invitrogen). qPCR was carried out at 60°C using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) on a QuantStudio3 (Applied Biosystems). Promoter sequence of Tgfb1 has been obtained at UCSC Genome Browser GRCm38/mm10 (chr7:25686954-25687013). Primer sequences are reported in Supplementary Table 2.
Protein extraction and western blot
Cells were lysed in cold RIPA buffer (ab156034, Abcam) supplemented with protease and phosphatase inhibitors and centrifuged at 20,000 × g for 20 min at 4 °C. Supernatants were collected and protein quantification was performed using the Pierce BCA Protein Assay kit (Thermofisher Scientific). Equal amounts of protein were resolved by SDS-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, and incubated for 1 h in Everyblot blocking buffer (Cat. No 12010020, Bio-Rad) at room temperature. Primary antibodies are reported in Supplementary Table 3. Blotted membranes were developed by using Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore) and imaged with UVITEC Alliance Q9 Advanced (UVITEC Cambridge).
Enzyme-linked immunosorbent assay (ELISA)
ELISA assays were performed with mouse Lif Quantikine ELISA Kit (Cat. no MLF00, R&D System), or mouse MIP1α ELISA Kit (Cat. no MMA00, R&D System). Plasma samples from mice were diluted before use following manufacturer’s instructions. The optical density was determined with microplate reader iMark (Bio-Rad) at 540 nm.
The multi-analyte immunoassay panel developed by Myriad RBM (Austin, Texas, USA) was used to assess CCL3 levels in plasma samples from patients14.
In vivo orthotopic transplantations
5–6 weeks old C57BL/6 J female with body weight ranging from 21 to 25 g were purchased from Charles River Laboratories. Murine PDAC cells were resuspended in a PBS:Matrigel solution (1:1) at the concentration of 2.5 × 105 cells per 40 μl/injection. On day 0, mice were anesthetized by exposure to isoflurane and injected orthotopically into the pancreas parenchyma. For in vivo treatments, mice bearing pancreatic cancer were randomly allocated into 8 group (n = 10) to receive galunisertib (100 mg/kg) via oral gavage twice/day, gemcitabine (75 mg/kg) intraperitoneally once/week, or their vehicles. During the treatment period, tumor growth was monitored by non-invasive imaging with ultrasound (Vevo System) in anesthesia. For Kaplan-Meier survival analysis, 6 mice from each group were euthanized by cervical dislocation when tumors reached the ethical cutoff volume 1500 mm3, and included in the survival curve. For flow-cytometry and ELISA, 4 mice from each group were euthanized by cervical dislocation at the end of 4-weeks treatment, peripheral blood was collected by retro-orbital sinus puncture, and tumors collected for ex vivo analysis. For in vivo experiments, mice were maintained at the animal facility of the University of Verona. Animals were maintained in a pathogen-free and temperature-controlled environment, with 12 h light and dark cycles, housed in plastics cages and were fed ad libitum. Research involving animals was in accordance with relevant guidelines and regulations, and has been approved by the Italian Ministry of Health (authorization no. 299/2022-PR from Ministry of Health, prot. C46F4.29).
Immunohistochemistry
Immunohistochemistry (IHC) and hematoxylin and eosin (H&E) were performed on murine pancreatic sections of formalin-fixed paraffin-embedded specimens. Sections at 5 μm interval were cut from each tissue and stained via IHC or with H&E (cat. no H-3502, Vector Laboratories). IHC was performed as described in23. After antigen retrieval with citrate buffer pH 6.0 (Leica Biosystems), tissues were blocked with 2.5% BSA blocking solution (Vector laboratories) for 1 h and incubated with primary antibodies against E-cadherin (#3195), p-Smad2 (#3108) (Cell Signaling Technology), Vimentin (#ab92547, Abcam), Gata-6 (#55435-1-AP, Proteintech) or pan-Cytokeratin (#NB600-579, Novus Biologicals) at 1:100–1:1000 overnight at 4 °C (Supplementary Table 3). Slides were developed using ImmPACT DAB substrate peroxidase HRP substrate (Vector Laboratories) following manufacturer’s instructions.
RNAscope® in situ hybridization (ISH)
RNAscope® in situ hybridization assay was performed according to manufacturer’s instructions (Advance Cell Diagnostics (ACD), Hayward, CA, USA). Briefly, slides were dehydrated in Xylene for 10 min followed by 100% ethanol for 5 min. The pretreat solution 1 (hydrogen peroxide reagent, Cat. #322330, ACD) was applied for 10 min at RT followed by two washes with ultra-pure water. Tissue sections were then boiled at 98 °C for 20 min in pretreat solution 2 (target retrieval reagent, Cat. #322000, ACD), rinsed in distilled water and dehydrated in 100% ethanol for 5 min. The pretreat solution 3 (protease reagent, Cat #322330, ACD) was applied for 15 min at 40 °C followed by two washes with ultra-pure water. All incubation periods were performed on the HybEz™ hybridization system (ACD). RNAscope® buffered Z probe Mm-CCL3 (Cat. #319471, ACD), the negative (Cat. #310043, ACD) and the positive (Cat. #313911, ACD) control probes were then applied and let hybridized for 2 h at 40 °C. The amplification steps were performed according to manufacturer’s instructions. Detection was performed using a chromogenic red-assay (RNAscope® 2.5 HD detection kit – RED, Cat. #322360, ACD). Sections were incubated for 10 min at RT, rinsed with ultra-pure water and then stained with hematoxylin (Cat. #H-3401-500, Vector Laboratories).
Flow cytometry
For flow cytometry analysis of macrophages population, tumor samples were dissociated using Tumor dissociation kit mouse (Cat. no 130-096-730, Miltenyi Biotec) according to manufacturer’s instructions. The Fixable die Viobility 405/520 was used to gate for live cells. Antibodies employed were from Miltenyi Biotec, and are reported in Supplementary Table 3. Acquisition was performed using Becton Dickinson LSR-Fortessa X-20, and samples were analyzed using FlowJo software (TreeStar Inc., Ashland, USA).
Statistical analysis
Data represent mean ± s.d. values calculated on at least three independent experiments. P values were calculated using Student’s t test (two groups) or one-way analysis of variance (ANOVA) with Dunnett’s or Tukey’s corrections (more than two groups). A two-tailed value of p < 0.05 was considered statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001. Overall survival (mOS) was estimated by using the Kaplan–Meier method and a log-rank test was used to compare the results between groups. Statistical analyses were performed using GraphPad Prism version 9 and R version 4.2.2 (R Foundation for Statistical Computing. URL https://www.R-project.org/).
Study approval
H9H-MC-JBAJ study was conducted according to the principles of good clinical practice, applicable laws and regulations, the Council for International Organizations of Medical Sciences International Ethical Guidelines, and the Declaration of Helsinki. The local ethical review board approved the study. Written informed consent was received prior to participation. All animal protocols were approved by local ethic authorities (Centro Interdipartimentale di Servizio alla Ricerca Sperimentale, CIRSAL) and by the Italian Ministry of Health and conducted in accordance with Italian Governing Law (D.lgs 26/2014) (authorization no 299/2022-PR, prot. C46F4.29).
Results
Ccl3 acts as a poor prognostic factor in PDAC by sustaining a mesenchymal/basal-like phenotype through an enrichment in M2-polarized TAMs
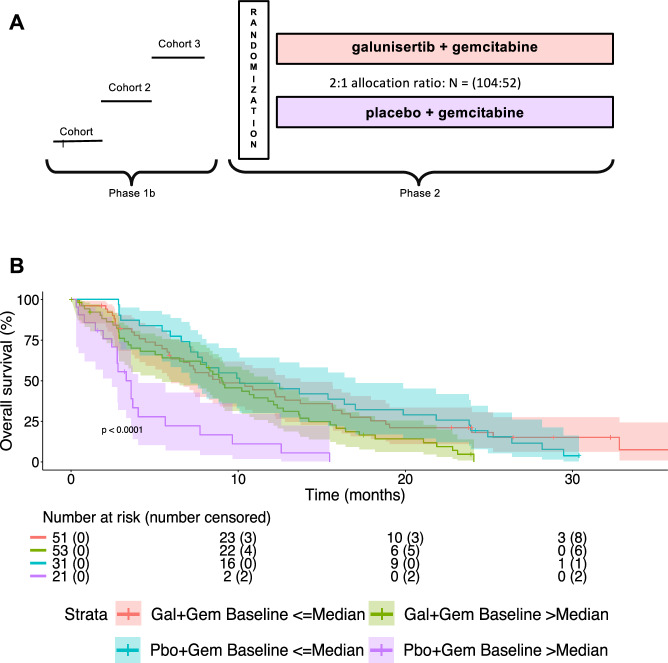
Initially, we confirmed the negative prognostic and the positive predictive role of CCL3 in patients enrolled in the H9H-MC-JBAJ study. Baseline clinical characteristics are described in Supplementary Table 1. With an updated median follow-up of 28.85 (95% CI: 24.21-NA) months, we measured a significantly different mOS in patients randomly allocated to receive placebo plus gemcitabine with basal levels of CCL3 higher or lower than the median value [mOS (95% CI), high vs low, 3.6 (2.5–4.0) vs 10.1 (7.2–17.0) months, HR (95% CI) = 3.93 (2.14–7.22); p < 0.01], confirming the negative prognostic value of this biomarker (Fig.1). Most importantly, we measured a significantly longer mOS in patients with basal levels of CCL3 higher than the median and allocated to receive galunisertib plus gemcitabine if compared with those allocated to receive placebo plus gemcitabine [mOS (95% CI), galunisertib+gemcitabine vs. placebo+gemcitabine, 9.2 (5.5–12.4) vs 3.6 (2.5–4.0) months, HR (95% CI) = 0.38 (0.22–0.66); p < 0.01], confirming the relevant positive predictive value of this cytokine.
Fig. 1. Plasma levels of CCL3 predict response to galunisertib plus chemotherapy in patients with advanced PDAC.
A Study design diagram. B Kaplan-Meier estimates of overall survival (OS) by plasma levels of CCL3, with 95% confidence bands and numbers at risk. Gal galunisertib, Gem gemcitabine, Pbo placebo.
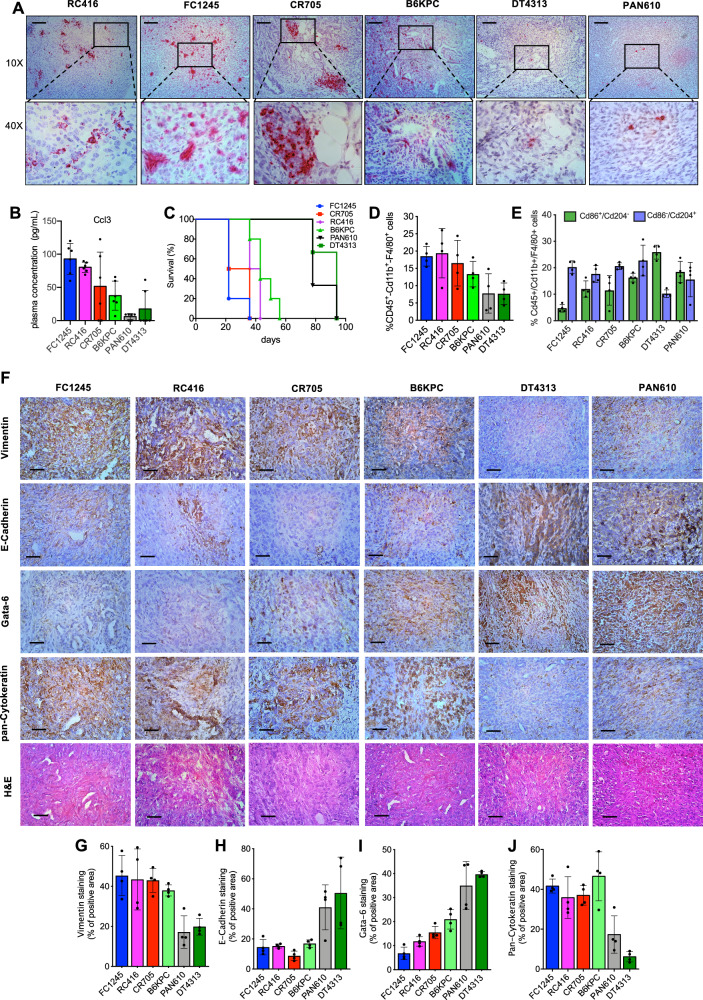
To delve into the mechanisms behind this exceptional response in PDACs with this prognostically negative ecotype sustained by CCL3, we initially analyzed six different murine PDAC cell lines established by tumors developed in LSL-KrasG12D/+; p53R172H/+; PdxCretg/+ (KPC) or LSL-KrasG12D/+; PdxCretg/+ (KC) mice. Based on their Ccl3 expression, we clustered two groups of models, FC1245, CR705, RC416 cell lines with a high expression, and DT4313, B6KPC, PAN610 cells with low expression of this cytokine (Supplementary Fig. 1A–C). Consistently, we measured a significantly different tumor expression (Fig. 2A, Supplementary Fig. 1D, E) and circulating plasma levels (p < 0.001) (Fig. 2B) of Ccl3 in mice bearing orthotopic tumors from these different cell lines.
Fig. 2. Characterization of orthotopic murine PDAC tumors.
A Representative RNA in situ hybridization (ISH) images of Ccl3 in tumor sections from mice bearing FC1245, RC416, CR705, B6KPC, DT4313 and PAN610 cells. Scale bar: 150 μm. B Plasma levels of Ccl3 were measured in C57BL/6 J mice bearing orthotopic PDAC tumors from 6 different murine PDAC cell lines using Enzyme-Linked Immunosorbent Assay (ELISA). Data are expressed as mean ± s.d. (n = 6). C Median overall survival (mOS) of C57BL/6 J mice bearing orthotopic PDAC tumors (n = 6). D, E Flow cytometry analysis of Cd45+/Cd11b+/F4/80+ tumor-associated macrophages (TAMs) (D) and their M2 (Cd45+/Cd11b+/F4/80+/Cd204+/Cd86−)/M1(Cd45+/Cd11b+/F4/80+/Cd204−/Cd86+) ratio (E) in FC1245, RC416, CR705, B6KPC, DT4313 and PAN610 tumor samples. Data are expressed as mean ± s.d. (n = 4). F Representative images of FC1245, RC416, CR705, B6KPC, DT4313 and PAN610 tumors stained with E-cadherin, vimentin, gata-6, pan-cytokeratin antibodies and H&E. Scale bar = 60 μm. G–J Quantification of paraffin sections from orthotopic murine PDAC tumors stained as indicated in (F). Data are expressed as mean ± s.d. (n = 4).
Consistently with the negative prognostic role of Ccl3 observed in PDAC patients, mOS in tumor-bearing mice with higher levels of plasma Ccl3 (FC1245, CR705, RC416) was significantly shorter (FC1245 = 22 days, CR705 = 29 days, RC416 = 39.5 days) than in those with undetectable levels of plasma Ccl3 (DT4313 = 94 days, PAN610 = 78 days) (Fig. 2C). Notably, we measured a significantly higher (p < 0.05) infiltration of Cd45+/Cd11b+/F4/80+ TAMs (Fig. 2D; Supplementary Fig. 2) and their M2(Cd204+/Cd86−)/M1(Cd204−/Cd86+) polarization ratio (Fig.2E; Supplementary Fig. 2) in high Ccl3 expressing FC1245, CR705, or RC416 tumors than in DT4313 or PAN610 ones. We histologically characterized these murine PDAC tumors into epithelial/classical or mesenchymal/basal-like phenotypes and correlated with their expression of Ccl3. We measured a high expression of epithelial/classical markers E-cadherin and Gata-6 and a low expression of the mesenchymal/basal-like markers vimentin and cytokeratins in Ccl3-low DT4313 and PAN610 tumors. Conversely, the Ccl3-high tumors FC1245, CR705 and RC416 had a clear mesenchymal/basal-like phenotype, as demonstrated by a high expression of vimentin and cytokeratins, and no expression of E-cadherin or Gata-6 (Fig. 2F–J).
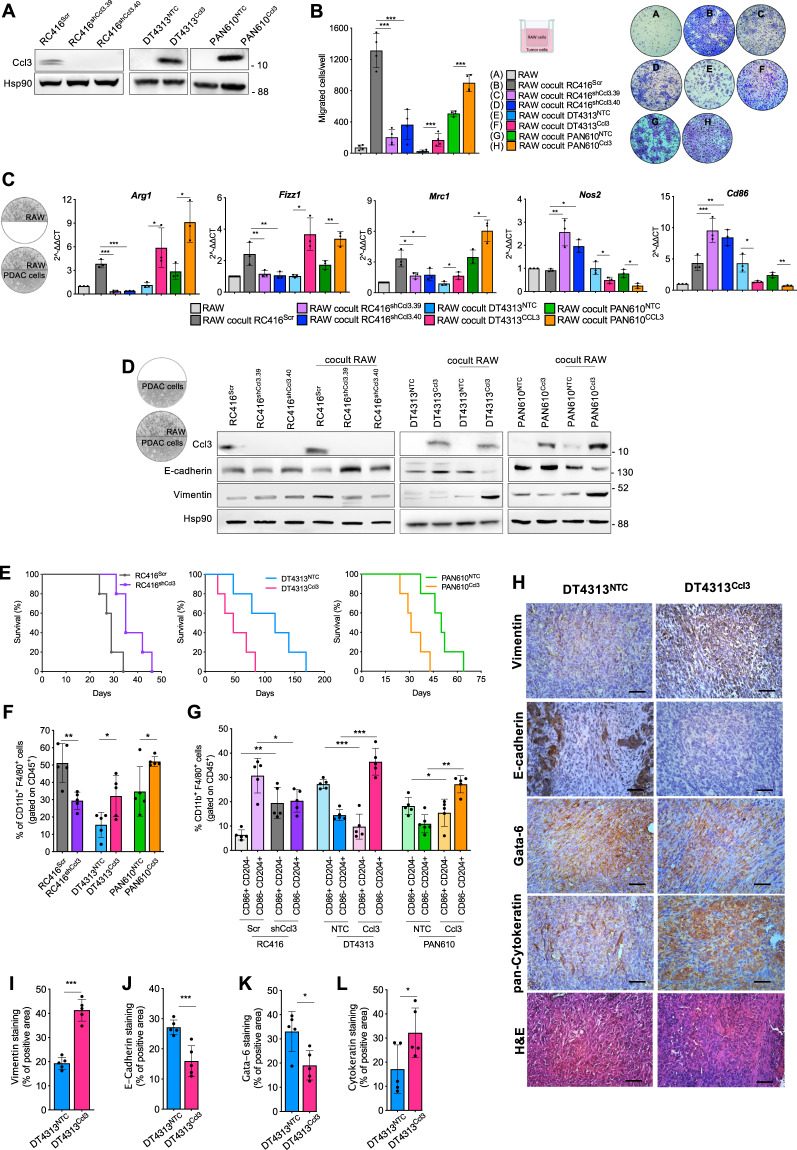
To dissect the molecular and cellular mechanisms acting within a high CCL3 ecotype, we knocked down Ccl3 expression in RC416 cells by using two different short-hairpin RNAs (RC416shCcl3.39 and RC416shCcl3.40), and stably overexpressed Ccl3 in DT4313 (DT4313Ccl3) and PAN610 (PAN610Ccl3) (Fig. 3A). We evaluated the role of Ccl3 in favoring the recruitment and polarization of TAMs by co-culturing RAW264.7 macrophages with tumor cells in a transwell system that allows them to migrate in presence or absence of stimuli from tumor cells. When RAW264.7 macrophages were co-cultured with Ccl3-high RC416SCR, DT4313 Ccl3 or PAN610Ccl3 cells, we observed a significant increase in their migration if compared with the co-culture with the corresponding Ccl3-low RC416shCcl3, DT4313NTC or PAN610NTC controls (Fig. 3B).
Fig. 3. Ccl3 favors recruitment and M2-polarization of TAMs that induce the acquisition of mesenchymal features in tumor cells.
A Representative western blot of Ccl3 in RC416, DT4313 and PAN610 cells transduced as indicated. Hsp90 was used as loading control. B Transwell migration assay of RAW264.7 macrophages as single culture or co-cultured as indicated. Results show mean ± s.d (n = 4). C qPCR analysis of M2 (Arg1, Fizz1, Mrc1) and M1 (Inos2, Cd86) markers in RAW264.7 cells as single cultures or co-cultured with RC416, DT4313 or PAN610 transduced as indicated. Data are shown as mean ± s.d (n = 3). A schematic representation of the co-culture technique used to culture RAW264.7 macrophages with PDAC cell lines is shown. D Representative western blot of Ccl3 and EMT markers in RC416, DT4313 and PAN610 transduced as indicated. Hsp90 was used as loading control. A schematic representation of the co-culture technique used to culture PDAC cell lines with RAW264.7 macrophages is shown. E mOS duration of C57BL6/J mice bearing orthotopic tumors from RC416, DT4313 or PAN610 transduced as indicated (n = 5). F, G Flow cytometry analysis showing increased Cd45+/Cd11b+/F4/80+ TAMs recruitment (F) and M1(Cd45+/Cd11b+/F4/80+/Cd204−/Cd86+)/M2(Cd45+/Cd11b+/F4/80+/ Cd204+/Cd86−) skewing (G) in Ccl3-high RC416Scr, DT4313Ccl3 and PAN610 Ccl3 tumors compared with Ccl3-low RC416shCcl3, DT4313NTC or PAN610NTC controls (n = 5). Data are expressed as mean ± s.d. P values were calculated by two-tailed unpaired Student’s t test (F, I, J, K, L), or ANOVA and Tukey’s test (B, C, G). H–L Representative images of paraffin sections from DT4313NTC and DT4313Ccl3 orthotopic tumors stained with vimentin, E-cadherin, Gata-6 or pan-cytokeratin antibodies (H) and relative quantifications (I–L). Scale bar = 60 μm. Data are expressed as mean ± s.d. (n = 5). *p < 0.05; **p < 0.01; ***p < 0.001.
To determine the effect of the paracrine crosstalk between TAMs and PDAC cells on their phenotype, we used co-culture models in which a hydrophobic barrier allows paracrine interactions between RAW264.7 and PDAC cells but prevents direct contact between the two cell types. In this model, RAW264.7 macrophages increased the expression of the M2 markers Arg1, Mrc1 and Fizz1, and decreased that of the M1 markers Nos2 and Cd86 only when co-cultured with Ccl3-high RC416SCR, DT4313Ccl3 or PAN610Ccl3 cells, but not with Ccl3-low RC416shCcl3, DT4313NTC or PAN610NTC controls (Fig. 3C). These results were reproduced by using a different co-culture model of bone marrow-derived macrophages (BMDMs) with PDAC cells (Supplementary Fig. 3A–E), establishing RAW264.7 cells as a valid model of in vitro murine macrophages.
Analyzing PDAC cells in the co-cultures, the overexpression of Ccl3 did not significantly affect their EMT phenotype as single cultures. Conversely, RC416Scr, DT4313Ccl3 and PAN610Ccl3 tumor cells co-cultured with RAW264.7 macrophages showed increased expression of the mesenchymal marker vimentin and decreased expression of the epithelial marker E-Cadherin (Fig. 3D) associated with increased migratory ability (Supplementary Fig. 4A–C), whereas RC416shCcl3.39/40, DT4313NTC and PAN610NTC respective controls did not. The role of macrophages in sustaining this phenotypic switch was corroborated by the evidence that the Ccl3 receptor Ccr5 was mainly expressed in RAW264.7 macrophages (Supplementary Fig. 1B). The negative prognostic role of Ccl3 was also confirmed in vivo. We measured a significantly shorter mOS duration in mice bearing RC416Scr, DT4313Ccl3 or PAN610Ccl3 orthotopic tumors than in those bearing RC416shCcl3, DT4313NTC or PAN610NTC tumors (mOS RC416Scr 29 days vs RC416shCcl3 35 days, HR = 3.85, 95% CI = 0.83–17.84, p = 0.0072; mOS DT4313Ccl3 47 days vs DT4313NTC 117 days, HR = 3.04, 95% CI = 0.72–12.76, p = 0.037; mOS PAN610Ccl3 31 days vs PAN610NTC 50 days, HR = 3.78, 95% CI = 0.82–17.37, p = 0.0088) (Fig. 3E).
We determined the role of Ccl3 in the recruitment and polarization of TAMs in these models. We measured a significantly higher (p < 0.05) infiltration of Cd11b+/F4/80+ TAMs in RC416Scr, DT4313Ccl3 or PAN610Ccl3 tumors if compared with the Ccl3-negative RC416shCcl3, DT4313NTC or PAN610NTC controls (Fig. 3F; Supplementary Fig. 5A, B). Most importantly, Ccl3-high tumors had a significantly lower (p < 0.05) infiltration of M1-polarized TAMs and a higher infiltration of M2-polarized TAMs than did their respective Ccl3-low controls (Fig.3G; Supplementary Fig. 5A, B). Histological examination revealed a shift toward a more mesenchymal/basal-like phenotype in DT4313Ccl3 tumors if compared with DT4313NTC controls (Fig. 3H–L).
These results indicate that tumor-derived Ccl3 acts as a poor prognostic factor in PDAC by recruiting and polarizing TAMs. M2-polarized TAMs sustain, in turn, a basal-like ecotype in the PDAC cells counterpart.
Ccl3 triggers autocrine TGFβ signaling in TAMs that, in turn, sustains EMT in PDAC cells through paracrine Lif
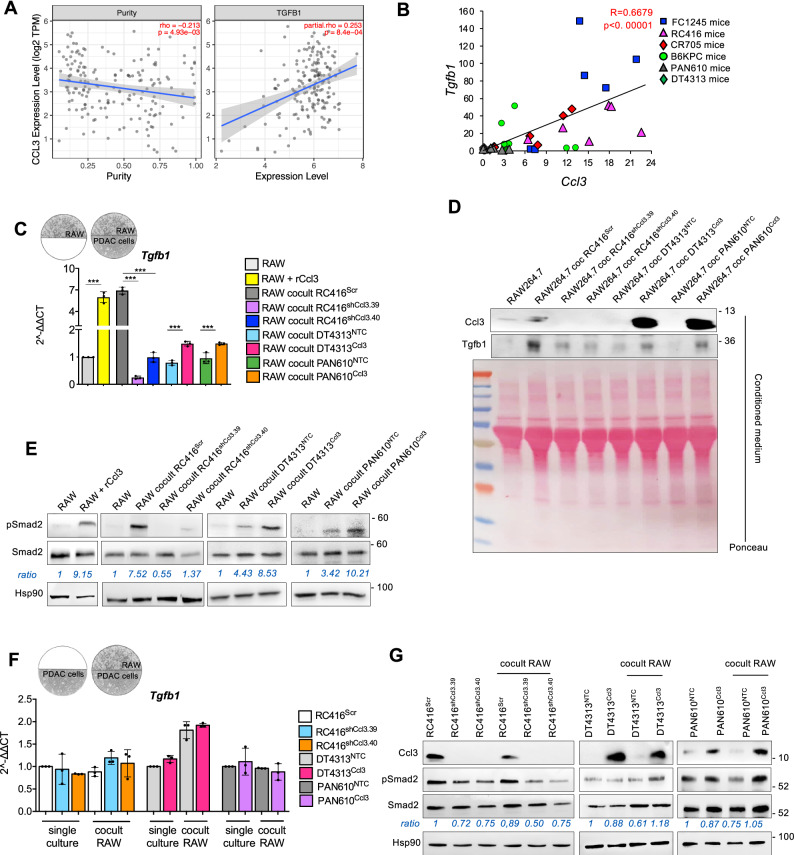
In order to investigate the role of TGFβ signaling pathway in this ecotype sustained by CCL3, we initially measured a significant (p = 0.00084) co-expression of CCL3 and TGFB1 mRNA transcripts in 179 PDAC patients from TCGA database (Fig. 4A). Consistently, we found a significant correlation between Ccl3 and Tgfb1 expression in our orthotopic PDAC models (Pearson score R = 0.6351; p < 0.00001) (Fig. 4B).
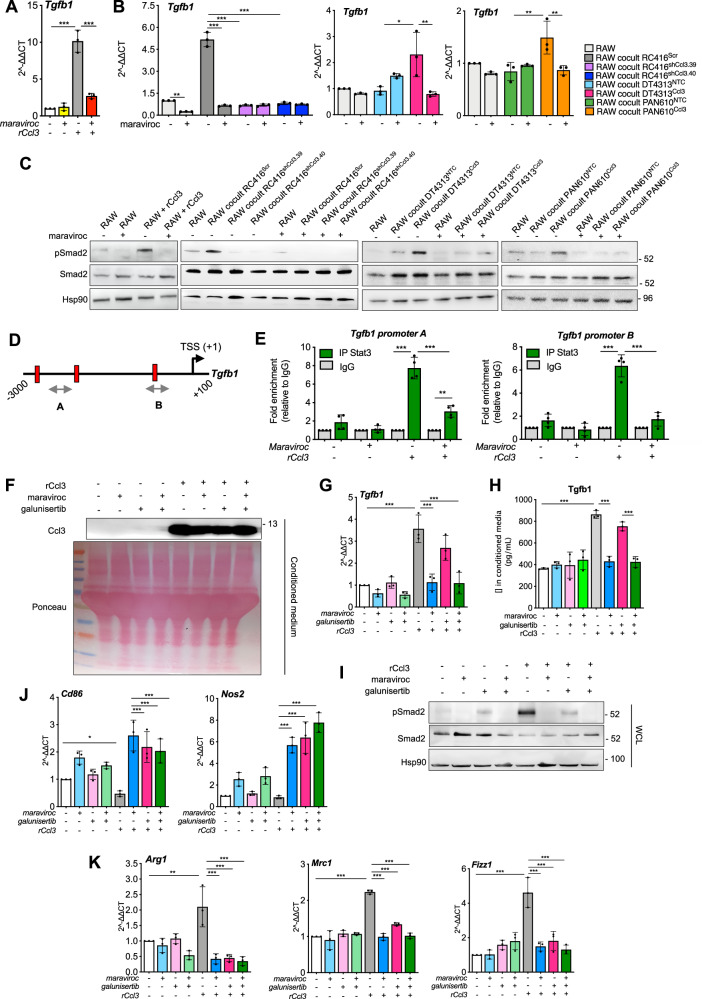
Fig. 4. Ccl3 activates TGFβ signaling in macrophages.
A Correlation of Ccl3 expression with tumor purity (left) and with TGFB1 expression (right) in PAAD patients from TIMER2.0 (n = 179). B Correlation between Tgfb1 and Ccl3 mRNA expression in tumors from C57BL6/J orthotopic PDAC models (n = 36). C qPCR of Tgfb1 in RAW264.7 unstimulated or stimulated with rCcl3, and as single cultures or co-cultured with RC416, DT4313 or PAN610 transduced as indicated. Results show mean ± s.d of 3 biological replicates. A schematic representation of the co-culture technique used to culture RAW264.7 macrophages with PDAC cell lines is shown. D Representative western blot of secreted Ccl3 and Tgfβ1 proteins in the conditioned medium of RAW264.7 macrophages as single cultures or cocultured with RC416, DT4313 and PAN610 transduced as indicated. Ponceau served as loading control. E Representative western blot of pSmad2 and Smad2 in RAW264.7 as single cultures or co-cultured as indicated. Hsp90 was used as loading control. Quantification of pSmad2/Smad2 is shown. F qPCR of Tgfb1 in RC416, DT4313 or PAN610 transduced as indicated, as single cultures or co-cultured with RAW264.7. Results show mean ± s.d of 3 biological replicates. A schematic representation of the co-culture technique used to culture PDAC cells with RAW264.7 macrophages is shown. P values in (C) and (F) were calculated by one-way ANOVA and Tukey’s test. G Representative western blot of pSmad2 and Smad2 in RC416, DT4313 or PAN610 transduced as indicated, as single cultures or co-cultured with RAW264.7. Hsp90 was used as loading control. Quantification of pSmad2/Smad2 is shown. *p < 0.05; **p < 0.01; ***p < 0.001.
We then used co-culture models to measure the activation of TGFβ signaling in PDAC cells or RAW264.7 macrophages upon modulation of Ccl3 expression by tumor cells. We measured a significantly (p < 0.001) higher expression (Fig. 4C) and secretion (Fig. 4D) of Tgfβ1 by RAW264.7 macrophages stimulated with rCcl3 or co-cultured with RC416Scr, DT4313Ccl3 or PAN610Ccl3 cells, if compared with unstimulated RAW264.7 or if co-cultured with the Ccl3-low controls RC416shCcl3, DT4313NTC or PAN610NTC. This was associated with a major increase of Smad2 phosphorylation in macrophages (Fig. 4E; Supplementary Fig. 3F, G). Conversely, we did not measure any significant increase in the expression of Tgfb1 upon modulation of Ccl3 in RC416, DT4313 or PAN610 tumor cells as single cultures or in coculture with RAW264.7 (Fig. 4F), nor in Smad2 phosphorylation (Fig. 4G).
By using maraviroc, a selective inhibitor of the Ccl3 receptor Ccr5, we measured a complete suppression of the increase of Tgfb1 (Fig. 5A, B) and Tgfb2 (Supplementary Fig. 6A–C) ligands, and of the consequent phosphorylation of Smad2 in RAW264.7 macrophages stimulated with rCcl3 or co-cultured with RC416Scr, DT4313Ccl3 or PAN610Ccl3 (Fig. 5C). To deepen into the mechanism underlying TGFβ regulation by the Ccl3/Ccr5 axis, we performed in silico analysis of Tgfb1 regulatory regions (obtained from the UCSC Genome Browser assembly ID: GRCm38/mm10) to identify putative transcription factors that could be involved in transcriptional regulation of Tgfb1. Consistently with the activation of Stat3 via Ccr5 signaling24, we identified three putative Stat3-binding sites (BS) possessing the consensus sequence TTC(N)2-4GAA within the Tgfb1 promoter (Fig. 5D). Chromatin immunoprecipitation (ChIP) assay of Stat3 using different sets of primers spanning the putative BS (regions A and B) revealed Stat3 enrichment at the Tgfb1 promoter in RAW264.7 macrophages stimulated with rCcl3, with a 5-to-9-fold enrichment in Stat3 signal over ChIP with a non-specific IgG (Fig. 5E). Most importantly, treatment with maraviroc almost completely abrogated the binding of Stat3 on Tgfb1 promoter sustained by Ccl3 (Fig. 5E). These results demonstrated the existence of an autocrine TGFβ signaling in macrophages, but not in tumor cells, triggered by tumor-derived Ccl3.
Fig. 5. The Ccr5 inhibitor maraviroc prevents the Ccl3-mediated activation of TGFβ signaling in macrophages.
A qPCR of Tgfb1 in RAW264.7 macrophages in presence or absence of rCcl3, after treatment with DMSO or 5 μM maraviroc. Results show mean ± s.d (n = 3). B qPCR of Tgfb1 in RAW as single cultures or co-cultured with RC416, DT4313 or PAN610 transduced as indicated, after treatment with DMSO or maraviroc (5 μM). Results show mean ± s.d (n = 3). C Representative western blot of pSmad2 and Smad2 in RAW264.7 in presence or absence of rCcl3, and as single cultures or in co-culture with RC416, DT4313 and PAN610 transduced as indicated, after treatment with DMSO or 5 μM maraviroc. Hsp90 was used as loading control. D Schematic representation of Tgfb1 promoter (available at UCSC Genome Browser GRCm38/mm10) with the position of ChIP probes (gray double arrowhead) and consensus Stat3 binding sites (TTC(N)2-4GAA) (vertical slashes) relative to the transcription starting site (TSS). E ChIP-qPCR of Stat3 occupancy at the Tgfb1 promoter in RAW264.7 macrophages upon stimulus with rCcl3, in the presence or absence of 5 μM maraviroc. The y-axis represents relative promoter enrichment, normalized on input material. IgG was set to 1. Data are represented as mean ± s.d. (n = 4). F Representative western blot of secreted Ccl3 in the conditioned media of RAW264.7 macrophages stimulated or not with rCcl3, and treated with 5 μM maraviroc, 5 μM galunisertib, their combination or DMSO as control. G qPCR of Tgfb1 in RAW264.7 macrophages in the presence or absence of rCcl3 and treated as indicated. Results show mean ± s.d (n = 3). H Secreted levels of Tgfβ1 ligand measured in conditioned medium of RAW264.7 macrophages treated as indicated using Enzyme-linked immunosorbent assay (ELISA). Data are expressed as mean ± s.d. (n = 3). I Representative western blot of pSmad2 and Smad2 in RAW264.7 macrophages treated as indicated. Hsp90 was used as loading control. J, K qPCR of M1 (J) and M2 (K) markers in RAW264.7 macrophages in presence or absence of rCcl3 and treated as indicated. Results show mean ± s.d (n = 3). P values in (A, B, E, G, H, J and K) were calculated by one-way ANOVA and Tukey’s test. *p < 0.05; **p < 0.01; ***p < 0.001.
To confirm TGFβ signaling as the mediator of the Ccl3/Ccr5 axis in sustaining macrophages phenotype, we first evaluated the effect of TGFβ receptor I (TGFβRI) inhibitor galunisertib on RAW264.7 macrophages. Pre-treatment of RAW264.7 with galunisertib followed by coculture with Ccl3-high and Ccl3-low PDAC cells, as well as addition of galunisertib to established co-cultures, almost completely suppressed the phosphorylation of Smad2 triggered by tumor-derived Ccl3 in macrophages (Supplementary Fig. 7A, and 8A, B), reverted their increase in migration (Supplementary Fig. 7B, C) and redirected their polarization toward a M1 phenotype (Supplementary Figs. 7D–G, and 8C–F). Most importantly, we observed a comparable modulation of macrophage phenotype achieved by either Ccr5 and TgfβRI inhibition in RAW264.7 stimulated with rCcl3, and their combination led to a similar M1-phenotype switch in these cells (Fig. 5F–K), supporting a direct effect of the Ccl3/Ccr5/TGFβ axis on macrophage phenotype and excluding any additional control from PDAC cells.
We, then, addressed the effect of TGFβRI inhibition on tumor cells EMT. We confirmed a significant increase of vimentin that was paralleled by a significant decrease of E-cadherin in RC416Scr, DT4313Ccl3 or PAN610Ccl3 cells only when co-cultured with RAW264.7 macrophages, and not in their corresponding controls RC416shCcl3, DT4313NTC or PAN610 NTC. This EMT happened despite only a minor modulation of Smad2 activation when Ccl3-high PDAC cells were co-cultured with RAW264.7 macrophages. Of note, galunisertib was able to fully revert the observed changes in the EMT phenotype (Fig. 6A). This discrepancy suggested the putative existence of additional paracrine signals from macrophages upon the activation of the TGFβ signaling sustained by Ccl3, which might be responsible for the acquisition of mesenchymal features in Ccl3-high tumor cells.
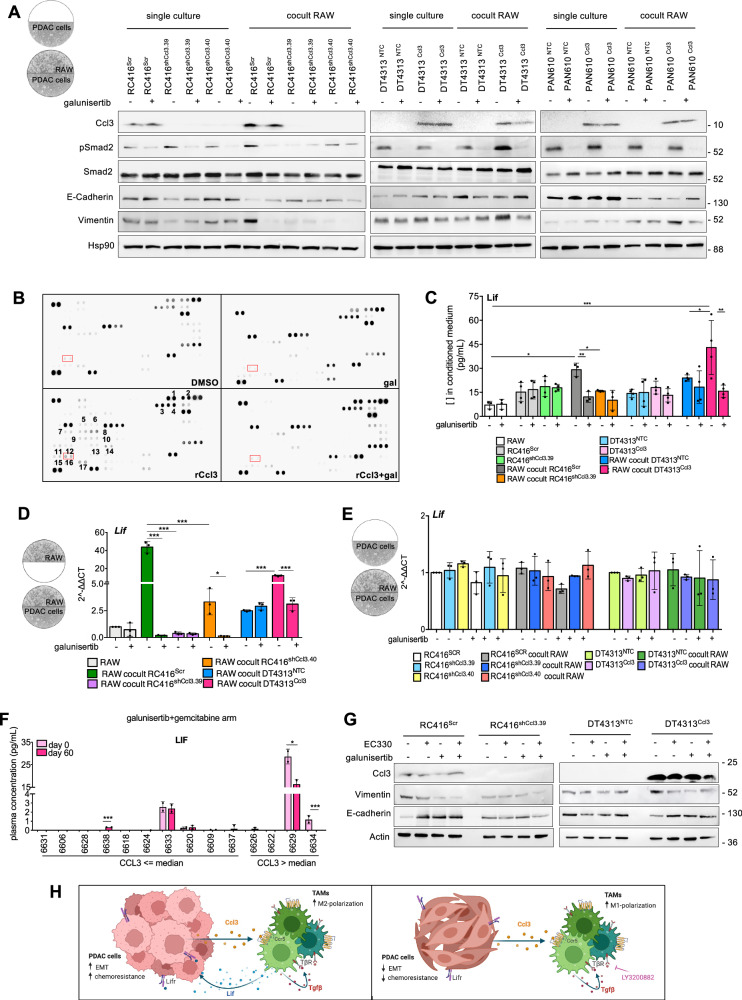
Fig. 6. TAM-derived Lif acts as a mediator of TGFβ signaling in sustaining EMT in PDAC cells.
A Representative western blot of pSmad2, Smad2, E-cadherin or vimentin in RC416, DT4313 or PAN610 transduced as indicated, as single culture or co-cultured with RAW264.7 and treated with DMSO or 5 μM galunisertib for 48 h. Hsp90 was used as loading control. A schematic representation of the co-culture technique used to culture PDAC cells with RAW264.7 macrophages is shown. B Representative cytokine array from the supernatants of RAW264.7 macrophages treated with DMSO or galunisertib, in presence or absence of recombinant Ccl3 (rCcl3). 1.Ccl3; 2.Ccl5; 3.Cd14; 4.Cd40; 5.Cxcl11; 6.Cxcl13; 7.Fgf21; 8.Gas6; 9.IGFBP-6; 10.Il1a; 11.Il10; 12.Il11; 13.Il13; 14.Il15; 15.Leptin; 16.Lif; 17.Pentraxin-3. C Lif concentration in the supernatants of RAW264.7, RC416Scr, RC416shCcl3, DT4313NTC or DT4313Ccl3 as single cultures or in co-culture, after treatment with 5 μM galunisertib or DMSO for 48 h, measured by ELISA. Data are expressed as mean ± s.d. (n = 4). D qPCR of Lif in RAW as single culture or co-cultured as indicated and treated with 5 μM galunisertib or DMSO for 48 h. Data are expressed as mean ± s.d. (n = 3). A schematic representation of the co-culture technique used to culture RAW264.7 macrophages with PDAC cells is shown. E qPCR of Lif in RC416Scr, RC416shCcl3, DT4313NTC or DT4313Ccl3 tumor cells as single cultures or in co-culture with RAW264.7 macrophages, treated with 5 μM galunisertib or DMSO for 48 h. Data are expressed as mean ± s.d. (n = 3). A schematic representation of the co-culture technique used to culture PDAC cells with RAW264.7 macrophages is shown. F Plasma levels of LIF at baseline (day 0) and after two cycles of treatment (day 60) in low- and high-CCL3 patients enrolled in the H9H-MC-JBAJ trial treated with galunisertib plus gemcitabine, measured by ELISA. P values were calculated by two-tailed unpaired Student's t test. G Representative western blot of vimentin and E-cadherin in RC416 and DT4313 transduced as indicated, co-cultured with RAW264.7 macrophages and treated with 1 μM of the Lif inhibitor EC330, 5 μM of the TGFβ inhibitor galunisertib, their double combination or DMSO as vehicle control. Actin was used as loading control. H Proposed mechanism of action of the Ccl3/Ccr5/Tgfβ/Lif axis. P values in (C–E) were calculated by one-way ANOVA and Tukey’s test. *p < 0.05; **p < 0.01; ***p < 0.001.
To identify these paracrine mediators, we performed a cytokine immunoassay. This assay enabled us to profile the relative levels of 111 different cytokines and chemokines in conditioned medium from RAW264.7 macrophages in basal conditions or upon stimulation with recombinant Ccl3 (rCcl3), in the presence or absence of the TGFβ receptor inhibitor galunisertib. We identified 17 cytokines whose expression was significantly increased in the conditioned medium of RAW264.7 cells upon stimulation with rCcl3. For 11 of them, the secretion was completely abrogated by treatment with galunisertib (Fig. 6B), including Lif and Il1α. While Il1α is known to support NF-κB activation in PDAC cells in an autocrine manner25, Lif is known to modulate EMT and chemoresistance in PDAC in a paracrine manner26. Thus, we evaluated by ELISA the amount of secreted Lif in the conditioned media of PDAC cells and RAW264.7 macrophages alone or in co-culture. We measured a significant increase in secreted Lif only when RAW264.7 macrophages were co-cultured with the Ccl3-high RC416Scr or DT4313Ccl3 PDAC cells if compared with single cultures or with co-culture with the Ccl3-low controls RC416shCcl3 and DT4313NTC cells. Galunisertib was able to significantly suppress Lif induction (Fig. 6C). qPCR analysis of co-cultures confirmed RAW264.7 macrophages as the cell type responsible for the secretion of Lif. Indeed, we measured increased Lif mRNA levels only in RAW264.7 macrophages when co-cultured with Ccl3-high PDAC cells (Fig. 6D), whereas its expression remained unaltered in PDAC cells both as single cultures and in co-culture with RAW264.7 (Fig. 6E). Administration of rCcl3 to RAW264.7 led to a similar upregulation. Most importantly, combined inhibition of Ccr5 and TgfβRI in these cells almost completely abrogated the increase of Lif mRNA induced by rCcl3 (Supplementary Fig. 9A), confirming the role of the Ccl3/Ccr5/Tgfβ axis in supporting the transcriptional regulation of Lif.
To strengthen our results, we measured circulating levels of LIF in plasma samples of 14 high-CCL3 patients collected at baseline and after 60 days of treatment with gemcitabine plus galunisertib. While we did not measure any significant difference in plasmatic level of LIF between day 0 and day 60 in patients with basal levels of CCL3 lower than the median, we observed a significant reduction in LIF circulating levels in those two out of four patients who expressed measurable levels of LIF at baseline (Fig. 6F).
To determine the role of macrophages-derived Lif in modulating EMT in PDAC cells, we used EC330, a small-molecule selective inhibitor of LIF receptor, as a single-agent treatment or in combination with galunisertib. We observed a comparable modulation of EMT achieved by either Lif or TGFβ inhibitor. Their combination did not obtain any additive effect (Fig. 6G), indicating a serial activation of these two pathways rather than a parallel and synergistic effect in sustaining EMT in tumor cells. Administration of EC330 was also able to revert EMT by decreasing the expression of vimentin while increasing that of E-cadherin in both high-Ccl3 PDAC cells wild type or knocked-down for TgfβrI (Supplementary Fig. 9B), confirming the importance of the Tgfβ/Lif paracrine axis in sustaining the EMT induced by Ccl3 in tumor cells.
Taken together, these data support a model in which tumor-derived Ccl3 induces a TGFβ autocrine loop in macrophages through Ccr5. This autocrine signal promotes a M2-polarized phenotype and triggers the secretion of Lif by macrophages, which then induces a poor prognostic mesenchymal/basal-like phenotype in tumor cells. Inhibitors of the TGFβ pathway act on macrophages by suppressing their TGFβ autocrine signaling, thus redirecting their polarization toward a M1 phenotype, and inhibiting their production of Lif (Fig. 6H).
Inhibition of TGFβ signaling in TAMs increases the response of CCL3-high PDAC to gemcitabine through a reversion toward a more epithelial/classical ecotype
To investigate the role of Ccl3 as a predictive marker for the combination of TGFβ receptor inhibition plus chemotherapy, we first evaluated the therapeutic efficacy of combined targeting of TGFβ with standard chemotherapy. In vitro, galunisertib led to a significant reduction in the EC50 values of gemcitabine only when RC416Scr, DT4313Ccl3 or PAN610Ccl3 were co-cultured with RAW264.7 macrophages (Supplementary Fig. 10A, B). Contrariwise, RC416shCcl3, DT4313NTC or PAN610NTC control cells were not affected (Supplementary Fig. 10C, D). Stimulation of co-cultures with rLif almost completely reverted the efficacy of galunisertib in increasing the sensitivity of Ccl3-high PDAC cells to gemcitabine (Supplementary Fig. 10E, F), suggesting that activation of the TGFβ/Lif signaling axis in macrophages could reduce antitumor activity of gemcitabine of Ccl3-high tumor cells.
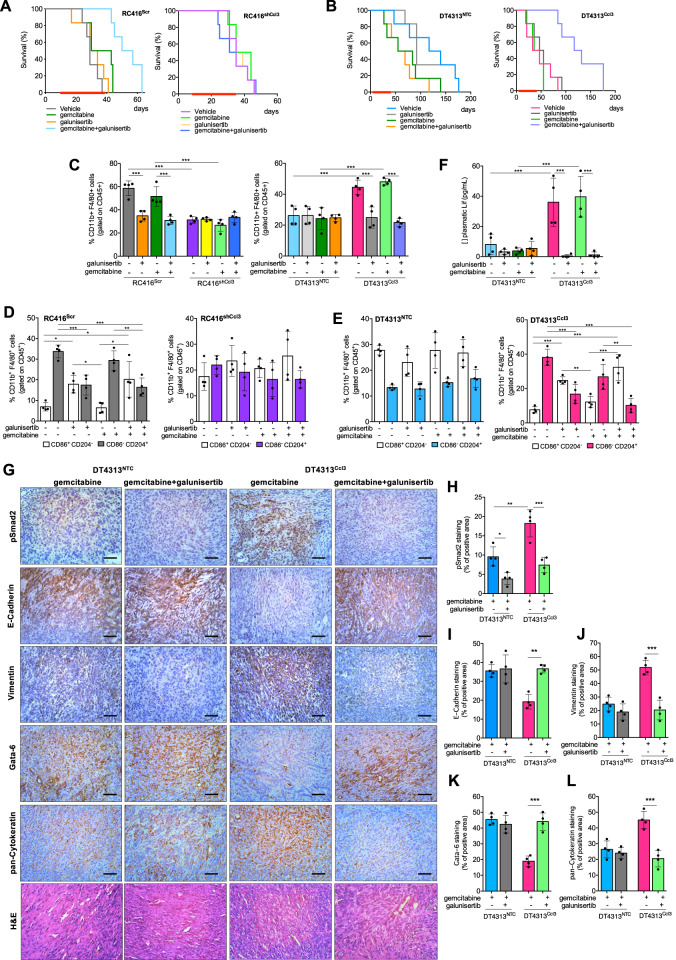
The in vitro efficacy of the combination of TGFβ receptor inhibition plus chemotherapy translated into a marked anti-tumor activity in orthotopic PDAC models. Mice bearing orthotopic RC416Scr and RC416shCcl3, or DT4313NTC and DT4313Ccl3 tumors were randomly assigned to be treated with gemcitabine, galunisertib, their double combinations, or their respective vehicles as control. Mice bearing Ccl3-high tumors receiving gemcitabine plus galunisertib had a statistically significantly longer mOS duration than those receiving gemcitabine alone (RC416Scr mOS: gemcitabine+galunisertib = 53.5 days vs gemcitabine + vehicle = 36.5 days, HR = 0.095, 95% CI = 0.01842–0.4913, p = 0.0005; RC416shCcl3 mOS: gemcitabine + galunisertib = 35 days vs gemcitabine + vehicle = 39.5 days, HR = 1.261, 95% CI = 0.354–4.496, p = 0.721; DT4313Ccl3, mOS: gemcitabine + galunisertib = 124.5 days vs gemcitabine + vehicle = 41.5 days, HR = 0.223, 95% CI = 0.012–0.221, p = 0.0008; DT4313NTC, mOS: gemcitabine + galunisertib = 62 days vs gemcitabine + vehicle = 69.5 days, HR = 1.381, 95% CI = 0.465–4.601, p = 0.546) (Fig. 7A, B). Ccl3-high tumors treated with the combination of gemcitabine plus galunisertib also had a significantly (p < 0.05) lower infiltration of Cd11b+/F4/80+/CD86−/CD204+ M2-TAMs, and a higher infiltration of Cd11b+/F4/80+/CD86+/CD204− M1-TAMs than did those treated with gemcitabine alone (Fig. 7C–E). Most importantly, galunisertib almost completely abrogated Lif secretion in the plasma of mice bearing Ccl3-high tumors (Fig. 7F). At immunohistochemical analysis, treatment with galunisertib was ineffective in DT4313NTC tumors, whereas it significantly reverted EMT in DT4313Ccl3 tumors, which showed a higher expression of the epithelial/classical markers E-cadherin and gata-6, and a lower expression of the mesenchymal markers vimentin and cytokeratin (Fig. 7G–L; Supplementary Fig. 11A–F). IHC of pSmad2 confirmed the efficacy of galunisertib in inhibiting TGFβ signaling in vivo (Fig. 7G, H; Supplementary Fig. 11A, B).
Fig. 7. Inhibition of TGFβ signaling improves the activity of gemcitabine in Ccl3-high PDAC tumors.
A mOS of mice bearing RC416Scr or RC416shCcl3 orthotopic tumors treated with gemcitabine, galunisertib, their double combinations or their respective vehicles as control (n = 6). B mOS of mice bearing DT4313NTC or DT4313Ccl3 orthotopic tumors treated with gemcitabine, galunisertib, their double combinations, or their respective vehicles as control (n = 6). C–E Flow cytometry analysis of Cd45+/Cd11b+/F4/80+ TAMs (C) and their M2 (Cd45+/Cd11b+/F4/80+/Cd204+/Cd86−)/M1(Cd45+/Cd11b+/F4/80+/Cd204−/Cd86+) ratio in tumors from mice bearing RC416 (D) or DT4313 (E) cells transduced as indicated and treated for 4 weeks with gemcitabine, galunisertib, their double combinations or their respective vehicles as control (n = 4). F Plasma levels of Lif measured by ELISA in DT4313NTC or DT4313Ccl3 orthotopic tumors treated as indicated (n = 4). G Representative images for DT4313NTC or DT4313Ccl3 tumors stained with pSmad2, E-cadherin, vimentin, Gata-6, pan-Cytokeratin antibodies, and H&E. Scale bar = 60 μm. H–L Quantification of paraffin sections from orthotopic murine PDAC tumors stained as indicated in (G). Data in (C–L) are expressed as mean ± s.d. (n = 4). P values in (C–F and H–L) were calculated by one-way ANOVA and Tukey’s test. *p < 0.05; **p < 0.01; ***p < 0.001.
Collectively, our results indicate that pharmacological inhibition of TGFβ signaling in TAMs modulates their M2 polarization and secretion of Lif. This leads to a reversion of the EMT toward a more epithelial phenotype of tumor cells and a classical ecotype of PDAC tissues. As a result, these CCL3-high tumors become significantly more sensitive to the cytotoxic effect of gemcitabine.
Discussion
TGFβ is recognized as one of the most relevant pathways to sustain PDAC progression. However, over the last decade, various agents targeting TGFβ ligands or receptors have been developed in unselected population of patients and, thus, failed to demonstrate large and significant efficacy. In our study, we demonstrated that inhibiting the TGFβ pathway could yield an exceptional response in patients affected by CCL3-high PDAC, and we defined the molecular and cellular mechanisms for this response. Our findings support a model where tumor-derived CCL3 promotes recruitment and induces a TGFβ pathway autocrine loop in macrophages. This autocrine signal induces an M2-polarized phenotype and triggers the secretion of Lif by macrophages. Lif derived from macrophages, in turn, acts on tumor cells inducing a poor prognostic mesenchymal/basal-like phenotype and resistance to classical chemotherapeutic agents. Inhibiting the TGFβ pathway acts on macrophages, suppressing their TGFβ autocrine signaling, redirecting their polarization toward an M1 phenotype, and inhibiting their production of LIF. Consequently, this restoration leads to a more epithelial/classical phenotype in PDAC cells, modulating their sensitivity to gemcitabine. This explains the exceptional response observed in patients affected by CCL3-high PDAC when treated with a combination of TGFβ pathway inhibition and chemotherapy.
The interaction between TAMs and cancer cells, as well as stromal cells, in the tumor microenvironment has been extensively demonstrated to facilitate and sustain most of the hallmarks of cancer27. In PDAC, particularly, thorough transcriptional network analyses have indicated that programs enriched with a macrophage-specific signature are the most significantly associated with poor survival in this disease7. More recently, a large-scale high-dimensional analysis of PDAC RNA sequencing data revealed that an ecotype consisting of aggressive basal-like malignant cells, tumor-promoting macrophages, and cancer-associated fibroblasts is associated with an especially poor prognosis28. In this present study, we identified CCL3, one of the most potent chemotactic cytokines for monocytes and macrophages, as a significant poorly prognostic factor in patients with metastatic PDAC. Importantly, we demonstrated that CCL3 is a robust predictive factor for using TGFβ inhibitors in this patient population, as treatment with galunisertib significantly improved the median overall survival from 3.6 to 9.2 months.
The TGFβ signaling pathway plays one of the most complex and pleiotropic roles in cancer. Tumor cells, as genetically unstable entities, have the capacity to corrupt the TGFβ signaling to promote tumor growth and metastasis. This effect is mediated by the activation of downstream intracellular pathways, both in tumor cells and in distinct mesenchymal or immune cell types of tumor microenvironment29. Over the last decade, various inhibitors of the TGFβ signaling have been developed, tested in preclinical murine models, and introduced into different clinical trials with the general assumption that tumor cells could be the most relevant target for these agents30. However, the specific cell type in which inhibiting the TGFβ pathway is most relevant for achieving a positive antitumor activity remained undetermined. In this study, we demonstrated for the first time that a significant portion of the therapeutic advantage obtained through the inhibition of TGFβ in PDAC patients could be attributable to the modulation of this signaling in macrophages rather than in tumor cells themselves. Ccl3 triggered an autocrine TGFβ signaling on TAMs but not on PDAC cells, and the inhibition of this autocrine loop suppressed the protumorigenic functions of TAMs and their secretion of factors such as LIF that acted paracrinally to sustain a mesenchymal phenotype in tumor cells. A similar mechanism has been recently identified in an in vivo screening for epigenetic mechanisms driving KRASG12D-independent PDAC recurrence. Paracrine TGFβ produced by CCL2-recruited TAMs emerged as critical signaling for bypassing KRASG12D dependency31. In this regard, the development of agents such as bispecific monoclonal antibodies, that could simultaneously bind TGFβ ligands or receptors and TAMs relevant antigens might obtain the largest advantage from this therapeutic strategy, inhibiting the TGFβ signaling where it is more significant and avoiding potential paradox effects or toxicities derived from its inhibition in different cell type in the tumor microenvironment where this effect is therapeutically irrelevant or, worse, detrimental.
LIF is the most pleiotropic member of the interleukin-6 family of cytokines. Its signaling is implicated in cancer progression, and its deregulation is observed in various solid cancers. Additionally, LIF signaling plays a role in modulating multiple immune cell types present in the tumor microenvironment32. The research group led by Joan Seoane extensively explored the role of LIF in different human tumors, revealing a significant correlation between LIF and TAMs across various tumor types, with PDAC expressing the highest level of LIF. In ovarian cancer, the LIF receptor was highly expressed within the macrophage compartment, and LIF treatment drove them to acquire immunosuppressive capacity. Targeting LIF regulated the expression of several protumoral cytokines in TAMs, including CCL2 and CCL333,34. In glioblastoma, TGFβ induced LIF, and LIF, in turn, promoted glioma-initiating cells self-renewal35. However, in models of PDAC, the research group led by Tony Hunter demonstrated LIF as a key paracrine factor from activated cancer-associated fibroblasts, acting on cancer cells, primarily by modulating cancer cell differentiation and EMT status26. Our present study contributes to this evidence by demonstrating that in our models of PDAC the main source of LIF was TAMs in response to an autocrine TGFβ signaling when cocultured with tumor cells expressing high levels of Ccl3. In this context, the inhibition of these two signaling pathways resulted in a similar modulation of EMT markers in PDAC cells. It is conceivable, however, that additional paracrine signals could meditate the crosstalk between TAMs and PDAC cells responsible for a mesenchymal/basal-like phenotype, and ongoing research is exploring these aspects.
In our present study, we elucidated these molecular and cellular mechanisms as responsible for the exceptional response of PDAC patients with high levels of CCL3 to the combination of TGFβ signaling inhibition and classical chemotherapeutic agents. However, it is conceivable that this model could be therapeutically relevant for the inhibition of different steps of the cascade that, from the overexpression of CCL3, lead to this mesenchymal/basal-like and poor prognostic ecotype. For instance, the inhibition of CCR5 with the highly selective receptor inhibitor maraviroc has been investigated in preclinical and clinical studies as a single-agent strategy36 or in combination with pembrolizumab in the treatment of MSS colorectal cancer but achieved only limited clinical activity37. Similarly, various approaches have also been developed to block LIF signaling for cancer treatment. A humanized monoclonal antibody that binds with high affinity to LIF—MSC-1 (AZD0171)—has been recently assessed in a phase I study, demonstrating a good safety profile and prolonged PFS in some patients also affected by PDAC38. Exploring these agents in the context of patients with CCL3-overexpressing tumors could have a unique rationale based on the role that CCR5 and LIF had in the model that we demonstrated in this present study.
In this present study, we established CCL3 as an intrinsic positive predictive biomarker for the inhibition of TGFβ signaling in PDAC patients. Nonetheless, we have recently identified additional stromal mechanisms of acquired resistance to this therapeutic strategy. We discovered that autotaxin, an enzyme secreted by inflammatory cancer-associated fibroblasts (CAFs) upon TGFβ signaling inhibition, activates a LPA-NF-κB axis in tumor cells that sustains their resistance to the combination of galunisertib plus gemcitabine, and that inhibition of autotaxin with the novel inhibitor IOA-289 overcomes this treatment resistance39. Overall, our previous and current studies provide a rationale to investigate IOA-289 plus a TGFβ inhibitor in combination with classic chemotherapy in CCL3-high PDAC patients, opening the path for the development of novel potential strategies to improve the therapeutic outcome of those patients selected for their high levels of CCL3. In this regard, we are currently conducting a phase 1b, open label, dose escalation study evaluating the safety and tolerability of escalating doses of IOA-289 in patients with metastatic PDAC in combination with standard chemotherapy consisting of gemcitabine and nab-paclitaxel (ClinicalTrials.gov Identifier: NCT05586516).
In recent years, several agents targeting TGFβ signaling have been discontinued in their clinical development. In September 2021, Merck announced a mutual decision with GSK to end their agreement on the development of bintrafusp alfa, a bifunctional fusion protein composed of the extracellular domain of the TGF-βRII receptor (a TGF-β “trap”) fused to a human IgG1 mAb blocking PD-L140. In February 2022, Sanofi terminated the clinical development program for SAR439459, a pan-TGFβ neutralizing antibody that inhibits all active isoforms of human TGFβ ligands41. In August 2023, Novartis discontinued the development of NIS793, a human IgG2 monoclonal antibody that binds to TGFβ1 and 2 ligands42, which was under investigation in a randomized, placebo-controlled phase 3 trial in combination with gemcitabine and nab-paclitaxel for the first-line treatment of patients with metastatic PDAC (NCT04935359). All these promising agents were, however, developed in unselected populations of patients. We believe that the current paradigm for the development of novel experimental therapeutics for PDAC should be challenged by giving growing importance to the fundamental step of early identification of those biological characteristics potentially useful as biomarkers for patient selection. By adopting this strategy, the most promising agents could be more considerately promoted to larger randomized phase 3 clinical trials, possibly explored in those biomarker-selected subpopulations of patients where their activity could have the highest rationale. Moreover, evidence emerging from these analyses would fuel a translational continuum in the laboratory, refining preclinical models to have those specific molecular characteristics to better dissect the real molecular and cellular mechanisms involved in the activity of a given agent in a biomarker-selected subtype of tumors. This present study follows this strategy as it strengthens its hypothesis about the role of CCL3 in predicting the benefit of inhibiting TGFβ signaling based on clinical evidence from the randomized H9H-MC-JBAJ study.
Novel experimental therapeutics are currently in active clinical development for the treatment of PDAC, including RO7496353, a humanized monoclonal IgG1 antibody that binds to latent TGF-β1 and inhibits TGF-β1 activation, and INCA33890, a human, Fc-silenced, IgG1-bispecific antibody that can simultaneously bind to both PD-1 and TGFβR2. Our results indicate that these agents should possibly be developed at the earliest possible lines of treatment. Since CCL3 is also a significant poor prognostic factor, evaluating these TGFβ-targeting agents in second or further lines of treatment could dilute the percentage of patients with this poor prognostic mesenchymal/basal-like phenotype, which, based on our results, could potentially derive the greatest advantage from this treatment strategy.
In conclusion, our study provides the rationale to explore TGFβ-targeting agents in the population of patients affected by PDAC with high levels of circulating CCL3 and a particularly poor prognosis due to their mesenchymal/basal-like PDAC ecotype. This strategic approach holds the potential to make a significant difference for those patients affected by one of the most challenging human solid tumors, PDAC, and characterized by the most profoundly adverse prognostic indicators.
Supplementary information
Acknowledgements
This study was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC) grant 23719 (D.M.), Italian Ministry of Health grant GR-2016-02361134 (DM), Italian Ministry of University and Research under PNRR -M4C2- I1.3 Project PE_00000019 “HEAL ITALIA” (DM), Associations “Nastro Viola” and “Voglio il Massimo” (DM). Part of the work was performed at the Laboratorio Universitario di Ricerca Medica (LURM) Research Center, University of Verona. We thank Dr. Hayley Louise Salt for data entry and administrative support.
Author contributions
D.M. and S.P. conceived and designed the studies; S.P., M.B., V.D.V. conducted experiments and acquired and analyzed data. C.F., F.S., F.F., E.S., D.M.1, S.C., C.Z., A.Q., V.M. generated data and samples. S.P., M.B., V.D.V., C.F. and F.F. analyzed data; D.M. provided financial support; D.M. and S.P. wrote the manuscript. All authors have read and approved the manuscript. The order of the co-first authors was determined by their efforts and contributions to the manuscript.
Data availability
All data supporting the graphs are provided in the Supplemental Supporting Data Values file. All other data are available upon request from the corresponding author.
Competing interests
DM received honoraria as an advisory board member or consultant from Servier, Incyte, Tahio, iOnctura, Eli Lilly, Evotec, Shire, Baxter; received institutional support for research project from Shire, Celgene, Incyte, iOnctura, Roche. The other authors have declared that no conflict of interest exists.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Silvia Pietrobono, Monica Bertolini.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-024-00742-3.
References
- 1.The Global Cancer Observatory. Pancreas, https://gco.iarc.fr/today/data/factsheets/cancers/13-Pancreas-fact-sheet.pdf (2020).
- 2.Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin.74, 12–49 (2024). [DOI] [PubMed] [Google Scholar]
- 3.Rahib, L., Wehner, M. R., Matrisian, L. M. & Nead, K. T. Estimated projection of US cancer incidence and death to 2040. JAMA Netw. Open4, e214708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamburrino, A., Piro, G., Carbone, C., Tortora, G. & Melisi, D. Mechanisms of resistance to chemotherapeutic and anti-angiogenic drugs as novel targets for pancreatic cancer therapy. Front. Pharm.4, 56 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collisson, E. A. et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med.17, 500–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffitt, R. A. et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet.47, 1168–1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Zhou, X. et al. Clinical impact of molecular subtyping of pancreatic cancer. Front. Cell Dev. Biol.9, 743908 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grunwald, B. T. et al. Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell184, 5577–5592.e5518 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Gaianigo, N., Melisi, D. & Carbone, C. EMT and treatment resistance in pancreatic cancer. Cancers9, 10.3390/cancers9090122 (2017). [DOI] [PMC free article] [PubMed]
- 11.Melisi, D. et al. Modulation of pancreatic cancer chemoresistance by inhibition of TAK1. J. Natl Cancer Inst.103, 1190–1204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melisi, D. et al. LY2109761, a novel transforming growth factor beta receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Mol. Cancer Ther.7, 829–840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melisi, D. et al. Galunisertib plus gemcitabine vs. gemcitabine for first-line treatment of patients with unresectable pancreatic cancer. Br. J. Cancer119, 1208–1214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melisi, D. et al. TGFbeta receptor inhibitor galunisertib is linked to inflammation- and remodeling-related proteins in patients with pancreatic cancer. Cancer Chemother. Pharmacol.83, 975–991 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Gueorguieva, I. et al. Population pharmacokinetics and exposure-overall survival analysis of the transforming growth factor-beta inhibitor galunisertib in patients with pancreatic cancer. Cancer Chemother. Pharmacol.10.1007/s00280-019-03931-1 (2019). [DOI] [PubMed]
- 16.Yap, T. A. et al. First-in-human phase I study of a next-generation, oral, TGFbeta Receptor 1 inhibitor, LY3200882, in patients with advanced cancer. Clin. Cancer Res.10.1158/1078-0432.CCR-21-1504 (2021). [DOI] [PMC free article] [PubMed]
- 17.Melisi, D. et al. Safety and activity of the TGFbeta receptor I kinase inhibitor galunisertib plus the anti-PD-L1 antibody durvalumab in metastatic pancreatic cancer. J Immunother. Cancer9, 10.1136/jitc-2020-002068 (2021). [DOI] [PMC free article] [PubMed]
- 18.Mantovani, A., Marchesi, F., Malesci, A., Laghi, L. & Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol.14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vayrynen, S. A. et al. Composition, spatial characteristics, and prognostic significance of myeloid cell infiltration in pancreatic cancer. Clin. Cancer Res.27, 1069–1081 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zucchetto, A. et al. Monocytes/macrophages but not T lymphocytes are the major targets of the CCL3/CCL4 chemokines produced by CD38(+)CD49d(+) chronic lymphocytic leukaemia cells. Br. J. Haematol.150, 111–113 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Menten, P., Wuyts, A. & Van Damme, J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev.13, 455–481 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Qin, R. et al. Role of chemokines in the crosstalk between tumor and tumor-associated macrophages. Clin. Exp. Med.10.1007/s10238-022-00888-z (2022). [DOI] [PMC free article] [PubMed]
- 23.Carbone, C. et al. Homeobox B9 mediates resistance to anti-VEGF therapy in colorectal cancer patients. Clin. Cancer Res.23, 4312–4322 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Wang, C. et al. Targeting a positive regulatory loop in the tumor-macrophage interaction impairs the progression of clear cell renal cell carcinoma. Cell Death Differ.28, 932–951 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melisi, D. et al. Secreted interleukin-1alpha induces a metastatic phenotype in pancreatic cancer by sustaining a constitutive activation of nuclear factor-kappaB. Mol. Cancer Res.7, 624–633 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi, Y. et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature569, 131–135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pittet, M. J., Michielin, O. & Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol.19, 402–421 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Storrs, E. P. et al. High-dimensional deconstruction of pancreatic cancer identifies tumor microenvironmental and developmental stemness features that predict survival. NPJ Precis Oncol.7, 105 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batlle, E. & Massague, J. Transforming growth factor-beta signaling in immunity and cancer. Immunity50, 924–940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabbadini, F. et al. The multifaceted role of TGF-beta in gastrointestinal tumors. Cancers13, 10.3390/cancers13163960 (2021). [DOI] [PMC free article] [PubMed]
- 31.Hou, P. et al. Tumor microenvironment remodeling enables bypass of oncogenic KRAS dependency in pancreatic cancer. Cancer Discov.10, 1058–1077 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viswanadhapalli, S., Dileep, K. V., Zhang, K. Y. J., Nair, H. B. & Vadlamudi, R. K. Targeting LIF/LIFR signaling in cancer. Genes Dis.9, 973–980 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pascual-Garcia, M. et al. LIF regulates CXCL9 in tumor-associated macrophages and prevents CD8(+) T cell tumor-infiltration impairing anti-PD1 therapy. Nat. Commun.10, 2416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hallett, R. M. et al. Therapeutic targeting of LIF overcomes macrophage-mediated immunosuppression of the local tumor microenvironment. Clin. Cancer Res.29, 791–804 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Penuelas, S. et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell15, 315–327 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Halama, N. et al. Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell29, 587–601 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Haag, G. M. et al. Pembrolizumab and maraviroc in refractory mismatch repair proficient/microsatellite-stable metastatic colorectal cancer - the PICCASSO phase I trial. Eur. J. Cancer167, 112–122 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Borazanci, E. et al. Phase I, first-in-human study of MSC-1 (AZD0171), a humanized anti-leukemia inhibitory factor monoclonal antibody, for advanced solid tumors. ESMO Open7, 100530 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietrobono, S. et al. Autotaxin secretion is a stromal mechanism of adaptive resistance to TGFbeta inhibition in pancreatic ductal adenocarcinoma. Cancer Res.10.1158/0008-5472.CAN-23-0104 (2023). [DOI] [PMC free article] [PubMed]
- 40.Gameiro, S. R., Strauss, J., Gulley, J. L. & Schlom, J. Preclinical and clinical studies of bintrafusp alfa, a novel bifunctional anti-PD-L1/TGFbetaRII agent: current status. Exp. Biol. Med.247, 1124–1134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greco, R. et al. Pan-TGFbeta inhibition by SAR439459 relieves immunosuppression and improves antitumor efficacy of PD-1 blockade. Oncoimmunology9, 1811605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer, T. M. et al. Phase I/Ib, open-label, multicenter, dose-escalation study of the anti-TGF-beta monoclonal antibody, NIS793, in combination with spartalizumab in adult patients with advanced tumors. J. Immunother. Cancer11, 10.1136/jitc-2023-007353 (2023). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the graphs are provided in the Supplemental Supporting Data Values file. All other data are available upon request from the corresponding author.