Abstract
The wide-spreading of multidrug resistance poses a significant threat to human and animal health. Although antimicrobial peptides (AMPs) show great potential application, their instability has severely limited their clinical application. Here, self-assembled AMPs composed of multiple modules based on the principle of associating natural marine peptide N6 with ß-sheet-forming peptide were designed. It is noteworthy that one of the designed peptides, FFN could self-assemble into nanoparticles at 35.46 µM and achieve a dynamic transformation from nanoparticles to nanofibers in the presence of bacteria, resulting in a significant increase in stability in trypsin and tissues by 1.72–57.5 times compared to that of N6. Additionally, FFN exhibits a broad spectrum of antibacterial activity against multidrug-resistant (MDR) gram-positive (G+) and gram-negative (G−) bacteria with Minimum inhibitory concentrations (MICs) as low as 2 µM by membrane destruction and complemented by nanofiber capture. In vivo mouse mastitis infection model further confirmed the therapeutic potential and promising biosafety of the self-assembled peptide FFN, which can effectively alleviate mastitis caused by MDR Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus), and eliminate pathogenic bacteria. In conclusion, the design of peptide-based nanomaterials presents a novel approach for the delivery and clinical translation of AMPs, promoting their application in medicine and animal husbandry.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12951-024-02896-5.
Keywords: Antimicrobial peptide N6, Self-assembly, Nanoparticles and nanofibers, Stability, Bacterial-capture, Bovine mastitis
Introduction
Antimicrobial resistance (AMR), as a "silent global pandemic", poses a significant peril to both human health and animal husbandry [1]. According to statistics, bacterial AMR was associated with 4.95 million deaths and directly caused 1.27 million deaths in 2019. Unfortunately, it is estimated that AMR could be accountable for approximately 10 million deaths by 2050 if no effective measures are taken [2]. It is worth noting that the widespread dissemination of AMR genes has resulted in the emergence of multidrug-resistant (MDR) bacteria, particularly Escherichia coli (E. coli) and Staphylococcus (S. aureus). The prevalence of AMR of S. aureus to clindamycin isolates from bovine mastitis increased 246-fold in 20142020 compared with 19992003 [3], and the prevalence of extended-spectrum ß-lactamase enzyme (ESBL) E. coli isolates has ranged from 0.4% to 25.4% [4, 5]. These two MDR bacteria have been included in the priority ESKAPE pathogens (Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) list, which urgently need for a novel antibiotic to be discovered by the World Health Organization (WHO) since 2017 [6, 7]. Therefore, developing novel antimicrobial agents to overcome MDR bacteria may become imperative for global security in the near future.
Antimicrobial peptides (AMPs), an integral part of the natural immune system, have attracted increasing attention due to their high antibacterial activity and low drug resistance [8, 9]. Most importantly, unlike traditional antibiotics focusing on bacterial protein receptors to disrupt metabolism and cell growth, AMPs mainly interact with negatively-charged bacterial cell membranes due to their positive-charge and hydrophobicity. Consequently, it is highly challenging to compel microbes to modify their membrane components to resist AMPs [10, 11]. Therefore, the development of AMPs represents an effective direction to solve the problem of traditional antibiotic resistance. However, only a few AMPs (like Dalbavancin, Daptomycin, and Nisin) have been put into practical application and marketing [12]. Studies have shown that one of the major obstacles hindering the clinical application of AMP is the stability problem. Typically, AMPs are cleared from the blood by enzymatic degradation, renal filtration, and uptake by the reticuloendothelial system [13, 14].
Hence, techniques are urgently required to enhance the stability and prolong the half-life of AMPs [15]. As one of the most promising approaches, nanotechnology has been recognized as the last push toward the clinical use of AMPs [16, 17]. Among various delivery systems, the self-assembly strategy represents one of the most effective and appealing methods for alleviating the short half-life in vivo of AMPs and enhancing their bioavailability [18,21]. Self-assembled AMPs can exert their functions on various microstructures (nanoparticles, nanofibers, nanotubes, or hydrogels). Nanoparticles exhibit enhanced penetration and retention (EPR) effects, and nanofibers can further enhance the accumulation and retention at target sites based on assembly induced retention (AIR) effect [22]. As previously stated, Qi et al. designed in situ self-assembled antimicrobial peptide CPC-1 with a fourfold prolongation of the half-life at the site of infection in comparison to self-assembled nanoparticles CPC-2 (Table 1) [23]. Meanwhile, the formed nanofibers could also trap bacteria, such as the peptide HDMP (Table 1) [24, 25], exhibiting a dual-action antibacterial mechanism. The marine peptide N6, previously investigated in our laboratory, exhibited excellent antibacterial activity. However, it was poorly resistant to enzymatic degradation, especially trypsin, showing a short half-life in vivo [26]. Self-assembly technology has been widely verified to improve the stability of AMPs [12]. By self-assembly technology, Chen et al. increased the stability of AMP PTP-7 in serum from 2 h to more than 5 h [27] (Table 1). Han et al. designed the self-assembled peptide A-10 compared to the parent peptide, whose peptide retention in proteinase K and serum was significantly increased to 43% and 56%, respectively [28]. So it is logical to propose that combining multiple strategies, including in vitro pre-assembly and in vivo self-assembly, could effectively improve the bioavailability and therapeutic effect of N6.
Table 1.
Summary of the design, structure and property of self-assembled peptide discussed in this study
| Peptide | Design | Structure | Improvements | Ref |
|---|---|---|---|---|
| W-4 | wkwkwNGwkwkw-NH2 | Nanofibers | Reduce toxicity and increase stability | 14 |
| CPC-1 | Chitosan-GPLGVRGC-PEG2000- CGGG(KLAKLAK)2 | Nanoparticle to nanofiber transition induced by enzyme | Improve the stability and prolong the half-retention time | 23 |
| HDMP | BP-KLVFF -RLYLRIGRR | Nanoparticle to nanofiber transition induced by LTA | LTA-induced in situ self-assembling nanofibers trap bacteria | 24 |
| PTP-7 | FLGALFRALSRLL | Nanofiber | Dramatically alter activity and stability in comparison with single molecule CL-1 | 27 |
| BET | BP-KFFVLK-RLKLILKSK | Nanoparticle to nanofiber transition induced by LPS | Transform in situ from nanoparticles to nanofibrous networks to trap bacteria and induce aggregation | 32 |
| BTT2 | LKLKLKVpPTKLKLKL-NH2 | LTA and LPS as nucleation sites, inducing BTT self-assembly into nanonet | Bacteria-induced in situ self-assembly form nanonet trap-and-kill bacteria and display robust stability against trypsin | 34 |
| FF | FF | Nanofiber | FF as the minimal model for antibacterial supramolecular polymers | 35 |
| NPs1 | C14-(PF)4P-K(PEG8-NH2)(KP)5-NH2 | Nanoparticle | Display broad-spectrum antibacterial activity and high stability | 38 |
| SAP | (WVHH)3PG(HHVV)3-NH2 | Nanospheres to nanofiber transition triggered by pH | Display the entrapment property and excellent biocompatibility | 51 |
| Assembling Peptide | C14-(HHHF)4HHH-K-(PEG8)-QRKLAAKLT-NH2 | Nanofiber to nanoparticle transition triggered by pH | Exhibit pH responsiveness and high biocompatibility | 53 |
| RW-1 | RRRRWWWW | Micelle | High and rapid bacteria-killing activity | 54 |
| Defensin | HD6 | Nanofiber | HD6 self-assembly to form fibrils and nanonets that surround and entangle bacteria | 56 |
| Z(WR)2 | WRWRCNSKSFCRWRW | Nanofiber | LPS and LTA-induced nanofibers to trap bacteria | 57 |
Here, we designed self-assembled AMPs to treat MDR bacterial infections according to the principles of precise module design and the structureactivity relationship of AMPs [11, 29]. AMPs are designed using the following specific modules: (i) the hydrophobic module, fluorenylmethy (Fmoc), is a commonly used hydrophobic motif that facilitates the assembly process by providing hydrophobic forces to form nanoparticles [30]; (ii) the hydrogen bond module: KFFK, which originates from the core sequence of Alzheimer's disease protein Aß40 (KLVFFAE), provides hydrogen bonding forces and - stacking forces for the transformation of nanofibers [31, 32]; (iii) the antibacterial module: N6, exhibits strong antibacterial activity and acts as a hydrophilic terminal to regulate the hydrophilic/lipophilic balance (HLB) of self-assembled peptides [26]. Based on the above design strategy, we screened highly effective in situ self-assembled AMPs, evaluated their self-assembly capability, antibacterial ability, biosafety, stability and bactericidal mechanism in vitro, and verified the therapeutic effect in vivo using mastitis models (Scheme 1).
Scheme 1.
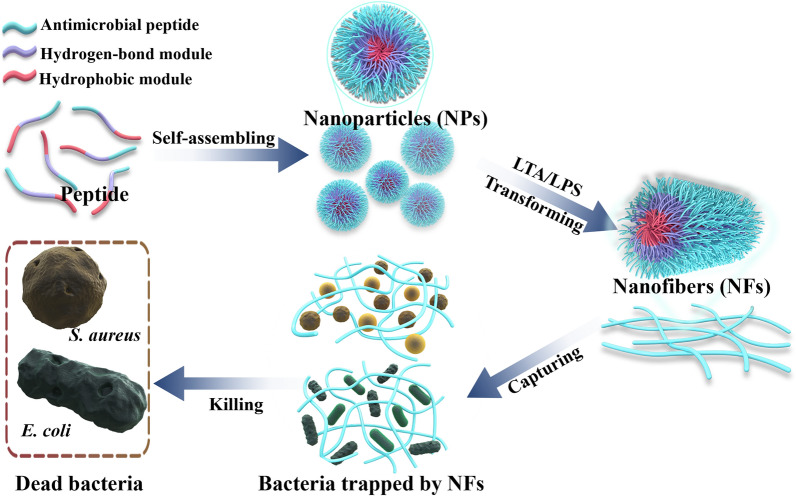
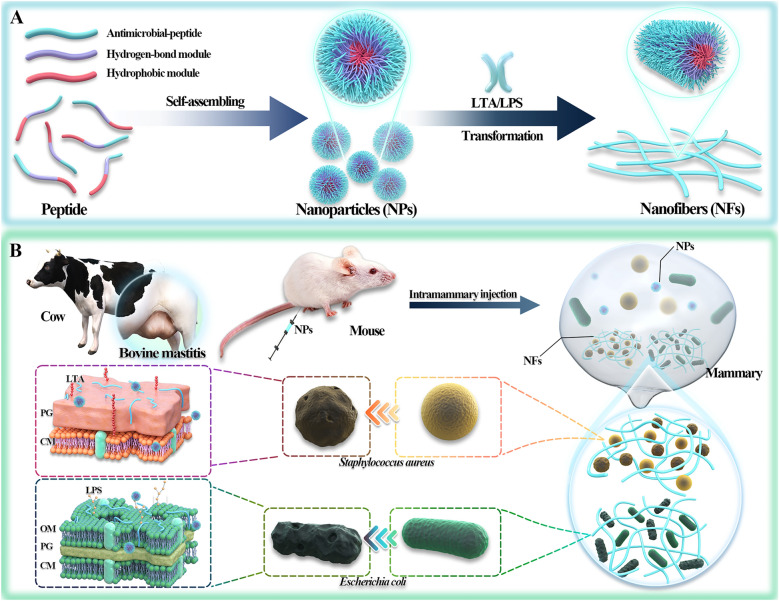
A Schematic illustration of the principle of bacteria-triggered transformation of self-assembled AMPs from nanoparticles to nanofibers. B Self-assembled peptides are administered via the mammary duct to treat mastitis caused by MDR S. aureus and E. coli. Bacteria-induced formation of nanofibers captures the bacteria and further kills the bacteria through membrane destruction mechanisms
Results and discussion
Design and self-assembled characterization of AMPs
Based on the comprehensive understanding of the precise module design theory of self-assembled peptide and antibacterial structurefunction relationships, the self-assembled AMPs were designed with different functional modules (hydrophobic, hydrogen-bonded self-assembly and antimicrobial modules) using the marine peptide N6 as a basis. Fluorescence spectroscopy, electron microscopy and circular dichroism (CD) were used to detect the self-assembly ability of the AMPs. Preliminary results indicate that hydrophobic forces, - stacking forces and hydrogen bonding, play important roles in facilitating self-assembly. The major bacterial cell wall components, lipopolysaccharides (LPS) and lipoteichoic acid (LTA) act as prominent amyloid core factors to promote the self-assembly of target peptides.
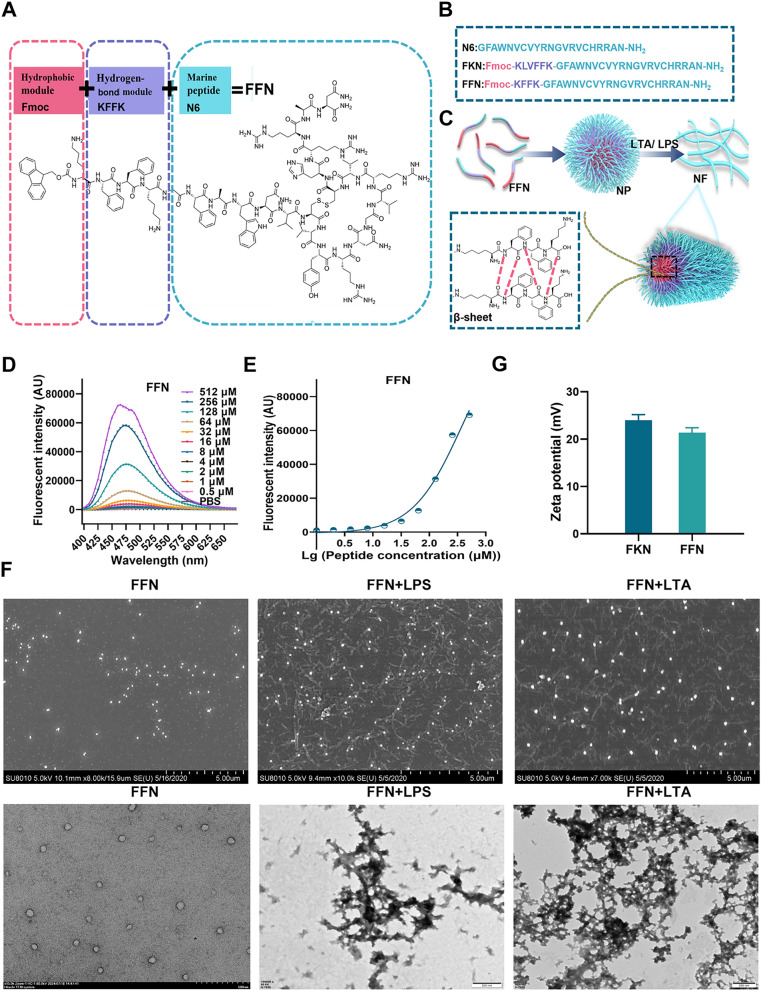
To investigate the molecular design and assembly behaviour, two AMPs on the basis of N6 (GFAWNVCVYRNGVRVCHRRAN-NH2) were modularly designed and solid-phase synthesized, which respectively were: FKN (Fmoc-KLVFFK-GFAWNVCVYRNGVRVCHRRAN-NH2, FKN) and FFN (Fmoc-KFFK-GFAWNVCVYRNGVRVCHRRAN-NH2, FFN) (Fig. 1A-C, Table S1). Their molecular weights were identified using matrix-associated laser desorption ionization time-of-flight mass spectrometry (MALDI-TOFMS) (Figure S1-S3 A). The final purities of N6, FKN and FFN were 95.0%, 95.2% and 96.2%, respectively. (Figure S1-S3 B).
Fig. 1.
Chemical structure and self-assembly properties of AMPs. A The molecular structure of peptides is designed as hydrophobic modules, hydrogen bond modules and antimicrobial peptide modules. B Amino acid composition of the designed peptides. C Diagram of the transition mode from nanoparticles to nanofibers. D The fluorescence spectra of FFN. E The CMC of FFN. F SEM and TEM images of self-assembled peptides only and self-assembled peptide FFN (80 µM) in LPS or LTA (1 M). G Zeta potential of self-assembled peptides
Firstly, the critical micelle concentration (CMC) of each AMP in PBS was determined using 1-phenylamino-8 sulfonate (ANS) fluorescent dye. As a hydrophobic fluorescent probe, ANS does not emit fluorescence in polar solvents. Still, its fluorescence intensity is notably enhanced in hydrophobic environments, making it suitable for determining the CMC of self-assembled AMPs. As shown in Fig. 1D, and Figure S4A, the fluorescence intensities of FKN and FFN at 480 nm exhibited a notable rise as the concentration rose. The CMCs of FKN and FFN were 7.94 and 35.46 µM, respectively, indicating that FKN and FFN can self-assemble (Fig. 1E and Figure S4B). Conversely, the fluorescence intensities of N6 exhibited a direct correlation with concentration, suggesting that N6 is incapable of self-assembling. To further monitor the structure of self-assembled AMPs, scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were utilized to evaluate the morphologies. As shown in Fig. 1F and Figure S4C, D, E, FKN and FFN self-assembled to form nanoparticles, which were characterized with particle sizes of 69.78±11.56 nm and 70.67±15.60 nm, respectively. The zeta-potentials of the FKN and FFN were 24 and 21.3 mV, respectively, suggesting that these AMPs could strongly bind to negatively charged bacterial membranes (Fig. 1G). Moreover, to verify the alteration in morphology of the self-assembled peptide following its interaction with components of the bacterial cell wall, self-assembled peptides (80 µM) were co-incubated with LPS or LTA. The fluorescence intensity of these AMPs, particularly FFN, was significantly increased upon their interaction with LPS or LTA (Figure S5A). This was further confirmed by SEM and TEM, FKN and FFN underwent a gradual transformation from nanoparticles to nanofibers when exposed to LPS or LTA for 6 h (Figure S4C and Fig. 1F). The morphology of FFN was gradually changed with LPS/LTA induction, multiple short nanofibers were visible at 12 h, and completely converted to nanofibers at 72 h (Figure S6). In contrast to N6, the hydrophobic heads of FKN and FFN aggregated to form nanoparticles and then gradually changed from hydrophobic to hydrogen bonding forces and -stacking forces. The morphology of self-assembled AMP FFN gradually changed from nanoparticles to nanofibers, which was basically consistent with the self-assembly behaviour of in situ self-assembling peptides HDMP [24] and BET [32], where HDMP completely transformed from nanoparticles to nanofibers after 48 h of LTA induction and BET completely transformed from nanoparticles to nanofibers after 72 h of LPS induction (Table 1). LPS/LTA, as prominent amyloid nucleators in the bacterial membrane, transforms the dominance of hydrophobic forces to hydrogen bonding forces and accelerates the conversion of nanoparticles to nanofibers in FKN and FFN [33, 34] (Table 1). Moreover, the significant disparity between the FKN and FFN underscores the substantial role played by the integrity of KLVFFK in the process of self-assembly. KLVFFK, derived from the core motif of Alzheimer's disease Aß40 (KLVFFAE), exhibits a stronger aggregation capacity than KFFK, mostly due to greater hydrogen bonding and -stacking force. The smallest nano-self-assembling motif, FF, has a significantly decreased self-assembly capacity than KLVFFK. This is attributed to the reduction of hydrogen bonding force, as shown by the CMC of 760 µg/mL [35] (Table 1).
Subsequently, the self-assembly behaviours of AMPs were characterized using ultraviolet spectroscopy (UVvis). As shown in Figure S5B, N6 presents UVvis absorption spectra in the range of 200300 nm, with a peak value of 275 nm. Compared to N6, FKN and FFN exhibit larger UVvis absorption peaks, broader spectral bands, and a blueshift at 275 nm, with a peak at 268 nm. The primary reason for this phenomenon is the hydrogen bonding forces of KLVFFK in FKN and KFFK in FFN. These forces shorten the intermolecular distance and enhance the intermolecular force, causing the absorption band of the molecule to shift towards shorter wavelengths [36]. Meanwhile, LPS slightly affected the UV spectra of all three peptides, but LTA had a greater effect on the spectra of FKN and FFN. Since the secondary structure affects AMP self-assembly and bioactivity, the CD and fourier transform infrared spectrometer (FTIR) analyses were performed to compare the secondary structure of each peptide. As shown in Figure S5C, the percentages of ß-sheet in N6, FKN and FFN were 35.24%, 40.45% and 38.82%, respectively. After incubation with LPS, the corresponding ß-sheet percentages of FKN and FFN increased to different extents, whereas the changes in N6 were negligible (Table S2). The confirmation of the ß-sheet structure of nanofibers was additionally validated using FTIR analysis. N6, FKN and FFN showed strong absorptions at approximately 1635 cm1 (C=O stretching vibration), 1529 cm1 (benzene skeleton vibration) and 3294 cm1 (OH and NH stretching vibration) (Figure S5D). The absorption at 1635 cm1 is specifically linked to the distinctive vibration of the - sheet structure [36]. However, N6 did not present a peak at 1194 cm1, possibly due to a deficiency of FF amino acids (Figure S5D). Furthermore, the affinity between the peptides and LPS or LTA was determined using the BC probe. The result has shown that the affinity of the self-assembled peptide FKN and FFN to LPS of E. coli and LTA of S. aureus exhibited a dose-dependent effect, which was even greater than that of N6 (Figure S5E and S5F), which is consistent with previous studies of SCPs and self-assembled peptide NPs-1 [37, 38] (Table 1).
2.2 Bioactivity of self-assembling AMPs in vitro
Bioactivity is an important property of self-assembled peptides. This part of the study analyzed the antibacterial activity and rate of self-assembled AMPs against G (E. coli) and G+ (S. aureus) by minimum inhibitory concentration (MIC), bactericidal kinetics (time and dose), mutant prevention concentrations (MPCs) and postantibiotic effect (PAE). The results demonstrated that FFN displayed superior antibacterial, a reduced MPC and an extended PAE, which can reduce the drug dose and prolong the dosing interval and be preliminarily used as a candidate therapeutic drug.
Firstly, to evaluate the in vitro bioactivity of the self-assembled AMPs, E. coli CGMCC 1.90026 and S. aureus CGMCC 1.90032 as test strains were used as test strains, and the standard strains E. coli ATCC 25922 and S. aureus ATCC 43300 were used as control strains. Initially, the clinical isolates were identified for drug resistance. The results revealed that E. coli CGMCC 1.90026 only exhibited sensitivity to polymyxin B, amikacin and developed the resistance genes gyrA, gyrB and parC (Table S3). S. aureus CGMCC 1.90032 developed resistance to penicillin, ampicillin, amoxicillin clindamycin, norfloxacin, sulfafurazole and bacitracin, and evolved the resistance genes blaZ, braRS and atpA (Table S4).
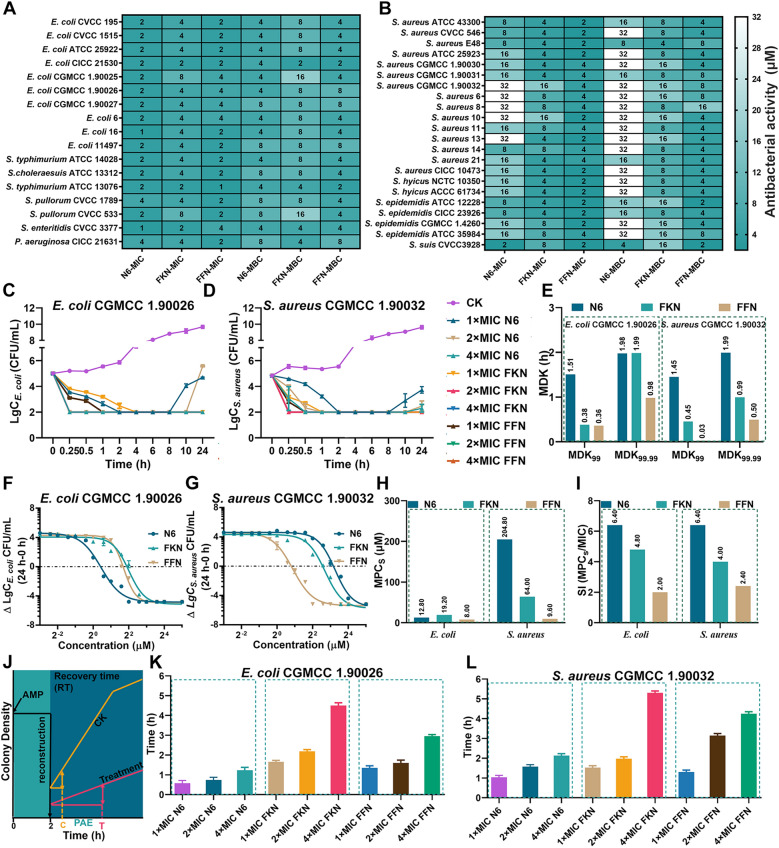
Secondly, the antibacterial efficacy of each AMP against G+ and G bacteria was evaluated by the MIC, minimum bactericidal concentration (MBC), time-kill kinetics and dose-kill kinetics. As shown in Fig. 2A, B and Table S5, in terms of antimicrobial activity, the geometric mean MICs of N6, FKN and FFN against all tested strains were 6.02, 4.78 and 2.52 µM, respectively. Specifically, the geometric mean of MICs of N6, FKN, and FFN against G being 2, 3.84 and 2.35 µM, respectively, and against G+ being 15.43, 5.98 and 2.78 µM, respectively. Since the antimicrobial activity and selectivity of AMPs are related to the ratio of cationic and hydrophobic, we inferred that the expansion of the antimicrobial spectrum of FKN and FFN was mainly attributed to the cationic properties of lysine (K) and the hydrophobicity of Fmoc, leucine (L), valine (V) and phenylalanine (F) [13]. The differences in antibacterial activity between FKN and FFN may be due to the alterations in secondary structure leading to intense aggregation [39, 40]. The time-antibacterial kinetics was determined to further compare the bactericidal rate and persistence of N6, FKN and FFN against S. aureus and E. coli (clinical isolates and standard strains) [41]. As depicted in Fig. 2C, E, after treatment with 1×, 2×or 4×N6, FKN or FFN, the E. coli CGMCC 1.90026 cells were completely eradicated within 4 h, as evidenced by the MDK99 and MDK99.99 ranging from 0.361.51 h and 0.981.99 h, respectively. Notably, N6 (1×and 2×MIC) exhibited a resurgence after 8 h. The time-kill curves of all AMPs toward E. coli ATCC 25922 showed similar effects, except that all AMPs demonstrated slightly accelerated rates of killing, as seen by MDK99 and MDK99.99 values falling within the range of 1.95 h (Figure S7A, C). This is possibly due to the greater resistance observed in clinical strains. The time-kill kinetics of AMPs against S. aureus CGMCC 1.90032 were shown in Fig. 2D, E, various concentrations of FKN and FFN could completely kill S. aureus CGMCC 1.90032 within 1 h, which was manifested by MDK99 and MDK99.99 range of 0.030.45 h and 0.500.99 h, respectively, and the effect was significantly greater than that of N6 (MDK99:1.45 h, MDK99.99:1.99 h). In contrast, they displayed a slower bactericidal rate against S. aureus ATCC 43300, with ranges of 0.291.95 h and 0.95 h for MDK99 and MDK99.99, respectively (Figure S7B, C). In this experiment, different concentrations (3/12816×MIC) of N6, FKN and FFN were added for 24 h to construct dose-dependent curves. The results showed that all the AMPs exhibited bactericidal effects in a concentration-dependent manner. Self-assembly did not affect the activity (Figure S7D, E, Figure S8A, B). Notably, FFN had a small EC50 against both E. coli and S. aureus (Table S6) and a relatively steep dose curve, which may imply that it is more difficult to evolve resistance to FFN [15].
Fig. 2.
In vitro antimicrobial activities of N6, FKN and FFN. A Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of N6, FKN and FFN against G bacteria (n=3). B The MICs and MBC of N6, FKN and FFN against G+ bacteria (n=3). C Time-kill curves of N6, FKN and FFN against clinical isolates E. coli CGMCC1.90026 (n=3), CK: PBS group. D Time-kill curves of N6, FKN and FFN against clinical isolates S. aureus CGMCC1.90032 (n=3), CK: PBS group. E The MDK99 and MDK99.99 of N6, FKN and FFN against clinical isolates E. coli CGMCC 1.90026 and S. aureus CGMCC 1.90032. F Schematic diagram of the test procedure for the postantibiotic effect. G Postantibiotic effects of 1×, 2×and 4×MIC of N6, FKN and FFN against clinical isolates E. coli CGMCC 1. 90026 (n=3). H Postantibiotic effects of 1×, 2×and 4×MIC of N6, FKN and FFN on clinical isolates S. aureus CGMCC 1.90032 (n=3). Results were given as mean±SD
Finally, the MPCs and PAE were determined to guide clinical drug dosing further. When directing clinical dosage, MPC is a crucial pharmacodynamic parameter [42]. To assess the persistence of AMPs resistance, the MPCs of AMPs were measured. As shown in Figure S8C, the MPCs of N6, FKN and FFN against the mutant E. coli CGMCC1.90026 ranged from 8.0 to 19.2 µM, which were comparable to those of the standard strain E. coli ATCC 25922 (4.825.6 µM) (Figure S7F). The MPCs of N6, FKN, and FFN against S. aureus CGMCC1.90032 ranged from 9.6204.8 M (Figure S8C), which were slightly greater than those of the standard strain S. aureus ATCC 43300 (6.451.2 µM) (Figure S7F). To further analyze the resistance mutation selection ability of all AMPs, the resistance selection index (SI) was calculated as the MPCs/MIC. The smaller the SI is, the stronger the resistance mutation selection ability of drugs to inhibit the development of bacterial resistance [42]. As shown in Figure S8D, the SI of FKN and FFN against E. coli CGMCC 1.90026 ranged from 2.0 to 4.8, slightly smaller than that of N6 (6.4). The SI of FKN and FFN against S. aureus CGMCC 1.90032 ranged from 2.4 to 4, somewhat smaller than that of N6 (6.4), suggesting that self-assembled AMPs might reduce the occurrence of drug-resistant bacteria. No significant difference was observed in SI indices between the standard and clinical isolates (Figure S7G). The PAE reflects the duration of the antibacterial effect on bacterial growth inhibition after a short period of contact with bacteria [43]. As shown in Fig. 2G, the PAE of FKN and FFN against E. coli CGMCC 1.90026 ranged from 1.35 to 4.50 h, similar to that of E. coli ATCC 25922 (1.334.47 h, Figure S7H). Similarly to those of S. aureus ATCC 43300, the PAE of FKN and FFN against S. aureus CGMCC 1.90032 ranged from 1.30 to 5.30 h, which were superior to the corresponding concentration of N6, (Fig. 2H, 1.844.41 h, Figure S7I). Based on the fact that PAE is associated with the binding kinetics and transport mechanism between antimicrobial drugs and their targets [44]. Here, it is reasonable to speculate that the prolongation of the self-assembled AMPs PAE is mainly due to the enhancement of its non-specific binding ability to bacteria, which is consistent with the results shown in Figure S5E, F.
Fig. 8.
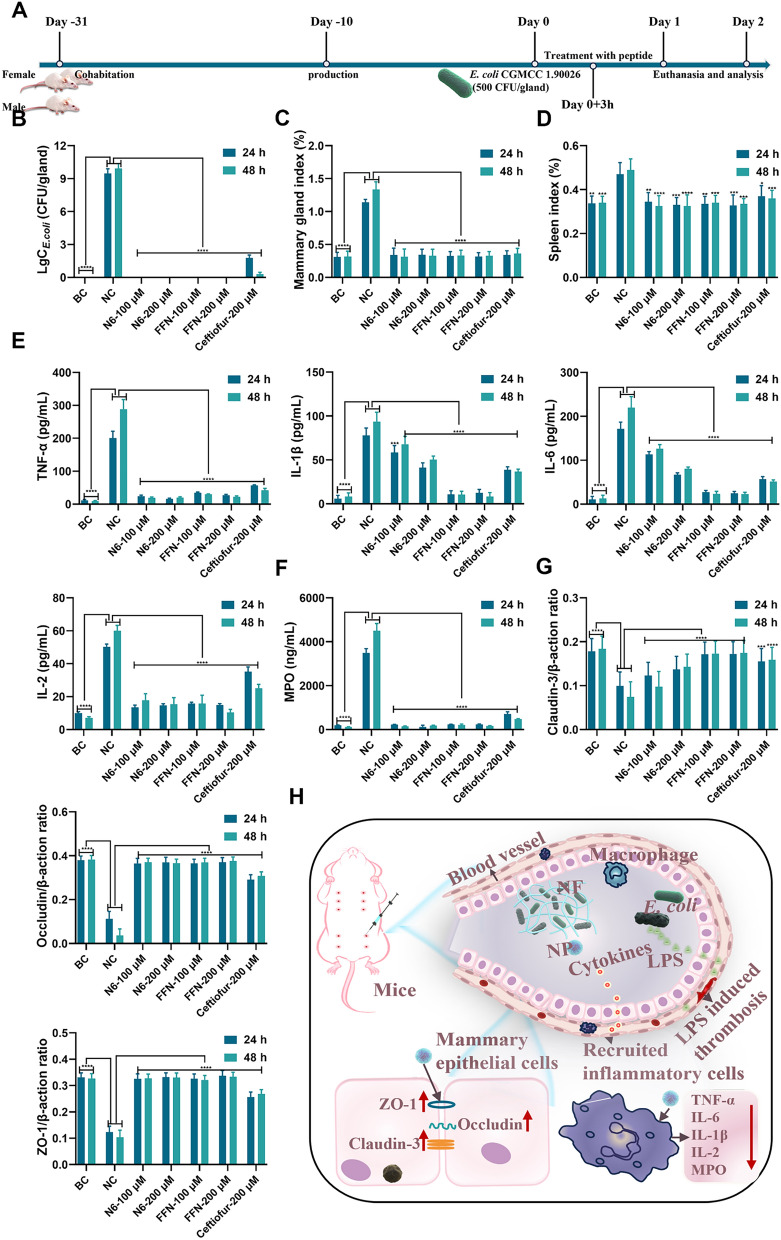
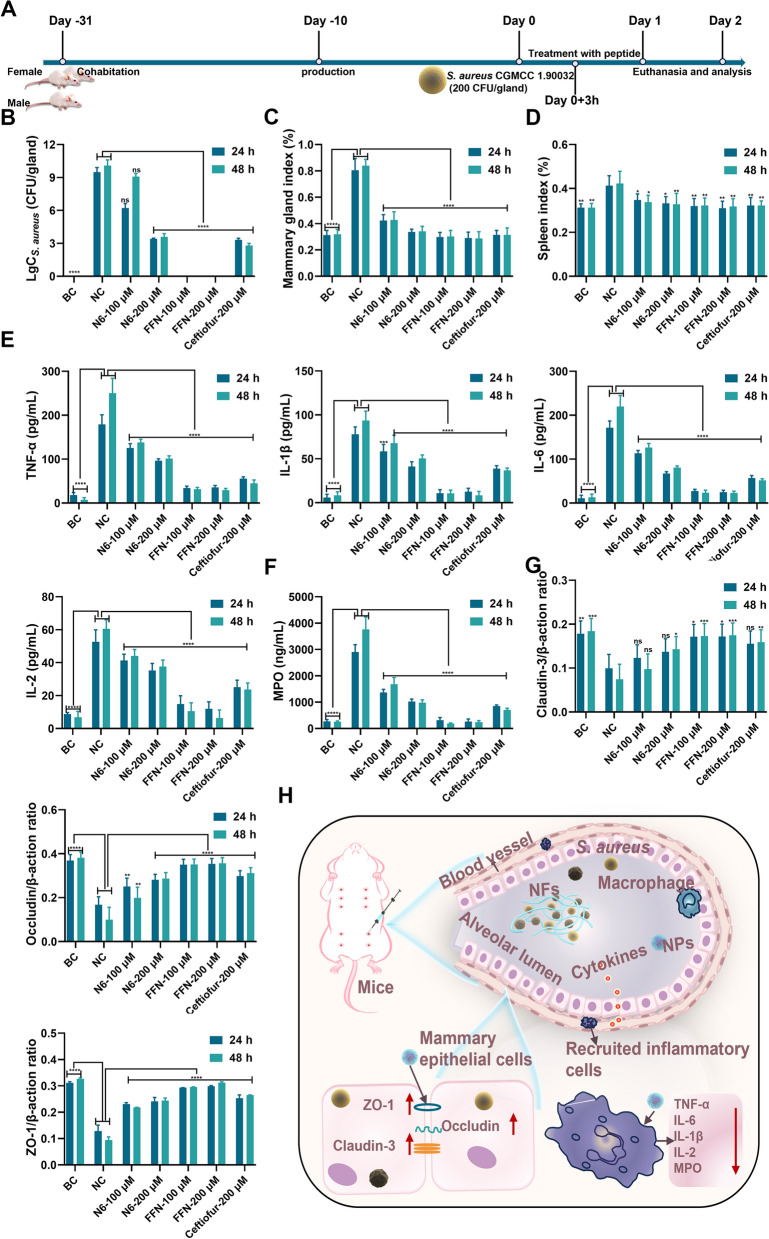
In vivo efficacy in mouse mastitis model caused by E. coli CGMCC 1.90026. A. Schematic diagram illustrating the experimental procedure for testing the treatment of E. coli CGMCC 1.90026 induced mastitis in mice. B. E. coli CGMCC 1.90026 bacterial load assessment in a murine mastitis model (n=8). C. Evaluation of mammary organ index in a mouse model of mastitis caused by E. coli CGMCC 1.90026 (n=8). D. Assessment of spleen organ index in a murine model infected with E. coli-induced mastitis (n=4). E. Inflammatory factors (TNF-, IL-1ß, IL-6 and IL-2) in the mammary tissue in a mouse mastitis model infected with E. coli (n=4). F. MPO in the mammary tissue in a mouse mastitis model infected with E. coli (n=4). G. Examination of tight junction protein expression in the mammary barrier in a murine model of mastitis induced by E. coli (n=4). H. Schematic representation depicting the treatment approach for E. coli-induced mastitis using self-assembling peptides. Blank control (BC): uninfected, negative control (NC): infected and untreated. Results were given as mean±SD. *: p<0.05, **: p<0.01, ***: p<0.0005, ****: p<0.0001
Preliminary biocompatibility evaluation of self-assembled antimicrobial peptides in vitro and in vivo
Biocompatibility intuitively reflects the toxic effects of materials on organisms, and this section demonstrates that self-assembled AMPs have good biosafety at the tested concentrations and can be used as safe antimicrobial candidates through in vitro hemolytic, cytotoxic and in vivo injection (intraperitoneal and mammary injections) tests.
Firstly, the hemolysis and cytotoxicity were determined to assess the biocompatibility of the self-assembled AMPs in vitro. As shown in Figure S9A, N6 (128 μM) showed no hemolysis, whereas FKN (128 M) and FFN (128 M) had hemolysis rates of 15.8% and 6.08%, respectively. This indicated that FKN was not suitable for intravenous administration. Subsequently, human keratinocytes (HaCaT), mouse mononuclear macrophage cells (RAW 264.7) and mammary epithelial cells (MAC-T) were used to evaluate the cytotoxicity of the self-assembled AMPs. In vitro compatibility tests revealed that N6 and FFN had the best biocompatibility, as evidenced by their high cell viability for HaCaT (Figure S9B), RAW 264.7 (Figure S9C) and MAC-T cells (Figure S9D) (>90%), and almost no cytotoxicity. In contrast, the viability of FKN lagged, with a corresponding cell viability range of 83.74%-89.6%. The therapeutic index of each peptide was utilized to evaluate the antibacterial activity and hemolysis comprehensively, and the results revealed that the therapeutic index of N6, FKN and FFN were 42.52, 26.78 and 101.59, respectively, and FFN displayed the best therapeutic potential (Table S5). Such a large discrepancy in biocompatibility between the FKN, FFN and N6 is attributed to the hydrophobic molecule Fmoc. Increased hydrophobicity inevitably leads to increased hemolysis and cytotoxicity, as the hydrophobic motif indiscriminately kills mammalian and bacterial cells, resulting in high toxicity to the host [45]. However, increasing hydrophobicity is an important strategy to optimize antimicrobial activity and self-assembly capacity [46]. Therefore, choosing the appropriate cationic/hydrophobicity ratio is challenging for a delicate balance of antimicrobial activity, hemolysis and self-assembly ability in the design of self-assembled AMPs.
Afterwards, to further assess the in vivo biocompatibility of the self-assembled AMPs, we performed intraperitoneal and mammary injections to evaluate the systemic and local safety of the self-assembled AMPs in mice. The clinical symptoms were rated according to pain scoring criteria (Table S7). Only the FKN group (10 mg/kg) showed a transient back arch after administration. In contrast, the other groups showed no symptoms of discomfort or pain during the entire administration cycle (Table S8). None of the groups showed significant weight changes compared to the BC group (Figure S9E). After three days of continuous administration of two different doses of AMPs (5 mg/kg or 10 mg/kg), the whole blood cell spectrum and serum biochemical indices of mice in each treatment group were within the normal range, with no significant difference when compared to those in the BC group (Table S9). This observation indicated that the tested AMPs doses did not cause acute hepatic and renal injury, as evidenced by liver and kidney tissue sections (Figure S10). The results of the local safety test revealed no alterations in clinical symptoms, serum biochemical indices and whole blood cell spectra after 3 days of continuous AMP administration (Figure S9F, Figure S10, Table S10). This showed that all AMPs were nontoxic at the tested concentrations, and even FKN did not exhibit strong toxicity. Further systematic studies are required to validate the safety of FKN's intravenous administration in the future.
Stability evaluation of self-assembled AMPs
Stability is the greatest challenge limiting the application of AMPs. In this section, the stability of self-assembled AMPs was assessed by enzyme stability, and metabolism time in tissues, and it was confirmed that self-assembly can be used as an effective strategy to improve the AMPs stability and the assembly-induced retention (AIR) effect of nanofibers can further improve the stability compared to nanoparticles.
The stability of AMPs is a critical factor restricting the clinical translation of peptide drugs. Here, the improvement in the stability of AMPs was evaluated. As shown in Figure S11A, the stability of all AMPs in various physiological salt environments revealed that monovalent cations (Na+ and K+) had no significant effect on the MICs of all AMPs, whereas Mg2+ and Ca2+ elevated the MICs of all AMPs by 1.344 times. This could be due to the competitive binding of hypervalent cations to LPS or LTA, which increases the hardness of the cell membrane and, ultimately, reduces antimicrobial activity [47]. After that, the serum stability of all AMPs was evaluated, and it found that 12.5% and 25% serum had less effect on AMPs MICs, with all AMPs having a stable MIC after incubated with 12.5% serum for 8 h, FKN and FFN even a stable MICs but N6 increased their MIC by twofold after 8 h of incubation in 25% serum. After incubation in 50% serum for 8 h, the MIC of each peptide increased by 13.33 times (Figure S11B).
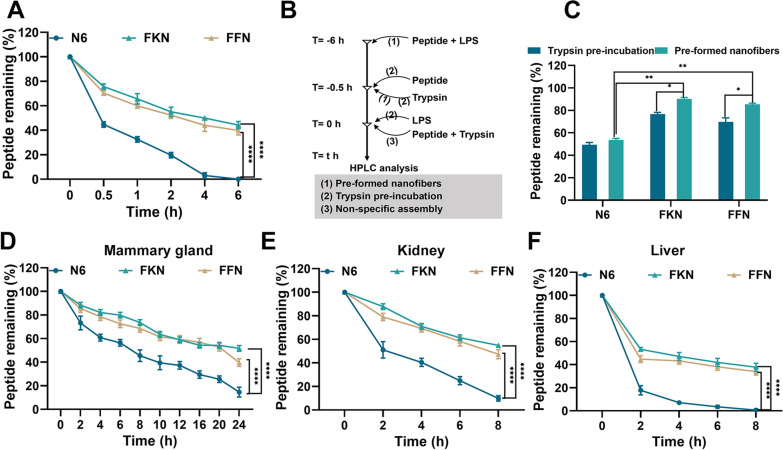
Trypsin is the primary digestive enzyme in vivo, and its breakdown of AMPs significantly reduces their bioavailability [48]. To evaluate the anti-tryptic hydrolysis potential of the self-assembled AMPs, we monitored the time-dependent degradation curves by HPLC. As shown in Fig. 3A and Table S12, after co-incubation with trypsin for 6 h, the peptide retention rates of FKN and FFN were 44.2% and 39.8%, respectively. In contrast, N6 was rapidly degraded by more than 50% after 0.5 h of co-incubation and completely degraded after 4 h. This observation supported that self-assembly of peptides significantly improved their anti-proteolytic ability, possibly due to the steric hindrance generated by self-assembly effectively preventing trypsin from cutting Arg and Lys [49]. To quantify the anti-tryptic hydrolysis ability of the nanofibers and nanoparticles, the peptide retention rates of the LPS-induced preformed nanofibers and self-assembled peptides after 0.5 h of trypsin incubation using HPLC (Fig. 3B). As shown in Fig. 3C, after the addition of LPS, the protein retention rates of FKN and FFN nanofibers were 90.1% and 85.3% (0.5 h), respectively, which were substantially greater than those of FKN and FFN nanoparticles (76.63% and 69.78%, respectively). This suggested that the nanofibers were more resistant to tryptic degradation than nanoparticles, possibly due to the assembly-induced retention effect of nanofibers enhancing their stability [14, 23, 34] (Table 1), which is consistent with previously reported results on the tryptic stability of the self-assemble AMP BTT2, where the peptide retention of BTT2 in trypsin was maintained in the range of 90100% by LPS-induced pre-assembly [34]. To evaluate the therapeutic potential of self-assembled AMPs, we assessed the metabolic stability of AMPs in mouse mammary glands, liver and kidney and monitored the time-dependent metabolic curve of peptides in tissue homogenates by HPLC. As shown in Fig. 3D, E and F and Table S13-S15, the self-assembled peptides FKN and FFN had great metabolic stability, as evidenced by peptide retention rates in the mammary, kidney and liver tissues being 39.65% and 51.61% (24 h), 47.37% and 54.88% (8 h), 33.93% and 37.74% (8 h), respectively. While the retention rate of N6 in the mammary maintained below 14.5%, N6 was virtually metabolized in the metabolic organs kidney and liver, with a retention rate of less than 9.76%. In summary, as previously reported [23, 29, 34] (Table 1), the self-assembly approach can successfully confer metabolic stability to AMPs, prolonging their half-lives by 57.564.07 times (Fig. 3F, 8 h).
Fig. 3.
Stability evaluation of self-assembled AMPs. A Stability of N6, FKN and FFN against 1 w/v% trypsin was monitored by HPLC (n=3). B The experimental protocol describes the time points at which different components (80 M peptide, 1 M LPS or 10 M trypsin) were added to the incubation mixtures under different conditions: (1) LPS was added to co-incubate with the AMP to generate pre-formed nanofibers before the addition of trypsin; (2) the peptide was pre-incubated with trypsin before the addition of LPS; and (3) non-specific assemblies were recorded in terms of the ratio of (1) or (2) to (3) to record peptide retention. C Peptide retention of N6, FKN and FFN under different conditions (n=3). D-F Temporal metabolic curves of N6, FKN and FFN in mammary /kidney/liver tissue (50 mg/mL), respectively (n=3). Results were given as mean±SD. *: p<0.05, **: p<0.01, ****: p<0.0001
Based on the comprehensive evaluation of AMP activity, biocompatibility and stability, we selected self-assembled AMP FFN for subsequent studies, with N6 serving as the AMP control.
Mechanism of self-assembled AMPs
Since AMPs have a variety of bactericidal mechanisms, this section explores the bactericidal mechanism of self-assembled AMPs through electron microscopy, fluorescence observation and membrane fluidity tests. The analysis showed that the self-assembled AMPs were triggered by LPS/LTA in the E. coli/S. aureus cell wall, formed nanofibers and captured bacteria, and further inserted into the cell membrane to cause leakage of contents and bacterial death.
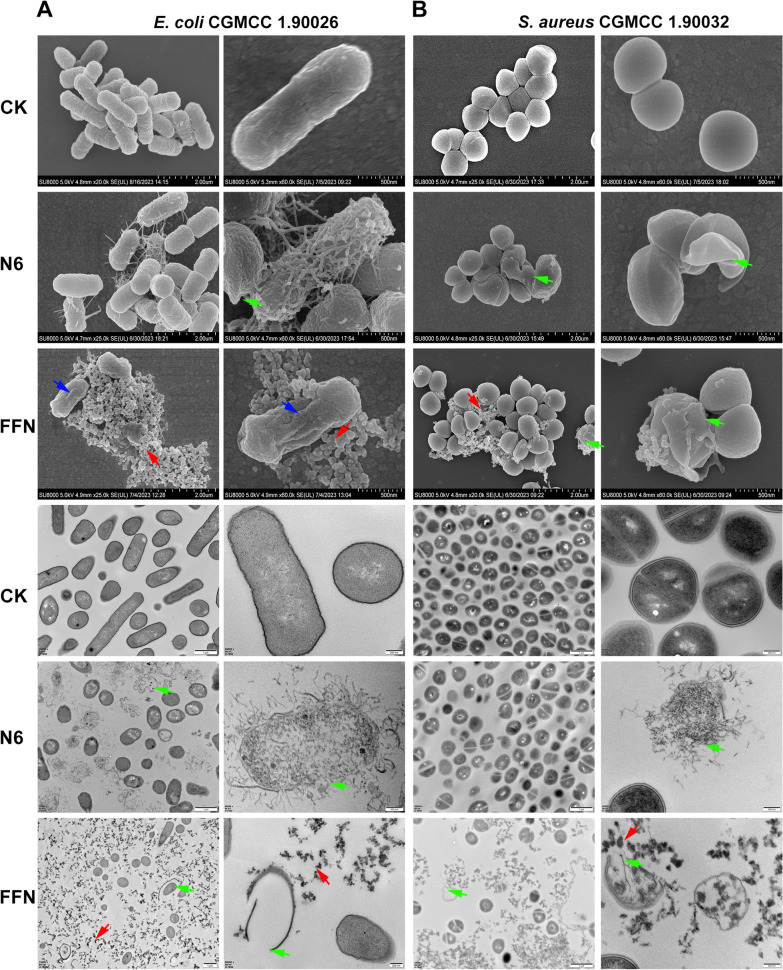
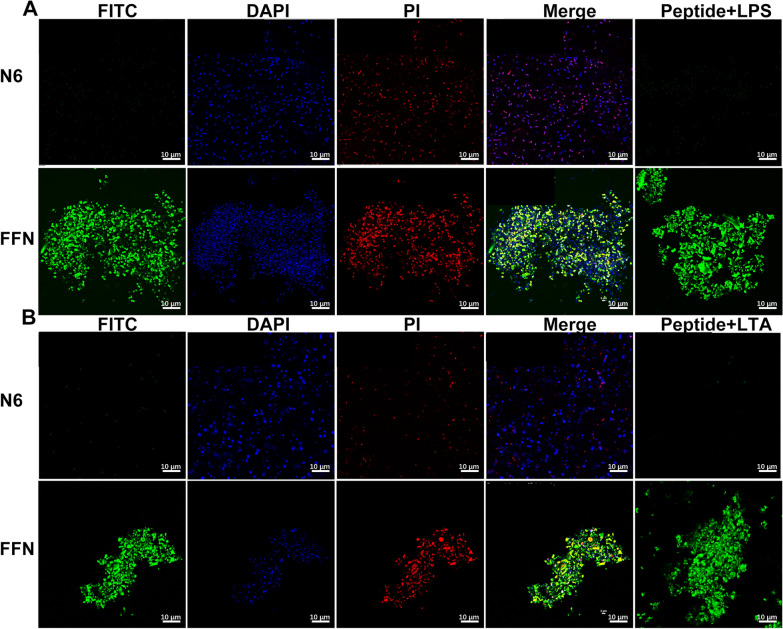
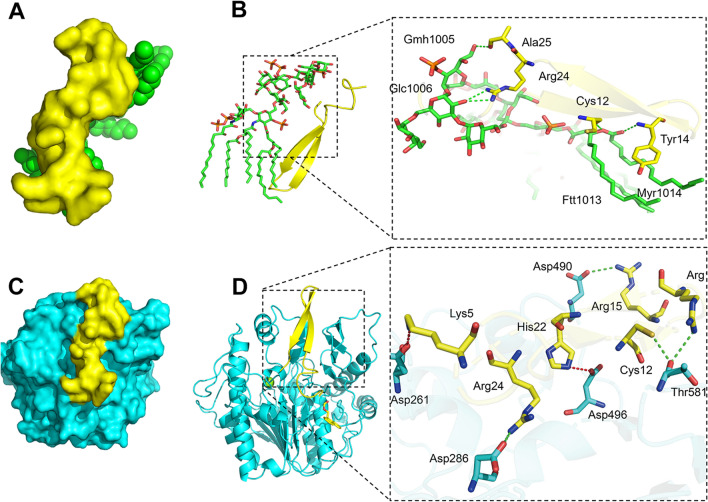
The mechanism of AMPs mainly focuses on their interaction with anionic bacterial cell membranes. Therefore, we first observed the effect of self-assembled AMPs on the structure of bacterial cell membranes by SEM and TEM. After treatment with N6 or FFN for 2 h, G bacteria (E. coli CGMCC1.90026 and E. coli ATCC 25922) mainly exhibited cell membrane crumpling, extensive membrane fusion and rupture (Fig. 4A, Figure S12A), and G+ bacteria (S. aureus CGMCC 1.90032 and S. aureus ATCC 43300) showed characteristic cell membrane rupture, and content leakage, but N6 was less effective against G+ bacteria (Fig. 4B, Figure S12B). In addition, it is worth noting that FFN successfully formed bacterial-trapping nanofiber networks when incubated with E. coli and S. aureus (red arrow), while N6 did not form nanofibers, which is consistent with self-assembled HDMP [24, 50] (Table 1). To further validate that the nanofibers were formed from self-assembling AMPs rather than bacterial secretions (biofilms), we used live/dead cell staining methods for confocal laser scanning microscope (CLSM) observation. As shown in Fig. 5 and Figure S13, after incubation with E. coli and S. aureus for 2 h, the FITC-labelled FFN formed distinct green clusters, which was consistent with what was observed for the FITC-labelled FFN after incubation with LPS or LTA. The cells co-located with the green nanofibers had live (blue) and dead (red) cells, indicating that the self-assembled AMPs could successfully capture and kill the bacteria. The capture of bacteria by the nanofibers was further demonstrated in the bacterial agglutination assay (Figure S14), consistent with the bacterial capture function of the self-assembling peptide SAP and NATs [51, 52] (Table 1), where 250 g/mL SAP formed a distinct precipitate after 8 h of co-incubation with E. coli ATCC 25922. After incubation with FFN for 4 h, no viable bacteria were detected in E. coli and S. aureus supernatant. Still, viable bacteria were detected in the precipitation, indicating that the nanofibers effectively trapped bacteria. In contrast, after incubation for 12 h, no viable bacteria were detected in the supernatant and precipitation, indicating that the nanofibers effectively killed bacteria. At the same time, compared with FFN, N6 only showed antibacterial activity (Table S16). In this study, the interaction modes of FFN with LPS and LTA were further simulated by molecular docking. As shown in Fig. 6, hydrogen bonds were formed between FFN and LPS at Cys12-FTT1013 (A), Tyr14-MYR1014 (A), Arg24-GLC1006 (A) and Ala25-GMH1005 (A) (Fig. 6B). Hydrogen bonds were formed between FFN and LTA at Arg24-Asp286, Arg19-Thr581, Cys12-Thr581 and Arg15-Asp490, and salt bridges were formed between Lys5-Asp261 and His22-Asp496 (Fig. 6D). The increase in hydrogen bonding forces promoted the formation of nanofibers, which is consistent with previous studies [25].
Fig. 4.
Observation of the effect of N6 and FFN on the bacterial cell membrane and morphology. A SEM and TEM images of E. coli CGMCC 1.90026 after treatment with N6 or FFN (80 μM) for 2 h. B SEM and TEM images of S. aureus CGMCC 1.90032 after treatment with N6 or FFN (80 M) for 2 h. The arrows indicate specific symptoms. Red arrow: nanofibers; blue arrow: cell shrinkage; green arrow: cell membrane rupture. CK: PBS group
Fig. 5.
Staining of live/dead bacteria cells tripped by self-assembled nanofibers. A Confocal microscope images of clinically isolated E. coli CGMCC1.90026 (1×109 CFU/mL) or LPS (1 µM) co-incubated with FITC-labelled N6 and FFN (80 µM). B Confocal microscope images of clinically isolated S. aureus CGMCC 1.90032 (1×109 CFU/mL) or LTA (1 µM) co-incubated with FITC-labelled N6 and FFN (80 µM)
Fig. 6.
Molecular docking (MD) of the self-assembled antimicrobial peptide FFN with LPS/LTA. A: Complex structure of FFN and LPS molecules. B: 3D mode of action between FFN and LPS. C: Complex structure of the FFN with LTA molecules. D: 3D mode of action between FFN and LTA. Green dashed line: hydrogen bond; Red dotted line: Salt bridge
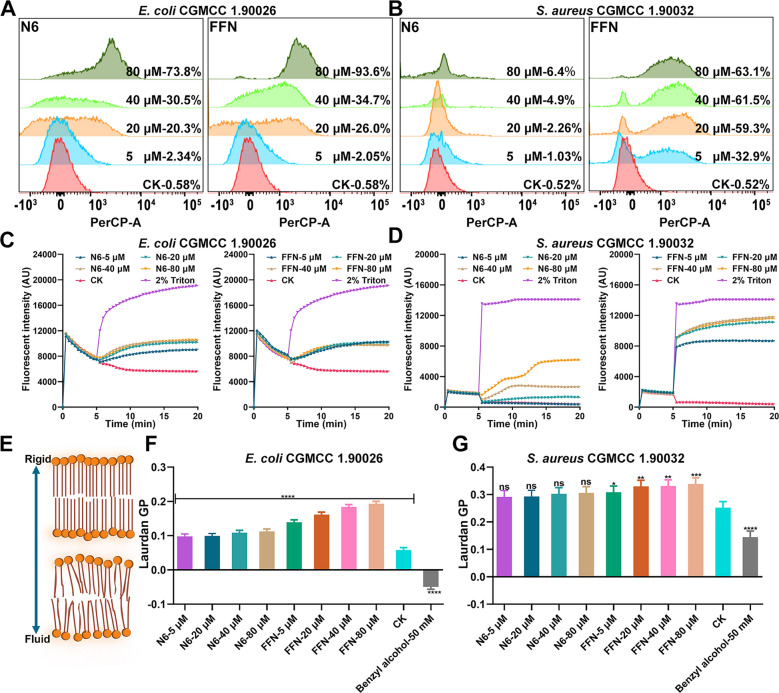
The above results indicate that each AMP treatment induces substantial perturbations and severe morphological disruptions, suggesting that bacterial membranes are the primary target of self-assembling AMPs. Therefore, we qualitatively and quantitatively analyzed the membrane damage and depolarization capacity induced by AMPs using the fluorescent dyes PI and DiSC3-5 and subsequently revealed their mode of action. The membrane penetration rates of FFN toward E. coli and S. aureus ranged from 2.05 to 97.3% and 0.30 to 63.1%, respectively, which were better than those of N6 (2.34 -80.7% and 0.166.40%, respectively), probably due to the strong penetration of the nanoparticles (Fig. 7A, B, Figure S15A, B), which was in line with the properties of the nanoparticle reported previously, where the penetration rate of nanoparticles to Pseudomonas aeruginosa was as high as 97.38% [53] (Table 1). To detect the interference effect of each self-assembled AMP on the potential of the bacterial plasma membrane, the membrane potential dye DiSC3-5 was used to detect the potential change in the bacterial plasma membrane. After treatment with N6 or FFN, the plasma membranes of S. aureus rapidly depolarized within 50 s in a dose-dependent manner (Fig. 7D, Figure S15D). Whereas the plasma membranes of E. coli underwent depolarization after N6 or FFN treatment, but curiously, their perturbing effect on the plasma membrane did not show a dose-dependent effect, which may be due to the outer membrane and LPS of E. coli and needs to be verified by further preparation of protoplasts (Fig. 7C, Figure S15C) [54] (Table 1). The insertion of AMPs into lipid bilayers usually leads to drastic changes in membrane fluidity, so we further assessed the changes in E. coli liposomes and S. aureus after incubation with various AMPs using Laurdan dye. The Laurdan GP increased, indicating membrane sclerosis, while the GP decreased, indicating enhanced membrane fluidity (Fig. 7E) [55]. N6 or FFN induced a significant increase in Laurdan GP in a concentration-dependent manner after interaction with E. coli liposomes (Fig. 7F, Figure S15E), and the GP of the corresponding S. aureus also increased (Fig. 7G, Figure S15F). However, the interference effect was weaker than that of E. coli. Overall, N6 only exhibited membrane disruption mechanisms, whereas the capture mechanisms of FFN included: (1) physical capture by cross-linked nanofibers such as peptide HD6, HDMP, BET and Z(WR)2 peptide [24,32, 56, 58] (Table 1), (2) as monomers or small oligomers inserted into the membrane to change the membrane potential and fluidity, and destroy the structure of the bacterial membrane.
Fig. 7.
Membrane destruction mechanism of N6 and FFN. A-B PI-positive cells of clinically isolated E. coli CGMCC1.90026 and S. aureus CGMCC 1.90032 were treated with N6 or FFN (5, 20, 40, 80 µM) by flow cytometry, respectively. C-D The inner potential changes of clinically isolated E. coli CGMCC1.90026 and S. aureus CGMCC 1.90032 were induced by N6 or FFN, respectively. E Cell membrane hardness model diagram characterized by Laurdan (F-G) Membrane fluidity of clinically isolated E. coli CGMCC1.90026 and S. aureus CGMCC 1.90032 after treatment with N6, or FFN (5, 20, 40, 80 μM). Membrane fluidizing agent benzyl alcohol (50 mM) as a positive control (n=3), CK: PBS group. Results were given as mean±SD. *: p<0.05, **: p<0.01, ***: p<0.0005, ****: p<0.0001
In vivo efficacy in mouse mastitis mode
Self-assembled AMPs (FFN) displayed effective bactericidal effects against E. coli and S. aureus in vitro. In contrast, in vivo bactericidal efficacy was assessed by the effectiveness of treating E. coli/S. aureus-induced mastitis in mice. The self-assembled AMPs could effectively kill pathogenic bacteria in the mammary gland, alleviate the symptoms of mastitis, inhibit the inflammatory storm by decreasing the expression of inflammatory factors (TNF-, IL-1ß, IL-6 and IL-2) and chemokines (MPO), and increase the expression of tight junction proteins (Claudin-3, Occludin, and ZO-1), thus inhibiting the destructive effect of bacteria on the blood-milk barrier and alleviating the destruction of mammary tissue structure.
Firstly, the therapeutic effects of N6 and FFN against E. coli CGMCC 1.90026-induced mastitis in mice were verified (Fig. 8A). After treatment for 24 h, the anatomical results of the negative control (NC) mice showed obvious symptoms of acute mastitis. Moreover, visible blood clots appeared in mammary tissue after E. coli infection, possibly due to the expression and activation of tissue factor (TF) caused by LPS in E. coli cell wall, thereby accelerating the initiation of coagulation reactions to form blood clots [59, 60]. The N6 and FFN treatment groups all completely inhibited the proliferation of E. coli CGMCC 1.90026 in the mammary glands and did not relapse at 48 h, which was better than those of the NC group (9.47 LgCE.coli CFU/gland) and ceftiofur sodium group (1.77 LgCE.coli CFU/gland) at the same dose (Fig. 8B). The mammary gland index of mice indicated that the mammary glands of mice were enlarged after E. coli infection, as evidenced by the mammary indices of mice in the NC group being 1.14% (24 h) and 1.34% (48 h), which were significantly greater than those of the other treatment groups (0.31%~0.36%) (Fig. 8C). Similarly, the spleen index in the NC group was 0.46% (24 h) and 0.47% (48 h), while those in the other treatment groups ranged from 0.30%-0.35%, which were significantly lower than those of the NC group (Fig. 8D). We further assessed the immunological protective effects of FFN against mastitis in mice by quantifying the levels of four inflammatory factors (TNF-, IL-1ß, IL-6, and IL-2) and myeloperoxidase (MPO), which can be secreted by various immune cells and play a pivotal role in the inflammatory response of mammary gland [61]. As shown in Fig. 8E, compared with the BC group, the levels of mammary gland tissue inflammatory factors TNF- (200.5 pg/mL), IL-1ß (90.43 pg/mL), IL-6 (200.54 pg/mL), and IL-2 (50.28 pg/mL) were significantly increased in the NC group. In contrast, after treatment with 100 or 200 µM N6 or FFN, the levels of inflammatory factors (TNF-, IL-1ß, IL-6 and IL-2) were significantly reduced. The effect was better than that of ceftiofur sodium at the same concentration (p<0.001). Similarly, the decrease in MPO (3492.70 ng/mL, Fig. 8F) was more significant. This suggests that N6 and FFN may modulate immune function, consistent with previous reports [49], while this effect is due to a decrease in bacterial load or direct immunomodulatory effects that need further verification. The blood-milk barrier, as a crucial physiological defence mechanism, plays a pivotal role in maintaining the health of mammalian mammary glands [62]. Quantitative PCR analysis revealed that the expression levels of the tight junction proteins Claudin-3, Occludin, and ZO-1 in mammary tissue of the NC group were significantly reduced than those of the BC group. Conversely, all treatment groups exhibited significant increases (Fig. 8G). These findings indicate that E. coli CGMCC 1.90026 infection leads to increased permeability of the blood-milk barrier, subsequently resulting in lower migration thresholds for inflammatory cells and neutrophils across the blood-milk barrier, causing an inflammatory storm (Fig. 8H) [63]. N6 and FFN can significantly alleviate the damage caused by E. coli to the blood-milk barrier. Histopathological changes in the mammary glands of mice were monitored by HE staining. Compared with those in the BC group, the mammary gland acinar structure in the NC group exhibited severe abnormalities with necrotic acinar epithelial cells and inflammation involving the entire mammary acinar, interlobular connective tissues and congestion of mammary interstitial blood vessels. Treatment with N6 and ceftiofur resulted in clear outlines of the mammary tissue acini and significant improvement in inflammatory symptoms. However, there was still scattered infiltration of inflammatory cells in the local interstitium and mammary gland acini, while the infiltration in the FFN group returned to normal (Figure S16).
Moreover, we further evaluated the therapeutic potential of FFN and N6 against G+ bacteria in vivo by S. aureus CGMCC 1.90032-induced mastitis (Fig. 9). After infection with S. aureus CGMCC 1.90032 for 27 h, the mammary gland exhibited erythema and oedema with tissue adhesion. The logarithmic values (Lg CFU/gland) of the bacterial loads of N6 and FFN treatment groups were 6.21, 3.40, 0 and 0 at 24 h, respectively. The corresponding 48 h bacterial loads were 9.07, 3.57, 0 and 0, respectively, among which FFN were better than the same concentration of ceftiofur sodium (3.31 and 2.78) and NC group (9.48 and 10.08) (Fig. 9B). The indices of the mammary gland and spleen also significantly decreased in the N6 and FFN groups compared with the NC group (Fig. 9C, D). Meanwhile, in line with the inflammatory response of E. coli CGMCC 1.90026, FFN exhibited varying degrees of reduction in the levels of inflammatory factors, surpassing the corresponding dose of ceftiofur sodium and N6 (Fig. 9E, F). In comparison to the S. aureus CGMCC 1.90032-NC group, the corresponding E. coli CGMCC 1.90026-NC group demonstrated a more robust inflammatory storm (Fig. 8E, F), which was triggered by LPS-driven IB/NF-B signalling fully activating its immune defences [64]. In contrast, S. aureus tended to show unbalanced immunosuppression rather than inflammation. The qPCR results showed that FFN could significantly increase the expression of tight junction protein (Claudin-3, Occludin, and ZO-1) to support the reconstruction of the integrity of the blood-milk barrier (Fig. 9G, H). Additionally, the tissue sections provided visual evidence for the protective effect of FFN against S. aureus-induced mastitis in mice, which surpassed that of both N6 and ceftiofur sodium (Figure S16). The results indicate that FFN exhibited superior therapeutic potential against mouse mastitis caused by gram-positive and gram-negative bacteria in terms of its in vivo therapeutic efficacy.
Fig. 9.
In vivo efficacy in mouse mastitis model caused by S. aureus CGMCC 1.90032. A Schematic diagram illustrating the experimental procedure for testing the treatment of S. aureus CGMCC 1.90032 induced mastitis in mice (n=8). B. S. aureus CGMCC 1.90032 bacterial load assessment in a murine model of mastitis. C. Evaluation of mammary organ index in a mouse model of mastitis caused by S. aureus CGMCC 1.90032 (n=8). D. Assessment of spleen organ index in a murine model infected with S. aureus CGMCC 1.90032 induced mastitis (n=4). E. Inflammatory factors (TNF-, IL-1ß, IL-6 and IL-2) in the mammary tissue in a mouse mastitis model infected with S. aureus (n=4). F. MPO in the mammary tissue in a mouse mastitis model infected with S. aureus (n=4). G. Examination of tight junction protein expression in the mammary barrier in a murine model of mastitis induced by S. aureus (n=4). H. Schematic representation depicting the treatment approach for S. aureus-induced mastitis using self-assembling peptides. Blank control (BC): uninfected, negative control (NC): infected and untreated. Results were given as mean±SD. *: p<0.05, **: p<0.01, ***: p<0.0005, ****: p<0.0001
Conclusion
Infections caused by MDR bacteria seriously affect human and animal health. In this study, based on a comprehensive understanding of the precise module design theory of self-assembled peptides and antibacterial structurefunction relationships, we successfully constructed a bacterial-responsive self-assembly platform consisting of hydrophobic, hydrogen-bonding and antimicrobial modules and screened a potent antimicrobial, highly stable and low-toxicity in situ self-assembled AMP FFN. FFN self-assembled into nanoparticles in vitro and initiated nanoparticle-to-nanofiber conversion under the action of bacterial cell wall LTA or LPS nucleation sites, thereby achieving bacterial capture and further disrupting the cell membrane lipid bilayer arrangement of the bacterial cell membrane and causing bacterial death. The in vivo mouse mastitis infection model further confirmed its therapeutic potential. In conclusion, this study provides a theoretical basis for the design of peptide-based nanomaterials based on AMPs, a technological means to overcome the poor stability of AMPs, and an alternative to the shortage of antibiotics for the MDR bacteria E. coli and S. aureus.
Experiment section
Self-assembly properties of AMPs
Self-assembled AMPs synthesis
The hydrophobic module fluorenylmethy (Fmoc), the self-assembling module (KLVFFK or KFFK) and the antimicrobial module N6 of AMPs FKN, FFN and fluorescein isothiocyanate (FITC)-labelled AMPs were covalently linked with amide bonds and synthesized by MIMOTOPES (Wuxi, China) using solid-phase synthesis method. All synthetic AMPs were purified by RP-HPLC to a purity above 95%. The molecular masses of AMPs are characterized by MALDI-TOFMS. Use ProtParam (ExPASy Proteomics Server: http://www.expasy.org/tools/protparam.html) to predict all AMPs' molecular weight, isoelectric point and hydrophobicity.
Fluorescence spectroscopy and CMC determination
As previous study reported, the hydrophobic fluorescent probe ANS was utilized to calculate the CMC of self-assembled AMPs [34]. All AMPs were diluted with PBS to a final concentration of 0.5512 µM. ANS was then added to the peptide solution at a final concentration of 25.6 µM and incubated for 1 h. The fluorescence spectra of peptide solution were determined by a microplate reader (ex: 360 nm, em: 400670 nm). Simultaneously, to assess the effect of LPS or LTA on peptide self-assembly ability, LPS or LTA was added to the 80 µM peptide at a final concentration of 1µM for fluorescence spectrometry. Finally, the crossing point of fluorescence intensity (480 nm) and the logarithm of peptide concentration were set as CMC.
SEM and TEM observation of self-assembled AMPs
The morphology of the self-assembled AMPs was observed using SEM and TEM. (1) Sample preparation: the AMP was diluted to 80 µM for use. At the same time, LPS or LTA at a final concentration of 1 µM was added to the 80 µM peptide to evaluate changes in the morphology of AMPs after interaction with LPS or LTA. The samples were equilibrated at 37 for 6 h for use. (2) SEM observation: 5 µL of the prepared samples were placed on silicon wafers, left at room temperature for 12 h, dried naturally, sprayed with gold for 120 s (Feica, EM ACE600) and then observed by SEM (Hitachi SU8000, Japan) [65]. (3) TEM observation: 5 µM of the above samples were dropped on the 100-mesh copper-plated grid, precipitated for 10 min, residual solution absorbed with filter paper, stained with 3% uranyl acetate dihydrate for 30 s, rinsed 3 times with ultrapure water, dried and observed by TEM (Hitachi H7650B, Japan).
Circular dichroism assay
CD spectroscopy was utilized to determine the secondary structure of AMPs (80 µM) in PBS (pH 7.4) and to examine the effect of LTA or LPS on the secondary structure of AMPs. The secondary structure of AMPs was determined in the presence of a final concentration of 1 µM of either LPS or LTA [30]. CD spectra from 197 to 280 nm were taken using a CD Spectrometer (Applied Photophysics, Chirascan V100) and then expressed as the average of three scans. All measurements were performed at room temperature. The structural analysis was performed using CDNN software.
In vitro antimicrobial assay
Determination of MIC and MBC
The MIC of all the AMPs were determined according to the broth microdilution method of the Clinical Laboratory Standards Institute (CLSI) [64]. In short, bacteria (mid-log phase) were diluted to 1×105 CFU/mL with fresh MHB medium. 90 µL of cell suspension and 10 µL of continuously twofold diluted AMP (0.25128 µM) were added to the 96-well plate and then incubated at 37 °C for 18 h. The MIC was determined as the lowest peptide concentration at which no bacterial growth was observed. MBC was determined as the lowest peptide concentration without CFU agar. All experiments were performed in triplicate.
Time-dependent sterilization curves
The killing kinetics assay is based on the previous method [26]. Briefly, the tested strains (mid-log phase) were diluted to 1×105 CFU/mL and mixed with different concentrations of AMPs (1×2×and 4×MIC), respectively, and then incubated at 37 . After incubation for 0, 0.5, 1, 2, 4, 6, 8, 10 and 24 h, samples were taken for colony counting on MHA plates. Each concentration was tested in triplicate. The MDK99 and MDK99.99 were defined as the minimum times required for achieving sterilization rates of 99% and 99.99%, respectively.
Biosafety of peptide
Hemolysis assay
The hemolysis activities of all AMPs were measured by the amount of haemoglobin released by the lysis of red blood cells, as described previously [50, 66]. Briefly, the peptide at a final concentration of 1128 µM was mixed with 8% mouse erythrocytes in equal volumes, incubated at 37 for 1 h, centrifuged at 1500 rpm for 5 min and collected supernatant. OD values were measured at 540 nm absorbance. PBS and 0.1% Tritalon X-100 were used as negative controls (A0) and positive controls (A100), respectively. Hemolysis rate (%)=((Apeptide-A0) / (A100-A0))×100%.
Cytotoxicity analysis
The cytotoxicity of all AMPs was determined using the CCK-8 assay [67]. The procedures were as follows: HaCaT, RAW 264.7 and MAC-T cells at a cell density of 1×105 cells/mL were inoculated into 96-well cell culture plates for 24 h. After removal of the medium, an equal volume of peptide at a final concentration of 1128 µM was added to the 96-well plate and incubated for 12 h. Then 10% CCK-8 (100 µL) was added, continually incubated for 2 h and then the absorbance (OD value) was detected at a wavelength of 450 nm. The assay was repeated at least three times. Cell viability=(As-Ab)/(Ac-Ab)×100%. (As: Absorbance value of experimental group Ac: Absorbance value of PBS group Ab: Absorbance value of the blank group).
Stability of peptide
Proteolytic stability test
The time-dependent proteolytic curves of self-assembled AMPs in trypsin buffer were quantitatively calculated using HPLC. The specific procedures were as follows: 0.5 mL of AMP (2560 µM) was incubated with trypsin (1 w/v%) at 37 , and 100 µL of samples were collected for each incubation time (0.5, 1, 2, 4, 6 h), diluted with 0.1% TFA/ddH2O to a final concentration of 256 µM and then quantified by HPLC [68]. Simultaneously, to further compare nanofiber and nanoparticle resistance trypsin, trypsin (10 µM) was added to an incubation mixture containing peptide (80 µM) and LPS (1 µM) at different times, and then samples were taken for HPLC analysis. Further details of the experimental protocol are shown in Fig. 3B [30]. The salt stability, serum stability and metabolic stability were in the "1.5 Stability of peptide" of Supporting Information.
Antibacterial mechanism
SEM and TEM observations
SEM. Mid-log phase tested bacteria (1×109 CFU/mL) were treated with peptide (80 µM) for 2 h at 37 °C, and then fixed with 2.5% glutaraldehyde, dehydrated with gradient ethanol series, and dried with CO2. Finally, the samples were sputtered with goldpalladium and observed with SEM (Hitachi SU8000, Japan). TEM. The morphology and intracellular changes were further observed by TEM. The bacterial samples were processed as described above, followed by fixationdehydrationembeddingsectioningstaining before TEM microscopy (Hitachi H7650B, Japan).
Confocal laser scanning microscope (CLSM) observation
The sample was prepared as described in 4.5.2. Tested bacteria were stained with DAPI (20 µg /mL) and PI (20 µg/mL) for 2 h. The DAPI-stained cells, PI-stained cells and FITC-labelled AMPs were visualized by CLSM (Leica TCS SP 5). At the same time, FITC-labelled AMPs were observed by CLSM after co-incubation with LPS or LTA [30].
Molecular docking
After incubation with LTA and LPS, the morphology of FFN changed from nanoparticles to nanofibers. To further study the binding region and interaction modes of FFN with LTA and LPS. In this study, the HDCOK program, a professional proteinprotein and protein-DNA/RNA docking program, was used to dock FFN with LTA and LPS [69,71]. The structure with the best docking score was selected as the standard result for further interaction analysis. The docking score was calculated based on the iterative scoring function of ITScorePP or ITScorePR. The lower the docking score, the greater the possibility of binding between the two molecules and the stronger the interaction. The docking score of proteinprotein/RNA/DNA complexes in PDB is usually around -200, and the confidence score is calculated according to the following formula to simulate the possibility of binding between two molecules.
In vivo efficacy in mouse mastitis model
Design of mice experiments
To investigate the in vivo efficacy of self-assembled FFN, a mouse mastitis model was selected for subsequent verification. The construction of the mouse mastitis model was described as follows [72]: i) Preparation of bacterial solution: E. coli CGMCC1.90026 and S. aureus CGMCC1.90032 (mid-log phase) were washed with PBS and diluted to final concentrations of 5×103 CFU/mL and 1×103 CFU/mL, respectively; ii) Establishment of a mastitis model: 8-week-old ICR female mice and male mice were caged (2:1) until pregnancy, post-partum female mice (10 d) were selected for subsequent model construction, 54 female mice were randomly divided into 7 groups, blank control (BC) (uninfected), negative control (NC) (untreated), N6 (100 µM and 200 µM), FFN (100 µM and 200 µM) and ceftiofur sodium (200 µM). Mice were separated from their offspring before 2 h, fixed in a supine position under a microscope, anaesthetized and subtracted 0.5 mm from the tip of the fourth pair of nipple ducts, and inoculated with 100 µL of the above bacterial solution through the nipple ducts using a micro syringe; (3) Mastitis treatment: after 3 h of infection, AMP was administered via the mammary duct according to the above groups.
Supplementary Information
Acknowledgements
We acknowledge Chunli Li from the Core Facility at the Institute of Microbiology at the Chinese Academy of Sciences (CAS) for his technical support with SEM, and Tong Zhao for her technical support with FACS analysis.
Author contribution
X.M., N.Y., J.W., D.T. and Y.H. (Yinhua Huang): conception and experiments design. X.M., Y. L.: experiments operation. X.M., N.Y., R.M., D.T., Y. G and Y.H (Ya Hao): methodology and data analysis. X.M.: writingoriginal draft. J.W., N.Y., R.M., Y.H. (Ya Hao), Y.H. (Yinhua Huang) and D.T.: writingreview and editing. J.W. and N.Y.: contributed to funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31872393). National Key Research and Development PlanHigh Expression of Thiopeptides and their Analogs (Grant No. 2022YFC2105000-03, 20222026). The Innovation Program of Agricultural Science and Technology (ASTIP) in Chinese Academy of Agricultural Sciences (CAAS) (Grant No. CAAS-ASTIP-2017-FRI-02) and its key projects (Grant No. CAAS-ZDRW202111 and Grant No. CAAS-ZDXT 201808).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
The mouse experiment was performed according to the Animal Care and Use Committee of the Feed Research Institute of the Chinese Academy of Agricultural Sciences (CAAS) and approved by the Laboratory Animal Ethical Committee and its Inspection of the Feed Research Institute of CAAS (IFR-CAAS20230818).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuanxuan Ma and Na Yang contributed equally to this work.
Contributor Information
Da Teng, Email: tengda@caas.cn.
Yinhua Huang, Email: cauhyh@cau.edu.cn.
Jianhua Wang, Email: wangjianhua@caas.cn, Email: wangjianhua.peking@qq.com.
References
- 1.Holmes AH, Moore LS, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387(10014):17687. 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 2.Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):62955. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molineri AI, Camussone C, Zbrun MV, Suárez Archilla G, Cristiani M, Neder V, et al. Antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis: systematic review and meta-analysis. Prev Vet Med. 2021;188: 105261. 10.1016/j.prevetmed.2021.105261. [DOI] [PubMed] [Google Scholar]
- 4.Goulart DB, Mellata M. Escherichiacoli mastitis in dairy cattle: etiology, diagnosis, and treatment challenges. Front Microbiol. 2022;13: 928346. 10.3389/fmicb.2022.928346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Jiang Y, Xia X, Wu J, Almeida R, Eda S, et al. An on-site, highly specific immunosensor for Escherichiacoli detection in field milk samples from mastitis-affected dairy cattle. Biosens Bioelectron. 2020;165: 112366. 10.1016/j.bios.2020.112366. [DOI] [PubMed] [Google Scholar]
- 6.De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, et al. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev. 2020;33(3):e00181-e219. 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):31827. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 8.Zhang QY, Yan ZB, Meng YM, Hong XY, Shao G, Ma JJ, et al. Antimicrobial peptides: mechanism of action, activity and clinical potential. Mil Med Res. 2021;8(1):48. 10.1186/s40779-021-00343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Y, Chen Y, Song Z, Tan Z, Cheng J. Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation. Adv Drug Deliv Rev. 2021;170:26180. 10.1016/j.addr.2020.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Magana M, Pushpanathan M, Santos AL, Leanse L, Fernandez M, Ioannidis A, et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect Dis. 2020;20(9):e21630. 10.1016/S1473-3099(20)30327-3. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Wang Y, Xue Z, Jia Y, Li R, He C, et al. The structure-mechanism relationship and mode of actions of antimicrobial peptides: a review. Trends Food Sci Tech. 2021;109:103. 10.1016/j.tifs.2021.01.005. [Google Scholar]
- 12.Yan Y, Li Y, Zhang Z, Wang X, Niu Y, Zhang S, et al. Advances of peptides for antibacterial applications. Colloids Surf B Biointerfaces. 2021;202: 111682. 10.1016/j.colsurfb.2021.111682. [DOI] [PubMed] [Google Scholar]
- 13.Lazzaro BP, Zasloff M, Rolff J. Antimicrobial peptides: application informed by evolution. Science. 2020. 10.1126/science.aau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Ouyang X, Liu Y, Ba Z, Yang Y, Zhang J, et al. Novel ß-hairpin antimicrobial peptide containing the -turn sequence of ß-NG- and the tryptophan zippers facilitate self-assembly into nanofibers, exhibiting excellent antimicrobial performance. J Med Chem. 2024;67(8):636583. 10.1021/acs.jmedchem.3c02339. [DOI] [PubMed] [Google Scholar]
- 15.Xuan J, Feng W, Wang J, Wang R, Zhang B, Bo L, et al. Antimicrobial peptides for combating drug-resistant bacterial infections. Drug Resist Updat. 2023;68: 100954. 10.1016/j.drup.2023.100954. [DOI] [PubMed] [Google Scholar]
- 16.Carratalá JV, Serna N, Villaverde A, Vázquez E, Ferrer-Miralles N. Nanostructured antimicrobial peptides: the last push towards clinics. Biotechnol Adv. 2020;44: 107603. 10.1016/j.biotechadv.2020.107603. [DOI] [PubMed] [Google Scholar]
- 17.Dzuvor CKO. Toward clinical applications: transforming nonantibiotic antibacterials into effective next-generation supramolecular therapeutics. ACS Nano. 2024;18(4):256477. 10.1021/acsnano.3c11045. [DOI] [PubMed] [Google Scholar]
- 18.Wang C, Hong T, Cui P, Wang J, Xia J. Antimicrobial peptides towards clinical application: delivery and formulation. Adv Drug Deliv Rev. 2021;175: 113818. 10.1016/j.addr.2021.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Mu R, Zhu D, Abdulmalik S, Wijekoon S, Wei G, Kumbar SG. Stimuli-responsive peptide assemblies: design, self-assembly, modulation, and biomedical applications. Bioact Mater. 2024;35:181207. 10.1016/j.bioactmat.2024.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batra R, Loeffler TD, Chan H, Srinivasan S, Cui H, Korendovych IV, Nanda V, Palmer LC, Solomon LA, Fry HC, Sankaranarayanan SKRS. Machine learning overcomes human bias in the discovery of self-assembling peptides. Nat Chem. 2022;14(12):142735. 10.1038/s41557-022-01055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbas M, Ovais M, Atiq A, Ansari T, Xing R, Spruijt E, et al. Tailoring supramolecular short peptide nanomaterials for antibacterial applications. Coord Chem Rev. 2022;460: 214481. 10.1016/j.ccr.2022.214481. [Google Scholar]
- 22.Mamuti M, Zheng R, An HW, Wang H. Invivo self-assembled nanomedicine. Nano Today. 2021;36: 101036. 10.1016/j.nantod.2020.101036. [Google Scholar]
- 23.Qi GB, Zhang D, Liu FH, Qiao ZY, Wang H. An, On-site transformation strategy for treatment of bacterial infection. Adv Mater. 2017;29(36):1703461. 10.1002/adma.201703461. [DOI] [PubMed] [Google Scholar]
- 24.Fan Y, Li XD, He PP, Hu XX, Zhang K, Fan JQ, et al. A biomimetic peptide recognizes and traps bacteria invivo as human defensin-6. Sci Adv. 2020. 10.1126/sciadv.aaz4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo X, Zhang Z, Song J, Wang Y, Yu Z. Self-assembly of peptides in living cells for disease theranostics. J Mater Chem B. 2024;12(18):4289306. 10.1039/d4tb00365a. [DOI] [PubMed] [Google Scholar]
- 26.Li T, Yang N, Teng D, Mao R, Hao Y, Wang X, et al. C-terminal mini-PEGylation of a marine peptide N6 had potent antibacterial and anti-inflammatory properties against Escherichiacoli and Salmonella strains invitro and invivo. BMC Microbiol. 2022;22(1):128. 10.1186/s12866-022-02534-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Liang JF. Peptide fibrils with altered stability, activity, and cell selectivity. Biomacromol. 2013;14(7):232631. 10.1021/bm400618m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Z, Feng D, Wang W, Wang Y, Cheng M, Yang H, et al. Influence of fatty acid modification on the anticancer activity of the antimicrobial peptide Figainin 1. ACS Omega. 2023;8(44):4187684. 10.1021/acsomega.3c06806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li LL, Qiao ZY, Wang L, Wang H. Programmable construction of peptide-based materials in living subjects: from modular design and morphological control to theranostics. Adv Mater. 2019;31(45): e1804971. 10.1002/adma.201804971. [DOI] [PubMed] [Google Scholar]
- 30.Debnath S, Shome A, Das D, Das PK. Hydrogelation through self-assembly of fmoc-peptide functionalized cationic amphiphiles: potent antibacterial agent. J Phys Chem B. 2010;114(13):440715. 10.1021/jp909520w. [DOI] [PubMed] [Google Scholar]
- 31.Yang PP, Luo Q, Qi GB, Gao YJ, Li BN, Zhang JP, et al. Host materials transformable in tumor microenvironment for homing theranostics. Adv Mater. 2017;29(15):1605869. 10.1002/adma.201605869. [DOI] [PubMed] [Google Scholar]
- 32.Sha XL, Lv GT, Chen QH, Cui X, Wang L, Cui X. A peptide selectively recognizes gram-negative bacteria and forms a bacterial extracellular trap (BET) through interfacial self-assembly. J Mater Chem B. 2024;12(15):367685. 10.1039/d3tb02559d. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Liu K, Xing R, Yan X. Peptide self-assembly: thermodynamics and kinetics. Chem Soc Rev. 2016;45(20):5589604. 10.1039/c6cs00176a. [DOI] [PubMed] [Google Scholar]
- 34.Tram N, Xu J, Mukherjee D, Obanel AE, Mayandi V, Selvarajan V, et al. Bacteria-responsive self-assembly of antimicrobial peptide nanonets for trap-and-kill of antibiotic-resistant strains. Adv Funct Mater. 2022;33:2210858. 10.1002/adfm.202210858. [Google Scholar]
- 35.Schnaider L, Brahmachari S, Schmidt NW, Mensa B, Shaham-Niv S, Bychenko D, et al. Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. Nat Commun. 2017;8(1):1365. 10.1038/s41467-017-01447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Gao H, Zhang C, Li D, Zhang L, Huang L, et al. Influence of acetylation on the mechanism of action of antimicrobial peptide L163. Int J Pept Res Ther. 2022;28(3):78. 10.1007/s10989-022-10387-0. [Google Scholar]
- 37.Wang Z, Liu X, Teng Da, Mao R, Hao Y, Yang N, et al. Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichiacoli. Commun Biol. 2020;3(1):41. 10.1038/s42003-020-0761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan P, Tang Q, Xu S, Zhang Y, Fu H, Ma X. Designing self-assembling chimeric peptide nanoparticles with high stability for combating piglet bacterial infections. Adv Sci. 2022;9(14):2105955. 10.1002/advs.202105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Häffner SM, Malmsten M. Influence of self-assembly on the performance of antimicrobial peptides. Curr Opin in Colloid In. 2018;38:56. 10.1016/j.cocis.2018.09.002. [Google Scholar]
- 40.Yang Y, Cai Z, Huang Z, Tang X, Zhang X. Antimicrobial cationic polymers: from structural design to functional control. Polym J. 2017;50:33. 10.1038/pj.2017.72. [Google Scholar]
- 41.Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol.2016;14(5):320. https://www.nature.com/articles/nrmicro.2016.34 [DOI] [PubMed] [Google Scholar]
- 42.Cantón R, Morosini MI. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev. 2011;35(5):977. 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 43.Srimani JK, Huang S, Lopatkin AJ, You L. Drug detoxification dynamics explain the postantibiotic effect. Mol Syst Biol. 2017;13(10):948. 10.15252/msb.20177723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abel Zur Wiesch P, Abel S, Gkotzis S, Ocampo P, Engelstädter J, Hinkley T, et al. Classic reaction kinetics can explain complex patterns of antibiotic action. Sci Transl Med. 2015. 10.1126/scitranslmed.aaa8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phuong PT, Oliver S, He J, Wong EHH, Mathers RT, Boyer C. Effect of hydrophobic groups on antimicrobial and hemolytic activity: developing a predictive tool for ternary antimicrobial polymers. Biomacromol. 2020;21(12):524155. 10.1021/acs.biomac.0c01320. [DOI] [PubMed] [Google Scholar]
- 46.Cao F, Ma G, Song M, Zhu G, Mei L, Qin Q. Evaluating the effects of hydrophobic and cationic residues on antimicrobial peptide self-assembly. Soft Matter. 2021;17(16):444551. 10.1039/d1sm00096a. [DOI] [PubMed] [Google Scholar]
- 47.Dou X, Zhu X, Wang J, Dong N, Shan A. Novel design of heptad amphiphiles to enhance cell selectivity, salt resistance, antibiofilm properties and their membrane-disruptive mechanism. J Med Chem. 2017;60(6):225770. 10.1021/acs.jmedchem.6b01457. [DOI] [PubMed] [Google Scholar]
- 48.Zuroff L, Daley D, Black KL, Koronyo-Hamaoui M. Clearance of cerebral Aß in Alzheimers disease: reassessing the role of microglia and monocytes. Cell Mol Life Sci. 2017;74(12):2167201. 10.1007/s00018-017-2463-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan P, Fu H, Ma X. Design, optimization, and nanotechnology of antimicrobial peptides: from exploration to applications. Nano Today. 2021;39: 101229. 10.1016/j.nantod.2021.101229. [Google Scholar]
- 50.El Battioui K, Chakraborty S, Wacha A, Molnár D, Quemé-Peña M, Szigyártó IC, et al. Insitu captured antibacterial action of membrane-incising peptide lamellae. Nat Commun. 2024;15(1):3424. 10.1038/s41467-024-47708-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Q, Li J, Yu W, Wang Z, Li J, Feng X, et al. De novo design of a pH-triggered self-assembled -hairpin nanopeptide with the dual biological functions for antibacterial and entrapment. J Nanobiotechnology. 2021;19(1):183. 10.1186/s12951-021-00927-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J, Zhang X, Zou P, Yao J, Liu L, Cai Y, et al. Peptide-based nano-antibiotic transformers with antibiotic adjuvant effect for multidrug resistant bacterial pneumonia therapy. Nano Today. 2022. 10.1016/j.nantod.2022.101505.37034182 [Google Scholar]
- 53.Tan P, Wu C, Tang Q, Wang T, Zhou C, Ding Y, et al. pH-triggered size-transformable and bioactivity-switchable self-assembling chimeric peptide nanoassemblies for combating drug-resistant bacteria and biofilms. Adv Mater. 2023;35(29): e2210766. 10.1002/adma.202210766. [DOI] [PubMed] [Google Scholar]
- 54.Fang Y, Zhu Y, Li L, Lai Z, Dong N, Shan A. Biomaterial-interrelated bacterial sweeper: simplified self-assembled octapeptides with double-layered trp zipper induces membrane destabilization and bacterial apoptosis-like death. Small Methods. 2021;5(12): e2101304. 10.1002/smtd.202101304. [DOI] [PubMed] [Google Scholar]
- 55.Wenzel M, Vischer NOE, Strahl H, Hamoen HLW, et al. Assessing membrane fluidity and visualizing fluid membrane domains in bacteria using fluorescent membrane dyes. Bio Protoc. 2018;8(20): e3063. 10.21769/bioprotoc.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu H, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, et al. Human -defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337(6093):47781. 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang H, Wang J, Wang X, Wang S, Xu J, Shan Q, et al. Nanofiber peptides for bacterial trapping: a novel approach to antibiotic alternatives in wound infections. Adv Healthc Mater. 2024;12: e2304657. 10.1002/adhm.202304657. [DOI] [PubMed] [Google Scholar]
- 58.Parkin HC, Street STG, Gowen B, Da-Silva-Correa LH, Hof R, Buckley HL, et al. Mechanism of action and design of potent antibacterial block copolymer nanoparticles. J Am Chem Soc. 2024;146(8):512841. 10.1021/jacs.3c09033. [DOI] [PubMed] [Google Scholar]
- 59.Yang X, Cheng X, Tang Y, Qiu X, Wang Y, Kang H, et al. Bacterial endotoxin activates the coagulation cascade through gasdermin D-dependent phosphatidylserine exposure. Immunity. 2019;51(6):983-996.e6. 10.1016/j.immuni.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 60.Johnzon CF, Dahlberg J, Gustafson AM, Waern I, Moazzami AA, Östensson K, et al. The effect of lipopolysaccharide-induced experimental bovine mastitis on clinical parameters, inflammatory markers, and the metabolome: a kinetic approach. Front Immunol. 2018;9:1487. 10.3389/fimmu.2018.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu X, Guo J, Zhao C, Jiang P, Maimai T, Yanyi L, et al. The gut microbiota contributes to the development of Staphylococcusaureus-induced mastitis in mice. ISME J. 2020;14(7):1897910. 10.1038/s41396-020-0651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wellnitz O, Zbinden C, Huang X, Bruckmaier RM. Short communication: Differential loss of bovine mammary epithelial barrier integrity in response to lipopolysaccharide and lipoteichoic acid. J Dairy Sci. 2016;99(6):48516. 10.3168/jds.2016-10927. [DOI] [PubMed] [Google Scholar]
- 63.Xu T, Dong Z, Wang X, Qi S, Li X, Cheng R, et al. IL-1 induces increased tight junction permeability in bovine mammary epithelial cells via the IL-1-ERK1/2-MLCK axis upon blood-milk barrier damage. J Cell Biochem. 2018;119(11):902841. 10.1002/jcb.27160. [DOI] [PubMed] [Google Scholar]
- 64.Günther J, Petzl W, Bauer I, Ponsuksili S, Zerbe H, Schuberth HJ, et al. Differentiating Staphylococcus aureus from Escherichia coli mastitis: S.aureus triggers unbalanced immune-dampening and host cell invasion immediately after udder infection. Sci Rep. 2017;7(1):4811. 10.1038/s41598-017-05107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zou P, Liu J, Li X, Yaseen M, Yao J, Liu L, et al. A membrane curvature modulated lipopeptide to broadly combat multidrug-resistant bacterial pneumonia with low resistance risk. ACS Nano. 2022;16(12):20545. 10.1021/acsnano.2c07251. [DOI] [PubMed] [Google Scholar]
- 66.Song M, Liu Y, Huang X, Ding S, Wang Y, Shen J, et al. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant gram-negative pathogens. Nat Microbiol. 2020;5(8):104050. 10.1038/s41564-020-0723-z. [DOI] [PubMed] [Google Scholar]
- 67.Ma Z, Liu X, Nie J, Zhao H, Li W. Nano-antimicrobial peptides based on constitutional isomerism-dictated self-assembly. Biomacromol. 2022;23(3):130213. 10.1021/acs.biomac.1c01532. [DOI] [PubMed] [Google Scholar]
- 68.Wang Z, Teng D, Mao R, Hao Y, Yang N, Wang X, et al. A cleavable chimeric peptide with targeting and killing domains enhances LPS neutralization and antibacterial properties against multidrug resistant E. coli. Commun Biol. 2023;6(1):1170. 10.1038/s42003-023-05528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):5839. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feng Y, Yan Y, He J, Tao H, Wu Q, et al. Docking and scoring for nucleic acid-ligand interactions: principles and current status. Drug Discov Today. 2022;27(3):83847. 10.1016/j.drudis.2021.10.013. [DOI] [PubMed] [Google Scholar]
- 71.Feng Y, Zhang K, Wu Q, Huang SY. NLDock: a fast nucleic acid-ligand docking algorithm for modeling RNA/DNA-ligand complexes. J Chem Inf Model. 2021;61(9):477182. 10.1021/acs.jcim.1c00341. [DOI] [PubMed] [Google Scholar]
- 72.Zheng X, Yang N, Mao R, Hao Y, Teng D, Wang J. Pharmacokinetics and pharmacodynamics of antibacterial peptide NZX in Staphylococcusaureus mastitis mouse model. Appl Microbiol Biotechnol. 2024;108(1):260. 10.1007/s00253-024-13101-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.