Abstract
Liquid-liquid phase separation (LLPS) in living cells provides innovative pathways for synthetic compartmentalized catalytic systems. While LLPS has been explored for enhancing enzyme catalysis, its potential application to catalytic peptides remains unexplored. Here, we demonstrate the use of coacervation, a key LLPS feature, to constrain the conformational flexibility of catalytic peptides, resulting in structured domains that enhance peptide catalysis. Using the flexible catalytic peptide P7 as a model, we induce reversible biomolecular coacervates with structured peptide domains proficient in hydrolyzing phosphate ester molecules and selectively sequestering phosphorylated proteins. Remarkably, these coacervate-based microreactors exhibit a 15,000-fold increase in catalytic efficiency compared to soluble peptides. Our findings highlight the potential of a single peptide to induce coacervate formation, selectively recruit substrates, and mediate catalysis, enabling a simple design for low-complexity, single peptide-based compartments with broad implications. Moreover, LLPS emerges as a fundamental mechanism in the evolution of chemical functions, effectively managing conformational heterogeneity in short peptides and providing valuable insights into the evolution of enzyme activity and catalysis in prebiotic chemistry.
Subject terms: Biocatalysis, NMR spectroscopy, Intrinsically disordered proteins, Phase transitions and critical phenomena
This study shows that liquid-liquid phase separation enhances the catalytic efficiency of peptides by up to 15,000-fold through the formation of peptide coacervates. These microreactors can also selectively recruit phosphorylated proteins, providing insights into the evolution of enzymatic activity.
Introduction
Compartmentalization has played a critical role in the emergence of catalysis and in the development of more complex biological systems1. For example, it is hypothesized that the first cells emerged from simple lipid vesicles, which provided a protective environment for the chemical reactions that gave rise to the first self-replicating molecules2,3. Membrane confinement, however, is not the only compartmentalization strategy available in nature. Several organisms use liquid-liquid phase separation (LLPS) to concentrate protein and nucleic acid molecules into membrane-less compartments, which enables the organization of cellular functionalities and complex biochemical reactions4–7. LLPS has been shown to promote the nucleation and assembly of actin filaments8,9, and to enhance the enzymatic efficiency of β-carboxysomes8,10 and multi-enzyme complexes11,12. These LLPS-derived biomolecular condensates, or coacervates, provide spatial organization and facilitate efficient substrate transfer between enzymes, leading to an overall increase in the efficiency of enzymatic reactions11,12.
The importance of LLPS in living systems has also fueled its use to create synthetic catalytic systems. Previous studies have used LLPS to confine enzymes in artificial cells via the use of polymers, or to confine substrates in peptide-based condensates13,14. While the impact of phase transitions on enzyme activity is complex and not fully understood, LLPS can create microenvironments that concentrate substrates and enzymes, thus increasing the local substrate concentration and influencing enzyme activity and selectivity7.
Though LLPS has been explored as a strategy to increase the catalytic rates of enzymes in artificial cells, its use for potentiating the activity of catalytic peptides remains unexplored. These catalytic peptides, which are for example widely used as chiral catalysts in organic reactions15, present substantial limitations when used in aqueous reactions as their conformational flexibility strongly limits catalytic efficiency15,16. However, the compartmentalization of catalytically active peptides in coacervates presents an enticing avenue to bolster their efficiency by confining their conformational flexibility into more densely packed environments, which may lead to the formation of structured peptide domains with improved catalytic activity. Furthermore, catalytic peptides offer an exciting opportunity as core components of coacervates, owing to their remarkable programmability and potential to simultaneously provide structural organization and catalytic functionality17. Despite this potential, the systematic investigation of coacervate designs incorporating catalytic peptides that inherently fuse condensate organization with functionality is still largely unexplored. Notably, the encapsulation of the L-dipeptide (Ser-His) within lipid-based vesicles represents the only example of compartmentalization involving catalytic peptides18, and this system was mainly explored as a model for the evolutionary competition between protocells.
While the mechanisms by which LLPS enables membraneless organelle formation are still being elucidated, previous studies have highlighted the importance of uncharged polar side chains, charged amino acids, and aromatic residues located in the repetitive low-complexity regions of intrinsically disordered proteins for coacervation1,3,4. Exploring this mechanism further, minimal model systems of self-coacervation have been developed using specifically designed peptides that adhere to these principles14,19–23. One of the major challenges in developing coacervates from simple building blocks that simultaneously govern condensate organization and mediate predictable functionality remains largely unaddressed24.
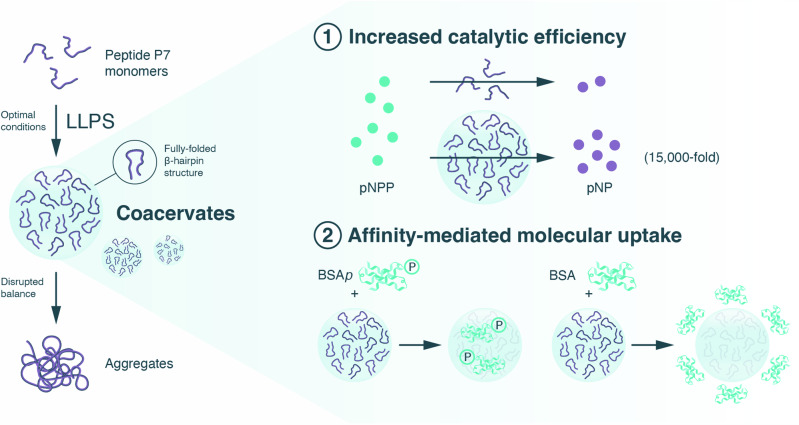
Here we show the impact of LLPS in peptide catalysis and explore LLPS as a strategy for creating single peptide-based coacervates with structured domains that result in improved catalytic function. To accomplish this, we utilized the hairpin-like catalytic peptide P7 (KVYFSIPWRVPM-NH2) as a model system, due to its ability to hydrolyze phosphate ester molecules and its high affinity for phosphorylated assemblies25 (Fig. 1). Moreover, P7 contains several residues predicted to mediate phase separation (Arg/R, Lys/K, Ser/S, Pro/P)1,3,4, enhancing its propensity for undergoing LLPS. First, we conduct a screening of peptide concentration under different environmental conditions (e.g., NaCl concentrations) and identify optimal parameters that induce formation of P7 coacervates. Then, we demonstrate that coacervation reduces the conformational flexibility of the peptide, stabilizing a fully folded β-hairpin structure. Furthermore, we show that these peptide-based coacervates effectively sequester a variety of guest molecules via charge and hydrophobic interactions, and can selectively sequester ordered entities, such as Bovine Serum Albumin (BSA), human microtubule-associated Tau protein, and Bacillus subtilis spore surface protein B (CotB), to a significantly greater extent in their phosphorylated form compared to the non-phosphorylated form. Finally, we demonstrate that these coacervate-based microreactors substantially enhance peptide catalysis, resulting in a 15,000-fold increase in catalytic efficiency of P7 for standard phosphatase substrates compared to the soluble peptide in solution. Our findings demonstrate that a single peptide can be used to induce compartmentalization, regulate substrate recruitment, and mediate catalysis, providing proof-of-concept for the establishment of low-complexity, single peptide-based compartments with a range of potential applications in areas such as drug delivery, sensing, and bioelectronic applications. As such, our findings represent a significant advance in the field of aqueous catalysis by using short oligomers such as catalytic peptides. Furthermore, the demonstration that LLPS-induced compartmentalization can manage the conformational heterogeneity in short peptides furthers our understanding of the evolution of enzymatic complexity, supporting previous suggestions that LLPS is a fundamental mechanism influencing the evolution of chemical functions and the emergence of catalysis in prebiotic chemistry.
Fig. 1. Illustration of the strategy used in this work, harnessing liquid-liquid phase separation (LLPS) to enhance peptide catalysis.
Depicted are biomolecular coacervates composed of a single peptide (P7), with densely packed peptide domains (where the fully folded ß-hairpin structure is stabilized), and which are proficient at (1) hydrolyzing phosphate ester molecules (e.g., pNPP, p-nitrophenyl phosphate with a 15,000-fold catalytic efficiency increase over soluble peptides; and 2) selectively sequestering phosphoryl assemblies (e.g., BSAp, phosphorylated Bovine Serum Albumin) through affinity interactions.
Results and discussion
LLPS can induce reversible self-coacervation of peptides with conformational flexibility
The catalytic peptide P7 (KVYFSIPWRVPM-NH2) has a primary sequence rich in residues recognized for their role in triggering LLPS, including R, K, S and P. Therefore, P7 is ideally placed to test our hypothesis that catalytic peptides can be used as a single component that merges functionality and structural organization within coacervates, through LLPS. To test this, we undertook a systematic high-throughput approach, screening how different NaCl and peptide concentrations influence P7 coacervation. The resulting samples were characterized using relative turbidity measurements and optical brightfield microscopy (Fig. 2).
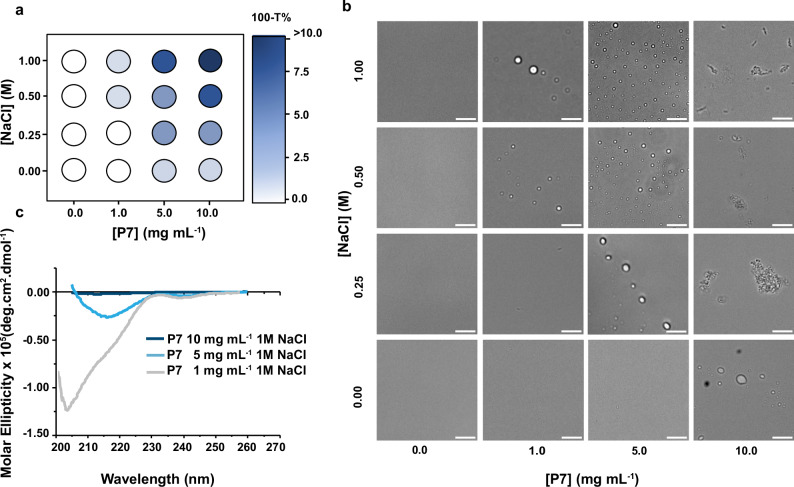
Fig. 2. Liquid-liquid phase separation of the catalytic peptide P7 leads to formation of coacervates with structured peptide domains.
a Phase diagrams illustrating the effect of different concentrations of NaCl on the liquid-liquid phase separation (LLPS) of P7, measured by the relative turbidity (100-T%). Different blue shades represent different relative turbidity levels. White circles indicate no LLPS (no relative turbidity), blue circles indicate the presence of coacervates (1 < 100-T% < 10), and the darkest blue indicates the co-existence of peptide aggregates (100-T% > 10). Source data are provided as a Source Data file (b), Bright-field microscope images depicting peptide-induced coacervation and aggregation at various NaCl and P7 concentrations. Scale bar: 10 μm. c Conformational analysis of P7 in different states using circular dichroism: soluble with low turbidity (gray line, stock P7 concentration: 1 mg mL−1), undergoing LLPS (light blue, stock P7 concentration: 5 mg mL−1), and in the aggregated form (dark blue, stock P7 concentration: 10 mg mL−1). Source data are provided as a Source Data file.
The turbidity values consistently increased with increasing concentrations of peptide and NaCl (Fig. 2a). For each peptide concentration tested, turbidity also increased as the NaCl concentration increased, suggesting peptide-driven coacervation (Fig. 2a, b, Supplementary Fig. 1). We confirmed the presence of coacervates by optical brightfield microscopy, observing increased abundance of coacervate with 1 M NaCl and at peptide concentrations from 1 to 5 mg mL−1, with the highest coacervate formation observed at 5 mg mL−1 (Fig. 2b). Interestingly, we found that higher peptide concentrations (10 mg mL−1) led to the formation of both coacervates and peptide aggregates (Fig. 2a, b). At this high peptide concentration, increasing NaCl concentrations increased the quantity and size of aggregates but did not result in coacervates formation.
These findings highlight the intricate interplay between coacervation and aggregation, with the peptide and salt concentrations serving as key regulators. The concentration of salts plays a crucial role in driving the phase separation process, underscoring the significant contribution of hydrophobic interactions to coacervate formation, which are enhanced in the presence of salt. This observation aligns with similar phenomena observed in other proteins undergoing LLPS26. However, it is important to emphasize that, at the highest concentrations of peptides and salts, the prevailing salting-out effect leads to the precipitation of peptides.
Our finding that higher peptide concentrations led to aggregate formation prompted us to test whether the aggregate to coacervate equilibrium could be tuned by altering the environmental conditions (Supplementary Information section 2.1). Indeed, when we diluted the aggregate rich solution to lower the peptide concentration from 10 to 5 mg mL−1 (while retaining 1.00 M NaCl), we observed the disappearance of aggregates and coacervate formation, highlighting the reversible nature of coacervation (Supplementary Fig. 2).
To further elucidate this process, we investigated the impact of temperature on P7 coacervation under optimal conditions (stock peptide concentration of 5 mg mL−1 in 100 mM sodium phosphate buffer, 1.00 M NaCl, pH 8), using turbidity measurements and optical brightfield microscopy (Supplementary Information section 2.2). We assessed different temperatures ranging from 23 °C–37 °C. Coacervate formation was observed within 1 h of incubation, with temperatures above 27 °C showing higher turbidity values (Supplementary Fig. 3), favoring coacervate formation compared to temperatures below 25 °C (Supplementary Fig. 4). These data are consistent with previous studies on similar systems where coacervation is primarily driven by hydrophobic interactions, which are known to be temperature-sensitive25. Similar to the effects of higher peptide concentrations, higher coacervation temperatures (>35 °C) also led to aggregation.
Furthermore, we also characterized the temporal dynamics of the coacervation process through time-course analysis. The samples were initially incubated at 27 °C to induce coacervation and subsequently maintained at room temperature (23 °C ± 2 °C) for a total duration of 6 h. This analysis revealed that while the coacervates had a larger size at 1-h coacervation, their size gradually decreased over time due to the formation of liquid bridges, leading to an increased number of coacervates (Supplementary Fig. 5). This phenomenon is characteristic of liquid-like condensates, which can fuse, deform or, as in this case, split into two coacervates. This cooling process from 27 °C to room temperature seems to impact coacervate dynamics. These temperature-dependent changes appear to contribute significantly to the observed division and size decrease of the coacervates. The observed coacervate size range was consistent with values reported in the literature, typically 1–10 μm18. This size range of the coacervates also corroborated by dynamic light scattering (DLS) (Supplementary Fig. 6). Having demonstrated that P7 can form coacervates, we then used circular dichroism (CD) analysis to examine the influence of LLPS on the conformational constraints of this flexible catalytic peptide during phase separation. The secondary structure of the peptide was analyzed in three distinct assembly states: soluble (any of the conditions 1 mg mL−1 with no salt, 5 mg mL−1 with no salt or 1 mg mL−1 with 1 M NaCl showing absence or low turbidity), undergoing LLPS (5 mg mL −1 1 M NaCl), and in the aggregate (10 mg ml−1 1 M NaCl) (Supplementary Information section 2.3). We found that the LLPS state of the peptide exhibits a CD signature, with a minimum ellipticity observed around 213–219 nm and a maximum at 232 nm (Fig. 2c), characteristic of a fully folded β-hairpin conformation (typified by a broad negative band at ~217 nm23,24). Our results suggest that LLPS induces the stabilization of the secondary structure of P7, transforming a flexible hairpin-like peptide in solution into a fully folded β-hairpin structure in the LLPS state. The CD analysis also shows that in the aggregated state, the peptide loses its conformational order. Further supporting this conformational equilibrium, NMR analysis for the soluble and undergoing LLPS assembly states corroborate these findings. The secondary structures propensities calculated from the assigned chemical shifts (Supplementary Table 1) revealed a beta-hairpin in LLPS state compared with a flexible hairpin-like peptide structure in the soluble state as showed by the presence of a turn within two random coil stretches (Supplementary Fig. 7).
Fourier-transform infrared (FTIR) spectroscopy was employed as a complementary method to identify the secondary structure of the P7 peptide in various assembly states. The FTIR spectra, shown in Supplementary Fig. 8 reveal distinct differences in the amide I band region, indicative of secondary structure variations. Deconvolution of the amide I band in the soluble state (Supplementary Fig. 9a and Supplementary Table 2) primarily indicates the presence of a random coil conformation (band at 1640 cm−1), with a minor band at 1685 cm−1 attributed to a β-turn27, reinforcing the presence of a hairpin-like peptide structure.
In the P7 coacervates, the β-sheet conformation was significantly higher, accounting for ~70% of the structure (Supplementary Fig. 9a–d and Supplementary Table 2). This was evidenced by the contributions of β-sheet bands at 1623 cm−1 and 1663 cm−1, along with a β-turn band at 1683 cm−127,28, thus reinforcing the presence of a β-hairpin structure in the LLPS state. Conversely, in the aggregated assembly state (Supplementary Fig 9c and Supplementary Table 2), there is a significant decrease in β-sheet content (down to 20%) (Supplementary Fig. 9d) compared to the P7 coacervates, with a more pronounced random coil conformation, corroborating a loss of conformational order.
These FTIR findings are consistent with the results obtained from CD and NMR analyses, indicating an enhanced content of β-sheet and higher structural organization in P7 coacervates compared to the soluble and aggregated states, Collectively, these structural investigations underscore the critical role of LLPS in modulating the conformational flexibility of the P7 peptide, stabilizing structured domains.
These findings also underscore the delicate interplay between LLPS and aggregation, which is governed by the environmental conditions utilized to induce LLPS and is contingent upon the β-hairpin conformation. Thus, our findings demonstrate that P7 undergoes a transition from a flexible hairpin-like peptide to a coacervate-forming peptide with a structured domain exhibiting a fully folded β-hairpin structure, highlighting the potential importance of such structural plasticity in the evolution of chemical function.
P7 coacervates selectively control the partitioning of guest molecules
Coacervates are dynamic compartments, with the capacity to partake in high internal mixing and rapid exchange with the external environment29–34. Thus, we explored the potential to sequester and partition specific molecules inside P7 coacervates. We first investigated the concentration ability of the P7-based coacervates for the aromatic dye molecules (9-(2-Carboxyphenyl)−6-(diethylamino)-N,N-diethyl-3H-xanthen-3-iminium chloride (rhodamine), Fluorescein 5-isothiocyanate (FITC), and 4′,6-diamidino-2-phenylindole (DAPI)) and for Green Fluorescent Protein (GFP). We assessed the partitioning of these molecules under optimized conditions, quantified their encapsulation efficiency (%EE = (c solution – c diluted phase)/csolution)20 and imaged the resulting coacervates by laser scanning confocal microscopy (Fig. 3).
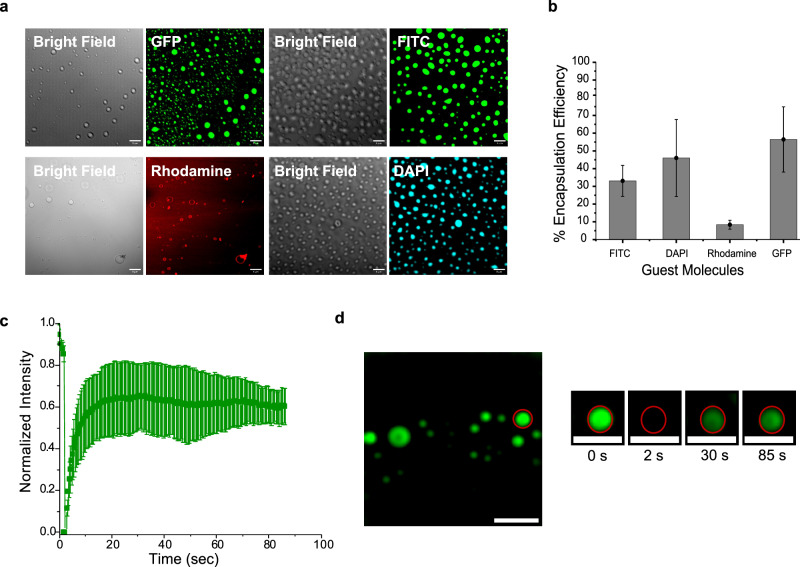
Fig. 3. Partitioning behavior of guest molecules and dynamics of P7 coacervates.
a Confocal microscopy images showing P7 coacervates encapsulating GFP, FITC, Rhodamine and DAPI. b Encapsulation efficiency analysis of the P7 coacervates calculated from fluorescence measurements of bulk solutions and diluted phases. Data are presented as mean values ± SD (n = 3) from three independent experiments. Source data are provided as a Source Data file (c), FRAP analysis of coacervates using 1 mM FITC. Data are presented as mean values ± SD (n = 3 coacervates). Source data are provided as a Source Data file (d), Representative confocal images illustrating the FRAP process in an individual coacervate at different time points. All scale bars: 10 μm.
At pH 8.0, the P7 coacervates present a net positive surface charge of +9.4 ± 3.2 mV as determined by dynamic light scattering (DLS) measurements. Local apparent pH inside coacervate droplets and the dilute phase was measured using 5-(and 6)-carboxy SNARF-1 (SNARF), a pH-sensitive indicator dye, as described by the Keating group35,36. By partitioning SNARF-1, we were able to measure the local apparent pH of both the coacervate phase and the dilute phase, which were estimated to be ~7.3 and 7.4, respectively (Supplementary Information Section 3—Supplementary Figs. 10 and 11). The slightly positive charge of P7 is predicted to promote the segregation of negatively charged molecules inside the coacervates through electrostatic interactions. Indeed, we found that the negatively charged molecule GFP exhibits a higher encapsulation efficiency than neutral dye molecules (Fig. 3a, b, Supplementary Fig. 6a). Our analysis also shows that for neutral dyes, hydrophobicity seems to be a dominant factor affecting partitioning, with encapsulation efficiency increasing as hydrophobicity decreases (Supplementary Information section 3.1, Supplementary Fig. 6b). Less hydrophobic molecules (with a lower octanol-water partition coefficient, logP), such as FITC and DAPI, preferentially partition into the coacervates, while dyes with a higher hydrophobic character (rhodamine) accumulate at the coacervate boundaries (Fig. 3a). Different ratios of the peptide P7-to-guest molecule ratio were used (Supplementary Fig. 8). Our findings reveal that for hydrophobic molecules such as FITC and Rhodamine, the partitioning is largely independent of concentration, suggesting that hydrophobic interactions, governed by the P7 peptide sequence, play a dominant role. For less hydrophobic molecules like DAPI, the partitioning efficiency increases with higher peptide-to-guest molecule ratios. This indicates that additional interactions, such as hydrogen bonding, may be involved.
We then used fluorescence recovery after photobleaching (FRAP) to characterize the dynamics of coacervates, with FITC as a representative guest molecule (Fig. 3c, d). Our results demonstrate a significant (~60%) fluorescence recovery in the coacervates, affirming their liquid-like nature.
Collectively, these observations demonstrate that P7 coacervates selectively control the dynamic partitioning of guest molecule and suggest that π-π interactions between the aromatic groups of the dye molecules and the aromatic residues of the P7 peptide together with hydrogen bonding between the peptide and less hydrophobic guest molecules mediate partitioning within the coacervates. Therefore, the functional properties of peptide coacervates are intricately tied to the encoded peptide sequence, which influences the partitioning of molecules within the coacervates.
P7 coacervates selectively recruit phosphorylated molecules
We then expanded our investigation to explore the capability of peptide-based coacervates to selectively recruit and uptake phosphorylated molecules in comparison to their non-phosphorylated counterparts. We capitalized on the fact that the P7 peptide exhibits a dissociation constant in the micromolar range specifically towards phosphorylated assemblies25. Leveraging this property, we incubated the P7 coacervates with different fluorophore-labeled phosphorylated proteins and their non-phosphorylated counterparts to explore the applicability of the P7 coacervates. Three distinct proteins were evaluated: phosphorylated bovine serum albumin (BSAp), its non-phosphorylated counterpart (BSA), phosphorylated human microtubule-associated Tau protein (Taup) along with its non-phosphorylated form (Tau), and Bacillus subtilis spore surface protein B (CotB) and its phosphorylated variant (CotBp)37. BSA is a versatile model protein to study, suitable for a broad range of applications. Tau is an intrinsically disordered protein vital to neuronal structure, which undergoes a pathological transformation when excessively phosphorylated, leading to brain dysfunction38. CotB is an abundant bacterial spore protein that is hyperphosphorylated by a kinase, CotH in the presence of a facilitator protein, CotG, the three proteins forming a phosphorylation module that patterns the spore surface layers37. BSA, Tau, and all their phosphorylated versions are intrinsically negative due to their isoelectric points and degrees of phosphorylation. In contrast, CotB is intrinsically positive (Table S3).
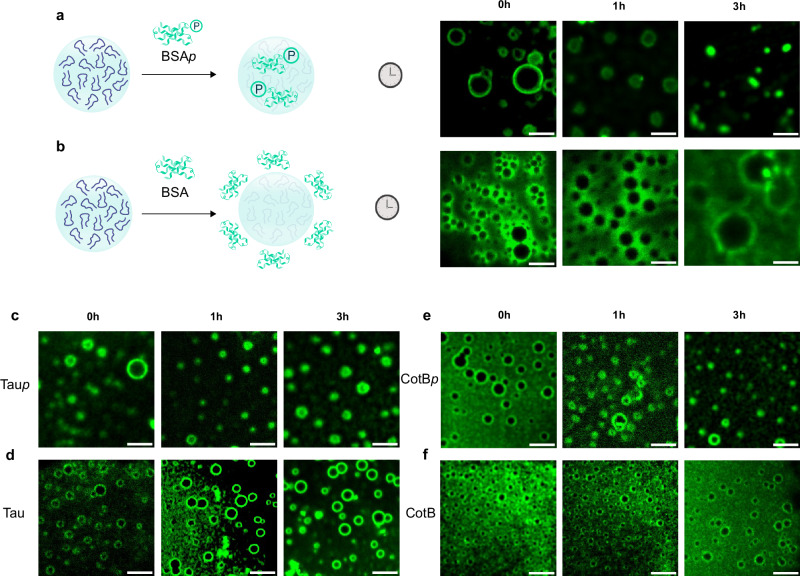
We incubated the P7 coacervates with FITC/Alexa-labeled phosphorylated proteins or FITC/Alexa-labeled non-phosphorylated proteins, monitoring their uptake by confocal microscopy over a 3-h period. Interestingly, we observed coacervate internalization of BSAp/ Taup/ CotBp (Supplementary Information section 3.2 and 3.3, Fig. 4), but not of non-phosphorylated BSA, Tau and CotB which is excluded and remains confined to the boundaries of the coacervates even after the 3-h period (Fig. 4).
Fig. 4. Affinity-mediated molecular uptake within P7 coacervates.
Time-lapse microscopy captures of the uptake of phosphorylated assemblies inside P7 coacervates. FITC-labeled BSAp and BSA, Alexa-labeled Taup and Tau, and Alexa-labeled CotBp and CotB were incubated with P7 coacervates for 3 h. Representative fluorescence confocal images highlight the molecular uptake within P7 coacervates (a) for FITC-labeled BSAp (c) Alexa-labeled Taup (e) Alexa-labeled CotBp and the exclusion of (b) BSA, (d) Tau, and (f) CotB at the boundaries of P7 coacervates. Scale bar set to 10μm. Full field of view in Supplementary Figs. 8–12.
This selective uptake is particularly noteworthy given that BSA and Tau are intrinsically negatively charged proteins, which might be expected to interact electrostatically with the positively charged coacervate surface. These findings strongly suggest that electrostatic interactions are not the primary driving force for protein sequestration within the coacervates. Instead, our results indicate that the selective binding properties of the P7 peptide sequence towards phosphorylated assemblies25 play a crucial role in the uptake mechanism, as demonstrated by the preferential uptake of phosphorylated proteins over their non-phosphorylated counterpart25. This phosphate-specific recognition suggests that it may be possible to incorporate other molecular recognition capabilities towards specific targets within the sequences of phase-separating peptides, paving the way for the creation of a class of functional biomolecular coacervates capable of mediating selective molecular uptake, resembling the processes observed in cellular functions.
Similarly, van Veldhuisen et al. have shown the selective recruitment of phosphorylated binding partners into coacervates, highlighting the role of phosphorylation in mediating specific protein-protein interactions within coacervates39.
This discriminative capacity of the P7- coacervates of selectively detecting phosphorylation levels in Tau over non-phosphorylated Tau holds promise for future diagnostic tools. The detection of phosphorylation in tauopathies is paramount for diagnosis, comprehending disease mechanisms, and developing treatments. Detection of phosphorylated CotB is a proxy for proper functioning of the CotB/CotG/CotH phosphorlytaion module and thus for the proper structural organization of the spore surface layers, which in turn is important for the environmental persistence of spores and their interaction with host cells and abiotic surfaces37.
Enhanced catalytic efficiency in peptide-based coacervates
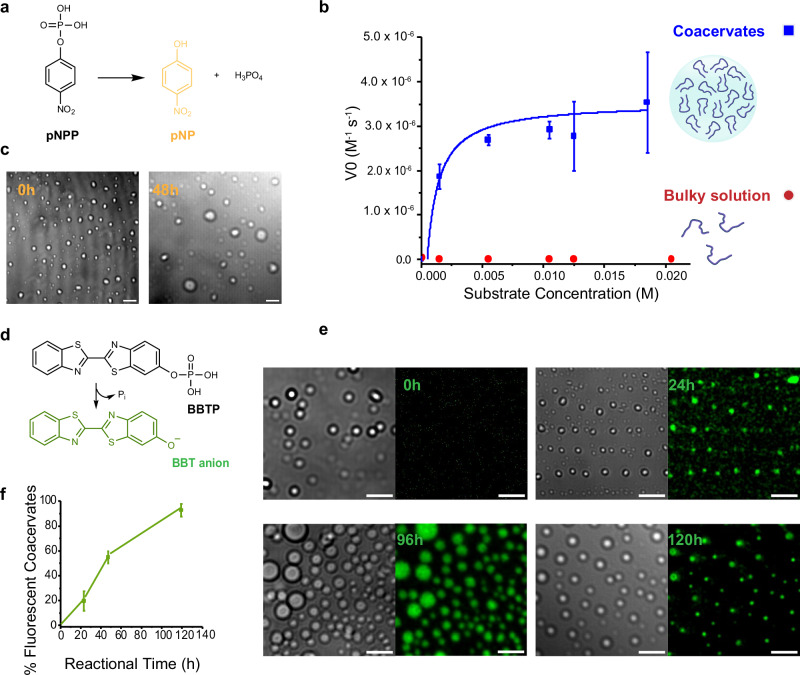
Having shown that LLPS can induce the formation of P7 coacervates and lead to the emergence of structured domains within the coacervates, we then explored the impact of constraining peptide conformation through LLPS on P7 catalytic efficiency. We first used a standard colorimetric phosphate ester hydrolysis reaction employing p-nitrophenyl phosphate (pNPP) as the substrate (Fig. 5a). We assessed the catalytic activity of P7 under LLPS conditions (5 mg mL−1 stock concentration peptide 100 mM sodium phosphate pH 8.0, 1 M NaCl), both with and without triggering coacervate formation. To induce coacervation, we combined the peptide and buffer solution at a ratio of 1:5 (peptide:buffer) and incubated the mixture at 27 °C. This resulted in the formation of coacervates. Subsequently, we introduced the substrate into the coacervation phase at 27 °C to initiate the catalytic reaction. To better estimate pNPP sequestration within the coacervates, the encapsulation efficiency of pNPP was determined, showing an efficiency ranging from 8–18%, depending on the pNPP concentration (Supplementary Figs. 21 and 22). These results indicate that pNPP sequestration inside the coacervates is not significantly higher than in the bulk solution. In the condition without triggering coacervation, we dissolved the peptide in the same buffer solution to achieve the same final concentration of 1 mg mL−1. This concentration was selected to correspond with the final peptide concentration in the coacervate samples, while maintaining the peptide in a soluble state and preventing phase separation. The substrate was then added to the resulting solution, and the catalytic reaction was also conducted at 27 °C (Fig. 5b). Without coacervation, no observable hydrolysis of pNPP occurred even after 90 h. However, under LLPS conditions, the initial rate of the reaction displayed substrate concentration dependency consistent with the Michaelis-Menten model (Fig. 5b and Supplementary Fig. 13, Supplementary Information Section 4). The catalytic parameters obtained were: kcat of (4.9 ± 0.6) x 10-3 s−1, KM of (8.2 ± 3.2) x 10-4 M and a catalytic efficiency, kcat/KM, of 5.9 ± 0.2 (Fig. 5b, Table 1). Additionally, the stability of the coacervates was evident even after 48 h of incubation (Fig. 5c). The comparison between the kinetic parameters obtained with P7 peptide-based coacervates to those previously determined for the peptide P7 in a bulk solution under catalytic optimal conditions (i.e., P7 final concentration of 50 µM in 100 mM sodium phosphate pH 8.0 with no salt, and catalytic reaction temperature 25 °C),25 show that the coacervates exhibit a 20-fold decrease in KM, a 500-fold increase in kcat, and a 15,000-fold improvement in catalytic efficiency (Table 1). Thus, catalysis is substantially improved in peptide-based coacervates.
Fig. 5. Enhanced catalytic efficiency and stability of P7 coacervates over time.
a Reaction scheme depicting the hydrolysis of p-nitrophenyl phosphate (pNPP) (b) kinetics of P7 peptide-based coacervates (in blue squares) and P7 in bulk solution (red dots) towards the substrate pNPP. The data fitting followed the Michaelis-Menten equation . Data are presented as mean values ± SD (n = 3) from three independent experiments. In the bulky solution the errors bars are not visible (c) Representative confocal microscopy images showing stability of P7-based coacervates during the hydrolysis of pNPP, at 0 h vs 48 h. Source data are provided as a Source Data file (d), Reaction scheme illustrating the hydrolysis of 2´-[2-benzothiazoyl]−6´-hydroxybenzothiazole phosphate (BBTP). e Representative confocal images showing the progression of BBTP hydrolysis over time. f Percentage of coacervates that are fluorescent due to the formation of the BBT product during BBTP hydrolysis over time. Data are presented as mean values ± SD (n = 5) of five independent samples. Source data are provided as a Source Data file. All scale bars: 10 μm.
Table 1.
Kinetic parameters of the peptide P7 under bulky solution and upon LLPS
| Catalyst | kcat (s−1) | KM (M) | kcat / KM (M−1 s−1) | Ref. |
|---|---|---|---|---|
| P7-based peptide coacervates | (4.9 ± 0.4) x 10-3 | (8.2 ± 3.2) x 10-4 | (5.9 ± 0.2) | This work |
| P7 peptide in solution | (1.0 ± 0.0) x 10-5 | (1.4 ± 0.4) x 10-2 | (4.0 ± 0.3) x 10-4 | 25 |
To confirm that the phosphate ester hydrolysis reaction occurred within the P7 coacervates, we employed a different substrate, 2´-[2-benzothiazoyl]-6´-hydroxybenzothiazole phosphate (BBTP), which, upon cleavage by a phosphatase biocatalyst, produces inorganic phosphate and the fluorescent alcohol, 2´-[2-benzothiazoyl]-6´-hydroxybenzothiazole (BBT) (Fig. 5d). Analysis of this reaction using confocal microscopy showed that the number of coacervates exhibiting fluorescence increases over time, reaching a maximum after 120 h of reaction, further highlighting the ability of peptide-based coacervates to hydrolyze phosphate ester molecules (Fig. 5e, f). Remarkably, these coacervates remain stably dispersed in the catalytic environment for the entire 120-h duration of the reaction (Fig. 5e and Supplementary Fig. 14).
Collectively, these data demonstrate that LLPS-mediated compartmentalization can enhance the catalytic efficiency of P7, as a result of the conformational stabilization of the peptide within coacervates. Through precise control of peptide conformation via LLPS, we have now achieved catalytic efficiencies that rival those observed in phosphatase enzymes40. Furthermore, unlike typical coacervates that often require surrounding membrane layers to prevent droplet coalescence and maintain stability for extended periods41, the P7 peptide-based coacervates demonstrate exceptional stability without the need for such additional structures. Our results demonstrate that peptide-based coacervates can not only serve as efficient microreactors but also undergo in situ conversions into condensed structures with improved stability.
We present a pioneering proof-of-concept for a unique class of coacervate protocells, in which a single phase-separating catalytic peptide can be used to induce coacervate formation, selectively recruit substrates and mediate catalysis. Our findings also highlight the significant impact of LLPS on modulating the flexibility of the peptides, with the ability of LLPS to facilitate the formation of crowded structured domains within coacervates playing a crucial role in enhancing enzymatic efficiency through precise control over the catalytic structure. By effectively managing the conformational heterogeneity in short peptides, LLPS emerges as a fundamental mechanism influencing the evolution of chemical functions, providing valuable insights into the emergence of catalysis in prebiotic chemistry and the origin of life. In fact, enzymes have been hypothesized to have evolved from much simpler functional precursors, such as peptides42, and coacervates have been proposed as potentially associated with the origin of life1,43. Coacervates provided mechanisms for shielding oligomers and concentrating biomolecules from bulk solutions, thus facilitating a variety of processes important for the evolution of catalytic function.34,43–45 Therefore, coacervate-based compartmentalization offers an attractive solution to the challenge of explaining how simple chemical reactions could have given rise to the complex biochemical processes that underpin life.
Moreover, our findings pave the way of the encoded peptide sequences in regulating the function of coacervates. In our model P7, the peptide sequence not only enables catalysis but also regulates the selective partitioning and uptake of target molecules into coacervates through affinity-based processes. The integration of LLPS with the affinity of P7 towards assemblies bearing phosphoryl groups represents an alternative approach for the specific sequestration of phosphoryl-containing proteins within condensates. This substrate specificity, combined with the microreactor functionality of the coacervates, eliminates the need for surrounding membranes while still facilitating solute uptake. This innovative feature not only simplifies the design of coacervates but also allows for precise control over solute uptake, laying the foundation for the development of low-complexity, peptide-based compartments with the ability to target other molecules of interest.
This work introduces a concept of designing complexity within protocells by engineering the sequences of phase-separating catalytic peptides, leading to fundamental changes in coacervate-based compartments. The integration of molecular-level complexity and functionality into the design of coacervates holds far-reaching implications, particularly in diagnostics requiring the recognition of specific molecules and precise responses. Looking ahead, the creation of dual-function coacervates capable of substrate-specific recognition and catalysis holds immense potential in various applications, including in medicine and biosensing. The selection of the target biomolecule to be sequestered can be tailored to the specific objectives of materials-based coacervates, offering versatility and adaptability. Leveraging the unique properties of these coacervates will drive transformative advancements in diverse fields.
In summary, this study not only demonstrates the remarkable capabilities of coacervate protocells but also provides valuable insights into LLPS-driven catalysis, shedding light on the fundamental processes underlying the evolution of chemical functions and the origin of life. The findings presented herein have potential broad applications in the development of innovative biotechnological solutions and transformative breakthroughs in healthcare and bioengineering.
Methods
Trifluoroacetic acid (TFA) removal of P7 peptide
Caslo’s P7 peptide was subjected to TFA removal using the following procedure: the peptide was dissolved in a 10 mM HCl solution, incubated for 30 min, and subsequently lyophilized using a LabConco Freeze-Dryer. This process was repeated twice to ensure efficient removal of TFA. The final TFA content was verified to be below 1%.
Coacervation
The lyophilized powder of P7 peptide was dissolved in 1 mL of distilled water and vortexed until a transparent solution with the desired stock concentration was obtained. The stock concentrations of P7 peptide used were 1 mg mL−1, 5 mg mL−1, and 10 mg mL−1.
Coacervation was induced by mixing 60 μL of the P7 stock peptide solution 5 mg mL−1 (or 1 mg mL−1 and 10 mg mL−1) with 240 μL of a 100 mM phosphate buffer solution containing 1 M NaCl at pH 8 (or any other buffer used in the phase diagram) in a 1:5 ratio of peptide to buffer. The samples were incubated for 1 h at 27 °C ( ± 1 °C) and then left at room temperature (23 °C ± 2 °C). For brightfield optical microscopy, 10 μL of the sample was analyzed using a 63x oil immersion lens on a glass slide covered by a functionalized coverslip with 1% Pluronics.
Turbidity measurements
Turbidity measurements were conducted using a Tecan Infinite M Nano instrument. Absorbance at 600 nm was recorded every 30 s for a total duration of 50 min. All measurements were carried out at a temperature of 27 °C ( ± 1 °C). The relative turbidity values reported represent triplicate measurements and were calculated using the formula , where A600 is the absorbance at 600 nm46 by using Microsoft Excel 2019 HSO. A well containing an equivalent volume of buffer solution served as the blank46.
Coacervates imaging
For imaging samples from the phase diagrams, optical brightfield microscopy was used with a Leica DM6000B upright microscope and for the for the remaining experiments, a Zeiss LSM 880 point laser scanning confocal microscope was used.
Cover slides functionalization
The cover slides were functionalized following the protocol by Pereira, PM et al.47. Firstly, the cover slides were immersed in absolute ethanol, swirled repeatedly, and then rinsed three times with MilliQ water. This washing procedure was repeated five times. Subsequently, the cover slides were washed with absolute anhydrous acetone, rinsed, and washed three times with MilliQ water. This washing step was also repeated five times. The cover slides were then immersed in 1 M KOH and sonicated for 45 min. Afterward, they were rinsed ten times with MilliQ water and immersed in a 1% (w/v) solution of pluronics for 1 h. Finally, the cover slides were rinsed ten times with MilliQ water to remove any residual solution.
Optical brightfield microscopy
For imaging samples from the phase diagrams, a Leica DM6000B upright microscope equipped with an Andor iXon 885 EMCCD camera was used. The MetaMorph V5.8 software was employed to control the microscope, and the images were acquired using a 63 × 1.4 NA oil immersion phase-contrast objective (Leica). Image analysis was conducted using ImageJ/FIJI 1.54 f48.
Laser scanning confocal microscopy
Confocal images were acquired using a Zeiss LSM 880 point scanning confocal microscope equipped with photomultiplier tube detectors (PMTs) and a gallium arsenide phosphide (GaAsP) detector. To visualize individual coacervates, 10 μL of the coacervate mixture was added to Pluronic-functionalized glass slides. Coacervates were imaged using a 63x Plan-Apochromat 1.4 NA DIC oil immersion objective (Zeiss) with laser lines at 405 nm, 488 nm, and 561 nm, and appropriate spectral separation for DAPI, FITC, GFP, and rhodamine§ respectively. The Zeiss Zen 2.3 (black edition) software was used to control the microscope, adjust spectral detection for the excitation/emission of the fluorophores used (following manufactures recommendations). Imaging was performed with 2% laser intensity for all lasers and a gain between 500 and 650. Image analysis was conducted using Zeiss Zen 2.3 (black edition) software and ImageJ/FIJI 1.54 f48.
Circular dichroism analysis
CD was employed to investigate the secondary structure of the P7 peptide in different forms: soluble, undergoing liquid-liquid phase separation (LLPS), and as peptide aggregates. The following samples were analyzed: (1) P7 dissolved peptide at a stock concentration of 1 mg mL−1 in 100 mM sodium phosphate buffer with 1 M NaCl (P7 completely soluble), (2) stock P7 peptide at a concentration of 5 mg mL−1 in 100 mM sodium phosphate buffer with 1 M NaCl at pH 8.0 (undergoing LLPS), and (3) P7 peptide at a stock concentration of 10 mg mL−1 in 100 mM sodium phosphate buffer at pH 8.0 with 1 M NaCl (peptide aggregates). All samples were prepared following the coacervation protocol.
For the CD measurements, quartz cuvettes with a path length of 0.1 cm were used for the P7 peptide stock solution at 1 mg mL−1, while cuvettes with a path length of 0.01 cm were used for the 5 mg mL−1 and 10 mg mL−1 P7 peptide stock solutions. Far-UV CD spectra in the range of 260 to 205–200 nm were acquired using a J-815 CD spectrometer from Jasco at a temperature of 25 °C. The measurements were performed in the appropriate cuvette under a nitrogen flow, with a scanning interval of 1 nm, a spectral bandwidth of 2 nm, and a scanning velocity of 50 nm/min. Each spectrum represents an average of three scans, and buffer signals were subtracted from each scan to obtain the final spectra, and the ellipticity was calculated according to literature49 by using in Microsoft Excel 2019 HSO.
NMR samples preparation and experiments
For NMR studies the P7 peptide samples were prepared according to the coacervation protocol described as in supplementary information note 2 in soluble and undergoing LLPS states. The conditions prepared were P7 peptide in a stock concentration of 5 mg mL−1 in 100 mM sodium phosphate buffer with no salt (soluble) and 1 M of NaCl at pH 8.0 (undergoing LLPS) in 92%H2O/8%D2O.
NMR experiments were acquired either at FCT-NOVA or ITQB in Bruker Avance Neo 500 MHz spectrometers equipped with a 5 mM triple resonance Prodigy cryoprobe (TCI). 2D 1H TOCSY (40 ms, dipsi2esgpph)50,51, 2D 1H ROESY (250 ms, roesyesgpph)51,52 and 2D 1H,13C HSQC (hsqcetgpsisp2)53–55 spectra were acquired at 23 °C with a spectral width of 13 ppm for proton and 165 ppm for carbon, processed using TopSpin 4.0.7 (Bruker Biospin) and analyzed with Poky56. The proton chemical shifts are referenced to 2,2-dimethyl-2-silapentane-5-sulfonate at 0 ppm by using the residual water signal57 the carbon shifts were calibrated through indirect referencing58. Secondary structure elements were determined with the CSI 3.0 web server59.
FTIR spectroscopy
Attenuated total reflection (ATR)-FTIR measurements were performed using a zinc-selenide crystal with a 45° angle of incidence. Samples of P7 peptide were prepared according to the coacervation protocol at three concentrations: 1 mg mL−1 in 100 mM sodium phosphate buffer pH 8.0 without salt (soluble), 5 mg mL−1 in the same buffer with 1 M NaCl (undergoing LLPS), and 10 mg mL−1 in the same buffer with 1 M NaCl. 10 µL of each sample was analyzed. ATR-FTIR spectra were recorded in the amide I band (1550–1720 cm−1) with a spectral resolution of 4 cm−1 using a Bruker INVENIO R spectrometer equipped with a DTGS detector. Measurements were conducted at room temperature while purging the sample compartment with dry air. Each spectrum comprised 64 scans, was baseline corrected, and the amide I band was deconvoluted using OriginPro 2024b software.
Dynamic light scattering
The size of coacervates and their zeta potential was measured by dynamic laser scattering (DLS) (Malvern Zeta-sizer Nano ZS). For size measurements, P7 samples with the different assembly states (soluble with low turbidity, undergoing LLPS and aggregated state) were induced for a final volume of 2 mL. Furthermore, 500 uL of each sample was transferred to 1.5 mL SARSTEDT disposable cuvettes and proceeded to size measurements. Lastly, for the measurement of the zeta potential, we transferred 700 uL from the stock solutions to ZETASIZER NANO SERIES disposable folded capillary cells. The experiments were performed in triplicates.
Partitioning experiments
To investigate the encapsulation of guest molecules within the coacervates, we performed partitioning experiments. The guest molecules used were DAPI (4’,6-diamidino-2-phenylindole), FITC (3’,6’-dihydroxy-6-isothiocyanatospiro[2-benzofuran-3,9’-xanthene]-1-one), GFP (Green Fluorescent Protein), and TMR (tetramethylrhodamine). Firstly, 5 mg of the lyophilized P7 peptide powder was dissolved in 1 mL of distilled water and vortexed until a transparent solution was obtained. Coacervation was induced by mixing 60 μL of the P7 peptide solution with 240 μL of 100 mM phosphate buffer solution containing 1 M NaCl at pH 8.0. This mixture was incubated for 5 min at 27 °C (±1 °C) in the microplate reader INFINITE M NANO + TECAN, resulting in the formation of milky-colored peptide coacervates. Next, 25 μL of the guest molecule solution (1 mM fluorophores and 0.1 mM GFP) was added to the coacervate mixture in a 1:12 ratio, resulting in a final volume of 325 μL. Different ratios of 1:5 and 1:20 were also tested. The samples were then incubated for 1 h at 27 °C (±1 °C). Then samples of partitioning were centrifuged at room temperature for 30 min, and 10 μL of the supernatant was mixed with 190 μL of 100 mM sodium phosphate buffer with 1 M NaCl at pH 8.0 for fluorescence measurement. The gain of the guest molecules was measured using the microplate reader INFINITE M NANO + TECAN. The guest molecule samples were diluted (1:20) into 100 mM sodium phosphate buffer with 1 M NaCl at pH 8.0, resulting in a final volume of 200 μL. The percentage of encapsulation (%EE) was calculated in Microsoft Excel 2019 HSO by using the following equation, where CT represents the total concentration of the fluorophore and Csup represents the concentration in the diluted phase: .
Fluorescence recovery after photobleaching (FRAP)
FRAP experiments were performed on a Zeiss LSM 880 confocal microscope. Sample preparation was performed as described above in the “Confocal Microscopy” section of supplementary information note 2. For fluorescence recovery dynamics assessment, a pre-bleached image of the coacervates was acquired using 488 nm laser line excitation and emission collected with a GaAsP detector. Subsequently, the target coacervates were bleached using the 488 nm laser line at maximum power for 5 s. The subsequent recovery of the bleached area was recorded using the same acquisition parameters as the pre-bleached image, with a total recovery time of 87 s. The final FRAP recovery curves represent the average of three recovery curves collected from n = 3 separate droplets. Correction for photobleaching, normalization, and averaging were performed using ImageJ/FIJI 1.54 f60.
Affinity-mediated molecular uptake within P7 coacervates
Labeling of the phosphorylated and non-phosphorylated proteins
All the proteins were commercially available except for CotBp and CotB, which were produced in Escherichia coli and purified by chromatographic techniques according to previous work37. BSAp and BSA were labeled with FITC by using the SIGMA FluoroTagTM FITC conjugation kit according to the supplier’s instructions. The sample of FITC-labeled BSAp was quantified using the QuantiProTM BCA Assay Kit, following the manufacturer’s instructions. Taup, Tau, CotBp and CotB were fluorescently labeled using the Alexa 488 Microscale Protein Labeling Kit and further quantified also according to the kit supplier’s instructions.
Binding experiments
For the binding experiments, P7 peptide coacervation was induced according to the coacervation protocol, using a 100 mM phosphate buffer solution containing 1 M NaCl at pH 8.0. After the designated incubation time, the P7 coacervates were transferred to IBIDI µ-Slide 8-well glass bottom previously coated with 1% (w/v) solution of pluronics. A solution of 3 µM BSAp-FITC, Taup-Alexa, CotBp- Alexa or 10 µM BSA-FITC, Tau-Alexa, CotB-Alexa (control samples) was added to the coacervate solution, with a volume of 25 μL. The binding experiments were conducted at room temperature for a total of 3 h.
Visualization of binding and internalization
To observe the binding and internalization of the labeled-phosphorylated and non-phosphorylated proteins into the P7 peptide coacervates, each well of the IBIDI µ-Slide 8-well glass bottom, containing the P7 peptide coacervates, was imaged using a Zeiss LSM 800 confocal microscope with Airyscan, equipped with a 63x oil immersion lens and a FITC and Alexa filter. Imaging was performed to capture fluorescence signals indicating the presence of the labeled molecules within the coacervates for a total period of 3 h. All acquired images were subsequently processed using the ImageJ/FIJI 1.54 f software for further analysis and visualization.
Kinetic experiments of P7 coacervates towards the p-nitrophenyl phosphate (pNPP)
The kinetics of P7 coacervates towards p-nitrophenyl phosphate (pNPP) were investigated using the following protocol. To prepare the P7 coacervates encapsulated with pNPP, a final volume of 1 mL was used, following the partitioning protocol. The peptide stock solution of 5 mg mL−1 and the coacervation condition of 100 mM sodium phosphate buffer with 1 M NaCl at pH 8.0 were utilized. In a cuvette, 186.4 μL of the 5 mg mL−1 P7 peptide solution was mixed with 738.46 μL of buffer and incubated for 5 min at 27 °C ( ± 1 °C) to induce coacervation. Subsequently, 76.923 μL of pNPP substrate at different concentrations (1, 5, 10, 12, 18 mM) were added to a final volume of 1 mL, maintaining the ratios specified in the partitioning protocol. The samples were then incubated for 1 h at 27 °C ( ± 1 °C). The formation of p-nitrophenol (pNP) was monitored by measuring the absorbance at 405 nm. The analysis of kinetic experiment data conducted with P7 coacervates involved two key corrections: (1) to account for the turbidity interference caused by coacervates, the absorbances of P7 coacervates without pNPP were subtracted from the experimental readings, and (2) to account for the autohydrolysis of the substrate in bulk solution, the absorbances of pNPP over time (without coacervates) were also subtracted from the experimental readings. These corrections allowed for a more accurate assessment of the catalytic activity of P7 coacervates by isolating the pNPP hydrolysis specifically caused by the peptide-based coacervates.
The initial velocity rate of each substrate concentration catalyzed by the P7 coacervates was determined by analyzing the linear phase of pNP formation over time (90 min). The kinetic parameters, namely Vmax and KM, were determined using Origin 2018 software by fitting the data to the Michaelis-Menten equation . The rate of catalysis was calculated using the equation The experiments were performed in triplicates, and the obtained kinetic parameters were compared with those reported by Pina et al.25 for P7 in bulky solution under optimal conditions.
Kinetic experiments of P7 coacervates towards the 2´-[2-benzothiazoyl]-6´-hydroxybenzothiazole phosphate (BBTP)
For the kinetic experiments of P7 coacervates with BBTP substrate, the commercially available AttoPhos® AP Fluorescent Substrate System from Promega was used. A 1 mM solution of AttoPhos was prepared by dissolving the contents of the attophos vial (36 mg) in 60 mL of AttoPhos® Buffer. To prepare the P7 coacervates encapsulated with BBTP substrate, the partitioning protocol was followed with a final volume of 1 mL. The peptide stock solution of 5 mg mL-1 and the coacervation condition of 100 mM sodium phosphate buffer with 1 M NaCl at pH 8.0 were utilized. In a cuvette, 186.4 μL of the 5 mg mL-1 P7 peptide solution was mixed with 738.46 μL of 100 mM sodium phosphate buffer with 1 M NaCl at pH 8.0, followed by incubation for 5 min at 27 °C ( ± 1 °C) to induce coacervation. Subsequently, 76.923 μL of the BBTP substrate at a concentration of 1 mM was added, and the mixture was further incubated for 1 h at 27 °C ( ± 1 °C). The formation of the alcohol product, 2´-[2-benzothiazoyl]-6´-hydroxybenzothiazole (BBT anion), was monitored after 5 d (120 h) using confocal microscopy with the FITC filter. The fluorescence gain was set to 650, and the PMT gain was set to 500. The laser intensity was maintained at 2.0 for all experiments.
All acquired images were processed using the ImageJ/FIJI 1.54 f software for analysis and quantification.
Statistics and reproducibility
The turbidity measurements of P7 samples varying P7 and salt concentration were repeated at least two times with similar results, and similar coacervates were obtained within each condition (Fig. 2). Guest molecule encapsulation experiments (Fig. 3b), FRAP analysis (Fig. 3c) and kinetic assays towards the substrate pNPP (Fig. 5b) were repeated independently three times with similar results.
Microscopy images shown in peptide-induced coacervation experiments (Fig. 2), guest molecule encapsulation (Fig. 3a), FRAP analysis (Fig. 3d), molecular uptake of phosphorylated and non-phosphorylated proteins within P7 coacervates (Fig. 4), and stability of the P7 coacervates during hydrolysis (Fig. 5c) and regarding the BBTP hydrolysis (Fig. 5e, f) are representative of at least three independent analyses with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
The authors thank Dr. José Rino from Instituto de Medicina Moleular da Universidade de Lisboa for the help with FRAP experiment design and analysis and for helpful discussions. D.Q.P.R., S.P., A.P.R., J.C., P.M.P., A. S. P. acknowledge support from Fundação para a Ciência e Tecnologia (FCT), through MOSTMICRO-ITQB R&D Unit (UIDB/04612/2020, UIDP/04612/2020), LS4FUTURE Associated Laboratory (LA/P/0087/2020), 2021.01283.CEECIND/CP1657/CT0004 for A.S.P., UI/BD/154577/2022 for J.C. and PRT/BD/154753/2023 for D.Q.P.R. This work was partially supported by PPBI - Portuguese Platform of BioImaging (PPBI-POCI-01-0145-FEDER-022122) co-funded by national funds from OE - “Orçamento de Estado” and by european funds from FEDER - “Fundo Europeu de Desenvolvimento Regional. P.M.P acknowledges support from FCT project grant (PTDC/BIA MIC/2422/2020), a La Caixa Junior Leader Fellowship (LCF/BQ/PI20/11760012) financed by”la Caixa” Foundation (ID 100010434) and by European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 847648, and a Maratona da Saúde award. L.M. acknowledge the support from FCT in the scope of 2021.02185.CEECIND/CP1657/CT0008 and the projects (i) UIDP/04378/2020 and UIDB/04378/2020 (Research Unit on Applied Molecular Biosciences—UCIBIO) and (ii) LA/P/0140/2020 (Associate Laboratory Institute for Health and Bioeconomy—i4HB). The NMR spectrometers at CERMAX, ITQB-NOVA, Oeiras are funded by FCT through project AAC 01/SAICT/2016, while those from FCT-NOVA are part of the National NMR Network and are supported by FCT (ROTEIRO/0031/2013 and PINFRA/22161/2016) cofounded by FEDER through COMPETE 2020, POCI, PORL and FCT through PIDDAC.
Author contributions
A.S.P. conceived the idea. D.Q.P.R., L.M., P.M.P. and A.S.P. designed the experiments. D.Q.P.R., S.P. and A.P.R. performed the microscopy experiments. S.P. performed the CD experiments. D.Q.P.R. and P.M.P performed the microscopy analysis. D.Q.P.R. and L.M performed the NMR experiments and the respective analysis. M.S. and A.O.H performed the experiments related with protein expression and purification. D.Q.P.R., J.C. and A.S.P. performed the catalytic experiments and the respective data analysis. All authors discussed the results and interpretations. D.Q.P.R., S.V., P.M.P. and A.S.P. wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Vladimir Uversky and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data are available from the corresponding author. The data generated in this study are provided in the Supplementary Information/Source Data file. Source data are provided with this paper as a Source Data file. Source data are provided with this paper.
Competing interests
D.Q.P.R., J.C., P.M.P., L.M., and A.S.P. are named as inventors on a pending patent (PT119800) filed by Universidade NOVA de Lisboa describing the catalytic peptide-based coacervates mentioned in this manuscript. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/22/2024
A Correction to this paper has been published: 10.1038/s41467-024-54617-z
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53699-z.
References
- 1.Mann, S. Life as a nanoscale phenomenon. Angew. Chem. Int. Ed.47, 5306–5320 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Schrum, J. P., Zhu, T. F. & Szostak, J. W. The origins of cellular life. Cold Spring Harb. Perspect. Biol. 2, a002212 (2010). [DOI] [PMC free article] [PubMed]
- 3.Szostak, J. W., Bartel, D. P. & Luisi, P. L. Synthesizing life. Nature409, 387–390 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Rumyantsev, A. M., Jackson, N. E. & de Pablo, J. J. Polyelectrolyte complex coacervates: recent developments and new frontiers. Ann. Rev. Condens. Matter Phys.12, 155–176 (2021). [Google Scholar]
- 5.Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular Biochem. Nat. Rev. Mol. Cell Biol.18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol.28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bracha, D., Walls, M. T. & Brangwynne, C. P. Probing and engineering liquid-phase organelles. Nat. Biotechnol.37, 1435–1445 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su, X. et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, H. et al. Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature566, 131–135 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Peeples, W. & Rosen, M. K. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol.17, 693–702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, Y. et al. Phase-separated multienzyme compartmentalization for terpene biosynthesis in a prokaryote. Angew. Chem. Int. Ed.61, e202203909 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Song, S. et al. Engineering transient dynamics of artificial cells by stochastic distribution of enzymes. Nat. Commun.12, 6897 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbas, M., Lipiński, W. P., Nakashima, K. K., Huck, W. T. S. & Spruijt, E. A short peptide synthon for liquid–liquid phase separation. Nat. Chem.13, 1046–1054 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Metrano, A. J. & Miller, S. J. Peptide-based catalysts reach the outer sphere through remote desymmetrization and atroposelectivity. Acc. Chem. Res.52, 199–215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho, S., Peralta Reis, D. Q., Pereira, S. V., Kalafatovic, D. & Pina, A. S. Catalytic peptides: the challenge between simplicity and functionality. Isr. J. Chem.62, e202200029 (2022). [Google Scholar]
- 17.Abbas, M., Lipiński, W. P., Wang, J. & Spruijt, E. Peptide-based coacervates as biomimetic protocells. Chem. Soc. Rev.50, 3690–3705 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Adamala, K. & Szostak, J. W. Competition between model protocells driven by an encapsulated catalyst. Nat. Chem.5, 495–501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun, Y. et al. Phase-separating peptides for direct cytosolic delivery and redox-activated release of macromolecular therapeutics. Nat. Chem.14, 274–283 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Baruch Leshem, A. et al. Biomolecular condensates formed by designer minimalistic peptides. Nat. Commun.14, 421 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang, Y. et al. Prediction and characterization of liquid-liquid phase separation of minimalistic peptides. Cell Rep. Phys. Sci.2, 100579 (2021). [Google Scholar]
- 22.Fisher, R. S. & Elbaum-Garfinkle, S. Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat. Commun.11, 4628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sementa, D. et al. Sequence-tunable phase behavior and intrinsic fluorescence in dynamically interacting peptides. Angew. Chem. Int. Ed.62, e202311479 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Dai, Y. et al. Programmable synthetic biomolecular condensates for cellular control. Nat. Chem. Biol.19, 518–528 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pina, A. S. et al. Discovery of phosphotyrosine-binding oligopeptides with supramolecular target selectivity. Chem. Sci.13, 210–217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krainer, G. et al. Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun.12, 1085 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagińska, K., Makowska, J., Wiczk, W., Kasprzykowski, F. & ChmurzyńSKI, L. Conformational studies of alanine-rich peptide using CD and FTIR spectroscopy. J. Pept. Sci.14, 283–289 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Barreto, M. S. C., Elzinga, E. J. & Alleoni, L. R. F. The molecular insights into protein adsorption on hematite surface disclosed by in-situ ATR-FTIR/2D-COS study. Sci. Rep.10, 13441 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Booth, R., Qiao, Y., Li, M. & Mann, S. Spatial positioning and chemical coupling in coacervate-in-proteinosome protocells. Angew. Chem. Int. Ed.58, 9120–9124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Küchler, A., Yoshimoto, M., Luginbühl, S., Mavelli, F. & Walde, P. Enzymatic reactions in confined environments. Nat. Nanotechnol.11, 409–420 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Schoonen, L. & van Hest, J. C. M. Compartmentalization approaches in soft matter science: from nanoreactor development to organelle mimics. Adv. Mater.28, 1109–1128 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Love, C. et al. Reversible pH-responsive coacervate formation in lipid vesicles activates dormant enzymatic reactions. Angew. Chem. Int. Ed.59, 5950–5957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garenne, D. et al. Sequestration of proteins by fatty acid coacervates for their encapsulation within vesicles. Angew. Chem. Int. Ed.55, 13475–13479 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Hammes, G. G. & Wu, C.-W. Kinetics of allosteric enzymes. Ann. Rev. Biophys. Bioeng.3, 1–33 (1974). [DOI] [PubMed] [Google Scholar]
- 35.Cakmak, F. P., Choi, S., Meyer, M. O., Bevilacqua, P. C. & Keating, C. D. Prebiotically-relevant low polyion multivalency can improve functionality of membraneless compartments. Nat. Commun.11, 5949 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi, S., Knoerdel, A. R., Sing, C. E. & Keating, C. D. Effect of polypeptide complex coacervate microenvironment on protonation of a guest molecule. J. Phys. Chem. B127, 5978–5991 (2023). [DOI] [PubMed] [Google Scholar]
- 37.Freitas, C. et al. A protein phosphorylation module patterns the Bacillus subtilis spore outer coat. Mol. Microbiol.114, 934–951 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansson, O. Biomarkers for neurodegenerative diseases. Nat. Med.27, 954–963 (2021). [DOI] [PubMed] [Google Scholar]
- 39.van Veldhuisen, T. W. et al. Enzymatic regulation of protein–protein interactions in artificial cells. Adv. Mater.35, 2300947 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Pabis, A., Duarte, F. & Kamerlin, S. C. L. Promiscuity in the enzymatic catalysis of phosphate and sulfate transfer. Biochem55, 3061–3081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao, N. & Mann, S. Membranized coacervate microdroplets: from versatile protocell models to cytomimetic materials. Acc. Chem. Res.56, 297–307 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frenkel-Pinter, M., Samanta, M., Ashkenasy, G. & Leman, L. J. Prebiotic peptides: molecular hubs in the origin of life. Chem. Rev.120, 4707–4765 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Koga, S., Williams, D. S., Perriman, A. W. & Mann, S. Peptide–nucleotide microdroplets as a step towards a membrane-free protocell model. Nat. Chem.3, 720–724 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Crowe, C. D. & Keating, C. D. Liquid–liquid phase separation in artificial cells. Interface Focus8, 20180032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keating, C. D. Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc. Chem. Res.45, 2114–2124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakashima, K. K., André, A. A. M. & Spruijt, E. in Methods in Enzymology Vol. 646 (ed Christine D. Keating) 353–389 (Academic Press, 2021). [DOI] [PubMed]
- 47.Pereira, P. M., Almada, P. & Henriques, R. in Methods Cell Biol, Vol. 125 (ed Ewa K. Paluch) 95−117 (Academic Press, 2015). [DOI] [PubMed]
- 48.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenfield, N. J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc.1, 2876–2890 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaka, A. J., Lee, C. J. & Pines, A. Iterative schemes for bilinear operators; application to spin decoupling. J. Magn. Reson.77, 274–293 (1988). [Google Scholar]
- 51.Hwang, T. L. & Shaka, A. J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A112, 275–279 (1995). [Google Scholar]
- 52.Bax, A. & Davis, D. G. Practical aspects of two-dimensional transverse NOE spectroscopy. J. Magn. Reson.63, 207–213 (1985). [Google Scholar]
- 53.Palmer, A. G., Cavanagh, J., Wright, P. E. & Rance, M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J. Magn. Reson.93, 151–170 (1991). [Google Scholar]
- 54.Schleucher, J. et al. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR4, 301–306 (1994). [DOI] [PubMed] [Google Scholar]
- 55.Kay, L. E., Keifer, P. & Saarinen, T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc.114, 10663–10665 (1992). [Google Scholar]
- 56.Lee, W., Rahimi, M., Lee, Y. & Chiu, A. POKY: a software suite for multidimensional NMR and 3D structure calculation of biomolecules. Bioinformatics37, 3041–3042 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pierattelli, R., Banci, L. & Turner, D. L. Indirect determination of magnetic susceptibility tensors in peroxidases: a novel approach to structure elucidation by NMR. JBIC J. Biol. Inorg. Chem.1, 320–329 (1996). [Google Scholar]
- 58.Wishart, D. S. et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR6, 135–140 (1995). [DOI] [PubMed] [Google Scholar]
- 59.Hafsa, N. E., Arndt, D. & Wishart, D. S. CSI 3.0: a web server for identifying secondary and super-secondary structure in proteins using NMR chemical shifts. Nucleic Acids Res.43, W370–W377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rino, J., Martin, R. M., Carvalho, T. & Carmo-Fonseca, M. Imaging dynamic interactions between spliceosomal proteins and pre-mRNA in living cells. Methods65, 359–366, (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author. The data generated in this study are provided in the Supplementary Information/Source Data file. Source data are provided with this paper as a Source Data file. Source data are provided with this paper.