Abstract
The interactive pathway of the gut-liver axis underscores the significance of microbiome modulation in the pathogenesis and progression of various liver diseases, including hepatocellular carcinoma (HCC). This study aims to investigate the disparities in the composition and functionality of the hepatic microbiota between tumor tissues and adjacent normal liver tissues, and their implications in the etiology of HCC. We conducted a comparative analysis of the hepatic microbiome between adjacent normal liver tissues and tumor tissues from HCC patients. Samples were categorized according to the modified Union for International Cancer Control (mUICC) staging system into Non-tumor, mUICC stage I, mUICC stage II, and mUICC stage III groups. Microbial richness and community composition were analyzed, and phylogenetic profiles were examined to identify significantly altered microbial taxa among the groups. Predicted metabolic pathways were analyzed using PICRUSt2. Our analysis did not reveal significant differences in microbial richness and community composition with the development of HCC. However, phylogenetic profiling identified significantly altered microbial taxa among the groups. Sphingobium, known for degrading polychlorinated biphenyls (PCBs), exhibited a significantly negative correlation with clinical indices in HCC patients. Conversely, Sphingomonas, a gut bacterium associated with various liver diseases, showed a positive correlation. Predicted metabolic pathways suggested a correlation between atrazine degradation and valine, leucine, and isoleucine biosynthesis with mUICC stage and tumor size. Our results underscore the critical link between hepatic microbial composition and function and the HCC tumor stage, suggesting a potentially pivotal role in the development of HCC. These findings highlight the importance of targeting the hepatic microbiome for therapeutic strategies in HCC.
Keywords: Hepatocellular carcinoma, Phylogenetic analysis, Taxa associations, Tumor tissues, Gut-liver axis
Subject terms: Cancer, Cell biology, Microbiology
Introduction
Globally, hepatocellular carcinoma (HCC) ranks as the third leading cause of death related to cancer and stands as the seventh most prevalent cancer in the Republic of Korea1. The general progression and outlook for patients with HCC continue to be unfavorable, and the World Health Organization predicts that the death toll among individuals with HCC will rise, surpassing one million by the year 20302. In the array of treatments for HCC intended for cure, surgical removal offers the broadest spectrum of application compared to other methods like liver transplantation and local ablation treatments3. Surgical resection remains the primary treatment method for early-stage HCC4. However, the 5-year recurrence rate after the surgical resection of HCC is reported to be approximately 43.7–77.0%, and the 5-year survival rate is reported to be approximately 23.0–56.3%5,6. The outlook for HCC depends on various elements, including the stage of the tumor, the number and size of tumors, the characteristics of the tumor as indicated by blood markers (such as alpha-fetoprotein and prothrombin induced by vitamin K absence or antagonist-II), the presence of microvascular invasion, the tumor’s histological grade, and the fundamental health attributes of the individual patient3. The baseline factors of each patient may vary depending on the etiology of HCC, ethnicity, sex, liver inflammatory activity, and microbiome. The gut microbiome, an intricate assembly of diverse microorganisms, has a significant influence on the regulation of human health and the manifestation of diseases7. Imbalances and functional disturbances in the microbial community can result in numerous diseases, especially when a person’s gut microbiome undergoes metabolic disruption8. Furthermore, the microbiome is recognized as a major factor in the development of various liver conditions, such as alcoholic liver disease and fatty liver disease8–13. The gut microbiome can be viewed as a new type of ‘organ’ that contains a gene pool vastly surpassing the host’s in number and is subject to influences from both genetic factors and environmental conditions14. The gut-liver axis describes the interactive connection between the gut and its microbiome and the liver, forming a pathway that links these two critical components of the digestive system15. Changes in the microbial community, the movement of bacteria due to enhanced intestinal permeability, and the direct impact of bacterial by-products on the liver are considered fundamental microbial contributors to liver disease progression. Several research studies have highlighted the microbiome’s possible influence on the onset of HCC and its utility in forecasting the disease’s prognosis16,17. Therefore, our study was designed to identify differences in microbial composition among HCC patient groups classified by tumor stage, while simultaneously comparing the microbial makeup found in surrounding non-tumor tissues. Our findings indicate a potentially crucial role in the development of HCC by highlighting the vital relationship between the composition and function of the hepatic microbiota and the HCC tumor stage.
Material and methods
Patients and HCC staging
This study involved the participation of eight patients (from a total of 20 samples), who underwent surgical removal of HCC at Chonnam National University Hospital between April 2023 and September 2023. We excluded patients with a different type of tumor than HCC, those who had undergone previous treatments, and anyone who had taken antibiotics within four weeks prior to surgery. The diagnosis of HCC conformed to the guidelines established by the Korean Liver Cancer Study Group and the National Cancer Center, with confirmation through pathological examination post-surgery4. The staging of HCC at diagnosis utilized the modified Union for International Cancer Control (mUICC) staging system and the Barcelona Clinic Liver Cancer (BCLC) classification18,19. Tumor size was determined via pathological review, and the histological grading of HCC followed the Edmondson and Steiner criteria. Macrovascular invasion was identified through CT or MRI evidence of vascular involvement by HCC, while microvascular invasion was defined based on microscopic evaluation of the vascular structures in the excised tumor samples. Liver tissues were harvested, immediately preserved by freezing, and stored at − 80 °C until further analysis.
Statement of ethics
This study was approved by the Institutional Review Board of the Chonnam National University Hospital (IRB No. CNUH-2023-087). The study’s protocol followed the Declaration of Helsinki. The data and safety monitoring board provided insights. Informed consent was obtained from all the patients according to the respective institutional guidelines.
Bacterial culture
Liver tissues were homogenized in Dulbecco’s phosphate-buffered saline (DPBS; WELEGENE, Republic of Korea). The homogenized lysate was spread onto Tryptic Soy Blood Agar (TSA-B; BD, New Jersey, USA) under both aerobic and anaerobic conditions (BD GasPak™ EX pouch system, New Jersy, USA) at 35 °C. Single colonies were picked and cultured as they grew (cultured for at least 48 h) on TSA agar, and 16S rRNA sequencing was subsequently performed. Bacteria were identified through the use of the NCBI BLAST tool (available at https://blast.ncbi.nlm.nih.gov/).
Real-time quantitative PCR to estimate bacterial load
Total liver RNA was purified using a commercially available preparation, Hybrid-RTM (GeneAll Biotechnology Co., Ltd., Republic of Korea). A constant amount of 500 ng of RNA was transcribed into cDNA using the TOPscript™ cDNA Synthesis Kit (Enzynomics, Republic of Korea), following the manufacturer’s instructions. Real-time quantitative PCR (qPCR) was performed to estimate the bacterial load in the liver tissues. Exactly 200 ng of the synthesized cDNA was added to a mixture of 10 µl of TOPreal™ SYBR Green qPCR PreMIX (Enzynomics, Republic of Korea), 3 µl of RNAse-free distilled water, and 2 µl of a primer mixture containing a forward primer (515F, 5′-GTGCCAGCMGCCGCGGTAA-3′) and a reverse primer (806R, 5’-GGACTACVSGGGTATCTAAT-3’) for the 16S rRNA gene V3-V4 region. The qPCR was performed on a CFX96 Real-Time PCR Detection System (BIO-RAD, California, USA) at 98 °C for 15 min with 39 cycles at 95 °C for 10 s, 57 °C for 15 s, and 72 °C for 15 s. Then, a melting curve analysis was conducted.
Process of 16S rRNA amplicon sequencing
Bacterial genomic DNA was extracted from liver specimens using the Fast DNA Spin Kit for Soil, following the manufacturer’s protocol (MP Biomedicals GmbH, Heidelberg, Germany). DNA samples were processed for 16S rRNA gene sequencing using a 300 bp paired-end protocol on the Illumina Miseq system (Illumina, Inc., San Diago, USA). The V3-V4 region of the bacterial 16S rRNA gene was amplified using the barcoded primers (forward primer sequence: 5’-CCTACGGGNGGCWGCAG-3’, reverse primer sequence: 5’-GACTACHVGGGTATCTAATCC-3’). 16S rRNA gene amplicon sequencing was performed at Macrogen (Macrogen, Inc., Seoul, Republic of Korea).
Bioinformatics pipelines for microbiota analyses
Paired-end raw sequence data were imported as QIIME2 (version 2023.2) artifacts, and these sequences were demultiplexed using the QIIME2 plugin. We then filtered out noisy sequences and joined the denoised paired-end reads using the DADA2 denoising method. Amplicon sequencing variants (ASVs) were produced using these quality control methods. After the quality filtering step was completed, alpha- and beta-diversity analyses were performed using q2-diversity plugins. Alpha diversity was calculated using the observed ASVs, Faith’s Phylogenetic Diversity (Faith’s PD), and Shannon and Pielou’s evenness metrics. A rarefied table was created by computing the features observed at multiple sequencing depths. Principal coordinate analysis (PCoA) plots were generated using Emperor for each beta-diversity metric (Weighted UniFrac, Unweighted UniFrac, Bray–Curtis, and Jaccard). The ASVs were assigned to the SILVA-138.1 database using a Naïve Bayes classifier and q2-feature-classifier plugin.
Taxonomic composition analyses
A table was created for each taxonomic level (Domain, Phylum, Class, Order, Family, Genus, and Species). In each table, sequences that could not be allocated to the corresponding taxonomic level (but were allocated to any higher level) were accumulated into a new category name “unclassified (_uc).”
The linear discriminant analysis (LDA) effect size (LEfSe) method was used to identify discriminative taxonomic biomarkers using an LDA log score cut-off of 2.0, followed by the Kruskal–Wallis test with a Wilcoxon test cut-off of p < 0.05. The implementation of the LEfSe method incorporated into the Galaxy framework is available online at http://galaxy.biobakery.org/20.
Data availability statement
Publicly available datasets were analyzed in this study. The dataset can be found in the NCBI BioProject, and its accession number was PRJNA1094649.
PICRUSt2 prediction analysis
The phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2, version 2.0) Python script was used to predict the functional metabolic pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database from the 16S rRNA gene sequencing data21.
Statistical analyses
Graph plots were created using GraphPad Prism v.7.05 (GraphPad Inc., USA) and OriginPro® 2024 (OriginLab Corporation, USA). The statistical significance of the alpha- and beta-diversity were obtained from QIIME2 plugins (q2-alpha- and q2-beta-group-significance). To assess the statistical significance of the alpha-diversity, a one-tailed Student’s t-test was employed, and beta-diversity metrics were assessed using the permutation multivariate analysis of variance (PERMANOVA) function with 999 permutations. The normalized values at the genus and species levels for each group were calculated based on the relative abundance of taxonomic composition using the normalization method provided by GraphPad Prism. The statistical significance of these normalized values was assessed using Tukey’s multiple comparison test. The correlation of the relative abundance of selected species was assessed using the one-tailed Spearman’s correlation test adjusted P-value. All values are expressed as the mean ± standard deviation (SD). Values of p < 0.05 were considered statistically significant.
Results
Clinical characteristics
The fundamental characteristics of the study cohort are detailed in Table 1. The median age of the participants was 63 years, with predominant male representation (87.5%). The leading cause of liver disease was hepatitis B virus (HBV) infection (37.5%), followed by alcohol-related liver disease (25%), hepatitis C virus (HCV) infection (25%), and combined causes (12.5%). The median level of alpha-fetoprotein (AFP) was found to be 20.69 IU/mL, with an IQR of 4.7 to 370.3 ng/mL. Based on the mUICC staging system, the distribution of patients was roughly equal among stages I and II (37.5% each), with 25% in stage III. Using the BCLC staging system, the most frequent stage was A (50%), then stage 0 (37.5%), and stage B (12.5%). Single tumors were present in 61.4% of cases, with a median largest tumor size of 3.1 cm (IQR, 1.48 to 4.7 cm). The comprehensive analysis of the liver specimens resected identified mUICC stage I (n = 5), stage II (n = 6), stage III (n = 4), and normal adjacent non-tumor tissues (n = 5). Hematoxylin and eosin (H&E) staining was performed on the tumor and adjacent non-tumor tissues to assess their characteristic features in Fig. 1A.
Table 1.
Baseline characteristics of enrolled patients.
| Total (n = 8) | |
|---|---|
| Age (years) | 63.43 ± 9.32 |
| Male, n (%) | 7 (87.5) |
| Body mass index (kg/m2) | 25.46 ± 1.61 |
| Etiology of hepatocellular carcinoma, n (%) | |
| Alcohol / HBV | 2(25) / 3(37.5) |
| HCV / Combined | 2(25) / 1(12.5) |
| Platelet (× 103/μL) | 175.25 ± 52.61 |
| AST (IU/mL) | 30.63 ± 7.60 |
| ALT (IU/mL) | 29.13 ± 17.06 |
| Albumin (mg/dL) | 4.41 ± 0.27 |
| Total bilirubin (mg/dL) | 0.83 ± 0.20 |
| Liver cirrhosis, n (%) | 6 (75) |
| Tumor size (cm) | 3.10 ± 1.62 |
| Tumor numbers | 1.25 ± 0.71 |
| BCLC stage, n (%) | |
| 0 / A / B | 3(37.5) / 4(50) / 1(12.5) |
| Pathologic mUICC stage, n (%) | |
| I / II / III | 3(37.5) / 3(37.5) / 2(25) |
| Edmonson-Steiner grade, n (%) | |
| Worst, 2 / 3 /4 | 1(12.5) / 6(75) / 1(12.5) |
| Major, 2 / 3/ 4 | 6(75) / 2(25) / 0(0) |
| ICG-R15 | 18.95 ± 6.54 |
| Child–Pugh score, score (%) | 5 (100) |
| Serum AFP (IU/mL) | 20.69 ± 22.63 |
| PIVKA-II (mAU/mL) | 95.43 ± 132.11 |
| Follow-up duration, days (median, range) | 171 (134–245) |
Values are presented as mean ± SD. SD, standard deviation; HBV, hepatitis B virus; HCV, hepatitis C virus; AST, aspartate transaminase; ALT, alanine transaminase; BCLC, Barcelona Clinic Liver Cancer; mUICC, modified Union for International Cancer Control; ICG-R15, indocyanine green retention test at 15 min; AFP, alpha-fetoprotein; PIVKA-II, protein induced by vitamin K absence or antagonist-II
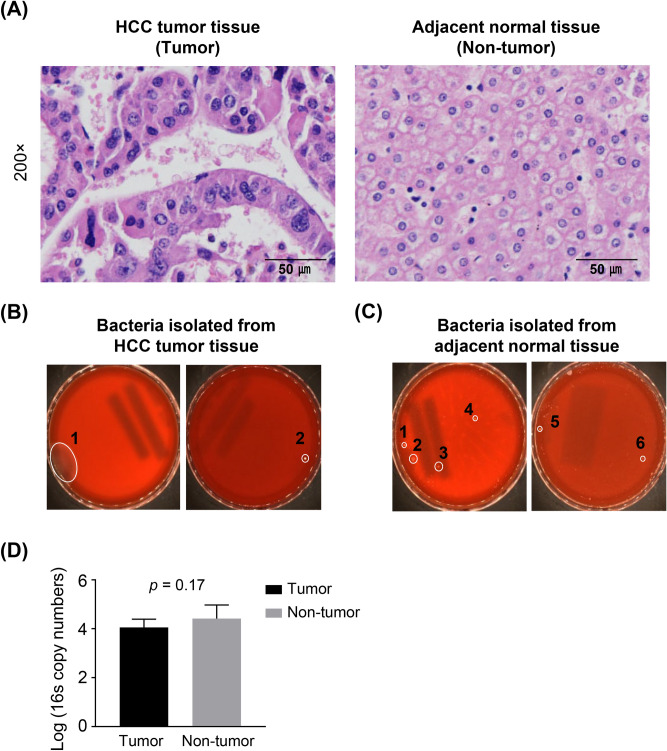
Fig. 1.
HCC tissues in presence of bacteria. (A) Representative hematoxylin and eosin (H&E) staining images from different regions of HCC tumor tissues and adjacent normal tissues. Scale bar: 50 μm. (B, C) Culture-based assay of liver samples in tumor tissues (B) and adjacent normal tissues (C). BLAST results of 16S rRNA sequences of colonies selected from HCC tumor tissues. (D) Bacterial load in the tumor and non-tumor samples determined by quantitative PCR of the 16S rRNA gene. Data are presented as mean ± standard deviation (SD).
Bacteria existed in both HCC and adjacent non-tumor liver tissues
For visualizing and confirming the existence of live bacteria within liver samples, we cultured bacteria from liver specimens in both aerobic and anaerobic environments on a TSA-B agar (Fig. 1B). We identified two bacterial colonies in the HCC tissues, which were classified as Bacillus oceanisediminis (Fig. 1B). More colonies were observed in the adjacent normal liver tissues and were identified as belonging to various species: Micromonospora terminaliae, Caldibacillus thermoamylovorans, Paenibacillus barengoltzii, Effusibacillus consociatus, Paenibacillus stellifer, Micromonospora globbae, and Bacillus thuringiensis (Fig. 1C). To measure the amount of bacterial DNA, quantitative PCR (qPCR) was conducted on liver tissue samples using primers that target a broad range of bacteria. While the difference in the number of copies of the 16S rRNA gene was not statistically significant, bacteria were indeed identified in the liver tissues of individuals diagnosed with HCC, as shown in Fig. 1D. These results, derived from all liver samples from the patients, underscore the microbial diversity present within both tumor and adjacent non-tumor liver tissues, verifying the existence of bacteria in these different tissue environments and indicating a varied microbiome presence.
Diversity analysis of the hepatic microbiome in patients with HCC
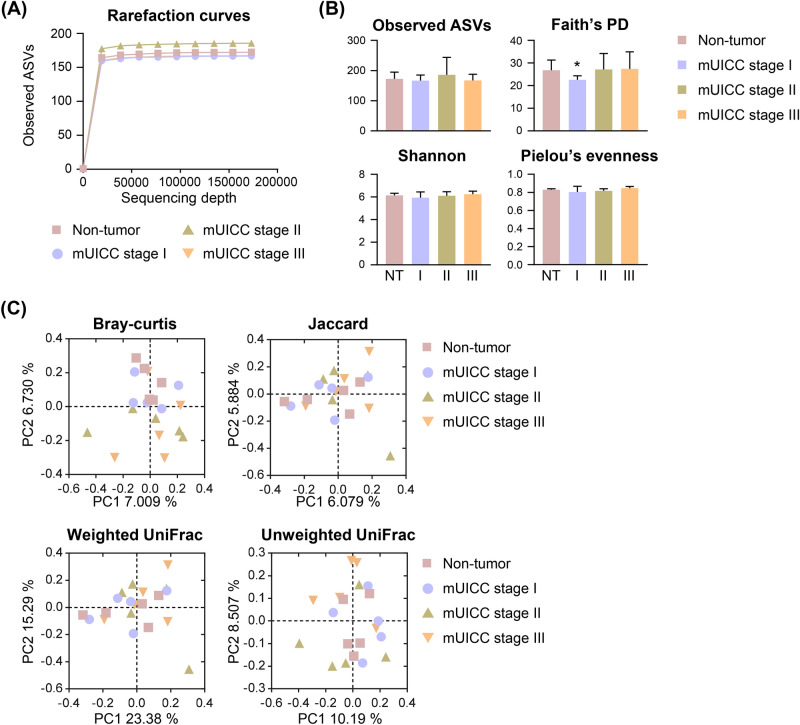
On average, 674 241 sequences of the 16S rRNA gene were utilized for our analysis. After analyzing the 16S rRNA amplicon sequencing data, we identified 3,479 Amplicon Sequence Variants (ASVs), encompassing 28 phyla, 78 classes, 167 orders, 275 families, 537 genera, and 851 species. The leveling off of the rarefaction curves (Fig. 2A) suggested that the sequence richness was adequately captured in further analyses. We applied various metrics to assess the microbial alpha-diversity, such as the count of observed ASVs, Faith’s Phylogenetic Diversity (PD), and indices for Shannon diversity and Pielou’s evenness (Fig. 2B). The Faith’s PD index indicated a reduction in species richness at the mUICC stage I compared to the non-tumor (NT) tissue group (p = 0.044). Yet, no statistically significant variance was detected across the groups for the other diversity indices, including observed ASVs and indices for Shannon diversity and Pielou’s evenness (Supplementary Table 1).
Fig. 2.
Microbial diversity in the liver of patients with HCC. (A) Rarefaction curves of the four groups demonstrating saturation: Non-tumor (NT), mUICC stage I (I), mUICC stage II (II), and mUICC stage III (III). The x-axis represents the number of sampled sequences, and y-axis represents the observed amplicon sequencing variants (ASVs). (B) Alpha diversity boxplots are illustrated for the observed ASVs, Faith’s phylogenetic diversity (Faith’s PD), Shannon, and Pielou’s evenness indices. Data are presented as mean ± SD. Statistical analysis was conducted using a one-tailed Student’s t-test (*p < 0.05) compared to the NT group. (C) Principal coordinates analysis (PCoA) plots generated based on Bray–Curtis distances, Jaccard, weighted-UniFrac, and unweighted-UniFrac indices among samples of the four groups. No significant differences were observed among the groups as PERMANOVA.
Differences in the community composition between samples (‘beta-diversity’) were evaluated using Bray–Curtis dissimilarity and Jaccard indices, as well as abundance-weighted and unweighted UniFrac distances (Fig. 2C). PERMANOVA analysis revealed no significant disparities among the groups (Supplementary Table 2). These results emphasize the subtle distinctions observed in the microbiota composition between hepatocellular carcinoma (HCC) tumor tissues and their adjacent normal counterparts. Although no statistical significance was found in the comparisons between groups, we observed a trend towards variability in the microbiota composition across samples, particularly with increasing mUICC stage.
Differences in hepatic microbial communities according to the HCC stage
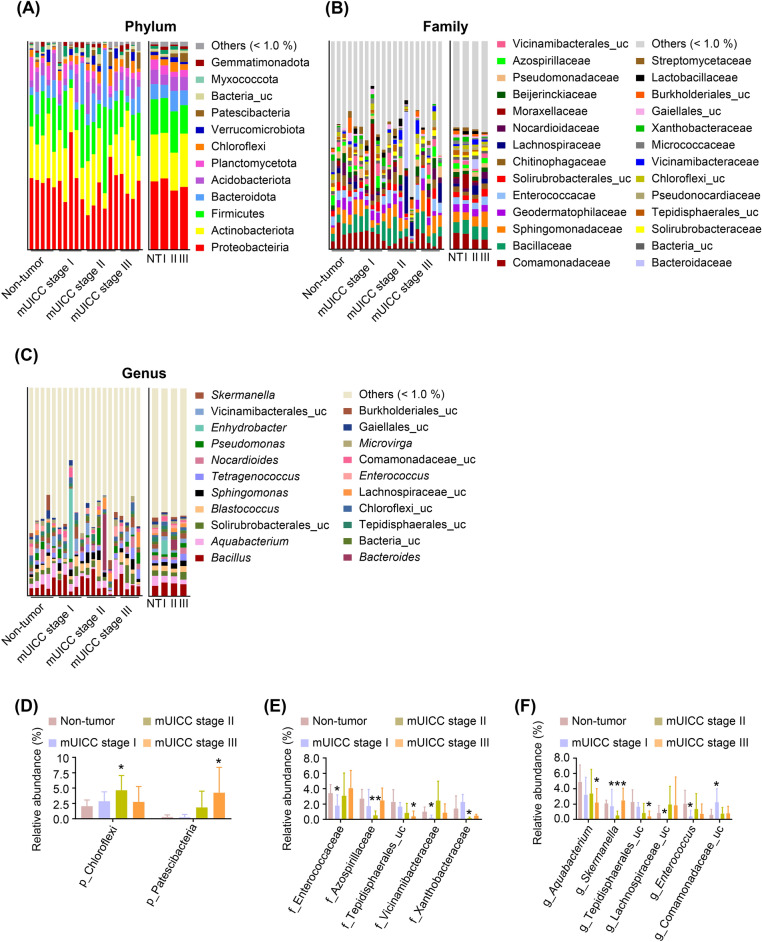
To address the hepatic microbial differences associated with HCC progression, we performed phylogenetic analyses at the phylum (Fig. 3A), family (Fig. 3B), and genus (Fig. 3C) levels. We identified the significantly enriched or depleted phyla, families, and genera in these groups. ‘Others’ represented the sum of phyla and families with an average of less than 1% relative abundance. As shown in Fig. 3D, there were two significantly enriched major phyla: Chloroflexi (p = 0.027, enriched in mUICC stage II) and Patescibacteria (p = 0.034, enriched in mUICC stage III) (Supplementary Table 3). As shown in Fig. 3E, At the family level, the five families, including Enterococcaceae (p = 0.037, decreased in mUICC stage I), Azospirillaceae (p = 0.002, decreased in mUICC stage II), unclassified Tepidisphaerales (p = 0.035, decreased in mUICC stage III), Vicinamibacteraceae (p = 0.019, decreased in mUICC stage I), and Xanthobacteraceae (p = 0.049, decreased in mUICC stage II), were significantly different among the groups (Supplementary Table 3). Furthermore, six genera were significantly different among the groups compared to the NT group (Fig. 3F): Aquabacterium (p = 0.046, decreased in mUICC stage III), Skermanella (p = 0.0004, decreased in mUICC stage II), unclassified Tepidisphaerales (p = 0.035, decreased in mUICC stage III), unclassified Lachnospiraceae (p = 0.041, decreased in mUICC stage I), Enterococcus (p = 0.048, decreased in mUICC stage I), and unclassified Comamonadaceae (p = 0.045, enriched in mUICC stage I) (Supplementary Table 3).
Fig. 3.
Phylogenetic profiles of the microbes at different HCC mUICC stages. Taxonomic composition of hepatic microbiota of patients with HCC at the phylum (A), family (B), and genus (C) levels; colors indicate the corresponding percentages of different taxa. The plots depict the relative abundance of two phyla (D), five families (E), and six genera (F), that exhibited statistically significant differences among the groups. Statistical analysis was conducted using a one-tailed Student’s t-test (*p < 0.05, **p < 0.01) compared to the non-tumor group. uc, unclassified taxa; others, sum of the taxa with averages of less than 1% relative abundance; NT, Non-tumor group; I, mUICC stage I group; II, mUICC stage II group; III, mUICC stage III group.
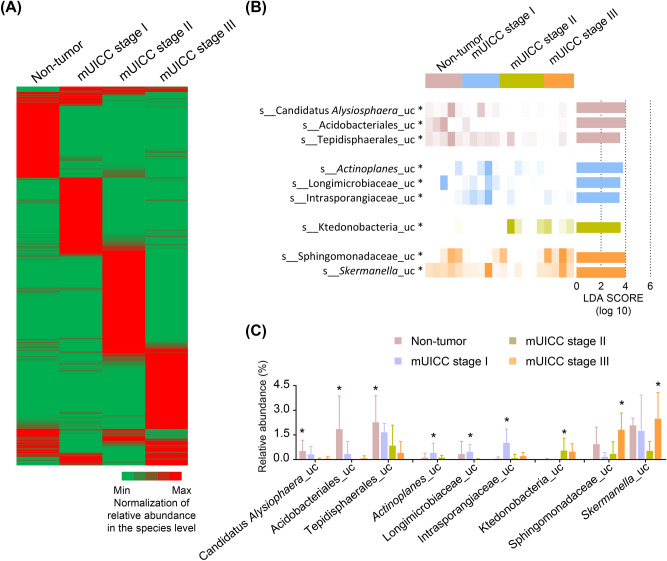
Following the normalization of species abundance for each group, the heatmap revealed distinct taxonomic patterns associated with HCC progression (Fig. 4A). The LEfSe method was employed to robustly identify abundant microbial taxa with log LDA scores above 2.0, revealing statistically significant differences in the phylogenetic analysis within this study. The LEfSe analysis revealed nine enriched species in each group (Fig. 4B). Three taxa, including unclassified Candidatus Alsiophaera (p = 0.029), unclassified Acidobacteriales (p = 0.036), and unclassified Tepidisphaerales (p = 0.034), were enriched in the NT group. In the mUICC stage I group, unclassified Actinoplanes (p = 0.049), unclassified Longimicrobiaceae (p = 0.020), and unclassified Intrasporangiaceae (p = 0.021) exhibited enrichment. The mUICC stage II group showed enrichment in one taxon (unclassified Ktedonobacteria, p = 0.046), and the mUICC stage III group displayed significant abundance in two taxa [unclassified Sphingomonadaceae (p = 0.047) and unclassified Skermanella (p = 0.014)] (Fig. 4C). This phylogenetic analysis revealed distinct bacterial compositions between cancerous liver tissues and adjacent normal tissues within the same patients with HCC, suggesting a potential correlation between HCC progression and the hepatic microbiota.
Fig. 4.
Differential composition in the hepatic microbiome across HCC mUICC stages. (A) Normalized heatmap of bacterial abundance at the species level in the Non-tumor, mUICC stage I, mUICC stage II, and mUICC stage III groups. (B) Linear discriminant analysis (LDA) score and p-value computed from differentially abundant bacteria among the groups at the species level, using the linear discriminant analysis effect size (LEfSe) algorithm. Statistical analysis was performed using LEfSe. Features with an LDA score > 2 were assessed, followed by Kruskal–Wallis test with a Wilcoxon test cut-off of p < 0.05 (*p < 0.05). (C) Relative abundance of significantly increased taxa in each group, followed by Kruskal–Wallis test with a Wilcoxon test cut-off of p < 0.05 (*p < 0.05). Values are expressed as mean ± SD. uc, unclassified taxa.
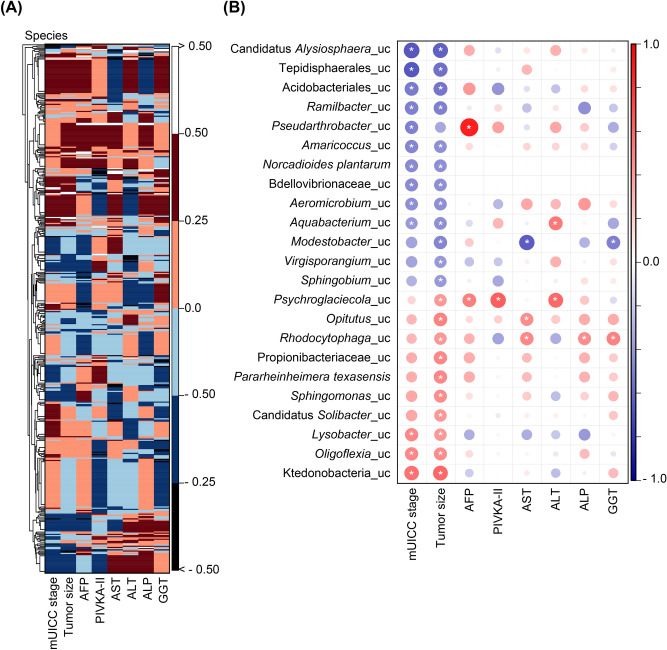
Relationship between liver microbiome and clinical indicators in individuals with hepatocellular carcinoma
To investigate the dynamic interaction between hepatic microbiota and the progression of HCC, a comprehensive analysis was undertaken employing Spearman’s correlation matrix to evaluate the associations between the abundance of hepatic bacteria and various clinical parameters. The generation of heatmaps facilitated the visualization of these associations, incorporating the species detected through co-occurrence patterns in conjunction with variables such as mUICC stage, tumor size, levels of alpha-fetoprotein (AFP), protein induced by vitamin K absence or antagonist-II (PIVKA-II), aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), and gamma-glutamyltransferase (GGT), as depicted in Fig. 5A. Subsequent analysis pinpointed specific taxa that demonstrated significant correlations with clinical indicators pertinent to HCC progression, highlighted in Fig. 5B. Remarkably, thirteen bacterial species showed a negative correlation with advancing stages of HCC, among which unclassified species within the genera Modestobacter, Virgisporangium, and Sphingobium were identified as displaying a pronounced inverse relationship with serum biochemical markers indicative of HCC pathology (AFP, PIVKA-II, AST, ALT, ALP, and GGT). In contrast, ten species exhibited a positive correlation with the progression of HCC, where species such as unclassified Psychroglaciecola, unclassified Opitutus, unclassified Rhodocytophaga, unclassified Propionibacteriaceae, Pararheinheimera texasensis, and unclassified Sphingomonas were noted for their direct correlation with clinical indices relevant to HCC, as detailed in Supplementary Table 4. This detailed inquiry underscores the intricate link between hepatic microbiota composition and key clinical indices associated with HCC, shedding light on potential microbial signatures reflective of disease progression.
Fig. 5.
Correlation between the hepatic microbiome and HCC progression. (A) Heatmap showing Spearman’s correlation coefficient of pairwise comparison based on bacterial abundance (at the species level) and clinical indices. (B) The corrplot illustrates statistically significant associations between the hepatic microbiota and clinical indices. Positive correlations are displayed in red, and negative correlations are in blue (the darker the color and the larger the graph, the more significant the correlation). The correlation results were considered statistically significant at *p < 0.05. AFP, alpha-fetoprotein; PIVKA-II, prothrombin-induced by vitamin K absence or antagonist-II; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALP, alkaline phosphatase; GGT, gamma-glutamyl peptidase.
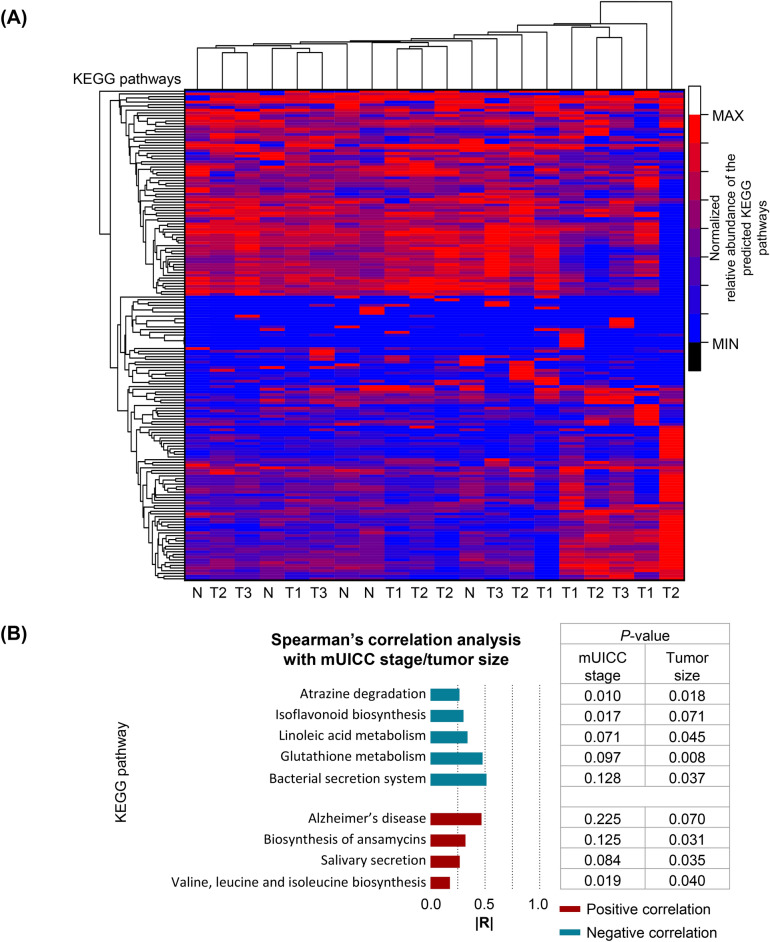
Analysis of functional correlations between pathways related to the staging of hepatocellular carcinoma (HCC) and tumor size dimensions
To elucidate the metabolic pathways linked with hepatic taxa exhibiting differential abundance in relation to the progression of hepatocellular carcinoma (HCC), we utilized Predictive Insights into Microbial Ecology (PICRUSt2) in conjunction with Spearman’s correlation analysis. This approach facilitated the alignment of microbial 16S rRNA gene sequences with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database, allowing for the prediction of microbial involvement in metabolic functions. Our analysis identified a comprehensive list of 181 KEGG categories deemed relevant to the study (Fig. 6A). Among these, nine metabolic pathways demonstrated significant correlations with both the modified Union for International Cancer Control (mUICC) stage and the dimensional attributes of the tumor (Fig. 6B). Notably, pathways associated with atrazine degradation, isoflavonoid biosynthesis, linoleic acid metabolism, glutathione metabolism, and the bacterial secretion system were found to have inverse correlations with both mUICC stage and tumor dimensions, suggesting a potential protective or diminishing role in HCC progression. In contrast, metabolic pathways linked to Alzheimer’s disease, ansamycin biosynthesis, salivary secretion, and the biosynthesis of branched-chain amino acids (valine, leucine, and isoleucine) exhibited positive correlations, indicating an enhancement or contributory role in the advancement of HCC. This intricate analysis underscores the multifaceted relationship between specific microbial metabolic activities and key clinical parameters of HCC, providing insight into potential microbial-driven mechanisms influencing the disease’s progression. Our findings highlight the critical link between hepatic microbial composition and function with the HCC tumor stage, suggesting a potentially pivotal role in HCC development.
Fig. 6.
Predictive functional profiling of microbial communities in the liver tissues of patients with HCC. (A) Normalized heatmap of predicted KEGG pathways in HCC and adjacent normal tissues. N, Non-tumor group; T1, mUICC stage I; T2, mUICC stage II; T3, mUICC stage III. (B) Absolute value of Spearman’s correlation coefficient (|R|) between the hepatic microbial metabolic pathway, mUICC stage, and tumor size. The correlation results were deemed statistically significant at p < 0.05. Positive correlations are displayed in red, and negative correlations are in blue.
Discussion
In this investigation, our premise posited that variances in the composition and functional capabilities of the microbial communities residing in hepatic environments—specifically, between neoplastic liver tissues and their adjacent non-neoplastic counterparts—play a contributory role in the pathogenesis of HCC. Unlike previous studies that primarily focused on analyzing the hepatic microbiome based on the presence or absence of tumors or disease etiology, our hypothesis is predicated on the understanding that the liver’s microbial ecosystem—through its diversity and metabolic functions, may influence tumorigenic processes, thereby implicating these microbial disparities in the initiation and progression of HCC.
Liver tissue samples obtained from hospitalized patients diagnosed with HCC were categorized based on the modified Union for International Cancer Control (mUICC) staging system. Subsequent analysis of these samples unveiled distinct variations in the abundance patterns of several microbial species across different mUICC stages of HCC. Notably, these species exhibited correlations with the associated risk levels of HCC development. We observed a reduction in the prevalence of unidentified Sphingobium species concurrent with the progression of HCC. Sphingobium species are known for their resilience to heavy metals and their capability to break down a wide array of environmental contaminants, including polychlorinated biphenyls (PCBs) and atrazine22. PCBs are highly carcinogenic chemical compounds that increase the risk of developing HCC23. A previous case–control study conducted in a PCB-polluted area in northern Italy found a potential association between elevated serum PCB levels and an increased risk of HCC23. Moreover, treatment with atrazine promotes the proliferation of mouse hepatocellular carcinoma cells and increases tumor volume in HCC-bearing C57BL/6 mice24. Thus, the abundant presence of Sphingobium and its increased ability to degrade atrazine in adjacent normal tissues may play an anticancer role.
Sphingomonas, a genus of bacteria prevalent within the gastrointestinal microbiome, has been identified as a key microbial agent implicated in the pathogenesis of a spectrum of hepatobiliary disorders. These disorders include, but are not limited to, primary biliary cholangitis (PBC)25, primary sclerosing cholangitis (PSC)26, liver fibrosis 27, liver cirrhosis, and HCC28. The association between Sphingomonas and these liver diseases underscores the bacterium’s potential role in modulating liver function and structure, possibly through mechanisms involving immune response dysregulation, bile acid metabolism disruption, and the induction of fibrogenic pathways29. This linkage highlights the intricate interplay between gut microbiota and liver health, suggesting that Sphingomonas may contribute to the initiation and progression of liver pathology through multifaceted interactions within the host’s biological systems. The expression of amine oxidase by Sphingomonas, a bacterium residing within the gut microbiota, has been correlated with the dysregulated migration of gut-derived lymphocytes to hepatic tissue, a phenomenon characterized as aberrant homing. This enzymatic activity is proposed to play a pivotal role in altering the chemotactic environment within the gut-liver axis, thereby facilitating the translocation of lymphocytes from the intestinal mucosa to the liver. The presence of these immune cells in hepatic tissue is believed to contribute to an inflammatory cascade, potentially exacerbating hepatic injury and fibrogenesis. Over time, this sustained inflammatory state may lay the groundwork for oncogenic transformations within hepatic cells, thereby elevating the risk of HCC development. This association underscores the intricate biochemical interactions mediated by microbial metabolites, such as those produced by Sphingomonas, in modulating immune cell trafficking and the subsequent implications for liver pathology.
A study conducted by Xue et al. demonstrated that the abundance of Sphingomonas and tumor-promoting sphingolipid, such as phosphoethanolamine was increased in HCC tissues through metabolomic analysis. Additionally, transcriptome analysis revealed an upregulation of amino acid metabolism in HCC tissues30,31. Consistent with these findings, our study also observed an increase in valine, leucine, and isoleucine biosynthesis as the mUICC stage progressed. Valine, leucine, and isoleucine, collectively classified as branched-chain amino acids (BCAAs), are fundamental components necessary for the biosynthesis of proteins and the facilitation of energy production within cellular systems32. These essential amino acids are critical for a multitude of cellular functions, including the regulation of protein synthesis pathways, the modulation of metabolic processes, and the maintenance of nitrogen balance33. Their roles extend beyond mere substrates for protein construction, as they are also involved in signaling pathways that influence overall metabolic homeostasis and cellular health. In the clinical context of HCC, a notable increase in the plasma concentrations of these amino acids has been documented among patients, indicating a potential aberration in metabolic regulation associated with the disease state34. This upregulation of BCAAs contributes to the pathogenesis and progression of pancreatic tumors through a mechanism involving the augmented phosphorylation of the mammalian target of rapamycin (mTOR)35. The mTOR pathway serves as a pivotal controller of cellular growth, proliferation, and viability, with its phosphorylation-induced activation representing a fundamental step in the initiation of tumorigenesis36. The increased availability of valine, leucine, and isoleucine in the tumor microenvironment facilitates this process, thereby enhancing the growth and aggressiveness of neoplasms. The interplay between elevated BCAA levels and mTOR signaling underscores the intricate relationship between metabolic dysregulation and cancer progression, highlighting the potential of these metabolic pathways as targets for therapeutic intervention in HCC and possibly other cancers characterized by similar metabolic anomalies.
Another interesting observation is the negative correlation between the tumor size and linoleic acid metabolism. Dysregulation of lipid metabolism is a well-known contributor to various liver diseases and liver cancer37. Consistent with this, downregulation of linoleic acid metabolism in HCC tissues has been reported31, along with a quantitative reduction in alpha-linoleic acid38. These results can be explained by the fact that alpha-linoleic acid inhibits tumor cell proliferation, migration, and invasion through modulation of the Farnesoid X receptor/β-catenin signaling pathway38.
This study presents several limitations. Firstly, the small sample size of enrolled patients restricted the ability to identify significant differences in microbial richness and community composition. Moreover, comparisons of microbial and metabolic profiles across various etiologies of HCC were not feasible due to this limited cohort. Secondly, the duration of follow-up was insufficient for a comprehensive assessment of recurrence-free survival and overall survival rates. Lastly, the absence of stool samples prevented the analysis of microbial and metabolic profiles in fecal matter. Future prospective studies with larger cohorts, incorporating a broader range of etiologies and tumor stages, as well as including stool analysis and extended follow-up, are necessary to enhance our understanding of these relationships.
The innovative findings from this investigation elucidate the significant impact of microbial and metabolic variances on the pathogenesis and progression of HCC, offering profound insights into the interplay between hepatic microenvironments and tumor development. The delineation of distinct microbial communities and metabolic profiles in neoplastic versus adjacent non-neoplastic liver tissues provides a pivotal understanding of how alterations in the liver’s microbiome and metabolic milieu may contribute to HCC pathophysiology. This investigation into the microbial and metabolic underpinnings of HCC not only advances our understanding of cancer biology but also opens new horizons for the development of innovative therapeutic strategies. Despite the introduction of novel therapies and advancements in treatment technologies, the prognosis for patients with HCC remains poor, and significant unmet clinical needs persist. These findings may offer new perspectives on HCC treatment through the exploration of microbial and metabolic components, thereby enhancing our understanding of the pathogenesis of HCC development and informing strategies for primary and secondary prevention. However, the translation of these findings into clinical practice will require further validation through studies with a larger sample size, along with mechanistic studies and clinical trials, to overcome the study’s limitations and fully harness the potential of microbial and metabolic modulation in cancer therapy.
Supplementary Information
Acknowledgements
This work was supported by the World Institute of Kimchi funded by the Ministry of Science and ICT, Republic of Korea (grant number: KE2403-1), National Research Foundation of Korea (NRF) grants funded by Korea government (MSIT) (grant numbers: NRF-2020R1C1C1015024, NRF-2021R1F1A1061719), and Chonnam National University Hospital Biomedical Research Institute (grant number: BCRI24041).
Author contributions
Misun Yun, Jae Hyun Yoon and Jae-Ho Jeong contributed to the study conception and design. Material preparation, data collection and analysis were performed by Hee Eun Jo, Sophallika Khom, Sumi Lee, Su Hyeon Cho, Shin Young Park, Ga Ram You, Hyosin Kim, and Nah Ihm Kim. The first draft of the manuscript was written by Hee Eun Jo, Jae Hyun Yoon and Misun Yun and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data availability
Publicly available datasets were analyzed in this study. The dataset can be found in the NCBI BioProject, and its accession number was PRJNA1094649.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jae-Ho Jeong, Email: jeongjaeho@chonnam.ac.kr.
Jae Hyun Yoon, Email: zenmake14@gmail.com.
Misun Yun, Email: misun.yun@wikim.re.kr.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-77260-6.
References
- 1.Chon, Y. E., Jeong, S. W. & Jun, D. W. Hepatocellular carcinoma statistics in South Korea. Clin. Mol. Hepatol.27(3), 512–514 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med.380(15), 1450–1462 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Yoon, J. H. & Choi, S. K. Management of early-stage hepatocellular carcinoma: challenges and strategies for optimal outcomes. J. Liver Cancer23(2), 300–315 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korean Liver Cancer, A., and National Cancer Center, K 2022 KLCA-NCC Korea practice guidelines for the management of hepatocellular carcinoma. Clin Mol Hepatol, 28(4), 583–705 (2022). [DOI] [PMC free article] [PubMed]
- 5.Chan, A. W. H. et al. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J. Hepatol.69(6), 1284–1293 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kim, J. et al. Substantial risk of recurrence even after 5 recurrence-free years in early-stage hepatocellular carcinoma patients. Clin. Mol. Hepatol.26(4), 516–528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heintz-Buschart, A. & Wilmes, P. Human gut microbiome: Function matters. Trends Microbiol.26(7), 563–574 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Fan, Y. & Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol.19(1), 55–71 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Jennison, E. & Byrne, C. D. The role of the gut microbiome and diet in the pathogenesis of non-alcoholic fatty liver disease. Clin. Mol. Hepatol.27(1), 22–43 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park, G., Jung, S., Wellen, K. E. & Jang, C. The interaction between the gut microbiota and dietary carbohydrates in nonalcoholic fatty liver disease. Exp. Mol. Med.53(5), 809–822 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganesan, R. & Suk, K. T. Microbiome and metabolomics in alcoholic liver disease. Clin. Mol. Hepatol.28(3), 580–582 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han, G. & Vaishnava, S. Microbial underdogs: exploring the significance of low-abundance commensals in host-microbe interactions. Exp. Mol. Med.55(12), 2498–2507 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo, D. O. & Holtzman, D. M. Current understanding of the Alzheimer’s disease-associated microbiome and therapeutic strategies. Exp. Mol. Med.56(1), 86–94 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchesi, J. R. et al. The gut microbiota and host health: A new clinical frontier. Gut65(2), 330–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albillos, A., de Gottardi, A. & Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol.72(3), 558–577 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Schwabe, R. F. & Greten, T. F. Gut microbiome in HCC - Mechanisms, diagnosis and therapy. J. Hepatol.72(2), 230–238 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Ma, J. et al. Association of gut microbiome and primary liver cancer: A two-sample Mendelian randomization and case-control study. Liver Int.43(1), 221–233 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Kudo, M., Kitano, M., Sakurai, T. & Nishida, N. General rules for the clinical and pathological study of primary liver cancer, nationwide follow-up survey and clinical practice guidelines: The outstanding achievements of the liver cancer study group of Japan. Dig. Dis.33(6), 765–770 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Reig, M. et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol.76(3), 681–693 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol.10.1186/gb-2011-12-6-r60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglas, G. M. et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol.38(6), 685–688 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, J. et al. Sphingobium fuliginis HC3: A novel and robust isolated biphenyl- and polychlorinated biphenyls-degrading bacterium without dead-end intermediates accumulation. PLoS One10(4), e0122740 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donato, F. et al. Polychlorinated biphenyls and risk of hepatocellular carcinoma in the population living in a highly polluted area in Italy. Sci. Rep.11(1), 3064 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian, Y. et al. Atrazine exposure improves the proliferation of H22 cells in vitro and in vivo. RSC Adv.8(39), 21759–21767 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitahata, S. et al. Ileal mucosa-associated microbiota overgrowth associated with pathogenesis of primary biliary cholangitis. Sci. Rep.11(1), 19705 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quraishi, M. N. et al. A pilot integrative analysis of colonic gene expression, gut microbiota, and immune infiltration in primary sclerosing cholangitis-inflammatory bowel disease: Association of disease with bile acid pathways. J. Crohns Colitis14(7), 935–947 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lelouvier, B. et al. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: A pilot analysis. Hepatology64(6), 2015–2027 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Cho, E. J. et al. Circulating microbiota-based metagenomic signature for detection of hepatocellular carcinoma. Sci. Rep.9(1), 7536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trivedi, P. J. et al. Vascular adhesion protein-1 is elevated in primary sclerosing cholangitis, is predictive of clinical outcome and facilitates recruitment of gut-tropic lymphocytes to liver in a substrate-dependent manner. Gut67(6), 1135–1145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue, C. et al. Intratumoral bacteria interact with metabolites and genetic alterations in hepatocellular carcinoma. Sig. Transduct. Target Ther.7(1), 335 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He, Y., Zhang, Q., Yu, X., Zhang, S. & Guo, W. Overview of microbial profiles in human hepatocellular carcinoma and adjacent nontumor tissues. J. Transl. Med.21(1), 68 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimomura, Y. & Kitaura, Y. Physiological and pathological roles of branched-chain amino acids in the regulation of protein and energy metabolism and neurological functions. Pharmacol. Res.133, 215–217 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Ling, Z. N. et al. Amino acid metabolism in health and disease. Sig. Transduct. Target Ther.8(1), 345 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nezami Ranjbar, M. R. et al. GC-MS based plasma metabolomics for identification of candidate biomarkers for hepatocellular carcinoma in Egyptian Cohort. PLoS One10(6), e0127299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, K. A., Lashinger, L. M., Rasmussen, A. J. & Hursting, S. D. Leucine supplementation differentially enhances pancreatic cancer growth in lean and overweight mice. Cancer Metab.10.1186/2049-3002-2-6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou, Z., Tao, T., Li, H. & Zhu, X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci.10, 31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul, B., Lewinska, M. & Anderson, J. B. Lipid alterations in chronic liver disease and liver cancer. JHEP Rep.4(6), 100479 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng, S. et al. Alpha-linoleic acid inhibits hepatocellular carcinoma cell growth through Farnesoid X receptor/β-catenin signaling pathway. Nutr. Metab.19, 57 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. The dataset can be found in the NCBI BioProject, and its accession number was PRJNA1094649.
Publicly available datasets were analyzed in this study. The dataset can be found in the NCBI BioProject, and its accession number was PRJNA1094649.